Candida albicans Multilocus Sequence Typing Clade I Contributes to the Clinical Phenotype of Vulvovaginal Candidiasis Patients
- 1Department of Obstetrics and Gynecology, Peking University Shenzhen Hospital, Shenzhen, China
- 2Shenzhen PKU-HKUST Medical Center, Institute of Obstetrics and Gynecology, Shenzhen, China
- 3Shenzhen Key Laboratory on Technology for Early Diagnosis of Major Gynecological Disease, Shenzhen, China
- 4BGI-Shenzhen, Shenzhen, China
- 5Clinical College of Peking University Shenzhen Hospital, Anhui Medical University, Hefei, China
- 6Department of Laboratory Science, Peking University Shenzhen Hospital, Shenzhen, China
- 7Research Institute of Huazhong University of Science and Technology in Shenzhen, Shenzhen, China
Candida albicans is the most frequent fungal species responsible for vulvovaginal candidiasis (VVC), which exhibits distinct genetic diversity that is linked with the clinical phenotype. This study aimed to assess the genotypes and clinical characteristics of different C. albicans isolates from VVC patients. Based on multilocus sequence typing (MLST), clade 1 was identified as the largest C. albicans group, which appeared most frequently in recurrent VVC and treatment failure cases. Further study of antifungal susceptibility demonstrated that MLST clade 1 strains presented significantly higher drug resistance ability than non-clade 1 strains, which result from the overexpression of MDR1. The mRNA and protein expression levels of virulence-related genes were also significantly higher in clade 1 isolates than in non-clade 1 isolates. Proteomic analysis indicated that the protein stabilization pathway was significantly enriched in clade 1 strains and that RPS4 was a central regulator of proteins involved in stress resistance, adherence, and DNA repair, which all contribute to the resistance and virulence of MLST clade 1 strains. This study was the first attempt to compare the correlation mechanisms between C. albicans MLST clade 1 and non-clade 1 strains and the clinical phenotype, which is of great significance for VVC classification and treatment.
1. Introduction
Nearly three-quarters of healthy women experience at least one episode of vulvovaginal candidiasis (VVC) during their lifetimes, and 5–10% of women suffer from recurrent VVC (RVVC), defined as at least three episodes of VVC within 1 year (1). The high recurrence rate has been a difficult clinical problem for decades due to various complex factors (2). Invasive fungal pathogens, the root etiology of VVC, harbor different genotypes that lead to varying degrees of host immune responses and further determine different infection outcomes, antifungal drug efficacy and recurrence probability. Candida albicans is the most frequent fungal species responsible for VVC and exhibits distinct phenotypic and genetic diversity (3). The genotype distribution of C. albicans strains has been associated with different clinical conditions of VVC (4, 5). The genotypic differences of C. albicans strains retrieved from VVC patients and healthy women reveal that VVC-linked strains possess specific genotypes (6). Hence, studying the genotypes of different subspecies of C. albicans strains will contribute to understanding their pathogenic mechanisms, which is important for increasing the recovery rate and controlling the infection rate.
Different molecular methods have been applied to identify the genotypes of C. albicans strains, including non-DNA-based methods, DNA-based conventional typing methods and exact DNA-based techniques (7). Multilocus enzyme electrophoresis (MLEE), a classic non-DNA-based method, has been replaced by conventional DNA-based typing methods with higher resolution, including electrophoretic karyotyping (EK), random amplified polymorphic DNA (RAPD), restriction fragment length polymorphism (RFLP), and repetitive extragenic palindromic element PCR (REP-PCR) (3). Southern blot hybridization with the DNA fingerprinting probe Ca3 is a widely used technique for the exact typing of C. albicans isolates, but the experimental process is particularly complex, so it was replaced by microsatellite length polymorphism (MLMT) and multilocus sequence typing (MLST). The MLST method directly analyzes polymorphisms within seven housekeeping DNA markers and exhibits high discriminatory power and reproducibility (8). To date, a public database for MLST data for C. albicans has been established (http://calbicans.mlst.net) that can be used to evaluate the worldwide diversity of C. albicans. The population structure of C. albicans consists of 18 well-defined clades (9) and is associated with epidemiological characteristics, such as the infection's anatomical location, geographical distribution, and antifungal drug susceptibility (10, 11). However, new clades have been successively identified, with an increase in the number of isolates from the baseline, which has also resulted in a more diversified association. Therefore, additional isolates with detailed epidemiological characteristics are needed to provide a more accurate correlation between the genotype of C. albicans and the phenotype of VVC, which will be helpful for guiding clinical diagnosis and treatment.
The drug resistance and virulence factors of the strain are also closely associated with the clinical characteristics of VVC, which can be attributed to the phenotypic plasticity and high genetic diversity of strains. A previous study indicated that the reduction in the 5-fluorocytosine susceptibility of MLST clade 1 strains was linked to mutation of the FUR1 gene (12), and isolates with significantly lower levels of acid phosphatase activity were clustered in MLST clade 2, which resulted in a higher virulence phenotype of the strains, as well as a higher number of tandem repeat sequences in the hyphal regulator genes (HYR1 and HYR3) and the agglutinin-like sequence (ALS) cell surface protein-encoding genes (13). Therefore, the phenetic and genetic profile related to the fungal susceptibility and virulence properties of clinical C. albicans isolates is necessary for optimizing the treatment methods for VVC and controlling the transmission of drug resistance. Furthermore, comparative proteomic studies have been performed to explain the mechanism of drug resistance in untreated control cells and cells treated with antifungal drugs (14–16), as well as avirulent and virulent C. albicans strains (17). However, the proteomic profiles among different MLST genotype clusters remain unclear.
In the current study, we aimed to assess the relevance of the association between the clinical characteristics and the MLST phenotype of C. albicans isolates from VVC patients and explain the MLST clade-specific antifungal drug resistance and virulence properties. This information will greatly support the graded diagnosis and treatment strategy of VVC.
2. Materials and Methods
2.1. The Isolation of C. albicans Strains
This study was approved by the Medical Review Board of Peking University Shenzhen Hospital. All C. albicans were isolated from vaginal samples of VVC patients in the Department of Obstetrics and Gynecology, Peking University Shenzhen Hospital, between April 2003 and December 2016. All the clinical information of the participants, including age, pregnancy status, and treatment history and results, was recorded after a consent form was signed.
Vaginal samples from the lateral vaginal wall were collected with a sterile cotton-tipped swab. The swab was placed in a tube filled with saline for microscopic examination with 10% potassium hydroxide. Specimens were simultaneously inoculated on Sabouraud's dextrose agar (SDA) plates and CHROMagar Candida plates (Biocell Laboratory Ltd., Zhengzhou, China) for 24–48 h at 37°C in ambient air. Single colonies obtained from the SDA plates were identified using the standardized Candida API system (bioMerieux, Marcy l'Etoile, France) and stored in medium containing 2% glucose, 2% peptone, and 20% glycerol at −70°C.
2.2. DNA Extraction and MLST Genotyping of the Isolates
The DNA of the isolates was extracted by an EZNA TM Yeast DNA Kit (Omega Bio-Tek, USA) according to the manufacturer's instructions. For MLST identification, seven housekeeping genes (AAT1a, ACC1, ADP1, MPIb, SYA1, VPS13, and ZWF1b) were amplified and sequenced. The primers are shown in Table 1. The sequences were uploaded to the C. albicans MLST database (http://calbicans.mlst.net) for genotype identification. Then, DNA sequences of the seven marker genes were concatenated for cluster analysis and clade definition, and a phylogenetic tree was constructed by the unweighted-pair group method using average linkages (UPGMA) according to the p-distance model (p = 0.04) with 1,000 replicates for bootstrap testing (MEGA 6.0). Additionally, we identified the ABC genotype and mating type locus (MTL) homozygosity of each isolate according to previously described methods (18, 19).
2.3. RNA Extraction and Quantitative Real-Time PCR
The RNA of the isolates was extracted by an EZNA TM Yeast RNA Kit (Omega Bio-Tek, USA) and reverse transcribed to cDNA using a Takara PrimeScriptRT Reagent Kit with gDNA Eraser (Takara, Japan) according to the manufacturer's instructions. Quantitative real-time PCR analysis of the genes related to drug resistance, including CDR1, CDR2, MDR1, and ERG11, and virulence factors, including ALS1-7, ALS9, HWP1, SAP1-SAP10, PLB1-3, and PLB5, was performed as described previously (22, 23, 25). The ACT1 gene was used as a housekeeping gene. Data for each target gene were calculated as the fold change in comparison with the reference gene ACT1 using the ΔΔCt quantification method. The primers used for quantitative real-time PCR analysis are shown in Table 1.
2.4. Antifungal Susceptibility Testing
The susceptibilities of the isolates to antifungal drugs were assayed by using the broth microdilution method according to the proposed Clinical and Laboratory Standards Institute M27-A3 and M60 standard guidelines (26, 27). In total, 14 antifungal drugs were selected for susceptibility testing, and they were grouped into five categories: imidazoles, triazoles, polyenes, echinocandins, and others. The fat-soluble drugs, including itraconazole (ITC), miconazole (MCA), clotrimazole (CLO), voriconazole (VRC), terconazole (TEC), butoconazole (BUC), amphotericin B (AMB), terbinafine (TRB), caspofungin (CFG), micafungin (MFG), and anidulafungin (AFG), were dissolved in DMSO, and the storage concentration was at least 100 times greater than the test concentration to avoid the influence of DMSO. Water-soluble drugs, including fluconazole (FLC), nystatin (NYS), and 5-fluorocytosine (5FC), were dissolved in sterile normal saline solution. The test concentration range of FLC, NYS, and 5FC was 0.125–64 μg/ml, and their storage concentration was 1,280 μg/ml. The test concentration range of ITC, MCA, CLO, VRC, TEC, BUC, AMB, and TRB was 0.0313–16 μg/ml, and their storage concentration was 3,200 μg/ml. The test concentration range of CFG, MFG, and AFG was 0.015–8 μg/ml, and their storage concentration was 1,600 μg/ml. For susceptibility testing, 5 single colonies with a diameter of ~1 mm were picked and suspended in 5 ml of sterile normal saline solution. After vortexing, the bacterial suspension was diluted to 1 × 103 CFU/ml −5 × 103 CFU/ml with RPMI1640 medium. Then, a mixture of 100 μl bacterial suspension and 100 μl drug solution was added to 96-well plates and incubated at 35°C. The minimum inhibitory concentration (MIC) values for AMB and NYS were determined as the minimum concentrations required for completely clear culture after 48 h of incubation. The MIC values for imidazoles, triazoles, and 5FC were determined as the minimum concentrations required for the culture to have a 50% turbidity decrease after 48 h of incubation. The MIC values for echinocandins were determined as the minimum concentrations required for the culture to have a 50% turbidity decrease after 24 h of incubation. The MIC of each isolate was recorded, and MIC90, which is the minimum inhibitory concentration required to inhibit the growth of 90% of isolates in each clade, was calculated.
2.5. Extracellular Enzyme Assays
The production of extracellular enzymes, such as esterase, hemolysis enzyme, and phospholipase, was measured to reflect the virulence of the strains. The esterase activity and hemolysis enzyme production were evaluated using the plate assays described by Pakshir et al. (28), while the phospholipase activity was determined using base medium modified by adding 0.15% potassium tellurite to the egg yolk emulsion to improve the visualization of the halos of precipitation and the accuracy of the measurements (29).
2.6. Quantitative Proteomic Analysis
In total, six strains were randomly selected for the proteomic study, including three strains from MLST clade 1 (2 isolates in this study and the type strain C. albicans ATCC90028) and three non-clade 1 strains. Proteomic analysis was carried out using liquid chromatography-tandem mass spectrometry (LC-MS/MS; SWATH-MS) by Wuhan GeneCreate Biological Engineering Co., Ltd, China (http://www.genecreate.com/). Total proteins were extracted using the ultrasonic crushing method (30). An AB SCIEX nano LC-MS/MS (Triple TOF 5600 plus) system was used for liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) analysis. Spectral library generation and SWATH data processing were performed using Skyline version 2.5 software (Department of Genome Sciences, University of Washington). The interactions among proteins with significantly different abundances were analyzed and visualized by Cytoscape (version 3.8.2).
2.7. Statistical Analysis
The statistical significance of the results was determined by the Wilcoxon test. The results were considered statistically significant when the p-values were <0.05.
3. Results
3.1. MLST Clade 1 Dominates the C. albicans Isolate Genotype Distribution
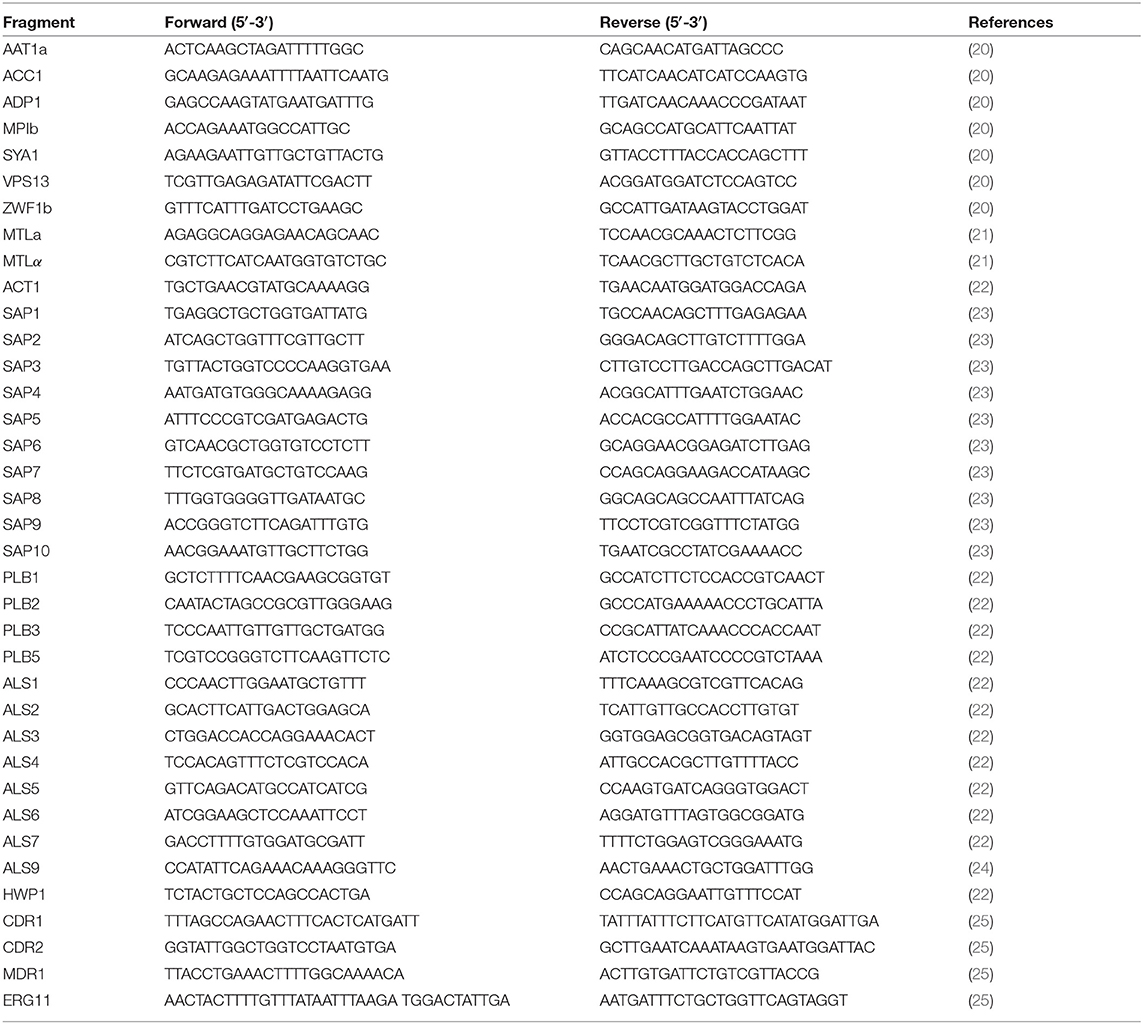
A total of 306 C. albicans isolates were analyzed by MLST (Supplementary Table 1). Single nucleotide polymorphisms (SNPs) of housekeeping genes were found after alignment, and the VPS13 and MPIb genes showed the highest rates of variation. Based on MLST database annotation, 76 diploid sequence types (DSTs) were identified, of which DST79 and DST1867 were the most common, including 79 strains and 71 strains, respectively. Moreover, 28 novel DSTs (DST3115-DST3148) were discovered. A dendrogram demonstrated that the 306 C. albicans isolates were clustered into 14 clades, including 11 known clades reported by Odds et al. (31) and 3 novel clades (Figure 1). Clade 1 was the largest group, accounting for 66% of the total isolates (202/306), followed by clade 8 (22/306), clade 14 (18/306), clade 17 (13/306), clade 15 (9/306), clade 11 (8/306), clade 4 (7/306), clade 3 (6/306), clade 12 (4/306), clade 6 (3/306), and clade 9 (1/306). The three novel clades discovered in this study were formed by three strains (E173B, 9028C, and 8633), which each constituted a single clade and were designated clade 101, clade 102, and clade 103, respectively. In addition, type A and heterozygotes were the main groups identified by ABC and MTL genotyping, respectively (Supplementary Table 1).
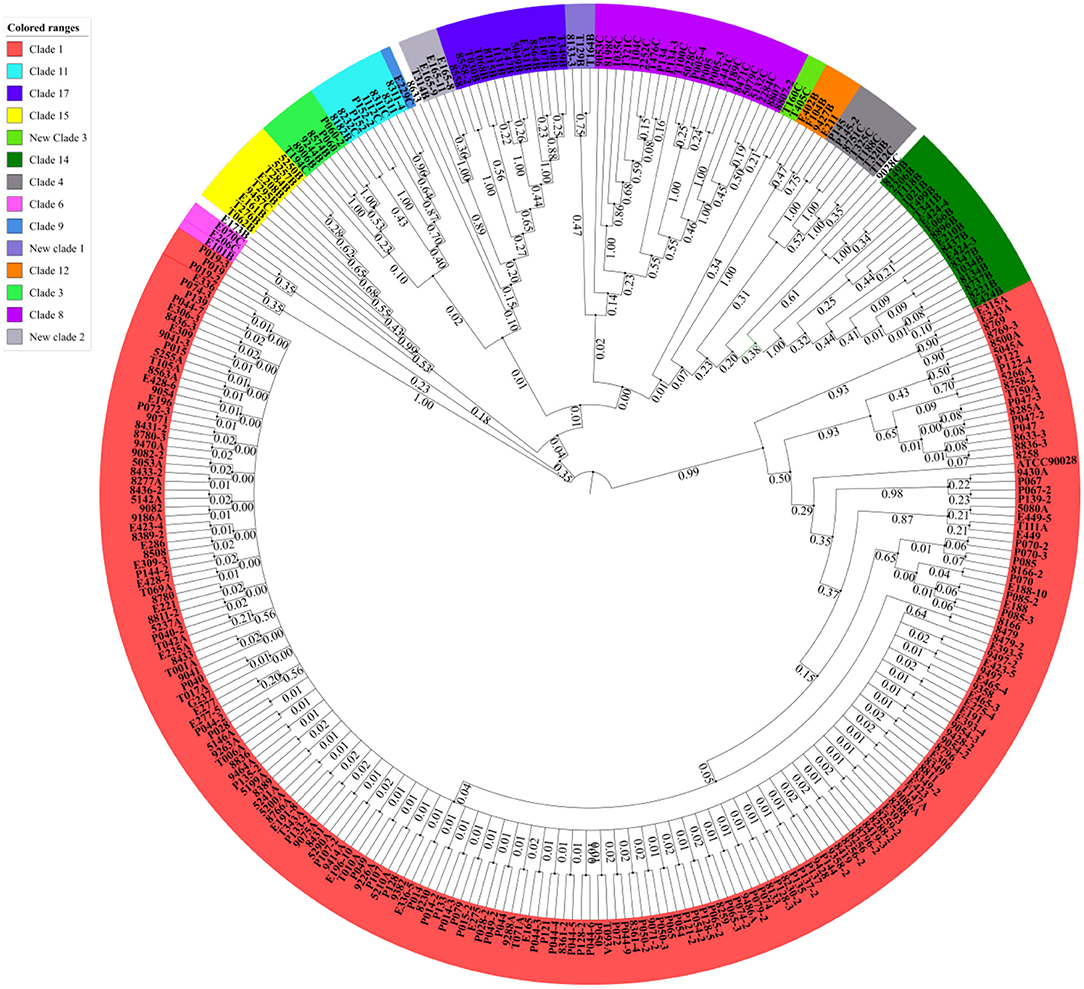
Figure 1. Candida albicans isolated from VVC patients were mainly clustered in MLST clade 1. The MLST phylogenetic tree was constructed by UPGMA according to the p-distance model (p = 0.04) with 1,000 replicates for bootstrap testing. In total, 306 strains were clustered into 14 clades and three singletons, including 11 known clades and three novel clades. Clade 1 was the largest group, accounting for 66% of the total isolates.
3.2. Worse Clinical Presentation of VVC Associated With C. albicans MLST Clade 1 Strains
A total of 306 C. albicans strains were isolated from VVC patients with different clinical characteristics, and the relationship between the clinical characteristics and strains is shown in Figure 2. Among the strains, 79 were from pregnant patients, and they were mostly from MLST clades 1, 3, 4, 8, and 9; this distribution was not significantly different from that of strains from nonpregnant patients. In addition, isolates from MLST clade 1 occurred most frequently in RVVC cases and treatment failure cases, as well as genotype A isolates (Supplementary Figure 1). Moreover, for the 44 treatment failure cases, the strains isolated from their subsequent samples were all identified as having the same MLST genotype, which mostly belonged to clade 1 (Supplementary Table 2). These results indicated that VVC caused by invasive C. albicans belonging to clade 1 was more difficult to cure and more likely to relapse than VVC caused by non-clade 1 C. albicans. In addition, the severity of VVC symptoms at the time of the patient's visit was diagnosed according to a published Chinese specification (32) and recorded. Although severe cases accounted for the highest percentage within MLST clade 14, the cure rate reached 77.8% (14/18). This result revealed that the strains in clade 14 had higher virulence but lower drug resistance. The same phenotype was present in genotype B and homozygous isolates (Supplementary Figure 1).
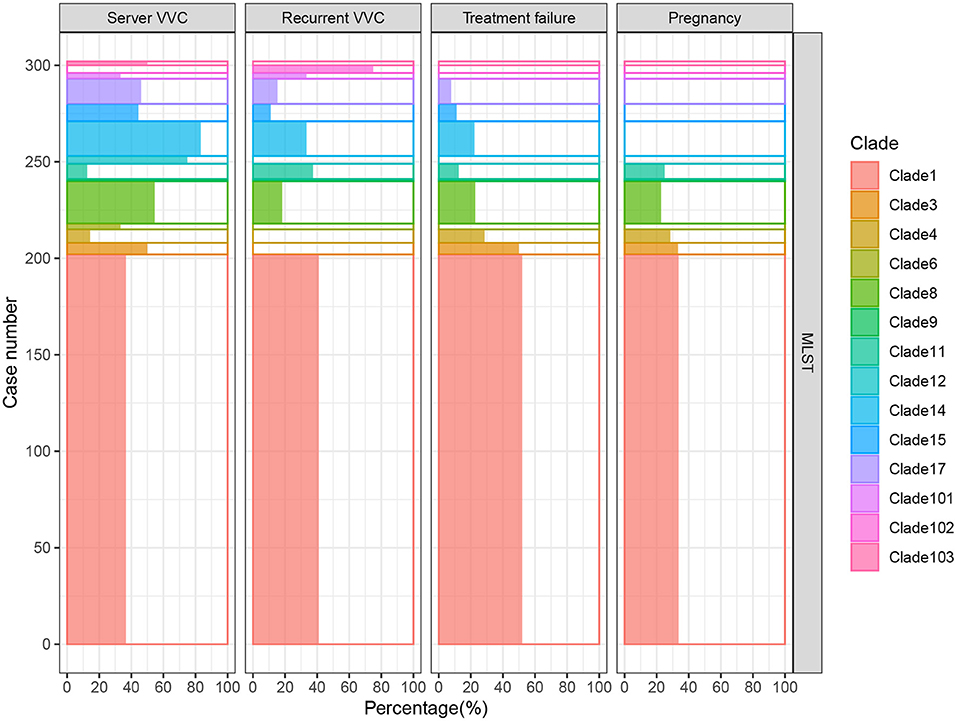
Figure 2. RVVC cases and VVC cases with treatment failure were mainly caused by C. albicans in MLST clade 1. In total, 306 samples were analyzed, and the 14 detected clades are presented in different colors. The percentage of clinical characteristics, including severe VVC, RVVC, treatment failure, and pregnancy cases, in each group is shown on the X axis. Clade 1 was dominant in RVVC and treatment failure cases, which demonstrated that VVC caused by invasive clade 1 C. albicans was more difficult to cure and relapsed more easily. Although isolates of clade 14 accounted for the highest proportion of severe VVC cases, the cure rate reached 77.8% (14/18). This result revealed that the strains in clade 14 had higher virulence but lower drug resistance.
3.3. Lower Antifungal Susceptibility and Gene/Protein Expression in MLST Clade 1
The MICs of 14 different drugs were determined for each C. albicans isolate (Supplementary Table 3). The MIC90 for each MLST clade was calculated and is shown in Figure 3. In general, all of the strains were sensitive to AFG but resistant to TRB. Although some non-clade 1 strains presented the highest resistance to several antifungal drugs, such as the clade 3 resistance to VRC and clades 12 and 15 resistance to 5FC, MLST clade 1 isolates showed the broadest spectrum of antifungal drug resistance. In addition, two of the three new clades discovered in this study, clades 101 and 102, showed the highest tolerance to NYS and FLC, respectively. This result indicates that the C. albicans in these two new clades not only had distinct genotypes but also exhibited different phenotypes. Furthermore, the difference in antifungal susceptibility was calculated between MLST clade 1 and non-clade 1 isolates, and the MIC values of clade 1 for all of the examined drugs were significantly higher than those of non-clade 1 strains, except that for MFG (Figure 3B). In addition, the MIC values of C. albicans strains in each MLST clade for the antifungal drugs are presented in Supplementary Figure 2 and Supplementary Table 3, and the statistical analysis based on Wilcoxon tests between clades also showed significantly different resistance abilities in clade 1. Interestingly, the new clade 102 manifested significantly higher VRC and FLC resistance than the other clades.
Figure 3. The susceptibility of C. albicans isolates in MLST clade 1 to antifungal drugs was significantly lower than that of non-clade 1 isolates. The MIC90 of each clade for 14 different antifungal drugs is presented (A), revealing that the drug resistance of isolates from clade 1 was higher than that of other isolates. The Wilcoxon test was performed based on the MICs of each strain between clade 1 and non-clade 1 (B). The resistance to all the antifungal drugs of the strains in clade 1 was significantly higher than that of non-clade 1 strains, except for resistance to MFG.
The mRNA expression of antifungal resistance genes was determined in this study to explore the mechanisms of the molecular resistance of C. albicans in different genotypes, as well as the expression of the proteins (Supplementary Figures 3A,B). Overall, the mRNA expression levels of resistance genes varied among different clades. The expression levels of the MDR1 and ERG11 genes were significantly higher (p < 0.0001) in MLST clade 1 strains than in non-clade 1 strains, while genes belonging to the CDR family showed no significant difference between clade 1 and other clades (Figure 4A). Proteins encoded by the MDR, ERG, and CDR gene families were also annotated by proteomic analysis, while only the protein amount of MDR1 was significantly different between MLST clade 1 strains and non-clade 1 strains, which is consistent with the corresponding transcriptional results (Figure 4B).
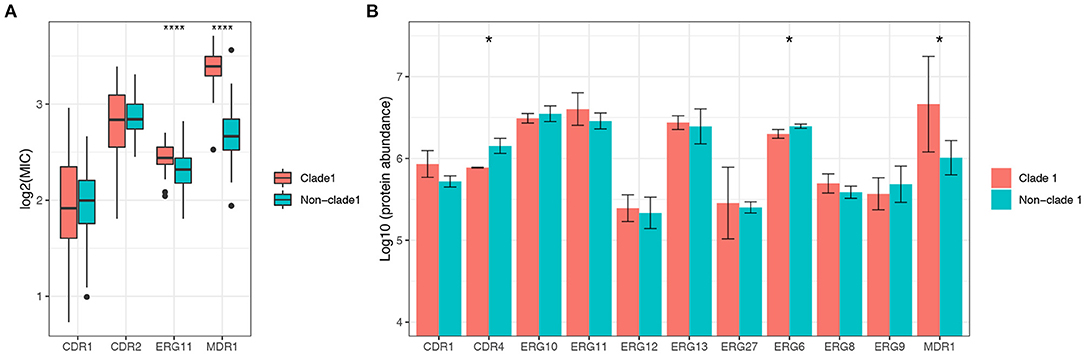
Figure 4. The expression level of the multidrug resistance gene MDR1 of C. albicans in MLST clade 1 isolates was significantly higher than that in non-clade 1 isolates. The mRNA expression levels of ERG11, CDR1, CDR2, and MDR1 were determined in 306 isolates (A), while ERG11 and MDR1 exhibited significantly higher expressions in clade 1 than in other clades (***p < 0.0001). Consistently, the proteomics results (B) also showed that the expression of MDR1 protein was significantly higher in clade 1 (*p < 0.05). The CDR4 and ERG6 proteins also presented significant differences in expression between clade 1 and non-clade 1 isolates.
3.4. Higher Virulence Characteristics Exhibited by MLST Clade 1 Isolates
The expression levels of virulence-related genes from the secretory aspartyl proteinase (SAP), hyphal wall protein (HWP), PLB, and ALS families were determined, and the results indicated that the virulence transcriptional pattern varied among different clades (Supplementary Figures 3C,D), as 10 out of 23 genes showed significantly different mRNA expression levels between clade 1 and non-clade 1 (Figure 5A). However, only seven proteins from these four virulence gene families were detected by proteomic analysis (Figure 5B), and the statistical analysis results were different. The ALS1 protein was expressed at non-significantly higher levels in clade 1, but its mRNA expression was significantly higher in clade 1 strains. The ALS2 protein was expressed at significantly higher levels in clade 1, but no significant difference was shown in mRNA expression. Moreover, all of the tested C. albicans strains were positive for phospholipase, esterase and hemolysis enzyme production, whereas there were no statistically significant differences in extracellular enzymatic activity among the different genotypes of C. albicans (Supplementary Figure 4 and Supplementary Table 4).
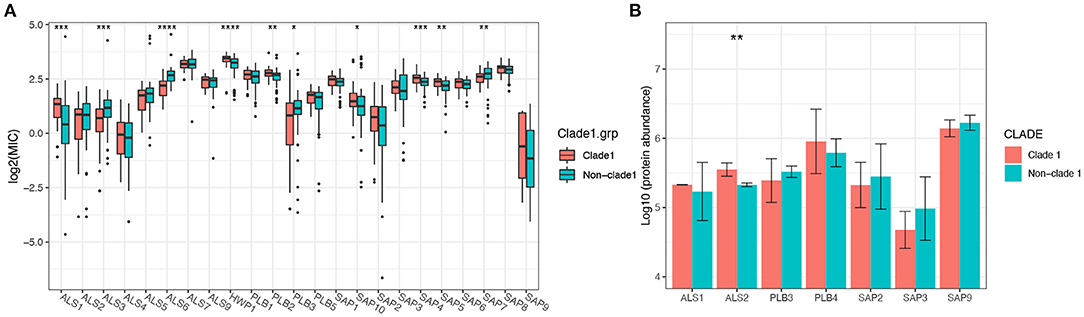
Figure 5. The mRNA expression levels of the virulence genes of C. albicans isolates varied significantly between MLST clade 1 and non-clade 1, while only ALS2 presented a significantly higher protein expression level. In total, 10 out of 23 genes from four virulence families, including SAP, HWP, PLB, and ALS, showed significantly different mRNA expression levels between clade 1 and non-clade 1 (A). However, only seven proteins from these four virulence gene families were detected by proteomic analysis (B), and ALS2 showed a significant difference between clade 1 and non-clade 1. *p < 0.05. **p < 0.01. ***p < 0.001. ****p < 0.0001.
3.5. Differences in the Proteomic Expression Profile Between Clade 1 and Non-clade 1 C. albicans Isolates
In total, 1,884 proteins were identified by proteomic analysis, and 1,677 proteins were subcategorized based on the GO classification biological process (23), cellular component (10), and molecular function (12). After statistical analysis, 41 proteins were found to be significantly differentially expressed between C. albicans MLST clade 1 isolates and non-clade 1 isolates (fold change > 2, p < 0.01, false discovery rate (FDR) < 0.01), among which 21 proteins were expressed at significantly higher levels in clade 1 isolates. The differentially expressed proteins were significantly enriched in five metabolic pathways, protein stabilization, the regulation of biological quality, poly(A)+ mRNA export from the nucleus, cytokinesis, and carbon metabolism (Figure 6A), which are pathways that contribute to tolerance, adaptation, and proliferation. The relationship between the differentially expressed proteins was analyzed, and the protein encoded by the RPS4 gene was discovered to be a central regulator that correlated with the proteins involved in the properties that influenced the resistance and virulence of the strain, such as stress resistance (HSP90, SGT1, SOD1, STI1, and YPD1), adherence (ALS2, PFK1, and SPA2), and DNA repair (HAT2, RFC1, and HTA2) (Figures 6B,C).
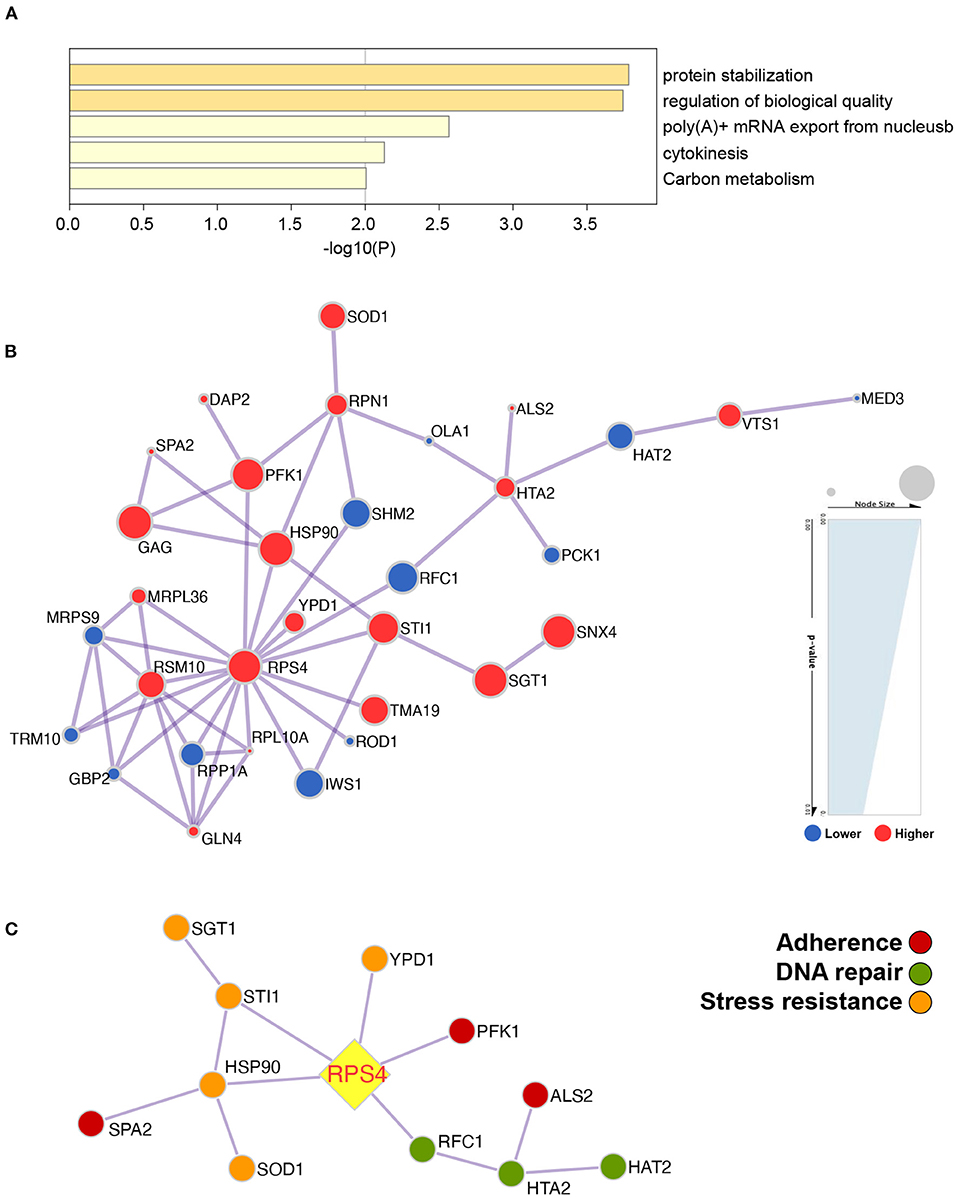
Figure 6. The protein profile demonstrated that the metabolic pathways were significantly different between C. albicans clade 1 isolates and non-clade 1 isolates and that RPS4 was the key gene linked to the proteins influencing the pathogenicity of C. albicans. The differentially expressed proteins were enriched in the pathways of protein stabilization, regulation of biological quality, poly(A)+ mRNA export from the nucleus, cytokinesis, and carbon metabolism (A). The network of the proteins that were expressed significantly differently between clade 1 and non-clade 1 isolates was analyzed (B), and RPS4 was the key protein associated with the stress resistance (HSP90, SGT1, SOD1, STI1, and YPD1), adherence (ALS2, PFK1, and SPA2), and DNA repair (HAT2, RFC1, and HTA2) of C. albicans (C).
4. Discussion
The MLST used in this study is based on nucleotide sequence analysis of the internal regions of multiple independent housekeeping genes and has proven to be a useful method for epidemiological investigation of clinical isolates of C. albicans. MLST studies have been conducted on C. albicans isolates from the digestive tract, blood infections, oral leukoplakia, and vaginal secretions (33–37). In addition, MLST has been applied widely to study nosocomial infection, which is important for the prevention and control of infection (38, 39). Therefore, fine genotyping for clinically pathogenic C. albicans has important implications for the epidemiology of candidal infections, especially for infections with high incidence, such as VVC. The genotype of C. albicans isolates collected from the vaginal samples of Chinese VVC patients and asymptomatic volunteers was explored in a previous study, and the results showed that 71.6% of the VVC isolates were classified in MLST clade 1, while a significantly lower proportion (40.6%) of the vaginal isolates from asymptomatic volunteers were classified in clade 1 (5). In our current study, MLST clade 1 was also the most common group in isolates from patients with VVC, which indicated that additional attention should be given to the difference between C. albicans MLST clade 1 and non-clade 1 strains. In contrast to previous studies, the clinical characteristics of VVC patients were examined in this study. We found that clade 1 isolates accounted for the highest percentage of the RVVC and treatment failure cases, which demonstrated that VVC caused by invasive C. albicans belonging to clade 1 was more difficult to cure and more likely to relapse. This finding suggests that novel treatment methods need to be proposed to prevent VVC recurrence, such as vaginal microbiota regulation or immunotherapies.
The C. albicans strains of VVC treatment failure cases were also evaluated in this study. The MLST genotypes of the strains isolated from samples from recurrent cases were the same as those of the original infecting strains, which were mostly from MLST clade 1. Recurrence could be caused by uneradicated candida or repeated infection of candida from the same living and working environment due to its conserved genetic signatures and strong infection ability. Moorhouse et al. performed MLST on 42 C. albicans strains from six patients with chronic cutaneous candida disease at multiple body sites across multiple time points over 17 months and found that the genotype of strains isolated from each individual patient was stable (40). It has also been confirmed that C. albicans can stay in one location for a long time and occasionally show microvariation (41). Notably, this genotype characteristic was more likely to occur in MLST clade 1 strains, which indicated that the strains may have higher resistance and growth abilities with stable inheritance.
The development of antifungal resistance in C. albicans is one of the crucial reasons for treatment failure and recurrence. Hence, it is necessary for clinicians to understand the local distribution of C. albicans susceptibility to antifungal drugs prior to treatment initiation. It has been reported that the decreasing cure rate of VVC was closely related to the increasing use of azoles (42, 43), and the susceptibility profiles have varied in different cohorts (43, 44). Moreover, the genotype of C. albicans was also associated with susceptibility. For instance, resistance to FLC and TRB is associated with MTL homozygosity and MLST clade 1 (45). In this study, C. albicans MLST clade 1 strains had a distinct and broader spectrum of resistance to antifungal drugs, with MIC90 values of all drugs that were significantly higher than those of non-clade 1 strains, except for MFG. This result suggested that MFG may be a better choice for the treatment of clade 1 strain infection. Furthermore, taking the molecular mechanisms of antifungal resistance into consideration, the ABC transporter CDR1/CDR2 and multidrug transporter MDR1, which are responsible for reducing intracellular drug levels via efflux pump activity, and one of the drug targets, ergosterol synthase ERG11, were selected to evaluate their mRNA expression levels in strains with different genotypes and susceptibilities, as well as the levels of proteins from the same families. MDR1 was found to be significantly more highly expressed in clade 1 strains than in non-clade 1 strains at both the transcriptional and translational levels. Therefore, the multidrug transporter could be the key mechanism for the resistance of MLST clade 1 strains.
The virulence factors of C. albicans contribute to VVC clinical characteristics by regulating adhesion, invasion and damage to host epithelial cells. Many proteins, including ALS proteins, phospholipase B (PLB), SAPs, and HWPs, mediate pathogen virulence (46), as well as some extracellular enzyme production (47). Our study showed that the transcriptional levels of SAP4, SAP5, SAP7, and SAP10 were significantly different in MLST clade 1 strains. Among them, SAP4 and SAP5 are involved in adherence, mucosal damage and the immune response of the host, while SAP10 maintains cell wall integrity. Moreover, in C. albicans MLST clade 1 strains, ALS1, ALS3, and ALS6 presented significantly higher mRNA expression levels, and ALS2 presented significantly higher protein expression levels. Genes of the ALS family participate in the pathogenic process of C. albicans by adhering to the host (48), and HWP1 expression also appeared to be significantly different. Although PLB2 and PLB3, which are involved in phospholipase synthesis, were overexpressed in clade 1 strains, their production showed no significant difference among the genotypes of C. albicans.
To further understand the mechanisms that drive the higher resistance and virulence of C. albicans in MLST clade 1, proteomic analysis was applied in this study. Proteins involved in the protein stabilization pathway were determined to be enriched in clade 1 strains, which should contribute to strain resistance against stress and the maintenance of regular growth and virulence. Based on the interaction network analysis of the differentially expressed proteins, a key gene, RPS4, was found to be linked with the proteins influencing the resistance and virulence of C. albicans. RPS4 is a gene encoding a ribosomal protein responsible for cellular protein biosynthesis. However, little is known about how the ribosome operates to mediate microbial pathogenesis. Recently, a pre-rRNA processing factor was verified to be a regulator of fungal development and pathogenesis by mediating the production of reactive oxygen species (49). Moreover, the expression of RPS4 has more than a twofold difference during biofilm development in C. albicans (50). In the current study, RPS4 was associated with the expression of HSP90, SGT1, STI1, SOD1, and YPD1, which are all involved in stress resistance. HSP90 is a heat shock protein that mediates the drug resistance pathways of C. albicans via various signaling pathways by binding with different cochaperones, such as SGT1 and STI, as discovered in this study (51–53). SOD1 and YPD1 have been reported to protect C. albicans against oxidative stress and maintain the virulence of strains (54, 55). In addition, the ALS2, SPA2, and PFK1 proteins that influence the adherence of the strains have been linked with RPS4 directly or indirectly (56–58). Furthermore, DNA repair proteins (HAT2, RFC1, and HTA2) might also be regulated by RPS4 in MLST clade 1 C. albicans and affect the growth stability of the strains.
5. Conclusion
The MLST of C. albicans is closely associated with clinical phenotypes. Among the 306 isolated strains from VVC patients, MLST clade 1 was the most common group and accounted for the highest proportion of RVVC and treatment failure cases. In addition, significantly higher antifungal resistance was shown in MLST clade 1 isolates. The mRNA of the genes and encoded proteins related to drug resistance and virulence were also expressed at significantly higher levels in clade 1 strains, indicating a higher pathogenicity, which corresponded with the clinical phenotype. The proteomic analysis indicated that these properties of C. albicans MLST clade 1 were associated with the protein stabilization pathway. Furthermore, RSP4 is a central regulator that influences the pathogenicity of clade 1 strains by linking to genes involved in stress resistance, adherence and DNA repair.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.iprox.cn/page/project.html?id=IPX0003855000.
Ethics Statement
The studies involving human participants were reviewed and approved by Medical Review Board of Peking University Shenzhen Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
SF and XZ: conceptualization and investigation. YZhu, CF, YShi, XiaL, and XZ: data curation. YZhu, CF, and XZ: formal analysis. SF: funding acquisition and supervision. YZhu, YSha, YL, LH, XinL and CL: methodology. YZhu and SF: project administration. YZhu, YSha, YL, LH, and SF: resources. YZhu and CF: software. YShi, XiaL, YL, and YZha: validation. CF: visualization. YZhu and XZ: writing—original draft. CF and XZ: writing—review and editing.
Funding
This research was funded by the Science and Technology Planning Project of Shenzhen Municipality (Grant Numbers JCYJ20180228162311024 and JCYJ20200109140614667) and the National Natural Science Foundation of China (Grant Number 82171676).
Conflict of Interest
CF was employed by BGI-Shenzhen.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.837536/full#supplementary-material
Abbreviations
VVC, vulvovaginal candidiasis; RVVC, recurrent vulvovaginal candidiasis; MLST, multilocus sequence typing; MTL, mating type locus; DST, diploid sequence type; UPGMA, unweighted-pair group method using average linkages; AAT1a, aspartate aminotransferase; ACC1, acetyl-coenzyme A carboxylase; ADP1, ATP-dependent permease; MPIb, mannose phosphate isomerase; SYA1, alanyl-RNA synthetase; VPS13, vacuolar protein sorting protein; ZWF1b, glucose-6-phosphate dehydrogenase; CDR, ATP-binding cassette multidrug transporter; MDR, multidrug resistance gene; ERG, ergosterol biosynthesis gene; ALS, agglutinin-like sequence; HWP, hyphal wall protein; SAP, secreted aspartyl protease; PLB, phospholipase B; ITC, itraconazole; MCA, miconazole; CLO, clotrimazole; VRC, voriconazole; TEC, terconazole; BUC, butoconazole; AMB, amphotericin B; TRB, terbinafine; CFG, caspofungin; MFG, micafungin; AFG, anidulafungin; FLC, fluconazole; NYS, nystatin; 5FC, 5-fluorocytosine; MIC, minimum inhibitory concentration.
References
1. Tian JY, Yang YG, Chen S, Teng Y, Li XZ. Genetic diversity and molecular epidemiology of Candida albicans from vulvovaginal candidiasis patients. Infect Genet Evol. (2021) 92:104893. doi: 10.1016/j.meegid.2021.104893
2. Willems HM, Ahmed SS, Liu J, Xu Z, Peters BM. Vulvovaginal candidiasis: a current understanding and burning questions. J Fungi. (2020) 6:27. doi: 10.3390/jof6010027
3. Dadar M, Tiwari R, Karthik K, Chakraborty S, Shahali Y, Dhama K. Candida albicans-Biology, molecular characterization, pathogenicity, and advances in diagnosis and control-An update. Microb Pathogen. (2018) 117:128–38. doi: 10.1016/j.micpath.2018.02.028
4. Wu YM, Lee CH, Cheng YC, Lu JJ, Wang SH. Association between CAI microsatellite, multilocus sequence typing, and clinical significance within Candida albicans isolates. Med Mycol. (2021) 59:498–504. doi: 10.1093/mmy/myaa090
5. Ge SH, Xie J, Xu J, Li J, Li DM, Zong LL, et al. Prevalence of specific and phylogenetically closely related genotypes in the population of Candida albicans associated with genital candidiasis in China. Fungal Genet Biol. (2012) 49:86–93. doi: 10.1016/j.fgb.2011.10.006
6. Wang M, Cao Y, Xia M, Al'Hatmi AM, Ou W, Wang Y, et al. Virulence and antifungal susceptibility of microsatellite genotypes of Candida albicans from superficial and deep locations. Yeast. (2019) 36:363–73. doi: 10.1002/yea.3397
7. Saghrouni F, Ben Abdeljelil J, Boukadida J, Ben Said M. Molecular methods for strain typing of C andida albicans: a review. J Appl Microbiol. (2013) 114:1559–74. doi: 10.1111/jam.12132
8. Bougnoux ME, Morand S, d'Enfert C. Usefulness of multilocus sequence typing for characterization of clinical isolates of Candida albicans. J Clin Microbiol. (2002) 40:1290–7. doi: 10.1128/JCM.40.4.1290-1297.2002
9. Odds FC. Molecular phylogenetics and epidemiology of Candida albicans. Fut Microbiol. (2010) 5:67–79. doi: 10.2217/fmb.09.113
10. Hong N, Lei Y, Chen H, Chen X, Tsui KM, Hu D, et al. Genotyping and drug resistance profile of clinical isolates of Candida albicans from vulvovaginal candidiasis in the Eastern China. Mycopathologia. (2022). doi: 10.1007/s11046-022-00616-x. [Epub ahead of print].
11. Bougnoux ME, Aanensen DM, Morand S, Théraud M, Spratt BG, d'Enfert C. Multilocus sequence typing of Candida albicans: strategies, data exchange and applications. Infect Genet Evol. (2004) 4:243–52. doi: 10.1016/j.meegid.2004.06.002
12. Dodgson AR, Dodgson KJ, Pujol C, Pfaller MA, Soll DR. Clade-specific flucytosine resistance is due to a single nucleotide change in the FUR1 gene of Candida albicans. Antimicrob Agents Chemother. (2004) 48:2223–7. doi: 10.1128/AAC.48.6.2223-2227.2004
13. MacCallum DM, Castillo L, Nather K, Munro CA, Brown AJ, Gow NA, et al. Property differences among the four major Candida albicans strain clades. Eukaryot Cell. (2009) 8:373–87. doi: 10.1128/EC.00387-08
14. Owens RA, Doyle S. Effects of antifungal agents on the fungal proteome: informing on mechanisms of sensitivity and resistance. Expert Rev Proteomics. (2021) 18:185–99. doi: 10.1080/14789450.2021.1912601
15. Kusch H, Biswas K, Schwanfelder S, Engelmann S, Rogers P, Hecker M, et al. A proteomic approach to understanding the development of multidrug-resistant Candida albicans strains. Mol Genet Genomics. (2004) 271:554–65. doi: 10.1007/s00438-004-0984-x
16. Hoehamer CF, Cummings ED, Hilliard GM, Rogers PD. Changes in the proteome of Candida albicans in response to azole, polyene, and echinocandin antifungal agents. Antimicrob Agents Chemother. (2010) 54:1655–64. doi: 10.1128/AAC.00756-09
17. Aoki W, Tatsukami Y, Kitahara N, Matsui K, Morisaka H, Kuroda K, et al. Elucidation of potentially virulent factors of Candida albicans during serum adaptation by using quantitative time-course proteomics. J Proteomics. (2013) 91:417–29. doi: 10.1016/j.jprot.2013.07.031
18. McCullough MJ, Clemons KV, Stevens DA. Molecular and phenotypic characterization of genotypic Candida albicans subgroups and comparison with Candida dubliniensis and Candida stellatoidea. J Clin Microbiol. (1999) 37:417–21. doi: 10.1128/JCM.37.2.417-421.1999
19. Hull CM, Raisner RM, Johnson AD. Evidence for mating of the "asexual" yeast Candida albicans in a mammalian host. Science. (2000) 289:307–10. doi: 10.1126/science.289.5477.307
20. Bougnoux ME, Tavanti A, Bouchier C, Gow NA, Magnier A, Davidson AD, et al. Collaborative consensus for optimized multilocus sequence typing of Candida albicans. J Clin Microbiol. (2003) 41:5265–6. doi: 10.1128/JCM.41.11.5265-5266.2003
21. Hull CM, Johnson AD. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science. (1999) 285:1271–5. doi: 10.1126/science.285.5431.1271
22. Alves CT, Wei XQ, Silva S, Azeredo J, Henriques M, Williams DW. Candida albican promotes invasion and colonisation of Candida glabrata in a reconstituted human vaginal epithelium. J Infect. (2014) 69:396–407. doi: 10.1016/j.jinf.2014.06.002
23. Correia A, Lermann U, Teixeira L, Cerca F, Botelho S, Gil da Costa RM, et al. Limited role of secreted aspartyl proteinases Sap1 to Sap6 in Candida albicans virulence and host immune response in murine hematogenously disseminated candidiasis. Infect Immun. (2010) 78:4839–49. doi: 10.1128/IAI.00248-10
24. Green CB, Cheng G, Chandra J, Mukherjee P, Ghannoum MA, Hoyer LL. RT-PCR detection of Candida albicans ALS gene expression in the reconstituted human epithelium (RHE) model of oral candidiasis and in model biofilms. Microbiology (Reading). (2004) 150:267–75. doi: 10.1099/mic.0.26699-0
25. Chau AS, Mendrick CA, Sabatelli FJ, Loebenberg D, McNicholas PM. Application of real-time quantitative PCR to molecular analysis of Candida albicans strains exhibiting reduced susceptibility to azoles. Antimicrob Agents Chemother. (2004) 48:2124–31. doi: 10.1128/AAC.48.6.2124-2131.2004
26. CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. 3rd Edn. CLSI document M27-A3. Wayne, PA: Clinical and Laboratory Standards Institute (2008).
27. CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests of Yeasts. 1st Edn. CLSI Supplement M60. Wayne, PA: Clinical and Laboratory Standards Institute (2017).
28. Pakshir K, Zomorodian K, Karamitalab M, Jafari M, Taraz H, Ebrahimi H. Phospholipase, esterase and hemolytic activities of Candida spp. isolated from onychomycosis and oral lichen planus lesions. J Mycol Méd. (2013) 23:113–8. doi: 10.1016/j.mycmed.2013.04.007
29. Sanitá PV, Zago CE, de Oliveira Mima EG, Pavarina AC, Jorge JH, Machado AL, et al. In vitro evaluation of the enzymatic activity profile of non-albicans Candida species isolated from patients with oral candidiasis with or without diabetes. Oral Surg Oral Med Oral Pathol Oral Radiol. (2014) 118:84–91. doi: 10.1016/j.oooo.2014.03.020
30. Jia H, Sun W, Li M, Zhang Z. Integrated analysis of protein abundance, transcript level, and tissue diversity to reveal developmental regulation of maize. J Proteome Res. (2018) 17:822–33. doi: 10.1021/acs.jproteome.7b00586
31. Odds FC, Bougnoux ME, Shaw DJ, Bain JM, Davidson AD, Diogo D, et al. Molecular phylogenetics of Candida albicans. Eukaryot Cell. (2007) 6:1041–52. doi: 10.1128/EC.00041-07
32. Liu Z, Liao Q. Specification for diagnosis and treatment of vulvovaginal candidiasis (VVC). Chinese J Pract Gynecol Obstetr. (2012) 6:401–2.
33. Tan J, Song Y, Liu W, Wang X, Zhang J, Chen W, et al. Molecular genotyping of Candida albicans isolated from different sites may trace the etiological infection routes: observation in China. Mycoses. (2021) 64:841–50. doi: 10.1111/myc.13327
34. Tsai MH, Wang SH, Hsu JF, Lin LC, Chu SM, Huang HR, et al. Clinical and molecular characteristics of bloodstream infections caused by Candida albicans in children from 2003 to 2011. Clin Microbiol Infect. (2015) 21:1018.e1–8. doi: 10.1016/j.cmi.2015.06.024
35. Gong YB, Zheng JL, Jin B, Zhuo DX, Huang ZQ, Qi H, et al. Particular Candida albicans strains in the digestive tract of dyspeptic patients, identified by multilocus sequence typing. PLoS ONE. (2012) 7:e35311. doi: 10.1371/journal.pone.0035311
36. Tavanti A, Davidson AD, Fordyce MJ, Gow NA, Maiden MC, Odds FC. Population structure and properties of Candida albicans, as determined by multilocus sequence typing. J Clin Microbiol. (2005) 43:5601–13. doi: 10.1128/JCM.43.11.5601-5613.2005
37. Abdulrahim MH, McManus BA, Flint SR, Coleman DC. Genotyping Candida albicans from Candida leukoplakia and non-Candida leukoplakia shows no enrichment of multilocus sequence typing clades but enrichment of ABC genotype C in Candida leukoplakia. PLoS ONE. (2013) 8:e73738. doi: 10.1371/journal.pone.0073738
38. Song N, Kan S, Pang Q, Mei H, Zheng H, Li D, et al. A prospective study on vulvovaginal candidiasis: multicentre molecular epidemiology of pathogenic yeasts in China. J Eur Acad Dermatol Venereol. (2021). doi: 10.1111/jdv.17874
39. Afsarian MH, Badali H, Boekhout T, Shokohi T, Katiraee F. Multilocus sequence typing of Candida albicans isolates from a burn intensive care unit in Iran. J Med Microbiol. (2015) 64:248–53. doi: 10.1099/jmm.0.000015
40. Moorhouse AJ, Rennison C, Raza M, Lilic D, Gow NA. Clonal strain persistence of Candida albicans isolates from chronic mucocutaneous candidiasis patients. PLoS ONE. (2016) 11:e0145888. doi: 10.1371/journal.pone.0145888
41. Odds FC, Davidson AD, Jacobsen MD, Tavanti A, Whyte J, Kibbler C, et al. Candida albicans strain maintenance, replacement, and microvariation demonstrated by multilocus sequence typing. J Clin Microbiol. (2006) 44:3647–58. doi: 10.1128/JCM.00934-06
42. Bhattacharya S, Sae-Tia S, Fries BC. Candidiasis and mechanisms of antifungal resistance. Antibiotics. (2020) 9:312. doi: 10.3390/antibiotics9060312
43. Kalkanci A, Güzel AB, Khalil IIJ, Aydin M, Ilkit M, Kuştimur S. Yeast vaginitis during pregnancy: susceptibility testing of 13 antifungal drugs and boric acid and the detection of four virulence factors. Med Mycol. (2012) 50:585–93. doi: 10.3109/13693786.2012.662597
44. Richter SS, Galask RP, Messer SA, Hollis RJ, Diekema DJ, Pfaller MA. Antifungal susceptibilities of Candida species causing vulvovaginitis and epidemiology of recurrent cases. J Clin Microbiol. (2005) 43:2155–62. doi: 10.1128/JCM.43.5.2155-2162.2005
45. Odds FC. Candida albicans, resistance to flucytosine and terbinafine is linked to MAT locus homozygosity and multilocus sequence typing clade 1. FEMS Yeast Res. (2009) 9:1091–101. doi: 10.1111/j.1567-1364.2009.00577.x
46. Talapko J, Juzbasic M, Matijevic T, Pustijanac E, Bekic S, Kotris I, et al. Candida albicans-the virulence factors and clinical manifestations of infection. J Fungi. (2021) 7:79. doi: 10.3390/jof7020079
47. Rapala-Kozik M, Bochenska O, Zajac D, Karkowska-Kuleta J, Gogol M, Zawrotniak M, et al. Extracellular proteinases of Candida species pathogenic yeasts. Mol Oral Microbiol. (2018) 33:113–24. doi: 10.1111/omi.12206
48. Hoyer LL. The ALS gene family of Candida albicans. Trends Microbiol. (2001) 9:176–80. doi: 10.1016/S0966-842X(01)01984-9
49. Cao S, Yuan Y, Qin Y, Zhang M, de Figueiredo P, Li G, et al. The pre-rRNA processing factor Nop53 regulates fungal development and pathogenesis via mediating production of reactive oxygen species. Environ Microbiol. (2018) 20:1531–49. doi: 10.1111/1462-2920.14082
50. Yeater KM, Chandra J, Cheng G, Mukherjee PK, Zhao X, Rodriguez-Zas SL, et al. Temporal analysis of Candida albicans gene expression during biofilm development. Microbiology. (2007) 153:2373–85. doi: 10.1099/mic.0.2007/006163-0
51. Shapiro RS, Uppuluri P, Zaas AK, Collins C, Senn H, Perfect JR, et al. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr Biol. (2009) 19:621–9. doi: 10.1016/j.cub.2009.03.017
52. Shapiro RS, Zaas AK, Betancourt-Quiroz M, Perfect JR, Cowen LE. The Hsp90 co-chaperone Sgt1 governs Candida albicans morphogenesis and drug resistance. PLoS ONE. (2012) 7:e44734. doi: 10.1371/journal.pone.0044734
53. Gu X, Xue W, Yin Y, Liu H, Li S, Sun X. The Hsp90 co-chaperones Sti1, Aha1, and P23 regulate adaptive responses to antifungal azoles. Front Microbiol. (2016) 7:1571. doi: 10.3389/fmicb.2016.01571
54. Hwang CS, Rhie Ge, Oh JH, Huh WK, Yim HS, Kang SO. Copper-and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology. (2002) 148:3705–13. doi: 10.1099/00221287-148-11-3705
55. Day AM, Smith DA, Ikeh MA, Haider M, Herrero-de Dios CM, Brown AJ, et al. Blocking two-component signalling enhances Candida albicans virulence and reveals adaptive mechanisms that counteract sustained SAPK activation. PLoS Pathog. (2017) 13:e1006131. doi: 10.1371/journal.ppat.1006131
56. Kumar R, Maulik M, Pathirana RU, Cullen PJ, Edgerton M. Sho1p Connects glycolysis to Ras1-cAMP Signaling and Is Required for Microcolony Formation in Candida albicans. Msphere. (2020) 5:e00366–20. doi: 10.1128/mSphere.00366-20
57. Zhao X, Oh SH, Yeater K, Hoyer LL. Analysis of the Candida albicans Als2p and Als4p adhesins suggests the potential for compensatory function within the Als family. Microbiology. (2005) 151(Pt 5):1619. doi: 10.1099/mic.0.27763-0
Keywords: multilocus sequence typing, Candida albicans, clinical characterization, antifungal resistance, virulence
Citation: Zhu Y, Fang C, Shi Y, Shan Y, Liu X, Liang Y, Huang L, Liu X, Liu C, Zhao Y, Fan S and Zhang X (2022) Candida albicans Multilocus Sequence Typing Clade I Contributes to the Clinical Phenotype of Vulvovaginal Candidiasis Patients. Front. Med. 9:837536. doi: 10.3389/fmed.2022.837536
Received: 05 January 2022; Accepted: 18 February 2022;
Published: 01 April 2022.
Edited by:
Hector Mora Montes, University of Guanajuato, MexicoReviewed by:
Samira Salari, Kerman University of Medical Sciences, IranJavad Javidnia, Mazandaran University of Medical Sciences, Iran
Copyright © 2022 Zhu, Fang, Shi, Shan, Liu, Liang, Huang, Liu, Liu, Zhao, Fan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaowei Zhang, zhangivy726@163.com; Shangrong Fan, fanshangrong@21cn.com
†These authors have contributed equally to this work and share first authorship
 Yuxia Zhu
Yuxia Zhu Chao Fang
Chao Fang Yu Shi5
Yu Shi5  Xinyang Liu
Xinyang Liu Chunfeng Liu
Chunfeng Liu Shangrong Fan
Shangrong Fan Xiaowei Zhang
Xiaowei Zhang