- 1University of Iowa Carver College of Medicine, Iowa City, IA, United States
- 2Department of Dermatology, University of Iowa, University of Iowa Hospitals and Clinics, Iowa City, IA, United States
- 3Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, MO, United States
- 4Cancer Biology Graduate Program, University of Iowa Carver College of Medicine, Iowa City, IA, United States
- 5Department of Hematology and Oncology, University of Iowa Hospitals and Clinics, Iowa City, IA, United States
- 6Department of Pathology, University of Iowa Hospitals and Clinics, Iowa City, IA, United States
- 7Iowa City Veterans Affairs Medical Center, Iowa City, IA, United States
Cutaneous 5T cell lymphoma (CTCL), characterized by malignant T cells infiltrating the skin with potential for dissemination, remains a challenging disease to diagnose and treat due to disease heterogeneity, treatment resistance, and lack of effective and standardized diagnostic and prognostic clinical tools. Currently, diagnosis of CTCL practically relies on clinical presentation, histopathology, and immunohistochemistry. These methods are collectively fraught with limitations in sensitivity and specificity. Fortunately, recent advances in flow cytometry, polymerase chain reaction, high throughput sequencing, and other molecular techniques have shown promise in improving diagnosis and treatment of CTCL. Examples of these advances include T cell receptor clonotyping via sequencing to detect CTCL earlier in the disease course and single-cell RNA sequencing to identify gene expression patterns that commonly drive CTCL pathogenesis. Experience with these techniques has afforded novel insights which may translate into enhanced diagnostic and therapeutic approaches for CTCL.
Introduction
Cutaneous lymphomas are a heterogeneous assortment of clonal T-, B-, and natural killer (NK) cell malignancies that localize to the skin but may additionally involve or disseminate to the blood, lymph nodes, and viscera. Cutaneous T cell lymphomas (CTCL) comprise most cases, with neoplastic cells commonly exhibiting a CD4+CD8- immunophenotype. The most common type of CTCL is mycosis fungoides (MF), characterized by malignant cells localizing to the dermal-epidermal junction and the epidermis as single cells and as collections (1, 2). MF, typified by pruritic patches, plaques, and tumors classically distributed over the trunk, proximal extremities, and waist and buttock region, often behaves indolently. However, a subset of patients with MF will experience a more rapidly progressive, treatment-resistant disease course (1, 3). The leukemic form of MF, Sézary syndrome (SS), most often presents with erythroderma with pruritus and lymphadenopathy (4). Because SS imparts significant morbidity and mortality (3), early, accurate diagnosis and an appreciation of factors indicative of disease progression should optimize management.
CTCL tumor cell populations typically exhibit a dominant T cell clone that shares common rearranged TCRγδ or TCRαβ chains, although CTCL cases with multiple clones have been reported (5, 6). Malignant cells also frequently exhibit genomic alterations, including the more common gene copy number alterations and less common somatic mutations (7). These mutations can result in aberrant protein expression and function such as increased JAK-STAT signaling that promotes tumor cell proliferation (8). The expansion of malignant CD4 T cells results in increased CD4:CD8 T cell ratios and the presence of atypical tumor cells in the blood (9).
CTCL management is frequently complicated by delayed diagnosis, in part because MF mimics inflammatory dermatoses such as atopic dermatitis and psoriasis at a clinical and histopathological level (9, 10). Diagnosing CTCL with traditional diagnostic tools, including clinical presentation, immunohistochemistry, and histopathology, is imperfect, resulting in a median delay in diagnosis of forty eight months in patients who presented with only cutaneous disease (11).
The accuracy and efficacy of clinical tools also remain limited when assessing individualized risk for disease progression and determining optimal targeted therapies. Diagnostic delays and prognostic errors may result in inappropriate treatment of CTCL and disease progression. Therefore, there is a clear need for new tools that the clinician can deploy in the care of CTCL patients. Scientists and clinicians previously validated a set of clinical variables to propose prediction models for disease progression (12), termed the Cutaneous Lymphoma International Prognostic Index (13). These risk factors for progression in patients are based on the physical exam, including total body surface area involvement and skin lesion morphology, e.g., the presence of skin plaques versus patches. Reliance on clinical staging is, however, of limited use in the initial stages of disease (14).
Current therapies for advanced CTCL include chemotherapy, radiotherapy, and immunomodulatory medications; however, allogeneic hematopoietic stem cell transplant in the setting of low disease burden is the only potentially curative treatment currently available (15, 16). Chemotherapy and non-chemotherapeutic therapies are frequently unable to provide durable disease control in MF/SS (17). Therefore, there is a need for novel therapies that will provide symptom relief, disease control, and disease cure. Identifying novel therapeutic targets can be challenging, in part because prominent levels of tumor heterogeneity exist within and between patients (18–20). Recent and emerging studies dissecting mutated pathways in CTCL continue to identify novel potential therapeutic targets. These include targets related to signaling pathways, cell surface proteins, and host immune responses (21).
Assessment of parameters such as tumor T cell receptor (TCR) clone frequency and CTCL somatic mutation burden, the latter of which is primarily driven by copy number alteration, offer the ability to more accurately diagnose CTCL, determine the optimal treatment of the disease, and monitor treatment response. Molecular techniques can assess these parameters. Techniques include flow cytometry, polymerase chain reaction (PCR), high throughput sequencing (HTS), and single-cell RNA sequencing (scRNA-seq) performed on patient tissue samples. In this review, we discuss how clinicians can leverage molecular techniques to aid in CTCL diagnosis, prognosis, and monitoring of treatment response. We also highlight how researchers utilize molecular techniques to increase understanding of CTCL pathogenesis and potentially identify future therapeutic targets.
Diagnostic and prognostic evaluation of CTCL
Diagnosis
Molecular techniques, including flow cytometry, PCR, quantitative PCR, HTS, scRNA-seq, and proteomic analysis, have shown promise in identifying CTCL tumor cells from the skin and/or blood to facilitate quicker and more dependable CTCL diagnosis as compared to clinical tools. These techniques distinguish CTCL from benign inflammatory dermatoses by exploiting tumor characteristics, including signatures of TCR clonality, gene expression, miRNA expression, and protein composition Table 1. It is important to consider the unique strengths and weaknesses of each approach such as sensitivity, specificity, and cost.
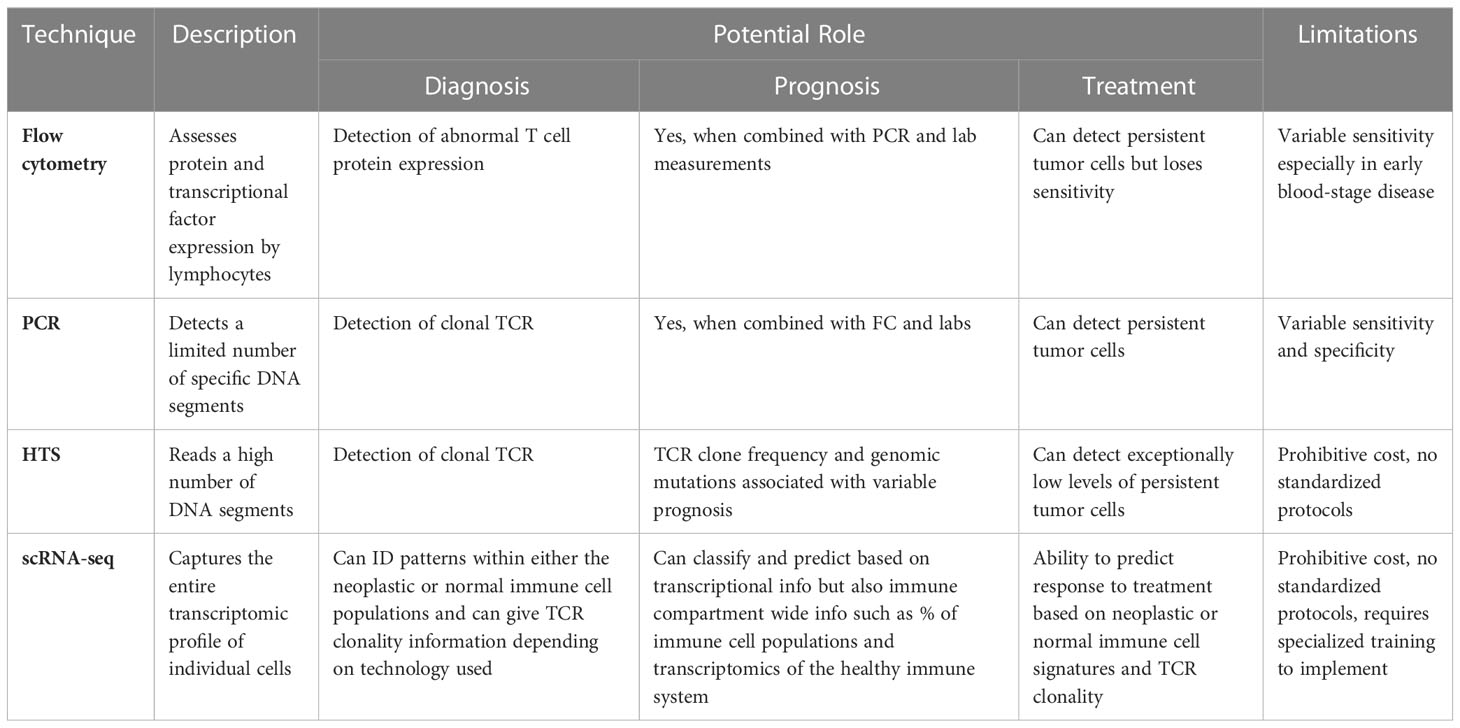
Table 1 Novel Molecular Tools with Potential for Enhancing Diagnosis, Prognosis, and Management of CTCL.
Flow cytometry is often used to diagnose the presence of a malignant cell population in peripheral blood and stage the disease. Well-established guidelines inform this. Updated WHO-EORTC criteria define significant (B2) blood involvement as ≥1000/ml aberrant T cells typically with a CD4+/CD7- or CD4+/CD26- immunophenotype (22). However, benign dermatoses may also have associated abnormal CD4 T cell populations, which may limit the specificity of these markers (23, 24). Proposed alternative parameters include the presence of clonal TCRVβ chains; gating on CD4+CD26- populations appears to increase the sensitivity of TCR-Vβ testing (25, 26).
CTCL diagnosis may be made by PCR detection of malignant TCR δ or β chains, another WHO-EORTC criteria (22, 27). Clonal PCR may facilitate the detection of early-stage MF when traditional diagnostic techniques lack sufficient sensitivity (7, 28). Strengths of PCR include higher sensitivity than flow cytometry for diagnosis and the ability to track disease from diagnosis and treatment, whereas treatment can reduce the reliability of flow cytometry and RNA based disease markers (29).
PCR is limited, however, by factors including pre-test probability, sample quality, disease stage, assay method, and primer design; as a result, it is estimated that the sensitivity of PCR among institutions varies from 50-90% (7). There is also not a clear consensus on where the cutoffs for PCR monoclonality should be set, as increasing the specificity of a PCR assay may compromise the sensitivity to an unacceptable level (30). For these reasons, it is recommended that standardized PCR assays, such as the PCR BIOMED-2 kit, be used in favor of non-standardized PCR kits to identify mutations more accurately (31). A proposed solution is to combine TCRβ and TCRγ clonality PCR assays to improve the sensitivity when detecting early MF (32). PCR specificity may be limited by the fact that autoimmune and inflammatory conditions sometimes exhibit dominant T cell clones (32). This may be addressed by determining if a shared dominant TCR clone is detected in multiple skin lesions, which is unique to CTCL (33).
HTS of malignant TCR clones holds advantages over PCR, including increased sensitivity and specificity, earlier disease and disease recurrence detection, and independence from diagnostic biases such as primer selection. HTS of the TCRγ and TCRβ genes exhibited 100% sensitivity versus 70% sensitivity for PCR analysis of paired samples in MF (34). In the same study, malignant T cells could be isolated from the blood of patients with new skin lesions but no clinical involvement of the peripheral blood, highlighting that early blood involvement may be detected by HTS. Numerous other studies have demonstrated that HTS has superior sensitivity and/or specificity as compared to PCR for detection of malignant cells in the skin and or blood (35–38).
While TCR sequencing is a powerful tool for the diagnosis of CTCL, it may sometimes be insufficient for this purpose such as is the case when CTCLs exhibit polyclonal TCR distributions. Two case reports identified patients with clinical features of CTCL but without dominant TCR clones on PCR or HTS. The researchers instead utilized somatic mutation profiling to identify tumor cell mutations in PIKC3D or TP53 that were consistent with CTCL (39). Single-cell RNA sequencing determined that skin-localized CTCL cells exhibit increased expression of TOX, CCR4, and STAT5 and decreased expression of PSORS1C2, among others, when compared with benign inflammatory dermatoses (40).
Prognosis
Accurate prognostic information in CTCL is critical because it guides decisions about how to best deploy resource-intensive and potentially risky therapeutic interventions. One technique that may be complementary to clinical staging for prognosis is the assessment of multiple parameters including patient age, lactate dehydrogenase (LDH) levels, and the presence of a dominant T cell clone. Newer methods to assess progression risk include HTS to determine tumor clone frequency and identifying unique tumor cell gene expression patterns through transcriptional profiling. These techniques often provide more reliable prognostic value than clinical staging does (7).
Integrating results from flow cytometry, TCR clonotyping, and lab measurements can predict disease progression risk and overall survival in MF. Overall patient survival was predicted via a Prognostic Index Model using the four risk factors of clinical stage IV, age greater than 60 years, elevated LDH, and large-cell transformation in the skin (39). Peripheral blood TCR clonality, flow cytometry, and LDH predicted progression to advanced stage MF (41). A dominant T cell clone in peripheral blood was associated with a shorter time to systemic treatment in patients with Stage IB MF (42). Low-level blood involvement, determined by positive TCR gene rearrangement and abnormal T cell population identification on flow cytometry, was associated with decreased overall survival in patients with stage IA to IIA CTCL (43).
HTS has the potential for more accurate identification of patients with early-stage disease who are at significant risk of disease progression. Tumor clone frequency (TCF) in the skin of patients with MF, as measured by HTS of TCRB, is predictive of progression-free and overall survival in patients with CTCL (44). A TCF of greater than 25% was associated with a 92% positive predictive value and 83% negative predictive value for 5-year disease progression or death. The prognostic value of TCF analysis was superior to other prognostic techniques, including clinical disease staging. Higher TCF was associated with increased somatic mutation burden in CTCL cells, which may explain why increasing TCF frequency correlates with increased disease risk (44).
Transcriptional profiling of CTCL cells holds the potential for predicting disease progression and overall survival based on gene expression patterns. One study performed microarray analysis of skin tumor cells and identified three distinct gene expression clusters that predicted favorable, intermediate, and poor disease prognosis. Certain genes were preferentially expressed in favorable, such as WIF-1, versus poor, such as IL-17F, prognosis clusters (45). RNA sequencing of skin samples from patients with early-stage MF determined that downregulation of genes such as CXCR4 and CD69 and upregulation of genes including HSPA1A and IL7R was associated with tumor progression (46). Malignant cells from clinically unaffected skin showed similar differential gene expression, suggesting that early disease spread may be clinically undetectable. In patients with advanced, disseminated disease, as defined by SS, IL32 overexpression portended poorer survival (2). TOX is overexpressed in early-stage MF, and TOX upregulation correlates with an increased risk of disease progression and decreased survival (47, 48). Whether these gene expression patterns correlate with advanced disease or are true drivers of progression remains unknown.
Advancing understanding of CTCL pathogenesis
Heterogeneity in T cell clones, tumor somatic mutations, and gene expression are key features of CTCL biology. Genetic alterations in SS include copy number alterations, DNA rearrangements and fusion transcripts, single nucleotide variation, and epigenetic changes (49). Genetic alterations in MF include DNA rearrangements, copy number alterations, fusion transcripts, gene mutations, and epigenetic alterations. This heterogeneity presents a challenge to researchers and clinicians who attempt to better understand CTCL pathogenesis. Indeed, one study determined that no one dominant mutation was shared between different patients’ tumor cells (21). Recent efforts by researchers using molecular techniques such as HTS and scRNA-seq are identifying pathways that drive CTCL pathogenesis and identifying potential therapeutic targets. Understanding these is important because it offers a glimpse into the future of CTCL treatment.
A multitude of studies have deployed qRT-PCR, scRNA-seq, and HTS of isolated tumor cells to identify genes associated with CTCL disease progression and response to treatment. Numerous differentially regulated genes have been identified; however, significant heterogeneity exists in mutations found in cells from different patients with the same type of CTCL and even within different tumor cells from the same patient (50, 51). This heterogeneity complicates efforts to identify shared drivers of CTCL pathogenesis, and it may be that currently available sequencing technologies limit researchers’ abilities to identify all mutations. Mutation patterns have been identified despite these challenges. One notable signaling pathway that is aberrantly upregulated in MF and SS is JAK-STAT signaling, which appears to drive increased proliferation and activation of malignant T cells and mediate disease progression in a mouse model (52). Loss of SOCS1 expression, which normally inhibits JAK activity, appears to drive increased JAK signaling in MF. In contrast, in SS copy number variants of the STAT3 and STAT5B genes or gain of function mutations in JAK and STAT genes drive aberrant JAK-STAT signaling.
Several additional mutated genes have been linked to CTCL pathogenesis. A genomic analysis of published CTCL genomic databases identified RLTPR as commonly mutated in tumors, which may drive NF-κB activation and IL-2 production (53). Massive parallel sequencing determined that some CTCL samples exhibited PLCG1 mutations, which was linked with increased NFAT activation that appeared to drive CTCL proliferation and cell survival (54). scRNA-seq determined that tumor cell gene signatures could accurately predict disease stage and that FOXP3 overexpression was a primary factor that could predict early disease in SS (55). In a separate study of patients with SS, tumor cells commonly exhibited activating mutations in CCR4 and CARD1, and ZEB1 was deleted in over half of the patients (2). AIRE, which encodes for a protein that functions in central immune tolerance, was upregulated in 58% of malignant cells in SS versus 8.7% of non-malignant cells in one study (56).
Differential mutational burdens may exist between skin resident and circulating tumor cells from the same patient, influencing disease progression. Single-cell sequencing of tumor cells from matched blood and skin samples from patients with L-CTCL revealed that distinct transcriptional signatures exist based on the tissue from which the cells were recovered (57). Expression of genes such as PDCD1 and NR4A1 were upregulated in cells isolated from the skin, whereas KLF2, TCF7, and SELL were upregulated in cells isolated from the blood. It is unclear if tissue localization drove epigenetic changes or vice versa. Interestingly, skin-localized tumor cells had higher proliferative activity ex vivo as compared to blood-localized cells, which suggested that the skin microenvironment promoted more aggressive malignant expansion than the blood.
Individualizing CTCL treatment
HTS and scRNA-seq characterization of unique gene expression patterns in CTCL to detect vulnerabilities allow for targeted therapies that minimize morbidity. Targetable tumor biology includes aberrant intracellular signaling pathways and altered protein expression. The high level of heterogeneity of the CTCL transcriptional landscape suggests that personalizing treatment to a patient’s unique tumor biology and treatment response monitoring are important components of high-quality care.
Some gene mutation patterns, most commonly TP53 loss of function or deletion mutations, are frequently associated with CTCL. Signaling pathway mutations that promote increased T cell survival and activation include increased TCR/CD28 signaling via decreased PTEN expression or increased expression of PLCG1, CD28, and/or RLTPR, increased JAK/STAT signaling via increased JAK1 and STAT3/STAT6 signaling (58, 59), and increased NFκB signaling via increased expression of CARD11 and IRF4 (53), among other genes. Researchers have documented epigenetic dysregulation with increased gene methylation (8). Ruxolitinib, a JAK1/JAK2 inhibitor, appears to provide treatment benefit to patients with CTCL-containing mutations promoting JAK/STAT signaling (60).
Malignant CTCL cells often overexpress surface proteins. These include the co-stimulatory receptor CD30 (55), the chemokine receptor CCR4 (2), and the IL-2 receptor CD25, among others. Drugs such as brentuximab vedotin, an antibody-drug conjugate, have been successfully utilized to treat CTCL patients with CD30+ tumor cells (61, 62).
Host immune targets offer therapeutic targets. CTCL tumor cells express higher levels of PD1, and anti-PD1 therapies have shown efficacy in treating this. However, in advanced CTCL, tumor cells may lose PD1 expression via PDCD1 gene deletion, a process associated with worse patient survival (63). There is no evidence to date, however, that PD1 blockade can induce accelerated progression of CTCL, even in patients with PDCD1 deletion (64). Blockade of CD47, a protein that suppresses phagocytosis, within skin localized CTCL lesions has shown promise in CTCL treatment (65).
Personalizing CTCL treatments based on tumor mutations and protein expression remains limited by the frequent failure of therapies targeted toward mutated genes. For example, patient responses to brentuximab vedotin are often discordant in relation to measured tumor CD30 expression. Additionally, only a minority of SS patients were found to respond to HDAC inhibitors despite many patients exhibiting HDAC-targetable mutations (66–68). CTCL tumor cells appear to diverge into transcriptionally distinct subsets after HDAC inhibitor therapy (56). These findings may be due to intra-tumoral heterogeneity that permits a fraction of the tumor cells to resist targeted therapies, and so treatment response monitoring is critical.
Accurate monitoring of CTCL treatment response is important to guide clinical decision-making, including determining the appropriateness of de-escalating therapy in patients with excellent disease control or considering the risk/benefit balance of allogeneic stem cell transplant. Tumor cell clone frequency and MF recurrence have been detected via HTS of TCR clones (35). Decreasing numbers and frequencies of clones, as determined by HTS, is associated with effective treatment response, including response to topical and systemic chemotherapy and radiotherapy (69–71).
Clinical adoption of molecular techniques
Widespread implementation of molecular techniques in CTCL treatment awaits resolution of challenging obstacles, including lack of standardization in assay design, differences in interpretation guidelines, and prohibitive cost. The only widely used molecular technique by clinicians in CTCL treatment may be flow cytometry and PCR-based TCR clonality assays which aid in CTCL diagnosis. HTS and NGS offer increased sensitivity and specificity but remain limited in use to a few academic centers.
Flow cytometry and PCR-based TCR clonality assays may, however, suffer from lower specificity despite offering high sensitivity. PCR-based TCR clonality assay costs are variable. For example, one 2018 study reported a range of $75-450 across four academic medical institutions (72). This study also surveyed dermatopathologists and determined that (1) most respondents had some training in clonality assays and were familiar with CTCL diagnosis and workup, (2), clonality assays were commonly used in the evaluation and diagnosis of CTCL, and (3), clonality assays still play an adjunctive role in CTCL diagnosis and management, with clinical, histopathologic, and immunophenotypic features being the primary determinants that clinicians employ. The authors concluded that this might be due to uncertainty regarding the accuracy of clonality assays.
Researchers have demonstrated that HTS and NGS offer superior sensitivity and specificity compared to PCR in TCR clonality (37, 44). This approach can also identify disease across time. One study estimated that, in the authors’ hands, the cost of HTS of CTCL TCR was about three to four times more expensive than PCR-based assays (37). However, implementing these techniques in patient care remains limited by financial and logistical complications to a few academic medical centers.
Conclusion
The numerous challenges to diagnosis and therapy posed by CTCL compels continuous exploration of innovative approaches and techniques to optimize management. Improving molecular techniques offer promise for earlier diagnosis, more accurate prognosis, and more effective individualized therapy for affected patients. The development of standardized protocols to inform how to implement and interpret such tests is critical to enable clinicians to utilize these tests in the care of CTCL patients. Decreasing costs and/or increasing reimbursement for the use of molecular techniques is also important for these goals. Finally, deployment of these techniques in research is crucial to improving understanding of CTCL biology and pathogenesis. A promising future for CTCL treatment awaits if these goals are met.
Author contributions
ML, NB, EM, VL, and AJ were responsible for the conception of the manuscript and reviewed the literature. ML was primarily responsible for drafting the manuscript. ML, NB, RR, EM, VL and AJ revised it critically for intellectual content. All authors approved the manuscript.
Funding
AJ receives funding from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR077194) and the Department of Veterans Affairs (I01BX004907). ML and RR are members of the NIH-funded University of Iowa Medical Scientist Training Program (T32GM139776).
Acknowledgments
We would like to thank the University of Iowa Department of Dermatology and the Medical Scientist Training Program for providing a training environment that is conducive to biomedical research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Campbell JJ, Clark RA, Watanabe R, Kupper TS. Sezary syndrome and mycosis fungoides arise from distinct T-cell subsets: a biologic rationale for their distinct clinical behaviors. Blood (2010) 116(5):767–71. doi: 10.1182/blood-2009-11-251926
2. Wang L, Ni X, Covington KR, Yang BY, Shiu J, Zhang X, et al. Genomic profiling of Sezary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet (2015) 47(12):1426–34. doi: 10.1038/ng.3444
3. Agar NS, Wedgeworth E, Crichton S, Mitchell TJ, Cox M, Ferreira S, et al. Survival outcomes and prognostic factors in mycosis fungoides/Sezary syndrome: validation of the revised International Society for Cutaneous Lymphomas/European Organisation for Research and Treatment of Cancer staging proposal. J Clin Oncol (2010) 28(31):4730–9. doi: 10.1200/JCO.2009.27.7665
5. Dereure O, Portales P, Clot J, Guilhou JJ. Biclonal Sezary syndrome with capillary leak syndrome. Dermatology (1994) 188(2):152–6. doi: 10.1159/000247123
6. Iyer A, Hennessey D, O'Keefe S, Patterson J, Wang W, Wong GK, et al. Skin colonization by circulating neoplastic clones in cutaneous T-cell lymphoma. Blood (2019) 134(18):1517–27. doi: 10.1182/blood.2019002516
7. Walia R, Yeung CCS. An update on molecular biology of cutaneous T cell lymphoma. Front Oncol (2019) 9:1558. doi: 10.3389/fonc.2019.01558
8. Lai P, Wang Y. Epigenetics of cutaneous T-cell lymphoma: biomarkers and therapeutic potentials. Cancer Biol Med (2021) 18(1):34–51. doi: 10.20892/j.issn.2095-3941.2020.0216
9. Baum C, Liu V. Inflammatory mimics of mycosis fungoides. AJSP: Rev Rep (2011) 16(1):45–51. doi: 10.1097/PCR.0b013e318202d439
10. Jawed SI, Myskowski PL, Horwitz S, Moskowitz A, Querfeld C. Primary cutaneous T-cell lymphoma (mycosis fungoides and Sezary syndrome): part I. Diagnosis: clinical and histopathologic features and new molecular and biologic markers. J Am Acad Dermatol (2014) 70(2):205.e1–16; quiz 21-2. doi: 10.1016/j.jaad.2013.07.049
11. van Doorn R, Van Haselen CW, van Voorst Vader PC, Geerts ML, Heule F, de Rie M, et al. Mycosis fungoides: disease evolution and prognosis of 309 Dutch patients. Arch Dermatol (2000) 136(4):504–10. doi: 10.1001/archderm.136.4.504
12. Olsen E, Vonderheid E, Pimpinelli N, Willemze R, Kim Y, Knobler R, et al. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC). Blood (2007) 110(6):1713–22. doi: 10.1182/blood-2007-03-055749
13. Benton EC, Crichton S, Talpur R, Agar NS, Fields PA, Wedgeworth E, et al. A cutaneous lymphoma international prognostic index (CLIPi) for mycosis fungoides and Sezary syndrome. Eur J Cancer (2013) 49(13):2859–68. doi: 10.1016/j.ejca.2013.04.018
14. Sanz-Bueno J, Lora D, Monsalvez V, Maronas-Jimenez L, Postigo C, Rodriguez-Peralto JL, et al. The new Cutaneous Lymphoma International Prognostic index (CLIPi) for early mycosis fungoides failed to identify prognostic groups in a cohort of Spanish patients. Br J Dermatol (2016) 175(4):794–6. doi: 10.1111/bjd.14559
15. de Masson A, Beylot-Barry M, Bouaziz JD, Peffault de Latour R, Aubin F, Garciaz S, et al. Allogeneic stem cell transplantation for advanced cutaneous T-cell lymphomas: a study from the French Society of Bone Marrow Transplantation and French Study Group on Cutaneous Lymphomas. Haematologica (2014) 99(3):527–34. doi: 10.3324/haematol.2013.098145
16. Virmani P, Zain J, Rosen ST, Myskowski PL, Querfeld C. Hematopoietic stem cell transplant for mycosis fungoides and Sezary syndrome. Dermatol Clin (2015) 33(4):807–18. doi: 10.1016/j.det.2015.05.014
17. Hughes CF, Khot A, McCormack C, Lade S, Westerman DA, Twigger R, et al. Lack of durable disease control with chemotherapy for mycosis fungoides and Sezary syndrome: a comparative study of systemic therapy. Blood (2015) 125(1):71–81. doi: 10.1182/blood-2014-07-588236
18. Khodadoust MS, Mou E, Kim YH. Integrating novel agents into the treatment of advanced mycosis fungoides and Sezary syndrome. Blood (2022) 141(7):695–703. doi: 10.1182/blood.2020008241
19. Liu X, Jin S, Hu S, Li R, Pan H, Liu Y, et al. Single-cell transcriptomics links Malignant T cells to the tumor immune landscape in cutaneous T cell lymphoma. Nat Commun (2022) 13(1):1158. doi: 10.1038/s41467-022-28799-3
20. Buus TB, Willerslev-Olsen A, Fredholm S, Blumel E, Nastasi C, Gluud M, et al. Single-cell heterogeneity in Sezary syndrome. Blood Adv (2018) 2(16):2115–26. doi: 10.1182/bloodadvances.2018022608
21. Iyer A, Hennessey D, O'Keefe S, Patterson J, Wang W, Wong GK, et al. Branched evolution and genomic intratumor heterogeneity in the pathogenesis of cutaneous T-cell lymphoma. Blood Adv (2020) 4(11):2489–500. doi: 10.1182/bloodadvances.2020001441
22. Olsen EA, Whittaker S, Willemze R, Pinter-Brown L, Foss F, Geskin L, et al. Primary cutaneous lymphoma: recommendations for clinical trial design and staging update from the ISCL, USCLC, and EORTC. Blood (2022) 140(5):419–37. doi: 10.1182/blood.2021012057
23. Harmon CB, Witzig TE, Katzmann JA, Pittelkow MR. Detection of circulating T cells with CD4+CD7- immunophenotype in patients with benign and Malignant lymphoproliferative dermatoses. J Am Acad Dermatol (1996) 35(3 Pt 1):404–10. doi: 10.1016/S0190-9622(96)90605-2
24. Vonderheid EC, Bernengo MG. The Sezary syndrome: hematologic criteria. Hematol Oncol Clin North Am (2003) 17(6):1367–89. doi: 10.1016/S0889-8588(03)00120-5
25. Gibson JF, Huang J, Liu KJ, Carlson KR, Foss F, Choi J, et al. Cutaneous T-cell lymphoma (CTCL): Current practices in blood assessment and the utility of T-cell receptor (TCR)-Vbeta chain restriction. J Am Acad Dermatol (2016) 74(5):870–7. doi: 10.1016/j.jaad.2015.12.018
26. Feng B, Jorgensen JL, Jones D, Chen SS, Hu Y, Medeiros LJ, et al. Flow cytometric detection of peripheral blood involvement by mycosis fungoides and Sezary syndrome using T-cell receptor Vbeta chain antibodies and its application in blood staging. Mod Pathol (2010) 23(2):284–95. doi: 10.1038/modpathol.2009.175
27. Willemze R, Cerroni L, Kempf W, Berti E, Facchetti F, Swerdlow SH, et al. The 2018 update of the WHO-EORTC classification for primary cutaneous lymphomas. Blood (2019) 133(16):1703–14. doi: 10.1182/blood-2018-11-881268
28. Tang MB, Chong TK, Tan ES, Sun YJ, Tan SH. A comparative study of polymerase chain reaction detection of clonal T-cell receptor gamma chain gene rearrangements using polyacrylamide gel electrophoresis versus fluorescence capillary electrophoresis. Ann Acad Med Singap (2008) 37(1):27–31. doi: 10.47102/annals-acadmedsg.V37N1p27
29. Muche JM, Lukowsky A, Asadullah K, Gellrich S, Sterry W. Demonstration of frequent occurrence of clonal T cells in the peripheral blood of patients with primary cutaneous T-cell lymphoma. Blood (1997) 90(4):1636–42. doi: 10.1182/blood.V90.4.1636.1636_1636_1642
30. Moczko A, Dimitriou F, Kresbach H, Amarov B, Hoetzenecker W, Pascolo S, et al. Sensitivity and specificity of T-cell receptor PCR BIOMED-2 clonality analysis for the diagnosis of cutaneous T-cell lymphoma. Eur J Dermatol (2020) 30(1):12–5. doi: 10.1684/ejd.2020.3698
31. Lukowsky A, Muche JM, Mobs M, Assaf C, Humme D, Hummel M, et al. Evaluation of T-cell clonality in archival skin biopsy samples of cutaneous T-cell lymphomas using the biomed-2 PCR protocol. Diagn Mol Pathol (2010) 19(2):70–7. doi: 10.1097/PDM.0b013e3181b2a1b7
32. Plaza JA, Morrison C, Magro CM. Assessment of TCR-beta clonality in a diverse group of cutaneous T-Cell infiltrates. J Cutan Pathol (2008) 35(4):358–65. doi: 10.1111/j.1600-0560.2007.00813.x
33. Thurber SE, Zhang B, Kim YH, Schrijver I, Zehnder J, Kohler S. T-cell clonality analysis in biopsy specimens from two different skin sites shows high specificity in the diagnosis of patients with suggested mycosis fungoides. J Am Acad Dermatol (2007) 57(5):782–90. doi: 10.1016/j.jaad.2007.06.004
34. Kirsch IR, Watanabe R, O'Malley JT, Williamson DW, Scott LL, Elco CP, et al. TCR sequencing facilitates diagnosis and identifies mature T cells as the cell of origin in CTCL. Sci Transl Med (2015) 7(308):308ra158. doi: 10.1126/scitranslmed.aaa9122
35. Sufficool KE, Lockwood CM, Abel HJ, Hagemann IS, Schumacher JA, Kelley TW, et al. T-cell clonality assessment by next-generation sequencing improves detection sensitivity in mycosis fungoides. J Am Acad Dermatol (2015) 73(2):228–36.e2. doi: 10.1016/j.jaad.2015.04.030
36. Rea B, Haun P, Emerson R, Vignali M, Farooqi M, Samimi S, et al. Role of high-throughput sequencing in the diagnosis of cutaneous T-cell lymphoma. J Clin Pathol (2018) 71(9):814–20. doi: 10.1136/jclinpath-2018-205004
37. Schumacher JA, Duncavage EJ, Mosbruger TL, Szankasi PM, Kelley TW. A comparison of deep sequencing of TCRG rearrangements vs traditional capillary electrophoresis for assessment of clonality in T-Cell lymphoproliferative disorders. Am J Clin Pathol (2014) 141(3):348–59. doi: 10.1309/AJCP5TYGBVW4ZITR
38. Hussaini MO, Srivastava J, Lee LW, Nishihori T, Shah BD, Alsina M, et al. Assessment of clonotypic rearrangements and minimal residual disease in lymphoid Malignancies. Arch Pathol Lab Med (2022) 146(4):485–93. doi: 10.5858/arpa.2020-0457-OA
39. Scarisbrick JJ, Prince HM, Vermeer MH, Quaglino P, Horwitz S, Porcu P, et al. Cutaneous lymphoma international consortium study of outcome in advanced stages of mycosis fungoides and Sezary syndrome: effect of specific prognostic markers on survival and development of a prognostic model. J Clin Oncol (2015) 33(32):3766–73. doi: 10.1200/JCO.2015.61.7142
40. Litvinov IV, Tetzlaff MT, Thibault P, Gangar P, Moreau L, Watters AK, et al. Gene expression analysis in Cutaneous T-Cell Lymphomas (CTCL) highlights disease heterogeneity and potential diagnostic and prognostic indicators. Oncoimmunology (2017) 6(5):e1306618. doi: 10.1080/2162402X.2017.1306618
41. Allen PB, McCook-Veal AA, Switchenko JM, Paulino DM, Niyogusaba T, Baird KM, et al. Staging lymph nodes and blood at diagnosis in mycosis fungoides identifies patients at increased risk of progression to advanced stage: A retrospective cohort study. Cancer (2023) 129(4):541–50. doi: 10.1002/cncr.34579
42. Raychaudhuri S, Charli-Joseph Y, Huang CY, Mintz MA, Pincus LB, Ai WZ. Association of a dominant T-cell clone in peripheral blood with time to systemic treatment in patients with stage IB mycosis fungoides. JAMA Dermatol (2022) 158(8):954–6. doi: 10.1001/jamadermatol.2022.2100
43. Marks JA, Switchenko JM, Martini DJ, Tarabadkar ES, Khan MK, Lechowicz MJ, et al. T-cell receptor gene rearrangement clonality, flow cytometry status, and associated outcomes in early-stage cutaneous T-cell lymphoma. JAMA Dermatol (2021) 157(8):954–62. doi: 10.1001/jamadermatol.2021.2191
44. de Masson A, O'Malley JT, Elco CP, Garcia SS, Divito SJ, Lowry EL, et al. High-throughput sequencing of the T cell receptor beta gene identifies aggressive early-stage mycosis fungoides. Sci Transl Med (2018) 10(440):1–13. doi: 10.1126/scitranslmed.aar5894
45. Litvinov IV, Jones DA, Sasseville D, Kupper TS. Transcriptional profiles predict disease outcome in patients with cutaneous T-cell lymphoma. Clin Cancer Res (2010) 16(7):2106–14. doi: 10.1158/1078-0432.CCR-09-2879
46. Rindler K, Jonak C, Alkon N, Thaler FM, Kurz H, Shaw LE, et al. Single-cell RNA sequencing reveals markers of disease progression in primary cutaneous T-cell lymphoma. Mol Cancer (2021) 20(1):124. doi: 10.1186/s12943-021-01419-2
47. Huang Y, Litvinov IV, Wang Y, Su MW, Tu P, Jiang X, et al. Thymocyte selection-associated high mobility group box gene (TOX) is aberrantly over-expressed in mycosis fungoides and correlates with poor prognosis. Oncotarget (2014) 5(12):4418–25. doi: 10.18632/oncotarget.2031
48. Lefrancois P, Xie P, Wang L, Tetzlaff MT, Moreau L, Watters AK, et al. Gene expression profiling and immune cell-type deconvolution highlight robust disease progression and survival markers in multiple cohorts of CTCL patients. Oncoimmunology (2018) 7(8):e1467856. doi: 10.1080/2162402X.2018.1467856
49. Tensen CP, Quint KD, Vermeer MH. Genetic and epigenetic insights into cutaneous T-cell lymphoma. Blood (2022) 139(1):15–33. doi: 10.1182/blood.2019004256
50. Gaydosik AM, Stonesifer CJ, Khaleel AE, Geskin LJ, Fuschiotti P. Single-cell RNA sequencing unveils the clonal and transcriptional landscape of cutaneous T-cell lymphomas. Clin Cancer Res (2022) 28(12):2610–22. doi: 10.1158/1078-0432.CCR-21-4437
51. Roelens M, Delord M, Ram-Wolff C, Marie-Cardine A, Alberdi A, Maki G, et al. Circulating and skin-derived Sezary cells: clonal but with phenotypic plasticity. Blood (2017) 130(12):1468–71. doi: 10.1182/blood-2017-03-772996
52. Fanok MH, Sun A, Fogli LK, Narendran V, Eckstein M, Kannan K, et al. Role of dysregulated cytokine signaling and bacterial triggers in the pathogenesis of cutaneous T-cell lymphoma. J Invest Dermatol (2018) 138(5):1116–25. doi: 10.1016/j.jid.2017.10.028
53. Park J, Yang J, Wenzel AT, Ramachandran A, Lee WJ, Daniels JC, et al. Genomic analysis of 220 CTCLs identifies a novel recurrent gain-of-function alteration in RLTPR (p.Q575E). Blood (2017) 130(12):1430–40. doi: 10.1182/blood-2017-02-768234
54. Vaque JP, Gomez-Lopez G, Monsalvez V, Varela I, Martinez N, Perez C, et al. PLCG1 mutations in cutaneous T-cell lymphomas. Blood (2014) 123(13):2034–43. doi: 10.1182/blood-2013-05-504308
55. Borcherding N, Voigt AP, Liu V, Link BK, Zhang W, Jabbari A. Single-cell profiling of cutaneous T-cell lymphoma reveals underlying heterogeneity associated with disease progression. Clin Cancer Res (2019) 25(10):2996–3005. doi: 10.1158/1078-0432.CCR-18-3309
56. Borcherding N, Severson KJ, Henderson N, Ortolan LS, Rosenthal AC, Bellizzi AM, et al. Single-cell analysis of Sezary syndrome reveals novel markers and shifting gene profiles associated with treatment. Blood Adv (2023) 7(3):321–35. doi: 10.1182/bloodadvances.2021005991
57. Herrera A, Cheng A, Mimitou EP, Seffens A, George D, Bar-Natan M, et al. Multimodal single-cell analysis of cutaneous T-cell lymphoma reveals distinct subclonal tissue-dependent signatures. Blood (2021) 138(16):1456–64. doi: 10.1182/blood.2020009346
58. Lee K, Evans MG, Yang L, Ng S, Snowden C, Khodadoust M, et al. Primary cytotoxic T-cell lymphomas harbor recurrent targetable alterations in the JAK-STAT pathway. Blood (2021) 138(23):2435–40. doi: 10.1182/blood.2021012536
59. Gaydosik AM, Queen DS, Trager MH, Akilov OE, Geskin LJ, Fuschiotti P. Genome-wide transcriptome analysis of the STAT6-regulated genes in advanced-stage cutaneous T-cell lymphoma. Blood (2020) 136(15):1748–59. doi: 10.1182/blood.2019004725
60. Moskowitz AJ, Ghione P, Jacobsen E, Ruan J, Schatz JH, Noor S, et al. A phase 2 biomarker-driven study of ruxolitinib demonstrates effectiveness of JAK/STAT targeting in T-cell lymphomas. Blood (2021) 138(26):2828–37. doi: 10.1182/blood.2021013379
61. Kim YH, Tavallaee M, Sundram U, Salva KA, Wood GS, Li S, et al. Phase II investigator-initiated study of brentuximab vedotin in mycosis fungoides and sezary syndrome with variable CD30 expression level: A multi-institution collaborative project. J Clin Oncol (2015) 33(32):3750–8. doi: 10.1200/JCO.2014.60.3969
62. Papadavid E, Kapniari E, Pappa V, Nikolaou V, Iliakis T, Dalamaga M, et al. Multicentric EORTC retrospective study shows efficacy of brentuximab vedotin in patients who have mycosis fungoides and Sezary syndrome with variable CD30 positivity. Br J Dermatol (2021) 185(5):1035–44. doi: 10.1111/bjd.20588
63. Park J, Daniels J, Wartewig T, Ringbloom KG, Martinez-Escala ME, Choi S, et al. Integrated genomic analyses of cutaneous T-cell lymphomas reveal the molecular bases for disease heterogeneity. Blood (2021) 138(14):1225–36. doi: 10.1182/blood.2020009655
64. Khodadoust MS, Rook AH, Porcu P, Foss F, Moskowitz AJ, Shustov A, et al. Pembrolizumab in relapsed and refractory mycosis fungoides and sezary syndrome: A multicenter phase II study. J Clin Oncol (2020) 38(1):20–8. doi: 10.1200/JCO.19.01056
65. Ansell SM, Maris MB, Lesokhin AM, Chen RW, Flinn IW, Sawas A, et al. Phase I study of the CD47 blocker TTI-621 in patients with relapsed or refractory hematologic Malignancies. Clin Cancer Res (2021) 27(8):2190–9. doi: 10.1158/1078-0432.CCR-20-3706
66. Kim EJ, Kim YH, Rook AH, Lerner A, Duvic M, Reddy S, et al. Clinically significant responses achieved with romidepsin across disease compartments in patients with cutaneous T-cell lymphoma. Leuk Lymphoma (2015) 56(10):2847–54. doi: 10.3109/10428194.2015.1014360
67. Kim YH, Bagot M, Pinter-Brown L, Rook AH, Porcu P, Horwitz SM, et al. Mogamulizumab versus vorinostat in previously treated cutaneous T-cell lymphoma (MAVORIC): an international, open-label, randomised, controlled phase 3 trial. Lancet Oncol (2018) 19(9):1192–204. doi: 10.1016/S1470-2045(18)30379-6
68. Kiel MJ, Sahasrabuddhe AA, Rolland DCM, Velusamy T, Chung F, Schaller M, et al. Genomic analyses reveal recurrent mutations in epigenetic modifiers and the JAK-STAT pathway in Sezary syndrome. Nat Commun (2015) 6:8470. doi: 10.1038/ncomms9470
69. Weng WK, Armstrong R, Arai S, Desmarais C, Hoppe R, Kim YH. Minimal residual disease monitoring with high-throughput sequencing of T cell receptors in cutaneous T cell lymphoma. Sci Transl Med (2013) 5(214):214ra171. doi: 10.1126/scitranslmed.3007420
70. Rook AH, Gelfand JM, Wysocka M, Troxel AB, Benoit B, Surber C, et al. Topical resiquimod can induce disease regression and enhance T-cell effector functions in cutaneous T-cell lymphoma. Blood (2015) 126(12):1452–61. doi: 10.1182/blood-2015-02-630335
71. O'Malley JT, de Masson A, Lowry EL, Giobbie-Hurder A, LeBoeuf NR, Larocca C, et al. Radiotherapy eradicates Malignant T cells and is associated with improved survival in early-stage mycosis fungoides. Clin Cancer Res (2020) 26(2):408–18. doi: 10.1158/1078-0432.CCR-18-4147
Keywords: cutaneous T cell lymphoma, flow cytometry, RNA sequencing, high throughput sequencing, TCR - T cell receptor
Citation: Lefebvre MN, Borcherding N, Reis RJ, Mou E, Liu V and Jabbari A (2023) Molecular techniques drive cutting edge advancements in management of cutaneous T cell lymphoma. Front. Immunol. 14:1228563. doi: 10.3389/fimmu.2023.1228563
Received: 25 May 2023; Accepted: 27 July 2023;
Published: 15 August 2023.
Edited by:
Patrizia Fuschiotti, University of Pittsburgh, United StatesReviewed by:
Zhiming Li, Sun Yat-sen University Cancer Center (SYSUCC), ChinaCopyright © 2023 Lefebvre, Borcherding, Reis, Mou, Liu and Jabbari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ali Jabbari, ali-jabbari@uiowa.edu
 Mitchell N. Lefebvre
Mitchell N. Lefebvre Nicholas Borcherding3
Nicholas Borcherding3 Ali Jabbari
Ali Jabbari