- 1College of Animal Science and Technology, Gansu Agricultural University, Lanzhou, China
- 2Tibet Academy of Agricultural and Animal Husbandry Science, Lasa, China
- 3Gansu Research Center for Swine Production Engineering and Technology, Lanzhou, China
Clostridium perfringens type C (Cp) is one of the principal microorganisms responsible for bacterial diarrhea in neonatal and pre-weaning piglets. To better understand the molecular effects of Cp infection, we performed a genome-wide comparison of the changes in DNA methylation and gene expression in Cp infected resistant and susceptible piglets. We characterized the pattern of changes in methylation and found 6485, 5968, and 6472 differentially methylated regions (DMRs) of piglets infected with Cp in IR vs. IC, IS vs. IC, and IS vs. IR groups, respectively. These methylation changes for genes mainly involved in immune and inflammatory responses, cell adhesion, and activation of transcription factors. Gene ontology and KEGG pathway analyses showed that the differentially methylated genes (DMGs) were associated with negative regulation of transcription, apoptotic processes, protein binding, and kinase activity. In addition, they were enriched in immunity-related pathways, such as MAPK signaling pathway, Toll-like receptor signaling pathway, and NF-kappa B signaling pathway. Integrative analysis identified 168, 198, and 7 mRNAs showing inverse correlations between methylation and expression with Cp infection. Altered DNA methylation and expression of various genes suggested their roles and potential functional interactions upon Cp infection, 14 immune-associated mRNAs with differential methylation and transcriptional repression were identified in IS vs. IR, commonly revealing that the differentially expressed genes (DEGs) LBP, TBX21, and LCN2 were likely involved in the piglets against Cp infection. The present results provide further insight into the DNA methylation epigenetic alterations of C. perfringens type C infected piglet ileum tissues, and may advance the identification of biomarkers and drug targets for predicting susceptibility to and controlling C. perfringens type C-induced piglet diarrhea.
Introduction
Clostridium perfringens (C. perfringens) type C (Cp) frequently causes the severe, acute, and lethal necrotic enteritis (NE) in humans and livestock (Rood and Cole 1991; Lyras and Rood 2014), such as calves, sheep, goats, and pigs (Songer and Uzal 2005), especially in newborn piglets (Petit et al., 1999; Rood et al., 2018). Newborn piglets from the birthday until 3 weeks of age are highly susceptible to the clostridia because of their incompletely developed intestinal immune system, leading to mortality rates up to 100%. The Cp infection spreads rapidly and becomes an important problem worldwide (Posthaus et al., 2020).
C. perfringens type C beta (CPB) toxin is the essential virulence factor (Sayeed et al., 2008; Vidal et al., 2008; Uzal et al., 2009). Usually, the colonization and rapid proliferation of Cp intruded into the piglet’s incompletely developed intestine forebode the start of NE disease. Due to trypsin inhibitors in the prevention of degradation of CPB, secretions of CPB toxin leads to initial epithelial damage or irritation, the toxin-induced intestinal damages rapidly cause an increase in permeability of vessels in the lamina propria, even the epithelial barrier further disrupted. The change of luminal environment causes acceleration of toxin production, which is absorbed into the systemic circulation and further causes hemorrhage, tissue necrosis, and even enterotoxemia (Posthaus et al., 2020). The Cp diseases can occur in acute and subacute-to-chronic forms (Hogh 1967; Chean et al., 2014). Piglets with acute disease usually appear with several symptoms such as depression, hemorrhagic diarrhea, and death mainly within the first 3 d after birth (Songer and Uzal 2005). Piglets with more protracted subacute-to-chronic clinical features almost have non-hemorrhagic diarrhea, and appear with hypo-immunity, growth reduction, and emaciation (Songer and Uzal 2005). Stoy et al. found that C. perfringens type A infection led to the increased expressions of inflammatory related genes (CCL5, NFKBIA, IL8, IL1RN, and TNFAIP3) of IPEC-J2 cells, and total count and densities of bacteria were markedly high in pigs NE, showing the significant positive correlation with disease severity (Stoy et al., 2015).
DNA methylation is one of the central epigenetic modifications; in mammalian genomes, it mainly occurs on cytosines at position C5 in CpG dinucleotides (Wang et al., 2019). DNA methylation always participates in numerous immunity and physiology processes, such as genomic imprinting, transcriptional regulation, growth, and developmental, immune, and inflammation regulation (Schübeler 2015). The methylation state normally dynamically changes and serves to regulate expressions of the responsive genes during host responses to environmental stimuli of pathogen infection, drug treatment, pollutants, and immune and inflammatory diseases (Kiga et al., 2014; Jiang et al., 2018; Swathy et al., 2018; Chen et al., 2019). Generally, the DNA occurrence of promoter methylation is often accompanied by transcription inhibition (Lee and Bartolomei 2013). Hypomethylation can promote the increase of transcriptional activity (Lluis and Cosma 2013), abnormal methylation can cause many diseases. Research has reported that bacterial endotoxins have profound impacts on gene expression in intestinal epithelial cells through DNA methylation modifications. The expressions of FUT1 (Dai et al., 2017) and FUT2 (Wu et al., 2018) were epigenetically modulated by DNA methylation of their promoters, regulating ETEC F18 resistance in weanling piglets. Other studies also addressed the impact of infection and LPS on the DNA methylation status of immune cells. In human macrophages, LPS-induced specific methylation changes lead to inactivation of pro-inflammatory pathways (Novakovic et al., 2016). However, systematic investigations on the global DNA methylation changes induced by C. perfringens type C infection and the methylation pattern of responsive genes are still scant.
This study aimed to explore the genomic regions and distribution of DNA methylation in piglet ileum tissues exposed to Cp infection and screened the potential DNA methylation targets for piglets against Cp infection, combined with RNAseq data of our previous study (Huang X. et al., 2019). This study comprehensively analyzed the effects of RRBS DNA methylome level and transcriptome level, and provided new insights in Cp-induced piglet diarrhea disease, which may contribute to the identification of biomarkers for diarrhea resistance against Cp infection.
Materials and Methods
Animal Experiment
Bacterial culture, feeding, and management of piglets were in accordance with the description of Huang et al. (Huang X. et al., 2019; Huang X. Y. et al., 2019); the details were as follows: Cp strain (CVCC 2032; China Veterinary Culture Collection Center) was anaerobic shaking cultured 16 h at 37°C in the bouillon culture-medium (HopeBio, Qingdao, China), and an expected concentration of 1 × 109 CFU/ml Cp medium was used to inoculate piglets orally.
The 30 7-day-old experimental piglets (Landrace male × Yorkshire female) tested seronegative for Escherichia coli (E. coli), Salmonella, and C. perfringens by commercial enzyme-linked immunosorbent assay (ELISA) kits (Jiancheng Bioengineering Institute, Nanjing, China) from Dingxi City, Gansu Province of China. Then 25 piglets were randomly orally challenged with 1 ml 1 × 109 CFU/ml Cp medium for 5 consecutive days, the 5 remaining piglets were the control group (IC), and all piglets were housed separately and isolated in climate-controlled and fully air-conditioned, receiving water and diets ad libitum (Huang X. et al., 2019; Huang X. Y. et al., 2019).
During Cp infection, summing and ranking total scores of each piglet, according to fecal consistency from 0 to 3 grade (Kelly et al., 1990), meanwhile combining with the clinical signs, the top 5 piglets with the highest and lowest fecal scores were designated as susceptibility (IS) and resistance (IR) groups, respectively. Piglets of IR, IS, and IC groups were humanly slaughtered under barbiturate anesthesia. The ileum tissues were collected to extract DNA (Supplementary Figure S1).
Nucleic Acid Isolation
According to the manufacturer’s instruction, genomic DNA and total RNA were exacted by using the QIAamp Fast DNA Tissue Kit (Qiagen, Dusseldorf, Germany) and TRIzol reagent (Invitrogen, United States). Qualities and integrities of RNA extracts were assessed using the NanoPhotometer® spectrophotometer (Thermo Scientific, United States) and by 1% agarose gel electrophoresis with RNA Nano6000 Assay Kit of the Bionalyzer 2100 system (Agilent Technologies, United States), which were used for library preparation and subsequent analysis.
Library Preparation and RRBS Sequencing
The 200 to 1,000 bp in length fragmented DNA samples by MspI enzyme (NEB, United States) were then subjected to bisulfite conversion for converting any unmethylated cytosine to Uracil by EZ DNA Methylation-Gold™ Kit (Zymo Research, United States). Further, the Accel-NGS Methyl-Seq DNA Library Kit (Swift, MI) was utilized for attaching adapters to single-stranded DNA fragments. Bead-based SPRI clean-ups were used to remove both oligonucleotides and small fragments, as well as changing enzymatic buffer composition. Finally, we performed the pair-end 2 × 150 bp sequencing on an Illumina Hiseq 4000 platform housed in the LC Sciences (Hangzhou, China).
Data Normalization
Sequencing reads that contained adapter contamination, low quality bases, and undetermined bases were removed using Cutadapt and perl scripts. Quality control was verified using FastQC v0.11.4 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) (Cock et al., 2010), then reads were mapped and aligned to Sus scrofa 11.1 reference genome using Bismark v0.22.1 (Krueger and Andrews 2011). Further, for each cytosine site (or guanine corresponding to a cytosine on the opposite strand) in the reference genome sequence, the DNA methylation levels were determined by the ratio of numbers of reads supporting C (methylated) to that of total reads (methylated and unmethylated) using per scripts in house and MethPipe (Song et al., 2013). Analysis of differentially methylated regions (DMRs) was calculated by R package-MethylKit (Akalin et al., 2012) with default parameters (1000 bp slide windows, 500 bp overlap, p value < 0.05).
The Conjunctive Analysis of RRBS and RNA-Seq
Gene promoter DNA methylation usually inhibits gene expression. To explore the effect of DNA methylation on gene expression during piglets suffering from Cp infection, we also had conjunctively analyzed the negative relationship between promoter DMGs and differentially expressed genes (DEGs). Overlapping analysis was performed for DMGs and DEGs, screening negative correlation between gene expression levels and methylation levels by Pearson correlation analysis. The methylation level of DMGs (|ΔMethylation| > 5%) and expression level of DEGs (|Δexpression log2FC| > 1) both with the p < 0.05 were selected to perform the conjoint analysis.
Bioinformatics Analyses
Gene ontology (GO) analysis of DMG-associated genes was performed using DAVID functional annotation tool (Huang et al., 2009). All annotated genes in Sus scrofa genome were used as background for GO analysis. Pathway enrichment analysis was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database available within the DAVID platform, and with WikiPathways database (https://www.wikipathways.org/) (Martens et al., 2021).
Data Statistics and Analysis
The t-test analysis and one-way ANOVA analysis were used to analyze the significance of diarrhea scores and fecal bacteria shedding of piglets in the IR, IS, and IC groups through SPSS 21.0 software. All values were expressed by mean ± standard error (M±SE), the p < 0.05 means the significant difference, p < 0.01 means the obviously significant difference.
Results
Diarrhea Scores and Fecal Bacteria Shedding of Piglets in the IR, IS, and IC Groups
The average diarrhea score and total diarrhea score of piglets among the IR, IS, and IC groups were statistically analyzed, and the results are shown in Table 1. The average diarrhea scores of piglets in the IR and IS groups were 2.01 ± 0.26 and 2.68 ± 0.04, respectively, which were significantly higher than that in the IC group (0.61 ± 0.02) (p < 0.01). The total diarrhea scores of piglets in the IR and IS groups were 37.60 ± 0.87 and 67.6 ± 1.21, respectively, which were significantly higher than that in the IC group (11.40 ± 0.51, p < 0.01). These results suggested that the average diarrhea scores and total diarrhea scores of the IR and IS groups were significantly increased after Cp infection (p < 0.01).
The numbers of piglet feces Cp in the IR, IS, and IC groups were also counted, which are shown in Figure 1. As increasing times of Cp infection, the numbers of Cp in feces of piglets in the IR and IS groups were significantly increased, meanwhile, the numbers of feces Cp of piglets in the IS group were also significantly higher than those in the IR group (p < 0.05), which were both significantly higher than those in the IC group (p < 0.01). The results showed that Cp infection led to increasing numbers of feces bacteria of piglets, the tolerant piglets in the IR group had stronger resistance to bacterial infection, showing numbers of feces bacteria shedding obviously less than those in the IS group.
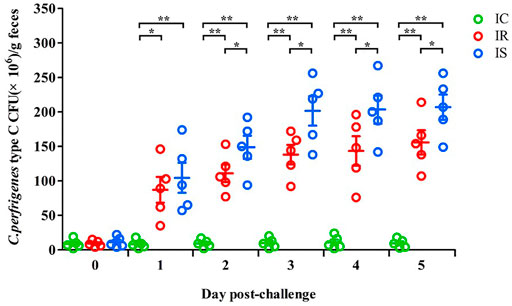
FIGURE 1. The fecal shedding levels of piglets in the IR, IS, and IC groups after Cp infection. Note: Fecal CFUs were determined by plate count method. The horizontal line represents the mean. Green circles represent the IC group, red circles represent the IR group, and blue circles represent the IS group. An asterisk denotes a significant difference (*p < 0.05, **p < 0.01).
DNA Methylation Profiles of RRBS
Genome DNA methylation profiles of quintuplicate samples of IR, IS, and IC groups were analyzed. Overall, RRBS yielded an amount of 39–49 million reads per sample. After quality filtering, ranging from 59.6–65.63%, average 61.23% of reads were successfully aligned to the Sus scrofa 11 reference genomes. In total, we had identified 1.7–2.2 million CpG sites per sample, of which, the average 1.96 million were covered in all samples, representing 7.9% of total numbers of CpGs in the Sus scrofa genome. Raw sequencing data and mapping statistics are summarized in Supplementary Table S1.
To detect the DNA methylome changes induced by Cp infection, we compared Cp infected and uninfected piglets to identify methylated enrichment peaks (MEPs) in genomic DNA (Supplementary Table S2). Statistically, compared to the IC group, there were 157,833 MEPs identified in the IR group (p < 0.05), of which 110,874 were hypermethylated and 46,959 were hypomethylated (Supplementary Table S3), and 150,128 MEPs were identified in the IS group (p < 0.05), of which 79,818 were hypermethylated and 70,310 were hypomethylated (Supplementary Table S4). Mentionable, compared to the IR group, 160,738 MEPs were identified in the IS group (p < 0.05), of which 52,995 were hypermethylated and 107,743 were hypomethylated (Supplementary Table S5).
The chromosomal distribution of the methylated peaks was determined to assess whether methylation was associated with specific chromosomal features. As shown in the methylation map (Figure 2), the distribution of identified methylations almost covered all chromosomal, the methylation density in these regions was distinct among the chromosomes, chromosome MT (mitochondria) in particular, contained a relatively large unmethylated region among samples, which was related to the different degrees and correlation between methylation profiles of the infected IR and IS groups and the uninfected IC group.
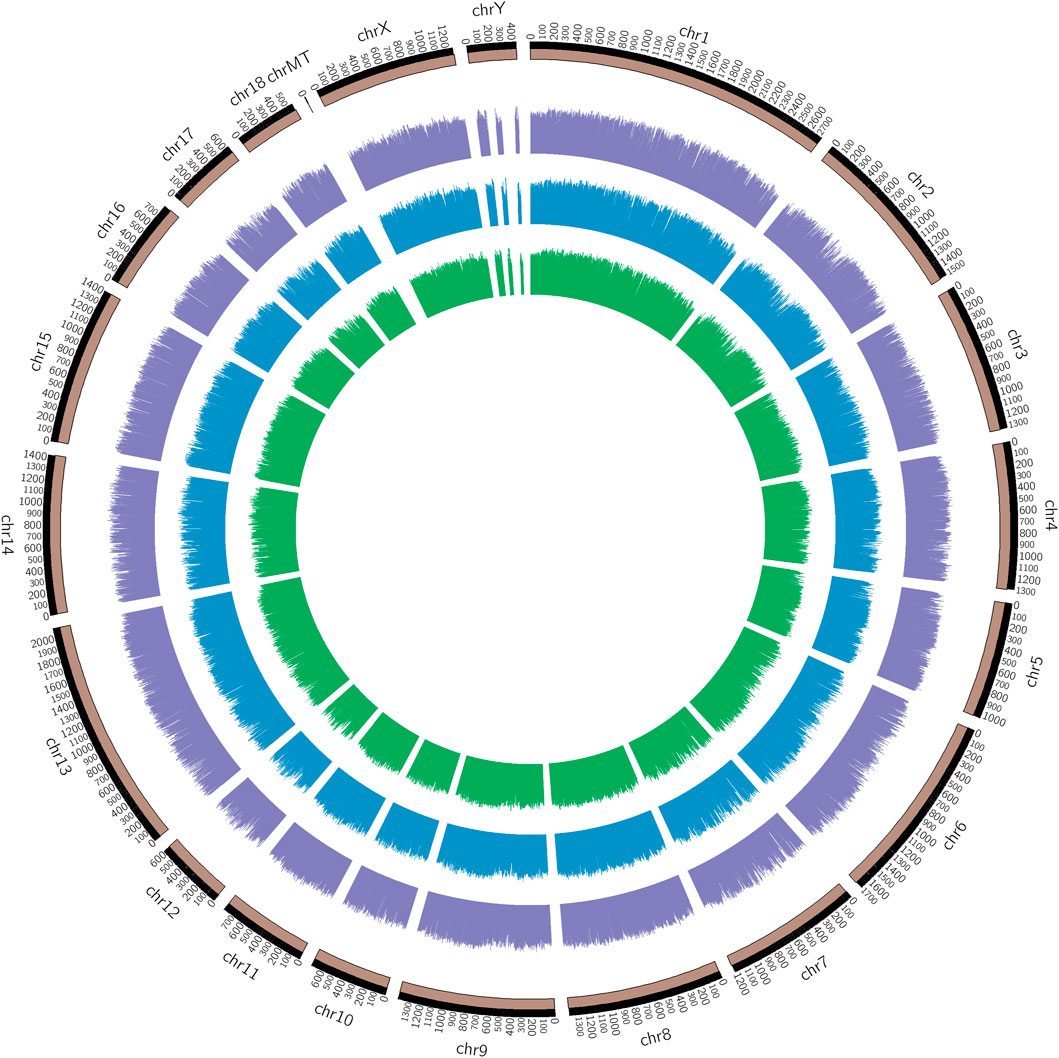
FIGURE 2. Genomic distribution of DMRs among Cp infected and uninfected piglets. Note: From the inner circle to the outer circle represents the IR vs. IC, IS vs. IC, and IS vs. IR groups.
Based on the CpG ratio, GC content, and length of the CpG-rich region, we divided gene promoters into three types: High-density CpG promoter (HCP), low-density CpG promoter (LCP), and intermediate density CpG promoter (ICP) (Yu et al., 2013). The information of MEPs in three comparative groups were shown and revealed the relatively uneven distribution across the genome. The majority of MEPs were in the intergenic regions, following in the exon and intron, lowest in the promoter (Table 2).
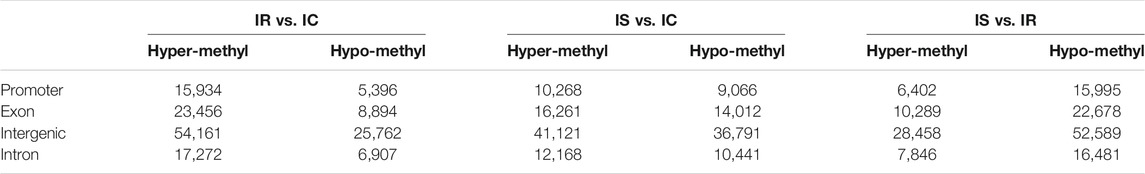
TABLE 2. Analysis and distribution of the significant methylated genes of the IR, IS, and IC groups.
Then distribution of the MEPs in three types of promoter were analyzed. We found that the numbers of MEPs were obviously increased in the IR and IS groups after C. perfringens type C infection, which included MEP in LCP types, followed by in HCPs and ICPs. It is worth attention that in promoter CGIs, HCPs had more MEPs in the IS vs. IR group than ICPs or LCPs (Table 2).
Methylation Status of Genome CpG Islands
CpG islands (CGIs) obtained particular attention and interest for their role in controlling gene expression through epigenetic modification. We grouped the CGIs into four classes according to their distance to the RefSeq genes: promoter CGIs [from about −2 kb to +0.5 kb around the transcription start site (TSS)], exon and intron CGIs [from +0.5 kb around the TSS to the transcription terminal site (TTS)], and intergenic CGIs (about 2 kb to those that do not fall into either the promoter or the intragenic group) (Figure 3A).
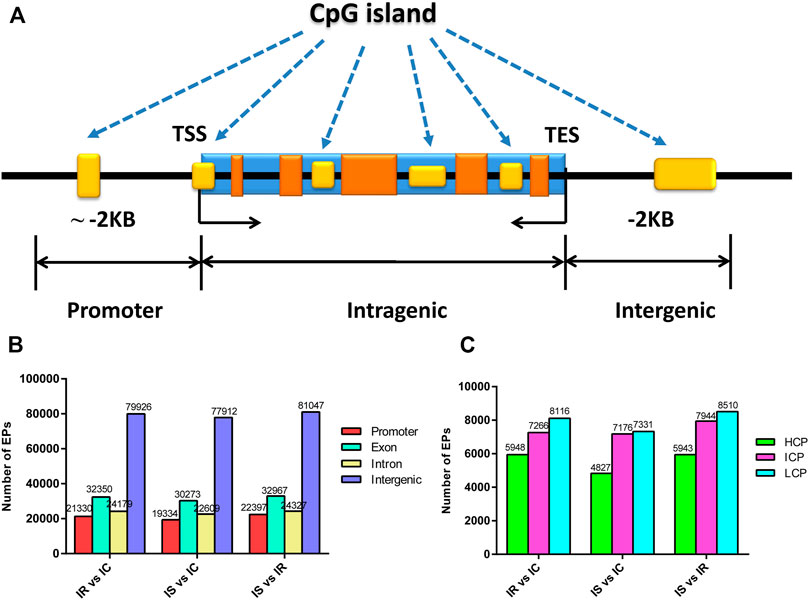
FIGURE 3. Distribution of DNA methylation enrichment peaks in piglet ileum infected with C. perfringens type C infection. (A) Generic diagram showing CpG islands (CGIs) relative to gene transcript regions. (B) Numbers of MEPs in the CGI region of IR, IS, and IC groups. (C) Numbers of MEPs in the promoter CGIs. Note: HCP represents high CpG density promoter; ICP represents intermediate CpG density promoter; LCP represents low CpG density promoter.
We also analyzed the CpG methylation status of different gene segments in the piglet ileum after Cp infection. The numbers of MEPs in the four classes of CGIs and CGIs shore among IR, IS, and IC groups are shown in Figures 3B,C. Most MEPs were distributed in the intergenic CGIs among IR, IS, and IC comparative groups. It was worth mentioning that more CGI methylations happened in the IR vs. IC group (Supplementary Table S6) and the IS vs. IC group (Supplementary Table S7) than in the IS vs. IR group (Supplementary Table S8) (p < 0.05). Importantly, for the promoter CGIs, intergenic CGIs and 3′ transcript CGI, the numbers of hyper MEPs were also higher than hypo MEPs in the IR vs. IC group and the IS vs. IC group, which was opposite to those in the IS vs. IR group (Figure 3B; Table 3).
Identification of Differentially Methylated Genes
To explore the DMGs of piglet ileum tissues induced by Cp infection, we subsequently mapped all DMRs to their nearest genomic features and analyzed the DMGs located in promoter and CGI regions. Compared to the IC group, there were 6,485 DMGs having one or more DMRs including 5,186 mRNAs, 14 miRNA, and 989 pseudogenes in the IR group (Supplementary Table S6), 5,968 DMGs including 4,819 mRNAs, 6 miRNA, and 886 pseudogenes in the IS group (Supplementary Table S7), meanwhile, compared to the IR group, 6,472 DMGs including 5,214 mRNAs, 14 miRNA, and 958 pseudogenes in the IS group (Supplementary Table S8). Except for miRNA and pseudogenes, DMGs almost contained an average of 4 DMRs in the gene body and promoter regions, mainly distributed in CGI and CGI shore (Figure 4). In total, 2607 DMGs were detected among the IR vs. IC, IS vs. IC, and IS vs. IR comparative groups (Figure 5).
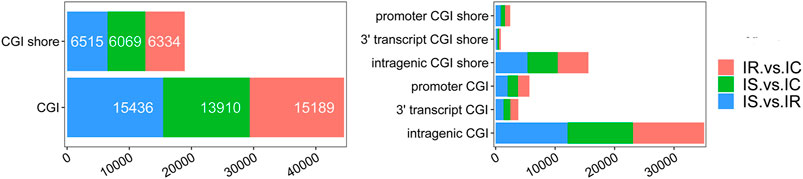
FIGURE 4. The details of CGIs and CGI shores distributed in the IR vs. IC, IS vs. IC, and IS vs. IR groups.
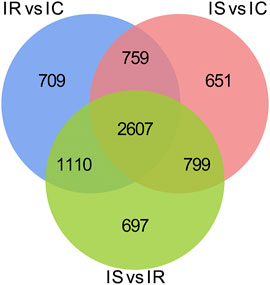
FIGURE 5. Venn distribution of the differentially methylated mRNAs with CGIs in the IR vs. IC, IS vs. IC, and IS vs. IR groups. GO and KEGG signaling pathway analyses.
In order to further characterize DMGs, gene ontology and KEGG signaling pathway analyses were carried out. GO analysis revealed 46, 35, and 38 GO terms significantly enriched in the IR vs. IC, IS vs. IC, and IS vs. IR groups (Supplementary Table S9), respectively. We found that Cp infection caused the DMGs mainly enriched in the biological and molecular functions, such as signal transduction, ion channel activity, nucleotide binding, protein transferase activity, immune response, cytoskeleton, GTPase activator activity, histone deacetylase complex, inactivation of MAPK activity, and nucleotide immunoglobulin production (Figure 6).
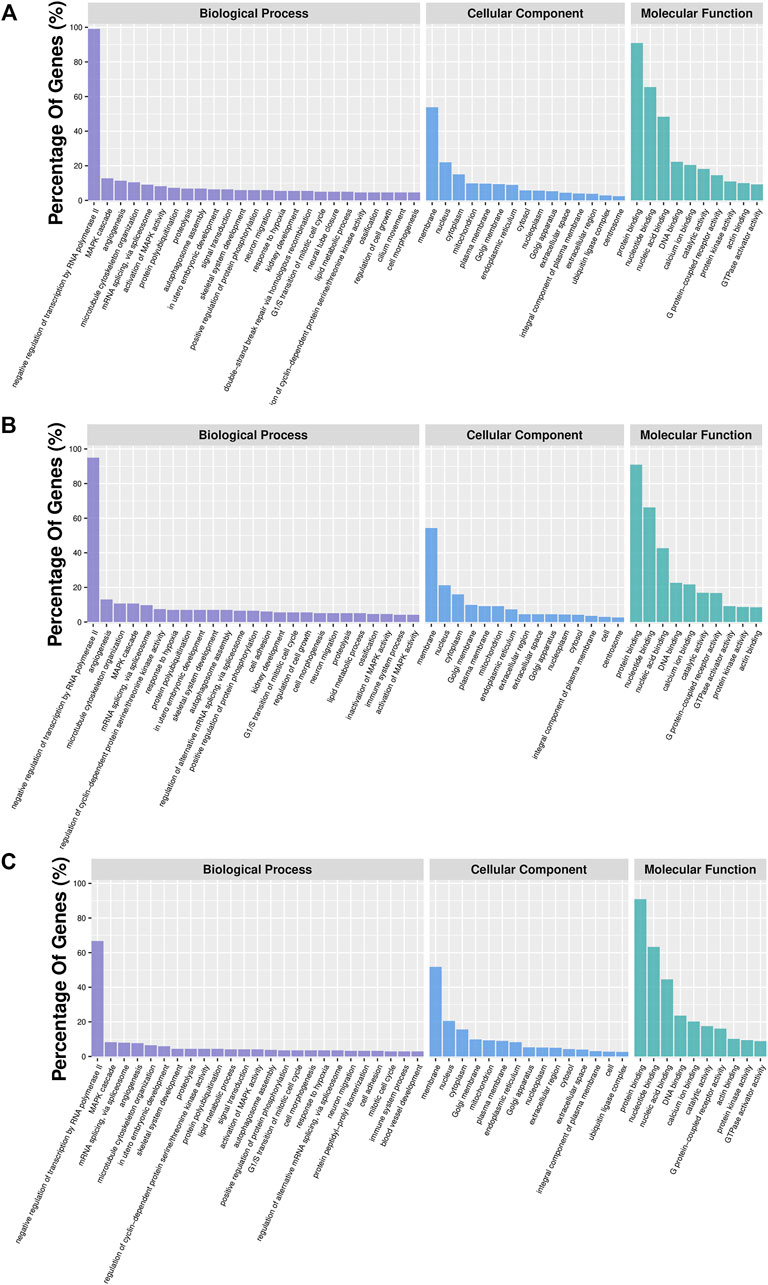
FIGURE 6. GO analyses of DMGs located in the vicinity of significant differentially methylated regions (DMRs) of the IR vs. IC (A), IS vs. IC (B), and IS vs. IR groups (C). Bar plots display enriched GO terms. The plots show significantly enriched degrees (p < 0.05).
Among the KEGG pathway enrichment, a total of 23, 25, and 29 pathways were significantly enriched in the IR vs. IC, IS vs. IC, and IS vs. IR groups (Supplementary Table S10), respectively. The DMRs were enriched by MAPK signaling pathway, NF-kappa B signaling pathway, tight junction, chemokine signaling pathway, calcium signaling pathway, lysosome, PI3K-Akt signaling pathway, autophagy, Toll-like receptor signaling pathway, ECM-receptor interaction, and several metabolism pathways (Figure 7).
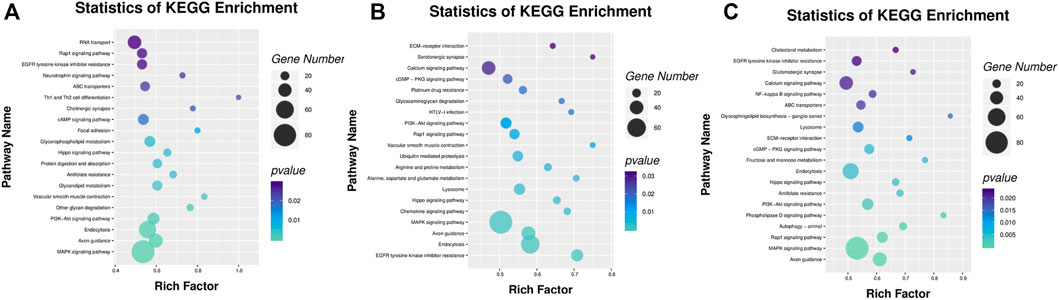
FIGURE 7. KEGG signal pathway analyses of DMGs located in the vicinity of significant differentially methylated regions (DMRs) of the IR vs. IC (A), IS vs. IC (B), and IS vs. IR groups (C). Bar plots display enriched KEGG pathways. The plots show significantly enriched degrees (p < 0.05).
Correlation Analysis Between DNA Methylation and Gene Expression
DNA methylation of gene promoters is usually involved in inhibiting the expression level of the corresponding genes (Jones 2012). The potential effects of DNA methylation on gene expression were characterized by comparing methylation and RNA expression data (Guo et al., 2019). By analyzing a wide association between transcriptome gene expression and epigenome DNA methylation (in promoter or body), we explored the relationship between methylation changes at the promoter regions and gene expression changes (our previous study) of the IR, IS, and IC groups (Huang X. et al., 2019; Huang X. Y. et al., 2019).
Usually, there is the significant negative association between mean methylation levels of promoter regions and expressions of mRNAs. According to methylation levels of DMGs (|ΔMethylation| > 5%) and expression levels of DEGs (|Δexpression log2FC| > 1) both with the p < 0.05, there were 168 mRNAs, 198 mRNAs, and 7 mRNAs screened in the IR vs. IC, IS vs. IC, and IS vs. IR groups, respectively (Supplementary Table S11). Importantly, to further reveal the functions of epigenetics and transcriptomics during piglets’ resistance to Cp infection, we also analyzed the immune-associated mRNAs in the IS vs. IR group and constructed a differential methylation and transcription network (Figure 8). In this network, the screened mRNAs mainly belonged to T cell receptor, Toll like receptor, NF-κB, MAPK, JAK-STAT, IL-17, Th1, and Th2 cell differentiation signaling pathways, such as NFKB1, MAPK14, TRIM25, TLR6, IL21R, LBP, IRF7, TBX21 and so on. Moreover, there were 14 mRNAs, which promoter methylations were obviously inversely related to transcriptional repressions, commonly identified (Table 4), in which LCN2 with down-expressed and hypomethylated, AKT2 with up-expressed and hypermethylated, were also identified in the IR vs. IC and IS vs. IC groups.
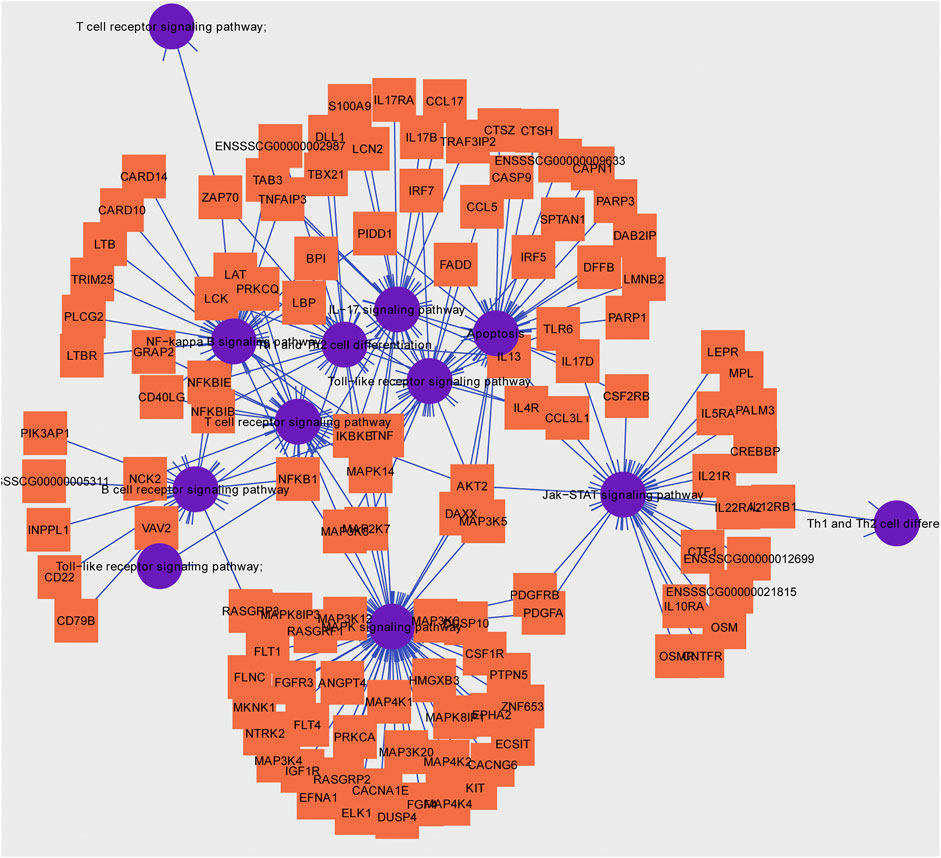
FIGURE 8. Hub pathways and associated genes in piglets of the IS vs. IR group suffering from C. perfringens type C infection.
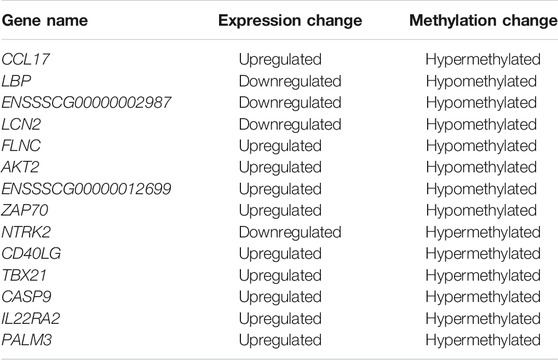
TABLE 4. List of differentially methylated and expressed mRNAs in C. perfringens type C infected piglets of the IR vs. IS group.
Discussion
In this study, we have revealed the epigenetic alterations in piglet ileum tissues due to infection by Cp from a genome-wide comparative methylome analysis. However, it may only provide limited insights into the biological mechanisms of piglet diarrhea induced by Cp infection. Generally, complementary effects and synergistic interactions between omics in life science can be captured by integrative studies of multiple molecular layers. Building upon the successes in single-omics research, a better understanding of the molecular functions and disease etiologies by multi-omics integrated approaches from different omic levels (e.g., genetics, epigenetics, mRNA transcripts, proteins, and metabolites), as well as their interrelations and combined influences on the disease processes (Sun and Hu 2016). Therefore, we further conducted an integrated analysis of RRBS and RNA sequencing data and identified a subset of genes that was implicated in the piglet response to Cp infection.
The host immune response is crucial for defense against microbial pathogens, it is not different to that found in the complex process of host-pathogen interactions, the genomic expression pattern and program of host reflects responses to pathogens and virulence (Boldrick et al., 2002). Recently, a study reported that epigenetic modulations such as host DNA methylation could be manipulated to influence the host’s gene expressions in response to defensing pathogens infection (Paschos and Allday, 2010). In addition, the changes and differences of DNA methylation presumably largely reflected the abilities of host epigenetic responses involved in the immune system against or triggers by pathogens (Tarakhovsky 2010). In this study, we have revealed the epigenetic alterations in piglet ileum tissues due to infection by Cp from a genome-wide comparative methylome analysis.
Our study found that the methylated peaks DNA methylation almost covered most chromosomal regions and presented obviously distinct methylation densities (Figure 2), indicating that Cp infection could trigger changes of DNA methylation of candidates. These changes may be correlated with piglets responding to Cp infection. Analysis of differential methylation genes revealed that 6635 DMGs were identified in piglets after Cp infection, in which 3,366 DMGs were identified both in the IR and IS groups (Figure 5), suggesting that the common DMGs identified in piglets of the IR and IS groups had an amount of similar methylation patterns during Cp infection, and further altered expressions of immune-related genes. Meanwhile, 697 DMGs especially identified in the IS vs. IR group (Figure 5), illustrating Cp-infected tolerant and sensitive piglets appeared with different methylation patterns induced by the Cp strain. These identified genes involved in the immune system and bacterial infection have been well studied. For example, JAK1, JAK2, and JAK3, found in three comparable groups, were the prototypical members of the JAK family and could play an essential signaling role for cytokines and interferons involved in immunity and antiviral responses (Ferrao et al., 2018). RASGRF1, found in IS vs. IC and IS vs. IR comparative groups, mainly participated in the Ras signaling pathway (Manyes et al., 2014). AIRE was one of the important regulators of peripheral T cell homeostasis, and played a certain role in control of intestinal tolerance (Kekalainen et al., 2015). Activation of AKT2 was considered to protect mice from defense against Salmonella enterica Typhimurium infection (Kum et al., 2011), even blocking the development of intestinal inflammatory disease (Liu et al., 2019). These differential DMGs may be the crucial resistance candidates of piglet diarrhea.
The candidate mRNAs modified by differential DNA methylation merit greater attention. Further, combining the promoter methylation gene and its mRNA expression level, we had constructed the networks with differential methylations of DMGs and differential expressions of DEGs (Supplementary Table S11), and screened many immune related mRNAs, S100A9, TARBP2, TRIM25, TBX21, LBP, TLR6, involving the piglets resistant to Cp-induced diarrhea disease through defense-related signaling pathways of T cell receptor, Toll-like receptor, NF-κB, MAPK, and so on. Furthermore, these screened mRNAs’ methylation of their promoters was inversely related to transcriptional repression in piglets after Cp infection. S100A9 gene is considered to play crucial functions in participating innate immunity and mediating the inflammatory response during infection-induced host inflammation (Ometto et al., 2017). Researchers found S100A8/A9 recombinant attenuated bacterial adherence and invasion (Wang et al., 2018), and inhibited growth of multiple species, including E. coli, S. aureus, Salmonella typhimurium, and C. perfringens type C (Wang et al., 2018; Huang X. et al., 2019; Wang et al., 2020). However, the expression of S100A9 gene was actually controlled by the methylation status of its promoter (Chandra et al., 2018). In our study, S100A9 gene was down-expressed in transcription and hypermethylated in methylation, and was identified in the Cp infected IR and IS groups, hinting that the hypermethylation S100A9 gene caused the down expression, which may play some roles in protecting piglets from Cp-induced diarrhea resistance.
Transactivation response element RNA-binding protein TARBP2 inhibits the catalytic activity of interferon-induced double-stranded RNA (dsRNA)-activated protein kinase (PKR) to regulate stress-induced signaling pathways during viral infections and cell stress, and TARBP2 is also a regulator of microRNA biogenesis and cellular stress response (Ukhueduan et al., 2021). TARBP2 has been characterized as a key regulator for promoting or inhibiting cell proliferation and invasion. Research has shown that the TARBP2 suppresses IFN-beta production and the innate antiviral response, especially in regulating the antiviral signaling pathways of the innate immune system (Ling et al., 2019). TARBP2 gene, differential expressed and differential DNA methylated, was identified in Cp infected piglets of the IR and IS groups, which may play some important roles in piglet diarrhea resistance.
To further explore the susceptibility and tolerance of piglet resistance to Cp-induced diarrhea, we screened the potential resistant candidates from the IR and IS groups by integrating data of DNA methylation and mRNA expression (Figure 8). There were 14 mRNAs for which methylation of their promoters obviously inversely related to transcriptional repression were identified (Table 4), such as LCN2, TBX21, and LBP. LCN2 with up-expressed and hypermethylated was identified both in the IR and IS groups after Cp infection. The changes in its promoter methylation and gene expression indicated that LCN2 may participate in immune response to C. perfringens type C stimulation. We also constructed a network of immune-associated mRNAs both different changes in methylation and transcription in the IS vs. IR group (Figure 8), we speculated that these genes may be functionally linked and regulated by promoter methylation level of piglets in response to C. perfringens type Cp-induced diarrhea.
TBX21 is an important transcription factor of adaptive immunity that regulates the Th1/Th2 balance and increasing evidence has pointed to the critical roles in regulating innate immunity, cytokine balance, immune dysregulation, and bacterial infection. A study reported that pneumococcus upregulated TBX21 expression in the respiratory epithelium, knocking down TBX21 suppressed pneumococcus-induced TLR2 expression (Woo et al., 2014), and a change of TBX21 may lead to the dysregulation of type I interferon pathways and T cell pathways (Gourh et al., 2009). These studies have proved the adaptive immune regulator TBX21 participated in regulating host innate immune responses during pathogenic bacterial infections. In this study, differential methylated TBX21 was identified in piglets in the IS and IR groups after Cp infection, and it was also found that up-regulated and hypermethylated TBX21 in the IS vs. IR comparative group suggests that hypermethylated TBX21 gene may cause the down expression in the transcriptional level, while the up-expression of TBX21 gene may be regulated by other factors, which commonly participate in the process of piglet resistance to Cp-induced diarrhea.
LPS-binding protein (LBP) is a plasma protein that transfers LPS to the cell surface CD14 presented on the myeloid lineage, playing the crucial roles in the host innate immune response during the development of inflammatory and infectious-related diseases (Meng et al., 2021). LBP is also essential to control bacterial infection. Recently, LBP has been shown to potentiate the host immune responses to LPS invade, relieving Salmonella typhimurium or E. coli induced generation of reactive oxygen species in host macrophages (Sclutt et al., 1999). The LPS-LBP complex can bind to a receptor complex (CD14, MD2, and TLR4) for initiating signal cascade and triggering the secretion of pro-inflammatory cytokines (Gabarin et al., 2021; Won et al., 2021). While accomplishing either blocking LBP binding to LPS or binding LPS/LBP complexes to CD14 can protect the host from LPS-induced toxicity (Le Roy et al., 1999). Therefore, in this study, the hyper methylation of LBP was significantly negative regulation with its down expression in transcription in susceptible piglets for Cp infection, while the tolerance piglets with high expression may maintain balance of the inflammatory response induced by Cp infection. Considering the functions and expression characteristic of these genes, we believe that LBP and TBX21 are strongly associated with piglet diarrhea induced by Cp infection. We speculated that these genes may be functionally linked and regulated by promoter methylation level of piglets in response to Cp-induced diarrhea, which may play some certain roles in protecting piglet resistance of diarrhea caused by bacterial infection.
In conclusion, we have profiled the landscape of DNA methylation of piglets in response to Cp infection, and analyzed the methylation and transcriptome data to further reveal the potential candidates implicated in the piglet immune response. Our findings have provided insight into the molecular effects of DNA methylation of piglet resistance to C. perfringens type C infection, which may contribute to the selection and breeding of piglet diarrhea resistant to C. perfringens type C infection.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Ethics Committee of Gansu Agricultural University and Gansu Research Center for Swine Production Engineering and Technology. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
XH and SG conceived of this study and wrote the manuscript. XH, HS and ZY performed the experiments. QY, PW and ZY performed the data analyses. JL and XS participated in the analysis and interpretation of data. All the authors read and approved the manuscript. All authors reviewed and approved the final manuscript.
Funding
This research was supported by the Young Science Fund, Gansu Agricultural University (GAU-DK-QNJJ-202112).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.803477/full#supplementary-material
Abbreviations
DMRs, differentially methylated regions; E. coli, Escherichia coli; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; mRNA, messenger RNA; RNAseq, RNA sequencing; RRBS, reduced representation bisulfite sequencing.
References
Akalin, A., Kormaksson, M., Li, S., Garrett-Bakelman, F. E., Figueroa, M. E., Melnick, A., et al. (2012). methylKit: a Comprehensive R Package for the Analysis of Genome-wide DNA Methylation Profiles. Genome Biol. 13, R87. doi:10.1186/gb-2012-13-10-R87
Boldrick, J. C., Alizadeh, A. A., Diehn, M., Dudoit, S., Liu, C. L., Belcher, C. E., et al. (2002). Stereotyped and Specific Gene Expression Programs in Human Innate Immune Responses to Bacteria. Proc. Natl. Acad. Sci. 99, 972–977. doi:10.1073/pnas.231625398
Chandra, A., Senapati, S., Roy, S., Chatterjee, G., and Chatterjee, R. (2018). Epigenome-wide DNA Methylation Regulates Cardinal Pathological Features of Psoriasis. Clin. Epigenet 10, 16. doi:10.1186/s13148-018-0541-9
Chean, R., Kotsanas, D., Francis, M. J., Palombo, E. A., Jadhav, S. R., Awad, M. M., et al. (2014). Comparing the Identification of Clostridium Spp. By Two Matrix-Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) Mass Spectrometry Platforms to 16S rRNA PCR Sequencing as a Reference Standard: A Detailed Analysis of Age of Culture and Sample Preparation. Anaerobe 30, 85–89. doi:10.1016/j.anaerobe.2014.09.007
Chen, J., Wu, Y., Sun, Y., Dong, X., Wang, Z., Zhang, Z., et al. (2019). Bacterial Lipopolysaccharide Induced Alterations of Genome-wide DNA Methylation and Promoter Methylation of Lactation-Related Genes in Bovine Mammary Epithelial Cells. Toxins 11, 298. doi:10.3390/toxins11050298
Cock, P. J. A., Fields, C. J., Goto, N., Heuer, M. L., and Rice, P. M. (2010). The Sanger FASTQ File Format for Sequences with Quality Scores, and the Solexa/Illumina FASTQ Variants. Nucleic Acids Res. 38, 1767–1771. doi:10.1093/nar/gkp1137
Dai, C., Sun, L., Xia, R., Sun, S., Zhu, G., Wu, S., et al. (2017). Correlation between the Methylation of the FUT1 Promoter Region and FUT1 Expression in the Duodenum of Piglets from Newborn to Weaning. 3 Biotech. 7, 247. doi:10.1007/s13205-017-0880-9
Ferrao, R. D., Wallweber, H. J., and Lupardus, P. J. (2018). Receptor-mediated Dimerization of JAK2 FERM Domains Is Required for JAK2 Activation. Elife 7, e38089. doi:10.7554/eLife.38089
Gabarin, R. S., Li, M., Zimmel, P. A., Marshall, J. C., Li, Y., and Zhang, H. (2021). Intracellular and Extracellular Lipopolysaccharide Signaling in Sepsis: Avenues for Novel Therapeutic Strategies. J. Innate Immun. 13, 323–332. doi:10.1159/000515740
Gourh, P., Agarwal, S. K., Divecha, D., Assassi, S., Paz, G., Arora-Singh, R. K., et al. (2009). Polymorphisms inTBX21andSTAT4increase the Risk of Systemic Sclerosis: Evidence of Possible Geneâ€"gene Interaction and Alterations in Th1/Th2 Cytokines. Arthritis Rheum. 60, 3794–3806. doi:10.1002/art.24958
Guo, Y., van Schaik, T., Jhamat, N., Niazi, A., Chanrot, M., Charpigny, G., et al. (2019). Differential Gene Expression in Bovine Endometrial Epithelial Cells after challenge with LPS; Specific Implications for Genes Involved in Embryo Maternal Interactions. Plos One 14, e0222081. doi:10.1371/journal.pone.0222081
Hogh, P. (1967). Necrotizing Infectious Enteritis in Piglets, Caused by Clostridium perfringens Type C. II. Incidence and Clinical Features. Acta Vet. Scand. 8, 301–323.
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 4, 44–57. doi:10.1038/nprot.2008.211
Huang, X. Y., Sun, W. Y., Yan, Z. Q., Shi, H. R., Yang, Q. L., Wang, P. F., et al. (2019a). Integrative Analyses of Long Non-coding RNA and mRNA Involved in Piglet Ileum Immune Response to Clostridium perfringens Type C Infection. Front. Cel. Infect. Microbiol. 9, 130. doi:10.3389/fcimb.2019.00130
Huang, X. Y., Sun, W. Y., Yan, Z. Q., Shi, H. R., Yang, Q. L., Wang, P. F., et al. (2019b). Novel Insights Reveal Anti-microbial Gene Regulation of Piglet Intestine Immune in Response to Clostridium perfringens Infection. Sci. Rep. 9, 1963. doi:10.1038/s41598-018-37898-5
Jiang, S., Yan, K., Sun, B., Gao, S., Yang, X., Ni, Y., et al. (2018). Long-Term High-Fat Diet Decreases Hepatic Iron Storage Associated with Suppressing TFR2 and ZIP14 Expression in Rats. J. Agric. Food Chem. 66, 11612–11621. doi:10.1021/acs.jafc.8b02974
Jones, P. A. (2012). Functions of DNA Methylation: Islands, Start Sites, Gene Bodies and beyond. Nat. Rev. Genet. 13, 484–492. doi:10.1038/nrg3230
Kekäläinen, E., Lehto, M.-K., Smeds, E., Pöntynen, N., Pekkarinen, P. T., Ulmanen, I., et al. (2015). Lymphopenia-induced Proliferation in the Absence of Functional Autoimmune Regulator (Aire) Induces Colitis in Mice. Immunol. Lett. 167, 17–22. doi:10.1016/j.imlet.2015.06.010
Kelly, D., O'Brien, J. J., and Mccracken, K. J. (1990). Effect of Creep Feeding on the Incidence, Duration and Severity of post-weaning Diarrhoea in Pigs. Res. Vet. Sci. 49, 223–228. doi:10.1016/s0034-5288(18)31082-8
Kiga, K., Mimuro, H., Suzuki, M., Shinozaki-Ushiku, A., Kobayashi, T., Sanada, T., et al. (2014). Epigenetic Silencing of miR-210 Increases the Proliferation of Gastric Epithelium during Chronic Helicobacter pylori Infection. Nat. Commun. 5, 4497. doi:10.1038/ncomms5497
Krueger, F., and Andrews, S. R. (2011). Bismark: a Flexible Aligner and Methylation Caller for Bisulfite-Seq Applications. Bioinformatics 27, 1571–1572. doi:10.1093/bioinformatics/btr167
Kum, W. W. S., Lo, B. C., Yu, H. B., and Finlay, B. B. (2011). Protective Role of Akt2 in Salmonella enterica Serovar Typhimurium-Induced Gastroenterocolitis. Infect. Immun. 79, 2554–2566. doi:10.1128/iai.01235-10
Le Roy, D., Di Padova, F., Tees, R., Lengacher, S., Landmann, R., Glauser, M. P., et al. (1999). Monoclonal Antibodies to Murine Lipopolysaccharide (LPS)-binding Protein (LBP) Protect Mice from Lethal Endotoxemia by Blocking Either the Binding of LPS to LBP or the Presentation of LPS/LBP Complexes to CD14. J. Immunol. 162, 7454–7460.
Lee, J. T., and Bartolomei, M. S. (2013). X-inactivation, Imprinting, and Long Noncoding RNAs in Health and Disease. Cell 152, 1308–1323. doi:10.1016/j.cell.2013.02.016
Ling, T., Weng, G.-X., Li, J., Li, C., Wang, W., Cao, L., et al. (2019). TARBP2 Inhibits IRF7 Activation by Suppressing TRAF6-Mediated K63-Linked Ubiquitination of IRF7. Mol. Immunol. 109, 116–125. doi:10.1016/j.molimm.2019.02.019
Liu, L., Liang, L., Liang, H., Wang, M., Lu, B., Xue, M., et al. (2019). Fusobacterium Nucleatum Aggravates the Progression of Colitis by Regulating M1 Macrophage Polarization via AKT2 Pathway. Front. Immunol. 10, 15. doi:10.3389/fimmu.2019.01324
Lluis, F., and Cosma, M. P. (2013). Resetting Epigenetic Signatures to Induce Somatic Cell Reprogramming. Cell. Mol. Life Sci. 70, 1413–1424. doi:10.1007/s00018-012-1137-8
Lyras, D., and Rood, J. I. (2014). Preface: ClostPath 2013 Meeting on the Molecular Biology and Pathogenesis of the Clostridia Special Issue. Anaerobe 30, 183. doi:10.1016/j.anaerobe.2014.11.006
Manyes, L., Arribas, M., Gomez, C., Calzada, N., Fernandez-Medarde, A., and Santos, E. (2014). Transcriptional Profiling Reveals Functional Links between RasGrf1 and Pttg1 in Pancreatic Beta Cells. Bmc Genomics 15, 1019. doi:10.1186/1471-2164-15-1019
Martens, M., Ammar, A., Riutta, A., Waagmeester, A., Slenter, D. N., Hanspers, K., et al. (2021). WikiPathways: Connecting Communities. Nucleic Acids Res. 49, D613–D621. doi:10.1093/nar/gkaa1024
Meng, L., Song, Z., Liu, A., Dahmen, U., Yang, X., and Fang, H. (2021). Effects of Lipopolysaccharide-Binding Protein (LBP) Single Nucleotide Polymorphism (SNP) in Infections, Inflammatory Diseases, Metabolic Disorders and Cancers. Front. Immunol. 12, 12. doi:10.3389/fimmu.2021.681810
Novakovic, B., Habibi, E., Wang, S.-Y., Arts, R. J. W., Davar, R., Megchelenbrink, W., et al. (2016). β-Glucan Reverses the Epigenetic State of LPS-Induced Immunological Tolerance. Cell 167, 1354–1368. e1314. doi:10.1016/j.cell.2016.09.034
Ometto, F., Friso, L., Astorri, D., Botsios, C., Raffeiner, B., Punzi, L., et al. (2017). Calprotectin in Rheumatic Diseases. Exp. Biol. Med. (Maywood) 242, 859–873. doi:10.1177/1535370216681551
Paschos, K., and Allday, M. J. (2010). Epigenetic Reprogramming of Host Genes in Viral and Microbial Pathogenesis. Trends Microbiology 18, 439–447. doi:10.1016/j.tim.2010.07.003
Petit, L., Gibert, M., and Popoff, M. R. (1999). Clostridium perfringens: Toxinotype and Genotype. Trends Microbiology 7, 104–110. doi:10.1016/s0966-842x(98)01430-9
Posthaus, H., Kittl, S., Tarek, B., and Bruggisser, J. (2020). Clostridium Perfringenstype C Necrotic Enteritis in Pigs: Diagnosis, Pathogenesis, and Prevention. J. VET. Diagn. Invest. 32, 203–212. doi:10.1177/1040638719900180
Rood, J. I., Adams, V., Lacey, J., Lyras, D., McClane, B. A., Melville, S. B., et al. (2018). Expansion of the Clostridium perfringens Toxin-Based Typing Scheme. Anaerobe 53, 5–10. doi:10.1016/j.anaerobe.2018.04.011
Rood, J. I., and Cole, S. T. (1991). Molecular Genetics and Pathogenesis of Clostridium perfringens. Microbiol. Rev. 55, 621–648. doi:10.1128/mmbr.55.4.621-648.1991
Sayeed, S., Uzal, F. A., Fisher, D. J., Saputo, J., Vidal, J. E., Chen, Y., et al. (2008). Beta Toxin Is Essential for the Intestinal Virulence of Clostridium perfringens Type C Disease Isolate CN3685 in a Rabbit Ileal Loop Model. Mol. Microbiol. 67, 15–30. doi:10.1111/j.1365-2958.2007.06007.x
Schutt, C. (1999). Fighting Infection: the Role of Lipopolysaccharide Binding Proteins CD14 and LBP. Pathobiology 67, 227–229. doi:10.1159/000028097
Schübeler, D. (2015). Function and Information Content of DNA Methylation. Nature 517, 321–326. doi:10.1038/nature14192
Song, Q., Decato, B., Hong, E. E., Zhou, M., Fang, F., Qu, J., et al. (2013). A Reference Methylome Database and Analysis Pipeline to Facilitate Integrative and Comparative Epigenomics. Plos One 8, e81148. doi:10.1371/journal.pone.0081148
Songer, J. G., and Uzal, F. A. (2005). Clostridial Enteric Infections in Pigs. J. VET. Diagn. Invest. 17, 528–536. doi:10.1177/104063870501700602
Støy, A. C. F., Mølbak, L., Delègue, C. L., Thymann, T., Sangild, P. T., Heegaard, P. M. H., et al. (2015). Necrotizing Enterocolitis in Preterm Pigs Is Associated with Increased Density of Intestinal Mucosa-Associated Bacteria Including Clostridium perfringens. Neonatology 108, 188–195. doi:10.1159/000431280
Sun, Y. V., and Hu, Y.-J. (2016). Integrative Analysis of Multi-Omics Data for Discovery and Functional Studies of Complex Human Diseases. Adv. Genet. Vol. 93, 147–190. doi:10.1016/bs.adgen.2015.11.004
Swathy, B., Saradalekshmi, K. R., Nair, I. V., Nair, C., and Banerjee, M. (2018). Understanding the Influence of Antipsychotic Drugs on Global Methylation Events and its Relevance in Treatment Response. Epigenomics 10, 233–247. doi:10.2217/epi-2017-0086
Tarakhovsky, A. (2010). Tools and Landscapes of Epigenetics. Nat. Immunol. 11, 565–568. doi:10.1038/ni0710-565
Ukhueduan, B., Chukwurah, E., and Patel, R. C. (2021). Regulation of PKR Activation and Apoptosis during Oxidative Stress by TRBP Phosphorylation. Int. J. Biochem. Cel Biol. 137, 106030. doi:10.1016/j.biocel.2021.106030
Uzal, F. A., Saputo, J., Sayeed, S., Vidal, J. E., Fisher, D. J., Poon, R., et al. (2009). Development and Application of New Mouse Models to Study the Pathogenesis of Clostridium perfringens Type C Enterotoxemias. Infect. Immun. 77, 5291–5299. doi:10.1128/iai.00825-09
Vidal, J. E., McClane, B. A., Saputo, J., Parker, J., and Uzal, F. A. (2008). Effects of Clostridium perfringens Beta-Toxin on the Rabbit Small Intestine and colon. Infect. Immun. 76, 4396–4404. doi:10.1128/iai.00547-08
Wang, H., Zong, Q., Wang, S., Zhao, C., Wu, S., and Bao, W. (2019). Genome-Wide DNA Methylome and Transcriptome Analysis of Porcine Intestinal Epithelial Cells upon Deoxynivalenol Exposure. J. Agric. Food Chem. 67, 6423–6431. doi:10.1021/acs.jafc.9b00613
Wang, S., Song, R., Wang, Z., Jing, Z., Wang, S., and Ma, J. (2018). S100A8/A9 in Inflammation. Front. Immunol. 9, 14. doi:10.3389/fimmu.2018.01298
Wang, W., Zhou, C., Tang, H., Yu, Y., and Zhang, Q. (2020). Combined Analysis of DNA Methylome and Transcriptome Reveal Novel Candidate Genes Related to Porcine Escherichia coli F4ab/ac-Induced Diarrhea. Front. Cel. Infect. Microbiol. 10, 15. doi:10.3389/fcimb.2020.00250
Won, Y., Yang, J. I., Park, S., and Chun, J. S. (2021). Lipopolysaccharide Binding Protein and CD14, Cofactors of Toll‐like Receptors, Are Essential for Low‐Grade Inflammation-Induced Exacerbation of Cartilage Damage in Mouse Models of Posttraumatic Osteoarthritis. Arthritis Rheumatol. 73, 1451–1460. doi:10.1002/art.41679
Woo, C. H., Shin, S. G., Koh, S. H., and Lim, J. H. (2014). TBX21 Participates in Innate Immune Response by Regulating Toll-like Receptor 2 Expression inStreptococcus Pneumoniaeinfections. Mol. Oral Microbiol. 29, 233–243. doi:10.1111/omi.12061
Wu, Z., Feng, H., Cao, Y., Huang, Y., Dai, C., Wu, S., et al. (2018). New Insight into the Molecular Mechanism of the FUT2 Regulating Escherichia coli F18 Resistance in Weaned Piglets. Ijms 19, 3301. doi:10.3390/ijms19113301
Keywords: DNA methylome, transcriptome, Clostridium perfringens type C, diarrhea, pig, resistance, susceptibility
Citation: Huang X, Yang Q, Yan Z, Wang P, Shi H, Li J, Shang X and Gun S (2022) Combined Analysis of RRBS DNA Methylome and Transcriptome Reveal Novel Candidate Genes Related to Porcine Clostridium perfringens Type C-Induced Diarrhea. Front. Genet. 13:803477. doi: 10.3389/fgene.2022.803477
Received: 28 October 2021; Accepted: 24 February 2022;
Published: 25 March 2022.
Edited by:
Sundararajan Jayaraman, University of Illinois, United StatesReviewed by:
Mengjin Zhu, Huazhong Agricultural University, ChinaXiangdong Ding, China Agricultural University, China
Copyright © 2022 Huang, Yang, Yan, Wang, Shi, Li, Shang and Gun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuangbao Gun, gunsbao056@126.com
 Xiaoyu Huang
Xiaoyu Huang Qiaoli Yang
Qiaoli Yang Zunqiang Yan1
Zunqiang Yan1 Hairen Shi
Hairen Shi Shuangbao Gun
Shuangbao Gun
