Processing of Naturally Sourced Macroalgal- and Coral-Dissolved Organic Matter (DOM) by High and Low Microbial Abundance Encrusting Sponges
- 1Department of Freshwater and Marine Ecology, Institute for Biodiversity and Ecosystem Dynamics, University of Amsterdam, Amsterdam, Netherlands
- 2Department of Earth Sciences, Faculty of Geosciences, Utrecht University, Utrecht, Netherlands
- 3Mote’s Elizabeth Moore International Center for Coral Reef Research & Restoration, Summerland Key, FL, United States
- 4Department of Marine Microbiology & Biogeochemistry, NIOZ Royal Netherlands Institute for Sea Research, Texel, Netherlands
- 5CARMABI Foundation, Willemstad, Curaçao
Sponges play a key role in (re)cycling of dissolved organic matter (DOM) and inorganic nutrients in coral reef ecosystems. Macroalgae and corals release different quantities of DOM and at different bioavailabilities to sponges and their microbiome. Given the current coral- to algal-dominance shift on coral reefs, we assessed the differential processing of macroalgal- and coral-DOM by three high and three low microbial abundance (HMA and LMA) encrusting sponge species. We followed the assimilation of naturally sourced 13C- and 15N-enriched macroalgal- and coral-DOM into bulk tissue and into host- versus bacteria-specific phospholipid fatty acids (PLFAs). Additionally, we compared sponge-processing of the two natural DOM sources with 13C- and 15N-enriched laboratory-made diatom-DOM. All investigated sponges utilized all DOM sources, with higher assimilation rates in LMA compared to HMA sponges. No difference was found in carbon assimilation of coral- versus macroalgal-DOM into bulk tissue and host- versus bacteria-specific PLFAs, but macroalgal nitrogen was assimilated into bulk tissue up to eight times faster compared to the other sources, indicating its higher bioavailability to the sponges. Additionally, LMA sponges released significantly more inorganic nitrogen after feeding on macroalgal-DOM. Therefore, we hypothesize that, depending on the abundance and composition of the sponge community, sponges could catalyze reef eutrophication through increased turnover of nitrogen under coral-to-algal regime shifts.
Introduction
Coral reefs are marine ecosystems thriving in oligotrophic waters. Despite being characterized by low nutrient concentrations, these ecosystems are highly productive and hotspots of biodiversity (Hatcher, 1990; Knowlton et al., 2010). The largest organic resource on coral reefs is dissolved organic matter (DOM) (Atkinson and Falter, 2003; de Goeij and van Duyl, 2007; Tanaka et al., 2011), which is mainly released by benthic primary producers, such as corals and algae (Naumann et al., 2010; Haas et al., 2011; Mueller et al., 2014b). The processing of DOM by bacterioplankton and its subsequent recycling through the microbial loop is well established (Azam et al., 1983; Haas et al., 2011; Nelson and Carlson, 2012), but in recent years, sponges have additionally been shown to play a major role in DOM recycling on coral reefs via the so-called “sponge loop” pathway (de Goeij et al., 2013). Sponges convert DOM—which is generally inaccessible to most heterotrophic reef organisms—into particulate organic matter (POM), which is then consumed by higher trophic levels (de Goeij et al., 2013; Rix et al., 2016; Rix et al., 2018). Conversion of DOM into detrital POM is mainly observed in encrusting sponge species (Alexander et al., 2014) that dominate the benthos of the cryptic habitats of reefs (e.g., overhangs, crevices, cavities), but also appear on exposed surfaces (Richter et al., 2001; Scheffers et al., 2005). By contrast, upward growing, non-encrusting sponges (e.g., massive, branching), which predominantly appear on exposed surfaces, are hypothesized to cycle DOM through a predatory sponge loop pathway, by investing more resources in net growth and then being preyed upon by spongivores (McMurray et al., 2018; Pawlik and McMurray, 2020).
Hurricanes, diseases, and coral bleaching events in combination with local (e.g., eutrophication and overfishing) and global (e.g. increased seawater temperatures) disturbances have resulted in considerable changes in benthic community compositions worldwide, including a shift from coral- to algal-dominance on many coral reefs (Hughes, 1994; McManus and Polsenberg, 2004; Hoegh-Guldberg et al., 2007; Mumby and Steneck, 2008). The loss of coral cover and the coral-to-algal community shift has been extensively described in terms of the decreased ability of reefs to build structure/habitat and maintain biodiversity (Hughes et al., 2003). But only recently has the biogeochemical role of DOM been incorporated into studies on overall reef functioning with changing benthic communities (Haas et al., 2016; Pawlik et al., 2016; de Goeij et al., 2017; Silveira et al., 2017; Silva et al., 2021). Algae release DOM at higher quantities per surface area and biomass than corals (Haas et al., 2011; Mueller et al., 2014b). Additionally, algal-DOM contains more labile components (e.g. sugars) than coral-DOM, consequently causing higher bacterioplankton production (Nelson et al., 2013), and fueling potentially pathogenic bacterial communities (Haas et al., 2011, 2016; Morrow et al., 2013; Cárdenas et al., 2018). Increased microbial respiration and other pathogenic mechanisms at the coral-algae interface can weaken or even kill corals, which frees space for further algal growth (Smith et al., 2006; Kline et al., 2006; Roach et al., 2020). This DDAM (dissolved organic carbon, disease, algae, microorganisms) feedback loop can thereby catalyze reef destruction (Haas et al., 2016) as reefs shift from coral to algal dominance (Rohwer et al., 2010; Dinsdale and Rohwer, 2011; Barott and Rohwer, 2012). By converting DOM into detritus and dissolved inorganic nutrients, sponges have been proposed to additionally fertilize algal growth, and thereby further accelerate the coral to algal shift (de Goeij et al., 2017), causing a feedback loop mechanism that may lock a reef in a degraded state (Pawlik et al., 2016).
The influence of sponge functional diversity on DOM-processing remains poorly understood. Different functional traits, including shape, microbial abundance, microbiome composition, filtering capacity, or pumping rates, have been proposed to influence the ability of sponges to process DOM (de Goeij et al., 2017; Gantt et al., 2019; Pawlik and McMurray, 2020). Sponges contain a highly diverse microbiome (Schmitt et al., 2011) and they are roughly classified into two main groups: high microbial abundance (HMA) species, in which bacteria can constitute up to 40% of their biomass (108–10 bacteria gsponge–1), and low microbial abundance (LMA) species, whose bacterial concentrations are similar to the surrounding seawater (105–6 bacteria gsponge–1) (Weisz et al., 2008; Hentschel et al., 2012). Due to their high abundances of microbial symbionts, HMA sponges are considered better adapted to utilize DOM (Reiswig, 1981; Ribes et al., 1999; Morganti et al., 2017; Hoer et al., 2018; Wooster et al., 2019). However, there is an increasing number of studies showing that LMA species take up DOM at rates equaling, or even higher than, those of HMA species. This was initially found in shallow water encrusting and excavating sponge species (de Goeij et al., 2013, 2017; Mueller et al., 2014a; Rix et al., 2017, 2020), but now also in deep-sea sponges with encrusting and massive growth forms (Bart et al., 2020, 2021).
First experimental assessment of differential processing of naturally sourced 13C/15N-enriched macroalgal- and coral-DOM was conducted on three common encrusting Red Sea sponge species (Rix et al., 2017). In both LMA (Hemimycale arabica and Mycale fistulifera) and HMA (Chondrilla sacciformis) species, macroalgal-DOM was processed at higher rates than coral-DOM and was preferentially incorporated into phospholipid-derived fatty acids (PLFAs) that are specific for sponge-associate bacteria, while coral-DOM was preferentially incorporated into sponge-host-specific PLFAs. Furthermore, the two LMA species were found to process coral-dissolved organic carbon (DOC) at higher rates than the HMA Chondrilla sacciformis, but no difference between LMA and HMA macroalgal-DOC processing rates were found. However, this is the only documented study on differential processing of naturally sourced coral- and macroalgal-DOM by sponges from one geographical area. Additionally, due to the low sample sizes and a limited number of species tested, the effect of sponge microbial abundance on the processing of coral- versus macroalgal-DOM sources is not yet established.
The Caribbean fringing reefs of Curaçao show a high variability in coral-to-algal abundance ratios, thereby providing a good experimental setting to study the ecological role of sponges in DOM cycling on coral-to-algal regime shifted reefs. On these reefs, encrusting sponges represent up to 35% of the reef benthic biomass (i.e., in ash-free dry weight) (Kornder et al., in press). As a first step to assess the role of sponges in the cycling of different naturally available DOM sources, we studied the differential processing of macroalgal- and coral-DOM by LMA and HMA encrusting sponge holobionts (i.e., host and microbial symbionts). Therefore, we compared the processing of different DOM sources by six encrusting sponges: three LMA and three HMA species. We followed the processing of two naturally sourced 13C- and 15N-enriched DOM food sources (macroalgal-DOM and coral-DOM) into bulk tissue of the sponge holobionts and into host- versus bacteria-specific PLFA biomarkers, and compared it with processing of 13C- and 15N-enriched laboratory-made diatom-DOM. Additionally, we measured inorganic nutrient fluxes relating to the processing of the two naturally sourced DOM types by the sponge holobionts.
Materials and Methods
Study Site, Organism Collection and Maintenance
All experimental work was conducted at the CARMABI Research Station over two fieldwork periods, in October–December 2017 and May–August 2018. Collections and experiments were performed under the research permit (#2012/48584) issued by the Curaçaoan Ministry of Health, Environment and Nature (GMN) to the CARMABI foundation.
All organisms, except the coral species Acropora cervicornis, were collected on the fringing reef close to Piscadera Bay on Curaçao (12°12′ N, 68°96′ W), between 5–30 m (i.e., macroalgae and corals between 5 and 15 m and sponges between 10 and 30 m water depth) by SCUBA. We collected macroalgae of the genus Dictyota and the scleractinian coral species Colpophyllia natans, Diploria labyrinthiformis, and Diploria strigosa (boulder brain corals). Fragments of the staghorn coral A. cervicornis grown in coral nurseries at Stella Maris reef (12°05′ N, 68°53′ W) were collected in collaboration with the Coral Restoration Curaçao. Dictyota spp. are common macroalgae on the reefs of Curaçao (i.e., 45% of total benthic macroalgal communities on the leeward reefs of Curaçao) (Institute Waitt, 2017; Kornder et al., in press). The four coral species were chosen because of their high mucus release rates, the minimal physical damage provoked by collection, and their common occurrence on reefs of Curaçao (i.e., C. natans, D. labyrinthiformis, and D. strigosa represent 3 of the 10 most abundant scleractinian coral species on Curaçao’s leeward reefs) (Institute Waitt, 2017). Corals and macroalgae were immediately transported without air exposure to the CARMABI running-seawater aquarium facilities and kept in 100 L flow-through aquaria, with a water exchange rate of 3 L min–1, supplied with seawater pumped directly from the reef at 10 m water depth. After an acclimatization period of at least 3 days, corals and algae were used for the production of labeled DOM sources. Six common coral reef encrusting sponge species (Porifera)—i.e., encrusting growth forms mainly follow the contours of the reef surface as opposed to massive growth forms that display emergent, upward growth forming a separate shape independent of the reef contour—were collected: low microbial abundance (LMA) sponge species Halisarca caerulea (Demospongiae; 1–2 mm thick sheet), Haliclona (Halichoclona) vansoesti (Demospongiae; 0.5–3 cm thick conulose), and Scopalina ruetzleri (Demospongiae; 0.5–4 cm thick conulose), and HMA species Chondrilla caribensis (Demospongiae; 3–8 mm thick sheet), Ectyoplasia ferox (Demospongiae; 2–3 cm thick conulose), and Plakortis angulospiculatus (Homoscleromorpha; 1–4 cm thick lobate/ficiform) (Figure 1). After collection, sponges were trimmed to a size between 10 and 30 cm2 and cleared of epibionts (de Goeij et al., 2008a). Trimmed specimens were allowed to recover at the collection site for 3–4 weeks to ensure full recovery from collection and handling (Alexander et al., 2015a). Only visually healthy sponges (no tissue damage, open oscula) were used in the experiments.
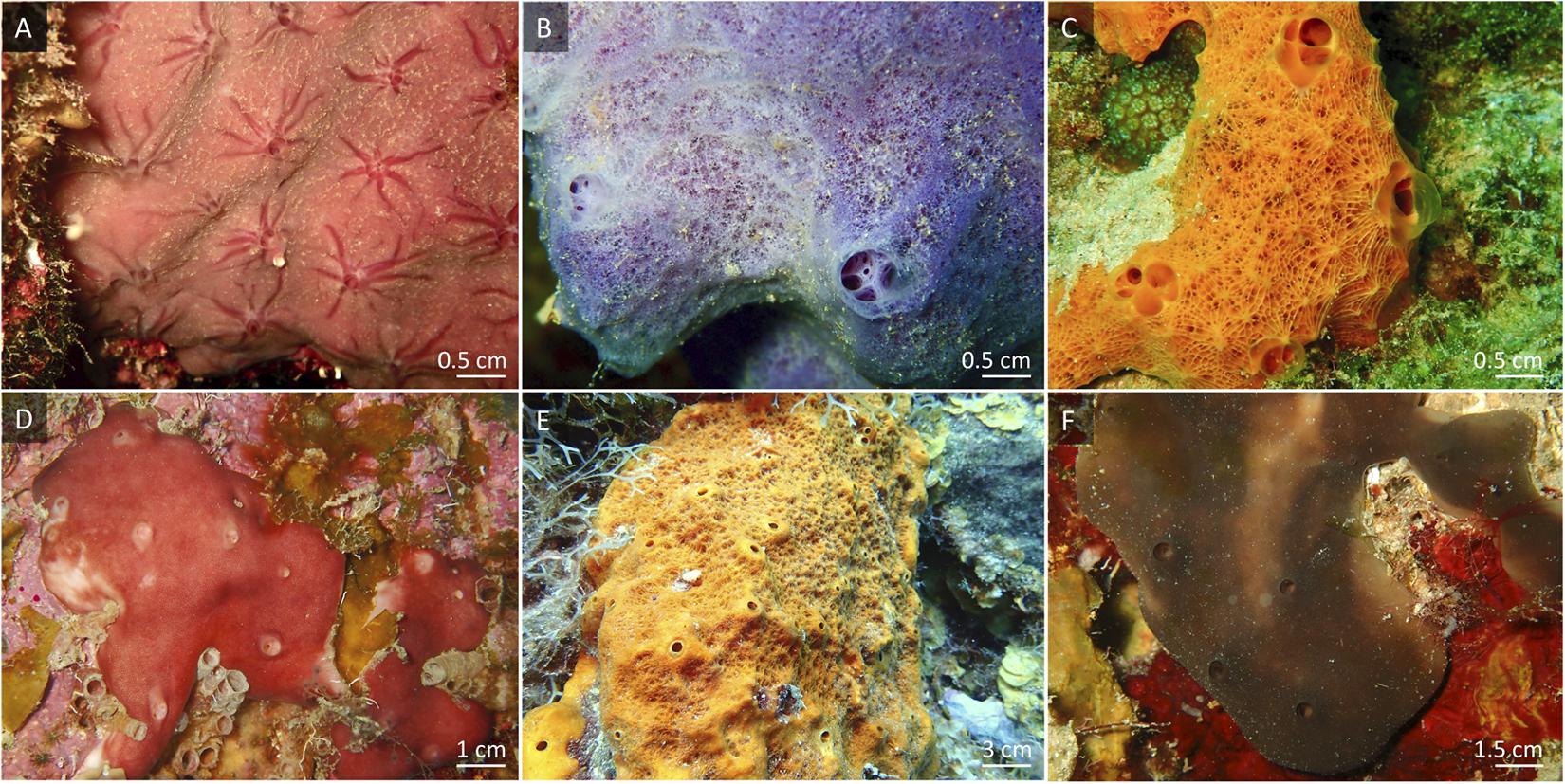
Figure 1. In situ views of the sponge species used in this experiment. (A) Halisarca caerulea, (B) Haliclona vansoesti, (C) Scopalina ruetzleri, (D) Chondrilla caribensis, (E) Ectyoplasia ferox and (F) Plakortis angulospiculatus. (A–C) Low microbial abundance (LMA) sponge species. (D–F) High microbial abundance (HMA) sponge species.
Production of Labeled DOM Sources
13C- and 15N-enriched DOM was naturally produced from macroalgae and corals, whereas the diatom-DOM source was laboratory-made. The latter served as a highly labeled control to our natural DOM sources and was produced according to established methodology (de Goeij et al., 2013; Rix et al., 2020), using axenic batch cultures of the cosmopolitan marine diatom Phaeodactylum tricornutum (details in the Supplementary Material). This diatom-DOM source contains total dissolved substances from lysed diatoms and may therefore represent naturally available DOM to only a limited extent. Macroalgal- and coral-DOM were naturally sourced to better reflect the composition of DOM exudates released into the environment. For the initial labeling phase, macroalgae (25 specimens of Dictyota, Supplementary Figure 1A) and corals (10 specimens of A. cervicornis and a total of 15 specimens of the brain corals, Supplementary Figures 1B,C) were separately incubated for 8 h per day over the course of five (algae) and seven (coral) consecutive days. Macroalgae and corals were incubated in 20-L aquaria filled with carbon-free artificial seawater (CFASW; see Supplementary Material for full description) with the addition of a final concentration of 1 mg L–1 Na15NO3 and 36 mg L–1 NaH13CO3 in the first fieldwork period and 14 mg L–1 Na15NO3 and 50 mg L–1 NaH13CO3 in the second fieldwork period (Eurisotop, Cambridge Isotope Laboratories). CFASW was used to maximize the relative abundance of 13C-labeled dissolved inorganic carbon (DI13C) in the seawater, in order to achieve the highest labeling of coral- and algal-DOM. Aquaria were placed in a running-seawater water bath and water circulation inside the 20-L aquaria was maintained with aquaria recirculating pumps. Temperature and pH of the aquaria were monitored with a pH/temperature meter (Hanna Instruments, United States). Light and temperature levels in the aquaria were adjusted to mimic in situ conditions at the average collection depths of our DOM-producing organisms (macroalgae and corals) at 10 m water depth (1000–20000 Lux and 29–31°C in 2017 and 27–29°C in 2018; HOBO pendant logger) using layers of mesh to shade the aquaria. The initial 5/7-day labeling phase was followed by a 2-day DOM-production phase. In the mornings, corals and macroalgae were again incubated with 13C and 15N for 4 h to maximize the 13C/15N-enrichment of the organisms. Corals and macroalgae were then thoroughly rinsed in non-labeled seawater for 20 min to remove any label adhering to the organisms. Meanwhile, the aquaria were also carefully rinsed and re-filled with fresh non-labeled seawater. Macroalgae and corals were finally returned to the label-free aquaria and incubated for 5 h in non-labeled seawater to produce 13C- and 15N-labeled DOM. The water containing the labeled DOM exudates was filtered through a 0.7 μm GF/F filter (45 mm, Whatman, Kent, United Kingdom), using a peristaltic pump. The resulting filtered seawater containing only the dissolved organic fractions of macroalgal and coral exudates (from here on referred to as ‘macroalgal-DOM’ and ‘coral-DOM’; NB: exudates from the different species of coral were pooled) were used in subsequent sponge incubations. Concentrations of dissolved organic C and (in)organic N (DOC, DON, NO2–, NO3–, NH4+, and PO43–) were determined during the DOM-production before (t = –1) and directly after (t = 0) the addition of the macroalgae and corals, as well as at the end of the 5 h incubation (t = 5). Macroalgae and corals were returned to their collection sites at the end of the experiments. While mucus production due to air exposure is a common phenomenon on reefs with a high tidal amplitude, this rarely occurs in the Caribbean. Thus, the here described incubation technique to produce coral-DOM is a more suitable method when working with Caribbean species, as opposed to previous protocols where (labeled) coral mucus was obtained via ‘milking’ methods (Nakajima et al., 2016; Rix et al., 2017).
Sponge Incubations
All sponge species (n = 3–5 per species) and controls without sponges (n = 4–7) were incubated with the three DOM sources for 3–6 h, depending on sponge biomass (de Goeij et al., 2013) (see Supplementary Table 1). Sponge individuals were transferred, without air exposure, to air-tight, stirred, incubation chambers (Supplementary Figure 2), which were filled with macroalgal- or coral-DOM, or with 0.7-μm filtered seawater (FSW) to which the resuspended diatom-DOM was added at the beginning of the incubation using a sterile syringe (to a final concentration of 100 μmol L–1 DOC and 16-20 μmol L–1 DON). Only sponges with open oscula (indicating actively pumping specimens) were used for incubations. All incubations were conducted in the dark and dissolved oxygen concentration was monitored continuously with an optical probe (OXY-4, PreSens, Regensburg, Germany). Incubation chambers were placed in a flow-through aquarium to ensure near in situ temperature. Water samples for inorganic nutrients, DOC, total dissolved nitrogen and dissolved inorganic 13carbon (DI13C) quantification (see full description of water sample analysis in Supplementary Material) were collected at the beginning and end of each incubation, in acid-washed (0.4 mol L–1 HCl) 100-mL polycarbonate syringes. At the end of each incubation, sponges were rinsed in non-labeled fresh seawater and dipped in Milli-Q to remove salts before sampling for later 13C and 15N bulk tissue and PLFA stable isotope analysis. Bulk tissue samples were used to calculate source DOC and DON assimilation by the sponges, whereas PLFA stable isotope analysis allowed us to distinguish source DOC incorporation into sponge-host versus associated-bacteria specific PLFAs (for full description of sample analysis and rates calculation see Supplementary Material). Using the carbon assimilation rates (obtained from bulk tissue stable isotope analysis) and the carbon respiration rates (obtained from dissolved inorganic carbon stable isotopes analysis, DI13C), we calculated the assimilation-to-respiration efficiency as the percentage of: Cassimilated/(Cassimilated + Crespired).
Statistical Data Analysis
Statistical analyses were performed in PRIMER-E version 7 (Clarke and Gorley, 2015) with the PERMANOVA + add-on (Anderson et al., 2008). Differences between species within LMA and HMA sponge types were tested using individual one-factor PERMANOVAs based on Euclidean distance matrices with type III (partial) sum of squares and unrestricted permutation of raw data (9999 permutations). Where ‘species’ was not a significant factor, species were pooled within the LMA or HMA category. Individual two-factor PERMANOVAs based on Euclidean distance matrices were used with type III (partial) sum of squares and permutation of residuals under a reduced model (9999 permutations) to test for possible differences between: (A) the effect of DOM source (macroalgal- versus coral- versus diatom-DOM) and sponge type (LMA versus HMA) on DOC and DON assimilation rates, (B) the effect of sponge type on assimialtion-to-respiration efficiencies, and, (C) the effect of DOM source and sponge type on the DOC incorporation rates into total PLFAs, and into bacteria and sponge specific PLFAs. Post hoc pairwise tests were carried out to identify the significant pairs when one of the factors returned significant. Results were considered significant at the level P(perm) < 0.05. To further investigate possible differences between all sponge species, we performed a parallel set of statistical analysis using sponge species instead of sponge type as a factor in the aforementioned two-factor PERMANOVAs design (these results are presented in Supplementary File 1).
Results
Production of Labeled DOM Sources
All produced DOM sources were enriched in both 13C and 15N (Table 1). The enrichment of the laboratory-made diatom-DOM was higher in both 13C and 15N compared to natural DOM sources due to the different preparation methodologies. The concentrations of macroalgal- and coral-DOC and -DON at the start of the sponge incubations ranged between 101 and 151 μmol L–1 and 7–31 μmol L–1 respectively, and dissolved inorganic nitrogen (DIN = NO2– + NO3– + NH4+) ranged between 1.4–1.8 μmol L–1 (Table 1).
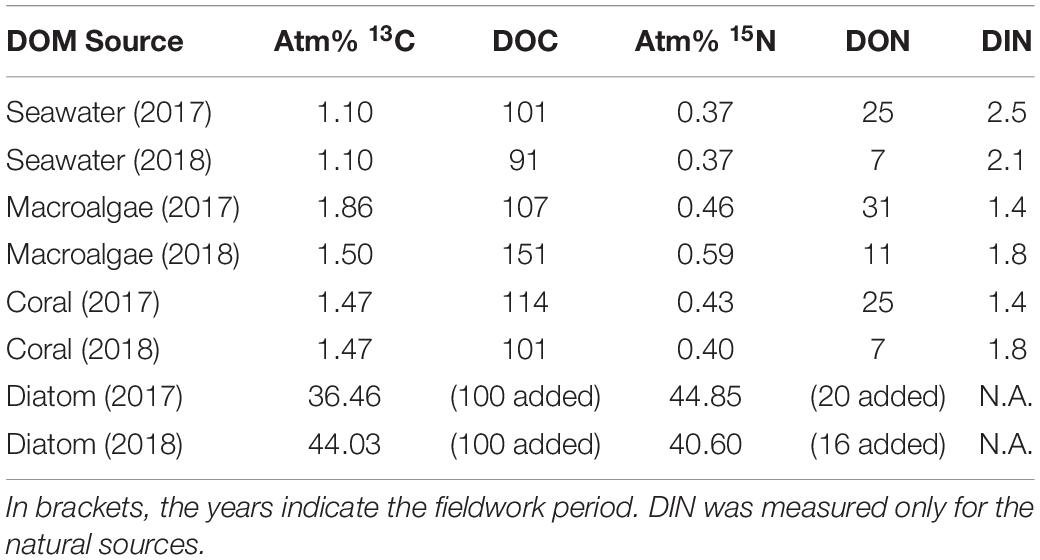
Table 1. Enrichment of macroalgal-, coral-, and diatom-DOM and average concentration of DOC, DON, and DIN (μmol C or N L–1) at the start of the sponge incubations.
Bulk DOM Assimilation and Respiration Rates
All the investigated sponges assimilated all three DOM sources in their bulk tissue (Table 2 and Figure 2). LMA sponges assimilated DOC and DON of all three DOM sources at a significantly higher rate than HMA sponges, except for laboratory-made diatom-DON (Table 3 and Supplementary Table 2). There was no significant difference between the assimilation rates of the natural macroalgal- and coral-DOC sources (LMA: 0.8 ± 0.1 and 0.8 ± 0.3 and HMA: 0.5 ± 0.2 and 0.5 ± 0.1 μmol Ctracer mmol Csponge–1 h–1, respectively; mean ± 95% confidence intervals throughout text), but both were assimilated at significantly lower rates than the laboratory-made diatom-DOC (LMA: 1.1 ± 0.3 and HMA: 0.6 ± 0.3) (Figure 2A and Supplementary Table 2). The largest variation in differential processing between sponge type and DOM source type was found for DON (Figure 2B). The difference in DON assimilation between LMA and HMA sponges was highest for sponges fed with macroalgal-DON (LMA: 8.7 ± 2.5 versus HMA: 4.3 ± 1.7 6 μmol Ntracer mmol Nsponge–1 h–1) and macroalgal-DON was assimilated 3–8 times faster than coral- and diatom-DON (LMA: 2.4 ± 0.6 and 1.0 ± 0.3 and HMA: 1.3 ± 0.6 and 0.8 ± 0.4, respectively).
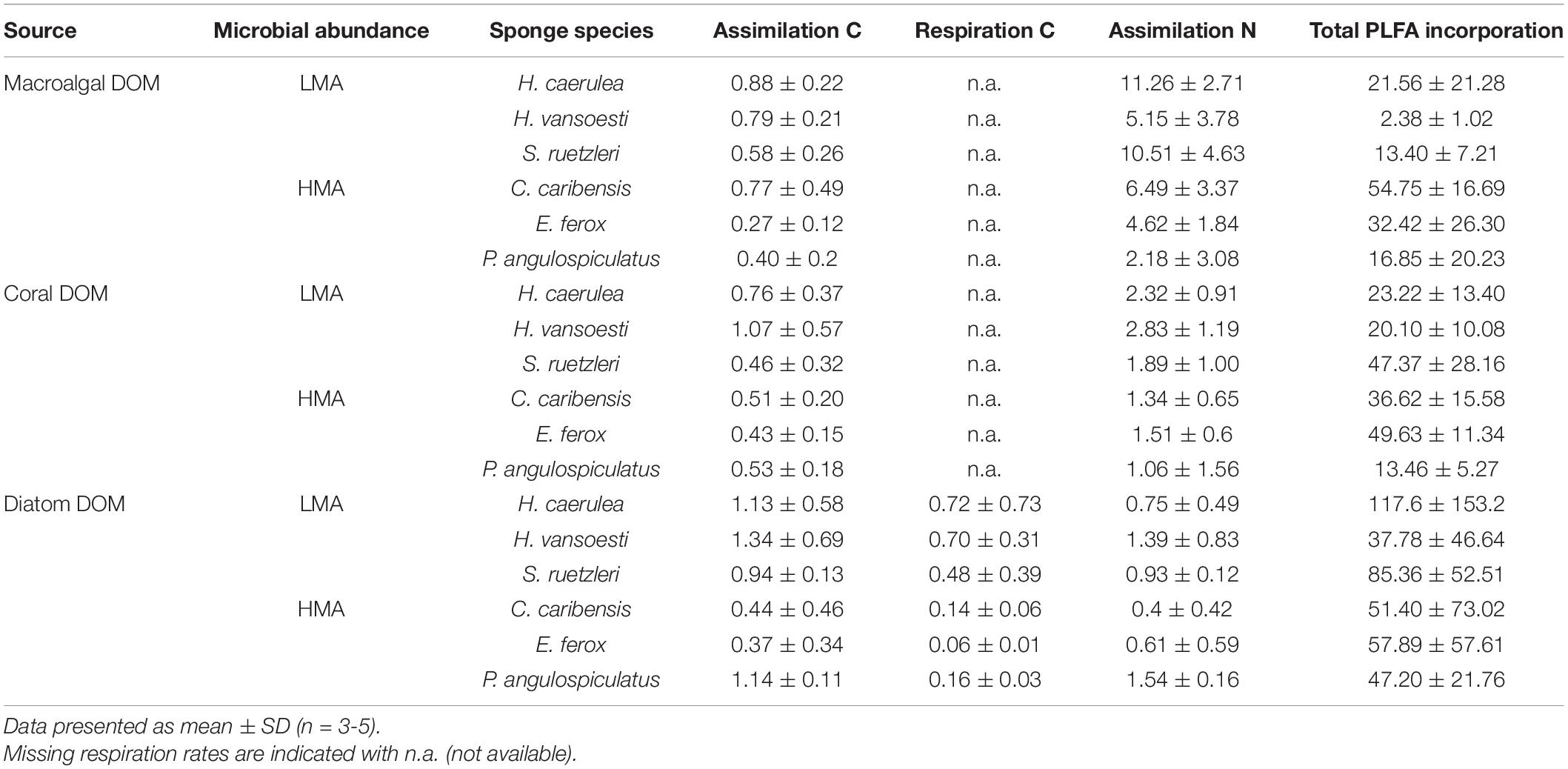
Table 2. Bulk tissue assimilation and respiration rates in μmol C or Ntracer mmol C or Nsponge–1 h–1 and total PLFA incorporation rates in nmol Ctracer mmol Csponge–1 12 h–1 of three tracer DOM sources per sponge species.
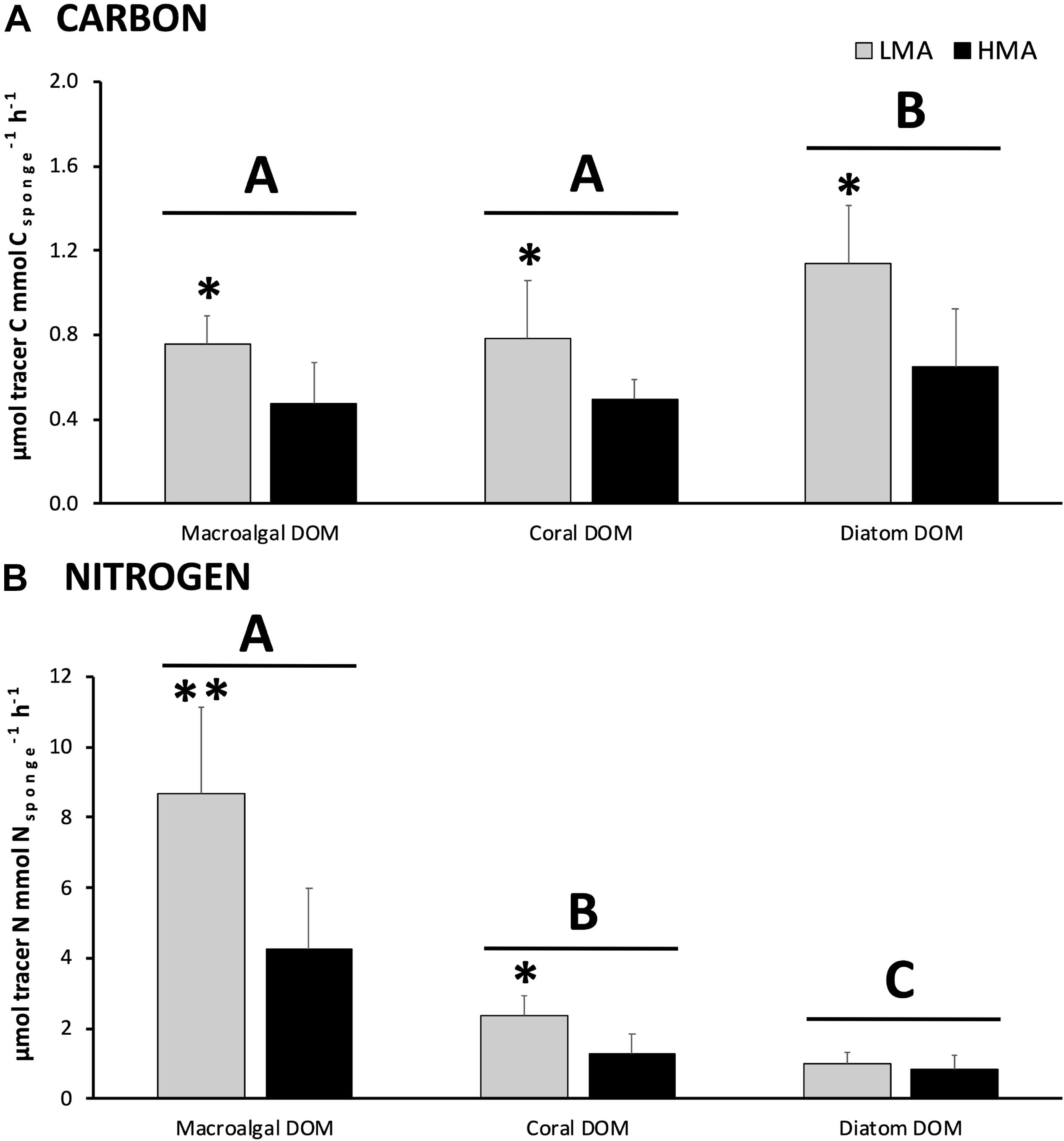
Figure 2. DOM assimilation into bulk sponge tissue. Assimilation rates of (A) carbon and (B) nitrogen from each 13C/15N-enriched DOM source by sponge type (in gray LMA species and in black HMA species). Note the different scales for carbon and nitrogen. Data presented as mean assimilation rates in μmol C or Ntracer mmol C or Nsponge–1 h–1 ± 95%-CI (n = 13). Letters (A, B, and C) indicate significant differences between the sources and asterisks (*) indicate significant differences between LMA and HMA types within each source. Signif. codes: ∗∗p < 0.01, ∗p < 0.05.
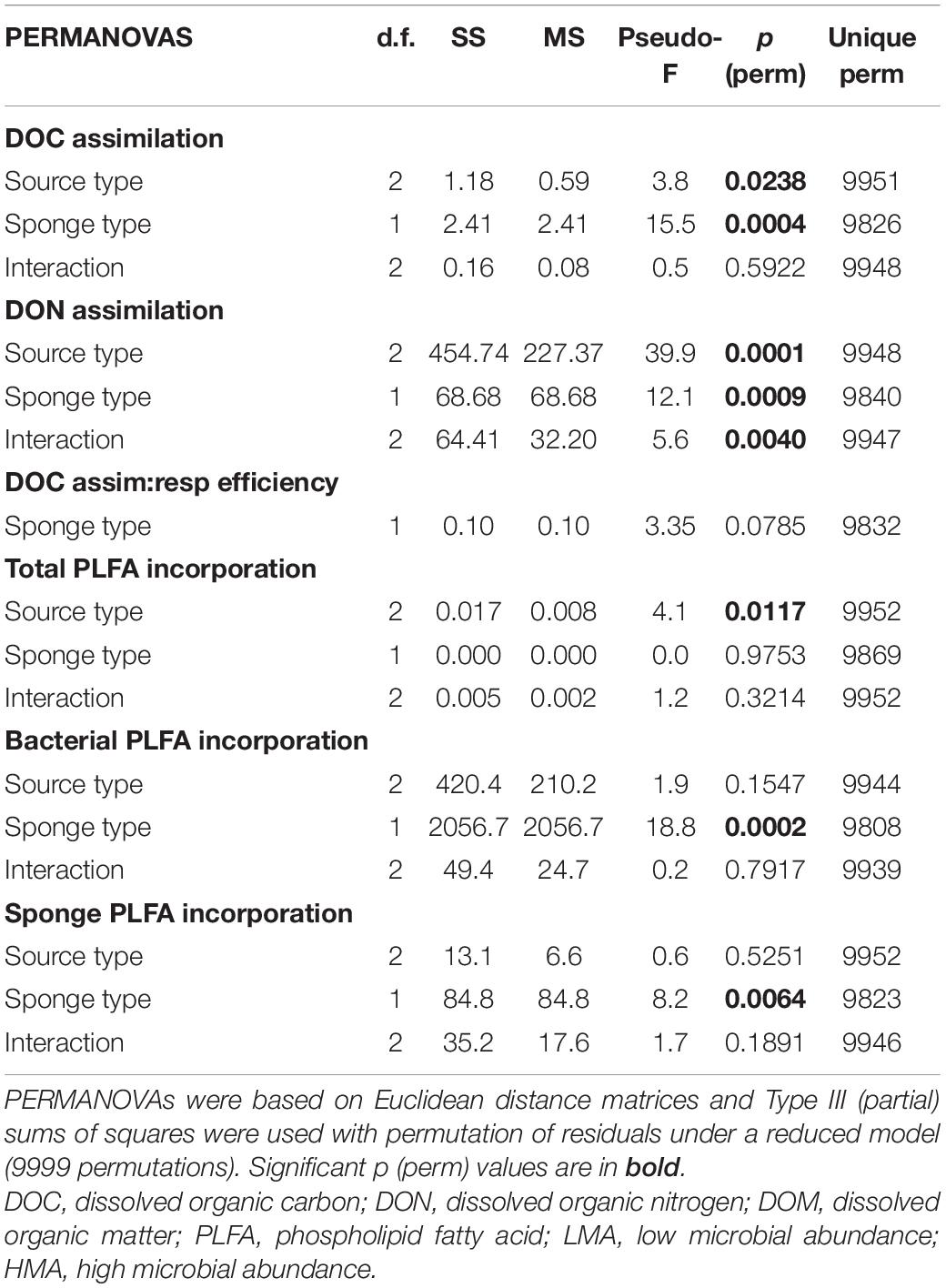
Table 3. Results of individual PERMANOVAs testing for differences in (A) carbon and nitrogen (DOC and DON) assimilation rates into bulk tissue, (B) assimilation-to-respiration efficiencies of diatom-DOM, and (C) carbon assimilation rates into PLFAs (total, sponge-, and bacteria-specific) by tracer DOM source type (macroalgal-, coral-, diatom-DOM), sponge type (LMA and HMA) and their interaction.
Respiration rates were only calculated for the diatom-DOM source (Table 2), due to very high background DI13C levels of the two natural sources. During isotope-labeling of the sources, macroalgae, coral, and diatoms respire a substantial part of the incorporated 13C. This background DI13C is removed during the collection step for laboratory made diatom-DOM, but not for the natural sources. Therefore, the assimilation-to-respiration efficiency was only estimated for the diatom-DOM source for LMA and HMA species. Despite lower assimilation rates, HMA species showed higher mean assimilation-to-respiration efficiencies (76 ± 10%) than the LMA Species (64 ± 9%), but this difference was not significant (Table 3).
PLFA Composition of Sponges and DOM Sources
Each sponge species exhibited a distinct PLFA profile, composed of bacterial-, sponge-specific and unspecific (referred to as ‘other’) biomarkers (Figure 3 and Supplementary Figure 3). Characteristic bacterial biomarkers included iso-, anteiso, methyl-branched and cyclic PLFAs, while sponge biomarkers comprised several long-chain PLFAs (>C24:0) with characteristic demosponge Δ5,9 poly unsaturation (Koopmans et al., 2015). Except for the homoscleromorph species P. angulospiculatus, all demosponges species exhibited one sponge-specific PLFA of a relatively high concentration, namely C30:3(5,9,23) in H. caerulea and C. caribensis, C32:2(5,9) in H. vansoesti, C28:2(5,9) in S. ruetzleri, and C27:2(5,9) in E. ferox (Figure 3 and Supplementary Figure 3). Common unspecific PLFAs present in all species include C14:0, C16:0, C18:1ω9, C22:0, and C24:0 (Figure 3). The overall average PLFA composition in LMA sponges was characterized by a higher contribution of sponge-specific (32.7 ± 9.1%) over bacteria-specific (14.7 ± 8.5%) PLFAs. For HMA species, a higher proportion of bacteria-specific PLFAs (37.0 ± 25.7%) was found compared to sponge-specific PLFAs (28.2 ± 25.1%) (Figure 3).
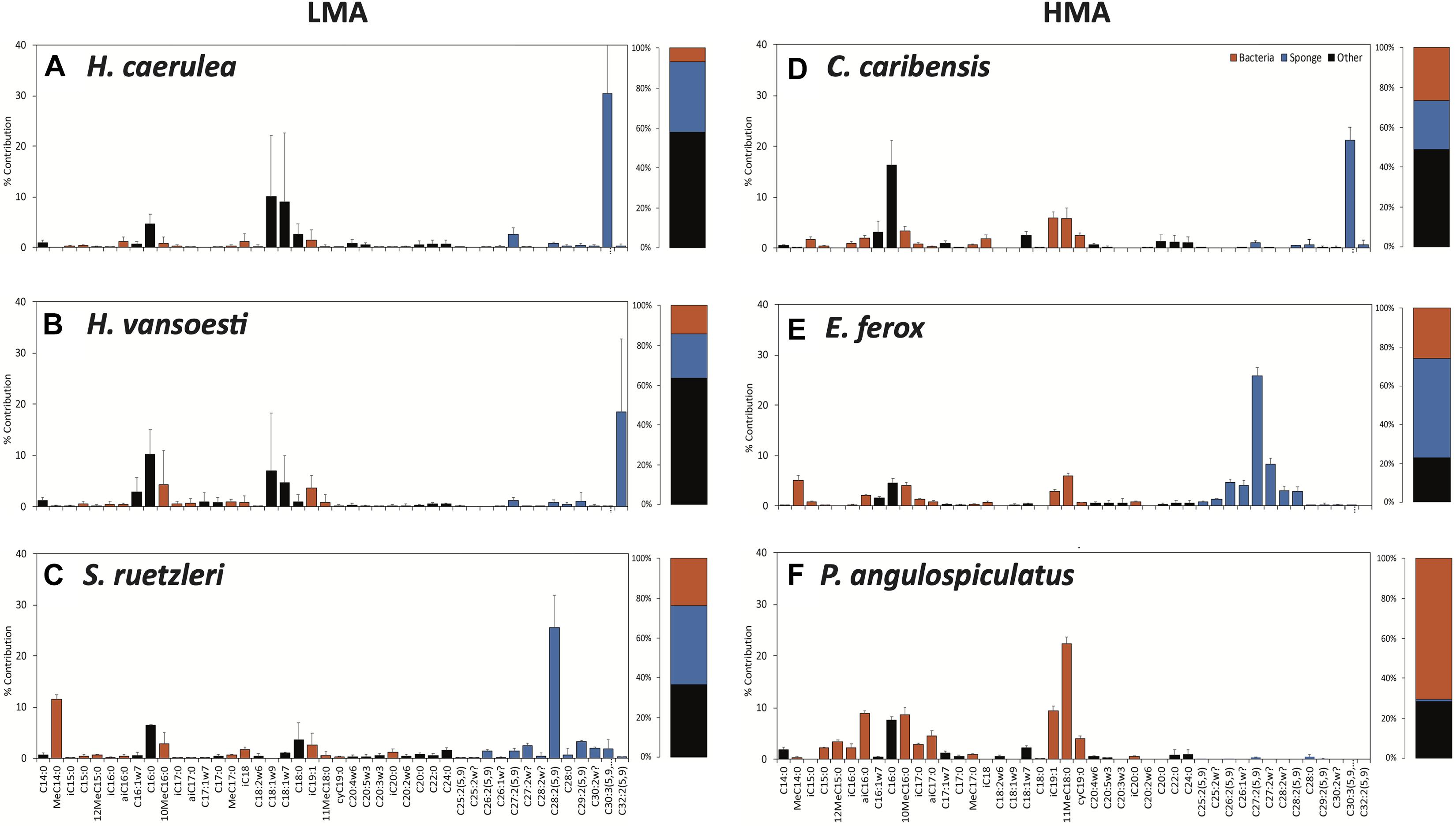
Figure 3. Phospholipid fatty acid (PLFA) profiles of the sponge species used in this experiment. (A) Halisarca caerulea, (B) Haliclona vansoesti, (C) Scopalina ruetzleri, (D) Chondrilla caribensis, (E) Ectyoplasia ferox, and (F) Plakortis angulospiculatus. LMA species on the left-hand side and HMA species on the right-hand side. Values presented as the percentage contribution to the total PLFA composition. Stacked barplots depict the total percentages of bacteria- (red), sponge-specific (blue) and other (black) PLFAs within the PLFA profiles of each species. Data presented as mean (%) ± SD (n = 3).
The three DOM sources were characterized by distinct PLFA profiles (Supplementary Figure 4). The PLFA compositions of macroalgal- and diatom-DOM were more similar than the composition of coral-DOM. C20:5ω3, a known algal biomarker, was present in higher concentrations in macroalgal- and diatom- DOM compared to coral-DOM. In macroalgal-DOM the most abundant fatty acid was C18:1ω9 (32%), while in diatom-DOM it was C16:0 (36%). Coral DOM was characterized by a greater contribution of longer fatty acids, such as C20:0 (18%), C22:0 (23%), and C24:0 (23%).
Incorporation of Different DOM Sources Into Host and Bacterial Symbiont PLFAs
All sponge species incorporated all DOM sources into host and bacterial symbiont PLFAs (Table 2 and Supplementary Table 3). There was no significant difference in total PLFA incorporation rates between LMA and HMA sponges (Table 3). Average incorporation of laboratory-made diatom-DOC into total PLFAs (66.21 ± 45.26 nmol Ctracer mmol Csponge–1 12 h–1; mean ± standard deviation) was higher than macroalgal- and coral-DOC incorporation (23.56 ± 15.46 and 31.73 ± 7.77, respectively), but it was significantly different only between diatom- and macroalgal-DOC (Table 3 and Supplementary Table 2). LMA and HMA sponges directly incorporated PLFAs from all DOM sources, as reflected by the presence of PLFAs in both source and sponge incorporation profiles (arrows in Figure 4). The relative incorporation into bacteria- and sponge- specific PLFAs did not differ between the three DOM sources per sponge type but was significantly different between LMA and HMA sponges (Supplementary Table 2). HMA species incorporated twice as much DOC into bacterial biomarkers (29.4 ± 8.0%) compared to LMA species (16.8 ± 8.0%), but three times less DOC into sponge biomarkers (1.4 ± 1.3%) compared to LMA species (3.8 ± 2.4%). DOC was used 4–21 times more to synthesize de novo bacteria-specific PLFAs than de novo sponge-specific PLFAs in LMA and HMA sponges, respectively. All species displayed high uptake of all the sources into common unspecific biomarkers (including C14:0, C16:1ω7, C16:0, C18:1ω9, C20:5ω3), which were also present in the sources and are used as precursors for further PLFAs synthesis (Supplementary Table 3).
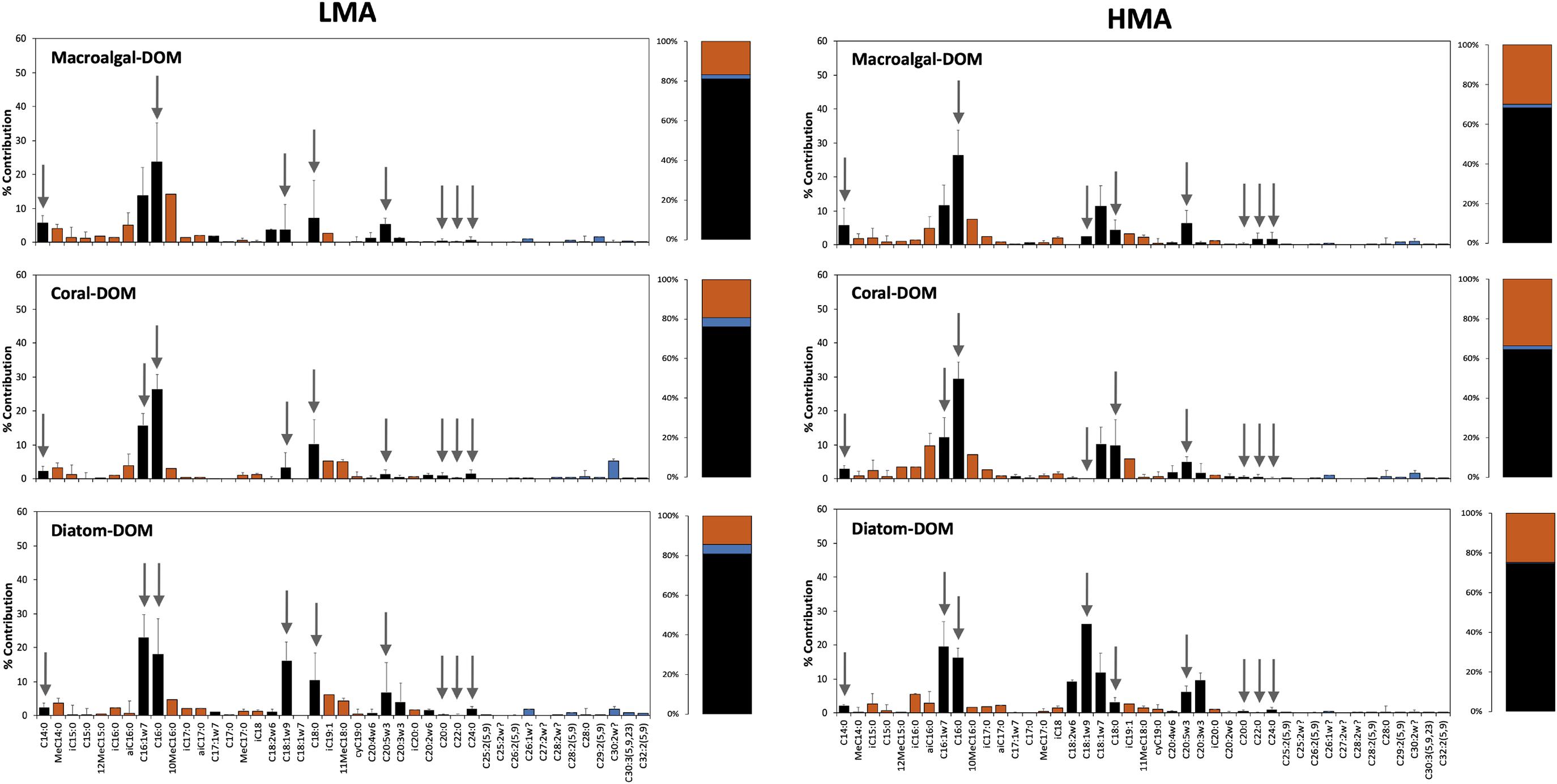
Figure 4. Assimilation of tracer DOM sources into phospholipid fatty acids (PLFAs). Incorporation of tracer DOM into bacteria- (red), sponge-specific (blue), and other (black) PLFAs. LMA species on the left-hand side and HMA species on the right-hand side; upper row sponges were fed with macroalgal-DOM, central row with coral-DOM and lower row with diatom-DOM. Gray arrows indicate PLFAs that were also present in the source (direct uptake), all other PLFAs are de novo synthesized. Values depicted as the percentage contribution to the total DOC incorporation for each individual PLFA. Stacked barplots depict the total percentages of bacterial, sponge and other PLFAs. Data presented as mean (%) ± SD (n = 3).
Inorganic Nutrient Fluxes With Natural DOM Sources
The release rate of DIN was significantly higher for LMA species (3.06 ± 0.88 μmol N g DWsponge h–1; mean ± 95% confidence intervals) compared to HMA species (0.96 ± 0.44) (PERMANOVA, Pseudo-F = 18.53 p = 0.0002), but for both LMA and HMA species, no significant difference was found between sponges fed with macroalgal- versus coral-DOM (Figure 5A and see Supplementary Figure 5 for all individual species). Taking a closer look at the different N-compounds within the DIN pool, in LMA species, macroalgal-DOM feeding induced a significantly higher release of nitrite + nitrate (NOx) (PERMANOVA pair-wise test, t = 2.69 p = 0.013) in combination with significantly lower release of ammonia (NH4+) (PERMANOVA pair-wise test, t = 2.31 p = 0.035) compared to coral-DOM (Figure 5B). A similar trend of higher release of NOx in combination with lower release (to even net uptake) of NH4+ was observed in HMA species fed with macroalgal- compared to coral-DOM (Figure 5C), however, only the NH4+ release rates were significantly different between the food sources (PERMANOVA pair-wise test, NOx: t = 1.99, p = 0.064 and NH4+: t = 3.07, p = 0.009). The mean release rates of phosphate (PO43–) were higher in LMA compared to HMA sponges (Figure 5D), however, pair-wise comparisons showed that this was only significant in coral-DOM fed sponges (PERMANOVA pair-wise test, t = 2.72, p = 0.016). Within the LMA sponges, PO43– release rates were significantly higher in sponges fed with coral-DOM (0.17 ± 0.07 μmol P g DWsponge h–1) compared to macroalgal-DOM (0.07 ± 0.05) (PERMANOVA pair-wise test, t = 2.35, p = 0.030).
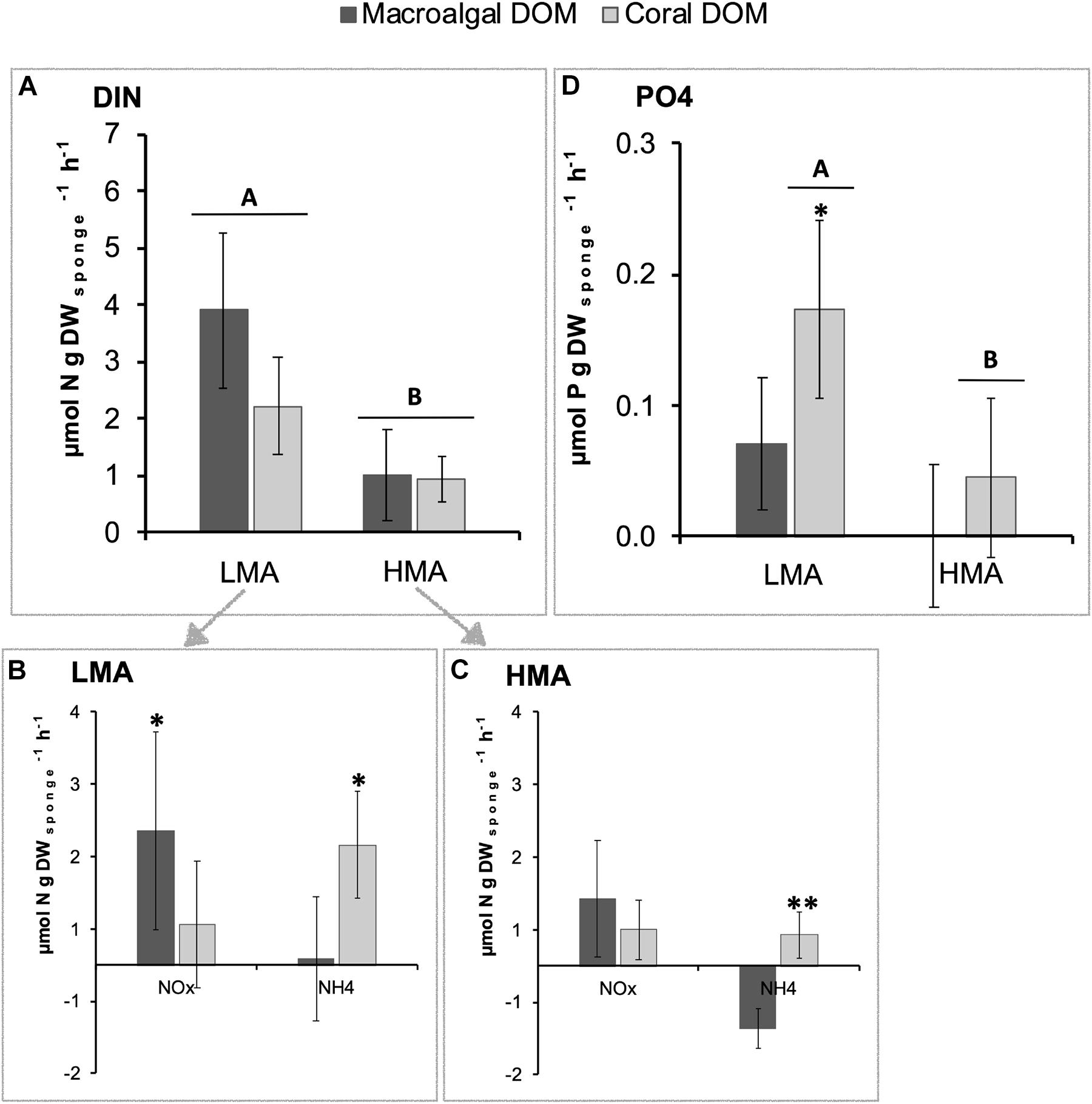
Figure 5. Average net inorganic nutrient fluxes of sponges fed with naturally sourced macroalgal- and coral-DOM. Positive values indicate net release and negative values indicate net uptake. In dark gray macroalgal-DOM and in light gray coral-DOM. Shown are (A) dissolved inorganic nitrogen (DIN) fluxes of LMA sponges and HMA sponges, (B) NOx and NH4 fluxes of LMA sponges, (C) NOx and NH4 fluxes of HMA sponges, (D) PO4 fluxes of LMA sponges and HMA sponges. Data presented as mean rate in μmol N or P g DWsponge h–1) ± 95%-CI (n = 10). Letters (A and B) indicate significant differences between LMA and HMA sponges and asterisks (∗) indicate significant differences between the sources per sponge type (LMA/HMA). Signif. codes: ∗∗p < 0.01, ∗p < 0.05.
Discussion
Bulk Processing of Macroalgal- and Coral-DOM by LMA and HMA Sponges
This study shows the differential processing of two natural DOM sources derived from macroalgae and corals by six Caribbean encrusting sponge species with different microbial abundances. All six species incorporated coral- and macroalgal-DOM into their tissue, with highest assimilation rates found for the species with low abundances of microbial symbionts (LMA). Contrary to the prevailing notion that HMA sponges are more prolific DOM feeders than LMA sponges, these findings add to an increasing body of evidence that encrusting sponges do not rely on microbial symbionts for DOM-processing (de Goeij et al., 2008a; Rix et al., 2017; Bart et al., 2020, 2021). The low volume-to-surface area morphology (e.g., sheet-, conulose-shapes) of encrusting sponges is suggested to aid their ability to process DOM (de Goeij et al., 2017). Massive sponge species (e.g., ball-, barrel-shapes) show a stronger positive correlation between high microbial abundances and their ability to utilize DOM (Morganti et al., 2017; Hoer et al., 2018; McMurray et al., 2018), although net DOM-feeding by massive LMA species is also found (Gantt et al., 2019; Bart et al., 2020, 2021). LMA and HMA massive sponges have recently been show to exhibit very different life strategies in relation to DOM-feeding: HMA sponges primarily rely on their microbial symbionts to process DOM (60–87%), whereas LMA sponges process DOM predominantly through their host sponge cells (>95%), with their filter cells (‘choanocytes’) as main DOM-processors (Rix et al., 2020).
The largest difference in macroalgal- versus coral-DOM processing among sponge holobionts was the significantly higher assimilation of nitrogen (N) from the macroalgal DOM-source. This corroborates the findings of Rix and colleagues (2017) who found preferential uptake of macroalgal-DOM by three encrusting Red Sea sponge species. In contrast to their study, we did not find preferential uptake of carbon from macroalgal-DOM compared with coral-DOM. Differences in preferential uptake of carbon and nitrogen between studies could be caused by overall differences in water quality (e.g., carbon and nitrogen availability) between the distinct geographical Caribbean and Red Sea waters. For example, the Gulf of Aqaba shows lower dissolved organic carbon concentrations than Curaçaon waters (65–96 versus 94–160 μmol L–1; de Goeij et al., 2008b; Naumann et al., 2012), which could indicate that carbon is less limiting for Curaçaon sponges. Also, the Red Sea has a much stronger seasonal fluctuation in (in)organic nutrient supply than the Caribbean, which also directly affects organic matter release and carbon and nitrogen composition by corals and algae (Wild et al., 2010). In addition, sponge functional traits, e.g., the composition of their microbiome, could have caused differences in preferential uptake of carbon and nitrogen from DOM. However, a recent study suggested that differences in fluxes of organic carbon and nitrogen were not related to the composition of the microbiomes of two distinct Caribbean populations of reef sponges (Gantt et al., 2019). In addition, the microbial community of the Red Sea sponge species used in the study by Rix et al. (2017) is, to our knowledge, not yet known. Our findings suggest that of the Red Sea and Caribbean sponges tested, macroalgal-DOM, at least in terms of N, is more bioavailable or ‘labile’ to sponges than coral-DOM. In addition to the concentration of DOM, the bioavailability of DOM is driven by its chemical composition. For example, it is known that coral- and macroalgal-DOM vary in their neutral sugar composition, which affects the lability of the exudates, and the latter has been shown to stimulate faster bacterioplankton growth compared to the coral-DOM (Haas et al., 2011, 2013, 2016; Nelson et al., 2013). However, the exact composition of the DOM pool (from any source) is still largely a black box. New untargeted metabolomic approaches (Hartmann et al., 2017; Petras et al., 2017) may identify the role that different components released by corals and macroalgae from different geographical regions have on sponge metabolism, and how sponges themselves change the chemical composition of the DOM pool by selectively processing (i.e., uptake and release) different components from the available sources (Fiore et al., 2017; Letourneau et al., 2020).
The assimilation-to-respiration efficiencies of DOM by encrusting sponges observed in our study (57–88%) are comparable to previously published values (55–80%) for four out of the six species used here (LMA H. caerulea, H. vansoesti, S. ruetzleri and HMA C. caribensis) (de Goeij et al., 2013). Quantifying respiration rates of naturally sourced 13C-enriched DOM has proved unfeasible using current methodology, since we were unable to remove the high background DI13C values caused by the respiration of the initial 13C label by corals and macroalgae, see also Rix et al. (2017). At present, only laboratory-made DOM from lysed algal sources, whereby respired H13CO3 is removed by collecting the algal cells on a filter, can be used to estimate sponge respiration rates and assimilation efficiencies after DOM processing. We are confident that the assimilation efficiencies of diatom-DOM presented here provide at least a qualitative approach to estimate the utilization of DOM by different sponge types (e.g., LMA, HMA), since overall diatom-DOM assimilation rates are similar to the two natural DOM sources (factor 1.2–1.3 difference in carbon utilization), despite higher initial labeling of the diatom-DOM source.
Macroalgal- and Coral-DOM Incorporation Into Sponge Host- and Bacterial Symbiont-PLFAs
The enrichment of bacterial- and sponge-specific PLFAs absent from the provided DOM sources suggests de novo synthesis and confirms DOM processing by the sponge host and its microbiome (Figure 4). There was no difference in the total PLFA incorporation rates between LMA and HMA species, but processing of DOM into bacteria-specific PLFAs compared to sponge-specific PLFAs was relatively higher in HMA compared to LMA species, as previously reported by Rix et al. (2017). Similar to bulk tissue carbon assimilation rates (Figure 2A), there was no difference in macroalgal- versus coral-DOM processing into PLFAs. This finding contrasts with the observation made by Rix et al. (2017), who found preferential incorporation of macroalgal-DOM into bacterial PLFAs and coral-DOM into sponge PLFAs, as well as overall higher incorporation rates of macroalgal-DOM. As aforementioned, this could be due to differences in ambient nutrient availability between the distinct Red Sea and Caribbean ocean basins. But, it cannot be excluded that differences in the production of the coral-DOM source (milking versus natural DOM release) resulted in considerable differences in the composition of the coral-DOM (Crossland, 1987) and thereby its processing by the sponge holobionts. In addition, the discrepancy could be caused by the shorter incubation periods (by a factor of 2–3) in our study than in Rix et al. (2017). This likely underrepresented the contribution of (very) long-chained sponge-specific PLFAs, since more time is required for their biosynthesis (Hahn et al., 1988). Recent nanoscale secondary ion mass spectrometry (NanoSIMS) assessment of the here used HMA P. angulospiculatus and LMA H. caerulea confirmed that both sponge host cells and microbial symbionts rapidly take up DOM (within 15 min after administration of the labeled food source), (Hudspith et al., 2021). However, longer time may be needed for the DOM-derived C and N to enter anabolic pathways and be incorporated into the sctructures of the sponge cells, including PLFAs. Therefore, despite the very low assimilation rates of DOM into sponge-specific PLFAs, it cannot be concluded that DOM is mainly processed by bacterial symbionts, since most (70–80%) DOM assimilation into PLFAs cannot be specifically linked to host or bacterial symbionts. The relative contribution of bacterial biomarkers as estimated through PLFA incorporation can be high, but the actual quantity of C-processing by bacterial symbionts can result in much lower contributions (Rix et al., 2020). The contribution of host cell PLFA incorporation of DOM can be further underestimated by a loss through a high cell turnover of the choanocyte cell compartment (de Goeij et al., 2009; Alexander et al., 2015a,b). In conclusion, PLFA biomarkers are certainly an important tool to identify processing of different food sources by host and microbial symbionts, but need additional methods (e.g., NanoSIMS, host–microbe cell separation, metatranscriptomics) to estimate actual host–symbiont contributions.
Inorganic Nutrient Fluxes With Natural DOM Sources
All sponge species acted as a net sources of inorganic nutrients. The fluxes of sponges fed with macroalgal- and coral-DOM showed a trend in both LMA and HMA species: sponges fed with macroalgal-DOM showed a higher net released of NOx (nitrite and nitrate) and a concomitant lower release or net uptake of ammonia compared to coral-DOM fed sponges. These fluxes can be attributed to nitrification processes carried out by the bacterial symbionts (Fiore et al., 2010; Schläppy et al., 2010; Maldonado et al., 2012; Zhang et al., 2019). Contrary, the net lower NOx release and higher release of ammonia by coral-DOM fed sponges are typical of heterotrophic metazoan metabolism and suggest a greater contribution of the sponge host. In combination, these results may indicate higher microbial processing of macroalgal-DOM versus higher sponge-host processing of coral-DOM, as previously hypothesized by Rix et al. (2017). The observed difference in DIN fluxes related to coral- and algal-DOM may also partly explain why sponges have been described as both sinks and sources of inorganic nitrogen (Fiore et al., 2010), depending on the relative concentration and processing of different ambient DOM sources. All sponge species acted as net sources of phosphate. Particularly, LMA sponges further showed a significant increase in PO43– release after being fed with coral-DOM compared to macroalgal-DOM. This suggests that sponges exposed to ambient coral-DOM stimulate inorganic phosphorous release compared with macroalgal-DOM, which could support reef primary production. PO43– fluxes may seem low compared to inorganic N fluxes (Figure 5), but P is considered the ultimate limiting macronutrient on coral reefs (Larned, 1998; Ferrier-Pagès et al., 2016), and marine oligotrophic environments in general (Tyrrell, 1999), and small changes in availability may provide competitive advantages to users of inorganic P (e.g., corals, algae, bacteria). Given the high abundance of encrusting sponges on Caribbean reefs (Kornder et al., in press), this sponge-mediated flux from DOM into inorganic P could be crucial to support reef primary production under pristine conditions (Colman, 2015).
Species Level Response to Macroalgal- and Coral-DOM
We assessed the relation between processing of DOM from different sources and the abundance of microbial symbionts (according to the LMA versus HMA dichotomy) as a functional trait. Yet, sponges are likely equipped with many more functional traits, including morphology, microbiome composition, and host phylogeny, which have been shown to influence DOM and POM processing by deep-sea sponges (Bart et al., 2020) and inorganic nutrient release by shallow-water sponges (Gantt et al., 2019). We show that species-level effects in DOM processing indeed transcend the HMA/LMA dichotomy (results presented in Supplementary File 1), but note that many differences found only appeared in macroalgal-DOM fed sponges. For example, S. ruetzleri released the highest amount of NO2 compared to all other LMA and HMA species, H. caerulea had the highest NH4 release rates, and overall DIN fluxes were lowest in P. angulospiculatus, indicating distinct microbially mediated metabolic pathways (Supplementary Figure 5). We acknowledge that microbial abundance is merely one functional trait affecting the processing of metabolic resources. Full integration of the host-microbe metabolic network, as was recently modeled for the common deep-sea species Geodia barretti (de Kluijver et al., 2021), may improve our understanding of how sponge holobionts drive nutrient fluxes within their ecosystem.
Ecological Implications of Macroalgal- Versus Coral-DOM Processing by LMA and HMA Sponges
This work represents only the second record of differential processing of naturally sourced macroalgal- and coral-DOM by sponges. Despite the fact that we mimicked the release of macroalgal- and coral-DOM in a more natural manner than in previous studies, both sources were produced from ‘representative’ macroalgal and coral species, making it difficult to draw definite conclusions on the ecological impact of the processing of both sources by sponges on coral reefs. Nevertheless, based on our results, we can hypothesize on possible ecological implications. It seems plausible that DOM source type (e.g., macroalgal, coral) affects (1) the metabolism of encrusting sponge types (i.e., LMA, HMA), but also (2) the release of inorganic nitrogen (N) and phosphorus (P), and thus the availability of these resources to other reef inhabitants. For example, under the current shift from coral- to algal-dominance, sponges could catalyze eutrophication of reefs through increased turnover of nitrogen. Furthermore, our findings suggest that LMA species have a greater influence on nutrient fluxes compared to HMA species. Therefore, it is important to quantify the biomass of LMA and HMA encrusting sponges on current reefs. Furthermore, understanding how these sponge types are affected by environmental change will help us better predict their relative abundance on future reefs and the concomitant effects on reef biogeochemical cycles.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
SC, MH, BM, and JG conceived the ideas and designed the methodology. SC, MH, DL, and CD performed the experiments and collected the data. SC, DL, AK, JS, SA, MM, and JG analyzed the data. SC, BM, and JG led the writing of the manuscript. All authors reviewed the manuscript.
Funding
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Program (grant agreement #715513; personal grant to JG).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Niklas Kornder, Mischa Streekstra, Mark Vermeij, and the staff of CARMABI for fieldwork and logistical support. A special thanks to Pol Bosch and the Coral Restoration Curaçao for their constant collaboration and availability. Thanks to Emiel van Loon for help with statistical analysis; Ronald van Bommel for 13C DOC analysis at NIOZ; Arnold van Dijk for DI13C analysis at the University of Utrecht and Eva de Rijke, Rutger van Hall, Pieter Slot, and the analytical laboratory of IBED for help with sample analysis at the University of Amsterdam. Many thanks also to Sean Hoetjes and Martijn Bart for helping with fatty acids extractions and analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.640583/full#supplementary-material
References
Alexander, B. E., Achlatis, M., Osinga, R., van der Geest, H. G., Cleutjens, J. P. M., Schutte, B., et al. (2015a). Cell kinetics during regeneration in the sponge Halisarca caerulea: how local is the response to tissue damage? PeerJ 3:e820. doi: 10.7717/peerj.820
Alexander, B. E., Liebrand, K., Osinga, R., van der Geest, H. G., Admiraal, W., Cleutjens, J. P. M., et al. (2014). Cell turnover and detritus production in marine sponges from tropical and temperate benthic ecosystems. PLoS One 9:e109486. doi: 10.1371/journal.pone.0109486
Alexander, B. E., Mueller, B., Vermeij, M. J. A., van der Geest, H. G., and de Goeij, J. M. (2015b). Biofouling of inlet pipes affects water quality in running seawater aquaria and compromises sponge cell proliferation. PeerJ 3:e1430. doi: 10.7717/peerj.1430
Anderson, M. J., Gorley, R. N., and Clarke, K. R. (2008). PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods. Plymouth: Primer-E Ltd.
Atkinson, M. J., and Falter, J. (2003). “Biogeochemistry of coral reefs,” in Biogeochemistry of Marine Ecosystems, eds K. Black and G. Shimmield (Boca Raton, FA: CRC Press), 40–64. doi: 10.1201/9780367812423-2
Azam, F., Fenchel, T., Field, J. G., Gray, J. S., Meyer-Reil, L. A., and Thingstad, F. (1983). The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263. doi: 10.3354/meps010257
Barott, K. L., and Rohwer, F. L. (2012). Unseen players shape benthic competition on coral reefs. Trends Microbiol. 20, 621–628. doi: 10.1016/j.tim.2012.08.004
Bart, M. C., de Kluijver, A., Hoetjes, S., Absalah, S., Mueller, B., Kenchington, E., et al. (2020). Differential processing of dissolved and particulate organic matter by deep-sea sponges and their microbial symbionts. Sci. Rep. 10:17515.
Bart, M. C., Mueller, B., Rombouts, T., van de Ven, C., Tompkins, G. J., Osinga, R., et al. (2021). Dissolved organic carbon (DOC) is essential to balance the metabolic demands of four dominant North-Atlantic deep-sea sponges. Limnol. Oceanogr. 66, 925–938. doi: 10.1002/lno.11652
Cárdenas, A., Neave, M. J., Haroon, M. F., Pogoreutz, C., Rädecker, N., Wild, C., et al. (2018). Excess labile carbon promotes the expression of virulence factors in coral reef bacterioplankton. ISME J. 12, 59–76. doi: 10.1038/ismej.2017.142
Colman, A. S. (2015). Sponge symbionts and the marine P cycle. Proc. Natl. Acad. Sci. U.S.A. 112, 4191–4192. doi: 10.1073/pnas.1502763112
Crossland, C. J. (1987). In situ release of mucus and DOC-lipid from the corals Acropora variabilis and Stylophora pistillata in different light regimes. Coral Reefs 61, 35–42. doi: 10.1007/bf00302210
de Goeij, J. M., and van Duyl, F. C. (2007). Coral cavities are sinks of dissolved organic carbon (DOC). Limnol. Oceanogr. 52, 2608–2617. doi: 10.4319/lo.2007.52.6.2608
de Goeij, J. M., De Kluijver, A., Van Duyl, F. C., Vacelet, J., Wijffels, R. H., De Goeij, A. F. P. M., et al. (2009). Cell kinetics of the marine sponge Halisarca caerulea reveal rapid cell turnover and shedding. J. Exp. Biol. 212(Pt 23), 3892–3900. doi: 10.1242/jeb.034561
de Goeij, J. M., Lesser, M. P., and Pawlik, J. R. (2017). “Nutrient fluxes and ecological functions of coral reef sponges in a changing ocean,” in Climate Change, Ocean Acidification and Sponges: Impacts Across Multiple Levels of Organization, eds J. L. Carballo and J. J. Bell (Cham: Springer International Publishing), 373–410. doi: 10.1007/978-3-319-59008-0_8
de Goeij, J. M., Moodley, L., Houtekamer, M., Carballeira, N. M., and van Duyl, F. C. (2008a). Tracing 13C-enriched dissolved and particulate organic carbon in the bacteria-containing coral reef sponge Halisarca caerulea: evidence for DOM feeding. Limnol. Oceanogr. 53, 1376–1386.
de Goeij, J. M., Van den Berg, H., Van Oostveen, M., Epping, E., and Van Duyl, F. (2008b). Major bulk dissolved organic carbon (DOC) removal by encrusting coral reef cavity sponges. Mar. Ecol. Prog. Ser. 357, 139–151. doi: 10.3354/meps07403
de Goeij, J. M., van Oevelen, D., Vermeij, M. J. A., Osinga, R., Middelburg, J. J., de Goeij Anton, F. P. M., et al. (2013). Surviving in a marine desert: the sponge loop retains resources within coral reefs. Science 342, 108–110. doi: 10.1126/science.1241981
de Kluijver, A., Bart, M., Van Oevelen, D., De Goeij, J., Leys, S., Maier, S., et al. (2021). An integrative model of carbon and nitrogen metabolism in a common deep-sea sponge. Front. Mar. Sci. 7:1131. doi: 10.3389/fmars.2020.596251
Dinsdale, E. A., and Rohwer, F. (2011). “Fish or germs? Microbial dynamics associated with changing trophic structures on coral reefs,” in Coral Reefs: An Ecosystem in Transition, eds Z. Dubinsky and N. Stambler (Dordrecht: Springer Netherlands), 231–240. doi: 10.1007/978-94-007-0114-4_16
Ferrier-Pagès, C., Godinot, C., D’ Angelo, C., Wiedenmann, J. Ö, and Grover, R. (2016). Phosphorus metabolism of reef organisms with algal symbionts. Ecol. Monogr. 863, 262–277. doi: 10.1002/ecm.1217
Fiore, C. L., Freeman, C. J., and Kujawinski, E. B. (2017). Sponge exhalent seawater contains a unique chemical profile of dissolved organic matter. PeerJ 5:e2870. doi: 10.7717/peerj.2870
Fiore, C. L., Jarett, J. K., Olson, N. D., and Lesser, M. P. (2010). Nitrogen fixation and nitrogen transformations in marine symbioses. Trends Microbiol. 18, 455–463. doi: 10.1016/j.tim.2010.07.001
Gantt, S. E., McMurray, S. E., Stubler, A. D., Finelli, C. M., Pawlik, J. R., and Erwin, P. M. (2019). Testing the relationship between microbiome composition and flux of carbon and nutrients in Caribbean coral reef sponges. Microbiome 71:124.
Haas, A. F., Fairoz, M. F. M., Kelly, L. W., Nelson, C. E., Dinsdale, E. A., Edwards, R. A., et al. (2016). Global microbialization of coral reefs. Nat. Microbiol. 1:16042.
Haas, A. F., Nelson, C. E., Rohwer, F., Wegley-Kelly, L., Quistad, S. D., Carlson, C. A., et al. (2013). Influence of coral and algal exudates on microbially mediated reef metabolism. PeerJ 1:e108. doi: 10.7717/peerj.108
Haas, A. F., Nelson, C. E., Wegley Kelly, L., Carlson, C. A., Rohwer, F., Leichter, J. J., et al. (2011). Effects of coral reef benthic primary producers on dissolved organic carbon and microbial activity. PLoS One 611:e27973. doi: 10.1371/journal.pone.0027973
Hahn, S., Stoilov, I. L., Ha, T. B. T., Raederstorff, D., Doss, G. A., Li, H. T., et al. (1988). Biosynthetic studies of marine lipids. The course of chain elongation and desaturation in long-chain fatty acids of marine sponges. J. Am. Chem. Soc. 110, 8117–8124. doi: 10.1021/ja00232a025
Hartmann, A. C., Petras, D., Quinn, R. A., Protsyuk, I., Archer, F. I., Ransome, E., et al. (2017). Meta-mass shift chemical profiling of metabolomes from coral reefs. Proc. Natl. Acad. Sci. U.S.A. 114, 11685–11690. doi: 10.1073/pnas.1710248114
Hatcher, B. G. (1990). Coral reef primary productivity. A hierarchy of pattern and process. Trends Ecol. Evol. 5, 149–155. doi: 10.1016/0169-5347(90)90221-X
Hentschel, U., Piel, J., Degnan, S. M., and Taylor, M. W. (2012). Genomic insights into the marine sponge microbiome. Nat. Rev. 109, 641–654. doi: 10.1038/nrmicro2839
Hoegh-Guldberg, O., Mumby, P. J., Hooten, A. J., Steneck, R. S., Greenfield, P., Gomez, E., et al. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742.
Hoer, D. R., Gibson, P. J., Tommerdahl, J. P., Lindquist, N. L., and Martens, C. S. (2018). Consumption of dissolved organic carbon by Caribbean reef sponges. Limnol. Oceanogr. 63, 337–351. doi: 10.1002/lno.10634
Hudspith, M., Rix, L., Achlatis, M., Bougoure, J., Guagliardo, P., Clode, P. L., et al. (2021). Subcellular view of host–microbiome nutrient exchange in sponges: insights into the ecological success of an early metazoan–microbe symbiosis. Microbiome 9:44.
Hughes, T. P. (1994). Catastrophes, phase shifts, and large-scale degradation of a Caribbean coral reef. Science 265, 1547–1551. doi: 10.1126/science.265.5178.1547
Hughes, T. P., Baird, A. H., Bellwood, D. R., Card, M., Connolly, S. R., Folke, C., et al. (2003). Climate change, human impacts, and the resilience of coral reefs. Science 301, 929–933. doi: 10.1126/science.1085046
Kline, D. I., Kuntz, N. M., Breitbart, M., Knowlton, N., and Rohwer, F. (2006). Role of elevated organic carbon levels and microbial activity in coral mortality. Mar. Ecol. Prog. Ser. 314, 119–125. doi: 10.3354/meps314119
Knowlton, N., Brainard, R. E., Fisher, R., Moews, M., Plaisance, L., and Caley, M. J. (2010). “Coral reef biodiversity,” in Life in the World’s Oceans, ed. A. D. McIntyre (New York, NY: Wiley), 65–78.
Koopmans, M., van Rijswijk, P., Boschker, H. T. S., Marco, H., Martens, D., and Wijffels, R. H. (2015). Seasonal variation of fatty acids and stable carbon isotopes in sponges as indicators for nutrition: biomarkers in sponges identified. Mar. Biotechnol. 171, 43–54. doi: 10.1007/s10126-014-9594-8
Kornder, N.A., Cappelletto, J., Mueller, B., Zalm, M.J.L., Martinez, S.J., Vermeij, M.J.A., Huisman, J., and de Goeij, J.M. (in press). The inclusion of habitat complexity and cryptic habitats greatly alters relative abundance estimates of benthic reef organisms. Coral Reefs
Larned, S. T. (1998). Nitrogen- versus phosphorus-limited growth and sources of nutrients for coral reef macroalgae. Mar. Biol. 13, 409–421. doi: 10.1007/s002270050407
Letourneau, M. L., Hopkinson, B. M., Fitt, W. K., and Medeiros, P. M. (2020). Molecular composition and biodegradation of loggerhead sponge Spheciospongia vesparium exhalent dissolved organic matter. Mar. Environ. Res. 162:105130. doi: 10.1016/j.marenvres.2020.105130
Maldonado, M., Ribes, M., and van Duyl, F. C. (2012). “Nutrient fluxes through sponges: biology, budgets, and ecological implications,” in Advances in Sponge Science: Physiology, Chemical and Microbial Diversity, Biotechnology, eds M. A. Becerro, M. J. Uriz, M. Maldonado, and X. Turon (Cambridge, MA: Academic Press), 113–182. doi: 10.1016/b978-0-12-394283-8.00003-5
McManus, J. W., and Polsenberg, J. F. (2004). Coral–algal phase shifts on coral reefs: ecological and environmental aspects. Prog. Oceanogr. 60, 263–279. doi: 10.1016/j.pocean.2004.02.014
McMurray, S. E., Stubler, A. D., Erwin, P. M., Finelli, C. M., and Pawlik, J. R. (2018). A test of the sponge-loop hypothesis for emergent Caribbean reef sponges. Mar. Ecol. Prog. Ser. 588, 1–14. doi: 10.3354/meps12466
Morganti, T., Coma, R., Yahel, G., and Ribes, M. (2017). Trophic niche separation that facilitates co-existence of high and low microbial abundance sponges is revealed by in situ study of carbon and nitrogen fluxes. Limnol. Oceanogr. 625, 1963–1983. doi: 10.1002/lno.10546
Morrow, K. M., Liles, M. R., Paul, V. J., Moss, A., and Chadwick, N. E. (2013). Bacterial shifts associated with coral-macroalgal competition in the Caribbean Sea. Mar. Ecol. Prog. Ser. 488, 103–117. doi: 10.3354/meps10394
Mueller, B., de Goeij, J. M., Vermeij, M. J. A., Mulders, Y., van der Ent, E., Ribes, M., et al. (2014a). Natural diet of coral-excavating sponges consists mainly of dissolved organic carbon (DOC). PLoS One 92:e90152. doi: 10.1371/journal.pone.0090152
Mueller, B., van der Zande, R. M., van Leent, P. J. M., Meesters, E. H., Vermeij, M., and van Duyl, F. C. (2014b). Effect of light availability on dissolved organic carbon release by Caribbean reef algae and corals. Bull. Mar. Sci. 903, 875–893. doi: 10.5343/bms.2013.1062
Mumby, P., and Steneck, R. (2008). Coral reef management and conservation in light of rapidly evolving ecological paradigms. Trends Ecol. Evol. 23, 555–563. doi: 10.1016/j.tree.2008.06.011
Nakajima, R., Nakatomi, N., Kurihara, H., Fox, D. M., Smith, E. J., and Okaji, K. (2016). Crown-of-thorns starfish larvae can feed on organic matter released from corals. Diversity 8:18. doi: 10.3390/d8040018
Naumann, M. S., Haas, A., Struck, U., Mayr, C., El-Zibdah, M., and Wild, C. (2010). Organic matter release by dominant hermatypic corals of the Northern Red Sea. Coral Reefs 293, 649–659. doi: 10.1007/s00338-010-0612-7
Naumann, M., Richter, C., Mott, C., El-Zibdah, M., Manasrah, R., and Wild, C. (2012). Budget of coral-derived organic carbon in a fringing coral reef of the Gulf of Aqaba, Red Sea. J. Mar. Syst. 105-108, 20–29. doi: 10.1016/j.jmarsys.2012.05.007
Nelson, C. E., and Carlson, C. A. (2012). Tracking differential incorporation of dissolved organic carbon types among diverse lineages of Sargasso Sea bacterioplankton. Environ. Microbiol. 146, 1500–1516. doi: 10.1111/j.1462-2920.2012.02738.x
Nelson, C. E., Goldberg, S. J., Wegley Kelly, L., Haas, A. F., Smith, J. E., Rohwer, F., et al. (2013). Coral and macroalgal exudates vary in neutral sugar composition and differentially enrich reef bacterioplankton lineages. ISME J. 7:962. doi: 10.1038/ismej.2012.161
Pawlik, J. R., and McMurray, S. E. (2020). The emerging ecological and biogeochemical importance of sponges on coral reefs. Annu. Rev. Mar. Sci. 121, 315–337. doi: 10.1146/annurev-marine-010419-010807
Pawlik, J. R., Burkepile, D. E., and Thurber, R. V. (2016). A vicious circle? Altered carbon and nutrient cycling may explain the low resilience of Caribbean coral reefs. Bioscience 66, 470–476. doi: 10.1093/biosci/biw047
Petras, D., Jarmusch, A. K., and Dorrestein, P. C. (2017). From single cells to our planet—recent advances in using mass spectrometry for spatially resolved metabolomics. Curr. Opin. Chem. Biol. 36, 24–31. doi: 10.1016/j.cbpa.2016.12.018
Reiswig, H. M. (1981). Partial carbon and energy budgets of the bacteriosponge Verohgia fistularis (Porifera: Demospongiae) in Barbados. Mar. Ecol. 24, 273–293. doi: 10.1111/j.1439-0485.1981.tb00271.x
Ribes, M., Coma, R., and Gili, J. (1999). Natural diet and grazing rate of the temperate sponge Dysidea avara (Demospongiae, Dendroceratida) throughout an annual cycle. Mar. Ecol. Prog. Ser. 176, 179–190. doi: 10.3354/meps176179
Richter, C., Wunsch, M., Rasheed, M., Kötter, I., and Badran, M. I. (2001). Endoscopic exploration of Red Sea coral reefs reveals dense populations of cavity-dwelling sponges. Nature 413, 726–730. doi: 10.1038/35099547
Rix, L., de Goeij, J. M., Mueller, C. E., Struck, U., Middelburg, J. J., van Duyl, F. C., et al. (2016). Coral mucus fuels the sponge loop in warm- and cold-water coral reef ecosystems. Sci. Rep. 6:18715.
Rix, L., de Goeij, J. M., van Oevelen, D., Struck, U., Al-Horani, F. A., Wild, C., et al. (2018). Reef sponges facilitate the transfer of coral-derived organic matter to their associated fauna via the sponge loop. Mar. Ecol. Prog. Ser. 589, 85–96. doi: 10.3354/meps12443
Rix, L., de Goeij, J. M., van Oevelen, D., Struck, U., Al-Horani, F. A., Wild, C., et al. (2017). Differential recycling of coral and algal dissolved organic matter via the sponge loop. Funct. Ecol. 313, 778–789. doi: 10.1111/1365-2435.12758
Rix, L., Ribes, M., Coma, R., Jahn, M. T., de Goeij, J. M., van Oevelen, D., et al. (2020). Heterotrophy in the earliest gut: a single-cell view of heterotrophic carbon and nitrogen assimilation in sponge-microbe symbioses. ISME J. 14, 2554–2567. doi: 10.1038/s41396-020-0706-3
Roach, T. N. F., Little, M., Arts, M. G. I., Huckeba, J., Haas, A. F., George, E. E., et al. (2020). A multiomic analysis of in situ coral–turf algal interactions.ons. Proc. Natl. Acad. Sci. U.S.A. 117, 13588–13595. doi: 10.1073/pnas.1915455117
Rohwer, F., Youle, M., and Vosten, D. (2010). Coral Reefs in the Microbial Sea. San Francisco, CA: Plaid Press United States.
Scheffers, S. R., Bak, R. P. M., and van Duyl, F. C. (2005). Why is bacterioplankton growth in coral reef framework cavities enhanced? Mar. Ecol. Prog. Ser. 299, 89–99. doi: 10.3354/meps299089
Schläppy, M., Schöttner, S., Lavik, G., Kuypers, M. M. M., de Beer, D., and Hoffmann, F. (2010). Evidence of nitrification and denitrification in high and low microbial abundance sponges. Mar. Biol. 157, 593–602. doi: 10.1007/s00227-009-1344-5
Schmitt, S., Tsai, P., Bell, J., Fromont, J., Ilan, M., Lindquist, N., et al. (2011). Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J. 63:564. doi: 10.1038/ismej.2011.116
Silva, L., Calleja, M. L., Ivetic, S., Huete-Stauffer, T., Roth, F., Carvalho, S., et al. (2021). Heterotrophic bacterioplankton responses in coral- and algae-dominated Red Sea reefs show they might benefit from future regime shift. Sci. Total Environ. 751:141628. doi: 10.1016/j.scitotenv.2020.141628
Silveira, C. B., Cavalcanti, G. S., Walter, J. M., Silva-Lima, A., Dinsdale, E. A., Bourne, D. G., et al. (2017). Microbial processes driving coral reef organic carbon flow. FEMS Microbiol. Rev. 414, 575–595. doi: 10.1093/femsre/fux018
Smith, J. E., Shaw, M., Edwards, R. A., Obura, D., Pantos, O., Sala, E., et al. (2006). Indirect effects of algae on coral: algae-mediated, microbe-induced coral mortality. Ecol. Lett. 97, 835–845. doi: 10.1111/j.1461-0248.2006.00937.x
Tanaka, Y., Miyajima, T., Watanabe, A., Nadaoka, K., Yamamoto, T., and Ogawa, H. (2011). Distribution of dissolved organic carbon and nitrogen in a coral reef. Coral Reefs 302, 533–541. doi: 10.1007/s00338-011-0735-5
Tyrrell, T. (1999). The relative influences of nitrogen and phosphorus on oceanic primary production. Nature 400, 525–531. doi: 10.1038/22941
Weisz, J. B., Lindquist, N., and Martens, C. S. (2008). Do associated microbial abundances impact marine demosponge pumping rates and tissue densities? Oecologia 155, 367–376. doi: 10.1007/s00442-007-0910-0
Wild, C., Niggl, W., Naumann, M., and Haas, A. (2010). Organic matter release by Red Sea coral reef organisms—potential effects on microbial activity and in situ O2 availability. Mar. Ecol. Prog. Ser. 411, 61–71. doi: 10.3354/meps08653
Wooster, M. K., McMurray, S. E., Pawlik, J. R., Morán, X. A. G., and Berumen, M. L. (2019). Feeding and respiration by giant barrel sponges across a gradient of food abundance in the Red Sea. Limnol. Oceanogr. 64, 1790–1801. doi: 10.1002/lno.11151
Keywords: dissolved organic matter (DOM), encrusting sponges, phospholipid fatty acids (PLFA), coral, macroalgae, coral reefs, nutrient cycling, coral-algae shifts
Citation: Campana S, Hudspith M, Lankes D, de Kluijver A, Demey C, Schoorl J, Absalah S, van der Meer MTJ, Mueller B and de Goeij JM (2021) Processing of Naturally Sourced Macroalgal- and Coral-Dissolved Organic Matter (DOM) by High and Low Microbial Abundance Encrusting Sponges. Front. Mar. Sci. 8:640583. doi: 10.3389/fmars.2021.640583
Received: 11 December 2020; Accepted: 06 April 2021;
Published: 04 May 2021.
Edited by:
Linda Wegley Kelly, University of California, San Diego, United StatesReviewed by:
Claudia Pogoreutz, University of Konstanz, GermanyCole G. Easson, Middle Tennessee State University, United States
Copyright © 2021 Campana, Hudspith, Lankes, de Kluijver, Demey, Schoorl, Absalah, van der Meer, Mueller and de Goeij. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sara Campana, s.campana@uva.nl
 Sara Campana
Sara Campana Meggie Hudspith
Meggie Hudspith David Lankes2,3
David Lankes2,3  Anna de Kluijver
Anna de Kluijver Celine Demey
Celine Demey Marcel T. J. van der Meer
Marcel T. J. van der Meer Benjamin Mueller
Benjamin Mueller Jasper M. de Goeij
Jasper M. de Goeij