- 1Department of Endocrinology and Metabolism, Cancer Center West China Hospital, Sichuan University, Chengdu, Sichuan, China
- 2Department of Endocrinology and Metabolism, West China Hospital, West China Medical School, Sichuan University, Chengdu, Sichuan, China
Hepatocellular carcinoma (HCC) is one of the most common causes of cancer-related death worldwide. Both genetic and environmental factors through a multitude of underlying molecular mechanisms participate in the pathogenesis of HCC. Recently, numerous studies have shown that circular RNAs (circRNAs), an emerging class of non-coding RNAs characterized by the presence of covalent bonds linking 3’ and 5’ ends, play an important role in the initiation and progression of cancers, including HCC. In this review, we outline the current status of the field of circRNAs, with an emphasis on the functions and mechanisms of circRNAs in HCC and its microenvironment. We also summarize and discuss recent advances of circRNAs as biomarkers and therapeutic targets. These efforts are anticipated to throw new insights into future perspectives about circRNAs in basic, translational and clinical research, eventually advancing the diagnosis, prevention and treatment of HCC.
1 Introduction
Liver cancer, the leading cause of cancer-related deaths in many countries, remains a global health challenge (Akinyemiju et al., 2017), and its prevalence continues to increase with an estimated incidence of >1 million cases by 2025 (Llovet J. et al., 2021). Hepatocellular carcinoma (HCC) represents the most frequent primary liver cancer. Most HCC patients are usually diagnosed at an advanced stage and accompanied by metastasis. In this regard, early detection is an effective option to attenuate HCC-related death and achieve long-term disease-free survival. Unfortunately, the only blood-based biomarker currently validated for HCC surveillance is α-fetoprotein. Furthermore, the principal cancer diagnosis methods, including imaging examination and histopathology (Yang and Heimbach, 2020), are difficult to detect concealed early symptoms of HCC. Therefore, early diagnosis of HCC remains a challenge that needs to be solved. On the other hand, researchers have been committed to finding effective systemic management for HCC for the past 70 years. Although the treatment of HCC is becoming more reasonable, the curative effect has not reached the ideal impact. For example, Sorafenib and Lenvatinib, two representative first-line drugs for advanced HCC, are just effective in a subset of patients. In addition, locoregional therapies play a substantial role in the management of 50%–60% HCC, but molecular therapies dominate the adjuvant trials after curative therapies (Llovet et al., 2021b). Thus, the molecular mechanisms underlying HCC pathogenesis, especially the identification of new druggable targets, are of significance for the prevention and treatment of the HCC pandemic.
CircRNAs, as the name suggests, are a class of single-stranded closed circle molecules that lack 5’ and 3’ ends and poly (A) tails, which make them resistant to RNase R and more stable than linear RNAs (Liu and Chen, 2022). When first discovered in the early 1970 s, circRNAs were thought to be byproducts of splicing without meritorious biological functions (Hsu and Coca-Prados, 1979). However, with the emergence of high-throughput sequencing technologies and bioinformatics approaches, many differentially expressed circRNAs have been identified in viruses, insects, plants and mammals (Tholken et al., 2019; Zhang P. et al., 2020; Espindola et al., 2021; Jiang et al., 2021). Moreover, numerous studies have indicated the potential of circRNAs as a promising disease biomarker. A common experimental standard for identifying, isolating, analyzing circRNAs is essential for circRNA biology. The improvement of practice processes and standards also provides convenience for circRNA research (Nielsen et al., 2022) (Figure 1). Certainly, increasing evidence suggests that circRNAs are involved in the occurrence and development of various diseases (Gomes et al., 2020; Stoll et al., 2020). In particular, most studies have focused on the role of circRNAs in cancers and demonstrated their pivotal roles in the pathogenesis of cancer (Wang Y. et al., 2020; Wang Y. et al., 2022), including HCC (Yang X. et al., 2021; Chen and Shan, 2021). Indeed, many classical tumor-related factors and signaling pathways, such as c-Myc, MAPK and Hedgehog, are mediated by circRNAs in HCC (Hu et al., 2020; Gu et al., 2021b; Chen et al., 2022b). These results indicate that circRNAs are emerging as novel cancer biomarkers and therapeutic targets.
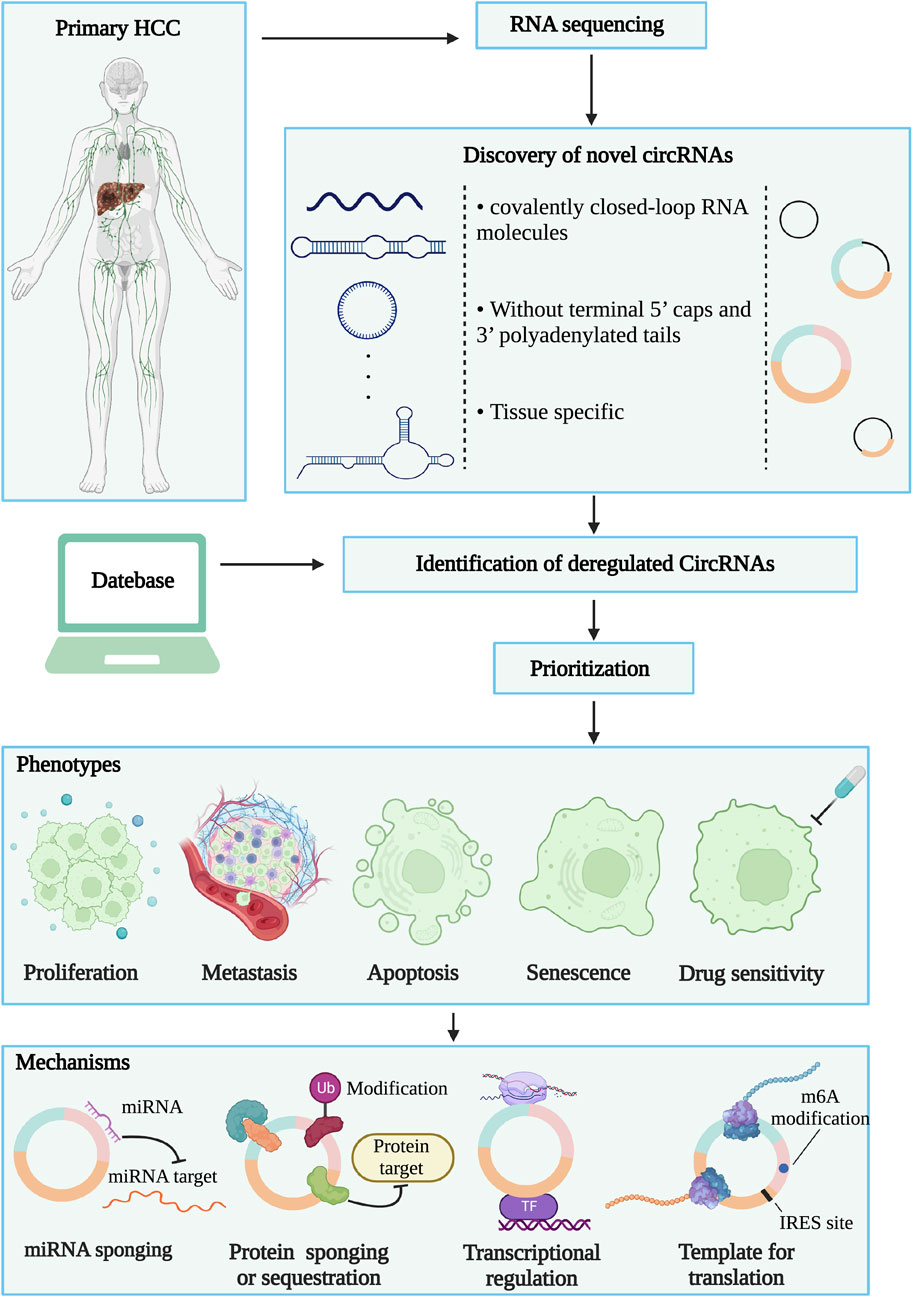
FIGURE 1. Identification and research strategies of circRNAs. Firstly, RNA sequencing is performed between hepatocellular carcinoma tissue and normal liver tissue. By comparing the two sets of sequencing data, novel or deregulated circRNAs could be found. Then, the technology of omics data integration and bioinformatics analysis prioritized circRNAs candidates. Finally, the screened circRNAs will be disclosed the function and mechanism characterization of liver cancer through biological experimental technology.
In recent years, tremendous efforts have been devoted to unraveling the involvement of circRNAs in HCC pathology. These exciting advances had brought new insights into tumorigenesis, and provided alternative tools for diagnosis and potential therapeutic approaches in clinic for HCC. In this review, we summarized the characterization, roles, and underlying mechanisms of circRNAs in HCC, and also discussed the current dilemma and future development direction of circRNAs.
2 Non-coding RNAs
In the nucleus of human cells, approximately 90% of the genomic sequence is transcribed into RNA, but less than 2% can be translated into functional proteins. The rest untranslated portion is traditionally regarded as “dark matter” that contains a vast portion of functional non-coding RNAs (ncRNAs), as demonstrated by the ENCODE project (Doolittle, 2013). NcRNAs can be grouped into several classes by size. Small ncRNAs (sncRNAs), including microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs) and tRNA-derived small RNAs (tsRNAs), play a key role in carcinogenesis. Long ncRNAs (lncRNAs), comprising ncRNAs longer than 200 nucleotides (nt), include pseudogenes and circRNAs (Slack and Chinnaiyan, 2019). NcRNAs are involved in numerous physiological and pathological processes, and thus modulate disease progression (Fu et al., 2015; Tian et al., 2018; Ma et al., 2019; Zhang W. et al., 2021). Accordingly, the recent decades have witnessed remarkable advances of ncRNAs, particularly miRNAs and lncRNAs, in both physiology and pathology (Liu G. et al., 2020; Du W. et al., 2022; Zou et al., 2023a). Although most of the molecular mechanisms are incompletely clear, circRNAs have shown universal and critical roles in regulating cellular processes of various diseases including cancers, which emphasizes the importance of circRNAs (Li et al., 2020b).
3 CircRNAs
CircRNAs, a class of ncRNAs, are usually generated from fragments of linear pre-messenger RNAs or other linear RNA species (Liu and Chen, 2022). CircRNAs are a kind of tissue-specific and covalently closed-loop RNA molecules, which are different from other ncRNAs (Liu and Chen, 2022). CircRNAs, which are without terminal 5’ caps and 3’ polyadenylated tails, are more stable than other ncRNAs. This high stability may be due to its covalently circular structure that avoids exonuclease-mediated degradation (Hou et al., 2023). The mechanisms underlying circRNA biosynthesis remain unclear. Currently, there are two patterns of circRNAs formation, namely, the ‘direct back-splicing’ model and the ‘lariat intermediate’ model (Chen L., 2020). According to the biogenesis mechanisms, four types of circRNAs have been classified: exonic circRNAs (ecircRNAs), intronic circRNAs (ciRNAs), exon-intron circRNAs (EIciRNAs), and intergenic circRNAs (icircRNAs). EcircRNAs are composed of one or several exons of cyclization, whereas ciRNAs contain only introns. Exons and introns cross cyclization are the existence form of EIciRNAs. Approximately 84% of circRNAs are derived from protein-coding genes (van Zonneveld et al., 2021), however, the region of icircRNAs sequency between two protein-coding genes. EcircRNAs and intergenic circRNAs are located in the cytoplasm, while ciRNAs and EIciRNAs generally localize to the nucleus. By the way, ecircRNAs make up the majority of circRNAs and are more conserved than other types of circRNAs (Liu and Chen, 2022). Recently, Shan et al. discovered a new class of circRNAs, named mecciRNAs, which are encoded by the mitochondrial genome (Liu et al., 2020c). MecciRNAs can facilitate the mitochondrial entry of nuclear-encoded proteins by serving as molecular chaperones. Further studies found that circRNAs are involved in immune responses, cell proliferation and transformation, and neuronal functions (Yan and Chen, 2020; D'Anca et al., 2022). In addition, dysregulation of circRNAs is associated with a wide range of diseases and can have phenotypes in animal models.
3.1 Biogenesis and regulation of circRNAs
The formation of circRNAs is different from other RNAs. CircRNAs are derived from canonical 3’ and 5’ splice sites (Vo et al., 2019). High-throughput analyses of genomic characterization of circRNAs exons combined with mutational analyses in circRNA expression vectors showed that the canonical spliceosomal machinery carries out a particular type of alternative splicing, named back-splicing (Starke et al., 2015). CircRNAs are generated from linear pre-messenger RNAs and other linear RNA species through back-splicing. This particular type of splicing connects the RNA downstream 5’ splice-donor (SD) site and an upstream 3’ splice-acceptor (SA) site to form a covalently closed loop (Dragomir and Calin, 2018; Chen L. L., 2020). CircRNAs usually have long introns on the flanks of exons, and back-splicing mediates the base-pairing of introns by inverted repeat elements (such as Alu elements) and looping formation (Jeck et al., 2013; Zhang X. O. et al., 2014). The base-pairing between inverted repeat elements recruits a downstream SD site into proximity with an upstream SA site, then an upstream branch point (BP) attacks a downstream SD site. After 5’ end of a downstream SD site breaks away from pre-mRNA, the downstream SD site attacks an upstream SA site and binds closely (Kristensen et al., 2019; Long et al., 2021). This connection results in the formation of EIciRNAs or ecircRNAs (the internal intron is spliced out). In addition, the dimerization of RNA-binding proteins (RBP) such as protein quaking, FUS, HNRNPL and MBL, has been shown to regulate certain circRNAs biogenesis by binding to specific elements in flanking introns (Yang T. et al., 2021; Ho et al., 2021; Han J. et al., 2022; Pamudurti et al., 2022).
In addition to the biogenesis model mentioned above, a ‘lariat intermediate’ model has been purposed, in which canonical linear splicing happens first to produce a linear mRNA and long intron lariat (Liu and Chen, 2022). CiRNAs could derive from intron lariat and escape the debranching step of linear splicing (Zhang et al., 2013). More importantly, exon-skipping events usually occur during linear splicing and produce a long intron lariat. The long intron lariat containing the skipped exon can mediate the formation of circRNA by internal back-splicing (Kelly et al., 2015; Eger et al., 2018). Notably, not every exon-skipping event can trigger back-splicing to produce circRNAs, but skipping exons that undergo back-splicing are less present in mature mRNAs (Kelly et al., 2015). Although the above mechanism hypotheses have been widely studied, the precise mechanisms of circRNA biogenesis are still obscure.
Both models rely on back-splicing, which is accomplished by the canonical splicing machinery. In this regard, anything modulating the spliceosome might logically influence the formation of circRNAs. Using RNAi screening in Drosophila melanogaster, Liang et al. found that depletion of SF3b or SF3a, components of the U2 small nuclear ribonucleoprotein particle (snRNP) that are required for pre-spliceosome assembly, resulted in obviously increased in circRNAs levels, accompanied by decreased linear RNA levels (Liang et al., 2017). This phenomenon suggests that back-splicing becomes a preferred pathway of pre-mRNA processing when canonical pre-mRNA processing events are impaired (Liang et al., 2017). Furthermore, some spliceosomal factors are involved in circRNA formation through directly or indirectly binding to specific RNA motifs. For example, Quaking (QKI) strengthens circRNA formation by binding to specific sites in flanking introns (Conn et al., 2015). The YY1 complex could bind to the super-enhancer and promoter of QKI to increase the formation of certain circRNAs (Han J. et al., 2022). Kramer et al. found that the combination of cis-acting elements and trans-acting splicing factors, including hnRNP (heterogeneous nuclear ribonucleoprotein) and SR (serine–arginine) proteins, regulates the biogenesis of Laccase circRNA (Kramer et al., 2015). RBPs, except core spliceosomal factors, can modulate back-splicing in trans-acting by binding the intronic complementary sequence (ICS). NF90 and NF110, both of which are protein products of interleukin enhancer-binding factors 3 (ILF3) and involved in the host antiviral system, are able to promote circRNA production by acting on inverted-repeat Alu elements and stabilizing intronic RNA pairs in the nucleus (Li et al., 2017; Shang et al., 2022). In contrast to NF90 and NF110, double-stranded RNA-specific adenosine deaminase 1 (ADAR1) prevents the looping of flanking intron by performing adenosine to inosine editing. Accordingly, the destruction of inverted-repeat Alu elements is capable of impairing the complementarity and stability of intronic RNA pairs (Ivanov et al., 2015; Eisenberg and Levanon, 2018). Similar as ADAR1, DHX9 is an ATP-dependent nuclear RNA helicase and can suppress circRNA biogenesis by binding to inverted-repeat Alu elements and unwinding RNA pairs that flank circularized exons (Aktas et al., 2017). Interestingly, DHX9 is remarkably upregulated in HCC tissues and regulates the expression of circDLC1, which inhibits liver cancer progression (Liu H. et al., 2021). In addition, Zhang et al. found that back-splicing is correlated with RNA Polymerase II transcription elongation rate (TER). The faster TER of genes results in a higher probability and production of circRNAs (Zhang et al., 2016), which probably due to the fact that a higher TER could transcribe more downstream ICS, thereby increasing the possibility of ICS pairing across exons (Chen L. L., 2020).
3.2 Biological functions of circRNAs
Truth will not be changed by misunderstanding of people. CircRNAs initially existed in the world as by-products of splicing, nevertheless, accumulating data demonstrate that circRNAs have diverse biological functions, such as miRNAs sponge and proteins scaffold. Figure 2 shows the function of circRNAs, and the further details are described as follows.
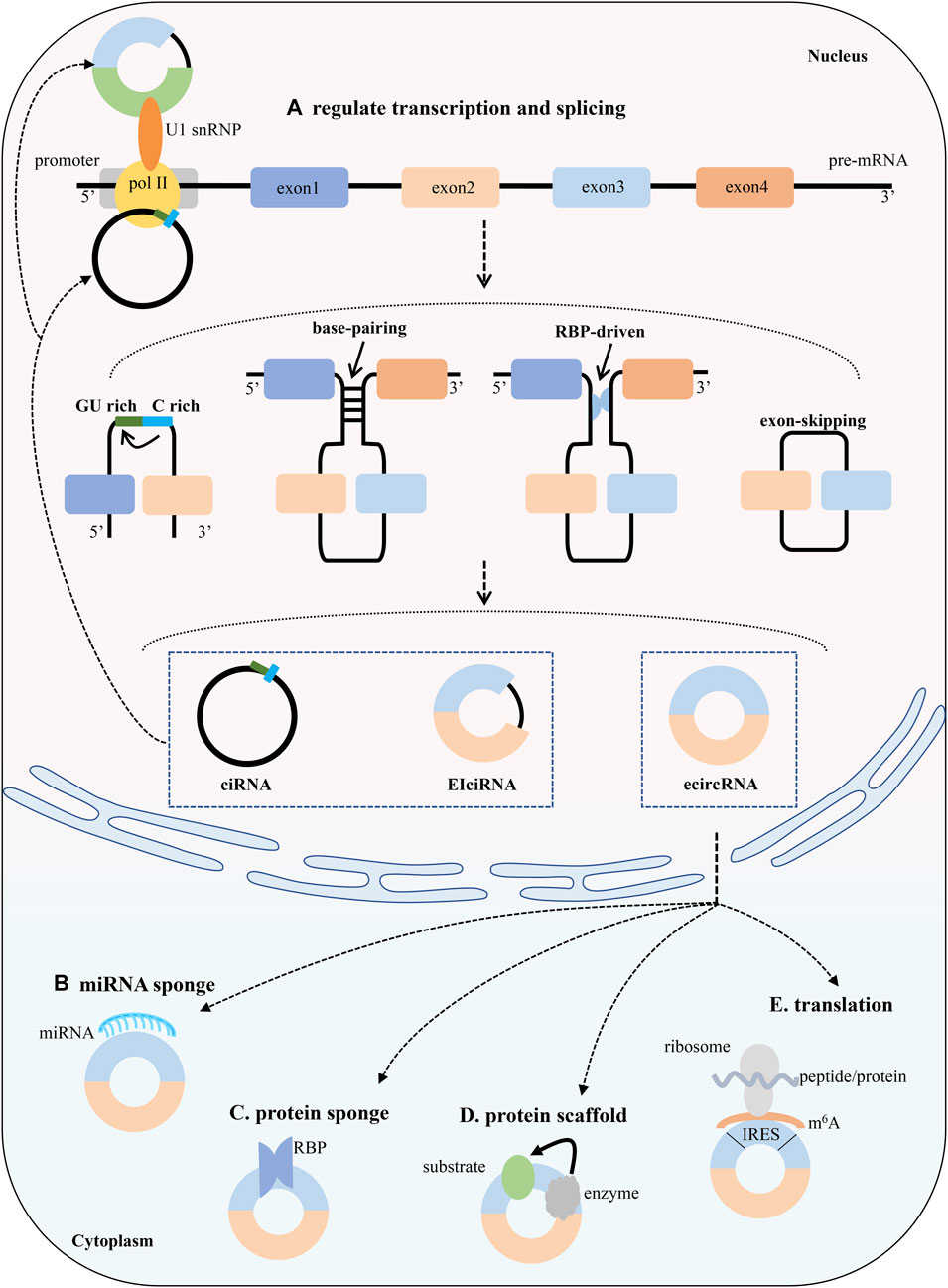
FIGURE 2. Biogenesis and functions of circRNAs. The canonical back-splicing generates three types of circRNAs: ciRNAs, EIciRNAs and ecircRNAs. CircRNAs enhance the transcription and splicing of their parental genes by interacting with RNA pol II or U1 small nuclear snRNP. Functions (A) Competition with splicing and regulation of parental gene transcription (B) miRNA sponges. CircRNAs affects the expression of miRNA and downstream target genes by adsorbing miRNAs (C) Protein sponges. CircRNAs regulates target proteins by binding to them (D) Prontein scaffolds. CircRNAs facilitates the formation of a complex (E) Translation. CircRNAs can be translated into peptides or proteins.
3.2.1 CircRNAs can act as miRNA sponges
The great majority of circRNAs are predominantly localized in the cytoplasm and contain miRNA-response elements. Consequently, circRNAs can function as competing for endogenous RNAs (ceRNAs) or miRNA sponges to sequester miRNA, and thus prevent the interactions between miRNA and mRNA (Cui et al., 2021). CDR1as (cerebellar degeneration-related protein 1 antisense transcript; also known as ciRS-7) is the most representative circRNA as yet, containing more than 70 conversed binding sites for miR-7. Thus, CDR1as can function as a negative regulator of miR-7, leading to an increase in the expression of miR-7 target genes (Guo et al., 2020). Intriguingly, CDR1as and miR-7 are enriched in brain tissues (Memczak et al., 2013), indicating that CDR1as may be involved in the development and function of the nervous system. Indeed, CDR1as-knockouted mice exhibited neuropsychiatric disorders associated with a strong deficit in prepulse inhibition of the startle response (Piwecka et al., 2017). In addition to its neuron-modulating function, CDR1as has been recently shown to regulate tumorigenesis, including HCC, through sponging miR-7. For example, Yang et al. found that CDR1as promotes HCC cell proliferation at least in part through the regulation of the expression of epidermal growth factor receptor (EGFR), a target gene of miR-7 (Yang X. et al., 2017). Other miR-7 target genes, such as phosphoinositide 3-kinase catalytic subunit delta (PIK3CD) and cyclin E1 (CCNE1), may also attribute to the oncogenic role of CDR1as in HCC progression (Fang et al., 2012; Zhang X. et al., 2014). Consistently, CDR1as is also upregulated and facilitates cell proliferation by activating miR-7 target genes in esophageal cancer, osteosarcoma, and nasopharyngeal carcinoma (Li R. et al., 2018; Xu et al., 2018; Zhong et al., 2019). Beyond CDR1as, CircZNF91 is regarded as another representative miRNA sponge. It contains 24 target sites for miR-23b-3p, a crucial regulator of many physiological processes, providing an attractive target for stem cell biology (Zeng Z. et al., 2021). The mouse circSry, derived from sex-determining region Y, contains 16 target sites for miR-138-5p and is involved in the development of testis (Song et al., 2023). These studies suggest that there is a competitive relationship between circRNAs and mRNA in binding miRNA. Of note, unlike the above-mentioned circRNAs that can act as a miRNA sponge, the vast majority of circRNAs do not contain more miRNA binding sites than expected by chance (Guo et al., 2014). Thus, when identifying the sponge role of circRNAs, it is necessary to consider the stoichiometric relationship between the miRNA binding sites of the circRNA and the mRNA target sites of the miRNA (Jarlstad Olesen and Lasse, 2021).
3.2.2 CircRNAs interact with RBPs
CircRNAs can also interact with RBPs to modulate the function and location of associated proteins, functioning as either protein scaffolds or protein sponges. Du et al. conducted a series of studies on circ-Foxo 3, a circular RNA generated from a member of the forkhead transcription factors family (Du et al., 2016). These studies demonstrate a role of Circ-Foxo 3 in cell cycle progression. Specifically, ectopic expression of circ-Foxo3 in the cytoplasm can repress cell cycle progression by binding to 2 cell cycle proteins, cyclin-dependent kinase 2 (CDK2) and cyclin-dependent kinase inhibitor 1 (p21), resulting in the formation of a ternary complex that prevents disease progression, such as HCC. Cellular senescence is defined as the arrest of cell cycle progression. Interestingly, circ-Foxo 3 can interact with several proteins, including the anti-senescent protein ID-1, the transcription factor E2F1 and the anti-stress proteins FAK and HIF1α, which subsequently prevents FAK to locate to mitochondria or HIF1α to translocate to the nucleus, exacerbating cardiac cell senescence under stressed conditions (Du et al., 2017b). In addition, circ-Foxo 3 promotes the mouse double-minute 2 (MDM2)-mediated ubiquitylation of mutant p53, and facilitates the proteasome-meditated mutant p53 degradation in breast carcinoma (Du et al., 2017a).
It has shown that a number of circRNAs could bind to HuR, a crucial RBP regulator for protein translation, and thus play a role in tumorigenesis. For instance, circPPFIA1s are generated from PPFIA1 with different length, including circPPFIA1-L (long) and -S (short). CircPPFIA1s binds to HuR and destroys the mRNA stability effect of HuR on oncogenic RAB36, thus decreasing RAB36 expression and inhibiting the liver metastasis of colorectal cancer (Ji et al., 2022). CircBACH1 is able to directly interact with HuR and facilitate HuR translocation to the cytoplasm, resulting in decreased p27 translation and increased HCC cell proliferation (Liu B. et al., 2020). Interestingly, recent studies suggest that circRNAs might serve as an entry point for tumor immunotherapy. It has shown that endogenous circRNAs tend to form a 16–26 bp imperfect RNA duplexes and bind to double-stranded RNA (dsRNA)-activated protein kinase (PKR), and thus prevent aberrant activation of innate immunity (Liu C. X. et al., 2019). In response to poly (I:C) stimulation or viral infection, circRNAs are globally degraded by RNase L, and PKR molecules are released to activate the immune response. It is of interest for future investigation to delineate the interplay of circRNAs and their associated proteins in immunity and tumorigenesis.
3.2.3 CircRNAs regulate transcription and splicing
A few circRNAs are localized in the nucleus and play a role in transcription regulation. CircRNAs in the nucleus, mainly CiRNA and EIciRNA, are involved in gene expression at the transcription level (Zhang et al., 2013; Li Z. et al., 2015). CircRIG-I binds to DDX3X and activates MAVS/TRAF5/TBK1 signaling cascade, which are crucial for activation of host innate immunity. TBK activates the transcriptional factor IRF3 through phosphorylation to form homodimers, eventually initiating the transcription of type I IFNs and various antiviral genes via binding to the target genome region (Song et al., 2022). On the contrary, several circRNAs can inhibit the transcription of their target genes. Circ-HuR directly interacts with CCHC-type zinc finger nucleic acid binding protein (CNBP) and acts as an inhibitor to restrain the binding of CNBP to HuR promoter, resulting in HuR repression (Yang F. et al., 2019). Notably, recent studies have indicated that several cytoplasmic circRNAs are also involved in the modulation of transcription. For example, circSLC25A16 in the cytoplasm can promote the transcription of lactate dehydrogenase A (LDHA) by interacting with miR-488-3p and hypoxia-inducible factor 1-alpha (HIF-1α) (Shangguan et al., 2020). CircPPKAA1 recruits mSREBP-1 and Ku80/Ku70 to form a ternary complex in HCC, thus facilitating the stability of mSREBP-1, a transcription factor to drive expression of essential fatty acid synthesis enzymes. Of note, the circPPKAA/Ku80/Ku70/mSREBP-1 ternary complex locates to the promoters of the fatty acid biosynthesis genes, and upregulates their transcription, including ACC1, ACLY, FASN and SCD1 (Li Q. et al., 2022). Although a growing number of circRNAs have been identified to be involved in the transcription of their host genes, the relevant mechanisms of action remain unclear.
Notably, circRNAs also influence the biogenesis of itself through affecting the canonical splicing and host gene transcription. Working in Drosophila indicated a genome-wide competition between canonical splicing and circRNAs generation. For example, MBL promotes the circularization of its own exon 2 to the detriment of mRNA production (Ashwal-Fluss et al., 2014). Another study, by exhausting or inhibiting the spliceosome components, found that the steady-state levels of circular RNAs were increased while the expression of their associated linear mRNAs concomitantly decreased (Liang et al., 2017). Back-splicing and canonical splicing use the same 3’ and 5’ splice sites may be part of the reasons for the negative correlation between mRNA production rates and circRNA biogenesis rates.
3.2.4 CircRNAs can translate proteins or peptides
CircRNAs were initially considered as a kind of endogenous ncRNA, since they lack the typical 5’ cap and 3’ poly(A) tail that are the essential elements for cap-dependent translation initiation. Nevertheless, a vast part of circRNAs are mainly located in the cytoplasm, the location of translation, and derived from the middle exons of pre-mRNA. More importantly, cap-independent translation pattern has been discovered (Wang J. et al., 2022). This indicates translation may be the potential function of circRNAs. As expected, a growing body of research has shown that translation can also proceed on circRNAs through cap-independent way, namely, internal ribosome entry sites (IRESs) or N6-methyladenosine (m6A)-dependent translation (Wen et al., 2022). Circular E-cadherin (circ-E-Cad) RNA, which is obviously enriched in glioblastoma, contains a potential IRES. IRES may have the potential possibility to mediate cross-junction and multiple-round ORF (open reading frame) to encode a 254-amino acid protein, named circRNA-encoded E-cadherin (C-E-Cad) (Gao et al., 2021). In addition, circMbl in Drosophila head extracts, circZNF609 in myogenesis, circPINTexon2, circSHPRH and circFBXW7 in glioma tumorigenesis, all of them are IRES-mediated protein-coding circRNAs (Legnini et al., 2017; Zhang et al., 2018a; Zhang et al., 2018b; Yang et al., 2018; Pamudurti et al., 2022). Of course, IRES-driven protein-coding circRNAs also exist in liver cancer. For example, circβ-catenin encodes a new 370-amino acid isform of β-catenin depending on its own existence IRES sequence (Liang et al., 2019). The β-catenin isoform binds to GSK3β protein kinase, thereby antagonizing the GSK3β-mediated ubiquitination degradation of β-catenin and activating the Wnt pathway. Notably, several IRES-activated protein-coding circRNA, such as circZNF609, at the same time with high m6A methylation levels, which indicates that there may be an unclear association between the two cap-independent translation ways (Zhao et al., 2014; Legnini et al., 2017). However, whether there are other cap-independent ways to initiate circRNA translation remains unclear. Given that proteins are the exerciser and undertakers of cell activities, the next step is still essential to determine the function of the encoded proteins. Indeed, several studies have found that cells undergoing heat stock and starvation conditions can alter circRNA translation, indicating that circRNA-encoded proteins may play roles in stress response (Pamudurti et al., 2017).
4 CircRNAs in HCC
The liver is the largest solid organ in mammals, and is considered to be the busiest organ in the body due to its many functions. Of note, the function of the liver in regulating energy homeostasis is particularly important, which executes its anabolic or catabolic programs to balance nutrient metabolism. The stability of liver metabolic function is essential for body homeostasis, and dysfunction of metabolism will lead to the occurrence of liver disease, including HCC (Ma et al., 2022). As a rising star for the past few years, circRNAs have been proven to be involved in the physiological function of liver. High-throughput sequencing has identified many circRNAs, such as circ_0001452, circ_0001453 and circ_0001454, which participate in hepatic lipid metabolism (Xie et al., 2022). Glucose metabolism is also an important manifestation of liver function. For example, circ_0000660 targets lgfbp1, an energy metabolism-related gene, and regulates glucose and lipid metabolism (Zeng Y. et al., 2021). Thus, circRNAs are closely related to the functional homeostasis of the liver, and the dysregulation of circRNAs may lead to liver disease.
In fact, increasing evidence indicates that evolutionarily conserved ncRNAs, including circRNAs, play an important role in various pathological and physiological processes. To date, some circRNAs have been implicated in several hallmarks of cancer, such as cell death and survival, invasion, metastasis, and angiogenesis. Various validation tests, such as Northern blot, dot-blotting, RNA-seq and circRNA-specific microarrays, demonstrate that many key circRNAs are significantly dysregulated in HCC cells (Figure 3), tissues, blood and exosomes. Despite these recent progresses, the role of circRNAs in the occurrence and development of HCC is still in their infancy. There is no doubt that better understanding of circRNAs biology is valuable to further deepen the pathogenesis of HCC and develop novel targeted treatments.
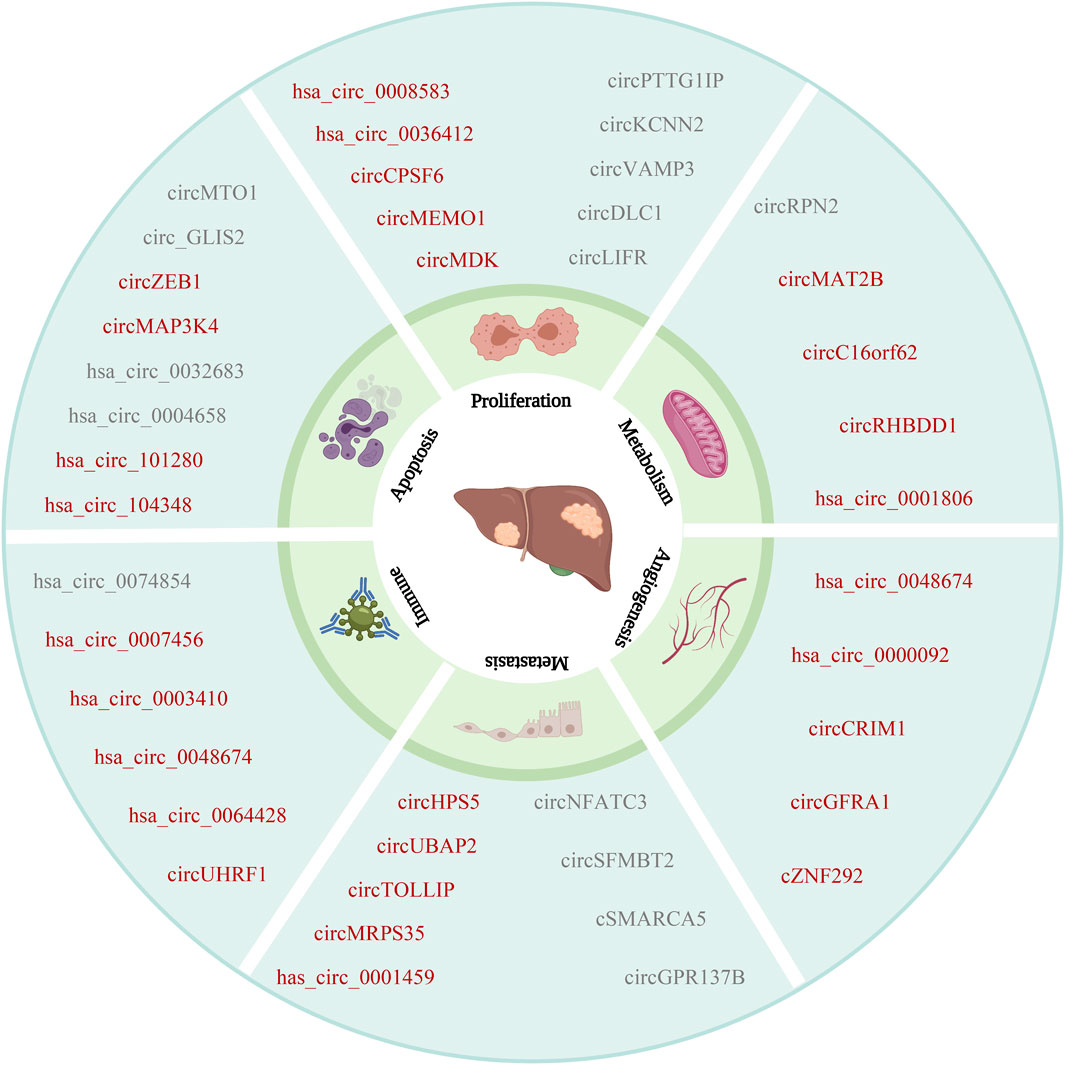
FIGURE 3. The relationship between circRNAs and HCC. CircRNAs are associated with the hallmarks of HCC. Including proliferation, metabolism, angiogenesis, metastasis, immune and apoptosis (The red font indicates oncogene, and the gray font indicates suppressor).
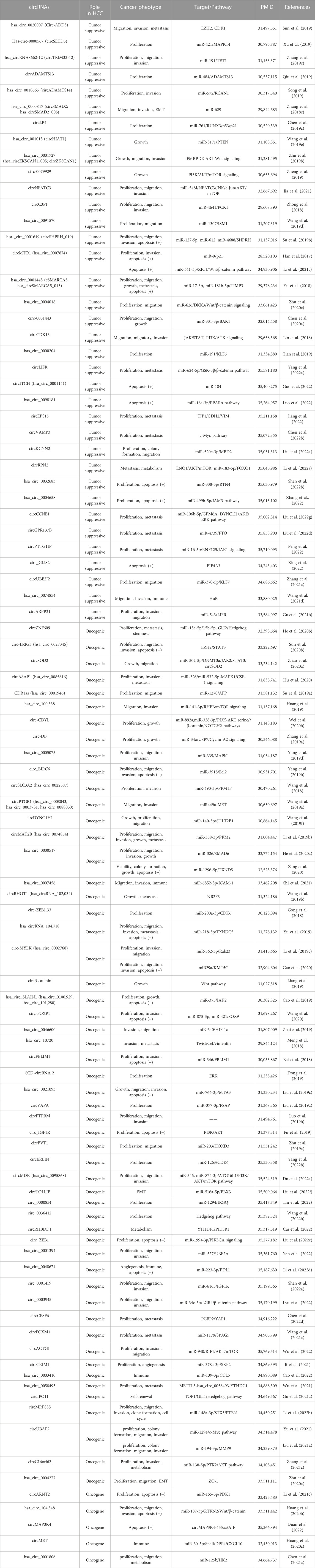
4.1 CircRNA expression profile in HCC
With the development of high-throughput sequencing technology, genome-wide circRNA expression profiles in liver cancer have long been revealed. Sunagawa et al. found that seven candidate circRNAs were downregulated in HCC tumors compared with normal liver tissues (Sunagawa et al., 2021). By sequencing mononuclear cells from the peripheral blood of healthy people and HCC, a total of 58 circRNAs were found to be significantly changed (≥2 or ≤ 0.5-fold) (Lei et al., 2019). In addition, multiple circRNA-related databases also showed that circRNA expressions are substantially different between human HCC and cancer-free liver tissues, suggesting that the dysregulation of circRNAs may be involved in HCC (Rebolledo et al., 2023) (Table 1). Both circHIPK3 and circTEME45A are significantly upregulated in HCC cells and enhance cell proliferation, migration, and invasion (Zheng et al., 2016; Zhang T. et al., 2020). Of note, the same circRNA may have different roles in distinct cancers. For example, ciRS-7 promotes the proliferation and invasion of HCC cells by targeting miR-7, yet acts as a tumor suppressor in ovarian and bladder cancer (Yang X. et al., 2017; Chen et al., 2019b). The dysregulation of circRNAs suggests that they have emerged as a potential target for diagnosing HCC and therapeutic intervention.
4.2 CircRNAs regulate HCC cell proliferation
To some extent, the ability to maintain chronic proliferation is the most fundamental characteristic of cancer cells. Cancer cells manipulate this ability by activating multiple oncogenic signaling pathways excessively. In mammals, many circRNAs promote HCC cell proliferation through activating oncogenic pathways, such as Wnt (Liang et al., 2019; Wei et al., 2020a), signal transducer and activator of transcription 3 (STAT3) (Chen L. et al., 2020; Sun et al., 2020b; Zhao Z. et al., 2020) and NOTCH (Wang L. et al., 2019; Wei Y. et al., 2020) signaling pathways. Deregulation of Wnt signaling has been linked to a number of human diseases, especially cancers. Circβ-catenin promotes HCC cell proliferation by encoding a 370-amino acid β-catenin isoform that can activate the Wnt pathway (Liang et al., 2019). Circ-DENND4C, an oncogene identified in breast cancer, was also overexpressed in HCC cells. It mediated malignant phenotypes in HCC cells through sequestering miR-195-5p, and thus augmented transcription factor 4 (TCF4) expression, leading to the activation of the Wnt/β-catenin signaling pathway (Liu et al., 2020d). Furthermore, several β-catenin downstream targets are also upregulated in liver cancer, such as C-MYC (Liang et al., 2019). Indeed, C-MYC often appears in the process of tumor occurrence and development (Dang, 2012). Hsa_circ_0091581 is derived from the GPC3 gene locus, and can regulate HCC progression by binding to molecules such as Wnt signaling proteins (Zhou F. et al., 2018). In addition, hsa_circ_0091581 promoted HCC cell proliferation via the hsa_circ_0091581/miR-526b/c-Myc axis (Wei et al., 2020a). The Janus kinase (JAK)/STAT signaling pathway is also involved in cancer cell migration, growth and differentiation (Leonard and O'Shea, 1998). CircSOD2, a circular RNA derived from SOD2 gene, promoted HCC cell growth by sponging miR-502-5p, thereby activating the DNMT3a/JAK2/STAT3 signaling axis (Zhao Z. et al., 2020). Circ-LRIG3, a novel covalently closed circRNA originated from the back-splicing of LRIG3 exon 2–11, can interact with EZH2 and STAT3 to form a ternary complex. The complex facilitates EZH2-induced STAT3 methylation and subsequent phosphorylation, and then activates STAT3 signaling and promotes HCC proliferation (Sun et al., 2020b). Of note, several circRNAs could maintain a specific cell state by multiple patterns. For example, circ-MALAT1, derived from lncRNA MALAT1, functions as a brake to form a complex with both ribosomes and mRNAs of paired box 5 (PAX5), a well-known tumor suppressor (Chen L. et al., 2020). The resultant tertiary complex retards PAX5 translation and promotes the self-renewal of HCC stem cells. In addition, circ-MALAT1 acts as a miR-6887-3p sponge to activate the JAK2/STAT3 signaling pathway, thus promoting HCC stem cell self-renewal. Circ-CDYL activates the PI3K-AKT-mTORC1/β-catenin and NOTCH2 pathways by sponging miR-892a and miR-328-3p respectively, which promote the translation of stem-cell–associated genes C-MYC and SURVIVIN and facilitate HCC tumorigenesis (Wei Y. et al., 2020). CircRHOT1 can accelerate the progression of HCC by recruiting TIP60 to the NR2F6 promoter, initiating NR2F6 transcription and eventually activating the NOTCH signaling pathway (Wang L. et al., 2019). NR2F6, a negative regulator of T cell development, is involved in human disease such as leukaemia and cancer (Klepsch et al., 2018; Malouf et al., 2021), albeit its involvement in HCC remains elusive. There is no doubt that many other signaling pathways might be involved in circRNA-mediated HCC cell proliferation. It is of great interest to expand the repertoire of circRNA-modulating signaling pathways in HCC tumorigenesis in the future.
In contrast, some circRNAs may inhibit HCC progression through inhibiting oncogenic pathways, such as Wnt (Zhang et al., 2019c) and STAT3 (Yu et al., 2018). TET1 is a member of the ten elven translocation (TET) family of methylcytosine dioxygenases, which participate in DNA demethylation by converting 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) (Tahiliani et al., 2009; Fu et al., 2013). Downregulated 5hmC/TET1 or upregulated 5 mC has been shown to be related with HCC progression (Chuang et al., 2015). Intriguingly, circMEMO1 is significantly reduced in HCC tissue and its expression is reversely associated with poor patient prognosis. Mechanistically, circMEMO1 could act as a sponge for miR-106b-5p and then modulate the promoter methylation and gene expression of TCF21, which targets the TET family members and increases genome-wide 5hmC levels, thus inhibiting HCC progression (Dong et al., 2021). More importantly, circMEMO1 and its downstream target miR-106b-5p can increase the sensitivity of HCC cells to sorafenib treatment. CircTRIM33-12, an independent risk factor for the overall survival (OS) and recurrence-free survival (RFS) of patients with HCC after surgery, upregulates TET1 expression by sponging oncogenic miR-191 (Zhang et al., 2019c), and it has shown that TET1 can suppress the Wnt signaling pathway and repress HCC cell proliferation and invasion (Fan J. et al., 2018; Zhang et al., 2019c). Interestingly, the expression of miR-191 is regulated by DNA methylation, adding another layer of complexity to the miR-191/Tet1 regulatory axis. Additionally, miR-191 can aggravate HCC progression by directly targeting TIMP metallopeptidase inhibitor 3 (TIMP3) (He et al., 2011). MiR-17-3p and miR-181b-5p may also enhance HCC progression through repressing TIMP3, a well-known tumor suppressor of the oncogenic STAT3 signaling pathway (Shen et al., 2016; Yu et al., 2018; Zou et al., 2020; Tian et al., 2022). Interestingly, there is a concomitant reduction of TIMP3 and cSMARCA5 expression in HCC cells (Yu et al., 2018). Further mechanistic investigations revealed that cSMARCA5 can inhibit the TIMP3-STAT3 pathway by sponging miR-17-3p/miR-181b-5p, thereby suppressing the growth and metastasis of HCC (Shen et al., 2016; Yu et al., 2018). Circ-102,166, a circRNA encoded by the TEX2 gene locus, inhibited HCC development by binding with miR-182 and miR-184 to regulate the expression of their downstream targets, including FOXO3a, MTSS1, SOX7, p-RB and c-MYC (Li R. et al., 2020). CircLIFR has also been identified to function as a tumor suppressor, suppressing the proliferation and invasion of HCC cells through the miR-624-5p/GSK-3β axis (Yang L. et al., 2022).
Due to the considerable bioenergy, biosynthesis and redox requirements caused by rapid proliferation, cancer cells need to reprogram pathways of nutrient acquisition and metabolism. According to the Warburg effect, HCC cells converts aerobic oxidation to glycolysis through metabolic reprogramming, thereby supporting rapid proliferation (Liberti and Locasale, 2016). PI3K/AKT/mTOR-related circRNAs exert their essential functions in this process. As a downstream target of the PI3K-Akt-mTOR signaling pathway, pyruvate kinase (PK) participates in the final rate-limiting step of glycolysis by catalyzing phosphoenolpyruvate (PEP) and ADP to pyruvate and ATP. Especially, PKM2, one of PK isoforms, is overexpressed in malignant cells and enhances tumor progression through modulating metabolic reprogramming and other processes (Wei et al., 2016). The miR-338-8p degrades PKM2 mRNA by targeting its 3’-UTR. Li et al. demonstrated that circMAT2B acts as a miR-338-3p sponge to upregulate the expression level of PKM2, thus promoting glycolysis and glycolysis-related cell proliferation of HCC (Li Q. et al., 2019). CircRPN2 also regulates aerobic glycolysis reprogramming in HCC cells (Li J. et al., 2022). Enolase 1 (ENO1) promotes AKT activation to exert its metabolic effects (Dai et al., 2018). CircRPN2 decreases the level of phosphorylated AKT and mTOR by accelerating ubiquitin/proteasome-dependent ENO1 degradation, thus inhibiting HCC glycolysis and progression. On the other hand, circRPN2 suppresses HCC glycolysis, glucose uptake, ATP levels, and lactate production via the miR-183-5p/FOXO1 axis.
Moreover, several circRNAs can also modulate cell cycle and apoptosis to regulate tumor cell growth and proliferation. Cyclins and CDKs are two classes of regulators for cell cycle progression and always exert functions by binding to each other. Therefore, CDK inhibitors, such as P21 and P27, can regulate the cell cycle by inhibiting the function of CDKs (Levkau et al., 1998). For example, circ-CSPP1 was upregulated in HCC and increased the expression of cyclin E2 (CCNE2), which regulates the G1-to-S phase transition via the CCNE2/CDK2 axis (Kanska et al., 2016; Sun Q. et al., 2020). Hsa_circ_0000517 downregulation induced cell cycle arrest in HCC cells and decreased the expression of P21 and cyclin D1 (He S. et al., 2020). P21 can bind CDK2, thus regulating the cell cycle progression of G1 to S phase (Brugarolas et al., 1999). As reported by Du et al., circ-Foxo3 forms a circ-Foxo3-p21-CDK2 ternary complex with p21 and CDK2, which hijacks CDK2 together with p21, and inhibits the cyclin E/CDK2 complex formation, thus arresting the process from G1 to S phase (Du et al., 2016). At the same time, the ternary complex can also block the progression of the cell cycle in S phase by abolishing the inhibitory effect of p21 on the cyclin A/CDK2 complex (Du et al., 2016). Similar to the p21, p27 is known to play a key role in regulating CDK2 activity (Sherr and Roberts, 1995). Liu et al. demonstrated that circBACH1 represses p27 mRNA translation by gathering HuR, and consequently accelerates the entrance of HCC cells from G1 to S phase (Liu B. et al., 2020). Undoubtedly, cell proliferation is closely related to senescence, and circRNAs may be involved in this regulation. It has been proposed that circLARP4 could sponge miR761 and upregulate RUNX3 expression, thus activating the downstream p53/p21 signaling and arresting the G1/S cell cycle of HCC cells (Chen et al., 2019b). Not only cell cycle, but apoptosis is also regulated by p53, p21 and RUNX3 (Hotta et al., 1998; Dotto, 2000; Vousden and Lu, 2002). Therefore, it is reasonable to speculate that the effect of circRNAs on apoptosis is associated with HCC development. Indeed, circ-LRIG3 has been demonstrated to reduce apoptosis by forming a ternary complex with EZH2 and STAT3 (Sun et al., 2020b). Hsa_circRNA_104,348 directly targeted miR-187-3p, and thus suppressed apoptosis of HCC cells (Huang G. et al., 2020). These studies collectively suggest that the majority of circRNAs may function as miRNA sponges, which subsequently regulate downstream genes and signaling pathways associated with proliferation, senescence, and apoptosis of HCC cells.
4.3 CircRNAs regulate HCC invasion, migration and metastasis
Cancer-related mortality is predominantly due to the metastasis of primary tumor and subsequent recrudescence at the distinct site. CircRNAs can regulate tumor cell invasion and metastasis through multiple signaling pathways, such as MAPK (including extracellular regulated kinase) (Wang et al., 2018; Hu et al., 2020), TGF-β (Meng et al., 2018; Song et al., 2020), and AKT (Huang et al., 2020c). Protein phosphatase, Mg2+/Mn2+ dependent 1 F (PPM1F), a member of the PP2C family of Ser/Thr protein phosphatases, plays a vital role in HCC progression. PPM1F facilitates cell motility and invasiveness by inactivating p53 signaling and promoting tumor metastasis via the MAPK signaling and exosomal cytokine secretion (Susila et al., 2010; Zhang et al., 2017). CircSLC3A2 upregulates PPM1F expression by sponging miR-490-3p, activating the MAPK signaling pathway in HCC (Wang et al., 2018). Hu et al. determined that circASAP1 promotes HCC cell proliferation and metastasis by sponging miR-326 and miR-532-5p to enhance the expression of colony-stimulating factor-1 (CSF-1) and MAPK1, which are direct targets of miR-326 and miR-532-5p (Hu et al., 2020). The extracellular regulated kinase (ERK) 1/2 signaling pathway is highly involved in tumor growth and metastasis, and MAPK1 acts as a tumor promoter that exerts its function through the ERK1/2 pathway (Han et al., 2021). Circ_0001175 is increased in HCC tumors and facilitates HCC cell proliferation, migration, invasion, epithelial-mesenchymal transition (EMT) and lung metastasis through enhancing the expression of SNX5, a target of miR-130a-5p, resulted in activation of the EGFR-ERK1/2 pathway (Li L. et al., 2020). It has been fairly elucidated that EMT functions as a prominent step in the transition of early-stage HCC to aggressive and metastatic malignancies, increasing cancer cell motility (Brabletz et al., 2018). It has shown that elevated circMET expression induces EMT by modulating associated signaling pathways, such as the chemokine pathway, TNF pathway, and PI3K-Akt pathway (Huang et al., 2020c). CD44v6 and FOSL2 is EMT-stimulated genes that trigger the TGF-β signaling pathway and promote EMT. Circ0003998 can act as a ceRNA of miRNA-143-3p to enhance FOSL2 expression, while circ0003998 could bind with PCBP1-poly (rC) binding protein 1 (PCBP1) to increase the expression level of CD44v6 (Tripathi et al., 2016; Sun et al., 2018; Song et al., 2020), exemplifying the coordination of two circRNAs in HCC EMT regulation.
The EMT process is controlled by many protein-coding factors, such as ZIC1 and Twist, which may be modulated by ncRNAs including circRNAs. For example, circMTO1 can competitively bind to miR-541-50 and upregulate the downstream target ZIC1 expression, thereby suppressing HCC EMT (Li D. et al., 2021). ZIC1 is a zinc-finger transcription factor and has a major impact on the Wnt/β-catenin and EMT signaling networks. Briefly, ZIC1 attenuates the activation of Wnt/β-catenin signaling, consequently decreases the expression of mesenchymal marker proteins, including MMP2, N-cadherin and Vimentin. Vimentin, a significant constituent of intermediate filament proteins, functions as a potent enhancer of cell migration in cancerous tissues and facilitates EMT process (Ridge et al., 2022). As one of the EMT-inducing transcription factors, Twist 1 regulates the transcription of EMT-associated genes, including Vimentin. As reported by Meng et al. (Meng et al., 2018), Twist1 upregulates the level of Cul2 circular RNA (circ-10720), which sponges miRNAs targeting Vimentin and promotes EMT in HCC. Intriguingly, circ-10720-overexpressing cells is enriched in gene sets associated with VEGF and TGF-β receptor signaling pathway, suggesting a broad role of this circRNA in HCC EMT.
CircRNAs may also modulate HCC metastasis through regulating certain oncogenes and anti-oncogenes. CircPTTG1IP blocked miR-16-5p and subsequently upregulated its target RNF125, leading to ubiquitination and degradation of JAK1, and eventually reduced HCC invasion and metastasis (Peng et al., 2022). CircZNF566 promoted HCC tumorigenesis and metastasis by sponging miR-4738-3p to increase its target TDO2, an essential enzyme of Try metabolism, albeit the exact mechanisms remains unclear (Opitz et al., 2011; Li S. et al., 2020). The histone methyltransferase EZH2 is a key metastasis-related gene that can physically bind to a large amount of ncRNAs. Sun et al. found that circ-ADD3 inhibits HCC cell migration and invasion, as well as lung metastasis, by potentiating CDK1-mediated ubiquitination and degradation of EZH2 (Benetatos et al., 2013). Subsequently, the decrease of EZH2 significantly increases the expression of anti-metastatic genes, including circ-ADD3, thereby forming a regulatory loop to inhibit the metastasis of HCC (Sun et al., 2019). In addition, circRNAs also exert function by binding with RBPs directly. For example, circDLC1 interacts with the RNA-binding protein HuR and impairs its binding to matrix metallopeptidases 1 (MMP1) mRNAs, blocking the translation of MMP1 (Liu H. et al., 2021), a known repressor of HCC cell proliferation and invasion (Kim et al., 2018).
Angiogenesis can sustain the growing malignant tissues to obtain nutrients and oxygen and remove metabolic wastes continuously. Undoubtedly, the tumor-associated neovascularization system plays critical roles in tumor dissemination and metastasis. Increasing evidence has revealed that circRNAs can act as metastasis regulators by managing angiogenesis (Zhao et al., 2022). Interestingly, it has been demonstrated that circRNAs possess the ability to promote angiogenesis under the special condition, hypoxia, a typical characteristic of the tumor microenvironment (Boeckel et al., 2015). SRY (sex determining region Y)-box 9 (SOX9), can interact with β-catenin to inhibit the activation of Wnt/β-catenin signaling pathway which is involved in vasculogenic mimicry. Yang et al. discovered that cZNF292 interacts with SOX9 protein and blocks its nuclear translocation, which subsequently stimulates the Wnt/β-catenin signaling pathway activity, thereby promoting angiogenesis in human hepatoma SMMC7721 cells under hypoxia (Yang W. et al., 2019). CircGFRA1/miR-149 also plays a vital role in the angiogenesis of HCC, probaly through activating multiple miR-149 targets, such as PPM1F, AKT1, and LRIG2 (Zhang Y. et al., 2014; Ji et al., 2019; Yu Y. et al., 2020). Therefore, targeting circRNAs to disrupt the angiogenesis of HCC may be a hopeful therapeutic strategy.
5 CircRNA as a modulator of HCC microenvironment
The tumor microenvironment (TME) is essential in the pathobiology of cancer. The interaction between malignant and surrounding cells creates the TME, which plays a crucial role in all stages of cancer progression, metastasis and treatment.
5.1 CircRNAs regulate tumor immune microenvironment
Immune cells are the most abundant cellular components in TME and function as a double-edged sword in cancer-protecting the host and promoting tumor proliferation. Previous studies have shown that foreign circRNAs potently stimulate immune signaling and inhibit RNA virus infection (Chen et al., 2017) (Figure 4). Tumor-infiltrating lymphocytes (TILs) are an important part of the immune system and are positively correlated with patient’s survival, tumor size and metastasis (Weng et al., 2019). Through performing global circRNA microarray screening and qRT-PCR validations, Weng et al. found that hsa_circ_0064428 was significantly reduced in HCC patients with high TILs and negatively correlated with overall survival and metastasis (Weng et al., 2019). Among the plethora of lymphocytes, natural killer (NK) cells and cytotoxic CD8+ T (CTLs) cells are deemed as the ‘professional killers’ because of their involvement in the direct killing of pathogens (Trinchieri, 1989). Interestingly, NK cells can directly kill the target cells without prior exposure to antigen. Moreover, NK cells are considered the first line of defense for host immune surveillance and play a vital role in tumor immune microenvironment. In cellular stress conditions, active ligands on tumor cell surface are upregulated and interact with activating receptors on NK cells. In the end, both the NK cell-mediated cytotoxicity and their secreted pro-inflammatory cytokines cause the death of the target cells (Chan et al., 2014). Recent studies elucidated that circRNAs may participate in HCC immunity in this way. For example, circARSP91 upregulates the expression of UL16 binding protein 1 (ULBP1). Notably, ULBP1 serves as a killer cell lectin to facilitate NK cells to recognize and attack HCC cells. At the same time, hsa_circ_0007456 increases the susceptibility of HCC cells to NK cell cytotoxicity (Shi et al., 2021), indicating a vital role in tumor immunity. CTLs differentiate into effector cytotoxic CTLs and bind to cancer cells through the interaction between their T cell receptors and the specific neo-antigen-MHC class I complex, then killing the malignant cells by the Fas/FasL/caspase cascade pathway or directly secreting effector cytokines and cytotoxic granules (Halle et al., 2017). In a pan-cancer analysis, Zou et al. discovered that CDR1as might play a specific role in immune and stromal cell infiltration in tumor tissues, especially those of CD8+T cells and activated NK cells (Zou et al., 2019). Cancer cells secret chemokines or cytokines, such as CCL2 and CCL5, thus recruiting macrophages to the TME and polarizing to a tumor-associated macrophage (TAM) phenotype (Christofides et al., 2022). CircASAP1 promotes TAM infiltration by sponging miR-326 and miR-532-5p, and that CSF-1 is a common target of those two miRNAs (Hu et al., 2020). Hsa_circ_0110,102, was significantly downregulated in HCC, and inhibited macrophage activation and HCC progression. C-C chemokine ligand 2 (CCL2) was overexpressed in HCC by the activation of PPARα and interacted with C-C motif chemokine receptor 2 to regulate the chemotaxis of TAMs, contributing to cancer progression in HCC TME. Hsa_circ_0110,102 inhibits the expression and release of CCL2 into the microenvironment through the miR-580-5p/PPARα pathway, thereby alleviating macrophage recruitment and TAM (Wang X. et al., 2021). Furthermore, through spearman correlation analysis, Zhou et al. found that at least six circRNAs are related to macrophage infiltration in HCC, including hsa_circ_0091570, hsa_circ_0072088, hsa_circ_0004913, hsa_circ_0002980, hsa_circ_0001955 and hsa_circ_0000520 (Zhou et al., 2020).
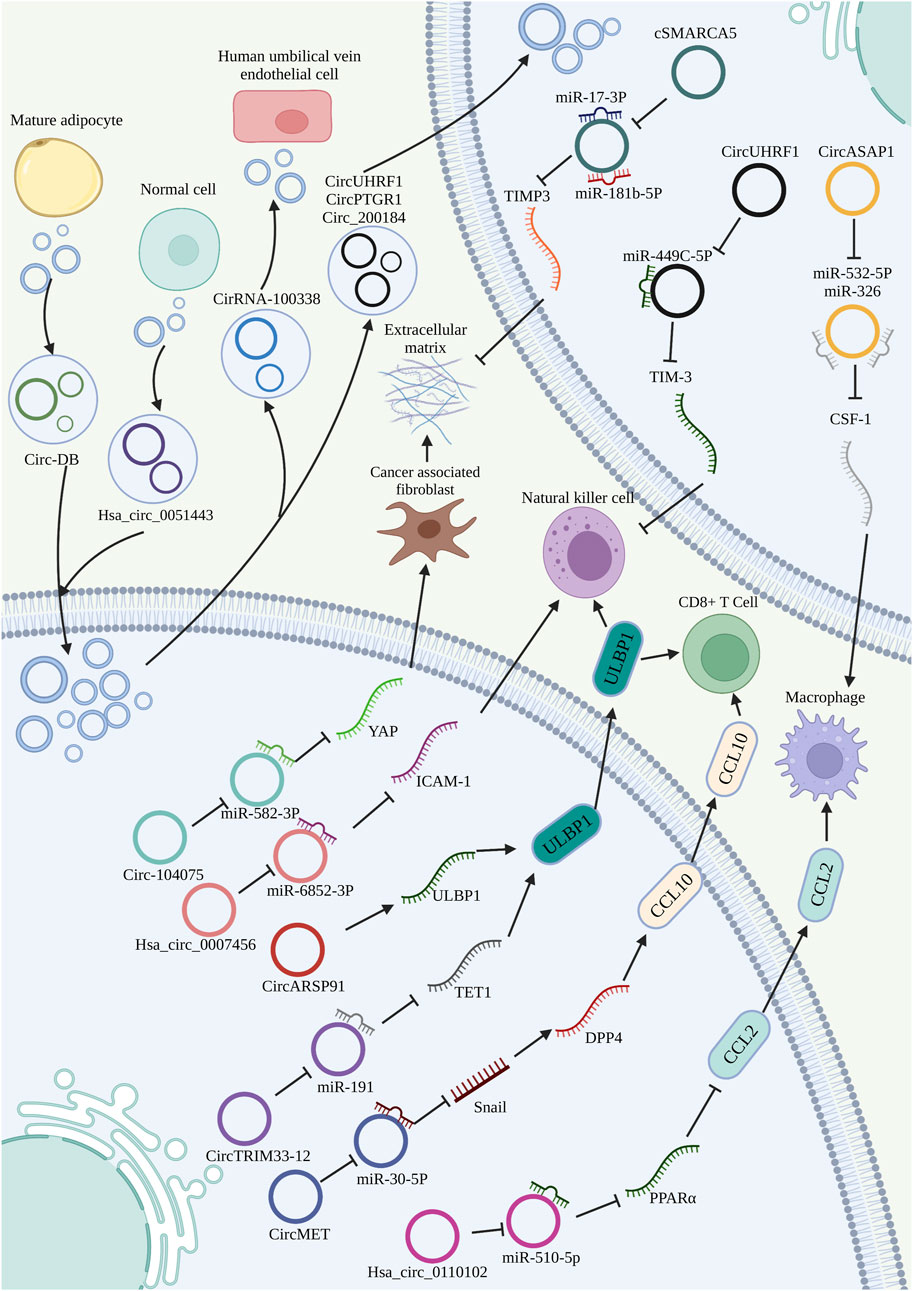
FIGURE 4. Role of circRNAs in the TME of HCC. CircRNAs promote or inhibit immune cell activity, and participate in ECM remodeling by regulating fibroblast recruitment and activation. Exosome-encapsulated circRNAs as modulators in intercellular communication of HCC.
5.2 CircRNAs regulate CAFs and ECM
Cancer-associated fibroblasts (CAFs) are involved in TME construction and promote cancer proliferation and metastasis. Interestingly, CDR1as was found to participate in regulating CAFs and extracellular matrix (ECM). Zou et al. identified that CDR1as expression is positively correlated with the infiltrating level of CAFs (Zou et al., 2019). This idea is backed up by another study (Du et al., 2016). Compare with cancer cell lines, the level of circ-Foxo3 was significantly reduced in fibroblast cells, such as NIH3T3 and mouse embryo fibroblasts (Du et al., 2016). In HCC tissues, HNF4a, a well-acknowledged HCC-promoting transcription factor, stimulates the expression of circ_104,075 by binding with its promoter. In addition, circ_104,075 can act as a ceRNA to upregulate YAP expression by absorbing miR-582-3p, which may cause the remodel of ECM by inducing CAFs, and promote the proliferation of HCC (Zhang et al., 2018d). ECM, consisting of collagens, proteoglycans, elastin, fibronectin, laminins, and several other glycoproteins, is one of the significant components of TME and continuously undergoes remodeling. cSMARCA5, whose expression is reduced in HCC, prevents degradation of the ECM of HCC by inhibiting the activity of MMPs (Yu et al., 2018). Several circRNAs are reported to be involved in ECM remodeling in other cancers, and it is of interest to explore their roles in HCC TME.
5.3 Exosomal circRNAs in HCC microenvironment
Exosomes are small bioactive vesicles produced by endocytosis in the cell membrane, and serve as a nanoscale messenger shuttling between cells to transfer information and cargos. The peculiarity that exosomes are secreted into surrounding body fluids make them play a vital role in various pathophysiological changes. CircRNA-sequencing on exosomes derived from liver cancer cells showed that circRNAs were enriched in exosomes compared with normal cells. Meanwhile, the enrichment degree of tumor circRNAs in serum was even related to tumor size (Li Y. et al., 2015). This phenomenon suggests that exosomal circRNAs are likely involved in the proliferation and metastasis of HCC (Figure 4). Several circRNAs are secreted into the TME by HCC cells. CircUHRF1, an exosomal circRNA secreted by HCC cells, can sponge miR-449c-5p to promote the expression of TIM-3, a receptor on NK cell surface, resulting in the loss of NK cell activity (Vivier et al., 2011). Thus, exosomal cireUHRF1 inhibits NK cell activity and facilitates HCC progression (Zhang P. F. et al., 2020). Huang et al. found that exosomal circRNA-100338 enhances HCC cell proliferation, invasion and angiogenesis via the mTOR signaling pathway. The HCC-derived exosomal circRNA-100338 can affect the proliferation of human umbilical vein endothelial cells (Huang et al., 2020c). Exosomal circPTGR1 from HCC-LM3 cells enhances cell migration and invasion through the miR449a-MET pathway. Acting as messengers to enhance intercellular communication is an important function of exosomes. Interestingly, exosomal circRNAs derived from normal tissues may be also involved in HCC progression. For example, exosomal circRNA hsa_circ_0051443, originated from normal cells, suppresses the progression of HCC via the miR-331-3p/BAK1 axis (Chen L. et al., 2020). Ubiquitin-specific protease 7 (USP7), a deubiquitinating enzyme, is associated with the proliferation and invasion of cancers, including HCC (Novellasdemunt et al., 2017). Circ-DB, an exosomal circRNA derived from the adipose tissue, can promote the growth of HCC by regulating the miR-34a/USP7 axis and deubiquitination (Zhang H. et al., 2019). Of note, exosomes are broadly existed in surrounding body fluids, as mentioned above, suggesting that exosoaml cirRNAs may be used as potential biomarkers for cancerdiagnosis.
6 CircRNAs in HCC-related diseases
Chronic infection with hepatitis viruses is one of the main risk factors for HCC, especially hepatitis B virus (HBV) and hepatitis C virus (HCV). HBV infection is the most prominent risk factor for HCC development, accounting for ∼50% of cases (Akinyemiju et al., 2017). Interestingly, a novel circRNA HBV_circ_1 produced by HBV pgRNA/pcRNA, rather than host mRNAs, was identified in HBV-positive HepG2 cells and HBV-related HCC tissues (Zhu et al., 2021). HBV_circ_1 upregulates CDK1 expression by directly binding to CDK1 in vitro and in vivo, and CDK1 then binds to cyclins and activates cell cycle progression. These results suggest that the interaction of HBV_circ_1 with CDK1 may promote HCC development, although the specific mechanism remains further studied. As to HCV, unique to this virus is a dependence on the liver-specific miR-122. The binding of miR-122 to two binding sites in the 5’ UTR of HCV RNA is critical for viral replication by moderately stimulating viral protein translation (Henke et al., 2008) and protecting the uncapped HCV RNA genome from exoribonuclease Xrn2 degradation (Sedano and Sarnow, 2014). This provides a new approach for HCV treatment by competitively inhibiting miR-122 binding to HCV, thus restraining its replication and translation. Based on the property of circRNAs as miRNA sponges, Jots et al. produced artificial circRNA sponges in vitro that efficiently sequester miR-122, thereby inhibiting viral protein in an HCV cell culture system (Jost et al., 2018). The stability of circRNAs is a precious advantage in scientific research and clinical application.
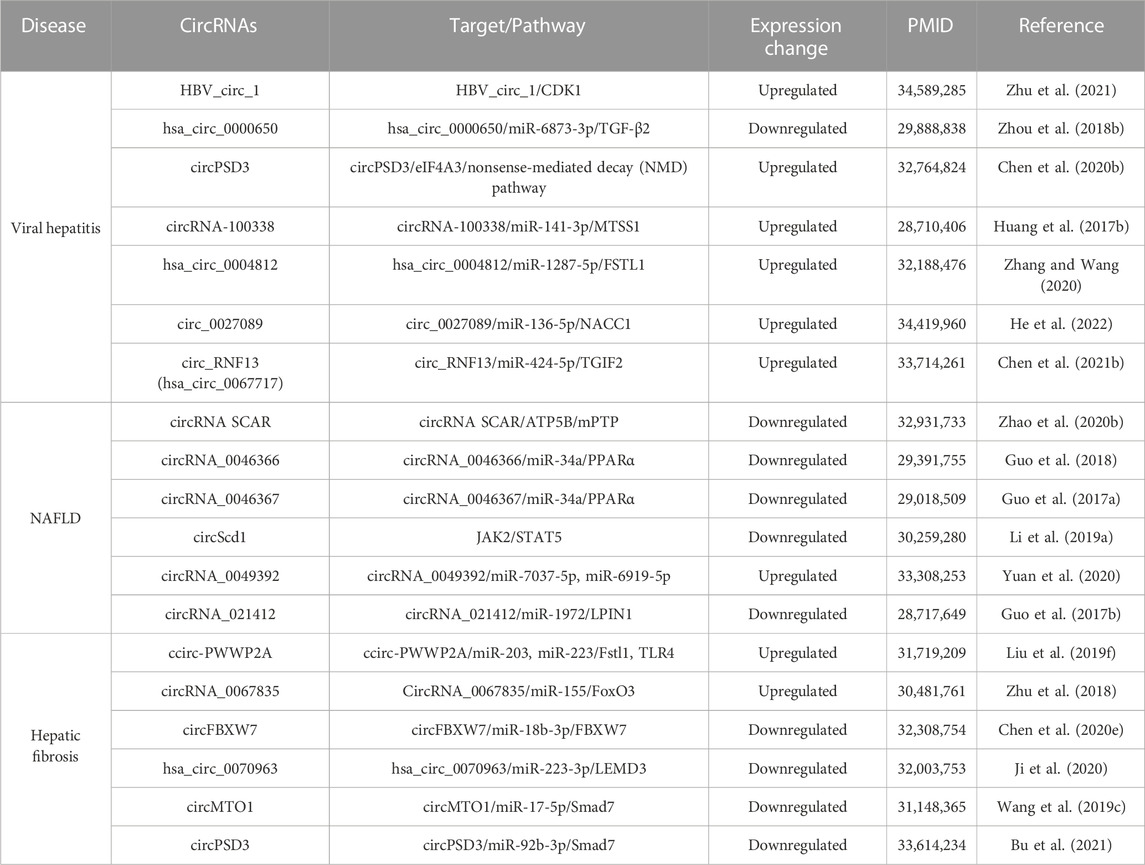
Non-alcoholic fatty liver disease (NAFLD) is the leading etiology of non-cirrhotic HCC (Ioannou, 2021). NAFLD is emerging as the most common cause of chronic liver disease worldwide. It is a multifaceted disorder that ranges from the simple accumulation of triglycerides in hepatocytes (hepatic steatosis) to steatosis with inflammation, non-alcoholic steatohepatitis (NASH), fibrosis and cirrhosis, which may evolve towards HCC. Many molecules play important roles in both NAFLD/NASH and HCC, including ncRNAs (Fu et al., 2014; Xu et al., 2021; Tian et al., 2022). Emerging evidence illustrates that oxidative stress caused by impaired mitochondrial function is involved in the pathogenesis of NAFLD, including NASH (Sunny et al., 2017). Mitochondrial dysfunction leads to an excessive production of reactive oxygen species (ROS), which is released from mitochondria into the cytoplasm and activates human liver fibroblasts, thus promoting the fibrosis process of NAFLD/NASH (Zhao Q. et al., 2020). Interestingly, circRNAs are the crucial mediators in inhibiting ROS leakage from mitochondria and blocking fibroblast activation. The circRNA, steatohepatitis-associated circRNA ATP synthase subunit b (ATP5B) regulator (SCAR), originates from the circularization of the second exon of its host gene (JA760602) and localizes in the mitochondria matrix. SCAR directly binds to ATP5B, a regulator of mitochondrial permeability transition pore (mPTP), leading to the closure of mPTP. mPTP is an indispensable channel for mitochondrial ROS to enter the cytoplasm. Accordingly, SCAR can modulate mitochondrial function by controlling the release of mROS and the activation of liver fibroblasts to regulate the progression of steatosis to NASH (Zhao Q. et al., 2020). It is well known that PPARα and associated signaling pathways are involved in the pathogenesis of NAFLD, and the expression level of PPARα is inversely proportional to the severity of NASH (Francque et al., 2015; Huang K. et al., 2017). CircRNA_0046366, circRNA_0046367 and PPARα shared the complementary sequences with miR-34a. Further studies found that circRNA_0046366 and circRNA_0046367 can serve as the endogenous sponges of miR-34a and then activate PPARα. Activation of hepatic PPARα attenuated lipid peroxidation and mitochondrial injury by promoting the transcriptional activation of multiple genes for lipid metabolism, such as CPT1A, SLC27A, CPT2 and ACBD3 (Guo et al., 2017a; Guo et al., 2018). Currently, several circRNAs have been identified in HCC-related diseases, but their relationship to disease transformation remains unclear (Table 2).
7 CircRNAs as diagnostic and prognositic biomarkers for HCC
Diagnosis at early stages is crucial for successful curative treatment. Still, the early diagnosis of HCC remains a challenge due to the scarcity of symptoms until progression to late-stage. Meanwhile, the prognosis for HCC remains dismal because of its aggressiveness and high recurrence rate. Therefore, effective biomarkers are required to improve early diagnosis and prognosis analysis. CircRNAs are shielded from exonuclease destruction because of their circular form, and thus have a substantially longer half-life than parent mRNAs—about 48 h as opposed to 10 h (Kristensen et al., 2019). Additionally, circRNAs exhibit cell- and tissue-specific expression patterns and are extraordinarily stable and preserved. Importantly, circRNAs have been found in bodily tissues and fluids, rais the possibility that they might be used as prognostic and diagnostic biomarkers at the molecular levels (Figure 5).
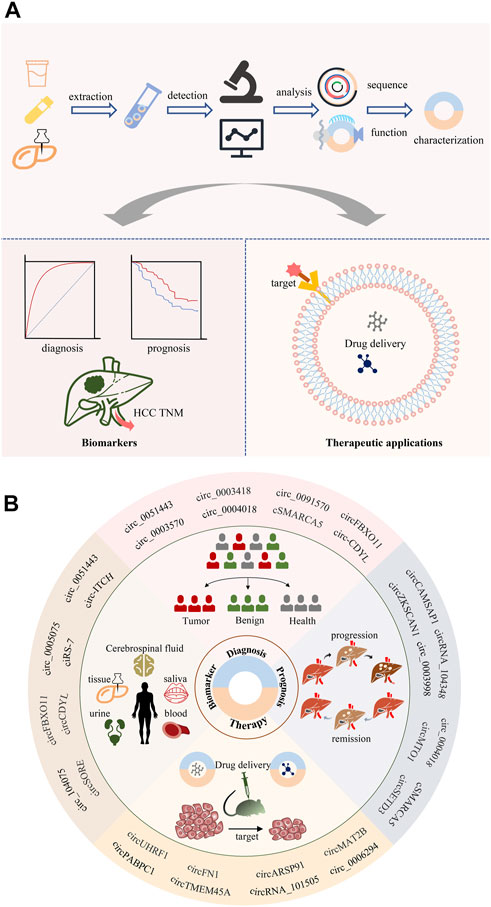
FIGURE 5. Clinical application of circRNAs. (A) CircRNA biomarkers can be isolated from blood, as well as from in the body fluids and tissues. (B) CircRNAs may serve as potential biomarkers for diagnosis, prognosis, and therapy selection of cancer.
Serum alpha-fetoprotein (AFP) is still the most common traditional serum biomarker for HCC screening and diagnosis. With the research on circRNAs, its application may be challenged. It has been shown that the area under the ROC curve (AUC-ROC: 0.973), a sensitivity (SE) of 96.0% and a specificity (SP) of 98.3% indicated that circ_104,075 might be a better serum biomarker for the diagnosis for HCC compared to the AFP (AUC-ROC: 0.750, SE69.3%, SP68.3%) (Zhang et al., 2018d). Furthermore, the use of AFP alone is constrained since AFP may be high in cirrhosis or hepatitis and not all HCC cells release AFP (Wang and Zhang, 2020). In comparison to the single standard biomarker, combinations of circRNAs and AFP may show greater diagnostic or prognostic accuracy. For example, the plasma level of hsa_circ_0003998 in HCC patients was significantly higher than in hepatitis B patients and healthy controls. Its level was significantly reduced after the operation, with an AUC value of 0.892 alone and 0.947 in combination with AFP for discriminating between HCC and healthy controls.
Recent studies have conducted that exosomes may serve as promising candidates for liquid biopsy, particularly for monitoring and forecasting tumor occurrence and metastasis (Kalluri and LeBleu, 2020). Exosomal circRNAs may be a unique and promising type of biomarkers for the early detection of cancer since they are numerous and stable in exosomes (Wang S. et al., 2021). For example, circ_0051443 has been found to have a significantly lower expression level in plasma exosomes from patients than in those from healthy controls. This biomarker can be used for both prediction and diagnosis (Chen W. et al., 2020).
Significantly, with the development of high-throughput sequencing techniques and public genetic databases, many circRNA-based prognostic models for HCC have been established. Zhang et al. employed The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO), and constructed a prognostic signature with seven target mRNAs by univariate, lasso and multivariate Cox regression analyses. According to the targeting relationship between 7 hub mRNAs and other RNAs, the circRNA-lncRNA-miRNA-mRNA ceRNA prognostic network was constructed, comprising of 21 circRNAs (Zhang et al., 2021b). Meanwhile, Huang et al. obtained an alternative prognostic model of HCC. Differentially expressed circRNAs and mRNAs in normal liver tissues and HCC were constructed into the ceRNA network, Significant hub nodes in the ceRNA network were 5 circRNAs, including hsa_circ_0004662, hsa_circ_0005735, hsa_circ_0006990, hsa_circ_0018403 and hsa_circ_0100,609. In addition, according to the mRNA expression level difference in this ceRNA network, a prognostic risk assessment tool, based on the expressions of 7 genes (PLOD2, TARS, RNF19B, CCT2, RAN, C5orf30 and MCM10), has been developed (Han L. et al., 2022). To sum up, the ceRNA network is the main form of prognostic model of HCC, which has a promising clinical application.
In total, the above examples demonstrated that circRNAs could be considered as promising diagnostic and prognostic biomarkers in different body fluids for HCC patients. However, the research of circRNAs as cancer biomarkers is still in its infancy, and some challenges remains on their clinical application. Confirmation of a marker as an indicator requires retrospective and prospective analyses of a large-scale sample to make statistics. In addition, the normalization of circRNAs requires repeated tests to determine the cut-off value and reference range. Because of the substantial ongoing improvements, we suppose that most of these queries will be overcome and that more circRNAs will be detected and applied in the clinic in the future. Table 3 list some circRNAs in application in HCC.
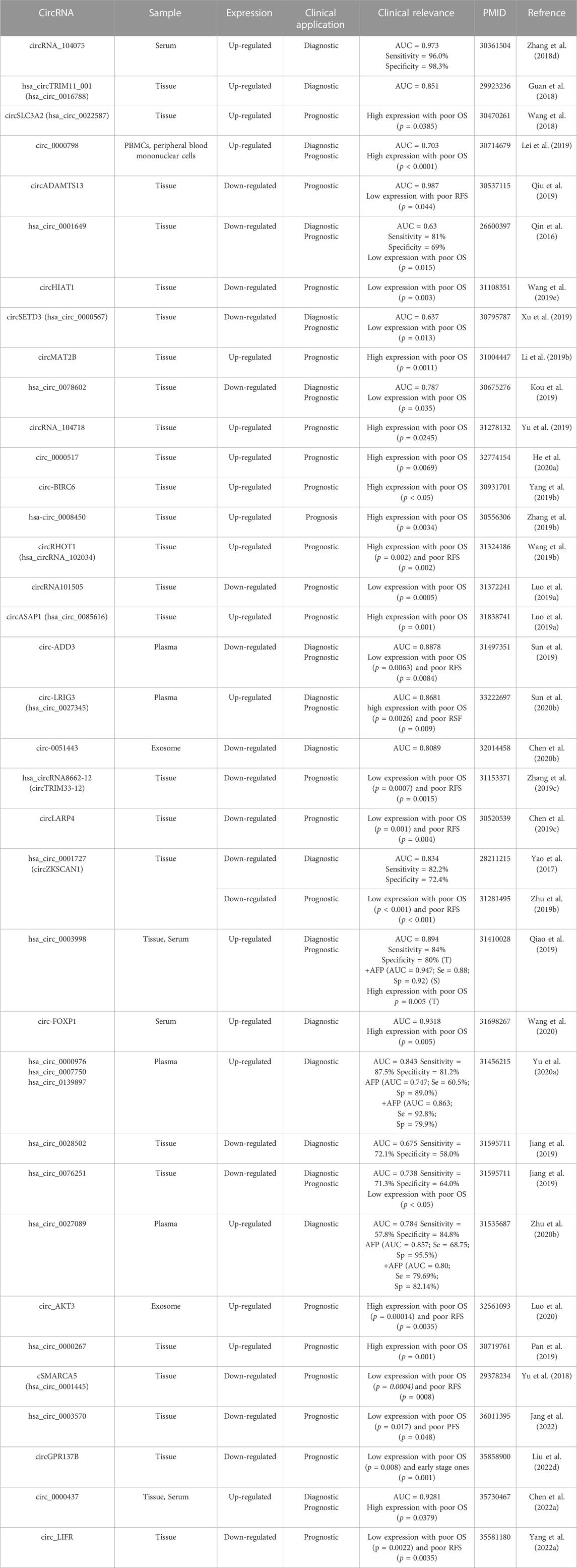
TABLE 3. Dysregulated circRNAs in HCC act as novel potential diagnostic and/or prognostic biomarkers.
8 CircRNAs as therapeutic agents or targets
Advances in surgery and chemotherapy have improved the prognosis of patients with early-stage HCC (Peng et al., 2021). The majority of patients are diagnosed at the advanced stages of the disease and thus have to undergo chemotherapy (Vasan et al., 2019). The important emerging role of circRNAs in the initiation and progression of HCC makes them an attractive therapeutic target option. Several strategies based on the functions of circRNAs have been proposed to treat HCC. For instance, circARSP91 could function as a progression inhibitor to prevent androgen receptor (AR)-induced HCC cell migration and invasion. Targeting the AR/ADAR1/circARSP91 axis could be a potential new route for the development of therapies for HCC (Shi et al., 2017). Meanwhile, several intracellular expression systems, such as the Tornado which transforms the RNA into a loop by modifying its two ends, have been developed to efficiently increase circRNA expression in cells (Litke and Jaffrey, 2019). Another effective and convenient strategy, synthetic (artificial) circRNAs, also has potential therapeutic application in human patients. As one of the main functions of circRNAs is to sponge miRNAs, synthetic circRNAs containing miRNA binding sites were proved to achieve targeted loss of miRNA function and suppress cancer development (Loan Young et al., 2023). Significantly, synthetic circRNAs display excellent capability in terms of both quantity of protein produced and stability of production (Wesselhoeft et al., 2018). In addition, modified exosomes might be an effective approach safely delivering synthetic circRNAs in vivo because (Li C. et al., 2021).
Additionally, a number of techniques may be used to target circRNAs that are favorably linked to carcinogenesis or HCC development in order to influence therapy or enhance prognosis. To downregulate the production of circRNA, siRNA or shRNA usually target the particular backspliced sequence. Similarly, to counteract the effects of circRNAs, antisense oligonucleotides and the CRISPR/Cas9 system might be utilized (Piwecka et al., 2017). A recent study employed DNAzymes, which had circRNA-cleaving properties, to disable circRNAs sequences implicated in HCC and inhibit its cancer promoting ability (Ding et al., 2019).
Although HCC is one of the most frequent cancers, there have been very few drugs available that improve survival, and the efficiency of HCC chemotherapy is usually limited by intrinsic and acquired resistance. Sorafenib, one of the most widely used drugs for patients with non-resectable tumors, inhibits tumor cell proliferation and angiogenesis and promotes tumor cell apoptosis by targeting multiple receptor tyrosine kinases (RTKs), representing an effective first-line therapy for HCC (Llovet et al., 2022). However, the development of its resistance greatly minimizes its therapeutic benefits (Schlacher, 2018). Therefore, elucidating the molecular mechanism of drug resistance is important for cancer treatment. Given the broad involvement of miRNAs in cancer chemotherapy resistance, there is no doubt that circRNAs play a variety of roles in HCC drug resistance. For instance, circFN1, which is overexpressed in HCC cells, contributes to sorafenib resistance by regulating the miR-1205/E2F1 signaling pathway, which may be targeted to improve sensitivity to the multi-kinase inhibitor (Yang et al., 2020). Thus, circRNAs may be applied as targets for drug sensitization since a rising number of drug-resistant circRNAs associated with HCC have been found (Table 4).
9 Conclusion and perspectives
Nearly two-thirds of individuals with HCC have advanced stages of the disease when they are first identified, making HCC one of the most prevalent malignancies and the second leading cause of cancer mortality globally (Llovet et al., 2021c). Thousands of highly conserved endogenous circRNAs have been found in mammalian cells, despite previously being assumed to be a “mistake” in RNA splicing. Numerous research has demonstrated the extent to which circRNAs is systematically changed in cancer, and the significance of circRNAs in different cellular and biological processes, as well as tumorigenesis and metastasis, in HCC has been thoroughly documented. In this review, we briefly summarized the biological functions of circRNAs and their implications in HCC. The broad modification of circRNAs will open up a plethora of new avenues for diagnosis, therapy, and prognosis, all of which are critical issues of HCC care.
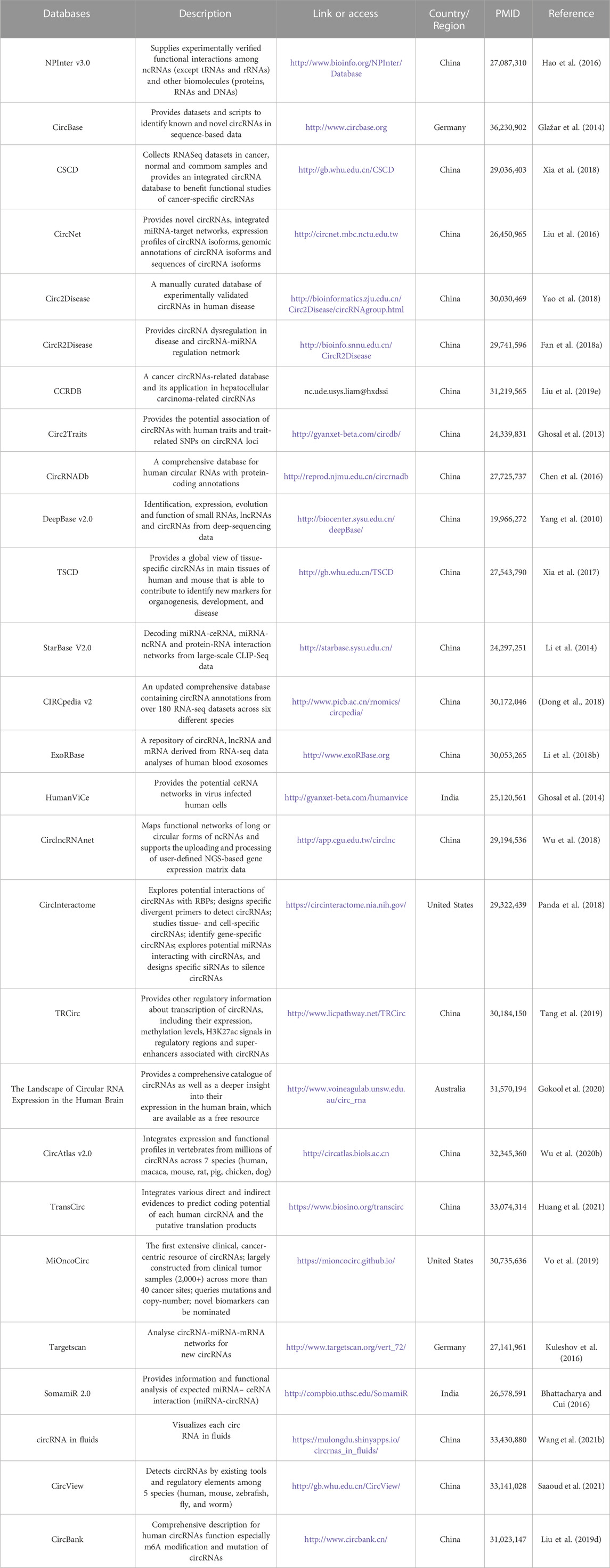
However, there are still many questions that need to be clarified. Firstly, at present, the nomenclature of circRNAs has not been unified. The chances are that the same molecule may be under research simultaneously while in different names, especially when multiple databases have been developed for circRNA analysis in this field (Table 5). Through these databases, researchers can search basic information about circRNAs, predict interactions of circRNAs with target molecules and their translation potential, and evaluate their relationships with diseases. Therefore, it is urgent to establish the uniformed name rule of circRNAs. Secondly, the studies discussed above have confirmed that circRNAs display altered expression patterns in HCC, their cell-type origins are not yet fully explored. This is very challenging because HCC is a complicated condition, especially the TME and the limitation of probe design targeting the back-spliced junction. Even though the effects of circRNAs on the TME have been extensively studied, it remains unclear how TME might affect the machinery of circRNAs biogenesis, secretion, transfer, and mode of action. Therefore, discriminating the cellular origins of secreted circRNAs to identify those involved in tissue function in health and dysfunction in disease; identifying potential circRNAs mediating inter-cell communications as well as those that may impact homeostasis in case of organ failure, and characterizing natural circRNAs carriers to reach distant organs may be part of the solution to these hurdles. Thirdly, the potential role of exosomal circRNAs in the development of HCC remains largely unknown even some investigations have shown their potential correlations. Exosomal circRNAs have been suggested as circulating biomarkers for cancer diagnosis as they have been shown to discriminate patients with cancer from healthy controls as shown above (Chen W. et al., 2020; Liu J. et al., 2022). However, effectively extracting circRNAs with low abundance from exosomes is a challenge, for instance how many copies of circRNA molecules in exosomes need to be evaluated. While concerning circRNAs found in adipose tissues, they may also be released to the circulation inside macrovesicles and have functions in target organs. Therefore, more in vitro and in vivo modeling of exosome-mediated circRNA communications, along with development of sensitive bioinformatic methods and mathematical mass-action models to capture all circRNA–target interactions, will undoubtedly provide insight into the function of these exosomal RNA species. Fourthly, functional peptides produced by circRNAs have emerged into scientific attention. As described above, β-catenin-370aa, encoded by circβ-catenin, activates the Wnt/β-catenin pathway and promotes liver cancer growth (Liang et al., 2019). Evidence has certificated the existence and function of circRNA-encoded peptides. However, high-throughput analytical and systematic detection methods such as ribosome profiling have technical challenges to such peptides. The similarity between circRNA-encoded peptides and the counterpart of mRNA makes them hard to distinguish. The identification of small peptides requires specific biochemical and bioinformatics methods seldom applied in genome-wide characterization. Moreover, cell- and tissue-specific expression complicate these assays. Thus, further stoichiometric analysis is required to explore these peptides’ functions better. Notably, it is also of interest to determine if circRNAs could be translated in the nucleus (Zou et al., 2023b). Fifth, it is speculated that the aberrant expression of circRNAs observed in cancer might be affected by epigenetic changes. Many circRNAs have protein-coding potential, and the translation process can be driven by m6A (Yang Y. et al., 2017). Meanwhile, m6A can affect the function of circRNA via changing the methylation state of downstream molecules. As one of the most abundant RNA modifications, m6A provides us with an intermediate mechanism by which circRNAs are regulated by upstream molecules and allows us to predict and interfere with disease progression caused by the dysregulation of circRNAs. There is no doubt that it would greatly expand our understanding on circRNAs and accelerate their applications. Further, up to date, no specific biological functions have been detected in the majority of already discovered circRNAs, which is also one of the reasons that circRNAs were regarded as by-products of splicing when first discovered (Nigro et al., 1991). Considering the ubiquitous m6A modification in annotated functional circRNAs, it is possible that the role of circRNA might be related to their tissue and developmental stage specificity. Several circRNAs displaying differential expression in specific tissues, developmental stages and subcellular locations may attribute to certain epigenetic and environmental mechanisms (Wu et al., 2019; Tang et al., 2023). In addition, recent evidence suggests an important role of certain RNA secondary structures, such as RNA G-quadruplex, in gene expression (Liu G. et al., 2020; Varshney et al., 2020; Liu G. et al., 2022; Tong et al., 2023). However, the implications of these regulatory modes in circRNA expression await further investigation. Sixthly, it is too soon to speculate on the viability of these molecules as biomarkers and therapeutic disease targets in a clinical HCC scenario. CircRNAs are being investigated for their potential value in the diagnosis, prognosis, and treatments of HCC. The creation of these methods is mostly based on the expression levels of particular circRNAs, although it is not known whether certain biomarkers are unique to HCC or prevalent in other malignancies. The efficacy of circRNAs has not been investigated in sizable, clinically monitored, and definitive cohorts, nor have their mechanisms of action been thoroughly investigated. Additionally, there are still questions regarding possible toxicity, pharmacological reaction, immunologic rejection, accumulation in other tissues, and sustained long-term effects. But studies have shown that exosomes could significantly broaden the uses of circRNAs (Dai et al., 2020). Nevertheless, a better understanding of these issues is required to see circRNAs clinical translation becoming a reality. Last but not least, given the complexity of cancer pathomechanisms, relying on the pattern of a single circRNA paradigm to diagnose and treat HCC may not be biologically maximally relevant and specific enough to provide clinical utility, even studies combining circRNAs with traditional cancer biomarkers for diagnosis have been conducted as mentioned above. Rather, one should consider a multi-markers approach that combines candidate circRNAs/signatures, miRNAs, RBPs, traditional cancer biomarkers, which may be highly discriminative, accurate, and efficient in predicting HCC.
Taken all, circRNAs are a new type of ncRNAs that have a variety of activities. Experimental studies conducted in vivo and in vitro have shown evidence that circRNAs have a role in the control of HCC. To fully comprehend their importance, effectiveness, generalizability, security, and reliability, tremendous investigations are required. It is advisable to employ multiple tools for analysis because different tools have varying degrees of efficiency. Moreover, the improvement in the high-throughput RNA sequencing and the cost reduction, coupled with the establishment of an unbiased bioinformatics analysis, will lead to the identification of a large number of novel circRNAs, which will accelerate the clinical application of circRNAs in the comprehensive management of HCC.
Author contributions
YZ, XF, and YT conceived the project. GR, XP, YT, YZ, and XF wrote the manuscript. GR and XP drew the figures and tables. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (ZYJC18049).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akinyemiju, T., Abera, S., Ahmed, M., Alam, N., Alemayohu, M., Allen, C., et al. (2017). The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: Results from the global burden of disease study 2015. JAMA Oncol. 3 (12), 1683–1691. doi:10.1001/jamaoncol.2017.3055
Aktas, T., Avsar Ilik, I., Maticzka, D., Bhardwaj, V., Pessoa Rodrigues, C., Mittler, G., et al. (2017). DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544 (7648), 115–119. doi:10.1038/nature21715
Ashwal-Fluss, R., Meyer, M., Pamudurti, N. R., Ivanov, A., Bartok, O., Hanan, M., et al. (2014). circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56 (1), 55–66. doi:10.1016/j.molcel.2014.08.019
Bai, N., Peng, E., Qiu, X., Lyu, N., Zhang, Z., Tao, Y., et al. (2018). circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J. Exp. Clin. Cancer Res. 37 (1), 172. doi:10.1186/s13046-018-0838-8
Benetatos, L., Voulgaris, E., Vartholomatos, G., and Hatzimichael, E. (2013). Non-coding RNAs and EZH2 interactions in cancer: Long and short tales from the transcriptome. Int. J. Cancer 133 (2), 267–274. doi:10.1002/ijc.27859
Bhattacharya, A., and Cui, Y. (2016). SomamiR 2.0: A database of cancer somatic mutations altering microRNA-ceRNA interactions. Nucleic Acids Res. 44 (D1), D1005–D1010. doi:10.1093/nar/gkv1220
Boeckel, J. N., Jae, N., Heumuller, A. W., Chen, W., Boon, R. A., Stellos, K., et al. (2015). Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ. Res. 117 (10), 884–890. doi:10.1161/CIRCRESAHA.115.306319
Brabletz, T., Kalluri, R., Nieto, M. A., and Weinberg, R. A. (2018). EMT in cancer. Nat. Rev. Cancer 18 (2), 128–134. doi:10.1038/nrc.2017.118
Brugarolas, J., Moberg, K., Boyd, S., Taya, Y., Jacks, T., and Lees, J. (1999). Inhibition of cyclin-dependent kinase 2 by p21 is necessary for retinoblastoma protein-mediated G1 arrest after gamma-irradiation. Proc. Natl. Acad. Sci. U. S. A. 96 (3), 1002–1007. doi:10.1073/pnas.96.3.1002
Bu, F. T., Zhu, Y., Chen, X., Wang, A., Zhang, Y. F., You, H. M., et al. (2021). Circular RNA circPSD3 alleviates hepatic fibrogenesis by regulating the miR-92b-3p/Smad7 axis. Mol. Ther. Nucleic Acids 23, 847–862. doi:10.1016/j.omtn.2021.01.007
Cai, J., Chen, Z., Zhang, Y., Wang, J., Zhang, Z., Wu, J., et al. (2022). CircRHBDD1 augments metabolic rewiring and restricts immunotherapy efficacy via m(6)A modification in hepatocellular carcinoma. Mol. Ther. Oncolytics 24, 755–771. doi:10.1016/j.omto.2022.02.021
Cao, P., Ma, B., Sun, D., Zhang, W., Qiu, J., Qin, L., et al. (2022). hsa_circ_0003410 promotes hepatocellular carcinoma progression by increasing the ratio of M2/M1 macrophages through the miR-139-3p/CCL5 axis. Cancer Sci. 113 (2), 634–647. doi:10.1111/cas.15238
Cao, S., Wang, G., Wang, J., Li, C., and Zhang, L. (2019). Hsa_circ_101280 promotes hepatocellular carcinoma by regulating miR-375/JAK2. Immunol. Cell Biol. 97 (2), 218–228. doi:10.1111/imcb.12213
Chan, C., Smyth, M., and Martinet, L. (2014). Molecular mechanisms of natural killer cell activation in response to cellular stress. Cell Death Differ. 21 (1), 5–14. doi:10.1038/cdd.2013.26
Chen, G., Xie, D., Zhang, P., and Zhou, H. (2022a). Circular RNA hsa_circ_0000437 may be used as a new indicator for the diagnosis and prognosis of hepatocellular carcinoma. Bioengineered 13 (6), 14118–14124. doi:10.1080/21655979.2022.2081458
Chen, H., Liu, S., Li, M., Huang, P., and Li, X. (2019a). circ_0003418 inhibits tumorigenesis and cisplatin chemoresistance through wnt/β-catenin pathway in hepatocellular carcinoma. Onco Targets Ther. 12, 9539–9549. doi:10.2147/ott.S229507
Chen, H., Mao, M., Jiang, J., Zhu, D., and Li, P. (2019b). Circular RNA CDR1as acts as a sponge of miR-135b-5p to suppress ovarian cancer progression. OncoTargets Ther. 12, 3869–3879. doi:10.2147/ott.S207938
Chen, L., Kong, R., Wu, C., Wang, S., Liu, Z., Liu, S., et al. (2020a). Circ-MALAT1 functions as both an mRNA translation brake and a microRNA sponge to promote self-renewal of hepatocellular cancer stem cells. Adv. Sci. (Weinh) 7 (4), 1900949. doi:10.1002/advs.201900949
Chen, L. L. (2020b). The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 21 (8), 475–490. doi:10.1038/s41580-020-0243-y
Chen, L., and Shan, G. (2021). CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Lett. 505, 49–57. doi:10.1016/j.canlet.2021.02.004
Chen, L. (2020a). The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 21 (8), 475–490. doi:10.1038/s41580-020-0243-y
Chen, S., Cao, X., Zhang, J., Wu, W., Zhang, B., and Zhao, F. (2022b). circVAMP3 drives CAPRIN1 phase separation and inhibits hepatocellular carcinoma by suppressing c-myc translation. Adv. Sci. (Weinh) 9 (8), e2103817. doi:10.1002/advs.202103817
Chen, T. C., Tallo-Parra, M., Cao, Q. M., Kadener, S., Böttcher, R., Pérez-Vilaró, G., et al. (2020b). Host-derived circular RNAs display proviral activities in Hepatitis C virus-infected cells. PLoS Pathog. 16 (8), e1008346. doi:10.1371/journal.ppat.1008346
Chen, W., Quan, Y., Fan, S., Wang, H., Liang, J., Huang, L., et al. (2020c). Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 475, 119–128. doi:10.1016/j.canlet.2020.01.022
Chen, X., Han, P., Zhou, T., Guo, X., Song, X., and Li, Y. (2016). circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep. 6, 34985. doi:10.1038/srep34985
Chen, X., Li, H. D., Bu, F. T., Li, X. F., Chen, Y., Zhu, S., et al. (2020e). Circular RNA circFBXW4 suppresses hepatic fibrosis via targeting the miR-18b-3p/FBXW7 axis. Theranostics 10 (11), 4851–4870. doi:10.7150/thno.42423
Chen, X., She, P., Wang, C., Shi, L., Zhang, T., Wang, Y., et al. (2021a). Hsa_circ_0001806 promotes glycolysis and cell progression in hepatocellular carcinoma through miR-125b/HK2. J. Clin. Lab. Anal. 35 (12), e23991. doi:10.1002/jcla.23991
Chen, Y. G., Kim, M. V., Chen, X., Batista, P. J., Aoyama, S., Wilusz, J. E., et al. (2017). Sensing self and foreign circular RNAs by intron identity. Mol. Cell 67 (2), 228–238.e225. doi:10.1016/j.molcel.2017.05.022
Chen, Y., Li, S., Wei, Y., Xu, Z., and Wu, X. (2021b). Circ-RNF13, as an oncogene, regulates malignant progression of HBV-associated hepatocellular carcinoma cells and HBV infection through ceRNA pathway of circ-RNF13/miR-424-5p/TGIF2. Bosn. J. Basic Med. Sci. 21 (5), 555–568. doi:10.17305/bjbms.2020.5266
Chen, Y., Ling, Z., Cai, X., Xu, Y., Lv, Z., Man, D., et al. (2022d). Activation of YAP1 by N6-methyladenosine-modified circCPSF6 drives malignancy in hepatocellular carcinoma. Cancer Res. 82 (4), 599–614. doi:10.1158/0008-5472.Can-21-1628
Chen, Z., Zuo, X., Pu, L., Zhang, Y., Han, G., Zhang, L., et al. (2019c). circLARP4 induces cellular senescence through regulating miR-761/RUNX3/p53/p21 signaling in hepatocellular carcinoma. Cancer Sci. 110 (2), 568–581. doi:10.1111/cas.13901
Christofides, A., Strauss, L., Yeo, A., Cao, C., Charest, A., and Boussiotis, V. (2022). The complex role of tumor-infiltrating macrophages. Nat. Immunol. 23 (8), 1148–1156. doi:10.1038/s41590-022-01267-2
Chuang, K., Whitney-Miller, C., Chu, C., Zhou, Z., Dokus, M., Schmit, S., et al. (2015). MicroRNA-494 is a master epigenetic regulator of multiple invasion-suppressor microRNAs by targeting ten eleven translocation 1 in invasive human hepatocellular carcinoma tumors. Hepatology. 62 (2), 466–480. doi:10.1002/hep.27816
Conn, S. J., Pillman, K. A., Toubia, J., Conn, V. M., Salmanidis, M., Phillips, C. A., et al. (2015). The RNA binding protein quaking regulates formation of circRNAs. Cell 160 (6), 1125–1134. doi:10.1016/j.cell.2015.02.014
Cui, W., Dang, Q., Chen, C., Yuan, W., and Sun, Z. (2021). Roles of circRNAs on tumor autophagy. Mol. Ther. Nucleic Acids 23, 918–929. doi:10.1016/j.omtn.2021.01.002
D'Anca, M., Buccellato, F. R., Fenoglio, C., and Galimberti, D. (2022). Circular RNAs: Emblematic players of neurogenesis and neurodegeneration. Int. J. Mol. Sci. 23 (8). doi:10.3390/ijms23084134
Dai, J., Su, Y., Zhong, S., Cong, L., Liu, B., Yang, J., et al. (2020). Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 5 (1), 145. doi:10.1038/s41392-020-00261-0
Dai, J., Zhou, Q., Chen, J., Rexius-Hall, M. L., Rehman, J., and Zhou, G. (2018). Alpha-enolase regulates the malignant phenotype of pulmonary artery smooth muscle cells via the AMPK-Akt pathway. Nat. Commun. 9 (1), 3850. doi:10.1038/s41467-018-06376-x
Ding, J., Zhou, W., Li, X., Sun, M., Ding, J., and Zhu, Q. (2019). Tandem DNAzyme for double digestion: A new tool for circRNA suppression. Biol. Chem. 400 (2), 247–253. doi:10.1515/hsz-2018-0232
Dong, R., Ma, X. K., Li, G. W., and Yang, L. (2018). CIRCpedia v2: An updated database for comprehensive circular RNA annotation and expression comparison. Genomics Proteomics Bioinforma. 16 (4), 226–233. doi:10.1016/j.gpb.2018.08.001
Dong, W., Dai, Z. H., Liu, F. C., Guo, X. G., Ge, C. M., Ding, J., et al. (2019). The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine 45, 155–167. doi:10.1016/j.ebiom.2019.06.030
Dong, Z. R., Ke, A. W., Li, T., Cai, J. B., Yang, Y. F., Zhou, W., et al. (2021). CircMEMO1 modulates the promoter methylation and expression of TCF21 to regulate hepatocellular carcinoma progression and sorafenib treatment sensitivity. Mol. Cancer 20 (1), 75. doi:10.1186/s12943-021-01361-3
Doolittle, W. (2013). Is junk DNA bunk? A critique of encode. Proc. Natl. Acad. Sci. U. S. A. 110 (14), 5294–5300. doi:10.1073/pnas.1221376110
Dotto, G. P. (2000). p21(WAF1/Cip1): more than a break to the cell cycle? Biochim. Biophys. Acta 1471 (1), M43–M56. doi:10.1016/s0304-419x(00)00019-6
Dragomir, M., and Calin, G. A. (2018). Circular RNAs in cancer - lessons learned from microRNAs. Front. Oncol. 8, 179. doi:10.3389/fonc.2018.00179
Du, A., Li, S., Zhou, Y., Disoma, C., Liao, Y., Zhang, Y., et al. (2022a). M6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol. Cancer 21 (1), 109. doi:10.1186/s12943-022-01575-z
Du, W., Liu, G., Shi, N., Tang, D., Ferdek, P., Jakubowska, M., et al. (2022b). A microRNA checkpoint for Ca signaling and overload in acute pancreatitis. Mol. Ther. 30 (4), 1754–1774. doi:10.1016/j.ymthe.2022.01.033
Du, W. W., Fang, L., Yang, W., Wu, N., Awan, F. M., Yang, Z., et al. (2017a). Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 24 (2), 357–370. doi:10.1038/cdd.2016.133
Du, W. W., Yang, W., Chen, Y., Wu, Z. K., Foster, F. S., Yang, Z., et al. (2017b). Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 38 (18), 1402–1412. doi:10.1093/eurheartj/ehw001
Du, W. W., Yang, W., Liu, E., Yang, Z., Dhaliwal, P., and Yang, B. B. (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44 (6), 2846–2858. doi:10.1093/nar/gkw027
Duan, J. L., Chen, W., Xie, J. J., Zhang, M. L., Nie, R. C., Liang, H., et al. (2022). A novel peptide encoded by N6-methyladenosine modified circMAP3K4 prevents apoptosis in hepatocellular carcinoma. Mol. Cancer 21 (1), 93. doi:10.1186/s12943-022-01537-5
Eger, N., Schoppe, L., Schuster, S., Laufs, U., and Boeckel, J. N. (2018). Circular RNA splicing. Adv. Exp. Med. Biol. 1087, 41–52. doi:10.1007/978-981-13-1426-1_4
Eisenberg, E., and Levanon, E. Y. (2018). A-to-I RNA editing - immune protector and transcriptome diversifier. Nat. Rev. Genet. 19 (8), 473–490. doi:10.1038/s41576-018-0006-1
Espindola, A. S., Sempertegui-Bayas, D., Bravo-Padilla, D. F., Freire-Zapata, V., Ochoa-Corona, F., and Cardwell, K. F. (2021). Taspert: Target-specific reverse transcript pools to improve HTS plant virus diagnostics. Viruses 13 (7). doi:10.3390/v13071223
Fan, C., Lei, X., Fang, Z., Jiang, Q., and Wu, F. X. (2018a). CircR2Disease: A manually curated database for experimentally supported circular RNAs associated with various diseases. Database (Oxford) 2018, bay044. doi:10.1093/database/bay044
Fan, J., Zhang, Y., Mu, J., He, X., Shao, B., Zhou, D., et al. (2018b). TET1 exerts its anti-tumor functions via demethylating DACT2 and SFRP2 to antagonize Wnt/β-catenin signaling pathway in nasopharyngeal carcinoma cells. Clin. Epigenetics 10 (1), 103. doi:10.1186/s13148-018-0535-7
Fan, W., Chen, L., Wu, X., and Zhang, T. (2021). Circ_0031242 silencing mitigates the progression and drug resistance in DDP-resistant hepatoma cells by the miR-924/POU3F2 Axis. Cancer Manag. Res. 13, 743–755. doi:10.2147/cmar.S272851
Fang, Y., Xue, J., Shen, Q., Chen, J., and Tian, L. (2012). MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 55 (6), 1852–1862. doi:10.1002/hep.25576
Francque, S., Verrijken, A., Caron, S., Prawitt, J., Paumelle, R., Derudas, B., et al. (2015). PPARalpha gene expression correlates with severity and histological treatment response in patients with non-alcoholic steatohepatitis. J. Hepatol. 63 (1), 164–173. doi:10.1016/j.jhep.2015.02.019
Fu, H. W., Lin, X., Zhu, Y. X., Lan, X., Kuang, Y., Wang, Y. Z., et al. (2019). Circ-IGF1R has pro-proliferative and anti-apoptotic effects in HCC by activating the PI3K/AKT pathway. Gene 716, 144031. doi:10.1016/j.gene.2019.144031
Fu, X., Dong, B., Tian, Y., Lefebvre, P., Meng, Z., Wang, X., et al. (2015). MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. J. Clin. Invest 125 (6), 2497–2509. doi:10.1172/jci75438
Fu, X., Jin, L., Wang, X., Luo, A., Hu, J., Zheng, X., et al. (2013). MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proc. Natl. Acad. Sci. U. S. A. 110 (44), 17892–17897. doi:10.1073/pnas.1317397110
Fu, X., Meng, Z., Liang, W., Tian, Y., Wang, X., Han, W., et al. (2014). miR-26a enhances miRNA biogenesis by targeting Lin28B and Zcchc11 to suppress tumor growth and metastasis. Oncogene 33 (34), 4296–4306. doi:10.1038/onc.2013.385
Gao, J., Li, E., Liu, W., Yang, Q., Xie, C., Ai, J., et al. (2020). Circular RNA MYLK promotes hepatocellular carcinoma progression through the miR29a/KMT5C signaling pathway. Onco Targets Ther. 13, 8615–8627. doi:10.2147/ott.S258715
Gao, X., Xia, X., Li, F., Zhang, M., Zhou, H., Wu, X., et al. (2021). Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat. Cell Biol. 23, 278–291. doi:10.1038/s41556-021-00639-4
Ghosal, S., Das, S., Sen, R., Basak, P., and Chakrabarti, J. (2013). Circ2Traits: A comprehensive database for circular RNA potentially associated with disease and traits. Front. Genet. 4, 283. doi:10.3389/fgene.2013.00283
Ghosal, S., Das, S., Sen, R., and Chakrabarti, J. (2014). HumanViCe: Host ceRNA network in virus infected cells in human. Front. Genet. 5, 249. doi:10.3389/fgene.2014.00249
Glažar, P., Papavasileiou, P., and Rajewsky, N. (2014). circBase: a database for circular RNAs. RNA (New York, N.Y.) 20 (11), 1666–1670. doi:10.1261/rna.043687.113
Gokool, A., Anwar, F., and Voineagu, I. (2020). The landscape of circular RNA expression in the human brain. Biol. Psychiatry 87 (3), 294–304. doi:10.1016/j.biopsych.2019.07.029
Gomes, C., Schroen, B., Kuster, G., Robinson, E., Ford, K., Squire, I., et al. (2020). Regulatory RNAs in heart failure. Circulation 141 (4), 313–328. doi:10.1161/circulationaha.119.042474
Gong, Y., Mao, J., Wu, D., Wang, X., Li, L., Zhu, L., et al. (2018). Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int. 18, 116. doi:10.1186/s12935-018-0602-3
Gu, Y., Wang, Y., He, L., Zhang, J., Zhu, X., Liu, N., et al. (2021a). Circular RNA circIPO11 drives self-renewal of liver cancer initiating cells via Hedgehog signaling. Mol. Cancer 20 (1), 132. doi:10.1186/s12943-021-01435-2
Gu, Y., Wu, F., Wang, H., Chang, J., Wang, Y., and Li, X. (2021b). Circular RNA circARPP21 acts as a sponge of miR-543 to suppress hepatocellular carcinoma by regulating LIFR. Onco Targets Ther. 14, 879–890. doi:10.2147/ott.S283026
Guan, Y., Zhang, Y., Hao, L., and Nie, Z. (2020). CircRNA_102272 promotes cisplatin-resistance in hepatocellular carcinoma by decreasing MiR-326 targeting of RUNX2. Cancer Manag. Res. 12, 12527–12534. doi:10.2147/cmar.S258230
Guan, Z., Tan, J., Gao, W., Li, X., Yang, Y., Li, X., et al. (2018). Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J. Cell Physiol. 234 (1), 500–508. doi:10.1002/jcp.26612
Guo, J., Agarwal, V., Guo, H., and Bartel, D. (2014). Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 15 (7), 409. doi:10.1186/s13059-014-0409-z
Guo, X., Wang, Z., Deng, X., Lu, Y., Huang, X., Lin, J., et al. (2022). Circular RNA CircITCH (has-circ-0001141) suppresses hepatocellular carcinoma (HCC) progression by sponging miR-184. Cell Cycle 21 (15), 1557–1577. doi:10.1080/15384101.2022.2057633
Guo, X. Y., Chen, J. N., Sun, F., Wang, Y. Q., Pan, Q., and Fan, J. G. (2017a). circRNA_0046367 prevents hepatoxicity of lipid peroxidation: An inhibitory role against hepatic steatosis. Oxid. Med. Cell Longev. 2017, 3960197. doi:10.1155/2017/3960197
Guo, X. Y., He, C. X., Wang, Y. Q., Sun, C., Li, G. M., Su, Q., et al. (2017b). Circular RNA profiling and bioinformatic modeling identify its regulatory role in hepatic steatosis. Biomed. Res. Int. 2017, 5936171. doi:10.1155/2017/5936171
Guo, X. Y., Sun, F., Chen, J. N., Wang, Y. Q., Pan, Q., and Fan, J. G. (2018). circRNA_0046366 inhibits hepatocellular steatosis by normalization of PPAR signaling. World J. Gastroenterol. 24 (3), 323–337. doi:10.3748/wjg.v24.i3.323
Guo, Z., Cao, Q., Zhao, Z., and Song, C. (2020). Biogenesis, features, functions, and disease relationships of a specific circular RNA: CDR1as. Aging Dis. 11 (4), 1009–1020. doi:10.14336/AD.2019.0920
Halle, S., Halle, O., and Förster, R. (2017). Mechanisms and dynamics of T cell-mediated cytotoxicity in vivo. Trends Immunol. 38 (6), 432–443. doi:10.1016/j.it.2017.04.002
Han, D., Li, J., Wang, H., Su, X., Hou, J., Gu, Y., et al. (2017). Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 66 (4), 1151–1164. doi:10.1002/hep.29270
Han, J., Liu, Y., Yang, S., Wu, X., Li, H., and Wang, Q. (2021). MEK inhibitors for the treatment of non-small cell lung cancer. J. Hematol. Oncol. 14 (1), 1. doi:10.1186/s13045-020-01025-7
Han, J., Meng, J., Chen, S., Wang, X., Yin, S., Zhang, Q., et al. (2022a). Correction: YY1 complex promotes quaking expression via super-enhancer binding during EMT of hepatocellular carcinoma. Cancer Res. 82 (24), 4694. doi:10.1158/0008-5472.Can-22-3444
Han, L., Wang, M., Yang, Y., Xu, H., Wei, L., and Huang, X. (2022b). Detection of prognostic biomarkers for hepatocellular carcinoma through CircRNA-associated CeRNA analysis. J. Clin. Transl. Hepatol. 10 (1), 80–89. doi:10.14218/jcth.2020.00144
Hao, Y., Wu, W., Li, H., Yuan, J., Luo, J., Zhao, Y., et al. (2016). NPInter v3.0: An upgraded database of noncoding RNA-associated interactions. Database (Oxford) 2016, baw057. doi:10.1093/database/baw057
He, S., Guo, Z., Kang, Q., Wang, X., and Han, X. (2020a). Circular RNA hsa_circ_0000517 modulates hepatocellular carcinoma advancement via the miR-326/SMAD6 axis. Cancer Cell Int. 20, 360. doi:10.1186/s12935-020-01447-w
He, W., Zhu, X., Tang, X., Xiang, X., Yu, J., and Sun, H. (2022). Circ_0027089 regulates NACC1 by targeting miR-136-5p to aggravate the development of Hepatitis B virus-related hepatocellular carcinoma. Anticancer Drugs 33 (1), e336–e348. doi:10.1097/cad.0000000000001211
He, Y., Cui, Y., Wang, W., Gu, J., Guo, S., Ma, K., et al. (2011). Hypomethylation of the hsa-miR-191 locus causes high expression of hsa-mir-191 and promotes the epithelial-to-mesenchymal transition in hepatocellular carcinoma. Neoplasia 13 (9), 841–853. doi:10.1593/neo.11698
He, Y., Huang, H., Jin, L., Zhang, F., Zeng, M., Wei, L., et al. (2020b). CircZNF609 enhances hepatocellular carcinoma cell proliferation, metastasis, and stemness by activating the Hedgehog pathway through the regulation of miR-15a-5p/15b-5p and GLI2 expressions. Cell Death Dis. 11 (5), 358. doi:10.1038/s41419-020-2441-0
Henke, J., Goergen, D., Zheng, J., Song, Y., Schüttler, C., Fehr, C., et al. (2008). microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 27 (24), 3300–3310. doi:10.1038/emboj.2008.244
Ho, J. S., Di Tullio, F., Schwarz, M., Low, D., Incarnato, D., Gay, F., et al. (2021). HNRNPM controls circRNA biogenesis and splicing fidelity to sustain cancer cell fitness. Elife 10. doi:10.7554/eLife.59654
Hotta, M., Tashiro, F., Ikegami, H., Niwa, H., Ogihara, T., Yodoi, J., et al. (1998). Pancreatic beta cell-specific expression of thioredoxin, an antioxidative and antiapoptotic protein, prevents autoimmune and Streptozotocin-induced diabetes. J. Exp. Med. 188(8), 1445–1451. doi:10.1084/jem.188.8.1445
Hou, L., Zhang, J., and Zhao, F. (2023). Full-length circular RNA profiling by nanopore sequencing with CIRI-long. Nat. Protoc. 18 (6), 1795–1813. doi:10.1038/s41596-023-00815-w
Hsu, M. T., and Coca-Prados, M. (1979). Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280 (5720), 339–340. doi:10.1038/280339a0
Hu, Z. Q., Zhou, S. L., Li, J., Zhou, Z. J., Wang, P. C., Xin, H. Y., et al. (2020). Circular RNA sequencing identifies CircASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology 72 (3), 906–922. doi:10.1002/hep.31068
Huang, G., Liang, M., Liu, H., Huang, J., Li, P., Wang, C., et al. (2020a). CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/beta-catenin pathway. Cell Death Dis. 11 (12), 1065. doi:10.1038/s41419-020-03276-1
Huang, K., Du, M., Tan, X., Yang, L., Li, X., Jiang, Y., et al. (2017a). PARP1-mediated PPARalpha poly(ADP-ribosyl)ation suppresses fatty acid oxidation in non-alcoholic fatty liver disease. J. Hepatol. 66 (5), 962–977. doi:10.1016/j.jhep.2016.11.020
Huang, W., Ling, Y., Zhang, S., Xia, Q., Cao, R., Fan, X., et al. (2021). TransCirc: An interactive database for translatable circular RNAs based on multi-omics evidence. Nucleic Acids Res. 49 (D1), D236–d242. doi:10.1093/nar/gkaa823
Huang, X. Y., Huang, Z. L., Huang, J., Xu, B., Huang, X. Y., Xu, Y. H., et al. (2020b). Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J. Exp. Clin. Cancer Res. 39 (1), 20. doi:10.1186/s13046-020-1529-9
Huang, X. Y., Huang, Z. L., Xu, Y. H., Zheng, Q., Chen, Z., Song, W., et al. (2017b). Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in Hepatitis B-related hepatocellular carcinoma. Sci. Rep. 7 (1), 5428. doi:10.1038/s41598-017-05432-8
Huang, X. Y., Huang, Z. L., Zhang, P. B., Huang, X. Y., Huang, J., Wang, H. C., et al. (2019). CircRNA-100338 is associated with mTOR signaling pathway and poor prognosis in hepatocellular carcinoma. Front. Oncol. 9, 392. doi:10.3389/fonc.2019.00392
Huang, X. Y., Zhang, P. F., Wei, C. Y., Peng, R., Lu, J. C., Gao, C., et al. (2020c). Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol. Cancer 19 (1), 92. doi:10.1186/s12943-020-01213-6
Ioannou, G. N. (2021). Epidemiology and risk-stratification of NAFLD-associated HCC. J. Hepatol. 75 (6), 1476–1484. doi:10.1016/j.jhep.2021.08.012
Ivanov, A., Memczak, S., Wyler, E., Torti, F., Porath, H. T., Orejuela, M. R., et al. (2015). Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 10 (2), 170–177. doi:10.1016/j.celrep.2014.12.019
Jang, S., Kim, G., Tak, W., Kweon, Y., Lee, Y., Han, Y., et al. (2022). Circular noncoding RNA hsa_circ_0003570 as a prognostic biomarker for hepatocellular carcinoma. Genes 13 (8). doi:10.3390/genes13081484
Jarlstad Olesen, M. T., and Lasse, S. K. (2021). Circular RNAs as microRNA sponges: Evidence and controversies. Essays Biochem. 65 (4), 685–696. doi:10.1042/EBC20200060
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19 (2), 141–157. doi:10.1261/rna.035667.112
Ji, D., Chen, G. F., Wang, J. C., Ji, S. H., Wu, X. W., Lu, X. J., et al. (2020). Hsa_circ_0070963 inhibits liver fibrosis via regulation of miR-223-3p and LEMD3. Aging 12 (2), 1643–1655. doi:10.18632/aging.102705
Ji, D., Wang, Y., Li, H., Sun, B., and Luo, X. (2019). Long non-coding RNA LINC00461/miR-149-5p/LRIG2 axis regulates hepatocellular carcinoma progression. Biochem. Biophys. Res. Commun. 512 (2), 176–181. doi:10.1016/j.bbrc.2019.03.049
Ji, H., Kim, T., Lee, W., Jeong, S., Cho, Y., and Kim, H. (2022). Two circPPFIA1s negatively regulate liver metastasis of colon cancer via miR-155-5p/CDX1 and HuR/RAB36. Mol. Cancer 21 (1), 197. doi:10.1186/s12943-022-01667-w
Ji, Y., Yang, S., Yan, X., Zhu, L., Yang, W., Yang, X., et al. (2021). CircCRIM1 promotes hepatocellular carcinoma proliferation and angiogenesis by sponging miR-378a-3p and regulating SKP2 expression. Front. Cell Dev. Biol. 9, 796686. doi:10.3389/fcell.2021.796686
Jia, C., Yao, Z., Lin, Z., Zhao, L., Cai, X., Chen, S., et al. (2021). circNFATC3 sponges miR-548I acts as a ceRNA to protect NFATC3 itself and suppressed hepatocellular carcinoma progression. J. Cell. Physiol. 236 (2), 1252–1269. doi:10.1002/jcp.29931
Jiang, B., Tian, M., Li, G., Sadula, A., Xiu, D., Yuan, C., et al. (2022). circEPS15 overexpression in hepatocellular carcinoma modulates tumor invasion and migration. Front. Genet. 13, 804848. doi:10.3389/fgene.2022.804848
Jiang, M., Chen, H., Du, Q., Wang, L., Liu, X., and Liu, C. (2021). Genome-wide identification of circular RNAs potentially involved in the biosynthesis of secondary metabolites in salvia miltiorrhiza. Front. Genet. 12, 645115. doi:10.3389/fgene.2021.645115
Jiang, Z., Shen, L., Wang, S., Wu, S., Hu, Y., Guo, J., et al. (2019). Hsa_circ_0028502 and hsa_circ_0076251 are potential novel biomarkers for hepatocellular carcinoma. Cancer Med. 8 (17), 7278–7287. doi:10.1002/cam4.2584
Jost, I., Shalamova, L. A., Gerresheim, G. K., Niepmann, M., Bindereif, A., and Rossbach, O. (2018). Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol. 15 (8), 1032–1039. doi:10.1080/15476286.2018.1435248
Kalluri, R., and LeBleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Science 367 (6478). doi:10.1126/science.aau6977
Kanska, J., Zakhour, M., Taylor-Harding, B., Karlan, B., and Wiedemeyer, W. (2016). Cyclin E as a potential therapeutic target in high grade serous ovarian cancer. Gynecol. Oncol. 143 (1), 152–158. doi:10.1016/j.ygyno.2016.07.111
Kelly, S., Greenman, C., Cook, P. R., and Papantonis, A. (2015). Exon skipping is correlated with exon circularization. J. Mol. Biol. 427 (15), 2414–2417. doi:10.1016/j.jmb.2015.02.018
Kim, E., Kim, D., Lee, J., Yoe, J., Park, J., Kim, C., et al. (2018). Capicua suppresses hepatocellular carcinoma progression by controlling the ETV4-MMP1 axis. Hepatology. 67 (6), 2287–2301. doi:10.1002/hep.29738
Klepsch, V., Hermann-Kleiter, N., Do-Dinh, P., Jakic, B., Offermann, A., Efremova, M., et al. (2018). Nuclear receptor NR2F6 inhibition potentiates responses to PD-L1/PD-1 cancer immune checkpoint blockade. Nat. Commun. 9 (1), 1538. doi:10.1038/s41467-018-04004-2
Kou, P., Zhang, C., Lin, J., and Wang, H. (2019). Circular RNA hsa_circ_0078602 may have potential as a prognostic biomarker for patients with hepatocellular carcinoma. Oncol. Lett. 17 (2), 2091–2098. doi:10.3892/ol.2018.9863
Kramer, M. C., Liang, D., Tatomer, D. C., Gold, B., March, Z. M., Cherry, S., et al. (2015). Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 29 (20), 2168–2182. doi:10.1101/gad.270421.115
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20 (11), 675–691. doi:10.1038/s41576-019-0158-7
Kuleshov, M. V., Jones, M. R., Rouillard, A. D., Fernandez, N. F., Duan, Q., Wang, Z., et al. (2016). Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44 (W1), W90–W97. doi:10.1093/nar/gkw377
Legnini, I., Di Timoteo, G., Rossi, F., Morlando, M., Briganti, F., Sthandier, O., et al. (2017). Circ-ZNF609 is a circular RNA that can Be translated and functions in myogenesis. Mol. Cell 66 (1), 22–37. doi:10.1016/j.molcel.2017.02.017
Lei, B., Zhou, J., Xuan, X., Tian, Z., Zhang, M., Gao, W., et al. (2019). Circular RNA expression profiles of peripheral blood mononuclear cells in hepatocellular carcinoma patients by sequence analysis. Cancer Med. 8 (4), 1423–1433. doi:10.1002/cam4.2010
Leonard, W., and O'Shea, J. (1998). Jaks and STATs: Biological implications. Annu. Rev. Immunol. 16, 293–322. doi:10.1146/annurev.immunol.16.1.293
Levkau, B., Koyama, H., Raines, E., Clurman, B., Herren, B., Orth, K., et al. (1998). Cleavage of p21Cip1/waf1 and p27Kip1 mediates apoptosis in endothelial cells through activation of Cdk2: Role of a caspase cascade. Mol. Cell 1 (4), 553–563. doi:10.1016/s1097-2765(00)80055-6
Li, C., Ni, Y. Q., Xu, H., Xiang, Q. Y., Zhao, Y., Zhan, J. K., et al. (2021a). Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct. Target Ther. 6 (1), 383. doi:10.1038/s41392-021-00779-x
Li, D., Zhang, J., Yang, J., Wang, J., Zhang, R., Li, J., et al. (2021b). CircMTO1 suppresses hepatocellular carcinoma progression via the miR-541-5p/ZIC1 axis by regulating Wnt/β-catenin signaling pathway and epithelial-to-mesenchymal transition. Cell Death Dis. 13 (1), 12. doi:10.1038/s41419-021-04464-3
Li, J. H., Liu, S., Zhou, H., Qu, L. H., and Yang, J. H. (2014). starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42, D92–D97. doi:10.1093/nar/gkt1248
Li, J., Hu, Z. Q., Yu, S. Y., Mao, L., Zhou, Z. J., Wang, P. C., et al. (2022a). CircRPN2 inhibits aerobic glycolysis and metastasis in hepatocellular carcinoma. Cancer Res. 82 (6), 1055–1069. doi:10.1158/0008-5472.CAN-21-1259
Li, J., Qin, X., Wu, R., Wan, L., Zhang, L., and Liu, R. (2020a). Circular RNA circFBXO11 modulates hepatocellular carcinoma progress and oxaliplatin resistance through miR-605/FOXO3/ABCB1 axis. J. Cell Mol. Med. 24 (9), 5152–5161. doi:10.1111/jcmm.15162
Li, J., Sun, D., Pu, W., Wang, J., and Peng, Y. (2020b). Circular RNAs in cancer: Biogenesis, function, and clinical significance. Trends Cancer 6 (4), 319–336. doi:10.1016/j.trecan.2020.01.012
Li, L., He, K., Chen, S., Wei, W., Tian, Z., Tang, Y., et al. (2020c). Circ_0001175 promotes hepatocellular carcinoma cell proliferation and metastasis by regulating miR-130a-5p. Onco Targets Ther. 13, 13315–13327. doi:10.2147/OTT.S262408
Li, P., Shan, K., Liu, Y., Zhang, Y., Xu, L., and Xu, L. (2019a). CircScd1 promotes fatty liver disease via the janus kinase 2/signal transducer and activator of transcription 5 pathway. Dig. Dis. Sci. 64 (1), 113–122. doi:10.1007/s10620-018-5290-2
Li, P., Song, R., Yin, F., Liu, M., Liu, H., Ma, S., et al. (2022b). circMRPS35 promotes malignant progression and cisplatin resistance in hepatocellular carcinoma. Mol. Ther. 30 (1), 431–447. doi:10.1016/j.ymthe.2021.08.027
Li, Q., Pan, X., Zhu, D., Deng, Z., Jiang, R., and Wang, X. (2019b). Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 Axis under hypoxic stress. Hepatology 70 (4), 1298–1316. doi:10.1002/hep.30671
Li, Q., Yao, H., Wang, Y., Wu, Y., Thorne, R., Zhu, Y., et al. (2022c). circPRKAA1 activates a Ku80/Ku70/SREBP-1 axis driving de novo fatty acid synthesis in cancer cells. Cell Rep. 41 (8), 111707. doi:10.1016/j.celrep.2022.111707
Li, R., Deng, Y., Liang, J., Hu, Z., Li, X., Liu, H., et al. (2020d). Circular RNA circ-102,166 acts as a sponge of miR-182 and miR-184 to suppress hepatocellular carcinoma proliferation and invasion. Cell Oncol. (Dordr) 44 (2), 279–295. doi:10.1007/s13402-020-00564-y
Li, R., Ke, S., Meng, F., Lu, J., Zou, X., He, Z., et al. (2018a). CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 9 (8), 838. doi:10.1038/s41419-018-0852-y
Li, S., Chen, Z., Zhou, R., Wang, S., Wang, W., Liu, D., et al. (2022d). Hsa_circ_0048674 facilitates hepatocellular carcinoma progression and natural killer cell exhaustion depending on the regulation of miR-223-3p/PDL1. Histol. Histopathol. 37 (12), 1185–1199. doi:10.14670/hh-18-440
Li, S., Li, Y., Chen, B., Zhao, J., Yu, S., Tang, Y., et al. (2018b). exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 46 (D1), D106–d112. doi:10.1093/nar/gkx891
Li, S., Weng, J., Song, F., Li, L., Xiao, C., Yang, W., et al. (2020e). Circular RNA circZNF566 promotes hepatocellular carcinoma progression by sponging miR-4738-3p and regulating TDO2 expression. Cell Death Dis. 11 (6), 452. doi:10.1038/s41419-020-2616-8
Li, X., Liu, C. X., Xue, W., Zhang, Y., Jiang, S., Yin, Q. F., et al. (2017). Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. Cell 67 (2), 214–227 e217. doi:10.1016/j.molcel.2017.05.023
Li, Y., Zhang, Y., Zhang, S., Huang, D., Li, B., Liang, G., et al. (2021c). circRNA circARNT2 suppressed the sensitivity of hepatocellular carcinoma cells to cisplatin by targeting the miR-155-5p/PDK1 Axis. Mol. Ther. Nucleic Acids 23, 244–254. doi:10.1016/j.omtn.2020.08.037
Li, Y., Zheng, Q., Bao, C., Li, S., Guo, W., Zhao, J., et al. (2015a). Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 25 (8), 981–984. doi:10.1038/cr.2015.82
Li, Z., Hu, Y., Zeng, Q., Wang, H., Yan, J., Li, H., et al. (2019c). Circular RNA MYLK promotes hepatocellular carcinoma progression by increasing Rab23 expression by sponging miR-362-3p. Cancer Cell Int. 19, 211. doi:10.1186/s12935-019-0926-7
Li, Z., Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., et al. (2015b). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22 (3), 256–264. doi:10.1038/nsmb.2959
Liang, D., Tatomer, D. C., Luo, Z., Wu, H., Yang, L., Chen, L. L., et al. (2017). The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol. Cell 68 (5), 940–954 e943. doi:10.1016/j.molcel.2017.10.034
Liang, W. C., Wong, C. W., Liang, P. P., Shi, M., Cao, Y., Rao, S. T., et al. (2019). Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 20 (1), 84. doi:10.1186/s13059-019-1685-4
Liberti, M. V., and Locasale, J. W. (2016). The Warburg effect: How does it benefit cancer cells? Trends Biochem. Sci. 41 (3), 211–218. doi:10.1016/j.tibs.2015.12.001
Lin, G., Li, J., Chen, K., Wang, A., and Guo, C. (2022). Circ_0000854 regulates the progression of hepatocellular carcinoma through miR-1294/IRGQ axis. Clin. Immunol. 238, 109007. doi:10.1016/j.clim.2022.109007
Lin, Q., Ling, Y. B., Chen, J. W., Zhou, C. R., Chen, J., Li, X., et al. (2018). Circular RNA circCDK13 suppresses cell proliferation, migration and invasion by modulating the JAK/STAT and PI3K/AKT pathways in liver cancer. Int. J. Oncol. 53 (1), 246–256. doi:10.3892/ijo.2018.4371
Litke, J., and Jaffrey, S. (2019). Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol. 37 (6), 667–675. doi:10.1038/s41587-019-0090-6
Liu, B., Tian, Y., Chen, M., Shen, H., Xia, J., Nan, J., et al. (2021a). CircUBAP2 promotes MMP9-mediated oncogenic effect via sponging miR-194-3p in hepatocellular carcinoma. Front. Cell Dev. Biol. 9, 675043. doi:10.3389/fcell.2021.675043
Liu, B., Yang, G., Wang, X., Liu, J., Lu, Z., Wang, Q., et al. (2020a). CircBACH1 (hsa_circ_0061395) promotes hepatocellular carcinoma growth by regulating p27 repression via HuR. J. Cell Physiol. 235 (10), 6929–6941. doi:10.1002/jcp.29589
Liu, C., and Chen, L. (2022). Circular RNAs: Characterization, cellular roles, and applications. Cell 185 (12), 2016–2034. doi:10.1016/j.cell.2022.04.021
Liu, C. X., Li, X., Nan, F., Jiang, S., Gao, X., Guo, S. K., et al. (2019b). Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177 (4), 865–880 e821. doi:10.1016/j.cell.2019.03.046
Liu, C., Zhong, X., Li, J., and Xu, F. (2019a). Circular RNA circVAPA promotes cell proliferation in hepatocellular carcinoma. Hum. Gene Ther. Clin. Dev. 30 (4), 152–159. doi:10.1089/humc.2019.079
Liu, D., Liu, W., Chen, X., Yin, J., Ma, L., Liu, M., et al. (2022a). circKCNN2 suppresses the recurrence of hepatocellular carcinoma at least partially via regulating miR-520c-3p/methyl-DNA-binding domain protein 2 axis. Clin. Transl. Med. 12 (1), e662. doi:10.1002/ctm2.662
Liu, G., Du, W., Sang, X., Tong, Q., Wang, Y., Chen, G., et al. (2022b). RNA G-quadruplex in TMPRSS2 reduces SARS-CoV-2 infection. Nat. Commun. 13 (1), 1444. doi:10.1038/s41467-022-29135-5
Liu, G., Du, W., Xu, H., Sun, Q., Tang, D., Zou, S., et al. (2020b). RNA G-quadruplex regulates microRNA-26a biogenesis and function. J. Hepatol. 73 (2), 371–382. doi:10.1016/j.jhep.2020.02.032
Liu, H., Lan, T., Li, H., Xu, L., Chen, X., Liao, H., et al. (2021b). Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR. Theranostics 11 (3), 1396–1411. doi:10.7150/thno.53227
Liu, J., Zhang, Y., Tian, Y., Huang, W., Tong, N., and Fu, X. (2022c). Integrative biology of extracellular vesicles in diabetes mellitus and diabetic complications. Theranostics 12 (3), 1342–1372. doi:10.7150/thno.65778
Liu, L., Gu, M., Ma, J., Wang, Y., Li, M., Wang, H., et al. (2022d). CircGPR137B/miR-4739/FTO feedback loop suppresses tumorigenesis and metastasis of hepatocellular carcinoma. Mol. Cancer 21 (1), 149. doi:10.1186/s12943-022-01619-4
Liu, L., Qi, X., Gui, Y., Huo, H., Yang, X., and Yang, L. (2019c). Overexpression of circ_0021093 circular RNA forecasts an unfavorable prognosis and facilitates cell progression by targeting the miR-766-3p/MTA3 pathway in hepatocellular carcinoma. Gene 714, 143992. doi:10.1016/j.gene.2019.143992
Liu, M., Wang, Q., Shen, J., Yang, B. B., and Ding, X. (2019d). Circbank: A comprehensive database for circRNA with standard nomenclature. RNA Biol. 16 (7), 899–905. doi:10.1080/15476286.2019.1600395
Liu, Q., Cai, Y., Xiong, H., Deng, Y., and Dai, X. (2019e). CCRDB: A cancer circRNAs-related database and its application in hepatocellular carcinoma-related circRNAs. Database (Oxford) 2019, baz063. doi:10.1093/database/baz063
Liu, W., Feng, R., Li, X., Li, D., and Zhai, W. (2019f). TGF-β- and lipopolysaccharide-induced upregulation of circular RNA PWWP2A promotes hepatic fibrosis via sponging miR-203 and miR-223. Aging 11 (21), 9569–9580. doi:10.18632/aging.102405
Liu, W., Zheng, L., Zhang, R., Hou, P., Wang, J., Wu, L., et al. (2022e). Circ-ZEB1 promotes PIK3CA expression by silencing miR-199a-3p and affects the proliferation and apoptosis of hepatocellular carcinoma. Mol. Cancer 21 (1), 72. doi:10.1186/s12943-022-01529-5
Liu, X., Wang, X., Li, J., Hu, S., Deng, Y., Yin, H., et al. (2020c). Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci. China Life Sci. 63 (10), 1429–1449. doi:10.1007/s11427-020-1631-9
Liu, X., Yang, L., Jiang, D., Lu, W., and Zhang, Y. (2020d). Circ-DENND4C up-regulates TCF4 expression to modulate hepatocellular carcinoma cell proliferation and apoptosis via activating Wnt/β-catenin signal pathway. Cancer Cell Int. 20 (1), 295. doi:10.1186/s12935-020-01346-0
Liu, Y. C., Li, J. R., Sun, C. H., Andrews, E., Chao, R. F., Lin, F. M., et al. (2016). CircNet: A database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 44 (D1), D209–D215. doi:10.1093/nar/gkv940
Liu, Y. M., Cao, Y., Zhao, P. S., Wu, L. Y., Lu, Y. M., Wang, Y. L., et al. (2022g). CircCCNB1 silencing acting as a miR-106b-5p sponge inhibited GPM6A expression to promote HCC progression by enhancing DYNC1I1 expression and activating the AKT/ERK signaling pathway. Int. J. Biol. Sci. 18 (2), 637–651. doi:10.7150/ijbs.66915
Liu, Y., Song, J., Zhang, H., Liao, Z., Liu, F., Su, C., et al. (2022f). EIF4A3-induced circTOLLIP promotes the progression of hepatocellular carcinoma via the miR-516a-5p/PBX3/EMT pathway. J. Exp. Clin. Cancer Res. 41 (1), 164. doi:10.1186/s13046-022-02378-2
Llovet, J., Kelley, R., Villanueva, A., Singal, A., Pikarsky, E., Roayaie, S., et al. (2021a). Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 7 (1), 6. doi:10.1038/s41572-020-00240-3
Llovet, J. M., Castet, F., Heikenwalder, M., Maini, M. K., Mazzaferro, V., Pinato, D. J., et al. (2022). Immunotherapies for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 19 (3), 151–172. doi:10.1038/s41571-021-00573-2
Llovet, J. M., De Baere, T., Kulik, L., Haber, P. K., Greten, T. F., Meyer, T., et al. (2021b). Locoregional therapies in the era of molecular and immune treatments for hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 18 (5), 293–313. doi:10.1038/s41575-020-00395-0
Llovet, J. M., Kelley, R. K., Villanueva, A., Singal, A. G., Pikarsky, E., Roayaie, S., et al. (2021c). Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 7 (1), 6. doi:10.1038/s41572-020-00240-3
Loan Young, T., Chang Wang, K., James Varley, A., and Li, B. (2023). Clinical delivery of circular RNA: Lessons learned from RNA drug development. Adv. drug Deliv. Rev. 197, 114826. doi:10.1016/j.addr.2023.114826
Long, F., Lin, Z., Li, L., Ma, M., Lu, Z., Jing, L., et al. (2021). Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Mol. Cancer 20 (1), 26. doi:10.1186/s12943-021-01318-6
Luo, Y., Fu, Y., Huang, R., Gao, M., Liu, F., Gui, R., et al. (2019a). CircRNA_101505 sensitizes hepatocellular carcinoma cells to cisplatin by sponging miR-103 and promotes oxidored-nitro domain-containing protein 1 expression. Cell Death Dis. 5, 121. doi:10.1038/s41420-019-0202-6
Luo, Y., Liu, F., and Gui, R. (2020). High expression of circulating exosomal circAKT3 is associated with higher recurrence in HCC patients undergoing surgical treatment. Surg. Oncol. 33, 276–281. doi:10.1016/j.suronc.2020.04.021
Luo, Y. Y., Tao, K. G., Lu, Y. T., Li, B. B., Wu, K. M., Ding, C. H., et al. (2022). Hsa_Circ_0098181 suppresses hepatocellular carcinoma by sponging miR-18a-3p and targeting PPARA. Front. Pharmacol. 13, 819735. doi:10.3389/fphar.2022.819735
Luo, Z., Mao, X., and Cui, W. (2019b). Circular RNA expression and circPTPRM promotes proliferation and migration in hepatocellular carcinoma. Med. Oncol. 36 (10), 86. doi:10.1007/s12032-019-1311-z
Lyu, L. H., Zhang, C. Y., Yang, W. J., Jin, A. L., Zhu, J., Wang, H., et al. (2022). Hsa_circ_0003945 promotes progression of hepatocellular carcinoma by mediating miR-34c-5p/LGR4/β-catenin axis activity. J. Cell Mol. Med. 26 (8), 2218–2229. doi:10.1111/jcmm.17243
Ma, M., Xu, H., Liu, G., Wu, J., Li, C., Wang, X., et al. (2019). Metabolism-induced tumor activator 1 (MITA1), an energy stress-inducible long noncoding RNA, promotes hepatocellular carcinoma metastasis. Hepatology. 70 (1), 215–230. doi:10.1002/hep.30602
Ma, M., Zhang, C., Cao, R., Tang, D., Sang, X., Zou, S., et al. (2022). UBE2O promotes lipid metabolic reprogramming and liver cancer progression by mediating HADHA ubiquitination. Oncogene 41 (48), 5199–5213. doi:10.1038/s41388-022-02509-1
Malouf, C., Antunes, E., O'Dwyer, M., Jakobczyk, H., Sahm, F., Landua, S., et al. (2021). miR-130b and miR-128a are essential lineage-specific codrivers of t(4;11) MLL-AF4 acute leukemia. Blood 138 (21), 2066–2092. doi:10.1182/blood.2020006610
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495 (7441), 333–338. doi:10.1038/nature11928
Meng, J., Chen, S., Han, J. X., Qian, B., Wang, X. R., Zhong, W. L., et al. (2018). Twist1 regulates vimentin through Cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res. 78 (15), 4150–4162. doi:10.1158/0008-5472.CAN-17-3009
Nielsen, A. F., Bindereif, A., Bozzoni, I., Hanan, M., Hansen, T. B., Irimia, M., et al. (2022). Best practice standards for circular RNA research. Nat. Methods 19 (10), 1208–1220. doi:10.1038/s41592-022-01487-2
Nigro, J. M., Cho, K. R., Fearon, E. R., Kern, S. E., Ruppert, J. M., Oliner, J. D., et al. (1991). Scrambled exons. Cell 64 (3), 607–613. doi:10.1016/0092-8674(91)90244-s
Novellasdemunt, L., Foglizzo, V., Cuadrado, L., Antas, P., Kucharska, A., Encheva, V., et al. (2017). USP7 is a tumor-specific WNT activator for APC-mutated colorectal cancer by mediating β-catenin deubiquitination. Cell Rep. 21 (3), 612–627. doi:10.1016/j.celrep.2017.09.072
Opitz, C. A., Litzenburger, U. M., Sahm, F., Ott, M., Tritschler, I., Trump, S., et al. (2011). An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478 (7368), 197–203. doi:10.1038/nature10491
Pamudurti, N., Bartok, O., Jens, M., Ashwal-Fluss, R., Stottmeister, C., Ruhe, L., et al. (2017). Translation of CircRNAs. Mol. Cell 66 (1), 9–21.e27. doi:10.1016/j.molcel.2017.02.021
Pamudurti, N. R., Patop, I. L., Krishnamoorthy, A., Bartok, O., Maya, R., Lerner, N., et al. (2022). circMbl functions in cis and in trans to regulate gene expression and physiology in a tissue-specific fashion. Cell Rep. 39 (4), 110740. doi:10.1016/j.celrep.2022.110740
Pan, H., Tang, L., Jiang, H., Li, X., Wang, R., Gao, J., et al. (2019). Enhanced expression of circ_0000267 in hepatocellular carcinoma indicates poor prognosis and facilitates cell progression by sponging miR-646. J. Cell. Biochem. 120 (7), 11350–11357. doi:10.1002/jcb.28411
Panda, A. C., Dudekula, D. B., Abdelmohsen, K., and Gorospe, M. (2018). Analysis of circular RNAs using the web tool CircInteractome. Methods Mol. Biol. 1724, 43–56. doi:10.1007/978-1-4939-7562-4_4
Peng, B., Hu, J., and Fu, X. (2021). ELANE: An emerging lane to selective anticancer therapy. Signal Transduct. Target Ther. 6 (1), 358. doi:10.1038/s41392-021-00766-2
Peng, R., Cao, J., Su, B., Bai, X., Jin, X., Wang, A., et al. (2022). Down-regulation of circPTTG1IP induces hepatocellular carcinoma development via miR-16-5p/RNF125/JAK1 axis. Cancer Lett. 543, 215778. doi:10.1016/j.canlet.2022.215778
Piwecka, M., Glažar, P., Hernandez-Miranda, L., Memczak, S., Wolf, S., Rybak-Wolf, A., et al. (2017). Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357 (6357). doi:10.1126/science.aam8526
Qiao, G., Chen, L., Jiang, W., Yang, C., Yang, C., Song, L., et al. (2019). Hsa_circ_0003998 may be used as a new biomarker for the diagnosis and prognosis of hepatocellular carcinoma. Onco Targets Ther. 12, 5849–5860. doi:10.2147/ott.S210363
Qin, L., Zhan, Z., Wei, C., Li, X., Zhang, T., and Li, J. (2021). Hsa-circRNA-G004213 promotes cisplatin sensitivity by regulating miR-513b-5p/PRPF39 in liver cancer. Mol. Med. Rep. 23 (6). doi:10.3892/mmr.2021.12060
Qin, M., Liu, G., Huo, X., Tao, X., Sun, X., Ge, Z., et al. (2016). Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 16 (1), 161–169. doi:10.3233/cbm-150552
Qiu, L., Huang, Y., Li, Z., Dong, X., Chen, G., Xu, H., et al. (2019). Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol. Oncol. 13 (2), 441–455. doi:10.1002/1878-0261.12424
Rebolledo, C., Silva, J., Saavedra, N., and Maracaja-Coutinho, V. (2023). Computational approaches for circRNAs prediction and in silico characterization. Briefings Bioinforma. 24 (3). doi:10.1093/bib/bbad154
Ridge, K. M., Eriksson, J. E., Pekny, M., and Goldman, R. D. (2022). Roles of vimentin in health and disease. Genes Dev. 36 (7-8), 391–407. doi:10.1101/gad.349358.122
Saaoud, F., Drummer, I. V. C., Shao, Y., Sun, Y., Lu, Y., Xu, K., et al. (2021). Circular RNAs are a novel type of non-coding RNAs in ROS regulation, cardiovascular metabolic inflammations and cancers. Pharmacol. Ther. 220, 107715. doi:10.1016/j.pharmthera.2020.107715
Schlacher, K. (2018). A new road to cancer-drug resistance. Nature 563 (7732), 478–480. doi:10.1038/d41586-018-07188-1
Sedano, C., and Sarnow, P. (2014). Hepatitis C virus subverts liver-specific miR-122 to protect the viral genome from exoribonuclease Xrn2. Cell Host Microbe 16 (2), 257–264. doi:10.1016/j.chom.2014.07.006
Shang, R., Kretov, D., Adamson, S., Treiber, T., Treiber, N., Vedanayagam, J., et al. (2022). Regulated dicing of pre-mir-144 via reshaping of its terminal loop. Nucleic Acids Res. 50 (13), 7637–7654. doi:10.1093/nar/gkac568
Shangguan, H., Feng, H., Lv, D., Wang, J., Tian, T., and Wang, X. (2020). Circular RNA circSLC25A16 contributes to the glycolysis of non-small-cell lung cancer through epigenetic modification. Cell Death Dis. 11 (6), 437. doi:10.1038/s41419-020-2635-5
Shen, B., Jiang, Y., Chen, Y., Zheng, H., Zeng, W., Li, Y., et al. (2016). Expression and inhibitory role of TIMP-3 in hepatocellular carcinoma. Oncol. Rep. 36 (1), 494–502. doi:10.3892/or.2016.4818
Shen, D., Zhao, H., Zeng, P., Ge, M., Shrestha, S., and Zhao, W. (2022a). Circular RNA circ_0001459 accelerates hepatocellular carcinoma progression via the miR-6165/IGF1R axis. Ann. N. Y. Acad. Sci. 1512 (1), 46–60. doi:10.1111/nyas.14753
Shen, H., Li, H., and Zhou, J. (2022b). Circular RNA hsa_circ_0032683 inhibits the progression of hepatocellular carcinoma by sponging microRNA-338-5p. Bioengineered 13 (2), 2321–2335. doi:10.1080/21655979.2021.2024961
Sherr, C., and Roberts, J. (1995). Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 9 (10), 1149–1163. doi:10.1101/gad.9.10.1149
Shi, L., Yan, P., Liang, Y., Sun, Y., Shen, J., Zhou, S., et al. (2017). Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis. 8 (11), e3171. doi:10.1038/cddis.2017.556
Shi, M., Li, Z., Zhang, L., Wu, X., Xiang, S., Wang, Y., et al. (2021). Hsa_circ_0007456 regulates the natural killer cell-mediated cytotoxicity toward hepatocellular carcinoma via the miR-6852-3p/ICAM-1 axis. Cell Death Dis. 12 (1), 94. doi:10.1038/s41419-020-03334-8
Slack, F., and Chinnaiyan, A. (2019). The role of non-coding RNAs in oncology. Cell 179 (5), 1033–1055. doi:10.1016/j.cell.2019.10.017
Song, C., Li, D., Liu, H., Sun, H., Liu, Z., Zhang, L., et al. (2019). The competing endogenous circular RNA ADAMTS14 suppressed hepatocellular carcinoma progression through regulating microRNA-572/regulator of calcineurin 1. J. Cell. Physiol. 234 (3), 2460–2470. doi:10.1002/jcp.26764
Song, J., Zhao, W., Zhang, X., Tian, W., Zhao, X., Ma, L., et al. (2022). Mutant RIG-I enhances cancer-related inflammation through activation of circRIG-I signaling. Nat. Commun. 13 (1), 7096. doi:10.1038/s41467-022-34885-3
Song, L. N., Qiao, G. L., Yu, J., Yang, C. M., Chen, Y., Deng, Z. F., et al. (2020). Hsa_circ_0003998 promotes epithelial to mesenchymal transition of hepatocellular carcinoma by sponging miR-143-3p and PCBP1. J. Exp. Clin. Cancer Res. 39 (1), 114. doi:10.1186/s13046-020-01576-0
Song, Y., Chen, M., Zhang, Y., Li, J., Liu, B., Li, N., et al. (2023). Loss of circSRY reduces γH2AX level in germ cells and impairs mouse spermatogenesis. Life Sci. Alliance 6 (2). doi:10.26508/lsa.202201617
Starke, S., Jost, I., Rossbach, O., Schneider, T., Schreiner, S., Hung, L. H., et al. (2015). Exon circularization requires canonical splice signals. Cell Rep. 10 (1), 103–111. doi:10.1016/j.celrep.2014.12.002
Stoll, L., Rodríguez-Trejo, A., Guay, C., Brozzi, F., Bayazit, M., Gattesco, S., et al. (2020). A circular RNA generated from an intron of the insulin gene controls insulin secretion. Nat. Commun. 11 (1), 5611. doi:10.1038/s41467-020-19381-w
Su, Y., Lv, X., Yin, W., Zhou, L., Hu, Y., Zhou, A., et al. (2019a). CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging 11 (19), 8183–8203. doi:10.18632/aging.102312
Su, Y., Xu, C., Liu, Y., Hu, Y., and Wu, H. (2019b). Circular RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression via multiple miRNAs sponge. Aging 11 (10), 3362–3375. doi:10.18632/aging.101988
Sun, L., Guo, Z., Sun, J., Li, J., Dong, Z., Zhang, Y., et al. (2018). MiR-133a acts as an anti-oncogene in Hepatocellular carcinoma by inhibiting FOSL2 through TGF-β/Smad3 signaling pathway. Biomed. Pharmacother. 107, 168–176. doi:10.1016/j.biopha.2018.07.151
Sun, Q., Yu, R., Wang, C., Yao, J., and Zhang, L. (2020a). Circular RNA circ-CSPP1 regulates CCNE2 to facilitate hepatocellular carcinoma cell growth via sponging miR-577. Cancer Cell Int. 20, 202. doi:10.1186/s12935-020-01287-8
Sun, S., Gao, J., Zhou, S., Li, Y., Wang, Y., Jin, L., et al. (2020b). A novel circular RNA circ-LRIG3 facilitates the malignant progression of hepatocellular carcinoma by modulating the EZH2/STAT3 signaling. J. Exp. Clin. Cancer Res. 39 (1), 252. doi:10.1186/s13046-020-01779-5
Sun, S., Wang, W., Luo, X., Li, Y., Liu, B., Li, X., et al. (2019). Circular RNA circ-ADD3 inhibits hepatocellular carcinoma metastasis through facilitating EZH2 degradation via CDK1-mediated ubiquitination. Am. J. Cancer Res. 9 (8), 1695–1707.
Sunagawa, Y., Yamada, S., Sonohara, F., Kurimoto, K., Tanaka, N., Suzuki, Y., et al. (2021). Genome-wide identification and characterization of circular RNA in resected hepatocellular carcinoma and background liver tissue. Sci. Rep. 11 (1), 6016. doi:10.1038/s41598-021-85237-y
Sunny, N. E., Bril, F., and Cusi, K. (2017). Mitochondrial adaptation in nonalcoholic fatty liver disease: Novel mechanisms and treatment strategies. Trends Endocrinol. Metab. 28 (4), 250–260. doi:10.1016/j.tem.2016.11.006
Susila, A., Chan, H., Loh, A., Phang, H., Wong, E., Tergaonkar, V., et al. (2010). The POPX2 phosphatase regulates cancer cell motility and invasiveness. Cell Cycle 9 (1), 179–187. doi:10.4161/cc.9.1.10406
Tahiliani, M., Koh, K., Shen, Y., Pastor, W., Bandukwala, H., Brudno, Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324 (5929), 930–935. doi:10.1126/science.1170116
Tang, D., Tang, Q., Huang, W., Zhang, Y., Tian, Y., and Fu, X. (2023). Fasting: From physiology to pathology. Adv. Sci. (Weinh) 10 (9), e2204487. doi:10.1002/advs.202204487
Tang, Z., Li, X., Zhao, J., Qian, F., Feng, C., Li, Y., et al. (2019). TRCirc: A resource for transcriptional regulation information of circRNAs. Brief. Bioinform 20 (6), 2327–2333. doi:10.1093/bib/bby083
Tholken, C., Thamm, M., Erbacher, C., and Lechner, M. (2019). Sequence and structural properties of circular RNAs in the brain of nurse and forager honeybees (Apis mellifera). BMC Genomics 20 (1), 88. doi:10.1186/s12864-018-5402-6
Tian, F., Yu, C., Wu, M., Wu, X., Wan, L., and Zhu, X. (2019). MicroRNA-191 promotes hepatocellular carcinoma cell proliferation by has_circ_0000204/miR-191/KLF6 axis. Cell Prolif. 52 (5), e12635. doi:10.1111/cpr.12635
Tian, Y., Xu, J., Du, X., and Fu, X. (2018). The interplay between noncoding RNAs and insulin in diabetes. Cancer Lett. 419, 53–63. doi:10.1016/j.canlet.2018.01.038
Tian, Y., Zhang, M., Fan, M., Xu, H., Wu, S., Zou, S., et al. (2022). A miRNA-mediated attenuation of hepatocarcinogenesis in both hepatocytes and Kupffer cells. Mol. Ther. Nucleic Acids 30, 1–12. doi:10.1016/j.omtn.2022.08.036
Tong, Q., Liu, G., Sang, X., Zhu, X., Fu, X., Dou, C., et al. (2023). Targeting RNA G-quadruplex with repurposed drugs blocks SARS-CoV-2 entry. PLoS Pathog. 19 (1), e1011131. doi:10.1371/journal.ppat.1011131
Trinchieri, G. (1989). Biology of natural killer cells. Adv. Immunol. 47, 187–376. doi:10.1016/s0065-2776(08)60664-1
Tripathi, V., Sixt, K., Gao, S., Xu, X., Huang, J., Weigert, R., et al. (2016). Direct regulation of alternative splicing by SMAD3 through PCBP1 is essential to the tumor-promoting role of TGF-β. Mol. Cell 64 (3), 549–564. doi:10.1016/j.molcel.2016.09.013
van Zonneveld, A., Kölling, M., Bijkerk, R., and Lorenzen, J. (2021). Circular RNAs in kidney disease and cancer. Nat. Rev. Nephrol. 17 (12), 814–826. doi:10.1038/s41581-021-00465-9
Varshney, D., Spiegel, J., Zyner, K., Tannahill, D., and Balasubramanian, S. (2020). The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 21 (8), 459–474. doi:10.1038/s41580-020-0236-x
Vasan, N., Baselga, J., and Hyman, D. (2019). A view on drug resistance in cancer. Nature 575 (7782), 299–309. doi:10.1038/s41586-019-1730-1
Vivier, E., Raulet, D., Moretta, A., Caligiuri, M., Zitvogel, L., Lanier, L., et al. (2011). Innate or adaptive immunity? The example of natural killer cells. Science 331 (6013), 44–49. doi:10.1126/science.1198687
Vo, J. N., Cieslik, M., Zhang, Y., Shukla, S., Xiao, L., Zhang, Y., et al. (2019). The landscape of circular RNA in cancer. Cell 176 (4), 869–881 e813. doi:10.1016/j.cell.2018.12.021
Vousden, K. H., and Lu, X. (2002). Live or let die: The cell's response to p53. Nat. Rev. Cancer 2 (8), 594–604. doi:10.1038/nrc864
Wang, G., Jiang, Y., Lu, C., Jiang, W., Wu, S., and Hua, Y. (2021a). CircFOXM1 promotes proliferation and metastasis of hepatocellular carcinoma via regulating miR-1179/SPAG5 axis. Sci. Rep. 11 (1), 23890. doi:10.1038/s41598-021-03285-w
Wang, G., Liu, W., Zou, Y., Wang, G., Deng, Y., Luo, J., et al. (2019a). Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine 40, 432–445. doi:10.1016/j.ebiom.2018.12.062
Wang, H., Chen, W., Jin, M., Hou, L., Chen, X., Zhang, R., et al. (2018). CircSLC3A2 functions as an oncogenic factor in hepatocellular carcinoma by sponging miR-490-3p and regulating PPM1F expression. Mol. Cancer 17 (1), 165. doi:10.1186/s12943-018-0909-7
Wang, J., Zhang, X., Greene, G., Xu, G., and Dong, X. (2022a). PABP/purine-rich motif as an initiation module for cap-independent translation in pattern-triggered immunity. Cell 185 (17), 3186–3200.e3117. doi:10.1016/j.cell.2022.06.037
Wang, L., Li, B., Yi, X., Xiao, X., Zheng, Q., and Ma, L. (2022b). Circ_0036412 affects the proliferation and cell cycle of hepatocellular carcinoma via hedgehog signaling pathway. J. Transl. Med. 20 (1), 154. doi:10.1186/s12967-022-03305-x
Wang, L., Long, H., Zheng, Q., Bo, X., Xiao, X., and Li, B. (2019b). Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol. Cancer 18 (1), 119. doi:10.1186/s12943-019-1046-7
Wang, S., Liu, D., Wei, H., Hua, Y., Shi, G., and Qiao, J. (2022c). The hsa_circRNA_102049 mediates the sorafenib sensitivity of hepatocellular carcinoma cells by regulating Reelin gene expression. Bioengineered 13 (2), 2272–2284. doi:10.1080/21655979.2021.2024332
Wang, S., Zhang, K., Tan, S., Xin, J., Yuan, Q., Xu, H., et al. (2021b). Circular RNAs in body fluids as cancer biomarkers: The new frontier of liquid biopsies. Mol. Cancer 20 (1), 13. doi:10.1186/s12943-020-01298-z
Wang, T., and Zhang, K. (2020). New blood biomarkers for the diagnosis of AFP-negative hepatocellular carcinoma. Front. Oncol. 10, 1316. doi:10.3389/fonc.2020.01316
Wang, W., Dong, R., Guo, Y., He, J., Shao, C., Yi, P., et al. (2019c). CircMTO1 inhibits liver fibrosis via regulation of miR-17-5p and Smad7. J. Cell Mol. Med. 23 (8), 5486–5496. doi:10.1111/jcmm.14432
Wang, W., Li, Y., Li, X., Liu, B., Han, S., Li, X., et al. (2020). Circular RNA circ-FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR-875-3p and miR-421. Biomed. Pharmacother. 121, 109517. doi:10.1016/j.biopha.2019.109517
Wang, X., Sheng, W., Xu, T., Xu, J., Gao, R., and Zhang, Z. (2021c). CircRNA hsa_circ_0110102 inhibited macrophage activation and hepatocellular carcinoma progression via miR-580-5p/PPARα/CCL2 pathway. Aging 13. doi:10.18632/aging.202900
Wang, Y., Gao, R., Li, J., Tang, S., Li, S., Tong, Q., et al. (2021d). Downregulation of hsa_circ_0074854 suppresses the migration and invasion in hepatocellular carcinoma via interacting with HuR and via suppressing exosomes-mediated macrophage M2 polarization. Int. J. Nanomedicine 16, 2803–2818. doi:10.2147/ijn.S284560
Wang, Y. G., Wang, T., Ding, M., Xiang, S. H., Shi, M., and Zhai, B. (2019d). hsa_circ_0091570 acts as a ceRNA to suppress hepatocellular cancer progression by sponging hsa-miR-1307. Cancer Lett. 460, 128–138. doi:10.1016/j.canlet.2019.06.007
Wang, Y., Zhang, Y., Su, X., Qiu, Q., Yuan, Y., Weng, C., et al. (2022d). Circular RNA circDVL1 inhibits clear cell renal cell carcinoma progression through the miR-412-3p/PCDH7 axis. Int. J. Biol. Sci. 18 (4), 1491–1507. doi:10.7150/ijbs.69351
Wang, Y., Zhang, Y., Wang, P., Fu, X., and Lin, W. (2020c). Circular RNAs in renal cell carcinoma: Implications for tumorigenesis, diagnosis, and therapy. Mol. cancer 19 (1), 149. doi:10.1186/s12943-020-01266-7
Wang, Z., Zhao, Y., Wang, Y., and Jin, C. (2019e). Circular RNA circHIAT1 inhibits cell growth in hepatocellular carcinoma by regulating miR-3171/PTEN axis. Biomed. Pharmacother. 116, 108932. doi:10.1016/j.biopha.2019.108932
Wang, Z. Y., Zhu, Z., Wang, H. F., Qin, B., Liu, J., Yao, X. H., et al. (2019f). Downregulation of circDYNC1H1 exhibits inhibitor effect on cell proliferation and migration in hepatocellular carcinoma through miR-140-5p. J. Cell Physiol. 234 (10), 17775–17785. doi:10.1002/jcp.28403
Wei, L., Li, K., Pang, X., Guo, B., Su, M., Huang, Y., et al. (2016). Leptin promotes epithelial-mesenchymal transition of breast cancer via the upregulation of pyruvate kinase M2. J. Exp. Clin. Cancer Res. 35 (1), 166. doi:10.1186/s13046-016-0446-4
Wei, X., Zheng, W., Tian, P., He, Y., Liu, H., Peng, M., et al. (2020a). Oncogenic hsa_circ_0091581 promotes the malignancy of HCC cell through blocking miR-526b from degrading c-MYC mRNA. Cell Cycle 19 (7), 817–824. doi:10.1080/15384101.2020.1731945
Wei, Y., Chen, X., Liang, C., Ling, Y., Yang, X., Ye, X., et al. (2020b). A noncoding regulatory RNAs network driven by circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology 71 (1), 130–147. doi:10.1002/hep.30795
Wen, S. Y., Qadir, J., and Yang, B. B. (2022). Circular RNA translation: Novel protein isoforms and clinical significance. Trends Mol. Med. 28 (5), 405–420. doi:10.1016/j.molmed.2022.03.003
Weng, H., Zeng, L., Cao, L., Chen, T., Li, Y., Xu, Y., et al. (2021). circFOXM1 contributes to sorafenib resistance of hepatocellular carcinoma cells by regulating MECP2 via miR-1324. Mol. Ther. Nucleic Acids 23, 811–820. doi:10.1016/j.omtn.2020.12.019
Weng, Q., Chen, M., Li, M., Zheng, Y., Shao, G., Fan, W., et al. (2019). hsa_circ_0064428Global microarray profiling identified as a potential immune-associated prognosis biomarker for hepatocellular carcinoma. J. Med. Genet. 56 (1), 32–38. doi:10.1136/jmedgenet-2018-105440
Wesselhoeft, R., Kowalski, P., and Anderson, D. (2018). Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 9 (1), 2629. doi:10.1038/s41467-018-05096-6
Wu, A., Hu, Y., Xu, Y., Xu, J., Wang, X., Cai, A., et al. (2021). Methyltransferase-like 3-mediated m6A methylation of Hsa_circ_0058493 accelerates hepatocellular carcinoma progression by binding to YTH domain-containing protein 1. Front. Cell Dev. Biol. 9, 762588. doi:10.3389/fcell.2021.762588
Wu, C., Tian, S., Guo, Y., Yu, C., Lei, L., Zheng, D., et al. (2022). circACTG1 promotes hepatocellular carcinoma progression by regulating miR-940/RIF1 Axis and activating AKT/mTOR pathway. J. Immunol. Res. 2022, 8649386. doi:10.1155/2022/8649386
Wu, J., Qi, X., Liu, L., Hu, X., Liu, J., Yang, J., et al. (2019). Emerging epigenetic regulation of circular RNAs in human cancer. Mol. Ther. Nucleic Acids 16, 589–596. doi:10.1016/j.omtn.2019.04.011
Wu, M. Y., Tang, Y. P., Liu, J. J., Liang, R., and Luo, X. L. (2020a). Global transcriptomic study of circRNAs expression profile in sorafenib resistant hepatocellular carcinoma cells. J. Cancer 11 (10), 2993–3001. doi:10.7150/jca.39854
Wu, S. M., Liu, H., Huang, P. J., Chang, I. Y., Lee, C. C., Yang, C. Y., et al. (2018). circlncRNAnet: an integrated web-based resource for mapping functional networks of long or circular forms of noncoding RNAs. Gigascience 7 (1), 1–10. doi:10.1093/gigascience/gix118
Wu, W., Ji, P., and Zhao, F. (2020b). CircAtlas: An integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 21 (1), 101. doi:10.1186/s13059-020-02018-y
Xia, S., Feng, J., Chen, K., Ma, Y., Gong, J., Cai, F., et al. (2018). CSCD: A database for cancer-specific circular RNAs. Nucleic Acids Res. 46, D925–D929. doi:10.1093/nar/gkx863
Xia, S., Feng, J., Lei, L., Hu, J., Xia, L., Wang, J., et al. (2017). Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Briefings Bioinforma. 18 (6), 984–992. doi:10.1093/bib/bbw081
Xie, Y., Cao, Y., Guo, C., Guo, X., He, Y., Xu, Q., et al. (2022). Profile analysis and functional modeling identify circular RNAs in nonalcoholic fatty liver disease as regulators of hepatic lipid metabolism. Front. Genet. 13, 884037. doi:10.3389/fgene.2022.884037
Xing, W., Zhou, P., Zhang, H., Chen, L., Zhou, Y., Cui, X., et al. (2022). Circular RNA circ_GLIS2 suppresses hepatocellular carcinoma growth and metastasis. Liver Int. official J. Int. Assoc. Study Liver 42 (3), 682–695. doi:10.1111/liv.15097
Xiong, D. D., Feng, Z. B., Lai, Z. F., Qin, Y., Liu, L. M., Fu, H. X., et al. (2019). High throughput circRNA sequencing analysis reveals novel insights into the mechanism of nitidine chloride against hepatocellular carcinoma. Cell Death Dis. 10 (9), 658. doi:10.1038/s41419-019-1890-9
Xu, B., Yang, T., Wang, Z., Zhang, Y., Liu, S., and Shen, M. (2018). CircRNA CDR1as/miR-7 signals promote tumor growth of osteosarcoma with a potential therapeutic and diagnostic value. Cancer Manag. Res. 10, 4871–4880. doi:10.2147/cmar.S178213
Xu, H., Tian, Y., Tang, D., Zou, S., Liu, G., Song, J., et al. (2021). An endoplasmic reticulum stress-MicroRNA-26a feedback circuit in NAFLD. Hepatology. 73 (4), 1327–1345. doi:10.1002/hep.31428
Xu, J., Ji, L., Liang, Y., Wan, Z., Zheng, W., Song, X., et al. (2020a). CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct. Target Ther. 5 (1), 298. doi:10.1038/s41392-020-00375-5
Xu, J., Wan, Z., Tang, M., Lin, Z., Jiang, S., Ji, L., et al. (2020b). N(6)-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol. Cancer 19 (1), 163. doi:10.1186/s12943-020-01281-8
Xu, L., Feng, X., Hao, X., Wang, P., Zhang, Y., Zheng, X., et al. (2019). CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J. Exp. Clin. Cancer Res. CR 38 (1), 98. doi:10.1186/s13046-019-1041-2
Yan, L., and Chen, Y. G. (2020). Circular RNAs in immune response and viral infection. Trends Biochem. Sci. 45 (12), 1022–1034. doi:10.1016/j.tibs.2020.08.006
Yan, Y., Nie, Y., Peng, C., Xing, F., Ji, S., Liu, H., et al. (2022). Correction to: The circular RNA hsa_circ_0001394 promotes hepatocellular carcinoma progression by targeting the miR-527/UBE2A axis. Cell Death Dis. 8 (1), 145. doi:10.1038/s41420-022-00955-0
Yang, C., Dong, Z., Hong, H., Dai, B., Song, F., Geng, L., et al. (2020). circFN1 mediates sorafenib resistance of hepatocellular carcinoma cells by sponging miR-1205 and regulating E2F1 expression. Mol. Ther. Nucleic Acids 22, 421–433. doi:10.1016/j.omtn.2020.08.039
Yang, F., Hu, A., Li, D., Wang, J., Guo, Y., Liu, Y., et al. (2019a). Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol. Cancer 18 (1), 158. doi:10.1186/s12943-019-1094-z
Yang, G., Wang, X., Liu, B., Lu, Z., Xu, Z., Xiu, P., et al. (2019b). circ-BIRC6, a circular RNA, promotes hepatocellular carcinoma progression by targeting the miR-3918/Bcl2 axis. Cell Cycle 18 (9), 976–989. doi:10.1080/15384101.2019.1601477
Yang, J., and Heimbach, J. (2020). New advances in the diagnosis and management of hepatocellular carcinoma. BMJ. 371, m3544. doi:10.1136/bmj.m3544
Yang, J., Shao, P., Zhou, H., Chen, Y., and Qu, L. (2010). deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res. 38, D123–D130. doi:10.1093/nar/gkp943
Yang, L., Tan, W., Wei, Y., Xie, Z., Li, W., Ma, X., et al. (2022a). CircLIFR suppresses hepatocellular carcinoma progression by sponging miR-624-5p and inactivating the GSK-3β/β-catenin signaling pathway. Cell Death Dis. 13 (5), 464. doi:10.1038/s41419-022-04887-6
Yang, S., Yu, F., Ji, Y., Shen, Y., Lu, H., Gao, Y., et al. (2022b). Circular RNA ERBIN promotes proliferation of hepatocellular carcinoma via the miR-1263/CDK6 Axis. Front. Oncol. 12, 878513. doi:10.3389/fonc.2022.878513
Yang, T., Shen, P., Chen, Q., Wu, P., Yuan, H., Ge, W., et al. (2021a). FUS-induced circRHOBTB3 facilitates cell proliferation via miR-600/NACC1 mediated autophagy response in pancreatic ductal adenocarcinoma. J. Exp. Clin. Cancer Res. 40 (1), 261. doi:10.1186/s13046-021-02063-w
Yang, W., Liu, Y., Gao, R., Xiu, Z., and Sun, T. (2019c). Knockdown of cZNF292 suppressed hypoxic human hepatoma SMMC7721 cell proliferation, vasculogenic mimicry, and radioresistance. Cell Signal 60, 122–135. doi:10.1016/j.cellsig.2019.04.011
Yang, X., Song, H., Zi, Z., Kou, J., Chen, S., Dai, Y., et al. (2019d). Circ_0005075 promotes hepatocellular carcinoma progression by suppression of microRNA-335. J. Cell Physiol. 234 (12), 21937–21946. doi:10.1002/jcp.28757
Yang, X., Xiong, Q., Wu, Y., Li, S., and Ge, F. (2017a). Quantitative proteomics reveals the regulatory networks of circular RNA CDR1as in hepatocellular carcinoma cells. J. Proteome Res. 16 (10), 3891–3902. doi:10.1021/acs.jproteome.7b00519
Yang, X., Ye, T., Liu, H., Lv, P., Duan, C., Wu, X., et al. (2021b). Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer. Mol. Cancer 20 (1), 4. doi:10.1186/s12943-020-01300-8
Yang, Y., Fan, X., Mao, M., Song, X., Wu, P., Zhang, Y., et al. (2017b). Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 27 (5), 626–641. doi:10.1038/cr.2017.31
Yang, Y., Gao, X., Zhang, M., Yan, S., Sun, C., Xiao, F., et al. (2018). Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 110 (3). doi:10.1093/jnci/djx166
Yao, D., Zhang, L., Zheng, M., Sun, X., Lu, Y., and Liu, P. (2018). Circ2Disease: A manually curated database of experimentally validated circRNAs in human disease. Sci. Rep. 8 (1), 11018. doi:10.1038/s41598-018-29360-3
Yao, Z., Luo, J., Hu, K., Lin, J., Huang, H., Wang, Q., et al. (2017). ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol. Oncol. 11 (4), 422–437. doi:10.1002/1878-0261.12045
Ye, R., Lu, X., Liu, J., Duan, Q., Xiao, J., Duan, X., et al. (2022). CircSOD2 contributes to tumor progression, immune evasion and anti-PD-1 resistance in hepatocellular carcinoma by targeting miR-497-5p/ANXA11 Axis. Biochem. Genet. 61 (2), 597–614. doi:10.1007/s10528-022-10273-w
Yu, J., Ding, W., Wang, M., Guo, X., Xu, J., Xu, Q., et al. (2020a). Plasma circular RNA panel to diagnose Hepatitis B virus-related hepatocellular carcinoma: A large-scale, multicenter study. Int. J. Cancer 146 (6), 1754–1763. doi:10.1002/ijc.32647
Yu, J., Xu, Q. G., Wang, Z. G., Yang, Y., Zhang, L., Ma, J. Z., et al. (2018). Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol. 68 (6), 1214–1227. doi:10.1016/j.jhep.2018.01.012
Yu, J., Yang, M., Zhou, B., Luo, J., Zhang, Z., Zhang, W., et al. (2019). CircRNA-104718 acts as competing endogenous RNA and promotes hepatocellular carcinoma progression through microRNA-218-5p/TXNDC5 signaling pathway. Clin. Sci. (Lond) 133 (13), 1487–1503. doi:10.1042/cs20190394
Yu, M. C., Ding, G. Y., Ma, P., Chen, Y. D., Zhu, X. D., Cai, J. B., et al. (2021). CircRNA UBAP2 serves as a sponge of miR-1294 to increase tumorigenesis in hepatocellular carcinoma through regulating c-Myc expression. Carcinogenesis 42 (10), 1293–1303. doi:10.1093/carcin/bgab068
Yu, Y., Ge, T., and Zhang, P. (2020b). Circular RNA circGFRA1 promotes angiogenesis, cell proliferation and migration of hepatocellular carcinoma by combining with miR-149. Eur. Rev. Med. Pharmacol. Sci. 24 (21), 11058–11064. doi:10.26355/eurrev_202011_23591
Yuan, X., Diao, J., Du, A., Wen, S., Zhou, L., and Pan, Y. (2020). Circular RNA expression profiles and features in NAFLD mice: A study using RNA-seq data. J. Transl. Med. 18 (1), 476. doi:10.1186/s12967-020-02637-w
Zang, H., Li, Y., Zhang, X., and Huang, G. (2020). Circ_0000517 contributes to hepatocellular carcinoma progression by upregulating TXNDC5 via sponging miR-1296-5p. Cancer Manag. Res. 12, 3457–3468. doi:10.2147/cmar.S244024
Zeng, Y., Zheng, Z., Liu, F., and Yi, G. (2021a). Circular RNAs in metabolism and metabolic disorders. Obes. Rev. official J. Int. Assoc. Study Obes. 22 (7), e13220. doi:10.1111/obr.13220
Zeng, Z., Zhao, Y., Chen, Q., Zhu, S., Niu, Y., Ye, Z., et al. (2021b). Hypoxic exosomal HIF-1α-stabilizing circZNF91 promotes chemoresistance of normoxic pancreatic cancer cells via enhancing glycolysis. Oncogene 40 (36), 5505–5517. doi:10.1038/s41388-021-01960-w
Zhai, Z., Fu, Q., Liu, C., Zhang, X., Jia, P., Xia, P., et al. (2019). Emerging roles of hsa-circ-0046600 targeting the miR-640/HIF-1α signalling pathway in the progression of HCC. Onco Targets Ther. 12, 9291–9302. doi:10.2147/ott.S229514
Zhang, H., Deng, T., Ge, S., Liu, Y., Bai, M., Zhu, K., et al. (2019a). Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene 38 (15), 2844–2859. doi:10.1038/s41388-018-0619-z
Zhang, J., Chang, Y., Xu, L., and Qin, L. (2019b). Elevated expression of circular RNA circ_0008450 predicts dismal prognosis in hepatocellular carcinoma and regulates cell proliferation, apoptosis, and invasion via sponging miR-548p. J. Cell Biochem. 120 (6), 9487–9494. doi:10.1002/jcb.28224
Zhang, L., Liu, Y., Tao, H., Zhu, H., Pan, Y., Li, P., et al. (2021a). Circular RNA circUBE2J2 acts as the sponge of microRNA-370-5P to suppress hepatocellular carcinoma progression. Cell Death Dis. 12 (11), 985. doi:10.1038/s41419-021-04269-4
Zhang, L., Tao, H., Li, J., Zhang, E., Liang, H., and Zhang, B. (2021b). Comprehensive analysis of the competing endogenous circRNA-lncRNA-miRNA-mRNA network and identification of a novel potential biomarker for hepatocellular carcinoma. Aging 13 (12), 15990–16008. doi:10.18632/aging.203056
Zhang, L., and Wang, Z. (2020). Circular RNA hsa_circ_0004812 impairs IFN-induced immune response by sponging miR-1287-5p to regulate FSTL1 in chronic Hepatitis B. Virol. J. 17 (1), 40. doi:10.1186/s12985-020-01314-0
Zhang, L., Zhang, J., Li, P., Li, T., Zhou, Z., and Wu, H. (2022). Exosomal hsa_circ_0004658 derived from RBPJ overexpressed-macrophages inhibits hepatocellular carcinoma progression via miR-499b-5p/JAM3. Cell Death Dis. 13 (1), 32. doi:10.1038/s41419-021-04345-9
Zhang, M., Huang, N., Yang, X., Luo, J., Yan, S., Xiao, F., et al. (2018a). A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37 (13), 1805–1814. doi:10.1038/s41388-017-0019-9
Zhang, M., Zhao, K., Xu, X., Yang, Y., Yan, S., Wei, P., et al. (2018b). A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 9 (1), 4475. doi:10.1038/s41467-018-06862-2
Zhang, P. F., Gao, C., Huang, X. Y., Lu, J. C., Guo, X. J., Shi, G. M., et al. (2020b). Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer 19 (1), 110. doi:10.1186/s12943-020-01222-5
Zhang, P., Wei, C., Huang, X., Peng, R., Yang, X., Lu, J., et al. (2019c). Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol. Cancer 18 (1), 105. doi:10.1186/s12943-019-1031-1
Zhang, P., Zhang, X. O., Jiang, T., Cai, L., Huang, X., Liu, Q., et al. (2020a). Comprehensive identification of alternative back-splicing in human tissue transcriptomes. Nucleic Acids Res. 48 (4), 1779–1789. doi:10.1093/nar/gkaa005
Zhang, S., Lu, Y., Jiang, H. Y., Cheng, Z. M., Wei, Z. J., Wei, Y. H., et al. (2021c). CircC16orf62 promotes hepatocellular carcinoma progression through the miR-138-5p/PTK2/AKT axis. Cell Death Dis. 12 (6), 597. doi:10.1038/s41419-021-03866-7
Zhang, S., Weng, T., Cheruba, E., Guo, T., Chan, H., Sze, S., et al. (2017). Phosphatase POPX2 exhibits dual regulatory functions in cancer metastasis. J. Proteome Res. 16 (2), 698–711. doi:10.1021/acs.jproteome.6b00748
Zhang, T., Jing, B., Bai, Y., Zhang, Y., and Yu, H. (2020c). Circular RNA circTMEM45A acts as the sponge of MicroRNA-665 to promote hepatocellular carcinoma progression. Mol. Ther. Nucleic Acids 22, 285–297. doi:10.1016/j.omtn.2020.08.011
Zhang, W., Fu, X., Xie, J., Pan, H., Han, W., and Huang, W. (2021d). miR-26a attenuates colitis and colitis-associated cancer by targeting the multiple intestinal inflammatory pathways. Mol. Ther. Nucleic Acids 24, 264–273. doi:10.1016/j.omtn.2021.02.029
Zhang, X., Hu, S., Zhang, X., Wang, L., Zhang, X., Yan, B., et al. (2014a). MicroRNA-7 arrests cell cycle in G1 phase by directly targeting CCNE1 in human hepatocellular carcinoma cells. Biochem. Biophys. Res. Commun. 443 (3), 1078–1084. doi:10.1016/j.bbrc.2013.12.095
Zhang, X., Luo, P., Jing, W., Zhou, H., Liang, C., and Tu, J. (2018c). circSMAD2 inhibits the epithelial-mesenchymal transition by targeting miR-629 in hepatocellular carcinoma. Onco Targets Ther. 11, 2853–2863. doi:10.2147/ott.S158008
Zhang, X. O., Wang, H. B., Zhang, Y., Lu, X., Chen, L. L., and Yang, L. (2014b). Complementary sequence-mediated exon circularization. Cell 159 (1), 134–147. doi:10.1016/j.cell.2014.09.001
Zhang, X., Xu, Y., Qian, Z., Zheng, W., Wu, Q., Chen, Y., et al. (2018d). circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 9 (11), 1091. doi:10.1038/s41419-018-1132-6
Zhang, Y., Guo, X., Xiong, L., Yu, L., Li, Z., Guo, Q., et al. (2014c). Comprehensive analysis of microRNA-regulated protein interaction network reveals the tumor suppressive role of microRNA-149 in human hepatocellular carcinoma via targeting AKT-mTOR pathway. Mol. Cancer 13, 253. doi:10.1186/1476-4598-13-253
Zhang, Y., Xue, W., Li, X., Zhang, J., Chen, S., Zhang, J. L., et al. (2016). The biogenesis of nascent circular RNAs. Cell Rep. 15 (3), 611–624. doi:10.1016/j.celrep.2016.03.058
Zhang, Y., Zhang, X. O., Chen, T., Xiang, J. F., Yin, Q. F., Xing, Y. H., et al. (2013). Circular intronic long noncoding RNAs. Mol. Cell 51 (6), 792–806. doi:10.1016/j.molcel.2013.08.017
Zhao, L., Guo, Y., Guo, Y., Ji, X., Fan, D., Chen, C., et al. (2022). Effect and mechanism of circRNAs in tumor angiogenesis and clinical application. Int. J. Cancer 150 (8), 1223–1232. doi:10.1002/ijc.33863
Zhao, Q., Liu, J., Deng, H., Ma, R., Liao, J. Y., Liang, H., et al. (2020a). Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell 183 (1), 76–93. doi:10.1016/j.cell.2020.08.009
Zhao, X., Yang, Y., Sun, B., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24 (12), 1403–1419. doi:10.1038/cr.2014.151
Zhao, Z., Song, J., Tang, B., Fang, S., Zhang, D., Zheng, L., et al. (2020b). CircSOD2 induced epigenetic alteration drives hepatocellular carcinoma progression through activating JAK2/STAT3 signaling pathway. J. Exp. Clin. Cancer Res. 39 (1), 259. doi:10.1186/s13046-020-01769-7
Zheng, H., Chen, T., Li, C., Xu, C., Ding, C., Chen, J., et al. (2019). A circular RNA hsa_circ_0079929 inhibits tumor growth in hepatocellular carcinoma. Cancer Manag. Res. 11, 443–454. doi:10.2147/cmar.S189338
Zheng, Q., Bao, C., Guo, W., Li, S., Chen, J., Chen, B., et al. (2016). Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 7, 11215. doi:10.1038/ncomms11215
Zhong, L., Wang, Y., Cheng, Y., Wang, W., Lu, B., Zhu, L., et al. (2018). Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem. Biophys. Res. Commun. 499 (4), 1044–1049. doi:10.1016/j.bbrc.2018.03.221
Zhong, Q., Huang, J., Wei, J., and Wu, R. (2019). Circular RNA CDR1as sponges miR-7-5p to enhance E2F3 stability and promote the growth of nasopharyngeal carcinoma. Cancer Cell Int. 19, 252. doi:10.1186/s12935-019-0959-y
Zhou, F., Shang, W., Yu, X., and Tian, J. (2018a). Glypican-3: A promising biomarker for hepatocellular carcinoma diagnosis and treatment. Med. Res. Rev. 38 (2), 741–767. doi:10.1002/med.21455
Zhou, P., Zheng, G., Li, Y., Wu, D., and Chen, Y. (2020). Construction of a circRNA-miRNA-mRNA network related to macrophage infiltration in hepatocellular carcinoma. Front. Genet. 11, 1026. doi:10.3389/fgene.2020.01026
Zhou, T. C., Li, X., Chen, L. J., Fan, J. H., Lai, X., Tang, Y., et al. (2018b). Differential expression profile of hepatic circular RNAs in chronic Hepatitis B. J. Viral Hepat. 25 (11), 1341–1351. doi:10.1111/jvh.12944
Zhu, C., Su, Y., Liu, L., Wang, S., Liu, Y., and Wu, J. (2020a). Circular RNA hsa_circ_0004277 stimulates malignant phenotype of hepatocellular carcinoma and epithelial-mesenchymal transition of peripheral cells. Front. Cell Dev. Biol. 8, 585565. doi:10.3389/fcell.2020.585565
Zhu, K., Zhan, H., Peng, Y., Yang, L., Gao, Q., Jia, H., et al. (2020b). Plasma hsa_circ_0027089 is a diagnostic biomarker for Hepatitis B virus-related hepatocellular carcinoma. Carcinogenesis 41 (3), 296–302. doi:10.1093/carcin/bgz154
Zhu, L., Ren, T., Zhu, Z., Cheng, M., Mou, Q., Mu, M., et al. (2018). Thymosin-β4 mediates hepatic stellate cell activation by interfering with CircRNA-0067835/miR-155/FoxO3 signaling pathway. Cell Physiol. Biochem. 51 (3), 1389–1398. doi:10.1159/000495556
Zhu, M., Liang, Z., Pan, J., Zhang, X., Xue, R., Cao, G., et al. (2021). Hepatocellular carcinoma progression mediated by Hepatitis B virus-encoded circRNA HBV_circ_1 through interaction with CDK1. Mol. Ther. Nucleic Acids 25, 668–682. doi:10.1016/j.omtn.2021.08.011
Zhu, P., Liang, H., Huang, X., Zeng, Q., Liu, Y., Lv, J., et al. (2020c). Circular RNA Hsa_circ_0004018 inhibits wnt/β-catenin signaling pathway by targeting microRNA-626/DKK3 in hepatocellular carcinoma. Onco Targets Ther. 13, 9351–9364. doi:10.2147/ott.S254997
Zhu, Y., Liu, Y., Xiao, B., Cai, H., Liu, M., Ma, L., et al. (2019a). The circular RNA PVT1/miR-203/HOXD3 pathway promotes the progression of human hepatocellular carcinoma. Biol. Open 8 (9). doi:10.1242/bio.043687
Zhu, Y., Zheng, B., Luo, G., Ma, X., Lu, X., Lin, X., et al. (2019b). Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics 9 (12), 3526–3540. doi:10.7150/thno.32796
Zou, S., Chen, S., Rao, G., Zhang, G., Ma, M., Peng, B., et al. (2023a). Extrachromosomal circular MiR-17-92 amplicon promotes hepatocellular carcinoma. Hepatology. doi:10.1097/hep.0000000000000435
Zou, S., Kim, B., Tian, Y., Liu, G., Zhang, J., Zerda, R., et al. (2023b). Enhanced nuclear translation is associated with proliferation and progression across multiple cancers. MedComm 4 (3), e248. doi:10.1002/mco2.248
Zou, S., Tong, Q., Liu, B., Huang, W., Tian, Y., and Fu, X. (2020). Targeting STAT3 in cancer immunotherapy. Mol. Cancer 19 (1), 145. doi:10.1186/s12943-020-01258-7
Keywords: HCC, circRNA, miRNA sponge, tumor microenvironment, biomarker, drug resistance
Citation: Rao G, Peng X, Tian Y, Fu X and Zhang Y (2023) Circular RNAs in hepatocellular carcinoma: biogenesis, function, and pathology. Front. Genet. 14:1106665. doi: 10.3389/fgene.2023.1106665
Received: 25 November 2022; Accepted: 16 June 2023;
Published: 07 July 2023.
Edited by:
William C. Cho, QEH, Hong Kong SAR, ChinaReviewed by:
Jianwei Li, Hebei University of Technology, ChinaZeng-Hong Wu, Huazhong University of Science and Technology, China
Copyright © 2023 Rao, Peng, Tian, Fu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianghui Fu, xfu@scu.edu.cn; Yuwei Zhang, doczhangyw@scu.edu.cn
†These authors have contributed equally to this work and share first authorship
 Guocheng Rao1,2†
Guocheng Rao1,2† Xi Peng
Xi Peng Xianghui Fu
Xianghui Fu