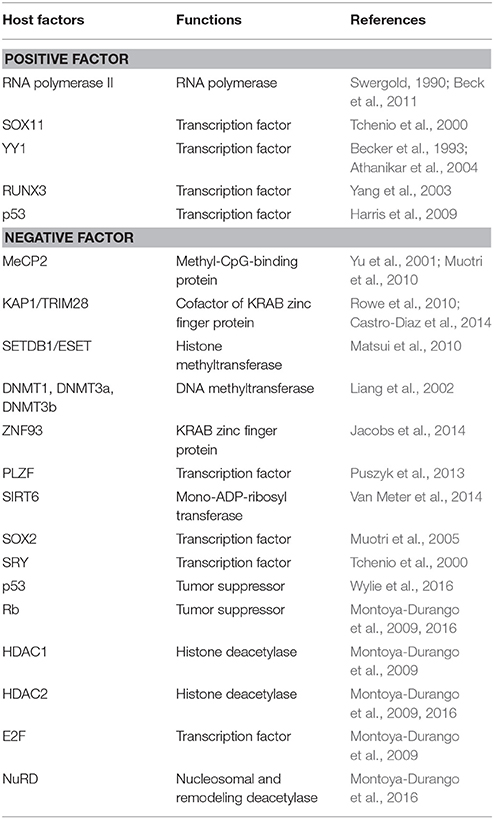
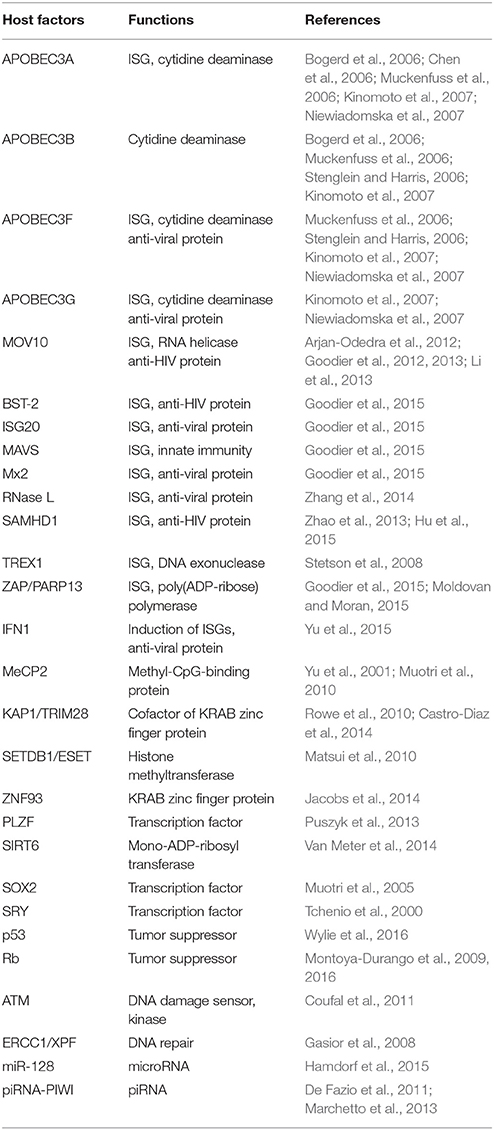
Guardian of the Human Genome: Host Defense Mechanisms against LINE-1 Retrotransposition
- Ariumi Project Laboratory, Center for AIDS Research and International Research Center for Medical Sciences, Kumamoto University, Kumamoto, Japan
Long interspersed element type 1 (LINE-1, L1) is a mobile genetic element comprising about 17% of the human genome, encoding a newly identified ORF0 with unknown function, ORF1p with RNA-binding activity and ORF2p with endonuclease and reverse transcriptase activities required for L1 retrotransposition. L1 utilizes an endonuclease (EN) to insert L1 cDNA into target DNA, which induces DNA double-strand breaks (DSBs). The ataxia-telangiectasia mutated (ATM) is activated by DSBs and subsequently the ATM-signaling pathway plays a role in regulating L1 retrotransposition. In addition, the host DNA repair machinery such as non-homologous end-joining (NHEJ) repair pathway is also involved in L1 retrotransposition. On the other hand, L1 is an insertional mutagenic agent, which contributes to genetic change, genomic instability, and tumorigenesis. Indeed, high-throughput sequencing-based approaches identified numerous tumor-specific somatic L1 insertions in variety of cancers, such as colon cancer, breast cancer, and hepatocellular carcinoma (HCC). In fact, L1 retrotransposition seems to be a potential factor to reduce the tumor suppressive property in HCC. Furthermore, recent study demonstrated that a specific viral-human chimeric transcript, HBx-L1, contributes to hepatitis B virus (HBV)-associated HCC. In contrast, host cells have evolved several defense mechanisms protecting cells against retrotransposition including epigenetic regulation through DNA methylation and host defense factors, such as APOBEC3, MOV10, and SAMHD1, which restrict L1 mobility as a guardian of the human genome. In this review, I focus on somatic L1 insertions into the human genome in cancers and host defense mechanisms against deleterious L1 insertions.
Introduction
Long interspersed element type 1 (LINE-1, L1) is an active and autonomous non-long terminal repeat (LTR) retrotransposon composing about 17% of the human genome and L1 is an essential evolutionary force (DeBerardinis et al., 1998; Ostertag and Kazazian, 2001; Cordaux and Batzer, 2009; Hancks and Kazazian, 2012). However, only 100 copies out of ~500,000 copies still remain active (Brouha et al., 2003; Mills et al., 2007; Beck et al., 2010). The remaining L1s are 5′ truncated and defective. Furthermore, L1 provides the trans-acting functions required for the retrotransposition of non-autonomous retrotransposons such as short interspersed element (SINE), which includes Alu repeats in humans, SINE-VNTR-Alu (SVA), and processed pseudogenes (Esnault et al., 2000; Dewannieux et al., 2003; Hancks et al., 2011).
L1 encodes three open reading frames, a newly identified ORF0 with unknown function, ORF1p with RNA-binding and nucleic acid chaperon activities, and ORF2p with AP-like endonuclease (EN) and reverse transcriptase (RT) activities required for L1 retrotransposition (Mathias et al., 1991; Martin and Bushman, 2001; Ostertag and Kazazian, 2001; Hancks and Kazazian, 2012; Denli et al., 2015). ORF0 is the primate-specific ORF in the anti-sense 5′ untranslated region (UTR) of L1 (Denli et al., 2015). ORF0 predominantly localizes in nuclear PML-adjacent foci and enhances L1 mobility. ORF1p and ORF2p preferentially assemble with L1 RNA and form a ribonucleoprotein (RNP) in the cytoplasmic foci (Goodier et al., 2007; Doucet et al., 2010). Although retroviruses and LTR-retrotransposons utilize a long terminal repeat (LTR) to synthesize full-length transcripts, L1 instead utilizes an internal promoter in the 5′UTR of L1 (Swergold, 1990). Several transcription factors including SOX11 (Tchenio et al., 2000), YY1 (Becker et al., 1993; Athanikar et al., 2004), RUNX3 (Yang et al., 2003), and p53 (Harris et al., 2009) positively regulate the L1 transcription. On the other hand, SOX2 (Muotri et al., 2005) and SRY (Tchenio et al., 2000) as well as several epigenetic factors negatively regulate the L1 transcription (Table 1).
L1 integrates into the genome by target-primed reverse transcription (TPRT) (Luan et al., 1993) after the L1-RNP complex enters the nucleus. During TPRT, the L1 EN creates a nicked DNA that serves as a primer for reverse transcription of L1 RNA, leading to integration of L1 cDNA into the human genome (Feng et al., 1996). A typical L1 EN cleavage site is 5′-TTTT/AA-3′ (Feng et al., 1996; Cost and Boeke, 1998). Thus, L1 insertion generates DNA double-strand breaks (DSBs) as well as L1 structural hallmarks such as frequent 5′ truncations, 3′ poly(A) tails and variable length target site duplications (TSDs) in the target DNA. L1 can alter the mammalian genome in many ways upon retrotransposition, since the insertion of L1 into the human genome may cause genomic instability, genetic disorders, and cancers through insertional mutagenesis (Kazazian et al., 1988; Morse et al., 1988; Miki et al., 1992; Narita et al., 1993; Holmes et al., 1994; Gilbert et al., 2002; Morrish et al., 2002; Symer et al., 2002; Belancio et al., 2008; Beck et al., 2011; Hancks and Kazazian, 2012; Bundo et al., 2014; Kines et al., 2014; Figure 1). So far, >100 disease-causing retrotransposon insertions have been identified in humans [26 L1, 61 Alu, 12 SVA, 4 poly(A)] (Figure 1).
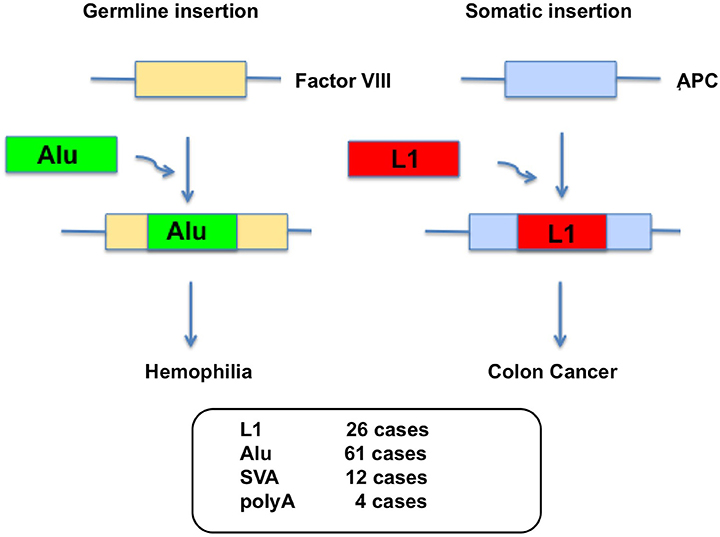
Figure 1. Germline Alu insertion in the human genome causes hemophilia and somatic L1 insertion causes colon cancer. L1 insertions have also been observed in Factor VIII and this causes hemophilia A. Disease causing L1 and Alu insertions are also found in genes that are X-linked inherited. >100 disease-causing retrotransposon insertions have been identified in humans [26 L1, 61 Alu, 12 SVA, 4 poly(A)].
L1-Mediated DSBs Induction and DNA Repair Machinery
L1 is known to induce DSBs in target DNA by L1 EN activity (Gasior et al., 2006). The ataxia-telangiectasia mutated (ATM) is activated by DSBs and subsequently phosphorylates downstream substrates including p53, Chk2, BRCA1 and the MRE11-Rad50-NBS1 (MRN) complex, resulting in the activation of DNA damage checkpoint and cell cycle arrest (Harper and Elledge, 2007; Ciccia and Elledge, 2010; Shiloh, 2014). Accordingly, L1 retrotransposition was increased in ATM-deficient cells, indicating ATM signaling pathway modulates L1 retrotransposition (Coufal et al., 2011). In contrast, the E6 protein from β-human papillomavirus (β-HPV 5 and 8) reduces ATM protein levels and attenuates L1 retrotransposition, suggesting that ATM is needed for efficient L1 retrotransposition (Wallace et al., 2013). Thus, the DNA damage response may modulate L1 retrotransposition. Notably, L1 can integrate into preformed DSBs generated independently of L1 EN, resulting in retrotransposon-mediated DNA repair (Morrish et al., 2002). Furthermore, host DNA repair machinery may also impact L1 retrotransposition. Gasior et al reported that DNA repair enzyme ERCC1/XPF heterodimer limits L1 retrotransposition (Gasior et al., 2008). Importantly, deficiencies of non-homologous end-joining (NHEJ) repair pathway such as Ku70, Artemis, and DNA ligase IV (LigIV) decrease retrotransposition frequencies of human L1 in chicken DT40 cells, suggesting that the NHEJ repair pathway is required for efficient L1 retrotransposition (Suzuki et al., 2009).
L1 Retrotransposition in Cancers
Somatic L1 insertions are seldom observed in normal tissues except hippocampus (Baillie et al., 2011; Evrony et al., 2012; Upton et al., 2015). Although most L1 retrotransposition was thought to occur in the germline, somatic L1 insertions were also found to occur in variety of tumors, including breast cancer, colon cancer, hepatocellular carcinoma (HCC), and lung cancer (Miki et al., 1992; Liu et al., 1997; Iskow et al., 2010; Lee et al., 2012; Solyom et al., 2012; Shukla et al., 2013; Carreira et al., 2014; Helman et al., 2014; Ewing et al., 2015; Table 2). First, three L1 insertion candidates were reported in human tumors (Morse et al., 1988; Miki et al., 1992; Liu et al., 1997). However, two insertions described by Liu et al. (1997) and Morse et al. (1988) lack all of the hallmark features of a true somatic retrotransposition event, such as L1 endonuclease cleavage site, the presence of L1 poly(A) tail, target-site duplication (TSD), 5′ truncation and inversion, and 3′ transduction (Holmes et al., 1994; Moran et al., 1999; Goodier et al., 2000; Pickeral et al., 2000; Szak et al., 2002; Table 2). These insertions may be derived from recombination events, L1 EN-independent insertions (Morrish et al., 2002), or other atypical integration mechanisms of L1 retrotransposition. Indeed, an L1 insertion disrupts the adenomatous polyposis coli (APC) gene in a colon cancer, indicating the disruption of a tumor suppressor gene caused by somatic L1 insertion (Miki et al., 1992). Accordingly, a recent study identified a novel somatic insertion in the APC gene and a hot spot for L1 insertion on Chromosome 17, suggesting that the L1 insertion initiates colorectal cancer (CRC) by mutating the APC gene through the classic two-hit CRC pathway (Scott et al., 2016). Furthermore, high-throughput sequencing-based approaches identified numerous somatic tumor-specific insertions in cancers (Miki et al., 1992; Liu et al., 1997; Iskow et al., 2010; Lee et al., 2012; Solyom et al., 2012; Shukla et al., 2013; Carreira et al., 2014; Helman et al., 2014; Tubio et al., 2014). Indeed, Lee et al. identified the L1 insertions in cadherin-12 (CDH12), roundabout, axon guidance receptor, homolog 2 (ROBO2), NRXN3, FPR2, COL11A1, NEGR1, NTM, and CTNNA2 (Lee et al., 2012). As well, Solyom et al. identified several tumor-specific insertions in colorectal tumors including odd Oz/ten-m homolog 3 (ODZ3), ROBO2, protein tyrosine phosphatase, receptor type, M (PTPRM), pericentriolar material 1 (PCM1), CDH11, and runt-related transcription factor 1 (RUNX1T1) of colorectal cancers (Solyom et al., 2012). All insertions were severely 5′ truncated. Interestingly, these genes are associated with cell-adhesion functions and both groups could identify the L1 insertions in the same ROBO2 genes, suggesting the potential role of cell-adhesion genes in L1 insertion-mediated colorectal tumorigenesis. In addition, Tubio et al. analyzed the somatic L1 retrotransposition activities in 290 cancers and noticed insertions occurring during cancer development. 53% of the patients have at least one somatic L1 retrotransposition event, of which 24% were 3′ transductions, most frequently colorectal cancers (93%) and lung cancers (75%), suggesting that 3′ transduction are potentially mutagenic. Somatic L1 retrotranspositions tend to insert in intergenic or heterochromatin regions of the cancer genome (Tubio et al., 2014). Furthermore, somatic L1 insertions participate in the dynamics of many tumor genomes and lead to driver mutations. Surprisingly, L1 insertion was reported in colonic adenoma, a known cancer precursor, suggesting that widespread somatic L1 retrotransposition occurs early during development of gastrointestinal (GI) tumors, probably before dysplastic growth (Ewing et al., 2015). Similarly, a recent study demonstrated that L1 retrotransposition is active in esophageal adenocarcinoma and its precursor, Barrett's esophagus (BE), indicating that somatic L1 insertions occur early in BE and esophageal adenocarcinoma. Notably, two L1 insertions were detected in normal esophagus, indicating that some L1 insertions may occur in normal squamous epithelium cells (Doucet-O'Hare et al., 2015). In this regard, most of the new somatic insertions are truncated, and would not mobilize again. So mutations arising from insertions in the normal precursor esophageal or benign BE would be contributing to tumorigenesis. Otherwise, only a rare full-length somatic insertion has the potential to contribute to mutation during the various stages of transition to tumorigenesis. In addition, L1 insertions in pancreatic ductal adenocarcinoma (PDAC) were reported with discordant rate of retrotransposition between primary and metastatic sites, suggesting that L1 insertions in gastrointestinal neoplasms occur discontinuously. Thus, somatic L1 insertions contribute to genetic and phenotypic heterogeneity in PDAC (Rodić et al., 2015). Interestingly, somatic insertions were identified in epithelial tumors but not in blood or brain cancers (Lee et al., 2012). However, we raise awareness regarding the following limitations of this study. For example, the sample size was small and the normal tissue was not from the same patient. In addition, in this study they only examined multiple myeloma and did not look at the entire spectrum of blood based cancers. In this regard, ten-eleven-translocation (TET) 2, a DNA demethylation-related protein, is frequently mutated in myeloid and lymphoid tumors (Ko et al., 2015). The TET family that oxidizes 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) in DNA, leads to the DNA demethylation. Since DNA methylation has a pivotal regulatory role in L1 silencing, TET2 may impact L1 mobility. Therefore, L1 insertions may be suppressed in such hematological cancers. Intriguingly, several somatic insertions occur in genes that are commonly mutated in cancers such as tumor suppressor gene. These insertions disrupt the expression of target genes, and are biased toward regions of cancer-specific DNA hypomethylation (Lee et al., 2012). Indeed, recent studies identified somatic L1 insertion in tumor suppressor genes, such as APC and PTEN (Miki et al., 1992; Helman et al., 2014). As well, the first case of familial retinoblastoma (Rb) caused by a de novo insertion of a full-length L1 into intron 14 of the Rb gene, resulting in the aberrant and non-canonical mRNA splicing of the Rb gene, was reported (Rodríguez-Martín et al., 2016). Furthermore, 18 retrotransposon insertions [14 Alu, 3 L1, and 1 poly(A)] were identified in neurofibromatosis type 1 (NF1) gene (Wimmer et al., 2011).
Although still debated, cell division seems to be required for efficient L1 retrotransposition (Shi et al., 2007; Xie et al., 2013). In fact, retrotransposition was strongly inhibited in the cells arrested in the G1, S, G2, or M phase of cell cycle. The reduction in L1 transcript abundance limits retrotransposition in non-dividing cells, suggesting that inhibition of retrotransposition in non-dividing cells protects somatic cells from accumulation of deleterious mutations caused by L1 insertions (Shi et al., 2007). In contrast, there is an opposite report that L1 retrotransposition was detected in non-dividing and primary human somatic cells using adenovirus-L1 hybrid vector, even though they detected L1 retrotransposition in G1/S- but not in G0-arrested cells (Kubo et al., 2006). In addition, retrotransposition was also inhibited during cellular senescence in primary human fibroblasts. So far, several biomarkers of cellular senescence have been identified such as senescence-associated β-galactosidase (SA-β-Gal), p53/p21, p16INK4a, senescence-associated heterochromatin foci (SAHF), senescence-associated secretory phenotype (SASP), autophagy, telomere-induced foci/DNA damage response (DDR), and cell cycle arrest (Kuilman et al., 2010) and the reduction in L1 retrotransposition may be a biomarker of cellular senescence. Thus, cell cycle may affect L1 retrotransposition.
L1 protein expression is a common feature of many types of high-grade malignant tumor, yet is rarely detected in early stage of tumorigenesis (Rodić et al., 2014). L1 promoter is normally silenced by methylation in normal somatic cells (Woodcock et al., 1997; Schulz et al., 2006). In contrast, L1 promoter is hypomethylated (Baba et al., 2014), and expression of L1 is elevated in many tumors. In fact, L1 expression was detected in human breast carcinomas and testicular cancers (Bratthauer and Fanning, 1992; Bratthauer et al., 1994; Nangia-Makker et al., 1998). L1 ORF1p protein is detected in a variety of tumor cells including breast cancer, colon cancer, pancreatic ductal adenocarcinoma, and HCC but not in normal somatic cells (Bratthauer et al., 1994; Asch et al., 1996; Rodić et al., 2014). Thus, L1 ORF1p expression seems to be a hallmark of many human cancers as a highly specific tumor marker.
In addition to expression of L1, a hallmark of tumor cells is an activated telomere maintenance mechanism that allows prolonged survival of the malignant tumor cells. In more than 80% of tumors, telomeres are typically maintained by telomerase. Notably, the reduced length of telomeres was reported in the L1 knockdown cells, indicating that L1 is involved in telomere maintenance in telomerase positive tumor cells (Aschacher et al., 2016). Accordingly, L1 involves in a transcriptional regulation of hTERT and upregulation of its transcription factors c-Myc and KLF-4 (Aschacher et al., 2016). Thus, L1 may contribute to the development of cancers. However, these studies were not done in alternative lengthening of telomeres (ALT)-positive tumors or telomerase negative tumors. Consequently, it is uncertain if L1 is directly contributing to telomere maintenance or if the reduction in telomere length is contributed to the reduction in telomerase levels. Indeed, the stoichiometry of telomerase is important for maintaining telomere length (Armanios et al., 2005; Goldman et al., 2005).
Chronic infection with hepatitis B virus (HBV) is a major risk for the development of HCC. HBV integration into the human genome was found in most HBV-related HCC and it has been implicated in the development of HCC. An initial study proposed that HBV integration occurs randomly without preferred integration site (Matsubara and Tokino, 1990). However, high-throughput sequencing-based approaches identified recurrent integration sites in HCC (Ding et al., 2012). HBV integration favored chromosome 17 and preferentially integrated into human transcript units. At least, telomerase reverse transcriptase (TERT) and fibronectin 1 (FN1) genes were identified as the recurrent HBV integration sites. Furthermore, seven integrations were found in the repeat regions including L1, LTR/ERV1, and SINE/Alu (Ding et al., 2012). Similarly, a recent transcriptome sequencing study of HBV-positive HCC cell lines discovered that HBV integrates into L1 (Lau et al., 2014). Insertion of the gene encoding hepatitis B virus x (HBx) into L1 on chromosome 8p11 produces an oncogenic HBx-LINE1 chimeric RNA transcript (Lau et al., 2014; Figure 2). The HBx-LINE1 RNA transcript was detected in 23.3% of HCC, suggesting that HBx-LINE1 is selected for in HCC oncogenesis. The long non-coding RNA (lncRNA)-like HBx-LINE1 transcript confers cancer-promoting properties through activation of Wnt/β-catenin signaling pathway (Lau et al., 2014).
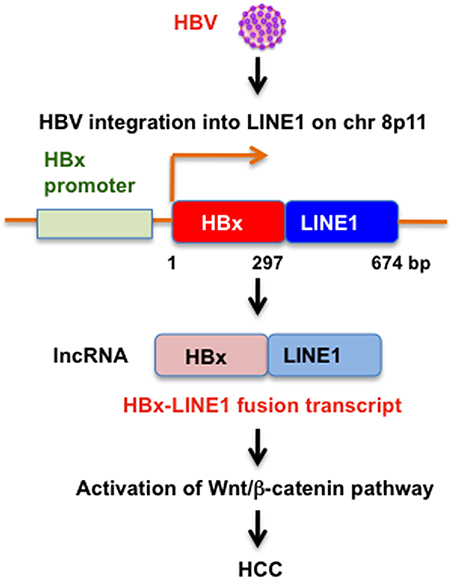
Figure 2. HBV integration into LINE1 in HCC. Schematic representation of the HBx-LINE1 fusion RNA transcript and potential role of oncogenic HBx-LINE1 fusion RNA transcript in HCC.
Furthermore, endogenous L1-mediated retrotransposition was identified in the germline and somatic cells of HCC patients (Shukla et al., 2013). The germline L1 insertion in the tumor suppressor mutated in colorectal cancers (MCC) was detected in 21.1% of HCC, resulting in the aberrant expression of MCC. Moreover, suppression of tumorigenicity 18 (ST18) was activated by a tumor-specific somatic L1 insertion (Shukla et al., 2013). Thus, L1-mediated retrotransposition seems to be a potential etiological factor in HCC.
Guardian of the Human Genome: Host Defense Mechanisms Against L1 Retrotransposition
Since insertion of L1 into the human genome may cause human genetic disorders and cancer, retrotransposition must be silenced under normal conditions. To restrict deleterious retrotransposition, host cells have evolved several defense mechanisms protecting cells against retrotransposition including epigenetic regulation through DNA methylation (Burden et al., 2005; Trono, 2015), RNA silencing by RNA interference (Soifer et al., 2005; Yang and Kazazian, 2006), PIWI-interacting RNA (piRNA)-PIWI system (Aravin et al., 2007a,b; Kuramochi-Miyagawa et al., 2008; De Fazio et al., 2011; Marchetto et al., 2013), microRNA (Hamdorf et al., 2015), and host restriction factors, such as apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3 (APOBEC3), Moloney leukemia virus 10 (MOV10), and SAM domain and HD domain containing protein 1 (SAMHD1) (Table 3).
DNA methylation within the 5′UTR promoter of L1 is essential for maintaining transcriptional inactivation and for inhibiting L1 retrotransposition (Woodcock et al., 1997; Liang et al., 2002; Burden et al., 2005). L1 is highly active during early embryogenesis, while L1 is silenced early in development through epigenetic mechanisms (Table 1). Indeed, methylation of the L1 promoter is maintained by DNA metyltransferases (DNMTs), including DNMT1, DNMT3a, and DNMT3b (Liang et al., 2002). L1 retrotransposition is negatively regulated by methyl-CpG-binding protein 2 (MeCP2)-mediated DNA methylation (Yu et al., 2001; Muotri et al., 2010). In addition, nucleosomal and remodeling deacetylase (NuRD) multiprotein complex specifically enriches the L1 promoter. Rb and E2F recruit to the L1 promoter along with histone deacetylase (HDAC), including HDAC1 and HDAC2 (Montoya-Durango et al., 2009, 2016). Furthermore, KRAB-associated protein1 (KAP1, also known as TRIM28) mediates transcriptional silencing of endogenous retroelements (EREs) including L1, Alu, SVA, and human endogenous retrovirus-K (HERV-K) as well as exogenous retrovirus mouse leukemia virus (MLV) in embryonic stem (ES) cells (Wolf and Goff, 2007, 2009; Matsui et al., 2010; Rowe et al., 2010; Castro-Diaz et al., 2014; Turelli et al., 2014; Trono, 2015). Krüppel-associated box (KRAB)-containing zinc-finger protein (KRAB-ZFP/ZNF), a large family of tetrapod-restricted transcription factors, and a cofactor KAP1 serve as a scaffold for a heterochromatin complex comprising the SETDB1 (also known as ESET) histone methyltransferase, histone deacetylase, nucleosome remodeling, and DNMT activities (Trono, 2015; Figure 3). Furthermore, the protein deacylase and mono-ADP ribosyltransferase Sirtuin 6 (SIRT6) represses L1 mobility by ribosylating KAP1 (Van Meter et al., 2014). SIRT6 binds to the 5′UTR of L1 and ribosylates KAP1, resulting in facilitation of KAP1 interaction with the heterochromatin factor HP1α, thereby contributing to the packaging of L1 into transcriptionally repressive heterochromatin. Moreover, promyelocytic leukemia zinc finger (PLZF) protein, a member of the POK (POZ and Kruppel zinc finger) family of transcription factors that acts as an epigenetic regulator of stem cell maintenance in germ cells and haematopoietic stem cells, represses L1 retrotransposition in germ and progenitor cells (Puszyk et al., 2013). PLZF-mediated DNA methylation induces silencing of the L1 gene, resulting in inhibition of L1 retrotransposition. Species-specific KZNFs might recruit KAP1 to species-specific retrotransposon classes that recently invaded the host genome. In this regard, Jacobs et al. recently reported that two primate-specific ZNF91 and ZNF93 repress SVA and L1 retrotransposons, respectively (Jacobs et al., 2014). ZNF93 evolved earlier to repress the primate L1 lineage until ~12.5 million years ago when the L1PA3 subfamily escaped ZNF93-mediated restriction through the removal of the ZNF93-binding site, suggesting an evolutionary arms race between KRAB-ZNFs and retrotransposons (Jacobs et al., 2014).
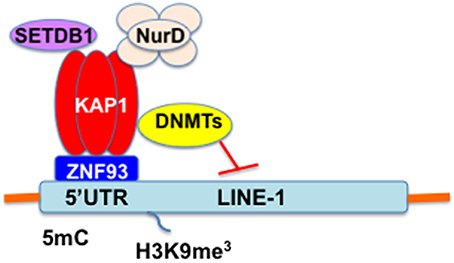
Figure 3. The KRAB/KAP1 system controls the transcriptional activity of L1 in ES cells. KRAB/KAP1 serves as a scaffold for a heterochromatin complex comprising the SETDB1 and DNMT.
Post-translational modification and subcellular localization of L1 protein seem to be important for modulation of L1 mobility. In fact, phosphorylation of ORF1p is required for L1 retrotransposition (Cook et al., 2015). L1 ORF1p contains four conserved proline-directed protein kinase (PDPKs) target sites. PDPK mutations in ORF1p could inactivate L1 mobility (Cook et al., 2015). The PDPK family includes mitogen-activated protein kinases (MAPKs) and cyclin-dependent kinases (CDKs). Although nuclear localization of L1 ORF1p and ORF2p is essential for L1 retrotransposition, L1 ORF1p predominantly localizes in punctate cytoplasmic foci in most of cases (Goodier et al., 2007; Harris et al., 2010; Chen et al., 2012). However, in several breast cancers, LI ORF1p and ORF2p were also detected in the nucleus (Harris et al., 2010; Chen et al., 2012). Indeed, the expression of L1 is elevated in breast cancers.
Recently, APOBEC3 family of cytidine deaminases, MOV10, and SAMHD1 have been identified as restriction factors for human immunodeficiency virus type 1 (HIV-1) (Sheehy et al., 2002; Harris et al., 2003; Mangeat et al., 2003; Zhang et al., 2003; Burdick et al., 2010; Furtak et al., 2010; Wang et al., 2010; Hrecka et al., 2011; Laguette et al., 2011). APOBEC3A, APOBEC3B, and APOBEC3F but not APOBEC3G inhibit L1 retrotransposition by a DNA deaminase-independent manner, indicating a novel anti-L1 retrotransposition mechanism (Turelli et al., 2004; Bogerd et al., 2006; Chen et al., 2006; Muckenfuss et al., 2006; Stenglein and Harris, 2006; Hulme et al., 2007; Kinomoto et al., 2007; Niewiadomska et al., 2007; Schumann, 2007; Arias et al., 2012). In contrast, Kinomoto et al. and Niewiadomska et al. reported that APOBEC3G could inhibit L1 retrotransposition by a DNA deamination-independent manner (Kinomoto et al., 2007; Niewiadomska et al., 2007). Furthermore, APOBEC3G inhibits Alu retrotransposition by a DNA deaminase-independent manner (Chiu et al., 2006; Hulme et al., 2007; Bulliard et al., 2009). MOV10 RNA helicase also inhibits L1 and Alu retrotransposition (Arjan-Odedra et al., 2012; Goodier et al., 2012, 2013; Li et al., 2013). Similarly, SAMHD1 inhibits LINE-1 and Alu/SVA retrotransposition (Zhao et al., 2013). SAMHD1 inhibits L1 retrotransposition through promoting the sequestration of L1 RNP within stress granules (Hu et al., 2015). Similarly, the zinc-finger antiviral protein (ZAP) also known as PARP13, a member of poly(ADP-ribose) polymerase (PARP) family, inhibits the retrotransposition of L1, Alu, and intracisternal A particle (IAP) retrotransposons (Goodier et al., 2015; Moldovan and Moran, 2015). ZAP interacts with L1 RNA and L1 ORF1p and co-localizes with stress granules.
Type I interferons (IFN 1) including IFNα and IFNβ have been involved in innate immunity against viruses. In this regard, a recent study reported that L1 induces IFN1 and IFN1, in turn, inhibits L1 retrotransposition, suggesting that IFN1 controls propagation of L1 as well as maintenance of genomic integrity (Yu et al., 2015). Accordingly, several interferon-stimulated genes (ISGs), including APOBEC3, MOV10, BST-2, ISG20, MAVS, MX2, RNase L, SAMHD1, TREX1, and ZAP restrict L1 retrotransposition, indicating that ISGs are key players of the type I interferon anti-retroelement response (Turelli et al., 2004; Bogerd et al., 2006; Chen et al., 2006; Muckenfuss et al., 2006; Stenglein and Harris, 2006; Hulme et al., 2007; Kinomoto et al., 2007; Niewiadomska et al., 2007; Schumann, 2007; Stetson et al., 2008; Arias et al., 2012; Zhao et al., 2013; Zhang et al., 2014; Goodier et al., 2015; Hu et al., 2015; Table 3).
Small RNAs have been implicated in the regulation of L1 mobility. Piwi proteins and Piwi-interacting RNAs (piRNA) silence L1 during genome reprogramming in the embryonic male germ line (De Fazio et al., 2011; Marchetto et al., 2013). Notably, Hamdorf et al. uncovered a new mechanism in which microRNAs restrict L1 mobilization and L1-associated mutations in cancer cells, cancer-initiating cells and iPS cells (Hamdorf et al., 2015). Indeed, miR-128 represses L1 retrotransposition by binding directly to L1 RNA, suggesting a new function of microRNAs in mediating genomic stability by suppressing the mobility of endogenous retrotransposons.
Tumor suppressor p53 mutations occur in most of human cancers, however, precisely how p53 functions to mediate tumor suppression is not well understood. In this regard, p53 was reported to restrict L1 mobility and suggested that p53 restricts oncogenesis in part by restricting transposon mobility (Wylie et al., 2016). Although normal human p53 suppressed transposons, mutant p53 from cancer patients could not. In contrast, L1 activity was elevated in p53 negative human cancers. Thus, ancestral function of p53 may be associated with transposon control as a guardian of human genome.
Conclusion
L1 has successfully propagated and composed 17% of the human genome, resulting in evolutionary force. Activation of the normally silent L1 is associated with a high level of cancer-associated DNA damage and genomic instability. Indeed, L1 insertions into the human genome may cause cancers through insertional mutagenesis. In fact, recent high-throughput sequencing-based approaches could identify numerous somatic tumor-specific L1 insertions in a variety of cancers (Iskow et al., 2010; Lee et al., 2012; Solyom et al., 2012; Shukla et al., 2013; Helman et al., 2014), however there is no sufficient evidence. Therefore, it should clarify the role of L1-mediated retrotransposition in human cancers. Indeed, the implication of L1 insertion events as either passenger or driver mutations with a causative role in tumorigenesis still remains to be clarified (Rodić and Burns, 2013). Intriguingly, somatic insertions were only identified in epithelial tumors (Lee et al., 2012). Accordingly, epithelial cells can be transformed to cancer stem cells (Wang et al., 2013) and metastasis is more prevalent in epithelial tumors (Gotzmann et al., 2004). Thus, epithelial cells seem to be plastic (Carreira et al., 2014). Cancer stem cells are defined as rare cells with indefinite potential for self-renewal that drive tumorigenesis (Reya et al., 2001). However, it remains to be clarified the role of L1 mobility in cancer stem cells. Recent studies focus on the relationship among L1 mobility, reprogramming, and differentiation. Indeed, reprogramming somatic cells into iPS cells activates L1 mobility (Wissing et al., 2012; Friedli et al., 2014; Klawitter et al., 2016). On the other hand, L1 mobility is enhanced in tumor cells. In this regard, the elevation of L1 protein or RNA expression levels may be useful as a diagnostic hallmark of many human cancers and as a tumor specific marker, metastasis, and prognosis. Furthermore, recent advances in single cell analysis will be useful for comparison of the L1 mobility and the integration site of L1 at a single cell level in human cancers.
Finally, tumor suppressor proteins may be associated with transposon control to restrict deleterious retrotransposition as a guardian of the human genome. Wild-type p53 suppresses transposon mobility in normal cells, while mutant p53 in cancer cells could not, resulting in the activation of L1 mobility in cancer cells (Wylie et al., 2016). Furthermore, recent studies identified somatic L1 insertion in tumor suppressor genes, such as APC, PTEN, NF1, and Rb (Miki et al., 1992; Wimmer et al., 2011; Helman et al., 2014; Rodríguez-Martín et al., 2016). Thus, L1 insertions in the tumor suppressor genes may disrupt their functions and be associated with tumorigenesis. Altogether, host cells have evolved several defense mechanisms protecting cells against retrotransposition.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Funding
This work was supported by a Lateral Research from the Japan Society for the Promotion of Science (JSPS), by the Research Program on Hepatitis from Japan Agency for Medical Research and Development, AMED, and by Takeda Science Foundation.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I thank Ms. Kazumi Tsuruhara for secretarial assistance and her kind encouragement.
References
Aravin, A. A., Hannon, G. J., and Brennecke, J. (2007a). The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318, 761–764. doi: 10.1126/science.1146484
Aravin, A. A., Sachidanandam, R., Girard, A., Fejes-Toth, K., and Hannon, G. J. (2007b). Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744–747. doi: 10.1126/science.1142612
Arias, J. F., Koyama, T., Kinomoto, M., and Tokunaga, K. (2012). Retroelements versus APOBEC3 family members: no great escape from the magnificent seven. Front. Microbiol. 3:275. doi: 10.3389/fmicb.2012.00275
Arjan-Odedra, S., Swanson, C. M., Sheree, N. M., Wolinsky, S. M., and Malim, M. H. (2012). Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication if exogenous retroviruses. Retrovirology 9:53. doi: 10.1186/1742-4690-9-53
Armanios, M., Chen, J. L., Chang, Y. P., Brodsky, R. A., Hawkins, A., Griffin, C. A., et al. (2005). Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenital. Proc. Natl. Acad. Sci. U.S.A. 102, 15960–15964. doi: 10.1073/pnas.0508124102
Asch, H. L., Eliacin, E., Fanning, T. G., Connolly, J. L., Bratthauer, G., and Asch, B. B. (1996). Comparative expression of the LINE-1 p40 protein in human breast carcinomas and normal breast tissues. Oncol. Res. 8, 239–247.
Aschacher, T., Wolf, B., Enzmann, F., Kienzl, P., Messner, B., Sampl, S., et al. (2016). LINE-1 induces hTERT and ensures telomere maintenance in tumour cell lines. Oncogene 35, 94–104. doi: 10.1038/onc.2015.65
Athanikar, J. N., Badge, R. M., and Moran, J. V. (2004). A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 32, 3846–3855. doi: 10.1093/nar/gkh698
Baba, Y., Murata, A., Watanabe, M., and Baba, H. (2014). Clinical implications of the LINE-1 methylation levels in patients with gastrointestinal cancer. Surg. Today 44, 1807–1816. doi: 10.1007/s00595-013-0763-6
Baillie, J. K., Barrett, M. W., Upton, K. R., Gerhardt, D. J., Richmond, T. A., De Sapio, F., et al. (2011). Somatic retrotransposition alters the genetic landscape of the human brain. Nature 479, 534–537. doi: 10.1038/nature10531
Beck, C. R., Collier, P., Macfariane, C., Malig, M., Kidd, J. M., Eichler, E. E., et al. (2010). LINE-1 retrotransposition activity in human genomes. Cell 141, 1159–1170. doi: 10.1016/j.cell.2010.05.021
Beck, C. R., Garcia-Perez, J. L., Badge, R. M., and Moran, J. V. (2011). LINE-1 elements in structural variation and disease. Ann. Rev. Genomics Hum. Genet. 12, 187–215. doi: 10.1146/annurev-genom-082509-141802
Becker, K. G., Swergold, G. D., Ozato, K., and Thayer, R. E. (1993). Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum. Mol. Genet. 2, 1697–1702. doi: 10.1093/hmg/2.10.1697
Belancio, V. P., Hedges, D. J., and Deininger, P. (2008). Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 18, 343–358. doi: 10.1101/gr.5558208
Bogerd, H. P., Wiegrand, H. L., Hulme, A. E., Garcua-Perez, J. L., O'Shea, K. S., and Moran, J. V. (2006). Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 103, 8780–8785. doi: 10.1073/pnas.0603313103
Bratthauer, G. L., Cardiff, R. D., and Fanning, T. G. (1994). Expression of LINE-1 retrotransposons in human breast cancer. Cancer 73, 2333–2336.
Bratthauer, G. L., and Fanning, T. G. (1992). Active LINE-1 retrotransposons in human testicular cancer. Oncogene 7, 507–510.
Brouha, B., Schustak, J., Badge, R. M., Lutz-Prigge, S., Farley, A. H., Moran, J. V., et al. (2003). Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. U.S.A. 100, 5280–5285. doi: 10.1073/pnas.0831042100
Bulliard, Y., Turelli, P., Röhrig, U. F., Zoete, V., Mangeat, B., Michielin, O., et al. (2009). Functional analysis and structural modeling of human APOBEC3G reveal the role of evolutionarily conserved elements in the inhibition of human immunodeficiency virus type 1 infection and Alu transposition. J. Virol. 83, 12611–12621. doi: 10.1128/JVI.01491-09
Bundo, M., Toyoshima, M., Okada, Y., Akamatsu, W., Ueda, J., Nemoto-Miyauchi, T., et al. (2014). Increased l1 retrotransposition in the neuronal genome in schizophrenia. Neuron 81, 306–313. doi: 10.1016/j.neuron.2013.10.053
Burden, A. F., Manley, N. C., Clark, A. D., Gartler, S. M., Laird, C. D., and Hansen, R. S. (2005). Hemimethylation and non-CpG methylation levels in a promoter region of human LINE-1 (L1) repeated elements. J. Biol. Chem. 280, 14413–14419. doi: 10.1074/jbc.M413836200
Burdick, R., Smith, J. L., Chaipan, C., Friew, Y., Chen, J., Venkatachari, N. J., et al. (2010). P-body-associated protein Mov10 inhibits HIV-1 replication at multiple stages. J. Virol. 84, 10241–10253. doi: 10.1128/JVI.00585-10
Carreira, P. E., Richardson, S. R., and Faulkner, G. J. (2014). L1 retrotransposons, cancer stem cells and oncogenesis. FEBS J. 281, 63–73. doi: 10.1111/febs.12601
Castro-Diaz, N., Ecco, G., Coluccio, A., Kapopoulou, A., Yazdanpanah, B., Friedli, M., et al. (2014). Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev. 28, 1397–1409. doi: 10.1101/gad.241661.114
Chen, H., Lilley, C. E., Yu, Q., Lee, D. V., Chou, J., Narvaiza, I., et al. (2006). APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 16, 480–485. doi: 10.1016/j.cub.2006.01.031
Chen, L., Dahlstrom, J. E., Chandra, A., Board, P., and Rangasamy, D. (2012). Prognostic value of LINE-1 retrotransposon expression and its subcellular localization in breast cancer. Breast Cancer Res. Treat. 136, 129–142. doi: 10.1007/s10549-012-2246-7
Chiu, Y. L., Witkowska, H. E., Hall, S. C., Santiago, M., Soros, V. B., Esnault, C., et al. (2006). High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 103, 15588–15593. doi: 10.1073/pnas.0604524103
Ciccia, A., and Elledge, S. J. (2010). The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204. doi: 10.1016/j.molcel.2010.09.019
Cook, P. R., Jones, C. E., and Furano, A. V. (2015). Phosphorylation of ORF1p is required for L1 retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 112, 4298–4303. doi: 10.1073/pnas.1416869112
Cordaux, R., and Batzer, M. A. (2009). The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 10, 691–703. doi: 10.1038/nrg2640
Cost, G. J., and Boeke, J. D. (1998). Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry 37, 18081–18093. doi: 10.1021/bi981858s
Coufal, N. G., Garcia-Perez, J. L., Peng, G. E., Marchetto, M. C., Muotri, A. R., Mu, Y., et al. (2011). Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc. Natl. Acad. Sci. U.S.A. 108, 20382–20387. doi: 10.1073/pnas.1100273108
DeBerardinis, R. J., Goodier, J. L., Ostertag, E. M., and Kazazian, H. H. Jr. (1998). Rapid amplification of a retrotransposon subfamily is evolving the mouse genome. Nat. Genet. 20, 288–290. doi: 10.1038/3104
De Fazio, S., Bartonicek, N., Di Giacomo, M., Abreu-Goodger, C., Sankar, A., Funaya, C., et al. (2011). The endonuclease activity of Mili fuels piRNA amplification that silences LINE-1 elements. Nature 480, 259–263. doi: 10.1038/nature10547
Denli, A. M., Narvaiza, I., Kerman, B. E., Pena, M., Benner, C., Marchetto, M. C. N., et al. (2015). Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell 163, 583–593. doi: 10.1016/j.cell.2015.09.025
Dewannieux, M., Esnault, C., and Heidmann, T. (2003). LINE-1-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35, 41–48. doi: 10.1038/ng1223
Ding, D., Lou, X., Hua, D., Yu, W., Li, L., Wang, J., et al. (2012). Recurrent targeted genes of hepatitis B virus in the liver cancer genomes identified by a next-generation sequencing-based approach. PLoS Genet. 8:e1003065. doi: 10.1371/journal.pgen.1003065
Doucet, A. J., Hulme, A. E., Sahinovic, E., Kulpa, D. A., Moldovan, J. B., Kopera, H. C., et al. (2010). Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 6:e1001150. doi: 10.1371/journal.pgen.1001150
Doucet-O'Hare, T. T., Rodić, N., Sharma, R., Darbari, I., Abril, G., Choi, J. A., et al. (2015). LINE-1 expression and retrotransposition in Barrett's esophagus and esophageal carcinoma. Proc. Natl. Acad. Sci. U.S.A. 112, E4894–E4900. doi: 10.1073/pnas.1502474112
Esnault, C., Maestre, J., and Heidmann, T. (2000). Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24, 363–367. doi: 10.1038/74184
Evrony, G. D., Cal, X., Lee, E., Hills, L. B., Elhosary, P. C., Lehmann, H. S., et al. (2012). Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell 151, 483–496. doi: 10.1016/j.cell.2012.09.035
Ewing, A. D., Gacita, A., Wood, L. D., Ma, F., Xing, D., Kim, M. S., et al. (2015). Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res. 25, 1536–1545. doi: 10.1101/gr.196238.115
Feng, Q., Moran, J. V., Kazazian, H. H. Jr., and Boeke, J. D. (1996). Human L1 retrotranspon encodes a conserved endonuclease required for retrotransposition. Cell 87, 905–916. doi: 10.1016/S0092-8674(00)81997-2
Friedli, M., Turelli, P., Kapopoulou, A., Rauwel, B., Castro-Diaz, N., Rowe, H. M., et al. (2014). Loss of transcriptional control over endogenous retroelements during reprogramming to pluripotency. Genome Res. 24, 1251–1259. doi: 10.1101/gr.172809.114
Furtak, V., Mulky, A., Rawlings, S. A., Kozhaya, L., Lee, K., Kewalramani, V. N., et al. (2010). Perturbation of the P-body component Mov10 inhibits HIV-1 infectivity. PLoS ONE 5:e9081. doi: 10.1371/journal.pone.0009081
Gasior, S. L., Roy-Engel, A. M., and Deininger, P. L. (2008). ERCC1/XPF limits L1 retrotransposition. DNA Repair (Amst) 7, 983–989. doi: 10.1016/j.dnarep.2008.02.006
Gasior, S. L., Wakeman, T. P., Xu, B., and Deininger, P. L. (2006). The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 357, 1383–1393. doi: 10.1016/j.jmb.2006.01.089
Gilbert, N., Lutz-Prigge, S., and Moran, J. V. (2002). Genomic deletions created upon LINE-1 retrotransposition. Cell 110, 315–325. doi: 10.1016/S0092-8674(02)00828-0
Goldman, F., Bouarich, R., Kulkarni, S., Freeman, S., Du, H.-Y., Harrington, L., et al. (2005). The effect of TERC haploinsufficiency on the inheritance of telomere length. Proc. Natl. Acad. Sci. U.S.A. 102, 17119–17124. doi: 10.1073/pnas.0505318102
Goodier, J. L., Cheung, L. E., and Kazazian, H. H. Jr. (2012). MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 8:e1002941. doi: 10.1371/journal.pgen.1002941
Goodier, J. L., Cheung, L. E., and Kazazian, H. H. Jr. (2013). Mapping the LINE-1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res. 41, 7401–7419. doi: 10.1093/nar/gkt512
Goodier, J. L., Ostertag, E. M., and Kazazian, H. H. Jr. (2000). Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum. Mol. Genet. 9, 653–657. doi: 10.1093/hmg/9.4.653
Goodier, J. L., Pereira, G. C., Cheung, L. E., Rose, R. J., and Kazazian, H. H. Jr. (2015). The broad-spectrum antiviral protein ZAP restricts human retrotransposition. PLoS Genet. 11:e1005252. doi: 10.1371/journal.pgen.1005252
Goodier, J. L., Zhang, L., Vetter, M. R., and Kazazian, H. H. Jr. (2007). LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 27, 6469–6483. doi: 10.1128/MCB.00332-07
Gotzmann, J., Mikula, M., Eger, A., Schulte-Hermann, R., Foisner, R., Beug, H., et al. (2004). Molecular aspects of epithelial cell plasticity: implications for local tumor invasion and metastasis. Mutat. Res. 566, 9–20. doi: 10.1016/S1383-5742(03)00033-4
Hamdorf, M., Idica, A., Zisoulis, D. G., Gamelin, L., Martin, C., Sanders, K. J., et al. (2015). miR-128 represses L1 retrotransposition by binding directly to L1 RNA. Nat. Struct. Mol. Biol. 22, 824–831. doi: 10.1038/nsmb.3090
Hancks, D. C., Goodier, J. L., Mandal, P. K., Cheung, L. E., and Kazazian, H. H. Jr. (2011). Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum. Mol. Genet. 20, 3386–3400. doi: 10.1093/hmg/ddr245
Hancks, D. C., and Kazazian, H. H. Jr. (2012). Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 22, 191–203. doi: 10.1016/j.gde.2012.02.006
Harper, J. W., and Elledge, S. J. (2007). The DNA damage response: ten years after. Mol. Cell 28, 739–745. doi: 10.1016/j.molcel.2007.11.015
Harris, C. R., Dewan, A., Zupnick, A., Normart, R., Gabriel, A., Prives, C., et al. (2009). p53 responsive elements in human retrotransposons. Oncogene 28, 3857–3865. doi: 10.1038/onc.2009.246
Harris, C. R., Normart, R., Yang, Q., Stevenson, E., Haffty, B. G., Ganesan, S., et al. (2010). Association of nuclear localization of a long interspersed nuclear element-1 protein in breast tumors with poor prognostic outcomes. Genes Cancer 1, 115–124. doi: 10.1177/1947601909360812
Harris, R. S., Bishop, K. N., Sheehy, A. M., Craig, H. M., Petersen-Mahrt, S. K., Watt, I. N., et al. (2003). DNA deamination mediates innate immunity to retroviral infection. Cell 113, 803–809. doi: 10.1016/S0092-8674(03)00423-9
Helman, E., Lawrence, M. S., Stewart, C., Sougnez, C., Getz, G., and Meyerson, M. (2014). Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 24, 1053–1063. doi: 10.1101/gr.163659.113
Holmes, S. E., Dombrowski, B. A., Krebs, C. M., Boehm, C. D., and Kazazian, H. H. Jr. (1994). A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat. Genet. 7, 143–148. doi: 10.1038/ng0694-143
Hrecka, K., Hao, C., Gierszewska, M., Swanson, S. K., Kesik-Brodacka, M., Srivastava, S., et al. (2011). Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661. doi: 10.1038/nature10195
Hu, S., Li, J., Xu, F., Mei, S., Duff, Y. L., Yin, L., et al. (2015). SAMHD1 inhibits LINE-1 retrotransposition by promoting stress granule formation. PLoS Genet. 11:e1005367. doi: 10.1371/journal.pgen.1005367
Hulme, A. E., Bogerd, H. P., Cullen, B. R., and Moran, J. V. (2007). Selective inhibition of Alu retrotransposition by APOBEC3G. Gene 390, 199–205. doi: 10.1016/j.gene.2006.08.032
Iskow, R. C., McCabe, M. T., Mills, R. E., Torene, S., Pittard, W. S., Neuwald, A. F., et al. (2010). Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141, 1253–1261. doi: 10.1016/j.cell.2010.05.020
Jacobs, F. M., Greenberg, D., Nguyen, N., Haeussler, M., Ewing, A. D., Katzman, S., et al. (2014). An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516, 242–245. doi: 10.1038/nature13760
Kazazian, H. H. Jr., Wong, C., Youssoufian, H., Scott, A. F., Philips, D. G., and Antonarakis, S. E. (1988). Haemophilia A resulting from de novo insertion of L1 sequence represents a novel mechanism for mutation in man. Nature 332, 164–166. doi: 10.1038/332164a0
Kines, K. J., Sokolowski, M., deHaro, D. L., Christian, C. M., and Belancio, V. P. (2014). Potential for genomic instability associated with retrotranspositionally-incompetent L1 loci. Nucleic Acids Res. 42, 10488–10502. doi: 10.1093/nar/gku687
Kinomoto, M., Kanno, T., Shimura, M., Ishizaka, Y., Kojima, A., Kurata, T., et al. (2007). ALL APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 35, 2955–2964. doi: 10.1093/nar/gkm181
Klawitter, S., Fuchs, N. V., Upton, K. R., Muñoz-Lopez, M., Shukla, R., Wang, J., et al. (2016). Reprogramming triggers endogenous L1 and Alu retrotransposition in human induced pluripotent stem cells. Nat. Commun. 7:10286. doi: 10.1038/ncomms10286
Ko, M., An, J., Pastor, W. A., Koralov, S. B., Rajewsky, K., and Rao, A. (2015). TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol. Rev. 263, 6–21. doi: 10.1111/imr.12239
Kubo, S., Seleme, M. C., Soifer, H. S., Perez, J. L., Moran, J. V., Kazazian, H. H. Jr., et al. (2006). L1 retrotransposition in nondividing and primary human somatic cells. Proc. Natl. Acad. Sci. U.S.A. 103, 8036–8041. doi: 10.1073/pnas.0601954103
Kuilman, T., Michaloglou, C., Mooi, W. J., and Peeper, D. S. (2010). The essence of senescence. Genes Dev. 24, 2463–2479. doi: 10.1101/gad.1971610
Kuramochi-Miyagawa, S., Watanabe, T., Gotoh, K., Totoki, Y., Toyoda, A., Ikawa, M., et al. (2008). DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fet al testes. Genes Dev. 22, 908–917. doi: 10.1101/gad.1640708
Laguette, N., Sobhian, B., Casartelli, N., Ringeard, M., Chable-Bessia, C., Segeral, E., et al. (2011). SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657. doi: 10.1038/nature10117
Lau, C. C., Sun, T., Ching, A. K., He, M., Li, J. W., Wong, A. M., et al. (2014). Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell 25, 335–349. doi: 10.1016/j.ccr.2014.01.030
Lee, E., Iskow, R., Yang, L., Gokcumen, O., Haseley, P., Luquette, L. J. III, et al. (2012). Landcape of somatic retrotransposition in human cancers. Science 337, 967–971. doi: 10.1126/science.1222077
Li, X., Zhang, J., Jia, R., Cheng, V., Xu, X., Qiao, W., et al. (2013). The MOV10 helicase inhibits LINE-1 mobility. J. Biol. Chem. 288, 21146–21160. doi: 10.1074/jbc.m113.465856
Liang, G., Chan, M. F., Tomigahara, Y., Tsai, Y. C., Gonzales, F. A., Li, E., et al. (2002). Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 22, 480–491. doi: 10.1128/MCB.22.2.480-491.2002
Liu, J., Nau, M. M., Zucman-Rossi, J., Powell, J. I., Allegra, C. J., and Wright, J. J. (1997). LINE-1 element insertion at the t(11;22) translocation breakpoint of a desmoplastic small round cell tumor. Genes Chromosomes Cancer 18, 232–239.
Luan, D. D., Korman, M. H., Jakubczak, J. L., and Eickbush, T. H. (1993). Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72, 595–605. doi: 10.1016/0092-8674(93)90078-5
Mangeat, B., Turelli, P., Caron, G., Friedli, M., Perrin, L., and Trono, D. (2003). Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103. doi: 10.1038/nature01709
Marchetto, M. C. N., Narvaiza, I., Denli, A. M., Benner, C., Lazzarini, T. A., Nathanson, J. L., et al. (2013). Differential L1 regulation in pluripotent stem cells of humans and apes. Nature 503, 525–529. doi: 10.1038/nature12686
Martin, S. L., and Bushman, F. D. (2001). Nucleic acid chaperon activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell Biol. 21, 467–475. doi: 10.1128/MCB.21.2.467-475.2001
Mathias, S. L., Scott, A. F., Kazazian, H. H. Jr., Boeke, J. D., and Gabriel, A. (1991). Reverse transcriptase encoded by a human transposable element. Science 254, 1808–1810. doi: 10.1126/science.1722352
Matsubara, K., and Tokino, T. (1990). Integration of hepatitis B virus DNA and its implications for hepatocarcinogenesis. Mol. Biol. Med. 7, 243–260.
Matsui, T., Leung, D., Miyashita, H., Maksakova, I. A., Miyachi, H., Kimura, H., et al. (2010). Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931. doi: 10.1038/nature08858
Miki, Y., Nishisho, I., Horii, A., Miyoshi, Y., Utsunomiya, J., Kinzler, K. W., et al. (1992). Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 52, 643–645.
Mills, R. E., Bennett, E. A., Iskow, R. C., and Devine, S. E. (2007). Which transposable elements are active in the human genome? Trends Genet. 23, 183–191. doi: 10.1016/j.tig.2007.02.006
Moldovan, J. B., and Moran, J. V. (2015). The zinc-finger antiviral protein ZAP inhibits LINE and Alu retrotransposition. PLoS Genet. 11:e1005121. doi: 10.1371/journal.pgen.1005121
Montoya-Durango, D. E., Liu, Y., Teneng, I., Kalbfleisch, T., Lacy, M. E., Steffen, M. C., et al. (2009). Epigenetic control of mammalian LINE-1 retrotransposition by retinoblastoma proteins. Mutat. Res. 665, 20–28. doi: 10.1016/j.mrfmmm.2009.02.011
Montoya-Durango, D. E., Ramos, K. A., Bojang, P., Ruiz, L., Ramos, I. N., and Ramos, K. S. (2016). LINE-1 silencing by retinoblastoma proteins is effected through the nucleosomal and remodeling deacetylase multiprotein complex. BMC Cancer 16:38. doi: 10.1186/s12885-016-2068-9
Moran, J. V., DeBerardinis, R. J., and Kazazian, H. H. Jr. (1999). Exon shuffling by L1 retrotransposition. Science 283, 1530–1534. doi: 10.1126/science.283.5407.1530
Morrish, T. A., Gilbert, N., Myers, J. S., Vincent, B. J., Stamato, T. D., Taccioli, G. E., et al. (2002). DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 31, 159–165. doi: 10.1038/ng898
Morse, B., Rotherg, P. G., South, V. J., Spandorfer, J. M., and Astrin, S. M. (1988). Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma. Nature 333, 87–90. doi: 10.1038/333087a0
Muckenfuss, H., Hamdorf, M., Held, U., Perkovic, M., Löwer, J., Cichutek, K., et al. (2006). APOBEC3 prteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 281, 22161–22172. doi: 10.1074/jbc.M601716200
Muotri, A. R., Chu, V. T., Marchetto, M. C., Deng, W., Moran, J. V., and Gage, F. H. (2005). Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435, 903–910. doi: 10.1038/nature03663
Muotri, A. R., Marchetto, M. C., Coufal, N. G., Oefner, R., Yeo, G., Nakashima, K., et al. (2010). L1 retrotransposition in neurons is modulated by MeCP2. Nature 468, 443–446. doi: 10.1038/nature09544
Nangia-Makker, P., Sarvis, R., Visscher, D. W., Bailey-Penrod, J., Raz, A., and Sarkar, F. H. (1998). Galectin-3 and L1 retrotransposons in human breast carcinomas. Breast Cancer Res. Treat. 49, 171–183. doi: 10.1023/A:1005913810250
Narita, N., Nishio, H., Kitoh, Y., Ishikawa, Y., Minami, R., Nakamura, H., et al. (1993). Insertion of a 5′ truncated L1 element into the 3′ end of exon 44 of the dystrophin gene resulted in skipping of the exon during splicing in a case of Duchenne muscular dystrophy. J. Clin. Invest. 91, 1862–1867. doi: 10.1172/JCI116402
Niewiadomska, A. M., Tian, C., Tan, L., Wang, T., Sarkis, P. T. N., and Yu, X. F. (2007). Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J. Virol. 81, 9577–9583. doi: 10.1128/JVI.02800-06
Ostertag, E. M., and Kazazian, H. H. Jr. (2001). Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35, 501–538. doi: 10.1146/annurev.genet.35.102401.091032
Pickeral, O. K., Makalowski, W., Boguski, M. S., and Boeke, J. D. (2000). Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 10, 411–415. doi: 10.1101/gr.10.4.411
Puszyk, W., Down, T., Grimwade, D., Chromienne, C., Oakey, R. J., Solomon, E., et al. (2013). The epigenetic regulator PLZF represses L1 retrotransposition in germ and progenitor cells. EMBO J. 32, 1941–1952. doi: 10.1038/emboj.2013.118
Reya, T., Morrison, S. J., Clarke, M. F., and Weissman, I. L. (2001). Stem cells, cancer, and cancer stem cells. Nature 414, 105–111. doi: 10.1038/35102167
Rodić, N., and Burns, K. H. (2013). Long interspersed element-1 (LINE-1): passenger or driver in human neoplasm? PLoS Genet. 9:e1003402. doi: 10.1371/journal.pgen.1003402
Rodić, N., Sharma, R., Sharma, R., Zampella, J., Dai, L., Taylor, M. S., et al. (2014). Long interspersed element-1 protein expression is a hallmark of many human cancers. Am. J. Pathol. 184, 1280–1286. doi: 10.1016/j.ajpath.2014.01.007
Rodić, N., Steranka, J. P., Makohon-Moore, A., Moyer, A., Shen, P., Sharma, R., et al. (2015). Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat. Med. 21, 1060–1064. doi: 10.1038/nm.3919
Rodríguez-Martín, C., Cidre, F., Fernández-Teijeiro, A., Gómez-Mariano, G., de la Vega, L., Ramos, P., et al. (2016). Familial retinoblastoma due to intronic LINE-1 insertion causes aberrant and noncanonical mRNA splicing of the RB1 gene. J. Hum. Genet. 61, 463–466. doi: 10.1038/jhg.2015.173
Rowe, H. M., Jakobsson, J., Mesnard, D., Rougemont, J., Reynard, S., Aktas, T., et al. (2010). KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463, 237–240. doi: 10.1038/nature08674
Schulz, W. A., Steinhoff, C., and Flori, A. R. (2006). Methylation of endogenous human retroelements in health and disease. Curr. Top. Microbiol. Immunol. 310, 211–250. doi: 10.1007/3-540-31181-5_11
Schumann, G. G. (2007). APOBEC3 proteins: major players in intracellular defence against LINE-1-mediated retrotransposition. Biochem. Soc. Trans. 35, 637–642. doi: 10.1042/BST0350637
Scott, E. C., Gardner, E. J., Masood, A., Chuang, N. T., Vertino, P. M., and Devine, S. E. (2016). A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 26, 745–755. doi: 10.1101/gr.201814.115
Sheehy, A. M., Gaddis, N. C., Choi, J. D., and Malim, M. H. (2002). Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650. doi: 10.1038/nature00939
Shi, X., Seluanov, A., and Gorbunova, V. (2007). Cell divisions are required for L1 retrotransposition. Mol. Cell. Biol. 27, 1264–1270. doi: 10.1128/MCB.01888-06
Shiloh, Y. (2014). ATM, expanding roles as a chief guardian of genome stability. Exp. Cell Res. 329, 154–161. doi: 10.1016/j.yexcr.2014.09.002
Shukla, R., Upton, K. R., Munoz-Lopez, M., Gerhardt, D. J., Fisher, M. E., Nguyen, T., et al. (2013). Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 153, 101–111. doi: 10.1016/j.cell.2013.02.032
Soifer, H. S., Zaragoza, A., Peyvan, M., Behlke, M. A., and Rossi, J. J. (2005). A potential role for RNA interference in controlling the activity of the human LINE-1 retrotransposon. Nucleic Acids Res. 33, 846–856. doi: 10.1093/nar/gki223
Solyom, S., Ewing, A. D., Rahrmann, E. P., Doucet, T., Nelson, H. H., Burns, M. B., et al. (2012). Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 22, 2328–2338. doi: 10.1101/gr.145235.112
Stenglein, M. D., and Harris, R. S. (2006). APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 281, 16837–16841. doi: 10.1074/jbc.M602367200
Stetson, D. B., Ko, J. S., Heidmann, T., and Medzhitov, R. (2008). Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598. doi: 10.1016/j.cell.2008.06.032
Suzuki, J., Yamaguchi, K., Kajikawa, M., Ichiyanagi, K., Adachi, N., Koyama, H., et al. (2009). Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS Genet. 5:e1000461. doi: 10.1371/journal.pgen.1000461
Swergold, G. D. (1990). Identification. Characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell. Biol. 10, 6718–6729. doi: 10.1128/MCB.10.12.6718
Symer, D. E., Connelly, C., Szak, S. T., Caputo, E. M., Cost, G. J., Parmigiani, G., et al. (2002). Human L1 retrotransposition is associated with genetic instability in vivo. Cell 110, 327–338. doi: 10.1016/S0092-8674(02)00839-5
Szak, S. T., Pickeral, O. K., Makalowski, W., Boguski, M. S., Landsman, D., and Boeke, J. D. (2002). Molecular archeology of L1 insertions in the human genome. Genome Biol. 3:research0052. doi: 10.1186/gb-2002-3-10-research0052
Tchenio, T., Casella, J. F., and Heidmann, T. (2000). Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 28, 411–415. doi: 10.1093/nar/28.2.411
Trono, D. (2015). Transposable elements, polydactyl proteins, and the genesis of human-specific transcription networks. Cold Spring Harb. Symp. Quant. Biol. 80, 281–288. doi: 10.1101/sqb.2015.80.027573
Tubio, J. C., Li, Y., Ju, Y. S., Martincorena, I., Cooke, S. L., Tojo, M., et al. (2014). Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 345:1251343. doi: 10.1126/science.1251343
Turelli, P., Castro-Diaz, N., Marzetta, F., Kapopoulou, A., Raclot, C., Duc, J., et al. (2014). Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res. 24, 1260–1270. doi: 10.1101/gr.172833.114
Turelli, P., Vianin, S., and Trono, D. (2004). The innate antiretroviral factor APOBEC3G does not affect human LINE-1 retrotransposition in a cell culture assay. J. Biol. Chem. 279, 43371–43373. doi: 10.1074/jbc.C400334200
Upton, K. R., Gerhardt, D. J., Jesuadian, J. S., Richardson, S. R., Sánchez-Luque, F. J., Bodea, G. O., et al. (2015). Ubiquitous L1 mosaicism in hippocampal neurons. Cell 161, 228–239. doi: 10.1016/j.cell.2015.03.026
Van Meter, M., Kashyap, M., Rezazadeh, S., Geneva, A. J., Morello, T. D., Seluanov, A., et al. (2014). SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 5, 5011. doi: 10.1038/ncomms6011
Wallace, N. A., Gasior, S. L., Faber, Z. J., Howie, H. L., Deininger, P. L., and Galloway, D. A. (2013). HPV 5 and 8 E6 expression reduces ATM protein levels and attenuates LINE-1 retrotransposition. Virology 443, 69–79. doi: 10.1016/j.virol.2013.04.022
Wang, X., Han, Y., Dang, Y., Fu, W., Zhou, T., Ptak, R. G., et al. (2010). Moloney leukemia virus 10 (MOV10) protein inhibits retrovirus replication. J. Biol. Chem. 285, 14346–14355. doi: 10.1074/jbc.M110.109314
Wang, Z. A., Mitrofanova, A., Bergren, S. K., Abate-Shen, C., Cardiff, R. D., Califano, A., et al. (2013). Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat. Cell Biol. 15, 274–283. doi: 10.1038/ncb2697
Wimmer, K., Callens, T., Wernstedt, A., and Messiaen, L. (2011). The NF1 gene contains hotspots for L1 endonuclease-dependent de novo insertion. PLoS Genet. 7:e1002371. doi: 10.1371/journal.pgen.1002371
Wissing, S., Muñoz-Lopez, M., Macia, A., Yang, Z., Montano, M., Collins, W., et al. (2012). Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Hum. Mol. Genet. 21, 208–218. doi: 10.1093/hmg/ddr455
Wolf, D., and Goff, S. P. (2007). TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131, 46–57. doi: 10.1016/j.cell.2007.07.026
Wolf, D., and Goff, S. P. (2009). Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458, 1201–1204. doi: 10.1038/nature07844
Woodcock, D. M., Lawler, C. B., Linsenmeyer, M. E., Doherty, J. P., and Warren, W. D. (1997). Asymmetric methylation in the hypermethylated CpG promoter region of the human L1 retrotransposon. J. Biol. Chem. 272, 7810–7816. doi: 10.1074/jbc.272.12.7810
Wylie, A., Jones, A. E., D'Brot, A., Lu, W. J., Kurtz, P., Moran, J. V., et al. (2016). P53 genes function to restrain mobile elements. Genes Dev. 30, 64–77. doi: 10.1101/gad.266098.115
Xie, Y., Mates, L., Ivics, Z., Izsvák, Z., Martin, S. L., and An, W. (2013). Cell division promotes efficient retrotransposition in a stable L1 reporter cell line. Mob. DNA 4:10. doi: 10.1186/1759-8753-4-10
Yang, N., and Kazazian, H. H. Jr. (2006). L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 9, 763–771. doi: 10.1038/nsmb1141
Yang, N., Zhang, L., Zhang, Y., and Kazazian, H. H. Jr. (2003). An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 31, 4929–4940. doi: 10.1093/nar/gkg663
Yu, F., Zingler, N., Schumann, G., and Strätling, W. H. (2001). Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res. 29, 4493–4501. doi: 10.1093/nar/29.21.4493
Yu, Q., Carbone, C. J., Katlinskaya, Y. V., Zheng, H., Zheng, K., Luo, M., et al. (2015). Type I interferon controls propagation of long interspersed element-1. J. Biol. Chem. 290, 10191–10199. doi: 10.1074/jbc.M114.612374
Zhang, A., Dong, B., Doucet, A., Moldovan, J. B., Moran, J. V., and Silverman, R. H. (2014). RNase L restricts the mobility of engineered retrotransposons in cultured human cells. Nucleic Acids Res. 42, 3803–3820. doi: 10.1093/nar/gkt1308
Zhang, H., Yang, B., Pomerantz, R. J., Zhang, C., Arunachalam, S. C., and Gao, L. (2003). The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424, 94–98. doi: 10.1038/nature01707
Keywords: LINE-1, retrotransposition, DNA double-strand breaks (DSBs), DNA repair, tumor suppressor, HBV, epigenetic regulation, somatic insertion
Citation: Ariumi Y (2016) Guardian of the Human Genome: Host Defense Mechanisms against LINE-1 Retrotransposition. Front. Chem. 4:28. doi: 10.3389/fchem.2016.00028
Received: 14 February 2016; Accepted: 14 June 2016;
Published: 28 June 2016.
Edited by:
Tammy A. Morrish, Formerly affiliated with University of Toledo, USAReviewed by:
Geoff Faulkner, Mater Medical Research Institute, AustraliaNemanja Rodic, Yale University, USA
Copyright © 2016 Ariumi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasuo Ariumi, ariumi@kumamoto-u.ac.jp
 Yasuo Ariumi
Yasuo Ariumi