Mesenchymal Stromal Cells as a Cellular Target in Myeloid Malignancy: Chances and Challenges in the Genome Editing of Stromal Alterations
- 1Department of Hematology, Erasmus Medical Center Cancer Institute, Rotterdam, Netherlands
- 2Oncode Institute, Erasmus Medical Center Cancer Institute, Rotterdam, Netherlands
- 3Department of Cell Biology, Faculty of Medicine, Institute for Biomedical Engineering, Rheinisch-Westfälische Technische Hochschule (RWTH) Aachen University, Aachen, Germany
The contribution of bone marrow stromal cells to the pathogenesis and therapy response of myeloid malignancies has gained significant attention over the last decade. Evidence suggests that the bone marrow stroma should not be neglected in the design of novel, targeted-therapies. In terms of gene-editing, the focus of gene therapies has mainly been on correcting mutations in hematopoietic cells. Here, we outline why alterations in the stroma should also be taken into consideration in the design of novel therapeutic strategies but also outline the challenges in specifically targeting mesenchymal stromal cells in myeloid malignancies caused by somatic and germline mutations.
Introduction
Under physiological conditions, hematopoietic stem cells (HSCs) are regulated by their bone marrow microenvironment (BMM) through cellular interactions and secreted factors to maintain a continuous pool of hematopoietic cells (Morrison and Scadden, 2014; Pinho and Frenette, 2019). This crosstalk between the hematopoietic system with its surroundings is essential for the proper functioning of HSCs throughout life and becomes deregulated in hematological malignancies. The main constituents of the BMM are bone marrow mesenchymal stromal cells (MSCs), osteolineage cells (OLCs), endothelial cells, amongst various other cells including adipocytes, neural, and hematopoietic cells (Pinho and Frenette, 2019; Méndez-Ferrer et al., 2020). MSCs are a heterogenous group of non-hematopoietic cells that express key hematopoiesis-supporting factors such as stem cell factor (SCF) and CXC motif ligand (CXCL)-12. In humans the surface markers CD271 and CD146 have been shown to enrich for cells that can form fibroblast colonies (CFU-F) (Kfoury and Scadden, 2015). MSCs have been described in mouse models using numerous Cre-drivers and surface markers outlined in Figure 1.
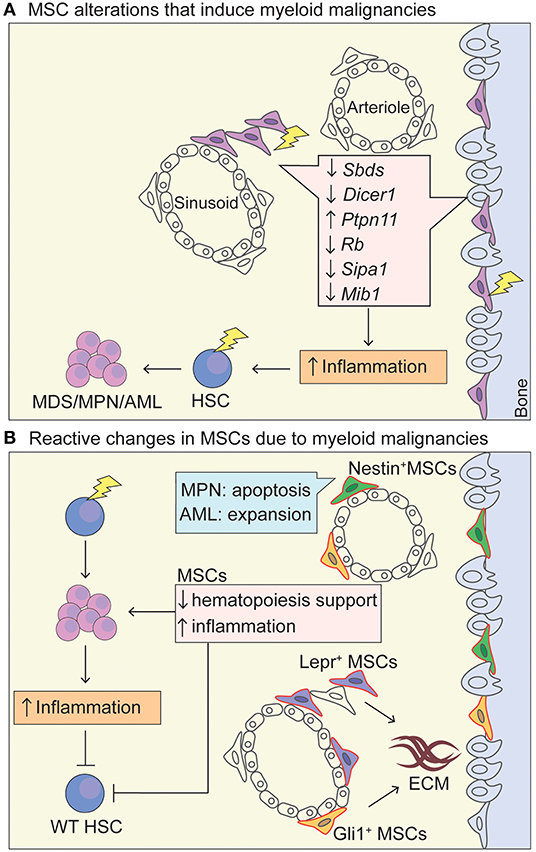
Figure 1. MSC niche in myeloid malignancies. (A) Schematic representation of MSC niche alterations that promote the leukemic transformation. Lightning bolt indicates genetic lesion in MSC-like cells that have been shown to promote oncogenesis through increased inflammation and increased (genotoxic) stress of HSCs. Deletion of Dicer1 and Sbds in Osterix+ osteoprogenitor MSCs leads to a MDS phenotype with sporadic AML upon Dicer1 deletion (Raaijmakers et al., 2010; Zambetti et al., 2016). Activating mutations in tyrosine-protein phosphatase non-receptor type 11 (Ptpn11) in Nestin-Cre+ cells induces a MPN phenotype. A MDS/MPN phenotype was also seen in activating Ptpn11 in Mx1-Cre+, Prx1-Cre+, Lepr-Cre+, Osx1-Cre+ cell-type specific knock-in mice, highlighting that MSCs and osteoprogenitors can induce MPN and that there is probably overlap in cells populations targeted by the Cre-drivers, whereas differentiated osteoblasts (Oc-Cre+) and endothelial cells (VE-Cadherin-Cre+-ERT2) could not induce MPN (Dong et al., 2016). Rb (encodes RB protein) (Walkley et al., 2007) and Mib1 (encodes mind bomb 1 protein) (Kim et al., 2008) promote an MPN-like phenotype in a Mx1-Cre+ driver. The Mx1-Cre+ driver traces MSC-like cells that are located within the bone marrow but also at the periosteum (Ortinau et al., 2019), and have limited in vivo adipogenic differentiation potential, making them more osteoprogenitor-like (Park et al., 2012). Sipa1 expression is most abundant in CD31+ BM endothelial cells, but also found in MSCs (CD45−Lin−CD31−CD51+Sca1+) (Xiao et al., 2018). Deletion of Sipa1 results in the development of MDS/MPN and Sipa1−/− MSCs cultured in vitro show increased adipogenic and chrondrogenic differentiation potential, but impaired osteogenic differentiation (Xiao et al., 2018). (B) Upon exposure of a mutated hematopoietic cell within the niche, MSCs are functionally altered through cytokine stimulation, direct cell-cell contact and activation of inflammatory pathways, promoting the survival of the mutant HSC in favor of WT HSCs and increase in inflammatory signatures. Specifically, Lepr-Cre+ (Decker et al., 2017) and Gli1-Cre+ (Schneider et al., 2017) cells expand and proliferate in an MPN setting and produce extracellular matrix. Nestin-Cre+ cells proliferate in AML and provide chemotherapy resistance (Forte et al., 2020), whereas they become apoptotic in MPN disease due to neural damage and Schwann cell death triggered by interleukin-1β production by the mutated HSC. AML cells seem to induce osteogenic differentiation and block adipogenesis of MSCs, as well as blocking maturation of osteolineage-MSCs into mature osteoblasts (Battula et al., 2017; Pievani et al., 2020b). MSCs, mesenchymal stem cells; HSC, hematopoietic stem cells; AML, acute myeloid leukemia; MPN, myeloproliferative neoplasm; MDS, myelodysplastic syndrome; ECM, extracellular matrix; Lepr, Leptin receptor; Prx1, Paired related homeobox 1; Mx-1, Myxovirus Resistance-1.
Historically, the development of myeloid malignancies was considered to be HSC-intrinsic, be it driven by germline or somatic mutation. The BMM can either facilitate oncogenesis by supporting the expansion of malignant cells, and suppressing normal hematopoiesis, or induce oncogenesis by acquiring mutations or functional alterations that pre-dispose for oncogenesis. These two theories are not mutually exclusive, as is amply exemplified in the pathogenesis of myeloid malignancies including myeloproliferative neoplasms (MPN), myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML) (Medyouf, 2017; Fathi et al., 2019; Behrmann et al., 2020). Thus, the mutual interaction between mutated HSCs and the BMM has further evolved as an attractive novel therapeutic target.
In this article, we will outline the role of stromal cells (specifically BM MSCs) in myeloid malignancies in somatic disease, as well as germline conditions, and describe recent progress in dissecting the HSC-stroma crosstalk. Finally, we discuss possible application of the established murine disease models and future challenges in developing genetically targeted therapies for the BM stroma.
The Role of the Stroma in Leukemia Pre-Disposition Syndromes
The World Health Organization (WHO) introduced a new category of “myeloid malignancies with germline predisposition” to the 2016 Classification of hematopoietic tumors. Given that these “rare” mutations are only coming to light with increased use of parallel sequencing platforms in population and family studies (Porter, 2016; Miller et al., 2018; Kim et al., 2020), it can be speculated that germline stromal mutations exist which have yet to be discovered. An exemplary disease is Shwachman-Diamond syndrome (SDS); a rare autosomal recessive bone marrow failure disorder caused by mutation in the SBDS gene with a cumulative probability of leukemic progression of >30% at the age of 30 years (Dale et al., 2006; Nelson and Myers, 2018). Hematopoietic cell specific deletion of Sbds did not result in MDS or AML in two murine disease models (Rawls et al., 2007; Zambetti et al., 2015), whereas exposure of wildtype HSCs to Sbds-deficient osteolineage MSCs led to an MDS phenotype and genotoxic stress in HSCs (Zambetti et al., 2016). The prominent clinical feature of skeletal abnormalities in SDS patients was recapitulated through the niche-specific deletion of Sbds (Zambetti et al., 2016). Importantly, the alarmin heterocomplex S100A8/9 secreted by the niche was identified as a candidate driver of inflammatory stress in HSCs, highlighting that the crosstalk between stroma and HSCs is of particular interest as a possible therapy target. Targeted deletion of Dicer1 in osteolineage MSCs resulted in reduced expression of Sbds in transplanted WT HSCs (Raaijmakers et al., 2010). The resulting phenotype displayed key features of human MDS and a tendency to develop AML; clearly showing that alterations in BM stromal cells can induce malignancy and stress in HSCs (Raaijmakers et al., 2010). Intriguing case studies of donor cell-derived leukemia (DCL) development upon allogeneic HSC transplantation in humans have brought about the possibility of oncogenesis driven by the diseased recipient BMM (Berger et al., 2016; Engel et al., 2018).
In line, numerous genetic modifications (deletions) in non-hematopoietic, stromal cells were reported to give rise to a myeloproliferative phenotype in vitro and in vivo (Rupec et al., 2005; Walkley et al., 2007; Xiao et al., 2018), but also activation of e.g., Notch signaling (Kim et al., 2008; Dong et al., 2016). Inflammation seems to play an important role in the pathogenesis of these myeloid malignancies. As an example, IL-1B propagates an inflammatory BMM as it activates HSCs to differentiate toward myeloid cells and monocytes (Rupec et al., 2005). Early stages of MPN disease are also characterized by increased IL-1β expression, which triggers pro-inflammatory damage to the BMM and advances disease progression (Arranz et al., 2014). This showcases that stromal drivers influence the hematopoietic system and can result in secondary neoplasms (schematically depicted in Figure 1A).
Crosstalk Between Non-Mutated Stroma and Hematopoietic Cells With Somatic Mutations
It is becoming evident that the BMM is functionally altered by exposure to hematopoietic cells harboring somatic mutations, creating a proinflammatory environment that seems to propagate leukemic disease development and supresses normal hematopoiesis (Figure 1B). Overall differences in MSC compartments have been noted in myeloid malignancies compared to normal bone marrow. In AML, there is generally a reduction of bulk MSCs. However, Nestin+ cells, as well-documented MSCs in the BM (Mendez-Ferrer et al., 2010), have been shown to be 4–5-fold more abundant in human AML patients, in line with expansion of Nestin+ cells in the murine iMLL-AF9 AML model (Hanoun et al., 2014; Forte et al., 2020). This is in striking contrast to the decrease in Nestin+ cells in murine models and human MPN (Arranz et al., 2014; Drexler et al., 2019), suggesting that the same group of niche cells can behave differently in various myeloid malignancies and/or stages of leukemic disease. Conditional depletion of Nestin+ cells upon AML development in iMLL-AF9 mice lead to a significantly extended mouse survival, suggesting that Nestin+ cells promote leukemogenesis in vivo (Forte et al., 2020). Importantly, in a competitive transplant setting, depletion of Nestin+ cells during AML development selectively diminished the number of leukemic cells, while leaving normal hematopoiesis unaffected (Forte et al., 2020), which is one of the major challenges in the treatment of AML.
The direct effect of a mutated hematopoietic clone on the bone microenvironment is evidently illustrated in murine models but also patient samples with bone marrow fibrosis. In a murine model of CML, endosteal OLCs expanded upon expression of BCR/ABL in the hematopoietic compartment leading to deposition of extracellular matrix (Schepers et al., 2013). In response to MPN clones, Gli1+ stromal cells are activated from their normal endosteal and perivascular niches and significantly expand in murine models and patient samples (Schneider et al., 2017). Importantly, their genetic ablation ameliorates fibrosis, proving functional proof that they play a central role in the fibrotic transformation. Another stromal subset of Lepr+ MSCs has been shown to expand in fibrosis (Decker et al., 2017). Conditional deletion of platelet-derived growth factor receptor a (Pdgfra) from Lepr+ cells or the administration of the tyrosine kinase inhibitor imatinib suppressed Lepr+ cell expansion and mitigated fibrosis. There seems to be a common initial pro-inflammatory stromal response to the malignant MPN clone that poises the stroma to become pro-fibrotic (Gleitz et al., 2020; Leimkuhler et al., 2020). This is in line with the observation of a diseased niche characterized by cellular stress and an increased inflammatory signature in bulk RNA-sequencing of prospectively sorted mesenchymal cells from human low-risk MDS patients (Chen et al., 2016). Additionally, human MDS MSCs produce inflammatory cytokines (IL-1β, IL-6, and TNFα) compared to control in vitro cultured MSCs (Flores-Figueroa et al., 2002, 2008). Notably, IL-6 knockout in the BM reduces fibrosis in a MPN setting (Gleitz et al., 2020). Likewise, our group demonstrated increased expression of the inflammatory S100A8 alarmin in the stromal niche in murine models and patient samples of del(5q) MDS (Ribezzo et al., 2018). This increased expression of S100A8 in MSCs resulted in decreased hematopoiesis-support in vitro, indicating that mutated hematopoietic cells can initiate a vicious cycle of inflammation in the niche, leading to decreased support of normal hematopoiesis and fuelling the progression of haematopoietic malignancy. The common denominator in hematological malignancies driven by somatic or germline mutations thus seems to be an inflammatory “mutagenic” microenvironment that precedes malignant transformation and disease progression (Craver et al., 2018; Gleitz et al., 2018; Leimkühler and Schneider, 2019; Pronk and Raaijmakers, 2019).
Challenges Of Genetic Editing in the Bone Marrow Stroma
As outlined, the bone marrow stroma seems to play a significant role in the initiation, maintenance and progression of myeloid malignancies and murine models indicate that MSCs are a highly attractive therapeutic target. The importance of targeting the stroma is highlighted by the fact that despite improvements in the treatment of AML, long-term survival is <30% in adults (Ferrara and Schiff, 2013). In murine models, specific subsets of stromal cells can be modified by using stromal Cre-drivers. The correlate to this procedure in the human setting would optimally be genome editing of stromal cells. Nuclease-based site-specific genome editing has provided an unprecedented opportunity to artificially modify genetic information within mammalian cells (Romito et al., 2019). The clustered regularly interspersed short palindromic repeats (CRISPR)/Cas9 system has been used to create germline and somatic mouse models, and has the benefits of relatively easy design and high mutational efficiency (Mou et al., 2015; González-Romero et al., 2019; Broeders et al., 2020; Lee et al., 2020). The HSC has been the most relevant cell type to edit, with major advances in Cas9 clinical translation made, particularly in the monogenetic disorders sickle cell disease and β-thalassemia (Dever and Porteus, 2017).
In this section we highlight some of the key challenges hampering the development of targeted genetic therapy of BM stromal populations: (1) identification of specific MSC population to edit, (2) targeting MSCs in their in situ location vs. ex vivo, (3) indirect targeting of MSC function in vivo through genome editing of hematopoietic cells and cell-to-cell interactions, and (4) in vitro functional characterization of MSCs and potential therapeutic targets through CRISPR screens and 3D models.
Identification and Targeting MSCs In Situ – Direct Vs. Indirect Strategies
Much of our understanding of the BM MSCs has originated from genetic-fate tracing mouse models in which MSC populations have been labeled via a stromal Cre-driver (Kfoury and Scadden, 2015). Functionality of these Cre-drivers has been shown by conditional deletion using diphteria-toxin receptor based mechanisms (Schneider et al., 2017; Pinho and Frenette, 2019). Additionally, Cre-drivers of MSC populations provide spatial information when combined with a fluorescent-reporter. Nevertheless, the current widely-used Cre-drivers likely label heterogeneous groups of MSCs, outlined in a recent review (Al-Sabah et al., 2020). The recombination efficacy in Cre-drivers has resulted in variable results (Chen et al., 2017), while conditional Cre-lines result in higher specificity compared to constitutive Cre-lines and allow fate-tracing experiments in health and disease setting (Méndez-Ferrer et al., 2020). Recent advancements in single-cell RNA sequencing (scRNAseq) have for the first time allowed us to zoom in on heterogeneous populations within the murine BMM (Schroeder et al., 2016; Baryawno et al., 2019; Tikhonova et al., 2019; Wolock et al., 2019; Baccin et al., 2020; Leimkuhler et al., 2020). Tikhonova et al. have shown that the Lepr+ Cre-driver previously studied as one MSC population, contains four subclusters of MSCs, with functional differences between them as current evidence suggests. As we gain knowledge of functionally distinct MSCs and their possibly common progenitors, it will become possible to target them. Importantly, the location of MSCs in relation to (mutant) HSPCs seems to predict biological functionality and these sinusoidal and CXCL12 niches will need to be further investigated (Gomariz et al., 2018; Baccin et al., 2020; Kokkaliaris et al., 2020). Perhaps the use of multiplexed imaging (Kokkaliaris et al., 2020) in combination with laser-capture techniques to isolate specific BM populations (Baccin et al., 2020) can aid in inferring spatial and signaling relationships between cells from single cell transcriptomic data.
Specifically, these new techniques can help identify new druggable pathways through, for example, ligand-receptor analysis between mutated hematopoietic cells and the stromal counterpart in myeloid malignancies (Efremova et al., 2020). This method was very recently employed in the unbiased scRNAseq paper showing populations of murine and human MSCs interacting with hematopoietic populations in MPN (Leimkuhler et al., 2020). A druggable alarmin axis was identified in the fibrotic transformation both in murine models and patients and treatment with Tasquinimod, inhibiting the binding of the alarmins S100A8/S100A9 to TLR4, ameliorated the MPN phenotype in mice.
Due to the lack of evident genetic modifications and a prominent cell of origin, a clear-cut molecular target for BM MSCs is not apparent. It is possible, however, to target HSPCs as they are relatively easily accessible for genome editing. The use of gene therapy for neurometabolic disorders using HSPC transplantation has shown that overexpression of therapeutic proteins has cross-correction capacity as also non-hematopoietic cells are being exposed to the therapeutic effect (Ferrari et al., 2020). This could be useful if loss-of-function mutations are found in MSCs.
One could imagine that mutated hematopoietic cells can be examined for specific receptors that are not vital for their physiological function but are unique for their malignant interaction with stroma (Kokkaliaris and Scadden, 2020; Pievani et al., 2020a). The α4β1 integrin–VCAM1 axis between stroma and the AML mutant cell aids in chemoresistance (Jacamo et al., 2014; Carter et al., 2016). AML chemo-resistant cells also have high expression of very late antigen 4 (VLA4) which facilitates adherence to the stroma through VCAM1 activated NF-kB signaling (Jacamo et al., 2014). Indeed, patients with VLA-4-negative AML have a more favorable prognosis, highlighting the role of stroma-HSCPs cross-talk (Matsunaga et al., 2003). Within the CXCL12-CXCR4 axis, CXCL12 is expressed by MSCs and interacts with HSCs via the binding to CXCR4, regulating their mobilization (Greenbaum et al., 2013). Blockade of this axis can release leukemic cells from their chemoprotective niches (Nervi et al., 2009). Recently, an elegant in vivo pooled CRISPR screen targeting selected cell surface genes was performed in murine MLL-AF9 AML cells and identified CXCR4 as a positive regulator of leukemic cells, indispensable for their growth and survival in vivo (Ramakrishnan et al., 2020). CXCR4 is essential for the development of AML independently of its interaction with CXCL12 on MSCs or endothelial cells. In contrast, Cxcr4−/− normal HSCs are capable of long-term hematopoiesis (Nie et al., 2008), highlighting the different biology in homeostasis and malignant disease and possible targeting avenues.
As an example, inflammation within the BM niche, specifically the erythroblastic niche, can be targeted by genetically editing the hematopoietic cell. In our previous work, we applied CRISPR-Cas9 technology in a murine MDS model to genetically inactivate S100a8 and improve the defective erythropoiesis characteristic for the disease. Compared to control non-targeting sgRNA, CRISPR-mediated inactivation of S100a8 in MDS cells restored erythropoiesis and restored a normal erythroid niche by interrupting the cycle of inflammation (Schneider et al., 2016).
Ideally, the complex interplay between the hematopoietic system and the stroma could be modeled more efficiently with CRISPR-Cas9 based techniques in mice (Heckl et al., 2014; Tothova et al., 2017). With advancements in deep-sequencing, novel germline/somatic mutations in stroma of patients might be identified. More complex models could then be made to mimic the different mutations identified in the hematopoietic and the stromal compartment in mice, to search for druggable targets. The genome-editing efficiency is also consistently being improved, with DNA-free systems being developed that are more suitable for human trials as there is no risk for random insertional mutagenesis (Shapiro et al., 2020).
Genetic Editing of Stromal Cells Ex Vivo: Feasibility of Delivery
A commonly used CRISPR/Cas9- based technique for gene editing ex vivo is the isolation of the target cell and delivery of the gene-editing machinery via electroporation, microinjection, or virus-based vehicles before injecting the corrected cells back into patients or mice (Broeders et al., 2020). MSCs in general have been widely investigated for use in multiple diseases due to the ease of their isolation (plastic adherence and self-renewal properties), their low immunity potential, and their ability to secrete factors (Kean et al., 2013). The production of inflammatory cytokines such as PDGF, TNFa, CCR8, and CCR2 within the solid organ tumor microenvironment, has been shown to enhance homing of MSCs to the tumor location (Marofi et al., 2017). Primary MSCs can express CRISPR/Cas9 proteins through nucleofection, lentivirus, and non-integrating adeno-associated virus (Golchin et al., 2020). However, the homing of edited MSCs to the bone marrow niche has not been formally tested yet.
A possible technique by which CRISPR-based strategies on the BM stroma could be performed, is by injecting complete CRISPR-proteins through intrafemoral injections. Intrafemoral injection has been used to model osteosarcoma in orthotopic mouse models (Sasaki et al., 2016). The only downside is that off-target effects on surrounding (hematopoietic) cells can occur. To circumvent this, a possibility could be to expand MSCs ex vivo and genetically alter them using CRISPR in vitro, and then inject them back through an intrafemoral injection. It has been shown that donor MSCs injected via intramarrow injection also contribute to the reconstitution of the stromal niche in the ablated bone marrow of recipient mice (Muguruma et al., 2006; Ahn et al., 2010; Zhou et al., 2014). Intraosseal therapy could pose clinical challenges, with an invasive procedure that has an increased chance of complications (in particular infections), compared to intravenous or intraarterial administration. However, the intravenously administered MSCs easily get trapped in the lung circulation and have limited engraftment of about a week, whereas arterially administered MSCs seem to engraftment better at the site of injury, e.g., hind leg bone irradiation in mice (Kean et al., 2013). First, proof of principle studies using intrafemoral/intraosseal injections need to be performed where candidate genes can be knocked out or mutations introduced within the mouse or even specifically in the stroma by using floxed Cas9 mice crossed to specific stromal Cre-drivers. The beauty of this method in mice is additionally that one leg can be edited while one leg serves as a non-targeted control. A major point to consider, however, is the determination of recombination efficiency within the bone marrow stroma as MSCs are difficult to obtain as single-cell suspension cells. A possible read-out here could be in situ hybridization of mRNA of the targeted genes in a multiplex imaging set-up.
Cas proteins need specially designed delivery vehicles for tissue-specific delivery as they cannot cross biological barriers themselves and have a high positive charge and molecular mass. Extracellular vesicles (EVs) are used as possible packing devices for sgRNA:Cas9 ribonucleoprotein complexes. It has been shown however that EVs are mainly taken up by the liver (~84%), whereas roughly 1.6% are found back in the bone marrow 4 h upon systemic administration, making delivery to the bone marrow quite challenging (Kostyushev et al., 2020). Progress is made on engineering functionalized exosomes (M-CRISPR-Cas9 exosome) which encapsulate CRISPR-Cas9 components more efficiently (Ye et al., 2020). Recently, the interest for vesicle nanoparticles containing the Cas9 machinery has been growing. While traditionally nanoparticles can mainly be found in the liver and lung after injection, a recent breakthrough study (Krohn-Grimberghe et al., 2020), reported the design and in vivo performance of systemically injected lipid–polymer nanoparticles encapsulating small interfering RNA (siRNA), for the silencing of genes specifically in bone-marrow endothelial cells. Using nanoparticle enabled RNAi, the group targeted stromal-derived factor 1 (Sdf1) resulting in stem cell liberation into the blood, and monocyte chemotactic protein 1 (Mcp1) whose silencing retained monocytes in the BM. These modified nanoparticles lay the ground for editing non-hematopoietic cells in the bone marrow with a high efficacy and show that HSPCs biology can be altered through stroma alterations.
Functional Testing of Genetic Modification of MSCs In Vitro and In Vivo
Isolation of MSCs directly from the BM remains a challenge as stromal cells are closely associated with extracellular matrix within the marrow and single-cell suspensions are difficult to obtain even after digestion of bone (Gomariz et al., 2018). Most often, MSCs are left to grow out from bone chips or human aspirates and selected for on the basis of their plastic adherence. Cultured human MSCs are minimally characterized by their trilineage differentiation potential, expression of surface markers that enhance CFU-F potential, and plastic adherence in vitro (Dominici et al., 2006; Kfoury and Scadden, 2015; Agha et al., 2017). Murine MSCs are often identified by a panel of typical surface markers and have a less stringent definition (Agha et al., 2017). Nevertheless, functional characterization of in vitro isolated cells still needs to be optimized, as even a short-term (passage 0) ex vivo culturing environment greatly reprograms MSCs compared to direct sorting of primary cells for microarray analysis (Ghazanfari et al., 2017). Despite retaining their in vitro clonogenicity and tri-lineage differentiation potential (Pevsner-Fischer et al., 2011), culture-induced gene expression changes are present and raise the question of comparability of primary and cultured cells, as well as the possibility that only specific subsets of MSCs are selected for in adherent culture (Tormin et al., 2009). BM MSCs cultured as non-adherent 3D sphere colonies termed mesenspheres, have been reported to retain MSC surface markers, tri-lineage potential, and to have an increased self-renewal potential in serial transplantations into immunodeficient NOD scid gamma mice compared to adherent cultured cells (Ghazanfari et al., 2016). Gene expression in cultured MSC mesenspheres was still altered compared to primary sorted MSCs, but 3D cultured cells had more osteogenic and adipogenic transcription factor expression compared to 2D adherent cells (Ghazanfari et al., 2017). This difference in culturing conditions might be confounding as Forte et al. have shown that in MSCs derived from the same AML donors, only mesenspheres provided enhanced chemoprotection of human AML blasts, whereas plastic-adherent MSCs did not (Forte et al., 2020). The improved fitness of 3D cultured MSCs advocates for its use. Ideally, there will be a standardized protocol for the isolation of murine and human BM MSCs so that results from different groups can be compared easily (Stroncek et al., 2020).
Patient derived cultured MSCs however, in 2D but also 3D cultures, can serve as a platform for personalized screening approaches to detect alterations which hamper therapy or find potential targets. As an example, a genome-scale CRISPR knock-out screen was used to uncover imatinib-sensitizing genes in vitro on K562 cells (Lewis et al., 2020). Although this was performed on cell lines, one can imagine broadening the application and do similar tests in smaller format (due to the high cell number needed) on patient derived cells.
These methods could be used as proof-of-principle platforms to identify candidate proteins for genome editing. Recently, human mesenchymal stromal cells were shown to endure nucleofection with Cas9-adeno-associated virus serotype 6 (AAV-6) and genome-editing including gene disruption and targeted integration of up to 3.2 kb of DNA with stable transgene expression, while retaining their in vitro tri-lineage differentiation potential and phenotypical signature (Srifa et al., 2020). Through integration of PDGF-BB, VEGFA, and IL-10 transgenes at the HBB locus they successfully created hypersecreting hMSC which actively improved wound healing in diabetic wounds of mice. Specifically the combination of scaffolds coated with human MSCs could be modified with the Cas9-AAV-6 system to model normal and malignant human hematopoiesis by subcutaneous implantation in immunodeficient mice (Vaiselbuh et al., 2010; Abarrategi et al., 2017; Passaro et al., 2017). The benefit of such a system is that patient-derived leukemic cells can grow in the hMSC scaffolds as they form ectopic humanized BMM and can be followed up for long periods of time in an in vivo setting. Similarly, human femur-derived bone fragments from AML patients were transplanted into NSG mice using Matrigel as a carrier and were vascularized 4 weeks post implantation (Battula et al., 2017). These systems could allow for easily-accessible and controllable in vivo gene-editing of multiple relevant human BMM populations in the presence of clonal xenografted AML cells.
Future Outlook
As we gain knowledge of the different functional subcomponents of the bone marrow niche, the disease model of myeloid malignancies will become more complex. It is evident that oncogenesis can arise from two non-mutually exclusive theories: niche-induced and niche-facilitated. In patients, we envision a future of personalized medicine in which the stroma can be pharmacologically targeted in combination with a hematopoietic cell-based therapy. We can use the accumulating knowledge with genome editing by (1) generation of murine disease models in vivo on the basis of new possible germline/somatic mutations within the niche found with targeted sequencing in human disease to study disease pathogenesis, (2) targeting MSCs in vivo directly through MSC/EV-based approaches, (3) indirectly through modulation of hematopoietic cells, (4) modeling of the human hematopoietic niche using ossified scaffolds in xenotransplantations, and (5) in vitro Cas9-based screening methods. Targeted genome-editing will most likely become more feasible as we characterize the true MSCs as the target cell and improve engineering of carriers which will deliver the sgRNA:Cas9 cargo with high efficacy to the bone marrow.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
RS was supported by grants from the MPN foundation (2017 MPNRF/LLS Award), a KWF Kankerbestrijding young investigator grant (11031/2017-1, Bas Mulder Award; Dutch Cancer Foundation), and a grant of the European Research Council (ERC) (deFIBER; ERC-StG 757339).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abarrategi, A., Foster, K., Hamilton, A., Mian, S. A., Passaro, D., Gribben, J., et al. (2017). Versatile humanized niche model enables study of normal and malignant human hematopoiesis. J. Clin. Invest. 127, 543–548. doi: 10.1172/JCI89364
Agha, E., El, Kramann, R., Schneider, R. K., Li, X., Seeger, W., and Humphreys, B. D. (2017). Mesenchymal stem cells in fibrotic disease. Stem Cell 21, 166–177. doi: 10.1016/j.stem.2017.07.011
Ahn, J., Park, G., and Shim, J. (2010). Intramarrow injection of β -catenin-activated, but not naïve mesenchymal stromal cells stimulates self-renewal of hematopoietic stem cells in bone marrow. Exp. Mol. Med. 42, 122–131. doi: 10.3858/emm.2010.42.2.014
Al-Sabah, J., Baccin, C., and Haas, S. (2020). Single-cell and spatial transcriptomics approaches of the bone marrow microenvironment. Curr. Opin. Oncol. 32, 146–153. doi: 10.1097/CCO.0000000000000602
Arranz, L., Sanchez-Aguilera, A., Martin-Perez, D., Isern, J., Langa, X., Tzankov, A., et al. (2014). Neuropathy of haematopoietic stem cell niche is essential for myeloproliferative neoplasms. Nature 512, 78–81. doi: 10.1038/nature13383
Baccin, C., Al-sabah, J., Velten, L., Helbling, P. M., Grünschläger, F., Hernández-Malmierca, P., et al. (2020). Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat. Cell Biol. 22, 38–48. doi: 10.1038/s41556-019-0439-6
Baryawno, N., Przybylski, D., Kowalczyk, M. S., Kfoury, Y., Severe, N., Gustafsson, K., et al. (2019). A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell 177, 1915–1932.e16. doi: 10.1016/j.cell.2019.04.040
Battula, V. L., Le, P. M., Sun, J. C., Nguyen, K., Yuan, B., Zhou, X., et al. (2017). AML-induced osteogenic differentiation in mesenchymal stromal cells supports leukemia growth. JCI Insight 2:e90036. doi: 10.1172/jci.insight.90036
Behrmann, L., Wellbrock, J., and Fiedler, W. (2020). The bone marrow stromal niche: a therapeutic target of hematological myeloid malignancies. Expert Opin. Ther. Targets 24, 451–462. doi: 10.1080/14728222.2020.1744850
Berger, G., Berg, E., Van Den Abbott, K. M., Sinke, R. J., Bungener, L. B., Mulder, A. B., et al. (2016). Re-emergence of acute myeloid leukemia in donor cells following allogeneic transplantation in a family with a germline DDX41 mutation. Leukemia 31, 520–522. doi: 10.1038/leu.2016.310
Broeders, M., Herrero-hernandez, P., Ernst, M. P. T., and Van Der Ploeg, A. T. (2020). Sharpening the molecular scissors : advances in gene-editing technology. iScience 23:100789. doi: 10.1016/j.isci.2019.100789
Carter, B. Z., Mak, P. Y., Chen, Y., Mak, D. H., Mu, H., Ruvolo, V., et al. (2016). Anti-apoptotic ARC protein confers chemoresistance by controlling leukemia-microenvironment interactions through a NFκB/IL1β signaling network. Oncotarget 7:7911. doi: 10.18632/oncotarget.7911
Chen, K. G., Johnson, K. R., and Robey, P. G. (2017). Mouse genetic analysis of bone marrow stem cell niches: technological pitfalls, challenges, and translational considerations. Stem Cell Rep. 9, 1343–1358. doi: 10.1016/j.stemcr.2017.09.014
Chen, S., Zambetti, N. A., Bindels, E. M. J., Kenswil, K. J. G., Mylona, M. A., Adisty, N., et al. (2016). Massive parallel RNA sequencing of highly purified mesenchymal elements in low-risk MDS reveals tissue- context-dependent activation of inflammatory programs. Leukemia 30, 1938–1942. doi: 10.1038/leu.2016.91
Craver, B. M., Alaoui, K., El Scherber, R. M., and Fleischman, A. G. (2018). The critical role of inflammation in the pathogenesis and progression of myeloid malignancies. Cancers 10:104. doi: 10.3390/cancers10040104
Dale, D. C., Bolyard, A. A., Schwinzer, B. G., Pracht, G., Bonilla, M. A., Boxer, L., et al. (2006). The severe chronic neutropenia international registry : 10-year follow-up report. Support. Cancer Ther. 3, 220–231. doi: 10.3816/SCT.2006.n.020
Decker, M., Martinez-morentin, L., Wang, G., Lee, Y., Liu, Q., Leslie, J., et al. (2017). Leptin receptor-expressing bone marrow stromal cells are myofibroblasts in primary myelofibrosis. Nat. Cell. Biol. 19, 677–688. doi: 10.1038/ncb3530
Dever, D. P., and Porteus, M. H. (2017). The changing landscape of gene editing in hematopoietic stem cells: a step towards cas9 clinical translation daniel. Curr. Opin. Hematol. 24, 481–488. doi: 10.1097/MOH.0000000000000385
Dominici, M., Blanc, K., Le Mueller, I., Marini, F. C., Krause, D. S., Deans, R. J., et al. (2006). Minimal criteria for defining multipotent mesenchymal stromal cells. International society for cellular therapy position statement. Cytotherapy 8, 315–317. doi: 10.1080/14653240600855905
Dong, L., Yu, W., Zheng, H., Loh, M. L., Bunting, S. T., Pauly, M., et al. (2016). Leukaemogenic effects of Ptpn11 activating mutations in the stem cell microenvironment. Nat. Publ. Gr. 539, 304–308. doi: 10.1038/nature20131
Drexler, B., Passweg, J. R., Tzankov, A., Bigler, M., Theocharides, A. P. A., Cantoni, N., et al. (2019). The sympathomimetic agonist mirabegron did not lower JAK2-V617F allele burden, but restored nestin-positive cells and reduced reticulin fibrosis in patients with myeloproliferative neoplasms: results of phase II study SAKK 33/14. Haematologica 104, 710–716. doi: 10.3324/haematol.2018.200014
Efremova, M., Vento-tormo, M., and Teichmann, S. A. (2020). CellPhoneDB : inferring cell – cell communication from combined expression of multi-subunit ligand – receptor complexes. Nat. Protoc. 15, 1484–1506. doi: 10.1038/s41596-020-0292-x
Engel, N., Rovo, A., Badoglio, M., Labopin, M., Basak, G. W., Beguin, Y., et al. (2018). European experience and risk factor analysis of donor cell-derived leukaemias / MDS following haematopoietic cell transplantation. Leukemia 33, 508–517. doi: 10.1038/s41375-018-0218-6
Fathi, E., Sanaat, Z., and Farahzadi, R. (2019). Mesenchymal stem cells in acute myeloid leukemia: a focus on mechanisms involved and therapeutic concepts. Blood Res. 54, 165–174. doi: 10.5045/br.2019.54.3.165
Ferrara, F., and Schiff, C. A. (2013). Acute myeloid leukaemia in adults. Lancet 381, 484–495. doi: 10.1016/S0140-6736(12)61727-9
Ferrari, G., Thrasher, A. J., and Aiuti, A. (2020). Gene therapy using haematopoietic stem and progenitor cells. Nat. Rev. Genet. doi: 10.1038/s41576-020-00298-5. [Epub ahead of print].
Flores-Figueroa, E., Gutiérrez-Espindola, G., José Montesinos, J., Maria Arana-Trejo, R., and Mayani, H. (2002). In vitro characterization of hematopoietic microenvironment cells from patients with myelodysplastic syndrome. Leuk. Res. 26, 677–686. doi: 10.1016/S0145-2126(01)00193-X
Flores-Figueroa, E., Montesinos, J. J., Flores-guzm, P., Espindola, G. G., Arana-Trejo, R. M., Hernandez-Estevez, E., et al. (2008). Functional analysis of myelodysplastic syndromes-derived mesenchymal stem cells. Leuk. Res. 32, 1407–1416. doi: 10.1016/j.leukres.2008.02.013
Forte, D., Fernandez, M. G., Sanchez-Aguilera, A., Stavropoulou, V., Fielding, C., Martin-Perez, D., et al. (2020). Bone marrow mesenchymal stem cells support acute myeloid leukemia bioenergetics and enhance antioxidant defense and escape from chemotherapy. Cell Metab. 32, 829–843.e9. doi: 10.1016/j.cmet.2020.09.001
Ghazanfari, R., Li, H., Zacharaki, D., Lim, H. C., and Scheding, S. (2016). Human non-hematopoietic CD271pos/CD140alow/neg bone marrow stroma cells fulfill stringent stem cell criteria in serial transplantations. Stem Cells Dev. 25, 1652–1658. doi: 10.1089/scd.2016.0169
Ghazanfari, R., Zacharaki, D., Li, H., Lim, H. C., Soneji, S., and Scheding, S. (2017). Human primary bone marrow mesenchymal stromal cells and their in vitro progenies display distinct transcriptional profile signatures. Sci. Rep. 7:10338. doi: 10.1038/s41598-017-09449-x
Gleitz, H., Dugourd, A. J. F., Leimkühler, N. B., Snoeren, I. A. M., Fuchs, S. N. R., Menzel, S., et al. (2020). Increased CXCL4 expression in hematopoietic cells links inflammation and progression of bone marrow fibrosis in MPN. Blood 136, 2051–2064. doi: 10.1182/blood.2019004095
Gleitz, H. F. E., Kramann, R., and Schneider, R. K. (2018). Understanding deregulated cellular and molecular dynamics in the haematopoietic stem cell niche to develop novel therapeutics for bone marrow fibrosis. J. Pathol. 245, 138–146. doi: 10.1002/path.5078
Golchin, A., Shams, F., and Karami, F. (2020). Advancing mesenchymal stem cell therapy with CRISPR/Cas9 for clinical trial studies. Adv. Exp. Med. Biol. 1247, 89–100. doi: 10.1007/5584_2019_459
Gomariz, A., Helbling, P. M., Isringhausen, S., Suessbier, U., Becker, A., Boss, A., et al. (2018). Quantitative spatial analysis of haematopoiesis-regulating stromal cells in the bone marrow microenvironment by 3D microscopy. Nat. Commun. 9:2532. doi: 10.1038/s41467-018-04770-z
González-Romero, E., Martínez-Valiente, C., García-Ruiz, C., Vázquez-Manrique, R. P., Cervera, J., and Sanjuan-Pla, A. (2019). CRISPR to fix bad blood : a new tool in basic and clinical hematology. Haematologica 104, 881–893. doi: 10.3324/haematol.2018.211359
Greenbaum, A., Hsu, Y.-M. S., Day, R. B., Schuettpelz, L. G., Christopher, M. J., Borgerding, J. N., et al. (2013). CXCL12 production by early mesenchymal progenitors is required. Nature 495, 227–230. doi: 10.1038/nature11926
Hanoun, M., Zhang, D., Mizoguchi, T., Pinho, S., Pierce, H., Kunisaki, Y., et al. (2014). Acute myelogenous leukemia-inducedsympathetic neuropathy promotes malignancyin an altered hematopoietic stem cell niche. Cell Stem Cell 15, 365–375. doi: 10.1016/j.stem.2014.06.020
Heckl, D., Kowalczyk, M. S., Yudovich, D., Belizaire, R., Puram, R. V., Mcconkey, M. E., et al. (2014). Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol. 32, 941–946. doi: 10.1038/nbt.2951
Jacamo, R., Chen, Y., Wang, Z., Wencai, M., Zhang, M., Spaeth, E. L., et al. (2014). Reciprocal leukemia-stroma VCAM-1/VLA-4-dependent activation of NF-κB mediates chemoresistance. Blood 123, 2691–2702. doi: 10.1182/blood-2013-06-511527
Kean, T. J., Lin, P., Caplan, A. I., and Dennis, J. E. (2013). MSCs : delivery routes and engraftment, cell-targeting strategies, and immune modulation. Stem Cells Int. 2013:732742. doi: 10.1155/2013/732742
Kfoury, Y., and Scadden, D. T. (2015). Mesenchymal cell contributions to the stem cell niche. Stem Cell 16, 239–253. doi: 10.1016/j.stem.2015.02.019
Kim, B., Yun, W., Lee, S. T., Choi, J. R., Yoo, K. H., Koo, H. H., et al. (2020). Prevalence and clinical implications of germline predisposition gene mutations in patients with acute myeloid leukemia. Sci. Rep. 10:14297. doi: 10.1038/s41598-020-71386-z
Kim, Y. W., Koo, B. K., Jeong, H. W., Yoon, M. J., Song, R., Shin, J., et al. (2008). Defective notch activation in microenvironment leads to myeloproliferative disease. Blood 112, 4628–4638. doi: 10.1182/blood-2008-03-148999
Kokkaliaris, K. D., Kunz, L., Cabezas-Wallscheid, N., Christodoulou, C., Renders, S., Camargo, F., et al. (2020). Adult blood stem cell localization reflects the abundance of reported bone marrow niche cell types and their combinations. Blood 136, 2296–2307. doi: 10.1182/blood.2020006574
Kokkaliaris, K. D., and Scadden, D. T. (2020). Cell interactions in the bone marrow microenvironment affecting myeloid malignancies. Blood Adv. 4, 3795–3803. doi: 10.1182/bloodadvances.2020002127
Kostyushev, D., Kostyusheva, A., Brezgin, S., Smirnov, V., Volchkova, E., Lukashev, A., et al. (2020). Gene editing by extracellular vesicles. Int. J. Mol. Sci. 21, 1–34. doi: 10.3390/ijms21197362
Krohn-Grimberghe, M., Mitchell, M. J., Schloss, M. J., Khan, O. F., Courties, G., Guimaraes, P. P. G., et al. (2020). Nanoparticle-encapsulated siRNAs for gene silencing in the haematopoietic stem-cell niche. Nat. Biomed. Eng. 4, 1076–1089. doi: 10.1038/s41551-020-00623-7
Lee, J., Bayarsaikhan, D., Bayarsaikhan, G., Kim, J., Schwarzbach, E., and Lee, B. (2020). Pharmacology & therapeutics recent advances in genome editing of stem cells for drug discovery and therapeutic application. Pharmacol. Ther. 209:107501. doi: 10.1016/j.pharmthera.2020.107501
Leimkuhler, N. B., Gleitz, H., Ronghui, L., Snoeren, I. A. M., Fuchs, S. N. R., Nagai, J. S., et al. (2020). Heterogeneous bone-marrow stromal progenitors drive myelofibrosis via a druggable alarmin axis. Cell Stem Cell 28, 1–16. doi: 10.1016/j.stem.2020.11.004
Leimkühler, N. B., and Schneider, R. K. (2019). Inflammatory bone marrow microenvironment. Hematol. Am. Soc. Hematol. Educ. Program 2019, 294–302. doi: 10.1182/hematology.2019000045
Lewis, M., Florence, V. P., Iggo, R., Turcq, B., Richard, E., Iggo, R., et al. (2020). A genome-scale CRISPR knock-out screen in chronic myeloid leukemia identifies novel drug resistance mechanisms along with intrinsic apoptosis and MAPK signaling. Cancer Med. 9, 6739–6751. doi: 10.1002/cam4.3231
Marofi, F., Vahedi, G., Biglari, A., Esmaeilzadeh, A., and Athari, S. S. (2017). Mesenchymal stromal / stem cells : a new era in the cell-based targeted gene therapy of cancer. Front. Immunol. 8:1770. doi: 10.3389/fimmu.2017.01770
Matsunaga, T., Takemoto, N., Sato, T., Takimoto, R., Tanaka, I., Fujimi, A., et al. (2003). Interaction between leukemic-cell VLA-4 and stromal fibronectin is a decisive factor for minimal residual disease of acute myelogenous leukemia. Nat. Med. 9, 1158–1165. doi: 10.1038/nm909
Medyouf, H. (2017). The microenvironment in human myeloid malignancies: emerging concepts and therapeutic implications. Blood 129, 1617–1626. doi: 10.1182/blood-2016-11-696070
Méndez-Ferrer, S., Bonnet, D., Steensma, D. P., Hasserjian, R. P., Ghobrial, I. M., Gribben, J. G., et al. (2020). Bone marrow niches in haematological malignancies. Nat. Rev. Cancer 20, 285–298. doi: 10.1038/s41568-020-0245-2
Mendez-Ferrer, S., Michurina, T. V., Ferraro, F., Mazloom, A. R., Macarthur, B. D., Lira, S. A., et al. (2010). Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466, 829–834. doi: 10.1038/nature09262
Miller, L. H., Qu, C. K., and Pauly, M. (2018). Germline mutations in the bone marrow microenvironment and dysregulated hematopoiesis. Exp. Hematol. 66, 17–26. doi: 10.1016/j.exphem.2018.07.001
Morrison, S. J., and Scadden, D. T. (2014). The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334. doi: 10.1038/nature12984
Mou, H., Kennedy, Z., Anderson, D. G., Yin, H., and Xue, W. (2015). Precision cancer mouse models through genome editing with CRISPR-Cas9. Genome Med. 7:53. doi: 10.1186/s13073-015-0178-7
Muguruma, Y., Yahata, T., Miyatake, H., Sato, T., Uno, T., Itoh, J., et al. (2006). Reconstitution of the functional human hematopoietic microenvironment derived from human mesenchymal stem cells in the murine bone marrow compartment. Blood 107, 1878–1887. doi: 10.1182/blood-2005-06-2211
Nelson, A. S., and Myers, K. C. (2018). Diagnosis, treatment, and molecular pathology of shwachman-diamond syndrome. Hematol. Oncol. Clin. North Am. 32, 687–700. doi: 10.1016/j.hoc.2018.04.006
Nervi, B., Ramirez, P., Rettig, M. P., Uy, G. L., Holt, M. S., Ritchey, J. K., et al. (2009). Chemosensitization of acute myeloid leukemia (AML) following mobilization by the CXCR4 antagonist AMD3100. Blood 113, 6206–6214. doi: 10.1182/blood-2008-06-162123
Nie, Y., Han, Y. C., and Zou, Y. R. (2008). CXCR4 is required for the quiescence of primitive hematopoietic cells. J. Exp. Med. 205, 777–783. doi: 10.1084/jem.20072513
Ortinau, L. C., Wang, H., Lei, K., Deveza, L., Jeong, Y., Hara, Y., et al. (2019). Identification of functionally distinct Mx1+αSMA+ periosteal skeletal stem cells. Cell Stem Cell 25, 784–796.e5. doi: 10.1016/j.stem.2019.11.003
Park, D., Spencer, J. A., Koh, B. I., Kobayashi, T., Fujisaki, J., Clemens, T. L., et al. (2012). Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell 10, 259–272. doi: 10.1016/j.stem.2012.02.003
Passaro, D., Abarrategi, A., Foster, K., Ariza-McNaughton, L., and Bonnet, D. (2017). Bioengineering of humanized bone marrow microenvironments in mouse and their visualization by live imaging. J. Vis. Exp. 2017:55914. doi: 10.3791/55914
Pevsner-Fischer, M., Levin, S., and Zipori, D. (2011). The origins of mesenchymal stromal cell heterogeneity. Stem Cell Rev. Rep. 7, 560–568. doi: 10.1007/s12015-011-9229-7
Pievani, A., Biondi, M., Tomasoni, C., Biondi, A., and Serafini, M. (2020a). Location first: targeting acute myeloid leukemia within its niche. J. Clin. Med. 9:1513. doi: 10.3390/jcm9051513
Pievani, A., Donsante, S., Tomasoni, C., Corsi, A., Dazzi, F., Biondi, A., et al. (2020b). Acute myeloid leukemia shapes the bone marrow stromal niche in vivo. Haematologica 2020:247205. doi: 10.3324/haematol.2020.247205
Pinho, S., and Frenette, P. S. (2019). Haematopoietic stem cell activity and interactions with the niche. Nat. Rev. Mol. Cell Biol. 20, 303–320. doi: 10.1038/s41580-019-0103-9
Porter, C. C. (2016). Germ line mutations associated with leukemias. Hematol. Am. Soc. Hematol. Educ. Program 2016, 302–308. doi: 10.1182/asheducation-2016.1.302
Pronk, E., and Raaijmakers, M. H. G. P. (2019). The mesenchymal niche in MDS. Blood 133, 1031–1038. doi: 10.1182/blood-2018-10-844639
Raaijmakers, M. H. G. P., Mukherjee, S., Guo, S., Zhang, S., Kobayashi, T., Schoonmaker, J. A., et al. (2010). Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature 464, 852–857. doi: 10.1038/nature08851
Ramakrishnan, R., Peña-Martínez, P., Agarwal, P., Rodriguez-Zabala, M., Chapellier, M., Högberg, C., et al. (2020). CXCR4 signaling has a CXCL12-independent essential role in murine MLL-AF9-driven acute myeloid leukemia. Cell Rep. 31:107684. doi: 10.1016/j.celrep.2020.107684
Rawls, A. S., Gregory, A. D., Woloszynek, J. R., Liu, F., and Link, D. C. (2007). Lentiviral-mediated RNAi inhibition of Sbds in murine hematopoietic progenitors impairs their hematopoietic potential. Blood 110, 2414–2422. doi: 10.1182/blood-2006-03-007112
Ribezzo, F., Snoeren, I. A. M., Ziegler, S., Stoelben, J., Olofsen, P. A., Henic, A., et al. (2018). Rps14, Csnk1a1 and miRNA145/miRNA146a deficiency cooperate in the clinical phenotype and activation of the innate immune system in the 5q- syndrome. Leukemia 33, 1759–1772. doi: 10.1038/s41375-018-0350-3
Romito, M., Rai, R., Thrasher, A. J., and Cavazza, A. (2019). Genome editing for blood disorders : state of the art and recent advances. Emerg. Top. Life Sci. 3, 289–299. doi: 10.1042/ETLS20180147
Rupec, R. A., Jundt, F., Rebholz, B., Eckelt, B., Weindl, G., Herzinger, T., et al. (2005). Stroma-mediated dysregulation of myelopoiesis in mice lacking IκBα. Immunity 22, 479–491. doi: 10.1016/j.immuni.2005.02.009
Sasaki, H., Iyer, S. V., Sasaki, K., Tawfik, O. W., Iwakuma, T., City, K., et al. (2016). An improved intrafemoral injection with minimized leakage as an orthotopic mouse model of osteosarcoma. Anal. Biochem. 486, 70–74. doi: 10.1016/j.ab.2015.06.030
Schepers, K., Pietras, E. M., Reynaud, D., Flach, J., Binnewies, M., Garg, T., et al. (2013). Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self-reinforcing leukemic niche. Cell Stem Cell 13, 285–299. doi: 10.1016/j.stem.2013.06.009
Schneider, R. K., Mullally, A., Dugourd, A., Peisker, F., Hoogenboezem, R., Van Strien, P. M. H., et al. (2017). Gli1+ mesenchymal stromal cells are a key driver of bone marrow fibrosis and an important cellular therapeutic target. Cell Stem Cell 20, 785–800.e8. doi: 10.1016/j.stem.2017.03.008
Schneider, R. K., Schenone, M., Ferreira, M. V., Kramann, R., Joyce, C. E., Hartigan, C., et al. (2016). Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat. Med. 22, 288–297. doi: 10.1038/nm.4047
Schroeder, T., Geyh, S., Germing, U., and Haas, R. (2016). Mesenchymal stromal cells in myeloid malignancies. Blood Res. 51, 225–232. doi: 10.5045/br.2016.51.4.225
Shapiro, J., Iancu, O., Jacobi, A. M., McNeill, M. S., Turk, R., Rettig, G. R., et al. (2020). Increasing CRISPR efficiency and measuring its specificity in HSPCs using a clinically relevant system. Mol. Ther. Methods Clin. Dev. 17, 1097–1107. doi: 10.1016/j.omtm.2020.04.027
Srifa, W., Kosaric, N., Amorin, A., Jadi, O., Camarena, J., Gurtner, G. C., et al. (2020). Cas9-AAV6-engineered human mesenchymal stromal cells improved cutaneous wound healing in diabetic mice. Nat. Commun. 11:2470. doi: 10.1038/s41467-020-16065-3
Stroncek, D. F., Jin, P., McKenna, D. H., Takanashi, M., Fontaine, M. J., Pati, S., et al. (2020). Human mesenchymal stromal cell (MSC) characteristics vary among laboratories when manufactured from the same source material : a report by the cellular therapy team of the biomedical excellence for safer transfusion (BEST) collaborative. Front. Cell Dev. Biol. 8:458. doi: 10.3389/fcell.2020.00458
Tikhonova, A. N., Dolgalev, I., Hu, H., Sivaraj, K. K., Hoxha, E., Cuesta-domínguez, Á., et al. (2019). The bone marrow microenvironment at single-cell resolution. Nature 569, 222–228. doi: 10.1038/s41586-019-1104-8
Tormin, A., Brune, J. C., Olsson, E., Valcich, J., Neuman, U., Olofsson, T., et al. (2009). Characterization of bone marrow-derived mesenchymal stromal cells (MSC) based on gene expression profiling of functionally defined MSC subsets. Cytotherapy 11, 114–128. doi: 10.1080/14653240802716590
Tothova, Z., Krill-burger, J. M., Popova, K. D., Landers, C. C., Sievers, Q. L., Yudovich, D., et al. (2017). Multiplex CRISPR-Cas9 based genome editing in human hematopoietic stem cells models clonal hematopoiesis and myeloid neoplasia. Cell Stem Cell 21, 547–555. doi: 10.1016/j.stem.2017.07.015
Vaiselbuh, S. R., Edelman, M., Lipton, J. M., and Liu, J. M. (2010). Ectopic human mesenchymal stem cell-coated scaffolds in NOD/SCID mice: an in vivo model of the leukemia niche. Tissue Eng. C Methods 16, 1523–1531. doi: 10.1089/ten.tec.2010.0179
Walkley, C. R., Shea, J. M., Sims, N. A., Purton, L. E., and Orkin, S. H. (2007). Rb regulates interactions between hematopoietic stem cells and their bone marrow microenvironment. Cell 129, 1081–1095. doi: 10.1016/j.cell.2007.03.055
Wolock, S. L., Krishnan, I., Tenen, D. E., Tenen, D. G., Klein, A. M., Welner, R. S., et al. (2019). Mapping distinct bone marrow niche populations and their differentiation paths. Cell Reports 28, 302–311.e5. doi: 10.1016/j.celrep.2019.06.031
Xiao, P., Dolinska, M., Sandhow, L., Kondo, M., Johansson, A. S., Bouderlique, T., et al. (2018). Sipa1 deficiency-induced bone marrow niche alterations lead to the initiation of myeloproliferative neoplasm. Blood Adv. 2, 534–548. doi: 10.1182/bloodadvances.2017013599
Ye, Y., Zhang, X., Xie, F., Xu, B., Xie, P., Yang, T., et al. (2020). An engineered exosome for delivering sgRNA:Cas9 ribonucleoprotein complex and genome editing in recipient cells. Biomater. Sci. 8, 2966–2976. doi: 10.1039/D0BM00427H
Zambetti, N. A., Bindels, E. M. J., Strien, P. M. H., Van Valkhof, M. G., Adisty, M. N., Remco, M., et al. (2015). Deficiency of the ribosome biogenesis gene Sbds in hematopoietic stem and progenitor cells causes neutropenia in mice by attenuating lineage progression in myelocytes. Haematologica 100, 1285–1293. doi: 10.3324/haematol.2015.131573
Zambetti, N. A., Ping, Z., Chen, S., Loosdrecht, A. A., Van De Vogl, T., Raaijmakers, M. H. G. P., et al. (2016). Mesenchymal inflammation drives genotoxic stress in hematopoietic stem cells and predicts disease evolution in human pre-leukemia. Stem Cell 19, 613–627. doi: 10.1016/j.stem.2016.08.021
Keywords: genome-editing, BM MSCS, myeloid maliganancies, stromal alterations, BM niche
Citation: Banjanin B and Schneider RK (2021) Mesenchymal Stromal Cells as a Cellular Target in Myeloid Malignancy: Chances and Challenges in the Genome Editing of Stromal Alterations. Front. Genome Ed. 2:618308. doi: 10.3389/fgeed.2020.618308
Received: 16 October 2020; Accepted: 22 December 2020;
Published: 22 January 2021.
Edited by:
Pietro Genovese, Boston Children's Hospital and Harvard Medical School, United StatesReviewed by:
Andreas Reinisch, Medizinische Universität Graz, AustriaMaria Rosa Lidonnici, San Raffaele Telethon Institute for Gene Therapy (SR-Tiget), Italy
Copyright © 2021 Banjanin and Schneider. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rebekka K. Schneider, reschneider@ukaachen.de
 Bella Banjanin
Bella Banjanin Rebekka K. Schneider
Rebekka K. Schneider