- 1Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, TX, United States
- 2Center for RNA Interference and Non-Coding RNAs, The University of Texas MD Anderson Cancer Center, Houston, TX, United States
- 3Department of Leukemia, The University of Texas MD Anderson Cancer Center, Houston, TX, United States
MicroRNAs (miRNAs) are critical mediators of posttranscriptional regulation via their targeting of the imperfect antisense complementary regions of coding and non-coding transcripts. Recently, researchers have shown that miRNAs play roles in many aspects of regulation of immune cell function by targeting of inflammation-associated genes, including Toll-like receptors (TLRs). Besides this indirect regulatory role of miRNAs, they can also act as physiological ligands of specific TLRs and initiate the signaling cascade of immune response. In this review, we summarize the potential roles of miRNAs in regulation of TLR gene expression and TLR signaling, with a focus on the ability of miRNAs bind to TLRs.
Introduction
An efficient immune system is required for all multicellular organisms to detect and respond to pathogenic microorganisms or cells and/or tissue damage (1, 2). After discovery of the Toll-like receptor (TLR) family of pattern recognition receptors in the late 1990s, investigators showed that they recognize specific and distinct conserved endogenous and exogenous molecular patterns (3, 4). TLRs are crucial in recognition of microbial products, release of inflammatory mediators through the induction of transcription factors in immune response and inflammation and control of adaptive immune responses (5, 6). Emerging evidence has demonstrated that non-coding RNAs such as microRNAs (miRNAs) are involved in almost all known cellular processes, including innate, and adaptive immune responses, via modulation of gene expression (7–11). MiRNAs are secreted by several cell types, including tumor cells and macrophages within extracellular vesicles, such as exosomes and microvesicles; act as cell-to-cell communication vectors; and are taken up by recipient cells (12–17). Moreover, several miRNAs can bind to TLRs and initiate immune response by inducing immune and inflammatory gene expression. This review focuses on the inflammation-related miRNAs in the let-7 family, miR-21, miR-146b, and miR-155 and their involvement in TLR signaling pathways via regulation of TLRs and/or TLR signaling expression and binding to TLRs.
TLRs and TLR Signaling
TLRs are evolutionarily conserved molecules that initiate the signaling cascade of immune response against a wide variety of pathogens (18). Moreover, TLRs are type I integral membrane proteins consisting of 10–30 leucine-rich repeats in the N-terminal portion of TLRs that participate in ligand recognition and a cytoplasmic domain of the Toll/interleukin (IL)-1 receptor (TIR) in the C-terminal portion of TLRs that is responsible for activation of downstream signaling. Both pathogen-associated and damage-associated molecular patterns can be recognized by different TLRs and subsequently trigger signaling transduction pathways through adaptor molecules (19, 20). Damage-associated molecular patterns are endogenous molecules released from stressed or dying cells. Depending on the type of cells and tissues damaged, they can be classified as protein damage-associated molecular patterns, such as heat shock proteins, high-mobility group box 1 protein, or non-protein damage-associated molecular patterns, such RNA and DNA (3, 21, 22). Pathogen associated-molecular patterns are exogenous molecules derived from pathogens such as bacteria, fungi, parasites, and viruses and can be recognized by TLRs, leading to activation of the TLR signaling cascade, which regulates the expression of inflammation-related genes such as IL-1 receptor-associated kinase (IRAK1), tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6), and type I interferon (IFN) (3, 21, 22). Various TLRs are primarily or selectively expressed in specific cell types, including immune cells such as lymphocytes, dendritic cells (DCs), macrophages, and neutrophils and non-immune cells such as epithelial cells and fibroblasts (23–26). Recent studies identified that TLRs are also expressed in tumor cells and their microenvironments that composed cancer-associated fibroblasts, tumor-associated macrophages, marrow-derived suppressive cells, and regulatory T cells, adipocytes, and immune cells (23, 27). In mammals, the TLR protein family currently comprises 13 members (humans, TLR1-10; mice, TLR1-9, and TLR11-13), with the TLRs in humans and mice having some functional differences (28–31). Based on the subcellular localization of TLRs, they can be broadly divided into two subgroups. Those in the first group, including TLR1, TLR2, TLR4, TLR5, TLR6, and TLR10, are expressed on the surface of cells and recognize microbially derived ligands. TLRs in the second group, including TLR3, TLR7, TLR8, and TLR9, are expressed intracellularly in vesicles such as endosomes and lysosomes and recognize microbial nucleic acids (32, 33). In contrast, TLR3 can be localized both on cell surfaces and in intracellular vesicles (34).
Upon activation of TLR signaling transduction pathways, TLRs interact with several TIR-containing intracellular adaptor molecules, including myeloid differentiation primary response gene 88 (MYD88), sterile alpha and TIR motif-containing protein 1, TIR domain-containing adaptor protein, TIR domain-containing adapter-inducing IFN-β (TRIF), TIR domain-containing adapter molecule 1 (TICAM1), and TICAM2, leading to transcription factor activation and ultimately causing the release of various proinflammatory cytokines, chemokines, and IFNs and activation of the adaptive immune system (35, 36). Depending on the adaptor protein recruited, TLR signaling can be activated via the MyD88-dependent pathway that leads to release of proinflammatory cytokines and a TRIF-dependent (MyD88-independent) pathway associated with production of IFN-β (37–40). TLR1, TLR2, TLR5, TLR6, TLR7, TLR8, and TLR9 signaling are activated by the MyD88-dependent pathway, which typically leads to activation of nuclear factor (NF)-κB, whereas TLR3 signaling is activated by the TRIF-dependent pathway. In contrast, TLR4 is activated by both pathways simultaneously (37–40).
The Effects of miRNAs on TLR Expression and Signaling
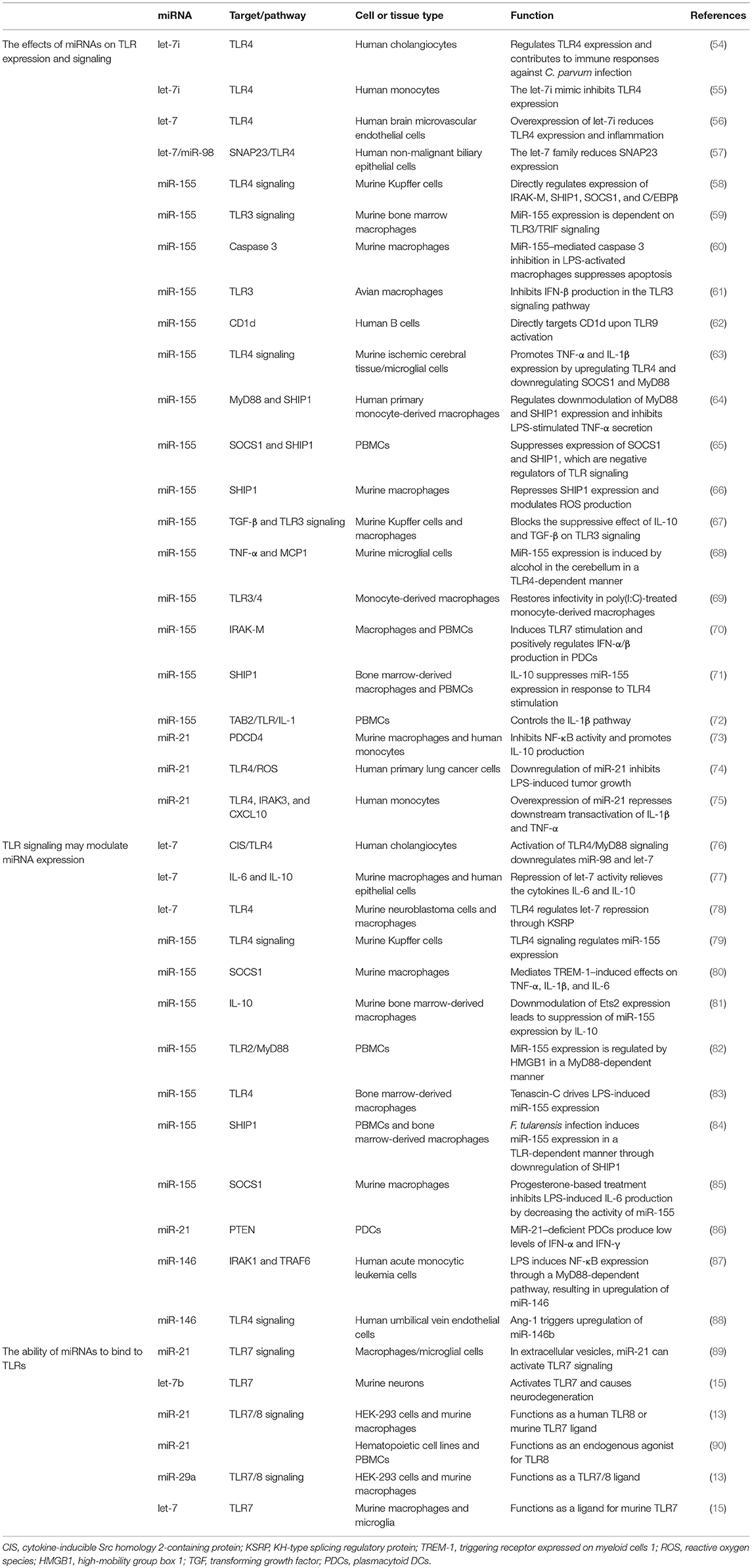
A growing number of reports have stated that specific epigenetic processes such as histone modifications, DNA methylation, and non-coding RNAs may regulate the transcriptional responses of TLRs (41–43). MiRNAs make up one of the well-characterized non-coding RNA families that generally bind to the 3' untranslated regions of their target messenger RNAs (mRNAs) to suppress translation or degradation of the mRNAs (44, 45). MiRNAs have a fundamental role in many biological processes, including apoptotic cell death, cell-cycle, tumorigenesis, and inflammation. Also, dysregulation of miRNAs has been associated with prognosis for and progression of multiple human diseases, including cancer (46–51). An increasing number of studies have demonstrated that several miRNAs, including miR-21, miR-146, miR-155, and let-7 family, target TLRs or proteins in TLR signaling pathways (Figure 1) that are involved in the regulation of various processes, such as inflammation, T-cell activation, cellular infiltration, and immunity development (52, 53). We have selectively listed recent miRNAs and their regulator roles on TLRs in Table 1.
Figure 1. Schematic of the regulatory mechanism of miRNAs in TLR signaling. Cell surface and cytoplasmic TLRs can be regulated by several miRNAs, including let-7 family members, miR-21, miR-146, and miR-155. First, miRNAs can bind directly to 3′untranslated region of TLRs or TLR-related genes, leading to modulated expression of TLRs through posttranscriptional regulation of TLR signaling. Second, miRNAs serve as physiological ligands of TLRs, such as miR-21, let-7 family members, and miR-29a, which can activate TLR signaling and stimulate the release of inflammatory cytokines and IFN genes in some cell types. Functional studies have demonstrated that these miRNAs may participate in activation of TLR signaling through regulating the NF-κB pathway and the production of inflammatory cytokines, which are shown here.
In one of the first studies demonstrating that miRNAs regulate immune response, researchers found that let-7i binds directly to TLR4 and regulates its expression in human cholangiocytes (54). In that study, infection of cultured cholangiocytes with Cryptosporidium parvum and lipopolysaccharide (LPS) stimulation of the cholangiocytes led to decreased let-7 expression via a MyD88/NF-κB–dependent mechanism, and low expression of let-7 was associated with upregulation of TLR4 in cholangiocytes. In concordance with this, upon C. parvum infection in non-malignant human biliary epithelial cells, inhibits expression of let-7 family miRNAs, including let-7i, let-7d, let-7f, let-7e, and miR-98, whereas induces the protein content of total SNAP23 and enhances phosphorylation of SNAP23. Activation of TLR4 signaling may induce SNAP23 protein expression by modulation of let-7-mediated gene regulation (57). Subsequently, investigators showed that let-7 and miR-98 target the 3' untranslated region of the cytokine-inducible Src homology 2-containing protein, resulting in translational repression of this protein in cholangiocytes, and that this may be associated with modulation of inflammatory responses in epithelial cells during microbial infection (76). In addition to this regulatory role of let-7 regarding TLR4 activation the inflammation-associated transcription factors NF-κB p50 and C/EBPβ can interact with the let-7 promoter region and repress transcription following microbial stimulus in human cholangiocytes (91).
MiR-155 has a well-characterized oncogenic role in tumorigenesis (92, 93), and aberrant expression and function of miR-155 have been associated with inflammation and affect immune cell functions at various levels by targeting inflammation-related genes, including TLRs (87, 94). This miRNA suppresses the expression of the adaptor protein TAB2 in the TLR/IL-1 signaling cascade, thereby regulating the feedback mechanism of IL-1β and other inflammatory cytokines produced during LPS-mediated DC activation (72). MiR-155 can also target suppressor of cytokine signaling 1 (SOCS1) and consequently modulate transcriptional expression of SOCS1 in LPS-activated Akt1−/− murine macrophages (95). In addition, miR-155 and miR-M4 (virally encoded functional orthologs of miR-155) may target coding sequences of the TLR3 gene and regulate TLR3 expression in macrophages (61). In line with this, inhibition of miR-155 by antagomirs markedly increased TLR3 expression, whereas ectopic overexpression of miR-155 decreased IFN-β production in primary chicken embryo fibroblast cells (61). In another study, researchers found not only that miR-155 regulates TLR expression but also that miR-155 and caspase 3 mRNA can interact with AGO2 in LPS-activated murine macrophages (60).
MiR-21 is one of the multifunctional miRNAs and is mainly characterized by overexpression in many inflamed states, including lung inflammation in LPS-treated mice, allergic airway inflammation, and osteoarthritis (73, 96–98). Moreover, researchers detected high miR-21 expression in extracellular vesicles during simian immunodeficiency virus pathogenesis and increased miR-21 expression in mouse hippocampal neurons associated with neurotoxicity due to neuronal TLR7 expression (89). Furthermore, investigators showed that miR-21 expression was induced in murine macrophages by treatment with LPS, whereas proinflammatory protein PDCD4 expression was downregulated in these cells due to induction of miR-21 expression via the adaptor proteins MyD88 and NF-κB (73). In another study, miR-21 expression decreased in patients with primary graft dysfunction after lung transplantation, and incubation of human monocytes with bronchoalveolar lavage fluid obtained from patients with primary graft dysfunction induced miR-21 expression, suggesting that dysregulation of miR-21expression is a novel regulator of TLR signaling during development of lung injury (75). Another study demonstrated that activation of TLR4 by treatment with LPS induced miR-21expression in primary human lung cancer cells and reactive oxygen species production by these cells (74). A more recent study demonstrated that miR-21 was upregulated in plasmacytoid DCs and that miR-21 deficiency significantly impaired production of IFN-γ and IFN-α in response to HSV-1 infection through targeting of the phosphoinositide 3-kinase/Akt/mammalian target of rapamycin signaling pathway in miR-21–knockout mice (86).
Importantly, an expression signature analysis of 200 miRNAs demonstrated that miR-146a/b, miR-132, and miR-155 were highly expressed in the acute monocytic leukemia cell line THP-1 after treatment with LPS as well as other microbial components and proinflammatory mediators (87). This finding suggests that miR-146 directly targets IRAK1 and TRAF6, which are key adapter molecules in the TLR4/NF-κB pathway (87). Furthermore, researchers found that miR-146 was significantly upregulated in hepatic stellate cells in mice infected with Schistosoma japonicum (99), is a negative regulator of NF-κB signaling in hepatic stellate cells, and acts by targeting TRAF6. Moreover, ectopic overexpression of the miR-146b-5p mimic significantly attenuated LPS-induced inflammatory responses and IRAK1 and TRAF6 expression in human umbilical vein endothelial cells (88).
Furthermore, miR-195 can regulate TLR2 expression through an indirect mechanism, as TLR2 expression was significantly reduced in miR-195–transfected THP-1 macrophages polarized toward the M1 phenotype (100). Treatment with LPS, synthetic lipid A, IL-2, IL-15, IL-1β, IFN-γ, and TNF-α similarly induced TLR2 gene expression in murine macrophages (101, 102), and acts is a key player in inflammation and atherosclerosis progression (103). Besides the role of cellular miRNAs in TLR signaling, authors recently reported that multiple viral miRNAs can activate production of proinflammatory mediators. For instance, Kaposi sarcoma herpes virus miRNAs such as miR-K-10b and miR-K12-12* are involved in sepsis as agonists of TLR8 through secretion of IL-6 and IL-10 (104, 105). Furthermore, Epstein-Barr virus miRNAs such as BHRF1-1 are expressed at higher levels in patients with chronic lymphocytic leukemia than in healthy individuals, and these viral miRNAs can serve as prognostic biomarkers for cancer (106, 107). Overall, these studies highlight miRNAs as central drivers of the TLR expression through transcriptional regulation of them, as indicated in Table 1.
How TLR Signaling may Modulate miRNA Expression
Initiation of the signaling cascade of immune response induced by TLR signaling can drive transcription of miRNAs during infection and inflammation. This is demonstrated by the fact that aberrant activation of TLR signaling after infection with microbial pathogens leads to dysregulation of miRNAs. Researchers have shown that infection of human peripheral blood monocytes (PBMCs) with Francisella tularensis, which is a highly pathogenic gram-negative bacterium that infects macrophages, induces expression of miR-155 in a TLR-dependent manner through downregulation of Src homology 2 domain-containing inositol 5-phosphatase 1 (SHIP1) (84). In concordance with this, authors reported significant differential expression of several miRNAs, including miR-155, after F. tularensis infection in primary human monocyte-derived macrophages and that F. tularensis infection leads to downmodulation of MyD88 and SHIP1 through an miR-155–dependent mechanism (64). Moreover, Leishmania RNA virus 1 was recognized by TLR3, and Leishmania infection induced miR-155 expression in murine bone-marrow macrophages (59). Concurrently, in that study, the pathogenesis of LRV1+ Leishmania infection decreased drastically in miR-155–deficient mice. In another study, let-7 and miR-98 were downregulated in murine macrophages upon Salmonella infection, whereas miR-155, miR-146a, and miR-21 were upregulated (77).
The immune-regulatory cytokine IL-10 may regulate transcription of miR-155 from the BIC gene in a signal transducer and activator of transcription 3-dependent manner in immortalized bone marrow-derived macrophages, and downmodulation of miR-155 expression leads to increased expression of SHIP1, which is one of the targets of miR-155 (71). Furthermore, investigators found that Ets2 is a critical transcription factor for the induction of miR-155 expression by LPS, and downmodulation of Ets2 leads to suppression of miR-155 by IL-10 (81). Another example miRNA signature is involved in human plasmacytoid DC activation (70), and miR-155 and its star form, miR-155*, were the most upregulated miRNAs in in primary human plasmacytoid DCs after TLR7 stimulation. MiR-155* induced IFN-α/β expression by suppressing IRAK-M expression, whereas miR-155 suppressed TAB2 expression (70). TLR3-dependent antiviral as well as inflammatory activity can be regulated by IL-10, transforming growth factor-β, and miR-155 in non-parenchymal liver cells in vitro (67). Other studies of macrophages demonstrated that chronic alcohol exposure induces TNF-α secretion through increased miR-155 expression both in vitro and in vivo (108), and miR-155 deficiency can protect against alcohol-induced liver injury, oxidative stress, steatosis, and inflammation in miR-155–knockout mice (79). In a similar study, after chronic ethanol feeding, miR-155 induced TNF-α and MCP1 expression in the cerebellum in a TLR4-dependent manner (68). Researchers have also observed aberrant expression of miR-155 in macrophages after stimulation by poly(I:C) and IFN-β and that miR-155 expression is induced by other TLR ligands through MyD88- or TRIF-dependent signaling pathways (109). However, investigators found TLR-independent upregulation of mature miR-155 in the murine macrophage cell line J774A and murine primary bone marrow-derived macrophages during Helicobacter pylori infection (110). Authors reported that treatment with progesterone augmented LPS- and poly(I:C)-induced miR-155 expression in macrophages through inhibition of NF-κB activation and led to downmodulation of IL-6 and IFN-β production in TLR-activated macrophages by increasing SOCS1 expression (85). A recent study identified that miR-155 expression increased in monocyte-derived macrophages upon TLR3/4 but not TLR7 stimulation and that inhibition of miR-155 expression partially restored infectivity in poly(I:C)-treated monocyte-derived macrophages (69). Furthermore, miR-155 in PBMCs of systemic lupus erythematosus patients was specifically upregulated by high-mobility group box 1 protein in a MyD88-dependent manner during induction of anti-double-stranded DNA antibody, which is the central pathogenic autoantibody involved in pathogenesis of systemic lupus erythematosus (82). Authors reported that cold exposure (32°C) induced miR-155 expression in human monocytes and that increased miR-155 expression was associated with suppressed SOCS1 and SHIP1expression (65). Additionally, miR-155 was upregulated in ischemic cerebral tissue and promoted TNF-α and IL-1β expression by upregulating TLR4 and downregulating SOCS1 and MyD88 (63). Negative regulator proteins for the TLR4 pathway (IRAK-M, SHIP1, and SOCS1) were upregulated in Kupffer cells isolated from miR-155–deficient mice (58). Researchers have shown that miR-155-3p and miR-155-5p (the two mature miRNAs processed from the precursor miR-155 transcript) were highly expressed in mice after treatment with LPS, whereas expression of both was decreased in the lungs of triggering receptor expressed on myeloid cells 1- (TREM-1) knockout mice. Deficiency of TREM-1 significantly inhibited neutrophils and proinflammatory chemokines and cytokines, particularly IL-1β, TNF-α, and IL-6 (80). Researchers also identified that protein kinase Akt1 activated by LPS positively regulates the expression of let-7e and miR-181c but negatively regulates that of miR-155 and miR-125b revealing that let-7e inhibits the expression of TLR4, whereas miR-155 inhibits the expression of SOCS1; both proteins TLR4 and SOCS1 are critical for TLR signaling after LPS stimulation (95). In another study, investigators showed that exposure to angiopoietin-1 significantly decreased IRAK1 and TRAF6 protein expression but did not affect TLR4, MYD88, IRAK4, or TAK1 expression in human umbilical vein endothelial cells (88).
The Ability of miRNAs to Bind to TLRs
MiRNAs may bind to TLRs and activate TLRs involved in intercellular communication in the tumor microenvironment (12, 13, 111). Authors reported that guanosine- and uridine-rich single-stranded RNA oligonucleotides derived from HIV-1 and influenza virus are recognized by murine TLR7 and human TLR8. Subsequently, activation of DCs and macrophages lead to the production of proinflammatory mediators such as IFN-α and cytokines (112, 113). Recently reported evidence demonstrated that extracellular vesicles such as exosomes and shed microvesicles isolated from different cell types may be novel mediators of cell-cell communication these vesicles can contain mRNAs, miRNAs, long non-coding RNAs, lipids, and DNA fragments. These active cargo molecules are packaged and released in exosome-derived cells and taken up by neighbor cells, where they are functionally active (114–116). MiRNAs are ubiquitously expressed in exosomes and are involved in modulation of the host immune response, expression of some activated molecules, enhanced tumor cell invasion, and mediation of intercellular communication (12, 117).
In 2012, researchers discovered that tumor-secreted exosomes in supernatants of lung cancer cells and exosomes loaded with miRNAs are physiological ligands for TLR7 and TLR8 (9, 12, 13). Expression of miR-21, miR-27b, and miR-29a was higher in exosomes derived from lung cancer cells than in those derived from HEK-293 cells (13). Upon co-culture of HEK-293 and RAW macrophages in vitro, labeled exosomes released from HEK-293 cells were incorporated with RAW macrophages, and miR-29a co-localized with TLR7 and TLR8 in the RAW macrophages (9, 12, 13). In addition, these investigators reported that cancer cell-derived exosomal miRNAs can bind to and activate TLR8 in macrophages and stimulate TLR8-mediated activation of NF-κB and NF-κB–mediated release of the proinflammatory and prometastatic cytokines IL-6 and TNF-α (13). Therefore, malignant cells release signals via exosomes loaded with miRNAs to the surrounding cells in their microenvironments that promote tumorigenesis and dissemination by different TLRs in humans (TLR8) and mice (TLR7) (13, 118). In our recent study, we demonstrated that exosomal miR-1246 released in abundance from ovarian cancer cells and miR-1246 transmit molecular signals to M2-type macrophages but not M0-type macrophages by shuttling exosomes (119).
Moreover, treatment with liposome-encapsulated miR-21 significantly induced human TLR8 expression in hematopoietic cell lines and PBMCs obtained from patients with systemic lupus erythematosus (90). Single-stranded RNAs containing twenty-nucleotide guanosine- and uridine-rich regions derived from human immunodeficiency virus and the influenza virus are physiological ligands for TLR7 and TLR8 that bind to TLR and activate TLR signaling (113, 120). Furthermore, the let-7 family contains a specific GU-rich motif GUUGUGU, which is present in the core of single-stranded RNA40 and responsible for murine TLR7 activation (15, 113, 120). Researchers recently discovered that let-7 may interact with TLR7 and activate TLR signaling in murine macrophages and microglia (15). They found that six nucleotide exchanges in the seed sequence of let-7b dramatically diminished induction of TNF-α expression in microglia and macrophages. In addition to let-7b, let-7a, −7c, and −7g induced a dose- and time-dependent cytokine response in wild-type immortalized bone marrow-derived macrophages but not TLR7-deficient cells (15). Consistent with these findings in macrophages, these investigators showed that neuronal loss was induced by let-7a, let-7c, let-7g, and miR-599 through TLR7. To further understand the role of let-7 in neurodegeneration in vivo, Lehmann et al. (15) showed that treatment with let-7b significantly induced marked axonal injury and neuronal loss in wild-type mice, whereas mutant let-7b rescued this phenotype. In contrast, TLR7 deficiency in Tlr7 knockout mice can protect against let-7b's induced neurotoxic effects. These findings suggest that endogenous miRNAs such as let-7b can be released during neuroinflammation and may cause further spread of central nervous system damage in patients with neurodegenerative disorders such as Alzheimer disease (15, 121).
Conclusion and Future Perspectives
MiRNAs are strongly implicated to have roles in the development and progression of inflammation-related diseases. Increasing numbers of studies have identified that miRNAs can act as physiological ligands for TLRs. The next challenges are to understand the complex mechanisms behind these integrated networks of interactions and, more importantly, determine whether therapeutic modulation of TLR-regulated and TLR-regulating miRNAs is beneficial for patients with cancer or inflammatory diseases. At present, MRG-106 therapy, is an oligonucleotide inhibitor of miR-155, for being tested in phase 1 clinical studies in Cutaneous T-cell Lymphoma, Mycosis Fungoides, Chronic Lymphocytic Leukemia, Diffuse Large B-Cell Lymphoma and Adult T-Cell Leukemia/Lymphoma (clinicaltrials.gov identifier NCT02580552). Therefore, researchers expect more preclinical and clinical studies regarding this new therapeutic avenue (93, 122, 123).
Author Contributions
RB, MB, and GC wrote the first draft of the manuscript and contributed to the writing of the manuscript.
Funding
GC is the Felix L. Haas Endowed Professor in basic science. Work in his laboratory is supported by NIH/NCATS grant UH3TR00943-01 through the NIH Common Fund, the Office of Strategic Coordination, NCI grants 1R01 CA182905-01 and 1R01CA222007-01A1, NIGMS grant 1R01GM122775-01, NIH U54 grant CA096297/CA096300, a 2016 University of Puerto Rico/The University of Texas MD Anderson Cancer Center Partnership for Excellence in Cancer Research Pilot Project, Team Department of Defense grant CA160445P1, an MD Anderson Cancer Center Chronic Lymphocytic Leukemia Moon Shot Flagship project, a 2017 Sister Institution Network Fund grant, and the estate of C.G. Johnson, Jr.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We also want to thank Donald R. Norwood, Scientific Editor Department of Scientific Publications, The University of Texas MD Anderson Cancer Center for critical reading and English editing of the review.
Abbreviations
AGO2, argonaute RISC catalytic component 2; CIS, cytokine-inducible Src homology 2-containing protein; DCs, dendritic cells; HMGB1, high-mobility group box 1; HSV-1, herpes simplex virus type 1; IFNs, interferons; IL, interleukin; IRAK1, interleukin 1 receptor associated kinase 1; IRF, interferon-regulatory factor; KSRP, KH-type splicing regulatory protein; LPS, lipopolysaccharide; MCP1, monocyte chemoattractant protein-1; miRNA, microRNA; mRNA, messenger RNA; MYD88, myeloid differentiation primary response protein; NF-κB, nuclear factor κB; PBMC, peripheral blood mononuclear cells; PDCD4, programmed cell death 4; PDCs, plasmacytoid dendritic cells; ROS, reactive oxygen species; SHIP1, inositol polyphosphate-5-phosphatase D; SNAP23, synaptosome associated protein 23; SOCS1, suppressor of cytokine signaling 1; TAB2, TGF-beta activated kinase 1 binding protein 2; TGF, transforming growth factor; TICAM, TIR domain-containing adapter molecule; TIR, Toll/interleukin-1 receptor; TLRs, Toll-like receptors; TNF, tumor necrosis factor.
References
1. Heneka MT, McManus RM, Latz E. Inflammasome signalling in brain function and neurodegenerative disease. Nat Rev Neurosci. (2018) 19:610–21. doi: 10.1038/s41583-018-0055-7
2. Kotas ME, Medzhitov R. Homeostasis, inflammation, and disease susceptibility. Cell. (2015) 160:816–27. doi: 10.1016/j.cell.2015.02.010
3. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. (2010) 11:373–84. doi: 10.1038/ni.1863
4. Miyake Y, Yamasaki S. Sensing necrotic cells. Adv Exp Med Biol. (2012) 738:144–52. doi: 10.1007/978-1-4614-1680-7_9
5. Carpenter S, O'Neill LA. How important are Toll-like receptors for antimicrobial responses? Cell Microbiol. (2007) 9:1891–901. doi: 10.1111/j.1462-5822.2007.00965.x
6. Kanzler H, Barrat FJ, Hessel EM, Coffman RL. Therapeutic targeting of innate immunity with Toll-like receptor agonists and antagonists. Nat Med. (2007) 13:552–9. doi: 10.1038/nm1589
7. Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. (2004) 101:11755–60. doi: 10.1073/pnas.0404432101
8. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. (2014) 9:287–314. doi: 10.1146/annurev-pathol-012513-104715
9. Fabbri M, Paone A, Calore F, Galli R, Croce CM. A new role for microRNAs, as ligands of Toll-like receptors. RNA Biol. (2013) 10:169–74. doi: 10.4161/rna.23144
10. Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol. (2016) 16:279–94. doi: 10.1038/nri.2016.40
11. van Leva E, Olson EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat Rev Drug Discov. (2012) 11:860–72. doi: 10.1038/nrd3864
12. Bayraktar R, Van Roosbroeck K, Calin GA. Cell-to-cell communication: microRNAs as hormones. Mol Oncol. (2017) 11:1673–86. doi: 10.1002/1878-0261.12144
13. Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. (2012) 109:E2110–6. doi: 10.1073/pnas.1209414109
14. Gonzalez-Villasana V, Rashed MH, Gonzalez-Cantú Y, Bayraktar R, Menchaca-Arredondo JL, Vazquez-Guillen JM, et al. Presence of circulating miR-145, miR-155, and miR-382 in exosomes isolated from serum of breast cancer patients and healthy donors. Dis Markers. (2019) 2019:6852917. doi: 10.1155/2019/6852917
15. Lehmann SM, Krüger C, Park B, Derkow K, Rosenberger K, Baumgart J, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. (2012) 15:827–35. doi: 10.1038/nn.3113
16. Rashed MH, Kanlikilicer P, Rodriguez-Aguayo C, Pichler M, Bayraktar R, Bayraktar E, et al. Exosomal miR-940 maintains SRC-mediated oncogenic activity in cancer cells: a possible role for exosomal disposal of tumor suppressor miRNAs. Oncotarget. (2017) 8:20145–64. doi: 10.18632/oncotarget.15525
17. Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, Lyle R, et al. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. (2014) 8:1432–46. doi: 10.1016/j.celrep.2014.07.035
18. Takeda K, Akira S. TLR signaling pathways. Semin Immunol. (2004) 16:3–9. doi: 10.1016/j.smim.2003.10.003
19. Bell JK, Mullen GE, Leifer CA, Mazzoni A, Davies DR, Segal DM. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. (2003) 24:528–33. doi: 10.1016/S1471-4906(03)00242-4
20. Matsushima N, Tanaka T, Enkhbayar P, Mikami T, Taga M, Yamada K, et al. Comparative sequence analysis of leucine-rich repeats. (LRRs) within vertebrate toll-like receptors. BMC Genomics. (2007) 8:124. doi: 10.1186/1471-2164-8-124
21. Fischer M, Ehlers M. Toll-like receptors in autoimmunity. Ann N Y Acad Sci. (2008) 1143:21–34. doi: 10.1196/annals.1443.012
22. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. (2010) 140:805–20. doi: 10.1016/j.cell.2010.01.022
23. Cen X, Liu S, Cheng K. The role of toll-like receptor in inflammation and tumor immunity. Front Pharmacol. (2018) 9:878. doi: 10.3389/fphar.2018.00878
24. González-Reyes S, Fernández JM, González LO, Aguirre A, Suárez A, González JM, et al. Study of TLR3, TLR4, and TLR9 in prostate carcinomas and their association with biochemical recurrence. Cancer Immunol Immunother. (2011) 60:217–26. doi: 10.1007/s00262-010-0931-0
25. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. (2010) 140:883–99. doi: 10.1016/j.cell.2010.01.025
26. Sato Y, Goto Y, Narita N, Hoon DS. Cancer cells expressing toll-like receptors and the tumor microenvironment. Cancer Microenviron. (2009) 2 (Suppl. 1):205–14. doi: 10.1007/s12307-009-0022-y
27. González-Reyes S, Marín L, González L, González LO, del Casar JM, Lamelas ML, et al. Study of TLR3, TLR4 and TLR9 in breast carcinomas and their association with metastasis. BMC Cancer. (2010) 10:665. doi: 10.1186/1471-2407-10-665
28. Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. (2011) 34:637–50. doi: 10.1016/j.immuni.2011.05.006
29. Lester SN, Li K. Toll-like receptors in antiviral innate immunity. J Mol Biol. (2014) 426:1246–64. doi: 10.1016/j.jmb.2013.11.024
30. Palsson-McDermott EM, O'Neill LA. Building an immune system from nine domains. Biochem Soc Trans. (2007) 35(Pt 6):1437–44. doi: 10.1042/BST0351437
31. Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. (2003) 21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126
32. McGettrick AF, O'Neill LA. Localisation and trafficking of Toll-like receptors: an important mode of regulation. Curr Opin Immunol. (2010) 22:20–7. doi: 10.1016/j.coi.2009.12.002
33. Oosenbrug T, van de Graaff MJ, Ressing ME, van Kasteren SI. Chemical tools for studying TLR signaling dynamics. Cell Chem Biol. (2017) 24:801–12. doi: 10.1016/j.chembiol.2017.05.022
34. Matsumoto M, Seya T. TLR3: interferon induction by double-stranded RNA including poly(I:C) Adv Drug Deliv Rev. (2008) 60:805–12. doi: 10.1016/j.addr.2007.11.005
35. Carty M, Goodbody R, Schröder M, Stack J, Moynagh PN, Bowie AG. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol. (2006) 7:1074–81. doi: 10.1038/ni1382
36. Kenny EF, O'Neill LA. Signalling adaptors used by Toll-like receptors: an update. Cytokine. (2008) 43:342–9. doi: 10.1016/j.cyto.2008.07.010
37. Barbalat R, Lau L, Locksley RM, Barton GM. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol. (2009) 10:1200–7. doi: 10.1038/ni.1792
38. Faure E, Equils O, Sieling PA, Thomas L, Zhang FX, Kirschning CJ, et al. Bacterial lipopolysaccharide activates NF-kappaB through toll-like receptor 4. (TLR-4) in cultured human dermal endothelial cells. Differential expression of TLR-4 and TLR-2 in endothelial cells. J Biol Chem. (2000) 275:11058–63. doi: 10.1074/jbc.275.15.11058
39. Janeway CA Jr, Medzhitov R Innate immune recognition. Annu Rev Immunol. (2002) 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359
40. Jin MS, Lee JO. Structures of the toll-like receptor family and its ligand complexes. Immunity. (2008) 29:182–91. doi: 10.1016/j.immuni.2008.07.007
41. Liew FY, Xu D, Brint EK, O'Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. (2005) 5:446–58. doi: 10.1038/nri1630
42. Liu Y, Yin H, Zhao M, Lu Q. TLR2 and TLR4 in autoimmune diseases: a comprehensive review. Clin Rev Allergy Immunol. (2014) 47:136–47. doi: 10.1007/s12016-013-8402-y
43. Qian C, Cao X. Regulation of Toll-like receptor signaling pathways in innate immune responses. Ann N Y Acad Sci. (2013) 1283:67–74. doi: 10.1111/j.1749-6632.2012.06786.x
44. Bayraktar R, Ivan C, Bayraktar E, Kanlikilicer P, Kabil NN, Kahraman N, et al. Dual suppressive effect of miR-34a on the FOXM1/eEF2-kinase axis regulates triple-negative breast cancer growth and invasion. Clin Cancer Res. (2018) 24:4225–41. doi: 10.1158/1078-0432.CCR-17-1959
45. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. (2004) 101:2999–3004. doi: 10.1073/pnas.0307323101
46. Bayraktar R, Pichler M, Kanlikilicer P, Ivan C, Bayraktar E, Kahraman N, et al. MicroRNA 603 acts as a tumor suppressor and inhibits triple-negative breast cancer tumorigenesis by targeting elongation factor 2 kinase. Oncotarget. (2017) 8:11641–58. doi: 10.18632/oncotarget.14264
47. Bullrich F, Fujii H, Calin G, Mabuchi H, Negrini M, Pekarsky Y, et al. Characterization of the 13q14 tumor suppressor locus in CLL: identification of ALT1, an alternative splice variant of the LEU2 gene. Cancer Res. (2001) 61:6640–8. Available online at: http://cancerres.aacrjournals.org/content/61/18/6640.full-text.pdf
48. Fabbri M, Garzon R, Andreeff M, Kantarjian HM, Garcia-Manero G, Calin GA. MicroRNAs and noncoding RNAs in hematological malignancies: molecular, clinical and therapeutic implications. Leukemia. (2008) 22:1095–105. doi: 10.1038/leu.2008.30
49. Kanlikilicer P, Rashed MH, Bayraktar R, Mitra R, Ivan C, Aslan B, et al. Ubiquitous release of exosomal tumor suppressor miR-6126 from ovarian cancer cells. Cancer Res. (2016) 76:7194–207. doi: 10.1158/0008-5472.CAN-16-0714
50. Munker R, Calin GA. MicroRNA profiling in cancer. Clin Sci. (2011) 121:141–58. doi: 10.1042/CS20110005
51. Sevignani C, Calin GA, Nnadi SC, Shimizu M, Davuluri RV, Hyslop T, et al. MicroRNA genes are frequently located near mouse cancer susceptibility loci. Proc Natl Acad Sci USA. (2007) 104:8017–22. doi: 10.1073/pnas.0702177104
52. O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. (2010) 10:111–22. doi: 10.1038/nri2708
53. O'Neill LA, Sheedy FJ, McCoy CE MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol. (2011) 11:163–75. doi: 10.1038/nri2957
54. Chen XM, Splinter PL, O'Hara SP, LaRusso NF. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. (2007) 282:28929–38. doi: 10.1074/jbc.M702633200
55. Satoh M, Tabuchi T, Minami Y, Takahashi Y, Itoh T, Nakamura M. Expression of let-7i is associated with Toll-like receptor 4 signal in coronary artery disease: effect of statins on let-7i and Toll-like receptor 4 signal. Immunobiology. (2012) 217:533–9. doi: 10.1016/j.imbio.2011.08.005
56. Xiang W, Tian C, Peng S, Zhou L, Pan S, Deng Z. Let-7i attenuates human brain microvascular endothelial cell damage in oxygen glucose deprivation model by decreasing toll-like receptor 4 expression. Biochem Biophys Res Commun. (2017) 493:788–93. doi: 10.1016/j.bbrc.2017.08.093
57. Hu G, Gong AY, Roth AL, Huang BQ, Ward HD, Zhu G, et al. Release of luminal exosomes contributes to TLR4-mediated epithelial antimicrobial defense. PLoS Pathog. (2013) 9:e1003261. doi: 10.1371/journal.ppat.1003261
58. Bala S, Csak T, Kodys K, Catalano D, Ambade A, Furi I, et al. Alcohol-induced miR-155 and HDAC11 inhibit negative regulators of the TLR4 pathway and lead to increased LPS responsiveness of Kupffer cells in alcoholic liver disease. J Leukoc Biol. (2017) 102:487–98. doi: 10.1189/jlb.3A0716-310R
59. Eren RO, Reverte M, Rossi M, Hartley MA, Castiglioni P, Prevel F, et al. Mammalian innate immune response to a leishmania-resident RNA virus increases macrophage survival to promote parasite persistence. Cell Host Microbe. (2016) 20:318–28. doi: 10.1016/j.chom.2016.08.001
60. De Santis R, Liepelt A, Mossanen JC, Dueck A, Simons N, Mohs A, et al. miR-155 targets Caspase-3 mRNA in activated macrophages. RNA Biol. (2016) 13:43–58. doi: 10.1080/15476286.2015.1109768
61. Hu X, Ye J, Qin A, Zou H, Shao H, Qian K. Both MicroRNA-155 and virus-encoded MiR-155 ortholog regulate TLR3 expression. PLoS ONE. (2015) 10:e0126012. doi: 10.1371/journal.pone.0126012
62. Liu F, Fan H, Ren D, Dong G, Hu E, Ji J, et al. TLR9-induced miR-155 and Ets-1 decrease expression of CD1d on B cells in SLE. Eur J Immunol. (2015) 45:1934–45. doi: 10.1002/eji.201445286
63. Wen Y, Zhang X, Dong L, Zhao J, Zhang C, Zhu C. Acetylbritannilactone modulates MicroRNA-155-mediated inflammatory response in ischemic cerebral tissues. Mol Med. (2015) 21:197–209. doi: 10.2119/molmed.2014.00199
64. Meyre D, Froguel P, Horber FF, Kral JG, Bandyopadhyay S, Long ME, Allen LA Differential expression of microRNAs in Francisella tularensis-infected human macrophages: miR-155-dependent downregulation of MyD88 inhibits the inflammatory response. PLoS ONE. (2014) 9:e109525. doi: 10.1371/journal.pone.0109525
65. Billeter AT, Hellmann J, Roberts H, Druen D, Gardner SA, Sarojini H, et al. MicroRNA-155 potentiates the inflammatory response in hypothermia by suppressing IL-10 production. FASEB J. (2014) 28:5322–36. doi: 10.1096/fj.14-258335
66. Wang J, Wu M, Wen J, Yang K, Li M, Zhan X, et al. MicroRNA-155 induction by Mycobacterium bovis BCG enhances ROS production through targeting SHIP1. Mol Immunol. (2014) 62:29–36. doi: 10.1016/j.molimm.2014.05.012
67. Jiang M, Broering R, Trippler M, Wu J, Zhang E, Zhang X, et al. MicroRNA-155 controls Toll-like receptor 3- and hepatitis C virus-induced immune responses in the liver. J Viral Hepat. (2014) 21:99–110. doi: 10.1111/jvh.12126
68. Lippai D, Bala S, Csak T, Kurt-Jones EA, Szabo G. Chronic alcohol-induced microRNA-155 contributes to neuroinflammation in a TLR4-dependent manner in mice. PLoS ONE. (2013) 8:e70945. doi: 10.1371/journal.pone.0070945
69. Swaminathan G, Rossi F, Sierra LJ, Gupta A, Navas-Martín S, Martín-García J. A role for microRNA-155 modulation in the anti-HIV-1 effects of Toll-like receptor 3 stimulation in macrophages. PLoS Pathog. (2012) 8:e1002937. doi: 10.1371/journal.ppat.1002937
70. Zhou H, Huang X, Cui H, Luo X, Tang Y, Chen S, et al. miR-155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells. Blood. (2010) 116:5885–94. doi: 10.1182/blood-2010-04-280156
71. McCoy CE, Sheedy FJ, Qualls JE, Doyle SL, Quinn SR, Murray PJ, et al. IL-10 inhibits miR-155 induction by toll-like receptors. J Biol Chem. (2010) 285:20492–8. doi: 10.1074/jbc.M110.102111
72. Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, et al. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci USA. (2009) 106:2735–40. doi: 10.1073/pnas.0811073106
73. Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. (2010) 11:141–7. doi: 10.1038/ni.1828
74. Zhang X, Wang C, Shan S, Liu X, Jiang Z, Ren T. TLR4/ROS/miRNA-21 pathway underlies lipopolysaccharide instructed primary tumor outgrowth in lung cancer patients. Oncotarget. (2016) 7:42172–82. doi: 10.18632/oncotarget.9902
75. Xu Z, Sharma M, Gelman A, Hachem R, Mohanakumar T. Significant role for microRNA-21 affecting toll-like receptor pathway in primary graft dysfunction after human lung transplantation. J Heart Lung Transplant. (2017) 36:331–9. doi: 10.1016/j.healun.2016.08.028
76. Hu G, Zhou R, Liu J, Gong AY, Eischeid AN, Dittman JW, et al. MicroRNA-98 and let-7 confer cholangiocyte expression of cytokine-inducible Src homology 2-containing protein in response to microbial challenge. J Immunol. (2009) 183:1617–24. doi: 10.4049/jimmunol.0804362
77. Schulte LN, Eulalio A, Mollenkopf HJ, Reinhardt R, Vogel J. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. (2011) 30:1977–89. doi: 10.1038/emboj.2011.94
78. Mueller M, Zhou J, Yang L, Gao Y, Wu F, Schoeberlein A, et al. PreImplantation factor promotes neuroprotection by targeting microRNA let-7. Proc Natl Acad Sci USA. (2014) 111:13882–7. doi: 10.1073/pnas.1411674111
79. Bala S, Csak T, Saha B, Zatsiorsky J, Kodys K, Catalano D, et al. The pro-inflammatory effects of miR-155 promote liver fibrosis and alcohol-induced steatohepatitis. J Hepatol. (2016) 64:1378–87. doi: 10.1016/j.jhep.2016.01.035
80. Yuan Z, Syed M, Panchal D, Joo M, Bedi C, Lim S, et al. TREM-1-accentuated lung injury via miR-155 is inhibited by LP17 nanomedicine. Am J Physiol Lung Cell Mol Physiol. (2016) 310:L426–38. doi: 10.1152/ajplung.00195.2015
81. Quinn SR, Mangan NE, Caffrey BE, Gantier MP, Williams BR, Hertzog PJ, et al. The role of Ets2 transcription factor in the induction of microRNA-155. (miR-155) by lipopolysaccharide and its targeting by interleukin-10. J Biol Chem. (2014) 289:4316–25. doi: 10.1074/jbc.M113.522730
82. Wen Z, Xu L, Chen X, Xu W, Yin Z, Gao X, et al. Autoantibody induction by DNA-containing immune complexes requires HMGB1 with the TLR2/microRNA-155 pathway. J Immunol. (2013) 190:5411–22. doi: 10.4049/jimmunol.1203301
83. Piccinini AM, Midwood KS, Piccinini AM, Midwood KS Endogenous control of immunity against infection: tenascin-C regulates TLR4-mediated inflammation via microRNA-155. Cell Rep. (2012) 2:914–26. doi: 10.1016/j.celrep.2012.09.005
84. Cremer TJ, Ravneberg DH, Clay CD, Piper-Hunter MG, Marsh CB, Elton TS, et al. MiR-155 induction by F. novicida but not the virulent F. tularensis results in SHIP down-regulation and enhanced pro-inflammatory cytokine response. PLoS ONE. (2009) 4:e8508. doi: 10.1371/journal.pone.0008508
85. Sun Y, Cai J, Ma F, Lü P, Huang H, Zhou J. miR-155 mediates suppressive effect of progesterone on TLR3, TLR4-triggered immune response. Immunol Lett. (2012) 146:25–30. doi: 10.1016/j.imlet.2012.04.007
86. Liu F, Liu C, Hu X, Shang Y, Wu L. MicroRNA-21: a positive regulator for optimal production of type i and type iii interferon by plasmacytoid dendritic cells. Front Immunol. (2017) 8:947. doi: 10.3389/fimmu.2017.00947
87. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. (2006) 103:12481–6. doi: 10.1073/pnas.0605298103
88. Echavarria R, Mayaki D, Neel JC, Harel S, Sanchez V, Hussain SN. Angiopoietin-1 inhibits toll-like receptor 4 signalling in cultured endothelial cells: role of miR-146b-5p. Cardiovasc Res. (2015) 106:465–77. doi: 10.1093/cvr/cvv120
89. Yelamanchili SV, Lamberty BG, Rennard DA, Morsey BM, Hochfelder CG, Meays BM, et al. MiR-21 in extracellular vesicles leads to neurotoxicity via TLR7 signaling in SIV neurological disease. PLoS Pathog. (2015) 11:e1005032. doi: 10.1371/journal.ppat.1005032
90. Young NA, Valiente GR, Hampton JM, Wu LC, Burd CJ, Willis WL, et al. Estrogen-regulated STAT1 activation promotes TLR8 expression to facilitate signaling via microRNA-21 in systemic lupus erythematosus. Clin Immunol. (2017) 176:12–22. doi: 10.1016/j.clim.2016.12.005
91. O'Hara SP, Splinter PL, Gajdos GB, Trussoni CE, Fernandez-Zapico ME, Chen XM, et al. NFkappaB p50-CCAAT/enhancer-binding protein beta. (C/EBPbeta)-mediated transcriptional repression of microRNA let-7i following microbial infection. J Biol Chem. (2010) 285:216–25. doi: 10.1074/jbc.M109.041640
92. Ferrajoli A, Shanafelt TD, Ivan C, Shimizu M, Rabe KG, Nouraee N, et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood. (2013) 122:1891–9. doi: 10.1182/blood-2013-01-478222
93. Van Roosbroeck K, Fanini F, Setoyama T, Ivan C, Rodriguez-Aguayo C, Fuentes-Mattei E, et al. Combining Anti-Mir-155 with chemotherapy for the treatment of lung cancers. Clin Cancer Res. (2017) 23:2891–2904. doi: 10.1158/1078-0432.CCR-16-1025
94. Bayraktar R, Van Roosbroeck K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev. (2018) 37:33–44. doi: 10.1007/s10555-017-9724-7
95. Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. (2009) 31:220–31. doi: 10.1016/j.immuni.2009.06.024
96. Iliopoulos D, Malizos KN, Oikonomou P, Tsezou A. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS ONE. (2008) 3:e3740. doi: 10.1371/journal.pone.0003740
97. Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. (2009) 182:4994–5002. doi: 10.4049/jimmunol.0803560
98. Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics. (2007) 8:240. doi: 10.1186/1471-2164-8-240
99. Cai P, Piao X, Liu S, Hou N, Wang H, Chen Q. MicroRNA-gene expression network in murine liver during Schistosoma japonicum infection. PLoS ONE. (2013) 8:e67037. doi: 10.1371/journal.pone.0067037
100. Bras JP, Silva AM, Calin GA, Barbosa MA, Santos SG, Almeida MI. miR-195 inhibits macrophages pro-inflammatory profile and impacts the crosstalk with smooth muscle cells. PLoS ONE. (2017) 12:e0188530. doi: 10.1371/journal.pone.0188530
101. Liu Y, Wang Y, Yamakuchi M, Isowaki S, Nagata E, Kanmura Y, et al. Upregulation of toll-like receptor 2 gene expression in macrophage response to peptidoglycan and high concentration of lipopolysaccharide is involved in NF-kappa b activation. Infect Immun. (2001) 69:2788–96. doi: 10.1128/IAI.69.5.2788-2796.2001
102. Matsuguchi T, Musikacharoen T, Ogawa T, Yoshikai Y. Gene expressions of Toll-like receptor 2, but not Toll-like receptor 4, is induced by LPS and inflammatory cytokines in mouse macrophages. J Immunol. (2000) 165:5767–72. doi: 10.4049/jimmunol.165.10.5767
103. Chávez-Sánchez L, Garza-Reyes MG, Espinosa-Luna JE, Chávez-Rueda K, Legorreta-Haquet MV, Blanco-Favela F. The role of TLR2, TLR4 and CD36 in macrophage activation and foam cell formation in response to oxLDL in humans. Hum Immunol. (2014) 75:322–9. doi: 10.1016/j.humimm.2014.01.012
104. Giza DE, Fuentes-Mattei E, Bullock MD, Tudor S, Goblirsch MJ, Fabbri M, et al. Cellular and viral microRNAs in sepsis: mechanisms of action and clinical applications. Cell Death Differ. (2016) 23:1906–18. doi: 10.1038/cdd.2016.94
105. Tudor S, Giza DE, Lin HY, Fabris L, Yoshiaki K, D'Abundo L, et al. Cellular and Kaposi's sarcoma-associated herpes virus microRNAs in sepsis and surgical trauma. Cell Death Dis. (2014) 5:e1559. doi: 10.1038/cddis.2014.515
106. Van Roosbroeck K, Bayraktar R, Calin S, Bloehdorn J, Dragomir MP, Okubo K, et al. miRNAs involvement in the pathogenesis of Richter's syndrome. Haematologica. (2019) 104:1004–15. doi: 10.3324/haematol.2018.203828
107. Van Roosbroeck K, Calin GA. When kissing. (disease) counts. Blood. (2016) 127:1947–8. doi: 10.1182/blood-2016-01-692087
108. Bala S, Marcos M, Kodys K, Csak T, Catalano D, Mandrekar P, et al. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor {alpha}. (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem. (2011) 286:1436–44. doi: 10.1074/jbc.M110.145870
109. O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. (2007) 104:1604–9. doi: 10.1073/pnas.0610731104
110. Koch M, Mollenkopf HJ, Klemm U, Meyer TF. Induction of microRNA-155 is TLR- and type IV secretion system-dependent in macrophages and inhibits DNA-damage induced apoptosis. Proc Natl Acad Sci USA. (2012) 109:E1153–62. doi: 10.1073/pnas.1116125109
111. Bosisio D, Gianello V, Salvi V, Sozzani S. Extracellular miRNAs as activators of innate immune receptors. Cancer Lett. (2019) 452:59–65. doi: 10.1016/j.canlet.2019.03.021
112. Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. (2004) 303:1529–31. doi: 10.1126/science.1093616
113. Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. (2004) 303:1526–9. doi: 10.1126/science.1093620
114. Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. (2005) 132:4653–62. doi: 10.1242/dev.02073
115. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. (2007) 9:654–9. doi: 10.1038/ncb1596
116. Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. (2005) 579:5911–22. doi: 10.1016/j.febslet.2005.07.070
117. Rashed HM, Bayraktar E, K Helal G, Abd-Ellah MF, Amero P, Chavez-Reyes A, et al. Exosomes: from garbage bins to promising therapeutic targets. Int J Mol Sci. (2017) 18:E538. doi: 10.3390/ijms18030538
118. Challagundla KB, Wise PM, Neviani P, Chava H, Murtadha M, Xu T, et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J Natl Cancer Inst. (2015) 107:djv135. doi: 10.1093/jnci/djv135
119. Kanlikilicer P, Bayraktar R, Denizli M, Rashed MH, Ivan C, Aslan B, et al. Exosomal miRNA confers chemo resistance via targeting Cav1/p-gp/M2-type macrophage axis in ovarian cancer. EBioMedicine. (2018) 38:100–12. doi: 10.1016/j.ebiom.2018.11.004
120. Forsbach A, Nemorin JG, Montino C, Müller C, Samulowitz U, Vicari AP, et al. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J Immunol. (2008) 180:3729–38. doi: 10.4049/jimmunol.180.6.3729
121. Paschon V, Takada SH, Ikebara JM, Sousa E, Raeisossadati R, Ulrich H, et al. Interplay between exosomes, microRNAs and toll-like receptors in brain disorders. Mol Neurobiol. (2016) 53:2016–28. doi: 10.1007/s12035-015-9142-1
122. Pichler M, Calin GA. MicroRNAs in cancer: from developmental genes in worms to their clinical application in patients. Br J Cancer. (2015) 113:569–73. doi: 10.1038/bjc.2015.253
Keywords: microRNAs, Toll-like receptors, inflammation, TLR, TLR ligands
Citation: Bayraktar R, Bertilaccio MTS and Calin GA (2019) The Interaction Between Two Worlds: MicroRNAs and Toll-Like Receptors. Front. Immunol. 10:1053. doi: 10.3389/fimmu.2019.01053
Received: 21 November 2018; Accepted: 24 April 2019;
Published: 14 May 2019.
Edited by:
Daniela Bosisio, University of Brescia, ItalyReviewed by:
Fausto Almeida, University of São Paulo, BrazilAlessandra Zingoni, Sapienza University of Rome, Italy
Copyright © 2019 Bayraktar, Bertilaccio and Calin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: George A. Calin, Gcalin@mdanderson.org
 Recep Bayraktar1
Recep Bayraktar1 Maria Teresa Sabrina Bertilaccio
Maria Teresa Sabrina Bertilaccio George A. Calin
George A. Calin