Surveillance of Antimalarial Drug-Resistance Genes in Imported Plasmodium falciparum Isolates From Nigeria in Henan, China, 2012–2019
- Department of Parasite Disease Control and Prevention, Henan Provincial Center for Disease Control and Prevention, Henan Key Laboratory of Infectious Disease Microbiology, Zhengzhou, China
Malaria remains a major public health issue in Nigeria, and Nigeria is one of the main sources of imported malaria in China. Antimalarial drug resistance is a significant obstacle to the control and prevention of malaria globally. The molecular markers associated with antimalarial drug resistance can provide early warnings about the emergence of resistance. The prevalence of antimalarial drug resistant genes and mutants, including PfK13, Pfcrt, Pfmdr1, Pfdhfr, and Pfdhps, was evaluated among the imported Plasmodium falciparum isolates from Nigeria in Henan, China, from 2012 to 2019. Among the 167 imported P. falciparum isolates, the wild-type frequency of PfK13, Pfcrt, Pfmdr1, Pfdhfr, and Pfdhps was 98.7, 63.9, 34.8, 3.1, and 3.1%, respectively. The mutation of PfK13 was rare, with just two nonsynonymous (S693F and Q613H) and two synonymous mutations (C469C and G496G) identified from four isolates. The prevalence of Pfcrt mutation at codon 74–76 decreased year-by-year, while the prevalence of pfmdr1 86Y also decreased significantly with time. The prevalence of Pfdhfr and Pfdhps mutants was high. Combined mutations of Pfdhfr and Pfdhps had a high prevalence of the quadruple mutant I51R59N108-G437 (39.0%), followed by the octal mutant I51R59N108-V431A436G437G581S613 (17.0%). These molecular findings update the known data on antimalarial drug-resistance genes and provide supplemental information for Nigeria.
Introduction
Globally, malaria incidence and mortality have substantially reduced since 2010, and an increasing number of countries are moving toward malaria elimination. In 2016, the World Health Organization (WHO) identified 21 countries that had the potential to eliminate malaria by 2020, including China (WHO, 2018). In Henan Province, there has been no local malaria case since 2012 (Liu et al., 2014), while in 2017, no indigenous malaria cases were reported in China for the first time (Zhang et al., 2018). However, malaria remains the major public problem in sub-Saharan Africa, and Nigeria had the greatest burden of global malaria cases (27%) and malaria deaths (23%) worldwide in 2019 (WHO, 2020).
Efficacious antimalarial medicines are critical to malaria control and elimination. However, the emergence of antimalarial drug resistance increases the burden of malaria, and is one of the recurring challenges in the global fight against malaria. Monitoring antimalarial drug efficacy is necessary to provide information for treatment policies, as well as to mitigate the impact of resistance and prevent its spread. Therapeutic efficacy studies (TESs) and integrated drug efficacy surveillance (iDES) are common and reliable measures to obtain data on treatment efficacy, and the molecular markers associated with parasite resistance can provide supplemental information for TESs and iDES.
Chloroquine (CQ), as the first-line therapy for Plasmodium falciparum malaria, was the most common antimalarial in Africa from the 1940s to the 2000s (Nuwaha, 2001). CQ was replaced by artemisinin-based combination therapies (ACTs) in most African countries from the late 1990s to 2000s with the spread of chloroquine resistance (Flegg et al., 2013). However, CQ remains the first-line treatment for P. vivax in most endemic countries (WHO, 2019), while ACTs have been recommended by the WHO as the first-line treatment for uncomplicated P. falciparum malaria in nearly all areas, as well as for chloroquine-resistant (CQR) P. vivax malaria (WHO, 2015). Artemether-lumefantrine (AL), artesunate-amodiaquine (AS-AQ), and dihydroartemisinin-piperaquine (DHA-PPQ) are used as the first-line treatment for P. falciparum in most African countries (WHO, 2019). Sulfadoxine-pyrimethamine (SP) is recommended for the intermittent preventive treatment of malaria in pregnant women and infants (IPTp and IPTi) by the WHO (WHO, 2010).
CQR was first reported at the Thailand-Cambodia border in 1957 (Mehlotra et al., 2001; Ridley, 2002) and then confirmed in Africa in 1979 (Campbell et al., 1979). The emergence of artemisinin resistance (ART-R) in P. falciparum was first reported in Cambodia and later became widespread in the Greater Mekong sub-region (GMS) (Noedl et al., 2008; Dondorp et al., 2009; Yeung et al., 2009). Currently, resistance to the partner drugs of artemisinin (ART) is also common in the GMS, which affects the efficacy of ACTs. The P. falciparum chloroquine resistance transporter gene (Pfcrt) is the most reliable molecular marker of CQR, and has also been considered to be associated with resistance to ACT partner drugs, such as piperaquine (Ross et al., 2018), amodiaquine (Sá et al., 2009), lumefantrine (Sisowath et al., 2009), and mefloquine (Picot et al., 2009). The polymorphism of the Kelch 13 (K13) propeller domain in P. falciparum has been identified as a molecular marker of partial ART resistance (Ariey et al., 2014). Many non-synonymous mutations in PfK13 have been identified (WHO, 2018); however, only a few PfK13 mutations have been validated, and all of them have been identified in the GMS in Southeast Asia (Ménard et al., 2016). Some mutations in PfK13 were also identified in Africa, although these have not been validated in vivo or in vitro (Taylor et al., 2014). A recent study confirmed the de novo emergence and clonal expansion of an ART-R PfK13 R561H lineage in Rwanda (Uwimana et al., 2020). These findings have substantial implications for the treatment and control of malaria in Africa. P. falciparum multidrug resistance 1 gene (Pfmdr1) has been linked with the efficacy of chloroquine, mefloquine, amodiaquine, lumefantrine, artemisinin, and others (Humphreys et al., 2007; Somé et al., 2010). The polymorphisms of Pfmdr1 at codons N86Y, Y184F, S1034C, N1042D, and D1246Y have also been shown to be linked with antimalarial drug resistance (Picot et al., 2009; Kamugisha et al., 2012). The Pfmdr1 mutations N86Y and D1246Y, together with the Pfcrt mutations, can reduce the efficacy of CQ (Sá et al., 2009). The mutations of P. falciparum dihydrofolate reductase (Pfdhfr) at codons A16V, N51I, C59R, S108N/T, and I164L have been reported to be related with pyrimethamine resistance (McCollum et al., 2006). Moreover, polymorphisms of P. falciparum dihydropteroate synthase (Pfdhps) at codons S436A/F, A437G, K540E, A581G, and A613S/T are considered to be associated with sulphadoxine resistance (Vinayak et al., 2010). The quintuple mutation composed of the pfdhfr triple mutant 51I59R108N and the Pfdhps double mutant 437G540E has been reported to reduce the efficacy of SP in IPTp and IPTi (Gosling et al., 2009; Nankabirwa et al., 2010).
In Nigeria, ACTs have been recommended by the National malaria drug policy since 2005 because of the failure of CQ treatment (FMoH, 2005). However, CQ is still used to treat malaria because it is both accessible and cheap. Currently, AL and AS-AQ, as the recommended ART-based combinations, are adopted for the treatment of uncomplicated malaria in Nigeria, and the most recent TES showed that the cure rate was more than 95% (Sowunmi et al., 2017). At present, pregnant Nigerian women are also recommended to receive 3+ doses of SP to prevent malaria (NPC and ICF, 2019).
Nigeria is one of the main sources of imported malaria in China. It is important to understand the efficacy of antimalarial drugs for these imported cases. Therefore, in this study, we performed molecular surveillance of antimalarial drug-resistant genes in P. falciparum isolates imported from Nigeria in Henan Province, China, to determine their haplotypes and prevalence.
Materials and Methods
Sample Characteristics
All of the information on imported malaria cases in Henan Province was collected from the Disease Surveillance Information Report Management system of China Center for Disease Control and Prevention. A total of 1,522 imported malaria cases were reported in Henan Province during 2012–2019, with no indigenous cases. Nigeria is one of the main sources of imported malaria in Henan Province, and 201 of the total cases originated from here. Among these 201 cases, 167 cases were infected with P. falciparum. With the exception of one patient who was Nigerian working in China, the other 200 cases were Chinese people who traveled to Nigeria and returned with malaria infection. One case was female, and the others were male. The youngest was 19 years old and the oldest was 62 years old. The average age was 38.11 ± 9.188 years, and most cases were concentrated in the 21–50 years age group (86.6%, 174/201). Most of the patients were migrant workers (84.6%, 170/201).
DNA Extraction and Amplification
The patients were diagnosed with malaria initially by blood smear microscopy and/or rapid diagnosis test (RDT) in the local hospital or County Centers for Disease Control and Prevention. The whole blood and blood smear samples of the cases were collected before antimalarial treatment and deposited in the Sample Resource Library in Henan Provincial Reference Laboratory for Malaria Diagnosis. All of the cases were confirmed to be infected with malaria parasite species using nested polymerase chain reaction (PCR) and blood smear microscopy performed at the Henan Provincial Reference Laboratory for Malaria Diagnosis (Yin et al., 2015). The genomic DNA was extracted from blood samples using QIAamp DNA Blood Mini Kits (Qiagen, USA) following the manufacturer’s instructions.
The target genes of PfK13, Pfcrt, Pfmdr1, Pfdhfr, and Pfdhps were amplified using nested PCR previously described (Zhou et al., 2016; Zhou et al., 2019). The primer sequences and conditions are shown in Supplementary Material. The amplification of the Pfcrt gene amplified covered the codons C72S, V73V, M74I, N75E, and K76T. The Pfmdr1 gene contained the codons N86Y, Y184F, S1034C, N1042D, and D1246Y. The Pfdhfr gene was amplified to identify the polymorphism at codons A16S, N51I, C59R, S108N, and I164L. The Pfdhps gene covered the codons I431V, S436A, A437G, K540E, A581G, and A613S. Bidirectional sequencing of the secondary PCR products was performed by Sangon Biotech Co Ltd (Shanghai, China).
Data Analysis
ChromasPro software v. 1.5 (https://technelysium.com.au/wp/chromaspro/) was used to assemble the forward and reverse sequences of the genes. MEGA7 (Molecular Evolutionary Genetics Analysis, https://www.megasoftware.net/show_eua) software was used to identify the mutations by comparing with their reference genomes. The reference genomes of PfK13, Pfcrt, Pfmdr1, Pfdhfr, and Pfdhps were from the P. falciparum 3D7 strain obtained from Genbank (Genbank ID: Pf3D7_1343700, Pf3D7_0709000, Pf3D7_0523000, PF3D7_0417200, and PF3D7_1324800, respectively). The data were analyzed using SSPS v.21.0 (Statistical Product and Service Solutions). The difference was compared using Chi-square or Fisher’s exact test, and a two-sided p value of <0.05 was considered statistically significant.
Results
K13-Propeller
The 850 bp fragments of the K13-propeller domain were successfully sequenced from 157 samples among 167 P. falciparum isolates. Two non-synonymous and two synonymous mutations were identified from four isolates: S693F, Q613H, C469C, and G496G. The total mutant prevalence was 2.5% (4/157). Two synonymous mutations were detected in 2014 and 2016, and one non-synonymous mutation was identified in 2017 and 2019.
Pfcrt
A total of 158 out of 167 isolates were successfully sequenced on Pfcrt. The codons 72 and 73 were all wild type. The mutant prevalence of codons 74, 75, and 76 was 36.1% (57/158), 36.1% (57/158), and 35.4% (56/158), respectively, during 2012–2019 (Table 1). Four haplotypes of Pfcrt were identified, wild-type CVMNK (63.9%, 101/158), CVIET (32.3%, 51/158), CVIEK (0.6%, 1/158), and CV M/I N/E K/T (3.2%, 5/158) (Table 2). The mutant prevalence of Pfcrt 74I, 75E, and 76T decreased with time, and the differences showed statistical significance (74I and 75E: χ2 = 9.837, p = 0.020; 76T: χ2 = 8.260, p = 0.041) (Figure 1A).
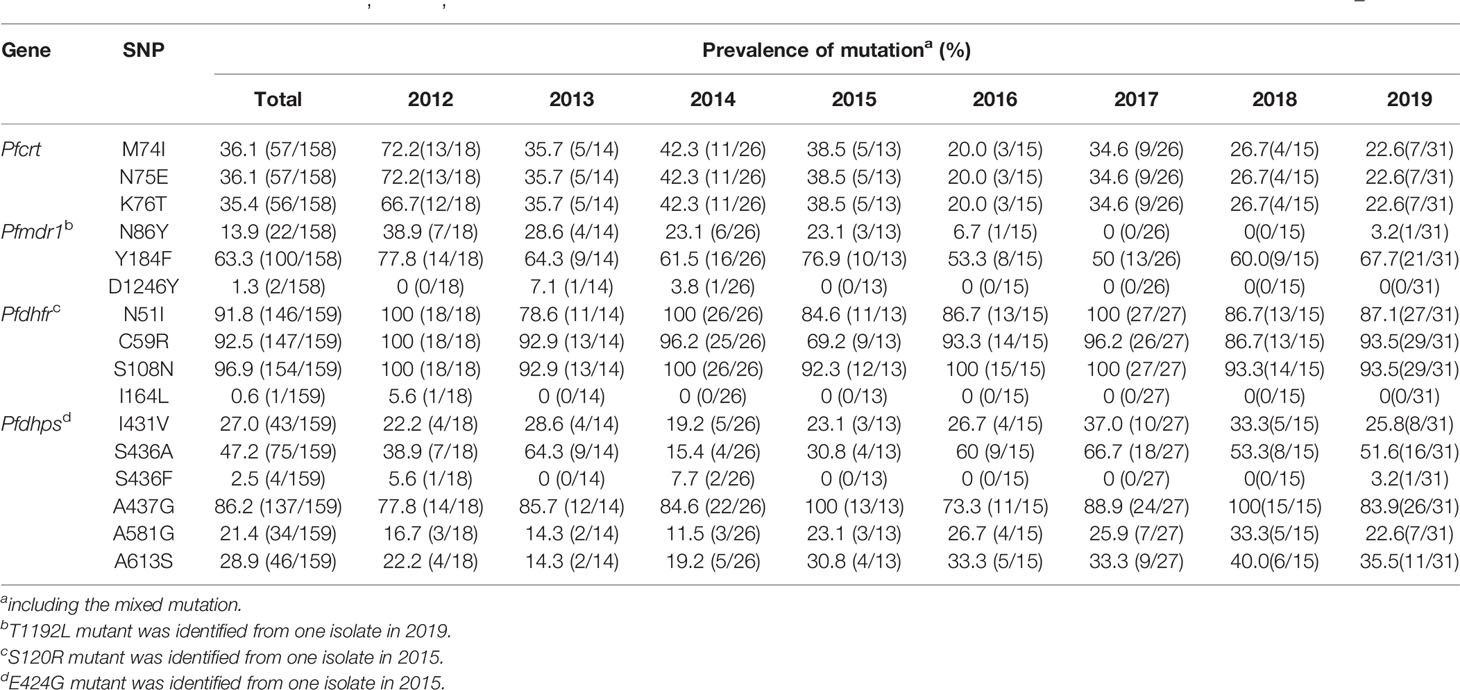
Table 1 Mutant prevalence of the Pfcrt, Pfmdr1, Pfdhfr, and Pfdhps genes detected in Plasmodium falciparum isolates returned from Nigeria during 2012–2019.

Table 2 Haplotypes of Pfcrt, Pfmdr1, Pfdhfr, and Pfdhps genes detected in Plasmodium falciparum isolates returned from Nigeria during 2012–2019.
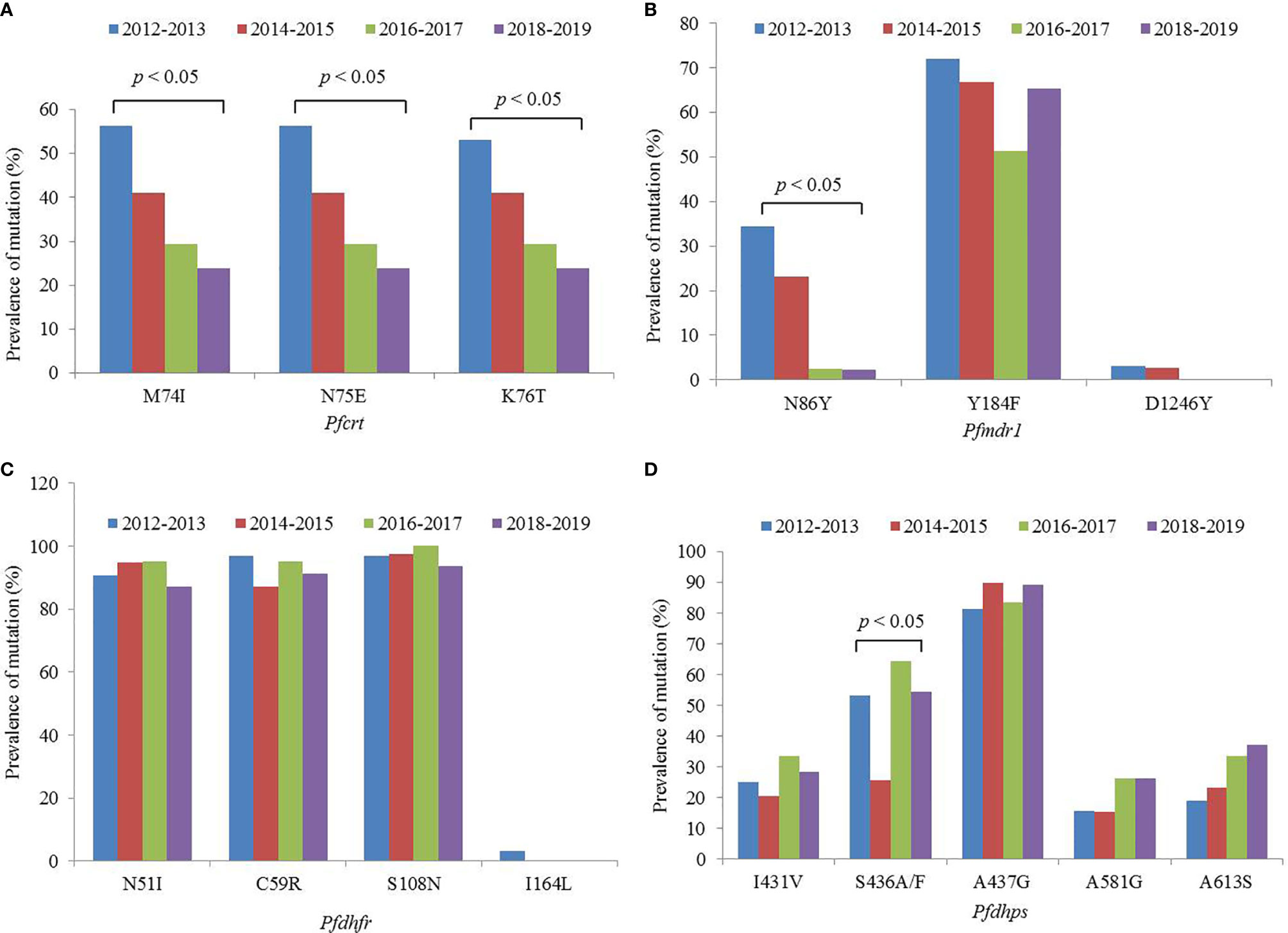
Figure 1 Mutant prevalence of the (A) Pfcrt, (B) Pfmdr1, (C) Pfdhfr, and (D) Pfdhps genes detected in Plasmodium falciparum isolates returned from Nigeria, 2012–2019.
Pfmdr1
The fragments of Pfmdr1 were successfully obtained from 158 isolates. No mutation was found at codons 1034 and 1042, and only two isolates had mutation at codon D1246Y. One T1192L mutant was identified for the first time in our study. The mutant prevalence of N86Y and Y184F was 13.9% (22/158) and 63.3% (100/158), respectively, during 2012–2019 (Table 1). Five haplotypes of Pfmdr1 were found, including wild-type NYSND, two single mutant haplotypes NFSND and NYSND-T1192L, and two double mutant haplotypes YFSNLD and YYSNY (Table 2). The mutant prevalence of Pfmdr1 86Y decreased significantly with time (χ2 = 23.704, p = 0.000) (Figure 1B).
Pfdhfr
Pfdhfr was successfully amplified from 159 P. falciparum isolates. Only five isolates were wild type, and the other 154 isolates had mutations among five codons N51I, C59R, S108N, S120R, and I164L. The mutations at codons N51I, C59R, and S108N were common, accounting for 91.8% (146/159), 92.5% (147/159), and 96.9% (154/159), respectively (Table 1). The Pfdhfr S120R and I164L mutants were identified from just one isolate each. The Pfdhfr S120R mutant was newly identified in this study. No Pfdhfr A16V mutant was found, although there was no statistical difference in the mutant prevalence among these codons during 2012–2019 (Figure 1C). Seven haplotypes of Pfdhfr were found, including wild-type, single, double, triple, and quadruple mutant haplotypes. However, the triple mutation, I51R59N108 haplotype, was the most common, accounting for 88.1% (140/159) (Table 2).
Pfdhps
Among the 159 successfully sequenced samples, five isolates were free of mutations, and the other 154 isolates had mutations among six codons E424G, I431V, S436A/F, A437G, A581G, and A613S. There were two types of mutations at codon 436, S436A (75 isolates), and S436F (4 isolates). The mutant S436F always appeared with Pfdhps 613S. No Pfdhps K540E mutant was found. The mutation at codon A437G (86.2%, 137/159) was the most prevalent, followed by S436A/F (49.7%, 79/159), A613S (28.9%, 46/159), I431V (27.0%, 43/159), and A581G (21.4%, 34/159) (Table 1). Only one E424G mutant was found and was newly identified in this study. There was statistical difference about the mutant prevalence at codon S436A/F during 2012–2019 (χ2 = 13.152, p = 0.004) (Figure 1D). The prevalence of Pfdhps A613S increased yearly, but there was no significant difference (χ2 = 4.100, p = 0.251). In addition to the wild type (ISAKAA), there were 14 mutant types, including two single mutant haplotypes (ISGKAA, IAAKAA), four double mutant haplotypes (IAGKAA, VAAKAA, IFAKAS, ISGKGA), four triple mutant haplotypes (VAGKAA, VSGKGA, IAGKAS, ISGKGA-E424G), three quadruple mutant haplotypes (VAGKGA, VAGKAS, VSGKGS), and one quintuple mutant haplotype (VAGKGS). The single mutant haplotype ISGKAA (40.0%) was the most common, followed by the quintuple mutant haplotype VAGKGS (17.0%), and the triple mutant haplotype IAGKAS (7.5%) (Table 2).
Combined Haplotypes of Pfdhfr and Pfdhps
Among the 167 P. falciparum isolates, 159 samples were successfully sequenced for Pfdhfr and Pfdhps. The results of sequencing demonstrated that only one isolate was free of mutations, and eight isolates had single-gene mutations, Pfdhfr or Pfdhps. The other 150 isolates (94.3%) had mutations in two genes simultaneously. The most frequent mutation was the quadruple mutant I51R59N108- G437, accounting for 39.0%, followed by I51R59N108- V431A436G437G581S613 (17.0%) and I51R59N108- A436 (6.9%). As no Pfdhps K540E mutants were detected, the combination of the Pfdhfr triple mutant I51R59N108 and the Pfdhps double mutant G437E540 was not observed (Table 3).

Table 3 Combination of the Pfdhfr and Pfdhps genes detected from Plasmodium falciparum isolates returned from Nigeria during 2012–2019.
Discussion
ACTs are currently considered the most effective treatment for uncomplicated falciparum malaria globally. However, the emergence and radical spread of ACT resistance represents a significant threat to malaria control and elimination. Until now, previously validated PfK13 mutants, including F446I, N485Y, M476I, Y493H, R539T, I543T, P553L, R561H, and C580Y (the most common), have been mainly identified in Southeast Asia (WHO, 2018). Meanwhile, a few PfK13 mutations have been reported in African P. falciparum isolates, including A557S, V566I, A567T, S576L, A578S, and L589I; however, none of them conferred the ART-R in vivo or in vitro (Taylor et al., 2014). The confirmation of PfK13 R561H in Rwanda would have an important impact on the ART-R in Africa (Uwimana et al., 2020). Moreover, delayed clearance of ACTs has been reported among a few cases in Nigeria (Ajayi and Ukwaja, 2013; Wundermann and Osiki, 2017). Several mutations of PfK13 have also been described in Nigerian isolates, including one non-synonymous mutation G665C discovered in southwestern Nigeria (Oboh et al., 2018), and six mutations (E433G, F434I, F434S, I684N, I684T, and E688K) identified in northern Nigeria, among which E433G and E688K were identified from isolates with the delayed clearance (Abubakar et al., 2020). The study performed in southwestern Nigeria in 2014 identified eight non-synonymous mutations in PfK13, including G496S, R539F, I543V, V566K, D584I, C580Y, and a deletion variant A557; the C580Y mutant was suspected by allelic discrimination in two samples with mixed genotypes (Tola et al., 2020). In this study, two non-synonymous mutations, S693F and Q613H, were identified in two isolates. However, none of these mutations detected in Nigerian isolates has been fully validated in vivo or in vitro for resistance to ART. Given the cases with delayed clearance to ACTs, and the fact that the PfK13 C580Y mutation has been reported in Nigeria, urgent monitoring of the efficacy of antimalarial drugs is required to obtain an early warning signal, update the treatment policy and stop the spread of ACTs-resistance.
An increasing number of studies have shown that CQ sensitivity is recovered as a consequence of CQ withdrawal (Mwanza et al., 2016; Lu et al., 2017; Ndam et al., 2017). In Nigeria, CQ was replaced with ACTs in 2005 (FMoH, 2005). In this study, the prevalence of the Pfcrt mutation reduced from 72.2% in 2012 to 22.6% in 2019, and decreased steadily, and significantly year-by-year (Figure 1A). The study of Tola et al. reported that the prevalence of mutant Pfcrt (CVIET) was 45% in 2014 in southwestern Nigeria (Tola et al., 2020), and Lu et al. reported that the prevalence of Pfcrt 76T was 46.9% in Nigeria during 2011–2014 (Lu et al., 2017), and the prevalence of mutant Pfcrt was 41.9% in Nigeria during 2012–2015 in our published study (Zhou et al., 2016), the results of the three studies performed in Nigeria were similar. However, there are few recent data on the prevalence in Nigeria. In this study, the molecular monitoring of Pfcrt took place over an 8-year period, from 2012 to 2019, and provided sufficient information to observe the reversal of CQR in Nigeria. It is possible that the mutant prevalence of Pfcrt will reduce further, leading to full recovery of the sensitivity to CQ. Moreover, the dynamics of population genetics may also account for the recovered sensitivity of CQ, in that as the corresponding antimalarial drug is withdrawn, the wild type gene might segregate and increase in population.
Pfmdr1 gene is considered to be associated with the efficacy of multiple antimalarial drugs. Meanwhile, Pfmdr1 and Pfcrt are assumed to be associated with resistance to ACT partner drugs, such as amodiaquine, lumefantrine, and mefloquine (Otienoburu et al., 2019). Pfmdr1 has also been found to be closely associated with chloroquine resistance, especially between the Pfmdr1 N86Y and Pfcrt K76T (Duraisingh and Cowman, 2005), and the prevalence of Pfmdr1 86Y reduced with the withdrawal of chloroquine (Gupta et al., 2018). Indeed, in this study, a significant decrease in the prevalence of Pfmdr1 86Y was observed from 38.9% in 2012 to 3.2% in 2019. The literature review also reported that the prevalence of Pfmdr1 86Y reduced significantly in all of the studied countries (Otienoburu et al., 2019), which was confirmed by the current study. The prevalence of Pfmdr1 184F was high in this study, at 63.3% during 2012–2019. The study performed in 2007–2008 showed that the frequency of 184F was 69.0% in Nigeria (Oladipo et al., 2015). Therefore, it can be considered that the prevalence of Pfmdr1 184F might be maintained at a certain level for a long time in Nigeria. The prevalence of Pfmdr1 86Y and 184F obtained from isolates imported from Angola was 11.7 and 30.9%, respectively, during 2012–2017 (Zhou et al., 2019). In Mozambique, the frequency of Pfmdr1 86Y and 184F was 3.1 and 46.7%, respectively, in 2015 (Gupta et al., 2018). In other countries, the prevalence of Pfmdr1 86Y has been shown to be lower, whereas that of Pfmdr1 184F has been found to vary considerably; these observations may be related to the medication strategy of individual countries.
Pfdhfr and Pfdhps were used to monitor resistance of SP, and the level of SP resistance was considered to be related to the number of combined mutations within the two genes. The level of resistance can be divided into three groups: the quadruple mutant, Pfdhfr I51R59N108-Pfdhps G437, is considered to be “partially resistant,” the quintuple mutant, Pfdhfr I51R59N108-Pfdhps G437E540, is considered to be “fully resistant,” and the sextuple mutant, Pfdhfr I51R59N108-G437E540G581/S613, is considered to be “super resistant” (Naidoo et al., 2013). In this study, Pfdhfr I51R59N108 was very common (88.1%); meanwhile, the Pfdhps gene had 15 haplotypes, among which the single mutant G437 had the highest prevalence, followed by the quintuple mutant V431A436G437G581S613. No Pfdhps K540E mutants were found. The combined mutants of the two genes showed that the quadruple mutant, Pfdhfr I51R59N108-Pfdhps G437, was common (39.0%) and classified as “partially resistant.” Because the Pfdhps K540E mutant was not identified in this study, the quintuple mutant Pfdhfr I51R59N108-Pfdhps G437E540 was not identified. However, 17.0% of isolates in Nigeria comprised the octal mutant Pfdhfr I51R59N108-Pfdhps V431A436G437G581S613. Moreover, the Pfdhps I431V mutant was discovered recently and it has been identified in Nigeria, Cameroon, and Equatorial Guinea (Chauvin et al., 2015; Oguike et al., 2016). It has also been reported that the prevalence of Pfdhps 431V is 8.3, 16.7, and 6.3% in Nigeria, Cameroon, and Equatorial Guinea, respectively (Xu et al., 2019). However, the frequency of Pfdhps 431V reached 27.0% in this study. Pfdhps 431V has always been found together with other mutants, among which Pfdhps V431A436G437G581S613 was the most common. The effects of Pfdhps 431V on SP resistance need further study. Although no data were obtained from pregnant women or children, our findings provide supplementary information for SP resistance in Nigeria. It is necessary to monitor SP resistance continuously using the two genes to guide the IPT strategy.
There are some limitations to this study. First, the samples were passively obtained from migrants returning from Nigeria; thus, the sampling was not planned, and the sample size was not controlled. Second, the exact information about which part of Nigeria these individuals worked/lived in was unavailable. Third, although the individuals were all cured, detailed information about the treatment process and the use of antimalarial drugs was not incomplete. Fourth, the individuals were almost all Chinese people who returned from Nigeria with malaria; therefore, the prevalence of mutations among them might differ from those among native Nigerians.
This study evaluated polymorphisms and prevalence of antimalarial drug-resistance genes in imported P. falciparum cases from Nigeria to Henan Province, China. The mutation of Pf K13, associated with ACTs, was rare, and no validated mutation was found. The prevalence of Pfcrt and Pfmdr1 mutants associated with the resistance of ACT partner drugs reduced gradually. Moreover, the prevalence of Pfdhfr and Pfdhps mutations was high. At present, ACTs are still effective for those returning from Nigeria with P. falciparum malaria infected in Henan Province. However, the validated PfK13 R561H mutation recently observed in Rwanda has substantial implications for ART-R in Africa (Uwimana et al., 2020). The routine molecular surveillance of antimalarial drugs is more important and imperative for the imported malaria cases, especially those from Africa, and will be helpful to rationalize drug guidance for local authorities in China.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethical Committee of Henan Province Center for Disease Control and Prevention. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
DZ, RZ, and YZ conceived and designed the study. HZ, SL, and CY performed the experiments. DZ was responsible for the data analysis and drafted the manuscript. SL and YL participated in the sample collection. PJ and DQ contributed to the data collection. RZ and YZ revised the manuscript. YD, HW, and DL provided the administrative coordination. All authors contributed to the article and approved the submitted version.
Funding
The study was supported by Science and Technology Project of Henan Province (No. 182102310631) and Henan Provincial Medical Science and Technology Project (No. 2018020515, No. 2018020509). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank all the patients who participated in this study and the staff of the hospitals and centers for disease control and prevention in Henan Province.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.644576/full#supplementary-material
References
Abubakar U. F., Adam R., Mukhtar M. M., Muhammad A., Yahuza A. A., Ibrahim S. S. (2020). Identification of Mutations in Antimalarial Resistance Gene Kelch13 From Plasmodium Falciparum Isolates in Kano, Nigeria. Trop. Med. Infect. Dis. 5 (2), 85. doi: 10.3390/tropicalmed5020085
Ajayi N. A., Ukwaja K. N. (2013). Possible Artemisinin-Based Combination Therapy-Resistant Malaria in Nigeria: A Report of Three Cases. Rev. Da Sociedade Bras. Medicina Tropical. 46 (4), 525–527. doi: 10.1590/0037-8682-0098-2013
Ariey F., Witkowski B., Amaratunga C., Beghain J., Langlois A.-C., Khim N., et al. (2014). A Molecular Marker of Artemisinin-Resistant Plasmodium Falciparum Malaria. Nature 505 (7481), 50–55. doi: 10.1038/nature12876
Campbell C. C., Chin W., Collins W. E., Teutsch S. M., Moss D. M. (1979). Chloroquine-Resistant Plasmodium Falciparum From East Africa: Cultivation and Drug Sensitivity of the Tanzanian I/CDC Strain From an American Tourist. Lancet 2 (8153), 1151–1154. doi: 10.1016/s0140-6736(79)92383-3
Chauvin P., Menard S., Iriart X., Nsango S. E., Tchioffo M. T., Abate L., et al. (2015). Prevalence of Plasmodium Falciparum Parasites Resistant to Sulfadoxine/Pyrimethamine in Pregnant Women in Yaoundé, Cameroon: Emergence of Highly Resistant Pfdhfr/ Pfdhps Alleles. J. Antimicrob. Chemother. 70 (9), 2566–2571. doi: 10.1093/jac/dkv160
Dondorp A. M., Nosten F., Yi P., Das D., Phyo A. P., Tarning J., et al. (2009). Artemisinin Resistance in Plasmodium Falciparum Malaria. N Engl. J. Med. 361 (5), 455–467. doi: 10.1056/NEJMoa0808859
Duraisingh M. T., Cowman A. F. (2005). Contribution of the Pfmdr1 Gene to Antimalarial Drug-Resistance. Acta Trop. 94, 181–190. doi: 10.1016/j.actatropica.2005.04.008
Flegg J. A., Metcalf C. J. E., Gharbi M., Venkatesan M., Shewchuk T., Sibley C. H., et al. (2013). Trends in Antimalatial Drug Use in Africa. Am. J. Trop. Med. Hyg. 89 (5), 857–865. doi: 10.4269/ajtmh.13-0129
FMoH (2005). National Antimalarial Treatment Guidelines Policy Federal Ministry of Health, National Malaria and Vector Control Division (Abuja-Nigeria, Nigeria: Federal Ministry of Health).
Gosling R. D., Gesase S., Mosha J. F., Carneiro I., Hashim R., Lemnge M., et al. (2009). Protective Efficacy and Safety of Three Antimalarial Regimens for Intermittent Preventive Treatment for Malaria in Infants: A Randomised, Double-Blind, Placebo-Controlled Trial. Lancet 374 (9700), 1521–1532. doi: 10.1016/S0140-6736(09)60997-1
Gupta H., Macete E., Bulo H., Salvador C., Warsame M., Carvalho E., et al. (2018). Drug-Resistant Polymorphisms and Copy Numbers in Plasmodium Falciparum, Mozambiqu. Emerg. Infect. Dis. 24 (1), 40–48. doi: 10.3201/eid2401.170864
Humphreys G. S., Merinopoulos I., Ahmed J., Whitty C. J. M., Mutabingwa T. K., Sutherland C. J., et al. (2007). Amodiaquine and Artemether-Lumefantrine Select Distinct Alleles of the Plasmodium Falciparum Mdr1 Gene in Tanzanian Children Treated for Uncomplicated Malaria. Antimicrob. Agents Chemother. 51 (3), 991–997. doi: 10.1128/AAC.00875-06
Kamugisha E., Jing S., Minde M., Kataraihya J., Kongola G., Kironde F., et al. (2012). Efficacy of Artemether-Lumefantrine in Treatment of Malaria Among Under-Fives and Prevalence of Drug Resistance Markers in Igombe-Mwanza, North-Western Tanzania. Malaria J. 11, 58. doi: 10.1186/1475-2875-11-58
Liu Y., Zhou R., Qian D., Yang C., Zhang H. (2014). Analysis of Malaria Epidemiological Characteristics in Henan Province From 2005 to 2013. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi 32, 419–422. (in Chinese).
Lu F., Zhang M., Culleton R. L., Xu S., Tang J., Zhou H., et al. (2017). Return of Chloroquine Sensitivity to Africa? Surveillance of African Plasmodium Falciparum Chloroquine Resistance Through Malaria Imported to China. Parasites Vectors 10 (1), 355. doi: 10.1186/s13071-017-2298-y
McCollum A. M., Poe A. C., Hamel M., Huber C., Zhou Z., Shi Y. P., et al. (2006). Antifolate Resistance in Plasmodium Falciparum: Multiple Origins and Identification of Novel Dhfr Alleles. J. Infect. Dis. 194 (2), 189–197. doi: 10.1086/504687
Mehlotra R. K., Fujioka H., Roepe P. D., Janneh O., Ursos L. M., Jacobs-Lorena V., et al. (2001). Evolution of a Unique Plasmodium Falciparum Chloroquine-Resistance Phenotype in Association With Pfcrt Polymorphism in Papua New Guinea and South America. Proc. Natl. Acad. Sci. U. S. A. 98 (22), 12689–12694. doi: 10.1073/pnas.221440898
Ménard D., Khim N., Beghain J., Adegnika A. A., Shafiul-Alam M., Amodu O., et al. (2016). A Worldwide Map of Plasmodium Falciparum K13-Propeller Polymorphisms. N Engl. J. Med. 374 (25), 2453–2464. doi: 10.1056/NEJMoa1513137
Mwanza S., Joshi S., Nambozi M., Chileshe J., Malunga P., Kabuya J.-B., et al. (2016). The Return of Chloroquine-Susceptible Plasmodium Falciparum Malaria in Zambia. Malar. J. 15 (1), 584. doi: 10.1186/s12936-016-1637-3
Naidoo I., Roper C. (2013). Mapping ‘Partially Resistant’, ‘Fully Resistant’, and ‘Super Resistant’ Malaria. Trends Parasitol. 29 (10), 505–515. doi: 10.1016/j.pt.2013.08.002
Nankabirwa J., Cundill B., Clarke S., Kabatereine N., Rosenthal P. J., Dorsey G., et al. (2010). Efficacy, Safety, and Tolerability of Three Regimens for Prevention of Malaria: A Randomized, Placebo-Controlled Trial in Ugandan Schoolchildren. PloS One 5 (10), e13438. doi: 10.1371/journal.pone.0013438
National Population Commission (NPC) [Nigeria] and ICF (2019). The 2018 Nigeria Demographic and Health Survey Key Findings (Abuja, Nigeria and Rockville, Maryland, USA: NPC and ICF).
Ndam N. T., Basco L. K., Ngane V. F., Ayouba A., Ngolle E. M., Deloron P., et al. (2017). Reemergence of Chloroquine-Sensitive Pfcrt K76 Plasmodium Falciparum Genotype in Southeastern Cameroon. Malar. J. 16 (1), 130. doi: 10.1186/s12936-017-1783-2
Noedl H., Se Y., Schaecher K., Smith B. L., Socheat D., Fukuda M. M. (2008). Evidence of Artemisinin-Resistant Malaria in Western Cambodia. N Engl. J. Med. 359 (24), 2619–2620. doi: 10.1056/NEJMc0805011
Nuwaha F. (2001). The Challenge of Chloroquine-Resistant Malaria in Sub-Saharan Africa. Health Policy Plann. 16 (1), 1–12. doi: 10.1093/heapol/16.1.1
Oboh M. A., Ndiaye D., Antony H. A., Badiane A. S., Singh U., Ali N. A., et al. (2018). Status of Artemisinin Resistance in Malaria Parasite Plasmodium Falciparum From Molecular Analyses of the Kelch13 Gene in Southwestern Nigeria. BioMed. Res. Int. 2018, 2305062. doi: 10.1155/2018/2305062
Oguike M. C., Falade C. O., Shu E., Enato I. G., Watila I., Baba E. S., et al. (2016). Molecular Determinants of Sulfadoxinepyrimethamine Resistance in Plasmodium Falciparum in Nigeria and the Regional Emergence of Dhps 431V. Int. J. Parasitol. Drugs Drug Resist. 6 (3), 220–229. doi: 10.1016/j.ijpddr.2016.08.004
Oladipo O. O., Wellington O. A., Sutherland C. J. (2015). Persistence of Chloroquine-Resistant Haplotypes of Plasmodium Falciparum in Children With Uncomplicated Malaria in Lagos, Nigeria, Four Years After Change of Chloroquine as First-Line Antimalarial Medicine. Diagn. Pathol. 10, 41. doi: 10.1186/s13000-015-0276-2
Otienoburu S. D., Suay I., Garcia S., Thomas N. V., Srisutham S., Björkman A., et al. (2019). An Online Mapping Database of Molecular Markers of Drug Resistance in Plasmodium Falciparum: The ACT Partner Drug Molecular Surveyor. Malar. J. 18 (1), 12. doi: 10.1186/s12936-019-2645-x
Picot S., Olliaro P., de Monbrison F., Bienvenu A.-L., Price R. N., Ringwald P. (2009). A Systematic Review and Meta-Analysis of Evidence for Correlation Between Molecular Markers of Parasite Resistance and Treatment Outcome in Falciparum Malaria. Malar. J. 8, 89. doi: 10.1186/1475-2875-8-89
Ridley R. G. (2002). Medical Need, Scientific Opportunity and the Drive for Antimalarial Drugs. Nature 415 (6872), 686–693. doi: 10.1038/415686a
Ross L. S., Dhingra S. K., Mok S., Yeo T., Wicht K. J., Kümpornsin K., et al. (2018). Emerging Southeast Asian Pfcrt Mutations Confer Plasmodium Falciparum Resistance to the First-Line Antimalarial Piperaquine. Nat. Commun. 9 (1), 3314. doi: 10.1038/s41467-018-05652-0
Sá J. M., Twu O., Hayton K., Reyes S., Fay M. P., Ringwald P., et al. (2009). Geographic Patterns of Plasmodium Falciparum Drug Resistance Distinguished by Differential Responses to Amodiaquine and Chloroquine. Proc. Natl. Acad. Sci. U. S. A. 106 (45), 18883–18889. doi: 10.1073/pnas.0911317106
Sisowath C., Petersen I., Veiga M. I., Mårtensson A., Premji Z., Björkman A., et al. (2009). In Vivo Selection of Plasmodium Falciparum Parasites Carrying the Chloroquine-Susceptible Pfcrt K76 Allele After Treatment With Artemether-Lumefantrine in Africa. J. Infect. Dis. 199 (5), 750–757. doi: 10.1086/596738
Somé A. F., Seré Y. Y., Dokomajilar C., Zongo I., Rouamba N., Greenhouse B., et al. (2010). Selection of Known Plasmodium Falciparum Resistance-Mediating Polymorphisms by Artemether-Lumefantrine and Amodiaquine-Sulfadoxine-Pyrimethamine But Not Dihydroartemisinin-Piperaquine in Burkina Faso. Antimicrob. Agents Chemother. 54 (5), 1949–1954. doi: 10.1128/AAC.01413-09
Sowunmi A., Akano K., Ntadom G., Ayede A. I., Ibironke F. O., Aderoyeje T., et al. (2017). Therapeutic Efficacy and Effects of Artemisinin-Based Combination Treatments on Uncomplicated Plasmodium Falciparum Malaria-Associated Anaemia in Nigerian Children During Seven Years of Adoption as First-Line Treatments. Infect. Dis. Poverty 6 (1), 36. doi: 10.1186/s40249-016-0217-7
Taylor S. M., Parobek C. M., DeConti D. K., Kayentao K., Coulibaly S. O., Greenwood B. M., et al. (2014). Absence of Putative Artemisinin Resistance Mutations Among Plasmodium Falciparum in Sub-Saharan Africa: A Molecular Epidemiologic Study. J. Infect. Dis. 211 (5), 680–688. doi: 10.1093/infdis/jiu467
Tola M., Ajibola O., Idowu E. T., Omidiji O., Awolola S. T., Amambua-Ngwa A. (2020). Molecular Detection of Drug Resistant Polymorphisms in Plasmodium Falciparum Isolates From Southwest, Nigeria. BMC Res. Notes 13 (1), 497. doi: 10.1186/s13104-020-05334-5
Uwimana A., Legrand E., Stokes B. H., Ndikumana J. M., Warsame M., Umulisa N., et al. (2020). Emergence and Clonal Expansion of In Vitro Artemisinin-Resistant Plasmodium Falciparum Kelch13 R561H Mutant Parasites in Rwanda. Nat. Med. 26, 1602–1608. doi: 10.1038/s41591-020-1005-2
Vinayak S., Alam M. T., Mixson-Hayden T., McCollum A. M., Sem R., Shah N. K., et al. (2010). Origin and Evolution of Sulfadoxine Resistant Plasmodium Falciparum. PloS Pathog. 6 (3), e1000830. doi: 10.1371/journal.ppat.1000830
World Health Organization (2010). Global Report on Antimalarial Drug Efficacy and Drug Resistance: 2000-2010. Available at: https://www.who.int/publications/i/item/9789241500470.
World Health Organization (2018). Artemisinin Resistance and Artemisininbased Combination Therapy Efficacy. Available at: https://www.who.int/publications/i/item/status-report-on-artemisinin-resistance-and-act-efficacy.
World Health Organization (2018). World Malaria Report 2017. Available at: https://www.who.int/publications/i/item/9789241565523.
World Health Organization (2019). Artemisinin Resistance and Artemisinin-Based Combination Therapy Efficacy. Available at: https://www.who.int/publications/i/item/status-report-on-artemisinin-resistance-and-act-efficacy.
World Health Organization (2020). World Malaria Report 2020. Available at: https://www.who.int/publications/i/item/9789240015791.
Wundermann G., Osiki A. (2017). Currently Observed Trend in the Resistance of Malaria to Artemisinin Based Combination Therapy in Nigeria – a Report of 5 Cases. Int. J. Trop. Dis. Health 21 (2), 1–5. doi: 10.9734/IJTDH/2017/30914
Xu C., Sun H., Wei Q., Li J., Xiao T., Kong X., et al. (2019). Mutation Profile of Pfdhfr and Pfdhps in Plasmodium Falciparum Among Returned Chinese Migrant Workers From Africa. Antimicrob. Agents Chemother. 63 (5), e01927–e01918. doi: 10.1128/AAC.01927-18
Yeung S., Socheat D., Moorthy V. S., Mills A. J. (2009). Artemisinin Resistance on the Thai–Cambodian Border. Lancet 374 (9699), 1418–1419. doi: 10.1016/S0140-6736(09)61856-0
Yin J., Yan H., Huang F., Li M., Xiao H., Zhou S., et al. (2015). Establishing a China Malaria Diagnosis Reference Laboratory Network for Malaria Elimination. Malar. J. 14, 40. doi: 10.1186/s12936-015-0556-z
Zhang L., Feng J., Zhang S., Xia Z., Zhou S. (2018). The Progress of National Malaria Elimination and Epidemiological Characteristics of Malaria in China in 2017. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi 36, 201–209. (in Chinese).
Zhou R., Yang C., Li S., Zhao Y., Liu Y., Qian D., et al. (2019). Molecular Surveillance of Drug Resistance of Plasmodium Falciparum Isolates Imported From Angola in Henan Province, China. Antimicrob. Agents Chemother. 63 (10), e00552–e00519. doi: 10.1128/AAC.00552-19
Keywords: Plasmodium falciparum, drug resistance, PfK13, Pfcrt, Pfmdr1, Pfdhfr, Pfdhps, Nigeria
Citation: Zhao D, Zhang H, Ji P, Li S, Yang C, Liu Y, Qian D, Deng Y, Wang H, Lu D, Zhou R and Zhao Y (2021) Surveillance of Antimalarial Drug-Resistance Genes in Imported Plasmodium falciparum Isolates From Nigeria in Henan, China, 2012–2019. Front. Cell. Infect. Microbiol. 11:644576. doi: 10.3389/fcimb.2021.644576
Received: 21 December 2020; Accepted: 31 March 2021;
Published: 23 April 2021.
Edited by:
Jun Feng, National Institute of Parasitic Diseases, ChinaReviewed by:
Carlo Severini, National Institute of Health (ISS), ItalyDidier Menard, Institut Pasteur, France
Yaming Cao, China Medical University, China
Olusola Ojurongbe, Ladoke Akintola University of Technology, Nigeria
Copyright © 2021 Zhao, Zhang, Ji, Li, Yang, Liu, Qian, Deng, Wang, Lu, Zhou and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruimin Zhou, zrm920@126.com; Yuling Zhao, 819482937@qq.com
†These authors have contributed equally to this work
 Dongyang Zhao†
Dongyang Zhao†  Hongwei Zhang
Hongwei Zhang Ying Liu
Ying Liu Ruimin Zhou
Ruimin Zhou