How does heat stress affect sponge microbiomes? Structure and resilience of microbial communities of marine sponges from different habitats
- 1Departament de Genètica, Microbiologia i Estadística, Universitat de Barcelona (UB), Barcelona, Spain
- 2Institut de Recerca de la Biodiversitat (IRBio), Universitat de Barcelona, Barcelona, Spain
- 3Departament de Biologia Evolutiva, Ecologia i Ciències Ambientals, Facultat de Biologia, Universitat de Barcelona (UB), Barcelona, Spain
- 4Red Sea Research Center, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia
- 5Division of Aquatic & Wildlife Resources, Guam Department of Agriculture, Mangilao, GU, United States
Introduction: Sponges are key components of marine benthic communities, providing many ecosystem functions and establishing close relationships with microorganisms, conforming the holobiont. These symbiotic microbiotas seem to be host species-specific and highly diverse, playing key roles in their sponge host. The effects of elevated seawater temperature on sponges and their microbiota are still poorly known, and whether sponges from polar areas are more sensitive to these impacts respect to temperate and tropical species is totally unknown.
Methods: We analyzed the microbiomes of different sponge species in their natural habitat and after exposure to heat stress in aquaria by 16S rRNA amplicon sequencing to (1) characterize the sponge microbiota covering a latitudinal gradient (polar, temperate and tropical environments), and (2) asses the effects of thermal stress on their microbial communities.
Results: Bacterial communities’ structure was different in the different sponge species and also respect the surrounding seawater. The core microbiome is maintained in most sponge species after a heat stress, although whether they would recover to the normal conditions previous to the stress remains yet to be further investigated. We observed increased abundances of transient bacteria from unknown origin in sponge species exposed to heat stress.
Discussion: Some of the transient bacteria may be opportunistic bacteria that may benefit from the heat stress-associated dysregulation in the sponge by occupying new niches in the holobiont. According to our results, sponges from Antarctic waters could be more resilient than tropical and temperate sponges. Both the microbiome composition and the changes produced by the heat stress seem to be quite host species-specific, and thus, depend on the sponge species. Under a global change scenario, the microbiomes of the tropical and temperate sponges will probably be those suffering the most the heat stress, and therefore the effects of global change may be dramatic for benthic ecosystems since sponges are a fundamental part of them.
1 Introduction
Sponges (phylum Porifera) are among the most ancient groups of metazoans (Bengtson, 1998; Feuda et al., 2017; Simion et al., 2017), and are widely distributed along tropical, temperate, and polar aquatic environments (Hooper and van Soest, 2002). These organisms are key components of marine benthic communities, providing ecosystem functions such as the creation of three-dimensional habitats to a variety of animals, giving shelter from predators, being source of food for spongivorous species, and contributing to the nutrient cycling (through the so-called benthic-pelagic coupling) due to their filter feeding nature (Bell, 2008; Southwell et al., 2008; de Goeij et al., 2013; Maldonado et al., 2015). Sponges often establish close relationships with diverse groups of microorganisms, including archaea, bacteria, microalgae, fungi, and viruses, that live within them, forming a complex structured ecosystem, called the sponge holobiont (Webster and Taylor, 2012; Thomas et al., 2016; Dittami et al., 2021; Leray et al., 2021). This symbiotic microbiota plays key roles in nutrient assimilation, waste metabolism, vitamin synthesis, or production of compounds with antifouling and defense properties, among many other aspects of the sponge’s physiology and ecology (Taylor et al., 2007; Hentschel et al., 2012).
Marine sponge-associated microbial community has been reported to be host species-specific and highly diverse, harboring over 60 bacterial and four archaeal phyla (Lee et al., 2011; Jackson et al., 2012; Schmitt et al., 2012a; Cleary et al., 2013; Kennedy et al., 2014; Reveillaud et al., 2014; Thomas et al., 2016; Moitinho-Silva et al., 2017a; Freeman et al., 2020). The sponge microbiome generally consists of a core microbiome (microbial species or amplicon sequence variants (ASVs) present in all specimens of the sponge species) and a variable part (ASVs not present in all specimens or present but at very different abundances) (Schmitt et al., 2012b; Blanquer et al., 2013; Astudillo-García et al., 2017). Despite the continuous flow-through of seawater within sponges, they manage to maintain a specific microbiome that is different and often more diverse than the surrounding seawater (Taylor et al., 2004; Taylor et al., 2007; Webster et al., 2010; Schmitt et al., 2012b; Taylor et al., 2013; Horn et al., 2016; Thomas et al., 2016). Although how the sponge selects and keeps certain microbial populations remains still unclear, some studies suggest that a very specific recognition system to discriminate symbiotic microbes may exist, with the host’s immune system being involved (Hentschel et al., 2012; Riesgo et al., 2014; Pita et al., 2018).
In terms of microbiota’s contribution to the sponge biomass, sponges have been classified as either high microbial abundance (HMA) or low microbial abundance (LMA) (Hentschel et al., 2003; Gloeckner et al., 2014). HMA sponges harbor microbes’ densities 2-4 orders to magnitude higher than LMA sponges (Webster et al., 2001; Hentschel et al., 2006; Hentschel et al., 2012). The microbiome of HMA sponges is richer and more diverse than that of LMA sponges, which is usually dominated by a few taxa (Björk et al., 2013; Erwin et al., 2015; Moitinho-Silva et al., 2017b). The dominant bacterial groups are also different for LMA and HMA sponges. While LMA sponges are usually dominated by Proteobacteria or Cyanobacteria, HMA sponges are enriched in Chloroflexi, Actinobacteria, or Acidobacteria, and usually harbor members of the phylum Poribacteria (Giles et al., 2013; Simister et al., 2013; Moitinho-Silva et al., 2017b). Despite the differences between HMA and LMA sponges’ microbiomes, the core microbial functions in the holobiont seem to be conserved independently of the HMA or LMA nature of the sponge (Thomas et al., 2010; Fan et al., 2012).
Since the development of next generation sequencing technologies in the late 1990s and early 2000s, the characterization of the microbiome diversity has grown exponentially (Taylor et al., 2007; Thomas et al., 2016; Webster and Thomas, 2016; Moitinho-Silva et al., 2017a; Stevens et al., 2017; Pita et al., 2018) The most common technique used to characterize microbial communities’ composition (e.g., Earth Microbiome Project) is 16S rRNA gene amplicon sequencing. Even though sponges’ microbiomes have been studied worldwide, most studies have focused on tropical and temperate sponges, and less attention has been paid to polar species. This is reflected in “The sponge microbiome project” (Moitinho-Silva et al., 2017a), where the microbiomes of 268 temperate and tropical sponge species were analyzed, but no polar species were included. However, in recent years, the microbiota of 31 Antarctic sponge species (or probably less since some of them were only identified to genus level) have been studied using 16S rRNA amplicon sequencing analysis (Webster et al., 2004; Rodríguez-Marconi et al., 2015; Cárdenas et al., 2018; Cárdenas et al., 2019; Lo Giudice et al., 2019; Steinert et al., 2019; Díez-Vives et al., 2020; Moreno-Pino et al., 2020; Papale et al., 2020; Sacristán-Soriano et al., 2020; Ruocco et al., 2021; Cristi et al., 2022; Happel et al., 2022). This is still a very small amount, considering that the most recent estimates of sponge species richness in the Southern Ocean and neighboring oceanographic regions were of 400 species as published in the Biogeographic Atlas of the Southern Ocean (Janussen and Downey, 2014).
Anthropogenic climate change has for long been known to have detrimental effects on marine environments (Smale et al., 2019; Cooley et al., 2022). For the last decades, along gradual ocean warming, ocean acidification, deoxygenation, and sea level rise, extreme events like heat waves have increased in frequency, duration, intensity, and extension (Oliver et al., 2018; Collins et al., 2019; Cooley et al., 2022). There is evidence that these heat waves have caused massive mortality events on marine benthic environments (Garrabou et al., 2009; Hereu and Kersting, 2016; Rubio-Portillo et al., 2016; Hughes et al., 2017; Ereskovsky et al., 2019; Garrabou et al., 2022). Although the sponge microbiome is in general stable across geographical and temporal scales (Erwin et al., 2012; Erwin et al., 2015; Cárdenas et al., 2019; Happel et al., 2022), it can also be influenced by environmental perturbations (e.g. Webster et al., 2008; Lesser et al., 2016; McDevitt-Irwin et al., 2017). Some studies have addressed the effect of elevated seawater temperature particularly on sponges, both on their physiology and on their microbiota (Simister et al., 2012b; Simister et al., 2012a; Vargas et al., 2021). While some sponges exposed to thermal stress suffered from tissue necrosis and bleaching (e.g. Bennett et al., 2017; Perkins et al., 2022), others seem to be less vulnerable (e.g. González-Aravena et al., 2019). Similarly, while warming produced changes and disruption on the microbiota of some sponge species (e.g. Simister et al., 2012a; Fan et al., 2013; Blanquer et al., 2016; Ramsby et al., 2018; Rondon et al., 2020), others remained unaffected (Webster et al., 2008; Erwin et al., 2012; Simister et al., 2012b; Strand et al., 2017). Mostly, however, the effect of the temperature increase in the microbiota of the sponges remains to be further investigated. Moreover, whether sponges from polar areas are more sensitive to these impacts respect to temperate and tropical species is totally unknown. The water temperature in Antarctic environments is very stable all year round, whereas temperate and tropical regions experience larger variations in seawater temperature (Gonzalez-Acosta et al., 2006; Thébault et al., 2007; Clarke et al., 2008; Lie and Lee, 2010). The adaptation of organisms to the particular environmental conditions of their region have led to organisms in the Antarctic regions to survive in a narrow temperature range (stenothermal organisms), compared to temperate and tropical organisms (Peck and Conway, 2000).
Here, the microbiomes of different sponge species from their natural habitat along with that of sponge specimens exposed to heat stress experiments in aquaria were assessed by 16S rRNA amplicon sequencing to (1) characterize the sponge species microbiota covering a latitudinal gradient (polar, temperate and tropical environments), and (2) asses the effects of thermal stress on their microbial communities.
2 Materials and methods
2.1 Sample collection
A total of 88 specimens belonging to four demosponge species living in similar shallow rocky benthic environments were collected at three regions representative of polar, temperate, and tropical coastal environments. Mycale acerata (LMA) (Sacristán-Soriano et al., 2020; Happel et al., 2022) and Dendrilla antarctica (LMA) (Koutsouveli et al., 2018; Díez-Vives et al., 2020), common yellow sponges in hard bottom benthic environments from polar (Antarctic) waters; and Agelas oroides (HMA) (Vacelet and Donadey, 1977; Blanquer et al., 2013), a lobed orange sponge present in temperate (Mediterranean Sea and Eastern Atlantic) waters; and Acanthella cavernosa (LMA) (Coelho et al., 2018; Cleary et al., 2019), a red spiky sponge present in tropical (Pacific Ocean) waters. Both Antarctic sponges, M. acerata (n=28) and D. antarctica (n=28) were collected at four different stations on Deception and Livingston Islands (South Shetland Islands) (Figure 1 and Table 1). The temperate species, A. oroides (n=25) was collected at two different stations from the Western Mediterranean coast (Costa Brava, Catalonia), while the Indo-pacific sponge, A. cavernosa (n=25) was collected at two different stations from Guam (Mariana Islands). Whole healthy sponge specimens and 2 L of seawater adjacent to the animals were collected manually by scuba diving at 10-25 m depths. Sponge specimens were kept in independent plastic zip bags or screw cap plastic containers filled with seawater and transported to the lab in liquid N2 or ice, within 1-2 h. Sampling took place in Antarctica (polar site) during the austral Summer of 2018, and 2019, while sampling in the Mediterranean Sea (temperate site) took place in May 2018 and sampling in Guam (tropical site) took place in April 2019 (Figure 1). Seawater temperatures at the moment of collection were -1 to 4°C in the South Shetland Islands, 14-15°C in the Mediterranean coast, and 28-29°C in Guam Island. A total of 29 specimens were used to assess the bacterial composition of the holobiont and 59 were used to simulate the effect of a heatwave in the microbiome. A summary of the number of samples used in the different experiments are shown in Table 1.
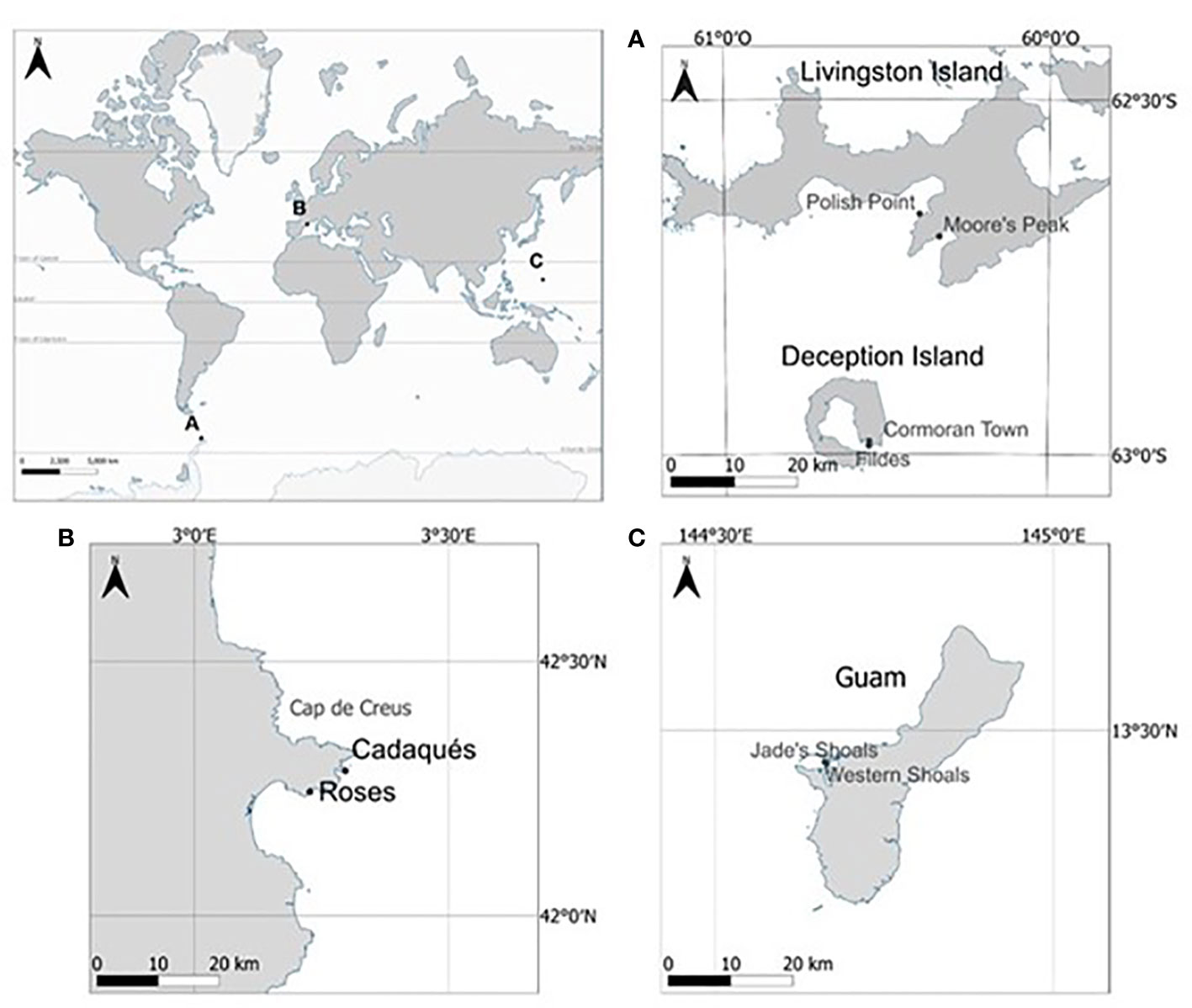
Figure 1 Sampling sites of the different regions. (A) Antarctic region (South Shetland Islands). (B) Mediterranean region. (C) sampling sites in Guam Island (Mariana Islands). Sampling locations are marked with black dots at each sampling region.
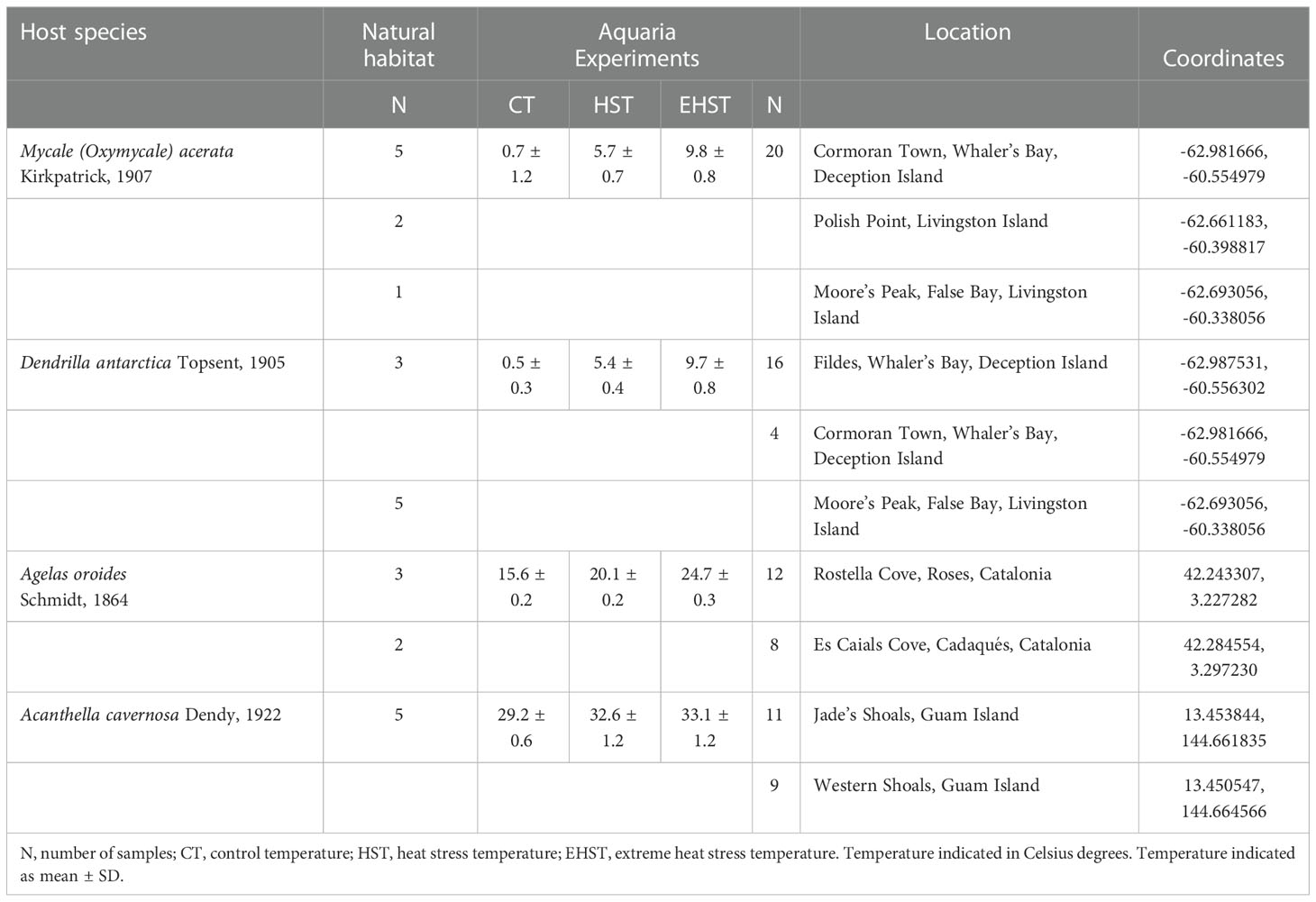
Table 1 Samples, number of sponge specimens, experimental conditions and sampling sites coordinates of the different specimens.
2.2 Heat stress experimental design
For the temperature experiments, at each locality, 20 different individuals of each species (M. acerata, D. antarctica, A. cavernosa, and A. oroides) were placed in aquaria at three different temperatures, including local seawater temperature, to be used as control temperature (CT), and at two higher temperatures [heat stress temperature (HST) and extreme heat stress temperature (EHST), around 4-5°C and 6-10°C higher than the CT, respectively]. Temperatures were chosen on each site according to the increase predicted by the IPCC and other reports, by duplicating the expected values and higher, and to the aquarium possibilities available for us to carry out the experiments on each place. Five more specimens of each species were placed in the aquaria at the beginning of the experiment and processed after 24 h to control for changes in the microbiota due to the manipulation, transport, and placement in the aquaria (aquaria adaptation control group). The experimental temperatures are summarized in Table 1. Water temperature was measured and controlled using a digital controller (Aqua Medic T controller twin) connected to heating (Sera 50 W or 150 W) and/or cooling (Aqua Medic Titan 150) units. The system was kept steady all along the experiment. Sponge specimens were kept in compartmented tanks (volumes ranging between 24-112.5 and 480 L according to the organism sizes), with seawater circulating through all the compartments, and were incubated for approximately three weeks for the Antarctic, two weeks for Mediterranean and one week for tropical sponge species. Samples were incubated at different times because temperature affects metabolism, and this is much fastest in the tropics and slowest in Antarctica. Therefore, the experimental times were adjusted to these different metabolisms.
2.3 Total DNA extraction and PCR amplification of 16S rRNA
All sponge specimens were washed with sterile seawater. An amount of 250 mg of each sponge specimen was homogenized to small pieces in a glass Petri dish with sterile scissors and blade. DNA was extracted using FastDNA™ SPIN Kit for Soil (MP Biomedicals, Illkirch, France). Extraction was performed following manufacturer’s instructions. Isolation of bacterial DNA from seawater collected from the surrounding environment was performed using a concentration-filtration method. One liter of each sample was filtered by vacuum filtration through a 0.22 μm pore-size mixed ester cellulose membrane. DNA from filter-retained bacteria was extracted using the same DNA extraction kit. Each filter was fitted in a 5 ml flask so that the bacteria retaining side was exposed to buffers and the beads in the first step of the kit’s protocol. The DNA concentration was quantified by Qubit fluorometer (Invitrogen). A negative control for all the extractions were performed.
2.4 Illumina 16S rRNA amplicon sequencing
Negative controls (blanks from the DNA extraction process, as well as from the DNA amplification, and sterile seawater used to wash the animals) as well as two different positive controls (ABRF-MGRG 10 Strain Staggered Mix Genomic Material (ATCC MSA-3002), and ZymoBIOMICS Microbial Community DNA Standard (D6306) were included in the 16S rRNA amplicon sequencing. Sample sequencing was performed in three runs using the Illumina MiSeq platform at the Genomics Unit of Centre for Genomic Regulation Core Facilities (CRG, Barcelona). The data is available at Mendely Data public repository (DOI: 10.17632/9c6y62nv9z.1, DOI: 10.17632/d67pjpc47g.1, DOI: 10.17632/fnjhzsybxj.1).
The V4 region was amplified from DNA sample extracts using the primers from the Earth Microbiome Project [515F (Parada et al., 2016) (5’- GTGYCAGCMGCCGCGGTAA-3’) and 806R (Apprill et al., 2015) (5’-GGACTACNVGGGTWTCTAAT-3’)] The PCR was performed in 25 μl volume with 0.2 μM primer concentration and KAPA HiFi HotStart ReadyMix (Roche). Cycling conditions were initial denaturation of 3 min at 95°C followed by 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, ending with a final elongation step of 5 min at 72°C. After this first PCR step, water was added to a total volume of 50 μl and reactions were purified using AgenCourt AMPure XP beads (Beckman Coulter). The first PCR primers contain overhangs allowing the addition of full-length Nextera adapters with barcodes for multiplex sequencing in a second PCR step, resulting in sequencing ready libraries with approximately 450 bp insert sizes. To do so, five μl of the first amplification were used as template for the second PCR with Nextera XT v2 adaptor primers in a final volume of 50 μl using the same PCR mix and thermal profile as for the first PCR but only 8 cycles. After the second PCR, 25 μl of the final product was used for purification and normalization with SequalPrep normalization kit (ThermoFisher Scientific), according to manufacturer’s protocol. Libraries were eluted and pooled for sequencing. Final pool libraries were analyzed using Agilent Bioanalyzer or Fragment analyzer High Sensitivity assay to estimate the quantity and check size distribution, and were then quantified by qPCR using the KAPA Library Quantification Kit (KapaBiosystems) prior to sequencing with Illumina’s Miseq 2x300bp.
2.5 Bioinformatic analyses
Cutadapt was used to trim adapters, primers, barcodes and leading Ns from sequencing reads. Sequences were processed to amplicon sequence variants (ASV) using the default parameters of the Dada2 workflow (Callahan et al., 2016). Firstly, quality filtering and the trimming of sequences was set to 180 bp (for forward reads) and 150 bp (for reverse reads) with a maximum number of expected errors allowed per read set at two (EE = 2). This parameter has been shown to be a better filter than simply averaging quality scores (Edgar and Flyvbjerg, 2015). Filtered sequences were dereplicated, the forward and reverse reads were aligned and merged. The sequences from the three runs were merged, chimeras were removed and an amplicon sequence variant (ASV) table was obtained. Taxonomy was assigned to the resulting ASVs using the SILVA SSU 138 reference database and was imported to the phyloseq R package for microbiome analyses. To obtain more accurate profiling of microbial communities, the decontam (Davis et al., 2018) R package was used to remove sequences derived from contaminating DNA present in extraction or sequencing reagents. In addition, chloroplast and mitochondrial reads were removed. The sequences of most abundant ASV that could not be identified at genus level were searched against GenBank database (rRNA database for Archaea and Bacteria) using Blast tool from NCBI (https://blast.ncbi.nlm.nih.gov/).
2.6 Data analyses
Alpha and beta diversity were analysed using the Phyloseq (McMurdie and Holmes, 2013) and vegan (Oksanen et al., 2022) R packages. For alpha diversity analysis, estimates of richness (namely Chao1 index, which is an estimate based on the abundance but affected by the number of ASVs appearing few times), and diversity indices (Shannon, which indicates both the richness but also considers the abundance of ASV, and Inverse Simpson, which is mostly affected by the dominance of certain ASVs in the sample) were calculated after rarefying the ASV table. In temperature experiments, rarefaction was performed to the different experimental groups ASV table for each target species. One-way analyses of variance (ANOVA) were used to assess differences in the alpha diversity indices across species (Mycale acerata, Dendrilla antarctica, Acanthella cavernosa, Agelas oroides), followed by HSD Tukey post-hoc test for significant ANOVAs. Student’s t-test was used to detect differences in the alpha diversity indices between the sponge specimens of the two Antarctic locations (Deception Island and Livingston Island) for M. acerata and D. antarctica, and between the sponge specimens of the two Mediterranean locations (Roses and Cadaqués) for A. oroides. One-way analyses of variance (ANOVA) were used to assess differences in the alpha diversity indices across aquaria experimental groups (natural habitat, adaptation group, control temperature, heat stress temperature, and high heat stress temperature) within each sponge species, followed by HSD Tukey post-hoc test for significant ANOVAs. The univariate statistics were performed using SPSS Statistics v27 (IBM Corporation). For beta diversity analysis, the number of reads of each ASV was previously transformed to relative abundance, the Bray Curtis distance was calculated, and samples were ordinated by non-linear multidimensional scaling (nMDS). Microbial core communities were determined at the sponge species level, defined as taxa shared (ASVs) by 100% of the sponge specimens. Additionally, for those sponges collected at different locations a separate core community was also determined for each location.
Furthermore, SourceTracker2 package (Knights et al., 2011) was used to determine the source of ASVs of sponge specimens from different experimental groups. For this, the natural habitat group, seawater, and aquarium water from the end of the aquaria experiments were treated as “source”, whereas the aquaria adaptation control group, control temperature (CT), heat stress temperature (HST) and extreme heat stress temperature (EHST) groups from each sponge species were treated as “sink” samples.
3 Results
3.1 Diversity of the sponge-associated bacterial communities in their natural habitat
A total of 2,947,270 reads were obtained after denoising and quality filtering of the raw sequencing data of sponge specimens collected from their natural habitat and their surrounding water. Among these, after chloroplast and mitochondria removal, 2,583,832 reads (88%) affiliated to Bacteria, representing 5,183 ASVs, with an average of 80,744 reads per sample (ranging from 9,681 to 180,387 reads) (Table 2). A total of 689 ASVs were recovered from M. acerata, 936 ASVs from D. antarctica, 2,071 ASVs from A. cavernosa, and 550 ASVs from A. oroides. Samples of seawater recovered in the same area as the sponge specimens had 480 ASVs on average. The sponge species were in all cases enriched for ASVs in relation to the surrounding seawater with the exception of the Mediterranean environment, from where 755 ASVs were recovered from the water whereas 550 ASVs were recovered from A. oroides.
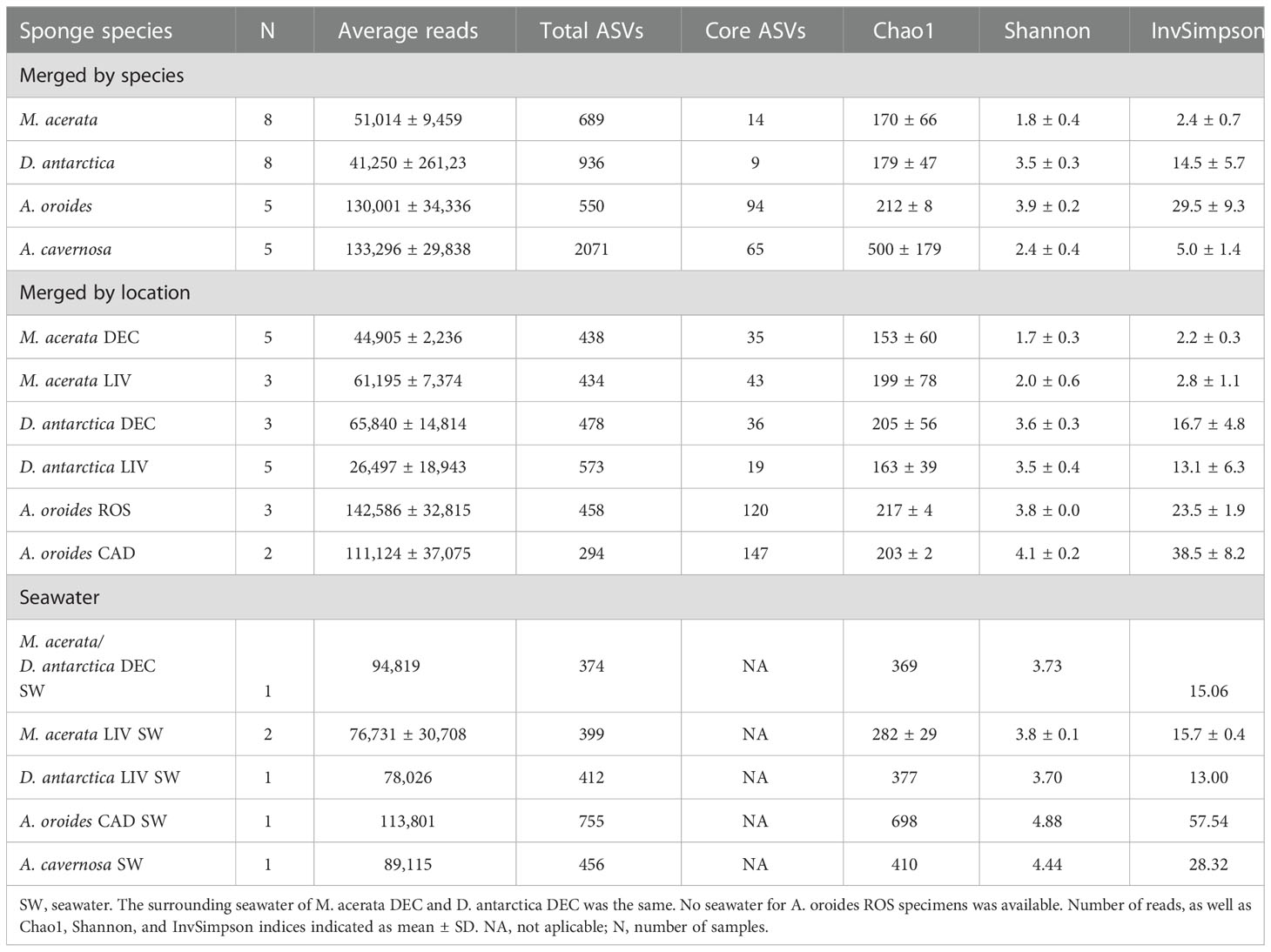
Table 2 Summary of reads, ASVs, and diversity of sponge species and water of the different geographic areas.
Around 3% of the reads affiliated to domain Archaea (ranging from 3 to 25,329 reads per sample), showing an uneven distribution between the different sponge species. In fact, only two of the studied species (A. oroides and D. antarctica) and their corresponding seawater samples harbored Archaea at an abundance higher than 1% of the total reads [A. oroides (8.08-18.84% and D. antarctica from Livingston (1.12-2.22%)]. In both cases, the archaeal community was dominated with 99-100% abundance by a unique genus, Cenarchaeum in A. oroides and “Candidatus Nitrosopumilus” in D. antarctica, respectively. Both genera belong to the Nitrososphaeria class in the Crenarchaeota phylum. Seawater associated to A. oroides were dominated by Marine Group II archaea (52%), “Candidatus Nitrosopumilus” (28%), “Candidatus Nitrosopelagicus” (11%) and Cenarchaeum (6%) genus, and Marine Group III (4%). On the other hand, seawater associated to A. cavernosa was dominated by the Marine Group II (98%). Archaea were not included in further analyses.
The richness was significantly higher in the Tropical sponge A. cavernosa (observed ASVs, 2071; and Chao 1 index, 500) than in the other sponge species (p<0.001) (Table 2). It has to be noted that more than 50% of these ASVs were found less than 10 times. A lower number of ASVs was observed for D. antarctica (936), followed by M. (689), and A. oroides (550). A. oroides and D. antarctica had the highest Shannon diversity index (3.9 and 3.4, respectively), followed by A. cavernosa (2.4) and M. acerata (1.8) (Table 2). Statistically significant differences were observed between M. acerata and the sponges D. antarctica, A. cavernosa and A. oroides, and between A. cavernosa and the sponges M. acerata, D. antarctica and A. oroides (p<0.001). This index indicates both the richness but also considers the proportion of ASV. In the case of the Inverse Simpson index, which is mostly affected by the dominance of certain ASVs in the sample, a similar trend was observed with the highest average value Obtained for A. oroides (29.5), followed by D. antarctica (14.5), A. cavernosa (5.0) and M. acerata (2.4) (Table 2). Statistically significant differences were observed between M. acerata and the sponges D. antarctica and A. oroides, between D. antarctica and the sponge A. oroides, and between A. cavernosa and the sponges D. antarctica and A. oroides. The effect of the sampling location on the richness and diversity indices was also investigated showing no statistically significant differences among the different sampling locations of the same species. (p>0.05). The exception was the Chao1 index in A. oroides, which was higher in the sponge specimens from Roses than in the sponge specimens from Cadaqués (p=0.025) (Table 2).
3.2 Taxonomic composition and structure of the sponge-associated bacterial communities in their natural habitat
The taxonomic composition of the bacterial communities at phylum level is shown in Figure 2 and Supplementary Table 1. The bacterial communities of M. acerata were dominated by Proteobacteria (86±7%), specifically class Gammaproteobacteria (79±9%), independently of the sampling location. D. antarctica was also dominated by Proteobacteria (62±13%), with Alphaproteobacteria constituting 35±13% of the bacterial community, followed by Bacteroidota (29±11%) and Gammaproteobacteria (27±15%), also independently of the sampling location. A. oroides was dominated by the phylum Chloroflexi (37±9%), which was practically absent in the other sponge species and the surrounding seawater, followed by Gammaproteobacteria (22±4%), in both sampling locations. Finally, A. cavernosa was dominated by Cyanobacteria (47±14%), followed by Planctomycetota (27±7%), which was present at <1% on average in the other sponge species and seawater, and Gammaproteobacteria (20±22%).
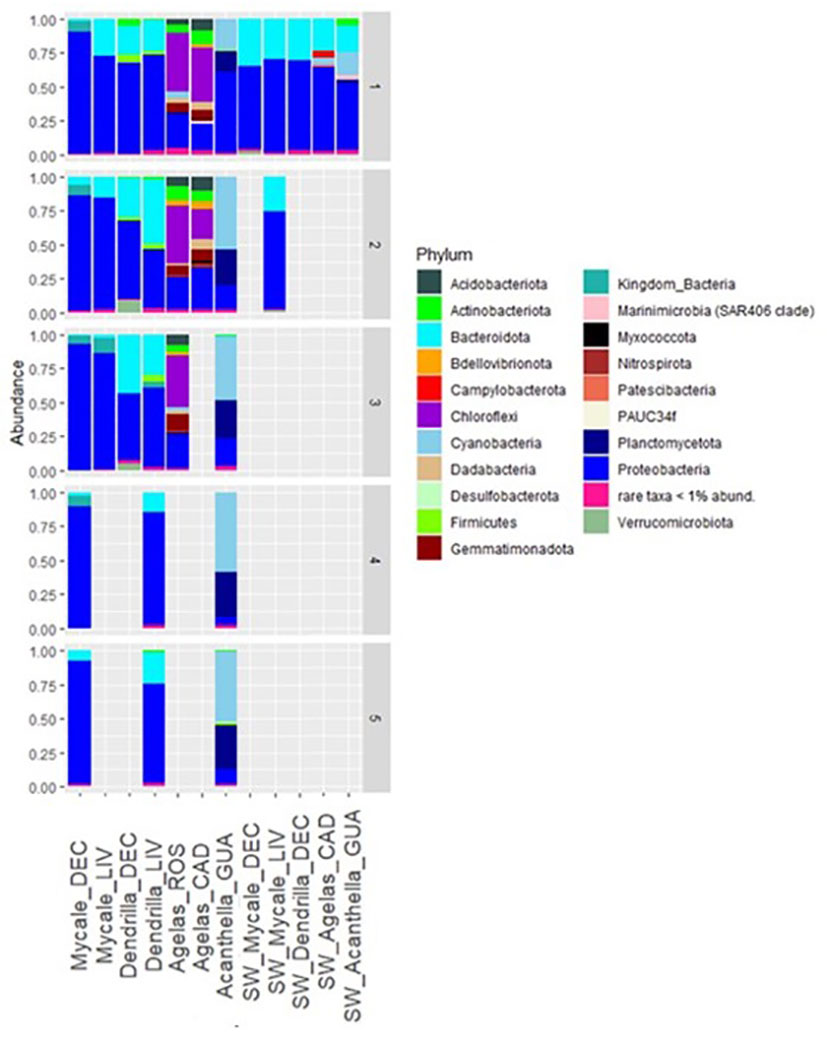
Figure 2 Taxonomic affiliation of ASVs detected in sponge species and seawater in their natural habitat at Phylum level. DEC, Deception Island; LIV, Livingston Island; ROS, Roses; CAD, Cadaqués; GUA, Guam; SW, seawater. Numbers on the right axis indicate sample replicates.
The bacterial community of the seawater from the different locations where the sponges were collected was dominated by Proteobacteria (63±8%), followed by Bacteroidota (27±6%) (Figure 2). Class Alphaproteobacteria was in all seawater samples equal or more abundant than class Gammaproteobacteria. Specifically, seawater surrounding M. acerata hosted a bacterial community dominated by Proteobacteria (67±6%) and Bacteroidota (30±5%), independently of the sampling location. The seawater surrounding D. antarctica in Livingston Island had a community dominated by Proteobacteria (66%) and Bacteroidota (31%). Seawater surrounding A. cavernosa specimens hosted a bacterial community dominated by Proteobacteria (50%) (with Alphaproteobacteria as the most abundant (37%) versus Bacteroidota (20%), Cyanobacteria (17%), and Gammaproteobacteria (12%). Finally, seawater around A. oroides (temperate sampling region), was enriched in Proteobacteria (61%), Bacteroidota (24%), Cyanobacteria (5%) and Campylobacterota (5%).
The majority of ASVs could not be affiliated to the genus taxonomic rank, and therefore they were affiliated at a higher taxonomic rank. The complete taxonomic affiliation from genus to class is shown in Supplementary Figures 3. Notably, the ten most abundant ASVs from each sponge species were distributed unevenly in the other sponges (Figure 3). For example, M. acerata most abundant ASVs were affiliated to the family EC94 with an abundance of 69% altogether. In this species, 4% of the reads could not be affiliated at a lower taxonomic rank below Bacteria kingdom. In D. antarctica, the most abundant ASVs were an unidentified ASV affiliated to an unidentified genus of class Alphaproteobacteria (>13%), Polaribacter (>12%), an unidentified ASV affiliated to the family SAR116 clade (10%), followed by Burkholderia-Caballeronia-Paraburkholderia (3%), two unidentified ASVs affiliated to family Nitrincolaceae and family S25-593, respectively, Kistimonas and the NS4 marine group (all of them at relative abundance <3%). Among the ten most abundant ASVs detected in A. cavernosa Cyanobium was the most abundant (40%), followed by an unidentified ASVs affiliated to family Pirellulaceae (23%), and ASVs affiliated to Ralstonia (13%) and Synechococcus (6%). In A. oroides, again the most abundant ASVs at a relative abundance higher than 5% were not shared with the other species or were practically absent. These included mostly unidentified genera from different families or classes (family A4b (>14%), Sva0996 marine group (6%), clade KI89A (5%), class TK17 (7%), PAUC43f marine benthic group (3%), class TK30 (5%), and family Caldilineaceae (2.5%)).
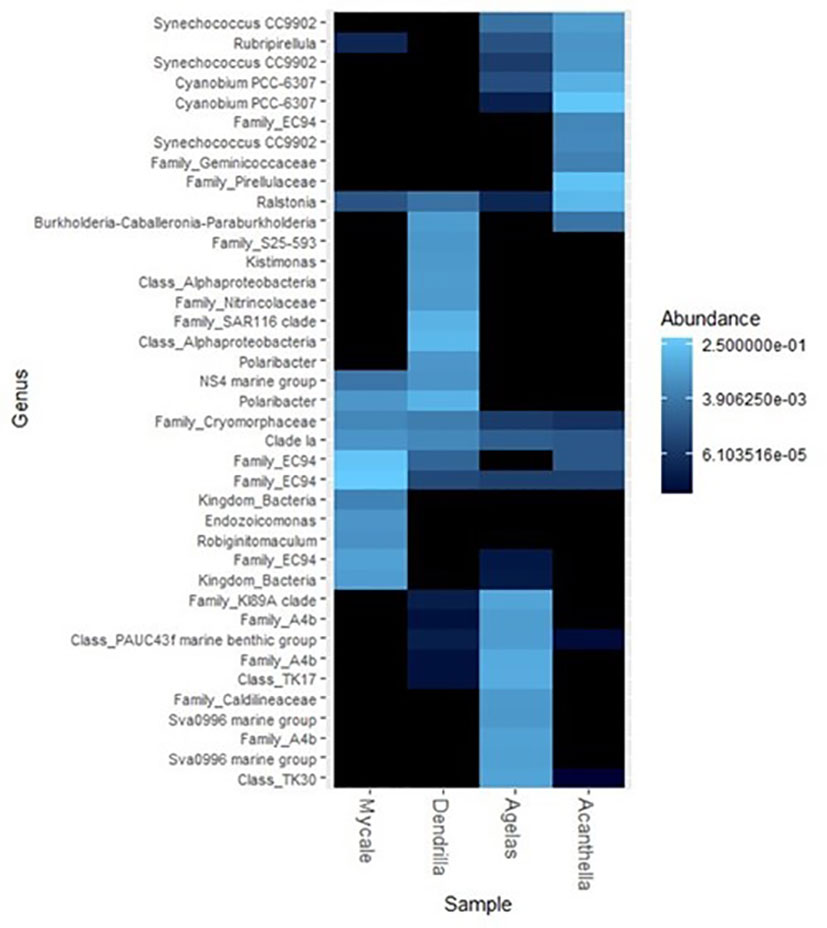
Figure 3 Heatmap representation of the abundance of the 10 most abundant ASVs of the different sponge species with its taxonomic affiliation at the lowest taxonomic rank. Their presence in all sponge species is shown.
This distinct bacterial communities’ structure observed between the different sponge species and between the surrounding water and the corresponding sponges was also visualized by non-metric multidimensional scaling (nMDS) of Beta-diversity (Bray-Curtis) coefficients (Figure 4). The samples clustered together according to the sponge species. The Antarctic sponges M. acerata and D. antarctica were closer between them than the other sponge species, but without overlapping each other. All sponge species were distant from seawater in the multidimensional space, except from M. acerata specimens from Livingston, that exhibited overlap with the group formed by Antarctic seawater samples.
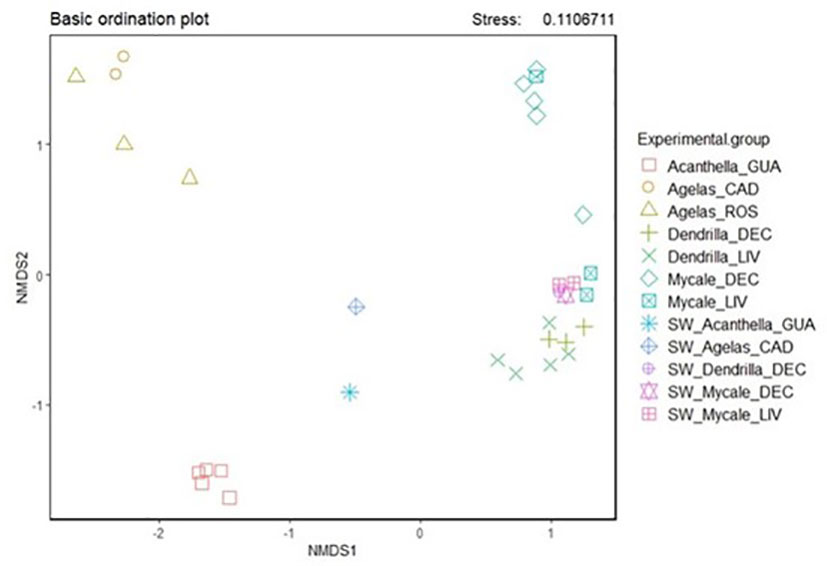
Figure 4 Non-linear multidimensional scaling representation of the different sponge species from the different geographic areas and sampling locations and their surrounding water. DEC, Deception Island; LIV, Livingston Island; GUA, Guam; ROS, Roses; CAD, Cadaqués; SW, seawater.
3.3 Core bacterial communities of the sponges in their natural habitat
The core bacterial community of M. acerata from Deception Island was composed of 35 ASVs, and that from Livingston Island of 43 ASVs (Table 2). Considering all replicates from both locations together, the core community of M. acerata was restricted to 14 ASVs. It must be noted that although the most abundant bacterial taxa in all M. acerata replicates was a unidentified genus from the EC94 family, with relative abundance ranging 68-78%, this genus was represented by a total of 27 ASVs, none of which was shared by 100% of the replicates. ASVs 1 and 3 (both affiliated to the EC94 family) were the two most abundant ASVs in M. acerata specimens in average. These ASVs showed the highest identity to different Nitrosomonas spp using Blast against GenBank rRNA database [Supplementary Table 3 (Table 1)]. All M. acerata replicates had at least one of these ASVs. ASVs 1 and 3 were also present in seawater samples, whereas ASV47 (also affiliated to the EC94 family) was only present in one seawater sample. Nevertheless, they were up to 1000-2000 times more abundant in the sponge specimens than in seawater. The most abundant ASVs shared by 100% of the replicates of the sponge were affiliated to SAR11 Ia clade and Polaribacter (around 2% on average). Both were more abundant in seawater’s community than in the sponges’ communities: around ten times more abundant for the genus affiliated to SAR11 Ia clade, and around two times for Polaribacter. The other ASVs in M. acerata core community were less abundant than 1% on average.
The core bacterial community of D. antarctica from Deception Island included 36 ASVs, and that from Livingston Island, 19 ASVs. Considering all specimens from both locations, the core community of D. antarctica was composed of 9 ASVs. The five most abundant ASVs on average were affiliated to a unidentified genus of the SAR116 clade, Polaribacter, a unidentified genus of the NS4 marine group, a unidentified genus of the OM43 clade, and a unidentified genus of the Nitrincolaceae family. These unidentified ASVs showed the highest identity to genera Thalassobaculum, Kordia, Rivicola, and Maribrevibacterium from GenBank DB, respectively [Supplementary Table 3 (Table 1)]. SAR116 clade was not present in the seawater samples, the ASVs associated to a unidentified genus of the Nitrincolaceae family was equally abundant in the sponge specimens and seawater (~1% on average), and the other three were six to nine times more abundant in the sponge specimens compared to seawater.
The core bacterial community of the tropical sponge A. cavernosa included 65 ASVs. The five more abundant ASVs in the core bacterial community represented on average around 80% of the whole bacterial community. These ASVs affiliated to the genus Cyanobium, a unidentified genus of the Pirellulaceae family (showing the highest identity to genus Botrimarina from GenBank DB) [Supplementary Table 3 (Table 1)], Ralstonia genus and Synechococcus genus. All these five ASVs were present in the seawater community, with relative abundances under 0.5%, except for ASV34 (Synechococcus genus), which represented the 14% of the seawater community.
The core bacterial community of the temperate sponge A. oroides included 94 ASVs. Four out of the five more abundant ASVs on average (with abundances 4-7% of the community) belonged to bacteria in the Chloroflexi phylum. Specifically, these bacteria belonged either to class TK17, family A4b or class TK30 (showing the highest identity to genera Litorilinea, Ornatilinea and Ammoniphilus, respectively, from GenBank DB) [Supplementary Table 3 (Table 1)]. The third most abundant bacterial taxa in the core community of A. oroides was the KI89A clade (4.7% average relative abundance), belonging to the oligotrophic marine Gammaproteobacteria (OMG) group. These ASVs were either absent in the seawater bacterial community (class TK30, KI89A clade), or were 1000-2000 times more concentrated in the sponge specimens than in the seawater (class TK17, family A4b).
There were four ASVs (ASV 1, 2, 18, 112) that were shared among the four sponge species studied although not present in 100% of the replicates. The taxa assigned to them were a unidentified genus of the EC94 family, a unidentified genus of the SAR11 Ia clade, Ralstonia, and a unidentified genus of family Cryomorphaceae. These unidentified ASVs showed the highest identity to genera Nitrosomonas, Pelagibacter and Phaeocystidibacter, from GenBank DB) (Supplementary Table 1). The ASVs relative frequencies varied among the different sponge species being ASV 1 dominant in M. acerata (39%), with respect to the other sponge species (0.01-0.02%). The second most abundant shared ASV was ASV 18 in A. cavernosa (14%), being detected in the other sponges at lower relative abundances (with up to 4000 times lower relative abundance in A. oroides). The other left ASVs were present at lower frequencies (<2% in all of the sponge species). All except ASV1 were found ubiquitously in the surrounding seawaters, except ASV1, which was only found in the seawater sample from Deception Island at very low relative abundance (0.03%).
3.4 Effect of heat stress
The results of the effect of a heat wave on the bacterial communities of the sponge species are shown in Table 3 and Supplementary Table 2. In all four species, the reads from the adaptation control specimens mainly matched those of the natural habitat group (M. acerata 91.8%, A. cavernosa 61.6%, and A. oroides 82.8%), with lower percentage in D. antarctica, 46.6% (Table 3). At the end of the experiments, the percentage of sequences that were tracked to the natural habitat specimens had decreased in all cases. In the specimens exposed to extreme heat stress temperature, the source of the highest percentage of reads was the aquarium seawater in all cases. The experimental groups maintained at CT were those with a higher percentage of sequencing tracking the natural habitat specimens, although only M. acerata retained most sequences of the natural habitat specimens at CT condition. Taking into consideration the increasing temperatures, the percentages of shared sequences decreased as the temperature increased (p<0.05 in all comparisons of HST vs. CT, and EHST vs. CT, except for D. antarctica HST vs. CT).
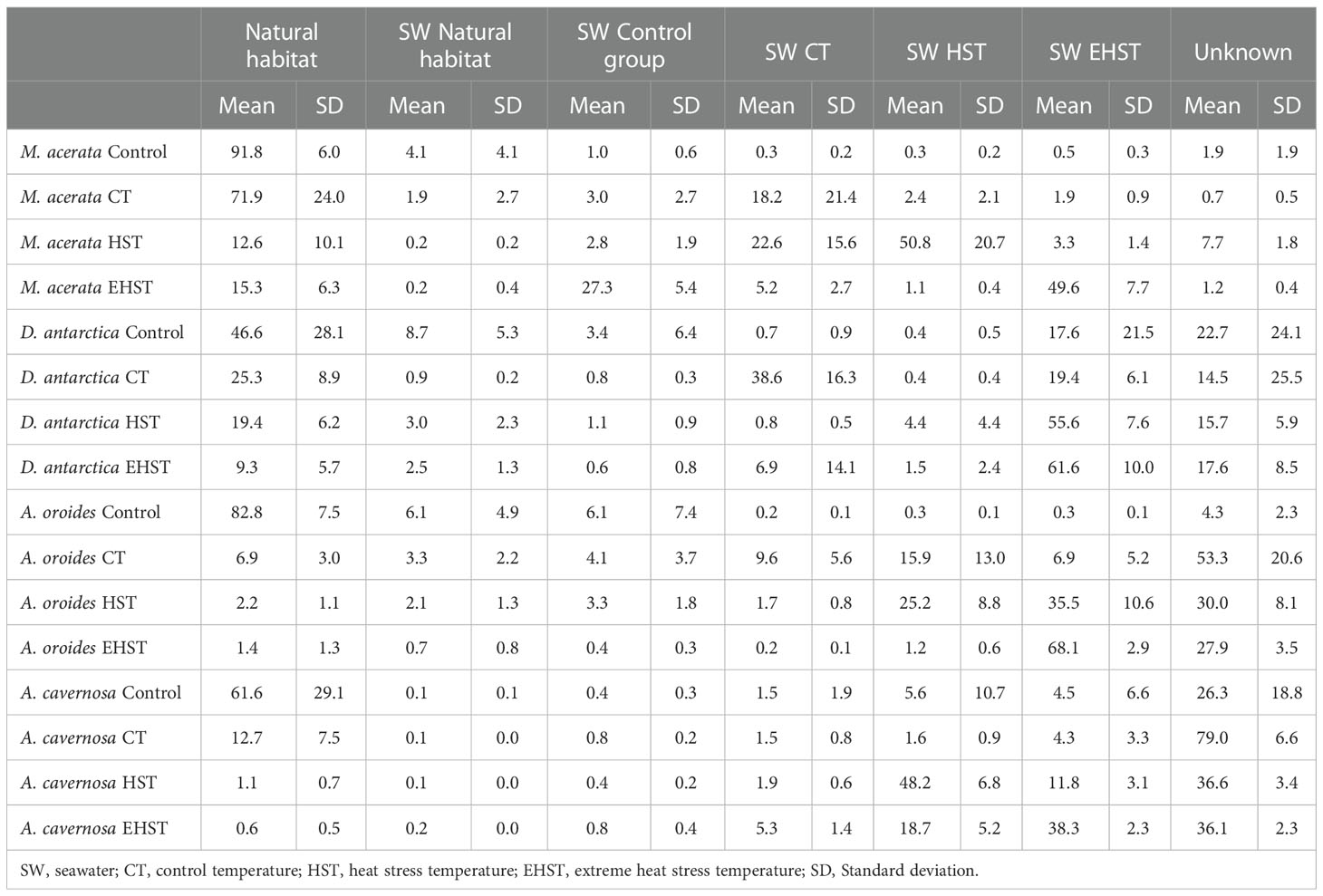
Table 3 Average contribution of reads (%) of the possible sources to the microbiome of the different sponge species experimental groups detected in the aquaria exposed at different temperatures.
In order to assess the effect of temperature in the communities’ biodiversity, the richness of observed species and Shannon index were analyzed (Figure 5). A general decrease trend in the observed species richness and Shannon diversity index with the increasing temperature was observed for the tropical A. cavernosa and the temperate A. oroides. This trend was reversed for the Antarctic sponges, although in the case of M. acerata the diversity of the EHST experimental group was between HST and CT experimental groups. The diversity of the sponge specimens in their natural habitat was higher than the corresponding experimental group mimicking the natural conditions (CT) in the case of D. antarctica (p<0.05 for Shannon index difference) and lower in the case of A. cavernosa (p<0.05 for Shannon index difference). The diversity in natural habitat and CT sponge specimens was comparable in M. acerata (p>0.05 for Shannon index difference) and the same was observed in A. oroides (p>0.05 for Shannon index difference).
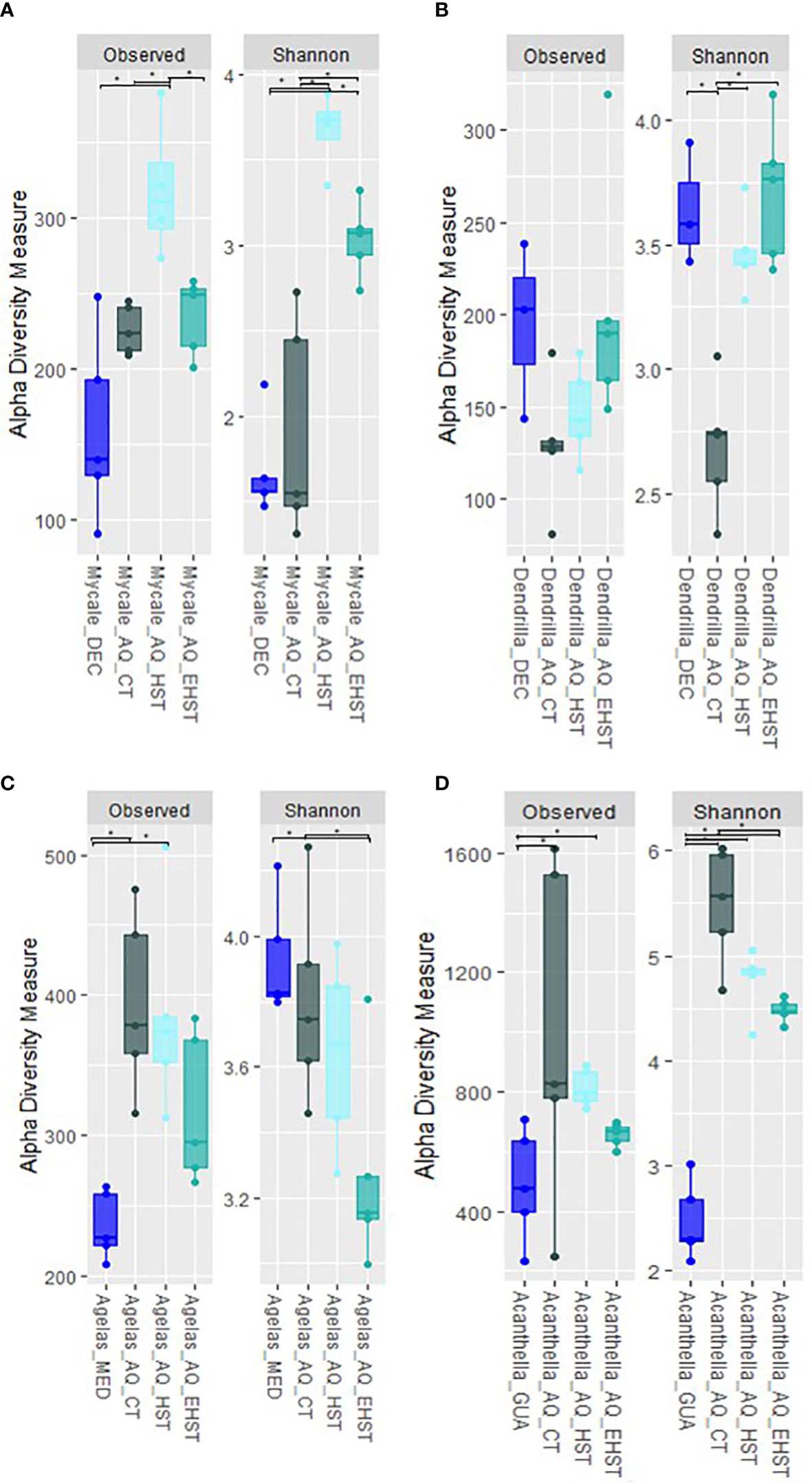
Figure 5 Alpha-diversity measurements of the bacterial communities of sponge specimens in aquaria experiments at the different experimental temperatures (CT, control temperature, HST, heat stress temperature, EHST, extreme heat stress temperature) compared to the natural habitat specimens: (A) M. acerata from Deception Island; (B) D. antarctica from Deception Island; (C) A. oroides from Cadaqués and Roses; (D) A. cavernosa from Guam. AQ, aquarium experiments. * p-value <0.05.
The diversity changes observed in the different experimental conditions were also assessed taxonomically (Figure 6). Changes were observed in the most abundant phyla among the different experimental groups. The phylum Proteobacteria decreased in M. acerata exposed to low heat stress (HST) compared to the other experimental groups. Instead, Bacteroidota increased in M. acerata exposed to HST (28±9%) compared to the natural habitat group, and slightly decreased at the extreme heat stress (EHST). Additionally, an enrichment in Campylobacterales was observed (14±2%), compared to the other experimental groups, where it was practically absent. Cyanobacteria decreased in M. acerata exposed to experimental temperatures and was almost absent in the EHST group.
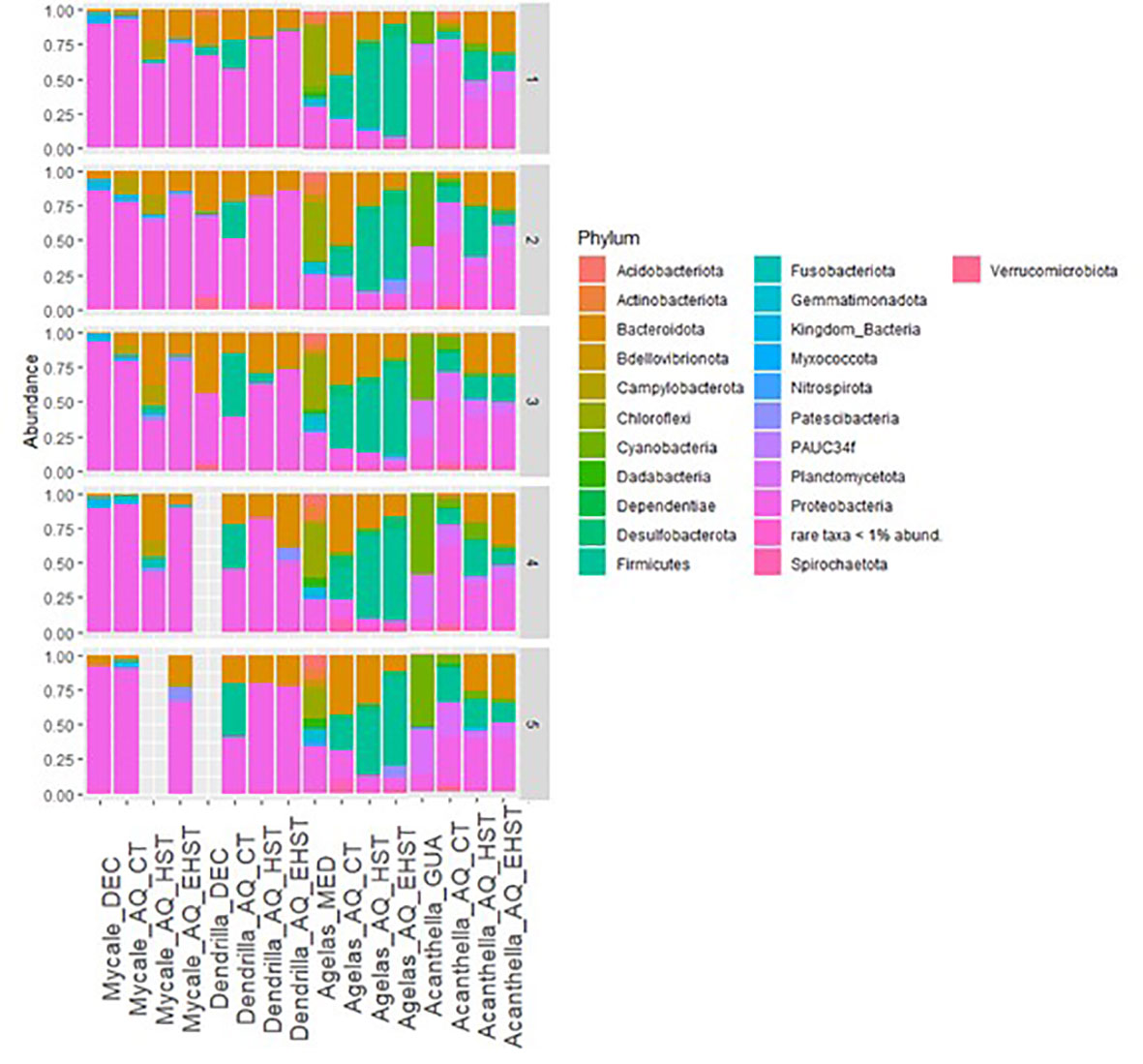
Figure 6 Taxonomic affiliation of ASVs at Phylum level in sponge species in their natural habitat and in the aquaria exposed at different temperatures (CT, control temperature, HST, heat stress temperature, EHST, extreme heat stress temperature). DEC, Deception Island; MED, Roses and Cadaqués origin; GUA, Guam; SW, seawater. Numbers on the right axis indicate sample replicates. AQ, aquarium experiments.
In D. antarctica exposed to both heat stresses the abundance of Proteobacteria increased compared to the natural habitat group. Additionally, the proportion of Gamma- versus Alphaproteobacteria was inversed in all experimental groups compared to the natural habitat group. Bacteroidota was less abundant in the three experimental groups than in the natural habitat group. Firmicutes represented 25±17% of the bacterial community on average in CT sponge specimens but was practically absent in the other groups’ communities.
In A. oroides, Proteobacteria phylum was less abundant in the sponge specimens of the heat stress experiment than in the natural habitat group. Moreover, while in the natural habitat communities’ class Gammaproteobacteria was more abundant than class Alphaproteobacteria, this relationship was reversed in the communities of the three experimental groups. The three experimental groups were enriched in Bacteroidota compared to the natural habitat group, but HST and EHST sponge specimens were less enriched than CT. The phylum Chloroflexi represented 37±9% of the bacterial community of the natural habitat group in A. oroides but was practically absent in the communities of the experimental groups. Firmicutes was more abundant in the CT group than in the natural habitat group. HST group was more enriched in bacteria from this phylum than CT, and in EHST group, the abundance of Firmicutes was even higher than in HST.
In A. cavernosa, the communities from the groups exposed to the three experimental temperatures were enriched in Proteobacteria compared to the natural habitat sponge specimens’ communities. Also, while in the natural habitat communities, class Gammaproteobacteria was more abundant than class Alphaproteobacteria, this relationship was reversed in the communities of the three experimental groups in A. cavernosa. Bacteroidota showed and increase trend with the heat stress from lower than 1% in CT and natural habitat up to 38% in the EHST experimental group. Cyanobacteria was markedly less abundant in A. cavernosa in aquaria experiments compared to the natural habitat, being almost absent at A. cavernosa EHST group. The bacteria belonging to Planctomycetota were less abundant in A. cavernosa HST experimental group compared to the other groups. Firmicutes was more abundant in the CT group than in the natural habitat group and even more in the HST group. Nevertheless, in EHST group, the presence of Firmicutes was similar to CT.
Other phyla that were present in the natural habitat group but were not in the aquaria experimental groups were Acidobacteriota, Gemmatimonadota, and Actinobacteriota. Conversely, Desulfobacterota was more abundant in the communities of CT than natural habitat group, and its presence was higher in HST and even higher in EHST experimental groups. At genus level changes were also observed between the different experimental groups. If we focus on the core communities of the sponge species, the most abundant core taxa were maintained in the different experimental groups in the Antarctic sponges although with changing relative proportions (Figure 7). Nevertheless, some genera were practically lost from the core communities in all the experimental conditions such as Oceanicoccus, SAR92 clade, Sulfitobacter, Rubritalea, in D. antarctica, or Arenicella, Rubidimonas, Ralstonia, Lutibacter and Winogradskyella in M. acerata, among others.
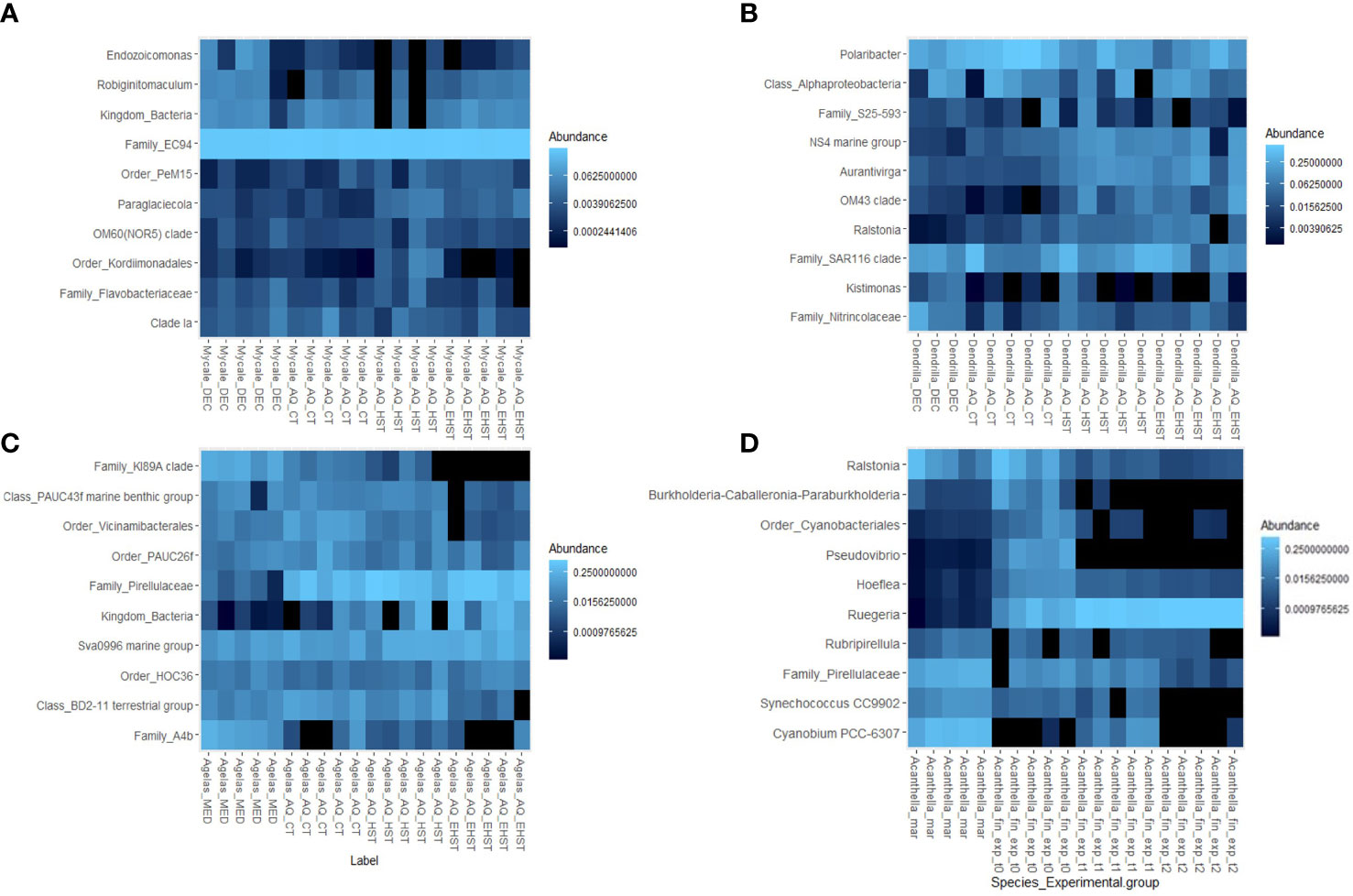
Figure 7 Heatmap representation of the abundance of the 10 most abundant genera from the core sponge communities of each sponge species in the different aquaria experimental groups exposed to different temperatures (CT, control temperature, HST, heat stress temperature, EHST, extreme heat stress temperature) and natural habitat: (A) M. acerata from Deception Island; (B) D. antarctica from Deception Island; (C) A. oroides from Cadaqués and Roses; (D) A. cavernosa from Guam. AQ, aquarium experiments.
The dominant taxa in the microbial communities of M. acerata natural habitat group were also hosted by the sponge specimens exposed to CT, HST and EHST (Figure 7A). The mean relative abundances in the 5 CT replicates were as follows: unidentified genus of family EC94 (67±22%), genus Endozoicomonas (2±5%). The unidentified genus of Family EC94 and genus Endozoicomonas were less abundant in CT sponge specimens with respect to natural habitat sponge specimens. In HST experimental group, family EC94 represented 12±9% of the bacterial community, while genus Endozoicomonas was 2±1%. Regarding EHST experimental group, unidentified genus of family EC94 represented 16±7% of the bacterial community, and Endozoicomonas, 0.1±0.1%.
Concerning the dominant taxa in M. acerata exposed to HST the dominant taxa were, after family EC94, genus Pseudarcobacter (8±4%), genus Colwellia (8±3%), genus Crocinitomix (7±4%), family Marinifilaceae (6±6%), and genus Psychrobium (5±5%). All these taxa were present in the aquarium water, being 2-3 times less abundant in the water, except for genus Crocinitomix, which was almost equally abundant. In the EHST group bacterial communities, the most abundant taxa were genus Colwellia (27±3%), which was also the dominant taxa in the EHST aquarium water, family Rhodobacteraceae (15±7%), genus Pseudoalteromonas (5±2%), and genus Polaribacter (5±2%). All these taxa were practically absent in the natural habitat group sponges.
Concerning the most abundant taxa, the range of abundances in D. antarctica in the 5 CT replicates were as follows: genus Polaribacter (14±2%), unidentified genus of class Alphaproteobacteria (7±7%), clade SAR116 (6±8%), and unidentified genus of family Nitrincolaceae (1±1%) (Figure 7B). Genus Polaribacter was less abundant in CT sponge specimens with respect to natural habitat sponge specimens. Class Alphaproteobacteria and SAR116 clade had half relative abundance in CT sponge specimens than in the natural habitat sponge specimens. Family Nitrincolaceae was more than 10 times less abundant in CT sponge specimens than in natural habitat sponge specimens. Similarly, these taxa were present in the HST sponge specimens’ communities, with similar abundances to CT sponge specimens, with the exception of Polaribacter (6±2%). The abundances of these taxa in the microbial communities of the sponge specimens exposed to EHST were lower than in HST sponge specimens, except for family Nitrincolaceae, which was more abundant in EHST sponge specimens.
The dominant taxon in D. antarctica HST group communities was group Burkholderia-Caballeronia-Paraburkholderia (34±5%), almost absent in the aquarium water. In the EHST group bacterial communities, the most abundant taxon was also group Burkholderia-Caballeronia-Paraburkholderia (29±17%). The relative abundance of Burkholderia-Caballeronia-Paraburkholderia was similar in the aquarium water communities and in EHST sponges. Genus Burkholderia-Caballeronia-Paraburkholderia were practically absent in the natural habitat group sponges.
In A. oroides, some genera were found in all experimental groups such as the class TK17 and Sva0996 marine group (Figure 7D). HST sponge specimens kept core taxa such as family A4b, class TK30, or clades KI89A and SAR202 in most of the specimens, which were lost in EHST sponge specimens. The mean abundances in the 5 CT replicates were as follows: family A4b (0.2±0.2%), KI89A clade (0.2-0.1%), class TK17 (0.04±0.03%), and class TK30 (0.1±0.041%). These had lower abundances in the microbial communities of the sponge specimens exposed to HST, and even lower in EHST.
The dominant taxa in A. oroides HST group communities were genus Fusibacter (23±6%), family Clostridiaceae (15±5%), family Marinifilaceae (13±7%), order Peptostreptococcales-Tissierellales (5±2%), and genus Halodesulfovibrio (4±1%). In the EHST group bacterial communities, the most abundant taxa were family Clostridiaceae (40±7%), genus Halodesulfovibrio (7±3%), order Peptostreptococcales-Tissierellales (6±2%), and group Clostridia vadinBB60 (5±0.6%). Only two of the dominant taxa of the bacterial communities in HST and EHST experimental groups were present in the natural habitat group: family Clostridiaceae and genus Halodesulfovibrio, with abundances <0.01% on average.
In the tropical sponge A. cavernosa, most taxa were still present in CT sponge specimens although at lower abundance, but lost in HST and EHST groups such as the unidentified genus of Family EC94, Filomicrobium or Blastopirellula, or lost mostly at the highest temperatures (in EHST group), such as Synecococcus and Cyanobium. Still, the dominant taxa of the sponge species’ core microbiota were present in HST sponge specimens (Figure 7C). The range of abundances in the 5 CT replicates were as follows: genus Cyanobium, 1±1%, family Pirellulaceae, 2±1%, genus Ralstonia, 5±5%, and genus Synechococcus, 0.3±0.1%. These had lower abundances in the microbial communities of the sponge specimens exposed to HST, and even lower in EHST.
The dominant taxa in A. cavernosa HST group communities were family Clostridiaceae (14±6%), family Flavobacteriaceae (7±2%), genus Ruegeria (6±2%), genus Winogradskyella (6±1%) and family Rhodobacteraceae (5±0.6%). In the EHST group bacterial communities, the most abundant taxa were family Flavobacteriaceae (17±3%), genus Ruegeria (15±2%), family Clostridiaceae (8±3%) and genus Muricauda (3±2%). All the dominant taxa of the bacterial communities in HST and EHST experimental groups were present in the natural habitat group, except for genera Winogradskyella and Muricauda. The abundances in the natural habitat group of the experimental groups dominant taxa were <0.7%.
The number of ASVs shared between the sponge specimens from natural habitat and those exposed to CT, HST and EHST for each sponge species are shown in Table 4. As shown, the number of ASVs shared between natural habitat specimens and those exposed to HST and EHST decreased notably with the increasing temperature specially for the temperate A. oroides and the tropical A. cavernosa sponge species.
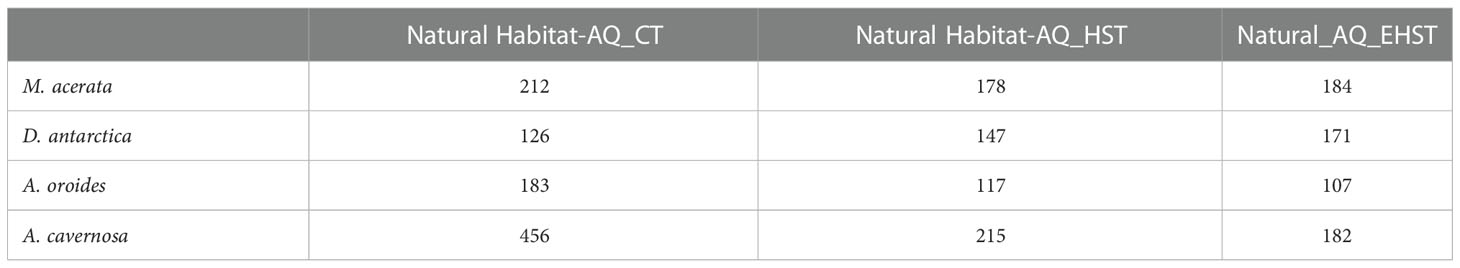
Table 4 Number of ASVs shared between natural habitat sponge specimens and those from aquaria exposed to control temperature (CT), heat stress temperature (HST) and extreme heat stress temperature (EHST) in the different sponge species.
4 Discussion
Sponges, which are important components of benthic environments both in biomass and in abundance, host species-specific microbial communities that can also be very abundant and diverse. In this study, we have described the microbiome of four sponge species from three different environments (polar, tropical and temperate) and examined the heat stress effect on the abundances and diversities of their bacterial communities. The effects of warming on the microbiome of sponges have been previously reported to be different for different sponge species (e.g. Fan et al., 2013; Lesser et al., 2016; Ramsby et al., 2018).
The microbiome of the four sponge species studied here has been analyzed also in previous works, although using different methodologies. The diversity and composition of the microbiota of the four studied species are comparable to those previously reported in the literature (Webster et al., 2004; Gerçe et al., 2011; Ribes et al., 2012; Björk et al., 2013; Blanquer et al., 2013; Ribes et al., 2015; Erwin et al., 2015; Ribes et al., 2016; Steinert et al., 2016; Cárdenas et al., 2018; Coelho et al., 2018; Cárdenas et al., 2019; Cleary et al., 2019; Díez-Vives et al., 2020; Papale et al., 2020; Sacristán-Soriano et al., 2020; Ruocco et al., 2021; Happel et al., 2022). Nevertheless, there are some remarkable differences that must be noted. Similar Shannon and Inverse of Simpson indices have been reported for M. acerata from other Antarctic Peninsula spots (Cárdenas et al., 2019; Sacristán-Soriano et al., 2020), while a higher Shannon index was reported for the microbiome of M. acerata from the distant Terra Nova Bay (Ross Sea, Antarctica) (Papale et al., 2020). However, Papale and colleagues amplified V1-V2 hypervariable regions from 16S rRNA gene, while our study and the other works cited amplified V4 or V4-V5 regions. Recent work showed high variability in the Shannon diversity index of M. acerata’s microbiome along the Western Antarctic Peninsula (0.65-3.79; Happel et al., 2022). The diversity found here in D. antarctica is comparable to that found previously (Díez-Vives et al., 2020), both regarding observed ASVs and Shannon index in Deception Island, but they found variability in diversity, with higher values of Shannon index (up to 5) in specimens from the neighbor King George Island (South Shetland Islands, Antarctica). In our study we have not detected significant changes in M. acerata or D. antarctica collected from either Livingston Island or Deception Island. The microbiome of A. oroides from our study harbored less bacterial richness (ASVs) than reported before from a nearby location (243±28 vs. 521±275), but similar Shannon indices (3.9±0.2 vs. 4.3±0.4) (Ribes et al., 2015). Concerning A. cavernosa, the bacterial community in our specimens was richer compared to previously reported results of the same species from Taiwan (Cleary et al., 2019), although our specimens had a similar Shannon index (2.2-3.2 vs. 2.9-4.3). Slight variations may be caused by the amplification of different 16S hypervariable regions. While we amplified the V4 hypervariable region, they amplified V3-V4 region.
The composition of the microbial communities observed in our specimens agrees with their previous HMA-LMA classification. The notably higher diversity indices (particularly the Inverse Simpson index) in A. oroides with respect to the other sponge species, agrees with the classification of this species as HMA. Moreover, according to (Giles et al., 2013; Simister et al., 2013), LMA sponges host microbiomes dominated by Proteobacteria or Cyanobacteria, while HMA sponges host Chloroflexi or other phyla as dominant (Moitinho-Silva et al., 2017b). In our work, among the LMA sponges, M. acerata, and D. antarctica were dominated by Proteobacteria, A. cavernosa by Cyanobacteria, while the HMA A. oroides was dominated by Chloroflexi. The presence of bacteria affiliated to phylum Poribacteria only in A. oroides microbiome and its absence in the other sponge species or the seawater samples adds evidence to the assignment of HMA status to this sponge species (Moitinho-Silva et al., 2017b). An unidentified genus affiliated to the gammaproteobacterial family EC94 was the dominant taxon of M. acerata’s bacterial microbiome, confirming what was observed in previous studies (Cárdenas et al., 2019; Sacristán-Soriano et al., 2020; Ruocco et al., 2021; Happel et al., 2022). This bacterial group has been reported to be dominant in the microbiome of other LMA sponges (Jackson et al., 2013; Jeong et al., 2015; Cárdenas et al., 2019; Happel et al., 2022) and of some corals (Yang et al., 2013; Gonzalez-Zapata et al., 2018). Recent research of the EC94 group suggested that these bacteria became sponge symbionts quite early in the evolutionary history and re-classified them as a new order, “Candidatus Tethybacterales” (Taylor et al., 2020). These EC94 bacteria are heterotrophs and can use various carbon, nitrogen, and sulfur sources (Taylor et al., 2020).
In a previous study in which the composition of the microbiota of four Antarctic sponges (including D. antarctica) was analyzed, a larger species intra-variability was found in the microbiome composition compared to the other sponge species studied (Sacristán-Soriano et al., 2020). Our results agree with this large inter-individual variability. The presence of the Phyla Proteobacteria and Bacteroidetes, accounting for most of the bacterial community in this species, has been reported before (Díez-Vives et al., 2020; Sacristán-Soriano et al., 2020). The most abundant ASV on average were an unidentified genus and one affiliated to family SAR116 clade, both affiliated to class Alphaproteobacteria. The role of the SAR116 clade in sponges is still unknown with the small number of cultured representatives but a significant role in the sulfur cycle has been reported (Roda-Garcia et al., 2021).
The microbiome of the temperate sponge A. oroides has been described as dominated by Chloroflexi phylum (Ribes et al., 2012; Björk et al., 2013; Blanquer et al., 2013; Ribes et al., 2016), except for one study that found Proteobacteria as the main phylum (Ribes et al., 2015). Bacteria belonging to the dominant Chloroflexi clades in A. oroides microbiome (Anaerolineae, Caldilineales, and SAR202) are involved in dissolved organic matter degradation. Specifically, clades Anaerolineae and Caldilineales have a wide gene repertoire in carbohydrates degradation, and SAR202 in amino acid degradation. Bacteria from SAR202 are also capable of cofactor production and therefore this HMA sponges characteristic phylum is thought to provide nutrients to the host, and also participate in nutrient cycling in the benthic ecosystem (Bayer et al., 2018; Campana et al., 2021). It is worth noting that while A. oroides studied in “The sponge microbiome project” had SAR202 as the most abundant Chloroflexi clade (Moitinho-Silva et al., 2017a), bacteria from class Anaerolineae were the dominant Chloroflexi in our A. oroides specimens.
The microbiome of A. cavernosa was dominated by one Cyanobacteria genus (Cyanobium), except for one specimen, enriched in a Gammaproteobacteria genus (Ralstonia). The microbiome of this tropical sponge has been reported before to be dominated by Proteobacteria or Acidobacteriota (Steinert et al., 2016; Coelho et al., 2018; Cleary et al., 2019). Differences in irradiance due to seasonal change or depth have been suggested to influence sponge-associated microbial communities, with some phototrophic symbionts being less abundant with depth (Morrow et al., 2016) or being absent in winter (Alex et al., 2012). Cyanobacteria from the Cyanobium genus have been reported from other marine sponges (Regueiras et al., 2017; Pagliara et al., 2021), but not from previously studied A. cavernosa specimens. However, bacteria from the same order, Synechococcales, were reported (Steinert et al., 2016; Coelho et al., 2018). Chemical extracts from Cyanobium strains have been reported to display cytotoxicity (Costa et al., 2015; Pagliara et al., 2021). Bacteria from genus Ralstonia have been found to be autotrophic ammonia oxidizers (Calvó et al., 2004). This genus has been previously reported in other sponges and macroalgae too (Florez et al., 2019).
Gradual ocean warming and more frequent heat wave episodes, along with other phenomena associated to anthropogenic climate change, have detrimental effects on benthic communities (Smale et al., 2019; Cooley et al., 2022). Environmental stress may cause imbalance of the microbial communities of sponges and other benthic components, disrupting or changing the holobiont’s functions, and ultimately affecting the whole ecosystem (Pita et al., 2018; Ramsby et al., 2018; Rondon et al., 2020; Rubio-Portillo et al., 2021; Vargas et al., 2021). Since responses to heat stress seem to be related to the sponge host species, we assessed the effect of heat stress and extreme heat stress in aquarium experiments on the four sponge species analyzed previously, which are important components of the benthos at different latitudes, and therefore, adapted to different climate conditions. According to our results, the microbiome of the sponges was quite stable despite the manipulation of the sponge specimens and after 24 h in the aquaria. The exception was D. antarctica, with its microbiome being the most altered at control adaptation aquarium group specimens. An “aquarium effect”, described as the changes in the microbiome of the Control Temperature (CT) experimental group compared to the natural habitat sponge specimens, was observed for all sponge species with varying degrees, being the microbiome of M. acerata the most conserved, with more than 70% of the sequences of the natural habitat control group, followed by D. antarctica (25%).
Differences were also found in the response of each sponge species’ microbiome to the heat stress experiments, being again the microbiomes of the Antarctic sponges, M. acerata and D. antarctica, those maintaining a higher percentage of reads when exposed to increasing temperatures (up to 20% for D. antarctica) compared to the tropical species. The microbiome of D. antarctica was highly altered at control specimens, but a fifth of the microbiome exposed to heat stress seemed to be maintained through the experiment, including the most abundant core taxa from the natural habitat sponge specimens. Both for the tropical A. cavernosa and the Mediterranean, A. oroides (up to 2% for A. oroides), the SourceTracker results suggest there was a shift in the microbiomes by the end of control temperature exposure in the aquarium experiment, with most sequences coming from unknown sources. The microbiomes exposed to heat stress also experienced a shift, modifying the proportions of rare or less abundant bacteria, but mostly becoming more similar to the microbiome found in the water in A. oroides. Concerning bacterial diversity, the microbiomes of the tropical and temperate sponge species lost diversity when exposed to a heat stress. However, while in A. oroides the highest diversity is found in the natural habitat specimens, in A. cavernosa, the diversity in the natural habitat group is lower than in the experimental groups (Figure 5). Contrastingly, the bacterial community associated with M. acerata became more diverse after heat stress exposure. We suggest warming boosted the activity of previously scarce or inactive bacteria that could not be detected previously due to the detection limit of the technique. Extreme heat stress seemed to dysregulate the microbiome leading to dysbiosis, and diversity was lost. The diversity in D. antarctica microbiome was similar in the sponges from natural habitat, HST and EHST, but was significantly lower in CT sponge specimens (p<0.05).
The changes in diversity were also reflected in changes in taxonomy. A shift in A. oroides microbiome occurred at sponge specimens exposed to CT even at phylum level (Figure 6). Changes at phylum level could also be observed in D. antarctica and A. cavernosa exposed to CT (“aquarium effect”), but not in M. acerata (Figure 6). Phyla Bacteroidota and Firmicutes were more abundant in the heat stress specimens in all sponge species, except for D. antarctica. Firmicutes was reported to also increased in diseased deep water Geodia barretti (Luter et al., 2017) and Bacteroidota abundance increased in heat stressed Antarctic sponge Isodictya kerguelenensis (Rondon et al., 2020) and diseased Lubomirskia baicalensis (Kulakova et al., 2018). Proteobacteria was more abundant after heat stress in D. antarctica and A. cavernosa, and less abundant after heat stress in M. acerata and A. oroides. Campylobacterota were enriched in HST M. acerata. Cyanobacteria was less abundant in A. cavernosa. Planctomycetota was less abundant after HST in A. cavernosa. Chloroflexi was practically absent in A. oroides exposed to HST.
The dominant taxa of the core microbiota were still present in the sponge specimens exposed to heat stress in all the four studied species. Nevertheless, different responses were observed among them. Both Antarctic sponges (M. acerata and D. antarctica) seem to host a microbiome that is more resistant to heat stress. After exposure to heat and extreme heat stress, not only the dominant core taxa were still present, but their relative abundance was notable. In A. cavernosa and A. oroides, the dominant taxa were present at very low abundances in heat stress exposed sponge specimens and were virtually absent in extreme heat stress sponge specimens. The exposure to heat stress caused important changes in the microbiomes at phylum level in many studies although not in all of them. Examples of sponge holobionts that have reported to be resistant to heat stress are found among tropical sponges (e.g. Simister et al., 2012a; Lesser et al., 2016) but there are also evidences of resistance in a polar deep sea sponge (Strand et al., 2017).
The most abundant taxon in the microbiome of the Antarctic sponge M. acerata, the EC94 group, was still present at the specimens at the end of both the heat stress and extreme heat stress experiments. We can presumably argue that the functions that this group develops within the holobiont may also be maintained even when exposed to stressful conditions. The microbiome of sponge specimens exposed to heat or extreme heat stress experienced a shift in composition, that was more or less prominent depending on the sponge species. There were some taxa, presumably opportunistic, that seemed to benefit from the heat stress-associated dysregulation in the sponge by occupying new niches in the holobiont, and other taxa already present at natural habitat sponge specimens that increased in abundance with temperature. In general, the most abundant taxa in the microbiome of heat stress sponge specimens were different for each sponge species. However, some taxa were shared among stressed species, such as bacteria from the family Marinifilaceae, more abundant in heat stressed M. acerata and A. oroides, or the family Clostridiaceae, more abundant in the microbiome of A. oroides and A. cavernosa. Genus Marinifilum (Bacteroidota, family Marinifilaceae), and also genus Fusibacter (Firmicutes), which was also more abundant in heat stressed A. oroides specimens, were among the dominant taxa in the microbiome of necrotic Rhopaloeides odorabile (Fan et al., 2013). An ASV affiliated to family Marinifilaceae increased in abundance in the sponge Leucetta chagosensis exposed to pH 7.8, 30°C in comparison to those specimens exposed to present day conditions (Posadas et al., 2021). In our experiment with M. acerata, the specimens exposed to heat stress had higher abundances of genera Colwellia, Crocinitomix, Pseudarcobacter, and Psychrobium. Genus Colwellia (Gammaproteobacteria) was found to be much more abundant in injured specimens than it was in healthy specimens (Rondon et al., 2020). This genus was present in healthy M. acerata in previous studies (Ruocco et al., 2021). It is a denitrifying bacterium and produces natural compounds (Ruocco et al., 2021). Members from families Colwelliaceae and Flavobacteriaceae (Bacteroidota) were more abundant in injured specimens (Rondon et al., 2020), and were reported to be disease-related in necrotic and stressed sponge specimens (Simister et al., 2012a; Fan et al., 2013). They were associated with disease and stress in corals (Gajigan et al., 2017). Genera Crocinitomix (Flavobacteriales, Bacteroidota) and Colwellia were reported from other Antarctic benthic invertebrates such as sea anemones or soft corals (Webster and Bourne, 2007; Murray et al., 2016). Genus Psychrobium increased in abundance in the microbiota of marine bivalves after a viral infection (de Lorgeril et al., 2018) and after exposure to nanoplastics (Auguste et al., 2020). One bacterial genus that was more abundant in the M. acerata exposed to extreme heat stress than in the other groups in our study was genus Pseudoalteromonas. Strains from this genus have been found in other species of Mycale (Lau et al., 2005), and have been reported to have a great biotechnological potential, specially regarding cold-active enzymes (Borchert et al., 2017). On the other hand, a strain from Pseudoalteromonas agarivorans has been reported to be a pathogen of the tropical sponge R. odorabile (Choudhury et al., 2015).
In heat stress exposed D. antarctica, we observed an increase in the group Burkholderia-Caballeronia-Paraburkholderia (Gammaproteobacteria). ASVs affiliated to this group were among the most abundant in the microbiome of the coral Acropora hyacinthus (Kriefall et al., 2022). Contrastingly, in heat stressed A. cavernosa, we observed an increase in members of several bacterial families (Clostridiaceae, Flavobacteriaceae, Rhodobacteraceae) and two genera (Ruegeria and Winogradskyella). Genus Clostridium (family Clostridiaceae, Firmicutes) can be found in healthy Xestospongia muta specimens (Leal et al., 2022), and order Clostridiales in Leucetta chagosensis after experimental exposition to “present day” seawater temperature and pH conditions (Posadas et al., 2021). Moreover, class Clostridia was more abundant in diseased than in healthy corals (reviewed in Mouchka et al., 2010). Phyla Firmicutes and Bacteroidota were reported to be more abundant in diseased sponge specimens (Luter et al., 2017). Orders Flavobacteriales (Bacteroidota) and Rhodobacterales (Proteobacteria) were reported to be more abundant in various coral species exposed to stressors (reviewed in McDevitt-Irwin et al., 2017), as well as in sponge species such as Isodictya kerguelenensis (Rondon et al., 2020) and Lubomirskia baicalensis (Kulakova et al., 2018). Previous literature has found an important contribution of Operational Taxonomic Units (OTUs) from order Flavobacteriales to the pool of OTUs associated to coral diseases and stress (Gignoux-Wolfsohn and Vollmer, 2015). Bacteria from order Rhodobacterales (Alphaproteobacteria) have been reported to be fast growing and opportunistic (Teeling et al., 2012), have been found in both healthy and stressed corals (Meron et al., 2010; Sharp and Ritchie, 2012) and have the potential to bloom when new niches are available under stress periods (Welsh et al., 2017). Order Rhodobacterales was more abundant in the microbiome of the sponge Leucetta chagosensis experimentally exposed to warming and acidification scenarios (Posadas et al., 2021). Photoheterotrophic members of the Rhodobacteraceae family were more abundant in Neopetrosia compacta exposed to acidification and warming scenario. Those were suggested to provide heat tolerance to the sponge (Posadas et al., 2021). It also was more abundant in Isodictya kerguelenensis exposed to heat stress (Rondon et al., 2020). Genus Rhodobacter (family Rhodobacteraceae) was more abundant in diseased than in healthy corals (reviewed in (Mouchka et al., 2010)). Similarly, the Rhodobacteraceae family increased fourfold in coral white syndrome lesions compared to healthy tissues (Pollock et al., 2017; Rosales et al., 2020) and are implicated in Stony Coral Tissue Loss Disease that is currently affecting Caribbean reefs. Bacteria affiliated to genus Winogradskyella has been described from various sponge species. Species of this genus have been reported to produce antibiofilm compounds (Pinnaka et al., 2013; Schellenberg et al., 2017; Rizzo et al., 2021). Ruegeria (family Rhodobacteraceae) species were found to increase in abundance in diseased tissues of several Mediterranean corals collected during a marine heat wave (Rubio-Portillo et al., 2021). Ruegeria sp. was found to increase in corals exposed to stress conditions and was proposed to be a microbial bio-indicator of marine invertebrate diseases. This bacterium may be able to protect corals from pathogenic Vibrio spp. by growth inhibition (Miura et al., 2019; Rosado et al., 2019). Apart from genus Fusibacter and family Marinifilaceae, the microbiome of heat stressed A. oroides was enriched in family Clostridiaceae, order Peptostreptococcales-Tissierellales, and genera Halodesulfovibio. Genus Halodesulfovibrio has been found in corals and species from this genus seem to have antimicrobial properties (Shivani et al., 2017; Chen et al., 2021).
Despite all the differences observed between studies it must be noted that observed differences may be influenced by the use of different methodologies, e.g. DNA extraction method, amplification primers targeting different 16S rRNA regions, sequencing platform, or bioinformatic pipeline. Sampling and experimental design should also be considered at comparisons.
Overall, we can say that the core microbiome is maintained in most sponge species after a heat stress, although whether they would recover to the normal conditions previous to the stress remains yet to be further investigated. Although the core bacteria may or may not change, it is clear that there is a change in transient bacteria appearing new species from unknown origin, some of which may be opportunistic pathogens finding this opportunity to grow after a dysbiosis occurs. According to our results, sponge species from Antarctic waters could be more resilient than tropical and temperate sponge species, and perhaps their recovery after the heat stress would be possible. Changes produced by an extreme heat stress however do not seem to be reversible for the sponge microbiomes in any of the sponge species tested. Remarkably, both the microbiome composition and the changes produced by the heat stress seem to be quite host species-specific, and thus, depend on the sponge species, being always different from the seawater. Therefore, we may conclude that although there are some general trends, heat stress will affect differently the different sponge species, as well as the different bacterium species, and thus we may see winners and losers when temperature increases high enough in the near future. Global change is impacting significantly in marine environments and the effects are particularly evident for sessile invertebrate species, which often exhibit narrow ranges of thermal tolerance. For all these reasons, tropical and temperate sponge species will probably be those suffering the most the heat stress. Consequently, the effects of global change may be dramatic for benthic ecosystems since sponges are a fundamental part of them.
Data availability statement
The data presented in the study are deposited in the Mendely Data public repository (DOI: 10.17632/9c6y62nv9z.1, DOI: 10.17632/d67pjpc47g.1, DOI: 10.17632/fnjhzsybxj.1). Further queries should be directed to the corresponding author.
Author contributions
Idea and conceptualization, PC-F, CA, EB, CG-A. Methodology, PC-F, CA, EB, CG-A. Investigation, PC-F, CA, EB, CG-A, JB, CA-P. Resources, CA, CG-A, EB. Sample collection: PC-F, CA, CA-P, JB. Data curation, PC-F, CA, EB, CG-A. Writing—original draft preparation, PC-F, CG-A. Writing—review and editing, PC-F, CA, EB, CG-A, CA-P. Supervision and project administration, CA, CG-A, EB. Funding acquisition, CA, CG-A, EB. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the BLUEBIO (CTM2016-78901/ANT) and CHALLENGE (PID2019-107979RB-I00/ANT) research grants to C.A. by the Spanish Government and the SGR (Research Quality Group) funding of the Generalitat de Catalunya (2014SGR336; 2017SGR1120). P.C.F. acknowledges financial support from an FPU Grant (2016-02877) of the MEFP (Spain) and the Water Research Institute (IdRA).
Acknowledgments
Thanks are due to all the researchers of the BLUEBIO and CHALLENGE projects for their support during sample collection and aquaria experiments in the Antarctic and Mediterranean sites, and A. Townsend (University of Guam Marine Laboratory) in Guam. Thanks are also due to the technicians from the Unitat de Tecnologia Marina (UTM-CSIC) for their support in the Antarctic sampling seasons. This work is a contribution to the Integrated Science to Inform Antarctic and Southern Ocean Conservation (Ant-ICON) Scientific Research Programme of the Scientific Committee on Antarctic Research (SCAR). We acknowledge the QGIS Development Team and the Norwegian Polar Institute’s Quantarctica package.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1072696/full#supplementary-material
References
Alex A., Vasconcelos V., Tamagnini P., Santos A., Antunes A. (2012). Unusual symbiotic cyanobacteria association in the genetically diverse intertidal marine sponge hymeniacidon perlevis (Demospongiae, halichondrida). PloS One 7 (12), e51834. doi: 10.1371/journal.pone.0051834
Apprill A., McNally S., Parsons R., Weber L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75 (2), 129–137. doi: 10.3354/ame01753
Astudillo-García C., Bell J. J., Webster N. S., Glasl B., Jompa J., Montoya J. M., et al. (2017). Evaluating the core microbiota in complex communities: a systematic investigation. Environ. Microbiol. 19 (4), 1450–1462. doi: 10.1111/1462-2920.13647
Auguste M., Lasa A., Balbi T., Pallavicini A., Vezzulli L., Canesi L. (2020). Impact of nanoplastics on hemolymph immune parameters and microbiota composition in mytilus galloprovincialis. Mar. Environ. Res. 159, 105017. doi: 10.1016/J.MARENVRES.2020.105017
Bayer K., Jahn M. T., Slaby B. M., Moitinho-Silva L., Hentschel U. (2018). Marine sponges as chloroflexi hot spots: Genomic insights and high-resolution visualization of an abundant and diverse symbiotic clade. mSystems 3 (6), e00150–e00118. doi: 10.1128/msystems.00150-18
Bell J. J. (2008). The functional roles of marine sponges. Estuar. Coast. Shelf Sci. 79 (3), 341–353. doi: 10.1016/j.ecss.2008.05.002
Bennett H. M., Altenrath C., Woods L., Davy S. K., Webster N. S., Bell J. J. (2017). Interactive effects of temperature and pCO2 on sponges: from the cradle to the grave. Glob. Chang. Biol. 23 (5), 2031–2046. doi: 10.1111/GCB.13474
Björk J. R., Díez-Vives C., Coma R., Ribes M., Montoya J. M. (2013). Specificity and temporal dynamics of complex bacteria-sponge symbiotic interactions. Ecology 94 (12), 2781–2791. doi: 10.1890/13-0557.1
Blanquer A., Uriz M. J., Galand P. E. (2013). Removing environmental sources of variation to gain insight on symbionts vs. transient microbes in high and low microbial abundance sponges. Environ. Microbiol. 15 (11), 3008–3019. doi: 10.1111/1462-2920.12261
Blanquer A., Uriz M. J., Cebrian E., Galand P. E. (2016). Snapshot of a bacterial microbiome shift during the early symptoms of a massive sponge die-offin the western Mediterranean. Front. Microbiol. 7, 752. doi: 10.3389/FMICB.2016.00752/BIBTEX
Borchert E., Knobloch S., Dwyer E., Flynn S., Jackson S. A., Jóhannsson R., et al. (2017). Biotechnological potential of cold adapted pseudoalteromonas spp. isolated from ‘deep sea’ sponges. Mar. Drugs 15 (6), 184. doi: 10.3390/MD15060184
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13 (7), 518–583. doi: 10.1038/nmeth.3869
Calvó L., Vila X., Abella C. A., Garcia-Gil L. J. (2004). Use of the ammonia-oxidizing bacterial-specific phylogenetic probe Nso1225 as a primer for fingerprint analysis of ammonia-oxidizer communities. Appl. Microbiol. Biotechnol. 63 (6), 715–721. doi: 10.1007/s00253-003-1433-1
Campana S., Busch K., Hentschel U., Muyzer G., de Goeij J. M. (2021). DNA-Stable isotope probing (DNA-SIP) identifies marine sponge-associated bacteria actively utilizing dissolved organic matter (DOM). Environ. Microbiol. 23 (8), 4489–4504. doi: 10.1111/1462-2920.15642
Cárdenas C. A., Font A., Steinert G., Rondon R., González-Aravena M. (2019). Temporal stability of bacterial communities in Antarctic sponges. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02699
Cárdenas C. A., González-Aravena M., Font A., Hestetun J. T., Hajdu E., Trefault N., et al. (2018). High similarity in the microbiota of cold- water sponges of the genus mycale from two different geographical areas. PeerJ 6, e4935. doi: 10.7717/peerj.4935
Chen Y. H., Yang S. H., Tandon K., Lu C. Y., Chen H. J., Shih C. J., et al. (2021). Potential syntrophic relationship between coral-associated prosthecochloris and its companion sulfate-reducing bacterium unveiled by genomic analysis. Microb. Genom. 7 (5), 574. doi: 10.1099/mgen.0.000574
Choudhury J. D., Pramanik A., Webster N. S., Llewellyn L. E., Gachhui R., Mukherjee J. (2015). The pathogen of the great barrier reef sponge rhopaloeides odorabile is a new strain of pseudoalteromonas agarivorans containing abundant and diverse virulence-related genes. Mar. Biotechnol. 17 (4), 463–478. doi: 10.1007/S10126-015-9627-Y
Clarke A., Meredith M. P., Wallace M. I., Brandon M. A., Thomas D. N. (2008). Seasonal and interannual variability in temperature, chlorophyll and macronutrients in northern Marguerite bay, Antarctica. Deep-Sea Res. Part II-Top. Stud. Oceanogr. 55 (18-19), 1988–2006. doi: 10.1016/j.dsr2.2008.04.035
Cleary D. F. R., Becking L. E., de Voogd N. J., Pires A. C. C., Polónia A. R. M., Egas C., et al. (2013). Habitat- and host-related variation in sponge bacterial symbiont communities in Indonesian waters. FEMS Microbiol. Ecol. 85 (3), 465–482. doi: 10.1111/1574-6941.12135
Cleary D. F. R., Polónia A. R. M., Huang Y. M., Putchakarn S., Gomes N. C. M., de Voogd N. J. (2019). A comparison of prokaryote communities inhabiting sponges, bacterial mats, sediment and seawater in southeast Asian coral reefs. FEMS Microbiol. Ecol. 95 (12), fiz169. doi: 10.1093/femsec/fiz169
Coelho F. J. R. C., Cleary D. F. R., Gomes N. C. M., Pólonia A. R. M., Huang Y. M., Liu L. L., et al. (2018). Sponge prokaryote communities in Taiwanese coral reef and shallow hydrothermal vent ecosystems. Microb. Ecol. 75 (1), 239–254. doi: 10.1007/s00248-017-1023-x
Collins M., Sutherland M., Bouwer L., Cheong S.-M., Frölicher T., Jacot Des Combes H., et al. (2019). “Extremes, abrupt changes and managing risks,” in IPCC special report on the ocean and cryosphere in a changing climate. Eds. Pörtner H.-O., Roberts D. C., Masson-Delmotte V., Zhai P., Tignor M., Poloczanska E., et al (Cambridge, UK and New York: Cambridge University Press), 589–655. doi: 10.1017/9781009157964.008
Cooley S., Schoeman D., Bopp L., Boyd P., Donner S., Ghebrehiwet D. Y., et al. (2022). “Oceans and coastal ecosystems and their services,” in Climate change 2022: Impacts, adaptation and vulnerability. Eds. Pörtner H.-O., Roberts D. C., Tignor M., Poloczanska E. S., Mintenbeck K., Alegría A., et al (Cambridge, UK and New York: Cambridge University Press), 379–550. doi: 10.1017/9781009325844.005
Costa M. S., Costa M., Ramos V., Leão P. N., Barreiro A., Vasconcelos V., et al. (2015). Picocyanobacteria from a clade of marine cyanobium revealed bioactive potential against microalgae, bacteria, and marine invertebrates. J. Toxicol. Env. Health Part A 78 (7), 432–442. doi: 10.1080/15287394.2014.991466
Cristi A., Parada-Pozo G., Morales-Vicencio F., Cárdenas C. A., Trefault N. (2022). Variability in host specificity and functional potential of Antarctic sponge-associated bacterial communities. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.771589
Davis N. M., Proctor D. M., Holmes S. P., Relman D. A., Callahan B. J. (2018). Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6 (1), 1–14. doi: 10.1186/s40168-018-0605-2
de Goeij J. M., van Oevelen D., Vermeij M. J. A., Osinga R., Middelburg J. J., de Goeij A. F. P. M., et al. (2013). Surviving in a marine desert: The sponge loop retains resources within coral reefs. Science 342 (6154), 108–110. doi: 10.1126/science.1241981
de Lorgeril J., Lucasson A., Petton B., Toulza E., Montagnani C., Clerissi C., et al. (2018). Immune-suppression by OsHV-1 viral infection causes fatal bacteraemia in pacific oysters. Nat. Commun. 9, 4215. doi: 10.1038/s41467-018-06659-3
Díez-Vives C., Taboada S., Leiva C., Busch K., Hentschel U., Riesgo A. (2020). On the way to specificity-microbiome reflects sponge genetic cluster primarily in highly structured populations. Mol. Ecol. 29 (22), 4412–4427. doi: 10.1111/mec.15635
Dittami S. M., Arboleda E., Auguet J. C., Bigalke A., Briand E., Cárdenas P., et al. (2021). A community perspective on the concept of marine holobionts: Current status, challenges, and future directions. PeerJ 9, e10911. doi: 10.7717/peerj.10911
Edgar R. C., Flyvbjerg H. (2015). Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31 (21), 3476–3482. doi: 10.1093/BIOINFORMATICS/BTV401
Ereskovsky A., Ozerov D. A., Pantyulin A. N., Tzetlin A. B. (2019). Mass mortality event of white Sea sponges as the result of high temperature in summer 2018. Polar Biol. 42 (12), 2313–2318. doi: 10.1007/S00300-019-02606-0
Erwin P. M., Coma R., López-Sendino P., Serrano E., Ribes M. (2015). Stable symbionts across the HMA-LMA dichotomy: Low seasonal and interannual variation in sponge-associated bacteria from taxonomically diverse hosts. FEMS Microbiol. Ecol. 91 (10), fiv115. doi: 10.1093/femsec/fiv115
Erwin P. M., Pita L., López-Legentil S., Turon X. (2012). Stability of sponge-associated bacteria over large seasonal shifts in temperature and irradiance. Appl. Environ. Microbiol. 78 (20), 7358–7368. doi: 10.1128/aem.02035-12
Fan L., Liu M., Simister R., Webster N. S., Thomas T. (2013). Marine microbial symbiosis heats up: The phylogenetic and functional response of a sponge holobiont to thermal stress. ISME J. 7 (5), 991–1002. doi: 10.1038/ismej.2012.165
Fan L., Reynolds D., Liu M., Stark M., Kjelleberg S., Webster N. S., et al. (2012). Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc. Natl. Acad. Sci. U.S.A. 109 (27), E1878–E1887. doi: 10.1073/pnas.1203287109
Feuda R., Dohrmann M., Pett W., Philippe H., Rota-Stabelli O., Lartillot N., et al. (2017). Improved modeling of compositional heterogeneity supports sponges as sister to all other animals. Curr. Biol. 27 (24), 3864–3870.e4. doi: 10.1016/j.cub.2017.11.008
Flores J. Z., Camus C., Hengst M. B., Marchant F., Bushmann A. H. (2019). Structure of the epiphytic bacterial communities of Macrocystis pyrifera in localities with contrasting nitrogen concentrations and temperature. Algal Res. 44, 101706. doi: 10.1016/J.ALGAL.2019.101706
Freeman C. J., Easson C. G., Matterson K. O., Thacker R. W., Baker D. M., Paul V. J. (2020). Microbial symbionts and ecological divergence of Caribbean sponges: A new perspective on an ancient association. ISME J. 14 (6), 1571–1583. doi: 10.1038/s41396-020-0625-3
Gajigan A. P., Diaz L. A., Conaco C. (2017). Resilience of the prokaryotic microbial community of acropora digitifera to elevated temperature. MicrobiologyOpen 6 (4), e00478. doi: 10.1002/mbo3.478
Garrabou J., Coma R., Bensoussan N., Bally M., Chevaldonné P., Cigliano M., et al. (2009). Mass mortality in northwestern Mediterranean rocky benthic communities: Effects of the 2003 heat wave. Glob. Change Biol. 15 (5), 1090–1103. doi: 10.1111/j.1365-2486.2008.01823.x
Garrabou J., Gómez-Gras D., Medrano A., Cerrano C., Ponti M., Schlegel R., et al. (2022). Marine heatwaves drive recurrent mass mortalities in the Mediterranean Sea. Glob. Change Biol. 28 (19), 5708–5725. doi: 10.1111/gcb.16301
Gerçe B., Schwartz T., Syldatk C., Hausmann R. (2011). Differences between bacterial communities associated with the surface or tissue of Mediterranean sponge species. Microb. Ecol. 61 (4), 769–782. doi: 10.1007/s00248-011-9802-2
Gignoux-Wolfsohn S. A., Vollmer S. V. (2015). Identification of candidate coral pathogens on white band disease-infected staghorn coral. PloS One 10 (8), e0134416. doi: 10.1371/journal.pone.0134416
Giles E. C., Kamke J., Moitinho-Silva L., Taylor M. W., Hentschel U., Ravasi T., et al. (2013). Bacterial community profiles in low microbial abundance sponges. FEMS Microbiol. Ecol. 83 (1), 232–241. doi: 10.1111/j.1574-6941.2012.01467.x
Gloeckner V., Wehrl M., Moitinho-Silva L., Gernert C., Hentschel U., Schupp P., et al. (2014). The HMA-LMA dichotomy revisited: An electron microscopical survey of 56 sponge species. Biol. Bull. 227 (1), 78–88. doi: 10.1086/BBLv227n1p78
Gonzalez-Acosta B., Bashan Y., Hernandez-Saavedra N. Y., Ascencio F., de la Cruz-Agüero G. (2006). Seasonal seawater temperature as the major determinant for populations of culturable bacteria in the sediments of an intact mangrove in an arid region. FEMS Microbiol. Ecol. 55 (2), 311–321. doi: 10.1111/j.1574-6941.2005.00019.x
Gonzalez-Zapata F. L., Bongaerts P., Ramírez-Portilla C., Adu-Oppong B., Walljasper G., Reyes A., et al. (2018). Holobiont diversity in a reef-building coral over its entire depth range in the mesophotic zone. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00029
González-Aravena M., Kenny N. J., Osorio M., Font A., Riesgo A. H., Cárdenas C. A. (2019). Warm temperatures, cool sponges: The effect of increased temperatures on the Antarctic sponge Isodictya sp. PeerJ 7, e8088. doi: 10.7717/peerj.8088
Happel L., Rondon R., Font A., González-Aravena M., Cárdenas C. A. (2022). Stability of the microbiome of the sponge mycale (Oxymycale) acerata in the Western Antarctic peninsula. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.827863
Hentschel U., Fieseler L., Wehrl M., Gernert C., Steinert M., Hacker J., et al. (2003). “Microbial diversity of marine sponges,” in Progress in molecular and subcellular biology, vol. 37 . Ed. Müller W. E. G. (Berlin, Heidelberg: Springer), 59–88. doi: 10.1007/978-3-642-55519-0_3
Hentschel U., Piel J., Degnan S. M., Taylor M. W. (2012). Genomic insights into the marine sponge microbiome. Nat. Rev. Microbiol. 10 (9), 641–654. doi: 10.1038/nrmicro2839
Hentschel U., Usher K. M., Taylor M. W. (2006). Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 55 (2), 167–177. doi: 10.1111/j.1574-6941.2005.00046.x
Hereu B., Kersting D. K. (2016). Diseases of coralline algae in the Mediterranean Sea. Coral Reefs 35 (2), 713. doi: 10.1007/s00338-016-1428-x
Hooper J. N. A., van Soest R. W. M. (2002). “Systema Porifera: A guide to the classification of sponges,” in Systema Porifera. Eds. Hooper J. N. A., Van Soest R. W. M., Willenz P. (Boston, MA: Springer), 1–7. doi: 10.1007/978-1-4615-0747-5_1
Horn H., Slaby B. M., Jahn M. T., Bayer K., Moitinho-Silva L., Förster F., et al. (2016). An enrichment of CRISPR and other defense-related features in marine sponge-associated microbial metagenomes. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01751
Hughes T. P., Kerry J. T., Álvarez-Noriega M., Álvarez-Romero J. G., Anderson K. D., Baird A. H., et al. (2017). Global warming and recurrent mass bleaching of corals. Nature 543 (7645), 373–377. doi: 10.1038/nature21707
Jackson S. A., Flemer B., McCann A., Kennedy J., Morrissey J. P., O’Gara F., et al. (2013). Archaea appear to dominate the microbiome of inflatella pellicula deep sea sponges. PloS One 8 (12), e84438. doi: 10.1371/journal.pone.0084438
Jackson S. A., Kennedy J., Morrissey J. P., O’Gara F., Dobson A. D. W. (2012). Pyrosequencing reveals diverse and distinct sponge-specific microbial communities in sponges from a single geographical location in Irish waters. Microb. Ecol. 64 (1), 105–116. doi: 10.1007/s00248-011-0002-x
Janussen D., Downey R. V. (2014). “Chapter 5.5. Porifera,” in Biogeographic atlas of the southern ocean. Eds. de Broyer C., Koubbi P., Griffiths H. J., Raymond B., d’Udekem d’Acoz C., Van de Putte A., et al (Cambridge: Scientific Committee on Antarctic Research), 94–102.
Jeong J. B., Kim K. H., Park J. S. (2015). Sponge-specific unknown bacterial groups detected in marine sponges collected from Korea through barcoded pyrosequencing. J. Microbiol. Biotechnol. 25 (1), 1–10. doi: 10.4014/jmb.1406.06041
Kennedy J., Flemer B., Jackson S. A., Morrissey J. P., O’Gara F., Dobson A. D. W. (2014). Evidence of a putative deep sea specific microbiome in marine sponges. PloS One 9 (3), e91092. doi: 10.1371/journal.pone.0091092
Knights D., Kuczynski J., Charlson E. S., Zaneveld J., Mozer M. C., Collman R. G., et al. (2011). Bayesian Community-wide culture-independent microbial source tracking. Nat. Methods 8 (9), 761–763. doi: 10.1038/nmeth.1650
Koutsouveli V., Taboada S., Moles J., Cristobo J., Rı P., Bertran A., et al. (2018). Insights into the reproduction of some Antarctic dendroceratid, poecilosclerid, and haplosclerid demosponges. PloS One 13 (2), e0192267. doi: 10.1371/journal.pone.0192267
Kriefall N. G., Kanke M. R., Aglyamova G. V., Davies S. W. (2022). Reef environments shape microbial partners in a highly connected coral population. Proc. R. Soc B-Biol. Sci. 289 (1967), 20212459. doi: 10.1098/rspb.2021.2459
Kulakova N. V., Sakirko M. V., Adelshin R. V., Khanaev I. V., Nebesnykh I. A., Pérez T. (2018). Brown rot syndrome and changes in the bacterial сommunity of the baikal sponge lubomirskia baicalensis. Microb. Ecol. 75 (4), 1024–1034. doi: 10.1007/S00248-017-1097-5
Lau S. C. K., Tsoi M. M. Y., Li X., Dobretsov S., Plakhotnikova Y., Wong P. K., et al. (2005). Pseudoalteromonas spongiae sp. nov., a novel member of the γ-proteobacteria isolated from the sponge mycale adhaerens in Hong Kong waters. Int. J. Syst. Evol. Microbiol. 55 (4), 1593–1596. doi: 10.1099/IJS.0.63638-0
Leal C. V., Avelino-Alves D., Salazar V., Omachi C., Thompson C., Berlinck R. G. S., et al. (2022). Sponges present a core prokaryotic community stable across tropical Western Atlantic. Sci. Total Environ. 835, 155145. doi: 10.1016/j.scitotenv.2022.155145
Lee O. O., Wang Y., Yang J., Lafi F. F., Al-Suwailem A., Qian P. Y. (2011). Pyrosequencing reveals highly diverse and species-specific microbial communities in sponges from the red Sea. ISME J. 5 (4), 650–664. doi: 10.1038/ismej.2010.165
Leray M., Wilkins L. G. E., Apprill A., Bik H. M., Clever F., Connolly S. R., et al. (2021). Natural experiments and long-term monitoring are critical to understand and predict marine host-microbe ecology and evolution. PloS Biol. 19 (8), e3001322. doi: 10.1371/journal.pbio.3001322
Lesser M. P., Fiore C., Slattery M., Zaneveld J. (2016). Climate change stressors destabilize the microbiome of the Caribbean barrel sponge. Exp. Mar. Biol. Ecol. 475, 11–18. doi: 10.1016/j.jembe.2015.11.004
Lie H. J., Lee S. (2010). Reconstruction of regular time series from bi-monthly temperature data in the yellow Sea and the northwestern East China Sea. Ocean Sci. J. 45 (2), 139–149. doi: 10.1007/S12601-010-0012-5
Lo Giudice A., Azzaro M., Schiaparelli S. (2019). “Microbial symbionts of Antarctic marine benthic invertebrates,” in The ecological role of micro-organisms in the Antarctic environment. Ed. Castro-Sowinski S. (Cham: Springer), 277–296. doi: 10.1007/978-3-030-02786-5_13
Luter H. M., Bannister R. J., Whalan S., Kutti T., Pineda M. C., Webster N. S. (2017). Microbiome analysis of a disease affecting the deep-sea sponge geodia barretti. FEMS Microbiol. Ecol. 93 (6), fix074. doi: 10.1093/femsec/fix074
Maldonado M., Aguilar R., Bannister R. J., Bell J. J., Conway K. W., Dayton P. K., et al. (2015). “Sponge grounds as key marine habitats: a synthetic review of types, structure, functional roles, and conservation concerns,” in Marine animal forests. Eds. Rossi S., Bramanti L., Gori A., Orejas C., Saco del Valle C. (Cham: Springer), 1–39. doi: 10.1007/978-3-319-17001-5_24-1
McDevitt-Irwin J. M., Baum J. K., Garren M., Vega Thurber R. L. (2017). Responses of coral-associated bacterial communities to local and global stressors. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00262
McMurdie P. J., Holmes S. (2013). Phyloseq: An r package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8 (4), e61217. doi: 10.1371/journal.pone.0061217
Meron D., Atias E., Iasur Kruh L., Elifantz H., Minz D., Fine M., et al (2010). The impact of reduced pH on the microbial community of the coral Acropora eurystoma. ISME J. 5 (1), 51–60. doi: 10.1038/ismej.2010.102
Miura N., Motone K., Takagi T., Aburaya S., Watanabe S., Aoki W., et al. (2019). Ruegeria sp. strains isolated from the reef-building coral galaxea fascicularis inhibit growth of the temperature-dependent pathogen vibrio coralliilyticus. Mar. Biotechnol. 21 (1), 1–8. doi: 10.1007/S10126-018-9853-1
Moitinho-Silva L., Nielsen S., Amir A., Gonzalez A., Ackermann G. L., Cerrano C., et al. (2017a). The sponge microbiome project. GigaScience 6 (10), gix077. doi: 10.1093/gigascience/gix077
Moitinho-Silva L., Steinert G., Nielsen S., Hardoim C. C. P., Wu Y. C., McCormack G. P., et al. (2017b). Predicting the HMA-LMA status in marine sponges by machine learning. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00752
Moreno-Pino M., Cristi A., Gillooly J. F., Trefault N. (2020). Characterizing the microbiomes of Antarctic sponges: A functional metagenomic approach. Sci. Rep. 10 (1), 1–12. doi: 10.1038/s41598-020-57464-2
Morrow K. M., Fiore C. L., Lesser M. P. (2016). Environmental drivers of microbial community shifts in the giant barrel sponge, xestospongia muta, over a shallow to mesophotic depth gradient. Environ. Microbiol. 18 (6), 2025–2038. doi: 10.1111/1462-2920.13226
Mouchka M. E., Hewson I., Harvell C. D. (2010). Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 50 (4), 662–674. doi: 10.1093/icb/icq061
Murray A. E., Rack F. R., Zook R., Williams M. J. M., Higham M. L., Broe M., et al. (2016). Microbiome composition and diversity of the ice-dwelling sea anemone, edwardsiella andrillae. Integr. Comp. Biol. 56 (4), 542–555. doi: 10.1093/icb/icw095
Oksanen J., Simpson G., Blanchet F., Kindt R., Legendre P., Minchin P., et al. (2022). “Vegan: Community ecology package,” in R package version 2.6-2. Available at: https://CRAN.R-project.org/package=vegan.
Oliver E. C. J., Donat M. G., Burrows M. T., Moore P. J., Smale D. A., Alexander L., et al. (2018). Longer and more frequent marine heatwaves over the past century. Nat. Commun. 9 (1), 1–12. doi: 10.1038/s41467-018-03732-9
Pagliara P., de Benedetto G. E., Francavilla M., Barca A., Caroppo C. (2021). Bioactive potential of two marine picocyanobacteria belonging to cyanobium and synechococcus genera. Microorganisms 9 (10), 2048. doi: 10.3390/microorganisms9102048
Papale M., Rizzo C., Fani R., Bertolino M., Costa G., Paytuví-Gallart A., et al. (2020). Exploring the diversity and metabolic profiles of bacterial communities associated with Antarctic sponges (Terra Nova bay, Ross Sea). Front. Ecol. Evol. 8. doi: 10.3389/fevo.2020.00268
Parada A. E., Needham D. M., Fuhrman J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18 (5), 1403–1414. doi: 10.1111/1462-2920.13023
Peck L. S., Conway L. Z. (2000). The myth of metabolic cold adaptation: Oxygen consumption in stenothermal Antarctic bivalves. Geol. Soc Spec. Publ. 177 (1), 441–450. doi: 10.1144/GSL.SP.2000.177.01.29
Perkins N. R., Monk J., Soler G., Gallagher P., Barrett N. S. (2022). Bleaching in sponges on temperate mesophotic reefs observed following marine heatwave events. Clim. Change Ecol. 3, 100046. doi: 10.1016/J.ECOCHG.2021.100046
Pinnaka A. K., Ara S., Singh A., Shivaji S. (2013). Draft genome sequence of winogradskyella psychrotolerans rs-3t, isolated from the marine transect of kongsfjorden, ny-Ålesund, Svalbard, Arctic ocean. Genome Announc. 1 (4), e00630–e00613. doi: 10.1128/genomea.00630-13
Pita L., Rix L., Slaby B. M., Franke A., Hentschel U. (2018). The sponge holobiont in a changing ocean: From microbes to ecosystems. Microbiome 6 (1), 46. doi: 10.1186/s40168-018-0428-1
Pollocks J. F., Katz S. M., van de Water J. A. J. M., Davies S. W., Hein M., Torda G. (2017). Coral larvae for restoration and research: A large-scale method for rearing Acropora millepora larvae, inducing settlement, and establishing symbiosis. PeerJ 3, e3732. doi: 10.7717/PEERJ.3732
Posadas N., Baquiran J. I. P., Nada M. A. L., Kelly M., Conaco C. (2021). Microbiome diversity and host immune functions influence survivorship of sponge holobionts under future ocean conditions. ISME J. 16 (1), 58–67. doi: 10.1038/S41396-021-01050-5
Ramsby B. D., Hoogenboom M. O., Whalan S., Webster N. S. (2018). Elevated seawater temperature disrupts the microbiome of an ecologically important bioeroding sponge. Mol. Ecol. 27 (8), 2124–2137. doi: 10.1111/mec.14544
Regueiras A., Alex A., Pereira S., Costa M. S., Antunes A., Vasconcelos V. (2017). Cyanobacterial diversity in the marine sponge hymeniacidon perlevis from a temperate region (Portuguese coast, northeast Atlantic). Aquat. Microb. Ecol. 79 (3), 259–272. doi: 10.3354/ame01830
Reveillaud J., Maignien L., Eren M. A., Huber J. A., Apprill A., Sogin M. L., et al. (2014). Host-specificity among abundant and rare taxa in the sponge microbiome. ISME J. 8 (6), 1198–1209. doi: 10.1038/ismej.2013.227
Ribes M., Calvo E., Movilla J., Logares R., Coma R., Pelejero C. (2016). Restructuring of the sponge microbiome favors tolerance to ocean acidification. Environ. Microbiol. Rep. 8 (4), 536–544. doi: 10.1111/1758-2229.12430
Ribes M., Dziallas C., Coma R., Riemann L. (2015). Microbial diversity and putative diazotrophy in high- and low-Microbial-Abundance Mediterranean sponges. Appl. Environ. Microbiol. 81 (17), 5683–5693. doi: 10.1128/aem.01320-15
Ribes M., Jiménez E., Yahel G., López-Sendino P., Diez B., Massana R., et al. (2012). Functional convergence of microbes associated with temperate marine sponges. Environ. Microbiol. 14 (5), 1224–1239. doi: 10.1111/j.1462-2920.2012.02701.x
Riesgo A., Farrar N., Windsor P. J., Giribet G., Leys S. P. (2014). The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol. Biol. Evol. 31 (5), 1102–1120. doi: 10.1093/molbev/msu057
Rizzo C., Zammuto V., Lo Giudice A., Rizzo M. G., Spanò A., Laganà P., et al. (2021). Antibiofilm activity of Antarctic sponge-associated bacteria against pseudomonas aeruginosa and staphylococcus aureus. J. Mar. Sci. Eng. 9 (3), 243. doi: 10.3390/jmse9030243
Roda-Garcia J. J., Haro-Moreno J. M., Huschet L. A., Rodriguez-Valera F., López-Pérez M. (2021). Phylogenomics of SAR116 clade reveals two subclades with different evolutionary trajectories and an important role in the ocean sulfur cycle. mSystems 6 (5), e00944–e00921. doi: 10.1128/msystems.00944-21
Rodríguez-Marconi S., de la Iglesia R., Díez B., Fonseca C. A., Hajdu E., Trefault N., et al. (2015). Characterization of bacterial, archaeal and eukaryote symbionts from Antarctic sponges reveals a high diversity at a three-domain level and a particular signature for this ecosystem. PloS One 10 (9), e0138837. doi: 10.1371/journal.pone.0138837
Rondon R., González-Aravena M., Font A., Osorio M., Cárdenas C. A. (2020). Effects of climate change stressors on the prokaryotic communities of the Antarctic sponge isodictya kerguelenensis. Front. Ecol. Evol. 8. doi: 10.3389/fevo.2020.00262
Rosado P. M., Leite D. C. A., Duarte G. A. S., Chaloub R. M., Jospin G., Nunes da Rocha U., et al. (2019). Marine probiotics: Increasing coral resistance to bleaching through microbiome manipulation. ISME J. 13 (4), 921–936. doi: 10.1038/s41396-018-0323-6
Rosales S. M., Clark A. S., Huebner L. K., Ruzicka R. R., Muller E. M. (2017). Rhodobacterales and Rhizobiales are associated with stony coral tissue loss disease and its suspected sources of transmission. Front. Microbiol. 11, 681. doi: 10.3389/FMICB.2020.00681
Rubio-Portillo E., Izquierdo-Muñoz A., Gago J. F., Rosselló-Mora R., Antón J., Ramos-Esplá A. A. (2016). Effects of the 2015 heat wave on benthic invertebrates in the tabarca marine protected area (southeast Spain). Mar. Environ. Res. 122, 135–142. doi: 10.1016/j.marenvres.2016.10.004
Rubio-Portillo E., Ramos-Esplá A. A., Antón J. (2021). Shifts in marine invertebrate bacterial assemblages associated with tissue necrosis during a heat wave. Coral Reefs 40 (2), 395–404. doi: 10.1007/s00338
Ruocco N., Esposito R., Bertolino M., Zazo G., Sonnessa M., Andreani F., et al. (2021). A metataxonomic approach reveals diversified bacterial communities in Antarctic sponges. Mar. Drugs 19 (3), 173. doi: 10.3390/md19030173
Sacristán-Soriano O., Pérez Criado N., Avila C. (2020). Host species determines symbiotic community composition in Antarctic sponges (Porifera: Demospongiae). Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00474
Schellenberg J., Busse H. J., Hardt M., Schubert P., Wilke T., Kämpfer P., et al. (2017). Winogradskyella haliclonae sp. nov., isolated from a marine sponge of the genus haliclona. Int. J. Syst. Evol. Microbiol. 67 (12), 4902–4910. doi: 10.1099/ijsem.0.002192
Schmitt S., Hentschel U., Taylor M. W. (2012a). Deep sequencing reveals diversity and community structure of complex microbiota in five Mediterranean sponges. Hydrobiologia 687, 341–351. doi: 10.1007/s10750-011-0799-9
Schmitt S., Tsai P., Bell J., Fromont J., Ilan M., Lindquist N., et al. (2012b). Assessing the complex sponge microbiota: Core, variable and species-specific bacterial communities in marine sponges. ISME J. 6 (3), 564–576. doi: 10.1038/ismej.2011.116
Sharp K. H., Ritchie K. B. (2021). Multi-partner interactions in corals in the face of climate change. Biol. Bull 223 (1), 66–77. doi: 10.1086/BBLV223N1P66
Shivani Y., Subhash Y., Sasikala C., Ramana C. H. V. (2017). Halodesulfovibrio spirochaetisodalis gen. nov. sp. nov. and reclassification of four desulfovibrio spp. Int. J. Syst. Evol. Microbiol. 67 (1), 87–93. doi: 10.1099/ijsem.0.001574
Simion P., Philippe H., Baurain D., Jager M., Richter D. J., di Franco A., et al. (2017). A large and consistent phylogenomic dataset supports sponges as the sister group to all other animals. Curr. Biol. 27 (7), 958–967. doi: 10.1016/j.cub.2017.02.031
Simister R., Taylor M. W., Rogers K. M., Schupp P. J., Deines P. (2013). Temporal molecular and isotopic analysis of active bacterial communities in two new Zealand sponges. FEMS Microbiol. Ecol. 85 (1), 195–205. doi: 10.1111/1574-6941.12109
Simister R., Taylor M. W., Tsai P., Fan L., Bruxner T. J., Crowe M. L., et al. (2012a). Thermal stress responses in the bacterial biosphere of the great barrier reef sponge, rhopaloeides odorabile. Environ. Microbiol. 14 (12), 3232–3246. doi: 10.1111/1462-2920.12010
Simister R., Taylor M. W., Tsai P., Webster N. (2012b). Sponge-microbe associations survive high nutrients and temperatures. PloS One 7 (12), e52220. doi: 10.1371/journal.pone.0052220
Smale D. A., Wernberg T., Oliver E. C. J., Thomsen M., Harvey B. P., Straub S. C., et al. (2019). Marine heatwaves threaten global biodiversity and the provision of ecosystem services. Nat. Clim. Change 9 (4), 306–312. doi: 10.1038/s41558-019-0412-1
Southwell M. W., Weisz J. B., Martens C. S., Lindquist N. (2008). In situ fluxes of dissolved inorganic nitrogen from the sponge community on conch reef, key largo, Florida. Limnol. Oceanogr. 53 (3), 986–996. doi: 10.4319/LO.2008.53.3.0986
Steinert G., Taylor M. W., Deines P., Simister R. L., de Voogd N. J., Hoggard M., et al. (2016). In four shallow and mesophotic tropical reef sponges from Guam the microbial community largely depends on host identity. PeerJ 4, e1936. doi: 10.7717/peerj.1936
Steinert G., Wemheuer B., Janussen D., Erpenbeck D., Daniel R., Simon M., et al. (2019). Prokaryotic diversity and community patterns in Antarctic continental shelf sponges. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00297
Stevens M. A., Riebesell U., McDevitt-Irwin J. M., Karlsson R., Kanbar J., Gifford I., et al. (2017). A communal catalogue reveals earth’s multiscale microbial diversity. Nature 551 (7681), 457–463. doi: 10.1038/nature24621
Strand R., Whalan S., Webster N. S., Kutti T., Fang J. K. H., Luter H. M., et al. (2017). The response of a boreal deep-sea sponge holobiont to acute thermal stress. Sci. Rep. 7 (1), 1–12. doi: 10.1038/s41598-017-01091-x
Taylor M. W., Radax R., Steger D., Wagner M. (2007). Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71 (2), 295–347. doi: 10.1128/mmbr.00040-06
Taylor M. W., Schupp P. J., Dahllöf I., Kjelleberg S., Steinberg P. D. (2004). Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ. Microbiol. 6 (2), 121–130. doi: 10.1046/J.1462-2920.2003.00545.x
Taylor M. W., Tsai P., Simister R. L., Deines P., Botte E., Ericson G., et al. (2013). ‘Sponge-specific’ bacteria are widespread (but rare) in diverse marine environments. ISME J. 7 (2), 438–443. doi: 10.1038/ismej.2012.111
Taylor J. A., Palladino G., Wemheuer B., Steinert G., Sipkema D., Williams T. J. (2020). Phylogeny resolved, metabolism revealed: functional radiation within a widespread and divergent clade of sponge symbionts. ISME J. 15 (2), 503–519. doi: 10.1038/s41396-020-00791-z
Teeling H., Fuchs B. M., Becher D., Klockow C., Gardebrecht A., Bennke C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336 (6081), 608–611. doi: 10.1126/science.1218344
Thébault J., Chauvaud L., Clavier J., Guarini J., Dunbar R. B., Fichez R., et al. (2007). Reconstruction of seasonal temperature variability in the tropical pacific ocean from the shell of the scallop, comptopallium radula. Geochim. Cosmochim. Acta 71 (4), 918–928. doi: 10.1016/j.gca.2006.10.017
Thomas T., Moitinho-Silva L., Lurgi M., Björk J. R., Easson C., Astudillo-García C., et al. (2016). Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 7 (1), 1–12. doi: 10.1038/ncomms11870
Thomas T., Rusch D., DeMaere M. Z., Yung P. Y., Lewis M., Halpern A., et al. (2010). Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J. 4 (12), 1557–1567. doi: 10.1038/ismej.2010.74
Vacelet J., Donadey C. (1977). Electron microscope study of the association between some sponges and bacteria. J. Exp. Mar. Biol. Ecol. 30 (3), 301–314. doi: 10.1016/0022-0981(77)90038-7
Vargas S., Leiva L., Wörheide G. (2021). Short-term exposure to high-temperature water causes a shift in the microbiome of the common aquarium sponge lendenfeldia chondrodes. Microb. Ecol. 81 (1), 213–222. doi: 10.1007/s00248-020-01556-z
Webster N., Bourne D. (2007). Bacterial community structure associated with the Antarctic soft coral, alcyonium antarcticum. FEMS Microbiol. Ecol. 59 (1), 81–94. doi: 10.1111/j.1574-6941.2006.00195.x
Webster N. S., Cobb R. E., Negri A. P. (2008). Temperature thresholds for bacterial symbiosis with a sponge. ISME J. 2 (8), 830–842. doi: 10.1038/ismej.2008.42
Webster N. S., Negri A. P., Munro M. M. H. G., Battershill C. N. (2004). Diverse microbial communities inhabit Antarctic sponges. Environ. Microbiol. 6 (3), 288–300. doi: 10.1111/j.1462-2920.2004.00570.x
Webster N. S., Taylor M. W. (2012). Marine sponges and their microbial symbionts: love and other relationships. Environ. Microbiol. 14 (2), 335–346. doi: 10.1111/j.1462-2920.2011.02460.x
Webster N. S., Taylor M. W., Behnam F., Lücker S., Rattei T., Whalan S., et al. (2010). Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol. 12 (8), 2070–2082. doi: 10.1111/j.1462-2920.2009.02065.x
Webster N. S., Thomas T. (2016). The sponge hologenome. mBio 7 (2), e00135–e00116. doi: 10.1128/mBio.00135-16
Webster N. S., Webb R. I., Ridd M. J., Hill R. T., Negri A. P. (2001). The effects of copper on the microbial community of a coral reef sponge. Environ. Microbiol. 3 (1), 19–31. doi: 10.1046/j.1462-2920.2001.00155.x
Welsh R. M., Rosales S. M., Zaneveld J. R., Payet J. P., McMinds R., Hubbs S. L., et al. (2017). Alien vs. predator: bacterial challenge alters coral microbiomes unless controlled by halobacteriovorax predators. PeerJ 5, e3315. doi: 10.7717/peerj.3315
Keywords: holobiont, heat shock, global warming, high microbial abundance, low microbial abundance, metabarcoding
Citation: De Castro-Fernández P, Ballesté E, Angulo-Preckler C, Biggs J, Avila C and García-Aljaro C (2023) How does heat stress affect sponge microbiomes? Structure and resilience of microbial communities of marine sponges from different habitats. Front. Mar. Sci. 9:1072696. doi: 10.3389/fmars.2022.1072696
Received: 17 October 2022; Accepted: 16 December 2022;
Published: 19 January 2023.
Edited by:
Nicole Trefault, Universidad Mayor, ChileReviewed by:
Angelina Lo Giudice, National Research Council (CNR), ItalyMario Moreno-Pino, Universidad Mayor, Chile
Copyright © 2023 De Castro-Fernández, Ballesté, Angulo-Preckler, Biggs, Avila and García-Aljaro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cristina García-Aljaro, crgarcia@ub.edu
 Paula De Castro-Fernández
Paula De Castro-Fernández Elisenda Ballesté
Elisenda Ballesté Carlos Angulo-Preckler
Carlos Angulo-Preckler Jason Biggs
Jason Biggs Conxita Avila
Conxita Avila Cristina García-Aljaro
Cristina García-Aljaro