- Department of Clinical Laboratory Center, Beijing Youan Hospital, Capital Medical University, Beijing, China
Background: Clinically, some patients whose HBsAg becomes negative owing to antiviral therapy or spontaneously still show a low level of HBV DNA persistence in serum. T-lymphocyte subsets, cytokine levels and HBV S gene sequences were analyzed in this study.
Methods: A total of 52 HBsAg-negative and HBV DNA-positive patients(HBsAg-/HBV DNA+ patients), 52 persistently HBsAg-positive patients(HBsAg+/HBV DNA+ patients) and 16 healthy people were evaluated. T-lymphocyte subsets of these patients were detected by flow cytometry, serum cytokines and chemokines were detected by the Luminex technique, and the HBV S region was evaluated by Sanger sequencing. T%, T-lymphocyte, CD8+ and CD4+T lymphocyte were lower in the HBsAg-negative group than in the HC group. Compared with the HBsAg-positive group, the HBsAg-negative group had lower levels in T lymphocyte %, CD8+T lymphocyte %, CD8+T lymphocyte and CD4/CD8. These difference were statistically significant (P<0.05). Serum IFN-γ, IFN-α and FLT-3L levels were significantly higher in the HBsAg-negative group than in the HBsAg-positive group (P<0.05). However, levels of many cytokines related to inflammation (i.e., IL-6, IL-8, IL10, IL-12, IL-17A) were lower in the HBsAg-negative group. Fifty-two HBsAg-negative samples were sequenced, revealing high-frequency amino acid substitution sites in the HBV S protein, including immune escape mutations (i.e., Y100C, S114T, C124Y, P127L, G130R, T131N, M133T, C137S, G145A) and TMD region substitutions (i.e., E2K/R/D, G7D/R, G10D, A17R, F20L/S, L21V, L22V).
Conclusions: According to the results of T-lymphocyte subsets and serum cytokines, it can be deduced that the cellular immune function of HBsAg-negative patients is superior to that of HBsAg-positive patients, with attenuation of liver inflammation. HBsAg-negative patients may show a variety of mutations and amino acid replacement sites at high frequency in the HBV S region, and these mutations may lead to undetectable HBsAg, HBsAg antigenic changes or secretion inhibition.
Hepatitis B virus (HBV) infection is a major global health problem. According to the latest data released by the WHO in 2020, there are approximately 257 million cases of chronic HBV infection worldwide. Moreover, approximately 887,000 people die each year due to uncompensated liver cirrhosis, liver failure, primary liver cancer and other end-stage liver diseases caused by HBV infection, indicating that the HBV infection rate has not decreased (1). In hepatitis B virus infection, the most representative serological marker is hepatitis B surface antigen (HBsAg), but some patients infected with hepatitis B virus infection may be negative for HBsAg. Occult HBV infection (OBI) is described as having HBV DNA in the blood, and the current detection methods do not include hepatitis B surface antigen (HBsAg) (2). Loss of HBsAg and anti-HBs seroconversion are considered signs of hepatitis B virus (HBV) elimination. However, serum/intrahepatic HBV DNA can be found in some patients who are negative for serum HBsAg. This status is described as OBI (1, 2). OBI may lead to progression of liver cirrhosis and eventually become an important risk factor for hepatocellular carcinoma (3). Cases of OBI arise from a variety of sources, including patients who are clinically cured of chronic hepatitis B, with HBsAg being serologically cleared but with a small amount of HBV remaining in the liver, or patients infected with mutant HBV (3–5). In addition, it is important to emphasize that the window of acute hepatitis B virus infection is not OBI patients. At present, the mechanism of OBI and how it participates in the occurrence of liver injury remain uncertain, and more studies and long-term clinical follow-up are necessary to better understand the viral and host biological mechanisms and clinical significance of OBI. This study included patients with HBsAg-negative and HBV DNA-positive states in the clinic.
There are several mechanisms that might explain such a state of infection, including the following: mutations in HBV genes, particularly HBsAg escape mutations that are undetectable by commercial HBsAg assays; host mechanisms that lead to strong suppression of HBV replication and transcription; host immune surveillance; coinfection with other viruses, such as hepatitis C virus (HCV); and epigenetic mechanisms (6). The HBV S region can be divided into three sections: N-terminal (aa 1-99), major hydrophilic region (MHR) (aa 100-169) and C-terminal (aa 170-226) regions (7, 8). The ‘α’ antigen determinant (aa 124-147 or 149) in the major hydrophilic region (MHR) is the primary target by which HBsAg is recognized by anti-HBs and immune cells (9). There are four transmembrane domains of HBsAg (TMDs): TMD1 (aa 4-24), TMD2 (aa 80-98), TMD3 (aa 160-193) and TMD4 (aa 202-222). Amino acid substitution in and around the ‘α’ antigen determinant may change the conformation of HBsAg and T-cell epitopes, altering immunogenicity (10). HBV S gene mutations are known to influence the occurrence of HBsAg-negative and HBV DNA-positive cases. These mutations are called immune escape-associated mutations, and they are mainly located in the MHR. A variety of mutations associated with HBsAg-negative and HBV DNA-positive cases have been discovered. The most well-known immune escape mutant is G145R, which impairs HBsAg secretion, antigenicity and anti-HBs binding (11).
It is generally believed that HBV infection can cause disruption of the function and proliferation of various immune cells involved in innate and adaptive immune responses. In general, the result of HBV infection is determined by the interaction between the virus and host (12). HBV itself is a noncytopathogenic virus, and liver damage is mainly attributed to the host's immune response. During HBV infection, the host immune response is a double-edged sword that defends against infection by destroying virus-infected cells while inducing liver inflammation and aggravating liver injury (13, 14). Cellular immunity is crucial in the occurrence and progression of hepatitis B disease. T helper cells (Th cells) are classified into Th1 and Th2 subsets according to the different cytokines produced, and the Th1/Th2 balance is critical for maintaining immune function. In chronic HBV infection, Th cells secrete high levels of Th2-type cytokines, inducing the cellular immune balance toward Th2 cells, high expression of HBsAg in serum and liver tissues, a decrease in the number of T lymphocyte subsets, significantly reduced cytokine production ability, and functional exhaustion (15–17).
At present, clinical research on HBsAg-negative and HBV DNA-positive patients is still lacking. Based on a large amount of clinical data, serum HBsAg clearance after nucleotide antiviral treatment or spontaneously may endure for more than 6 months in a small number of HBV-infected patients while serum HBV DNA remains present at a low level in serum. Nevertheless, the majority of HBV-infected patients remain positive for HBsAg after antiviral treatment, and HBV DNA can become negative or be present in serum to varying degrees. The aim of this study was to elucidate the characteristics of T lymphocyte subsets, serum cytokines and HBV S gene status in HBsAg-negative and HBV DNA-positive patients.
Materials and methods
Population studied
This is a cross-sectional study. Between May 2019 and May 2022, 279 patients who were HBsAg negative(<0.05 IU/mL, S/CO<1) and HBV DNA positive(≥10 IU/mL) were screened by hospital data systems. Only 52 of the 279 patients had persistent HBsAg-/HBV DNA+ serological status(>3 months). They had all been diagnosed with chronic hepatitis B in the past. 52 HBsAg+/HBV DNA+ patients who were diagnosed with chronic hepatitis B were matched by sex, age, and disease type. At the same time, clinical residual serum samples from 16 healthy people were collected as the control group. The criteria for healthy subjects were: healthy, no liver disease, no other diseases, and normal laboratory tests. The HBsAg-positive patient samples came from the patient's first blood draw after admission, before antiviral treatment. The diagnostic criteria are based on WHO guidelines for the prevention and treatment of chronic hepatitis B (2019 version) (12). The exclusion criteria were as follows: HBV infection window period; HCV infection not combined with other viral infections; alcoholic liver disease; streptocarpus; drug-induced liver injury; autoimmune diseases, hematological tumors and other serious genetic and metabolic diseases; use of immunosuppressants and hormones within the past three months; and post chemotherapy. The study was approved by the Medical Ethics Committee of Capital Medical University, Beijing Youan Hospital. Because the study used clinical residual serum samples, it was exempt from informed consent.
Laboratory assays
Roche automatic immaterialize was routinely used to detect serum HBsAg, anti-HBs, HBeAg, anti-HBe and anti-HBc at the hospital clinical examination center. HBsAg titer<0.05 IU/mL and anti-HBs<10 IU/L, HBeAg S/CO<1, or anti-HBe and anti-HBc S/CO>1 was considered negative. Quantitative detection of serum HBV DNA was carried out by Abbott Real-Time fluorescence quantitative PCR. The lower limit of quantification was 10 IU/mL (34.1 HBV DNA copies/mL) in the 0.5 mL sample preparation protocol. The results of the assay were expressed as undetected, detectable but below the lower limit of quantification (i.e., HBV DNA detected but not quantifiable), and calculated results in IU/mL within the linear range of the assay. The Siemens Aptio automatic biochemical assembly pipeline was used to detect liver function indexes, such as serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin (TBIL), and direct bilirubin (DBIL).
Detection of peripheral blood T-lymphocyte subsets
Whole-blood samples were analyzed using a BECKMAN flow cytometer (Coulter, USA). Lymphocytes were analyzed using a gate set on a forward scatter vs. side scatter, and three-color flow cytometry was applied to combine CD3, CD4 and CD8. The peripheral blood T-lymphocyte subsets detected in each sample were analyzed using CellQuest software (Coulter, USA).
Cytokine and chemokine profiles
We use MILLIPLEX MAP Human Cytokine/Chemokine/Growth Factor Panel A 48 Plex Premixed Magnetic Bead Panel Kit (Merck, USA) following the manufacturer’s recommendations and a Luminex MAGPIX Instrument System, as used for simultaneous quantification of any or all of the following analytes in human tissue/cell lysate and culture supernatant samples and serum or plasma samples: Soluble Cluster of Differentiation 40 Ligand (sCD40L), epidermal growth factor (EGF), Eotaxin, fibroblast growth factor 2 (FGF-2), Fms-related tyrosine kinase 3 ligand (FLT-3L), Fractalkine, granulocyte-stimulating factor(G-CSF), granulocyte macrophage colony-stimulating factor (GM-CSF), growth-regulated oncogene-α (GRO-α), interferon-α2 (IFN-α2), interferon-γ (IFN-γ), IL-1α, IL-1β, interleukin 1 receptor antagonist (IL-1RA), IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12 (p40), IL-12(p70), IL-13, IL-15, IL-17A, IL-17E/IL-25, IL-17F, IL-18, IL-22, IL-27, interferon gamma-induced protein 10 (IP-10), monocyte chemoattractant protein 1 (MCP-1/CCL2), MCP-3, macrophage-stimulating factor(M-CSF), macrophage-derived chemokine (MDC), monokine induced by IFN-γ (MIG/CXCL9), macrophage inflammatory protein 1α (MIP-1α/CCL3), MIP-1β/CCL4, platelet-derived growth factor AA (PDGF-AA), PDGF-AB/BB, regulated upon activation normal T-Cell expressed and presumably secreted (RANTES/CCL5), TGF-α, tumor necrosis factor α (TNF-α), TNF-β/lymphotoxin alpha (LTA), and vascular endothelial growth factor A (VEGF-A). Serum cytokine profiles were quantitatively measured in 52 HBsAg-negative and HBV DNA-positive patients, 52 HBsAg-positive patients and 16 healthy subjects. MILLIPLEX® products are based on Luminex xMAP technology. Cytokines with more than 50% missing data were not analyzed. Cytokine concentrations have an intra-assay coefficient of variation within 15%. As 4 of the 48 cytokines were not detectable in more than 50% of the samples (GM-CSF, IL-3, IL-7, IL-22), these four cytokines were not analyzed. A variety of cytokine levels are shown in the Supplementary Materials.
Sequencing analysis of the HBV S region
HBV DNA was extracted from 1.0 ml of serum using the silica gel membrane centrifugal column method. First-round and second-round PCR were performed to amplify the HBV S region using PrimeSTAR MAX DNA polymerase. The primers used for both rounds of PCR were PF (5’-TTCCTGC TGGTGGCTCCAGTTC-3’, nt54-75) and PR (5’-TTCCGCAGTATGGATCGGCAG-3’, nt1258-1278), amplifying a 1224-bp fragment. PCR amplicons were assessed by 1% agarose gel electrophoresis (200 V for 20 min), and positive amplicons were purified. Both strands of purified amplification products were sequenced directly using ABI 3730xl DNA Analyzer. Homology evaluations were performed with the GenBank database using BLAST analysis at https://www.ncbi.nlm.nih.gov. Nucleic and amino acid sequences were analyzed using MEGA 7.0 software. HBV was genotyped based on the full sequence of the S gene using an online prediction tool (https://hbv.geno2pheno.org/index.php). Amino acid substitutions were determined by comparing specimen sequences to the genotype consensus sequence from the alignment of genotype sequences downloaded from https://www.ncbi.nlm.nih.gov. The immune escape mutation of the HBV S protein was determined by the Geno-2-pheno-hbv tool (https://hbv.geno2pheno.org/index.php).
Statistical analysis
Statistical analysis was performed using SPSS 26.0. Single-factor ANOVA was employed for comparison of continuous variables following a normal distribution and the SNK-q test for comparisons between groups. A t test was used to compare two groups of continuous variables. The Kruskal-Wallis H test was used to compare continuous variables with a nonnormal distribution between groups, and the Mann-Whitney U test was used to compare the two groups. Two groups of classifying variables were compared using the X2 test. Continuous variables are expressed as (±s) or the median (P25-P75), and a difference was considered to be statistically significant at P< 0.05. GraphPad Prism 8 was utilized for the statistical analysis.
Results
Basic clinical data and laboratory test results
The HBsAg-negative patients were mainly male, at 76.92% (40/52), and the mean age was 53 years old. Most of the HBsAg-negative patients had received antiviral therapy with nucleotide analogs and/or interferon according to their clinical history. The positive rates of anti-HBs and anti-HBe in HBsAg-negative patients were significantly higher than those among HBsAg-positive patients[51.92%(27/52) vs. 0.00%(0/52), 80.77% (42/52) vs. 57.69%(30/52)] (P<0.05). A total of 80.77% of HBsAg-negative patients had HBV DNA below 200 IU/mL, and the median value of HBV DNA load was significantly lower than that of HBsAg-positive patients (log, 1.76 vs. 5.77 IU/mL) (P<0.05). Compared with HBsAg-positive patients, median levels of ALT, AST and TBA in HBsAg-negative patients were lower (36 vs. 103 U/L, 38 vs. 92 U/L and 12.40 vs. 120.00 μmol/L), and the differences were significant (P<0.05) (Table 1).
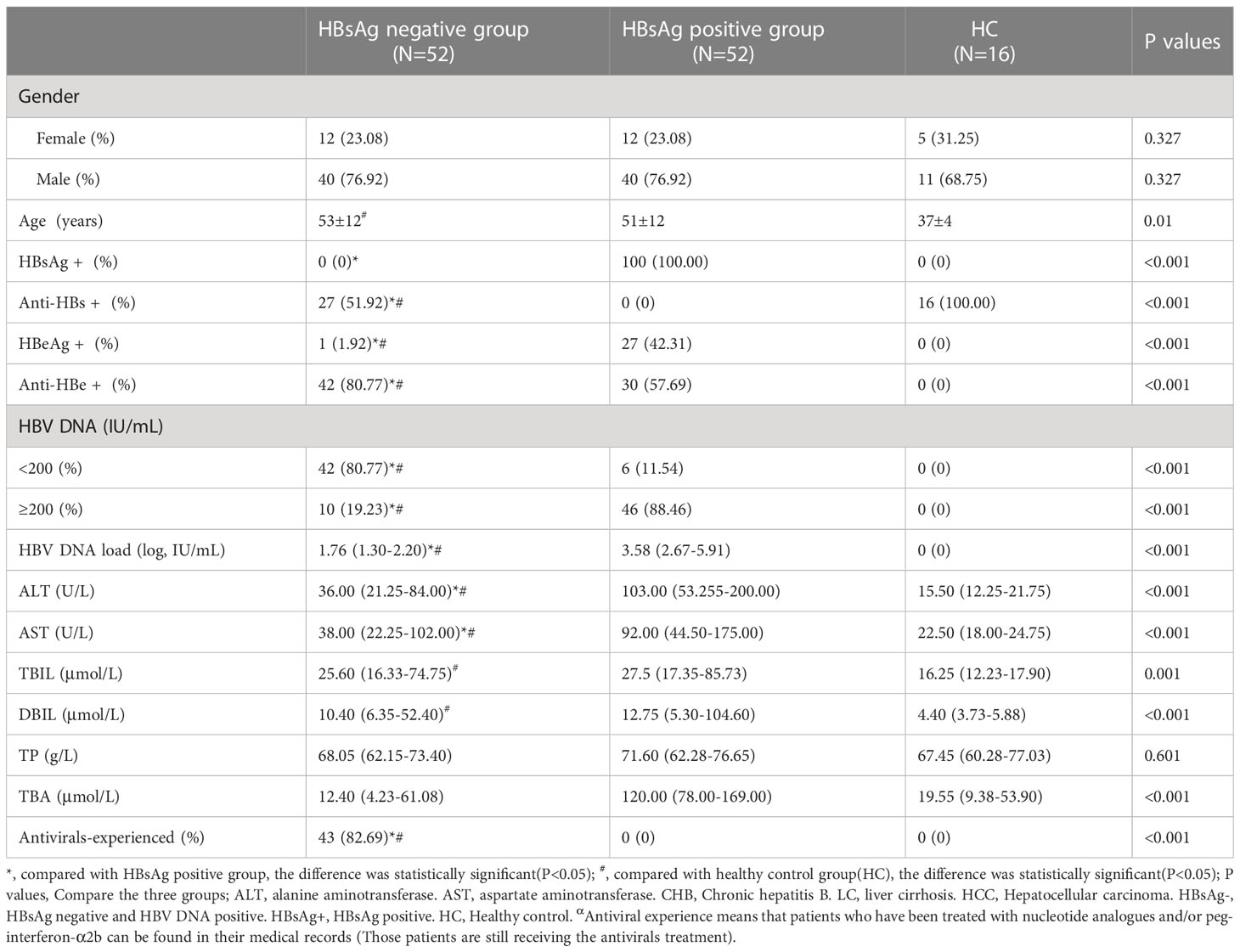
Table 1 General clinical information of HBsAg negative group, HBsAg positive group, and healthy control group.
Characteristics of peripheral blood T-lymphocyte subsets
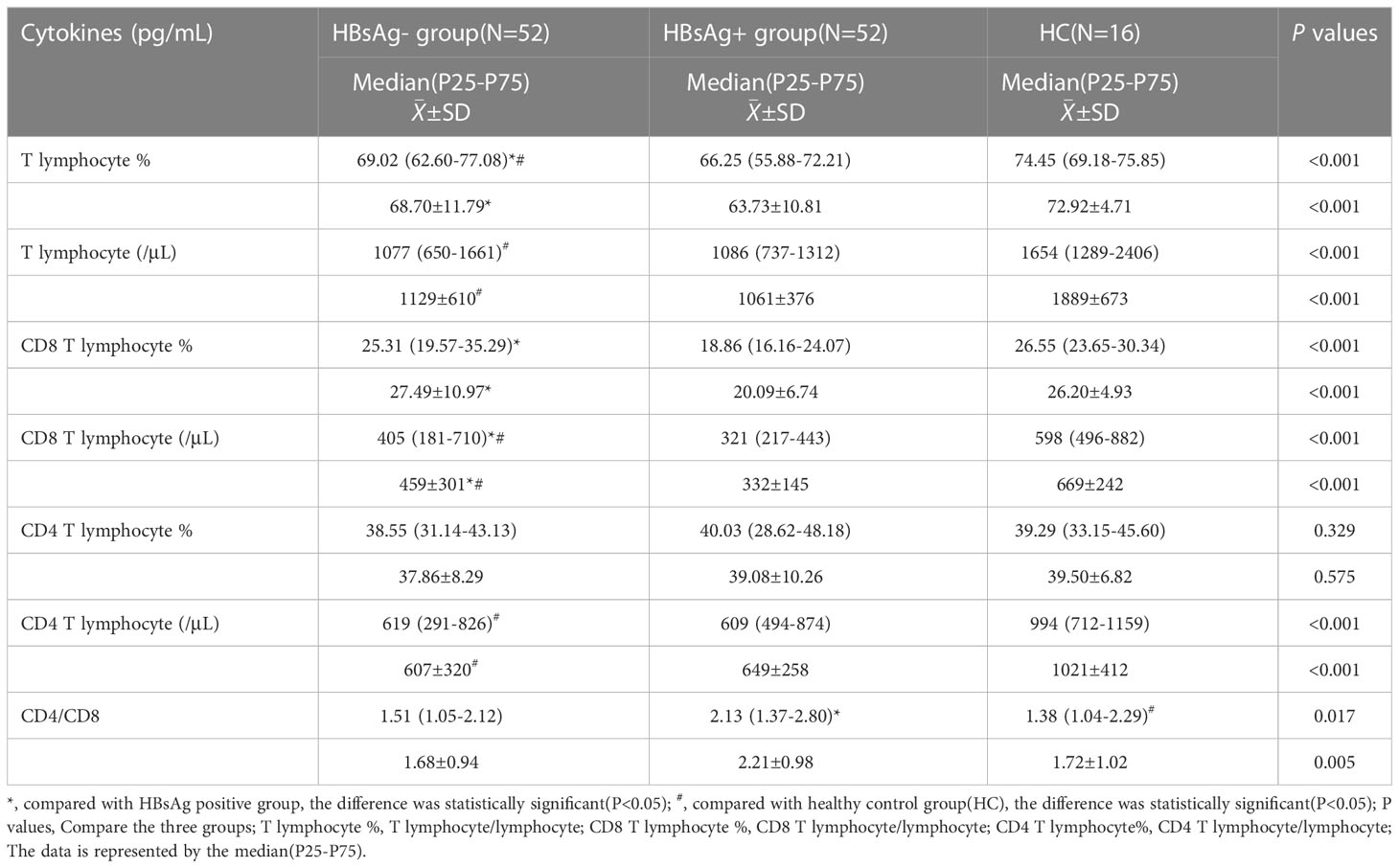
The number of T lymphocytes and CD8+T lymphocytes decreased successively in the healthy control group, HBsAg-negative group and HBsAg-positive group. Each group of data was compared with the mean and median(Table 2). Levels of T lymphocytes and CD8+ T lymphocytes in HBsAg-negative group were higher than HBsAg-positive group, and the difference was statistically significant (P<0.05). The number of T-lymphocyte subsets in the HBsAg negative group and HBsAg positive group was lower than that in the healthy control group. Ratios of T lymphocytes/lymphocytes(T lymphocyte%), T lymphocytes, CD8+T lymphocytes/ lymphocytes(CD8+T lymphocyte%) and CD8+T lymphocytes in the HBsAg-negative group were significantly lower than those in the HBsAg-positive group(P<0.05). However, the CD4/CD8 ratio was the highest in HBsAg-positive group, and the difference was statistically significant compared with the other two groups(P<0.05). There was no difference in the CD4/CD8 ratio between HBsAg-negative group and healthy control group (Figure 1).
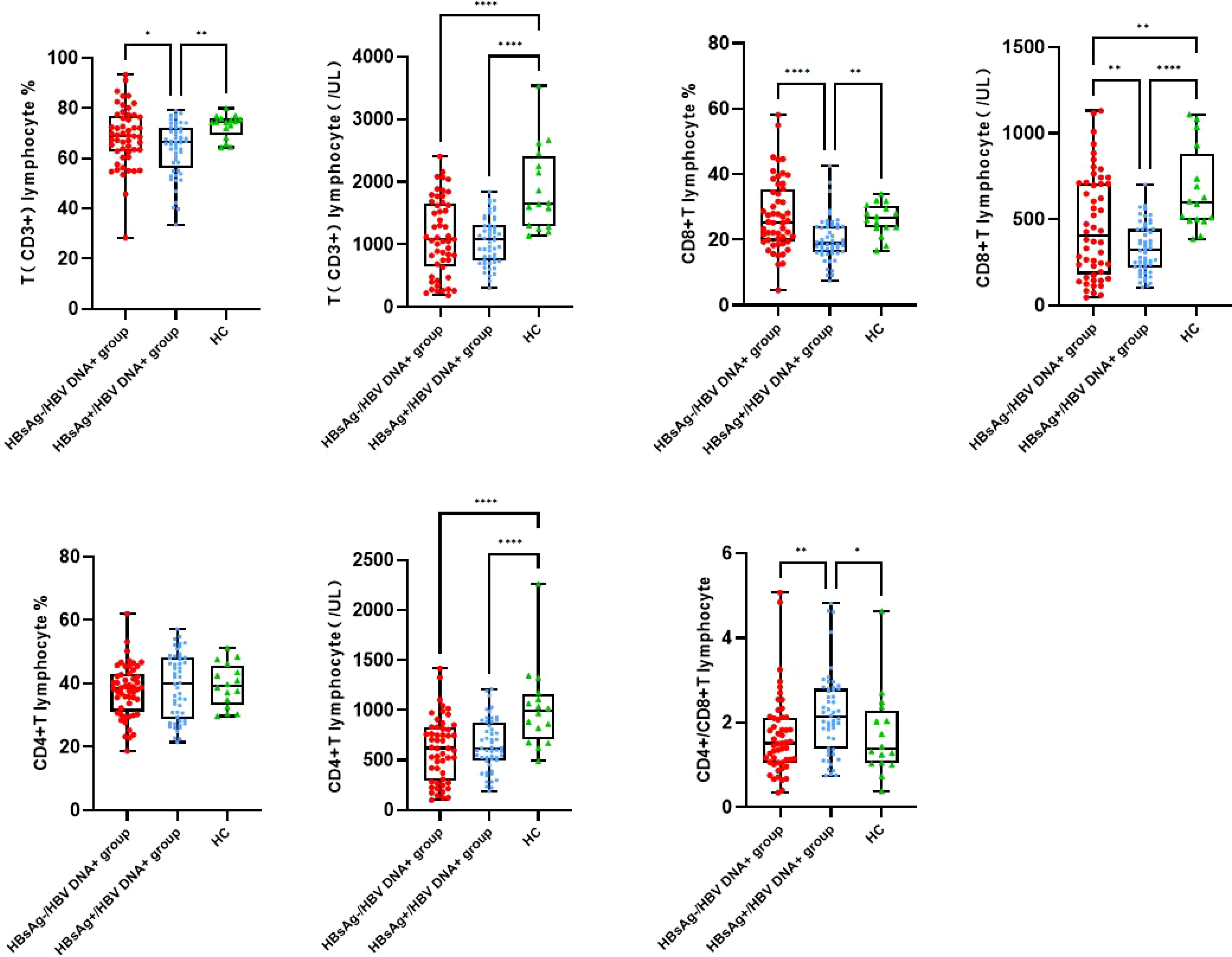
Figure 1 Peripheral blood T lymphocyte subsets among the three groups. Comparison of the mean of T lymphocyte, CD4 and CD8 T lymphocyte and CD4/CD8 ratio. HBsAg-, HBsAg negative and HBV DNA positive. HBsAg+, HBsAg positive. *P<0.05, **P<0.01, ****P<0.0001.
Levels of serum cytokines and chemokines
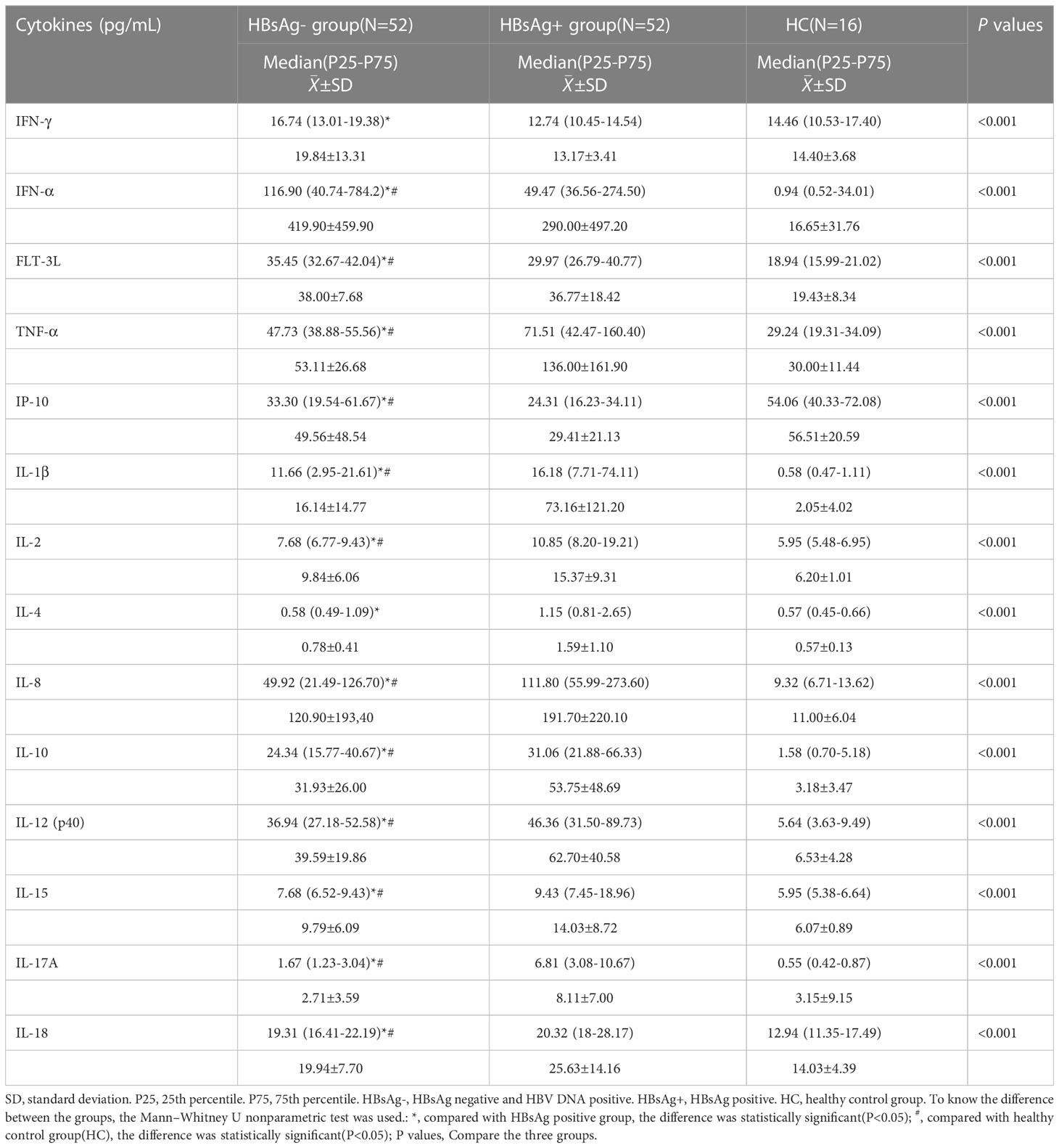
By comparing median levels of cytokines and chemokines between the HBsAg-negative group and HBsAg-positive groups, significant differences in IFN-γ, IFN-α, FLT-3L, IP-10, TNF-α, IL-1β, IL-2, IL-4, IL-8, IL-10, IL-12, IL-15 IL-17A and IL-18 were found (P<0.05). Additionally, higher median values of IFN-γ, IFN-α and FLT-3L were detected in the HBsAg-negative group and higher median levels of IL-1β, IL-2, IL-4, IL-8, IL-10, IL-12, IL-15, IL-17A and IL-18 were detected in the HBsAg-positive group (Figure 2, Table 3). Among the 48 cytokines and chemokines detected, 14 each showed significant differences between the two groups, with the other cytokines having no significant difference. Furthermore, median levels of IFN-γ and FLT-3 L in the HBsAg-positive group were lower than those in the HBsAg-negative group and healthy controls, with significant differences (P<0.05); median values of IFN-α were highest in the HBsAg-negative group, with significant differences between groups (P<0.05). Moreover, median levels of IFN-α, IP-10, TNF-α, IL-1β, IL-2, IL-8, IL-10, IL-12, IL-15, IL-17A and IL-18 were higher in the HBsAg-negative and HBsAg-positive groups than in the healthy controls, a statistically significant difference (P<0.05). The median (P25-P75) and differences in cytokines in the three groups are shown in Figure 2 and Table 3.
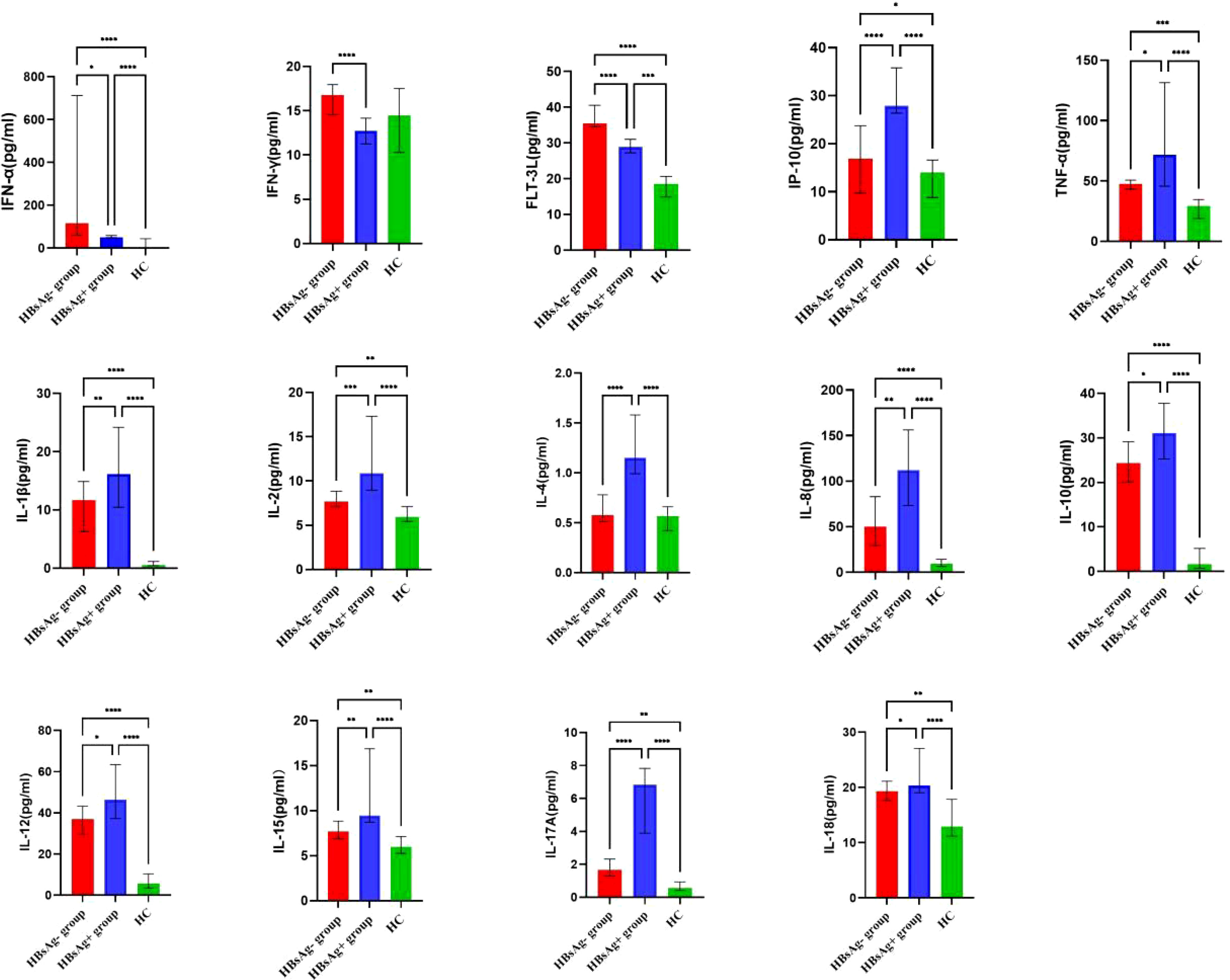
Figure 2 Comparison of the median levels of serum cytokine in the HBsAg negative, HBsAg positive and healthy control group. The figure shows the median with 95%CI of cytokines in three groups patients. Kruskal-Wallis H test was used for comparison among the three groups, and Benjamini test was used for pairwise comparison between groups. HBsAg-, HBsAg negative and HBV DNA positive. HBsAg+, HBsAg positive. *P<0.05. **P<0.01. ***P<0.001, ****P<0.0001.
HBV S region mutation and amino acid substitution in HBsAg-negative and HBV DNA-positive patients
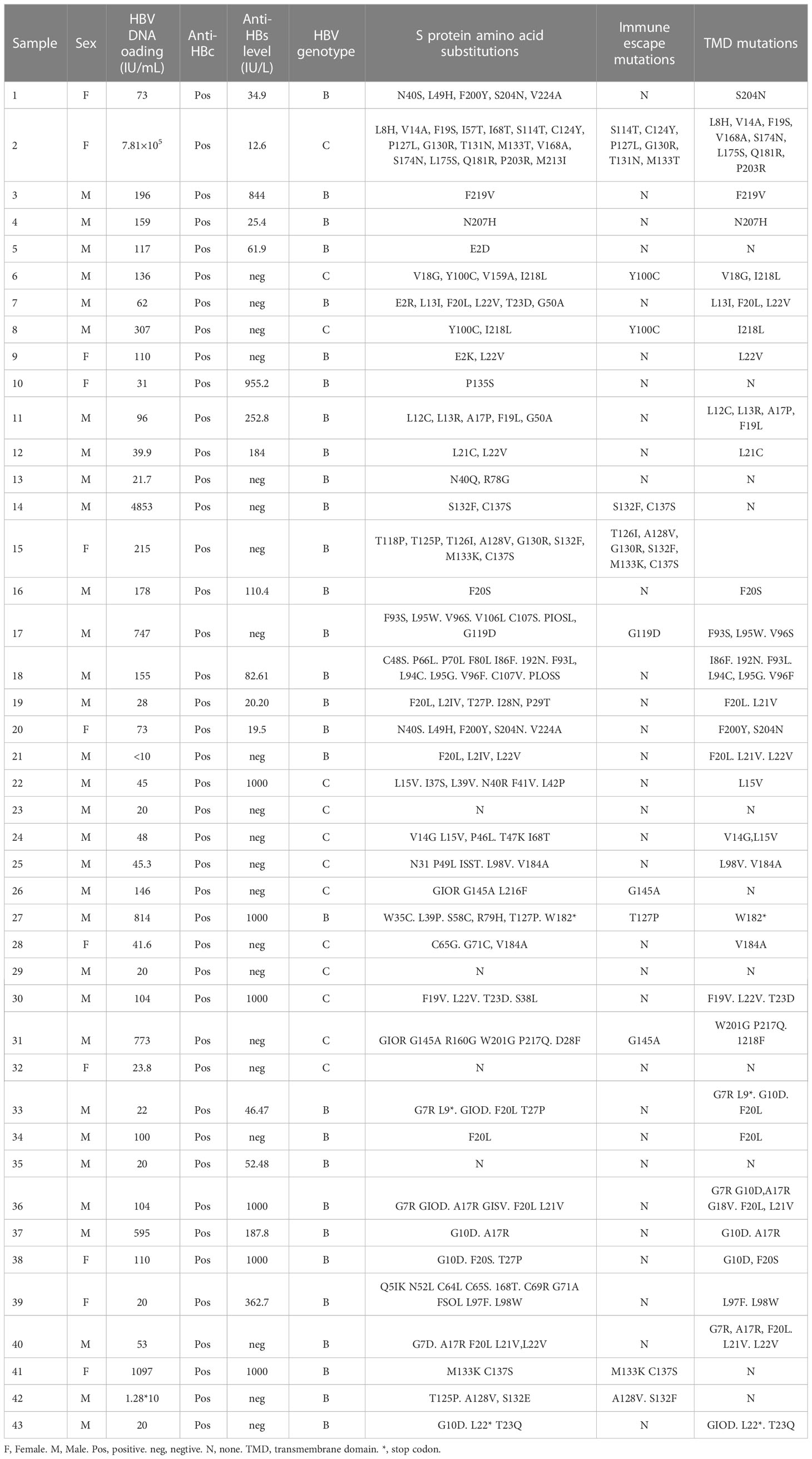
Only 43 of 52 HBsAg-negative samples were successfully sequenced; thus, 43 HBV S gene sequences were analyzed. Genotypes of HBV were determined according to the S gene sequence. Of the 30 HBV S region fragments identified in HBsAg-negative strains, 30 were classified as genotype B (30/43, 69.77%) and 13 strains as genotype C (13/43, 31.23%). DNA sequencing revealed an amino acid substitution in the HBV S protein in 39 of the 43 (90.70%) HBsAg-negative and HBV DNA-positive patients. E kinds of immune escape mutation sites were detected in the genotype B HBV S protein, namely, G119D, T126I, T127P, A128V, G130R, S132F, M133K and C137S, in six HBsAg-negative and HBV DNA-positive patients (6/43, 13.95%). Additionally, 8 kinds of immune escape mutation sites were detected in the genotype C HBV S protein in five HBsAg-negative and HBV DNA-positive patients: Y100C, S114T, C124Y, P127L, G130R, T131N, M133T and G145A (5/43, 11.63%). Amino acid substitution in the transmembrane domain (TMD) region was observed in 31 HBsAg-negative patients (31/43, 72.09%), but only 8 HBsAg-negative samples showed an amino acid substitution in the "α" determinant (amino acids 124-147) of the S protein (Table 4). One female patient’s HBV DNA load was as high as 7.81×105 IU/mL, but HBsAg was negative after antiviral treatment with entecavir. In her sample, we detected many HBV S region mutations resulting in multiple amino acid replacement sites, including immune escape mutations (S114T, C124Y, P127L, G130R, T131N and M133T) and TMD region mutations (L8H, V14A, F19S, V168A, S174N, L175S, Q181R and P203R). There were also three patients with high HBV DNA loads at more than 1×104 IU/mL, and they also showed many HBV S region mutations (Table 4).
Amino acid substitution frequency in HBV S protein
A total of 43 samples from HBsAg-negative patients and 49 samples from HBsAg-positive patients were successfully sequenced. Among 43 HBV S-region sequences in HBsAg-negative patients, 30 (30/43, 69.77%) were genotype B and 13 (13/43, 31.23%) genotype C; 34 of 49 (34/49, 69.39%) HBV S-region sequences in HBsAg-positive patients were genotype B and 15 (15/49, 31.23%) genotype C. The mutation rate of the HBV S protein in HBsAg-negative patients was 90.70% (39/43) and that in HBsAg-positive patients was 28.57% (14/49), significantly higher in HBsAg-negative patients. F20L/S (9/30, 30.00%) displayed the highest occurrence frequency in HBsAg-negative patients with genotype B, followed by L22V (5/30, 16.67%) and G10D (5/30, 16.67%). The frequency of multiple immune escape mutations in HBsAg-negative patients with the B and C genotypes was higher than that in HBsAg-positive patients. A variety of amino acid substitution sites also occurred at high frequency in the TMD region of the S protein in HBsAg-negative patients. The details of amino acid substitution frequencies for HBsAg-negative and HBsAg-positive patients are provided in Table 5.
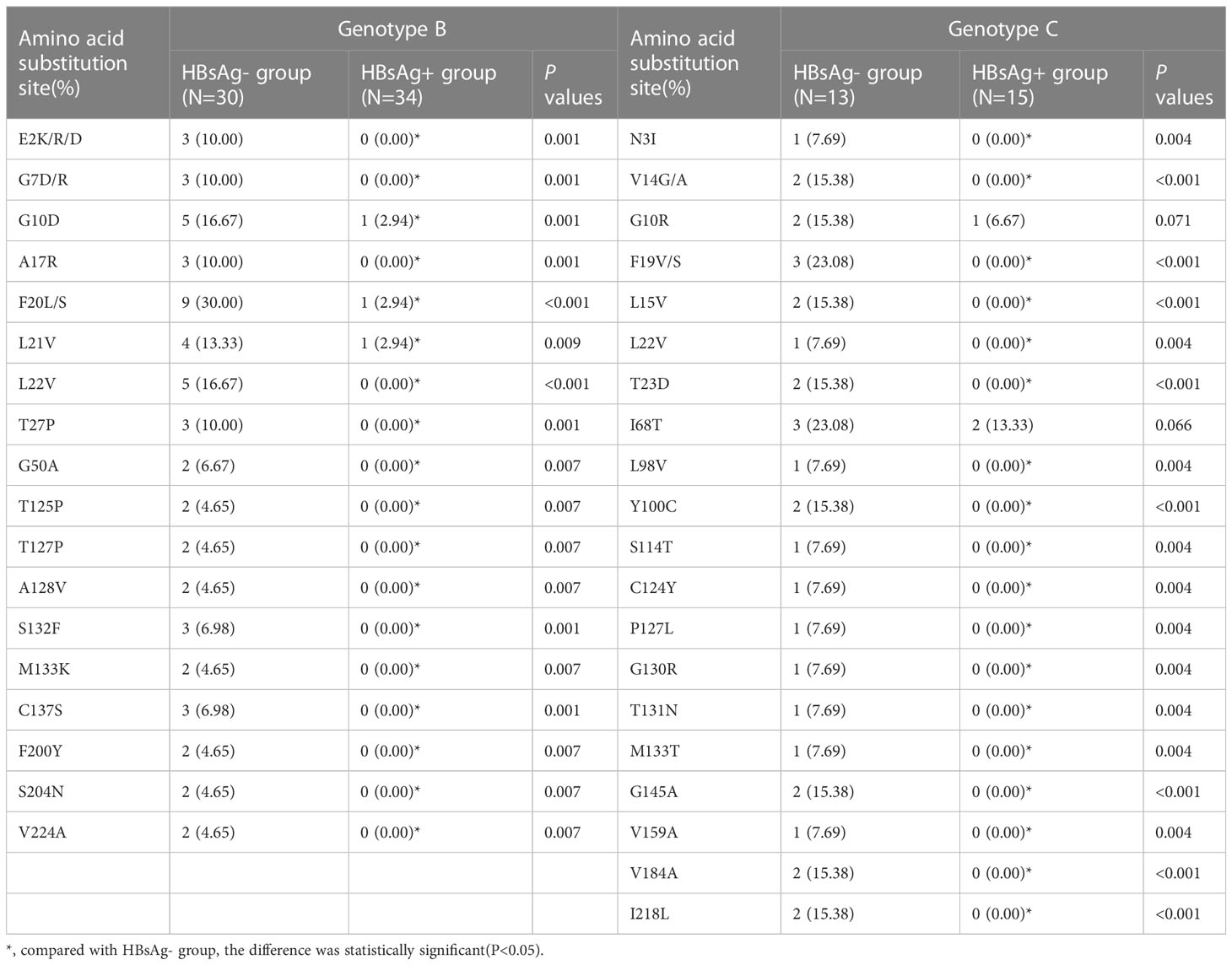
Table 5 Frequency of amino acid substitution of HBV S protein in HBsAg negative and HBsAg positive patients.
Discussion
In this study, HBsAg-negative patients were treated with antiviral therapy or spontaneously experienced a gradual decrease in HBsAg titer until it disappeared; the HBV DNA load also decreased but remained at a low level in serum. This paper describes T-lymphocyte subsets, various cytokine levels and HBV S gene mutations in HBsAg-negative and HBV DNA-positive patients. The subjects of this study were clinically HBsAg-negative and HBV DNA-positive patients, and their serological characteristics and viral load were the same as those of OBI patients among blood donors in previous studies (18–20). Hence, regardless of whether HBsAg-negative cases are caused by antiviral therapy or naturally occurring OBI, clinical indicator characteristics are the same.
HBeAg is a marker of active HBV DNA replication in hepatocytes. However, most HBsAg-negative patients are negative for HBeAg, suggesting low viral replication in these patients. The positive rates of anti-HBs and anti-HBe in HBsAg-negative and HBV DNA-positive patients in our study were 51.92% and 80.77%, respectively, significantly higher than those in HBsAg-positive patients. Thus, most HBsAg-negative patients have undergone serological conversion, with low levels of viral replication and protein expression. To explore the host immune function and inflammatory state in HBsAg-negative patients, levels of T lymphocyte subsets and serum cytokines were analyzed in this study; the HBV S gene was also sequenced and analyzed. Despite suppression of HBV DNA replication and protein expression, the virus may not be completely eliminated. In fact, a small amount of HBV cccDNA remains in liver tissue, with the potential to progress to cirrhosis and hepatocellular carcinoma. As it is considered a risk factor for hepatocellular carcinoma, it cannot be ignored (21–23).
Some studies have suggested that HBsAg-negative and HBV DNA-positive cases are related to viral genome mutations, that is, HBsAg antigenicity changes in HBsAg-negative and HBV DNA-positive patients with mutations in the HBV PreS/S region. Mutations located in the immunodominant "α" determinant region (aa124-147 or 149) can impair the efficiency of HBsAg detection reagents by reducing the binding affinity of the HBsAg capture antibody (24). A single mutation in the "α" determinant in the main hydrophilic region (MHR) of the HBV S region can also inhibit HBsAg secretion (25). Additionally, amino acid substitution in the reverse transcriptase (RT) domain may lead to low levels of HBV DNA replication and HBsAg synthesis, resulting in HBsAg-negative and HBV DNA-positive cases (26). The substitutions M133T and T131N generate an extra N-linked glycosylation site that reportedly does not affect HBsAg secretion but may mask antigenic sites affecting detection (27). Moreover, the mutants Y100C and P127L may affect HBsAg expression and secretion as well as anti-HBs binding (28).
Among the 43 successfully sequenced HBsAg-negative samples, 39 HBV S region fragments showed mutations; among HBsAg-positive samples, 49 samples were sequenced successfully, and 14 HBV S region fragments showed mutations. The mutation rate of the HBV S gene in HBsAg-negative patients (39/43, 90.70%) was significantly higher than that in HBsAg-positive patients (14/49, 28.57%) in our study. Six HBsAg-negative samples showed immune escape mutation sites in the genotype B HBV S region, G119D, T126I, T127P, A128V, G130R, S132F, M133K and C137S (29), with immune escape mutation sites in the genotype C HBV S region in five HBsAg-negative samples, Y100C, S114T, C124Y, P127L, G130R, T131N, M133T and G145A (29). No immune escape mutation was detected in HBsAg-positive patients. These HBsAg immune escape mutants may have arisen as a result of specific selection, such as through the host immune system due to vaccination or antiviral selective pressure attenuating production of HBsAg and resulting in low plasma HBV DNA levels (30). Furthermore, these mutations may be responsible for virus detection failure in routine screening (25). Indeed, this is common in the clinical setting in which patients may have HBsAg-negative results but classical symptoms and, if tested for HBV DNA, will show high HBV DNA levels. In general, detection of some of these emerging mutants has become a major challenge for commercially available immunoassays. Thirty-one HBsAg-negative samples in the TMD region exhibited multiple amino acid substitution sites, many of which were newly discovered, such as E2K/R/D, G10D, A17R, F20L/S, L21V, L22V, F200Y, and S204N. E2 site mutations in the S protein are confirmed to impair secretion of HBsAg, which significantly affected detection of HBsAg (31). For genotype B, the highest mutation frequency of the S protein in HBsAg-negative patients was sF20L/S; regarding genotype C, sF19V/S had the highest frequency. Both mutations occur in the TMD region of the S protein. sF20L/S and sF19V/S are high-frequency mutations associated with HBsAg negativity, which may be a result of the pressure of antiviral therapy and have certain effects on HBsAg production or secretion. The truncation mutant sW182* was present in one HBsAg-negative patient in the study of Pollicino et al (32). This mutation induces retention of the truncated S protein in the perinuclear endoplasmic reticulum (ER) and is associated with lower HBV transcript levels due to decreased stability (33). Large-scale studies and in vitro experiments are needed to determine the significance of newly detected mutations.
Studies have shown that the immune system of HBsAg-negative and HBV DNA-positive patients strongly inhibits viral replication and gene expression (34–37). The ability of CD8 T lymphocytes to proliferate and secrete cytokines (e.g., IFN-γ, IL-2) is weakened in chronic HBV infection (38), and T-lymphocyte subsets can be used to assess the immune function of the host. As cytokines can alter the process of infection and affect the tendency and progression of chronic hepatitis B, they are worthy of study.
In this study, the number of CD4+ T lymphocytes, CD8+ T lymphocytes, CD4+ T lymphocytes/lymphocytes (CD4+ T%) and CD8+ T lymphocytes/lymphocytes (CD8+ T%) was lowest in the HBsAg-positive group, which may be related to the low immune function caused by chronic HBV infection. The numbers of CD4+ T cells and CD8+ T cells in the HBsAg-negative group were higher than those in the HBsAg-positive group. Levels of a variety of cytokines and chemokines (IFN-γ, IFN-α, FLT-3 L) related to immune regulation and virus clearance were also higher in the HBsAg-negative group. Therefore, the immune function of HBsAg-negative patients is enhanced compared with HBsAg-positive patients, which may be related to virus suppression in the former after antiviral treatment. The immune function of HBV-infected patients treated with antiviral interferon may also be enhanced, which can increase the clearance rate of HBsAg. Because this study was a cross-sectional study and we could only determine T lymphocyte subsets and serum cytokine levels after HBsAg clearance, it is unclear whether enhanced host immune function occurred before or after HBsAg clearance. Regardless, it can be confirmed that HBsAg-negative patients have strong host immune function. Although the number of CD4 and CD8 T cells in the HBsAg-negative patients was higher than that in the HBsAg-positive patients, it was lower than that in the healthy controls. Therefore, the immune function of HBsAg-negative patients did not completely recover to the normal level, which corresponds to the low level of serum HBV DNA in HBsAg-negative patients. A high level of HBsAg is associated with impairment of anti-HBV-specific T and B-cell immune function. Reducing the HBsAg level should promote recovery of specific immune function and in turn promote clearance of HBsAg (39, 40). In this study, we only analyzed the increase in overall T-lymphocyte subsets in HBsAg-negative patients and did not detect the immune function of HBV-specific T lymphocytes. Further research is needed.
Cytokines represent a large family of molecules, including the following: Th1-associated cytokines [e.g., IL-2 and IFN-γ], which have a functional contribution to cellular immune responses; Th2-associated cytokines (e.g., IL-4, IL-6, and IL-10), which have roles in humoral immune responses; regulatory T-cell (Treg)-associated cytokines [e.g., tumor growth factor-beta (TGF-β) and IL-10], which have been associated with immunomodulation and immunosuppression; and Th17-associated cytokines (e.g., IL-17, IL-22, and IL-23), which play critical roles in mediating inflammation (41). There are few studies on the production profile of cytokines and chemokines in HBsAg-negative and HBV DNA-positive patients, and the mechanism of liver injury is still unclear, but some studies have shown that persistence and transcription of HBV cccDNA in hepatocytes can stimulate production of cytokines, such as TNF-α and IFN-γ, resulting in hepatocyte injury (17). We conclude that levels of TNF-α and IFN-γ in HBsAg-negative patients are significantly higher than those in healthy subjects, which may be related to a small amount of HBV in the body. The persistence and transcription of HBV cccDNA in hepatocytes can lead to the production of cytokines, such as TNF-α and INF-γ (38). Cytokines and chemokines are essential effector molecules in the HBV-related inflammatory response. CD4+ T cells, CD8+ T cells, NK cells, DC cells and their related cytokines participate in the immune injury process of chronic HBV infection (42, 43).
In addition, IFN-γ, IFN-α and FLT-3 L levels in HBsAg-negative patients were significantly higher than those in HBsAg-positive patients in our study. High HBV DNA load and high HBeAg and HBsAg levels may inhibit immune cell function, leading to a reduction in FLT-3L, IFN-γ, and other cytokines with virus clearance effects and to an increase in the level of the most important cytokine for immunosuppression: IL-10 (43–45). HBsAg induces depletion phenotypes and dysfunction of T and B cells, leading to innate and adaptive immune deficiencies, and lowering serum HBsAg contributes to recovery of the host immune response (46). The increase in IFN-γ and FLT-3 L in the HBsAg-negative patients in the present study may be due to the decrease in HBV DNA and clearance of HBsAg after the use of nucleotide analog antiviral drugs. HBV DNA reduction and HBsAg clearance in HBsAg-negative patients after antiviral therapy may promote recovery of cellular immune function, thus leading to increased serum IFN-γ and FLT-3L levels. In general, levels of various proinflammatory cytokines (IL-2, IL-4, IL-8, IL-10, IL-12, IL-15, IL17A, IL-18) in HBsAg-positive patients were higher than those in HBsAg-negative patients and healthy people. Compared with HBsAg-positive patients, the inflammatory reaction of HBsAg-negative patients was reduced and the immune response function enhanced, but they were still in a state of HBV infection. Overall, a mild inflammatory response compared to healthy people was observed.
Conclusion
High HBsAg serum levels lead to failure of the host immune system, preventing an effective antiviral response. Levels of HBsAg and virus decrease after antiviral treatment, and the inhibitory effect on host cellular immune function is weakened. Cellular immune function is gradually enhanced, which further increases the virus clearance effect. However, HBV S gene mutation may occur in HBsAg-negative patients during antiviral therapy, which leads to amino acid substitutions in the S protein. If the mutation occurs in the main hydrophilic region, the antigenicity of the surface antigen would be changed, rendering commercial detection reagents unable to detect it. If the mutation occurs in the TMD region and changes the S protein conformation, it may affect production and secretion of HBsAg and result in a concentration of serum HBsAg lower than the limit of detection. The decrease in HBsAg secretion is beneficial to host cellular immune function, and antiviral immune responses are continuously stimulated by persistent/intermittent low levels of HBV antigens. This study found a variety of new high-frequency mutation sites in clinically HBsAg-negative and HBV DNA-positive patients.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://doi.org/10.6084/m9.figshare.23903928.v1.
Ethics statement
The study was approved by the Medical Ethics Committee of Capital Medical University, Beijing Youan Hospital. Because the study used clinical residual serum samples, it was exempt from informed consent.
Author contributions
XL and S-XC are joint first authors(Equal & First authors). XL and S-XC are responsible for designing research plans, collecting clinical data, analyzing data, and writing first drafts of papers. HL participate in the review and editing of the first draft. J-LL participates in the acquisition of research funds and the review and revision of papers. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Beijing Outstanding Talents Training fund young backbone individual project (grant no. 2018000021469G287).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1211980/full#supplementary-material
Glossary
References
1. Stasi C, Silvestri C, Voller F. Hepatitis B vaccination and immunotherapies: an update. Clin Exp Vaccine Res (2020) 9(1):1–7. doi: 10.7774/cevr.2020.9.1.1
2. European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol (2017) 67(2):370–98. doi: 10.1016/j.jhep.2017.03.021
3. Locarnini S, Raimondo G. How infectious is the hepatitis B virus? Readings from the occult. Gut (2019) 68(2):182–3. doi: 10.1136/gutjnl-2018-316900
4. Hoofnagle JH, Seeff LB, Bales ZB, Zimmerman HJ. Type B hepatitis after transfusion with blood containing antibody to hepatitis B core antigen. N Engl J Med (1978) 298(25):1379–83. doi: 10.1056/NEJM197806222982502
5. Cho HJ, Cheong JY. Role of immune cells in patients with hepatitis B virus-related hepatocellular carcinoma. Int J Mol Sci (2021) 22(15):8011. doi: 10.3390/ijms22158011
6. Bréchot C, Hadchouel M, Scotto J, Fonck M, Potet F, Vyas GN, et al. State of hepatitis B virus DNA in hepatocytes of patients with hepatitis B surface antigen-positive and negative liver diseases. Proc Natl Acad (1981) 78:3906–10. doi: 10.1073/pnas.78.6.3906
7. Jiang X, Chang L, Yan Y, Wang L. Paradoxical HBsAg and anti-HBs coexistence among chronic HBV infections:causes and consequences. Int J Biol Sci (2021) 17(4):1125–37. doi: 10.7150/ijbs.55724
8. Wu Y, Zhu Z, Wu J, Bi W, Xu W, Xia X, et al. Evolutionary analysis of pre-S/S mutations in HBeAg-negative chronic hepatitis B with HBsAg< 100 IU/ml. Front Public Health (2021) 9:633792. doi: 10.3389/fpubh.2021.633792
9. Yong-Lin Y, Qiang F, Ming-Shun Z, Jie C, Gui-Ming M, Zu-Hu H, et al. Hepatitis B surface antigen variants in voluntary blood donors in Nanjing, China. Virol J (2012) 9:82. doi: 10.1186/1743-422X-9-82
10. Zhu HL, Li X, Li J, Zhang ZH. Genetic variation of occult hepatitis B virus infection. World J Gastroenterol (2016) 22(13):3531–46. doi: 10.3748/wjg.v22.i13.3531
11. Xue Y, Wang MJ, Yang ZT, Yu DM, Han Y, Huang D, et al. Clinical features and viral quasispecies characteristics associated with infection by the hepatitis B virus G145R immune escape mutant. Emerg Microbes Infect (2017) 6(3):e15. doi: 10.1038/emi.2017.2
12. Wu J, Han M, Li J, Yang X, Yang D. Immunopathogenesis of HBV infection. Adv Exp Med Biol (2020) 1179:71–107. doi: 10.1007/978-981-13-9151-4_4
13. Khanam A, Chua JV, Kottilil S. Immunopathology of chronic hepatitis B infection: role of innate and adaptive immune response in disease progression. Int J Mol Sci (2021) 22(11):5497. doi: 10.3390/ijms22115497
14. Nguyen LN, Nguyen L, Zhao J, Schank M, Dang X, Cao D, et al. Immune activation induces telomeric DNA damage and promotes short-lived effector T cell differentiation in chronic HCV infection. Hepatology (2021) 74(5):2380–94. doi: 10.1002/hep.32008
15. Bertoletti A, Ferrari C. Innate and adaptive immune responses in chronic hepatitis B virus infections: towards restoration of immune control of viral infection. Gut (2012) 61(12):1754–64. doi: 10.1136/gutjnl-2011-301073
16. Martinot-Peignoux M, Lapalus M, Asselah T, Marcellin P. The role of HBsAg quantification for monitoring natural history and treatment outcome. Liver Int (2013) 33(Suppl 1):125–32. doi: 10.1111/liv.12075
17. Ribeiro CR, de Almeida NA, Martinelli KG, Pires MA, Mello CE, Barros JJ, et al. Cytokine profile during occult hepatitis B virus infection in chronic hepatitis C patients. Virol J (2021) 18(1):15. doi: 10.1186/s12985-021-01487-2
18. Liu CJ, Chen DS, Chen PJ. Epidemiology of HBV infection in Asian blood donors: emphasis on occult HBV infection and the role of NAT. J Clin Virol (2006) 36(Suppl 1):S33–44. doi: 10.1016/S1386-6532(06)80007-7
19. Ye X, Liu L, Chen L, Nie X, Huang L, Ye D, et al. High-frequency notable HBV mutations identified in blood donors with occult hepatitis B infection from heyuan city of southern China. Front Immunol (2022) 13:754383. doi: 10.3389/fimmu.2022.754383
20. Fopa D, Candotti D, Tagny CT, Doux C, Mbanya D, Murphy EL, et al. Occult hepatitis B infection among blood donors from Yaoundé, Cameroon. Blood Transfus (2019) 17(6):403–8. doi: 10.2450/2019.0182-19
21. Wang J, Liu Y, Liao H, Liu L, Chen R, Si L, et al. The sK122R mutation of hepatitis B virus (HBV) is associated with occult HBV infection: Analysis of a large cohort of Chinese patients. J Clin Virol (2020) 130:104564. doi: 10.1016/j.jcv.2020.104564
22. Simonetti J, Bulkow L, McMahon BJ, HOman C, Snowball M, Negus S, et al. Clearance of hepatitis B surface antigen and risk of hepatocellular carcinoma in a cohort chronically infected with hepatitis B virus. Hepatology (2010) 51(5):1531–7. doi: 10.1002/hep.23464
23. Chen L, Zhao H, Yang X, Gao JY, Cheng J. HBsAg-negative hepatitis B virus infection and hepatocellular carcinoma. Discovery Med (2014) 18(99):189–93.
24. Cao GW. Clinical relevance and public health significance of hepatitis B virus genomic variations. World J Gastroenterol (2009) 15(46):5761–9. doi: 10.3748/wjg.15.5761
25. Salpini R, Colagrossi L, Bellocchi MC, Surdo M, Becker C, Alteri C, et al. Hepatitis B surface antigen genetic elements critical for immune escape correlate with hepatitis B virus reactivation upon immunosuppression. Hepatology (2015) 61(3):823–33. doi: 10.1002/hep.27604
26. Li S, Zhao K, Liu S, Wu C, Yao Y, Cao L, et al. HBsAg sT123N mutation induces stronger antibody responses to HBsAg and HBcAg and accelerates in vivo HBsAg clearance. Virus Res (2015) 210:119–25. doi: 10.1016/j.virusres.2015.08.004
27. Bi X, Tong S. Impact of immune escape mutations and N-linked glycosylation on the secretion of hepatitis B virus virions andsubviral particles: Role of the small envelope protein. Virology (2018) 518:358–68. doi: 10.1016/j.virol.2018.03.011
28. Taffon S, Genovese D, Blasi M, Pierotti P, Degli Esposti A, Catone S, et al. HBV whole-genome mutation profile in HIV-1/HBV coinfected patients in a long-term follow-up study. Infection (2014) 42(4):675–87. doi: 10.1007/s15010-014-0616-2
29. Kim MH, Kang SY, Lee WI. Occult HBV among anti-HBc alone: mutation analysis of an HBV surface gene and pre-S gene. Yonsei Med J (2017) 58(3):557–63. doi: 10.3349/ymj.2017.58.3.557
30. Liu Y, Wang CM, Cheng J, Liang ZL, Zhong YW, Ren XQ, et al. Hepatitis B virus in tenofovir-naive Chinese patients with chronic hepatitis B contains no mutation of rtA194T conferring a reduced tenofovir susceptibility. Chin Med J (Engl) (2009) 122:1585–6.
31. Wang H, Liao F, Xie J, Gao W, Wang M, Huang J, et al. E2 site mutations in S protein strongly affect hepatitis B surface antigen detection in the occult hepatitis B virus. Front Microbiol (2021) 12:664833. doi: 10.3389/fmicb.2021.664833
32. Pollicino T, Amaddeo G, Restuccia A, Raffa G, Alibrandi A, Cutroneo G, et al. Impact of hepatitis B virus (HBV) preS/S genomic variability on HBV surface antigen and HBV DNA serum levels. Hepatology (2012) 56:434–43. doi: 10.1002/hep.25592
33. Xiang KH, Michailidis E, Ding H, Peng YQ, Su MZ, Li Y, et al. Effects of amino acid substitutions in hepatitis B virus surface protein on virion secretion, antigenicity, HBsAg and viral DNA. J Hepatol (2017) 66(2):288–96. doi: 10.1016/j.jhep.2016.09.005
34. Pollicino T, Raffa G, Costantino L, Lisa A, Campello C, Squadrito G, et al. Molecular and functional analysis of occult hepatitis B virus isolates from patients with hepatocellular carcinoma. Hepatology (2007) 45(2):277–85. doi: 10.1002/hep.21529
35. Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, et al. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology (2006) 130(3):823–37. doi: 10.1053/j.gastro.2006.01.001
36. Toscanini F, De Leo P, Calcagno G, Malfatti F, Grasso A, Anselmo M. Hepatitis B reactivation in a HBsAg-negative, HBcAb-positive patient receiving fludarabine for the treatment of chronic lymphocytic leukaemia. Case Rep Hepatol (2011) 2011:258791. doi: 10.1155/2011/258791
37. Hoofnagle JH. Reactivation of hepatitis B. Hepatology (2009) 49(5 Suppl):S156–65. doi: 10.1002/hep.22945
38. Xiong S, Zhu D, Liang B, Li M, Pan W, He J, et al. Longitudinal characterization of phenotypic profile of T cells in chronic hepatitis B identifies immune markers associated with HBsAg loss. EBioMedicine (2021) 69:103464. doi: 10.1016/j.ebiom.2021.103464
39. Kondo Y, Ninomiya M, Kakazu E, Kimura O, Shimosegawa T. Hepatitis B surface antigen could contribute to the immunopathogenesis of hepatitis B virus infection. ISRN Gastroenterol (2013) 2013:935295. doi: 10.1155/2013/935295
40. Tsai KN, Kuo CF, Ou JJ. Mechanisms of hepatitis B virus persistence. Trends Microbiol (2018) 26(1):33–42. doi: 10.1016/j.tim.2017.07.006
41. Huang Z, van Velkinburgh JC, Ni B, Wu Y. Pivotal roles of the interleukin-23/T helper 17 cell axis in hepatitis B. Liver Int (2012) 32(6):894–901. doi: 10.1111/j.1478-3231.2012.02764.x
42. Oh IS, Park SH. Immune-mediated liver injury in hepatitis B virus infection. Immune Netw (2015) 15(4):191–8. doi: 10.4110/in.2015.15.4.191
43. Li M, Zhang L, Xie S, Sun F, Zeng Z, Deng W, et al. Dynamic changes of cytokine profiles and virological markers associated with HBsAg loss during peginterferon alpha-2a treatment in HBeAg-positive chronic hepatitis B patients. Front Immunol (2022) 13:892031. doi: 10.3389/fimmu.2022.892031
44. Li MH, Zhang D, Zhang L, Qu XJ, Lu Y, Shen G, et al. Ratios of T-helper 2 cells to T-helper 1 cells and cytokine levels in. Chin Med J (2017) 130(15):1810–5. doi: 10.4103/0366-6999.211541
45. Li Y, Fan W, Link F, Wang S, Dooley S. Transforming growth factor β latency: A mechanism of cytokine storage and signalling regulation in liver homeostasis and disease. JHEP Rep (2021) 4(2):100397. doi: 10.1016/j.jhepr.2021.100397
Keywords: T lymphocyte, cytokine, HBsAg negative, HBV DNA, Occult hepatitis B virus infection
Citation: Liu X, Chen S-x, Liu H and Lou J-l (2023) Host immunity and HBV S gene mutation in HBsAg-negative HBV-infected patients. Front. Immunol. 14:1211980. doi: 10.3389/fimmu.2023.1211980
Received: 25 April 2023; Accepted: 20 July 2023;
Published: 14 August 2023.
Edited by:
Rongtuan Lin, McGill University, CanadaReviewed by:
Juandy Jo, University of Pelita Harapan, IndonesiaChuanwu Zhu, Fifth People's Hospital of Suzhou, China
Copyright © 2023 Liu, Chen, Liu and Lou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-li Lou, loujinli@ccmu.edu.cn
†These authors share first authorship
 Xin Liu
Xin Liu Shu-xiang Chen
Shu-xiang Chen Hui Liu
Hui Liu