- 1Key Laboratory of Horticultural Plant Biology (Ministry of Education), College of Horticulture and Forestry Science, Huazhong Agricultural University, Wuhan, China
- 2Department of Botany, National Museum of Natural History, MRC166, Smithsonian Institution, Washington, DC, United States
- 3Alpha Genomics Private Limited, Islamabad, Pakistan
- 4Department of Biochemistry, Faculty of Biological Sciences, Quaid-i-Azam University, Islamabad, Pakistan
- 5Southwest Engineering Technology and Research Center of Landscape Architecture, State Forestry Administration, Southwest Forestry University, Kunming, China
- 6Key Laboratory of Plant Resources Conservation and Utilization, College of Biology and Environmental Sciences, Jishou University, Jishou, China
Chimonanthus of Calycanthaceae is a small endemic genus in China, with unusual winter-blooming sweet flowers widely cultivated for ornamentals and medicinal uses. The evolution of Chimonanthus plastomes and its phylogenetic relationships remain unresolved due to limited availability of genetic resources. Here, we report fully assembled and annotated chloroplast genomes of five Chimonanthus species. The chloroplast genomes of the genus (size range 153,010 – 153,299 bp) reveal high similarities in gene content, gene order, GC content, codon usage, amino acid frequency, simple sequence repeats, oligonucleotide repeats, synonymous and non-synonymous substitutions, and transition and transversion substitutions. Signatures of positive selection are detected in atpF and rpoB genes in C. campanulatus. The correlations among substitutions, InDels, and oligonucleotide repeats reveal weak to strong correlations in distantly related species at the intergeneric levels, and very weak to weak correlations among closely related Chimonanthus species. Chloroplast genomes are used to reconstruct a well-resolved phylogenetic tree, which supports the monophyly of Chimonanthus. Within Chimonanthus, C. praecox and C. campanulatus form one clade, while C. grammatus, C. salicifolius, C. zhejiangensis, and C. nitens constitute another clade. Chimonanthus nitens appears paraphyletic and is closely related to C. salicifolius and C. zhejiangensis, suggesting the need to reevaluate the species delimitation of C. nitens. Chimonanthus and Calycanthus diverged in mid-Oligocene; the radiation of extant Chimonanthus species was dated to the mid-Miocene, while C. grammatus diverged from other Chimonanthus species in the late Miocene. C. salicifolius, C. nitens (a), and C. zhejiangensis are inferred to have diverged in the Pleistocene of the Quaternary period, suggesting recent speciation of a relict lineage in the subtropical forest regions in eastern China. This study provides important insights into the chloroplast genome features and evolutionary history of Chimonanthus and family Calycanthaceae.
Introduction
Due to its uniparental inheritance, moderate evolutionary rate, and highly conserved genome structure and gene content in most land plants, the chloroplast genome has been widely used in phylogenetic studies (Jansen et al., 2007; Daniell et al., 2016; Valcárcel and Wen, 2019; Wang Y. B. et al., 2020; Mehmood et al., 2020c), DNA barcoding (Ahmed et al., 2013; Bi et al., 2018; Abdullah et al., 2020e), dating divergence times (Zhang et al., 2019; Ahmed et al., 2020; Mehmood et al., 2020c), and studying the origins of horticulturally important plants, e.g., Chrysanthemum cultivars (Ma et al., 2020). With the rapid advancements in next-generation sequencing, the availability of cp genome sequences has dramatically increased for flowering plants, offering opportunities for comparative evolutionary and systematic studies, and improvement of horticultural plant breeding (Sonah et al., 2011; Ahmed, 2015; Xiong et al., 2015; Wen et al., 2018; Thode et al., 2020).
The Calycanthaceae (sweetshrubs or spicebushes) are a small family of flowering plants, which is sister to the remaining six families of the order Laurales of the Magnoliids (Renner, 1999; Chase et al., 2016). The family contains 10 known species in three genera (Zhou et al., 2006; Christenhusz and Byng, 2016). Calycanthus comprises three species, Cal. floridus L. and Cal. occidentalis Hook. and Arn. in North America and Cal. chinensis W.C. Cheng and S.Y. Chang in eastern China. The latter species was sometimes recognized as a member of the monotypic genus Sinocalycanthus, based on floral morphology (Cheng and Chang, 1964). However, the chloroplast DNA analyses supported Sinocalycanthus as part of Calycanthus (Wen et al., 1996; Wen, 1999). The Chinese endemic genus Chimonanthus consists of six species: C. praecox L., C. campanulatus R.H. Chang and C.S., C. nitens Oliv., C. salicifolius S.Y. Hu., C. grammatus M.C. Liu, and C. zhejiangensis M.C. Liu. The phylogenetic relationships among the latter four species remain poorly resolved [see (Zhou et al., 2006) for details]. Chimonanthus species are widely cultivated as ornamental plants with high economic value, and several species of the genus are also utilized for their medicinal properties (Sun et al., 2017; Zhang X. et al., 2017, Zhang et al., 2017 S.). Idiospermum is a highly distinctive monotypic genus from Queensland, Australia, consisting of Idiospermum australiense (Diels) S.T. Blake (Blake, 1972; Carlquist, 1983). Based on DNA markers and morphological characters, Idiospermum is sister to the clade of Chimonanthus and Calycanthus (Li and Li, 2000; Li et al., 2004; Zhou et al., 2006).
The previous molecular phylogenetic studies have resolved C. praecox and C. campanulatus as a clade, while phylogenetic relationships among the rest of Chimonanthus species remain poorly resolved (Zhou et al., 2006), partly due to lack of informative characters for all species. In the context of inferring the temporal diversification of Calycanthaceae, Zhou et al. (2006) only used a few markers with inconsistent age estimates. Therefore, it is important to use a large region of the chloroplast genomes to provide more precise age estimates (Nikiforova et al., 2013; Middleton et al., 2014; Wen et al., 2018; Zhang et al., 2019). So far, only two chloroplast genomes of Chimonanthus have been reported (Dong et al., 2020).
We herein newly report fully assembled and annotated chloroplast genomes of four species: C. campanulatus, C. grammatus, C. salicifolius, and C. zhejiangensis. We have also assembled the chloroplast genome of one additional accession of C. nitens collected from Jiangxi, China, and included two previously published cp genomes in the analyses. The objectives of our study are to characterize and perform comparative analyses of the chloroplast genomes, evaluate correlations among mutational events, infer the phylogenetic relationships within Chimonanthus with all six recognized species of the genus sampled, and estimate their divergence times to explore the evolutionary diversification of the endemic genus from China. Our assemblies and data will serve as useful genetic resources for future research in this horticulturally unique and systematically important genus Chimonanthus and the family Calycanthaceae of the Magnoliids.
Materials and Methods
Taxon Sampling and Sequencing
Fresh and healthy leaves of C. grammatus, C. salicifolius, and C. nitens were collected from natural populations located at Anyuan (25.28°N, 115.45°E), Yushan (28.77°N, 118.12°E), and Guixi (27.90°N, 117.20°E) in Jiangxi Province, China, respectively. C. campanulatus leaves were collected from the Ornamental Horticulture Nursery, Huazhong Agricultural University, Wuhan (30.47°N, 114.36°E), China, while those of C. zhejiangensis were sampled from its native habitat at Longquan (28.13°N, 119.03°E) in Zhejiang Province, China (Supplementary Table S1). The voucher specimens were deposited at the Herbarium of the Key Laboratory of Horticultural Plant Biology (Ministry of Education), College of Horticulture and Forestry Science, Huazhong Agricultural University (China). No specific permissions were required for all the samples, which are neither protected nor privately owned, and the field study did not involve protected or endangered species.
Total genomic DNA was isolated from fresh or silica gel-dried leaves of the five Chimonanthus species (including one accession of C. nitens) using the CTAB method (Doyle and Doyle, 1987). Electrophoresis with 1% agarose gel was used to evaluate the DNA quality. DNA samples were sent to Novogene (Beijing, China) for whole-genome sequencing on Illumina HiSeq platform, with the PE 150 run.
Genome Assembly and Annotation
The quality of the generated short read data was assessed using FastQC (Andrews et al., 2015) with default parameters. Chimonanthus praecox cp genome (NCBI accession number MH377057) was used for initial mapping of the short reads using the Burrows–Wheeler Alignment (BWA) algorithm (Li and Durbin, 2009). The alignment file (SAM format) was visualized in Tablet (Milne et al., 2009) to check for presence of short reads coming from the chloroplast genomes and to obtain an estimate of average coverage depth, a statistical measure needed for the downstream assembly process. The genomes were assembled following the methodology reported earlier (Ahmed et al., 2012; Iram et al., 2019; Abdullah et al., 2020b). Briefly, the chloroplast genomes were assembled using the Velvet (v.1.2.10) tool (Zerbino and Birney, 2008) to form contigs. Assembly process was reiterated thrice for each individual samples by selecting different kmer sizes, including 71, 91, and 111 kmer lengths, and selecting coverage cutoff option of 100 during the Velvet assembly process. The assembled contigs were imported in Geneious 8.1 (Kearse et al., 2012) and further assembled from large scaffolds into full-length genomes. The boundaries of large single copy (LSC), small single copy (SSC), and the two inverted repeats (IRa and IRb) were visually curated. The assembled genomes were curated by mapping the original short reads to their respective assembled genomes using BWA mapping and visualizing in Tablet as given above. Average coverage depth statistics were visually recorded in Tablet for all the genomes. The assembled genomes were annotated using online annotations GeSeq (Tillich et al., 2017), whereas tRNA genes were further verified by tRNAscan-SE v.2.0.3 (Schattner et al., 2005) using default parameters. The annotations of each protein-coding gene were further validated with homologous genes using BLAST of the National Center for Biotechnology Information. Moreover, the annotations of GeSeq were also checked by comparing with previously reported genome of C. praecox using the Multiple Alignment Fast Fourier Transform (MAFFT) alignment (Katoh et al., 2005). The assembled chloroplast genomes were circularized using OGDRAW (Greiner et al., 2019). The fully assembled and annotated cp genomes of the five species were submitted to the GenBank database of NCBI and given accession numbers MW166216 through MW166220.
Comparative Analyses of Chloroplast Genomes
We used Geneious R8.1 (Kearse et al., 2012) to compare genomic features, including sizes of LSC, SSC, and IRs, and determine amino acid frequency and codon usage. The junctions of single copy and inverted repeat regions were also analyzed in Geneious R8.1. We analyzed microsatellite repeats using MISA-web (Beier et al., 2017) with repeat units as follows: mononucleotide repeats ≥ 10, dinucleotide ≥ 5, trinucleotide ≥ 4, tetranucleotide, pentanucleotide, and hexanucleotide ≥ 3 (Mehmood et al., 2020a; Shahzadi et al., 2020). In addition, the oligonucleotide repeats were analyzed using the REPuter (Kurtz et al., 2001) tool by selecting a repeat length of 30 bp and determining all four types of repeats (forward, reverse, complimentary, and palindromic) with minimum 90% identity (Abdullah et al., 2019a).
The contraction and expansion of the IR regions at the junctions of four main parts (LSC/IRb/SSC/IRa) of the cp genomes were inspected and plotted with IRscope (Amiryousefi et al., 2018a). The possibility of large-scale genomic rearrangements among the studied Chimonanthus cp genomes was also investigated by generating a multiple sequence alignment of the six chloroplast genomes using MAUVE multiple sequence alignment (Darling et al., 2004) embedded in Geneious R8.1. One IR region (IRa) was excluded in the alignment for this comparison. Large-scale contraction and expansion in inverted repeats at the junctions of inverted repeats with single copies were compared in Geneious 8.1.
We determined transition (Ts), transversion (Tv) substitutions, and their ratio (Ts/Tv) in 78 protein-coding genes. For this purpose, we concatenated the protein-coding genes of all five species. The sequences of the concatenated protein-coding genes of these species were pairwise aligned to C. praecox by MAFFT (Katoh et al., 2005) plugin provided in Geneious 8.1 (Kearse et al., 2012). The substitution types were determined from each alignment in Geneious R8.1. We also determined the rate of synonymous (Ks), non-synonymous (Ka) substitutions, and their ratio (Ka/Ks) in 78 protein-coding genes. We extracted and aligned protein-coding genes from all five species. The chloroplast genome of C. praecox was used as a reference, and the rates of evolution of protein-coding genes were recorded. The data were interpreted in terms of purifying selection (Ka/Ks < 1), neutral evolution (Ka/Ks = 1), and positive selection (Ka/Ks > 1) (Henriquez et al., 2020b).
Correlations Among Mutational Events in Calycanthaceae
We determined correlations among substitutions, InDels, and oligonucleotide repeats at the family and genus levels in pairwise comparison using the same approach used previously (Abdullah et al., 2020a). The chloroplast genome of Idiospermum australiense was used as reference for family-level comparison. The cp genome of Calycanthus chinensis was used as reference in comparison of two species of Calycanthus, whereas the cp genome of Chimonanthus campanulatus was used as reference for all the species of genus Chimonanthus. We also expressed the strength of the correlations as negligible or very weak (0.1–0.19), weak (0.20–0.29), moderate (0.30–0.39), strong (0.4–0.69), very strong (0.70–0.99), and perfect (1.0) following studies by Abdullah et al. (2020d, 2020a).
Phylogenetic Inference, Bayesian Dating, and Fossil Calibration
A phylogenetic analysis was performed with the maximum-likelihood method (Stamatakis, 2014) using the 10 cp genomes in the family, comprising seven Chimonanthus samples representing all six species (two cp genomes for C. nitens), two Calycanthus species, and one Idiospermum species. Chloroplast genomes of Cal. chinensis, Cal. floridus, C. praecox, C. nitens (a), and Idiospermum australiense were selected from NCBI (Goremykin et al., 2003; Dong et al., 2020). Full-length cp genomes of all these species were aligned using MAFFT (Katoh et al., 2005) after removal of one IR. We removed indels from the alignment to construct the phylogenetic relationships based only on substitution mutations (Ahmed et al., 2012; Henriquez et al., 2020b). JModelTest2 (Darriba et al., 2012) was used to infer the best fit model on the alignment. A maximum likelihood tree was built with 100 bootstrap replicates, using IQ-tree (Nguyen et al., 2015), and the tree was refined using TreeDyn (Chevenet et al., 2006).
To estimate divergence times for taxa in Calycanthaceae, the maximum likelihood tree obtained from whole chloroplast genomes was used as input to perform the Bayesian dating analyses in the BEAST v1.7.5 package using uncorrelated relaxed clock model (Drummond et al., 2006; Drummond and Rambaut, 2007). Rate variation among sites was modeled using a gamma distribution with four rate categories in the GTR model with a birth–death speciation tree prior and an uncorrelated lognormal distributed relaxed clock model (Drummond et al., 2006). Two independent MCMC runs were performed for 10 million generations each and sampling for every 1,000 generations. Samples from the two chains, which yielded similar results, were combined, and convergence of the chains was checked using the program Tracer version 1.7.1 (Rambaut et al., 2018). The effective sample size (ESS) values of all parameters were greater than 200, indicating a sufficient level of sampling. After the discarding of ca. 15% burn-in, the rest sampled posterior trees were summarized to generate a maximum clade credibility tree using the program TreeAnnotator 1.8.5 (Drummond and Rambaut, 2007) with a PP limit of 0.5 and mean node heights. The means and 95% higher posterior densities (HPD) of age estimates were obtained from the combined outputs using Tracer. FigTree version 1.4.0 (Rambaut, 2012) was used to view the BEAST tree.
The oldest fossil record of Calycanthaceae was reported from the Potomac group in the Early Cretaceous (Early or Middle Albian), based on charcoalified flower Virginianthus calycanthoides (Friis et al., 1994). Cladistic analyses by Crepet et al. (2005) suggested that Virginianthus may lie on the Laurales stem lineage rather than Calycanthaceae and instead placed a new fossil Jeresyanthus calycanthoides described from the Late Cretaceous (∼90 Ma) in New Jersey within Calycanthcaeae. Other fossils belonging to Calycanthaceae, include Araripia florifera described from the Early Cretaceous Crato Formation in Brazil (Mohr and Eklund, 2003), and a potential Miocene (∼16 Ma) fossil fruit from Germany (Mai, 1987). Araripia may represent an extinct lineage within or close to the Laurales, as it shares features not only with Calycanthaceae but also with several other families of Laurales including Hernandiaceae and Lauraceae (Mohr and Eklund, 2003). Based on these fossils, the emergence and early diversification of Calycanthaceae might have occurred between 90 and 110 Ma in the Cretaceous.
We used two approaches to calibrate the divergence estimates. First, the minimum age for Calycanthaceae was set to 108 Mya, based on secondary calibration (Dong et al., 2020) with normal prior mean = 108 and sigma = 5.0. This age is also in accordance with the fossil Araripia florifera representing Calycanthaceae (Mohr and Eklund, 2003). Second, the minimum age for Calycanthaceae was set to 90 Mya, based on the fossil record of Jereysanthus calycanthoides (Crepet et al., 2005) with lognormal mean = 1.0, standard = 1.25, and offset = 90.0.
Results
Comparative Analyses of the Chloroplast Genomes
The number of short reads and data produced per sample are summarized in Supplementary Table S2. The table also gives the average coverage depth information of the short reads when mapped to their respective assembled cp genomes. For the paired-end 150 bp run, the total data obtained ranged from 7.1 to 9.5 Gb, and the number of short reads ranged from 19.21 to 25.87 million per sample. The phred quality of the data for all samples remained above 35. Very high average coverage depth (ranging from 1,429× to 6,270×) ensures the accuracy of the assembled genomes.
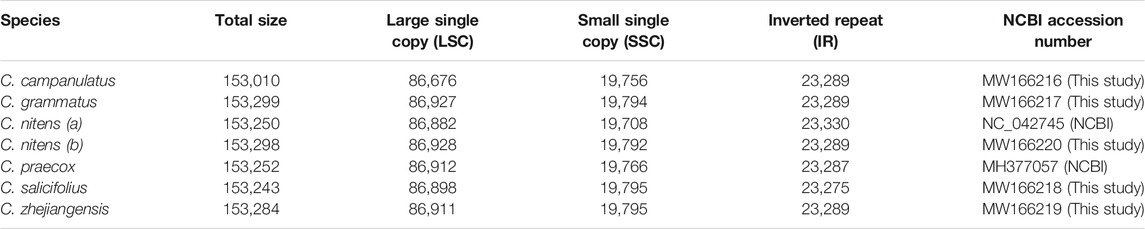
The sizes of the cp genomes within Chimonanthus did not show large variation and ranged from 153,010 bp (C. campanulatus) to 153,299 bp (C. grammatus). The LSC regions ranged from 86,676 bp (C. campanulatus) to 86,928 bp (C. nitens); SSC from 19,756 bp (C. campanulatus) to 19,795 bp (C. salicifolius and C. zhejiangensis); and IRs from 23,275 bp (C. salicifolius) to 23, 289 bp (all other species, except C. praecox) (Table 1). Notable size differences for the LSC, SSC, and IR regions were observed between cp genomes of the two C. nitens accessions (Table 1).
A representative circular map of the chloroplast genomes of the five Chimonanthus species assembled in this study is given in Figure 1. All five species were found to be highly conserved in terms of gene organization, gene content, and intron content. All species exhibited 113 unique genes, including 79 protein-coding, 30 tRNA, and four rRNA genes (Supplementary Table S3). We recorded 15 duplicated genes in the IRs of all genomes. Among these duplicated genes, four were protein coding, seven encoded tRNAs, and four encoded rRNA genes. Hence, the total number of genes in each of the Chimonanthus species was 128. We detected two introns in clpP, rps12, and ycf3 genes. A complete open reading frame (ORF) of the infA gene was present in all species (Supplementary Table S3). The guanine–cytosine (GC) content of the complete chloroplast genomes and of all regions showed high similarities among the species, whereas fluctuation in GC content was observed within the different regions of the same chloroplast genome. The GC content of coding regions, rRNAs, and tRNAs also showed high similarities among the species (Supplementary Table S4).
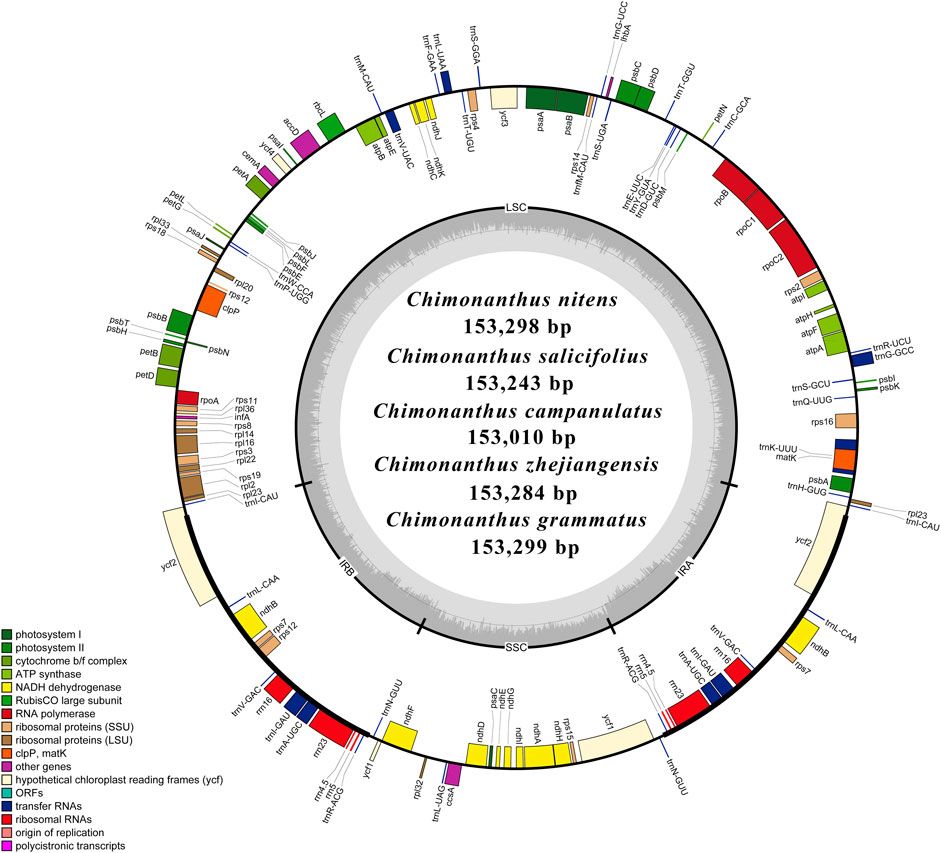
FIGURE 1. Schematic representation of the de novo assembled circular chloroplast genomes of five Chimonanthus species. Genes present inside the circle are transcribed counterclockwise, whereas genes present outside the circle are transcribed clockwise. Genes are color-coded based on their role in different pathways, as given in the key. Large single copy (LSC), inverted repeat b (IRb), small single copy (SSC), and inverted repeat a (IRa) of the inner circle represent quadripartite structure of cp genomes. Dark grey area in the inner circle represents average guanine–cytosine (GC) content, while light grey represents AT content.
Dynamics at the Boundaries of Inverted Repeats and Single Copy Regions
The chloroplast genomes showed overall similarities at all four junctions of the single copy and inverted repeat regions in all six Chimonanthus species. No significant differences were observed across all six species at these junctions (Figure 2B). At the JLA (junction of large single copy with IRa), trnH gene was found in LSC 11–14 bases downstream at the start of the LSC in different species, while rpl23 gene was present 27–30 bases prior to the IRa end in different species. At the JLB (LSC–IRb junction), rpl2 gene started two to four bases inside IRb and extended in LSC. At the JSA (SSC-IRa junction), ycf1 gene was found to extend from IRa to SSC; the first 266 bases of this gene were found in all the six chloroplast genomes. This resulted in a truncated copy (or a pseudogene) of ycf1 gene at the JSB (SSC-IRb junction) in IRb, whereas ndhF gene was found completely in the SSC region, ending exactly at the junction. This conserved structure was also confirmed using colinear block analyses of Mauve (Figure 2A).
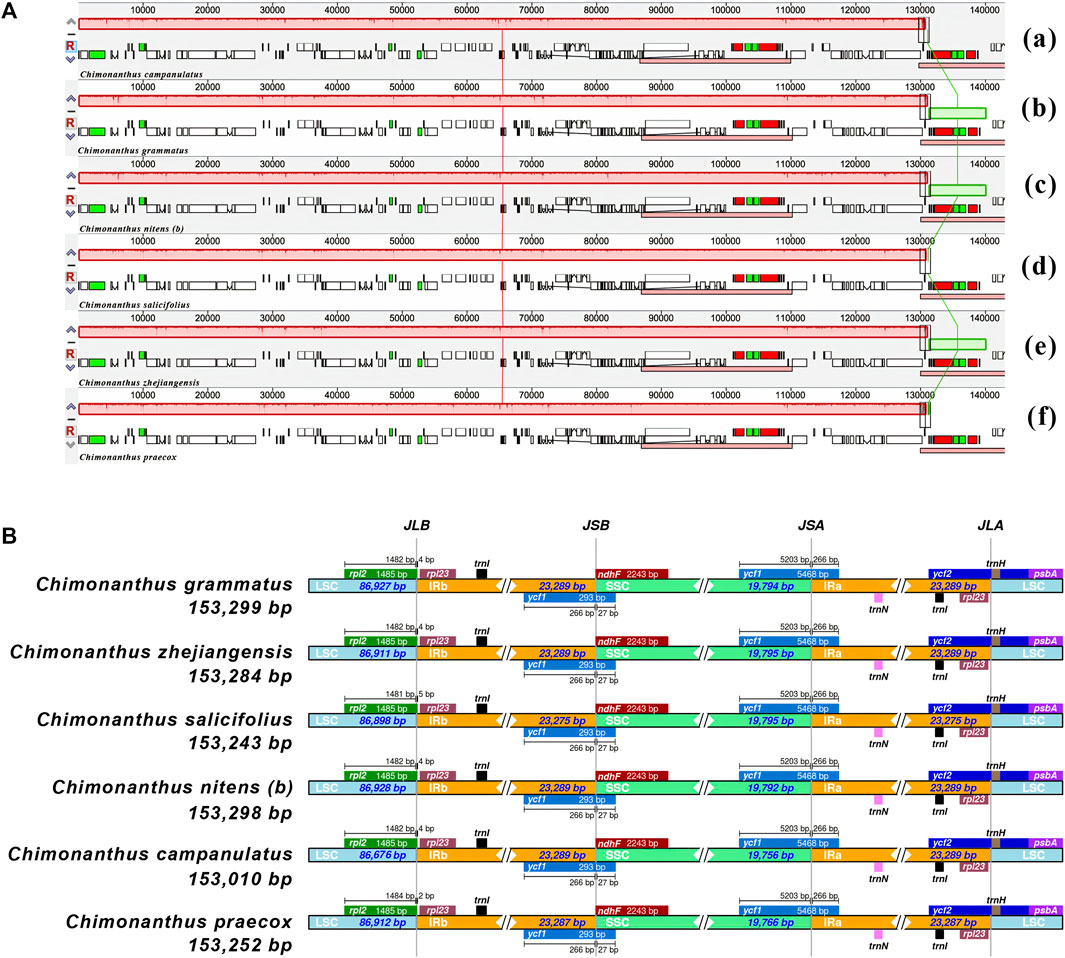
FIGURE 2. Comparative analysis of gene order and junction sites in Chimonanthus chloroplast genomes. (A) Mauve alignment of six Chimonanthus chloroplast genomes. Within each of the alignments, local collinear blocks are represented by blocks of the same color connected by lines. (a) C. campanulatus, (b) C. grammatus, (c) C. nitens (b), (d) C. salicifolius, (e) C. zhejiangensis, and (f) C. praecox. (B) The IRscope based analyses provide insight into similarities and variations at junction sites of cp genomes. For each species, genes present on the top of their corresponding track transcribed in positive strand from right to left direction whereas genes present below their respective track transcribe on negative strand from left to right direction. JLA, JLB, JSA, and JSB denote corresponding junctions between the LSC, SSC, and IR regions. The genes extending from one region to another region of chloroplast genomes are shown with the T bar above or below the solid lines. The size of T bars corresponds to the length of the genes or gene fragments present in the different regions. The plotted genes and distances in the vicinity of the junction sites are the scaled projection of the genome.
Analyses of Codon Usage and Amino Acid Frequency
The codon usage analyses revealed high encoding efficacy for those codons that end with A/T at the 3′ end as opposed to codons that end with C/G at the 3′. The amino acid frequency analyses revealed that leucine was the highest (>10%) encoded amino acid followed by iso-leucine (>8%), whereas cysteine (1) and tyrosine (1.5%) were among the least encoded amino acids (Supplementary Table S4).
Repeats Analyses
The analyses of microsatellites revealed 49–54 repeats in the six cp genomes (Supplementary Table S5). Most of the repeats existed in the non-coding regions of LSC, followed by SSC, and then IR (Supplementary Figure S1A). Mononucleotide A/T repeats were most abundant in all species, especially in C. campanulatus, followed by dinucleotide and trinucleotide repeats. Notably, mononucleotide G/C repeats were only found once or twice in the genomes. Most of the microsatellites were in A/T rich regions (Supplementary Table S5). The analyses of oligonucleotide repeats (30 nucleotides or longer in length) showed that most of the repeat types present in the chloroplast genomes were forward repeats, followed by palindromic repeats. Very few reverse repeats were found only in two cp genomes, and complimentary repeats were not found in the genomes (Supplementary Figure S1B).
Analyses of Substitution Types
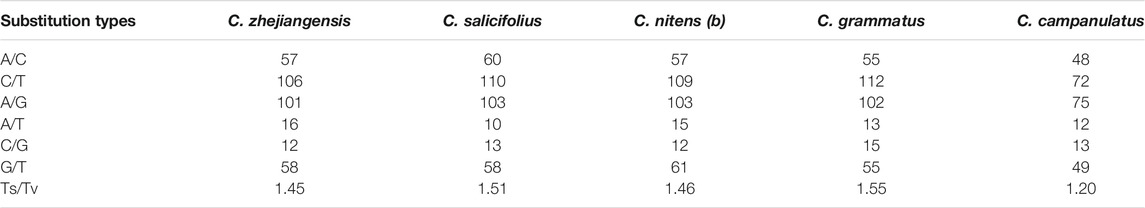
We recorded a higher number of transition (Ts) than transversion (Tv) substitutions. The ratios of Ts/Tv ranged from 1.2 to 1.55 (Table 2). The majority of Ts substitutions were promoted by C/T mutations except in C. campanulatus where A/G mutations were slightly higher than C/T mutations. Tv substitutions were found to be related to A/C and G/T rather than to A/T and C/G (Table 2). For Ks and Ka, we found a higher average of Ks than Ka on 78 protein-coding genes in the Chimonanthus species. Only atpF and rpoB genes showed positive selection (Ka/Ks ratio of 1.92 and 1.23, respectively) in C. campanulatus chloroplast genome. The rbcL gene in C. grammatus showed neutral selection (Ka/Ks ratio of 0.96), while the matK gene in C. grammatus, C. nitens, and C. zhejiangensis showed approximate neutral selection (Ka/Ks ratio of 0.86). All other genes in all species showed strong purifying selection (Supplementary Table S6).
Quantitative and Qualitative Analyses of Correlations Among Mutational Events
We performed four comparisons at the family level and six comparisons at the genus level to unravel the quantitative and qualitative correlations among the species of Calycanthaceae.
At the family level, we found strong correlations between substitutions and InDels in all four comparisons with an average of 0.43. The correlations between InDels, and repeats were moderate in two comparisons while near to strong in two comparisons. The average of correlations was found at 0.39. Correlations between substitutions and repeats were very weak in two comparisons, whereas weak correlations were observed in two comparisons. The average of correlations was found at 0.195 (Figure 3A).
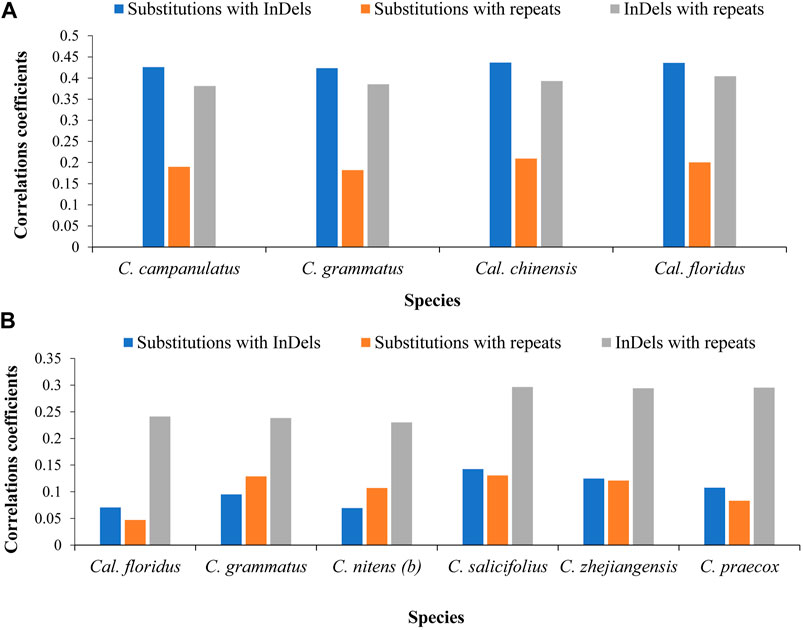
FIGURE 3. Correlations among mutational events at family and genus levels. (A) Correlations among distinctly related species of family level. The chloroplast genome of Idiospermum australiense was used as reference for all comparisons. (B) Correlations among closely related species at genus level. The Calycanthus chinensis was used as reference in comparison of two species of Calycanthus, whereas the cp genome of Chimonanthus campanulatus was used as reference for all species of Chimonanthus. The x-axis shows name of the species, whereas the y-axis shows the value of correlations coefficient.
At the genus level, we observed negligible/very weak or weak correlations. We recorded negligible correlations between substitutions and InDels (0.07–0.14) and between substitutions and repeats (0.04–0.13) in pairwise comparisons of all species, whereas we recorded weak correlations between InDels and repeats (0.23–0.29). Here, our results revealed weak correlations between closely related species (Figure 3B).
Qualitative analyses revealed that, at the family level, 98.36–98.90% of InDels co-occurred with substitutions in the same bins, whereas 95.30–97.01% of repeats co-occurred with substitutions, and 56.41–58.97% of repeats co-occurred with InDels in the same bins. At the genus level, 53.23–69.57% of InDels coexisted with substitutions, whereas 8.93–17.85% of repeats coexisted with InDels, and 41.96–56.25% of repeats coexisted with substitutions (Table 3). The distributions of substitutions, InDels, and repeats in 250-bp bins are shown in Supplementary Table S7.
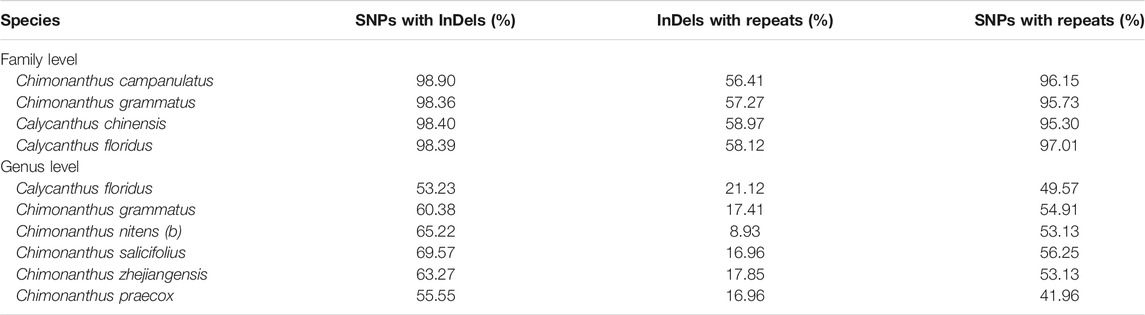
TABLE 3. The co-occurrence of InDels with substitutions, and of repeats with substitutions and InDels in family Calycanthaceae.
Phylogenetic Inference and Estimation of Divergence Times
A maximum likelihood tree was reconstructed using 10 chloroplast genomes in the family Calycanthaceae from all three genera. These included all six recognized species of Chimonanthus, two of the three Calycanthus species, and the only Idiospermum species. The C. nitens chloroplast genome was represented twice in this analysis: “a” was taken from NCBI (Dong et al., 2020), and “b” was assembled in this study (LIU2047, Supplementary Table S1). The multiple sequence alignment excluded one IR copy (IRb) and indels in the alignment, and was 127,010 nucleotides long, wherein 123,302 nucleotides (97.1%) were invariant sites. Best fit model on the data was found to be TVM + I + G4. The resulting phylogeny shows the monophyly of Chimonanthus that is sister to Calycanthus. However, Chimonanthus nitens is paraphyletic with accession (b) forming a clade with C. zhejiangensis but distinct from C. nitens (a) (Figure 4). The Bayesian inference (BI) using BEAST (Drummond and Rambaut, 2007) generated the same topology, and Figure 4 also shows the posterior probabilities of each node/clade.
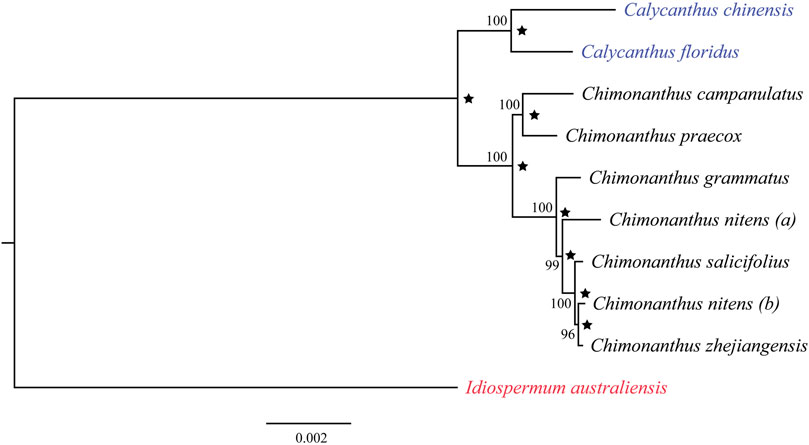
FIGURE 4. Phylogenetic relationships among nine species in the family Calycanthaceae based on the maximum-likelihood analyses and Bayesian inference. Idiospermum australiense was used as the outgroup. Numbers at each node are bootstrap support values in 100 replicates. Star indicates Bayesian posterior probabilities above 0.95 from the BEAST analyses.
Based on whole cp genome sequences, the BEAST analyses (Drummond and Rambaut, 2007) estimated the divergence between Chimonanthus and Calycanthus to be 29.88 Ma (95% HPD: 17.19–45.50), and that between Calycanthus chinensis and Calycanthus floridus as 17.27 Ma (95% HPD: 9.31–29.50; Figure 5). Within Chimonanthus, the divergence between the C. praecox-campanulatus clade and the clade of the rest of the four species of Chimonanthus was 15.20 Ma (95% HPD: 8.82–24.96), that between C. praecox and C. campanulatus was 11.89 Ma (node 1; 95% HPD: 6.04–19.80). The split of C. grammatus and the clade of C. nitens, C. zhejiangensis, and C. salicifolius occurred in the late Miocene, with an estimated age of 7.01 Ma (node 2; 95% HPD: 3.89–11.74). C. nitens (a) diverged from C. salicifolius, C. zhejiangensis, and C. nitens (b) at 5.54 Ma (node 3; 95% HPD: 2.92–9.26). The divergence time of C. salicifolius and that of C. zhejiangensis and C. nitens (b) occurred in the Pleistocene, with an estimated age of 2.13 Ma (node 4; 95% HPD: 1.04–3.76) and 1.38 Ma (node 5; 95% HPD: 0.56–2.54), respectively (Figure 5) [Supplementary Table S8 (A)]. The estimates for the divergence times of the different nodes through fossil record calibration showed slightly more recent estimates for each node [Supplementary Table S8 (B)].
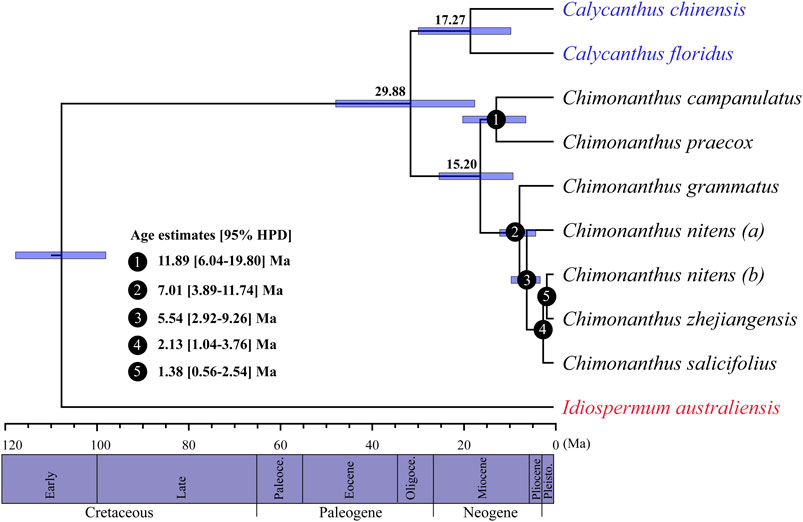
FIGURE 5. BEAST-derived chronograms of Calycanthaceae based on whole chloroplast genome sequences. Blue bars indicate the 95% highest posterior density (HPD) credibility intervals for node ages (Ma). Estimated divergence times of Chimonanthus species are indicated at the nodes (nodes 1–5).
Discussion
In this study, we have assembled chloroplast genomes for five Chimonanthus species, and compared them with the two previously reported species of the genus (Dong et al., 2020). The chloroplast genomes of Chimonanthus species range in size from 153,010 to 153,299 bp with no obvious structural variation among the taxa. The organization and gene content of chloroplast genomes are similar among the studied species, consistent with previously reported genomes of Calycanthaceae (Goremykin et al., 2003; Dong et al., 2020). We have also evaluated correlations among mutational events, performed phylogenetic analyses in Calycanthaceae based on whole cp genomes, and explored their divergence history. These comparative chloroplast genomic analyses are important to provide insights into the genome organization and evolutionary history of species within the genus Chimonanthus and family Calycanthaceae.
Chloroplast Genome Evolution of Chimonanthus
The infA gene (translation initiation factor 1), which has been independently lost multiple times during land plant evolution (Millen et al., 2001), is present in all Chimonanthus species in this study. This gene was also found in one species of Calycanthus (Goremykin et al., 2003) suggesting that the presence of infA in the chloroplast genome might be an ancestral condition in the family Calycanthaceae.
Chloroplast genomes are conserved in most plant lineages, but the expansion and contraction of the border regions between SC and IR regions contribute to variation in chloroplast genome lengths (Li et al., 2013; Dong et al., 2016; Menezes et al., 2018; Henriquez et al., 2020a; Abdullah et al., 2020c). Higher variation at the boundaries of single copy and inverted repeats is found in comparisons of deeply diverged lineages; conversely, those species that evolved recently have less variation at the boundary regions (Abdullah S. et al., 2019; Iram et al., 2019; Henriquez et al., 2020b; Shahzadi et al., 2020). In the current study, Chimonanthus species showed resemblance of this variational pattern at the junctions of single copy and inverted repeats. This might be indicative of recent evolutionary divergences among Chimonanthus species.
Codon usage analysis is essential to understand genome structure, evolutionary processes, and selection pressure on the genes (Morton, 1998; Goodarzi et al., 2008). The degenerative property of the genetic code shows that more than one codon can encode a single amino acid. Codons that encode a single amino acid are known as synonymous codons. Codon usage analysis in Chimonanthus species showed a bias for A/T-ending codons at 3′ end. This phenomenon has been mostly observed in many sequenced chloroplast genomes of land plants and may be due to A/T-rich chloroplast genome content (Amiryousefi et al., 2018b; Abdullah I. et al., 2019, Abdullah et al., 2019 S.; Mehmood et al., 2020a, 2020b). The high similarity exists in the codon usage of closely related species or of the same lineage, which provides insight into the evolution of plants, and the species of the same genus have high similarity compared with family-level comparison (Abdullah et al., 2020e; Abdullah et al., 2021). Hence, the high similarity in codon usage of Chimonanthus species showed their close evolutionary relationships.
Simple sequence repeats (SSRs) in the chloroplast genome can serve as highly informative genetic markers and are therefore often used in analysis of genetic variation, taxonomy, parentage analysis, functional diversity, linkage and comparative mapping, and evolutionary studies in various plant species (McCouch et al., 2002; Shirasawa et al., 2013; Xu et al., 2013; Ren et al., 2014; George et al., 2015; Yang et al., 2016; Zhu et al., 2016). We observed an abundance of mononucleotide repeats with A/T motif and of dinucleotide repeats with AT/AT motif in the chloroplast genome of Chimonanthus species. A similar pattern of SSR distribution was also reported in chloroplast genomes of other plant lineages (Du et al., 2017; Huo et al., 2019). The presence of abundant SSR loci in the genome suggests the potential utility for future population genetic work.
The Ka/Ks is important in evolutionary studies, as it reveals the selection pressure on protein-coding genes (Nazareno et al., 2015). The Ka/Ks < 1 describes purifying selection, Ka/Ks = 1 reveals neutral selection, and Ka/Ks > 1 shows positive selection (Lawrie et al., 2013). Due to higher Ks than Ka, we observed Ka/Ks < 1 for most of the protein-coding genes in Chimonanthus species. These results are consistent with previous studies of angiosperm chloroplast genomes, as purifying selection pressure mostly acts on the genes of chloroplast genomes (Cheng et al., 2017; Abdullah et al., 2020e; Shahzadi et al., 2020). The genes that showed higher Ka/Ks value are rpoB and atpF in C. campanulatus. This positive selection might confer some selective advantage to C. campanulatus.
The weak to strong correlations among mutational events including substitutions, InDels, and oligonucleotide repeats have been reported in the family Araceae (Ahmed et al., 2012; Abdullah et al., 2020a), Malvaceae (Abdullah et al., 2020d), between two species of Cephalotaxus (Cephalotaxaceae) (Yi et al., 2013), and in various species of Dendrobium (Orchidaceae) (Li et al., 2020). The strong correlations were observed at the family and subfamily levels (Abdullah et al., 2020d; 2020a), whereas weak correlations were observed among closely related species such as in pairwise comparisons of Symplocarpus (Abdullah et al., 2020a), Theobroma (Abdullah et al., 2020d), and Cephalotaxus (Yi et al., 2013). In the current study, the phylogenetic analyses revealed very close relationships among the species within Calycanthus and Chimonanthus. Therefore, we were interested in seeing how the mutational events are correlated. Our study again confirms that weak correlations exist among the closely related species. Previously, it was suggested that existence of fewer mutations between the compared species and incomplete lineage sorting may lead to low correlation coefficient (Abdullah et al., 2020a). Hence, the low correlations can be expected in the closely related species with very short divergence events due to availability of less time to InDels and repeats to generate mutations, as the mutations arise by errors in replications due to the presence of InDels or existence of a high number of repeats in regions of the genome (Tian et al., 2008; McDonald et al., 2011).
Phylogenetic Relationships
Our phylogenetic analyses of Calycanthaceae (Figure 4) support the monophyly of both Calycanthus and Chimonanthus. Calycanthus floridus is sister to Calycanthus chinensis; however, the inclusion of Calycanthus occidentalis is needed in future studies to test the relationships within Calycanthus. Although Calycanthus chinensis endemic to Zhejiang, China, is considered morphologically highly distinct from the two North American species (Cheng and Chang, 1964), we have detected that it shares unusually similar dark brownish color trichomes with Calycanthus floridus, on the abaxial surface of the leaf midvein. On the other hand, species in Chimonanthus, e.g., C. praecox (other species not shown), share transparent trichomes distributed on the midvein of the abaxial leaf surface (Figure 6). The dark brownish color of trichomes may be due to the presence of biominerals (Ensikat et al., 2017), which may also provide phylogenetic signals (Mustafa et al., 2018). However, the confirmation and detailed patterns of biomineralization particularly in Calycanthus will require further investigation.
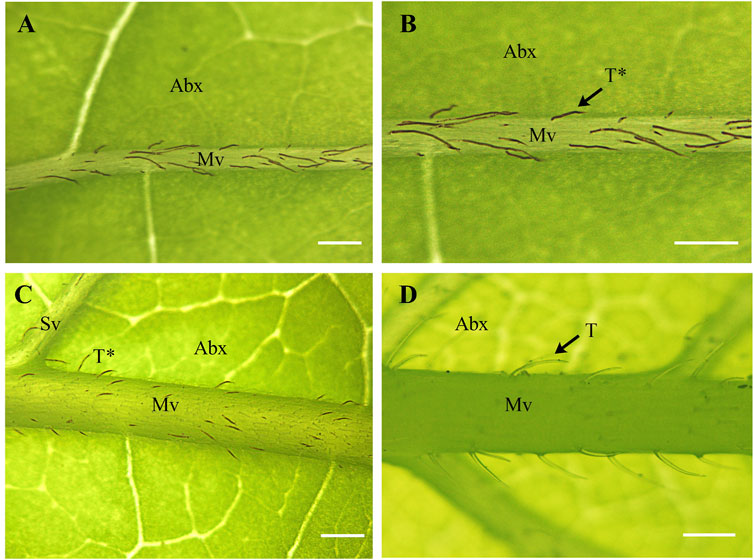
FIGURE 6. Structure and distribution of unicellular trichomes in Calycanthaceae. (A,B) Calycanthus chinenses (C) Calycanthus floridus; and (D) Chimonanthus praecox. Abx, abaxial; Mv, midvein; Sv, secondary vein; T, transparent trichomes; T*, dark brownish (non-transparent) trichomes. Scale bars (A–D) = 200 µm. Abaxial leaf surfaces were examined using a Nikon SMZ800 stereomicroscope.
Within Chimonanthus, C. praecox and C. campanulatus form a clade, consistent with previous studies (Zhou et al., 2006, Zhou et al., 2007). The Chimonanthus praecox–C. campanulatus clade is sister to the clade of the remaining four species of the genus. Chimonanthus grammatus is sister to the clade of C. salicifolius, C. zhejiangensis, and C. nitens. Chimonanthus nitens is paraphyletic and it is closely related to C. salicifolius and C. zhejiangensis. In the previous study by Zhou et al. (2006), the relationships among the four species C. salicifolius, C. grammatus, C. zhejiangensis, and C. nitens were difficult to resolve. These species show high similarities in morphology and have been difficult to distinguish based on morphological characters (Chen, 1998; Dai et al., 2012). We sampled C. nitens (b) from the natural population located in Jiangxi, China (Supplementary Table S1), and it may represent an ecotype. Based on electrochemical fingerprint data, Xu et al. (2020) suggested that even C. zhejiangensis and C. grammatus may represent two ecotypes or varieties of C. nitens, and the small morphological variations between these three species may be due to environmental factors and may not be used for distinguishing the species. Chloroplast sequence data have also been used to suggest a particular species (Colocasia formosana) as an ecotype of another species (Colocasia esculenta) (Ahmed et al., 2020). However, confirmation of ecotypes requires reciprocal transplant experiments to link their establishment with local adaptation (Lowry, 2012). The complex species relationship in Chimonanthus and paraphyly of C. nitens in particular may have also arisen because of interspecific hybridization and introgression (Rieseberg and Brouillet, 1994; Hegarty and Hiscock, 2005; Mallet, 2005; Vriesendorp and Bakker, 2005). The reticulate phylogenetic relationships of closely related species caused by these naturally occurring processes present challenges to species delimitation (Mallet et al., 2016; Pennisi, 2016). Another cause of complex phylogenetic relationships within a group is incomplete lineage sorting, which cannot be ruled out in our case. Both incomplete lineage sorting and interspecific hybridization/introgression may have resulted in shared genetic variation (De Queiroz, 2007; Zhou et al., 2017). Both phenomena are observed frequently in taxa that are products of incipient radiation (Gavrilets and Losos, 2009; Goetze et al., 2017). Further studies at the population and species levels using both nuclear and chloroplast markers are required to test the alternate hypotheses of hybridization among distinct species and incomplete lineage sorting due to incipient radiation of C. nitens and its close relatives, eventually leading to the well-tested species delimitations in Chimonanthus.
Diversification History of Chimonanthus in Eastern Asia
Overall, the species of Chimonanthus (except Chimonanthus praecox) are restricted to the subtropical regions of China, and our analyses indicate that early diversifications of most extant Chimonanthus species might have occurred during the middle and late Miocene (Figure 5, node 1, node 2, and node 3). This period corresponds to the intensification of the Eastern Asian monsoon and climate changes (Zhisheng et al., 2001; Sun and Wang, 2005), which might have contributed to the rapid radiation of many Eastern Asian plants, including subtropical lineages. For example, the initial diversification of subtropical Cyclocarya paliurus was associated with the intensification of Eastern Asian monsoon in the middle Miocene, which provided suitable climatic conditions that facilitated its survival in Southwestern China (Kou et al., 2016). Similarly, climatic fluctuations during late Miocene might have triggered the rapid radiation of Quercus arbutifolia (Fagaceae) inhabiting subtropical montane cloud forests (MCFs) in Southern China (Xu et al., 2016). Speciation event in the genus Cercidiphyllum at the Miocene/Pliocene boundary (Qi et al., 2012) and divergence of Tetracentron sinense by late Miocene global cooling (Sun et al., 2014) also suggest that pre-Quaternary climate changes might have contributed to the diversification of temperate and subtropical plants in Eastern Asia.
Zhou et al. (2006) reported that several species within Chimonanthus diverged recently approximately 1–2 Ma ago. Here we used whole cp genomes sequences to enhance the phylogenetic resolution and estimate the divergence times of Chimonanthus species, with all species sampled. The three species, C. salicifolius, C. nitens (b), and C. zhejiangensis were found to have diverged relatively recently, with C. salicifolius diverging from the other two roughly 2.13–1.88 Ma (node 4 and node 5 in Figure 5), in the Pleistocene, which also agrees with the estimates of Zhou et al. (2006). Our results, hence, support active recent speciation events of an ancient lineage in the subtropical forest biome in Eastern China. The subtropical forests in Eastern China have been well known as an important refugia for the Eastern Asian flora (Wu, 1980) and for many Eastern Asian–North American disjunct lineages (Wen, 1999). Our results strongly showcase the importance of the subtropical forests as recent diversification centers for endemic elements of the Eastern Asian flora (see also Qiu et al., 2011; Wang H. X. et al., 2020).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article. Moreover, additional data generated and analyzed are provided as Supplementary Material.
Author Contributions
X-QL, AJ, and L-QC designed the experiment and contributed to the sampling. AJ performed the experiments and drafted the manuscript. AJ, A, and IA analyzed the data. IA, JW, X-QL, and Z-YM revised the manuscript. All authors approved the final version of the manuscript.
Funding
This project was supported by the Fundamental Research Funds for the Central Universities (Grant No. 510320059).
Conflict of Interest
IA was employed by the Alpha Genomics Private Limited.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.709996/full#supplementary-material
Supplementary Figure S1 | Distribution of repeats in the six Chimonanthus chloroplast genomes. (A) Represents distribution of microsatellite repeats in different regions of the chloroplast genomes. (B) Reputer results showing the number of forward, reverse, complimentary and palindromic oligonucleotide repeats in Chimonanthus chloroplast genomes.
Supplementary Table 1 | Collection location and voucher specimen number of the studied Chimonanthus species.
Supplementary Table 2 | Summary statistics of the data. Average coverage depth obtained after mapping the short reads to their respective assembled chloroplast genomes using BWA and visualizing in Tablet.
Supplementary Table 3 | List of genes found in the chloroplast genomes of Chimonanthus species.
Supplementary Table 4 | Frequencies and percentages of nucleotides and amino acids, and codon usage for the six Chimonanthus chloroplast genomes.
Supplementary Table 5 | Distribution of microsatellite repeats in the six Chimonanthus chloroplast genomes.
Supplementary Table 6 | Non-synonymous (Ka) versus synonymous (Ks) substitutions of 78 protein coding genes. Cc: C. campanulatus; Cg: C. grammatus; Cn: C. nitens (b); Cs: C. salicifolius; Cz: C. zhejiangensis.
Supplementary Table 7 | Distributions of substitutions, InDels, and oligonucleotide repeats in bins of 250 bp in pairwise comparisons at family and genus level.
Supplementary Table 8 | Divergence time estimates (in million years) for Calycanthaceae with the highest posterior density (HPD) interval limits. (A) The minimum age of Calycanthaceae was set to minimally 108 Mya (secondary calibration). (B) The minimum age of Calycanthaceae was set to minimally 90 Mya (fossil record).
References
Abdullah, , Henriquez, C. L., Croat, T. B., Poczai, P., and Ahmed, I. (2020a). Mutational Dynamics of Aroid Chloroplast Genomes II. Front. Genet. 11, 1562. doi:10.3389/FGENE.2020.610838
Abdullah, , Henriquez, C. L., Mehmood, F., Carlsen, M. M., Islam, M., et al. (2020b). Complete Chloroplast Genomes of Anthurium Huixtlense and Pothos Scandens (Pothoideae, Araceae): Unique Inverted Repeat Expansion and Contraction Affect Rate of Evolution. J. Mol. Evol. 88, 562–574. doi:10.1007/s00239-020-09958-w
Abdullah, , Henriquez, C. L., Mehmood, F., Shahzadi, I., Ali, Z., et al. (2020c). Comparison of Chloroplast Genomes Among Species of Unisexual and Bisexual Clades of the Monocot Family Araceae. Plants 9, 737. doi:10.3390/plants9060737
Abdullah, , Mehmood, F., Rahim, A., Heidari, P., Ahmed, I., and Poczai, P. (2021). Comparative Plastome Analysis of Blumea , with Implications for Genome Evolution and Phylogeny of Asteroideae. Ecol. Evol. 11, 7810–7826. doi:10.1002/ece3.7614
Abdullah, , Mehmood, F., Shahzadi, I., Ali, Z., Islam, M., Naeem, M., et al. (2020d). Correlations Among Oligonucleotide Repeats, Nucleotide Substitutions, and Insertion-Deletion Mutations in Chloroplast Genomes of Plant Family Malvaceae. J. Syst. Evol. 59, 388–402. doi:10.1111/jse.12585
Abdullah, , Mehmood, F., Shahzadi, I., Waseem, S., Mirza, B., Ahmed, I., et al. (2020e). Chloroplast Genome of Hibiscus rosa-sinensis (Malvaceae): Comparative Analyses and Identification of Mutational Hotspots. Genomics 112, 581–591. doi:10.1016/j.ygeno.2019.04.010
Abdullah, , Shahzadi, I., Shahzadi, I., Mehmood, F., Ali, Z., Malik, M. S., et al. (2019a). Comparative Analyses of Chloroplast Genomes Among Three Firmiana Species: Identification of Mutational Hotspots and Phylogenetic Relationship with Other Species of Malvaceae. Plant Gene 19, 100199. doi:10.1016/J.PLGENE.2019.100199
Abdullah, , Waseem, S., Waseem, S., Mirza, B., Ahmed, I., and Waheed, M. T. (2019b). Comparative Analyses of Chloroplast Genomes of Theobroma cacao and Theobroma grandiflorum. Biologia 75, 761–771. doi:10.2478/s11756-019-00388-8
Ahmed, I., Biggs, P. J., Matthews, P. J., Collins, L. J., Hendy, M. D., and Lockhart, P. J. (2012). Mutational Dynamics of Aroid Chloroplast Genomes. Genome Biol. Evol. 4, 1316–1323. doi:10.1093/gbe/evs110
Ahmed, I. (2015). Chloroplast Genome Sequencing: Some Reflections. Next Generat. Sequenc. Applic. 02, 119. doi:10.4172/2469-9853.1000119
Ahmed, I., Lockhart, P. J., Agoo, E. M. G., Naing, K. W., Nguyen, D. V., Medhi, D. K., et al. (2020). Evolutionary Origins of Taro (Colocasia esculenta) in Southeast Asia. Ecol. Evol. 10, 13530–13543. doi:10.1002/ece3.6958
Ahmed, I., Matthews, P. J., Biggs, P. J., Naeem, M., Mclenachan, P. A., and Lockhart, P. J. (2013). Identification of Chloroplast Genome Loci Suitable for High‐resolution Phylogeographic Studies of Colocasia esculenta (L.) Schott (Araceae) and Closely Related Taxa. Mol. Ecol. Resour. 13, 929–937. doi:10.1111/1755-0998.12128
Amiryousefi, A., Hyvönen, J., and Poczai, P. (2018a). IRscope: an Online Program to Visualize the junction Sites of Chloroplast Genomes. Bioinformatics 34, 3030–3031. doi:10.1093/bioinformatics/bty220
Amiryousefi, A., Hyvönen, J., and Poczai, P. (2018b). The Chloroplast Genome Sequence of Bittersweet (Solanum dulcamara): Plastid Genome Structure Evolution in Solanaceae. PLoS One 13, e0196069. doi:10.1371/journal.pone.0196069
Andrews, S., Krueger, F., Seconds-Pichon, A., Biggins, F., and Wingett, S. (2015). FastQC. A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinforma. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Beier, S., Thiel, T., Münch, T., Scholz, U., and Mascher, M. (2017). MISA-web: a Web Server for Microsatellite Prediction. Bioinformatics 33, 2583–2585. doi:10.1093/bioinformatics/btx198
Bi, Y., Zhang, M.-F., Xue, J., Dong, R., Du, Y.-P., and Zhang, X.-H. (2018). Chloroplast Genomic Resources for Phylogeny and DNA Barcoding: A Case Study on Fritillaria. Sci. Rep. 8, 1–12. doi:10.1038/s41598-018-19591-9
Blake, S. T. (1972). Idiospermum (Idiospermaceae), a New Genus and Family for Calycanthus australiensis. Contrib. Queensl. Herb. 12, 1–37.
Carlquist, S. (1983). Wood Anatomy of Calycanthaceae. Aliso 10, 427–441. doi:10.5642/aliso.19831003.06
Chase, M. W., Christenhusz, M. J. M., Fay, M. F., Byng, J. W., Judd, W. S., Soltis, D. E., et al. (2016). An Update of the Angiosperm Phylogeny Group Classification for the Orders and Families of Flowering Plants: APG IV. Bot. J. Linn. Soc. 181, 1–20. doi:10.1111/boj.12385
Chen, L. Q. (1998). Studies on Biosystematics of Calycanthaceae L. Ph.D. Dissertation. Beijing: Beijing Forestry University.
Cheng, H., Li, J., Zhang, H., Cai, B., Gao, Z., Qiao, Y., et al. (2017). The Complete Chloroplast Genome Sequence of Strawberry (Fragaria × ananassa Duch.) and Comparison with Related Species of Rosaceae. PeerJ 5, e3919. doi:10.7717/peerj.3919
Cheng, W. C., and Chang, S. Y. (1964). Genus Novum Calycanthacearum Chinae Orientalis. Acta Phytotaxon. Sin. 9, 135–138.
Chevenet, F., Brun, C., Bañuls, A.-L., Jacq, B., and Christen, R. (2006). TreeDyn: Towards Dynamic Graphics and Annotations for Analyses of Trees. BMC Bioinformatics 7, 439. doi:10.1186/1471-2105-7-439
Christenhusz, M. J. M., and Byng, J. W. (2016). The Number of Known Plants Species in the World and its Annual Increase. Phytotaxa 261, 201. doi:10.11646/phytotaxa.261.3.1
Crepet, W. L., Nixon, K. C., and Gandolfo, M. A. (2005). An Extinct Calycanthoid Taxon, Jerseyanthus calycanthoides, from the Late Cretaceous of New Jersey. Am. J. Bot. 92, 1475–1485. doi:10.3732/ajb.92.9.1475
Dai, P. F., Yang, J., Zhou, T. H., Huang, Z. H., Feng, L., Su, H. L., et al. (2012). Genetic Diversity and Differentiation in Chimonanthus praecox and Ch. salicifolius (Calycanthaceae) as Revealed by Inter-simple Sequence Repeat (ISSR) Markers. Biochem. Syst. Ecol. 44, 149–156. doi:10.1016/j.bse.2012.04.014
Daniell, H., Lin, C. S., Yu, M., and Chang, W. J. (2016). Chloroplast Genomes: Diversity, Evolution, and Applications in Genetic Engineering. Genome Biol. 17, 134. doi:10.1186/s13059-016-1004-2
Darling, A. C. E., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: Multiple Alignment of Conserved Genomic Sequence with Rearrangements. Genome Res. 14, 1394–1403. doi:10.1101/gr.2289704
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). JModelTest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 9, 772. doi:10.1038/nmeth.2109
De Queiroz, K. (2007). Species Concepts and Species Delimitation. Syst. Biol. 56, 879–886. doi:10.1080/10635150701701083
Dong, W., Xu, C., Li, D., Jin, X., Li, R., Lu, Q., et al. (2016). Comparative Analysis of the Complete Chloroplast Genome Sequences in psammophytic Haloxylon species (Amaranthaceae). PeerJ 4, e2699. doi:10.7717/peerj.2699
Dong, W., Xu, C., Wen, J., and Zhou, S. (2020). Evolutionary Directions of Single Nucleotide Substitutions and Structural Mutations in the Chloroplast Genomes of the Family Calycanthaceae. BMC Evol. Biol. 20, 96. doi:10.1186/s12862-020-01661-0
Doyle, J., and Doyle, J. (1987). A Rapid Isolation Procedure for Small Amounts of Leaf Tissue. Phytochem. Bull. 19 (1), 11–15. doi:10.2307/4119796
Drummond, A. J., Ho, S. Y. W., Phillips, M. J., and Rambaut, A. (2006). Relaxed Phylogenetics and Dating with Confidence. Plos Biol. 4, e88. doi:10.1371/journal.pbio.0040088
Drummond, A. J., and Rambaut, A. (2007). BEAST: Bayesian Evolutionary Analysis by Sampling Trees. BMC Evol. Biol. 7, 214–218. doi:10.1186/1471-2148-7-214
Du, Y. P., Bi, Y., Yang, F. P., Zhang, M. F., Chen, X. Q., Xue, J., et al. (2017). Complete Chloroplast Genome Sequences of Lilium: Insights into Evolutionary Dynamics and Phylogenetic Analyses. Sci. Rep. 7, 5751. doi:10.1038/s41598-017-06210-2
Ensikat, H. J., Mustafa, A., and Weigend, M. (2017). Complex Patterns of Multiple Biomineralization in Single-Celled Plant Trichomes of the Loasaceae. Am. J. Bot. 104, 195–206. doi:10.3732/ajb.1600331
Friis, E. M., Eklund, H., Pedersen, K. R., and Crane, P. R. (1994). Virginianthus calycanthoides gen. et sp. nov.-A Calycanthaceous Flower from the Potomac Group (Early Cretaceous) of Eastern North America. Int. J. Plant Sci. 155, 772–785. doi:10.1086/297217
Gavrilets, S., and Losos, J. B. (2009). Adaptive Radiation: Contrasting Theory with Data. Science 323, 732–737. doi:10.1126/science.1157966
George, B., Bhatt, B. S., Awasthi, M., George, B., and Singh, A. K. (2015). Comparative Analysis of Microsatellites in Chloroplast Genomes of Lower and Higher Plants. Curr. Genet. 61, 665–677. doi:10.1007/s00294-015-0495-9
Goetze, M., Zanella, C. M., Palma-Silva, C., Büttow, M. V., and Bered, F. (2017). Incomplete Lineage Sorting and Hybridization in the Evolutionary History of Closely Related, Endemic Yellow-Flowered Aechmea Species of Subgenus Ortgiesia (Bromeliaceae). Am. J. Bot. 104, 1073–1087. doi:10.3732/ajb.1700103
Goodarzi, H., Torabi, N., Najafabadi, H. S., and Archetti, M. (2008). Amino Acid and Codon Usage Profiles: Adaptive Changes in the Frequency of Amino Acids and Codons. Gene 407, 30–41. doi:10.1016/j.gene.2007.09.020
Goremykin, V., Hirsch-Ernst, K. I., Wölfl, S., and Hellwig, F. H. (2003). The Chloroplast Genome of the “Basal” Angiosperm Calycanthus fertilis - Structural and Phylogenetic Analyses. Plant Syst. Evol. 242, 119–135. doi:10.1007/s00606-003-0056-4
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) Version 1.3.1: Expanded Toolkit for the Graphical Visualization of Organellar Genomes. Nucleic Acids Res. 47, W59–W64. doi:10.1093/nar/gkz238
Hegarty, M. J., and Hiscock, S. J. (2005). Hybrid Speciation in Plants: New Insights from Molecular Studies. New Phytol. 165, 411–423. doi:10.1111/j.1469-8137.2004.01253.x
Henriquez, C. L., Abdullah, , Ahmed, I., Carlsen, M. M., Zuluaga, A., Croat, T. B., et al. (2020a). Evolutionary Dynamics of Chloroplast Genomes in Subfamily Aroideae (Araceae). Genomics 112, 2349–2360. doi:10.1016/j.ygeno.2020.01.006
Henriquez, C. L., Abdullah, , Ahmed, I., Carlsen, M. M., Zuluaga, A., Croat, T. B., et al. (2020b). Molecular Evolution of Chloroplast Genomes in Monsteroideae (Araceae). Planta 251, 72. doi:10.1007/s00425-020-03365-7
Huo, Y., Gao, L., Liu, B., Yang, Y., Kong, S., Sun, Y., et al. (2019). Complete Chloroplast Genome Sequences of Four Allium Species: Comparative and Phylogenetic Analyses. Sci. Rep. 9, 12250. doi:10.1038/s41598-019-48708-x
Iram, S., Hayat, M. Q., Tahir, M., Gul, A., Abdullah, , , and Ahmed, I. (2019). Chloroplast Genome Sequence of Artemisia scoparia: Comparative Analyses and Screening of Mutational Hotspots. Plants 8, 476. doi:10.3390/plants8110476
Jansen, R. K., Cai, Z., Raubeson, L. A., Daniell, H., Depamphilis, C. W., Leebens-Mack, J., et al. (2007). Analysis of 81 Genes from 64 Plastid Genomes Resolves Relationships in Angiosperms and Identifies Genome-Scale Evolutionary Patterns. Proc. Natl. Acad. Sci. 104, 19369–19374. doi:10.1073/pnas.0709121104
Katoh, K., Kuma, K. I., Toh, H., and Miyata, T. (2005). MAFFT Version 5: Improvement in Accuracy of Multiple Sequence Alignment. Nucleic Acids Res. 33, 511–518. doi:10.1093/nar/gki198
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 28, 1647–1649. doi:10.1093/bioinformatics/bts199
Kou, Y., Cheng, S., Tian, S., Li, B., Fan, D., Chen, Y., et al. (2016). The Antiquity of Cyclocarya paliurus (Juglandaceae) Provides New Insights into the Evolution of Relict Plants in Subtropical China since the Late Early Miocene. J. Biogeogr. 43, 351–360. doi:10.1111/jbi.12635
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., and Giegerich, R. (2001). REPuter: the Manifold Applications of Repeat Analysis on a Genomic Scale. Nucleic Acids Res. 29, 4633–4642. doi:10.1093/nar/29.22.4633
Lawrie, D. S., Messer, P. W., Hershberg, R., and Petrov, D. A. (2013). Strong Purifying Selection at Synonymous Sites in D. melanogaster. Plos Genet. 9, e1003527. doi:10.1371/journal.pgen.1003527
Li, H., and Durbin, R. (2009). Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 25, 1754–1760. doi:10.1093/bioinformatics/btp324
Li, Y., and Li, P. T. (2000). Cladistic Analysis of Calycanthaceae. J. Trop. Subtrop. Bot. 8, 275–281. doi:10.1007/978-3-662-10777-5_49
Li, J., Ledger, J., Ward, T., and Tredici, P. del. (2004). Phylogenetics of Calycanthaceae Based on Molecular and Morphological Data, with a Special Reference to Divergent Paralogues of the nrDNA ITS Region. Harv. Pap. Bot. 9, 69–82. Available at: http://www.jstor.org/stable/41761683.
Li, L., Jiang, Y., Liu, Y., Niu, Z., Xue, Q., Liu, W., et al. (2020). The Large Single-Copy (LSC) Region Functions as a Highly Effective and Efficient Molecular Marker for Accurate Authentication of Medicinal Dendrobium Species. Acta Pharmaceutica Sinica B. 10, 1989–2001. doi:10.1016/j.apsb.2020.01.012
Li, R., Ma, P. F., Wen, J., and Yi, T. S. (2013). Complete Sequencing of Five Araliaceae Chloroplast Genomes and the Phylogenetic Implications. PLoS One 8, e78568. doi:10.1371/journal.pone.0078568
Lowry, D. B. (2012). Ecotypes and the Controversy over Stages in the Formation of New Species. Biol. J. Linn. Soc. 106, 241–257. doi:10.1111/j.1095-8312.2012.01867.x
Ma, Y. P., Zhao, L., Zhang, W. J., Zhang, Y. H., Xing, X., Duan, X. X., et al. (2020). Origins of Cultivars of Chrysanthemum -Evidence from the Chloroplast Genome and Nuclear LFY Gene. J. Syst. Evol. 58, 925–944. doi:10.1111/jse.12682
Mai, D. H. (1987). Neue Arten nach Früchten und Samen aus dem Tertiär von Nordwestsachsen und der Lausitz. Feddes Repert 98, 105–126. doi:10.1002/fedr.4910980106
Mallet, J., Besansky, N., and Hahn, M. W. (2016). How Reticulated Are Species. BioEssays 38, 140–149. doi:10.1002/bies.201500149
Mallet, J. (2005). Hybridization as an Invasion of the Genome. Trends Ecol. Evol. 20, 229–237. doi:10.1016/j.tree.2005.02.010
McCouch, S. R., Teytelman, L., Xu, Y., Lobos, K. B., Clare, K., Walton, M., et al. (2002). Development and Mapping of 2240 New SSR Markers for rice (Oryza sativa L.). DNA Res. 9, 199–207. doi:10.1093/dnares/9.6.199
McDonald, M. J., Wang, W. C., Huang, H. D., and Leu, J. Y. (2011). Clusters of Nucleotide Substitutions and Insertion/deletion Mutations Are Associated with Repeat Sequences. Plos Biol. 9, e1000622. doi:10.1371/journal.pbio.1000622
Mehmood, F., Abdullah, , Shahzadi, I., Ahmed, I., Waheed, M. T., and Mirza, B. (2020a). Characterization of Withania somnifera Chloroplast Genome and its Comparison with Other Selected Species of Solanaceae. Genomics 112, 1522–1530. doi:10.1016/j.ygeno.2019.08.024
Mehmood, F., Abdullah, , Ubaid, Z., Bao, Y., Poczai, P., and Mirza, B. (2020b). Comparative Plastomics of Ashwagandha (Withania, Solanaceae) and Identification of Mutational Hotspots for Barcoding Medicinal Plants. Plants 9, 752. doi:10.3390/plants9060752
Mehmood, F., Abdullah, , Ubaid, Z., Shahzadi, I., Ahmed, I., Waheed, M. T., et al. (2020c). Plastid Genomics of Nicotiana (Solanaceae): Insights into Molecular Evolution, Positive Selection and the Origin of the Maternal Genome of Aztec Tobacco (Nicotiana rustica). PeerJ 8, e9552. doi:10.1101/2020.01.13.90515810.7717/peerj.9552
Menezes, A. P. A., Resende-Moreira, L. C., Buzatti, R. S. O., Nazareno, A. G., Carlsen, M., Lobo, F. P., et al. (2018). Chloroplast Genomes of Byrsonima Species (Malpighiaceae): Comparative Analysis and Screening of High Divergence Sequences. Sci. Rep. 8, 1–12. doi:10.1038/s41598-018-20189-4
Middleton, C. P., Senerchia, N., Stein, N., Akhunov, E. D., Keller, B., Wicker, T., et al. (2014). Sequencing of Chloroplast Genomes from Wheat, Barley, rye and their Relatives Provides a Detailed Insight into the Evolution of the Triticeae Tribe. PLoS One 9, e85761. doi:10.1371/journal.pone.0085761
Millen, R. S., Olmstead, R. G., Adams, K. L., Palmer, J. D., Lao, N. T., Heggie, L., et al. (2001). Many Parallel Losses of Infa from Chloroplast Dna during Angiosperm Evolution with Multiple Independent Transfers to the Nucleus. Plant Cell 13, 645–658. doi:10.1105/tpc.13.3.645
Milne, I., Bayer, M., Cardle, L., Shaw, P., Stephen, G., Wright, F., et al. (2009). Tablet--next Generation Sequence Assembly Visualization. Bioinformatics 26, 401–402. doi:10.1093/bioinformatics/btp666
Mohr, B. A. R., and Eklund, H. (2003). Araripia florifera, a Magnoliid Angiosperm from the Lower Cretaceous Crato Formation (Brazil). Rev. Palaeobotany Palynology 126, 279–292. doi:10.1016/S0034-6667(03)00092-7
Morton, B. R. (1998). Selection on the Codon Bias of Chloroplast and Cyanelle Genes in Different Plant and Algal Lineages. J. Mol. Evol. 46, 449–459. doi:10.1007/PL00006325
Mustafa, A., Ensikat, H. J., and Weigend, M. (2018). Mineralized Trichomes in Boraginales: Complex Microscale Heterogeneity and Simple Phylogenetic Patterns. Ann. Bot. 121, 741–751. doi:10.1093/aob/mcx191
Nazareno, A. G., Carlsen, M., and Lohmann, L. G. (2015). Complete Chloroplast Genome of Tanaecium tetragonolobum: The First Bignoniaceae Plastome. PLoS One 10, e0129930. doi:10.1371/journal.pone.0129930
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 32, 268–274. doi:10.1093/molbev/msu300
Nikiforova, S. V., Cavalieri, D., Velasco, R., and Goremykin, V. (2013). Phylogenetic Analysis of 47 Chloroplast Genomes Clarifies the Contribution of Wild Species to the Domesticated Apple Maternal Line. Mol. Biol. Evol. 30, 1751–1760. doi:10.1093/molbev/mst092
Pennisi, E. (2016). Shaking up the Tree of Life. Science 354, 817–821. doi:10.1126/science.354.6314.817
Qi, X. S., Chen, C., Comes, H. P., Sakaguchi, S., Liu, Y. H., Tanaka, N., et al. (2012). Molecular Data and Ecological Niche Modelling Reveal a Highly Dynamic Evolutionary History of the East Asian Tertiary Relict Cercidiphyllum (Cercidiphyllaceae). New Phytol. 196, 617–630. doi:10.1111/j.1469-8137.2012.04242.x
Qiu, Y. X., Fu, C. X., and Comes, H. P. (2011). Plant Molecular Phylogeography in China and Adjacent Regions: Tracing the Genetic Imprints of Quaternary Climate and Environmental Change in the World's Most Diverse Temperate flora. Mol. Phylogenet. Evol. 59, 225–244. doi:10.1016/j.ympev.2011.01.012
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 67, 901–904. doi:10.1093/sysbio/syy032
Rambaut, A. (2012). FigTree v1.4.0. Available at: http//tree.bio.ed.ac.uk/software/figtree/ (Accessed January 17, 2021).
Ren, X., Jiang, H., Yan, Z., Chen, Y., Zhou, X., Huang, L., et al. (2014). Genetic Diversity and Population Structure of the Major Peanut (Arachis hypogaea L.) Cultivars Grown in China by SSR Markers. PLoS One 9, e88091. doi:10.1371/journal.pone.0088091
Renner, S. S. (1999). Circumscription and Phylogeny of the Laurales: Evidence from Molecular and Morphological Data. Am. J. Bot. 86, 1301–1315. doi:10.2307/2656778
Rieseberg, L. H., and Brouillet, L. (1994). Are many Plant Species Paraphyletic. Taxon 43, 21–32. doi:10.2307/1223457
Schattner, P., Brooks, A. N., and Lowe, T. M. (2005). The tRNAscan-SE, Snoscan and snoGPS Web Servers for the Detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, W686–W689. doi:10.1093/nar/gki366
Shahzadi, I., Abdullah, Mehmood, F., Mehmood, F., Ali, Z., Ahmed, I., and Mirza, B. (2020). Chloroplast Genome Sequences of Artemisia maritima and Artemisia absinthium: Comparative Analyses, Mutational Hotspots in Genus Artemisia and Phylogeny in Family Asteraceae. Genomics 112, 1454–1463. doi:10.1016/j.ygeno.2019.08.016
Shirasawa, K., Ishii, K., Kim, C., Ban, T., Suzuki, M., Ito, T., et al. (2013). Development of Capsicum EST-SSR Markers for Species Identification and In Silico Mapping onto the Tomato Genome Sequence. Mol. Breed. 31, 101–110. doi:10.1007/s11032-012-9774-z
Sonah, H., Deshmukh, R. K., Singh, V. P., Gupta, D. K., Singh, N. K., and Sharma, T. R. (2011). Genomic Resources in Horticultural Crops: Status, Utility and Challenges. Biotechnol. Adv. 29, 199–209. doi:10.1016/j.biotechadv.2010.11.002
Stamatakis, A. (2014). RAxML Version 8: a Tool for Phylogenetic Analysis and post-analysis of Large Phylogenies. Bioinformatics 30, 1312–1313. doi:10.1093/bioinformatics/btu033
Sun, Q., Zhu, J., Cao, F., and Chen, F. (2017). Anti-inflammatory Properties of Extracts from Chimonanthus nitens Oliv. Leaf. PLoS One 12, e0181094. doi:10.1371/journal.pone.0181094
Sun, X., and Wang, P. (2005). How Old Is the Asian Monsoon System?Palaeobotanical Records from China. Palaeogeogr. Palaeoclimatol. Palaeoecol. 222, 181–222. doi:10.1016/j.palaeo.2005.03.005
Sun, Y., Moore, M. J., Yue, L., Feng, T., Chu, H., Chen, S., et al. (2014). Chloroplast Phylogeography of the East Asian Arcto-Tertiary Relict Tetracentron sinense (Trochodendraceae). J. Biogeogr. 41, 1721–1732. doi:10.1111/jbi.12323
Thode, V. A., Lohmann, L. G., and Sanmartín, I. (2020). Evaluating Character Partitioning and Molecular Models in Plastid Phylogenomics at Low Taxonomic Levels: A Case Study using Amphilophium (Bignonieae, Bignoniaceae). J. Syst. Evol. 58, 1071–1089. doi:10.1111/jse.12579
Tian, D., Wang, Q., Zhang, P., Araki, H., Yang, S., Kreitman, M., et al. (2008). Single-nucleotide Mutation Rate Increases Close to Insertions/deletions in Eukaryotes. Nature 455, 105–108. doi:10.1038/nature07175
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq - Versatile and Accurate Annotation of Organelle Genomes. Nucleic Acids Res. 45, W6–W11. doi:10.1093/nar/gkx391
Valcárcel, V., and Wen, J. (2019). Chloroplast Phylogenomic Data Support Eocene Amphi‐Pacific Early Radiation for the Asian Palmate Core Araliaceae. J. Syst. Evol. 57, 547–560. doi:10.1111/jse.12522
Vriesendorp, B., and Bakker, F. T. (2005). Reconstructing Patterns of Reticulate Evolution in Angiosperms: What Can We Do. Taxon 54, 593–604. doi:10.2307/25065417
Wang, H. X., Moore, M. J., Barrett, R. L., Landrein, S., Sakaguchi, S., Maki, M., et al. (2020a). Plastome Phylogenomic Insights into the Sino‐Japanese Biogeography of Diabelia (Caprifoliaceae). J. Syst. Evol. 58, 972–987. doi:10.1111/jse.12560
Wang, Y. B., Liu, B. B., Nie, Z. L., Chen, H. F., Chen, F. J., Figlar, R. B., et al. (2020b). Major Clades and a Revised Classification of Magnolia and Magnoliaceae Based on Whole Plastid Genome Sequences via Genome Skimming. J. Syst. Evol. 58, 673–695. doi:10.1111/jse.12588
Wen, J. (1999). Evolution of Eastern Asian and Eastern North American Disjunct Distributions in Flowering Plants. Annu. Rev. Ecol. Syst. 30, 421–455. doi:10.1146/annurev.ecolsys.30.1.421
Wen, J., Harris, A., Kalburgi, Y., Zhang, N., Xu, Y., Zheng, W., et al. (2018). Chloroplast Phylogenomics of the New World Grape Species (Vitis, Vitaceae). J. Syst. Evol. 56, 297–308. doi:10.1111/jse.12447
Wen, J., Jansen, R. K., and Zimmer, E. A. (1996). “Phylogenetic Relationships and DNA Sequence Divergence of Eastern Asian and Eastern North American Disjunct Plants,” in Current Topics on Molecular Evolution ( Nei, M., and Takahata, N., eds). Pennsylvania State University, USA and Graduate School for Advance Studies, Hayama, Japan.
Xiong, J. S., Ding, J., and Li, Y. (2015). Genome-editing Technologies and Their Potential Application in Horticultural Crop Breeding. Hortic. Res. 2, 15019. doi:10.1038/hortres.2015.19
Xu, J., Jiang, X. L., Deng, M., Westwood, M., Song, Y. G., and Zheng, S. S. (2016). Conservation Genetics of Rare Trees Restricted to Subtropical Montane Cloud Forests in Southern China: a Case Study from Quercus arbutifolia (Fagaceae). Tree Genet. Genomes 12, 1–16. doi:10.1007/s11295-016-1048-1
Xu, J., Liu, L., Xu, Y., Chen, C., Rong, T., Ali, F., et al. (2013). Development and Characterization of Simple Sequence Repeat Markers Providing Genome-wide Coverage and High Resolution in maize. DNA Res. 20, 497–509. doi:10.1093/dnares/dst026
Xu, Y., Lu, Y., Zhang, P., Wang, Y., Zheng, Y., Fu, L., et al. (2020). Infrageneric Phylogenetics Investigation of Chimonanthus Based on Electroactive Compound Profiles. Bioelectrochemistry 133, 107455. doi:10.1016/j.bioelechem.2020.107455
Yang, Y., Zhou, T., Duan, D., Yang, J., Feng, L., and Zhao, G. (2016). Comparative Analysis of the Complete Chloroplast Genomes of Five Quercus Species. Front. Plant Sci. 07, 959. doi:10.3389/fpls.2016.00959
Yi, X., Gao, L., Wang, B., Su, Y.-J., and Wang, T. (2013). The Complete Chloroplast Genome Sequence of Cephalotaxus oliveri (Cephalotaxaceae): Evolutionary Comparison of Cephalotaxus Chloroplast DNAs and Insights into the Loss of Inverted Repeat Copies in Gymnosperms. Genome Biol. Evol. 5, 688–698. doi:10.1093/gbe/evt042
Zerbino, D. R., and Birney, E. (2008). Velvet: Algorithms for De Novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829. doi:10.1101/gr.074492.107
Zhang, S., Zhang, H., Xu, Z., Wu, M., Xia, W., and Zhang, W. (2017a). Chimonanthus praecox Extract/cyclodextrin Inclusion Complexes: Selective Inclusion, Enhancement of Antioxidant Activity and thermal Stability. Ind. Crops Prod. 95, 60–65. doi:10.1016/j.indcrop.2016.09.033
Zhang, X., Xu, M., Zhang, J., Wu, L., Liu, J., and Si, J. (2017b). Identification and Evaluation of Antioxidant Components in the Flowers of Five Chimonanthus Species. Ind. Crops Prod. 102, 164–172. doi:10.1016/j.indcrop.2017.03.014
Zhang, X., Zhang, H. J., Landis, J. B., Deng, T., Meng, A. P., Sun, H., et al. (2019). Plastome Phylogenomic Analysis of Torreya (Taxaceae). Jnl Sytematics Evol. 57, 607–615. doi:10.1111/jse.12482
Zhisheng, A., Kutzbach, J. E., Prell, W. L., and Porter, S. C. (2001). Evolution of Asian Monsoons and Phased Uplift of the Himalaya-Tibetan Plateau since Late Miocene Times. Nature 411, 62–66. doi:10.1038/35075035
Zhou, M. Q., Zhao, K. G., and Chen, L. Q. (2007). Genetic Diversity of Calycanthaceae Accessions Estimated Using AFLP Markers. Scientia Horticulturae 112, 331–338. doi:10.1016/j.scienta.2006.12.033
Zhou, S., Renner, S. S., and Wen, J. (2006). Molecular Phylogeny and Intra- and Intercontinental Biogeography of Calycanthaceae. Mol. Phylogenet. Evol. 39, 1–15. doi:10.1016/j.ympev.2006.01.015
Zhou, Y., Duvaux, L., Ren, G., Zhang, L., Savolainen, O., and Liu, J. (2017). Importance of Incomplete Lineage Sorting and Introgression in the Origin of Shared Genetic Variation between Two Closely Related Pines with Overlapping Distributions. Heredity 118, 211–220. doi:10.1038/hdy.2016.72
Keywords: chloroplast genomics, correlations, phylogeny, incipient radiation, divergence time, subtropical forest biome assembly
Citation: Jamal A, Wen J, Ma Z-Y, Ahmed I, Abdullah , Chen L-Q, Nie Z-L and Liu X-Q (2021) Comparative Chloroplast Genome Analyses of the Winter-Blooming Eastern Asian Endemic Genus Chimonanthus (Calycanthaceae) With Implications For Its Phylogeny and Diversification. Front. Genet. 12:709996. doi: 10.3389/fgene.2021.709996
Received: 14 May 2021; Accepted: 18 October 2021;
Published: 30 November 2021.
Edited by:
Mahendar Thudi, Shandong Academy of Agricultural Sciences (SAAS), ChinaReviewed by:
Tao Deng, Kunming Institute of Botany (CAS), ChinaDeng-Feng Xie, Sichuan University, China
Copyright © 2021 Jamal, Wen, Ma, Ahmed, Abdullah, Chen, Nie and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiu-Qun Liu, liu_xiuqun@sina.com
†ORCID ID: Abbas Jamal, orcid.org/0000-0003-0607-3705; Abdullah, orcid.org/0000-0003-1628-8478
 Abbas Jamal
Abbas Jamal Jun Wen
Jun Wen Zhi-Yao Ma2
Zhi-Yao Ma2 Ibrar Ahmed
Ibrar Ahmed Abdullah
Abdullah Xiu-Qun Liu
Xiu-Qun Liu