- 1Center for Interdisciplinary Research in Health, Universidade Católica Portuguesa, Lisboa, Portugal
- 2Faculty of Medicine, Universidade Católica Portuguesa, Rio de Mouro, Portugal
The success of the first licensed mRNA-based vaccines against COVID-19 has created a widespread interest on mRNA technology for vaccinology. As expected, the number of mRNA vaccines in preclinical and clinical development increased exponentially since 2020, including numerous improvements in mRNA formulation design, delivery methods and manufacturing processes. However, the technology faces challenges such as the cost of raw materials, the lack of standardization, and delivery optimization. MRNA technology may provide a solution to some of the emerging infectious diseases as well as the deadliest hard-to-treat infectious diseases malaria, tuberculosis, and human immunodeficiency virus/acquired immunodeficiency syndrome (HIV/AIDS), for which an effective vaccine, easily deployable to endemic areas is urgently needed. In this review, we discuss the functional structure, design, manufacturing processes and delivery methods of mRNA vaccines. We provide an up-to-date overview of the preclinical and clinical development of mRNA vaccines against infectious diseases, and discuss the immunogenicity, efficacy and correlates of protection of mRNA vaccines, with particular focus on research and development of mRNA vaccines against malaria, tuberculosis and HIV.
Introduction
From historical empirical immunization methods or “variolation” against smallpox, and the development of the first live-attenuated whole pathogen vaccines in early 1900’s, the field of vaccinology has been constantly improving. While live-attenuated vaccines are still widely used, the current arsenal of vaccines includes killed whole organisms (inactivated vaccines), toxoid, subunit vaccines (purified proteins and peptides, polysaccharides), conjugated vaccines (protein-polysaccharide conjugate), virus-like particles, outer membrane vesicles, viral vectored, and nucleic acid vaccines, including mRNA vaccines (1, 2).
Vaccination is undoubtedly amongst the greatest advances of medicine, preventing millions of deaths to infectious diseases each year. National and international vaccination programs and mass vaccination campaigns have been responsible for a significant decrease in mortality, particularly during childhood (3). Importantly, vaccination led to the complete eradication of smallpox (4). However, despite this success, current technologies have failed to provide truly effective protection against human immunodeficiency virus (HIV), tuberculosis (TB) and malaria, which remain among the main causes of death, particularly in low and middle-income countries (LMIC). Indeed, the only available vaccine against TB, a live-attenuated strain of Mycobacterium bovis, bacillus Calmette-Guérin (BCG), has inconsistent efficacy, does not prevent transmission, and no vaccine against HIV or Malaria has been licensed to date.
The recent COVID-19 pandemic and the quick development of mRNA-based vaccines have drawn attention to this technology. These vaccines are safe, efficient, rapid and relatively simple to produce, and may quickly respond to the needs in emergency settings such as a pandemic caused by emerging pathogens (5). The research on mRNA vaccines has grown exponentially, and vaccine candidates against a great variety of infections have entered clinical trials, including HIV, TB and malaria. This brings hope that mRNA technologies could offer a solution to prevent these deadly diseases, but also for other applications such as cancer treatment and protein-replacement therapies for genetic disorders (6).
mRNA vaccine technology presents several advantages compared to other types of vaccines. They are safer and generally well-tolerated in healthy patients. Live attenuated vaccines, present a risk of uncontrolled replication in immunocompromised subjects, which does not occur with mRNA vaccines. Inactivated vaccines induce essentially humoral response, while mRNA vaccines activate both humoral and cellular responses. In addition, unlike recombinant protein-based vaccines which often need adjuvants to improve their immunogenicity, mRNA vaccine has intrinsic adjuvant activity (5). Furthermore, mRNA in vitro synthesis and purification processes are cell-free, quick and easily scalable, in contrast with the fastidious and time-consuming production of conventional vaccines (7).
In this review we focus on the mRNA vaccine formulation design, delivery methods and manufacturing processes. We describe an overview on the preclinical and clinical development of mRNA vaccines against infectious diseases, and we discuss their immunogenicity, efficacy, and correlates of protection, with particular focus on the three major killers, malaria, TB and HIV.
mRNA vaccine design, production and optimization
Structure and biological function
Messenger RNA (mRNA) is a single strand nucleic acid molecule encoding genetic information of one or several genes. Endogenous mRNA is transcribed in the nucleus of cells from genomic DNA by the RNA polymerase, either constitutively or under the control of transcription factors that activate or repress gene expression. mRNA is then transported to the cytoplasm, where it is translated by the ribosomal translation machinery into the encoded protein.
Endogenous eucaryotic mRNA is composed of an open reading frame (ORF), which contains the sequence of the encoded genes. This ORF is flanked by 5’ and 3’ untranslated regions (UTRs), that are important for the regulation of translation, as well as for the stability of the molecule. Additional essential structures are the cap, consisting of a N7-methylated guanosine residue (m (7)Gppp) at the 5’ end, and the poly(A) tail at the 3’ end, composed of approximately 250 to 300 adenosine residues (5, 8) (Figure 1A). The 5’ cap is important for translation initiation, through binding to the cap recognition protein eIF4E, which is engaged in the preinitiation complex eIF4F, facilitating ribosome binding and initiation of translation (9). The 2’ O-methylation of the cap (cap-1) is a characteristic of endogenous mRNA, allowing discrimination of self vs exogenous RNA (such as viral RNA), preventing inappropriate immune activation through detection by pattern recognition receptors (PRRs) (10). Indeed, this detection induces type I interferon (IFN) production and IFN-dependent signalling pathways that degrade and block the translation of RNA lacking 2′O methylation (cap-0), thus preventing translation of viral RNA (11). The poly(A) tail results from a nuclear polyadenylation, and its length is tightly regulated both in the nucleus and cytoplasm, notably through binding to the polyadenylate-binding protein (PABP), which plays dual functions: protecting and stabilizing the poly(A) tail, but also facilitating the deadenylation process through recruitment of deadenylases, under certain conditions that are not fully elucidated (12). Beside its importance for the protection and stability of mRNA, the poly(A) tail, in combination with PABP, participates in the stabilisation of the 5’ cap binding to the translation initiation complex, which is an important target of translation initiation regulation (8, 12, 13). While it has long been postulated that a positive correlation exists between poly(A) tail length and mRNA stability, recent findings also suggest that short poly(A) tails are a feature of highly expressed mRNA, while longer tails are associated with transcripts of lower abundance and poor translation (12, 14).
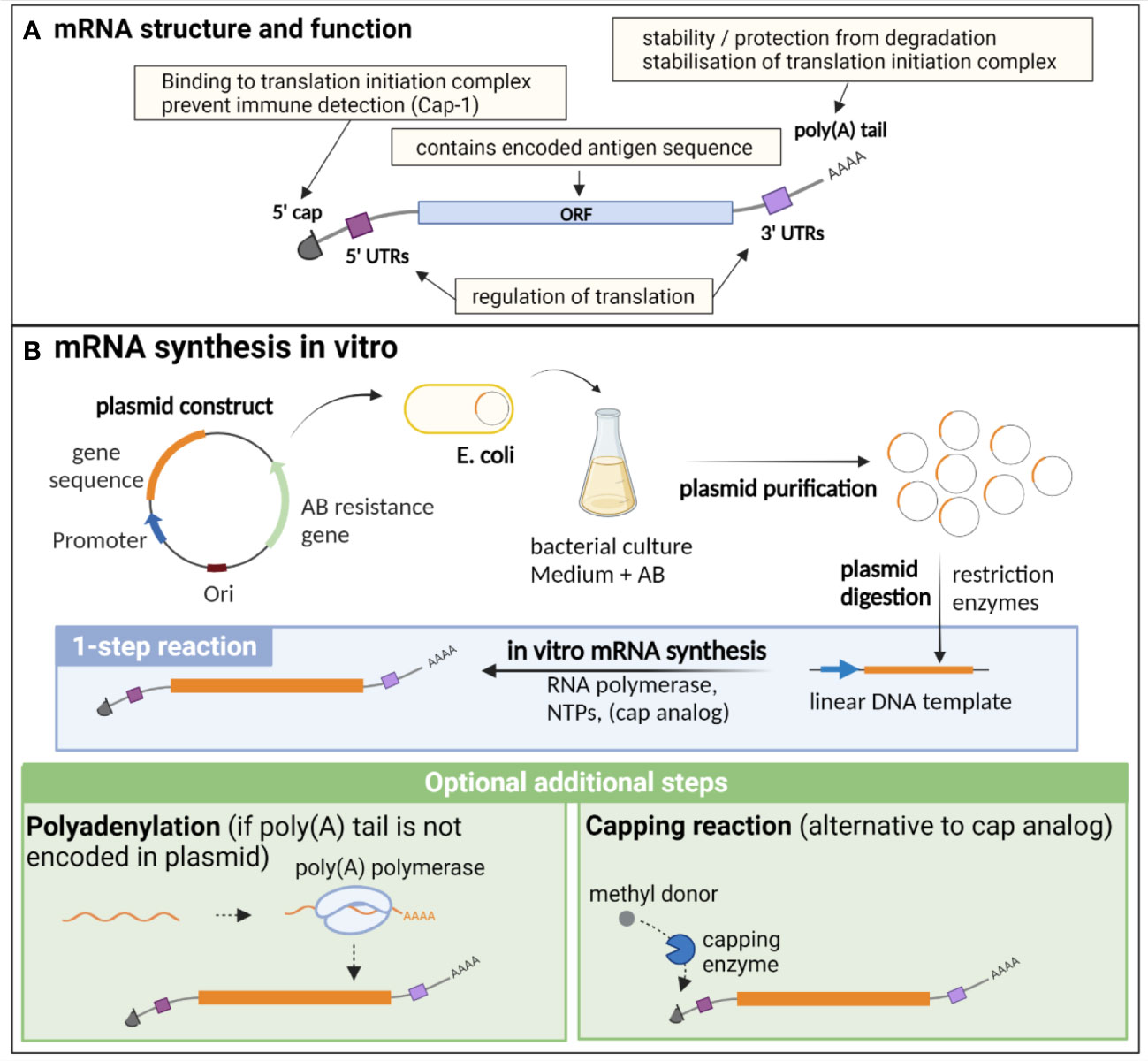
Figure 1 Structure, function and in vitro synthesis of vaccine mRNA. (A) mRNA are single stranded nucleic acids composed of an open reading frame (ORF) encoding the gene of interest, flanked by untranslated regions (UTRs) implicated in translation regulation, a cap at the 5’ end consisting of a N7-methylated guanosine residue, important for translation initiation and immune detection, and a poly(A) tail at the 3’ end, participating in the stability of the mRNA, as well as the stabilisation of the translation initiation complex. (B) In vitro synthesis of mRNA is often performed from a linearised plasmid template. The gene of interest is encoded in the plasmid template downstream of a promoter sequence. E. coli are transformed with the plasmid and cultured in liquid medium containing an antibiotic for which the plasmid encodes a resistance gene, thereby allowing the selection of bacteria that express the plasmid. The plasmid is then purified from the culture and digested using restriction enzymes to obtain a linear DNA template. In vitro transcription of mRNA is performed in the presence of the DNA template, an RNA polymerase and nucleotides triphosphates (NTPs). The capping can be performed by directly adding a cap analogue in the IVT reaction mix (1-step reaction), or alternatively by an enzymatic capping reaction after the IVT. If the poly(A) tail is not encoded in the plasmid, an additional step of polyadenylation is required. Created with BioRender.com.
Of note, beside conventional mRNA vaccines, another type of mRNA, self-amplifying mRNA (saRNA), has been widely studied as a vaccine platform against infectious diseases, particularly for viral infections (15–20). saRNA contains a replicase gene derived from a viral replicon, usually from alphaviruses or flaviviruses, conferring them an auto-replicative activity. This results in higher antigen production in host cells, allowing to lower the dose of mRNA needed for vaccination and thus lowering the manufacturing costs (15, 21).
In vitro synthesis of mRNA and optimization for vaccine development
mRNA vaccine production relies entirely on an in vitro, cell-free process, and is thus safe, quick and can be easily standardised (Figure 1B). It necessitates a DNA template (usually a linearized plasmid), in which the antigen and promoter sequences are encoded. In vitro transcription (IVT) of mRNA is then performed using an RNA polymerase from bacteriophage origin (such as T7, T3 or SP6) and nucleotides triphosphates (NTPs) under the control of the promoter, which is located upstream of the antigen sequence in the DNA template. Virtually any antigen sequence or multiple antigens can be transcribed with this method, and the DNA template can encode all the elements of functional mRNA except the 5’ cap, which needs to be added during the manufacturing process (6). Two methods are used for this purpose: a synthetic cap analogue (such as the CleanCap developed by TriLink) added during the IVT reaction to be directly incorporated in the synthesised mRNA, or alternatively a capping enzyme, such as the vaccinia capping enzyme, which will add the cap during an additional step after the transcription, in the presence of a methyl donor substrate. The poly(A) tail can either be encoded directly in the DNA template sequence or be added enzymatically with the recombinant poly(A) polymerase after IVT. However, this enzymatic process generates mRNAs with poly(A) tails of different lengths and thus direct encoding of poly(A) tail is often preferred for clinical applications (22).
mRNA instability, as well as its recognition by the innate immune system, are major obstacles for their use in therapeutic applications. Thus, several methods have been developed to increase the stability, translation efficiency and immune profile of therapeutic mRNA, such as codon optimisation, nucleotide modification, and selection of efficient purification processes (6). The codon optimization, consisting of the replacement of rarely used codons by synonymous, frequently used codons in humans, is commonly performed in order to increase the translation efficiency of the mRNA in the target cells. Indeed, due to species-specific differences in codon usage and abundance of transfer RNAs (tRNAs), which transport the corresponding amino acids to the ribosomes, this approach may increase the protein elongation rate, yet the importance of codon optimisation for therapeutic mRNA design is still debated as it may affect mRNA secondary structure, translation dynamics and protein conformation (23, 24).
Nucleoside modification, notably the replacement of uridines by pseudouridine and derivatives, has been widely used as a strategy to avoid recognition by innate immune receptors (detailed below), thus decreasing unwanted immune activation and side effects. Preclinical studies demonstrated that nucleoside-modified mRNAs induced increased translation efficiency, prolonged detectability, and increased immunogenicity of mRNA vaccines in animal models (25–27). Pfizer-BioNTech and Moderna have both adopted this strategy, as uridines are replaced by N1-methylpseudouridine in both licensed mRNA-based COVID-19 vaccines, BNT162b2 and mRNA-1273. Of note, while CureVac has always defended the interest of using non-modified mRNA despite the mitigated results of its COVID-19 candidate vaccine CVnCoV, they recently reported preliminary data of a study comparing two next generation nucleoside-modified versus unmodified COVID-19 mRNA vaccines (28). The results indicate that the modified mRNA induces similar antibody levels than the unmodified vaccine at the same dose, but with many fewer side effects, allowing to safely increase the dosage to achieve maximal protection efficacy. As a consequence, CureVac announced they will now focus their development activity on high dose modified mRNA vaccines for both its COVID-19 and flu vaccine programmes (28).
Regulatory regions in the 5’ and 3’ UTRs can also be modulated to increase mRNA stability and translation efficiency (29). For example, the COVID-19 vaccine BNT162b2 (Pfizer-BioNTech) incorporates the 5′-UTR of the highly expressed human α-globin gene, and the 3’-UTR contains regulatory elements of the α- and β-globin genes as well as other segments from the human mitochondrial 12S rRNA and AES/TLE5 gene (30). Interestingly, a recent study in rhesus macaques comparing the immunogenicity and efficacy of the LNP-mRNA vaccine candidate CVnCoV (Curevac) and a similar construct with optimized UTRs (CV2CoV), showed that the CV2CoV elicited higher neutralising antibodies titres and memory B and T cell responses, correlating with higher protective efficacy than CVnCoV for the same dose, highlighting the importance of UTRs optimisation for vaccine design (31).
The purity of mRNA is essential for its therapeutic use, as DNA template, enzymes, residual NTPs or dsRNA contaminants generated during IVT can significantly impact on the translation efficiency and immunogenicity profile of the vaccine, due to innate immune activation (5). The choice of the purification methods depends on the production scale and application (i.e from laboratory research and clinical trials to mass industrial production of licensed products). At the laboratory scale, frequently used methods include DNase treatment, lithium-chloride (LiCL) precipitation, cellulose-based chromatography or the use of commercially available purification kits (32, 33). For large scale, GMP-compliant production, the purification usually relies on advanced chromatography technics, such as ion pair reverse phase chromatography (IPC), ion exchange chromatography (IEC), diafiltration using tangential flow filtration (TFF) and affinity chromatography, which are more efficient in removing short abortive mRNA and dsRNA (5, 34).
Delivery of mRNA vaccines
mRNA vaccines need to reach the target cells and cross the cell membrane to enter the cytosol, where they use the cell machinery to be translated into the encoded antigen. However, naked mRNA is rapidly degraded by RNases in the extracellular space, and its negative charge prevents cell membrane crossing due to electrostatic repulsion. Thus, one of the main challenges is the development of efficient delivery systems to carry mRNA to the cytosol, while protecting it from degradation. Lipid-based carriers, such as liposomes or lipid nanoparticles (LNPs) are the most widely used for delivery of mRNA vaccines, but several other systems have been explored: polymers and polyplexes, such as polyethylenimine (PEI) or poly(amidoamine)s, which form complexes with nucleic acids and efficiently deliver mRNA to the cytoplasm, but can present a significant toxicity due to their positive charge; pH-responsive polymers, protonated at acidic pH in endosomes and presenting a lower toxicity profile; peptides and cell-penetrating peptides (CPPs) such as protamine that electrostatically bind to mRNA and form nanocomplexes due to their cationic or amphipathic amine groups; or squalene-based cationic nanoemulsions that adsorb mRNA on their surface (5, 22, 35).
Cationic liposomes such as N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium (DOTMA) and 1,2-dioleoyloxy-3-trimethylammoniumpropane (DOTAP) have been used successfully for in vitro transfection of mRNA to mammalian cells (Malone et al., (36); Zohra et al., (37)). However, the positively charged cationic lipids tend to aggregate with negatively charges serum proteins, increasing the clearance rate, and have a cytotoxic effect, limiting their clinical applications (35, 38). Incorporation of hydrophilic polymers such as polyethylene glycol (PEG) reduces the toxicity of cationic liposomes but can generate anti-PEG antibodies upon repeated administrations, increasing their clearance rate (39).
LNPs are among the most efficient and widely used mRNA delivery systems, and notably enter in the formulation of the COVID-19 vaccines developed by Moderna and Pfizer/BioNTech. LNPs are typically composed of cationic or ionisable lipids, and structural lipids including sterols, helper phospholipids and PEGylated lipids, and encapsulate mRNA in their core (35, 40). Various parameters influence LNP properties, and depend on the nature and proportion of the different lipids in LNP formulation: the size impacts on biodistribution and internalisation (with an optimal size usually comprised between 20 and 200 nm), the charge is important for cell uptake, cytotoxicity, encapsulation efficiency and organ targeting, while membrane hydration affects the fluidity, deformability, membrane fusion and responsiveness to pH, which is important for mRNA release in the acidic lysosome environment. Cationic and ionisable lipids are composed of 3 parts: the positively charged headgroup entraps the nucleic acid, interacts with the cell membrane and facilitates endosomal escape; the hydrophobic tails (typically 1 to 4) are saturated or unsaturated and impact the lipophilicity, fluidity and fusogenicity of LNPs; and the linker (composed of esters, amides or thiols), impacts on the stability, biodegradability, cytotoxicity and transfection efficiency of LNPs (40). Ionisable lipids are preferred over cationic lipids, as they have a neutral charge in biologic fluids and acquire their positive charge only at acidic pH, i.e in endolysosomes, while the constant positive charge of cationic lipids can lead to cytotoxicity by causing a destabilization of the cell membrane (38). Phospholipids, such as the 1,2-Distearoyl-sn-glycero-3-phosphocholine (DSPC) used in COVID-19 vaccines or the 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE)) are located at the periphery of LNP and organise into a bilayer structure, influencing the fluidity, membrane fusion, as well as the biodistribution of LNPs, as they can be designed for organ-specific targeting (41–43). Sterols (such as cholesterol, oxidised cholesterol derivatives and phytosterols) are the most abundant lipids in LNP formulation (20-50% of total lipids). They are essential for surface organisation through stabilisation of the lipid bilayer, shielding the positive charge, and influence the fluidity of the nanoparticles, protein fusion and endosomal escape (44, 45). PEG-anchored lipids (2-5% of total lipids) are composed of the hydrophilic polyethylene glycol conjugated to an anchoring phospholipid. The amount, length and type of PEG-lipids regulate the size of LNPs by limiting lipid diffusion and protect them from aggregation and undesired interactions with biological environments, such as serum protein and macrophage phagocytosis. However, an excess of PEG lipids can reduce receptor-mediated cell uptake and transfection of LNPs, thus reducing mRNA delivery. This can be overcome by using a PEGylating agent with weak bilayer anchor or a cleavable linker, resulting in a progressive loss of PEGylation in biologic fluids (35, 43). They can also be used to functionalise the surface of LNPs to enable bioconjugation with ligands and macromolecules, for organ/tumour targeting for example (46).
Immunological bases of mRNA vaccines
Innate immune detection of exogenous RNAs
Exogenous nucleic acids are detected by the innate immune system, as a coevolutionary mechanism of eucaryotes against invading pathogens. Exogenous RNAs are recognised by Immune-sensing receptors localised at the cellular or endolysosomal membrane, such as toll-like receptor (TLR)3, TLR7 and TLR8, or by cytoplasmic receptors, including the Retinoic Acid Inducible Gene-I (RIG-I), melanoma differentiation-associated protein 5 (MDA5) or nucleotide-binding oligomerization domain (NOD)-like receptors (NLR) Family Pyrin Domain Containing 1 (NLRP1). Activation of these receptors induces expression of pro-inflammatory genes, particularly type-I and type III IFNs, and IFN-stimulatory genes (ISGs), triggering various defence mechanisms to eliminate the pathogens (Figure 2). Additional mechanisms, such as antiviral restriction factors and RNases participate in exogenous RNA neutralisation and degradation (48). RNA degradation by RNases, such as RNase T2 and RNase 2, which occurs at sites containing uridine, generates by-products, that are further detected by TLR8, amplifying the immune response (49–51). Indeed, TLR7 and TLR8 have been shown to recognise guanosine- and uridine-rich ssRNA (51–53). Activation of this receptors induce cytokine production, particularly type I IFN, which promotes expression of RNA sensors, leading to a reduction of mRNA translation and increase inflammation (54). Thus, as previously mentioned, mRNA vaccine design often includes uridine replacement by methylpseudouridine, in order to avoid this mechanism.
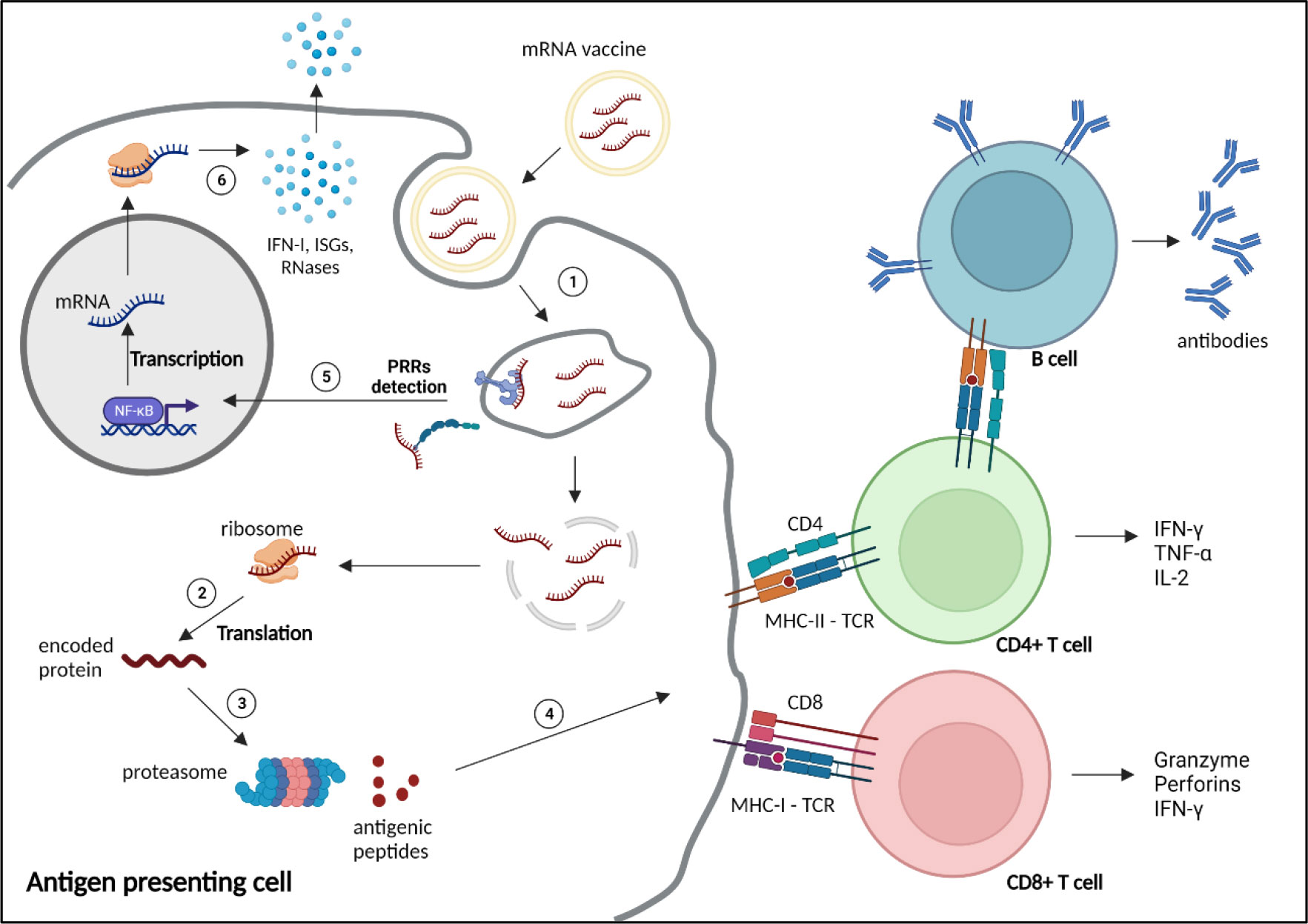
Figure 2 Mechanism of action and immune response induced by mRNA vaccines. 1) mRNA vaccines enter the cells through different mechanisms depending on the nature and size of the nanoparticles, such as clathrin-, caveolin- and receptor-mediated endocytosis, micropinocytosis, phagocytosis or diffusion across the cell membrane (47). 2) After reaching the cytoplasm, mRNAs are translated by the ribosomes into the encoded protein. 3) The protein is processed by the proteasome into small antigenic peptides. 4) The peptides are presented at the surface of the antigen presenting cell by major histocompatibility complex (MHC) molecules to prime CD4+ and CD8+ T cells through, respectively, MHC-II or MHC-I interaction with the T cell receptor (TCR), to activate humoral and cellular adaptive responses. 5) Exogenous mRNAs can be detected by the innate immune system through binding to pattern recognition receptors (PRRs) localised at the endosomal membrane or in the cytosol, inducing the transcription and translation (6) of proinflammatory factors, such as type 1 interferons (IFN-I), IFN-stimulated genes (ISGs) and RNases. NF-κB, nuclear factor κB. Created with BioRender.com.
The cap is also important in this regard, as the 5’-triphosphate end of RNA (m (7)GpppN, cap-0) can be detected by RIG-I, triggering type I IFN immune response, whereas N1 methylation of the 5’ cap (m (7)GpppNm, cap-1), which is a feature of self-RNA, prevents this recognition (55, 56). Similarly, the human IFN-induced protein with tetratricopeptide repeats 1 protein (IFIT1) binds to Cap-0, competing with the translation initiation factor complex eIF4F, thus reducing mRNA translation (57–59).
In addition, IVT mRNA production can generate double-stranded (ds) RNA by-products during the synthesis process, which can drive inflammatory and translation inhibitory pathways through TLR3, RIG-I, or MDA5 activation, as well as through detection by restriction factors such as the Protein kinase R (PKR), Oligoadenylate synthetase (OAS), and Adenosine deaminase acting on RNA (ADAR1) (60).
On the other hand, the production of inflammatory cytokines in antigen presenting cells (APCs) participates in the activation of adaptive immune response through enhancement of antigen presentation to T cells (61). These mechanisms are important considerations when designing and producing mRNA vaccines, and several optimisation and purification methods have been developed to limit the innate immune recognition to generate a balanced innate response, for efficient activation of APCs, while limiting mRNA degradation which reduces translation efficiency (5, 32, 61). In addition, high innate immune activation is responsible of acute post-vaccination symptoms, including injection-site pain, fever, chill, headache and fatigue, limiting the maximal dose that can be administered (62–64).
Immunogenicity of mRNA vaccines and correlates of protection
There are several immunologic aspects to consider when developing a vaccine: the innate immune stimulation or “adjuvant effect”, and the adaptive response, which comprises humoral and cellular responses, as well as the generation of memory cells. The adjuvant effect is essential to the recruitment and activation of APCs and priming of T cells to induce adaptive responses. Therefore, it is often necessary to add adjuvant molecules to the vaccine formulation in order to increase its immunogenicity, particularly in the case of subunit vaccines (5). The humoral and cellular adaptive responses, relying on antibody production and cytotoxic T lymphocytes (CTL) respectively, are the two main effector mechanisms of acquired, -natural or vaccine-induced-, protection against pathogens. While live attenuated vaccines stimulate both responses, inactivated and subunit vaccines, although having a better stability and safety profile, often induce only humoral response (5). This is of importance since, while antibody titres are usually the main correlate of protection for most available vaccines (65), cellular immunity may participate to the protection against intracellular pathogens such as M. tuberculosis (66–71) or influenza (72–74). Finally, as antibody titres fade over time after immunisation, the generation of long-lasting memory B cells and T cells, which can rapidly respond, upon new encounter with the pathogen, by clonal expansion and massive antibody production, is crucial for long-term vaccinal protection, particularly for diseases with long incubation time (65).
Since the first demonstration by Wolff and colleagues in 1990 of the ability of mRNA to induce protein expression after direct in vivo administration (75), several studies reported the production of specific antibodies and induction of CTL-mediated cellular immunity, highlighting its potential for vaccination against infectious diseases as well as for targeted cancer therapy (27, 76–80) (Figure 2). However, our understanding of the exact contribution of these mechanisms to the protection against infection is still incomplete and pathogen-dependent (54).
The innate response and the role of APCs have been investigated in a recent preclinical study with the nucleoside-modified mRNA-LNP COVID-19 vaccine BNT162b2 (Pfizer/BioNTech). The authors reported an increased activation of monocytes, macrophages and DCs in dLN, lung and spleen as well as increased cytokine and chemokine levels (including MCP1, Mip1b, CXCL10, IL-6, IFN-α and IFN-γ) in serum in the first hours/days following intramuscular vaccination (81). Consistent with observations in vaccinated patients (82, 83), this innate response was enhanced in the mouse model following the boost dose compared to the first dose, and was accompanied by a higher magnitude of IFN-γ-secreting CD4+ and CD8+ T cells in draining lymph nodes (dLNs), lung and spleen. By contrast, NK cells appeared to be the major source of IFN-γ after the first dose (81). Similar observations were made with an LNP-mRNA vaccine coding for influenza haemagglutinin H10 in rhesus macaques, with rapid and transient infiltration of neutrophils, monocytes and DCs, associated with type I IFN-inducible gene stimulation in dLNs, which correlated with priming of specific CD4+ T cells (84). However, while particle uptake by DCs and monocytes/macrophages resulted in high transcript translation, poor protein production is observed in neutrophils, suggesting that those cells may rather have a competitive role for particle uptake by APCs. In line with this, a study showed that, following intradermal administration of influenza vaccine, DC and langherans cells contribute to optimal T cell activation in dLNs, while neutrophils are dispensable (85). Of note, transgenic mice were used to demonstrate that MDA5 and Interferon-alpha/beta receptor alpha chain (IFNAR1) (but not TLR2,3,4,5,7, inflammasome or STING-cGAS) signalling pathways are essential for innate immune activation and induction of specific CD8+ T cell response following BNT162b2 vaccination, but have only a modest role in inducing specific antibodies, as IgG titres were only mildly decreased in deficient mice (81).
LNP-mRNA vaccines against SARS-Cov-2, HIV, influenza or Zika were shown to elicit durable antibody and Th1 cell responses in animals, associated with strong activation of antigen-specific T follicular helper (Tfh) and germinal centre B cells in dLNs (27, 86–89). The central role of rapidly -induced CD4+ T cells in orchestrating humoral and cellular responses during a prime-boost vaccination protocol against SARS-CoV-2 was highlighted in a longitudinal cohort analysis, where CD4+ T cells induced by the prime vaccination correlated with CD8+ T cells and antibody titres induced following the second dose in naïve healthy individuals (83). In addition, the prime-boost vaccination protocol with SARS-CoV-2 vaccines induces durable memory B cells and memory CD4+ and CD8+ T cells (detectable 6 months post-immunisation), similar to those induced by SARS-CoV-2 natural infection (83, 90, 91).
A large body of evidence points towards collaborative roles of humoral and CD8+ T cell-mediated responses in the protection induced by mRNA COVID-19 vaccines. In a phase 3 clinical trial, the titres of binding and neutralising antibodies against the viral spike protein were indeed correlated with protection against COVID-19 disease in individuals vaccinated with mRNA-1273 vaccine (Moderna) (92). The protective and therapeutic role of neutralising antibodies was further demonstrated in a study showing that the adoptive transfer of IgG from convalescent rhesus macaques to naïve animals protected recipient animals against SARS-CoV-2 intranasal challenge in a dose-dependent manner (93). Beside this, CD8+ T cell depletion partially abrogated the protection against rechallenge with SARS-CoV-2 in convalescent macaques with declining antibody titres (4 to 7 weeks post-primary infection (93). Another study points out a potential role of spike-specific CD8+ T cell response in early protection following prime vaccination with bnt162b2, when neutralising antibodies are hardly detectable (94). Interestingly, some observations suggest that memory T cells are less affected by immune escape observed with mutation variants of the virus than antibody-mediated responses and may thus be important for protection against SARS-CoV-2 variants (95, 96).
Of note, it is now recognised that LNPs, which enter in the composition of the commercialised COVID-19 vaccines and are the most widely used carriers in mRNA vaccine development, exert a strong adjuvant activity. LNPs containing ionizable lipids (iLNPs) were shown to induce a strong chemokine and cytokine production (including IFN-α, IL-6, IL-1b and GM-CSF), immune cell infiltration at the injection site and in dLNs, and promote DC maturation and monocytes and DC activation (27, 85, 97, 98). Moreover, iLNPs were shown to play an essential role in the stimulation of Tfh cells and germinal centre B cells, inducing their differentiation into long lived plasma cells and memory B cells, associated with a durable protective antibody response. This effect was mediated by IL-6 signalling pathway, and dependant on ionisable lipids (97). iLNPs have thus been successfully used as adjuvants in sub-unit vaccines against dengue and hepatitis B, VZV, influenza and SARS-CoV-2 (97, 99–101). In addition, several studies demonstrated that cationic lipid-based nanoparticles are detected by immune sensors, such as TLR4, TLR2, TLR3, NLRP3 or STING and enhance vaccine immunogenicity (102–105). IL-1 and IL-1ra are strongly produced by immune cells in vitro and in vivo upon administration of liposome-encapsulated mRNA vaccine and are important regulators of the inflammatory response induced by the vaccine in a lipid-dependent manner (106). Overall, these studies bring some clarification about the key innate pathways implicated in the induction of protective, specific immunity by mRNA-LNPs vaccines, even though complementary analyses are still required to better understand the contribution of memory B and T cells, particularly regarding strain-diversity and protection against reinfection with different variants in the case of SARS-CoV-2.
Pre-clinical and clinical development of mRNA vaccines against infectious diseases
If the research on mRNA vaccine against infectious diseases have truly exploded since 2020, driven by the success of COVID-19 vaccines, first preclinical evidence of the potential of RNA-based vaccines for tumours and infections have emerged since the 90s. In 1993, mice immunization with liposome-encapsulated IVT mRNA coding for the influenza virus nucleoprotein was shown to induce virus-specific CTL (107). Of note, many of early preclinical reports on the induction of protective immunity against infectious diseases were conducted with self-amplifying RNA replicons derived from alphaviruses or flaviviruses, packed in virus-like particles (108). They targeted mostly viruses, such as influenza (16, 109, 110), vaccinia virus (111), parainfluenza virus type 3 (20), tick-borne encephalitis virus (16, 112), HIV (19, 113, 114), herpes simplex virus (115), or Ebola (18, 116). However, viral vectors present a risk due to anti-vector immunity, have limited loading capacity, and their production is more fastidious (108). Nonetheless, the undeniable advantages of mRNA over other vaccine platforms as a safe, highly adaptable and easily produced template for in vivo protein expression, with intrinsic adjuvant properties and no risk of insertional mutagenesis (unlike DNA), kept the interest of the scientific community despite the absence of clinical validation.
The use of lipid-based nanocarriers, formulated to protect and deliver small molecules such as nucleic acids to the cells, have significantly contributed to the mRNA vaccine field (40, 97). In parallel, important advances have been made in the design of mRNA, with modifications by genetic engineering to improve their stability, translation efficiency and immunogenicity/safety profile (117). These efforts culminated in the development and approval of two COVID-19 vaccines in a record time. As a result of this success, modified or unmodified mRNA formulated with LNPs are now predominant in both preclinical and clinical development of mRNA-based vaccines against infectious diseases (40) (Table 1).
Influenza vaccine research, for example, have largely beneficiated from these technologic advances. Indeed, the high mutation rate of Influenza viruses means that the vaccine formulation needs to be constantly adapted to the last circulating strains, which is difficult to manage due to the long production time of conventional influenza vaccines, and results in variable effectiveness. mRNA technology could help in improving antigen design, or ideally developing a universal, cross-reactive vaccine, as well as ease the production to better respond to seasonal epidemics and pandemics. Preclinical studies showed that multitargeting mRNA-LNP vaccines elicit broad protective immunity in animal models against multiple Influenza virus strains (118–122). Several influenza vaccines have reached clinical trials, and Moderna (mRNA-1010) and Pfizer (qIRV) vaccine candidates have recently entered phase 3 of clinical trials (Table 1).
In recent years, nucleotide-modified or unmodified IVT mRNA vaccines formulated with lipid-based vectors have demonstrated efficacy in animal models against various infectious diseases, such as RSV (123), rabbies (80, 124), malaria (125), HIV (126–128), Ebola (129), Zika virus (89) and Cytomegalovirus (130). Clinical trials, active or completed, evaluating mRNA vaccines for infectious diseases, to the exclusion of COVID-19, are listed in Table 1. Of note, mRNA vaccines against RSV (mRNA-1345) and CMV (mRNA-1647), both developed by Moderna, have also entered phase 3 clinical trials.
Interest of mRNA vaccine technology for TB, HIV and malaria
TB, malaria and acquired immunodeficiency syndrome (AIDS), remain among the leading causes of death by infectious diseases, particularly in low-income countries (WHO, top 10 causes of death). Despite considerable efforts, the development of safe, efficient, and cost-effective vaccine easily deployable in endemic areas of these diseases remains a global health priority. The path opened by COVID-19 mRNA vaccines thus raises a new hope for the fight against these infections.
Tuberculosis
TB is caused by a pulmonary bacterium, Mycobacterium tuberculosis (M.tb). The disease classically progresses from latent TB infection (LTBI), where the bacteria are contained by the host immune system inside lung granulomas, to active TB disease if the pathogen is not eliminated and escapes immune containment. While reactivation of LTBI is the major source of TB disease patients, some patients develop active TB disease soon after M.tb exposure, within months or few years (131). During active TB, the patient is symptomatic (the main symptoms are fever, cough, weight loss, haemoptysis) and contagious. Of note, HIV infection is the strongest risk factor for TB disease, accounting for around 25% of all TB-related deaths (131). An antibiotic regimen against TB exists, which classically combines four molecules for several months, in order to target all populations of bacteria and avoid emergence of resistance. However, poor access to diagnostic, treatment, and healthcare support in the regions where TB is the most prevalent favours the emergence and rapid spread of antibiotic resistant M.tb strains, further aggravating the situation (Global Tuberculosis Report (132), WHO).
Thus, an efficient and cost-effective vaccination program appears as the best strategy to face this situation. Despite BCG being the oldest licensed vaccine still in use, with high coverage throughout the world, TB remains the second leading cause of death by an infectious disease after COVID-19, killing around 1.6 million people per year essentially in low and middle-income countries (132–134).
Macrophages, dendritic cells and T cells are implicated in the control of bacterial growth in granulomas, preventing the spread of bacteria in blood circulation and progression to active TB (131). Of note, HIV/AIDS, characterised by an impairment of CD4+ T cells, is a major risk factor for progression to active TB disease. Furthermore, the lung and spleen protection following vaccination of mice correlates with the magnitude and quality of multi-functional CD4 T cells expressing IFN-γ, TNF-α, and IL-2 (135). While the mechanisms of natural and BCG-induced protective immunity against mycobacteria are incompletely elucidated, this suggest a role for cellular immunity for the control of infection and for vaccine-induced protection against M.tb (136). Overall, the complexity of M.tb culture and difficulty to establish a relevant animal model for TB infection, as well as the lack of clearly defined correlates of protection both in preclinical animal models and in clinical trials, are still major challenges for researchers (131).
Mycobacteria are complex microorganisms, encoding about 4000 genes, and the identification of immunodominant and protective antigenic targets have been a major focus for the design of effective protein subunit or nucleic acid-based vaccine against TB, even though whole pathogen vaccines, attenuated or inactivated, and viral vector-based vaccines are also being investigated (137–139). Immunopeptidomics, based on mass spectrometry identification of MHC-bound peptides from infected cells, has been demonstrated as a useful approach for the identification of novel antigenic peptides for vaccine development (140). Furthermore, Immunoinformatic approaches provide useful tools for in silico modelling of mRNA vaccines against TB, combining epitope identification, mRNA construction and optimisation and immune simulation to guide further in vivo evaluation (141–143).
Interestingly, the first proof of concept of an mRNA vaccine against TB has been reported in 2004. The authors demonstrated that immunisation of mice with naked IVT mRNA coding for the immunodominant antigen MPT83 (four injections at 3-weeks intervals) induced specific humoral and cellular responses and conferred a modest but significant protection against TB challenge (144). Intranasal immunisation with naked mRNA coding for Hsp65 protein also demonstrated significant protection against M.tb in mouse model (145). More recently, a replicating mRNA-based vaccine coding for a fusion protein comprising 4 M.tb Antigens (Rv3619, Rv2389, Rv3478, Rv1886) formulated with a lipid nanocarrier induced cellular immune response and protection against M.tb and M. avium challenge in mice in a heterologous RNA-prime and protein-boost vaccination protocol (146, 147). Of note, a phase I clinical trial evaluating two investigational RNA-based vaccines against TB in BCG-vaccinated volunteers have been launched by BioNTech (Table 1).
HIV
In 2021, 650 000 persons died from HIV/AIDS-related cause, 1.5 million were newly infected, while around 38.4 million people were living with HIV (www.unaids.org). In the absence of a licensed vaccine, the arsenal against HIV/AIDS still relies on information and screening campaigns and antiretroviral therapies (ART). Even though ART have been significantly improved over the years, with stronger efficacy and less side effects than in the past, they remain expensive and require strict observance. Thus, poor access to diagnostic and treatment in low and middle-income countries with high prevalence (particularly Sub-Saharan Africa) is a major issue (148). Despite the dedication of governments, health organisms and the scientific community, no vaccine against HIV has been licensed to date. Indeed, in the last decades, numerous vaccine candidates have reached phase III efficacy trials, but none of them demonstrated sufficient efficacy (149, 150).
The proposed correlates of protection, identified in animal models and clinical studies, are neutralizing antibodies targeting HIV envelope (Env) glycoprotein epitopes, preventing the entry of virions in CD4+ T cells, and antibody-dependent cellular cytotoxicity (ADCC) (relying on Fc-mediated effector functions of non-neutralising antibodies), to eliminate infected cells and prevent virus reactivation from reservoir cells (151–153). However, targeting the highly glycosylated Env glycoprotein is hampered by the high mutation rate of the virus, resulting in high viral diversity (pseudoviruses), while broad neutralizing B cell precursor development is difficult to elicit and limited by immune tolerance mechanisms such as auto- or polyreactivity (150, 152, 154). Thus, vaccine design needs to be optimized to target a combination of multiple conserved epitopes from Env glycoprotein, eliciting broad neutralizing antibodies, as well as T cell epitopes on other viral proteins, such as Gag, Pol and Nef, to elicit CD8+ T cell-mediated responses (148, 153, 155). mRNA technology, combined with efficient delivery system, may hold the potential to overcome the challenges faced in HIV vaccine development.
Recent preclinical studies of mRNA-based vaccines in mouse and non-human primate models are indeed encouraging. Intradermal immunization of rabbits and rhesus macaques with mRNA-lipid nanoparticle (mRNA-LNP) vaccines encoding the clade C transmitted/founder HIV-1 Env 1086C elicited high levels of gp120-specific antibodies, with significant but transient neutralising activity, as well as ADCC activity in serum (126). In macaques, an HIV-1 Env-encoding mRNA-LNP elicited comparable titres and functions of neutralising and non-neutralising antibody than adjuvanted Env recombinant protein (156). In addition, Immunisation with polymer-formulated self-amplifying mRNA encoding mosaic Gag-Pol epitopes induced potent specific CD8+ T cell response in mice (17). Furthermore, an mRNA-LNP vaccine encoding Gag conserved elements induced potent humoral and cellular responses in macaques in a prime-boost protocol with Gag DNA vaccine (128). Interestingly, the authors report that higher dose of mRNA vaccine increased T cell, but not humoral response. In another study, mRNA-LNP vaccine encoding HIV-1 Env and simian immunodeficiency virus (SIV) Gag proteins to generate virus-like particles, induced broad neutralising antibodies and reduced the risk of infection in rhesus macaques immunized through a prime-boosts protocol with autologous and mixed heterologous Env challenged with repeated low doses of heterologous SHIV (127).
The HIV mRNA vaccine eOD-GT8 60mer (mRNA-1644), developed by the International AIDS Vaccine Initiative (IAVI), in collaboration with Moderna, is in phase I clinical trial to evaluate the safety and immunogenicity in healthy adult volunteers (Table 1). This promising candidate vaccine was designed to target germinal centre’s naïve progenitor naive B cells to produce broad neutralising antibodies (press release, First-in-human clinical trial confirms novel HIV vaccine approach (iavi.org)). The National Institute of Allergy and Infectious Diseases (NIAID) is also evaluating the safety and immunogenicity of three HIV trimer mRNA vaccines (BG505 MD39.3, BG505 MD39.3 gp151, and BG505 MD39.3 gp151 CD4KO) in healthy individuals (Table 1).
Malaria
Malaria, caused by Plasmodium falciparum parasites transmitted by female anopheline mosquitoes, is responsible for 200 million infections and 400 000 deaths per year, particularly in young infants in endemic areas, such as sub-Saharan Africa (157). Anti-malaria drugs, while effective to prevent and treat the infection, are poorly accessible to populations living in endemic areas in LMIC, and the progression of parasitic resistance to available molecules compromises their efficacy. In this context, the development of a cost-effective vaccine is a highly relevant strategy to reduce the global burden of the disease.
The parasite has a complex life cycle with multiple development stages in human and mosquitoes. The sexual cycle occurs in the mosquito, which transmits the sporozoites through the skin. Sporozoites then migrate to the liver, where they invade hepatocytes and develop into merozoites, that are released in the blood circulation and invade erythrocytes, causing repeated cycles of erythrocytes invasion and lysis, responsible for the fever characteristic of malaria. Some merozoites develop into gametocytes, which are transmitted to the mosquito during the blood meal.
One of the first vaccination strategies investigated, consisted of the administration of attenuated sporozoites, the infective form of the parasite (158). However, injection of purified, radiation-attenuated whole sporozoites (PfSPZ), despite excellent protection in non-human primates and rodents, as well as in naïve adults when administered intravenously (but not via subcutaneous or intramuscular route) (159, 160), failed to efficiently protect previously exposed adults and infants (161, 162).
The immune mechanisms underlying protection are not completely elucidated, but the role of liver CD8+ T cells have been demonstrated in animal models, and a role for γδT cell priming and Fc-dependent antibody effector functions have been suggested (157, 163). Of note, the circumsporozoite protein (CSP), expressed at the surface of sporozoites of different Plasmodium species, has been identified as a major immunogen eliciting binding antibodies that prevent infection of hepatocytes, leading to the development of vaccines targeting the CSP (164, 165). Among those, the adjuvanted subunit vaccine RTS,S/AS01 (Mosquirix™) developed by GSK, composed of CSP repeats (R) and C-terminal T-cell epitopes (T) recombinantly fused to HBsAg, demonstrated modest but significant protection against P. falciparum infection in children in endemic areas (but not in adults), and the World Health Organization (WHO) has recommended its widespread use among children living in malaria endemic areas (164). However, the protection wanes over time, correlating with decreased anti-CSP antibody levels, and the vaccine does not prevent the infection of mosquitoes by gametocytes from infected individuals, and thus does not decrease the circulation and transmission of the parasite (164). Of note, the next generation R21-matrix M vaccine, composed of a fusion of CPS and HBsAg that aggregates as virus-like particles and formulated with matrix M adjuvant, demonstrated 77% efficacy in children aged 5-17 months in a phase 1/2b clinical trial in Burkina Faso over 6 months after 3 doses, and a booster dose 1 year after initial vaccination allow to maintain high protection efficacy in those children (165–167). Despite these encouraging results, a malaria vaccine eliciting long-lasting, high-level protection in all subgroups and which could block the transmission in endemic areas has yet to be developed.
So far, preclinical studies on mRNA vaccines against malaria are still scarce. A mRNA-LNP vaccine coding for the major sporozoite-targeting antigen PfCSP demonstrated potent antibody and cytokine responses in mice, and protected the animals in a dose-dependent manner against P. burghei infection (168). In another study, the authors evaluated two mRNA-LNP vaccine candidates encoding PfCSP and Pfs25, a protein expressed by ookinetes, essential for oocyst development (125). These vaccines thus block different stages of the parasite cycle, i.e the liver stage (prevention of hepatocyte invasion by sporozoites), and sexual stage (disruption of sexual cycle and transmission by mosquitoes), respectively. Both vaccines were highly immunogenic following single or co-immunisation, eliciting high, dose-dependent antibody titres and B and T cell responses in mice. The Pfs25 mRNA-LNP elicited potent mosquito transmission-blocking activity, while PfCPS mRNA-LNP alone or in combination with Pfs25 mRNA-LNP conferred significant protection against sporozoite infection challenge (125). This study supports the evidence that multiple stage targeting mRNA vaccine platforms may provide an effective strategy to contribute to decrease malaria transmission. Additional potential antigenic targets for vaccines were identified in preclinical studies, such as the protein PfGARP, expressed on the exofacial surface of erythrocytes infected by early-to-late-trophozoite-stage parasites. Cohort studies on Tanzanian children and male adults showed that detection of naturally-acquired anti-PfGARP-A antibodies in the blood was associated with a significantly lower risk of severe malaria and lower parasitemia (169). Anti-PfGARP antibodies demonstrated potent parasite killing activity in vitro, and immunisation with PfGARP-A-mRNA LNPs of Aotus monkeys challenged with P. falciparum resulted in significantly lower parasitemia than control monkeys (169). The Cell-Traversal protein for Ookinetes and Sporozoites (CelTOS), a secreted protein playing a role both in mosquito transmission and hepatocyte invasion, has also been identified as a potent vaccine target in preclinical settings (170). The authors demonstrated that CelTOS-targeting mRNA vaccine can induce potent humoral and cellular response in mice and highlighted the importance of careful vaccine design. However, further investigations are required to state on the protective efficacy of this vaccine platform and the roles of humoral and cellular responses in the protection. Of note, a phase 1 clinical trial is evaluating the safety and immune response of an mRNA-based vaccine targeting PfCSP, developed by BioNTech (Table 1).
Conclusions
The success of mRNA vaccines against COVD-19 inspired scientist throughout the world to develop new mRNA vaccines against transmissible and non-transmissible diseases. Twenty-seven mRNA vaccine candidates against infectious diseases are in clinical trials and hundreds are currently being developed in pre-clinical studies. The number of mRNA vaccine candidates in clinical trials is expected to increase in the coming years. Moreover, the mRNA vaccine technology is being improved with new discoveries and developments at a daily basis.
Many complex diseases, particularly those that depend heavily on T cells for protection, lack the identification of the most protective antigens and the corresponding correlates of protection. In recent years, immunopeptidomics has been applied to identify antigens presented by MHC-I and MHC-II in an unbiased way, leading to the identification of antigens for vaccine development (140, 171). The combination of the most relevant antigenic targets identified by immunopeptidomics with the simplicity of the mRNA vaccine technology, allows for the development of vaccines encoding multiple antigens, or antigens to prevent multiple diseases, and paves the way for a new generation of more efficacious and effective vaccines against some of the most dramatic infectious diseases that depend on T cells for protection, such as malaria, TB and HIV.
The challenges faced by the mRNA vaccine technology includes the cost of raw materials, the lack of standardization, and delivery optimization. The high cost of raw materials and the cold-chain requirements for mRNA vaccines since manufacture to the moment of vaccination, will have an important impact on the vaccine cost-effectiveness. Moreover, the mRNA purity can vary significantly between different purification processes, which may contribute to the lack of process standardization. Importantly, the Intellectual Property landscape and the difficulty of access to methods and technologies, particularly to reagents and manufacturing processes required for vaccine formulation development, may delay the progress of new mRNA vaccines against the deadliest hard-to-treat infectious diseases malaria, TB and HIV.
Thus, we should expect future mRNA vaccines designed and optimised to induce the right type of immune response against each disease for maximal efficacy, improved stability of formulations, as well as formulations adapted to long term storage at room temperature, leading to a reduction and eventual elimination of cold-chain supply, ease of distribution, and above all, reduced cost. Of particular relevance is the difficulty of equitable distribution of mRNA vaccines to LMICs and remote regions. To overcome this difficulty, BioNtech have implemented modular mRNA manufacturing facilities to produce vaccines locally, being already installed in Rwanda (press release, BioNTech Starts Construction of First mRNA Vaccine Manufacturing Facility in Africa | BioNTech).
The mRNA vaccines represent a new generation of vaccines against transmissible and non-transmissible diseases. We are witnessing a scientific revolution, with direct consequences to improving global health. The success of the mRNA vaccines will change the history of medicine.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by internal funding of the Faculty of Medicine, Universidade Católica Portuguesa, and external funding from Fundação para a Ciência e a Tecnologia (FCT), under the projects UIDP/04279/2020 and UIDB/04279/2020.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
2. Pollard AJ, Bijker EM. A guide to vaccinology: from basic principles to new developments. Nat Rev Immunol (2021) 21:83–100. doi: 10.1038/s41577-020-00479-7
3. Younger DS, Younger APJ, Guttmacher S. Childhood vaccination: Implications for global and domestic public health. Neurol Clin (2016) 34:1035–47. doi: 10.1016/j.ncl.2016.05.004
5. Rosa SS, Prazeres DMF, Azevedo AM, Marques MPC. mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine (2021) 39:2190–200. doi: 10.1016/j.vaccine.2021.03.038
6. Sahin U, Karikó K, Türeci Ö. mRNA-based therapeutics–developing a new class of drugs. Nat Rev Drug Discovery (2014) 13:759–80. doi: 10.1038/nrd4278
7. Ghattas M, Dwivedi G, Lavertu M, Alameh M-G. Vaccine technologies and platforms for infectious diseases: Current progress, challenges, and opportunities. Vaccines (Basel) (2021) 9:1490. doi: 10.3390/vaccines9121490
8. Weill L, Belloc E, Bava F-A, Méndez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol (2012) 19:577–85. doi: 10.1038/nsmb.2311
9. Batool A, Aashaq S, Andrabi KI. Eukaryotic initiation factor 4E (eIF4E): A recap of the cap-binding protein. J Cell Biochem (2019) 120:14201–12. doi: 10.1002/jcb.28851
10. Ramanathan A, Robb GB, Chan S-H. mRNA capping: biological functions and applications. Nucleic Acids Res (2016) 44:7511–26. doi: 10.1093/nar/gkw551
11. Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, et al. 2’-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature (2010) 468:452–6. doi: 10.1038/nature09489
12. Nicholson AL, Pasquinelli AE. Tales of detailed Poly(A) tails. Trends Cell Biol (2019) 29:191–200. doi: 10.1016/j.tcb.2018.11.002
13. Munroe D, Jacobson A. mRNA poly(A) tail, a 3’ enhancer of translational initiation. Mol Cell Biol (1990) 10:3441–55. doi: 10.1128/mcb.10.7.3441-3455.1990
14. Lima SA, Chipman LB, Nicholson AL, Chen Y-H, Yee BA, Yeo GW, et al. Short poly(A) tails are a conserved feature of highly expressed genes. Nat Struct Mol Biol (2017) 24:1057–63. doi: 10.1038/nsmb.3499
15. Vogel AB, Lambert L, Kinnear E, Busse D, Erbar S, Reuter KC, et al. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol Ther (2018) 26:446–55. doi: 10.1016/j.ymthe.2017.11.017
16. Fleeton MN, Chen M, Berglund P, Rhodes G, Parker SE, Murphy M, et al. Self-replicative RNA vaccines elicit protection against influenza a virus, respiratory syncytial virus, and a tickborne encephalitis virus. J Infect Dis (2001) 183:1395–8. doi: 10.1086/319857
17. Moyo N, Vogel AB, Buus S, Erbar S, Wee EG, Sahin U, et al. Efficient induction of T cells against conserved HIV-1 regions by mosaic vaccines delivered as self-amplifying mRNA. Mol Ther Methods Clin Dev (2019) 12:32–46. doi: 10.1016/j.omtm.2018.10.010
18. Pushko P, Bray M, Ludwig GV, Parker M, Schmaljohn A, Sanchez A, et al. Recombinant RNA replicons derived from attenuated Venezuelan equine encephalitis virus protect guinea pigs and mice from Ebola hemorrhagic fever virus. Vaccine (2000) 19:142–53. doi: 10.1016/S0264-410X(00)00113-4
19. Ajbani SP, Velhal SM, Kadam RB, Patel VV, Lundstrom K, Bandivdekar AH, et al. Immunogenicity of virus-like semliki forest virus replicon particles expressing Indian HIV-1C gag, env and polRT genes. Immunol Lett (2017) 190:221–32. doi: 10.1016/j.imlet.2017.08.019
20. Greer CE, Zhou F, Legg HS, Tang Z, Perri S, Sloan BA, et al. A chimeric alphavirus RNA replicon gene-based vaccine for human parainfluenza virus type 3 induces protective immunity against intranasal virus challenge. Vaccine (2007) 25:481–9. doi: 10.1016/j.vaccine.2006.07.048
21. Blakney AK, Ip S, Geall AJ. An update on self-amplifying mRNA vaccine development. Vaccines (Basel) (2021) 9:97. doi: 10.3390/vaccines9020097
22. Yang L, Tang L, Zhang M, Liu C. Recent advances in the molecular design and delivery technology of mRNA for vaccination against infectious diseases. Front Immunol (2022) 13. doi: 10.3389/fimmu.2022.896958
23. Cannarozzi G, Schraudolph NN, Faty M, Rohr Pv, Friberg MT, Roth AC, et al. A role for codon order in translation dynamics. Cell (2010) 141:355–67. doi: 10.1016/j.cell.2010.02.036
24. Mauro VP. Codon optimization in the production of recombinant biotherapeutics: Potential risks and considerations. BioDrugs (2018) 32:69–81. doi: 10.1007/s40259-018-0261-x
25. Anderson BR, Muramatsu H, Nallagatla SR, Bevilacqua PC, Sansing LH, Weissman D, et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res (2010) 38:5884–92. doi: 10.1093/nar/gkq347
26. Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity (2005) 23:165–75. doi: 10.1016/j.immuni.2005.06.008
27. Pardi N, Hogan MJ, Naradikian MS, Parkhouse K, Cain DW, Jones L, et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center b cell responses. J Exp Med (2018) 215:1571–88. doi: 10.1084/jem.20171450
28. Dolgin E. Trial settles debate over best design for mRNA in COVID vaccines. Nature (2023) 613:419–20. doi: 10.1038/d41586-023-00042-z
29. Zhuang X, Qi Y, Wang M, Yu N, Nan F, Zhang H, et al. mRNA vaccines encoding the HA protein of influenza a H1N1 virus delivered by cationic lipid nanoparticles induce protective immune responses in mice. Vaccines (Basel) (2020) 8:123. doi: 10.3390/vaccines8010123
30. Xia X. Detailed dissection and critical evaluation of the Pfizer/BioNTech and moderna mRNA vaccines. Vaccines (Basel) (2021) 9:734. doi: 10.3390/vaccines9070734
31. Gebre MS, Rauch S, Roth N, Yu J, Chandrashekar A, Mercado NB, et al. Optimization of non-coding regions for a non-modified mRNA COVID-19 vaccine. Nature (2022) 601:410–4. doi: 10.1038/s41586-021-04231-6
32. Baiersdörfer M, Boros G, Muramatsu H, Mahiny A, Vlatkovic I, Sahin U, et al. A facile method for the removal of dsRNA contaminant from In vitro-transcribed mRNA. Mol Ther Nucleic Acids (2019) 15:26–35. doi: 10.1016/j.omtn.2019.02.018
33. Karikó K, Muramatsu H, Ludwig J, Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res (2011) 39:e142. doi: 10.1093/nar/gkr695
34. Ouranidis A, Vavilis T, Mandala E, Davidopoulou C, Stamoula E, Markopoulou CK, et al. mRNA therapeutic modalities design, formulation and manufacturing under pharma 4.0 principles. Biomedicines (2021) 10:50. doi: 10.3390/biomedicines10010050
35. Chaudhary N, Weissman D, Whitehead KA. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat Rev Drug Discovery (2021) 20:817–38. doi: 10.1038/s41573-021-00283-5
36. Malone RW, Felgner PL, Verma IM. Cationic liposome-mediated RNA transfection. Proc Natl Acad Sci (1989) 86:6077–81. doi: 10.1073/pnas.86.16.6077
37. Zohra FT, Chowdhury EH, Nagaoka M, Akaike T. Drastic effect of nanoapatite particles on liposome-mediated mRNA delivery to mammalian cells. Anal Biochem (2005) 345:164–6. doi: 10.1016/j.ab.2005.06.031
38. Cui S, Wang Y, Gong Y, Lin X, Zhao Y, Zhi D, et al. Correlation of the cytotoxic effects of cationic lipids with their headgroups. Toxicol Res (Camb) (2018) 7:473–9. doi: 10.1039/C8TX00005K
39. Ishida T, Kiwada H. Accelerated blood clearance (ABC) phenomenon upon repeated injection of PEGylated liposomes. Int J Pharm (2008) 354:56–62. doi: 10.1016/j.ijpharm.2007.11.005
40. Taina-González L, de la Fuente M. The potential of nanomedicine to unlock the limitless applications of mRNA. Pharmaceutics (2022) 14:460. doi: 10.3390/pharmaceutics14020460
41. Liu S, Cheng Q, Wei T, Yu X, Johnson LT, Farbiak L, et al. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR-cas gene editing. Nat Mater (2021) 20:701–10. doi: 10.1038/s41563-020-00886-0
42. Cheng Q, Wei T, Farbiak L, Johnson LT, Dilliard SA, Siegwart DJ, et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-cas gene editing. Nat Nanotechnol (2020) 15:313–20. doi: 10.1038/s41565-020-0669-6
43. Cheng X, Lee RJ. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv Drug Delivery Rev (2016) 99:129–37. doi: 10.1016/j.addr.2016.01.022
44. Yang S-T, Kreutzberger AJB, Lee J, Kiessling V, Tamm LK. The role of cholesterol in membrane fusion. Chem Phys Lipids (2016) 199:136–43. doi: 10.1016/j.chemphyslip.2016.05.003
45. Patel S, Ashwanikumar N, Robinson E, Xia Y, Mihai C, Griffith JP 3rd, et al. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat Commun (2020) 11:983. doi: 10.1038/s41467-020-14527-2
46. Eygeris Y, Gupta M, Kim J, Sahay G. Chemistry of lipid nanoparticles for RNA delivery. Acc Chem Res (2022) 55:2–12. doi: 10.1021/acs.accounts.1c00544
47. Zhang S, Gao H, Bao G. Physical principles of nanoparticle cellular endocytosis. ACS Nano (2015) 9:8655–71. doi: 10.1021/acsnano.5b03184
48. Wuebben C, Bartok E, Hartmann G. Innate sensing of mRNA vaccines. Curr Opin Immunol (2022) 79:102249. doi: 10.1016/j.coi.2022.102249
49. Ostendorf T, Zillinger T, Andryka K, Schlee-Guimaraes TM, Schmitz S, Marx S, et al. Immune sensing of synthetic, bacterial, and protozoan RNA by toll-like receptor 8 requires coordinated processing by RNase T2 and RNase 2. Immunity (2020) 52:591–605.e6. doi: 10.1016/j.immuni.2020.03.009
50. Greulich W, Wagner M, Gaidt MM, Stafford C, Cheng Y, Linder A, et al. TLR8 is a sensor of RNase T2 degradation products. Cell (2019) 179:1264–1275.e13. doi: 10.1016/j.cell.2019.11.001
51. Tanji H, Ohto U, Shibata T, Taoka M, Yamauchi Y, Isobe T, et al. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat Struct Mol Biol (2015) 22:109–15. doi: 10.1038/nsmb.2943
52. Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science (2004) 303:1526–9. doi: 10.1126/science.1093620
53. Zhang Z, Ohto U, Shibata T, Krayukhina E, Taoka M, Yamauchi Y, et al. Structural analysis reveals that toll-like receptor 7 is a dual receptor for guanosine and single-stranded RNA. Immunity (2016) 45:737–48. doi: 10.1016/j.immuni.2016.09.011
54. Verbeke R, Hogan MJ, Loré K, Pardi N. Innate immune mechanisms of mRNA vaccines. Immunity (2022) 55:1993–2005. doi: 10.1016/j.immuni.2022.10.014
55. Devarkar SC, Wang C, Miller MT, Ramanathan A, Jiang F, Khan AG, et al. Structural basis for m7G recognition and 2’-o-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc Natl Acad Sci U.S.A. (2016) 113:596–601. doi: 10.1073/pnas.1515152113
56. Schuberth-Wagner C, Ludwig J, Bruder AK, Herzner A-M, Zillinger T, Goldeck M, et al. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2’O-Methylated self RNA. Immunity (2015) 43:41–51. doi: 10.1016/j.immuni.2015.06.015
57. Hyde JL, Diamond MS. Innate immune restriction and antagonism of viral RNA lacking 2'-O methylation. Virology (2015) 479–480:66–74. doi: 10.1016/j.virol.2015.01.019
58. Abbas YM, Laudenbach BT, Martínez-Montero S, Cencic R, Habjan M, Pichlmair A, et al. Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2’-O methylations. Proc Natl Acad Sci U.S.A. (2017) 114:E2106–15. doi: 10.1073/pnas.1612444114
59. Leung DW, Amarasinghe GK. When your cap matters: structural insights into self vs non-self recognition of 5’ RNA by immunomodulatory host proteins. Curr Opin Struct Biol (2016) 36:133–41. doi: 10.1016/j.sbi.2016.02.001
60. Hartmann G. Nucleic acid immunity. Adv Immunol (2017) 133:121–69. doi: 10.1016/bs.ai.2016.11.001
61. Linares-Fernández S, Lacroix C, Exposito J-Y, Verrier B. Tailoring mRNA vaccine to balance Innate/Adaptive immune response. Trends Mol Med (2020) 26:311–23. doi: 10.1016/j.molmed.2019.10.002
62. Sharif N, Alzahrani KJ, Ahmed SN, Dey SK. Efficacy, immunogenicity and safety of COVID-19 vaccines: A systematic review and meta-analysis. Front Immunol (2021) 12:714170. doi: 10.3389/fimmu.2021.714170
63. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N Engl J Med (2020) 383:2603–15. doi: 10.1056/NEJMoa2034577
64. Kremsner PG, Guerrero RA, Arana-Arri E, Martinez GJA, Bonten M, Chandler R, et al. Efficacy and safety of the CVnCoV SARS-CoV-2 mRNA vaccine candidate in ten countries in Europe and Latin America (HERALD): a randomised, observer-blinded, placebo-controlled, phase 2b/3 trial. Lancet Infect Dis (2022) 22:329–40. doi: 10.1016/S1473-3099(21)00677-0
65. Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol (2010) 17:1055–65. doi: 10.1128/CVI.00131-10
66. Morgan J, Muskat K, Tippalagama R, Sette A, Burel J, Arlehamn CSL, et al. Classical CD4 T cells as the cornerstone of antimycobacterial immunity. Immunol Rev (2021) 301:10–29. doi: 10.1111/imr.12963
67. Srivastava S, Ernst JD. Cutting edge: Direct recognition of infected cells by CD4 T cells is required for control of intracellular mycobacterium tuberculosis in vivo. J Immunol (2013) 191:1016–20. doi: 10.4049/jimmunol.1301236
68. Green AM, DiFazio R, Flynn JL. IFN-γ from CD4 T cells is essential for host survival and enhances CD8 T cell function during mycobacterium tuberculosis infection. J Immunol (2013) 190:270–7. doi: 10.4049/jimmunol.1200061
69. Bian Y, Shang S, Siddiqui S, Zhao J, Joosten SA, Ottenhoff THM, et al. MHC ib molecule qa-1 presents mycobacterium tuberculosis peptide antigens to CD8+ T cells and contributes to protection against infection. PloS Pathog (2017) 13:e1006384. doi: 10.1371/journal.ppat.1006384
70. Shang S, Siddiqui S, Bian Y, Zhao J, Wang C-R. Nonclassical MHC ib-restricted CD8+ T cells recognize mycobacterium tuberculosis-derived protein antigens and contribute to protection against infection. PloS Pathog (2016) 12:e1005688. doi: 10.1371/journal.ppat.1005688
71. Behar SM. Antigen-specific CD8(+) T cells and protective immunity to tuberculosis. Adv Exp Med Biol (2013) 783:141–63. doi: 10.1007/978-1-4614-6111-1_8
72. Sridhar S, Begom S, Bermingham A, Hoschler K, Adamson W, Carman W, et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med (2013) 19:1305–12. doi: 10.1038/nm.3350
73. McElhaney JE, Xie D, Hager WD, Barry MB, Wang Y, Kleppinger A, et al. T Cell responses are better correlates of vaccine protection in the elderly. J Immunol (2006) 176:6333–9. doi: 10.4049/jimmunol.176.10.6333
74. Forrest BD, Pride MW, Dunning AJ, Capeding MRZ, Chotpitayasunondh T, Tam JS, et al. Correlation of cellular immune responses with protection against culture-confirmed influenza virus in young children. Clin Vaccine Immunol (2008) 15:1042–53. doi: 10.1128/CVI.00397-07
75. Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, et al. Direct gene transfer into mouse muscle in vivo. Science (1990) 247:1465–8. doi: 10.1126/science.1690918
76. Hoerr I, Obst R, Rammensee HG, Jung G. In vivo application of RNA leads to induction of specific cytotoxic T lymphocytes and antibodies. Eur J Immunol (2000) 30:1–7. doi: 10.1002/1521-4141(200001)30:1<1::AID-IMMU1>3.0.CO;2-#
77. Qiu P, Ziegelhoffer P, Sun J, Yang NS. Gene gun delivery of mRNA in situ results in efficient transgene expression and genetic immunization. Gene Ther (1996) 3:262–8.
78. Conry RM, LoBuglio AF, Wright M, Sumerel L, Pike MJ, Johanning F, et al. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res (1995) 55:1397–400.
79. Bertoletti A, le Bert N, Tan AT. SARS-CoV-2-specific T cells in the changing landscape of the COVID-19 pandemic. Immunity (2022) 55:1764–78. doi: 10.1016/j.immuni.2022.08.008
80. Li J, Liu Q, Liu J, Wu X, Lei Y, Li S, et al. An mRNA-based rabies vaccine induces strong protective immune responses in mice and dogs. Virol J (2022) 19:184. doi: 10.1186/s12985-022-01919-7
81. Li C, Lee A, Grigoryan L, Arunachalam PS, Scott MKD, Trisal M, et al. Mechanisms of innate and adaptive immunity to the pfizer-BioNTech BNT162b2 vaccine. Nat Immunol (2022) 23:543–55. doi: 10.1038/s41590-022-01163-9
82. Arunachalam PS, Scott MKD, Hagan T, Li C, Feng Y, Wimmers F, et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature (2021) 596:410–6. doi: 10.1038/s41586-021-03791-x
83. Painter MM, Mathew D, Goel RR, Apostolidis SA, Pattekar A, Kuthuru O, et al. Rapid induction of antigen-specific CD4+ T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity (2021) 54:2133–2142.e3. doi: 10.1016/j.immuni.2021.08.001
84. Liang F, Lindgren G, Lin A, Thompson EA, Ols S, Röhss J, et al. Efficient targeting and activation of antigen-presenting cells In vivo after modified mRNA vaccine administration in rhesus macaques. Mol Ther (2017) 25:2635–47. doi: 10.1016/j.ymthe.2017.08.006
85. Ndeupen S, Bouteau A, Herbst C, Qin Z, Jacobsen S, Powers NE, et al. Langerhans cells and cDC1s play redundant roles in mRNA-LNP induced protective anti-influenza and anti-SARS-CoV-2 immune responses. PloS Pathog (2022) 18:e1010255. doi: 10.1371/journal.ppat.1010255
86. Kim W, Zhou JQ, Horvath SC, Schmitz AJ, Sturtz AJ, Lei T, et al. Germinal centre-driven maturation of b cell response to mRNA vaccination. Nature (2022) 604:141–5. doi: 10.1038/s41586-022-04527-1
87. Turner JS, O'Halloran JA, Kalaidina E, Kim W, Schmitz AJ, Zhou JQ, et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature (2021) 596:109–13. doi: 10.1038/s41586-021-03738-2
88. Mudd PA, Minervina AA, Pogorelyy MV, Turner JS, Kim W, Kalaidina E, et al. SARS-CoV-2 mRNA vaccination elicits a robust and persistent T follicular helper cell response in humans. Cell (2022) 185:603–613.e15. doi: 10.1016/j.cell.2021.12.026
89. Pardi N, Hogan MJ, Pelc RS, Muramatsu H, Andersen H, DeMaso CR, et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature (2017) 543:248–51. doi: 10.1038/nature21428
90. Zhang Z, Mateus J, Coelho CH, Dan JM, Moderbacher CR, Gálvez RI, et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell (2022) 185:2434–2451.e17. doi: 10.1016/j.cell.2022.05.022
91. Mateus J, Dan JM, Zhang Z, Moderbacher CR, Lammers M, Goodwin B, et al. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Sci (1979) (2021) 374:374. doi: 10.1126/science.abj9853
92. Gilbert PB, Montefiori DC, McDermott AB, Fong Y, Benkeser D, Deng W, et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science (2022) 375:43–50. doi: 10.1126/science.abm3425
93. McMahan K, Yu J, Mercado NB, Loos C, Tostanoski LH, Chandrashekar A, et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature (2021) 590:630–4. doi: 10.1038/s41586-020-03041-6
94. Oberhardt V, Luxenburger H, Kemming J, Schulien I, Ciminski K, Giese S, et al. Rapid and stable mobilization of CD8+ T cells by SARS-CoV-2 mRNA vaccine. Nature (2021) 597:268–73. doi: 10.1038/s41586-021-03841-4
95. Tarke A, Sidney J, Methot N, Yu ED, Zhang Y, Dan JM, et al. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep Med (2021) 2:100355. doi: 10.1016/j.xcrm.2021.100355
96. Geers D, Shamier MC, Bogers S, Hartog Gd, Gommers L, Nieuwkoop NN, et al. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci Immunol (2021) 6. doi: 10.1126/sciimmunol.abj1750
97. Alameh M-G, Tombácz I, Bettini E, Lederer K, Sittplangkoon C, Wilmore JR, et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity (2021) 54:2877–2892.e7. doi: 10.1016/j.immuni.2021.11.001
98. Connors J, Joyner D, Mege NJ, Cusimano GM, Bell MR, Marcy J, et al. Lipid nanoparticles (LNP) induce activation and maturation of antigen presenting cells in young and aged individuals. Res Sq (2022) 6(1):188. doi: 10.21203/rs.3.rs-2199652/v1
99. Luan N, Cao H, Wang Y, Lin K, Liu C. Ionizable lipid nanoparticles enhanced the synergistic adjuvant effect of CpG ODNs and QS21 in a varicella zoster virus glycoprotein e subunit vaccine. Pharmaceutics (2022) 14. doi: 10.3390/pharmaceutics14050973
100. Swaminathan G, Thoryk EA, Cox KS, Meschino S, Dubey SA, Vora KA, et al. A novel lipid nanoparticle adjuvant significantly enhances b cell and T cell responses to sub-unit vaccine antigens. Vaccine (2016) 34:110–9. doi: 10.1016/j.vaccine.2015.10.132
101. Swaminathan G, Thoryk EA, Cox KS, Smith JS, Wolf JJ, Gindy ME, et al. A tetravalent Sub-unit dengue vaccine formulated with ionizable cationic lipid nanoparticle induces significant immune responses in rodents and non-human primates. Sci Rep (2016) 6:34215. doi: 10.1038/srep34215
102. Miao L, Li L, Huang Y, Delcassian D, Chahal J, Han J, et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat Biotechnol (2019) 37:1174–85. doi: 10.1038/s41587-019-0247-3
103. Hartikka J, Bozoukova V, Yang CK, Ye M, Rusalov D, Shlapobersky M, et al. Vaxfectin, a cationic lipid-based adjuvant for protein-based influenza vaccines. Vaccine (2009) 27:6399–403. doi: 10.1016/j.vaccine.2009.06.014
104. Lonez C, Bessodes M, Scherman D, Vandenbranden M, Escriou V, Ruysschaert J-M, et al. Cationic lipid nanocarriers activate toll-like receptor 2 and NLRP3 inflammasome pathways. Nanomedicine (2014) 10:775–82. doi: 10.1016/j.nano.2013.12.003
105. Tanaka T, Legat A, Adam E, Steuve J, Gatot J, Vandenbranden M, et al. DiC14-amidine cationic liposomes stimulate myeloid dendritic cells through toll-like receptor 4. Eur J Immunol (2008) 38:1351–7. doi: 10.1002/eji.200737998
106. Tahtinen S, Tong A-J, Himmels P, Oh J, Paler-Martinez A, Kim L, et al. IL-1 and IL-1ra are key regulators of the inflammatory response to RNA vaccines. Nat Immunol (2022) 23:532–42. doi: 10.1038/s41590-022-01160-y
107. Martinon F, Krishnan S, Lenzen G, Magné R, Gomard E, Guillet JG, et al. Induction of virus-specific cytotoxic T lymphocytesin vivo by liposome-entrapped mRNA. Eur J Immunol (1993) 23:1719–22. doi: 10.1002/eji.1830230749
108. Lundstrom K. Self-amplifying RNA viruses as RNA vaccines. Int J Mol Sci (2020) 21:5130. doi: 10.3390/ijms21145130
109. Schultz-Cherry S, Dybing JK, Davis NL, Williamson C, Suarez DL, Johnston R, et al. Influenza virus (A/HK/156/97) hemagglutinin expressed by an alphavirus replicon system protects chickens against lethal infection with Hong Kong-origin H5N1 viruses. Virology (2000) 278:55–9. doi: 10.1006/viro.2000.0635
110. Petsch B, Schnee M, Vogel AB, Lange E, Hoffmann B, Voss D, et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza a virus infection. Nat Biotechnol (2012) 30:1210–6. doi: 10.1038/nbt.2436
111. Anraku I, Harvey TJ, Linedale R, Gardner J, Harrich D, Suhrbier A, et al. Kunjin virus replicon vaccine vectors induce protective CD8+ T-cell immunity. J Virol (2002) 76:3791–9. doi: 10.1128/JVI.76.8.3791-3799.2002
112. Aberle JH, Aberle SW, Kofler RM, Mandl CW. Humoral and cellular immune response to RNA immunization with flavivirus replicons derived from tick-borne encephalitis virus. J Virol (2005) 79:15107–13. doi: 10.1128/JVI.79.24.15107-15113.2005
113. Berglund P, Quesada-Rolander M, Putkonen P, Biberfeld G, Thorstensson R, Liljeström P, et al. Outcome of immunization of cynomolgus monkeys with recombinant semliki forest virus encoding human immunodeficiency virus type 1 envelope protein and challenge with a high dose of SHIV-4 virus. AIDS Res Hum Retroviruses (1997) 13:1487–95. doi: 10.1089/aid.1997.13.1487
114. Gupta S, Janani R, Bin Q, Luciw P, Greer C, Perri S, et al. Characterization of human immunodeficiency virus gag-specific gamma interferon-expressing cells following protective mucosal immunization with alphavirus replicon particles. J Virol (2005) 79:7135–45. doi: 10.1128/JVI.79.11.7135-7145.2005
115. Polo JM, Belli BA, Driver DA, Frolov I, Sherrill S, Hariharan MJ, et al. Stable alphavirus packaging cell lines for sindbis virus and semliki forest virus-derived vectors. Proc Natl Acad Sci USA (1999) 96:4598–603. doi: 10.1073/pnas.96.8.4598
116. Pyankov O, Bodnev SA, Pyankova OG, Solodkyi VV, Pyankov SA, Setoh YX, et al. A kunjin replicon virus-like particle vaccine provides protection against Ebola virus infection in nonhuman primates. J Infect Dis (2015) 212 Suppl 2:S368–71. doi: 10.1093/infdis/jiv019
117. Barbier AJ, Jiang AY, Zhang P, Wooster R, Anderson DG. The clinical progress of mRNA vaccines and immunotherapies. Nat Biotechnol (2022) 40:840–54. doi: 10.1038/s41587-022-01294-2
118. Freyn AW, da Silva JR, Rosado VC, Bliss CM, Pine M, Mui BL, et al. A multi-targeting, nucleoside-modified mRNA influenza virus vaccine provides broad protection in mice. Mol Ther (2020) 28:1569–84. doi: 10.1016/j.ymthe.2020.04.018
119. Chivukula S, Plitnik T, Tibbitts T, Karve S, Dias A, Zhang D, et al. Development of multivalent mRNA vaccine candidates for seasonal or pandemic influenza. NPJ Vaccines (2021) 6:153. doi: 10.1038/s41541-021-00420-6
120. McMahon M, O'Dell G, Tan J, Sárközy A, Vadovics M, Carreño JM, et al. Assessment of a quadrivalent nucleoside-modified mRNA vaccine that protects against group 2 influenza viruses. Proc Natl Acad Sci (2022) 119. doi: 10.1073/pnas.2206333119
121. Arevalo CP, Bolton MJ, Le Sage V, Ye N, Furey C, Muramatsu H, et al. A multivalent nucleoside-modified mRNA vaccine against all known influenza virus subtypes. Sci (1979) (2022) 378:899–904. doi: 10.1126/science.abm0271
122. Pardi N, Carreño JM, O'Dell G, Tan J, Bajusz C, Muramatsu H, et al. Development of a pentavalent broadly protective nucleoside-modified mRNA vaccine against influenza b viruses. Nat Commun (2022) 13:4677. doi: 10.1038/s41467-022-32149-8
123. Espeseth AS, Cejas PJ, Citron MP, Wang D, DiStefano DJ, Callahan C, et al. Modified mRNA/lipid nanoparticle-based vaccines expressing respiratory syncytial virus f protein variants are immunogenic and protective in rodent models of RSV infection. NPJ Vaccines (2020) 5:16. doi: 10.1038/s41541-020-0163-z
124. Schnee M, Vogel AB, Voss D, Petsch B, Baumhof P, Kramps T, et al. An mRNA vaccine encoding rabies virus glycoprotein induces protection against lethal infection in mice and correlates of protection in adult and newborn pigs. PloS Negl Trop Dis (2016) 10:e0004746. doi: 10.1371/journal.pntd.0004746
125. Hayashi CTH, Cao Y, Clark LC, Tripathi AK, Zavala F, Dwivedi G, et al. mRNA-LNP expressing PfCSP and Pfs25 vaccine candidates targeting infection and transmission of plasmodium falciparum. NPJ Vaccines (2022) 7:155. doi: 10.1038/s41541-022-00577-8
126. Pardi N, LaBranche CC, Ferrari G, Cain DW, Tombácz I, Parks RJ, et al. Characterization of HIV-1 nucleoside-modified mRNA vaccines in rabbits and rhesus macaques. Mol Ther Nucleic Acids (2019) 15:36–47. doi: 10.1016/j.omtn.2019.03.003
127. Zhang P, Narayanan E, Liu Q, Tsybovsky Y, Boswell K, Ding S, et al. A multiclade env–gag VLP mRNA vaccine elicits tier-2 HIV-1-neutralizing antibodies and reduces the risk of heterologous SHIV infection in macaques. Nat Med (2021) 27:2234–45. doi: 10.1038/s41591-021-01574-5
128. Valentin A, Bergamaschi C, Rosati M, Angel M, Burns R, Agarwal M, et al. Comparative immunogenicity of an mRNA/LNP and a DNA vaccine targeting HIV gag conserved elements in macaques. Front Immunol (2022) 13. doi: 10.3389/fimmu.2022.945706
129. Chahal JS, Khan OF, Cooper CL, McPartlan JS, Tsosie JK, Tilley LD, et al. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and toxoplasma gondii challenges with a single dose. Proc Natl Acad Sci (2016) 113:E4133–42. doi: 10.1073/pnas.1600299113
130. John S, Yuzhakov O, Woods A, Deterling J, Hassett K, Shaw CA, et al. Multi-antigenic human cytomegalovirus mRNA vaccines that elicit potent humoral and cell-mediated immunity. Vaccine (2018) 36:1689–99. doi: 10.1016/j.vaccine.2018.01.029
131. Pai M, Behr MA, Dowdy D, Dheda K, Divangahi M, Boehme CC, et al. Tuberculosis. Nat Rev Dis Primers (2016) 2:16076. doi: 10.1038/nrdp.2016.76
133. Bettencourt PJG. The 100 th anniversary of bacille calmette-guérin (BCG) and the latest vaccines against COVID-19. Int J Tuberculosis Lung Dis (2021) 25:611–3. doi: 10.5588/ijtld.21.0372
134. Bettencourt PJG, Joosten SA, Arlehamn CSL, Behr MA, Locht C, Neyrolles O, et al. 100 years of the bacillus calmette-guérin vaccine. Vaccine (2021) 39:7221–2. doi: 10.1016/j.vaccine.2021.11.038
135. Derrick SC, Yabe IM, Yang A, Morris SL. Vaccine-induced anti-tuberculosis protective immunity in mice correlates with the magnitude and quality of multifunctional CD4 T cells. Vaccine (2011) 29:2902–9. doi: 10.1016/j.vaccine.2011.02.010
136. Seder RA, Darrah PA, Roederer M. T-Cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol (2008) 8:247–58. doi: 10.1038/nri2274
137. Whitlow E, Mustafa AS, Hanif SNM. An overview of the development of new vaccines for tuberculosis. Vaccines (Basel) (2020) 8:586. doi: 10.3390/vaccines8040586
138. Fan X-Y, Lowrie DB. Where are the RNA vaccines for TB? Emerg Microbes Infect (2021) 10:1217–8. doi: 10.1080/22221751.2021.1935328
139. Stylianou E, Harrington-Kandt R, Beglov J, Bull N, Pinpathomrat N, Swarbrick GM, et al. Identification and evaluation of novel protective antigens for the development of a candidate tuberculosis subunit vaccine. Infect Immun (2018) 86. doi: 10.1128/IAI.00014-18
140. Bettencourt P, Müller J, Nicastri A, Cantillon D, Madhavan M, Charles PD, et al. Identification of antigens presented by MHC for vaccines against tuberculosis. NPJ Vaccines (2020) 5:2. doi: 10.1038/s41541-019-0148-y
141. Sharma R, Rajput VS, Jamal S, Grover A, Grover S. An immunoinformatics approach to design a multi-epitope vaccine against mycobacterium tuberculosis exploiting secreted exosome proteins. Sci Rep (2021) 11:13836. doi: 10.1038/s41598-021-93266-w
142. al Tbeishat H. Novel in silico mRNA vaccine design exploiting proteins of m. tuberculosis that modulates host immune responses by inducing epigenetic modifications. Sci Rep (2022) 12:4645. doi: 10.1038/s41598-022-08506-4
143. Shahrear S, Islam ABMM. Modeling of MT. P495, an mRNA-based vaccine against the phosphate-binding protein PstS1 of mycobacterium tuberculosis. Mol Divers (2022) 1–20. doi: 10.1007/s11030-022-10515-4
144. Xue T, Stavropoulos E, Yang M, Ragno S, Vordermeier M, Chambers M, et al. RNA Encoding the MPT83 antigen induces protective immune responses against mycobacterium tuberculosis infection. Infect Immun (2004) 72:6324–9. doi: 10.1128/IAI.72.11.6324-6329.2004
145. Lorenzi JC, Trombone APF, Rocha CD, Almeida LP, Lousada RL, Malardo T, et al. Intranasal vaccination with messenger RNA as a new approach in gene therapy: Use against tuberculosis. BMC Biotechnol (2010) 10:77. doi: 10.1186/1472-6750-10-77
146. Larsen SE, Erasmus JH, Reese VA, Pecor T, Archer J, Kandahar A, et al. An RNA-based vaccine platform for use against mycobacterium tuberculosis. Vaccines (Basel) (2023) 11:130. doi: 10.3390/vaccines11010130
147. Rais M, Abdelaal H, Reese VA, Ferede D, Larsen SE, Pecor T, et al. Immunogenicity and protection against mycobacterium avium with a heterologous RNA prime and protein boost vaccine regimen. Tuberculosis (2023) 138:102302. doi: 10.1016/j.tube.2022.102302
148. Mu Z, Haynes BF, Cain DW. HIV mRNA vaccines–progress and future paths. Vaccines (Basel) (2021) 9:134. doi: 10.3390/vaccines9020134
149. Robinson HL. HIV / AIDS vaccines: 2018. Clin Pharmacol Ther (2018) 104:1062–73. doi: 10.1002/cpt.1208
150. Haynes BF, Burton DR. Developing an HIV vaccine. Sci (1979) (2017) 355:1129–30. doi: 10.1126/science.aan0662
151. Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunol Rev (2013) 254:225–44. doi: 10.1111/imr.12075
152. Sliepen K, Sanders RW. HIV-1 envelope glycoprotein immunogens to induce broadly neutralizing antibodies. Expert Rev Vaccines (2016) 15:349–65. doi: 10.1586/14760584.2016.1129905
153. Suryawanshi P, Bagul R, Shete A, Thakar M. Anti-HIV-1 ADCC and HIV-1 env can be partners in reducing latent HIV reservoir. Front Immunol (2021) 12. doi: 10.3389/fimmu.2021.663919
154. Finney J, Kelsoe G. Poly- and autoreactivity of HIV-1 bNAbs: implications for vaccine design. Retrovirology (2018) 15:53. doi: 10.1186/s12977-018-0435-0
155. Karpenko LI, Bazhan SI, Antonets DV, Belyakov IM. Novel approaches in polyepitope T-cell vaccine development against HIV-1. Expert Rev Vaccines (2014) 13:155–73. doi: 10.1586/14760584.2014.861748
156. Saunders KO, Pardi N, Parks R, Santra S, Mu Z, Sutherland L, et al. Lipid nanoparticle encapsulated nucleoside-modified mRNA vaccines elicit polyfunctional HIV-1 antibodies comparable to proteins in nonhuman primates. NPJ Vaccines (2021) 6:50. doi: 10.1038/s41541-021-00307-6
157. Nkumama IN, Osier FHA. Malaria vaccine roller coaster. Nat Microbiol (2021) 6:1345–6. doi: 10.1038/s41564-021-00982-0
158. Nussenzweig RS, Vanderberg J, Most H, Orton C. Protective immunity produced by the injection of X-irradiated sporozoites of plasmodium berghei. Nature (1967) 216:160–2. doi: 10.1038/216160a0
159. Seder RA, Chang L-J, Enama ME, Zephir KL, Sarwar UN, Gordon IJ, et al. Protection against malaria by intravenous immunization with a nonreplicating sporozoite vaccine. Sci (1979) (2013) 341:1359–65. doi: 10.1126/science.1241800
160. Epstein JE, Tewari K, Lyke KE, Sim BKL, Billingsley PF, Laurens MB, et al. Live attenuated malaria vaccine designed to protect through hepatic CD8+ T cell immunity. Sci (1979) (2011) 334:475–80. doi: 10.1126/science.1211548
161. Oneko M, Steinhardt LC, Yego R, Wiegand RE, Swanson PA, Kc N, et al. Safety, immunogenicity and efficacy of PfSPZ vaccine against malaria in infants in western Kenya: a double-blind, randomized, placebo-controlled phase 2 trial. Nat Med (2021) 27:1636–45. doi: 10.1038/s41591-021-01470-y
162. Sissoko MS, Healy SA, Katile A, Omaswa F, Zaidi I, Gabriel EE, et al. Safety and efficacy of PfSPZ vaccine against plasmodium falciparum via direct venous inoculation in healthy malaria-exposed adults in Mali: a randomised, double-blind phase 1 trial. Lancet Infect Dis (2017) 17:498–509. doi: 10.1016/S1473-3099(17)30104-4
163. Stanisic DI, McCall MBB. Correlates of malaria vaccine efficacy. Expert Rev Vaccines (2021) 20:143–61. doi: 10.1080/14760584.2021.1882309
165. Datoo MS, Natama MH, Somé A, Traoré O, Rouamba T, Bellamy D, et al. Efficacy of a low-dose candidate malaria vaccine, R21 in adjuvant matrix-m, with seasonal administration to children in Burkina Faso: a randomised controlled trial. Lancet (2021) 397:1809–18. doi: 10.1016/S0140-6736(21)00943-0
166. Daubenberger CA, Moncunill G. Next-generation malaria subunit vaccines to reduce disease burden in African children. Lancet Infect Dis (2022) 22:1655–6. doi: 10.1016/S1473-3099(22)00523-0
167. Datoo MS, Natama HM, Somé A, Bellamy D, Traoré O, Rouamba T, et al. Efficacy and immunogenicity of R21/Matrix-m vaccine against clinical malaria after 2 years’ follow-up in children in Burkina Faso: a phase 1/2b randomised controlled trial. Lancet Infect Dis (2022) 22:1728–36. doi: 10.1016/S1473-3099(22)00442-X
168. Mallory KL, Taylor JA, Zou X, Waghela IN, Schneider CG, Sibilo MQ, et al. Messenger RNA expressing PfCSP induces functional, protective immune responses against malaria in mice. NPJ Vaccines (2021) 6:84. doi: 10.1038/s41541-021-00345-0
169. Raj DK, Mohapatra AD, Jnawali A, Zuromski J, Jha A, Cham-Kpu G, et al. Anti-PfGARP activates programmed cell death of parasites and reduces severe malaria. Nature (2020) 582:104–8. doi: 10.1038/s41586-020-2220-1
170. Waghela IN, Mallory KL, Taylor JA, Schneider CG, Savransky T, Janse CJ, et al. Exploring in vitro expression and immune potency in mice using mRNA encoding the plasmodium falciparum malaria antigen, CelTOS. Front Immunol (2022) 13. doi: 10.3389/fimmu.2022.1026052
Keywords: RNA vaccines, malaria, tuberculosis, HIV, infectious diseases
Citation: Matarazzo L and Bettencourt PJG (2023) mRNA vaccines: a new opportunity for malaria, tuberculosis and HIV. Front. Immunol. 14:1172691. doi: 10.3389/fimmu.2023.1172691
Received: 23 February 2023; Accepted: 10 April 2023;
Published: 24 April 2023.
Edited by:
Selidji Todagbe Agnandji, Centre de Recherche Médicales de Lambaréné, GabonReviewed by:
Evelina Angov, Walter Reed Army Institute of Research, United StatesZhidong Hu, Shanghai Public Health Clinical Center, Fudan University, China
Copyright © 2023 Matarazzo and Bettencourt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paulo J. G. Bettencourt, pbettencourt@ucp.pt
 Laura Matarazzo
Laura Matarazzo Paulo J. G. Bettencourt
Paulo J. G. Bettencourt