Genetic evidence of differential dispersal pattern in the Asiatic wild dog: Comparing two populations with different pack sizes
- Wildlife Institute of India, Dehradun, India
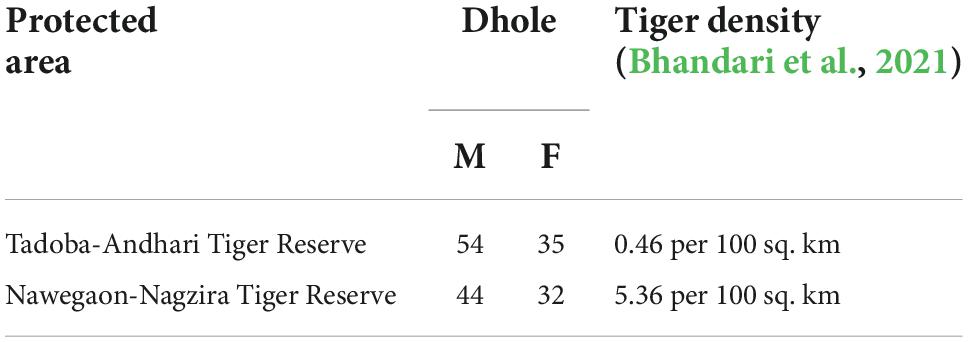
Introduction: Dispersal is a multi-causal, crucial life-history event in shaping the genetic and behavioral structure of mammals. We assessed the dispersal pattern of dholes aka Asiatic wild dog (Cuon alpinus), a social monogamous mammal at two tiger reserves of Maharashtra with different degrees of pack size and competition with tigers i.e. Tadoba-Andhari (TATR, smaller pack size, higher tiger density) and Nawegaon-Nagzira (NNTR, larger pack size, lower tiger density).
Methods: We used the microsatellite data of 174 individual genotypes (98 males and 67 females) to assess the dispersal pattern of dholes from two populations with varying pack size, tiger density, and landscape connectivity using gene flow as a proxy. We compared the population structure, pairwise F statistics, assignment index, and relatedness across a spatial scale.
Results and discussion: Overall, the results suggested a difference in sex-bias dispersal pattern for the two sub-populations, exhibiting significant results for female-biased dispersal in the TATR population with a smaller pack size and higher tiger density. Our study highlights the variability in sex-biased dispersal patterns in two different populations which could be the consequence of different variables such as pack size, tiger density, and geographical scale. The study warrants further quantitative investigation including several factors such as individual behavior, pack composition, pack size, tiger density, etc. In the present Anthropocene era, determining the sex bias in dispersal patterns for a short-range, pack-living carnivore will help in devising an effective conservation management plan for their long-term survival.
Introduction
Dispersal of individuals from the natal group to other breeding sites is one of the most important aspects of an organism’s life history and governs a range of ecological and evolutionary processes (Lawson Handley and Perrin, 2007). Effective dispersal is the key process for the maintenance of population genetic connectivity and persistence across the fragmented landscape (Bowler and Benton, 2005; Lowe and Allendorf, 2010) and affects the genetic structure across the populations or social groups (Ross, 2001). It is a costly but inevitable decision with risks of injury and mortality, depending on the dispersal distance, habitat quality and connectivity, mating system, and individual’s personality as well as sex (van Overveld et al., 2014; Burgess et al., 2016). The three major drivers to induce individuals to disperse have been proposed as follows: resource competition, inbreeding avoidance, and competition with the kin (Pusey, 1987; Dieckmann et al., 1999; Gandon, 1999; Perrin and Mazalov, 2000; Szulkin and Sheldon, 2008). Species, populations, and individuals do not always react similarly to the cues that trigger dispersal, which sometimes results in contrasting dispersal strategies between individuals (Baguette et al., 2013) governed by external information and internal state of individuals resulting in variable dispersal patterns such as personality dependent or sex-biased dispersal patterns.
In the Anthropocene era with limited resources and habitat patches, there are a few other factors that can play a decisive role in an individual’s dispersal such as landscape connectivity and interspecific as well as intraguild competition. Interspecific competition affects virtually all species and has long been recognized as an important force structuring large carnivore guilds (Palomares and Caro, 1999; Courchamp and Macdonald, 2001).
Tiger (Panthera tigris), leopard (Panthera leo), and the Asiatic wild dog (Cuon alpinus) represent the major carnivore guilds in the Indian sub-continent with the tiger as the dominant predator. Of the three, the Asiatic wild dog is the least studied group; they are socially monogamous canid exhibiting communal breeding, paternal care, and large packs often with multiple breeding females in a group (Fox, 1984; Johnsingh, 1985). Dholes are cooperative breeders where the sub-ordinate females of the group provide parental care to the offsprings of the breeding alpha female and show a highly organized social dominance hierarchy with an alpha male and female as main breeders. The species mainly thrives in deep forests and comes under the endangered category by the International Union for Conservation of Nature (IUCN) (Kamler et al., 2015). Preference for deeply forested habitat and home range of 58.67 sq. km (±4.08) (Habib et al., 2021a), smaller as compared to other large carnivores, makes this species vulnerable to habitat fragmentation and disrupted gene flow which may result in reproductive isolation, decrease in fitness, and reduced genetic diversity in the population, posing major concerns for their long-term survival (Pavlova et al., 2017). Previous studies have suggested three genetically differentiated major dhole populations in India: Western and Eastern Ghats (WEG), Central Indian Landscape (CIL), and North-East India (NEI) (Rodrigues et al., 2022).
Previous telemetry, behavioral, and genetic studies on dholes have suggested a bias toward female dispersal due to the majority of male-biased packs (Johnsingh, 1982; Venkataraman, 1998; Iyengar et al., 2005), but, to date, no extensive study has been done, comprehending the effects of competition, group size, density, and spatial heterogeneity on dhole dispersal (Miyamoto et al., 2013; Groom et al., 2017). Though the male-biased sex ratio in dhole packs possibly be a consequence of female-biased nature of dhole dispersal (Venkataraman, 1998; Modi et al., 2018), the dispersal pattern of a pack-living animal is further complicated by other variables. Dholes are socially monogamous, which contributes to the male-biased packs and female dispersal due to delayed dispersal in males, resource competition, and biparental care (Iyengar et al., 2005; Randall et al., 2007). Female dholes dispersal can be favorable to the sub-ordinate female by enhancing the possibilities of breeding and resources in other packs and reducing the likelihood of inbreeding with closely related kin (Sterck et al., 1997). A study done on red colobus monkeys found that female dispersal is more frequent in larger groups to avoid the increased competition for food resources within the group (Miyamoto et al., 2013). In the case of dholes, there is also a significant difference in the average pack size: a larger pack in Nawegaon-Nagzira Tiger Reserve (NNTR) and a smaller pack size in Tadoba-Andhari Tiger Reserve (TATR) within the landscape (Bhandari et al., 2021). The wide-scale assessment of local patterns of variation in pack size showed a significant inverse relationship between tiger density and pack size of dhole while accounting for the variability in resources and habitat heterogeneity (Bhandari et al., 2021). High intraguild competition from larger predators is known to result in lower group sizes and low recruitment rates in sub-ordinate social carnivores (Groom et al., 2017).
To understand the evolutionary pressures giving rise to asymmetric dispersal owing to the difference in sex and pack size and its genetic and ecological consequences, we describe the dispersal pattern of this group-living canid by using sex-specific population structure as a proxy for dispersal. Changes in dispersal patterns influence the geneflow and are predicted to affect a population’s genetic composition and conservation needs (Radespiel and Bruford, 2014). This information on dispersal patterns will also provide a baseline for delineating dispersal corridors and managing land use in the areas surrounding the dhole habitat. However, determining sex-biased dispersal in the wild can be difficult for elusive species like dholes. The direct methods based on field observations using mark-recapture and telemetry (Cozzi et al., 2020; Woodroffe et al., 2020) can be impractical, time-consuming, and expensive to apply considering the need for the large sample size for such experiments (Goudet et al., 2002; Solberg et al., 2006). Most importantly, the data obtained from the direct research methods to deduce dispersal patterns refer to the present migration status but do not reveal the effective dispersal followed by genetic exchange and reduced genetic differentiation between subpopulations. Advancements in non-invasive techniques and population genetics have provided tools for inferring dispersal patterns via indirect estimates coupled with faster, more accessible, and wide sampling coverage with the use of biparental markers (Goudet et al., 2002; Andrew et al., 2013).
In this study, we hypothesized that dispersal could be sex-biased in dholes due to their complex social hierarchy and cooperative breeding. The male-biased packs in dholes could be an outcome of female-biased dispersal. We tested whether there is any relationship between pack size and sex-biased dispersal pattern between two populations with varying tiger densities and habitat contiguity. To test our hypothesis, we used the microsatellite dataset of two different populations (NNTR and TATR) having different average pack sizes, which is an outcome of different top predator and prey densities between the two populations (Bhandari et al., 2021). We expected that the bias in the dispersal pattern, if there is any, would be more evident in the TATR population compared to the NNTR population. We found a strong genetic differentiation with five genetic clusters across the major protected areas of Maharashtra and each sub-population exhibiting its own signature in our previous study on dhole population genetics (Modi et al., 2021). Based on the findings, we expected the nature of dispersal to be short and within the sub-population.
Materials and methods
Study area
The study was conducted in two tiger reserves, namely TATR and NNTR, of Eastern Vidarbha Landscape which holds a major part of the Asiatic wild dog population in Maharashtra. Both parks have different topographic and climatic conditions where TATR has dry-deciduous forest and plain terrain while NNTR possesses moist deciduous forest and undulating terrain. Both the protected areas are separated by an average distance of 200 kms and interspersed by human habitation and roads possibly acting as a barrier for gene flow. Apart from physical differences, both tiger reserves exhibit an interesting sympatric predator dynamic due to comparative differences in tiger population and pack size variation (Bhandari et al., 2021).
Sampling
The sampling for dhole scats was conducted in the two tiger reserves of Maharashtra comprising an almost similar number of dhole individuals and a significant difference in their average pack size (Bhandari et al., 2021), i.e., TATR (area—1727.59 sq. km; smaller packs—6.4 ± 1.3; tiger density—5.36/100 km2) and NNTR (area—1894.94 sq. km; larger packs—16.8 ± 3.1; tiger density—0.46/100 km2). Our study primarily focuses on the dispersal bias within the population at the inter-pack level. Extensive sampling was conducted in both areas during the winter months of November and December (2018–2019) looking for possible dhole scats, focusing mainly on the junction of the road and trails where dholes generally defecate. We conducted opportunistic sampling and collected a total of 180 scats from TATR, while 194 scats from NNTR along with their geographic location. Only the scats with large bolus were collected to target the adult dholes (Keiter et al., 2016). The samples were collected in butter paper with separate zip-lock bags and temporarily preserved with silica gel (Sigma-Aldrich, Cat:85340) and further stored at −20°C for an average period of 20–25 days in the Conservation Genetics lab, Wildlife Institute of India, until processing.
DNA extraction, microsatellite loci genotyping, and molecular sexing
DNA extraction was performed using the QIAamp DNA Tissue Kit (QIAGEN Inc., Hilden, Germany), following a modified approach by Ball et al. (2007) by replacing the swabbing technique with the scraping method. Negative controls were maintained during all the steps of extraction to detect any contamination. Scats were confirmed to be from dholes using the DNA barcoding technique with the help of species-specific primers (DholespID-F/R) targeted at the ND4 gene (Modi et al., 2018). Species ascertainment was done through the visualization of dhole-specific bands (236 bp) and comparison with a positive dhole blood sample in a 2% agarose gel. The confirmed dhole scats were further analyzed using a cross-species microsatellite panel of 12 markers (Modi et al., 2019). To obtain reliable consensus data for analysis, we used the multiple tube strategy paired with the quality index protocol established by Modi et al. (2019) and repeated each locus genotyping four times. Only genotypes that yielded data for at least seven of the 12 loci in the consensus (Goudet et al., 2002) were included. For the samples to be eligible for downstream analysis, a quality index threshold of 0.66 per locus was set, with a mean quality index of 0.75 across loci. We utilized GIMLET’s v 1.3.3 genotyping error (Valière, 2002) estimate module to assess overall genotyping error rates and MICRO CHECKER v 2.2.3 (Van Oosterhout et al., 2004) to determine large allele dropouts. We used GENEPOP v 4.7 and ARLEQUIN v 3.1 to examine deviations from Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD) (Excoffier et al., 2005).
The probability of identity (pID) and probability of identity among sibs (pIDsibs) (Waits et al., 2001) were computed for both populations separately. The genetic recaptures were determined using the identity analysis module implemented in CERVUS v 3.0.7 (Kalinowski et al., 2007). The sex of the confirmed individual genotypes was determined using a multiplex sexing approach (Modi et al., 2018). The sexing multiplex PCR resulted in a three-band pattern (112, 190, and 199 bp from DBY, AHTX-40, and SRY genes, respectively) for males and a single band (190 bp from AHTX-40 gene) for females. We conducted three independent PCR repeats for the sex identification of all the individuals identified. The amplification was confirmed by electrophoresis at 90 V followed by visualization on 2% agarose gel.
Data analyses
Fine-scale genetic structure
To develop a broad understanding of the population structure at the reserve level, we used the Bayesian clustering method implemented in STRUCTURE v 2.3.4 (Pritchard et al., 2000). The analysis was conducted first on NNTR and TATR populations as a whole, and then separately on TATR and NNTR, males and females to understand their assignment within the population. The dispersing sex should be less genetically structured. Ten independent runs for k = 1 to 10 were performed using an admixture model with 100,000 burn in steps and 1,000,000 Markov Chain Monte Carlo (MCMC) repetitions to ensure chain stabilization. We used the LOCPRIOR model to improve the genetic assignment in case of a weak structure within the population. The optimum k (the number of clusters) was determined with the help of the highest estimated log-likelihood, the ad hoc ΔK (Evanno’s method) using STRUCTURE HARVESTER v 6.8 (Evanno et al., 2005; Earl and VonHoldt, 2012). We also used a Bayesian approach implemented in BAYESASS ver. 3.0.3 (Wilson and Rannala, 2003) to infer the directional contemporary migration rate (m) between the two populations.
Sex-biased dispersal
To understand the dispersal pattern of dholes, we compared different genetic indices between males and females for both reserves based on biparentally inherited markers (Goudet et al., 2002).
Genetic diversity, inbreeding coefficient, and relatedness
We calculated the observed (Ho) and expected (He) heterozygosity per locus, H–W equilibrium, and linkage disequilibrium using FSTAT v2.9.4 and Arlequin 3.1 for both populations (Excoffier et al., 2005). To determine the dispersing bias, we calculated the genetic diversity measured by allelic richness per locus, observed heterozygosity (Ho), expected heterozygosity (He), and inbreeding coefficient (F) of males and females to determine whether there was a significant deviation of the genotype frequencies of an individual from the H–W equilibrium (Goudet et al., 2002). The evaluation was done using the demerelate function for F calculation incorporated in the r package “Demerelate” (Kraemer and Gerlach, 2017) with R version 1.1.453 (R Core Team, 2018). F-value should be higher, and the pool of genotypes will be deficient for the dispersing sex as compared to the philopatric sex as it will be a mixture of two populations, residents, and immigrants. Due to the Wahlund effect, the sample should have a heterozygote deficit and a positive Fis. In general, members of the dispersing sex should, therefore, display a higher Fis than the more philopatric sex (Goudet et al., 2002). We used the most common and unbiased method (Weir and Cockerham, 1984) for the determination of the F-value.
We estimated the relatedness within male and female groups of NNTR and TATR with all the 12 microsatellites using the Queller–Goodnight index (Queller and Goodnight, 1989) incorporated in GenAlEx v 6.502 (Peakall and Smouse, 2006). This index calculates the relatedness between any two individuals by comparing the alleles shared by these individuals with the allele frequency of the group and relatedness coefficients value R ranging from −1 to 1 (Queller and Goodnight, 1989) with jackknifing over the loci to estimate the standard error. We estimated the relatedness in three categories males (M–M), females (F–F), and female–male (F–M) for the two populations. In the context of sex-biased dispersal, the dispersing sex is expected to have lower mean relatedness than the philopatric sex and between opposite-sex individuals. If there is bisexual philopatry, there will be no substantial difference in mean relatedness (Queller and Goodnight, 1989).
Spatial autocorrelation analyses
GenAlEx v 6.502 was used to compare the spatial genetic structure between the sexes referred to as spatial genetic autocorrelation (Smouse and Peakall, 1999). It uses the matrices of pairwise genetic and geographic distance to determine the spatial autocorrelation coefficient r. We first calculated the pairwise codominant genetic distance and linear genetic distance for male and female individuals separately. Geographical distances were calculated using the GPS coordinates of the samples registered in the UTM (Universal Transversal Mercator) coordinate system and translated to km using GenAlEx.
The autocorrelation coefficient r (−1 to 1) measures the genetic similarity (r > 0) or dissimilarity (r < 0) between pairs of individuals grouped in distance classes. Significant spatial structure (P < 0.05) is present when r lies outside a 95% upper and lower confidence interval, obtained by permutation with 9,999 replicates. Samples were distributed in fourteen distance classes of 5-km intervals based on the maximum geographic distance between two samples in a particular area. The maximum Euclidean distance between two sampling points was used to consider the maximum distance for dispersal within the protected area. We used the squared paired sample test and omega test to compare the heterogeneity between correlograms. If there is a sex-biased dispersal, the correlograms of males and females are expected to be distinct, and the philopatric sex should display significant positive genetic structure at shorter distances and significant negative genetic structure at longer distances (Smouse and Peakall, 1999). Spatial autocorrelation has proved to be a powerful and flexible tool to detect fine-scale genetic structure in animals, and this method does not make any assumption about the relationship between geographic and genetic distance (Banks and Peakall, 2012).
Assignment index
Program FSTAT v 2.9.4 was used to compare the assignment index of males and females in a population. This statistic was first introduced by Paetkau et al. (1995) and modified later by Favre et al. (1997) to differentiate the dispersers from residents in a population. We calculated the corrected assignment index, mAIC, and vAIC to determine the bias in the dispersal of dholes. The distribution of the AIC is centered on zero, and a positive value indicates a genotype more likely than average to occur in the sub-population in which the individual was sampled (probably resident), while a negative value indicates a potential immigrant (Goudet et al., 2002). This implies that the dispersing sex would show a lower mean of the AIC (mAIC) and a higher variance of AIC (vAIC) compared to the more philopatric sex.
Results
Genotyping and sex determination
Of the total samples collected, 84 were individually identified from TATR and 90 were identified from the NNTR population. The probability of identity for the NNTR and TATR population using the set of 12 markers was 5.98 × 10–8 and 4.52 × 10–8, while the probability of identity sibs was 8.52 × 10–4 and 6.16 × 10–4, respectively. We removed 12 genotypes from TATR and 15 from the NNTR population as recaptures. The panel calculated from three genotyping repeats had a low genotyping error rate, with a mean allelic dropout rate of 0.15 per allele per locus, a mean false allele frequency of 0.14 per allele per locus, and a null allele frequency of 0.01. There was no evidence of strong linkage disequilibrium between any pair of loci, according to the panel. However, few loci from the panel were found to be out of HW equilibrium in each population, but no loci were found to be out of HW equilibrium in all populations (Supplementary Table 1). All genotype datasets are deposited at Dryad.1
Molecular sexing confirmed 44 males and 32 females (90% success rate) in the TATR population, while 54 males and 35 females (98% success rate) in the NNTR population (Figure 1 and Table 1). The remaining individuals for which the sex was not confirmed were removed from the final dataset. The number of males is proportionately higher than females in both populations.
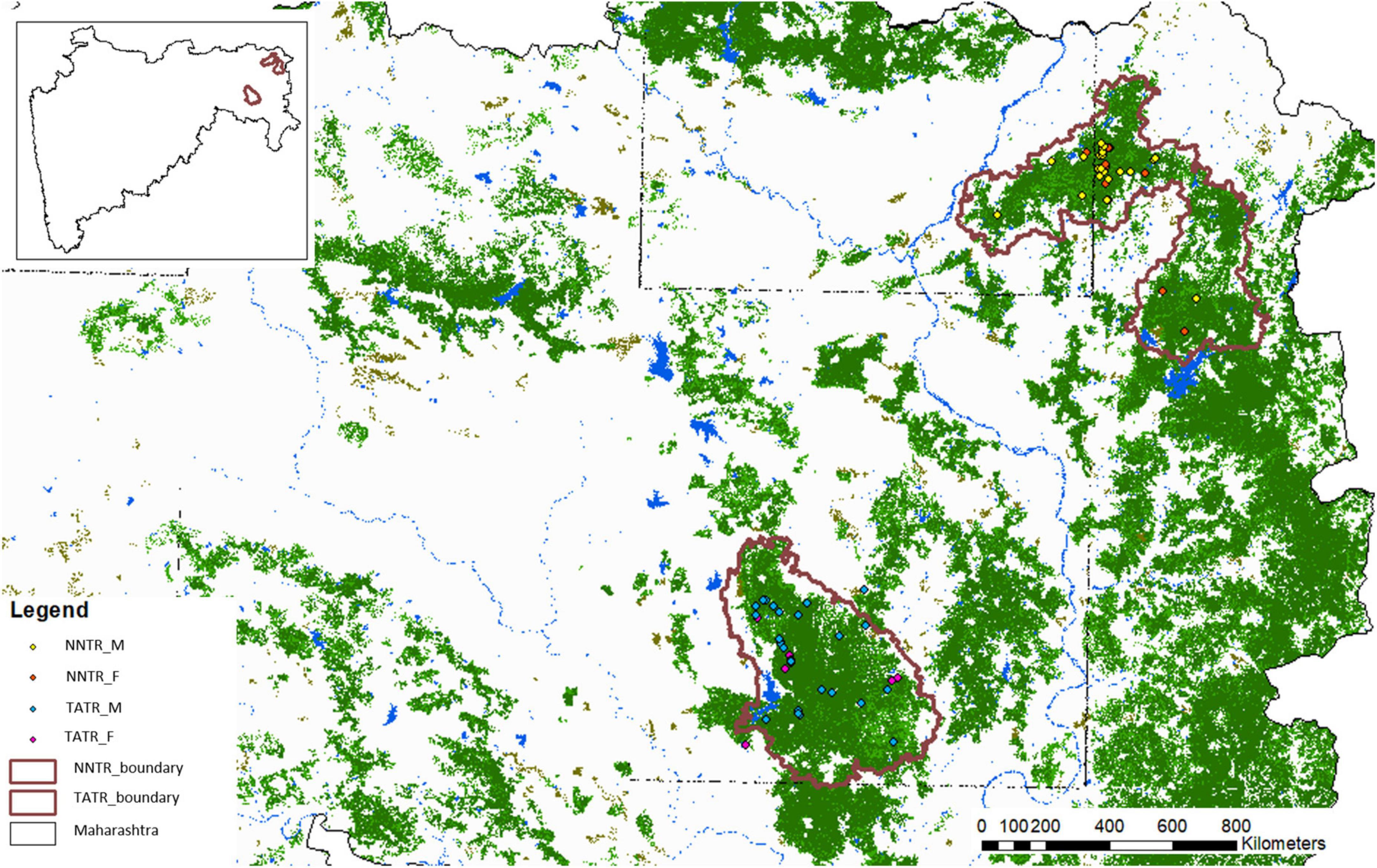
Figure 1. Study area map with locations of identified male and female individuals in Nawegaon-Nagzira Tiger Reserve (NNTR) and Tadoba-Andhari Tiger Reserve (TATR). The inset map represents the geographical location of the tiger reserves in the state of Maharashtra.
Genetic diversity
The genetic diversity of NNTR was found to be three to 12 alleles per locus with expected heterozygosity of He = 0.51 and observed heterozygosity of Ho = 0.32, while the genetic diversity of TATR was three to eight alleles per locus with expected and observed heterozygosity of He = 0.54 and Ho = 0.42, respectively. One locus was found to be out of HW equilibrium. The details of genetic diversity for males and females from each protected area are shown in Table 2.
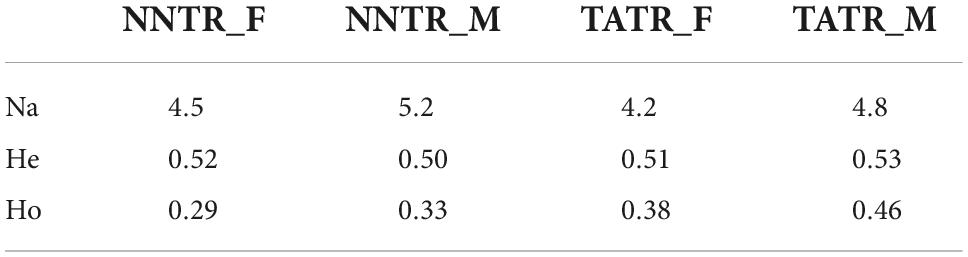
Table 2. Detailed estimates of genetic diversity (number of alleles per locus, expected (He), and observed (Ho) heterozygosity for males and females).
Fine-scale genetic structure
The structure results for both populations were found to be k = 2, suggesting that NNTR and TATR are two genetically differentiated populations (Supplementary Figure 1 and Supplementary Table 2). A minimal amount of gene flow was found between the two populations (Supplementary Table 3). The k-value obtained by running the STRUCTURE for NNTR (n = 76) and TATR (n = 84) individuals suggests k = 3 and k = 2, respectively, as per the largest log-likelihood and delta k-values. The plot for ranked average partial membership q for each cluster shows q > 0.75 for most individuals with only 10 and 12 as admixed individuals (0 < q < 0.75) for NNTR and TATR, respectively. Males and females did not show any distinct genetic clustering for both protected areas.
When STRUCTURE was run for males and females separately, the log-likelihood values for TATR_M suggest k = 4, while the log-likelihood values for TATR_F suggest k = 2. In the case of NNTR, both males and females showed the same value of log-likelihood (k = 2) (Figure 2 and Supplementary Figure 2).
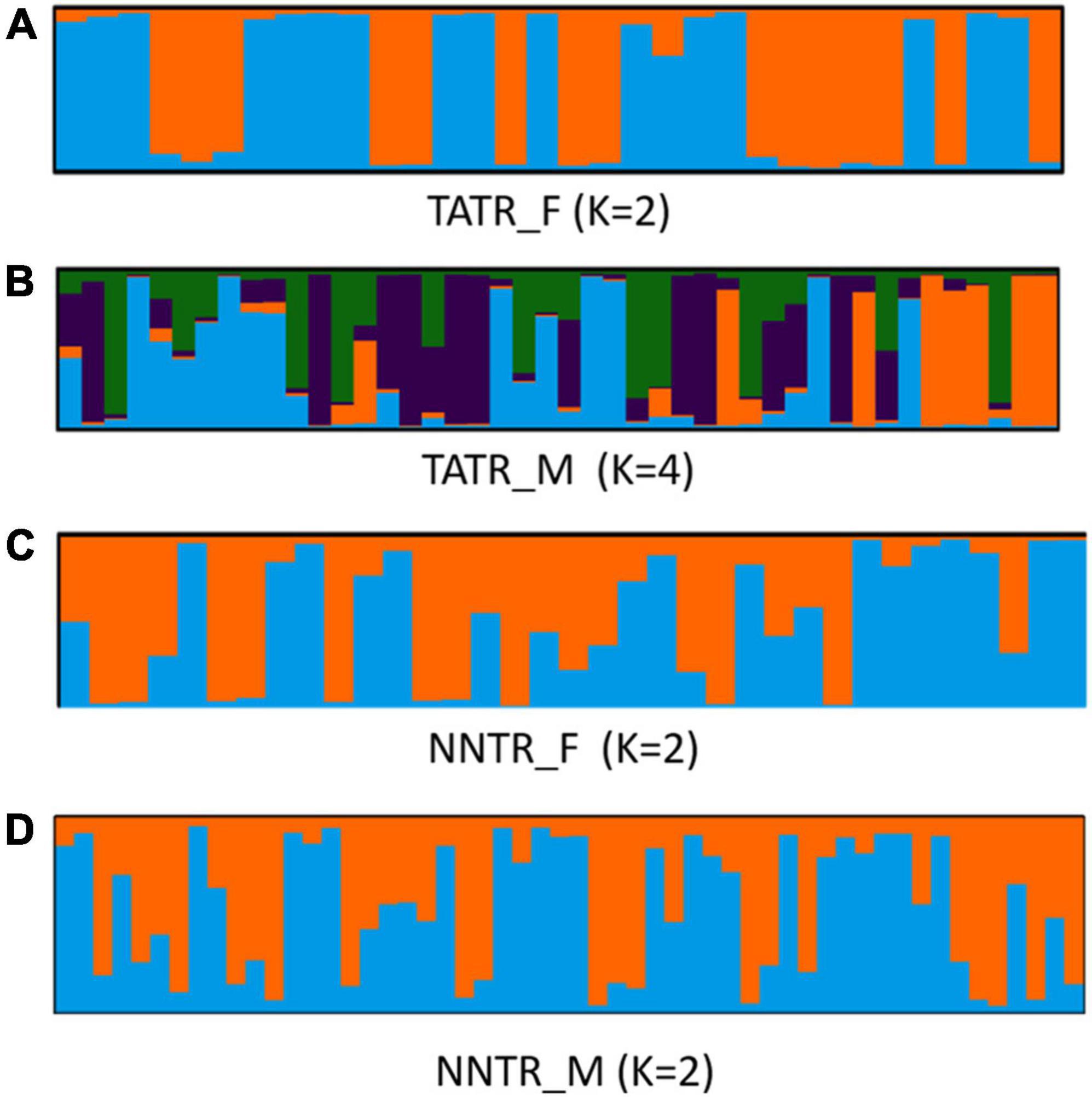
Figure 2. Bar-plot graph of the estimated membership coefficient of dhole male and female genotypes separately using STRUCTURE. The log-likelihood value confirms the number of clusters (A) TATR_F (K = 2), (B) TATR_M (K = 4), (C) NNTR_F (K = 2), and (D) NNTR_M (K = 2).
Sex-biased dispersal
The F-value for NNTR males is significantly lower than the NNTR females. Similarly, the F-value for TATR males is lower than the TATR females, but with no significant difference (Table 3). Relatedness coefficient r estimated as Queller–Goodnight mean showed no difference between the relatedness value of NNTR and TATR males and females with an insignificant p-value (Table 4) which indicates that the individuals are mostly unrelated.

Table 3. Test for the difference between sexes in the F-values within Nawegaon-Nagzira Tiger Reserve (NNTR) and Tadoba-Andhari Tiger Reserve (TATR) populations with significant values (P < 0.05).
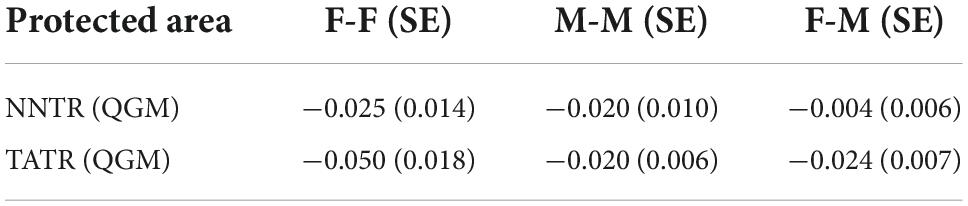
Table 4. Mean relatedness between the same-sex and opposite-sex pairs of individual dhole genotypes of Nawegaon-Nagzira Tiger Reserve (NNTR) and Tadoba-Andhari Tiger Reserve (TATR) using Queller–Goodnight index.
Nawegaon-Nagzira Tiger Reserve males for neighboring pairwise comparisons were found to be positively autocorrelated at a distance of 0–10 km, P = 0.001, while they were found to be negatively autocorrelated at 10–15 km and 35–40 km, P = 0.001 and 0.014, respectively, while for NNTR females, positive significant autocorrelation was found only at one distance class at 40–45 km, P = 0.009. The rest of the distance classes did not show any significant results for NNTR males and females (Figures 3A,B).
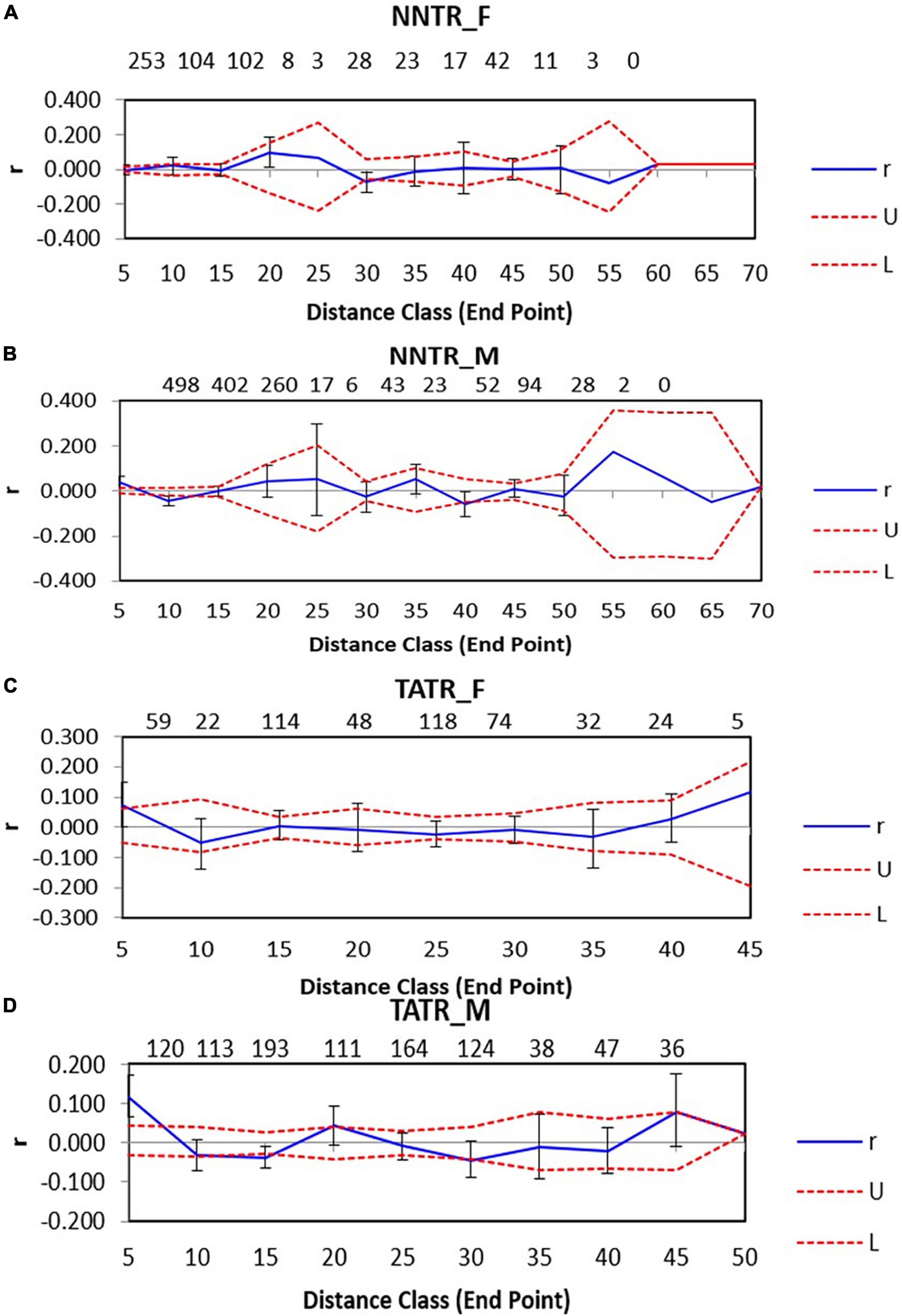
Figure 3. Spatial genetic structure correlograms for the two dhole populations Nawegaon-Nagzira Tiger Reserve (NNTR) and Tadoba-Andhari Tiger Reserve (TATR). The sample size at each distance class is provided above each correlogram. Spatial genetic structure correlograms for (A) NNTR females; (B) NNTR males; (C) TATR females; (D) TATR males.
For TATR males, the spatial correlogram was found to be significant with a positive autocorrelation at 0–10 and 20–25 km, P = 0.001 and 0.003 and negative autocorrelation at 15–20 and 30–35 km, P = 0.006 and 0.005, respectively. While for TATR females, we found no significant autocorrelation across the distance classes (Figures 3C,D). We also found a significant difference in the correlogram of males and females for both the populations [(NNTR–omega = 61.552, p-value = 0.001) and (TATR–omega = 55.44, p-value = 0.001)] (Figure 4).
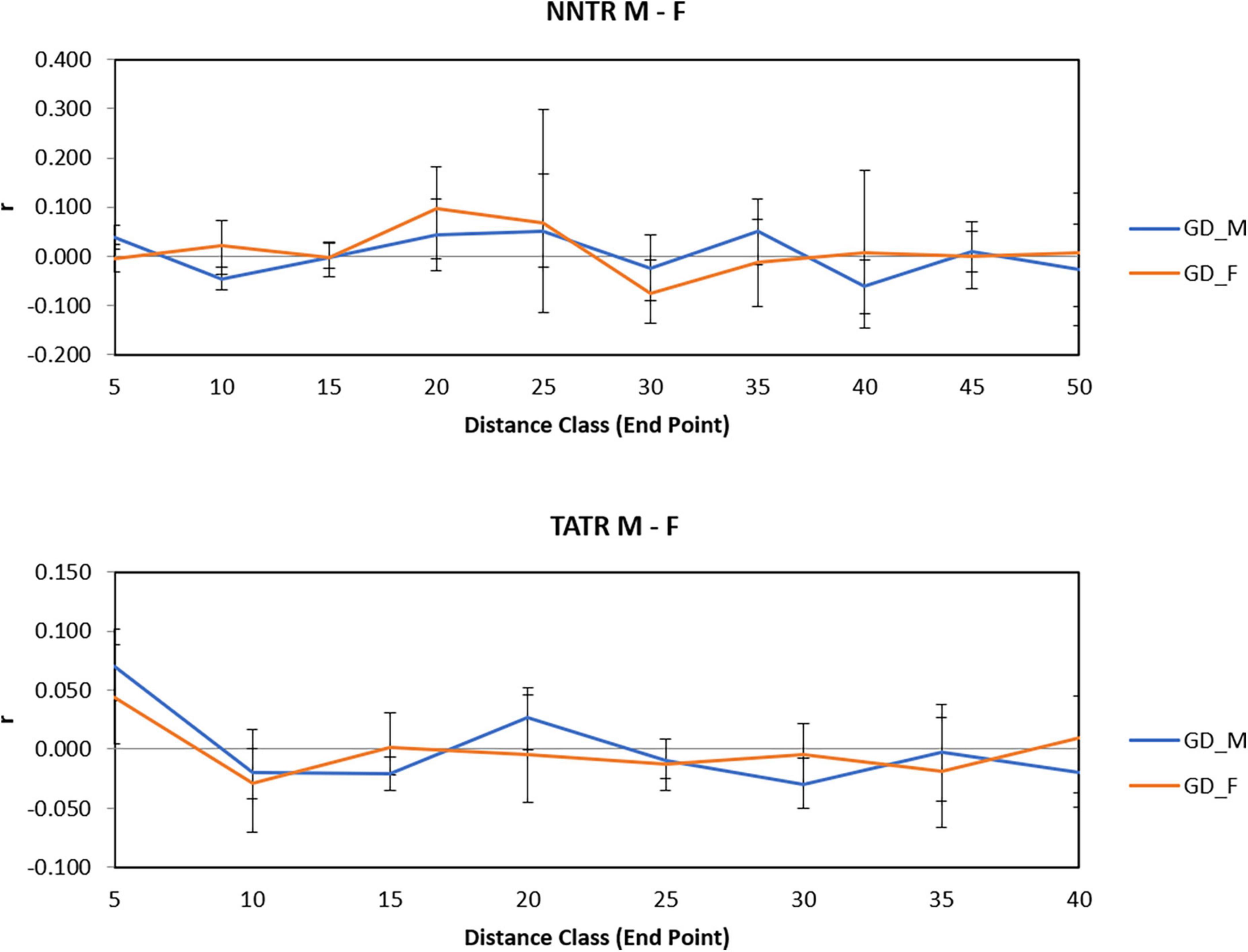
Figure 4. Spatial genetic structure correlograms for Nawegaon-Nagzira Tiger Reserve (NNTR) M and F and Tadoba-Andhari Tiger Reserve (TATR) M and F.
The assignment test conducted for NNTR showed a lower mean AIC and higher variance for females as compared to males, while in the case of TATR, females showed a higher mAIC and lower variance than males, but no significant differences were found. The t-test conducted to compare the mean and variance of males and females were also found to be non-significant (Table 5). Therefore, the assignment test could not be used to make any further conclusion regarding the dispersal bias in dholes.
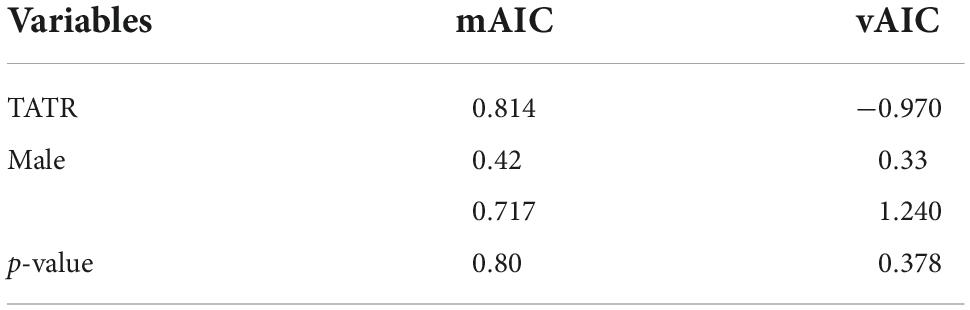
Table 5. Test for the differences between sexes in the mean (mAIC) and variance (vAIC) of the corrected assignment index with significant values (P < 0.05).
Discussion
Our results demonstrated the inter-population level variation in the sex-biased dispersal pattern of dholes using gene flow as a proxy from two genetically differentiated populations of NNTR and TATR at different tiger densities and different pack sizes. Our results suggest that males are more philopatric sex than females in TATR, whereas the NNTR population does not show any sex-biased dispersal. We have not directly tested the effect of tiger densities on the pack size of dholes, but we have used the inference from the previous study which shows the role of interspecific competition in modulating the dhole pack size which may consequently impact the dispersal decisions (Miyamoto et al., 2013; Bhandari et al., 2021). The genotyping error rate of the panel used for individual identification is less than the 20% recommended for non-invasive population level studies (Smith and Wang, 2014). The PID values for both populations indicate a statistically robust value for differentiating dhole individuals within a population. Multiple genetic analyses conducted to determine the dispersal pattern of dholes indicate two possibilities, either it is female-biased or there is no bias in dispersal. Our results suggest that males are more structured in TATR than females, indicating female dispersal (Ferreira da Silva et al., 2018), whereas males and females in NNTR showed a similar level of structure with the same number of clusters (k = 2). This indicates that males are more philopatric sex than females in TATR, while in the case of NNTR, both sexes do not show any significant difference in structure patterns which can be a possibility when both sexes disperse.
Tadoba-Andhari Tiger Reserve has sufficiently good connectivity with the adjacent Brahmapuri Forest Division, which can act as a dispersal escape, while in the case of NNTR, the connectivity to the nearest protected area is very patchy (Habib et al., 2021a). A tentative reason behind the high genetic structure in TATR males is that, due to better geographical connectivity, males disperse to greater distances, whereas females disperse to shorter distances within the park. Such dispersal pattern has been observed in African wild dogs, where males disperse less frequently but traverse longer distances, while females travel shorter distances but disperse more frequently (McNutt and Silk, 2008). A similar pattern was also observed in dholes in a previous study (Iyengar et al., 2005) where females disperse close to their natal packs while males travel longer distances, although we did not find any significant results supporting this hypothesis. Other corresponding analyses for sex-biased dispersal also revealed a significant difference between males and females of TATR, though the differences were not very profound for NNTR. We conducted four sex-biased dispersal tests based on the relatedness index, assignment values, F-values, and spatial autocorrelation between males and females in the two populations. The relatedness index shows negative values of the r coefficient among the male and female individuals of the respective populations, implying that the individuals as mainly unrelated. The lack of relatedness among males could be due to the frequent introduction of new alleles by immigrant females, which could be enough to prevent elevated levels of allele sharing resulting in related male individuals (Goudet et al., 2002). We did not find any significant difference between the dispersal patterns through the mean and variance of the assignment test also, but it gives a hint that males are more related than females, suggesting that females have a higher likelihood of being immigrants to the population sampled from TATR.
The other two sex-biased dispersal tests conducted using F-value and spatial autocorrelation for dholes showed a significant difference, indicating the biased nature of dispersal. We found a lower F-value and a positive spatial autocorrelation at a shorter distance for males of both populations. This suggests that the males are more philopatric sex than females due to the Wahlund effect resulting in higher homozygosity in males. Both results indicate that males are more philopatric than females. The spatial autocorrelation results show that males are generally more similar at shorter distances, suggesting that males could be more philopatric than females. We did not use the Fst index as indicated by Goudet et al. (2002) because dholes are considered to have a short dispersal distance with a home range of 58.67 sq. km (±4.08) (Habib et al., 2021b). Contrary to their African counterparts, we did not find any significant inter-dispersal events between these two populations.
Dhole is a socially monogamous canid that breeds cooperatively, often with multiple breeding females in a pack (Fox, 1984; Johnsingh, 1985). The pack is composed of an alpha-breeding pair and shows a dominant hierarchy (Serfass et al., 1998). Therefore, one major reason for highly structured male-biased packs in TATR can be the intra-specific competition due to which females disperse more, while males remain in the packs as there are higher chances of females getting accepted in a new pack. Another major reason for such a pattern can be the higher tiger density in TATR giving rise to a high turnover rate. Contrary to this in NNTR, there is no interspecific competition pressure due to lower tiger density and the larger pack size provides similar opportunities to both males and females. Differences in the turnover rate of any species can influence the genetic structure of the population (Williams et al., 2003), for example, higher competition in TATR leads to a larger turnover of male members of the pack, resulting in a more structured male population (Groom et al., 2017). The tiger population modulates the pack size of dholes in both the tiger reserves with larger pack size in NNTR due to an extremely low number of tigers and smaller pack size in TATR for better avoidance strategy (McNutt, 1996; Bhandari et al., 2021) in high-density tiger areas. This can be due to the reason that tiger presence is solely correlated with prey-rich sites, while the presence probability of dhole was a trade-off between prey availability and active spatial avoidance of tigers (Steinmetz et al., 2013).
The male-biased sex ratio in both populations indicates the possibility of female-biased dispersal, which is further supported by previous ecological studies (Johnsingh, 1982; Venkataraman, 1998). The probability of female-biased dispersal is suggested by the male-biased sex ratio in both populations, which is further confirmed by earlier ecological studies (Johnsingh, 1982; Venkataraman, 1998). According to the local resource enhancement (LRE) paradigm, research on species that breed cooperatively in hierarchies also shows that the pack tends to favor the more helpful sex (Silk and Brown, 2008). The present genetic study revealed a differential dispersal pattern in the case of TATR, but due to larger pack size and less competition in NNTR, dispersal bias may not be evident. Despite being non-significant, the sex-biased dispersal test points to the female-biased nature of dispersal in the case of NNTR as well.
Combining the knowledge of ecology and the findings obtained during this study, we suggest that dispersal is a complex phenomenon governed by the local dynamics, competition, and landscape connectivity of a population. This study adds a piece of empirical evidence to the aspect of inter-population variability in dispersal studies. Though African wild dogs being the social canid with a rigid structure of dominant hierarchies and sharing a sister lineage with dholes often found to have female-biased dispersal, it cannot be generalized in the case of dholes owing to various ecological, environmental, and geographical local factors. We have used the biparental markers to delineate the dispersal pattern by comparing the genetic diversity of males and females after dispersal and before reproduction. This approach is consistent with our sampling strategy for just one generation and provides the advantage of using the genetic diversity of both sexes (Prugnolle and De Mee^us, 2002).
Most solitary carnivores are reported to have a male-biased dispersal pattern (Biek et al., 2006; Janečka et al., 2007; Gour et al., 2013) and are generally characterized by uniparental care. As found through our analysis, the biased dispersal can be because of the biparental care and pack-living nature of socially structured dholes (Stockley and Hobson, 2016). Our study has paved the way to develop a fundamental understanding of future research on dholes’ dispersal. This is the first and foremost effort to define the dispersal of a lesser-studied social pack-living endangered canid that thrives in Indian forests with varying pack sizes and interspecific competitions. This has contributed to the idea that, for pack-living social species like the dhole, dispersion decisions which are typically influenced by the search for a mate and territory can be either female-biased or completely neutral. Future investigations in this field are needed to delineate a detailed pattern of dhole dispersal with broader coverage at the landscape level, the inclusion of interspecific competition, pack size, and other landscape variables as predictor variables using a multi-disciplinary approach, that is, camera trapping and telemetry.
Conclusion
Understanding the dispersal pattern of this endangered monophyletic species is critical in developing a better framework for the long-term persistence of populations in this anthropogenic era. Dispersal can be a combined outcome of several ecological and geographical variables and an individual’s life-history decisions. Our results validate that the Asiatic wild dogs are one of the genera among canids, where dispersal decision could be affected by pack size, top-predator density, and geographical connectivity. This will further help in developing focused conservation strategies for short-ranging species with lesser-known ecology. We support the fact that a comprehensive knowledge of dispersal patterns is required to develop any conservation management plan. Dholes have short dispersal ranges, and a lack of sex-biased dispersal can increase the risk of inbreeding within the packs and population. Further research including the different geographical and anthropogenic variables can help in deriving the requirements for sex-specific corridors for a pack-living species.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was reviewed and approved by Maharashtra State Forest Department (Permit No. 09/2016). This work did not require any approval from the ethical committee due to its non-invasive nature and does not involve any animal directly.
Author contributions
BH and ShM: conceptualization. ShM: methodology, software, formal analysis, investigation, data curation, writing original draft preparation, and visualization. BH, ShM, and SaM: validation. BH: resources, project administration, and funding acquisition. BH, ShM, SaM, PN, and PG: writing – review and editing. BH, SaM, and PN: supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Maharashtra State Forest Department. ShM was supported by a CSIR-SRF grant from the Government of India.
Acknowledgments
We are grateful for the support provided by the CWLW Maharashtra as well as the Field Directors and Deputy Directors of respective tiger reserves. We acknowledged A. Madhanraj and Ketaki Dadhe for their assistance in laboratory work. We are grateful to DWII, Dean and Research Coordinator for their encouragement and support. We also thank all the members of BH Lab.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.993851/full#supplementary-material
Footnotes
References
Andrew, R. L., Bernatchez, L., Bonin, A., Buerkle, C. A., Carstens, B. C., Emerson, B. C., et al. (2013). A road map for molecular ecology. Mol. Ecol. 22, 2605–2626. doi: 10.1111/mec.12319
Baguette, M., Blanchet, S., Legrand, D., Stevens, V. M., and Turlure, C. (2013). Individual dispersal, landscape connectivity and ecological networks. Biol. Rev. 88, 310–326. doi: 10.1111/brv.12000
Ball, M. C., Pither, R., Manseau, M., Clark, J., Petersen, S. D., Kingston, S., et al. (2007). Characterization of target nuclear DNA from faeces reduces technical issues associated with the assumptions of low-quality and quantity template. Conserv. Genet. 8, 577–586. doi: 10.1007/s10592-006-9193-y
Banks, S. C., and Peakall, R. O. D. (2012). Genetic spatial autocorrelation can readily detect sex-biased dispersal. Mol. Ecol. 21, 2092–2105. doi: 10.1111/j.1365-294X.2012.05485.x
Bhandari, A., Ghaskadbi, P., Nigam, P., and Habib, B. (2021). Dhole pack size variation: Assessing the effect of prey availability and apex predator. Ecol. Evol. 11, 4774–4785. doi: 10.1002/ece3.7380
Biek, R., Akamine, N., Schwartz, M. K., Ruth, T. K., Murphy, K. M., and Poss, M. (2006). Genetic consequences of sex-biased dispersal in a solitary carnivore: Yellowstone cougars. Biol. Lett. 2, 312–315. doi: 10.1098/rsbl.2005.0437
Bowler, D. E., and Benton, T. G. (2005). Causes and consequences of animal dispersal strategies: Relating individual behaviour to spatial dynamics. Biol. Rev. 80, 205–225. doi: 10.1017/S1464793104006645
Burgess, S. C., Baskett, M. L., Grosberg, R. K., Morgan, S. G., and Strathmann, R. R. (2016). When is dispersal for dispersal? Unifying marine and terrestrial perspectives. Biol. Rev. 91, 867–882. doi: 10.1111/brv.12198
Courchamp, F., and Macdonald, D. W. (2001). “Crucial importance of pack size in the African wild dog Lycaon pictus,” in Animal conservation forum, Vol. 4, (Cambridge: Cambridge University Press), 169–174. doi: 10.1017/S1367943001001196
Cozzi, G., Behr, D. M., Webster, H. S., Claase, M., Bryce, C. M., Modise, B., et al. (2020). African wild dog dispersal and implications for management. J. Wildlife Manag. 84, 614–621. doi: 10.1002/jwmg.21841
Dieckmann, U., O’Hara, B., and Weisser, W. (1999). The evolutionary ecology of dispersal. Trends Ecol. Evol. 14, 88–90. doi: 10.1016/S0169-5347(98)01571-7
Earl, D. A., and VonHoldt, B. M. (2012). STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier, L., Laval, G., and Schneider, S. (2005). Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinform. 1, 47–50. doi: 10.1177/117693430500100003
Favre, L., Balloux, F., Goudet, J., and Perrin, N. (1997). Female-biased dispersal in the monogamous mammal Crocidura russula: Evidence from field data and microsatellite patterns. Proc. R. Soc. Lond. Ser. B Biol. Sci. 264, 127–132. doi: 10.1098/rspb.1997.0019
Ferreira da Silva, M. J., Kopp, G. H., Casanova, C., Godinho, R., Minhos, T., Sa, R., et al. (2018). Disrupted dispersal and its genetic consequences: Comparing protected and threatened baboon populations (Papio papio) in West Africa. PLoS One 13:e0194189. doi: 10.1371/journal.pone.0194189
Fox, M. W. (1984). The whistling hunters: Field studies of the Asiatic wild dog (Cuon alpinus). Albany, NY: SUNY Press.
Gandon, S. (1999). Kin competition, the cost of inbreeding and the evolution of dispersal. J. Theor. Biol. 200, 345–364. doi: 10.1006/jtbi.1999.0994
Goudet, J., Perrin, N., and Waser, P. (2002). Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Mol. Ecol. 11, 1103–1114. doi: 10.1046/j.1365-294X.2002.01496.x
Gour, D. S., Bhagavatula, J., Bhavanishankar, M., Reddy, P. A., Gupta, J. A., Sarkar, M. S., et al. (2013). Philopatry and dispersal patterns in tiger (Panthera tigris). PLoS One 8:e66956. doi: 10.1371/journal.pone.0066956
Groom, R. J., Lannas, K., and Jackson, C. R. (2017). The impact of lions on the demography and ecology of endangered African wild dogs. Anim. Conserv. 20, 382–390. doi: 10.1111/acv.12328
Habib, B., Nigam, P., Mondal, I., Hussain, Z., Ghaskadbi, P., Govekar, R., et al. (2021a). Telemetry based tiger corridors of Vidarbha Landscape, Maharashtra, India. Technical Report. Dehradun: Wildlife Institute of India, 39.
Habib, B., Ghaskadbi, P., Khan, S., Hussain, Z., and Nigam, P. (2021b). Not a cakewalk: Insights into movement of large carnivores in human-dominated landscapes in India. Ecol. Evol. 11, 1653–1666. doi: 10.1002/ece3.7156
Iyengar, A., Babu, V. N., Hedges, S., Venkataraman, A. B., Maclean, N., and Morin, P. A. (2005). Phylogeography, genetic structure, and diversity in the dhole (Cuon alpinus). Mol. Ecol. 14, 2281–2297. doi: 10.1111/j.1365-294X.2005.02582.x
Janečka, J. E., Blankenship, T. L., Hirth, D. H., William Kilpatrick, C., Tewes, M. E., and Grassman, L. I. Jr. (2007). Evidence for male-biased dispersal in bobcats Lynx rufus using relatedness analysis. Wildlife Biol. 13, 38–47. doi: 10.2981/0909-6396(2007)13[38:EFMDIB]2.0.CO;2
Johnsingh, A. J. T. (1982). Reproductive and social behaviour of the dhole, Cuon alpinus (Canidae). J. Zool. 198, 443–463. doi: 10.1111/jzo.1982.198.4.443
Johnsingh, A. J. T. (1985). Distribution and status of dhole Cuon alpinus Pallas, 1811 in South Asia. Mammalia 49, 203–208. doi: 10.1515/mamm.1985.49.2.203
Kalinowski, S. T., Taper, M. L., and Marshall, T. C. (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x
Kamler, J. F., Songsasen, N., Jenks, K., Srivathsa, A., Sheng, L., and Kunkel, K. (2015). Cuon alpinus. The IUCN Red List of Threatened Species 2015: e.T5953A72477893. doi: 10.2305/IUCN.UK.2015-4.RLTS.T5953A72477893.en
Keiter, D. A., Cunningham, F. L., Rhodes, O. E. Jr., Irwin, B. J., and Beasley, J. C. (2016). Optimization of scat detection methods for a social ungulate, the wild pig, and experimental evaluation of factors affecting detection of scat. PLoS One 11:e0155615. doi: 10.1371/journal.pone.0155615
Kraemer, P., and Gerlach, G. (2017). Demerelate: Calculating interindividual relatedness for kinship analysis based on codominant diploid genetic markers using R. Mol. Ecol. Resour. 17, 1371–1377. doi: 10.1111/1755-0998.12666
Lawson Handley, L. J., and Perrin, N. (2007). Advances in our understanding of mammalian sex-biased dispersal. Mol. Ecol. 16, 1559–1578. doi: 10.1111/j.1365-294X.2006.03152.x
Lowe, W. H., and Allendorf, F. W. (2010). What can genetics tell us about population connectivity? Mol. Ecol. 19, 3038–3051. doi: 10.1111/j.1365-294X.2010.04688.x
McNutt, J. W. (1996). Sex-biased dispersal in African wild dogs, Lycaon pictus. Anim. Behav. 52, 1067–1077. doi: 10.1006/anbe.1996.0254
McNutt, J. W., and Silk, J. B. (2008). Pup production, sex ratios, and survivorship in African wild dogs, Lycaon pictus. Behav. Ecol. Sociobiol. 62, 1061–1067. doi: 10.1007/s00265-007-0533-9
Miyamoto, M. M., Allen, J. M., Gogarten, J. F., and Chapman, C. A. (2013). Microsatellite DNA suggests that group size affects sex-biased dispersal patterns in Red Colobus monkeys. Am. J. Primatol. 75, 478–490. doi: 10.1002/ajp.22124
Modi, S., Habib, B., Ghaskadbi, P., Nigam, P., and Mondol, S. (2019). Standardization and validation of a panel of cross-species microsatellites to individually identify the Asiatic wild dog (Cuon alpinus). PeerJ 7:e7453. doi: 10.7717/peerj.7453
Modi, S., Mondol, S., Ghaskadbi, P., Hussain, Z., Nigam, P., and Habib, B. (2018). Noninvasive DNA-based species and sex identification of Asiatic wild dog. J. Genet. 97, 1457–1461. doi: 10.1007/s12041-018-1017-6
Modi, S., Mondol, S., Nigam, P., and Habib, B. (2021). Genetic analyses reveal demographic decline and population differentiation in an endangered social carnivore, Asiatic wild dog. Sci. Rep. 11:16371. doi: 10.1038/s41598-021-95918-3
Paetkau, D., Calvert, W., Stirling, I., and Strobeck, C. (1995). Microsatellite analysis of population structure in Canadian polar bears. Mol. Ecol. 4, 347–354. doi: 10.1111/j.1365-294X.1995.tb00227.x
Palomares, F., and Caro, T. M. (1999). Interspecific killing among mammalian carnivores. Am. Nat. 153, 492–508. doi: 10.1086/303189
Pavlova, A., Beheregaray, L. B., Coleman, R., Gilligan, D., Harrisson, K. A., Ingram, B. A., et al. (2017). Severe consequences of habitat fragmentation on genetic diversity of an endangered Australian freshwater fish: A call for assisted gene flow. Evol. Appl. 10, 531–550. doi: 10.1111/eva.12484
Peakall, R. O. D., and Smouse, P. E. (2006). GENALEX 6: Genetic analysis in excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295. doi: 10.1111/j.1471-8286.2005.01155.x
Perrin, N., and Mazalov, V. (2000). Local competition, inbreeding, and the evolution of sex-biased dispersal. Am. Nat. 155, 116–127. doi: 10.1086/303296
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Prugnolle, F., and De Mee^us, T. (2002). Inferring sex-biased dispersal from population genetic tools: A review. Heredity 88, 161–165. doi: 10.1038/sj.hdy.6800060
Pusey, A. E. (1987). Sex-biased dispersal and inbreeding avoidance in birds and mammals. Trends Ecol. Evol. 2, 295–299. doi: 10.1016/0169-5347(87)90081-4
Queller, D. C., and Goodnight, K. F. (1989). Estimating relatedness using genetic markers. Evolution 43, 258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x
Radespiel, U., and Bruford, M. W. (2014). Fragmentation genetics of rainforest animals: Insights from recent studies. Conserv. Genet. 15, 245–260. doi: 10.1007/s10592-013-0550-3
Randall, D. A., Pollinger, J. P., Wayne, R. K., Tallents, L. A., Johnson, P. J., and Macdonald, D. W. (2007). Inbreeding is reduced by female-biased dispersal and mating behavior in Ethiopian wolves. Behav. Ecol. 18, 579–589. doi: 10.1093/beheco/arm010
R Core Team (2018). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. Available online at: http://www.R-project.org/
Rodrigues, R. G., Srivathsa, A., and Vasudev, D. (2022). Dog in the matrix: Envisioning countrywide connectivity conservation for an endangered carnivore. J. Appl. Ecol. 59, 223–237. doi: 10.1111/1365-2664.14048
Ross, K. G. (2001). Molecular ecology of social behaviour: Analyses of breeding systems and genetic structure. Mol. Ecol. 10, 265–284. doi: 10.1046/j.1365-294x.2001.01191.x
Serfass, T. L., Brooks, R. P., Novak, J. M., Johns, P. E., and Rhodes, O. E. Jr. (1998). Genetic variation among populations of river otters in North America: Considerations for reintroduction projects. J. Mammal. 79, 736–746. doi: 10.2307/1383084
Silk, J. B., and Brown, G. R. (2008). Local resource competition and local resource enhancement shape primate birth sex ratios. Proc. R. Soc. B Biol. Sci. 275, 1761–1765. doi: 10.1098/rspb.2008.0340
Smith, O., and Wang, J. (2014). When can noninvasive samples provide sufficient information in conservation genetics studies? Mol. Ecol. Resour. 14, 1011–1023. doi: 10.1111/1755-0998.12250
Smouse, P. E., and Peakall, R. O. D. (1999). Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 82, 561–573. doi: 10.1038/sj.hdy.6885180
Solberg, K. H., Bellemain, E., Drageset, O. M., Taberlet, P., and Swenson, J. E. (2006). An evaluation of field and non-invasive genetic methods to estimate brown bear (Ursus arctos) population size. Biol. Conserv. 128, 158–168. doi: 10.1016/j.biocon.2005.09.025
Steinmetz, R., Seuaturien, N., and Chutipong, W. (2013). Tigers, leopards, and dholes in a half-empty forest: Assessing species interactions in a guild of threatened carnivores. Biol. Conserv. 163, 68–78. doi: 10.1016/j.biocon.2012.12.016
Sterck, E. H., Watts, D. P., and Van Schaik, C. P. (1997). The evolution of female social relationships in nonhuman primates. Behav. Ecol. Sociobiol. 41, 291–309. doi: 10.1007/s002650050390
Stockley, P., and Hobson, L. (2016). Paternal care and litter size coevolution in mammals. Proc. R. Soc. B Biol. Sci. 283:20160140. doi: 10.1098/rspb.2016.0140
Szulkin, M., and Sheldon, B. C. (2008). Dispersal as a means of inbreeding avoidance in a wild bird population. Proc. R. Soc. B Biol. Sci. 275, 703–711. doi: 10.1098/rspb.2007.0989
Valière, N. (2002). GIMLET: A computer program for analysing genetic individual identification data. Mol. Ecol. Notes 2, 377–379. doi: 10.1046/j.1471-8286.2002.00228.x-i2
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P., and Shipley, P. (2004). MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
van Overveld, T., Careau, V., Adriaensen, F., and Matthysen, E. (2014). Seasonal and sex-specific correlations between dispersal and exploratory behaviour in the great tit. Oecologia 174, 109–120. doi: 10.1007/s00442-013-2762-0
Venkataraman, A. B. (1998). Male-biased adult sex ratios and their significance for cooperative breeding in dhole, Cuon alpinus, packs. Ethology 104, 671–684. doi: 10.1111/j.1439-0310.1998.tb00101.x
Waits, L. P., Luikart, G., and Taberlet, P. (2001). Estimating the probability of identity among genotypes in natural populations: Cautions and guidelines. Mol. Ecol. 10, 249–256. doi: 10.1046/j.1365-294X.2001.01185.x
Weir, B. S., and Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x
Williams, C. L., Blejwas, K., Johnston, J. J., and Jaeger, M. M. (2003). Temporal genetic variation in a coyote (Canis latrans) population experiencing high turnover. J. Mammal. 84, 177–184. doi: 10.1644/1545-1542(2003)084<0177:TGVIAC>2.0.CO;2
Wilson, G. A., and Rannala, B. (2003). Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163, 1177–1191. doi: 10.1093/genetics/163.3.1177
Keywords: sex-biased dispersal, tiger density, dhole, conservation genetics, predator interaction
Citation: Modi S, Mondol S, Ghaskadbi P, Nigam P and Habib B (2022) Genetic evidence of differential dispersal pattern in the Asiatic wild dog: Comparing two populations with different pack sizes. Front. Ecol. Evol. 10:993851. doi: 10.3389/fevo.2022.993851
Received: 14 July 2022; Accepted: 14 November 2022;
Published: 13 December 2022.
Edited by:
Izeni Pires Farias, Federal University of Amazonas, BrazilReviewed by:
Kanchan Thapa, World Wildlife Fund Nepal, NepalThomas Clifford Wood, George Mason University, United States
Copyright © 2022 Modi, Mondol, Ghaskadbi, Nigam and Habib. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bilal Habib, bh@wii.gov.in
 Shrushti Modi
Shrushti Modi Samrat Mondol
Samrat Mondol Pallavi Ghaskadbi
Pallavi Ghaskadbi  Parag Nigam
Parag Nigam Bilal Habib
Bilal Habib