- 1Noguchi Memorial Institute for Medical Research, University of Ghana, Accra, Ghana
- 2West African Centre for Cell Biology of Infectious Pathogens, University of Ghana, Accra, Ghana
- 3Department of Biochemistry, Cell, and Molecular Biology, University of Ghana, Accra, Ghana
- 4Division of Human Genetics, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa
- 5Department of Chest Diseases, Korle-Bu Teaching Hospital Korle-Bu, Accra, Ghana
Despite advancements made toward diagnostics, tuberculosis caused by Mycobacterium africanum (Maf) and Mycobacterium tuberculosis sensu stricto (Mtbss) remains a major public health issue. Human host factors are key players in tuberculosis (TB) outcomes and treatment. Research is required to probe the interplay between host and bacterial genomes. Here, we explored the association between selected human/host genomic variants and TB disease in Ghana. Paired host genotype datum and infecting bacterial isolate information were analyzed for associations using a multinomial logistic regression. Mycobacterium tuberculosis complex (MTBC) isolates were obtained from 191 TB patients and genotyped into different phylogenetic lineages by standard methods. Two hundred and thirty-five (235) nondisease participants were used as healthy controls. A selection of 29 SNPs from TB disease-associated genes with high frequency among African populations was assayed using a TaqMan® SNP Genotyping Assay and iPLEX Gold Sequenom Mass Genotyping Array. Using 26 high-quality SNPs across 326 case-control samples in an association analysis, we found a protective variant, rs955263, in the SORBS2 gene against both Maf and Mtb infections (PBH = 0.05; OR = 0.33; 95% CI = 0.32–0.34). A relatively uncommon variant, rs17235409 in the SLC11A1 gene was observed with an even stronger protective effect against Mtb infection (MAF = 0.06; PBH = 0.04; OR = 0.05; 95% CI = 0.04–0.05). These findings suggest SLC11A1 and SORBS2 as a potential protective gene of substantial interest for TB, which is an important pathogen in West Africa, and highlight the need for in-depth host-pathogen studies in West Africa.
Introduction
Pulmonary tuberculosis (TB), a disease of public health importance, is caused by pathogenic members of the Mycobacterium tuberculosis complex (MTBC), such as M. africanum (Maf) and M. tuberculosis sensu stricto (Mtbss). Each species comprising several clades of specific strains show variable virulence and disease-causing mechanisms (Yeboah-Manu et al., 2011; Delogu et al., 2013; Reiling et al., 2013; Brites and Gagneux, 2015). While Maf is restricted to West African countries including Ghana, Mtb is responsible for TB cases worldwide (Gagneux and Small, 2007). As a disease transmitted through aerosol, (Delogu et al., 2013), it is expected that susceptible immunocompetent individuals who come into contact with aerosol(s) containing viable bacteria will be equally infected. However, in about 90% of infected individuals, the host immune system can wall off the site of infection in a granuloma (Ghon complex) (Malik and Godfrey-Faussett, 2005; Gagneux et al., 2008; Yeboah-Manu et al., 2011). However, with the remaining 10%, only 5% will potentially develop active TB disease within 2–3 years of infection, while the remaining 5% may develop TB later in their life (Sakamoto, 2012; Fattorini et al., 2013; Narasimhan et al., 2013). This unique differential infection propensity depends on the interplay between the genetic makeup of the human host (Thye et al., 2010; Oki et al., 2011; Thye et al., 2012; Chimusa et al., 2014; Kinnear et al., 2017), the environment, and socioeconomic factors (Cheng et al., 2017; Mohidem et al., 2018).
In recent years, genetic association studies between important genes and susceptibility to infectious diseases such as TB, Malaria, and HIV using linkage and genome-wide association (GWAS) have been reported (Comstock, 1978; Pydi et al., 2013; McHenry et al., 2020). With these studies, several genes have been reported to be associated with TB, including Toll-like receptors (TLRs), cytokines/chemokines, and their receptors leukocyte antigens (HLAs), SLC11A1, major histocompatibility complex-(MHC), mannose-binding lectin (MBL), vitamin D Receptor (VDR), etc. (Søborg et al., 2003; Salie et al., 2013). For instance, in 2012, Pydi et al. reported on inhibitory genes, such as KIR3DL1 and KIR2DL3, that confer susceptibility toward TB in individuals.
Although these studies provide useful insights into the disease etiology, very few of these studies investigated the association between these gene variants and specific MTBC strains (McHenry et al., 2020). For example, in a cohort study of 1,916 sputum-positive Ghanaian TB patients genotyped for the ALOX5 g.760G > A variant, individuals with heterozygous alleles were found to be at increased risk for developing TB (Herb et al., 2008). Through stratification by MTBC lineage, this association was mainly driven by patients harboring the exonic variant (g.760A) and infected with Maf [OR = 1.70 (95% CI: 1.2–2.6)] (Herb et al., 2008). Conversely, a protective association [OR = 0.60 (95% CI: 0.4–0.9)] was identified among the occurrence of TB caused by Maf but not Mtb and the mannose-binding lectin (MBL2) G57E variant in another cohort of Ghanaian patients (Thye et al., 2011). Moreover, this latter study also found that Maf binds human recombinant MBL more efficiently, perhaps leading to an improved uptake of Maf by macrophages and selection of deficient MBL variants among human populations exposed to Maf.
Comparative analysis of HLA alleles and the Mtb strains isolated from pulmonary TB patients in a South African study found a significant association of HLA-B27 allele with decreased risk of TB caused by the Beijing strain (Salie et al., 2013). Using a candidate gene approach, from a cohort of 237 adult Vietnamese TB patients, Caws et al. concluded that individuals carrying the C allele of the Toll-like receptor-2 (TLR2) T597C polymorphism were significantly more likely to develop TB caused by mycobacteria belonging to the East-Asian/Beijing strain family [OR = 1.57 (95% CI 1.15–2.15)] (Caws et al., 2008). Similar studies have been conducted to understand the coevolution of MTBC and its human host, with regard to the association between different MTBC strains and disease severity. Findings from one such study in Uganda reported an interaction between a single-nucleotide polymorphism (SNP) in SLC11A1 and the lineage (L) L4 of Mtb with several IL12B polymorphisms associated with disease severity (McHenry et al., 2020).
Even though the Sub-Saharan African continent is home to the highest diversity of humans as well as the MTBC, studies to explore the potential association of specific host gene variants and susceptibility to strains of the MTBC are limited (Mboowa, 2014; Omae et al., 2017; McHenry et al., 2020). Thus, host genetics and susceptibility to distinct MTBC lineages cannot be overlooked. Findings from two independent molecular epidemiological studies by our group and other groups (Asante-Poku et al., 2015; Asante-Poku et al., 2016) showed a strong association between Maf (driven by lineage 5) and a native West African ethnic group. To improve our understanding of the genetic susceptibility to the MTBC clades, this current study thence explored genotyping data from the host and pathogen to screen for clade-specific genetic associations in cohorts originating from Ghana.
Materials and Methods
Ethical Approval
The study was performed in accordance with the Declaration of Helsinki. Ethical approval for the study was obtained from the Noguchi Memorial Institute for Medical Research Institutional Review Board (NMIMR-IRB 097/15–16). Written and signed informed consent was obtained from participants before enrolment into the study.
Study Population
From July 1, 2016 to July 31, 2018, this case-control study enrolled participants. The study population was in two categories: One group was made up of only newly diagnosed TB patients (cases), and the other group was made up of healthy individuals serving as the control group (NTB). For the case group, only newly diagnosed sputum smear-positive adult TB cases, registered at the Department of Chest Disease, Korle-Bu Teaching Hospital in Ghana, were recruited into the study before the commencement of TB treatment. All patients were unrelated. Sample size for the study was computed using the EPITOOLS webtool (https://epitools.ausvet.com.au/casecontrolss) for case-control studies based on odds ratios. The parameters furnished to the algorithm included the expected proportion exposed in controls (5%), assumed (minimum) odds ratio, confidence interval (95%), and desired power (80%). Based on these values, a sample size of 348 was estimated, 174 for each group. Therefore, 191 cases and 235 controls were recruited in anticipation of quality filters.
For the control group, five sites of the community by the national TB control program (NTP) were selected. These sites included Korle-gonno (Ablekuma south sub metro), Bukom (Ashiedu Keteke sub metro), Abossey-okai (Okaikoi south), Glefe (Ablekuma West), and Amanfrom (Ga West District). Medical outreaches were conducted in these sites and with a new structured questionnaire, clinical characteristics [diabetes (American Diabetes Association, 2016), HIV, hypertension], and demography and epidemiological data were obtained from each participant using a structure questionnaire. Only patients with no history of TB were recruited into the study. Participants with elevated stage 1 or 2 hypertension were transported to the nearest hospital for immediate medical attention and excluded from the study.
Sample Collection
Diagnosed Tuberculosis Patients Group
To confirm the initial diagnosis at the health facility and to identify the infecting mycobacterial species, a sputum specimen was collected from each TB study participant, following the NTP guidelines. Samples were taken only after participants’ written or thumb-printed consent. Clinical characteristics (previous history of TB, diabetes, BCG vaccination, HIV) as well demography and epidemiological data including age, sex, substance abuse, education, education and ethnic origin were obtained from each participant using a structured questionnaire.
Non-tuberculosis Patients Control Group and Chest X-Ray Screening
The screening of eligible control individuals was done in a stepwise manner. First, adults (>18 years) presenting at any of the outreach centers were screened for diabetes and hypertension. Individuals with no evidence of high body temperature (38°C or above) diabetes or elevated hypertension were then taken through chest x-ray (CXR) screening using CAD4TB (version 3.07, Diagnostic Image Analysis Group, The Netherlands) for abnormalities suggestive of pulmonary TB. The software has two abnormality detection systems (textural abnormality and shape abnormality systems), which analyze abnormalities in the unobscured lung fields that have been segmented automatically (Zaidi et al., 2018). A higher score is suggestive of TB. A CAD4TB threshold score of 60 was used for this population determined using previously collected CXR data in a similar population. Sputum samples were collected from all individuals with high CAD4TB scores (60 or greater) and transported to the laboratory for further analysis. Individuals with high CAD4TB scores were referred to the hospitals for further clinical evaluation. Whole blood (5 ml) was collected from each TB patient for host genetics analysis.
Laboratory Analysis
Isolation and Characterization of Mycobacterium spp.
Sputum samples obtained were decontaminated using 5% oxalic acid (Yeboah-Manu et al., 2001) and inoculated on two pairs of Lowenstein–Jensen (LJ) slants: one supplemented with 0.4% sodium pyruvate to enhance the isolation of Maf and M. bovis, and the other with glycerol for the growth of Mtb. The cultures were incubated at 37°C and were observed weekly for growth for a maximal duration of 16 weeks. MTBC strains were identified by PCR detection of insertion sequence IS6110 as previously described (Yeboah-Manu et al., 2004). Colonies from positive cultures were purified and stored at −80°C in 2 ml of Middlebrook 7H9 supplemented with ADC enrichment media until use. Pure bacteria DNA was extracted for genotyping using a modified protocol (Otchere et al., 2016) and stored at −20°C until further use.
All MTBC isolates were further typed by spoligotyping (Kamerbeek et al., 1997). This was performed according to the instructions of the manufacturer, using commercially available kits (Isogen Bioscience BV Maarssen, The Netherlands). Briefly, The DR-containing region was amplified by PCR using primers DRa and DRb (GGTTTTGGGTCTGACGAC and CCGAGGGGACGGAAAC). The amplified products were hybridized to a set of 43 oligonucleotides each corresponding to one spacer, immobilized on a nylon membrane. Detection of hybridization was achieved using chemiluminescent ECL (Amersham) liquid followed by x-ray exposure. The spoligotyping patterns obtained were defined according to the SITVITWEB database (http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE). SITVITWEB-assigned shared type numbers were used whenever a spoligotyping pattern was found in the database, while families and subfamilies were assigned based on the MIRU-VNTRplus database (http://www.miru-vntrplus) (http://www. miru-vntrplus.org). Shared types were defined as patterns common to at least two or more isolates. All patterns that could not be assigned were considered orphan spoligotypes.
Host DNA Isolation
DNA was extracted within 24 h from peripheral blood, following instructions on the available commercial kit Gentra Puregene Blood Kit (QIAGEN) in accordance with the recommendations of the manufacturer. All DNA samples were stored at 80°C prior to genotyping. DNA quality was evaluated according to the 260/280 ratio with a Nanodrop 2000 spectrophotometer (Thermo Scientific). In total, 792 samples with concentrations of 5–851.87 µg/µl were selected and sent for single-nucleotide polymorphism (SNP) typing.
Host Genetic Analysis: Genotyping of Targeted Single-Nucleotide Polymorphisms in Tuberculosis Disease-Related Host Genes
Targeted genotyping was conducted in the Division of Human Genetics Laboratory, Faculty of Health Sciences, at the University of Cape Town, South Africa. Genetic variants that are potentially associated with MTB were identified from the recent literature. Once the SNPs of interests were identified, we investigated their allele frequencies in African populations present in the 1000 Genomes project (http://www.internationalgenome.org/home) and further narrowed the selection to SNPs that showed high frequency among African populations. This resulted in the selection of 29 SNPs from MTBC-related genes that were investigated in the present study (Supplementary Table S1). SNPs were genotyped using a TaqMan® SNP Genotyping Assay and TaqMan® Universal Master Mix (Life Technologies, Carlsbad, CA, USA), at the Division of Human Genetics, Faculty of Health Sciences, University of Cape Town and by iPLEX Gold Sequenom Mass Genotyping Array (Inqaba Biotec, Pretoria, South Africa). Validation was done in a subset of samples (10%) by Sanger sequencing using BigDye terminator mix (Promega, Madison, WI, USA).
Quality Assessment
The genotype dataset was captured in Excel and transformed to PLINK ped + map format using a custom script. SNP coordinates were obtained by querying the ENSEMBL GRCh38 web browser. PLINK binary format files (bed + bim + fam) were then generated using PLINK2, while aligning against the human reference genome in GRCh38 coordinate. Quality checks involved removal of samples for which one or both parents were non-Ghanaians, SNPs with genotyping call rate <90%, individuals with missing genotype >10%, and SNPs with minor allele frequency <0.01.
Data Analysis
Determination of Axes of Genetic Variation
Epidemiological data for this study was double entered into Microsoft© Access and validated to remove duplicates. To determine whether there were significant genetic differences among the different ethnicities based on the typed SNPs that could impair association analysis, 10 (Kinnear et al., 2017) principal components (axes of genetic variation) were computed using PLINK2. A generalized linear model (GLM) was then run using the R statistical package to determine which axes of genetic variation were significantly associated with case-control status (i.e., presence or absence of TB infection). A GLM of case-control status against age, height, and weight was also run to determine their influence on association analysis.
Association Analysis
Association tests of SNPs with case-control status were run using PLINK1.9 with the maximum permutation test procedure used to correct for multiple testing and to determine empirical significance levels. The --logistic command of PLINK was used with 100,000 maximum permutations to test for significant SNPs under different models of inheritance/association (i.e., additive, dominant, recessive, heterozygote/homozygote, and genotypic), while adjusting for sex, age, weight, and height (the variable that showed significant association with case-control status by the GLM test). The association analyses were run on three separate datasets: all controls versus all cases irrespective of the infecting MTBC genotype, all controls versus cases with Maf, and all controls versus cases with Mtb TB. Association testing was run on each chromosome separately. The quality of the variants was assessed, and association analysis was run with 26 high-quality variants for the controls against all the cases put together. A total of 326 participants (145 cases; 181 controls), of which 156 were males and 170 were females, were included in the association analysis.
Results
Characteristics of Study Cohorts
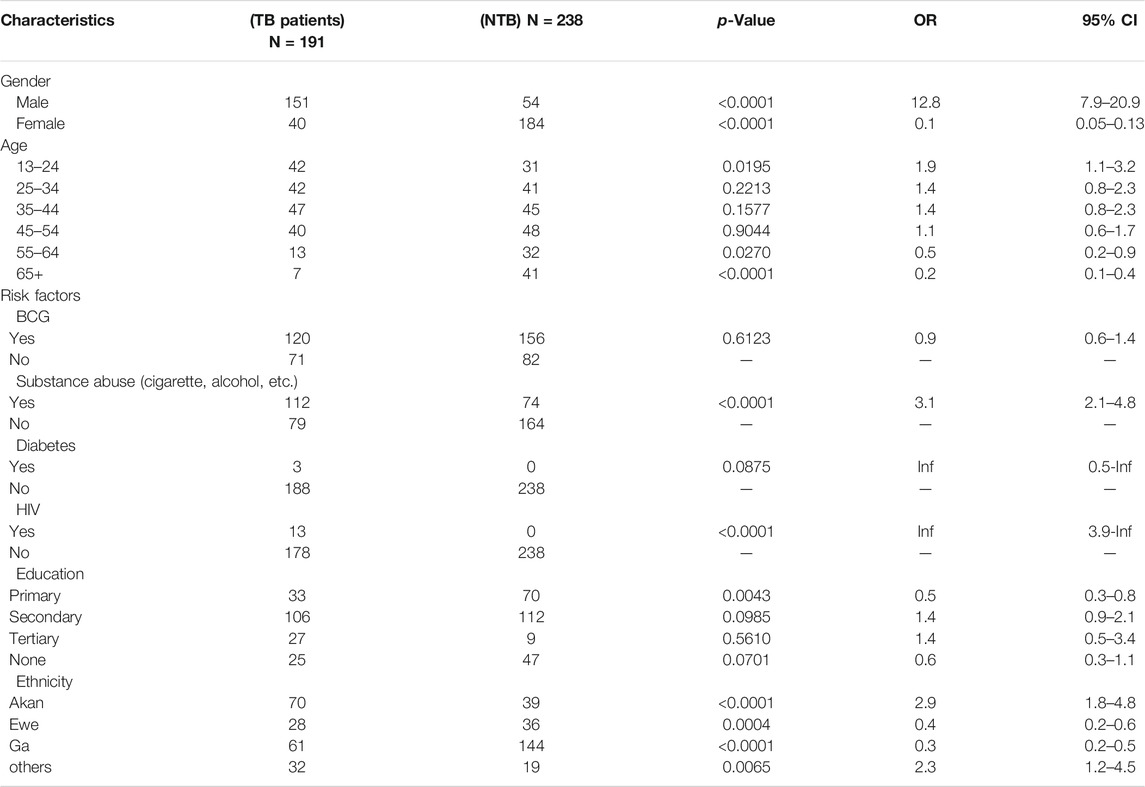
The study population consisted of 426 individuals (191 TB individuals and 235 controls). The odds of any gender being associated with active TB in univariate logistics regression was greater for young male patients (less than 24 years; n = 73) and patients older than 65 years (n = 7) [odds ratio (OR) 12.8, 95% confidence interval (CI). 7.9–20.9, p = 0.001], (Table 1). Analysis of risk factors revealed that 34%, 11.6%, and 5.8% of the patients were registered as alcohol abusers, cigarette users, and substances abusers, respectively. Participants who have a pattern of drinking that interferes with their day-to-day activities were considered alcohol abusers, cigarette users were people who smoke at least once daily, and substances abusers were people who use legal and illegal drugs. Among the TB patients with available clinical data (185/191), the vast majority had pulmonary TB only (97.0%) followed by pulmonary TB with anemia (2.0%) and TB with lobar pneumonia (1.0%). At the time of sampling, 16.7% (n = 32) of the patients were hospitalized, and 10.9% of the patients (n = 21) had a previous history of TB. Analysis of risk factors revealed that 1.6% of the patients were registered diabetic TB patients. HIV-positive patients accounted for 6.8% of the study population, and 9.4% were notified as suffering from malnutrition. Sputum bacillary load at presentation was significantly higher in the Mtb group with n = 90 patients having sputum grades of 2+ compared with (n = 28) patients in the Maf group (p < 0.001.). Analysis of the cohort showed contributing ethnicities from the Akan, Ga-Adangbe, Ewe, and several other ethnic groups from northern Ghana.
Genetic Structure of Pathogen Lineages and Correlation of Participants’ Demographic Variables
The 191 TB isolates after spoligotyping (S1) were identified as 78.5% (n = 150) Mtb and 21.5% (n = 41) Maf. Through stratifying by lineages (L), 86.6% (n = 130/150) of the Mtbss strains belonged to the Euro-American lineage (L4), with sublineages Cameroon (72.3%), Ghana (15.4%), and Haarlem (4.6%), with Uganda 11 (4.6%) being the most prevalent followed by Maf, L5 (58.5%) (n = 24/41). A total of 66 spoligotypes were detected; 170/191 isolates (89.0%) had previously defined shared type number (SIT). The remaining 21 isolates could not be defined by the SITVIT database and, thus, were defined as “orphan.” There was a significant correlation between case-control status and each of the demographic variables age, height, and weight-based on a generalized linear model (Table 1). None of the 10 (Kinnear et al., 2017) principal components computed showed a significant correlation with cases-control status (Table 2). On the other hand, age, height, weight, and sex showed statistically significant associations with case-control status and were, therefore, included as covariates in association tests.
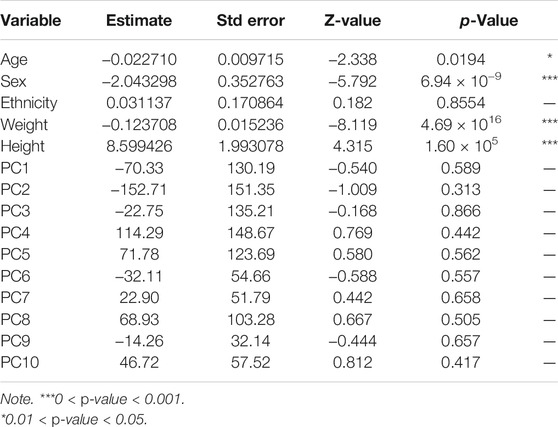
TABLE 2. Generalized linear model (GLM) of case-control status against age, ethnicity, weight, height, sex, and principal components.
Genetic Association Between Tuberculosis Cases and Controls
Of the 426 samples, 10 were removed due to a questionable ethnicity status (i.e., either father or mother, or both were non-Ghanaians). Using PLINK for quality assessment of the remaining 416 samples and 29 variants (SNPs), 90 samples were removed due to missing genotype data (--mind 0.105), 1 variant (rs1616723) was removed due to missing genotype data (--geno 0.105), and 2 variants were removed due to the Hardy–Weinberg exact test—performed on control samples only (--hwe 0.001). No SNPs were removed as a result of minor allele frequency threshold (--maf 0.01). Upon alignment to the human reference genome (build 38 - GRCh38), 9 variants were changed due to allele mismatch, while the remaining 14 variants were validated for reference/alternate allele assignment. A total of 326 samples (145 cases and 181 controls; 170 females, 156 males) and 26 SNPs were retained for further analysis with a total genotyping rate of 99%.
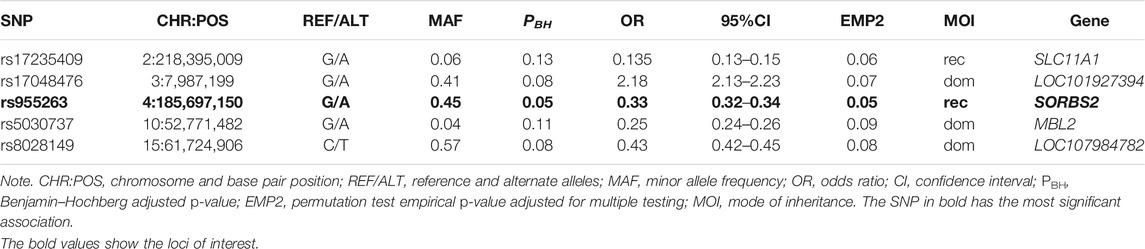
Table 3 shows association tests results after adjustment for sex, age, height, and weight, and correction for multiple testing. All p-values reported are Benjamin–Hochberg adjusted (PBH) after 100,000 permutations. SNPs located on chromosome 4 (rs955263) in the SORBS2 gene and on chromosome 2 (rs17235409) in SLC11A1 encoding mannose-binding lectin were found tending to confer apparent protection (PBH = 0.05) and (PBH = 0.06), respectively, against TB infection irrespective of the genotype of the infecting bacteria under a recessive model of inheritance. Additionally, whereas SNP rs17048476 (on chromosome 3) tends to increase the risk of TB infection by twofold (PBH = 0.07), SNPs rs8028149 (on chromosome 15) and rs5030737 decreased the risk of TB infection under a dominant model of inheritance (Table 3).
Association Analysis Between Mycobacterium tuberculosis sensu stricto Lineages and Controls
Similar association test procedures were performed for Mtb cases only and controls as described with 289 samples (108 cases and 181 controls) and 26 variants that passed all the aforementioned filters. Table 4 shows the association test results when considering TB caused by Mtbss. A single significant missense variant, rs17235409 in the SLC11A1 gene, under a homozygous recessive model of inheritance appeared to confer protection against TB infection (PBH = 0.04), whereas another SNP (rs8028149), under a dominant model of inheritance, was found tending to decrease the risk of Mtbss infection (PBH = 0.09).

TABLE 4. Association tests results for controls versus Mycobacterium tuberculosis sensu stricto (Mtbss) cases.
Association Analysis Between Mycobacterium africanum Lineages and Controls
A single-variant rs2681052 in the gene THSD7A, which encodes the transmembrane protein called thrombospondin type 1 domain-containing 7A, was found tending to confer marginal protection against TB caused by Maf (EMP2 = 0.09) under a heterozygote model of inheritance.
Discussion
Host factors are increasingly being recognized as critical for TB control considering the diversity of the outcome of interaction between MTBC and distinct human host populations. This study sought to explore potential host genetic factor (s) that may confer susceptibility or protection to distinct MTBC lineages, toward understanding crucial mechanisms of host–pathogen interaction. From this study, the most significant findings of the association analysis were two statistically significant protective SNPs: rs17235409 in SLC11A1 against Mtb only and rs955263 in SORBS2 against Mtb and Maf together. In addition, we found multiple marginally significant protective variants including in SLC11A1 and MBL2, and younger patients <35 years and patients >65 years are associated with active TB in Ghana.
Solute carrier family 11A Member 1 (SLC11A1) (OMIM: 600266), formerly NRAMP1, a bivalent antiporter located on chromosome 2q35, delivers metal cations from the cytosol into acidic endosomal and lysosomal compartments where Fenon and Haber–Weiss reaction generates toxic antimicrobial radicals for direct antimicrobial activity against infectious microorganisms such as mycobacteria (Mulero et al., 2002; Sobota et al., 2016). SLC11A1 also appears to have multiple functions, having a role in both the resolution of infections; thus, any polymorphism in the gene may influence the function of the gene, primarily affecting the survival of the TB pathogen after phagocytosis (O'Brien et al., 2008). Of the most assessed SLC11A1 variants, the polymorphic (GT)n microsatellite repeat has been shown to alter the level of SLC11A1 expression and is, therefore, a strong candidate for influencing disease incidence. Several alleles of different repeat lengths have been identified, with (GT)n allele 2 conferring lower SLC11A1 expression compared with the more commonly occurring (GT)n allele 3 (Zaahl et al., 2004).
Earlier studies analyzing (GT)n allele 2 in humans with TB in West Africans have primarily focused on four or five polymorphisms distributed across the gene: A GTn repeat in the 5′ promoter region, a four base-pair (TGTG) insertion/deletion (rs17235416) in the 3′ untranslated region (UTR), and two single-nucleotide polymorphisms (SNPs) in intron 4 (rs3731865). These mutations were found to be significantly associated with pulmonary TB. This association has been replicated in studies from Guinea-Conakry (Cervino et al., 2000) and Gambia (Yakubu et al., 2014). Bellamy et al. (Bellamy, 1998; Taype et al., 2006) found a significant association between heterozygous GC, GA, and +del genotypes, of INT4, D543N, and 3′UTR, respectively, and pulmonary TB patients, in a case-control study in Gambia. Similarly, case-control studies found an association between GA and +del genotypes, of D543N, and 3′UTR, and lung TB in Cambodia. All these genotypes were associated with resistance to TB. However, in contrast, this study identified a protective association with the more commonly occurring (GT)n allele 3, exon 15 (rs17235409, D543N).
Although there are several studies on the rs17235409 variant, the role of the variant in the pathobiology of TB remains unclear. A study by Liu et al. associated the variant with treatment failure of pulmonary TB in the Chinese (Yajie et al., 2021) and Mexican populations (Salinas-Delgado et al., 2015). Among the Tibetan population in Qinghai, the variant has been associated with TB susceptibility (Xiying et al., 2016). The variant was not only associated with TB but also was implicated in other diseases as well. The rs17235409 variant was among the list of host factors associated with susceptibility to leishmaniasis (Ates et al., 2009) and rheumatoid arthritis (Niño-Moreno et al., 2017; Braliou et al., 2019). Our analysis shows that rs17235409 might be a promotor/repression gene mutation, which could lead to significant phenotypic consequences. Mutations in this gene might eventually make the phagocytic cells less toxic, thus, providing the patients with more protection from infections by Mtb. It has, therefore, been hypothesized that allele 3 would provide protection against infectious disease by driving high SLC11A1 expression and resulting in Th1-mediated immune response. It was also suspected that the intronic position of this polymorphism might affect posttranscriptional modification of the affected gene, hence, potentially affecting the resulting SLC11A1 protein. Further analysis of the mechanism of action of NRAMP1 and its genetic variants may lead to new approaches in controlling tuberculosis, which kills more people than any other disease caused by an infectious pathogen.
Sorbin and sh3 domain-containing protein 2 (SORBS2) gene (OMIM: 616,349) located on chromosome 4q35.1 is responsible for the regulation of many cell signaling pathways. It is involved in several biological processes such as apoptosis, mitochondrial dysfunction, and innate immunity. Our study found a protective association of rs955263 in SORBS2 against Mtb and Maf together. Contrary to the protective value of rs955263 observed within the Ghanaian population, the variant was associated with active TB in a combined cohort from Uganda and Tanzania (36). Similar to our finding, a study from China reported an inconsistent risk association between SNPs in the SORBS2 gene and active TB cases with respect to the data published from Uganda and Tanzania (Qi et al., 2017). The inconsistency maybe explained by different host factors that may modify the activity of the gene as well as genetic diversity observed in the different populations. There seems to be some level of population specificity, which needs to be investigated from different populations to fully understand the role of SORBS2 gene variants in the development of active TB.
Associations between MTBC lineages and human ethnicities have been reported. Indeed, lineages 1, 2, and 4 are reported to be strongly associated with Filipino, Chinese, and “white” ethnicities, respectively (Gagneux et al., 2006; Asante-Poku et al., 2016). Likewise, in China, Hui ethnicity was found to be associated with the Beijing family of MTBC (Pang et al., 2012). Indeed, human genetic diversity has been linked to an increased or reduced susceptibility to TB. Recent studies have reported human genetic polymorphisms that influence the susceptibility to TB caused by Maf but Mtbss or vice versa. These studies indicate that human genetic susceptibility to TB is further influenced by the MTBC genotype. Conversely, a human polymorphism that reported recently on mannose-binding lectin (MBL) was associated with protection against TB caused by Maf (23). Moreover, this latter study also found Maf to bind to human recombinant MBL more efficiently, perhaps leading to an improved uptake by macrophages and selection of deficient MBL variants among human populations exposed to Maf. Although our study did not find a significant association between ethnicity and MTBC lineages, our study suggests that host genetics play an important role in TB pathogenesis, hence, the need for newer approaches to TB therapy such as host-directed immune-therapy, which have the potential to shorten the TB treatment and prevent resistance by promoting autophagy.
Conclusion
In conclusion, we found SLC11A1 as a potential susceptibility gene of substantial interest for TB caused by M. africanum, which is an important pathogen in West Africa. Given the odds ratio specified in the sample size calculation method (which effectively powers the study to detect loci that increase TB susceptibility), it implies that further studies on larger case-control sample sizes are needed to confirm the resistance loci observed in this study.
Data Availability Statement
The datasets presented in this article are not readily available because data cannot be shared publicly because of patient confidentiality. Data are available from the Noguchi Memorial Institutional Data Access/Ethics Committee (contact via GWemakor@noguchi.ug.edu.gh) for researchers who meet the criteria for access to confidential data. Requests to access the datasets should be directed to GWemakor@noguchi.ug.edu.gh.
Ethics Statement
The studies involving human participants were reviewed and approved by the Noguchi Memorial Institute for Medical Research Institutional Review Board with federal assurance number FWA00001824. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
AA-P, DY-M, and AW conceptualized the study. AA-P, PM, PA, KM, SA, IO, SO-W, SA, and KM developed the methodology. AA-P, PA, KM, IO, SA, SA, KE, KM, and GM performed the formal analysis. AA-P, PM, KM, SA, SA, and KM investigated the study. AA-P, KM, SA, and KM were in charge of the data curation. AA-P, DY-M, and AW wrote and prepared the original draft. AA-P, PM, PA, KM, SA, IO, SO-W, SA, KM, KE, DY-M, and AW wrote, reviewed, and edited the manuscript. AA-P, DY-M, and AW was in charge of the project administration. AA-P acquired the funding. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the WACCBIP Postdoctoral Fellowship funds to AA-P from a DELTAS Africa grant (DEL-15-007: Awandare). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS) Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (107755/Z/15/Z: Awandare) and the United Kingdom government. The views expressed in this publication are those of the author(s) and not necessarily those of the AAS, NEPAD Agency, Wellcome Trust, or the United Kingdom government.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.729737/full#supplementary-material
Supplementary Table S1 | Descriptive statistics of the allelic frequencies of SNPs in different human populations.
References
American Diabetes Association (2016). Erratum. Classification and Diagnosis of Diabetes. Sec. 2. In Standards of Medical Care in Diabetes-2016. Diabetes Care 2016;39(Suppl. 1):S13-S22. Diabetes Care 39 (1), 1653–S22. doi:10.2337/dc16-er09
Asante-Poku, A., Otchere, I. D., Osei-Wusu, S., Sarpong, E., Baddoo, A., Forson, A., et al. (2016). Molecular Epidemiology of Mycobacterium Africanum in Ghana. BMC Infect. Dis. 16, 385. doi:10.1186/s12879-016-1725
Asante-Poku, A., Yeboah-Manu, D., Otchere, I. D., Aboagye, S. Y., Stucki, D., Hattendorf, J., et al. (2015). Mycobacterium Africanum Is Associated with Patient Ethnicity in Ghana. Plos Negl. Trop. Dis. 9 (1), e3370. doi:10.1371/journal.pntd.0003370
Ates, O., Dalyan, L., Musellim, B., Hatemi, G., Turker, H., Ongen, G., et al. (2009). NRAMP1 (SLC11A1) Gene Polymorphisms that Correlate with Autoimmune versus Infectious Disease Susceptibility in Tuberculosis and Rheumatoid Arthritis. Int. J. immunogentics. doi:10.1111/j.1744-313X.2008.00814.x
Bellamy, R. (1998). Genetics and Pulmonary Medicine Bullet 3: Genetic Susceptibility to Tuberculosis in Human Populations. Thorax 53, 588–593. doi:10.1136/thx.53.7.588
Braliou, G. G., Kontou, P. I., and Boleti, H. (2019). Susceptibility to Leishmaniasis Is Affected by Host SLC11A1 Gene Polymorphisms: a Systematic Review and Meta-Analysis. Parasitol Res 118, 2329–2342. doi:10.1007/s00436-019-06374-y
Brites, D., and Gagneux, S. (2015). Co‐evolution of M Ycobacterium Tuberculosis and H Omo Sapiens. Immunol. Rev. 264 (1), 6–24. doi:10.1111/imr.12264
Caws, M., Thwaites, G., Dunstan, S., Hawn, T. R., Thi Ngoc Lan, N., Thuong, N. T. T., et al. (2008). The Influence of Host and Bacterial Genotype on the Development of Disseminated Disease with Mycobacterium tuberculosis. Plos Pathog. 4 (3), e1000034. doi:10.1371/journal.ppat.1000034
Cervino, A. C. L., Lakiss, S., Sow, O., and Hill, A. V. S. (2000). Allelic Association between the NRAMP1 Gene and Susceptibility to Tuberculosis in Guinea-Conakrygene and Susceptibility to Tuberculosis in Guinea-Conakry. Ann. Hum. Genet 64, 507–512. doi:10.1046/j.1469-1809.2000.6460507.x
Cheng, M. P., Abou Chakra, C. N., Yansouni, C. P., Cnossen, S., Shrier, I., Menzies, D., et al. (2017). Risk of Active Tuberculosis in Patients with Cancer: A Systematic Review and Meta-Analysis. Clin. Infect. Dis. 64 (5), 635–644. doi:10.1093/cid/ciw838
Chimusa, E. R., Zaitlen, N., Daya, M., Möller, M., van Helden, P. D., Mulder, N. J., et al. (2014). Genome-wide Association Study of Ancestry-specific TB Risk in the South African Coloured Population. Hum. Mol. Genet. 23 (1), 796–809. doi:10.1093/hmg/ddt462
Comstock, G. W. (1978). Tuberculosis in Twins: a Re-analysis of the Prophit Survey. Am. Rev. Respir. Dis. 117, 621–624. doi:10.1164/arrd.1978.117.4.621
Delogu, G., Sali, M., and Fadda, G. (2013). The Biology of mycobacterium Tuberculosis Infection. Mediterr. J. Hematol. Infect. Dis. 5 (1), e2013070. doi:10.4084/mjhid.2013.070
Fattorini, L., Piccaro, G., Mustazzolu, A., and Giannoni, F. (2013). Targeting Dormant Bacilli to Fight Tuberculosis. Mediterr. J. Hematol. Infect. Dis. 5(1), e2013072. doi:10.4084/mjhid.2013.072
Gagneux, S., and Small, P. M. “Molecular Evolution of Mycobacteria. (2008),” in Handbook of Tuberculosis. Editors S.H. Kaufmann, and E. Rubin, Wiley VCH, Weinheim, Vol. 1.
Gagneux, S., DeRiemer, K., Van, T., Kato-Maeda, M., de Jong, B. C., Narayanan, S., et al. (2006). Variable Host-Pathogen Compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. 103, 2869–2873. doi:10.1073/pnas.0511240103
Herb, F., Thye, T., Niemann, S., Browne, E. N., Chinbuah, M. A., Gyapong, J., et al. (2008). ALOX5 Variants Associated with Susceptibility to Human Pulmonary Tuberculosis. Hum. Mol. Genet. 17 (7), 1052–1060. doi:10.1093/hmg/ddm378
Kamerbeek, J., Schouls, L., Kolk, A., van Agterveld, M., van Soolingen, D., Kuijper, S., et al. (1997). Simultaneous Detection and Strain Differentiation of Mycobacterium tuberculosis for Diagnosis and Epidemiology. J. Clin. Microbiol. 35, 907–914. doi:10.1128/jcm.35.4.907-914.1997
Kinnear, C., Hoal, E. G., Schurz, H., van Helden, P. D., and Möller, M. (2017). The Role of Human Host Genetics in Tuberculosis Resistance. Expert Rev. Respir. Med. 11 (9), 721–737. doi:10.1080/17476348.201710.1080/17476348.2017.1354700
Malik, A. N., and Godfrey-Faussett, P. (2005). Effects of Genetic Variability of Mycobacterium tuberculosis Strains on the Presentation of Disease. Lancet Infect. Dis. 5 (3), 174–183. doi:10.1016/s1473-3099(05)01310-1
Mboowa, G. (2014). Tuberculosis and Genetics of Sub-saharan Africa Human Population. J Mycobac Dis 4 (5), 5. doi:10.4172/2161-1068.1000164
McHenry, M. L., Bartlett, J., Igo, R. P., Wampande, E. M., Benchek, P., Mayanja-Kizza, H., et al. (2020). Interaction between Host Genes and Mycobacterium tuberculosis Lineage Can Affect Tuberculosis Severity: Evidence for Coevolution. Plos Genet. 16 (4), e1008728. doi:10.1371/journal.pgen.1008728
Mohidem, N. A., Hashim, Z., Osman, M., Shaharudin, R., Muharam, F. M., and Makeswaran, P. (2018). Demographic, Socio-Economic and Behavior as Risk Factors of Tuberculosis in Malaysia: a Systematic Review of the Literature. Rev. Environ. Health 33 (4), 407–421. doi:10.1515/reveh-2018-0026
Mulero, V., Searle, S., Blackwell, J. M., and Brock, J. H. (2002). Solute Carrier 11a1 (Slc11a1; Formerly Nramp1) Regulates Metabolism and Release of Iron Acquired by Phagocytic, but Not Transferrin-Receptor-Mediated, Iron Uptake. Biochem. J. 363 (Pt 1), 89–94. doi:10.1042/0264-6021:3630089
Narasimhan, P., Wood, J., MacIntyre, C. R., and Mathai, D. (2013) “Risk Factors for Tuberculosis,” Pulmonary. Med. 11.
Niño-Moreno, P., Turrubiartes-Martínez, E., Oceguera-Maldonado, B., Baltazar-Benítez, N., Negrete-González, C., Oliva-Ramírez, B., et al. (2017). The Role of NRAMP1/SLC11A1 Gene Variant D543N (1730G/A) in the Genetic Susceptibility to Develop Rheumatoid Arthritis in the Mexican Mestizo Population. Rev. Invest. Clin. 69 (1), 5–10. doi:10.24875/ric.17002152
O'Brien, B. A., Archer, N. S., Simpson, A. M., Torpy, F. R., and Nassif, N. T. (2008). Association of SLC11A1 Promoter Polymorphisms with the Incidence of Autoimmune and Inflammatory Diseases: A Meta-Analysis. J. Autoimmun. 31, 42–51. doi:10.1016/j.jaut.2008.02.002
Oki, N. O., Motsinger-Reif, A. A., Antas, P. R., Levy, S., Holland, S. M., and Sterling, T. R. (2011). Novel Human Genetic Variants Associated with Extrapulmonary Tuberculosis: a Pilot Genome Wide Association Study. BMC Res. Notes 4 (4), 28. doi:10.1186/1756-0500-4-28
Omae, Y., Toyo-Oka, L., Yanai, H., Nedsuwan, S., Wattanapokayakit, S., Satproedprai, N., et al. (2017). Pathogen Lineage-Based Genome-wide Association Study Identified CD53 as Susceptible Locus in Tuberculosis. J. Hum. Genet. 62 (12), 1015–1022. doi:10.1038/jhg.2017.82
Otchere, I. D., Asante-Poku, A., Osei-Wusu, S., Baddoo, A., Sarpong, E., Ganiyu, A. H., et al. (2016). Detection and Characterization of Drug-Resistant Conferring Genes in Mycobacterium tuberculosis Complex Strains: A Prospective Study in Two Distant Regions of Ghana. Tuberculosis 99, 147–154. doi:10.1016/j.tube.2016.05.014
Pang, Y., Song, Y., Xia, H., Zhou, Y., Zhao, B., and Zhao, Y. (2012). Risk Factors and Clinical Phenotypes of Beijing Genotype Strains in Tuberculosis Patients in China. BMC Infect. Dis. 12, 354. doi:10.1186/1471-2334-12-354
Pydi, S. S., Sunder, S. R., Venkatasubramanian, S., Kovvali, S., Jonnalagada, S., Valluri, V. L., et al. (2013). Killer Cell Immunoglobulin like Receptor Gene Association with Tuberculosis. Hum. Immunol. 74, 85–92. doi:10.1016/j.humimm.2012.10.006
Qi, H., Zhang, Y. B., Sun, L., Chen, C., Xu, B., Xu, F., et al. (2017). Discovery of Susceptibility Loci Associated with Tuberculosis in Han Chinese. Hum. Mol. Genet. 26 (23), 4752–4763. doi:10.1093/hmg/ddx365
Reiling, N., Homolka, S., Walter, K., Brandenburg, J., Niwinski, L., Ernst, M., et al. (2013). Clade-Specific Virulence Patterns of Mycobacterium tuberculosis Complex Strains in Human Primary Macrophages and Aerogenically Infected MiceMycobacterium tuberculosis Complex Strains in Human Primary Macrophages and Aerogenically Infected Mice. mBio 4 (4), e00250–13. doi:10.1128/mBio.00250-13
Sakamoto, K. (2012). The Pathology of Mycobacterium tuberculosis Infection. Vet. Pathol. 49 (3), 423–439. doi:10.1177/0300985811429313
Salie, M., van der Merwe, L., Möller, M., Daya, M., van der Spuy, G. D., van Helden, P. D., et al. (2013). Associations between Human Leukocyte Antigen Class I Variants and the Mycobacterium tuberculosis Subtypes Causing DiseaseAssociations between Human Leukocyte Antigen Class I Variants and the Mycobacterium tuberculosis Subtypes Causing Disease. J Infect Dis. 15 209 (2), 216–223. doi:10.1093/infdis/jit443
Salinas-Delgado, Y., Galaviz-Hernández, C., Toral, R. G., Ávila Rejón, C. A., Reyes-Lopez, M. A., Martínez, A. R., et al. (2015). The D543N Polymorphism of the SLC11A1/NRAMP1 Gene Is Associated with Treatment Failure in Male Patients with Pulmonary Tuberculosis. Drug Metab. Pers Ther. 30 (3), 211–214. doi:10.1515/dmpt-2015-0019
Søborg, C., Madsen, H. O., Andersen, A. B., Lillebaek, T., Kok-Jensen, A., and Garred, P. (2003). Mannose-binding Lectin Polymorphisms in Clinical Tuberculosis. J. Infect. Dis. 188, 777–782. doi:10.1086/377183
Sobota, R. S., Stein, C. M., Kodaman, N., Scheinfeldt, L. B., Maro, I., Wieland-Alter, W., et al. (2016). A Locus at 5q33.3 Confers Resistance to Tuberculosis in Highly Susceptible Individuals. Am. J. Hum. Genet. 98 (3), 514–524. doi:10.1016/j.ajhg.2016.01.015
Taype, C. A., Castro, J. C., Accinelli, R. A., Herrera-Velit, P., Shaw, M. A., and Espinoza, J. R. (2006). Association between SLC11A1 Polymorphisms and Susceptibility to Different Clinical Forms of Tuberculosis in the Peruvian Population. Infect. Genet. Evol. 6 (5), 361–367. doi:10.1016/j.meegid.2006.01.002
Thye, T., Niemann, S., Walter, K., Homolka, S., Intemann, C. D., Chinbuah, M. A., et al. (2011). Variant G57E of Mannose Binding Lectin Associated with protection against Tuberculosis Caused by Mycobacterium Africanum but Not by M. tuberculosis. PLoS One 6 (6), e20908. doi:10.1371/journal.pone.0020908
Thye, T., Owusu-Dabo, E., Vannberg, F. O., van Crevel, R., Curtis, J., Sahiratmadja, E., et al. (2012). Common Variants at 11p13 Are Associated with Susceptibility to Tuberculosis. Nat. Genet. 44 (3), 257–259. doi:10.1038/ng.1080
Thye, T., Vannberg, F. O., Vannberg, F. O., Wong, S. H., Owusu-Dabo, E., Osei, I., et al. (2010). Genome-wide Association Analyses Identifies a Susceptibility Locus for Tuberculosis on Chromosome 18q11.2. Nat. Genet. 42 (9), 739–741. doi:10.1038/ng.639
Xiying, F., Wenwen, L. I., Zhenzhong,Wei, B. A. I., ,Tai,Jiu., G., Mao ,Hua., , et al. (2016). Study of the Correlation between NRAMP1 Gene Polymorphisms and Susceptibility to Tuberculosis in Tibetan People in Qinghai. Chin. J. Microbiol. Immunol. 36 (7), 494–499.
Yajie, L., Zhang, Y., Chen, Y., Zhang, W., Ma, L., Cao, M., et al. (2021). The Association Between SLC11A1 Gene Polymorphism and Treatment Failure of Pulmonary Tuberculosis. J. Prev. Med. (12), 563–567.
Yakubu, A., Alade, D., and Dim, N. (2014). Molecular Analysis of Solute Carrier Family 11 Member A1 (Slc11a1) Gene in Ruminants and Non-ruminants Using Computational Method. Genetika 46, 925–934. doi:10.2298/gensr1403925y
Yeboah-Manu, D., Asante-Poku, A., Bodmer, T., Stucki, D., Koram, K., Bonsu, F., et al. (2011). Genotypic Diversity and Drug Susceptibility Patterns Among M. tuberculosis Complex Isolates from South-Western Ghana. PLoS One 6 (7), e21906. doi:10.1371/journal.pone.0021906
Yeboah-Manu, D., Bodmer, T., Mensah-Quainoo, E., Owusu, S., Ofori-Adjei, D., and Pluschke, G. (2004). Evaluation of Decontamination Methods and Growth Media for Primary Isolation of Mycobacterium Ulcerans from Surgical Specimens. J. Clin. Microbiol. 42 (12), 5875–5876. doi:10.1128/JCM.42.12.5875-5876.2004
Yeboah-Manu, D., Yates, M. D., and Stuart, M. (2001). Wilson Application of a Simple Multiplex Polymerase Chain Reaction to Aid in the Routine Work of the mycobacterium Reference Laboratory. J. Clin. Microbiol. 2001 (39), 4166–4168. doi:10.1128/jcm.39.11.4166-4168.2001
Zaahl, M. G., Robson, K. J. H., Warnich, L., and Kotze, M. J. (2004). Expression of the SLC11A1 (NRAMP1) 5′-(GT)n Repeat: Opposite Effect in the Presence of −237C→T. Blood Cell Mol. Dis. 33 (1), 45–50. doi:10.1016/j.bcmd.2004.04.003
Zaidi, S. M. A., Habib, S. S., Van Ginneken, B., Ferrand, R. A., Creswell, J., Khowaja, S., et al. (2018). Evaluation of the Diagnostic Accuracy of Computer-Aided Detection of Tuberculosis on Chest Radiography Among Private Sector Patients in Pakistan. Sci. Rep. 8 (1), 12339. doi:10.1038/s41598-018-30810-1
Keywords: tuberculosis, SLC11A1, Sorbs2, Ghana, genomics
Citation: Asante-Poku A, Morgan P, Osei-Wusu S, Aboagye SY, Asare P, Otchere ID, Adadey SM, Mnika K, Esoh K, Mawuta KH, Arthur N, Forson A, Mazandu GK, Wonkam A and Yeboah-Manu D (2022) Genetic Analysis of TB Susceptibility Variants in Ghana Reveals Candidate Protective Loci in SORBS2 and SCL11A1 Genes. Front. Genet. 12:729737. doi: 10.3389/fgene.2021.729737
Received: 23 June 2021; Accepted: 08 December 2021;
Published: 15 February 2022.
Edited by:
Dana C. Crawford, Case Western Reserve University, United StatesReviewed by:
Stephanie Julia Müller, Stellenbosch University, South AfricaSundeep Chaitanya Vedithi, University of Cambridge, United Kingdom
Michael McHenry, Case Western Reserve University, United States
Copyright © 2022 Asante-Poku, Morgan, Osei-Wusu, Aboagye, Asare, Otchere, Adadey, Mnika, Esoh, Mawuta, Arthur, Forson, Mazandu, Wonkam and Yeboah-Manu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adwoa Asante-Poku, aasante-poku@noguchi.ug.edu.gh
 Adwoa Asante-Poku
Adwoa Asante-Poku Portia Morgan
Portia Morgan Stephen Osei-Wusu
Stephen Osei-Wusu Samuel Yaw Aboagye
Samuel Yaw Aboagye Prince Asare
Prince Asare Isaac Darko Otchere
Isaac Darko Otchere Samuel Mawuli Adadey
Samuel Mawuli Adadey Khuthala Mnika
Khuthala Mnika Kevin Esoh
Kevin Esoh Kenneth Hayibor Mawuta
Kenneth Hayibor Mawuta Nelly Arthur
Nelly Arthur Audrey Forson
Audrey Forson Gaston Kuzamunu Mazandu
Gaston Kuzamunu Mazandu Ambroise Wonkam
Ambroise Wonkam Dorothy Yeboah-Manu
Dorothy Yeboah-Manu