- 1Key Lab for Zoonoses Research, Ministry of Education, Jilin Provincial Key Laboratory of Animal Embryo Engineering, College of Animal Sciences, Jilin University, Changchun, China
- 2Research Unit of Key Technologies for Prevention and Control of Virus Zoonoses, Chinese Academy of Medical Sciences, Academy of Military Medical Sciences, Changchun, China
- 3Department of Infectious Diseases, Shenzhen People's Hospital, Second Clinical Hospital of Jinan University, Shenzhen, China
Host antiviral factor interferon-induced transmembrane proteins (IFITMs) are a kind of small-molecule transmembrane proteins induced by interferon. Their broad-spectrum antiviral activity and unique ability to inhibit viral invasion have made them a hot molecule in antiviral research in recent years. Since the first demonstration of their natural ability to resist viral infection in 1996, IFITMs have been reported to limit a variety of viral infections, including some major pathogens that seriously endanger human health and social stability, such as influenza A, Ebol, severe acute respiratory syndrome, AIDS, and Zika viruses, etc. Studies show that IFITMs mainly exert antiviral activity during virus entry, specifically interfering with the fusion of the envelope and the endosome membrane or forming fusion micropores to block the virus from entering the cytoplasm. However, their specific mechanism is still unclear. This article mainly reviews the research progress in the structure, evolution, function, and mechanism of IFITMs, which may provide a theoretical basis for clarifying the molecular mechanism of interaction between the molecules and viruses and the research and development of new antiviral drugs based on IFITMs.
Introduction
Human interferon-induced transmembrane proteins (IFITMs), first reported in 1984, are proteins that can be induced by interferon (IFN). Twelve years later, Alber et al. discovered that these proteins promote antiviral activity (1), providing clues for studying their role in the host antiviral response. In 2009, Brass and colleagues also found that IFITMs were effective limiting factors for the influenza A virus (IAV), further confirming the IFITM antiviral function (2). Since then, research on the interaction between IFITMs and viruses has rapidly become a research hot spot in related fields.
It is reported that IFITM proteins could significantly inhibit IAV, West Nile virus (WNV), Ebola virus (EBOV), SARS coronavirus (SARS-CoV), vesicular pharyngitis virus (VSV), Rift Valley fever virus (RVFV), dengue virus (DENV), Semliki forest virus (SFV), Zika virus, Respiratory syncytial virus (RSV), human immunodeficiency virus-1 (HIV-1), hepatitis C virus (HCV), Reovirus, and other capsular or noncapsular RNA viruses (2–10). Besides this, IFITM proteins also exhibit antiviral activity against individual DNA viruses (5, 11, 12). For example, IFITM1 inhibits frog iridovirus (RGV) replication by preventing the virus from entering cells (11). IFITM1, 2, and 3 can inhibit the early stage of African swine fever virus (ASFV) infection (5). We previously found that IFITM3 protein restricts vaccinia virus (VACV) infection by interfering with virus binding and entry in a low pH-dependent manner, and VACV can also inhibit IFITM3 translation (12).
To date, the antiviral spectrum of this kind of protein involves more than 20 viruses from 12 families. IFITMs, as a branch of the large family of interferon stimulating genes (ISGs), have become star molecules in antiviral immune responses, especially in the study of ISGs, in recent years due to their broad-spectrum antiviral activity and unique ability to inhibit virus invasion. Numerous groups have continued to reveal that IFITMs have a restrictive effect on many viruses and made progress in their antiviral mechanism. In this article, we aim to review the latest research progress on the structure, localization, function, and mechanism of IFITM proteins, providing a reference for the further exploration of the mechanism of IFITMs and the development of their clinical application.
Structure and Cellular Location of IFITMs
IFITMs mainly include the IFITM1, IFITM2, IFITM3, IFITM5, and IFITM10 genes in humans, and these are located on human chromosome 11 (Figure 1) (13–16). IFITM1, IFITM2, and IFITM3 are expressed at low levels in a variety of human tissues, playing roles in embryonic development, cell adhesion, tumorigenesis, and signal transduction as well as antiviral activities (13–15). IFITM5 is only expressed in osteoblasts and participates in bone mineralization and maturation, and the function of IFITM10 is still unknown (17, 18). It was found that the homologous IFITMs also exist in several animals (13–15). For example, five IFITM genes of chickens, including ifitm1, ifitm2, ifitm3, ifitm5, and ifitm10 genes, are located on chicken chromosome 5 (13–15). There are seven ifitm genes in mice, six of which are located on chromosome 7, and the ifitm7 gene is on chromosome 16, which is probably reverse-transcribed from the IFITM1 gene (13, 15, 16). Except for the ifitm7 gene, all IFITM proteins in humans and mice include one intron and two exons.
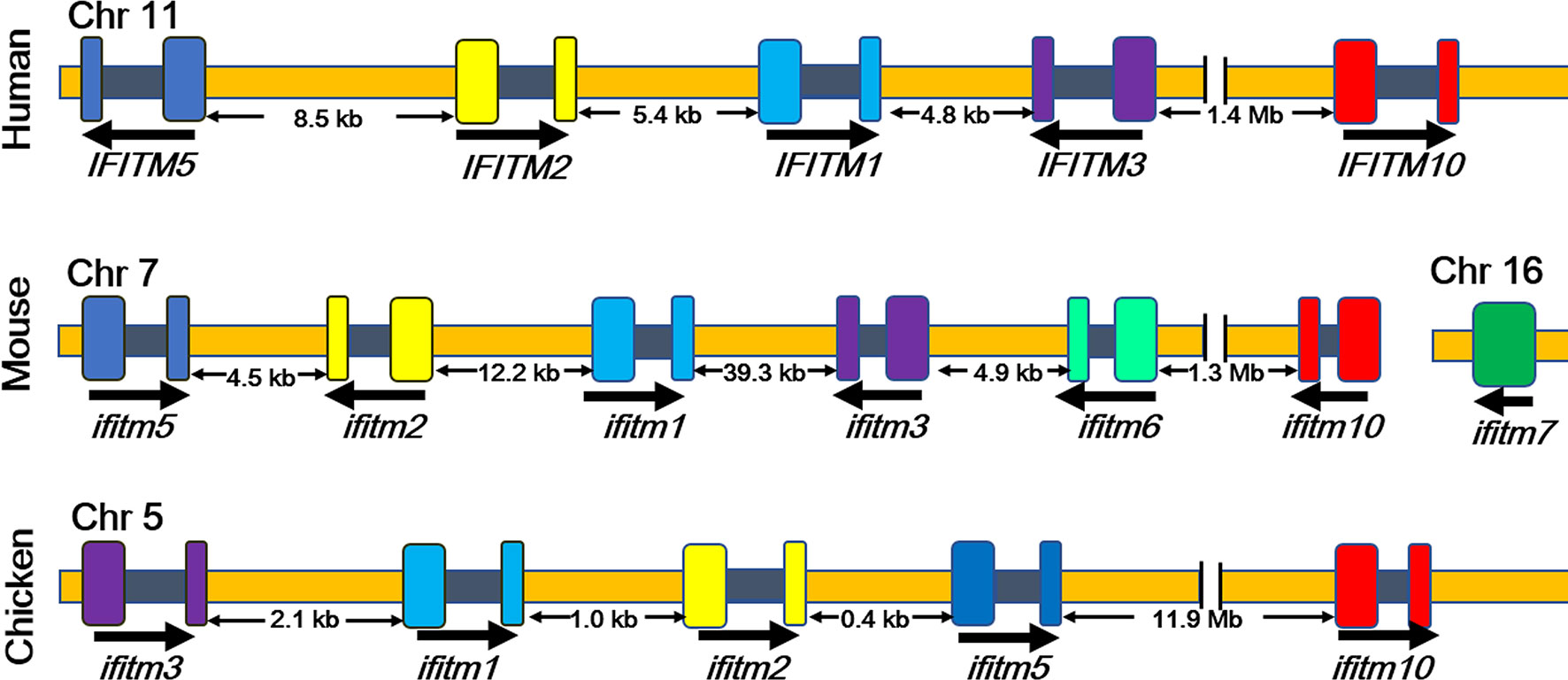
Figure 1 Localization of ifitm genes in different species (13). Human IFITM1, IFITM2, IFITM3, IFITM5, and IFITM10 genes are located on human chromosome 11. Seven ifitm genes were found in mice, six of which are located on chromosome 7, and the ifitm7 gene is on chromosome 16. Chicken ifitm1, ifitm2, ifitm3, ifitm5, and ifitm10 genes are located on chromosome 5 of the chicken. Arrows indicate the direction of expression. Exons are expressed in color, and the intron is blank. Chr, chromosome.
IFITMs, as transmembrane proteins, can be divided into five domains according to their structural characteristics (Figure 2) (6, 15). Human IFITM3 contains a variable and hydrophobic N-terminal domain (NTD, 1-57 aa), a conservative and hydrophobic transmembrane domain (IMD, also known as IM1 or TM1, 58-80 aa), a conservative intracellular cyclic structure (CIL, 81-104 aa), a variable and hydrophobic transmembrane domain (TMD, also known as IM2 or TM2, 105-126 aa), and a short and highly variable C-terminal domain (CTD, 127-133 aa) (6).
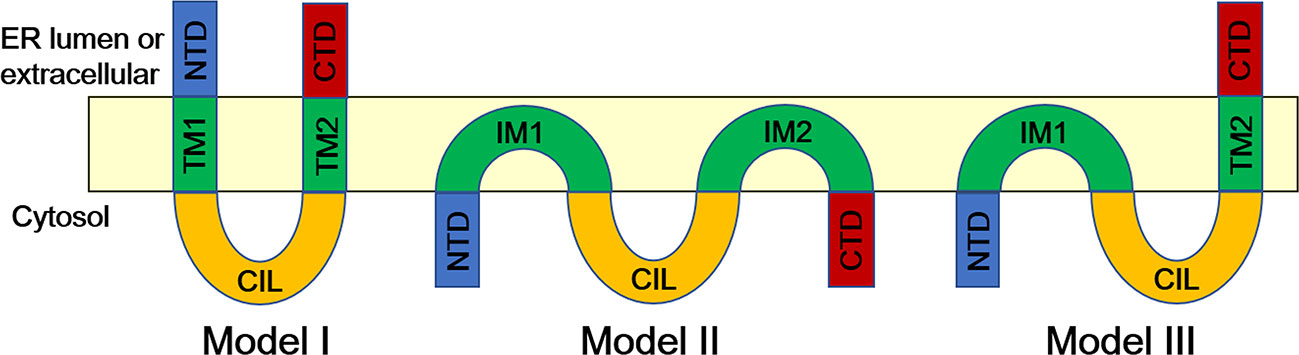
Figure 2 Topological Structure of IFITM Proteins (13). In Model I, IFITMs are U-shaped, two-transmembrane structures with N- and C-terminals either facing the endoplasmic reticulum cavity or extracellular. Another model (Model II) suggests that NTD, CTD, and CIL of IFITM proteins are in the cytoplasm, and IM1 and IM2 are inserted into the lipid bilayer but do not pass through the lipid layer (model II). In the third model (Model III), both NTD and CIL are located in the cytoplasm, and CTD is located in the endoplasmic reticulum cavity. CIL, conserved intracellular loop; CTD, C-terminal domain; IM, intramembrane domain; NTD, N-terminal domain; TM, transmembrane domain.
To date, there is still controversy about the topological structure of IFITM proteins on the cell membrane, which mainly focuses on the direction of the N- and C-terminals. According to the earliest model, IFITMs have a U-shaped, two-transmembrane structure with N- and C-terminals either facing the endoplasmic reticulum cavity or extracellular (Figure 2, Model I) (19). However, Yount et al. proposed another topology structure according to their research results (20). The model suggested that the NTD, CTD, and CIL of IFITM proteins were in the cytoplasm, and IM1 and IM2 were inserted into the lipid bilayer but did not pass through the lipid layer (Figure 2, Model II) (20). Bailey et al. reexamined the topological structure of IFITM3 in mice and proved that both NTD and CIL are located in the cytoplasm and CTD is located in the endoplasmic reticulum cavity, thus proposing a structural model of IFITM3 in which NTD is intracellular and CTD is extracellular (Figure 2, Model III) (21). Recently, Tian and colleagues further confirmed the Type II transmembrane protein model of IFITM3 by electron paramagnetic resonance (EPR) and nuclear magnetic resonance (NMR) technology and found that IFITM3 contains a C-terminal transmembrane α helix and two N-terminal short intramembrane α helices (22). However, what kind of structure the IFITM protein has remains to be further studied through crystal structure analysis, which helps explore the targets or cofactors that interact with IFITMs, thus explaining their antiviral mechanism.
Evolution of IFITMs
IFITM proteins belong to the Dispanin protein family, which first appeared as the common ancestor of Choanoflagellates and Metazoa and then evolved and formed four subfamilies (DSPA to DSPD) in the vertebrates, among which human IFITM1, 2, and 3 with antiviral activity belong to the DSPA subfamily (23).
According to the similarity of the IFITM sequence and its presumed function, it can be further divided into 3 clades. The first clade includes human IFITM1, 2, and 3, which are immune-related IFITMs, and mouse IFITM6 and IFITM7, encoded by intron-free inverse genes derived from the IFITM1 gene. IFITM2 and IFITM3 are highly homologous, and the IFITM1 protein is slightly different. Moreover, many mammals and poultry also have homologs of IFITM1, 2, and 3 although the IFITM2/3-like gene has low homology with human IFITM2 or 3 in other species, such as rhesus monkey. In contrast, clades 2 and 3 consist of IFITM5 and IFITM10, respectively. Although IFITM5 and IFITM10 genes are very close to the sites of human IFITM1, 2, and 3, neither of them can be induced by IFN nor do they have antiviral activity. Therefore, it is assumed that IFITM5 and IFITM10 are reflections of evolution but not positive selection. IFITM5 and IFITM10 also have homologs in many other mammals.
The Antiviral Spectrum of IFITMs
In 1996, Alber and Staeheli first reported that overexpression of IFITM1 inhibited VSV replication (1). Although this inhibitory effect is not as strong as IFN-induced MxA protein (1), it suggests that IFITM proteins may have an inhibitory effect on viral infection. Besides this, mouse cells overexpressing human IFITM1 are more resistant to VSV infection than control cells, but the effect is not obvious in IAV infection (7). Although this result is different from the current progress (IFITMs have a good inhibitory effect on IAV), it is the first time the antiviral activity of IFITM proteins were described and reported.
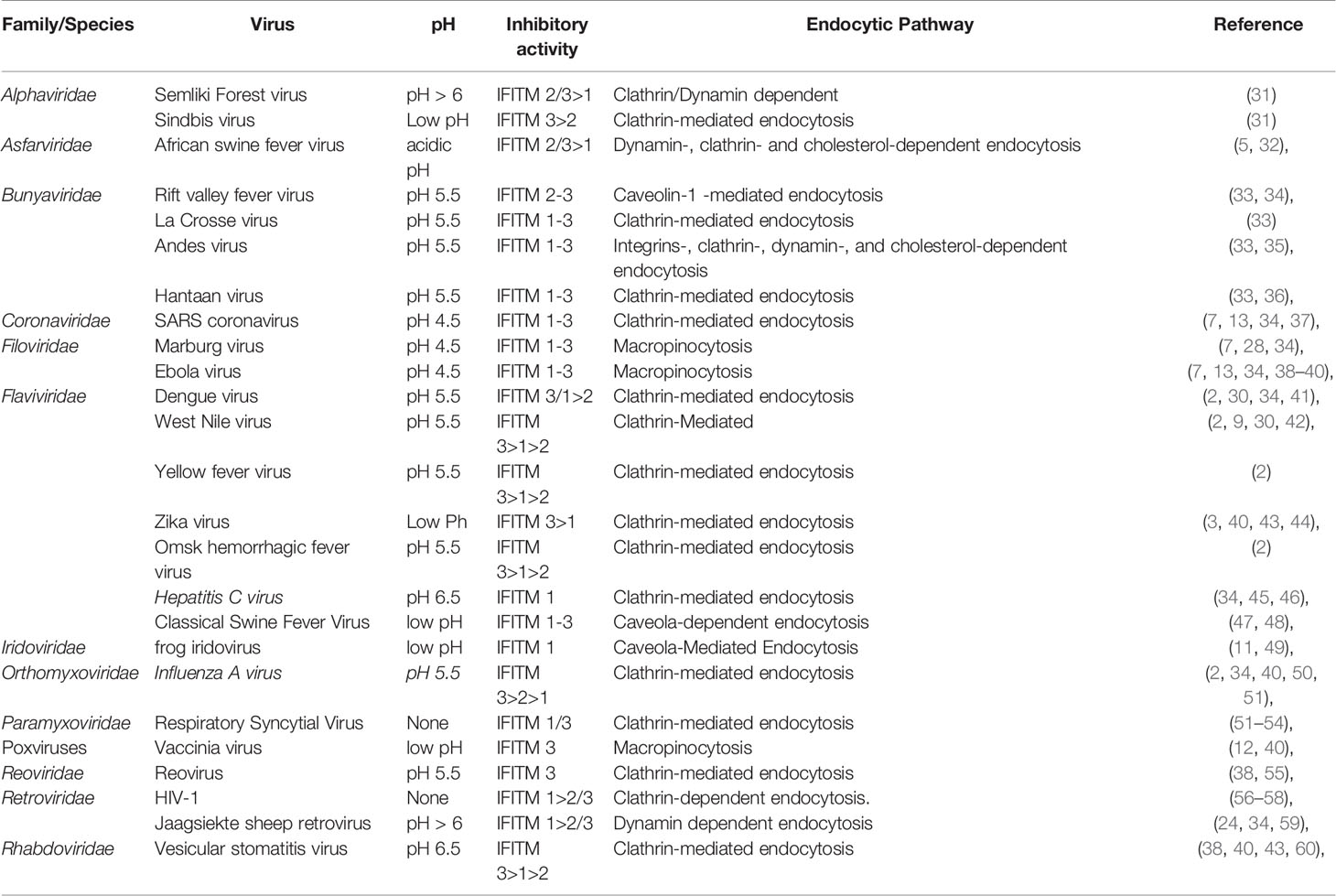
In 2009, Brass et al. systematically analyzed and confirmed IFITM3 as a significant limiting factor for IAV infection (2). Further research shows that silencing IFITM3 of U2OS cells can strongly enhance the replication of IAV H1N1 (A/PR/8/34), and siRNA targeting IFITM3 has a significant effect on eliminating IFN-γ mediated virus inhibition (2). We previously found that IFITM3 can suppress H5N1 replication in the early stage of the infection (10). Moreover, overexpression of human IFITM1, 2, or 3 inhibits replication of an IAV H1N1 subtype (A/PR/8/34) and H3N2 subtype (A/Udorn/72) (2, 24, 25). Embryonic fibroblasts (MEFs) from ifitmDel-/- mice are more sensitive to IAV than MEFs from wild-type mice, and the anti-IAV effect of type I and II IFN is weakened in ifitmDel-/- mice (26). Moreover, the infection of pseudo-retroviruses with different hemagglutinin proteins (H1, H3, H5, and H7) as outer membranes can be effectively inhibited by IFITM1, 2, and 3 (7, 24, 27–29). Studies have shown that arboviruses, including DENV and WNV, have similar sensitivity to IFITM-mediated restriction (30). Before now, it has been found that human IFITMs have potential inhibitory effects on more than 20 viruses in 10 families, mainly inhibiting the entry of viruses. The virus and virus entry characteristics inhibited by IFITMs are shown in Table 1. In addition to viruses, IFITM3 can also inhibit Mycobacterium tuberculosis invasion (61).
Unfortunately, IFITMs are not universal antiviral proteins that can resist all viruses. Studies have shown that IFITM proteins have no inhibitory effect on the infection of some viruses, including murine leukemia virus (MLV), arenavirus (LASV), lymphocytic choroid plexus meningitis virus (LCMV), and Crimean-Congo hemorrhagic fever (CCHFV) (7, 25, 62). Warren et al. found that IFITM1, 2, and 3 have no inhibitory effect on human papillomavirus (HPV), cytomegalovirus (HCMV), and adenovirus (Ad5) infected cells (4). Zhao et al. found IFITM proteins promote human coronavirus OC43 infection (63). HCMV utilizes IFITM proteins to facilitate virion assembly compartment formation during infection in MRC5 cells (62). Besides this, Ifitm3-/- mice have no obvious susceptibility to bacterial and protozoan pathogens, including Citrobacter, Salmonella typhimurium, and Plasmodium berghei, compared with wild mice (52). Although the reason for these results is still unclear, we can find that most viruses inhibited by IFITMs are enveloped RNA viruses, which mainly rely on low pH to enter cells. Furthermore, the inhibitory activity of IFITMs also has cell and virus specificity.
Besides this, there is still controversy about the effect of IFITM proteins on the alphavirus. It was reported that IFITM3 protein expressed in vitro had no obvious inhibitory effect on the Chikungunya virus (CHIKV) and Venezuelan equine encephalitis virus (VEEV) (64). However, recent studies show that IFITM2 and IFITM3 proteins can significantly inhibit SFV and Sindbis virus (SINV) although IFITM1 does not affect these viruses (31). Ifitm3–/– mice are more sensitive to multiple alphaviruses, including CHIKV, VEEV, SFV, SINV, and O’nyong-nyong virus (ONNV), than wild-type mice, and higher viral loads can be detected in multiple organs of ifitm3–/– mice, indicating that IFITM3 can limit the infection and pathogenicity of alphavirus (65).
Antiviral mechanism of IFITMs
Antiviral Effect of IFITMs Regulated by Post-Translation Modifications
In recent years, progress has been made in the research on the antiviral spectrum, intracellular localization, protein post-translational modification [phosphorylation (66, 67), ubiquitination (21, 68), palmitoylation (21, 69–71) and methylation (66, 72)], and upstream signal pathways generated by IFITMs.
Topological studies of IFITMs indicate that the N-terminal domains of IFITM2 and IFITM3 contain 20 and 21 amino acid residues, respectively, which are crucial for their transport (73). The N-terminal region includes important tyrosine (Tyr, Y), among which Y20 seems to control the cellular distribution of IFITMs (74). It is reported that its antiviral effect decreases or even disappears if the N-terminal of IFITM2 and IFITM3 is deleted or its key amino acid is mutated (74). However, the N-terminal of IFITM1 lacks this domain. Further studies show that IFITM1 is mainly located on the cell membrane and early endosome, and IFITM2 and IFITM3 are mainly located in the endosomal and lysosomal compartments of the endoplasmic reticulum (74), suggesting that the different cell localization of IFITMs determines their antiviral spectrum. Therefore, IFITM1 mainly limits the viruses that fuse to and penetrate the cell surface. For example, IFITM1 is more effective than IFITM2 and IFITM3 in inhibiting Jaagsiekte sheep retrovirus (JSRV) (24). However, IFITM2 and IFITM3 mainly restrict those viruses that enter through the endocytosis as IFITM3 has a stronger inhibition of viral entry by IAV, EBOV, SARS-CoV, etc. (7, 75). Additionally, the cysteine (C71, C72, C105) in IFITM3 can be palmitoylated (69, 70), and the lysine (K24, K83, K88, K104) in its N-terminal and conserved intracellular ring region (CIL) can also be ubiquitinated (20, 56). Codon 70 within the conserved CD225 domain of IFITMs plays a functionally important role in restricting virus entry (70). If this post-translational modification is removed, its antiviral activity is greatly affected (20, 56, 69).
Possible Antiviral Mechanism of IFITMs
To date, IFITM proteins are believed to exert antiviral activity by blocking the fusion of viral membranes, but the research on the molecular mechanism of IFITMs is relatively slow, and the mechanism of IFITMs inhibiting virus entry has not yet been determined. Based on previous studies, there are mainly three possible mechanisms (Figure 3).
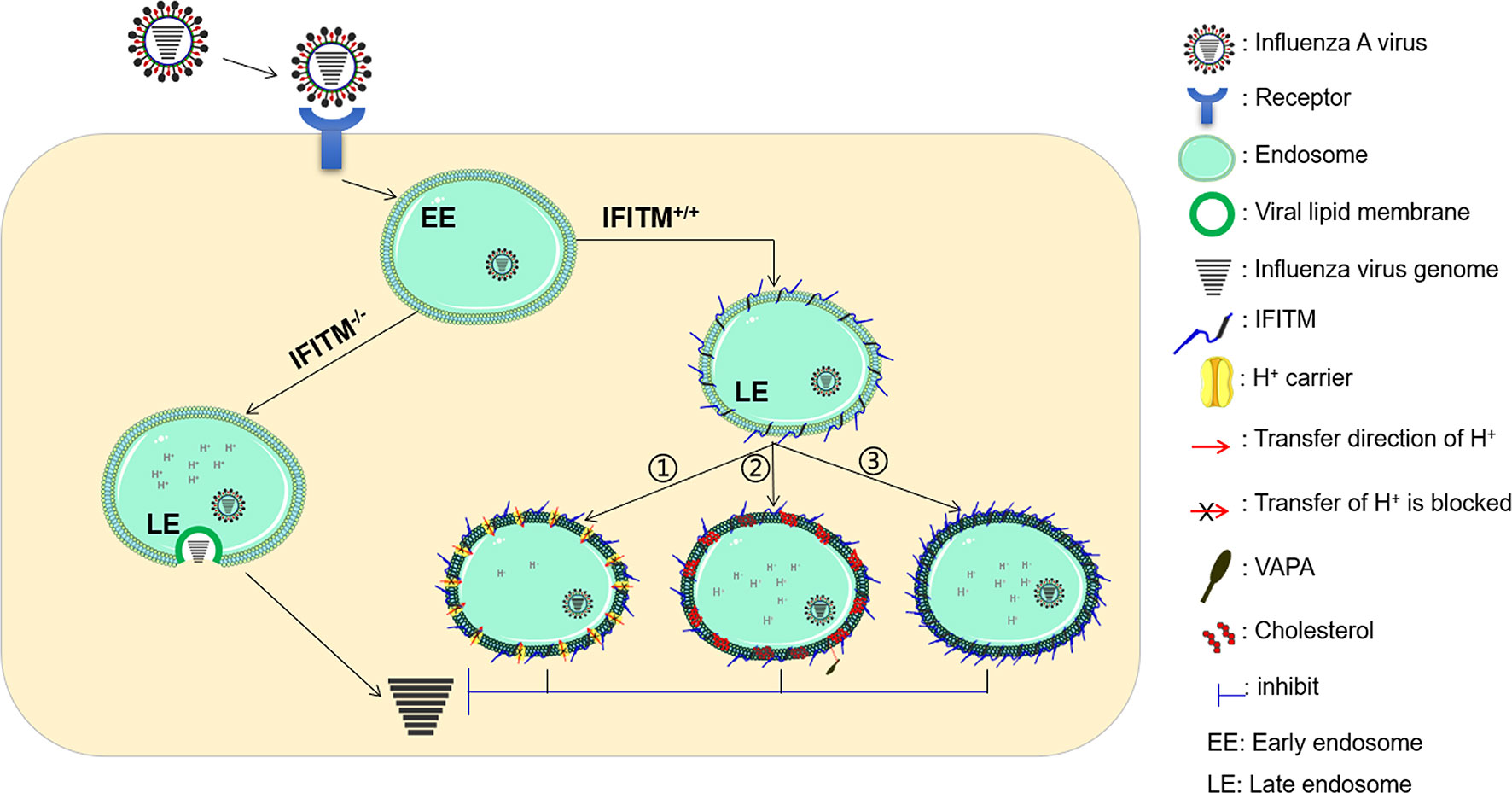
Figure 3 Possible antiviral mechanism of IFITMs. There are mainly three possible antiviral mechanisms. The first possible mechanism is that IFITMs may change the characteristics of the endosomal/lysosomal cavity, making these structures unfavorable for virus fusion (①). Another possible mechanism is that IFITM proteins block the formation of fusion pores following virus-endosome hemifusion by changing the physical properties of cell membranes (②). Besides this, IFITM proteins may also function independently by affecting the cell membrane structure or stimulating effective immune responses and cytokine signaling (③). VAPA, Vesicle-membrane-protein-associated protein A.
The first possible mechanism is that IFITMs may change the lysosomes’ characteristics, making these structures unfavorable for virus fusion (13). This mechanism is attractive because it can explain the difficulty of IFITM proteins in inhibiting virus entry in the cytoplasmic membrane. This mechanism is further supported by the results reported by Bailey and colleagues (21). When expressing IFITM3 with N- and C-terminal double tags in cells, it was found that IFITM3 C-terminal tags were cleaved in most cytoplasmic vesicles, but most tags remained in perinuclear vesicles, where the IFITM protein was overexpressed (21). A highly conserved, short amphipathic helix within a hydrophobic region of IFITM3 plays a critical role in IFITM3-dependent inhibition of IAV, Zika virus, VSV, EBOV, HIV, and SARS-CoV-2 infections (43, 71). IFITM3 may accumulate and locate in endosomal vesicles during IAV infection and eventually coat the IAV-containing endosomal vesicles (76). Moreover, IFITM3 can fuse with the incoming virus and enhance the trafficking of the IFITM-virus cargo to the lysosomes for degradation via specific S-palmitoylation (76, 77).
Another possible mechanism is that IFITM proteins block the formation of fusion pores following virus-endosome hemifusion by changing the cell membranes’ physical properties (8, 78). Li et al. found that overexpression of IFITM proteins resulted in decreased fluidity of the host cell membranes (24). These results were further confirmed as IFITM1 decreases host-membrane fluidity (79). It was found that IFITM3 interacts with vesicle membrane protein associated protein A (VAPA) and prevents its association with oxysterol-binding protein (OSBP), thereby disrupting intracellular cholesterol homeostasis and inhibiting viral entry (80).
However, other researchers believe that IFITM proteins may also function independently. It is reported that two phenylalanines within IM1 (F75 and F78) of IFITM3 mediate a physical association between IFITM proteins, and the loss of this interaction decreases IFITM3-mediated restriction (41). Further studies show that multiple residues in the NTD and CIL of IFITM3 are required to restrict both IAV and DENV (41). These results suggest that IFITM3 inhibits virus infection by affecting the cell membrane structure (13).
Besides this, IFITM1-3, especially IFITM3, is also expressed in T cells and lymphocytes, which can protect immune cells and the lungs, airway, spleen, skin, and brain from viral infection by stimulating effective immune responses (15). IFITM proteins also affect Th differentiation and are involved in regulating cytokine signaling (15).
In Vivo Function of IFITM3
Researchers infected ifitm3–/– mice with IAV and found that ifitm3–/– mice showed higher sensitivity to the virus than wild-type mice with increased lung viral load, aggravated pathological reaction, and decreased CD4+ and CD8+ T cells and activated NK cells as well as aberrant cardiac electrical activity, increased activation of fibrotic pathways, and fibrotic lesions in the heart (26, 81–83). Wakim et al. found that CD8+ resident memory T (TRM) cells deposited in the lungs after IAV infection can selectively express IFITM3 protein, facilitating their survival and protection from viral infection during subsequent exposures (84). Everitt et al. found that the viral load of ifitm3–/– mice infected with RSV was higher than wild-type mice (52). Diamond and colleagues found that ifitm3–/– mice showed greater joint swelling accompanied by higher levels of pro-inflammatory cytokines (such as TNF-α, IL-1β, etc.) and viral load during CHIKV infection (65). Besides this, they also found that IFITM3 protein plays a critical role in restricting VEEV and WNV infection and disease progression in vivo as ifitm3–/– mice were more susceptible to VEEV and WNV infection, resulting in greater virus accumulation in peripheral organs and central nervous system tissues (9, 65). The total number of B cells, CD4+ T cells, and antigen-specific CD8+ T cells were decreased in WNV-infected ifitm3-/- mice compared with that of wild-type mice (9). It is confirmed that the antiviral effect of IFITM3 does not depend on the participation of other IFITMs. These results prove that IFITM3 may limit virus infection through multiple mechanisms, which is of great significance in the occurrence and development of virus-induced diseases.
Conclusion and Perspective
Viruses must enter cells and use cell components for replication and proliferation to survive, resulting in disease. On the contrary, the host has also evolved mechanisms to prevent virus infection by blocking the virus from entering the host cells. During the last few years, IFITM proteins have been proven to be important proteins for endogenous cell defense against various pathogenic virus infections by blocking virus entry. In particular, IFITM3 protein and its homologs play a direct role in controlling infection of IAV, RSV, and WNV in mice. In vitro, experimental data show certain differences in virus types and antiviral activities of IFITM1, IFITM2, and IFITM3 proteins. Therefore, IFITM proteins may inhibit virus infection through various ways or mechanisms that still need to be elucidated. However, the question that cannot be ignored is, how do viruses that are not restricted by IFITMs escape the inhibition of IFITMs, whether they can inhibit IFITMs, and why can viruses that are inhibited by IFITMs not similarly evade this inhibition? Besides this, what is the molecular mechanism of IFITMs inhibiting the virus? What role does the network formed by host IFITMs and viruses play in the interaction between virus and cell, and what specific effects does the network have on the host’s natural immunity and disease process, etc. These mysteries have become the focus of antiviral research, but the ultimate challenge for ISG research, such as that on IFITM proteins, is transforming it into a new strategy for the prevention or treatment of viral diseases.
Author Contributions
Conceptualization: RL and LC. Writing—original draft preparation: DS and XW. Writing—review, RL, LT, and WS. Figures: DS and RL. Supervision: RL, LC, and JN. Funding acquisition: RL and LC. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by National Natural Science Foundation of China [No 31972719 31702210 31772747], Jilin University Science and Technology Innovative Research Team [JLU-STIRT No. 2017TD-05], the Jilin Province Science and Technology Development Projects [No. 20200402043NC], the Science and Technology Research Program during the 13th Five-year Plan Period of Jilin Educational Committee [No. JJKH20190172KJ].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Alber D, Staeheli P. Partial inhibition of vesicular stomatitis virus by the interferon-induced human 9-27 protein. J Interferon Cytokine Res (1996) 16:375–80. doi: 10.1089/jir.1996.16.375
2. Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, et al. The IFITM Proteins Mediate Cellular Resistance to Influenza A H1N1 Virus, West Nile Virus, and Dengue Virus. Cell (2009) 139:1243–54. doi: 10.1016/j.cell.2009.12.017
3. Savidis G, Perreira JM, Portmann JM, Meraner P, Guo Z, Green S, et al. The IFITMs Inhibit Zika Virus Replication. Cell Rep (2016) 15:2323–30. doi: 10.1016/j.celrep.2016.05.074
4. Jin D, Warren CJ, Griffin LM, Little AS, Huang IC, Farzan M, et al. The Antiviral Restriction Factors IFITM1, 2 and 3 Do Not Inhibit Infection of Human Papillomavirus, Cytomegalovirus and Adenovirus. PloS One (2014) 9:e96579. doi: 10.1371/journal.pone.0096579
5. Beer M, Muñoz-Moreno R, Cuesta-Geijo MÁ, Martínez-Romero C, Barrado-Gil L, Galindo I, et al. Antiviral Role of IFITM Proteins in African Swine Fever Virus Infection. PloS One (2016) 11:e0154366. doi: 10.1371/journal.pone.0154366
6. Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol (2012) 13:46–57. doi: 10.1038/nri3344
7. Baric RS, Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, et al. Distinct Patterns of IFITM-Mediated Restriction of Filoviruses, SARS Coronavirus, and Influenza A Virus. PloS Pathog (2011) 7:e1001258. doi: 10.1371/journal.ppat.1001258
8. Basler CF, Desai TM, Marin M, Chin CR, Savidis G, Brass AL, et al. IFITM3 Restricts Influenza A Virus Entry by Blocking the Formation of Fusion Pores following Virus-Endosome Hemifusion. PloS Pathog (2014) 10:e1004048. doi: 10.1371/journal.ppat.1004048
9. Gorman MJ, Poddar S, Farzan M, Diamond MS, Frueh K. The Interferon-Stimulated GeneIfitm3Restricts West Nile Virus Infection and Pathogenesis. J Virol (2016) 90:8212–25. doi: 10.1128/jvi.00581-16
10. Du S, Jiang Y, Xu W, Bai J, Tian M, Wang M, et al. Construction, expression and antiviral activity analysis of recombinant adenovirus expressing human IFITM3 in vitro. Int J Biol Macromol (2019) 131:925–32. doi: 10.1016/j.ijbiomac.2019.03.161
11. Zhu R, Wang J, Lei X-Y, Gui J-F, Zhang Q-Y. Evidence for Paralichthys olivaceus IFITM1 antiviral effect by impeding viral entry into target cells. Fish Shellfish Immunol (2013) 35:918–26. doi: 10.1016/j.fsi.2013.07.002
12. Li C, Du S, Tian M, Wang Y, Bai J, Tan P, et al. The Host Restriction Factor Interferon-Inducible Transmembrane Protein 3 Inhibits Vaccinia Virus Infection. Front Immunol (2018) 9:228:228. doi: 10.3389/fimmu.2018.00228
13. Yánez DC, Ross S, Crompton T. The IFITM protein family in adaptive immunity. Immunology (2019) 159:365–72. doi: 10.1111/imm.13163
14. Siegrist F, Ebeling M, Certa U. The Small Interferon-Induced Transmembrane Genes and Proteins. J Interferon Cytokine Res (2011) 31:183–97. doi: 10.1089/jir.2010.0112
15. Bailey CC, Zhong G, Huang IC, Farzan M. IFITM-Family Proteins: The Cell’s First Line of Antiviral Defense. Annu Rev Virol (2014) 1:261–83. doi: 10.1146/annurev-virology-031413-085537
16. Robinson-Rechavi M, Zhang Z, Liu J, Li M, Yang H, Zhang C. Evolutionary Dynamics of the Interferon-Induced Transmembrane Gene Family in Vertebrates. PloS One (2012) 7:e49265. doi: 10.1371/journal.pone.0049265
17. Moffatt P, Gaumond M-H, Salois P, Sellin K, Bessette M-C, Godin É, et al. Bril: A Novel Bone-Specific Modulator of Mineralization. J Bone Mineral Res (2008) 23:1497–508. doi: 10.1359/jbmr.080412
18. Hanagata N, Li X, Morita H, Takemura T, Li J, Minowa T. Characterization of the osteoblast-specific transmembrane protein IFITM5 and analysis of IFITM5-deficient mice. J Bone Miner Metab (2010) 29:279–90. doi: 10.1007/s00774-010-0221-0
19. Smith RA, Young J, Weis JJ, Weis JH. Expression of the mouse fragilis gene products in immune cells and association with receptor signaling complexes. Genes Immun (2006) 7:113–21. doi: 10.1038/sj.gene.6364278
20. Yount JS, Karssemeijer RA, Hang HC. S-Palmitoylation and Ubiquitination Differentially Regulate Interferon-induced Transmembrane Protein 3 (IFITM3)-mediated Resistance to Influenza Virus. J Biol Chem (2012) 287:19631–41. doi: 10.1074/jbc.M112.362095
21. Bailey CC, Kondur HR, Huang IC, Farzan M. Interferon-induced Transmembrane Protein 3 Is a Type II Transmembrane Protein. J Biol Chem (2013) 288:32184–93. doi: 10.1074/jbc.M113.514356
22. Ling S, Zhang C, Wang W, Cai X, Yu L, Wu F, et al. Combined approaches of EPR and NMR illustrate only one transmembrane helix in the human IFITM3. Sci Rep (2016) 6:24029. doi: 10.1038/srep24029
23. Uversky VN, Sällman Almén M, Bringeland N, Fredriksson R, Schiöth HB. The Dispanins: A Novel Gene Family of Ancient Origin That Contains 14 Human Members. PloS One (2012) 7:e31961. doi: 10.1371/journal.pone.0031961
24. Emerman M, Li K, Markosyan RM, Zheng Y-M, Golfetto O, Bungart B, et al. IFITM Proteins Restrict Viral Membrane Hemifusion. PloS Pathog (2013) 9:e1003124. doi: 10.1371/journal.ppat.1003124
25. Smith SE, Weston S, Kellam P, Marsh M. IFITM proteins—cellular inhibitors of viral entry. Curr Opin Virol (2014) 4:71–7. doi: 10.1016/j.coviro.2013.11.004
26. Basler CF, Bailey CC, Huang IC, Kam C, Farzan M. Ifitm3 Limits the Severity of Acute Influenza in Mice. PloS Pathog (2012) 8:e1002909. doi: 10.1371/journal.ppat.1002909
27. Smith J, Smith N, Yu L, Paton IR, Gutowska MW, Forrest HL, et al. A comparative analysis of host responses to avian influenza infection in ducks and chickens highlights a role for the interferon-induced transmembrane proteins in viral resistance. BMC Genomics (2015) 16:574. doi: 10.1186/s12864-015-1778-8
28. Gerlach T, Hensen L, Matrosovich T, Bergmann J, Winkler M, Peteranderl C, et al. pH Optimum of Hemagglutinin-Mediated Membrane Fusion Determines Sensitivity of Influenza A Viruses to the Interferon-Induced Antiviral State and IFITMs. J Virol (2017) 91:e00246-17. doi: 10.1128/jvi.00246-17
29. Hensen L, Matrosovich T, Roth K, Klenk H-D, Matrosovich M, García-Sastre A. HA-Dependent Tropism of H5N1 and H7N9 Influenza Viruses to Human Endothelial Cells Is Determined by Reduced Stability of the HA, Which Allows the Virus To Cope with Inefficient Endosomal Acidification and Constitutively Expressed IFITM3. J Virol (2019) 94:e01223-19. doi: 10.1128/jvi.01223-19
30. Jiang D, Weidner JM, Qing M, Pan X-B, Guo H, Xu C, et al. Identification of Five Interferon-Induced Cellular Proteins That Inhibit West Nile Virus and Dengue Virus Infections. J Virol (2010) 84:8332–41. doi: 10.1128/jvi.02199-09
31. Weston S, Czieso S, White IJ, Smith SE, Wash RS, Diaz-Soria C, et al. Alphavirus Restriction by IFITM Proteins. Traffic (2016) 17:997–1013. doi: 10.1111/tra.12416
32. Galindo I, Cuesta-Geijo MA, Hlavova K, Muñoz-Moreno R, Barrado-Gil L, Dominguez J, et al. African swine fever virus infects macrophages, the natural host cells, via clathrin- and cholesterol-dependent endocytosis. Virus Res (2015) 200:45–55. doi: 10.1016/j.virusres.2015.01.022
33. Mudhasani R, Tran JP, Retterer C, Radoshitzky SR, Kota KP, Altamura LA, et al. IFITM-2 and IFITM-3 but Not IFITM-1 Restrict Rift Valley Fever Virus. J Virol (2013) 87:8451–64. doi: 10.1128/jvi.03382-12
34. Perreira JM, Chin CR, Feeley EM, Brass AL. IFITMs Restrict the Replication of Multiple Pathogenic Viruses. J Mol Biol (2013) 425:4937–55. doi: 10.1016/j.jmb.2013.09.024
35. Kuhn JH, Chiang C-F, Flint M, Lin J-MS, Spiropoulou CF. Endocytic Pathways Used by Andes Virus to Enter Primary Human Lung Endothelial Cells. PloS One (2016) 11:e0164768. doi: 10.1371/journal.pone.0164768
36. Zheng X, Bian P, Ye C, Ye W, Ma H, Tang K, et al. Interferon-Induced Transmembrane Protein 3 Inhibits Hantaan Virus Infection, and Its Single Nucleotide Polymorphism rs12252 Influences the Severity of Hemorrhagic Fever with Renal Syndrome. Front Immunol (2016) 7:535:535. doi: 10.3389/fimmu.2016.00535
37. Zhao X, Sehgal M, Hou Z, Cheng J, Shu S, Wu S, et al. Identification of Residues Controlling Restriction versus Enhancing Activities of IFITM Proteins on Entry of Human Coronaviruses. J Virol (2018) 92:e01535-17. doi: 10.1128/jvi.01535-17
38. Ahi YS, Yimer D, Shi G, Majdoul S, Rahman K, Rein A, et al. IFITM3 Reduces Retroviral Envelope Abundance and Function and Is Counteracted by glycoGag. mBio (2020) 11:e03088-19. doi: 10.1128/mBio.03088-19
39. Wrensch F, Karsten CB, Gnirß K, Hoffmann M, Lu K, Takada A, et al. Interferon-Induced Transmembrane Protein–Mediated Inhibition of Host Cell Entry of Ebolaviruses. J Infect Dis (2015) 212:S210–S8. doi: 10.1093/infdis/jiv255
40. Dorf ME, Li S, Wang L, Fu B. ZMPSTE24 defends against influenza and other pathogenic viruses. J Exp Med (2017) 214:919–29. doi: 10.1084/jem.20161270
41. John SP, Chin CR, Perreira JM, Feeley EM, Aker AM, Savidis G, et al. The CD225 Domain of IFITM3 Is Required for both IFITM Protein Association and Inhibition of Influenza A Virus and Dengue Virus Replication. J Virol (2013) 87:7837–52. doi: 10.1128/jvi.00481-13
42. Chu JJH, Ng ML. Infectious Entry of West Nile Virus Occurs through a Clathrin-Mediated Endocytic Pathway. J Virol (2004) 78:10543–55. doi: 10.1128/jvi.78.19.10543-10555.2004
43. Chesarino NM, Compton AA, McMichael TM, Kenney AD, Zhang L, Soewarna V, et al. IFITM 3 requires an amphipathic helix for antiviral activity. EMBO Rep (2017) 18:1740–51. doi: 10.15252/embr.201744100
44. Ganapathiraju MK, Karunakaran KB, Correa-Menéndez J. Predicted protein interactions of IFITMs may shed light on mechanisms of Zika virus-induced microcephaly and host invasion. F1000Research (2017) 5:1919. doi: 10.12688/f1000research.9364.2
45. Narayana SK, Helbig KJ, McCartney EM, Eyre NS, Bull RA, Eltahla A, et al. The Interferon-induced Transmembrane Proteins, IFITM1, IFITM2, and IFITM3 Inhibit Hepatitis C Virus Entry. J Biol Chem (2015) 290:25946–59. doi: 10.1074/jbc.M115.657346
46. Wrensch F, Ligat G, Heydmann L, Schuster C, Zeisel MB, Pessaux P, et al. Interferon-Induced Transmembrane Proteins Mediate Viral Evasion in Acute and Chronic Hepatitis C Virus Infection. Hepatology (2019) 70:1506–20. doi: 10.1002/hep.30699
47. Li C, Zheng H, Wang Y, Dong W, Liu Y, Zhang L, et al. Antiviral Role of IFITM Proteins in Classical Swine Fever Virus Infection. Viruses (2019) 11:126. doi: 10.3390/v11020126
48. Zhang Y-N, Liu Y-Y, Xiao F-C, Liu C-C, Liang X-D, Chen J, et al. Rab5, Rab7, and Rab11 Are Required for Caveola-Dependent Endocytosis of Classical Swine Fever Virus in Porcine Alveolar Macrophages. J Virol (2018) 92:e00797-18. doi: 10.1128/jvi.00797-18
49. Guo CJ, Liu D, Wu YY, Yang XB, Yang LS, Mi S, et al. Entry of Tiger Frog Virus (an Iridovirus) into HepG2 Cells via a pH-Dependent, Atypical, Caveola-Mediated Endocytosis Pathway. J Virol (2011) 85:6416–26. doi: 10.1128/jvi.01500-10
50. Zhao X, Li J, Winkler CA, An P, Guo J-T. IFITM Genes, Variants, and Their Roles in the Control and Pathogenesis of Viral Infections. Front Microbiol (2019) 9:3228:3228. doi: 10.3389/fmicb.2018.03228
51. Zani A, Yount JS. Antiviral Protection by IFITM3 In Vivo. Curr Clin Microbiol Rep (2018) 5:229–37. doi: 10.1007/s40588-018-0103-0
52. Ikeda Y, Everitt AR, Clare S, McDonald JU, Kane L, Harcourt K, et al. Defining the Range of Pathogens Susceptible to Ifitm3 Restriction Using a Knockout Mouse Model. PloS One (2013) 8:e80723. doi: 10.1371/journal.pone.0080723
53. Smith SE, Busse DC, Binter S, Weston S, Diaz Soria C, Laksono BM, et al. Interferon-Induced Transmembrane Protein 1 Restricts Replication of Viruses That Enter Cells via the Plasma Membrane. J Virol (2019) 93:e02003-18. doi: 10.1128/jvi.02003-18
54. Zhang W, Zhang L, Zan Y, Du N, Yang Y, Tien P. Human respiratory syncytial virus infection is inhibited by IFN-induced transmembrane proteins. J Gen Virol (2015) 96:170–82. doi: 10.1099/vir.0.066555-0
55. Anafu AA, Bowen CH, Chin CR, Brass AL, Holm GH. Interferon-inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry. J Biol Chem (2013) 288:17261–71. doi: 10.1074/jbc.M112.438515
56. Chutiwitoonchai N, Hiyoshi M, Hiyoshi-Yoshidomi Y, Hashimoto M, Tokunaga K, Suzu S. Characteristics of IFITM, the newly identified IFN-inducible anti-HIV-1 family proteins. Microbes Infect (2013) 15:280–90. doi: 10.1016/j.micinf.2012.12.003
57. Lee W-YJ, Fu RM, Liang C, Sloan RD. IFITM proteins inhibit HIV-1 protein synthesis. Sci Rep (2018) 8:14551. doi: 10.1038/s41598-018-32785-5
58. Yu J, Li M, Wilkins J, Ding S, Swartz TH, Esposito AM, et al. IFITM Proteins Restrict HIV-1 Infection by Antagonizing the Envelope Glycoprotein. Cell Rep (2015) 13:145–56. doi: 10.1016/j.celrep.2015.08.055
59. Li K, Jia R, Li M, Zheng Y-M, Miao C, Yao Y, et al. A Sorting Signal Suppresses IFITM1 Restriction of Viral Entry. J Biol Chem (2015) 290:4248–59. doi: 10.1074/jbc.M114.630780
60. Hornick AL, Li N, Oakland M, McCray PB, Sinn PL. Human, Pig, and Mouse Interferon-Induced Transmembrane Proteins Partially Restrict Pseudotyped Lentiviral Vectors. Hum Gene Ther (2016) 27:354–62. doi: 10.1089/hum.2015.156
61. Ranjbar S, Haridas V, Jasenosky LD, Falvo JV, Goldfeld AE. A Role for IFITM Proteins in Restriction of Mycobacterium tuberculosis Infection. Cell Rep (2015) 13:874–83. doi: 10.1016/j.celrep.2015.09.048
62. Xie M, Xuan B, Shan J, Pan D, Sun Y, Shan Z, et al. Human Cytomegalovirus Exploits Interferon-Induced Transmembrane Proteins To Facilitate Morphogenesis of the Virion Assembly Compartment. J Virol (2015) 89:3049–61. doi: 10.1128/jvi.03416-14
63. Zhao X, Guo F, Liu F, Cuconati A, Chang J, Block TM, et al. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc Natl Acad Sci (2014) 111:6756–61. doi: 10.1073/pnas.1320856111
64. Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature (2011) 472:481–5. doi: 10.1038/nature09907
65. Poddar S, Hyde JL, Gorman MJ, Farzan M, Diamond MS, García-Sastre A. The Interferon-Stimulated Gene IFITM3 Restricts Infection and Pathogenesis of Arthritogenic and Encephalitic Alphaviruses. J Virol (2016) 90:8780–94. doi: 10.1128/jvi.00655-16
66. Chesarino NM, McMichael TM, Yount JS. Regulation of the trafficking and antiviral activity of IFITM3 by post-translational modifications. Future Microbiol (2014) 9:1151–63. doi: 10.2217/fmb.14.65
67. Chesarino NM, McMichael TM, Hach JC, Yount JS. Phosphorylation of the Antiviral Protein Interferon-inducible Transmembrane Protein 3 (IFITM3) Dually Regulates Its Endocytosis and Ubiquitination. J Biol Chem (2014) 289:11986–92. doi: 10.1074/jbc.M114.557694
68. Ashour J, Chesarino NM, McMichael TM, Yount JS. E3 Ubiquitin Ligase NEDD4 Promotes Influenza Virus Infection by Decreasing Levels of the Antiviral Protein IFITM3. PloS Pathog (2015) 11:e1005095. doi: 10.1371/journal.ppat.1005095
69. Yount JS, Moltedo B, Yang Y-Y, Charron G, Moran TM, López CB, et al. Palmitoylome profiling reveals S-palmitoylation–dependent antiviral activity of IFITM3. Nat Chem Biol (2010) 6:610–4. doi: 10.1038/nchembio.405
70. Benfield CTO, MacKenzie F, Ritzefeld M, Mazzon M, Weston S, Tate E, et al. Bat IFITM3 restriction depends on S-palmitoylation and a polymorphic site within the CD225 domain. Life Sci Alliance (2019) 3:e201900542. doi: 10.26508/lsa.201900542
71. Shi G, Kenney AD, Kudryashova E, Zhang L, Hall-Stoodley L, Robinson RT, et al. Opposing activities of IFITM proteins in SARS-CoV-2 infection. bioRxiv (2020). doi: 10.1101/2020.08.11.246678
72. Shan Z, Han Q, Nie J, Cao X, Chen Z, Yin S, et al. Negative Regulation of Interferon-induced Transmembrane Protein 3 by SET7-mediated Lysine Monomethylation. J Biol Chem (2013) 288:35093–103. doi: 10.1074/jbc.M113.511949
73. Jia R, Xu F, Qian J, Yao Y, Miao C, Zheng Y-M, et al. Identification of an endocytic signal essential for the antiviral action of IFITM3. Cell Microbiol (2014) 16:1080–93. doi: 10.1111/cmi.12262
74. Johannes L, Weston S, Czieso S, White IJ, Smith SE, Kellam P, et al. A Membrane Topology Model for Human Interferon Inducible Transmembrane Protein 1. PloS One (2014) 9:e104341. doi: 10.1371/journal.pone.0104341
75. Diamond MS, Feeley EM, Sims JS, John SP, Chin CR, Pertel T, et al. IFITM3 Inhibits Influenza A Virus Infection by Preventing Cytosolic Entry. PloS Pathog (2011) 7:e1002337. doi: 10.1371/journal.ppat.1002337
76. Kummer S, Avinoam O, Krausslich HG. IFITM3 Clusters on Virus Containing Endosomes and Lysosomes Early in the Influenza A Infection of Human Airway Epithelial Cells. Viruses (2019) 11:548. doi: 10.3390/v11060548
77. Spence JS, He R, Hoffmann H-H, Das T, Thinon E, Rice CM, et al. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat Chem Biol (2019) 15:259–68. doi: 10.1038/s41589-018-0213-2
78. Whelan SPJ, Suddala KC, Lee CC, Meraner P, Marin M, Markosyan RM, et al. Interferon-induced transmembrane protein 3 blocks fusion of sensitive but not resistant viruses by partitioning into virus-carrying endosomes. PloS Pathog (2019) 15:e1007532. doi: 10.1371/journal.ppat.1007532
79. Lin T-Y, Chin CR, Everitt AR, Clare S, Perreira JM, Savidis G, et al. Amphotericin B Increases Influenza A Virus Infection by Preventing IFITM3-Mediated Restriction. Cell Rep (2013) 5:895–908. doi: 10.1016/j.celrep.2013.10.033
80. Amini-Bavil-Olyaee S, Choi YJ, Lee JH, Shi M, Huang IC, Farzan M, et al. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe (2013) 13:452–64. doi: 10.1016/j.chom.2013.03.006
81. Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature (2012) 484:519–23. doi: 10.1038/nature10921
82. Kenney AD, McMichael TM, Imas A, Chesarino NM, Zhang L, Dorn LE, et al. IFITM3 protects the heart during influenza virus infection. Proc Natl Acad Sci (2019) 116:18607–12. doi: 10.1073/pnas.1900784116
83. Sun Q, Lei N, Lu J, Gao RB, Li Z, Liu LQ, et al. Interferon-induced Transmembrane Protein 3 Prevents Acute Influenza Pathogenesis in Mice. BioMed Environ Sci (2020) 33:295–305. doi: 10.3967/bes2020.041
Keywords: interferon-inducible transmembrane proteins, entry, virus, interaction, host antiviral factor
Citation: Ren L, Du S, Xu W, Li T, Wu S, Jin N and Li C (2020) Current Progress on Host Antiviral Factor IFITMs. Front. Immunol. 11:543444. doi: 10.3389/fimmu.2020.543444
Received: 04 May 2020; Accepted: 26 October 2020;
Published: 30 November 2020.
Edited by:
Uday Kishore, Brunel University London, United KingdomReviewed by:
Chunfu Zheng, Fujian Medical University, ChinaHelene Minyi Liu, National Taiwan University, Taiwan
Copyright © 2020 Ren, Du, Xu, Li, Wu, Jin and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linzhu Ren, renlz@jlu.edu.cn; Chang Li, lichang78@163.com
†These authors have contributed equally to this work
 Linzhu Ren
Linzhu Ren Shouwen Du
Shouwen Du Wang Xu
Wang Xu Tiyuan Li
Tiyuan Li Shipin Wu
Shipin Wu Ningyi Jin1,2
Ningyi Jin1,2 Chang Li
Chang Li