- 1Laboratory of Molecular Immunology and Immunotherapy, Blood Research Institute, Blood Center of Wisconsin, Milwaukee, WI, United States
- 2Department of Microbiology and Immunology, Medical College of Wisconsin, Milwaukee, WI, United States
- 3Department of Pediatrics, Medical College of Wisconsin, Milwaukee, WI, United States
- 4Department of Medicine, Medical College of Wisconsin, Milwaukee, WI, United States
- 5Center of Excellence in Prostate Cancer, Medical College of Wisconsin, Milwaukee, WI, United States
Natural killer (NK) cells are the predominant innate lymphocyte subsets that mediate anti-tumor and anti-viral responses, and therefore possess promising clinical utilization. NK cells do not express polymorphic clonotypic receptors and utilize inhibitory receptors (killer immunoglobulin-like receptor and Ly49) to develop, mature, and recognize “self” from “non-self.” The essential roles of common gamma cytokines such as interleukin (IL)-2, IL-7, and IL-15 in the commitment and development of NK cells are well established. However, the critical functions of pro-inflammatory cytokines IL-12, IL-18, IL-27, and IL-35 in the transcriptional-priming of NK cells are only starting to emerge. Recent studies have highlighted multiple shared characteristics between NK cells the adaptive immune lymphocytes. NK cells utilize unique signaling pathways that offer exclusive ways to genetically manipulate to improve their effector functions. Here, we summarize the recent advances made in the understanding of how NK cells develop, mature, and their potential translational use in the clinic.
Introduction
Experiments aimed at characterizing T cell-mediated cytotoxicity inadvertently uncovered the existence of a naturally occurring cytotoxic lymphocyte with intrinsic and innate anti-tumor properties (1). These original observations were made in the 1960s (2, 3) and, within 10 years, researchers began to explore a previously uncharacterized innate lymphocyte population known today as natural killer (NK) cells (4–7). As their name suggests, NK cells are “naturally” cytotoxic and, in contrast to cytotoxic T cells, do not require prior antigen exposure to mediate their anti-tumor effects (4, 7). NK cell activity was first observed in human peripheral blood mononuclear cells (8, 9) and rodent splenocytes (5, 6); however, these large granular lymphocytes are known to reside in multiple lymphoid and non-lymphoid tissues including the bone marrow (BM), lymph nodes (LNs), skin, gut, tonsils, liver, and lungs (10). In this review, we summarize the established and emerging themes of NK cells related to their development, maturation, effector functions such as cytokine production and anti-tumor cytotoxicity, role in the clearance of viral and bacterial infections, and the clinical utilization of donor-derived or genetically modified NK cells.
Development and Functional Maturation of NK Cells
Natural killer cells were initially thought to develop exclusively in the BM. However, recent evidence in humans and mice suggests that they can also develop and mature in secondary lymphoid tissues (SLTs) including tonsils, spleen, and LNs (11). The cellular progenitors and intermediate populations that give rise to NK cells are defined by the differential expression of lineage-specific surface markers (12). Although these markers are often different between humans and mice, the developmentally regulated expression of critical transcription factors, such as the T-box transcription factors T-bet and Eomesodermin, control NK cell-specific qualities in both species (13).
Natural killer cells represent 5–20% of circulating lymphocytes in humans (14). The percentages of NK cells among lymphocytes ranges between about 2–5% in the spleens and BMs of inbred laboratory mice (15) and about twice that number in wild-caught mice (16). They are distinguished by their unique functions and expression of surface antigens. NK cells lack the clonotypic T cell receptor (TCR) of T and NKT cells and its associated signal-transducing adaptor, CD3ε. In humans, subsets of NK cells express the activating Fc receptor, CD16 and most express CD56 [neural cell adhesion molecule (NCAM) or Leu-19] (17, 18). In C57BL/6 mice, NK cells are identified by the presence of NK1.1 (NKR-P1C) and NCR1 (NKp46/CD335), as well as CD49b (DX5, Integrin VLA-2α), are common NK cell markers in other mouse backgrounds (19, 20). NK cells are most similar to a group of lymphocytes known as innate lymphoid cells (ILCs) (21). ILCs are further categorized into three distinct groups and are present in both humans and mice (11, 21). NK cells are related to group 1 ILCs as both produce interferon-gamma (IFN-γ) and tumor necrosis factor (TNF)-α upon stimulation (22). However, unlike Group 1 ILCs, NK cells have cytolytic functions that resemble those of CD8+ cytotoxic T lymphocytes (22).
Developmental Stages of Murine and Human NK Cells
In mice, the NK cells develop in specialized BM niches (Figure 1). The hematopoietic niche is most often localized in the perivascular regions proximal to sinusoidal vessels. The multipotent self-renewing hematopoietic stem cells (HSCs) are regulated by an integrated cytokine milieu as part of the endocrine, autocrine, and paracrine signaling. HSCs contain transient self-renewing and long-term quiescent populations. HSCs give rise to all leukocytes and red blood cells. A branch of which constitutes the common lymphoid progenitor (CLP). CLPs give rise to Pro-B, Pre-T, innate lymphoid cells (ILCs), lymphoid tissue inducers, and CD122+ Pre-T/early NKP lineages. The cellular origin of NK cells in humans and mice can be traced back to oligopotent CLP (23). Expression of interleukin (IL)-7 receptor-alpha (IL-7Rα, CD127) in Lin−CD244+ cells mark the earliest step in the transition of CLPs into the lymphoid lineage. A subset of this early progenitor defined as pre-NK cell precursors (Pre-NKPs) expresses the IL-2 receptor β chain (CD122) to become NKPs (24) (Figure 2).
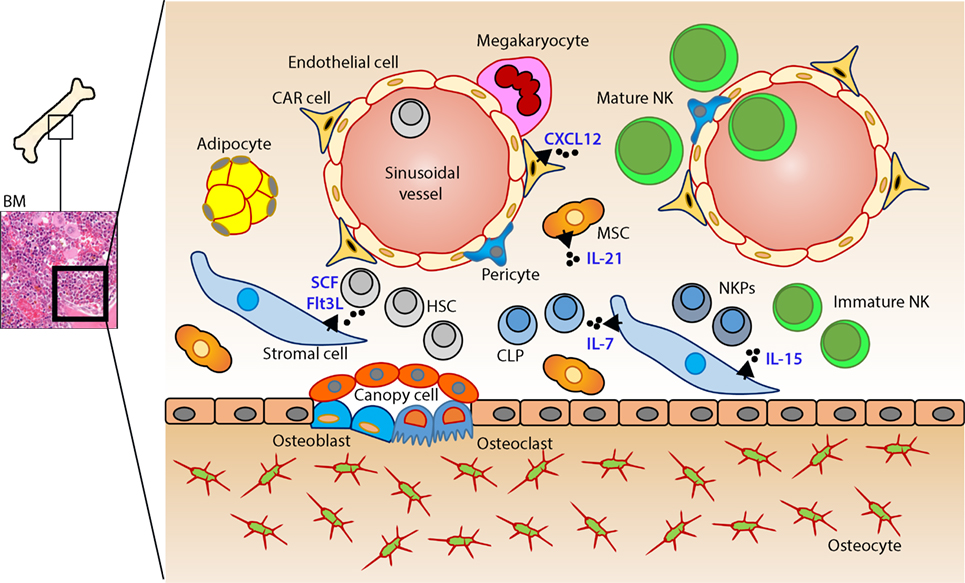
Figure 1. Murine bone marrow niche where natural killer (NK) cells develop. Quiescent hematopoietic stem cells (HSCs) from a hypoxic microenvironment, within the perivascular region proximal to sinusoidal vessels, are induced by hormonal and cytokine cues. Upon unique stimulations [such as stem cell factor (SCF); Fms-like tyrosine kinase-3 ligand (Flt3L)], the self-renewing multipotent HSCs commit to becoming common lymphoid progenitors (CLPs). Non-hematopoietic stromal cells [mesenchymal stromal cells (MSCs), fibroblastic reticular cells] that produce interleukin (IL)-7 or IL-15 play pleiotropic roles in programming CLPs into distinct lymphoid lineages including NK cell progenitors (NKPs). MSCs also produce another common gamma chain receptor (γcR)-binding cytokine, IL-21 that may help with the expansion of the NKPs. CXCL12-abundant reticular (CAR) cells generate CXCL12, which stimulates NKPs via CXCR4 to functionally mature the NKPs or immature NK cells (iNKs) into established Mature NK (mNK) cells subsets. mNK cells traffic to secondary lymphoid organs via the sinusoidal blood vessels. Other cell types, pericytes, megakaryocytes, adipocytes, canopy cells, osteoblasts, osteoclasts, and osteocytes help form the niche and other supporting systems.
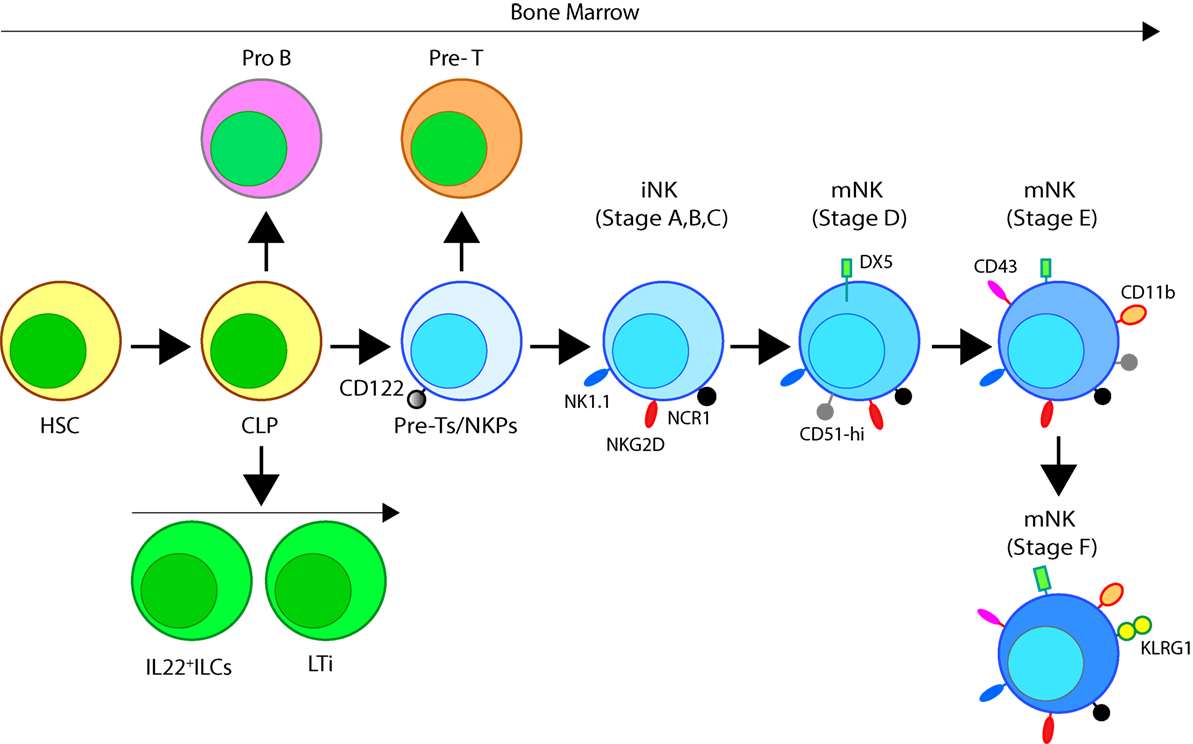
Figure 2. Developmental origin of murine natural killer (NK) cells in the bone marrow (BM). Murine NK cells develop in the BM. A subset of multipotent HSCs commits to becoming oligopotent common lymphoid progenitors (CLPs). CLPs give rise to Pro-B, Pre-T, innate lymphoid cells (ILCs), lymphoid tissue inducers, and CD122+ Pre-T/early NK cell progenitor (NKP) lineages. Expression of NKG2D by the CD122+ NKPs mark the earliest transition of NKPs into committed immature NK cells (iNK, Stage A). This is followed by the expression of NK1.1 and NCR1 (Stages B and C). Expression of CD51 (Integrin αV) and CD49b (DX5, Integrin VLA-2α) defines the initial stage of mature NK (mNK) cells. Expression of CD43 (Leukosialin), CD11b (Mac-1), and the acquisition of distinct sets of Ly49s define the terminal stage of mNK cells (Stage E). mNK cells migrate into secondary lymphoid organs following the expression of Killer cell Lectin-like Receptor G1 (KLRG1) (Stage F) at least in part by a subset. Additional functional classifications of mNK cells are made using CD27 and CD11b.
Expression of the activation receptor complex NKG2D/DNAX-activating protein of 10 kDa (DAP10) defines Stage A (Figure 3) of immature NK (iNK) population (25, 26). NKP maintenance and progression to the iNK cell stage requires the activation of transcription factors including an inhibitor of DNA binding 2 (Id2) (27–29) and E4-binding protein 4 (30, 31). By the iNK stage, NK cells express receptors including, NKG2A, DNAM-1 (CD226), NK1.1 (Stage B), and NCR1 (Stage C) as well as the cell adhesion molecules, L-selectin (CD62L) and Leukosialin (CD43) (32). Expression of CD51 (Integrin αV) and CD49b (DX5, Integrin VLA-2α) defines the initial stage (Stage D) of mature NK (mNK) cells. Terminally mNK cells are identified based on the expression of CD43 (Leukosialin) and CD11b (Mac-1). The acquisition of distinct sets of Ly49 receptors also define mNK cells (Stage E) that are functionally licensed (33). In C57BL/6 mice, these inhibitory or activating Ly49s include Ly49A, Ly49C/I, Ly49G or Ly49D, and Ly49H, respectively. mNK cells migrate into secondary lymphoid organs following the expression of Killer cell Lectin-like Receptor G1 (KLRG1) (Stage F) at least in part by a subset (10, 34). NK cells that have reached terminal maturation are fully functional; however, evidence suggests that their capabilities with regards to anti-tumor cytotoxicity and inflammatory cytokine production may not be acquired equally (35, 36).
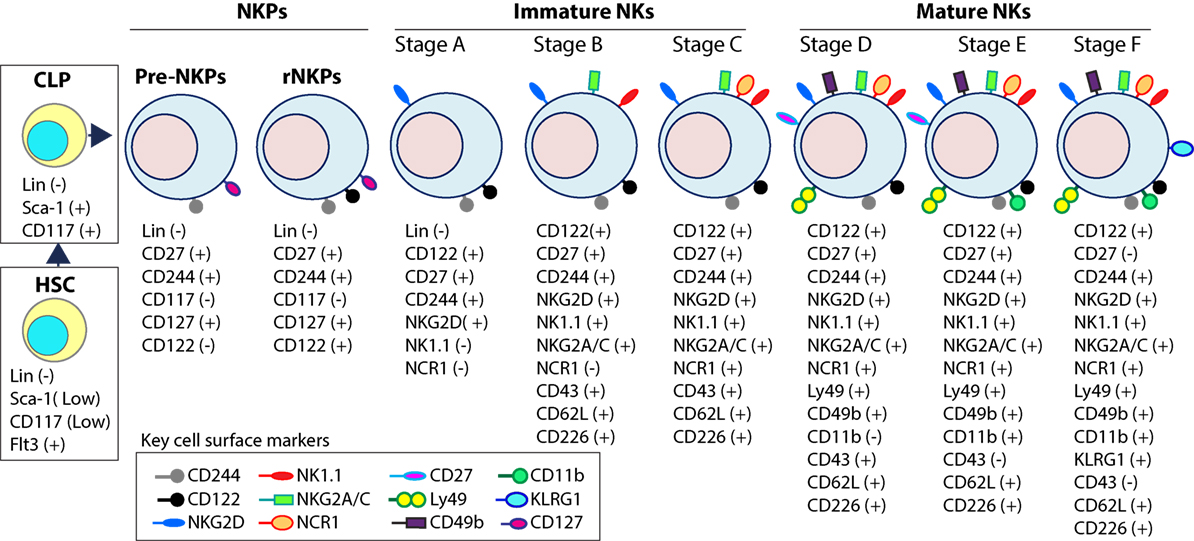
Figure 3. Distinct developmental stages of murine NK cell progenitors (NKPs), immature NK cells (iNKs), and mature NKs (mNKs). Lineage negative (Lin−) Sca+CD117+ hematopoietic stem cells (HSCs) differentiate into common lymphoid progenitors (CLPs) (Lin−ScaLowCD117LowFlt3+). Expression of IL-7 receptor-alpha (IL-7Rα) (CD127), CD27, and CD244 mark the full commitment of CLPs into pre-NK cell precursors (Pre-NKPs). Committed NKPs transition from Pre-NKPs to refined-NKPs (rNKPs) by expressing IL-2Rβ (CD122). Expression of NKG2D marks the conversion of rNKPs into iNK cells. Natural killer (NK) cells progressing through the iNK stages express NK1.1 and NKG2A/C followed by NCR1 (Stage A through C). Terminal maturation of iNK cells into mNK cells is defined by the acquisition of distinct sets of Ly49s that help to identify distinct subsets (Stage D). NK cells that have reached terminal maturation downregulate CD27 and express CD11b (Stage E) followed by Killer cell Lectin-like Receptor G1 (KLRG1) (Stage F) by a subset of matured NK cells.
Functional NK cell maturation can be defined by the differential surface expression of CD27 and CD11b (Mac-1) whereby NK cells develop consecutively through a three-stage program (37). NK cells begin expressing neither receptor, known as the double-negative population, and progress to CD27+CD11b− (Stages B, C, and D), double-positive (DP, Stages E), and the CD27−CD11b+ (Stage F) NK cells, which are considered the most mature (33, 37). Lack of signaling molecule PLC-g2 but not PLC-g1 significantly reduced the terminal maturation of NK cells (38). mNK cells express the activation receptor, CD49b (33), and acquire KLRG1, an inhibitory receptor and marker of terminal maturation (39, 40). Interestingly, DP NK cells have increased effector responses compared to CD27−CD11b+ NK cells, which suggests the acquisition of regulatory mechanisms during the NK cell maturation process (36).
Human NK cells have been shown to mature in the BM and secondary lymphoid organs such as LNs (11, 41). Lin−CD34+CD133+CD244+ HSCs differentiate into CD45RA+ lymphoid-primed multipotential progenitor in Stage 1 (LMPP, Figure 4). CD34 is a highly glycosylated cell membrane protein and a marker for stemness that facilitates the adhesion of stem cells to the extracellular matrix (42). CD133 is a glycoprotein known as Prominin-1 (43, 44) and CD244 (2B4) is a SLAM family member (45). By expressing CD38 (cyclic ADP ribose hydrolase) (46), CD7 (Ig family, co-stimulatory molecule) (47), CD10 (neutral endopeptidase) (48), and the cytokine receptor CD127 (IL-7Rα), LMPPs transition into CLPs with potential to make lineage commitments into Pro-B, Pre-T, NKPs, or other innate lymphoid cells (ILCs) (49). Expression of CD122 (IL-2Rβ) marks the irreversible fate decision of CLPs into NK lineage. The appearance of CD56 (NCAM) indicates a final transition of iNK into mNK cells. It is also suggested that iNK cells can directly give rise to CD56dim population (dotted arrow) that is yet to be validated (50) (Figure 4).
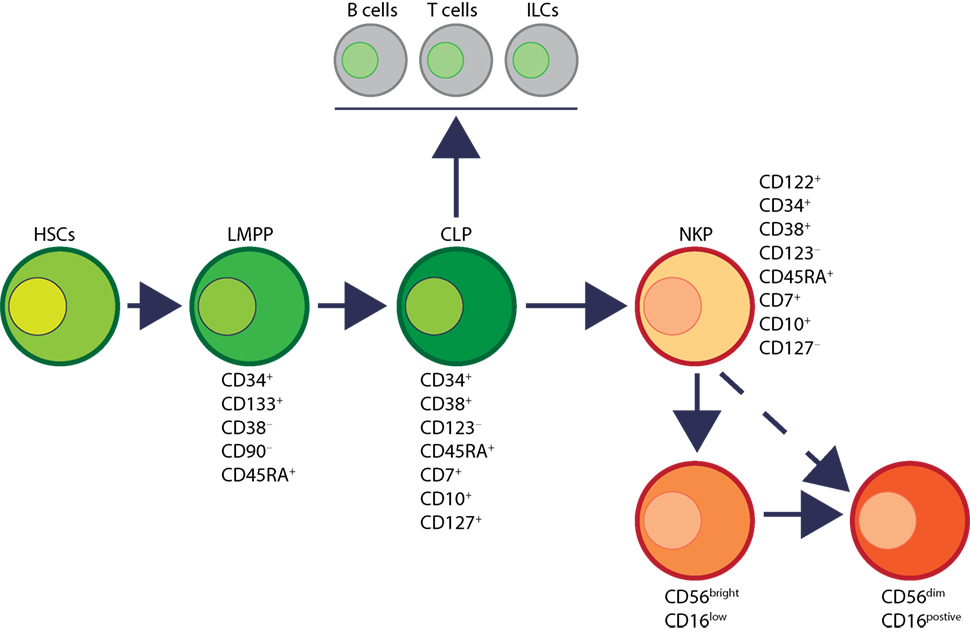
Figure 4. Developmental origin of human natural killer (NK) cells. In human, the primary organ where NK cells mature is still under active investigation. There is ample evidence that NK cells can mature from the lymph nodes (LNs). Lin−CD34+ hematopoietic stem cells (HSCs) differentiate into CD45RA+ lymphoid-primed multipotential progenitor (LMPP). By expressing CD38, CD7, CD10, and the cytokine receptor CD127 (IL-7 receptor-alpha), LMPPs transition into common lymphoid progenitors (CLPs) that have the potential to make lineage commitment into Pro-B, Pre-T, NK cell progenitors (NKPs), or other innate innate lymphoid cells. Expression of CD122 (IL-2Rβ) marks the irreversible fate decision of CLPs into NK lineage. The appearance of CD56 (neural cell adhesion molecule) indicates a final transition of immature NK cell (iNK) into mature NK cells. Most of the iNK cells transition into a minor CD56bright population (~5%) that convert into major CD56dim (>90%) population. It is also suggested that iNK cells can directly give rise to CD56dim population (dotted arrow) that is yet to be validated.
Distinct stages through which human NK cells develop are less understood compared to that of the murine counterparts (51). Recent work has helped to demarcate a total of six stages of human NK cell development (Figure 5) based on their both BM and LN development (11, 41). CD3ε−CD7+CD127+ cells mark the earliest stage of committed NKPs (Stage 2a). CD7, whose expression persists throughout development and in mNK cells is a cell membrane protein that recruits PI(3)K via a YEDM motif in its cytoplasmic tail (52). Although discrete subsets of CD7-expressing (low and high) CD8+ T cells (53) have been described, similar distinctions are yet to be identified in NK cells. Expression of IL-1R, a receptor for IL-1β defines Stage 2b. Expression of activation receptors including NKG2D (CD314, C-type lectin-like, KLRK1), CD335 (Natural cytotoxicity receptor, NCR1, NKp46), and CD337 (NCR3, NKp30) marks the transition of NK cells from Stage 2b to Stage 3. Human NKG2D uses only DAP10 adaptor protein, compared to mouse NKG2D that uses both DAP10 and DNAX-activating protein of 12 kDa (DAP12). NCR1 uses CD3ζ and FcεRγ while NCR3 utilizes CD3ζ as their adaptor complexes. Stage 4 of human NK cell development is sub-divided into two parts based on the expression of the activating receptor NKP80 (KLRF1, type II transmembrane protein) (54, 55). The primary distinction of NK cells in the Stage 4a is that they express abundant amounts of CD56 (CD56bright). These NK cells are NKP80− and express the maximal levels of NKG2D, CD335, CD337, inhibitory NKG2A [CD159a, contains two immunoreceptor-based tyrosine inhibitory motifs (ITIMs)] and CD161 (NK1.1, KLRB1, NKR-P1A). At Stage 4b, human NK cells become positive for NKP80 and maintain their CD56bright status.
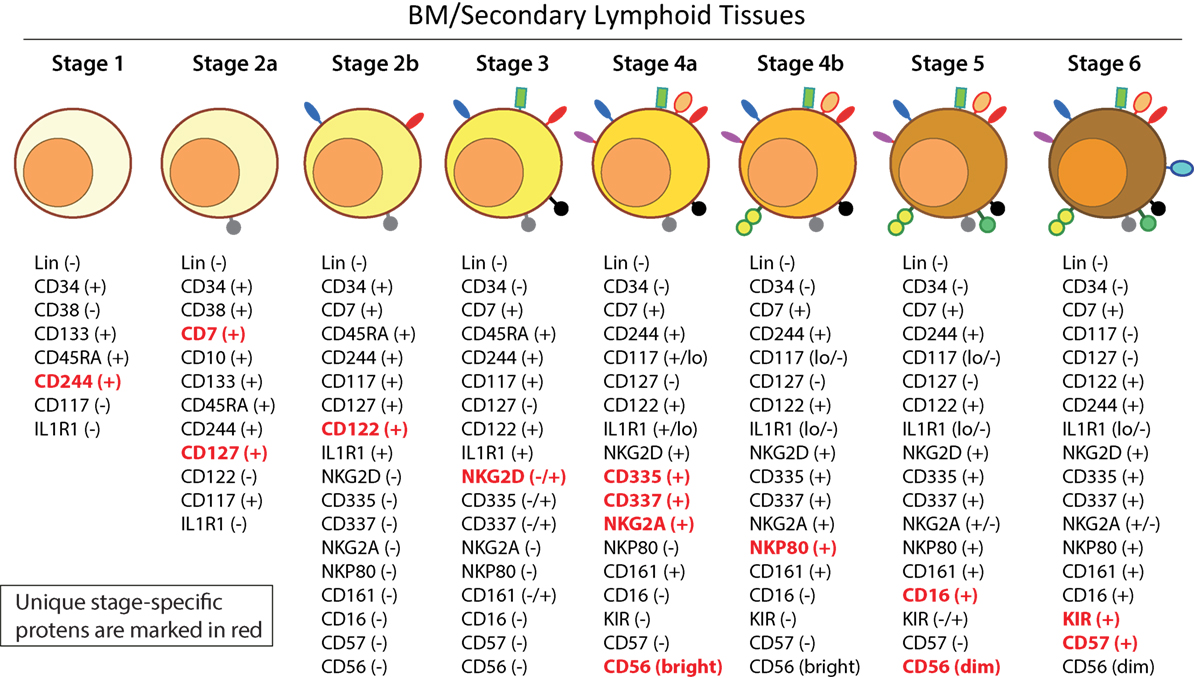
Figure 5. A common schema of human natural killer (NK) cell development in the bone marrow and lymph nodes. A total of six distinct developmental stages have been described with Stages 2 and 4 having additional bifurcations. Similar to the mouse, human NK cells express CD244 (2B4) throughout the developmental process starting at Stage 1 (pre-NK cell precursors). CD117 (c-Kit) and the low levels of interleukin (IL)-1R1 expressions define the Stage 2a and Stage 2b, respectively (NK cell progenitors). A higher expression of IL-1R1 defines the Stage 3 [immature NK cell (iNK)], and the expressions of NKG2D, CD335 (NKp46), CD337 (NKp30), and CD161 (NK1.1) are initiated. Stages 4a and 4b defines an entry of iNKs into mature nks, and are differentiated by the expression of NKp80 at the Stage 4b. Expressions of NKG2D, CD335, CD337, and CD161 reach their maximal levels at Stage 4. Most important of all, CD56 expression peaks (CD56bright). Significant differences between Stage 4b and Stage 5 are defined by a decrease in the expression of CD56 (CD56dim) in most and initiation of the expression of CD16 (FcγRIIIA) and killer immunoglobulin-like receptor (KIR) (CD158) in a subset of NK cells. Stage 6 defines the generation of “adaptive” or “memory-like” NK cells following “antigen” exposure, and it is identified by the high levels of NKG2C.
Downregulation of CD56bright expression to become CD56dim in most and the expression of immunoglobulin superfamily member CD16 (FcγRIII) in a subset of NK cells defines Stage 5 (Figure 5). Similar to the CD27/CD11b classification in mouse, expression levels of CD56 provides a functional classification of human NK cells. Most human NK cells in the peripheral blood are CD56dim (56). CD56bright NK cells are considered less mature and reside primarily in SLTs while the CD56dim subset represents the majority of NK cells in circulation (57). Most of the iNK cells transition into a minor CD56bright population (~5%) that convert into major CD56dim (>90%) population. The downregulation of CD56 during human NK cell maturation is strongly associated with the acquisition of anti-tumor cytotoxicity as CD56bright NK cells are potent producers of inflammatory cytokines, while the cytolytic function of human NK cells resides primarily in the CD56dim population (58, 59). Terminal maturation (Stage 6) of CD56dim NK cells are defined by the expression of CD57 (HNK-1, Leu-7). Additional classification such as “antigen-experienced” or “adaptive” CD2+ NK cells is defined by a higher expression of NKG2C (KLRC2, CD159c) (60–63).
Role of Common Gamma Chain Cytokines in the Development of NK Cells
Cytokines are essential inflammatory mediators that control multiple aspects of NK cell biology. NK cells express cytokine receptors early in their development (26) and require signaling through the common gamma (γc) chain for their development, homeostasis, and function (64). The γc chain (CD132) is a 40 kDa type I transmembrane glycoprotein that serves as the signaling subunit for IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21 (65). Although these cytokines display some functional redundancy, their cell-specific functions during an immune response are determined by the expression of distinct receptor complexes (Figure 6). For instance, IL-4, IL-7, IL-9, and IL-21, bind to high-affinity receptor complexes consisting of a cytokine-specific alpha-chain and the γc (64). These receptors have no intrinsic kinase activity, so signal transduction in response to γc cytokines is initiated by receptor-associated Janus kinases (JAKs) which phosphorylate different STAT molecules in a cytokine-dependent manner (66, 67).
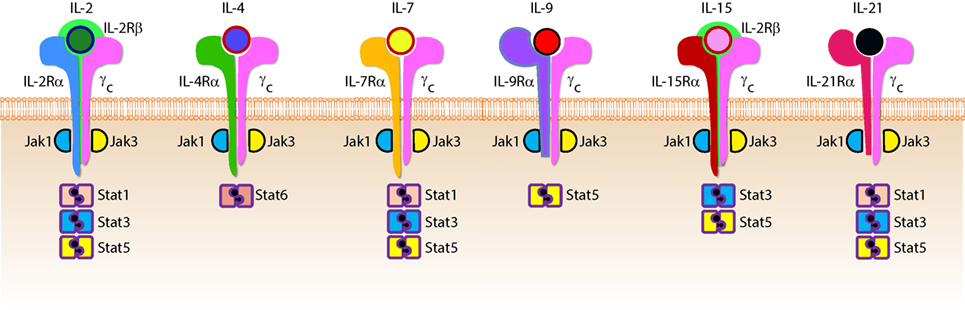
Figure 6. Role of common gamma-containing receptors in natural killer (NK) cell development. The common gamma chain-containing receptor family consists of six members, interleukin (IL)-2R, IL-4R, IL-7R, IL-9R, IL-15R, and IL-21R. Each one of them is distinguished from others by their unique α-chains. IL-2Rβ is shared by IL-2R and IL-15R complexes. In mouse, NK cell progenitors (NKPs) utilize IL-7R early during their transition from Pre-NKPs into refined-NKPs. In human, apart from its role in the early development, IL-7 also regulates the survival and expansion of mature CD56bright NK cells. IL-15R and IL-21R are required by NK cells to initiate and sustain their proliferation. Although it has been widely used to expand human and mouse NK cells ex vivo, the in vivo role of IL-2 that is primarily produced by CD4+ T cells is yet to be better understood. Role of IL-4 and IL-9 in NK cell development is less explored. Distinct sets of Janus kinases (JAK) and signal transducers and activators of transcription (STAT) associate and transmit the signaling from the common gamma chain-associated cytokine receptors.
Interleukin-2 and IL-15 are functionally related members the γc family of cytokines with respect to their receptor interactions as both can signal through complexes consisting of the γc and IL-2Rβ chains (68) resulting in the activation of STAT1 and STAT5 via JAK-1 and JAK-3, respectively (69). However, cellular affinity for either IL-2 or IL-15 is altered by the expression of high-affinity heterotrimeric complexes containing IL-2 or IL-15-specific alpha subunits (64). IL-2Rα (CD25) is expressed on activated NK cells and substantially increases their affinity for IL-2 which drives their proliferation and production of lytic molecules such as perforin and Granzyme B (70). Given that NK cells are found near T cell areas in SLTs (10), T cell-derived IL-2 may facilitate a vital functional crosstalk between innate and adaptive lymphocytes during an infection (71).
Although NK cells require γc signaling, as evidenced by the significant reduction in NK cell number and functional impairment in mice lacking the γc chain (γc−/−) (72, 73), IL-15 is unique in this regard. Mice lacking IL-15, IL-15Rα, or IL-2Rβ have similar phenotypes to γc−/− mice with respect to NK cell deficiencies (74–76), and transgenic overexpression of IL-15 in mice results in increased NK cell generation (77). It was determined that IL-15-mediated proliferation of mouse T cells was dependent on the presence of IL-15Rα on surrounding cells (78) which revealed a trans-presentation mechanism that is not required for IL-2-mediated proliferation. For this to occur, soluble IL-15 binds to IL-15Rα on the surface presenting cells which trans-present this complex to apposing NK cells expressing IL2-Rβ/γc heterodimers (79). IL-15 can be trans-presented by dendritic cells (DCs) and macrophages as well as non-hematopoietic cells including stromal cells and epithelial cells (80). The importance of IL-15 trans-presentation for NK cell survival in vivo was demonstrated with adoptive transfer experiments that showed normal NK cells were unable to survive in IL-15Rα-deficient mice while NK cells lacking IL-15Rα persist in IL-15Rα-sufficient recipients (81). IL-21R utilizes IL-21Rα and the γc (82). IL-21 synergizes with IL-2 to augment the expression of NKG2A, CD25, CD86, CD69, Perforin, and Granzyme B and thereby augmented cytotoxicity (83). These cytokines that use the γc-based receptors are the obligatory link between NK cells and the cells that produce them. For example, T helper cells that produce IL-21 can regulate the expression levels of activation receptors or cytolytic contents in NK cells. Similarly, DCs that produce IL-15 plays an essential role in the proliferation and priming of NK cells (discussed in detail elsewhere in this review).
Educating NK Cells to Distinguish “Self” From “Non-Self”
Functional differences between NK cells is also a consequence of the NK cell education process through which NK cells interact with self-major histocompatibility complex (MHC)-I (84). Initial observations concerning hybrid resistance to NK cell-mediated transplant rejection demonstrated that F1 hybrid mice reject transplanted BM from either parent while they do not reject transplants from other F1 mice (85, 86). These studies, along with others utilizing β2-microglobulin-deficient mice, further revealed that the underlying mechanistic basis of this rejection was dependent on MHC-I surface expression (87). The NK cell receptors that interact with MHC-I belong primarily to the killer immunoglobulin-like receptor (KIR) family in humans and the lectin-like homodimeric Ly49 receptor family in mice, and it is through these receptors that MHC-I regulates NK cell function (84). The molecular basis of NK cell education is still under debate and, based on the “missing-self” hypothesis of NK cell activation, it was initially thought that self-tolerance was exclusively due to inhibitory receptor signaling upon MHC-I engagement when interacting with normal cells (88). However, there exists a relatively small population of NK cells that do not express self-reactive inhibitory receptors under normal conditions, and these cells are hypofunctional upon stimulation (89).
The use of transgenic mouse models has led to the prevailing theories that attempt to explain the NK cell education process. In 2005, Yokoyama and colleagues termed the widely accepted model of NK cell education as “licensing” (90) which proposes that phosphatase activation in response to the ITIMs found in inhibitory receptors ultimately controls NK cell responsiveness. Thus, licensed NK cells are deemed functionally competent and are self-tolerant due to the interaction between inhibitory receptors and MHC-I while unlicensed NK cells, represented by those that do not express self-MHC-I-specific inhibitory receptors, are tolerant because they are functionally incompetent (84).
To further explain how NK cells become educated or “licensed,” Raulet and Vance proposed the NK cell “arming” and “disarming” models (91). In the “arming” model of NK cell education, NK cells are deemed functionally mature through self-MHC-I-specific inhibitory receptor interactions which are sufficient to drive the NK cell education process. This may seem counterintuitive given that these receptors are known to be exclusively inhibitory; however, their designation as such was described with respect to NK cell effector functions (91). Thus, inhibitory receptors may possess alternative functions in terms of NK cell education, and it has been demonstrated that signaling through these receptors is likely more complicated than previously appreciated (92). The “disarming” model proposes that chronic stimulation of NK cells that lack self-MHC-I inhibitory receptors are rendered hyporesponsive to stimulatory receptor activation potentially through a process similar to anergy in T or B cells (91). While these processes are thought to control NK cell responsiveness primarily during development, new interpretations of these models suggest that they may be altered under disease conditions and function as a rheostat to set the threshold of NK cell activation in the periphery (93, 94). Overall, the molecular mechanisms that regulate NK cell education have yet to be described though it is clear that the NK cell education process dictates their functional capabilities.
Signaling in NK Cells: Role of Germline-Encoded Activation Receptors
Natural killer cells do not express clonotypic receptors. However, they mediate strong anti-tumor cytotoxicity and generate significant quantities of pro-inflammatory cytokines (95). Lack of variable clonotypic receptors is compensated by multiple germline-encoded NK cell activation receptors (NKRs) such as NKG2D, NCR1, NCR2, NCR3, NKG2C, CD244, Ly49D, and Ly49H. Expression of more than one NKR that recognize self or pathogen-derived ligands endows NK cells with inherent, innate abilities to mediate effector functions. Due to the expression of multiple activation receptors, NK cells have to follow a distinct developmental program to obviate misrecognition of “self” leading to autoimmune responses. The varied nature of NKRs and the absence of signaling domains in their cytoplasmic tails necessitates the association and recruitment of receptor-associated adaptor molecules for signal transduction (96). The adaptor molecules that propagate NKR signaling includes FcεRIγ, CD3ζ, and the DAP12 which signal via immunoreceptor tyrosine-based activation motifs (ITAMs) contained within their cytoplasmic domains. NKRs that utilize these signaling adaptors include CD16, NCR1, Ly49D, Ly49H, and NKG2D (97–101). However, Ly49H and NKG2D can also signal via the YINM motif present within the adaptor, DAP10 (101–103). NK cell activation through these receptors occurs by interacting with distinct cellular and foreign ligands present on diseased cells and form the basis for the NK cell-mediated immune response in multiple contexts.
NKG2D is a homodimer forming C-type lectin-like type II transmembrane glycoprotein that is highly conserved from mice to humans (104). NKG2D is constitutively expressed on NK cells (105) and recognizes stress-inducible ligands that are structurally related to MHC-I (104). These ligands include ULBPs (106–108), MIC-A (109), and MIC-B (110, 111) in humans, and H60 (112–115) (a, b, and c), Rae-1 (α-ε) (115–117), and Mult1 (118, 119) in mice (120). NKG2D signaling is mediated through DAP10 and DAP12 via YINM and ITAM tyrosine-based signaling motifs, respectively. DAP10 recruits and activates the p85α subunit of PI(3)K (121) and recruits Grb2 (105) while DAP12 recruits ZAP70 and Syk to initiate NKG2D-mediated NK cell activation (105, 122).
These receptor-proximal signaling molecules activate the CBM signalosome containing Carma1, Bcl10, and Malt1, as well as Akt and the MAPKs, Erk1/2, Jnk1/2, and p38 (123–125). NK cell activation through NKG2D results in the mobilization of lytic granules as well as cytokine production via activation of transcription factors including activator protein-1 (AP-1) and NF-κB (123, 124). Pharmacological or genetic inhibition of these pathways causes deficiencies in NK cell-mediated cytotoxicity and pro-inflammatory cytokine production (126, 127). Pro-inflammatory cytokine production from NK cells expressing a catalytically inactive form of PI(3)K-p110δD910A, was significantly reduced while anti-tumor cytotoxicity was only moderately impaired (128–131). This finding substantiates the notion that the signaling molecules required for NK cell effector functions are not mutually exclusive (124) and further investigation is required to fully elucidate the molecular mechanisms that regulate NK cell effector functions in response to NKG2D-mediated stimulation.
NK Cell Effector Functions
Natural killer cells mediate their immunomodulatory effects through two critical effector functions. First, NK cells are cytotoxic lymphocytes that can directly lyse cells that have undergone a malignant transformation or have become infected with a virus or other intracellular pathogen (22). The cytolytic function of NK cells can initiate through a variety of processes, including degranulation and death receptor ligation, and is critical for the clearance of diseased and dysfunctional cells (132, 133). Second, NK cells can produce a variety of inflammatory cytokines in response to activation receptor stimulation as well as inflammatory cytokine-induced activation signaling (134, 135). These NK cell effector functions are essential components of the immune response and are the primary mechanisms through which NK cells mediate protective immunity.
The Mechanisms That Facilitate NK Cell Cytotoxicity
The molecular mechanisms that regulate NK cell cytotoxicity have been well described and can be divided into three main processes: (1) target cell recognition, (2) target cell contact and immunological synapse (IS) formation, and (3) NK cell-induced target cell death. Distinct mechanisms have been described for how target cells are recognized by NK cells and how they deem diseased cells appropriate for destruction (Figure 7). Once recognized, NK cells directly interact with the target cell of interest through the formation of a lytic IS which facilitates NK cell-induced target cell death through two essential mechanisms (136).
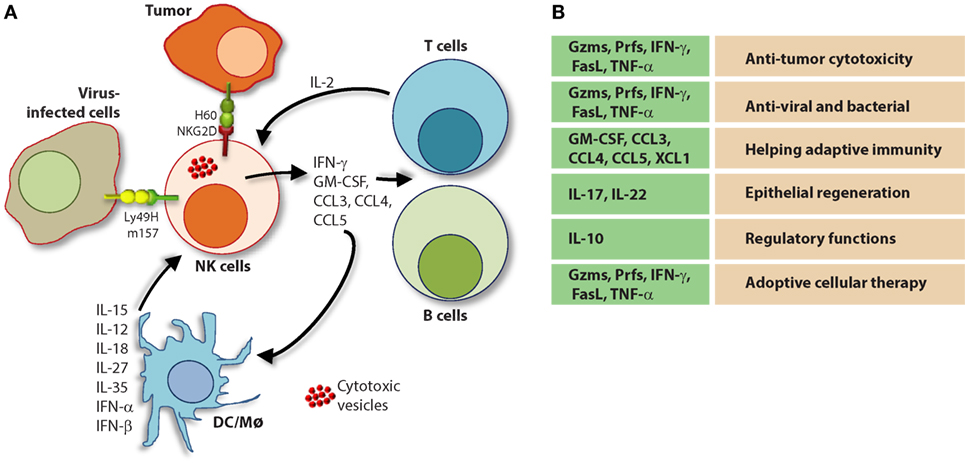
Figure 7. Role of a “third signal” in natural killer (NK) cell activation. (A) A brief description of the significant interactions between NK and myeloid cells. NK cells possess inherent abilities to mediate cytotoxicity and produce inflammatory cytokines and chemokines. Myeloid cell-derived cytokines play a central role in regulating the effector functions of NK cells. Interactions between the innate NK cells and the primary arms of the adaptive immunity (T and B cells) are less explored. Stimulation through activation receptors (i.e., NKG2D or Ly49H) help recognize tumor (H60) or infected target cells (murine cytomegalovirus-derived m157). (B) A summary of major soluble factors produced by NK cells and their intended functions.
The first mechanism involves the activation of death receptors present on the surface of the target cell which initiates the extrinsic apoptotic pathway (137). These receptors include TNF-related apoptosis-inducing ligand-receptor (TRAIL-R) and Fas (CD95) which are activated by their cognate ligands, Fas ligand (FasL) (CD95L) and TRAIL, present on NK cells (133). The surface expression of death receptors can be induced on target cells by NK cell-derived IFN-γ (138), and their activation initiates many pro-apoptotic signaling programs (139, 140). The death receptor superfamily is characterized by the utilization of a cytoplasmic death domain which enables these receptors to activate the apoptotic machinery including initiator caspases-8 and 10 (141, 142). Initiator caspases promote a cascade of IL1β-converting enzyme (ICE) superfamily proteases, including caspase-3 (143), and induce mitochondrial damage and cytochrome C release resulting in the formation of the apoptosome (144). The apoptosome amplifies initiator caspase-mediated substrate cleavage and, along with caspase-3-induced DNA fragmentation via caspase-activated DNase activation (145), results in cell death via apoptosis (146).
The primary mechanism of NK cell-mediated cytotoxicity involves the directed release of lytic molecules to the target cell (147). NK cells store these molecules in cytolytic granules that are delivered to the target cell through membrane fusion at the IS (136). This process requires cytoskeletal reorganization events including actin polymerization at the IS (148, 149) as well as polarization of the microtubule organizing center toward the target cell (150). Polarized lytic granules travel along microtubules and, once at the IS, fuse with the target cell membrane and release enzymes that facilitate that activation of the intrinsic apoptosis program within the target cell (136, 151). The molecules contained within lytic granules include the 60–70-kDa pore-forming glycoprotein, perforin (152), class of serine proteases known as granzymes (133), FasL (CD178), TRAIL (CD253), and granulysin (153). Granzyme B and perforin are a critical component of NK cell lytic granules and is classified as an apase that cleaves peptides after aspartic acid residues (133). Once inside the target cell, Granzyme B can trigger apoptosis through caspase-dependent and independent mechanisms. Granzyme B activates caspase-dependent apoptosis at multiple points in the apoptotic pathway by directly cleaving the apoptotic initiator caspase-8 as well as caspase-3 (154, 155). Granzyme B can also induce apoptosis in a caspase-independent manner and induce cytochrome C release from the mitochondria through the proteolytic cleavage of the pro-apoptotic protein, Bid (156).
NK Cell-Mediated Pro-Inflammatory Cytokine Production
Natural killer cells are potent producers of pro-inflammatory and immunosuppressive cytokines. However, the release of inflammatory cytokines is distinct from cytotoxic granule secretion (157) and NK cells utilize activation-induced signaling components to differentially regulate these two functions (124). Although NK cells can produce a wide-range of cytokines depending on the inflammatory environment (158, 159), NK cells primarily produce Th1-type cytokines when responding to tumor ligands and intracellular pathogens (160, 161). These include IFN-γ, TNF, and granulocyte/monocyte colony-stimulating factor (GM-CSF) which facilitate the activation of T cells as well as other innate immune mediators such as DCs, macrophages, and neutrophils (162, 163). NK cells also produce chemotactic cytokines (chemokines) including CCL3 (MIP-1α), CCl4 (MIP-1β), CCL5 (RANTES), XCL1 (lymphotoxin), and CXCL8 (IL-8) which can attract effector lymphocytes and myeloid cells to inflamed tissues (164).
Transcriptional activation of cytolytic molecules and inflammatory cytokines is a highly regulated process mediated by a variety of transcriptional regulators in NK cells. Many of these transcription factors, such as T-bet, are lineage defining and become activated early in NK cell development (13). Cytokine-induced activation of transcription factors, such as signal Transducers and Activators of Transcription (STAT) 4 and 5, occurs in response to IL-12 and IL-2 + IL-15 signaling, respectively (165). NKRs also initiate inflammatory transcriptional programs upon activation. These include the c-Fos and c-Jun heterodimer, AP-1, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), and nuclear factor of activated T cells (124, 166, 167) which bind promotor regions and promote inflammatory cytokine gene transcription (168, 169).
Role of Pro-Inflammatory Cytokines that Provide a “Third Signal” to NK Cells
A variety of cells generate a number of inflammatory mediators to sensitize and prime NK cells. Among these DCs play a central role (170). A complex interplay between DCs and NK cells is defined as one of the critical steps for the sensitization of NK cells (171). Given DC generate critical cytokines such as IL-15, IL-12, IL-23, IL-27, and IL-18, the crosstalk with NK cells determines the pathophysiological outcome of an ongoing immune response (172). Priming with type-1 IFN-α/IFN-β results in the expression of IL-15Rα and generation of IL-15 from plasmacytoid DCs (171). Multiple cell types including NK cells produce type-1 IFNs by which they can prime DCs (173). The trans-presentation of IL-15 by IL-15Rα to IL-15Rα/IL-2Rβ/IL-2Rγ complex on NK cells initiates multiple cellular tasks including proliferation and transcriptional reprogramming (81, 174). The IL-12 family of heterodimeric cytokines includes IL-12, IL-23, IL-27, and IL-35 which mediate diverse functions in NK cells (Figure 7) (175). IL-27 has both activating and inhibitory functions (176, 177) and IL-35 is an immunosuppressive cytokine produced exclusively by regulatory T cells (178). IL-12 and IL-23 are both produced by pathogen-activated macrophages and DCs and share a common component of their heterodimeric receptors, IL-12Rβ1 (175). Although the function of IL-23 in NK cells remains under debate, the role of IL-12 in NK cell activation is well established (175). IL-12 is a combination of the p40 and p35 alpha and beta subunits, respectively, and binds the IL-12 receptor (IL-12R) complex, IL-12Rβ1/IL-12Rβ2 (179). IL-12R signaling is propagated by Tyrosine kinase 2/JAK-2 and activates the transcriptional regulator, STAT4 (180).
Interleukin-12 signaling synergizes with those of other cytokines, including IL-2, IL-15, and IL-18 significantly enhances IFN-γ production by NK cells (181). IL-18 is a member of the IL-1 cytokine family and signals via the IL-18 receptor (IL-18R) through the signaling adaptors, myeloid differentiation primary response 88, and IL-1R-associated kinase (182, 183). IL-18 alone is not sufficient to induce IFN-γ production; however, the expression of IL-18R is induced by IL-12-mediated activation in lymphocytes (184) and IL-18 signaling synergizes with IL-12-mediated stimulation. Specifically, STAT4 activation by IL-12 enhances Ifng gene transcription while IL-18R signaling simultaneously induces the promoter binding activity of AP-1 and activates p38 MAPK to promote Ifng transcript stability and IFN-γ protein production (185, 186).
NK Cells in Health and Disease
To date, the diverse functions of NK cells in mammalian immunity is not fully understood. However, accumulating data collected from patients with rare disorders characterized by NK cell deficiency have shed light on their relevance to human health (187) and studies using genetically modified mouse models have generated intriguing ideas with regards to their pro-inflammatory and immunosuppressive functions (188). NK cells produce and respond to inflammatory stimuli and are most well known for their roles in anti-viral immunity and tumor immunosurveillance; however, NK cells are also involved in a variety of autoimmune disorders as drivers of pathologic inflammation (189). Emerging evidence also demonstrates that NK cells can regulate anti-inflammatory programs, such as tissue repair (190, 191). Whether NK cells act as primary innate effectors or accessory cells as part of the adaptive immune response appears to be context-dependent, but their contribution as first-line responders and essential inflammatory mediators is well established. Importantly, how the crosstalk between NK cells and lymphocytes (αβ-TCR+ T, γδ-TCR+ T, NKT, and B cells), myeloid cells (monocytes, macrophages, and DCs), or non-immune cells (epithelial or endothelial cells) enumerate a productive immune response is far from fully understood.
NK Cell Functions During Viral and Bacterial Infections
Natural killer cells are critical for defense against a wide variety of pathogens. Pattern recognition receptors (PRRs) recognize pathogen-associated molecular patterns and are essential components of the NK cell-mediated innate immune response (192). Activation of NK cells through PRRs elicit the production of TNF and IFN-γ which contribute to antibacterial defense (192, 193). NK cells also contribute to antifungal immunity by direct and indirect mechanisms (194). First, NK cells can directly damage fungal membranes through the targeted release of cytotoxic granules containing the membrane disrupting protein, perforin (195). They can also facilitate the antifungal host response through direct phagocytosis as well as the production of inflammatory mediators (196). Specifically, the production of GM-CSF by NK cells is critical for controlling C. albicans infection by promoting the fungicidal activity of neutrophils (197). However, the direct contribution of NK cells to microbial immunity has best been described with regards to their discrete actions against intracellular pathogens.
Intracellular pathogens have evolved a variety of mechanisms to evade the host immune response including subversion of the MHC immunosurveillance system (198). MHC molecules are highly polymorphic within a population and are encoded by human leukocyte antigen (HLA) genes in humans and, H-2 in mice (199). MHC molecules can be divided into two major classes, MHC class I (MHC-I) and MHC class II (MHC-II). MHC-I molecules bind, and present endogenous peptides to cytotoxic CD8+ T cells and subversion of this immunosurveillance mechanism results in an insufficient adaptive immune response (200). MHC-II is abundantly expressed on antigen-presenting cells (APCs) and facilitates the presentation of exogenous peptides to CD4+ helper T cells (201). Nearly all somatic cells express endogenous peptides on their surface in the context of MHC-I, and this allows the immune system to sample the intracellular environment (201). The peptide–MHC-I complex also defines the immunological “self” condition and maintenance of this system is essential for both immune tolerances as well as the rejection of “non-self” cells (Figure 8) and tissues that express distinct MHC-I haplotypes (202).
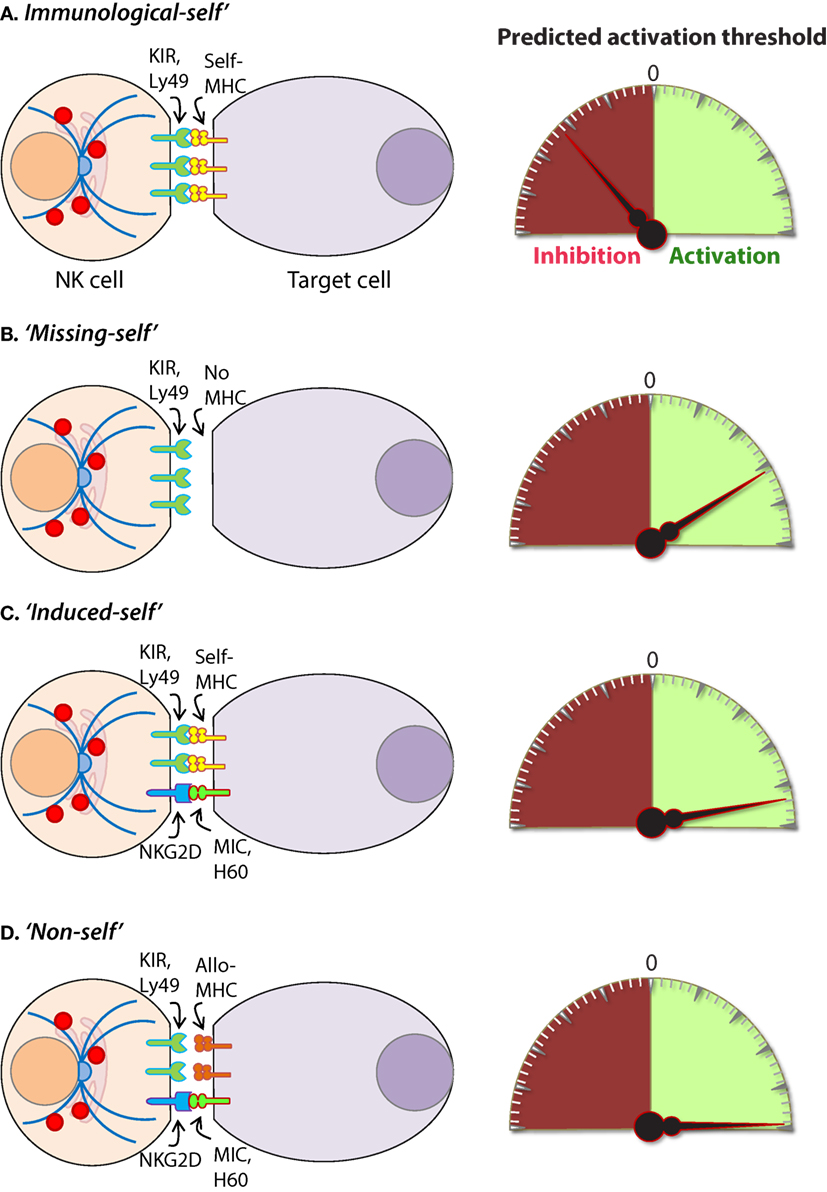
Figure 8. Mechanisms of target cell recognition by natural killer (NK) cells. NK cells lack clonotypic receptors and rely on germline-encoded activation and inhibitory receptors to recognize other cells around them. The following are some of the primary mechanisms by which NK cells perceive target cells. (A) “Immunological Self”: recognition of autologous MHC class I (MHC-I) (human leukocyte antigen (HLA)) or histocompatibility antigen-2 (H2, mouse) by inhibitory receptors [killer cell immunoglobulin-like receptor (KIR) or Ly49] let the NK cells know that they are interacting with normal cells and contain their activation. (B) “Missing-self”: recognition of target cells that either does not express MHC-I or reduce them below optimal levels can induce NK cell activation. (C) “Induced-self”: recognition of activating ligands that are expressed on target cells by the germline-encoded receptors such as NKG2D (H60, mouse; MIC-A/B, human), Ly49H (murine cytomegalovirus-derived m157, mouse), NCR1 (a number of viral proteins) can overcome MHC-I-mediated inhibitory signaling resulting in NK cell activation. (D) “Non-self”: recognition of transplanted tissue by NK cells, where the donor tissue expresses either allogeneic or haploidentical MHC-I.
Natural killer cells possess unique mechanisms to contain intracellular pathogens including viruses and some species of bacteria by lysing infected cells, releasing them and exposing them to adaptive cell-mediated immunity (203, 204). NK cells also produce inflammatory cytokines, such as IFN-γ to contain viral or bacterial growth (205–207). For example, hemagglutinin, a sialic acid receptor expressed by the influenza virus, serves as an activating ligand for NCR1 (208, 209). The murine cytomegalovirus (MCMV)-encoded membrane glycoprotein, m157, is recognized by the Ly49H receptor expressed in C57BL/6-derived NK cells (210). NK cells from other mouse backgrounds, such as 129/SvJ and BALB/c, do not express Ly49H, or another resistance factor, which renders them susceptible to MCMV as they are unable to mount a specific NK cell-mediated immune response to the virus (211–213). NKG2D has also involved in NK cell-mediated anti-viral immunity as evidenced by multiple observations in which human and mouse CMV proteins downregulate cellular stress ligands that activate NK cells through this receptor (214–217).
Natural killer cells have the unique ability to identify infected cells without direct engagement of the MHC-I complex (12, 218). Therefore, intracellular pathogens that evade CD8+ T cells by interfering with MHC-I surface expression remain vulnerable to NK cell-mediated immunity (219). In terms of anti-viral immunity, NK cells and CD8+ T cells have long been considered to represent the innate and adaptive arms of the immune response, respectively (220). However, the separation of these cells with regards to their contributions to adaptive immunity has recently been reconsidered due to the discovery of NK cells that exhibit immunological memory (160, 221). Although they do not utilize clonotypic receptors, such as the TCR, a relatively small population of memory NK cells has been described as long-lived effectors capable of rapid recall responses (222).
The formation of memory NK cells has been extensively investigated in mice infected with MCMV and studies using this system have been critical in defining the molecules that mediate this phenomenon (222–225). A vaccination study using antigens from viruses including, influenza, vesicular stomatitis virus, and human immunodeficiency virus type 1 also showed memory-like NK cell responses in mice (226) and NK cells exhibited enhanced protection against secondary infections with vaccinia virus and herpes simplex virus type 2 (227, 228). Collectively, these studies provide compelling evidence demonstrating the functional relevance of NK cell memory as a universal anti-viral immune mechanism. Observations in humans have also suggested the ability of human NK cells to form memory (229, 230); however, the full contribution of memory NK cells to anti-viral immunity and potential implications this may have on vaccine development has yet to be determined.
Natural killer cells also recognize bacteria and bacterial products either directly or from infected cells and professional APCs (Figure 9) (231). Recent work has shown that NK cells can directly release granzymes proteases to initiate disruption of electron transport, generate superoxide anion, and inactivate bacterial oxidative defenses causing the death of Listeria monocytogenes, Escherichia coli, and Mycobacteria tuberculosis (232–234). In addition, NK cells using Granzyme B mediated the killing of facultative anaerobic bacteria such as L. monocytogenes by cleaving essential proteins that are required for protein translation (aminoacyl tRNA synthetases and ribosomal proteins), folding (protein chaperones), and protein degradation (Clp system) (235). Indirect killing and containment of L. monocytogenes (236, 237), Staphylococcus aureus (238), Lactobacillus johnsonii (239), Mycobacterium tuberculosis (240), and Mycobacterium bovis bacille Calmette-Guérin (241) by NK cells have been described. Mechanisms by which NK cells mediate indirect clearance of bacteria are complex. Substantial evidence suggests that interleukins including IL-12 and IL-18 from monocytes and DCs play a central role (242–244). Role of other inflammatory cytokines such as IL-27 and its cooperation with IL-18, IL-6, and IL-12 during the clearance of bacterial infections have been identified; however, the precise mechanisms by which NK cells evoke the anti-microbial responses are yet to be elucidated (245, 246).
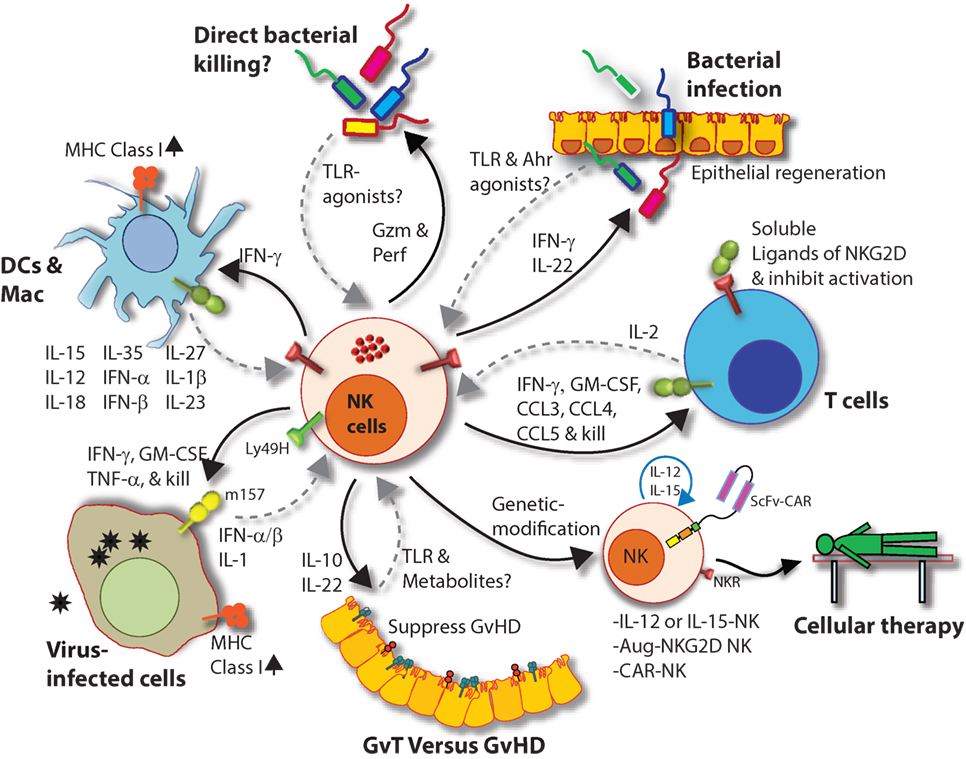
Figure 9. Natural killer (NK) cells in health and disease. As the largest lymphocyte population representing innate immunity, NK cells perform diverse functions. Through their ability to mediate killing and to produce soluble factors, NK cells perform multitudes of immunological functions. Counter-clockwise: bidirectional interactions between NK cell and dendritic cells (DCs)/macrophages result in priming. Activated DCs and macrophages generate interleukin (IL)-15, IL-12, IL-18, IL-35, IFN-α, IFN-β, IL-27, IL-1β, and IL-23. These, in turn, activate NK cells to be primed, proliferate, and to produce inflammatory factors and chemokines such as interferon-gamma (IFN-γ), granulocyte/monocyte colony-stimulating factor (GM-CSF), tumor necrosis factor (TNF)-α, CCL3, CCL4, and CCL5. In addition, IFN-γ from NK cells can increase the MHC class I expression and the transcription of genes encoding immuno-proteasomal subunits in these professional antigen-presenting cells and thereby augmenting T cell priming and activation. Similarly, virus-infected cells produce IFN-α, IFN-β, and IL-1β and present either “stress-induced” self-ligands or viral proteins on the cell surface that activate NK cells. A reduction in graft-versus-host disease (GvHD) is mediated through the production of IL-10 by the CD56brightCD16Neg NK cell subset and augmentation of GvT is potentiated via direct tumor killing by CD56dimCD16Pos NK cell subset. In addition, production of IL-22 by NK subsets may help the regeneration of epithelial cells in the mucosal tissues. Irrespective of these observations, the mechanisms by which NK cells are activated to respond during active GvHD/GvT is not fully understood. Genetic manipulation of NK cells has helped to improve the effector functionality and the longevity of human NK cells in vivo. Stable integration of gene encoding IL-15 into the genome of NK cells promotes sustained proliferation via an artificial autocrine loop. Similarly, integration of gene encoding IL-12 makes this cytokine abundantly available within the microenvironmental milieu and thereby augment the effector functions of NK cells, specifically, the production of IFN-γ. Augmented expression of NK cell activation receptors (NKRs) including NKG2D and NCR1 by genetic engineering increases the anti-tumor cytotoxicity of NK cells. Other studies have shown the expression of single chain variable fragment that forms the core ectodomain of chimeric antigen receptor (CAR) to augments the tumor-targeted killing of NK cells. These genetically modified NK cells provide exciting newer opportunities for cell-based therapies. The bidirectional interaction between NK and T cells results in the regulation of adaptive immunity. IL-2 produced by CD4+ Th1 cells play a vital role in the proliferation and expansion of NK cells. Although in vitro experiments consistently have provided support toward this notion, the in vivo evidence is far from convincing. However, the inflammatory factors produced by NK cells have a significant impact on both CD8+ and CD4+ T cells. Expression of “self” ligands for NKG2D by T cells results in the recognition and killing of T cells by NK cells during GvHD and anti-viral responses. In addition, a cleaved soluble form of these ligands (MIC-A/B) is present in the serum of cancer patients. This, in turn, plays an important role in containing the effector functions of T cells via direct binding to the NKG2D receptor expressed on T cells. NK cells recognize bacteria-infected cells (such as epithelial cells) either using toll-like receptors (TLR) or by activated through soluble factors including aryl hydrocarbon receptor (Ahr). This results in the production of IFN-γ and IL-22 that helps with the reduction in bacterial load and regeneration of epithelial cells, respectively. NK cells can also directly mediate the lysis of bacteria using granzymes and perforin.
Anti-Tumor Functions and the Clinical Utilization of NK Cells
The vital role of NK cells in tumor immunosurveillance was recognized soon after their initial characterization (247, 248). NK cells can detect changes in surface expression of self-MHC-I molecules on autologous cells which distinctively qualifies them to detect cells that have undergone malignant transformation (Figure 8) (218, 248). Genomic mutations that arise during the transformation process are reflected by a variety of phenotypic changes which alter the expression of cell surface molecules, including downregulation of the inhibitory “self” MHC-I (200, 249). The activity of NK cells against this “missing-self” condition has been well described (250, 251) and serves as a critical mechanism through which NK cells facilitate anti-tumor immunity. Transformed cells also express increased numbers of stress-induced molecules on their surface which can be recognized by specific NK cell receptors, such as NKG2D (120, 252). This concept, known as “induced self” (Figure 8) recognition (253, 254), explains why NK cell does not kill normal cells, such as erythrocytes, that do not express MHC-I on their surface but retain cytotoxic activity against MHC-I sufficient tumors (255). Elicitation of NK cell function is determined by the relative strength of activating and inhibitory receptor signaling and this concept, known as “altered balance,” ultimately controls NK cell activity under normal and disease conditions (256).
Decades of research in rodents have demonstrated the importance of NK cells in tumor clearance (14, 117, 247, 248). In humans, an 11-year follow-up study showed that low NK cell cytotoxic activity was correlated to an increased risk of cancer (257) and the presence of tumor-infiltrating NK cells is a positive prognostic marker for multiple malignancies including colorectal carcinoma (258), gastric carcinoma (259), and squamous cell lung cancer (260). Results from multiple studies demonstrate that NK cells have promise as a cancer immunotherapeutic for the treatment of hematological malignancies including acute myeloid leukemia and acute lymphoblastic leukemia (261–263). Allogenic NK cell therapy has proven effective in the clinic and, unlike T cell-based interventions, NK cell transfusion carries a relatively low risk of adverse off-tumor effects such as graft-versus-host disease (GvHD) (264).
Autologous NK cells may be inhibited by “self” MHC-I, thus limiting GvT effects in the absence of exogenous cytokines or antibodies (265, 266). Therefore, allogeneic NK cells along with hematopoietic stem cell transplant has been explored as a potential treatment for patients with high-risk solid tumors (263, 267, 268). Using non-myeloablative conditioning regimens to provide potent immune suppression without toxicity, the burden of cure then relies on the ability of transplanted donor cells to provide a GvT effect. Precedence in using low-intensity conditioning before transplanting allogeneic stem cells has been reported in Ewing sarcoma (269–271), osteosarcoma (272, 273), germ cell tumors (274), rhabdomyosarcoma (275–277), neuroblastoma (278–280), Wilms tumor (281), and CNS tumors (282), suggesting that alloreactive donor NK cells infiltrate heterogeneous solid tumors and cross the blood–brain barrier. A sizeable reduction in tumor burden has been observed (269). Using HLA-haploidentical family donors (parents and siblings), matched by only one HLA haplotype to the patient, have not only shown favorable outcomes in patients with solid tumors (263, 267, 283) but are also readily available and highly motivated donor sources. Thus, using HLA-haploidentical donors to augment GvT may be an effective strategy in patients undergoing allogeneic hematopoietic stem cell transplantation (HCT) for treatment of solid tumors (263, 284).
Regulatory Functions of NK Cells
Most functions of NK cells are analogous to either CD8+ T or Th1 cells, including the production of pro-inflammatory cytokines (IFN-γ, TNF-α, and GM-CSF) and mediating cytotoxicity against infected or tumor cells (95). However, in addition to these, recent reports suggest NK cells also play regulatory functions (285, 286). NK cells mediate regulatory functions of other cell types including myeloid [DC (246, 287–290), monocytes (291–293), and macrophages (246, 294–296)] or lymphoid [T (297, 298) and B (299–301) cells] via cytokines production or through direct cell–cell contact in a receptor–ligand interaction-dependent manner. As part of the innate immune responses, effector functions of NK cells during the early phase is expected to dictate the threshold, direction, and the outcome of an immune response. These NK cell-mediated regulatory functions are predicted to occur during viral, bacterial, or protozoan infections, anti-tumor immune responses, unexpected immuno-pathological outcomes such as GvHD, and autoimmune diseases (302). Few of the examples are described below. A unique innate immunoregulatory function for the smaller CD56bright subset of human NK cells (CD56brightCD16dimNKG2A+KIR−) was proposed due to their inherent ability to produce significant amounts of IL-10, and IL-13 along with IFN-γ, TNF-α, and GM-CSF compared to that of the more substantial CD56dimCD16+ subset (58). An Il-27-stimulated CD56brightCD16dimNKG2A+KIR− subset was able to suppress the proliferation of autologous CD4+ T cells in patients with multiple sclerosis through a cytotoxic mechanism involving perforin (303) or by the release of Granzymes (304, 305). Importantly, CD56brightCD16dimNKG2A+KIR− subset through their ability to produce adenosine and by the restricted expression of the ecto-nucleotide pyrophosphatase/phosphodiesterase 1 (CD203a/PC-1) and the nucleotide-metabolizing ectoenzyme CD38 (an NAD+ nucleosidase) was able to inhibit the proliferation of autologous CD4+ T cells (306).
Regulatory role of NK cells during GvHD is highly controversial (307). GvHD is one of the major complications and limiting factor in allogeneic HCT (308). Studies in both mouse and human lead to either suppressing or promoting rejection of HCT by NK cells. Furthermore, persistence or expansion of NK cells following HCT resulted in rejection and severe GvHD (309) while allograft-derived donor NK cells helped the engraftment of HCT by suppressing GvHD (310–313). Mechanistically, NK cells can help to contain GvHD through distinct mechanisms including the killing of professional APCs and thereby controlling the proliferation and expansion of graft-specific T cell (314, 315). In addition, NK cells were able to directly lyse graft-specific T cells following the expression of activating ligands of NKG2D on these T cell (316, 317). Expression of both mouse (316, 318) (H60a, H60b, H60c, Rae-1, and Mult-1) and human (319–321) (MIC-A, MIC-B, and ULBPs) activating ligands of NKG2D on stimulated T cells has been reported in a number of models. Also, shedding of these murine and human activating ligands has been demonstrated to employ a critical negative regulatory function on both T (322–324) and NK (325, 326) cells. These findings provide an exciting new avenue in understanding an inherent regulatory interaction between NK cell and APCs or T cells and thereby potential clinical utilization. Irrespective of the recent advances, the precise functions and associated mechanisms by which NK cells contribute to an immune-suppressive or immune-sufficient tumor microenvironment is far from fully defined. Similarly, the complex interplay of cytokines and ILs that are derived from and regulating the functions of NK and professional APCs during viral or bacterial infections is yet to be fully appreciated. Furthermore, defining the interactions between conventional NK cells (ILC1) and ILC2 or ILC3 can help to formulate better immunotherapeutic approaches to infections associated with mucosal tissues.
NK Cells and CAR Therapy
Recent efforts to improve the clinical efficacy of NK cell immunotherapy has led to the development of genetically engineered NK cells that express a chimeric antigen receptor (CAR). Primary NK cells and NK cell lines can be engineered to express CARs which redirect the anti-tumor specificity of NK cells on an antigen-dependent basis (327). Through the manipulation of signaling motifs critical for lymphocyte activation, CARs are also designed to utilize specific intracellular signaling molecules which can further refine NK cell function and optimize their therapeutic potential (328, 329). Interestingly, the use of a clonal cell line derived from a human NK cell leukemia, known as NK-92, has been genetically modified to express fully functional CARs and these cells have shown great promise with regards to their safety and efficacy in recent clinical trials (327, 330, 331). Moreover, the use of irradiated cell lines may provide a fast and affordable off-the-shelf option for a personalized cellular immunotherapy treatment (332, 333) and are quickly rising to the forefront of cell-based cancer immunotherapies (Figure 9).
Summary and Future Outlook
Natural killer cells possess promising potentials as a therapeutic tool to treat a number of maladies including malignancies (334). However, irrespective of their comparable ability in mediating anti-tumor cytotoxicity to that of CD8+ T cells, the clinical utilization of NK cells remains far from practical. In-depth understanding of NK cells at the single-cell transcriptomic landscape, methods to expand them in vitro without phenotypic and functional skewing, and detailed analyses of their in vivo longevity are central to facilitate the clinical utilization. NK cells regulate their effector functions utilizing both activating and inhibitory receptors (335, 336). Irrespective of our decades-long understanding, the precise intracellular signaling mechanisms by which NK cells discriminate the “self” from “missing-self” or “non-self” are still elusive. Emerging evidence suggests that mNK cells possess the ability to produce both pro-inflammatory to anti-inflammatory cytokines (159). However, the temporal regulation of these discrete functions is not yet fully understood. NK cells can be primed in response to a wide panel of ILs and other immunomodulatory factors (132, 158, 337). Our knowledge related to transcriptomic definitions of priming for an individual or combination of these priming factors is limited. NK cell subsets are comprised of a highly heterogeneous population (338). A pioneering study utilizing a novel technique known as mass-cytometry (CyTOF) determined that there are between 6,000 and 30,000 distinct NK cell phenotypes within a given individual based on unique combinations of 35 cell surface antigens (339). Studies on the genome-wide chromatin accessibility for regulomes provided similarities in regulatory circuitries of transcriptional programs between ILCs (ILC1 includes conventional NK cells) and CD4+ T helper subsets (340). However, the functional plasticity of subsets of NK cells yet to be fully appreciated. Controversies related to “adaptive” and “memory” characteristics of NK cells should be resolved by defining transcriptomic, genetic, and epigenetic alterations between naïve and “antigen-experienced” NK cells. Collectively, the future holds promising challenges to decipher new knowledge which will facilitate the utilization of NK cells for better therapeutic outcomes.
Author Contributions
AA conceived and wrote the manuscript. CY contributed to the writing. MT edited the text. SM conceived, wrote, and edited the text and generated all the figures for the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Lucia Sammarco and her Lulu’s Lemonade Stand for inspiration, motivation, and support. This work was supported in part by NIH R01 AI102893 and NCI R01 CA179363 (SM); Alex Lemonade Stand Foundation (SM); HRHM Program of MACC Fund (SM), Nicholas Family Foundation (SM); Gardetto Family (SM); Hyundai Scholars Program (MT); Pavlove Foundation (MT); Rebecca Jean Slye Endowment (MT); MCW-Cancer Center-Large Seed Grant (SM and MT); MACC Fund (MT and SM); Ann’s Hope Melanoma Foundation (SM and MT); and Advancing Healthier Wisconsin (SM).
References
1. Oldham RK. Natural killer cells: artifact to reality: an odyssey in biology. Cancer Metastasis Rev (1983) 2:323–36. doi:10.1007/BF00048565
2. Rosenau W, Moon HD. Lysis of homologous cells by sensitized lymphocytes in tissue culture. J Natl Cancer Inst (1961) 27:471–83.
3. Smith HJ. Antigenicity of carcinogen-induced and spontaneous tumours in inbred mice. Br J Cancer (1966) 20:831–7. doi:10.1038/bjc.1966.95
4. Kiessling R, Klein E, Pross H, Wigzell H. “Natural” killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur J Immunol (1975) 5:117–21. doi:10.1002/eji.1830050209
5. Herberman RB, Nunn ME, Holden HT, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. II. Characterization of effector cells. Int J Cancer (1975) 16:230–9.
6. Herberman RB, Nunn ME, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic acid allogeneic tumors. I. Distribution of reactivity and specificity. Int J Cancer (1975) 16:216–29. doi:10.1002/ijc.2910160204
7. Kiessling R, Klein E, Wigzell H. “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol (1975) 5:112–7. doi:10.1002/eji.1830050208
8. Oldham RK, Siwarski D, McCoy JL, Plata EJ, Herberman RB. Evaluation of a cell-mediated cytotoxicity assay utilizing 125 iododeoxyuridine-labeled tissue-culture target cells. Natl Cancer Inst Monogr (1973) 37:49–58.
9. Pross HF, Jondal M. Cytotoxic lymphocytes from normal donors. A functional marker of human non-T lymphocytes. Clin Exp Immunol (1975) 21:226–35.
10. Carrega P, Ferlazzo G. Natural killer cell distribution and trafficking in human tissues. Front Immunol (2012) 3:347. doi:10.3389/fimmu.2012.00347
11. Scoville SD, Freud AG, Caligiuri MA. Modeling human natural killer cell development in the era of innate lymphoid cells. Front Immunol (2017) 8:360. doi:10.3389/fimmu.2017.00360
12. Sun JC, Lanier LL. NK cell development, homeostasis and function: parallels with CD8(+) T cells. Nat Rev Immunol (2011) 11:645–57. doi:10.1038/nri3044
13. Simonetta F, Pradier A, Roosnek E. T-bet and eomesodermin in NK cell development, maturation, and function. Front Immunol (2016) 7:241. doi:10.3389/fimmu.2016.00241
14. Langers I, Renoux VM, Thiry M, Delvenne P, Jacobs N. Natural killer cells: role in local tumor growth and metastasis. Biologics (2012) 6:73–82. doi:10.2147/BTT.S23976
15. Jiao Y, Huntington ND, Belz GT, Seillet C. Type 1 innate lymphoid cell biology: lessons learnt from natural killer cells. Front Immunol (2016) 7:426. doi:10.3389/fimmu.2016.00426
16. Abolins S, King EC, Lazarou L, Weldon L, Hughes L, Drescher P, et al. The comparative immunology of wild and laboratory mice, Mus musculus domesticus. Nat Commun (2017) 8:14811. doi:10.1038/ncomms14811
17. Lanier LL, Testi R, Bindl J, Phillips JH. Identity of Leu-19 (CD56) leukocyte differentiation antigen and neural cell adhesion molecule. J Exp Med (1989) 169:2233–8. doi:10.1084/jem.169.6.2233
18. Lanier LL, Phillips JH, Hackett J Jr, Tutt M, Kumar V. Natural killer cells: definition of a cell type rather than a function. J Immunol (1986) 137:2735–9.
19. Tsukerman P, Stern-Ginossar N, Yamin R, Ophir Y, Stanietsky AM, Mandelboim O. Expansion of CD16 positive and negative human NK cells in response to tumor stimulation. Eur J Immunol (2014) 44:1517–25. doi:10.1002/eji.201344170
20. Walzer T, Bléry M, Chaix J, Fuseri N, Chasson L, Robbins SH, et al. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc Natl Acad Sci U S A (2007) 104:3384–9. doi:10.1073/pnas.0609692104
21. Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells – a proposal for uniform nomenclature. Nat Rev Immunol (2013) 13:145–9. doi:10.1038/nri3365
22. Zhang Y, Huang B. The development and diversity of ILCs, NK cells and their relevance in health and diseases. Adv Exp Med Biol (2017) 1024:225–44. doi:10.1007/978-981-10-5987-2_11
23. Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell (1997) 91:661–72. doi:10.1016/S0092-8674(00)80453-5
24. Male V, Nisoli I, Kostrzewski T, Allan DS, Carlyle JR, Lord GM, et al. The transcription factor E4bp4/Nfil3 controls commitment to the NK lineage and directly regulates Eomes and Id2 expression. J Exp Med (2014) 211:635–42. doi:10.1084/jem.20132398
25. Carotta S, Pang SH, Nutt SL, Belz GT. Identification of the earliest NK-cell precursor in the mouse BM. Blood (2011) 117:5449–52. doi:10.1182/blood-2010-11-318956
26. Rosmaraki EE, Douagi I, Roth C, Colucci F, Cumano A, Di Santo JP. Identification of committed NK cell progenitors in adult murine bone marrow. Eur J Immunol (2001) 31:1900–9. doi:10.1002/1521-4141(200106)31:6<1900::AID-IMMU1900>3.0.CO;2-M
27. Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med (2007) 204:1119–30. doi:10.1084/jem.20061959
28. Ikawa T, Fujimoto S, Kawamoto H, Katsura Y, Yokota Y. Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc Natl Acad Sci U S A (2001) 98:5164–9. doi:10.1073/pnas.091537598
29. Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature (1999) 397:702–6. doi:10.1038/17812
30. Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol (2009) 10:1118–24. doi:10.1038/ni.1787
31. Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, Grosveld G, et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J Exp Med (2009) 206:2977–86. doi:10.1084/jem.20092176
32. Goh W, Huntington ND. Regulation of murine natural killer cell development. Front Immunol (2017) 8:130. doi:10.3389/fimmu.2017.00130
33. Kim S, Iizuka K, Kang HS, Dokun A, French AR, Greco S, et al. In vivo developmental stages in murine natural killer cell maturation. Nat Immunol (2002) 3:523–8. doi:10.1038/ni796
34. Ito M, Maruyama T, Saito N, Koganei S, Yamamoto K, Matsumoto N. Killer cell lectin-like receptor G1 binds three members of the classical cadherin family to inhibit NK cell cytotoxicity. J Exp Med (2006) 203:289–95. doi:10.1084/jem.20051986
35. Di Santo JP. Natural killer cell developmental pathways: a question of balance. Annu Rev Immunol (2006) 24:257–86. doi:10.1146/annurev.immunol.24.021605.090700
36. Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J Immunol (2006) 176:1517–24. doi:10.4049/jimmunol.176.3.1517
37. Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T. Maturation of mouse NK cells is a 4-stage developmental program. Blood (2009) 113:5488–96. doi:10.1182/blood-2008-10-187179
38. Regunathan J, Chen Y, Kutlesa S, Dai X, Bai L, Wen R, et al. Differential and nonredundant roles of phospholipase Cgamma2 and phospholipase Cgamma1 in the terminal maturation of NK cells. J Immunol (2006) 177:5365–76. doi:10.4049/jimmunol.177.8.5365
39. Huntington ND, Tabarias H, Fairfax K, Brady J, Hayakawa Y, Degli-Esposti MA, et al. NK cell maturation and peripheral homeostasis is associated with KLRG1 up-regulation. J Immunol (2007) 178:4764–70. doi:10.4049/jimmunol.178.8.4764
40. Müller-Durovic B, Lanna A, Covre LP, Mills RS, Henson SM, Akbar AN. Killer cell lectin-like receptor G1 inhibits NK cell function through activation of adenosine 5’-monophosphate-activated protein kinase. J Immunol (2016) 197:2891–9. doi:10.4049/jimmunol.1600590
41. Yu J, Freud AG, Caligiuri MA. Location and cellular stages of natural killer cell development. Trends Immunol (2013) 34:573–82. doi:10.1016/j.it.2013.07.005
42. Akashi K, Traver D, Kondo M, Weissman IL. Lymphoid development from hematopoietic stem cells. Int J Hematol (1999) 69:217–26.
43. Miraglia S, Godfrey W, Yin AH, Atkins K, Warnke R, Holden JT, et al. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood (1997) 90:5013–21.
44. Li Z. CD133: a stem cell biomarker and beyond. Exp Hematol Oncol (2013) 2:17. doi:10.1186/2162-3619-2-17
45. Brown MH, Boles K, van der Merwe PA, Kumar V, Mathew PA, Barclay AN. 2B4, the natural killer and T cell immunoglobulin superfamily surface protein, is a ligand for CD48. J Exp Med (1998) 188:2083–90. doi:10.1084/jem.188.11.2083
46. Higuchi Y, Zeng H, Ogawa M. CD38 expression by hematopoietic stem cells of newborn and juvenile mice. Leukemia (2003) 17:171–4. doi:10.1038/sj.leu.2402785
47. Rabinowich H, Pricop L, Herberman RB, Whiteside TL. Expression and function of CD7 molecule on human natural killer cells. J Immunol (1994) 152:517–26.
48. Luetke-Eversloh M, Killig M, Romagnani C. Signatures of human NK cell development and terminal differentiation. Front Immunol (2013) 4:499. doi:10.3389/fimmu.2013.00499
49. Renoux VM, Zriwil A, Peitzsch C, Michaëlsson J, Friberg D, Soneji S, et al. Identification of a human natural killer cell lineage-restricted progenitor in fetal and adult tissues. Immunity (2015) 43:394–407. doi:10.1016/j.immuni.2015.07.011
50. Mace EM, Hsu AP, Monaco-Shawver L, Makedonas G, Rosen JB, Dropulic L, et al. Mutations in GATA2 cause human NK cell deficiency with specific loss of the CD56(bright) subset. Blood (2013) 121:2669–77. doi:10.1182/blood-2012-09-453969
51. Marcenaro E, Notarangelo LD, Orange JS, Vivier E. Editorial: NK cell subsets in health and disease: new developments. Front Immunol (2017) 8:1363. doi:10.3389/fimmu.2017.01363
52. Subrahmanyam G, Rudd CE, Schneider H. Association of T cell antigen CD7 with type II phosphatidylinositol-4 kinase, a key component in pathways of inositol phosphate turnover. Eur J Immunol (2003) 33:46–52. doi:10.1002/immu.200390006
53. Aandahl EM, Sandberg JK, Beckerman KP, Taskén K, Moretto WJ, Nixon DF. CD7 is a differentiation marker that identifies multiple CD8 T cell effector subsets. J Immunol (2003) 170:2349–55. doi:10.4049/jimmunol.170.5.2349
54. Vitale M, Falco M, Castriconi R, Parolini S, Zambello R, Semenzato G, et al. Identification of NKp80, a novel triggering molecule expressed by human NK cells. Eur J Immunol (2001) 31:233–42. doi:10.1002/1521-4141(200101)31:1<233::AID-IMMU233>3.0.CO;2-4
55. Freud AG, Keller KA, Scoville SD, Mundy-Bosse BL, Cheng S, Youssef Y, et al. NKp80 defines a critical step during human natural killer cell development. Cell Rep (2016) 16:379–91. doi:10.1016/j.celrep.2016.05.095
56. Farag SS, Caligiuri MA. Human natural killer cell development and biology. Blood Rev (2006) 20:123–37. doi:10.1016/j.blre.2005.10.001
57. Poli A, Michel T, Thérésine M, Andrès E, Hentges F, Zimmer J. CD56bright natural killer (NK) cells: an important NK cell subset. Immunology (2009) 126:458–65. doi:10.1111/j.1365-2567.2008.03027.x
58. Cooper MA, Fehniger TA, Turner SC, Chen KS, Ghaheri BA, Ghayur T, et al. Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset. Blood (2001) 97:3146–51. doi:10.1182/blood.V97.10.3146
59. Jacobs R, Hintzen G, Kemper A, Beul K, Kempf S, Behrens G, et al. CD56bright cells differ in their KIR repertoire and cytotoxic features from CD56dim NK cells. Eur J Immunol (2001) 31:3121–7. doi:10.1002/1521-4141(2001010)31:10<3121::AID-IMMU3121>3.0.CO;2-4
60. Wagner JA, Fehniger TA. Human adaptive natural killer cells: beyond NKG2C. Trends Immunol (2016) 37:351–3. doi:10.1016/j.it.2016.05.001
61. Pupuleku A, Costa-García M, Farré D, Hengel H, Angulo A, Muntasell A, et al. Elusive role of the CD94/NKG2C NK cell receptor in the response to cytomegalovirus: novel experimental observations in a reporter cell system. Front Immunol (2017) 8:1317. doi:10.3389/fimmu.2017.01317
62. Redondo-Pachón D, Crespo M, Yélamos J, Muntasell A, Pérez-Sáez MJ, Pérez-Fernández S, et al. Adaptive NKG2C+ NK cell response and the risk of cytomegalovirus infection in kidney transplant recipients. J Immunol (2017) 198:94–101. doi:10.4049/jimmunol.1601236
63. Kared H, Martelli S, Wen Tan S, Simoni Y, Li Chong M, Hwei Yap S, et al. Adaptive NKG2C(+)CD57(+) natural killer cell and Tim-3 expression during viral infections. Front Immunol (2018) 9:686. doi:10.3389/fimmu.2018.00686
64. Meazza R, Azzarone B, Orengo AM, Ferrini S. Role of common-gamma chain cytokines in NK cell development and function: perspectives for immunotherapy. J Biomed Biotechnol (2011) 2011:861920. doi:10.1155/2011/861920
65. Boulanger MJ, Garcia KC. Shared cytokine signaling receptors: structural insights from the gp130 system. Adv Protein Chem (2004) 68:107–46. doi:10.1016/S0065-3233(04)68004-1
66. Ghoreschi K, Laurence A, O’Shea JJ. Janus kinases in immune cell signaling. Immunol Rev (2009) 228:273–87. doi:10.1111/j.1600-065X.2008.00754.x
67. Suzuki K, Nakajima H, Saito Y, Saito T, Leonard WJ, Iwamoto I. Janus kinase 3 (Jak3) is essential for common cytokine receptor gamma chain (gamma(c))-dependent signaling: comparative analysis of gamma(c), Jak3, and gamma(c) and Jak3 double-deficient mice. Int Immunol (2000) 12:123–32. doi:10.1093/intimm/12.2.123
68. Bamford RN, Grant AJ, Burton JD, Peters C, Kurys G, Goldman CK, et al. The interleukin (IL) 2 receptor beta chain is shared by IL-2 and a cytokine, provisionally designated IL-T, that stimulates T-cell proliferation and the induction of lymphokine-activated killer cells. Proc Natl Acad Sci U S A (1994) 91:4940–4. doi:10.1073/pnas.91.11.4940
69. Smith GA, Uchida K, Weiss A, Taunton J. Essential biphasic role for JAK3 catalytic activity in IL-2 receptor signaling. Nat Chem Biol (2016) 12:373–9. doi:10.1038/nchembio.2056
70. Gasteiger G, Hemmers S, Firth MA, Le Floc’h A, Huse M, Sun JC, et al. IL-2-dependent tuning of NK cell sensitivity for target cells is controlled by regulatory T cells. J Exp Med (2013) 210:1167–78. doi:10.1084/jem.20122462
71. Wu Z, Frascaroli G, Bayer C, Schmal T, Mertens T. Interleukin-2 from adaptive T cells enhances natural killer cell activity against human cytomegalovirus-infected macrophages. J Virol (2015) 89:6435–41. doi:10.1128/JVI.00435-15
72. Williams NS, Klem J, Puzanov IJ, Sivakumar PV, Schatzle JD, Bennett M, et al. Natural killer cell differentiation: insights from knockout and transgenic mouse models and in vitro systems. Immunol Rev (1998) 165:47–61. doi:10.1111/j.1600-065X.1998.tb01229.x
73. Vosshenrich CA, Ranson T, Samson SI, Corcuff E, Colucci F, Rosmaraki EE, et al. Roles for common cytokine receptor gamma-chain-dependent cytokines in the generation, differentiation, and maturation of NK cell precursors and peripheral NK cells in vivo. J Immunol (2005) 174:1213–21. doi:10.4049/jimmunol.174.3.1213
74. Gilmour KC, Fujii H, Cranston T, Davies EG, Kinnon C, Gaspar HB. Defective expression of the interleukin-2/interleukin-15 receptor beta subunit leads to a natural killer cell-deficient form of severe combined immunodeficiency. Blood (2001) 98:877–9. doi:10.1182/blood.V98.3.877
75. Kennedy MK, Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med (2000) 191:771–80. doi:10.1084/jem.191.5.771
76. Lodolce JP, Boone DL, Chai S, Swain RE, Dassopoulos T, Trettin S, et al. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity (1998) 9:669–76. doi:10.1016/S1074-7613(00)80664-0
77. Fehniger TA, Suzuki K, Ponnappan A, VanDeusen JB, Cooper MA, Florea SM, et al. Fatal leukemia in interleukin 15 transgenic mice follows early expansions in natural killer and memory phenotype CD8+ T cells. J Exp Med (2001) 193:219–31. doi:10.1084/jem.193.2.219
78. Stonier SW, Schluns KS. Trans-presentation: a novel mechanism regulating IL-15 delivery and responses. Immunol Lett (2010) 127:85–92. doi:10.1016/j.imlet.2009.09.009
79. Dubois S, Mariner J, Waldmann TA, Tagaya Y. IL-15Ralpha recycles and presents IL-15 In trans to neighboring cells. Immunity (2002) 17:537–47. doi:10.1016/S1074-7613(02)00429-6
80. Mortier E, Bernard J, Plet A, Jacques Y. Natural, proteolytic release of a soluble form of human IL-15 receptor alpha-chain that behaves as a specific, high affinity IL-15 antagonist. J Immunol (2004) 173:1681–8. doi:10.4049/jimmunol.173.3.1681
81. Koka R, Burkett PR, Chien M, Chai S, Chan F, Lodolce JP, et al. Interleukin (IL)-15R[alpha]-deficient natural killer cells survive in normal but not IL-15R[alpha]-deficient mice. J Exp Med (2003) 197:977–84. doi:10.1084/jem.20021836
82. Vosshenrich CA, Di Santo JP. Cytokines: IL-21 joins the gamma(c)-dependent network? Curr Biol (2003) 11:R175–7. doi:10.1016/S0960-9822(01)00087-2
83. Skak K, Frederiksen KS, Lundsgaard D. Interleukin-21 activates human natural killer cells and modulates their surface receptor expression. Immunology (2008) 123:575–83. doi:10.1111/j.1365-2567.2007.02730.x
84. Elliott JM, Yokoyama WM. Unifying concepts of MHC-dependent natural killer cell education. Trends Immunol (2011) 32:364–72. doi:10.1016/j.it.2011.06.001
85. Ohlén C, Kling G, Höglund P, Hansson M, Scangos G, Bieberich C, et al. Prevention of allogeneic bone marrow graft rejection by H-2 transgene in donor mice. Science (1989) 246:666–8. doi:10.1126/science.2814488
86. Yu YY, Kumar V, Bennett M. Murine natural killer cells and marrow graft rejection. Annu Rev Immunol (1992) 10:189–213. doi:10.1146/annurev.iy.10.040192.001201
87. Bix M, Liao NS, Zijlstra M, Loring J, Jaenisch R, Raulet D. Rejection of class I MHC-deficient haemopoietic cells by irradiated MHC-matched mice. Nature (1991) 349:329–31. doi:10.1038/349329a0
88. Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D’Andrea A, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity (1997) 7:739–51. doi:10.1016/S1074-7613(00)80393-3
89. Fernandez NC, Treiner E, Vance RE, Jamieson AM, Lemieux S, Raulet DH. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood (2005) 105:4416–23. doi:10.1182/blood-2004-08-3156
90. Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature (2005) 436:709–13. doi:10.1038/nature03847
91. Raulet DH, Vance RE. Self-tolerance of natural killer cells. Nat Rev Immunol (2006) 6:520–31. doi:10.1038/nri1863
92. Peterson ME, Long EO. Inhibitory receptor signaling via tyrosine phosphorylation of the adaptor Crk. Immunity (2008) 29:578–88. doi:10.1016/j.immuni.2008.07.014
93. Brodin P, Hoglund P. Beyond licensing and disarming: a quantitative view on NK-cell education. Eur J Immunol (2008) 38:2934–7. doi:10.1002/eji.200838760
94. Brodin P, Karre K, Hoglund P. NK cell education: not an on-off switch but a tunable rheostat. Trends Immunol (2009) 30:143–9. doi:10.1016/j.it.2009.01.006
95. Rajasekaran K, Riese MJ, Rao S, Wang L, Thakar MS, Sentman CL, et al. Signaling in Effector Lymphocytes: insights toward Safer Immunotherapy. Front Immunol (2016) 7:176. doi:10.3389/fimmu.2016.00176
96. Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol (2008) 9:495–502. doi:10.1038/ni1581
97. Arase H, Suenaga T, Arase N, Kimura Y, Ito K, Shiina R, et al. Negative regulation of expression and function of Fc gamma RIII by CD3 zeta in murine NK cells. J Immunol (2001) 166:21–5. doi:10.4049/jimmunol.166.1.21
98. Augugliaro R, Parolini S, Castriconi R, Marcenaro E, Cantoni C, Nanni M, et al. Selective cross-talk among natural cytotoxicity receptors in human natural killer cells. Eur J Immunol (2003) 33:1235–41. doi:10.1002/eji.200323896
99. May RM, Okumura M, Hsu CJ, Bassiri H, Yang E, Rak G, et al. Murine natural killer immunoreceptors use distinct proximal signaling complexes to direct cell function. Blood (2013) 121:3135–46. doi:10.1182/blood-2012-12-474361
100. Rosen DB, Araki M, Hamerman JA, Chen T, Yamamura T, Lanier LL. A structural basis for the association of DAP12 with mouse, but not human, NKG2D. J Immunol (2004) 173:2470–8. doi:10.4049/jimmunol.173.4.2470
101. Smith KM, Wu J, Bakker AB, Phillips JH, Lanier LL. Ly-49D and Ly-49H associate with mouse DAP12 and form activating receptors. J Immunol (1998) 161:7–10.
102. Lanier LL. DAP10- and DAP12-associated receptors in innate immunity. Immunol Rev (2009) 227:150–60. doi:10.1111/j.1600-065X.2008.00720.x
103. Orr MT, Sun JC, Hesslein DG, Arase H, Phillips JH, Takai T, et al. Ly49H signaling through DAP10 is essential for optimal natural killer cell responses to mouse cytomegalovirus infection. J Exp Med (2009) 206:807–17. doi:10.1084/jem.20090168
104. Lopez-Soto A, Huergo-Zapico L, Acebes-Huerta A, Villa-Alvarez M, Gonzalez S. NKG2D signaling in cancer immunosurveillance. Int J Cancer (2015) 136:1741–50. doi:10.1002/ijc.28775
105. Spear P, Wu MR, Sentman ML, Sentman CL. NKG2D ligands as therapeutic targets. Cancer Immun (2013) 13:8.
106. Sutherland CL, Chalupny NJ, Cosman D. The UL16-binding proteins, a novel family of MHC class I-related ligands for NKG2D, activate natural killer cell functions. Immunol Rev (2001) 181:185–92. doi:10.1034/j.1600-065X.2001.1810115.x
107. Cosman D, Müllberg J, Sutherland CL, Chin W, Armitage R, Fanslow W, et al. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity (2001) 14:123–33. doi:10.1016/S1074-7613(01)00095-4
108. Kubin M, Cassiano L, Chalupny J, Chin W, Cosman D, Fanslow W, et al. ULBP1, 2, 3: novel MHC class I-related molecules that bind to human cytomegalovirus glycoprotein UL16, activate NK cells. Eur J Immunol (2001) 31:1428–37. doi:10.1002/1521-4141(200105)31:5<1428::AID-IMMU1428>3.0.CO;2-4
109. Steinle A, Li P, Morris DL, Groh V, Lanier LL, Strong RK, et al. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics (2001) 53:279–87. doi:10.1007/s002510100325
110. Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci U S A (1996) 93:12445–50. doi:10.1073/pnas.93.22.12445
111. Groh V, Steinle A, Bauer S, Spies T. Recognition of stress-induced MHC molecules by intestinal epithelial gammadelta T cells. Science (1998) 279:1737–40. doi:10.1126/science.279.5357.1737
112. Malarkannan S, Shih PP, Eden PA, Horng T, Zuberi AR, Christianson G, et al. The molecular and functional characterization of a dominant minor H antigen, H60. J Immunol (1998) 161:3501–9.
113. Malarkannan S, Horng T, Eden P, Gonzalez F, Shih P, Brouwenstijn N, et al. Differences that matter: major cytotoxic T cell-stimulating minor histocompatibility antigens. Immunity (2000) 13:333–44. doi:10.1016/S1074-7613(00)00033-9
114. Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat Immunol (2000) 1:119–26. doi:10.1038/77793
115. Cerwenka A, Bakker AB, McClanahan T, Wagner J, Wu J, Phillips JH, et al. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity (2000) 12:721–7. doi:10.1016/S1074-7613(00)80222-8
116. O’Callaghan CA, Cerwenka A, Willcox BE, Lanier LL, Bjorkman PJ. Molecular competition for NKG2D: H60 and RAE1 compete unequally for NKG2D with dominance of H60. Immunity (2001) 15:201–11. doi:10.1016/S1074-7613(01)00187-X
117. Diefenbach A, Jensen ER, Jamieson AM, Raulet DH. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature (2001) 413:165–71. doi:10.1038/35093109
118. Carayannopoulos LN, Naidenko OV, Fremont DH, Yokoyama WM. Cutting edge: murine UL16-binding protein-like transcript 1: a newly described transcript encoding a high-affinity ligand for murine NKG2D. J Immunol (2002) 169:4079–83. doi:10.4049/jimmunol.169.8.4079
119. Diefenbach A, Hsia JK, Hsiung MY, Raulet DH. A novel ligand for the NKG2D receptor activates NK cells and macrophages and induces tumor immunity. Eur J Immunol (2003) 33:381–91. doi:10.1002/immu.200310012
120. Samarakoon A, Chu H, Malarkannan S. Murine NKG2D ligands: "double, double toil and trouble". Mol Immunol (2009) 46:1011–9. doi:10.1016/j.molimm.2008.09.035
121. Awasthi A, Samarakoon A, Dai X, Wen R, Wang D, Malarkannan S. Deletion of PI3K-p85alpha gene impairs lineage commitment, terminal maturation, cytokine generation and cytotoxicity of NK cells. Genes Immun (2008) 9:522–35. doi:10.1038/gene.2008.45
122. Upshaw JL, Schoon RA, Dick CJ, Billadeau DD, Leibson PJ. The isoforms of phospholipase C-gamma are differentially used by distinct human NK activating receptors. J Immunol (2005) 175:213–8. doi:10.4049/jimmunol.175.1.213
123. Kwon HJ, Choi GE, Ryu S, Kwon SJ, Kim SC, Booth C, et al. Stepwise phosphorylation of p65 promotes NF-kappaB activation and NK cell responses during target cell recognition. Nat Commun (2016) 7:11686. doi:10.1038/ncomms11686
124. Rajasekaran K, Kumar P, Schuldt KM, Peterson EJ, Vanhaesebroeck B, Dixit V, et al. Signaling by Fyn-ADAP via the Carma1-Bcl-10-MAP3K7 signalosome exclusively regulates inflammatory cytokine production in NK cells. Nat Immunol (2013) 14:1127–36. doi:10.1038/ni.2708
125. Rajasekaran K, Chu H, Kumar P, Xiao Y, Tinguely M, Samarakoon A, et al. Transforming growth factor-{beta}-activated kinase 1 regulates natural killer cell-mediated cytotoxicity and cytokine production. J Biol Chem (2011) 286:31213–24. doi:10.1074/jbc.M111.261917
126. Cella M, Fujikawa K, Tassi I, Kim S, Latinis K, Nishi S, et al. Differential requirements for Vav proteins in DAP10- and ITAM-mediated NK cell cytotoxicity. J Exp Med (2004) 200:817–23. doi:10.1084/jem.20031847
127. Sutherland CL, Chalupny NJ, Schooley K, VandenBos T, Kubin M, Cosman D. UL16-binding proteins, novel MHC class I-related proteins, bind to NKG2D and activate multiple signaling pathways in primary NK cells. J Immunol (2002) 168:671–9. doi:10.4049/jimmunol.168.2.671
128. Guo H, Samarakoon A, Vanhaesebroeck B, Malarkannan S. The p110 delta of PI3K plays a critical role in NK cell terminal maturation and cytokine/chemokine generation. J Exp Med (2008) 205:2419–35. doi:10.1084/jem.20072327
129. Kim N, Saudemont A, Webb L, Camps M, Ruckle T, Hirsch E, et al. The p110delta catalytic isoform of PI3K is a key player in NK-cell development and cytokine secretion. Blood (2007) 110:3202–8. doi:10.1182/blood-2007-02-075366
130. Tassi I, Cella M, Gilfillan S, Turnbull I, Diacovo TG, Penninger JM, et al. p110gamma and p110delta phosphoinositide 3-kinase signaling pathways synergize to control development and functions of murine NK cells. Immunity (2007) 27:214–27. doi:10.1016/j.immuni.2007.07.014
131. Giurisato E, Cella M, Takai T, Kurosaki T, Feng Y, Longmore GD, et al. Phosphatidylinositol 3-kinase activation is required to form the NKG2D immunological synapse. Mol Cell Biol (2007) 27:8583–99. doi:10.1128/MCB.01477-07
132. Stabile H, Fionda C, Gismondi A, Santoni A. Role of distinct natural killer cell subsets in anticancer response. Front Immunol (2017) 8:293. doi:10.3389/fimmu.2017.00293
133. Smyth MJ, Cretney E, Kelly JM, Westwood JA, Street SE, Yagita H, et al. Activation of NK cell cytotoxicity. Mol Immunol (2005) 42:501–10. doi:10.1016/j.molimm.2004.07.034
134. Fauriat C, Long EO, Ljunggren HG, Bryceson YT. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood (2010) 115:2167–76. doi:10.1182/blood-2009-08-238469
135. Freeman BE, Raue HP, Hill AB, Slifka MK. Cytokine-mediated activation of NK cells during viral infection. J Virol (2015) 89:7922–31. doi:10.1128/JVI.00199-15
136. Krzewski K, Strominger JL. The killer’s kiss: the many functions of NK cell immunological synapses. Curr Opin Cell Biol (2008) 20:597–605. doi:10.1016/j.ceb.2008.05.006
137. Khosravi-Far R, Esposti MD. Death receptor signals to mitochondria. Cancer Biol Ther (2004) 3:1051–7. doi:10.4161/cbt.3.11.1173
138. Screpanti V, Wallin RP, Ljunggren HG, Grandien A. A central role for death receptor-mediated apoptosis in the rejection of tumors by NK cells. J Immunol (2001) 167:2068–73. doi:10.4049/jimmunol.167.4.2068
139. Nagata S, Golstein P. The Fas death factor. Science (1995) 267:1449–56. doi:10.1126/science.7533326
140. Degli-Esposti M. To die or not to die – the quest of the TRAIL receptors. J Leukoc Biol (1999) 65:535–42. doi:10.1002/jlb.65.5.535
141. Guicciardi ME, Gores GJ. Life and death by death receptors. FASEB J (2009) 23:1625–37. doi:10.1096/fj.08-111005
142. Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science (1998) 281:1305–8. doi:10.1126/science.281.5381.1305
144. Crowder RN, El-Deiry WS. Caspase-8 regulation of TRAIL-mediated cell death. Exp Oncol (2012) 34:160–4.
145. Wolf BB, Schuler M, Echeverri F, Green DR. Caspase-3 is the primary activator of apoptotic DNA fragmentation via DNA fragmentation factor-45/inhibitor of caspase-activated DNase inactivation. J Biol Chem (1999) 274:30651–6. doi:10.1074/jbc.274.43.30651
146. Bao Q, Shi Y. Apoptosome: a platform for the activation of initiator caspases. Cell Death Differ (2007) 14:56–65. doi:10.1038/sj.cdd.4402028
147. Bhat R, Watzl C. Serial killing of tumor cells by human natural killer cells – enhancement by therapeutic antibodies. PLoS One (2007) 2:e326. doi:10.1371/journal.pone.0000326
148. Mace EM, Wu WW, Ho T, Mann SS, Hsu HT, Orange JS. NK cell lytic granules are highly motile at the immunological synapse and require F-actin for post-degranulation persistence. J Immunol (2012) 189:4870–80. doi:10.4049/jimmunol.1201296
149. Mizesko MC, Banerjee PP, Monaco-Shawver L, Mace EM, Bernal WE, Sawalle-Belohradsky J, et al. Defective actin accumulation impairs human natural killer cell function in patients with dedicator of cytokinesis 8 deficiency. J Allergy Clin Immunol (2013) 131:840–8. doi:10.1016/j.jaci.2012.12.1568
150. Chen X, Trivedi PP, Ge B, Krzewski K, Strominger JL. Many NK cell receptors activate ERK2 and JNK1 to trigger microtubule organizing center and granule polarization and cytotoxicity. Proc Natl Acad Sci U S A (2007) 104:6329–34. doi:10.1073/pnas.0611655104
151. Stinchcombe JC, Majorovits E, Bossi G, Fuller S, Griffiths GM. Centrosome polarization delivers secretory granules to the immunological synapse. Nature (2006) 443:462–5. doi:10.1038/nature05071
152. Osinska I, Popko K, Demkow U. Perforin: an important player in immune response. Cent Eur J Immunol (2014) 39:109–15. doi:10.5114/ceji.2014.42135
153. Gwalani LA, Orange JS. Single degranulations in NK cells can mediate target cell killing. J Immunol (2018) 200:3231–43. doi:10.4049/jimmunol.1701500
154. Atkinson EA, Barry M, Darmon AJ, Shostak I, Turner PC, Moyer RW, et al. Cytotoxic T lymphocyte-assisted suicide. Caspase 3 activation is primarily the result of the direct action of granzyme B. J Biol Chem (1998) 273:21261–6. doi:10.1074/jbc.273.33.21261
155. Barry M, Heibein JA, Pinkoski MJ, Lee SF, Moyer RW, Green DR, et al. Granzyme B short-circuits the need for caspase 8 activity during granule-mediated cytotoxic T-lymphocyte killing by directly cleaving Bid. Mol Cell Biol (2000) 20:3781–94. doi:10.1128/MCB.20.11.3781-3794.2000
156. Pinkoski MJ, Waterhouse NJ, Heibein JA, Wolf BB, Kuwana T, Goldstein JC, et al. Granzyme B-mediated apoptosis proceeds predominantly through a Bcl-2-inhibitable mitochondrial pathway. J Biol Chem (2001) 276:12060–7. doi:10.1074/jbc.M009038200
157. Reefman E, Kay JG, Wood SM, Offenhäuser C, Brown DL, Roy S, et al. Cytokine secretion is distinct from secretion of cytotoxic granules in NK cells. J Immunol (2001) 184:4852–62. doi:10.4049/jimmunol.0803954
158. Kiniwa T, Enomoto Y, Terazawa N, Omi A, Miyata N, Ishiwata K, et al. NK cells activated by Interleukin-4 in cooperation with Interleukin-15 exhibit distinctive characteristics. Proc Natl Acad Sci U S A (2016) 113:10139–44. doi:10.1073/pnas.1600112113
159. Vivier E, Ugolini S. Regulatory natural killer cells: new players in the IL-10 anti-inflammatory response. Cell Host Microbe (2009) 6:493–5. doi:10.1016/j.chom.2009.12.001
160. Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? The example of natural killer cells. Science (2011) 331:44–9. doi:10.1126/science.1198687
161. Cook KD, Waggoner SN, Whitmire JK. NK cells and their ability to modulate T cells during virus infections. Crit Rev Immunol (2014) 34:359–88. doi:10.1615/CritRevImmunol.2014010604
162. Blanchard DK, Michelini-Norris MB, Djeu JY. Production of granulocyte-macrophage colony-stimulating factor by large granular lymphocytes stimulated with Candida albicans: role in activation of human neutrophil function. Blood (1991) 77:2259–65.
163. van den Bosch G, Preijers F, Vreugdenhil A, Hendriks J, Maas F, De Witte T. Granulocyte-macrophage colony-stimulating factor (GM-CSF) counteracts the inhibiting effect of monocytes on natural killer (NK) cells. Clin Exp Immunol (1995) 101:515–20. doi:10.1111/j.1365-2249.1995.tb03143.x
164. Walzer T, Dalod M, Robbins SH, Zitvogel L, Vivier E. Natural-killer cells and dendritic cells: "l’union fait la force". Blood (2005) 106:2252–8. doi:10.1182/blood-2005-03-1154
165. Gotthardt D, Sexl V. STATs in NK-cells: the good, the bad, and the ugly. Front Immunol (2017) 7:694. doi:10.3389/fimmu.2016.00694
166. Tato CM, Villarino A, Caamano JH, Boothby M, Hunter CA. Inhibition of NF-kappa B activity in T and NK cells results in defective effector cell expansion and production of IFN-gamma required for resistance to Toxoplasma gondii. J Immunol (2003) 170:3139–46. doi:10.4049/jimmunol.170.6.3139
167. Tassi I, Cella M, Presti R, Colucci A, Gilfillan S, Littman DR, et al. NK cell-activating receptors require PKC-theta for sustained signaling, transcriptional activation, and IFN-gamma secretion. Blood (2008) 112:4109–16. doi:10.1182/blood-2008-02-139527
168. Sica A, Dorman L, Viggiano V, Cippitelli M, Ghosh P, Rice N, et al. Interaction of NF-kappaB and NFAT with the interferon-gamma promoter. J Biol Chem (1997) 272:30412–20. doi:10.1074/jbc.272.48.30412
169. Zhang F, Wang DZ, Boothby M, Penix L, Flavell RA, Aune TM. Regulation of the activity of IFN-gamma promoter elements during Th cell differentiation. J Immunol (1998) 161:6105–12.
170. Long EO. Ready for prime time: NK cell priming by dendritic cells. Immunity (2007) 26:385–7. doi:10.1016/j.immuni.2007.04.001
171. Lucas M, Schachterle W, Oberle K, Aichele P, Diefenbach A. Dendritic cells prime natural killer cells by trans-presenting interleukin 15. Immunity (2007) 26:503–17. doi:10.1016/j.immuni.2007.03.006
172. Degli-Esposti MA, Smyth MJ. Close encounters of different kinds: dendritic cells and NK cells take centre stage. Nat Rev Immunol (2005) 5:112–24. doi:10.1038/nri1549
173. Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol (1999) 17:189–220. doi:10.1146/annurev.immunol.17.1.189
174. Koka R, Burkett P, Chien M, Chai S, Boone DL, Ma A. Cutting edge: murine dendritic cells require IL-15R alpha to prime NK cells. J Immunol (2004) 173:3594–8. doi:10.4049/jimmunol.173.6.3594
175. Vignali DA, Kuchroo VK. IL-12 family cytokines: immunological playmakers. Nat Immunol (2012) 13:722–8. doi:10.1038/ni.2366
176. Cox JH, Kljavin NM, Ramamoorthi N, Diehl L, Batten M, Ghilardi N. IL-27 promotes T cell-dependent colitis through multiple mechanisms. J Exp Med (2011) 208:115–23. doi:10.1084/jem.20100410
177. Hunter CA. New IL-12-family members: IL-23 and IL-27, cytokines with divergent functions. Nat Rev Immunol (2005) 5:521–31. doi:10.1038/nri1648
178. Collison LW, Vignali DA. Interleukin-35: odd one out or part of the family? Immunol Rev (2008) 226:248–62. doi:10.1111/j.1600-065X.2008.00704.x
179. Presky DH, Yang H, Minetti LJ, Chua AO, Nabavi N, Wu CY, et al. A functional interleukin 12 receptor complex is composed of two beta-type cytokine receptor subunits. Proc Natl Acad Sci U S A (1996) 93:14002–7. doi:10.1073/pnas.93.24.14002
180. Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature (1996) 382:171–4. doi:10.1038/382171a0
181. Lusty E, Poznanski SM, Kwofie K, Mandur TS, Lee DA, Richards CD, et al. IL-18/IL-15/IL-12 synergy induces elevated and prolonged IFN-gamma production by ex vivo expanded NK cells which is not due to enhanced STAT4 activation. Mol Immunol (2017) 88:138–47. doi:10.1016/j.molimm.2017.06.025
182. Lee JK, Kim SH, Lewis EC, Azam T, Reznikov LL, Dinarello CA. Differences in signaling pathways by IL-1beta and IL-18. Proc Natl Acad Sci U S A (2004) 101:8815–20. doi:10.1073/pnas.0402800101
183. Klekotka PA, Yang L, Yokoyama WM. Contrasting roles of the IL-1 and IL-18 receptors in MyD88-dependent contact hypersensitivity. J Invest Dermatol (2010) 130:184–91. doi:10.1038/jid.2009.242
184. Yoshimoto T, Takeda K, Tanaka T, Ohkusu K, Kashiwamura S, Okamura H, et al. IL-12 up-regulates IL-18 receptor expression on T cells, Th1 cells, and B cells: synergism with IL-18 for IFN-gamma production. J Immunol (1998) 161:3400–7.
185. Mavropoulos A, Sully G, Cope AP, Clark AR. Stabilization of IFN-gamma mRNA by MAPK p38 in IL-12- and IL-18-stimulated human NK cells. Blood (2005) 105:282–8. doi:10.1182/blood-2004-07-2782
186. Nakahira M, Ahn HJ, Park WR, Gao P, Tomura M, Park CS, et al. Synergy of IL-12 and IL-18 for IFN-gamma gene expression: IL-12-induced STAT4 contributes to IFN-gamma promoter activation by up-regulating the binding activity of IL-18-induced activator protein 1. J Immunol (2002) 168:1146–53. doi:10.4049/jimmunol.168.3.1146
187. Orange JS. Natural killer cell deficiency. J Allergy Clin Immunol (2013) 132:515–25. doi:10.1016/j.jaci.2013.07.020
188. Zamora AE, Grossenbacher SK, Aguilar EG, Murphy WJ. Models to study NK cell biology and possible clinical application. Curr Protoc Immunol (2015) 110:14.37.1–14. doi:10.1002/0471142735.im1437s110
189. Poggi A, Zocchi MR. NK cell autoreactivity and autoimmune diseases. Front Immunol (2014) 5:27. doi:10.3389/fimmu.2014.00027
190. Jewett A, Man YG, Tseng HC. Dual functions of natural killer cells in selection and differentiation of stem cells; role in regulation of inflammation and regeneration of tissues. J Cancer (2013) 4:12–24. doi:10.7150/jca.5519
191. Kumar P, Thakar MS, Ouyang W, Malarkannan S. IL-22 from conventional NK cells is epithelial regenerative and inflammation protective during influenza infection. Mucosal Immunol (2013) 6:69–82. doi:10.1038/mi.2012.49
192. Souza-Fonseca-Guimaraes F, Adib-Conquy M, Cavaillon JM. Natural killer (NK) cells in antibacterial innate immunity: angels or devils? Mol Med (2012) 18:270–85. doi:10.2119/molmed.2011.00201
193. Dillon SM, Lee EJ, Bramante JM, Barker E, Wilson CC. The natural killer cell interferon-gamma response to bacteria is diminished in untreated HIV-1 infection and defects persist despite viral suppression. J Acquir Immune Defic Syndr (2014) 65:259–67. doi:10.1097/01.qai.0000435603.50598.2b
194. Schmidt S, Zimmermann SY, Tramsen L, Koehl U, Lehrnbecher T. Natural killer cells and antifungal host response. Clin Vaccine Immunol (2013) 20:452–8. doi:10.1128/CVI.00606-12
195. Ma LL, Wang CL, Neely GG, Epelman S, Krensky AM, Mody CH. NK cells use perforin rather than granulysin for anticryptococcal activity. J Immunol (2004) 173:3357–65. doi:10.4049/jimmunol.173.5.3357
196. Voigt J, Hünniger K, Bouzani M, Jacobsen ID, Barz D, Hube B, et al. Human natural killer cells acting as phagocytes against Candida albicans and mounting an inflammatory response that modulates neutrophil antifungal activity. J Infect Dis (2014) 209:616–26. doi:10.1093/infdis/jit574
197. Bar E, Whitney PG, Moor K, Reis e Sousa C, LeibundGut-Landmann S. IL-17 regulates systemic fungal immunity by controlling the functional competence of NK cells. Immunity (2014) 40:117–27. doi:10.1016/j.immuni.2013.12.002
198. Antoniou AN, Powis SJ. Pathogen evasion strategies for the major histocompatibility complex class I assembly pathway. Immunology (2008) 124:1–12. doi:10.1111/j.1365-2567.2008.02804.x
199. Pratheek BM, Nayak TK, Sahoo SS, Mohanty PK, Chattopadhyay S, Chakraborty NG, et al. Mammalian non-classical major histocompatibility complex I and its receptors: important contexts of gene, evolution, and immunity. Indian J Hum Genet (2014) 20:129–41. doi:10.4103/0971-6866.142855
200. Khanna R. Tumour surveillance: missing peptides and MHC molecules. Immunol Cell Biol (1998) 76:20–6. doi:10.1046/j.1440-1711.1998.00717.x
201. Rock KL, Reits E, Neefjes J. Present yourself! By MHC class I and MHC class II molecules. Trends Immunol (2016) 37:724–37. doi:10.1016/j.it.2016.08.010
202. Schwartz RH. Historical overview of immunological tolerance. Cold Spring Harb Perspect Biol (2012) 4:a006908. doi:10.1101/cshperspect.a006908
203. Shegarfi H, Sydnes K, Løvik M, Inngjerdingen M, Rolstad B, Naper C. The role of natural killer cells in resistance to the intracellular bacterium Listeria monocytogenes in rats. Scand J Immunol (2009) 70:238–44. doi:10.1111/j.1365-3083.2009.02292.x
204. Unanue ER. Studies in listeriosis show the strong symbiosis between the innate cellular system and the T-cell response. Immunol Rev (1997) 158:11–25. doi:10.1111/j.1600-065X.1997.tb00988.x
205. Tay CH, Welsh RM. Distinct organ-dependent mechanisms for the control of murine cytomegalovirus infection by natural killer cells. J Virol (1997) 71:267–75.
206. Ge MQ, Ho AW, Tang Y, Wong KH, Chua BY, Gasser S, et al. NK cells regulate CD8+ T cell priming and dendritic cell migration during influenza A infection by IFN-gamma and perforin-dependent mechanisms. J Immunol (2012) 189:2099–109. doi:10.4049/jimmunol.1103474
207. Werner JM, Serti E, Chepa-Lotrea X, Stoltzfus J, Ahlenstiel G, Noureddin M, et al. Ribavirin improves the IFN-gamma response of natural killer cells to IFN-based therapy of hepatitis C virus infection. Hepatology (2014) 60:1160–9. doi:10.1002/hep.27092
208. Gazit R, Gruda R, Elboim M, Arnon TI, Katz G, Achdout H, et al. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol (2006) 7:517–23. doi:10.1038/ni1322
209. Glasner A, Zurunic A, Meningher T, Lenac Rovis T, Tsukerman P, Bar-On Y, et al. Elucidating the mechanisms of influenza virus recognition by Ncr1. PLoS One (2012) 7:e36837. doi:10.1371/journal.pone.0036837
210. Adams EJ, Juo ZS, Venook RT, Boulanger MJ, Arase H, Lanier LL, et al. Structural elucidation of the m157 mouse cytomegalovirus ligand for Ly49 natural killer cell receptors. Proc Natl Acad Sci U S A (2007) 104:10128–33. doi:10.1073/pnas.0703735104
211. Bubic I, Wagner M, Krmpotić A, Saulig T, Kim S, Yokoyama WM, et al. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J Virol (2004) 78:7536–44. doi:10.1128/JVI.78.14.7536-7544.2004
212. Cheng TP, French AR, Plougastel BF, Pingel JT, Orihuela MM, Buller ML, et al. Ly49h is necessary for genetic resistance to murine cytomegalovirus. Immunogenetics (2008) 60:565–73. doi:10.1007/s00251-008-0313-3
213. Fodil-Cornu N, Lee SH, Belanger S, Makrigiannis AP, Biron CA, Buller RM, et al. Ly49h-deficient C57BL/6 mice: a new mouse cytomegalovirus-susceptible model remains resistant to unrelated pathogens controlled by the NK gene complex. J Immunol (2008) 181:6394–405. doi:10.4049/jimmunol.181.9.6394
214. Bahram S, Inoko H, Shiina T, Radosavljevic M. MIC and other NKG2D ligands: from none to too many. Curr Opin Immunol (2005) 17:505–9. doi:10.1016/j.coi.2005.07.016
215. Dunn C, Chalupny NJ, Sutherland CL, Dosch S, Sivakumar PV, Johnson DC, et al. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J Exp Med (2005) 197:1427–39. doi:10.1084/jem.20022059
216. Welte SA, Sinzger C, Lutz SZ, Singh-Jasuja H, Sampaio KL, Eknigk U, et al. Selective intracellular retention of virally induced NKG2D ligands by the human cytomegalovirus UL16 glycoprotein. Eur J Immunol (2003) 33:194–203. doi:10.1002/immu.200390022
217. Wu J, Chalupny NJ, Manley TJ, Riddell SR, Cosman D, Spies T. Intracellular retention of the MHC class I-related chain B ligand of NKG2D by the human cytomegalovirus UL16 glycoprotein. J Immunol (2003) 170:4196–200. doi:10.4049/jimmunol.170.8.4196
218. Ljunggren HG, Karre K. In search of the ’missing self’: MHC molecules and NK cell recognition. Immunol Today (1990) 11:237–44. doi:10.1016/0167-5699(90)90097-S
219. Jonjic S, Babic M, Polic B, Krmpotic A. Immune evasion of natural killer cells by viruses. Curr Opin Immunol (2008) 20:30–8. doi:10.1016/j.coi.2007.11.002
220. Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature (2009) 457:557–61. doi:10.1038/nature07665
221. O’Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol (2006) 7:507–16. doi:10.1038/ni1332
222. Sun JC, Beilke JN, Lanier LL. Immune memory redefined: characterizing the longevity of natural killer cells. Immunol Rev (2010) 236:83–94. doi:10.1111/j.1600-065X.2010.00900.x
223. Kamimura Y, Lanier LL. Homeostatic control of memory cell progenitors in the natural killer cell lineage. Cell Rep (2015) 10:280–91. doi:10.1016/j.celrep.2014.12.025
224. O’Sullivan TE, Sun JC, Lanier LL. Natural killer cell memory. Immunity (2015) 43:634–45. doi:10.1016/j.immuni.2015.09.013
225. Nabekura T, Lanier LL. Tracking the fate of antigen-specific versus cytokine-activated natural killer cells after cytomegalovirus infection. J Exp Med (2016) 213:2745–58. doi:10.1084/jem.20160726
226. Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B, et al. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol (2010) 11:1127–35. doi:10.1038/ni.1953
227. Abdul-Careem MF, Lee AJ, Pek EA, Gill N, Gillgrass AE, Chew MV, et al. Genital HSV-2 infection induces short-term NK cell memory. PLoS One (2012) 7:e32821. doi:10.1371/journal.pone.0032821
228. Gillard GO, Bivas-Benita M, Hovav AH, Grandpre LE, Panas MW, Seaman MS, et al. Thy1+ NK [corrected] cells from vaccinia virus-primed mice confer protection against vaccinia virus challenge in the absence of adaptive lymphocytes. PLoS Pathog (2011) 7:e1002141. doi:10.1371/journal.ppat.1002141
229. Muccio L, Bertaina A, Falco M, Pende D, Meazza R, Lopez-Botet M, et al. Analysis of memory-like natural killer cells in human cytomegalovirus-infected children undergoing alphabeta+T and B cell-depleted hematopoietic stem cell transplantation for hematological malignancies. Haematologica (2016) 101:371–81. doi:10.3324/haematol.2015.134155
230. Newhook N, Fudge N, Grant M. NK cells generate memory-type responses to human cytomegalovirus-infected fibroblasts. Eur J Immunol (2017) 47:1032–9. doi:10.1002/eji.201646819
231. Horowitz A, Stegmann KA, Riley EM. Activation of natural killer cells during microbial infections. Front Immunol (2012) 2:88. doi:10.3389/fimmu.2011.00088
232. Chowdhury D, Lieberman J. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu Rev Immunol (2008) 26:389–420. doi:10.1146/annurev.immunol.26.021607.090404
233. Walch M, Dotiwala F, Mulik S, Thiery J, Kirchhausen T, Clayberger C, et al. Cytotoxic cells kill intracellular bacteria through granulysin-mediated delivery of granzymes. Cell (2015) 161:1229. doi:10.1016/j.cell.2015.05.021
234. Jacquemin G, Margiotta D, Kasahara A, Bassoy EY, Walch M, Thiery J, et al. Granzyme B-induced mitochondrial ROS are required for apoptosis. Cell Death Differ (2015) 22:862–74. doi:10.1038/cdd.2014.180
235. Dotiwala F, Sen Santara S, Binker-Cosen AA, Li B, Chandrasekaran S, Lieberman J. Granzyme B disrupts central metabolism and protein synthesis in bacteria to promote an immune cell death program. Cell (2017) 171:1125–37.e11. doi:10.1016/j.cell.2017.10.004
236. Teixeira HC, Kaufmann SH. Role of NK1.1+ cells in experimental listeriosis. NK1+ cells are early IFN-gamma producers but impair resistance to Listeria monocytogenes infection. J Immunol (1994) 152:1873–82.
237. Thale C, Kiderlen AF. Sources of interferon-gamma (IFN-gamma) in early immune response to Listeria monocytogenes. Immunobiology (2005) 210:673–83. doi:10.1016/j.imbio.2005.07.003
238. Walch M, Dotiwala F, Mulik S, Thiery J, Kirchhausen T, Clayberger C, et al. Cytotoxic cells kill intracellular bacteria through granulysin-mediated delivery of granzymes. Cell (2014) 157:1309–23. doi:10.1016/j.cell.2014.03.062
239. Haller D, Serrant P, Granato D, Schiffrin EJ, Blum S. Activation of human NK cells by staphylococci and lactobacilli requires cell contact-dependent costimulation by autologous monocytes. Clin Diagn Lab Immunol (2002) 9:649–57.
240. Feng CG, Kaviratne M, Rothfuchs AG, Cheever A, Hieny S, Young HA, et al. NK cell-derived IFN-gamma differentially regulates innate resistance and neutrophil response in T cell-deficient hosts infected with Mycobacterium tuberculosis. J Immunol (2006) 177:7086–93. doi:10.4049/jimmunol.177.10.7086
241. Esin S, Batoni G, Pardini M, Favilli F, Bottai D, Maisetta G, et al. Functional characterization of human natural killer cells responding to Mycobacterium bovis bacille Calmette-Guerin. Immunology (2004) 112:143–52. doi:10.1111/j.1365-2567.2004.01858.x
242. van de WD, De Paus RA, van Dissel JT, van de Vosse E. Salmonella induced IL-23 and IL-1beta allow for IL-12 production by monocytes and Mphi1 through induction of IFN-gamma in CD56 NK/NK-like T cells. PLoS One (2009) 4:e8396. doi:10.1371/journal.pone.0008396
243. Humann J, Lenz LL. Activation of naive NK cells in response to Listeria monocytogenes requires IL-18 and contact with infected dendritic cells. J Immunol (2010) 184:5172–8. doi:10.4049/jimmunol.0903759
244. Klezovich-Bénard M, Corre JP, Jusforgues-Saklani H, Fiole D, Burjek N, Tournier JN, et al. Mechanisms of NK cell-macrophage Bacillus anthracis crosstalk: a balance between stimulation by spores and differential disruption by toxins. PLoS Pathog (2012) 8:e1002481. doi:10.1371/journal.ppat.1002481
245. Ziblat A, Domaica CI, Spallanzani RG, Iraolagoitia XL, Rossi LE, Avila DE, et al. IL-27 stimulates human NK-cell effector functions and primes NK cells for IL-18 responsiveness. Eur J Immunol (2015) 45:192–202. doi:10.1002/eji.201444699
246. Zwirner NW, Ziblat A. Regulation of NK cell activation and effector functions by the IL-12 family of cytokines: the case of IL-27. Front Immunol (2017) 8:25. doi:10.3389/fimmu.2017.00025
247. Ljunggren HG, Karre K. Host resistance directed selectively against H-2-deficient lymphoma variants. Analysis of the mechanism. J Exp Med (1985) 162:1745–59. doi:10.1084/jem.162.6.1745
248. Seaman WE, Sleisenger M, Eriksson E, Koo GC. Depletion of natural killer cells in mice by monoclonal antibody to NK-1.1. Reduction in host defense against malignancy without loss of cellular or humoral immunity. J Immunol (1987) 138:4539–44.
249. Heerboth S, Housman G, Leary M, Longacre M, Byler S, Lapinska K, et al. EMT and tumor metastasis. Clin Transl Med (2015) 4:6. doi:10.1186/s40169-015-0048-3
250. Algarra I, Cabrera T, Garrido F. The HLA crossroad in tumor immunology. Hum Immunol (2000) 61:65–73. doi:10.1016/S0198-8859(99)00156-1
251. French AR, Yokoyama WM. Natural killer cells and viral infections. Curr Opin Immunol (2003) 15:45–51. doi:10.1016/S095279150200002X
252. Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science (1999) 285:727–9. doi:10.1126/science.285.5428.727
253. Diefenbach A, Raulet DH. Strategies for target cell recognition by natural killer cells. Immunol Rev (2001) 181:170–84. doi:10.1034/j.1600-065X.2001.1810114.x
254. Raulet DH. Interplay of natural killer cells and their receptors with the adaptive immune response. Nat Immunol (2004) 5:996–1002. doi:10.1038/ni1114
255. Waldhauer I, Steinle A. NK cells and cancer immunosurveillance. Oncogene (2008) 27:5932–43. doi:10.1038/onc.2008.267
256. Malarkannan S. The balancing act: inhibitory Ly49 regulate NKG2D-mediated NK cell functions. Semin Immunol (2006) 18:186–92. doi:10.1016/j.smim.2006.04.002
257. Imai K, Matsuyama S, Miyake S, Suga K, Nakachi K. Natural cytotoxic activity of peripheral-blood lymphocytes and cancer incidence: an 11-year follow-up study of a general population. Lancet (2000) 356:1795–9. doi:10.1016/S0140-6736(00)03231-1
258. Coca S, Perez-Piqueras J, Martinez D, Colmenarejo A, Saez MA, Vallejo C, et al. The prognostic significance of intratumoral natural killer cells in patients with colorectal carcinoma. Cancer (1997) 79:2320–8. doi:10.1002/(SICI)1097-0142(19970615)79:12<2320::AID-CNCR5>3.0.CO;2-P
259. Ishigami S, Natsugoe S, Tokuda K, Nakajo A, Che X, Iwashige H, et al. Prognostic value of intratumoral natural killer cells in gastric carcinoma. Cancer (2000) 88:577–83. doi:10.1002/(SICI)1097-0142(20000201)88:3<577::AID-CNCR13>3.0.CO;2-V
260. Villegas FR, Coca S, Villarrubia VG, Jiménez R, Chillón MJ, Jareño J, et al. Prognostic significance of tumor infiltrating natural killer cells subset CD57 in patients with squamous cell lung cancer. Lung Cancer (2002) 35:23–8. doi:10.1016/S0169-5002(01)00292-6
261. Torelli GF, Peragine N, Raponi S, Pagliara D, De Propris MS, Vitale A, et al. Recognition of adult and pediatric acute lymphoblastic leukemia blasts by natural killer cells. Haematologica (2014) 99:1248–54. doi:10.3324/haematol.2013.101931
262. Handgretinger R, Lang P, Andre MC. Exploitation of natural killer cells for the treatment of acute leukemia. Blood (2016) 127:3341–9. doi:10.1182/blood-2015-12-629055
263. Mehta RS, Randolph B, Daher M, Rezvani K. NK cell therapy for hematologic malignancies. Int J Hematol (2018) 107:262–70. doi:10.1007/s12185-018-2407-5
264. Lim O, Jung MY, Hwang YK, Shin EC. Present and future of allogeneic natural killer cell therapy. Front Immunol (2015) 6:286. doi:10.3389/fimmu.2015.00286
265. Margolin KA, Negrin RS, Wong KK, Chatterjee S, Wright C, Forman SJ. Cellular immunotherapy and autologous transplantation for hematologic malignancy. Immunol Rev (1997) 157:231–40. doi:10.1111/j.1600-065X.1997.tb00986.x
266. Parkhurst MR, Riley JP, Dudley ME, Rosenberg SA. Adoptive transfer of autologous natural killer cells leads to high levels of circulating natural killer cells but does not mediate tumor regression. Clin Cancer Res (2011) 17(19):6287–97. doi:10.1158/1078-0432.CCR-11-1347
267. Björklund AT, Carlsten M, Sohlberg E, Liu LL, Clancy T, Karimi M, et al. Complete remission with reduction of high-risk clones following haploidentical NK-cell therapy against MDS and AML. Clin Cancer Res (2018) 24(8):1834–44. doi:10.1158/1078-0432.CCR-17-3196
268. Van Elssen C, Ciurea SO. NK cell therapy after hematopoietic stem cell transplantation: can we improve anti-tumor effect? Int J Hematol (2018) 107:151–6. doi:10.1007/s12185-017-2379-x
269. Hosono A, Makimoto A, Kawai A, Takaue Y. Segregated graft-versus-tumor effect between CNS and non-CNS lesions of Ewing’s sarcoma family of tumors. Bone Marrow Transplant (2008) 41:1067–8. doi:10.1038/bmt.2008.26
270. Lucas KG, Schwartz C, Kaplan J. Allogeneic stem cell transplantation in a patient with relapsed Ewing sarcoma. Pediatr Blood Cancer (2008) 51:142–4. doi:10.1002/pbc.21503
271. Burdach S, van Kaick B, Laws HJ, Ahrens S, Haase R, Körholz D, et al. Allogeneic and autologous stem-cell transplantation in advanced Ewing tumors. An update after long-term follow-up from two centers of the European Intergroup study EICESS. Stem-cell transplant programs at Dusseldorf university medical center, Germany and St. Anna Kinderspital, Vienna, Austria. Ann Oncol (2000) 11:1451–62.
272. Goi K, Sugita K, Tezuka T, Sato H, Uno K, Inukai T, et al. A successful case of allogeneic bone marrow transplantation for osteosarcoma with multiple metastases of lung and bone. Bone Marrow Transplant (2006) 37:115–6. doi:10.1038/sj.bmt.1705209
273. Fagioli F, Berger M, Brach del Prever A, Lioji S, Aglietta M, Ferrari S, et al. Regression of metastatic osteosarcoma following non-myeloablative stem cell transplantation. A case report. Haematologica (2003) 88:ECR16.
274. Goodwin A, Gurney H, Gottlieb D. Allogeneic bone marrow transplant for refractory mediastinal germ cell tumour: possible evidence of graft-versus-tumour effect. Intern Med J (2007) 37:127–9. doi:10.1111/j.1445-5994.2007.01244.x
275. Doelken R, Weigel S, Schueler F, Doelken G, Beck JF. Poor outcome of two children with relapsed state stage IV alveolar rhabdomyosarcoma after allogeneic stem cell transplantation. Pediatr Hematol Oncol (2005) 22:699–703. doi:10.1080/08880010500278806
276. Misawa A, Hosoi H, Tsuchiya K, Iehara T, Sawada T, Sugimoto T. Regression of refractory rhabdomyosarcoma after allogeneic stem-cell transplantation. Pediatr Hematol Oncol (2003) 20:151–5. doi:10.1080/0880010390158658
277. Ohta H, Hashii Y, Yoshida H, Kusuki S, Tokimasa S, Yoneda A, et al. Allogeneic hematopoietic stem cell transplantation against recurrent rhabdomyosarcoma. J Pediatr Hematol Oncol (2011) 33:e35–8. doi:10.1097/MPH.0b013e3181e7ddc5
278. Sung KW, Park JE, Chueh HW, Lee SH, Yoo KH, Koo HH, et al. Reduced-intensity allogeneic stem cell transplantation for children with neuroblastoma who failed tandem autologous stem cell transplantation. Pediatr Blood Cancer (2011) 57:660–5. doi:10.1002/pbc.23035
279. Jubert C, Wall DA, Grimley M, Champagne MA, Duval M. Engraftment of unrelated cord blood after reduced-intensity conditioning regimen in children with refractory neuroblastoma: a feasibility trial. Bone Marrow Transplant (2011) 46:232–7. doi:10.1038/bmt.2010.107
280. Ash S, Gigi V, Askenasy N, Fabian I, Stein J, Yaniv I. Graft versus neuroblastoma reaction is efficiently elicited by allogeneic bone marrow transplantation through cytolytic activity in the absence of GVHD. Cancer Immunol Immunother (2009) 58:2073–84. doi:10.1007/s00262-009-0715-6
281. Lucas KG, Shapiro T, Freiberg A, Frauenhoffer E. Matched unrelated umbilical cord blood transplantation for a patient with chemotherapy resistant Wilms tumor. Pediatr Blood Cancer (2010) 55:763–5. doi:10.1002/pbc.22635
282. Aoyama Y, Yamamura R, Shima E, Nakamae H, Makita K, Kho G, et al. Successful treatment with reduced-intensity stem cell transplantation in a case of relapsed refractory central nervous system lymphoma. Ann Hematol (2003) 82:371–3. doi:10.1007/s00277-003-0651-z
283. Ichise H, Nagano S, Maeda T, Miyazaki M, Miyazaki Y, Kojima H, et al. NK cell alloreactivity against KIR-ligand-mismatched HLA-haploidentical tissue derived from HLA haplotype-homozygous iPSCs. Stem Cell Reports (2017) 9:853–67. doi:10.1016/j.stemcr.2017.07.020
284. Fang F, Xiao W, Tian Z. NK cell-based immunotherapy for cancer. Semin Immunol (2017) 31:37–54. doi:10.1016/j.smim.2017.07.009
285. Schuster IS, Coudert JD, Andoniou CE, Degli-Esposti MA. “Natural regulators”: NK cells as modulators of T cell immunity. Front Immunol (2016) 7:235. doi:10.3389/fimmu.2016.00235
286. Cichicki F, Schlums H, Theorell J, Tesi B, Miller JS, Ljunggren HG, et al. Diversification and functional specialization of human NK cell subsets. Curr Top Microbiol Immunol (2016) 395:63–94. doi:10.1007/82_2015_487
287. Fernandez NC, Lozier A, Flament C, Ricciardi-Castagnoli P, Bellet D, Suter M, et al. Dendritic cells directly trigger NK cell functions: cross-talk relevant in innate anti-tumor immune responses in vivo. Nat Med (1999) 5:405–11. doi:10.1038/7403
288. Moretta A. Natural killer cells and dendritic cells: rendezvous in abused tissues. Nat Rev Immunol (2002) 2:957–64. doi:10.1038/nri956
289. Fernandez NC, Flament C, Crépineau F, Angevin E, Vivier E, Zitvogel L. Dendritic cells (DC) promote natural killer (NK) cell functions: dynamics of the human DC/NK cell cross talk. Eur Cytokine Netw (2002) 13:17–27.
290. Cooper MA, Fehniger TA, Fuchs A, Colonna M, Caligiuri MA. NK cell and DC interactions. Trends Immunol (2004) 25:47–52. doi:10.1016/j.it.2003.10.012
291. Newman KC, Riley EM. Whatever turns you on: accessory-cell-dependent activation of NK cells by pathogens. Nat Rev Immunol (2007) 7:279–91. doi:10.1038/nri2057
292. Kloss M, Decker P, Baltz KM, Baessler T, Jung G, Rammensee HG, et al. Interaction of monocytes with NK cells upon toll-like receptor-induced expression of the NKG2D ligand MICA. J Immunol (2008) 181:6711–9. doi:10.4049/jimmunol.181.10.6711
293. Knorr M, Munzel T, Wenzel P. Interplay of NK cells and monocytes in vascular inflammation and myocardial infarction. Front Physiol (2014) 5:295. doi:10.3389/fphys.2014.00295
294. Chiche L, Forel JM, Thomas G, Farnarier C, Vely F, Bléry M, et al. The role of natural killer cells in sepsis. J Biomed Biotechnol (2011) 2011:986491. doi:10.1155/2011/986491
295. Bosmann M, Ward PA. Modulation of inflammation by interleukin-27. J Leukoc Biol (2013) 94:1159–65. doi:10.1189/jlb.0213107
296. Tosello-Trampont A, Surette FA, Ewald SE, Hahn YS. Immunoregulatory role of NK cells in tissue inflammation and regeneration. Front Immunol (2017) 8:301. doi:10.3389/fimmu.2017.00301
297. Laouar Y, Sutterwala FS, Gorelik L, Flavell RA. Transforming growth factor-beta controls T helper type 1 cell development through regulation of natural killer cell interferon-gamma. Nat Immunol (2005) 6:600–7. doi:10.1038/ni1197
298. Krebs P, Barnes MJ, Lampe K, Whitley K, Bahjat KS, Beutler B, et al. NK-cell-mediated killing of target cells triggers robust antigen-specific T-cell-mediated and humoral responses. Blood (2009) 113:6593–602. doi:10.1182/blood-2009-01-201467
299. Gao N, Dang T, Dunnick WA, Collins JT, Blazar BR, Yuan D. Receptors and counterreceptors involved in NK-B cell interactions. J Immunol (2005) 174:4113–9. doi:10.4049/jimmunol.174.7.4113
300. Gao N, Schwartzberg P, Wilder JA, Blazar BR, Yuan D. B cell induction of IL-13 expression in NK cells: role of CD244 and SLAM-associated protein. J Immunol (2006) 176:2758–64. doi:10.4049/jimmunol.176.5.2758
301. Blanca IR, Bere EW, Young HA, Ortaldo JR. Human B cell activation by autologous NK cells is regulated by CD40-CD40 ligand interaction: role of memory B cells and CD5+ B cells. J Immunol (2001) 167:6132–9. doi:10.4049/jimmunol.167.11.6132
302. Pallmer K, Oxenius A. Recognition and regulation of T cells by NK cells. Front Immunol (2016) 7:251. doi:10.3389/fimmu.2016.00251
303. Laroni A, Gandhi R, Beynon V, Weiner HL. IL-27 imparts immunoregulatory function to human NK cell subsets. PLoS One (2011) 6:e26173. doi:10.1371/journal.pone.0026173
304. Gross CC, Schulte-Mecklenbeck A, Wiendl H, Marcenaro E, Kerlero de Rosbo N, Uccelli A, et al. Regulatory functions of natural killer cells in multiple sclerosis. Front Immunol (2016) 7:606. doi:10.3389/fimmu.2016.00606
305. Jiang W, Chai NR, Maric D, Bielekova B. Unexpected role for granzyme K in CD56bright NK cell-mediated immunoregulation of multiple sclerosis. J Immunol (2011) 187:781–90. doi:10.4049/jimmunol.1100789
306. Morandi F, Horenstein AL, Chillemi A, Quarona V, Chiesa S, Imperatori A, et al. CD56brightCD16- NK cells produce adenosine through a CD38-mediated pathway and act as regulatory cells inhibiting autologous CD4+ T cell proliferation. J Immunol (2015) 195:965–72. doi:10.4049/jimmunol.1500591
307. Simonetta F, Alvarez M, Negrin RS. Natural killer cells in graft-versus-host-disease after allogeneic hematopoietic cell transplantation. Front Immunol (2017) 8:465. doi:10.3389/fimmu.2017.00465
308. Verneris MR, Ito M, Baker J, Arshi A, Negrin RS, Shizuru JA. Engineering hematopoietic grafts: purified allogeneic hematopoietic stem cells plus expanded CD8+ NK-T cells in the treatment of lymphoma. Biol Blood Marrow Transplant (2001) 7:532–42. doi:10.1016/S1083-8791(01)70014-6
309. Shah NN, Baird K, Delbrook CP, Fleisher TA, Kohler ME, Rampertaap S, et al. Acute GVHD in patients receiving IL-15/4-1BBL activated NK cells following T-cell-depleted stem cell transplantation. Blood (2015) 125:784–92. doi:10.1182/blood-2014-07-592881
310. Baron F, Petersdorf EW, Gooley T, Sandmaier BM, Malkki M, Chauncey TR, et al. What is the role for donor natural killer cells after nonmyeloablative conditioning? Biol Blood Marrow Transplant (2009) 15:580–8. doi:10.1016/j.bbmt.2009.01.018
311. Cooley S, Trachtenberg E, Bergemann TL, Saeteurn K, Klein J, Le CT, et al. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood (2009) 113:726–32. doi:10.1182/blood-2008-07-171926
312. Leveson-Gower DB, Olson JA, Sega EI, Luong RH, Baker J, Zeiser R, et al. Low doses of natural killer T cells provide protection from acute graft-versus-host disease via an IL-4-dependent mechanism. Blood (2011) 117:3220–9. doi:10.1182/blood-2010-08-303008
313. Chan YLT, Zuo J, Inman C, Croft W, Begum J, Croudace J, et al. NK cells produce high levels of IL-10 early after allogeneic stem cell transplantation and suppress development of acute GVHD. Eur J Immunol (2018) 48:316–29. doi:10.1002/eji.201747134
314. Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science (2002) 295:2097–100. doi:10.1126/science.1068440
315. Marcenaro E, Carlomagno S, Pesce S, Della Chiesa M, Moretta A, Sivori S. Role of alloreactive KIR2DS1(+) NK cells in haploidentical hematopoietic stem cell transplantation. J Leukoc Biol (2011) 90:661–7. doi:10.1189/jlb.0311137
316. Rabinovich BA, Li J, Shannon J, Hurren R, Chalupny J, Cosman D, et al. Activated, but not resting, T cells can be recognized and killed by syngeneic NK cells. J Immunol (2003) 170:3572–6. doi:10.4049/jimmunol.170.7.3572
317. Cerboni C, Zingoni A, Cippitelli M, Piccoli M, Frati L, Santoni A. Antigen-activated human T lymphocytes express cell-surface NKG2D ligands via an ATM/ATR-dependent mechanism and become susceptible to autologous NK-cell lysis. Blood (2007) 110:606–15. doi:10.1182/blood-2006-10-052720
318. Ogasawara K, Benjamin J, Takaki R, Phillips JH, Lanier LL. Function of NKG2D in natural killer cell-mediated rejection of mouse bone marrow grafts. Nat Immunol (2005) 6(9):938–45. doi:10.1038/ni1236
319. Zwirner NW, Fernandez-Vina MA, Stastny P. MICA, a new polymorphic HLA-related antigen, is expressed mainly by keratinocytes, endothelial cells, and monocytes. Immunogenetics (1998) 47:139–48. doi:10.1007/s002510050339
320. Ward J, Bonaparte M, Sacks J, Guterman J, Fogli M, Mavilio D, et al. HIV modulates the expression of ligands important in triggering natural killer cell cytotoxic responses on infected primary T-cell blasts. Blood (2007) 110:1207–14. doi:10.1182/blood-2006-06-028175
321. Yang D, Tian Z, Zhang M, Yang W, Tang J, Wu Y, et al. NKG2D(+)CD4(+) T cells kill regulatory T cells in a NKG2D-NKG2D ligand-dependent manner in systemic lupus erythematosus. Sci Rep (2017) 7:1288. doi:10.1038/s41598-017-01379-y
322. Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature (2002) 419:734–8. doi:10.1038/nature01112
323. El-Gazzar A, Groh V, Spies T. Immunobiology and conflicting roles of the human NKG2D lymphocyte receptor and its ligands in cancer. J Immunol (2013) 191:1509–15. doi:10.4049/jimmunol.1301071
324. Hamada S, Caballero-Benitez A, Duran KL, Stevens AM, Spies T, Groh V. Soluble MICB in plasma and urine explains population expansions of NKG2D(+)CD4 T cells inpatients with juvenile-onset systemic lupus erythematosus. Open J Immunol (2017) 7:1–17. doi:10.4236/oji.2017.71001
325. Ferrari de Andrade L, Tay RE, Pan D, Luoma AM, Ito Y, Badrinath S, et al. Antibody-mediated inhibition of MICA and MICB shedding promotes NK cell-driven tumor immunity. Science (2018) 359:1537–42. doi:10.1126/science.aao0505
326. Deng W, Gowen BG, Zhang L, Wang L, Lau S, Iannello A, et al. Antitumor immunity. A shed NKG2D ligand that promotes natural killer cell activation and tumor rejection. Science (2015) 348:136–9. doi:10.1126/science.1258867
327. Rezvani K, Rouce R, Liu E, Shpall E. Engineering natural killer cells for cancer immunotherapy. Mol Ther (2017) 25:1769–81. doi:10.1016/j.ymthe.2017.06.012
328. Sadelain M, Brentjens R, Riviere I. The basic principles of chimeric antigen receptor design. Cancer Discov (2013) 3:388–98. doi:10.1158/2159-8290.CD-12-0548
329. Srivastava S, Riddell SR. Engineering CAR-T cells: design concepts. Trends Immunol (2015) 36:494–502. doi:10.1016/j.it.2015.06.004
330. Tonn T, Schwabe D, Klingemann HG, Becker S, Esser R, Koehl U, et al. Treatment of patients with advanced cancer with the natural killer cell line NK-92. Cytotherapy (2013) 15:1563–70. doi:10.1016/j.jcyt.2013.06.017
331. Chen KH, Wada M, Pinz KG, Liu H, Lin KW, Jares A, et al. Preclinical targeting of aggressive T-cell malignancies using anti-CD5 chimeric antigen receptor. Leukemia (2017) 31:2151–60. doi:10.1038/leu.2017.8
332. Zhang C, Oberoi P, Oelsner S, Waldmann A, Lindner A, Tonn T, et al. Chimeric antigen receptor-engineered NK-92 cells: an off-the-shelf cellular therapeutic for targeted elimination of cancer cells and induction of protective antitumor immunity. Front Immunol (2017) 8:533. doi:10.3389/fimmu.2017.00533
333. Klingemann H, Boissel L, Toneguzzo F. Natural killer cells for immunotherapy – advantages of the NK-92 cell line over blood NK cells. Front Immunol (2016) 7:91. doi:10.3389/fimmu.2016.00091
334. Palmer JM, Rajasekaran K, Thakar MS, Malarkannan S. Clinical relevance of natural killer cells following hematopoietic stem cell transplantation. J Cancer (2013) 4:25–35. doi:10.7150/jca.5049
335. Raulet DH, Vance RE, McMahon CW. Regulation of the natural killer cell receptor repertoire. Annu Rev Immunol (2001) 19:291–330. doi:10.1146/annurev.immunol.19.1.291
336. Huntington ND, Vosshenrich CA, Di Santo JP. Developmental pathways that generate natural-killer-cell diversity in mice and humans. Nat Rev Immunol (2007) 7:703–14. doi:10.1038/nri2154
337. Brady J, Carotta S, Thong RP, Chan CJ, Hayakawa Y, Smyth MJ, et al. The interactions of multiple cytokines control NK cell maturation. J Immunol (2010) 185:6679–88. doi:10.4049/jimmunol.0903354
338. Leavy O. Natural killer cells: a virtual pick and mix. Nat Rev Immunol (2013) 13:844–5. doi:10.1038/nri3566
339. Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med (2013) 5:208ra145. doi:10.1126/scitranslmed.3006702
Keywords: developmental stages, human, mouse, natural killer cells, effector functions
Citation: Abel AM, Yang C, Thakar MS and Malarkannan S (2018) Natural Killer Cells: Development, Maturation, and Clinical Utilization. Front. Immunol. 9:1869. doi: 10.3389/fimmu.2018.01869
Received: 22 May 2018; Accepted: 30 July 2018;
Published: 13 August 2018
Edited by:
Laurent Brossay, Brown University, United StatesReviewed by:
Michael G. Brown, University of Virginia, United StatesStephen Noel Waggoner, Cincinnati Children’s Hospital Medical Center, United States
Copyright: © 2018 Abel, Yang, Thakar and Malarkannan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Subramaniam Malarkannan, subra.malar@bcw.edu
 Alex M. Abel
Alex M. Abel Chao Yang1,2
Chao Yang1,2 Monica S. Thakar
Monica S. Thakar Subramaniam Malarkannan
Subramaniam Malarkannan