- 1National Center of Technology Innovation for Saline-Alkali Tolerant Rice, College of Coastal Agricultural Sciences, Guangdong Ocean University, Zhanjiang, China
- 2Key Laboratory for Quality Regulation of Tropical Horticultural Crops of Hainan Province, School of Horticulture, Hainan University, Haikou, China
- 3Xiangyang Academy of Agricultural Sciences, Xiangyang, China
Background: Nitrogen (N), absorbed primarily as ammonium (NH4+) from soil by plant, is a necessary macronutrient in plant growth and development. Ammonium transporter (AMT) plays a vital role in the absorption and transport of ammonium (NH4+). Cassava (Manihot esculenta Crantz) has a strong adaptability to nitrogen deprivation. However, little is known about the functions of ammonium transporter AMT2 in cassava.
Methods: The cassava AMT2-type genes were identified and their characteristics were analyzed using bioinformatic techniques. The spatial expression patterns were analyzed based on the public RNA-seq data and their expression profiles under low ammonium treatment were studied using Real-time quantitative PCR (RT-qPCR) method. The cassava AMT2 genes were transformed into yeast mutant strain TM31019b by PEG/LiAc method to investigate their functions.
Results: Seven AMT2-type genes (MeAMT2.1-2.7) were identified in cassava and they were distributed on 6 chromosomes and included two segmental duplication events (MeAMT2.2/MeAMT2.4 and MeAMT2.3/MeAMT2.5). Based on their amino acid sequences, seven MeAMT2 were further divided into four subgroups, and each subgroup contained similar motif constitution and protein structure. Synteny analysis showed that two and four MeAMT2 genes in cassava were collinear with those in the Arabidopsis and soybean genomes, respectively. Sixteen types of cis-elements were identified in the MeAMT2 promoters, and they were related to light-, hormone-, stress-, and plant growth and development-responsive elements, respectively. Most of the MeAMT2 genes displayed tissue-specific expression patterns according to the RNA-seq data, of them, three MeAMT2 (MeAMT2.3, MeAMT2.5, and MeATM2.6) expressions were up-regulated under ammonium deficiency. Complementation experiments showed that yeast mutant strain TM31019b transformed with MeAMT2.3, MeAMT2.5, or MeATM2.6 grew better than untransgenic yeast cells under ammonium deficiency, suggesting that MeAMT2.3, MeAMT2.5, and MeATM2.6 might be the main contributors in response to ammonium deficiency in cassava.
Conclusion: This study provides a basis for further study of nitrogen efficient utilization in cassava.
Introduction
Plants utilize nutrients to maintain their normal growth and development. Nitrogen (N), mainly absorbed from soil, is an essential nutrient in growth and development of plants. N sources in soil include inorganic nitrogen (nitrate and ammonium), organic compounds and amino acids (Williams and Miller, 2001). Nitrate nitrogen and ammonium nitrogen are considered as main plant nitrogen sources. Lack of nitrogen will affect the synthesis of organic substances in plants, resulting in growth suppression, leaf yellowing and low yields (Tabata et al., 2014). When nitrogen is deficient, plants prefer absorbing ammonium nitrogen to nitrate nitrogen because absorption and assimilation of ammonium nitrogen consume less energy than that of nitrate nitrogen (Gazzarrini et al., 1999; Xuan et al., 2017). However, excessive ammonium is toxic to plants, therefore, its absorption and assimilation must be accurately regulated. The ammonium transport and homeostasis are controlled by unsaturated low-affinity absorption system (LATS, i.e., aquaporins or cation channels) and saturated high-affinity system (HATS, i.e., ammonium transporters) (Sohlenkamp et al., 2000). When the concentration of [NH4+]ext is lower than 1 mM, HATS is mainly responsible for absorbing NH4+, and the absorption approaches Michaelis-Menten kinetics. Whereas when the concentration is higher than 1 mM, LATS is activated and plays a role in absorbing NH4+, with a linear unsaturated characteristic (Kronzucker et al., 1996; Wang et al., 1996).
Ammonium transporter (AMT) proteins, which are encoded by a small multigene family, play critical roles in regulating ammonium absorption and transport. There are 10 and six AMTs in rice and Arabidopsis, respectively (Gazzarrini et al., 1999; Sonoda et al., 2003). According to their sequences and phylogenetic relationships, AMT proteins in plant cells can be divided into two categories: AMT1 and AMT2 (Loqué and von Wirén, 2004). Many AMT1s have been reported in different plants (Ninnemann et al., 1994; Ranathunge et al., 2014; Li et al., 2015; Filiz and Akbudak, 2020; Yang et al., 2021), and it has been found that these AMT1 proteins function as high-affinity ammonium transporters (McDonald et al., 2012). Ammonium transport mediated by AMT1s is regulated by phosphorylation and dephosphorylation of the transporters (Loque et al., 2007; Neuhauser et al., 2007). It has recently been reported that CIPK23, a Ser/Thr kinase, can negatively impact the NH4+ transport activities of AtAMT1; 1 and AtAMT1; 2 by direct interactions with their C-termini (Straub et al., 2017). Compared with the studies on functional and regulation mechanisms of AMT1s, the reports for expression patterns and physiological functions of AMT2s are little, due to the large number of AMT2 family members and complex expression patterns (Sohlenkamp et al., 2002; Neuhäuser et al., 2009).
AMT2 ammonium transporters belong to the family of methylamine permease (MEP) as AMT1s. The primary structure of AMT2 proteins is quite different from that of AMT1 proteins, but their advanced structure is similar as that of AMT1 proteins (Howitt and Udvardi, 2000). AMT2s have a high transport capacity for NH4+ (Neuhäuser et al., 2009), so they play a main role in distribution and regulation of NH4+ between the above-ground and underground parts of plants (Sohlenkamp et al., 2000; Giehl et al., 2017). Evolutionary analysis showed the close correlation between AMT2 proteins and ammonium transporters in many thermophilic bacteria, which may be related to the horizontal transfer of MEP genes in prokaryotes (Howitt and Udvardi, 2000; Zheng et al., 2004; McDonald et al., 2012). Arabidopsis thaliana AMT2 family contains only one member (AtAMT2.1) (Yuan et al., 2007), while the AMT2 family in many other plants, including rice, can be divided into AMT2, AMT3, AMT4, AMT5 and other groups according to different clades in phylogenetic analysis (Li et al., 2009). AtAMT2.1 is a high-affinity ammonium transporter with a Km value of about 20 μM (Sohlenkamp et al., 2000; Sohlenkamp et al., 2002), but most of the AMT2 members are low-affinity ammonium transporters. Compared with AMT1 genes that are mainly expressed in roots, AMT2 proteins distribute in different plant organs. For example, AtAMT2.1 is widely expressed in roots, leaves, and other tissues of Arabidopsis plants, and play a critical role in the reuse of ammonium ions (Sohlenkamp et al., 2000). CsAMT3.1 is a type of AMT2-type gene and has a high expression level in roots and green tissues, with the highest expression in mature leaves. Its function is related to transport and distribution of NH4+ in various tissues of tea plants (Zhang et al., 2018).
Cassava (Manihot esculenta Crantz) is an important raw material crop for food and energy (Ou et al., 2018). As a safe and nutritious high quality food, cassava has gradually been favored by more and more people. Cassava has a wide and strong adaptability and adverse resistance, especially to nitrogen deprivation (Xu et al., 2013). In our previous study, six cassava AMT1 genes were identified, and MeAMT1; 1 was found to play a vital role in response to ammonium deficiency (Xia et al., 2022). However, little is known about the MeAMT2 gene family. Herein, seven cassava AMT2 genes (MeAMT2.1-2.7) were found from the cassava genome. The phylogeny, gene structures, genomic locations, and cis-elements, the expression patterns of these MeAMT2 genes in different tissues and their expression profiles at low NH4+ level were investigated. Moreover, the functions of three MeAMT2 genes (MeAMT2.3, MeAMT2.5, and MeATM2.6) were explored using a yeast complementation system. The present results may provide valuable information for further studies on AMT2 functions and nitrogen efficient utilization in cassava.
Materials and methods
Plant materials and treatments
The plant material of cassava cultivar ‘SC8’ (Manihot esculenta Crantz cv. SC8) was grown in a greenhouse with conditions: temperature, 28°C; light intensity, 300 μmol m−2·s−1; relative humidity, 60%, light/darkness, 16 h/8 h (Xia et al., 2022). The seedlings were subjected to a 1-week culture in Afdaling nutrient solution (Hu et al., 2016), followed by low ammonium treatment in medium containing 0.05 mM NH4Cl for 6, 12, 24 and 48 h, and the leaves, roots, and stems were collected at each time point, all samples were frozen in liquid nitrogen and stored at −80°C for further use.
Identification and characterization of AMT2 family members in cassava
To identify the AMT2 transporters in cassava, the AMT2 protein sequences from A. thaliana (Sohlenkamp et al., 2000) were utilized as query sequences with E-value cutoff set as 1e-5 to perform a local BLASTP against the whole-genome data of cassava (Manihot esculenta v6.1) (phytozome v11.0, https://phytozome.jgi.doe.gov/pz/portal.html). The gene location information was obtained. The theoretical isoelectric point (pI) and molecular weight (MW) of the MeAMT2 proteins were obtained using the ExPASy online software (http://web.expasy.org/protparam/). PSORT instrument (https://www.genscript.com/) was used to characterize the subcellular localization of MeAMT2 proteins.
Analysis of phylogenetic, conserved motifs and gene structure
In order to understand the phylogenetic relationships of AMT2 proteins in different plant species, the sequences of AMT2 proteins from A. thaliana (Gazzarrini et al., 1999), rice (Oryza sativa) (Sonoda et al., 2003), soybean (Glycine max) (Kobae et al., 2010), maize (Zea mays) (Hui et al., 2022), and Lotus japonicus (Wang et al., 2022) were obtained from the previous study, and the sequences from Ricinus communis, belonging to Euphorbiaceae, were identified using the identification method for the cassava AMT2 gene family. Multiple sequence alignment using MUSCLE and default parameters was applied to evaluate the evolutionary relationship of MeAMT2s proteins in cassava (Edgar, 2004), and a neighbor-joining (NJ) phylogenetic tree was established based on the alignment utilizing the MEGA version X software (Kumar et al., 2018) with 1000 bootstrap replications. The online software Multi Em for Motif Elicitation (MEME Suite 4.12.0) (http://meme-suite.org/tools/meme) was used to analyze the conserved motifs of MeAMT2 proteins with optimization parameters: maximum number of motifs, 10; optimum width, 6–50; number of repetitions, any; as well as other default parameters. The MeAMT2 gene structures were analyzed using Gene Structure Display Server (GSDS) (http://gsds.cbi.pku.edu.cn/).
Analysis of gene collinearity and chromosomal distribution
The MeAMT2 genes were mapped to the chromosomes utilizing TBtools software (Chen et al., 2020) according to the information obtained from the cassava genome database. Simultaneously, gene duplication of MeAMT2 genes was analyzed utilizing MCScanX software (Wang et al., 2012) and illustrated with TBtools. The nucleotide substitution parameters Ks (synonymous) and Ka (non-synonymous) of the duplicated genes were assessed using TBtools, and then the Ka/Ks ratio was calculated. In addition, the gene duplication information from cassava, soybean, and A. thaliana was analyzed using MCscanX software, followed by integral visualization of synteny with TBtools software (Wang et al., 2012; Chen et al., 2020).
Analysis of cis-elements in MeAMT2 promoters
The 2.0-kb upstream sequences of the MeAMT2 genes were acted as the promoters and submitted to PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The cis-regulatory elements were identified and visualized by TBtools.
Spatial expression profiles of MeAMT2s based on RNA-seq data
The Transcripts Per Million (TPM) values of the MeAMT2 genes obtained from the RNA-seq data in the SRA dataset (https://www.ncbi.nlm.nih.gov/sra) (SRP076160) (Wilson et al., 2017) were utilized to examine the expression profiles of MeAMT2 genes in 11 selected cassava tissues including leaf, root, petiole, stem, midevein, fibrous root, lateral bud, middle storage root, early storage root, storage root and last storage root. The heatmap of MeAMT2 genes was constructed by TBtools.
Real-time quantitative PCR (RT-qPCR) analysis of MeAMT2 genes
The total RNA of cassava roots, stems and leaves was extracted according to the instructions of the Plant Total RNA Extraction Kit (TIANGEN, China, DP437). The first-strand cDNA was synthesized using the FastKing cDNA First Strand Synthesis Kit (TIANGEN, China, KR116-02). The transcript levels of MeAMT2 genes were analyzed using RT-qPCR on an ABI 7900HT RT-PCR system (TaKaRa, Japan) using SuperReal PreMix Plus (SYBR Green I) fluorescence quantification kit (TIANGEN, China, FP205-02) with Actin gene as internal reference control (Mo et al., 2018). The PCR amplification conditions were referred to the previous study (Mo et al., 2018). The experiments were repeated three times, and the relative expression of target genes was determined using the 2−ΔΔCt method (Livak and Schmittgen, 2001), where ΔΔCt = (Cttarget gene—Ctinternal reference gene)experimental group—(Cttarget gene—Ctinternal reference gene)control group. Three independent biological replicates were conducted. The primers used are designed using Primer Premier five software and shown in Supplementary Table S1.
Construction of yeast expression vector and functional analysis
To investigate the functions of the MeAMT2 genes, these genes were cloned using the cDNA as templates and inserted into the yeast expression vector p416. The restriction sites for construction could be seen in the primer sequences. After digestion and sequencing, the recombinant plasmids were confirmed. The plasmids p416 or p416-MeAMT2s were then transformed into the Saccharomyces cerevisiae mutant strain TM31019b (MATα ura3 mep1Δ mep2Δ:LEU2 mep3Δ:KanMX2) (Marini et al., 1997) using the PEG/LiAc method (Yin et al., 2020). The TM31019b strains were then grown on a SD/-ura solid medium to obtain positive clones (Li et al., 2017). The successful transformants were then grown on yeast nitrogen base medium containing 1 mM arginine (control) or low ammonium (different concentrations of NH4Cl) for 3–5 days at 30°C. The primers used are designed using Primer Premier five software and shown in Supplementary Table S1.
Results
Identification of the AMT2 gene family in cassava
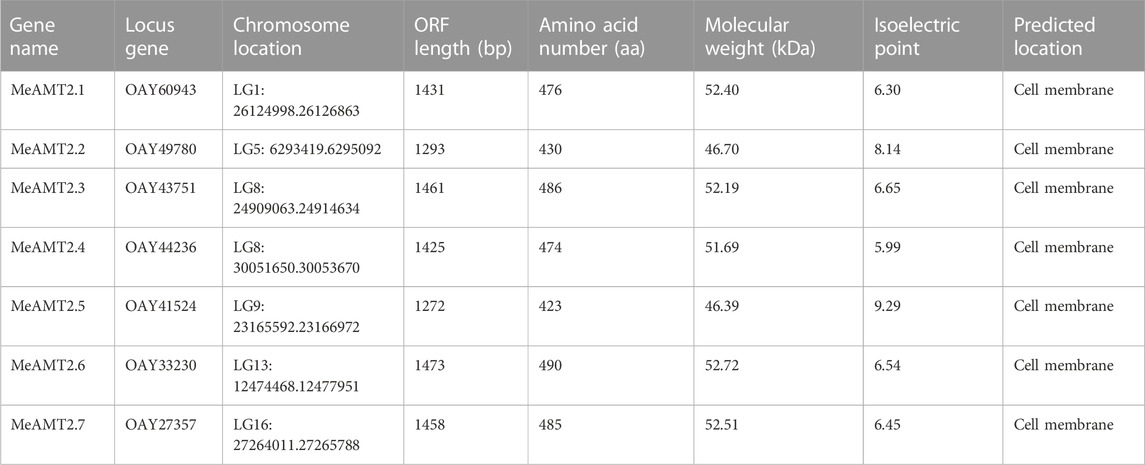
Through a BLASTP search using Arabidopsis AMT2 protein, seven AMT2 genes were identified in the cassava genome. These seven MeAMT2 genes distributed on six chromosomes (LG1, LG5, LG8, LG9, LG13, and LG16) were named MeAMT2.1 to MeAMT2.7 accordingly (Figure 1; Table 1). The physicochemical characteristics of the MeAMT2 genes are summarized in Table 1, with coding sequences (CDSs) ranging from 1272 (MeAMT2.5) to 1473 (MeAMT2.6) bp, and the length, MW and pI of the predicted proteins ranging from 423 (MeAMT2.5) to 490 (MeAMT2.6) amino acids, from 46.39 (MeAMT2.5) to 52.72 (MeAMT2.6) kDa, and from 5.99 (MeAMT2.4) to 9.29 (MeAMT2.5), average of 7.05, respectively. Furthermore, all the proteins were predicted to be located in cell membrane.
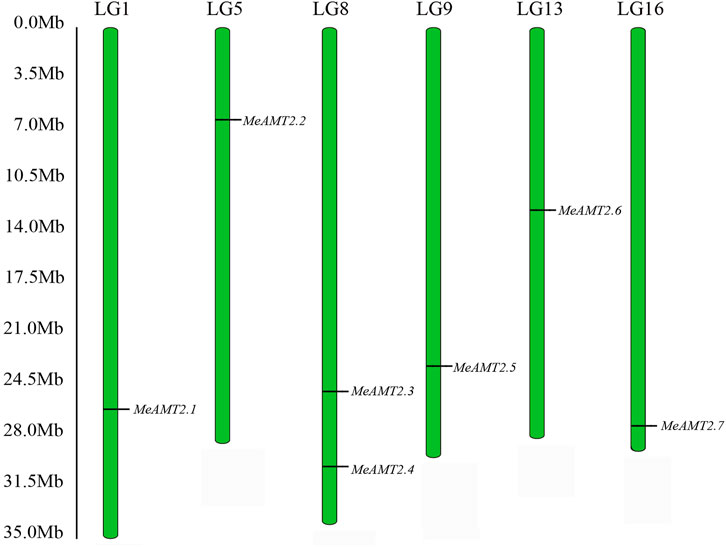
FIGURE 1. Chromosomal distribution of MeAMT2 genes. The chromosome number is listed above each chromosome while the numbers on the left represent the locations of the MeAMT2 genes.
Phylogenetic analysis of cassava AMT2 proteins
The evolutionary relationship between cassava AMT2 proteins and those from other plant species was explored next. A total of 37 protein sequences from cassava, R. communis, G. max, L. japonicus, Z. mays, O. sativa, and A. thaliana were used to construct a NJ phylogenetic tree. These proteins were clustered into five subgroups: subgroup 1 consisted of two MeAMT2s (MeAMT2.3 and MeAMT2.5), one RcAMT2 (RcAMT2.5), three GmAMT2s (GmAMT2.1, GmAMT2.2 and GmAMT2.3), three OsAMT2s (OsAMT2.1, OsAMT2.2 and OsAMT2.3), one LjAMT2 (LjAMT2.1), one ZmAMT2 (ZmAMT2.1), and one AtAMT2 (AtAMT2.1); subgroup 2 consisted of one MeAMT2 (MeAMT2.6), one RcAMT2 (RcAMT2.6), one GmAMT2 (GmAMT3.1), three OsAMT2s (OsAMT3.1, OsAMT3.2 and OsAMT3.3), and three ZmAMT2s (ZmAMT3.1, ZmAMT3.2 and ZmAMT3.3); subgroup 3 consisted of three MeAMT2s (MeAMT2.2, MeAMT2.4 and MeAMT2.7), two RcAMT2s (RcAMT2.2 and RcAMT2.7), three GmAMT2s (GmAMT4.1, GmAMT4.3 and GmAMT4.5), and one LjAMT2 (LjAMT2.2); subgroup 4 contained one MeAMT2 (MeAMT2), one RcAMT2 (RcAMT2.1), and one GmAMT2 (GmAMT4.4); subgroup 5 contained four RcAMT2s .1 (RcAMT2.3, RcAMT2.4, RcAMT2.8 and RcAMT2.9). The Euphorbiaceae crops, cassava and R. communis, had similar distribution in subgroups 1 to 4. Differently, only R. communis AMT2 proteins were present in subgroup 5 (Figure 2).
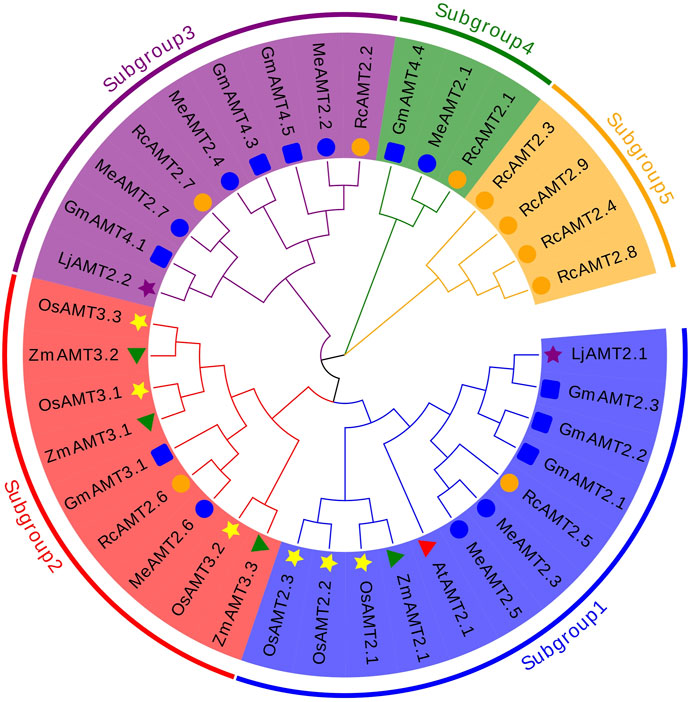
FIGURE 2. Phylogenetic analyses of AMT2 proteins. The phylogenetic tree of the AMT2 proteins was constructed using MEGA-X software with default parameters. The five subgroups are marked with different colors. The blue circles represent MeAMT2s, The orange circles represent RcAMT2s, the red triangles represent AtAMT2s, the yellow stars represent OsAMT2s, the purple stars represent LjAMT2s, the green triangles represent ZmAMT2s, and the blue squares represent GmAMT2s.
Analysis of conserved motifs and gene structure of the cassava AMT2 genes
To further characterize structural features of the AMT2 genes, the gene structures and distribution of conserved motifs were analyzed using the MEME program. As shown in Figure 3B, 10 putative conserved motifs (named motifs 1–10) were detected in most of the MeAMT2 protein sequences with length ranging from 11 to 50 amino acid residues (Supplementary Figure S1). All the MeAMT2 proteins except MeAMT2.1 (lacking motif two in N-terminal region) and MeAMT2.5 (lacking motif five and six in C-terminal region) contained the 10 motifs (Figure 3B). The structure of the MeAMT2 genes contained three exons separated by two introns except for MeAMT2.3 and MeAMT2.6 (Figure 3C), which contained four exons and three introns. Moreover, most of the MeAMT2 genes in the same subgroups shared similar gene structures and motif distributions (Figure 3).
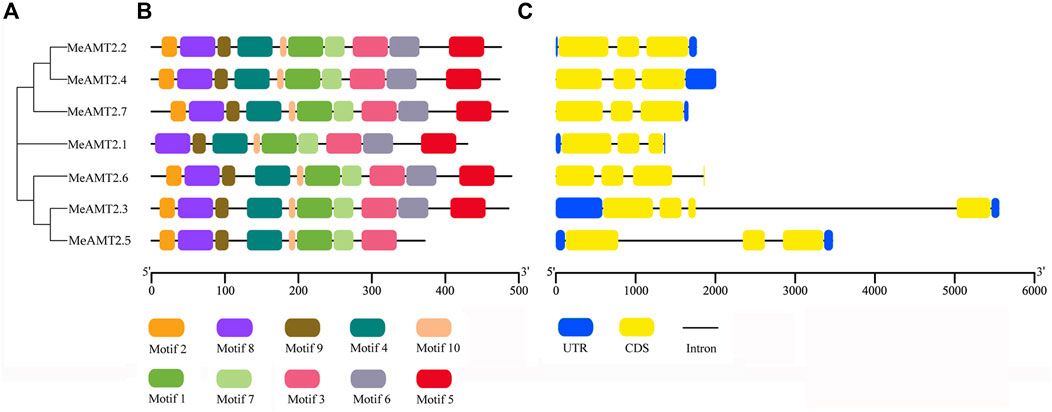
FIGURE 3. Motif compositions and gene structures of MeAMT2s (A) Phylogenetic analyses of MeAMT2 proteins (B) Distribution of conserved motifs in MeAMT2 proteins. The different colored boxes represent different motifs and their positions in each MeNRT2 protein sequence. Scale bar indicates number of amino acids (C) Exon-intron structures of MeAMT2 genes. The introns and exons are shown with black lines and yellow boxes, respectively. Green boxes represent the untranslated regions. Scale bar indicates number of nucleic acids (bp).
Analysis of gene duplication and synteny of MeAMT2s
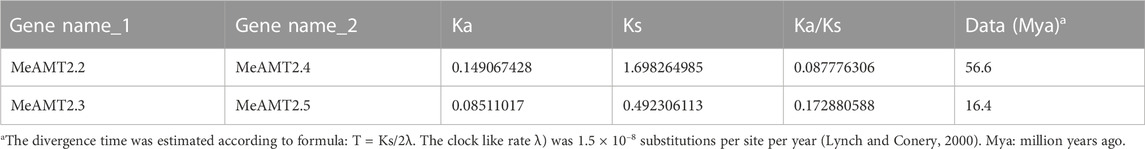
Figure 4 shows the gene duplication events. It can be seen that the MeAMT2 members were unevenly distributed in six among the 18 cassava chromosomes with only segmental duplication but no tandem duplication genes were detected. Two pairs of observed genes, MeAMT2.2/MeAMT2.4 and MeAMT2.3/MeAMT2.5, were regarded as segmental duplication genes (Figure 4), suggesting the critical roles of segmental duplication in the expansion of MeAMT2 family. The value of Ka/Ks can reflect the pressure of selection for a gene during evolution. The Ka/Ks values of the duplicated genes were both less than 1, suggesting that these MeAMT2 genes were influenced by purifying selection during evolution. The estimated time of duplication for paralogous genes indicated that all paralogs were ancient (from 16.4 to 56.6 Mya) (Table 2).
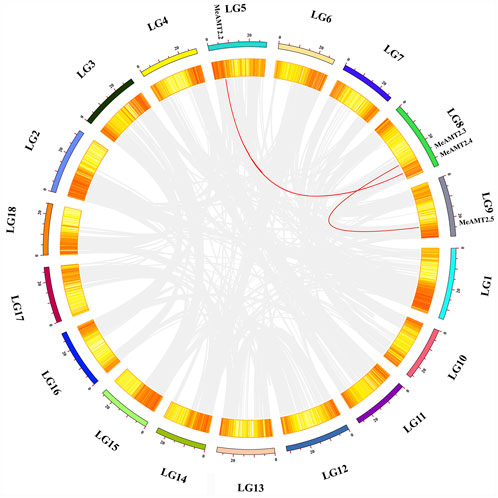
FIGURE 4. Schematic representations of the chromosomal distribution and interchromosomal relationships among cassava AMT2 genes. Chromosomes are represented in different colors with the chromosomal number indicated below each chromosome. Gray lines indicate synteny blocks within the cassava genome and red lines between MeAMT2 genes represent segmental duplication events that occurred in the cassava AMT2 gene family.
The comparative syntenic map was then constructed to show the associations between cassava and Arabidopsis, rice and soybean. As shown in Figure 5, four MeAMT2 genes showed a syntenic correlation with genes in soybean, and two MeAMT2 genes exhibited a syntenic correlation with genes in Arabidopsis, while no associations were found between MeAMT2s and OsAMT2s (data not shown). MeAMT2.3 and MeAMT2.5 were found to have syntenic relationships with AtAMT2.1, MeAMT2.2 had syntenic relationship with GmAMT3.1, MeAMT2.3 had syntenic relationships with GmAMT2.1, GmAMT2.2, and GmAMT2.3, MeAMT2.4 had syntenic relationship with GmAMT4.3, and MeAMT2.5 had syntenic relationships with GmAMT2.1, GmAMT2.2, and GmAMT2.3 (Figure 5, Supplementary Table S2).
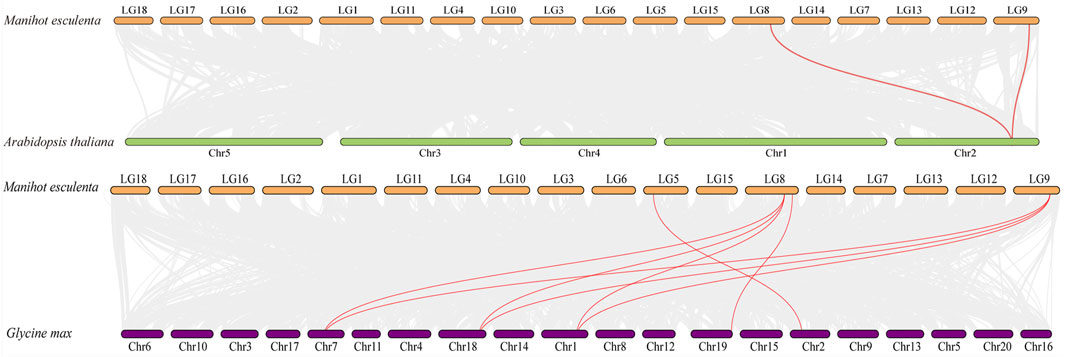
FIGURE 5. Synteny analyses of AMT2 genes between cassava, Arabidopsis and soybean. Gray lines in the background indicate the collinear blocks within cassava and other plant genomes, while the red lines highlight the syntenic AMT2 gene pairs.
Analysis of cis-elements in the promoter regions of MeAMT2 genes
The 2000 bp upstream sequences of start codons of the MeAMT2 genes were exacted as promoter sequences and used to analyze their cis-acting elements with PlantCARE software. Based on their biological functions, these 16 types of cis-acting elements were classified into four categories: 1) the light-responsive elements, the most numerous categories. The MeAMT2.1 and MeAMT2.5 promoter regions had the smallest and largest number of light-responsive elements, respectively (5, 23). 2) The hormone response related elements, including salicylic acid responsiveness element (SARE), abscisic acid responsiveness element (ABRE), gibberellin-responsive element, MeJA-responsiveness element, and auxin-responsive element (ARE). The ABRE only existed in the MeAMT2.5 and MeAMT2.6 promoter regions, indicating these two genes may be involved in ABA signaling pathway. The ARE only existed in the MeAMT2.7 promoter region, indicating this gene may be involved in auxin signaling pathway. The MeJA-responsiveness element existed in most promoter regions except for MeAMT2.3 and MeAMT2.5.3) The cis-acting elements associated with plant development. For example, the MeAMT2.6 promoter contained seed-specific regulation elements. 4) The stress-related element, which was uncommon in the MeAMT2 promoter region. The promoter region of MeAMT2.4 had a defense and stress responsiveness element, and the drought-inducibility element existed in the promoter regions of MeAMT2.2, MeAMT2.3, MeAMT2.4, MeAMT2.5 and MeAMT2.7. The low-temperature responsiveness element (LTR) existed in the promoter regions of MeAMT2.1, MeAMT2.2, MeAMT2.4 and MeAMT2.5 (Figure 6).
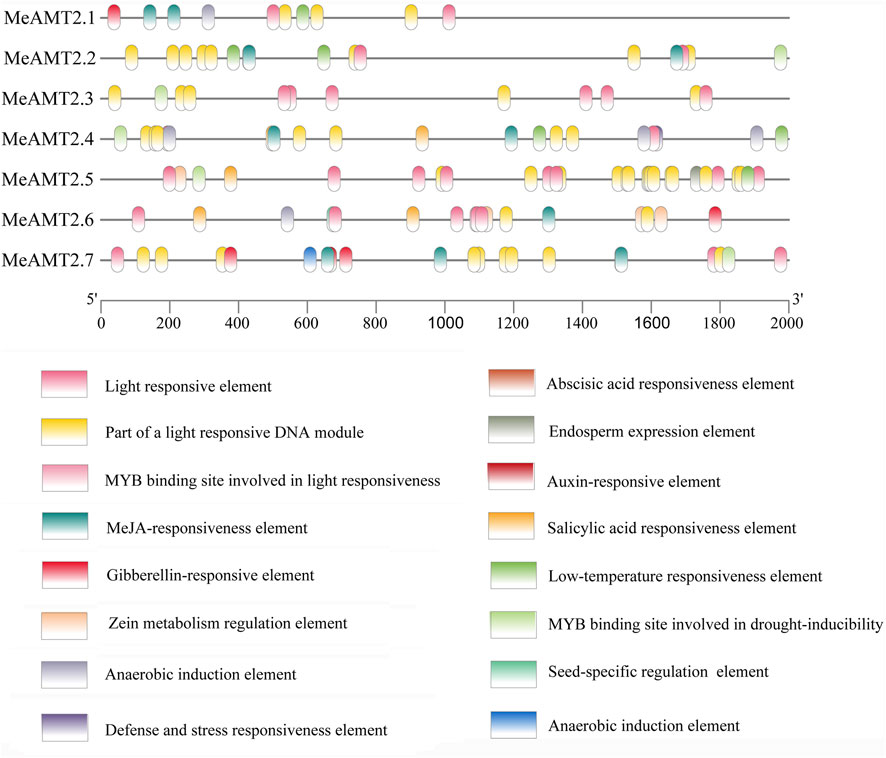
FIGURE 6. Analysis of cis-elements in the promoter regions of MeAMT2 genes. The potential elements in the promoter regions 2-kb upstream of the MeAMT2 genes were analyzed by PlantCARE. The upstream length to the translation start site can be inferred according to the scale at the bottom. Different color boxes represent different functions as described.
Expression profiles of AMT2 genes in different tissues of cassava based on RNA-seq
The spatial expression profiles of MeAMT2 genes in 11 cassava tissues including root, leaf, stem, storage root, fibrous root, midvein, petiole, lateral buds, early, medium and late storage roots were analyzed using RNA-seq data (SRP076160) of cassava from the public SRA database (Wilson et al., 2017) (Figure 7, Supplementary Table S3). The three genes, MeAMT2.3, MeAMT2.5 and MeAMT2.6, exhibited higher expression than other genes in most of the tested tissues. MeAMT2.3, MeAMT2.5 and MeAMT2.6 were highly expressed in leaves, lateral buds, and stems, respectively. MeAMT2.1 gene was also expressed in all tissues with a much lower level than the above three genes. The remaining three genes (MeAMT2.2, MeAMT2.4 and MeAMT2.7) were only expressed in some tissues at low levels.
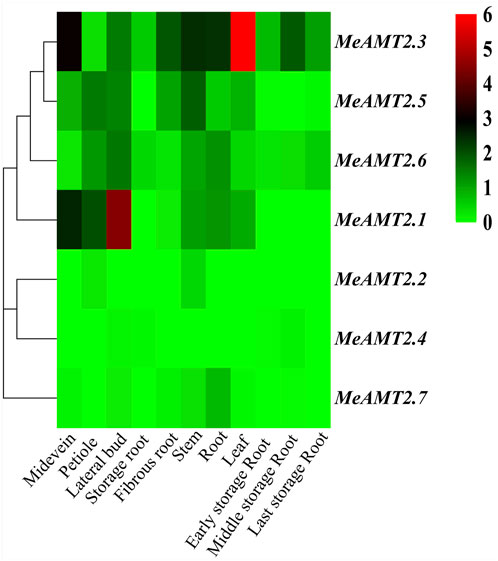
FIGURE 7. Hierachical clustering of MeAMT2 gene expression profiles across different cassava tissues. Data were normalized relative to each gene’s mean expression value across all tissues and log2-transformed. TPM (transcripts per kilobase million) values were used to create heat map showing the expression of MeAMT2 genes in different tissues. The expression level ranges from low expression (green) to high expression (red).
Expression patterns of cassava AMT2 genes under low ammonium conditions
To access whether ammonium deficiency influences the expression of MeAMT2 genes, we treated cassava seedlings with 0.05 mM NH4Cl for different periods. The results of RNA detection (Supplementary Figure S2A) and cDNA amplification (Supplementary Figure S2B) showed that the qualities were good for subsequent analysis. The three MeAMT2 genes (MeAMT2.3, MeAMT2.5 and MeAMT2.6) with higher spatial expression levels were selected for analysis in roots, stems and leaves under low ammonium conditions using RT-qPCR. Results showed that these three genes were up-regulated in different degrees under the condition of ammonium deficiency (Figure 8). The expression of MeAMT2.3 in leaves, roots, and stems was similarly slightly up-regulated after low ammonium treatment. The maximum expression level of MeAMT2.3 in roots, stems and leaves was reached at 48, 24 and 48 h after low ammonium treatment, respectively, which was about 3.10, 4.79, and 5.95 times of that at 0 h, respectively. The maximum expression level of MeAMT2.5 in roots, stems and leaves was reached at 48, 12 and 6 h after low ammonium treatment, respectively, which was about 2.60, 2.58, and 2.57 times of that at 0 h, respectively. The expression level of MeAMT2.6 in leaves did not change significantly after low ammonium treatment, but it was obviously up-regulated in roots and stems. MeAMT2.6 reached the highest expression level in roots and stems at 12 and 6 h after low ammonium treatment, which was about 10.59 and 15 times of that at 0 h, respectively.
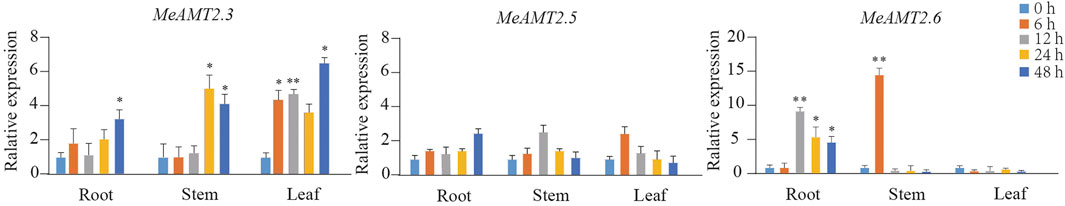
FIGURE 8. Expression profiles of MeAMT2 genes under ammonium deficiency. Data were analyzed by real-time quantitative PCR (RT-qPCR) using Actin gene as an internal cotrol. The unstressed level (0 h) was regarded as a standard. Values are the mean ± SE, n = 3. Vertical bars indicate the standard deviation. *p < 0.05 and **p < 0.01.
Functional analysis of MeAMT2s in transgenic yeast
The three genes (MeAMT2.3, MeAMT2.5 and MeAMT2.6) were cloned (Supplementary Figure S3A) and inserted into the yeast expression vector p416 (Supplementary Figure S3B) and then transformed into TM31019b mutant strain to further investigate the functions of MeAMT2 genes in response to low ammonium. Results showed that the growth of yeast cells transformed with either empty vector or MeAMT2 genes was similar under normal condition with 2 mM Arg. However, yeast cells transformed with empty vector p416 grew poorly with decreasing concentration of ammonium, and could not grow on SD/-ura medium containing 5 mM NH4Cl. In contrast, the MeAMT2.3-, MeAMT2.5-, or MeAMT2.6-transgenic TM31019b still grew well with decreasing concentration of NH4Cl, and could grow even under 0.05 mM NH4Cl treatment (Figure 9).
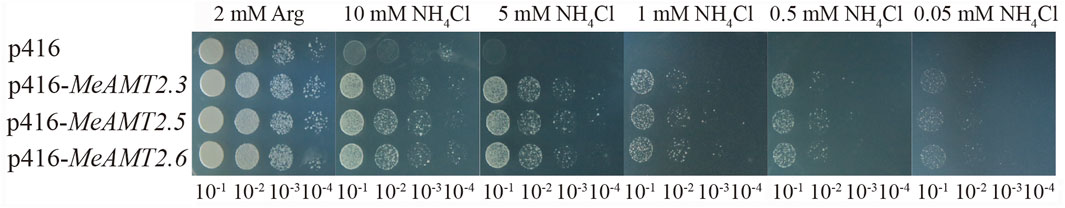
FIGURE 9. The effect of low ammonium treatment on yeast growth. p416-MeAMT2.3, p416-MeAMT2.5, p416-MeAMT2.6, and p416 were transformed into the TM31019b mutant yeast strain, respectively. The transgenic yeast cells were pre-cultured to saturation, and serial tenfold dilutions of the yeast cells were spotted on SD/-ura plates as previously described. After 3–5 days, the growth of the yeast cells on the plates was recorded. SD/-ura supplemented with 2 mM Arg was used as a control.
Discussion
Nitrogen is a plant-demanded mineral element necessary for plant growth (Williams and Miller, 2001). Nitrogen plays an extremely important role in crop growth, development, yield and quality. However, excessive nitrogen fertilizer application is not only a waste, but can also negatively affect crop growth and cause serious environmental pollution (Liu et al., 2013; Zhang et al., 2016). Plants can absorb both organic nitrogen (including peptides, amino acids, etc.) and inorganic nitrogen (including ammonium and nitrate nitrogen) from soil with nitrate nitrogen (NO3−) and ammonium nitrogen (NH4+) as the main forms (Xu et al., 2012). In agricultural production, regardless of soil type, the application of nitrogen fertilizer will lead to NH4+ becoming the main form in a short period of time (Xu and Takahashi, 2020). Since NH4+ consumes less energy than NO3− in the process of assimilation and utilization, NH4+ is considered the dominant nitrogen source (Socci and Templer, 2011). However, excessive absorption of NH4+ can result in toxic effects on crops, inducing leaf yellowing and growth inhibition. Therefore, maintaining an appropriate range of NH4+ absorption from the soil is critical for plant growth.
Plants absorb and transport NH4+ mainly through ammonium transporters (Yuan et al., 2007; Hao et al., 2020). The AMT gene family has been divided into two subfamilies: AMT1 subfamily (AMT1 cluster) and AMT2 subfamily (AMT2/3/4 cluster) (Koegel et al., 2013; Hao et al., 2020). At present, the AMT family genes in Arabidopsis have been well studied. The AtAMT family has been divided into two subfamilies: AtAMT1 and AtAMT2. The AtAMT1 subfamily contains five members: AtAMT1; 1∼AtAMT1; 5, and the AtAMT2 subfamily contains only AtAMT2; 1 (Gazzarrini et al., 1999). Cassava, as a tropical economic crop, is highly adaptable and can achieve high yield even in arid and barren mountainous areas. However, few studies have been on nitrogen utilization of cassava thus far. We have identified the cassava NRT2 and AMT1 gene families, and studied their response to low nitrogen stress, and found that MeNRT2.2 and MeAMT1; 1 could improve the plant growth of transgenic A. thaliana under low nitrate or ammonium stress (Xia et al., 2022; You et al., 2022). Our results suggest that these two genes play important roles in cassava responding to low nitrogen stress. In this study, we identified seven AMT2 genes from cassava genome, similar to those of rice (at least seven OsAMT2 members) (Suenaga et al., 2003), but more than that of Arabidopsis (only one AtAMT2 member) (Sohlenkamp et al., 2002), indicative of a possible gene gain event in the evolutionary process.
Phylogenetic analysis showed that the seven MeAMT2 and AMT2 proteins from Ricinus communis, Lotus japonicus, rice, soybean, maize and Arabidopsis could be divided into five subgroups (Figure 2). Cassava AMT2 is closely correlated with Ricinus communis AMT2 protein, both of the plants belonging to Euphorbiaceae. For example, MeAMT2.3 and MeAMT2.5 are highly similar to RcAMT2.5, MeAMT2.1/2.2/2.6/2.7 are highly similar to RcAMT2.1/2.2/2.6/2.7, respectively (Figure 2). In addition, the MeAMT2.4 was found to be closed to soybean GmAMT4.3. Synteny analysis also showed that MeAMT2.4 was correlated with GmAMT4.3 (Figure 5, Supplementary Table S2). Furthermore, MeAMT2.3 and MeAMT2.5 were found to be correlated with multiple genes from Arabidopsis and G. max, both of which were correlated with AtAMT2.1, GmAMT2.1, GmAMT2.2 and GmAMT2.3 (Figure 5, Supplementary Table S2), suggesting that MeAMT2.3 and MeAMT2.5 might have multiple functions. However, we did not find collinear pairs between monocot rice and cassava (data not shown), suggesting these orthologous pairs might be formed after differentiation of dicot and monocot plants (You et al., 2022). Analysis of conserved motifs showed that almost all MeAMT2 proteins, except MeAMT2.1 and MeAMT2.5, contained these ten motifs in a consistent order. The difference is that MeAMT2.1 lacked motif two in the N-terminus, while MeAMT2.5 lacked motif five and six in the C-terminus, suggesting loss of C- or N-terminus might have happened in some MeAMT2 members during evolution (Liu et al., 2018). Except for MeAMT2.3 and MeAMT2.6, which contained four exons and three introns, most of the MeAMT2 genes contained three exons and two introns (Figure 3). Similarly, three exons were found in Pyrus betulaefolia (Li et al., 2016), and five exons were found in Chinese cabbage AMT2 genes (Zhu et al., 2018). Moreover, we found segmental duplicated genes (MeAMT2.2/MeAMT2.4, MeAMT2.3/MeAMT2.5) in synteny analysis (Figure 4). The intron loss rate has been shown to be faster than the intron gain rate after segmental duplication (Nuruzzaman et al., 2008). Based on this, MeAMT2.5 (containing two introns) may be younger than MeAMT2.3 (containing three introns) (Figure 3C; Figure 4), which might be diverged at 16.4 Mya (Table 2). In contrast, the cassava AMT1 genes contain fewer exons; only MeAMT1;2 gene contains three exons, and all the other MeAMT1 genes contain no introns (Xia et al., 2022). This indicates that there are differences in evolution between AMT1 and AMT2 gene families in cassava, and AMT2 gene family may have more intron insertion during evolution.
In addition, various light responsive, hormone responsive (e.g., ARE, ABRE, SARE, etc.), stress-responsive (drought-responsive and LTR) and plant growth and development-related elements were found in the MeAMT2 promoter regions (Figure 6). But different cis-acting elements were found in each MeAMT2 promoter, indicating that cassava MeAMT2 genes exert multiple or specific functions. Previous studies showed that besides contributing to absorption of ammonium in roots, AtAMT2;1 is mainly responsible for transport of ammonium from roots to shoots (Sohlenkamp et al., 2000; Giehl et al., 2017). The expression level of OsAMT3.1 is relatively low, while OsAMT2.1 is constitutively expressed in both shoots and roots and is not regulated by nitrogen sources (Suenaga et al., 2003). Our study observed tissue specificity for the expression of MeAMT2 genes in cassava (Figure 7), indicating they might have different functions. GmAMT4.1 showed specific expression in arbusculated cortical cells and localized on the periarbuscular membrane (Kobae et al., 2010). The LjAMT2.2 gene was significantly upregulated when inoculating with arbuscular mycorrhizal fungi (AMF) (Wang et al., 2022). Phylogenetic relationship analysis showed that MeAMT2.2, MeAMT2.4, MeAMT2.7, GmAMT4.1, and LjAMT2.2 were all assigned to subgroup 3 (Figure 2). These results suggested that these cassava AMT2 proteins might be responsible for ammonium uptake in arbuscular mycorrhiza symbiosis. In this study, we selected MeAMT2.3, MeAMT2.5 and MeAMT2.6 for further analysis due to higher spatial expression levels. The expression patterns of the three genes were similar under low ammonium stress, especially the MeAMT2.6 gene, which exhibited a significant up-regulation in stems and roots (Figure 8). The function investigation showed that MeAMT2.3, MeAMT2.5 and MeAMT2.6 all could enhance the growth of transgenic yeast under low ammonium condition (Figure 9), suggesting that they might be involved in response to ammonium deficiency.
Conclusion
In conclusion, seven MeAMT2 genes were identified from cassava, and the corresponding proteins were divided into four subgroups based on the phylogeny with other plant species. The gene location on chromosomes, physicochemical property, conserved motifs and gene structures were further analyzed. Genes in the same subgroup exhibited similar structures and properties. Two pairs of segmental duplicated genes were identified in the MeAMT2 family. Spatial expression analysis showed tissue-specific expression for these genes. RT-qPCR analysis showed up-regulation of MeAMT2.3, MeAMT2.5, and MeAMT2.6 genes under ammonium deficiency. Yeast strains overexpressing the above genes grew well under ammonium deficiency. This study provides substantial valuable reference information for further study of nitrogen efficient utilization in cassava.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
YZ and XJ conceived and designed the study and prepared the manuscript. JX, YW, and TZ performed the experiments. JX, YW, TZ, CP, and YJ assisted with the analysis and interpretation of the data. YZ and TZ drafted the manuscript. YZ and XJ participated in the design of the experiments and provided a critical review. All authors have read, edited, and approved the current version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (2018YFD1000500, 2018YFE0207203-2), High-level Talents Project of Basic and Applied Basic Research Program of Hainan Province (in Natural Science) (2019RC046), the Education Department of Hainan Province (Hnky 2021–19), and the Startup funding from Hainan University (KYQD (ZR)1845).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1145735/full#supplementary-material
References
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Edgar, R. C. (2004). Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi:10.1093/nar/gkh340
Filiz, E., and Akbudak, M. A. (2020). Ammonium transporter 1 (AMT1) gene family in tomato (Solanum lycopersicum L.): Bioinformatics, physiological and expression analyses under drought and salt stresses. Genomics 112 (5), 3773–3782. doi:10.1016/j.ygeno.2020.04.009
Gazzarrini, S., Lejay, L., Gojon, A., Ninnemann, O., Frommer, W. B., and von Wiren, N. (1999). Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into arabidopsis roots. Plant Cell. 11, 937–948. doi:10.1105/tpc.11.5.937
Giehl, R. F. H., Laginha, A. M., Duan, F., Rentsch, D., Yuan, L., and von Wirén, N. (2017). A critical role of AMT2;1 in root-to-shoot translocation of ammonium in Arabidopsis. Mol. Plant 10 (11), 1449–1460. doi:10.1016/j.molp.2017.10.001
Hao, D. L., Zhou, J. Y., Yang, S. Y., Qi, W., Yang, K. J., and Su, Y. H. (2020). Function and regulation of ammonium transporters in plants. Int. J. Mol. Sci. 21 (10), 3557. doi:10.3390/ijms21103557
Howitt, S. M., and Udvardi, M. K. (2000). Structure, function and regulation of ammonium transporters in plants. Biochim. Biophys. Acta 1465 (1-2), 152–170. doi:10.1016/S0005-2736(00)00136-X
Hu, M., Hu, W., Xia, Z., Zhou, X., and Wang, W. (2016). Validation of reference genes for relative quantitative gene expression studies in cassava (Manihot esculenta Crantz) by using quantitative real-time PCR. Front. Plant Sci. 7, 680. doi:10.3389/fpls.2016.00680
Hui, J., An, X., Li, Z., Neuhauser, B., Ludewig, U., Wu, X., et al. (2022). The mycorrhiza-specific ammonium transporter ZmAMT3;1 mediates mycorrhiza-dependent nitrogen uptake in maize roots. Plant Cell. 34 (10), 4066–4087. doi:10.1093/plcell/koac225
Kobae, Y., Tamura, Y., Takai, S., Banba, M., and Hata, S. (2010). Localized expression of arbuscular mycorrhiza-inducible ammonium transporters in soybean. Plant Cell. Physiol. 51 (9), 1411–1415. doi:10.1093/pcp/pcq099
Koegel, S., Ait Lahmidi, N., Arnould, C., Chatagnier, O., Walder, F., Ineichen, K., et al. (2013). The family of ammonium transporters (AMT) in Sorghum bicolor: Two AMT members are induced locally, but not systemically in roots colonized by arbuscular mycorrhizal fungi. New Phytol. 198 (3), 853–865. doi:10.1111/nph.12199
Kronzucker, H. J., Siddiqi, M. Y., and Glass, A. D. M. (1996). Kinetics of NH4+ influx in spruce. Plant Physiol. 110, 773–779. doi:10.1104/pp.110.3.773
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). Mega X: Molecular evolutionary Genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Li, B. Z., Merrick, M., Li, S. M., Li, H. Y., Zhu, S. W., Shi, W. M., et al. (2009). Molecular basis and regulation of ammonium transporter in rice. Rice Sci. 16 (4), 314–322. doi:10.1016/S1672-6308(08)60096-7
Li, H., Cong, Y., Lin, J., and Chang, Y. H. (2015). Molecular cloning and identification of an ammonium transporter gene from pear. Plant Cell. Tiss. Organ Cult. 120, 441–451. doi:10.1007/s11240-014-0611-4
Li, H., Cong, Y., Chang, Y., and Lin, J. (2016). Two AMT2-type ammonium transporters from Pyrus betulaefolia demonstrate distinct expression characteristics. Plant Mol. Biol. Rep. 34, 707–719. doi:10.1007/s11105-015-0957-8
Li, T., Liao, K., Xu, X., Gao, Y., Wang, Z., Zhu, X., et al. (2017). Wheat ammonium transporter (AMT) gene family: Diversity and possible role in host-pathogen interaction with stem rust. Front. Plant Sci. 8, 1637. doi:10.3389/fpls.2017.01637
Liu, X. J., Zhang, Y., Han, W. X., Tang, A. H., Shen, J. L., Cui, Z. L., et al. (2013). Enhanced nitrogen deposition over China. Nature 494 (7438), 459–462. doi:10.1038/nature11917
Liu, Q., Dang, H., Chen, Z., Wu, J., Chen, Y., Chen, S., et al. (2018). Genome-wide identification, expression, and functional analysis of the sugar transporter gene family in cassava (Manihot esculenta). Int. J. Mol. Sci. 19, 987. doi:10.3390/ijms19040987
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Loqué, D., and von Wirén, N. (2004). Regulatory levels for the transport of ammonium in plant roots. J. Exp. Bot. 55, 1293–1305. doi:10.1093/jxb/erh147
Loque, D., Lalonde, S., Looger, L. L., von Wiren, N., and Frommer, W. B. (2007). A cytosolic trans-activation domain essential for ammonium uptake. Nature 446, 195–198. doi:10.1038/nature05579
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290 (5494), 1151–1155. doi:10.1126/science.290.5494.1151
Marini, A. M., Soussi-Boudekou, S., Vissers, S., and Andre, B. (1997). A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 4282–4293. doi:10.1128/MCB.17.8.4282
McDonald, T. R., Dietrich, F. S., and Lutzoni, F. (2012). Multiple horizontal gene transfers of ammonium transporters/ammonia permeases from prokaryotes to eukaryotes: Toward a new functional and evolutionary classification. Mol. Biol. Evol. 29, 51–60. doi:10.1093/molbev/msr123
Mo, C., Wan, S., Xia, X., Ren, N., Zhou, Y., and Jiang, X. Y. (2018). Expression patterns and identified protein-protein interactions suggest that cassava CBL-CIPK signal networks function in responses to abiotic stresses. Front. Plant Sci. 9, 269. doi:10.3389/fpls.2018.00269
Neuhauser, B., Dynowski, M., Mayer, M., and Ludewig, U. (2007). Regulation of NH4+ transport by essential cross talk between AMT monomers through the carboxyl tails. Plant Physiol 143, 1651–1659. doi:10.1104/pp.106.094243
Neuhäuser, B., Ludewig, U., and Dynowski, M. (2009). Channel-like NH3 flux by ammonium transporter AtAMT2. FEBS Lett. 583, 2833–2838. doi:10.1016/j.febslet.2009.07.039
Ninnemann, O., Jauniaux, J., and Frommer, W. (1994). Identification of a high affinity NH4+ transporter from plants. EMBO J. 13 (15), 3464–3471. doi:10.1002/j.1460-2075.1994.tb06652.x
Nuruzzaman, M., Gupta, M., Zhang, C., Wang, L., Xie, W., Xiong, L., et al. (2008). Sequence and expression analysis of the thioredoxin protein gene family in rice. Mol. Gen. Genomics 280, 139–151. doi:10.1007/s00438-008-0351-4
Ou, W., Mao, X., Huang, C., Tie, W., Yan, Y., Ding, Z., et al. (2018). Genome-wide identification and expression analysis of the KUP family under abiotic stress in cassava (Manihot esculenta Crantz). Front. Physiol. 9, 17. doi:10.3389/fphys.2018.00017
Ranathunge, K., Ei-kereamy, A., Gidda, S., Bi, Y. M., and Rothstein, S. J. (2014). AMT1;1 transgenic rice plants with enhanced NH4+ permeability show superior growth and higher yield under optimal and suboptimal NH4+ conditions. J. Exp. Bot. 65 (4), 965–979. doi:10.1093/jxb/ert458
Socci, A. M., and Templer, P. H. (2011). Temporal patterns of inorganic nitrogen uptake by mature sugar maple (Acer saccharum Marsh.) and red spruce (Picea rubens Sarg.) trees using two common approaches. Plant Ecol. divers. 4 (2), 141–152. doi:10.1080/17550874.2011.624557
Sohlenkamp, C., Shelden, M., Howitt, S., and Udvardi, M. (2000). Characterization of Arabidopsis AtAMT2, a novel ammonium transporter in plants. FEBS Lett. 467, 273–278. doi:10.1016/S0014-5793(00)01153-4
Sohlenkamp, C., Wood, C. C., Roeb, G. W., and Udvardi, M. K. (2002). Characterization of Arabidopsis AtAMT2, a high-affinity ammonium transporter of the plasma membrane. Plant Physiol. 130 (4), 1788–1796. doi:10.1104/pp.008599
Sonoda, Y., Ikeda, A., Saiki, S., Yamaya, T., and Yamaguchi, J. (2003). Feedback regulation of the ammonium transporter gene family AMT1 by glutamine in rice. Plant Cell. Physiol. 44, 1396–1402. doi:10.1093/pcp/pcg169
Straub, T., Ludewig, U., and Neuhäuser, B. (2017). The kinase CIPK23 inhibits ammonium transport in Arabidopsis thaliana. Plant Cell 29, 409–422. doi:10.1105/tpc.16.00806
Suenaga, A., Moriya, K., Sonoda, Y., Ikeda, A., von Wiren, N., Hayakawa, T., et al. (2003). Constitutive expression of a novel-type ammonium transporter OsAMT2 in rice plants. Plant Cell. Physiol. 44, 206–211. doi:10.1093/pcp/pcg017
Tabata, R., Sumida, K., Yoshii, T., Ohyama, K., Shinohara, H., and Matsubayashi, Y. (2014). Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346, 343–346. doi:10.1126/science.1257800
Wang, M. Y., Siddiqi, M. Y., and Glass, A. D. M. (1996). Interactions between K+ and NH4+: Effects on ion uptake by rice roots. Plant Cell. Environ. 19, 1037–1046. doi:10.1111/j.1365-3040.1996.tb00210.x
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49. doi:10.1093/nar/gkr1293
Wang, Y., Zhou, W., Wu, J., Xie, K., and Li, X. (2022). LjAMT2;2 promotes ammonium nitrogen transport during arbuscular mycorrhizal fungi symbiosis in Lotus japonicus. Lotus Jpn. Int. J. Mol. Sci. 23 (17), 9522. doi:10.3390/ijms23179522
Williams, L. E., and Miller, A. J. (2001). Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu. Rev. Plant Phys. 52, 659–688. doi:10.1146/annurev.arplant.52.1.659
Wilson, M. C., Mutka, A. M., Hummel, A. W., Berry, J., Chauhan, R. D., Vijayaraghavan, A., et al. (2017). Gene expression atlas for the food security crop cassava. New Phytol 213 (4), 1632–1641. doi:10.1111/nph.14443
Xia, Y., Liu, Y., Zhang, T., Wang, Y., Jiang, X. Y., and Zhou, Y. (2022). Genome-wide identification and expression analysis of ammonium transporter 1 (AMT1) gene family in cassava (Manihot esculenta Crantz) and functional analysis of MeAMT1;1 in transgenic Arabidopsis. 3 Biotech. 12, 4. doi:10.1007/s13205-021-03070-6
Xu, G. H., and Takahashi, H. (2020). Improving nitrogen use efficiency: From cells to plant systems. J. Exp. Bot. 71 (15), 4359–4364. doi:10.1093/jxb/eraa309
Xu, G., Fan, X., and Miller, A. J. (2012). Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63, 153–182. doi:10.1146/annurev-arplant-042811-105532
Xu, J., Duan, X., Yang, J., Beeching, J. R., and Zhang, P. (2013). Enhanced reactive oxygen species scavenging by overproduction of superoxide dismutase and catalase delays postharvest physiological deterioration of cassava storage roots. Plant Physiol. 161, 1517–1528. doi:10.1104/pp.112.212803
Xuan, W., Beeckman, T., and Xu, G. (2017). Plant nitrogen nutrition: Sensing and signaling. Curr. Opin. Plant Biol. 39, 57–65. doi:10.1016/j.pbi.2017.05.010
Yang, S., Yuan, D. P., Zhang, Y., Sun, Q., and Xuan, Y. H. (2021). BZR1 regulates brassinosteroid-mediated activation of AMT1;2 in rice. Front. Plant Sci. 12, 665883. doi:10.3389/fpls.2021.665883
Yin, X. C., Xia, Y. Q., Xie, Q., Cao, Y. X., Wang, Z. Y., Hao, G. P., et al. (2020). The protein kinase complex CBL10-CIPK8-SOS1 functions in Arabidopsis to regulate salt tolerance. J. Exp. Bot. 71 (6), 1801–1814. doi:10.1093/jxb/erz549
You, L. L., Wang, Y., Zhang, T. T., Zhu, Y. F., Ren, N., Jiang, X. Y., et al. (2022). Genome-wide identification of nitrate transporter 2 (NRT2) gene family and functional analysis of MeNRT2.2 in cassava (Manihot esculenta Crantz). Gene 809, 146038. doi:10.1016/j.gene.2021.146038
Yuan, L., Loque, D., Kojima, S., Rauch, S., Ishiyama, K., Inoue, E., et al. (2007). The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1 type transporters. Plant Cell. 19 (8), 2636–2652. doi:10.1105/tpc.107.052134
Zhang, W. F., Cao, G. X., Li, X. L., Zhang, H. Y., Wang, C., Liu, Q. Q., et al. (2016). Closing yield gaps in China by empowering smallholder farmers. Nature 537 (7622), 671–674. doi:10.1038/nature19368
Zhang, F., Liu, Y., Wang, L. Y., Bai, P. X., Ruan, L., Zhang, C. C., et al. (2018). Molecular cloning and expression analysis of ammonium transporters in tea plants (Camellia sinensis (L.) O. Kuntze) under different nitrogen treatments. Gene 658, 136–145. doi:10.1016/j.gene.2018.03.024
Zheng, L., Kostrewa, D., Bernèche, S., Winkler, F. K., and Li, X. D. (2004). The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 101 (49), 17090–17095. doi:10.1073/pnas.0406475101
Keywords: ammonium uptake, ammonium deficiency, ammonium transporter 2, gene expression, cassava
Citation: Xia J, Wang Y, Zhang T, Pan C, Ji Y, Zhou Y and Jiang X (2023) Genome-wide identification, expression profiling, and functional analysis of ammonium transporter 2 (AMT2) gene family in cassava (Manihot esculenta crantz). Front. Genet. 14:1145735. doi: 10.3389/fgene.2023.1145735
Received: 16 January 2023; Accepted: 13 February 2023;
Published: 22 February 2023.
Edited by:
Vijay Gahlaut, Chandigarh University, IndiaReviewed by:
Harinder Vishwakarma, Indian Council of Agricultural Research (ICAR), IndiaLi Song, Yangzhou University, China
Copyright © 2023 Xia, Wang, Zhang, Pan, Ji, Zhou and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Zhou, zhouyang@hainanu.edu.cn; Xingyu Jiang, jiangxingyuhu@163.com
†These authors have contributed equally to this work
 Jinze Xia1†
Jinze Xia1† Tingting Zhang
Tingting Zhang Yang Zhou
Yang Zhou Xingyu Jiang
Xingyu Jiang