- 1Department of Respiratory and Critical Care Medicine, Med-X Center for Manufacturing, Frontiers Science Center for Disease-Related Molecular Network, West China Hospital, West China School of Medicine, Sichuan University, Chengdu, China
- 2Artificial Intelligence (AI) Lab, Deepwise Healthcare, Beijing, China
- 3Department of Integrated Traditional Chinese and Western Medicine, West China Hospital, Sichuan University, Chengdu, China
Tuberculosis, caused by Mycobacterium tuberculosis, engenders an onerous burden on public hygiene. Congenital and adaptive immunity in the human body act as robust defenses against the pathogens. However, in coevolution with humans, this microbe has gained multiple lines of mechanisms to circumvent the immune response to sustain its intracellular persistence and long-term survival inside a host. Moreover, emerging evidence has revealed that this stealthy bacterium can alter the expression of demic noncoding RNAs (ncRNAs), leading to dysregulated biological processes subsequently, which may be the rationale behind the pathogenesis of tuberculosis. Meanwhile, the differential accumulation in clinical samples endows them with the capacity to be indicators in the time of tuberculosis suffering. In this article, we reviewed the nearest insights into the impact of ncRNAs during Mycobacterium tuberculosis infection as realized via immune response modulation and their potential as biomarkers for the diagnosis, drug resistance identification, treatment evaluation, and adverse drug reaction prediction of tuberculosis, aiming to inspire novel and precise therapy development to combat this pathogen in the future.
Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis (M. tuberculosis), poses a severe threat to public health, with approximately 10 million new cases and 1.5 million deaths reported in 2020 according to the World Health Organization (WHO). Much effort has been devoted to preventing the spread of the contagious disease. Nevertheless, the emergence of coronavirus disease 2019 (COVID-19) reversed the death toll of TB back to the level equivalent to that in 2017 (1). In addition, the proportion of drug-resistant tuberculosis (DR-TB) has steadily increased, resulting in an exacerbated challenge to TB supervision (2). Thus, precise indicators for the diagnosis and remedy direction of TB are urgently needed. The exploitation of novel biomarkers and host-directed therapy (HDT) may provide opportunities to surmount these conundrums.
M. tuberculosis, an intracellular pathogen, has evolved ingenious strategies to evade host immune defenses, armed with a tenacious power to proliferate in innate immune cells (3). Macrophages constitute the predominant niche of resident M. tuberculosis while functioning as the first line of self-defense, inducing innate immune cell responses such as cytokine secretion, autophagy, and apoptosis, and participating in acquired immunological reactions to eradicate this cunning microbial enemy (4).
Up to 90% of the human genome is transcribed into ribose nucleic acids (RNAs). However, only 1.5%–2.0% of them possess the ability to manufacture particular protein production (5). Recently, accumulating evidence has led to a reevaluation of the perception that the nonprotein-coding section of the genome is merely spurious transcriptional noise. In fact, some sequences in this proverbial “dark matter” can encode specific functional noncoding RNAs (ncRNAs) that play pivotal roles in biological regulation including immunology (6–9). Advances in RNA sequencing have led to the discovery of a multitude of ncRNAs including microRNAs (miRNAs), long noncoding RNAs (lncRNAs), circular RNAs (circRNAs), and PIWI-interacting RNAs (piRNAs) (10–12). Currently, the roles played by ncRNAs in eukaryotic cellular process mediation, ranging from gene regulation on a molecular scale to macroscopic manipulation of disease-inducing mechanisms, have been illuminated (13, 14). For instance, ncRNAs significantly influence bacterial and viral infections (15, 16). Specifically, in immunology, numerous studies have resolved the mystique of ncRNAs, which are implicated in multifarious events such as immune cell development and immune escape (17, 18). Furthermore, the contribution of ncRNAs to the underlying pathogenesis of M. tuberculosis in hosts has also been enumerated (19). The significant discovery of the host-pathogen interactions has ushered in an inspiring time for HDT exploration (20). Moreover, previous effort has revealed the feasibility of using these seemingly inconspicuous molecules as biomarkers of TB (21, 22).
In this review, we elaborated on the promising roles of ncRNAs, focusing on miRNAs, lncRNAs, and circRNAs, in TB. We described the regulatory impact of the three ncRNAs on immune function in disparate parts. In each section, we offered a brief introduction to the foundational structure and function of the highlighted ncRNAs, followed by a concrete explanation of their effects on M. tuberculosis infection. Then, we narrated their potential as biomarkers for TB diagnosis, DR-TB identification, and treatment monitoring. Finally, we summarized the included studies with achievements and existing challenges for the purpose of utilizing ncRNAs for TB management, including rapid diagnosis and precise treatment. The major content of the study is illustrated succinctly in Figure 1.
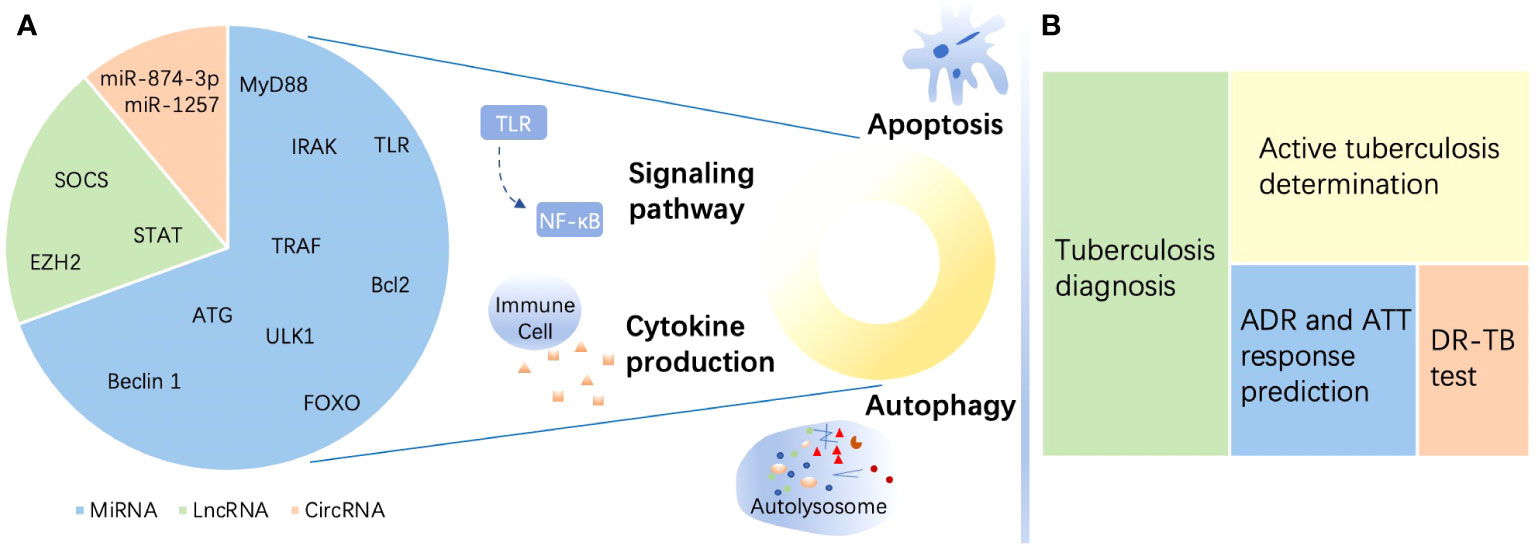
Figure 1 A brief summary of the roles of noncoding RNAs in tuberculosis. (A) Targets of noncoding RNAs (ncRNAs) in immune regulation in tuberculosis (TB) and estimated proportion of studies directed to the three ncRNAs enumerated in this review. MicroRNAs (miRNAs) mostly target myeloid differentiation factor 88 (MyD88), Toll-like receptor (TLR), tumor necrosis factor (TNF) receptor-associated factor (TRAF), and interleukin-1R-associated kinase (IRAK) to regulate signaling pathways and target Unc-51-like autophagy-activating kinase 1 (ULK1), autophagy-related gene (ATG), and beclin 1 to affect autophagy. Moreover, miRNAs target the Bcl-2 family and Forkhead box transcription factor class O (FOXO) to modulate apoptosis during TB. Long noncoding RNAs (lncRNAs) regulate the immune response in the host by targeting enhancer of zeste homolog (EZH) 2, suppressor of cytokine signaling (SOCS), signal transducer and activator of transcription (STAT), etc. Circular RNAs (circRNAs) perform immunity functions by acting as miRNA sponges. (B) ncRNAs serve as biomarkers in TB. ncRNAs show the potential to be biomarkers of different functions including diagnosis, active individual determination, adverse drug reaction (ADR) and response to antituberculosis therapy (ATT) monitoring, and drug resistance prediction of TB. DR-TB, drug-resistant tuberculosis.
Immune regulation of miRNAs in tuberculosis
A concise description of miRNAs
miRNAs, approximately 22 nucleotides in length, are endogenous single-stranded small ncRNAs with highly conserved structure, emerging as posttranscriptional epigenetic modulators after binding to the 3′ untranslated regions (3′ UTRs) of targeting messenger RNAs (mRNAs), resulting in mRNA decay or destabilization (23–25). Numerous studies have authenticated the polytropic roles of miRNAs in multiple biological processes, from cell differentiation to disease occurrence (26, 27). In addition, miRNAs exert a crucial influence on immune response regulation (28, 29). When certain morbific microorganisms stealthily enter the human body, miRNAs will respond to modulate the immune response in various ways (30). Focused on M. tuberculosis, previous studies have revealed that this crafty pathogen can alter the expression of host miRNAs, enabling this microorganism to evade immune clearance and achieve long-term dormancy inside the body (31). In innate immune reactions, macrophages serve as the first-line defense in the face of the complex enemy microorganism invasion (32), and the aftermath of the war between M. tuberculosis and macrophages determines the infection outcome-latent or active TB. Moreover, M. tuberculosis has developed various strategies to subvert sthe host immune response, such as autophagy (33). Hence, we enumerated the miRNA modulation caused by M. tuberculosis and the subsequent influence of this regulatory effect on antimicrobial immunity.
miRNA-regulated signaling pathways in M. tuberculosis infection
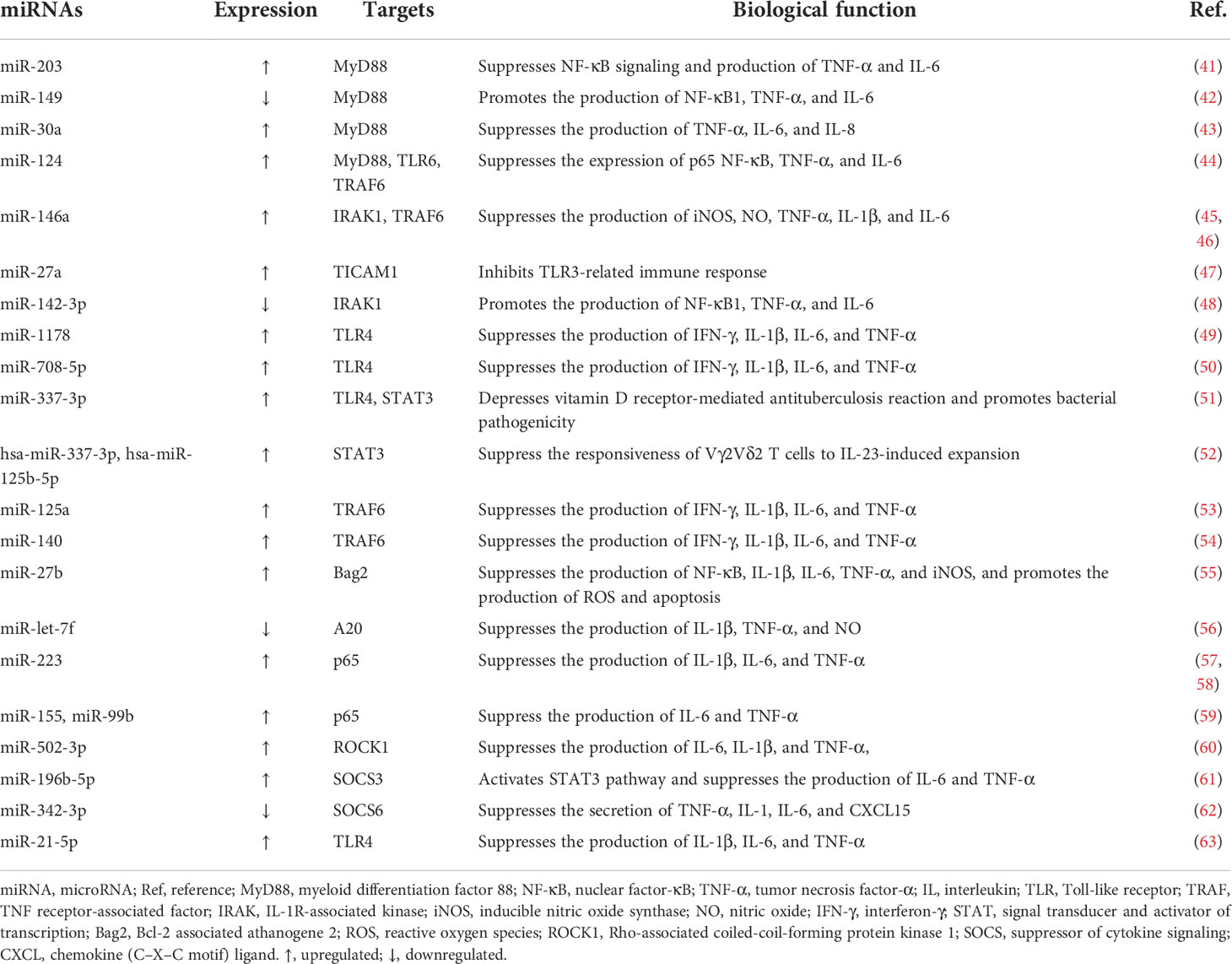
Toll-like receptors (TLRs), a family of pattern recognition receptors (PRRs) that reside on the plasma membrane of macrophages and other tissues involved in immunity, sense pathogen-associated molecular patterns (PAMPs) such as lipopolysaccharide (LPS) in Gram-negative bacteria and single-stranded RNAs. Subsequently, adaptor proteins including myeloid differentiation factor 88 (MyD88), Toll-interleukin (IL)-1-resistance (TIR) domain-containing adaptor-inducing interferon-β (TRIF), TRIF‐related adaptor molecule (TRAM), and TIR‐containing adaptor protein (TIRAP) (34–36), are engaged by TIRs, cytoplasmic domains of TLRs, after which IL-1R-associated kinase (IRAK), tumor necrosis factor (TNF) receptor-associated factor (TRAF), and IκB kinase (IKK) complex are recruited by appropriate signals in turn. The activation of IKK leads to the phosphorylation and degradation of IκB in the canonical nuclear factor-κB (NF-κB) pathway, releasing NF-κB, a two-subunit nuclear transcription factor that dimerizes and is further activated through different posttranscriptional modifications. NF-κB is then translocated into the nucleus to regulate corresponding gene expression by binding to specific DNA sequences, ultimately eliciting inflammatory cytokine secretion to eradicate invading pathogens (37–40). The role of miRNAs in modulating signaling pathway cascades, including the TLR/NF-κB pathway during M. tuberculosis infection, which results in the suppression or enhancement of the immune response, has been explored as follows (Figure 2; Table 1).
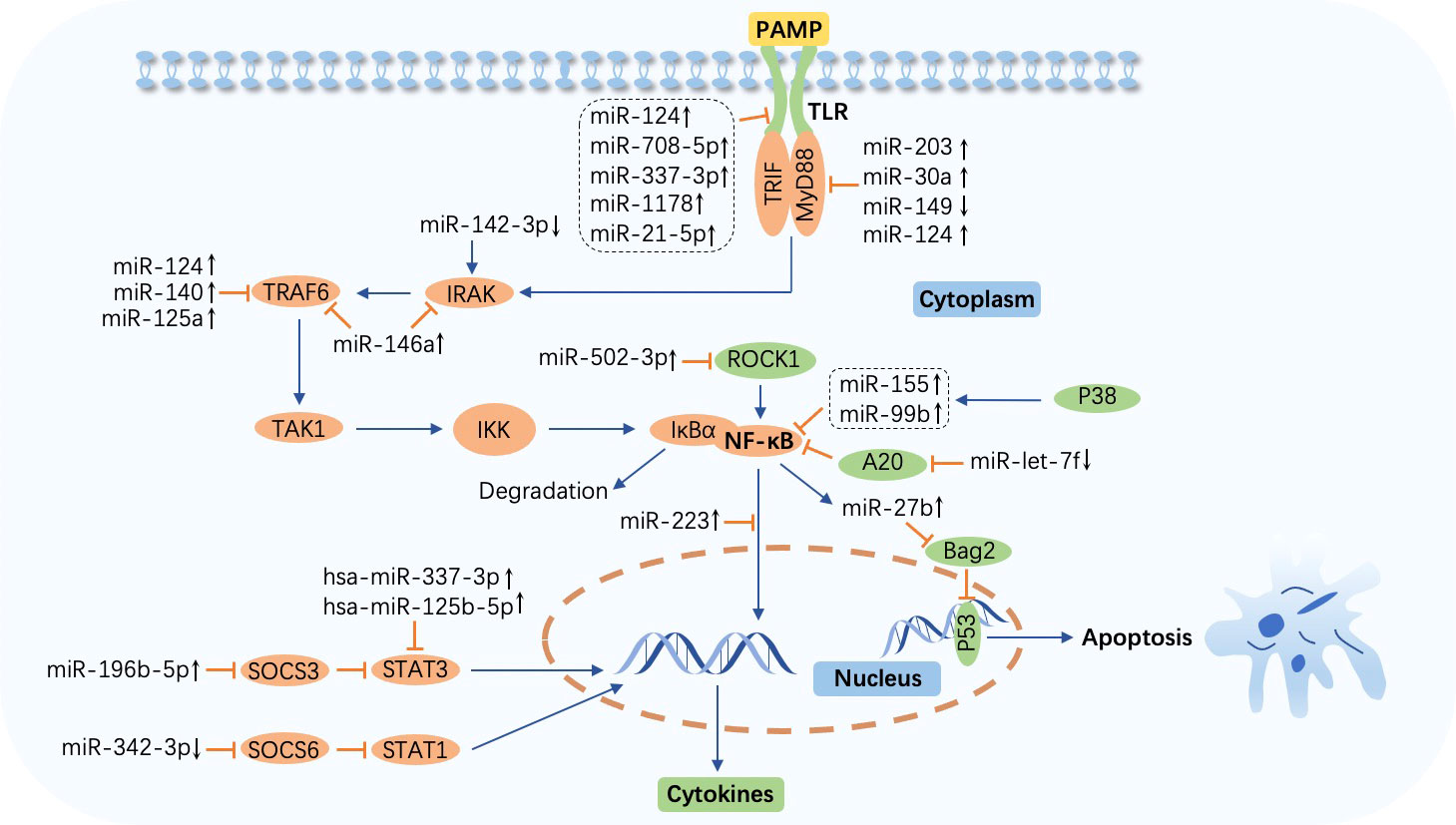
Figure 2 MicroRNA-regulated signaling pathway during Mycobacterium tuberculosis infection. The Toll-like receptor (TLR)/nuclear factor-κB (NF-κB) pathway is activated by the combination of TLRs and pathogen-associated molecular patterns (PAMPs) and impacts cytokine production. Abundant molecules such as TAK1 and IκB kinase (IKK) participate in this process. IκBα is degraded after the release of NF-κB from IKK. Overexpressed or downregulated microRNAs (miRNAs) target myeloid differentiation factor 88 (MyD88), Toll-interleukin (IL)-1-resistance (TIR) domain-containing adaptor inducing interferon-β (TRIF), IL-1R-associated kinase (IRAK), tumor necrosis factor (TNF) receptor-associated factor (TRAF), NF-κB, suppressor of cytokine signaling (SOCS), signal transducer and activator of transcription (STAT), and p53 to affect host immune cell response in tuberculosis. miR-27b is induced by NF-κB and triggers p53-dependent apoptosis by targeting Bcl-2 associated athanogene 2 (Bag2). ↑, upregulated; ↓, downregulated; ⟶, stimulate; ⟞, inhibit.
In Bacille Calmette–Guérin (BCG)-infected macrophages, miR-203 inhibits NF-κB signaling pathway activation, decreasing the expression of IL-6 and TNF-α by targeting MyD88 (41). Similarly, miR-149 and miR-30a also target MyD88 and suppress the downstream secretion of cytokines including TNF-α, IL-6, and IL-8, inhibiting pathogen clearance (42, 43). miR-124 is highly expressed via MyD88 activation in M. tuberculosis-infected alveolar macrophages (AMs), and in response, this miRNA targets MyD88, TRAF6, and TLR6 to alleviate inflammation triggered by the microorganism invasion (44). Another study demonstrated that miR-146a suppresses the inflammatory response by negatively modulating TRAF6 and IRAK1 expression in NF-κB signaling pathway cascades, repressing the generation of proinflammatory factors such as inducible nitric oxide synthase (iNOS), nitric oxide (NO), IL-6, TNF-α, and IL-1β to facilitate replication and survival of M. tuberculosis in macrophages (45, 46). Targeting TIR domain-containing adaptor molecule 1 (TICAM1), a TLR3 adaptor, the engagement of miR-27a stifles TLR3-related innate immune response to promote M. tuberculosis survival. Bone morphogenesis protein (BMP), regulated by c-Abl, is the primary molecule that induces miR-27a overexpression(47). Furthermore, a study discovered that miR-142-3p downregulates IRAK1 and suppresses the action of inflammatory mediators including TNF-α, IL-6, and NF-κB1 (a subunit of NF-κB) (48). The time- and concentrate-dependent upregulation of miR-1178 and miR-708-5p attenuates the accumulation of proinflammatory factors such as IFN-γ, TNF-α, IL-1β, and IL-6 by targeting TLR4 and subsequently enhances microorganism vitality in M. tuberculosis-infected macrophages (49, 50). Similarly, overexpression of miR-337-3p is implicated in enhanced M. tuberculosis pathogenicity and impaired vitamin D receptor (VDR)-mediated antibacterial response by depressing the TLR4 and the signal transducer and activator of transcription 3 (STAT3, a transcription activator that plays a key role in multiple biological processes such as apoptosis and immune regulation by modulating the expression of various genes) signaling pathway (51, 64, 65). Meanwhile, another study pointed out a marked increase in hsa-miR-337-3p and hsa-miR-125b-5p in patients with TB is causative of impaired STAT3 function, which results in the abrogation of IL-23-mediated expansion of Vγ2Vδ2 T cells and weakens the ability of these cells to produce anti-TB factors (52).
Overexpression of miR-125a and miR-140 inhibits NF-κB signaling and secretion of proinflammatory cytokines such as IFN-γ and IL-1β during M. tuberculosis infection by targeting TRAF6 directly, leading to immune response suppression and increased microbe viability. In addition, the miR-125a level is increased in a TLR4-dependent manner (53, 54). Moreover, miR-27b is induced via TLR2/MyD88/NF-κB cascades in M. tuberculosis-infected murine lungs and spleens, subsequently targeting Bcl-2-associated athanogene 2 (Bag2) and elevating p53-dependent apoptosis and reactive oxygen species (ROS) secretion rates to eliminate intracellular pathogens while forming a negative feed loop that prevents excessive inflammation induced by NF-κB (55). A20, a feedback inhibitor of the NF-κB signaling pathway, is upregulated via the early secreted antigenic target of 6 kDa (ESAT-6)-dependent suppression of miR-let-7f, which leads to diminished NF-κB signaling and cytokine production, resulting in pathogen maintenance in the macrophages of M. tuberculosis-infected mice (56). miR-223 is abundantly expressed in monocytes and monocyte-derived macrophages (MDMs) from TB patients and attenuates nuclear translocation and p65 (a subunit of NF-κB) (66) phosphorylation, resulting in suppression of NF-κB activation and inhibition of cytokine secretion, hindering M. tuberculosis eradication (57). Moreover, miR-223 regulates the engagement of myeloid cells by targeting chemokine (C-X-C motif) ligand (CXCL) 2, chemokine (C-C motif) ligand (CCL) 3, and IL-6, and TB infection has been shown to be exacerbated in miR-233−/− animals (58). p38 is promoted by M. tuberculosis Rv2346c, a member of ESAT6, which induces the overexpression of miR-155 and miR-99b, leading to the inhibition of both the activation of NF-κB and secretion of cytokines including IL-6 and TNF-α, ultimately enhancing bacillary persistence and restraining the proliferation of BCG-infected macrophages (59). Targeting Rho-associated coiled-coil-forming protein kinase 1 (ROCK1), induction of miR-502-3p by M. tuberculosis decreases the production of TNF-α, IL-6, and IL-1β via the inhibition of the TLR4/NF-κB pathway, promoting pathogen survival (60).
In addition, the STAT pathway participates in inflammatory mediator production in M. tuberculosis infection as well. The expression of miR-196-5p is elevated in monocytes from smoking patients with TB. By repressing the suppressor of cytokine signaling (SOCS) 3 and activating the STAT3 pathway, miR-196-5p inhibits proinflammatory cytokine production and bacterial uptake (61). However, miR-342-3p facilitates the production of cytokines and chemokines such as TNF-α, IL-1, IL-6, and CXCL15 via SOCS6 suppression and subsequent STAT1 activation and switches the death modality from necrosis to apoptosis in M. tuberculosis-infected macrophages, enhancing the anti-TB immune response. Moreover, mice with higher expression of miR-342-3p are more resistant to M. tuberculosis (62).
The TLR/NF-κB signaling pathway is affected most frequently by miRNAs during M. tuberculosis infection, which results in inflammatory cytokine regulation, ultimately resulting in the prevention or facilitation of microorganism eradication. Nevertheless, studies related to the regulatory function of other signaling pathways in bacterial infection have rarely been performed, and more research is yet to be carried out. Focusing on the likely fundamental mechanisms, scientists are called to ceaselessly pursue more novel and precise miRNA-based therapies for TB in clinical practice.
miRNA-regulated cytokine production in M. tuberculosis infection
Different from those focused on signaling pathways, several studies have revealed a more direct modulatory impact of miRNAs on the expression of inflammatory cytokines, as presented in brief in Supplementary Table S1. For instance, miR-29 is observed to inhibit interferon (IFN)-γ production to negatively control immune reactions, while in mice, competitive sponging of miR-29 leads to a higher concentration of IFN-γ and renders the mice more resistant to M. tuberculosis infection (67). Meanwhile, another study pointed out that in patients with active tuberculosis (ATB), upregulated miR-29 may attenuate the response of CD4+ T cells to M. tuberculosis (68). However, the relationship between the differential expression of miR-29a, a member of the miR-29 family, and IFN-γ has not yet constituted a plausible line of inquiry (69). Likewise, M. tuberculosis restrains the immune response by inducing miR-132 and miR-26a, which target p300, a transcriptional coactivator of IFN-γ signaling, and lead to lower IFN-γ expression and reduce responsiveness of macrophages to this lymphokine, permitting the pathogen persistence (70). miR-144* enables the reduction in TNF-α and IFN-γ secretion and T-cell proliferation, which hinders the clearance of M. tuberculosis (71). Lower production of TNF occurs along with overexpression of miR-125b and downregulation of miR-155 in macrophages incubated with M. tuberculosis lipomannan, promoting bacterial survival inside the host (72). Another study revealed that ESAT-6-induced miR-155 inhibits the expression of SH2-containing inositol 5’-phosphatase (SHIP1) and BTB and CNC homology 1 (Bach1), a repressor of haem oxygenase-1 (HO-1), and reduces IL-6 and cyclooxygenase-2 (Cox-2) production, finally facilitating M. tuberculosis existence in macrophages (73). Downregulating CCAAT/enhancer binding protein β (C/EBPβ), a positive transcriptional modulator of nitric oxide synthase (NOS) 2, miR-155 further decreases the synthesis of NO, thus preventing the microbe killing. When transfected with anti-miR-155, IFN-γ-activated macrophages exhibit a higher level of NO and a reduced M. tuberculosis burden (74).
Follistatin-like protein 1 (FSTL1) gene expression is triggered by TLR4 signaling and certain proinflammatory cytokines. In return, FSTL1 activates macrophages to promote proinflammatory cytokine and chemokine expression (75–78). In the duration of M. tuberculosis infection, greatly enhanced miR-32-5p targets FSTL1 and significantly attenuates the secretion of certain cytokines such as IL-1β and IL-6, which promotes inflammatory reactions, finally increasing the intracellular survival rate of M. tuberculosis (79). Rab10, a member of the Ras oncogene family, plays a vital role in macrophage activation. Suppressed miR-378d, which is associated with the activation of NF-κB signaling, increases the production of cytokines, including IL-1β, IL-6, and TNF-α, mediated via Rab10 in M. tuberculosis-infected macrophages and facilitates the clearance of the microbe (80). In contrast, activation of the NF-κB pathway after BCG infection induces the upregulation of miR-21, which decreases the expression of IL-12 by targeting IL-12p35 and promotes dendritic cell (DC) apoptosis by targeting Bcl2, impairing the anti-mycobacterial reactions (81). Moreover, miR-206 decreases the expression of tissue inhibitor of matrix metalloproteinase 3 (TIMP3) to elevate inflammatory cytokine secretion in THP-1 macrophages infected by M. tuberculosis, facilitating immune reactions against the pathogen (82). EAST6-inhibited miR-222-3p suppresses the production of proinflammatory cytokines such as IL-6, IL-1β, and TNF-α by promoting the expression of phosphatase and tensin (PTEN), which ultimately benefits microbial replication (83). In addition, BCG/H37Rv-downregulated miR-495 enables intracellular bacterial survival as a result of a superoxide dismutase 2 (SOD2)-promoted decrease in reactive oxygen species (ROS) (84).
Through various molecule targeting, miRNAs modulate the secretion of cytokines and chemokines to regulate the anti-mycobacterial response in a more straightforward manner, which gives us a hint to explore relevant therapeutic plans based on these theories.
miRNA-regulated autophagy in M. tuberculosis infection
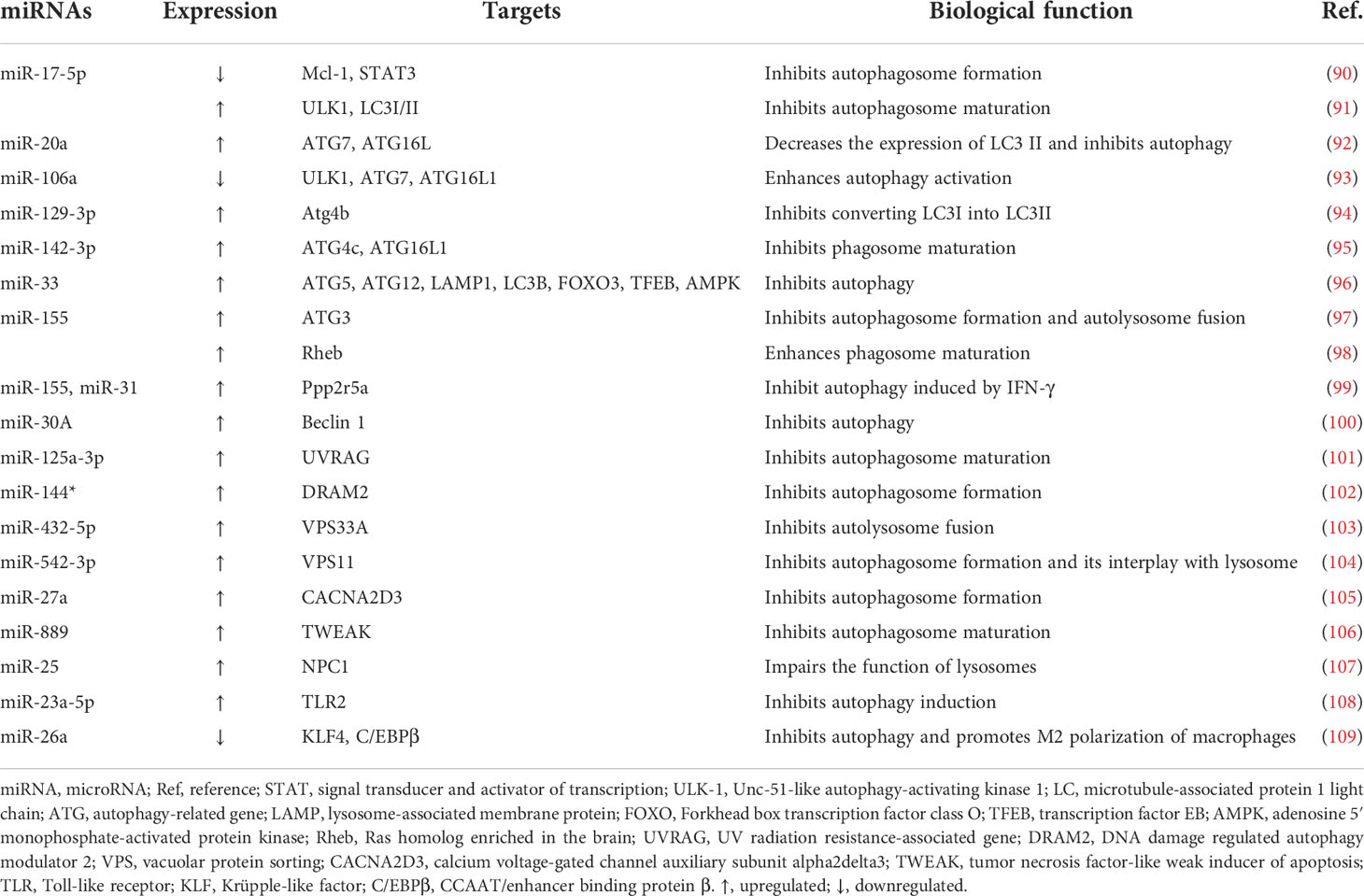
Autophagy, an evolutionarily conserved catabolic pathway, degrades macromolecules and specific organelles through the fusion between autophagosomes and lysosomes following the entrapment of superfluous intracellular substances (85). When the bacteria invade, autophagy is mobilized to resist the pathogenic infection, playing a critical role in the innate immune response. Autophagy disruption can lead to multiple diseases (86). Moreover, research has suggested that intracellular M. tuberculosis elimination may be associated with autophagy (87) and that miRNAs exert a crucial impact on this cellular cargo obliteration tactic (88). Recently, mounting evidence has discovered that the contagious agent M. tuberculosis can combat host autophagy by interacting with autophagic components such as beclin 1, autophagy-related genes (ATGs), and microtubule‐associated protein 1 light chain 3 (LC3), which contribute significantly to essential autophagic processes including membrane nucleation and autophagosome formation and subsequent fusion with lysosomes (89). The primary miRNAs involved in the biological process and their expression and impacts are displayed in Figure 3 and Table 2.
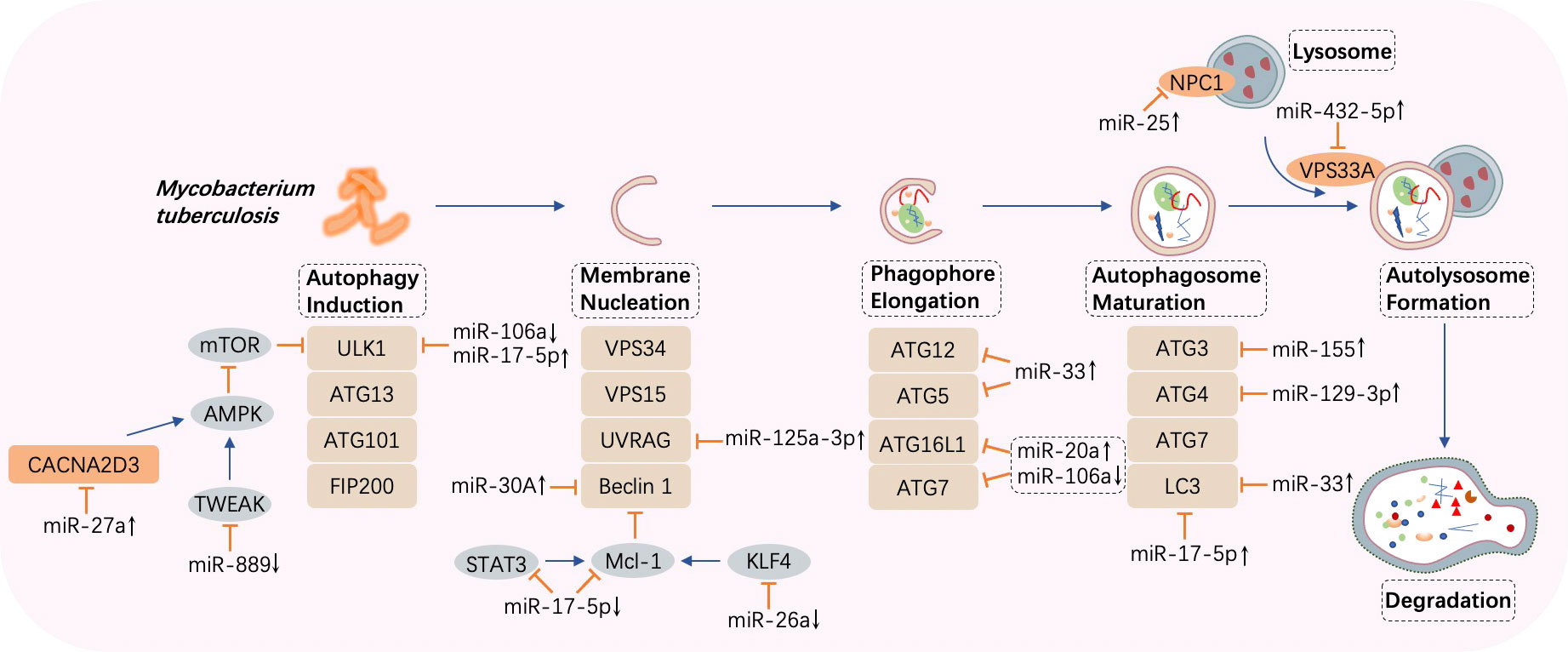
Figure 3 MicroRNA-regulated autophagy during Mycobacterium tuberculosis infection. Once autophagy is induced by Mycobacterium tuberculosis, several processes such as membrane nucleation to form the phagophore, elongation to realize autophagosome maturation, and the fusion between autophagosomes and lysosomes take place in turn. MicroRNAs (miRNAs) are differentially expressed and target crucial components in autophagy such as autophagy-related genes (ATGs), UV radiation resistance-associated gene (UVRAG), beclin 1, and microtubule‐associated protein 1 light chain 3 (LC3). CACNA2D3, calcium voltage-gated channel auxiliary subunit alpha2delta3; AMPK, adenosine 5’ monophosphate-activated protein kinase; TWEAK, tumor necrosis factor-like weak inducer of apoptosis; ULK1, Unc-51 like autophagy activating kinase 1; VPS, vacuolar protein sorting; STAT, signal transducer and activator of transcription; KLF, Krüpple-like factor. ↑, upregulated; ↓, downregulated; ⟶, stimulate; ⟞, inhibit.
The targets of miR-17-5p in M. tuberculosis-infected macrophages are verified to be Mcl-1 and STAT3 (a transcriptional activator of Mcl-1). Forced expression of miR-17-5p suppresses the interaction between Mcl-1 and beclin 1, resulting in the promotion of autophagy and acceleration of microbe killing. Furthermore, phosphorylation of protein kinase C (PKC) δ accelerates autophagy, and this effect is diminished by miR-17-5p (90). In BCG-infected RAW264.7 cells, enhanced expression of miR-17-5p prevents the maturation of phagosomes through the downregulation of Unc-51-like autophagy-activating kinase 1 (ULK1), an initiator of autophagy, and autophagosome-related protein LC3I/II, thereby increasing BCG propagation (91). Moreover, by targeting ATG7 and ATG16L, upregulated miR-20a attenuates autophagy and favors pathogen growth in BCG-infected macrophages (92). On the contrary, in H37Ra-infected THP-1 macrophages, downregulation of miR-106a suppresses bacterial proliferation as a result of promoted autophagy via increased levels of ULK1, ATG7, and ATG16L, which are important factors in autophagy (93). A study showed that overexpression of miR-129-3p induced by BCG in RAW264.7 cells is capable of attenuating M. tuberculosis killing via autophagy inhibition by targeting Atg4b, an ATG that contributes to the autophagosome formation step in which LC3I is converted into LC3II (94, 110). Furthermore, upregulation of miR-142-3p significantly abates H37Ra-induced autophagy by negatively controlling the expression of ATG16L and ATG4c, leading to the promotion of intracellular survival of the pathogen (95). M. tuberculosis replication is supported by overexpression of miR-33 and its passenger strand miR-33*, which target critical effectors that participate in autophagy such as ATG5, ATG12, LC3B, and lysosome-associated membrane protein 1 (LAMP1). Moreover, transcriptional regulators, including Forkhead box transcription factor class O (FOXO3) (111) and transcription factor EB (TFEB) (112), which promote the expression of genes implicated in autophagy biogenesis, are simultaneously repressed, resulting in impaired lipid catabolism (96).
Targeting ATG3, an E2-ubiquitin-like-conjugating enzyme participating in LC3-lipidation and autophagosome formation, virulent M. tuberculosis-induced miR-155 contributes to autophagy subversion to maintain bacterial survival (97). Meanwhile, miR-155 and miR-31 limit IFN-γ-induced autophagy by posttranscriptionally downregulating Ppp2r5a (99). In contrast, combined with the 3′ UTR of Ras homolog enriched in the brain (Rheb), overexpression of miR-155 accelerates the maturation of phagosomes and enhances autophagy in macrophages to eliminate intracellular M. tuberculosis (98).
Inversely correlated with beclin 1, miR-30A is overexpressed in THP-1 cells and AMs from bronchoalveolar lavage, inhibiting autophagy and thus permitting the immune escape of M. tuberculosis, but is expressed at a lower level after anti-TB treatment, indicating that miR-30A is a potential target and biomarker for treatment (100). UV radiation resistance-associated gene (UVRAG) can induce autophagosome formation in conjunction with beclin 1 (113). A profound increase in miR-125a-3p in M. tuberculosis-infected macrophages decreases the UVRAG protein level, inhibiting autophagosome maturation and prolonging intracellular pathogen survival (101). Interacting with UVRAG and LAMP1, DNA damage-regulated autophagy modulator 2 (DRAM2), an initiator of autophagy, is downregulated by miR-144* overexpression in human monocytes and macrophages after M. tuberculosis invasion, blocking autophagosome formation and inhibiting subsequent antimicrobial responses (102, 114). In addition, taking vacuolar protein sorting 33A (VPS33A) as the target, upregulated miR-432-5p suppresses the fusion between autophagosomes and lysosomes, playing a potentially critical role in the occurrence of ATB (103). Similarly, in the duration of phagolysosome biogenesis, miR-30a-3p and miR-30a-5p levels are elevated by recombinant ESAT-6 inc-treated differentiated THP-1 cells, impeding IL-18-mediated fusion between phagosomes and lysosomes and augmenting the survival of internalized M. tuberculosis (115). Furthermore, the promotion of miR-542-3p attenuates autophagy during M. tuberculosis infection by downregulating VPS11, resulting in the pathogen persistence in macrophages, whereas upregulation of VPS11 can counteract this effect (104).
Abundant expression of miR-27a in peripheral blood mononuclear cells (PBMCs) from ATB patients and primary peritoneal macrophages from H37Rv-infected mice is observed to promote M. tuberculosis intracellular residence. The rationale behind this effect is explained by the targeting of calcium voltage-gated channel auxiliary subunit alpha2delta3 (CACNA2D3), a transporter of Ca2+ located in the endoplasmic reticulum (ER), by miR-27a, which inhibits autophagy initiation (105). In addition, TNF-like weak inducer of apoptosis (TWEAK) is a target of miR-889, and an increased miR-889 level in latent tuberculosis infection (LTBI) individuals is related to autophagy suppression and M. tuberculosis maintenance in granulomas (106). Moreover, a team discovered that miR-25 is upregulated in M. tuberculosis- or BCG-infected macrophages, resulting in autophagy impairment and prolonged pathogen survival by blunting the function of the NPC1 protein, a cholesterol transporter located on the lysosomal membrane and involved in autophagolysosome formation (107). Furthermore, upregulation of miR-23a-5p in RAW264.7 cells and bone marrow-derived macrophages (BMDMs) prevents the induction of autophagy by targeting TLR2, thus conferring persistent M. tuberculosis existence in macrophages (108).
As previously stated, weakened autophagy resulting from M. tuberculosis infection is a considerable obstacle to clearing the microorganisms via the acid hydrolases deposited in the lysosomes, and may enable long-term bacterial survival inside the body, placing a heavy burden on the host immune system. Thus, the abovementioned studies on miRNAs may suggest promising therapeutic targets to counter autophagy subversion and opportunities to eliminate the pathogens.
miRNA-regulated apoptosis in M. tuberculosis infection
Apoptosis, a programmed cell demise mechanism that is triggered by an internal or external cellular stimulus, follows two patterns, intrinsic and extrinsic apoptosis, with both culminating in the activation of cysteine-aspartic proteases (caspases), enabling cell structure degradation and death. Cytochrome c (Cyt c), released into the cytosol from mitochondria through mitochondrial outer membrane permeability (MOMP), induces downstream caspase 9 to activate caspases 3 and 7, precursors in cell death via the intrinsic apoptosis pathway and the extrinsic pathway is initiated by the binding of Fas and FasL (116), which activates caspase 8. Bcl-2 family proteins have been identified as key modulators of Cyt c release into the cytoplasm during apoptosis and comprise anti-apoptotic (Bcl-2, Mcl-1, Bcl-XL, Bcl-W, etc.) and proapoptotic (Bim, Bid, Bax, Bak, PUMA, etc.) members (117, 118). Once the war between the host and M. tuberculosis begins, macrophages play a dual role by not only killing the pathogen via apoptosis but also by providing a natural niche for the invading microbe (119, 120). Moreover, the cunning bacteria can induce abnormal expression of host miRNAs to modulate Bcl-2 family activity, thereby either enhancing or inhibiting apoptosis (121) (Figure 4; Supplementary Table S2).
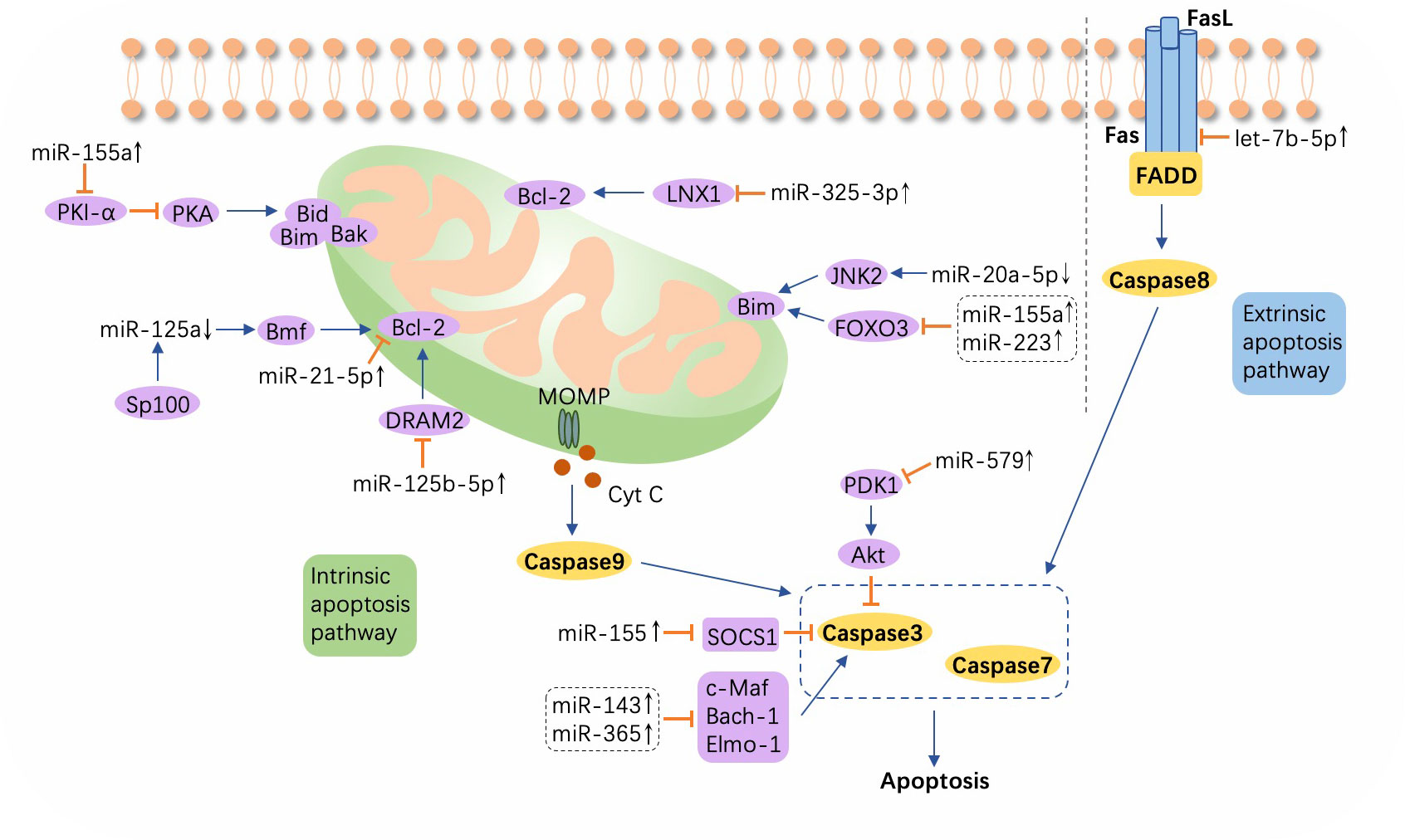
Figure 4 A schematic showing apoptosis and relevant microRNAs during Mycobacterium tuberculosis infection. Extrinsic apoptosis is activated by the combination of Fas and FasL, with the engagement of Fas-associated protein with DD (FADD) and the subsequent activation of caspase 8. In intrinsic apoptosis, cytochrome c (Cyt c) is released into the cytosol from mitochondria through mitochondrial outer membrane permeability (MOMP), which process is regulated by the Bcl-2 family. Altered microRNAs (miRNAs) target the Bcl-2 family or other molecules such as ligand of numb-protein X1 (LNX1), Forkhead box transcription factor class O (FOXO), DNA damage regulated autophagy modulator 2 (DRAM2), suppressor of cytokine signaling (SOCS), c-Maf, BTB and CNC homology 1 (Bach-1), and Engulfment and cell motility protein 1 (Elmo-1). ↑, upregulated; ↓, downregulated; ⟶, stimulate; ⟞, inhibit.
The expression of Jun N-terminal kinase 2 (JNK2), an upstream signaling gene that induces Bim expression, is observed to concomitantly increase with the downregulation of miR-20a-5p in M. tuberculosis-infected macrophages, resulting in the promotion of apoptosis and a higher rate of bacteria clearance (122). An increase in miR-21-5p, which targets Bcl-2 and TLR4, accelerates the apoptosis of macrophages and suppresses the secretion of inflammatory cytokines including IL-1β, IL-6, and TNF-α, which subsequently enhances M. tuberculosis survival (63). Overexpression of miR-125b-5p is associated with DRAM2 downregulation in H37Rv-infected macrophages, and an increase in apoptosis and a reduction in inflammation can protect macrophages when miR-125b-5p is forced into silencing, resulting in accelerated M. tuberculosis killing and suggesting a novel molecular therapeutic strategy for TB (123). Sp100, a nuclear body protein, suppresses miR-125a to increase the positive regulator of apoptosis, the Bcl2 modifying factor (Bmf), which promotes the elimination of the pathogen (124). Furthermore, when Mycobacterium bovis BCG attacks, upregulation of miR-155 targets protein kinase inhibitor-α (PKI-α) and activates the protein kinase A (PKA) signaling pathway to enhance proapoptotic genes, positively mediating macrophage apoptosis (125). ESAT6 can also induce overexpression of miR-155 in a TLR2-dependent manner, which inhibits SOCS1 expression while increasing caspase 3 activity and promoting macrophage apoptosis (126). Both of the aforementioned studies have verified apoptosis promotion and M. tuberculosis clearance facilitation induced by miR-155. However, miR-155 may act as a double-edged sword in the immune modulation, as monocyte apoptosis is inhibited by miR-155 via the suppressed expression of FOXO3, one of the transcription factors that induce apoptosis, leading to the subsistence of M. tuberculosis in PBMCs from ATB patients (127, 128).
In addition, elevated hsa-let-7b-5p and miR-223 inhibit apoptosis and enable enhanced M. tuberculosis survival in macrophages by downregulating Fas and FOXO3, respectively (129, 130). Similarly, in ATB patients, miR-582-5p is found to be upregulated and to suppress monocyte apoptosis by inhibiting FOXO1, attenuating bacteria elimination (131). miR-20b-5p carried in exosomes derived from M. tuberculosis-infected macrophages presents at a low level, inhibiting apoptosis by upregulating Mcl-1 and leading to acceleration of M. tuberculosis proliferation (132). The ligand of numb-protein X 1 (LNX1), an E3 ubiquitin ligase of NIMA-related expressed kinase 6 (NEK6), is directly targeted by miR-325-5p, which inhibits macrophage apoptosis through the activation of STAT3, a signaling pathway triggered by the accumulation of NEK6, ultimately leading to immune escape of M. tuberculosis (121). miR-143 and miR-365, highly overexpressed during M. tuberculosis infection, inhibit apoptosis by differentially downregulating c-Maf, Bach-1, and Engulfment and cell motility protein 1 (Elmo-1) in macrophages alternatively activated by IL-4/13. And the knockdown of these two miRNAs eases the M. tuberculosis burden and decreases chemokine CCL5 and IL-6 production (133). Moreover, ectopic overexpression of miR-579 in M. tuberculosis-infected macrophages intensifies cell apoptosis by negatively modulating phosphoinositide-dependent protein kinase 1 (PDK1) and sirtuin 1 (SIRT1), and this effect can be reversed by the accumulation of cPWWP2A, an endogenous sponge of miR-579 (134).
In addition to apoptosis and autophagy, necrosis, a biological process in the context of microbial invasion that is mediated by molecules such as cyclophilin-D (CypD) and p53, contributes to M. tuberculosis-mediated cytotoxicity in macrophages (119, 135). Virulent M. tuberculosis strains not only evade apoptosis but also induce necrotic cell death (136). However, miR-1281 can prevent trauma to macrophages from undergoing bacteria-induced necrosis and apoptosis by downregulating CypD expression (137).
Moreover, macrophages are capable of acquiring one of two phenotypes, the M1 and M2 phenotypes. When macrophages are polarized to the M1 type, they produce NO and inflammatory cytokines like TNF-α and IL-6, which promote an antibacterial response, whereas alternative activation triggers M2 polarization, which facilitates the production of anti-inflammatory cytokines such as IL-10 and arginase (138). As a member of the Krüpple-like factor (KLF) family, KLF4 activates M2 polarization and inhibits M1 polarization (139). C/EBPβ also plays a pivotal role in driving M2 polarization (140). Nucleotide-binding oligomerization domain-like receptor pyrin domain-containing protein 3 (NLPR3), a pivotal modulator in the inflammatory process, triggers the innate immune response with activation of caspase 1 and secretion of IL-1β and IL-18 (141). In a TB mouse model, miR-20b, which directly binds to NLRP3, is suppressed. After a miR-20b mimic is injected into TB mice, macrophages polarize into the M2 type, alleviating inflammation via the suppression of the NLRP3/caspase 1/IL-1β pathway, which may reflect a novel molecule-based therapeutic strategy (142). miR-26a, which has been observed to be downregulated during M. tuberculosis infection, elicits the upregulation of KLF4 and C/EBPβ, leading to microbe survival by inducing M2 polarization and repressing M. tuberculosis trafficking to lysosomes (109).
Taken together, the abovementioned studies showed that M. tuberculosis-induced alteration in the expression of host miRNAs facilitates the virulence of the bacteria and hinders their elimination. These effects are realized through diverse molecular mechanisms including regulation of signaling pathways, cytokine production, autophagy, and apoptosis. Therefore, pathogenesis of TB is likely based on various miRNAs, which may open up new exploratory avenues for developing innovative immunological therapies to eradicate this fierce bacterial foe.
The role of lncRNAs in immune regulation in tuberculosis
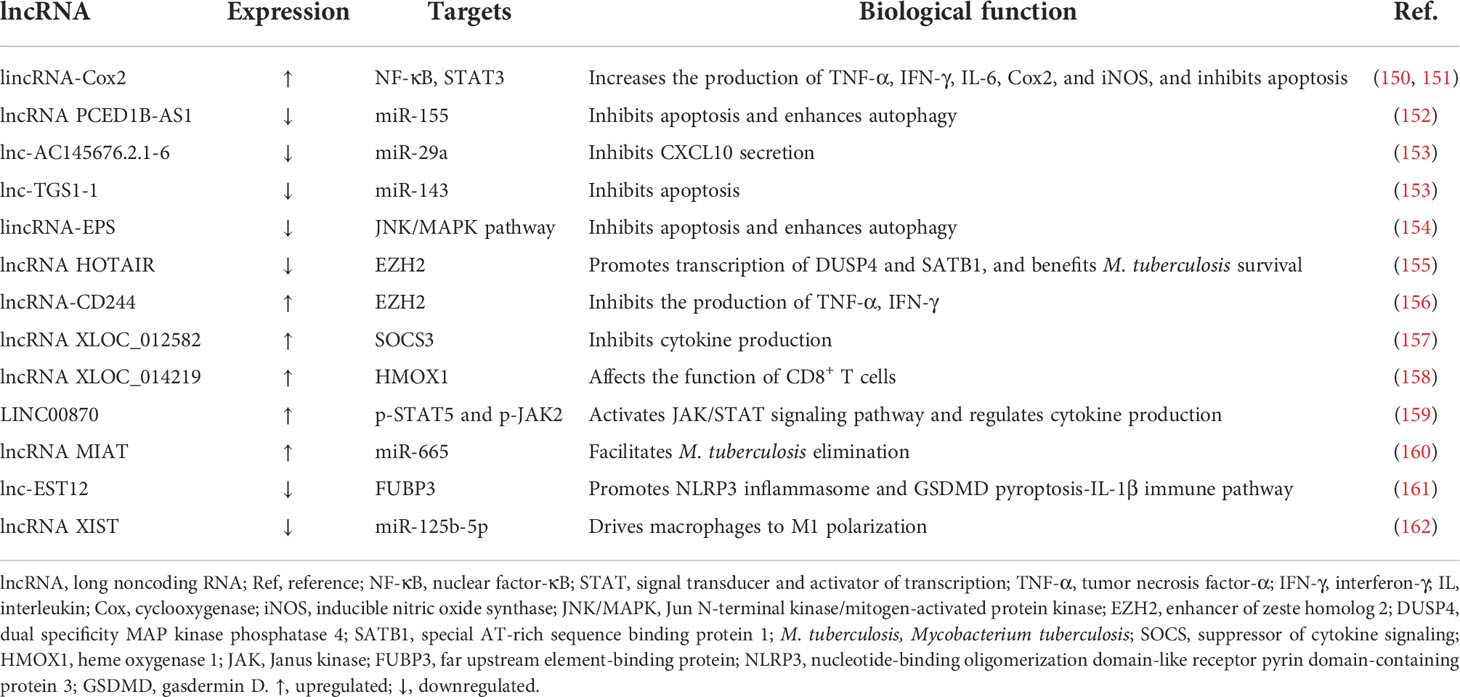
Constituting a class of transcripts longer than 200 nucleotides, lncRNAs, the structures of which do not demonstrate high interspecies conservation but follow a tissue-specific expression pattern, directly modulate cellular processes but not by encoding proteins (10, 143, 144). In the human transcriptome, lncRNAs can be divided into several categories, such as intragenic and intergenic lncRNAs (145). It has been proven that lncRNAs play crucial regulatory roles in diverse biological processes, including cancer metastasis, through multiple mechanisms. For instance, certain lncRNA can sponge a miRNA by competing for binding sites in the miRNA, resulting in corresponding miRNA availability alterations and reducing its regulatory effect on specific mRNAs. Other mechanisms of lncRNA function in gene regulation, including modulating chromatin interactions and affecting gene expression via regulatory complex recruitment, have also been proposed (146–148). In terms of immunology, emerging evidence has confirmed the character of lncRNAs in modulating the mammalian immune response in host–pathogen interactions (149). For M. tuberculosis infection, we concluded relevant lncRNAs which are implicated in innate immune regulation and elaborated their potential roles in the pathogenetic process herein (Table 3).
Induced by the M. tuberculosis H37Ra strain in macrophages, long intergenic noncoding RNA (lincRNA)-Cox2 is revealed to regulate inflammatory reactions in a broad-acting manner, with the promotion of TNF-α, IFN-γ, IL-6, Cox2, and iNOS production (150). Meanwhile, the knockdown of lincRNA-Cox2 inhibits the inflammatory response, promotes the apoptotic rate of H37Ra-infected macrophages, and facilitates pathogen proliferation via the suppression of the NF-κB and STAT3 signaling pathways (151). lncRNA PCED1B-AS1, which is downregulated in CD14+ monocytes from ATB patients, attenuates apoptosis and enhances autophagy by acting as an endogenous sponge of miR-155, leading to impeded M. tuberculosis elimination. FOXO3 and Rheb, genes targeted by miR-155, can reverse the impact of PCED1B-AS1 (152). miR-29a harbors a binding site for certain lncRNAs and has been verified to repress the secretion of CXCL10, negatively affecting T-cell recruitment after M. tuberculosis infection (163). Compared with the levels in healthy control (HC) groups, lnc-AC145676.2.1-6 and lnc-TGS1-1 are significantly decreased in TB patients, suppressing immune function via the elevation of miR-29a and miR-143, respectively, the functions of which have been enumerated above. Furthermore, the downregulation of lnc-TGS-1 is related to thrombocytopenia during TB treatment (153).
Downregulation of lincRNA-erythroid prosurvival (lincRNA-EPS) enhances autophagy by promoting LC3 and suppresses apoptosis, facilitating the eradication of the pathogen in BCG-infected RAW264.7 macrophages via the activation of JNK/mitogen-activated protein kinase (MAPK) signaling, which plays a key role in controlling the balance between apoptosis and autophagy (154). The expression of lncRNA HOX transcript antisense RNA (HOTAIR) has been found to be completely opposite in different M. tuberculosis strain-infected macrophages; it is upregulated in H37Ra-resided cells and suppressed in H37Rv-infected cells. Downregulation of lncRNA HOTAIR facilitates intracellular M. tuberculosis persistence due to the increased transcription of dual specificity MAP kinase phosphatase 4 (DUSP4) and special AT-rich sequence binding protein 1 (SATB1) by targeting enhancer of zeste homolog (EZH) 2 (155). Likewise, taking EZH2 as the target, lncRNA-CD244, which is induced by the T-cell inhibitory molecule CD244, suppresses the production of IFN-γ and TNF-α in M. tuberculosis-infected CD8+ T cells and attenuates the protective immunity of T cells to combat the invasion of the microorganisms (156).
The differential expression of 844 lncRNAs in B cells between individuals with or without TB was reported by Fu and his colleagues. Among these deregulated lncRNAs, lncRNA XLOC_012582 is highly expressed in B cells from TB patients, along with SOCS3 promotion, an inhibitor of cytokine secretion. However, the relationship between the altered expression of XLOC_012582 and SOCS3 and the particular impact of this change on M. tuberculosis infection remains to be further explored (157). Meanwhile, the team revealed the upregulation of lncRNA XLOC_014219 and a decrease in heme oxygenase 1 (HMOX1) in CD8+ T cells from ATB individuals. Nevertheless, whether this phenomenon is involved in the dysfunction of CD8+ T cells has not been clarified (158). LINC00870, which is significantly induced by M. tuberculosis in PBMCs, mediates the immune response against the bacteria by suppressing the ability of Th1 cells to produce cytokines such as IL-2 and IFN-γ and promoting the expression of cytokines generated by Th2 cells including IL-4 and IL-10. These impacts may be attributable to the activation of the Janus kinase (JAK)/STAT signaling pathway via the acceleration of the expression of p-STAT5 and p-JAK2 mediated by LINC00870 (159). Another study illuminated that the expression level of lncRNA MIAT is heightened, which accelerates autophagy and apoptosis in BCG-infected THP-1 macrophages by binding to miR-665 to enhance ULK1 activation, ultimately inhibiting intracellular pathogen maintenance (160).
Inhibited by the M. tuberculosis Rv1579c via the activation of the JAK2-STAT5a pathway, lnc-EST12 decreases the production of IL-1β, IL-6, and CCL5/8. Moreover, it suppresses the NLRP3 and gasdermin D (GSDMD) pyroptosis-IL-1β pathway by binding too far upstream element-binding protein (FUBP) 3 to suppress innate immunity toward M. tuberculosis (161). Negative pressure therapy, promoting wound healing via vacuum dressings, has been utilized as a TB treatment and has shown promising effects. Upregulation of lncRNA XIST and downregulation of miR-125b-5p during the infection can be reversed by this treatment strategy. Through modulation of the lncRNA XIST/miR-125b-5p/A20/NF-κB axis, which ultimately increases the activity of NF-κB p65, this regimen facilitates the polarization of macrophages to the M1 phenotype, enhancing the inflammatory response to reduce M. tuberculosis survival (162).
As discussed above, lncRNAs regulate the innate immune response to M. tuberculosis in various ways, including serving as sponges of miRNAs and regulating signaling pathways, which implies that we should undertake related research for the exploration of novel therapies. However, existing studies are not sufficiently systematic for organized analysis, and certain results remain unverified. Therefore, more investigations are yet to be done.
The role of circRNAs in immune regulation in tuberculosis
Due to the noncanonical splicing process, back-splicing, the structure of circRNAs is thoroughly distinct from that of other RNA molecules. They are covalently closed, without typical components such as 5′ capping and 3′ polyadenylation (164). Viroids were the first circRNAs to be discovered (165). With considerable progress in high-throughput RNA sequencing technology and bioinformatics, thousands of circRNAs have been identified in eukaryotes (166), demonstrating a tissue- and cell-specific expression pattern (167, 168). Acting as miRNA sponges, the more binding sites circRNAs contain, the more competitive rivalry there will be (169, 170). Furthermore, circRNAs can enhance the function of certain proteins by interacting with RNA-binding proteins (RBPs) (171) and serve as scaffolds in various processes involving enzymes and their substrates (172). Continuous evidence has confirmed that circRNAs exert a nonnegligible influence on numerous cellular processes, from cell cycle control to cancer development (173, 174).
With respect to infection, previous studies have found that circRNAs perform flexible roles when viruses invade (175). Several studies on TB have been carried out and have revealed the impact of circRNAs on the modulation of the host immune response (Supplementary Table S3). For example, circAGFG1, which is upregulated in TB patients, promotes autophagy and inhibits apoptosis by targeting miR-1257 in macrophages, which subsequently increases the Notch 2 level (176). Another study found that circTRAPPC6B is downregulated during M. tuberculosis infection. Furthermore, forced expression of this circRNA antagonizes the capacity of miR-874-3p to inhibit autophagy by targeting ATG16L, thereby increasing autophagy sequestration and restricting intracellular pathogen growth (177). Moreover, circRNA-0003528 enhances M. tuberculosis-related macrophage polarization, which is mediated by the promotion of CTLA4 via the inhibition of miR-224-5p, miR-324-5p, and miR-488-5p (178). Deng et al. discovered that circ_0001490 suppresses M. tuberculosis survival and promotes the viability of host macrophages by sponging miR-579-3p to increase the expression of FSTL1 (179). Similarly, hsa_circ_0045474 is downregulated and plays a positive role in autophagy induction by promoting the expression of miR-582-5p and suppressing the expression of the downstream target TNKS2 in M. tuberculosis-infected macrophages, facilitating the bacterial clearance in the end (180).
As demonstrated, the rarity and unconfirmed results of relevant studies lead to a vague understanding of circRNAs’ function in TB. Because circRNAs present peculiar structures, a cyclic construction formed through covalent bonds, they are highly stable and resistant to degradation mediated by exonucleases (164). Therefore, circRNAs may be ideal biomarkers for TB detection and prognosis prediction due to their aberrant expression (181), and this possibility is discussed, along with other potential ncRNA biomarkers, in the following section.
Noncoding RNAs as biomarkers in tuberculosis
On account of the huge burden of the disease, accurate diagnosis of TB is important but remains a formidable challenge. Conventional methods based on sputum microscopy or culture are time- consuming and resource-limited. Moreover, there is a grim fact that the incidence of DR-TB continues to increase, leading to lower treatment effectiveness (2). In addition, adverse drug reactions (ADRs) to anti-TB chemotherapy complicate disease management (182, 183). Thus, novel, precise, and efficient indicators for diagnosis, drug resistance prediction, and treatment monitoring of TB are urgently required. Based on differentially expressed ncRNAs, related studies have identified numerous biomarkers with various functions as follows.
Diagnostic biomarkers of pulmonary tuberculosis
As mentioned above, miR-432-5p, upregulated during M. tuberculosis infection, inhibits the fusion between autophagosomes and lysosomes by targeting VPS33A. In the meantime, the expression of miR-17-5p and miR-20b-5p is significantly elevated in the serum of TB patients. Therefore, one group developed a diagnostic model for TB via the combination of the three miRNAs, achieving an area under the curve (AUC) of 0.908 (103). As previously mentioned, miR-889 inhibits autophagy by targeting TWEAK and can serve as a biomarker for LBTI and a potential therapeutic target due to its high expression in patients, which is decreased after prophylactic therapy (106). Another study noticed that miR-29a and miR-99b are upregulated while miR-21, miR-146a, and miR-652 are decreased in the plasma of TB patients. Therefore, the team constructed a model based on these five miRNAs to determine M. tuberculosis infection, showing an AUC of 0.976 (184). After obtaining this result, they utilized a combination of proteins and miRNAs to further improve the model (185). Another group combined miR-142-3p with electronic health record (EHR) data to detect TB, and the AUC reached 0.94 (186). Compared with LTBI patients, hsa-miR-1246, hsa-miR-2110, hsa-miR-370-3p, hsa-miR-193b-5p, and hsa-miR-28-3p are specifically expressed in TB cohorts (187). In addition, miR-185-5p is more highly expressed in plasma exosomes obtained from TB patients than in HCs, demonstrating a diagnostic potential with an AUC of 0.75 (21).
Regarding lncRNAs, considerable effort has also been directed to explore their capacity for TB detection. Consisting of four lncRNAs including NR_038221, NR_003142, ENST00000570366, and ENST00000422183, the AUC of a diagnostic model was reported to be 0.845 (188). Hu and his team identified that ENST00000497872, n333737, and n335265 are differentially expressed in clinically diagnosed TB patients and established a nomogram to predict the infection by taking advantage of the data on these three lncRNAs and six clinical covariates such as age and hemoglobin, with an AUC equal to 0.89 (189). Similarly, lncRNAs TCONS_00001838 and n406498 are significantly differentially expressed in TB patients. Combining the two lncRNA loci with eight EHR indicators through logistic regression, the model achieved a decent predictive value of TB with an AUC of 0.86 (190). Another study discovered that lncRNA CCAT1 is upregulated in TB patients, concomitant with high mortality rates, and is negatively correlated with IL-10 (191). Furthermore, the level of LINC00870 is higher in the plasma and sputum obtained from TB and LTBI patients and decreases after 3 months of anti-TB therapy (ATT), showing the potential of being a biomarker for the diagnosis and treatment evaluation of TB (159).
In terms of circRNAs, the significant dysregulation of hsa_circ_0043497 and hsa_circ_0001204 in PBMCs from TB patients indicated their diagnostic capacity, as their AUCs achieved at 0.860 and 0.848, respectively (192). Another group validated the overexpression of circRNA_051239, circRNA_029965, and circRNA_404022 in the serum, and constructed a panel of the three circRNAs for use in TB diagnosis, reaching an AUC of 0.992 (193). Moreover, hsa_circRNA_001937 is significantly increased in PBMCs from TB patients compared to patients with lung cancer, pneumonia, or chronic obstructive pulmonary disease, demonstrating the potential of this circRNA to be a diagnostic biomarker with an AUC of 0.873, and it is deemed a candidate molecule for measuring TB severity (194).
As mentioned above, utilizing a combination of various ncRNAs, several diagnostic models have shown favorable performance, inspiring us to develop creative approaches based on these molecules to detect TB. Rather than waiting for the test results of pathogens, physicians can rapidly determine M. tuberculosis infection through fast evaluation of blood or other conveniently obtained clinical samples (Table 4).
Biomarkers for early detection of active tuberculosis
A study discovered that 24 miRNAs are upregulated and 6 miRNAs are downregulated in ATB patients, among which the significantly overexpressed hsa-miR-196b and hsa-miR-376c demonstrate the greatest potential to be ATB indicators (195). Presenting a higher fold change in ATB patients than in HCs, miR-155* and miR-155 can also serve as diagnostic biomarkers for ATB, with respective AUC of 0.7945 and 0.8972 (196). Moreover, a nested case-control study revealed that, combined with body mass index (BMI) and TB history, downregulated hsa-miR-16-5p and hsa-miR-451a can contribute to the prediction of developing ATB from LBTI, with AUCs of 0.84 and 0.85, respectively (197). Shown to be increased in the serum of ATB patients, miR-96, miR-425, and miR-484 are also diagnostic candidates, showing moderate performance with AUC values ranging from 0.62 to 0.72 (198).
Notably, after synthesizing analysis of the NONCODE database and GEO dataset, Fang and his colleagues discovered the ability of four lncRNAs, NONHSAT101518.2, NONHSAT067134.2, NONHSAT148822.1, and NONHSAT078957.2, to discriminate ATB patients from HCs via plasma samples, with AUCs ranging from 0.7080 to 0.9502 (199).
In addition, overexpressed circRNA_103017, circRNA_059914, and circRNA_101128 are confirmed to be increased by M. tuberculosis, while circRNA_062400 is downregulated in PBMCs from ATB individuals. Among these circRNAs, circRNA_103017 shows the maximum potential as an indicator for ATB diagnosis, with an AUC of 0.870 (200). Furthermore, the upregulation of hsa_circ_002883 in PBMCs endows it with the power to be a candidate biomarker for ATB determination, showing significant discrimination efficiency with an AUC of 0.773 (201). In contrast, hsa_circRNA_103571 expression is obviously decreased in plasma from ATB patients, with a diagnostic ability for identifying ATB (AUC 0.838) (202). Moreover, the lower expression of hsa_circ_0005836 in PBMCs from the ATB cohort is verified, indicating that it may serve as a novel biomarker for M. tuberculosis infection (22) (Supplementary Table S4).
To control the spread of TB, proactively recognizing and ameliorating active individuals is of imperative priority. Although the abovementioned studies demonstrate decent performance, the clinical practicality of using these molecules for diagnosis remains to be verified in realistic medical settings. After being strictly tested, these methods may be novel diagnostic decision-making tools, providing opportunities to carry out proper treatment against ATB as early as possible.
Biomarkers for prediction of tuberculosis drug resistance
Considering the roles of ncRNAs in identifying TB drug resistance, a few studies have also been carried out to analyze their potential in this daunting task. For example, in DR-TB patients, miR-197-3p and miR-223-3p are downregulated, while miR-let-7e-5p is increased, and a multivariate analysis based on these three miRNAs was performed to distinguish resistant individuals from HCs. Finally, the diagnostic model achieved an AUC of 0.96 for DR-TB recognition. A favorable outcome showing an AUC of 0.95 was observed for multidrug-resistant tuberculosis (MDR-TB) subgroup identification (203). Moreover, the expression of miR-378 in the serum is associated with adverse therapeutic consequences since it presented a higher level in MDR-TB than in single-drug-resistant tuberculosis (SDR-TB). With an AUC of 0.767, this miRNA can also distinguish the ATB population from the LBTI patients (204). Furthermore, lncRNA n335659 shows a statistically significant downward trend in the MDR-TB group compared with the HC cohort; hence, it may act as a latent biomarker for TB drug resistance prediction as well (205) (Supplementary Table S5).
Nonetheless, predicting drug resistance in TB through differentially expressed ncRNAs is still a challenging task, as there are not abundant or sufficiently comprehensive studies that have been validated in actual clinical environments. Therefore, we need to continue to explore the hidden potential of ncRNAs in DR-TB recognition to realize early diagnosis of drug resistance and carry out effective treatment.
Biomarkers for treatment evaluation of tuberculosis
During anti-TB treatment, drug-induced liver injury (DILI) is the most common severe adverse drug response, with an approximate incidence of 2%–28% (206). miR-122 and miR-192 are significantly decreased in the serum of TB patients with DILI and could be used to predict this serious complication in TB (207). Likewise, another study revealed the value of upregulated circMARS in identifying TB patients suffering from anti-TB drug-induced liver injury (ADLI), with an AUC of 0.80 (208). Moreover, the lower expression of lnc-TGS1-1 is capable of indicating the development of thrombocytopenia after treatment (153).
Used for treatment response monitoring, the expression of hsa-miR-346 is significantly increased in the supernatant of macrophages and serum during M. tuberculosis infection but declines after two months of ATT (209). Moreover, the upregulation of miR-29a and miR-99b in patients with TB is attenuated, reaching levels equal to those in HCs after the completion of treatment (184). Similarly, LINC00870 is overexpressed in both sputum and plasma samples from TB or LBTI patients, but its expression is reduced after three months of ATT (159). In addition, the expression of lncRNA CCAT1 also decreases during ATT, and a higher level of CCAT1 is correlated with high mortality (191), as briefly described in Supplementary Table S5.
The treatment monitoring capacity of ncRNAs in response evaluation and side effect prediction of anti-TB chemotherapy has provided researchers with a promising prospect to precisely assess the curative effect in patients with TB and to adjust therapy regimens on an individual basis.
Conclusion remarks
In summary, we concluded the latest released original studies investigating the promising roles of ncRNAs in M. tuberculosis infection, elucidating their functions, which vary in pathogenesis and as biomarkers. The research referenced may shed light on the principles behind novel TB therapeutic schemes based on those seemingly negligible but indeed crucial elements. Moreover, owing to the differential expression of ncRNAs in the clinical samples of patients with different statuses, a new chapter is added to the diagnosis, drug resistance prediction, and treatment monitoring of TB, with the further purpose of making appropriate clinical decisions.
Despite the attainment obtained in the field of ncRNAs in TB, pitfalls remain in the path of integrating these critical molecules into a realistic clinical scenario. First, since previous studies have linked various ncRNAs to the pathogenesis of TB, which may be instructive for HDT development, novel remedies for TB based on these ncRNAs are promising to explore, but relevant investigations are lacking, making their use in treatment infeasible in practice. Second, a single ncRNA can interfere with the expression of diverse genes, but the complete targets of ncRNAs have not been explored thoroughly yet (210). Thus, it is rational that one ncRNA may lead to paradoxical effects in M. tuberculosis infection, making determining a therapeutic strategy complicated and confusing. Third, although the parameters of the diagnostic models based on ncRNAs seem to be promising with decent AUCs, rigorous and prospective trials of these ncRNAs tested in substantial samples are required to verify their true potential as biomarkers in TB. Another considerable issue is that a large proportion of the studies to date have focused on miRNAs, with research advancements made with lncRNAs, circRNAs, and other ncRNAs being relatively rare. Prominent advances in RNA sequencing have opened the gate to an extensive domain from which to determine their function, and related work in the future is warranted.
In conclusion, the research field of ncRNAs in M. tuberculosis infection has met a certain degree of success in pathogenesis and biomarker exploration and holds broad promise. However, several challenges still need to be addressed before these molecules can be seamlessly integrated into clinical practice to enrich personalized and creative diagnostic strategies (211, 212), and further direct treatment decision-making in TB, especially for the management of drug-resistant patients.
Author contributions
WL and CW designed the conception of the manuscript. SL, JM, JS, JL and YZ drafted the original version of the manuscript and drew the figures and tables. ZW, HG and CW revised the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The study was supported by the National Natural Science Foundation of China (82100119, 92159302), the Science and Technology Project of Sichuan (2022ZDZX0018, 2020YFG0473), the Chinese Postdoctoral Science Foundation (2022T150451, 2021M692309), and the Postdoctoral Program of Sichuan University (2021SCU12018).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.987018/full#supplementary-material
References
2. Lange C, Dheda K, Chesov D, Mandalakas AM, Udwadia Z, Horsburgh CR Jr. Management of drug-resistant tuberculosis. Lancet (2019) 394(10202):953–66. doi: 10.1016/s0140-6736(19)31882-3
3. Divangahi M, Chen M, Gan H, Desjardins D, Hickman TT, Lee DM, et al. Mycobacterium tuberculosis evades macrophage defenses by inhibiting plasma membrane repair. Nat Immunol (2009) 10(8):899–906. doi: 10.1038/ni.1758
4. Rajaram MV, Ni B, Dodd CE, Schlesinger LS. Macrophage immunoregulatory pathways in tuberculosis. Semin Immunol (2014) 26(6):471–85. doi: 10.1016/j.smim.2014.09.010
5. Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non-coding regions of the genome. Nat Rev Genet (2010) 11(8):559–71. doi: 10.1038/nrg2814
6. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet (2009) 10(3):155–9. doi: 10.1038/nrg2521
7. McCabe EM, Rasmussen TP. LncRNA involvement in cancer stem cell function and epithelial-mesenchymal transitions. Semin Cancer Biol (2021) 75:38–48. doi: 10.1016/j.semcancer.2020.12.012
8. Huang T, Alvarez A, Hu B, Cheng SY. Noncoding RNAs in cancer and cancer stem cells. Chin J Cancer (2013) 32(11):582–93. doi: 10.5732/cjc.013.10170
9. Zhou M, Zhao H, Xu W, Bao S, Cheng L, Sun J. Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large b cell lymphoma. Mol Cancer (2017) 16(1):16. doi: 10.1186/s12943-017-0580-4
10. Johnsson P, Lipovich L, Grandér D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta (2014) 1840(3):1063–71. doi: 10.1016/j.bbagen.2013.10.035
11. Iwasaki YW, Siomi MC, Siomi H. PIWI-interacting RNA: Its biogenesis and functions. Annu Rev Biochem (2015) 84:405–33. doi: 10.1146/annurev-biochem-060614-034258
12. Hombach S, Kretz M. Non-coding RNAs: Classification, biology and functioning. Adv Exp Med Biol (2016) 937:3–17. doi: 10.1007/978-3-319-42059-2_1
13. Krol J, Loedige I, Filipowicz W. The widespread regulation of MicroRNA biogenesis, function and decay. Nat Rev Genet (2010) 11(9):597–610. doi: 10.1038/nrg2843
14. Anastasiadou E, Jacob L, Slack F. Non-coding RNA networks in cancer. Nat Rev Cancer (2018) 18(1):5–18. doi: 10.1038/nrc.2017.99
15. Zeng J, Dong S, Luo Z, Xie X, Fu B, Li P, et al. The zika virus capsid disrupts corticogenesis by suppressing dicer activity and miRNA biogenesis. Cell Stem Cell (2020) 27(4):618–32.e9. doi: 10.1016/j.stem.2020.07.012
16. Lu M, Zhang PJ, Li CH, Lv ZM, Zhang WW, Jin CH. MiRNA-133 augments coelomocyte phagocytosis in bacteria-challenged apostichopus japonicus via targeting the TLR component of IRAK-1 in vitro and in vivo. Sci Rep (2015) 5:12608. doi: 10.1038/srep12608
17. Huang M, Peng X, Yang L, Yang S, Li X, Tang S, et al. Non-coding RNA derived from extracellular vesicles in cancer immune escape: Biological functions and potential clinical applications. Cancer Lett (2021) 501:234–46. doi: 10.1016/j.canlet.2020.11.005
18. Wang JK, Wang Z, Li G. MicroRNA-125 in immunity and cancer. Cancer Lett (2019) 454:134–45. doi: 10.1016/j.canlet.2019.04.015
19. Arnvig K, Young D. Non-coding RNA and its potential role in mycobacterium tuberculosis pathogenesis. RNA Biol (2012) 9(4):427–36. doi: 10.4161/rna.20105
20. Stanley SA, Cox JS. Host-pathogen interactions during mycobacterium tuberculosis infections. Curr Top Microbiol Immunol (2013) 374:211–41. doi: 10.1007/82_2013_332
21. Kaushik AC, Wu Q, Lin L, Li H, Zhao L, Wen Z, et al. Exosomal ncRNAs profiling of mycobacterial infection identified miRNA-185-5p as a novel biomarker for tuberculosis. Brief Bioinform (2021) 22(6):bbab210. doi: 10.1093/bib/bbab210
22. Zhuang ZG, Zhang JA, Luo HL, Liu GB, Lu YB, Ge NH, et al. The circular RNA of peripheral blood mononuclear cells: Hsa_circ_0005836 as a new diagnostic biomarker and therapeutic target of active pulmonary tuberculosis. Mol Immunol (2017) 90:264–72. doi: 10.1016/j.molimm.2017.08.008
23. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of MicroRNAs. Genome Res (2009) 19(1):92–105. doi: 10.1101/gr.082701.108
24. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell (2004) 116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5
25. Rajewsky N. MicroRNA target predictions in animals. Nat Genet (2006) 38 Suppl:S8–13. doi: 10.1038/ng1798
26. Shenoy A, Blelloch RH. Regulation of MicroRNA function in somatic stem cell proliferation and differentiation. Nat Rev Mol Cell Biol (2014) 15(9):565–76. doi: 10.1038/nrm3854
27. Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol (2009) 4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222
28. Wu CJ, Lu LF. MicroRNA in immune regulation. Curr Top Microbiol Immunol (2017) 410:249–67. doi: 10.1007/82_2017_65
29. Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by MicroRNAs. Nat Rev Immunol (2008) 8(2):120–30. doi: 10.1038/nri2252
30. Lu LF, Liston A. MicroRNA in the immune system, MicroRNA as an immune system. Immunology (2009) 127(3):291–8. doi: 10.1111/j.1365-2567.2009.03092.x
31. Sinigaglia A, Peta E, Riccetti S, Venkateswaran S, Manganelli R, Barzon L. Tuberculosis-associated MicroRNAs: From pathogenesis to disease biomarkers. Cells (2020) 9(10):2160. doi: 10.3390/cells9102160
32. Simmons JD, Stein CM, Seshadri C, Campo M, Alter G, Fortune S, et al. Immunological mechanisms of human resistance to persistent mycobacterium tuberculosis infection. Nat Rev Immunol (2018) 18(9):575–89. doi: 10.1038/s41577-018-0025-3
33. Shariq M, Quadir N, Alam A, Zarin S, Sheikh JA, Sharma N, et al. The exploitation of host autophagy and ubiquitin machinery by mycobacterium tuberculosis in shaping immune responses and host defense during infection. Autophagy (2022) 1-21. doi: 10.1080/15548627.2021.2021495
34. Horng T, Barton GM, Flavell RA, Medzhitov R. The adaptor molecule TIRAP provides signalling specificity for toll-like receptors. Nature (2002) 420(6913):329–33. doi: 10.1038/nature01180
35. Marongiu L, Gornati L, Artuso I, Zanoni I, Granucci F. Below the surface: The inner lives of TLR4 and TLR9. J Leukoc Biol (2019) 106(1):147–60. doi: 10.1002/jlb.3mir1218-483rr
36. Ciesielska A, Matyjek M, Kwiatkowska K. TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell Mol Life Sci (2021) 78(4):1233–61. doi: 10.1007/s00018-020-03656-y
37. Hayden MS, Ghosh S. Shared principles in NF-KappaB signaling. Cell (2008) 132(3):344–62. doi: 10.1016/j.cell.2008.01.020
38. Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol (2014) 5:461. doi: 10.3389/fimmu.2014.00461
39. Hayden MS, Ghosh S. NF-κB, the first quarter-century: Remarkable progress and outstanding questions. Genes Dev (2012) 26(3):203–34. doi: 10.1101/gad.183434.111
40. Chen ZJ. Ubiquitin signalling in the NF-KappaB pathway. Nat Cell Biol (2005) 7(8):758–65. doi: 10.1038/ncb0805-758
41. Wei J, Huang X, Zhang Z, Jia W, Zhao Z, Zhang Y, et al. MyD88 as a target of MicroRNA-203 in regulation of lipopolysaccharide or bacille calmette-guerin induced inflammatory response of macrophage RAW264.7 cells. Mol Immunol (2013) 55(3-4):303–9. doi: 10.1016/j.molimm.2013.03.004
42. Xu G, Zhang Z, Xing Y, Wei J, Ge Z, Liu X, et al. MicroRNA-149 negatively regulates TLR-triggered inflammatory response in macrophages by targeting MyD88. J Cell Biochem (2014) 115(5):919–27. doi: 10.1002/jcb.24734
43. Wu Y, Sun Q, Dai L. Immune regulation of miR-30 on the mycobacterium tuberculosis-induced TLR/MyD88 signaling pathway in THP-1 cells. Exp Ther Med (2017) 14(4):3299–303. doi: 10.3892/etm.2017.4872
44. Ma C, Li Y, Li M, Deng G, Wu X, Zeng J, et al. MicroRNA-124 negatively regulates TLR signaling in alveolar macrophages in response to mycobacterial infection. Mol Immunol (2014) 62(1):150–8. doi: 10.1016/j.molimm.2014.06.014
45. Li M, Wang J, Fang Y, Gong S, Li M, Wu M, et al. MicroRNA-146a promotes mycobacterial survival in macrophages through suppressing nitric oxide production. Sci Rep (2016) 6:23351. doi: 10.1038/srep23351
46. Li S, Yue Y, Xu W, Xiong S. MicroRNA-146a represses mycobacteria-induced inflammatory response and facilitates bacterial replication via targeting IRAK-1 and TRAF-6. PloS One (2013) 8(12):e81438. doi: 10.1371/journal.pone.0081438
47. Mahadik K, Prakhar P, Rajmani RS, Singh A, Balaji KN. C-Abl-TWIST1 epigenetically dysregulate inflammatory responses during mycobacterial infection by co-regulating bone morphogenesis protein and miR27a. Front Immunol (2018) 9:85. doi: 10.3389/fimmu.2018.00085
48. Xu G, Zhang Z, Wei J, Zhang Y, Zhang Y, Guo L, et al. MicroR-142-3p down-regulates IRAK-1 in response to mycobacterium bovis BCG infection in macrophages. Tuberculosis (Edinb) (2013) 93(6):606–11. doi: 10.1016/j.tube.2013.08.006
49. Li WT, Zhang Q. MicroRNA-708-5p regulates mycobacterial vitality and the secretion of inflammatory factors in mycobacterium tuberculosis-infected macrophages by targeting TLR4. Eur Rev Med Pharmacol Sci (2019) 23(18):8028–38. doi: 10.26355/eurrev_201909_19019
50. Shi G, Mao G, Xie K, Wu D, Wang W. MiR-1178 regulates mycobacterial survival and inflammatory responses in mycobacterium tuberculosis-infected macrophages partly via TLR4. J Cell Biochem (2018) 119(9):7449–57. doi: 10.1002/jcb.27054
51. Liang S, Huang G, Wu T, Peng Y, Liu X, Ji X, et al. MIR337-3p enhances mycobacterial pathogenicity involving TLR4/MYD88 and STAT3 signals, impairing VDR antimicrobial response and fast-acting immunity. Front Immunol (2021) 12:739219. doi: 10.3389/fimmu.2021.739219
52. Shen H, Gu J, Xiao H, Liang S, Yang E, Yang R, et al. Selective destruction of interleukin 23-induced expansion of a major antigen-specific γδ T-cell subset in patients with tuberculosis. J Infect Dis (2017) 215(3):420–30. doi: 10.1093/infdis/jiw511
53. Niu W, Sun B, Li M, Cui J, Huang J, Zhang L. TLR-4/MicroRNA-125a/NF-κB signaling modulates the immune response to mycobacterium tuberculosis infection. Cell Cycle (2018) 17(15):1931–45. doi: 10.1080/15384101.2018.1509636
54. Li X, Huang S, Yu T, Liang G, Liu H, Pu D, et al. MiR-140 modulates the inflammatory responses of mycobacterium tuberculosis-infected macrophages by targeting TRAF6. J Cell Mol Med (2019) 23(8):5642–53. doi: 10.1111/jcmm.14472
55. Liang S, Song Z, Wu Y, Gao Y, Gao M, Liu F, et al. MicroRNA-27b modulates inflammatory response and apoptosis during mycobacterium tuberculosis infection. J Immunol (2018) 200(10):3506–18. doi: 10.4049/jimmunol.1701448
56. Kumar M, Sahu SK, Kumar R, Subuddhi A, Maji RK, Jana K, et al. MicroRNA let-7 modulates the immune response to mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-κB pathway. Cell Host Microbe (2015) 17(3):345–56. doi: 10.1016/j.chom.2015.01.007
57. Liu Y, Wang R, Jiang J, Yang B, Cao Z, Cheng X. MiR-223 is upregulated in monocytes from patients with tuberculosis and regulates function of monocyte-derived macrophages. Mol Immunol (2015) 67(2 Pt B):475–81. doi: 10.1016/j.molimm.2015.08.006
58. Dorhoi A, Iannaccone M, Farinacci M, Faé KC, Schreiber J, Moura-Alves P, et al. MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J Clin Invest (2013) 123(11):4836–48. doi: 10.1172/jci67604
59. Yao J, Du X, Chen S, Shao Y, Deng K, Jiang M, et al. Rv2346c enhances mycobacterial survival within macrophages by inhibiting TNF-α and IL-6 production via the p38/miRNA/NF-κB pathway. Emerg Microbes Infect (2018) 7(1):158. doi: 10.1038/s41426-018-0162-6
60. Liu F, Dong Z, Lin Y, Yang H, Wang P, Zhang Y. MicroRNA−502−3p promotes mycobacterium tuberculosis survival in macrophages by modulating the inflammatory response by targeting ROCK1. Mol Med Rep (2021) 24(5):753. doi: 10.3892/mmr.2021.12393
61. Yuan Y, Lin D, Feng L, Huang M, Yan H, Li Y, et al. Upregulation of miR-196b-5p attenuates BCG uptake via targeting SOCS3 and activating STAT3 in macrophages from patients with long-term cigarette smoking-related active pulmonary tuberculosis. J Transl Med (2018) 16(1):284. doi: 10.1186/s12967-018-1654-9
62. Fu B, Lin X, Tan S, Zhang R, Xue W, Zhang H, et al. MiR-342 controls mycobacterium tuberculosis susceptibility by modulating inflammation and cell death. EMBO Rep (2021) 22(9):e52252. doi: 10.15252/embr.202052252
63. Zhao Z, Hao J, Li X, Chen Y, Qi X. MiR-21-5p regulates mycobacterial survival and inflammatory responses by targeting bcl-2 and TLR4 in mycobacterium tuberculosis-infected macrophages. FEBS Lett (2019) 593(12):1326–35. doi: 10.1002/1873-3468.13438
64. Kogan D, Grabner A, Yanucil C, Faul C, Ulaganathan VK. STAT3-enhancing germline mutations contribute to tumor-extrinsic immune evasion. J Clin Invest (2018) 128(5):1867–72. doi: 10.1172/jci96708
65. Weng Q, Zhao M, Zheng J, Yang L, Xu Z, Zhang Z, et al. STAT3 dictates β-cell apoptosis by modulating PTEN in streptozocin-induced hyperglycemia. Cell Death Differ (2020) 27(1):130–45. doi: 10.1038/s41418-019-0344-3
66. Gasparini C, Feldmann M. NF-κB as a target for modulating inflammatory responses. Curr Pharm Des (2012) 18(35):5735–45. doi: 10.2174/138161212803530763
67. Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, et al. The MicroRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol (2011) 12(9):861–9. doi: 10.1038/ni.2073
68. Fu Y, Yi Z, Li J, Li R. Deregulated MicroRNAs in CD4+ T cells from individuals with latent tuberculosis versus active tuberculosis. J Cell Mol Med (2014) 18(3):503–13. doi: 10.1111/jcmm.12205
69. Afum-Adjei Awuah A, Ueberberg B, Owusu-Dabo E, Frempong M, Jacobsen M. Dynamics of T-cell IFN-γ and miR-29a expression during active pulmonary tuberculosis. Int Immunol (2014) 26(10):579–82. doi: 10.1093/intimm/dxu068
70. Ni B, Rajaram MV, Lafuse WP, Landes MB, Schlesinger LS. Mycobacterium tuberculosis decreases human macrophage IFN-γ responsiveness through miR-132 and miR-26a. J Immunol (2014) 193(9):4537–47. doi: 10.4049/jimmunol.1400124
71. Liu Y, Wang X, Jiang J, Cao Z, Yang B, Cheng X. Modulation of T cell cytokine production by miR-144* with elevated expression in patients with pulmonary tuberculosis. Mol Immunol (2011) 48(9-10):1084–90. doi: 10.1016/j.molimm.2011.02.001
72. Rajaram MV, Ni B, Morris JD, Brooks MN, Carlson TK, Bakthavachalu B, et al. Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and MicroRNA miR-125b. Proc Natl Acad Sci U.S.A. (2011) 108(42):17408–13. doi: 10.1073/pnas.1112660108
73. Kumar R, Halder P, Sahu SK, Kumar M, Kumari M, Jana K, et al. Identification of a novel role of ESAT-6-dependent miR-155 induction during infection of macrophages with mycobacterium tuberculosis. Cell Microbiol (2012) 14(10):1620–31. doi: 10.1111/j.1462-5822.2012.01827.x
74. Qin Y, Wang Q, Zhou Y, Duan Y, Gao Q. Inhibition of IFN-γ-induced nitric oxide dependent antimycobacterial activity by miR-155 and C/EBPβ. Int J Mol Sci (2016) 17(4):535. doi: 10.3390/ijms17040535
75. Wilson DC, Marinov AD, Blair HC, Bushnell DS, Thompson SD, Chaly Y, et al. Follistatin-like protein 1 is a mesenchyme-derived inflammatory protein and may represent a biomarker for systemic-onset juvenile rheumatoid arthritis. Arthritis Rheum (2010) 62(8):2510–6. doi: 10.1002/art.27485
76. Clutter SD, Wilson DC, Marinov AD, Hirsch R. Follistatin-like protein 1 promotes arthritis by up-regulating IFN-gamma. J Immunol (2009) 182(1):234–9. doi: 10.4049/jimmunol.182.1.234
77. Miyamae T, Marinov AD, Sowders D, Wilson DC, Devlin J, Boudreau R, et al. Follistatin-like protein-1 is a novel proinflammatory molecule. J Immunol (2006) 177(7):4758–62. doi: 10.4049/jimmunol.177.7.4758
78. Chaly Y, Marinov AD, Oxburgh L, Bushnell DS, Hirsch R. FSTL1 promotes arthritis in mice by enhancing inflammatory cytokine/chemokine expression. Arthritis Rheum (2012) 64(4):1082–8. doi: 10.1002/art.33422
79. Zhang ZM, Zhang AR, Xu M, Lou J, Qiu WQ. TLR-4/miRNA-32-5p/FSTL1 signaling regulates mycobacterial survival and inflammatory responses in mycobacterium tuberculosis-infected macrophages. Exp Cell Res (2017) 352(2):313–21. doi: 10.1016/j.yexcr.2017.02.025
80. Zhu Y, Xiao Y, Kong D, Liu H, Chen X, Chen Y, et al. Down-regulation of miR-378d increased Rab10 expression to help clearance of mycobacterium tuberculosis in macrophages. Front Cell Infect Microbiol (2020) 10:108. doi: 10.3389/fcimb.2020.00108
81. Wu Z, Lu H, Sheng J, Li L. Inductive MicroRNA-21 impairs anti-mycobacterial responses by targeting IL-12 and bcl-2. FEBS Lett (2012) 586(16):2459–67. doi: 10.1016/j.febslet.2012.06.004
82. Fu X, Zeng L, Liu Z, Ke X, Lei L, Li G. MicroRNA-206 regulates the secretion of inflammatory cytokines and MMP9 expression by targeting TIMP3 in mycobacterium tuberculosis-infected THP-1 human macrophages. Biochem Biophys Res Commun (2016) 477(2):167–73. doi: 10.1016/j.bbrc.2016.06.038
83. Chen Z, Luo T, Ma P, Ge L, Zhao R, Wang X, et al. Mycobacterium tuberculosis ESAT6 modulates host innate immunity by downregulating miR-222-3p target PTEN. Biochim Biophys Acta Mol Basis Dis (2022) 1868(1):166292. doi: 10.1016/j.bbadis.2021.166292
84. Ren X, Dong W, Feng J, Li P, Zheng Y, Wang G, et al. MiR-495 regulates cellular reactive oxygen species levels by targeting sod2 to inhibit intracellular survival of mycobacterium tuberculosis in macrophages. Infect Immun (2021) 89(12):e0031521. doi: 10.1128/iai.00315-21
85. Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell (2008) 132(1):27–42. doi: 10.1016/j.cell.2007.12.018
86. Deretic V. Autophagy in inflammation, infection, and immunometabolism. Immunity (2021) 54(3):437–53. doi: 10.1016/j.immuni.2021.01.018
87. Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and mycobacterium tuberculosis survival in infected macrophages. Cell (2004) 119(6):753–66. doi: 10.1016/j.cell.2004.11.038
88. Xu J, Wang Y, Tan X, Jing H. MicroRNAs in autophagy and their emerging roles in crosstalk with apoptosis. Autophagy (2012) 8(6):873–82. doi: 10.4161/auto.19629
89. Huang J, Brumell JH. Bacteria-autophagy interplay: A battle for survival. Nat Rev Microbiol (2014) 12(2):101–14. doi: 10.1038/nrmicro3160
90. Kumar R, Sahu SK, Kumar M, Jana K, Gupta P, Gupta UD, et al. MicroRNA 17-5p regulates autophagy in mycobacterium tuberculosis-infected macrophages by targeting mcl-1 and STAT3. Cell Microbiol (2016) 18(5):679–91. doi: 10.1111/cmi.12540
91. Duan X, Zhang T, Ding S, Wei J, Su C, Liu H, et al. MicroRNA-17-5p modulates bacille calmette-guerin growth in RAW264.7 cells by targeting ULK1. PloS One (2015) 10(9):e0138011. doi: 10.1371/journal.pone.0138011
92. Guo L, Zhao J, Qu Y, Yin R, Gao Q, Ding S, et al. MicroRNA-20a inhibits autophagic process by targeting ATG7 and ATG16L1 and favors mycobacterial survival in macrophage cells. Front Cell Infect Microbiol (2016) 6:134. doi: 10.3389/fcimb.2016.00134
93. Liu K, Hong D, Zhang F, Li X, He M, Han X, et al. MicroRNA-106a inhibits autophagy process and antimicrobial responses by targeting ULK1, ATG7, and ATG16L1 during mycobacterial infection. Front Immunol (2020) 11:610021. doi: 10.3389/fimmu.2020.610021
94. Qu Y, Ding S, Ma Z, Jiang D, Xu X, Zhang Y, et al. MiR-129-3p favors intracellular BCG survival in RAW264.7 cells by inhibiting autophagy via Atg4b. Cell Immunol (2019) 337:22–32. doi: 10.1016/j.cellimm.2019.01.004
95. Qu Y, Gao Q, Wu S, Xu T, Jiang D, Xu G. MicroRNA-142-3p inhibits autophagy and promotes intracellular survival of mycobacterium tuberculosis by targeting ATG16L1 and ATG4c. Int Immunopharmacol (2021) 101(Pt A):108202. doi: 10.1016/j.intimp.2021.108202
96. Ouimet M, Koster S, Sakowski E, Ramkhelawon B, van Solingen C, Oldebeken S, et al. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat Immunol (2016) 17(6):677–86. doi: 10.1038/ni.3434
97. Etna MP, Sinigaglia A, Grassi A, Giacomini E, Romagnoli A, Pardini M, et al. Mycobacterium tuberculosis-induced miR-155 subverts autophagy by targeting ATG3 in human dendritic cells. PloS Pathog (2018) 14(1):e1006790. doi: 10.1371/journal.ppat.1006790
98. Wang J, Yang K, Zhou L, Minhaowu, Wu Y, Zhu M, et al. MicroRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting rheb. PloS Pathog (2013) 9(10):e1003697. doi: 10.1371/journal.ppat.1003697
99. Holla S, Kurowska-Stolarska M, Bayry J, Balaji KN. Selective inhibition of IFNG-induced autophagy by miR155- and miR31-responsive WNT5A and SHH signaling. Autophagy (2014) 10(2):311–30. doi: 10.4161/auto.27225
100. Chen Z, Wang T, Liu Z, Zhang G, Wang J, Feng S, et al. Inhibition of autophagy by miR-30A induced by mycobacteria tuberculosis as a possible mechanism of immune escape in human macrophages. Jpn J Infect Dis (2015) 68(5):420–4. doi: 10.7883/yoken.JJID.2014.466
101. Kim JK, Yuk JM, Kim SY, Kim TS, Jin HS, Yang CS, et al. MicroRNA-125a inhibits autophagy activation and antimicrobial responses during mycobacterial infection. J Immunol (2015) 194(11):5355–65. doi: 10.4049/jimmunol.1402557
102. Kim JK, Lee HM, Park KS, Shin DM, Kim TS, Kim YS, et al. MIR144* inhibits antimicrobial responses against mycobacterium tuberculosis in human monocytes and macrophages by targeting the autophagy protein DRAM2. Autophagy (2017) 13(2):423–41. doi: 10.1080/15548627.2016.1241922
103. Tu H, Yang S, Jiang T, Wei L, Shi L, Liu C, et al. Elevated pulmonary tuberculosis biomarker miR-423-5p plays critical role in the occurrence of active TB by inhibiting autophagosome-lysosome fusion. Emerg Microbes Infect (2019) 8(1):448–60. doi: 10.1080/22221751.2019.1590129
104. Luo D, Wu J, Liu Y, Li P, Liang X, Xiao S, et al. Overexpression of VPS11 antagonizes the promoting effect of miR-542-3p on mycobacterium tuberculosis survival in macrophages by regulating autophagy. Microb Pathog (2022) 169:105609. doi: 10.1016/j.micpath.2022.105609
105. Liu F, Chen J, Wang P, Li H, Zhou Y, Liu H, et al. MicroRNA-27a controls the intracellular survival of mycobacterium tuberculosis by regulating calcium-associated autophagy. Nat Commun (2018) 9(1):4295. doi: 10.1038/s41467-018-06836-4
106. Chen DY, Chen YM, Lin CF, Lo CM, Liu HJ, Liao TL. MicroRNA-889 inhibits autophagy to maintain mycobacterial survival in patients with latent tuberculosis infection by targeting TWEAK. mBio (2020) 11(1):e03045–19. doi: 10.1128/mBio.03045-19
107. Dong W, Wang G, Feng J, Li P, Wang R, Lu H, et al. MiR-25 blunts autophagy and promotes the survival of mycobacterium tuberculosis by regulating NPC1. iScience (2022) 25(5):104279. doi: 10.1016/j.isci.2022.104279
108. Gu X, Gao Y, Mu DG, Fu EQ. MiR-23a-5p modulates mycobacterial survival and autophagy during mycobacterium tuberculosis infection through TLR2/MyD88/NF-κB pathway by targeting TLR2. Exp Cell Res (2017) 354(2):71–7. doi: 10.1016/j.yexcr.2017.03.039
109. Sahu SK, Kumar M, Chakraborty S, Banerjee SK, Kumar R, Gupta P, et al. MicroRNA 26a (miR-26a)/KLF4 and CREB-C/EBPβ regulate innate immune signaling, the polarization of macrophages and the trafficking of mycobacterium tuberculosis to lysosomes during infection. PloS Pathog (2017) 13(5):e1006410. doi: 10.1371/journal.ppat.1006410
110. Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, et al. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell (2008) 19(11):4651–9. doi: 10.1091/mbc.e08-03-0312
111. He C, Klionsky D. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet (2009) 43:67–93. doi: 10.1146/annurev-genet-102808-114910
112. Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, et al. A gene network regulating lysosomal biogenesis and function. Science (2009) 325(5939):473–7. doi: 10.1126/science.1174447
113. Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol (2006) 8(7):688–99. doi: 10.1038/ncb1426
114. Yoon JH, Her S, Kim M, Jang IS, Park J. The expression of damage-regulated autophagy modulator 2 (DRAM2) contributes to autophagy induction. Mol Biol Rep (2012) 39(2):1087–93. doi: 10.1007/s11033-011-0835-x
115. Behura A, Das M, Kumar A, Naik L, Mishra A, Manna D, et al. ESAT-6 impedes IL-18 mediated phagosome lysosome fusion via MicroRNA-30a upon calcimycin treatment in mycobacteria infected macrophages. Int Immunopharmacol (2021) 101(Pt A):108319. doi: 10.1016/j.intimp.2021.108319
116. Ehrenschwender M, Wajant H. The role of FasL and Fas in health and disease. Adv Exp Med Biol (2009) 647:64–93. doi: 10.1007/978-0-387-89520-8_5
117. Singh R, Letai A, Sarosiek K. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol (2019) 20(3):175–93. doi: 10.1038/s41580-018-0089-8
118. Youle RJ, Strasser A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat Rev Mol Cell Biol (2008) 9(1):47–59. doi: 10.1038/nrm2308
119. Lamkanfi M, Dixit VM. Manipulation of host cell death pathways during microbial infections. Cell Host Microbe (2010) 8(1):44–54. doi: 10.1016/j.chom.2010.06.007
120. Hmama Z, Peña-Díaz S, Joseph S, Av-Gay Y. Immunoevasion and immunosuppression of the macrophage by mycobacterium tuberculosis. Immunol Rev (2015) 264(1):220–32. doi: 10.1111/imr.12268
121. Fu B, Xue W, Zhang H, Zhang R, Feldman K, Zhao Q, et al. MicroRNA-325-3p facilitates immune escape of mycobacterium tuberculosis through targeting LNX1 via NEK6 accumulation to promote anti-apoptotic STAT3 signaling. mBio (2020) 11(3):e00557–20. doi: 10.1128/mBio.00557-20
122. Zhang G, Liu X, Wang W, Cai Y, Li S, Chen Q, et al. Down-regulation of miR-20a-5p triggers cell apoptosis to facilitate mycobacterial clearance through targeting JNK2 in human macrophages. Cell Cycle (2016) 15(18):2527–38. doi: 10.1080/15384101.2016.1215386
123. Liu G, Wan Q, Li J, Hu X, Gu X, Xu S. Silencing miR-125b-5p attenuates inflammatory response and apoptosis inhibition in mycobacterium tuberculosis-infected human macrophages by targeting DNA damage-regulated autophagy modulator 2 (DRAM2). Cell Cycle (2020) 19(22):3182–94. doi: 10.1080/15384101.2020.1838792
124. Wu Y, Guo Z, Yao K, Miao Y, Liang S, Liu F, et al. The transcriptional foundations of Sp110-mediated macrophage (RAW264.7) resistance to mycobacterium tuberculosis H37Ra. Sci Rep (2016) 6:22041. doi: 10.1038/srep22041
125. Ghorpade DS, Leyland R, Kurowska-Stolarska M, Patil SA, Balaji KN. MicroRNA-155 is required for mycobacterium bovis BCG-mediated apoptosis of macrophages. Mol Cell Biol (2012) 32(12):2239–53. doi: 10.1128/mcb.06597-11
126. Yang S, Li F, Jia S, Zhang K, Jiang W, Shang Y, et al. Early secreted antigen ESAT-6 of mycobacterium tuberculosis promotes apoptosis of macrophages via targeting the MicroRNA155-SOCS1 interaction. Cell Physiol Biochem (2015) 35(4):1276–88. doi: 10.1159/000373950
127. Huang J, Jiao J, Xu W, Zhao H, Zhang C, Shi Y, et al. MiR-155 is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO3. Mol Med Rep (2015) 12(5):7102–8. doi: 10.3892/mmr.2015.4250
128. Zhang X, Tang N, Hadden TJ, Rishi AK. Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta (2011) 1813(11):1978–86. doi: 10.1016/j.bbamcr.2011.03.010
129. Tripathi A, Srivastava V, Singh B. Hsa-let-7b-5p facilitates mycobacterium tuberculosis survival in THP-1 human macrophages by fas downregulation. FEMS Microbiol Lett (2018) 365(7). doi: 10.1093/femsle/fny040
130. Xi X, Zhang C, Han W, Zhao H, Zhang H, Jiao J. MicroRNA-223 is upregulated in active tuberculosis patients and inhibits apoptosis of macrophages by targeting FOXO3. Genet Test Mol Biomarkers (2015) 19(12):650–6. doi: 10.1089/gtmb.2015.0090
131. Liu Y, Jiang J, Wang X, Zhai F, Cheng X. MiR-582-5p is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO1. PloS One (2013) 8(10):e78381. doi: 10.1371/journal.pone.0078381
132. Zhang D, Yi Z, Fu Y. Downregulation of miR-20b-5p facilitates mycobacterium tuberculosis survival in RAW 264.7 macrophages via attenuating the cell apoptosis by mcl-1 upregulation. J Cell Biochem (2019) 120(4):5889–96. doi: 10.1002/jcb.27874
133. Tamgue O, Gcanga L, Ozturk M, Whitehead L, Pillay S, Jacobs R, et al. Differential targeting of c-maf, Bach-1, and Elmo-1 by MicroRNA-143 and MicroRNA-365 promotes the intracellular growth of mycobacterium tuberculosis in alternatively IL-4/IL-13 activated macrophages. Front Immunol (2019) 10:421. doi: 10.3389/fimmu.2019.00421
134. Ma J, Chen XL, Sun Q. MicroRNA-579 upregulation mediates death of human macrophages with mycobacterium tuberculosis infection. Biochem Biophys Res Commun (2019) 518(2):219–26. doi: 10.1016/j.bbrc.2019.08.035
135. Qin LS, Jia PF, Zhang ZQ, Zhang SM. ROS-p53-cyclophilin-D signaling mediates salinomycin-induced glioma cell necrosis. J Exp Clin Cancer Res (2015) 34(1):57. doi: 10.1186/s13046-015-0174-1
136. Park JS, Tamayo MH, Gonzalez-Juarrero M, Orme IM, Ordway DJ. Virulent clinical isolates of mycobacterium tuberculosis grow rapidly and induce cellular necrosis but minimal apoptosis in murine macrophages. J Leukoc Biol (2006) 79(1):80–6. doi: 10.1189/jlb.0505250
137. Sun Q, Shen X, Wang P, Ma J, Sha W. Targeting cyclophilin-d by miR-1281 protects human macrophages from mycobacterium tuberculosis-induced programmed necrosis and apoptosis. Aging (Albany NY) (2019) 11(24):12661–73. doi: 10.18632/aging.102593
138. Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity (2014) 41(1):14–20. doi: 10.1016/j.immuni.2014.06.008
139. Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, et al. Krüppel-like factor 4 regulates macrophage polarization. J Clin Invest (2011) 121(7):2736–49. doi: 10.1172/jci45444
140. Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez RG, Rosenthal N, et al. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci U.S.A. (2009) 106(41):17475–80. doi: 10.1073/pnas.0908641106
141. Mishra BB, Rathinam VA, Martens GW, Martinot AJ, Kornfeld H, Fitzgerald KA, et al. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1β. Nat Immunol (2013) 14(1):52–60. doi: 10.1038/ni.2474
142. Lou J, Wang Y, Zhang Z, Qiu W. MiR-20b inhibits mycobacterium tuberculosis induced inflammation in the lung of mice through targeting NLRP3. Exp Cell Res (2017) 358(2):120–8. doi: 10.1016/j.yexcr.2017.06.007
143. Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell (2013) 154(1):240–51. doi: 10.1016/j.cell.2013.06.009
144. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE V7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res (2012) 22(9):1775–89. doi: 10.1101/gr.132159.111
145. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet (2015) 47(3):199–208. doi: 10.1038/ng.3192
146. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell (2007) 129(7):1311–23. doi: 10.1016/j.cell.2007.05.022
147. Nojima T, Proudfoot NJ. Mechanisms of LncRNA biogenesis as revealed by nascent transcriptomics. Nat Rev Mol Cell Biol (2022) 23(6):389–406. doi: 10.1038/s41580-021-00447-6
148. Liu SJ, Dang HX, Lim DA, Feng FY, Maher CA. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer (2021) 21(7):446–60. doi: 10.1038/s41568-021-00353-1
149. Agliano F, Rathinam VA, Medvedev AE, Vanaja SK, Vella AT. Long noncoding RNAs in host-pathogen interactions. Trends Immunol (2019) 40(6):492–510. doi: 10.1016/j.it.2019.04.001
150. Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science (2013) 341(6147):789–92. doi: 10.1126/science.1240925
151. Li D, Gao C, Zhao L, Zhang Y. Inflammatory response is modulated by lincRNACox2 via the NF−κB pathway in macrophages infected by mycobacterium tuberculosis. Mol Med Rep (2020) 21(6):2513–21. doi: 10.3892/mmr.2020.11053
152. Li M, Cui J, Niu W, Huang J, Feng T, Sun B, et al. Long non-coding PCED1B-AS1 regulates macrophage apoptosis and autophagy by sponging miR-155 in active tuberculosis. Biochem Biophys Res Commun (2019) 509(3):803–9. doi: 10.1016/j.bbrc.2019.01.005
153. Bai H, Wu Q, Hu X, Wu T, Song J, Liu T, et al. Clinical significance of lnc-AC145676.2.1-6 and lnc-TGS1-1 and their variants in western Chinese tuberculosis patients. Int J Infect Dis (2019) 84:8–14. doi: 10.1016/j.ijid.2019.04.018
154. Ke Z, Lu J, Zhu J, Yang Z, Jin Z, Yuan L. Down-regulation of lincRNA-EPS regulates apoptosis and autophagy in BCG-infected RAW264.7 macrophages via JNK/MAPK signaling pathway. Infect Genet Evol (2020) 77:104077. doi: 10.1016/j.meegid.2019.104077
155. Subuddhi A, Kumar M, Majumder D, Sarkar A, Ghosh Z, Vasudevan M, et al. Unraveling the role of H3K4 trimethylation and lncRNA HOTAIR in SATB1 and DUSP4-dependent survival of virulent mycobacterium tuberculosis in macrophages. Tuberculosis (Edinb) (2020) 120:101897. doi: 10.1016/j.tube.2019.101897
156. Wang Y, Zhong H, Xie X, Chen CY, Huang D, Shen L, et al. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc Natl Acad Sci U.S.A. (2015) 112(29):E3883–92. doi: 10.1073/pnas.1501662112
157. Fu Y, Xu X, Xue J, Duan W, Yi Z. Deregulated lncRNAs in b cells from patients with active tuberculosis. PloS One (2017) 12(1):e0170712. doi: 10.1371/journal.pone.0170712
158. Fu Y, Gao K, Tao E, Li R, Yi Z. Aberrantly expressed long non-coding RNAs in CD8+ T cells response to active tuberculosis. J Cell Biochem (2017) 118(12):4275–84. doi: 10.1002/jcb.26078
159. Wang J, Wang H, Kang L. LINC00870 regulates Th1/Th2 via the JAK/STAT pathway in peripheral blood mononuclear cells infected with mycobacterium tuberculosis. Int Immunopharmacol (2022) 102:107188. doi: 10.1016/j.intimp.2020.107188
160. Jiang F, Lou J, Zheng XM, Yang XY. LncRNA MIAT regulates autophagy and apoptosis of macrophage infected by mycobacterium tuberculosis through the miR-665/ULK1 signaling axis. Mol Immunol (2021) 139:42–9. doi: 10.1016/j.molimm.2021.07.023
161. Yao Q, Xie Y, Xu D, Qu Z, Wu J, Zhou Y, et al. Lnc-EST12, which is negatively regulated by mycobacterial EST12, suppresses antimycobacterial innate immunity through its interaction with FUBP3. Cell Mol Immunol (2022) 19(8):883–97. doi: 10.1038/s41423-022-00878-x
162. Luo XB, Li LT, Xi JC, Liu HT, Liu Z, Yu L, et al. Negative pressure promotes macrophage M1 polarization after mycobacterium tuberculosis infection via the lncRNA XIST/MicroRNA-125b-5p/A20/NF-κB axis. Ann N Y Acad Sci (2022) 1514(1):116–131. doi: 10.1111/nyas.14781
163. Siddle KJ, Deschamps M, Tailleux L, Nédélec Y, Pothlichet J, Lugo-Villarino G, et al. A genomic portrait of the genetic architecture and regulatory impact of MicroRNA expression in response to infection. Genome Res (2014) 24(5):850–9. doi: 10.1101/gr.161471.113
164. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet (2019) 20(11):675–91. doi: 10.1038/s41576-019-0158-7
165. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U.S.A. (1976) 73(11):3852–6. doi: 10.1073/pnas.73.11.3852
166. Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, et al. Circular RNA is expressed across the eukaryotic tree of life. PloS One (2014) 9(6):e90859. doi: 10.1371/journal.pone.0090859
167. Maass PG, Glažar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl) (2017) 95(11):1179–89. doi: 10.1007/s00109-017-1582-9
168. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PloS Genet (2013) 9(9):e1003777. doi: 10.1371/journal.pgen.1003777
169. Thomson DW, Dinger ME. Endogenous MicroRNA sponges: Evidence and controversy. Nat Rev Genet (2016) 17(5):272–83. doi: 10.1038/nrg.2016.20
170. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient MicroRNA sponges. Nature (2013) 495(7441):384–8. doi: 10.1038/nature11993
171. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol (2015) 22(3):256–64. doi: 10.1038/nsmb.2959
172. Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics (2017) 7(16):3842–55. doi: 10.7150/thno.19764
173. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res (2016) 44(6):2846–58. doi: 10.1093/nar/gkw027
174. Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol (2022) 19(3):188–206. doi: 10.1038/s41571-021-00585-y
175. Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell (2017) 67(2):214–27.e7. doi: 10.1016/j.molcel.2017.05.023
176. Shi Q, Wang J, Yang Z, Liu Y. CircAGFG1modulates autophagy and apoptosis of macrophages infected by mycobacterium tuberculosis via the notch signaling pathway. Ann Transl Med (2020) 8(10):645. doi: 10.21037/atm.2020-20-3048
177. Luo HL, Pi J, Zhang JA, Yang EZ, Xu H, Luo H, et al. Circular RNA TRAPPC6B inhibits intracellular mycobacterium tuberculosis growth while inducing autophagy in macrophages by targeting MicroRNA-874-3p. Clin Transl Immunol (2021) 10(2):e1254. doi: 10.1002/cti2.1254
178. Huang Z, Yao F, Liu J, Xu J, Guo Y, Su R, et al. Up-regulation of circRNA-0003528 promotes mycobacterium tuberculosis associated macrophage polarization via down-regulating miR-224-5p, miR-324-5p and miR-488-5p and up-regulating CTLA4. Aging (Albany NY) (2020) 12(24):25658–72. doi: 10.18632/aging.104175
179. Deng Q, Huang J, Yan J, Mao E, Chen H, Wang C. Circ_0001490/miR-579-3p/FSTL1 axis modulates the survival of mycobacteria and the viability, apoptosis and inflammatory response in mycobacterium tuberculosis-infected macrophages. Tuberculosis (Edinb) (2021) 131:102123. doi: 10.1016/j.tube.2021.102123
180. Wu M, Liu Z, Zhang S. Down-regulation of hsa_circ_0045474 induces macrophage autophagy in tuberculosis via miR-582-5p/TNKS2 axis. Innate Immun (2022) 28(1):11–8. doi: 10.1177/17534259211064285
181. Zhang Z, Yang T, Xiao J. Circular RNAs: Promising biomarkers for human diseases. EBioMedicine (2018) 34:267–74. doi: 10.1016/j.ebiom.2018.07.036
182. Campbell J, Trajman A, Cook V, Johnston J, Adjobimey M, Ruslami R, et al. Adverse events in adults with latent tuberculosis infection receiving daily rifampicin or isoniazid: Post-hoc safety analysis of two randomised controlled trials. Lancet Infect Dis (2020) 20(3):318–29. doi: 10.1016/s1473-3099(19)30575-4
183. Lan Z, Ahmad N, Baghaei P, Barkane L, Benedetti A, Brode S, et al. Drug-associated adverse events in the treatment of multidrug-resistant tuberculosis: an individual patient data meta-analysis. Lancet Respir Med (2020) 8(4):383–94. doi: 10.1016/s2213-2600(20)30047-3
184. Barry SE, Ellis M, Yang Y, Guan G, Wang X, Britton WJ, et al. Identification of a plasma MicroRNA profile in untreated pulmonary tuberculosis patients that is modulated by anti-mycobacterial therapy. J Infect (2018) 77(4):341–8. doi: 10.1016/j.jinf.2018.03.006
185. Pedersen JL, Barry SE, Bokil NJ, Ellis M, Yang Y, Guan G, et al. High sensitivity and specificity of a 5-analyte protein and MicroRNA biosignature for identification of active tuberculosis. Clin Transl Immunol (2021) 10(6):e1298. doi: 10.1002/cti2.1298
186. Gao SH, Chen CG, Zhuang CB, Zeng YL, Zeng ZZ, Wen PH, et al. Integrating serum MicroRNAs and electronic health records improved the diagnosis of tuberculosis. J Clin Lab Anal (2021) 35(8):e23871. doi: 10.1002/jcla.23871
187. Lyu L, Zhang X, Li C, Yang T, Wang J, Pan L, et al. Small RNA profiles of serum exosomes derived from individuals with latent and active tuberculosis. Front Microbiol (2019) 10:1174. doi: 10.3389/fmicb.2019.01174
188. Chen ZL, Wei LL, Shi LY, Li M, Jiang TT, Chen J, et al. Screening and identification of lncRNAs as potential biomarkers for pulmonary tuberculosis. Sci Rep (2017) 7(1):16751. doi: 10.1038/s41598-017-17146-y
189. Hu X, Liao S, Bai H, Gupta S, Zhou Y, Zhou J, et al. Long noncoding RNA and predictive model to improve diagnosis of clinically diagnosed pulmonary tuberculosis. J Clin Microbiol (2020) 58(7):e01973–19. doi: 10.1128/jcm.01973-19
190. Chen J, Wu L, Lv Y, Liu T, Guo W, Song J, et al. Screening of long non-coding RNAs biomarkers for the diagnosis of tuberculosis and preliminary construction of a clinical diagnosis model. Front Microbiol (2022) 13:774663. doi: 10.3389/fmicb.2022.774663
191. Ye T, Zhang J, Zeng X, Xu Y, Li J. LncRNA CCAT1 is overexpressed in tuberculosis patients and predicts their survival. Immun Inflammation Dis (2022) 10(2):218–24. doi: 10.1002/iid3.565
192. Huang Z, Su R, Deng Z, Xu J, Peng Y, Luo Q, et al. Identification of differentially expressed circular RNAs in human monocyte derived macrophages response to mycobacterium tuberculosis infection. Sci Rep (2017) 7(1):13673. doi: 10.1038/s41598-017-13885-0
193. Liu H, Lu G, Wang W, Jiang X, Gu S, Wang J, et al. A panel of circRNAs in the serum serves as biomarkers for mycobacterium tuberculosis infection. Front Microbiol (2020) 11:1215. doi: 10.3389/fmicb.2020.01215
194. Huang ZK, Yao FY, Xu JQ, Deng Z, Su RG, Peng YP, et al. Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from active tuberculosis patients. Cell Physiol Biochem (2018) 45(3):1230–40. doi: 10.1159/000487454
195. Zhang H, Sun Z, Wei W, Liu Z, Fleming J, Zhang S, et al. Identification of serum MicroRNA biomarkers for tuberculosis using RNA-seq. PloS One (2014) 9(2):e88909. doi: 10.1371/journal.pone.0088909
196. Wu J, Lu C, Diao N, Zhang S, Wang S, Wang F, et al. Analysis of MicroRNA expression profiling identifies miR-155 and miR-155* as potential diagnostic markers for active tuberculosis: a preliminary study. Hum Immunol (2012) 73(1):31–7. doi: 10.1016/j.humimm.2011.10.003
197. Xin H, Cao X, Du Y, Yan J, He R, Liu Z, et al. The association between circulating MicroRNAs and the risk of active disease development from latent tuberculosis infection: a nested case-control study. Microbiol Spectr (2022) 10(3):e0262521. doi: 10.1128/spectrum.02625-21
198. Alipoor SD, Tabarsi P, Varahram M, Movassaghi M, Dizaji MK, Folkerts G, et al. Serum exosomal miRNAs are associated with active pulmonary tuberculosis. Dis Markers (2019) 2019:1907426. doi: 10.1155/2019/1907426
199. Fang Y, Zhao J, Wang X, Wang X, Wang L, Liu L, et al. Identification of differentially expressed lncRNAs as potential plasma biomarkers for active tuberculosis. Tuberculosis (Edinb) (2021) 128:102065. doi: 10.1016/j.tube.2021.102065
200. Fu Y, Wang J, Qiao J, Yi Z. Signature of circular RNAs in peripheral blood mononuclear cells from patients with active tuberculosis. J Cell Mol Med (2019) 23(3):1917–25. doi: 10.1111/jcmm.14093
201. Zhang X, Zhang Q, Wu Q, Tang H, Ye L, Zhang Q, et al. Integrated analyses reveal hsa_circ_0028883 as a diagnostic biomarker in active tuberculosis. Infect Genet Evol (2020) 83:104323. doi: 10.1016/j.meegid.2020.104323
202. Yi Z, Gao K, Li R, Fu Y. Dysregulated circRNAs in plasma from active tuberculosis patients. J Cell Mol Med (2018) 22(9):4076–84. doi: 10.1111/jcmm.13684
203. Carranza C, Herrera MT, Guzmán-Beltrán S, Salgado-Cantú MG, Salido-Guadarrama I, Santiago E, et al. A dual marker for monitoring MDR-TB treatment: Host-derived miRNAs and m. tuberculosis-derived RNA sequences in serum. Front Immunol (2021) 12:760468. doi: 10.3389/fimmu.2021.760468
204. Sun X, Liu K, Zhao Y, Zhang T. High miRNA-378 expression has high diagnostic values for pulmonary tuberculosis and predicts adverse outcomes. BMC Mol Cell Biol (2022) 23(1):14. doi: 10.1186/s12860-022-00413-w
205. Zhao J, Gao S, Chen C, Li H, Wang S, Yu Y, et al. Screening and identification of differentially expressed long non-coding RNAs in multidrug-resistant tuberculosis. PeerJ (2022) 10:e12776. doi: 10.7717/peerj.12776
206. Tostmann A, Boeree MJ, Aarnoutse RE, de Lange WC, van der Ven AJ, Dekhuijzen R. Antituberculosis drug-induced hepatotoxicity: Concise up-to-date review. J Gastroenterol Hepatol (2008) 23(2):192–202. doi: 10.1111/j.1440-1746.2007.05207.x
207. Bakshi S, Kaur M, Saini N, Mir AA, Duseja A, Sinha SK, et al. Altered expressions of circulating MicroRNAs 122 and 192 during antitubercular drug induced liver injury indicating their role as potential biomarkers. Hum Exp Toxicol (2021) 40(9):1474–84. doi: 10.1177/0960327121997975
208. Li B, Ren Q, Li Y, Tian S, Chong Y, Sun S, et al. Screening differential circular RNA expression profiles reveals the regulatory role of circMARS in anti-tuberculosis drug-induced liver injury. J Cell Mol Med (2022) 26(4):1050–9. doi: 10.1111/jcmm.17157
209. Uno S, Nishimura T, Nishio K, Kohsaka A, Tamizu E, Nakano Y, et al. Potential biomarker enhancing the activity of tuberculosis, hsa-miR-346. Tuberculosis (Edinb) (2021) 129:102101. doi: 10.1016/j.tube.2021.102101
210. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol (2018) 141(4):1202–7. doi: 10.1016/j.jaci.2017.08.034
211. Chen X, Hu TY. Strategies for advanced personalized tuberculosis diagnosis: Current technologies and clinical approaches. Precis Clin Med (2021) 4(1):35–44. doi: 10.1093/pcmedi/pbaa041
Keywords: Mycobacterium tuberculosis, microRNA, long noncoding RNA, circular RNA, immune regulation, biomarker
Citation: Liang S, Ma J, Gong H, Shao J, Li J, Zhan Y, Wang Z, Wang C and Li W (2022) Immune regulation and emerging roles of noncoding RNAs in Mycobacterium tuberculosis infection. Front. Immunol. 13:987018. doi: 10.3389/fimmu.2022.987018
Received: 05 July 2022; Accepted: 29 August 2022;
Published: 13 October 2022.
Edited by:
Hongbo Shen, Tongji University, ChinaReviewed by:
Hui Wang, University of Pennsylvania, United StatesBingdong Zhu, Lanzhou University, China
Copyright © 2022 Liang, Ma, Gong, Shao, Li, Zhan, Wang, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weimin Li, weimi003@scu.edu.cn; Chengdi Wang, chengdi_wang@scu.edu.cn; Zhoufeng Wang, wangzhoufeng@wchscu.cn
†These authors have contributed equally to this work and share first authorship
 Shufan Liang
Shufan Liang Jiechao Ma
Jiechao Ma Hanlin Gong3†
Hanlin Gong3† Jun Shao
Jun Shao Jingwei Li
Jingwei Li Yuejuan Zhan
Yuejuan Zhan Zhoufeng Wang
Zhoufeng Wang Chengdi Wang
Chengdi Wang Weimin Li
Weimin Li