- 1Department of Biological Sciences, Faculty of Science, Kuwait University, Kuwait City, Kuwait
- 2Department of Medical Microbiology, Faculty of Medical Laboratory Sciences, University of Khartoum, Khartoum North, Sudan
- 3Centre for Tropical Livestock Genetics and Health, The Roslin Institute, Edinburgh, United Kingdom
- 4Department of Animal Production, Faculty of Agricultural and Environmental Sciences, University of Gadarif, Gadarif State, Sudan
- 5Department of Public Health, College of Veterinary Medicine, King Faisal University, Al-Hasa, Saudi Arabia
- 6LiveGene, International Livestock Research Institute (ILRI), Addis Ababa, Ethiopia
Dromedary camels (Camelus dromedarius) are single-humped animals found throughout the deserts of Africa, the Arabian Peninsula, and the southwest of Asia. This well-adapted species is mainly used for milk and meat production, although some specific types exhibit superior running performance and are used in racing competitions. However, neither performance nor production camels are bred under intensive genomic selection programs with specific aims to improve these traits. In this study, the full genome sequence data of six camels from the Arabian Peninsula and the genotyping-by-sequencing data of 44 camels (29 packing and 15 racing) from Sudan were analyzed to assess their genome diversities, relationships, and candidate signatures of positive selection. Genome ADMIXTURE and principle component analyses indicate clear geographic separation between the Sudanese and the Arabian Peninsula camels, but with no population-specific genetic distinction within populations. Camel samples from the Arabian Peninsula show higher mean heterozygosity (0.560 ± 0.003) than those from Sudan (0.347 ± 0.003). Analyses of signatures of selection, using pooled heterozygosity (Hp) approach, in the Sudanese camels revealed 176, 189, and 308 candidate regions under positive selection in the combined and packing and racing camel populations, respectively. These regions host genes that might be associated with adaptation to arid environment, dairy traits, energy homeostasis, and chondrogenesis. Eight regions show high genetic differentiation, based on Fst analysis, between the Sudanese packing and racing camel types. Genes associated with chondrogenesis, energy balance, and urinary system development were found within these regions. Our results advocate for further detailed investigation of the genome of the dromedary camel to identify and characterize genes and variants associated with their valuable phenotypic traits. The results of which may support the development of breeding programs to improve the production and performance traits of this unique domesticated species.
Introduction
The Camelidae family is divided into two tribes, the New World camel (Lamini) and the Old World camel (Camelini). Within the Camelini, there are two domesticated species, the two-humped Bactrian camel (Camelus bactrianus) and the single-humped dromedary camel (Camelus dromedarius), in addition to the critically endangered two-humped wild Bactrian camel (Camelus ferus) found in northern China and southern Mongolia (Wilson, 1998).
Out of approximately 35 million camel heads worldwide (FAO, 2019), the majority (95%) are of the dromedary type (Hashim et al., 2015). Unlike the Bactrian camels, which are distributed throughout central and eastern Asia, dromedary camels mainly populate the desert and semi-desert areas across Africa, the Arabian Peninsula, and southwest of Asia (Wilson, 1998). They are highly adapted to the harsh desert environment, which is characterized by high temperatures and scarcity of food and water. Dromedary camels are tolerant to temperatures in excess of 40°C and can survive for up to 20 to 35 days without water, losing up to 25% of their body weight (Schmidt-Nielsen, 1959; Musa et al., 2006).
Archaeological evidence indicates dromedary camels to have been domesticated in the southeast of the Arabian Peninsula in the late second millennium BC (Magee, 2015), following which they spread to the northern part of the Arabian Peninsula and Africa via ancient trading routes. Being highly adapted to the desert environment, dromedary camels were historically the preferred means of transporting people and goods along routes such as the “incense road”—the trans-Arabian trading route used to transport valuable spices and perfumes from southern to northern areas of the Arabian Peninsula. The connection between the Arabian Peninsula and Africa was first established in East of Africa, through the Islands of Socotra, in association with sea-borne incense trading during the 1st millennium BC (Epstein and Mason, 1971). In parallel, luxury goods, such as ivory, wools, and skins, were also transported from Africa to the Arabian Peninsula through Aden (Andree-Salvini, 2010). By the 7th century, dromedary camels were widespread throughout the African Sahara employed in the transport of goods (Gauthier-Pilters and Anne, 1983). The spread of Islam to Africa at that time also contributed to the spread of dromedary between Africa and the Arabian Peninsula, as they were used to transport people to Makkah in the Arabian Peninsula during the annual pilgrimage “hajj” (Wilson, 1998).
The majority of dromedary, around 80%, are found in Africa. Sudan accommodates around 4.8 million heads, second in size to Somalia (Ali et al., 2019), representing around 22% of the animal biomass in the country (Eisa and Mustafa, 2011). The history of dromedary camel in Sudan can be traced to 15–25 years BC following human migrations from Egypt (Eisa and Mustafa, 2011; Hashim et al., 2015). Camels inhabit an area in Sudan characterized by arid conditions and erratic periods of rainfall. The area is flanked by the Butana plains and Red Sea hills in the East, and the Darfur and Kordofan States in the West (Eisa and Mustafa, 2011). Here, camels are classified as either “packing” or “racing” with the former type being used for milk and meat production, while the latter is typically used in racing competitions. Packing camel populations, such as the Arabi, Kenanai, Lahawi, and Rashaydi, constitute around 80% of a nomad’s herd and are characterized by their heavy weight and for having a well-developed hump (Hashim et al., 2015). Milk production in these camels is in the range of 820–2,400 liters per lactation period, which typically lasts for 12–18 months. The level of productivity depends mainly on the season and the farming management system (Faye et al., 2011). Other camel populations in Sudan, such as the Anafi and Bishari, which are famous for their racing performance, are lighter than the pack camels and are generally found in the northeast of Sudan and the River Nile State (Wardeh, 2004; Hashim et al., 2015). Populations of each of these camel types in Sudan are named after the ethnic groups that rear them.
Dromedaries in the Arabian Peninsula follow a similar classification system to that in Africa (Wardeh, 2004) but are further classified according to their coat color. Black dromedary camels are called Majaheem, whereas white and brown camels are called Wodh and Shual, respectively. A further type also characterized by a brown coat, but darker than Shual on the hump and tail, is called Sofor (Al-Swailem et al., 2007; Almathen et al., 2018). Each of these types are considered to be packing animals used for meat and milk production, with the Majaheem type being the most popular (Wardeh, 2004). Other popular types include the Omani originating from Oman, and the Hura from Saudi Arabia, which are both considered high-performance racing camels (Wardeh, 2004). Despite the different classifications and livestock functions, no selection programs aimed at improving the different types are in place.
Until recently, genetic analyses of dromedary camel have been largely restricted to studies involving autosomal microsatellite markers (Mburu et al., 2003; Almathen et al., 2016), partial mitochondrial DNA (mtDNA) sequences (Babar et al., 2015; Almathen et al., 2016) and candidate gene sequencing (Pauciullo et al., 2013; Shuiep et al., 2013; Almathen et al., 2018). These tools have mainly been employed to assess the genetic diversity and structure of dromedary camel populations from different geographical locations, e.g., Kenya (Mburu et al., 2003), the Arabian Peninsula (Almathen et al., 2016), and Pakistan (Babar et al., 2015). Efforts to link genotypes to phenotypes have also been undertaken—for example, Almathen et al. (2018) linked an arginine to cysteine missense mutation at protein position 301 in the melanocortin 1 receptor (MC1R) gene to white coat color. In the same study, a 1-bp deletion (23del T/T) and a SNP (25G/A) in exon 2 of the agouti signaling protein (ASIP) gene were linked to the black and dark-brown coat color phenotypes of Saudi Arabian dromedary. Recently, Khalkhali-Evrigh et al. (2018) analyzed the full genomes of two Iranian dromedary camels and reported non-synonymous variants in genes related to adaptation to the desert environment.
Signatures of selection analyses in African indigenous livestock have been conducted in different breeds or populations for a number of species, including, for instance, cattle (Bahbahani et al., 2015; Bahbahani et al., 2017; Kim et al., 2017a; Bahbahani et al., 2018a; Bahbahani et al., 2018b), goat (Kim et al., 2016; Onzima et al., 2018), and sheep (Kim et al., 2016), but to date, nothing has been reported for dromedary. Reference genomes of dromedary camels from the Arabian Peninsula and North Africa were published in 2014 and 2015, respectively (Wu et al., 2014; Fitak et al., 2015); however, neither is very contiguous, being assembled into 32,573 and 35,752 scaffolds, respectively. These assemblies are a fundamental first-step toward facilitating genome-wide analyses for signatures of selection. For example, Wu et al. (2014) analysis of the Arabian Peninsula dromedary genome revealed accelerated evolution and positive selection in genes related to fat metabolism, heat stress response, and salt metabolism.
Although the costs of whole-genome sequencing (WGS) have declined significantly in recent years, it remains an expensive endeavour to sequence large numbers of individuals, with large and complex genomes, to enable genome-wide studies. For some livestock species such as cattle and sheep, high-density genotyping microarrays are available enabling hundreds of thousands of genomic positions to be genotyped at a fraction of the cost of WGS (Rincon et al., 2011; Kijas et al., 2014). Where these tools are unavailable, typically in the case of non-model species with poor genomic resources, an alternative approach is to employ a genotyping-by-sequencing (GBS) strategy. GBS is a high-throughput multiplex sequencing system based on constructing reduced representative libraries for subsequent sequencing (Elshire et al., 2011). As with genotype microarrays, GBS can be performed at a fraction of the cost of WGS and does not suffer the ascertainment bias associated with array-based genotyping (Poland and Rife, 2012). The main disadvantage of this approach, however, is that the distribution of coverage throughout the genome is generally not uniform, and as a result, there is typically extensive between-sample variation in the genomic regions sequenced (Beissinger et al., 2013). GBS has been widely used in plant breeding for genome-wide association analysis, genomic diversity studies, and genomic selection (reviewed in He et al. (2014)). In addition, Wang et al. (2018) employed GBS to analyze the genomes of Chinese Landrace and Yorkshire pigs, identifying candidate signatures of selection in genes related to fatty acid biosynthesis, animal growth and development, and immune responses.
The availability of reference genome assemblies coupled with GBS enables us to explore in greater detail than ever before the genomes of different dromedary populations and to seek to identify genetic associations with different phenotypic traits. The aim of this study is to exploit these resources to assess the genomic diversity and relationship of dromedary camel populations from Sudan and the Arabian Peninsula and to undertake signatures of selection analyses to identify genes that might be associated with environmental adaptation, milk production, and racing performance in the Sudanese camels.
Materials and Methods
Animal Resources
Five Sudanese indigenous camel populations were sampled for this study. These included three packing camel populations: Arabi from western Sudan (n = 9), Kenani from central Sudan (n = 10), and Lahawi from eastern Sudan (n = 10) and two racing camel populations from eastern Sudan: Bishari (n = 7) and Anafi (n = 8) (Table 1 and Supplementary Figure S1). Six camel samples from the Arabian Peninsula were also included in this study: three Majaheem, two Omani, and one Sofor. Two Majaheem samples were collected from the Conservation and Genetic Improvement Centre at Al-Kharj in Saudi Arabia, while one Majaheem and one Sofor were sampled from Alwatani camel dairy farm in Saudi Arabia. The Omani samples were from the King Fahad camel herd (Table 1).
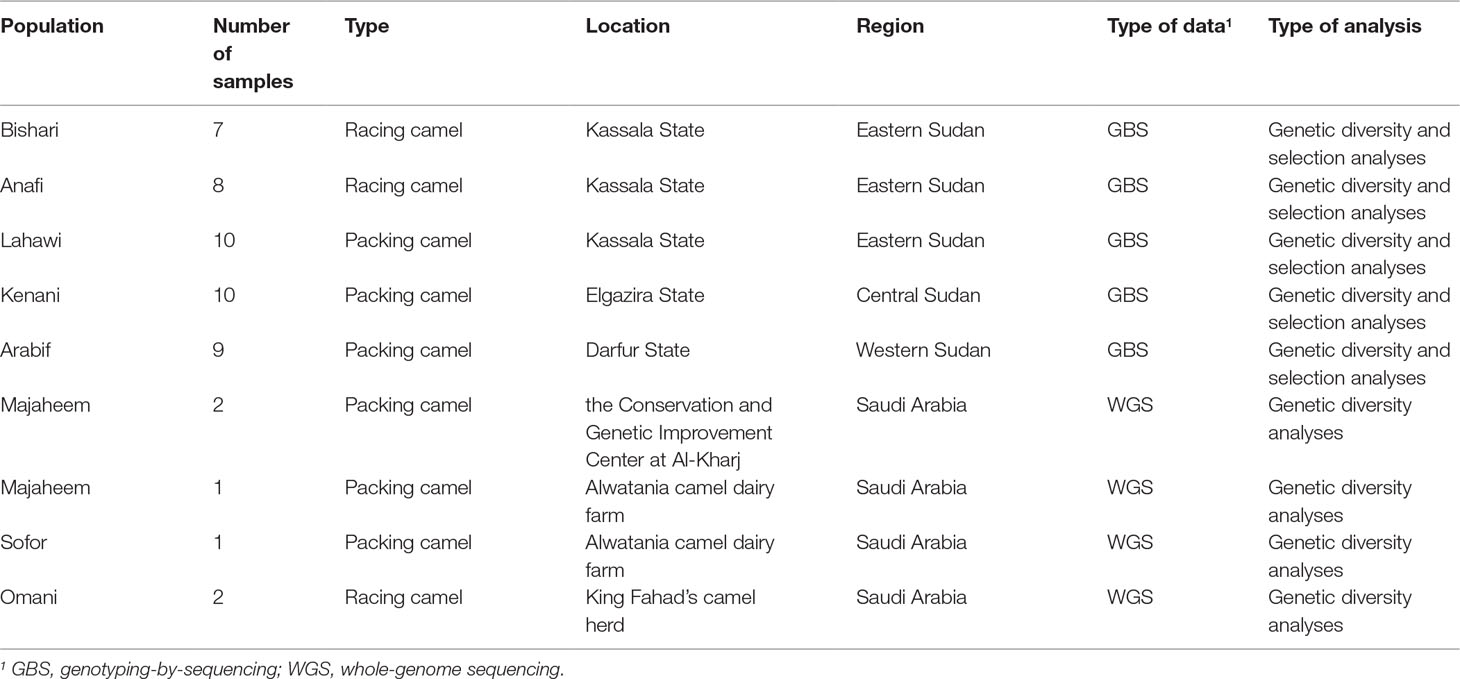
Table 1 Sampling summary for the different Sudanese and Arabian Peninsula camel populations included in this study.
Genotyping-by-Sequencing (GBS) Data
Ten milliliters of whole blood were collected from each Sudanese sample using EDTA VACUETTE® tubes. Genomic DNA was extracted from these whole-blood samples using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s protocol. The quality and quantity of the DNA were evaluated using a NanoDrop Spectrophotometer (NanoDrop Technologies, USA) and by gel electrophoresis. The extracted DNA samples were genotyped using pair-end GBS technology incorporating two restriction enzymes, MseI and EcoRI, sequenced on the Illumina HiSeq 2000 platform by Novogene (Novogene Co., Ltd., Tianjin, China). Novogene trimmed adapters from the sequence reads and discarded raw read pairs if (1) they were contaminated with adapter sequences, (2) uncertain nucleotides constituted more than 10% of either read, or (3) if low-quality nucleotides (base Qphred ≤ 5) constituted more than 50% of either read.
Whole-Genome Sequence (WGS) Data
Genomic DNA of the six Arabian Peninsula dromedary samples was extracted from 5 ml whole blood from each sample using the DNeasy® Blood and Tissue Kit (Qiagen) according to the manufacturer’s protocol. The extracted genomic DNA was sequenced using pair-end libraries on the Illumina HiSeq 2000 platform (Beijing Genomics Institute (BGI), China). BGI trimmed adapters from the sequence reads and discarded raw read pairs if low-quality nucleotides (base Qphred ≤ 5) constituted more than 50% of either read.
Processing of Sequence Data
The clean raw sequence reads of both the GBS and WGS data were mapped to the Arabian dromedary camel reference genome assembly (GCF_000767585.1) (Wu et al., 2014) using the BWA-MEM algorithm of Burrows–Wheeler Aligner (BWA) version 0.7.5a (Li and Durbin, 2010). This aligner employs local alignment which results in the ends of reads being soft-clipped if they fail to map sufficiently well. Picard tools version 1.119 (http://broadinstitute.github.io/picard/index.html) was used to sort the reads by coordinate and mark PCR duplicates. Marking duplicate reads results in those reads being ignored in downstream analyses both by the Genome Analysis Toolkit GATK version 4.1 (Mckenna et al., 2010) and SAMtools version 1.3.1 (Li, 2011). Mapped reads with insertions or deletions (indels) were realigned using GATK’s IndelRealigner.
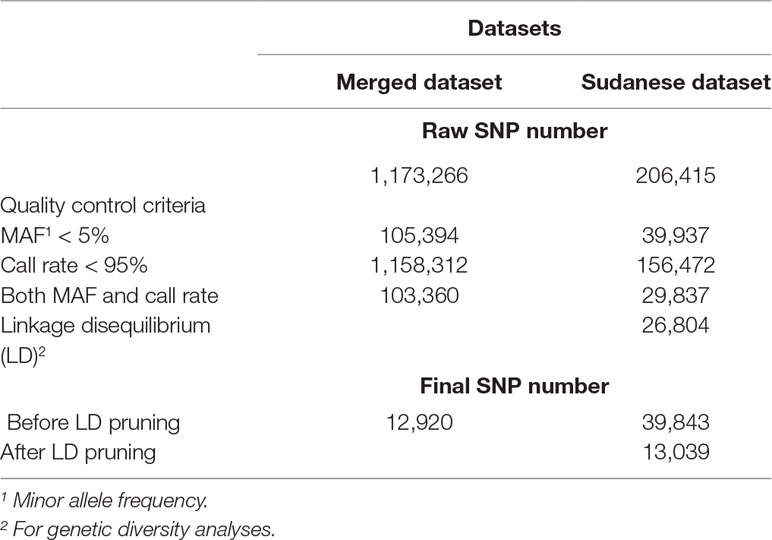
Single-nucleotide polymorphisms (SNPs) were called across the WGS (Arabian Peninsula) and GBS (Sudan) data, separately, using GATK’s HaplotypeCaller and SAMtools mpileup. HaplotypeCaller employs a number of default read filters; these include: removing reads that fail platform/vendor checks, unmapped reads, secondary alignments, reads that are not well-formed, reads that fail the minimum mapping quality (20), reads marked as duplicates, and reads with a bad CIGAR string (for further details, refer to the GATK documentation available at https://software.broadinstitute.org/gatk). Variants identified by SAMtools mpileup were required to have a minimum read mapping quality of 20 (MAPQ20) and a minimum base quality of 20. For each dataset, the genotypes of SNPs that were identified by both variant detection algorithms were retrieved from SAMtools, resulting in a total of 1,065,798 SNPs in Arabian Peninsula samples and 402,077 SNPs in the Sudanese populations. These variants were further hard-filtered using the VariantFiltration tool of GATK. This included removing variants with low quality by depth (QD < 2) to normalize variant quality and avoid inflation in the presence of deep coverage, removing variants indicating a high probability of strand bias (FS > 60), removing variants with a low root mean score mapping quality (MQ < 40), removing variants with low variant site quality (QUAL < 30), retaining variants where the mapping qualities of reads supporting the reference and alternate allele did not exhibit a bias for either allele (MQRankSum < −12.5), and retaining variants where their positions did not exhibit a bias toward the ends of reads (ReadPosRankSum < −8). The remaining variants were subsequently filtered using bcftools version 1.6 (Li, 2011) to retain those with a depth of coverage (DP) ≥ 5, and with a per-SNP DP within three standard deviations (SD) of the mean DP across all samples for a given dataset. The transition/transversion (Ts/Tv) ratio was calculated before and after filtering using bcftools stats. Variants on the mtDNA (scaffold NC_009849.1), indels, and SNPs that were not bi-allelic were also removed. A total of 206,415 and 1,028,936 SNPs were retained for the GBS and WGS data, respectively (Supplementary Table S1).
The filtered genotype data from the WGS and GBS datasets were merged, retaining a total of 1,173,266 SNPs common to both, and is herein referred to as the merged dataset. The filtered genotype data from the GBS dataset, independent of the WGS dataset, is herein referred to as the Sudanese dataset. A quality control (QC) steps was performed using the check.marker function implemented in the GenABEL package (Aulchenko et al., 2007) for R software version 2.15.1 (R Development Core Team, 2012). SNPs with minor allele frequency (MAF) less than 5% and call rate less than 95% were excluded from each dataset. A breakdown of the SNPs failing QC in each dataset is provided in Table 2. The final numbers of SNPs after QC were 12,920 and 39,843 in the merged and Sudanese datasets, respectively. For the genetic diversity analyses, SNPs with high linkage disequilibrium (LD) (r2 > 0.1) were filtered out using the indep-pairwise tool (–indep-pairwise 50 10 0.1) in PLINK version 1.9 (Purcell et al., 2007). A total of 7,273 and 26,804 SNPs were removed from the merged and Sudanese datasets, respectively. For the pooled heterozygosity (Hp) analyses of the Sudanese dataset, the MAF criteria were not applied as the statistic is specifically testing for large deviations in heterozygosity levels and as such is sensitive to MAF. The LD filter was also excluded, as regions under selection are expected to accommodate SNPs in LD, and so removing these reduces the power of the analysis. The QC filtering was applied to the combined Sudanese, packing, and racing camel subsets independently, resulting in 49,943 SNPs in the combined Sudanese, 48,808 SNPs in packing and 46,535 SNPs in racing camels. None of the samples from either QC-filtered dataset (merged and Sudanese) exhibited a genotyping call rate <95%, or an identity by state (IBS) ≥ 90% with any other sample.
Genetic Diversity Analyses
The mean heterozygosity and the inbreeding coefficient (Fis) of the different camel populations were computed from the merged dataset using the hom function implemented in GenABEL (Aulchenko et al., 2007). The two-sample Mann–Whitney U test was used to test for statistically significant differences in the heterozygosity and Fis values between the Arabian Peninsula and Sudanese camels, and between the different Sudanese populations. The one-sample Mann–Whitney U test was used to check if the Fis values of each of the distinct Sudanese populations, the combined Sudanese populations, and the Arabian Peninsula camels were significantly different from zero. Principle component analyses (PCA) were conducted in R using the prcomp function on both datasets to determine the genomic relationship between Arabian Peninsula and Sudanese camels, and separately among the Sudanese camels only.
To assess levels of genetic ADMIXTURE, analyses were performed using ADMIXTURE 1.23 (Alexander et al., 2009) on each dataset, assuming a number of clusters ranging from 1 to the number of populations sampled in each dataset (merged = 8; Sudanese = 5). In each case, 200 bootstrap iterations were performed. The optimal number of clusters was determined following Evanno et al. (2005) by calculating the second order rate of change of the likelihood for each K value. Figures of the ancestry assignments were plotted using the ggplot2 package (Wickham, 2009) for R.
The relatedness of samples was evaluated in PLINK using the –make-rel tool. This tool estimates genetic relatedness among genome-wide SNPs based on the unadjusted Ajk statistic described in Yang et al. (2011), which ranges from 0 for an unrelated pair of individuals to 1 when comparing an individual to itself. Yang et al. (2011) consider a pair of individuals to be related where Ajk > 0.025. The relationship matrix was plotted using the ggplot2 package for R.
Signatures of Selection Analyses
Pooled Heterozygosity (Hp) Analysis
Hp values were calculated using the formula described by Rubin et al. (2010): Hp = 2 ∑ nMAJ ∑ nMIN/(∑ nMAJ + ∑ nMIN)2. The analysis was performed in 50-kb sliding windows with a 25-kb step on all of the Sudanese camels, and independently for the Sudanese packing and racing types. After discarding windows supported by only a single SNP, the major and minor allele counts for each SNP were recorded (nMAJ and nMIN, respectively), and Hp values of each 50-kb window were calculated, which were subsequently Z-transformed: ZHp = Hp – median Hp/SD Hp. Values in the extreme lower 0.1% tail of the empirical distribution of each analysis were considered significant (all Sudanese Zhp = −2.51; packing Zhp = −2.45; racing Zhp = −2.22) (Supplementary Figure S2). Windows with significant Zhp values that overlapped were merged into single regions.
Fixation Index (Fst) Analysis
Fixation index (Fst) analysis (Weir and Cockerham, 1984) was calculated between Sudanese packing and racing camels in 50-kb windows with a 25-kb step using VCFtools version 0.1.13 (Danecek et al., 2011). As with the Hp analyses, windows comprising only a single SNP were discarded. Values in the extreme upper 0.1% tail of the Fst distribution were considered significant (Fst = 0.206) (Supplementary Figure S2). Windows with significant Fst values that overlapped were merged into single regions.
Functional Characterization of Candidate Regions of Selection
The positions of the significant windows from each analyses were cross-referenced against the genes annotated in the Arabian Peninsula dromedary camel reference genome assembly using the intersectBed function from the BedTools software (Quinlan and Hall, 2010). From each analysis, a background gene set was also identified by retrieving all genes that intersect with all windows tested. Gene-set over-representation analyses were performed using the ConsensusPathDB-human (CPDB) online tool (http://cpdb.molgen.mpg.de/CPDB). From each analysis, the significant and background gene lists were analyzed, and the significantly enriched biological pathways and gene ontology (GO) terms were recorded. Pathways and GO terms that remained significant after false discovery rate (FDR) correction (q-value < 0.05) were considered for discussion. A review of the literature for the genes identified from each analysis was also conducted to evaluate their relevance to adaptation to the desert environment, production, and performance traits.
Results
Genetic Diversity and Relationship Among the Camel Populations
The mean observed heterozygosity in Arabian Peninsula (0.560 ± 0.003) was significantly higher than within Sudanese camel populations (0.347 ± 0.003) (P-value < 0.0001). Among Sudanese camels, heterozygosity values were not significantly different (P-values > 0.05) ranging from 0.341 in Anafi to 0.353 in Bishari (Table 3). The mean Fis value of Arabian Peninsula samples (−0.814 ± 0.01) was significantly lower than in Sudanese camel populations (−0.123 ± 0.033) (P-value < 0.0001). As with the heterozygosity values, the Fis values among the Sudanese camel populations were not significantly different from each other (P-value > 0.05) (Table 3). Fis values of the Arabian Peninsula and Sudanese dromedary populations were all significantly different from zero (P-value < 0.05) (Supplementary Table S2). The camels sampled from Arabian Peninsula exhibited higher relatedness (median Ajk = 0.347 ± 0.093) than those sampled from Sudan (median Ajk = −0.007 ± 0.018) (Supplementary Figure S3, Supplementary Figure S4 and Supplementary Table S3).
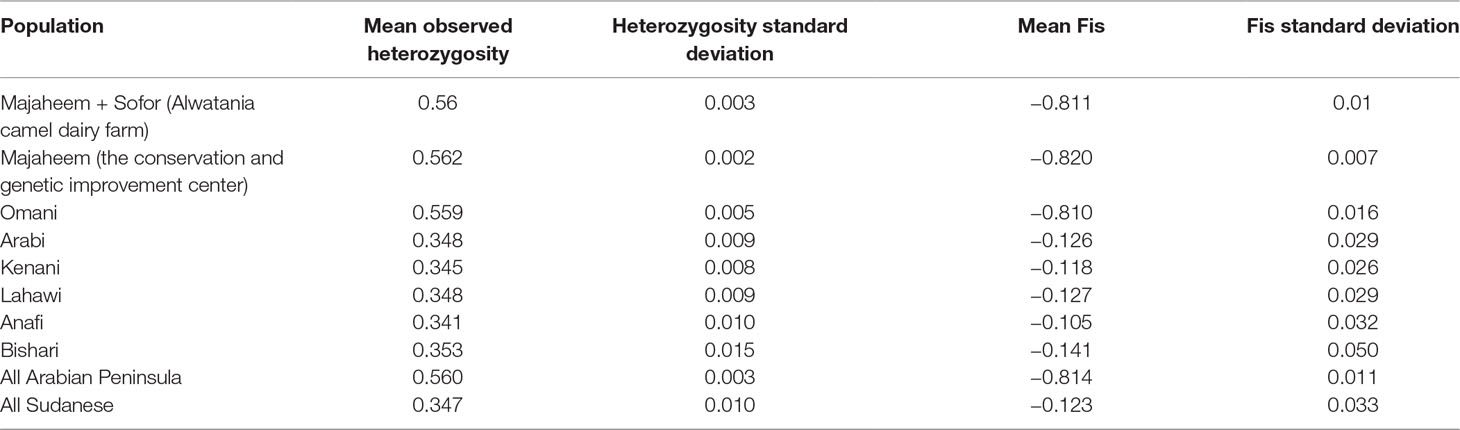
Table 3 Summary of observed heterozygosity and inbreeding coefficient (Fis) for each camel population.
Principal component analysis (PCA) of the merged dataset differentiates the camel populations of Sudan from the Arabian Peninsula along the first principal component (PC1), which explains 9.2% of the total variation. PC2, which accounts for 3.2% of the total variation, distinguishes the two Majaheem from Al-Kharj from the other Arabian Peninsula dromedary (Figure 1A), while PC3, which explains 2.8% of the total variation, separates the Omani camels from the other Arabian Peninsula dromedary (Supplementary Figure S5). Analysis of the Sudanese camels alone reveals PC1 and PC2 to explain only 3.4 and 3.2% of the total variation, respectively. This indicates that the populations sampled are relatively homogeneous. Nonetheless, the variance of data along these components indicates two Bishari and three Anafi to diverge from the origin for PC1 and PC2, respectively (Figure 1B).
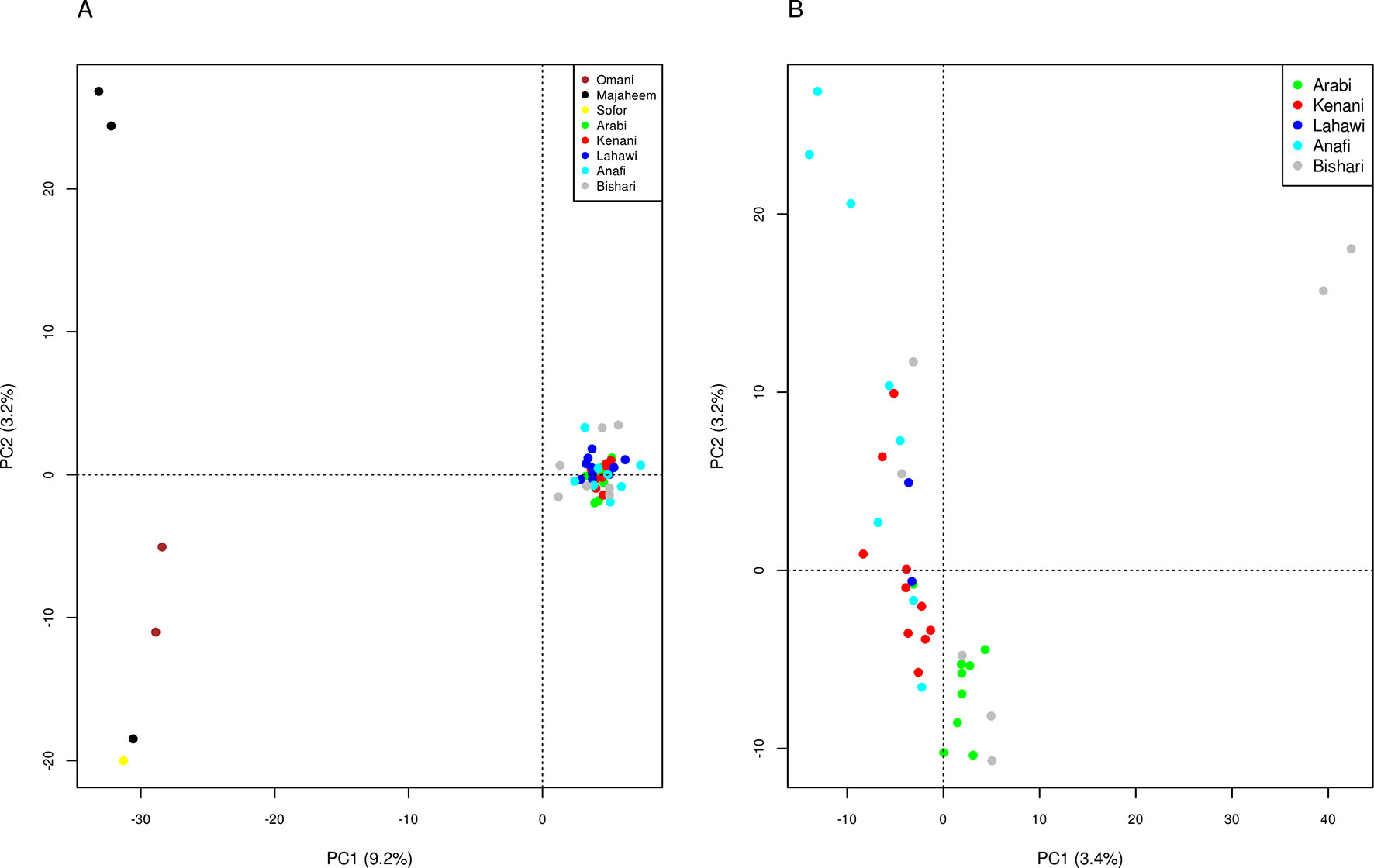
Figure 1 Principle components analysis, components 1 and 2 for (A) camel populations from Sudan and the Arabian Peninsula and (B) camel populations from Sudan only.
We applied the ΔK approach of Evanno et al. (2005) to identify the optimal number of genetic backgrounds from the results of the ADMIXTURE analyses of the merged dataset and the Sudanese dataset. In both cases, the optimal number of genetic backgrounds is found to be two (Supplementary Figure S6). This is not surprising in the merged dataset, given the differentiation of samples collected from Sudan and the Arabian Peninsula by PCA (Figure 1A). At K = 3, a degree of ADMIXTURE is observed among the Sudanese camels, with two samples from Bishari providing the signal for the 3rd genetic background (Figure 2). This pattern of ADMIXTURE is broadly observed at K = 2 in the independent analysis of Sudanese camels (Sudanese dataset; Supplementary Figure S7), although the extent of ADMIXTURE is slightly larger. At K < 4, the camels sampled from the Arabian Peninsula exhibit a more homogenous genetic background, distinct from the Sudanese camels with traces of Sudanese ancestry found in Omani camels (Figure 2). This distinction can also be observed at K values from 5 to 7 (Supplementary Figure S8). At K = 4 and 8, the two Majaheem samples from the Conservation and Genetic Improvement Centre provide a separate genetic background, while a single Majaheem camel retains a background shared with the Sofor sample which is also the dominant background in the Omani camels (Figure 2 and Supplementary Figure S8).
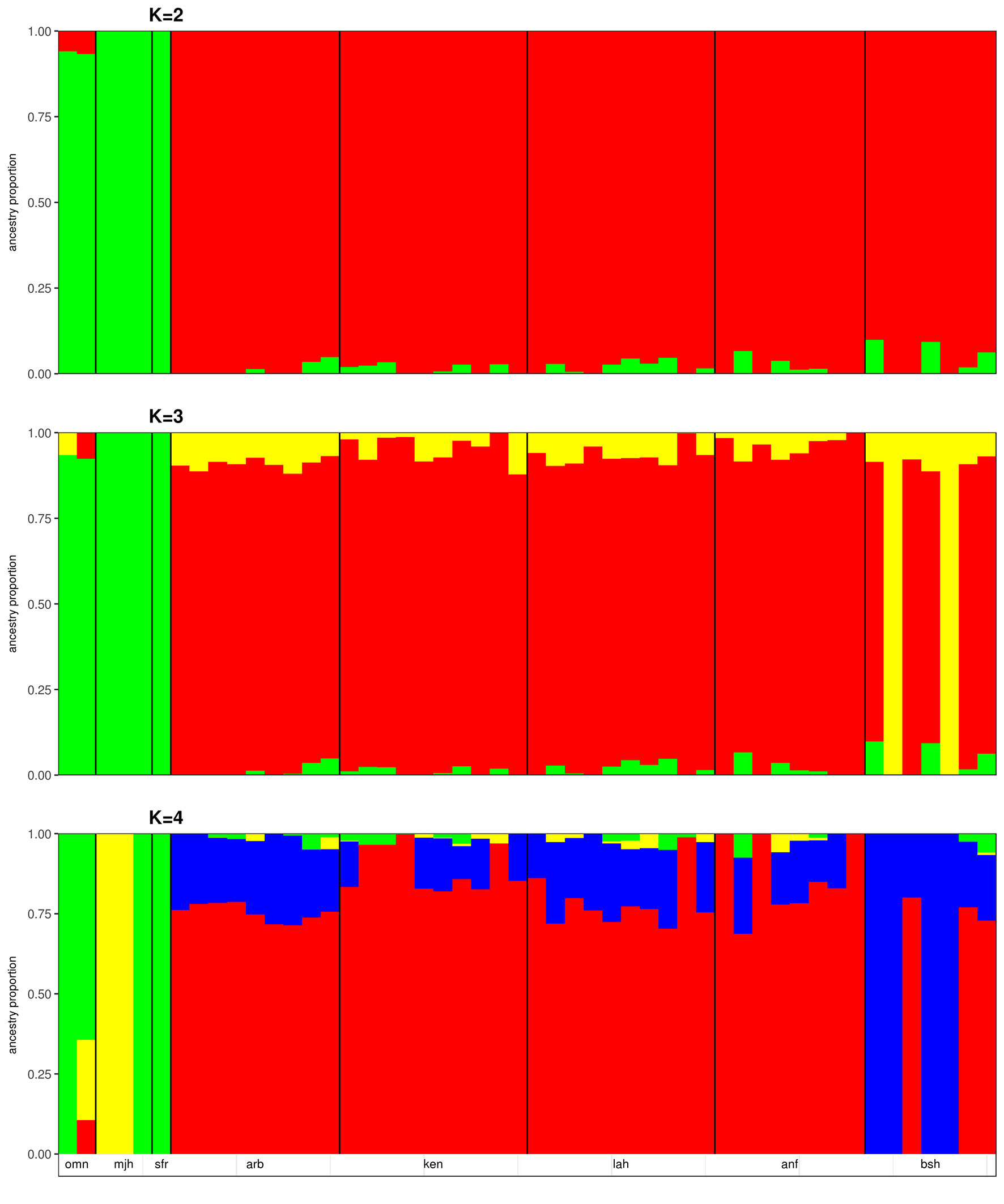
Figure 2 ADMIXTURE plots of Sudanese and Arabian Peninsula camels for cluster values (K) from 2 to 4. omn, Omani; mjh, Majaheem; sfr, Sofor; arb, Arabi; ken, Kenani; lah, Lahawi; anf, Anafi; bsh, Bishari.
Candidate Signatures of Positive Selection
The mean Hp value across all sliding windows analyzed for the three Sudanese camel subsets (all Sudanese, packing, racing) was 0.24 ± 0.09. The analysis of all Sudanese camels returned 267 significant windows representing 176 regions. The subsets of packing and racing camels returned 281 and 472 significant windows representing 189 and 308 regions, respectively (Table 4 and Supplementary Table S4), of which 133 were common to both camel types (Supplementary Table S5). A total of 159 and 148 regions identified in packing and racing camels, respectively, overlapped with the significant regions identified across all Sudanese camels. In total, 132 regions were found to overlap across each analysis (Supplementary Table S5).
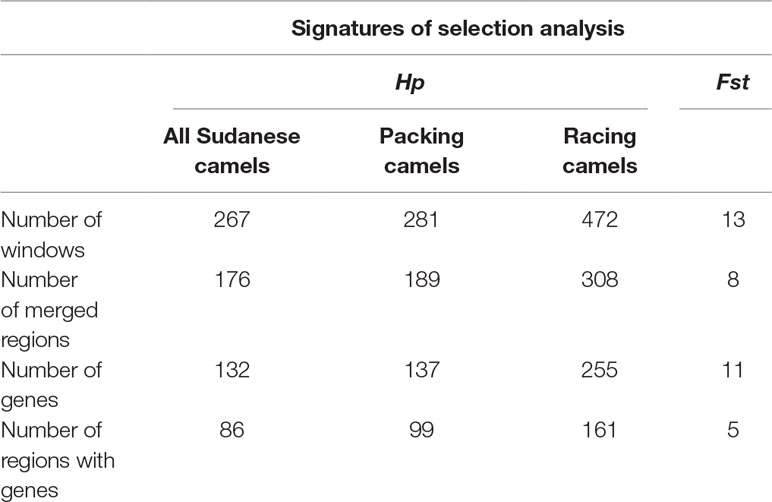
Table 4 Summary of windows, regions, and genes identified in signatures of selection analyses in Sudanese camel.
The Fst analysis revealed 13 out of 13,097 windows to exhibit high genetic differentiation (Fst > 0.206) between racing and packing camels from Sudan. These windows represented eight distinct regions (Table 4 and Supplementary Table S6).
Functional Characterization of Candidate Regions of Selection
The Hp analysis identified 132, 137, and 255 genes to intersect with 86, 99, and 161 regions in all Sudanese, packing, and racing camels, respectively (Table 4 and Supplementary Table S7). While the Fst analysis identified 11 genes to overlap with five genetically differentiated regions between Sudanese packing and racing camels (Table 4 and Supplementary Table S8). These genes are associated with different biological pathways, including immune response, fertility, milk content, energy homeostasis, and chondrogenesis (Table 5). No biological pathways were found to be significantly over-represented in the genes identified from the analyses of all Sudanese and packing camels. While a single GO term (monocarboxylic acid transmembrane transporter activity) was found to be significantly enriched in the analysis of all Sudanese camels. Moreover, a single biological pathway associated with liver kinase B1 (LKB-1) signaling was found to be significantly over-represented in Sudanese racing camels (Table 6 and Supplementary Table S9). In total, four biological pathways and five GO terms are significantly enriched among the genes identified from the Fst analysis between Sudanese racing and packing camels (Table 6 and Supplementary Table S9).
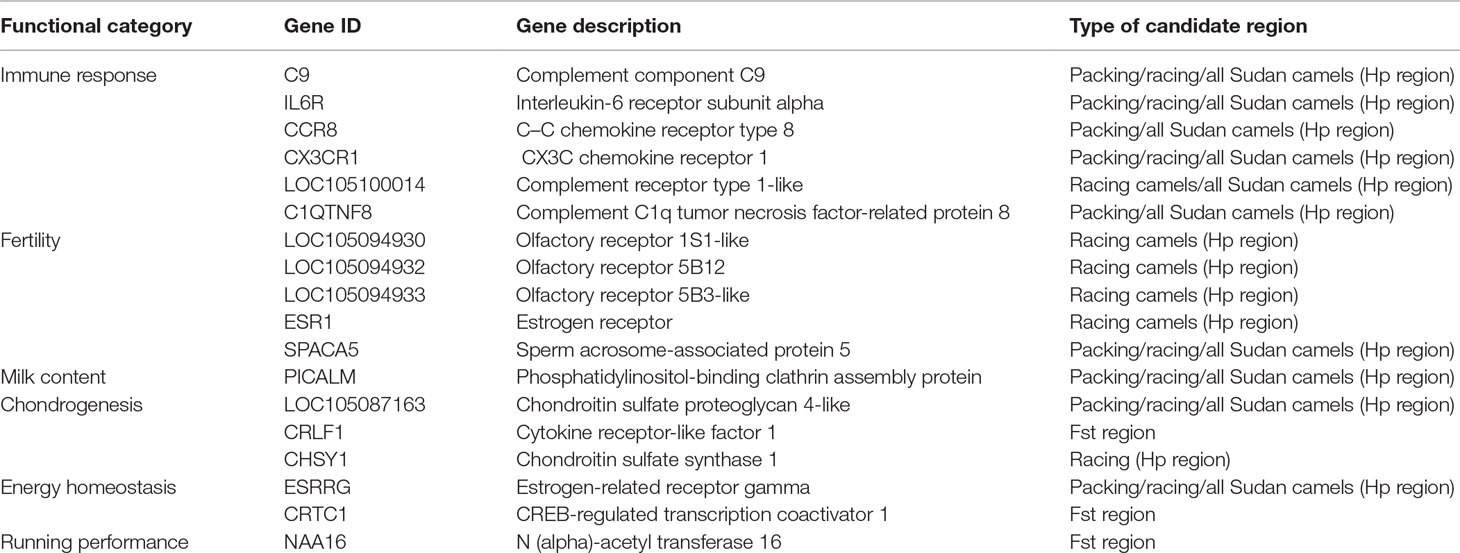
Table 5 Genes intersecting the pooled heterozygosity (Hp) and genetic differentiation (Fst) candidate regions.
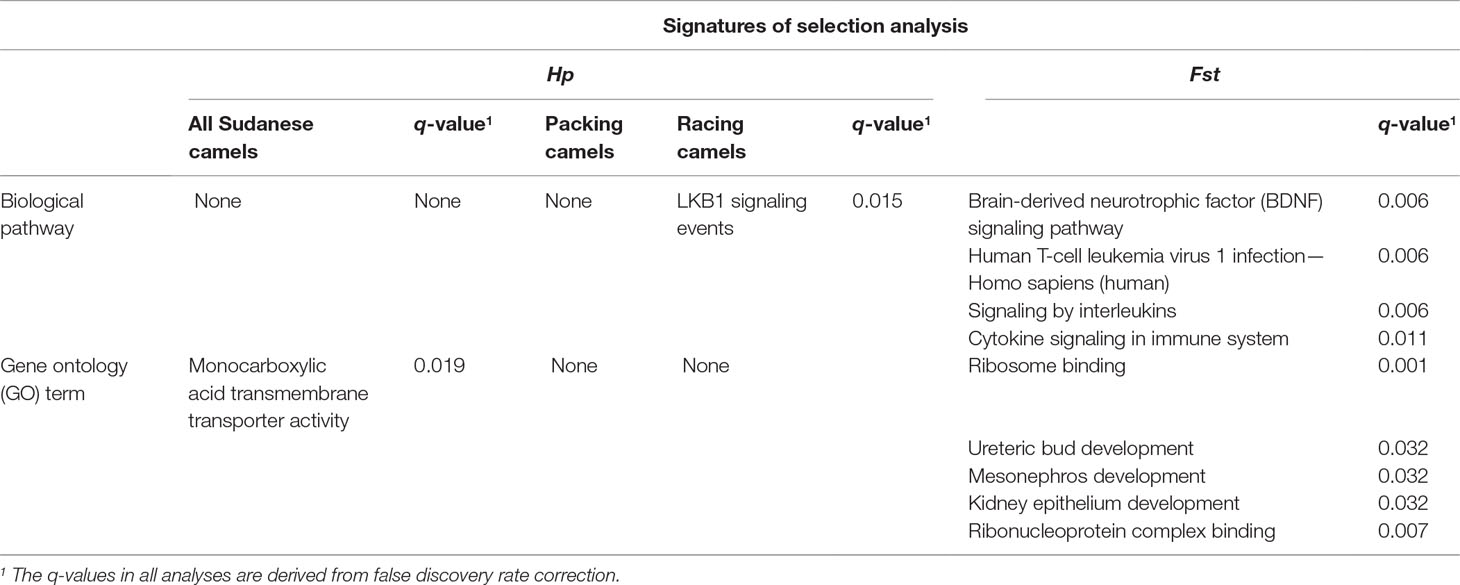
Table 6 Summary of significantly enriched biological pathways and gene ontology (GO) terms following gene-set over-representation analyses.
Discussion
In this study, we investigated the genetic diversity and relationship between camels sampled from Sudan and the Arabian Peninsula using genotype data derived from GBS and WGS. The migration of camels to Sudan from their putative center of domestication in the Arabian Peninsula may have resulted in a founder effect coupled with genetic drift that can explain the reduced heterozygosity observed in the Sudanese camels we studied. Populations that are closer to their centers of domestication are expected to exhibit higher levels of heterozygosity than more distant populations. Thus, the higher level of heterozygosity observed in the camels sampled from the Arabian Peninsula compared to those from Sudan offers some support for the southeast of the Arabian Peninsula as a center of domestication. Investigating a greater breadth and depth of sampling throughout Africa and the Arabian Peninsula will provide further context for these interpretations.
The Arabian Peninsula and Sudanese camel populations both exhibit mean negative Fis values significantly different from zero, which may be suggestive of low levels of inbreeding among these populations as a consequence of their historical use in transportation and trading—which might be associated with continuous interbreeding and gene flow. We sought to evaluate the relatedness of samples using the Ajk statistic, which indicated the majority of Sudanese camels to be unrelated, whereas all of the camels from the Arabian Peninsula returned high values (Ajk > 0.025). These high values observed across the Arabian Peninsula camels are likely an artifact of the small sample size that can result in severely biased estimates (Wang, 2017).
PCA and ADMIXTURE analyses revealed a phylogeographic distinction between the Arabian Peninsula and Sudanese camels. This might be attributed to the geographical isolation of these two populations by the Red Sea, hindering gene flow between them. Such genetic distinction has been observed between camel populations from Kenya, the Arabian Peninsula, and Pakistan (Mburu et al., 2003). We also observed traces of Sudanese ancestry in Omani camels, which might possibly result from historical interbreeding while following the trading routes connecting Africa with the south of Arabian Peninsula (Andree-Salvini, 2010). These results imply that the history of dromedary camel migration and interbreeding is highly complex and requires further investigation. A recent study by Almathen et al. (2016) showed that Sudanese camels are genetically distinct to camels from the south of the Arabian Peninsula but not the north. The discrepancy between that study and our results is likely to be due to our limited sampling of the Arabian Peninsula, which did not adequately capture the broader diversity of camel populations present. Furthermore, Almathen et al. (2016) employed autosomal microsatellite markers, whereas our genotypic data was derived from next-generation sequencing and is potentially more informative.
We observed no clear genetic structuring of the different Sudanese camel populations studied, providing further support of continuous gene flow. The two Bishari and three Anafi divergent camels exhibited marginally higher relatedness with one another than with other camels sampled from the same regions (Supplementary Figure S3). This likely accounts for their divergence in the PCA (Figure 1B). The lack of genetic distinction between the packing and the racing camels reflects a common practice in Sudan of breeding racing camels from East Sudan with packing camels in the West to improve the performance of packing camels. This is also reflected in the mean negative Fis values in the Sudanese dromedary camel populations analyzed.
The camels from the Arabian Peninsula were broadly found to be homogeneous in the ADMIXTURE analyses. PCA differentiated Omani samples from the rest of the Arabian Peninsula along the 3rd principal component. The 2nd principle component, although explaining little variation, differentiated the Majaheem and Sofor samples from the Alwatania camel dairy farm from the two Majaheem samples from the conservation and genetic improvement center. This was also observed in the ADMIXTURE analysis at K = 4 and K = 8. This suggests that camels from the Arabian Peninsula might exhibit a degree of genetic structure based on their breeding location and/or geographical origin. This observation however, which agrees with the findings of Almathen et al. (2016), requires further investigation using a larger and more diverse sample of camels from the Arabian Peninsula.
We further investigated the Sudanese camels for signatures of positive selection. A number of genomic regions exhibiting reduced heterozygosity were identified across the Sudanese camels, and within the subsets of packing and racing camels (Table 4). The Fst analysis, however, revealed few genetically differentiated regions between racing and packing camels. There was no overlap between the regions identified by the Fst analysis and those identified by the Hp analysis. This is not entirely unexpected as the two analyses are better-suited to different selection time frames, whereby Fst focuses on more recent selection than the Hp statistic (Oleksyk et al., 2010).
The regions identified from these analyses host a number of genes, for which gene set over-representation analyses and extensive literature review revealed a number to be of functional interest. In particular, we identified genes linked to the immune response, fertility, milk content, chondrogenesis, energy homeostasis, and running performance (Table 5). Whether or not these genes are linked to the domestication of dromedary camels from their wild ancestors, as found in sheep and goats (Alberto et al., 2018), requires further validation using genomic data from the wild ancestor of dromedary camels.
Examples of genes involved in the immune response include members of the complement system (C9, LOC105100014, and C1QTNF8), CX3C chemokine receptor 1 (CX3CR1), and interleukin-6 receptor subunit alpha (IL6R) genes, both of which were identified in packing and racing camels. The complement system links the innate and adaptive immune responses, mediating responses to inflammatory triggers through a co-ordinated enzyme cascade (Nesargikar et al., 2012). CX3CR1 and interleukin-6 (IL-6) are both involved in inducing inflammation in response to infection and tissue injuries (Ishida et al., 2008; Tanaka et al., 2014), while IL-6 is also involved in native T-cell differentiation (Tanaka et al., 2014). The chemokine receptor type 8 (CCR8) gene, identified in packing camels, plays a role in regulating the immune system by controlling regulatory T-cell activity (Coghill et al., 2013). Genes in this category have also been identified to be under adaptive evolution in dromedary and Bactrian camels in a study by Wu et al. (2014).
Genes involved in fertility have been identified in both packing and racing Sudanese dromedary camels. These include the ESR1 gene which encodes the estrogen receptor alpha that mediates estrogen action on target tissues. Mice whose estrogen receptor-alpha gene has been knocked out demonstrate complete infertility (Matthews and Gustafsson, 2003) and impaired spermatogenesis (Couse et al., 2001). Olfactory receptors have been shown to be expressed in mature sperm (Parmentier et al., 1992; Vanderhaeghen et al., 1993) and to play a role in oocyte fertilization upon interaction with chemo-attractants secreted from oocyte-cumulus cells (Spehr et al., 2003; Fukuda et al., 2004). The sperm acrosome-associated protein 5 gene (SPACA5) plays a role in acrosome reaction and the fertilization process (Agarwal et al., 2015). Fertility-associated genes have also been found to be under selection in indigenous African zebu cattle (Bahbahani et al., 2018a; Bahbahani et al., 2018b) in order to maintain fertility in the harsh environment (Skinner and Louw, 1966; Hansen, 2004).
An example of a gene likely to be a feature of adaptation to the desert environment is ESRRG. This gene, which is in candidate regions identified in both camel types, is related to energy homeostasis. ESRRG encodes the estrogen-related receptor gamma protein, which is involved in regulating metabolism and energy production in cells (Eichner and Giguere, 2011). A transition polymorphism (rs1890552 A > G) in this gene has been found to be associated with decreased levels of fasting glucose in humans (Kim et al., 2017b). The gene-set over-representation analysis in racing camel also identified the over-representation of genes associated with LKB1 signaling pathway, which is associated with regulating cellular energy metabolism in eukaryotic cells (Alessi et al., 2006). Genes in this category have also been found to be under adaptive evolution in dromedary and Bactrian camels (Wu et al., 2014).
A gene which encodes the phosphatidylinositol-binding clathrin assembly protein (PICALM), which is involved in regulating milk content, was identified in both racing and packing camel types. Variants in PICALM have been associated with αS1-casein content in cattle milk (Sanchez et al., 2017). This gene is therefore of potential future interest to improve the productivity of dromedary camels through genomic selection breeding programs (Al Abri and Faye, 2019).
A number of genes identified in candidate regions in packing and racing camels were found to be associated with chondrogenesis. These include the LOC105087163 gene which encodes the chondroitin sulfate proteoglycan 4 protein. Members of this protein family, such as aggrecan and versican, have been found to be involved in cartilage and limb joint formation (Watanabe et al., 1994; Choocheep et al., 2010). Another chondrogenesis-related gene identified, CHSY1, encodes the chondroitin sulfate synthase 1 protein, which, when knocked-out in mice, results in cartilage impairment, aberrant joint formation, and decreased bone density (Wilson et al., 2012). The identification of genes associated with chondrogenesis in packing dromedary camels might be a reflection of the historical use of camels in trading and transportation, which likely placed chronic physical demands on these animals similar to other livestock employed in the provision of draught power. The signatures of selection identified for genes associated with dairy traits in racing camels might also be a historical reflection of the general use of camels by nomads in the provision of milk.
A number of genes associated with energy homeostasis, chondrogenesis, and running performance were identified within the genetically differentiated regions between racing and packing camels. One such example is CREB-regulated transcription coactivator 1 (CRTC1). This gene is linked to energy homeostasis, which is likely to be an important function in racing camels in order to maintain their stamina during racing competitions. Studies in mice support this, indicating that the gene is associated with energy balance (Altarejos et al., 2008). Another candidate gene identified was cytokine receptor-like factor 1 (CRLF1), which plays a role in chondrogenesis. The expression of this gene has been shown to be up-regulated in mice chondrocytes upon stimulation by TGF-beta factor which, together with its co-factor cardiotrophin-like cytokine factor 1 (CLCF1), induces proliferation of chondrocyte precursors (Stefanovic and Stefanovic, 2012). A final example, associated with running performance, is the N(alpha)-acetyl transferase 16 (NAA16) gene, which has been correlated with average running speed in mice (Kelly et al., 2014). Interestingly, three out of the five significantly enriched gene ontology terms in the Fst analysis genes are associated urinary tract and kidney development, which might be linked with the running performance of racing camels (Table 6). A study by Leikis et al. (2006) has revealed reduced exercise performance in patients with chronic kidney disease at stages 3 and 4. This reduction in exercise capacity, and associated muscle strength, was also been linked with renal function failure. Moreover, the impairment in muscle K+ level regulation, which is associated with renal function failure (Bergstrom et al., 1983), has also been found to be linked with muscle fatigue (Sejersted and Sjogaard, 2000). This requires further investigations through detailed physiological studies on racing and packing dromedary camels.
To further our understanding of the genetic mechanisms underlying phenotypic traits of importance in dromedary camel, a number of actions are required. In the first instance, the reference genome is currently at the scaffold stage and would greatly benefit from the application of long-read single-molecule sequencing technologies coupled with optical mapping to improve its quality (Mostovoy et al., 2016; Weissensteiner et al., 2017). Improving the assembly of the reference genome to reach the chromosome stage is essential to facilitate detailed analyses for signatures of selection. The recent construction of two whole-genome radiation hybrid panels for dromedary camels by Perelman et al. (2018) is a promising first step toward improving the assembly. Secondly, in parallel to work improving the contiguity of the reference genome is the need for functional analyses on the annotated genes. Improving our understanding of the function of genes provides greater context toward interpreting genotype–phenotype associations. Thirdly, the detailed characterization of camels using standard phenotypic and morphometric parameters is required to more accurately classify the different camel types. Using WGS data in future studies instead of GBS will provide greater resolution to analyses, and less redundancy of data, while increasing the breadth and depth of sampling will enable more reliable calculations of genetic diversity parameters.
Conclusion
We have reported here for the first time the phylogeographic classification between the Arabian Peninsula and Sudanese dromedary camel populations using WGS and GBS data. We identified a number of genomic regions under positive selection hosting genes of putative relevance to Sudanese packing and racing camels, including genes potentially associated with dairy traits and running performance. The results of this study call for further investigation of the genome of dromedary camel using larger and more diverse populations to better identify variants associated with their important phenotypes. The results of which can support the development of informed breeding programs with the aim to improve the productivity and performance of dromedary camels.
Data Availability
The Arabian Peninsula camel full genome sequence data and the genotyping-by-sequencing data of Sudanese camels are publicly available from the European Nucleotide Archive (ENA) with the Bioproject accession number PRJEB32117.
Ethics Statement
Standard techniques were used to collect blood. The procedure was reviewed and approved by Faculty of Veterinary Science, University of Nyala, Sudan.
Author Contributions
HB, HM and OH conceived and designed the project. ES collected the dromedary blood samples from Sudan. HM and FA contributed sequence data for the Sudanese and Arabian Peninsula samples, respectively. HB and DW performed bioinformatic analysis. HB, DW and OH wrote the manuscript. All authors have commented upon and agreed on the contents of the manuscript.
Funding
This work is part of the project “Agricultural growth, capacity building for scientific preservation of livestock breeds in Sudan.” The project was supported by a grant from the Korea- Africa Economic Cooperation Trust Fund through the African Development Bank and co-funded by the livestock CGIAR-CRP. We would like to extend our gratitude to King Faisal University (Saudi Arabia) for their research award (Grant F289) to sequence the genomes of the six Arabian Peninsula dromedary camel samples.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor and reviewer PO-TW declared their involvement as co-editors in the Research Topic.
Acknowledgments
We would like to extend our sincere gratitude to the undersecretary of the Ministry of Finance and National Economy, Sudan, and the resident representative(s) of the African Development Bank (Khartoum, Sudan) for their support in the approval and implementation of the project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00893/full#supplementary-material
References
Al Abri, M. A., Faye, B. (2019). Genetic improvement in dromedary camels: challenges and opportunities. Front. Genet. 10, 167. doi: 10.3389/fgene.2019.00167
Agarwal, A., Sharma, R., Durairajanayagam, D., Ayaz, A., Cui, Z., Willard, B., et al. (2015). Major protein alterations in spermatozoa from infertile men with unilateral varicocele. Reprod. Biol. Endocrinol.: RB&E 13, 8–8. doi: 10.1186/s12958-015-0007-2
Al-Swailem, A. M., Al-Busadah, K. A., Shehata, M. M., Al-Anazi, I. O., Askari, E. (2007). Classification of Saudi Arabian camel (Camelus dromedarius) subtypes based on RAPD technique. J. Food Agric. Environ. 5, 143. doi: 10.1234/4.2007.749
Alberto, F. J., Boyer, F., Orozco-Terwengel, P., Streeter, I., Servin, B., De Villemereuil, P., et al. (2018). Convergent genomic signatures of domestication in sheep and goats. Nat. Commun. 9, 813. doi: 10.1038/s41467-018-03206-y
Alessi, D. R., Sakamoto, K., Bayascas, J. R. (2006). LKB1-dependent signaling pathways. Annu. Rev. Biochem. 75, 137–163. doi: 10.1146/annurev.biochem.75.103004.142702
Alexander, D. H., Novembre, J., Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
Ali, A., Baby, B., Vijayan, R. (2019). From desert to medicine: a review of camel genomics and therapeutic products. Front. Genet. 10, 17. doi: 10.3389/fgene.2019.00017
Almathen, F., Charruau, P., Mohandesan, E., Mwacharo, J. M., Orozco-Terwengel, P., Pitt, D., et al. (2016). Ancient and modern DNA reveal dynamics of domestication and cross-continental dispersal of the dromedary. Proc. Natl. Acad. Sci. U. S. A. 113, 6707–6712. doi: 10.1073/pnas.1519508113
Almathen, F., Elbir, H., Bahbahani, H., Mwacharo, J., Hanotte, O. (2018). Polymorphisms in MC1R and ASIP genes are associated with coat colour variation in the Arabian camel. J. Hered. 109, 700–706. doi: 10.1093/jhered/esy024
Andree-Salvini, B. (2010). Roads of Arabia: archeology and history of the Kingdom of Saudi Arabia. Paris, France: Somogy Editions d’Art.
Altarejos, J. Y., Goebel, N., Conkright, M. D., Inoue, H., Xie, J., Arias, C. M., et al. (2008). The Creb1 coactivator Crtc1 is required for energy balance and fertility. Nat. Med. 14, 1112–1117. doi: 10.1038/nm.1866
Aulchenko, Y. S., Ripke, S., Isaacs, A., Van Duijn, C. M. (2007). GenABEL: an R library for genome-wide association analysis. Bioinformatics 23, 1294–1296. doi: 10.1093/bioinformatics/btm108
Babar, M. E., Hussain, T., Wajid, A., Nawaz, A., Nadeem, A., Shah, S. A., et al. (2015). Mitochondrial cytochrome-b and D-loop sequence based genetic diversity in Mareecha and Bareela camel breeds of Pakistan. J. Anim. Plant Sci. 25, 591–594. doi: 10.1111/age.12158
Bahbahani, H., Afana, A., Wragg, D. (2018a). Genomic signatures of adaptive introgression and environmental adaptation in the Sheko cattle of southwest Ethiopia. PLoS One 13, e0202479. doi: 10.1371/journal.pone.0202479
Bahbahani, H., Clifford, H., Wragg, D., Mbole-Kariuki, M. N., Van Tassell, C., Sonstegard, T., et al. (2015). Signatures of positive selection in East African Shorthorn Zebu: a genome-wide single nucleotide polymorphism analysis. Sci. Rep. 5, 11729. doi: 10.1038/srep11729
Bahbahani, H., Salim, B., Almathen, F., Al Enezi, F., Mwacharo, J. M., Hanotte, O. (2018b). Signatures of positive selection in African Butana and Kenana dairy zebu cattle. PLoS One 13, e0190446. doi: 10.1371/journal.pone.0190446
Bahbahani, H., Tiijani, A., Mukasa, C., Wragg, D., Almathen, F., Nash, O., et al. (2017). Signature of selection for environmental adaptation and zebu x taurine hybrid fitness in East African Shorthorn Zebu. Front. Genet. 8, 1–20. doi: 10.3389/fgene.2017.00068
Beissinger, T. M., Hirsch, C. N., Sekhon, R. S., Foerster, J. M., Johnson, J. M., Muttoni, G., et al. (2013). Marker density and read depth for genotyping populations using genotyping-by-sequencing. Genetics 193, 1073–1081. doi: 10.1534/genetics.112.147710
Bergstrom, J., Alvestrand, A., Furst, P., Hultman, E., Widstam-Attorps, U. (1983). Muscle intracellular electrolytes in patients with chronic uremia. Kidney Int. Suppl. 16, S153–S160.
Choocheep, K., Hatano, S., Takagi, H., Watanabe, H., Kimata, K., Kongtawelert, P., et al. (2010). Versican facilitates chondrocyte differentiation and regulates joint morphogenesis. J. Biol. Chem. 285, 21114–21125. doi: 10.1074/jbc.M109.096479
Coghill, J. M., Fowler, K. A., West, M. L., Fulton, L. M., Van Deventer, H., Mckinnon, K. P., et al. (2013). CC chemokine receptor 8 potentiates donor Treg survival and is critical for the prevention of murine graft-versus-host disease. Blood 122, 825–836. doi: 10.1182/blood-2012-06-435735
Couse, J. E., Mahato, D., Eddy, E. M., Korach, K. S. (2001). Molecular mechanism of estrogen action in the male: insights from the estrogen receptor null mice. Reprod. Fertil. Dev. 13, 211–219. doi: 10.1071/RD00128
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., Depristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Eichner, L. J., Giguere, V. (2011). Estrogen related receptors (ERRs): a new dawn in transcriptional control of mitochondrial gene networks. Mitochondrion 11, 544–552. doi: 10.1016/j.mito.2011.03.121
Eisa, M. O., Mustafa, A. (2011). Production systems and dairy production of Sudan Camel (Camelus dromedarius): a review. Middle-East J. Sci. Res. 7, 132–135. doi: 10.12895/jaeid.20132.166
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6, e19379. doi: 10.1371/journal.pone.0019379
Epstein, H., Mason, I. L. (1971). The origin of the domestic animals of Africa. New York: Africana publishing corporation.
Evanno, G., Regnaut, S., Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Faye, B., Abdelhadi, O. M. A., Ahmed, A. I., Bakheit, S. A. (2011). Camel in Sudan: future prospects. Livestock Res. Rural Dev. 23, 219.
Fitak, R. R., Mohandesan, E., Corander, J., Burger, P. A. (2015). The de novo genome assembly and annotation of a female domestic dromedary of North African origin. Mol. Ecol. Resour. 16, 314–324. doi: 10.1111/1755-0998.12443
Fukuda, N., Yomogida, K., Okabe, M., Touhara, K. (2004). Functional characterization of a mouse testicular olfactory receptor and its role in chemosensing and in regulation of sperm motility. J. Cell Sci. 117, 5835–5845. doi: 10.1242/jcs.01507
Gauthier-Pilters, H. D., Anne, I. (1983). The camel: its evolution, ecology, behavior, and relationship to man. Chicago: University of Chicago Press.
Hansen, P. J. (2004). Physiological and cellular adaptations of zebu cattle to thermal stress. Anim. Reprod. Sci. 82-83, 349–360. doi: 10.1016/j.anireprosci.2004.04.011
Hashim, M. W., Galal, Y. M., Ali, M. A., Khalafalla, A. I., Hamid, A. S., et al. (2015). Dromedary camels in Sudan, types and sub types, distribution and movement. Int. J. Pharm. Res. Anal. 5, 8–12.
He, J., Zhao, X., Laroche, A., Lu, Z. X., Liu, H., Li, Z. (2014). Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front. Plant Sci. 5, 484. doi: 10.3389/fpls.2014.00484
Ishida, Y., Gao, J. L., Murphy, P. M. (2008). Chemokine receptor CX3CR1 mediates skin wound healing by promoting macrophage and fibroblast accumulation and function. J. Immunol. 180, 569–579. doi: 10.4049/jimmunol.180.1.569
Kelly, S. A., Nehrenberg, D. L., Hua, K., Garland, T., Jr., Pomp, D. (2014). Quantitative genomics of voluntary exercise in mice: transcriptional analysis and mapping of expression QTL in muscle. Physiol. Genomics 46, 593–601. doi: 10.1152/physiolgenomics.00023.2014
Khalkhali-Evrigh, R., Hafezian, S. H., Hedayat-Evrigh, N., Farhadi, A., Bakhtiarizadeh, M. R. (2018). Genetic variants analysis of three dromedary camels using whole genome sequencing data. PLoS One 13, e0204028. doi: 10.1371/journal.pone.0204028
Kijas, J. W., Porto-Neto, L., Dominik, S., Reverter, A., Bunch, R., Mcculloch, R., et al. (2014). Linkage disequilibrium over short physical distances measured in sheep using a high-density SNP chip. Anim. Genet. 45, 754–757. doi: 10.1111/age.12197
Kim, E. S., Elbeltagy, A. R., Aboul-Naga, A. M., Rischkowsky, B., Sayre, B., Mwacharo, J. M., et al. (2016). Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment. Heredity (Edinb.) 116, 255–264. doi: 10.1038/hdy.2015.94
Kim, J., Hanotte, O., Mwai, O. A., Dessie, T., Bashir, S., Diallo, B. (2017a). The genome landscape of indigenous African cattle. Genome Biol. 18, 34. doi: 10.1186/s13059-017-1153-y
Kim, M., Yoo, H. J., Kim, M., Seo, H., Chae, J. S., Lee, S. H., et al. (2017b). Influence of estrogen-related receptor gamma (ESRRG) rs1890552 A > G polymorphism on changes in fasting glucose and arterial stiffness. Sci. Rep. 7, 9787. doi: 10.1038/s41598-017-10192-6
Leikis, M. J., McKenna, M. J., Petersen, A. C., Kent, A. B., Murphy, K. T., Leppik, J. A., et al. (2006). Exercise performance falls over time in patients with chronic kidney disease despite maintenance of hemoglobin concentration. Clin. J. Am. Soc. Nephrol. 1, 488–495. doi: 10.2215/CJN.01501005
Li, H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. doi: 10.1093/bioinformatics/btr509
Li, H., Durbin, R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Magee, P. (2015). When was the dromedary domesticated in the ancient Near East? Z. Orient-Archäologie 8, 253–278.
Matthews, J., Gustafsson, J. A. (2003). Estrogen signaling: a subtle balance between ER alpha and ER beta. Mol. Interv. 3, 281–292. doi: 10.1124/mi.3.5.281
Mburu, D. N., Ochieng, J. W., Kuria, S. G., Jianlin, H., Kaufmann, B., Rege, J. E., et al. (2003). Genetic diversity and relationships of indigenous Kenyan camel (Camelus dromedarius) populations: implications for their classification. Anim. Genet. 34, 26–32. doi: 10.1046/j.1365-2052.2003.00937.x
Mckenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Mostovoy, Y., Levy-Sakin, M., Lam, J., Lam, E. T., Hastie, A. R., Marks, P., et al. (2016). A hybrid approach for de novo human genome sequence assembly and phasing. Nat. Methods 13, 587. doi: 10.1038/nmeth.3865
Musa, H. H., Shuiep, E. S., El-Zubeir, I. (2006). Camel husbandry among pastoralists in Darfur, Western Sudan. Nomadic Peoples 10, 101–105. doi: 10.3167/082279406780246438
Nesargikar, P. N., Spiller, B., Chavez, R. (2012). The complement system: history, pathways, cascade and inhibitors. Eur. J. Microbiol. Immunol. (Bp) 2, 103–111. doi: 10.1556/EuJMI.2.2012.2.2
Oleksyk, T. K., Smith, M. W., O’brien, S. J. (2010). Genome-wide scans for footprints of natural selection. Philos. Trans. R Soc. Lond. B Biol. Sci. 365, 185–205. doi: 10.1098/rstb.2009.0219
Onzima, R. B., Upadhyay, M. R., Doekes, H. P., Brito, L. F., Bosse, M., Kanis, E., et al. (2018). Genome-wide characterization of selection signatures and runs of homozygosity in Ugandan Goat Breeds. Front. Genet. 9, 318. doi: 10.3389/fgene.2018.00318
Parmentier, M., Libert, F., Schurmans, S., Schiffmann, S., Lefort, A., Eggerickx, D., et al. (1992). Expression of members of the putative olfactory receptor gene family in mammalian germ cells. Nature 355, 453–455. doi: 10.1038/355453a0
Pauciullo, A., Shuiep, E. S., Cosenza, G., Ramunno, L., Erhardt, G. (2013). Molecular characterization and genetic variability at κ-casein gene (CSN3) in camels. Gene 513, 22–30. doi: 10.1016/j.gene.2012.10.083
Perelman, P. L., Pichler, R., Gaggl, A., Larkin, D. M., Raudsepp, T., Alshanbari, F., et al. (2018). Construction of two whole genome radiation hybrid panels for dromedary (Camelus dromedarius): 5000RAD and 15000RAD. Sci. Rep. 8, 1982. doi: 10.1038/s41598-018-20223-5
Poland, J. A., Rife, T. W. (2012). Genotyping-by-sequencing for plant breeding and genetics. Plant Genome 5, 92–102. doi: 10.3835/plantgenome2012.05.0005
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Quinlan, A. R., Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. doi: 10.1093/bioinformatics/btq033
R Development Core Team (2012). R: a language and environment for statistical computing. Vienna, Austria.
Rincon, G., Weber, K. L., Eenennaam, A. L., Golden, B. L., Medrano, J. F. (2011). Hot topic: performance of bovine high-density genotyping platforms in Holsteins and Jerseys. J. Dairy Sci. 94, 6116–6121. doi: 10.3168/jds.2011-4764
Rubin, C. J., Zody, M. C., Eriksson, J., Meadows, J. R., Sherwood, E., Webster, M. T., et al. (2010). Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 464, 587–591. doi: 10.1038/nature08832
Sanchez, M.-P., Govignon-Gion, A., Croiseau, P., Fritz, S., Hozé, C., Miranda, G., et al. (2017). Within-breed and multi-breed GWAS on imputed whole-genome sequence variants reveal candidate mutations affecting milk protein composition in dairy cattle. Genet. Sel. Evol. GSE 49, 68–68. doi: 10.1186/s12711-017-0344-z
Schmidt-Nielsen, K. (1959). The physiology of the camel. Sci. Am. 201, 140–151. doi: 10.1038/scientificamerican1259-140
Sejersted, O. M., Sjogaard, G. (2000). Dynamics and consequences of potassium shifts in skeletal muscle and heart during exercise. Physiol. Rev. 80, 1411–1481. doi: 10.1152/physrev.2000.80.4.1411
Skinner, J. D., Louw, G. N. (1966). Heat stress and spermatogenesis in Bos indicus and Bos taurus cattle. J. Appl. Physiol. 21, 1784–1790. doi: 10.1152/jappl.1966.21.6.1784
Shuiep, E. T. S., Giambra, I. J., El Zubeir, I. E. Y. M., Erhardt, G. (2013). Biochemical and molecular characterization of polymorphisms of αs1-casein in Sudanese camel (Camelus dromedarius) milk. Int. Dairy J. 28, 88–93. doi: 10.1016/j.idairyj.2012.09.002
Spehr, M., Gisselmann, G., Poplawski, A., Riffell, J. A., Wetzel, C. H., Zimmer, R. K., et al. (2003). Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science 299, 2054–2058. doi: 10.1126/science.1080376
Stefanovic, L., Stefanovic, B. (2012). Role of cytokine receptor-like factor 1 in hepatic stellate cells and fibrosis. World J. Hepatol. 4, 356–364. doi: 10.4254/wjh.v4.i12.356
Tanaka, T., Narazaki, M., Kishimoto, T. (2014). IL-6 in inflammation, immunity, and disease. Cold Spring Harbor Perspect. Biol. 6, a016295. doi: 10.1101/cshperspect.a016295
Vanderhaeghen, P., Schurmans, S., Vassart, G., Parmentier, M. (1993). Olfactory receptors are displayed on dog mature sperm cells. J. Cell Biol. 123, 1441–1452. doi: 10.1083/jcb.123.6.1441
Wang, K., Wu, P., Yang, Q., Chen, D., Zhou, J., Jiang, A., et al. (2018). Detection of selection signatures in Chinese Landrace and Yorkshire pigs based on genotyping-by-sequencing data. Front. Genet. 9, 119. doi: 10.3389/fgene.2018.00119
Wang, J. (2017). Estimating pairwise relatedness in a small sample of individuals. Heredity (Edinb.) 119, 302–313. doi: 10.1038/hdy.2017.52
Watanabe, H., Kimata, K., Line, S., Strong, D., Gao, L. Y., Kozak, C. A., et al. (1994). Mouse cartilage matrix deficiency (cmd) caused by a 7 bp deletion in the aggrecan gene. Nat. Genet. 7, 154–157. doi: 10.1038/ng0694-154
Weir, B. S., Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x
Weissensteiner, M. H., Pang, A. W. C., Bunikis, I., Hoijer, I., Vinnere-Petterson, O., Suh, A., et al. (2017). Combination of short-read, long-read, and optical mapping assemblies reveals large-scale tandem repeat arrays with population genetic implications. Genome Res. 27, 697–708. doi: 10.1101/gr.215095.116
Wickham, H. (2009). ggplot2: elegant graphics for data analysis. New York: Springer-Verlag New York. doi: 10.1007/978-0-387-98141-3
Wilson, D. G., Phamluong, K., Lin, W. Y., Barck, K., Carano, R. A., Diehl, L., et al. (2012). Chondroitin sulfate synthase 1 (Chsy1) is required for bone development and digit patterning. Dev. Biol. 363, 413–425. doi: 10.1016/j.ydbio.2012.01.005
Wilson, R. T. (1998). Camels the tropical agriculturalist. London, United Kingdom: Macmillan Education.
Wu, H., Guang, X., Al-Fageeh, M. B., Cao, J., Pan, S., Zhou, H., et al. (2014). Camelid genomes reveal evolution and adaptation to desert environments. Nat. Commun. 5, 5188. doi: 10.1038/ncomms6188
Keywords: genotype-by-sequence, positive selection, milk production, racing camels, Arabian Peninsula, Sudan
Citation: Bahbahani H, Musa HH, Wragg D, Shuiep ES, Almathen F and Hanotte O (2019) Genome Diversity and Signatures of Selection for Production and Performance Traits in Dromedary Camels. Front. Genet. 10:893. doi: 10.3389/fgene.2019.00893
Received: 17 January 2019; Accepted: 23 August 2019;
Published: 19 September 2019.
Edited by:
Stéphane Joost, École Polytechnique Fédérale de Lausanne, SwitzerlandReviewed by:
Filippo Biscarini, Italian National Research Council (CNR), ItalyPablo Orozco-terWengel, Cardiff University, United Kingdom
Copyright © 2019 Bahbahani, Musa, Wragg, Shuiep, Almathen and Hanotte. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hussain Bahbahani, hussain.bahbahani@ku.edu.kw; hussain.bahbahani@gmail.com; Hassan H. Musa, hassanhm@uofk.edu
 Hussain Bahbahani
Hussain Bahbahani Hassan H. Musa
Hassan H. Musa David Wragg3
David Wragg3 Faisal Almathen
Faisal Almathen Olivier Hanotte
Olivier Hanotte