- 1Jockey Club College of Veterinary Medicine and Life Sciences, City University of Hong Kong, Kowloon, Hong Kong SAR, China
- 2Department of Electrical Engineering, City University of Hong Kong, Hong Kong, Hong Kong SAR, China
- 3Department of Biomedical Sciences, City University of Hong Kong, Hong Kong, Hong Kong SAR, China
- 4Department of Surgery, The University of Hong Kong, Pokfulam, Hong Kong SAR, China
- 5Department of Computer Sciences, National University of Singapore, Singapore, Singapore
- 6Department of Epidemiology, Centre for Global Cardiometabolic Health, Brown University, Providence, RI, United States
Esophageal cancer (EC) remains a significant challenge globally, having the 8th highest incidence and 6th highest mortality worldwide. Esophageal squamous cell carcinoma (ESCC) is the most common form of EC in Asia. Crucially, more than 90% of EC cases in China are ESCC. The high mortality rate of EC is likely due to the limited number of effective therapeutic options. To increase patient survival, novel therapeutic strategies for EC patients must be devised. Unfortunately, the development of novel drugs also presents its own significant challenges as most novel drugs do not make it to market due to lack of efficacy or safety concerns. A more time and cost-effective strategy is to identify existing drugs, that have already been approved for treatment of other diseases, which can be repurposed to treat EC patients, with drug repositioning. This can be achieved by comparing the gene expression profiles of disease-states with the effect on gene-expression by a given drug. In our analysis, we used previously published microarray data and identified 167 differentially expressed genes (DEGs). Using weighted key driver analysis, 39 key driver genes were then identified. These driver genes were then used in Overlap Analysis and Network Analysis in Pharmomics. By extracting drugs common to both analyses, 24 drugs are predicted to demonstrate therapeutic effect in EC patients. Several of which have already been shown to demonstrate a therapeutic effect in EC, most notably Doxorubicin, which is commonly used to treat EC patients, and Ixazomib, which was recently shown to induce apoptosis and supress growth of EC cell lines. Additionally, our analysis predicts multiple psychiatric drugs, including Venlafaxine, as repositioned drugs. This is in line with recent research which suggests that psychiatric drugs should be investigated for use in gastrointestinal cancers such as EC. Our study shows that a drug repositioning approach is a feasible strategy for identifying novel ESCC therapies and can also improve the understanding of the mechanisms underlying the drug targets.
Introduction
There are two major subtypes of Esophageal cancer (EC), esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC) (Ye et al., 2021). In China, more than 90% of esophageal cancer cases are ESCC (Zhang X. et al., 2021). EC as a whole remains a significant challenge globally, having the 8th highest incidence and the 6th highest mortality worldwide killing over 500,000 people in 2020 (Sung et al., 2021). A major driver of the high mortality rate is likely due to the fact that there are very few effective therapeutic options for EC patients. In recent years, there has been a significant increase in survival for many cancers, largely due to the availability of targeted therapies. For EC, however, targeted therapies are yet to make a significant impact on patient survival. Consequently, patients are often relying on more traditional therapies such as chemotherapy and surgical resection. In-order-to increase patient survival, novel therapeutic strategies for EC patients must be devised. Unfortunately, the development of novel drugs also presents its own significant challenges as most novel drugs do not make it to market due to lack of efficacy or safety concerns. Therefore, it is more time and cost effective to identify existing drugs, that have already been approved for treatment of other diseases, which can be repurposed to treat EC patients. This can be achieved using a drugs gene signature, the alterations in gene expression as a result of exposure to the drug. The gene signature of a drug indicates the underlying biological pathways and mechanisms that are involved in the therapeutic effect of the drug. With this knowledge, we can then identify candidate drugs which have gene signatures capable of reversing aberrant gene expression patterns observed in disease-states to those observed in normal cells. This gene signature-based approach has been adopted by previous research to identify drugs that can be repositioned to treat a variety of diseases including, but not limited to, cancer, Alzheimer’s, hyperlipidaemia, hypertension, and inflammatory disease (Corbett et al., 2012; Hall et al., 2014; Guney et al., 2016; Subramanian et al., 2017; Cheng et al., 2018; Carvalho et al., 2021; Wu et al., 2022). To date, drug repositioning to target gene signatures has primarily involved identifying directly overlapping drug genes and disease genes (herein referred to as overlap analysis) (Subramanian et al., 2017; Wang et al., 2018; Chen et al., 2022). More recently, network analysis has been greatly employed in this area as it offers distinct advantages over more traditional statistical methods. This is due to the fact that the models that can be built with this methodology are an excellent way to capture a molecules relationship with other molecules. In particular, nodes can be used to represent multiple entities such as genes, molecules, proteins, etc, and the edges can also represent a vast array of information such as mode-of-actions (MoAs), underlying mechanisms, or functional similarities (Jarada et al., 2020) Hence, network-based methods can accurately represent the biological mechanisms which are driving diseases (Barabási et al., 2011). As a result, network-based drug repositioning can identify drugs which target the underlying biology of the disease. It is worth noting, however, that other methods of computational drug repositioning have also been adopted, such as Data Mining and Machine Learning. An excellent review of the different methodologies, as well as their advantages and disadvantages has recently been published (Jarada et al., 2020). Due to the success of drug repositioning overall, and the absence of effective treatments for ESCC, it has been proposed that this method be used to identify novel treatment strategies for ESCC. However, these studies have largely, though not completely, been limited to testing existing cancer drugs in vitro with drug screening methods (Xie et al., 2020; Li Y. et al., 2021). Herein, we adopt both a network-based and overlap-based drug repositioning methodology, to identify existing drugs that can specifically target the aberrant expression profile of ESCC and impede oncogenesis. To do this, we used previously published data for in-silico drug repositioning analysis utilising the PharmOmics webserver (Chen et al., 2022). The repositioning analysis consisted of two arms, the ‘overlap analysis’ arm and the ‘network analysis’ arm (Figure 1), which utilise two methods of drug repositioning.
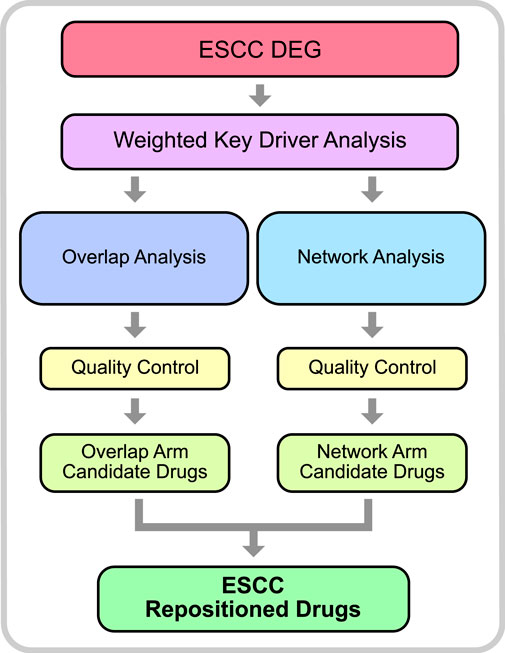
FIGURE 1. Drug Repositioning Analysis Methodology. The initial step of the analysis included differential gene expression on previously published array data from GEO (accession: GSE23400). DEGs were then used to identify key driver genes in a weighted key driver analysis. The key driver genes were then used as input in 2 arms; overlap analysis and network analysis. Qualify control was performed to filter out erroneous results and identify candidate drugs. Drugs which were common to both arms were considered robust and considered ESCC Repositioned Drugs.
Results
Identification of DEGs
The dataset GSE23400 was downloaded using the GEOquery R package function getGEO. In total, 167 DEGs were identified between ESC and normal samples (Details of the differential gene expression analysis can be found in the methods section). Of which, 65 were upregulated and 102 were downregulated. The top 5 most upregulated genes are MMP1, SPP1, POSTN, COL1A1, and JUP. The top 5 most downregulated genes are CRISP3, MAL, CRNN, SCEL, CLCA4. The top 25 up-regulated genes can be observed in Table 1, whereas the top 25 down-regulated genes can be observed in Table 2.
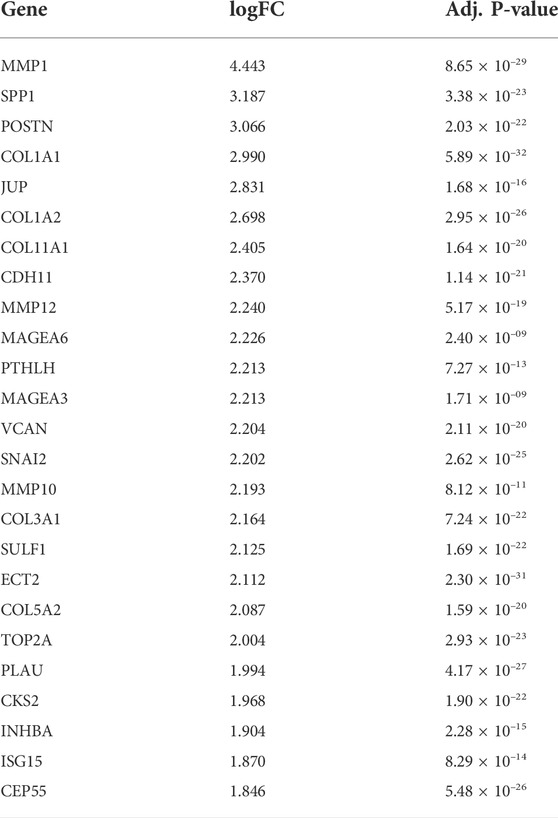
TABLE 1. Top 25 up-regulated genes in differential gene expression analysis comparing cancer tissue with adjacent tissue in ESCC patients.
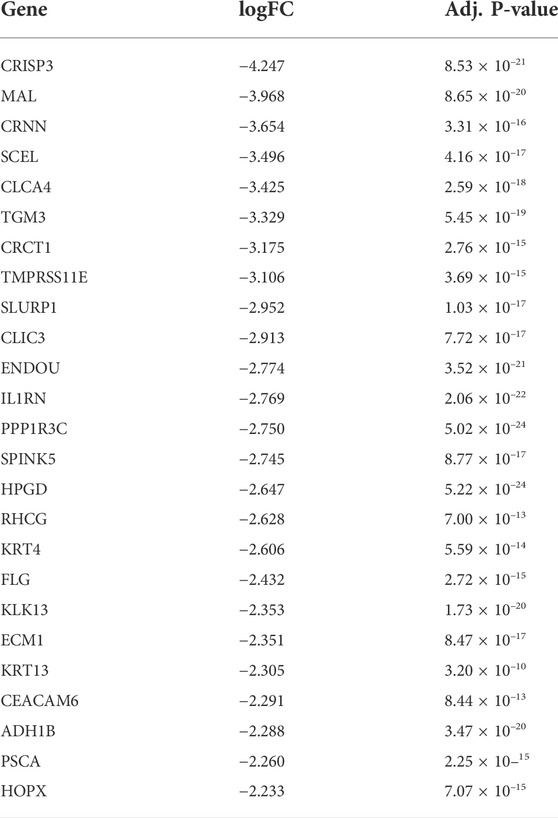
TABLE 2. Top 25 down-regulated genes in differential gene expression analysis comparing cancer tissue with adjacent tissue in ESCC patients.
Functional and pathway enrichment analyses
Functional and pathway enrichment analyses were performed used the ‘clusterProfiler’ R package. Gene Set Enrichment Analysis (GSEA) was performed with Gene Ontology (GO) (hereafter referred to as GSEA-GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway (hereafter referred to as GSEA-KEGG). GSEA-GO analysis was performed with the gene set categories Biological Process (BP), Cellular Component (CC), and Molecular Function (MF), which identified 253, 36, 25 enriched gene sets, respectively. Numerous BP gene sets identified by the analysis are related to extracellular matrix and cell differentiation. Ranking BP analysis by adjusted p-value, the top 5 most enriched gene sets are cellular component organization, cellular component organization or biogenesis, extracellular matrix organization, extracellular structure organization, multicellular organism development. According to adjusted p-value, the top 5 most enriched CC category are endoplasmic reticulum lumen, external encapsulating structure, extracellular matrix, fibrillar collagen trimer, banded collagen fibril. Furthermore, the top 5 categories in the MF analysis identified extracellular matrix structural constituent, extracellular matrix structural constituent conferring tensile strength, protein-containing complex binding, cell adhesion molecule binding, glycosaminoglycan binding. The full results for BP, CC, MF can be observed in Supplementary Tables S1–S3, respectively. Gene Set Enrichment Analysis of KEGG (GSEA-KEGG) identified 12 enriched gene sets (Supplementary Table S4), including those previously identified as ESCC-related, such as Focal adhesion, ECM-receptor interaction, PI3K-Akt signalling pathway.
Weighted key driver analysis
Weighted Key Driver Analysis (wKDA) was performed using Mergeomics webserver. In this analysis, genes which possess a local network neighbourhood that have a significant enrichment of genes that are ESCC-associated are considered key drivers (KDs) (Ding et al., 2021). The analysis identified 89 key driver genes which were then filtered to select those which possessed an FDR <0.05, ensuring that only the strongest KDs are used in subsequent analyses. This resulted in 39 key driver genes (Supplementary Table S5). The top 10 key driver genes are NCAPG, PLG, NUSAP1, COL17A1, ASPM, TOP2A, ITGB3, P4HB, TTK, and COL7A1.
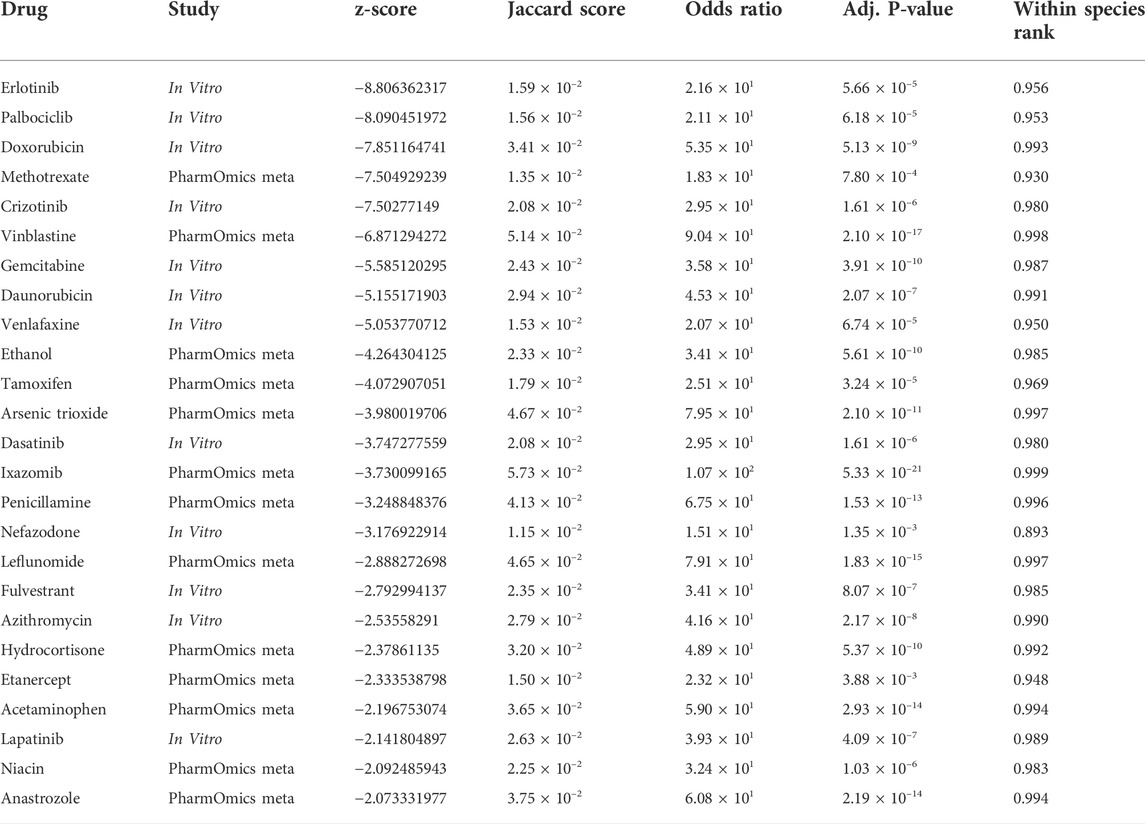
Repositioned drugs
Drug repositioning analysis was performed using both the Overlap Drug Repositioning and the Network Drug Repositioning modules from PharmOmics (Chen et al., 2022). The potential drugs from the analysis, were then filtered to identify robust ESCC repositioned drugs. The repositioning analysis identified 25 drugs that are strong candidates for ESCC treatment (Table 3). The top 10 repositioned drugs are Erlotinib, Palbociclib, Doxorubicin, Methotrexate, Crizotinib, Vinblastine, Gemcitabine, Daunorubicin, Venlafaxine, and Ethanol. We predicted that drugs which interact with EGFR (Erlotinib, Crizotinib, and Lapatinib), estrogen signalling (Tamoxifen, Fulvestrant, Hydrocortisone, and Anastrozole) and TRAIL-mediated apoptosis (Azithromycin and Anastrozole) pathways have potential for treating ESCC.
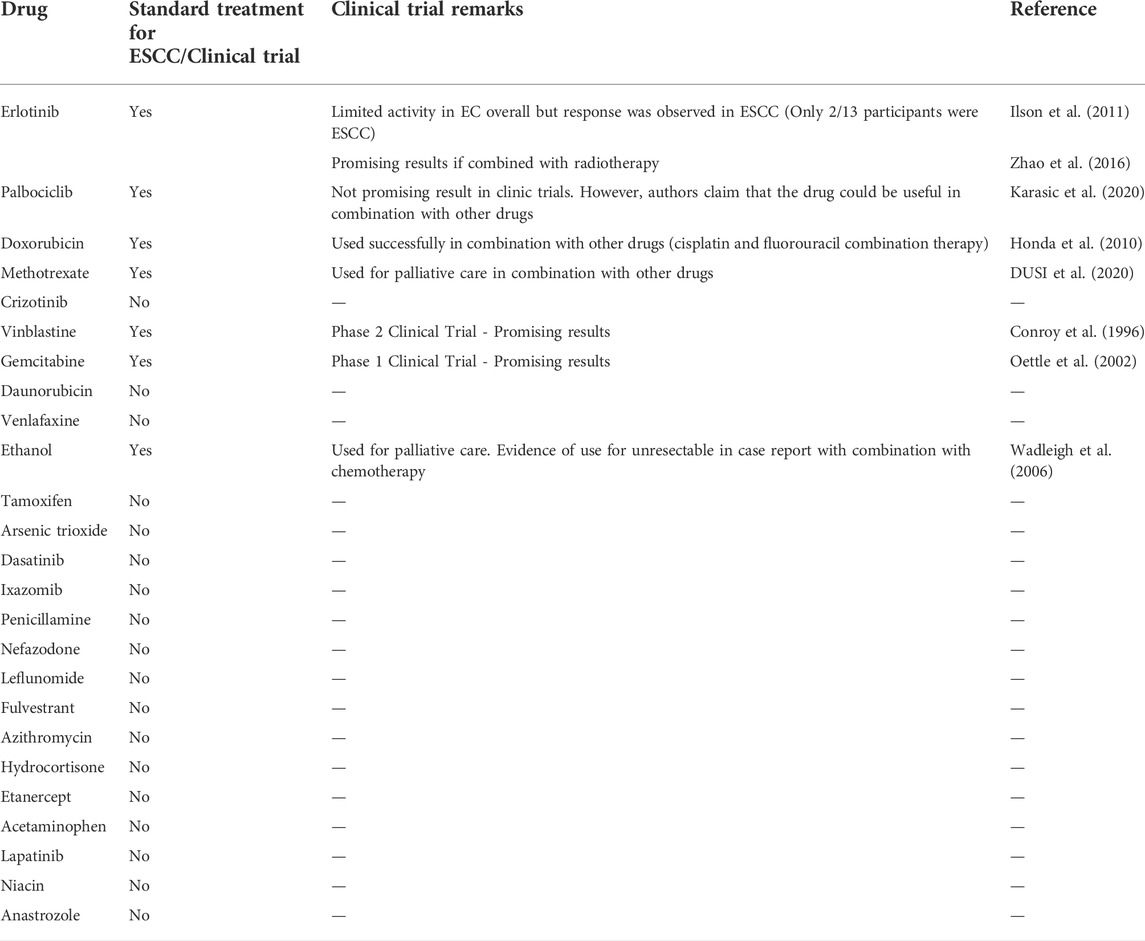
Drug validation
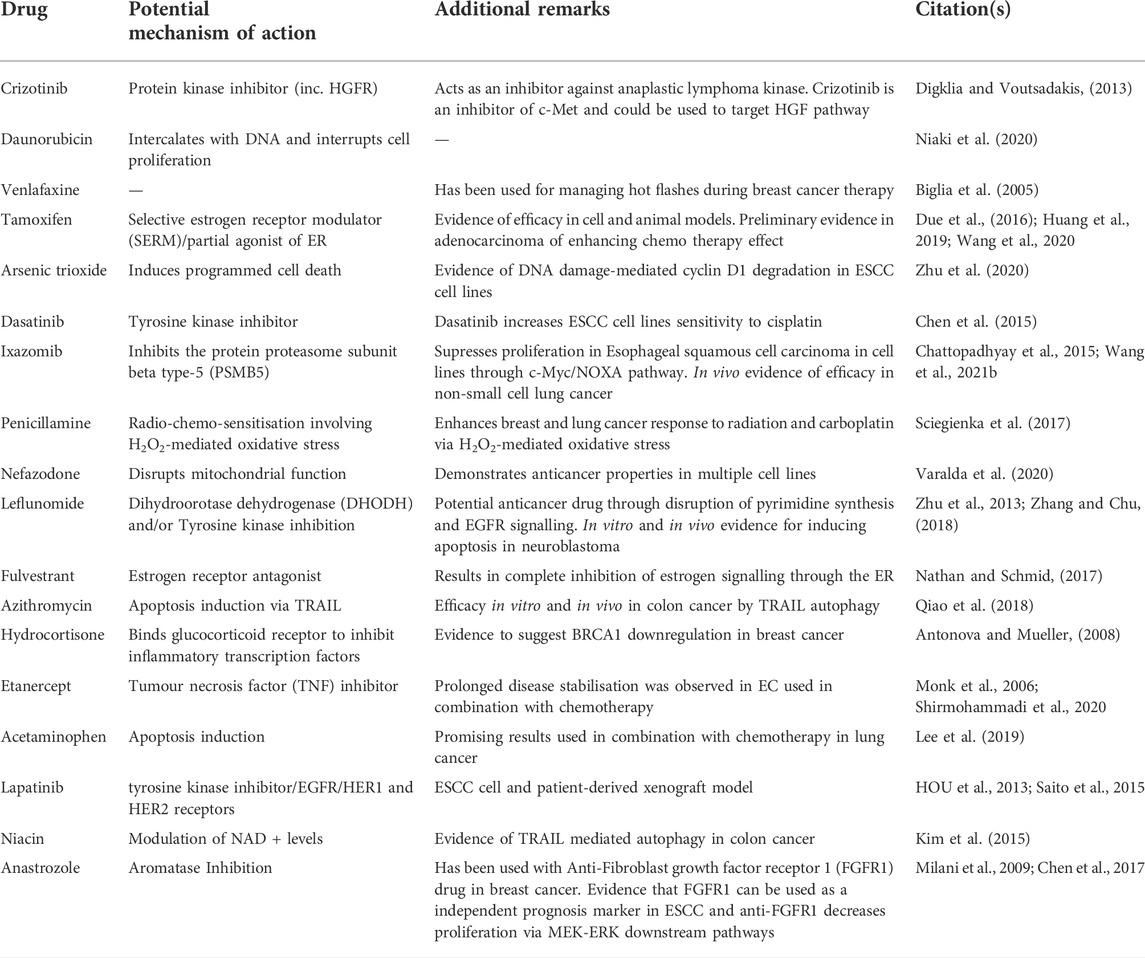
To validate our findings, we performed a literature search to determine whether any of the drugs identified by our analysis are currently used in ESCC treatment (Table 4). We found that 7 of the top 10 repositioned drugs, according to z-score, are already used to treat ESCC or have been shown to demonstrate efficacy in clinical trials. Candidate drugs were then validated using Binding DB (Gilson et al., 2016). Each drug was searched in the database to ascertain whether they bind to proteins known to be involved in ESCC. We found that 21 out of 25 repositioned drugs have a strong binding affinity to proteins that have been associated with ESCC in some manner previously (Table 5). Additionally, we performed a literature search to assess whether there is any biological evidence (in vitro or in vivo) that demonstrates efficacy or establishes a plausible mechanism by which the novel repositioned drugs could be beneficial for ESCC patients (Table 6). We found that all of our novel ESCC drugs, except for Venlafaxine, target pathways or proteins which have been demonstrated to drive oncogenesis in several cancers, including ESCC. Therefore, these drugs should be able to target the underlying biological processes driving oncogenesis in ESCC and inhibit proliferation and/or initiate apoptosis in ESCC.
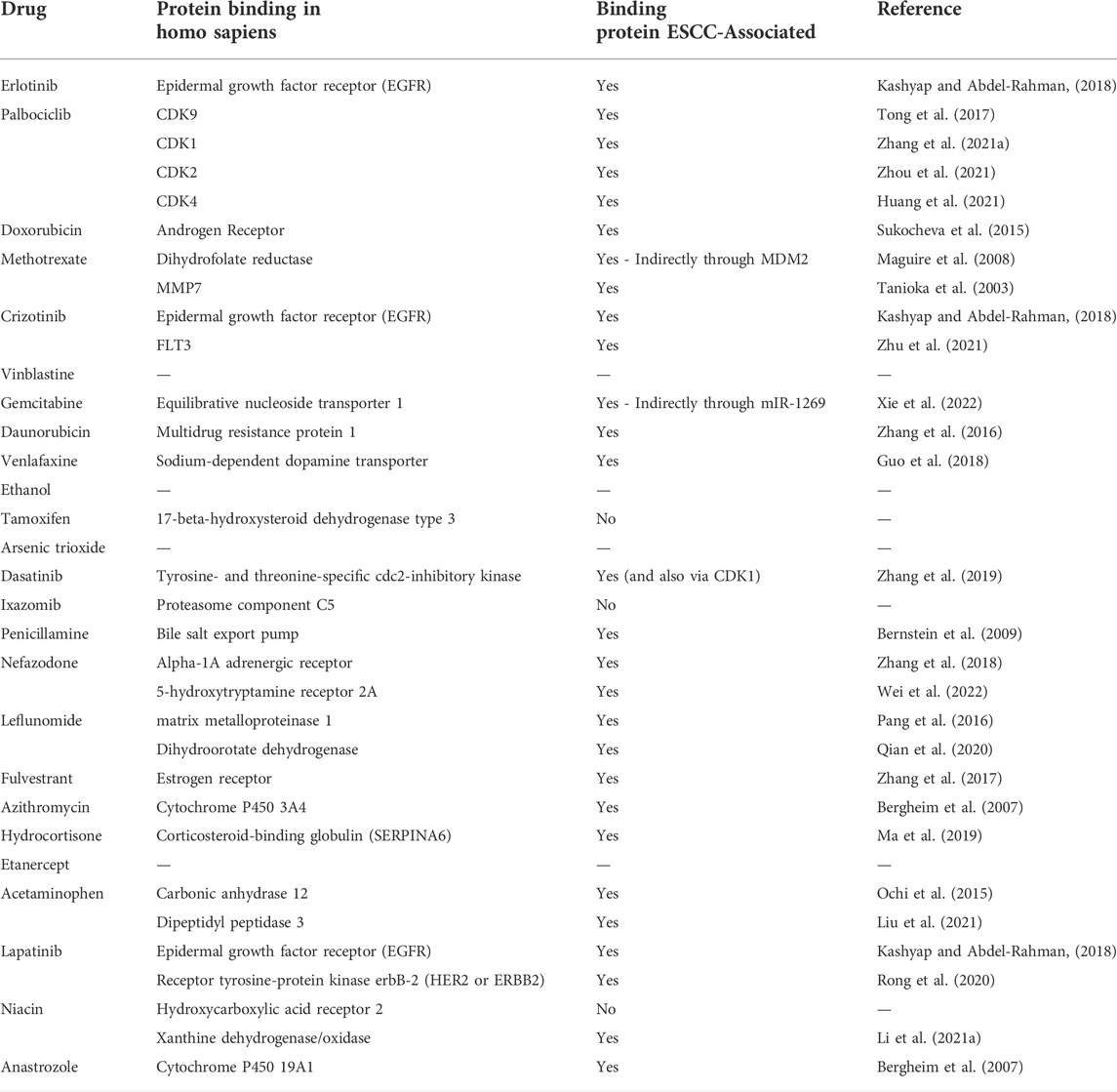
TABLE 5. Binding DB Target Validation. Repositioned drugs were investigated using Binding DB to determine whether the proteins that the drugs have strong affinity to have been previously shown to be associated with ESCC.
Discussion
ESCC is one of the most common malignancies and possess a significant mortality rate worldwide. This is largely due to late diagnosis and scarcity of efficacious treatment strategies upon being diagnosed (Feng et al., 2021). To address this, we performed a disease-based drug repositioning analysis with previously published ESCC gene expression data from paired patient samples. Differential gene expression analysis data identified 167 differentially expressed genes (DEGs) which were then used in wKDA and identified 39 key driver genes (KDGs). The genes with the highest absolute logFC identified by our differential gene expression analysis are MMP1, CRISP3, MAL, CRNN, SCEL. The most upregulated gene, MMP1, encodes a protein involved in the breakdown of the extracellular matrix (ECM) by cleaving collagens and other molecules. The most downregulated gene, CRISP3, encodes a protein located in the ECM and thought to be involved in cellular matrix remodelling (Ribeiro et al., 2011). The wKDA identified 39 significant driver genes for ESCC. Amongst the top 10 most significant KDGs, NUSAP1, COL17A1, ITGB3 and COL7A1 are involved in ECM maintenance. For example, the 4th most significant key driver gene, COL17A1, encodes a protein involved in cell-matrix adhesion (Jones et al., 2020). Taken together, these results suggest that alterations in ECM are an important driver of ESCC oncogenesis (Chen et al., 2016). Furthermore, KEGG analysis found both Focal adhesion and ECM-receptor interaction to be the 3rd and 4th most enriched term, respectively. This is in line with previous research that indicates higher levels of Serum human relaxin 2 (H2 RLN), a protein involved in ECM, collagen, and matrix metalloproteinase is associated with worse prognosis, including higher clinical stage and poorer survival (Ren et al., 2013; Napier et al., 2014).
Using 39 KDGs in an ESCC drug repositioning analysis, we identified 25 drugs that are predicted to have therapeutic effect in ESCC. Of which, 7 are either currently used in the clinic or have been used in clinical trials and 2 have shown efficacy in vitro or in vivo. Importantly, those which have been used in clinical trials have demonstrated efficacy particularly when used in combination with other drugs, such as chemotherapy. This is not surprising, however, as combination therapy has long been a standard practice in cancer therapy, including for ESCC where the current first-line treatment regimen is a combination of 5-fluorouracil and cisplatin (Hiramoto et al., 2018; Hirano and Kato, 2019). Each repositioned drug was validated in-silico using the drug binding database BindingDB, to identify which drug targets have previously been associated with ESCC (Table 5). Significantly, 21 of the 25 repositioned drugs have targets that have previously been associated with ESCC in some manner, which demonstrate the robustness of our findings. To further validate our findings, we performed a literature search on the novel repositioned drugs to examine whether there is an underlying biological mechanism which would justify the drugs appearance in the results (Table 6). We found that almost all of the repositioned drugs have been shown to demonstrate anti-cancer effects in multiple cancers, most notably breast cancers and non-small cell lung carcinoma (NSCLC). Interestingly, many of the repositioned drugs target specific pathways; EGFR (Erlotinib, Crizotinib, and Lapatinib), estrogen signalling (Tamoxifen, Fulvestrant, Hydrocortisone, and Anastrozole) and TRAIL-mediated apoptosis (Azithromycin and Anastrozole) pathways, suggesting that these pathways are key drivers of ESCC. This is in line with previous research which identified the EGFR AND ER pathways as drivers of ESCC oncogenesis and metastasis and have also been associated with patient outcome (Maron et al., 2020). Crucially, some of these drugs have been shown to have therapeutic potential in vitro. For example, Lapatinib, which acts through EGFR and HER2 has been shown to be efficacious in ESCC patient-derived xenografts (Rong et al., 2020). The potential mechanisms by which novel drugs identified by our study can be observed in Table 6. It is also worth noting that there are 2 anti-depressants present in our results, Venlafaxine and Nefazodone. These results are particularly interesting as it has recently been shown that psychiatric drugs offer potential as anti-cancer therapeutics (Loehr et al., 2021). Moreover, a recent review has specifically addressed the need to investigate psychiatric drugs for treatment of gastrointestinal cancers (Avendaño-Félix et al., 2020). We hypothesise that Crizotinib, Lapatinib, and Dasatinib are amongst the drugs with the most potential. Particularly Crizotinib and Lapatinib are of note as they target the EGFR pathway which is already targeted in ESCC treatment with Erlotinib. Dasatinib also has high potential due to targeting Tyrosine- and threonine-specific cdc2-inhibitory kinase and CDK1, proteins known to be involved in ESCC, and also due to displaying efficacy in cell lines (Chen et al., 2015; Zhang et al., 2019).
There are several limitations to our study, however, most notably that due to limited data availability, the sample size of patient samples was relatively small. We were unable to stratify patients according to subtype of ESCC. This means that the analysis is focussed on ESCC as whole and does not take into consideration specific subtypes. Moreover, as multiple drugs identified in our study are more efficacious when in combination with another drug, it would be beneficial to know what other drugs should be used in combination with the novel therapeutics identified. However, this analysis does not predict drug combinations that would be effective in treating ESCC.
On the other hand, our study has several strengths. To our knowledge, this is the first study to adopt a primarily computational approach to perform drug repositioning analysis in ESCC. Particularly, there are studies that have a computational component, but they do not use patient samples to identify drugs based on network analysis of differentially expressed genes (Li et al., 2022). Moreover, this is also the first to adopt Pharmomics unique network analysis to perform the analysis on ESCC. Furthermore, as Pharmomics contains >18000 species/tissue-specific gene signatures for 941 drugs and chemicals, it provides a larger scope of potential drugs compared to other studies in ESCC. Another strength of the study is that we used a two armed approach to ensure robust findings as each repositioning methodology has its own strengths. The overlap-based repositioning allows us to identify drugs which target the KDGs whereas the network-based repositioning allows for insights into the molecular and mechanistic therapeutic effects of the drugs. As this specific form of network-based repositioning is unique to PharmOmics, our study can provide valuable insights into the underlying molecular mechanisms driving ESCC. Another strength of our study is the consistency of our results with previously published literature. DEGs which displayed the highest absolute logFC were consistent with previously published literature including MMP1, SPP1, COL1A2, and COL1A1 amongst the top upregulated genes and CRISPR3, MAL, TMPRSS11E, and CRNN amongst the top downregulated genes (Feng et al., 2021; Song et al., 2021). Indeed, the gene with the highest absolute logFC, MMP1, is already known to be associated with ESCC oncogenesis (Chen et al., 2016). Additionally, higher MMP1 is associated with poorer prognosis (Feng et al., 2021). Moreover, the most significant key driver genes (KDGs) identified by our wKDA are consistent with previously published studies (Li et al., 2020; Yu-jing et al., 2020; Wang M. et al., 2021). Significantly, multiple drugs identified by our analysis target key pathways known to be involved in ESCC oncogenesis and metastasis.
Conclusion
Herein we utilised in silico disease-based drug repositioning to identify novel therapeutics for esophageal squamous cell carcinoma. Amongst 25 potential repositioned drugs identified in our study, 9 are currently used in the clinic or have shown promising results in clinical trials in combination with other treatments. Crucially, we identified 16 novel therapeutic strategies which possess a strong biological rationale for use in ESCC patients. Our study shows that drug repositioning approach is a feasible strategy in ESCC therapies and can improve the understanding of the mechanisms of the drug targets.
Materials and methodology
Data acquisition and identification of DEGs
Dataset was acquired from the Gene Expression Omnibus under accession code GSE23400 using the GEOquery R package function getGEO. The dataset consists of 53 paired patient samples from Esophageal squamous cell carcinoma (ESCC) patients. Additionally, 14,335 genes were in the dataset. Differentially expressed genes (DEGs) between paired tumour and non-tumour samples were identified using the limma package. The log-fold change (logFC) was calculated for DEGs. Genes with absolute logFC >1.5 and adjusted p-value < 0.01 were considered significant and used in subsequent analyses.
Functional and pathway enrichment analyses
The DEGs identified above were analysed using the clusterProfiler R package in order to identify biological annotations from the Gene ontology (GO) functional enrichment and Kyoto Encyclopaedia of Genes and Genomes (KEGG). The GO analysis was performed for biological process (BP), cellular component (CC) and molecular function (MF). An adjusted p-value < 0.05 was considered as statically significant for all analyses.
Weighted key driver analysis
Weighted key driver analysis (wKDA) was performed on DEGs using Mergeomics webserver to identify key driver genes (KDGs). wKDA has higher accuracy than standard key driver analysis as it considers edge weight information. The network used in the analysis was STRING PPI Network and default parameters were used (Search depth of 1, Undirected Edges, Min Hub Overlap of 0.33, and edge factor of 0.0). Genes which had an FDR <0.05 were considered as significant KDGs and used in subsequent analyses.
Drug repositioning analysis
By the Pharmomics webserver, Drug Repositioning Analyses was performed using genes obtained from the wKDA analysis. The analysis consisted of two arms: the Overlap Drug Repositioning and ADR Analysis (Overlap-DR) arm and the Meta-Signature Network Drug Repositioning and ADR Analysis (Meta-Net-DR) arm. The network analysis adopted by Pharmomics uses a network proximity measure between drug DEGs and disease-related genes that has been adopted previously for protein-network-based analysis. Specifically, tissue-specific Bayesian gene regulatory networks (BNs) are used and then the mean shortest distance between drug DEGs and disease genes are tested. Hence, it combines species and tissue specific in vivo drug signatures with gene networks to identify connections between disease genes and known drug targets. On the other hand, overlap analysis adopted by Pharmomics is largely similar to that which has been adopted previously, and assesses direct overlap between input genes and drug gene signatures. To do so, the Jaccard score, gene overlap fold enrichment, and Fisher’s exact test p values as measures of direct gene overlap are calculated. This analysis is based upon the premise that if disease and drug signatures target similar pathways then they would more than likely have gene overlaps and/or connect extensively in a gene network. Meta-Signature Network Drug Repositioning and ADR Analysis was performed using the multi-tissue network. In Overlap-DR, Jaccard score was used to measure the similarity between the 39 KDG’s gene networks and the drug target gene networks. In Meta-Net-DR, the connectivity of the gene network between drug signatures from PharmOmics and the KDs is used. The z-score of each drug is calculated which represents the distance between the KD network and the PharmOmics drug network. The smaller the z-score, the closer the distance between the networks. The output from these analyses were considered as possible repositioned drugs and were then filtered in Drug Candidate Selection to identify ESCC repositioned drugs.
Drug candidate selection
Repositioned drugs from both Pharmomics analyses were used as candidates to identify potential drugs for ESCC. Candidate drugs from Overlap-DR results were filtered to keep drugs with an adjusted P-value < 0.05, species equal to Homo sapiens, and a within species rank >0. The mean Jaccard score was then calculated and drugs with a Jaccard score less than the mean were removed. Subsequently, the drugs were sorted according to ‘Drug Name’, ‘Within Species Rank’, ‘Jaccard Score’, and ‘P-value’ and duplicate drugs were removed, keeping only the highest-ranking occurrence of each drug. Candidate drugs from Meta-Net-DR were filtered to keep drugs with adjusted p-value < 0.05. Candidate drugs were then sorted according to ‘Drug Name’ and ‘Rank’ and then duplicate drugs were removed, keeping only the highest-ranking occurrence of each drug. The filtered results from Overlap-DR and Meta-Net-DR were then compared to extract candidate drugs common to both arms of analysis. Drugs common to both arms were considered ESCC Repositioned Drugs. ‘Study’, ‘Jaccard Score’, ‘Odds Ratio’, ‘Adj. P-Value’, ‘Within Species Rank’ data from the Overlap-DR analysis and ‘z-score’ from Meta-Net-DR was used to construct the final ESCC Repositioned Drugs table. ESCC Repositioned Drugs were then sorted according to z-score (Table 3).
Drug candidate validation
In order to validate the repositioned drugs that were identified by the analysis, we performed a literature search to ascertain whether the drugs have previously been used in ESCC treatment (Table 4). Each drug was then investigated using the drug binding database Binding DB. For each ESCC Repositioned Drug, we identified which proteins they display a high binding affinity to. We then performed a literature search on these proteins, using Google Scholar and PubMed, to ascertain whether or not they have previously been shown to be ESCC-related in vitro or in vivo (Table 5). Finally, we performed a literature search on novel drugs identified by our analysis to elucidate the underlying biological processes and causal mechanisms which would explain why it is predicted to have therapeutic utility (Table 6).
Data availability Statement
Publicly available datasets were analyzed in this study. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving human participants were reviewed and approved by Human Subjects Ethics Sub-Committee at City University of Hong Kong (Reference number 2-11-201810_02). The patients/participants provided their written informed consent to participate in this study.
Author contributions
NL, W-KS, and KC envisioned and directed the project, assisted in writing manuscript, and performed data interpretation. AB and RH developed the analytical pipeline and interpreted data. AB performed the analysis and wrote the manuscript. QH assisted in analytical pipeline development and contributed towards the manuscript. KC coordinated the development of the analysis and revised the manuscript.
Funding
This project was funded by The Jockey Club College of Veterinary Medicine and Life Sciences (JCC) Interdisciplinary PhD Programme at City University of Hong Kong in Collaboration with Cornell University. This work was supported by the City University of Hong Kong New Research Initiatives/Infrastructure Support from Central (APRC; grant number 9610401).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.991842/full#supplementary-material
Supplementary Figure S1 | This figure shows the enriched gene set categories for the Gene Set Enrichment Analysis for Biological Processes.
Supplementary Figure S2 | This figure shows the enriched gene set categories for the Gene Set Enrichment Analysis for Cellular Component.
Supplementary Figure S3 | This figure shows the enriched gene set categories for the Gene Set Enrichment Analysis for Molecular Function.
References
Antonova, L., and Mueller, C. R. (2008). Hydrocortisone down‐regulates the tumor suppressor gene BRCA1 in mammary cells: A possible molecular link between stress and breast cancer. Genes Chromosom. Cancer 47, 341–352. doi:10.1002/gcc.20538
Avendaño-Félix, M., Aguilar-Medina, M., Bermudez, M., Lizárraga-Verdugo, E., López-Camarillo, C., and Ramos-Payán, R. (2020). Refocusing the use of psychiatric drugs for treatment of gastrointestinal cancers. Front. Oncol. 10, 1452. doi:10.3389/fonc.2020.01452
Barabási, A.-L., Gulbahce, N., and Loscalzo, J. (2011). Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 12, 56–68. doi:10.1038/nrg2918
Bergheim, I., Wolfgarten, E., Bollschweiler, E., Hölscher, A.-H., Bode, C., and Parlesak, A. (2007). Cytochrome P450 levels are altered in patients with esophageal squamous-cell carcinoma. World J. Gastroenterol. 13, 997–1002. doi:10.3748/wjg.v13.i7.997
Bernstein, H., Bernstein, C., Payne, C. M., and Dvorak, K. (2009). Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J. Gastroenterol. 15, 3329–3340. doi:10.3748/wjg.15.3329
Biglia, N., Torta, R., Roagna, R., Maggiorotto, F., Cacciari, F., Ponzone, R., et al. (2005). Evaluation of low-dose venlafaxine hydrochloride for the therapy of hot flushes in breast cancer survivors. Maturitas 52, 78–85. doi:10.1016/j.maturitas.2005.01.001
Carvalho, R. F., Canto, L. M. do, Cury, S. S., Hansen, T. F., Jensen, L. H., and Rogatto, S. R. (2021). Drug repositioning based on the reversal of gene expression signatures identifies TOP2A as a therapeutic target for rectal cancer. Cancers 13, 5492. doi:10.3390/cancers13215492
Chattopadhyay, N., Berger, A. J., Koenig, E., Bannerman, B., Garnsey, J., Bernard, H., et al. (2015). KRAS genotype correlates with proteasome inhibitor Ixazomib activity in preclinical in vivo models of colon and non-small cell lung cancer: Potential role of tumor metabolism. Plos One 10, e0144825. doi:10.1371/journal.pone.0144825
Chen, B., Liu, S., Gan, L., Wang, J., Hu, B., Xu, H., et al. (2017). FGFR1 signaling potentiates tumor growth and predicts poor prognosis in esophageal squamous cell carcinoma patients. Cancer Biol. Ther. 19, 76–86. doi:10.1080/15384047.2017.1394541
Chen, J., Lan, T., Zhang, W., Dong, L., Kang, N., Fu, M., et al. (2015). Dasatinib enhances cisplatin sensitivity in human esophageal squamous cell carcinoma (ESCC) cells via suppression of PI3K/AKT and Stat3 pathways. Arch. Biochem. Biophys. 575, 38–45. doi:10.1016/j.abb.2014.11.008
Chen, Y.-K., Tung, C.-W., Lee, J.-Y., Hung, Y.-C., Lee, C.-H., Chou, S.-H., et al. (2016). Plasma matrix metalloproteinase 1 improves the detection and survival prediction of esophageal squamous cell carcinoma. Sci. Rep. 6, 30057. doi:10.1038/srep30057
Chen, Y.-W., Diamante, G., Ding, J., Nghiem, T. X., Yang, J., Ha, S.-M., et al. (2022). PharmOmics: A species- and tissue-specific drug signature database and gene-network-based drug repositioning tool. Iscience 25, 104052. doi:10.1016/j.isci.2022.104052
Cheng, F., Desai, R. J., Handy, D. E., Wang, R., Schneeweiss, S., Barabási, A.-L., et al. (2018). Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun. 9, 2691. doi:10.1038/s41467-018-05116-5
Conroy, T., Etienne, P. L., Adenis, A., Wagener, D. J., Paillot, B., François, E., et al. (1996). Phase II trial of vinorelbine in metastatic squamous cell esophageal carcinoma. European organization for research and treatment of cancer gastrointestinal treat cancer cooperative group. J. Clin. Oncol. 14, 164–170. doi:10.1200/jco.1996.14.1.164
Corbett, A., Pickett, J., Burns, A., Corcoran, J., Dunnett, S. B., Edison, P., et al. (2012). Drug repositioning for Alzheimer’s disease. Nat. Rev. Drug Discov. 11, 833–846. doi:10.1038/nrd3869
Digklia, A., and Voutsadakis, I. A. (2013). Targeted treatments for metastatic esophageal squamous cell cancer. World J. Gastrointest. Oncol. 5, 88–96. doi:10.4251/wjgo.v5.i5.88
Ding, J., Blencowe, M., Nghiem, T., Ha, S., Chen, Y.-W., Li, G., et al. (2021). Mergeomics 2.0: A web server for multi-omics data integration to elucidate disease networks and predict therapeutics. Nucleic Acids Res. 49, W375–W387. doi:10.1093/nar/gkab405
Due, S. L., Watson, D. I., Bastian, I., Ding, G. Q., Sukocheva, O. A., Astill, D. St. J., et al. (2016). Tamoxifen enhances the cytotoxicity of conventional chemotherapy in esophageal adenocarcinoma cells. Surg. Oncol. 25, 269–277. doi:10.1016/j.suronc.2016.05.029
Dusi, V. S., C, O. R., Attili, S. V., and Palanki, S. D. (2020). Gefitinb along with methotrexate as palliative therapy in PS 3 and above in metastatic esophagus squamous cell carcinoma with focus on Q-TWIST. J. Clin. Oncol. 38, 355. doi:10.1200/jco.2020.38.4_suppl.355
Feng, Z., Qu, J., Liu, X., Liang, J., Li, Y., Jiang, J., et al. (2021). Integrated bioinformatics analysis of differentially expressed genes and immune cell infiltration characteristics in Esophageal Squamous cell carcinoma. Sci. Rep. 11, 16696. doi:10.1038/s41598-021-96274-y
Gilson, M. K., Liu, T., Baitaluk, M., Nicola, G., Hwang, L., and Chong, J. (2016). BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 44, D1045–D1053. doi:10.1093/nar/gkv1072
Guney, E., Menche, J., Vidal, M., and Barábasi, A.-L. (2016). Network-based in silico drug efficacy screening. Nat. Commun. 7, 10331. doi:10.1038/ncomms10331
Guo, J., Huang, J., Zhou, Y., Zhou, Y., Yu, L., Li, H., et al. (2018). Germline and somatic variations influence the somatic mutational signatures of esophageal squamous cell carcinomas in a Chinese population. Bmc Genomics 19, 538. doi:10.1186/s12864-018-4906-4
Hall, C. J., Wicker, S. M., Chien, A.-T., Tromp, A., Lawrence, L. M., Sun, X., et al. (2014). Repositioning drugs for inflammatory disease – fishing for new anti-inflammatory agents. Dis. Model. Mech. 7, 1069–1081. doi:10.1242/dmm.016873
Hiramoto, S., Kato, K., Shoji, H., Okita, N., Takashima, A., Honma, Y., et al. (2018). A retrospective analysis of 5-fluorouracil plus cisplatin as first-line chemotherapy in the recent treatment strategy for patients with metastatic or recurrent esophageal squamous cell carcinoma. Int. J. Clin. Oncol. 23, 466–472. doi:10.1007/s10147-018-1239-x
Hirano, H., and Kato, K. (2019). Systemic treatment of advanced esophageal squamous cell carcinoma: Chemotherapy, molecular-targeting therapy and immunotherapy. Jpn. J. Clin. Oncol. 49, 412–420. doi:10.1093/jjco/hyz034
Honda, M., Miura, A., Izumi, Y., Kato, T., Ryotokuji, T., Monma, K., et al. (2010). Doxorubicin, cisplatin, and fluorouracil combination therapy for metastatic esophageal squamous cell carcinoma. Dis. Esophagus 23, 641–645. doi:10.1111/j.1442-2050.2010.01070.x
Hou, W., Qin, X., Zhu, X., Fei, M., Liu, P., Liu, L., et al. (2013). Lapatinib inhibits the growth of esophageal squamous cell carcinoma and synergistically interacts with 5-fluorouracil in patient-derived xenograft models. Oncol. Rep. 30, 707–714. doi:10.3892/or.2013.2500
Huang, C.-M., Huang, C.-S., Hsu, T.-N., Huang, M.-S., Fong, I.-H., Lee, W.-H., et al. (2019). Disruption of cancer metabolic SREBP1/miR-142-5p suppresses epithelial–mesenchymal transition and stemness in esophageal carcinoma. Cells 9, 7. doi:10.3390/cells9010007
Huang, J., Wang, X., Zhang, X., Chen, W., Luan, L., Song, Q., et al. (2021). CDK4 amplification in esophageal squamous cell carcinoma associated with better patient outcome. Front. Genet. 12, 616110. doi:10.3389/fgene.2021.616110
Ilson, D. H., Kelsen, D., Shah, M., Schwartz, G., Levine, D. A., Boyd, J., et al. (2011). A phase 2 trial of erlotinib in patients with previously treated squamous cell and adenocarcinoma of the esophagus. Cancer 117, 1409–1414. doi:10.1002/cncr.25602
Jarada, T. N., Rokne, J. G., and Alhajj, R. (2020). A review of computational drug repositioning: Strategies, approaches, opportunities, challenges, and directions. J. Cheminform. 12, 46. doi:10.1186/s13321-020-00450-7
Jones, V. A., Patel, P. M., Gibson, F. T., Cordova, A., and Amber, K. T. (2020). The role of collagen XVII in cancer: Squamous cell carcinoma and beyond. Front. Oncol. 10, 352. doi:10.3389/fonc.2020.00352
Karasic, T. B., O’Hara, M. H., Teitelbaum, U. R., Damjanov, N., Giantonio, B. J., d’Entremont, T. S., et al. (2020). Phase II trial of Palbociclib in patients with advanced esophageal or gastric cancer. Oncologist 25, e1864–e1868. doi:10.1634/theoncologist.2020-0681
Kashyap, M. K., and Abdel-Rahman, O. (2018). Expression, regulation and targeting of receptor tyrosine kinases in esophageal squamous cell carcinoma. Mol. Cancer 17, 54. doi:10.1186/s12943-018-0790-4
Kim, S.-W., Lee, J.-H., Moon, J.-H., Nazim, U. M. D., Lee, Y.-J., Seol, J.-W., et al. (2015). Niacin alleviates TRAIL-mediated colon cancer cell death via autophagy flux activation. Oncotarget 7, 4356–4368. doi:10.18632/oncotarget.5374
Lee, S.-M., Park, J. S., and Kim, K.-S. (2019). Improving combination cancer therapy by acetaminophen and romidepsin in non-small cell lung cancer cells. Biomed. Sci. Lett. 25, 293–301. doi:10.15616/bsl.2019.25.4.293
Li, J., Xie, Y., Wang, X., Jiang, C., Yuan, X., Zhang, A., et al. (2020). Identification of hub genes associated with esophageal cancer progression using bioinformatics analysis. Oncol. Lett. 20, 214. doi:10.3892/ol.2020.12077
Li, X., Zhao, L., Wei, M., Lv, J., Sun, Y., Shen, X., et al. (2021a). Serum metabolomics analysis for the progression of esophageal squamous cell carcinoma. J. Cancer 12, 3190–3197. doi:10.7150/jca.54429
Li, Y., Xu, F., Chen, F., Chen, Y., Ge, D., Zhang, S., et al. (2021b). Transcriptomics based multi-dimensional characterization and drug screen in esophageal squamous cell carcinoma. Ebiomedicine 70, 103510. doi:10.1016/j.ebiom.2021.103510
Li, Z., Zou, L., Xiao, Z.-X., and Yang, J. (2022). Transcriptome-based drug repositioning identifies TPCA-1 as a potential selective inhibitor of esophagus squamous carcinoma cell viability. Int. J. Mol. Med. 49, 75. doi:10.3892/ijmm.2022.5131
Liu, J.-K., Abudula, A., Yang, H.-T., Xu, L.-X., Bai, G., Tulahong, A., et al. (2021). DPP3 expression promotes cell proliferation and migration in vitro and tumor growth in vivo that associates with poor prognosis of esophageal carcinoma. doi:10.21203/rs.3.rs-1078211/v1
Loehr, A. R., Pierpont, T. M., Gelsleichter, E., Galang, A. M. D., Fernandez, I. R., Moore, E. S., et al. (2021). Targeting cancer stem cells with differentiation agents as an alternative to genotoxic chemotherapy for the treatment of malignant testicular germ cell tumors. Cancers 13, 2045. doi:10.3390/cancers13092045
Ma, X.-L., Yao, H., Wang, X., Wei, Y., Cao, L.-Y., Zhang, Q., et al. (2019). ILK predicts the efficacy of chemoradiotherapy and the prognosis of patients with esophageal squamous cell carcinoma. Oncol. Lett. 18, 4114–4125. doi:10.3892/ol.2019.10768
Maguire, M., Nield, P. C., Devling, T., Jenkins, R. E., Park, B. K., Polański, R., et al. (2008). MDM2 regulates dihydrofolate reductase activity through monoubiquitination. Cancer Res. 68, 3232–3242. doi:10.1158/0008-5472.can-07-5271
Maron, S. B., Xu, J., and Janjigian, Y. Y. (2020). Targeting EGFR in esophagogastric cancer. Front. Oncol. 10, 553876. doi:10.3389/fonc.2020.553876
Milani, M., Jha, G., and Potter, D. A. (2009). Anastrozole use in early stage breast cancer of post-menopausal women. Clin. Med. Ther. 1, 141–156. doi:10.4137/cmt.s9
Monk, J. P., Phillips, G., Waite, R., Kuhn, J., Schaaf, L. J., Otterson, G. A., et al. (2006). Assessment of tumor necrosis factor Alpha blockade as an intervention to improve tolerability of dose-intensive chemotherapy in cancer patients. J. Clin. Oncol. 24, 1852–1859. doi:10.1200/jco.2005.04.2838
Napier, K. J., Scheerer, M., and Misra, S. (2014). Esophageal cancer: A review of epidemiology, pathogenesis, staging workup and treatment modalities. World J. Gastrointest. Oncol. 6, 112–120. doi:10.4251/wjgo.v6.i5.112
Nathan, M. R., and Schmid, P. (2017). A review of fulvestrant in breast cancer. Oncol. Ther. 5, 17–29. doi:10.1007/s40487-017-0046-2
Niaki, E. F., Acker, T. V., Imre, L., Nánási, P., Tarapcsák, S., Bacsó, Z., et al. (2020). Interactions of cisplatin and Daunorubicin at the chromatin level. Sci. Rep. 10, 1107. doi:10.1038/s41598-020-57702-7
Ochi, F., Shiozaki, A., Ichikawa, D., Fujiwara, H., Nakashima, S., Takemoto, K., et al. (2015). Carbonic anhydrase XII as an independent prognostic factor in advanced esophageal squamous cell carcinoma. J. Cancer 6, 922–929. doi:10.7150/jca.11269
Oettle, H., Arnold, D., Kern, M., Hoepffner, N., Settmacher, U., Neuhaus, P., et al. (2002). Phase I study of gemcitabine in combination with cisplatin, 5-fluorouracil and folinic acid in patients with advanced esophageal cancer. Anticancer. Drugs 13, 833–838. doi:10.1097/00001813-200209000-00008
Pang, L., Li, Q., Li, S., He, J., Cao, W., Lan, J., et al. (2016). Membrane type 1-matrix metalloproteinase induces epithelial-to-mesenchymal transition in esophageal squamous cell carcinoma: Observations from clinical and in vitro analyses. Sci. Rep. 6, 22179. doi:10.1038/srep22179
Qian, Y., Liang, X., Kong, P., Cheng, Y., Cui, H., Yan, T., et al. (2020). Elevated DHODH expression promotes cell proliferation via stabilizing β-catenin in esophageal squamous cell carcinoma. Cell Death Dis. 11, 862. doi:10.1038/s41419-020-03044-1
Qiao, X., Wang, X., Shang, Y., Li, Y., and Chen, S. (2018). Azithromycin enhances anticancer activity of TRAIL by inhibiting autophagy and up-regulating the protein levels of DR4/5 in colon cancer cells in vitro and in vivo. Cancer Commun. 38, 43. doi:10.1186/s40880-018-0309-9
Ren, P., Yu, Z.-T., Xiu, L., Wang, M., and Liu, H.-M. (2013). Elevated serum levels of human relaxin-2 in patients with esophageal squamous cell carcinoma. World J. Gastroenterol. 19, 2412–2418. doi:10.3748/wjg.v19.i15.2412
Ribeiro, F. R., Paulo, P., Costa, V. L., Barros-Silva, J. D., Ramalho-Carvalho, J., Jerónimo, C., et al. (2011). Cysteine-rich secretory protein-3 (CRISP3) is strongly up-regulated in prostate carcinomas with the TMPRSS2-ERG fusion gene. Plos One 6, e22317. doi:10.1371/journal.pone.0022317
Rong, L., Wang, B., Guo, L., Liu, X., Wang, B., Ying, J., et al. (2020). HER2 expression and relevant clinicopathological features in esophageal squamous cell carcinoma in a Chinese population. Diagn. Pathol. 15, 27. doi:10.1186/s13000-020-00950-y
Saito, S., Morishima, K., Ui, T., Hoshino, H., Matsubara, D., Ishikawa, S., et al. (2015). The role of HGF/MET and FGF/FGFR in fibroblast-derived growth stimulation and lapatinib-resistance of esophageal squamous cell carcinoma. Bmc Cancer 15, 82. doi:10.1186/s12885-015-1065-8
Sciegienka, S. J., Solst, S. R., Falls, K. C., Schoenfeld, J. D., Klinger, A. R., Ross, N. L., et al. (2017). D-penicillamine combined with inhibitors of hydroperoxide metabolism enhances lung and breast cancer cell responses to radiation and carboplatin via H2O2-mediated oxidative stress. Free Radic. Biol. Med. 108, 354–361. doi:10.1016/j.freeradbiomed.2017.04.001
Shirmohammadi, E., Ebrahimi, S.-E. S., Farshchi, A., and Salimi, M. (2020). The efficacy of etanercept as anti-breast cancer treatment is attenuated by residing macrophages. Bmc Cancer 20, 836. doi:10.1186/s12885-020-07228-y
Song, Y., Wang, X., Wang, F., Peng, X., Li, P., Liu, S., et al. (2021). Identification of four genes and biological characteristics of esophageal squamous cell carcinoma by integrated bioinformatics analysis. Cancer Cell Int. 21, 123. doi:10.1186/s12935-021-01814-1
Subramanian, A., Narayan, R., Corsello, S. M., Peck, D. D., Natoli, T. E., Lu, X., et al. (2017). A next generation connectivity map: L1000 platform and the first 1, 000, 000 profiles. Cell 171, 1437–1452. e17. doi:10.1016/j.cell.2017.10.049
Sukocheva, O. A., Li, B., Due, S. L., Hussey, D. J., and Watson, D. I. (2015). Androgens and esophageal cancer: What do we know? World J. Gastroenterol. 21, 6146–6156. doi:10.3748/wjg.v21.i20.6146
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Tanioka, Y., Yoshida, T., Yagawa, T., Saiki, Y., Takeo, S., Harada, T., et al. (2003). Matrix metalloproteinase-7 and matrix metalloproteinase-9 are associated with unfavourable prognosis in superficial oesophageal cancer. Br. J. Cancer 89, 2116–2121. doi:10.1038/sj.bjc.6601372
Tong, Z., Chatterjee, D., Deng, D., Veeranki, O., Mejia, A., Ajani, J. A., et al. (2017). Antitumor effects of cyclin dependent kinase 9 inhibition in esophageal adenocarcinoma. Oncotarget 8, 28696–28710. doi:10.18632/oncotarget.15645
Varalda, M., Antona, A., Bettio, V., Roy, K., Vachamaram, A., Yellenki, V., et al. (2020). Psychotropic drugs show anticancer activity by disrupting mitochondrial and lysosomal function. Front. Oncol. 10, 562196. doi:10.3389/fonc.2020.562196
Wadleigh, R. G., Abbasi, S., and Korman, L. (2006). Palliative ethanol injections of unresectable Advanced esophageal carcinoma combined with chemoradiation. Am. J. Med. Sci. 331, 110–112. doi:10.1097/00000441-200602000-00022
Wang, M., Liu, D., Huang, Y., Jiang, Z., Wu, F., Cen, Y., et al. (2021a). Identification of key genes related to the prognosis of esophageal squamous cell carcinoma based on chip Re-annotation. Appl. Sci. (Basel). 11, 3229. doi:10.3390/app11073229
Wang, T., Zhang, P., Chen, L., Qi, H., Chen, H., Zhu, Y., et al. (2021b). Ixazomib induces apoptosis and suppresses proliferation in esophageal squamous cell carcinoma through activation of the c-Myc/NOXA pathway. J. Pharmacol. Exp. Ther. 380, 15–25. JPET-AR-2021-000837. doi:10.1124/jpet.121.000837
Wang, X., Li, K., Cheng, M., Wang, G., Han, H., Chen, F., et al. (2020). Bmi1 severs as a potential tumor-initiating cell marker and therapeutic target in esophageal squamous cell carcinoma. Stem Cells Int. 2020, 8877577. doi:10.1155/2020/8877577
Wang, Z., Lachmann, A., Keenan, A. B., and Ma’ayan, A. (2018). L1000FWD: Fireworks visualization of drug-induced transcriptomic signatures. Bioinformatics 34, 2150–2152. doi:10.1093/bioinformatics/bty060
Wei, Y., Wu, W., Jiang, Y., Zhou, H., Yu, Y., Zhao, L., et al. (2022). Nuplazid suppresses esophageal squamous cell carcinoma growth in vitro and in vivo by targeting PAK4. Br. J. Cancer 126, 1037–1046. doi:10.1038/s41416-021-01651-z
Wu, P., Feng, Q., Kerchberger, V. E., Nelson, S. D., Chen, Q., Li, B., et al. (2022). Integrating gene expression and clinical data to identify drug repurposing candidates for hyperlipidemia and hypertension. Nat. Commun. 13, 46. doi:10.1038/s41467-021-27751-1
Xie, Y., Zhang, J., Lu, B., Bao, Z., Zhao, J., Lu, X., et al. (2020). Mefloquine inhibits esophageal squamous cell carcinoma tumor growth by inducing mitochondrial autophagy. Front. Oncol. 10, 1217. doi:10.3389/fonc.2020.01217
Xie, Z., Zhong, C., and Duan, S. (2022). miR-1269a and miR-1269b: Emerging carcinogenic genes of the miR-1269 family. Front. Cell Dev. Biol. 10, 809132. doi:10.3389/fcell.2022.809132
Ye, B., Fan, D., Xiong, W., Li, M., Yuan, J., Jiang, Q., et al. (2021). Oncogenic enhancers drive esophageal squamous cell carcinogenesis and metastasis. Nat. Commun. 12, 4457. doi:10.1038/s41467-021-24813-2
Yu-jing, T., Wen-jing, T., and Biao, T. (2020). Integrated analysis of hub genes and pathways in esophageal carcinoma based on NCBI’s gene expression Omnibus (GEO) database: A bioinformatics analysis. Med. Sci. Monit. 26, 9239344–e924020. doi:10.12659/msm.923934
Zhang, C., and Chu, M. (2018). Leflunomide: A promising drug with good antitumor potential. Biochem. Biophys. Res. Commun. 496, 726–730. doi:10.1016/j.bbrc.2018.01.107
Zhang, H., Chen, G., Chen, S., Fu, Z., Zhou, H., Feng, Z., et al. (2021a). Overexpression of cyclin‐dependent kinase 1 in esophageal squamous cell carcinoma and its clinical significance. Febs Open Bio 11, 3126–3141. doi:10.1002/2211-5463.13306
Zhang, Q., Zhao, X., Zhang, C., Wang, W., Li, F., Liu, D., et al. (2019). Overexpressed PKMYT1 promotes tumor progression and associates with poor survival in esophageal squamous cell carcinoma. Cancer Manag. Res. 11, 7813–7824. doi:10.2147/cmar.s214243
Zhang, S., Cao, W., Yue, M., Zheng, N., Hu, T., Yang, S., et al. (2016). Caveolin-1 affects tumor drug resistance in esophageal squamous cell carcinoma by regulating expressions of P-gp and MRP1. Tumour Biol. 37, 9189–9196. doi:10.1007/s13277-015-4778-z
Zhang, X., Peng, L., Luo, Y., Zhang, S., Pu, Y., Chen, Y., et al. (2021b). Dissecting esophageal squamous-cell carcinoma ecosystem by single-cell transcriptomic analysis. Nat. Commun. 12, 5291. doi:10.1038/s41467-021-25539-x
Zhang, Y., Xu, Y., Li, Z., Zhu, Y., Wen, S., Wang, M., et al. (2018). Identification of the key transcription factors in esophageal squamous cell carcinoma. J. Thorac. Dis. 10, 148–161. doi:10.21037/jtd.2017.12.27
Zhang, Z., He, Q., Fu, S., and Zheng, Z. (2017). Estrogen receptors in regulating cell proliferation of esophageal squamous cell carcinoma: Involvement of intracellular Ca2+ signaling. Pathol. Oncol. Res. 23, 329–334. doi:10.1007/s12253-016-0105-2
Zhao, C., Lin, L., Liu, J., Liu, R., Chen, Y., Ge, F., et al. (2016). A phase II study of concurrent chemoradiotherapy and erlotinib for inoperable esophageal squamous cell carcinoma. Oncotarget 7, 57310–57316. doi:10.18632/oncotarget.9809
Zhou, Y., He, X., Jiang, Y., Wang, Z., Yu, Y., Wu, W., et al. (2021). Benzydamine, a CDK2 kinase inhibitor, suppresses the growth of esophageal squamous cell carcinoma in vitro and in vivo. doi:10.21203/rs.3.rs-464145/v1
Zhu, G., Li, X., Li, J., Zhou, W., Chen, Z., Fan, Y., et al. (2020). Arsenic trioxide (ATO) induced degradation of Cyclin D1 sensitized PD-1/PD-L1 checkpoint inhibitor in oral and esophageal squamous cell carcinoma. J. Cancer 11, 6516–6529. doi:10.7150/jca.47111
Zhu, S., Yan, X., Xiang, Z., Ding, H.-F., and Cui, H. (2013). Leflunomide reduces proliferation and induces apoptosis in neuroblastoma cells in vitro and in vivo. Plos One 8, e71555. doi:10.1371/journal.pone.0071555
Keywords: drug repositioning, drug repurposing, esophageal squamous cell carcinoma, ESCC treatment, cancer biology
Citation: Bennett AN, Huang RX, He Q, Lee NP, Sung W-K and Chan KHK (2022) Drug repositioning for esophageal squamous cell carcinoma. Front. Genet. 13:991842. doi: 10.3389/fgene.2022.991842
Received: 12 July 2022; Accepted: 12 September 2022;
Published: 28 September 2022.
Edited by:
Marco Pellegrini, National Research Council (CNR), ItalyReviewed by:
Manuela Petti, Sapienza University of Rome, ItalyTeng Mao, Shanghai Jiao Tong University, China
Copyright © 2022 Bennett, Huang, He, Lee, Sung and Chan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kei Hang Katie Chan, katie.kh.chan@cityu.edu.hk
 Adam N. Bennett
Adam N. Bennett Rui Xuan Huang
Rui Xuan Huang Qian He3
Qian He3 Nikki P. Lee
Nikki P. Lee Wing-Kin Sung
Wing-Kin Sung Kei Hang Katie Chan
Kei Hang Katie Chan