- 1Department of Microbiology, Guangdong Provincial Key Laboratory of Tropical Disease Research, School of Public Health, Southern Medical University, Guangzhou, China
- 2Department of Infectious Disease, Jiangmen Central Hospital, Affiliated Jiangmen Hospital of Sun Yat-Sen University, Jiangmen, China
- 3Saban Research Institute, University of Southern California, Children’s Hospital Los Angeles, Los Angeles, CA, United States
- 4Department of Pediatrics, The Second Xiangya Hospital, Central South University, Changsha, China
- 5Kunming Key Laboratory of Children Infection and Immunity, Yunnan Institute of Pediatrics, Kunming Children’s Hospital, Kunming, China
Alpha 7 nicotinic acetylcholine receptor (α7 nAChR) is critical for the pathogenesis of Escherichia coli (E. coli) K1 meningitis, a severe central nervous system infection of the neonates. However, little is known about how E. coli K1 manipulates α7 nAChR signaling. Here, through employing immortalized cell lines, animal models, and human transcriptional analysis, we showed that E. coli K1 infection triggers releasing of secreted Ly6/Plaur domain containing 1 (SLURP1), an endogenous α7 nAChR ligand. Exogenous supplement of SLURP1, combined with SLURP1 knockdown or overexpression cell lines, showed that SLURP1 is required for E. coli K1 invasion and neutrophils migrating across the blood-brain barrier (BBB). Furthermore, we found that SLURP1 is required for E. coli K1-induced α7 nAChR activation. Finally, the promoting effects of SLURP1 on the pathogenesis of E. coli K1 meningitis was significantly abolished in the α7 nAChR knockout mice. These results reveal that E. coli K1 exploits SLURP1 to activate α7 nAChR and facilitate its pathogenesis, and blocking SLURP1-α7 nAChR interaction might represent a novel therapeutic strategy for E. coli K1 meningitis.
Introduction
Despite the widespread use of antibiotics, sepsis and meningitis remain to be severe complications in premature neonates, leading to high morbidity and mortality (1–3). Thus, developing more targeted therapeutic methods for meningitis is urgently needed. Escherichia coli K1 (E. coli K1), an opportunistic pathogen in the gut, accounts for 17.7% of meningitis patients and causes a mortality rate of 40%–58% in developing countries (4, 5). Recent studies from our group have revealed that alpha 7 nicotinic acetylcholine receptor (α7 nAChR) mediated the key E. coli K1 meningitis pathogenesis by promoting bacteria migrating across the blood-brain barrier (BBB). α7 nAChR-deficient mice had a higher survival rate, lower pathogen counts, and less inflammatory responses in the brain tissues than the wild-type littermates upon E. coli K1 infection (6–8). Furthermore, we found memantine, an antagonist of α7 nAChR approved by the FDA for therapy of dementia, could ameliorate E. coli K1 meningitis very efficiently (9, 10). While these studies revealed the critical role of α7 nAChR in E. coli K1 meningitis, little is known about how E. coli K1 manipulates α7 nAChR to facilitate its translocation into the central nervous system (CNS).
As a ligand-gated ion channel, α7 nAChR is abundantly expressed throughout the brain, including brain microvascular endothelium and astrocytes, which are major components of the BBB (11–13). The α7 nAChR is activated by many endogenous and exogenous ligands. Nicotine, a notable exogenous α7 nAChR agonist derived from tobacco, has been reported to impair the BBB permeability and host-microbial defense via stimulation of α7 nAChR (14–18). Many researchers have demonstrated that children under 5 years who had exposure to tobacco smoke in the air suffered from a higher risk of meningitis (19, 20). In contrast to nicotine, secreted Ly6/Plaur domain containing 1 (SLURP1) is an endogenous α7 nAChR ligand. SLURP1 serves as a positive allosteric modulator to potentiate α7 nAChR activity effectively (21, 22). However, whether this endogenous α7 nAChR ligand could dampen host defense against E. coli K1 to promote its penetration of the BBB is unclear. Thus, the present study aimed to explore the role of SLURP1 in the pathological process of E. coli K1 meningitis.
Materials and Methods
Ethics Approval
The Medical Ethics Committee of Southern Medical University approved all of the animal experiments (Protocol number: L2018018). All the experiments on mice were done according to the corresponding guidelines. Every attempt was taken to minimize the number and suffering of mice used. We purchased neonatal C57BL/6 mice (8 days old) from the Animal Experimental Center of Southern Medical University. The α7 nAChR heterozygous (A7R+/−) mice with C57BL/6J background were obtained from the Jackson Laboratory (B6.129S7-Chrna7tm1Bay/J, Stock No: 003232, Bar Harbor, ME). Littermate A7R−/− and A7R+/+ (wild-type) mice were generated from the heterozygous for the experiment. All animals were specific pathogen free and were kept on a 12-h light/dark cycle and free to get food and water.
Public Transcriptional Data and Analysis
Two transcriptional data of E. coli infection patients were retrieved from Gene Expression Omnibus database (GSE33341, GSE65088). The metadata and SLURP1 transcriptional levels, as measured by fragments per kilobase of exon per million reads mapped (FPKM), were directly extracted from the data sets.
Chemicals and Reagents
The chemicals and reagents used in this study were obtained as follows: Sigma-Aldrich (St. Louis, MO, USA) for bull serum albumin (BSA), Evans blue, Triton X-100, Tween-20, 4′,6-diamidino-2-phenylindole (DAPI), isopropyl-β-d-thiogalactoside (IPTG), Coomassie brilliant blue G 250, rifampin, kanamycin, gentamicin, and methyllycaconitine citrate (MLA); Thermo Fisher Scientific (Waltham, MA, USA) for fluorescent α-bungarotoxin conjugates; Gibco (Thermo Fisher Scientific, Inc., Waltham, MA, USA) for penicillin G, streptomycin, glutamine, and pyruvate; Abcam (Cambridge, UK) for antibodies against SLURP1, α7 nAChR or β-actin; Proteintech (Proteintech Group, Chicago, IL, USA) for enzyme-linked immunosorbent assay (ELISA) kits. The rest reagents were purchased from Beyotime Institute of Biotechnology, Shanghai, China.
Clone, Expression, and Purification of Recombinant SLURP1
In this study, the amino acid sequence of SLURP1 we used to construct recombinant was described by previously (23), which is secreted by N-terminal signal cleavage (23-103 aa, as showed in Supplementary Figure S1A). Total RNA was extracted and used to amplify the SLURP1 cDNA, using primers containing 5′ BamHI and 3′Not I restriction sites at their termini. The primers used in the study were: sense, 5′-CGGGATCCCTCAAGTGCTACACCTGCAA-3′, and antisense, 5′ TTGCGGCCGCTCAGAGTTCCGAGTTGCAGA-3′. The cDNA was ligated into BamHI- and NotI- digested pET-28a, and transformed into E. coli BL21(DE3). Bacteria was added into the LB broth (1:100, containing 50 μg/ml kanamycin) and incubated at 37°C for 3–4 h (OD≈1). Afterward, IPTG was added at the final concentration of 0.1 mM and incubated at 30°C for 8 h for induction of protein expression. The protein was expressed as inclusion body form and purified using His-Tagged Protein Purification Kit (KangWeiShiJi Inc, Beijing, China) at denatured condition according to the manufacturer’s instructions. Purified SLURP1 was refolded in a series of gradient solutions of urea containing 50 mM Tris-HCl (pH 7.0), 0.5 M L-arginine, 4 mM glutathione, 1 mM glutathione disulfide, and 20% glycerin. The potential endotoxin was removed by passing through a Detoxi-Gel Endotoxin Removing Gel (Pierce Biotechnology, Rockford, IL, USA).
Bacterial Strains, Cell Lines, Invasion, and PMN Transmigration Assay
E. coli K1 strain RS218 (O18:K1:H7) was isolated from the cerebrospinal fluid (CSF) of a meningitis neonate and showed rifampicin-resistant property (8, 24). The brain heart infusion broth was used to culture E. coli K1 at 37°C for 14 h, with supplementation of rifampin (100 μg/ml). The immortalized human brain microvascular endothelial cells (HBMEC) were isolated and cultured as described previously (8, 24, 25). RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used as basic medium, with the following supplementations: 10% fetal bovine serum from Gibco (Thermo Fisher Scientific, Inc., Waltham, MA, USA), 50 U/ml penicillin G, 50 μg/ml streptomycin, 2 mM glutamine, and 1 mM pyruvate according to previous studies (8, 25).
For invasion assays, HBMEC were cultured in 24-well plates and incubated with SLUPR1 (0.1–2 μg/ml) for 2 h, followed by infected with E. coli K1 [1 × 107 colony-forming unit (CFU)] for another 2 h. To kill the extracellular bacteria, the HBMEC was washed twice with sterile PBS and incubated with RPMI 1640 medium containing gentamicin (100 μg/ml) for 1 h. Then, the HBMEC were washed again and lysed using sterile water. Internalized bacteria were counted by plating the cell lysates on Luria-Bertani broth agar (containing 100 μg/ml rifampicin).
Polymorphonuclear leukocytes (PMN) transmigration experiments were carried out as previously (8, 14, 25). HBMEC monolayers on transwell filters (3 μm pore size, 6.5 mm diameter, Corning, product number 3415) were monitored by measuring trans-endothelial electrical resistance (TEER) changes across the endothelial cell monolayer using an End Ohm epithelial voltohmeter (World Precision Instruments, Sarasota, FL, USA). To exclude the possibility that the PMN migration elicited was due to destruction of HBMEC monolayer, the integrity of the monolayer was inspected by TEER and microscopy before the start of the PMN transmigration assay. Transwell filters with or without supplementation of SLUPR1 (0.1–2 μg/ml) were employed to culture fully confluent HBMEC monolayers for 2 h. Then E. coli K1 (1 × 105 CFU/ml) was added to the bottom of the Transwell filters and infected for another 2 h. Then PMNs were applied to the upper compartment at a concentration of 1 × 106 cells. The Transwell filters system was kept at 37°C, 5% CO2. After incubated for 4 h, the Transwell filters were removed and migrated PMNs in the bottom of 24-well plates were harvested and counted in a blinded manner.
Knockdown and Overexpression of SLURP1
SLURP1 expression was knockdown using RNA interference. In brief, predesigned siRNA specific for SLURP1 and nontargeting scrambled siRNA (control) were obtained from Santa Cruz Biotechnology (CA, USA). The Lipofectamine™3000 transfection reagents (Invitrogen, USA) were mixed with the siRNA solutions, applied to the HBMEC monolayers, and maintained at 37°C, 5% CO2 for 24 h. The transfected HBMECs were used for invasion and PMN transmigration assays as described above.
For overexpression of SLURP1, the full-length cDNA of SLURP1 was cloned into a pcDNA3.1(+) vector to construct the pcDNA3.1-SLURP1 expression plasmid. The control vector or pcDNA3.1-SLURP1 plasmid was transfected into HBMEC for 24 h using Lipofectamine™3000. The transfected HBMECs were used for invasion and PMN transmigration experiments as described above.
For the transfection in the Transwell filters, HBMECs were seeded onto Transwell filters and grown to confluency, then transfected with siRNA or SLURP1 expression vector as mentioned above. In order to ensure the barrier function remains comparable between different groups, HBMEC transfected with nontargeting scrambled siRNA or control vector were served as the scrambled control.
Mouse Model of E. coli K1 Meningitis
From postnatal days 8 to 10, neonatal C57BL/6 mice were intraperitoneally injected with SLURP1 or BSA daily at a dose of 0–100 mg/kg body weight. To establish an E. coli K1 meningitis model, mice were injected with E. coli K1 (1 × 106 CFU, in 20 μl PBS) intraperitoneally at day 10. Control mice were given 20 μl PBS using the same route of injection. After infection for 18 h, the blood samples were collected and plated on Luria-Bertani agar (containing 100 μg/ml rifampicin) plates. Puncture through cisterna magna were carried out to collect CSF samples, followed by inoculating into the Luria-Bertani agar plates (containing 100 μg/ml rifampicin). Mice were perfused with 30 ml sterile PBS by the intracardiac route. Then brain tissues were harvested under aseptic conditions and homogenized in saline. Serial tenfold dilutions of brain homogenates were carried out and plated on Luria-Bertani agar (containing 100 µg/ml rifampicin) for counting. CSF samples were stained with a FITC-Ly-6G (Gr-1) (ProteinTech Group, Chicago, IL, USA) antibody and counted under fluorescence microscopy for PMN counting. For Evans blue assay, mice were injected intraperitoneally with Evans blue at a concentration of 40 mg/kg body weight 3 h before sacrificing. After intracardiac perfused with 30 ml PBS, the brain tissues were harvested and immersed in formamide. The OD620 of the supernatant was measured using a spectrophotometry.
Immunohistochemical Staining
Brain tissues were kept in formalin and transported to histological examination. After cutting into 3 µm sections, the tissues were hematoxylin-eosin (H&E) stained to assess tissue inflammation and damage. For the immunohistochemical staining, xylene was first used to dewax paraffin sections for 10 min, followed by gradient alcohol to dehydrated and rinsing in distilled water. Then the sections were heated in citrate buffer solution at 100°C for 40 min to retrieve the antigen. Hydrogen peroxide/methanol (30%) was used to stop endogenous peroxidase activity (45 min at 25°C). One percent BSA was used to block tissue sections and then incubated with an antibody specific for rabbit anti-SLURP1 (1:200, Abcam) at 4°C for 12 h. After repetitive washing by PBS, the sections were incubated with peroxidase-conjugated antirabbit antibodies, followed by visualization using 3,3-diaminobenzidine with hematoxylin counterstain. Immunostaining was quantified by ImageJ software.
For the immunofluorescence staining, the deparaffinization and antigen retrieval of sections were done as described above. Three percent normal goat serum and 0.1% Triton X-100 were used to block sections. After washing, sections were incubated with antibody specific for SLURP1 (1:200, Abcam) and a7 nAChR (1:200, Abcam) at 4°C for 16 h. After washing and incubating with appropriate secondary antibodies and DAPI, the sections were observed using fluorescence microscopy. NIH image analysis software (ImageJ) was employed to quantify the results of immunofluorescence staining.
Fluorescent α-Bungarotoxin Binding
HBMEC cells cultured in 24-well tissue were treated with BSA (2 μg/ml), SLURP1 (2 μg/ml), E. coli K1 (1 × 105 CFU) or E. coli K1 (1 × 105 CFU) + anti-SLURP1 antibody (1 μg) at 37°C, 5% CO2 for 2 h. The SLURP1 antibody was added simultaneously with E. coli K1. After general washing, 4% paraformaldehyde were used to fix cells for 10 min and 5% BSA was employed to block cells for 30 min. Alexa Fluor 488-conjugated α-bungarotoxin was then added at a concentration of 1 μg/ml and kept for 6 h at room temperature. Followed by washing, the sections were observed using fluorescence microscopy. The relative fluorescence intensity was determined using the ImageJ software (NIH). Briefly, the Spot Enhancing Filter 2D plugin was used to amplify signals from the cells, and then threshold settings were used to specifically select the fluorescent regions. The selected regions were overlaid on the original images and analyzed for mean fluorescence intensity of the area.
Immunoblot Analysis
The culture supernatants of HBMEC infected with or without E. coli K1 were concentrated using ultrafiltration for immunoblot analysis. Cell lysates were prepared in RIPA buffer. SDS-polyacrylamide gel was used to separated protein (20–30 μg), followed by transferring onto polyvinylidene difluoride membranes (Millipore). Five percent of skim milk was used to block membranes for 1 h. Membranes were then incubated with rabbit-anti-SLURP1 antibody (1:1,000, Abcam) at 4°C overnight. β-Actin (1:20,000) or Coomassie staining of total proteins was employed as an internal control. SLURP1 expression was detected using goat antirabbit IgG antibody conjugated with horseradish peroxidase and enhanced chemiluminescence reagent kit.
ELISA
The proinflammatory cytokines TNF-α (ab208348, Abcam), MMP-9 (ab253227, Abcam), and ICAM-1 (ab100688, Abcam) from homogenized brain extracts were evaluated using ELISA kits according to the manufacturer’s instructions.
Statistical Analysis
Data are shown in mean ± standard error. All the analyses in this study were done by SPSS (v25.0). Group differences between two groups were analyzed using the Student’s t-test. Group differences between three or more groups were analyzed using the one-way ANOVA followed by Bonferroni post-hoc test. Survival rates comparations were analyzed with log-rank test. Two-side p-value less than 0.05 was considered significant and is represented as *p < 0.05, **p < 0.01, and **p < 0.001.
Results
E. coli K1 Infection Induces SLURP1 Secretion in Cell Lines, Murine Model, and Humans
Secretion of SLURP1 in the culture supernatants of E. coli K1-infected HBMEC were analyzed using immunoblot assay and ELISA. We found E. coli K1 infection enhanced the SLURP1 secretion in both time- and dose-dependent manner compared with uninfected HBMECs (Figures 1A, B). To confirm these results in vivo, we assessed the SLURP1 expression in brain sections of neonatal C57BL/6 mice infected with or without E. coli K1 by immunohistochemical detection. As shown in Figures 1C, D, SLURP1 protein expression from hippocampus areas was significantly increased in mice infected with E. coli K1 compared with that of control. Notably, we also found a lot of SLURP1 was specially gathered around the blood vessels in the cortex sections of mice infected with E. coli K1 (Figures 1E, F). ELISA showed that mice infected with E. coli K1 showed a higher concentration of SLURP1 both in the serum and CSF than that of control (Figures 1G, H). Pearson correlation analysis indicated that the concentrations of SLURP1 were positively correlated with E. coli K1 counts in the CSF (Figure 1I, r = 0.7635, p = 0.0167).
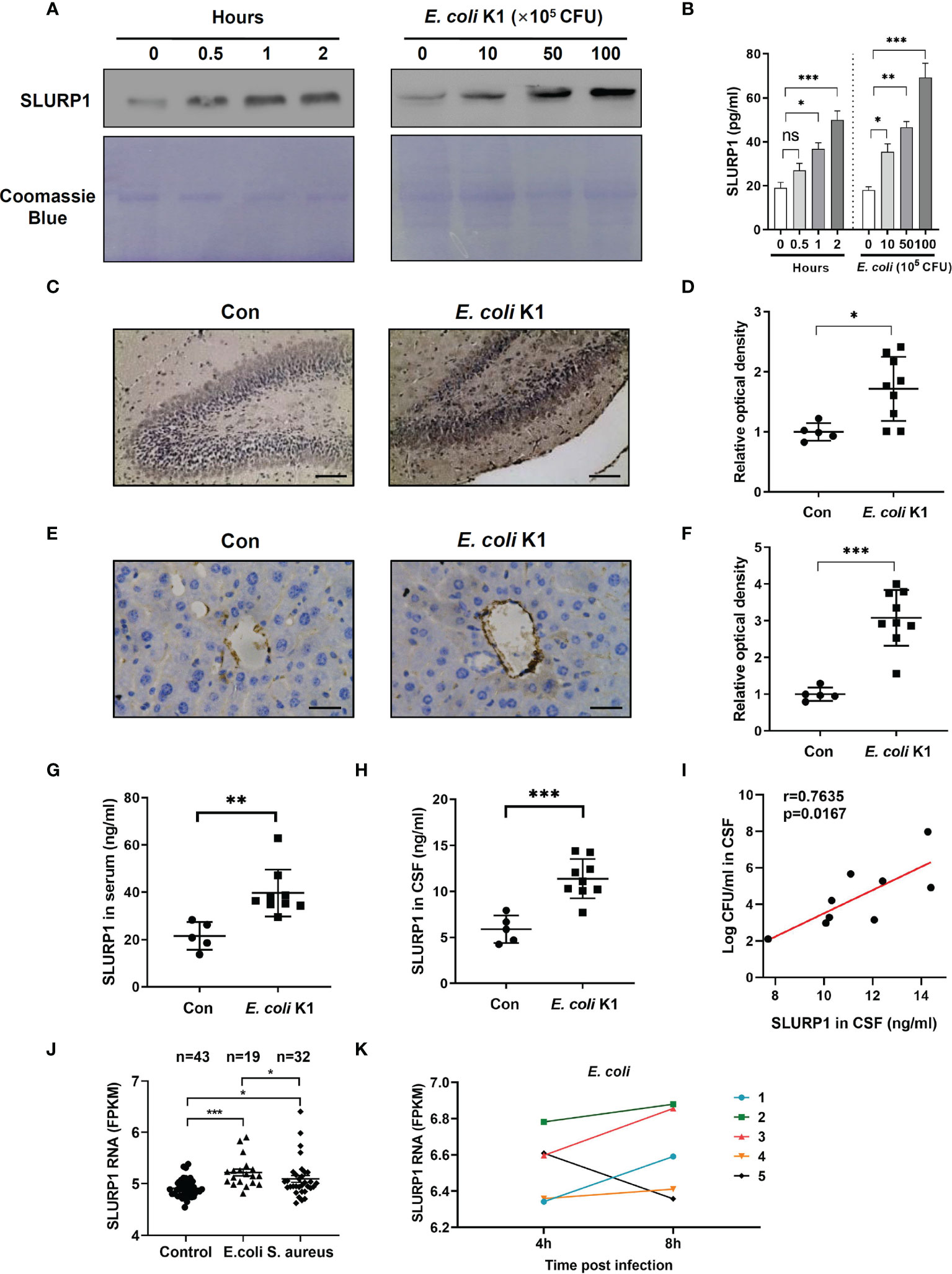
Figure 1 E coli K1 enhances SLURP1 secretion. (A, B) Immunoblot analysis (A) and ELISA (B) showed the SLURP1 release in the culture supernatant of HBMEC infected with E coli K1 (5 × 106 CFU) at different time intervals (0–2 h, left panel), or infected with various doses of E coli K1 (0–1 × 107 CFU) for 2 h (right). (C) Immunohistochemical staining of SLURP1 in the hippocampus sections of mice infected with or without E coli K1, scale bar = 200 μm. (D) The semiquantitative results of immunohistochemical staining of SLURP1, shown as fold change relative to control. (E) Immunohistochemical staining of SLURP1 in the cortex sections from mice challenged with or without E coli K1, scale bar = 40 μm. (F) Semiquantitative analysis of immunohistochemical staining of SLURP1, shown as fold change relative to control. (G, H) Quantification of SLURP1 secretion in the serum (G) or CSF (H) of mice infected with or without E coli K1. (I) Correlation analysis of SLURP1 levels and E coli K1 counts in the CSF. (J) Relative SLURP1 transcription levels (FPKM) among patients with E coli or S. aureus sepsis. (K) Change of relative SLURP1 transcription levels (FPKM) of five patients in 4- or 8-h post-E. coli infection. FPKM, fragments per kilobase of exon per million reads mapped. Data are presented as mean ± SEM. The immunoblots and immunohistochemical results are representative of two independent experiments (A, C, E). The data are displayed as the mean ± SEM from two independent experiments (B, D, F, G–I). *p < 0.05; **p < 0.01; ***p < 0.001 by one-way ANOVA followed by Bonferroni post-hoc test (B, J) and Student’s t-test (D, F–H). Correlation analysis was performed using Pearson and Spearman correlation tests. ns, not significant.
To identify if E. coli infection can upregulate SLURP1 transcription in humans, we analyzed one public human data set encompassed sepsis caused by E. coli or Staphylococcus aureus (S. aureus), and found that SLURP1 transcription level was higher in E. coli-infected patients, as compared with healthy controls or S. aureus-infected patients (Figure 1J). Furthermore, SLURP1 transcription levels get higher at 8 h postinfection than 4 h postinfection of E. coli (Figure 1K). Together, these results revealed that E. coli infection could induce SLURP1 secretion.
SLURP1 Promotes E. coli K1 Invasion and PMN Transmigration Across the BBB In Vitro
The pathogenesis of E. coli K1 meningitis required two key events: invasion of the brain microvascular endothelial cells by the bacteria and PMN transmigration across the BBB (8), we therefore next employed immortalized HBMEC monolayers to determine whether exogenous supplement of SLURP1 could promote E. coli K1 penetrating the endothelial cells, as well as support PMN transmigration across the BBB in vitro. SLURP1 was obtained recombinantly with a His-tag as described in the Material and Methods section. The amino acid sequence is shown in Supplementary Figure S1A. The results of double-digestion analysis, protein purity, and immunoblot detection, DNA sequencing are shown in Supplementary Figures S1B, C and Supplementary Material 1, respectively. As shown in Figures 2A, B, supplement with SLURP1 promoted E. coli K1 penetrating the endothelial cells, accompanied by enhancing E. coli K1-induced PMN transmigration across the HBMEC monolayers in a dose- and time-dependent manner. To explore the possibility that SLURP1 may promote E. coli K1 growth, we compared its growth on brain heart infusion broth in presence or absence of SLURP1. The result showed that SLURP1 has no obvious influence on E. coli K1 growth (Figure 2C), suggesting that the promotive effects of SLURP1 on E. coli K1 infection is not through promoting the growth of the pathogen. In order to further confirm the promotive role of SLURP1, we generated SLURP1 overexpression or knockdown HBMEC. Figures 2D, G showed the effects of SLUPR1 overexpression or knockdown, respectively. We found overexpression/knockdown of SLUPR1 significantly increased/decreased E. coli K1 penetrating the endothelial cells and PMN transmigration across the HBMEC monolayers, respectively (Figures 2E, F, H, I). Taken together, these results indicate that SLURP1 promotes E. coli K1 invasion of the endothelial cells, as well as enhances PMN transmigration across the BBB.
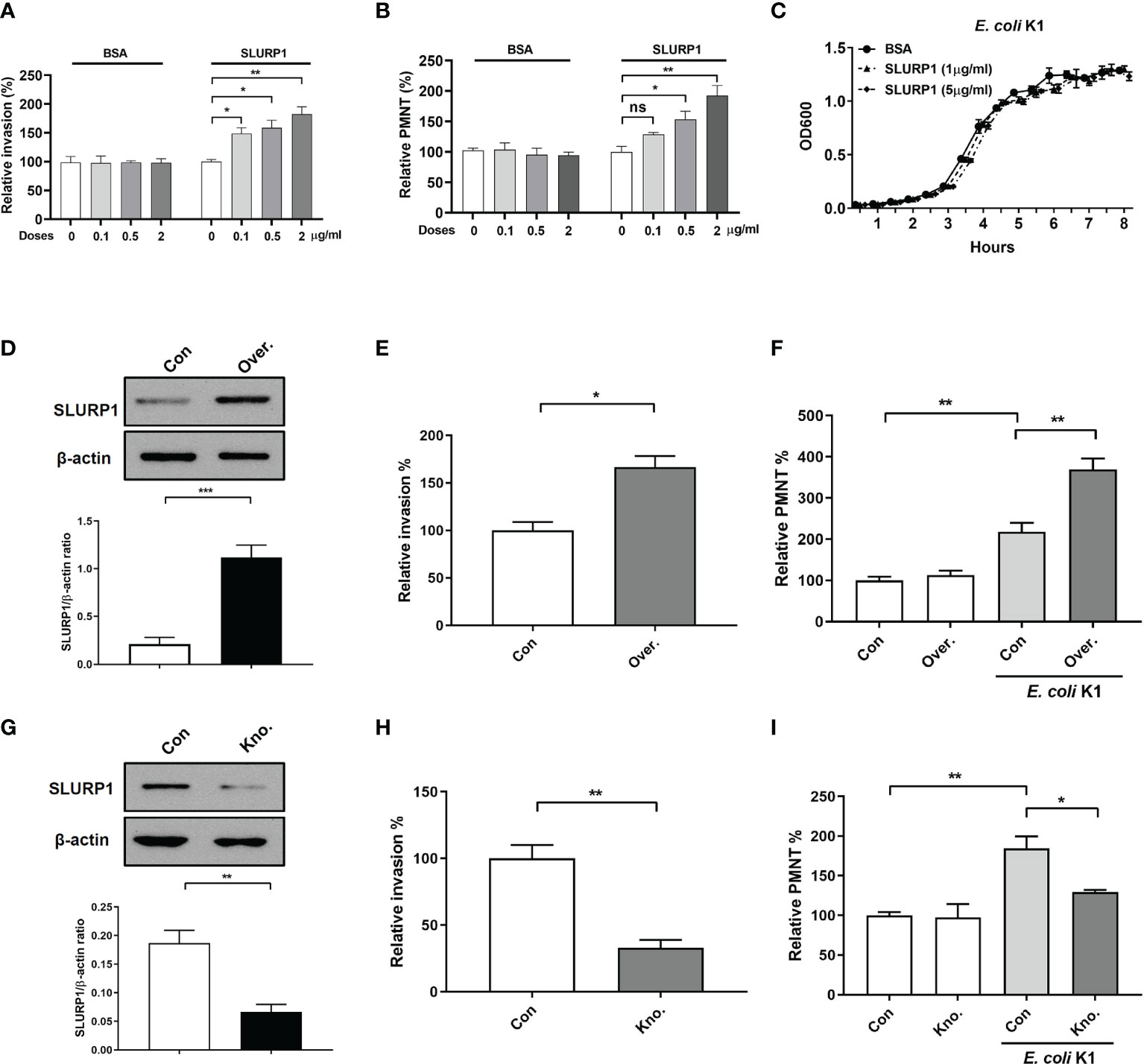
Figure 2 The effect of SLURP1 on E coli K1 penetration and PMN transmigration across the BBB in vitro. (A) The invasion of E coli K1 into HBMEC which pretreated with indicated doses of BSA or SLURP1. Data are presented as percent of the control values. (B) Transwell-cultured HBMEC monolayers were pre treatment with indicated doses of BSA or SLURP1 for 2 h, followed by incubated with E coli K1 in the bottom and PMN in the top of filter successively. PMN in the bottom of filter were harvested and counted. (C) The growth curve of E coli K1 in medium containing indicated doses of SLURP1. (D) The upregulation effect of SLURP1 in HBMEC which had been transfected with pCNDA3.1+-SLURP1. (E) The invasion of E coli K1 into HBMEC which had been transfected with pCNDA3.1+-SLURP1. Data are presented as percent of the control values. (F) Effect of SLURP1 upregulation on E coli K1-induced PMN transmigration across the HBMEC monolayers. Data are presented as percent of the control values. (G) The knockdown effect of SLURP1 in HBMEC transfected with siRNA. (H) The invasion of E coli K1 into HBMEC which had been transfected with siRNA. Data are presented as percent of the control values. (I) Effect of SLURP1 knockdown on E coli K1-induced PMN transmigration across the HBMEC monolayers. Data are presented as percent of the control values. Over., overexpression; Kno., knockdown. The immunoblots results are representative of three independent experiments (D, G). The data are displayed as the mean ± SEM from three independent experiments (A–I). *p < 0.05; **p < 0.01 by one-way ANOVA followed by Bonferroni post-hoc test (A, B, F, I) and Student’s t-test (D, E, G, H). ns, not significant.
SLURP1 Promotes the Pathogenesis Process of E. coli K1 Meningitis In Vivo
In order to confirm the biological significance of the in vitro findings described above, we further tested the effect of SLURP1 on E. coli K1 meningitis pathogenesis in the murine model. Neonatal mice were intraperitoneally administered with BSA or SLURP1 two consecutive days prior to E. coli K1 challenge. The survival rate, pathogen counts in CSF, PMN transmigration, and brain damage were detected as described in Materials and Methods. The results showed that only 20% of the SLURP1-pretreated mice infected with E. coli K1 survived within 60 h postinfection (Figure 3A), while the survival rate for E. coli K1-infected mice without SLURP1 supplementation reached almost 50%. Furthermore, we found administration with SLURP1 was able to markedly increase pathogen and PMN counts in the CSF (Figures 3B, C). Evans blue assay showed that SLURP1-pretreated mice have more severe BBB damage than the control group (Figure 3D). Notably, we found SLURP1 has no influence on the BBB integrity of uninfected mice. H&E staining of brain sections indicated that supplement with SLURP1 dramatically promotes neutrophil infiltration into the meninges and meningeal inflammation (Figures 3E, F). Additionally, we found exogenous supplement of SLURP1 could robustly enhance the levels of proinflammatory cytokines in brain homogenates (Figures 3G–I). Above all, these results suggested that SLURP1 promotes the pathogenesis process of E. coli K1 meningitis.
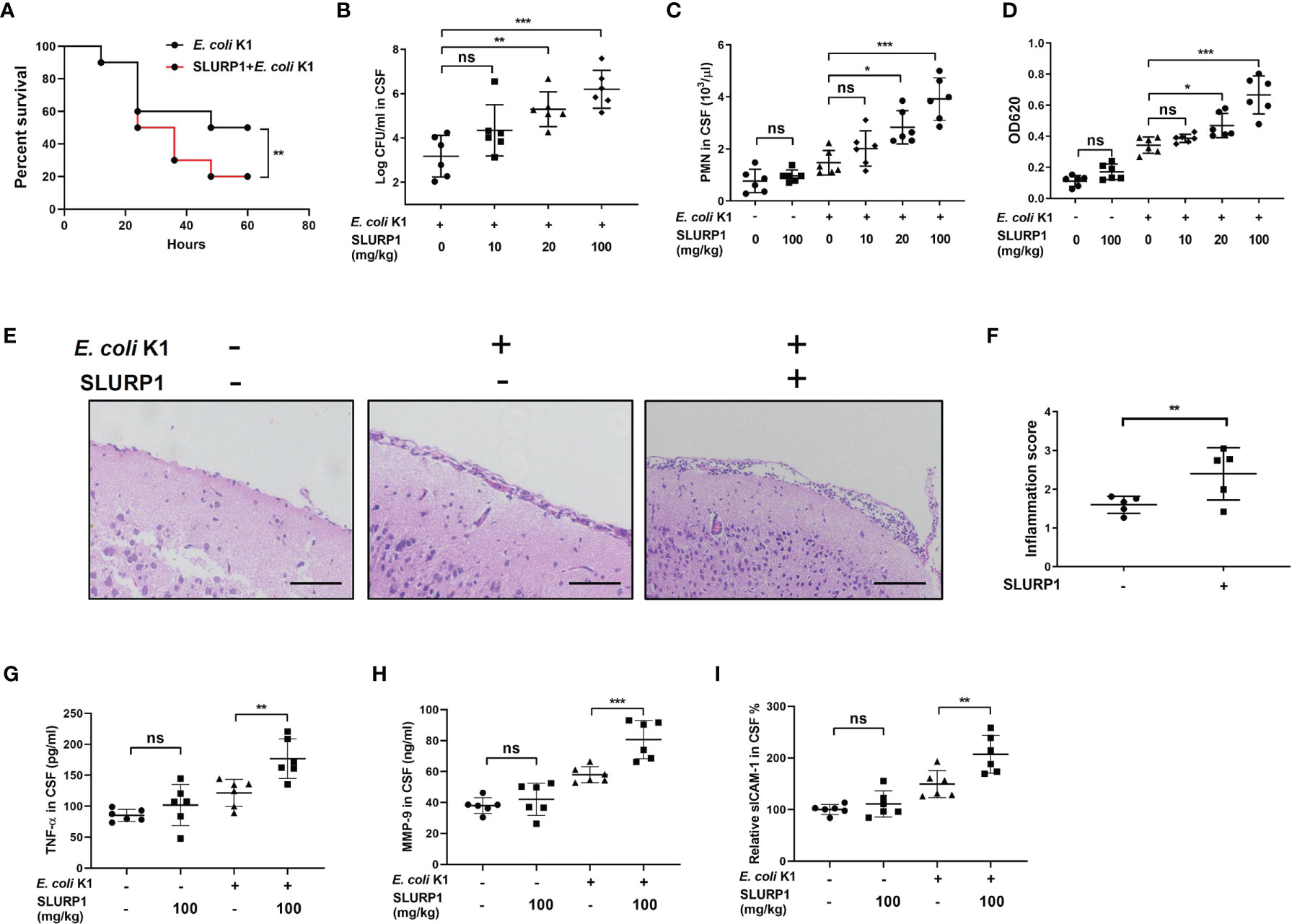
Figure 3 SLURP1 promotes the pathogenesis of E coli K1 meningitis in mice model. (A) Survival curve of C57BL/6 mice treated with SLURP1 (100 mg/kg body weight) + E. coli K1 (1 × 106) or treated with only E coli K1 (1 × 106). SLURP1 was intraperitoneally injected 2 days before E coli K1 challenge. n = 10 per group. (B, C) Pathogen (B) and PMN (C) counts in the CSF of E coli K1-infected mice pretreated with BSA or indicated doses of SLURP1. (D) The OD620 values of Evans blue extracted from the brain of E coli K1-infected mice pretreated with BSA or indicated doses of SLURP1. (E) Representative H&E staining of the cortex sections, scale bar = 200 μm; and (F) meningeal inflammation score. (G–I) The cytokines levels in the CSF were analyzed by ELISA: TNF-α (G), MMP-9 (H), and ICAM-1 (I). The H&E staining are representative of two independent experiments (E). Data are presented as mean ± SEM from two independent experiments. Each dot indicates an individual mouse (n = 5). *p < 0.05; **p < 0.01; ***p < 0.001 by log-rank test (A), Student’s t-test (F), and one-way ANOVA followed by Bonferroni post-hoc test (B–D, G–I). ns, not significant.
E. coli K1-Induced SLURP1 Activates α7 nAChR
We further confirmed if E. coli K1-induced SLURP1 activates α7 nAChR. Firstly, a fluorescent α-bungarotoxin (α-bgtx) binding assay was performed to detect the activity of α7 nAChR. HBMEC were cultured in 24-well plates, followed by incubating with SLURP1 or E. coli K1 for 2 h. Afterward, fluorescently labeled α-bgtx incubation was carried out to test α7 nAChR activity. As shown in Figures 4A, B, SLURP1 or E. coli K1-treated HBMEC showed brighter green fluorescence than control, indicating an increase in α7 nAChR activity. Notably, when added with SLURP1 antibody, the promotive effect of E. coli K1 on α7 nAChR activity was blocked, indicating the E. coli K1-induced SLURP1 was responsible for the α7 nAChR activation. To further confirm whether SLURP1 is directly linked to α7 nAChR activation, we analyzed the colocalization of fluorescently labeled SLURP1 with α7 nAChRs by immunofluorescence staining. The cortex sections of E. coli K1-treated mice showed colocalization of SLURP1 (green) and α7 nAChRs (red) (Figure 4C). What is more, with increasing doses of E. coli K1 challenge, the Pearson’s coefficient and overlap coefficient of colocalization also increased (Figure 4D). These results suggest that E. coli K1-induced SLURP1 is responsible for α7 nAChR activation.
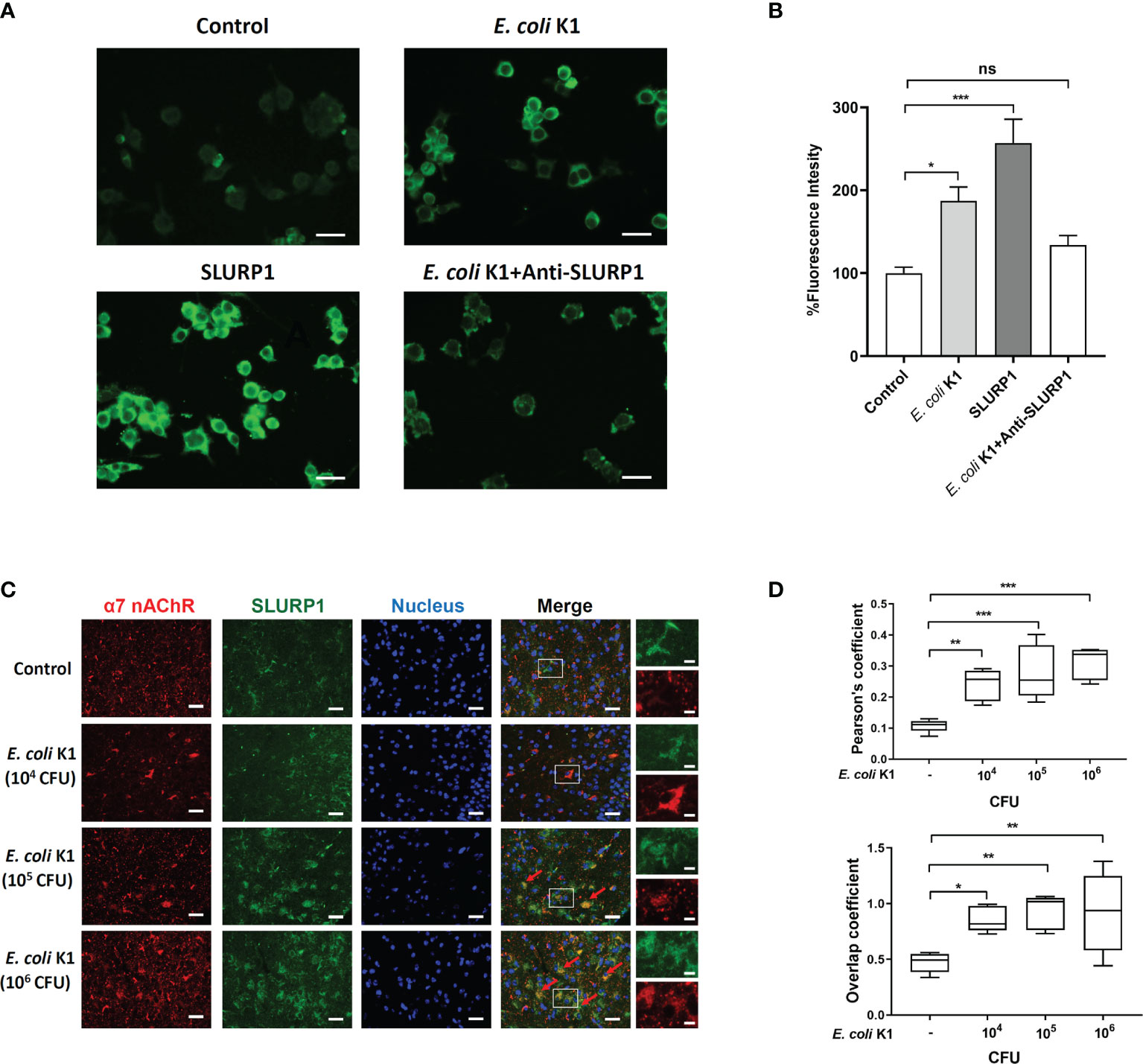
Figure 4 E coli K1-induced SLURP1 activates α7 nAChR. (A) The Alexa Fluor 488-conjugated α-bungarotoxin binding to HBMEC was detected upon treatment with BSA (control, 2 μg/ml), E coli K1 (1 × 105 CFU), SLURP1(2 μg/ml) or E coli K1 (1 × 105 CFU) + SLURP1 antibody (1 μg). SLURP1 antibody was added simultaneously with E coli K1. Scale bars = 40 μm. (B) Fluorescence intensity of Alexa Fluor 488-conjugated α-bungarotoxin binding to HBMEC. Data are presented as percent of the control values. (C) The cortex sections of neonatal mice infected with indicated doses of E coli K1 were permeabilized and immunofluorescence stained with either α7 nAChR and SLUPR1 antibodies. Nuclei were stained by DAPI. Arrows show the colonization of α7 nAChR (red) and SLUPR1 (green). Scale bars = 50 μm (left) or 10 μm (right). (D) Quantification of the Pearson’s correlation coefficient (upper panel) and overlap coefficient (bottom) for colonization of α7 nAChR and SLURP1. Data are presented as mean ± SEM from three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001 by one-way ANOVA followed by Bonferroni post-hoc test (B, D). ns, not significant.
Inhibition of α7 nAChR Blocks the Promotive Effects of SLURP1 in the Pathogenesis of E. coli K1 Meningitis
Finally, we determined whether α7 nAChR is necessary for SLURP1-enhanced E. coli K1 meningitis. We first used the MLA, an α7 nAChR inhibitor, to explore the role of α7 nAChR on the function of SLURP1 in vitro. As shown in Figures 5A, B, MLA inhibited the promotive effects of SLURP1 in a dose-dependent manner, including attenuating E. coli K1 invasion and PMN transmigration. Furthermore, we used α7 nAChR knockout (A7R−/−) mice to confirm these findings in vitro. Wild-type (A7R+/+) and A7R−/− mice were intraperitoneally injected with BSA or SLURP1 for 2 days, followed by challenge with E. coli K1 for 18 h. As shown in Figures 5C–F, SLURP1 treatment has enhanced the pathogen load, PMN transmigration and the BBB damage in the A7R+/+ mice, while all these promotive effects of SLURP1 were blocked in the A7R−/− mice. H&E staining of the brain sections showed that neutrophil recruitment and meningeal inflammation of SLURP1-treated A7R−/− mice was not significantly increased when compared with untreated A7R−/− mice (Figures 5G, H). Above all, these results indicated that SLURP1 acts through α7 nAChR to enhance the pathogenesis process of E. coli K1 meningitis.
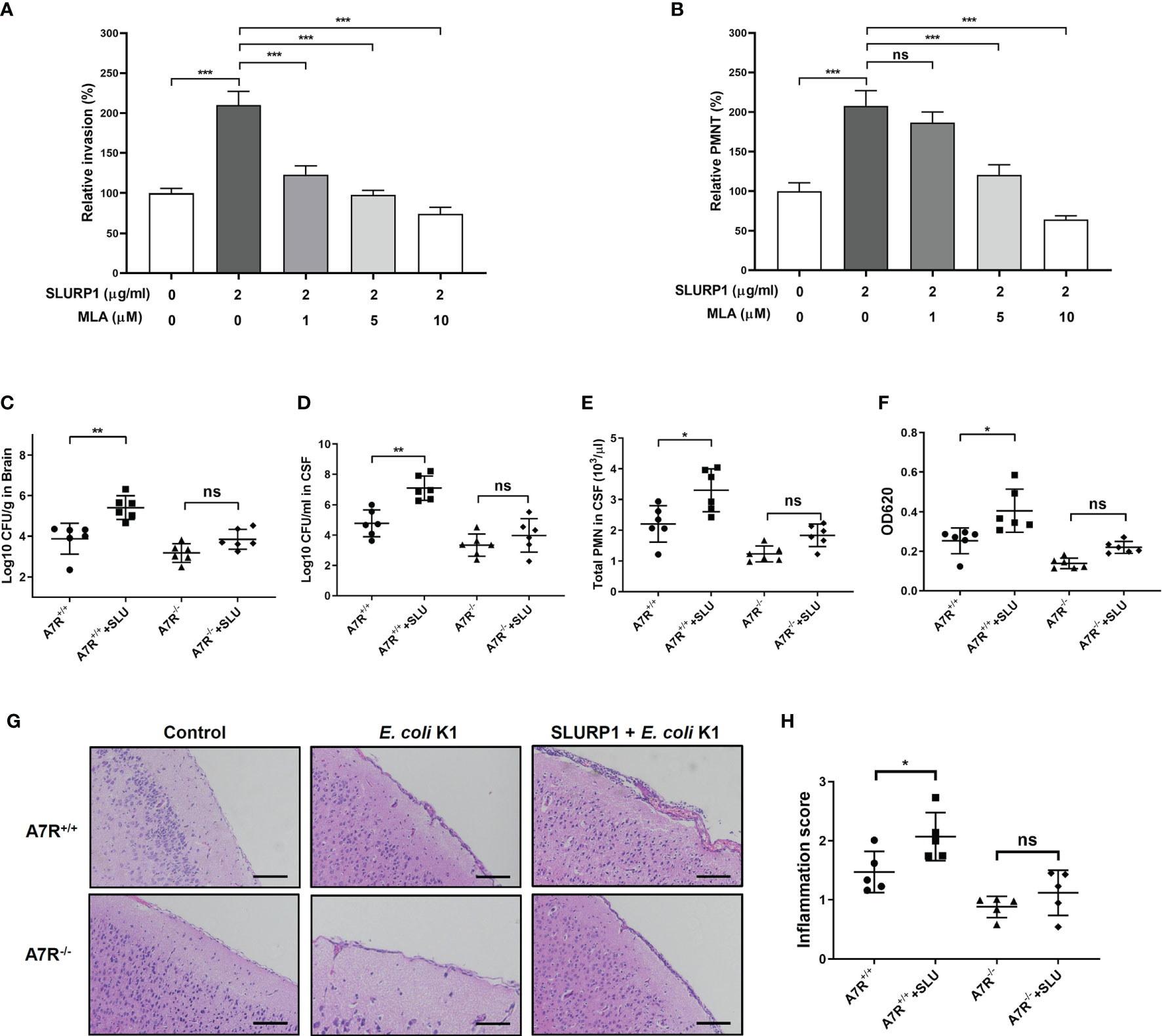
Figure 5 Effects of α7 nAChR knockout on SLURP1-enhanced E coli K1 entry into the CNS and PMN transmigration across the BBB. (A) The invasion of E coli K1 into HBMEC which pretreated with indicated doses of SLURP1 or SLURP1+α7 nAChR inhibitor MLA. Data are presented as percent of the control values. (B) E coli K1-induced PMN transmigration across the HBMEC monolayers which pretreated with indicated doses of SLURP1 or SLURP1+α7 nAChR inhibitor MLA. Data are presented as percent of the control values. (C, D) Pathogen counts in the brain tissues (C) or CSF (D) of E coli K1-infected wild type (A7R+/+) or α7 nAChR knockout (A7R−/−) mice pretreated with or without SLURP1 (100 mg/kg body weight). (E) PMN counts in the CSF of E coli K1-infected A7R+/+ or A7R−/− mice pretreated with or without SLURP1 (100 mg/kg body weight). (F) Measurement of the BBB permeability by Evans blue assay in E coli K1-infected A7R+/+ or A7R−/− mice administrated with or without SLURP1 (100 mg/kg body weight). (G) Representative H&E staining of the brain cortex sections, scale bar, 200 μm, and (H) meningeal inflammation score. The H&E staining are representative of two independent experiments (G). Data are presented as mean ± SEM from two independent experiments. Each dot indicates an individual mouse. *p < 0.05; **p < 0.01; ***p < 0.001 by one-way ANOVA followed by Bonferroni post-hoc test (A–F, H). ns, not significant.
Discussion
As an important cholinergic anti-inflammatory signaling, α7 nAChR has been intensively investigated in sterile inflammation over the last decades (26–30). However, there are few studies that focus on the role of α7 nAChR in the host immune response to microbial infection. A number of studies showed that activation of α7 nAChR impairs host defense to bacterial infections, indicating that the use of α7 nAChR ligands might not be a good strategy in treating infectious diseases (31–37). Ilona and coworkers have demonstrated that stimulates α7 nAChR promotes the development of E. coli peritonitis after intraperitoneal infection (37). Consistent with their study, our group recently revealed that α7 nAChR impaired the host defense against E. coli K1 infection in the CNS (7–9). In this report, we further found that E. coli K1 infection promotes SLURP1, an endogenous α7 nAChR ligand secretion, and supplement of SLURP1 could greatly facilitate E. coli K1 dissemination into the CNS. These findings expand our knowledge of the pathogenesis process of E. coli K1 infection and provide a new perspective on the establishment of the CNS infection.
SLURP1 is a secreted protein that has structural homology with three-finger snake α-neurotoxins, acts in a both autocrine and paracrine manner, to activate α7 nAChR and thus exert potent anti-inflammatory activity (38–43). Although the function of SLURP1 in modulated sterile inflammation has been extensively studied, its effects on the inflammation induced by microbial infection are largely unknown, especially on neuroinflammation. Taken into account that SLURP1 is highly expressed in the CNS, a study into its pathologist effects on the CNS infection constitutes a promising field for developing new therapeutic methods (44–46). To the best of our knowledge, this is the first study that reported that microbial infection could induce SLURP1 expression, and consequently stimulate α7 nAChR to establish infection. Whether the enhanced SLURP1 secretion is a universal phenomenon upon infection is very interesting and warrants further investigation.
How E. coli K1 promotes SLURP1 secretion needs to be addressed. Kruppel-like factor 4 (Klf4) is the first reported factor that regulates SLPRP1 expression (47, 48). Klf4 is a member of the Krüppel-like factor transcription factor family, which can stimulate microglial activation and induce neuroinflammation. As a zinc finger protein, Klf4 could effectively induce SLURP1 expression via binding to its promoter. Interestingly, two recent studies reported that Klf4 is robustly upregulated upon infection with pathogens like E. coli or Streptococcus pneumoniae (49, 50). Thus, we speculated that E. coli K1 might upregulate Klf4 expression to promote SLURP1 release. Another question that needs to be addressed is how SLURP1-α7 nAChR mediated the pathogenesis process of E. coli K1 meningitis. Previous studies by our groups and others have reported that nuclear factor-κB (NF-κB) is critical for E. coli K1 entry into the CNS (51, 52). Actually, Chernyavsky and coworkers have reported that SLURP1 can bind to α7 nAChR, activate the Raf-1/MEK1/ERK1/2 cascade to modulate NF-κB signaling (53). It has been demonstrated in our previous research that NF-κB modulation, CaMKII, ERK, and protein kinase C are involved in α7 nAChR-mediated signaling (7, 52, 54). It is most likely that the same pathway may contribute to SLURP1-mediated signaling as SLURP1 is an endogenous α7 nAChR ligand. It thus seems that SLPRP1-α7 nAChR-NF-κB cascade might be critical for E. coli K1 meningitis.
The limitation of the present study is that the recombinant SLURP1 used was not a native one. Recently, several studies reported the contradictory role of SLURP1 on α7 nAChR based on recombinant SLURP1 with N- and/or C-terminal extensions. In 2003 (22), it was demonstrated that a recombinant SLURP1 containing N-terminal hemagglutinin tag and C-terminal myc tag, could potentiate the α7 nAChRs-mediated responses, while a recent study by Lyukmanova et al. (38, 46) has reported the inhibitory role of recombinant SLURP1, which only added a Met residue in its N-terminal. However, this inhibitory effect was not observed in the case of a synthetic human SLURP1, which is identical with the amino acid sequence of the native source (55). These contradictory findings indicate that additional extensions may produce marked changes in the functional activity of SLURP1. To the best of our knowledge, we believe that SLURP1 may act as a positive modulator, because mutations in SLURP1 cause Mal de Meleda (an inflammatory palmoplantar hyperkeratosis), and α7 nAChR plays a central role in the differentiation of stratified squamous epithelium (22, 56, 57). In spite of this, the conclusion that SLURP1 facilitates E. coli K1 crossing the blood-brain barrier needed to be further verified by using the native SLURP1.
Taken together, the present study reveals that SLURP1, an endogenous α7nAChR ligand, is the key mediator for E. coli K1 meningitis pathogenesis. Blocking initial SLURP1-α7nAChR interaction would be an attractive strategy for preventing E. coli K1 meningitis.
Data Availability Statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by Medical Ethics Committee of Southern Medical University.
Author Contributions
HC, SH, TZ, and XH conceived and designed the experiment. XH, LW, FC, LQL, JG, YW, TH, and ZG performed the experiment. ZG, LW, XH, LL, PZ, TZ, JG, and FC analyzed the data. SH and PZ contributed reagents/materials/analysis tools. XH, HC, JG, BL, TH, and SH participated in its design and coordination and helped in drafting the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 81871198; 81801985; 81873762; 82060192), China Postdoctoral Science Foundation (No. 2020M682808; 2021T140299), the Key Research Program of Hunan Provincial Department of Science and Technology, China (No. 2022SK2032), Postdoctoral innovative practice project of Jiang Meng (No. JMBSH2020B04), and the Yunnan Key Laboratory of Children’s Major Disease Research (202005AG070073).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Prof. Bao Zhang for generously providing the vector pcDNA3.1+.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.745854/full#supplementary-material
References
1. Xu M, Hu L, Huang H, Wang L, Tan J, Zhang Y, et al. Etiology and Clinical Features of Full-Term Neonatal Bacterial Meningitis: A Multicenter Retrospective Cohort Study. Front Pediatr (2019) 7:31. doi: 10.3389/fped.2019.00031
2. Biondi EA, Lee B, Ralston SL, Winikor JM, Lynn JF, Dixon A, et al. Prevalence of Bacteremia and Bacterial Meningitis in Febrile Neonates and Infants in the Second Month of Life: A Systematic Review and Meta-Analysis. JAMA Netw Open (2019) 2(3):e190874–e190874. doi: 10.1001/jamanetworkopen.2019.0874
3. Zainel A, Mitchell H, Sadarangani M. Bacterial Meningitis in Children: Neurological Complications Associated Risk Factors and Prevention. Microorganisms (2021) 9(3):535. doi: 10.3390/microorganisms9030535
4. Sarowska J, Futoma-Koloch B, Jama-Kmiecik A, Frej-Madrzak M, Ksiazczyk M, Bugla-Ploskonska G, et al. Virulence Factors Prevalence and Potential Transmission of Extraintestinal Pathogenic Escherichia Coli Isolated From Different Sources: Recent Reports. Gut Pathog (2019) 11(1):1–16. doi: 10.1186/s13099-019-0290-0
5. Oordt-Speets AM, Bolijn R, van Hoorn RC, Bhavsar A, Kyaw MH. Global Etiology of Bacterial Meningitis: A Systematic Review and Meta-Analysis. PloS One (2018) 13(6):e0198772. doi: 10.1371/journal.pone.0198772
6. Liu R, Wu C, Li L, Chi F, Zhang T, Xu Y, et al. CD48 and α7 Nicotinic Acetylcholine Receptor Synergistically Regulate fimH-Mediated Escherichia Coli K1 Penetration and Neutrophil Transmigration Across Human Brain Microvascular Endothelial Cells. J Infect Dis (2019) 219(3):470–79. doi: 10.1093/infdis/jiy531
7. Chi F, Wang L, Zheng X, Jong A, Huang SH. Recruitment of α7 Nicotinic Acetylcholine Receptor to Caveolin-1-Enriched Lipid Rafts is Required for Nicotine-Enhanced Escherichia Coli K1 Entry Into Brain Endothelial Cells. Future Microbiol (2011) 6(8):953–66. doi: 10.2217/fmb.11.65
8. Chi F, Wang L, Zheng X, Wu CH, Jong A, Sheard MA, et al. Meningitic Escherichia Coli K1 Penetration and Neutrophil Transmigration Across the Blood–Brain Barrier are Modulated by Alpha7 Nicotinic Receptor. PloS One (2011) 6(9):e25016. doi: 10.1371/journal.pone.0025016
9. Yu JY, Zhang B, Peng L, Wu CH, Cao H, Zhong JF, et al. Repositioning of Memantine as a Potential Novel Therapeutic Agent Against Meningitic E Coli–induced Pathogenicities Through Disease-Associated Alpha7 Cholinergic Pathway and RNA Sequencing-Based Transcriptome Analysis of Host Inflammatory Responses. PloS One (2015) 10(5):e0121911. doi: 10.1371/journal.pone.0121911
10. Qiu HS, Li ZT, Lin QX, Cai YP, Lan J, Chen YL, et al. Memantine Attenuated E Coli K1-Induced Tight Junction Injuries Through Inhibition of α7 Nachr-Dependent Pathway In Vitro. Int J Infect Dis (2020) 101:132. doi: 10.1016/j.ijid.2020.09.360
11. Graham AJ, Martin-Ruiz CM, Teaktong T, Ray MA. Human Brain Nicotinic Receptors Their Distribution and Participation in Neuropsychiatric Disorders. Curr Drug Targets CNS Neurol Disord (2002) 1(4):387–97. doi: 10.2174/1568007023339283
12. Arvanitis CD, Ferraro GB, Jain RK. The Blood–Brain Barrier and Blood–Tumour Barrier in Brain Tumours and Metastases. Nat Rev Cancer (2020) 20(1):26–41. doi: 10.1038/s41568-019-0205-x
13. Profaci CP, Munji RN, Pulido RS, Daneman R. The Blood–Brain Barrier in Health and Disease: Important Unanswered Questions. J Exp Med (2020) 217(4):e20190062. doi: 10.1084/jem.20190062
14. Zhang B, Yu JY, Liu LQ, Peng L, Chi F, Wu CH, et al. Alpha7 Nicotinic Acetylcholine Receptor Is Required for Blood-Brain Barrier Injury-Related CNS Disorders Caused by Cryptococcus Neoformans and HIV-1 Associated Comorbidity Factors. BMC Infect Dis (2015) 15:352. doi: 10.1186/s12879-015-1075-9
15. Chen YH, Chen SHM, Jong A, Zhou ZY, Li W, Suzuki K, et al. Enhanced Escherichia Coli Invasion of Human Brain Microvascular Endothelial Cells is Associated With Alterations in Cytoskeleton Induced by Nicotine. Cell Microbiol (2002) 4(8):503–14. doi: 10.1046/j.1462-5822.2002.00209.x
16. Pimentel E, Sivalingam K, Doke M, Samikkannu T. Effects of Drugs of Abuse on the Blood-Brain Barrier: A Brief Overview. Front Neurosci (2020) 14:513. doi: 10.3389/fnins.2020.00513
17. Sajja RK, Rahman S, Cucullo L. Drugs of Abuse and Blood-Brain Barrier Endothelial Dysfunction: A Focus on the Role of Oxidative Stress. J Cereb Blood Flow Metab (2016) 36(3):539–54. doi: 10.1177/0271678X15616978
18. Wu C, Yang M, Liu R, Hu H, Ji L, Zhang X, et al. Nicotine Reduces Human Brain Microvascular Endothelial Cell Response to Escherichia Coli K1 Infection by Inhibiting Autophagy. Front Cell Infect Microbiol (2012) 10:484. doi: 10.3389/fcimb.2020.00484
19. Murray RL, Britton J, Leonardi-Bee J. Second Hand Smoke Exposure and the Risk of Invasive Meningococcal Disease in Children: Systematic Review and Meta-Analysis. BMC Public Health (2012) 12:1062. doi: 10.1186/1471-2458-12-1062
20. Bagaitkar J, Demuth DR, Scott DA. Tobacco Use Increases Susceptibility to Bacterial Infection. Tob Induc Dis (2008) 4(1):12. doi: 10.1186/1617-9625-4-12
21. Narumoto O, Niikura Y, Ishii S, Morihara H, Okashiro S, Nakahari T, et al. Effect of Secreted Lymphocyte Antigen-6/Urokinase-Type Plasminogen Activator Receptor-Related Peptide-1 (SLURP-1) on Airway Epithelial Cells. Biochem Biophys Res Commun (2013) 438(1):175–9. doi: 10.1016/j.bbrc.2013.07.048
22. Chimienti F, Hogg RC, Plantard L, Lehmann C, Brakch N, Fischer J, et al. Identification of SLURP-1 as an Epidermal Neuromodulator Explains the Clinical Phenotype of Mal De Meleda. Hum Mol Genet (2003) 12(22):3017–24. doi: 10.1093/hmg/ddg320
23. Adermann K, Wattler F, Wattler S, Heine G, Meyer M, Forssmann WG, et al. Structural and Phylogenetic Characterization of Human SLURP-1, the First Secreted Mammalian Member of the Ly-6/uPAR Protein Superfamily. Protein Sci (1999) 8(4):810–9. doi: 10.1110/ps.8.4.810
24. Stins MF, Gilles F, Kim KS. Selective Expression of Adhesion Molecules on Human Brain Microvascular Endothelial Cells. J Neuroimmunol (1997) 76(1-2):81–90. doi: 10.1016/s0165-5728(97)00036-2
25. He X, Shi X, Puthiyakunnon S, Zhang L, Zeng Q, Li Y, et al. CD44-Mediated Monocyte Transmigration Across Cryptococcus Neoformans-Infected Brain Microvascular Endothelial Cells is Enhanced by HIV-1 Gp41-I90 Ectodomain. J BioMed Sci (2016) 23:28. doi: 10.1186/s12929-016-0247-2
26. Hoover DB. Cholinergic Modulation of the Immune System Presents New Approaches for Treating Inflammation. Pharmacol Therapeut (2017) 179:1–16. doi: 10.1016/j.pharmthera.2017.05.002
27. Li DJ, Fu H, Tong J, Li YH, Qu LF, Wang P, et al. Cholinergic Anti-Inflammatory Pathway Inhibits Neointimal Hyperplasia by Suppressing Inflammation and Oxidative Stress. Redox Biol (2018) 15:22–33. doi: 10.1016/j.redox.2017.11.013
28. Andersson U. The Cholinergic Anti-Inflammatory Pathway Alleviates Acute Lung Injury. Mol Med (2020) 26(1):64. doi: 10.1186/s10020-020-00184-0
29. Benfante RD, Lascio S, Cardani S, Fornasari D. Acetylcholinesterase Inhibitors Targeting the Cholinergic Anti-Inflammatory Pathway: A New Therapeutic Perspective in Aging-Related Disorders. Aging Clin Exp Res (2021) 33(4):823–34. doi: 10.1007/s40520-019-01359-4
30. Xie H, Yepuri N, Meng Q, Dhawan R, Leech CA, Chepurny OG, et al. Therapeutic Potential of α7 Nicotinic Acetylcholine Receptor Agonists to Combat Obesity Diabetes and Inflammation. Rev Endocr Metab Disord (2020) 21(4):431–47. doi: 10.1007/s11154-020-09584-3
31. Giebelen IA, Leendertse M, Florquin S, van der Poll T. Stimulation of Acetylcholine Receptors Impairs Host Defence During Pneumococcal Pneumonia. Eur Respir J (2009) 33(2):375–81. doi: 10.1183/09031936.00103408
32. Sussan TE, Gajghate S, Thimmulappa RK, Ma J, Kim JH, Sudini K, et al. Exposure to Electronic Cigarettes Impairs Pulmonary Anti-Bacterial and Anti-Viral Defenses in a Mouse Model. PloS One (2015) 10(2):e0116861. doi: 10.1371/journal.pone.0116861
33. Su X, Matthay MA, Malik AB. Requisite Role of the Cholinergic α7 Nicotinic Acetylcholine Receptor Pathway in Suppressing Gram-Negative Sepsis-Induced Acute Lung Inflammatory Injury. J Immunol (2010) 184(1):401–10. doi: 10.4049/jimmunol.0901808
34. Radek KA, Elias PM, Taupenot L, Mahata SK, O'Connor DT, Gallo RL. Neuroendocrine Nicotinic Receptor Activation Increases Susceptibility to Bacterial Infections by Suppressing Antimicrobial Peptide Production. Cell Host Microbe (2010) 7(4):277–89. doi: 10.1016/j.chom.2010.03.009
35. Kanashiro A, Sônego F, Ferreira RG, Castanheira FV, Leite CA, Borges VF, et al. Therapeutic Potential and Limitations of Cholinergic Anti-Inflammatory Pathway in Sepsis. Pharmacol Res (2017) 117:1–8. doi: 10.1016/j.phrs.2016.12.014
36. Matsunaga K, Klein TW, Friedman H, Yamamoto Y. Involvement of Nicotinic Acetylcholine Receptors in Suppression of Antimicrobial Activity and Cytokine Responses of Alveolar Macrophages to Legionella Pneumophila Infection by Nicotine. J Immunol (2001) 167(11):6518–24. doi: 10.4049/jimmunol.167.11.6518
37. Giebelen IL, Moine A, van den Pangaart PS, Sadis C, Goldman M, Florquin S, et al. Deficiency of Alpha7 Cholinergic Receptors Facilitates Bacterial Clearance in Escherichia Coli Peritonitis. J Infect Dis (2008) 198(5):750–57. doi: 10.1086/590432
38. Tsetlin VI, Kasheverov IE, Utkin YN. Three-Finger Proteins From Snakes and Humans Acting on Nicotinic Receptors: Old and New. J Neurochem (2020) 158(6):1223–35. doi: 10.1111/jnc.15123
39. Swamynathan S, Tiwari A, Loughner C, Alexander N, Gnalian J, Swamynathan SK. Secreted Ly-6/uPAR Related Protein-1 (SLURP1) Modulates Inflammation by Influencing Both Neutrophils and Endothelial Cells. Invest Ophth Vis Sci (2018) 59(9):3325.
40. Campbell G, Swamynathan S, Tiwari A, Swamynathan SK. The Secreted Ly-6/uPAR Related Protein-1 (SLURP1) Stabilizes Epithelial Cell Junctions and Suppresses TNF-α-Induced Cytokine Production. Biochem Biophys Res Commun (2019) 517(4):729–34. doi: 10.1016/j.bbrc.2019.07.123
41. Ertle CM, Rommel FR, Tumala S, Moriwaki Y, Klein J, Kruse J, et al. New Pathways for the Skin's Stress Response: The Cholinergic Neuropeptide SLURP-1 Can Activate Mast Cells and Alter Cytokine Production in Mice. Front Immunol (2021) 12:631881. doi: 10.3389/fimmu.2021.631881
42. Swamynathan S, Tiwari A, Loughner CL, Gnalian J, Alexander N, Jhanji V, et al. The Secreted Ly6/uPAR-Related Protein-1 Suppresses Neutrophil Binding Chemotaxis and Transmigration Through Human Umbilical Vein Endothelial Cells. Sci Rep (2019) 9(1):5898. doi: 10.1038/s41598-019-42437-x
43. Chernyavsky AI, Galitovskiy V, Shchepotin IB, Grando SA. Anti-Inflammatory Effects of the Nicotinergic Peptides SLURP-1 and SLURP-2 on Human Intestinal Epithelial Cells and Immunocytes. BioMed Res Int (2014) 2014:609086. doi: 10.1155/2014/609086
44. Fujii T, Mashimo M, Moriwaki Y, Misawa H, Ono S, Horiguchi K, et al. Expression and Function of the Cholinergic System in Immune Cells. Front Immunol (2017) 8:1085. doi: 10.3389/fimmu.2017.01085
45. Moriwaki Y, Watanabe Y, Shinagawa T, Kai M, Miyazawa M, Okuda T, et al. Primary Sensory Neuronal Expression of SLURP-1 an Endogenous Nicotinic Acetylcholine Receptor Ligand. Neurosci Res (2009) 64(4):403–12. doi: 10.1016/j.neures.2009.04.014
46. Vasilyeva NA, Loktyushov EV, Bychkov ML, Shenkarev ZO, Lyukmanova EN. Three-Finger Proteins From the Ly6/uPAR Family: Functional Diversity Within One Structural Motif. Biochem (Moscow) (2017) 82(13):1702–15. doi: 10.1134/S0006297917130090
47. Swamynathan S, Buela KA, Kinchington P, Lathrop KL, Misawa H, Hendricks RL, et al. Klf4 Regulates the Expression of Slurp1 Which Functions as an Immunomodulatory Peptide in the Mouse Cornea. Invest Ophth Vis Sci (2012) 53(13):8433–46. doi: 10.1167/iovs.12-10759
48. Swamynathan SK, Piatigorsky J. Slurp-1 Promoter Activity Is Regulated by KLF4 in the Corneal Epithelial and Skin Keratinocytes. Invest Ophth Vis Sci (2008) 49(13):4298.
49. Bhattacharyya A, Herta T, Conrad C, Frey D, García P, Suttorp N, et al. Induction of Krueppel-Like Factor 4 Mediates Polymorphonuclear Neutrophil Activation in Streptococcus Pneumoniae Infection. Front Microbiol (2020) 11:582070. doi: 10.3389/fmicb.2020.582070
50. Shen Y, Hong H, Sangwung P, Lapping S, Nayak L, Zhang L, et al. Kruppel-Like Factor 4 Regulates Neutrophil Activation. Blood Adv (2017) 1(11):662–8. doi: 10.1182/bloodadvances
51. Chi F, Jong TD, Wang L, Ouyang Y, Wu C, Li W, et al. Vimentin-Mediated Signalling is Required for IbeA+ E Coli K1 Invasion of Human Brain Microvascular Endothelial Cells. Biochem J (2010) 427(1):79–90. doi: 10.1042/BJ20091097
52. Huang SH, Chi F, Peng L, Bo T, Zhang B, Liu LQ, et al. Vimentin, a Novel NF-κb Regulator is Required for Meningitic Escherichia Coli K1-Induced Pathogen Invasion and PMN Transmigration Across the Blood-Brain Barrier. PloS One (2016) 11(9):e0162641. doi: 10.1371/journal.pone.0162641
53. Chernyavsky AI, Arredondo J, Galitovskiy V, Qian J, Grando SA. Upregulation of Nuclear Factor-κb Expression by SLURP-1 is Mediated by α7-Nicotinic Acetylcholine Receptor and Involves Both Ionic Events and Activation of Protein Kinases. Am J Physiol Cell Physiol (2010) 299(5):C903–11. doi: 10.1152/ajpcell.00216.2010
54. Huang SH, Wang L, Chi F, Wu CH, Cao H, Zhang A, et al. Circulating Brain Microvascular Endothelial Cells (cBMECs) as Potential Biomarkers of the Blood-Brain Barrier Disorders Caused by Microbial and non-Microbial Factors. PloS One (2013) 8(4):e62164. doi: 10.1371/journal.pone.0062164
55. Durek T, Shelukhina IV, Tae HS, Thongyoo P, Spirova EN, Kudryavtsev DS, et al. Interaction of Synthetic Human SLURP-1 With the Nicotinic Acetylcholine Receptors. Sci Rep (2017) 7(1):16606. doi: 10.1038/s41598-017-16809-0
56. Favre B, Plantard L, Aeschbach L, Brakch N, Christen-Zaech S, de Viragh PA, et al. SLURP1 is a Late Marker of Epidermal Differentiation and is Absent in Mal De Meleda. J Invest Dermatol (2007) 127(2):301–8. doi: 10.1038/sj.jid.5700551
Keywords: SLURP1, E. coli K1 meningitis, blood-brain barrier, inflammation, α7 nAChR
Citation: He X, Wang L, Liu L, Gao J, Long B, Chi F, Hu T, Wan Y, Gong Z, Li L, Zhen P, Zhang T, Cao H and Huang S-H (2021) Endogenous α7 nAChR Agonist SLURP1 Facilitates Escherichia coli K1 Crossing the Blood-Brain Barrier. Front. Immunol. 12:745854. doi: 10.3389/fimmu.2021.745854
Received: 22 July 2021; Accepted: 13 September 2021;
Published: 14 October 2021.
Edited by:
Gee W. Lau, University of Illinois at Urbana-Champaign, United StatesReviewed by:
Jian Huang, Coriell Institute For Medical Research, United StatesAnirban Banerjee, Indian Institute of Technology Bombay, India
Christian Schwerk, University of Heidelberg, Germany
Copyright © 2021 He, Wang, Liu, Gao, Long, Chi, Hu, Wan, Gong, Li, Zhen, Zhang, Cao and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Cao, gzhcao@smu.edu.cn; Tiesong Zhang, zts68420@sina.com
†These authors have contributed equally to this work
 Xiaolong He
Xiaolong He Lei Wang
Lei Wang Liqun Liu
Liqun Liu Jie Gao
Jie Gao Beiguo Long1
Beiguo Long1 Tongtong Hu
Tongtong Hu Yu Wan
Yu Wan Zelong Gong
Zelong Gong Li Li
Li Li Hong Cao
Hong Cao Sheng-He Huang
Sheng-He Huang