- 1Key Laboratory of Exploration and Utilization of Aquatic Genetic Resources, Ministry of Education, Shanghai Ocean University, Shanghai, China
- 2International Research Center for Marine Biosciences, Ministry of Science and Technology, Shanghai Ocean University, Shanghai, China
- 3National Demonstration Center for Experimental Fisheries Science Education, Shanghai Ocean University, Shanghai, China
- 4Department of Microbiology and Immunology, University of Maryland School of Medicine, Baltimore, MD, United States
- 5Institute of Marine and Environmental Technology, Baltimore, MD, United States
- 6Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Introduction: Cartilaginous fishes are the most evolutionary-distant vertebrates from mammals and possess an immunoglobulin (Ig)- and T cell-mediated adaptive immunity. CD8 is the hallmark receptor of cytotoxic T cells and is required for the formation of T cell receptor-major histocompatibility complex (TCR-MHC) class I complexes.
Methods: RACE PCR was used to obtain gene sequences. Direct dilution was applied for the refolding of denatured recombinant CD8 protein. Hanging-drop vapor diffusion method was performed for protein crystallization.
Results: In this study, CD8α and CD8β orthologues (termed ScCD8α and ScCD8β) were identified in small-spotted catshark (Scyliorhinus canicula). Both ScCD8α and ScCD8β possess an extracellular immunoglobulin superfamily (IgSF) V domain as in previously identified CD8 proteins. The genes encoding CD8α and CD8β are tandemly linked in the genomes of all jawed vertebrates studied, suggesting that they were duplicated from a common ancestral gene before the divergence of cartilaginous fishes and other vertebrates. We determined the crystal structure of the ScCD8α ectodomain homodimer at a resolution of 1.35 Å and show that it exhibits the typical topological structure of CD8α from endotherms. As in mammals, the homodimer formation of ScCD8αα relies upon interactions within a hydrophobic core although this differs in position and amino acid composition. Importantly, ScCD8αα shares the canonical cavity required for interaction with peptide-loaded MHC I in mammals. Furthermore, it was found that ScCD8α can co-immunoprecipitate with ScCD8β, indicating that it can form both homodimeric and heterodimeric complexes.
Conclusion: Our results expand the current knowledge of vertebrate CD8 dimerization and the interaction between CD8α with p/MHC I from an evolutionary perspective.
Introduction
The divergence of jawless and jawed (gnathostome) vertebrates is a major event in evolution, which was accompanied by drastic morphological and physiological changes (1, 2). Of note, all gnathostomes are equipped with an antibody-based adaptive immune system orchestrated by immunoglobulins (Igs) and T cell receptors (TCRs), which are somatically rearranged by the recombination-activating genes (RAGs), and polymorphic/polygenic major histocompatibility complex (MHC) molecules, which are used to fend off pathogen evasion (3–5).
CD8+ T cytotoxic cells are key immune cells in the adaptive immune response and are vital for limiting the replication of intracellular pathogens such as viruses by killing the infected cells. These cells are activated by peptides presented by MHC class I molecules (p/MHC I) and secret cytotoxins such as perforin and granzymes to induce apoptosis in the target cells. CD8αα-p/MHC I complexes interact with TCR/CD3 complexes to transduce signals within the T cells. To ensure efficient T cell activation, CD8 acts as a co-receptor to stabilize the interaction of TCR with MHC I on the surface of antigen-presenting cells (APCs) (6, 7). The CD8 molecule is a type I membrane glycoprotein with two isoforms, CD8α and CD8β, which can form homodimeric (CD8αα) or heterodimeric (CD8αβ) complexes (8). Both CD8α and CD8β have an extracellular Ig superfamily (IgSF) V domain, a stalk region, a transmembrane domain and an intracellular cytoplasmic tail which contains a CxC/Cxx motif for binding to the Src kinase p56Lck to elicit cellular responses (9, 10). CD8α and CD8β are structurally related and the genes encoding them are tandemly linked in the genomes in both mammals and bony fishes (11). Although CD8αα and CD8αβ bind with similar affinities to p/MHC I and both can be recruited to the immunological synapse, their expression differs between immune cell types. CD8αα is found on γδ T cells, NK cells, subsets of dendritic cells and intestinal intraepithelial lymphocytes, whilst CD8αβ expression is limited to αβ T cells (6, 12). To date, the functions of CD8αα are still not fully characterized. However, it has been shown that CD8αα can bind non-classical MHC I molecules with greater affinity than classical MHC I molecules (13, 14), helping determine the differentiation of memory and mucosal T cell immune responses. Additionally, recent studies suggest that CD8αα may function as a negative regulator for T cell activation (15). The crystal structures of CD8αα homodimer and CD8αα/p/MHC I complex have been solved in mammals and birds (16–18). The CD8αα homodimer consists of two symmetric IgSF V domains, with several highly conserved residues being critical for CD8α dimerization and interaction between CD8α and p/MHC I molecule. Recently, we solved the crystal structure of grass carp CD8αα (19) and found that it has several unique topological features. For instance, the cavity of human CD8αα which is shaped by the CDR loops and forms the principal area of interaction with p/MHC I, is absent in grass carp CD8αα. However, this appears to have little impact upon the binding with peptide-Ctid-UAA-β2m (p/UAA-β2m) binding (20), suggesting that the mechanisms of interactions between CD8αα and p/MHC I in exothermic vertebrates may differ from that in endotherms.
The cartilaginous fishes (Chondrichthyes: sharks, skates, rays, and chimaeras) diverged from other jawed vertebrates approximately 450 million years ago and are the most ancient vertebrate lineage to possess a canonical adaptive immune system. It has been shown that the protein structure of elephant shark (Callorhinchus milii) MHC I molecule is structurally conserved (21). Analyses of the elephant shark draft genome assembly confirmed that the CD8α and CD8β genes are present in cartilaginous fishes (2, 22). In this study, we sequenced CD8α and CD8β from small-spotted catshark (Scyliorhinus canicular; Sc), and show that the encoded proteins possess an IgSF V domain and that CD8α could form homodimer and heterodimer with CD8β. Moreover, the structure of the ScCD8αα extracellular region was solved, revealing that CD8α IgSF V domain contains a canonical cavity which is present in the interface with p/MHC I in mammals. Our study provides the first evidence that cartilaginous fish CD8α can form homodimer through a conserved hydrophobic core, and that CD8α can interact with CD8β.
Materials and methods
Animals
Captive-bred, small-spotted catsharks of approximately three years of age, were maintained in artificial seawater at 8-12°C in indoor tanks at the University of Aberdeen, UK. Animals were overdosed in MS-222 prior to sacrifice and tissue harvest. All procedures were conducted in accordance with UK Home Office ‘Animals and Scientific Procedures Act 1986; Amendment Regulations 2012’ on animal care and use, with prior ethical approval from the University of Aberdeen’s Animal Welfare and Ethical Review Body (AWERB). Tissue samples were homogenized in TRIzol reagent (Sigma-Aldrich) using a TissueLyser II (Qiagen) and total RNA isolation performed according to standard protocols. RNA samples were quantified using the Agilent 2100 Bioanalyzer (Agilent) and were pooled for subsequent gene cloning (23).
Identification and cloning of ScCD8
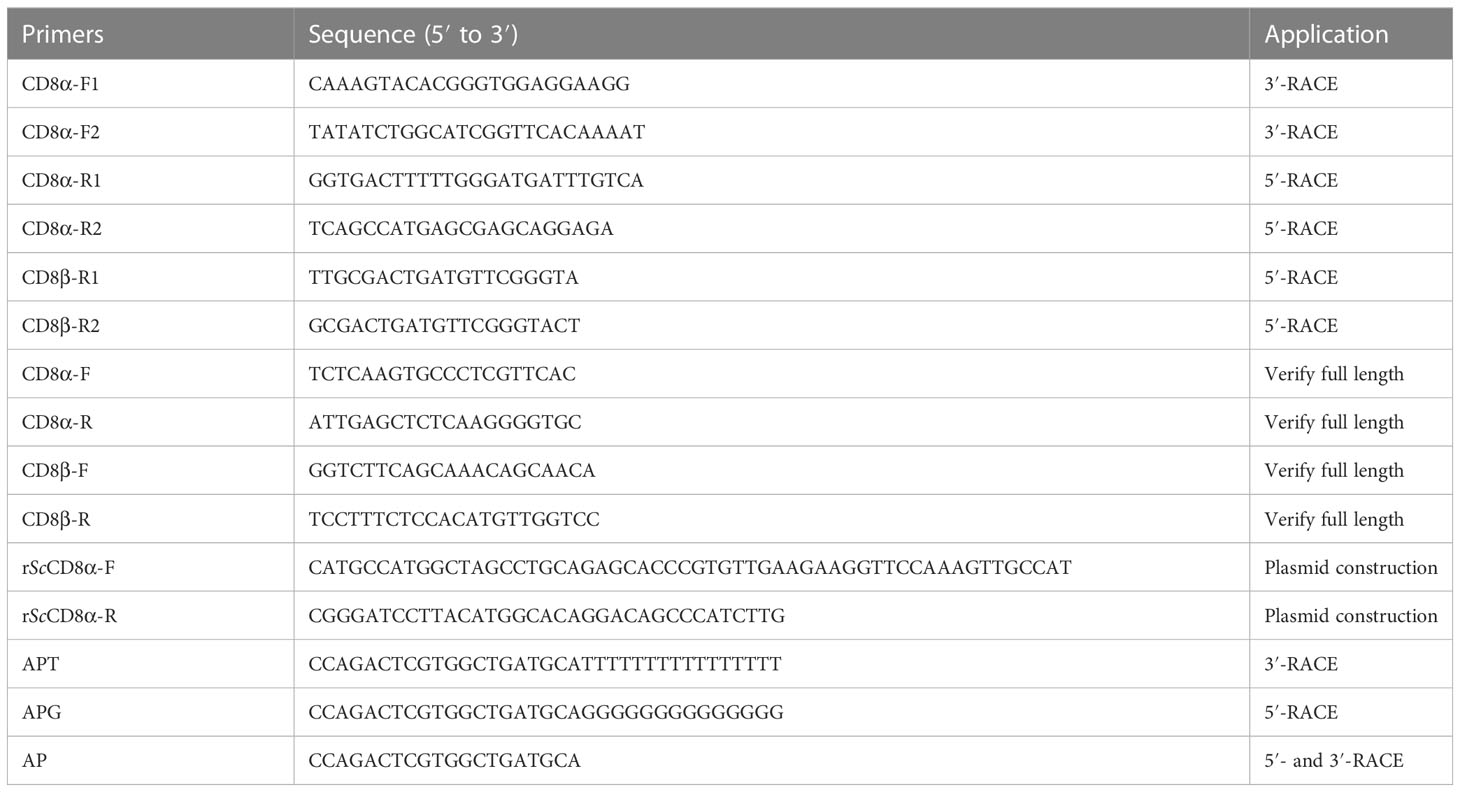
Partial cDNA sequences of ScCD8α and ScCD8β were obtained from the NCBI Scyliorhinus database (https://www.ncbi.nlm.nih.gov/) using the local BLAST search tool and the top hits used to design gene-specific primers (GSP) for cloning. The full-length cDNA sequences were obtained using GSP by rapid amplification of cDNA ends (RACE) PCR. For 5′ RACE, first strand cDNA was tailed with dCTPs and used as template. The first round PCR was performed using a Premix Ex Taq™ Hot Start Mix (TaKaRa) and primer pairs, 5R1/APG. Primers 5R2/AP were used for the second round PCR of 5′ RACE. For the 3’ RACE, primers 3F1/APT and 3F2/AP were used for the first and second round PCR, respectively (24). 5′ and 3′ RACE was performed as described previously (25). Briefly, the first round PCR was performed under the following conditions: 1 cycle of 95°C/3 min; 35 cycles of 98°C/10 s, 60°C/30 s, 72°C/1 min; 1 cycle of 72°C/7 min. The first round PCR products were diluted (1:50, v/v) and used as template for the second round PCR and cycling conditions are: 1 cycle of 95°C/3 min; 5 cycles of 98°C/10 s, 70°C/30 s, 72°C/1 min; 5 cycles of 98°C/10 s, 68°C/30 s, 72°C/1 min; 5 cycles of 98°C/10 s, 66°C/30 s, 72°C/1 min; 5 cycles of 98°C/10 s, 64°C/30 s, 72°C/1 min; 5 cycles of 98°C/10 s, 62°C/30 s, 72°C/1 min; 10 cycles of 98°C/10 s, 60°C/30 s, 72°C/1 min;1 cycle of 72°C/7 min.
PCR products were sequenced and assembled into the full-length cDNA sequences using the DNAMAN program (version 6.0). The complete coding sequences (CDS) were verified by sequencing the PCR products amplified using one pair of primers located at the 5’ and 3’ untranslated region (UTR). Primers are described in Table 1.
Sequence analysis of ScCD8α and ScCD8β
The cDNA and protein sequences of ScCD8α and ScCD8β were predicted using the Genetyx program. Signal peptides, molecular weight and isoelectric point (pI) were predicted using software available on the ExPasy server (https://www.expasy.org/). Known CD8α sequences were retrieved from the NCBI GenBank database. Amino acid (aa) identity between the sequences was determined using the EMBOSS Tools (https://www.ebi.ac.uk/Tools/psa/emboss_needle/). The phylogenetic tree was constructed using the Neighbor-Joining method of MEGA X program and repeated for 10,000 times to obtain the bootstrap scores. The signal peptide cleavage site was predicted using the SignalP 5.0 program (http://www.cbs.dtu.dk/services/SignalP/).
Production and purification of recombinant ScCD8α protein
The cDNA fragment encoding the extracellular IgSF V domain of ScCD8α (Ser20-Met163) was amplified and cloned into the expression vector pET21d between the Nco I and BamH I sites, with the codons for the first ten aa optimized for expression in the E. coli. The recombinant pET21d-ScCD8α plasmid was transformed into E. coli Rosetta (DE3) cells. The recombinant ScCD8α protein was expressed as inclusion bodies after induction with 0.5 mM IPTG. The inclusion bodies were collected as previously described (19), and denatured in 6 M guanidine hydrochloride buffer (6 M guanidine hydrochloride, 50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 100 mM NaCl, 10% glycerol (v/v) and 10 mM DTT) (19). The protein concentration was adjusted to 30 mg/ml prior to protein refolding.
Dilution renaturation was used to obtain soluble ScCD8α (19). Briefly, 5 ml of ScCD8α-containing denaturing solution (at 30 mg/ml) was slowly dropped into 500 ml of refolding buffer (100 mM Tris-HCl, 2 mM EDTA, 400 mM L-arginine-HCl, 0.5 mM oxidized glutathione and 5 mM reduced glutathione, pH 8.0) at 4°C and slowly stirred for 24 h. The resulting protein solution was then concentrated using a 10 kDa cutoff filter and buffer-exchanged into 10 mM Tris-HCl with 50 mM NaCl (pH 8.0). Concentrated protein was then subject to size exclusion chromatography over a Superdex 200 16/600 column (GE Healthcare) and polished by passage over a Resource Q anion-exchange chromatography column (GE Healthcare). Protein fractions were collected at 1 ml per tube and checked by SDS-PAGE. Purified ScCD8α protein was again buffer-exchanged into 20 mM Tris-HCl with 50 mM NaCl (pH 8.0). The recombinant ScCD8α was also checked by PAGE under non-reducing conditions. PAGE was run in Tris-Gly buffer (pH 4.4, WSHTBio, China) in an ice-water bath at 120 V for 2 h.
Co-immunoprecipitation
The coding sequences of ScCD8α and ScCD8β were synthesized with a C-terminal Myc-tag or HA-tag and cloned into pcDNA3.1 (GENEWIZ, China). HEK293 cells were grown in 25cm2 flasks to reach 80-90% confluence and transfected with 5 μg of each plasmid using the jetOPTIMUS® reagent (polyplus). After 24 h, the transfected HEK293 cells were collected and lysed in the radioimmunoprecipitation assay (RIPA) buffer (Beyotime, China) containing 1% phenylmethanesulfonyl fluoride (PMSF, Beyotime) on ice for 30 min. The lysed sample was centrifuged at 12,000 g for 15 min to pellet cellular debris and the supernatant collected into a clean 1.5 ml microtube. Following the removal of 50 μl for Western blot analysis, 40 μl of α-Myc-conjugated magnetic beads (Abmart) was added to the remaining supernatant and incubated overnight on a shaker. The magnetic beads were collected by centrifugation at 1,500 g for 3 min at 4°C, washed 3 times with ice-cold PBS, resuspended in 30 µl 2 × SDS loading buffer and prepared for Western blot analysis as detailed description previous (26). Briefly, Protein samples are separated by SDS-PAGE, then transferred to PVDF membranes using a semi-dry method. The membrane is blocked with a TBS buffer containing 5% non-fat milk for 1 h at room temperature. It is then incubated overnight at 4°C with the α-Myc or α-HA (1:1000, v/v, Huabio, China). After washing with TBS-T buffer 3 times, the membrane was incubated with a goat α-mouse IgG H&L antibody (IRDye® 680RD, 1: 10,000 dilution, v/v, Odyssey, USA), for 1 h in the dark. The membrane was washed as above then imaged using the Odyssey CLx image system (Odyssey, USA).
Protein crystallization
Purified ScCD8α protein was concentrated to 5 mg/ml or 10 mg/ml and an initial crystallization screen performed by the hanging-drop vapor diffusion method at 277 K. The Wizard™ Classic line of random sparse matrix screening (Rigaku Reagents) was used to establish crystallization conditions where the protein solution was mixed with reservoir buffer at a 1:1 ratio (v/v). The ScCD8α crystals formed in Wizard classic 1, Formulation 41 (30% (w/v) PEG3000 and 100 mM CHES/Sodium hydroxide pH 9.5) after 5 days at a protein concentration of 10 mg/ml.
Collection and processing of diffraction data
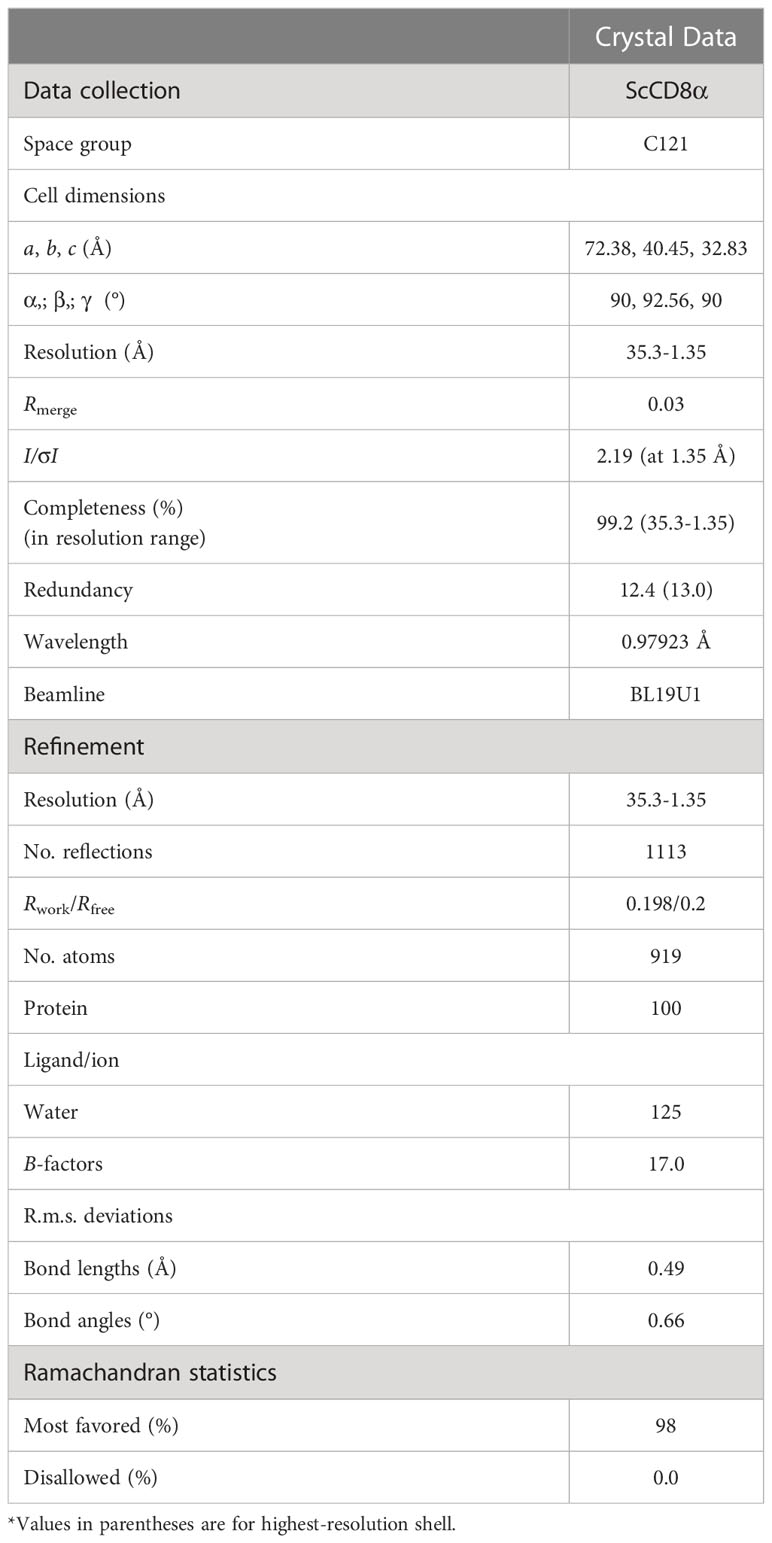
Diffraction data on the ScCD8α crystals were collected on beamline BL19U1 at the wavelength of 0.97923 Å, with an ADSC 315 CCD detector at the Shanghai Synchrotron Radiation Facility (SSRF), China. The crystals were soaked for several seconds in a reservoir solution containing 15% glycerol as a cryoprotectant and then flash-cooled in a stream of gaseous nitrogen at 100 K. The collected intensities were indexed, integrated, corrected for absorption, scaled and merged using HKL 3000.
Structural analysis
The structure of ScCD8α was determined by molecular replacement with the MOLREP and PHASER programmer using the chicken CD8α protein structure (Protein Data Bank [PDB] code: 5EB9) as a search model. A comprehensive model was built manually with COOT (27), and the structure was refined with the REFMAC 5 program. Refinement rounds were implemented using PHENIX as previously described (28), and the stereochemical quality of the final model was assessed by the PROCHECK program. Details of data collection and refinement are shown in Table 2. PyMOL was used to generate and visualize the structural figures. Multiple sequence alignment was performed by the Clustal Omega server (https://www.ebi.ac.uk/Tools/msa/clustalo/) and ESPript 3.0 (http://espript.ibcp.fr/ESPript/ESPript/). Scores for accessible surface area (ASA) and buried surface area (BSA) were calculated using the PDBePISA server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html).
Results
The CD8α and CD8β genes had diverged in cartilaginous fishes
In the present study, CD8α and CD8β were identified from small-spotted catshark. The full-length cDNA sequence of the ScCD8α (NCBI accession number: MT840192) is 917 bp with an open reading frame (ORF) of 657 bp and encodes a protein of 218 aa with a putative signal peptide of 19 aa. A CQH motif is present in the cytoplasmic tail of ScCD8α permitting its interaction with Lyk kinase. Three cysteine residues are present in the ectodomain, two of which are conserved in all previously characterized CD8α molecules (Supplementary Figure 1). The obtained cDNA sequence of ScCD8β (NCBI accession number: MW713126.1) is 726 bp containing an ORF of 648 bp and encodes a protein of 215 aa with a signal peptide of 19 aa. It has a 5′-UTR of 39 bp and a 3′-UTR of 42 bp (Supplementary Figure 2). Although ScCD8α and ScCD8β have low (20%-30%) aa identity with their orthologues from other species (Supplementary Table 1), they share a similar domain structure (i.e., IgSF V-transmembrane region-cytoplasmic tail).
Gene synteny for cartilaginous fish CD8α and CD8β genes was determined using the draft genomes for elephant shark, white shark (Carcharodon carcharias), and small-spotted catshark (Figure 1A). This showed that the CD8α and CD8β genes are tandemly linked in all three species and are sandwiched by the RMND5A and FOX12/13 genes. This synteny is conserved in chicken, mouse, and human (Figure 1A). The CD8 genes are also arranged in tandem in zebrafish, while the surrounding genes are not conserved. To determine the phylogenetic relationships of ScCD8α and ScCD8β, an unrooted Neighbor-Joining tree was constructed using CD8α and CD8β protein sequences from cartilaginous fishes, teleosts, amphibians, reptiles, birds and mammals. As shown in Figure 1B, the CD8α and CD8β clades are well separated, suggesting that the duplication event generating two CD8 genes occurred before the divergence of cartilaginous fishes and other vertebrates. In line with many other studies, cartilaginous fish CD8α and CD8β grouped with their tetrapod orthologs (node bootstrap values of 83% and 97% respectively), while teleost CD8αs and CD8βs form an independent clade. This is likely a consequence of the extra round(s) of genome wide duplication experienced by teleost fishes (29) and the higher evolutionary rate of both protein-coding and non-coding sequences in this lineage compared to other vertebrates (30).
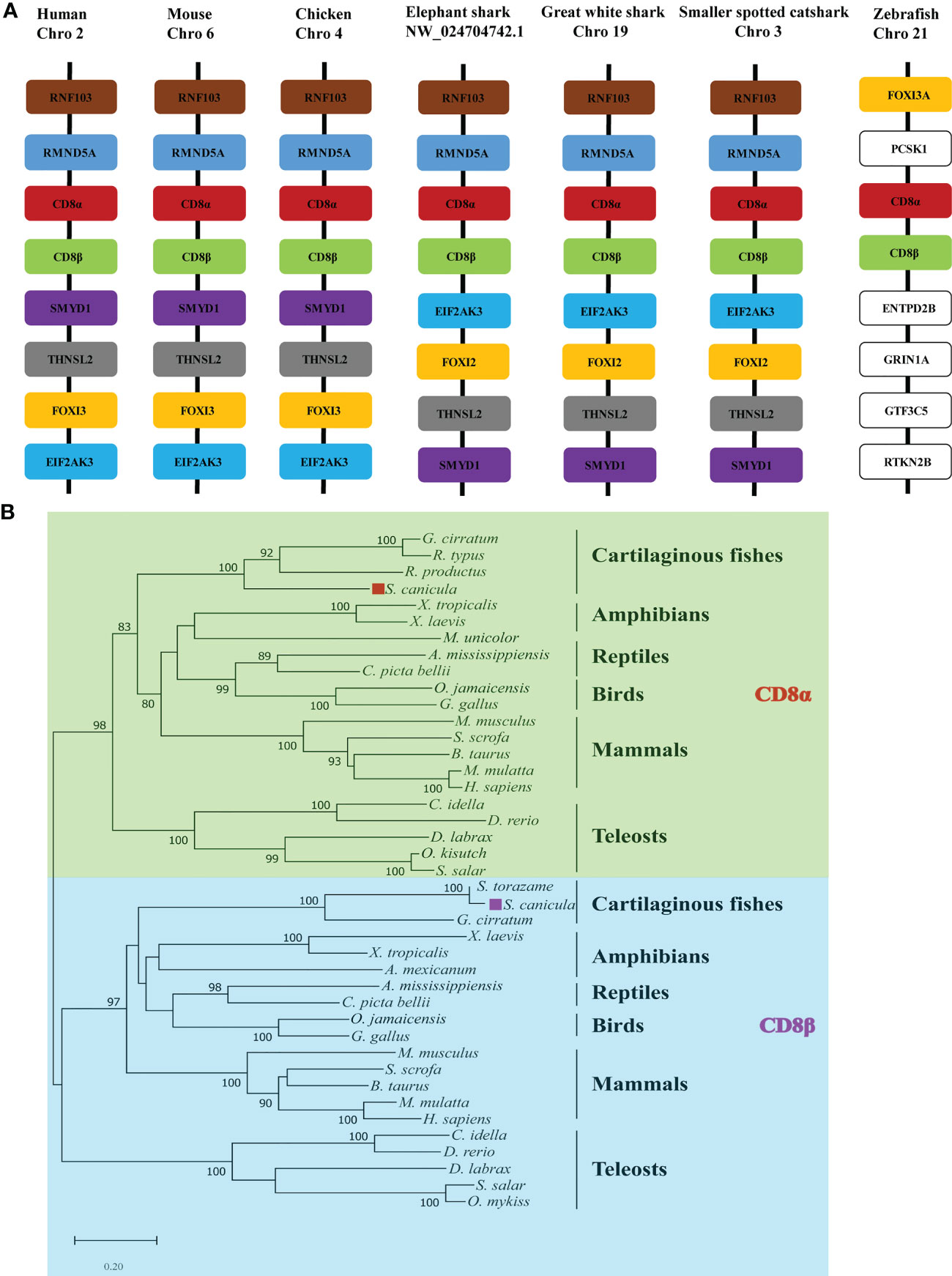
Figure 1 Analysis of ScCD8 genes. (A) Gene synteny analysis of CD8α, the synteny information for human, mouse, chicken, elephant shark, great white shark, zebrafish and small-spotted catshark, and spotted gar genes were obtained from the Ensembl database. (B) Neighbor-joining (N-J) phylogenetic tree of CD8 genes. The tree was constructed using the N-J method within the MEGA-X program with bootstrap values shown next to the branches based on 10,000 bootstrap replications. “◼” indicates ScCD8α (red) and ScCD8β (purple). The accession numbers of protein sequences are provided in Supplementary Table 1.
The topological structure of ScCD8αα
The recombinant protein of the ScCD8α extracellular region expressed in bacteria and purified (Supplementary Figure 3). SDS-PAGE analysis under reducing conditions revealed a single band of approx. 16 kDa. However, PAGE analysis under non-reducing conditions showed a protein band of approx. 32 kDa, indicating that it exists as a dimer (Supplementary Figure 3C). The structure of recombinant ScCD8α extracellular region (Supplementary Figure 2) was solved at a resolution of 1.35 Å with a space group of C121 and further refined to a Rwork/Rfree factor of 0.198/0.2 (Table 2). The structure is composed of two molecules arranged in the asymmetric unit. Despite low sequence homology with known CD8α molecules, ScCD8αα retains typical IgSF V architecture. Like its counterparts in higher vertebrates, the ScCD8α dimer consists of 2 anti-parallel β sheets containing 10 β strands (Figure 2A), labeled as A, A’, B, C, C’, D, E, F, G, G’. The front sheet is occupied by strands A’, D and E, while the back sheet contains strand A, B, C, C’ F, G and G’ (Figures 2A, B). Notably, the A strand is in the back β sheet, lying parallel to strand G’ (Figures 2C, D), an arrangement seen only in grass carp CD8α thus far. In contrast, the A strand of chicken and human CD8αs is in the front sheet and lies in parallel to the B strand (Figures 2E–G). Two cysteines (Cys19 and Cys84) form a disulphide bond linking the front and back sheet of ScCD8α and stabilizing the IgSF V architecture (Figure 2A). The RMSD values of ScCD8αα with known CD8αα from other species were determined, including human (1.63), monkey (1.70), mouse (1.70), bovine (2.56), swine (2.10) chicken (1.54), and grass carp (1.66) (Supplementary Figure 4).
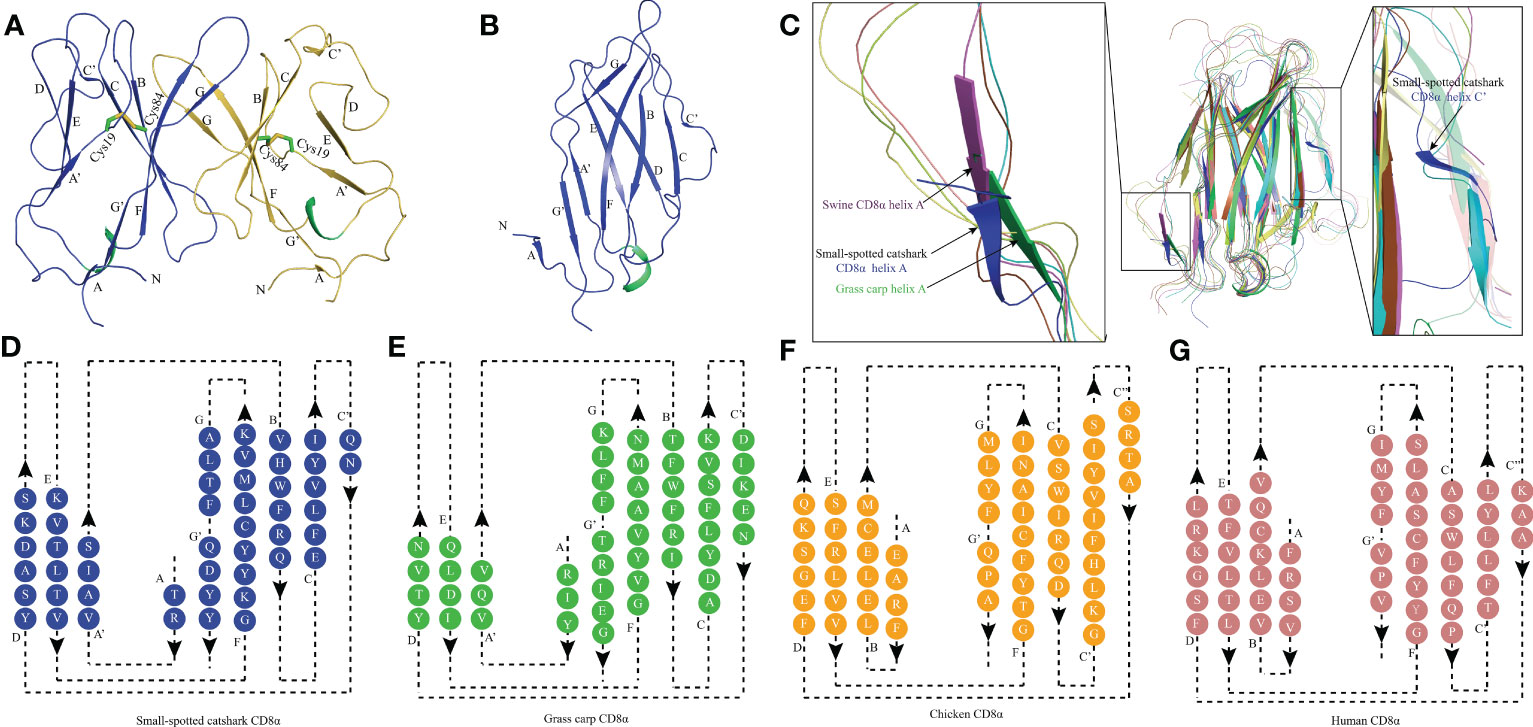
Figure 2 The topological structure of ScCD8αα extracellular region. (A) Cartoonstructure of the ScCD8αα homodimer, CD8α monomers are colored blue and orange. The β-strands are labelled. Disulfide bonds are shown as sticks and colored lime green. (B) The monomer structure of the ScCD8α extracellular region. (C) Comparison of ScCD8α extracellular region to that of other species. All the structures are shown in cartoon representation. The ScCD8α is colored blue, grass carp CD8α is lime green, chicken CD8α is yellow, bovine CD8α is cyan, human CD8α is pink, mouse CD8α is orange, monkey CD8α is limon, and swine CD8α is light magenta. Helix A of ScCD8α, swine CD8α, and grass carp CD8α are labeled. Helix C’ of ScCD8α is labeled in the righthand panel. PDB accession numbers for each structure are as follows; human CD8αα 1CD8, chicken CD8αα 5EB9, swine CD8αα 5EDX, grass carp CD8αα 5Z11, monkey CD8αα 2Q3A, mouse CD8αα 2ARJ and bovine CD8αα 5EBG. (D-G) Two-dimensional topology diagrams of vertebrate CD8αs. The strands form a sandwich of 2 sheets in the catshark, grass carp, chicken, and human, which are colored blue, lime green, orange, and pink respectively, β-strands are labeled.
To understand how CD8α has maintained core structural features during evolution, structure-based sequence alignment of selected vertebrate CD8α molecules was performed (Figure 3). Unsurprisingly, ScCD8α shares relatively higher aa identities with CD8α homologs from cartilaginous fish CD8α (48.1-49.1%) than with those from bony fish and tetrapods, for instance, 24.3% with human CD8α, 28.6% with chicken CD8α and 28.6% with grass carp CD8α. The disulfide bond in the IgSF V domain, known to be vital for maintaining the overall structure of CD8α, is conserved in all the species examined (Figures 2, 3). Also conserved are Trp30ScCD8α, Leu77ScCD8α, Tyr91ScCD8α and Phe104ScCD8α, which constitute the backbone of CD8α for stabilizing conformation.
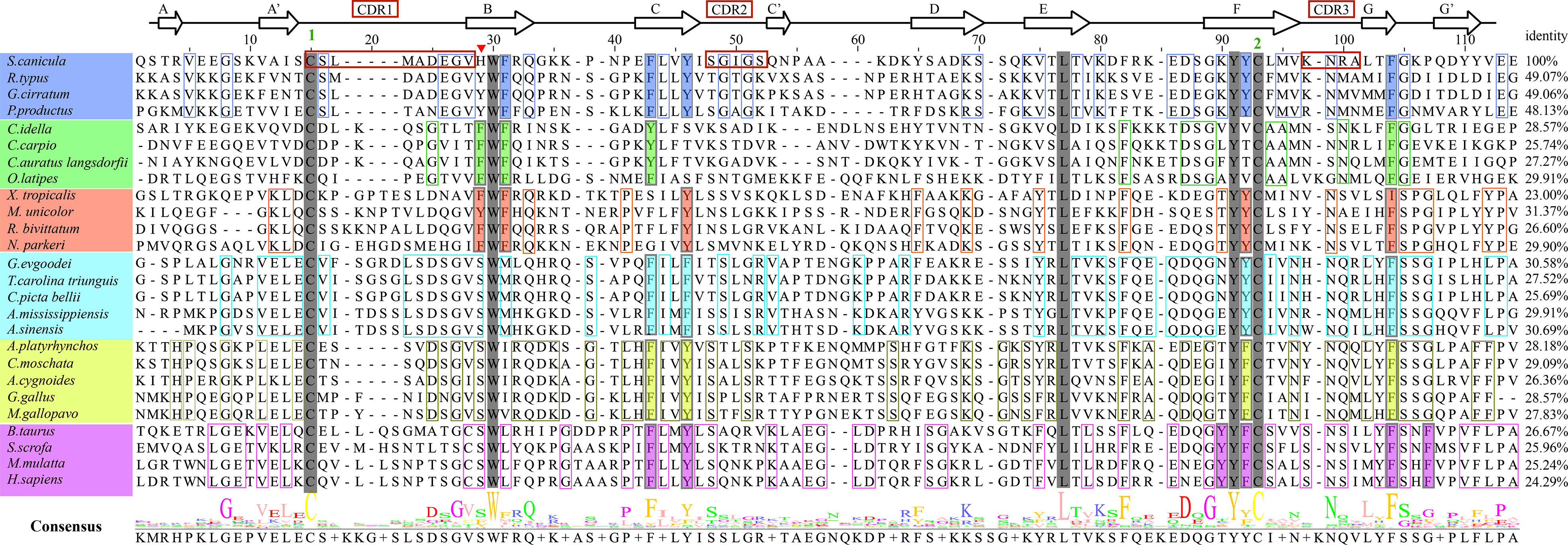
Figure 3 Amino acid sequence alignment of ScCD8α with CD8αs from different vertebrate species, with secondary structure elements indicated. Black arrows above the alignment indicate β-strands. Green numbers denote cysteines that form disulfide bonds, CDRs are outlined with red boxes. The residues conserved in the sampled species are shaded gray. The aa residues conserved in specific vertebrate lineages are marked with colored boxes. The hydrophobic core or predicted core are highlighted with colored rectangles. Amino acid identities between ScCD8α and the listed CD8α molecules are shown at the end of each sequence. The consensus sequence is shown at the bottom of the alignment. His28 in ScCD8α IgSF V domain is indicated by “▾”.
ScCD8α maintains dimerization through a distinctive hydrophobic core
Previous studies have shown that stabilization of the CD8αα homodimer relies upon a hydrophobic core which forms at the interface of the two CD8α monomers and is composed of several hydrophobic aromatic residues (18, 19). The ScCD8αα interface contains four inter-chain hydrogen bonds, which are formed by Gln32 (chain A) and Tyr83 (chain B), Asn51 (chain A) and Arg90 (chain B), Gln32 (chain B) and Tyr83 (chain A), Asn51 (chain B) and Arg90 (chain A), respectively (Figure 4A). Strands B, C, C’, F, G and G’ in the back sheet of ScCD8α are heavily involved in dimerization, with a buried surface area (BSA) of 853.3 Å2. In the ScCD8αα interface, 23 residues are involved in dimerization, 12 of which are hydrophobic, burying a surface area of 541.29 Å2 and making a major contribution (63.4%) to the van der Waals interactions (Figure 4B and Table 2). Five aromatic residues including Phe30 (strand B), Phe40 and Tyr43 (strand C), Tyr83 (strand F) and Phe94 (strand G) are located in the center of ScCD8αα dimer, forming a hydrophobic core with a BSA of 315.51 Å2 and contributing to 58.2% of the total hydrophobic force (Figures 4C, 5A). Interestingly, the composition and spatial distribution of these 5 aromatic residues are conserved in all cartilaginous fish CD8αs but not in CD8αs from bony fishes, birds, and mammals (Figures 5B–D).
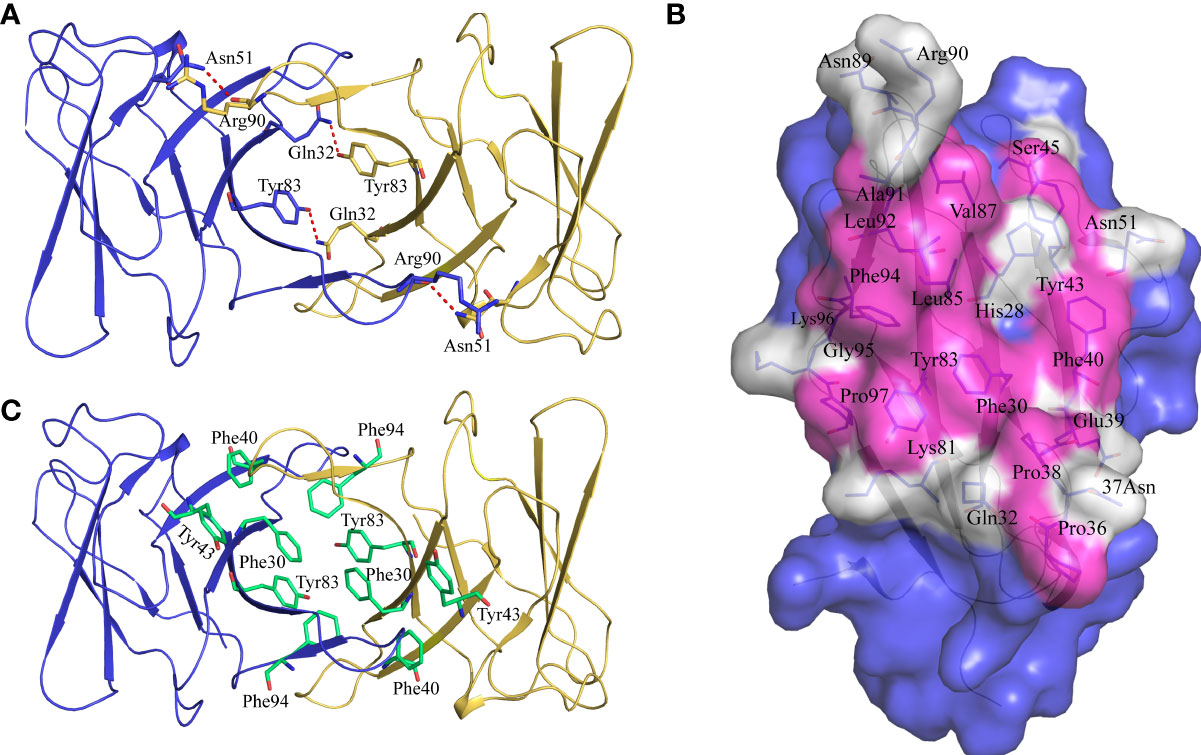
Figure 4 Characterization of the intermolecular interactions that maintain the dimerization of ScCD8αα. (A) The inter-chain hydrogen bonds of ScCD8αα. The ScCD8αα is shown in cartoon representation with the monomers colored blue and orange. Side chains that assist dimerization through the formation of hydrogen bonds are shown as stick models with their amino acid and position labeled. Hydrogen bonds are shown as red dashed lines. (B) The residues in the interface of the ScCD8αα. ScCD8α is surface rendered in blue. The residues forming the interface are shown as sticks and surface rendered in white while hydrophobic residues are surface rendered in magenta. Amino acid residues and their positions are labeled. (C) Side chains forming the hydrophobic core of ScCD8αα. The backbone of ScCD8αα is shown as a cartoon with the monomers colored blue and orange. The residues forming the hydrophobic core are shown as stick models and colored lime green. Amino acid residues and their positions are labeled.
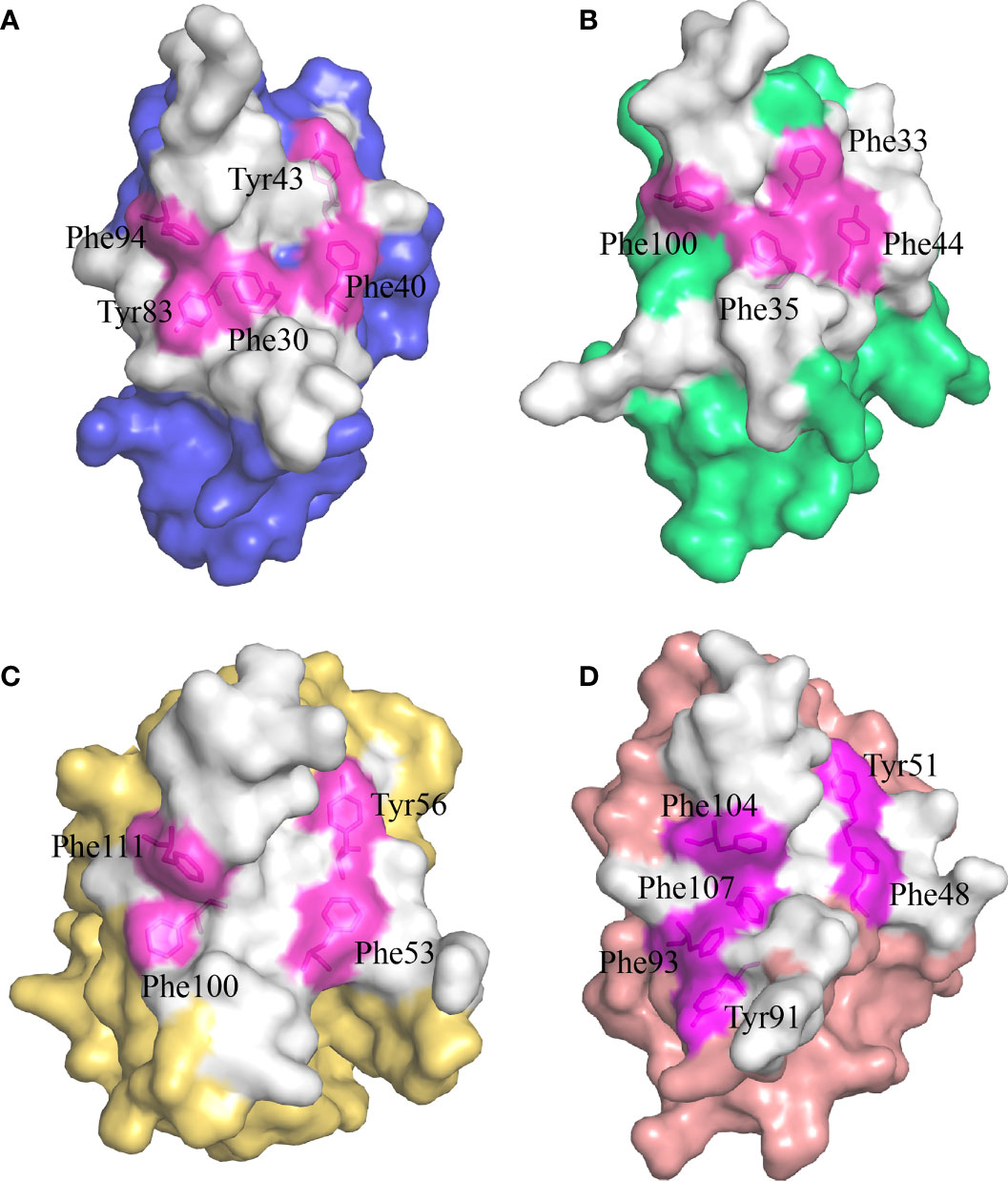
Figure 5 The hydrophobic core of the CD8αα homodimer in small-spotted catshark compared to other vertebrate species. The surface of each CD8α monomer is rendered in a different color while residues forming the hydrophobic core are shown in stick representation and surface rendered in magenta. (A) The hydrophobic core of ScCD8αα is formed by five hydrophobic residues (Phe30, Tyr43, Phe40, Tyr83 and Phe94). (B) The hydrophobic core of grass carp CD8αα is formed by four hydrophobic residues (Phe32, Phe35, Tyr44, and Phe100). (C) The hydrophobic core of chicken CD8αα is formed by four hydrophobic residues (Phe53, Tyr56, Phe100, and Phe111). (D) The hydrophobic core of human CD8αα is formed by six hydrophobic residues (Phe48, Tyr51, Tyr91, Phe93, Phe104 and Phe107).
ScCD8α can form a heterodimer with ScCD8β
To explore whether ScCD8α can also interact with ScCD8β, we co-transfected HEK293 cells with ScCD8α-Myc and ScCD8α-HA or ScCD8α-Myc and ScCD8β-HA then immunoprecipitated (IP) any complexes formed with an agarose bead conjugated α-Myc antibody. Our results show that both HA-tagged ScCD8α (Figure 6A) and HA-tagged ScCD8β (Figure 6B) can be immunoprecipitated with Myc-tagged ScCD8α using an α-Myc antibody. This indicates that both CD8αα homodimer and CD8αβ heterodimer can be formed in small-spotted catshark. To investigate this further, we modelled the 3D structure of ScCD8β extracellular region (Val22-Val132, without signal peptide) using human immunoglobulin lambda light chain (PDB code: 6ID4) as a search model, and showed that it also exhibits typical IgSF V architecture (Figure 6C).
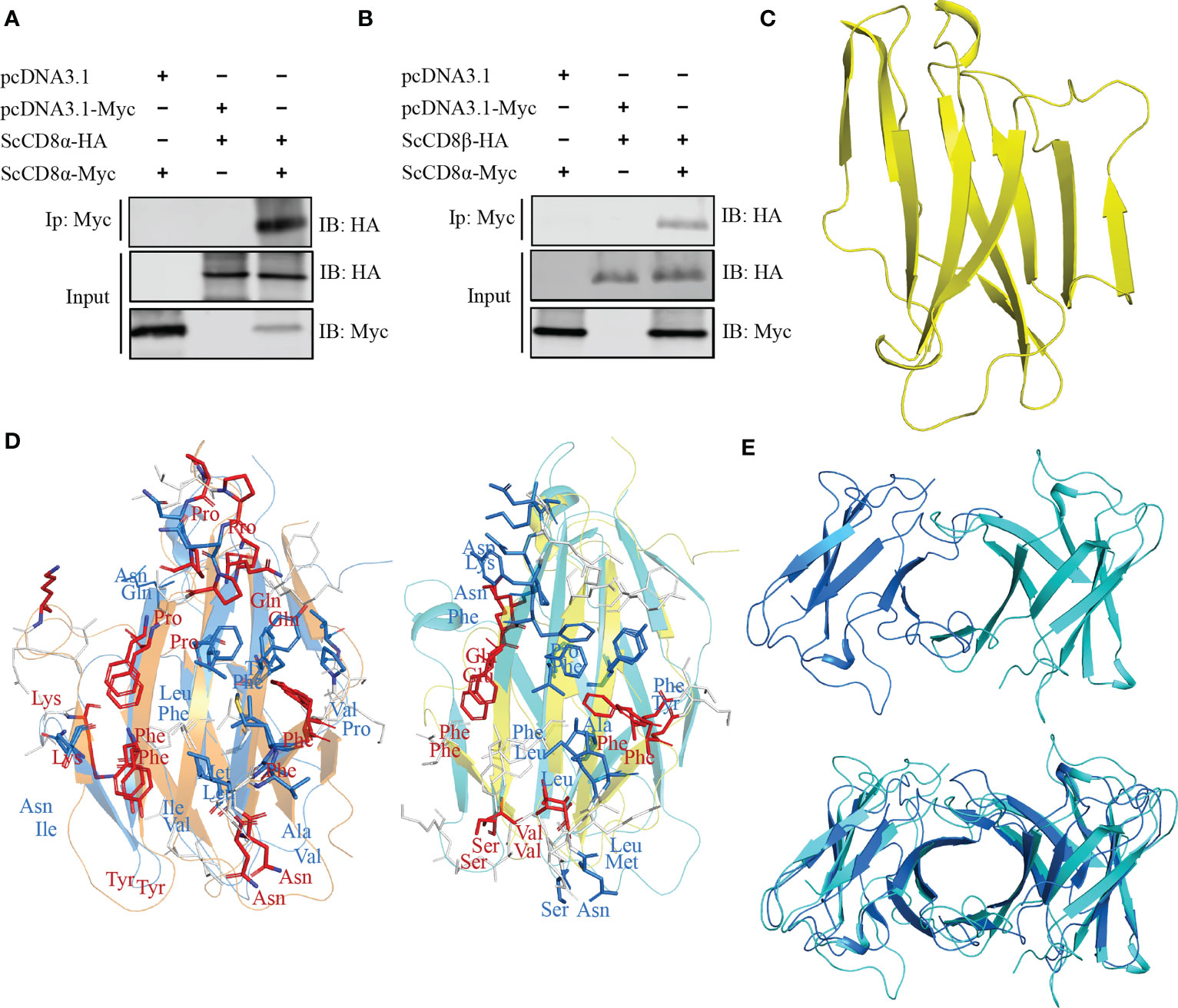
Figure 6 Interaction and modelling and of ScCD8α and ScCD8β. (A, B) Interaction of ScCD8α with ScCD8α (A) and ScCD8β (B). HEK293 cells were co-transfected with the indicated expression plasmids (5 μg each). After 24h, cell lysates were immunoprecipitated (IP) with α-Myc Ab conjugated agarose beads, and immunoblotted (IB) was detected with α-Myc or α-HA Abs. (C) Model of ScCD8β shown as a cartoon structure. (D) Predicted interface of CD8αβ with the ScCD8α monomer shown in marine, ScCD8β in cyan, mouse CD8α in orange, and CD8β in yellow. The side chains of key residues forming the CD8αβ interface are shown as sticks. Conserved residues are colored red, physiochemically-conserved residues are colored marine, other residues are shown in white. (E) The structural model of ScCD8αβ (top) and shown structurally aligned with ScCD8αα (bottom). ScCD8α is colored marine and ScCD8β cyan.
The structure of the mouse CD8αβ (mCD8αβ) heterodimer structure (PDB code: 2ATP) indicates that dimerization of CD8αβ is maintained by hydrophobic interactions as seen in CD8αα (10). We superposed the structures of ScCD8α and ScCD8β with the mouse CD8αβ structure (Figure 6D). We observed that 8 residues of CD8α within the interface are identical between the two structures and 8 residues are physico-chemically similar. Similarly, five identical residues and 8 physico-chemically conserved residues in the interface of CD8αβ structure can be identified in ScCD8β and mCD8β (Figure 6D). Furthermore, structural superposition revealed striking resemblance between ScCD8αα and ScCD8αβ (Figure 6E).
ScCD8αα is capable of interacting with ScMHC I
CD8α is a co-receptor of the TCR-MHC class I complex. Studies of human, mouse and chicken CD8α have shown that the cavity formed by the CDR loops of CD8α is the main area for interaction with p/MHC I. Similarly, ScCD8αα has a typical cavity, composed of 8 residues (Glu25, Gly26, His28, Tyr42, Leu85, Met86, Val87 and Lys88) symmetrically arranged in the CDR loops of the two monomers (Figure 7A), resulting in an accessible surface area (ASA) of 319.96Å2. This is a much smaller ASA than CD8ααs of other vertebrate species studied so far (e.g., chicken 641.36 Å2, swine 676.96 Å2, cow 585.2 Å2, mouse 727.85 Å2, monkey 526.42 Å2 and human 527.73 Å2) (Figure 7). Previous studies have shown that Ser34hCD8α is vital for maintaining the cavity while substitution of Ser34hCD8α by Phe33 results in disappearance of the cavity in grass carp CD8αα (12, 18). In cartilaginous fishes, the corresponding residue is occupied by histidine or tyrosine which, like phenylalanine, have large side chains. Structure alignment showed that His28ScCD8α is placed approximately 1.2Å away from the surface, which may explain the formation of the cavity (Figures 7E, F).
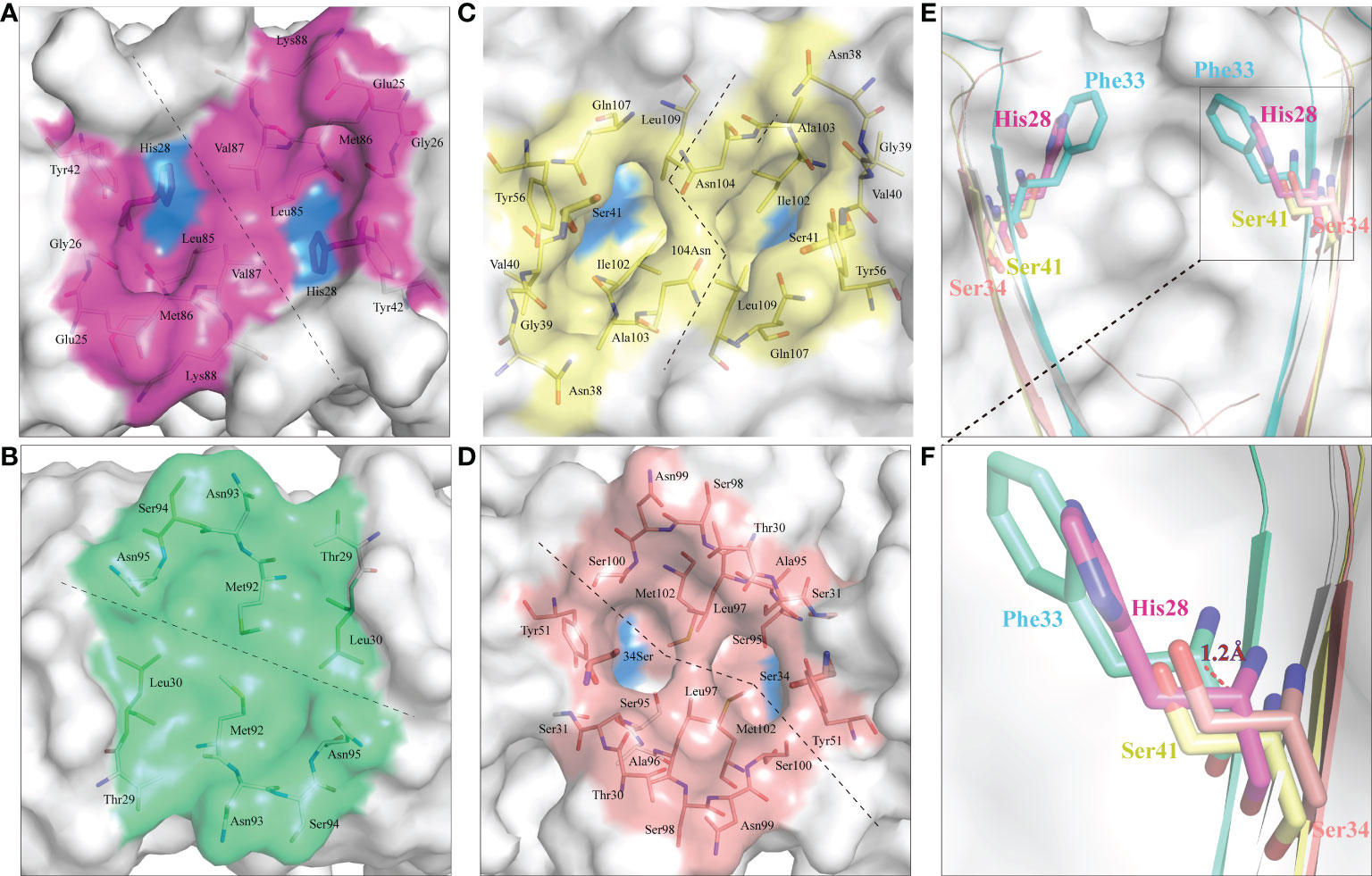
Figure 7 ScCD8αα possess a canonical binding cavity. The composition of the binding cavities in representative vertebrate CD8αα structures are shown. Molecular surfaces are rendered in white, with residues key to dimeric interaction highlighted blue. (A) The cavity of ScCD8αα, the side chains of cavity residues are shown as sticks and colored magenta. (B) The cavity of grass carp CD8αα, the side chains of cavity residues are shown as sticks and colored lime green. (C) The cavity of chicken CD8αα, the side chains of cavity residues are shown as sticks and colored yellow. (D) The cavity of human CD8αα, the side chains of cavity residues are shown as sticks and colored pink. (E, F) Comparison of key amino acids in the cavity of CD8αα structure in selected species, the residues and the distance between His28 and Phe33 are labeled.
Study of the human CD8αα-p/MHC I complex showed that three residues, Ser34hCD8α, Tyr51hCD8α and Asn99hCD8α form hydrogen bonds with the side chains of Gln226, Asp227 and Leu230 of CD loop and D strand of human MHC I α3 domain (16). Notably, the side chain of Gln226 from MHC I protrudes into the cavity of CD8α where it forms a bond with Ser34 in the C strand of CD8α (Figure 8A). This cavity and the key residues for interaction with MHC I are highly conserved in chickens and mammals (18, 31). While Tyr51hCD8α and Asn99hCD8α are conserved in cartilaginous fish CD8αs, a His residue is found in the position corresponding to Ser32hCD8α in ScCD8αα (Figure 3). Correspondingly, the Gln residue of MHC I that interacts with Ser32hCD8α in endothermic species is replaced by a negatively charged Glu in almost all ectotherms, potentially strengthening the interaction through formation of an ionic bond.
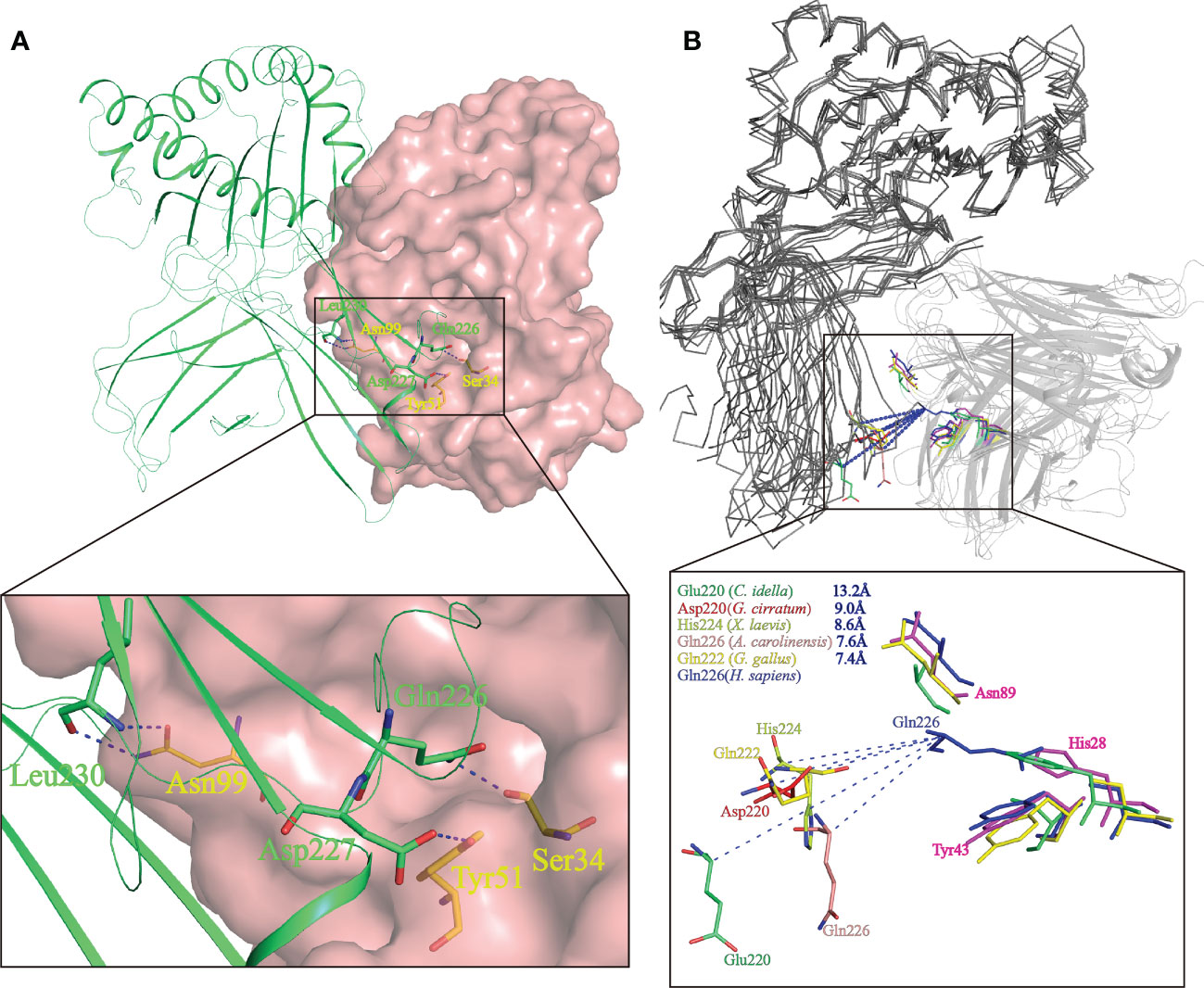
Figure 8 Prediction of the key intermolecular forces maintaining the interaction between ScCD8αα and p/MHC I molecules. (A) The interaction between human CD8αα and p/MHC I Human MHC is shown as a lime green cartoon structure while the surface of human CD8αα is rendered in pink. The key amino acid side chains of the interaction between CD8αα (yellow) and p/MHC I (green) are shown. Key hydrogen bonds at the interface of human CD8αα and p/MHC I (PDB ID: 1AKJ) are indicated by blue dashed lines. (B) The shift distance between the CD loops reveals a different binding mode of CD8 in non-mammals. The MHC I molecules are shown as gray ribbons and the CD8αα structure as cartoon with a transparency of 50%. The shift distance is indicated by blue dash lines. Key residues in the CD loop of MHC I and the corresponding residues interacting with the CD8αα molecules are shown as sticks. Residues of MHC I and CD8αα of grass carp (limegreen), nurse shark (red), African clawed frog (limon), chicken (yellow) and human (blue) are colored.
The variation and a shift in the spatial location of the key CD loop residues in the α3 domain of MHC I plays a critical role in maintaining the interaction with CD8. In human MHC I (PDB code: 1AKJ), the key amino acid in the CD loop is Gln226, whereas in MHC I of nurse shark (PDB code:6LUP), grass carp (PDB code:5Y91), African clawed frog (Xenopus laevis) (PDB code: 6A2B), Green Anole Lizard (PDB code:7CPO) and chicken (PDB code: 4E0R), it is Asp220, Glu219, His224, Gln226 and Gln222 respectively. Superposition of representative MHC I and CD8αα structures, as shown in Figure 8B, taking Gln226 of human MHC-I as a reference, show the distance from Gln226 to Asp220, Glu219, His224, Gln226 and Gln222 is ~9.0 Å, ~13.2 Å, ~8.6 Å, ~7.6 Å and ~7.4 Å respectively. Thus, distance between CD8αα and p/MHC I decreases from lower to higher vertebrates (Figure 8B). Grass carp CD8αα, of which the residue Ser32hCD8α is replaced by Phe33, still can interact with MHC I. Notably, residue Gln is responsible to interact with Ser in MHC I of most endotherm, while it is replaced by a negative charged Glu in almost all MHC I of ectotherm. Together, our data indicate that CD8αα is capable of interaction with MHC I and that the interaction between MHC I and CD8α differs between mammalian and non-mammalian species.
Discussion
The cartilaginous fishes, including sharks, are an ancient lineage which diverged from other vertebrates approx. 450 million years ago (2, 32). In addition to Igs, cartilaginous fishes possess a complete CD8/MHC I system. Recently, the structure of nurse shark MHC I was solved and showed many features, including the interactions between the MHC I heavy chain, β2-microglobulin (β2m) and peptide ligand (21), are similar to those of bony vertebrates. To investigate the interaction between CD8 and MHC I in cartilaginous fishes, CD8α and CD8β were cloned from small-spotted catshark and the structure of ScCD8αα solved. This is the first reported CD8αα structure from a cartilaginous fish.
Our gene synteny analysis shows that the CD8α and CD8β genes are tandemly linked from cartilaginous fishes to humans (31, 33), suggesting the duplication giving rise to these genes occurred in an ancestor of all jawed vertebrates. Despite low sequence identity (25.8%) between ScCD8α and ScCD8β, their IgSF V extracellular domains, which are vital for their interaction with MHC I (20, 34), share a similar domain architecture. However, the topology of ScCD8α displays significant differences with CD8αs previously characterized in endotherms. Most notably, the A strand of ScCD8α is located in the rear β sheet (Figure 2) but in the front sheet of chicken and human CD8α (16, 18), however, it should be noted that A strand exhibits considerable variations in all known structures of CD8α (17, 19). Further, an A’ strand is present only in the CD8α structures of catshark, grass carp and swine (17, 19). Therefore, we propose that the ancestral CD8α molecule likely adopted the topological conformation comprising A and A’ strands.
Our study also demonstrated that, as in mammals (16), cartilaginous fish CD8α can form a homodimer (Supplementary Figure 3). Our PAGE analysis under non-reducing conditions clearly showed a single band of approx. 32 kDa, matching the size of ScCD8α homodimer (32.1 kDa). Previous analyses have shown that a hydrophobic core, predominantly involving hydrophobic aromatic residues, serves as the key interaction force which maintains CD8α dimerization (19). A similar hydrophobic core can be found in the ScCD8αα structure (Figure 6). Moreover, the aromatic residues forming the hydrophobic core of ScCD8αα, including Phe30ScCD8α, Phe40ScCD8α, Tyr43ScCD8α, Tyr83ScCD8α and Phe94ScCD8α, are relatively well conserved in vertebrates (Figure 5). This suggests the mechanism of CD8αα-MHC I interaction is evolutionarily conserved from cartilaginous fishes through to mammals.
So far structures of the CD8αα-p/MHC I complex have been solved for humans, mice and chickens (16, 18, 31). In all these species the cavity formed by the CDR loops of CD8αα is the principal area of interaction with p/MHC I (16, 18, 31). In humans, Ser34, Tyr51 and Asn99 of CD8α are vital for connection with the side chains of Gln226, Asp227 and Leu230 of MHC I (16). Ma et al. (35) report that the distance between CD8α and pMHC I within the interface decreased from Xenopus to mammals (35), which is believed to contribute to the stronger interactions seen in mammals. The structure of nurse shark MHC I was recently solved (28). Structural modeling revealed that the CD loop in the α3 domain of nurse shark MHC I is protruded ~9.0 Å shorter in the cavity relative to that of human MHC I (Figure 8), suggesting that the distance between shark CD8α and pMHC I may be longer than that in humans and as a result, the interaction may be weaker.
The structure of ScCD8αα shows a canonical cavity is formed between the CDR loops of the two monomers. This suggests that the primordial CD8αα homodimer possesses the core structural unit to interact with p/MHC I. This cavity has been found in all known CD8αα structures except grass carp CD8αα, where the cavity is notably absent (16–19). Despite lack of the cavity, grass carp CD8αα can still bind to p/UAA-β2m (19). While this suggests that formation of the cavity is not essential for the interaction between CD8α and p/MHC I-β2m and that the interaction may be different in grass carp/bony fish relative to other vertebrates.
In summary, this study identified the CD8α and CD8β genes in small-spotted catshark and solved the structure of CD8αα. The ScCD8α and ScCD8β genes are found to be tandemly clustered in the genome and their encoded proteins share a characteristic IgSF V domain in their extracellular region. It was shown that ScCD8α can form a homodimer or a heterodimer with ScCD8β. Our structure revealed that ScCD8αα, like its mammalian counterparts, mainly relies upon a hydrophobic core to form a dimeric structure. Furthermore, ScCD8αα has a canonical cavity which mediates the interaction with p/MHC I in most species examined.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by University of Aberdeen’s Animal Welfare and Ethical Review Body (AWERB).
Author contributions
ZJ and JF: investigation, methodology, data curation, and writing original draft. HD: conceptualization, supervision and editing. JZ: conceptualization, funding acquisition, project administration, supervision and editing. JW: conceptualization, data curation, project administration, supervision and editing. All authors contributed to the article and approved the submitted version.
Funding
This work is funded by the National Natural Science Foundation of China (Grant numbers: 32030112 and U21A20268) and Hunan Provincial Science and Technology Program (Grant number: 2021NK2025), China.
Acknowledgments
We thank staff at the Shanghai Synchrotron Radiation Facility of China for technical support and Anthony K. Redmond from the University of Aberdeen for assistance with tissue sampling.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1156219/full#supplementary-material
References
1. Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: Genetic events and selective pressures. Nat Rev Genet (2010) 11(1):47–59. doi: 10.1038/nrg2703
2. Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature (2014) 505(7482):174–9. doi: 10.1038/nature12826
3. Flajnik MF, Kasahara M. Comparative genomics of the mhc glimpses into the evolution of the adaptive immune system. Immunity (2001) 15(3):351–62. doi: 10.1016/S1074-7613(01)00198-4
4. Flajnik MF, Du Pasquier L. Evolution of innate and adaptive immunity: Can we draw a line? Trends Immunol (2004) 25(12):640–4. doi: 10.1016/j.it.2004.10.001
5. Pancer Z, Cooper MD. The evolution of adaptive immunity. Annu Rev Immunol (2006) 24:497–518. doi: 10.1146/annurev.immunol.24.021605.090542
6. Holler PD, Kranz DM. Quantitative analysis of the contribution of Tcr/Pepmhc affinity and Cd8 to T cell activation. Immunity (2003) 18(2):255–64. doi: 10.1016/s1074-7613(03)00019-0
7. Wooldridge L, Clement M, Lissina A, Edwards ES, Ladell K, Ekeruche J, et al. Mhc class I molecules with superenhanced Cd8 binding properties bypass the requirement for cognate tcr recognition and nonspecifically activate ctls. J Immunol (2010) 184(7):3357–66. doi: 10.4049/jimmunol.0902398
8. Cole DK, Gao GF. Cd8: Adhesion molecule, Co-receptor and immuno-modulator. Cell Mol Immunol (2004) 1(2):81–8.
9. Gao GF, Jakobsen BK. Molecular interactions of coreceptor Cd8 and mhc class I: The molecular basis for functional coordination with the T-cell receptor. Immunol Today (2000) 21(12):630–6. doi: 10.1016/s0167-5699(00)01750-3
10. Chang HC, Tan K, Ouyang J, Parisini E, Liu JH, Le Y, et al. Structural and mutational analyses of a Cd8alphabeta heterodimer and comparison with the Cd8alphaalpha homodimer. Immunity (2005) 23(6):661–71. doi: 10.1016/j.immuni.2005.11.002
11. Nakanishi T, Shibasaki Y, Matsuura Y. T Cells in fish. Biol (Basel) (2015) 4(4):640–63. doi: 10.3390/biology4040640
12. Wang R, Natarajan K, Margulies DH. Structural basis of the Cd8 alpha Beta/Mhc class I interaction: Focused recognition orients Cd8 beta to a T cell proximal position. J Immunol (2009) 183(4):2554–64. doi: 10.4049/jimmunol.0901276
13. Liu Y, Xiong Y, Naidenko OV, Liu JH, Zhang R, Joachimiak A, et al. The crystal structure of a Tl/Cd8alphaalpha complex at 2.1 a resolution: Implications for modulation of T cell activation and memory. Immunity (2003) 18(2):205–15. doi: 10.1016/s1074-7613(03)00027-x
14. Olivares-Villagomez D, Van Kaer L. Tl and Cd8alphaalpha: Enigmatic partners in mucosal immunity. Immunol Lett (2010) 134(1):1–6. doi: 10.1016/j.imlet.2010.09.004
15. Janeway Jr. CA. How the immune system protects the host from infection. Microbes Infect (2001) 3(13):1167–71. doi: 10.1016/s1286-4579(01)01477-0
16. Gao GF, Tormo J, Gerth UC, Wyer JR, McMichael AJ, Stuart DI, et al. Crystal structure of the complex between human Cd8alpha(Alpha) and hla-A2. Nature (1997) 387(6633):630–4. doi: 10.1038/42523
17. Liu Y, Li X, Qi J, Zhang N, Xia C. The structural basis of chicken, swine and bovine Cd8alphaalpha dimers provides insight into the Co-evolution with mhc I in endotherm species. Sci Rep (2016) 6:24788. doi: 10.1038/srep24788
18. Liu Y, Chen R, Liang R, Sun B, Wu Y, Zhang L, et al. The combination of Cd8alphaalpha and peptide-Mhc-I in a face-to-Face mode promotes chicken gammadeltat cells response. Front Immunol (2020) 11:605085. doi: 10.3389/fimmu.2020.605085
19. Wang J, Zhang N, Wang Z, Yanan W, Zhang L, Xia C. Structural insights into the evolution feature of a bony fish Cd8alphaalpha homodimer. Mol Immunol (2018) 97:109–16. doi: 10.1016/j.molimm.2018.03.023
20. Chen Z, Zhang N, Qi J, Chen R, Dijkstra JM, Li X, et al. The structure of the mhc class I molecule of bony fishes provides insights into the conserved nature of the antigen-presenting system. J Immunol (2017) 199(10):3668–78. doi: 10.4049/jimmunol.1600229
21. Wu Y, Zhang N, Wei X, Lu S, Li S, Hashimoto K, et al. The structure of a peptide-loaded shark mhc class I molecule reveals features of the binding between Beta(2)-microglobulin and h chain conserved in evolution. J Immunol (2021) 207(1):308–21. doi: 10.4049/jimmunol.2001165
22. Criscitiello MF, Ohta Y, Saltis M, McKinney EC, Flajnik MF. Evolutionarily conserved tcr binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark. J Immunol (2010) 184(12):6950–60. doi: 10.4049/jimmunol.0902774
23. Li R, Wang T, Bird S, Zou J, Dooley H, Secombes CJ. B cell receptor accessory molecule Cd79alpha: Characterisation and expression analysis in a cartilaginous fish, the spiny dogfish (Squalus acanthias). Fish Shellfish Immunol (2013) 34(6):1404–15. doi: 10.1016/j.fsi.2013.02.015
24. Xue Y, Jiang X, Gao J, Li X, Xu J, Wang J, et al. Functional characterisation of interleukin 34 in grass carp Ctenopharyngodon idella. Fish Shellfish Immunol (2019) 92:91–100. doi: 10.1016/j.fsi.2019.05.059
25. Sun Z, Qin Y, Liu D, Wang B, Jia Z, Wang J, et al. The evolution and functional characterization of cxc chemokines and receptors in lamprey. Dev Comp Immunol (2021) 116:103905. doi: 10.1016/j.dci.2020.103905
26. Wang Z, Xu J, Feng J, Wu K, Chen K, Jia Z, et al. Structural and functional analyses of type I ifna shed light into its interaction with multiple receptors in fish. Front Immunol (2022) 13:862764. doi: 10.3389/fimmu.2022.862764
27. Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr (2004) 60(Pt 12 Pt 1):2126–32. doi: 10.1107/S0907444904019158
28. Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, et al. Phenix: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr (2002) 58(Pt 11):1948–54. doi: 10.1107/s0907444902016657
29. Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res (2003) 13(3):382–90. doi: 10.1101/gr.640303
30. Robinson-Rechavi VL M. Evolutionary rates of duplicate genes in fish and mammals. Mol Biol Evol (2001) 4):681–3. doi: 10.1093/oxfordjournals.molbev.a003849
31. Quiniou SM, Sahoo M, Edholm ES, Bengten E, Wilson M. Channel catfish Cd8alpha and Cd8beta Co-receptors: Characterization, expression and polymorphism. Fish Shellfish Immunol (2011) 30(3):894–901. doi: 10.1016/j.fsi.2011.01.011
32. Hara Y, Yamaguchi K, Onimaru K, Kadota M, Koyanagi M, Keeley SD, et al. Shark genomes provide insights into elasmobranch evolution and the origin of vertebrates. Nat Ecol Evol (2018) 2(11):1761–71. doi: 10.1038/s41559-018-0673-5
33. Guo Z, Wang GL, Fu JP, Nie P. Characterization and expression of Cd8 molecules in mandarin fish Siniperca chuatsi. J Fish Biol (2013) 82(1):189–205. doi: 10.1111/j.1095-8649.2012.03475.x
34. Wang Z, Chen R, Tariq M, Jiang B, Chen Z, Xia C. Complex assembly, crystallization and preliminary X-ray crystallographic analysis of the bovine Cd8alphaalpha-Bola-2*02201 complex. Acta Crystallogr F Struct Biol Commun (2014) 70(Pt 6):742–6. doi: 10.1107/S2053230X14008838
Keywords: cartilaginous fishes, shark, T cells, CD8, MHC, structure, evolution
Citation: Jia Z, Feng J, Dooley H, Zou J and Wang J (2023) The first crystal structure of CD8αα from a cartilaginous fish. Front. Immunol. 14:1156219. doi: 10.3389/fimmu.2023.1156219
Received: 01 February 2023; Accepted: 27 March 2023;
Published: 14 April 2023.
Edited by:
Maria Forlenza, Wageningen University and Research, NetherlandsReviewed by:
Fumio Takizawa, Fukui Prefectural University, JapanChun Xia, China Agricultural University, China
Copyright © 2023 Jia, Feng, Dooley, Zou and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junya Wang, jywang@shou.edu.cn
†These authors have contributed equally to this work
 Zhao Jia1,2,3†
Zhao Jia1,2,3† Jianhua Feng
Jianhua Feng Helen Dooley
Helen Dooley Jun Zou
Jun Zou Junya Wang
Junya Wang