The presence of Enterococcus faecalis in saliva as a risk factor for endodontic infection
- 1Unit of Periodontology, Endodontic and Restorative Dentistry, Department of Medical Biotechnology, University of Siena, Siena, Italy
- 2Department of Endodontics, Faculty of Dentistry, Mansoura University, Mansoura, Egypt
- 3Faculty of Dentistry, The University of Hong Kong, Sai Ying Pun, Hong Kong, Hong Kong SAR, China
Aim: The aim of the present study was to investigate and correlate the prevalence of Enterococcus faecalis in saliva and in root canals with different pulpal and periapical conditions.
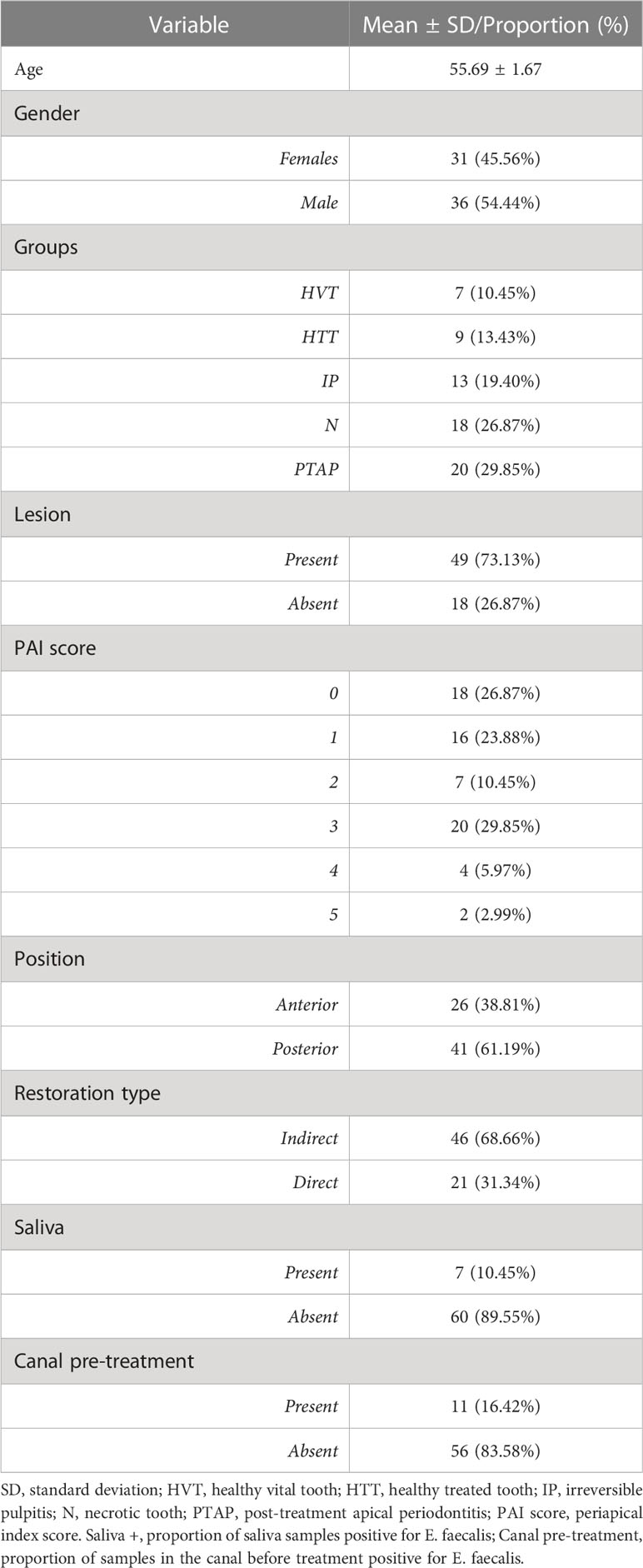
Methodology: Sixty-seven patients were divided into five groups based on pulpal and periapical tissue status: healthy vital teeth (HVT, n=7), healthy treated teeth without lesion (HTT, n=9), irreversible pulpitis (IP, n=13), necrosis (N, n=18), and post-treatment apical periodontitis (PTAP, n=20). Saliva, rubber dam, sterility control and pre-treatment root canal samples were collected and microbiologically processed by culture method. The phylogenetic relationship of E. faecalis isolates collected from root canals and saliva were investigated by whole genome sequencing. Fisher’s exact test was used to correlate the presence of E. faecalis in root canals or saliva with clinical and/or radiographic findings. Linear/logistic regression analyses were performed to establish the relationship between the presence of E. faecalis in root canals, saliva, and the status of periapical tissues.
Results: E. faecalis was found in 18 root canal and saliva samples. E. faecalis root canal isolates were recovered with the highest frequency from post-treatment apical periodontitis. The occurrence of E. faecalis in saliva was strongly associated with its detection in the root canals (P < 0.001). The pretreatment presence of E. faecalis in root canals was associated with significantly higher odds of having periapical lesions (OR=11.03; 95% CI, 1.27-95.70; p < 0.05). Saliva and root canal isolates from the same patient were highly correlated at the phylogenetic level (Jaccard index >0.95).
Conclusion: This pilot study confirms the role of E. faecalis in developing peri-radicular lesions in secondary endodontic infections and suggests that saliva could be the main source of infection. Further studies are needed to investigate the exact origin of this bacteria and its true role in the pathogenesis of secondary/persistent endodontic infections.
Introduction
The role of bacteria in the initiation and progression of apical periodontitis has been widely demonstrated (Kakehashi et al., 1965). Bacteria can invade the root canal system via different pathways such as carious, periodontal lesions and cracks (Siqueira and Rôças, 2009). Microbial profiling of endodontic infections revealed compositionally unspecific, yet differentially abundant microbiota depending on clinical diagnosis (Manoil et al., 2020). While primary infections are caused by microorganisms that initially invade and colonize necrotic root canals, secondary and persistent infections are caused by microorganisms that enter root canals as a result of professional intervention or survive the chemo-mechanical debridement and persist within the root canal environment (Wong et al., 2021). Enterococcus faecalis is a facultative anaerobic gram-positive bacterium which has been frequently recovered from secondary/persistent endodontic infections (Rôças et al., 2004; Sedgley et al., 2006b; Bouillaguet et al., 2018). The contribution of E. faecalis to endodontic treatment failures is attributed to its ability to withstand nutrient scarcity encountered in root-filled teeth (Evans et al., 2002; Stuart et al., 2006) and tolerance to antimicrobials employed during endodontic treatment (Ali et al., 2020a; Ali et al., 2020b). The ability of E. faecalis to form dense biofilms on root canal walls, by a biofilm-associated pili (Ebp) and its collagen-binding protein (Ace), make this microorganism able to invade dentinal tubules and root canal complexities and contributing to be recalcitrant to endodontic disinfectants and intracanal dressings (Zhang et al., 2015; Hahn and Hanford, 2021; Momenijavid et al., 2022). Also many other virulence factors and its predisposition to be resistant to some antibiotics contribute to persistence and recovery of E. faecalis from endodontic failures (Aas et al., 2005; Gomes et al., 2021). Given that endodontic microbiota is derived from oral microbiota under the influence of specific ecological conditions of root canal environment (Wong et al., 2021), and despite the recovery of E. faecalis from root-filled teeth with post-treatment diseases, E. faecalis is not a typical member of commensal oral microbiota (Aas et al., 2005). It is less likely that E. faecalis occurs in advanced carious lesions, and primary endodontic infections (Martin et al., 2002). Therefore, the origin of E. faecalis recovered from root canals has been questioned, and its association with their respective counterparts in saliva was studied (Zehnder and Guggenheim, 2009). A significant association was found between the presence of E. faecalis in saliva and root canals with post-treatment apical periodontitis (Wang et al., 2012). Similar genotype was detected in E. faecalis isolated from saliva and endodontically treated teeth (Delboni et al., 2017), while different genetic profiles were observed in salivary and root canals strains from the same patient (Zhu et al., 2010). Therefore, the origin of E. faecalis in endodontic treatment failures was proposed to be exogenous (Vidana et al., 2011). With such contradictory findings, the relationship between E. faecalis in saliva and root canals remains unsolved and additional evidence is warranted. Therefore, the aim of this study was to determine the prevalence of E. faecalis in root canals and saliva and to investigate whether its presence could influence the presence and dimension of periapical lesions.
Materials and methods
Study design
The present cohort study is reported following the Strengthening the Reporting of Observational studies in Epidemiology (STROBE) guidelines for cohort studies (Von Elm et al., 2008). The research protocol was approved by the local Ethics Committee (protocol number: 18202/2020) and was registered on Clinicaltrials.gov (NCT04637659).
Setting and participants
Sixty-seven patients were sequentially recruited among those attending the Unit of Endodontology and Restorative dentistry, School of Dentistry, University of Siena between July 2020 and November 2020 according to the following eligibility criteria:
- need for a root canal treatment or retreatment with previous therapy aging for at least five years.
- ability and willingness to give informed consent.
The exclusion criteria were:
- presence of periodontitis (Tonetti et al., 2018).
- impossibility to isolate the operating field.
- retreatment cases with missing or calcified canals, perforation and separated endodontic instruments in which was impossible to reach the apex.
- administration of antibiotics within the last 3 months.
- patients with diabetes, rheumatoid arthritis, and inflammatory bowel diseases.
The cohort of patients included in the present study was defined once all participants read and signed a written informed consent, according to the Declaration of Helsinki.
Variables
Clinical and radiographic assessment
For each participant, demographic characteristics (age, gender) as well as medical and dental history were collected. Tooth position (anterior/posterior) and type of coronal restoration (direct/indirect) were recorded during the clinical examination; The quality of each restoration was defined as proper or improper, according to the “Modified USPHS ‘‘ criteria (Bayne and Schmalz, 2005). A standardized periapical intraoral radiograph was performed to evaluate the status through the Periapical Index (PAI) score (Orstavik et al., 1986). Afterwards, the included teeth were categorized into five groups according to their pulpal and periapical status as determined by clinical and radiographic findings: (i) healthy vital tooth (HVT) group was represented by a clinical situation in which endodontic treatment is needed for prosthetic reasons despite the pulp not showing any sign of inflammation; (ii) healthy treated tooth (HTT) group included teeth in which the pre-existing endodontic filling material was exposed to oral cavity with no sign of periapical lesion; (iii) irreversible pulpitis (IP) diagnosed by sharp spontaneous pain and tenderness to percussion or pain exacerbated by lying down or cold test (Levin et al., 2009); (iv) pulp necrosis (N) group belonged to untreated teeth, negative to cold test, with and without apical periodontitis; post-treatment apical periodontitis (PTAP).
Sampling and clinical procedures
Root canal and saliva samples were collected as previously described (Sedgley et al., 2006a). Before isolation with the rubber dam, saliva samples from the floor of the mouth, dorsum of the tongue and the crown of the affected tooth were collected for each patient using three sterile ISO size 40 paper points (Dentsply-Maillefer, Ballaigues, Switzerland). The paper points were resuspended in 100 μl of PBS/10% glycerol and stored at -70°C until analysis. Plaque around the affected tooth was removed using scalers and the surfaces were brushed with pumice. Teeth were isolated with a rubber dam and disinfected with 30% hydrogen peroxide and 5.25% sodium hypochlorite (NaOCl), which is inactivated by sodium thiosulphate 5%. As a sterility control, three sterile paper points (Size 40) were rubbed on the crown of the tooth and on the surrounding areas. After access preparation, root canal patency was achieved with minimal instrumentation and without using hypochlorite irrigant. In case of retreatment, coronal gutta percha was removed by sterile Gates Glidden drills size 2 & 3 (Dentsply-Maillefer, Ballaigues, Switzerland), while the middle and apical gutta percha were removed with endodontic files without a chemical solvent. Irrigation was performed with sterile saline to remove any residual material before the collection of the intracanal sample. Once the working length was established, the pre-treatment sample was collected using ISO size 10 K-file (Dentsply-Maillefer, Ballaigues, Switzerland). An additional pretreatment sampling was performed by introducing two sterile paper points (ISO size 15) into the full working length kept for at least 60 seconds. The sample was then transferred to PBS/10% glycerol solution. When the canal was dry, a sterile paper point moistened with sterile saline was used to acquire the sample. In multi-rooted teeth, a single root canal was chosen, based on the presence of periapical radiolucency and/or exudation.
Laboratory assessment
Isolation and identification of Enterococci
Ten μl of PBS/10% glycerol from each sample were plated on Brain Heart Infusion (BHI) agar containing 5% horse blood. The plates were incubated in 5% CO2 at 37°C for 48 hours and monitored daily for the presence of microbial growth. Putative enterococcal colonies were isolated on BHI agar/blood and identified with a latex agglutination test (Oxoid™ Streptococcal Grouping Kit, Thermo Fisher, Hampshire, United Kingdom). Group D colonies were then identified on a MALDI Biotyper (Bruker Daltonics, Bremen, Germany) and by ribosomal RNA operon sequencing (Cuscó et al., 2018). Colonies identified as E. faecalis were frozen at -70°C in BHI/10% glycerol.
High molecular weight DNA extraction
E. faecalis strains were streak plated on BHI agar/blood, incubated overnight at 37°C and checked for purity. About ten single colonies were inoculated in BHI broth and the starter cultures of exponentially growing bacteria (OD590 of 0.3-0.4) were frozen at -70°C with 10% glycerol. Bacteria were inoculated 1:50 (vol:vol) from starter cultures in 10 ml of BHI broth and incubated at 37° C until an OD590 of 1.0 was reached. Samples were then centrifuged at 6600 x g for 5 minutes. Bacterial pellets were washed with 10 ml of sterile 1X TE buffer (Tris 10 mM-EDTA 1 mM) and resuspended in 7.5 ml of Raffinose buffer (50 mM Tris pH 8, 5 mM EDTA, 20% Raffinose). DNA extraction was carried out as described previously (Pinzauti et al., 2022). The DNA pellet was resuspended in 100 μl of saline. Genomic DNA was quantified using a Qubit 2.0 fluorometer (Invitrogen, Whaltan, Massachusetts, USA) and a NanoPhotometer device (Implen, Westlake Village, USA) before molecular analysis and whole genome sequencing.
Sequencing and bioinformatic analysis
Whole genome sequencing (WGS) was performed employing Oxford Nanopore technology. Following manufacturers’ instruction, the sequencing library was prepared using a ligation sequencing kit (SQK-LSK108) and barcode expansion kits (EXP-NBD104/114) for sample multiplexing. The sequencing run was performed on the GridION x5 platform (Oxford Nanopore Technologies). Nanopore reads were filtered using the tool Filtlong (v. 0.2.0) (https://github.com/rrwick/Filtlong) removing reads shorter than 1,000 bases (–min_length 1000) and getting rid of the 5% worst (low quality) reads (–keep_percentage 95). Samples were also sequenced with Illumina technology at MicrobesNG (Birmingham, UK) (https://microbesng.com/) which performed library preparation and sequencing of paired end 250 bp reads on a HiSeq2500. Raw Illumina reads were quality checked at MicrobesNG: reads were trimmed using Trimmomatic (v. 0.30) (Bolger et al., 2014) and analyzed with FastQC (v. 0.11.5) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc).
High quality complete genomes were de novo assembled using Unicycler (v 0.4.7) (Wick et al., 2017), with both Nanopore and Illumina reads as an input. Phylogenetic relationships among sequenced genomes were explored using PopPUNK (v. 2.4.0) using the ‘fit-model lineage’ parameter for data fitting (Lees et al., 2019). PopPUNK exploits the Jaccard index (J) to establish the similarity between k-mer data sets (oligonucleotide sequences of k length) of two genome sequences (0<J<1, with J=1 describing two genome sequences sharing the same k -mers) (De Giorgi et al., 2022).
Power analysis
The detection rate of E. faecalis in culture medium was reported to be 2% and 71% in primary and secondary endodontic infections respectively (Guo et al., 2011). Therefore, setting the level of significance at alpha=0.05, the power of the study resulted to be above 90%.
Statistical analysis
All analyses were performed using a statistical software (STATA BE, version 17.1, StataCorp LP, Texas, USA), setting the level of significance at 5%. Continuous variables were expressed as Mean (SD), while categorical variables were expressed as number of observations (percentage - %). Fisher’s exact test was used to investigate the association between clinical and microbiological variables. Simple linear/logistic multilevel regression models were built in order to evaluate the association between E. faecalis presence in the canal before treatment/E. faecalis presence in saliva and tooth vitality, presence of periapical lesion and PAI score, respectively. Multiple multilevel regression models were obtained by adjusting the crude estimates for confounders (i.e. proper/improper restoration, type of restoration, tooth position).
Results
Participants and samples
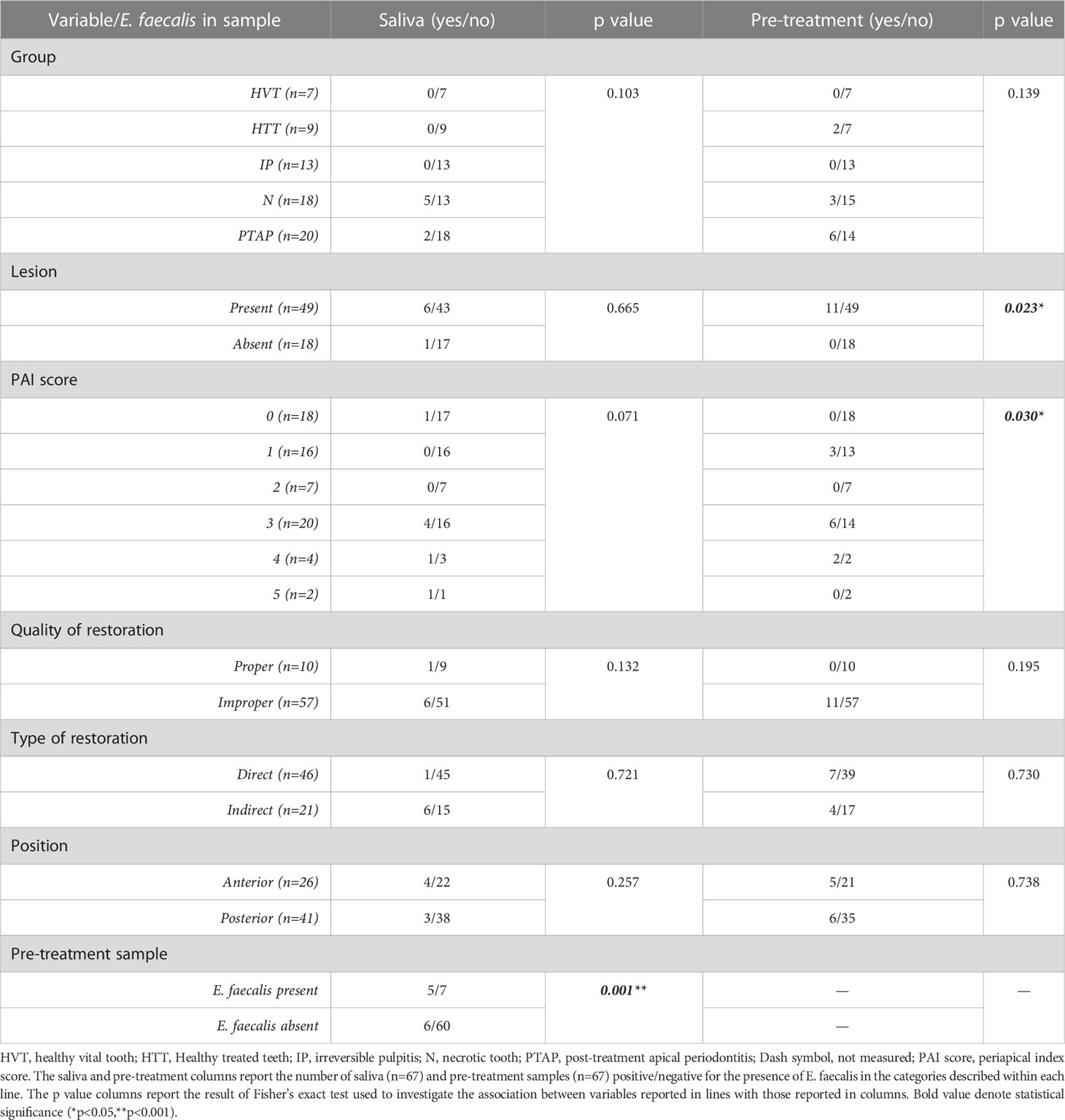
Sixty-seven patients (36 males and 31 females), aged from 26 to 90 (mean ± SD = 56 ± 1.67), were included in the study. A total of 79 teeth in the recruited patients were sampled. Eleven samples were discarded due to sampling or laboratory errors, and one saliva sample was repeated in the same patient after 4 months after endodontic therapy. Therefore, a total of 67 teeth were included in the final analysis. E. faecalis was recovered from 11 (16.42%) and 7 (10.45%) root canal and saliva samples, respectively. The highest frequency was from the PTAP group (30%), followed by N (22.2%) and HTT (16.6%) groups. E. faecalis was not detected in IP and HVT groups. Descriptive statistics of patients’ characteristics, and clinical and microbiological assessments are shown in Table 1.
Outcome data
Clinical and microbiological variables
Results of the association between clinical and microbiological variables are shown in Table 2. The presence of E. faecalis in root canals before the endodontic treatment (pre-treatment) was significantly associated with the presence in saliva (p<0.001) and with the presence of radiologically evident periapical lesions (p<0.05). The tooth position, type and quality of coronal restorations were not significantly associated with the presence of E. faecalis in any of the samples.
Linear/logistic regression analyses
The combined effect of the variables that were related to the presence of E. faecalis in pre-treatment samples were investigated using a logistic regression model (Table 3). The presence of E. faecalis in root canal samples before the treatment significantly increased the odds of having a secondary endodontic infections (OR=2.94; 95% CI [1.47, 11.59]; p<0.05) while its presence in saliva was associated with higher odds of identifying E. faecalis in root canals (OR=3.70; 95% CI [1.031, 19.229]; p<0.05) and to develop a secondary/persistent infection (OR=3.07; 95% CI [1.67, 6.88]; p<0.05). The presence of Enterococcus faecalis in root canals significantly increased the odds of periapical lesion (OR=11.03; 95% CI [1.273, 95.704]; p<0.05). However, this was not the case when E. faecalis was identified in saliva (OR=1.97; 95% CI [0.333, 11.674]; p<0.454). Finally, the presence of E. faecalis in pretreatment samples increases the odds of a higher PAI index score (MD=1.031; 95% CI[0.091, 1.971]; p<0.05).
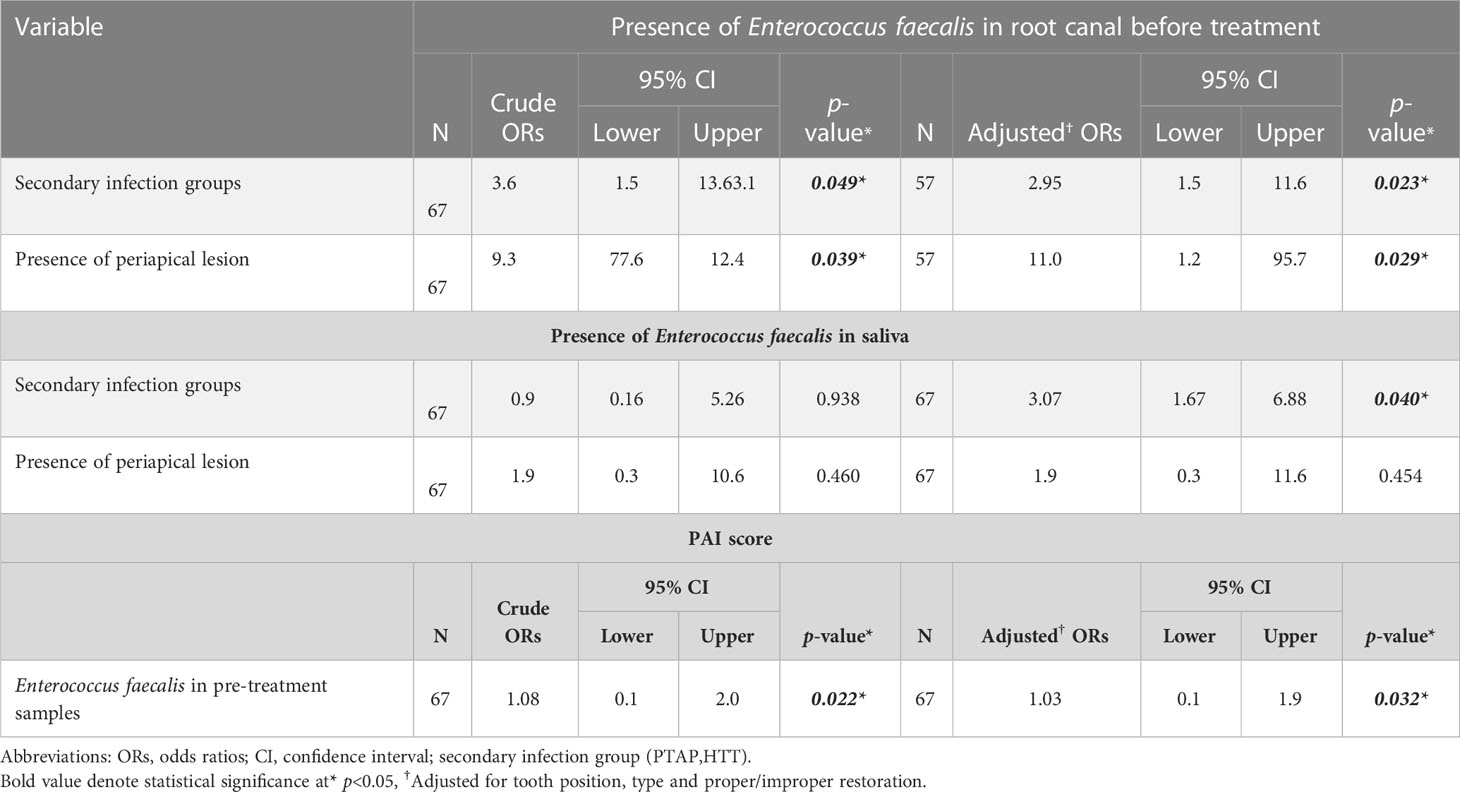
Table 3 Linear/logistic regression analyses for the association between E.faecalis in the root canal/saliva and PAI score with clinical variables, respectively.
Whole genome sequencing and phylogenetic relationships of E. faecalis isolates
Seventeen out of 18 E. faecalis strains were sequenced using both Illumina and Nanopore technologies (n= 17) or only with Nanopore (n=14). One strain was not vital and could not be sequenced. Genomes were assembled and the whole genome sequences were used to investigate the phylogenetic relationships among different isolates with PopPUNK (Figure 1). Core genome analysis and whole genome analysis identified five major clusters. Analysis of phylogenetic distances among isolates using the Jaccard index (J) with different k-mer lengths indicated that saliva and root canal isolates of E. faecalis retrieved from the same patient, BE15 and BE43, BE16 and BE17, BE7 and BE8, BE32 and BE33 were highly correlated (J>0.95). Moreover, the strains BE11 and BE32 were recovered from the saliva of the same subjects at 2 different visits 4 months apart, and share a J index of 0.987 (k=29). The strains BE5 and BE52 share a J index of 0.8 although they were recovered from root canals of different subjects.
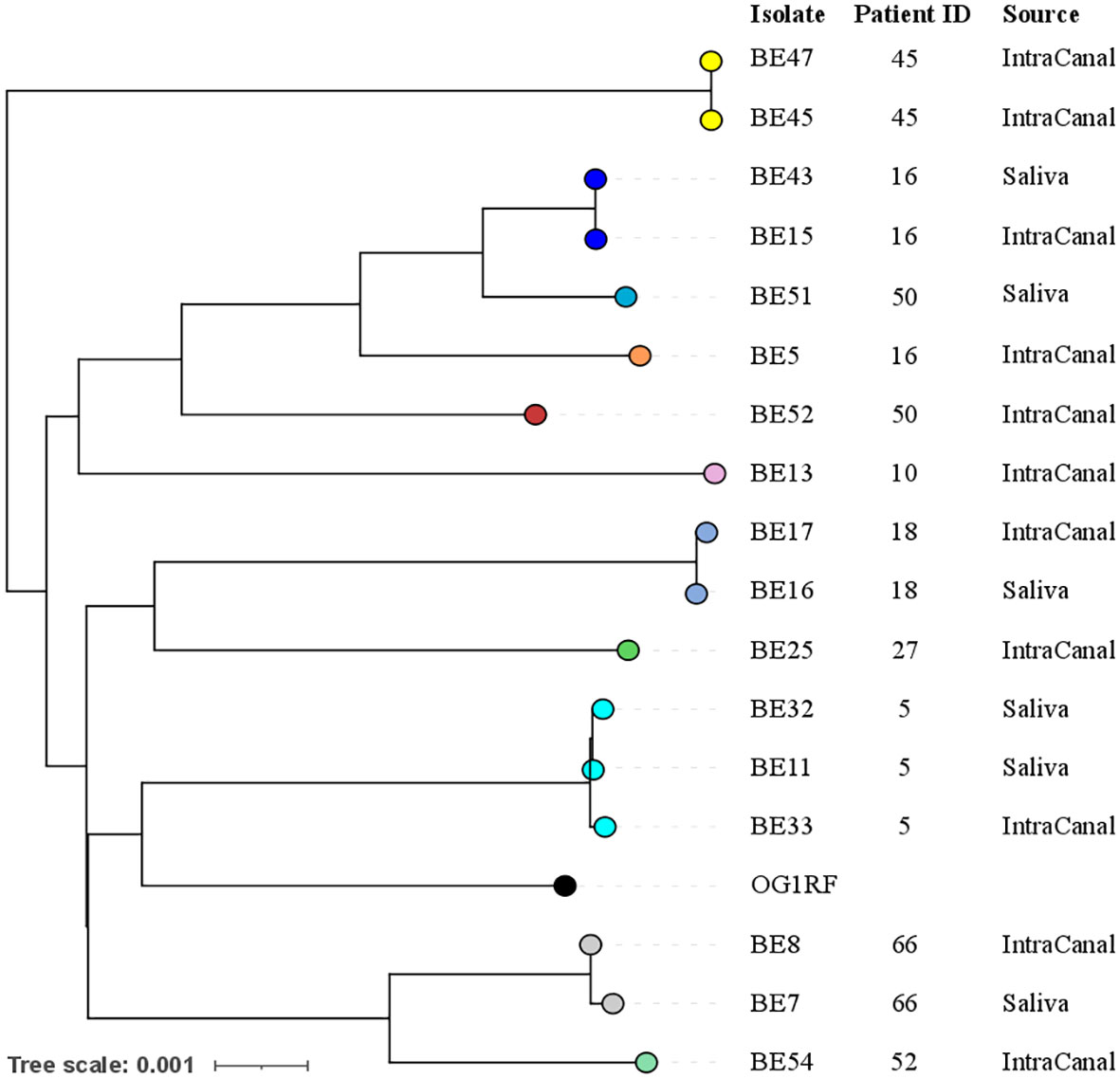
Figure 1 Phylogenetic relationships among E. faecalis isolates. Different colors indicate different genomic clusters: ten population clusters were identified using core genome sequences, 5 clusters contain a single genome, while another 5 contain either 2 or 3 genomes, notably all coming from the same patient. The phylogenetic tree was generated based on whole genome sequences with branch lengths indicating the number of nucleotide substitutions per site (scale bar). Patient ID and source of the sample are indicated on the right. The genome of the reference laboratory strain OG1RF was used as an outgroup.
Discussion
The present study aimed to assess the prevalence and correlation between E. faecalis isolates from root canals, with different pulpal and periapical conditions, and saliva to better understand the origin of E. faecalis in endodontic infections. This study analyzed the association of E. faecalis presence in root canals before treatment with (i) the status of periapical tissues, and (ii) clinical characteristics such as type, quality and location of restorations. Previous studies investigated the prevalence of E. faecalis in failed endodontic treatment and persistent infections (Zhu et al., 2010; Wang et al., 2012; Delboni et al., 2017), and recovered E. faecalis from primary endodontic infections (Rôças et al., 2004; Sedgley et al., 2006a; Guo et al., 2011). This study included clinical conditions ranging from healthy pulp to teeth with post-treatment apical periodontitis, classifying each condition into primary or secondary/persistent endodontic infection groups as previously established (Delboni et al., 2017). A cultural approach was used to isolate E. faecalis from saliva and endodontic samples, this allowed to recover strains for further molecular characterization and to avoid PCR-based techniques, which could be influenced by contamination and by the presence of extracellular DNA or DNA from dead bacterial cells (Siqueira, 2002; Gomes et al., 2015). Recently, Next Generation Sequencing (NGS)-based studies revealed an unspecific composition of endodontic microbiota (Wong et al., 2021), and challenged the role of E. faecalis in the etiology of persistent/secondary root canal infections (Rogers et al., 2010; Brundin et al., 2014) even if a recent 16S rRNA amplicon sequencing study detected high abundance of E. faecalis OTUs in secondary apical periodontitis (Bouillaguet et al., 2018). E. faecalis was not identified in root canals with healthy vital pulp or irreversible pulpitis, coherently with the reported absence of E. faecalis in carious lesions close to the pulp (Martin et al., 2002). On the other hand, a more recent NGS-based study identified the genus Enterococcus in the microbiome of root canals with irreversible pulpitis, albeit at a very low relative abundance (Siqueira et al., 2016). According to our study, the prevalence of E. faecalis in necrotic root canals was 22%. This percentage was essentially in line with a previous study, wherein the prevalence of E. faecalis was 26% and 32% when identified by culture- and PCR-based methods, respectively (Zandi et al., 2018). The prevalence of E. faecalis in primary root canal infections was even lower (7.5%) when investigated using the checkerboard DNA-DNA hybridization (Zahran et al., 2021). These findings collectively support a relatively low occurrence of enterococci in primary endodontic infections. This could be explained by the fact that enterococci are transient members of oral microbiota (Wang et al., 2012), given that endodontic microbiota are derived from oral microbiota influenced by the specific ecological conditions of root canal system (Cogulu et al., 2007). It is also possible that microbial species predominant in primary endodontic infections can inhibit the proliferation of E. faecalis, yet such assumption should be investigated in future studies. Our study revealed that E. faecalis was identified by 30% in secondary/persistent endodontic infections, which lies in agreement with previous sequencing-based studies, which reported a prevalence of E. faecalis equal or greater than 30% in these infections (Siqueira et al., 2002; Zehnder and Belibasakis, 2015). The higher prevalence of E. faecalis in secondary/persistent endodontic infections group compared to primary infections in this study (30% vs 22%) agrees with a previous systematic review, which significantly correlated E. faecalis with persistent infections (Zhang et al., 2015). Adaptation to environmental conditions of root-filled teeth and tolerance to intracanal disinfection could explain the higher occurrence of E. faecalis in post-treatment apical periodontitis (Evans et al., 2002). It has been demonstrated that mechanical instrumentation and exposure to endodontic irrigants increased the number and adhesion forces of E. faecalis to dentine and root canal filling materials respectively (Kishen et al., 2008; Vengerfeldt et al., 2014; Keskin et al., 2017). Our study reported a 10% prevalence of E. faecalis in saliva samples. Previous studies, both using cultural and molecular methods, reported similar values of prevalence ranging from 19 to 21% (Wang et al., 2012; Delboni et al., 2017). Isolation of E. faecalis from saliva samples could be also linked with the isolation of this pathogen from multiple oral sites (Delboni et al., 2017), which supports the assumption that oral cavity could be a potential reservoir of E. faecalis. In contrast to our and previous studies, E. faecalis was never identified in saliva of patients seeking endodontic retreatment (Zhu et al., 2010). In addition, a significant association was observed between the presence of E. faecalis in saliva and root canals, as demonstrated previously (Wang et al., 2012), while contradicting an earlier study (Vidana et al., 2011). The higher odds of identifying E. faecalis in root canals when it exists in saliva supports a possible role of E. faecalis in saliva as a risk factor for root canal infection with this pathogen. A higher prevalence of E. faecalis in saliva and subgingival samples from patients with chronic periodontitis compared to healthy subjects was reported (Xu et al., 2019), and suggests that periodontal infections could favor the colonization of E. faecalis as observed in endodontic-periodontal lesions (Guo et al., 2011). For this reason, in our study, subjects with periodontitis were excluded. Our study also agrees with the study by Wang et al. (2012), wherein tooth position, quality and type of restorations were not significantly associated with the presence of E. faecalis in root canals despite differences in the demographic characteristics of the investigated populations. Our results demonstrated that the odds of developing a periapical lesion were significantly increased when E. faecalis was detected in root canals. These results could be explained by several studies, which demonstrated the role of E. faecalis and its virulence factors (such as extracellular proteases and cytolysin) in local inflammation and alveolar bone destruction in apical periodontitis (Souto and Colombo, 2008; Guerreiro-Tanomaru et al., 2013). Our results correlate with the study by Molander et al., wherein enterococci were recovered from 32% of teeth with radiographically verified apical periodontitis versus only 5% in teeth with no apical periodontitis (Molander et al., 1998), while other studies revealed no significant association of enterococci with diseased periapical tissues (Kaufman et al., 2005; Zoletti et al., 2006). Although it is well-established that AP is of bacterial etiology, it is important to consider that multiple local and systemic factors predispose the incidence of periapical lesions and affect the healing of periapical tissues in endodontically-treated teeth (Kirkevang et al., 2007; Holland et al., 2017). Our pilot findings showed that two genetically related salivary isolates of E. faecalis were recovered from the same subjects at four months apart, which could support the assumption that this species could persist in the oral cavity for a long time frame, as observed in repeated oral rinses after the ingestion of enterococci-rich food and in mature biofilms recovered from intraoral dental splints (Razavi et al., 2007). We demonstrated genetic relatedness of four pairs of salivary and endodontic E. faecalis isolates from the same patient, this supports the hypothesis that E. faecalis in saliva could serve as a potential source of infecting root canals. A similar finding was also reported for E. faecalis strains isolated from saliva, pulp chamber and root canals of endodontic patients (Delboni et al., 2017). These findings can be explained by the possible transition of E. faecalis from oral cavity into root canals during or after endodontic treatment or less likely via carious lesions. We also found a pair of genetically different E. faecalis in saliva and root canals of the same patient (BE51 and BE52). Interestingly, strain BE52 was genetically related to BE5, which was isolated from the root canal of a different patient. These findings suggest that similar strains of E. faecalis can be present in different individuals as observed by Pinheiro et al (Pinheiro et al., 2006), which could be related to bacterial intake by exogenous sources such as food (Al-Ahmad et al., 2010). Future studies should be focused on investigating the genetic profiles of E. faecalis strains longitudinally collected from the same patient, and their association with food intake. It seems also worthy to explore the long-term occurrence of E. faecalis in the oral cavity in a larger cohort, and to investigate the factors which govern the long-term survival of E. faecalis and its integration into oral biofilms. The mechanisms which explore the role of E. faecalis in the pathogenesis of AP should also be investigated.
Conclusion
The findings of this study confirmed the presence of E. faecalis in saliva and root canals especially those with post-treatment apical periodontitis. The significant association and genetic relatedness of E. faecalis in saliva and root canals suggest that the presence of E. faecalis in saliva is a risk factor for root canal contamination with this pathogen. The latter could increase the risk of developing a periapical lesion. The present study shifts the focus back to the role of E. faecalis in the pathogenesis of endodontic infections.
Data availability statement
The data presented in the study are deposited in the NCBI bioprojects repository, accession number PRJNA891504. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA891504.
Ethics statement
The studies involving human participants were reviewed and approved by Comitato Etico Regionale per la Sperimentazione Clinica della Regione Toscana Sezione: AREA VASTA SUD EST Segreteria Tecnico Scientifica ubicata c/o: Farmacia Ospedaliera AOUS - Viale Bracci, 16 - 53100 Siena Telefono: 0577-586358 E-mail: c.etico@ao-siena.toscana.it. The patients/participants provided their written informed consent to participate in this study.
Author contributions
CG: study concept and design, data analysis, writing of original manuscript. CM: performed statistical analysis, study concept and design. IA: data analysis, writing of original manuscript. DP: laboratory experiments, sequencing, genomic and phylogenetic analysis. AF: study concept and sample collection. FS: microbiologic analyses, data analysis, review of manuscript. PN: review of manuscript. GP: study concept, review of manuscript. SG: study concept and design, data analysis, editing and review of manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aas, J. A., Paster, B. J., Stokes, L. N., Olsen, I., Dewhirst, F. E. (2005). Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43 (11), 5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005
Al-Ahmad, A., Maier, J., Follo, M., Spitzmüller, B., Wittmer, A., Hellwig, E., et al. (2010). Food-borne enterococci integrate into oral biofilm: An in vivo study. J. Endod. 36 (11), 1812–1819. doi: 10.1016/j.joen.2010.08.011
Ali, I. A. A., Cheung, B. P. K., Matinlinna, J., Lévesque, C. M., Neelakantan, P. (2020a). Trans-cinnamaldehyde potently kills enterococcus faecalis biofilm cells and prevents biofilm recovery. Microb. Pathog. 149, 104482. doi: 10.1016/j.micpath.2020.104482
Ali, I. A. A., Cheung, B. P. K., Yau, J. Y. Y., Matinlinna, J. P., Lévesque, C. M., Belibasakis, G. N., et al. (2020b). The influence of substrate surface conditioning and biofilm age on the composition of enterococcus faecalis biofilms. Int. Endod. J. 53 (1), 53–61. doi: 10.1111/iej.13202
Baik, J. E., Ryu, Y. H., Han, J. Y., Im, J., Kum, K. Y., Yun, C. H., et al. (2008). Lipoteichoic acid partially contributes to the inflammatory responses to enterococcus faecalis. J. Endod. 34 (8), 975–982. doi: 10.1016/j.joen.2008.05.005
Bayne, S. C., Schmalz, G. (2005). Reprinting the classic article on USPHS evaluation methods for measuring the clinical research performance of restorative materials. Clin. Oral. Investig. 9 (4), 209–214. doi: 10.1007/s00784-005-0017-0
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bouillaguet, S., Manoil, D., Girard, M., Louis, J., Gaïa, N., Leo, S., et al. (2018). Root microbiota in primary and secondary apical periodontitis. Front. Microbiol. 9 (9). doi: 10.3389/fmicb.2018.02374
Brundin, M., Figdor, D., Johansson, A., Sjögren, U. (2014). Preservation of bacterial DNA by human dentin. J. Endod. 40 (2), 241–245. doi: 10.1016/j.joen.2013.08.025
Cogulu, D., Uzel, A., Oncag, O., Aksoy, S. C., Eronat, C. (2007). Detection of enterococcus faecalis in necrotic teeth root canals by culture and polymerase chain reaction methods. Eur. J. Dent. 1 (4), 216–221. doi: 10.1055/s-0039-1698342
Cuscó, A., Catozzi, C., Viñes, J., Sanchez, A., Francino, O. (2018). Microbiota profiling with long amplicons using nanopore sequencing: full-length 16S rRNA gene and the 16S-ITS-23S of the rrn operon. F1000Res 7, 1755. doi: 10.12688/f1000research.16817.2
Dai, X., Ma, R., Jiang, W., Deng, Z., Chen, L., Liang, Y., et al. (2022). Enterococcus faecalis-induced macrophage necroptosis promotes refractory apical periodontitis. Microbiol. Spectr. 16, e0104522. doi: 10.1128/spectrum.01045-22
De Giorgi, S., Ricci, S., Colombini, L., Pinzauti, D., Santoro, F., Iannelli, F., et al. (2022). Genome sequence typing and antimicrobial susceptibility testing of infertility-associated enterococcus faecalis reveals clonality of aminoglycoside-resistant strains. J. Glob Antimicrob. Resist. 29, 194–196. doi: 10.1016/j.jgar.2022.03.017
Delboni, M. G., Gomes, B. P., Francisco, P. A., Teixeira, F. B., Drake, D. (2017). Diversity of enterococcus faecalis genotypes from multiple oral sites associated with endodontic failure using repetitive sequence-based polymerase chain reaction and arbitrarily primed polymerase chain reaction. J. Endod. 43 (3), 377–382. doi: 10.1016/j.joen.2016.10.042
Evans, M., Davies, J. K., Sundqvist, G., Figdor, D. (2002). Mechanisms involved in the resistance of enterococcus faecalis to calcium hydroxide. Int. Endod. J. 35 (3), 221–228. doi: 10.1046/j.1365-2591.2002.00504.x
Gomes, B. P., Berber, V. B., Kokaras, A. S., Chen, T., Paster, B. J. (2015). Microbiomes of endodontic-periodontal lesions before and after chemomechanical preparation. J. Endod. 41 (12), 1975–1984. doi: 10.1016/j.joen.2015.08.022
Gomes, B. P. F. A., Francisco, P. A., Godoi, E. P., Jr, Endo, M. S., Barbosa-Ribeiro, M., Delboni, M. G., et al. (2021). Identification of culturable and nonculturable microorganisms, lipopolysaccharides, and lipoteichoic acids from root canals of teeth with endodontic failure. J. Endod. 47 (7), 1075–1086. doi: 10.1016/j.joen.2021.04.011
Guerreiro-Tanomaru, J. M., de Faria-Júnior, N. B., Duarte, M. A., Ordinola-Zapata, R., Graeff, M. S., Tanomaru-Filho, M. (2013). Comparative analysis of enterococcus faecalis biofilm formation on different substrates. J. Endod. 39 (3), 346–350. doi: 10.1016/j.joen.2012.09.027
Guo, Hj, Yue, L., Gao, Y. (2011). Status of bacterial colonization in infected root canal. Beijing Da Xue Xue Bao Yi Xue Ban. 43 (1), 26–28.
Hahn, C. L., Hanford, K. (2021). An In vitro model to study the colonization and tubular invasion of enterococcus faecalis. J. Endod. 47 (3), 451–457. doi: 10.1016/j.joen.2020.12.004
Holland, R., Gomes, J. E. F., Cintra, L. T. A., Queiroz, ÍOA, Estrela, C. (2017). Factors affecting the periapical healing process of endodontically treated teeth. J. Appl. Oral. Sci. 25 (5), 465–476. doi: 10.1590/1678-7757-2016-0464
Kakehashi, S., Stanley, H. R., Fitzgerald, R. J. (1965). The effects of surgical exposures of dental pulps in germ-free and conventional laboratory rats. Oral. Surg. Oral. Med. Oral. Pathol. 20, 340–349. doi: 10.1016/0030-4220(65)90166-0
Kaufman, B., Spångberg, L., Barry, J., Fouad, A. F. (2005). Enterococcus spp. endodontically treated teeth without periradicular lesions. J. Endod. 31 (12), 851–856. doi: 10.1097/01.don.0000164133.04548.26
Keskin, C., Demiryürek, EÖ, Onuk, E. E. (2017). Pyrosequencing analysis of cryogenically ground samples from primary and Secondary/Persistent endodontic infections. J. Endod. 43 (8), 1309–1316. doi: 10.1016/j.joen.2017.03.019
Kirkevang, L. L., Vaeth, M., Hörsted-Bindslev, P., Bahrami, G., Wenzel, A. (2007). Risk factors for developing apical periodontitis in a general population. Int. Endod. J. 40 (4), 290–299. doi: 10.1111/j.1365-2591.2007.01224.x
Kishen, A., Sum, C. P., Mathew, S., Lim, C. T. (2008). Influence of irrigation regimens on the adherence of enterococcus faecalis to root canal dentin. J. Endod. 34 (7), 850–854. doi: 10.1016/j.joen.2008.04.006
Lees, J. A., Harris, S. R., Tonkin-Hill, G., Gladstone, R. A., Lo, S. W., Weiser, J. N., et al. (2019). Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Res. 29 (2), 304–316. doi: 10.1101/gr.241455.118
Levin, L. G., Law, A. S., Holland, G. R., Abbott, P. V., Roda, R. S. (2009). Identify and define all diagnostic terms for pulpal health and disease states. J. Endod. 35 (12), 1645–1657. doi: 10.1016/j.joen.2009.09.032
Manoil, D., Al-Manei, K., Belibasakis, G. N. (2020). A systematic review of the root canal microbiota associated with apical periodontitis: Lessons from next-generation sequencing. Proteomics Clin. Appl. 14 (3), e1900060. doi: 10.1002/prca.201900060
Martin, F. E., Nadkarni, M. A., Jacques, N. A., Hunter, N. (2002). Quantitative microbiological study of human carious dentine by culture and real-time PCR: association of anaerobes with histopathological changes in chronic pulpitis. J. Clin. Microbiol. 40 (5), 1698–1704. doi: 10.1128/JCM.40.5.1698-1704.2002
Molander, A., Reit, C., Dahlén, G., Kvist, T. (1998). Microbiological status of root-filled teeth with apical periodontitis. Int. Endod. J. 31 (1), 1–7. doi: 10.1046/j.1365-2591.1998.t01-1-00111.x
Momenijavid, M., Salimizand, H., Korani, A., Dianat, O., Nouri, B., Ramazanzadeh, R., et al. (2022). Effect of calcium hydroxide on morphology and physicochemical properties of enterococcus faecalis biofilm. Sci. Rep. 12 (1), 7595. doi: 10.1038/s41598-022-11780-x
Orstavik, D., Kerekes, K., Eriksen, H. M. (1986). The periapical index: a scoring system for radiographic assessment of apical periodontitis. Endod. Dent. Traumatol. 2 (1), 20–34. doi: 10.1111/j.1600-9657.1986.tb00119.x
Pinheiro, E. T., Anderson, M. J., Gomes, B. P., Drucker, D. B. (2006). Phenotypic and genotypic identification of enterococci isolated from canals of root-filled teeth with periapical lesions. Oral. Microbiol. Immunol. 21 (3), 137–144. doi: 10.1111/j.1399-302X.2006.00285.x
Pinzauti, D., Iannelli, F., Pozzi, G., Santoro, F. (2022). DNA Isolation methods for nanopore sequencing of the Streptococcus mitis genome. Microb. Genom. 8 (2), 764. doi: 10.1099/mgen.0.000764
Razavi, A., Gmür, R., Imfeld, T., Zehnder, M. (2007). Recovery of enterococcus faecalis from cheese in the oral cavity of healthy subjects. Oral. Microbiol. Immunol. 22 (4), 248–251. doi: 10.1111/j.1399-302X.2006.00349.x
Rôças, I. N., Siqueira, J. F., Jr, Santos, K. R. (2004). Association of enterococcus faecalis with different forms of periradicular diseases. J. Endod. 30 (5), 315–320. doi: 10.1097/00004770-200405000-00004
Rogers, G. B., Marsh, P., Stressmann, A. F., Allen, C. E., Daniels, T. V., Carroll, M. P., et al. (2010). The exclusion of dead bacterial cells is essential for accurate molecular analysis of clinical samples. Clin. Microbiol. Infect. 16 (11), 1656–1658. doi: 10.1111/j.1469-0691.2010.03189.x
Sedgley, C., Buck, G., Appelbe, O. (2006a). Prevalence of enterococcus faecalis at multiple oral sites in endodontic patients using culture and PCR. J. Endod. 32 (2), 104–109. doi: 10.1016/j.joen.2005.10.022
Sedgley, C., Nagel, A., Dahlén, G., Reit, C., Molander, A. (2006b). Real-time quantitative polymerase chain reaction and culture analyses of enterococcus faecalis in root canals. J. Endod. 32 (3), 173–177. doi: 10.1016/j.joen.2005.10.037
Siqueira, J. F., Jr. (2002). Endodontic infections: concepts, paradigms, and perspectives. Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. Endod. 94 (3), 281–293. doi: 10.1067/moe.2002.126163
Siqueira, J. F., Jr, Antunes, H. S., Rôças, I. N., Rachid, C. T., Alves, F. R. (2016). Microbiome in the apical root canal system of teeth with post-treatment apical periodontitis. PloS One 11 (9), e0162887. doi: 10.1371/journal.pone.0162887
Siqueira, J. F., Jr, Rôças, I. N. (2009). Diversity of endodontic microbiota revisited. J. Dent. Res. 88 (11), 969–981. doi: 10.1177/0022034509346549
Siqueira, J. F., Jr, Rôças, I. N., Souto, R., de Uzeda, M., Colombo, A. P. (2002). Actinomyces species, streptococci, and enterococcus faecalis in primary root canal infections. J. Endod. 28 (3), 168–172. doi: 10.1097/00004770-200203000-00006
Souto, R., Colombo, A. P. (2008). Prevalence of enterococcus faecalis in subgingival biofilm and saliva of subjects with chronic periodontal infection. Arch. Oral. Biol. 53 (2), 155–160. doi: 10.1016/j.archoralbio.2007.08.004
Stuart, C. H., Schwartz, S. A., Beeson, T. J., Owatz, C. B. (2006). Enterococcus faecalis: its role in root canal treatment failure and current concepts in retreatment. J. Endod. 32 (2), 93–98. doi: 10.1016/j.joen.2005.10.049
Tonetti, M. S., Greenwell, H., Kornman, K. S. (2018). Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Periodontol. 89 (Suppl 1), S159–S172. doi: 10.1002/JPER.18-0006
Vengerfeldt, V., Špilka, K., Saag, M., Preem, J. K., Oopkaup, K., Truu, J., et al. (2014). Highly diverse microbiota in dental root canals in cases of apical periodontitis (data of illumina sequencing). J. Endod. 40 (11), 1778–1783. doi: 10.1016/j.joen.2014.06.017
Vidana, R., Sullivan, A., Billström, H., Ahlquist, M., Lund, B. (2011). Enterococcus faecalis infection in root canals - host-derived or exogenous source? Lett. Appl. Microbiol. 52 (2), 109–115. doi: 10.1111/j.1472-765X.2010.02972.x
Von Elm, E., Altman, D. G., Egger, M., Pocock, S. J., Gøtzsche, P. C., Vandenbroucke, J. P., et al. (2008). The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. J. Clin. Epidemiol. 61 (4), 344–349. doi: 10.1016/j.jclinepi.2007.11.008
Wang, Q. Q., Zhang, C. F., Chu, C. H., Zhu, X. F. (2012). Prevalence of enterococcus faecalis in saliva and filled root canals of teeth associated with apical periodontitis. Int. J. Oral. Sci. 4 (1), 19–23. doi: 10.1038/ijos.2012.17
Wick, R. R., Judd, L. M., Gorrie, C. L., Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput. Biol. 13 (6), e1005595. doi: 10.1371/journal.pcbi.1005595
Wong, J., Manoil, D., Näsman, P., Belibasakis, G. N., Neelakantan, P. (2021). Microbiological aspects of root canal infections and disinfection strategies: An update review on the current knowledge and challenges. Front. Oral. Health 25 (2). doi: 10.3389/froh.2021.672887
Xu, J., He, J., Shen, Y., Zhou, X., Huang, D., Gao, Y., et al. (2019). Influence of endodontic procedure on the adherence of enterococcus faecalis. J. Endod. 45 (7), 943–949. doi: 10.1016/j.joen.2019.04.006
Zahran, S., Witherden, E., Mannocci, F., Koller, G. (2021). Characterization of root canal microbiota in teeth diagnosed with irreversible pulpitis. J. Endod. 47 (3), 415–423. doi: 10.1016/j.joen.2020.12.009
Zandi, H., Kristoffersen, A. K., Ørstavik, D., Rôças, I. N., Siqueira, J. F., Jr, Enersen, M. (2018). Microbial analysis of endodontic infections in root-filled teeth with apical periodontitis before and after irrigation using pyrosequencing. J. Endod. 44 (3), 372–378. doi: 10.1016/j.joen.2017.11.019
Zehnder, M., Belibasakis, G. N. (2015). On the dynamics of root canal infections-what we understand and what we don't. Virulence 6 (3), 216–222. doi: 10.4161/21505594.2014.984567
Zehnder, M., Guggenheim, B. (2009). The mysterious appearance of enterococci in filled root canals. Int. Endod. J. 42 (4), 277–287. doi: 10.1111/j.1365-2591.2008.01537.x
Zhang, C., Du, J., Peng, Z. (2015). Correlation between enterococcus faecalis and persistent intraradicular infection compared with primary intraradicular infection: A systematic review. J. Endod. 41 (8), 1207–1213. doi: 10.1016/j.joen.2015.04.008
Zhu, X., Wang, Q., Zhang, C., Cheung, G. S., Shen, Y. (2010). Prevalence, phenotype, and genotype of enterococcus faecalis isolated from saliva and root canals in patients with persistent apical periodontitis. J. Endod. 36 (12), 1950–1955. doi: 10.1016/j.joen.2010.08.053
Keywords: apical periodontitis, endodontic infections, enterococcus faecalis, phylogenetic analysis, risk factor, saliva, whole-genome sequencing (WGS)
Citation: Gaeta C, Marruganti C, Ali IAA, Fabbro A, Pinzauti D, Santoro F, Neelakantan P, Pozzi G and Grandini S (2023) The presence of Enterococcus faecalis in saliva as a risk factor for endodontic infection. Front. Cell. Infect. Microbiol. 13:1061645. doi: 10.3389/fcimb.2023.1061645
Received: 04 October 2022; Accepted: 02 March 2023;
Published: 06 April 2023.
Edited by:
Zhongchun Tong, Guanghua School of Stomatology, Sun Yat-sen University, ChinaReviewed by:
Daniel Manoil, University of Geneva, SwitzerlandJ. Christopher Fenno, University of Michigan, United States
Copyright © 2023 Gaeta, Marruganti, Ali, Fabbro, Pinzauti, Santoro, Neelakantan, Pozzi and Grandini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carlo Gaeta, c.gaeta@student.unisi.it
 Carlo Gaeta
Carlo Gaeta Crystal Marruganti
Crystal Marruganti Islam A. A. Ali
Islam A. A. Ali Andrea Fabbro
Andrea Fabbro David Pinzauti
David Pinzauti Francesco Santoro
Francesco Santoro Prasanna Neelakantan
Prasanna Neelakantan Gianni Pozzi
Gianni Pozzi Simone Grandini
Simone Grandini