- 1School of Physiology, Pharmacology and Neuroscience, Biomedical Sciences Building, University of Bristol, Bristol, United Kingdom
- 2Musculoskeletal Research Unit, Translational Health Sciences, Bristol Medical School, Southmead Hospital, University of Bristol, Bristol, United Kingdom
Osteoporosis is metabolic bone disease caused by an altered balance between bone anabolism and catabolism. This dysregulated balance is responsible for fragile bones that fracture easily after minor falls. With an aging population, the incidence is rising and as yet pharmaceutical options to restore this imbalance is limited, especially stimulating osteoblast bone-building activity. Excitingly, output from large genetic studies on people with high bone mass (HBM) cases and genome wide association studies (GWAS) on the population, yielded new insights into pathways containing osteo-anabolic players that have potential for drug target development. However, a bottleneck in development of new treatments targeting these putative osteo-anabolic genes is the lack of animal models for rapid and affordable testing to generate functional data and that simultaneously can be used as a compound testing platform. Zebrafish, a small teleost fish, are increasingly used in functional genomics and drug screening assays which resulted in new treatments in the clinic for other diseases. In this review we outline the zebrafish as a powerful model for osteoporosis research to validate potential therapeutic candidates, describe the tools and assays that can be used to study bone homeostasis, and affordable (semi-)high-throughput compound testing.
Introduction
Osteoporosis (OP) is a degenerative bone disease that affects around 27.6 million people over the age of 50 in the 27 European Union (EU27) countries alone (1). As average life expectancies increase, it is predicted that the annual cost of treating OP in the EU will rise from €37 billion in 2010 to €46.5 billion by 2025 (2). OP is characterized by a reduction in bone mineral density (BMD), reduction of bone mass (BM), and a decrease in the trabecular volume of long bones; resulting in brittle bones that are more prone to fracture (3). The underlying mechanism behind OP is a dysregulation of bone homeostasis; with decreased bone anabolism (decreased activity of osteoblasts and osteocytes) and increased catabolism (enhanced osteoclast activity). Successful treatment of OP should therefore increase bone anabolism and decrease catabolism to reinstate the equilibrium in bone homeostasis (4, 5). While therapeutic options are increasing, all but one available therapies aim to reduce bone resorption. However, as osteoclast and osteoblast activity are coupled, anti-resorptives can negatively affect anabolic osteoblast activity and may not fully restore bone architecture (6). The only injectable osteoanabolic compound, teriparatide, is an analog of the parathyroid hormone (7). However, it is not an ideal long-term therapy option as, not only is it expensive, long term exposures in rat increase susceptibility to osteosarcoma (8, 9) limiting treatment duration (currently 2-years) in OP patients (10). Thus, an ideal treatment plan should focus on both strengthening bones using an osteoanabolic compound, combined with use of an anti-resorptive treatment (also ideally non-invasive) to maintain bone integrity (5), few such options exist. Currently, a major bottleneck in the development of new pharmaceuticals is the collection of primary functional data on new biological drug targets with osteo-anabolic capacities.
The twinning of genetic information with mechanistic data is key for development of new treatments. For example, familial studies on high bone mass (HBM) cases led to the discovery of mutations in SOST (Sclerostin). Further mechanistic data generated in model systems showed that SOST acts negatively on the WNT signaling pathway and led to the development of a novel antibody treatment Romosozumab (approved in 2018 for clinical use), which blocks SOST activity (11–13). With the advent of genome-wide association studies (GWAS), and efficient whole-genome/exome sequencing (WGS/WES) data mapping there has been a sizeable increase in availability of human genetic data from cohort studies for musculoskeletal conditions including OP, high bone mass (HBM), and osteoarthritis (OA) (14–20). Recent large cohort studies, such as UK-Biobank, have identified many new loci that contain novel osteogenic factors. For example, the UK-Biobank (21) data yielded 518 loci associated with changes in BMD using heel ultrasound data (16, 19). Currently, there is a substantial gap in translating these human genetic findings to model systems (22) in which the mechanism by which these genes act on the skeleton can be defined, where hypotheses can be tested, and ultimately define new putative drug targets that can be assessed with pharmacological agents. Because the skeletal system involves complex interactions between different cell and tissue types, genes and mechanical stimuli it is difficult to recapitulate features of OP in a petri dish. However, traditional rodent models are expensive to genetically manipulate. Zebrafish (Danio rerio) could therefore bridge this gap by offering fast genetic manipulation and complex tissue interactions required to model complex diseases such as OP.
Zebrafish are vertebrates and show strong similarities in their skeletal physiology to mammals (23). They are highly fecund and a single pair of fish can lay up to 300 eggs a week, which develop externally and are translucent (24). They show conservation of 70% of all genes and 85% of disease genes with humans (25, 26). However, the main advantage of zebrafish for functional genetic studies is their genetic tractability, as constructs that modify the genome can be injected directly into embryos at the single cell stage. This has allowed the generation of transgenic lines that allow dynamic imaging of all the cells of the developing skeletal system in live larvae (27–29) (Table 1) and in more recent years allowed genome editing strategies to be employed. In this review we set-out these different approaches and how developing and adult zebrafish can be used to study bone mineralization, bone content formation, and osteoblast-osteoclast interactions in a whole animal context. We also discuss future prospects for drug screening pipelines in zebrafish which may confer advantages over other pre-clinical model systems.
Flexible Genetic Manipulation in the Zebrafish
Zebrafish are genetically high amenable and new ways to manipulate the genome are constantly being added to the zebrafish genetic toolbox, which includes knockout, knock-down and, DNA insertion strategies. The external development of the embryos allows tools targeting genes of interest to be microinjected directly in embryos at the 1-cell stage and hundreds of embryos can readily be injected in a morning. Acute knockdown of gene expression can be achieved either by targeting mRNA with antisense RNA morpholino (MO) molecules that stably bind the target mRNA to block translation or splicing through steric hindrance (41). MOs offer a rapid method to assess the phenotype of a gene of interest during early development. However, they can only be used to study developmental processes occurring over the first 4 or 5 days of development, which limits their utility in skeletal studies as mineralization occurs from 4 days of development. While concerns have been raised about MO veracity as morphants frequently show more severe phenotypes than stables mutants generated for the same gene (42, 43). This is due to a transcriptional compensation response for chronic loss of a gene as has been shown in mouse, cultured human cell lines, plants, and zebrafish models (44–51). Thus, while MOs have a role, their use has been largely supplanted by use of genome editing strategies.
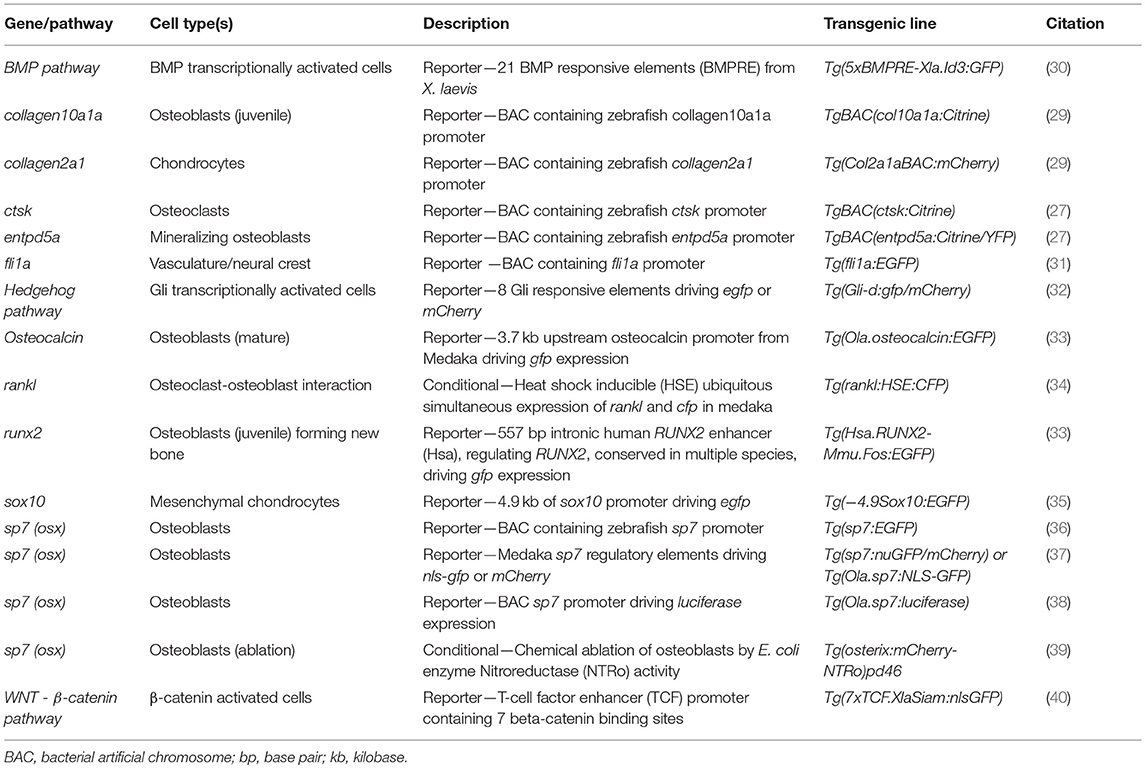
Traditionally, zebrafish mutant lines have been generated by forward genetic screening; using mutagens [e.g., N-ethyl-N-nitroso urea (ENU)] to induce random point mutations in offspring that were then screened for phenotypes of interest (52–56). The expansion of the zebrafish genetic toolkit with zinc-finger nucleases (ZFN), Transcription Activator-Like Effector Nucleases (TALEN) (57, 58), and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 (59) reverse genetic strategies, which, in combination with a fully sequenced genome (25), allow tailored gene-specific mutagenesis in the zebrafish. Gene function can be studied in genetic knockouts by generating insertion/deletion (indel) mutations leading to premature stop codons, deleting whole exons containing important protein domains and generate new stable mutant lines (Figure 1A). Moreover, the CRISPR/Cas9 protocol is so efficient that the F0 injected fish (crispants) can be used to study loss of gene function in these crispants, despite them carrying mosaic mutations (i.e., not every cell carries a mutation and more than one mutation may be present) (23, 60) (Figure 1B). Single base gene editing (knock ins) using modified Cas9 enzymes or supplying a DNA template for the endogenous homologous recombination machinery initiated by a double stranded break allows to introduce specific genetic changes to model specific human disease mutations in zebrafish orthologs (62, 63).
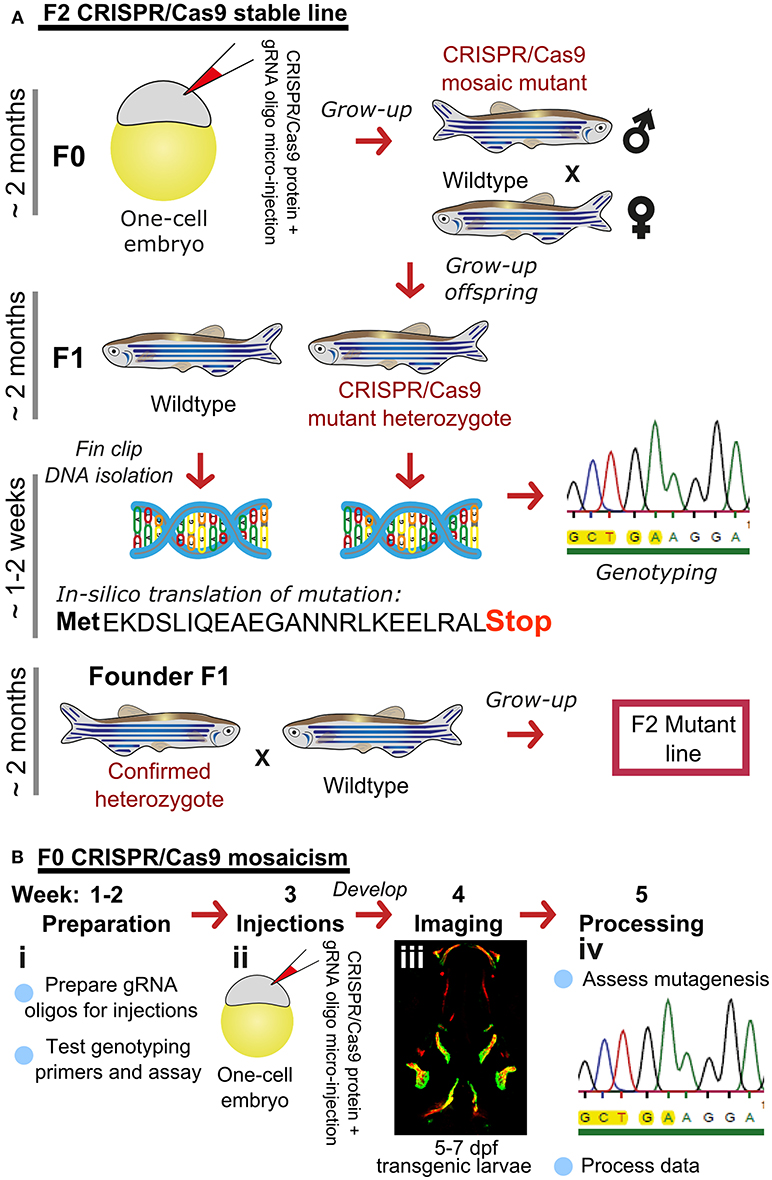
Figure 1. Rapid and efficient mutagenesis using CRISPR/Cas9 genome editing in zebrafish. (A) To generate a stable mutant line, F0 CRISPR/Cas9 injected individuals carrying mosaic mutations (defined by fin-clipping, B) should be outcrossed to wildtype fish to allow selection of a single germline mutation. Out-crossing the founder to wildtype will establish a stable F2 mutant line. Note that the F1 can have multiple founders with damaging mutations, incrossing these will result in F2 homozygotes (for recessive alleles) for functional analysis. When performing incrosses from F2, it will take another 2 months of breeding time. (B) This rapid protocol can be used to generate mutations in a gene of interest using CRISPR/Cas9 RNA or protein with gRNAs targeted against the gene from custom made gRNA oligos (i). Micro-injection of CRISPR/Cas9 RNA or protein and gRNAs specific to gene of interest into embryos at the single cell stage (ii) generating double stranded breaks during the first few rounds of cell divisions. The repair machinery is prone to errors and those cells will carry a different type of mutation giving a range of insertion and deletion (indel) mutations (spectrum of mutations, mosaicism). The overall mutagenic efficiency is typically high (around 80% with fragment analysis) allowing larval skeletal phenotypes to be assessed in the injected (F0) population (60). After imaging an Alizarin Red S (AR) stained individual in a transgenic background (here osteoblast marker sp7:gfp)(iii), mutagenesis assessment such as fragment analysis will determine a quantified mutagenesis rate (61) which can be correlated to a phenotype (iv). Note that mosaic mutants (crispants) can also be grown up to see the effect on the adult skeleton.
Simple Assessment of Zebrafish Bones During Development and Adulthood
Zebrafish in common with higher vertebrates, have both dermal/intramembranous ossification, in which bone is formed de novo directly by osteoblasts, and chondral/endochondral ossification in which bone forms by progressively replacing a cartilaginous template. Although zebrafish have thinner bones than terrestrial vertebrates, with fewer embedded osteocytes and little trabeculation, all of the relevant skeletal cell types and modes of regulation are conserved between zebrafish and higher vertebrates. This, importantly for the study of OP, includes osteoblast and osteoclast coupling and regulation of bone remodeling (64, 65).
A major advantage of using zebrafish to probe the mechanism of bone homeostasis is that cell behavior can be visualized dynamically in vivo. Zebrafish larvae are translucent and develop rapidly (24), and early skeletal processes can be dynamically visualized in the living fish through use of fluorescent transgenic reporter lines marking these cell types (see Table 1 for examples). Formation of the craniofacial skeleton occurs early, with the first cartilaginous structures of the jaw forming by 2 days post fertilization (dpf) (66), the first skeletal joints are formed and mobile by 3 dpf (60), by 5 dpf, hypertrophic chondrocytes, marked by col10a1a, are seen in some elements from 5 dpf (29), and first osteoblasts surrounding the cartilage and forming bone matrix by 7 dpf (67). The first intramembranous bones, such as the cleithrum, anterior notochord, and operculum, are visible in the craniofacial skeleton from 72 hpf (66). While skeletal development occurs early, true remodeling through the combined activity of osteoblasts and osteoclasts does not commence until the second week of development as osteoclasts (marked by Cathepsin-K (Ctsk) or TRAP) are not formed until day 10–12. Unlike mammals, mononucleated osteoclasts as well as multinucleated cells are present and actively resorb bone (65, 67).
There are many transgenic lines available to mark musculoskeletal tissues, these include reporter lines which label cells or signaling pathway activation by driving expression of proteins in the cytoplasm, targeted to the nucleus, or plasma membrane, and lines that tag proteins (28, 68). Reporter lines mark cell types by using a tissue specific promotor, responsive elements from a signaling pathway, or transcription factor binding sites controlling expression of a fluorescent protein (Table 1). For example, to study bone homeostasis, osteoblasts and osteoclasts can both be labeled in vivo, using osteoblast reporters such as sp7, and osteoclast reporters such as ctsk, so that their numbers, location and activity monitored in living bone tissue either longitudinally, in response to drug treatment, genetic mutation, or environmental stimuli (Table 1). Relevant to research into OP, osteoclasts can be specifically temporally activated by use of a heat shock promoter driving RANK ligand (rankl) expression; such that following a period of immersion in water at 39°C, osteoclast activity, labeled with the blue fluorescent protein (CFP), is increased resulting degradation of the bone matrix and in an osteoporotic phenotype of low BMD (34). A simple Alizarin Red S (AR) staining, which marks calcium phosphate crystals and fluoresces strongly in the red channel (580 nm wavelength), allows a rapid assessment of ossified elements in live or fixed fish. In combination with transgenic lines, endochondral ossification in the lower jaw (Figure 2A) and intramembranous bone formation in the operculum (Figure 2B) can be easily visualized compared to traditional rodent models.
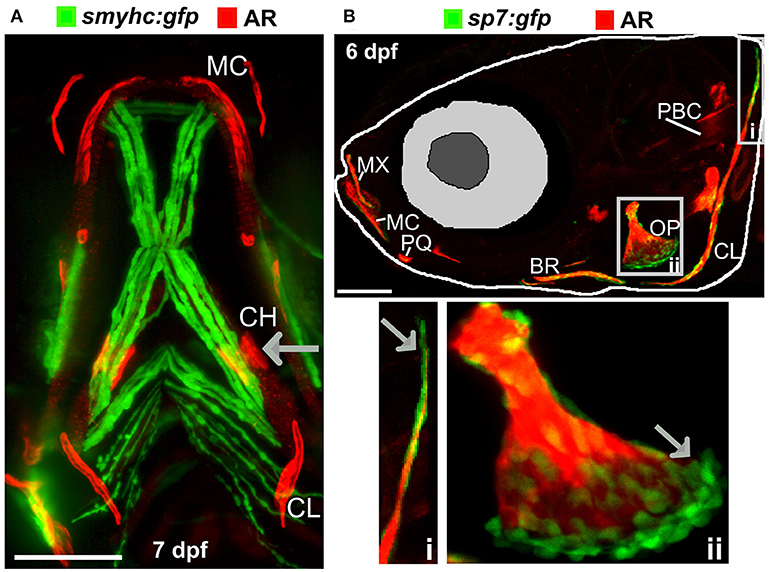
Figure 2. Ossified elements in the cranial region during early development. (A) Ventral view of a 7 days live Alizarin Red S (AR) labeled larval jaw showing dermal ossification of cleithrum (CL), and ossification of the cartilaginous ceratohyal (CH). Arrow indicates the CH which undergoes endochondral ossification. Slow muscle transgene reporter in green (smych:gfp). Image taken on a Leica lightsheet microscope. (B) Lateral view of a 6 days old larva live labeled with Alizarin Red S (red) and carrying GFP under the control of the osteoblast promoter s7/osterix (green; sp7:gfp) allowing visualization of mineralized elements (red) and osteoblasts (green) in a living individual. Insets show the cleithrum (i) and operculum (ii) with osteoblast enrichment at the distal ends of these elements (gray arrows). Image taken on a confocal microscope. Wildtype strains AB/TL in both panels. Ossified elements: BR, branchiostegal ray; CH, ceratohyal; CL, cleithrum; MC, Meckel's cartilage; MX, maxilla; OP, operculum; PBC, posterior basicranial commissure; PQ, palatoquadrate. Scale bars = 100 μm.
Imaging the Adult Skeleton for Assessing Mineralization
The zebrafish adult skeleton is relatively complex and once fully formed by around 2 months is composed of 74 ossified cranial elements (compared with 22 in humans), 28–31 vertebrae; 4 cervical, 10–11 thoracic vertebrae, and 15–16 separated vertebrae in the tail region and fins (pectoral, dorsal, anal (ventral), and caudal) (69). As in larvae, live AR and Calcein staining, or use of transgenic lines, allows easy detection of superficially located calcified elements in the skull, elasmoid scales, and fins using a simple fluorescent microscope. Deeper tissues can be imaged by multiphoton microscopy in small juveniles. However, bones located more internally (e.g., vertebrae and ribs) in large adults are difficult to visualize using this method. Post-mortem staining of bone (AR) and cartilage [Alcian blue (AB)] is a cost-effective way to analyse these structures for adult skeletal abnormalities (Figure 3A) and has been used in forward genetic screens to obtain detailed skeletal morphology information (56, 70, 71).
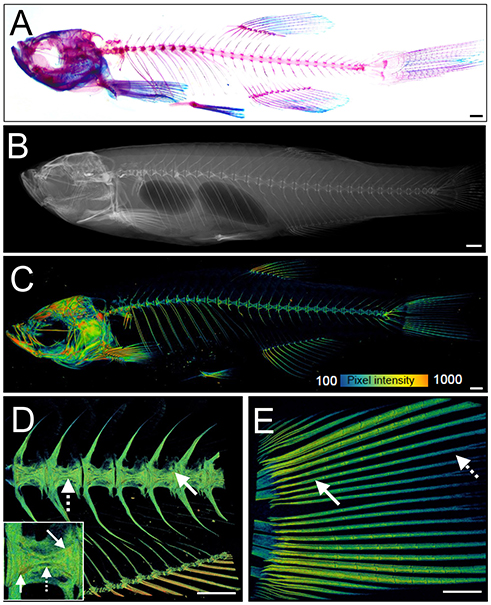
Figure 3. Examples of visualization and quantification of mineralized bone in zebrafish. (A) Wholemount Alizarin Red S (AR) and Alcian Blue staining of 3 months fixed fish. (B) Radiograph of 1-year old live fish showing whole body: endo- and exoskeleton. (C) Low resolution μCT images acquired with a 20 μm voxel size of a 3 months old fish. Note that pixel intensity can be used to determine BMD; represented on the color coded pixel intensity bar. (D,E) High resolution (5 μm voxel size) μCT images of vertebral column with anal fin rays (D) and caudal fin rays (E). Vertebral centrae have higher density at their edges (solid arrow) than the center (dashed arrow). In the fin rays, a higher density (solid arrow) is observed in older segments within the proximity to the body in comparison to younger segments located more caudally showing lower pixel intensity (dashed arrow). The same pixel intensity color coding as (C) applies. All fish and their insets are depicted from a lateral view in an anterior-posterior (left-right) orientation. Scale bars = 50 μm in (A,B); and 100 μm in (C–E).
Recent advances in X-ray based imaging: radiographs, micro-computed tomography (μCT), and synchrotron equipped μCT technologies (SR-μCT), and their subsequent downstream imaging processing, opened avenues to assess the adult zebrafish skeleton. The major advantage of using these X-ray imaging techniques is that they are non-destructive and can be used in the intact fish, allowing the samples to be used for other purposes, such as histology. Radiographs give two-dimensional (2D) images of the zebrafish skeleton at relatively low resolution (Figure 3B), permitting the visualization of bone elements and a broad evaluation of changes in the skeleton, radiographs can be used to image live anesthetized fish permitting longitudinal analysis of the skeleton over time (70). Higher resolution (2 μm voxel size) and three-dimensional (3D) assessment of the zebrafish skeleton can be achieved by using μCT (Figures 3C–E). As fish bone, like that of mammals, is composed of hydroxyapatite crystals, quantification of BMD can be performed by comparison to phantoms, which are samples of known hydroxyapatite content (72). Additionally, treatment with agents to improve contrast such as silver nitrate (AgNO3) or iodine, allow detection of juvenile (less dense) bone and of soft tissues such as muscle and cartilage (72).
Very detailed data on bone micro-architecture can be achieved with SR-μCT (73, 74). This technique can yield a spatial resolution of 100 nanometres on tissue samples and visualize fine bone structures at a cellular level including the vasculature in mineralized bone, osteoclast resorption pits, and osteocyte lacunae (75). As the size and resolution of data sets increase, the bottleneck in the process is frequently data analysis. Commercially available software packages such as “boneJ” are tailored for CT data analysis, and recently open source user friendly software have become available to process μCT data from zebrafish scans. For example, the “FishCut” software processes whole-body μCT scan datasets and applies semi-automated analysis algorithms. The current version segments the axial skeleton, then generates values for the surface area of vertebrae and centrae, and calculates BMD and mineralized thickness in a semi-automated fashion (76).
Zebrafish Mutants of Brittle or Thin Bones
An increasing number of zebrafish genetic mutants in skeletally relevant genes have been shown to recapitulate human bone disease. These have provided insight into the dynamic regulation of bone formation, mineralization, and remodeling. We have included a list of zebrafish skeletal mutants in Table 2. While there are currently few models for OP, there are various zebrafish mutant lines that accurately model human skeletal dysplasias, including collagenopathies and forms of osteogenesis imperfecta, which are characterized by brittle bones and frequent low-impact bone fractures. Autosomal dominant mutations COL1A1 and COL1A2 genes predominantly affect glycine-X-Y (Gly-X-Y) repeat domains that result in collagen α1(I) and α2(I) heterotrimer maturation defects (119), causing fragile bone matrix and insufficient mineralization (120). The Gly-X-Y mutations lead to impaired hydroxylation and defects in collagen maturation in the endoplasmic reticulum (ER), which is also conserved in zebrafish (121–123). The autosomal dominant chihuahua (chi) zebrafish mutant, was identified in a forward genetic screen using radiography (70). Linkage mapping identified a mutation causing a glycine to aspartate amino acid substitution in a conserved Gly-X-Y repeat of col1a1a (zebrafish col1a1 is duplicated). Note that in contrast to mammals, zebrafish type-I collagen is constituted by three different α chains [α1 (col1a1a), α3 (col1a1b), α2 (col1a2)] due to duplication (124). chi/+ zebrafish display phenotypes resembling those seen in humans, including a shortened axial skeleton, with irregular radiodensity, uneven mineralization, and brittle bones that fracture easily (especially ribs). Transmission electron microscopy revealed that chi/+ fish show signs of ER stress (70). The ER trapping of insufficiently hydroxylated oligotrimerized α1(I)/α2(I)/α3(I) collagen leads to lower extra-cellular collagen maturity, abnormally shaped and thinner vertebrae bodies, areas of higher calcium content, different local mechanical properties, and reduced osteocyte number (84). Osteogenesis imperfecta has a broad disease spectrum in the clinic, and recent comparative studies of multiple mutant alleles for col1a1a, col1a1b, col1a2, and also bmp1a (described later) and plod2 described a diversity of skeletal phenotypes (Table 2) with brittle bones as the common feature (85).
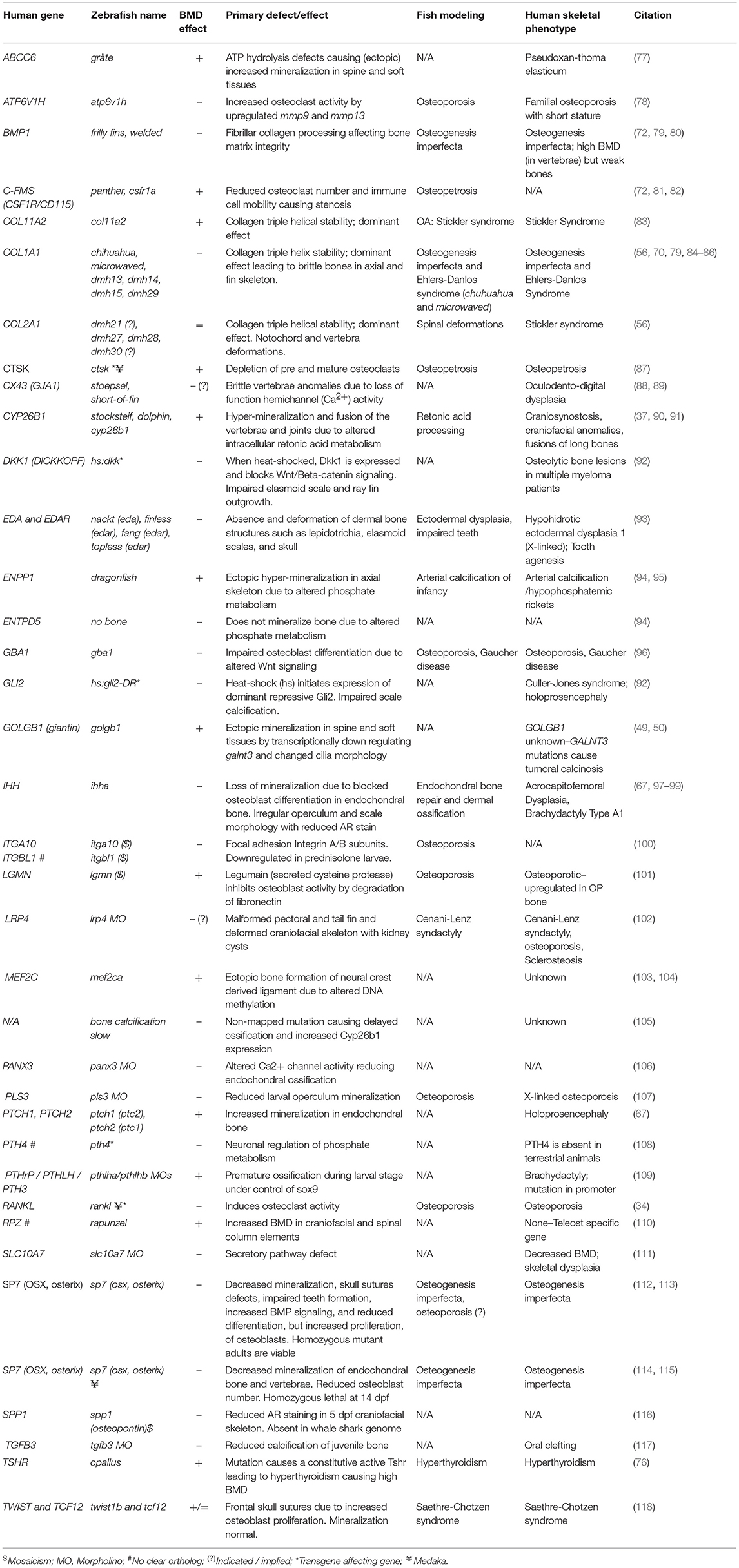
Table 2. Zebrafish mutants, transgene insertion mutants, and morphants showing altered skeletal mineralization.
The zebrafish sp7/osterix mutant has been shown to model human osteogenesis imperfecta caused by recessive damaging mutations in SP7 (125). This mutant showed uneven mineralization, severe fractures caused by minimal impact, and misshapen bones. Moreover, rare craniofacial characteristics caused by impaired SP7 function, such as wormian bones, reported in human patients carrying mutations in SP7 were also observed in zebrafish (112).
Another example of a zebrafish mutant that recapitulates patient phenotype is the bmp1a mutant frilly fins (frf). In humans a damaging missense mutation in the BMP1 signal peptide causes brittle bones in an osteogenesis imperfecta pedigree (79). frf mutants showed normal osteoblast number, but pericellular pro-collagen processing (C-pro-peptide removal) defect leading to mineralization defects in the axial skeleton and fin rays (79).
Collagenopathies, such as Stickler Syndrome, have also been successfully modeled in zebrafish. We have recently reported a col11a2 zebrafish mutant showing specific traits of the human disease which include thicker collagen fibers and degradation of type-II collagen in zebrafish larvae leading to compromised jaw shape, mechanical properties and movement of the jaw leading to premature OA (83). In many skeletal dysplasias zebrafish not only model the human condition but allow mechanistic insight into how genetic changes lead to the cellular changes that underpin the disease symptoms. As such zebrafish offer exciting prospects for delivering functional studies in new osteoporotic genetic loci.
Assays of Caudal Fin Regeneration and Fracture Repair to Asses de novo Bone Matrix Formation
Zebrafish are capable of regeneration many tissues and organs including the heart, lens, and pancreas. They also show regeneration of skeletal tissues following amputation of the tail fin (lepidotrichia) or removal of elasmoid scales (126, 127). As the fins and scales are translucent, and readily imaged they allow cells and their calcified matrix to be visualized in detail using standard fluorescent microscopes (Figure 4A). After amputation of a ray fin (typically a caudal fin), a wound healing response results in the formation of an epimorphic blastema which regenerates all affected tissues of the amputated organ, including bone, in a controlled fashion (128). Following this inflammation response, osteoblasts undergo dedifferentiation and proliferate to contribute to the blastema (33, 129). These juvenile osteoblasts then secrete matrix with intermediate properties between cartilage and bone and are later remodeled as mature bone by matured osteoblasts and recruited osteoclasts (Figure 4B) (33, 128). These fins can also be injured via cryo-injury by placing a −196°C knife perpendicularly to the caudal fin rays allowing to study the bone resorption response (130). These techniques offer great perspectives to compare bone formation and bone remodeling, in an adult context.
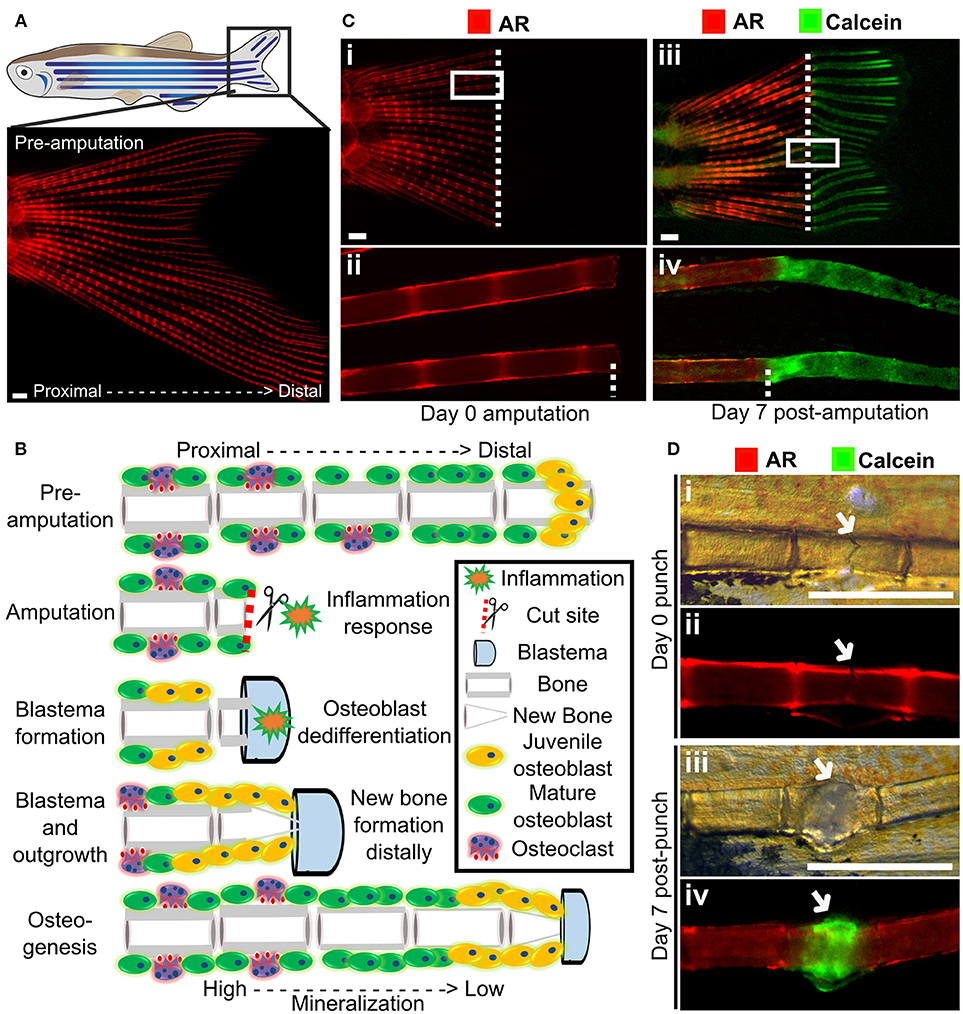
Figure 4. Fin regeneration and fracture assay to visualize and quantify live bone formation and repair. (A) Schematic representation of a zebrafish with a standard fluorescent stereomicroscope image of a live Alizarin red S (AR) pre-amputation caudal fin (inset). (B) Schematic representation of bone regeneration after fin amputation showing the (simplified) cascade of events that follow after fin amputation to regenerate bone (a single ray depicted here). This allows studying de novo bone formation by newly formed osteoblasts (orange cells) and differentiated osteoblasts (green cells) and subsequent remodeling by osteoblasts and osteoclasts (purple cells) in an adult fish. Note that during osteogenesis that there is a gradient of mineralization. (C) Live images of the tail fin labeled with Alizarin red (red) prior to amputation (i, ii) and Calcein (green) post-amputation (iii, iv) taken on a fluorescent dissecting microscope. All images in panel come from the same fish. Seven days post-amputation showing regrowth of new bone (green). Note that intense Calcein staining is visible distally from the amputation site (white dotted line). (D) The fracture healing assay involves applying pressure on a fin ray bone element to induce a small fracture to one segment of the fin ray (i), which is visible with life AR staining (ii). Green Calcein labels the new bone formed in the fracture callus by 7 days (iii and iv). The white arrow indicates the fracture site. Scale bars = 500 μm, 3 months old wildtype TL/EKK females.
Using transgenic lines and in vivo staining methods, such as AR (fluoresces red, 545 nm excitation, 580 nm emission) and Calcein (fluoresces green, 495 nm excitation, 515 nm emission), which binds to calcified matrix, the dynamics of bone formation can be visualized by using a fluorescent stereomicroscope in a regenerating caudal fin of a living fish. This allows longitudinal analysis by following regeneration rate and volume, since AR stains fully mineralized bone and Calcein binds to newly deposited bone matrix (Figure 4C).
The utility of fin regeneration assays to test bioactive compounds has been demonstrated by treating regenerating fins with the glucocorticoid prednisolone. Following treatment bone formation was reduced, and furthermore, both osteoblast number and subsequent bone deposition and osteoclast recruitment was reduced in these fins (131). Interestingly, skull injury repair is less affected following prednisolone treatment (131), this is similar to mammals. Treatment of fins with Botulinum toxin (Botox) leads to a reduction in bone mineralization and regeneration following amputation (132), comparable to the situation in mammals where fracture repair is impaired following Botox induced paralysis (133, 134).
A major issue with OP is increased fracture risk due to weaker bone structure, and therefore identification of therapeutics that can improve fracture healing is desirable. Zebrafish show a fracture healing response, including callus formation (Figure 4D), with strong similarities to that of mammals. Fractures can be induced in zebrafish fins using simple pressure applied externally to the fin (131, 135). As the fin has around 300 bony rays, multiple fractures can be induced in a single fin. A fracture callus is formed and de novo bone formation is initiated 2 days post-injury accompanied by an increased expression of osteoblast genes such as runx2 and sp7/osx (131, 135). As the fin is flat, the fracture repair process can be dynamically tracked at cellular resolution using transgenic lines (Table 1) or by labeling bone formation with AR and Calcein (Figure 4D). As for regeneration, it is possible to add pharmacological agents to the regenerating tissue (131), allowing potential osteoanabolic compounds to be tested for beneficial effects in fracture repair in vivo (136).
Skeletal Assays Using Elasmoid Scales
The body of zebrafish is covered with elasmoid scales made of calcified dermal bone harboring osteoblasts and osteoclasts (Figures 5A–C). The calcified matrix is composed of a plywood structure of collagen fibrils (137), which are easily visualized with second harmonics generation microscopy (Figure 5D). Scales are embedded in, and grow from, the dermis and shed and replace naturally throughout life of the fish (138). As scales are part of the exoskeleton they are easy to collect from an anesthetized fish. Each flat scale is subdivided in four regions by its morphology: anterior, lateral, central, and central with epidermis (Figure 5A) (139). The anterior region is attached to the skin and does not grow or form new bone. The lateral area is characterized by its curved ridges (circuli), whereas the central area has linear trenches. Within the lateral circuli and central grooves newly mineralized matrix is formed by osteoblasts (139) and degraded by osteoclasts (140). The posterior area has increased osteoblast number and bone is continuously deposited (Figures 5A,B). Osteoblasts in different regions of the scale express different markers of maturity (97). As the scale contains living cells, including nerve and vascular endothelial cells, their use offers an opportunity to study bone cell behavior in a mature context.
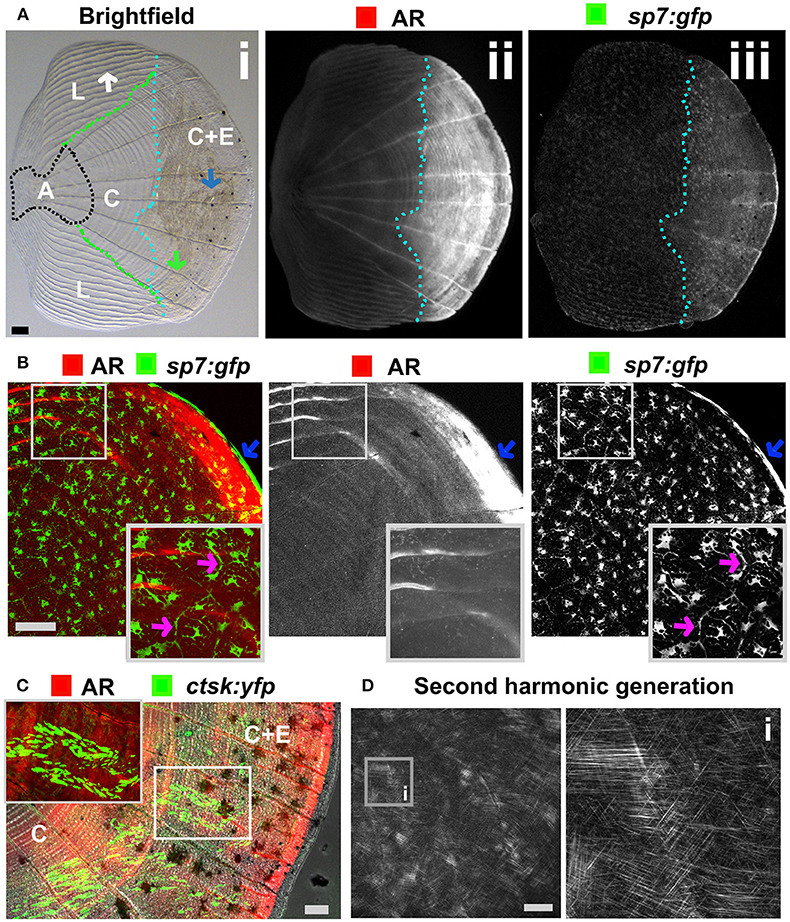
Figure 5. Zebrafish elasmoid scale structure and bone cell types. (A) Single scale from the flank of a 3 months old fish carrying the sp7:gfp osteoblast reporter transgene (green) and stained for Alizarin Red S (AR, red). Whole scale is shown in bright field (i) and gray scale images for AR (ii) and GFP (iii) in the top panels. The brightfield image (i) depicts the anterior anchor region (A, black dotted line boundaries), the lateral circuli (L, green dotted line boundaries, white arrow), central region (C, surrounded by black, green, and light blue dotted lines), and central region covered by epidermis (C+E, light blue dotted line, with grooves by green arrow) with enhanced mineralization. (B) Confocal images showing a merge image of osteoblasts (sp7:gfp transgenic fish, green) abundantly distributed over the freshly harvested scale and AR staining (red). Individual channels are depicted in gray scale images. Note increased mineralization at the edge of the scale corresponding increased GFP presence (blue arrows). Insets focus on the lateral circulus and note osteoblast cytoplasmic protrusions (pink arrows). (C) Confocal images visualizing osteoclasts with cathepsin K (ctsk) YFP reporter expression (green), mineralization by AR (red), and brightfield (gray). Note that YFP positive cells were predominantly seen in the central region with epidermis (C+E) and distal edges of the central region (C). (D) Multiphoton forward scattering (second harmonic generation (SHG), 880 nm wavelength) visualizes collagen fibrils in an ethanol fixed scale. Inset (i) shows the organization of collagen fibrils in a plywood structure. Wildtype strains (panel): TL/EKK (A), TL (B), AB/TL (C). Scale bars 100 μm.
Pharmacological Manipulation of Bone Tissue and Chemical Genetic Screening
As larvae are small and develop in water, it is possible to grow larvae in multi-well format with the addition of water-soluble compounds to their growth media for easy uptake. Zebrafish have been used extensively for high-throughput screening using larvae and now drugs are used in clinical studies that were first identified in zebrafish. A great example is the identification of the kinase inhibitor dorsomorphin (BMP type-1 receptor (BMP1R) antagonist) to treat lymphoma which was discovered in an early embryogenesis phenotype screen using 7,500 small-molecules (141, 142). Another example used semi-automated imaging strategy of Calcein stained larvae exposed to a small-compound library identifying 6 catabolic and 2 anabolic compounds that alter notochord mineralization (143) (Table 3). Thus, when fluorescent compounds are twinned with fluorescent reporters for osteoblasts (e.g., sp7:gfp with AR) (Figure 2B), it will allow assessment of osteoblast number and activity in a semi-high content setting using plate imaging microscopy (162). When these assays are combined with high efficiency CRISPR/Cas9 genome engineering strategies, it will open avenues to test compounds of interest that could alter disease causing mutations deteriorating effects. Thus, this comprehensive approach will also offer opportunities to develop compounds for personalized medicine. For OP research it may be more advantageous to focus on adult skeletal assays to allow assessment of osteoclast activity (bone catabolism) simultaneously with an assessment of osteoblasts (bone anabolism). An example of pharmaco-genetics improving brittle bones, is when type-I collagen secretion in the bone matrix is ameliorated by treating chi/+ mutants with 4 phenyl butyrate (4PBA) compound (86).
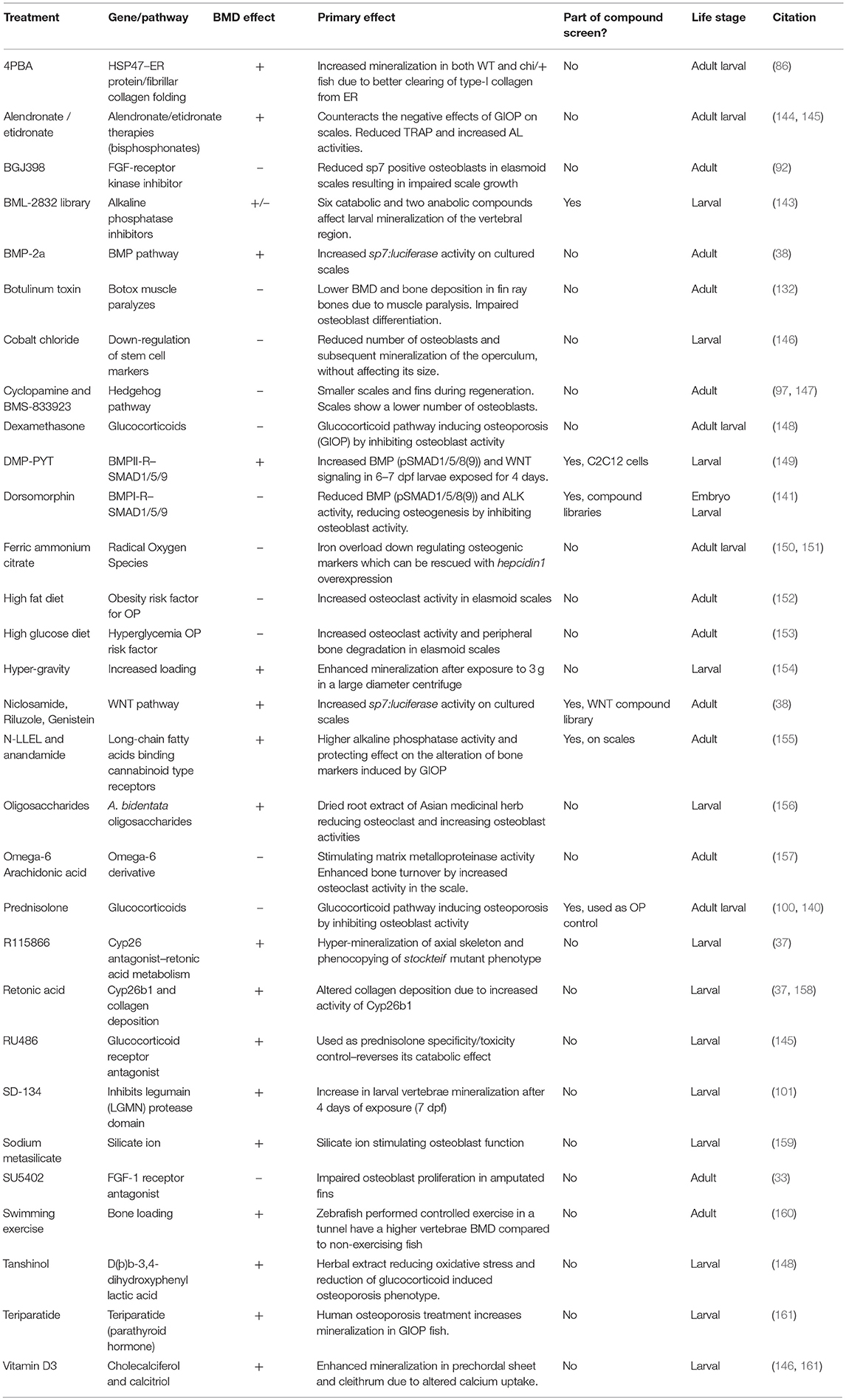
Table 3. List of compounds, diets, and exercise that alter ossification in zebrafish larvae, adults, and/or adult elasmoid scales.
Zebrafish elasmoid scales are bony plates that are small and contain bioactive osteoblasts and osteoclasts (Figures 5B,C). These therefore offer huge potential as a primary pharmacological screening tool for skeletal compounds. The scales can be cultured for 72-h post-harvesting during which they can arrayed in multi-well plates and exposed to pharmacological compounds. As the scale is thin, osteoblasts are accessible to osteogenic factors, and have been demonstrated to react in a dose dependent manner to BMP-2 (38). To model an OP-like phenotype, individuals can easily be exposed to prednisolone / dexamethasone (glucocorticoid pathway) (140, 148), ferric ammonium citrate (150, 151), or metabolically with a high fat or glucose diet (152, 153), see also Table 3. In the context of glucocorticoid induced OP (GIOP), the bisphosphonate Alendronate reverses the effects of prednisolone on ex vivo cultured elasmoid scale bone, which showed a reduction in osteoclast activity (measured by TRAP) and an increase in bone anabolism (measured by alkaline phosphatase activity) (144); the same response as in mammals (163, 164). As fat metabolism has been implicated with OP, a small fatty acid derivative library was used on GIOP adult fish. Biochemical assays on scales derived from these fish showed that cannaboid receptor 2 binding anandamide and N-linoleoylethanolamine (N-LLEL) fatty acids drive osteogenesis by stimulating alkaline phosphatase (ALK) activity (155).
A WNT-pathway compound library was tested to identify new osteo-anabolic compounds using an assay in which luciferase was expressed under control of the sp7 promoter allowing a quantitative readout of osteoblast activity (Figure 6). This screen identified three osteo-anabolic (Table 3) and 15 osteo-catabolic compounds from 85 trial compounds (38). This library contained five previously published compounds tested in vivo, and nine tested in vitro mammalian bone progenitor cell lines. Strikingly, this scale luciferase assay was able to reproduce the effect of all in vivo tested compounds and about half of all in vitro tested compounds (38). These studies demonstrate the exciting potential that scale assays represent for testing of skeletal compounds relevant to OP in a cost-effective manner.
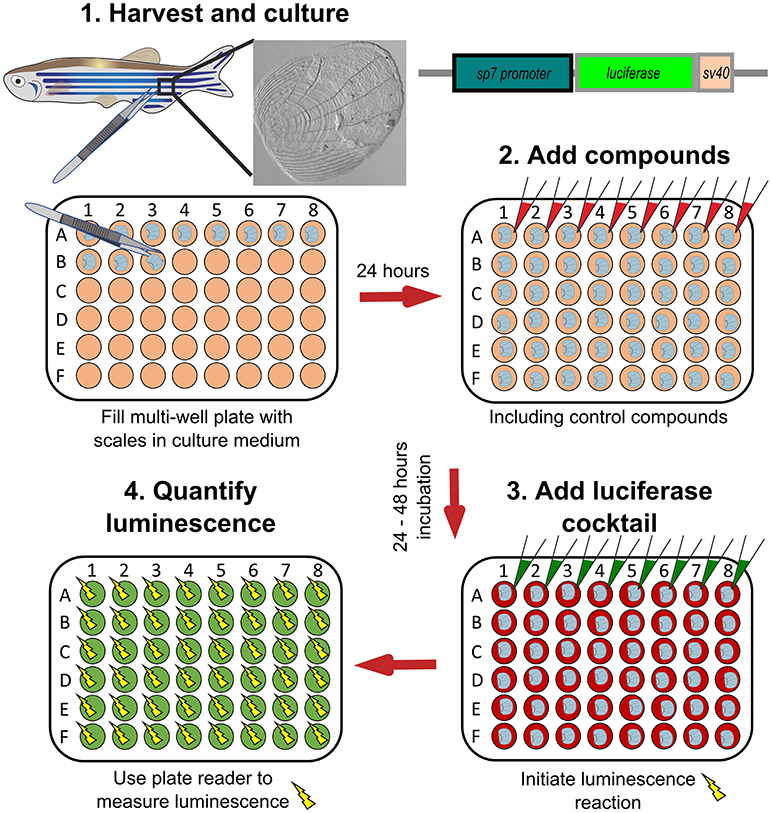
Figure 6. Schematic representation to show how osteoblast activity can be quantified from scales. Scales from sp7:luciferase transgenic reporter fish are harvested from the lateral flanks of a fish, then cultured in multi-well plates with DMEM culture medium (orange wells) at 28°C for 24 h. Compounds of interest can then be added (red wells) to the scales and incubated prior addition of a luciferin cocktail (green wells) and measurement of luciferase activity with a luminescent (yellow sparks) plate reader. Based on text from de Vrieze et al. (38).
Potential Drug Discovery Pipeline for Osteoporosis
Recently, there has been a substantial expansion in the quantity of high-quality genetic data from large-scale human genomic and transcriptomic studies that contain potential osteo-anabolic factors. Here we describe a potential screening pipeline that makes use of the genetic tractability and imaging in zebrafish to offer a relatively low cost, high-throughput option compared to traditional in vitro and in vivo models (Figure 7).
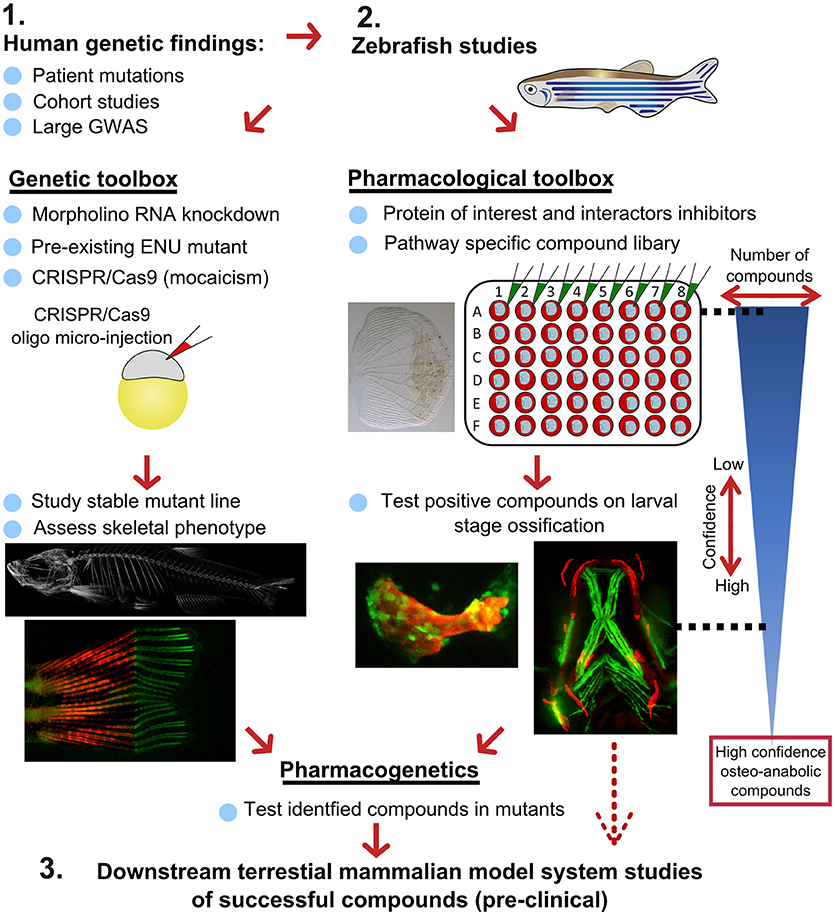
Figure 7. Proposed pipeline using zebrafish as a primary testing platform to address bottleneck for fast and affordable translation of human genetic findings. Two experimental arms using the genetic and pharmacological toolboxes allow simultaneous drug target validation. The blue reversed triangle depicts the reduction in number of putative osteo-anabolic compounds (along with an increase in confidence) when testing the compounds using the skeletal assays available.
After identification of several candidate genes/drug targets from human genetic studies, the pipeline consists of two experimental arms that can be carried out simultaneously to generate primary pre-clinical data to validate the putative drug targets. Using genome editing, loss-of function studies can be performed in transgenic backgrounds to test the effect of the gene of interest on the developing skeleton or on mineralization, and simultaneously allowing safety testing for deleterious effects on other tissues or organs. For example, using CRISPR/Cas9 editing, it is possible to generate hundreds of mosaic zebrafish mutants within 3–4 weeks (includes the generation of the targeting reagents), which is difficult to achieve in other available systems, such as cultured chondrocytes and osteoblasts (differentiation of these takes multiple weeks). With CRISPR/Cas9 editing it is also feasible to study the specific human disease mutation in zebrafish, as long as it is in a conserved coding region. These fish can be grown to adulthood and germline mutations identified allowing more detailed studies on the mature skeleton to be performed (Figure 7).
In addition to genetic studies, pharmacological assessment of the identified putative drug target can be performed. By using water-soluble compounds, or lipid soluble compounds dissolved in DMSO, screens in a multi-well format can be performed using ex vivo culture of elasmoid scales. As a single adult fish has around 200 scales (138), this assay allows testing of many compounds, including control compounds (e.g., osteo-anabolic (alendronate) and -catabolic (prednisolone), on scales harvested from a single individual, reducing intra-individual variation (38). Therefore, this technique offers a platform to generate a primary read-out of novel osteo-active compounds in the context of homeostasis in a mature tissue. Additionally, this ex vivo technique will reduce the number of (potentially harmful) compounds being exposed to living fish, therefore contributing to ethical refinement and reduction of experimental animal use, but also reducing associated costs. As this scale assay reduces the number of putative osteogenic compounds substantially, these positive compounds can be further validated (along with safety testing) on developing transgenic larvae. These larvae would be plated out at 3 larvae per well and the compounds added from 3 days of development, with high-content imaging used for preliminary assessment of the effects of each compound and more detailed analysis including dose response followed up for validated positive hits. Further downstream tests, such as fin regeneration or fracture assays, can further reduce the number of compounds as such that only high-confidence compounds will be assessed in tetrapod pre-clinical studies (Figure 7).
If desired, the two experimental arms can be performed simultaneously, so that stable mutants are being generated during the compound testing phase. This opens the possibility to perform pharmacogenetic experiments in a relatively short time frame to validate the effects of putative drugs on specific disease mutations to see if they can “rescue” the disease phenotype (Figure 7). Together, zebrafish offer the potential in future to bridge the gap between human genetic hits, and fast functional validation.
Prospects for Zebrafish in Osteoporosis Research
The zebrafish is a well-established increasingly used animal model for studying various diseases including (congenital) metabolic bone diseases (165). As zebrafish have historically been mainly used for its fast-embryonic development properties to better understand disease onset, zebrafish aging studies have only recently been conducted to model age-related diseases such as OA and OP. OP is an emerging field in zebrafish modeling and more research is needed to fully establish an OP-like phenotype as it was previously determined in its teleost cousin medaka (34). The advantageous properties as set-out in this review should be further exploited to benefit drug development for OP. Zebrafish show the appropriate response to increased mechanical loading, where the cellular (transcriptional) response initiates increased bone formation and mineralization in the loaded bone elements that are easily quantified (154, 160). However, since zebrafish and mammalian bone morphology show some differences (64), a pharmacological assay should particularly focus on the complex tissue and osteoblast-osteoclast interactions that underpin OP pathology. As traditional rodent and in vitro co-culture both have limitations to pursue large-scale drug discovery in a genetic context, zebrafish can take the place as a primary testing platform and therefore opening avenues to work toward gene specific compound discovery that have been identified as risk factors in human genetic studies. After primary safety testing, these identified compounds can be further tested in mammalian OP models to determine the effect on BMD, bone strength, and trabeculation. Fully exploiting these opportunities by using zebrafish as a primary screening model will open exciting avenues to perform pharmacogenetics for OP on a larger scale.
Ethics Statement
Zebrafish procedures were approved by the University of Bristol Animal Welfare and Ethical Review Body (AWERB) and performed in accordance with a UK Home Office project license.
Author Contributions
DB and EK generated the figures for the manuscript. DB, EK, and CH researched and drafted the manuscript together.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the Wolfson Bioimaging Facility for confocal microscope (Leica SP5II) access and imaging support; in particular Katy Jepson for assistance with lightsheet (Zeiss Z1 lightsheet funded by the Wellcome Trust) imaging, and Dom Alhibi and Dr. Nicola Stevenson for assistance with SHG imaging (acquired on Leica SP8 AOBS CLSM with multiphoton lasers funded by the MRC). Faxitron is acknowledged for trialing the digital X-ray machine (Multifocus). We gratefully acknowledge Versus Arthritis (formerly known as Arthritis Research UK) for their funding of CH, EK, and DB (21211, 21937, 19476), and The Elizabeth Blackwell Institute for Health Research (University of Bristol) and the Wellcome Trust Institutional Strategic Support Fund for the postgraduate extension (discipline hopping) fellowship (204813/Z/16/Z) that funded DB.
References
1. Hernlund E, Svedbom A, Ivergard M, Compston J, Cooper C, Stenmark J, et al. Osteoporosis in the European Union: medical management, epidemiology and economic burden. A report prepared in collaboration with the International Osteoporosis Foundation (IOF) and the European Federation of Pharmaceutical Industry Associations (EFPIA). Arch Osteoporos. (2013) 8:136. doi: 10.1007/s11657-013-0136-1
2. Svedbom A, Hernlund E, Ivergard M, Compston J, Cooper C, Stenmark J, et al. Osteoporosis in the European Union: a compendium of country-specific reports. Arch Osteoporos. (2013) 8:137. doi: 10.1007/s11657-013-0137-0
3. Kanis JA, McCloskey EV, Johansson H, Cooper C, Rizzoli R, Reginster JY, et al. European guidance for the diagnosis and management of osteoporosis in postmenopausal women. Osteoporos Int. (2013) 24:23–57. doi: 10.1007/s00198-012-2074-y
4. Baron R, Hesse E. Update on bone anabolics in osteoporosis treatment: rationale, current status, and perspectives. J Clin Endocrinol Metab. (2012) 97:311–25. doi: 10.1210/jc.2011-2332
5. Garber K. Two pioneering osteoporosis drugs finally approach approval. Nat Rev Drug Discov. (2016) 15:445. doi: 10.1038/nrd.2016.132
6. Khosla S, Hofbauer LC. Osteoporosis treatment: recent developments and ongoing challenges. Lancet Diabetes Endocrinol. (2017) 5:898–907. doi: 10.1016/S2213-8587(17)30188-2
7. Metcalf LM, Aspray TJ, McCloskey EV. The effects of parathyroid hormone peptides on the peripheral skeleton of postmenopausal women. Syst Rev Bone (2017) 99:39–46. doi: 10.1016/j.bone.2017.03.007
8. Vahle JL, Long GG, Sandusky G, Westmore M, Ma YL, Sato M. Bone neoplasms in F344 rats Given teriparatide [rhPTH(1-34)] are dependent on duration of treatment and dose. Toxicol Pathol. (2004) 32:426–38. doi: 10.1080/01926230490462138
9. Watanabe A, Yoneyama S, Nakajima M, Sato N, Takao-Kawabata R, Isogai Y, et al. Osteosarcoma in Sprague-Dawley rats after long-term treatment with teriparatide (human parathyroid hormone (1-34)). J Toxicol Sci. (2012) 37:617–29. doi: 10.2131/jts.37.617
10. Ishtiaq S, Fogelman I, Hampson G. Treatment of post-menopausal osteoporosis: beyond bisphosphonates. J Endocrinol Invest. (2015) 38:13–29. doi: 10.1007/s40618-014-0152-z
11. Balemans W, Ebeling M, Patel N, Van Hul E, Olson P, Dioszegi M, et al. Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST). Hum Mol Genet. (2001) 10:537–43. doi: 10.1093/hmg/10.5.537
12. McClung MR, Grauer A, Boonen S, Bolognese MA, Brown JP, Diez-Perez A, et al. Romosozumab in postmenopausal women with low bone mineral density. N Engl J Med. (2014) 370:412–20. doi: 10.1056/NEJMoa1305224
13. McClung MR. Romosozumab for the treatment of osteoporosis. Osteopor Sarcop. (2018) 4:11–5. doi: 10.1016/j.afos.2018.03.002
14. Gregson CL, Steel SA, O'Rourke KP, Allan K, Ayuk J, Bhalla A, et al. ‘Sink or swim': an evaluation of the clinical characteristics of individuals with high bone mass. Osteopor Int. (2012) 23:643–54. doi: 10.1007/s00198-011-1603-4
15. Gregson CL, Wheeler L, Hardcastle SA, Appleton LH, Addison KA, Brugmans M, et al. Mutations in known monogenic high bone mass loci only explain a small proportion of high bone mass cases. J Bone Min Res. (2016) 31:640–9. doi: 10.1002/jbmr.2706
16. Kemp JP, Morris JA, Medina-Gomez C, Forgetta V, Warrington NM, Youlten SE, et al. Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat Genet. (2017) 49:1468–75. doi: 10.1038/ng.3949
17. Gregson CL, Newell F, Leo PJ, Clark GR, Paternoster L, Marshall M, et al. Genome-wide association study of extreme high bone mass: contribution of common genetic variation to extreme BMD phenotypes and potential novel BMD-associated genes. Bone (2018) 114:62–71. doi: 10.1016/j.bone.2018.06.001
18. Kim SK. Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture. PLoS ONE (2018) 13:e0200785. doi: 10.1371/journal.pone.0200785
19. Morris JA, Kemp JP, Youlten SE, Laurent L, Logan JG, Chai R, et al. An atlas of human and murine genetic influences on osteoporosis. bioRxiv [preprint] (2018). doi: 10.1101/338863
20. Tachmazidou I, Hatzikotoulas K, Southam L, Esparza-Gordillo J, Haberland V, Zheng J, et al. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank. bioRxiv [preprint] (2018). doi: 10.1101/453530
21. Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. (2015) 12:e1001779. doi: 10.1371/journal.pmed.1001779
22. Cannon ME, Mohlke KL. Deciphering the emerging complexities of molecular mechanisms at GWAS loci. Am J Hum Gene (2018) 103:637–53. doi: 10.1016/j.ajhg.2018.10.001
23. Mackay EW, Apschner A, Schulte-Merker S. A bone to pick with zebrafish. Bonekey Rep. (2013) 2:445–445. doi: 10.1038/bonekey.2013.179
24. Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. (1995) 203:253–310. doi: 10.1002/aja.1002030302
25. Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature (2013) 496:498–503. doi: 10.1038/nature12111
26. Kettleborough RN, Busch-Nentwich EM, Harvey SA, Dooley CM, de Bruijn E, van Eeden F, et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature (2013) 496:494–7. doi: 10.1038/nature11992
27. Bussmann J, Schulte-Merker S. Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development (2011) 138:4327–32. doi: 10.1242/dev.068080
28. Hammond CL, Moro E. Using transgenic reporters to visualize bone and cartilage signaling during development in vivo. Front Endocrinol. (2012) 3:91. doi: 10.3389/fendo.2012.00091
29. Mitchell RE, Huitema LF, Skinner RE, Brunt LH, Severn C, Schulte-Merker S, et al. New tools for studying osteoarthritis genetics in zebrafish. Osteoarthr Cartil. (2013) 21:269–78. doi: 10.1016/j.joca.2012.11.004
30. Alexander C, Zuniga E, Blitz IL, Wada N, Le Pabic P, Javidan Y, et al. Combinatorial roles for BMPs and Endothelin 1 in patterning the dorsal-ventral axis of the craniofacial skeleton. Development (2011) 138:5135–46. doi: 10.1242/dev.067801
31. Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. (2002) 248:307–18. doi: 10.1006/dbio.2002.0711
32. Schwend T, Loucks EJ, Ahlgren SC. Visualization of Gli activity in craniofacial tissues of hedgehog-pathway reporter transgenic zebrafish. PLoS ONE (2010) 5:e14396. doi: 10.1371/journal.pone.0014396
33. Knopf F, Hammond C, Chekuru A, Kurth T, Hans S, Weber CW, et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell (2011) 20:713–24. doi: 10.1016/j.devcel.2011.04.014
34. To TT, Witten PE, Renn J, Bhattacharya D, Huysseune A, Winkler C. Rankl-induced osteoclastogenesis leads to loss of mineralization in a medaka osteoporosis model. Development (2011) 139:141–50. doi: 10.1242/dev.071035
35. Wada N, Javidan Y, Nelson S, Carney TJ, Kelsh RN, Schilling TF. Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development (2005) 132:3977–88. doi: 10.1242/dev.01943
36. DeLaurier A, Eames BF, Blanco-Sánchez B, Peng G, He X, Swartz ME, et al. Zebrafish sp7:EGFP: a transgenic for studying otic vesicle formation, skeletogenesis, and bone regeneration. Genesis (2010) 48:505–11. doi: 10.1002/dvg.20639
37. Spoorendonk KM, Peterson-Maduro J, Renn J, Trowe T, Kranenbarg S, Winkler C, et al. Retinoic acid and Cyp26b1 are critical regulators of osteogenesis in the axial skeleton. Development (2008) 135:3765–74. doi: 10.1242/dev.024034
38. de Vrieze E, Zethof J, Schulte-Merker S, Flik G, Metz JR. Identification of novel osteogenic compounds by an ex vivo sp7:luciferase zebrafish scale assay. Bone (2015) 74(Suppl. C):106–13. doi: 10.1016/j.bone.2015.01.006
39. Singh SP, Holdway JE, Poss KD. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell (2012) 22:879–86. doi: 10.1016/j.devcel.2012.03.006
40. Moro E, Ozhan-Kizil G, Mongera A, Beis D, Wierzbicki C, Young RM, et al. In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev Biol. (2012) 366:327–40. doi: 10.1016/j.ydbio.2012.03.023
41. Summerton J, Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. (1997) 7:187–95. doi: 10.1089/oli.1.1997.7.187
42. Schulte-Merker S, Stainier DY. Out with the old, in with the new: reassessing morpholino knockdowns in light of genome editing technology. Development (2014) 141:3103–4. doi: 10.1242/dev.112003
43. Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell (2015) 32:97–108. doi: 10.1016/j.devcel.2014.11.018
44. De Souza AT, Dai X, Spencer AG, Reppen T, Menzie A, Roesch PL, et al. Transcriptional and phenotypic comparisons of Ppara knockout and siRNA knockdown mice. Nucleic Acids Res. (2006) 34:4486–94. doi: 10.1093/nar/gkl609
45. Daude N, Wohlgemuth S, Brown R, Pitstick R, Gapeshina H, Yang J, et al. Knockout of the prion protein (PrP)-like Sprn gene does not produce embryonic lethality in combination with PrP(C)-deficiency. Proc Natl Acad Sci USA. (2012) 109:9035–40. doi: 10.1073/pnas.1202130109
46. Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci USA. (2015) 112:2275–80. doi: 10.1073/pnas.1500365112
47. Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature (2015) 524:230–3. doi: 10.1038/nature14580
48. Cerikan B, Shaheen R, Colo GP, Gläßer C, Hata S, Knobeloch K-P, et al. Cell-intrinsic adaptation arising from chronic ablation of a key Rho GTPase regulator. Dev Cell (2016) 39:28–43. doi: 10.1016/j.devcel.2016.08.020
49. Bergen DJM, Stevenson NL, Skinner REH, Stephens DJ, Hammond CL. The Golgi matrix protein giantin is required for normal cilia function in zebrafish. Biol Open (2017) 6:1180–9. doi: 10.1242/bio.025502
50. Stevenson NL, Bergen DJM, Skinner REH, Kague E, Martin-Silverstone E, Robson Brown KA, et al. Giantin knockout models reveal a feedback loop between Golgi function and glycosyltransferase expression. J Cell Sci. (2017) 130:4132–43. doi: 10.1242/jcs.212308
51. Stevenson NL, Bergen DJM, Xu A, Wyatt E, Henry F, McCaughey J, et al. Regulator of calcineurin-2 is a centriolar protein with a role in cilia length control. J Cell Sci. (2018) 131:jcs212258. doi: 10.1242/jcs.212258
52. Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development (1996) 123:1.
53. Odenthal J, Haffter P, Vogelsang E, Brand M, van Eeden FJ, Furutani-Seiki M, et al. Mutations affecting the formation of the notochord in the zebrafish, Danio rerio. Development (1996) 123:103–15.
54. Knapik EW. ENU mutagenesis in zebrafish–from genes to complex diseases. Mamm Genome (2000) 11:511–9. doi: 10.1007/s003350010098
55. Kettleborough RN, Bruijn E, Eeden F, Cuppen E, Stemple DL. High-throughput target-selected gene inactivation in zebrafish. Methods Cell Biol. (2011) 104:121–7. doi: 10.1016/B978-0-12-374814-0.00006-9
56. Henke K, Daane JM, Hawkins MB, Dooley CM, Busch-Nentwich EM, Stemple DL, et al. Genetic screen for post-embryonic development in the zebrafish (Danio rerio): dominant mutations affecting adult form. Genetics (2017) 207:609–23. doi: 10.1534/genetics.117.300187
57. Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG II, et al. In vivo genome editing using a high-efficiency TALEN system. Nature (2012) 491:114–8. doi: 10.1038/nature11537
58. Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, et al. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. (2012) 40:8001–10. doi: 10.1093/nar/gks518
59. Talbot JC, Amacher SL. A streamlined CRISPR pipeline to reliably generate zebrafish frameshifting alleles. Zebrafish (2014) 11:583–5. doi: 10.1089/zeb.2014.1047
60. Brunt LH, Begg K, Kague E, Cross S, Hammond CL. Wnt signalling controls the response to mechanical loading during zebrafish joint development. Development (2017) 144:2798–809. doi: 10.1242/dev.153528
61. Carrington B, Varshney GK, Burgess SM, Sood R. CRISPR-STAT: an easy and reliable PCR-based method to evaluate target-specific sgRNA activity. Nucleic Acids Res. (2015) 43:e157. doi: 10.1093/nar/gkv802
62. Rees HA, Komor AC, Yeh W-H, Caetano-Lopes J, Warman M, Edge ASB, et al. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat Commun. (2017) 8:15790. doi: 10.1038/ncomms15790
63. Prykhozhij SV, Berman JN. Zebrafish knock-ins swim into the mainstream. Dis Model Mech. (2018) 11:dmm037515. doi: 10.1242/dmm.037515
64. Weigele J, Franz-Odendaal TA. Functional bone histology of zebrafish reveals two types of endochondral ossification, different types of osteoblast clusters and a new bone type. J Anat. (2016) 229:92–103. doi: 10.1111/joa.12480
65. Witten PE, Harris MP, Huysseune A, Winkler C. Chapter 13 - Small teleost fish provide new insights into human skeletal diseases. Methods Cell Biol. (2017) 138:321–46. doi: 10.1016/bs.mcb.2016.09.001
66. Schilling TF, Kimmel CB. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development (1997) 124:2945–60.
67. Hammond CL, Schulte-Merker S. Two populations of endochondral osteoblasts with differential sensitivity to Hedgehog signalling. Development (2009) 136:3991–4000. doi: 10.1242/dev.042150
68. Borovina A, Superina S, Voskas D, Ciruna B. Vangl2 directs the posterior tilting and asymmetric localization of motile primary cilia. Nat Cell Biol. (2010) 12:407–12. doi: 10.1038/ncb2042
69. Bird NC, Mabee PM. Developmental morphology of the axial skeleton of the zebrafish, Danio rerio (Ostariophysi: Cyprinidae). Dev Dyn. (2003) 228:337–57. doi: 10.1002/dvdy.10387
70. Fisher S, Jagadeeswaran P, Halpern ME. Radiographic analysis of zebrafish skeletal defects. Dev Biol. (2003) 264:64–76. doi: 10.1016/S0012-1606(03)00399-3
71. Andreeva V, Connolly MH, Stewart-Swift C, Fraher D, Burt J, Cardarelli J, et al. Identification of adult mineralized tissue zebrafish mutants. Genesis (2011) 49:360–6. doi: 10.1002/dvg.20712
72. Charles JF, Sury M, Tsang K, Urso K, Henke K, Huang Y, et al. Utility of quantitative micro-computed tomographic analysis in zebrafish to define gene function during skeletogenesis. Bone (2017) 101:162–71. doi: 10.1016/j.bone.2017.05.001
73. Neldam CA, Pinholt EM. Synchrotron μCT imaging of bone, titanium implants and bone substitutes – A systematic review of the literature. J Cranio Maxillofac Surg. (2014) 42:801–5. doi: 10.1016/j.jcms.2013.11.015
74. Ding Y, Vanselow DJ, Yakovlev MA, Katz SR, Lin AY, Clark DP, et al. Three-dimensional histology of whole zebrafish by sub-micron synchrotron x-ray micro-tomography. bioRxiv [preprint] (2018). doi: 10.1101/392381
75. Matsumoto T, Ando N, Tomii T, Uesugi K. Three-dimensional cortical bone microstructure in a rat model of hypoxia-induced growth retardation. Calcif Tissue Int. (2011) 88:54–62. doi: 10.1007/s00223-010-9415-7
76. Hur M, Gistelinck CA, Huber P, Lee J, Thompson MH, Monstad-Rios AT, et al. MicroCT-based phenomics in the zebrafish skeleton reveals virtues of deep phenotyping in a distributed organ system. Elife (2017) 6:e26014. doi: 10.7554/eLife.26014
77. Mackay EW, Apschner A, Schulte-Merker S. Vitamin K reduces hypermineralisation in zebrafish models of PXE and GACI. Development (2015) 142:1095–101. doi: 10.1242/dev.113811
78. Barsh GS, Zhang Y, Huang H, Zhao G, Yokoyama T, Vega H, et al. ATP6V1H deficiency impairs bone development through activation of MMP9 and MMP13. PLoS Genet. (2017) 13:e1006481. doi: 10.1371/journal.pgen.1006481
79. Asharani PV, Keupp K, Semler O, Wang W, Li Y, Thiele H, et al. Attenuated BMP1 function compromises osteogenesis, leading to bone fragility in humans and zebrafish. Am J Hum Gene (2012) 90:661–74. doi: 10.1016/j.ajhg.2012.02.026
80. Bowen ME, Henke K, Siegfried KR, Warman ML, Harris MP. Efficient mapping and cloning of mutations in zebrafish by low-coverage whole-genome sequencing. Genetics (2012) 190:1017–24. doi: 10.1534/genetics.111.136069
81. Herbomel P, Thisse B, Thisse C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev Biol. (2001) 238:274–88. doi: 10.1006/dbio.2001.0393
82. Chatani M, Takano Y, Kudo A. Osteoclasts in bone modeling, as revealed by in vivo imaging, are essential for organogenesis in fish. Dev Biol. (2011) 360:96–109. doi: 10.1016/j.ydbio.2011.09.013
83. Lawrence EA, Kague E, Aggleton JA, Harniman RL, Roddy KA, Hammond CL. The mechanical impact of col11a2 loss on joints; col11a2 mutant zebrafish show changes to joint development and function, which leads to early-onset osteoarthritis. Philos Transact R Soc Lond B Biol Sci. (2018) 373:20170335. doi: 10.1098/rstb.2017.0335
84. Fiedler IAK, Schmidt FN, Wölfel EM, Plumeyer C, Milovanovic P, Gioia R, et al. Severely impaired bone material quality in chihuahua zebrafish resembles classical dominant human osteogenesis imperfecta. J Bone Miner Res. (2018) 33:1489–99. doi: 10.1002/jbmr.3445
85. Gistelinck C, Kwon RY, Malfait F, Symoens S, Harris MP, Henke K, et al. Zebrafish type I collagen mutants faithfully recapitulate human type I collagenopathies. Proc Natl Acad Sci USA. (2018) 115:E8037–46. doi: 10.1073/pnas.1722200115
86. Gioia R, Tonelli F, Ceppi I, Biggiogera M, Leikin S, Fisher S, et al. The chaperone activity of 4PBA ameliorates the skeletal phenotype of Chihuahua, a zebrafish model for dominant osteogenesis imperfecta. Hum Mol Genet. (2017) 26:2897–911. doi: 10.1093/hmg/ddx171
87. To TT, Witten PE, Huysseune A, Winkler C. An adult osteopetrosis model in medaka reveals the importance of osteoclast function for bone remodeling in teleost fish. Comp Biochem Physiol C Toxicol Pharmacol. (2015) 178:68–75. doi: 10.1016/j.cbpc.2015.08.007
88. Zhang Y, Cui FZ, Wang XM, Feng QL, Zhu XD. Mechanical properties of skeletal bone in gene-mutated stopsel(dtl28d) and wild-type zebrafish (Danio rerio) measured by atomic force microscopy-based nanoindentation. Bone (2002) 30:541–6. doi: 10.1016/S8756-3282(02)00676-2
89. Misu A, Yamanaka H, Aramaki T, Kondo S, Skerrett IM, Iovine MK, et al. Two different functions of connexin43 confer two different bone phenotypes in zebrafish. J Biol Chem. (2016) 291:12601–11. doi: 10.1074/jbc.M116.720110
90. Laue K, Janicke M, Plaster N, Sonntag C, Hammerschmidt M. Restriction of retinoic acid activity by Cyp26b1 is required for proper timing and patterning of osteogenesis during zebrafish development. Development (2008) 135:3775–87. doi: 10.1242/dev.021238
91. Laue K, Pogoda H-M, Daniel PB, van Haeringen A, Alanay Y, von Ameln S, et al. Craniosynostosis and multiple skeletal anomalies in humans and zebrafish result from a defect in the localized degradation of retinoic acid. Am J Hum Gene (2011) 89:595–606. doi: 10.1016/j.ajhg.2011.09.015
92. Aman AJ, Fulbright AN, Parichy DM. Wnt/β-catenin regulates an ancient signaling network during zebrafish scale development. Elife (2018) 7:e37001. doi: 10.7554/eLife.37001
93. Harris MP, Rohner N, Schwarz H, Perathoner S, Konstantinidis P, Nusslein-Volhard C. Zebrafish eda and edar mutants reveal conserved and ancestral roles of ectodysplasin signaling in vertebrates. PLoS Genet. (2008) 4:e1000206. doi: 10.1371/journal.pgen.1000206
94. Huitema LFA, Apschner A, Logister I, Spoorendonk KM, Bussmann J, Hammond CL, et al. Entpd5 is essential for skeletal mineralization and regulates phosphate homeostasis in zebrafish. Proc Natl Acad Sci USA. (2012) 109:21372–7. doi: 10.1073/pnas.1214231110
95. Apschner A, Huitema LFA, Ponsioen B, Peterson-Maduro J, Schulte-Merker S. Zebrafish enpp1 mutants exhibit pathological mineralization, mimicking features of generalized arterial calcification of infancy (GACI) and pseudoxanthoma elasticum (PXE). Dis Model Mech. (2014) 7:811–22. doi: 10.1242/dmm.015693
96. Zancan I, Bellesso S, Costa R, Salvalaio M, Stroppiano M, Hammond C, et al. Glucocerebrosidase deficiency in zebrafish affects primary bone ossification through increased oxidative stress and reduced Wnt/beta-catenin signaling. Hum Mol Genet. (2015) 24:1280–94. doi: 10.1093/hmg/ddu538
97. Iwasaki M, Kuroda J, Kawakami K, Wada H. Epidermal regulation of bone morphogenesis through the development and regeneration of osteoblasts in the zebrafish scale. Dev Biol. (2018) 437:105–19. doi: 10.1016/j.ydbio.2018.03.005
98. Huycke TR, Eames BF, Kimmel CB. Hedgehog-dependent proliferation drives modular growth during morphogenesis of a dermal bone. Development (2012) 139:2371–80. doi: 10.1242/dev.079806
99. Paul S, Schindler S, Giovannone D, de Millo Terrazzani A, Mariani FV, Crump JG. Ihha induces hybrid cartilage-bone cells during zebrafish jawbone regeneration. Development (2016) 143:2066–76. doi: 10.1242/dev.131292
100. Huo L, Wang L, Yang Z, Li P, Geng D, Xu Y. Prednisolone induces osteoporosis-like phenotypes via focal adhesion signaling pathway in zebrafish larvae. Biol. Open (2018) 7:bio029405. doi: 10.1242/bio.029405
101. Jafari A, Qanie D, Andersen TL, Zhang Y, Chen L, Postert B, et al. Legumain regulates differentiation fate of human bone marrow stromal cells and is altered in postmenopausal osteoporosis. Stem Cell Rep. (2017) 8:373–86. doi: 10.1016/j.stemcr.2017.01.003
102. Tian J, Shao J, Liu C, Hou H-Y, Chou C-W, Shboul M, et al. Deficiency of lrp4 in zebrafish and human LRP4 mutation induce aberrant activation of Jagged–Notch signaling in fin and limb development. Cell Mol Life Sci. (2019). 76:163–78. doi: 10.1007/s00018-018-2928-3
103. DeLaurier A, Huycke TR, Nichols JT, Swartz ME, Larsen A, Walker C, et al. Role of mef2ca in developmental buffering of the zebrafish larval hyoid dermal skeleton. Dev Biol. (2014) 385:189–99. doi: 10.1016/j.ydbio.2013.11.016
104. Nichols JT, Blanco-Sanchez B, Brooks EP, Parthasarathy R, Dowd J, Subramanian A, et al. Ligament versus bone cell identity in the zebrafish hyoid skeleton is regulated by mef2ca. Development (2016) 143:4430–40. doi: 10.1242/dev.141036
105. Xi Y, Chen D, Sun L, Li Y, Li L. Characterization of zebrafish mutants with defects in bone calcification during development. Biochem Biophys Res Commun. (2013) 440:132–6. doi: 10.1016/j.bbrc.2013.09.043
106. Oh SK, Shin JO, Baek JI, Lee J, Bae JW, Ankamerddy H, et al. Pannexin 3 is required for normal progression of skeletal development in vertebrates. FASEB J. (2015) 29:4473–84. doi: 10.1096/fj.15-273722
107. van Dijk FS, Zillikens MC, Micha D, Riessland M, Marcelis CLM, de Die-Smulders CE, et al. PLS3 mutations in X-linked osteoporosis with fractures. N Engl J Med. (2013) 369:1529–36. doi: 10.1056/NEJMoa1308223
108. Suarez-Bregua P, Torres-Nuñez E, Saxena A, Guerreiro P, Braasch I, Prober DA, et al. Pth4, an ancient parathyroid hormone lost in eutherian mammals, reveals a new brain-to-bone signaling pathway. FASEB J. (2017) 31:569–83. doi: 10.1096/fj.201600815R
109. Yan Y-L, Bhattacharya P, He XJ, Ponugoti B, Marquardt B, Layman J, et al. Duplicated zebrafish co-orthologs of parathyroid hormone-related peptide (PTHrP, Pthlh) play different roles in craniofacial skeletogenesis. J Endocrinol. (2012) 214:421–35. doi: 10.1530/JOE-12-0110
110. Green J, Taylor JJ, Hindes A, Johnson SL, Goldsmith MI. A gain of function mutation causing skeletal overgrowth in the rapunzel mutant. Dev Biol. (2009) 334:224–34. doi: 10.1016/j.ydbio.2009.07.025
111. Ashikov A, Abu Bakar N, Wen X-Y, Niemeijer M, Rodrigues Pinto Osorio G, Brand-Arzamendi K, et al. Integrating glycomics and genomics uncovers SLC10A7 as essential factor for bone mineralization by regulating post-Golgi protein transport and glycosylation. Hum Mol Genet. (2018) 27:3029–45. doi: 10.1093/hmg/ddy213
112. Kague E, Roy P, Asselin G, Hu G, Simonet J, Stanley A, et al. Osterix/Sp7 limits cranial bone initiation sites and is required for formation of sutures. Dev Biol. (2016) 413:160–72. doi: 10.1016/j.ydbio.2016.03.011
113. Kague E, Witten PE, Soenens M, Campos CL, Lubiana T, Fisher S, et al. Zebrafish sp7 mutants show tooth cycling independent of attachment, eruption and poor differentiation of teeth. Dev Biol. (2018) 435:176–84. doi: 10.1016/j.ydbio.2018.01.021
114. Azetsu Y, Inohaya K, Takano Y, Kinoshita M, Tasaki M, Kudo A. The sp7 gene is required for maturation of osteoblast-lineage cells in medaka (Oryzias latipes) vertebral column development. Dev Biol. (2017) 431:252–62. doi: 10.1016/j.ydbio.2017.09.010
115. Yu T, Graf M, Renn J, Schartl M, Larionova D, Huysseune A, et al. A vertebrate-specific and essential role for osterix in osteogenesis revealed by gene knockout in the teleost medaka. Development (2017) 144:265–71. doi: 10.1242/dev.139550
116. Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature (2014) 505:174–9. doi: 10.1038/nature12826
117. Cheah FS, Winkler C, Jabs EW, Chong SS. Tgfbeta3 regulation of chondrogenesis and osteogenesis in zebrafish is mediated through formation and survival of a subpopulation of the cranial neural crest. Mech Dev. (2010) 127:329–44. doi: 10.1016/j.mod.2010.04.003
118. Teng CS, Ting MC, Farmer DT, Brockop M, Maxson RE, Crump JG. Altered bone growth dynamics prefigure craniosynostosis in a zebrafish model of Saethre-Chotzen syndrome. Elife (2018) 7:37024. doi: 10.7554/eLife.37024
119. Bella J, Liu J, Kramer R, Brodsky B, Berman HM. Conformational effects of Gly-X-Gly interruptions in the collagen triple helix. J Mol Biol. (2006) 362:298–311. doi: 10.1016/j.jmb.2006.07.014
120. Eyre DR, Weis MA. Bone collagen: new clues to its mineralization mechanism from recessive osteogenesis imperfecta. Calcif Tissue Int. (2013) 93:338–47. doi: 10.1007/s00223-013-9723-9
121. Weis MA, Hudson DM, Kim L, Scott M, Wu JJ, Eyre DR. Location of 3-hydroxyproline residues in collagen types I, II, III, and V/XI implies a role in fibril supramolecular assembly. J Biol Chem. (2010) 285:2580–90. doi: 10.1074/jbc.M109.068726
122. Banerjee S, Isaacman-Beck J, Schneider VA, Granato M. A novel role for Lh3 dependent ECM modifications during neural crest cell migration in zebrafish. PLoS ONE (2013) 8:e54609. doi: 10.1371/journal.pone.0054609
123. Unlu G, Levic DS, Melville DB, Knapik EW. Trafficking mechanisms of extracellular matrix macromolecules: insights from vertebrate development and human diseases. Int J Biochem Cell Biol. (2014) 47:57–67. doi: 10.1016/j.biocel.2013.11.005
124. Gistelinck C, Gioia R, Gagliardi A, Tonelli F, Marchese L, Bianchi L, et al. Zebrafish collagen type i: molecular and biochemical characterization of the major structural protein in bone and skin. Sci Rep. (2016) 6:21540. doi: 10.1038/srep21540
125. Lapunzina P, Aglan M, Temtamy S, Caparrós-Martín JA, Valencia M, Letón R, et al. Identification of a frameshift mutation in osterix in a patient with recessive osteogenesis imperfecta. Am J Hum Gene (2010) 87:110–4. doi: 10.1016/j.ajhg.2010.05.016
126. Mariotti M, Carnovali M, Banfi G. Danio rerio: the Janus of the bone from embryo to scale. Clin Cases Miner Bone Metabol. (2015) 12:188–94. doi: 10.11138/ccmbm/2015.12.2.188
127. Roehl HH. Linking wound response and inflammation to regeneration in the zebrafish larval fin. Int J Dev Biol. (2018) 62:473–7. doi: 10.1387/ijdb.170331hr
128. Akimenko M-A, Marí-Beffa M, Becerra J, Géraudie J. Old questions, new tools, and some answers to the mystery of fin regeneration. Dev Dyn. (2003) 226:190–201. doi: 10.1002/dvdy.10248
129. Sousa S, Afonso N, Bensimon-Brito A, Fonseca M, Simoes M, Leon J, et al. Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development (2011) 138:3897–905. doi: 10.1242/dev.064717
130. Chassot B, Pury D, Jazwinska A. Zebrafish fin regeneration after cryoinjury-induced tissue damage. Biol Open (2016) 5:819–28. doi: 10.1242/bio.016865
131. Geurtzen K, Vernet A, Freidin A, Rauner M, Hofbauer LC, Schneider JE, et al. Immune suppressive and bone inhibitory effects of prednisolone in growing and regenerating zebrafish tissues. J Bone Miner Res. (2017) 32:2476–88. doi: 10.1002/jbmr.3231
132. Recidoro AM, Roof AC, Schmitt M, Worton LE, Petrie T, Strand N, et al. Botulinum toxin induces muscle paralysis and inhibits bone regeneration in zebrafish. J Bone Miner Res. (2014) 29:2346–56. doi: 10.1002/jbmr.2274
133. Kim HM, Galatz LM, Patel N, Das R, Thomopoulos S. Recovery potential after postnatal shoulder paralysis. J Bone Joint Surg. Am Vol. (2009) 91:879–91. doi: 10.2106/JBJS.H.00088
134. Hao Y, Ma Y, Wang X, Jin F, Ge S. Short-term muscle atrophy caused by botulinum toxin-A local injection impairs fracture healing in the rat femur. J Orthop Res. (2012) 30:574–80. doi: 10.1002/jor.21553
135. Sousa S, Valerio F, Jacinto A. A new zebrafish bone crush injury model. Biol Open (2012) 1:915–21. doi: 10.1242/bio.2012877
136. Monstad-Rios AT, Watson CJ, Kwon RY. Screencube: a 3D printed system for rapid and cost-effective chemical screening in adult zebrafish. Zebrafish (2017) 15:1–8. doi: 10.1089/zeb.2017.1488
137. Zylberberg L, Bereiter-Hahn J, Sire JY. Cytoskeletal organization and collagen orientation in the fish scales. Cell Tissue Res. (1988) 253:597–607. doi: 10.1007/BF00219750
138. Sire JY, Donoghue PC, Vickaryous MK. Origin and evolution of the integumentary skeleton in non-tetrapod vertebrates. J Anat. (2009) 214:409–40. doi: 10.1111/j.1469-7580.2009.01046.x
139. Pasqualetti S, Banfi G, Mariotti M. The zebrafish scale as model to study the bone mineralization process. J Mol Histol. (2012) 43:589–95. doi: 10.1007/s10735-012-9425-z
140. de Vrieze E, van Kessel MAHJ, Peters HM, Spanings FAT, Flik G, Metz JR. Prednisolone induces osteoporosis-like phenotype in regenerating zebrafish scales. Osteopor Int. (2014) 25:567–78. doi: 10.1007/s00198-013-2441-3
141. Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. (2008) 4:33–41. doi: 10.1038/nchembio.2007.54
142. Kithcart A, MacRae CA. Using zebrafish for high-throughput screening of novel cardiovascular drugs. JACC Basic Transl Sci. (2017) 2:1–12. doi: 10.1016/j.jacbts.2017.01.004
143. Chen JR, Lai YH, Tsai JJ, Hsiao CD. Live fluorescent staining platform for drug-screening and mechanism-analysis in zebrafish for bone mineralization. Molecules (2017) 22:12. doi: 10.3390/molecules22122068
144. Pasqualetti S, Congiu T, Banfi G, Mariotti M. Alendronate rescued osteoporotic phenotype in a model of glucocorticoid-induced osteoporosis in adult zebrafish scale. Int J Exp Pathol. (2015) 96:11–20. doi: 10.1111/iep.12106
145. Barrett R, Chappell C, Quick M, Fleming A. A rapid, high content, in vivo model of glucocorticoid-induced osteoporosis. Biotechnol J. (2006) 1:651–5. doi: 10.1002/biot.200600043
146. Tarasco M, Laize V, Cardeira J, Cancela ML, Gavaia PJ. The zebrafish operculum: a powerful system to assess osteogenic bioactivities of molecules with pharmacological and toxicological relevance. Comp Biochem Physiol C Toxicol Pharmacol. (2017) 197:45–52. doi: 10.1016/j.cbpc.2017.04.006
147. Armstrong BE, Henner A, Stewart S, Stankunas K. Shh promotes direct interactions between epidermal cells and osteoblast progenitors to shape regenerated zebrafish bone. Development (2017) 144:1165–76. doi: 10.1242/dev.143792
148. Luo S, Yang Y, Chen J, Zhong Z, Huang H, Zhang J, et al. Tanshinol stimulates bone formation and attenuates dexamethasone-induced inhibition of osteogenesis in larval zebrafish. J Orthopaed Transl. (2016) 4:35–45. doi: 10.1016/j.jot.2015.07.002
149. Bae SJ, Kim HJ, Won HY, Min YK, Hwang ES. Acceleration of osteoblast differentiation by a novel osteogenic compound, DMP-PYT, through activation of both the BMP and Wnt pathways. Sci Rep. (2017) 7:8455. doi: 10.1038/s41598-017-08190-9
150. Chen B, Yan Y-L, Liu C, Bo L, Li G-F, Wang H, et al. Therapeutic effect of deferoxamine on iron overload-induced inhibition of osteogenesis in a zebrafish model. Calcif Tissue Int. (2014) 94:353–60. doi: 10.1007/s00223-013-9817-4
151. Zhang W, Xu J, Qiu J, Xing C, Li X, Leng B, et al. Novel and rapid osteoporosis model established in zebrafish using high iron stress. Biochem Biophys Res Commun. (2018) 496:654–60. doi: 10.1016/j.bbrc.2017.12.172
152. Carnovali M, Luzi L, Terruzzi I, Banfi G, Mariotti M. Metabolic and bone effects of high-fat diet in adult zebrafish. Endocrine (2017) 61:317–26. doi: 10.1007/s12020-017-1494-z
153. Carnovali M, Luzi L, Banfi G, Mariotti M. Chronic hyperglycemia affects bone metabolism in adult zebrafish scale model. Endocrine (2016) 54:808–17. doi: 10.1007/s12020-016-1106-3
154. Aceto J, Nourizadeh-Lillabadi R, Maree R, Dardenne N, Jeanray N, Wehenkel L, et al. Zebrafish bone and general physiology are differently affected by hormones or changes in gravity. PLoS ONE (2015) 10:e0126928. doi: 10.1371/journal.pone.0126928
155. Carnovali M, Ottria R, Pasqualetti S, Banfi G, Ciuffreda P, Mariotti M. Effects of bioactive fatty acid amide derivatives in zebrafish scale model of bone metabolism and disease. Pharmacol Res. (2016) 104:1–8. doi: 10.1016/j.phrs.2015.12.009
156. Zhang S, Zhang Q, Zhang D, Wang C, Yan C. Anti-osteoporosis activity of a novel Achyranthes bidentata polysaccharide via stimulating bone formation. Carbohydr Polym. (2018) 184:288–98. doi: 10.1016/j.carbpol.2017.12.070
157. de Vrieze E, Moren M, Metz JR, Flik G, Lie KK. Arachidonic acid enhances turnover of the dermal skeleton: studies on zebrafish scales. PLoS ONE (2014) 9:e89347. doi: 10.1371/journal.pone.0089347
158. Pogoda HM, Riedl-Quinkertz I, Löhr H, Waxman JS, Dale RM, Topczewski J, et al. Direct activation of chordoblasts by retinoic acid is required for segmented centra mineralization during zebrafish spine development. Development (2018) 145:9. doi: 10.1242/dev.159418
159. Montazerolghaem M, Nyström L, Engqvist H, Karlsson Ott M. Zebrafish: a possible tool to evaluate bioactive ions. Acta Biomater. (2015) 19:10–4. doi: 10.1016/j.actbio.2015.03.010
160. Suniaga S, Rolvien T, vom Scheidt A, Fiedler IAK, Bale HA, Huysseune A, et al. Increased mechanical loading through controlled swimming exercise induces bone formation and mineralization in adult zebrafish. Sci Rep. (2018) 8:3646. doi: 10.1038/s41598-018-21776-1
161. Fleming A, Sato M, Goldsmith P. High-throughput in vivo screening for bone anabolic compounds with zebrafish. J Biomol Screen. (2005) 10:823–31. doi: 10.1177/1087057105279952
162. Wiley DS, Redfield SE, Zon LI. Chemical screening in zebrafish for novel biological and therapeutic discovery. Methods Cell Biol. (2017) 138:651–79. doi: 10.1016/bs.mcb.2016.10.004
163. Hassler N, Gamsjaeger S, Hofstetter B, Brozek W, Klaushofer K, Paschalis EP. Effects of long-term alendronate treatment on postmenopausal osteoporosis bone material properties. Osteopor Int. (2014) 26:339–52. doi: 10.1007/s00198-014-2929-5
164. Ma X, Xu Z, Ding S, Yi G, Wang Q. Alendronate promotes osteoblast differentiation and bone formation in ovariectomy-induced osteoporosis through interferon-β/signal transducer and activator of transcription 1 pathway. Exp Ther Med. (2017) 15:182–90. doi: 10.3892/etm.2017.5381
Keywords: zebrafish, screening, genetic mutants, osteoblast, osteoclast, osteoporosis, drug development, animal model
Citation: Bergen DJM, Kague E and Hammond CL (2019) Zebrafish as an Emerging Model for Osteoporosis: A Primary Testing Platform for Screening New Osteo-Active Compounds. Front. Endocrinol. 10:6. doi: 10.3389/fendo.2019.00006
Received: 07 November 2018; Accepted: 09 January 2019;
Published: 29 January 2019.
Edited by:
David Karasik, Bar-Ilan University, IsraelReviewed by:
Matthew Harris, Harvard Medical School, United StatesAntonella Forlino, University of Pavia, Italy
Copyright © 2019 Bergen, Kague and Hammond. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chrissy L. Hammond, chrissy.hammond@bristol.ac.uk
 Dylan J. M. Bergen
Dylan J. M. Bergen Erika Kague1
Erika Kague1 Chrissy L. Hammond
Chrissy L. Hammond