Subsurface Microbial Community Composition in Anchialine Environments Is Influenced by Original Organic Carbon Source at Time of Deposition
- 1Department of Marine Biology, Texas A&M University at Galveston, Galveston, TX, United States
- 2Département de sciences biologiques, Université de Montréal, Montréal, QC, Canada
- 3Department of Oceanography, Texas A&M University, College Station, TX, United States
- 4Department of Marine and Coastal Environmental Science, Texas A&M University at Galveston, Galveston, TX, United States
Prokaryotes constitute the majority of sedimentary biomass, where they cycle organic carbon and regulate organic matter transformation. The microbes inhabiting sediment are diverse and the factors controlling microbial community composition are not fully understood. Here, we characterized the prokaryotic community using 16S rRNA gene sequencing in 24 stratigraphic layers within a 89 cm (dated to ~1900 years old) sediment core from an anchialine sinkhole in the Bahamas with a stratified water column and anoxic bottom water. The microbial community was dominated by members of the Alphaproteobacteria, Dehalococcoidia, Gammaproteobacteria, Bathyarchaeota, and Campylobacter classes. Most interestingly, subsurface microbial community structure could be correlated to previous evidence for timewise changes in the main source of organic matter that was supplied to the sediment accumulating during the last 2000 years, which itself was caused by regional terrestrial vegetation changes. The C:N ratio was correlated to the relative abundance of the microbial classes, and the microbial communities followed three previously determined time periods based on the source of organic matter, which suggests that the carbon source at time of deposition influences the resultant subsurface microbial community composition. These results show that carbon source is a driver of the microbial community composition inhabiting anoxic sediment, which could have implications for improving understanding of carbon cycling in coastal sedimentary basins.
Introduction
Coastal sediment contains an estimated 103 to 109 prokaryotes/gram, and these microorganisms play pivotal roles in the global biogeochemical cycling of carbon (Jorgensen, 2012; Orsi, 2018), sulfur (Holmkvist et al., 2011), and nitrogen (Kuypers et al., 2003). Microbial community diversity in sediment has been characterized by sequencing of the 16S ribosomal RNA (rRNA) gene in coastal marine environments (Koretsky et al., 2005; Lenk et al., 2012; Ul-Hasan et al., 2019), freshwater lakes (Nold et al., 2010; Kojima et al., 2012), and the deep-sea (Reed et al., 2002; Engelen et al., 2008; Inagaki et al., 2015; Labonté et al., 2017). Metagenomics and transcriptomics analyses showed that coastal sediment microbes are physiologically versatile and functionally diverse (Dyksma et al., 2016; Chen et al., 2017; Yu C. et al., 2018). However, the factors influencing microbial community composition and distribution in the sediment below anoxic water columns remain understudied, and are mostly unknown in anchialine environments.
The subsurface mixing of rain and marine water on carbonate platforms creates karst subterranean estuaries (van Hengstum et al., 2019; Brankovits and Pohlman, 2020; Cresswell and van Hengstum, 2021), which are analogous to other coastal estuaries by having an upper meteoric water mass of varying salinity buoyed on a denser saline groundwater mass below. The steep environmental gradients created by these variable water mass conditions create subsurface habitats and environments frequently prefaced with the adjective ‘anchialine’ (Stock et al., 1986), especially in the Atlantic realm. Blue holes and sinkholes not only provide access points for human hydrographic observation of karst subterranean estuaries (Smart et al., 1988), they also enable terrigenous chemicals, nutrients and organic matter to enter the otherwise subsurface anchialine environments (Collins et al., 2015b), and exposure to sunlight also promotes organic matter production by photosynthesis (Tamalavage et al., 2018). Sinkholes and blue holes are abundant across the Bahamian archipelago, and they are located in both subaerial (‘sinkhole’) and shallow subtidal settings (‘blue hole’). Structurally, Bahamian carbonate platforms began forming in the Jurassic from the long-term accumulation of shallow water carbonates (Mullins and Lynts, 1977), with sinkholes and blue holes subsequently created by subsurface dissolution and collapse of limestone bedrock (Mylroie et al., 1995). Owing to the high primary porosity of the antecedent carbonate and the present elevation of relative sea level, sinkholes on the terrestrial landscape are now aquatic environments as their depth extends below the water table and into the local groundwater. With sufficient local rainfall and aquifer recharge, groundwater on low-lying Bahamian islands is stratified with an upper meteoric water mass (typically oxic, lower salinity) divided from a lower saline groundwater water mass (anoxic, seawater), both of which destabilize and mix with increasing proximity to the shoreline. Both terrestrial organic matter coming from runoff and organic matter produced in situ (primary productivity) contribute to the particulate organic matter pool that is ultimately deposited within a sinkhole basin (van Hengstum et al., 2010; Collins et al., 2015a; Tamalavage et al., 2018; van Hengstum et al., 2018).
While the drivers of subsurface microbial communities remain poorly understood, several hypotheses have been proposed, which included varying sediment composition, enzymatic activity, or organic matter bioavailability. For example, evidence has been provided that subsurface microbial community structure in deep-sea anoxic sediment from the Juan de Fuca ridge may be caused by variations in sediment chemistry and texture (e.g., clays, carbonate-content, and organic carbon), as opposed to porewater geochemistry (Labonté et al., 2017). Subsurface microbial community composition has also been linked to enzymatic activity (Böer et al., 2009) and labile organic matter (Franco et al., 2007). Variations within microbial community structure in subtidal sands significantly correlated to enzymatic activities, specifically chitobiase, α-glucosidase, and phosphatase activity, as well as microphytobenthic algae productivity (Böer et al., 2009). The seasonal variation of chlorophyll a, a proxy for the availability of labile organic matter, slightly shifted the subsurface microbial community structure within sediment from the North Sea (Franco et al., 2007). In riparian settings, organic matter provenance between different terrestrial organic matter (OM) pools (e.g., soil, vegetation) was shown to influence the subsurface microbial community (Fagervold et al., 2014).
In anchialine environments, microbial communities in the water columns vary greatly between sinkholes (Sahl et al., 2011; Kajan et al., 2022), and they tend to follow oxygen concentration and density stratification (Gonzalez et al., 2011; Davis and Garey, 2018; He et al., 2019). Other than site and the type of watershed, salinity is one of the strongest drivers of the microbial community composition (Hoffman et al., 2018). Inferred microbial functions showed high potential for sulfur oxidation and nitrate reduction in sinkholes, with nitrite and sulfite reduction associated with the photic zone (He et al., 2019). Moreover, there are striking differences between the microbial communities found in the water column and those found in microbial mats and surface sediment (Sahl et al., 2011). Surface sediment microbial communities also depend on shoreline proximity (i.e., coastal or inland) (Suárez-Moo et al., 2022). However, little is known regarding sediment microbial communities in anchialine environments, or the factors shaping their composition and distribution. In this study, we aimed to explore the subsurface prokaryotic community in Blackwood Sinkhole on Abaco Island in the northern Bahamian Archipelago. Based on work introduced above, we hypothesize that changes in the structure and microbial composition will be mainly related to organic matter provenance at time of deposition. Aside from this work, terrestrial landscape changes and productivity changes in the water column at Blackwood Sinkhole are well documented in previous studies (van Hengstum et al., 2016; Tamalavage et al., 2018; Tamalavage et al., 2020; Fall et al., 2021). To test these ideas, we (a) described the composition and structure of the microbial communities using high-throughput sequencing the V4-V5 hypervariable regions of the 16S rRNA gene, and (b) compared to results to known changes in sediment and organic matter supply in Blackwood Sinkhole over the last ~1900 years that occurred from known evidence of local landscape and sinkhole productivity changes (Tamalavage et al., 2018; Tamalavage et al., 2020).
Study Site
Blackwood Sinkhole is located ~220 m from the shoreline on the northeastern coast of the Great Abaco Island on the Little Bahama Bank (26.79, 77.42) (Figure 1). Blackwood Sinkhole is 32 m in diameter, and this groundwater-fed basin has a stratified water column that has remained stable during the last 3000 years (van Hengstum et al., 2016; Tamalavage et al., 2018; Fall et al., 2021). The sinkhole has an upper ~10 m of meteoric water (1.4 psu), which transitions to a mixing zone below (10-15 m below sea level), and this rests above anoxic saline groundwater (~15-40 m below sea level, 39.9 psu) (van Hengstum et al., 2016). The groundwater stratification and geomorphology of Blackwood Sinkhole has promoted excellent sediment preservation from a lack of vertical sediment mixing from either invertebrate bioturbation or waves. Basal sedimentary deposits in Blackwood Sinkhole are carbonate (karst) gravel, which abruptly transition to laminated sapropel (i.e., high organic matter) interbedded with carbonate horizons that have accumulated over the last 3000 years (van Hengstum et al., 2016). The laminated sapropel has a total organic carbon (TOC) content ~10% (Tamalavage et al., 2018). However, sedimentation rate and site of primary deposition has not been laterally or temporally uniform through time. This is because (a) the bottom geometry of the sinkhole basin is complex since it was inherited from the collapse of an original limestone cave, and (b) the sediment sources and delivery mechanisms during the last 3000 years are linked to changing primary productivity, differential erosion of the vertical sinkhole walls, and organic matter transport and deposition from adjacent wetland and surrounding forest habitats (Tamalavage et al., 2018). Using detailed radiocarbon dating on other core samples, constant sedimentation (~0.4 mm yr−1) was initiated 3000 Calibrated years Before Present (cal yrs BP) around the sinkhole periphery, with evidence of coarse-grained particle deposition during intense hurricane strikes on Abaco Island (van Hengstum et al., 2016). However, sedimentation in the center of the sinkhole was delayed until ~1900 cal yrs BP, with higher sedimentation rates (1.2 mm yr−1) than the periphery and far less erosional inputs from the vertical sinkhole walls (Tamalavage et al., 2018).
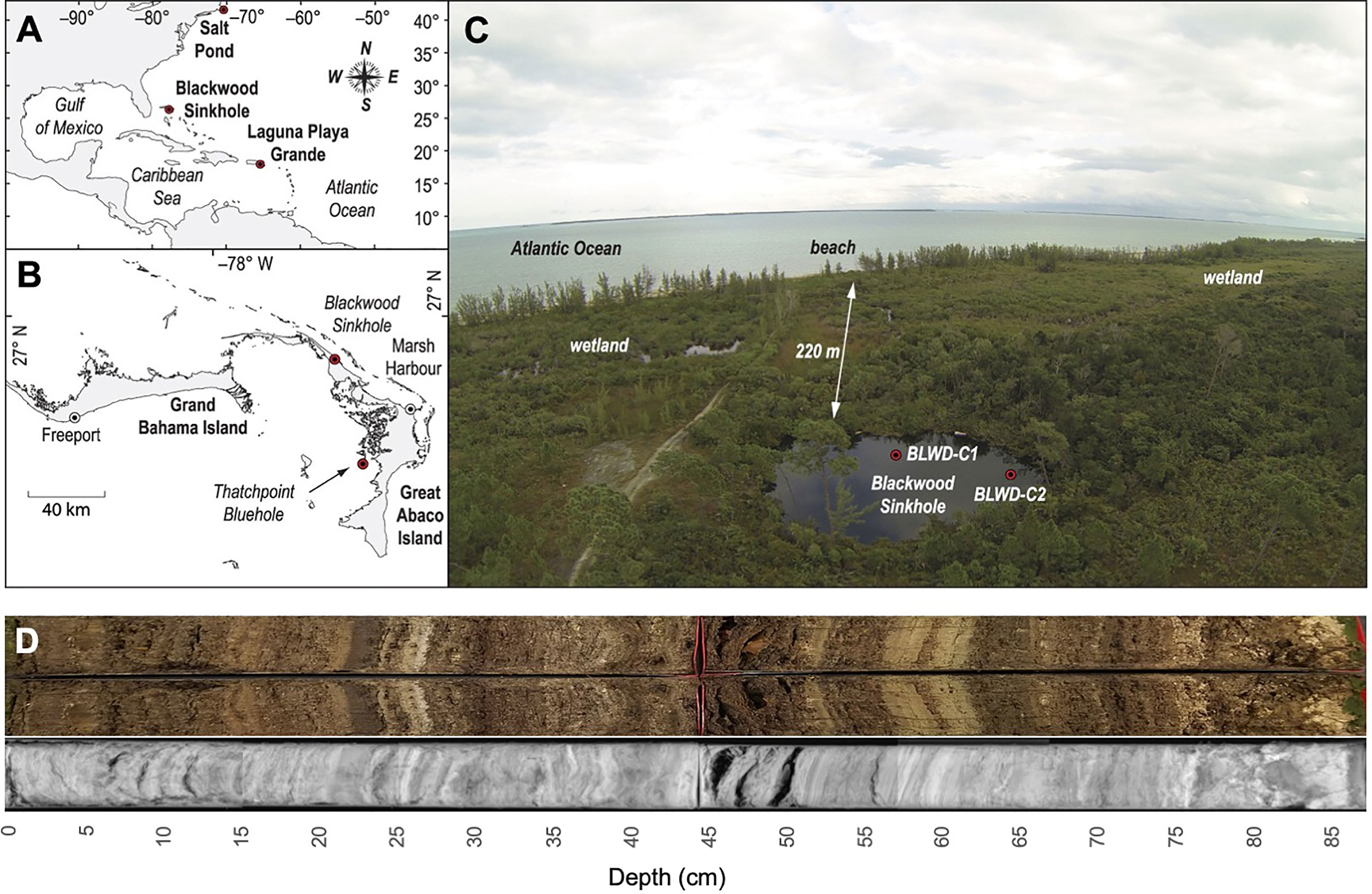
Figure 1 Map of Blackwood Sinkhole within the context of the Bahamas region. (A) The western North Atlantic region noting the location of the Blackwood Sinkhole on the Little Bahama Bank. (B) Islands on the Little Bahama Bank, and the position of Blackwood Sinkhole on Great Abaco Island. (C) Aerial photograph of Blackwood Sinkhole and the relation (220 m) to the Atlantic Ocean. Red dots indicate coring locations, including BLWD-C7, used in this study. Indicating the location of the core sampled in this study, BLWD-C7. (D) Radiograph and picture of core BLWD-C7. Figure adapted from (Tamalavage et al., 2018).
Based on a detailed analysis on the changes in organic matter (OM) provenance through time using stable carbon isotopes (δ13Corg) and the C:N ratio, Tamalavage et al. (2018) documented how the primary source of OM accumulating in Blackwood Sinkhole changed through time. While OM was always typically derived from primary productivity and erosion of terrigenous materials, the dominant source was not always constant. For example, OM was primarily derived from the terrestrial landscape from ~3000 to ~1500 cal yrs BP. At ~1500 years ago, however, OM deposition was dominated by constituents of authigenic OM produced in situ. Finally, during the last 1000 years, organic matter input into Blackwood Sinkhole was dominated by inputs from an adjacent wetland that colonized the epikarst surface at around the same time. These wetlands colonized the favorable habitat that was created when concomitant regional sea-level and groundwater-level rise flooded topographic lows adjacent to Blackwood Sinkhole (Tamalavage et al., 2018; Fall et al., 2021).
Material and Methods
Sample Collection and Handling
A 7.6 cm diameter push core (BLWD-C7; 26.79°N, 77.42°W, depth: 34 ± 1 m below sea level) was collected on 29 July 2018 with a polyvinyl chloride pipe, using advanced technical scuba diving procedures that followed guidelines established by the American Academy of Underwater Sciences. The periphery of the sinkhole bottom was targeted for core sampling in an attempt to re-collect the last 3000 years of sedimentary deposition (van Hengstum et al., 2016). In the lab, the 89-cm long core was sectioned lengthwise into a working and archive half (subsequently stored at 4°C), photographed, radiographed, and the lithology was qualitatively described (Schnurrenberger et al., 2003). The working half was split into 24 stratigraphic layers, based on sediment color and texture, and stored at –80°C for further analyses (Supplementary Table 1).
To validate the age of the core and compare to previous results, five terrestrial plant macrofossils (e.g., leaves, twigs) were sent for radiocarbon dating of at the National Ocean Sciences Accelerator Mass Spectrometry facility at Woods Hole Oceanographic Institution (Woods Hole, MA, USA, 20.1−22.2 cm, 32.9−35 cm, 52.1−54.2 cm, 64.5−67.7 cm, and 79.5−86.9 cm). Conventional radiocarbon ages were calibrated into Calendar years Before Present (cal yrs BP, present is considered 1950 Common Era) with IntCAL13 (Reimer et al., 2013). A final downcore Bayesian age model for BLWD-C7 was computed using the R program Bacon v2.2 (Blaauw and Christen, 2011) to provide probability estimates at each core depth (Supplementary Figure 1) using three of the five samples (32.9−35 cm, 52.1−54.2 cm, 64.5−67.7 cm), and the surface designated as −68 Cal yrs BP to mark the date of collection in the year 2018. Two samples dates were rejected from the final radiocarbon age model (Figure 2B; Supplementary Figure 1). The shallowest radiocarbon result from the base of the prominent sapropel horizon at 20.1 to 22.2 cm in BLWD-C7 was not used in the age model because it provided a calibrated age result that was too old relative to previous higher-resolution dating results (Supplementary Table 2). Our previous, more detailed, stratigraphic dating results indicate a much younger age for this sapropel horizon (291-223 Cal yrs BP, Tamalavage et al., 2018), so the dated terrestrial macrofossil must have been an older plant fragment that resided on the terrestrial surface prior to its erosion into Blackwood Sinkhole. Similarly, the deepest radiocarbon calibration result from 79.5−86.9 cm in BLWD-C7 was rejected because it caused significant change in sedimentation rate that was inconsistent with previous results (Supplementary Figure 1), and as such is also suspected to be an old, reworked terrestrial plant fragment.
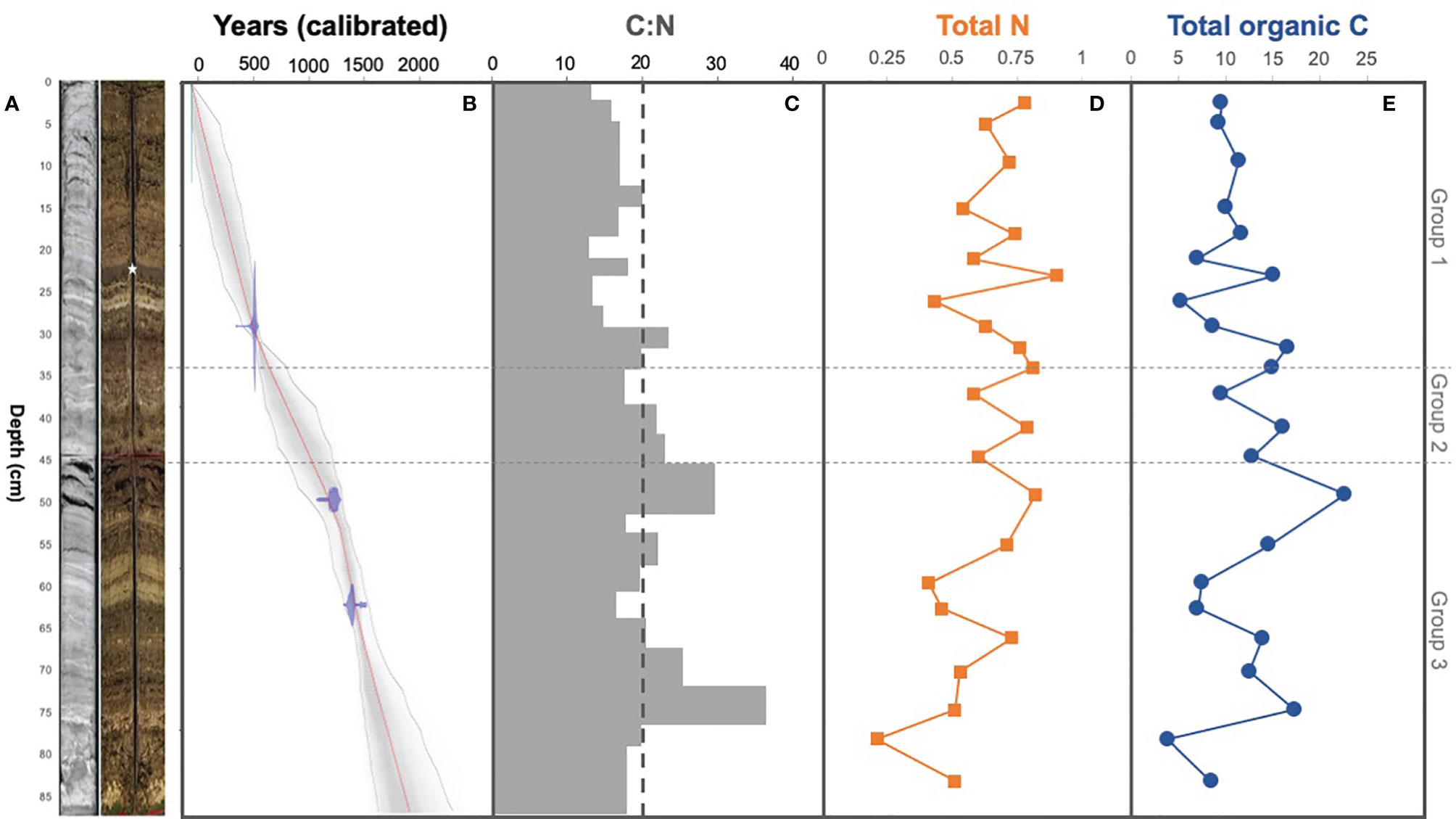
Figure 2 (A) Radiograph and picture of core BLWD-C7. The white star indicates the sapropel layer that is synchronous across all cores from Blackwood Sinkhole. (B) Downcore age model for BLWD-C7 showing core to be up to ~1900 years old. Radiocarbon results were calibrated into calendar years before present (present being 1950) with IntCAL13 and visualized using Bacon v2.2. The calibrated 14C dates are shown in the transparent blue regions, the grey coloring indicates more likely calendar ages, and the red line shows the best model based on the weighted mean age for each depth. (C) C:N ratio based on the percentage of (D) total nitrogen (%) and (E) total organic carbon (%), in accordance to depth in centimeters.
Measurement of Carbon and Nitrogen Content
Total carbon and nitrogen content was measured on 24 subsamples downcore from BLWD-C7 (Table 1) that were also targeted for 16S rRNA gene analysis. First, the subsamples were freeze dried overnight, homogenized, and 2 to 6 mg of ground sample were placed into tin capsules and measured on a Costech instruments ECS 4010 CHNSO Analyzer (Costech Analytical Technologies) to measure total carbon (TC) and total nitrogen (TN). Data calibration was determined relative to acetanilide and standard reference material for organics in marine sediment according to the National Institute of Standards and Technology (NIST). To measure organic carbon (with mass correction applied), the samples were acidified using 8 mL of 1M HCl for 24 h or until effervescence ceased, then desiccated at 60°C, and re-homogenized. The ground, acidified samples were weighed (0.5 to 1.8 mg) into silver capsules then processed on the CHNSO analyzer (Tamalavage et al., 2018). The potential loss of carbon from the direct acidification process was corrected by multiplying the percent of sample remaining (post-acidification weight subtracted from pre-acidification weight/pre acidification weight) (Tamalavage et al., 2018). The atomic C:N ratio was determined using the organic carbon (OC) acidified values divided by the unacidified Total Nitrogen (TN) values and multiplied by the molecular weight ratio (14.01/12.01) (Tamalavage et al., 2018). Precision on replicates measurements of C and N was within ± 2% weight percent.
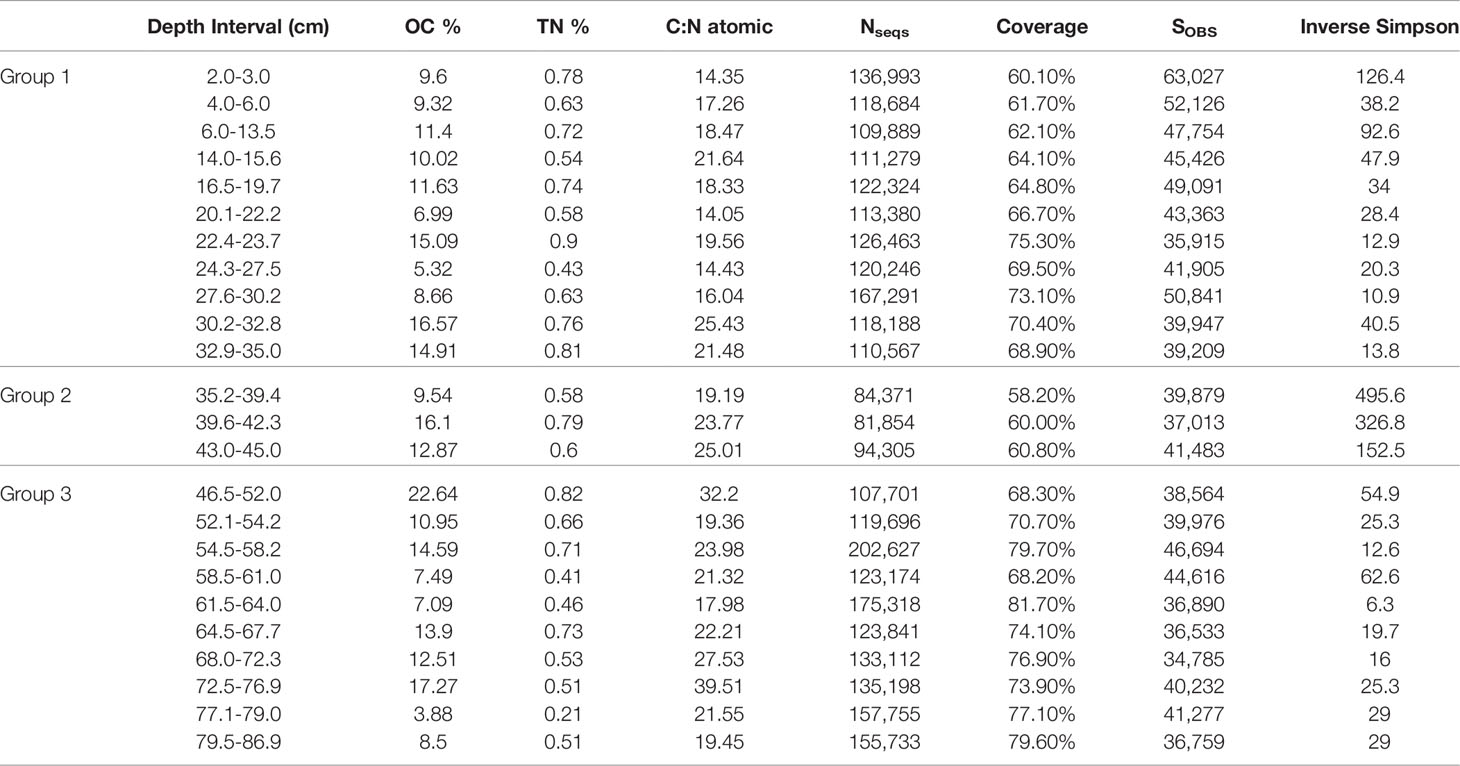
Table 1 Downcore organic matter analysis [OC% (organic carbon corrected to the initial volume of sediment), TN% (total nitrogen) and C:N atomic ratios] as well as sequence-based calculations [Number of sequences (Nseq), sequence coverage, observed richness (SOBS), and inverse Simpson index] for core BLWD C7.
16S rRNA Gene Sequencing and Analysis
Total DNA was extracted from separate sediment subsamples (1–2g) from each of the 24 horizons using the DNeasy PowerSoil kit (Qiagen, USA) (Supplementary Table 1) according to the manufacturer’s protocol, except more material was used than the recommended 0.25 g, and then stored at –20°C until PCR amplification. As recommended for sediment and low biomass samples (Sheik et al., 2018), a negative control, or extraction blank, using 0.5 mL of 0.2 µm filtered sterile MilliQ water was also extracted to identify possible contamination from the ambient laboratory and kit reagents. The Earth Microbiome Project PCR primers 515F-Y (5′-GTGYCAGCMGCCGCGGTAA-3′) and 926R (5′-CCGYCAATTYMTTTRAGTTT-3′) were used to target the V4-V5 region of the 16S rRNA gene (Parada et al., 2016). Thermal cycling was performed under the following conditions: initial preheating for 3 min at 94°C; 35 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 1 min 45 s; final extension at 72°C for 10 min. PCR reactions were completed in triplicates and the products were pooled and cleaned using the MinElute PCR Purification Kit (Qiagen, USA). Amplicons (100-200 ng) were sent to the Texas A&M AgriLife Bioinformatics and Genomics facility for library preparation with the Amplicon library preparation kit (Illumina) and sequencing with Illumina MiSeq with 250 bp paired-ends.
Analyses of the 16S rRNA gene amplicons were completed using the software mothur as presented in the MiSeq Standard Operating Procedure (SOP) tutorial (Kozich et al., 2013), which included reducing sequencing and PCR errors, processing the improved sequences, running an alignment using the reference SILVA alignment (v132), removing poorly aligned sequences and undesirables, and pre-clustering the sequences into amplicon sequence variants (ASVs). The remove.lineage command using all the lineages observed in the negative control at taxonomic level 6 (genus) was used to remove any potential contaminants from final analyses. For the non-metric multidimensional scaling (NMDS) analysis, the taxa (class level) abundances were square root transformed and the environmental variables (geochemical data) were z-transformed, which allowed the variables to be compared using a similar scale. The NMDS analysis was performed using the envfit function (permutations = 9999) available in the vegan R package (Oksanen et al., 2019) and plotted with ggplot2 (Wickham, 2016). The ANOVA tests were performed using the aov command to fit an analysis of variance model in R (RCore Team, 2017). Finally, stratigraphically-constrained (i.e., age constrained according to radiocarbon results) hierarchical clustering analysis was performed using the package ‘rioja’ in R, which can provide a semi-quantitative tool to assess intervals of time when the microbial community structure was similar (Juggins, 2017). The sequences are available in GenBank under the BioProject #PRJNA639820. Accession numbers for each sample are available in Supplementary Table 3.
Results
Sedimentation
Basal sediment in BLWD-C7 is carbonate gravel, above which is laminated algal sapropel interbedded with layers of calcium carbonate (Figure 2A). This general lithology is similar to previous results (van Hengstum et al., 2016; Tamalavage et al., 2018). Most notably, Blackwood Sinkhole has a prominent layer of non-laminated algal sapropel that is located near the top of the stratigraphic architecture (see photographs in Figure 2 of van Hengstum et al., 2016), which was identified in BLWD-C7 from 23–25 cm. Tamalavage et al. (2018) provided a more precise median age range for deposition of this non-laminated algal sapropel layer in an alternate core sample of 291-223 cal yrs BP (BLWD-C3). The 2σ uncertainty age envelope for this layer with the radiocarbon age results for BLWD-C7 (this work) is similar to previous age results. However, the new radiocarbon results indicate that the specific location where BLWD-C7 was collected only started accumulating sediment ~1900 years ago instead of ~3,000 yrs BP as observed in BLWD-C2 from Blackwood Sinkhole (Figure 2B). This result further underscores that bathymetric complexities created by the original cave collapse impacted subsequent sediment focusing and deposition (van Hengstum et al., 2016).
In core BLWD-C7 (this study), C and N contents show variation in accord with the primary mineralogical and organic matter variability in the stratigraphic layers (Table 1 and Figure 2). In general, sedimentary layers with high TOC content (%) also have a corresponding high C:N ratio (Table 1 and Figure 2). A high C:N ratio (>20) usually indicates that the source of carbon fluxed to the sediment was likely dominated by terrestrial organic carbon input, whereas a lower C:N ratio (<15) indicates a higher proportion of carbon from primary productivity [e.g., phytoplankton, (Stenuite et al., 2007)] (Lamb et al., 2006; Fuentes et al., 2016). The C:N ratio fluctuates in Blackwood Sinkhole, with the lowest value at horizon 20.1−22.2 cm (C:N ratio of 14.4) and the highest value at 72.5–76.9 cm (C:N ratio of 39.5) (Figure 2). The sapropel layer seen at approximately 23–25 cm contains 15% TOC and has a C:N ratio of 19, indicating that the organic matter has both terrestrial and primary productivity sources (Tamalavage et al., 2018).
Microbial Community Composition Analysis Using 16S rRNA Gene Sequencing
We sequenced on average 749,985 reads for each of the 24 samples, for a total of 20,669,626 raw reads. After trimming and deduplication, between 277,000-551,107 reads per sample, or a total of 13,907,092 reads remained (Supplementary Table 3). We ran a negative control, which returned 3,764 sequences, to identify possible extraction kit contamination (Supplementary Figure 2). Contaminants made up ~5% of the entire dataset, and they consisted of classes and species known to be common contaminants of DNA extraction and sequencing kits reagents (e.g., Ralstonia, unclassified Xanthobacteraceae, Afipia, Streptococcus, Sphingomonas, unclassified Enterobacteriaceae, Pseudomonas, Bradyrhizobium, Micrococcus, Cutibacterium, Paenibacillus, Mesorhizobium, Pelagibacterales Clade I, Marivivens, Lactococcus, unclassified Burkholderiaceae, unclassified Sphingomonadaceae, and unclassified Rhizobiaceae) (Salter et al., 2014; Glassing et al., 2016). Using a conservative approach, ASVs belonging to the same genus as the contaminants were removed. After removal of poorly aligned and chimeric sequences, and decontamination, we were left with a total of 4,055,715 ASVs for taxonomic assignment (Supplementary Table 3).
Main taxonomic classes observed in all samples are common in anoxic sediment, such as wetlands and marine sediment, and belong to Dehalococcoidia, Gammaproteobacteria, Bathyarchaeota, and Campylobacteria (Figure 3 and Supplementary Figure 3) (Wang et al., 2012; Biderre-Petit et al., 2016; Franco et al., 2017). In addition to Bathyarchaeota, a number of other archaeal phyla (e.g., including Euryarchaeota, Nanoarchaeota, and Crenarcheaota) were also present (Figure 3). When looking at a deeper taxonomic level, negligible changes were observed within the class Dehalococcoidia, except a slight increase in the class Sh765B-AG-111 was observed down core (Supplementary Figure 3).
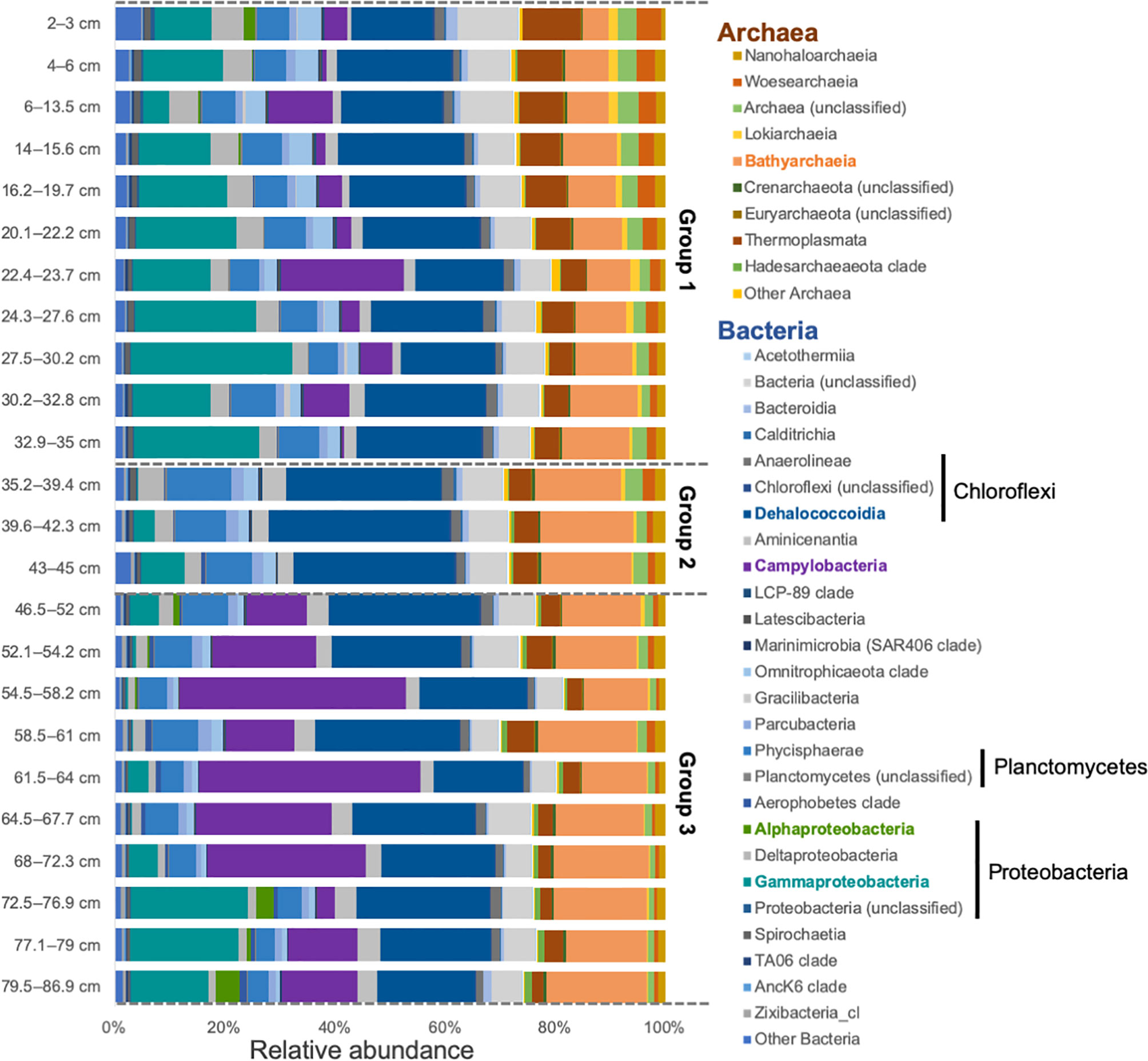
Figure 3 Class level prokaryotic relative abundance for each layer of the sediment core. Groups 1, 2, and 3 are divided by the dashed lines. Taxa highlighted in color are the ones considered in the discussion.
We also compared the samples using a stratigraphically-constrained cluster analysis, which provides a semi-quantitative time-wise (i.e., core depth) division of changes in the microbial community structure (Figure 4). Interestingly, the output dendrogram can be assessed in two different ways. First, three groups emerge on the dendrogram at Bray-Curtis Dissimilarity Index of 2.3: (a) group 1 (0-32.8 cm) spanning 545 cal yrs BP until present, (b) group 2 (32.9-42.3 cm) spanning 545-930 cal yrs B, and (c) group 3 (43 to 86.9 cm) spanning 930 to 1908 cal yrs BP. For example, Gammaproteobacteria was mainly present in Group 1 and Group 3. Gammaproteobacteria were mainly dominated by the order Thiomicrospirales in Group 1 and displayed greater taxonomic variations in Group 3, especially in the deeper part of the core (Supplementary Figure 3). Campylobacteria was mainly present in Group 3, where it was dominated by the families Sulfurovaceae and Arcobacteraceae. In Group 1, there were more unclassified Campylobacterales members and Thiovulaceae, which were absent from Group 3. Other taxa present showed no major affiliation with any group. Secondly, the dendrogram can be perceived slightly broader, and at a Bray-Curtis Dissimilarity Index of 2.75 two groups emerge: (a) group 1 (0-42.3 cm) spanning the last 1000 years, and (b) group 2 (43-86.9 cm) contains samples than are older than 1000 years.
To determine if the beta diversity, or the distribution of the microbial community, was related to carbon content, the relative abundance of the different taxonomic classes was analyzed in relation to total organic carbon content (%), total nitrogen (%), and C:N ratio in an MSDS (Figure 5). The samples clustered based on their age group. While total organic carbon (p = 0.737) and total nitrogen (p = 0.258) are not statistically correlated to the relative abundance of the microbial taxa, the C:N ratio is weakly statistically correlated to the relative abundance for the microbial taxa (p = 0.096) (Figure 5). Calculated diversity indices include both taxon richness and evenness, both of which varied throughout the core. The shallowest samples, BLWD-C7 2–3 cm, 6–13.5 cm, and the three samples belonging to Group 2, 35.2–39.4 cm, 39.6–42.3 cm, and 43–45 cm, displayed the highest diversity indices (Inverse Simpson), with 126.4, 92.6, 495.6, 326.8, and 152.5, respectively (Table 1). There was no correlation between the inverse Simpson diversity index and C:N ratio values (ANOVA p = 0.897).
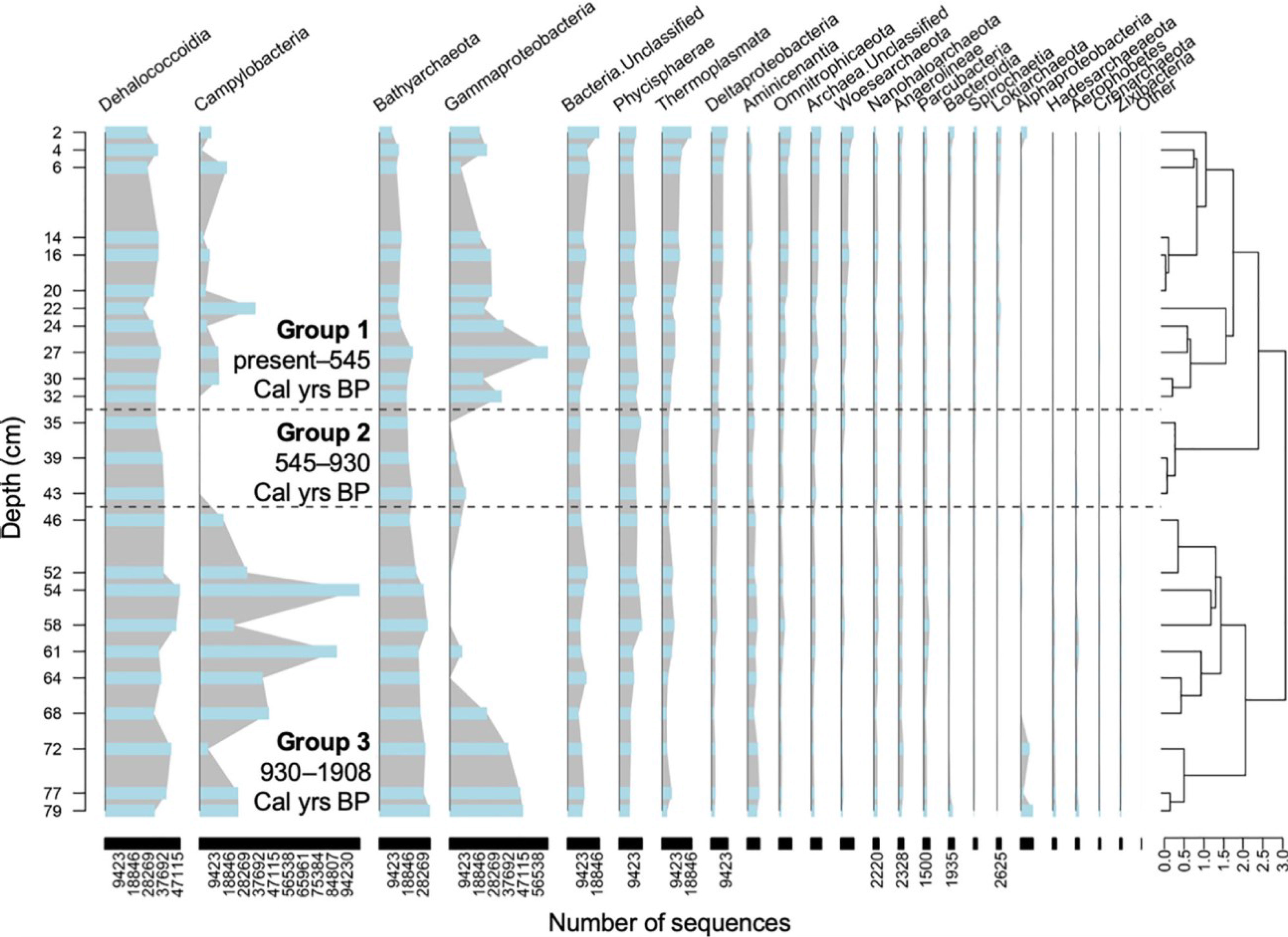
Figure 4 Stratigraphically-constrained hierarchical cluster analysis of microbial community downcore based on the three previously determined age groups based on source of organic matter. Blue bars, linked by a grey area line, represent the number of ASVs for each class.
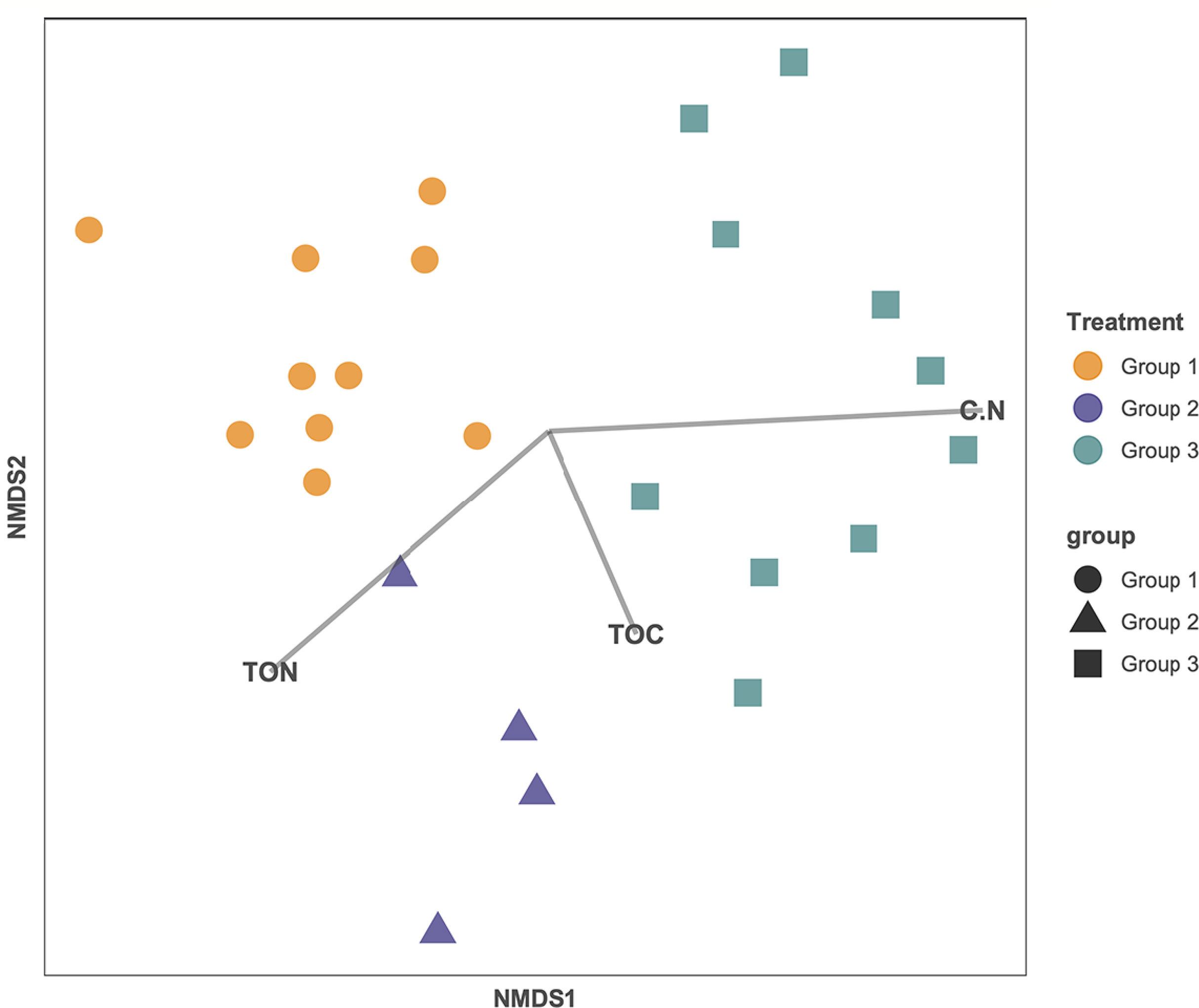
Figure 5 Non-metric multidimensional scaling (NMDS) ordination of each sample based on their microbial composition. Groups previously identified are labeled as orange circles (Group 1), blue triangles (Group 2), and teal squares (Group 3). Each line is correlated to the ordination and represents the direction and strength of the carbon influence (C:N ratio, OC, and TN). C:N ratio is significantly correlated (p = 0.096).
Discussion
The most interesting result is the timewise division of the subsurface prokaryotic community structure that generally correlates to previous evidence of changes in OM source and deposition in Blackwood Sinkhole. According to the 16S rRNA gene data, the most prevalent classes within the entire sediment core are Dehalococcoidia, Bathyarchaeota, Campylobacteria, and Gammaproteobacteria. These classes exhibit meaningful timewise distributional changes in the ~1900 year old succession of BLWD-C7, suggesting that the type of organic matter at time of burial provided resources for the enrichment of these microbial communities.
At the broadest level, the dendrogram can be divided at Bray-Curtis Dissimilarity Index of 2.75 (Figure 4), which creates two groups: (a) group 1 (0-42.3 cm) spanning the last 1000 years, and (b) group 2 (43-86.9 cm) contains samples than are older than 1000 years. In this greater division of the dendrogram, the older Group 2 has a higher proportion of Campylobacteria, whereas the younger Group 1 is characterized by increased proportion of Gammaproetobacteria. This bipartite division of the dendrogram compares well with the distinct intervals of OM sourcing in Blackwood Sinkhole determined from a multicore analysis of δ13C, δ15N, and C:N (Tamalavage et al., 2018). The oldest sediment deposited in Blackwood Sinkhole from 2990-1520 cal yrs BP had OM that was primarily derived from terrestrial sources. From 1502-1009 cal yrs BP, OM derived from freshwater primary productivity produced in situ in the upper part of the stratified water column. At 1300 years ago, however, a wetland began colonizing the topographic lows adjacent to Blackwood Sinkhole (van Hengstum et al., 2016). Pollen grains preserved in the stratigraphy of Blackwood Sinkhole indicate that new vegetation on the landscape which increased around ~1000 years ago included increased abundance of mangrove species (e.g., Lagunclularia, Avicennia) and emergent macrophytes (e.g., Typha, (Fall et al., 2021)). As a result of wetland expansion, by ~1000 years ago, organic matter produced within the wetland eroded and transported into Blackwood Sinkhole, and became the dominant source of OM into the system (Tamalavage et al., 2018). It would appear that increased delivery of wetland-derived organic matter at ~1000 cal yrs BP generally favored expansion of subsurface populations of Grammaproteobacteria, at the expense of Camphylobacteria. In addition, it seems that the expanded primary productivity in the upper water column from 1500 to 1000 years ago, which would provide more bioavailable organic carbon, was not as meaningful for the eventual subsurface prokaryotic community structure.
In balance, however, a bipartite division of the dendrogram does not completely explain the changes observed in the subsurface microbial community structure. If the dendrogram is alternatively divided at a Bray-Curtis Dissimilarity Index of 2.3, three different communities of prokaryotes emerge (as depicted in Figure 4): Group 1 (0-32.8 cm) spanning 545 cal yrs BP until present, (b) group 2 (32.9-42.3 cm) spanning 545-930 cal yrs B, and (c) group 3 (43 to 86.9 cm) spanning 930 to 1908 cal yrs BP. The numerical separation is most likely caused by the salient decrease in Camphylobacteria in sediment that was deposited from 545 to 930 cal yrs BP.
The relative abundance of Dehaloccoidia and Bathyarchaeota showed little variation throughout the core. However, studies completed in anoxic sediment elsewhere have often discovered an abundance of the class Dehalococcoidia (phylum Chloroflexi) (Cetecioğlu et al., 2009; Biderre-Petit et al., 2016). Specifically, members of the class Dehalococcoidia are known to only inhabit environments with strict anoxic conditions, such as river sediments, aquifers, and sludge (Löffler et al., 2013). Based on metabolic potential found in metagenome assembled genomes (MAGs), some strains are known to rely only on halogenated compounds for anaerobic respiration, while others are anaerobic acetate fermenters (Hug et al., 2013). Members of the phylum Chloroflexi are commonly found in anoxic environments, including within the water column (Cetecioğlu et al., 2009). Analysis of Chloroflexi MAGs from aquifer sediments (Hug et al., 2013) and a bioreactor (Kindaichi et al., 2012) also revealed that some strains may also have the ability to degrade cellulose, which is the main component of plant cell walls. Chloroflexi members may be thriving on the terrestrial input of organic matter in Blackwood Sinkhole. Bathyarchaeota are widespread in anoxic sediment in both marine and freshwater (Zhou et al., 2018). Their wide distribution may be caused by the fact that these microbes are metabolic generalists able to anaerobically utilize detritus (proteins and polymeric), aromatic compounds, methane and methylated compounds, and a wide range of organic matter (Zhou et al., 2018). Bathyarchaeota are able to break down carbohydrates of terrestrial origin (Lazar et al., 2016) and lignin (Yu T. et al., 2018), which are important sources of organic matter in Blackwood Sinkhole. Metagenomic analyses of the microbial communities inhabiting Blackwood Sinkhole may shed more light on the potential impact of the members of the Dehalococcoidia and Bathyarchaeota in this anoxic system influenced by both aquatic and terrestrial OM.
Campylobacteria was generally more abundant in Group 3, coeval with deposition of OM primarily from the terrestrial surface and primary productivity. Campylobacteria are nitrogen fixers and sulfate reducers commonly found in sulfur-rich environments such as hydrothermal vents, or salt marshes (Drexler et al., 2020), since Campylobacteria has been observed with the root system of Spartina (McClung and Patriquin, 1980; Miller et al., 2007). While we do not have companion measurements of porewater sulfate concentrations, sulfate has been found in higher concentrations in anoxic subsurface environments, and increasing with depth in another stratified anchialine sinkhole in a riverbed (Bottrell et al., 1991; Davis and Garey, 2018), suggesting that the higher prevalence of Campylobacteria in Group 3 may be influenced by sulfate concentrations. The higher relative abundance of Thiovulaceae (specifically members the family Sulfurimonas, a widespread sulfur-oxidizing species found in sediments, hydrothermal vent fields, aquifers and subsurface environments) in the upper part of the core in Group 1 also supports the idea that the sulfur cycle is a determining factor in shaping the microbial communities (Supplementary Figure 3).
Gammaproteobacteria was abundant in Group 1 and towards the end of Group 3, which were both characterized by OM originating from wetlands and mangroves, or terrestrial input, respectively, which are sources of carbon that are less labile, therefore not as easily degraded by microorganisms than carbon of photodetritus origin (Guillemette et al., 2013). Gammaproteobacteria, known to be facilitating carbon fixation (Dyksma et al., 2016), have also been found in high abundance in coastal sediment from sites in Western Europe, Germany, and Australia (Lenk et al., 2011; Dyksma et al., 2016). In the layers where carbon isn’t as labile, therefore less available for catabolism, Gammaproteobacteria could be playing a key role in the system by fixing CO2 to produce usable organic carbon compounds, but more studies are required to confirm.
Alphaproteobacteria were found only in the top horizon (2–3cm) and the bottom 20 cm (72.5–86.9 cm) of the core. These bacteria are known for a variety of metabolic abilities within anoxic sediment, including nitrogen fixation, ammonia oxidation, and methylotrophy (Acosta-González and Marqués, 2016; Williams et al., 2017). Specifically, the order Rhodobacteraceae within the Alphaproteobacteria class are associated with marine algae and seagrasses in coastal sediments (Raulf et al., 2015; Park et al., 2016). Alphaproteobacteria and other microbes, especially nitrate reducers, are commonly found in the water column. They are known to decrease with subsurface depth (Lloyd et al., 2020), which can explain their dominance only in the top most sediment. Our system is marine groundwater fed, as such, an influx of fresh nutrients at the base of the sediment could fuel bacteria such as Alphaproteobacteria. This groundwater input could explain why Alphaproteobacteria were seen at the bottom of the core as well as at its surface, but a detailed nutrient analysis would provide more information regarding Alphaproteobacteria distribution in the subsurface.
Conclusions
Overall, the entire core displayed variability in carbon source (terrestrial/aquatic derived), which promotes a diverse subsurface microbial community. The microbial communities were dominated by members of the Chloroflexi, Bathyarchaeota, Proteobacteria, and Campylobacteria phyla, all of which are commonly found in coastal anoxic sediment. The microbial communities varied based on landscape change and variability in in situ productivity, suggesting that the source of carbon at time of deposition is a driver of the microbial community’s presence within this anoxic environment at Blackwood sinkhole. Further exploration of microbial communities’ relationships to carbon in other sinkhole environments will expand our understanding of the role of microorganisms living in anoxic sediment within the carbon cycle.
Data Availability Statement
The data presented in the study are deposited in the NCBI repository, BioProject accession number PRJNA639820, available at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA639820.
Author Contributions
JL and PvH designed the study. CR generated the data, performed the analyses and wrote the first draft of the manuscript. AT helped with total C and N measurements, and manuscript preparation. PvH collected the sample and participated in the manuscript preparation. JL edited the manuscript, with contributions from all. All authors contributed to the article and approved the submitted version.
Funding
Funding for this research was provided by the Center for Dark Energy Biosphere Investigations (Research Grant to JL and PvH), Texas Sea Grant (Graduate student Grant in Aid to CR), and the National Sciences Foundation (EAR-1703087 to PvH).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This is C-DEBI contribution # 593.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.872789/full#supplementary-material
References
Acosta-González A., Marqués S. (2016). Bacterial Diversity in Oil-Polluted Marine Coastal Sediments. Curr. Opin. Biotechnol. 38, 24–32. doi: 10.1016/j.copbio.2015.12.010
Biddanda B. A., Nold S. C., Dick G. J., Kendall S. T., Vail J. H., Ruberg S. A., et al. (2012). Rock, Water, Microbes: Underwater Sinkholes in Lake Huron Are Habitats for Ancient Microbial Life. Nat. Educ. Knowl. 3, 13.
Biderre-Petit C., Dugat-Bony E., Mege M., Parisot N., Adrian L., Moné A., et al. (2016). Distribution of Dehalococcoidia in the Anaerobic Deep Water of a Remote Meromictic Crater Lake and Detection of Dehalococcoidia-Derived Reductive Dehalogenase Homologous Genes. PloS One 11, e0145558. doi: 10.1371/journal.pone.0145558
Blaauw M., Christen J. A. (2011). Flexible Paleoclimate Age-Depth Models Using an Autoregressive Gamma Process. Bayesian Anal. 6, 457–474. doi: 10.1214/ba/1339616472
Böer S. I., Hedtkamp S. I. C., Van Beusekom J. E. E., Fuhrman J. A., Boetius A., Ramette A. (2009). Time-And Sediment Depth-Related Variations in Bacterial Diversity and Community Structure in Subtidal Sands. ISME J. 3, 780. doi: 10.1038/ismej.2009.29
Bottrell S. H., Smart P. L., Whitaker F., Raiswell R. (1991). Geochemistry and Isotope Systematics of Sulphur in the Mixing Zone of Bahamian Blue Holes. Appl. Geochem. 6, 97–103. doi: 10.1016/0883-2927(91)90066-X
Brankovits D., Pohlman J. W. (2020). Methane Oxidation Dynamics in a Karst Subterranean Estuary. Geochim. Cosmochim. Acta 277, 320–333. doi: 10.1016/j.gca.2020.03.007
Cetecioğlu Z., Ince B. K., Kolukirik M., Ince O. (2009). Biogeographical Distribution and Diversity of Bacterial and Archaeal Communities Within Highly Polluted Anoxic Marine Sediments From the Marmara Sea. Mar. Pollut. Bull. 58, 384–395. doi: 10.1016/j.marpolbul.2008.10.009
Chen J., Hanke A., Tegetmeyer H. E., Kattelmann I., Sharma R., Hamann E., et al. (2017). Impacts of Chemical Gradients on Microbial Community Structure. ISME J. 11, 920–931. doi: 10.1038/ismej.2016.175
Collins S. V., Reinhardt E. G., Werner C. L., Le Maillot C., Devos F., Meacham S. S. (2015a). Regional Response of the Coastal Aquifer to Hurricane Ingrid and Sedimentation Flux in the Yax Chen Cave System (Ox Bel Ha) Yucatan, Mexico. Palaeogeogr. Palaeoclimatol. Palaeoecol. 438, 226–238. doi: 10.1016/j.palaeo.2015.07.030
Collins S. V., Reinhardt E. G., Werner C. L., Le Maillot C., Devos F., Rissolo D. (2015b). Late Holocene Mangrove Development and Onset of Sedimentation in the Yax Chen Cave System (Ox Bel Ha) Yucatan, Mexico: Implications for Using Cave Sediments as a Sea-Level Indicator. Palaeogeogr. Palaeoclimatol. Palaeoecol. 438, 124–134. doi: 10.1016/j.palaeo.2015.07.042
Cresswell J. N., van Hengstum P. J. (2021). Habitat Partitioning in the Marine Sector of Karst Subterranean Estuaries and Bermuda’s Marine Caves: Benthic Foraminiferal Evidence. Front. Environ. Sci. 8. doi: 10.3389/fenvs.2020.594554
Davis M. C., Garey J. R. (2018). Microbial Function and Hydrochemistry Within a Stratified Anchialine Sinkhole: A Window Into Coastal Aquifer Interactions. Water (Switzerland) 10, 1–17. doi: 10.3390/w10080972
Drexler J. Z., Davis M. J., Woo I., de la Cruz S. (2020). Carbon Sources in the Sediments of a Restoring vs. Historically Unaltered Salt Marsh. Estuaries Coasts. 43, 1345–1360 doi: 10.1007/s12237-020-00748-7
Dyksma S., Bischof K., Fuchs B. M., Hoffmann K., Meier D., Meyerdierks A., et al. (2016). Ubiquitous Gammaproteobacteria Dominate Dark Carbon Fixation in Coastal Sediments. ISME J. 10, 1939–1953. doi: 10.1038/ismej.2015.257
Engelen B., Ziegelmüller K., Wolf L., Köpke B., Gittel A., Cypionka H., et al. (2008). Fluids From the Oceanic Crust Support Microbial Activities Within the Deep Biosphere. Geomicrobiol. J. 25, 56–66. doi: 10.1080/01490450701829006
Fagervold S. K., Bourgeois S., Pruski A. M., Charles F., Kerherve P., Vetion G., et al. (2014). River Organic Matter Shapes Microbial Communities in the Sediment of the Rhone Prodelta. ISME J. 8, 2327–2338. doi: 10.1038/ismej.2014.86
Fall P. L., van Hengstum P. J., Lavold-Foote L., Donnelly J. P., Albury N. A., Tamalavage A. E. (2021). Human Arrival and Landscape Dynamics in the Northern Bahamas. Proc. Natl. Acad. Sci. 118, e2015764118. doi: 10.1073/pnas.2015764118
Franco M. A., De Mesel I., Diallo M. D., van der Gucht K., Van Gansbeke D., Van Rijswijk P., et al. (2007). Effect of Phytoplankton Bloom Deposition on Benthic Bacterial Communities in Two Contrasting Sediments in the Southern North Sea. Aquat. Microb. Ecol. 48, 241–254. doi: 10.3354/ame048241
Franco D. C., Signori C. N., Duarte R. T. D., Nakayama C. R., Campos L. S., Pellizari V. H. (2017). AndHigh Prevalence of Gammaproteobacteria in the Sediments of Admiralty Bay and North Bransfield Basin, Northwestern Antarctic Peninsula. Front. Microbiol. 8, 153. doi: 10.3389/fmicb.2017.00153
Fuentes J. L., Garbayo I., Cuaresma M., Montero Z., González-del-Valle M., Vílchez C. (2016). Impact of Microalgae-Bacteria Interactions on the Production of Algal Biomass and Associated Compounds. Mar. Drugs 14, 100. doi: 10.3390/md14050100
Glassing A., Dowd S. E., Galandiuk S., Davis B., Chiodini R. J. (2016). Inherent Bacterial DNA Contamination of Extraction and Sequencing Reagents may Affect Interpretation of Microbiota in Low Bacterial Biomass Samples. Gut Pathog. 8, 24. doi: 10.1186/s13099-016-0103-7
Gonzalez B. C., Iliffe T. M., Macalady J. L., Schaperdoth I., Kakuk B. (2011). Microbial Hotspots in Anchialine Blue Holes: Initial Discoveries From the Bahamas. Hydrobiologia 677, 149–156. doi: 10.1007/s10750-011-0932-9
Guillemette F., McCallister S. L., Del Giorgio P. A. (2013). Differentiating the Degradation Dynamics of Algal and Terrestrial Carbon Within Complex Natural Dissolved Organic Carbon in Temperate Lakes. J. Geophys. Res. Biogeosci. 118, 963–973. doi: 10.1002/jgrg.20077
He H., Fu L., Liu Q., Fu L., Bi N., Yang Z., et al. (2019). Community Structure, Abundance and Potential Functions of Bacteria and Archaea in the Sansha Yongle Blue Hole, Xisha, South China Sea. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02404
Hoffman S. K., Seitz K. W., Havird J. C., Weese D. A., Santos S. R. (2018). Diversity and the Environmental Drivers of Spatial Variation in Bacteria and Micro-Eukarya Communities From the Hawaiian Anchialine Ecosystem. Hydrobiologia 806, 265–282. doi: 10.1007/s10750-017-3365-2
Holmkvist L., Ferdelman T. G., Jørgensen B. B. (2011). A Cryptic Sulfur Cycle Driven by Iron in the Methane Zone of Marine Sediment (Aarhus Bay, Denmark). Geochim. Cosmochim. Acta 75, 3581–3599. doi: 10.1016/j.gca.2011.03.033
Hug L. A., Castelle C. J., Wrighton K. C., Thomas B. C., Sharon I., Frischkorn K. R., et al. (2013). Community Genomic Analyses Constrain the Distribution of Metabolic Traits Across the Chloroflexi Phylum and Indicate Roles in Sediment Carbon Cycling. Microbiome 1, 1–17. doi: 10.1186/2049-2618-1-22
Inagaki F., Kubo Y., Bowles M. W., Heuer V. B., Ijiri A., Imachi H., et al. (2015). Exploring Deep Microbial Life in Coal-Bearing Sediment Down to ~2.5 Km Below the Ocean Floor. Sci. (80-. ). 349, 420–424. doi: 10.1126/science.aaa6882
Jorgensen B. B. (2012). Shrinking Majority of the Deep Biosphere. Proc. Natl. Acad. Sci. 109, 15976–15977. doi: 10.1073/pnas.1213639109
Juggins S. (2017) Rioja: Analysis of Quaternary Science Data. Available at: http://www.staff.ncl.ac.uk/stephen.juggins/.
Kajan K., Cukrov N., Cukrov N., Bishop-Pierce R., Orlić S. (2022). Microeukaryotic and Prokaryotic Diversity of Anchialine Caves From Eastern Adriatic Sea Islands. Microb. Ecol. 83, 257–270. doi: 10.1007/s00248-021-01760-5
Kindaichi T., Yuri S., Ozaki N., Ohashi A. (2012). Ecophysiological Role and Function of Uncultured Chloroflexi in an Anammox Reactor. Water Sci. Technol. 66, 2556–2561. doi: 10.2166/wst.2012.479
Kojima H., Tsutsumi M., Ishikawa K., Iwata T., Mußmann M., Fukui M. (2012). Distribution of Putative Denitrifying Methane Oxidizing Bacteria in Sediment of a Freshwater Lake, Lake Biwa. Syst. Appl. Microbiol. 35, 233–238. doi: 10.1016/j.syapm.2012.03.005
Koretsky C. M., Van Cappellen P., DiChristina T. J., Kostka J. E., Lowe K. L., Moore C. M., et al. (2005). Salt Marsh Pore Water Geochemistry Does Not Correlate With Microbial Community Structure. Estuar. Coast. Shelf Sci. 62, 233–251. doi: 10.1016/j.ecss.2004.09.001
Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., Schloss P. D. (2013). Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Kuypers M. M. M., Sliekers A. O., Lavik G., Schmid M., Jørgensen B. B., Kuenen J. G., et al. (2003). Anaerobic Ammonium Oxidation by Anammox Bacteria in the Black Sea. Nature 422, 608. doi: 10.1038/nature01472
Labonté J. M., Lever M. A., Edwards K. J., Orcutt B. N. (2017). Influence of Igneous Basement on Deep Sediment Microbial Diversity on the Eastern Juan De Fuca Ridge Flank. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01434
Lamb A. L., Wilson G. P., Leng M. J. (2006). A Review of Coastal Palaeoclimate and Relative Sea-Level Reconstructions Using δ13c and C/N Ratios in Organic Material. Earth-Sci. Rev. 75, 29–57. doi: 10.1016/j.earscirev.2005.10.003
Lazar C. S., Baker B. J., Seitz K., Hyde A. S., Dick G. J., Hinrichs K., et al. (2016). Genomic Evidence for Distinct Carbon Substrate Preferences and Ecological Niches of B Athyarchaeota in Estuarine Sediments. Environ. Microbiol. 18, 1200–1211. doi: 10.1111/1462-2920.13142
Lenk S., Arnds J., Zerjatke K., Musat N., Amann R., Mußmann M. (2011). Novel Groups of Gammaproteobacteria Catalyse Sulfur Oxidation and Carbon Fixation in a Coastal, Intertidal Sediment. Environ. Microbiol. 13, 758–774. doi: 10.1111/j.1462-2920.2010.02380.x
Lenk S., Moraru C., Hahnke S., Arnds J., Richter M., Kube M., et al. (2012). Roseobacter Clade Bacteria are Abundant in Coastal Sediments and Encode a Novel Combination of Sulfur Oxidation Genes. ISME J. 6, 2178–2187. doi: 10.1038/ismej.2012.66
Lloyd K. G., Bird J. T., Buongiorno J., Deas E., Kevorkian R., Noordhoek T., et al. (2020). Erratum for Lloyd et al., “Evidence for a Growth Zone for Deep-Subsurface Microbial Clades in Near-Surface Anoxic Sediments”. Appl. Environ. Microbiol. 86, e00877–20. doi: 10.1128/AEM.02224-20
Löffler F. E., Yan J., Ritalahti K. M., Adrian L., Edwards E. A., Konstantinidis K. T., et al. (2013). Dehalococcoides Mccartyi Gen. Nov., Sp. Nov., Obligately Organohalide-Respiring Anaerobic Bacteria Relevant to Halogen Cycling and Bioremediation, Belong to a Novel Bacterial Class, Dehalococcoidia Classis Nov., Order Dehalococcoidales Ord. Nov. And Famil. Int. J. Syst. Evol. Microbiol. 63, 625–635. doi: 10.1099/ijs.0.034926-0
McClung C. R., Patriquin D. G. (1980). Isolation of a Nitrogen-Fixing Campylobacter Species From the Roots of Spartina Alterniflora Loisel. Can. J. Microbiol. 26, 881–886. doi: 10.1139/m80-153
Miller W. G., Parker C. T., Rubenfield M., Mendz G. L., Wösten M. M. S. M., Ussery D. W., et al. (2007). The Complete Genome Sequence and Analysis of the Epsilonproteobacterium Arcobacter Butzleri. PloS One 2, e1358. doi: 10.1371/journal.pone.0001358
Mullins H. T., Lynts G. W. (1977). Origin of the Northwestern Bahama Platform: Review and Reinterpretation. GSA Bull. 88, 1447–1461. doi: 10.1130/0016-7606(1977)88<1447:OOTNBP>2.0.CO;2
Mylroie J. E., Carew J. L., Vacher H. L. (1995). Karst Development in the Bahamas and Bermuda. Geol. Soc Am. Spec. Pap. 300, 251–267. doi: 10.1130/0-8137-2300-0.251
Nold S. C., Pangborn J. B., Zajack H. A., Kendall S. T., Rediske R. R., Biddanda B. A. (2010). Benthic Bacterial Diversity in Submerged Sinkhole Ecosystems. Appl. Environ. Microbiol. 76, 347–351. doi: 10.1128/AEM.01186-09
Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., et al (2019). vegan: Community Ecology Package. R package version 2.5-6. Available at: https://CRAN.R-project.org/package=vegan.
Orsi W. D. (2018). Ecology and Evolution of Seafloor and Subseafloor Microbial Communities. Nat. Rev. Microbiol. 16, 671–683 doi: 10.1038/s41579-018-0046-8
Parada A. E., Needham D. M., Fuhrman J. A. (2016). Every Base Matters: Assessing Small Subunit rRNA Primers for Marine Microbiomes With Mock Communities, Time Series and Global Field Samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Park S., Ha M. J., Jung Y. T., Yoon J. H. (2016). Litorisediminivivens Gilvus Gen. Nov., Sp. Nov.,Isolated From a Tidal Flat. Int. J. Syst. Evol. Microbiol. 66, 4681–4685. doi: 10.1099/ijsem.0.001410
Raulf F. F., Fabricius K., Uthicke S., de Beer D., Abed R. M. M., Ramette A. (2015). Changes in Microbial Communities in Coastal Sediments Along Natural CO2 Gradients at a Volcanic Vent in Papua New Guinea. Environ. Microbiol. 17, 3678–3691. doi: 10.1111/1462-2920.12729
RCore Team (2017). R: A Language and Environment for Statistical Computing. Available at: https://www.r-project.org/.
Reed D. W., Fujita Y., Delwiche M. E., Blackwelder D. B., Sheridan P. P., Uchida T., et al. (2002). Microbial Communities From Methane Hydrate-Bearing Deep Marine Sediments in a Forearc Basin. Appl. Environ. Microbiol. 68, 3759–3770. doi: 10.1128/AEM.68.8.3759-3770.2002
Reimer P. J., Bard E., Bayliss A., Beck J. W., Blackwell P. G., Ramsey C. B., et al. (2013). IntCal13 and Marine13 Radiocarbon Age Calibration Curves 0–50,000 Years Cal BP. Radiocarbon 55, 1869–1887. doi: 10.2458/azu_js_rc.55.16947
Sahl J. W., Gary M. O., Harris J. K., Spear J. R. (2011). A Comparative Molecular Analysis of Water-Filled Limestone Sinkholes in North-Eastern Mexico. Environ. Microbiol. 13, 226–240. doi: 10.1111/j.1462-2920.2010.02324.x
Salter S. J., Cox M. J., Turek E. M., Calus S. T., Cookson W. O., Moffatt M. F., et al. (2014). Reagent and Laboratory Contamination Can Critically Impact Sequence-Based Microbiome Analyses. BMC Biol. 12, 87. doi: 10.1186/s12915-014-0087-z
Schnurrenberger D., Russell J., Kelts K. (2003). Classification of Lacustrine Sediments Based on Sedimentary Components. J. Paleolimnol. 29, 141–154. doi: 10.1023/A:1023270324800
Sheik C. S., Reese B. K., Twing K. I., Sylvan J. B., Grim S. L., Schrenk M. O., et al. (2018). Identification and Removal of Contaminant Sequences From Ribosomal Gene Databases: Lessons From the Census of Deep Life. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00840
Smart P., Dawans J. M., Whitaker F. (1988). Carbonate Dissolution in a Modern Mixing Zone. Nature 335, 811–813. doi: 10.1038/335811a0
Stenuite S., Pirlot S., HARDY M., Sarmento H., TARBE A., Leporcq B., et al. (2007). Phytoplankton Production and Growth Rate in Lake Tanganyika: Evidence of a Decline in Primary Productivity in Recent Decades. Freshw. Biol. 52, 2226–2239. doi: 10.1111/j.1365-2427.2007.01829.x
Stock J. H., Iliffe T. M., Williams D. (1986). The Concept of “Anchialine” Reconsidered. Stygologia 2, 90–92.
Suárez-Moo P., Remes-Rodríguez C. A., Márquez-Velázquez N. A., Falcón L. I., García-Maldonado J. Q., Prieto-Davó A. (2022). Changes in the Sediment Microbial Community Structure of Coastal and Inland Sinkholes of a Karst Ecosystem From the Yucatan Peninsula. Sci. Rep. 12, 1–11. doi: 10.1038/s41598-022-05135-9
Tamalavage A. E., van Hengstum P. J., Louchouarn P., Fall P. L., Donnelly J. P., Albury N. A., et al. (2020). Plant Wax Evidence for Precipitation and Vegetation Change From a Coastal Sinkhole Lake in the Bahamas Spanning the Last 3000 Years. Org. Geochem. 150, 104120. doi: 10.1016/j.orggeochem.2020.104120
Tamalavage A. E., van Hengstum P. J., Louchouarn P., Molodtsov S., Kaiser K., Donnelly J. P., et al. (2018). Organic Matter Sources and Lateral Sedimentation in a Bahamian Karst Basin (Sinkhole) Over the Late Holocene: Influence of Local Vegetation and Climate. Palaeogeogr. Palaeoclimatol. Palaeoecol. 506, 70–83. doi: 10.1016/j.palaeo.2018.06.014
Ul-Hasan S., Bowers R. M., Figueroa-Montiel A., Licea-Navarro A. F., Beman J. M., Woyke T., et al. (2019). Community Ecology Across Bacteria, Archaea and Microbial Eukaryotes in the Sediment and Seawater of Coastal Puerto Nuevo, Baja California. PLoS One 14, 1–19. doi: 10.1371/journal.pone.0212355
van Hengstum P. J., Cresswell J. N., Milne G. A., Iliffe T. M. (2019). Development of Anchialine Cave Habitats and Karst Subterranean Estuaries Since the Last Ice Age. Sci. Rep. 9, 1–10. doi: 10.1038/s41598-019-48058-8
van Hengstum P. J., Donnelly J. P., Fall P. L., Toomey M. R., Albury N. A., Kakuk B. (2016). The Intertropical Convergence Zone Modulates Intense Hurricane Strikes on the Western North Atlantic Margin. Sci. Rep. 6, 21728. doi: 10.1038/srep21728
van Hengstum P. J., Maale G., Donnelly J. P., Albury N. A., Onac B. P., Sullivan R. M., et al. (2018). Drought in the Northern Bahamas From 3300 to 2500 Years Ago. Quat. Sci. Rev. 186, 169–185. doi: 10.1016/j.quascirev.2018.02.014
van Hengstum P. J., Reinhardt E. G., Beddows P. A., Gabriel J. J. (2010). Linkages Between Holocene Paleoclimate and Paleohydrogeology Preserved in a Yucatan Underwater Cave. Quat. Sci. Rev. 29, 2788–2798. doi: 10.1016/j.quascirev.2010.06.034
Wang Y., Sheng H. F., He Y., Wu J. Y., Jiang Y. X., Tam N. F. Y., et al. (2012). Comparison of the Levels of Bacterial Diversity in Freshwater, Intertidal Wetland, and Marine Sediments by Using Millions of Illumina Tags. Appl. Environ. Microbiol. 78, 8264–8271. doi: 10.1128/AEM.01821-12
Wickham H. (2016). Ggplot2: Elegant Graphics for Data Analysis (New York: Springer-Verlag). Available at: https://ggplot2.tidyverse.org.
Williams A. K., Bacosa H. P., Quigg A. (2017). The Impact of Dissolved Inorganic Nitrogen and Phosphorous on Responses of Microbial Plankton to the Texas City “Y” Oil Spill in Galveston Bay, Texas (USA). Mar. Pollut. Bull. 121, 32–44. doi: 10.1016/j.marpolbul.2017.05.033
Yu C., Hou L., Zheng Y., Liu M., Yin G., Gao J., et al. (2018). Evidence for Complete Nitrification in Enrichment Culture of Tidal Sediments and Diversity Analysis of Clade a Comammox Nitrospira in Natural Environments. Appl. Microbiol. Biotechnol. 102, 9363–9377. doi: 10.1007/s00253-018-9274-0
Yu T., Wu W., Liang W., Lever M. A., Hinrichs K. U., Wang F. (2018). Growth of Sedimentary Bathyarchaeota on Lignin as an Energy Source. Proc. Natl. Acad. Sci. U.S.A. 115, 6022–6027. doi: 10.1073/pnas.1718854115
Keywords: anchialine, microbial communities, organic matter, 16S rRNA gene, sediment
Citation: Risley CA, Tamalavage AE, van Hengstum PJ and Labonté JM (2022) Subsurface Microbial Community Composition in Anchialine Environments Is Influenced by Original Organic Carbon Source at Time of Deposition. Front. Mar. Sci. 9:872789. doi: 10.3389/fmars.2022.872789
Received: 10 February 2022; Accepted: 16 March 2022;
Published: 20 April 2022.
Edited by:
Efrain Miguel Chavez Solis, Autonomous University of Baja California, MexicoReviewed by:
Alejandra Prieto-Davó, Universidad Nacional Autónoma de México, MexicoSandi Orlic, Rudjer Boskovic Institute, Croatia
Copyright © 2022 Risley, Tamalavage, van Hengstum and Labonté. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jessica M. Labonté, labontej@tamug.edu
 Catherine A. Risley1
Catherine A. Risley1  Anne E. Tamalavage
Anne E. Tamalavage Jessica M. Labonté
Jessica M. Labonté