Associated Bacteria and Their Effects on Growth and Toxigenicity of the Dinoflagellate Prorocentrum lima Species Complex From Epibenthic Substrates Along Mexican Coasts
- 1Alfred-Wegener-Institut, Helmholtz-Zentrum für Polar-und Meeresforschung, Bremerhaven, Germany
- 2International Max Planck Research School, Max Planck Institute for Marine Microbiology, Bremen, Germany
- 3Laboratorio de Microbiología Molecular y Genómica Bacteriana, Universidad Científica del Sur, Lima, Peru
- 4Facultad de Ciencias, Universidad Nacional Autónoma de México, Mexico City, Mexico
- 5Instituto de Ciencias del Mar y Limnología, Universidad Nacional Autónoma de México, Mexico City, Mexico
- 6Laboratorio Nacional de Biodiversidad, Instituto de Biología, Universidad Nacional Autónoma de México, Mexico City, Mexico
- 7Instituto de Ciencias Marinas y Pesquerías, Universidad Veracruzana, Veracruz, Mexico
- 8CONACyT-Departamento de Biotecnología Marina, Centro de Investigación Científica y de Educación Superior de Ensenada, Benito Juarez, Mexico
The dinoflagellate genus Prorocentrum is globally represented by a wide variety of species found upon benthic and/or epiphytic substrates. Many epibenthic Prorocentrum species produce lipophilic polyether toxins, some of which act as potent protein phosphatase inhibitors and tumor-promoters associated with Diarrheic Shellfish Poisoning (DSP). Most members of the Prorocentrum lima species complex (PLSC) commonly found in the tropics and sub-tropics are toxigenic. Epiphytic and planktonic bacteria co-occur with toxigenic Prorocentrum but reciprocal allelochemical interactions are under-investigated. The aim of the present study was to identify the culturable bacteria collected together with isolates of the PLSC from seagrass (Thalassia testudinum) and macroalgae along tropical Atlantic coasts of Mexico, and to explore potential species interactions with selected isolates. Twenty-one bacterial genera belonging to Proteobacteria, Actinobacteria, and Bacteroidetes were identified by amplification of the 16S rRNA gene marker from nine clonal Prorocentrum cultures, with γ-proteobacteria comprising the dominant class. A positive correlation was found between the bacterial genera associated with two Prorocentrum clones and the esterified toxin analog DTX1a-D8, but there was no apparent correlation between the other PLSC clones and their associated bacteria with the other five DSP toxins detected. No bacteriostatic or allelochemical response was found for cell- and culture medium extracts of five Prorocentrum isolates assayed for bioactivity against Staphylococcus sp. DMBS2 and Vibrio sp. HEL66. Bulk cell-washing of Prorocentrum PA1, followed by growth with antibiotics, was only effective in reducing bacterial load in the initial growth stages, but did not yield axenic cultures or lower bacterial cell densities throughout the culture cycle. Antibiotic treatment did not impair growth or survival of the dinoflagellate, or apparently affect DSP toxin production. There was no significant correlation between Prorocentrum cell volume, growth rate, bacterial cell counts, or cellular toxin concentration over the entire time-series culture cycle. Benthic Prorocentrum and associated bacterial communities comprise highly diverse and characteristic microbiomes upon substrates, and among compartments in culture, but this study provides little evidence that allelochemical interactions among Prorocentrum cells and associated bacteria originating from epibenthic substrates play a definable role in growth and toxigenicity.
Introduction
Benthic dinoflagellates tend to flourish in shallow waters, where they grow attached to macroalgae, seagrass, or hard substrates by fibrillar components of extracellular mucilage (Honsell et al., 2013). The epiphytic relationship between the dinoflagellate and host, in the case of macroalgal substrates, can be species-specific or defined by the host morphology (Parsons and Preskitt, 2007). Benthic Prorocentrum species are typically rather sessile epiphytes but can also thrive and survive dispersal over long distances upon floating detritus (“rafting”) (Faust, 2004; Durán-Riveroll et al., 2019). Although ecophysiological information about benthic Prorocentrum species is limited from field populations, some physiological parameters and associated growth responses to key environmental factors, such as light availability, photoperiod, temperature, salinity, and nutrients, have been well described in laboratory studies (Morton et al., 1994; Pan et al., 1999; references cited in Hoppenrath et al., 2013).
The most commonly reported benthic Prorocentrum species is Prorocentrum lima (Ehrenberg) Stein (Hoppenrath et al., 2013), but recent studies have shown high variability among morphological features of the classically defined species. These morphological characteristics can vary among genetic lineages; hence, it is essential to determine the molecular identity of field specimens and cultured isolates for unambiguous taxonomic assignment (Nascimento et al., 2017; Durán-Riveroll et al., 2019). In the absence of comprehensive molecular genetic evidence, morphological “P. lima” can be interpreted as representing an unresolved species complex [P. lima species complex (PLSC)] (Zhang et al., 2015; Nishimura et al., 2019).
Members of the PLSC are globally distributed from high latitudes of the North Atlantic and Scandinavia to the tropics, but are more frequently found in higher cell abundances in tropical and sub-tropical coastal waters (Durán-Riveroll et al., 2019). Although these benthic dinoflagellates are only occasionally (and usually circumstantially) associated with Diarrheic Shellfish Poisoning (DSP) events after human consumption of contaminated shellfish (Lawrence et al., 1998; Foden et al., 2005), cultured isolates and natural populations are usually found to be toxigenic (citations in Durán-Riveroll et al., 2019). The polyether toxins produced among benthic Prorocentrum species, such as Prorocentrum arenarium, Prorocentrum concavum, Prorocentrum belizeanum, Prorocentrum faustiae, Prorocentrum hoffmannianum, P. lima, and Prorocentrum maculosum, include okadaic acid (OA) and at least two dozen dinophysistoxin (DTX) analogs, plus related polyketides of uncertain toxicity (Hu et al., 2010).
The physical contact and close association between epibenthic dinoflagellates and the often-organic host substrate provides ample opportunity for chemical ecological interactions. The substrate provides a surface platform for a complex microbiome. Host preferences for dinoflagellate colonization and growth can be determined by organic exudates, i.e., by providing concentrated fixed nutrients for heterotrophic growth, including vitamins. Alternatively, the substrate may yield inhibitory compounds, inhibiting growth of potential competitors for dinoflagellate colonization in two-dimensional space. Nutrient inputs may promote enhanced macroalgal growth and thereby provide additional substrate for epibenthic colonization (Parsons and Preskitt, 2007), but can also lead to shifts in the equilibrium of the dinoflagellates, microeukaryotic and metazoan competitors and predators, and the bacterial flora associated with this microbiome. Colonization and growth of the toxic dinoflagellate P. lima upon macroalgal biofouling attached to mussel aquaculture lines was shown not to be directly dependent on the stimulatory presence of live mussels; the cell density was a function of the magnitude and growth of the fouling biomass (Lawrence et al., 2000).
Marine bacteria are frequently found in association with epibenthic P. lima cell aggregations and the growth substrate (Basu et al., 2013; Park et al., 2018). Interactions between bacteria and benthic dinoflagellates occur within the phycosphere—a zone surrounding the dinoflagellate cell wherein leaked or excreted metabolites tend to be retained in higher concentrations and this can potentially enhance cell-to-cell chemical communication between adjacent bacteria and the dinoflagellate. Coexistence and allelochemical interactions between Prorocentrum cells and bacteria can occur at many levels, i.e., via extracellular-attached or free-living bacteria within the phycosphere, often including sticky mucopolysaccharides, or from intracellular bacteria (Park et al., 2018).
The extent to which associated bacteria affect growth and polyketide toxin production by epibenthic Prorocentrum in natural microbiomes or in monoclonal cultures remains a controversial issue. Accordingly, the current study aimed to determine the composition and diversity of the bacterial flora associated with unialgal cultures of the PLSC from natural populations and different substrates, i.e., from seagrass (Thalassia testudinum) and macroalgae from tropical Mexican reef systems. The second objective was to identify putative effects of associated bacteria on growth and cellular toxin content and composition throughout a culture cycle under controlled environmental conditions, and to evaluate potential allelochemical interactions between Prorocentrum and bacteria in culture.
Materials and Methods
Sample Collection and Initial Dinoflagellate Culture
For benthic dinoflagellate isolation, substrates of seagrass (T. testudinum), macroalgae (Ulva, Laurencia, Sargassum, and Padina) were collected, and inanimate surfaces such as buoys and ropes were sampled from the Veracruz Reef System (VRS) (19°11’54.10”N, 96° 4’0.70”W) and Puerto Morelos (Quintana Roo) (20°50’48.55”N, 86°52’30.53”W) (Table 1). Samples were collected from seagrass beds and ropes by snorkeling in shallow water along sandy shores and from macroalgae and buoys from rocky shores. Live samples were transported with site water in 50 mL conical plastic centrifuge tubes in ice chests with ice packs to maintain ambient temperature around 24°C during the 12 h surface transport to the laboratory at UNAM, Mexico City. Substrate specimens and surrounding medium were examined for colonizing dinoflagellates in Petri plates under a stereo-dissecting microscope (Discovery.V8, Zeiss, Göttingen, Germany). Substrates were gently brushed and single-cells of epibenthic dinoflagellates were isolated by micropipette into sterile 96-well microplates containing 300 μL 50%-strength GSe growth medium (modified without soil extract) (Blackburn et al., 2001) prepared from autoclaved (121°C, 15 min) seawater filtered through sand, activated carbon, and 1 μm-cartridge-filters from the Acuario de Veracruz. The growth medium, supplemented with GeO2 (final concentration: 2.5 mg L–1) (Markham and Hagmeier, 1982) to inhibit diatom growth, was prepared from heat-sterilized seawater stock at salinity 36. Clonal isolates were cultured by incubation at 25 ± 1°C on a 12:12 h light:dark cycle and illumination of 50 μmol photons m–2 s–1.
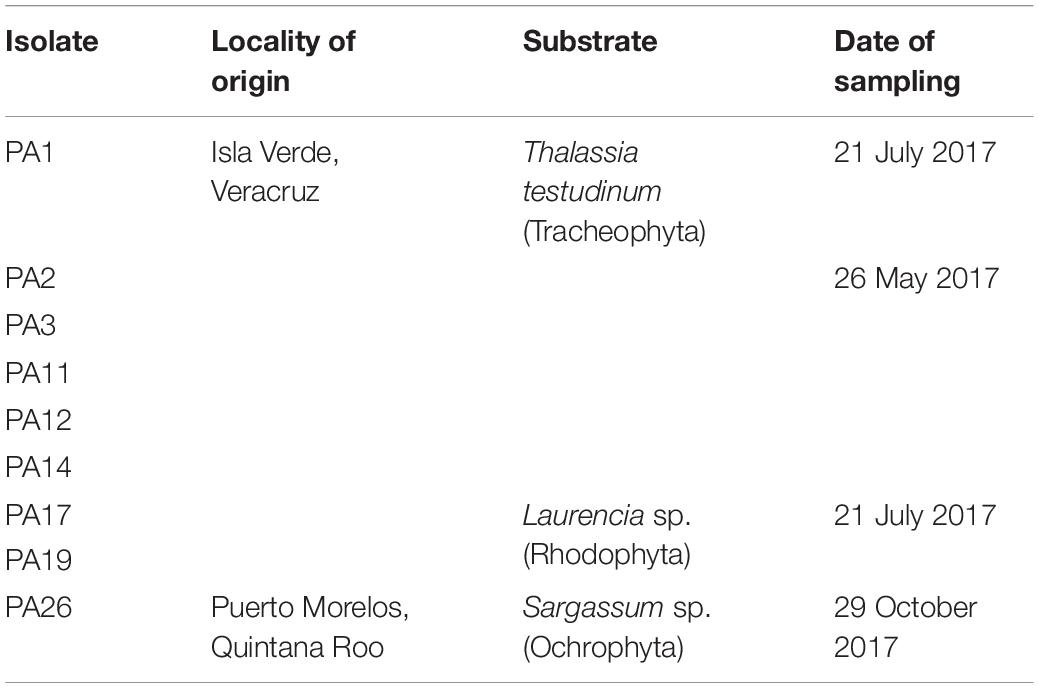
Table 1. Geographical origin of the monoclonal isolates of the Prorocentrum lima species complex (PLSC) obtained for this study.
After 10–12 days, once dinoflagellate cell division was observed (around 6–8 cells per well), preliminary cultures were transferred into 24-well microplates, each well containing 2 mL growth medium, and after subsequent growth, into 60 × 15 mm sterile plastic Petri plates with 15 mL 50% GSe seawater medium. Clonal isolates were maintained in 50% GSe medium for further analysis and as part of a benthic dinoflagellate reference collection. A sub-set of Prorocentrum isolates from this collection was selected for the following experiments (Table 1). Isolate PA14 from Isla Verde, Veracruz, was initially brought into culture and subjected to preliminary analysis, including of the associated bacterial community. Unfortunately, the culture soon exhibited heavy contamination by cyanobacteria, and subsequently died, and hence was excluded from further analysis of cell morphology, toxin content and composition, and bacterial interactions.
Sub-cultures of the Prorocentrum isolates maintained at UNAM were transferred to the culture facilities at the Alfred Wegener Institute (AWI), Bremerhaven, Germany, for further experimentation on growth and toxin production interactions between selected dinoflagellate isolates and associated bacteria in culture. These cultures were initially grown in plastic Petri plates on GSe medium as in Mexico, and then acclimated to grow on K medium (Keller et al., 1987) added as enrichment to 0.2-μm-filtered and autoclaved North Sea seawater at salinity 33. Cultures were maintained at 24 ± 1°C on a light:dark cycle of 14:10 h and illumination of 86 μmol photons m–2 s–1 measured with a LI-1000 light sensor (DataLogger LI-COR, Lincoln, NE, United States).
Morphological Analysis and Description of Prorocentrum Isolates and Associated Bacteria
Specimens of Prorocentrum isolates from stable cultures were analyzed in detail by light and scanning electronic microscopy (SEM) for morphological characteristics. Cells stained with 0.2% Calcofluor White M2R in aqueous solution (Fritz and Triemer, 1985) were studied under epifluorescence microscopy (Axio Scope.A1, Carl Zeiss, Oberkochen, Germany) with filter set 18 shift free EX BP 390-420 (excitation), BS FT 425 (optical divider), EM LP 450 (emission). Light micrographs were taken with an Axiocam 506 color digital camera (Zeiss, Göttingen, Germany).
Cultured dinoflagellate cells for SEM were fixed with 2% glutaraldehyde for 90 min. Specimens were washed in 1.5 mL distilled water (5°C) and centrifuged at 1200 × g at 5°C for 6 min. The wash procedure was repeated four times. Samples (1.5 mL each) underwent a graded ethanol dehydration series (10, 30, 50, 70, 90, and 99%), with centrifugation as for the sample wash at each dehydration step. Finally, a drop (250 μL) of hexamethyldisilazane (HDMS) air-drying agent was placed upon the sample on aluminum SEM stubs, and specimens were gold sputter-coated for 5 min.
Bacteria for SEM analysis were inoculated into marine broth (Difco, Detroit, MI, United States) and incubated for 6 days under the previously referred conditions. Each sample (1.5 mL) was fixed with 100 μL of 25% glutaraldehyde for 45 min. Specimens then were washed and dehydrated as described for Prorocentrum cells. After the 99% ethanol dehydration step, 1 mL of HDMS:ethanol 1:1 was added to the microtube for 3 min. After centrifugation, 1 mL of pure HDMS was added for 3 min; 10 μL of sample was pipetted onto round coverslips and gold sputter-coated as above.
Specimens of dinoflagellates and bacteria were observed with a JEOL JSM6360LV electron microscope equipped with a backscattered electron detector under 8 kV voltage acceleration and at 15 mm working distance.
Molecular Characterization of Bacteria From Epibenthic Prorocentrum
Nine Prorocentrum isolates from the VRS and Puerto Morelos sites (Table 1) were chosen for the isolation of dinoflagellate-associated bacteria. Bacteria were isolated from clustered Prorocentrum cells and the surrounding culture medium onto Marine and Luria Berthani agar (Difco, Detroit, MI, United States) supplemented with seawater salts. After 5–9 days of incubation at 25°C, 2023 colonies were harvested and characterized. Afterward, 45 representative bacterial morphotypes were chosen for DNA extraction with the 2% CTAB protocol, and amplification of the small-subunit ribosomal RNA (16S rRNA) with the primers 27F and 1492R (Lane, 1991). Purified PCR products were sequenced by the Sanger method (Sanger et al., 1977) at the Institute of Biology (LaNaBio, UNAM, Mexico). Sequence reactions were prepared with big dye (v3.1) (Applied Biosystems, Foster City, CA, United States) following manufacturer’s instructions, purified with Centrisep plates, and processed in a 3730xl DNA Analyzer (Applied Biosystems-Hitachi, Foster City, CA, United States).
After trimming and editing with the software Bioedit (Hall, 1999), sequences were aligned with the ClustalW algorithm, and taxonomically classified according to the Ribosomal Database Project (Cole et al., 2014). A neighbor-joining phylogenetic tree was constructed with the software Mega 7, using a bootstrap of 10,000 repetitions (Kumar et al., 2016). The resulting phylogenetic tree was edited with the online program Tree of Life (iTOL) (Ciccarelli et al., 2006). Sequences were deposited in GenBank with the accession numbers MN853604–MN853648.
The software R (R Core Team, 2018) was used to obtain the rarefaction curves to identify the sampling effort of bacterial genera among the isolates. A non-metric multidimensional scaling (NMDS) analysis, an indirect gradient approach for producing an ordination plot based upon a distance matrix, was performed upon the data. In this case, a Bray-Curtis similarity matrix was plotted to detect differences in the distribution of bacterial genera associated with the Prorocentrum isolates, excluding Pseudomonas and Microbacterium due to their cosmopolitan character. The identified toxins from each isolate were transformed and then fitted onto the NMDS ordination plot. Permutation tests (n = 1000) were used to test the significance of vector fits, and only significant vectors were depicted (p < 0.1).
Cell-Free Extracts for Toxin Analysis
Dense actively growing Prorocentrum cultures, cultivated in plastic Petri plates under the conditions detailed in Section “Sample Collection and Initial Dinoflagellate Culture,” were harvested with a 1 mL micropipette after sub-sampling 2 mL of stirred culture for cell counts in a Sedgewick Rafter cell-counting chamber under a light microscope (Olympus BH2, Tokyo, Japan) at 100X magnification. The remaining cells were transferred into 2 mL cryotubes and centrifuged at 5000 × g at 4°C for 5 min. The cell pellets were resuspended in 0.2 μm-filter-sterilized seawater (5°C) and centrifuged again under the same conditions. The supernatant was again decanted and the cryotubes were placed in a thermomixer at 100°C for 5 min to inactivate esterase enzymes that could modify the toxin profile. Cell pellets were stored frozen (−20°C) for later extraction.
Cell pellets were resuspended in 500 μL 50% methanol in FastPrep tubes. After adding 0.9 g FastPrep lysing matrix D (Thermo Savant, Illkirch, France), cells were homogenized by reciprocal shaking in a FastPrep FP120 (Bio 101, Thermo Savant, Illkirch, France) at 6.5 m s–1 for 45 s. Homogenized samples were centrifuged at 16,000 × g for 15 min at 11°C. The supernatant from each sample was transferred into a 0.45 μm pore-size spin-filter (Millipore Ultrafree, Eschborn, Germany) and centrifuged at 11,000 × g for 1.5 min at 11°C. Finally, filtrates were transferred into 2 mL LC-autosampler vials (Agilent, Waldbronn, Germany) for analysis by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS).
Toxin Analysis by LC-MS/MS
The analytical system consisted of an SCIEX- 4000 Q-Trap (Sciex, Darmstadt, Germany), triple quadrupole mass spetrometer equipped with a TurboSpray® interface coupled to an 1100 liquid chromatograph (LC) (Agilent, Waldbronn, Germany). The LC equipment included a solvent reservoir, in-line degasser (G1379A), binary pump (G1311A), refrigerated autosampler (G1329A/G1330B), and a temperature-controlled column oven (G1316A).
Separation of toxins was achieved following previous protocols (Krock et al., 2008; Nielsen et al., 2013) after injection of 5 μL extract onto a C8 analytical column packed with 3 mm HypercloneTM 3 μm BDS C8 130 Å 50 × 2 mm (Phenomenex, Aschaffenburg, Germany), maintained at 20°C. The flow rate was 0.2 mL min–1 and gradient elution was performed by eluents A (water, formic acid, and ammonium formate) and B (acetonitrile, formic acid, and ammonium formate). Initial conditions were 12 min column equilibration with 5% B, followed by a linear gradient to 100% B in 10 min and isocratic elution until 16 min with 100% B. The program was then returned to initial conditions until 19 min (total run time: 31 min).
Detection of DSP toxin analogs was performed by LC-MS/MS by selected reaction monitoring (SRM) experiments carried out in positive-ion mode with selected mass transitions (Supplementary Table S1). The following parameters were applied: curtain gas: 10 psi, CAD gas: medium, ion spray voltage: 5500 V, temperature: no heating, interface heater: On, declustering potential: 50 V, entrance potential: 10 V, exit potential: 15 V and dwell times of 100–200 ms per transition. Due to detection of a putative novel DTX1 isomer and associated diol-ester, provisionally dubbed DTX1a and DTX1a-D8, respectively, a collision-induced dissociation (CID) spectrum of DTX1 and the novel DTX1a compound was recorded in an enhanced product ion (EPI) mode of m/z 836 (mass range m/z 150–800) in the positive mode. Mass spectrometric parameters were the same as in the respective SRM experiments (Nielsen et al., 2013).
Analytical standards of OA (1 ng μL–1), DTX 1 (DTX1) (500 pg μL–1), and DTX 2 (DTX2) (500 pg μL–1) from the Institute for Marine Biosciences, National Research Council, Halifax, Canada, were used to identify and quantify DSP toxins in extracts of Prorocentrum cells. Due to lack of standards for derivatives such as OA diol-ester (OA-D8) and DTX 1-diol ester (DTX1-D8), cell quotas were expressed as OA and DTX1 equivalents, respectively, considering the following detection limits for OA (47 pg μL–1), DTX1 (35 pg μL–1), and DTX2 (25 pg μL–1). Data acquisition and processing were performed with the Analyst Software (Version 1.5, Sciex).
Disk Diffusion Assays With Prorocentrum Extracts
Three Prorocentrum isolates (PA1, PA2, and PA3) from the VRS, were selected for larger-volume batch cultures based on relative toxin composition, and high biomass yield per culture volume. These isolates were scaled-up into 2.8 L glass Fernbach flasks and harvested (total volume = 1 L) in active growth phase by centrifugation in 45 mL aliquots at 1600 × g at 10°C for 10 min (Eppendorf 5810R, Hamburg, Germany). Cell pellets were combined for each isolate into a single tube and freeze-dried. The dried pellets were sequentially extracted with hexane, ethyl acetate, ethanol, and ethanol:water (1:1) after ultrasonication (Sonorex Digitec, Bandelin, Berlin, Germany) for 10 min. After the pellet had precipitated at each step, the supernatant was transferred into a new scintillation vial and 2 mL of the next solvent was added. The supernatant was rotary-evaporated (Laborota 4002, Heidolph, Schwabach, Germany) to dry residue at 40°C. Crude residues were dissolved in their respective solvent (∼500 μL) and stored at −20°C prior to disk application.
The pooled supernatants for each isolate were passed through a solid phase extraction (SPE) cartridge (Bond Elut PPL, Agilent, Waldbronn, Germany) and eluted with methanol. The column was first conditioned with methanol, then after sample application, the column was washed with Milli Q deionized water to remove unbound compounds and then eluted with 1 mL methanol to release target molecules.
Cell pellet extracts and SPE-concentrated supernatants were tested by disk diffusion assay (Bauer et al., 1966) for their effect against bacterial growth. 15 μL of each extract [hexane, ethyl acetate, ethanol, ethanol:water (1:1), and methanol] was evaporated on sterile 8 mm paper (two replicates per extract) for ∼30 min at room temperature. Cell equivalents were calculated based on the number of harvested cells and the extract volume applied in the assay (PA1: 382.5, PA2: 645, PA3; 409 cells). Aliquots (60 μL) of overnight-grown (∼108 cells mL–1) bacterial target strains Staphylococcus sp. DMBS2 and Vibrio sp. HEL66 were spread over the surface of marine agar plates (Difco, Detroit, MI, United States) with a sterile glass spreader. The dried filters containing the extracts were placed symmetrically upon the agar surface (four filters per plate). Paper disks with each solvent served as controls. The agar plates were incubated at 20°C for 24 h and the inhibition zones recorded.
Effect of Bacterial Load on Prorocentrum Growth and Toxin Production
An early stationary growth phase culture of Prorocentrum PA1 was subjected to alternative treatments to reduce bacterial load in a time-series growth experiment on bacterial interactions. A stirred 6 L batch culture (initial cell concentration: 13.6 × 106 cells L–1) was first divided into three 2 L aliquots in glass bottles. The first treatment consisted of washing 2 L batch culture in small aliquots (∼200 mL) across a 30 μm nylon-mesh and then rinsing three times with 0.2 μm-filter seawater to remove associated bacteria. For each aliquot, retained Prorocentrum cells were back-washed from the mesh with sterile K-medium to a final 2 L culture volume (initial cell concentration: 2.8 × 105 cells L–1). The second treatment was identical, except that the sterile K-medium (initial cell concentration: 3.62 × 105 cells L–1) contained a cocktail of antibiotics (details in Supplementary Table S2) throughout the 47 day growth experiment. The final 2 L culture (cell concentration: 4.84 × 105 cells L–1) was retained untreated by either cell washing or antibiotics and served as a growth control of the initial conditions from the bulk culture.
The treated and control cultures were incubated in 5 L glass bottles for acclimation under the same conditions detailed in Section “Sample Collection and Initial Dinoflagellate Culture.” After 4 days, the content from each bottle was distributed into 150 × 12 mm plastic Petri dishes after stirring for 20 min for homogenization to yield approximately equal cell numbers among the plates. Replicate (n = 5) cultures from each treatment and control were harvested every 3–5 days from Day 0 to 35. The final harvest was extended to the beginning of senescence phase, but only two treated replicates were evaluated at Day 47 and no control sample was considered at this point.
An aliquot of 1.5 mL was pipetted from a well-mixed sample for Prorocentrum cell counts at each time-point for each replicate. Cells were immediately fixed with acidic Lugol’s iodine solution and counted in a Sedgewick-Rafter chamber with an inverted microscope (AxioVert.A1, Carl Zeiss, Oberkochen, Germany) at 100X magnification (according to Reguera et al., 2016). Prorocentrum cell size was measured by the software Capture Express with an HDMI 16MDPX camera (DeltaPix, Smorum, Denmark) and volume calculated by the following equation:
where a = length, b = width, and c = cross-section (height).
Associated bacteria in the well-mixed culture aliquots were fixed with 0.25 mL 20% phosphate-buffered saline (PBS)-formaldehyde and prepared for counting following the fluorescence method of Porter and Feig (1980). The bacterial cells were stained by adding 0.5 mL of preserved sample to 5 mL sterilized water and 25 μL of 4,6-diamidino-2-phenylindole (DAPI, 0.5 mg mL–1), with incubation (20 min) at 4°C in the dark. Stained cells were filtered onto a 25 mm, 0.2 μm pore size black polycarbonate Nucleopore Track-Etch filter (Sigma–Aldrich, Munich, Germany) mounted over a 25 mm Whatman GF/C glass microfiber filter (Sigma–Aldrich, Munich, Germany). Filters were placed on a glass slide and the bacteria counted by epifluorescence microscopy (Axioskop 2 plus, Carl Zeiss, Oberkochen, Germany) at 1000X.
Data Analysis
Statistical analysis was performed with the software Infostat V.2018 (Di Rienzo et al., 2018) with inclusion of mean values and standard deviations. An analysis of variance (two-way ANOVA) was followed by a post hoc Tukey’s HSD test for mutually significant differences in interactions among multiple variables; namely, in cell abundance, cell-size, bacterial-cell density, and toxin content among the treated (washed cells plus antibiotics and washed cells) and non-treated cultures. In the same manner, all these variables were analyzed and compared at each timepoint throughout the 47 day growth experiment. All data complied with the assumptions of normality and homoscedasticity. Pearson’s correlation coefficient (r) was determined to establish a linear correlation among the same variables.
Results
Morphological Description of Prorocentrum Isolates and Associated Bacteria
Specimens from cultured isolates from the Gulf of Mexico and Caribbean coast studied under epifluorescence microscopy after Calcofluor staining exhibit a wide variation in cell outline in valve view (Table 2). Nevertheless, among 20 cells measured from each isolate, a dominant cell shape could be easily distinguished. The dominant ovoid cell type corresponds well to the illustrations and description for Prorocentrum in both classical and recent literature (Stein, 1883, as Dinophysis laevis Stein; Schiller, 1933, as Exuviaella ostenfeldi Schiller; Dodge, 1975; Nagahama et al., 2011; Hoppenrath et al., 2013; Zhang et al., 2015; Luo et al., 2017).

Table 2. Morphometric characteristics (n = 20) of cultured cells of the Prorocentrum lima species complex (PLSC) from the Gulf of Mexico and Caribbean coasts of Mexico observed by epifluorescence microscopy after staining with Calcofluor White M2R.
The cells of all Prorocentrum cells observed have a smooth thecal surface, and a trichocyst pore pattern with a central area on each valve devoid of pores, but with scattered pores throughout the valve surface, and a marginal row of closely situated pores. Cells are characterized by a central pyrenoid and a wide V-shaped periflagellar area. Some cells have shapes distinct from the typical morphology, in that they are smaller, shorter, or asymmetrical along the longitudinal axis. In all cases, the thecal surface features, the pore pattern, and other characters remained consistent with the dominant cell type in each culture. In natural samples, under an inverted microscope, these atypical cells might be ascribed to species other than P. lima, but because they are from clonal cultures, there is no doubt about the infraspecific variation in cell shape within each culture. The morphological characters observed under epifluorescence microscopy ascribed all the studied cells to the PLSC (Aligizaki et al., 2009).
Scanning electron microscopy of cultured Prorocentrum showed some dinoflagellate cells with attached bacteria, but most bacteria appeared as free-living coccoid or rod-like forms (sometimes filaments) often clumped in mucilaginous aggregates in the culture medium (Figure 1).
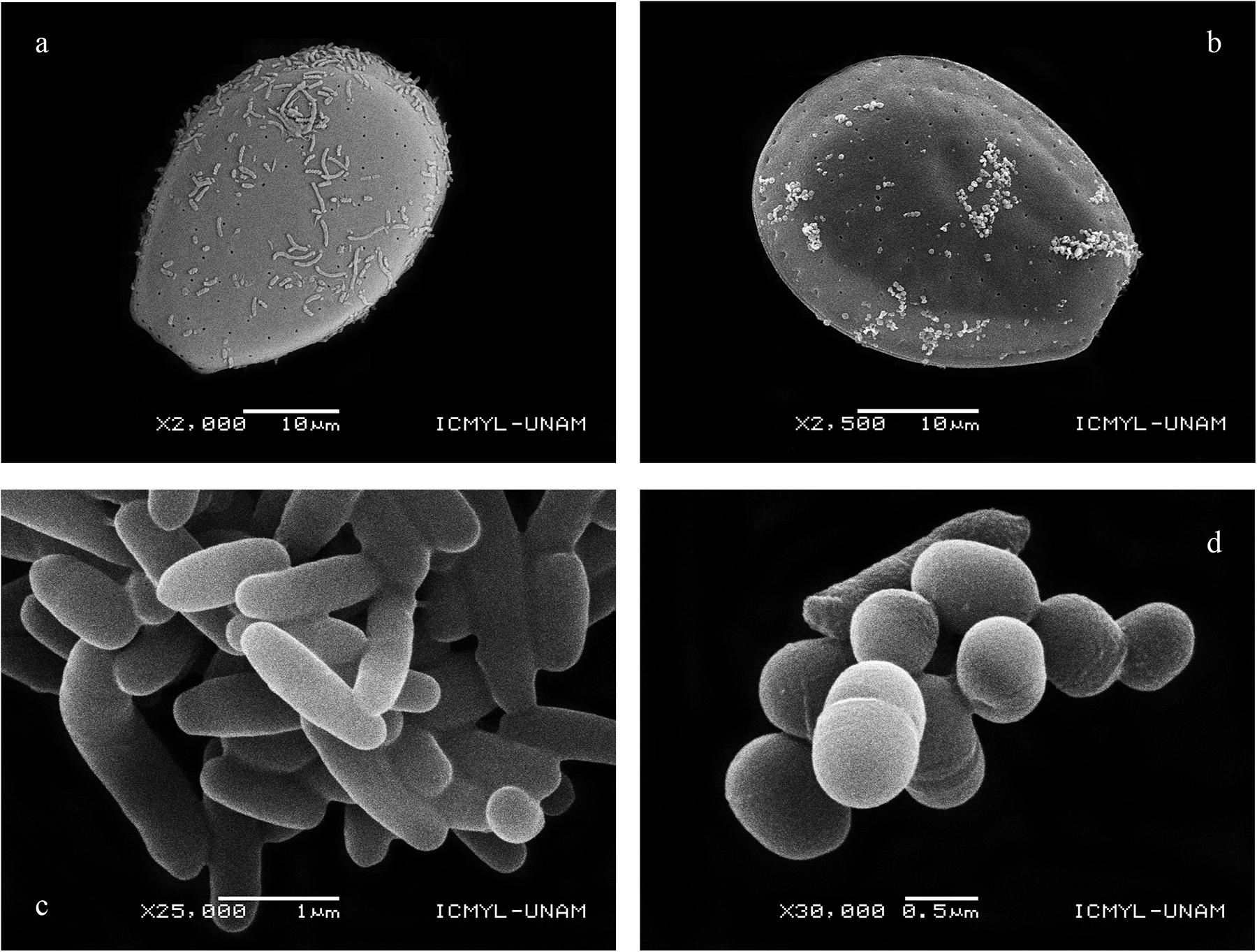
Figure 1. Bacteria attached to Prorocentrum cells: (a) PA3; (b) PA19, and isolated from cell aggregates versus culture medium: (c) Thalassospira sp. isolated exclusively from cells belonging to PA11; (d) Kocuria sp. isolated exclusively from the culture medium of PA11, PA12, and PA17.
Identification of Cultured Bacteria Associated With Epibenthic Prorocentrum
The phylogenetic tree of bacteria isolated from nine Prorocentrum cell cultures shows the phylogenetic association pattern among four dominant classes: Actinobacteria, Flavobacteriia, α-Proteobacteria, and γ-Proteobacteria (Figure 2). Class γ-Proteobacteria was the most abundant, with 11 genera within the orders Alteromonadales, Cellvibrionales, Oceanospirillales, and Pseudomonadales. Class α-Proteobacteria comprised five genera, within the orders Rhodobacterales, Rhodospirillales, and Sphingomonadales, whereas Actinobacteria presented four genera. Genus Euzebyella was the only representative of the class Flavobacteriia. Prorocentrum isolate PA17 showed the highest number of bacterial genera (13 of 21 identified genera), whereas PA1 exhibited the lowest abundance, represented by only three genera.
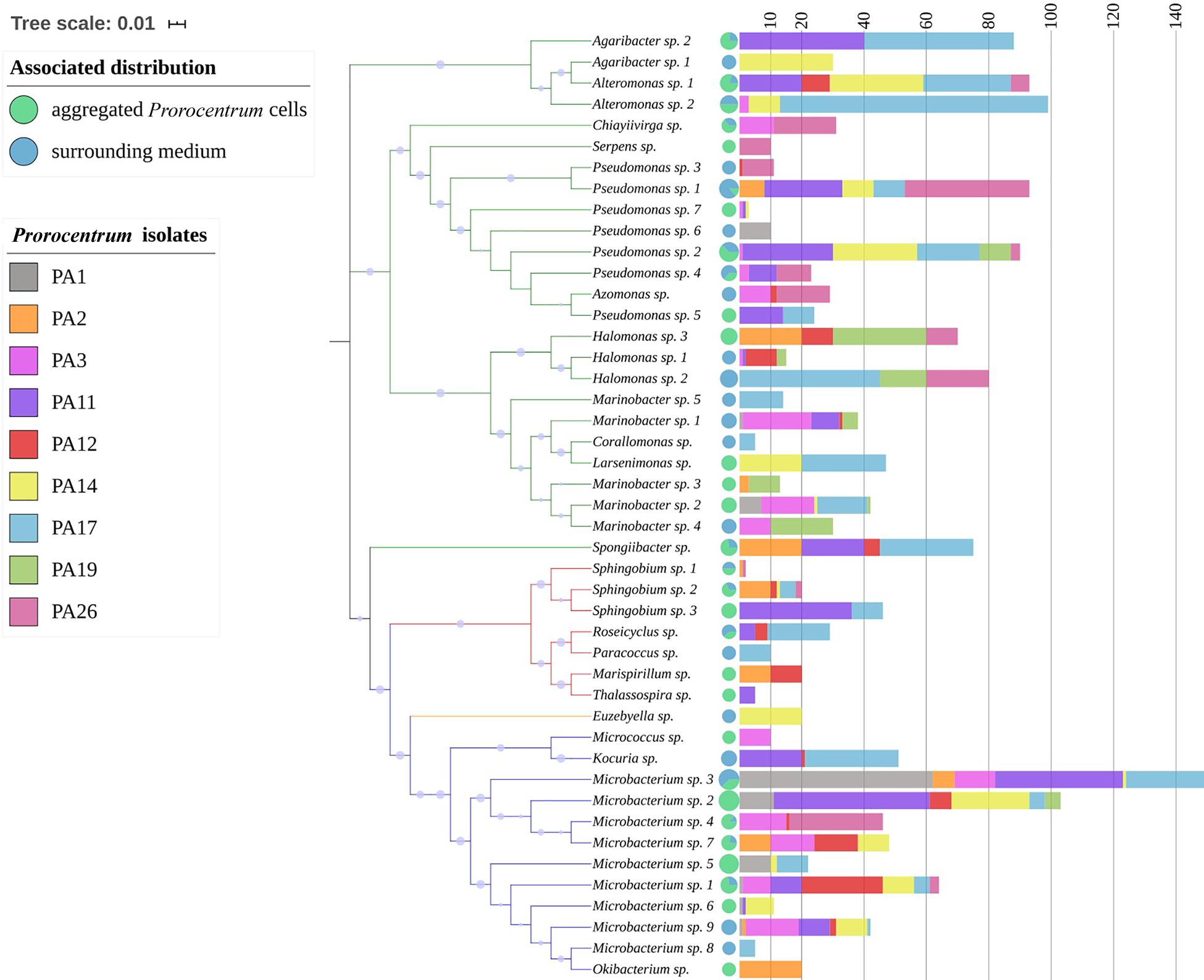
Figure 2. Neighbor-joining phylogenetic tree of bacterial isolates from Prorocentrum cell cultures from the Veracruz Reef System and the Mexican Caribbean coast. The tree is constructed from 16S rRNA sequences with classification according to the Ribosomal Database Project, and with 10,000 bootstrap repetitions (blue dots). The tree scale indicates the evolutionary distance among the isolates. The branch color denotes the corresponding bacterial class: Actinobacteria (blue); Flavobacteriia (orange); α-Proteobacteria (red); γ-Proteobacteria (green). On the right, the pie charts show the proportion of colonies of each isolate associated with Prorocentrum cell aggregates (D) versus the surrounding culture medium (CM), whereas the colored stacked histogram bars (non-cumulative) indicate the number of colonies of each bacterial species (isolate) found in the nine Prorocentrum cultures.
At the bacterial genus/species level, there was high variation among Prorocentrum isolates in cultured bacteria composition and even between those associated with dinoflagellate cell aggregates versus free in the medium (Figure 3A). More homogeneity was exhibited at the higher group level (phylum/class), with dominance by y-Proteobacteria and Actinobacteria, except often (but not always) more α-Proteobacteria were associated with the dinoflagellate cells than free in the culture medium among Prorocentrum isolates (Figure 3B).
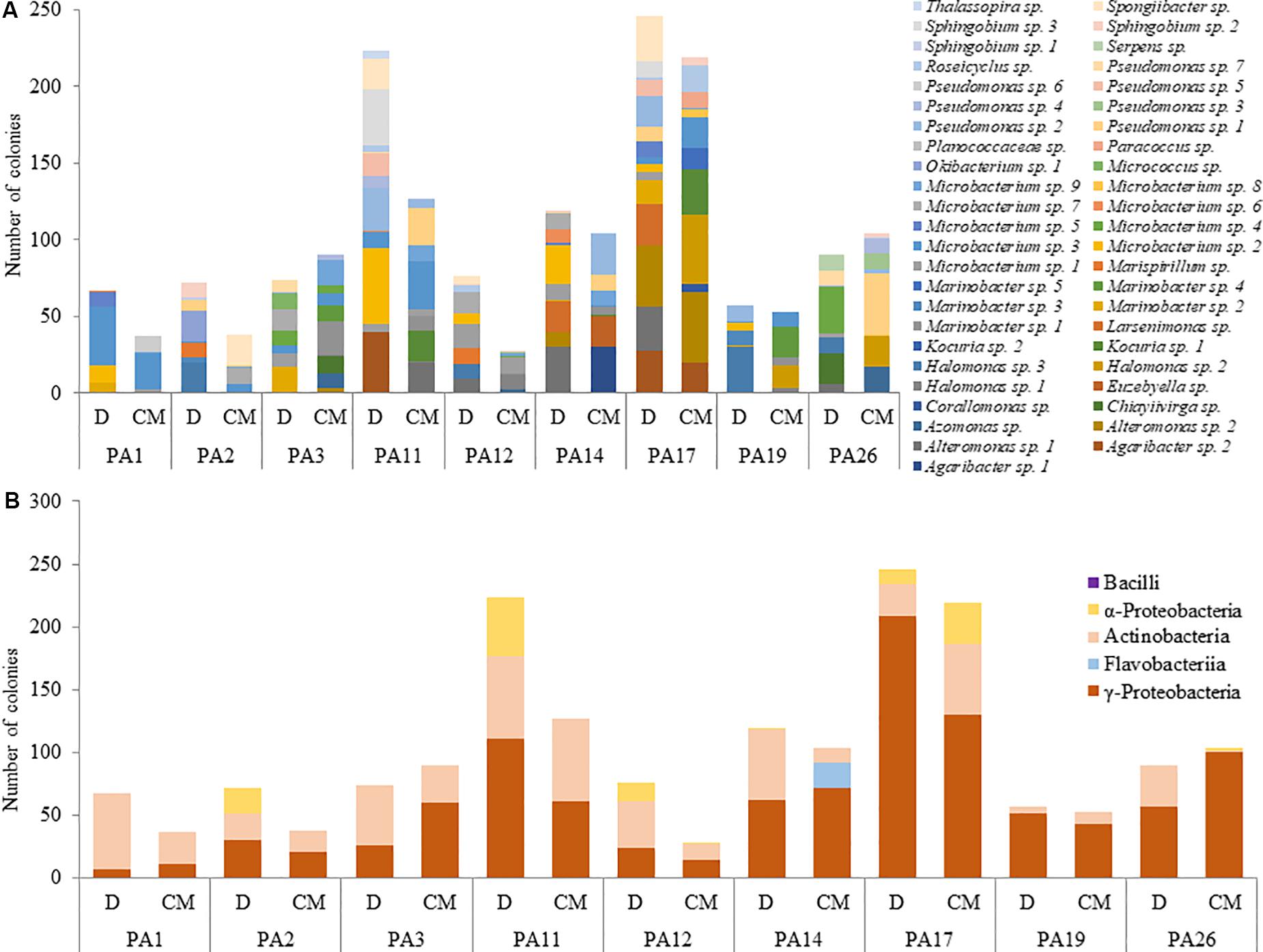
Figure 3. Abundance of cultured bacterial genera/species (A) and classes (B) from dinoflagellate cell-aggregates (D) and culture medium (CM).
Among the 45 bacterial strains isolated from nine cultured Prorocentrum clones, most rarefaction curves attained an asymptote, based upon amplified 16s rDNA sequences of cultured bacterial strains, identified among 21 bacterial genera, with respect to the isolated colonies (Supplementary Figure S1). This indicates that the sampling effort was sufficient to cover the diversity of the bacterial community, except from Prorocentrum clones PA1, PA12, and PA14. Even though most bacterial genera were identified in all Prorocentrum isolates, some were restricted to particular isolates (i.e., Serpens in PA26, Thalassospira in PA11, Micrococcus in PA3, Okibacterium in PA2). Certain genera were isolated exclusively from the dinoflagellate cell-aggregates or the surrounding medium (Table 3), i.e., Thalassospira was isolated exclusively from dinoflagellate cells, whereas Kocuria was isolated only from the culture medium. Microbacterium was the most abundant genus isolated from the cell-aggregates, but Pseudomonas was dominant from the surrounding medium. The distribution of Prorocentrum cultures in relation to the abundance of isolated bacterial genera and toxin production showed four groups in the NMDS plot (Figure 4). Prorocentrum isolates PA19 and PA26 were clustered mainly by the abundance of Halomonas and Marinobacter, while PA2 and PA12 were clustered by Marispirillum and Sphingobium. Isolates PA11 and PA17 were clustered primarily by the high abundance of Alteromonas, Kocuria, and Agaribacter. Marinobacter grouped isolates PA1 and PA3, which showed a positive correlation with DTX1a-D8. Other Prorocentrum clones and their associated bacterial isolates were not significantly correlated with the identified DSP toxins.
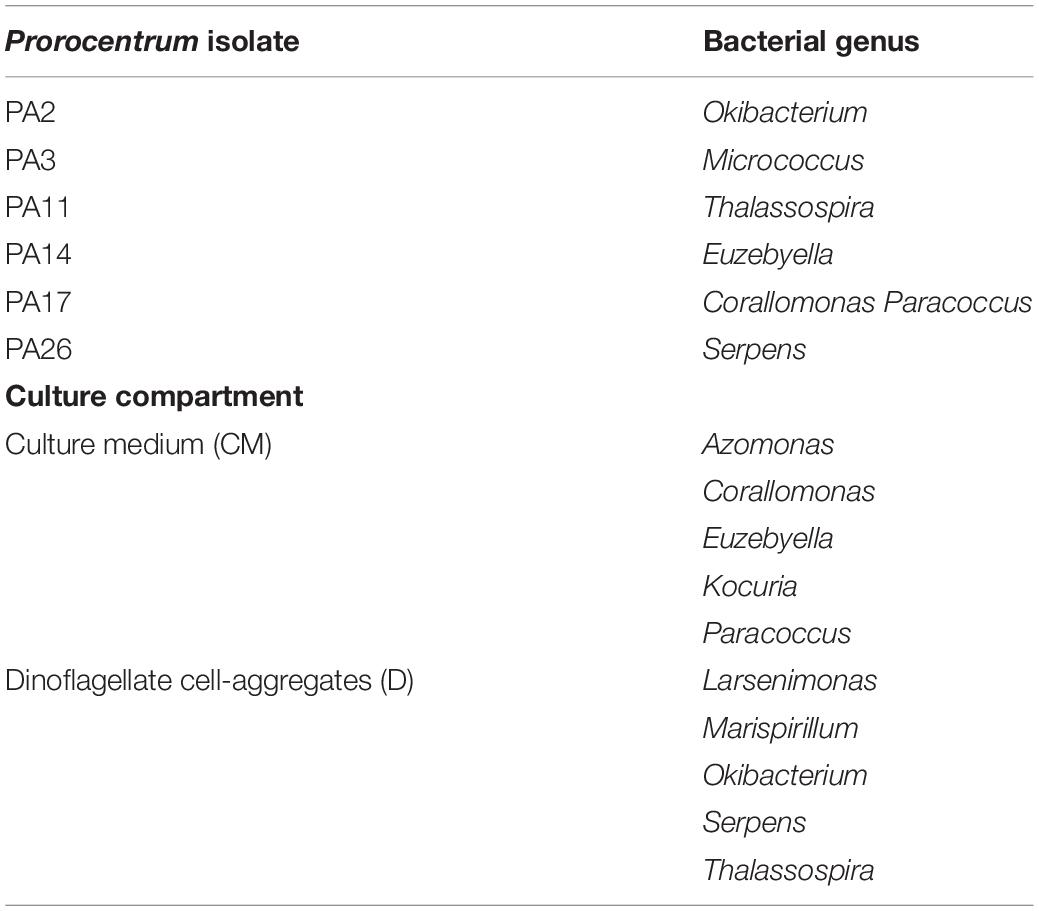
Table 3. Bacterial genera isolated exclusively from specific Prorocentrum isolates and culture compartments: Prorocentrum cell aggregates versus surrounding medium.
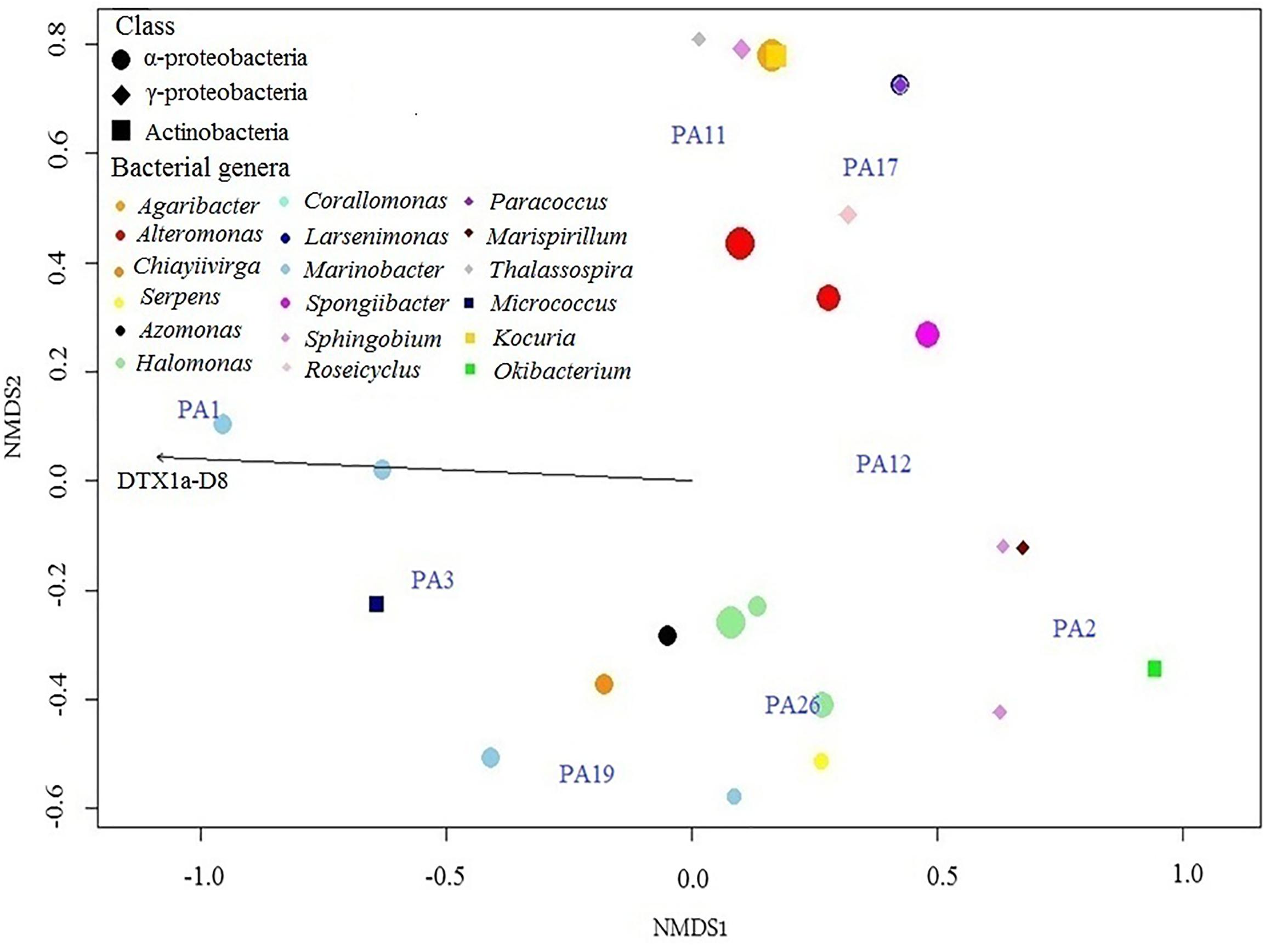
Figure 4. NMDS plot based upon a Bray–Curtis matrix of the distribution of clones of the Prorocentrum lima species complex (PLSC) in relation to the abundance of their associated isolated bacterial genera and DSP toxin composition in culture. The scale of the axes is arbitrary as is the orientation of the plot, with closeness of the clustered elements indicating the degree of similarity and association. The symbol shape denotes the bacterial class; color indicates genera; and size denotes abundance. Vector length indicates magnitude and correlation of the toxin with the Prorocentrum clones and their isolated associated bacteria.
Toxin Composition of Prorocentrum Isolates
Six main DSP toxin analogs were identified and quantified among the cultured PLSC clones from the Gulf of Mexico and Caribbean coasts of Mexico: OA, OA-D8, DTX1, DTX1-D8, DTX1a, and DTX1a-D8. The analyte transition m/z 836.6 > 237.1 eluted at two retention times: 12.41 and 12.83 min (standard value for DTX-1); this putative DTX1 isomer eluting at 12.41 min was provisionally named DTX1a but remains structurally uncharacterized. Novel isomers DTX1a and DTX1a-D8 were detected from most clones, but typically at relative concentrations <10%. The relative toxin composition (% total concentration) indicated OA and OA-D8 as co-dominant toxins among seven of eight evaluated clonal cell extracts, with OA-D8 absent from only PA2 (Figure 5).
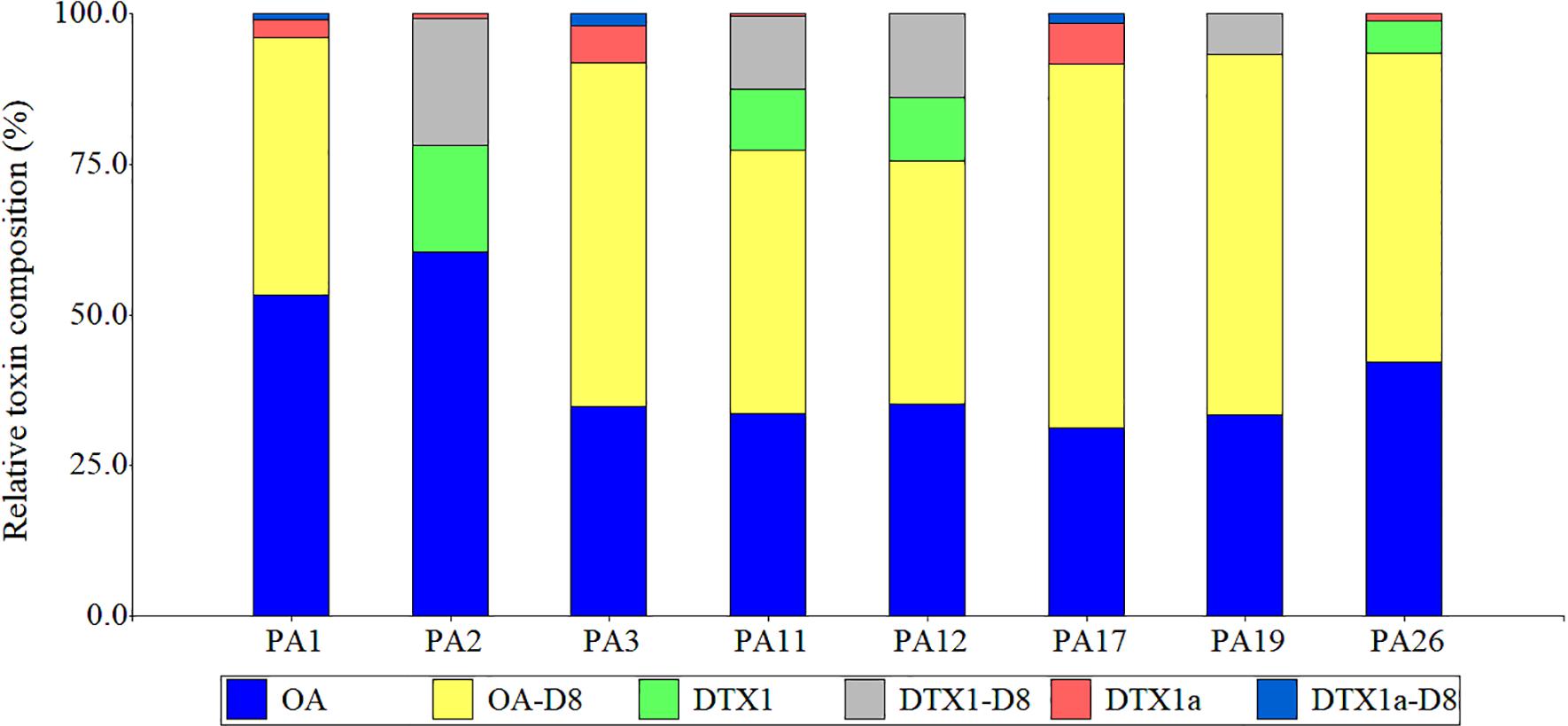
Figure 5. Relative toxin composition (%total concentration) of DSP toxins (OA and DTX analogs) from cultured benthic Prorocentrum isolates from the Veracruz Reef System and the Mexican Caribbean coast. OA, okadaic acid; OA-D8, okadaic acid diol-ester; DTX1, dinophysistoxin 1; DTX1a, undescribed DTX1 isomer; DTX1-D8, dinophysistoxin1diol-ester; DTX1a-D8, undescribed DTX1-D8 isomer.
Effect of Prorocentrum Cell and Supernatant Extracts on Cultured Bacterial Growth
The agar disk diffusion assay showed no inhibition zones nor apparent changes in growth of bacteria near the disk area for either Prorocentrum cell pellet extracts or supernatants. Solvent controls were similarly negative for growth effects. The first assay series did show an apparent reduction in the number of colonies of Staphylococcus sp. around the disks for the 50% ethanolic extract of Prorocentrum PA1 cells, but subsequent application of extracts in a dilution series (1:10), failed to demonstrate any effect. Since there was no apparent effect on bacterial growth, figures are not shown.
Growth Characteristics of Prorocentrum PA1 and Associated Bacteria
Bacterial cell shape was consistent among Prorocentrum PA1 cultures over time: long filaments of small individual cells (∼2.5 μm) and rod-shaped cells (∼1.6 μm) were most frequent. Aggregations of bacterial cells (clumps) were observed in all cultures. In washed and antibiotic-treated cultures, these aggregations became more abundant after Day 16 and 21, respectively, whereas, in the untreated control, there were small aggregates from the start of the experiment (Figure 6).
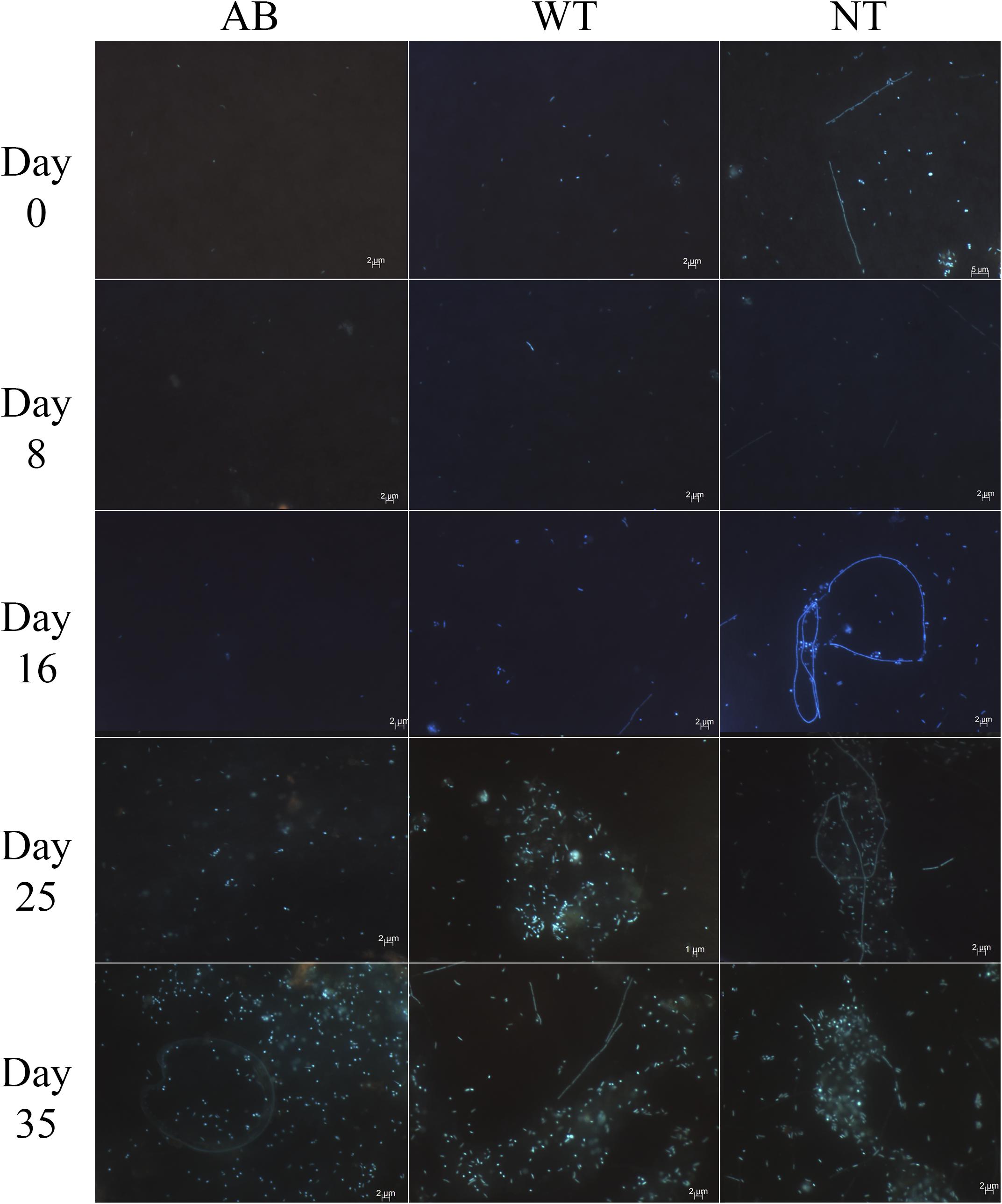
Figure 6. Epifluorescence microscopy after DAPI staining of bacteria in Prorocentrum PA1 cultures showing apparent increase in bacterial cell density over time in all treatments: AB, washed cells plus antibiotics; WT, washed cells; NT, non-treated culture (control). 1000X magnification.
Initial bacterial cell densities (n = 5 cell counts) in washed cell cultures without antibiotics (WT) (2.4 × 108 cells mL–1) and with antibiotics (AB) (1.5 × 108) compared to unwashed control cultures (NT) (7.5 × 108 cell mL–1) indicated substantial but not totally effective reduction in initial bacterial load by washing cells. After 1 week, the bacterial density in the washed cell culture was not significantly different from that of the untreated control, although the antibiotic-treated bacterial load did not begin to increase before Day 11 and remained significantly lower until Day 21 (Figure 7). Nevertheless, over the entire culture cycle, on a daily basis, there were no significant differences among treatments (two-way ANOVA, p < 0.05 and Tukey’s HSD test, n = 49).
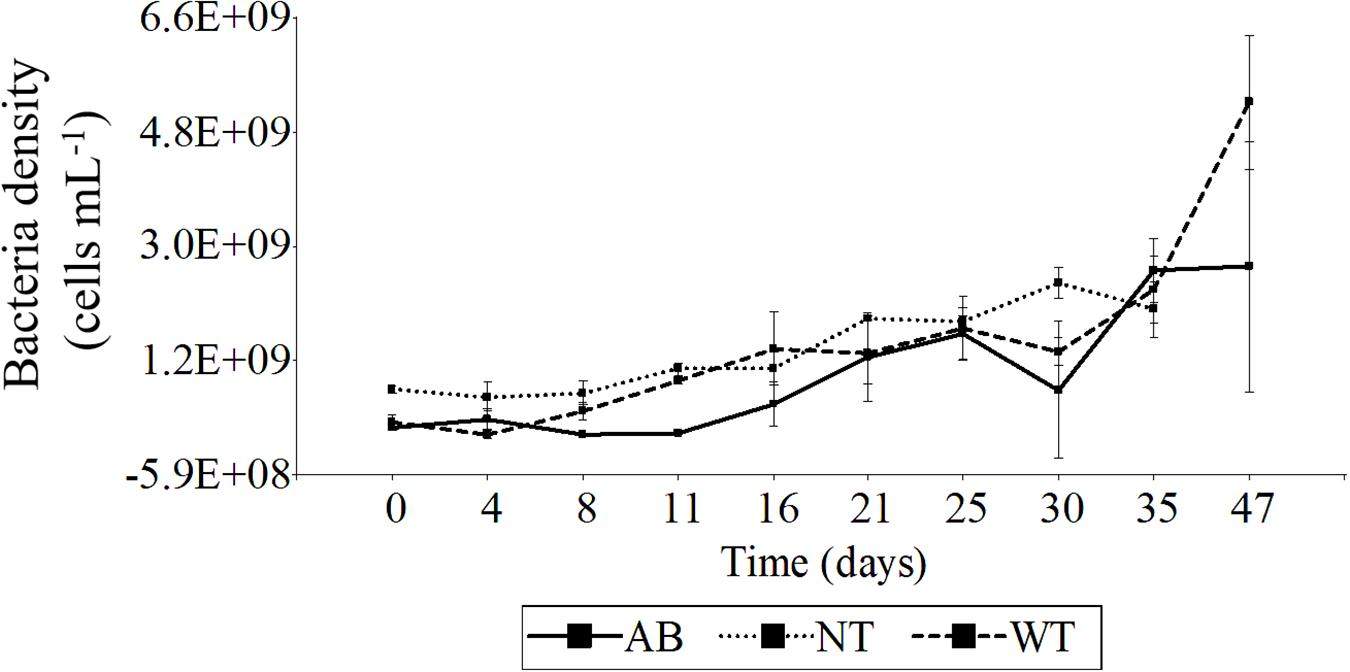
Figure 7. Time-series of bacterial cell density from Prorocentrum PA1 cultures under various treatments. AB, washed cells plus antibiotics; WT, washed cells; NT, non-treated culture (control). AB, WT, and NT: n = 3 from Day 0 to 35, AB and WT: n = 2 on Day 47. Error bars indicate mean ± standard deviation (SD) of replicate cell counts.
The increase in bacterial load tracked the increase in Prorocentrum cell density for the untreated control, but whereas bacterial growth remained partially suppressed for the first week with the antibiotic treatment, bacterial growth tracked the Prorocentrum density much earlier in the washed cell (WT) cultures (Figure 8). There was a moderate positive correlation between bacteria and Prorocentrum PA1 cell densities for all treatments—wash plus antibiotic-treated (AB), wash only (WT), and control (NT) cell cultures (ANOVA p < 0.05, Pearson correlation coefficient: 0.48, 0.67, 0.79, respectively). No significant differences in Prorocentrum growth rates were found among treatments and days of incubation during the initial phase (Day 0–4) (two-way ANOVA, p < 0.05 and Tukey’s HSD test, n = 49). During early exponential phase (i.e., Day 4–8), cultures exhibited a maximum mean growth rate (μmax): 0.22 ± 0.15, 0.35 ± 0.25, and 0.17 ± 0.09 div day–1 for the AB, WT, and NT cultures, respectively (Supplementary Figure S2). Prorocentrum cell volume remained constant through the growth cycle and among treatments, with only minor non-significant fluctuations on a daily basis among treatments (two-way ANOVA, p < 0.05 and Tukey’s HSD test, n = 49).
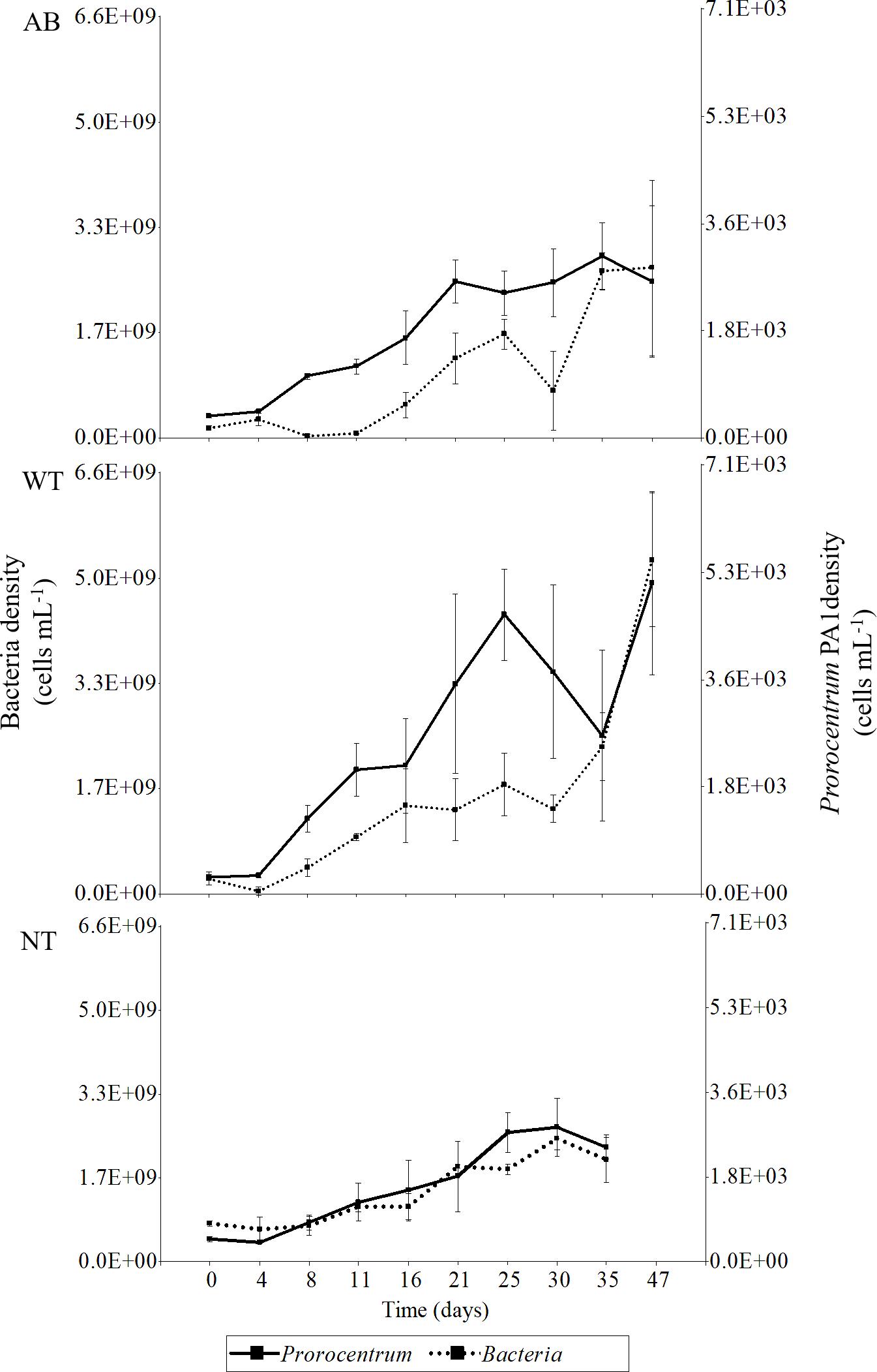
Figure 8. Time-series of bacterial cell density along the growth curve of Prorocentrum PA1 subjected to alternative treatments. AB, washed cells plus antibiotics; WT, washed cells; NT, non-treated cultures. AB, WT, and NT: n = 3 from Day 0 to 35, AB and WT: n = 2 on Day 47. Error bars indicate mean ± standard deviation (SD) of replicate cell counts.
Toxin Composition and Cell Toxin Quota of Prorocentrum PA1
The toxin profile of P. lima isolate PA1 comprised six major analogs: OA, OA-D8, DTX1, DTX1-D8, DTX1a, and DTX1a-D8. The relative abundance of six analogs was consistent and did not vary significantly among wash- (WT) and wash plus antibiotic- (AB) treatments or for the control (NT) over 47 days in batch culture (Supplementary Figure S3). The most relatively abundant toxin (>50%) was OA, followed by OA-D8, then DTX1a > DTX1-D8 > DTX1a-D8 > DTX1. No significant differences in toxin cell quota (two-way ANOVA, p < 0.05 and Tukey’s HSD test, n = 49) were found among different culture treatments evaluated over the entire 47 day culture cycle (Figure 9), but the toxin quota varied significantly during the first 16 days of the experiment. Toxin cell quotas in antibiotic-treated (AB) and washed cultures without antibiotics (WT) were substantially lower at Day 4 compared to initial values (Day 0), where the AB culture reached minimum quota (1.58 × 102 fmol cell–1). At Day 8, WT and NT control reached minimum values: 1.36 × 102 and 1.43 × 102 fmol cell–1, respectively. During late exponential to stationary growth phase from Day 16 to 47, cell toxin quota remained roughly stable, although AB-cultures reached the highest value (3.36 × 102 fmol cell–1) on Day 25.
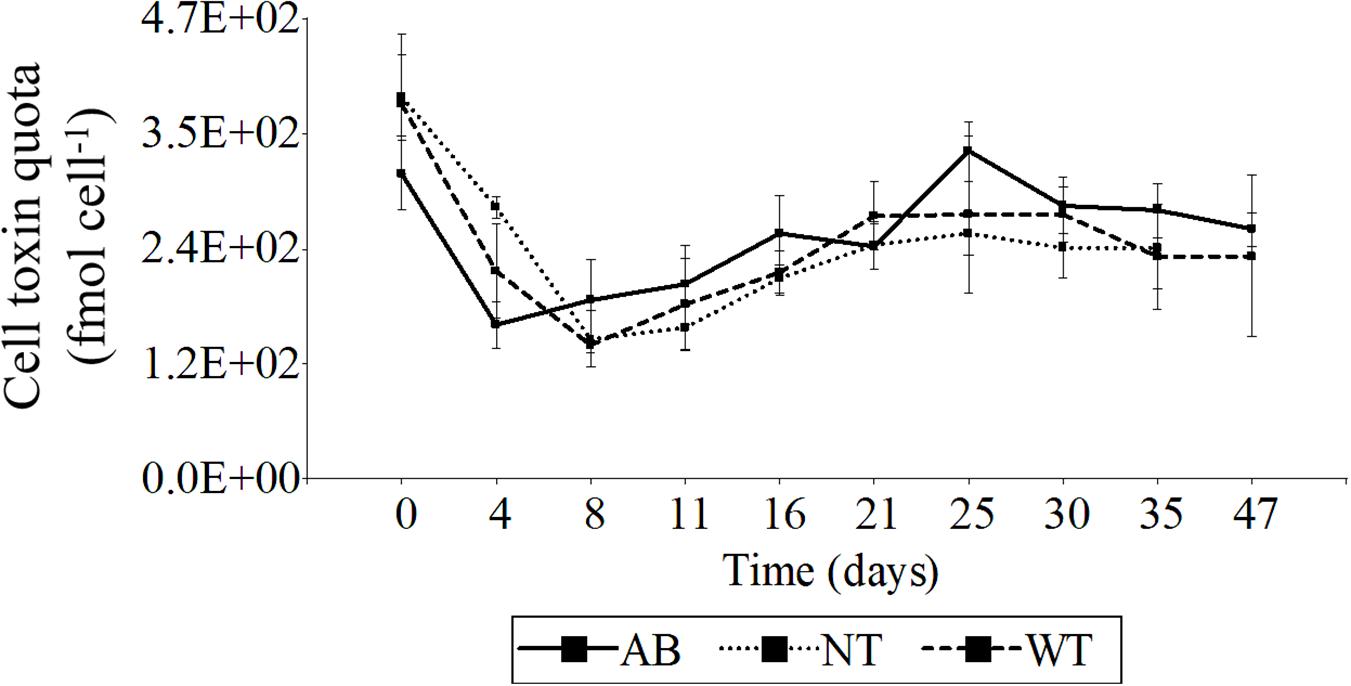
Figure 9. Variation of total DSP toxin cell quota over time-series growth subjected to alternative treatments. Total cell quota (fmol cell– 1) calculated as sum of all detectable DSP toxin analogs: OA, OA-D8, DTX1, DTX1-D8, DTX1A, and DTX1a-D8. AB, washed cells plus antibiotics; WT, washed cells; NT, non-treated culture (control). AB, WT, and NT: n = 3 from Day 0 to 35, AB and WT: n = 2 on Day 47. Error bars indicate mean ± standard deviation (SD) of replicate measurements.
Discussion
Bacterial Associations With Toxigenic Dinoflagellates
A few studies have explored the dynamic relationship between groups of bacteria and blooms of dinoflagellates in natural populations characterized as harmful algal blooms (HABs) (i.e., Park et al., 2015; Santi-Delia et al., 2015). Typically, α- and γ-Proteobacteria are the most widespread and diverse classes in marine environments, but can shift proportions during evolution of a HAB. Members of the γ-Proteobacteria are usually in low relative abundance during the initial and stationary growth phase of the bloom, while α-Proteobacteria decrease as the number of dinoflagellate cells increases (Park et al., 2015; Guidi et al., 2018). These generalities have been defined with respect to dense pelagic blooms in the water column, but it is not clear that such growth dynamics apply to consortia of bacteria–dinoflagellates in benthic systems, where the “blooms” of dinoflagellates and associated bacteria are more spatially constrained upon and adjacent to the substrates.
Many investigations on the molecular identity and phylogeny of marine bacteria, typically based upon 16S rRNA sequencing, have acknowledged the gross underestimation of bacterial diversity represented by the small fraction of total bacterial that can be successfully cultured (Vartoukian et al., 2010). Most marine bacteria from coral reefs do not exhibit favorable growth when cultured in nutrient-rich media (Giovannoni and Stingl, 2005), particularly those from oligotrophic tropical oceanic environments. Nevertheless, the high diversity of bacteria that can be cultured from diverse benthic assemblages and substrates in reef systems is indeed noteworthy. In the work reported here, the extracellular-attached and free-living bacteria isolated from benthic Prorocentrum cultures from the Gulf of Mexico and Caribbean coasts of Mexico are mostly γ-Proteobacteria, which together with α-Proteobacteria are the bacteria most commonly detected in microalgal cultures and natural blooms (Buchan et al., 2014; Park et al., 2018). Unfortunately, the high differential selection inherent in culture experiments means that the results from co-culture of bacterial assemblages and dinoflagellates isolates cannot be simply extrapolated back to natural benthic systems as representative of in situ bacterial composition and diversity.
Previous studies on cultured isolates of toxigenic benthic Prorocentrum also noted, as found herein for tropical Atlantic populations, that the microbiota from the bulk culture medium and the phycosphere could be considerably different (Prokic et al., 1998; Perez et al., 2008). Even though five bacterial genera were exclusively isolated from Prorocentrum cell aggregates among dinoflagellate clones from tropical coasts of Mexico, the distribution of the identified cultivable bacteria between cell-aggregates and surrounding medium was very similar. This is likely attributable to the unique and nutrient-rich microhabitat provided by Prorocentrum cells.
Variation in the microbiota between Prorocentrum clones recently isolated and from cultures long established in the laboratory may be due to microenvironmental selection, but apparent selection biases can also arise from differences in culture media, growth conditions, and molecular detection techniques employed among laboratories. Nevertheless, growth-dependent interactions among dinoflagellates and associated bacteria in culture can be identified with respect to specific bacterial taxa. For example, Rhodobacterales was the dominant order among bacterial isolates from Prorocentrum PA17, the clone with the highest diversity and abundance of bacterial isolates. Most known Rhodobacterales are capable of synthesizing vitamin B12, an essential nutrient for dinoflagellate growth (Sañudo-Wilhelmy et al., 2014). The most reported Rhodobacterales associated with P. lima is Roseobacter (De Traubenberg et al., 1995; Lafay et al., 1995; Prokic et al., 1998), but genus Roseobacter was not found among bacterial isolates in the current study.
Negative allelochemical interactions between dinoflagellates and bacterial growth are also well known. For instance, Micrococcus has algicidal effects against various dinoflagellates; the cosmopolites Alteromonas and Pseudomonas can synthesize hydrolytic components capable of destroying the cellular wall of dinoflagellates (Park and Lee, 1998; Mayali and Azam, 2004; Shi et al., 2018); and Thalassospira degrades polycyclic aromatic hydrocarbons that control growth of certain dinoflagellates (Kodama et al., 2008; Lu et al., 2016). These bacterial taxa that are putatively bioactive against dinoflagellate growth were isolated from various clonal cultures in the current experiments: Micrococcus from the cell-aggregate medium of Prorocentrum PA3; Alteromonas and Pseudomonas from most of the Prorocentrum clones; and Thalassospira from PA11. Euzebyella sp. was exclusively found in Prorocentrum PA14 and was the only representative of the Cytophaga-Flavobacteria-Bacteroidetes (CFB) group known to synthesize proteins with algicidal effects. For example, CFB bacteria have been reported during the peak and termination of HABs (Mayali and Doucette, 2002; Roth, 2005; Park et al., 2015). Isolate PA14 was heavily colonized by cyanobacteria shortly after bacterial isolation, and the dinoflagellate subsequently died, which may indicate the association of CFB-member Euzebyella with the senescence of this Prorocentrum in high cell density culture. Further experiments are required to determine the algicidal potency of specific bacterial isolates against benthic Prorocentrum clones.
The role (if any) of particular bacteria from various compartments (i.e., free-living, extracellular aggregates, externally cell-attached, endosymbiotic intracellular) on DSP toxin biosynthesis and biotransformation remains unresolved. Environmentally determined mechanisms controlling cell proliferation and the coupling of toxin biosynthesis within the cell division cycle of P. lima are defined under controlled laboratory conditions (Pan et al., 1999), but without reference to associated bacteria. Low levels of OA were found in the extracellular fraction (about 1% of the total toxin) in previous studies with bacteria in P. lima cultures (De Traubenberg, 1993; Lee et al., 2016); it was not clear if this represented toxin synthesized (and then leaked or excreted) by the dinoflagellates or associated bacteria. Perez et al. (2008) found that 50% of all rRNA bacterial sequences originating from toxic OA-producing strains of Prorocentrum belonged to genus Roseobacter, but were not present in non-toxigenic species Prorocentrum donghaiense or Prorocentrum micans. Phylogenetic analysis of the polyketide synthase (PKS) genes, essential for biosynthesis of the polyketide OA-analogs, showed that most PKS genes within a Prorocentrum-specific bacterial PKS clade were indeed non-modular bacterial type, but were unrelated to PKSs of Roseobacter or other α-proteobacteria. Despite some early evidence that certain bacteria may contain the PKS genes capable of synthesizing these polyketide toxins (Perez et al., 2008), it is now doubtful that bacteria alone can produce OA analogs, because unlike certain dinoflagellates, they do not possess the complete genetic machinery.
This lack of genetic capacity for DSP toxin biosynthesis does not rule out a potential role for associated bacteria as a modular or effector for toxin biosynthesis or release from dinoflagellate cells. The NMDS plot showed a positive correlation between Prorocentrum PA1 and PA3 with cellular DTX1a-D8. Neither of these Prorocentrum strains presented culturable members of α-proteobacteria, which may indicate they were in stationary-senescence growth phase and thus produced this esterified analog under unfavorable growth conditions (Hu et al., 2017). None of the other Prorocentrum strains and their isolated bacteria showed any correlation with the identified toxins in this study, implying that these bacteria do not interfere directly with toxin production or composition in the dinoflagellates. This provides further evidence that toxins are produced by Prorocentrum alone and not by their associated extracellular bacteria.
Prorocentrum Bioactivity Against Bacteria
The potent inhibitory activity of certain DSP toxins against ubiquitous serine-threonine protein phosphatases (PP1A and PP2A) led to the suggestion that these compounds may be produced by dinoflagellates as defensive bacteriostatic agents, particular for epibenthic dinoflagellates (Cembella, 2003). Literature regarding bioactivity of Prorocentrum species is scarce since most investigations have focused on the production and composition of their DSP toxins, without reference to toxin-related bioactivity. Nagai et al. (1990) demonstrated potent antifungal activity against Aspergillus niger, Penicillium funicullosum, and Candida rugosa from extracts containing OA and DTX1, but other antibacterial assays involving these compounds did not show significant results (Bajpai, 2016; Falaise et al., 2016). Antimicrobial assays based on the microdilution method revealed that methanolic extracts of P. hoffmannianum 1031 prepared from the cell-free culture medium, inhibited the growth of the bacterium Enterococcus faecalis and the fungus Candida albicans by 100 and 98%, respectively (De Vera et al., 2018). Other OA-producing Prorocentrum species were also tested under the same conditions, but no antimicrobial activity was found; these authors suggested the presence of specific antimicrobial compounds (presumably not DSP toxins) for P. hoffmannianum 1031.
Prorocentrum species can either inhibit or stimulate, or have no effect, on bacterial growth, depending on the target bacteria (De Vera et al., 2018). The bioactivity screening in the experiments reported here did not reveal any apparent allelochemical effect on the bacterial growth of Staphylococcus sp. DMBS2 or Vibrio sp. HEL66 from either cell-extracts or supernatants from Prorocentrum PA1. Initial results showed a slightly positive growth inhibition, but this was not sustained through the dilution series. Of course, it is possible that higher biomass equivalents in the cell extracts could yield bioactivity against bacteria from Prorocentrum metabolites, but not at environmentally realistic exposure concentrations. The bacterial isolates for the bioassays originated from the North Sea and were deliberately selected, rather than bacterial isolates from the Prorocentrum cultures from Mexico, to eliminate any conceivable possibility of resistance or co-evolution from prior exposure. Negative evidence of allelochemical interactions of Prorocentrum metabolites, i.e., antibiotic effect against these two naïve strains, does not preclude the possibility that allelochemicals and/or polyketide toxins could be effective growth inhibitors against other potentially competing bacteria in their own native benthic environment, and even against other strains of Staphylococcus and Vibrio. Further experiments considering extracts prepared from axenic versus non-axenic Prorocentrum cultures would help to resolve the issue of bioactivity against its naturally occurring associated bacteria (i.e., isolated from Prorocentrum cultures or directly from natural assemblages). Disk diffusion assays with purified DSP toxins would demonstrate the specific potency against bacteria, in the absence of potentially interfering components in crude extracts.
Time-Series Growth and Toxin Composition of Prorocentrum With Bacteria
The maximum growth rate among Prorocentrum PA1 cultures from tropical Mexican waters varied from 0.17 to 0.35 div day–1. Similar values, ranging from 0.06 to 0.3 div day–1 (Morton et al., 1992) and 0.2 to 0.35 div day–1 (Morton and Tindall, 1995) were recorded for cultured P. lima isolated from Florida, United States, and Heron Island, Australia, when grown in K medium and under similar temperature and light conditions to those reported herein. The length of active growth (i.e., until stationary phase) in the Prorocentrum PA1 experiments with alternative bacterial treatments is also roughly consistent with literature values. Exponential growth that exceeded 60 days in P. lima batch cultures has been reported (Pan et al., 1999; Varkitzi et al., 2010; Ben-Gharbia et al., 2016), but most active growth phases do not last longer than 16 (Vale et al., 2009) or 25 days (Nascimento et al., 2005). This corresponds with the exponential growth phases from PA1 of about 20–25 days under all treatments.
The most noteworthy finding in the time-series growth experiment with Prorocentrum PA1 was the inability of either cell washing or antibiotic treatment to maintain persistently reduced bacterial load over time, much less to produce axenic cultures. Even beginning with clonal isolates of Prorocentrum cells with sterile isolation techniques, it is difficult to achieve and maintain axenic cultures. Careful washing of bulk Prorocentrum cells in established bacterized batch cultures was only partially effective in reducing the bacterial load, and bacterial cell densities rapidly increased in subsequent culture, presumably due to the increasingly organic-rich environment provided by dinoflagellate growth. Cell washing of benthic dinoflagellates to remove bacteria was complicated by the aggregation of bacteria into mucilaginous clumps and the attachment of bacteria to the Prorocentrum cells. These dinoflagellates tend to grow together in cell aggregates and produce copious exopolysaccharides (EPS) (Vanucci et al., 2010), and this likely interferes with the exposure of these bacteria to the antibiotics. The selected antibiotic cocktail and final concentration were chosen with respect to conventional methods to achieve axenic cultures of phytoplankton but were ineffective for dense assemblages of benthic dinoflagellates in culture. In this context, it is appropriate to be somewhat skeptical of previous references to “axenic” cultures of benthic dinoflagellates, where convincing evidence of bacterial absence is not provided.
The cell toxin quota in strain PA1 decreased at the beginning of the culture cycle, i.e., during acclimation and lag growth phase, but then remained stable through the growth curve, while toxin composition remained stable. Similarly, in a previous study, no significant differences in toxin composition between axenic and non-axenic cultures were found for the closely related toxigenic benthic species P. hoffmannianum (Morton et al., 1994).
Both toxin cell quota and toxin composition for Prorocentrum PA1 were apparently independent of the effect of antibiotics or bacterial cell load throughout the culture cycle. Ben-Gharbia et al. (2016) also reported no variation in cell size between exponential and stationary growth phase from a thermophilic P. lima strain (Bizerte Bay, Tunisia). Conversely, the general tracking of the growth of bacteria with the cell increase in Prorocentrum does not indicate allelochemical suppression of bacteria by metabolites leaked or excreted from the dinoflagellate. No significant variations were noted in the Prorocentrum cell volume between antibiotic-treated and only washed cell cultures throughout the growth curve, further indicating lack of effect on growth and cell division. In any case, the changes in bacterial load among treatments over time provide little evidence that external bacteria directly control Prorocentrum cell division rate or affect cell size under optimal growth conditions. The negative data regarding the influence of bacteria on Prorocentrum growth may be attributable to the fact that dinoflagellate isolates were grown in nutrient-replete highly enriched seawater medium and hence were rather refractory to bacterial influence. This result does not preclude the possibility that bacteria do enhance Prorocentrum growth in natural benthic habitats, particularly when certain micronutrients are limiting, e.g., via nutrient re-cycling or provision of micronutrients such as vitamins or chelated trace metals. The only firm conclusion regarding growth that can be drawn from these laboratory experiments is that under optimal growth conditions, bacterial load and species composition did not influence growth. Factors such as nutrient availability and N:P ratios have been reported to play a major role in toxin production (Varkitzi et al., 2010). Further experiments considering N and/or P deprivation and micronutrient and trace element availability would provide a closer estimation of the potential interactions between Prorocentrum and associated bacteria in the natural benthic environment.
Conclusion and Perspectives
Association of specific bacterial assemblages with the evolution of HABs in the pelagic zone, in particular with the high abundance of γ-Proteobacteria, may also be represented in the benthic realm for dense aggregations of benthic dinoflagellates. Nevertheless, it is premature to conclude that the culturable bacterial community yields an appropriate insight into the bacteria–dinoflagellate interactions modulated in natural blooms. A metagenomic analysis of the bacterial and microeukaryotic assemblages (including the toxigenic dinoflagellates) is required as the first step for such a comparison.
High inorganic nutrient loads in culture medium promote optimal growth of cultured benthic dinoflagellates, which in turn yield enhanced organic substrates for associated bacterial growth. The high component of γ-Proteobacteria in the Prorocentrum cultures may indicate that this microcosm reflects a mature high cell density “bloom” with concomitant selection features. The different taxonomic distribution of cultivable bacteria among culture compartments, i.e., Prorocentrum cell aggregates versus the surrounding medium, does suggest that the phycosphere constitutes a unique microenvironment with high selective capacity for bacteria. The lack of bioactivity of cell extracts and culture supernatants against selected model bacteria does not rule out the possible production of allelochemical metabolites, but does not support arguments for high general potency against bacteria, i.e., related to phosphatase inhibition by DSP toxins. Production of large volume axenic dinoflagellate cultures is essential to unraveling the potential role of bacterial interactions in growth and toxin production. This approach would allow to assay the effect of taxonomically identified specific consortia of naturally co-occurring bacteria on axenic cultures of P. lima to determine effects on growth and toxin production. In any case, current studies provide little evidence that extracellular bacteria play a critical role in regulation of DSP toxin production in Prorocentrum or in the modulation of growth under non-nutrient limited conditions in culture.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
All authors contributed actively to the preparation of data content and writing of this manuscript. LD-R, AC, and YO conducted the dinoflagellate field sampling. LD-R and AC prepared preliminary isolates for culture. LD-R and YO performed the morphotaxonomic analysis of Prorocentrum isolates. LD-R, UT-J, and AC maintained the clonal strains at UNAM, Mexico and AWI, Germany, respectively. MP-Z and SP were responsible for bacterial identification and phylogenetic analysis, while LM-V sequenced the bacterial genes. UT-J and JT conducted the bioactivity assays against bacteria. UT-J performed the toxin extractions and data analysis for quantification of DSP toxins under supervision of BK and JT. UT-J and AC conceived and designed the time series experiment, implemented by UT-J. The coordination of the drafting of the manuscript was led by UT-J, AC, and LD-R.
Funding
Financial contribution for the preparation and publication of this manuscript and associated research activities was provided to AC, BK, and JT via the PACES II Research Program (Topic II Coast: WP3) of the Alfred-Wegener-Institut, Helmholtz-Zentrum für Polar- und Meeresforschung, Germany. The Deutscher Akademischer Austauschdienst (DAAD) made possible the research stay of LD-R at the AWI in order to complete this project on benthic dinoflagellates from Mexico.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Manuel Victoria-Muguira of Dorado Buceo for boat access and logistical support with field sampling, and Manuel Rodríguez-Gómez from the Acuario de Veracruz for donating filtered seawater. Laura Elena Gómez-Lizárraga from the Scanning Electron Microscopy Laboratory at Instituto de Ciencias del Mar y Limnología, UNAM, provided micrographs and Hugo Pérez-López assisted with SEM sample preparation. The help of Annegret Müller for technical assistance in the laboratory and Thomas Max for LC-MS/MS measurements of the DSP toxins at AWI is much appreciated. Jennifer Bergemann assisted with the antimicrobial assays within laboratory facilities graciously provided by Tilmann Harder, Marine Chemistry Group, Bremen University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00569/full#supplementary-material
References
Aligizaki, K., Nikolaidis, G., Katikou, P., Baxevanis, A. D., and Abatzopoulos, T. J. (2009). Potentially toxic epiphytic Prorocentrum (Dinophyceae) species in Greek coastal waters. Harmful Algae 8, 299–311. doi: 10.1016/j.hal.2008.07.002
Bajpai, V. K. (2016). Antimicrobial bioactive compounds from marine algae: a mini review. Indian J. Geo Mar. Sci. 45, 1076–1085.
Basu, S., Deobagkar, D., Matondkar, P., and Furtado, I. (2013). Culturable bacterial flora associated with the dinoflagellate green Noctiluca miliaris during active and declining bloom phases in Northen Arabian Sea. Microb. Ecol. 65, 934–954. doi: 10.1007/s00248-012-0148-1
Bauer, A. W., Kirby, W. M. M., Sherris, J. C., and Turck, M. (1966). Antibiotic suceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496. doi: 10.1093/ajcp/45.4_ts.493
Ben-Gharbia, H., Yahia, O. K. D., Amzil, Z., Chomerat, N., Abadie, E., Masseret, E., et al. (2016). Toxicity and growth assessments of three thermophilic benthic dinoflagellates (Ostreopsis cf. ovata, Prorocentrum lima and Coolia monotis) developing in the Southern Mediterranean basin. Toxins 8, 1–38. doi: 10.3390/toxins8100297
Blackburn, S. I., Bolch, C. J. S., Haskard, K. A., and Hallegraeff, G. M. (2001). Reproductive compatibility among four global populations of the toxic dinoflagellate Gymnodinium catenatum (Dinophyceae). Phycologia 40, 78–87. doi: 10.2216/i0031-8884-40-1-78.1
Buchan, A., LeCleir, G. R., Gulvik, C. A., and González, J. M. (2014). Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698. doi: 10.1038/nrmicro3326
Cembella, A. D. (2003). Chemical ecology of eukaryotic microalgae in marine ecosystems. Phycologia 42, 420–447. doi: 10.2216/i0031-8884-42-4-420.1
Ciccarelli, F. D., Doerks, T., Von Mering, C., Creevey, C. J., Snel, B., and Bork, P. (2006). Toward automatic reconstruction of a highly resolved Tree of Life. Science 311, 1283–1288. doi: 10.1126/science.1123061
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., et al. (2014). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, 633–642. doi: 10.1093/nar/gkt1244
De Traubenberg, C. R. (1993). Interactions Between a Dinoflagellate and its Associated Bacterial Microflora: Role of Bacteria in the Toxicity of Prorocentrum lima Ehrenberg (Dodge). Doctoral dissertation, University of Nantes, Nantes.
De Traubenberg, C. R., Soyer-Gobillard, M. O., Géraud, M. L., and Albert, M. (1995). The toxic dinoflagellate Prorocentrum lima and its associated bacteria. Eur. J. Protistol. 31, 383–388. doi: 10.1016/S0932-4739(11)80450-5
De Vera, C. R., Díaz-Crespín, G., Hernández-Daranas, A., Montalvão-Looga, S., Lillsunde, K.-E., Tammela, P., et al. (2018). Marine microalgae: promising source for new bioactive compounds. Mar. Drugs 16:317. doi: 10.3390/md16090317
Di Rienzo, J. A., Casanoves, F., Balzarini, M. G., Gonzalez, L., Tablada, M., and Robledo, C. W. (2018). Infostat. Universidad Nacional de Córdoba; Argentina: 2018. InfoStat, Versión 2018. London: FCA.
Dodge, J. D. (1975). The Prorocentrales (Dinophyceae). II. Revision of the taxonomy within the genus Prorocentrum. Bot. J. Linn. Soc. 71, 103–125. doi: 10.1111/j.1095-8339.1975.tb02449.x
Durán-Riveroll, L. M., Cembella, A. D., and Okolodkov, Y. B. (2019). A review on the biodiversity and biogeography of toxigenic benthic marine dinoflagellates of the coasts of Latin America. Front. Mar. Sci. 6:148. doi: 10.3389/fmars.2019.00148
Falaise, C., François, C., Travers, M.-A., Morga, B., Haure, J., Tremblay, R., et al. (2016). Antimicrobial compounds from eukaryotic microalgae against human pathogens and diseases in aquaculture. Mar. Drugs 14:159. doi: 10.3390/md14090159
Faust, M. A. (2004). The dinoflagellates of Twin Cays, Belize: biodiversity, distribution, and vulnerability. Atoll Res. Bull. 514, 1–20.
Foden, J., Purdie, D. A., Morris, S., and Nascimento, S. (2005). Epiphytic abundance and toxicity of Prorocentrum lima populations in the Fleet Lagoon, UK. Harmful Algae 4, 1063–1074. doi: 10.1016/j.hal.2005.03.004
Fritz, L., and Triemer, R. E. (1985). A rapid simple technique utilizaing calcofluor white M2R for the visualization of dinoflagellate thecal plates. J. Phycol. 21, 662–664. doi: 10.1111/j.0022-3646.1985.00662.x
Giovannoni, S. J., and Stingl, U. (2005). Molecular diversity and ecology of microbial plankton. Nature 437, 343–348. doi: 10.1038/nature04158
Guidi, F., Pezzolesi, L., and Vanucci, S. (2018). Microbial dynamics during harmful dinoflagellate Ostreopsis cf. ovata growth: bacterial succession and viral abundance pattern. MicrobiologyOpen 7, 1–15. doi: 10.1002/mbo3.584
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Honsell, G., Bonifacio, A., De Bortoli, M., Penna, A., Battocchi, C., Ciminiello, P., et al. (2013). New insights on cytological and metabolic features of Ostreopsis cf. ovata Fukuyo (Dinophyceae): a multidisciplinary approach. PLoS One 8:5729. doi: 10.1371/journal.pone.0057291
Hoppenrath, M., Chomérat, N., Horiguchi, T., Schweikert, M., Nagahama, Y., and Murray, S. (2013). Taxonomy and phylogeny of the benthic Prorocentrum species (Dinophyceae)-A proposal and review. Harmful Algae 27, 1–28. doi: 10.1016/j.hal.2013.03.006
Hu, T., LeBlanc, P., Burton, I. W., Walter, J. A., McCarron, P., Melanson, J. E., et al. (2017). Sulfated diesters of okadaic acid and DTX-1: self-protective precursors of diarrhetic shellfish poisoning (DSP) toxins. Harmful Algae 63, 85–93. doi: 10.1016/j.hal.2017.01.012
Hu, W., Xu, J., Sinkkonen, J., and Wu, J. (2010). Polyketides from marine dinoflagellates of the genus Prorocentrum, biosynthetic origin and bioactivity of their okadaic acid analogues. Mini. Rev. Med. Chem. 10, 51–61. doi: 10.2174/138955710791112541
Keller, M. D., Selvin, R. C., Claus, W., and Guillard, R. R. L. (1987). Media for the culture of oceanic ultraphytoplankton. J. Phycol. 23, 633–638. doi: 10.1111/j.1529-8817.1987.tb04217.x
Kodama, Y., Stiknowati, L. I., Ueki, A., Ueki, K., and Watanabe, K. (2008). Thalassospira tepidiphila sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from seawater. Int. J. Syst. Evol. Microbiol. 58, 711–715. doi: 10.1099/ijs.0.65476-0
Krock, B., Tillmann, U., John, U., and Cembella, A. (2008). LC-MS-MS aboard ship: tandem mass spectrometry in the search for phycotoxins and novel toxigenic plankton from the North Sea. Anal. Bioanal. Chem. 392, 797–803. doi: 10.1007/s00216-008-2221-7
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evo. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lafay, B., Ruimy, R., De Traubenberg, C., Breittmayer, V., Gauthier, M. J., and Christen, R. (1995). Roseobacter algicola sp. nov., a new marine bacterium isolated from the phycosphere of the toxin-producing dinoflagellate Prorocentrum lima. Int. J. Syst. Bacteriol. 45, 290–296. doi: 10.1055/s-0029-1193666
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (New York, NY: John Wiley & Sons), 115–175.
Lawrence, J. E., Bauder, A. G., Quilliam, M. A., and Cembella, A. D. (1998). “Prorocentrum lima, a putative link to diarrhetic shellfish poisoning in Nova Scotia, Canada,” in Harmful Algae, Proceedings of the VIII International Conference on Harmful Algae, eds B. Reguera, J. Blanco, M. Fernandez, and T. Wyatt (Vigo: Xunta de Galicia and IOC of UNESCO), 78–79.
Lawrence, J. E., Grant, J., Quilliam, M. A., Bauder, A. G., and Cembella, A. D. (2000). Colonization and growth of the toxic dinoflagellate Prorocentrum lima and associated fouling macroalgae on mussels in suspended culture. Mar. Ecol. Prog. Ser. 201, 147–154. doi: 10.3354/meps201147
Lee, T. C. H., Fong, F. L. Y., Ho, K. C., and Lee, F. W. F. (2016). The mechanism of diarrhetic shellfish poisoning toxin production in Prorocentrum spp.: physiological and molecular perspectives. Toxins 8:272. doi: 10.3390/toxins8100272
Lu, X., Zhou, B., Xu, L., Liu, L., Wang, G., Liu, X., et al. (2016). A marine algicidal Thalassospira and its active substance against the harmful algal bloom species Karenia mikimotoi. Appl. Microbiol. Biotechnol. 100, 5131–5139. doi: 10.1007/s00253-016-7352-8
Luo, Z., Zhang, H., Krock, B., Lu, S., Yang, W., and Gu, H. (2017). Morphology, molecular phylogeny and okadaic acid production of epibenthic Prorocentrum (Dinophyceae) species from the northern South China Sea. Algal Res. 22, 14–30. doi: 10.1016/j.algal.2016.11.020
Markham, J., and Hagmeier, E. (1982). Observations on the effects of germanium dioxide on the growth of macro-algae and diatoms. Phycologia 21, 125–130.
Mayali, X., and Azam, F. (2004). Algicidal bacteria in the sea and their impact on algal blooms. J. Eukaryot. Microbiol. 51, 139–144.
Mayali, X., and Doucette, G. J. (2002). Microbial community interactions and population dynamics of an algicidal bacterium active against Karenia brevis (Dinophyceae). Harmful Algae 1, 277–293.
Morton, S. L., Bomber, J. W., and Tindall, P. M. (1994). Environmental effects on the production of okadaic acid from Prorocentrum hoffmannianum Faust I. temperature, light, and salinity. J. Exp. Mar. Biol. Ecol. 178, 67–77. doi: 10.1016/0022-0981(94)90225-9
Morton, S. L., Norris, D. R., and Bomber, J. W. (1992). Effect of temperature, salinity and light intensity on the growth and seasonality of toxic dinoflagellates associated with ciguatera. J. Exp. Mar. Biol. Ecol. 157, 79–90. doi: 10.1016/0022-0981(92)90076-M
Morton, S. L., and Tindall, D. R. (1995). Morphological and biochemical variability of the toxic dinoflagellate Prorocentrum lima isolated from three locations at Heron Island. Australia. J. Phycol. 31, 914–921.
Nagahama, Y., Murrar, S., Tomaru, A., and Fukuyo, Y. (2011). Species boundaries in the toxic dinoflagellate Prorocentrum lima (Dinophyceae, Prorocentrales), based on morphological and phylogenetic characters. J. Phycol. 189, 178–189. doi: 10.1111/j.1529-8817.2010.00939.x
Nagai, H., Satake, M., and Yasumoto, T. (1990). Antimicrobial activities of polyether compounds of dinoflagellate origins. J. Appl. Phycol. 2, 305–308. doi: 10.1007/BF02180919
Nascimento, S. M., Mendes, M. C. Q., Menezes, M., Rodríguez, F., Alves-de-Souza, C., Branco, S., et al. (2017). Morphology and phylogeny of Prorocentrum caipirignum sp. nov. (Dinophyceae), a new tropical toxic benthic dinoflagellate. Harmful Algae 70, 73–89. doi: 10.1016/j.hal.2017.11.001
Nascimento, S. M., Purdie, D. A., and Morris, S. (2005). Morphology, toxin composition and pigment content of Prorocentrum lima strains isolated from a coastal lagoon in southern UK. Toxicon 45, 633–649. doi: 10.1016/j.toxicon.2004.12.023
Nielsen, L. T., Krock, B., and Hansen, P. J. (2013). Production and excretion of okadaic acid, pectenotoxin-2 and a novel dinophysistoxin from the DSP-causing marine dinoflagellate Dinophysis acuta - Effects of light, food availability and growth phase. Harmful Algae 23, 34–45. doi: 10.1016/j.hal.2012.12.004
Nishimura, T., Uchida, H., Noguchi, R., Oikawa, H., Suzuki, T., Funaki, H., et al. (2019). Abundance of the benthic dinoflagellate Prorocentrum and the diversity, distribution, and diarrhetic shellfish toxin production of Prorocentrum lima complex and P. caipirignum in Japan. Harmful Algae 96:101687. doi: 10.1016/j.hal.2019.101687
Pan, Y., Cembella, A. D., and Quilliam, M. A. (1999). Cell cycle and toxin production in the benthic dinoflagellate Prorocentrum lima. Mar. Biol. 134, 541–549. doi: 10.1007/s002270050569
Park, B. S., Guo, R., Lim, W.-A., and Jang, S.-K. (2018). Importance of free living and particle asssociated bacteria for the growth of the harmful dinoflagellate Prorocentrum minimum: evidence in culture stages. Mar. Freshw. Res. 69, 290–299.
Park, B. S., Kim, J. H., Kim, J. H., Gobler, C. J., Baek, S. H., and Han, M. S. (2015). Dynamics of bacterial community structure during blooms of Cochlodinium polykrikoides (Gymnodiniales, Dinophyceae) in Korean coastal waters. Harmful Algae 48, 44–54. doi: 10.1016/j.hal.2015.07.004
Park, Y. T., and Lee, W. J. (1998). Changes of bacterial population during the decomposition process of red tide dinoflagellate, Cochlodinium polykrikoides in the marine sediment addition of Yellow Loess. J. Korean Fish. Soc. 31, 920–926.
Parsons, M. L., and Preskitt, L. B. (2007). A survey of epiphytic dinoflagellates from the coastal waters of the island of Hawai’i. Harmful Algae 6, 658–669. doi: 10.1016/j.hal.2007.01.001
Perez, R., Liu, L., Lopez, J., An, T., and Rein, K. S. (2008). Diverse bacterial PKS sequences derived from okadaic acid-producing dinoflagellates. Mar. Drugs 6, 164–179. doi: 10.3390/md6020164
Porter, K. G., and Feig, S. Y. (1980). The use of DAPI for identifying aquatic microfloral. Limnol. Oceanogr. 25, 943–948.
Prokic, I., Brümmer, F., Brigge, T., Görtz, H. D., Gerdts, G., Schütt, C., et al. (1998). Bacteria of the genus Roseobacter associated with the toxic dinoflagellate Prorocentrum lima. Protist 149, 347–357. doi: 10.1016/S1434-4610(98)70041-0
R Core Team (2018). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Reguera, B., Alonso, R., Moreira, Á, Méndez, S., and Dechraoui Bottein, M. Y. (2016). Guide for Designing and Implementing a Plan to Monitor Toxin-Producing Microalgae. IOC Manuals and Guides. Paris: Intergovernmental Oceanographic Commission.
Roth, P. (2005). The Microbial Community Associated With the Florida Red Tide Dinoflagellate Karenia Brevis: Algicidal and Antagonistic Interactions. Doctoral thesis, College of Charleston, Charleston, SC.
Sanger, F., Nicklen, S., and Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 74, 5463–5467. doi: 10.1073/pnas.74.12.5463
Santi-Delia, A., Caruso, G., Melcarne, L., Caruso, G., Parisi, S., and Laganà, P. (2015). “Biological toxins from marine and freshwater microalgae,” in Histamine in Fish and Fishery Products, eds P. Laganà, G. Caruso, C. Barone, G. Caruso, S. Parisi, L. Melcarne, F. Mazzù, and A. Santi Delia (Berlin: Springer), 13–56.
Sañudo-Wilhelmy, S. A., Gómez-Consarnau, L., Suffridge, C., and Webb, E. A. (2014). The role of B vitamins in marine biogeochemistry. Annu. Rev. Mar. Sci. 6, 339–367. doi: 10.1146/annurev-marine-120710-100912
Schiller, J. (1933). Dinoflagellatae (Peridineae) in Monographischer Behandlung. New York, NY: Johnson Reprint Corporation.
Shi, X., Liu, L., Li, Y., Xiao, Y., Ding, G., Lin, S., et al. (2018). Isolation of an algicidal bacterium and its effects against the harmful-algal-bloom dinoflagellate Prorocentrum donghaiense (Dinophyceae). Harmful Algae 80, 72–79. doi: 10.1016/j.hal.2018.09.003
Stein, F. R. (1883). Der Organismus der Infusionsthiere. III Abt. Der Organismus der Arthodelen Flagellaten. II. Hälfte. Die Naturgeschichte der Arthrodelen Flagellaten.. Leipzig: Einleitung und Eklärung der Abbildungen, 25.
Vale, P., Veloso, V., and Amorim, A. (2009). Toxin composition of a Prorocentrum lima strain isolated from the Portuguese coast. Toxicon 54, 145–152. doi: 10.1016/j.toxicon.2009.03.026
Vanucci, S., Guerrini, F., Milandri, A., and Pistocchi, R. (2010). Effects of different levels of N- and P-deficiency on cell yield, okadaic acid, DTX-1, protein and carbohydrate dynamics in the benthic dinoflagellate Prorocentrum lima. Harmful Algae 9, 590–599. doi: 10.1016/j.hal.2010.04.009
Varkitzi, I., Pagou, K., Granéli, E., Hatzianestis, I., Pyrgaki, C., Pavlidou, A., et al. (2010). Unbalanced N:P ratios and nutrient stress controlling growth and toxin production of the harmful dinoflagellate Prorocentrum lima (Ehrenberg) Dodge. Harmful Algae 9, 304–311. doi: 10.1016/j.hal.2009.12.001
Vartoukian, S. R., Palmer, R. M., and Wade, W. G. (2010). Strategies for culture of “unculturable” bacteria. FEMS Microbiol. Lett. 309, 1–7. doi: 10.1111/j.1574-6968.2010.02000.x
Keywords: Prorocentrum, Diarrheic Shellfish Poisoning, bacteria, allelochemical, polyether toxins
Citation: Tarazona-Janampa UI, Cembella AD, Pelayo-Zárate MC, Pajares S, Márquez-Valdelamar LM, Okolodkov YB, Tebben J, Krock B and Durán-Riveroll LM (2020) Associated Bacteria and Their Effects on Growth and Toxigenicity of the Dinoflagellate Prorocentrum lima Species Complex From Epibenthic Substrates Along Mexican Coasts. Front. Mar. Sci. 7:569. doi: 10.3389/fmars.2020.00569
Received: 14 December 2019; Accepted: 22 June 2020;
Published: 10 July 2020.
Edited by:
Elisa Berdalet, Spanish National Research Council (CSIS), SpainReviewed by:
Sai Elangovan S., National Institute of Oceanography (CSIR), IndiaRichard Wayne Litaker, National Centers for Coastal Ocean Science (NOAA), United States
Copyright © 2020 Tarazona-Janampa, Cembella, Pelayo-Zárate, Pajares, Márquez-Valdelamar, Okolodkov, Tebben, Krock and Durán-Riveroll. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Allan D. Cembella, allan.cembella@awi.de; Lorena M. Durán-Riveroll, lorena.duran@awi.de; lduran@conacyt.mx
 Ulrike I. Tarazona-Janampa
Ulrike I. Tarazona-Janampa Allan D. Cembella
Allan D. Cembella María C. Pelayo-Zárate
María C. Pelayo-Zárate Silvia Pajares
Silvia Pajares Laura M. Márquez-Valdelamar6
Laura M. Márquez-Valdelamar6  Yuri B. Okolodkov
Yuri B. Okolodkov Bernd Krock
Bernd Krock Lorena M. Durán-Riveroll
Lorena M. Durán-Riveroll