- 1Department of Biochemistry, College of Natural Sciences, Kangwon National University, Chuncheon, South Korea
- 2Institute of New Frontier Research, College of Medicine, Hallym University, Chuncheon, South Korea
Tumor microenvironment consists of cancer cells and various stromal cells such as endothelial cells, cancer-associated fibroblasts (CAFs), myeloid-derived suppressor cells (MDSCs), neutrophils, macrophages, and other innate and adaptive immune cells. Of these innate immune cells, macrophages are an extremely heterogeneous population, and display both pro-inflammatory and anti-inflammatory functions. While M1 macrophages (classically activated macrophages) display anti-tumoral and pro-inflammatory functions, M2 macrophages display pro-tumoral and anti-inflammatory functions. Cellular interactions and molecular factors in the tumor microenvironment affect the polarization of macrophages. We review molecules and immune cells that influence the polarization status of macrophages. Tumor-associated macrophages (TAMs) generally express M2 phenotype, and mediate many processes that include tumor initiation, angiogenesis, and metastasis. A high number of TAMs has been associated with the poor prognosis of cancers. MicroRNAs (miRNAs) have been known to regulate cellular interactions that involve cancer cells and macrophages. Tumor-derived exosomes play critical roles in inducing the M1 or M2-like polarization of macrophages. The roles of exosomal miRNAs from tumor cells in the polarization of macrophages are also discussed and the targets of these miRNAs are presented. We review the effects of exosomal miRNAs from TAMs on cancer cell invasion, growth, and anti-cancer drug resistance. The relevance of exosomal microRNAs (miRNAs) as targets for the development of anti-cancer drugs is discussed. We review recent progress in the development of miRNA therapeutics aimed at elevating or decreasing levels of miRNAs.
Introduction
The Tumor Microenvironment Modulates Cancer Progression
The tumor microenvironment (TME) is the environment surrounding tumor. The tumor microenvironment is critical for the initiation and promotion of cancer (1). The tumor microenvironment consists of various components such as endothelial cells, cancer associated fibroblasts (CAFs), innate and adaptive immune cells, and extracellular matrix. These innate immune cells include dendritic cells (DC), macrophages, neutrophils, myeloid-derived suppressor cells (MDSC), mesenchymal stem cells (MSCs), and natural killer cells (NK). CAFs release various growth factors and cytokines and mediate cancer progression (2–4). MDSCs derived by C-X-C Motif Chemokine Ligand 17 (CXCL17) enhances the metastatic potential of breast cancer cells through platelet-derived growth factor (PDGF) (5). MDSCs suppresses cytolytic T lymphocytes (CTL) activity via interleukin 6 (IL6)/IL8-arginase (Arg) axis to promote gastric cancer cell progression (6). DCs stimulated by IL37 enhance the anti-tumor effect of CD8+ T cells by secreting cytokines such as secrete cytokines such as IL-2, IL-12, and interferon- γ (IFN-γ) (7). Neutrophils account for up to 70% of circulating leukocytes and are the first line of defense against pathogens. Neutrophils are classified into N1 neutrophils and N2 neutrophils. N1 neutrophils are inflammatory and anti-tumorigenic and N2 neutrophils are anti-inflammatory and tumorigenic, respectively. Neutrophils can promote or inhibit tumor progression by secretion of cytokines and growth factors. Neutrophils enhance extravasation of tumor cells through the secretion of IL1β and matrix metalloproteinases (MMPs) (8). Neutrophils mediate hepatocellular carcinoma progression by C-C Motif Chemokine Ligand 2 (CCL2) and CCL17 (9). Neutrophils suppress lung cancer cell proliferation by Fas Ligand (Fas L)/Fas pathway (10). Hepatocyte growth factor (HGF)/MET proto-oncogene- dependent nitric oxide (NO) release by neutrophils suppresses tumor growth (11). MSCs mediate gastric cancer progression by connective tissue growth factor (CTGF) (12). Human umbilical cord mesenchymal stem cells suppress cholangiocarcinoma cell growth by regulating Wingless-Type MMTV Integration Site Family (Wnt)/β-catenin and phosphoinositide-3 kinase (PI3K)/Akt signaling pathway (13). NK cells suppress tumor immune evasion by IFN-γ (14). NK cells suppress the metastatic potential of cancer cells by IL-33 (15). Tryptase released by mast cells enhances tumor cell metastasis through the janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathway (16). Human mast cells induce apoptosis of breast cancer cells through tumor necrosis factor-α (TNF-α) (17). Mast cell derived IL-6 suppresses proliferation of glioma cells by abolition of STAT3 signaling and the down-regulation of glycogen synthase kinase (GSK3β) (18). Figure 1 depicts tumor-stroma interactions that modulate cancer progression.
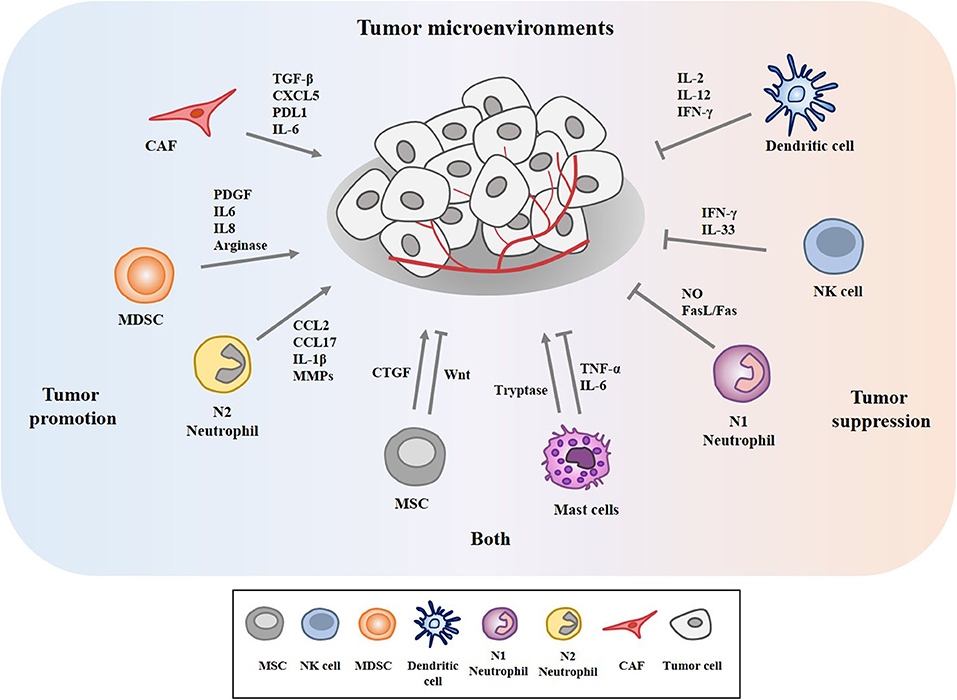
Figure 1. Tumor-stroma interactions in the tumor microenvironment modulate cancer progression. The tumor microenvironment comprises cancer cells, various stromal cells, and extracellular matrix. These stromal cells include endothelial cells, fibroblasts, and innate and adaptive immune cells. These cells can enhance or inhibit cancer progression by secreting cytokines, growth factors, and various other molecules. Mesenchymal stem cells (MSCs) and mast cells can enhance and inhibit cancer progression.
Macrophages in the Tumor Microenvironment
The most abundant stromal cells within the tumor microenvironment are macrophages. Macrophages modulate immune responses through antigen presentation and phagocytosis, and also play important roles in tissue repair and wound healing. Depletion of peritoneal macrophages suppresses the progression of ovarian tumor (19). Macrophages are highly plastic and classified into two distinct phenotypes, M1 and M2. Depending on the type of stimuli from their milieu, macrophages show either M1 or M2 phenotype (20, 21). M1 macrophages (classical) display pro-inflammatory and M2 macrophages display anti-inflammatory function, respectively. Circulating monocytes, recruited into the tumor sites by monocyte chemoattractant 1 (MCP1), are differentiated into tumor associated macrophages (TAMs). TAMs could represent up to 50% of the tumor mass, and play a key role in tumor development (22). TAMs comprise distinct populations that share features of both M1 and M2 macrophages; however, most studies have shown that TAMs are anti-inflammatory, and correlate with a poor prognosis of cancers (23–26).
M1 macrophages are activated by cytokines such as interferon γ (IFNγ) and produce interleukin-1 (IL-1), IL-6, IL-12, tumor necrosis factor (TNF)-a, reactive oxygen species (ROS), and nitric oxide (NO) (20, 23). Pattern recognition receptors (Toll-like receptors, C-type lectins, retinoic acid inducible gene-like receptors) are necessary for the M1 macrophages polarization. These receptors recognize molecular patterns such as lipopolysaccharide (LPS), and increase the production of pro-inflammatory cytokines (IL-6, IL-12, tumor necrosis factor- α (TNF-α), etc.). Classically activated M1 macrophages are also induced by IFN-γ, alone or in combination with cytokines [e.g., TNF-α and Granulocyte Macrophage Colony-Stimulating Factor (GM-CSF)] (27). M1 macrophages can enhance tumor specific antigen presentation by increasing the expression levels of the major histocompatibility complex (MHC) class I and II molecules (28). M1 macrophages convert L-arginine into citrulline and NO by inducible nitric oxide synthase (iNOS) (28).
T helper 2 (Th2) cytokines, such as IL-4, IL-10, and IL-13, prostaglandin E2 (PGE2), and transforming growth factor-β (TGF-β), induce the differentiation of monocytes into the alternative M2 macrophages which play essential roles in humoral immunity, wound healing, parasite clearance, dampening of inflammation, angiogenesis, and immunosuppression (29), and tissue remodeling (30). IL-33, a cytokine of the IL-1family, is associated with Th2 and M2 polarization (31). M2 macrophages produce and secrete IL-10 and TGF-β to inhibit M1 polarization of macrophages through the suppression of IL-12 (32). M2 macrophages convert L-arginine into polyamine and urea by Arg1 (28). Figure 2 shows the polarization of macrophages by T-helper cells and molecules that mediate various functions of the M1 and M2 macrophages.
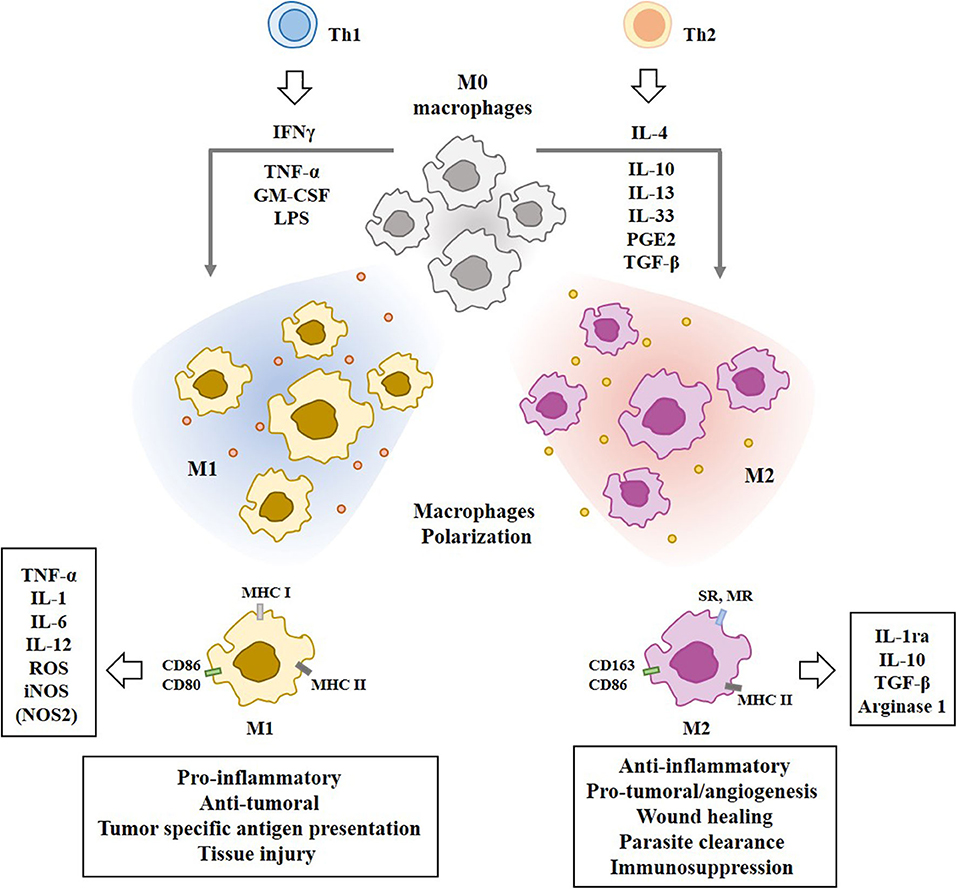
Figure 2. Polarization of macrophages by T-helper cells. Tumor associated macrophages (TAMs) are composed of pro-inflammatory M1 macrophages and anti-inflammatory M2 macrophages. IFN-γ, TNF-α, LPS, and GM-CSF induce M1 macrophages polarization. Th2 cytokines induce M2 macrophages polarization. Mo macrophages are circulating monocytes. M1 macrophages display higher expression level of MHC class I than M2 macrophages. M1 macrophages display higher expression level of iNOS than M2 macrophages. M2 macrophages display higher expression level of CD163 than M1 macrophages. Molecules that mediate effects of M1 macrophages or M2 macrophages are shown. SR denotes scavenger receptor. MR denotes mannose receptor. Nitric oxide synthase NOS2 (iNOS) in M1 macrophages converts L-arginine into citrulline and NO. Arginase 1 in M2 macrophages converts L-arginine into polyamine and urea.
Cellular Interactions in the Tumor Microenvironment
The phenotype of macrophages within the tumor microenvironment is significantly affected by cellular interactions that involve cancer cells, immune cells, endothelial cells, and fibroblasts. Vascular endothelial growth factor A (VEGFA) from hepatic cancer cells (HCC) activates VEGFR2 to enhance tube formation, and the invasion potential of vascular endothelial cells (33). CC chemokine ligand 2 (CCL2)-CCR2 signaling is necessary for the activation of endothelial cells by cancer cells (34). CCL5 derived from cancer cells is necessary for tumor angiogenesis and metastasis (35). Clinical studies have revealed interactions between cancer cells (including ovarian cancer) and TAMs (36). Tumor-derived CCL2 is a monocyte-chemotactic protein and its high level correlates with high numbers of TAMs in tumor tissue and a poor prognosis (37). Tumor-derived VEGF, platelet-derived growth factor (PDGF), macrophage colony stimulating factor (M-CSF), and interleukin IL-10 leads to the recruitment of macrophages (38, 39). Renal cell carcinoma-derived IL-10 induces the M2 polarization of macrophages (40). Nicotinamide adenine dinucleotide phosphate oxidase 4 (NADPH oxidase 4) recruits M2 macrophages via the rho-associated protein kinase (ROCK)/PI3 kinase-dependent production of cytokines, thus contributing to cancer cell growth (41). Human monocytes exposed to culture medium from thyroid tumor cell lines undergo M2-like polarization of macrophages, showing high cluster of differentiation 206 (CD206) and low MHC class II markers. Thyroid tumor-derived prostaglandin E2 (PGE2) induces the M2 polarization of macrophages (42). Recombinant endothelin-1, a 21-amino acid peptide, induces the M2 macrophages polarization and leads to the increased expression levels of CD204, CD206, CD163, IL-10, and CCL22, and the production of matrix metalloproteinase-9 (MMP-9) (43).
TAMs activate tumor stem cells and enhance the metastatic potential of cancer cells (44, 45). TAMs induce the vascularization of tumor tissue by producing VEGF, PDGF, and transforming growth factor (TGF)-β (46). TAMs enhance the tumorigenic potential of cancer cells and induce anti-cancer drug resistance through cathepsin β and milk-fat globule epidermal growth factor (EGF)-VIII (47–49). TAMs produce MMPs to induce lung cancer cell invasion (50). Intraperitoneal TAMs enhance the migration and invasion of gastric cancer cells via IL-6 (51). IL-10-derived from M2 macrophages enhances the growth potential of non-small cell lung cancer cells (NSCLCs) by activating cancer stemness via janus kinase (JAK)/signal transducer and activator of transcription 1 (STAT1)/nuclear factor-kappa B (NF-kB) pathway (52). TAMs secrete TGF-β1, and promotes the proliferation and invasion potential of colorectal cancer cells (53). TAMs secrete IL-35 to promote the metastatic potential of cancer cells by activating JAK2-STAT6-GATA-bnding protein 3 (GATA3) signaling (54).
Myeloid cells infiltrating into tumor stroma mostly express M2 markers play essential roles in immunosuppression, tumor angiogenesis, and tumor metastasis (23). IL-10, released by TAMs, increases the number of regulatory T (Treg) cells by activating Forkhead Box P3 (Foxp3) (55). TAMs-derived TGFβ induces Treg response by cellular interaction (56). TAMs induce immunosuppressive and tumorigenic functions, and express the enzyme Arg1. Anti-programmed cell death ligand 1 (PDL1) therapy decreases the number of TAMs with Arg1+ to suppress cancer progression (57). TAMs suppress CD8+T cell activity by releasing CCL17, CCL22, TGF-β, IL-10, Arg1, and galectin-3 (58). Genetic ablation of colony stimulating factor (CSF1) in colorectal cancer cells inhibits the influx of immunosuppressive CSF1R+ TAMs in tumors. This reduction results in CD8+ T cell attack on tumors, but a compensatory increase in Foxp3+ Treg cells limits its effect on tumor growth (59). Cancer cells and TAMs induce CD4+FoxP3- type I regulatory T cells (Tr1) cells in glioblastoma (GBM) patients. Co-culture of Tr1 with CD8+ T cells reduces the tumor-specific cytotoxic effects of CD8+T cells (60). TAMs-derived TGF-β induces the macrophage polarization of M1 into M2-like phenotype via the upregulation of SNAIL, and blockade of TGF-β/SNAIL signaling restores the production of pro-inflammatory cytokines (61). Figure 3 provides an overview of the cellular interactions involving cancer cells and macrophages within the tumor microenvironment.
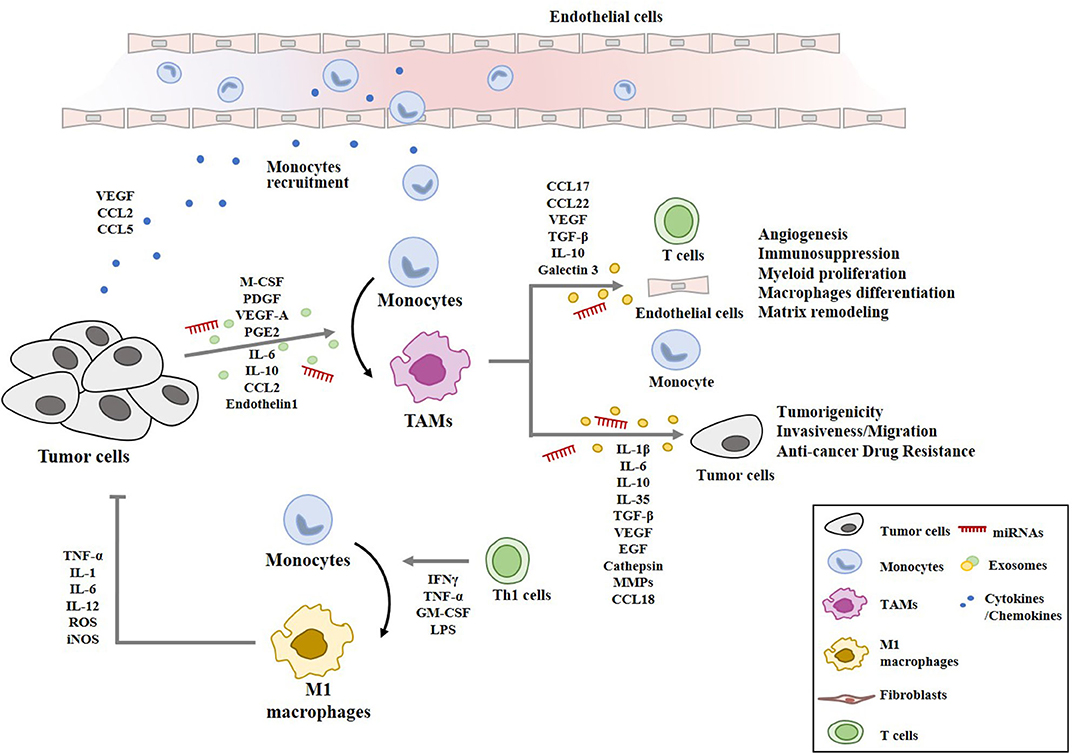
Figure 3. Cellular interactions involving macrophages in tumor microenvironment. Cancer cells secrete chemoattractants and growth factors (CCL2, CCL5, VEGF, M-CSF) to recruit blood monocytes. Tumor-derived M-CSF, PDGF, IL-6, IL-10, TGF-β, VEGF-A, CCL2, PGE2, and endothelin 1 induce the polarization into M2-like macrophages with pro-tumor functions (TAMs). TAMs-derived cytokines/growth factors (IL-1β, IL-6, IL-10, IL-35, TGF-β, VEGF, and EGF), cathepsin, and MMPs enhance tumor cell proliferation, invasion, anti-cancer drug-resistance, and remodeling of extracellular matrix. TAMs-derived chemokines (CCL17, CCL18, and CCL22) recruit naïve and Th2 lymphocytes, which results in ineffective anti-tumor immune response. miRNAs denote microRNAs. M1 macrophages are activated by Th1 cells. M1 macrophages display anti-tumoral functions through TNF-α, IL-1, IL-6, IL-12, ROS, and iNOS.
Tumor-Derived Exosomal miRNAs Regulate the Polarization of Macrophages
In the tumor microenvironment, cancer cells secrete exosomes. Exosomes are lipid bilayer membrane vesicles of (50–100) nm that are derived from the luminal membrane of multivesicular bodies, which are constitutively released by fusion with the cell membrane (62). Most eukaryotes release exosomes to various biological fluids such as blood, lymph, and milk. Exosomes are particularly enriched in various tumor microenvironment (63). Cancer cells generate more exosomes than normal cells, and circulating exosomes levels are increased in the blood of cancer patients when compared to healthy individuals (64). Exosomes can affect the phenotype of recipient cells as it can transfer their contents to recipient cells. Tumor-derived exosomes enhance tumorigenic potential, and metastatic potential by increasing the expression of MET oncogene in bone marrow progenitor cells from individuals with melanoma (65). Immunosuppressive exosomes of tumor can enhance tumor growth by inducing apoptosis of cytotoxic T cells (66). Exosomes from EGFR2-overexpressing anti-cancer drug resistant breast cancer cells contain immunosuppressive cytokine such as TGF-β and confers anti-cancer drug resistance in anti-cancer drug sensitive breast cancer cells (67).
miRNAs represent one of the predominant RNAs contained in exosomes (68, 69). miRNAs are short, non-coding RNAs that regulate the expression of complementary mRNAs (70). miRNAs function in RNA silencing and post-transcriptional regulation of gene expression (71). miRNAs are initially transcribed as long primary miRNAs (pre-miRNAs), and are subsequently processed by the microprocessor complex that contains the double strand RNA-specific endoribonuclease DROSHA and its RNA binding partner DiGeorge Syndrome critical region gene 8 (DGCR8) (72). Mature miRNAs bind to multiple targets (73). miRNAs are frequently deregulated in many types of cancer and regulate tumor progression, metastasis, and anti-cancer drug resistance (74–76). miRNAs paly critical roles in tumor immunity by regulating the polarization of macrophages (77).
Exosomes protect miRNAs from degradation, enabling them to be stably expressed in the extracellular space (78). Exosomal miRNAs derived from cancers act as mediators of interactions between cancer cells and the tumor microenvironment (79, 80). The exosomal miRNAs mediate communication between colon cancer cells and TAMs and reprograms macrophages to create a microenvironment for tumor growth; this microenvironment enables cancer cells to metastasize (81). Modification or inhibition of exosomal miRNAs might offer a valuable therapeutic strategy in cancer.
Tumor-derived exosomes (TDEs) deliver proteins, messenger RNAs (mRNAs), and miRNAs to various stromal cells in the tumor microenvironment. Epithelial ovarian cancer (EOC)-derived exosomal miR-222 activates STAT3 signaling and induces the M2 polarization of macrophages (82). Exosomal miRNAs (miR-21–3p, miR-125 b-5p, and miR-181 d-5p) induce the M2 polarization of macrophages (83). These miRNAs are induced by hypoxia via hypoxia-inducible factors (HIFs) (83). Exosomal miR-940 derived from EOC cells stimulates the M2 polarization of macrophages (84). Hypoxic conditions induce the expression of exosomal miR-940 in EOC cells (84). Exosomal miR-222 from EOC cells induces the M2 polarization of macrophages by targeting the suppressor of cytokine signaling 3 (SOCS3) (82, 85). Exosomal miRNAs (miR-25-3p, miR-130b-3p, miR-425-5p), upregulated in colorectal cancer cells by activation of the CXCL12/CXCR4 axis, induce the M2 polarization of macrophages by regulating the expression of phosphatase and tensin homolog (PTEN) through the activation of phosphatidyl inositol 3 kinase (PI3K)/Akt signaling pathway (74). Exosomal mR-203 from colorectal cancer cells induces the M2 polarization of macrophages and enhances the metastatic potential of colorectal cancer cells by increasing the expression levels of CD163 and STAT3 (86). Exosomal miR-145 from colorectal cancer cells induces the M2 polarization of macrophages by decreasing the expression of histone deacetylase 11 (HDAC11) (87). P53 is responsible for the increased expression of miR-145 (87). Exosomal miR-150 from hepatic cancer cells increases the secretion of VEGF from M2 macrophages (88). Exosomal miR-146a from hepatic cancer cells induces the M2 polarization of macrophages and suppresses the anti-tumor function of T cells by increasing the expression of programmed death-1 (PD-1) in T cells (89). Spalt Like Transcription Factor 4 (SALL-4) binds to the promoter sequences of miR-146a, and directly increases the expression of miR-146a (89). Endoplasmic reticulum (ER)-stressed hepatic cancer cells release exosomes to upregulate programmed death-ligand 1 (PD-L1) expression in macrophages, which subsequently inhibits anti-tumor T-cell function through an exosome miR-23a-3p-PTEN-AKT pathway (90). Exosomal miR-301a-3p from pancreatic cancer cells induces the M2 polarization of macrophages by targeting PTEN/PI3Kγ pathway (91). HIF-1a or HIF-2a is responsible for the increased expression level of exosomal miR-301a-3p (91).
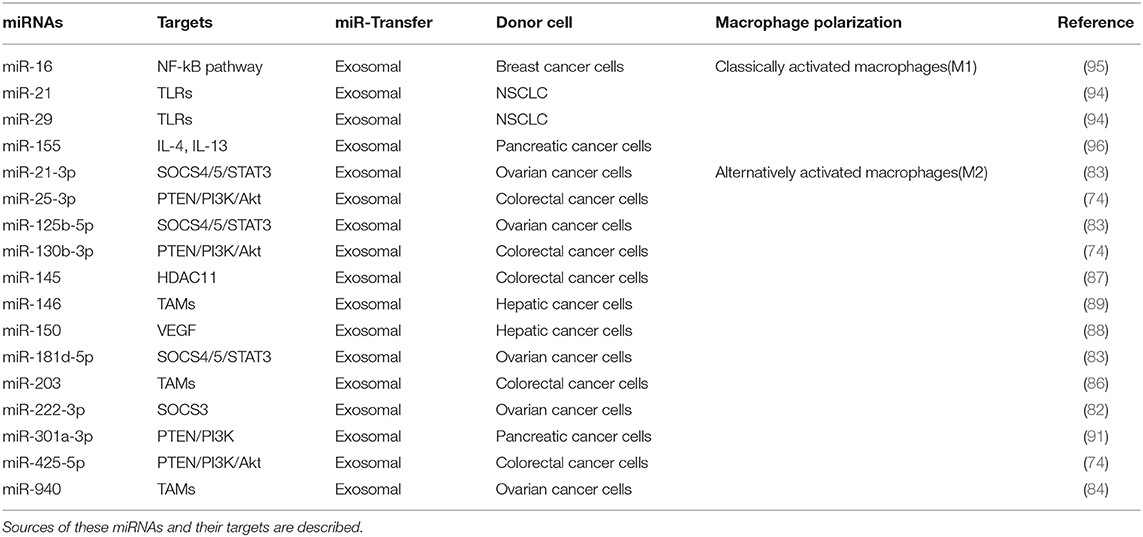
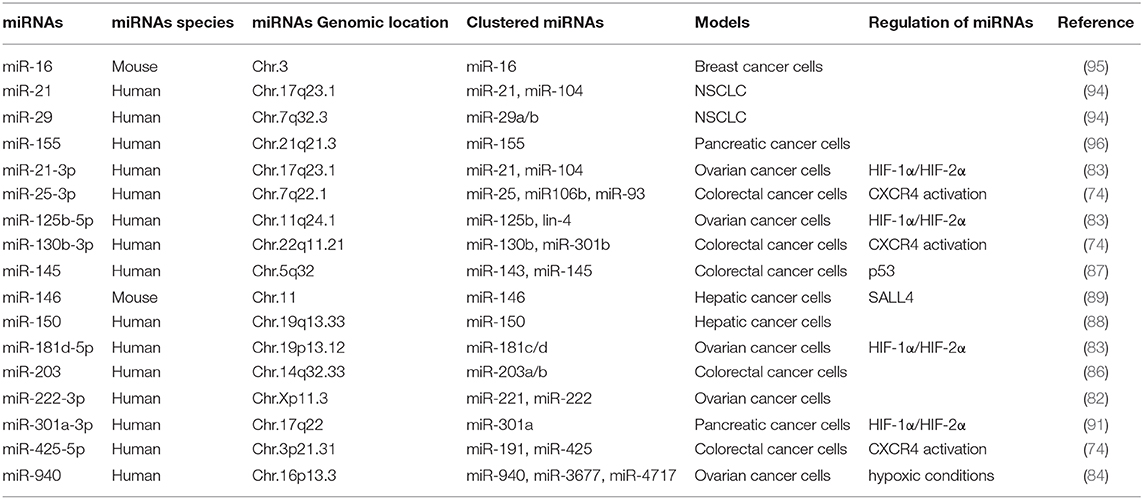
Exosomes of miR-125b-transsfectd lung cancer cells display reprogrammed miRNA expression profiles and the miRNAs in the exosomes regulate expression levels of apoptosis-related genes (92). Exosomal miR-19a-3p suppresses breast cancer progression by inducing M1 macrophages polarization by regulating the expression levels of Fos-related antigen 1 (Fra-1), VEGF, and signal transducer and activator of transcription 3 (STAT3) (93). Exosomal miR-21 and miR-29 from NSCLCs bind to toll-like receptors (TLRs), and induce the M1 polarization of macrophages (94). Epigallocatechin-3-gallate (EGCG) has anti-tumor and anti-inflammatory activities. Exosomes from EGCG-treated 4T1 (breast cancer) cells inhibit the infiltration of TAMs and differentiation into M2 macrophages. miR-16 expression is increased in both cancer cells and exosomes by ECGC treatment. miR-16 decreases I kappa B kinase α (IKKα) expression in TAMs, and subsequently inhibits NF-κB pathway, which is critical for TAMs infiltration and the M2 polarization of macrophages (95). Exosomal miR-155 and miR-125b-5p derived from pancreatic cancer cells induce repolarization of M2 macrophages into M1 macrophages by increasing expression levels of iNOS and TNF-α (96). Figure 4 shows the tumor-derived miRNAs that regulate the polarization of macrophages. Table 1 shows the targets of these miRNAs. Table 2 shows the chromosomal localizations of these miRNAs and factors regulating the expression levels of these miRNAs. Some of these miRNAs form cluster. The expression of clustered miRNAs is believed to be co-regulated and these miRNAs display similar expression patterns (97, 98). These miRNAs may be involved in common biological processes.
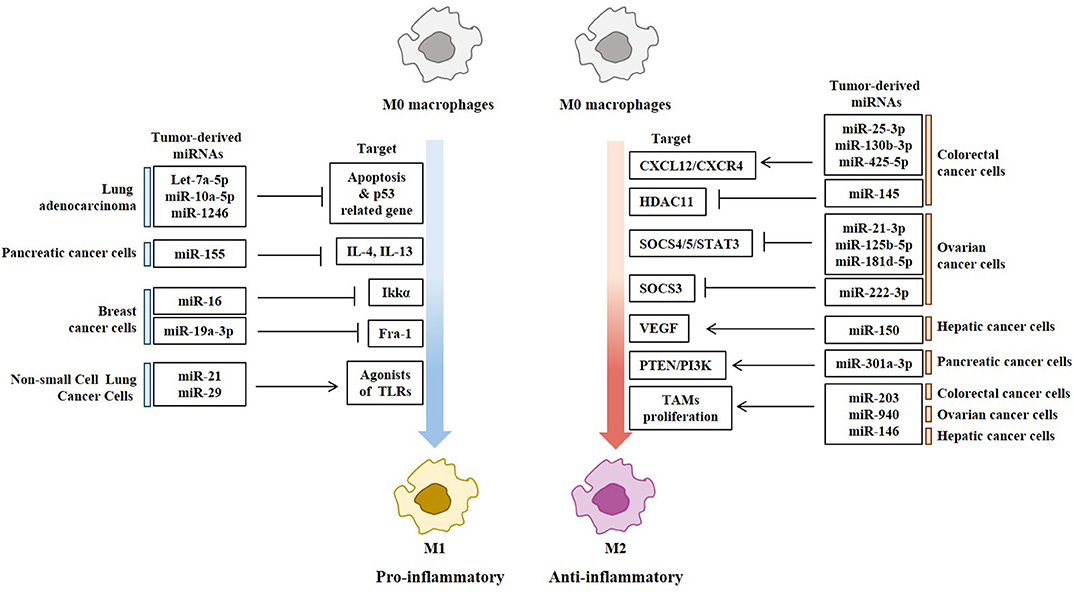
Figure 4. Tumor-derived miRNAs regulate polarization of macrophages. Tumor-derived miRNAs induce polarization of anti-tumoral and pro-inflammatory M1 macrophages or Pro-tumoral and anti-inflammatory M2 macrophages.
Exosomal miRNAs of Cancer Cells Regulate Anti-Cancer Drug Resistance
Accumulating evidence suggests that the tumor microenvironment plays a pivotal role in the development of anti-cancer drug resistance (99). Exosomes from cancer cells, cancer associated fibroblasts, or immune cells, carrying miRNAs have been shown to confer anti-cancer drug-resistance. Exosomal drug-efflux pumps and miRNAs can regulate anti-cancer drug resistance (100). miR-365a-3p enhance the migration and invasion of lung cancer cells by inhibiting ubiquitin specific peptidase 33 (USP33)/ slit guidance ligand 2 (SLIT2)/ roundabout guidance Receptor 1 (ROBO1) signaling pathway (101). Exosomal miR-365 from imatinib resistant chronic myeloid leukemia cells can confer drug resistance phenotypes in imatinib-sensitive chronic myeloid leukemia cells by decreasing apoptosis (102). Exosomes from anti-cancer drug-resistant breast cancer cells increase the levels of TGFβ1 and the lymphocyte activation inhibitor PD-L1 to confer resistance to anti-cancer drugs, such as trastuzumab (103). The down-regulation of miR-155 reduces the malignancy of chordoma cells, and enhances sensitivity to anti-cancer drugs by increasing the expression of PTEN and activating PI3K-Akt-mammalian target of rapamycin (mTOR) signaling (104). miR-155 overexpression suppresses suppressor oy cytokine signaling 1 (SOCS1) expression and induces the progression of Anaplastic thyroid cancer (ATC) (105). The inhibition of SOCS1 reverses the effects of mR-155 on cancer cell proliferation (105). Exosomal miR-155 of hepatocellular carcinoma cells (HCC) binds to the 3′-untranslated region (UTR) of PTEN, and stimulates the proliferation of HCC (106).
Exosomal miRNAs of Tams Regulate Tumor Growth and Anti-Cancer Drug Resistance
TAMs modulate various factors in the tumor microenvironment to facilitate tumor progression. TAMs secrete pro-tumorigenic factors that induce the gemcitabine resistance of pancreatic ductal adenocarcinoma (PDAC) cells (107). TAMs infiltrate solid tumors, and stimulate cell proliferation and angiogenesis (108, 109). TAMs mediate lung tumor progression enhanced by protein arginine methyl transferase 6 (PRMT6) (110). Exosomes derived from TAMs enhance the tumor growth and metastasis in mice (111). TAMs-derived exosomal miR-501-3p inhibits tumor suppressor transforming growth factor beta receptor 3 (TGFBR3) gene, and facilitates the development of PDAC (111). Exosomes derived from TAMs confer resistance to cisplatin in gastric cancer cells by activating PI3K/AKT signaling pathway (112). TAMs-derived exosomes (MDE) enhance the migration and invasion potential of colon cancer cells. In this, exosomal miR-21-5p and miR-155-5p bind to transcriptional activator brahma related gene 1(BRG1) and decreases the expression of BRG1 (113). BRG1 is a negative regulator of colorectal cancer metastasis. That study also showed that culture medium of M2 macrophages enhanced the motility, invasion, and metastasis of colorectal cancer cells. BRG1 mutation or lack of expression occurs in primary lung tumors and several cancers (114). Exosomal miR-223 from IL4-activated macrophages enhances the invasion potential of breast cancer cells by targeting myocyte enhancer factor 2 (MEF2)-β-catenin pathway (115). Overexpression of miR-223-3p enhances cell proliferation and metastasis by regulating the expression of solute carrier family 4 (SLC4A4) (116). miR-223 is upregulated in colon cancer and enhances colon cancer cell invasion and metastasis by decreasing the expression of p120 (117). Exosomal miR-223 derived from TAMs induces anti-cancer drug resistance in EOC cells by activating PTEN-PI3K/AKT pathway (118). miR-223 induces anti-cancer drug resistance in gastric cancer cells by regulating the expression of F-box and WD repeat domain-containing 7 (FBXW7) (119, 120). Adoptive transfer of miR-365 in TAMs confers resistance to gemcitabine in PDAC-bearing mice by upregulation of the triphospho-nucleotide pool in cancer cells (121). Cisplatin-stimulated TAMs enhance ovarian cancer cell migration by increasing the expression of CCL20 (122). Exosomes from plasma of EOC patients display higher expression level of miR-21, as compared to that of healthy women (123). The exosomal miR-21 confers resistance to cisplatin through the downregulation of PTEN, leading to the activation of PI3K/AKT pathway. The lack of miR-21 expression in TAMs results in anti-tumoral immune response involving the improvement of cytotoxic T-cell responses by macrophages through the induction of IL-12 and C-X-C motif chemokine 10 (124). miR-21-3p promotes proliferation and anti-apoptosis in esophageal squamous cell carcinoma (ESCC) by regulating tnf receptor associated factor 4 (TRAF4) (125). KRAS upregulates miR-21 and enhances cell migration/invasion by inhibiting tumor suppressor genes, such as neurofibromin 1 (NF1), ras p21 protein activator (RASA1), and Ras Association Domain-Containing Protein 8 (RASSF8) (126).
Exosomal miR-7 from TAMs suppresses the metastasis of EOC cells by inhibiting epidermal growth factor receptor (EGFR)/Akt/extracellular regulated kinase (ERK) pathway (127). Exosomal miR-142-3p from macrophages decreases the expression levels of stathmin-1 and insulin-like growth factor-1 receptor to inhibit the proliferation of HCCs (80).
Figure 5 provides an overview of exosomal miRNAs from TAMs that exert tumor or anti-tumoral functions.
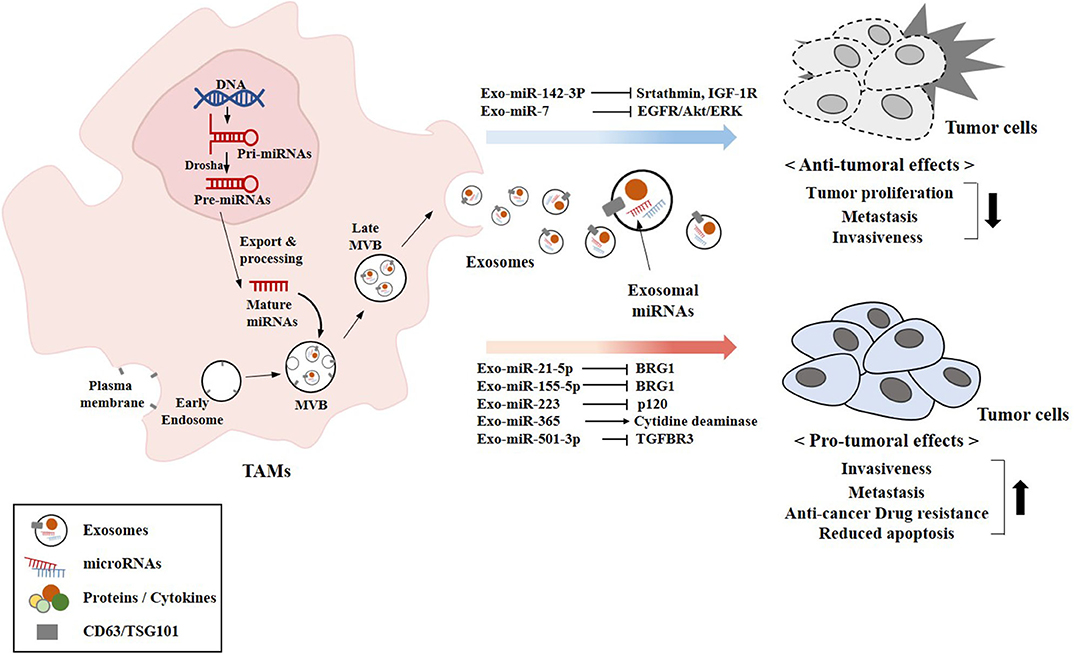
Figure 5. Exosomal miRNAs from TAMs exert pro-tumoral and anti-tumoral effects. The biogenesis of exosomes involves four different steps: (a) the membrane invagination; (b) endosome formation; (c) inward budding of endosomes to form multivesicular bodies (MVBs); and (d) the fusion of MVBs with the plasma membrane release the ILVs in the extracellular space by exocytosis and become exosomes. Exosomal miRNAs (miR-7 and miR-142-3p) that exert anti-tumoral effects are shown. Exosomal miRNAs (miR-21-5p and other miRNAs) that exert pro-tumoral effects are also shown. CD63 and TSG101 are surface markers of exosomes.
Current Progress in miRNA Therapeutics
miRNAs can serve as targets for the development of anti-cancer drugs based on their roles in tumorigenesis, cancer progression, and anti-cancer drug resistance. Resistance to standard anti-cancer drugs, such as Taxol, Doxorubicin, and Cisplatin, involves the deregulated expression of miRNAs in breast cancer (128). These miRNAs can also be employed as targets for the development of anti-cancer therapeutics. The close relationship between miRNA, tumor microenvironment, and the hallmarks of cancer makes miRNAs valuable targets for the development of therapeutic strategy to complement current cancer therapies. Small interfering RNA (siRNA), was granted FDA approval in 2018. However, miRNA drugs have not yet been granted FDA approval.
miRNA-based therapeutics that decrease the levels of oncogenic miRNAs (oncomiRs) or increase the levels of tumor suppressor miRNAs have great potential as cancer therapeutics. Clinical trials of miRNA-based drugs have shown promising results for the treatment of cancer (129). Since miRNAs have multiple targets, miRNA drugs might induce unwanted side effects. Systemic delivery methods of miRNA drugs involve injection or intravenous administration. Intratumoral injections of miRNA drugs can enhance target specificity and efficacy, and minimize side effects (130, 131). Intratumoral injection of cationic liposome/pVAX-miR-143 complex (CL-pVAX-miR-143) inhibits subcutaneous tumor growth and systemic injection inhibits tumor metastasis in a dose-dependent manner in early-stage experimental lung cancer metastasis models (132).
Clinical phase 1 trial employing “TargomiR” shows promising results in patients with malignant pleural mesothelioma or non-small cell lung cancer. TargomiR delivery vehicles comprise miRNA mimic, minicells, and a targeting moiety (a specific antibody that recognizes a target protein). MesomiR-1, the targomiR drug, comprises the miRNA mimic corresponding to tumor-suppressor miR-16 and an antibody to the EGFR (133, 134).
An antagomiR is a synthetic single-helix nucleic acid, consisting of DNA or analogs, peptide or locked nucleic acids (PNAs or LNAs, respectively), or miRNA sponges, perfectly complementary to a specific miRNA target. AntagomiRs can inhibit many different mRNAs. Many miRNA drugs are in the form of antagomiRs. Many approaches are now being employed to decrease the expression levels of miRNAs that display oncogenic potential (OncomiRs). For example, CRISPR/Cas-9 system is employed to decrease the expression levels of these oncomiRs (135). Putative miRNA drugs have exhibited significant efficacy in some cancers (miR-16 in lung cancer and miR-155 in T cell lymphoma), hepatitis C (miR-122), heart abnormalities (miR-92), and pathologic fibrosis (miR-29). Phase 1 clinical trial of MRG 110, a locked nucleic acid (LNA)-modified antisense oligonucleotide to inhibit the function of miR-92, is underway. Cobomarsen (MRG-106), a locked nucleic acid-modified oligonucleotide inhibitor of miR-155, inhibits cell proliferation and activates apoptosis in human lymphotropic virus type 1 (HTLV-1+) cutaneous T-cell lymphoma (CTCL) cell lines (136). A first-in-human phase 1 clinical trial of cobomarsen in patients with CTCL is currently underway. Identification of targets of miR-155 will be necessary for the rational design of cancer therapeutics. miR-10b is up-regulated in high-grade, and significantly down-regulated in low-grade gliomas (137). miR-10b targeting drug will be tried in patients with glioblastoma multiforme with a median survival of approximately 14.6 months.
Future Prospect
Cellular interactions within the microenvironment lead to tumor growth and progression. Understanding of cellular interactions within the tumor microenvironment is necessary for the development of anti-cancer therapeutics. TAMs play critical roles in tumorigenesis, tumor metastasis, and anti-cancer drug resistance. TAMs can therefore serve as targets for the development of anti-cancer therapeutics. Changing immunosuppressive TAMs into M1 macrophages can be used in combination with current anti-cancer immunotherapies (138). Specific deletion of TAMs might also be used in combination with current anti-cancer therapy. Inhibition of the infiltration of monocytes into the tumor site can be an effective anti-cancer therapy. Many reports suggest that circulating miRNAs can be employed as diagnostic markers for cancers (139). Identification of the signature miRNAs of each cancer will be helpful for the diagnosis of cancers, and the development of anti-cancer therapeutics. Identification of the target genes and their implicated functions, along with an in vitro and in vivo preclinical research models, has made it possible for the development of anti-cancer drugs employing miRNAs. miRNAs that induce the M2 polarization of macrophages can serve as targets for the development of anti-cancer drugs. Macrophages miRNAs that regulate ant-cancer drug-resistance, tumor growth, and tumor metastasis can also serve as targets for the development of anti-cancer drugs. miRNA therapeutics offer a valuable approach for the development of anti-cancer therapeutics.
Author Contributions
DJ wrote the manuscript. YKw and MK made the figures and tables. HJ helped in editing. YKi and HJ provided intellectual output in the manuscript.
Funding
This work was supported by National Research Foundation Grants (2020R1A2C1006996, 2017M3A9G7072417, and 2018R1D1A1B07043498), a grant from the BK21 plus Program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. (2011) 17:320–9. doi: 10.1038/nm.2328
2. Sun DY, Wu JQ, He ZH, He MF, Sun HB. Cancer-associated fibroblast regulate proliferation and migration of prostate cancer cells through TGF-β signaling pathway. Life Sci. (2019) 235:116791. doi: 10.1016/j.lfs.2019.116791
3. Li Z, Zhou J, Zhang J, Li S, Wang H, Du J. Cancer-associated fibroblasts promote PD-L1 expression in mice cancer cells via secreting CXCL5. Int J Cancer. (2019) 145:1946–57. doi: 10.1002/ijc.32278
4. Goulet CR, Champagne A, Bernard G, Vandal D, Chabaud S, Pouliot F, et al. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of bladder cancer cells through paracrine IL-6 signalling. BMC Cancer. (2019) 19:137. doi: 10.1186/s12885-019-5353-6
5. Hsu YL, Yen MC, Chang WA, Tsai PH, Pan YC, Liao SH, et al. CXCL17-derived CD11b+Gr-1+ myeloid-derived suppressor cells contribute to lung metastasis of breast cancer through platelet-derived growth factor-BB. Breast Cancer Res. (2019) 21:23. doi: 10.1186/s13058-019-1114-3
6. Mao FY, Zhao YL, Lv YP, Teng YS, Kong H, Liu YG, et al. CD45+CD33lowCD11bdim myeloid-derived suppressor cells suppress CD8+ T cell activity via the IL-6/IL-8-arginase I axis in human gastric cancer. Cell Death Dis. (2018) 9:763. doi: 10.1038/s41419-018-0803-7
7. Liu Y, Zhao JJ, Zhou ZQ, Pan QZ, Zhu Q, Tang Y, et al. IL-37 induces anti-tumor immunity by indirectly promoting dendritic cell recruitment and activation in hepatocellular carcinoma. Cancer Manag Res. (2019) 11:6691–702. doi: 10.2147/CMAR.S200627
8. Spiegel A, Brooks MW, Houshyar S, Reinhardt F, Ardolino M, Fessler E, et al. Neutrophils suppress intraluminal NK cell-mediated tumor cell clearance and enhance extravasation of disseminated carcinoma cells. Cancer Discov. (2016) 6:630–49. doi: 10.1158/2159-8290.CD-15-1157
9. Zhou SL, Zhou ZJ, Hu ZQ, Huang XW, Wang Z, Chen EB, et al. Tumor-associated neutrophils recruit macrophages and T-regulatory cells to promote progression of hepatocellular carcinoma and resistance to sorafenib. Gastroenterology. (2016) 150:1646–58.e17. doi: 10.1053/j.gastro.2016.02.040
10. Sun B, Qin W, Song M, Liu L, Yu Y, Qi X, et al. Neutrophil suppresses tumor cell proliferation via fas /fas ligand pathway mediated cell cycle arrested. Int J Biol Sci. (2018) 14:2103–13. doi: 10.7150/ijbs.29297
11. Finisguerra V, Di Conza G, Di Matteo M, Serneels J, Costa S, Thompson AA, et al. MET is required for the recruitment of anti-tumoural neutrophils. Nature. (2015) 522:349–53. doi: 10.1038/nature14407
12. Wu YL, Li HY, Zhao XP, Jiao JY, Tang DX, Yan LJ, et al. Mesenchymal stem cell-derived CCN2 promotes the proliferation, migration and invasion of human tongue squamous cell carcinoma cells. Cancer Sci. (2017) 108:897–909. doi: 10.1111/cas.13202
13. Liu J, Han G, Liu H, Qin C. Suppression of cholangiocarcinoma cell growth by human umbilical cord mesenchymal stem cells: a possible role of Wnt and Akt signaling. PLoS ONE. (2013) 8:e62844. doi: 10.1371/journal.pone.0062844
14. Freeman AJ, Vervoort SJ, Ramsbottom KM, Kelly MJ, Michie J, Pijpers L, et al. Natural killer cells suppress T cell-associated tumor immune evasion. Cell Rep. (2019) 28:2784–94.e5. doi: 10.1016/j.celrep.2019.08.017
15. Qi L, Zhang Q, Miao Y, Kang W, Tian Z, Xu D, et al. Interleukin-33 activates and recruits natural killer cells to inhibit pulmonary metastatic cancer development. Int J Cancer. (2020) 146:1421–34. doi: 10.1002/ijc.32779
16. Xiao H, He M, Xie G, Liu Y, Zhao Y, Ye X, et al. The release of tryptase from mast cells promote tumor cell metastasis via exosomes. BMC Cancer. (2019) 19:1015. doi: 10.1186/s12885-019-6203-2
17. Plotkin JD, Elias MG, Fereydouni M, Daniels-Wells TR, Dellinger AL, Penichet ML, et al. Human mast cells from adipose tissue target and induce apoptosis of breast cancer cells. Front Immunol. (2019) 10:138. doi: 10.3389/fimmu.2019.00138
18. Attarha S, Roy A, Westermark B, Tchougounova E. Mast cells modulate proliferation, migration and stemness of glioma cells through downregulation of GSK3beta expression and inhibition of STAT3 activation. Cell Signal. (2017) 37:81–92. doi: 10.1016/j.cellsig.2017.06.004
19. Robinson-Smith TM, Isaacsohn I, Mercer CA, Zhou M, Van Rooijen N, Husseinzadeh N, et al. Macrophages mediate inflammation-enhanced metastasis of ovarian tumors in mice. Cancer Res. (2007) 67:5708–16. doi: 10.1158/0008-5472.CAN-06-4375
20. Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. (2012) 122:787–95. doi: 10.1172/JCI59643
21. Brown JM, Recht L, Strober S. The promise of targeting macrophages in cancer therapy. Clin Cancer Res. (2017) 23:3241–50. doi: 10.1158/1078-0432.CCR-16-3122
22. Kim J, Bae JS. Tumor-associated macrophages and neutrophils in tumor microenvironment. Mediators Inflamm. (2016) 2016:6058147. doi: 10.1155/2016/6058147
23. Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. (2010) 141:39–51. doi: 10.1016/j.cell.2010.03.014
24. Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. (2014) 41:49–61. doi: 10.1016/j.immuni.2014.06.010
25. Chen J, Yao Y, Gong C, Yu F, Su S, Chen J, et al. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell. (2011) 19:541–55. doi: 10.1016/j.ccr.2011.02.006
26. Cardoso AP, Pinto ML, Pinto AT, Oliveira MI, Pinto MT, Gonçalves R, et al. Macrophages stimulate gastric and colorectal cancer invasion through EGFR Y (1086), c-Src, Erk1/2 and Akt phosphorylation and small GT Pase activity. Oncogene. (2014) 33:2123–33. doi: 10.1038/onc.2013.154
27. Sica A, Erreni M, Allavena P, Porta C. Macrophage polarization in pathology. Cell Mol Life Sci. (2015) 72:4111–26. doi: 10.1007/s00018-015-1995-y
28. Hao NB, Lü MH, Fan YH, Cao YL, Zhang ZR, Yang SM. Macrophages in tumor microenvironments and the progression of tumors. Clin Dev Immunol. (2012) 2012:948098. doi: 10.1155/2012/948098
29. Caronni N, Savino B, Bonecchi R. Myeloid cells in cancer-related inflammation. Immunobiology. (2015) 220:249–53. doi: 10.1016/j.imbio.2014.10.001
30. Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. (2010) 32:593–604. doi: 10.1016/j.immuni.2010.05.007
31. Gadani SP, Walsh JT, Smirnov I, Zheng J, Kipnis J. The glia-derived alarmin IL-33 orchestrates the immune response and promotes recovery following CNS injury. Neuron. (2015) 85:703–9. doi: 10.1016/j.neuron.2015.01.013
32. Sica A, Saccani A, Bottazzi B, Polentarutti N, Vecchi A, van Damme J, et al. Autocrine production of IL-10 mediates defective IL-12 production and NF-kappa B activation in tumor-associated macrophages. J Immunol. (2000) 164:762–7. doi: 10.4049/jimmunol.164.2.762
33. Yang W, Li Z, Qin R, Wang X, An H, Wang Y, et al. YY1 promotes endothelial cell-dependent tumor angiogenesis in hepatocellular carcinoma by transcriptionally activating VEGFA. Front Oncol. (2019) 9:1187. doi: 10.3389/fonc.2019.01187
34. Roblek M, Protsyuk D, Becker PF, Stefanescu C, Gorzelanny C, Glaus Garzon JF, et al. CCL2 is a vascular permeability factor inducing CCR2-dependent endothelial retraction during lung metastasis. Mol Cancer Res. (2019) 17:783–93. doi: 10.1158/1541-7786.MCR-18-0530
35. Sax MJ, Gasch C, Athota VR, Freeman R, Rasighaemi P, Westcott DE, et al. Cancer cell CCL5 mediates bone marrow independent angiogenesis in breast cancer. Oncotarget. (2016) 7:85437–49. doi: 10.18632/oncotarget.13387
36. Wen Z, Liu H, Li M, Li B, Gao W, Shao Q, et al. Increased metabolites of 5-lipoxygenase from hypoxic ovarian cancer cells promote tumor-associated macrophage infiltration. Oncogene. (2015) 34:1241–52. doi: 10.1038/onc.2014.85
37. Zou K, Wang Y, Hu Y, Zheng L, Xu W, Li G. Specific tumor-derived CCL2 mediated by pyruvate kinase M2 in colorectal cancer cells contributes to macrophage recruitment in tumor microenvironment. Tumour Biol. (2017) 39:1010428317695962. doi: 10.1177/1010428317695962
38. Allavena P, Sica A, Solinas G, Porta C, Mantovani A. The inflammatory micro-environment in tumor progression: the role of tumor-associated macrophages. Crit Rev Oncol Hematol. (2008) 66:1–9. doi: 10.1016/j.critrevonc.2007.07.004
39. Murdoch C, Muthana M, Coffelt SB, Lewis CE. The role of myeloid cells in the promotion of tumour angiogenesis. Nat Rev Cancer. (2008) 8:618–31. doi: 10.1038/nrc2444
40. Lee JH, Lee GT, Woo SH, Ha YS, Kwon SJ, Kim WJ, et al. BMP-6 in renal cell carcinoma promotes tumor proliferation through IL-10-Dependent M2 polarization of tumor-associated macrophages. Cancer Res. (2013) 73:3604–14. doi: 10.1158/0008-5472.CAN-12-4563
41. Zhang J, Li H, Wu Q, Chen Y, Deng Y, Yang Z, et al. Tumoral NOX4 recruits M2 tumor-associated macrophages via ROS/PI3K signaling-dependent various cytokine production to promote NSCLC growth. Redox Biol. (2019) 22:101116. doi: 10.1016/j.redox.2019.101116
42. Mazzoni M, Mauro G, Erreni M, Romeo P, Minna E, Vizioli MG, et al. Senescent thyrocytes and thyroid tumor cells induce M2-like macrophage polarization of human monocytes via a PGE2-dependent mechanism. J Exp Clin Cancer Res. (2019) 38:208. doi: 10.1186/s13046-019-1198-8
43. Soldano S, Pizzorni C, Paolino S, Trombetta AC, Montagna P, Brizzolara R, et al. Alternatively Activated (M2) M2. PLoS ONE. (2016) 11:e0166433. doi: 10.1371/journal.pone.0166433
44. Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. (2010) 22:231–7. doi: 10.1016/j.coi.2010.01.009
45. Ruffell B, Affara NI, Coussens LM. Differential macrophage programming in the tumor microenvironment. Trends Immunol. (2012) 33:119–26. doi: 10.1016/j.it.2011.12.001
46. Dirkx AE, Oude Egbrink MG, Wagstaff J, Griffioen AW. Monocyte/macrophage infiltration in tumors: modulators of angiogenesis. J Leukoc Biol. (2006) 80:1183–96. doi: 10.1189/jlb.0905495
47. Bruchard M, Mignot G, Derangère V, Chalmin F, Chevriaux A, Végran F, et al. Chemotherapy-triggered cathepsin B release in myeloid-derived suppressor cells activates the Nlrp3 inflammasome and promotes tumor growth. Nat Med. (2013) 19:57–64. doi: 10.1038/nm.2999
48. De Palma M, Lewis CE. Macrophage regulation of tumor responses to anticancer therapies. Cancer Cell. (2013) 23:277–86. doi: 10.1016/j.ccr.2013.02.013
49. Jinushi M, Chiba S, Yoshiyama H, Masutomi K, Kinoshita I, Dosaka-Akita H, et al. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc Natl Acad Sci USA. (2011) 108:12425–30. doi: 10.1073/pnas.1106645108
50. Han Y, Guo W, Ren T, Huang Y, Wang S, Liu K, et al. Tumor-associated macrophages promote lung metastasis and induce epithelial-mesenchymal transition in osteosarcoma by activating the COX-2/STAT3 axis. Cancer Lett. (2019) 440–441:116–25. doi: 10.1016/j.canlet.2018.10.011
51. Sakamoto S, Kagawa S, Kuwada K, Ito A, Kajioka H, Kakiuchi Y, et al. Intraperitoneal cancer-immune microenvironment promotes peritoneal dissemination of gastric cancer. Oncoimmunology. (2019) 8:e1671760. doi: 10.1080/2162402X.2019.1671760
52. Yang L, Dong Y, Li Y, Wang D, Liu S, Wang D, et al. IL-10 derived from M2 macrophage promotes cancer stemness via JAK1/STAT1/NF-κB/Notch1 pathway in non-small cell lung cancer. Int J Cancer. (2019) 145:1099–110. doi: 10.1002/ijc.32151
53. Zhang D, Qiu X, Li J, Zheng S, Li L, Zhao H. TGF-β secreted by tumor-associated macrophages promotes proliferation and invasion of colorectal cancer via miR-34a-VEGF axis. Cell Cycle. (2018) 17:2766–78. doi: 10.1080/15384101.2018.1556064
54. Lee CC, Lin JC, Hwang WL, Kuo YJ, Chen HK, Tai SK, et al. Macrophage-secreted interleukin-35 regulates cancer cell plasticity to facilitate metastatic colonization. Nat Commun. (2018) 9:3763. doi: 10.1038/s41467-018-06268-0
55. Zhu Q, Wu X, Wu Y, Wang X. Interaction between Treg cells and tumor-associated macrophages in the tumor microenvironment of epithelial ovarian cancer. Oncol Rep. (2016) 36:3472–8. doi: 10.3892/or.2016.5136
56. Savage ND, de Boer T, Walburg KV, Joosten SA, van Meijgaarden K, Geluk A, et al. Human anti-inflammatory macrophages induce Foxp3+ GITR+ CD25+ regulatory T cells, which suppress via membrane-bound TGFbeta-1. J Immunol. (2008) 181:2220–6. doi: 10.4049/jimmunol.181.3.2220
57. Arlauckas SP, Garren SB, Garris CS, Kohler RH, Oh J, Pittet MJ, et al. Arg1 expression defines immunosuppressive subsets of tumor-associated macrophages. Theranostics. (2018) 8:5842–54. doi: 10.7150/thno.26888
58. Sawa-Wejksza K, Kandefer-Szerszen M. Tumor-associated macrophages as target for antitumor therapy. Arch Immunol Ther Exp. (2018) 66:97–111. doi: 10.1007/s00005-017-0480-8
59. Gyori D, Lim EL, Grant FM, Spensberger D, Roychoudhuri R, Shuttleworth SJ, et al. Compensation between CSF1R+ macrophages and Foxp3+ Treg cells drives resistance to tumor immunotherapy. JCI Insight. (2018) 3:120631. doi: 10.1172/jci.insight.120631
60. Li Z, Liu X, Guo R, Wang P. CD4+ Foxp3- type 1 regulatory T cells in glioblastoma multiforme suppress T cell responses through multiple pathways and are regulated by tumor-associated macrophages. Int J Biochem Cell Biol. (2016) 81:1–9. doi: 10.1016/j.biocel.2016.09.013
61. Zhang F, Wang H, Wang X, Jiang G, Liu H, Zhang G, et al. TGF-β induces M2-like macrophage polarization via SNAIL-mediated suppression of a pro-inflammatory phenotype. Oncotarget. (2016) 7:52294–306. doi: 10.18632/oncotarget.10561
62. Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. (2019) 18:75. doi: 10.1186/s12943-019-0991-5
63. Pucci F, Pittet MJ. Molecular pathways: tumor-derived microvesicles and their interactions with immune cells in vivo. Clin Cancer Res. (2013) 19:2598–604. doi: 10.1158/1078-0432.CCR-12-0962
64. Melo SA, Sugimoto H, O'Connell JT, Kato N, Villanueva A, Vidal A, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. (2014) 26:707–21. doi: 10.1016/j.ccell.2014.09.005
65. Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. (2012) 18:883–91. doi: 10.1038/nm.2753
66. Taylor DD, Gercel-Taylor C. Exosomes/microvesicles: mediators of cancer associated immunosuppressive microenvironments. Semin Immunopathol. (2011) 33:441–54. doi: 10.1007/s00281-010-0234-8
67. Martinez VG, O'Neill S, Salimu J, Breslin S, Clayton A, Crown J, et al. Resistance to HER2-targeted anti-cancer drugs is associated with immune evasion in cancer cells and their derived extracellular vesicles. Oncoimmunology. (2017) 6:e1362530. doi: 10.1080/2162402X.2017.1362530
68. Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. (2013) 14:319. doi: 10.1186/1471-2164-14-319
69. Yuan T, Huang X, Woodcock M, Du M, Dittmar R, Wang Y, et al. Plasma extracellular RNA profiles in healthy and cancer patients. Sci Rep. (2016) 6:19413. doi: 10.1038/srep19413
70. Wang FF, Zhang XJ, Yan YR, Zhu XH, Yu J, Ding Y, et al. FBX8 is a metastasis suppressor downstream of miR-223 and targeting mTOR for degradation in colorectal carcinoma. Cancer Lett. (2017) 388:85–95. doi: 10.1016/j.canlet.2016.11.031
71. Graves P, Zeng Y. Biogenesis of mammalian microRNAs: a global view. Genom Proteom Bioinform. (2012) 10:239–45. doi: 10.1016/j.gpb.2012.06.004
72. Motiño O, Francés DE, Mayoral R, Castro-Sánchez L, Fernández-Velasco M, Boscá L, et al. Regulation of microRNA 183 by cyclooxygenase 2 in liver is dead-box helicase p68 (DDX5) dependent: role in insulin signaling. Mol Cell Biol. (2015) 35:2554–67. doi: 10.1128/MCB.00198-15
73. Kopkova A, Sana J, Machackova T, Vecera M, Radova L, Trachtova K, et al. Cerebrospinal fluid microRNA signatures as diagnostic biomarkers in brain tumors. Cancers. (2019) 11:E1546. doi: 10.3390/cancers11101546
74. Wang D, Wang X, Si M, Yang J, Sun S, Wu H, et al. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett. (2020) 474:36–52. doi: 10.1016/j.canlet.2020.01.005
75. Yeon M, Byun J, Kim H, Kim M, Jung HS, Jeon D, et al. CAGE binds to beclin1, regulates autophagic flux and CAGE-derived peptide confers sensitivity to anti-cancer drugs in non-small cell lung cancer cells. Front Oncol. (2018) 8:599. doi: 10.3389/fonc.2018.00599
76. Kwon Y, Kim Y, Jung HS, Jeoung D. Role of HDAC3-miRNA-CAGE network in anti-cancer drug-resistance. Int J Mol Sci. (2018) 20:E51. doi: 10.3390/ijms20010051
77. Frank AC, Ebersberger S, Fink AF, Lampe S, Weigert A, Schmid T, et al. Apoptotic tumor cell-derived microRNA-375 uses CD36 to alter the tumor-associated macrophage phenotype. Nat Commun. (2019) 10:1135. doi: 10.1038/s41467-019-08989-2
78. Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles. (2014) 3:24743. doi: 10.3402/jev.v3.23743
79. Lu FB, Chen DZ, Chen L, Hu ED, Wu JL, Li H, et al. Attenuation of experimental autoimmune hepatitis in mice with bone mesenchymal stem cell-derived exosomes carrying microRNA-223-3p. Mol Cells. (2019) 42:906–18. doi: 10.14348/molcells.2019.2283
80. Aucher A, Rudnicka D, Davis DM. MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation. J Immunol. (2013) 191:6250–60. doi: 10.4049/jimmunol.1301728
81. Szebeni GJ, Vizler C, Kitajka K, Puskas LG. Inflammation and cancer: extra and intracellular determinants of tumor-associated macrophages as tumor promoters. Mediators Inflamm. (2017) 2017:9294018. doi: 10.1155/2017/9294018
82. Ying X, Wu Q, Wu X, Zhu Q, Wang X, Jiang L, et al. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget. (2016) 7:43076–87. doi: 10.18632/oncotarget.9246
83. Chen X, Zhou J, Li X, Wang X, Lin Y, Wang X. Exosomes derived from hypoxic epithelial ovarian cancer cells deliver microRNAs to macrophages and elicit a tumor-promoted phenotype. Cancer Lett. (2018) 435:80–91. doi: 10.1016/j.canlet.2018.08.001
84. Chen X, Ying X, Wang X, Wu X, Zhu Q, Wang X. Exosomes derived from hypoxic epithelial ovarian cancer deliver microRNA-940 to induce macrophage M2 polarization. Oncol Rep. (2017) 38:522–8. doi: 10.3892/or.2017.5697
85. Moradi-Chaleshtori M, Hashemi SM, Soudi S, Bandehpour M, Mohammadi-Yeganeh S. Tumor-derived exosomal microRNAs and proteins as modulators of macrophage function. J Cell Physiol. (2019) 234:7970–82. doi: 10.1002/jcp.27552
86. Takano Y, Masuda T, Iinuma H, Yamaguchi R, Sato K, Tobo T, et al. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget. (2017) 8:78598–613. doi: 10.18632/oncotarget.20009
87. Shinohara H, Kuranaga Y, Kumazaki M, Sugito N, Yoshikawa Y, Takai T, et al. Regulated polarization of tumor-associated macrophages by mir-145 via colorectal cancer-derived extracellular vesicles. J Immunol. (2017) 199:1505–15. doi: 10.4049/jimmunol.1700167
88. Liu Y, Zhao L, Li D, Yin Y, Zhang CY, Li J, et al. Microvesicle-delivery miR-150 promotes tumorigenesis by up-regulating VEGF, and the neutralization of miR-150 attenuate tumor development. Protein Cell. (2013) 4:932–41. doi: 10.1007/s13238-013-3092-z
89. Yin C, Han Q, Xu D, Zheng B, Zhao X, Zhang J. SALL4-mediated upregulation of exosomal miR-146a-5p drives T-cell exhaustion by M2 tumor-associated macrophages in HCC. Oncoimmunology. (2019) 8:1601479. doi: 10.1080/2162402X.2019.1601479
90. Liu J, Fan L, Yu H, Zhang J, He Y, Feng D, et al. Endoplasmic reticulum stress causes liver cancer cells to release exosomal miR-23a-3p and up-regulate programmed death ligand 1 expression in macrophages. Hepatology. (2019) 70:241–58. doi: 10.1002/hep.30607
91. Wang X, Luo G, Zhang K, Cao J, Huang C, Jiang T, et al. Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kγ to promote pancreatic cancer metastasis. Cancer Res. (2018) 78:4586–98. doi: 10.1158/0008-5472.CAN-17-3841
92. Trivedi M, Talekar M, Shah P, Ouyang Q, Amiji M. Modification of tumor cell exosome content by transfection with wt-p53 and microRNA-125b expressing plasmid DNA and its effect on macrophage polarization. Oncogenesis. (2016) 5:e250. doi: 10.1038/oncsis.2016.52
93. Yang J, Zhang Z, Chen C, Liu Y, Si Q, Chuang TH, et al. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene. (2014) 33:3014–23. doi: 10.1038/onc.2013.258
94. Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. (2012) 109:E2110–16. doi: 10.1073/pnas.1209414109
95. Jang JY, Lee JK, Jeon YK, Kim CW. Exosome derived from epigallocatechin gallate treated breast cancer cells suppresses tumor growth by inhibiting tumor-associated macrophage infiltration and M2 polarization. BMC Cancer. (2013) 13:421. doi: 10.1186/1471-2407-13-421
96. Su MJ, Aldawsari H, Amiji M. Pancreatic cancer cell exosome-mediated macrophage reprogramming and the role of MicroRNAs 155 and 125b2 transfection using nanoparticle delivery systems. Sci Rep. (2016) 6:30110. doi: 10.1038/srep30110
97. Plum PS, Warnecke-Eberz U, Drebber U, Chon SH, Alakus H, Hölscher AH, et al. Upregulation of miR-17-92 cluster is associated with progression and lymph node metastasis in oesophageal adenocarcinoma. Sci Rep. (2019) 9:12113. doi: 10.1038/s41598-019-48624-0
98. Niveditha D, Jasoria M, Narayan J, Majumder S, Mukherjee S, Chowdhury R, et al. Common and unique microRNAs in multiple carcinomas regulate similar network of pathways to mediate cancer progression. Sci Rep. (2020) 10:2331. doi: 10.1038/s41598-020-59142-9
99. McMillin DW, Negri JM, Mitsiades CS. The role of tumour-stromal interactions in modifying drug response: challenges and opportunities. Nat. Rev. Drug Discov. (2013) 12:217–28. doi: 10.1038/nrd3870
100. Sousa D, Lima RT, Vasconcelos MH. Intercellular transfer of cancer drug resistance traits by extracellular vesicles. Trends Mol Med. (2015) 21:595–608. doi: 10.1016/j.molmed.2015.08.002
101. Wang Y, Zhang S, Bao H, Mu S, Zhang B, Ma H, et al. MicroRNA-365 promotes lung carcinogenesis by downregulating the USP33/SLIT2/ROBO1 signalling pathway. Cancer Cell Int. (2018) 18:64. doi: 10.1186/s12935-018-0563-6
102. Min QH, Wang XZ, Zhang J, Chen QG, Li SQ, Liu XQ, et al. Exosomes derived from imatinib-resistant chronic myeloid leukemia cells mediate a horizontal transfer of drug-resistant trait by delivering miR-365. Exp Cell Res. (2018) 362:386–93. doi: 10.1016/j.yexcr.2017.12.001
103. Ning S, Liu H, Gao B, Wei W, Yang A, Li J, et al. miR-155, miR-96 and miR-99a as potential diagnostic and prognostic tools for the clinical management of hepatocellular carcinoma. Oncol Lett. (2019) 18:3381–7. doi: 10.3892/ol.2019.10606
104. Zhao K, Li X, Chen X, Zhu Q, Yin F, Ruan Q, et al. Inhibition of miR-140-3p or miR-155-5p by antagomir treatment sensitize chordoma cells to chemotherapy drug treatment by increasing PTEN expression. Eur J Pharmacol. (2019) 854:298–306. doi: 10.1016/j.ejphar.2019.03.034
105. Zhang W, Ji W, Zhao X. MiR-155 promotes anaplastic thyroid cancer progression by directly targeting SOCS1. BMC Cancer. (2019) 19:1093. doi: 10.1186/s12885-019-6319-4
106. Sun JF, Zhang D, Gao CJ, Zhang YW, Dai QS. Exosome-Mediated MiR-155 transfer contributes to hepatocellular carcinoma cell proliferation by targeting PTEN. Med Sci Monit Basic Res. (2019) 25:218–28. doi: 10.12659/MSMBR.918134
107. Weizman N, Krelin Y, Shabtay-Orbach A, Amit M, Binenbaum Y, Wong RJ, et al. Macrophages mediate gemcitabine resistance of pancreatic adenocarcinoma by upregulating cytidine deaminase. Oncogene. (2014) 33:3812–9. doi: 10.1038/onc.2013.357
108. Fukuda K, Kobayashi A, Watabe K. The role of tumor-associated macrophage in tumor progression. Front Biosci. (2012) 4:787–98. doi: 10.2741/s299
109. Atanasov G, Pötner C, Aust G, Schierle K, Dietel C, Benzing C, et al. TIE2-expressing monocytes and M2-polarized macrophages impact survival and correlate with angiogenesis in adenocarcinoma of the pancreas. Oncotarget. (2018) 9:29715–26. doi: 10.18632/oncotarget.25690
110. Avasarala S, Wu PY, Khan SQ, Yanlin S, Van Scoyk M, Bao J, et al. PRMT6 promotes lung tumor progression via the alternate activation of tumor-associated macrophages. Mol Cancer Res. (2020) 18:166–78. doi: 10.1158/1541-7786.MCR-19-0204
111. Yin Z, Ma T, Huang B, Lin L, Zhou Y, Yan J, et al. Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-β signaling pathway. J Exp Clin Cancer Res. (2019) 38:310. doi: 10.1186/s13046-019-1313-x
112. Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie G, et al. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J Exp Clin Cancer Res. (2017) 36:53. doi: 10.1186/s13046-017-0528-y
113. Lan J, Sun L, Xu F, Liu L, Hu F, Song D, et al. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. (2019) 79:146–58. doi: 10.1158/0008-5472.CAN-18-0014
114. Medina PP, Romero OA, Kohno T, Montuenga LM, Pio R, Yokota J, et al. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat. (2008) 29:617–22. doi: 10.1002/humu.20730
115. Yang M, Chen J, Su F, Yu B, Su F, Lin L, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. (2011) 10:117. doi: 10.1186/1476-4598-10-117
116. Xiao W, Wang X, Wang T, Xing J. MiR-223-3p promotes cell proliferation and metastasis by downregulating SLC4A4 in clear cell renal cell carcinoma. Aging. (2019) 11:615–33. doi: 10.18632/aging.101763
117. Liu L, Zhang C, Li X, Sun W, Qin S, Qin L, et al. miR-223 promotes colon cancer by directly targeting p120 catenin. Oncotarget. (2017) 8:63764–79. doi: 10.18632/oncotarget.19541
118. Zhu X, Shen H, Yin X, Yang M, Wei H, Chen Q, et al. Macrophages derived exosomes deliver miR-223 to epithelial ovarian cancer cells to elicit a chemoresistant phenotype. J Exp Clin Cancer Res. (2019) 38:81. doi: 10.1186/s13046-019-1095-1
119. Eto K, Iwatsuki M, Watanabe M, Ishimoto T, Ida S, Imamura Y, et al. The sensitivity of gastric cancer to trastuzumab is regulated by the miR-223/FBXW7 pathway. Int J Cancer. (2015) 136:1537–45. doi: 10.1002/ijc.29168
120. Zhou X, Jin W, Jia H, Yan J, Zhang G. MiR-223 promotes the cisplatin resistance of human gastric cancer cells via regulating cell cycle by targeting FBXW7. J Exp Clin Cancer Res. (2015) 34:28. doi: 10.1186/s13046-015-0145-6
121. Binenbaum Y, Fridman E, Yaari Z, Milman N, Schroeder A, Ben David G, et al. Transfer of miRNA in macrophage-derived exosomes induces drug resistance in pancreatic adenocarcinoma. Cancer Res. (2018) 78:5287–99. doi: 10.1158/0008-5472.CAN-18-0124
122. Liu W, Wang W, Wang X, Xu C, Zhang N, Di W. Cisplatin-stimulated macrophages promote ovarian cancer migration via the CCL20-CCR6 axis. Cancer Lett. (2020) 472:59–69. doi: 10.1016/j.canlet.2019.12.024
123. Pan C, Stevic I, Müller V, Ni Q, Oliveira-Ferrer L, Pantel K, et al. Exosomal microRNAs as tumor markers in epithelial ovarian cancer. Mol Oncol. (2018) 12:1935–48. doi: 10.1002/1878-0261.12371
124. Sahraei M, Chaube B, Liu Y, Sun J, Kaplan A, Price NL, et al. miR-21Suppressing miR-21 activity in tumor-associated macrophages promotes an antitumor immune response. J Clin Invest. (2019) 129:5518–36. doi: 10.1172/JCI127125
125. Gao Z, Liu H, Shi Y, Yin L, Zhu Y, Liu R. Identification of cancer stem cell molecular markers and effects of hsa-miR-21-3p on stemness in esophageal squamous cell carcinoma. Cancers. (2019) 11:E518. doi: 10.3390/cancers11040518
126. Shi L, Middleton J, Jeon YJ, Magee P, Veneziano D, Laganà A, et al. KRAS induces lung tumorigenesis through microRNAs modulation. Cell Death Dis. (2018) 9:219. doi: 10.1038/s41419-017-0243-9
127. Hu Y, Li D, Wu A, Qiu X, Di W, Huang L, et al. TWEAK-stimulated macrophages inhibit metastasis of epithelial ovarian cancer via exosomal shuttling of microRNA. Cancer Lett. (2017) 393:60–7. doi: 10.1016/j.canlet.2017.02.009
128. Hu W, Tan C, He Y, Zhang G, Xu Y, Tang J. Functional miRNAs in breast cancer drug resistance. Onco Targets Ther. (2018) 11:1529–41. doi: 10.2147/OTT.S152462
129. Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol. (2019) 234:5451–65. doi: 10.1002/jcp.27486
130. Chen Y, Gao DY, Huang L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv Drug Deliv Rev. (2015) 81:128–41. doi: 10.1016/j.addr.2014.05.009
131. Bertucci A, Kim KH, Kang J, Zuidema JM, Lee SH, Kwon EJ, et al. Tumor-Targeting, microRNA-Silencing porous silicon nanoparticles for ovarian cancer therapy. ACS Appl Mater Interfaces. (2019) 11:23926–37. doi: 10.1021/acsami.9b07980
132. Jiang Q, Yuan Y, Gong Y, Luo X, Su X, Hu X, et al. Therapeutic delivery of microRNA-143 by cationic lipoplexes for non-small cell lung cancer treatment in vivo. J Cancer Res Clin Oncol. (2019) 145:2951–67. doi: 10.1007/s00432-019-03051-6
133. Reid G, Kao SC, Pavlakis N, Brahmbhatt H, MacDiarmid J, Clarke S, et al. Clinical development of TargomiRs, a miRNA mimic-based treatment for patients with recurrent thoracic cancer. Epigenomics. (2016) 8:1079–85. doi: 10.2217/epi-2016-0035
134. van Zandwijk N, Pavlakis N, Kao SC, Linton A, Boyer MJ, Clarke S, et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. (2017) 18:1386–96. doi: 10.1016/S1470-2045(17)30621-6
135. Chang H, Yi B, Ma R, Zhang X, Zhao H, Xi Y. CRISPR/cas9, a novel genomic tool to knockdown microRNA in vitro and in vivo. Sci Rep. (2016) 6:22312. doi: 10.1038/srep22312
136. Seto AG, Beatty X, Lynch JM, Hermreck M, Tetzlaff M, Duvic M, et al. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br J Haematol. (2018) 183:428–44. doi: 10.1111/bjh.15547
137. Visani M, de Biase D, Marucci G, Cerasoli S, Nigrisoli E, Bacchi Reggiani ML, et al. Expression of 19 microRNAs in glioblastoma and comparison with other brain neoplasia of grades I-III. Mol Oncol. (2014) 8:417–30. doi: 10.1016/j.molonc.2013.12.010
138. Xi J, Huang Q, Wang L, Ma X, Deng Q, Kumar M, et al. miR-21 depletion in macrophages promotes tumoricidal polarization and enhances PD-1 immunotherapy. Oncogene. (2018) 37:3151–65. doi: 10.1038/s41388-018-0178-3
Keywords: anti-cancer drugs, cancer cells, cellular interactions, exosomes, macrophages, microRNAs
Citation: Kwon Y, Kim M, Kim Y, Jung HS and Jeoung D (2020) Exosomal MicroRNAs as Mediators of Cellular Interactions Between Cancer Cells and Macrophages. Front. Immunol. 11:1167. doi: 10.3389/fimmu.2020.01167
Received: 31 January 2020; Accepted: 12 May 2020;
Published: 11 June 2020.
Edited by:
Luminita Aurelia Stanciu, Imperial College London, United KingdomReviewed by:
Chaofeng Han, Second Military Medical University, ChinaParamananda Saikia, Cleveland Clinic, United States
Copyright © 2020 Kwon, Kim, Kim, Jung and Jeoung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dooil Jeoung, jeoungd@kangwon.ac.kr
 Yoojung Kwon
Yoojung Kwon Misun Kim1
Misun Kim1 Youngmi Kim
Youngmi Kim Hyun Suk Jung
Hyun Suk Jung Dooil Jeoung
Dooil Jeoung