- 1Immunology Research Center, Mashhad University of Medical Sciences, Mashhad, Iran
- 2Department of Immunology and Allergy, School of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran
- 3Immunoregulation Research Center, Shahed University, Tehran, Iran
- 4Department of Immunology, Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran
- 5Noncommunicable Diseases Research Center, Fasa University of Medical Science, Fasa, Iran
- 6Department of Immunology, Shahed University, Tehran, Iran
Background: Since the beginning of the pandemic of coronavirus disease 2019 (COVID-19), many countries have experienced a considerable number of COVID-19 cases and deaths. The etiology of a broad spectrum of symptoms is still debated. Host genetic variants might also significantly influence the outcome of the disease. This study aimed to evaluate the association of angiotensin-converting enzyme (ACE1) gene Insertion/Deletion (I/D) polymorphism (rs1799752) and ACE2 gene rs1978124 single nucleotide polymorphism with the COVID-19 severity.
Methods: This study was conducted on 470 COVID-19 patients and a control group of 56 healthy individuals across several major cities in Iran. The blood sample and clinical data were collected from the participants, and their ACE1 I/D and ACE2 rs1978124 polymorphisms were determined using polymerase chain reaction and PCR-RFLP, respectively. Serum levels of C-reactive protein (CRP), interleukin 6 (IL-6), and ACE1 were measured in the blood samples.
Results: We found that the ACE1 DD genotype frequency was inversely correlated with the risk of intubation (p = 0.017) and mortality in COVID-19 patients (p = 0.049). Even after adjustment, logistic regression demonstrated that this significant inverse association remained constant for the above variables at odds ratios of (OR) = 0.35 and Odds Ratio = 0.49, respectively. Also, in the expired (p = 0.042) and intubated (p = 0.048) groups with II + ID genotypes, the mean level of CRP was significantly higher than in the DD genotype group. Furthermore, in both intubated and expired groups, the mean serum level of ACE1 was higher compared with non-intubated and survived groups with II or II + ID genotypes. The results also indicated that ACE2 rs1978124 TT + CT genotypes in females have a significant positive role in susceptibility to COVID-19; however, in females, the TT + CT genotypes had a protective effect (OR = 0.098) against the severity of COVID-19.
Conclusion: These findings suggest that ACE1 I/D and ACE2 rs1978124 polymorphism could potentially influence the outcome of COVID-19 in the Iranian population.
1 Introduction
The current pandemic results from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causing coronavirus disease 2019 (COVID-19). It has turned into a full-blown global crisis since its onset, prompting researchers worldwide to seek solutions for this problem. So far, COVID-19 has affected over 566 million individuals worldwide, resulting in 6.3 million deaths (WHO, 2022). The clinical spectrum of SARS-CoV-2 patients ranges from asymptomatic and mild flu-like symptoms to severe acute respiratory distress syndrome (ARDS). The prevalence and mortality rates of COVID-19 vary considerably from country to country, which raises the question of whether geographical origin and host genetic variations play a role in the severity and mortality of COVID-19 infection.
Angiotensin-converting enzyme-2 (ACE2) is the receptor of SARS-CoV-2, and transmembrane protease serine 2 (TMPRSS2) facilitates the virus entry. ACE2 is mainly expressed in the lung, intestine, cardiovascular system, kidney, adipose tissue, and central nervous system (Gheblawi et al., 2020). ACE1 converts Angiotensin I into Angiotensin II, which promotes inflammation, thrombosis, and vasoconstriction. ACE2 converts Angiotensin II into Angiotensin 1–7 and hence promotes vasodilation (D'Ardes et al., 2020). Downregulation of ACE2 expression due to SARS-CoV-2 infection may prevent viral infection. However, it also diminishes the beneficial impacts of ACE2 in the lungs and other organs (Gao et al., 2022). Therefore, COVID-19 may lead to ACE1/ACE2 imbalance and increase angiotensin II levels because it activates the renin-angiotensin-aldosterone system (RAAS) and thus the progression of COVID-19, especially in patients with underlying diseases such as high blood pressure (HTN), cardiovascular disease (CVD) and diabetes (DM) (Adamzik et al., 2007; Beyerstedt et al., 2021a). Moreover, factors like sex, age, smoking habit, obesity, blood group, HTN, DM, CVD, and genetics might be important in COVID-19 infection (Gard, 2010; Cai et al., 2020; Ejaz et al., 2020; Ovsyannikova et al., 2020; Sattar et al., 2020; Zeberg and Pääbo, 2020; Goel et al., 2021). On the other hand, genetic variation of a gene likely modifies the function and expression of an encoded product, which could be considered the interindividual differences in susceptibility to several infectious diseases. So far, few studies have shown the roles of angiotensin-converting enzyme 1 (ACE1), ACE2, and transmembrane protease serine 2 (TMPRSS2) gene variants in the COVID-19 severity (Gorbalenya et al., 2020; Hou et al., 2020; de Araújo et al., 2022; Gintoni et al., 2022).
1.1 ACE1
The ACE1 gene is located on chromosome 17q35 with 26 exons and 25 introns. The insertion/deletion (I/D) ACE1 polymorphism (rs1799752) is described by an insertion (allele I) or deletion (allele D) of a 287-base pair (bp) Alu repeat sequence in the 16th intron of the ACE1 gene, which accounts for most of the interindividual variability in circulating ACE activity and shows significant geographic variability. Therefore, I/D polymorphism has three different genotypes: II, ID, and DD (Rigat et al., 1990; Rieder et al., 1999; Sayed-Tabatabaei et al., 2006). Some recent studies suggest that the ACE1 ID polymorphism as a main geographical variation could be one of the genetic markers of susceptibility and pathogenicity of COVID-19 (Pati et al., 2020; Yamamoto et al., 2020). A review on I/D polymorphism suggested that the DD genotype in COVID-19 patients might cause severe lung injury (Zheng and Cao, 2020). However, a meta-analysis by Delanghe et al. demonstrated a negative association between COVID-19 mortality and D alleles frequency from an evaluation of 25 countries in the Middle East, North Africa, and Europe (Delanghe et al., 2020a). An ecological study demonstrated the distribution of II genotype is highest in Asian countries and lower among the African and European countries across 25 countries (Aung et al., 2020). A case-control study in one of the southeastern cities of Iran with a smaller sample size has shown that the II genotype decreases the risk of COVID-19 infection (Kouhpayeh et al., 2021).
1.2 ACE2
ACE2 rs1978124 SNP is located on chromosome Xp22 in intron one, suggesting that this variant may affect the expression of the ACE2 gene (Zhao et al., 2010; Patel et al., 2014). Also, some studies demonstrated that the ACE2 rs1978124 SNP was associated with the severity of COVID-19, the risk of diabetes-related left ventricular remodeling, and dyslipidemia (Liu et al., 2018; Sabater Molina et al., 2022).
The present study is the first to determine the potential role of ACE1/ACE2 gene polymorphisms in susceptibility to COVID-19 and the disease outcome of COVID-19 with a larger sample size compared to previous studies, representing the entire Iranian population.
2 Participants and methods
2.1 Study subjects
This case-control study was conducted on 470 patients with COVID-19 and 56 healthy controls referred to hospitals between 2020 and 2021 across several major cities in Iran. COVID-19 infections were confirmed by real-time reverse transcription-polymerase chain reaction (RT-PCR) or chest CT scan findings. COVID-19 severity was classified into mild, moderate, severe, and critical as defined by the World Health Organization (WHO) (WHO, 2021). The research protocol was approved by the National Institute for Medical Research Development (IR.NIMAD.REC.1399.041).
2.2 Data collection and blood sampling
The relevant personal information and medical history of most subjects, including their sex, age, smoking status, and comorbidities, were obtained by a patient checklist. Informed consent was acquired from all individuals or their family members before collecting blood samples. The selection method of patients was not probabilistic. At the beginning of hospitalization, all patients and healthy control individuals donated 5 ml blood samples collected in the clot activator tubes and tubes containing ethylene diamine tetraacetic acid (EDTA).
2.3 Interleukin-6, C-reactive protein, and ACE1 assessment
Interleukin-6 (IL-6) was measured in serum samples using an automated immunoassay (IMMULITE 2000; Siemens Healthcare Diagnostics, The United Kingdom). Serum level of C-reactive protein (CRP) and the quantitative enzymatic determination of ACE1 were done in serum samples with 7180 clinical analyzers (Hitachi, Japan) using ACE BIOLIS (Genbio, Ireland).
2.4 DNA extraction and genotyping
DNA was extracted from the buffy coat samples of all subjects using a spin column kit (GenAll Exgene Cell SV mini kit, GenAll Biotechnology, South Korea).
2.4.1 Detection of ACE I/D gene polymorphism
The ACE1 intronic Alu insertion (I) or deletion (D) polymorphism (rs1799752) was determined by polymerase chain reaction (PCR) and agarose gel electrophoresis methods with specific primers (forward primer- 5′ -CTGGAGACCACTCCCATCCTTTCT-3′ and reverse primer- 5′ -GATGTGGCCATCACATTCGTCAGAT-3′). PCR reactions were performed in a final volume of 25 μL comprising TEMPase Hot Start 2x Master Mix A (Ampliqon, Denmark), ten pmol of each primer (TAG Copenhagen A/S, Denmark), 20–100 ng genomic DNA, and distilled water. After the initial denaturation step at 95°C for 15 min, the reaction mixtures were subjected to 40 cycles of 95°C for 30 s, 58°C for 45 s, 72°C for 1 min, and a final extension at 72°C for 5 min. The PCR products were electrophoresed and visualized in 2% agarose gels containing DNA-safe stains. This technique provided amplification products of 191 bp for the DD genotype, 480 base pairs (bp) for the II genotype, and 480 bp + 191 bp for the ID genotype (Figure 1).
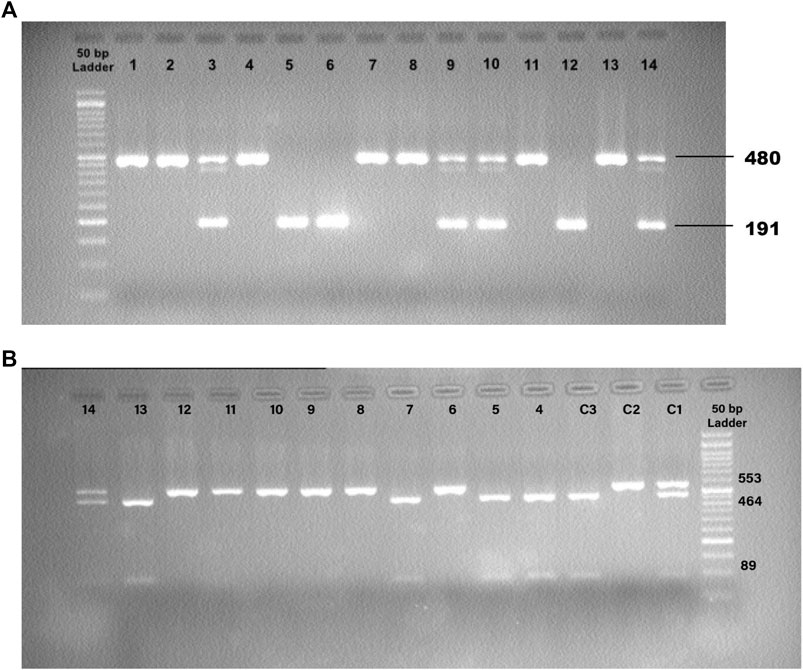
FIGURE 1. (A) Detection of the PCR Products for ACE1 Insertion/Deletion (I/D) Polymorphism. 12: Control, DD; 13: Control, II; 14: Control, ID; 1,2,4,7,8,11: II; 5,6: DD; 3,9,10: ID. (B) Detection of the PCR-RFLP Products for ACE2 rs1978124 SNP. C1: Control, CT; C2: Control, TT; C3: Control, CC; 4, 5, 7, 13: CC; 6, 8, 9, 10, 11, 12: TT; 14: CT.
2.4.2 Determination of rs1978124 SNP of the ACE2 gene
The ACE2 rs1978124 polymorphism was assessed by PCR–restriction fragment length polymorphism (PCR-RFLP). The PCR product of 553 bp was generated using the forward primer-5′- CAACCACACATACCACAAT-3′and reverse primer-5′- TTTCCTTTAGCCTACAATATCAAT -3′, and were incubated with 1 μL of Echo471 (Ava II) restriction enzyme at 37°C overnight. After digestion, two 464 bp and 89 bp products identify the C allele, and a 553 bp band identifies the T allele on agarose gel (Figure 2). About 10% of the samples were directly Sanger sequenced to ensure PCR-RFLP for SNP rs1978124. Using PCR primers, sanger sequencing was performed on 10% of the resulting samples. The sequencing result of rs1978124 SNP after alignment is shown in Supplementary Figure S1.
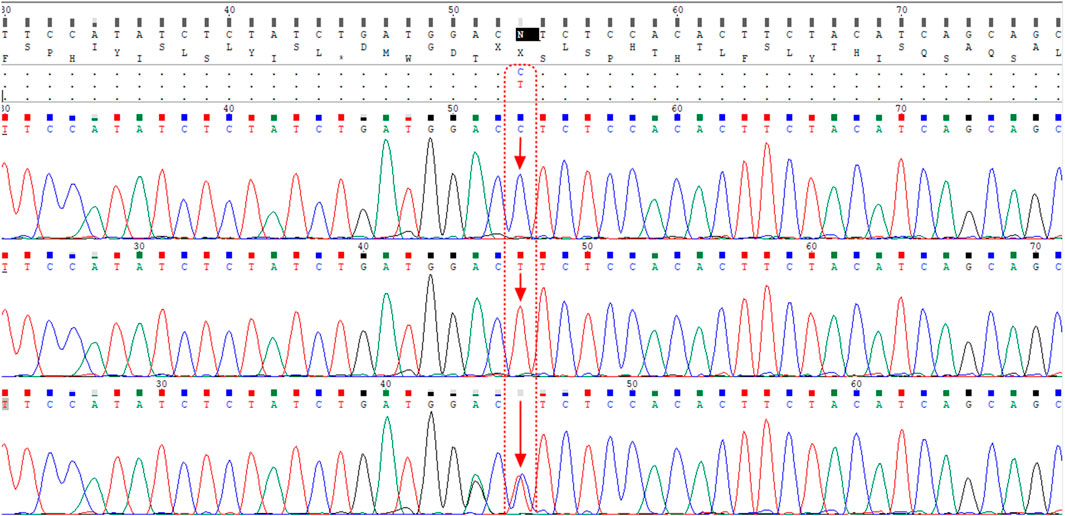
FIGURE 2. Alignment of the sequencing results of rs1978124 SNP located in ACE2 gene; the first, second, and third rows are related to the sample with CC genotype, TT genotype, and CT genotype, respectively. The presence of both peaks for both alleles is evident in the third row.
2.5 Statistical analysis
The numerical variables of each group were presented as mean ± standard deviation, and the Mann-Whitney test compared continuous data. The genotypes frequencies were reported as number (percentages) or n (%) in each group and assessed using the Chi-square. Also, Hardy–Weinberg equilibrium (HWE) was calculated and tested by the Chi-square test. The associations of ACE1 insertion/deletion polymorphism and ACE2 rs1978124 SNP with susceptibility and severity for SARS-CoV-2 infection at both the multiple and univariate levels were assessed by multinomial or binary logistic regressions to calculate odds ratios (ORs) (adjusted and unadjusted) with 95% confidence intervals (CI). Individuals were included in the comparison among groups after adjustment for sex, age, HTN, diabetes mellitus, CVD, renal disease, and cigarette smoking. For rs1978124, males and females were analyzed separately since the ACE2 gene is on the X chromosome. A p-value less than 0.05 was regarded to be significant.
3 Results
3.1 Demographic characteristics of the study on Iranian population
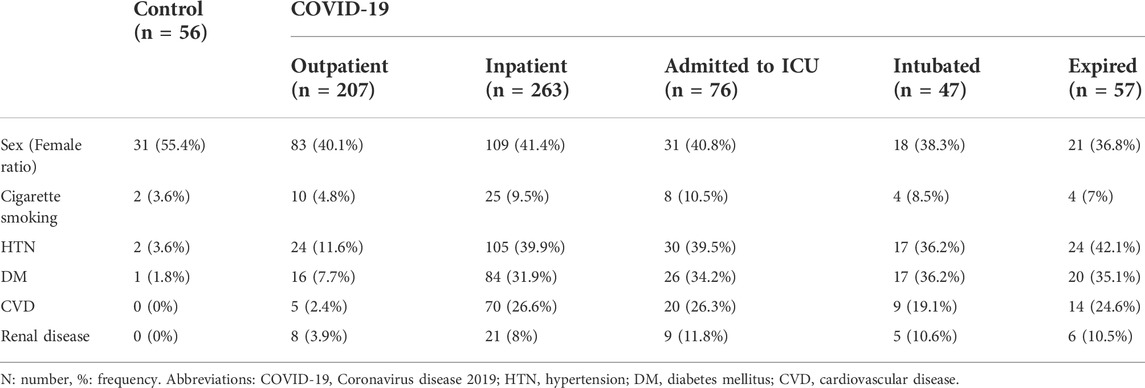
Patients were divided into groups based on the disease severity (COVID-19 Treatment Guidelines, 2019). The frequencies of comorbidities such as HTN, diabetes mellitus, CVD, renal disease, and cigarette smoking are presented in Table 1 and Supplementary Table S1. The results of this study demonstrated that HTN (p < 0.001), diabetes mellitus (p < 0.001), CVD (p < 0.001), and renal disease (p = 0.024) were significantly higher in COVID-19 patients compared to controls. Also, HTN (p = 0.008), diabetes mellitus (p = 0.007), and CVD (p = 0.009) were significantly correlated with COVID-19 mortality. There was no significant gender difference between the groups. The mean age was associated with increased severity and disease mortality in COVID-19. The mean age of healthy control, outpatients, inpatients, ICU admitted patients, intubated, and expired patients were 45.3 ± 13.3, 44.6 ± 14.1, 57.5 ± 16.5, 61.8 ± 17, 62 ± 14.9, and 65.8 ± 13.9 years, respectively.
3.2 ACE1 I/D and ACE2 rs1978124 genotypes
The genotype frequencies of the ACE1 I/D and ACE2 rs1978124 polymorphisms in the patients and control groups agreed with the Hardy-Weinberg equilibrium using the Chi-square analysis. (Supplementary Table S2).
3.3 Statistical comparisons between controls and patients (P value) for ACE1 and ACE2 genotypes/alleles
We found no significant relationship between different ACE1 I/D and ACE2 rs1978124 genotypes/alleles frequencies with comorbidities. The relevant statistical details are presented in Supplementary Table S3.
3.3.1 Susceptibility to COVID-19 infection
There was no significant association between ACE1 I/D (rs1799752) and ACE2 rs1978124 SNP genotypes/allele frequencies and susceptibility to COVID-19, but ACE2 rs1978124 T allele and TT+CT genotypes frequencies were higher in women with COVID-19 than in female controls. After adjusting for sex, age, HTN, diabetes mellitus, CVD, renal disease, and cigarette smoking, the comparison of 470 COVID-19 patients and 56 healthy controls through logistic regression revealed no significant association between genotypes of ACE1 and susceptibility to COVID-19 infection. However, for ACE2 CT + TT genotypes, this significant association remained even after adjustment (p = 0.008, 95%CI = 5.99) (Table 2).
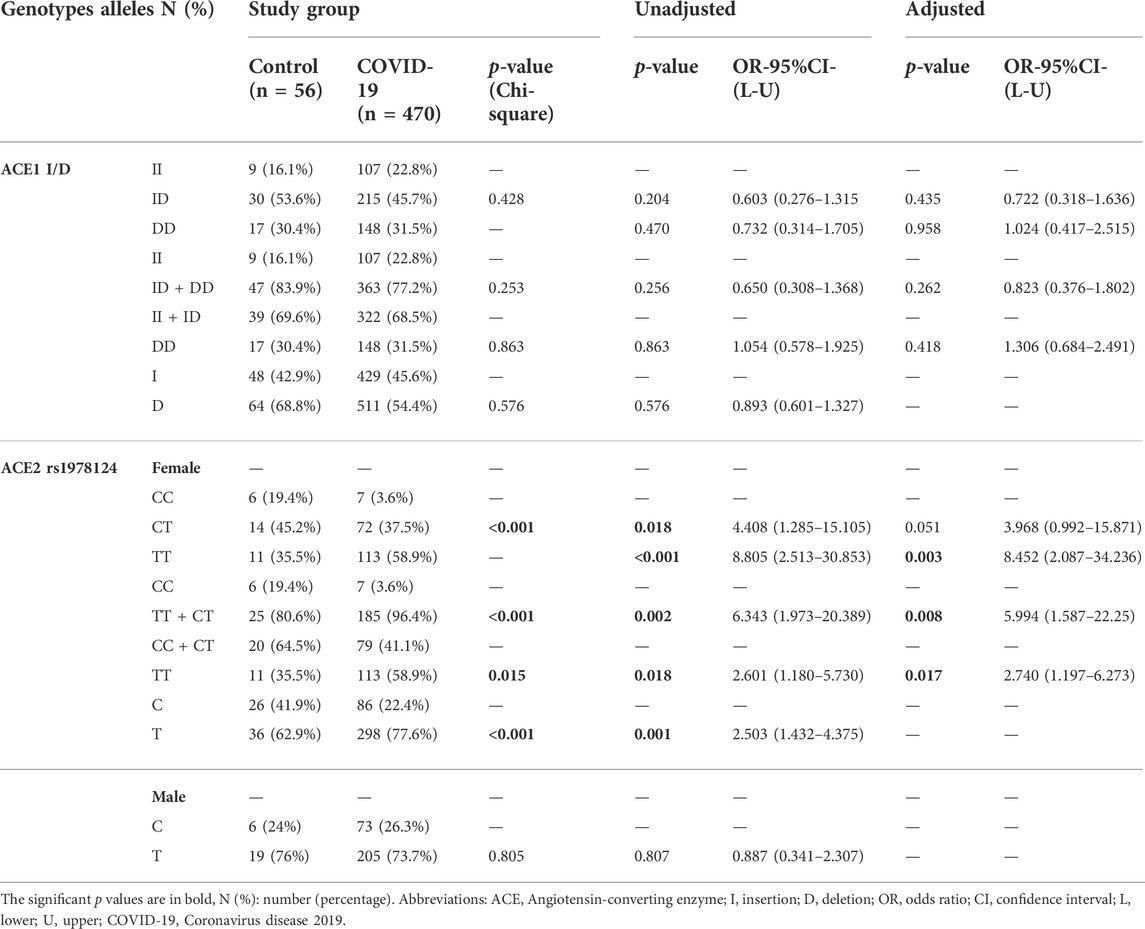
TABLE 2. Association of ACE1 I/D and ACE2 rs1978124 Genotypes/Alleles Distribution with Susceptibility to COVID-19, Adjusted by Age, Sex, Cigarette Smoking, Diabetes Mellitus, HTN, CVD, and renal diseases.
3.3.2 Severity of COVID-19
Different genotypes/alleles of ACE1 rs1799752 and ACE2 rs1978124 are not associated with COVID-19 hospitalization. After adjustment, binary logistic regression showed that patients with the TT + CT genotypes of rs1978124 carried a lower risk of hospitalization (94.5% vs 98.8%; p = 0.042, OR = 0.099; Supplementary Table S4).
Also, we observed a significant inverse association between the frequency of DD genotype and the risk for ICU admission (21.1% vs 33.2%, p = 0.049, OR = 0.53; Supplementary Table S5). Moreover, there was a significant inverse association between the frequency of DD genotype and intubation (84% vs 66.8, p = 0.017). While, the frequency of DD genotype and D allele significantly decreased in intubated patients (14.9% vs 32.9%, p = 0.014; 42.6% vs 55.1%, p = 0.027). After adjustment, patients with the DD genotype were at a reduced risk of intubation (OR = 0.35, p = 0.018; Table 3 and Figure 3).
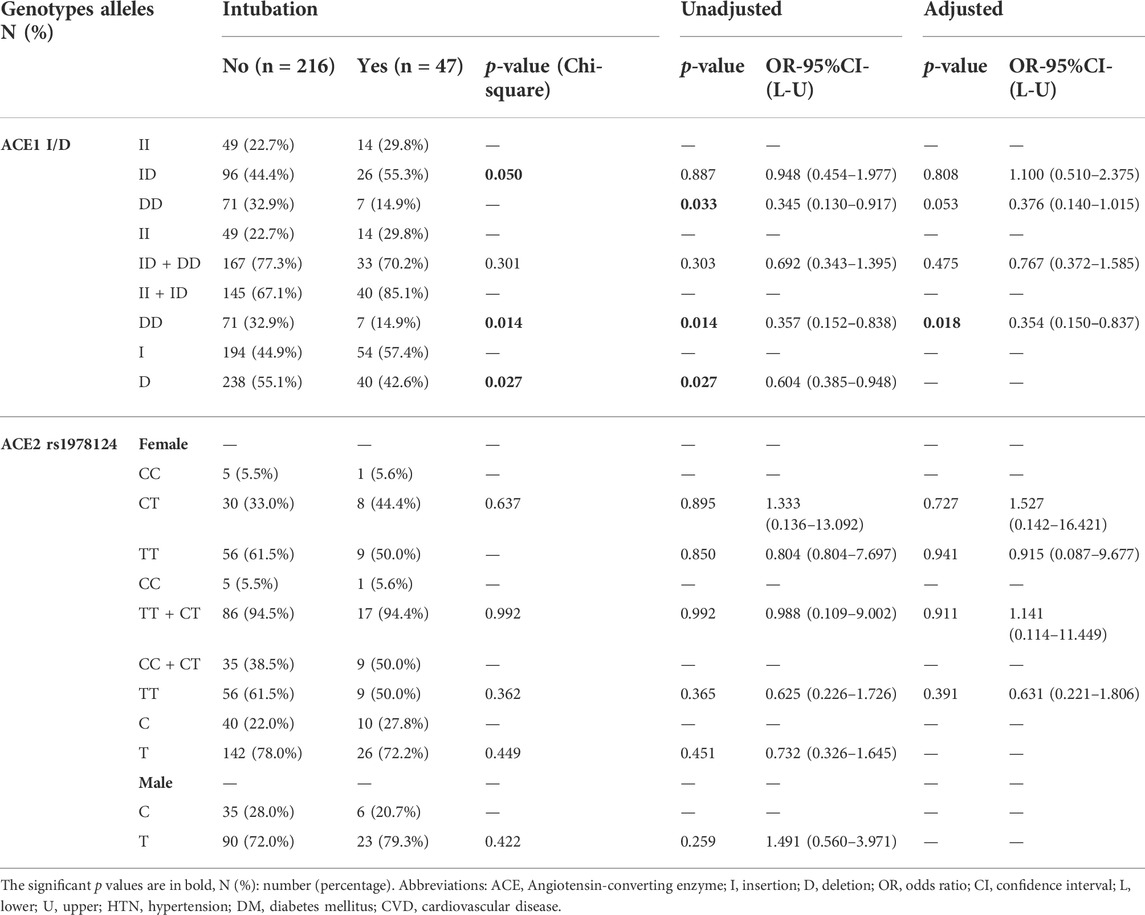
TABLE 3. Association of ACE1 Genotypes Distribution with Intubation of COVID-19 Patients, Adjusted by Age, Sex, Cigarette Smoking, DM, HTN, CVD, and renal diseases.
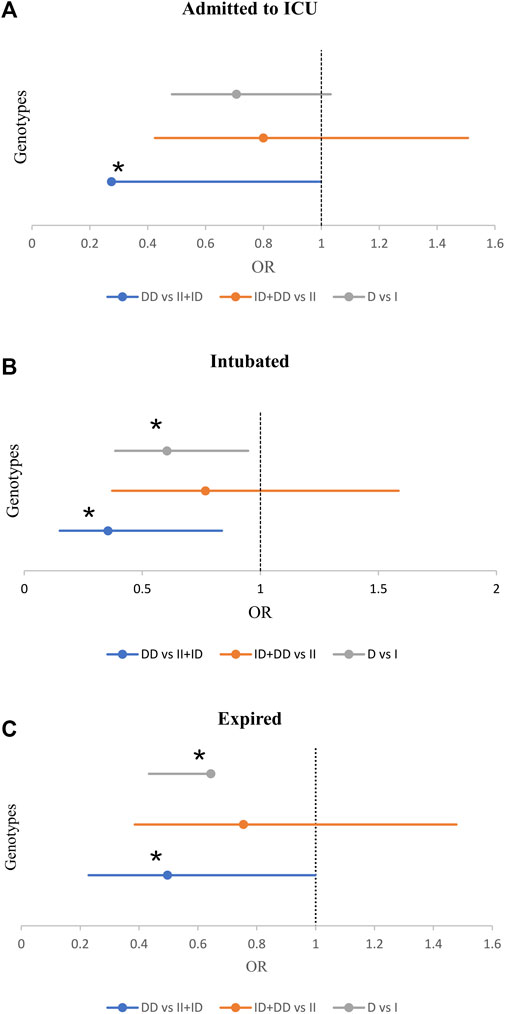
FIGURE 3. The Forest Plot displays Adjusted Odds Ratio (OR) ± 95% Confidence Intervals (CI) of ACE1 Polymorphism for ICU Admission (A), Intubated (B), and Expired (C) Groups; *p < 0.05. (Adjusted by Age, Sex, Cigarette Smoking, Diabetes Mellitus, Hypertension, Cardiovascular Disease, and Renal Diseases).
3.3.3 Mortality of COVID-19
Of the 470 patients, 413 (88.3%) survived, and 57 (12.1%) expired. The D allele and DD genotype frequency were lower in the expired group than in the surviving group (19.3% vs 32.3%, p = 0.035; 44.7% vs 55.7%, p = 0.028, respectively). After adjustment, patients with DD genotype were at a decreased risk of mortality (OR = 0.49; p = 0.049; Table 4 and Figure 3). However, there was no significant relationship between genotype/allele frequency of ACE2 rs1978124 and mortality, even after adjustment (Table 4).
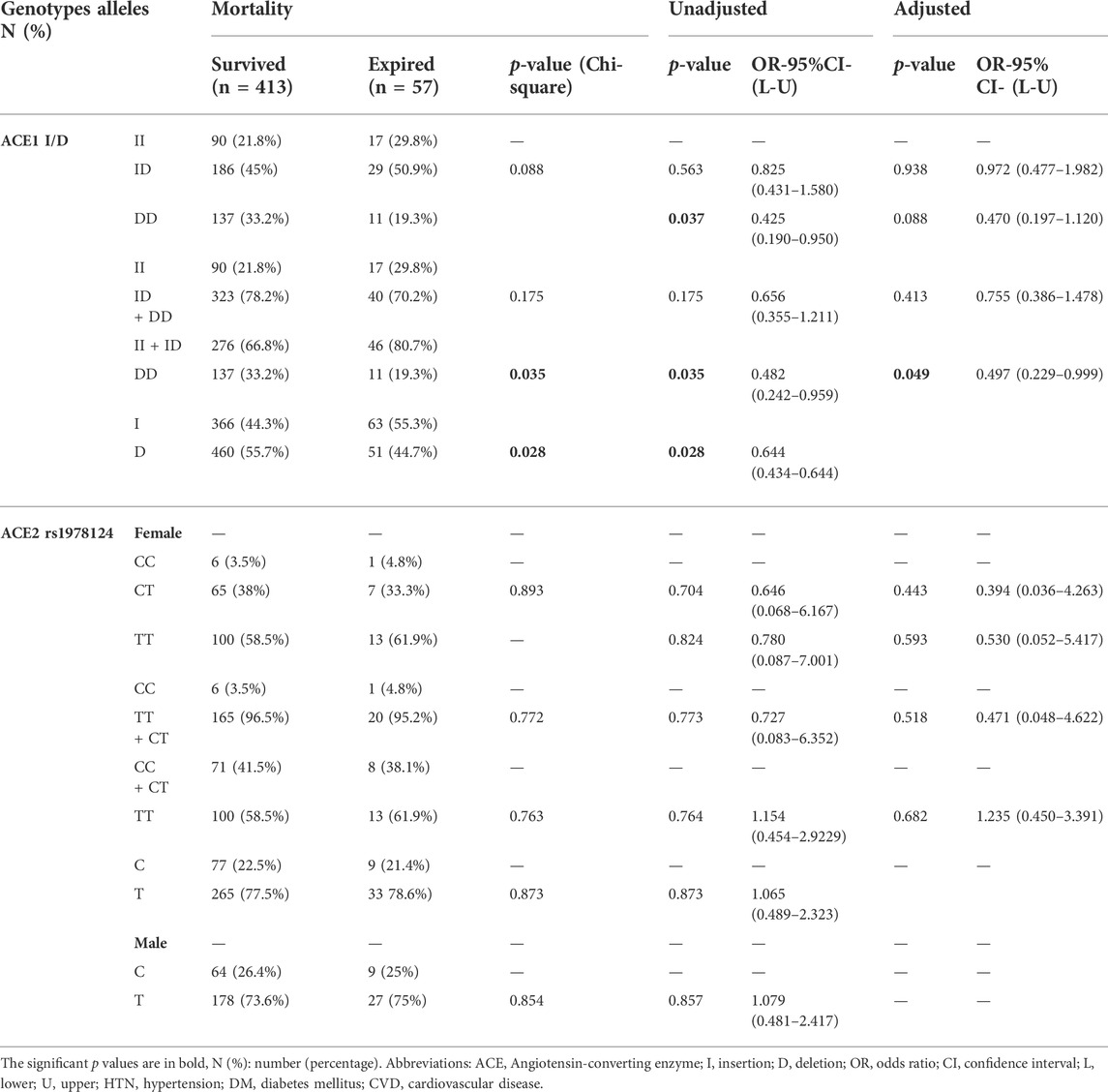
TABLE 4. Association of ACE1 Genotypes Distribution with COVID-19 Mortality, Adjusted by Age, Sex, Cigarette Smoking, DM, HTN, CVD, and renal diseases.
3.3.4 Clinical laboratory data
3.3.4.1 C-reactive protein
C-reactive protein (CRP) increased along with COVID-19 severity, as shown in Supplementary Tables S6-S8. The mean ± SD serum levels of CRP in intubated patients with DD and II + ID genotypes were 16.057 ± 7.69 and 23.91 ± 8.05, respectively. Also, it was observed that the mean ± SD serum level of CRP in the intubated groups with DD genotype was significantly lower than in the II + ID genotypes group. Also, the serum level of CRP decreased in the expired group with DD genotype compared to II + ID genotype carriers.
In comparing COVID-19 patients with the control group, the increase in the serum level of CRP in the ACE2 TT genotype was significant compared to CC + CT genotypes (12.05 ± 11.39 vs 3.14 ± 1.69; p = 0.029).
3.3.4.2 Interleukin-6
In all carriers of genotypes ACE1 rs1799752 and ACE2 rs1978124, the amount of IL-6 was noticeably increased in the control group compared with the COVID-19 group, as well as outpatients compared with inpatients, and in the survived compared with the expired (Supplementary Tables S9-S11). The IL-6 serum levels in patients admitted to the ICU as opposed to those admitted to the non-ICU ward were considerably increased only in groups with ID, ID + II, and ID + DD genotypes. This finding was also detected when comparing intubated with non-intubated patients. The serum level of IL-6 in all rs1978124 genotypes increased with disease severity, except for the CC genotype.
3.3.4.3 ACE1
The mean serum ACE1 level was significantly higher amongst DD and ID genotypes compared to II genotype in control, COVID-19, outpatients, inpatients, and survived groups. However, this association between ACE1 genotypes and serum ACE1 levels was not observed in ICU, intubated, and expired groups. Even in these groups, the level of ACE1 in the DD genotype was lower than II + ID. Notably, the serum level of ACE1 was significantly decreased in the intubated patients with DD genotype compared to the non-intubated patients with DD genotype. In addition, serum levels of ACE1 in patients with II or II + ID genotypes were increased in both intubated and expired groups compared to non-intubated and survived groups. Supplementary Table S12-S14 summarize the relationship between the serum level of ACE1 and COVID-19 severity.
4 Discussion
In this case-control study, the ACE1 I/D and ACE2 (rs1978124) genotypes were found to be distributed in accordance with the Hardy-Weinberg equilibrium, indicating that the chosen samples were representative of the society. This study is the first to evaluate the possible associations between these genetic factors (ACE1 rs1799752 and ACE2 rs1978124) and COVID-19 severity in several major cities in Iran.
The present study showed that HTN, DM, CVD, and renal disease were significantly associated with COVID-19 infection. Also, HTN, DM, and CVD were significantly correlated with the COVID-19 mortality rate. These findings confirm the results of previous studies (Docherty et al., 2020; Ejaz et al., 2020; Garg et al., 2020; Richardson et al., 2020).
Since ACE2 is the cellular receptor for SARS-COV-2 entry and a main component of the RAAS system, the downregulation of ACE2 expression following a viral infection promotes ACE1/ACE2 imbalance (Beyerstedt et al., 2021b). Therefore, studying ACE1 and ACE2 polymorphisms promises to be an effective way of regulating RAAS activity, which can improve the prognosis of COVID-19 patients. Accordingly, numerous studies have been conducted on the association of ACE1 I/D (rs1799752) and ACE2 rs1978124 polymorphisms with different diseases (Rigat et al., 1990; Tiret et al., 1992; Danser et al., 1995; Luo et al., 2019). However, the location of the ACE1 I/D rs1799752 polymorphism and ACE2 rs1978124 SNP in a non-coding region of the genes means they are unlikely to be functional variants. But previous studies have pointed to an association between serum and tissue levels of the ACE1 protein with rs1799752 polymorphism, which can affect the balance of ACE1/ACE2. The rs1799752 polymorphism can describe approximately 50% of ACE activity (Sayed-Tabatabaei et al., 2006). Higher activity of ACE can lead to an increase in the concentration of angiotensin II, which plays an essential role in inflammation (Osadnik et al., 2016).
A recent study showed the effect of a family history of hypertension and ACE1 I/D polymorphism (rs1799752) on cardiac autonomic modulation in adolescents. The D allele is a prognostic factor associated with increased serum ACE levels (Dias-Filho et al., 2021). Another study showed no association between the rs1799752 ACE I/D polymorphism and diabetic retinopathy (Li et al., 2013). The ACE2 rs1978124 SNP is a common genetic factor for cardiovascular disease (Yang et al., 2006; Palmer et al., 2008; Chaoxin et al., 2013), diabetes (Patel et al., 2012), and hypertension (Benjafield et al., 2004). Since the results of relevant studies in different populations display considerable variation, it is necessary that other ethnic groups should be evaluated separately. The present study showed that ACE1 I/D and ACE2 rs1978124 genotypes/alleles frequencies are not significantly correlated with diabetes mellitus, HTN, CVD, and renal disease.
That CRP and IL-6 levels reflect disease severity (Rostamian et al., 2020; Wang, 2020; Luan et al., 2021; Santa Cruz et al., 2021). Accordingly, we also observed that CRP levels were higher in intubated patients with genotype II + ID compared to those with genotype DD. Also, comparing the two groups of COVID-19 and control in terms of ACE2, the CRP level was found to be significantly higher in carriers of TT genotype, which is positively correlated with COVID-19 susceptibility.
Recent studies have demonstrated that ACE1 I/D polymorphism could have a significant role in the prognosis of COVID-19, but these results are controversial (Delanghe et al., 2020a; Bellone and Calvisi, 2020; Saadat, 2020; Yamamoto et al., 2020; Hubacek et al., 2021). Even though numerous studies have reported that the ACE1 DD genotype is associated with COVID-19 severity, they are mostly limited to epidemiological studies and in silico analyses (Gómez et al., 2020; Pati et al., 2020; Verma et al., 2021). Also, a study on the population of Turkey indicated that ACE1 I/D polymorphism was not associated with the severity of COVID-19 (Karakaş Çelik et al., 2021). Overall, we found no significant relationship between ACE1 I/D genotypes and the susceptibility to COVID-19.
Interestingly, we found that the frequencies of DD genotype and D allele were significantly low in the group of patients admitted to the ICU, intubated, and expired. Notably, this association remained significant, even after adjustment. Our results are consistent with the findings of Delanghe et al. They found a significant inverse association between the frequency of the D allele and mortality rate in a study that spanned more than 25 countries (Delanghe et al., 2020b; Delanghe et al., 2020c). Saad H et al. found a positive correlation between ACE1 II genotype and a heightened risk of contracting COVID-19 (Saad et al., 2021). In addition, Hubacek JA et al. showed that ACE1 II genotypes increased the risk of symptomatic COVID-19 in the Czech Republic (Hubacek et al., 2021). Furthermore, Jacobs found a significant elevation in the level of ACE2 protein in the alveolar epithelium cells when patients had II genotype of the rs1799752 polymorphism, as it can facilitate host cell entry of SARS-CoV-2 (Jacobs et al., 2021).
In the COVID-19 outpatients and healthy control, the serum level of ACE1 was significantly higher in DD compared with II + ID genotypes carriers, as expected. Furthermore, the ACE1 level was considerably higher in COVID-19 outpatients than in control subjects. Nevertheless, this significant association was not seen in patients admitted to ICU, intubated, and expired compared to those admitted to the non-ICU ward, not intubated, and who survived. In fact, in these groups, the ACE1 level had even decreased among the DD genotype and increased among II + ID genotypes. Therefore, the serum ACE1 level can be an influential factor in disease prognosis. Similar to our results, Annoni F et al. demonstrated that ACE1 levels were higher in non-survivors compared with survivors of ARDS (Annoni et al., 2019). Therefore, elevated ACE1 levels are associated with poor prognosis in ARDS patients (Sriram and Insel, 2020), which might point to endothelial activation and can prove to be a therapeutic target. Cambien F et al. demonstrated that plasma level of ACE was higher in myocardial infarction patients compared with the control group among subjects with II and ID genotypes (Cambien et al., 1994).
In conclusion, we observed that the ACE I/D polymorphism might alter ACE, IL-6, and CRP expression levels.
Previous studies have demonstrated a relation between ACE2 rs1978124 gene polymorphism and underlying comorbidities affecting the severity of COVID-19. A case-control study was conducted in West China to evaluate the association of rs1978124 ACE2 polymorphism with diabetes. It was revealed that there is found a significant relationship between the frequency of TT + CT genotypes and diabetes (OR = 2.2) (Liu et al., 2018). Another study in China showed a significant relationship between the TT + CT genotypes of rs1978124 polymorphism and dyslipidemia (Pan et al., 2018). Barry R. Palmer also found that the T allele of the ACE2 SNP rs1978124 was associated with higher mortality in an acute coronary syndrome cohort of European ancestry (Palmer et al., 2008).
A recent study on 318 Spanish COVID-19 patients aimed to evaluate the association of rs1978124 polymorphism with the severity of the disease. This study showed that CT genotype in women has a protective role (OR = 0.32) against the severity of COVID-19 (Sabater Molina et al., 2022). Our results also showed that the TT + CT genotypes have a protective effect (OR = 0.098) against the severity of COVID-19 in females. However, we observed that TT + CT genotypes in females have a significant positive role in the susceptibility to COVID-19 (infectivity), and even after adjusting for age and underlying diseases, this association remained significant. Also, the serum level of CRP was higher in patients with TT + CT genotypes compared to the control group. The functional mechanism by which rs1978124 SNP, a noncoding region of the ACE2 gene, affects the outcome of COVID-19 is unclear and needs further investigation. One possible explanation is that polymorphism affects the stability of ACE2 mRNA (i.e., splicing), post-transcriptional regulation by microRNA, and the efficiency of mRNA splicing (for example, silencing element or enhancer of intron splicing).
5 Study limitations
One of the limitations of this study was that the blood sample volume obtained from some patients was insufficient for laboratory tests, such as the ACE1, CRP, and IL-6 serum levels tests. Moreover, even though the assessment of angiotensin II and ACE2 serum levels can provide additional evidence for predicting the outcome of COVID-19, unfortunately, no blood sample was left to perform these tests.
6 Conclusion
We was demonstrated that the ACE1 I/D and ACE2 rs1978124 polymorphisms are relevant prognostic factors for the outcome of COVID-19. Patients with the II + ID genotype might have a significantly worse prognosis than those with the DD genotype. The T allele of SNP rs1978124 could affect susceptibility to COVID-19; ACE1/ACE2 polymorphism-mediated pathology is relevant, at least in the Iranian population. Consequently, further genetic studies on COVID-19 patients in different countries are required to clarify the mechanisms of lung injury and determine new therapeutic approaches.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving human participants were reviewed and approved by The research protocol was approved by the National Institute for Medical Research Development (IR.NIMAD.REC.1399.041). The patients/participants provided their written informed consent to participate in this study.
Author contributions
All authors contributed to the conception and design of this paper. MM and TH were responsible for the organizing and management of the project and also the final edition of the paper. AA, SH, MZ, and MN had an essential role in collecting blood samples and questionnaires, performing the experiments, analyzing the data, and writing papers. AH had a role in collecting blood samples and writing part of the draft article. All authors read and approved the final manuscript.
Funding
This work was supported by the Immunoregulation Research Center of Shahed University and the Ministry of Health and Medical Education of Iran.
Acknowledgments
The authors highly appreciate all the people for their participation in this project. The facilities provided by the universities of Mashhad, Tehran, Esfahan, Babol, and Zahedan are gratefully acknowledged.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.955965/full#supplementary-material
References
Adamzik, M., Frey, U., Sixt, S., Knemeyer, L., Beiderlinden, M., Peters, J., et al. (2007). ACE I/D but not AGT (-6)A/G polymorphism is a risk factor for mortality in ARDS. Eur. Respir. J. 29 (3), 482–488. doi:10.1183/09031936.00046106
Annoni, F., Orbegozo, D., Rahmania, L., Irazabal, M., Mendoza, M., De Backer, D., et al. (2019). Angiotensin-converting enzymes in acute respiratory distress syndrome. Intensive Care Med. 45 (8), 1159–1160. doi:10.1007/s00134-019-05600-6
Aung, A. K., Aitken, T., Teh, B. M., Yu, C., Ofori-Asenso, R., Chin, K. L., et al. (2020). Angiotensin converting enzyme genotypes and mortality from COVID-19: An ecological study. J. Infect. 81 (6), 961–965. doi:10.1016/j.jinf.2020.11.012
Bellone, M., and Calvisi, S. L. (2020). ACE polymorphisms and COVID-19-related mortality in Europe. J. Mol. Med. 98 (11), 1505–1509. doi:10.1007/s00109-020-01981-0
Benjafield, A. V., Wang, W. Y., and Morris, B. J. (2004). No association of angiotensin-converting enzyme 2 gene (ACE2) polymorphisms with essential hypertension. Am. J. Hypertens. 17 (7), 624–628. doi:10.1016/j.amjhyper.2004.02.022
Beyerstedt, S., Casaro, E. B., and Rangel É, B. (2021). COVID-19: Angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. 40 (5), 905–919. doi:10.1007/s10096-020-04138-6
Beyerstedt, S., Casaro, E. B., and Rangel É, B. (2021). COVID-19: Angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. 40 (5), 905–919. doi:10.1007/s10096-020-04138-6
Cai, G., Bossé, Y., Xiao, F., Kheradmand, F., and Amos, C. I. (2020). Tobacco smoking increases the lung gene expression of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 201 (12), 1557–1559. doi:10.1164/rccm.202003-0693LE
Cambien, F., Costerousse, O., Tiret, L., Poirier, O., Lecerf, L., Gonzales, M. F., et al. (1994). Plasma level and gene polymorphism of angiotensin-converting enzyme in relation to myocardial infarction. Circulation 90 (2), 669–676. doi:10.1161/01.cir.90.2.669
Chaoxin, J., Daili, S., Yanxin, H., Ruwei, G., Chenlong, W., and Yaobin, T. (2013). The influence of angiotensin-converting enzyme 2 gene polymorphisms on type 2 diabetes mellitus and coronary heart disease. Eur. Rev. Med. Pharmacol. Sci. 17 (19), 2654–2659.
COVID-19 Treatment Guidelines (2019). COVID-19 treatment Guidelines. Available at: https://www.covid19treatmentguidelines.nih.gov/.
D'Ardes, D., Boccatonda, A., Rossi, I., Guagnano, M. T., Santilli, F., Cipollone, F., et al. (2020). COVID-19 and RAS: Unravelling an unclear relationship. Int. J. Mol. Sci. 21 (8), E3003. doi:10.3390/ijms21083003
Danser, A. H., Schalekamp, M. A., Bax, W. A., van den Brink, A. M., Saxena, P. R., Riegger, G. A., et al. (1995). Angiotensin-converting enzyme in the human heart. Effect of the deletion/insertion polymorphism. Circulation 92 (6), 1387–1388. doi:10.1161/01.cir.92.6.1387
de Araújo, J. L. F., Menezes, D., Aguiar, R. S. D., and Souza, R. P. D. (2022). IFITM3, FURIN, ACE1, and TNF-α genetic association with COVID-19 outcomes: Systematic review and meta-analysis. Front. Genet. 13, 775246. doi:10.3389/fgene.2022.775246
Delanghe, J. R., Speeckaert, M. M., and De Buyzere, M. L. (2020). ACE polymorphism and COVID-19 outcome. Endocrine 70 (1), 13–14. doi:10.1007/s12020-020-02454-7
Delanghe, J. R., Speeckaert, M. M., and De Buyzere, M. L. (2020). COVID-19 infections are also affected by human ACE1 D/I polymorphism. Clin. Chem. Lab. Med. 58 (7), 1125–1126. doi:10.1515/cclm-2020-0425
Delanghe, J. R., Speeckaert, M. M., and De Buyzere, M. L. (2020). The host's angiotensin-converting enzyme polymorphism may explain epidemiological findings in COVID-19 infections. Clin. Chim. Acta. 505, 192–193. doi:10.1016/j.cca.2020.03.031
Dias-Filho, C. A. A., Soares, N. J. S. J., Bomfim, M. R. Q., José Dias, C., Vidal, F. C. B., Magalhães, B. C., et al. (2021). The effect of family history of hypertension and polymorphism of the ACE gene (rs1799752) on cardiac autonomic modulation in adolescents. Clin. Exp. Pharmacol. Physiol. 48 (2), 177–185. doi:10.1111/1440-1681.13426
Docherty, A. B., Harrison, E. M., Green, C. A., Hardwick, H. E., Pius, R., Norman, L., et al. (2020). Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO clinical characterisation protocol: Prospective observational cohort study. BMJ Clin. Res. ed) 369, m1985. doi:10.1136/bmj.m1985
Ejaz, H., Alsrhani, A., Zafar, A., Javed, H., Junaid, K., Abdalla, A. E., et al. (2020). COVID-19 and comorbidities: Deleterious impact on infected patients. J. Infect. Public Health 13 (12), 1833–1839. doi:10.1016/j.jiph.2020.07.014
Gao, X., Zhang, S., Gou, J., Wen, Y., Fan, L., Zhou, J., et al. (2022). Spike-mediated ACE2 down-regulation involved in the pathogenesis of SARS-CoV-2 infection. J. Infect. doi:10.1016/j.jinf.2022.06.030
Gard, P. R. (2010). Implications of the angiotensin converting enzyme gene insertion/deletion polymorphism in health and disease: A snapshot review. Int. J. Mol. Epidemiol. Genet. 1 (2), 145–157.
Garg, S., Kim, L., Whitaker, M., O'Halloran, A., Cummings, C., Holstein, R., et al. (2020). Hospitalization rates and Characteristics of patients hospitalized with laboratory-confirmed coronavirus disease 2019 - COVID-NET, 14 States, march 1-30, 2020. MMWR. Morb. Mortal. Wkly. Rep. 69 (15), 458–464. doi:10.15585/mmwr.mm6915e3
Gheblawi, M., Wang, K., Viveiros, A., Nguyen, Q., Zhong, J. C., Turner, A. J., et al. (2020). Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: Celebrating the 20th anniversary of the discovery of ACE2. Circ. Res. 126 (10), 1456–1474. doi:10.1161/CIRCRESAHA.120.317015
Gintoni, I., Adamopoulou, M., and Yapijakis, C. (2022). The impact of ACE and ACE2 gene polymorphisms in pulmonary diseases including COVID-19. Vivo 36 (1), 13–29. doi:10.21873/invivo.12672
Goel, R., Bloch, E. M., Pirenne, F., Al-Riyami, A. Z., Crowe, E., Dau, L., et al. (2021). ABO blood group and COVID-19: A review on behalf of the ISBT COVID-19 working group. Vox Sang. 116 (8), 849–861. doi:10.1111/vox.13076
Gómez, J., Albaiceta, G. M., García-Clemente, M., López-Larrea, C., Amado-Rodríguez, L., Lopez-Alonso, I., et al. (2020). Angiotensin-converting enzymes (ACE, ACE2) gene variants and COVID-19 outcome. Gene 762, 145102. doi:10.1016/j.gene.2020.145102
Gorbalenya, A. E., Baker, S. C., Baric, R. S., de Groot, R. J., Drosten, C., Gulyaeva, A. A., et al. (2020). The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5 (4), 536–544. doi:10.1038/s41564-020-0695-z
Hou, Y., Zhao, J., Martin, W., Kallianpur, A., Chung, M. K., Jehi, L., et al. (2020). New insights into genetic susceptibility of COVID-19: An ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 18 (1), 216. doi:10.1186/s12916-020-01673-z
Hubacek, J. A., Dusek, L., Majek, O., Adamek, V., Cervinkova, T., Dlouha, D., et al. (2021). ACE I/D polymorphism in Czech first-wave SARS-CoV-2-positive survivors. Clin. Chim. Acta. 519, 206–209. doi:10.1016/j.cca.2021.04.024
Jacobs, M., Lahousse, L., Van Eeckhoutte, H. P., Wijnant, S. R. A., Delanghe, J. R., Brusselle, G. G., et al. (2021). Effect of ACE1 polymorphism rs1799752 on protein levels of ACE2, the SARS-CoV-2 entry receptor, in alveolar lung epithelium. ERJ Open Res. 7 (2), 00940–02020. doi:10.1183/23120541.00940-2020
Karakaş Çelik, S., Çakmak Genç, G., Pişkin, N., Açikgöz, B., Altinsoy, B., Kurucu İşsiz, B., et al. (2021). Polymorphisms of ACE (I/D) and ACE2 receptor gene (Rs2106809, Rs2285666) are not related to the clinical course of COVID-19: A case study. J. Med. Virol. 93 (10), 5947–5952. doi:10.1002/jmv.27160
Kouhpayeh, H. R., Tabasi, F., Dehvari, M., Naderi, M., Bahari, G., Khalili, T., et al. (2021). Association between angiotensinogen (AGT), angiotensin-converting enzyme (ACE) and angiotensin-II receptor 1 (AGTR1) polymorphisms and COVID-19 infection in the southeast of Iran: A preliminary case-control study. Transl. Med. Commun. 6 (1), 26. doi:10.1186/s41231-021-00106-0
Li, N., Yang, X. F., Gu, H., Deng, Y., Xu, J., Ma, K., et al. (2013). Relationship of angiotensin converting enzyme gene polymorphism with diabetic retinopathy. Zhonghua. Yan Ke Za Zhi. 49 (1), 52–57.
Liu, C., Li, Y., Guan, T., Lai, Y., Shen, Y., Zeyaweiding, A., et al. (2018). ACE2 polymorphisms associated with cardiovascular risk in Uygurs with type 2 diabetes mellitus. Cardiovasc. Diabetol. 17 (1), 127. doi:10.1186/s12933-018-0771-3
Luan, Y-y., Yin, C-h., and Yao, Y-m. (2021). Update advances on C-reactive protein in COVID-19 and other viral infections. Front. Immunol. 12, 720363. doi:10.3389/fimmu.2021.720363
Luo, Y., Liu, C., Guan, T., Li, Y., Lai, Y., Li, F., et al. (2019). Association of ACE2 genetic polymorphisms withhypertension-related target organ damages in south Xinjiang. Hypertens. Res. 42 (5), 681–689. doi:10.1038/s41440-018-0166-6
Osadnik, T., Strzelczyk, J. K., Fronczek, M., Bujak, K., Reguła, R., Gonera, M., et al. (2016). Relationship of the rs1799752 polymorphism of the angiotensin-converting enzyme gene and the rs699 polymorphism of the angiotensinogen gene to the process of in-stent restenosis in a population of Polish patients with stable coronary artery disease. Adv. Med. Sci. 61 (2), 276–281. doi:10.1016/j.advms.2016.03.006
Ovsyannikova, I. G., Haralambieva, I. H., Crooke, S. N., Poland, G. A., and Kennedy, R. B. (2020). The role of host genetics in the immune response to SARS-CoV-2 and COVID-19 susceptibility and severity. Immunol. Rev. 296 (1), 205–219. doi:10.1111/imr.12897
Palmer, B. R., Jarvis, M. D., Pilbrow, A. P., Ellis, K. L., Frampton, C. M., Skelton, L., et al. (2008). Angiotensin-converting enzyme 2 A1075G polymorphism is associated with survival in an acute coronary syndromes cohort. Am. Heart J. 156 (4), 752–758. doi:10.1016/j.ahj.2008.06.013
Pan, Y., Wang, T., Li, Y., Guan, T., Lai, Y., Shen, Y., et al. (2018). Association of ACE2 polymorphisms with susceptibility to essential hypertension and dyslipidemia in Xinjiang, China. Lipids Health Dis. 17 (1), 241. doi:10.1186/s12944-018-0890-6
Patel, S. K., Velkoska, E., Freeman, M., Wai, B., Lancefield, T. F., and Burrell, L. M. (2014). From gene to protein-experimental and clinical studies of ACE2 in blood pressure control and arterial hypertension. Front. Physiol. 5, 227. doi:10.3389/fphys.2014.00227
Patel, S. K., Wai, B., Ord, M., MacIsaac, R. J., Grant, S., Velkoska, E., et al. (2012). Association of ACE2 genetic variants with blood pressure, left ventricular mass, and cardiac function in Caucasians with type 2 diabetes. Am. J. Hypertens. 25 (2), 216–222. doi:10.1038/ajh.2011.188
Pati, A., Mahto, H., Padhi, S., and Panda, A. K. (2020). ACE deletion allele is associated with susceptibility to SARS-CoV-2 infection and mortality rate: An epidemiological study in the Asian population. Clin. Chim. Acta. 510, 455–458. doi:10.1016/j.cca.2020.08.008
Richardson, S., Hirsch, J. S., Narasimhan, M., Crawford, J. M., McGinn, T., Davidson, K. W., et al. (2020). Presenting Characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York city area. Jama 323 (20), 2052–2059. doi:10.1001/jama.2020.6775
Rieder, M. J., Taylor, S. L., Clark, A. G., and Nickerson, D. A. (1999). Sequence variation in the human angiotensin converting enzyme. Nat. Genet. 22 (1), 59–62. doi:10.1038/8760
Rigat, B., Hubert, C., Alhenc-Gelas, F., Cambien, F., Corvol, P., and Soubrier, F. (1990). An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J. Clin. Invest. 86 (4), 1343–1346. doi:10.1172/JCI114844
Rostamian, A., Ghazanfari, T., Arabkheradmand, J., Edalatifard, M., Ghaffarpour, S., Salehi, M. R., et al. (2020). Interleukin-6 as a potential predictor of COVID-19 disease severity in hospitalized patients and its association with clinical laboratory routine tests. Immunoregulation 3 (1), 29–36. doi:10.32598/immunoregulation.3.1.4
Saad, H., Jabotian, K., Sakr, C., Mahfouz, R., Akl, I. B., and Zgheib, N. K. (2021). The role of angiotensin converting enzyme 1 insertion/deletion genetic polymorphism in the risk and severity of COVID-19 infection. Front. Med. 8, 798571. doi:10.3389/fmed.2021.798571
Saadat, M. (2020). No significant correlation between ACE Ins/Del genetic polymorphism and COVID-19 infection. Clin. Chem. Lab. Med. 58 (7), 1127–1128. doi:10.1515/cclm-2020-0577
Sabater Molina, M., Nicolás Rocamora, E., Bendicho, A. I., Vázquez, E. G., Zorio, E., Rodriguez, F. D., et al. (2022). Polymorphisms in ACE, ACE2, AGTR1 genes and severity of COVID-19 disease. PLoS One 17 (2), e0263140. doi:10.1371/journal.pone.0263140
Santa Cruz, A., Mendes-Frias, A., Oliveira, A. I., Dias, L., Matos, A. R., Carvalho, A., et al. (2021). Interleukin-6 is a biomarker for the development of fatal severe acute respiratory syndrome coronavirus 2 pneumonia. Front. Immunol. 12, 613422. doi:10.3389/fimmu.2021.613422
Sattar, N., McInnes, I. B., and McMurray, J. J. V. (2020). Obesity is a risk factor for severe COVID-19 infection: Multiple potential mechanisms. Circulation 142 (1), 4–6. doi:10.1161/CIRCULATIONAHA.120.047659
Sayed-Tabatabaei, F. A., Oostra, B. A., Isaacs, A., van Duijn, C. M., and Witteman, J. C. (2006). ACE polymorphisms. Circ. Res. 98 (9), 1123–1133. doi:10.1161/01.RES.0000223145.74217.e7
Sriram, K., and Insel, P. A. (2020). A hypothesis for pathobiology and treatment of COVID-19: The centrality of ACE1/ACE2 imbalance. Br. J. Pharmacol. 177 (21), 4825–4844. doi:10.1111/bph.15082
Tiret, L., Rigat, B., Visvikis, S., Breda, C., Corvol, P., Cambien, F., et al. (1992). Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am. J. Hum. Genet. 51 (1), 197–205.
Verma, S., Abbas, M., Verma, S., Khan, F. H., Raza, S. T., Siddiqi, Z., et al. (2021). Impact of I/D polymorphism of angiotensin-converting enzyme 1 (ACE1) gene on the severity of COVID-19 patients. Infect. Genet. Evol. 91, 104801. doi:10.1016/j.meegid.2021.104801
Wang, L. (2020). C-reactive protein levels in the early stage of COVID-19. Med. Mal. Infect. 50 (4), 332–334. doi:10.1016/j.medmal.2020.03.007
WHO (2022). Coronavirus (COVID-19) dashboard. Geneva: World Health Organization. Available at: https://covid19.who.int/.
WHO (2021). COVID-19 clinical management: Living guidance, 25 january 2021. Geneva: World Health Organization. Contract No.: WHO/2019-nCoV/clinical/2021.1.
Yamamoto, N., Ariumi, Y., Nishida, N., Yamamoto, R., Bauer, G., Gojobori, T., et al. (2020). SARS-CoV-2 infections and COVID-19 mortalities strongly correlate with ACE1 I/D genotype. Gene 758, 144944. doi:10.1016/j.gene.2020.144944
Yang, W., Huang, W., Su, S., Li, B., Zhao, W., Chen, S., et al. (2006). Association study of ACE2 (angiotensin I-converting enzyme 2) gene polymorphisms with coronary heart disease and myocardial infarction in a Chinese Han population. Clin. Sci. 111 (5), 333–340. doi:10.1042/CS20060020
Zeberg, H., and Pääbo, S. (2020). The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature 587 (7835), 610–612. doi:10.1038/s41586-020-2818-3
Zhao, Q., Gu, D., Kelly, T. N., Hixson, J. E., Rao, D. C., Jaquish, C. E., et al. (2010). Association of genetic variants in the apelin-APJ system and ACE2 with blood pressure responses to potassium supplementation: The GenSalt study. Am. J. Hypertens. 23 (6), 606–613. doi:10.1038/ajh.2010.36
Keywords: polymorphism, insertion/deletion, angiotensin-converting enzyme (ACE), coronavirus disease 2019 (COVID-19), severity, SNP
Citation: Faridzadeh A, Mahmoudi M, Ghaffarpour S, Zamani MS, Hoseinzadeh A, Naghizadeh MM and Ghazanfari T (2022) The role of ACE1 I/D and ACE2 polymorphism in the outcome of Iranian COVID-19 patients: A case-control study. Front. Genet. 13:955965. doi: 10.3389/fgene.2022.955965
Received: 29 May 2022; Accepted: 08 August 2022;
Published: 05 September 2022.
Edited by:
Zhongshan Cheng, St. Jude Children’s Research Hospital, United StatesReviewed by:
Elisa Zavattaro, University of Eastern Piedmont, ItalyYu-si Luo, Affiliated Hospital of Guizhou Medical University, China
Copyright © 2022 Faridzadeh, Mahmoudi, Ghaffarpour, Zamani, Hoseinzadeh, Naghizadeh and Ghazanfari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mahmoud Mahmoudi, mahmoudim@mums.ac.ir; Tooba Ghazanfari, Tghazanfari@yahoo.com
 Arezoo Faridzadeh
Arezoo Faridzadeh Mahmoud Mahmoudi
Mahmoud Mahmoudi Sara Ghaffarpour
Sara Ghaffarpour Mohammad Saber Zamani3
Mohammad Saber Zamani3 Akram Hoseinzadeh
Akram Hoseinzadeh Tooba Ghazanfari
Tooba Ghazanfari