Bibliometric and visual analysis of RAN methylation in cardiovascular disease
- 1Xiyuan Hospital, China Academy of Chinese Medical Sciences, Beijing, China
- 2Graduate School, Beijing University of Chinese Medicine, Beijing, China
- 3National Clinical Research Center for Chinese Medicine Cardiology, Beijing, China
Background: RNA methylation is associated with cardiovascular disease (CVD) occurrence and development. The purpose of this study is to visually analyze the results and research trends of global RNA methylation in CVD.
Methods: Articles and reviews on RNA methylation in CVD published before 6 November 2022 were searched in the Web of Science Core Collection. Visual and statistical analysis was performed using CiteSpace 1.6.R4 advanced and VOSviewer 1.6.18.
Results: There were 847 papers from 1,188 institutions and 63 countries/regions. Over approximately 30 years, there was a gradual increase in publications and citations on RNA methylation in CVD. America and China had the highest output (284 and 259 papers, respectively). Nine of the top 20 institutions that published articles were from China, among which Fudan University represented the most. The International Journal of Molecular Sciences was the journal with the most studies. Nature was the most co-cited journal. The most influential writers were Zhang and Wang from China and Mathiyalagan from the United States. After 2015, the primary keywords were cardiac development, heart, promoter methylation, RNA methylation, and N6-methyladenosine. Nuclear RNA, m6A methylation, inhibition, and myocardial infarction were the most common burst keywords from 2020 to the present.
Conclusions: A bibliometric analysis reveals research hotspots and trends of RNA methylation in CVD. The regulatory mechanisms of RNA methylation related to CVD and the clinical application of their results, especially m6A methylation, are likely to be the focus of future research.
1. Introduction
Cardiovascular diseases (CVD) are the leading cause of death among noncommunicable diseases worldwide. Ischemic heart disease is the highest risk factor for premature death in women in more than half of countries and men in more than three-quarters of countries (1). Common risk factors for CVD include diabetes, hypertension, obesity, and smoking (2). CVD has been extensively studied at the genetic and epigenetic levels (3–5). Due to the close relationship between epigenetic regulation and environmental adaptation, the former has attracted substantial attention. As new insights are gained, epigenomics is emerging. Epigenetic transcriptomics does not cover all epigenetic regulation but focuses on RNA-level modification (6). RNA methylation accounts for the most significantly proportion.
RNA methylation refers to the methylation modification of RNA molecules at various positions, affecting gene expression. The molecules involved in RNA methylation are classified as Writers, Erasers, and Readers. Taking N6-methyladenosine (m6A) methylation as an example, for Writers, the methylation modification is “written” into RNA; that is, the methylation modification process of RNA is mediated (7). The most common molecules are methyltransferase-like 3 (METTL3) and METTL14. Studies have found that MettL3/14 rapidly locates disease gene loci and enriches m6A, promoting the RNA methylation process (8). “Erasers” of the RNA methylation modification signal mediate RNA demethylation modifications. The most common are obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5), which remove methyl groups on RNA (9). Readers “read” the information on RNA methylation modification and participate in downstream RNA translation, degradation, and other processes. For example, proteins with the YTH domain family proteins recognize and bind m6A in mRNA, reducing the half-life of mRNA and promoting its degradation (10). Depending on the methyl modification positions, it can be roughly divided into m6A, N1-methyladenosine (m1A), and 5-methylcytosine (m5C). m6A is the most abundant form, distributed near the stop codon and the 3′UTR region, affecting RNA pairing, changing the secondary structure of RNA, or being directly recognized by proteins, thereby regulating mRNA maturation, variable splicing, stability, and translation processes (11, 12). m6A is catalyzed by methyltransferase complex and removed by demethylated transferase ALKBH5 or FTO (13); m1A is a reversible apparent transcription modification removed by the RNA demethylated transferase ALKBH3. No m1A modification enzyme or modification recognition protein has been found. The expression abundance of m1A is low and distributed in the 5′UTR region of mRNA, which may be involved in regulating translation initiation (14). m5C is widely distributed in tRNA and rRNA, stabilizing the secondary structure of tRNA, affecting the anticodon loop conformation, and maintaining the fidelity of rRNA translation and other functions. m5C can be catalyzed by theNOP2/Sun RNA methyltransferase 2 or tRNA aspartic acid methyltransferase 1 and oxidized by the dioxygenase ten-eleven translocation to form 5-hydroxymethylcytosine. m5C does not affect base pairing but may enhance base stacking and the hydrophobic interaction between RNA and protein (15).
Research into RNA methylation increases yearly. Napoli et al. focused on clinical trials of sensitive epigenetic drugs for the treatment of heart failure (HF) and outlined the discovery of epigenetic biomarkers and cardiac remodeling characteristics (16). METTL3 silencing inhibits m6A RNA hypermethylation, inflammatory responses, and monocyte adhesion induced by oxidative stress. It reverses the changes in the expression levels of nucleotide-binding domain leucine-rich repeat pyrin domain-containing 1 and Kruppel-like factor 4 in endothelial cells under oxidative stress conditions (17). These findings suggest that RNA methylation regulates the development of CVD; nevertheless, the hotspots and trends in RNA methylation in CVD research have not been analyzed.
The bibliometric analysis enables qualitative and quantitative analysis of published literature and is used to analyze disciplinary knowledge structures and explore development trends (18), which traditional meta-scores and systematic reviews cannot achieve. Bibliometric analysis reveals hotspots and trends in research fields and analyzes the information from countries, journals, authors, and keywords of published articles (19). This analysis method has been widely used in various fields.
This study used bibliometric analysis with CiteSpace 1.6.R4 advanced, VOSviewer 1.6.18, and Microsoft Excel 2016 software to explore the research hotspots and trends of RNA methylation in CVD since 1991 and create a visual knowledge map. We hope to help scholars understand this field's scientific basis and research trends.
2. Methods
2.1. Data collection
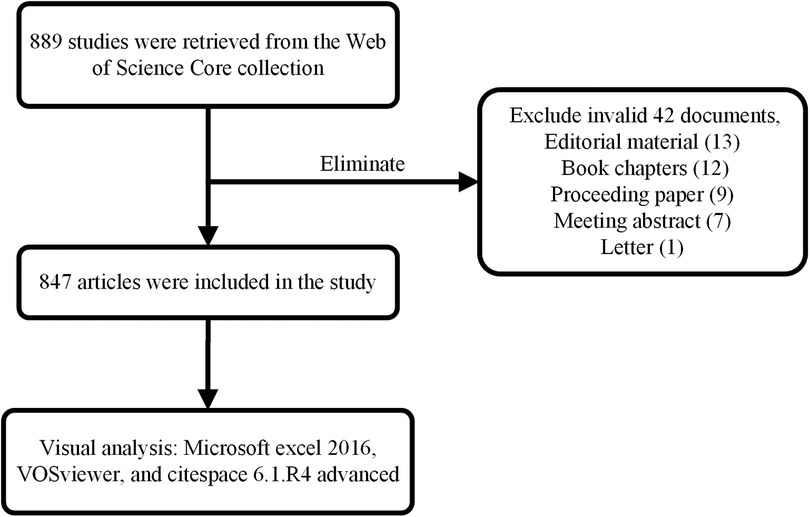
Articles were extracted from the Web of Science Core Collection (WoSCC) and were downloaded on November 06, 2022. The search terms were as follows: TS = (RNA methylation) AND (“circulation” OR “heart” OR “cardiovascular”), and the publication date: “1990-01-01” to “2022-11-06”, resulting in 889 records. We excluded invalid documents, including editorial material (13 articles), book chapters (12 articles), proceeding paper (nine articles), meeting abstract (seven articles), and letters (one article). Finally, 847 documents were included, and all records were exported and saved in plain text format. Figure 1 displays the flow chart.
2.2. Visual analytics
All valid documents retrieved from the WoSCC are exported into.txt files for visual analysis. We input the information into Microsoft Excel 2016 (Microsoft, United States) for the number of publications, the number of literature citations, and the analysis of research fields. CiteSpace 6.1.R4 advanced (CiteSpace for short, Drexel University, United States) and VOSviewer 1.6.18 (VOSviewer for short, the Centre for Science and Technology Studies, Holland) were used for literature clustering and visualization analysis. CiteSpace is a bibliometric analysis software developed by Chaomei Chen (20). It is a visual analytic tool for visualizing landmarks, critical paths, and emerging trends in a field of research based on relevant scholarly publications (21). We used CiteSpace to visualize the author and institutional collaboration, co-cited references and citation burst, keyword co-occurrence, and cluster and burst. The file was named starting with “download_” and imported into CiteSpace to remove duplicate files. There were no duplicate files. We imported the deduplicated files to CiteSpace for visual analysis. For institutional analysis, journal analysis, literature analysis, and author analysis, the parameter was set as “time slice 1 year” and “N = 50”; that is, the top 50 entries in each year were included in the analysis. The connection cable-cutting modes were pathfinder & pruning networks. To display image information more clearly, the time slice was set as two years in the keyword timeline view analysis, while other parameters remained unchanged. VOSviewer is an application for visual literature analysis, capable of coupling analysis of countries, journals, authors, citations, and more (22). We used VOSviwer to establish the item density of the countries. In the country analysis, the minimum number of articles per country was set at 1. In addition, the 2021 journal impact factors (IF) and JCR were obtained from the Web of Science.
3. Results
3.1. General analysis
3.1.1. Number of publications and citations over the years
The first RNA methylation article on CVD appeared in 1991. There was a small peak in 2008, and the number of published articles fell back in 2009. The number of published papers and citations increased steadily from 2010 to 2022. This finding indicates that RNA methylation has attracted the attention of researchers (Figure 2).
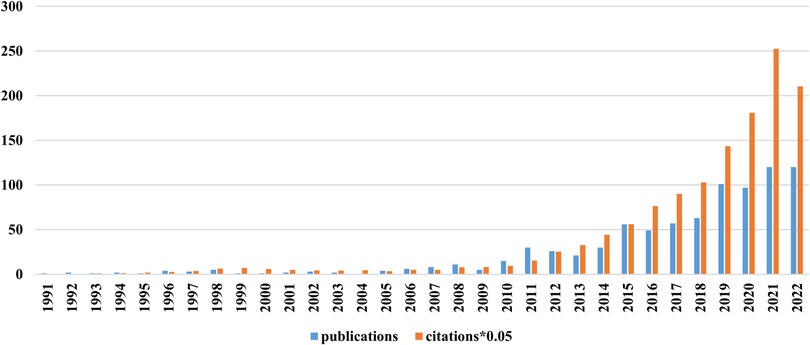
Figure 2. Annual publication and citation trends related to RNA methylation in CVD. Blue bars represent publications and yellow bars represent citations (*0.05).
3.1.2. Analysis of research areas
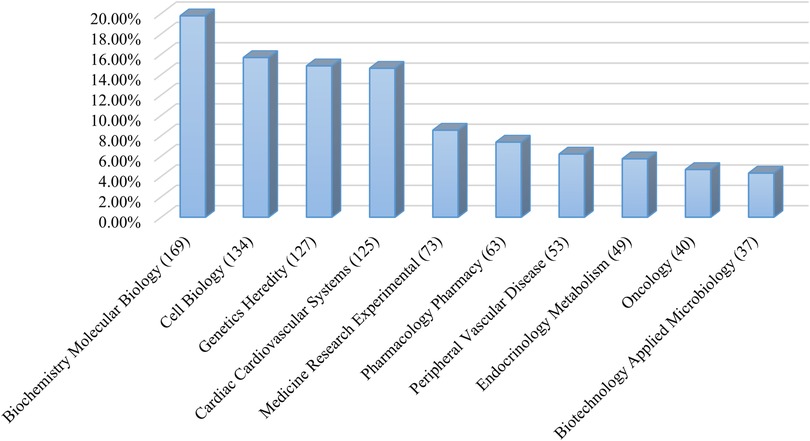
The 847 articles covered 50 fields, including biochemistry and molecular biology (167 articles), cell biology (132 articles), genetics and heredity (124 articles), and biochemistry and cardiac cardiovascular systems (122 articles) were most common. Figure 3 shows the top ten research areas in RNA methylation in CVD.
3.2. Countries/regions and institutional analysis
3.2.1. Analysis of countries/regions
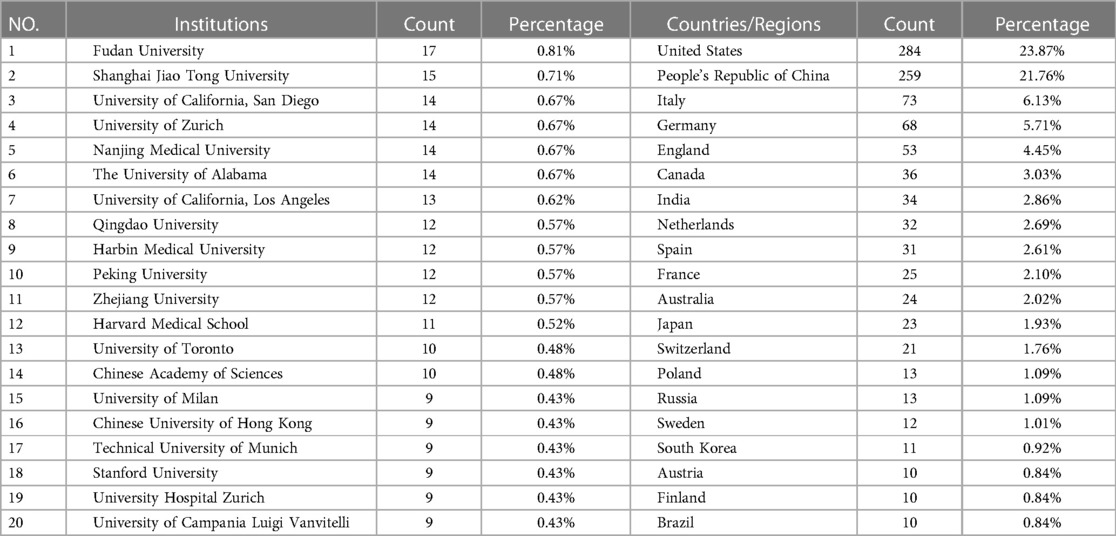
847 articles were published in 1,188 institutions from 63 countries and regions. Table 1 displays the top 20 countries with the most significant number of publications. The top five countries/regions were the United States (23.87%), China (21.76%), Italy (6.13%), Germany (5.71%), and the United Kingdom (4.45%), accounting for 61.93% of the total publications.
3.2.2. Item density
Figure 4 shows that more items near a node mean a higher weight of adjacent items; red indicates the highest item density. The opposite is closer to blue. Countries with more than two articles were analyzed (Figure 4). As can be seen from the picture, there was close cooperation between the United States, Italy, and Japan. Although China has many publications, it shows few connections with other countries.
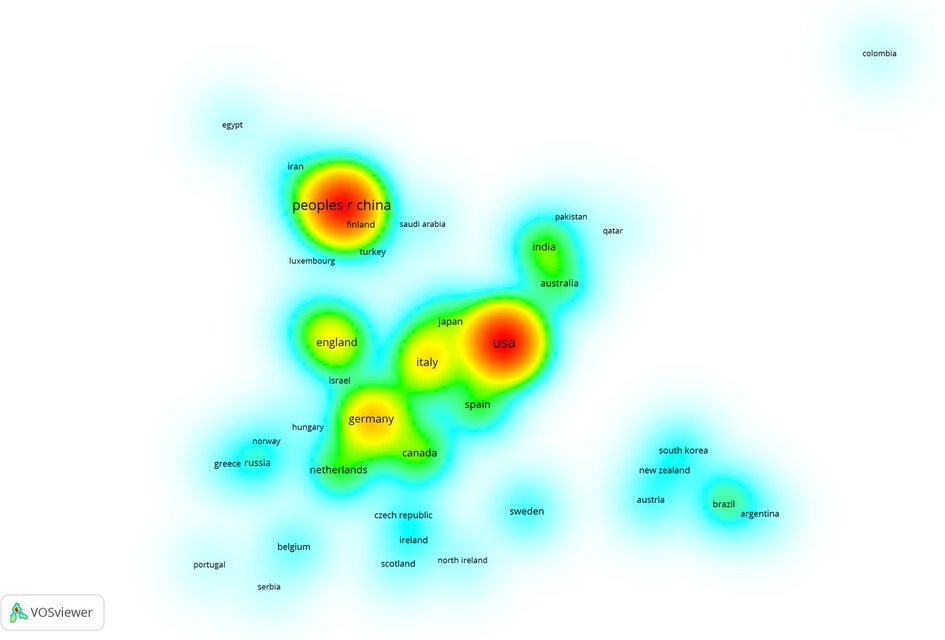
Figure 4. Item density of different countries in RNA methylation in CVD. The more items near a node, the higher the weight of adjacent items, and the closer the color of this point is to red.
3.2.3. Analysis of institutions
Table 2 shows the top 20 institutions in terms of publication volume among 1,188 research institutions, among which nine are Chinese research institutions. The top five were Fudan University (17 publications), Shanghai Jiao Tong University (15 publications), University of California, San Diego (14 publications), University of Zurich (14 publications), and Nanjing Medical University (14 publications). Figure 5 shows the top ten clusters (K = 10) established by CiteSpace analysis. Each cluster is displayed in a different color. Set parameters: The threshold is ten, and the node type is color ring history. The color of the node represents the year when the article was published. The reader should refer to the time color scale in the lower-left corner of the picture. The mechanisms at the ends of each line segment are cooperative relations. Modularity Q > 0.3 indicates significant modularity. Weighted Mean Silhouette S > 0.5 is reasonable clustering, and >0.7 means that the total cited clustering can clearly define each subdomain of clustering (23). Figure 5 contains 1,188 nodes and 3,445 lines, modularity Q = 0.8007, Weighted Mean Silhouette S = 0.9371, with high credibility. As shown in the figure, Fudan University, Nanjing Medical University, and Shanghai Jiao Tong University in China have close cooperation and are newly published; however, there was no evident cooperation relationship with institutions outside China. The University of California, San Diego, the Icahn School of Medicine at Mount Sinai, and the German Center for Cardiovascular Research work closely with Harvard Medical School but have not published recently.
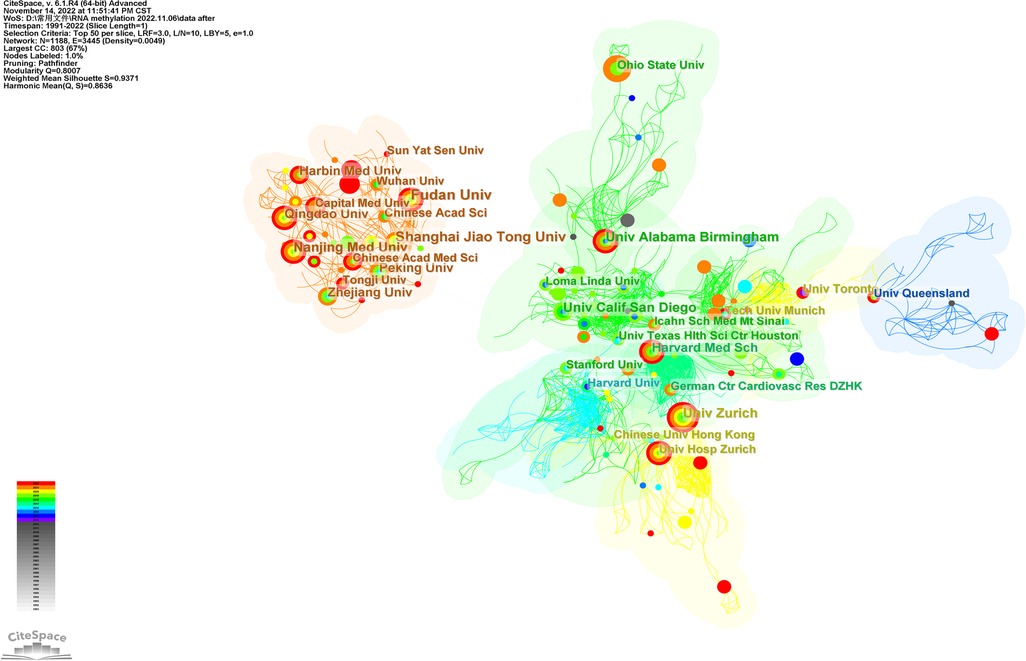
Figure 5. The visualization map of institutions involved in RNA methylation in CVD (K = 10). The size of nodes matches the publications of the institution. The links between nodes represent the institution collaborative relationships.
3.3. Journals and co-cited journals
3.3.1. Dual-map overlay
The dual-map overlay of journals shows the distribution of relationships between journals, citing journals on the left and cited journals on the right. The colored paths between them indicate the cited relationships. As shown in Figure 6, there are two primary citation paths, including one orange path and one green path. The two paths indicate that studies published in molecular/biology/immunology journals and medicine/medical/clinical journals cite from the studies in molecular/biology/immunology journals.
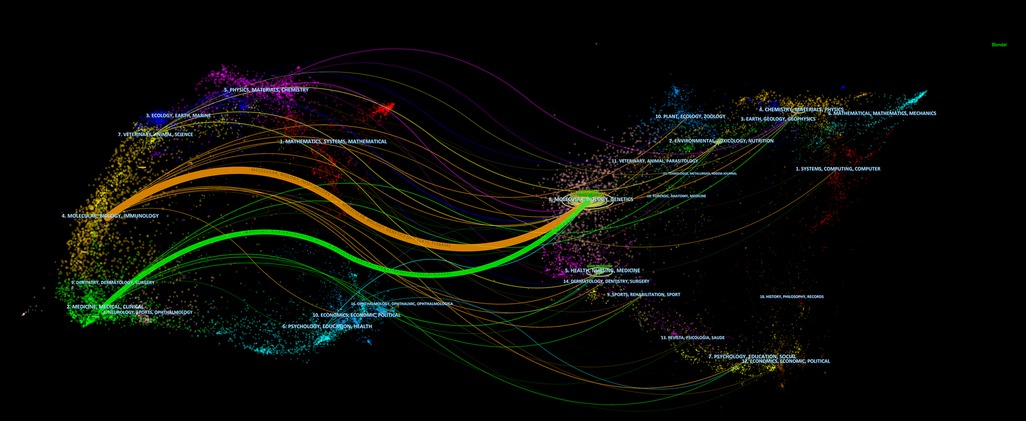
Figure 6. The dual-map overlay of journals on RNA methylation in CVD. Cited journals are on the left and cited journals are on the right. The link represents the cited path.
The dual-map overlay shows the citations between journals. As shown in Figure 6, cited journals are on the left, and cited journals are on the right. The colored curves between them indicate the relationship of citation. The orange and green curves represent the two primary reference paths, indicating that studies published in molecular/biology/immunology journals and medical/medical/clinical journals cited studies in molecular/biology/immunology journals.
3.3.2. Journal analysis
Since 1991, 847 articles on RNA methylation have been published in 317 academic journals, among which 35 journals had an IF more significant than five. The top five journals were the International Journal of Molecular Sciences (27 publications), Frontiers in Cardiovascular Medicine (22 publications), Circulation Research (15 publications), Frontiers in Cell and Developmental Biology (15 publications), and Clinical Epigenetics (14 publications). The IF of Circulation was the highest (39.922). Among the top ten journals, six are in JCR Q1: International Journal of Molecular Sciences (IF = 6.208), Circulation Research (IF = 23.218), Clinical Epigenetics (IF = 7.28), Circulation (IF = 39.922), Arteriosclerosis Thrombosis and Vascular Biology (IF = 10.514), and Frontiers in Genetics (IF = 4.772). Six journals had an IF more significant than five (Table 3).
3.3.3. Co-cited journals analysis
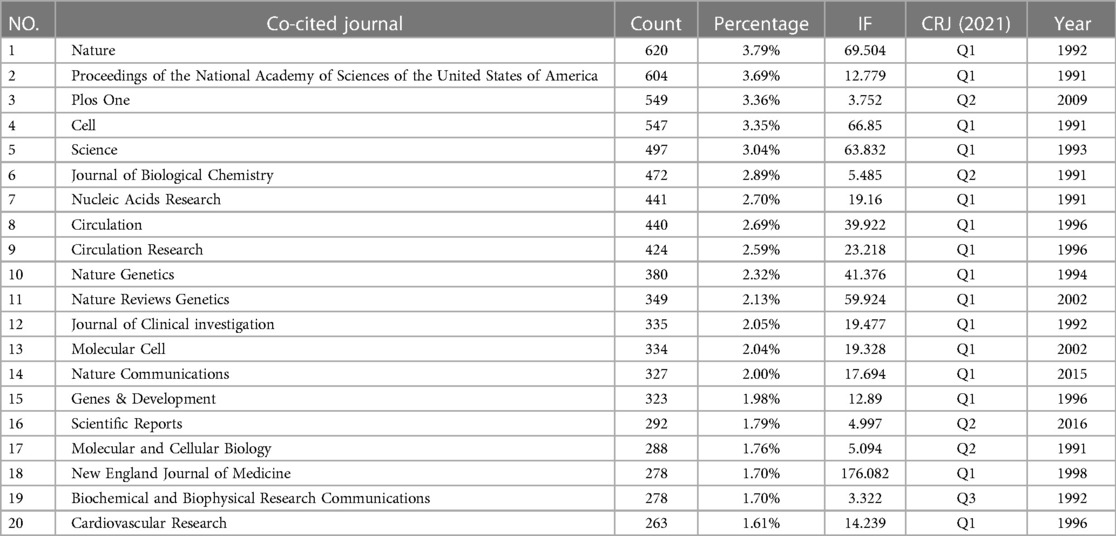
If papers are cited by one or more articles simultaneously, the two journals are said to have a co-citation relationship. The number of co-citations reflects the relevance of the literature in terms of content (24). CiteSpace was used for journal co-citation analysis, and there were 631 journals co-cited in this study. Thirty journals were cited more than 200 times, and nine were cited more than 400 times. As shown in Table 4, the top journals were Nature (620), Proceedings of the National Academy of Sciences of the United States of America (604), Plos One (549), Cell (547), Science (497), Journal of Biological Chemistry (472), Nucleic Acids Research (441), Circulation (440), Circulation Research (424), and Nature Genetics (380). Among the top ten cited journals, Nature had the highest IF of 69.504, followed by Cell (IF = 66.85), Science (IF = 63.832), and Circulation (IF = 29.69). Among the top 20 co-cited journals, 15 journals were CRJ Q1.
3.4. Co-cited reference and reference burst detection
3.4.1. Co-cited reference analysis
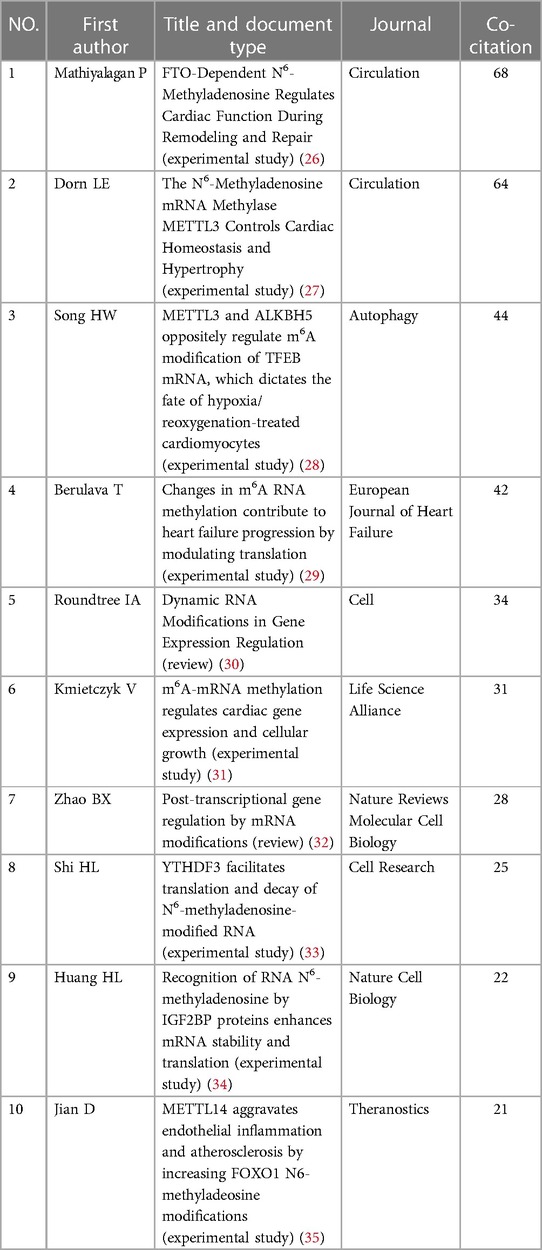
Co-citation refers to two or more papers being cited by one or more papers simultaneously (25). Table 5 displays the top ten co-cited references in which a co-citation occurred at least 20 times. “FTO-dependent N6-methyladenosine regulates cardiac function during remodeling and repair,” which was authored by Mathiyalagan and published in Circulation, was the most co-cited reference in RNA methylation in CVD (26), followed by the article entitled “The N6-methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy”. Nine of the top ten most highly co-cited papers were experimental studies, and one was a review article.
3.4.2. Co-cited references burst detection
Burst detection identifies events that occur unusually frequently or suddenly during a period (36). Co-cited references burst detection can detect significant changes in citations during a specific period. It is used to find the decline or rise of a specific co-cited study. As shown in Figure 7, CiteSpace detected 25 references with the most citation bursts. The earliest reference with citation bursts began in 2012, entitled “Distinct epigenomic features in end-stage failing human hearts” by Movassagh et al. and published in Circulation (37). Since 2012, the number of studies cited has increased yearly. Among the 25 studies, 14 had bursts in the previous three years and continued until 2022. All articles published after 2019 were published in CRJ Q1.
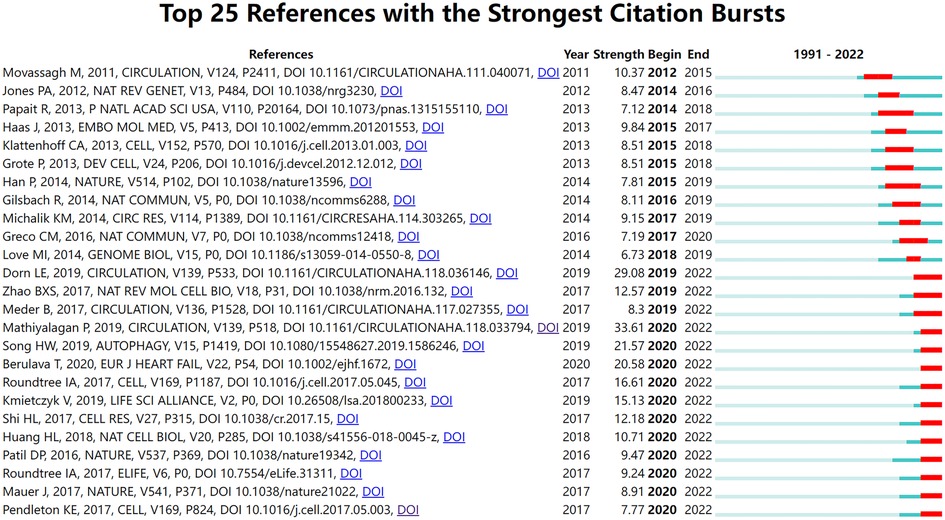
Figure 7. Top 25 references with the strongest citation bursts. The blue bar indicates that the reference has been published; the red bar means that the citation has exploded.
3.5. Authors and co-cited authors
3.5.1. Analysis of authors
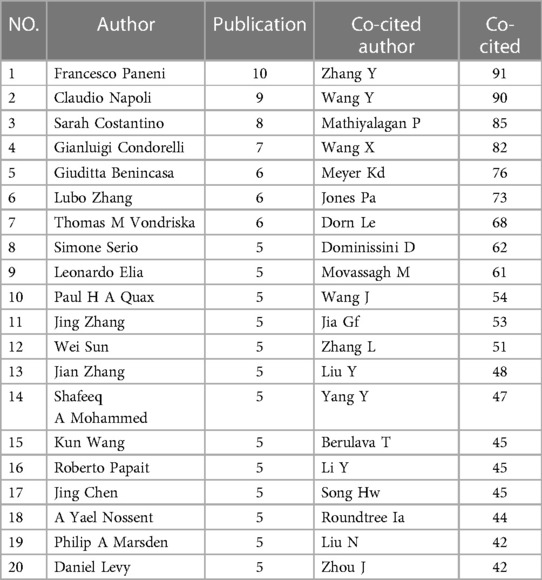
5,116 authors were included in 847 studies. Table 1 shows the top 20 authors in the number of publications. Francesco Paneni published the most articles (n = 10), followed by Claudio Napoli (n = 9), Sarah Costantino (n = 8), and Gianluigi Condorelli (n = 7).
3.5.2. Analysis of co-cited authors
If two (or more than one) authors were cited in one or more papers simultaneously, it is said that these two or more authors constitute a co-cited relationship. In this study, there were 2,809 co-cited authors, 12 of whom were cited more than 50 times, among which six were cited more than 70 times. Table 1 shows the top 20 authors with the most citations, of which the top five authors were Zhang (n = 91), Wang (n = 90), Mathiyalagan (n = 85), Wang (n = 82), and Meyer (n = 76). Figure 8 shows the top 10 clusters based on the keyword clustering for authors who co-cited over 30 times. The figure contains 2,809 nodes and 9,474 connections, Modularity Q = 0.8952, Weighted Mean Silhouette S = 0.8948. Each node represents an author, and more connections between nodes indicate closer author cooperation. The largest cluster in Figure 8 is 0#RNA methylation, containing 2,430 nodes, accounting for 86% of the total. Wang and Han, and Zhang Y and Zhang L showed close cooperation relationships.
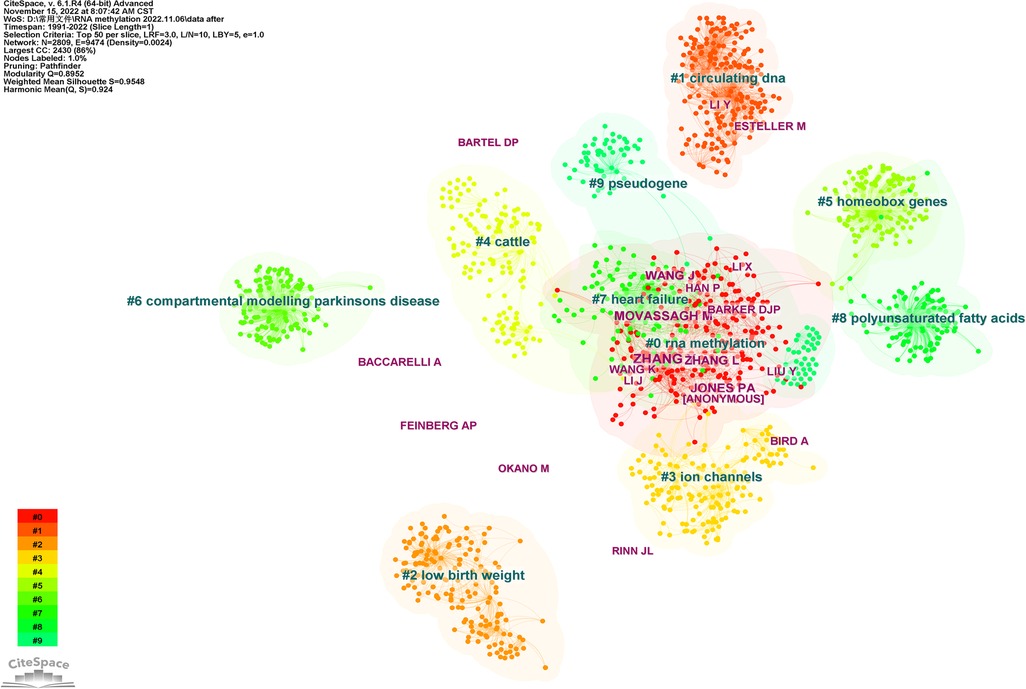
Figure 8. The visualization map of co-cited authors involved in RNA methylation in CVD (K = 10). The size of nodes matches the publications of the authors. Links between nodes represent co-citation relationships of authors.
3.6. Keyword co-occurrence and burst detection
3.6.1. Keywords timeline view
The keywords timeline view can reveal the period and research process of the development and evolution of various clusters. This research requires two years as a time slice to obtain the timeline view displaying keywords over time (Figure 9). Each node represents a keyword, and the node's size represents the cumulative use of the keyword from its appearance to the present. Keyword nodes appearing in different years show different tree ring colors. From 1991 to 2005, the top keywords were methylation, gene expression, DNA methylation, cell, messenger RNA, differentiation, transcription, and cgp island. From 1991 to 2005, chromatin, cardiovascular disease, ndothelial cells, coronary artery disease, oxidative stress, histone modification, and deacetylase were primary keywords. After 2015, the primary keywords were cardiac development, heart, promoter methylation, RNA methylation, and N6-methyladenosine.
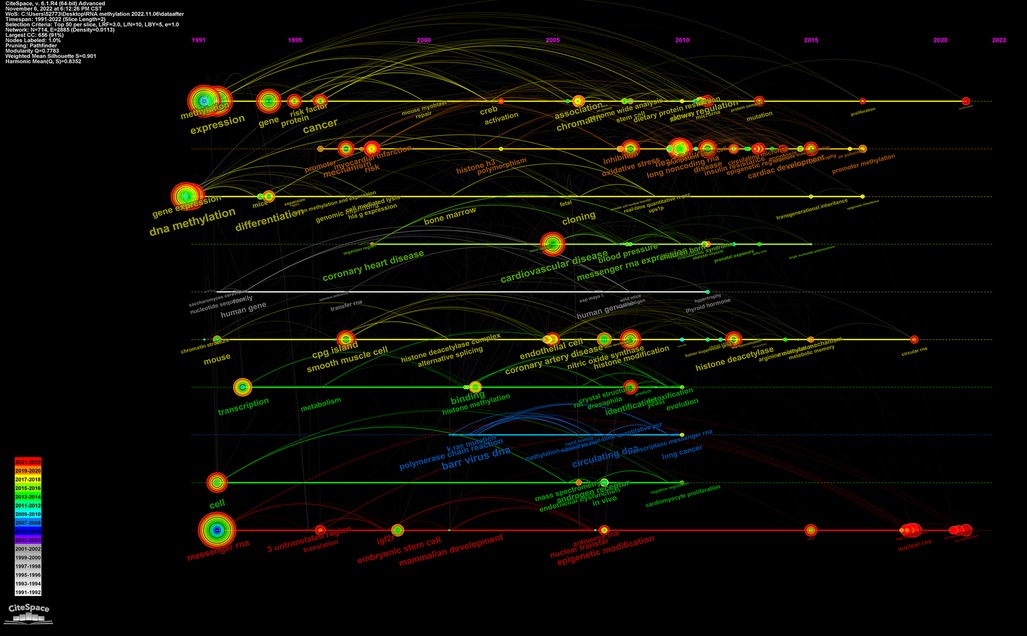
Figure 9. Timeline view of the CiteSpace visualization map relevant to RNA methylation in CVD. The horizontal lines of different colors represent the clusters formed by different keywords. The node on the horizontal line represents the keyword and its position represents the year in which the literature containing the keyword first appeared.
3.6.2. Keywords burst detection
In Figure 10, the green line segment's starting point represents the keyword's beginning and end. The red line segment represents the duration of keyword bursts. As we can see from the graph, there was no keyword burst before 2015. In 2016–2018, cardiac hypertrophy, coronary artery disease, DNA methylation, and histone methylation emerged as keywords. From 2016 to 2020, the number of keywords increased. These included inflammation, endothelial damage, RNA methylation, long noncoding RNA, proliferation, and others. Nuclear RNA, m6A methylation, inhibition, and myocardial infarction were new burst keywords from 2020 to the present.
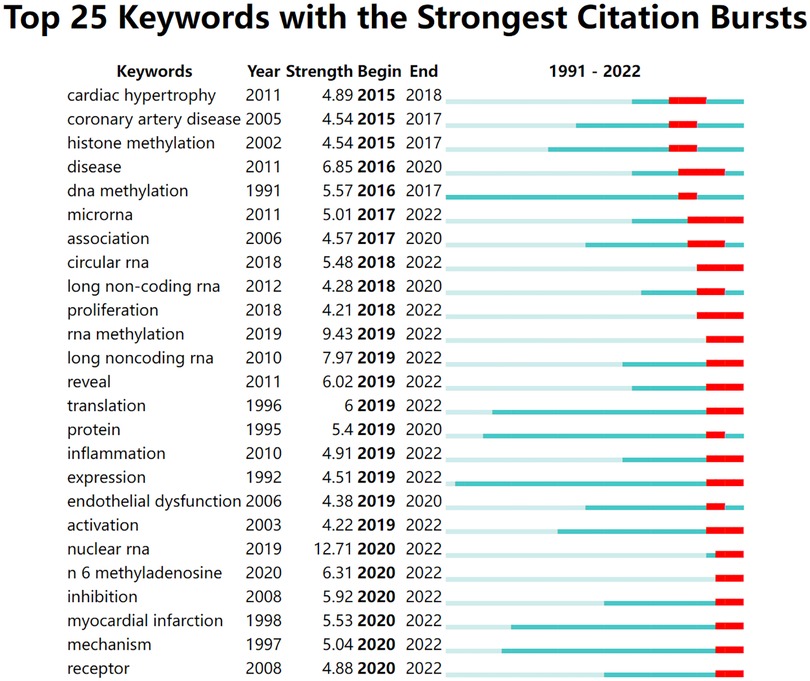
Figure 10. Top 25 keywords with the strongest citation bursts. The blue bar indicates that the keyword has been published; the red bar means that the keyword has burst.
According to the keywords burst detection, we can appreciate the rise of RNA methylation research from 2019. We identified articles after 2019. There were 83 articles with m6A as the topic, including 50 original studies, 13 articles with m5C as the topic (including four original studies), and only three review articles with m1A as the topic.
4. Discussion
4.1. General trend
The number of articles published in a field over the years can display the trend and popularity of research. Before 2010, the number of publications and citations related to RNA methylation in CVD was very small, with no papers in 2004 and only five in 2009. A turning point occurred in 2011, when 30 research papers were published, indicating that the field had captured the attention of researchers. Since then, the number of papers published has risen steadily. Since 2012, the number of citations of RNA methylation literature has increased rapidly, with a growth spurt from 2019 to 2021. Combined with the number of published articles and references cited, we infer that RNA methylation in CVD is rising rather than peaking. We expect that more articles will emerge in the next few years.
In national and institutional studies, the United States and China have an absolute advantage in the number of published articles; however, the institutions that publish articles are more scattered, and the number of published articles by a single research institution is not high. In contrast, though less than the United States in terms of the total number of articles published, China has the most among the top 20 institutions. This finding suggests that Chinese institutions such as Nanjing Medical University and Fudan University have a better research foundation and may make future breakthroughs in CVD RNA methylation.
In the journal analysis, the top ten journals were all in the CRJ Q1/Q2 area, suggesting that the outstanding journals have a high degree of recognition for the research results of RNA methylation. These results suggest that the study of RNA methylation in CVD has high credibility.
4.2. Research basis
The co-cited literature with high citations represents the research basis of a discipline. We interpreted the top five co-cited references.
The first co-cited paper was the experimental article published in Circulation by Mathiyalagan et al. (26). The functional roles of m6A and FTO in the heart and cardiomyocytes were investigated using clinical samples, pig and mouse models, and primary cardiomyocyte cultures. The authors found that the expression of the demethylation enzyme FTO was decreased in the HF model, and the increase of FTO expression in HF mice reduced the decline of systolic function induced by m6A. These findings suggest the functional importance of the FTO-dependent m6A methyl group in cardiac contraction in HF.
Second place went to Dorn et al. (27), published in Circulation. The authors showed that METTL3-mediated methylation of m6A mRNA is a dynamic modification. Enhanced m6A leads to compensatory myocardial hypertrophy, while inhibition of m6A leads to muscle cell remodeling and dysfunction, highlighting the importance of METTL3-mediated methylation of m6A mRNA for maintaining normal cardiac function.
Third place went to Song et al. (28) for an article published in Autophagy. This study found that METTL3 silencing enhanced autophagy and inhibited hypoxia/reoxygenation-induced apoptosis of cardiomyocytes. However, overexpression of METTL3 had the opposite effect, suggesting that METTL3 inhibits autophagy. The transcription factor EB activates its transcription by binding to the ALKBH5 promoter, while the inhibition of METTL3 downregulates the stability of mRNA and does not involve transcription inhibition. The essential function of RNA methylation in the bidirectional regulation of ischemic heart disease was revealed.
In fourth place was a study by Berulava et al. (29) published in the European Journal of Heart Failure. Changes in m6A RNA methylation outweighed changes in gene expression in mammalian models of HF. Moreover, changes in m6A methylation of mRNA affected the polymeric binding of corresponding transcripts in the heart, affecting protein balance in a transcription-independent manner.
Roundtree et al. (30) described RNA methylation with different modifications, including adenosine methylation and cytosine modifications. Adenosine methylation in studies includes m6A and m1A, while cytosine modifications include m5C and 5-hydroxymethylcytosine. This paper points out the technical difficulties of RNA methylation. Taking m6A methylation as an example, although m6A accounts for 0.2%–0.6% of all adenosine in mammalian mRNAs, only semi-quantitative detection can be performed within the transcriptome.
These studies explored the clinical, animal, and cellular levels. At present, there have been many studies on the methodology of RNA methylation. For example, XNA probe and CRISPR/Cas12a-powered flexible fluorescent and electrochemical dual-mode biosensor (38), mG-quant-seq (39), direct RNA sequencing (40), and other techniques provide technical support for the study of RNA methylation in CVD. We should also recognize the shortcomings of the research. The most prominent problems are the deviation of detection results due to low RNA methyl content (41, 42) and the lack of international standards for methylation studies. Although RNA methylation includes various types, current studies focus on the effects of m6A methylation on HF (43), ischemic cardiomyopathy (44, 45), and myocardial hypertrophy (46). Other diseases in CVD require further exploration.
4.3. Author analysis
Ten co-cited papers in the author's analysis were published by Francesco Paneni, assistant professor of cardiovascular epigenetics at the Schlieren Campus of the University of Zurich and team leader. A primary goal of Paneni's lab is to characterize the epigenetic mechanisms of the myocardium and vascular damage in the setting of cardiometabolic disturbances (47, 48).
Paneni's research team believes that epigenetic biomarkers should be regarded as the target of molecular intervention to treat diseases rather than blindly clinging to complex biological models of diseases (49). The aim is to apply epigenetic research to clinical applications rather than focusing on microscopic gene changes. One of these studies applied the central regulator SIRT1 to obese patients of different ages and found that SIRT1 affects the complex epigenetic control of p66sch or arginase II. NO availability and mitochondrial respiratory chain protein expression were affected to treat obesity and age-related early microvascular endothelial dysfunction (50). The team also reviewed drugs with known epigenetic effects and potential use in HF (51), including resveratrol (52), curcumin (53), and danshen (54).
The runner-up author, Claudio Napoli, published nine articles. Napoli's research on epigenetics is related to histones, microRNA, and DNA methylation, dedicated to the precise treatment of CVD and its high-risk pathogenic factors from the level of gene methylation (55–57). The Napoli group performed reduced-representation bisulfite sequencing and found that aortic endothelial cells exposed to high glucose had higher methylation differences of vascular endothelial growth factor (3.6 times), nitric oxide synthase 3 (1.6 times), and gene expression than normal aortic endothelial cells (58).
Third, Sarah Costantino suggested that studying cell-specific epigenetic information and developing chromatin modification drugs could provide a personalized risk assessment and treatment for patients with cardiometabolic disorders (51). The team examined the effects of hypoxia and hypertrophy stimulation on cardiomyocyte function and methylation levels and found that both resulted in epigenetic-driven upregulation of tenascin C and subsequent damage to cardiomyocyte energy metabolism, revealing the role of RNA methylation in the pathogenesis of CVD (59).
Gene-targeted therapy (60), precision medicine (61), and personalized therapy (62) were frequently used in co-cited literature. It is not difficult to see that the current research on RNA methylation in CVD is no longer limited to the simple mechanism study of disease but is committed to the clinical application of research results.
Co-cited author analysis can obtain high-influence authors in RNA methylation in CVD. We identified the authors with similar research topics, which authors created liaisons among topics, and which authors have multidisciplinary backgrounds or research fields with interdisciplinary attributes (63, 64). In the analysis of co-cited authors, Zhang, Wang, Mathiyalagan, and Wang were all cited more than 80 times. This finding suggests that their research results substantially influence this field.
4.4. Research hotspot and trend analysis
Using CiteSpace for burst detection analysis, we discovered the evolutionary trends of research from macro to micro, from single to diversified, reviewed the period when keywords become hot and predicted which keywords will continue the explosive trend in the future (65).
The keyword burst detection analysis showed no keyword emergence before 2015, suggesting that RNA methylation was an emerging field. In 2016–2018, investigators began to study the relationship between RNA methylation and specific types of CVD, including cardiac hypertrophy (66, 67); and coronary artery disease (68, 69), and began to explore mechanisms such as DNA methylation and histone methylation. From 2016 to 2020, RNA methylation-mediated molecular mechanisms, including inflammation and endothelial damage, attracted attention. Endothelial dysfunction and inflammatory infiltration are essential processes that promote the development of atherosclerosis. High blood lipids, smoking, high blood shear stress, and other factors lead to abnormal vascular endothelial function, release adhesion factors and chemokines, such that monocytes in blood migrate to the arterial subendothelial space, differentiate into macrophages to take up lipids, and convert into foam cells, promoting atherosclerosis (70). Studies found that FTO can promote the demethylation of m6A by yes-associated protein 1 and reduce apoptosis and inflammation of hypoxia/re-oxygenation-induced cardiomyocytes (71). Lysine methyltransferase Smyd1 reduces IL-6 expression by enhancing methylation of trimethylation of lysine-4 of histone-3 within the IL-6 promoter (72). After 2020, m6A RNA methylation, nuclear RNA, inhibition, and other microscopic precise regulation gradually attracted investigator attention (73, 74).
Co-citation burst detection reflects the research nodes in the development of a specific field and is suitable for investigating research hotspots and predicting development trends (75). There were four articles with a burst intensity >20 (Figure 7), all of which were experimental articles published in CRJ Q1 journals. An experimental article published by Mathiyalagan et al. in 2019 (strength = 34.78, Circulation) was also the highest co-citation. From clinical to animal to cell, from in vitro to in vivo research, this paper comprehensively and meticulously demonstrated the role of FTO de-m6A methylation. This article represents a turning point in research methods and content (26). The second, an experimental article by Dorn et al. published in 2019 (strength = 29.9, Circulation), inhibited m6A RNA methylation in vitro and in vivo, clarifying the importance of RNA methylation for maintaining cardiac homeostasis (27). The authors of the third paper, Song et al. (strength = 22.39, Autophagy), revealed a link between METTL3-ALKBH5 and autophagy, providing a scientific basis for the role of reversible mRNA m6A methylation in ischemic heart disease (28). A fourth, published in 2020 by Berulava et al. (strength = 21.37, European Journal of Heart Failure), found that m6A methylation is altered in cardiac hypertrophy and HF with corresponding protein abundance but independent of mRNA levels, revealing a novel transcription-independent translation regulatory mechanism (29).
In RNA methylation studies, m6A is the most common methylation modification in mRNA, with approximately 25% of mRNA carrying at least one m6A site (76), while m5C and m1A are found in tRNA and rRNA at relatively low levels (77, 78). m6A methylation is currently the focus of research on RNA methylation in CVD due to the differences in the content of different methylation types and the limited detection techniques.
Combined with citation bursts and keyword bursts, myocardial infarction, myocardial hypertrophy, and HF are current research hot spots. The exploration of methylation regulators related to these diseases and the clinical application of their results, especially the research related to m6A methylation, will become the goal of future research. The current RNA methylation detection methods are few and expensive, including liquid chromatography-mass spectrometry (79) and high-throughput sequencing (80). Dot blot represents a cost reduction for RNA methylation detection; however, the procedure is tedious and may result in false positives (81). Currently, the research on RNA methylation in the field of CVD is in its early stages, and optimizing the detection method of methylation to reduce the detection cost is a future direction of efforts.
5. Limitations
There are some shortcomings in this study. First, it was conducted on November 6, 2022, and covered all previous articles; however, some data may have been missed because the 2022 WoSCC database is still open to relevant documentation. Second, all data were downloaded from the WoSCC database, and studies not collected in the WoSCC were missed. WoSCC is the most used database in econometric analysis and contains the most information in the relevant core articles. Because the research data's quality may affect the knowledge graph's credibility, we chose WoSCC for this study. Other bibliometric studies also reported these limitations (82, 83).
6. Conclusion
We used CiteSpace and VOSviewer software visually to analyze RNA methylation in CVD and found that it has research value and application prospects in cardiovascular science. The leading countries studying this issue are the United States and China; however, other countries and institutions should strengthen cooperation. The increasing number of articles published in core international journals reflects the increasing emphasis of researchers in this field. The research focus of scholars shifted from basic to clinical research. Currently, research on RNA methylation in cardiovascular science focuses on the mechanism of ischemic heart disease, biomarkers, targeted therapy, and (especially) m6A methylation. These areas will be the focus of future research.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
BS: conceived the study. BX, ML and HL: collected the data. BS and HL: re-examined the data. BS and BX: analyzed the data. BS: wrote the manuscript. FZ and DS: reviewed and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Details of all funding sources will be provided, including grant numbers if applicable. This work was supported by the Natural Science Foundation of China (grant no. 81973674) and Beijing Municipal Natural Science Foundation (grant no. 7202175).
Acknowledgments
The authors thank Xiyuan Hospital, China Academy of Chinese Medical Sciences, for supporting this work.
Conflict of interest
The reviewer YZ declared a shared affiliation with the authors BS, BX, ML, DS, FZ to the handling editor at the time of review.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. NCD Countdown 2030 collaborators. NCD Countdown 2030: pathways to achieving sustainable development goal target 3.4. Lancet. (2020) 396(10255):918–34. doi: 10.1016/S0140-6736(20)31761-X
2. Francula-Zaninovic S, Nola IA. Management of measurable variable cardiovascular disease’ risk factors. Curr Cardiol Rev. (2018) 14:153–63. doi: 10.2174/1573403X14666180222102312
3. Ishikawa K, Weber T, Hajjar RJ. Human cardiac gene therapy. Circ Res. (2018) 123:601–13. doi: 10.1161/CIRCRESAHA.118.311587
4. Liu S, Li K, Wagner Florencio L, Tang L, Heallen TR, Leach JP, et al. Gene therapy knockdown of hippo signaling induces cardiomyocyte renewal in pigs after myocardial infarction. Sci Transl Med. (2021) 13:eabd6892. doi: 10.1126/scitranslmed.abd6892
5. Prasher D, Greenway SC, Singh RB. The impact of epigenetics on cardiovascular disease. Biochem Cell Biol. (2020) 98:12–22. doi: 10.1139/bcb-2019-0045
6. Mongelli A, Atlante S, Bachetti T, Martelli F, Farsetti A, Gaetano C. Epigenetic signaling and RNA regulation in cardiovascular diseases. Int J Mol Sci. (2020) 21:509. doi: 10.3390/ijms21020509
7. Menezo Y, Clement P, Clement A, Elder K. Methylation: an ineluctable biochemical and physiological process essential to the transmission of life. Int J Mol Sci. (2020) 21:9311. doi: 10.3390/ijms21239311
8. Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, et al. Corrigendum: RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature. (2017) 552:430. doi: 10.1038/nature24007
9. Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun. (2013) 4:1798. doi: 10.1038/ncomms2822
10. Chelmicki T, Roger E, Teissandier A, Dura M, Bonneville L, Rucli S, et al. m6A RNA methylation regulates the fate of endogenous retroviruses. Nature. (2021) 591:312–6. doi: 10.1038/s41586-020-03135-1
11. Zhang H, Shi X, Huang T, Zhao X, Chen W, Gu N, et al. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. (2020) 48:6251–64. doi: 10.1093/nar/gkaa347
12. Wu S, Zhang S, Wu X, Zhou X. m6A RNA methylation in cardiovascular diseases. Mol Ther. (2020) 28:2111–9. doi: 10.1016/j.ymthe.2020.08.010
13. Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. (2019) 74:640–50. doi: 10.1016/j.molcel.2019.04.025
14. Alriquet M, Calloni G, Martínez-Limón A, Delli Ponti R, Hanspach G, Hengesbach M, et al. The protective role of m1A during stress-induced granulation. J Mol Cell Biol. (2021) 12:870–80. doi: 10.1093/jmcb/mjaa023
15. Bohnsack KE, Höbartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m⁵C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes. (2019) 10:102. doi: 10.3390/genes10020102
16. Napoli C, Bontempo P, Palmieri V, Coscioni E, Maiello C, Donatelli F, et al. Epigenetic therapies for heart failure: current insights and future potential. Vasc Health Risk Manag. (2021) 17:247–54. doi: 10.2147/VHRM.S287082
17. Chien CS, Li JY, Chien Y, Wang ML, Yarmishyn AA, Tsai PH, et al. METTL3-dependent N-methyladenosine RNA modification mediates the atherogenic inflammatory cascades in vascular endothelium. Proc Natl Acad Sci U S A. (2021) 118:e2025070118. doi: 10.1073/pnas.2025070118
18. Ma D, Guan B, Song L, Liu Q, Fan Y, Zhao L, et al. A bibliometric analysis of exosomes in cardiovascular diseases from 2001 to 2021. Front Cardiovasc Med. (2021) 8:734514. doi: 10.3389/fcvm.2021.734514
19. Ma C, Su H, Li H. Global research trends on prostate diseases and erectile dysfunction: a bibliometric and visualized study. Front Oncol. (2021) 10:627891. doi: 10.3389/fonc.2020.627891
20. Chen C. Searching for intellectual turning points: progressive knowledge domain visualization. Proc Natl Acad Sci U S A. (2004) 101(Suppl 1):5303–10. doi: 10.1073/pnas.0307513100
21. Synnestvedt MB, Chen C, Holmes JH. Citespace II: visualization and knowledge discovery in bibliographic databases. AMIA Annu Symp Proc. (2005) 2005:724–8. 1560567.16779135
22. Van Eck N, Waltman L. Software survey: vOSviewer, a computer program for bibliometric mapping. Scientometrics. (2010) 84(2):523–38. doi: 10.1007/s11192-009-0146-3
23. Chen C. How to use citespace 6.1.R2. E. coli (2022). Available at: http://leanpub.com/howtousecitespace (Accessed June 12, 2022).
24. He J, Wu W. Comprehensive landscape and future perspectives of long noncoding RNAs (lncRNAs) in colorectal cancer (CRC): based on a bibliometric analysis. Noncoding RNA Res. (2022) 8(1):33–52. doi: 10.1016/j.ncrna.2022.10.001
25. Small H. Co-citation in the scientific literature: a new measure of the relationship between two documents. JASIST. (1973) 24:265–9. doi: 10.1002/asi.4630240406
26. Mathiyalagan P, Adamiak M, Mayourian J, Sassi Y, Liang Y, Agarwal N, et al. FTO-dependent N-methyladenosine regulates cardiac function during remodeling and repair. Circulation. (2019) 139:518–32. doi: 10.1161/CIRCULATIONAHA.118.033794
27. Dorn LE, Lasman L, Chen J, Xu X, Hund TJ, Medvedovic M, et al. The N-methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation. (2019) 139:533–45. doi: 10.1161/CIRCULATIONAHA.118.036146
28. Song H, Feng X, Zhang H, Luo Y, Huang J, Lin M, et al. METTL3 and ALKBH5 oppositely regulate mA modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. (2019) 15:1419–37. doi: 10.1080/15548627.2019.1586246
29. Berulava T, Buchholz E, Elerdashvili V, Pena T, Islam MR, Lbik D, et al. Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur J Heart Fail. (2020) 22:54–66. doi: 10.1002/ejhf.1672
30. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. (2017) 169:1187–200. doi: 10.1016/j.cell.2017.05.045
31. Kmietczyk V, Riechert E, Kalinski L, Boileau E, Malovrh E, Malone B, et al. m6A-mRNA methylation regulates cardiac gene expression and cellular growth. Life Sci Alliance. (2019) 2:e201800233. doi: 10.26508/lsa.201800233
32. Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. (2017) 18:31–42. doi: 10.1038/nrm.2016.132
33. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. (2017) 27:315–28. doi: 10.1038/cr.2017.15
34. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. (2018) 20:285–95. doi: 10.1038/s41556-018-0045-z
35. Jian D, Wang Y, Jian L, Tang H, Rao L, Chen K, et al. METTL14 aggravates endothelial inflammation and atherosclerosis by increasing FOXO1 N6-methyladeosine modifications. Theranostics. (2020) 10:8939–56. doi: 10.7150/thno.45178
36. Ye R, Cheng Y, Ge Y, Xu G, Tu W. A bibliometric analysis of the global trends and hotspots for the ketogenic diet based on CiteSpace. Medicine. (2023) 102(5):e32794. doi: 10.1097/MD.0000000000032794
37. Movassagh M, Choy MK, Knowles DA, Cordeddu L, Haider S, Down T, et al. Distinct epigenomic features in end-stage failing human hearts. Circulation. (2011) 124:2411–22. doi: 10.1161/CIRCULATIONAHA.111.040071
38. Pu Q, Ye Y, Hu J, Xie C, Zhou X, Yu H, et al. XNA probe and CRISPR/Cas12a-powered flexible fluorescent and electrochemical dual-mode biosensor for sensitive detection of m6A site-specific RNA modification. Talanta. (2023) 252:123754. doi: 10.1016/j.talanta.2022.123754
39. Zhang LS, Ju CW, Liu C, Wei J, Dai Q, Chen L, et al. m7G-quant-seq: quantitative detection of RNA internal N7-methylguanosine. ACS Chem Biol. (2022) 17(12):3306–12. doi: 10.1021/acschembio.2c00792
40. Hendra C, Pratanwanich PN, Wan YK, Goh WSS, Thiery A, Göke J. Detection of m6A from direct RNA sequencing using a multiple instance learning framework. Nat Methods. (2022) 19(12):1590–8. doi: 10.1038/s41592-022-01666-1
41. Chen K, Luo GZ, He C. High-resolution mapping of N⁶-methyladenosine in transcriptome and genome using a photo-crosslinking-assisted strategy. Methods Enzymol. (2015) 560:161–85. doi: 10.1016/bs.mie.2015.03.012
42. Clark WC, Evans ME, Dominissini D, Zheng G, Pan T. tRNA base methylation identification and quantification via high-throughput sequencing. RNA. (2016) 22:1771–84. doi: 10.1261/rna.056531.116
43. Vausort M, Niedolistek M, Lumley AI, Oknińska M, Paterek A, Mączewski M, et al. Regulation of N6-methyladenosine after myocardial infarction. Cells. (2022) 11:2271. doi: 10.3390/cells11152271
44. Wang J, Li Y, Zhang S. N(6)-methyladenosine modification: a vital role of programmed cell death in myocardial ischemia/reperfusion injury. Int J Cardiol. (2022) 367:11–9. doi: 10.1016/j.ijcard.2022.08.042
45. Wang T, Tian J, Jin Y. VCAM1 expression in the myocardium is associated with the risk of heart failure and immune cell infiltration in myocardium. Sci Rep. (2021) 11:19488. doi: 10.1038/s41598-021-98998-3
46. Li Y, Zhang W, Dai Y, Chen K. Identification and verification of IGFBP3 and YTHDC1 as biomarkers associated with immune infiltration and mitophagy in hypertrophic cardiomyopathy. Front Genet. (2022) 13:986995. doi: 10.3389/fgene.2022.986995
47. Ambrosini S, Mohammed SA, Costantino S, Paneni F. Disentangling the epigenetic landscape in cardiovascular patients: a path toward personalized medicine. Minerva Cardiol Angiol. (2021) 69:331–45. doi: 10.23736/S2724-5683.20.05326-8
48. Mohammed SA, Albiero M, Ambrosini S, Gorica E, Karsai G, Caravaggi CM, et al. The BET protein inhibitor apabetalone rescues diabetes-induced impairment of angiogenic response by epigenetic regulation of thrombospondin-1. Antioxid Redox Signal. (2022) 36:667–84. doi: 10.1089/ars.2021.0127
49. Khan AW, Paneni F, Jandeleit-Dahm KAM. Cell-specific epigenetic changes in atherosclerosis. Clin Sci. (2021) 135:1165–87. doi: 10.1042/CS20201066
50. Mengozzi A, Costantino S, Paneni F, Duranti E, Nannipieri M, Mancini R, et al. Targeting SIRT1 rescues age- and obesity-induced microvascular dysfunction in E×vivo human vessels. Circ Res. (2022) 131:476–91. doi: 10.1161/CIRCRESAHA.122.320888
51. Gorica E, Mohammed SA, Ambrosini S, Calderone V, Costantino S, Paneni F. Epi-drugs in heart failure. Front Cardiovasc Med. (2022) 9:923014. doi: 10.3389/fcvm.2022.923014
52. Bagul PK, Deepthi N, Sultana R, Banerjee SK. Resveratrol ameliorates cardiac oxidative stress in diabetes through deacetylation of NFkB-p65 and histone 3. J Nutr Biochem. (2015) 26:1298–307. doi: 10.1016/j.jnutbio.2015.06.006
53. Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, et al. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem. (2004) 279:51163–71. doi: 10.1074/jbc.M409024200
54. Barillas R, Friehs I, Cao-Danh H, Martinez JF, del Nido PJ. Inhibition of glycogen synthase kinase-3β improves tolerance to ischemia in hypertrophied hearts. Ann Thorac Surg. (2007) 84:126–33. doi: 10.1016/j.athoracsur.2007.02.015
55. Napoli C, Benincasa G, Donatelli F, Ambrosio G. Precision medicine in distinct heart failure phenotypes: focus on clinical epigenetics. Am Heart J. (2020) 224:113–28. doi: 10.1016/j.ahj.2020.03.007
56. Schiano C, Benincasa G, Franzese M, Della Mura N, Pane K, Salvatore M, et al. Epigenetic-sensitive pathways in personalized therapy of major cardiovascular diseases. Pharmacol Ther. (2020) 210:107514. doi: 10.1016/j.pharmthera.2020.107514
57. Benincasa G, Mansueto G, Napoli C. Fluid-based assays and precision medicine of cardiovascular diseases: the “hope” for pandora’s box? J Clin Pathol. (2019) 72:785–99. doi: 10.1136/jclinpath-2019-206178
58. Pepin ME, Schiano C, Miceli M, Benincasa G, Mansueto G, Grimaldi V, et al. The human aortic endothelium undergoes dose-dependent DNA methylation in response to transient hyperglycemia. Exp Cell Res. (2021) 400:112485. doi: 10.1016/j.yexcr.2021.112485
59. Hussain S, Khan AW, Akhmedov A, Suades R, Costantino S, Paneni F, et al. Hyperglycemia induces myocardial dysfunction via epigenetic regulation of JunD. Circ Res. (2020) 127:1261–73. doi: 10.1161/CIRCRESAHA.120.317132
60. Shi Y, Zhang H, Huang S, Yin L, Wang F, Luo P, et al. Epigenetic regulation in cardiovascular disease: mechanisms and advances in clinical trials. Signal Transduct Target Ther. (2022) 7:200. doi: 10.1038/s41392-022-01055-2
61. Wang L, Qiao C, Cao L, Cai S, Ma X, Song X, et al. Significance of HOXD transcription factors family in progression, migration and angiogenesis of cancer. Crit Rev Oncol Hematol. (2022) 179:103809. doi: 10.1016/j.critrevonc.2022.103809
62. Li H, An JR, Park M, Choi J, Heo R, Kang M, et al. The antidiabetic drug teneligliptin induces vasodilation via activation of PKG, Kv channels, and SERCA pumps in aortic smooth muscle. Eur J Pharmacol. (2022) 935:175305. doi: 10.1016/j.ejphar.2022.175305
63. Li Y, Ouyang Y, He C. Research trends on clinical fecal microbiota transplantation: a biliometric analysis from 2001 to 2021. Front Immunol. (2022) 13:991788. doi: 10.3389/fimmu.2022.991788
64. Zheng K, Wang X. Publications on the association between cognitive function and pain from 2000 to 2018: a bibliometric analysis using CiteSpace. Med Sci Monit. (2019) 25:8940–51. doi: 10.12659/MSM.917742
65. Ma D, Yang B, Guan B, Song L, Liu Q, Fan Y, et al. A bibliometric analysis of pyroptosis from 2001 to 2021. Front Immunol. (2021) 12:731933. doi: 10.3389/fimmu.2021.731933
66. Wang L, Wang J, Yu P, Feng J, Xu GE, Zhao X, et al. METTL14 is required for exercise-induced cardiac hypertrophy and protects against myocardial ischemia-reperfusion injury. Nat Commun. (2022) 13:6762. doi: 10.1038/s41467-022-34434-y
67. Fang M, Deng J, Zhou Q, Hu Z, Yang L. Maslinic acid protects against pressure- overload-induced cardiac hypertrophy by blocking METTL3-mediated m6A methylation. Aging (Albany NY). (2022) 14:2548–57. doi: 10.18632/aging.203860
68. Zhong N, Nong X, Diao J, Yang G. piRNA-6426 increases DNMT3B-mediated SOAT1 methylation and improves heart failure. Aging (Albany NY). (2022) 14:2678–94. doi: 10.18632/aging.203965
69. Sun Z, Chen W, Wang Z, Wang S, Zan J, Zheng L, et al. Matr3 reshapes m6A modification complex to alleviate macrophage inflammation during atherosclerosis. Clin Immunol. (2022) 245:109176. doi: 10.1016/j.clim.2022.109176
70. Duan Y, Zhang X, Zhang X, Lin J, Shu X, Man W, et al. Inhibition of macrophage-derived foam cells by Adipsin attenuates progression of atherosclerosis. Biochim Biophys Acta Mol Basis Dis. (2022) 1868:166533. doi: 10.1016/j.bbadis.2022.166533
71. Ke WL, Huang ZW, Peng CL, Ke YP. m6A demethylase FTO regulates the apoptosis and inflammation of cardiomyocytes via YAP1 in ischemia-reperfusion injury. Bioengineered. (2022) 13:5443–52. doi: 10.1080/21655979.2022.20305726
72. Shamloul A, Steinemann G, Roos K, Liem CH, Bernd J, Braun T, et al. The methyltransferase Smyd1 mediates LPS-triggered up-regulation of IL-6 in endothelial cells. Cells. (2021) 10:3515. doi: 10.3390/cells10123515
73. Wang X, Wu Y, Guo R, Zhao L, Yan J, Gao C. Comprehensive analysis of N6-methyladenosine RNA methylation regulators in the diagnosis and subtype classification of acute myocardial infarction. J Immunol Res. (2022) 2022:5173761. doi: 10.1155/2022/5173761
74. Chen J, Liu Z, Ma L, Gao S, Fu H, Wang C, et al. Targeting epigenetics and non-coding RNAs in myocardial infarction: from mechanisms to therapeutics. Front Genet. (2021) 12:780649. doi: 10.3389/fgene.2021.780649
75. Huang X, Yang Z, Zhang J, Wang R, Fan J, Zhang H, et al. A bibliometric analysis based on web of science: current perspectives and potential trends of SMAD7 in oncology. Front Cell Dev Biol. (2022) 9:712732. doi: 10.3389/fcell.2021.712732
76. Zhou W, Wang C, Chang J, Huang Y, Xue Q, Miao C, et al. RNA methylations in cardiovascular diseases, molecular structure, biological functions and regulatory roles in cardiovascular diseases. Front Pharmacol. (2021) 12:722728. doi: 10.3389/fphar.2021.722728
77. Yang T, Low JJA, Woon ECY. A general strategy exploiting m5C duplex-remodelling effect for selective detection of RNA and DNA m5C methyltransferase activity in cells. Nucleic Acids Res. (2020) 48(1):e5. doi: 10.1093/nar/gkz1047
78. Shima H, Igarashi K. N 1-methyladenosine (m1A) RNA modification: the key to ribosome control. J Biochem. (2020) 167(6):535–9. doi: 10.1093/jb/mvaa026
79. Marchand V, Motorin Y. Detection and functions of viral RNA modifications: perspectives in biology and medicine. Virologie. (2021) 25(2):5–20. doi: 10.1684/vir.2021.0884
80. Kisan A, Chhabra R. Modulation of gene expression by YTH domain family (YTHDF) proteins in human physiology and pathology. J Cell Physiol. (2023) 238(1):5–31. doi: 10.1002/jcp.30907
81. Zhu W, Wang JZ, Xu Z, Cao M, Hu Q, Pan C, et al. Detection of N6-methyladenosine modification residues (review). Int J Mol Med. (2019) 43(6):2267–78. doi: 10.3892/ijmm.2019.4169
82. Zhou R, Xiao L, Xiao W, Yi Y, Wen H, Wang H. Bibliometric review of 1992–2022 publications on acupuncture for cognitive impairment. Front Neurol. (2022) 13:1006830. doi: 10.3389/fneur.2022.1006830
Keywords: bibliometric analysis, RNA methylation, cardiovascular disease, visual analysis, citespace, VOSviewer
Citation: Song B, Xie B, Liu M, Li H, Shi D and Zhao F (2023) Bibliometric and visual analysis of RAN methylation in cardiovascular disease. Front. Cardiovasc. Med. 10:1110718. doi: 10.3389/fcvm.2023.1110718
Received: 29 November 2022; Accepted: 7 March 2023;
Published: 30 March 2023.
Edited by:
Jia Meng, Xi’an Jiaotong-Liverpool University, ChinaReviewed by:
Lingfeng Zha, Huazhong University of Science and Technology, ChinaYehao Zhang, China Academy of Chinese Medical Sciences, China
Qiuyue Ren, First Affiliated Hospital of Henan University of Traditional Chinese Medicine, China
Bai Chang, Tianjin Medical University, China
© 2023 Song, Xie, Liu, Li, Shi and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuhai Zhao 13911134962@163.com
Specialty Section: This article was submitted to General Cardiovascular Medicine, a section of the journal Frontiers in Cardiovascular Medicine
Abbreviations CVD, cardiovascular diseases; m6A, N6-methyladenosine; METTL3, methyltransferase-like 3; FTO, obesity-associated protein; ALKBH5, alkB homolog 5; m1A, N1-methyladenosine; m5C, 5-methylcytosine; IF, impact factor; HF, heart failure; WoSCC, web of science core collection.
 Boce Song
Boce Song Beili Xie1
Beili Xie1  Haohao Li
Haohao Li Dazhuo Shi
Dazhuo Shi