- 1Laboratory for Transcriptional Regulation, RIKEN Center for Integrative Medical Sciences, Yokohama, Kanagawa, Japan
- 2Department of Pediatrics and Developmental Biology, Graduate School of Medical and Dental Sciences, Tokyo Medical and Dental University (TMDU), Tokyo, Japan
- 3Laboratory for Structural Bioinformatics, RIKEN Center for Biosystems Dynamics Research, Yokohama, Kanagawa, Japan
AIOLOS, encoded by IKZF3, is a member of the IKZF family of proteins that plays an important role in regulating late B-cell differentiation. Human individuals heterozygous for the AIOLOS p.N160S variant displayed impaired humoral immune responses as well as impaired B and T cell development. We have previously reported that a mouse strain harboring an Ikzf3N159S allele that corresponds to human IKZF3N160S recapitulated immune-deficient phenotypes, such as impaired B cell development and loss of CD23 expression. In this study, we investigated the effect of the Ikzf3N159S variant and found that B1a cell development was impaired in Ikzf3N159S/N159S mice. In addition, CD62L expression was severely decreased in both B and T lymphocytes by the Ikzf3N159S mutation, in a dose-dependent manner. Mixed bone marrow chimera experiments have revealed that most immunodeficient phenotypes, including low CD62L expression, occur in intrinsic cells. Interestingly, while Ikzf3N159S/N159S lymphocytes were still present in the spleen, they were completely outcompeted by control cells in the lymph nodes, suggesting that the capacity for homing or retention in the lymph nodes was lost due to the Ikzf3N159S mutation. The homing assay confirmed severely decreased homing abilities to lymph nodes of Ikzf3N159S/N159S B and T lymphocytes but selective enrichment of CD62L expressing Ikzf3N159S/N159S lymphocytes in lymph nodes. This finding suggests that impaired CD62L expression is the major reason for the impaired homing capacity caused by the Ikzf3N159S mutation. Interestingly, an excess amount of Ikaros, but not Aiolos, restored CD62L expression in Ikzf3N159S/N159S B cells. Together with the loss of CD62L expression due to Ikaros deficiency, the AiolosN159S mutant protein likely interferes with Ikaros function through heterodimerization, at least in activating the Sell gene encoding CD62L expression. Thus, our results revealed that AiolosN159S causes some immunodeficient phenotypes via the pathogenesis referred to as the heterodimeric interference as observed for AiolosG158R variant.
Introduction
Inborn errors of immunity (IEI), also known as primary immunodeficiency disorders (PIDs), are characterized by defects in development or dysfunction of the immune system, resulting in increased susceptibility to infections or dysregulated immune responses (1). IEIs are a heterogeneous group of disorders characterized by a defect in the function of at least one, and often more, components of the immune system (2). Germline variants in single genes often caused IEI, and more than 450 genes have been identified to be related to IEIs (1). Some IEIs are caused by monoallelic variants through haploinsufficiency, negative dominance, or gain of function (GOF). Among these monoallelic variants, missense variants in the genes encoding transcriptional regulators have received attention. For instance, variants of STAT1, c-MYB, and BCL11B, have been isolated from human patients with IEI (3–6).
The IKAROS (IKZF) family members include IKZF1 (Ikaros), IKZF2 (Helios), IKZF3 (Aiolos), IKZF4 (Eos), and IKZF5 (Pegasus), which are critical regulators of hematopoiesis (7, 8). The IKZF family proteins function as either homo- or heterodimer and regulate gene expression in both positive and negative manners (9). Structurally, IKZF family proteins are characterized by four zinc fingers (ZFs) in the middle of the molecule for binding to DNA and two ZFs at the C-terminus for homo- and heterodimerization with IKZF members (10). IKAROS/IKZF1, the first and most studied member of the IKZF family, plays a key role in generating early hematopoietic progenitors and is necessary for lymphoid differentiation (10, 11). In a murine Ikzf1 mutant allele that removes ZF1, ZF2, and ZF3 that are responsible for DNA binding, a truncated Ikzf1 protein lacking DNA-binding capacity functions as a dominant negative (DN) form. In mice homozygous for this Ikzf1 mutant allele (Ikzf1DN/DN mice), the generation of lymphoid progenitors is severely damaged, thereby generating almost no mature B and T lymphocytes (7, 8). Natural killer (NK) cell development is also inhibited in Ikzf1DN/DN mice (12). Thus, studies in mice have revealed the critical roles of Ikzf1 in the hematopoiesis and lymphoid cell development (7, 8, 12–14). Moreover, heterozygous variants of IKZF1 have been reported from human patients with combined immunodeficiency (15–19). IKZF1 creates macromolecular complexes that produce different functional activities in the hematopoietic, lymphopoietic, and non-lymphoid systems (20–24). Among IKZF family proteins, AIOLOS/IKZF3 shows the highest homology to IKAROS and has been confirmed to make heterodimerize with IKAROS (20). However, contrary to IKZF1, IKZF3 is not expressed in hematopoietic stem or progenitor cells. Its expression in the bone marrow is first detected at low levels in pro-B cells and is upregulated as this progress to pre-B cells and mature peripheral B cells (16, 25). Mouse studies have shown that Aiolos promotes activation and maturation of B lymphocytes (25). Recently, a missense mutation in IKZF3, IKZF3G159R, has been identified in a human family with impaired B-cell development (16). Yamashita et al. proposed that B cell deficiency in IKZF3+/G159R patients is caused by hijacking IKZF1 function by the IKZF3G159R variant, a pathogenesis referred to as “heterodimeric interference” (16, 26). Another IKZF3 variant encoding AIOLOSN160S was identified in the patients presenting with hypogammaglobulinemia, susceptibility to Pneumocystis jirovecii pneumonia, and chronic lymphocytic leukemia (27). In mice harboring Ikzf3N159S corresponding to the human IKZF3N160S, certain immune phenotypes observed in human patients, such as loss of follicular T cells and decreased CD23 expression, were shown to be recapitulated (27).
In this study, we further characterized Ikzf3N159S mutant mice and found CD62L expression was severely reduced in both B and T lymphocytes by AiolosN159S in a dose-dependent manner. The homing assay confirmed the impaired homing ability of Ikzf3N159S/N159S T and B lymphocytes. Selective homing of CD62L expressing Ikzf3N159S/N159S cells into the lymph nodes (LNs) suggested that low CD62L expression is a major reason for reduced homing activity. Retroviral transduction of Ikaros, but not Aiolos, into Ikzf3N159S/N159S B cells restored CD62L expression, indicating that AiolosN159S reduces CD62L expression by disturbing the function of Ikaros.
Materials and methods
Mice
The Ikzf3N159S mutant mouse strain was generated by CRISPRCas9–mediated genome editing and has been reported (27). CD45.1 congenic mice and Rag1-deficient mice were obtained Jackson Laboratory. All mice were maintained in the animal facility at RIKEN Center for Integrative Medical Sciences, and all animal procedures were in accordance with institutional guidelines for animal care and the protocol approved by the Institutional Animal Care and Use Committee of RIKEN Yokohama Branch (2020–026).
Antibodies
Anti-Ikaros (MABE912) and anti-Aiolos (MABE911) purchased from Merck were used for western blot experiments. Antibodies and clone names used for flow cytometry were CD43 (S7), BP-1 (BP-1), IgM (R6-60.2), B220 (RA3-6B2), CD11b (M1/70), CD19 (1D3), CD24 (M1/69), CD93 (AA4.1), CD62L (MEL-14), CD44 (IM7), CD4 (RM4-5), CD8a (53-6.7), TCRβ (H57-597), CD3e (17A2), IgD (11-26c.2a), CD23 (B3B4), CD21 (7G6), CD5 (53-7.3), CD45.1 (A20), CD45.2 (104), Ly6G (1A8), Ly6C (HK1.4), CD11c (HL3), CD127 (A7R34) and NK1.1 (PK136) and were purchased from either BD Bioscience or BioLegend.
Flow cytometry and definition of cell types
Flow cytometry experiments and fluorescence-activated cell sorting were performed on FACSCanto™ II (BD Biosciences) and FACSAria (BD Biosciences), respectively, and were analyzed by FlowJo software (Tree Star). Cell types were defined as follows: B cells (B220+CD19+), B-1 cells (CD19hiB220lo), B-2 cells (CD19loB220hi), B-1a cells (CD19hiB220loCD5+), B-1b cells (CD19hiB220loCD5−), T cells (TCRβ+CD3+), naïve T cells (CD62L+CD44−), central memory cells (CD62L+CD44+), effector memory cells (CD62L−CD44+), FO B cells (CD19+CD21intCD23hi), MZ B cells (CD19+CD21hiCD23lo/-). B cells in bone marrow, Fr.A (B220+CD43+Bp-1−CD24−), Fr.B (B220+CD43+Bp-1−CD24+), Fr.C (B220+CD43+Bp-1+CD24+), Fr.D (B220+CD43−AA4.1+IgM−), Fr.E&F (B220+CD43−AA4.1+IgM+) and recirculating B cells (B220+CD43−AA4.1−IgM+),dendritic cells (B220−TCRβ−CD11b+CD11c+), natural killer cell (B220−TCRβ−CD11b+CD11c−NK1.1+Ly6G−), neutrophils (B220−TCRβ−CD11b+CD11c−NK1.1−Ly6G+), eosinophils (B220−TCRβ−CD11b+CD11c−NK1.1−Ly6G−Ly6Clo), monocytes/macrophages (B220−TCRβ−CD11b+CD11c−NK1.1−Ly6G−Ly6C−), inflammatory monocytes (B220−TCRβ−CD11b+CD11c−NK1.1−Ly6G−Ly6Chi).
RNA-Seq
RNA-seq was performed using RNA extracted from purified splenic T cells (TCRβ+CD4+CD44−CD127+) or naïve B cells (B220+CD19+) from Ikzf3+/+ (n=2 or 4), Ikzf3+/N159S (n=2) and Ikzf3N159S/N159S (n=2) mice. Total RNA was extracted by using Trizol reagent (Invitrogen). The mRNA was obtained by poly(A) selection with NEBNext Poly(A) mRNA Magnetic Isolation Module (E7490). RNA-seq libraries were prepared by NEBNext UltraIIRNA Library Prep Kit for Illumina (E7770) following the manufacturer’s protocol. Briefly, the first and second strands of cDNA were synthesized from mRNA, and were end-repaired, dA tailed and ligated with an adapter (E7335). The adapter-ligated DNA fragments were size selected by dual bead-based size selection using AMPure XP Beads (A63881). The size-selected DNA was amplified by PCR for 11–12 cycles. The purified DNA was diluted to 1 ng and was sequenced with a HiSeqX sequencer (Illumina) at Macrogen.
Trimmomatic is used to trim reads and removing adapter sequences. For analysis of differentially expressed genes (DEGs), RNA-seq reads were mapped to the murine genome (mm10) using HISAT2 (version 2.2.1) with default parameters, and read counts were calculated with HTseq (version2.0.2) using refGene as genome index files (28). Generated read counts were analyzed with the R package edgeR (version 3.38.4) (29). For gene ontology (GO) analysis, downregulated DEGs with false discovery rate (FDR) <0.05 in Ikzf3N159S/N159S T cells or B cells compared to Ikzf3+/+ controls were extracted and subjected to the analysis. GO enrichment analysis was performed with the R package clusterProfiler (version 4.4.4) (30).
And the ENCODE database showed the Ikaros protein binds to SELL gene in human B-Lymphocyte cell line (GM12878 cell line). And Harmonizome databases extract and organize the genes or proteins from publicly available resources (31). In Harmonizome databases, we search and confirm which down-regulated genes of RNA-seq data are the target genes of the IKZF1 transcription factor in ChIP-seq datasets, and then verified them in the ENCODE database.
Plasmid preparation
We constructed retroviral vectors for generating retroviruses to infect mouse B cells. The entire coding region of mouse Ikaros/Ikzf1 (NM_001025597.3) was amplified from cDNA prepared from wild-type splenic cells by RT-PCR and was cloned into pCR2.1®-TOPO vector by TOPO TA Cloning Kit (Invitrogen). After confirming the sequence, the cDNA fragment was cloned into the pMigR-DsRED plasmid, which was generated by ourselves via replacing the GFP sequence with DsRED in the pMigR1 plasmid (Addgene Plasmid #27490). Similarly, cDNA encoding mouse Aiolos/Ikzf3 (NM_011771.1) was amplified by RT-PCR and was cloned into pMigR1 plasmid. Primers sequences were following: Ikzf1-Forward : CCCAGATCTAGACAATGGATGTCG-ATGAGG, Ikzf1-Reverse : CCCCTCGAGGGTTTAGCTCAGGTGGTAAC, Ikzf3-Forward : CCCGCGGCCGCCGGCGACATGGAAGATATAC, Ikzf3-Reverse : CCCCTC GAGTGCTCACTTCAACATGGCTC
Retroviral transduction into B cells
Plate-E packaging cell line was cultured in DMEM medium (Gibco) supplemented with 10% FBS (Hyclone), 1% penicillin/streptomycin (Gibco), 10 µg/ml Blasticidin (Gibco) and 1 µg/ml Puromycin (Gibco) at 37°C with 5% CO2. Plate-E cells (2*105/ml in a 6-well dish) were transfected with 2 µg of the retroviral vector using a FuGENER HD Transfection Reagent (Promega) at a 3:1 ratio (FuGENER HD: DNA mass per well). After 48h, the supernatant was collected. B cells from WT and Ikzf3N159S/N159S mice were isolated using B cell Microbead (Miltenyi Biotec) with purity over 92%. 106 of these B cells were stimulated with 10 µg/ml LPS (Sigma) in RPMI medium (Gibco), supplemented with 10% FBS, 50 µmol 2-mercaptoethanol (Sigma), 1mM sodium pyruvate (Gibco), 1% penicillin/streptomycin (Gibco) and 1% MEM (Gibco). Two days after stimulation, viral supernatant was added with 20 mg/ml polybrene (Greiner bio-one) to the cells in a 24-well plate. To enhance transduction efficiency, cells were subjected to a round of centrifugation with vector-containing medium for 90min at 2000rpm and 32°C, separated by a 2-hour interval in which cells were cultured in medium without viral particles. Two days after infection, the cells were FACS analyzed for various purposes as indicated.
Generation of bone marrow chimeras
For the generation of bone marrow chimeras, bone marrow (BM) cells were prepared from CD45.2 WT or Ikzf3N159S/N159S mice and CD45.1 wild type mice. Those donor-derived CD45.2 bone marrow cells were mixed 1:1 with bone marrow cells from CD45.1+ wild type cells and were transferred intravenously into sub-lethally (950 rad) irradiated Rag1−/− recipients. The recipient mice were analyzed 8–12 weeks after bone marrow injection.
Homing assay
The homing assay was performed as described (32–35) with slight modifications. Single splenic cell suspensions were prepared from CD45.2 WT or Ikzf3N159S/N159S mice and CD45.1 WT mice. Equal numbers (3-5 × 107) of CD45.2 WT or Ikzf3N159S/N159S cells and CD45.1 WT cells were mixed and were intravenously co-injected into CD45.1/2 recipient mice. An aliquot of the injected cells mixture was analyzed by flow cytometry to confirm the injected ratio of CD45.1+ and CD45.2+ (Ri) in B220+CD19+ or TCRβ+CD3+ population. After 24h of migration, single-cell suspensions of blood and organs were prepared and were stained with antibodies. The ratio of CD45.1+CD45.2− and CD45.2−CD45.2+ (Ro) was determined by flow cytometry. The ratio of CD45.1+CD45.2− and CD45.2−CD45.2+ cells in B220+CD19+ or TCRβ+CD3+ population within individual injection mixture (Ri) and organs (Ro) was measured, and the homing assay results were presented as the ratio of Ro/Ri in each tissue.
Template selection and homology modeling
Given the absence of experimentally determined three-dimensional structure for human AIOLOS, a homology model of the four N-terminal zinc fingers was constructed. Initially, a template search was performed using BLASTP against the RCSB protein data bank (PDB) with the human AIOLOS sequence as the query, leading to the identification of potential template sequences. After careful evaluation, a template fragment of human PR/SET domain 9 (PRDM9c) with a PDB ID of 5V3G, encompassing six zinc fingers, was selected (36–38). The choice was based on its sequence identity of 38%, absence of gaps (0% gap), 55% positive residues, and a high BLAST bit score of 224.
To generate the homology models, MODELLERv9.17 was employed (39). Initially, the target and template sequences were aligned using MODELLER’s align2d command, which employs global dynamic programming with a linear gap penalty function to align the two profiles. Subsequently, 20 three-dimensional models of human AIOLOS with four N-terminal zinc fingers and DNA were built using MODELLER. Among the generated models, the one with the lowest DOPE (Discrete Optimized Protein Energy) score, as well as visual inspection, was chosen for further analysis (40). This selected model represents the complex of human AIOLOS with four N-terminal zinc fingers, cognate DNA sequence of AIOLOS, and zinc ions, and served as the starting structure for subsequent molecular dynamics simulations and analyses.
Molecular dynamics simulations and analyses
To investigate the behavior of the N160S as compared to wild-type (WT) and G159R-mutant structures, we first prepared the N160S-mutant by mutating the N160 to S160 in silico, while preserving the secondary structure. Subsequently, molecular dynamics (MD) simulations were performed. The initial step involved the addition of hydrogen atoms to all the systems. Subsequently, these structures were solvated using the three-point transferable intermolecular potential (TIP3P) water model in an octahedral box, while maintaining a distance of approximately 10 Å between the protein-DNA complex surface and the box boundary (41). The systems were then neutralized by introducing counter ions, and the simulations were performed using GROMACS 5.1.2 with the ff14SB forcefield (42, 43). The MD simulations commenced with an energy minimization phase, consisting of 50,000 steps, followed by a gradual heating process from 0 to 300 K over 500 ps. Subsequently, a constant temperature equilibration was performed for 3000 ps at 300 K. To ensure velocity stability, the Parrinello-Rahman barostat pressure coupling method was employed. The production runs for the WT, N160S and G159R mutants lasted 500 ns, employing a periodic boundary condition in the NPT ensemble. Berendsen temperature coupling with modifications was utilized to maintain a constant temperature of 300 K and a pressure of 1 atm. The leap-frog integrator and the Verlet cut-off schemes were employed during the simulations (44). To preserve bond lengths, the LINCS algorithm was used, while the Particle-mesh Ewald method was employed for the calculation of electrostatic forces. In these calculations, Fourier grid spacing and Coulomb radius values of 0.16 nm and 1.4 nm, respectively, were utilized. The standard van der Waals interactions were set to 1.4 nm. To analyze the MD simulated trajectories of the three systems, several key metrics were calculated. The root means square deviation (RMSD), root mean square fluctuation (RMSF), and radius of gyration (Rg) were determined, for which the GROMACS utilities, namely gmx rms, gmx rmsf, and gmx gyrate, were employed for these calculations respectively, providing essential insights into the structural stability and flexibility of the systems. The MD simulation trajectories were visualized using Visual Molecular Dynamics (VMD) (45). Molecular visualization and figure generation were accomplished using PyMOL (http://www.pymol.org).
Results
Impaired B1a cell development in AiolosN159S mutant mice
We have previously reported that the knock-in mouse model carrying the Ikzf3N159S mutant allele generating the AiolosN159S missense mutant protein, which corresponds to the human AIOLOSN160S variant, recapitulated the abnormal B and T lymphocyte development similarly to what was observed in the patients with IKZF3N160S variant. B cell frequencies in peripheral blood (PBL) was decreased in AiolosN159S/N159S mice (27). In addition, we showed that the expressions of IgM, IgD, and CD23 significantly decreased in B lymphocyte populations in both Ikzf3+/N159S and Ikzf3N159S/N159S mice (27). To clarify the developmental stages of B cell development impaired by the AiolosN159S mutation, we examined early B cell development in the bone marrow according to the criteria of Fractions A to G defined by Hardy (46). No significant differences were observed in the frequencies of fractions A-C among Ikzf3+/+, Ikzf3+/N159S and Ikzf3N159S/N159S mice (Figures 1A, B). However, in the CD11b−B220+CD43− population, AA4.1+IgM+ cells corresponded to Fr. E & F were increased in Ikzf3N159S/N159S mice with a decrease in recirculating B cells, which were defined as CD11b−B220+CD43−AA4.1−IgM+ (Figures 1A, B). In contrast, recirculating B cells increased in Ikzf3+/N159S mice (Figures 1A, B), which is consistent with the tendency of increased splenic B cell frequency in Ikzf3+/N159S mice (27). Therefore, an increase in Fr. E & F in Ikzf3N159S/N159S mice might reflect a relative increase due to the low frequency of recirculating B cells, rather than the accumulation of immature B cells by developmental blockade.
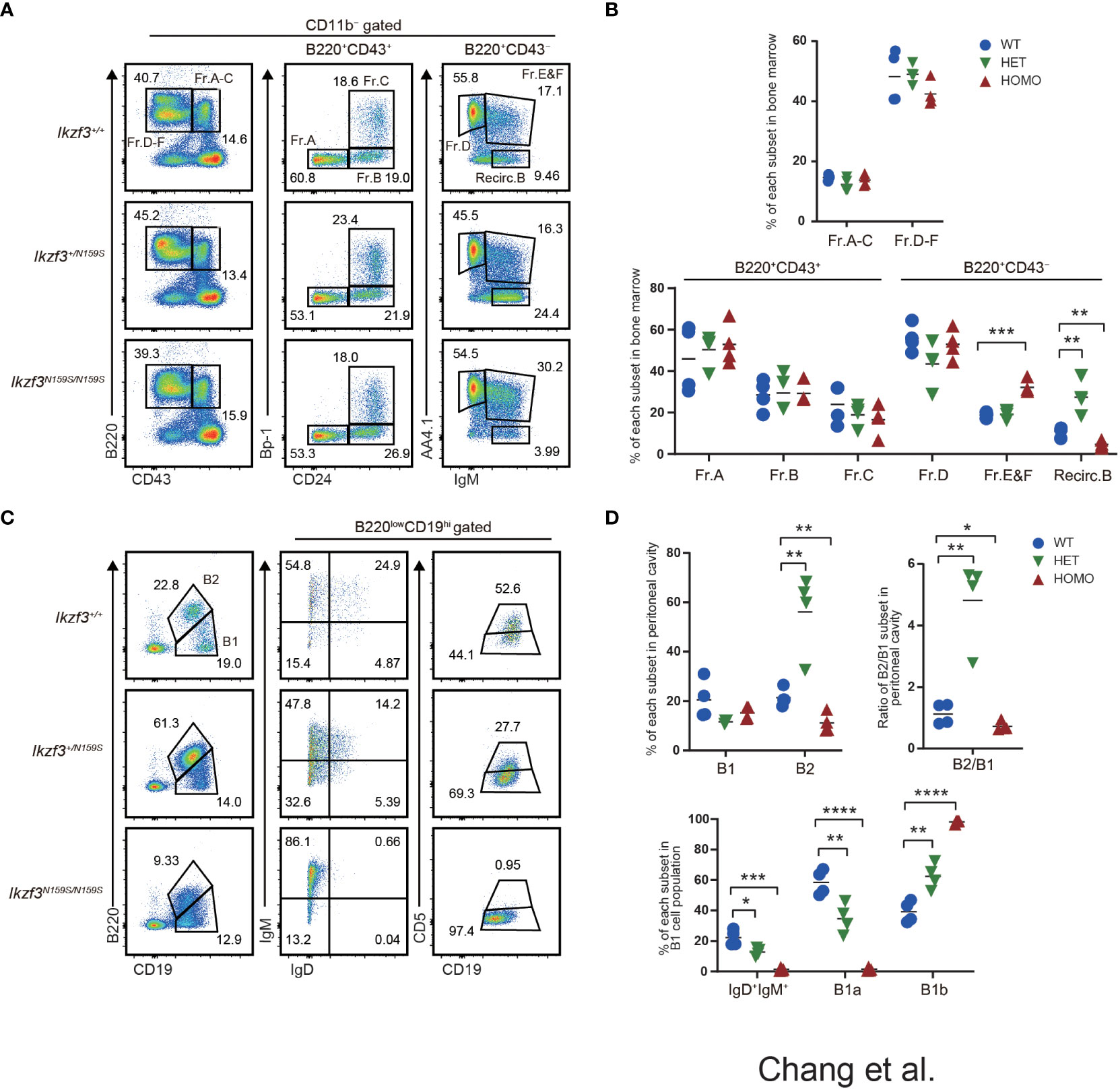
Figure 1 Impaired B1a cell development in Ikzf3N159S/N159S mice. (A) Flow cytometric analysis of bone marrow cells from 6-12 weeks age mice with indicated genotypes. CD11b−B220+ cells can be resolved into Fr.A-C (B220+CD43+) and Fr.D-E (B220+CD43−) subsets. Fr.A-C and Fr.D-F subsets are divided into six subsets: Fr.A, Bp1−CD24−; Fr.B, Bp-1−CD24+; Fr.C, Bp-1+CD24+; Fr.D, AA4.1+IgM−; Fr.E&F, AA4.1+IgM+; Recirc.B, AA4.1−IgM+. Numerical values in the plots indicate percentages of each subpopulation. (B) Graphs show the frequencies of B cell subsets in the bone marrow (n=3). The horizontal lines indicate the mean values from at least three different mice per group. (C) Flow cytometric analyzes of B1 (CD19hiB220lo) and B2 (CD19loB220hi) cell population in the peritoneal cavity, using the indicated markers. CD19hiB220loCD5+ (B1a) and CD19hiB220loCD5− (B1b) of the peritoneal cavity were stained. FACS analysis of B1 cell population from the peritoneal cavity stained for IgM and IgD. Numerical values in the plots indicate percentages of each subpopulation. (D) Frequencies of B1, B2 population (top-left) and the ratio of B2/B1 (top-right) in the peritoneal cavity from Ikzf3+/+, Ikzf3+/N159S, Ikzf3N159S/N159S mice are shown (n=3). Additionally, the graph below shows the frequencies of IgM+IgD+, B1a and B1b subpopulations from total B1 cells (n=3). Statistically significant differences (Student’s unpaired, two-tailed t test) between the groups are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
B-lymphocytes are divided into B1 and B2 subtypes. B1 cells are an important part of the innate immune response, and spontaneously secrete polyreactive IgM antibodies (47, 48). In mice, B1 cells differ from conventional B2 B cells in that they do not express CD21 and CD23 and express only low levels of B220 (B220loCD19hi) and IgD, but high levels of IgM. B1 cells are abundant in both peritoneal and pleural cavities and can be further divided into B1a (B220loCD19hiCD5+) and B1b (B220loCD19hiCD5−) cells (47, 48). CD5+ B1a cells are thought to be long-lived, self-renewing cells that predominantly reside in these pleural and peritoneal cavities, where they produce natural polyreactive IgM antibodies with a biased, autoreactive repertoire (49).
Given the reduction in B2 cell numbers in Ikzf3N159S/N159S mice, we examined whether B1 cell development was impaired by the Ikzf3N159S mutation. Consistent with the tendency of splenic B2 cell numbers (27), the frequency of B2 cells, defined as CD19loB220hi cells in the peritoneal cavity of Ikzf3+/N159S and Ikzf3N159S/N159S mice, increased and decreased, respectively, thereby increasing and decreasing the B2 to B1 ratio (Figures 1C, D). B2 cells in the peritoneal cavity of both mutants also showed decreased IgD and CD23 expression (Figure S1A), as observed in the spleen (27). In the CD19hiB220lo B1 cell population, the expression level of IgD was also decreased by the Ikzf3N159S mutation in a dose-dependent manner, and IgD+ cells were hardly detected in Ikzf3N159S/N159S mice (Figures 1C, D). B1a cells, defined as CD19hiB220loCD5+ cells, were significantly decreased in the peritoneal cavities of Ikzf3+/N159S and Ikzf3N159S/N159S mice (Figures 1C, D). Since CD5 is the only marker to distinguish B1a and B1b cells, loss of B1a cell population could be either caused by loss of CD5 expression from B1a cells or developmental block of B1a cells. In order to distinguish whether CD5 expression was lost in B1a cells or whether B1a cell development was inhibited by AiolosN159S, we examined CD5 expression in other cells such as T lymphocytes. In the spleen, the frequency of CD5+ T cells were decreased in Ikzf3+/N159S mice with severer reduction in CD8+ T cells (Figure S1B). Primary T cell development in the thymus is divided into four stages according to CD4 and CD8 co-receptor expression: most immature CD4−CD8− (DN) stage is flowed by CD4+CD8+ (DP) precursor stage, and DP precursor is maturated into CD4+CD8− (CD4 SP) or CD4−CD8+ (CD8 SP) stage after positive selection. In thymic T cells, the frequency of CD5 expressing cells was significantly decreased in the CD4+CD8+ (DP) and CD4−CD8+ (CD8 SP) subsets and increased in the CD4−CD8− (DN) subset (Figure S1C). This indicated that Ikzf3N159S mutation disturbed CD5 expression during the DN to DP stages in the thymus, specifically in the CD8 SP population. However, CD5 expression was not completely lost, suggesting that the loss of CD5+ B1a population was likely caused by developmental inhibition rather than a loss of CD5 expression.
Impaired CD62L expression on T and B cells by AiolosN159S variant
Although just from a single patient, RNA-seq analyses of naïve B cell from such a patient carrying IKZF3N160S variant showed lower expression of SELL gene which encodes CD62L (27). In Ikzf3N159S/N159S mice, circulating T cells in the peripheral blood, particularly CD8+ T cells, showed decreased CD62L expression (27). We then next examined the developmental stages that initiate low CD62L expression during T cell development. In primary T cell development of thymus, flow cytometry analyses detected low CD62L expression of thymocytes of Ikzf3+/N159S and Ikzf3N159S/N159S mice from CD4+CD8+ (DP) stage onward, while CD62L expression on CD4−CD8− DN thymocytes was comparable (Figures 2A, B). In mice, RNA-seq data available in the ImmGen database showed that the expression of Ikzf3 gene is initiated from the DN to DP transition, whereas the Ikzf1 gene is expressed earlier in the DN1 stage (Figure S2A). Thus, low CD62L and CD5 expression was correlated with the expression of Ikzf3N159S gene, suggesting that the AiolosN159S mutant protein quickly suppresses CD62L expression in a dominant-negative form. Given that CD62L is expressed in mature B cells, we also examined CD62L expression in splenic B220+ CD19+ B cells and found that expression of CD62L is heavily decreased in Ikzf3+/N159S and Ikzf3N159S/N159S mice (Figures 2C, D). Furthermore, B220+CD19+ cells in the peripheral blood and peripheral LN (pLN), and mesenteric LN (mLN), and B2 and B1 cells in the peritoneal cavity showed decreased CD62L expression in Ikzf3N159S/N159S mice (Figures S2B, C). Interestingly, the frequency of CD62L+ B cells varied in secondary lymphoid tissues of Ikzf3+/N159S mice.
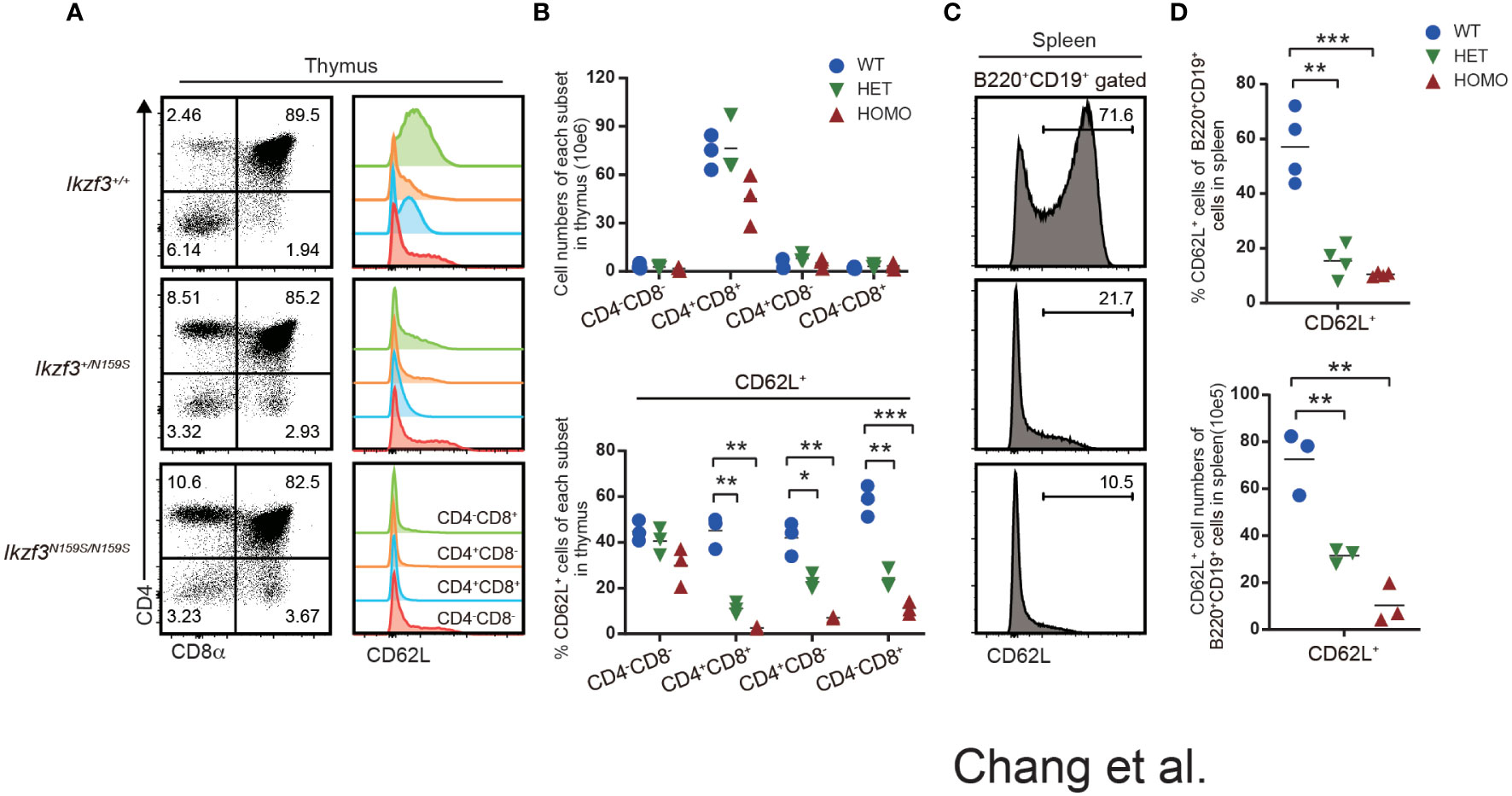
Figure 2 Low CD62L expression on T and B cells by AiolosN159S variant. (A) Flow cytometric analyses of thymocytes from Ikzf3+/+, Ikzf3+/N159S, Ikzf3N159S/N159S mice, at age of 6-12 weeks. CD4 and CD8a were used to stain DN (CD4−CD8−), DP (CD4+CD8+), CD4 SP (CD4+CD8−), CD8 SP (CD4−CD8+) thymocyte subsets. CD62L expression levels on each thymocyte subsets are adjacently shown as histograms. (B) Graphs show cell numbers of CD4−CD8−, CD4+CD8+, CD4+CD8− and CD4−CD8+ thymocytes (top) and the percentage of CD62L+ cells within each subset (bottom) (n=3). The horizontal lines indicate the mean values of at least three different mice per group. (C) Flow cytometric analyzes of CD62L expression on splenic B (B220+CD19+) cells. (D) Graphs show the frequency (top) and cell number (bottom) of CD62L+ cells from splenic B cell population from Ikzf3+/+, Ikzf3+/N159S, Ikzf3N159S/N159S mice (n=3). Statistically significant differences (Student’s unpaired, two-tailed t test) between the groups are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Cell intrinsic phenotypes of Ikzf3N159S/N159S cells assessed by mixed bone marrow chimera
We observed that the differentiation of T and B lymphocytes, NK1.1+ natural killer (NK) cells, and eosinophils (Eos) was impaired with the loss of certain markers such as CD23 and CD62L in Ikzf3N159S/N159S mice (27). To address whether these B, T, and myeloid cell phenotypes occur in a cell-intrinsic manner and to test the developmental potency of progenitors under competitive settings, we performed mixed bone marrow (BM) chimera experiments. CD45.1+ bone marrow cells were mixed with CD45.2+ bone marrow cells prepared from either Ikzf3+/+ or Ikzf3N159S/N159S mice in equal proportions and transplanted into sublethally irradiated Rag1 KO recipient mice (Figure S3A). Reconstitution of hematopoiesis in recipients was analyzed 8 weeks after transplantation using flow cytometry. In the total bone marrow cells, the CD45.2+/CD45.1+ ratio was close to 1:1 in recipients injected with CD45.1+ Ikzf3+/+ and CD45.2+ Ikzf3N159S/N159S BM cells (Figures 3A, B). However, during early B cell development, the percentage of CD45.2+ Ikzf3N159S/N159S cells increased in Fr. E and F (Figures S3B, C), suggesting that mature B cells in the bone marrow were retained or accumulated in the presence of AiolosN159S. In B220+CD19+ B cell population in peripheral lymphoid tissues, the percentage of CD45.2+ Ikzf3N159S/N159S cells was significantly decreased to approximately 10% in spleen and was further reduced to less than 1% in pLN, mLN, and peritoneal cavity, comparing with CD45.2+ Ikzf3+/+ cells (Figures 3A, B and S3D).
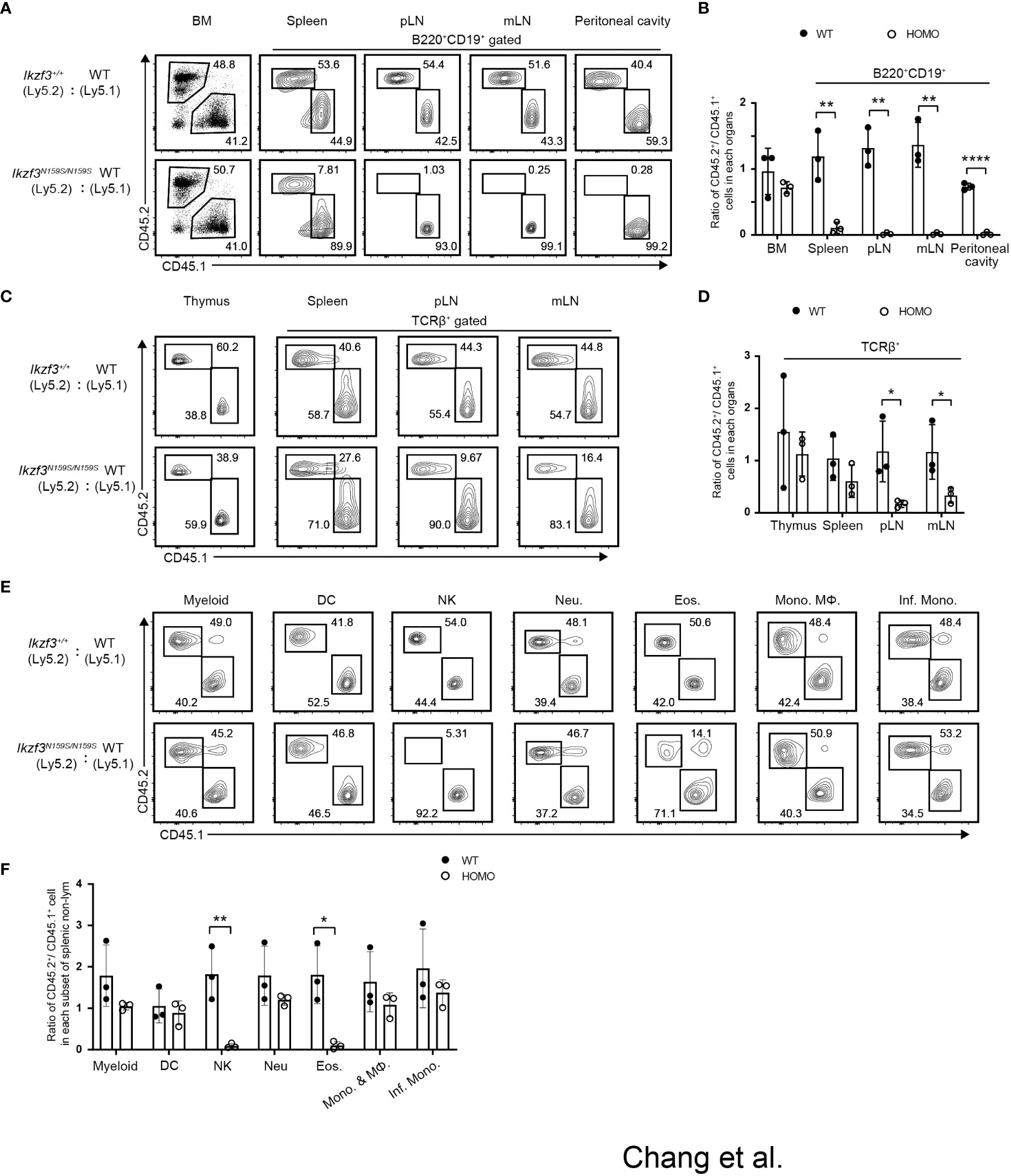
Figure 3 Cell intrinsic phenotypes by the AiolosN159S variant assessed by mixed bone marrow chimera experiment. (A) Flow cytometric analyses show the percentages of CD45.1+ (WT) versus CD45.2+ (Ikzf3+/+or Ikzf3N159S/N159S) donor cells in total bone marrow and in B cell population (B220+CD19+) from spleen, pLN, mLN and peritoneal cavity, after >8 weeks post-transplantation. (B) Ratios of CD45.2+/CD45.1+ cells in total bone marrow and B cell populations (B220+CD19+) from spleen, pLN and mLN, after >8 weeks post-transplantation (n=3). Each dot represents data from one recipient mouse. Data are presented as the mean ± SD, n = 3 per group. (C) Flow cytometric analyzes show the percentages of CD45.2 and CD45.1 donor cells in total cells of thymus and T cells (TCRβ+) from spleen, pLN and mLN, after >8 weeks post-transplantation. (D) Ratios of CD45.2+/CD45.1+ cells in total thymus and in T cells (TCRβ+) from spleen, pLN, mLN and peritoneal cavity, after >8 weeks post-transplantation. (E) Flow cytometric analyzes show percentage of CD45.2+ and CD45.1+ donor cells in splenic CD11b+CD11c− cell, dendritic cell (DC), natural killer cell (NK), neutrophil (Neu.), eosinophil (Eos.), Monocyte (Mono.), Macrophage (MΦ) and Inflammatory monocyte (Inf. Mono.) subsets, after >8 weeks post-transplantation. (F) Ratios of CD45.2+/CD45.1+ cells in each splenic myeloid cell subset (CD11b+CD11c−, DC, NK, Neu., Eos., Mono. & MΦ and Inf. Mono.) after >8 weeks post-transplantation (n=3). Statistically significant differences (Student’s unpaired, two-tailed t test) between the groups are shown. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
In the TCRβ+ T cell population, CD45.2+ Ikzf3N159S/N159S cells were significantly decreased in the peripheral and mesenteric lymph nodes compared to CD45.2+ Ikzf3+/+ cells (Figures 3C, D and S3E). In NK and myeloid lineage cells, the frequency of CD45.2+ Ikzf3N159S/N159S cells was lower, specifically in CD11b+NK1.1+ NK cells and CD11b+Ly6G−Ly6Clo Eos cells. (Figures 3E, F and S3F, G). These observations indicate that developmental defects of NK and Eos by the Ikzf3N159S mutation were caused in a cell-intrinsic manner and developmental potency to other myeloid-lineage cells, at least cell types we examined, were not affected by the Ikzf3N159S mutation.
Next, we examined whether the impaired expression of surface proteins caused by the Ikzf3N159S mutation occurs in a cell-intrinsic or cell-extrinsic manner. In recipients transplanted with CD45.1+ Ikzf3+/+ and CD45.2+ Ikzf3N159S/N159S BM cells, the expressions of IgD, CD23, and CD21 in the splenic B cell population were heavily reduced, specifically in CD45.2+ Ikzf3N159S/N159S cells (Figures 4A, B). Similarly, CD62L expression was significantly decreased, specifically in CD45.2+ Ikzf3N159S/N159S thymocytes (Figures 4C, D). These results indicated that the AiolosN159S mutant protein inhibits the expression of IgD, CD23, CD21, and CD62L in a cell-intrinsic manner. Interestingly, the frequency of CD62L+ cells in each thymocyte subset was higher in these recipients than in the non-irradiated control mice (Figures 2A, B and 4C, D). This suggests that the reconstitution of T cell development in the thymic environment in irradiated recipients may induce CD62L. It is noteworthy that the frequency of CD62L+ in CD4−CD8− DN subset was already lower in CD45.2+ Ikzf3N159S/N159S thymocytes compared with CD45.2+ Ikzf3+/+, suggesting that a small amount of AiolosN159S may shortly inhibit CD62L expression after induction in DN cells (Figures 4C, D). As expected, CD62L expression was severely inhibited in T cells from the DP thymocytes stage onward and in the spleen (Figures 4E, F). However, in pLNs, although the CD45.2+ Ikzf3N159S/N159S T cells comprised a small population, the frequency of CD62L+ cells was comparable to that of control cells, similar to CD45.2+ Ikzf3N159S/N159S CD4 T cells in mLN (Figures 4E, F and S4E, F).
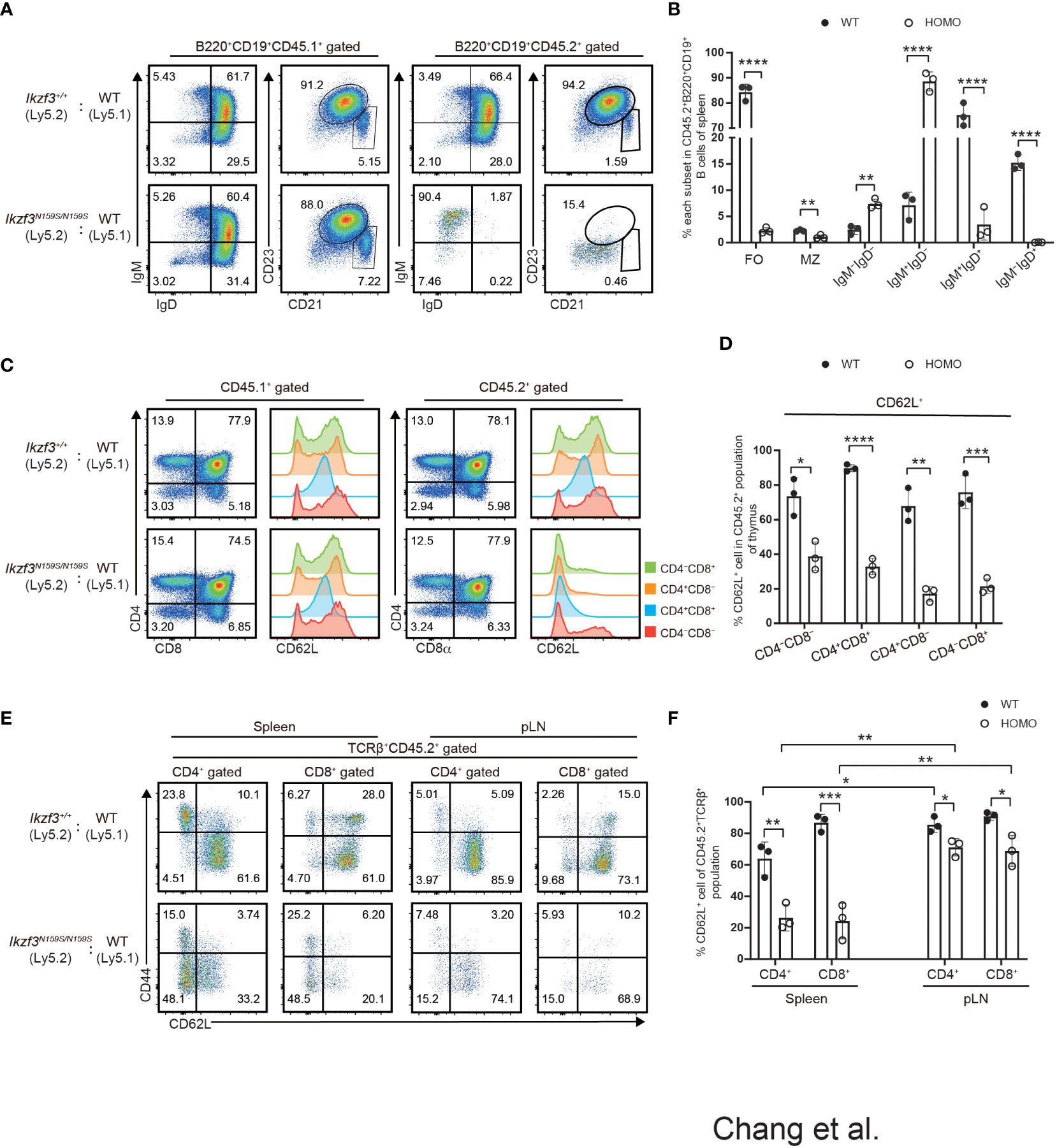
Figure 4 Cell intrinsic defect of CD62L and CD23 expression by the AiolosN159S variant. (A) Flow cytometric analyses show IgM, IgD, CD21 and CD23 expression level on B220+CD19+ splenic B cells from CD45.1+ (WT) versus CD45.2+ (Ikzf3+/+or Ikzf3N159S/N159S) donor cells, after >8 weeks post-transplantation. (B) Frequencies of splenic follicular B cell (CD21intCD23hi, FO), marginal zone B cell (CD21hiCD23lo/−, MZ), IgM−IgD−, IgM+IgD−, IgM+IgD+ and IgM−IgD+ subsets from CD45.2+ donor cells are shown (n=3). Each dot represents data from one recipient. Data are presented as the mean ± SD, n = 3 per group. (C) Flow cytometric analyses of CD4 and CD8a subsets of CD45.1+ or CD45.2+ donor cells from WT and Homo group, after >8 weeks post-transplantation. CD62L expression levels on each subset (CD4−CD8−, CD4+CD8+, CD4+CD8−, CD4−CD8+) are adjacently shown as histograms. (D) Graph shows the frequencies of CD62L+ of each thymocyte subset from CD45.2+ donor cells (n=3). (E) Flow cytometric analyses of CD44 and CD62L expression of CD45.2+ T cells in spleen and pLN (TCRβ+CD4+, TCRβ+CD8+), after >8 weeks post-transplantation. (F) Frequencies of CD62L+ cells from CD45.2+ T cell subsets in spleen and pLN are shown. Statistically significant differences (Student’s unpaired, two-tailed t test) between the groups are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Reduced homing ability of AiolosN159S/N159S T and B cells
Our mixed bone marrow chimera experiments clearly showed a greater reduction in the frequency of CD45.2+ Ikzf3N159S/N159S lymphocytes in the LNs than in the spleen, whereas the frequency of CD62L+ T cells increased in the LNs. Given the important role of CD62L in lymphocyte homing to lymphoid tissue, we compared the homing abilities of Ikzf3+/+ and Ikzf3N159S/N159S lymphocytes. Total splenocytes from CD45.2+ Ikzf3+/+ or CD45.2+ Ikzf3N159S/N159S mice and CD45.1+ control mice were mixed in a 1 to 1 ratio and intravenously injected into CD45.1+CD45.2+ recipient mice (Figure 5A). After 24 hours, the frequencies of CD45.1+ and CD45.2+ cells in the PBL, spleen, and LNs were analyzed by flow cytometry. The ratio of CD45.1+CD45.2− and CD45.2−CD45.2+ in B220+CD19+ or TCRβ+CD3+ population in recipient organs (Ro) and that in injection mixture (Ri) were used to calculate the homing index as the ratio of Ro/Ri (Figure 5A). In recipients injected with CD45.1+ control cells and CD45.2+ Ikzf3+/+, the homing index was approximately one in all tissues examined (Figures 5B–E and S5A). In contrast, in recipients injected with CD45.1+ control cells and CD45.2+ Ikzf3N159S/N159S cells, the homing index in the B cell compartment was lower than 0.2 in the PBL, although it remained around 0.5 in spleen, and CD45.2+ Ikzf3N159S/N159S B cells were hardly detected in the LNs (Figures 5B, C and S5A). In the T cell population of the above recipients, the homing index was decreased in the pLN and mLN, but not in the PBL and spleen (Figures 5D, E and S5A). These observations indicate that the homing ability to the LNs was impaired in Ikzf3N159S/N159S B and T lymphocytes. As expected from analyses of mixed bone marrow chimeras, the frequency of CD62L+ cells in Ikzf3N159S/N159S B and T cell populations was significantly increased in pLN and mLN from the input, although these were still lower than those in CD45.2+ Ikzf3+/+ cells (Figures 5B–E and S5A). These results revealed that Ikzf3N159S/N159S B and T lymphocytes were outcompeted by control cells in homing to the LNs. Selective accumulation of CD62L+ Ikzf3N159S/N159S lymphocytes in LNs revealed that Ikzf3N159S/N159S lymphocytes expressing CD62L retained their homing activity to some extent, suggesting that the AiolosN159S mutation impaired the homing ability of lymphocytes, mainly via disturbing the CD62L expression.
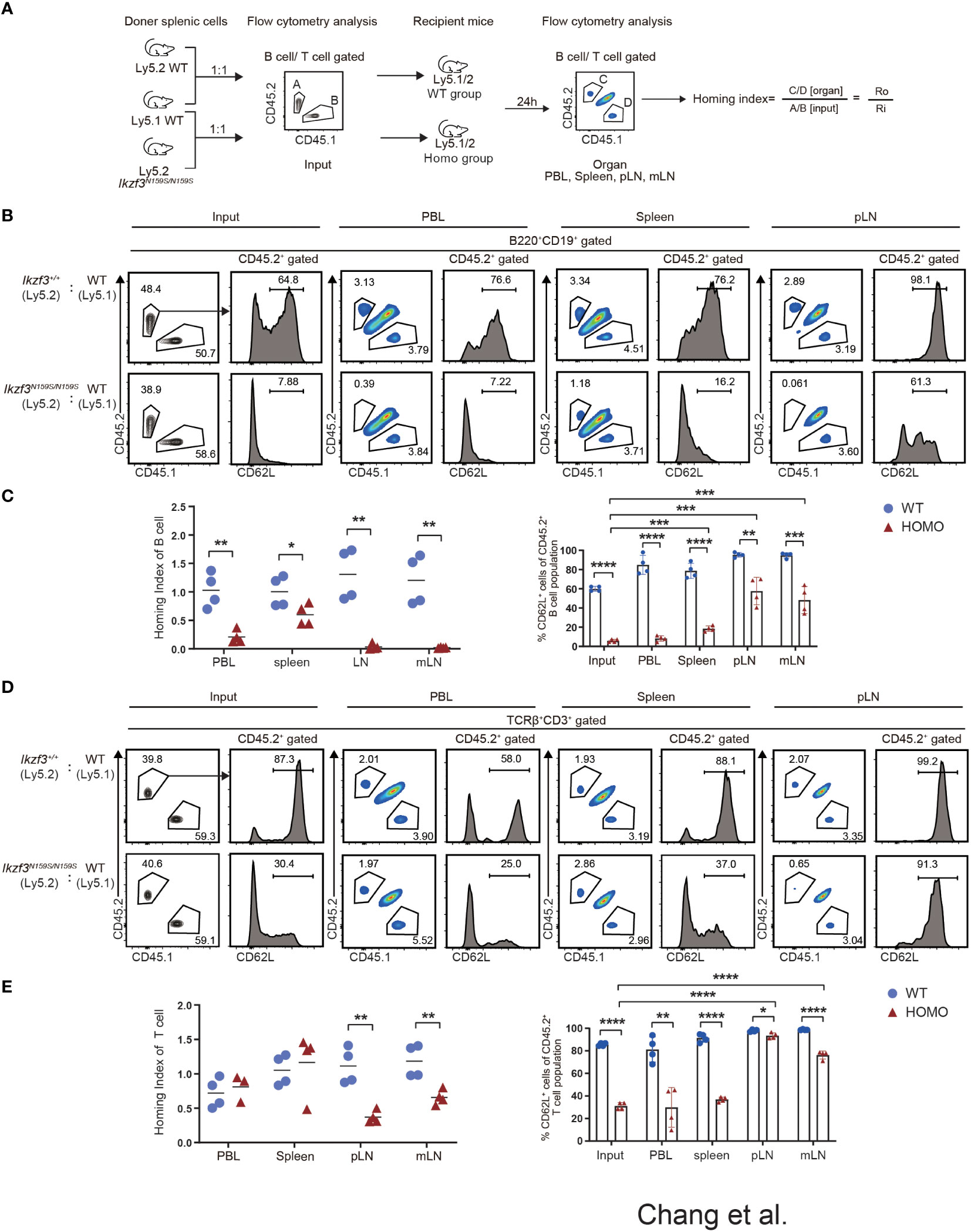
Figure 5 Impaired Homing ability of Ikzf3N159S/N159S lymphocytes. (A) Ly5.2 (CD45.2+) Ikzf3+/+ (WT group) or Ikzf3N159S/N159S (Homo group) splenic cells were mixed with Ly5.1 (CD45.1+) wild type (WT) splenic cells and injected into sub-lethally irradiated CD45.1+CD45.2+ recipient mice. Flow cytometry was used to assess the input ratio (Ri) of CD45.2+CD45.1− and CD45.2−CD45.1+ in B220+CD19+ or TCRβ+CD3+ population before injection. After 24h post-injection, flow cytometric analyzes shows ratios of CD45.2+CD45.1− and CD45.2−CD45.1+ donor cells in B or T cell populations from various organs (Ro). The ratio of Ro and Ri is analyzed as the homing index. (B) Flow cytometric analyzes shows ratios of CD45.2+ and CD45.1+ cells in B220+CD19+ B cells, from input and 24h post-transplanted organs (PBL, spleen, and pLN), alongside CD62L expression levels on CD45.2+ B cells. (C) B cell homing indexes of PBL, spleen, pLN, and mLN, from WT and Homo groups, are shown on the left. The horizontal lines indicate the mean values of at least three mice per group. The frequencies of CD62L+ cells in CD45.2+ B cell population from input and 24h post-transplanted organs (PBL, spleen, pLN, and mLN) are shown on the right (n=4). Each dot represents data from one recipient mouse. Data are presented as the mean ± SD, n = 4 per group (right). (D) Flow cytometry analyses show ratios of CD45.2+ and CD45.1+ cells in TCRβ+CD3+ T cells, from input and 24h post-transplanted organs (PBL, spleen, and pLN), alongside CD62L expression levels on CD45.2+ T cells. (E) T cell homing indexes of PBL, spleen, pLN, and mLN, from WT and Homo groups, are shown on the left. The frequencies of CD62L+ cells from CD45.2+ T cell population from input and 24h post-transplanted organs (PBL, spleen, pLN, and mLN) are shown on the right (n=4). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Low expression of putative IKZF1 target genes by AiolosN159S variant
To examine what genes were dysregulated in the presence of AiolosN159S variant, we conducted RNA-seq analyses. Considering low CD62L expression in T cells, we chose CD127 as a marker to isolate cells that would correspond to naïve T cells and purified splenic B220+CD19+ B cells and TCRβ+CD4+CD44−CD127+ T cells of Ikzf3+/+, Ikzf3+/N159S, and Ikzf3N159S/N159S mice. The results obtained from RNA-sequencing (RNA-seq) were analyzed using principal component analysis (PCA). In both B and T cells, PCA showed that Ikzf3+/+, Ikzf3+/N159S, and Ikzf3N159S/N159S cells were differentially clustered in terms of global gene expression (Figure 6A). Gene ontology (GO) analysis was subsequently performed using downregulated genes in Ikzf3N159S/N159S B and T cells. GO analysis indicated that the downregulated genes were enriched in lymphocyte activation, differentiation, and proliferation, leukocyte migration, and cell adhesion (Figure 6B). In the heat map, we defined the downregulated genes of leukocyte migration in Ikzf3N159S/N159S B cells, showing an intermediate phenotype in Ikzf3+/N159S B cells between WT and homo B cells in terms of gene expression (Figure 6C). In Ikzf3+/N159S T cells, most of genes in the leukocyte migration term also showed an intermediate phenotype, except for CD24a and Cnr2 genes (Figure 6D). This observation provides evidence of the gene dosage effect of the AiolosN159S variant on transcriptional dysregulation. We also examined our RNA-seq data for expression of genes encoding chemokine receptors and found that some chemokine receptors Ccr6, Ccr7, Ackr2 and Ccr1 were downregulated in Ikzf3N159S/N159S B cells, while only Cxcr1 showed a minimal increased expression of Ikzf3N159S/N159S T cells (Figure S6).
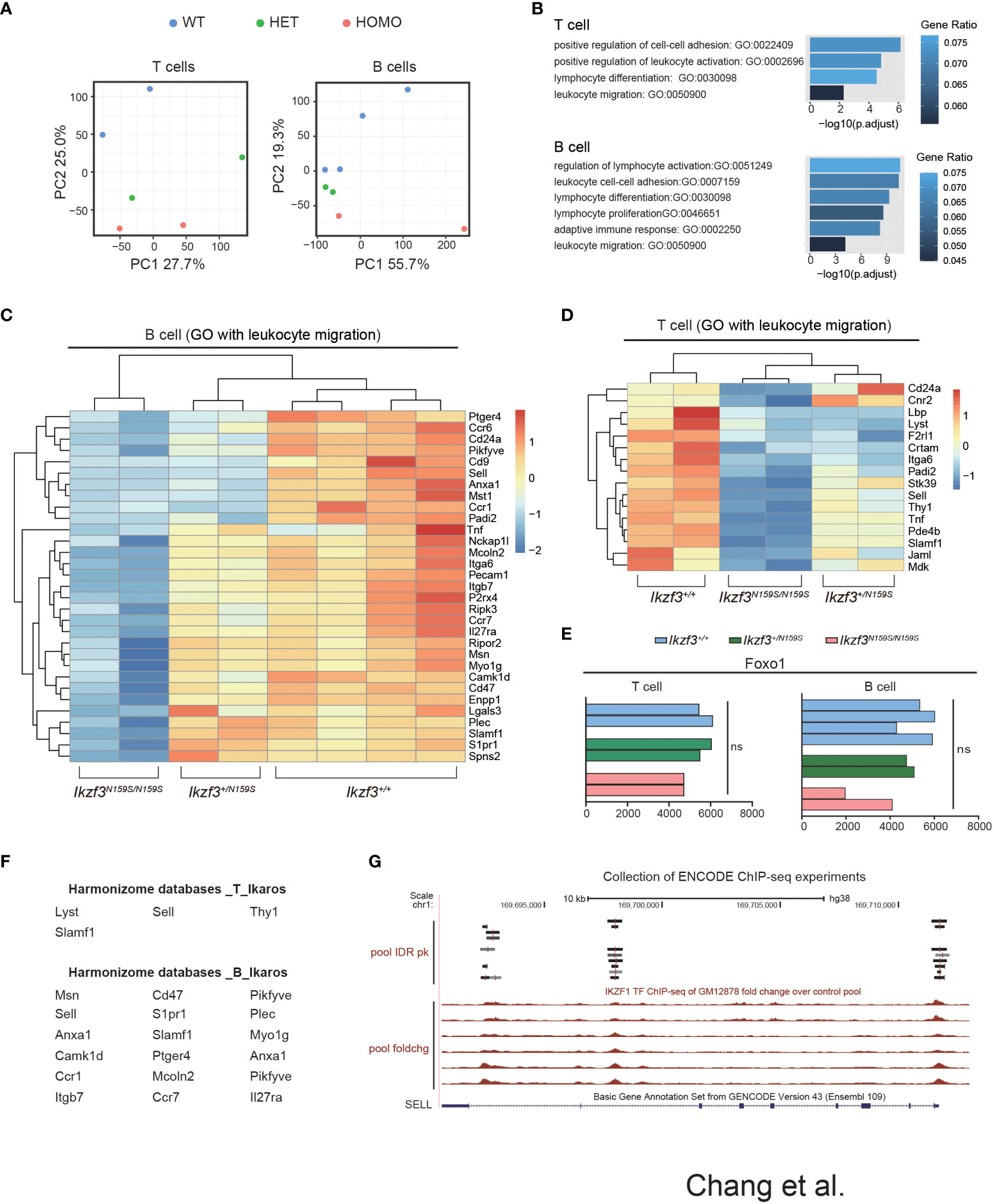
Figure 6 Dysregulated genes by AiolosN159S revealed by transcriptome analyses. (A) Principal component analysis (PCA) of the T and B cells transcriptomes from Ikzf3+/+, Ikzf3+/N159S and Ikzf3N159S/N159S mice (n = 2 or 4 per genotype). (B) GO analysis of downregulated genes in Ikzf3+/N159S and Ikzf3N159S/N159S T and B cells, compared to Ikzf3+/+ controls, with FDR <0.05. Adjusted p values are presented for each GO term. Colors indicate ratio of number of downregulated genes within total number of genes included for each GO term. (C, D) Z-score heat maps of downregulated genes in Ikzf3N159S/N159S B cells (C) and T cells (D), with the GO term of leukocyte migration included. (E) Read counts of Foxo1 in RNA-seq are shown. (F) Putative IKZF1 target genes, abstracted by Harmonizome database among the downregulated genes in Ikzf3N159S/N159S B and T cells. (G) UCSC Genome Browser track view of IKAROS ChIP-seq performed in GM12878 cell line (B-lymphocyte) in the ENCODE project, showing the binding by IKAROS around the SELL gene. Pool IDR pk is IKZF1 ChIP-seq of GM12878 irreproducible discovery rate (IDR) thresholded peaks pool. Pool foldchg is IKZF1 ChIP-seq of GM12878 fold change over control pool.
While CD62L expression in CD4+ T cells was not reduced by Aiolos deficiency (50), loss of Ikaros function led to reduced CD62L expression in naïve T cells due to low Foxo1 expression (51). However, Foxo1 expression was not reduced in B or T cells by the Ikzf3N159S mutation (Figure 6E). We have shown that another murine Aiolos variant, AiolosG158R, which has a G to R replacement next to the N159 residue, interferes with Ikaros function (16). To explore the possibility that the genes downregulated by AiolosN159S were correlated with Ikaros function, we utilized the Harmonizome databases, which is a collection of processed datasets gathered to serve and mine knowledge about genes and proteins from over 70 major online resources (31). Although the Harmonizome database does not have sufficient datasets to predict IKZF3/AIOLOS target genes, IKZF1/IKAROS target genes can be predicted using chromatin immunoprecipitation sequencing (ChIP-seq) datasets of IKZF1 from the ENCODE Transcription Factor Targets datasets. Our analyses of 16 and 31 dysregulated genes in T and B cells, respectively, found that four genes in T cells and more than half of the genes in B cells were associated with IKAROS protein binding (Figure 6F). The genome browser track of the ENCODE IKAROS ChIP-seq datasets in the human B cell line showed potential binding of IKAROS to the SELL locus encoding CD62L (Figure 6G). Thus, transcriptome analyses suggested that the downregulation of some genes by the AiolosN159S variant was caused by impaired Ikaros function.
Overexpression of Ikaros restored CD62L expression on AiolosN159S/N159S B cells
Given the gene dosage-dependent effect of Ikzf3N159S on CD62L expression, when low CD62L expression was caused by the interference of Ikaros by AiolosN159S, an increase in the ratio of Ikaros to AiolosN159S may overcome the dominant effect of the AiolosN159S variant. To test this possibility, we constructed a retroviral vector encoding either Ikaros with a DsRed marker or Aiolos with a GFP marker. After confirming the presence of Ikaros and Aiolos proteins in the packaging cell line Plat-E by western blotting (WB) (Figure 7A), we transduced splenic Ikzf3+/+ and Ikzf3N159S/N159S B cells by retroviral transduction. Induction with Ikaros significantly increased the frequency of the CD62L+ population in Ikzf3+/+ cells. However, Aiolos transduction decreased CD62L expression compared to the empty vector (EV) (Figures 7B, C). Importantly, CD62L expression in Ikaros transduced (DsRED+) Ikzf3N159S/N159S B cells was partially restored to approximately 20% (Figures 7B, C), although the CD62L expression remained lost after Aiolos transduction. These findings support the hypothesis that AiolosN159S interferes with Ikaros function, at least in the transcriptional regulating of CD62L expression.
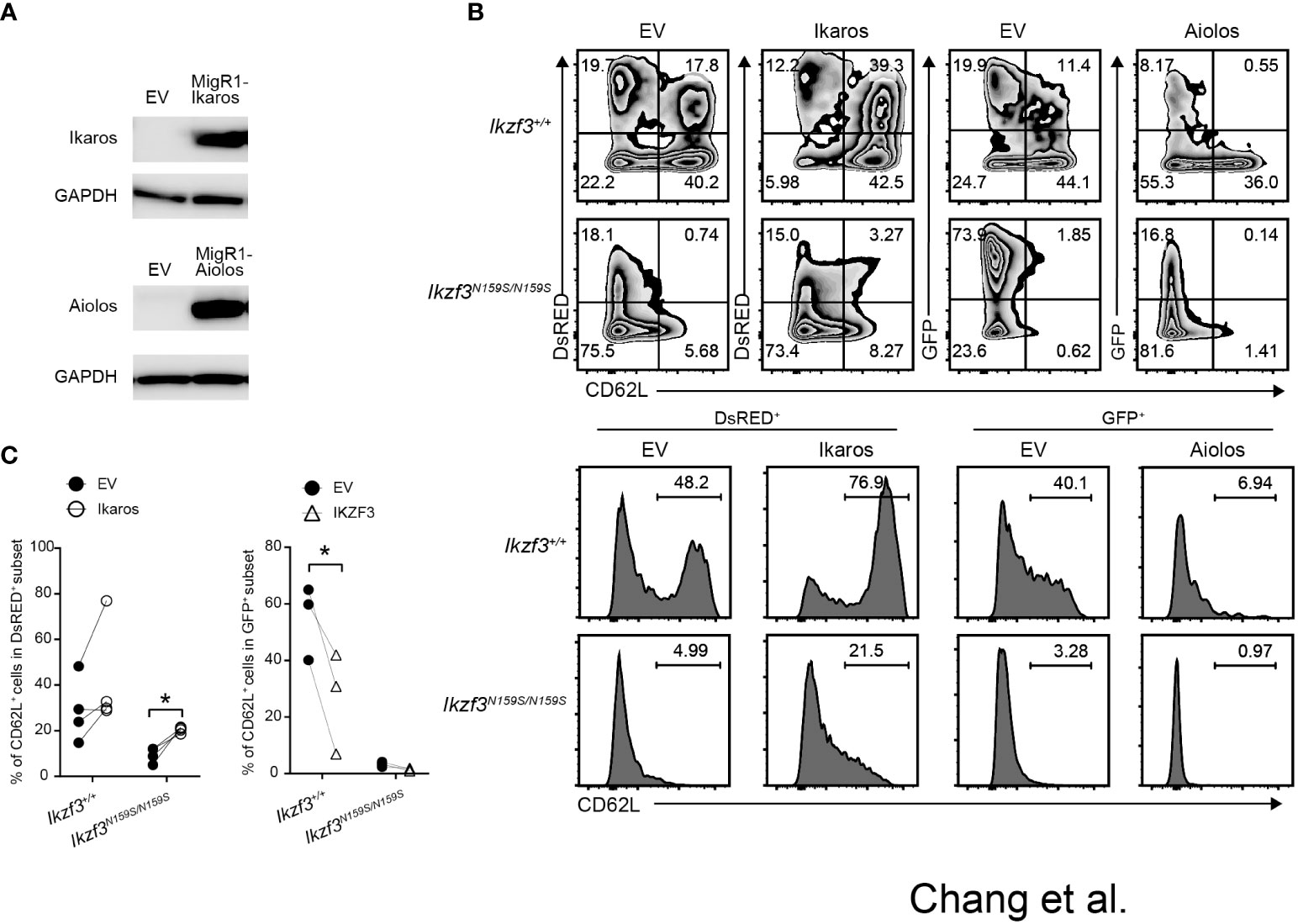
Figure 7 Overexpression of Ikaros restored CD62L expression in Ikzf3N159S/N159S B cells. (A) Immunoblot showing expression of Ikaros and Aiolos protein in packaging Plate E cell line transfected with empty vector (EV), pMigR1-Ikaros or pMigR1-Aiolos vector. (B) Zebra plots showing CD62L expression of retrovirally transduced Ikzf3+/+ and Ikzf3N159S/N159S B cells. DsRED and GFP fluorescent marker expression indicate Ikaros and Aiolos expression, respectively. Lower histograms showing CD62L expression on DsRED+ or GFP+ transduced cells. Numerical values in each histogram indicate percentages of each region. (C) Graphs show the percentage of CD62L+ in DsRED+ or GFP+ subset. Each circle represents data from one experiment. Data are presented as the mean ± SD, n = 3 per group. Student’ s unpaired, two-tailed t test). * P < 0.05.
Discussion
We have previously reported that the patient’s family carrying the heterozygous IKZF3 (c.479 A>G, p.N160S) variant showed impaired T and B cell development (27). A mouse model carrying Ikzf3N159S mutant allele corresponding to N160S in human AIOLOS showed a similar abnormal immune phenotype in B and T cells, confirming that the IKZF3N160S is the causal variant of the immune-deficient phenotype in these patients (27). In this study, we investigated the homing capacity of Ikzf3N159S/N159S lymphocytes and the mechanisms underlying low CD62L expression. Although our previous analyses did not reveal any dominant negative effects on IKAROS by AIOLOSN160S in terms of in vitro DNA binding and pericentromeric targeting in the cell line in previous analyses (27), this study revealed that some phenotypes, such as impaired CD62L expression observed in the Ikzf3N159S mouse model, were probably caused by hijacking of Ikaros function by the AiolosN159S variant.
Currently, the regulation of the Sell gene encoding CD62L remains elusive. Thereby, little is known about how the IKZF family proteins are involved in Sell gene regulation. Indu et al. reported that the dominant negative form (IK7) of the Ikaros protein, which lacks the N-terminal DNA-binding ZFs but retains the C-terminal dimerization zinc fingers, disturbs the CD62L expression (52). This suggests that Ik7 Ikaros possibly interferes with wild-type (wt) Ikaros protein function necessary for activating Sell gene (52). It is conceivable that Ik7 Ikaros influences the binding of Ikzf1 to the regulatory sequences of the Sell gene. In this study, we found that two regions in the SELL locus are bound by IKAROS in a human B cell line, suggesting that IKAROS directly activates the SELL gene. Future studies are required to elucidate the function of these IKAROS-bound regions. In contrast to Ikaros, there have been no reports demonstrating the involvement of Aiolos in CD62L expression. Aiolos-deficient mice showed normal CD62L expression in CD4+ T cells (50). Therefore, the AiolosN159S variant downregulates CD62L expression by interfering with protein functions other than those of Aiolos. Yamashita et al. reported that the AiolosG159R variant interferes with Ikaros function, causing developmental inhibition of B cells at an early stage in a gene dose-dependent manner (16). Although AiolosG159R can alter the nuclear localization pattern of Ikaros and in vitro binding to the canonical Ikaros site (16), AIOLOSN159S variant did not show such a capacity (27). However, the analyses in this study revealed that some phenotypes observed in Ikzf3N159S/N159S mice could be caused by interference with Ikaros function. For instance, our RNA-Seq of splenic B and T cells revealed that genes downregulated genes by AiolosN159S variant included putative targets of Ikaros, such as Sell, at least according to the Harmonizome databases. In addition, excessive exogenous Ikaros expression in AiolosN159S/N159S B cells restored CD62L expression. These findings indicate that the low CD62L expression caused by AiolosN159S is affected by disruption of Ikaros function. Further elucidation of how Ikaros regulates Sell should be conducted in future studies. To date, no IKZF protein variant, other than Ikzf3G158R and Ikzf3N159S, has been shown to interfere with other IKZF family members. Interestingly, IKZF1N159S and recently reported immunodeficiency and multiple system anomalies-causing IKZF2G153R variants were shown to act dominant-negative against IKZF1 and IKZF2, respectively (15, 53). Thus, it is possible that these pathogenic variants also act against other IKZF proteins, and interference of other IKZF proteins may be partially responsible for the pathogenesis of immunodeficiency caused by these variants.
Parul et al. proposed that the decreased expression of Sell due to Ikaros deficiency is mediated by the reduced expression of Foxo1 (51). Although Ikaros transduction of Ikaros-null CD4+ T cells upregulated Foxo1 level, CD62L expression remained low. However, Foxo1 transduction increased the expression of CD62L. Parul et al. discussed that increased Ikaros expression might interfere with Foxo1’s ability to activate the Sell locus (51). In contrast, in Ikzf3N159S/N159S B and T cells, Foxo1 level were not reduced, and the transduction of Ikaros into Ikzf3N159S/N159S B cells partially restored CD62L expression. This discrepancy between Ikaros-null and Ikzf3N159S/N159S cells suggests that the mechanisms underlying CD62L reduction are different. Further studies are required to address this issue. In contrast to Ikaros, overexpression of wild-type Aiolos over endogenous Ikaros inhibited CD62L expression. It is conceivable that excess Aiolos occupies the Ikaros-bound regions in the Sell locus. In this case, Sell gene was not activated by Aiolos, suggesting that there must be a functional difference between Ikaros and Aiolos in term of the activating of the Sell gene after binding to DNA. Both Ikaros and Aiolos are expressed in primary murine lymphocytes; however, the expression level of AiolosN159S was slightly higher than that of wt Aiolos (27). Therefore, it is possible that high amounts of AiolosN159S cause low CD62L expression, rather than functional differences between Aiolos and AiolosN159S. Interestingly, one among four patients harboring the same N-to-S substitution in the IKZF1/IKAROS protein (IKZF1N159S variant) showed low CD62L expression (15). Therefore, we speculate that the N to S substitution alters Aiolos function and make AiolosN159S variant to function as a dominant negative, at least in Sell gene regulation.
In mice harboring the Ikzf3G158R mutation, early B cell development in the bone marrow is inhibited at the pre-Pro B to Pre-B transition, at least in part, by interference with Ikaros function (16). In contrast, early B cell development was mostly normal in Ikzf3+/N159S and Ikzf3N159S/N159S mice, with an increased and decreased frequency of recirculating mature B cells in Ikzf3+/N159S and Ikzf3N159S/N159S mice, respectively. As Aiolos-deficient mice show a decrease in mature recirculating B cells in the bone marrow (25), the reduction in recirculating B cells in Ikzf3N159S/N159S mice likely stems from the loss of Aiolos function due to N159S replacement. Similar to recirculating B cells, the frequency of the peritoneal B2 cells was significantly increased in heterozygous mice, while it was decreased in homozygous mice. Thus, as far as these two types of B cells are concerned, Ikzf3+/N159S and Ikzf3N159S/N159S mice showed divergent phenotypes. Currently, the mechanisms causing this divergent and cell-specific phenotypes caused by Ikzf3N159S variant are not yet characterized and need further studies. In addition to the reduction in the splenic B220+ population, which consisted mainly of B2 cells, this study revealed that CD5+ B1a cells in the peritoneal cavity were nearly lost in Ikzf3N159S/N159S mice. Given the decrease in the B2/B1 ratio, the loss of CD5+ B1a cells likely stems from the developmental inhibition of B1a cells, although our results do not formally exclude the possibility that this is caused by the loss of CD5 expression from B1a cells. A previous study has shown that peritoneal B1a cells are severely reduced in Aiolos-deficient mice (25). In contrast, Ikaros-deficiency increases the number of B1 cells, suggesting a negative role in B1 cell development and function (54, 55). Thus, Ikaros and Aiolos function differently in regulating B1 cell development, suggesting that the loss of CD5+ B1a cells in Ikzf3N159S/N159S mice is likely caused by the loss of Aiolos function. In contrast, CD23 expression in follicular B cells was reduced in Ikaros-deficient mice (54, 56), whereas follicular B cells retained CD23 expression in Aiolos-deficient mice (25). Thus, the loss of CD23 expression in Ikzf3N159S/N159S splenic B cells was likely caused by the disruption of Ikaros function by AiolosN159S variant.
The migration of lymphocytes into tissues, a process referred to as lymphocyte homing, is regulated by several molecules. L-selectin (CD62L), which is encoded by Sell gene, is an adhesion molecule that binds to ligands expressed on high endothelial venules (HEVs) and plays an important role in maximizing lymphocyte trafficking across HEV (57–60). CD62L−/− lymphocytes failed to enter peripheral lymph nodes across HEV, resulting in a striking reduction in the lymphocyte cellularity in these tissues with a corresponding increase in their cellularity in spleen (57, 60, 61). L-selectin (CD62L) plays a key role in leukocyte migration into lymphoid tissues via the regulation of leukocyte rolling and entry into tissues (62). CD62L expression in Ikzf3N159S/N159S T and B cells was low, and BM chimera experiments showed that those Ikzf3N159S/N159S cells were outcompeted by control cells, particularly in LNs. These observations suggested that the homing abilities of Ikzf3N159S/N159S B and T cells were reduced. Indeed, the homing assay confirmed that the ability to enter LNs was weaker in Ikzf3N159S/N159S B cells than control cells. Unexpectedly, Ikzf3N159S/N159S cells were nearly undetectable in the bloodstream after injection. In Ikzf3N159S/N159S mice, the reduction in B cell frequency declined more in the peripheral blood than in the spleen (27). Thus, the reduction of circulating lymphocytes by AiolosN159S is mediated in part by intrinsic defect in staying in the bloodstream, although the underlying molecular mechanisms for this defect in Ikzf3N159S/N159S cells are unclear. RNA-seq of B cells detected a decrease in mRNAs-encoding molecules related to cell migration, such as Ccr1, Ccr7 and Slamf1, in Ikzf3N159S/N159S cells. It is possible that the reduction in these molecules is involved in the decreased frequency of Ikzf3N159S/N159S cells in the bloodstream. However, these factors are not primarily involved in the low homing capacity of Ikzf3N159S/N159S cells. Compared to the input Ikzf3N159S/N159S cells, the frequency of CD62L+ cells as well as CD62L expression level were higher in cells that migrated to LNs. This selective enrichment of CD62L+ cells in LNs not only confirms the importance of CD62L for lymphocyte homing but also indicates that Ikzf3N159S/N159S cells retain their homing capacity to LNs once sufficient levels of CD62L are expressed. Therefore, the reduced homing capacity of Ikzf3N159S/N159S cells is likely attributable to low CD62L expression, although our results did not formally exclude the involvement of other down-regulated molecules for impaired migration capacity of Ikzf3N159S/N159S cells.
In summary, using a mouse model, we found that Ikzf3N159S inhibited B1a cell differentiation, and most of immune phenotype occurred in a cell-intrinsic manner. Importantly, reduced CD62L expression by the AiolosN159S variant is likely caused by hijacking of Ikaros function. The AiolosG158R variant also interferes with Ikaros function (16), however, the phenotypes caused by Ikzf3G158R and Ikzf3N159S mutations are quite different. In terms of CD23 and CD62L expression, Ikzf3+/N159S cells showed a greater reduction than Ikzf3+/G158R cells (16, 27). The in silico simulation predicted that G159R replacement pushed the second zinc finger away from DNA (16). Comparison of G159R and N160S by the same in silico simulation predicted that that the N160S mutant experiences reduced stability, higher flexibility, and reduced compactness as a result of loss of key hydrogen bond interactions between the cognate DNA (Figure S7). These subtle differences in interactions in the structures might serve as the causative factor for the observed phenotypic effect of the AiolosN159S mutant as compared to the AiolosG158R mutant. Thus, amino acid replacement in the residue next to each other differentially affects Aiolos function, and even though these two variants potentially interfere with Ikaros function, the different phenotypes between the two mutant mouse lines suggest that the affected Ikaros target genes should be different. It would be interesting to examine whether the phenotypes observed with Ikzf3G158R or Ikzf3N159S were retained, restored, or synergized when the two mutations were combined on one allele by generating an Ikzf3G158R:N159S mutation. ChIP-seq or interactome studies of Ikzf3G158R or Ikzf3N159S variant would be other potential approaches to study differences in molecular pathogenesis between two variants. Currently, little is known about how homo- or heterodimer formation among Ikzf family proteins is controlled, and what target genes are regulated by homo- or heterodimers. To understand the mechanisms that regulate the diversity of Ikzf protein family functions, it is important to explore how homo- or heterodimer formation is regulated.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GSE234649 (GEO).
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee of RIKEN Yokohama Branch. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JC performed experiments, analyzed data, prepared figures, and wrote the manuscript. MY analyzed the RNA-seq data. AP and KZ performed structural analyses. IT supervised the research and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the primary immune deficient program by RIKEN IMS, was supported by the Grant-in-Aid for Scientific Research on Innovative Areas 19H04820 (to IT) and was supported by RIKEN international program associate (IPA) program (to JC).
Acknowledgments
We appreciate Animal Facility Group at RIKEN IMS, and thank members, in particular Chengcheng Zou and Kazuaki Matsumoto, of laboratory for transcriptional regulation for valuable discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1239779/full#supplementary-material
References
1. Tangye SG, Al-Herz W, Bousfiha A, Cunningham-Rundles C, Franco JL, Holland SM, et al. Human inborn errors of immunity: 2022 update on the classification from the international union of immunological societies expert committee. J Clin Immunol (2022) 42(7):1473–507. doi: 10.1007/s10875-022-01289-3
2. Tiri A, Masetti R, Conti F, Tignanelli A, Turrini E, Bertolini P, et al. Inborn Errors Immun Cancer Biol (Basel) (2021) 10(4). doi: 10.3390/biology10040313
3. Liu L, Okada S, Kong XF, Kreins AY, Cypowyj S, Abhyankar A, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med (2011) 208(8):1635–48. doi: 10.1084/jem.20110958
4. Smits BM, Hartley T, Dünnebach E, Bartels M, Boycott KM, Kernohan KD, et al. Heterozygous variants in the DNA-binding domain of c-Myb may affect normal B/T cell development. Hemasphere (2022) 6(10):e774. doi: 10.1097/HS9.0000000000000774
5. Punwani D, Zhang Y, Yu J, Cowan MJ, Rana S, Kwan A, et al. Multisystem AnoMalies in severe combined immunodeficiency with mutant BCL11B. N Engl J Med (2016) 375(22):2165–76. doi: 10.1056/NEJMoa1509164
6. Lessel D, Gehbauer C, Bramswig NC, Schluth-Bolard C, Venkataramanappa S, van Gassen KLI, et al. BCL11B mutations in patients affected by a neurodevelopmental disorder with reduced type 2 innate lymphoid cells. Brain J Neurol (2018) 141(8):2299–311. doi: 10.1093/brain/awy173
7. Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell (1995) 83(2):289–99. doi: 10.1016/0092-8674(95)90170-1
8. Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, et al. The Ikaros gene is required for the development of all lymphoid lineages. Cell (1994) 79(1):143–56. doi: 10.1016/0092-8674(94)90407-3
9. Holmes ML, Huntington ND, Thong RP, Brady J, Hayakawa Y, Andoniou CE, et al. Peripheral natural killer cell maturation depends on the transcription factor Aiolos. EMBO J (2014) 33(22):2721–34. doi: 10.15252/embj.201487900
10. John LB, Ward AC. The Ikaros gene family: transcriptional regulators of hematopoiesis and immunity. Mol Immunol (2011) 48(9-10):1272–8. doi: 10.1016/j.molimm.2011.03.006
11. Georgopoulos K. The making of a lymphocyte: the choice among disparate cell fates and the IKAROS enigma. Genes Dev (2017) 31(5):439–50. doi: 10.1101/gad.297002.117
12. Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity (1996) 5(6):537–49. doi: 10.1016/S1074-7613(00)80269-1
13. Sun L, Liu A, Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J (1996) 15(19):5358–69. doi: 10.1002/j.1460-2075.1996.tb00920.x
14. Nichogiannopoulou A, Trevisan M, Neben S, Friedrich C, Georgopoulos K. Defects in hemopoietic stem cell activity in Ikaros mutant mice. J Exp Med (1999) 190(9):1201–14. doi: 10.1084/jem.190.9.1201
15. Boutboul D, Kuehn HS, Van de Wyngaert Z, Niemela JE, Callebaut I, Stoddard J, et al. Dominant-negative IKZF1 mutations cause a T, B, and myeloid cell combined immunodeficiency. J Clin Invest (2018) 128(7):3071–87. doi: 10.1172/JCI98164
16. Yamashita M, Kuehn HS, Okuyama K, Okada S, Inoue Y, Mitsuiki N, et al. A variant in human AIOLOS impairs adaptive immunity by interfering with IKAROS. Nat Immunol (2021) 22(7):893–903. doi: 10.1038/s41590-021-00951-z
17. Yoshida N, Sakaguchi H, Muramatsu H, Okuno Y, Song C, Dovat S, et al. Germline IKAROS mutation associated with primary immunodeficiency that progressed to T-cell acute lymphoblastic leukemia. Leukemia (2017) 31(5):1221–3. doi: 10.1038/leu.2017.25
18. Hoshino A, Okada S, Yoshida K, Nishida N, Okuno Y, Ueno H, et al. Abnormal hematopoiesis and autoimmunity in human subjects with germline IKZF1 mutations. J Allergy Clin Immunol (2017) 140(1):223–31. doi: 10.1016/j.jaci.2016.09.029
19. Kuehn HS, Boast B, Rosenzweig SD. Inborn errors of human IKAROS: LOF and GOF variants associated with primary immunodeficiency. Clin Exp Immunol (2023) 212(2):129–36. doi: 10.1093/cei/uxac109
20. Morgan B, Sun L, Avitahl N, Andrikopoulos K, Ikeda T, Gonzales E, et al. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J (1997) 16(8):2004–13. doi: 10.1093/emboj/16.8.2004
21. Kelley CM, Ikeda T, Koipally J, Avitahl N, Wu L, Georgopoulos K, et al. Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Curr Biol (1998) 8(9):508–15. doi: 10.1016/S0960-9822(98)70202-7
22. Honma Y, Kiyosawa H, Mori T, Oguri A, Nikaido T, Kanazawa K, et al. Eos: a novel member of the Ikaros gene family expressed predominantly in the developing nervous system. FEBS Lett (1999) 447(1):76–80. doi: 10.1016/S0014-5793(99)00265-3
23. Perdomo J, Holmes M, Chong B, Crossley M. Eos and pegasus, two members of the Ikaros family of proteins with distinct DNA binding activities. J Biol Chem (2000) 275(49):38347–54. doi: 10.1074/jbc.M005457200
24. Boast B, Nunes-Santos CJ, Kuehn HS, Rosenzweig SD. Ikaros-associated diseases: from mice to humans and back again. Front Pediatr (2021) 9:705497. doi: 10.3389/fped.2021.705497
25. Wang JH, Avitahl N, Cariappa A, Friedrich C, Ikeda T, Renold A, et al. Aiolos regulates B cell activation and maturation to effector state. Immunity (1998) 9(4):543–53. doi: 10.1016/S1074-7613(00)80637-8
26. Yamashita M, Morio T. AIOLOS variants causing immunodeficiency in human and mice. Front Immunol (2022) 13:866582. doi: 10.3389/fimmu.2022.866582
27. Kuehn HS, Chang J, Yamashita M, Niemela JE, Zou C, Okuyama K, et al. T and B cell abnorMalities, pneumocystis pneumonia, and chronic lymphocytic leukemia associated with an AIOLOS defect in patients. J Exp Med (2021) 218(12):e20211118. doi: 10.1084/jem.20211118
28. Putri GH, Anders S, Pyl PT, Pimanda JE, Zanini F. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics (2022) 38(10):2943–5. doi: 10.1093/bioinformatics/btac166
29. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (2010) 26(1):139–40. doi: 10.1093/bioinformatics/btp616
30. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics (2012) 16(5):284–7. doi: 10.1089/omi.2011.0118
31. Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) (2016) 2016:1–16. doi: 10.1093/database/baw100
32. Zeng J, Eljalby M, Aryal RP, Lehoux S, Stavenhagen K, Kudelka MR, et al. Cosmc controls B cell homing. Nat Commun (2020) 11(1):3990. doi: 10.1038/s41467-020-17765-6
33. McHeik S, Van Eeckhout N, De Poorter C, Galés C, Parmentier M, Springael JY. Coexpression of CCR7 and CXCR4 during B cell development controls CXCR4 responsiveness and bone marrow homing. Front Immunol (2019) 10:2970. doi: 10.3389/fimmu.2019.02970
34. Hoggatt J, Singh P, Sampath J, Pelus LM. Prostaglandin E2 enhances hematopoietic stem cell homing, survival, and proliferation. Blood (2009) 113(22):5444–55. doi: 10.1182/blood-2009-01-201335
35. Liu Y, Ding L, Zhang B, Deng Z, Han Y, Wang S, et al. Thrombopoietin enhances hematopoietic stem and progenitor cell homing by impeding matrix metalloproteinase 9 expression. Stem Cells Transl Med (2020) 9(6):661–73. doi: 10.1002/sctm.19-0220
36. Patel A, Zhang X, Blumenthal RM, Cheng X. Structural basis of human PR/SET domain 9 (PRDM9) allele C-specific recognition of its cognate DNA sequence. J Biol Chem (2017) 292(39):15994–6002. doi: 10.1074/jbc.M117.805754
37. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Res (2000) 28(1):235–42. doi: 10.1093/nar/28.1.235
38. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol (1990) 215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2
39. Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol (1993) 234(3):779–815. doi: 10.1006/jmbi.1993.1626
40. Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci (2006) 15(11):2507–24. doi: 10.1110/ps.062416606
41. Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Physics (1983) 79:926–35. doi: 10.1063/1.445869
42. Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput (2015) 11(8):3696–713. doi: 10.1021/acs.jctc.5b00255
43. Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem (2005) 26(16):1701–18. doi: 10.1002/jcc.20291
44. Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys (2007) 126(1):014101. doi: 10.1063/1.2408420
45. Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph (1996) 14(1):33–8, 27-8. doi: 10.1016/0263-7855(96)00018-5
46. Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med (1991) 173(5):1213–25. doi: 10.1084/jem.173.5.1213
47. Manohar V, Brown E, Leiserson WM, Chused TM. Expression of Lyt-1 by a subset of B lymphocytes. J Immunol (1982) 129(2):532–8. doi: 10.4049/jimmunol.129.2.532
48. Hayakawa K, Hardy RR, Parks DR, Herzenberg LA. The "Ly-1 B" cell subpopulation in normal immunodefective, and autoimmune mice. J Exp Med (1983) 157(1):202–18. doi: 10.1084/jem.157.1.202
49. Wong JB, Hewitt SL, Heltemes-Harris LM, Mandal M, Johnson K, Rajewsky K, et al. B-1a cells acquire their unique characteristics by bypassing the pre-BCR selection stage. Nat Commun (2019) 10(1):4768. doi: 10.1038/s41467-019-12824-z
50. Quintana FJ, Jin H, Burns EJ, Nadeau M, Yeste A, Kumar D, et al. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat Immunol (2012) 13(8):770–7. doi: 10.1038/ni.2363
51. Agnihotri P, Robertson NM, Umetsu SE, Arakcheeva K, Winandy S. Lack of Ikaros cripples expression of Foxo1 and its targets in naive T cells. Immunology (2017) 152(3):494–506. doi: 10.1111/imm.12786
52. Christopherson I, Piechoki M, Liu G, Ratner S, Galy A. Regulation of L-selectin expression by a dominant negative Ikaros protein. J Leukoc Biol (2001) 69(4):675–83. doi: 10.1189/jlb.69.4.675
53. Mohajeri A, Vaseghi-Shanjani M, Rosenfeld JA, Yang GX, Lu H, Sharma M, et al. Dominant negative variants in IKZF2 cause ICHAD syndrome, a new disorder characterised by immunodysregulation, craniofacial anoMalies, hearing impairment, athelia and developmental delay. J Med Genet (2023). doi: 10.1016/j.clim.2023.109546
54. Macias-Garcia A, Heizmann B, Sellars M, Marchal P, Dali H, Pasquali JL, et al. Ikaros is a negative regulator of B1 cell development and function. J Biol Chem (2016) 291(17):9073–86. doi: 10.1074/jbc.M115.704239
55. Oliveira VC, Lacerda MP, Moraes BBM, Gomes CP, Maricato JT, Souza OF, et al. Deregulation of Ikaros expression in B-1 cells: New insights in the Malignant transformation to chronic lymphocytic leukemia. J Leukoc Biol (2019) 106(3):581–94. doi: 10.1002/JLB.MA1118-454R
56. Cariappa A, Tang M, Parng C, Nebelitskiy E, Carroll M, Georgopoulos K, et al. The follicular versus marginal zone B lymphocyte cell fate decision is regulated by Aiolos, Btk, and CD21. Immunity (2001) 14(5):603–15. doi: 10.1016/S1074-7613(01)00135-2
57. Arbonés ML, Ord DC, Ley K, Ratech H, Maynard-Curry C, Otten G, et al. Lymphocyte homing and leukocyte rolling and migration are impaired in L-selectin-deficient mice. Immunity (1994) 1(4):247–60. doi: 10.1016/1074-7613(94)90076-0
58. Tedder TF, Steeber DA, Chen A, Engel P. The selectins: vascular adhesion molecules. FASEB J (1995) 9(10):866–73. doi: 10.1096/fasebj.9.10.7542213
59. Springer TA. Traffic signals on endothelium for lymphocyte recirculation and leukocyte emigration. Annu Rev Physiol (1995) 57:827–72. doi: 10.1146/annurev.ph.57.030195.004143
60. Subramanian H, Grailer JJ, Ohlrich KC, Rymaszewski AL, Loppnow JJ, Kodera M, et al. Signaling through L-selectin mediates enhanced chemotaxis of lymphocyte subsets to secondary lymphoid tissue chemokine. J Immunol (2012) 188(7):3223–36. doi: 10.4049/jimmunol.1101032
61. Steeber DA, Tang ML, Zhang XQ, Müller W, Wagner N, Tedder TF. Efficient lymphocyte migration across high endothelial venules of mouse Peyer's patches requires overlapping expression of L-selectin and beta7 integrin. J Immunol (1998) 161(12):6638–47. doi: 10.4049/jimmunol.161.12.6638
Keywords: IKZF family proteins, lymphocyte homing, CD62L, immune deficiency, heterodimeric interference
Citation: Chang J, Yamashita M, Padhi AK, Zhang KYJ and Taniuchi I (2023) Impaired tissue homing by the Ikzf3N159S variant is mediated by interfering with Ikaros function. Front. Immunol. 14:1239779. doi: 10.3389/fimmu.2023.1239779
Received: 14 June 2023; Accepted: 31 July 2023;
Published: 17 August 2023.
Edited by:
Yoji Sasahara, Tohoku University, JapanReviewed by:
Hidenori Ohnishi, Gifu University, JapanMario Ernesto Cruz-Munoz, Autonomous University of the State of Morelos, Mexico
Copyright © 2023 Chang, Yamashita, Padhi, Zhang and Taniuchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ichiro Taniuchi, ichiro.taniuchi@riken.jp
†Present address: Aditya K. Padhi, Laboratory for Computational Biology & Biomolecular Design, School of Biochemical Engineering, Indian Institute of Technology (BHU), Varanasi, Uttar Pradesh, India
 Jingjie Chang
Jingjie Chang Motoi Yamashita
Motoi Yamashita Aditya K. Padhi
Aditya K. Padhi Kam Y. J. Zhang
Kam Y. J. Zhang Ichiro Taniuchi
Ichiro Taniuchi