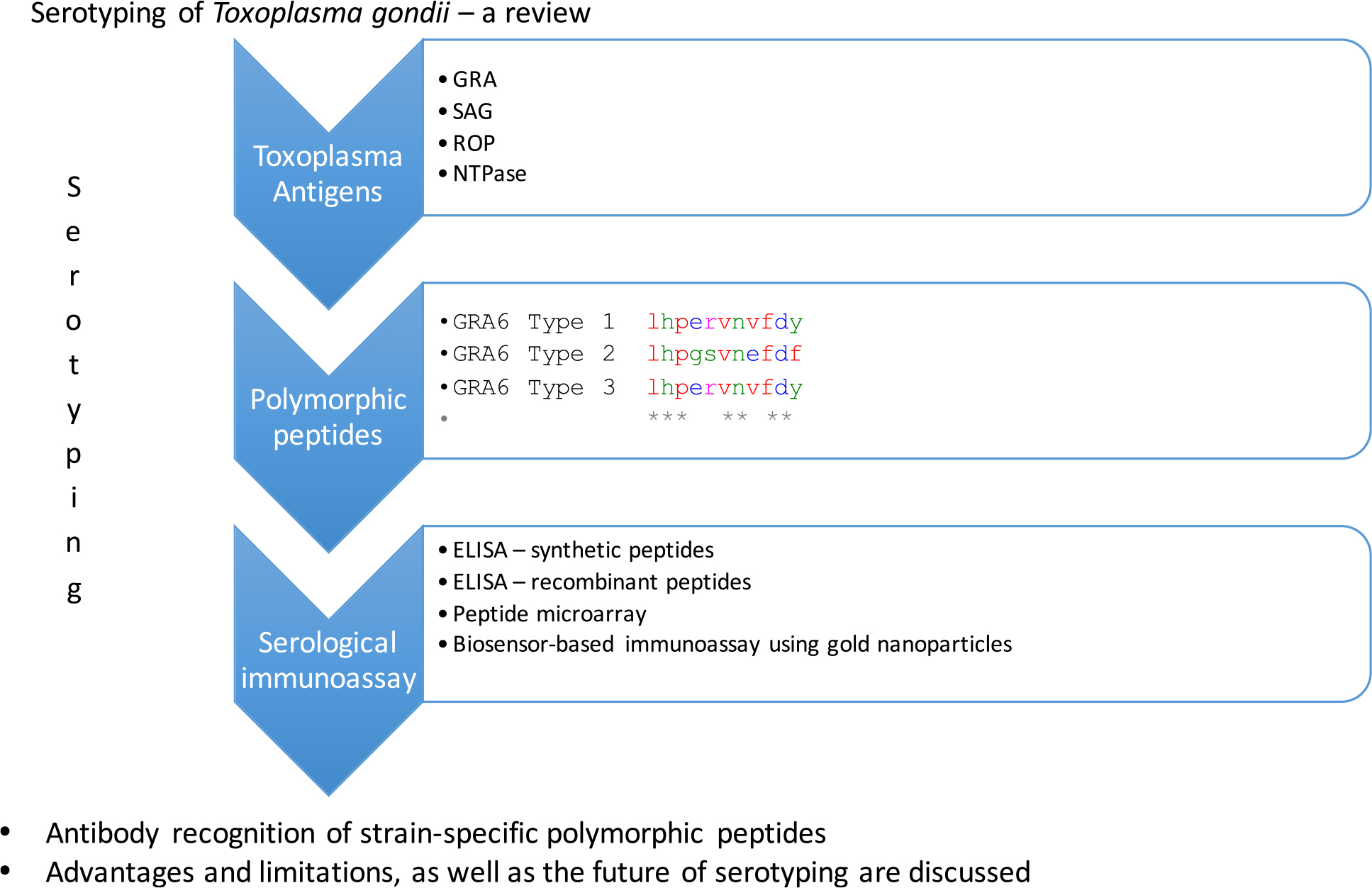
Serotyping, a challenging approach for Toxoplasma gondii typing
- 1Center for the Study of Animal Science (CECA), University of Porto, Porto, Portugal
- 2Department of Infectious Diseases, R&D Unit, National Health Institute Dr. Ricardo Jorge (INSA), Porto, Portugal
- 3Associate Laboratory for Animal and Veterinary Sciences (AL4AnimalS), University of Porto, Porto, Portugal
- 4Associated Laboratory for Green Chemistry (LAQV) of the Network of Chemistry and Technology (REQUIMTE), Department of Chemistry and Biochemistry, Faculty of Sciences, University of Porto, Porto, Portugal
- 5Center for the Study of Animal Science, University of Porto, Porto, Portugal
Genotype analysis has revealed a high genetic diversity in strains of Toxoplasma gondii, isolated from a wide range of intermediate hosts and different geographic origins. Diversity is notably striking for parasites from wild hosts in South America, generally referred as non-archetypal genotypes. Those genotypes are implicated in the etiology of severe clinical disease, multivisceral toxoplasmosis, associated with high rate of mortality in immunocompetent individuals. Can we accept specific antibodies produced during T. gondii infection as biomarkers to identify infecting genotypes? Scientific evidence supports a positive response to this question; however, the genetic diversity of T. gondii genotypes organized into 16 haplogroups and collectively defined in 6 major clades, provides a reminder of the complexity and difficulty for the purpose. This review discusses serological approaches to genotyping T. gondii.
1. Toxoplasma gondii and toxoplasmosis, an overview
Toxoplasmosis is the most reported parasitic zoonosis in Europe. The global prevalence of toxoplasmosis is estimated to be around 30% with 10 million clinical cases (1–4) and it is ranked as the third most important contributor to health burden caused by food-borne illness in Europe (4). Most infections in humans are asymptomatic. However, severe complications may occur during (i) congenital Toxoplasma infection, such as abortion, stillbirth, and hydrocephalus in newborns (1, 4, 5); (ii) ocular toxoplasmosis, with retinochoroidal lesions leading to chronic ocular disease (1, 2); (iii) encephalitis in immunosuppressed patients (1, 4, 5); and (iv) multivisceral toxoplasmosis due to atypical genotypes in South America (6–8). A possible implication of Toxoplasma gondii genetic diversity on the pathogenesis of toxoplasmosis has been postulated (9–11). However, a better comprehension of the population structure is crucial in order to predict the potential spread of pathogenicity (or virulent) determinants related with isolates of T. gondii, their geographic and host origin. This process must be performed based on the study of a large population and representative of the T. gondii gene pool circulating in nature. Carme et al. (12) postulated a novel scenario in French Guiana wherein atypical genotypes of T. gondii, were associated to severe toxoplasmosis acquired by immunocompetent adults. The investigators claim that “…this unusual form of toxoplasmosis is associated with an unusual genotype of T. gondii and may be linked to a neotropical forest based cycle…” and that “the “wild-type” strain of T. gondii should be isolated from neotropical mammals so that we can study its virulence and genetic characteristics….” There is some consensus that the population structure of Toxoplasma includes the three archetypal genotypes (I, II, and III), along with non-archetypal genotypes. Archetypal strains, particularly genotype II and III are the most prevalent in Europe. In addition, in North America, along with type II and III, strains belonging to haplogroup 12 has been described in the sylvatic reservoir; finally, specific clonal strains have been described in Africa, South America, and Asia, whereas non-archetypal genotypes prevail in South America (11).
Can we anticipate that pathogenesis of toxoplasmosis may reflect the genotype structure of infecting strains? A new area of investigation has focused on this issue: a systematic isolation of parasites by bioassay of T. gondii of livestock, wild animals and humans (13). In addition, multilocus genotyping approach of T. gondii isolates provides data related with genetic diversity, including those genotypes favoring virulence (13). Both approaches are crucial to comprehend the mechanisms implicated in host derived genotype selection, and the pathogenesis of the disease. Both approaches are labor intensive and biological samples for bioassay are not always available. Moreover, the requirement for the parasite isolates and/or DNAs in bioassays may favor the isolation of one strain, while other may be missed in cases of co-infections with different strains.
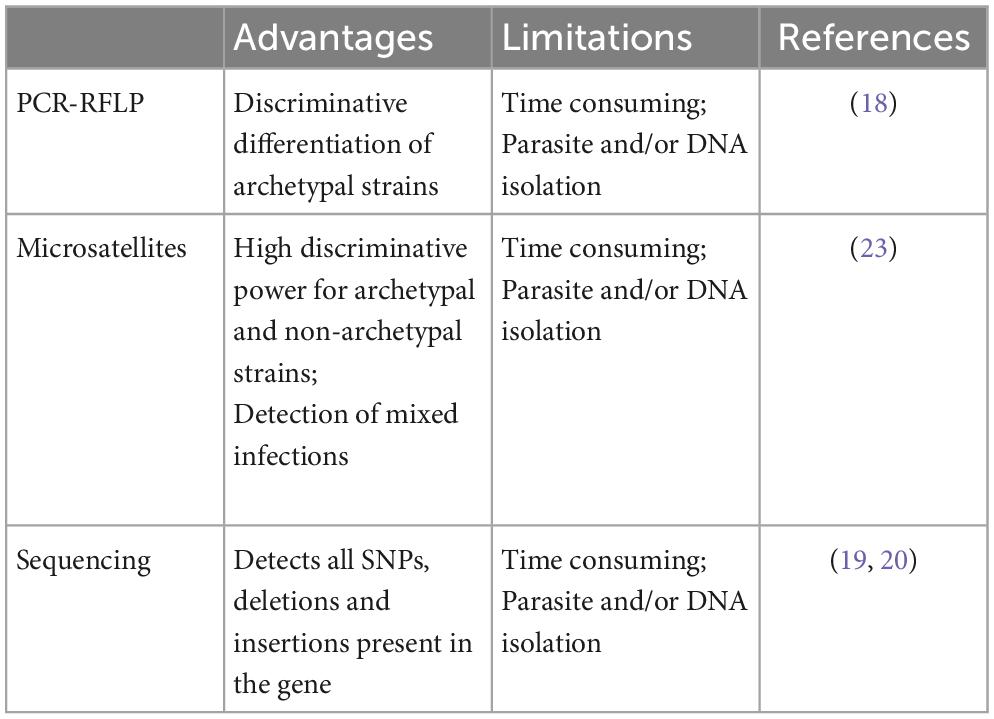
In addition, to achieve genotype analysis, different approaches have been proposed: (i) PCR-RFLP (14–18); (ii) gene sequencing (multilocus genes sequencing and whole genome sequencing (19, 20)); and (iii) length polymorphism of microsatellite sequences (21–24). Table 1 summarizes the advantages and limitations of various genotyping methods.
Herein, serotyping is our focus, including on the state of the art, and controversies around effectiveness of approaches, considering the spectrum of T. gondii strains.
2. Serotyping: The other way of typing Toxoplasma gondii
The isolation of T. gondii from human samples is complex. Serotyping appears to be an alternative method for typing T. gondii strains. The process does not require isolation of the parasite. Serotyping is based on the antibody recognition of strain-specific polymorphic peptides (25–29). Early findings indicated that serotyping was informative and a promising approach. It requires access to sera from both symptomatic and asymptomatic humans. Serotyping limitations are related with the sensitivity and specificity of peptides investigated. The critical limitations are related with difficulties in differentiating the clonal genotype I and III, and all the non-archetypal genotypes (28, 29).
3. Methods to serotype T. gondii
Different strategies have been described to serotype T. gondii. Immunoenzymatic assays (ELISA) using polymorphic synthetic peptides are widely accepted (25, 28, 29); small peptides with a cysteine residue added to either the C or the N terminus of each peptide and a carrier protein such as keyhole limpet hemocyanin (KLH) (25), or synthetic peptides used by Sousa et al. (28) with a triple repeats of the polymorphic sequence in a single synthetic peptide. The use of synthetic peptides as ELISA antigens can be problematic due to the small size of the peptides. Such peptides are difficult to attach to the plate and consequently the sensitivity of the assay will be lower. To cross this problem and to facilitate peptide attachment to the plate and consequently improve ELISA sensitivity, small peptides can be coupled to carrier proteins (25). As an alternative, another approach was proposed and consisted on a trimer of the chosen peptide (if smaller than 20 aa) (28). This would increase the antigen size, with the objective of facilitating attachment to microplates and the antibody recognition. Peptides were coupled to the Immobilizer Amino plates (Nunc). These plates are specially adapted for small molecules, like peptides. The use of these plates replaces the role played by the carrier proteins. In another approach, polymorphic recombinant peptides expressed as polypeptides fused to Escherichia coli glutathione S-transferase or to a His-tag were employed as the target antigen in enzyme-linked immunosorbent assays (26, 30). The advantage is that bigger sequences, with more SNPs, are expressed for genotype I, II, and III. However, these recombinant polymorphic peptides discriminate genotype II, but cross-reactivity was found between genotype I and III. Peptide microarray assays has been assessed (27, 31–33). The major advantage of these assays is screening large number of peptides from different antigens. Furthermore, they provide higher sensitivity than ELISA-based assays (27, 30, 32). Similar conclusions were obtained comparing these two methods on other infectious diseases (34, 35).
Finally, a novel gold nanoparticles (AuNPs) assay improved the sensitivity for genotype II. This procedure uses AuNPs conjugated with a polymorphic synthetic peptide previously identified (25, 28), and presents values suggesting higher sensitivity when compared with the classical ELISA approach (36).
4. Antigens and peptides: Which are the best targets for serotyping
If we accept that specific antibodies elicited during toxoplasmosis are useful as biomarkers for T. gondii genotypes, a new question emerges: which are the best targets for serotyping? Polymorphic regions from native Toxoplasma antigens can be identified by multiple sequence alignments of genotype specific target antigens (25, 26, 29). Polymorphic peptides can be predicted using a bioinformatics-based approach, and synthesized, according to these criteria:
1. The content of charged and/or hydrophilic amino acids,
2. a predicted α-helical structure,
3. presence of proline residues
4. the presence of short regions of typically 8–12 aa that are bounded by 2 cysteine residues (25, 32)
5. high antigenic index, and
6. high surface probability (32).
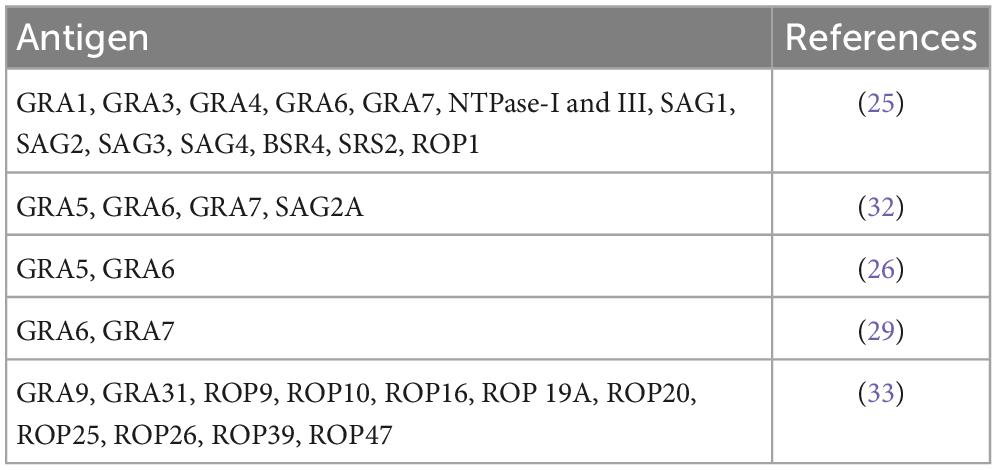
Different selected peptides derived from a variety of Toxoplasma antigens, such as granule dense proteins (GRA), surface antigens (SAG), rhoptry proteins (ROP), and NTPases, have been explored (25, 26, 29, 32, 33, 37). Table 2 provides global data. Curiously, a limited number of peptides were able to differentiate T. gondii serotypes, and we assumed that polymorphic site and length of peptides might play a pivotal role in serological typing (38).
Peptides derived from native SAG2, GRA3, GRA6, and GRA7 antigens proved ability to differentiate Toxoplasma strains in infected mice (25). However, these findings are not reproduced, completely, in humans. Meanwhile, GRA6 and GRA7 (Table 3) derived peptides evidences ability to differentiate genotype II from non-genotype II strains in human sera (25). The presence of a common epitope in the non-polymorphic amino acids of GRA6II peptide may explain this cross-reactivity. In order to verify this hypothesis, delimited versions of the GRA6II peptide were constructed, by truncation at the N terminus, that failed to improve the specificity of the original peptide, or deletion of the C-terminal residue, that eliminated false-positives, in mice (25).
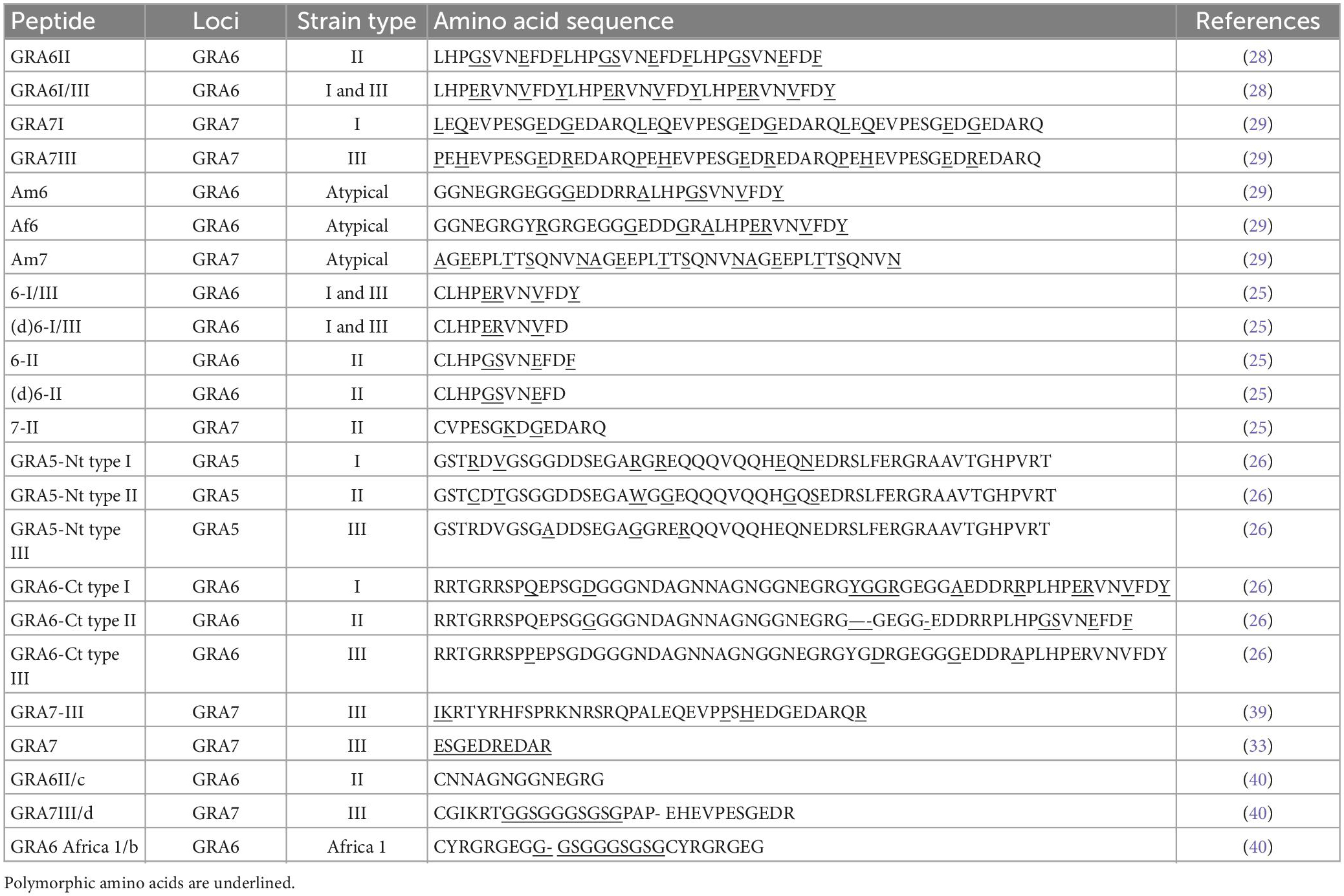
Table 3. Amino acid sequence of GRA5, GRA6, and GRA7 synthetic peptides that better discriminate the different serotypes.
Peyron et al. (26) proposed the use of recombinant polymorphic peptides of GRA5 and GRA6 antigens, specific for strains of T. gondii of genotypes I, II, and III (Table 3). The peptides were derived from the hydrophilic N-terminal region in native GRA5, the first 75 amino acids, and native GRA6 C-terminal region (172–230 amino acids). Derived peptides from GRA5 and GRA6 type 2 strains could differentiate genotype II strains from non-genotype II. Similar peptides derived from GRA5 and GRA6 genotypes I and III strains, failed to differentiate these two strains (30).
Sousa et al. (28) validated C-terminal GRA6-derived peptides for serotyping assay: peptide GRA6II, polymorphic specific for genotype II Toxoplasma strains, and the peptide GRA6I/III with polymorphisms specific for genotype I and III strains. Their findings were in accordance with genotyping results for 20 out of 23 cases associated with strains of the three main lineages. In addition, two peptides derived from the GRA7 locus with polymorphisms specific for genotype I (GRA7I) and genotype III (GRA7III) strains were investigated (29); peptide GRA7I provided only low sensitivity whereas peptide GRA7III appears to be informative for serotyping human infection with genotype III strains (29). A shorter version of the GRA7III C-terminal peptide was designed and specifically discriminates genotype III from non-genotype III infections in mice, rabbits, and humans (33). Xiao et al. (39) designed 12 peptides from GRA5, GRA6, and GRA7, specific for the archetypal genotypes I, II, and III. GRA5-derived peptides were from the N-terminal region and GRA6 and GRA7 from the C-terminus. One particular peptide, named GRA7-III distinguished genotype I from genotype III in infected mice with a specificity and sensitivity of 100% (39).
Xicotucal-Garcia et al. (37) designed new peptides of GRA6, GRA7, and SAG1 proteins. The peptides were designed with more SNPs, including linkers joining non-contiguous segments. Only with the GRA6 peptides it was possible to distinguish the three clonal genotypes.
5. Serotyping and non-archetypal genotypes
Non-archetypal strains, or atypical genotypes, exhibit, for the selected markers, a unique combination of alleles, discrete from the archetypal genotype, but also unique or different alleles. We hypothesize that infections with those atypical genotypes may induce a specific humoral response. Is it possible to provide serotyping assays for atypical genotypes using the polymorphic peptides defined so far, and based on archetypal strains? Mixed serum reactivity signatures, or reaction patterns exclusive of some non-archetypal strains, can be observed (28, 29, 38). Arranz-Solis et al. (38), using specific combinations of peptide ratios revealed unique profiles for the non-archetypal strains MAS (haplogroup 4), BOF (haplogroup 6), CAST (haplogroup 7), and Cougar (haplogroup 11). Furthermore, a distinct reactivity pattern using GRA6 derived peptides is able to distinguish haplogroup 11 and 12 from infections with archetypal North American/European strains (38).
Sequencing of loci GRA6 and GRA7 from 49 non-archetypal strains revealed 18 and 10 different amino acid sequences, respectively (29). Table 4 shows GRA6 polymorphisms of five non-archetypal strains, differing from clonal strains by one or two amino acids at the C-terminal region (SNP 223, 224, 227, and 230) (28). Three different sequences were described for the C-terminal region of those five atypical strains (Table 4). Strains GUY-2002-MAT and GUY-2002-KOE shared the same four SNPs as genotype I and genotype III strains, inducing the misclassification of the non-archetypal strains as genotype I/III. Strain GUY-2004-AKO differed from the peptide characterizing genotype I and III by a single amino acid. This single amino acid substitution was insufficient to distinguish Toxoplasma infections with strains harboring these alleles. Strains GUY-2003-BAS and GUY-2004-TER shared two SNPs with genotype II strains and two SNPs with genotype III strains. Strains that shared these polymorphisms displayed different reactivity profiles. The same atypical allele may induce a genotype II response, a genotype I/III response or a double response against both peptides.
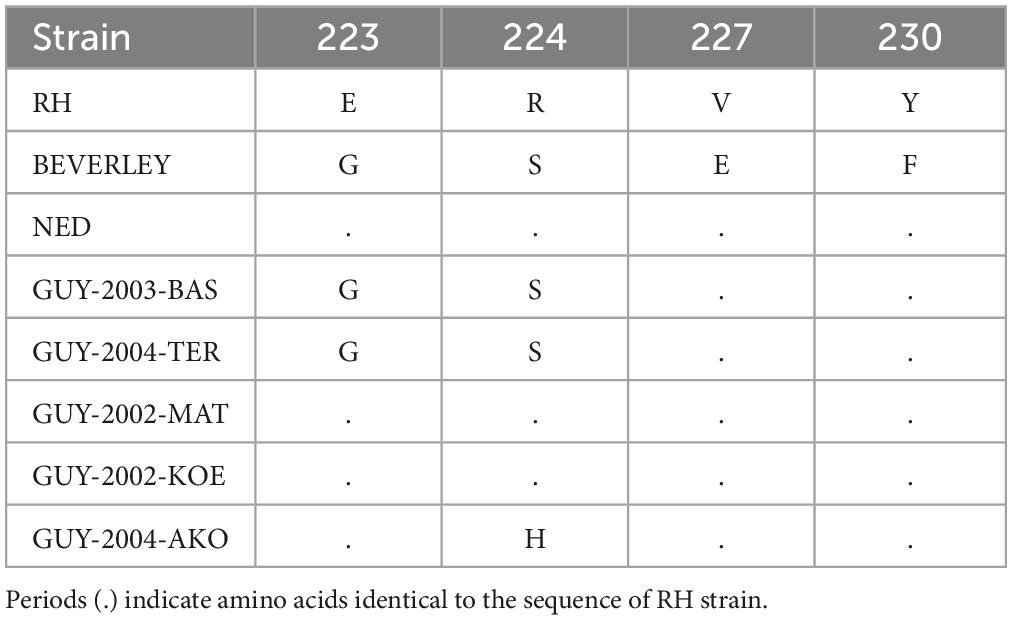
Table 4. C-terminal amino acid polymorphisms for GRA6 marker for genotype I (RH), genotype II (Beverley), genotype III (NED), and five non-archetypal strains (28).
Three peptides specific for some non-archetypal strains were selected from GRA6 and GRA7 antigens (29). From GRA7, peptide (Am7) contained polymorphisms specific for three haplotypes found in South America. Two other peptides were derived from GRA6, one specific for South American strains (peptide Am6) and the other specific for African strains (peptide Af6). These two peptides were selected from the most representative haplotypes among strains from Africa and South America (29). Peptides Am6 and Af6 had low specificities, since they react with sera from cases with strains belonging to the three archetypal lineages. Peptide Am7 was specific but poorly sensitive.
Another peptide from GRA6 locus, named GRA6 Africa 1/b was described for Africa 1 lineage (40). Peptide GRA6 Africa 1/b is derived from an insertion sequences found in genotype III and Africa 1 genotype. This epitope differs between genotype III and Africa 1 at position 206 involving a glycine to arginine substitution. This peptide had a sensitivity of 100% for Africa 1/b genotype, but cross-reacted with genotype II and III (40).
6. Serological signature, geographic origin, and disease outcome
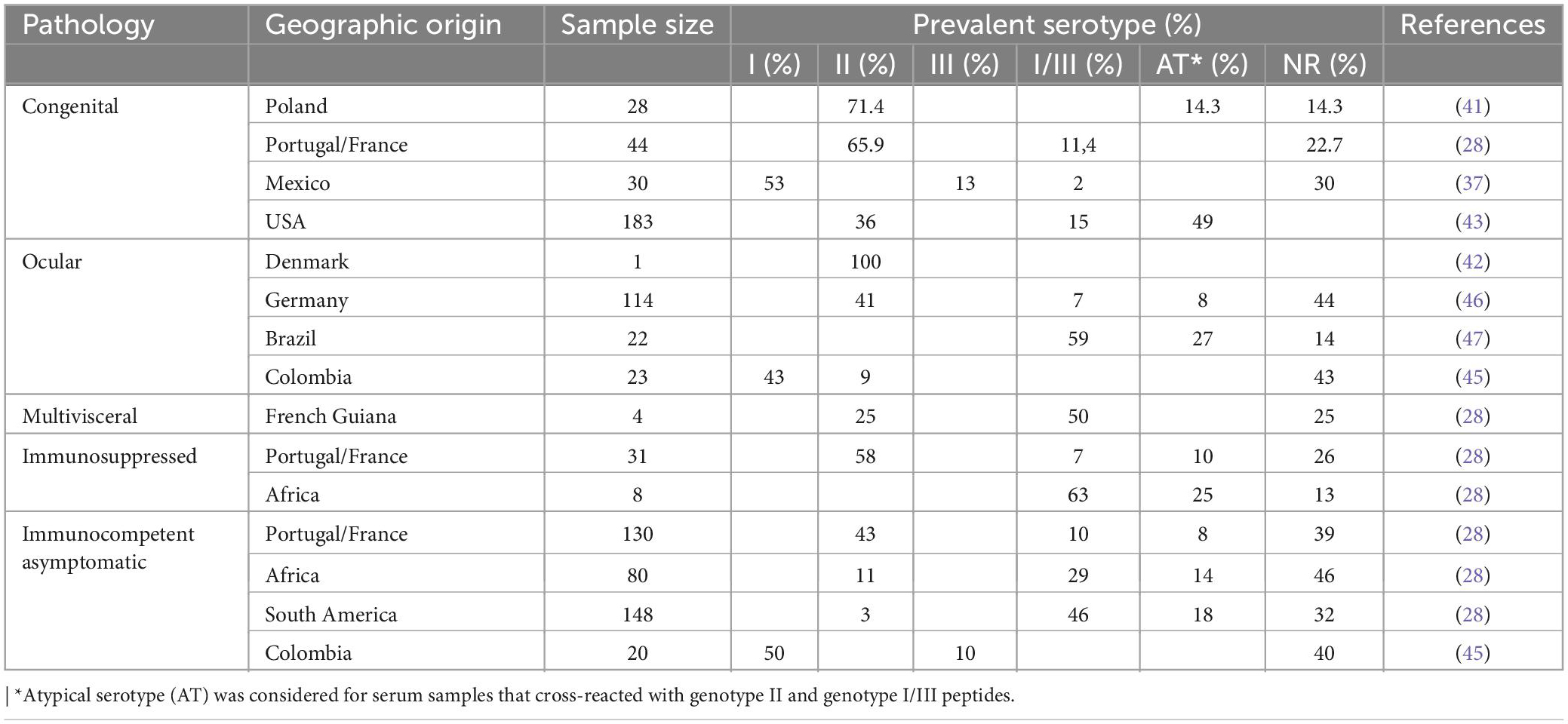
The serotype II profile is significantly more frequent in sera from Europeans, independent of clinical presentation of the disease (Table 5) (26, 28, 30, 40–43). In USA, serotype II was also associated with congenital toxoplasmosis (43). In contrast, serotypes I/III are more prevalent in South America, where non-archetypal genotypes prevail (26, 28, 30, 37, 44, 45). Few serotyping studies have been conducted in sera of African origin. However, as in South America, serotype I/III was also more prevalent than serotype II (28). Serotype I was associated with retinal lesions in patients from Colombia (45), and was the most frequent among congenital infections in Mexico (37).
Atypical serologic signatures with different reactivity against GRA6 and GRA7 type II and I/III were identified among Germans with ocular toxoplasmosis (46). Three of these atypical profiles had been identified in sera associated with a waterborne outbreak of human toxoplasmosis in Brazil (47). In addition, a novel non-reactive (NR) serotype was identified among Toxoplasma-infected uveitis cases in Germany who developed ocular toxoplasmosis (46). Non-archetypal strains typically possess entirely divergent polymorphic epitopes at GRA6 or GRA7, and these epitopes do not necessarily elicit antibodies capable of reacting with the diagnostic allelic peptide motifs. Alternatively, it is also possible that host genetic factors preclude the ability of ocular toxoplasmosis patients to produce allele-specific antibodies at GRA6 and GRA7 or that the polymorphic epitopes are not immunogenic in this patient cohort (48, 49). NR serotype patients with active ocular toxoplasmosis who have a higher risk of recurrences can be expected to receive appropriate management including preventive treatment.
7. Limitations, the future, and conclusion
Toxoplasma serotypes are defined by the serological profile based on the antibodies reaction to genotype-specific peptides. The peptides described thus far have limited utility. Those peptides only distinguished genotype II strains from non-genotype II. As a consequence, it is not possible to differentiate the clonal genotype III, and all the non-archetypal genotypes, as well as genotype I. The limited number of loci that have been investigated in depth raised the same problems found in single locus genotyping (28, 29). This is especially true in infections with non-archetypal strains (28). The epitope expressed by the infecting strain ultimately determined the reactivity against the diagnostic peptides, not the genotype of the infecting strain (47). For each lineage, peptides must be selected from as a high a number of antigens as possible. Therefore, in order to validate serotyping as an expedient method for genotyping and diagnosis, several peptides from different strain-specific markers need to be characterized (29).
In addition to specificity problems, the sensitivity of serotyping is a limitation given that some infections cannot presently be serotyped. Serotyping with a larger number of peptides, specific for both archetypal and non-archetypal strains can be anticipated to enhance the sensitivity of the method and consequently diminish the number of non-typed samples (29). For congenital toxoplasmosis and other T. gondii-related diseases (cerebral, pulmonary, and ocular) from Europe, the number of non-serotyped infections was 22.7 and 33.3%, respectively (28). Similar results were obtained for congenital infections from Poland (14.3%) by Nowakowska et al. (41). In addition, for immunocompetent asymptomatic cases from Europe and Africa serotyped with GRA6 archetypal peptides, 39.2 and 46.3%, respectively of the sera could not be serotyped (28). These non-reactive sera were later defined as a new serotype (46). Nevertheless, serotyping has clear potential in diagnosis. Given that it is non-invasive, serotyping can be readily deployed in large-scale epidemiological studies (32) with the objective of defining serotype distribution among the continents and in wild, feral, and domesticated hosts to understand whether strain selection is dependent on the host species. Regarding human infection, serotyping will enhance our understanding of the relationships between strain genotype and pathogenesis (28, 29, 46). Accomplishing these objectives will positively influence the diagnostic performance and treatment of the cases. Serotyping may be useful in the detection of a re-infection with a different strain, which may be particularly relevant in the follow-up of pregnant women. Last, since toxoplasmosis is a foodborne disease, serotyping can play a critical role for detection of T. gondii infections by non-archetypal strains in livestock and meat products destined for human consumption.
This review discusses the state of the art of the serological typing of T. gondii, known as serotyping. Many questions remain regarding host-parasite interaction, parasite biology and virulence factors. Understanding the role of strain-specific Toxoplasma antigens on infection and consequent host immune response underpins the selection of the best targets for in vitro genotype detection. Multilocus genotyping of T. gondii has revealed a markedly high genetic diversity. This population diversity is challenging to a serotyping approach, particularly for not clonal lineages. The many peptides suggested for genotype-focused serotyping, in tandem with the few only sensitive and specific peptides that have been reported reflects this difficulty. However, for clonal lineages, serotyping may become a routine. Selection of efficient markers is time and cost expensive, and there is still a long way to go. Nevertheless, we consider that this typing method offer promising advantages and a similar multilocus approach must be followed with peptides from different antigens. Indeed, we ourselves continue to investigate novel peptides for serotyping type II strains.
Author contributions
SS and MF wrote the manuscript. JC proposed the subject and was responsible for the critical review of the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by UIDB/00211/2020 and UIDP/00211/2020 with funding from FCT/MCTES through national funds.
Acknowledgments
The authors would like to thank Drs. Paul Brindley and Gabriel Rinaldi for English revision, critical suggestions, and helpful advices.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Dubey JP. Toxoplasmosis of Animals and Humans. 2nd ed. Boca Raton, FL: CRC Press, Taylor & Francis Group (2010).
2. Robert-Gangneux F, Dardé ML. Epidemiology of and diagnostic strategies for toxoplasmosis. Clin Microbiol Rev. (2012) 25:264–96. doi: 10.1128/CMR.05013-11
3. Montoya JG, Liesenfeld O. Toxoplasmosis. Lancet (2004) 363:1965–76. doi: 10.1016/S0140-6736(04)16412-X
5. Koutsoumanis K, Allende A, Alvarez-Ordóñez A, Bolton D, Bover-Cid S, Chemaly M, et al. Public health risks associated with food-borne parasites. EFSA J. (2018) 16:e05495. doi: 10.2903/j.efsa.2018.5495
6. Demar M, Ajzenberg D, Maubon D, Djossou F, Panchoe D, Punwasi W, et al. Fatal outbreak of human toxoplasmosis along the Maroni river: epidemiological, clinical, and parasitological aspects. Clin Infect Dis. (2007) 45:88–95. doi: 10.1086/521246
7. Demar M, Hommel D, Djossou F, Peneau C, Boukhari R, Louvel D, et al. Acute toxoplasmosis in immunocompetent patients hospitalized in an intensive care unit in French Guiana. Clin Microbiol Infect. (2012) 18:221–31. doi: 10.1111/j.1469-0691.2011.03648.x
8. Pomares C, Devillard S, Holmes TH, Olariu TR, Press CJ, Ramirez R, et al. Genetic characterization of Toxoplasma gondii DNA samples isolated from humans living in north America: an unexpected high prevalence of atypical genotypes. J Infect Dis. (2018) 218:1783–91. doi: 10.1093/infdis/jiy375
9. Boothroyd JC, Grigg ME. Population biology of Toxoplasma gondii and its relevance to human infection: do different strains cause different disease? Curr Opin Microbiol. (2002) 5:438–42. doi: 10.1016/S1369-5274(02)00349-1
10. Xiao J, Yolken RH. Strain hypothesis of Toxoplasma gondii infection on the outcome of human diseases. Acta Physiol. (2015) 213:828–45. doi: 10.1111/apha.12458
11. Galal L, Hamidović A, Dardé ML, Mercier M. Diversity of Toxoplasma gondii strains at the global level and its determinants. Food Waterborne Parasitol. (2019) 15:e00052. doi: 10.1016/j.fawpar.2019.e00052
12. Carme B, Bissuel F, Ajzenberg D, Bouyne R, Aznar C, Demar M, et al. Severe acquired toxoplasmosis in immunocompetent adult patients in French Guiana. J Clin Microbiol. (2002) 40:4037–44. doi: 10.1128/JCM.40.11.4037-4044.2002
13. Sibley LD, Khan A, Ajioka JW, Rosenthal BM. Genetic diversity of Toxoplasma gondii in animals and humans. Phil Trans R Soc B. (2009) 364:2749–61. doi: 10.1098/rstb.2009.0087
14. Howe DK, Sibley LD. Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J Infect Dis. (1995) 172:1561–6. doi: 10.1093/infdis/172.6.1561
15. Biñas M, Johnson AM. A polymorphism in a DNA polymerase α gene intron differentiates between murine virulent and avirulent strains of Toxoplasma gondii. Int J Parasitol. (1998) 28:1033–40. doi: 10.1016/S0020-7519(98)00052-6
16. Howe DK, Honoré S, Derouin F, Sibley D. Determination of genotypes of Toxoplasma gondii strains isolated from patients with toxoplasmosis. J Clin Microbiol. (1997) 35:1411–4. doi: 10.1128/jcm.35.6.1411-1414.1997
17. Sibley LD, Boothroyd JC. Virulent strains of Toxoplasma gondii comprise a single clonal lineage. Nature. (1992) 359:82–5.
18. Su C, Zhang X, Dubey JP. Genotyping Of Toxoplasma gondii By multilocus PCR-RFLP markers: a high resolution and simple method for identification of parasites. Int J Parasitol. (2006) 36:841–8. doi: 10.1016/j.ijpara.2006.03.003
19. Su C, Khan A, Zhou P, Majumdar D, Ajzenberg D, Dardé ML, et al. Globally diverse Toxoplasma gondii isolates comprise six major clades originating from a small number of distinct ancestral lineages. Proc Natl Acad Sci USA. (2012) 109:5844–9. doi: 10.1073/pnas.1203190109
20. Lorenzi H, Khan A, Behnke MS, Namasivayam S, Swapna LS, Hadjithomas M, et al. Local admixture of amplified and diversified secreted pathogenesis determinants shapes mosaic Toxoplasma gondii genomes. Nat Commun. (2016) 7:10147. doi: 10.1038/ncomms10147
21. Ajzenberg D, Bañuls AL, Dardé ML. Microsatellite analysis of Toxoplasma gondii shows considerable polymorphism structured into two main clonal groups. Int J Parasitol. (2002) 32:27–38. doi: 10.1016/S0020-7519(01)00301-0
22. Ajzenberg D, Dumètre A, Dardé ML. Multiplex PCR for typing strains of Toxoplasma gondii. J Clin Microbiol. (2005) 43:1940–3. doi: 10.1128/JCM.43.4.1940-1943.2005
23. Ajzenberg D, Collinet F, Mercier A, Vignoles P, Dardé ML. Genotyping of Toxoplasma gondii isolates with 15 microsatellite markers in a single multiplex PCR assay. J Clin Microbiol. (2010) 48:4641–5. doi: 10.1128/JCM.01152-10
24. Costa JM, Dardé ML, Assouline B, Vidaud M, Bretagne S. Microsatellite in the beta-tubulin gene of Toxoplasma gondii as a new genetic marker for use in direct screening of Amniotic fluids. J Clin Microbiol. (1997) 35:2542–5. doi: 10.1128/jcm.35.10.2542-2545.1997
25. Kong JT, Grigg ME, Uyetake L, Parmley S, Boothroyd JC. Serotyping of Toxoplasma gondii infections in humans using synthetic peptides. J Infect Dis. (2003) 187:1484–95. doi: 10.1086/374647
26. Peyron F, Lobry JR, Musset K, Ferrandiz J, Gomez-Marin JE, Petersen E, et al. Serotyping of Toxoplasma gondii in chronically infected pregnant women: predominance of type II in Europe and types I and III in Colombia (South America). Microbes Infect. (2006) 8:2333–40. doi: 10.1016/j.micinf.2006.03.023
27. Maksimov P, Zerweck J, Maksimov A, Hotop A, Gross U, Spekker K, et al. Analysis of clonal type-specific antibody reactions in Toxoplasma gondii seropositive humans from Germany by peptide-microarray. PLoS One. (2012) 7:e34212. doi: 10.1371/journal.pone.0034212
28. Sousa S, Ajzenberg D, Vilanova M, Costa J, Dardé ML. Use of GRA6-derived synthetic polymorphic peptides in an immunoenzymatic assay to serotype Toxoplasma gondii in human serum samples collected from three continents. Clin Vaccine Immunol. (2008) 15:1380–6. doi: 10.1128/CVI.00186-08
29. Sousa S, Ajzenberg D, Marle M, Aubert D, Villena I, Correia Da Costa J, et al. Selection of polymorphic peptides from GRA6 and GRA7 sequences of Toxoplasma gondii strains to be used in serotyping. Clin Vaccine Immunol. (2009) 16:1158–69. doi: 10.1128/CVI.00092-09
30. Morisset S, Peyron F, Lobry JR, Garweg J, Ferrandiz J, Musset K, et al. Serotyping of Toxoplasma gondii: striking homogeneous pattern between symptomatic and asymptomatic infections within Europe and South America. Microbes Infect. (2008) 10:742–7. doi: 10.1016/j.micinf.2008.04.001
31. Maksimov P, Zerweck J, Maksimov A, Hotop A, Gross U, Pleyer U, et al. Peptide microarray analysis of in silico-predicted epitopes for serological diagnosis of Toxoplasma gondii infection in humans. Clin Vaccine Immunol. (2012) 19:865–74. doi: 10.1128/CVI.00119-12
32. Maksimov P, Zerweck J, Dubey JP, Pantchev N, Frey CF, Maksimov A, et al. Serotyping of Toxoplasma gondii in cats (Felis domesticus) reveals predominance of type II infections in Germany. PLoS One. (2013) 8:e80213. doi: 10.1371/journal.pone.0080213
33. Arranz-Solis D, Cordeiro C, Young LH, Dardé ML, Commodaro AG, Grigg ME, et al. Serotyping of Toxoplasma gondii infection using peptide membrane arrays. Front Cell Infect Microbiol. (2019) 9:408. doi: 10.3389/fcimb.2019.00408
34. Melnyk O, Duburcq X, Olivier C, Urbes F, Auriault C, Gras-Masse H. Peptide arrays for highly sensitive and specific antibody-binding fluorescence assays. Bioconjug Chem. (2002) 13:713–20.
35. Mezzasoma L, Bacarese-Hamilton T, Di Cristina M, Rossi R, Bistoni F, Crisanti A. Antigen microarrays for serodiagnosis of infectious diseases. Clin Chem. (2002) 48:121–30.
36. Sousa S, Castro A, Correia da Costa J.M, Pereira E. Biosensor based immunoassay: a new approach for serotyping of Toxoplasma gondii. Nanomaterials. (2021) 11:2065.
37. Xicotucal-Garcia L, Enriquez-Flores S, Correa D. Testing new peptides from Toxoplasma gondii SAG1, GRA6, and GRA7 for serotypin: better definition using GRA6 in mother/newborns pairs with risk of congenital transmission in Mexico. Front Cell Infect Microbiol. (2019) 9:368. doi: 10.3389/fcimb.2019.00368
38. Arranz-Solis D, Carvalheiro CG, Zhang ER, Grigg ME, Saeij JPJ. Toxoplasma GRA peptide-specific serologic fingerprints discriminate among major strains causing toxoplasmosis. Front Cell Infect Microbiol. (2021) 11:621738. doi: 10.3389/fcimb.2021.621738
39. Xiao J, Buka SL, Cannon TD, Suzuki Y, Viscidi RP, Torrey EF, et al. Serological pattern consistent with infection with type I Toxoplasma gondii in mothers and risk of psychosis among adult offspring. Microbes Infect. (2009) 11:1011–8. doi: 10.1016/j.micinf.2009.07.007
40. Can H, Gokmen AA, Doskaya M, Alak SE, Doskaya AD, Karakavuk M, et al. Development of a new serotyping ELISA for Toxoplasma gondii type II, type III and Africa 1 lineages using in silico peptide discovery methods, well categorized feline and human outbreak serum samples. BMC Infect Dis. (2022) 22:110. doi: 10.1186/s12879-022-07088-w
41. Nowakowska D, Colon I, Remington JS, Grigg M, Golab E, Wilczynski J, et al. Genotyping of Toxoplasma gondii by multiplex PCR and peptide-based serological testing of samples from infants in Poland diagnosed with congenital toxoplasmosis. J Clin Microbiol. (2006) 44:1382–9.
42. Sandfeld L, Petersen E, Sousa S, Laessoe M, Milea D. Bilateral papillitis in ocular toxoplasmosis. Eye. (2010) 24:188–9. doi: 10.1038/eye.2009.55
43. McLeod R, Boyer KM, Lee D, Mui E, Wroblewski K, Karrison T, et al. Prematurity and severity are associated with Toxoplasma gondii alleles (NCCCTS, 1981-2009). Clin Infect Dis. (2012) 54:1595–605. doi: 10.1093/cid/cis258
44. de-la-Torre A, Sauer A, Pfaff AW, Bourcier T, Brunet J, Speeg-Schatz C, et al. Severe South American ocular toxoplasmosis is associated with decreased Ifn-γ/Il-17a and increased Il-6/Il-13 intraocular levels. PLoS Negl Trop Dis. (2013) 7:e2541. doi: 10.1371/journal.pntd.0002541
45. Mantilla-Muriel LE, Hernández-de-los-Ríos A, Rincón M, de-la-Torre A, Murillo-León M, Cardona N, et al. Serotyping, host genes and cytokines response in human ocular toxoplasmosis. Microb Pathog. (2020) 148:104465.
46. Shobab L, Pleyer U, Johnsen J, Metzner S, James ER, Torun N, et al. Toxoplasma serotype is associated with development of ocular toxoplasmosis. J Infect Dis. (2013) 208:1520–8. doi: 10.1093/infdis/jit313
47. Vaudaux JD, Muccioli C, James ER, Silveira C, Margargal SL, Jung C, et al. Identification of an atypical strain of Toxoplasma gondii as the cause of a waterborne outbreak of toxoplasmosis in Santa Isabel do Ivai. Brazil J Infect Dis. (2020) 202:1226–33. doi: 10.1086/656397
48. Alvarez C, de-la-Torre A, Vargas M, Herrera M, Uribe-Huertas LD, Lora F, et al. Striking divergence in Toxoplasma ROP16 nucleotide sequences from human and meat samples. J Infect Dis. (2015) 211:2006–13.
Keywords: serotype, Toxoplasma gondii, strain, antibodies, polymorphism, peptides
Citation: Sousa S, Fernandes M and Correia da Costa JM (2023) Serotyping, a challenging approach for Toxoplasma gondii typing. Front. Med. 10:1111509. doi: 10.3389/fmed.2023.1111509
Received: 29 November 2022; Accepted: 20 March 2023;
Published: 06 April 2023.
Edited by:
Jorge Enrique Gómez Marín, University of Quindío, ColombiaReviewed by:
Dolores Correa, Anahuac University of North Mexico, MexicoMarie-Laure Dardé, University of Limoges, France
Copyright © 2023 Sousa, Fernandes and Correia da Costa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Susana Sousa, adelaide.sousa@insa.min-saude.pt
 Susana Sousa
Susana Sousa Maria Fernandes5
Maria Fernandes5  José Manuel Correia da Costa
José Manuel Correia da Costa