- 1Istituto di Ricerca in Biomedicina (IRB), Università della Svizzera italiana, Bellinzona, Switzerland
- 2Euler institute, Università della Svizzera italiana, Lugano-Viganello, Switzerland
- 3Department of Information Technology and Electrical Engineering, Swiss Federal Institute of Technology Zurich (ETHZ) Zürich, Zürich, Switzerland
The migration of immune cells plays a key role in inflammation. This is evident in the fact that inflammatory stimuli elicit a broad range of migration patterns in immune cells. Since these patterns are pivotal for initiating the immune response, their dysregulation is associated with life-threatening conditions including organ failure, chronic inflammation, autoimmunity, and cancer, amongst others. Over the last two decades, thanks to advancements in the intravital microscopy technology, it has become possible to visualize cell migration in living organisms with unprecedented resolution, helping to deconstruct hitherto unexplored aspects of the immune response associated with the dynamism of cells. However, a comprehensive classification of the main motility patterns of immune cells observed in vivo, along with their relevance to the inflammatory process, is still lacking. In this review we defined cell actions as motility patterns displayed by immune cells, which are associated with a specific role during the immune response. In this regard, we summarize the main actions performed by immune cells during intravital microscopy studies. For each of these actions, we provide a consensus name, a definition based on morphodynamic properties, and the biological contexts in which it was reported. Moreover, we provide an overview of the computational methods that were employed for the quantification, fostering an interdisciplinary approach to study the immune system from imaging data.
Introduction
Inflammation is a highly dynamic process that involves changes in cell behavior both at the site of the insult as well as at distant organs (1, 2). Immune cells are key players in this process, as they relocate into inflamed tissues, and secrete mediators of inflammation that orchestrate a cascade of immune reactions (3–7).
Over the last two decades, intravital microscopy (MP-IVM) techniques have consolidated the in vivo analysis of the immune response. Videos acquired via MP-IVM capture the behavior of immune cells, including their migratory and interaction patterns, in organs of living organisms (8–11). However, the quantification of these videos remains challenging. This is due to a range of factors, such as the complexity of the in vivo environment, which includes a multitude of cell types and anatomical structures (12) or the high plasticity and dynamism of the migration patterns displayed by immune cells, which change over time. Moreover, numerous technical artifacts introduced by the intravital imaging procedure also affect the analysis to a large extent (13).
The recently established image-based systems biology paradigm offers a unique opportunity to study cell behavior in vivo, as it combines imaging data and computational methods (14). Analogously, recently developed computer vision methods for action recognition (AR) have enabled the analysis of the complex behavior of humans associated with specific actions such as walking, jumping, etc. (15). This is a particularly challenging task, as human actions may be hierarchical in their nature, composed of multiple actors, or captured by different imaging modalities (15). Interestingly, these three challenges are shared with the quantification of the immune cell behavior in intravital movies, as they display different morphodynamics, undergo cell-to-cell interactions, and can be imaged in different anatomical regions. Hence, in line with AR, we define cell actions as motility patterns associated with relevant biological functions to dissect leukocyte behavior.
To this end, we collected and reported from the literature a list of actions displayed by immune cells in different organs during key inflammatory processes. A summary of the diseases, organs, and studies included in this review is reported in Table 1. Moreover, we provide a consensus definition for each action and its biological relevance during inflammation. Lastly, we report the computational methods currently available for the detection and quantification of each reviewed action.
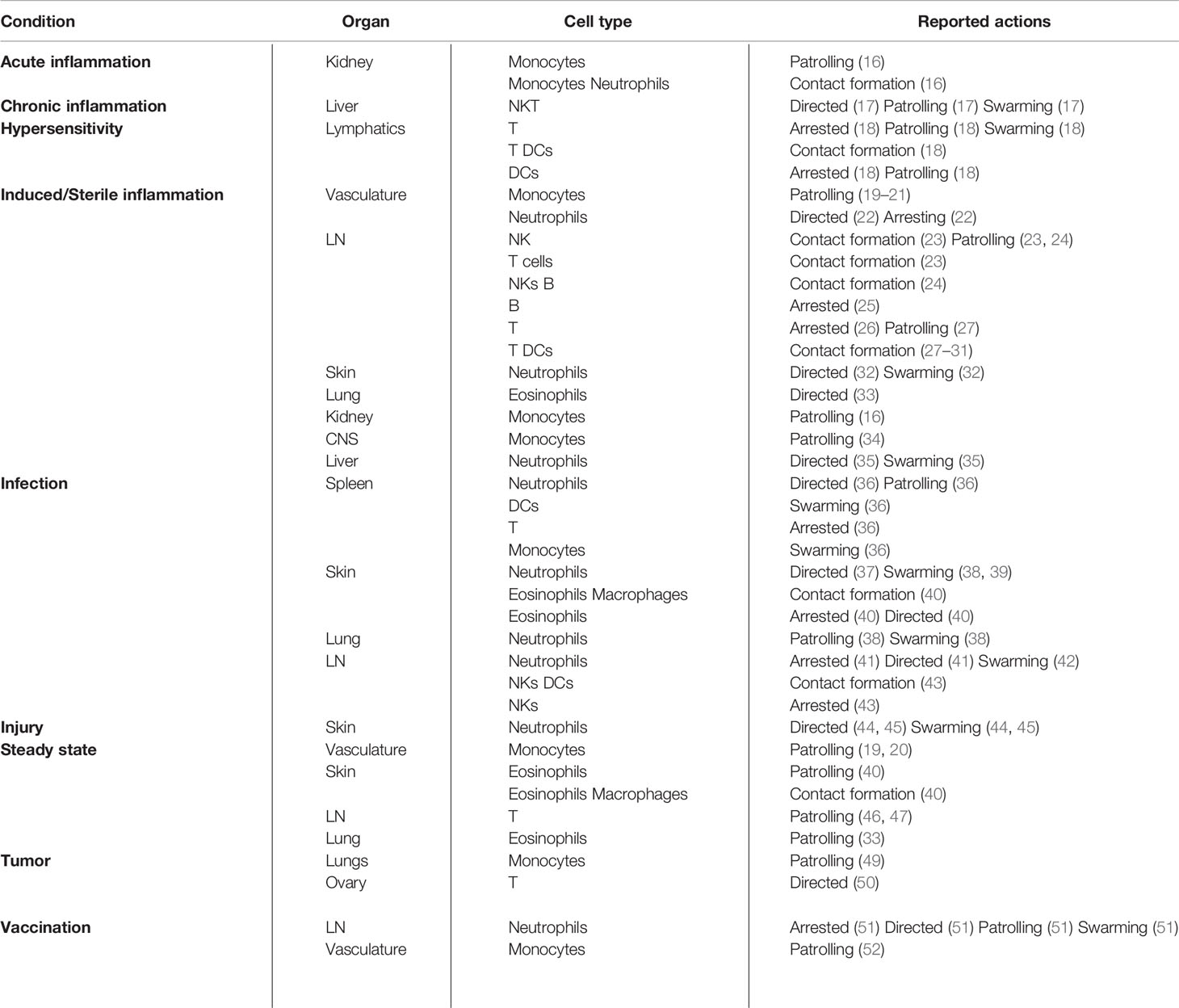
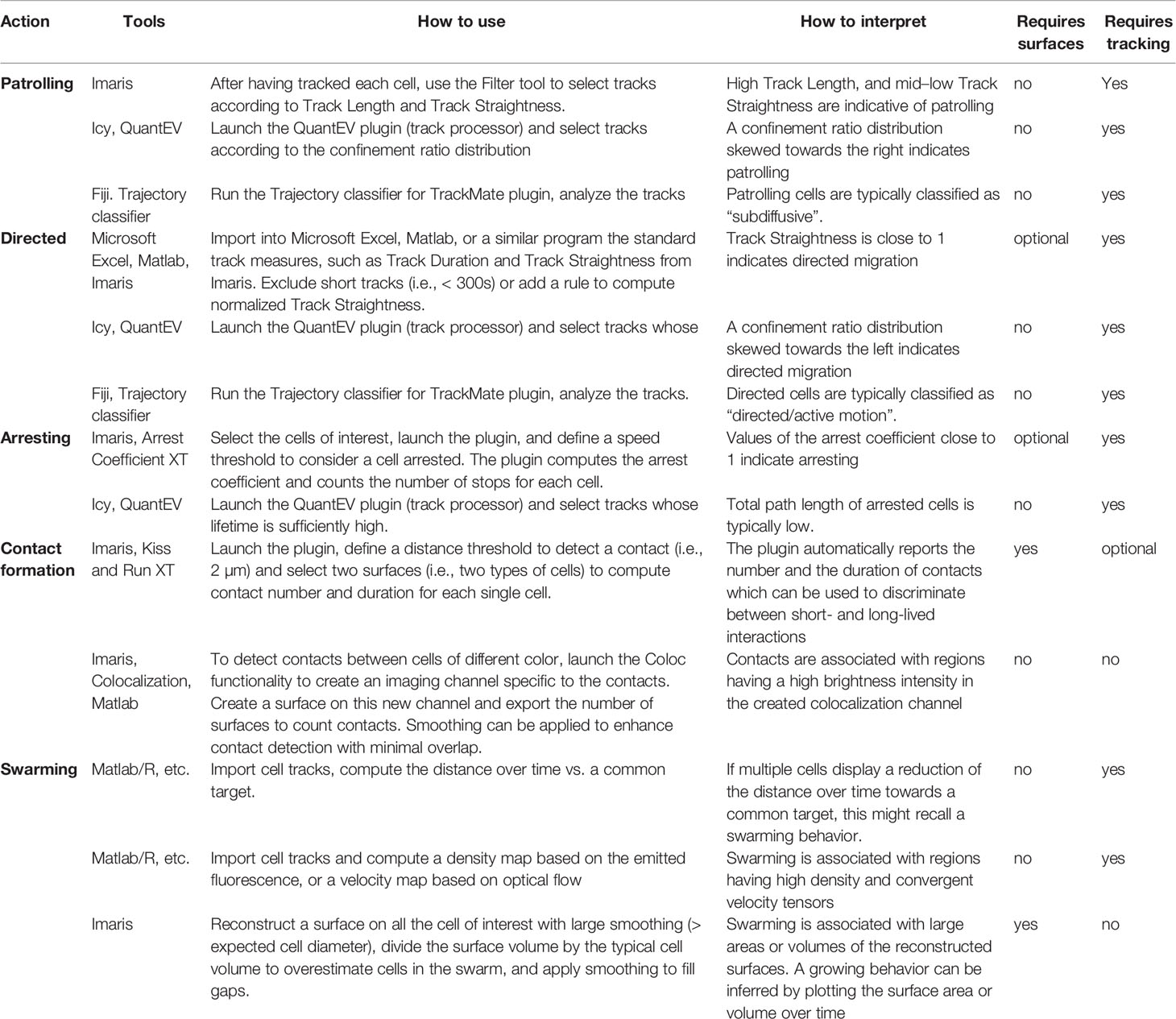
Table 1 Summary of the actions described in different inflammatory conditions, organs, and cell types.
Intravital Imaging Workflow
The application of MP-IVM for the imaging of multiple cells during the inflammatory process involves the following steps.
Cell labeling
Different methods are available, including the adoptive transfer of cells from transgenic animals expressing a fluorophore-tagged protein, in vitro labeling with fluorescent dies, or the injection of fluorescently labeled antibodies that specifically bind to the cells of interest. Available optical probes for MP-IVM and fluorescent proteins are comprehensively reviewed elsewhere (53, 54) and are beyond the scope of this work.
Surgery
The next step to perform a MP-IVM protocol is to select the proper surgical model, to enable the exposure and immobilization of the targeted organ (Figure 1A) (8, 9, 55). Although this typically requires minimally invasive surgery, more advanced surgical setups can be employed for long-term imaging of internal organs, including gut (56), brain and spinal cord (57), primary tumors and metastasis (58, 59) amongst others (60–63).
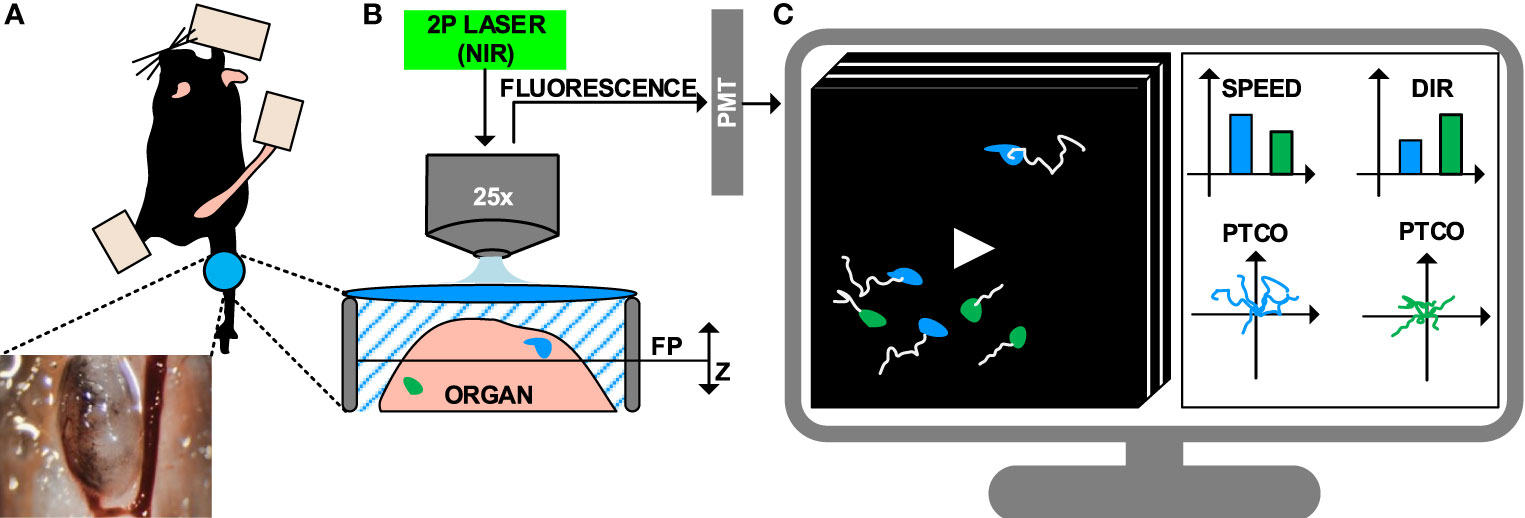
Figure 1 Intravital imaging of the immune system under inflammatory conditions. (A) Representation of the surgical model used to perform intravital imaging in the murine popliteal lymph node, including a minimally invasive surgery and imaging through a transparent window. (B) Example of intravital imaging setup based on 2-photon microscopy, including a pulsed laser with near-infrared (NIR) emission wavelengths and photomultipliers (PMT) for fluorescence detection. (C) 4D videos (3D z-stacks over time) capturing cell motility are acquired and visualized on a computer. Cells are tracked (white lines) to compute metrics such as speed, directionality (dir), and plotting of tracks with a common origin (PTCO).
Image Acquisition
Once surgery is completed, the anesthetized animal is transported to the microscope where image acquisition is performed. The fluorophores present in the sample are excited, and the resulting emitted fluorescence is acquired. A number of microscopy platforms are available for intravital imaging of the immune system (11, 64). Amongst these, multiphoton microscopy (MP-IVM) allows for deeper tissue penetration (by reducing scattering and autofluorescence) and prolonged acquisition time (by significantly reducing photodamage) (65, 66). This is achieved by employing a pulsed laser that emits excitation photons in the near-infrared range (NIR). The simultaneous absorption of multiple photons by a single fluorophore leads to the emission of one photon with higher energy. Finally, emitted photons are collected with detectors such as high-sensitivity photomultipliers (Figure 1B) (67).
4D imaging data (time lapses of 3D z-stacks) are obtained at different time points by sliding the excitation point throughout the sample on a focal plane and repeating this process by moving the focal plane along the z-axis. One drawback of this process is the reduced acquisition speed of MP-IVM. Conversely, other technologies such as resonant scanners or spinning disk confocal microscopy may be employed to capture rapid biological processes such as short-lived interactions or morphological changes (68).
Data Analysis
The standard pipeline to analyze IVM videos consists of tracking the cells in the field of view, then computing motility measures from the cell trajectories (Figure 1C) (69, 70). Computer vision stands as a promising approach to automatically performing cell tracking (71). However, to date, a series of limitations hamper the accuracy of state-of-the-art automatic tracking algorithms when applied to immune cells observed via MP-IVM. For example, the high plasticity of the immune cells might yield to double tracking errors (69). Additionally, the high cell density associated with biological processes such as swarm formation hinder the distinction of individual cells. Lastly, technical artifacts introduced by the intravital imaging, such as varying signal-to-noise ratio across space and time, might affect the overall experimental readout (72, 73).
Therefore, to obtain insightful results, manual tracking and editing of automatically generated tracks are still required. Indeed, manual tracking significantly minimizes tracking errors and improves the accuracy of motility measures used to quantify cell migration and interaction (13). However, these procedures are time-consuming and prone to bias from each individual researcher.
Common Measures of Cell Motility
A variety of measures formerly used to study particle dynamics in physics have been adopted by image analysts to study cell motility in different experimental setups, including intravital imaging (69, 70). Amongst these, speed and confinement ratio (also known as directionality, or meandering index) are the most common parameters when performing MP-IVM analysis of immune cells. Speed is defined as the ratio between the track length and the track duration, while the confinement ratio is defined as the distance between the first and the last point of the trajectory (displacement) divided by the total length of the track followed by a cell. This parameter tends to 1 for straight tracks, but decreases to 0 for circular tracks.
The aforementioned measures can be computed either on entire tracks (track-based) or on track fragments (step-based) (51). Track-based measures describe the overall motility of a cell for the entire period of observation. An important limitation in the application of these measures is that tracking errors can compromise the readout. Additionally, a cell whose behavior varies over time is represented by a single average value, yielding to an information loss. By contrast, step-based measures are computed amongst adjacent time points only, or on a temporal window, limiting the temporal propagation of errors. Moreover, step-based measures further allow the quantification of instantaneous changes in cell behavior, which may occur over time, rather than taking an average value of the entire track. If cells cannot be tracked for long periods of time, a step-based measure may represent the only possible choice for quantification.
More advanced measurements to evaluate the directionality of cells have also been defined. For example, the mean squared displacement analysis (MSD) evaluates the diffusivity of a particle by comparing it with the expected motion of a random walk. This measure can be represented by plotting the squared displacement over consecutive time steps, resulting in a straight line for a randomly diffusive process. Conversely, plots above this line refer to super diffusive, or directed, processes, while plots below it indicate a confined motion (74). The motility coefficient, expressed in µm2/min, is a diffusivity measure derived from the MSD (70). This coefficient considers the square of the cell displacement over time, which can be inferred as the slope of the MSD plot and can be used to compare migratory modes of different cells. In addition, the distribution of the turning angles can be evaluated to assess how much a cell deviates from its previous path. Following this analysis, narrow distribution centered on small angles are indicative of straight trajectories (75).
Actions Performed by Individual Cells
Patrolling
Patrolling, also referred as scanning (76) or stochastic migration (46, 77), is an action associated with random-like movement characterized by long tracks in a confined area, which results in low directionality (Figures 2A, B) (19). The speed of patrolling cells varies according to the cell type, conditions, and anatomical site. For instance, monocytes exhibited a speed of 36 µm/min in the endothelium of carotid arteries and 9 µm/min in the mesenteric venules (52), while B cells exhibited a speed of 6 um/min in the lymph node follicles.
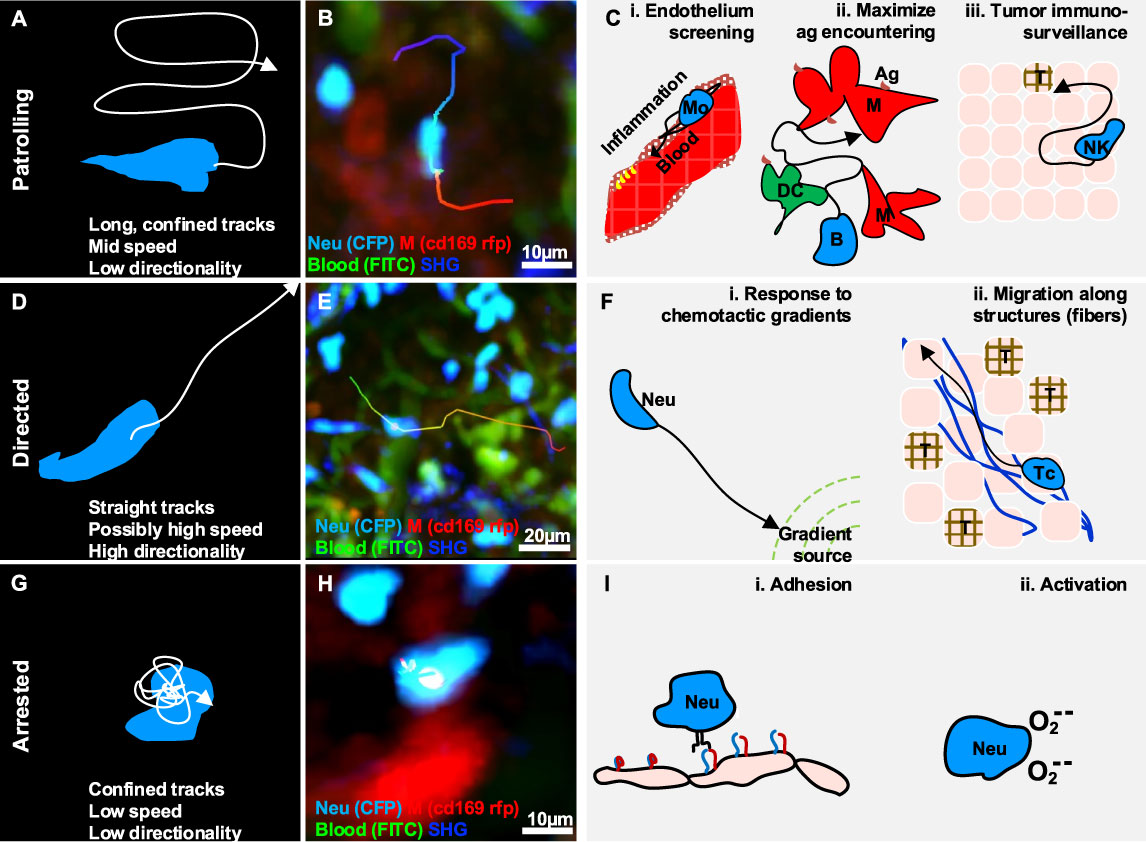
Figure 2 Gallery of actions displayed by individual immune cells. (A) Illustration of a patrolling cell, with the characteristic long track in a confined area, which is associated with mid-speed and low directionality (high confinement). (B) MP-IVM micrograph showing a patrolling neutrophil (light blue) migrating between macrophages (red) in the subcapsular sinus of a lymph node following infection. (C) Illustration of biological cases of patrolling behavior, including (i) a monocyte (Mo) screening the endothelium of blood vessels, (ii) a B cell surveying antigen-presenting cells in the lymph nodes (M: macrophages, DC: dendritic cells), and (iii) a natural killer (NK) cell during immune-surveillance in tumor microenvironments (T). (D) Illustration of a cell migrating directionally, with the characteristic straight tracks associated with high directionality and possibly high speed. (E) MP-IVM micrograph showing a neutrophil (light blue) exhibiting directed migration towards the subcapsular sinus area of a lymph node following infection. (F) Illustration of biological cases of directed migration including (i) a neutrophil (Neu) directed towards the source of a chemotactic gradient, and (ii) a T cell (Tc) moving with directed migration while following collagen fibers (blue structures) in the tumor microenvironment (T). (G) Illustration of an arrested cell with the characteristic folded track, which is associated with a low speed and high confinement. (H) MP-IVM micrograph showing a neutrophil (light blue) arresting in the proximity of a macrophage (red) in the subcapsular sinus area of a lymph node following infection. (I) Illustration of biological cases of arresting including (i) a neutrophil (Neu) during an adhesive interaction with an epithelial cell layer, and (ii) a neutrophil arresting during the production of reactive oxygen species.
Patrolling cells are found in different biological processes occurring both at steady state and under inflammatory conditions.
Maximization of antigen encountering in steady state conditions
Patrolling cells are capable of monitoring large areas and promptly responding to specific antigens. For example, monocytes display a patrolling behavior while monitoring the endothelium of blood vessels (Figures 2C, i) (19). Upon activation, these cells promote the recruitment of immune cells locally via paracrine secretion of proinflammatory cytokines (16, 19, 20, 34, 52), and transient interactions (21). Similarly, a population of neutrophils were described with a patrolling behavior within the lumen of blood vessels. This was and associated with an increased capacity of these cells for being recruited to the inflammation site (78, 79). More recently, tissue-resident eosinophils have also been reported to display a patrolling behavior in different organs (33, 40).
In the LN, patrolling B cells continuously survey subcapsular macrophages and follicular dendritic cells in order to identify antigens that are either presented on a cell surface or suspended in the environment (80). Moreover, within the germinal centers (GC), patrolling B cells exhibited a probing, dendritic morphology that conferred them a larger surface area and therefore a greater opportunity for antigen encountering (Figures 2C, ii) (80). In addition, patrolling of T cells was also reported as a strategy to maximize the encountering of antigen presenting cells (APCs) (46, 77, 81) and avoid obstacles in densely packed microenvironments (82). Finally, Bajenoff and colleagues reported that the apparently random movement associated with the patrolling of T cells in the LN is indeed reflecting the complex network of fibroblastic reticular cells (47).
Patrolling under inflammatory conditions
NK cells were reported to maintain a patrolling behavior during priming (26), and while searching for cognate targets and transformed cells (Figure 2C, iii) (24), suggesting that the patrolling pattern is an efficient strategy for sensing and integrating cytokine signals under inflammatory conditions (26). Similarly, T cells displayed a patrolling behavior in the LN, to integrate signals from multiple APCs. Upon the encountering of APCs this behavior was maintained if the affinity was low, or switched to the formation of local clusters in case of high affinity (83).
Moreover, within the tumor microenvironment, patrolling monocytes were also associated with immune surveillance, promptly detecting tumor material, establishing interactions with metastasizing cells, and promoting recruitment and activation of natural killer (NK) cells in lung carcinoma (49).
Directed Migration
Directed migration is associated with cells displacing along straight trajectories. These cells typically exhibit long tracks with high confinement ratio and possibly high speed (Figures 2D, E) depending on the cell type, the conditions, and the microenvironment.
In inflammatory contexts, cells undergo directional migration in response to chemotactic cues and inflammatory signals, as well as when influenced by anatomical structures. Generally, directional migration is described as a strategy to rapidly reach a specific target, which also plays important roles in recruitment, tissue repair, cleaning, and antigen presentation (84, 85). Amongst the different biological context where cells display this action we can find:
Response to Chemotactic Gradients
One of the best-characterized processes associated with directional migration is chemotaxis, which involves the polarization and displacement of cells towards the source of a chemotactic gradient (Figures 2F, i). For instance, neutrophils perform directed migration towards injured, infected, or inflamed areas (35, 37, 44, 86, 87), where their presence is relevant for tissue repairing, microbial clearing (88), amplification of the inflammatory response (89), and shaping of the adaptive immune response (90). In addition, macrophages perform directed migration in interstitial tissue in response to bacterial infection or tissue injury (91).
Influence of Anatomical Structures on the Directed Migration
Tissue architecture can influence cell movements, conferring properties of directed migration. The most compelling example is the transportation of cells via the bloodstream (92, 93). More recently, transportation of immune cells via lymphatics (94, 95) was also reported and associated with a strategy for rapidly reaching lymphoid tissues (84). Moreover, the architecture of the LN was reported to influence the recruitment of B and T cells, which displayed directional migration to relocate precisely in their respective areas (96). Finally, directed migration of immune cells was also associated with the architecture of tumor microenvironments. For instance, CD8+ T cells exhibited a directed migration pattern along collagen fibers in a model of ovarian carcinoma (50) (Figures 2F, ii).
Arresting
Arresting is an action associated with cells that typically display confined trajectories and a speed below a predefined threshold (96) (Figures 2G, H). However, the migration of immune cells typically involves alternating cycles of “stop-and-go” (97). Hence, to define a cell as arrested, we consider that it should be tracked for longer than the duration of the stop-and-go cycle.
During the inflammatory process, motile cells change their behavior to arresting in order to perform a variety of functions, including signaling, killing, and activation.
Cell Activation and Signaling
Effective intracellular communication requires arresting. Notably, both B cells and T cells undergo an arresting phase prior to interacting with DC during priming (98). This step is essential to maximize the contact duration and to induce signaling.
In neutrophils, arresting was associated with the oxidative burst (38), a state in which reactive oxygen species are generated. This occurs both during phagocytosis and in response to soluble antigens. In contrast, Beuneu and colleagues (26) reported that NK cells do not arrest while being activated by DC. However, NK cells were reported to arrest in the medullary part of the LN (99) following influenza vaccination. Although the arrested NK cells were forming stable contacts with macrophages, this behavior was not associated with NK-mediated lysis. Therefore, it may suggest an alternative activation pattern.
Killing
The formation of stable contacts between a cytotoxic cell and its target is one of the best-characterized biological processes during which cytotoxic cells arrest. For instance, CD8+ T cells arrest during the formation of the cytotoxic synapses with target cells and resume their migration after killing the target (48, 100).
Adhesive Interactions During Recruitment
During recruitment from the blood stream, several types of leukocytes form adhesive interactions with stromal cells, leading to a decrease in motility and eventually to their arrest (22, 96) (Figure 2I). This process has been extensively revised (4, 101) and coincides with findings showing, that T cells interacting with lymphatic capillaries were commonly arrested (18).
Actions Performed by Two or More Cells
Studies of cell migration have typically been performed by quantifying the motility of individual cells. Collective migration patterns, meanwhile, are more difficult to interpret (69) but they remain necessary for understanding complex biological processes such as inflammation. Indeed, Mayor and colleagues argue that considering cells as part of supracellular entities allows the quantification of migration at a higher scale (102).
Contact Formation
Contact formation is an action characterized by the absence of space between two or more cells (103) (Figures 3A, B). Indeed, during contact formation, the distance between membranes of cells decreases up to a distance of 15 nm to 100 nm (104). Cells forming contacts may exhibit an arrested behavior or maintain a patrolling behavior according to the duration and the type of contact.
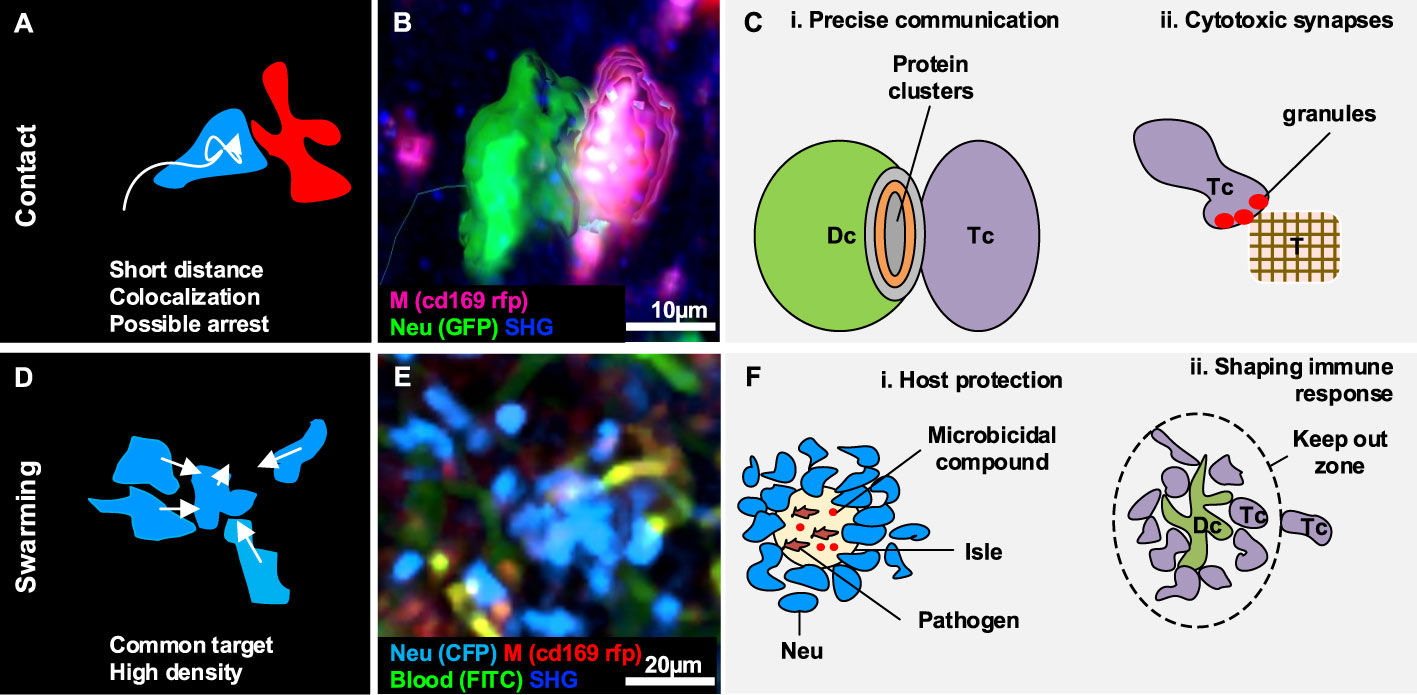
Figure 3 Gallery of actions displayed by two or a collectivity of cells. (A) Illustration of the morphodynamics of contact formation between two cells, characterized by a low distance and the possible overlap of colors. (B) MP-IVM micrograph showing a neutrophil (green) establishing contact with a macrophage (violet). 3D reconstructions are shown to highlight the shape of the cells during the formation of the contacts. (C) Illustration of biological cases of contact formation including (i) a T cell (Tc) forming an immunological synapse with a dendritic cell (Dc) with a cluster of proteins in the contact area, and (ii) a T cell (Tc) accumulating cytotoxic granules in contact with a tumor cell (T). (D) Illustration of the morphodynamics of swarm formation, characterized by cells moving towards a common target, resulting in the accumulation of cells in a confined area (high density). (E) MP-IVM micrograph showing a neutrophil swarm (light blue) following infection in the subcapsular area of a lymph node. (F) Illustration of biological cases including (i) a swarm of neutrophil (Neu) to contain pathogens in an isle enriched with microbicidal compounds, and (ii) a swarm of T cell (Tc) accumulating around an antigen-presenting dendritic cell (Dc) to prevent the other Tc from interacting with the Dc.
Cellular contacts are a form of cell-to-cell communication that enables the formation of clusters between the proteins on the surface of distinct cells (Figures 3A–C) (105, 106), and the delivery of highly localized signals (40). Although contacts are continuously formed and disrupted between migratory and resident cells in physiological conditions, certain contacts of immune cells are important for the inflammatory processes (99, 107) due to their involvement in the modulation of the immune response.
Immunological Synapses
One of the best-characterized cases of contact formation between immune cells is the immune synapse that occurs between DC and T cells (Figures 3C, i). This process can occur either in lymphoid organs such as the LN (28), or non-lymphoid organs such as the lymphatic capillaries of the ear skin (18), and is pivotal for immunity and tolerance (29). DC play a crucial role in initiating the immune cell response as they scan the surrounding environment in search of antigens to capture and present to naive T cells (108). The interaction between T cells and DC follow a series of steps characterized by a varying contact duration. At first, T cells engage many short-lived contacts with the surrounding DC, reducing their overall motility due to the multiple interactions (25). Upon successful encountering of a DC presenting the antigen specific for the T cell receptor, long-term stable contacts occur, and T cells remain arrested, which leads to their activation. Finally, the T cell recover its motility and proliferate. This process has been observed in an OT-I model, where a comparison between antigen-specific CD8+ T cells and polyclonal CD8+ T cells revealed that antigen-specific cells significantly decreased their speed in response to the formation of stable interactions with DC (109). By contrast, polyclonal CD8+ T cells maintained a constant speed (109). This finding is in agreement with the T-DC model, where different phases of the T cell-DC interaction were associated with different contact durations (28), and highlights the importance of contact duration for efficient cell activation (30).
Moreover, some studies reported that NK cells maintained a motile behavior during the formation of short-term (1 – 3 min) contacts with DC, by recognizing cytokines on the surface of DC in addition to soluble signals (23, 43). This suggested an efficient strategy to sense and integrate cytokine signals from multiple DC (23). However, other in vivo studies have reported the formation of stable contacts with macrophages during the activation of these cells (99), in accordance with previous studies performed in vitro (110, 111).
Cytotoxic Synapses and Lysis
Cytotoxic T leukocytes (CTL) can establish cytotoxic synapses with target cells, which eventually leads to the lysis of the target (112). Cytotoxic synapses formed by CD8+ T cells (Figures 3C, ii), rely on a shared molecular mechanism with CD4+ T cell immunological synapses (112). However, CD8+ T cell synapses appear to be more stable and efficient in killing the target (113). Two known killing mechanisms involve the binding of Fas death ligand to Fas death receptor, resulting in the induction of apoptotic death by caspase activation (114). The second mechanism involves calcium-dependent release of perforin and granzymes, yielding to the activation of alternative apoptotic pathways (114). The latter mechanism was reported to be faster since it does not require specific receptors to be activated (112). Common targets of CTL are virus-infected or transformed cells. Moreover, CTL killing efficiency was reported to be affected by the affinity for its ligand (25, 112).
NK cells are also able to form contacts to lyse target cells through degranulation of lytic enzymes. Within the LN, NK has been observed to form contacts with B cells to eliminate major histocompatibility complexes mismatched targets (24). Additionally, in the context of tumor microenvironment, NK-mediated lysis was reported to occur either by establishing contacts of long duration with a single NK or via multiple short contacts with several NK (26).
Swarming
Swarming is an action that involves a collectivity of cells clustering in a defined space or moving towards a common target in a coordinated manner, giving rise to a swarm (Figures 3D, E) (42). Swarms have been classified according to their size and duration (42). Transient swarms with fewer than 150 cells are reported to last up to 40 minutes. Larger swarms can include more than 300 cells and can persist for hours (42).
The swarming process has been primarily described in neutrophils, which form cell aggregates in inflamed and injured tissues. Notably, duration and swarm size were positively correlated with the severity of the tissue damage or infection, with extended lesions massively recruiting neutrophils involved in swarms that persisted for days (32, 115). Cell death, known to induce recruitment of phagocytic cells (51, 116, 117), is regarded as one of the triggers of swarming.
Swarming is associated with two key biological functions, host protection and tissue remodeling.
Host Protection
Swarm formation was reported in infection models as a strategy to contain pathogens and protect the host (118). To this end, swarms lead to the confinement of pathogens in isles where microbicidal compounds concentrate (Figures 3F, i) (115). Accordingly, neutrophil swarming was observed to contain bacteria spread (39) and limit the growth of fungi in vivo (119). Eosinophils were also observed performing swarms throughout the parenchyma in the lungs in different infection models. Amongst these, during parasitic infections, swarms of eosinophils were maintained for several days (120).
Tissue Remodeling and Shaping the Immune Response
Neutrophil swarming was also reported in the context of sterile inflammation. Sterile photo burning (87) and needle damage (44) caused neutrophils to form abrupt and long-lasting clusters of large dimensions, suggesting a role in tissue remodeling and repair.
Additionally, the formation of swarms can alter the cellular structure of immune organs. For instance, swarms formed by neutrophils were reported to disrupt the network of resident SCS macrophages in parasitic infection models (41, 115, 121). Considering that SCS macrophages are important for containing the spread of pathogens (122, 123) and for activating the adaptive immunity (86, 122), the alteration of this cell layer by swarms might influence the overall immune response. Interestingly, other cell types, such as NK cells, were also observed to form swarms in the SCS area of the LN and to interact with resident CD11b+ cells. The accumulation of NK cells in the SCS area was linked to the function of promoting self-activation by the encountering of specific APC (124). Other cell types such as T cells were reported to form swarms around APC following immunization. Since most of the interactions in the swarms were maintained over time (108), it has been proposed that swarms may keep newly arrived T cells at the boundaries of the swarm, limiting their interaction with DC (Figures 3F, ii) (108). Finally, swarming of invariant natural killer T cells was associated with a reduced level of fibrosis in a model of steatohepatitis (17), suggesting a further role of swarming in tissue remodeling under inflammatory conditions.
Methods to Detect and Quantify Cell Actions
Due to the difficulties associated with the processing and quantification of IVM movies (125), computational methods have become essential for the analysis of cell motility in vivo. A variety of software and methods were applied to detect and quantify cell actions. A summary of these methods, including how to use them and how to interpret the computed numerical values is presented in Table 2.
Quantification of Patrolling Cells
To quantify the patrolling behavior, coefficients that evaluate the displacement over time, such as the directionality and the motility index are typically used. These coefficients are larger than the ones displayed by arrested cells, but lower than the values displayed by directional cells (18, 126, 127). Additionally, the previously mentioned MSD analysis can be used to distinguish patrolling cells from arrested or directed cells, as they display a random-like migratory pattern. However, several studies demonstrated that the movement of cells in vivo is not stochastic, but rather influenced by alternative parameters such as the interaction with stromal cells, amongst others (70). Therefore, we suggest to complement the MSD analysis with other parameters, such as the angle or speed distribution, which would provide additional insights on the migratory mode.
Quantification of Directed Cells
Directional migration can be inferred by plotting the trajectories of the analyzed cells with a common origin, resulting in tracks with a strong preferential migration direction (45, 51, 63). More quantitatively, one of the parameters that better characterizes directed migration is the confinement ratio, which presents high values for highly directional cells (69). However, the confinement ratio of different tracks is comparable only if they have similar track durations. Otherwise, normalizing the trajectories for their duration is often required. Another measure typically used for predicting directed migration is the distribution of the turning angles (36). Following this analysis, a skewing toward small angles would indicate that a cell trajectory does not deviate abruptly from its established path. In addition, the MSD analysis could also indicate directional migration recalling super diffusivity (36, 44).
When analyzing the overall motility of a cell population, directional migration can be inferred by evaluating the distance over time of all the cells with respect to a reference point or region. The chosen reference point should ideally represent a common target (32, 41, 51, 115) towards which the distance decreases or increases.
Quantification of Arresting Cells
An arrested cell is typically detected by evaluating the arrest coefficient (69). This coefficient measures the amount of time in which a cell migrates with a speed below a defined threshold (typically 2 µm/min) (31). However, the value of the arrest coefficient depends on the track duration. Therefore, tracks (or track fragments) with similar duration should be compared; otherwise, normalization strategies are required for comparative studies (i.e., dividing the arrest coefficient by the track duration). The confinement ratio used to predict the directionality of a trajectory is also used to detect arrested cells, which typically display low values (31).
Quantification of Contacts
Contacts are typically detected by evaluating the distance between cells. Such a distance can be computed either between the centroids of the cells (75), or reconstructed surfaces (i.e. between the closest points of two cells) (13). In the first case, a contact is detected when the distance is less than a threshold, which is equal to the expected cell diameter. However, errors may be introduced when cells with a non-convex shape are analyzed. In the second case, the distance threshold is preferably small, up to the spatial resolution of the microscope (i.e., 1 µm). This allows to detect contacts between cells of arbitrary shapes. However, the distance between the membranes of two cells forming a contact is on average lower than the spatial resolution of fluorescent microscopes used in intravital imaging (0.2 µm – 0.3 µm) (128). Moreover, an accurate reconstruction of cell surfaces might be hampered by the presence of cell-to-cell contacts themselves (13). For these reasons, cell-to-cell contacts are still annotated manually (18). More robust approaches inferred contact formation from time series, such as the trajectories of the individual cells or the changes in cell speed (27). Moreover, spatial colocalization of two distinct fluorophores can be used to highlight overlapping cells without the need for surface reconstruction (27) nor the computation of spatial distances between cells.
Contact dynamics can be quantified by computing for each cell the contact duration and the number of contacts. This ultimately allows one to distinguish between transient and long-lasting contacts.
Quantification of Cell Swarms
To quantify swarm dynamics in the case of localized tissue damage or infection, the distance between the affected region and each cell at different time points can be computed. Cells whose distance over time falls below a defined threshold are considered part of a forming swarm (32, 41, 44, 51, 63).
Alternatively, when the swarm coordinates are not known, the increase of fluorescence intensity over time in different areas can be computed. In turn, surface and volumetric reconstruction enable the monitoring of the swarm growth over time by encompassing the fluorescence intensity emitted by the forming swarm (41, 45, 51). This provides insights into the different stages of the process, including initiation, growth, stabilization (41) and whether the swarm is transient or persistent (119). Furthermore, dividing the measured surface or volume by the mean volume or area of cells leads to an estimate of the number of swarming cells (41).
Swarming can be also inferred from the trajectories and speed of cells. Indeed, color coding the cell trajectories for their instantaneous cell speed can help to locate transient and persistent swarms (39). Similarly, representing a heatmap of the cell velocities and densities generates a spatiotemporal visualization that accounts for both migratory and clustering dynamics (38).
Concluding Remarks
In line with the computer vision community, we considered distinct motility patterns displayed by cells as elementary actions (129, 130), which are the building blocks of several biological processes. This approach is relevant to dissecting the complex dynamics of inflammation, as it provides a link between identifiable morpho-phenotypes and the underlying cellular function. Moreover, by detecting the occurrence of each action over time, it is possible to quantify the dynamic behavior of immune cells in response to different stimuli.
Another advantage of decomposing cell motility in elementary actions is that these can be quantified from tracks of short duration (tracklets) or short image sequences using instantaneous measures. However, a longer imaging time can lead to more accurate results for actions such as swarming, which can persist for up to several hours.
In this review, the visual approach adopted to classify each action further aims to facilitate the interpretation of intravital microscopy data for immunologists and imaging specialists. The described measurements and definitions are provided to help researchers in differentiating between distinct cellular actions from a motility perspective. These are, however, intended only as guidelines rather than absolute discriminatory factors, as no consensus definition and numerical characterization exists thus far. In fact, to identify cell actions, it might be necessary to adopt a gating strategy that considers the combination of several motility parameters (51). Future advancements in this direction will require further characterization of cell motility based on the function, cell type, and organ.
In conclusion, the development of computer vision methods for cellular action recognition represents a promising methodology for deciphering biological processes occurring in vivo from imaging data.
Inclusion Criteria
This review includes studies that reported specific motility patterns of immune cells, which were observed under inflammatory conditions in vivo. The included imaging modalities were MP-IVM, spinning disk, laser scanning confocal, and epifluorescence microscopy. All the definitions of cell actions used in this work were inferred from the original studies. In most cases, the authors explicitly named the migratory patterns displayed by the imaged cells. Indeed, directed migration, arresting, contact formation and swarming (or clustering) are well-characterized processes that were typically referred to using a direct name. In these cases, we did not perform re-analysis of the data. In the case of patrolling instead, studies referred to it either with the same term, or a similar nomenclature (i.e., scanning, undirected migration, random migration), or provided measurements whose values were indicative of this motility pattern.
Author Contributions
DP and AP reviewed the literature and wrote the manuscript. MT and RK corrected the manuscript. SG supervised and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Swiss National Foundation (SNF) grants, 176124, SystemsX.ch (2013/124), Biolink (189699).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Luster AD, Alon R, von Andrian UH. Immune Cell Migration in Inflammation: Present and Future Therapeutic Targets. Nat Immunol (2005) 6:1182–90. doi: 10.1038/ni1275
2. BB MD. Systemic Response to Inflammation. Nutr Rev (2008) 65:S170–2. doi: 10.1111/j.1753-4887.2007.tb00357.x
3. Voisin MB, Nourshargh S. Neutrophil Transmigration: Emergence of an Adhesive Cascade Within Venular Walls. J Innate Immun (2013) 5:336–47. doi: 10.1159/000346659
4. Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the Site of Inflammation: The Leukocyte Adhesion Cascade Updated. Nat Rev Immunol (2007) 7:678–89. doi: 10.1038/nri2156
5. Teixidó J, Hidalgo A, Fagerholm S. Editorial: Leukocyte Trafficking in Homeostasis and Disease. Front Immunol (2019) 10:2560. doi: 10.3389/fimmu.2019.02560
6. Nowarski R, Gagliani N, Huber S, Flavell RA. Innate Immune Cells in Inflammation and Cancer. Cancer Immunol Res (2013) 1:77–84. doi: 10.1158/2326-6066.CIR-13-0081
7. Barton GM. A Calculated Response: Control of Inflammation by the Innate Immune System. J Clin Invest (2008) 118:413–20. doi: 10.1172/JCI34431
8. Pittet MJ, Garris CS, Arlauckas SP, Weissleder R. Recording the Wild Lives of Immune Cells. Sci Immunol (2018) 3:eaaq0491. doi: 10.1126/sciimmunol.aaq0491
9. Stein JV, Gonzalez SF F, Gonzalez S, Gonzalez SF. Dynamic Intravital Imaging of Cell-Cell Interactions in the Lymph Node. J Allergy Clin Immunol (2017) 139:12–20. doi: 10.1016/j.jaci.2016.11.008
10. Cahalan MD, Parker I, Wei SH, Miller MJ. Two-Photon Tissue Imaging: Seeing the Immune System in a Fresh Light. Nat Rev Immunol (2002) 2:872–80. doi: 10.1038/nri935
11. Sumen C, Mempel TR, Mazo IB, Von Andrian UH. Intravital Microscopy: Visualizing Immunity in Context. Immunity (2004) 21:315–29. doi: 10.1016/j.immuni.2004.08.006
12. SenGupta S, Parent CA, Bear JE. The Principles of Directed Cell Migration. Nat Rev Mol Cell Biol (2021) 22. doi: 10.1038/s41580-021-00366-6
13. Pizzagalli DU, Farsakoglu Y, Palomino-Segura M, Palladino E, Sintes J, Marangoni F, et al. Leukocyte Tracking Database, a Collection of Immune Cell Tracks From Intravital 2-Photon Microscopy Videos. Sci Data (2018) 5:180129. doi: 10.1038/sdata.2018.129
14. Figge MT, Murphy RF. Image-Based Systems Biology. Cytometry A (2015) 87:459–61. doi: 10.1002/cyto.a.22663
15. Poppe R. A Survey on Vision-Based Human Action Recognition. Image Vis Comput (2010) 28:976–90. doi: 10.1016/j.imavis.2009.11.014
16. Finsterbusch M, Hall P, Li A, Devi S, Westhorpe CLV, Kitching AR, et al. Patrolling Monocytes Promote Intravascular Neutrophil Activation and Glomerular Injury in the Acutely Inflamed Glomerulus. Proc Natl Acad Sci (2016) 113:E5172–81. doi: 10.1073/pnas.1606253113
17. Wang H, Li L, Li Y, Li Y, Sha Y, Wen S, et al. Intravital Imaging of Interactions Between iNKT and Kupffer Cells to Clear Free Lipids During Steatohepatitis. Theranostics (2021) 11:2149–69. doi: 10.7150/thno.51369
18. Hunter MC, Teijeira A, Montecchi R, Russo E, Runge P, Kiefer F, et al. Dendritic Cells and T Cells Interact Within Murine Afferent Lymphatic Capillaries. Front Immunol (2019) 10:520. doi: 10.3389/fimmu.2019.00520
19. Auffray C, Fogg D, Garfa M, Elain G, Join-Lambert O, Kayal S, et al. Monitoring of Blood Vessels and Tissues by a Population of Monocytes With Patrolling Behavior. Science (80-) (2007) 317:666–70. doi: 10.1126/science.1142883
20. Carlin LM, Stamatiades EG, Auffray C, Hanna RN, Glover L, Vizcay-Barrena G, et al. Nr4a1-Dependent Ly6Clow Monocytes Monitor Endothelial Cells and Orchestrate Their Disposal. Cell (2013) 153:362–75. doi: 10.1016/j.cell.2013.03.010
21. Westhorpe CLV, Ursula Norman M, Hall P, Snelgrove SL, Finsterbusch M, Li A, et al. Effector CD4+ T Cells Recognize Intravascular Antigen Presented by Patrolling Monocytes. Nat Commun (2018) 9. doi: 10.1038/s41467-018-03181-4
22. Phillipson M, Heit B, Colarusso P, Liu L, Ballantyne CM, Kubes P. Intraluminal Crawling of Neutrophils to Emigration Sites: A Molecularly Distinct Process From Adhesion in the Recruitment Cascade. J Exp Med (2006) 203:2569–75. doi: 10.1084/jem.20060925
23. Deguine J, Bousso P. Dynamics of NK Cell Interactions In Vivo. Immunol Rev (2013) 251:154–9. doi: 10.1111/imr.12015
24. Garrod KR, Wei SH, Parker I, Cahalan MD. Natural Killer Cells Actively Patrol Peripheral Lymph Nodes Forming Stable Conjugates to Eliminate MHC-Mismatched Targets. Proc Natl Acad Sci (2007) 104:12081–6. doi: 10.1073/pnas.0702867104
25. Mempel TR, Pittet MJ, Khazaie K, Weninger W, Weissleder R, von Boehmer H, et al. Regulatory T Cells Reversibly Suppress Cytotoxic T Cell Function Independent of Effector Differentiation. Immunity (2006) 25:129–41. doi: 10.1016/j.immuni.2006.04.015
26. Beuneu H, Deguine J, Breart B, Mandelboim O, Di Santo JP, Bousso P. Dynamic Behavior of NK Cells During Activation in Lymph Nodes. Blood (2009) 114:3227–34. doi: 10.1182/blood-2009-06-228759
27. Miller MJ, Hejazi AS, Wei SH, Cahalan MD, Parker I. T Cell Repertoire Scanning is Promoted by Dynamic Dendritic Cell Behavior and Random T Cell Motility in the Lymph Node. Proc Natl Acad Sci USA (2004) 101:998–1003. doi: 10.1073/pnas.0306407101
28. Mempel TR, Henrickson SE, von Andrian UH. T-Cell Priming by Dendritic Cells in Lymph Nodes Occurs in Three Distinct Phases. Nature (2004) 427:154–9. doi: 10.1038/nature02238
29. Shakhar G, Lindquist RL, Skokos D, Dudziak D, Huang JH, Nussenzweig MC, et al. Stable T Cell-Dendritic Cell Interactions Precede the Development of Both Tolerance and Immunity In Vivo. Nat Immunol (2005) 6:707–14. doi: 10.1038/ni1210
30. Celli S, Lemaître F, Bousso P. Real-Time Manipulation of T Cell-Dendritic Cell Interactions In Vivo Reveals the Importance of Prolonged Contacts for CD4+ T Cell Activation. Immunity (2007) 27:625–34. doi: 10.1016/j.immuni.2007.08.018
31. Hugues S, Fetler L, Bonifaz L, Helft J, Amblard F, Amigorena S. Distinct T Cell Dynamics in Lymph Nodes During the Induction of Tolerance and Immunity. Nat Immunol (2004) 5:1235–42. doi: 10.1038/ni1134
32. Lämmermann T, Afonso PV, Angermann BR, Wang JM, Kastenmüller W, Parent CA, et al. Neutrophil Swarms Require LTB4 and Integrins at Sites of Cell Death In Vivo. Nature (2013) 498:371–5. doi: 10.1038/nature12175
33. Chojnacki A, Wojcik K, Petri B, Aulakh G, Jacobsen EA, LeSuer WE, et al. Intravital Imaging Allows Real-Time Characterization of Tissue Resident Eosinophils. Commun Biol (2019) 2:181. doi: 10.1038/S42003-019-0425-3
34. Audoy-Remus J, Richard J-F, Soulet D, Zhou H, Kubes P, Vallieres L. Rod-Shaped Monocytes Patrol the Brain Vasculature and Give Rise to Perivascular Macrophages Under the Influence of Proinflammatory Cytokines and Angiopoietin-2. J Neurosci (2008) 28:10187–99. doi: 10.1523/JNEUROSCI.3510-08.2008
35. McDonald B, Pittman K, Menezes GB, Hirota SA, Slaba I, Waterhouse CCM, et al. Intravascular Danger Signals Guide Neutrophils to Sites of Sterile Inflammation. Science (80-) (2010) 330:362–6. doi: 10.1126/science.1195491
36. Waite JC, Leiner I, Lauer P, Rae CS, Barbet G, Zheng H, et al. Dynamic Imaging of the Effector Immune Response to Listeria Infection In Vivo. PLoS Pathog (2011) 7:1–16. doi: 10.1371/journal.ppat.1001326
37. Peters NC, Egen JG, Secundino N, Debrabant A, Kimblin N, Kamhawi S, et al. In Vivo Imaging Reveals an Essential Role for Neutrophils in Leishmaniasis Transmitted by Sand Flies. Science (80-) (2008) 321:970–4. doi: 10.1126/science.1159194
38. Kreisel D, Nava RG, Li W, Zinselmeyer BH, Wang B, Lai J, et al. In Vivo Two-Photon Imaging Reveals Monocyte-Dependent Neutrophil Extravasation During Pulmonary Inflammation. Proc Natl Acad Sci (2010) 107:18073–8. doi: 10.1073/pnas.1008737107
39. Kienle K, Glaser KM, Eickhoff S, Mihlan M, Knöpper K, Reátegui E, et al. Neutrophils Self-Limit Swarming to Contain Bacterial Growth In Vivo. Science (80-) (2021) 372(6548):eabe7729. doi: 10.1126/science.abe7729
40. Lee SH, Chaves MM, Kamenyeva O, Gazzinelli-Guimaraes PH, Kang BH, Pessenda G, et al. M2-Like, Dermal Macrophages are Maintained via IL-4/CCL24 Mediated Cooperative Interaction With Eosinophils in Cutaneous Leishmaniasis. Sci Immunol (2020) 5(46):eaaz4415. doi: 10.1126/SCIIMMUNOL.AAZ4415
41. Chtanova T, Schaeffer M, Han S-J, van Dooren GG, Nollmann M, Herzmark P, et al. Dynamics of Neutrophil Migration in Lymph Nodes During Infection. Immunity (2008) 29:487–96. doi: 10.1016/j.immuni.2008.07.012
42. Chtanova T, Schaeffer M, Han SJ, van Dooren GG, Nollmann M, Herzmark P, et al. Dynamics of Neutrophil Migration in Lymph Nodes During Infection (DOI:10.1016/J.Immuni.2008.07.012). Immunity (2008) 29:487–96. doi: 10.1016/j.immuni.2008.09.007
43. Bajénoff M, Breart B, Huang AYC, Qi H, Cazareth J, Braud VM, et al. Natural Killer Cell Behavior in Lymph Nodes Revealed by Static and Real-Time Imaging. J Exp Med (2006) 203:619–31. doi: 10.1084/jem.20051474
44. Ng LG, Qin JS, Roediger B, Wang Y, Jain R, Cavanagh LL, et al. Visualizing the Neutrophil Response to Sterile Tissue Injury in Mouse Dermis Reveals a Three-Phase Cascade of Events. J Invest Dermatol (2011) 131:2058–68. doi: 10.1038/jid.2011.179
45. Park SA, Choe YH, Park E, Hyun YM. Real-Time Dynamics of Neutrophil Clustering in Response to Phototoxicity-Induced Cell Death and Tissue Damage in Mouse Ear Dermis. Cell Adhes Migr (2018) 6918:1–8. doi: 10.1080/19336918.2018.1471322
46. Miller MJ, Wei SH, Cahalan MD, Parker I. Autonomous T Cell Trafficking Examined In Vivo With Intravital Two-Photon Microscopy. Proc Natl Acad Sci USA (2003) 100:2604–9. doi: 10.1073/pnas.2628040100
47. Bajénoff M, Egen JG, Koo LY, Laugier JP, Brau F, Glaichenhaus N, et al. Stromal Cell Networks Regulate Lymphocyte Entry, Migration, and Territoriality in Lymph Nodes. Immunity (2006) 25:989–1001. doi: 10.1016/j.immuni.2006.10.011
48. Boissonnas A, Fetler L, Zeelenberg IS, Hugues S, Amigorena S. In Vivo Imaging of Cytotoxic T Cell Infiltration and Elimination of a Solid Tumor. J Exp Med (2007) 204:345–56. doi: 10.1084/jem.20061890
49. Hanna RN, Cekic C, Sag D, Tacke R, Thomas GD, Nowyhed H, et al. Patrolling Monocytes Control Tumor Metastasis to the Lung. Science (80-) (2015) 350:985–90. doi: 10.1126/science.aac9407
50. Bougherara H, Mansuet-Lupo A, Alifano M, Ngô C, Damotte D, Le Frère-Belda M-A, et al. Real-Time Imaging of Resident T Cells in Human Lung and Ovarian Carcinomas Reveals How Different Tumor Microenvironments Control T Lymphocyte Migration. Front Immunol (2015) 6:500. doi: 10.3389/fimmu.2015.00500
51. Pizzagalli DU, Latino I, Pulfer A, Palomino-Segura M, Virgilio T, Farsakoglu Y, et al. Characterization of the Dynamic Behavior of Neutrophils Following Influenza Vaccination. Front Immunol (2019) 10:2621. doi: 10.3389/fimmu.2019.02621
52. Buscher K, Marcovecchio P, Hedrick CC, Ley K. Patrolling Mechanics of Non-Classical Monocytes in Vascular Inflammation. Front Cardiovasc Med (2017) 4:80. doi: 10.3389/fcvm.2017.00080
53. Ricard C, Arroyo ED, He CX, Portera-Cailliau C, Lepousez G, Canepari M, et al. Two-Photon Probes for In Vivo Multicolor Microscopy of the Structure and Signals of Brain Cells. Brain Struct Funct (2018) 223:3011–43. doi: 10.1007/s00429-018-1678-1
54. Shaner NC, Steinbach PA, Tsien RY. A Guide to Choosing Fluorescent Proteins. Nat Methods (2005) 2:905–9. doi: 10.1038/nmeth819
55. Secklehner J, Celso C, Carlin LM. Intravital Microscopy in Historic and Contemporary Immunology. Immunol Cell Biol (2017) 95:506–13. doi: 10.1038/icb.2017.25
56. Rakhilin N, Garrett A, Eom CY, Chavez KR, Small DM, Daniel AR, et al. An Intravital Window to Image the Colon in Real Time. Nat Commun (2019) 10(1). doi: 10.1038/s41467-019-13699-w
57. Jahromi NH, Tardent H, Enzmann G, Deutsch U, Kawakami N, Bittner S, et al. A Novel Cervical Spinal Cord Window Preparation Allows for Two-Photon Imaging of T-Cell Interactions With the Cervical Spinal Cord Microvasculature During Experimental Autoimmune Encephalomyelitis. Front Immunol (2017) 8:406. doi: 10.3389/fimmu.2017.00406
58. Zhang W, Karschnia P, von Mücke-Heim IA, Mulazzani M, Zhou X, Blobner J, et al. In Vivo Two-Photon Characterization of Tumor-Associated Macrophages and Microglia (TAM/M) and CX3CR1 During Different Steps of Brain Metastasis Formation From Lung Cancer. Neoplasia (United States) (2021) 23:1089–100. doi: 10.1016/j.neo.2021.09.001
59. Mulazzani M, Fräßle SP, von Mücke-Heim I, Langer S, Zhou X, Ishikawa-Ankerhold H, et al. Long-Term In Vivo Microscopy of CAR T Cell Dynamics During Eradication of CNS Lymphoma in Mice. Proc Natl Acad Sci USA (2019) 116:24275–84. doi: 10.1073/pnas.1903854116
60. Dunn KW, Sutton TA. Functional Studies in Living Animals Using Multiphoton Microscopy. ILAR J (2008) 49:66–77. doi: 10.1093/ilar.49.1.66
61. Soulet D, Lamontagne-Proulx J, Aubé B, Davalos D. Multiphoton Intravital Microscopy in Small Animals: Motion Artefact Challenges and Technical Solutions. J Microsc (2020) 278:3–17. doi: 10.1111/jmi.12880
62. Vaghela R, Arkudas A, Horch RE, Hessenauer M. Actually Seeing What Is Going on – Intravital Microscopy in Tissue Engineering. Front Bioeng Biotechnol (2021) 9:627462. doi: 10.3389/fbioe.2021.627462
63. Phillipson M, Kubes P. The Neutrophil in Vascular Inflammation. Nat Med (2011) 17:1381–90. doi: 10.1038/nm.2514
64. Marques PE, Oliveira AG, Chang L, Paula-Neto HA, Menezes GB. Understanding Liver Immunology Using Intravital Microscopy. J Hepatol (2015) 63:733–42. doi: 10.1016/j.jhep.2015.05.027
65. Zipfel WR, Williams RM, Webb WW. Nonlinear Magic: Multiphoton Microscopy in the Biosciences. Nat Biotechnol (2003) 21:1369–77. doi: 10.1038/nbt899
66. Helmchen F, Denk W. Deep Tissue Two-Photon Microscopy. Nat Methods (2005) 2:932–40. doi: 10.1038/nmeth818
67. Modi MN, Daie K, Turner GC, Podgorski K. Two-Photon Imaging With Silicon Photomultipliers. Opt Express (2019) 27:35830. doi: 10.1364/OE.27.035830
68. Jorch SK, Deppermann C. Intravital Imaging Allows Organ-Specific Insights Into Immune Functions. Front Cell Dev Biol (2021) 9:623906. doi: 10.3389/fcell.2021.623906
69. Beltman JB, Marée AFM, de Boer RJ, M Marée AF, de Boer RJ, Marée AFM, et al. Analysing Immune Cell Migration. Nat Rev Immunol (2009) 9:789–98. doi: 10.1038/nri2638
70. Cahalan MD, Parker I. Choreography of Cell Motility and Interaction Dynamics Imaged by Two-Photon Microscopy in Lymphoid Organs. Annu Rev Immunol (2008) 26:585–626. doi: 10.1146/annurev.immunol.24.021605.090620
71. Ulman VV, Maška M, Magnusson KEGG, Ronneberger O, Haubold C, Harder N, et al. An Objective Comparison of Cell-Tracking Algorithms. Nat Methods (2017) 14:1141. doi: 10.1038/nmeth.4473
72. Vladymyrov M, Haghayegh Jahromi N, Kaba E, Engelhardt B, Ariga A. VivoFollow 2: Distortion-Free Multiphoton Intravital Imaging. Front Phys (2020) 7:222. doi: 10.3389/fphy.2019.00222
73. Pizzagalli DU, Thelen M, Gonzalez SF, Krause R. Semi-Supervised Machine Learning Facilitates Cell Colocalization and Tracking in Intravital Microscopy. BioRxiv (2019). doi: 10.1101/829838
74. Loosley AJ, O’Brien XM, Reichner JS, Tang JX. Describing Directional Cell Migration With a Characteristic Directionality Time. PloS One (2015) 10:e0127425. doi: 10.1371/journal.pone.0127425
75. Beltman JB, Henrickson SE, von Andrian UH, de Boer RJ, Marée AFM. Towards Estimating the True Duration of Dendritic Cell Interactions With T Cells. J Immunol Methods (2009) 347:54–69. doi: 10.1016/j.jim.2009.05.013
76. Schienstock D, Mueller SN. Moving Beyond Velocity: Opportunities and Challenges to Quantify Immune Cell Behavior. Immunol Rev (2021) 1–14. doi: 10.1111/imr.13038
77. Wei SH, Parker I, Miller MJ, Cahalan MD. A Stochastic View of Lymphocyte Motility and Trafficking Within the Lymph Node. Immunol Rev (2003) 195:136–59. doi: 10.1034/j.1600-065X.2003.00076.x
78. Kolaczkowska E, Kubes P. Neutrophil Recruitment and Function in Health and Inflammation. Nat Rev Immunol (2013) 13:159–75. doi: 10.1038/nri3399
79. Nourshargh S, Alon R. Leukocyte Migration Into Inflamed Tissues. Immunity (2014) 41:694–707. doi: 10.1016/j.immuni.2014.10.008
80. Cyster JG, Allen CDC. B Cell Responses: Cell Interaction Dynamics and Decisions. Cell (2019) 177:524–40. doi: 10.1016/j.cell.2019.03.016
81. Miller MJ, Wei SH, Parker I, Cahalan MD. Two-Photon Imaging of Lymphocyte Motility and Antigen Response in Intact Lymph Node. Science (80-) (2002) 296:1869–73. doi: 10.1126/science.1070051
82. Katakai T, Kinashi T. Microenvironmental Control of High-Speed Interstitial T Cell Migration in the Lymph Node. Front Immunol (2016) 7:194. doi: 10.3389/fimmu.2016.00194
83. Moreau HD, Bousso P. Visualizing How T Cells Collect Activation Signals In Vivo. Curr Opin Immunol (2014) 26:56–62. doi: 10.1016/j.coi.2013.10.013
84. Arasa J, Collado-Diaz V, Kritikos I, Medina-Sanchez JD, Friess MC, Sigmund EC, et al. Upregulation of VCAM-1 in Lymphatic Collectors Supports Dendritic Cell Entry and Rapid Migration to Lymph Nodes in Inflammation. J Exp Med (2021) 218:e20201413. doi: 10.1084/jem.20201413
85. Singer AJ, Clark RA. Cutaneous Wound Healing. N Engl J Med (1999) 341:738–46. doi: 10.1056/NEJM199909023411006
86. Chatziandreou N, Farsakoglu Y, Palomino-Segura M, D’Antuono R, Pizzagalli DU, Sallusto F, et al. Macrophage Death Following Influenza Vaccination Initiates the Inflammatory Response That Promotes Dendritic Cell Function in the Draining Lymph Node. Cell Rep (2017) 18:2427–40. doi: 10.1016/j.celrep.2017.02.026
87. Kamenyeva O, Boularan C, Kabat J, Cheung GYC, Cicala C, Yeh AJ, et al. Neutrophil Recruitment to Lymph Nodes Limits Local Humoral Response to Staphylococcus Aureus. PloS Pathog (2015) 11:e1004827. doi: 10.1371/journal.ppat.1004827
88. de Oliveira S, Rosowski EE, Huttenlocher A. Neutrophil Migration in Infection and Wound Repair: Going Forward in Reverse. Nat Rev Immunol (2016) 16:378–91. doi: 10.1038/nri.2016.49
89. Mayadas TN, Cullere X, Lowell CA. The Multifaceted Functions of Neutrophils. Annu Rev Pathol Mech Dis (2014) 9:181–218. doi: 10.1146/annurev-pathol-020712-164023
90. Leliefeld PHC, Koenderman L, Pillay J. How Neutrophils Shape Adaptive Immune Responses. Front Immunol (2015) 6:471. doi: 10.3389/fimmu.2015.00471
91. Zhang Y, Bai X-T, Zhu K-Y, Jin Y, Deng M, Le H-Y, et al. In Vivo Interstitial Migration of Primitive Macrophages Mediated by JNK-Matrix Metalloproteinase 13 Signaling in Response to Acute Injury. J Immunol (2008) 181:2155–64. doi: 10.4049/jimmunol.181.3.2155
92. Zinselmeyer BH, Lynch JN, Zhang X, Aoshi T, Miller MJ. Video-Rate Two-Photon Imaging of Mouse Footpad - A Promising Model for Studying Leukocyte Recruitment Dynamics During Inflammation. Inflamm Res (2008) 57:93–6. doi: 10.1007/s00011-007-7195-y
93. De Filippo K, Rankin SM. The Secretive Life of Neutrophils Revealed by Intravital Microscopy. Front Cell Dev Biol (2020) 8:603230. doi: 10.3389/fcell.2020.603230
94. Hampton HR, Chtanova T. Lymphatic Migration of Immune Cells. Front Immunol (2019) 10:1168. doi: 10.3389/fimmu.2019.01168
95. Lindquist RL, Shakhar G, Dudziak D, Wardemann H, Eisenreich T, Dustin ML, et al. Visualizing Dendritic Cell Networks In Vivo. Nat Immunol (2004) 5:1243–50. doi: 10.1038/ni1139
96. Friedl P, Weigelin B. Interstitial Leukocyte Migration and Immune Function. Nat Immunol (2008) 9:960–9. doi: 10.1038/ni.f.212
97. Hallett MB. editor. "Biophysics of leukocytes: neutrophil chemotaxis, characteristics, and mechanisms". In: The Neutrophil: Cellular Biochemistry and Physiology. Boston, MA: CRC Press (1989).
98. Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. SAP-Controlled T–B Cell Interactions Underlie Germinal Centre Formation. Nature (2008) 455:764–9. doi: 10.1038/nature07345
99. Farsakoglu Y, Palomino-Segura M, Latino I, Zanaga S, Chatziandreou N, Pizzagalli DU, et al. Influenza Vaccination Induces NK-Cell-Mediated Type-II IFN Response That Regulates Humoral Immunity in an IL-6-Dependent Manner. Cell Rep (2019) 26:2307–15.e5. doi: 10.1016/j.celrep.2019.01.104
100. Kawakami N, Flügel A. Knocking at the Brain’s Door: Intravital Two-Photon Imaging of Autoreactive T Cell Interactions With CNS Structures. Semin Immunopathol (2010) 32:275–87. doi: 10.1007/s00281-010-0216-x
101. Xu N, Lei X, Liu L. Tracking Neutrophil Intraluminal Crawling, Transendothelial Migration and Chemotaxis in Tissue by Intravital Video Microscopy. J Vis Exp (2011) 55. doi: 10.3791/3296
102. Shellard A, Mayor R. Supracellular Migration - Beyond Collective Cell Migration. J Cell Sci (2019) 132:345–56. doi: 10.1242/jcs.226142
103. Mrass P, Takano H, Ng LG, Daxini S, Lasaro MO, Iparraguirre A, et al. Random Migration Precedes Stable Target Cell Interactions of Tumor-Infiltrating T Cells. J Exp Med (2006) 203:2749–61. doi: 10.1084/jem.20060710
104. Krummel MF, Cahalan MD. The Immunological Synapse: A Dynamic Platform for Local Signaling. J Clin Immunol (2010) 30:364–72. doi: 10.1007/s10875-010-9393-6
105. Dustin ML, Chakraborty AK, Shaw AS. Understanding the Structure and Function of the Immunological Synapse. Cold Spring Harb Perspect Biol (2010) 2:a002311–a002311. doi: 10.1101/cshperspect.a002311
106. Monks CRF, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three-Dimensional Segregation of Supramolecular Activation Clusters in T Cells. Nature (1998) 395:82–6. doi: 10.1038/25764
107. Xie J, Tato CM, Davis MM. How the Immune System Talks to Itself: The Varied Role of Synapses. Immunol Rev (2013) 251:65–79. doi: 10.1111/imr.12017
108. Bousso P, Robey E. Dynamics of CD8+ T Cell Priming by Dendritic Cells in Intact Lymph Nodes. Nat Immunol (2003) 4:579–85. doi: 10.1038/ni928
109. Kitano M, Yamazaki C, Takumi A, Ikeno T, Hemmi H, Takahashi N, et al. Imaging of the Cross-Presenting Dendritic Cell Subsets in the Skin-Draining Lymph Node. Proc Natl Acad Sci (2016) 113:1044–9. doi: 10.1073/pnas.1513607113
110. Brilot F, Strowig T, Roberts SM, Arrey F, Münz C. NK Cell Survival Mediated Through the Regulatory Synapse With Human DCs Requires IL-15rα. J Clin Invest (2007) 117:3316–29. doi: 10.1172/JCI31751
111. Borg C, Jalil A, Laderach D, Maruyama K, Wakasugi H, Charrier S, et al. NK Cell Activation by Dendritic Cells (DCs) Requires the Formation of a Synapse Leading to IL-12 Polarization in DCs. Blood (2004) 104:3267–75. doi: 10.1182/blood-2004-01-0380
112. Dustin ML, Long EO. Cytotoxic Immunological Synapses. Immunol Rev (2010) 235:24–34. doi: 10.1111/j.0105-2896.2010.00904.x
113. Beal AM, Anikeeva N, Varma R, Cameron TO, Vasiliver-Shamis G, Norris PJ, et al. Kinetics of Early T Cell Receptor Signaling Regulate the Pathway of Lytic Granule Delivery to the Secretory Domain. Immunity (2009) 31:632–42. doi: 10.1016/j.immuni.2009.09.004
114. Trapani JA, Smyth MJ. Functional Significance of the Perforin/Granzyme Cell Death Pathway. Nat Rev Immunol (2002) 2:735–47. doi: 10.1038/nri911
115. Kienle K, Lämmermann T. Neutrophil Swarming: An Essential Process of the Neutrophil Tissue Response. Immunol Rev (2016) 273:76–93. doi: 10.1111/imr.12458
116. Gregory CD, Pound JD. Cell Death in the Neighbourhood: Direct Microenvironmental Effects of Apoptosis in Normal and Neoplastic Tissues. J Pathol (2011) 223:178–95. doi: 10.1002/path.2792
117. Ravichandran KS. Find-Me and Eat-Me Signals in Apoptotic Cell Clearance: Progress and Conundrums. J Exp Med (2010) 207:1807–17. doi: 10.1084/jem.20101157
118. Shannon JG, Bosio CF, Hinnebusch BJ. Dermal Neutrophil, Macrophage and Dendritic Cell Responses to Yersinia Pestis Transmitted by Fleas. PloS Pathog (2015) 11:1–19. doi: 10.1371/journal.ppat.1004734
119. Alex H, Scherer A, Kreuzburg S, Abers MS, Zerbe CS, Dinauer MC, et al. Neutrophil Swarming Delays the Growth of Clusters of Pathogenic Fungi. Nat Commun (2020) 11:2031. doi: 10.1038/s41467-020-15834-4
120. Nguyen WNT, Jacobsen EA, Finney CAM, Colarusso P, Patel KD. Intravital Imaging of Eosinophils: Unwrapping the Enigma. J Leukoc Biol (2020) 108:83–91. doi: 10.1002/JLB.3HR0220-396R
121. Coombes JL, Charsar BA, Han S-J, Halkias J, Chan SW, Koshy AA, et al. Motile Invaded Neutrophils in the Small Intestine of Toxoplasma Gondii-Infected Mice Reveal a Potential Mechanism for Parasite Spread. Proc Natl Acad Sci (2013) 110:E1913–22. doi: 10.1073/pnas.1220272110
122. Louie DAP, Liao S. Lymph Node Subcapsular Sinus Macrophages as the Frontline of Lymphatic Immune Defense. Front Immunol (2019) 10:347. doi: 10.3389/fimmu.2019.00347
123. Moseman EA, Iannacone M, Bosurgi L, Tonti E, Chevrier N, Tumanov A, et al. Von Andrian UH. B Cell Maintenance of Subcapsular Sinus Macrophages Protects Against a Fatal Viral Infection Independent of Adaptive Immunity. Immunity (2012) 36:415–26. doi: 10.1016/j.immuni.2012.01.013
124. Coombes JL, Han S-J, van Rooijen N, Raulet DH, Robey EA. Infection-Induced Regulation of Natural Killer Cells by Macrophages and Collagen at the Lymph Node Subcapsular Sinus. Cell Rep (2012) 2:124–35. doi: 10.1016/j.celrep.2012.06.001
125. Pizzagalli DU, Farsakoglu Y, Palomino-Segura M, Palladino E, Sintes J, Marangoni F, et al. Leukocyte Tracking Database, a Collection of Immune Cell Tracks From Intravital 2-Photon Microscopy Videos. Sci Data (2018) 5:1–13. doi: 10.1038/sdata.2018.129
126. Selmeczi D, Mosler S, Hagedorn PH, Larsen NB, Flyvbjerg H. Cell Motility as Persistent Random Motion: Theories From Experiments. Biophys J (2005) 89:912–31. doi: 10.1529/biophysj.105.061150
127. Teijeira A, Hunter MC, Russo E, Proulx ST, Frei T, Debes GF, et al. T Cell Migration From Inflamed Skin to Draining Lymph Nodes Requires Intralymphatic Crawling Supported by ICAM-1/LFA-1 Interactions. Cell Rep (2017) 18:857–65. doi: 10.1016/j.celrep.2016.12.078
128. Doi A, Oketani R, Nawa Y, Fujita K. High-Resolution Imaging in Two-Photon Excitation Microscopy Using In Situ Estimations of the Point Spread Function. BioMed Opt Express (2018) 9:202–13. doi: 10.1364/boe.9.000202
129. Nguyen NT, Phung DQ, Venkatesh S, Bui H. Learning and Detecting Activities From Movement Trajectories Using the Hierarchical Hidden Markov Model. In: Proceedings - 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, CVPR 2005. June 2005, San Diego, CA, USA (2005). doi: 10.1109/CVPR.2005.203
Keywords: cell actions, computer vision, inflammation, intravital imaging, leukocytes, motility patterns
Citation: Pizzagalli DU, Pulfer A, Thelen M, Krause R and Gonzalez SF (2022) In Vivo Motility Patterns Displayed by Immune Cells Under Inflammatory Conditions. Front. Immunol. 12:804159. doi: 10.3389/fimmu.2021.804159
Received: 28 October 2021; Accepted: 26 November 2021;
Published: 03 January 2022.
Edited by:
Marco Erreni, Humanitas Research Hospital, ItalyCopyright © 2022 Pizzagalli, Pulfer, Thelen, Krause and Gonzalez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Santiago F. Gonzalez, santiago.gonzalez@irb.usi.ch
†These authors have contributed equally to this work
 Diego Ulisse Pizzagalli
Diego Ulisse Pizzagalli Alain Pulfer
Alain Pulfer Marcus Thelen
Marcus Thelen Rolf Krause2
Rolf Krause2 Santiago F. Gonzalez
Santiago F. Gonzalez