- State Key Laboratory of Biocontrol, Institute of Aquatic Economic Animals and Guangdong Province Key Laboratory for Aquatic Economic Animals, Guangdong Provincial Engineering Technology Research Center of Healthy Breeding in Important Economic Fish, School of Life Sciences, Sun Yat-sen University, Guangzhou, China
Background: Myostatin (Mstn), a member of the TGF-β superfamily, is a negative regulator of skeletal muscle mass in mammals. Precise regulation of Mstn expression is important for somite growth in fish. MicroRNA (miRNA), a type of small non-coding RNA, regulates gene expression at the post-transcriptional level and participates in various physiological functions. A growing amount of evidence has emphasized the importance of miRNA in the development of skeletal muscle.
Aims: This study aims to study how miRNAs regulate myostatin b (mstnb) post-transcriptionally in tilapia.
Methods/Results: Mstnb 3′ UTR sequences were obtained, and the results of tissue distribution showed that mstnb was expressed in several tissues, including brain, white muscle, gut, and adipose tissue. A total of 1,992 miRNAs were predicted to target mstnb in tilapia using bioinformatics, and a dual-luciferase reporter experiment confirmed that miR-181a/b-5p, miR-30-3p, miR-200a, and miR-27a were the target miRNAs of mstnb. Mutagenesis of the miR-181b-5p binding sites of mstnb significantly increased the luciferase signal compared to the wild-type mstnb. In tilapia primary muscle cells, overexpression of miR-181b-5p led to the downregulation of MSTNb expression, and the inhibitory effect of MSTNb on the downstream genes was dismissed, while inhibition of miR-181b-5p could reverse these phenomena.
Conclusion: Taken together, our results suggested that miR-181b-5p could promote the growth of skeletal muscle by decreasing the MSTNb protein level in tilapia.
Introduction
Genetic Improvement of Farmed Tilapia (GIFT), a freshwater fish with a fast growth rate and high disease resistance, is a popular aquaculture fish worldwide that provides premium protein for people. Since the major edible part of the fish is skeletal muscle, the fish growth performance is mainly determined by the development of skeletal muscle. Skeletal muscle development is an accurate process that is regulated by positive factors, including muscle-specific myogenic regulatory transcription factors (MRFs) (1) and negative factors such as myostatin (Mstn).
Myostatin, a member of the transforming growth factor β (TGFβ) superfamily, is regarded as a specific muscle negative regulatory factor (2). Mstn, a secretory protein in skeletal muscle, is composed of 376 amino acids, including a signal peptide, an N-terminal precursor peptide, and a C-terminal mature peptide that contains nine conserved cysteine amino acids (3). In mammals, mstn is specifically expressed in skeletal muscle, while mstn is widely distributed in teleosts. In addition, mstn has several different types in teleosts as a result of gene duplication. According to the genome listed in the NCBI, for example, mstn-1 (or mstnb) and mstn-2 (or mstna) exist in Nile tilapia. Tissue distribution revealed that mstnb is mainly expressed in the brain, eye, gill, gut, and skeletal muscle in Nile tilapia (4). Mstn can inhibit the growth of skeletal muscle in mammals, but its functions in teleosts are not clear. In tilapia, scientists reported that prolonged fasting reduced the mRNA level of mstn, but short-term fasting elevated the mRNA level (5, 6). Moreover, the proliferation was inhibited and differentiation was consequently activated after MSTN-1 incubation of the myosatellite cells in rainbow trout (7). Regardless, Mstn is an important factor for skeletal muscle in teleosts.
Due to the strong inhibitory effect on muscle growth in mammals, it is particularly important to regulate the expression of Mstn. On the one hand, Mstn can be tightly regulated at the transcriptional level. E-Box sequence motifs, the canonical binding site for the basic Helix-Loop-Helix (bHLH) transcription factors (MyoD, Myogenic Differentiation Antigen; Myf5, Myogenic factor 5; and MyoG, Myogenin), were found in the mstn promoter (8, 9). The putative myocyte enhancer factor 2 (mef2) transcription factors binding motifs were also observed in the mstn promoter (9–12), and they were shown to increase Mstn expression in myoblasts (11). On the other hand, Mstn can be regulated at the post-transcriptional level. MiRNAs, a type of short non-coding RNA, inhibit translation or degrade the mRNA by binding to the 3′ UTR of targeted mRNAs (13). MiRNAs take part in numerous developmental processes, including the development of skeletal muscle (14, 15). Several muscle-specific miRNAs, including miR-1, miR-133a, miR-133b, and miR-206, were identified and shown to regulate myogenesis in mammals (16). For example, miR-1 and miR-206 affected muscularity by targeting mstn in Texel Sheep due to a mutation in the 3′ UTR (17). There is a complex regulatory network between miRNAs and genes; one gene can be regulated by several miRNAs and one miRNA can regulate multiple genes (18). In mammals, miR-27 was reported to regulate mstn expression by directly targeting the 3′ UTR (19–22). For example, MSTN could inhibit its own expression by upregulating miR-27 expression through a smad3-dependent mechanism (21). In teleosts, only miR-181a-5p was reported to target the mstn 3′ UTR in Siniperca chuatsi (23). MiRNAs regulating the expression of Mstn post-transcription levels have attracted more attention in recent years. However, it is unclear whether miRNA regulates Mstn in tilapia.
In our previous study, a deep sequencing of the Nile tilapia miRNA transcriptome was conducted in our lab (24). In this study, the candidate miRNAs that target mstn were predicted based on the miRNA transcriptome database. We screened the miRNAs that targeted mstn using the dual-luciferase reporter system and verified the regulation of miRNA on mstn in tilapia primary muscle cells. The objective of this study was to find miRNAs that target mstn and regulate the growth of tilapia. Clarifying the regulatory mechanism of mstn using miRNA for skeletal muscle growth would help deepen the understanding of tilapia growth. In addition, it is a new paradigm to study miRNA in fish with economic value. This could increase economic benefits and make an important contribution to the aquaculture industry.
Materials and Methods
Experimental Fish and Tissue Sample Preparation
Tilapia were obtained from the local farm of Guangdong Tilapia Breeding Farm. They were maintained in a water circulation system with water temperature at 28°C under a 12/12 h light/dark photoperiod. The fish were fed to satiety daily with commercial extruded feed (Tongwei, Foshan, China). The time of domestication was longer than 1 week. They were narcotized with eugenol before decollating. Skeletal muscle samples were collected from fish weighting 6–8 g.
Prediction of mstnb-Binding miRNAs
First, the sequences of the mstnb and mstna 3′ UTR were obtained using PCR with KOD neo plus (TOYOBO, Osaka, Japan). To predict miRNAs that potentially bind to mstnb, a tilapia miRNA transcriptome was conducted (data not shown) and the PITA targets (http://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html) were queried (24).
Luciferase Assay
A recombined psiCHECK2 vector (Promega, Madison, USA) containing the mstnb 3′ UTR downstream of the stop codon of the Ranilla luciferase gene was constructed, and the firefly luciferase was used as a reference gene. The mutant mstnb 3′-UTR reporters were created using site-directed mutagenesis at the binding sites of the predicted miR-181b-5p, and primers were designed using Primer X (http://www.bioinformatics.org/primerx/). These reporters and miRNA mimics (synthesized by GenePharma, Shanghai, China) were co-transfected into HEK 293T cells, and the relative luciferase activity was detected using the Luciferase Assay Systems kit (Promega, USA) according to the manufacturer's protocol. All primers are listed in Table S1.
Tissue Distribution of mstnb/a mRNA and miR-181b-5p in Tilapia
For miR-181b-5p cloning, the specific primers were designed according to the miRNA transcriptome, and the sequence of miR-181b-5p was further verified. For tissue distribution, total RNA was extracted from the tissue samples of the telencephalon, diencephalon, cerebellum, medulla oblongata, spinal cord, hypothalamus, pituitary, gill, heart, liver, spleen, stomach, foregut, midgut, hindgut, adipose tissue, red muscle, white muscle, testis, and kidney of three adult male tilapia (BW 150–180 g). All samples were snap-frozen in liquid nitrogen once removed, followed by storage at −80°C until RNA extraction. After RNA extraction and reverse transcription, the tissue distribution of mstnb/a and miR-181b-5p was assayed using real-time PCR.
Real-Time PCR
Total RNA from each well (n = 3–4) was extracted from primary muscle cells using Trizol reagent (Invitrogen, Carlsbad, CA, USA), and the RNA concentration was determined using a Nanodrop200C spectrophotometer (Thermo Scientific, Waltham, USA). A total of 1 μg of total RNA was reverse-transcribed into cDNA with the M-MLV Reverse Transcriptase (Life Technology, Carlsbad, USA), and the PCR reaction was amplified with the Thunderbird SYBR Green qPCR Mix (TOYOBO, Japan) according to the manufacturer's protocol. For mRNA quantitative analysis, β-actin was detected as the internal normalization control. Specifically, miRNA stem-loop primers were used in reverse transcription and U6 snoRNA was used as the internal control. Gene expression was normalized against the expression of the control using the comparative Ct method (25). Each experiment was repeated three times independently.
Preparation of the Polyclonal Antibody Against Recombinant MSTNb
First, the sequence of the tilapia mstnb ORF without the signal peptide was subcloned into the pET-32a vector. The recombinant expression vector pET-32a-rMSTNb was transformed into Escherichia coli BL21. The method of recombinant protein expression was based on previous reports (26). Briefly, the cells were induced using 1 mM IPTG for 4 h at 37°C when the optical density (OD 600) reached 0.5–0.6. RMSTNb (recombinant MSTNb) was purified by cutting the object tape in SDS-PAGE. Western blots targeting a 6 × His-tag were used to assess the production of the purified protein. Second, the purified rMSTNb was injected into a New Zealand White rabbit for 4 weeks to produce the polyclonal antibody. The serum was sampled and stored at −80°C after the final immunization. All of the procedures involving the polyclonal antibody and specificity determination were based on previous reports (26).
Immunoblotting
To detect the MSTN protein expression, rMSTNb protein was expressed using the prokaryotic expression system and a rabbit polyclonal antibody against rMSTNb was developed. Before protein extraction, PMSF (phenylmethanesulfonyl fluoride) and protease inhibitor (Beyotime, Nantong, China) were added to a cell lysis reagent radio immunoprecipitation assay buffer (Beyotime, China) at a ratio of 1:100. First, the concentration of protein was determined, and 10 μg total protein was separated on an SDS-PAGE and transferred to a polyvinylidene fluoride membrane (0.45 μm, Millipore, New York, USA). Next, 5% BSA (dissolved in TBST) was used to block the membrane for an hour at room temperature, and the specific antibody was incubated at 4°C overnight. The internal control was β-actin (Proteintech, Chicago, USA) or GAPDH (Cell Signaling Technology, Boston, USA). Third, HRP-conjugated goat anti-rabbit or anti-mouse IgG antibody (Boster, Wuhan, China) was incubated for an hour at room temperature after washing the membrane three times (10 min each). Finally, the membranes were washed, and the immunoreactivity was determined by an enhanced chemiluminescence ECL detection kit (Amersham, Buckinghamshire, UK). The gray intensity analysis was conducted using Image J 1.45 (NIH, Bethesda, USA).
Primary Muscle Cell Isolation and Identification
The primary muscle cell culture was conducted as previously described with some modifications (27). Briefly, white muscle was obtained from the latero-dorsal muscle of juvenile tilapia (6–8 g, n = 25–30) and collected in an ice-cold isolation medium (DMEM, 9 mM NaHCO3, 20 mM HEPES, 100 U/ml penicillin, 100 U/ml streptomycin, and 15% horse serum). After removing the remaining red muscle and skin, the samples were sliced and hydrolyzed with collagenase (2 mg/ml, Sigma-Aldrich, St. Louis, MO, USA) at 28°C for 20 min. After washing twice with washing medium (isolation medium without horse serum), trypsin (1 mg/ml, Sigma-Aldrich, USA) was used to digest the remaining sample at 28°C for 20 min. This mixture was diluted with additional isolation medium (1:4) to neutralize the digestion of trypsin. After centrifugation, the cells were resuspended with complete medium (washing medium with 10% FBS) and filtered through a sterile nylon sieve (100-, 200-, and 400-mesh). Then, the myosatellite cells were collected in complete medium, and seeded onto 24-well or 12-well plates (Corning, NY, USA) at a density of 4 × 105 cells/cm2 for different experiments. After 24 h, the adhered myosatellite cells were covered with fresh complete medium. To further verify the adhered myosatellite cells, an immunofluorescence experiment with MyoD (ab203383, Abcam, Cambridge, UK) and MyoG (M-225; Santa Cruz Biotechnology, Santa Cruz, USA) antibody was conducted as previously described (27). Both MyoD and MyoG are myogenic regulatory factors with dynamic expression in the process of muscle differentiation (28).
Regulation of miR-181b-5p in vitro
To detect the transfection efficiency, we transfected miR-181b-5p (with or without CY3-label, synthesized by GenePharma, China) into primary muscle cells using Lipofectamine3000 (Life Technologies, USA) at 80 nM for 24 h, and the inverted fluorescent microscope ECLIPSE Ti-E (Nikon, Tokyo, Japan) was used to observe the fluorescence. Transfection efficiency was defined as the ratio of cells in red (CY3-labeled) and number of cells in blue (DAPI stained). MiRNA mimics and antagomir (synthesized by GenePharma, China) were used in the overexpression and inhibition experiments. The negative control was a scrambled RNA duplex that was not homologous to the tilapia genome. All oligonucleotides were 2′-OMe modified, and the end of the antagomir was conjugated to cholesterol. MiRNA mimics were transfected into the dispersed cells at 80 nM for 24 h, while antagomir was incubated at 100 nM for 24 h, and cells were harvested for RNA extraction or western blot. Each experiment was repeated three times independently.
Statistical Analysis
Data are expressed as the means ± SEM unless otherwise stated. Statistical significance was assessed by one-way ANOVA followed by Bonferroni's multiple comparison tests. Statistical significance was defined as p < 0.05. p < 0.05 was noted with *p < 0.01 with **, and p < 0.001 with ***.
Results
Molecular Cloning and Tissue Distribution of Mstnb and Msnta in Tilapia
The 3′ UTRs of Mstna and Mstnb were cloned. The results showed that the Mstnb 3′ UTR was 1,307 bp, which was consistent with the Mstnb sequences in the Nile tilapia genome (Figure 1A and Data Sheet S1). The Mstna 3′ UTR was 879 bp, which differed from Mstna sequences in the Nile tilapia genome (Figure 1B and Data Sheet S1). QPCR analysis showed that Mstnb mRNA was abundantly expressed in the telencephalon, diencephalon, cerebellum, and white muscle, and weakly expressed in the pituitary and gut (Figure 1C). However, Mstna mRNA was only expressed in the brain of tilapia (Figure 1D). Therefore, our present study was focused on Mstnb.
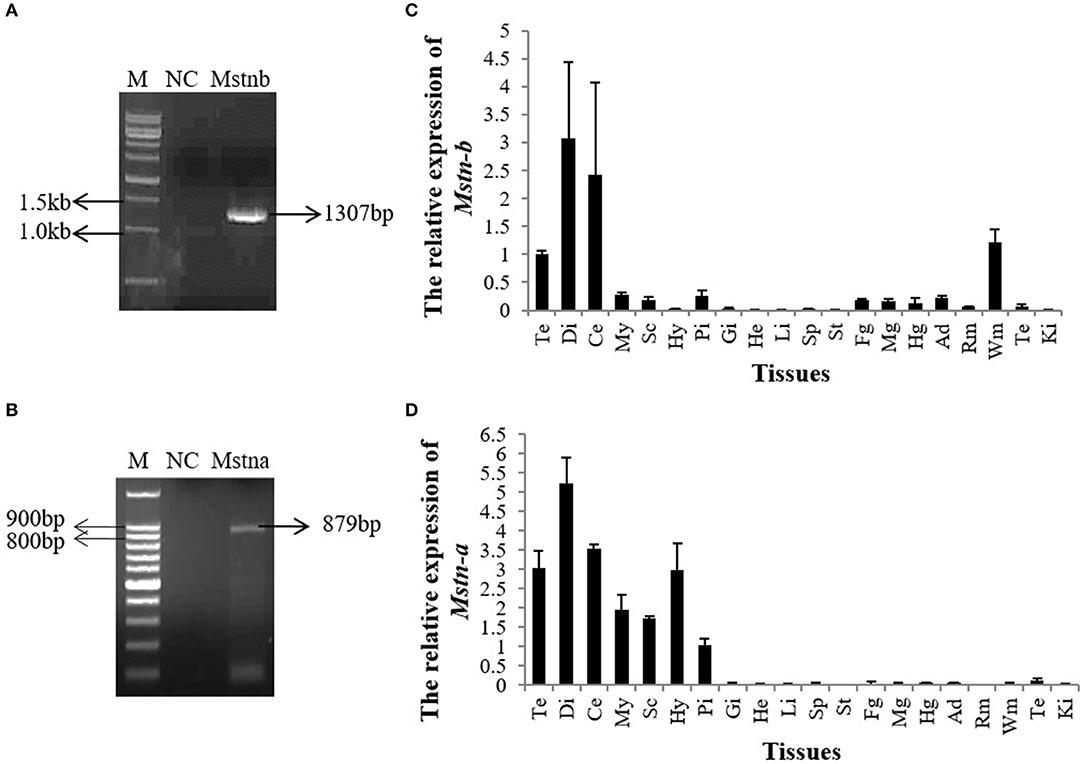
Figure 1. The expression pattern of tilapia mstnb and mstna by qPCR. Mstnb and mstna clones were identified by PCR in (A) tilapia brain and (B) white muscle. Relative mRNA expression of (C) mstnb and (D) mstna in the tissues of tilapia (n = 3, males). Te, telencephalon; Di, diencephalon; Ce, cerebellum; My, Myelencephalon; Sc, spinal cord; Hy, Hypothalamus; Pi, pituitary; Gi, gill; He, Heart; Li, Liver; Sp, spleen; St, stomach; Fg, foregut; Mg, midgut; Hg, hindgut; Ad, adipose tissue; Rm, red muscle; Wm, white muscle; Te, testis; Ki, Kidney. Bar: mean ± SEM.
Screening of miRNAs Targeting Mstnb
To predict miRNAs that potentially target Mstnb, the PITA prediction program was employed based on the database of the tilapia miRNA transcriptome. After the prediction, 1,992 miRNAs were predicted to target the 3′ UTR of Mstnb (data not shown). Subsequently, a preliminary screening was carried out with the principle of conservation, and, finally, 32 miRNAs with high scores were selected to conduct different experiments. The Mstnb 3′ UTR was subcloned into the psiCHECK2 reporter plasmid and used in a dual luciferase reporter experiment. The results showed that miR-30a-3p, miR-181a-5p, and miR-181b-5p downregulated the relative Renilla/Firefly luciferase ratio (Rluc/Fluc) of the Mstnb 3′ UTR (P < 0.001) (Figures 2A,D,E); miR-338, miR-455b, miR-200a, miR-27b, miR-27a, miR-31, miR-221, and miR-222 downregulated the Rluc/Fluc (P < 0.01) (Figures 2A–D); and miR-132-5p, miR-141-3p, miR-107, miR-22a, miR-27c, miR-27e, miR-72-5p, miR-25-5p, and miR-206-3p downregulated the Rluc/Fluc (P < 0.05) (Figures 2A–E). Specially, overexpression of miR-181a/b-5p and miR-30a-3p decreased the Rluc/Fluc by more than 50%. The other members of miR-181 and miR-30 were also included in the dual luciferase reporter experiment. The results showed that miR-181, miR-181a, miR-181b, and miR-30b-3p also downregulated the Rluc/Fluc (Figure 2F). Among these miRNAs, miR-181 was reported to be abundantly expressed in muscle and related to skeletal muscle growth in mammals (29). Thus, miR-181 was chosen for further study.
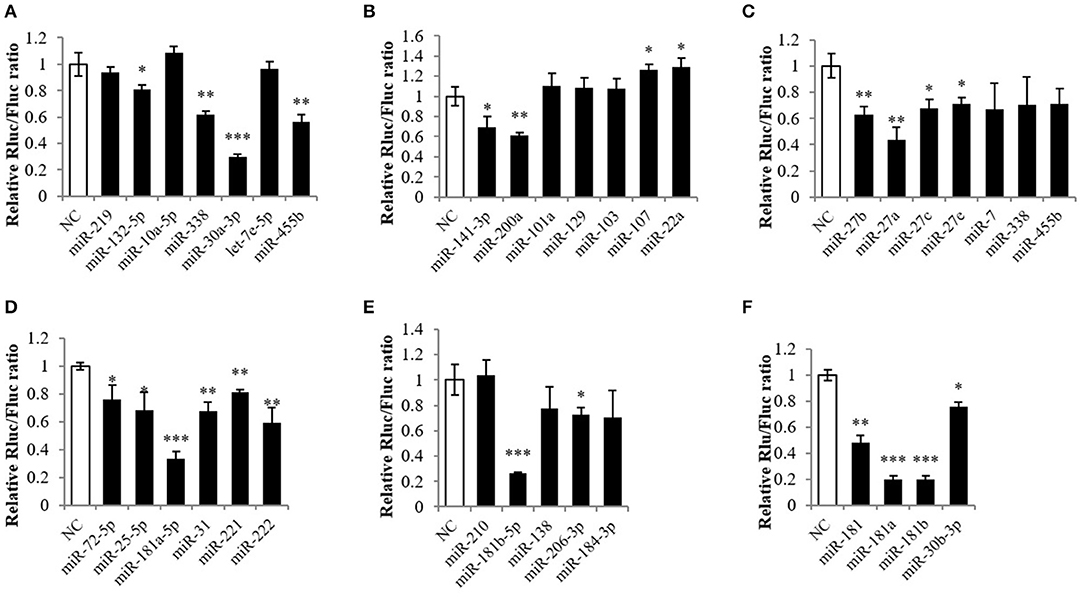
Figure 2. Screening the targeting miRNA of Mstnb. Dual luciferase assay with psiCHECK2-Mstnb and predicted target miRNA, and the y-axis is represented by the ratio of ranilla luciferase to firefly luciferase. (A–F) Showed different miRNA may target mstnb. (A) miR-338, miR-30a-3p, and miR-455b; (B) miR-141-3p and miR-200a; (C) miR-27b, miR-27a, and miR-27c; (D) miR-181a-5p, miR-31, miR-221, and miR- 222; (E) miR-181b-5p and miR-206-3p; (F) miR-181, miR-181a/b, and miR-30b-3p. Bar: mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 One-way ANOVA analysis, n = 3.
miR-181b-5p Directly Targets the Mstnb 3′ UTR
To further verify the miR-181 expression pattern, we attempted to clone the miR-181 family, but the results showed that only miR-181b-5p was cloned in tilapia (Figure 3A and Data Sheet S1). Therefore, miR-181b-5p was chosen for further study. Subsequently, the results of tissue distribution showed that miR-181b-5p had very high expression in the diencephalon and a lower expression in the telencephalon, spinal cord, adipose tissue, white muscle, and kidney (Figure 3B).
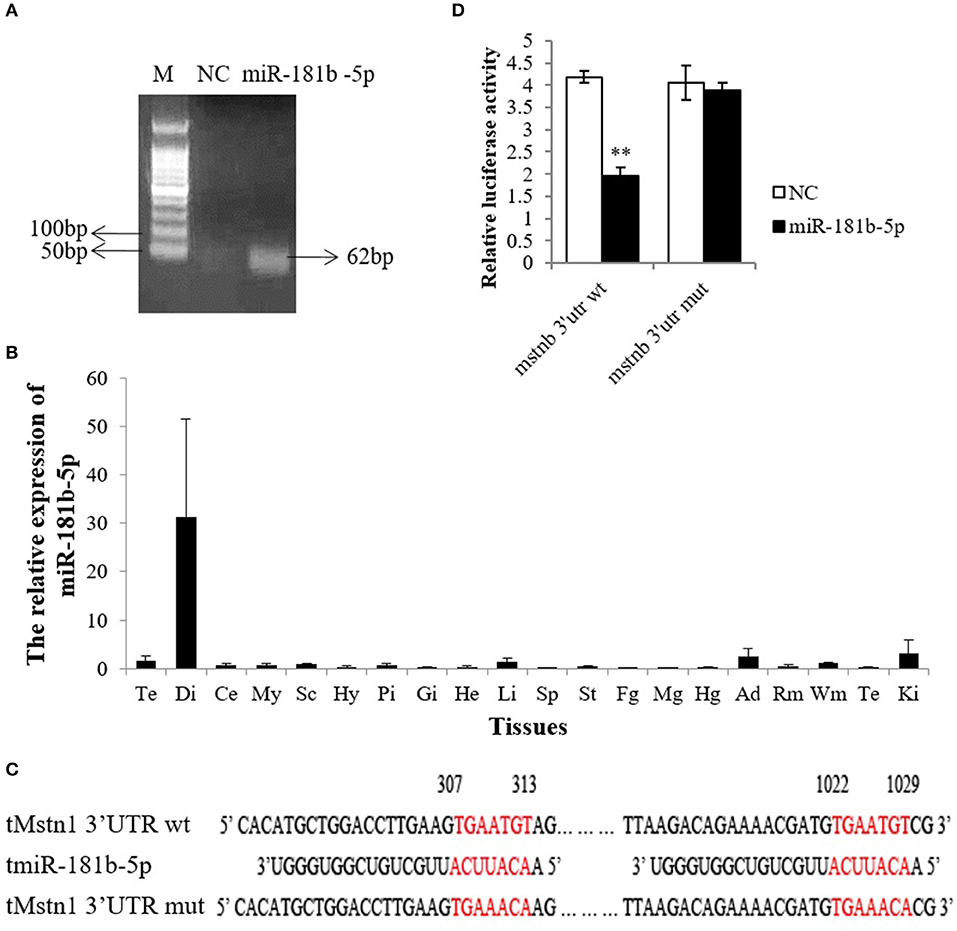
Figure 3. miR-181b-5p directly targeted Mstnb 3′UTR. (A) MiR-181b-5p clones were identified by PCR in tilapia brain. (B) Relative mRNA expression of miR-181b-5p in the tissues of tilapia (n = 3, males). Te, telencephalon; Di, diencephalon; Ce, cerebellum; My, Myelencephalon; Sc, spinal cord; Hy, Hypothalamus; Pi, pituitary; Gi, gill; He, Heart; Li, Liver; Sp, spleen; St, stomach; Fg, foregut; Mg, midgut; Hg, hindgut; Ad, adipose tissue; Rm, red muscle; Wm, white muscle; Te, testis; Ki, Kidney. (C) Complete complementarity between Mstnb 3′ UTR and miRNA (Mstnb 3′ UTR wt) and incomplete complementarity (Mstnb 3′ UTR mut). (D) Dual luciferase assay with psiCHECK2-Mstnb 3′ UTR wt or psiCHECK2-Mstnb 3′ UTR mut and miR-181b-5p. Bar: mean ± SEM. ***P < 0.01, One-way ANOVA analysis, n = 3–5.
As PITA predicted, the seed sequence of miR-181b-5p is completely complementary to the Mstnb 3′ UTR at 307–313 nt and 1022–1029 nt (Figure 3C). To determine whether miR-181b-5p directly binds to the 3′ UTR of Mstnb, nucleotides TGT were converted to ACA at both sites by site-directed mutagenesis (Figure 3C). Dual luciferase assay results showed that the wild-type Mstnb 3′ UTR dramatically downregulated the Rluc/Fluc, while mutagenesis of the Mstnb 3′ UTR could reverse miR-181b-5p-induced suppression (Figure 3D).
The Development of a Polyclonal Antibody Against rMSTNb
To detect the MSTNb protein level in the following experiment, an antibody against MSTNb was produced (Figure 4). First, the mstnb ORF was amplified by PCR and the sequence was confirmed by sequencing (Figure 4A and Data Sheet S1). Then, the amplified fragment was inserted between XhoI and BamHI sites in pET-32a to produce a C-terminal His-tagged rMSTNb protein (Figure 4B). SDS-PAGE revealed that the rMSTNb band with a molecular mass of 35 kDa was expressed successfully in IPTG-induced pET-32a-rMSTNb-transformed bacteria but not in the pET-32a-transformed control bacteria (Figure 4C and Data Sheet S1). After that, the rMSTNb was purified by incising the target strip in the albumen gel, and the results of SDS-PAGE and western blot showed that a 35 kDa protein band can be detected (Figures 4D,E and Data Sheet S1), which suggested that the rMSTNb protein was purified. The purified rMSTNb was used to induce production of polyclonal antibodies in serum of rabbit. To confirm the affinity, western blot was performed and showed that rMSTNb could be detected at the dilution of 1:10,000 (Figure 4F and Data Sheet S1).
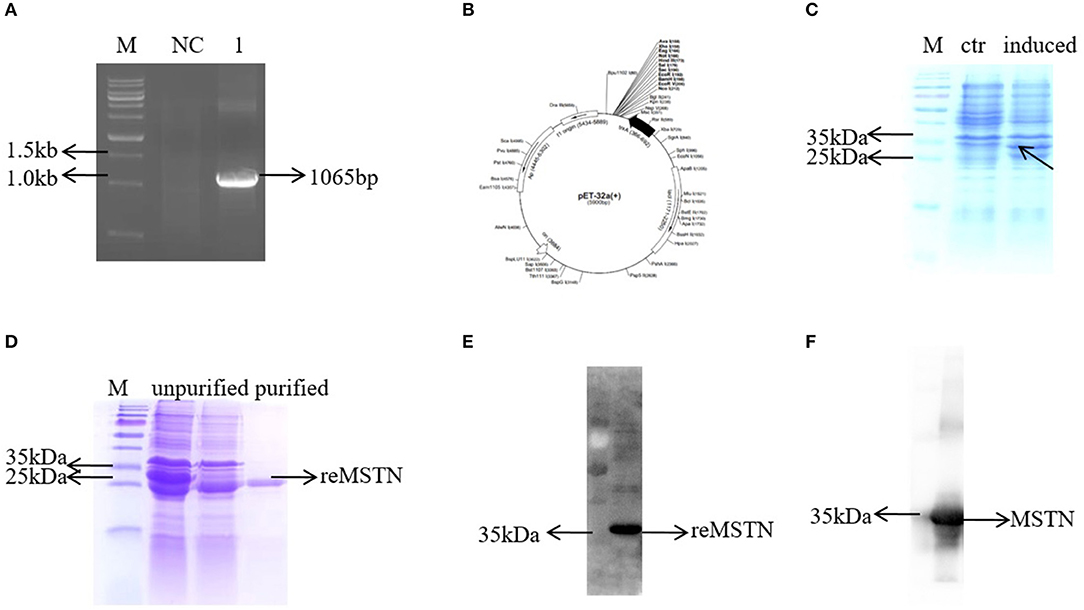
Figure 4. Development of the MSTNb anti-rabbit antibody. (A) The mstnb ORF was cloned by PCR. (B) Schematic diagram of recombinant plasmid pET32a-MSTNb. (C) The induced expression of recombinant protein. (D) Purification of recombinant protein. (E) The purified protein was verified by western blot. (F) Mstnb anti-serum was detected, dilution ratio was 1:10,000.
The Isolation and Identification of Primary Muscle Cells From Tilapia
To verify whether miR-181b-5p regulates the expression of Mstnb in vitro, a piece of technology for separating the tilapia primary muscle cells was established (Figure 5A). The myosatellite cells were separated and were round on day 1; they then differentiated into spindle-shaped cells called myoblasts on day 4, and they ultimately turned into myotubes on day 7 (Figure 5A). Additionally, the myosatellite cells were verified using immunofluorescence (Figure 5B). Myoblasts expressed MyoD1 during the proliferation phase (day 4), and differentiated myotubes expressed Myogenin at day 7 (Figure 5B).
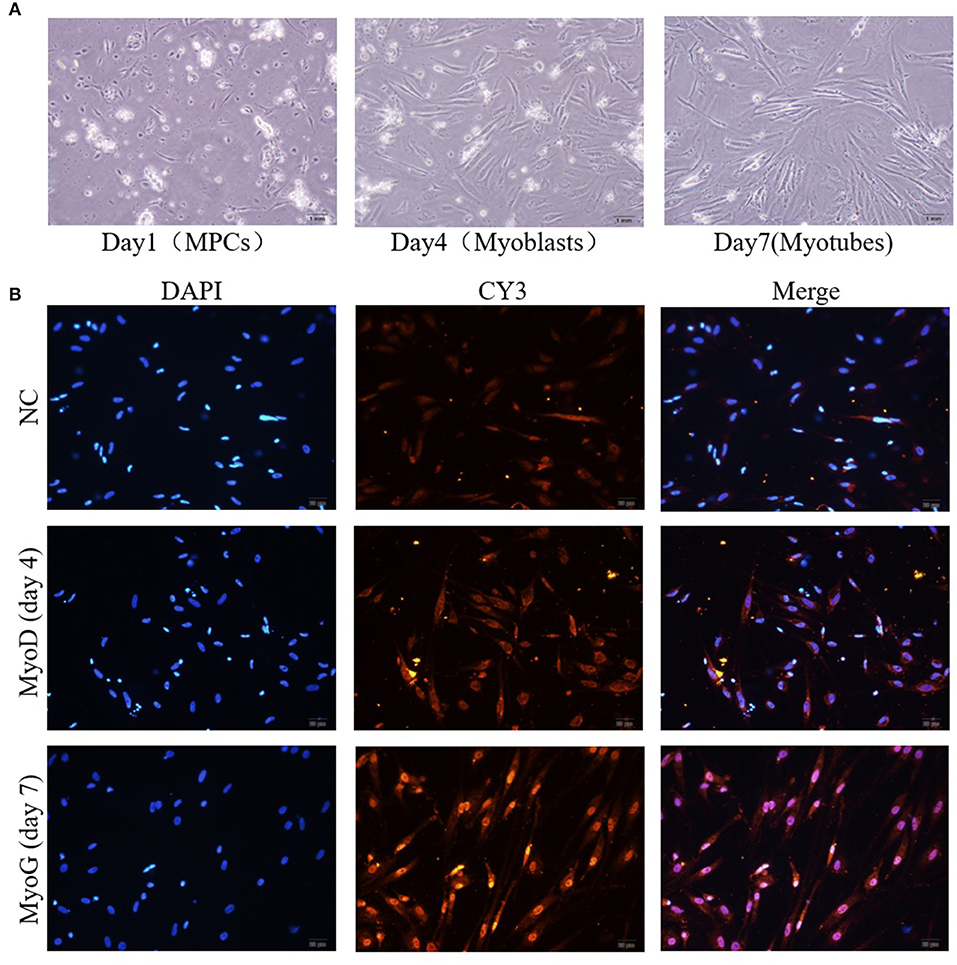
Figure 5. Isolation and identification of primary muscle cells from tilapia. (A) The culture of myosatellite cells (day 1), myoblasts (day 4), and myotubes (day 7). (B) Immunocytochemical staining of myoblasts (day 4) and myotubes (day 7).
miR-181b-5p Silencing Activates Mstnb and Inhibits Downstream Genes
As Mstnb was directly targeted by miR-181b-5p, we hypothesized that the upregulation of MSTNb by antagomir-181b-5p would inhibit the expression of MRFs. In tilapia primary muscle cells, knockdown of miR-181b-5p resulted in a decrease in miR-181b-5p expression at 24 h and 48 h (Figure 6A), while Mstnb mRNA was upregulated at 24 h but not at 48 h after the administration (Figure 6B). Knockdown of miR-181b-5p upregulated MSTNb protein levels with a significant difference at 24 h, but no significant difference was noted at 48 h (Figure 6C, Data Sheet S1, Figures S2–S4 and Presentation S1). Thus, 24 h was determined as the administration time. Meanwhile, the mRNA expression of MHC and Myf6 was downregulated by antagomir-181b-5p compared to antagomir-nc after 24 h. However, myf5, myoG, and myoD mRNA exhibited no change after the knockdown of miR-181b-5p (Figure 6D).
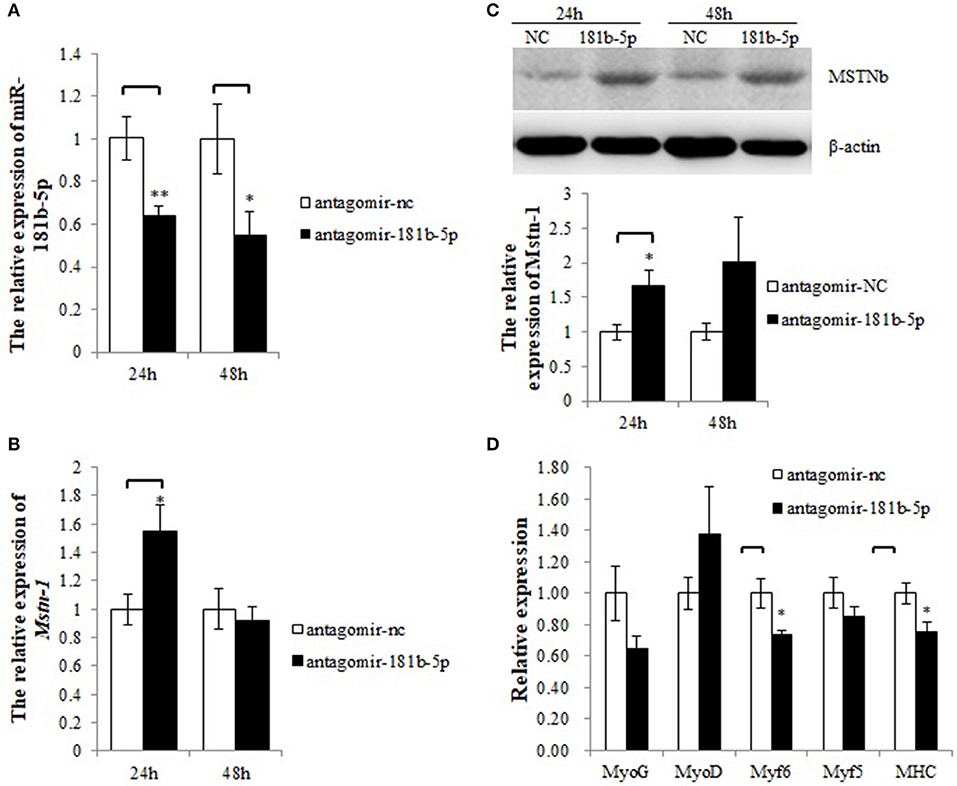
Figure 6. miR-181b-5p silencing influences the expression of Mstnb and its downstream genes. (A) The relative expression of miR-181b-5p. (B) The mRNA expression of Mstnb. (C) The protein level of Mstnb. (D) The mRNA expression of Mstnb downstream genes. Bar: mean ± SEM. *P < 0.05 and **P < 0.01, One-way ANOVA analysis, n = 3–4.
miR-181b-5p Overexpression Inhibits Mstnb and Activates Downstream Genes
To determine whether the miRNA mimics could be transfected into tilapia primary muscle cells, CY3-labeled miR-181b-5p was synthesized and the transfection efficiency was determined by a fluorescence microscope (Figure S1). A total of 59% of the dispersed muscle cells were transfected with the CY3-labeled miR-181b-5p, while the transfection efficiency of the control was 0%. The relative expression of miR-181b-5p was increased thousands of times after transfection (Figure 7A). Overexpression of miR-181b-5p did not affected the mRNA expression level of mstnb (Figure 7B), but it decreased MSTNb protein expression (Figure 7C, Data Sheet S1, Figures S5–S7 and Presentation S1). As expected, the mRNA expression of myf5, myoD, and myoG was upregulated by miR-181b-5p mimics, while myf6 and MHC mRNA expression was not affected (Figure 7D). This contrasted with the results of miR-181b-5p inhibition.
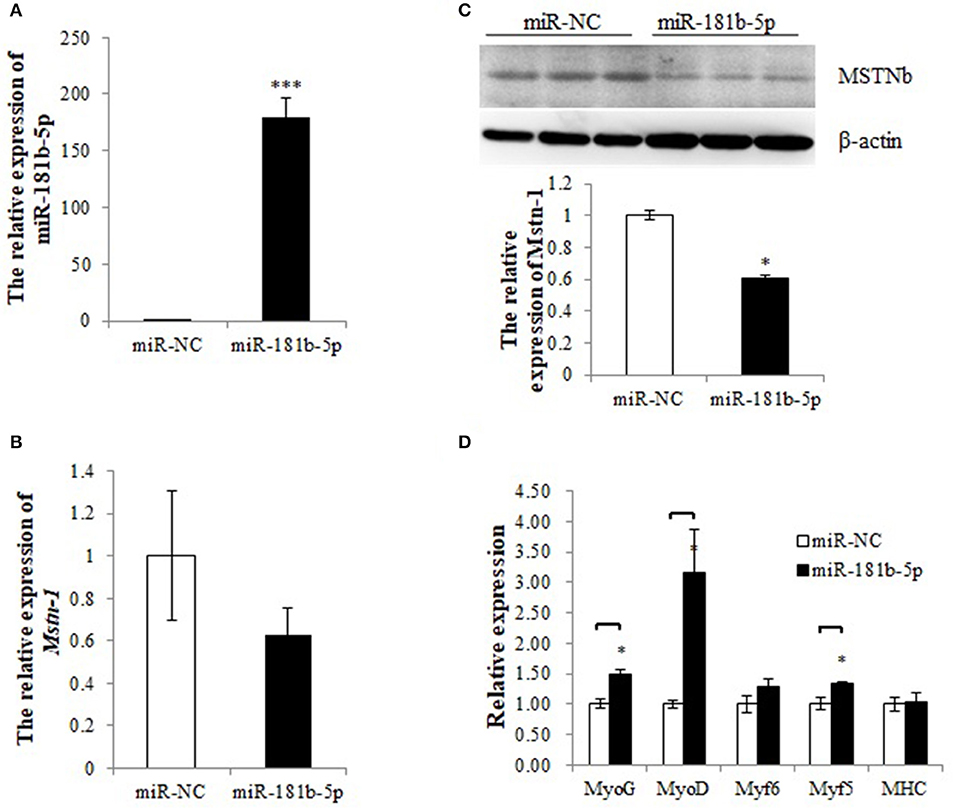
Figure 7. miR-181b-5p overexpression influences the expression of Mstnb and its downstream genes. (A) The relative expression of miR-181b-5p. (B) The mRNA expression of Mstnb. (C) The protein level of Mstnb. (D) The mRNA expression of Mstnb downstream genes. Bar: mean ± SEM. *P < 0.05 and ***P < 0.01, One-way ANOVA analysis, n = 3–4.
Discussion
Myostatin has been regarded as a muscle negative regulator in mammals since its discovery (2); its function in muscle development (2), adipogenesis (30), and insulin sensitivity (31) was revealed by several studies. Due to its importance in growth and metabolism, the precise regulation of Mstn protein levels is necessary. The regulation of Mstn at the transcriptional, post-transcriptional, and translational levels was documented widely. MiRNAs could directly degrade or inhibit the translation of targeted mRNAs at the post-transcriptional level, and they were found to take part in the development of skeletal muscle. In this study, we found that miR-181b-5p may regulate muscle growth of tilapia by targeting Mstnb.
In our study, the 3′ UTRs of mstna and mstnb were cloned, but only mstnb was chosen for further study as mstna mRNA was only expressed in the brain of tilapia whereas mstnb mRNA was expressed in the brain and skeletal muscle. The tissue distribution pattern of tilapia mstnb was similar to that of previous studies; RT-PCR results showed that mstn was expressed in the muscle, eye, gill, gonad, gut, and brain of Nile tilapia (4). In rainbow trout, both transcripts (mstn 1a and mstn 1b) were present in the muscle, testes, eye, brain, and spleen (32). In Trachidermus fasciatus, the mstn was highly expressed in the muscle and intestine and weakly expressed in the brain and liver (33). Although mstn is widely distributed in various tissues of teleosts, it is highly expressed in muscle (4, 34). We hypothesized that mstnb was important for the growth of skeletal muscle in tilapia.
The relative luciferase activity of mstnb was downregulated by several miRNAs, including miR-30a-3p, miR-181a/b-5p, miR-27a/b, miR-206-3p, miR-200a, and miR-455b. This is consistent with the fact that one gene may be targeted by multiple miRNAs and one miRNA may target several genes (35). In addition, the other members of the miR-181 family were also shown to target the mstnb 3′ UTR since their seed sequences are identical. These results suggested that miRNAs targeted mRNA by interacting with the seed sequences (36, 37). Although only miR-181b-5p was investigated in this study, other miRNAs might also play a role in regulating mstn at the post-transcriptional level. For example, miR-27a was reported to target mstn and induce the differentiation of C2C12 (20), and miR-27a/b regulates MSTN expression through negative feedback auto-regulation in mice (21). MiR-206, a miRNA that is abundantly expressed in skeletal muscle that modulates the development and disease of skeletal muscle in mammals (38), was also shown to decrease the luciferase activity of mstnb in tilapia. In addition, miR-206 was shown to target the 3′ UTR of mstn in Texel sheep, a sheep known for its double muscling (17). The above results suggested that mstnb could be regulated by several miRNAs in both mammals and teletosts.
Among those miRNAs, miR-181 was reported to associate with the TGF-β superfamily and regulate Hox-A11 expression in mammals (29). Meanwhile, only miR-181b-5p, a member of the miR-181 family from tilapia, was cloned. We focused on miR-181b-5p in the subsequent research. The results of tissue distribution showed that miR-181b-5p was expressed predominantly in the diencephalon, and smaller amounts in the telencephalon, liver, adipose, white muscle, and kidney, which was in line with the studies that showed that miR-181 was widely expressed across tissues and played a vital role in the immune system (39), skeletal muscle growth (29), hemopoiesis (40), brain ischemia (41), and so on. In addition, the expression profiles of miR-181 were correlated with the development stage (42) and nutrition status (43). Therefore, the development stage may be the cause of low abundance of miR-181b-5p expression in white muscle.
Furthermore, the results of our study showed that miR-181b-5p could target mstnb using a mutated reporter in the predicted target sites and a dual-luciferase assays. To further study the possible roles of miR-181b-5p in the post-transcriptional regulation of mstnb in tilapia, the primary muscle cells of tilapia were used as cell models to perform the experiments of miR-181b-5p knockdown and overexpression. In our study, double enzyme hydrolysis (collagenase IV and trypsin) was used to digest the muscle of tilapia. The research in fish cell culture developed quickly following Wolf establishing the RTG-2 cell line in rainbow trout (44). However, the culture of primary muscle cells was developed slowly because the myogenic precursor cells only proliferate under certain conditions, such as wound healing, exercise, and disease (45). A method to isolate and culture primary myogenic precursor cells was established in several fish species, including rainbow trout, salmon, and sea bream (46–48). We also cultured primary muscle cells in tilapia using the protocol described by Froehlich et al. (27). This study was the first paper to describe the technology of culturing primary muscle cells from tilapia.
In this study, knockdown of miR-181b-5p in primary muscle cells led to an upregulation of MSTNb protein, and overexpression caused a downregulation effect, indicating that miR-181b-5p might participate in the regulation of MSTNb expression levels in vitro. Similar results were obtained using deep sequencing and a dual-luciferase experiment in S. chuatsi (23). Meanwhile, the opposite expression pattern between miR-181a-5p and mstn in white and red muscle was observed (23). In our study, miR-181b-5p targeting mstnb was further confirmed in tilapia primary muscle cells. In addition, miR-181 was reported to regulate the differentiation of myoblasts in mice by targeting Hox-A11, an inhibitor of MyoD (29). Although miR-181 may target different genes in mammals and fish, the function of miR-181 in regulating muscle growth may be conserved in evolution.
Although MSTN was reported to inhibit the growth of skeletal muscle in mammals and some fishes, the regulatory role of MSTN on the growth of skeletal muscles in tilapia was not reported. In the present study, the mRNA expression of MRFs was detected after stimulating or inhibiting the expression of MSTNb by antagomir-181b-5p and miR-181b-5p. MRFs play an important role during the proliferation and differentiation of skeletal muscle cells. Lacking both MyoD and Myf5 in skeletal muscle stem cells caused the accumulation of satellite cell progeny in damaged muscle and blocked differentiation (49). MyoD and MyoG can induce the transcription of Myomaker and promote the fusion of myoblasts, which is an important step during skeletal muscle differentiation (50). It was also reported that miR-374b directly targeted Myf6 and inhibited the differentiation in C2C12 (51). It suggested that the expression of MRFs may reflect the differentiation of muscle cells. Except for MRFs, MSTN knockdown by small hairpin RNA (shRNAs) led to sustained cell proliferation of myoblasts and upregulated expression of Myf6 in goats (52). Our study showed that the mRNA expression of MRFs was decreased when MSTNb was promoted by antagomir-181b-5p and increased when MSTNb was inhibited by miR-181b-5p. These results suggested that MSTNb may regulate muscle growth by modulating the expression of MRFs in tilapia, which was consistent with those reported in mammals (53–56).
Since MSTN is a negative regulator, inhibition or mutation of this gene may be useful for muscle development (55). For instance, knocking down mstn using siRNA increased muscle mass in mice (57), and knocking out mstnb using TALEN (transcription activator-like effector nucleases) also induced muscle hyperplasia and body weight increase in zebrafish (58). In their study, the circumferences and body weights of mstnb-deficient zebrafish increased after 80 days post-fertilization (58). Similarly, primary muscle cells treated with human recombinant MSTN (huMSTN) resulted in a myotube diameter decreased of up to 20% (59). The mRNA expression of Myf5, MyoD, and MyoG was significantly increased after rainbow trout primary myosatellite cells were treated with MSTN-1 (7), which was inconsistent with our study. It is noteworthy that the treatments were different. In their study, primary myosatellite cells were incubated with MSTN-1 after culturing 72 h for 3 days or 7 days (7), while in our study, MSTNb was indirectly increased by antagomir after culturing over 1 day (for 24 h). Obviously, the mechanism of MSTN-1 and antagomir treatment of myosatellite cells is different. In addition, the expression of MRFs is dynamic during the differentiation of skeletal muscle. The expression of Myf5 and MyoD was increased in proliferating myoblasts, while MyoG and Myf6 increased in the terminally differentiated myotubes (28). It is hard to compare the two studies because the experimental conditions were different. Despite this, we hypothesized that mstnb participates in the regulation of muscle growth in tilapia in a similar way in fishes as in mammals.
In this study, miR-181b-5p overexpression inhibited MSTNb and activated downstream gene expression. Our results suggest that miR-181b-5p may regulate the muscle growth of tilapia by targeting myostatin b in tilapia.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Ethics Statement
The Tilapia used in this study were obtained from the local farm of Guangdong Tilapia Breeding Farm, Guangzhou, China. No specific permissions are required for the buying of fish. All animal experiments were performed with the approval of the Sun Yat-Sen University Animal Care and Use Committee and in full compliance with its ethical guidelines.
Author Contributions
WL and ZZ conceived and designed the experiments. ZZ, XY, JJ, and GY performed the experiments. ZZ and WL analyzed the data. ZZ and CS wrote the paper. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National Key R&D Program of China 2018YFD0900101, the China Agriculture Research System (CARS-46), the National Science Foundation of China (31472259 and 31272639), the Guangdong Provincial Science and Technology Program (2015A020216006), Guangdong Oceanic & Fishery Program (A201601C02), Science and Technology Planning Project of Guangzhou (201607020014), and the Program for Chinese Outstanding Talents in Agricultural Scientific Research (2016-2020) to WL.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2019.00812/full#supplementary-material
Figure S1. Transfection efficiency of tilapia muscle pituitary cells by CY3-labeled miR-181b-5p. DAPI, cells were dyed by DAPI and image taken with 358 nm exciting laser; CY3, image taken with 550 nm exciting laser; MERGE, image merged by the two images DAPI and CY3.
Figure S2. Western bolt original image of β-actin corresponding to Figure 6C.
Figure S3. Western bolt original image of Mstnb corresponding to Figure 6C.
Figure S4. Coomassie-blue staining of SDS-PAGE corresponding to Figure 6C.
Figure S5. Western bolt original image of Mstnb corresponding to Figure 7C.
Figure S6. Western bolt original image of β-actin corresponding to Figure 7C.
Figure S7. Coomassie-blue staining of SDS-PAGE corresponding to Figure 7C.
Table S1. Primers used in this research.
Data Sheet S1. Agarose gel original images corresponding to Figures 1A,B, 3A, 4A. Coomassie-blue original images corresponding to Figures 4C,D. Western bolt original images corresponding to Figures 4E,F, 6C, 7C.
Presentation S1. Coomassie-blue staining and Western bolt original images corresponding to Figures 6C, 7C.
References
1. Chen YH, Tsai HJ. Myogenic regulatory actors Myf5 and Mrf4 of fish: current status and perspective. J Fish Biol. (2008) 73:1872–90. doi: 10.1111/j.1095-8649.2008.01991.x
2. McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. (1997) 387:83–90. doi: 10.1038/387083a0
3. Sharma M, McFarlane C, Kambadur R, Kukreti H, Bonala S, Srinivasan S. Myostatin: expanding horizons. IUBMB Life. (2015) 67:589–600. doi: 10.1002/iub.1392
4. Rodgers BD, Weber GM, Sullivan CV, Levine MA. Isolation and characterization of myostatin complementary deoxyribonucleic acid clones from two commercially important fish: Oreochromis mossambicus and Morone chrysops. Endocrinology. (2001) 142:1412–8. doi: 10.1210/endo.142.4.8097
5. Rodgers BD, Weber GM, Kelley KM, Levine MA. Prolonged fasting and cortisol reduce myostatin mRNA levels in tilapia larvae; short-term fasting elevates. Am J Physiol Regul Integr Comp Physiol. (2003) 284:R1277–86. doi: 10.1152/ajpregu.00644.2002
6. Nebo C, Portella MC, Carani FR, de Almeida FL, Padovani CR, Carvalho RF, et al. Short periods of fasting followed by refeeding change the expression of muscle growth-related genes in juvenile Nile tilapia (Oreochromis niloticus). Comp Biochem Physiol B Biochem Mol Biol. (2013) 164:268–74. doi: 10.1016/j.cbpb.2013.02.003
7. Garikipati DK, Rodgers BD. Myostatin inhibits myosatellite cell proliferation and consequently activates differentiation: evidence for endocrine-regulated transcript processing. J Endocrinol. (2012) 215:177–87. doi: 10.1530/JOE-12-0260
8. Salerno MS, Thomas M, Forbes D, Watson T, Kambadur R, Sharma M. Molecular analysis of fiber type-specific expression of murine myostatin promoter. Am J Physiol Cell Physiol. (2004) 287:C1031–40. doi: 10.1152/ajpcell.00492.2003
9. Spiller MP, Kambadur R, Jeanplong F, Thomas M, Martyn JK, Bass JJ, et al. The myostatin gene is a downstream target gene of basic helix-loop-helix transcription factor MyoD. Mol Cell Biol. (2002) 22:7066–82. doi: 10.1128/MCB.22.20.7066-7082.2002
10. Du R, Chen YF, An XR, Yang XY, Ma Y, Zhang L, et al. Cloning and sequence analysis of myostatin promoter in sheep. DNA Sequence. (2005) 16:412–7. doi: 10.1080/10425170500226474
11. Li J, Deng J, Yu S, Zhang JL, Cheng D, Wang HY. The virtual element in proximal promoter of porcine myostatin is regulated by myocyte enhancer factor 2C. Biochem Biophys Res Commun. (2012) 419:175–81. doi: 10.1016/j.bbrc.2012.01.135
12. Ma K, Mallidis C, Artaza J, Taylor W, Gonzalez-Cadavid N, Bhasin S. Characterization of 5 '-regulatory region of human myostatin gene: regulation by dexamethasone in vitro. Am J Physiol-Endoc M. (2001) 281:E1128–36. doi: 10.1152/ajpendo.2001.281.6.E1128
13. Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. (2007) 23:243–9. doi: 10.1016/j.tig.2007.02.011
14. Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. (2006) 16:203–8. doi: 10.1016/j.gde.2006.02.012
15. Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. (2009) 136:642–55. doi: 10.1016/j.cell.2009.01.035
16. Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol. (2009) 21:461–9. doi: 10.1016/j.ceb.2009.01.029
17. Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. (2006) 38:813–8. doi: 10.1038/ng1810
18. Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: current status and prospective. Int J Cancer. (2007) 120:953–60. doi: 10.1002/ijc.22454
19. Allen DL, Loh AS. Posttranscriptional mechanisms involving microRNA-27a and b contribute to fast-specific and glucocorticoid-mediated myostatin expression in skeletal muscle. Am J Physiol-Cell. (2011) 300:C124–37. doi: 10.1152/ajpcell.00142.2010
20. Chen XL, Huang ZQ, Chen DW, Yang T, Liu GM. MicroRNA-27a is induced by leucine and contributes to leucine-induced proliferation promotion in C2C12 Cells. Int J Mol Sci. (2013) 14:14076–84. doi: 10.3390/ijms140714076
21. McFarlane C, Vajjala A, Arigela H, Lokireddy S, Ge XJ, Bonala S, et al. Negative auto-regulation of myostatin expression is mediated by Smad3 and MicroRNA-27. PLoS ONE. (2014) 9:e87687. doi: 10.1371/journal.pone.0087687
22. Miretti S, Martignani E, Accornero P, Baratta M. Functional effect of mir-27b on myostatin expression: a relationship in piedmontese cattle with double-muscled phenotype. BMC Genomics. (2013) 14:194. doi: 10.1186/1471-2164-14-194
23. Chu WY, Liu LS, Li YL, Chen L, Wang KZ, Li HH, et al. Systematic identification and differential expression profiling of MicroRNAs from white and red muscles of Siniperca chuatsi. Curr Mol Med. (2013) 13:1397–407. doi: 10.2174/15665240113139990059
24. Wang DF, Qin JK, Jia JR, Yan PP, Li WS. Pou1f1, the key transcription factor related to somatic growth in tilapia (Orechromis niloticus), is regulated by two independent post-transcriptional regulation mechanisms. Biochem Biophys Res Commun. (2017) 483:559–65. doi: 10.1016/j.bbrc.2016.12.106
25. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C-T method. Nat Protoc. (2008) 3:1101–8. doi: 10.1038/nprot.2008.73
26. Ma TY, Wu JY, Gao XK, Wang JY, Zhan XL, Li WS. Molecular cloning, functional identification and expressional analyses of FasL in Tilapia, Oreochromis niloticus. Dev Comp Immunol. (2014) 46:448–60. doi: 10.1016/j.dci.2014.06.003
27. Froehlich JM, Seiliez I, Gabillard JC, Biga PR. Preparation of primary myogenic precursor cell/myoblast cultures from basal vertebrate lineages. J Vis Exp. 86:e51354. (2014). doi: 10.3791/51354
28. Ferri P, Barbieri E, Burattini S, Guescini M, D'Emilio A, Biagiotti L, et al. Expression and subcellular localization of myogenic regulatory factors during the differentiation of skeletal muscle C2C12 myoblasts. J Cell Biochem. (2009) 108:1302–17. doi: 10.1002/jcb.22360
29. Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. (2006) 8:278–84. doi: 10.1038/ncb1373
30. McPherron AC, Lee SJ. Suppression of body fat accumulation in myostatin-deficient mice. J Clin Invest. (2002) 109:595–601. doi: 10.1172/JCI200213562
31. Guo TQ, Bond ND, Jou W, Gavrilova O, Portas J, McPherron AC. Myostatin inhibition prevents diabetes and hyperphagia in a mouse model of lipodystrophy. Diabetes. (2012) 61:2414–23. doi: 10.2337/db11-0915
32. Garikipati DK, Gahr SA, Rodgers BD. Identification, characterization, and quantitative expression analysis of rainbow trout myostatin-1a and myostatin-1b genes. J Endocrinol. (2006) 190:879–88. doi: 10.1677/joe.1.06866
33. Lu XY, Zhang Y, Wang XG, Zhang YP, Xu JR, Lai R, et al. [Identification and characterization of myostatin gene in rough-skinned sculp, Trachidermus fasciatus]. Zool Res. (2010) 31:387–94. doi: 10.3724/SP.J.1141.2010.04387
34. Ostbye TK, Galloway TF, Nielsen C, Gabestad I, Bardal T, Andersen O. The two myostatin genes of Atlantic salmon (Salmo salar) are expressed in a variety of tissues. Eur J Biochem. (2001) 268:5249–57. doi: 10.1046/j.0014-2956.2001.02456.x
35. Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. (2003) 115:787–98. doi: 10.1016/S0092-8674(03)01018-3
36. Wang XW. Composition of seed sequence is a major determinant of microRNA targeting patterns. Bioinformatics. (2014) 30:1377–83. doi: 10.1093/bioinformatics/btu045
37. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. (2005) 120:15–20. doi: 10.1016/j.cell.2004.12.035
38. Ma GD, Wang YJ, Li Y, Cui LL, Zhao YJ, Zhao B, et al. MiR-206, a key modulator of skeletal muscle development and disease. Int J Biol Sci. (2015) 11:345–352. doi: 10.7150/ijbs.10921
39. Sun X, Sit A, Feinberg MW. Role of miR-181 family in regulating vascular inflammation and immunity. Trends Cardiovasc Med. (2014) 24:105–12. doi: 10.1016/j.tcm.2013.09.002
40. Su R, Lin HS, Zhang XH, Yin XL, Ning HM, Liu B, et al. MiR-181 family: regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene. (2015) 34:3226–39. doi: 10.1038/onc.2014.274
41. Ouyang YB, Xu LJ, Yue SB, Liu SW, Giffard RG. Neuroprotection by astrocytes in brain ischemia: importance of microRNAs. Neurosci Lett. (2014) 565:53–8. doi: 10.1016/j.neulet.2013.11.015
42. Siengdee P, Trakooljul N, Murani E, Brand B, Schwerin M, Wimmers K, et al. Pre- and post-natal muscle microRNA expression profiles of two pig breeds differing in muscularity. Gene. (2015) 561:190–8. doi: 10.1016/j.gene.2015.02.035
43. Gabler J, Ruetze M, Kynast KL, Grossner T, Diederichs S, Richter W. Stage-specific miRs in chondrocyte maturation: differentiation-dependent and hypertrophy-related miR clusters and the miR-181 family. Tissue Eng Pt A. (2015) 21:2840–51. doi: 10.1089/ten.tea.2015.0352
44. Wolf K, Quimby MC. Established eurythermic line of fish cells in vitro. Science. (1962) 135:1065. doi: 10.1126/science.135.3508.1065
45. Wang YX, Rudnicki MA. Satellite cells, the engines of muscle repair. Nat Rev Mol Cell Biol. (2011) 13:127–33. doi: 10.1038/nrm3265
46. Castillo J, Codina M, Martinez ML, Navarro I, Gutierrez J. Metabolic and mitogenic effects of IGF-I and insulin on muscle cells of rainbow trout. Am J Physiol Regul Integr Comp Physiol. (2004) 286:R935–41. doi: 10.1152/ajpregu.00459.2003
47. Bower NI, Johnston IA. Paralogs of Atlantic salmon myoblast determination factor genes are distinctly regulated in proliferating and differentiating myogenic cells. Am J Physiol Regul Integr Comp Physiol. (2010) 298:R1615–26. doi: 10.1152/ajpregu.00114.2010
48. Funkenstein B, Balas V, Skopal T, Radaelli G, Rowlerson A. Long-term culture of muscle explants from Sparus aurata. Tissue Cell. (2006) 38:399–415. doi: 10.1016/j.tice.2006.09.003
49. Yamamoto M, Legendre NP, Biswas AA, Lawton A, Yamamoto S, Tajbakhsh S, et al. Loss of MyoD and Myf5 in skeletal muscle stem cells results in altered myogenic programming and failed regeneration. Stem Cell Rep. (2018) 10:956–69. doi: 10.1016/j.stemcr.2018.01.027
50. Luo W, Li E, Nie Q, Zhang X. Myomaker, Regulated by MYOD, MYOG and miR-140–3p, promotes chicken myoblast fusion. Int J Mol Sci. (2015) 16:26186–201. doi: 10.3390/ijms161125946
51. Ma Z, Sun X, Xu D, Xiong Y, Zuo B. MicroRNA, miR-374b, directly targets Myf6 and negatively regulates C2C12 myoblasts differentiation. Biochem Biophys Res Commun. (2015) 467:670–5. doi: 10.1016/j.bbrc.2015.10.086
52. Patel AK, Tripathi AK, Patel UA, Shah RK, Joshi CG. Myostatin knockdown and its effect on myogenic gene expression program in stably transfected goat myoblasts. In Vitro Cell Dev Biol Anim. (2014) 50:587–96. doi: 10.1007/s11626-014-9743-4
53. Collins-Hooper H, Sartori R, Macharia R, Visanuvimol K, Foster K, Matsakas A, et al. Propeptide-mediated inhibition of myostatin increases muscle mass through inhibiting proteolytic pathways in aged mice. J Gerontol Biol. (2014) 69:1049–59. doi: 10.1093/gerona/glt170
54. Mendias CL, Bakhurin KI, Gumucio JP, Shallal-Ayzin MV, Davis CS, Faulkner JA. Haploinsufficiency of myostatin protects against aging-related declines in muscle function and enhances the longevity of mice. Aging Cell. (2015) 14:704–6. doi: 10.1111/acel.12339
55. Qian LL, Tang MX, Yang JZ, Wang QQ, Cai CB, Jiang SW, et al. Targeted mutations in myostatin by zinc-finger nucleases result in double-muscled phenotype in Meishan pigs. Sci Rep. (2015) 5:14435. doi: 10.1038/srep14435
56. Kumar R, Singh SP, Kumari P, Mitra A. Small interfering RNA (siRNA)-mediated knockdown of myostatin influences the expression of myogenic regulatory factors in caprine foetal myoblasts. Appl Biochem Biotechnol. (2014) 172:1714–24. doi: 10.1007/s12010-013-0582-7
57. Kinouchi N, Ohsawa Y, Ishimaru N, Ohuchi H, Sunada Y, Hayashi Y, et al. Atelocollagen-mediated local and systemic applications of myostatin-targeting siRNA increase skeletal muscle mass. Gene Ther. (2008) 15:1126–30. doi: 10.1038/gt.2008.24
58. Gao YP, Dai ZR, Shi C, Zhai G, Jin X, He JY, et al. Depletion of Myostatin b promotes somatic growth and lipid metabolism in Zebrafish. Front Endocrinol. (2016) 7:88. doi: 10.3389/fendo.2016.00088
Keywords: myostatin, tilapia, microRNA, growth, muscle, primary muscle cells
Citation: Zhao Z, Yu X, Jia J, Yang G, Sun C and Li W (2019) miR-181b-5p May Regulate Muscle Growth in Tilapia by Targeting Myostatin b. Front. Endocrinol. 10:812. doi: 10.3389/fendo.2019.00812
Received: 15 July 2019; Accepted: 06 November 2019;
Published: 03 December 2019.
Edited by:
Chun Peng, York University, CanadaReviewed by:
Zhan Yin, Chinese Academy of Sciences, ChinaPeggy Biga, University of Alabama at Birmingham, United States
Copyright © 2019 Zhao, Yu, Jia, Yang, Sun and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wensheng Li, lsslws@mail.sysu.edu.cn
 Zaoya Zhao
Zaoya Zhao Xiaozheng Yu
Xiaozheng Yu Jirong Jia
Jirong Jia Guokun Yang
Guokun Yang Caiyun Sun
Caiyun Sun Wensheng Li
Wensheng Li