Identification of CHMP7 as a promising immunobiomarker for immunotherapy and chemotherapy and impact on prognosis of colorectal cancer patients
- 1Department of the General Surgery, The Second Hospital of Jilin University, Changchun, China
- 2Department of the Ridiotherapy, The Second Hospital of Jilin University, Changchun, China
Introduction: ESCRT is a molecular machine involved in various important physiological processes, such as the formation of multivesicular bodies, cellular autophagy, and cellular membrane repair. CHMP7 is a regulatory subunit of ESCRT-III and is necessary for the proper functioning of ESCRT. In this study, public databases were exploited to explore the role of CHMP7 in tumors.
Methods: The research on CHMP7 in oncology is rather limited. In this study, the differential expression of CHMP7 in multiple tumor tissues was analyzed with information from public databases and clinically collected colorectal cancer tissue samples. Subsequently, the mutational landscape of CHMP7, methylation levels, and the relationship between its expression levels and genomic instability were resolved. The immune microenvironment is a compelling emerging star in tumor research. The correlation of CHMP7 with various infiltrating immune cell types in TME was analyzed by online datasets and single-cell sequencing. In terms of clinical treatment, the impact of CHMP7 expression levels on chemotherapy and immunotherapy and the evaluation of small molecule drugs related to CHMP7 were assessed.
Results: CHMP7 has a predictive value for the prognosis of patients with tumors and is highly involved in tumor immunity. The downregulation of CHMP7 may lead to genomic instability. A strong correlation between CHMP7 and TME immune cell infiltration has been observed, participating in the formation of suppressive TME and promoting tumor progression. The expression level of CHMP7 is significantly lower in the non-responder group of multiple chemotherapeutic agents. CHMP7 can potentially serve as a new biomarker for predicting the efficacy of tumor chemotherapy and immunotherapy.
Conclusion: As a gene of interest, CHMP7 is expected to provide novel and promising targets for further treatment of patients with tumor.
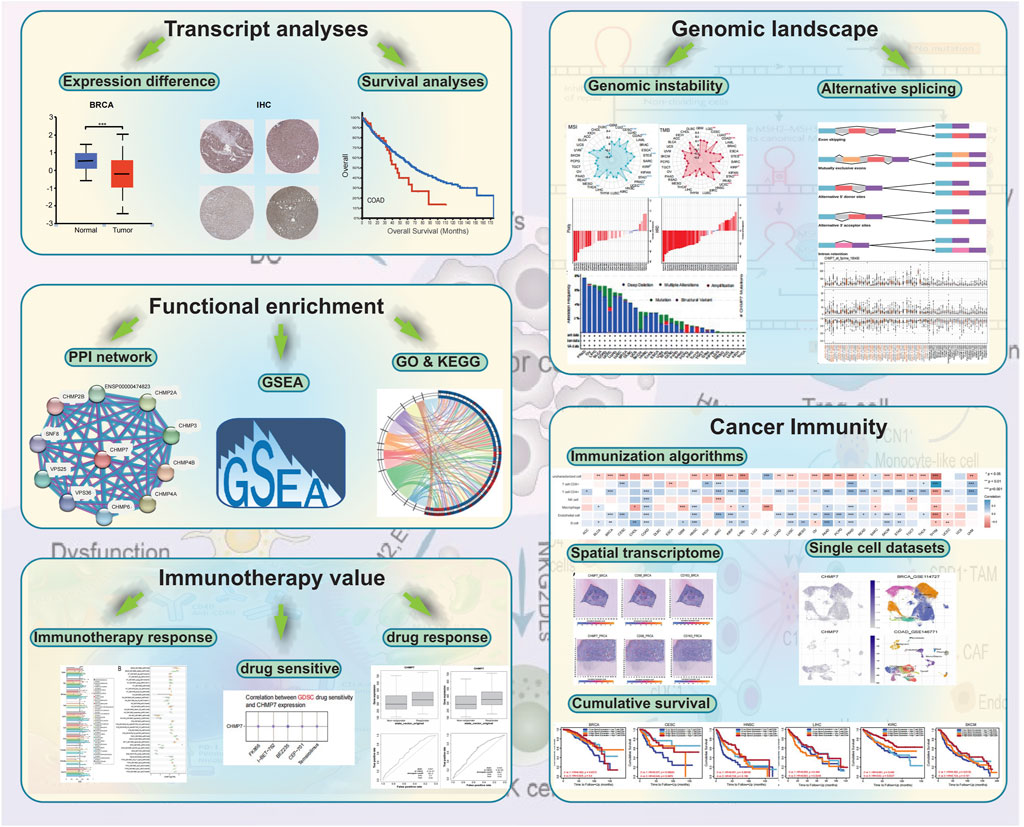
GRAPHICAL ABSTRACT | The main points of this study. The role of CHMP7 in COAD was analyzed in terms of differential expression, mutational analysis, functional enrichment analysis, and immune infiltration.
Introduction
The formation of malignant tumors is an extremely complicated process that typically takes decades. Tissue cells in normal individuals evolve and progressively develop into tumor cells with a malignant phenotype, a process called tumor progression, usually accompanied by multiple genetic alterations (Hausman, 2019). Tumor progression is attributed to the accumulation of random mutations and epigenetic alterations in DNA sequences that affect the proliferation of malignant cells associated with gene regulatory networks and other traits associated with the malignant phenotype (Vogelstein et al., 2013; Iranzo et al., 2018). Tumorigenesis is usually the result of the synergistic effect of multiple risk factors, including environmental chemical factors (e.g., atmospheric pollutants), physical factors (e.g., ionizing radiation), viral infections, adverse dietary habits, and pharmaceutical effects (Lewandowska et al., 2019; Sun et al., 2020). Current treatment modalities for malignancies include surgery, chemotherapy, targeted drug therapy, radiation therapy, hormone therapy, stem cell transplantation, and tumor immunotherapy. However, various genomic mutations and epigenetic modifications occur in tumor cell DNA during progression, leading to the emergence of malignant phenotypes, including abnormal metabolism, treatment resistance, unrestricted division, and weakened intercellular adhesion, subsequently limiting the effectiveness of various therapeutic modalities and affecting the prognosis of patient survival. Tumor heterogeneity affects the sensitivity of different patients with the same tumor to chemotherapeutic agents, radiation therapy, and targeted drugs (Lim and Ma, 2019). In addition to tumor cells, there are several kinds of infiltrating cells in tumor tissue, such as cancer-associated fibroblasts (CAFs), B cells, T cells, macrophages, and other immune cells, adipocytes, and endothelial cells of blood vessels, which together with the tumor extracellular matrix (ECM) constitute the tumor microenvironment (TME). Different immune cells may perform different roles in tumorigenesis by inhibiting or promoting tumorigenesis (Lee and Cheah, 2019; Bilotta et al., 2022). The tumor progression is frequently characterized by mutations in multiple genes, and the development of high-throughput sequencing technology has become a practical approach to unraveling the mystery of cancer genes.
CHMP7 (Charged multivesicular body protein 7) is essential in properly regulating the ESCRT (endosomal sorting complex required for transport). The ESCRT system is an integral molecular mechanism responsible for membrane protein sorting and membrane repair in eukaryotic cells, and it participates in several physiological processes, such as cell division and autophagy. Ubiquitin-tagged membrane proteins are primarily transported to the endosomal membrane through cytokinesis and then invaginated by the ESCRT system, which releases membrane components containing these proteins into the endosomal lumen to form intraluminal vesicles (ILVs). Subsequently, the ILVs and their membrane proteins are degraded through fusion with lysosomes, and the protein-bound ESCRT complex proteins are degraded and recycled (Korbei, 2022). For example, the epidermal growth factor receptor (EGFR) is degraded by this pathway. In addition to its involvement in the degradation of ubiquitinated proteins, the ESCRT system is also involved in the sorting and delivery of extracellular vehicles (EVs) (Wei et al., 2021) and in the repair of cell membranes to maintain cell integrity and normal function. It is also a vital component of membrane proteins in cellular life activities such as cell division and autophagy (Vietri et al., 2020; Ritter et al., 2022). All these physiological processes are involved in topological membrane remodeling. ESCRT-III is the key player in completing the critical step of the shearing of the budding body (Gatta and Carlton, 2019; Isono, 2021). The dysregulation of ESCRT function is highly related to tumor development, and CHMP7, an important regulatory subunit of ESCRT-III, has attracted our attention.
Aberrant expression of CHMP7 may lead to nuclear pore complex damage and TDP-43 dysfunction in amyotrophic lateral sclerosis/frontotemporal dementia (Coyne et al., 2021). Neurodegenerative diseases, such as Huntington’s disease and Parkinson’s disease, are often associated with the accumulation of intracellular ubiquitinated protein aggregates, with lesions that may be associated with the loss of ESCRT function (Coyne and Rothstein, 2022). Regarding oncology research, Ritter et al. discovered that ESCRT-mediated cell membrane repair mechanisms contribute to the immune escape of cancer cells from lethal attacks by cytotoxic T lymphocytes (CTL). Furthermore, inhibiting the ESCRT pathway significantly improves the killing efficiency of CTL (Ritter et al., 2022). Abnormal expression of several genes in the ESCRT system is also assumed to be associated with tumors. VPS4A is significantly overexpressed in liver cancer tissues and can promote tumor growth and invasion by affecting the sorting and delivery of exosomes (Han et al., 2019). However, conclusive studies on the role of CHMP7 in tumors are limited.
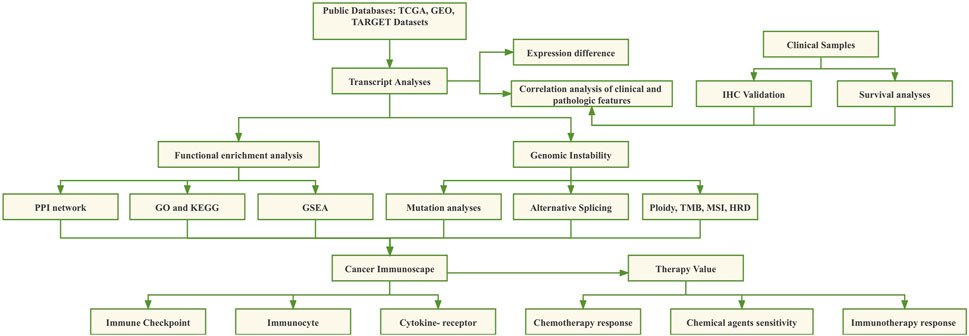
In this study, the differential expression of CHMP7 in tumor tissues and its prognostic impact were systematically analyzed with information from public databases such as TCGA, CCLE, and GTEX. Immunohistochemical (IHC) validation was performed on clinically collected colorectal cancer tissue samples. Subsequently, the mutational landscape of CHMP7, methylation levels, and the relationship between its expression levels and genomic instability were resolved. The co-expression of CHMP7 in various infiltrating immune cell types in TME was verified by an online dataset and single-cell sequencing analysis. The influence of CHMP7 on chemotherapy and immunotherapy was evaluated to predict the effect of immunotherapy and sensitive drugs against CHMP7 in these cancers (Figure 1).
Methods
Data source
The unified and standardized pan-cancer dataset was downloaded from the UCSC (https://xenabrowser.net/) database, Therapeutically Applicable Research to Generate Effective Treatments (TARGET; includes 7 pediatric cancers), and Genotype-Tissue Expression (GTEx; covers 54 normal tissues). Those samples with fewer than three tumor types were excluded from the analysis. Single-cell sequencing datasets were obtained from the Gene Expression Omnibus (GEO) database. They included breast invasive carcinoma (BRCA, GSE114727), colon adenocarcinoma (COAD, GSE146771), skin cutaneous melanoma (SKCM, GSE48190), liver hepatocellular carcinoma (LIHC, GSE98638), nasopharyngeal carcinoma (NPC, GSE150430), and kidney renal clear cell carcinoma (KIRC, GSE 111360).
Differential expression and prognostic analysis
Diseases associated with CHMP7 were analyzed with the Open Target website (https://www.opentargets.org/) and represented in bubble charts. The sample information obtained from UCSC was analyzed with R software (version 3.6.4) to calculate the difference in CHMP7 expression between normal and tumor samples in each tumor, and the significance of differences was performed using the unpaired Wilcoxon rank sum and signed rank tests. Protein level data were obtained from the UALCAN database (http://ualcan.path.uab.edu/) and further utilized to reveal the differential expression of CHMP7 in normal and tumors (Patra et al., 2021). The UALCAN database also facilitated further analysis of the correlation between CHMP7 expression levels and tumor stage (Patra et al., 2023). The SURVIVAL package from the R software was applied to analyze the association between CHMP7 expression and pan-cancer survival with risk tables.
IHC validation
IHC staining analysis was performed on fifty specimens of postoperative paraffin slides that underwent surgical treatment at the Department of Colorectal Surgery, Second Hospital of Jilin University, from January 2017 to December 2017, and whose postoperative pathology was confirmed as colorectal cancer. The experiment was approved by the ethics committee. The antibody was obtained from Proteintech under item number 16424-1-AP for IHC staining. Paraffin slides were stained by the Servicebio company. The slides were dried and observed with a light microscope, and two professional pathologists were invited to determine the intensity of the staining. Relevant clinical information of patients was collected and followed up to determine whether there was any significant difference in the effect of different CHMP7 expression intensities on the prognosis of patients.
Mutational analysis of CHMP7 and correlation with genomic instability
cBioPortal (https://www.cbioportal.org/) is a website that enables researchers to explore, visualize, and analyze multidimensional cancer genomic data. Using this website, we can easily explore genetic alterations in different tumor types, genes, and pathways. We queried the mutation characteristics of CHMP7 in different tumor types and further explored the mutation types of the genes. The prognostic impact of CHMP7 mutations on patients with tumors was evaluated through the “Comparison” module of the website.
To further analyze the correlation between CHMP7 and genomic stability, the R software was used to compute the correlation between tumor mutation burden (TMB), microsatellite instability (MSI), homologous recombination deficiency (HRD), and neoantigen (NEO) data for each tumor and CHMP7 expression levels. A heat map demonstrated the correlation between CHMP7 expression levels and five mismatch repair genes (MLH1, MSH2, MSH6, PMS2, and EPCAM) in each tumor type (Levy et al., 2017).
Analyses of tumor stemness and epigenetic modifications
The Stemness index is an indicator to assess the similarity of tumor cells to stem cells, which is associated with active biological processes in stem cells and a more advanced degree of tumor dedifferentiation. Wiznerowicz et al. constructed predictive models for multipotential stem cell samples from the PCBC dataset with the one-class logistic regression (OCLR) machine learning algorithm. The predictive model was subsequently applied to a training set such as TCGA to calculate the stemness score for each sample (Malta et al., 2018). The tumor stemness index calculated by mRNA expression and methylation signature was obtained from previous studies, and the stemness index and gene expression data of the samples and their correlation performed were further integrated (Kocaturk et al., 2019).
Homologous recombination repair (HRR), as one of the core DNA damage repair methods, is a DNA repair mechanism that maintains genome integrity to ensure high-fidelity transmission of genetic information. Mutation of related HRR genes or methylation of gene promoters can trigger HRR dysfunction, leading to genomic instability. Tumor cells tend to exploit HRR to save cells from apoptosis. HRR is a complex signaling pathway involving multiple steps, in which the most critical genes are BRCA1 and BRCA2, and other related genes include MLH1, MSH2, ATM, and TP53. The HRR-related genes analyzed in the present study were cited in the research of Liang et al. (Liang et al., 2022). The correlation between CHMP7 and HRR signature can be analyzed by applying the GEPIA2.0 online website (Ledermann et al., 2016). Heat maps were applied to visualize the correlation between CHMP7 and 44 RNA-modified genes.
Alternative splicing (AS) analysis
AS refers to the process of mRNA precursor to mature mRNA, in which various splicing methods can allow the same gene to produce multiple different mature mRNAs, resulting in the translation of different proteins (Ule and Blencowe, 2019). AS is a major mechanism for maintaining protein diversity (Li et al., 2017). It produces specific shear isoforms in certain tissues or conditions at different stages of development. The OncoSplicing website (http://www.oncosplicing.com/) was employed to explore SplAdder and SliceSeq projects that contain AS events for CHMP7. Differences in percent spliced-in (PSI) and AS events were further compared between TCGA tumor tissues and GTEx normal tissues. We also explored the impact of AS events of CHMP7 on patient prognosis in diverse tumors and confirmed shear isoforms of CHMP7 in pan-cancer.
Functional enrichment analysis
The protein-protein interaction (PPI) network of CHMP7 was explored using the STRING website (https://www.string-db.org/). Tumor development involves the aberrant activation of multiple critical pathways. Genes in the corresponding pathways were collected from TCGA and analyzed using the GSVA package of R software. The correlation between CHMP7 and pathway scores was analyzed using Spearman correlation analysis. The similar gene detection function in the GEPIA2.0 website helped us obtain genes that were the top 100 co-expressed with CHMP7 in tumor tissues and performed gene ontology (GO) functional analysis of the top 100 genes with the clusterProfiler package. Gene set enrichment analysis (GSEA) further explored the functional enrichment-related pathways CHMP7 may affect.
Immune infiltration analysis
The role of immune cells in TME has attracted much attention in recent years (Padma et al., 2023). Different immune cells perform various roles in tumorigenesis. The immune cells of tumor types are frequently quantified in studies of tumorigenesis, treatment, and other mechanisms. The correlation scores between CHMP7 and tumor tissue immune infiltration levels were calculated with the ESTIMATE package of R software, and the bar graphs present the stromal, immune, and ESTIMATE scores for each tumor (Yoshihara et al., 2013). The correlation between CHMP7 and immune checkpoints is also shown in a heat map.
The TISDB online website (http://cis.hku.hk/TISIDB/) was utilized to analyze the differential expression of CHMP7 in distinct immune subtypes of pan-cancerous tissues (Thorsson et al., 2018). The site was also exploited to construct heat maps exhibiting the correlation between CHMP7 and chemokines, chemokine receptors, immunostimulatory factors, and immunoinhibitory factors.
M2 macrophages are regarded to serve in pathogen clearance, anti-inflammatory response, and tumor progression (Biswas and Mantovani, 2010). The TIMER2.0 website was applied to analyze the relationship between CHMP7 and M2 macrophages according to different algorithms. Additionally, spatial transcripts in SpatialDB (https://www.spatialomics.org/SpatialDB/) were subjected to analysis of the overlapping levels of CHMP7 expression and spatial expression of M2 macrophage markers (CD68 and CD163). As for the TISCH website (http://tisch.comp-genomics.org/), single-cell data from multiple tumors were presented to compare the expression of CHMP7 in various cell subtypes of tumor tissues.
CTLs are one of the critical immune surveillance cells. A high abundance of CTL with a killing function in tumor tissues is a promising prognostic indicator, and increasing the proportion of CTL in patients’ tumor tissues can help inhibit tumor progression and eventual elimination (Mami-Chouaib et al., 2018). Multiple algorithms are available on the TIMER2.0 website to analyze the correlation between CHMP7 and CTL and the impact of CHMP7 expression levels and CTL infiltration levels on patient prognosis. Tumor immune dysfunction and exclusion (TIDE, a web tool) also enabled us to investigate the role of CHMP7 in T cell dysfunction and CTL-related prognosis in tumor tissues.
Drug sensitivity analysis
To elucidate the predictive value of CHMP7 in tumor immunotherapy, we explored it from public databases. The predictive role of CHMP7 as a new biomarker was compared with classical markers such as TMB at the TIDE website to calculate the predictive role of CHMP7. The TIDE website further enabled us to predict the therapeutic response of CHMP7 in the core dataset, immunotherapy dataset, CRISPR screening dataset, and mechanistic follow-up experiments of immunosuppressive cell types. And the differential expression of CHMP7 was compared between the responding and non-responding groups in 30 immunotherapy cohorts such as IMvigor210.
The ROC Plotter dataset (http://www.rocplot.org/site/index) was adopted to analyze the gene expression levels in the response and non-response groups of multiple chemotherapeutic agents. The RNAactDrug database (hrbmu.edu.cn) can facilitate our queries on the association between CHMP7 and drug sensitivity (Xie et al., 2022).
Statistical analysis
The overall survival (OS) differences between the high and low gene expression groups were explored using the log-rank test. The correlation coefficients were quantified using Spearman or Pearson. All data analyses included in this article, such as differential expression, gene interaction, immune infiltration, and drug sensitivity analyses, were considered significant only at p < 0.05.
Results
CHMP7 is aberrantly expressed in multiple tumor tissues and correlates with patient prognosis
The Open Target website presents diseases associated with CHMP7, and the bubble chart illustrates that CHMP7 may be associated with multiple tumors (Figure 2A). The differential expression of CHMP7 in tumor tissues and corresponding normal tissues were analyzed with TCGA, TARGET, and GTEx databases. The results indicated that CHMP7 expression levels were significantly upregulated in GBM, LGG, BRCA, KIPAN, STAD, HNSC, SKCM, PAAD, LAML, and CHOL tumor tissues, while markedly downregulated in ESCA, STES, KIRP, COAD, PRAD, LUSC, BLCA, THCA, READ, OV, TGCT, UCS, ACC, and KICH tumor tissues (Figure 2B). The UALCAN website analyzed the differences in CHMP7 protein levels, revealing that CHMP7 protein was downregulated in BRCA, COAD, and UCEC tumor tissues and upregulated in LUAD tumor tissues (Figure 2C). The expression level of CHMP7 is related to the stage of many tumors. For example, the more advanced the stage in COAD, the lower the CHMP7 expression level (Figure 2D). The TCGA data were employed to analyze the predictive value of CHMP7 on prognosis and demonstrated that low CHMP7 expression was associated with poorer OS in BRCA, ESCA, HNSC, KIRC, SARC, and SKCM, while in LAML, patients in the high CHMP7 expression group had a worse prognosis (Figure 2E).
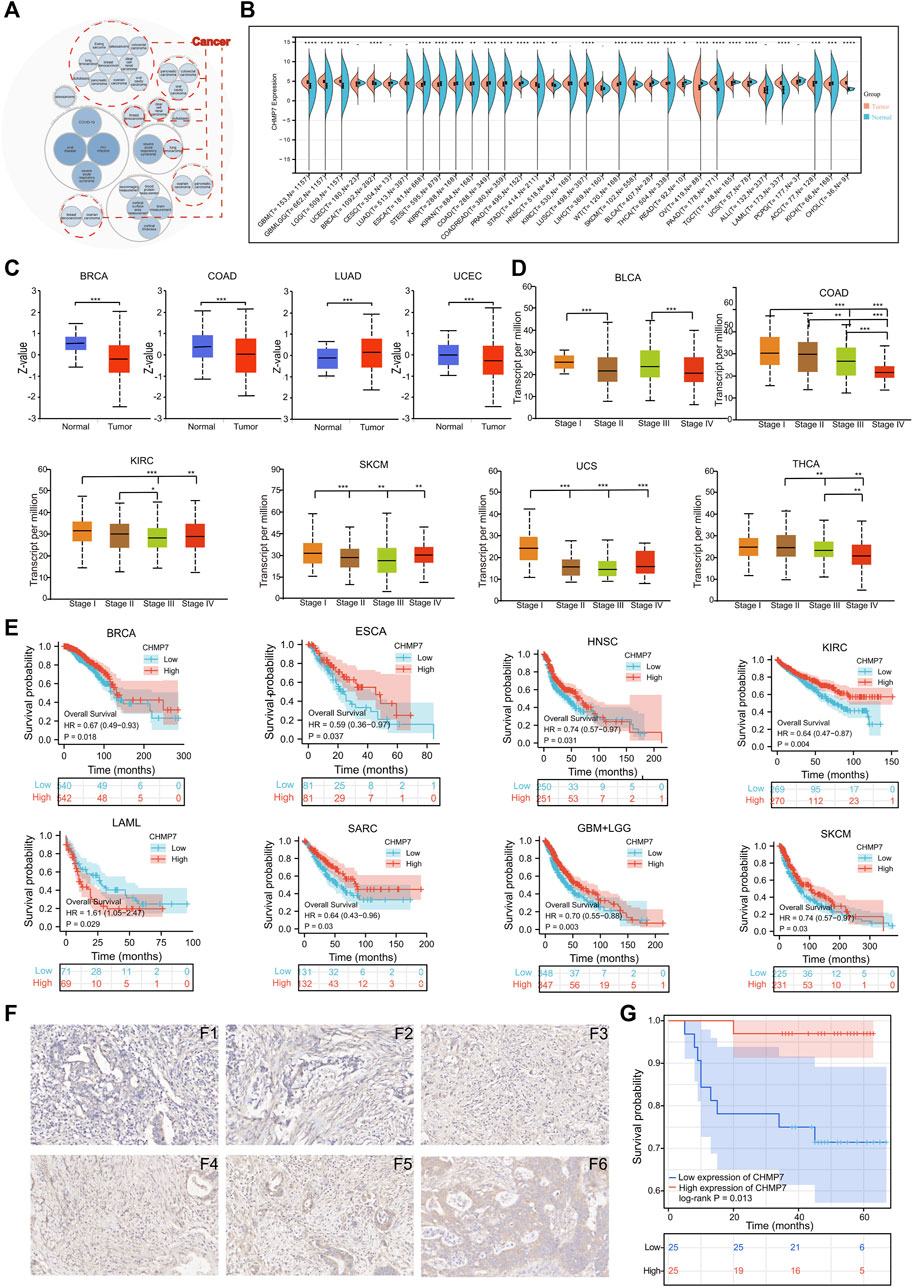
FIGURE 2. Differential expression and prognostic analysis. (A) Diseases associated with CHMP7 were analyzed with the openTarget website. And tumors that may be associated with CHMP7 are marked with red dotted lines. (B) Analysis of the TCGA, TARGET and GTEx databases for differential expression of CHMP7 in tumor tissues and corresponding normal tissues. (C) The UALCAN website facilitates the analysis of differential expression of CHMP7 in different tumor stages. (D) CHMP7 expression levels of multiple tumors at different stages were further analyzed on UALCAN. (E) Kaplan-Meier curves were plotted for the analysis of CHMP7 expression in relation to patient prognosis. (F) IHC staining maps of COAD tissues. F1-F3 is the low CHMP7 expression group and F4-F6 is the high group with a magnification of ×40. (G) The OS of COAD patients that performed IHC staining. (*: p < 0.05, **: p < 0.01 and ***: p < 0.001).
The IHC staining was performed on the collected 50 colorectal cancer paraffin slides. According to the staining intensity, specimens were divided into CHMP7 low expression group (Figures 2F1–3) and CHMP7 high expression group (Figures 2F4–6). The relevant information was collected into a baseline clinical information table (Table 1), and the collected data were processed and analyzed using R software. The results indicated that CHMP7 expression level was not significantly correlated with patients’ age, weight, gender, concomitant disease status (hypertension and diabetes), and T-stage, and the group with low CHMP7 expression had advanced N-stage and TNM-stage. The prognostic impact of CHMP7 on patients was analyzed by Kaplan-Meier survival analysis, which revealed that patients with low CHMP7 expression had a relatively poor prognosis (Figure 2G).
CHMP7 is associated with various pathways, such as angiogenesis and apoptosis
To explore the possible role of CHMP7 in tumor development in detail, the enrichment pathways of CHMP7 and its related genes were analyzed. The PPI network for CHMP7 was constructed with the STRING website (Figure 3A). The tumor data from the TCGA database were exploited to analyze the correlations between CHMP7 and multiple pathways, and the results were represented as scatter plots. CHMP7 and angiogenesis, apoptosis, inflammatory response, EMT markers, tumor proliferation signature, and tumor inflammation signature all demonstrated a significant positive correlation (Figure 3B).
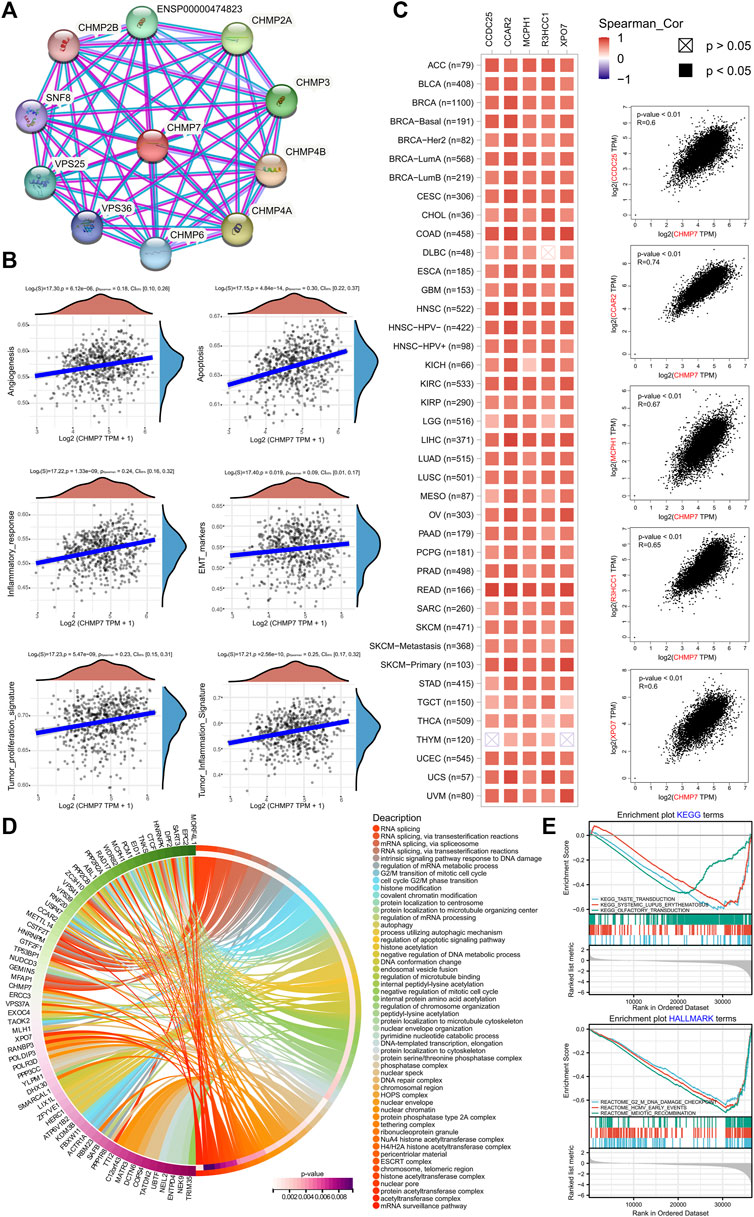
FIGURE 3. CHMP7 functional enrichment analysis. (A) The String site reveals proteins interacting with CHMP7. (B) CHMP7 expression levels correlate with the angiogenesis, apoptosis, inflammatory response, EMT markers, tumor proliferation signature, and tumor inflammation signature pathway. (C) The correlation between CHMP7 and the top 5 co-expressed genes identified on GEPIA2.0. Each cancer type is shown on the left, and all cancer samples are on the right. (D) GO enrichment analysis circle plot of the top 100 CHMP7 co-expressed genes identified on GEPIA2.0. (E) Enrichment plot of GSEA analysis of CHMP7 and its associated genes KEGG and HALLMARK terms.
The top 100 genes tightly associated with CHMP7 were explored via the GEPIA website, and heat maps were drawn to present the top 5 genes of relevance (Figure 3C; Supplementary Material S1). The genes of CHMP7 and its related top hundred were subjected to GO and KEGG enrichment analysis, revealing a strong association with RNA splicing, ESCRT complex, and DNA damage checkpoint (Figures 3D, E).
CHMP7 is altered in multiple tumor tissues and associated with genomic instability
Mutation analysis of CHMP7 in the TCGA database was performed on the cBioPortal website. The results reveal that the top five tumors with alterations are PRAD, OV, LIHC, BLCA, and COAD, with the main mutation type being deletion. The main loci of mutations are shown in Figure 4A. To further analyze the effect of CHMP7 mutations on patient prognosis, COAD and LUSC patients in the CHMP7-altered group had poorer disease-free survival (DFS; Figure 4B).
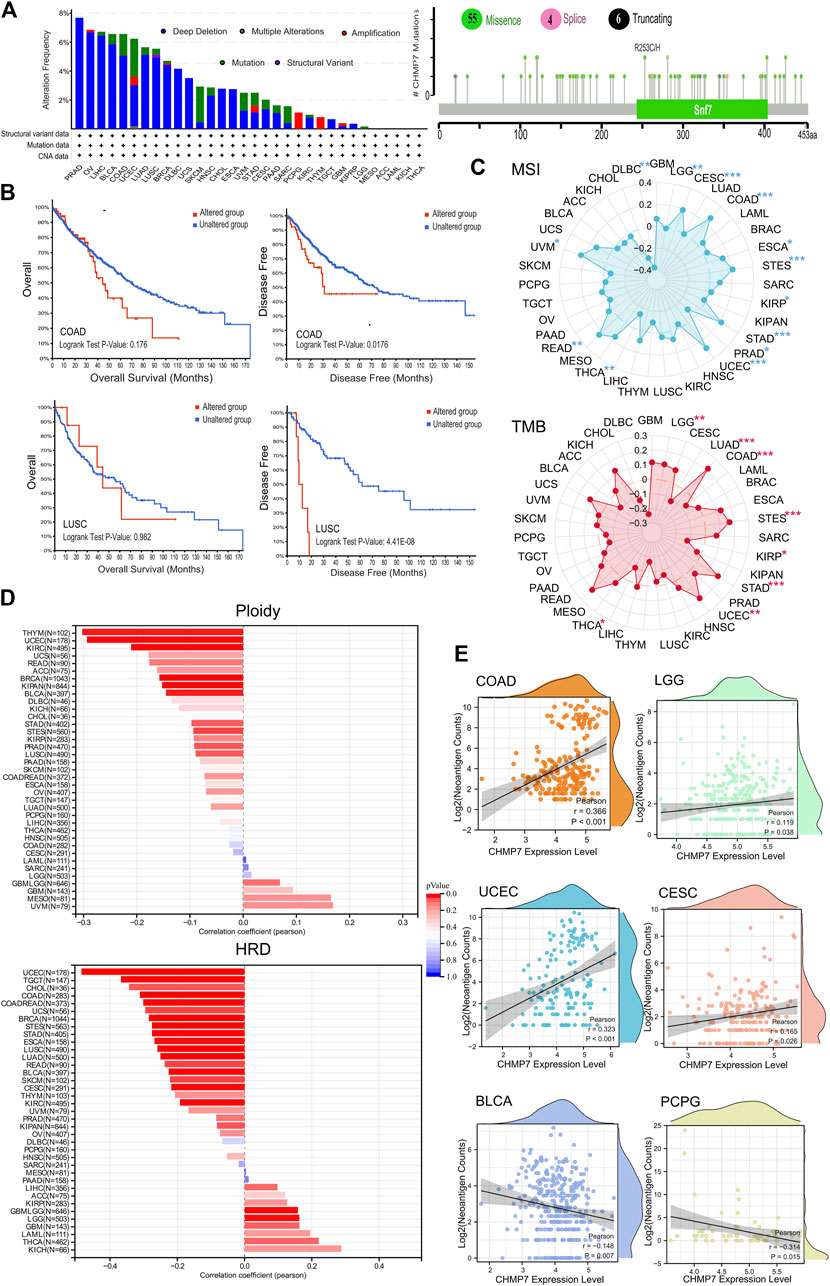
FIGURE 4. CHMP7 is correlated with genomic instability in tumor tissues. (A) Genomic alterations of CHMP7 in TCGA pan-cancer and the loci at which missense mutations, frameshift deletions and splicing occur. (B) Impact of CHMP7 mutations on patient prognosis, with poorer prognosis in the mutated group of COAD and LUSC patients. (C) Radar plot showing the association between CHMP7 expression levels and TMB (top) and MSI (bottom) in pan-cancer; correlation coefficients of 0 are indicated by dashed circles, points inside the dashed circles indicate negative correlation coefficients, those outside the circles indicate positive correlation coefficients, and * indicates the significance of the difference. (D) Correlation coefficients between CHMP7 expression levels and Ploidy or HRD. (E) Association between CHMP7 expression and neoantigen counts. The waves at the top and right side represent the distribution density of CHMP7 and neoantigen counts. (*: p < 0.05, **: p < 0.01 and ***: p < 0.001).
TMB and MSI are important clinical biomarkers that can be effectively predicted for tumor treatment (Baretti and Le, 2018; Zhang et al., 2022). Radar plots present the correlation between CHMP7 and TMB, and MSI in different tumor tissues (Figure 4C). CHMP7 is positively correlated with TMB in five tumors (LGG, COAD, STES, STAD, and UCEC) and significantly negatively correlated with TMB in three tumors (LUAD, KIRP, and THCA). As for MSI, CHMP7 is significantly positively correlated in CESC, COAD, ESCA, STES, STAD, UCEC, READ, and UVM and negatively correlated in PRAD, THCA, and DLBC.
Ploidy is tightly associated with chromosomal instability in tumor development (Müller et al., 2021). The bar chart demonstrates the correlation of CHMP7 with ploidy, which is significantly negatively correlated in nine types of tumors (BRCA, STES, KIPAN, PRAD, UCEC, KIRC, LUSC, THYM, and BLCA). CHMP7 is significantly negatively correlated with HRD in CESC, LUAD, COAD, BRCA, ESCA, STES, KIPAN, STAD, UCEC, KIRC, LUSC, READ, TGCT, SKCM, UCS, BLCA, and CHOL. It presents a positive correlation in LGG (Figure 4D). NEO is abundantly expressed in tumor cells with strong immunogenicity and tumor heterogeneity, making it an attractive target for tumor immunotherapy (Blass and Ott, 2021). Scatter plots indicate a significant positive correlation between CHMP7 and NEO in COAD, LGG, UCEC, and CESC, while a negative correlation in BLCA and PCPG (Figure 4E). All analyses suggest that CHMP7 is altered in various tumor tissues and associated with genomic instability.
CHMP7 is associated with gene repair in pan-cancerous tissues
The stability of the genome relies on the combined action of multiple repair mechanisms, such as MMR and HRR. The diagonal heat map demonstrates the correlation between CHMP7 and MMR-related genes (MLH1, MSH2, MSH6, PMS2, and EPCAM), exhibiting positive correlation in various tumor tissues, including ACC, BRCA, and KIRC (Figure 5A). The correlation between CHMP7 and cancer stemness was analyzed based on six tumor stemness indices calculated by mRNA expression and methylation signature, which were significantly positively correlated in HNSC and negatively correlated in LUAD, COAD, COADREAD, BRCA, THYM, PCPG, and BLCA (Figure 5B; Supplementary Material S2). A positive correlation between CHMP7 and HRR was found in multiple tumor tissues, indicating that CHMP7 may be associated with DNA repair (Figure 5C).
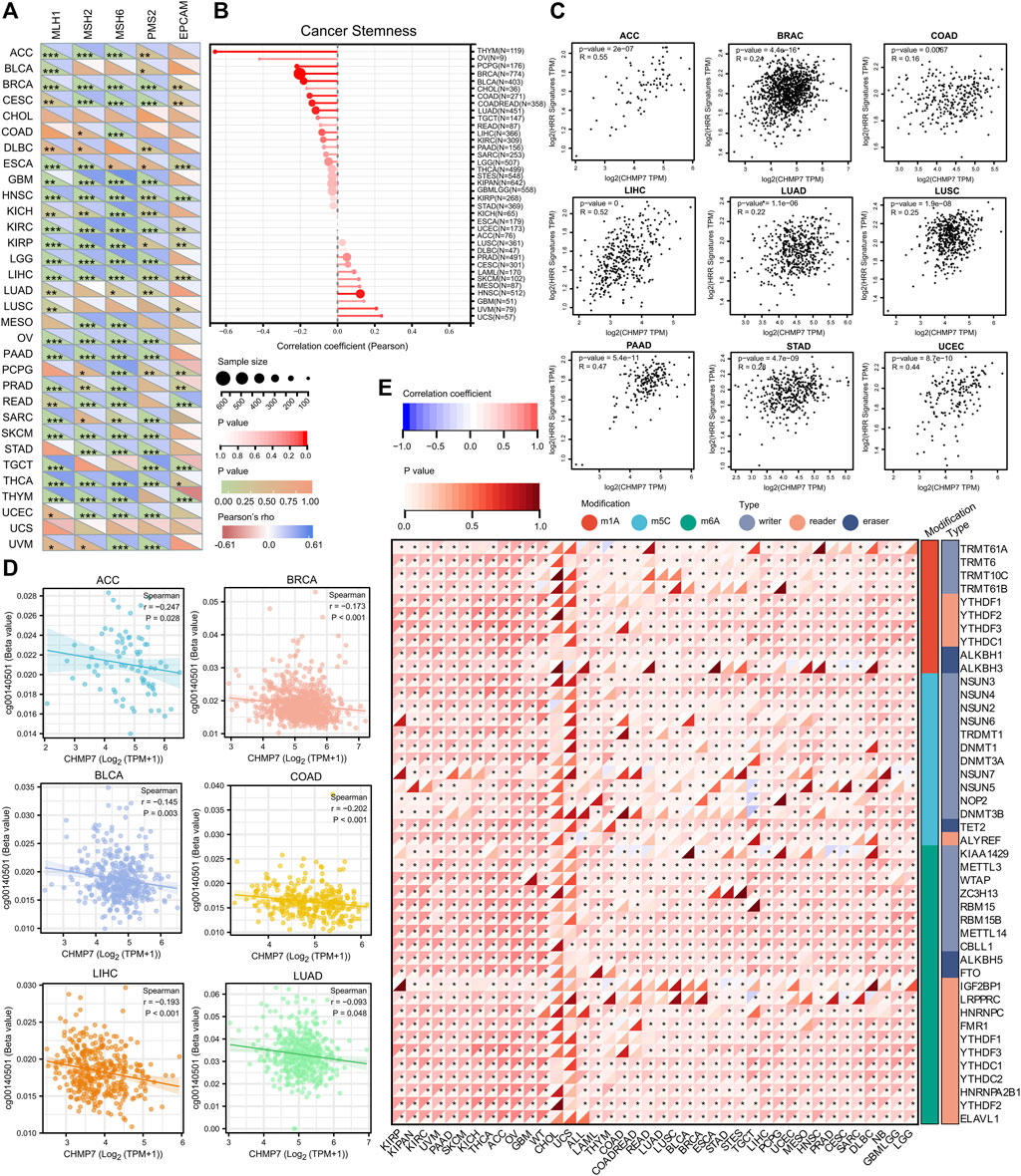
FIGURE 5. CHMP7 is involved in DNA mismatch repair, cancer stemness and epigenetic regulation. (A) Heat map demonstrating the correlation between CHMP7 and 5 MMR genes (MLH1, MSH2, MSH6, PMS2, EPCAM) in pan-cancer. (B) Lollipop plot reveals the association between CHMP7 expression levels and cancer stemness. The dot size indicates the number of samples and the color indicates the p-value. (C) Scatter plot depicting the correlation between CHMP7 expression levels and HRR characteristics. (D) CHMP7 DNA methylation was significantly negatively correlated with probe cg00140501 in ACC, BRCA, BLCA, COAD, LIHC and LUAD. (E) The correlation between CHMP7 expression levels and RNA regulation. (*: p < 0.05, **: p < 0.01 and ***: p < 0.001).
DNAss is a cancer stemness score based on the methylation profile calculated for each tumor and further analyzed for CHMP7 DNA methylation in the TCGA database (Malta et al., 2018). A significant negative correlation was observed between CHMP7 and DNA methylation probe cg00140501 in several tumor tissues of TCGA, including ACC, BRCA, BLCA, COAD, LIHC, and LUAD (Figure 5D). In addition to methylation analysis, the correlation between CHMP7 and RNA-modified genes was analyzed, revealing a heat map showing a positive correlation between CHMP7 and major RNA-modified genes (Figure 5E).
AS events in CHMP7 can contribute to predicting patient prognosis
AS is an essential mechanism for regulating gene expression and generating proteomic diversity, which has the potential to serve as a new biomarker in oncology and provide many new targets for drug development (Blencowe, 2006). The AS events for CHMP7 were evaluated at the OncoSplicing website, and alt_5prime_195490 is presented in Figure 6A and the rest in Supplementary Material S3. A higher PSI was observed in BRCA, CHOL, KIRP, and READ tumors. The difference in PSI between tumor tissues and adjacent/normal tissues was compared, and CHMP7_alt_5prime_195490 exhibited higher PSI in various tumor tissues, such as BRCA and STAD (Figure 6B). The predictive value of PSI values on the prognosis of patients with tumors was analyzed. The results suggested that high PSI values were associated with poorer OS in ESCA, COAD, HNSC, and UVM and poorer DFI in LIHC, SKCM, and TGCT (Figure 6C). Shear isoforms of CHMP7 in pan-cancer were also demonstrated (Figure 6D). All these results indicate that AS events of CHMP7 are critical in tumor research.
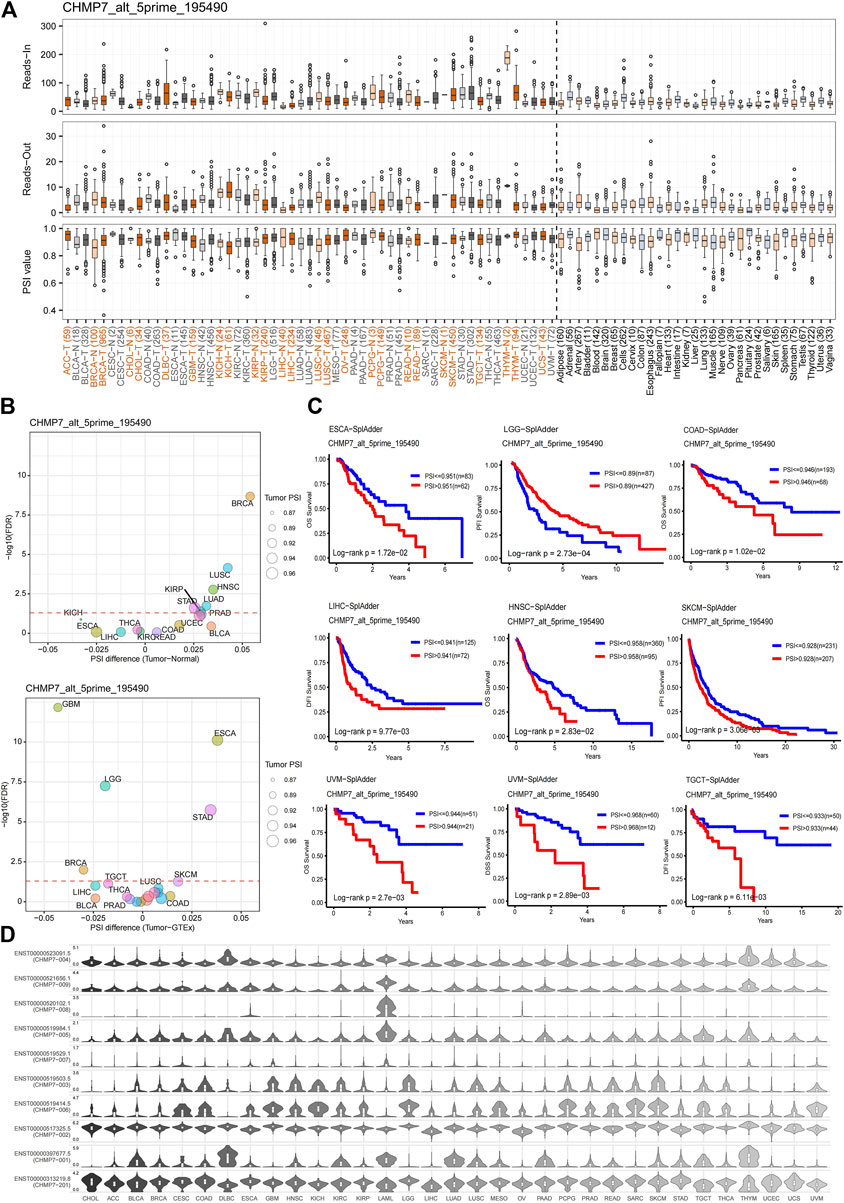
FIGURE 6. Alternative splicing analysis of CHMP7. (A) The read-in, read-out, and PSI values of CHMP7 _alt_5prime in tumor and normal tissues. (B) Differences in PSI value between tumor and adjacent normal tissue (top), and tumor and GTEx normal tissue (bottom); red dashed line refers to FDR of 0.05. (C) Kaplan-Meier curves present the effect of PSI values of CHMP7 _alt_5prime on patient OS, DSS, DFI, and PFI. (D) Isoform switch events of the CHMP7 gene in pan-cancer.
CHMP7 is engaged in tumor immune infiltration and regulation
To investigate the relationship between CHMP7 and immune infiltration in the tumor TME, we calculated the stromal and immune scores of tumor samples based on CHMP7 expression data using the ESTIMATE package of R software. The two scores were summed to obtain the ESTIMATE score, which can be used to estimate tumor purity. Significant correlation between CHMP7 expression and immune infiltration was observed in 20 cancer species, 12 of which were significantly positively correlated, including TCGA-BRCA, TCGA-STES, TCGA-KIPAN, TCGA-COAD, TCGA-COADREAD, TCGA-PRAD, TCGA-STAD, TCGA-HNSC, TCGA -KIRC, TCGA-BLCA, TCGA-PAAD, and TCGA-LAML. There were eight significant negative correlations, such as TCGA-GBM, TCGA-GBMLGG, TCGA-LGG, TCGA-SARC, TARGET-WT, TCGA-THCA, TARGET-NB, and TCGA-ACC (Figure 7A). CHMP7 was significantly positively correlated with most immune checkpoints and immunoregulatory genes in pan-cancerous tissues. Notably, CHMP7 was negatively correlated with most immune checkpoints and immunoregulatory genes in THYM (Figure 7B; Supplementary Material S4).
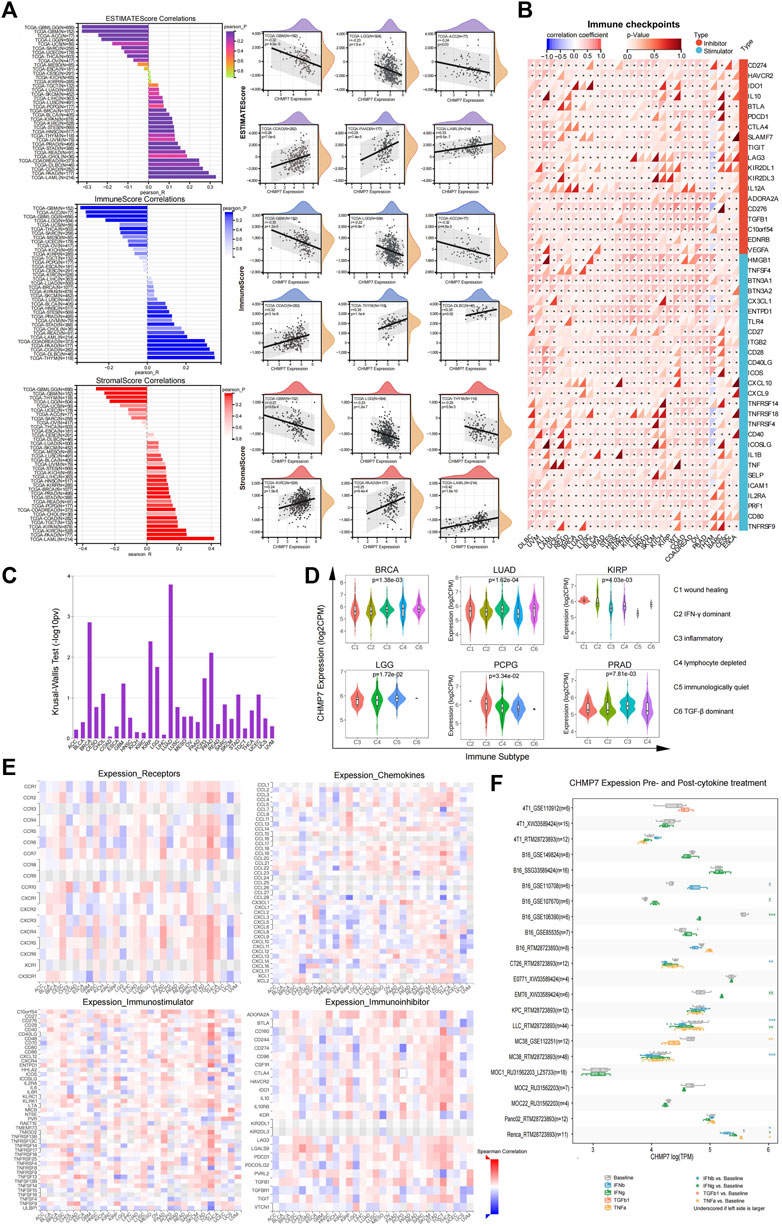
FIGURE 7. CHMP7 is correlated with immune infiltrates and immune checkpoints. (A) Bar graph displaying the correlation between CHMP7 and ESTIMATEScore, ImmuneScore and StromalScore immune infiltration scores, and scatter plot presenting the top 6 correlated cancers for each immune infiltration score. (B) Heat map of the association between immune checkpoint and CHMP7 expression levels. (C) The association between CHMP7 and immune subtypes was obtained by TSIDB online tool. (D) CHMP7 expression levels were correlated with immune subtypes. (E) Heat map of correlation between CHMP7 expression and receptors (top left), chemokines (bottom left), immunostimulatory factors (top right) immunosuppressive factors (bottom right). (F) Box line plot of changes in CHMP7 expression levels in tumor cell lines before and after cytokine treatment. (*: p < 0.05, **: p < 0.01 and ***: p < 0.001).
The TISIDB enabled us to investigate the differential expression of CHMP7 in different immune subtypes. The results revealed that CHMP7 expression was significantly elevated in the C4 subtype in BRCA, KIRP, and LGG, indicating that CHMP7 may be associated with lymphocyte function (Figures 7C, D). Heat maps demonstrate the correlation of CHMP7 with cytokines, cytokine receptors, immunostimulatory factors, and immunosuppressive factors (Figure 7E). The TISMO website was utilized to compare the changes in CHMP7 expression levels in tumor cell lines in both pre- and post-cytokine treatment (Figure 7F). CHMP7 expression levels increased in several cell lines after IFN-β treatment, and the same was observed in several cell lines treated with IFN-γ. The results suggest that the downregulation of our CHMP7 expression levels may lead to the suppression of immune checkpoint function, which is associated with suppressive TME.
CHMP7 is associated with M2 macrophage infiltration
The relationship between CHMP7 and immune cell infiltration in TME was analyzed using the EPIC algorithm (Sturm et al., 2019), and seven immune cells associated with CHMP7 expression were obtained (Figure 8A). The results indicate that CHMP7 negatively correlates with CD8+ T cells in CESC, KICH, KIRC, PRAD, THCA, THYM, and UVM. The negative correlation of CHMP7 with tumor-associated macrophages has been observed in BLCA, COAD, HNSC, LAML, PAAD, and PRAD, while the opposite has been observed in CHOL, GBM, KIRP, LIHC, and THYM. The relationship between CHMP7 and immune cells in COAD is shown in scatter plots (Figure 8B).
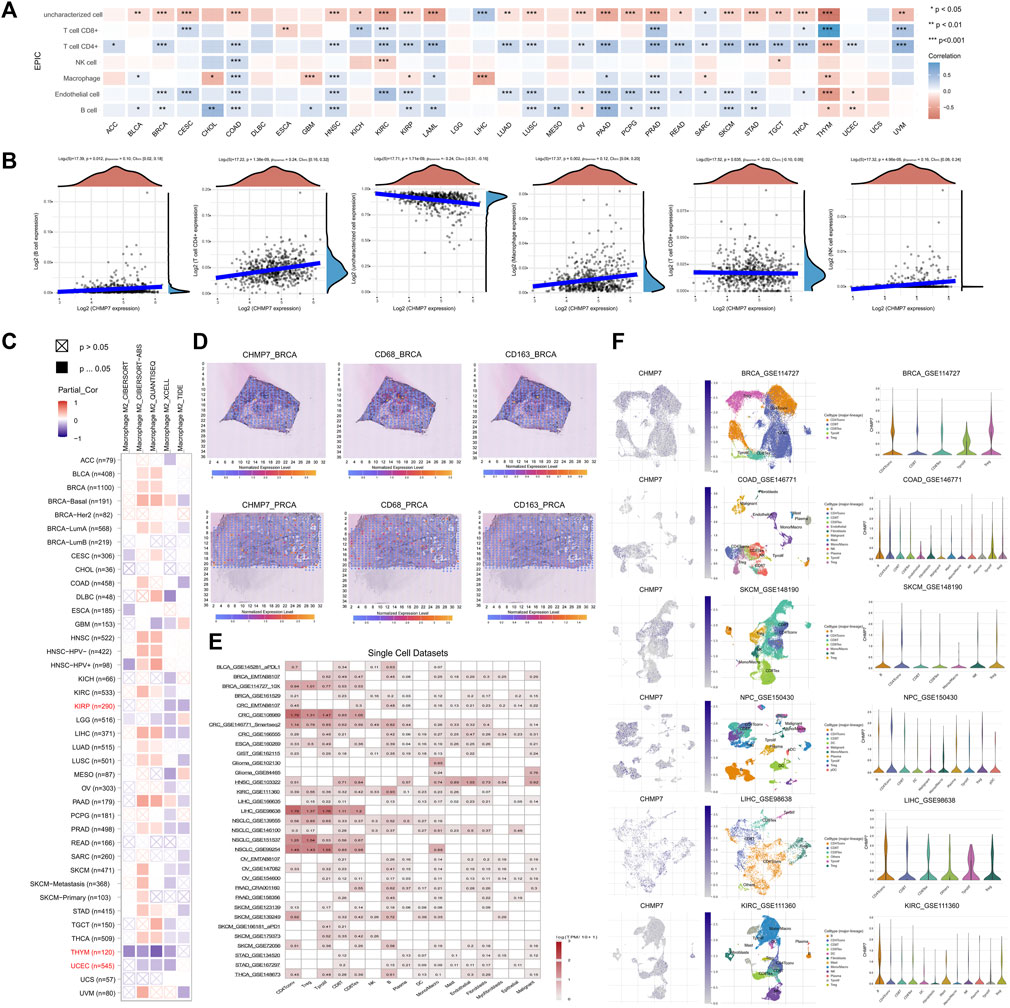
FIGURE 8. CHMP7 is associated with tumor tissue infiltration of immune cells. (A) EPIC algorithm for calculating CHMP7 correlation with immune cell infiltration. (B) Scatter plots representing CHMP7 correlation with B cell, CD4+ T cell, CD8+ T cell, uncharacterized cell, macrophage and NK cell infiltration. (C) Correlation of CHMP7 with M2 macrophage infiltration was calculated by TIMER2.0 with several algorithms. (D) Spatial transcriptional sections illustrated the spatial expression of CHMP7 in BRCA and PRCA tissues and CD68 and CD163 markers. (E) Expression of CHMP7 in cancer monocyte clusters correlates with immune cell infiltration. (F) Expression of CHMP7 in different immune cells in single cell clusters of multiple tumor cells. (*: p < 0.05, **: p < 0.01 and ***: p < 0.001).
M2 macrophages can secrete suppressive cytokines such as TGF-β and IL-10, producing an immunosuppressive TME that promotes tumor progression (Genin et al., 2015). The TIMER2.0 website facilitated us to analyze the correlation between CHMP7 and M2 macrophage infiltration levels with multiple algorithms, and consistent correlations were observed in KIRP, THYM, and UCEC, suggesting that downregulation of CHMP7 expression may be associated with M2 macrophage infiltration (Figure 8C). The correlation between CHMP7 and M2 macrophage markers (CD68, CD163) was analyzed at the spatial transcriptional level utilizing the SpatialDB, and CHMP7 and M2 macrophage biomarkers presented a similar spatial distribution (Figure 8D).
Subsequently, we investigated the correlation between CHMP7 and immune cell infiltration from the single-cell level. The relationship between CHMP7 and immune cell infiltration was investigated with the help of the TISCH (Figure 8E). The correlation between CHMP7 expression and immune cells in BRCA, COAD, SKCM, NPC, LIHC, and KIRC was analyzed, and there were differences in the expression levels of CHMP7 in various infiltrating immune cells (Figure 8F).
CHMP7 is associated with CTL dysfunction
In addition to M2 macrophages, the relevance of CHMP7 to CTL has also attracted our attention. Similar to M2 macrophages, we analyzed the correlation between CHMP7 and CTL with multiple immune infiltration algorithms in TIMER2.0 and expressed consistent significant positive correlations in BLCA, BRCA-Basal, BRCA-LumA, PAAD, PRAD, SARC, SKCM, STAD, THCA, and UVM (Figure 9A). Combined CHMP7 expression and CTL infiltration levels were analyzed for their predictive value for patient prognosis. The results reveal a poorer prognosis in the CHMP7 low expression and CTL low infiltration groups in BRCA, CSC, HNSC, LIHC, KIRC, and SKCM (Figure 9B).
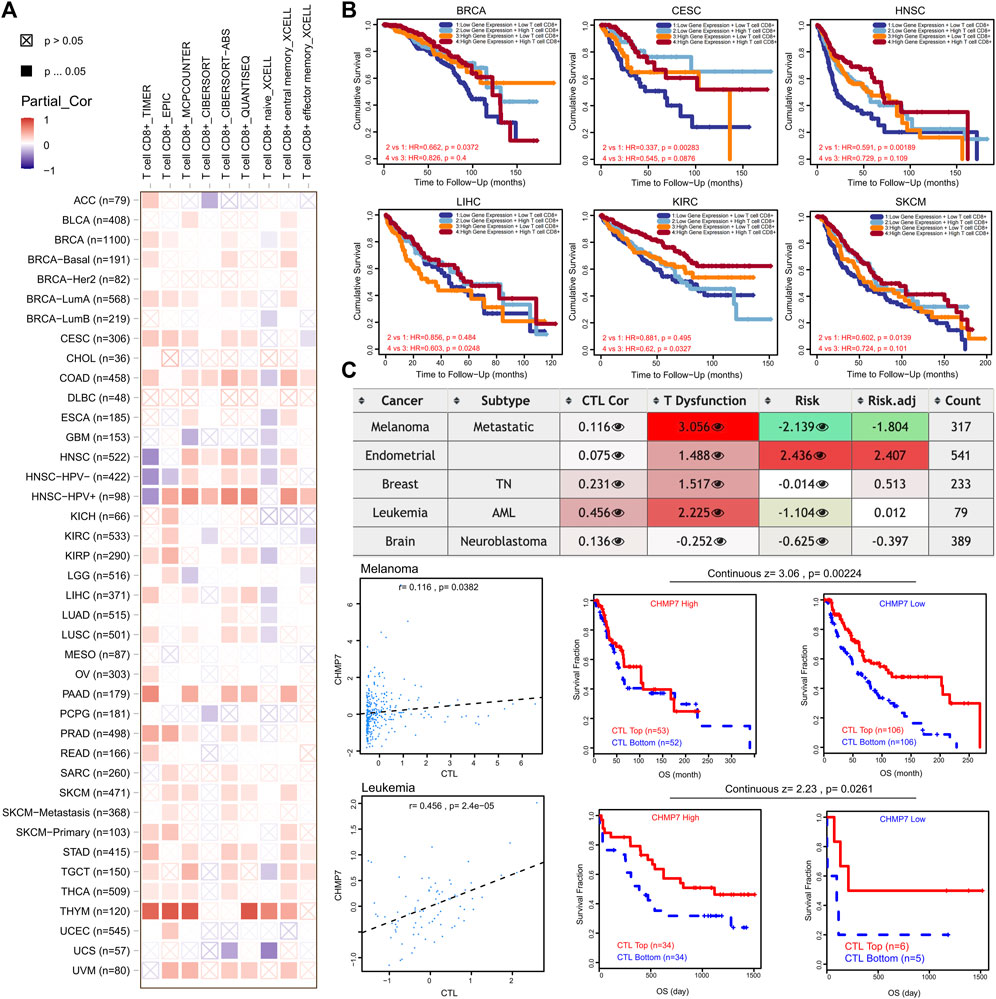
FIGURE 9. CHMP7 has been positively correlated with CD8+ T-cell infiltration. (A) Heat map of association between CHMP7 levels and CD8+ T-cell infiltration calculated by multiple algorithms. (B) Impact of CHMP7 expression levels and CD8+ T-cell infiltration levels on prognosis of tumor patients. (C) Table depicting the correlation between CHMP7 expression and CTL, CTL dysfunction and risk.
The correlation between CHMP7 and CTL dysfunction was observed in SKCM, UCEC, BRCA, and LAML through the TIDE website. A significant positive correlation between CHMP7 and CTL was demonstrated in SKCM and LAML (Figure 9C). The results suggest that CHMP7 is associated with many immune cells, with the possible involvement of killing tumor cells by targeting CTLs.
CHMP7 may assist in predicting the efficacy of chemotherapy and immunotherapy
To investigate the possible role of CHMP7 in tumor therapy, we investigated the predictive value of CHMP7 compared to classical biomarkers for immunotherapeutic response through the TIDE website (Figure 10A). The bar chart indicates that CHMP7 presents an AUC above 0.5 in 11 of the 25 immunotherapy cohorts, suggesting a predictive significance. In 11 cohorts, the predictive value of CHMP7 was superior to the MSI score of CHMP7. Higher predictive value of CHMP7 was observed in 14, seven, 18, 18, 16, nine, and six immunotherapy cohorts compared with TIDE, TMB, CD274, CD8, IFN, T. Clonality, T. Clonality, and Merck18, respectively. It was observed in the antiPD-L1 treatment group of the EMT6 mouse breast cancer cell line that CHMP7 expression was higher in the response group (Figure 10B). The TIDE website also assisted us in predicting the therapeutic response of CHMP7 in the core dataset, immunotherapy dataset, CRISPR screening dataset, and mechanistic follow-up experiments with immunosuppressive cell types (Figure 10C).
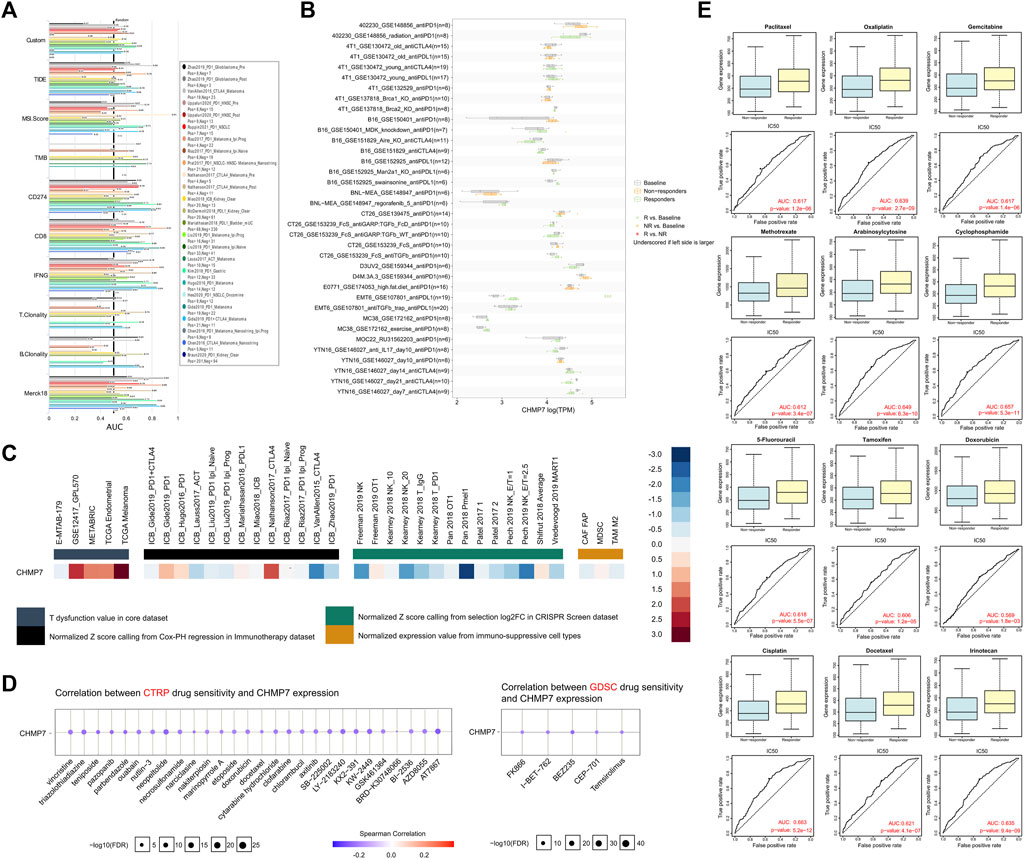
FIGURE 10. CHMP7 may predict immunotherapy and chemotherapy drug sensitivity. (A) CHMP7 correlates with immunotherapy response and (B) biomarkers in the immunotherapy cohort. (C) Therapeutic response of CHMP7 in mechanistic follow-up experiments. (D) Prediction of CHMP7 level-sensitive small compounds by CTRP and GDSC sites. (E) GDSC data-based analysis of differential CHMP7 expression in multiple chemotherapy drug response and non-response groups. (*: p < 0.05, **: p < 0.01 and ***: p < 0.001).
In addition to the predictive value of CHMP7 for immunotherapy efficacy, CHMP7-sensitive small molecule drugs were also analyzed using the CTRP and GDSC datasets (Figure 10D). It was further compared that CHMP7 was differentially expressed in the responding and non-responding groups of multiple immunotherapy cohorts. The results revealed that CHMP7 was differentially expressed in response and non-response groups in multiple immunotherapy cohorts including SRP094781, GSE67501, SRP230414, and IMvigor210 (Supplementary Material S5). The RNAact Drug website also facilitated our predictions (Supplementary Material S6). The results suggest that the top 5 sensitive drugs associated with CHMP7 expression based on the GDSC dataset are GSK429286A, KU-55933, BX-912, CCT007093, and Tretinoin (p < 0.05). Performing further exploration of the differential expression of CHMP7 in the response and non-response groups of multiple chemotherapeutic agents, we discovered that high CHMP7 expression may be associated with drug sensitivity to chemotherapeutic agents such as paclitaxel, oxaliplatin, gemcitabine, and methotrexate, which are now commonly recommended (Figure 10E).
Discussion
Tumor progression is attributed to the accumulation of random mutations and epigenetic alterations in DNA sequences that affect the proliferation of malignant cells associated with gene regulatory networks and other traits associated with the malignant phenotype (Nakagawa and Fujita, 2018). ESCRT is a molecular machine that participates in various essential physiological processes, such as the formation of multivesicular bodies, involvement in cellular autophagy, and repair of cellular membranes. The dysregulation of ESCRT function is highly related to tumor development, and CHMP7, an important regulatory subunit of ESCRT-III, has attracted our attention. The dysregulated function of ESCRT may affect the proliferation and migration capacity of tumor cells through the sorting and delivery of exosomes (Wei et al., 2015). In this study, CHMP7 has been comprehensively described with the help of public databases of tumor tissues. The differential expression of CHMP7 in tumor and normal tissues was first compared, and the predictive value of CHMP7 for patient OS was assessed further. The role of CHMP7 in tumor immunity is a central focus of our study and excels as a biomarker for predicting the efficacy of tumor immunotherapy and chemotherapy. The low CHMP7 expression group was associated with poor prognosis in BRCA, COAD, HNSC, and KIRC, and the level of CHMP7 expression decreased with a more advanced tumor stage. The results suggest that normal CHMP7 expression may be crucial for maintaining normal cellular function.
DNA integrity affected the accuracy of genetic information transmission in organisms, and the ESCRT system is thought to be related to cell division, where we further explored the correlation between CHMP7 and the DNA damage repair response. To maintain genome integrity, complex DNA repair systems are employed to counteract various forms of DNA damage, and these mechanisms are known as the DNA damage response (Jackson and Bartek, 2009). Cellular DNA damage can be classified into single-strand break (SSB) and double-strand break (DSB). SSBs mainly rely on poly ADP-ribose polymerase (PARP) for nucleotide excision repair (NER), base excision repair (BER), and MMR; while DSBs are repaired by HRR, non-homologous end joining (NHEJ), and microhomology-mediated end joining (MMEJ) pathways (Li et al., 2016). The inhibitor targeting PARP can promote the recruitment of DNA repair effector molecules and structural remodeling of chromatin around DNA damage sites, which can selectively kill tumor cells with HRD. Therefore, it is a promising therapeutic strategy for BRCA-mutated tumors (Slade, 2020). In this study, CHMP7 was significantly and positively correlated with MMR and HRR-related gene signatures in various tumor tissues. The results indicate that normal expression of CHMP7 is essential for cells to complete DNA repair through MMR and HRR pathways. Furthermore, CHMP7 was significantly and negatively correlated with Ploidy and HRD. The results show that the downregulation of CHMP7 may lead to chromosomal instability from another aspect. All these findings prompt us that CHMP7 may be intimately involved in the DNA repair process and deserves to be explored in depth.
The immune microenvironment and immunotherapy represent emerging trends in oncology research. In addition to tumor cells, TME includes infiltrating immune and inflammatory cells, CAFs, ECM, microvasculature, and various cytokines and chemokines (Bilotta et al., 2022). The heterogeneity of TME is inextricably linked to the different response rates of patients with tumors to immunotherapy. Low CHMP7 expression was significantly correlated with immunosuppressive TME, as evidenced by high infiltration of M2 macrophages and reduced CTL and cytokines. The analysis of the correlation between CHMP7 and immune cell infiltration levels shows that CHMP7 and M2 macrophages are significantly negatively correlated, which has been demonstrated with multiple algorithms and spatial transcriptional data. Significant elevations in CHMP7 expression were detected following multiple cytokine immunotherapy cohorts. As the immune cell that primarily kills tumor cells in TME, lower CHMP7 expression significantly inhibits the function of CTL, further leading to tumor progression and poor patient prognosis. Furthermore, CHMP7 was identified to perform superiorly as a biomarker for predicting the efficacy of immunotherapy and chemotherapy (Zheng et al., 2023). CHMP7 was superior to traditional biomarkers in several immunotherapy cohorts, such as TMB and MSI scores. CHMP7 levels were significantly lower in the non-responder group for several common chemotherapeutic agents. The results suggest that downregulated CHMP7 levels may lead to the occurrence of tumor drug resistance.
Conclusion
In this article, we utilized public databases such as TCGA, GTEx, TARGET, and single-cell sequencing data to provide a comprehensive and intensive analysis of the role of CHMP7 in tumor development and therapy. CHMP7 shows a predictive value for the prognosis of patients with tumors and is highly involved in tumor immunity. A strong correlation between CHMP7 and TME immune cell infiltration has been observed, which is involved in the formation of suppressive TME and promotes tumor progression. CHMP7 can potentially serve as a new biomarker for predicting the efficacy of chemotherapy and immunotherapy for tumors. As a gene of interest, CHMP7 is expected to provide novel and promising targets for further treatment of patients with tumors.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by The Ethics Committee of The Second Hospital of Jilin University. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from primarily isolated as part of your previous study for which ethical approval was obtained. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
YG and MW drafted the manuscript, and SW reviewed and revised the manuscript. FL has contributed to the article revision and checking process. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Science and Technology Department of Changchun (Grant No. 3D5220102429).
Acknowledgments
The author express gratitude to the public databases, websites, and software used in the paper. The data visualization in the manuscript obtained assistance from Xiantao Academic (https://www.xiantaozi.com/) and Sangerbox (http://vip.sangerbox.com/).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1211843/full#supplementary-material
SUPPLEMENTARY MATERIAL S1 | Correlation of CHMP7 expression levels with cancer stemness scores. (A) Cancer stemness index based on epigenetic regulation of DNA methylation signatures. (B) Cancer stemness index based on differential methylation probes. (C) Cancer stemness index based on epigenetically regulated RNA expression. (D) Cancer stemness index based on enhancer elements/DNA methylation. (E) Cancer stemness index based on RNA expression.
SUPPLEMENTARY MATERIAL S2 | AS events occurring with CHMP7 in pan-cancerous tissues. (A) The read-in, read-out, and PSI values of CHMP7 _alt_5prime_195490 in tumor and normal tissues. Differences in PSI value between tumor and adjacent normal tissue (left), and tumor and GTEx normal tissue (right); red dashed line refers to FDR of 0.05. (B) The read-in, read-out, and PSI values of CHMP7_exon_skip_482248 in tumor and normal tissues. Differences in PSI value between tumor and adjacent normal tissue (left), and tumor and GTEx normal tissue (right); red dashed line refers to FDR of 0.05. (C) PSI values of CHMP7_AD_83076 in tumor and normal tissues. (D) PSI values of CHMP7_ES_83072 in tumor and normal tissues. (E) PSI values of CHMP7_ES_83073 in tumor and normal tissues. (F) Differences in PSI value of CHMP7_AD_83076, CHMP7_ES_83072 and CHMP7_ES_83073 between tumor and adjacent normal tissue respectively. (AD, alternate donor site; ES, exon skipping).
SUPPLEMENTARY MATERIAL S3 | Top 100 genes associated with CHMP7 obtained from the GEPIA website.
SUPPLEMENTARY MATERIAL S4 | Correlation of CHMP7 expression levels and immune regulatory genes.
SUPPLEMENTARY MATERIAL S5 | CHMP7 expression for Response and Non-response based on Pre-treatment Samples in each dataset.
SUPPLEMENTARY MATERIAL S6 | Small molecule drugs related to CHMP7 as predicted by the RNAact Drug website.
Abbreviations
AS, alternative splicing; CAFs, cancer associated fibroblasts; CHMP7, charged multivesicular body protein 7; CTLs, cytotoxic T lymphocytes; ECM, extracellular matrix; TME, tumor microenvironment; DFS, disease-free survival; EGFR, epidermal growth factor receptor; ESCRT, endosomal sorting complex required for transport; EVs, extracellular vehicles; GEO, Gene Expression Omnibus database; GSEA, gene set enrichment analysis; HRD, homologous recombination deficiency; HRR, homologous recombination repair; ILVs, intraluminal vesicles; MSI, microsatellite instability; NEO, neoantigen; OS, overall survival; PPI, protein-protein interaction; PSI, percent spliced-in; TARGET, Therapeutically Applicable Research to Generate Effective Treatments; TCGA, the Cancer Genome Atlas; TMB, tumor mutation burden.
References
Baretti, M., and Le, D. T. (2018). DNA mismatch repair in cancer. Pharmacol. Ther. 189, 45–62. doi:10.1016/j.pharmthera.2018.04.004
Bilotta, M. T., Antignani, A., and Fitzgerald, D. J. (2022). Managing the TME to improve the efficacy of cancer therapy. Front. Immunol. 13, 954992. doi:10.3389/fimmu.2022.954992
Biswas, S. K., and Mantovani, A. (2010). Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol. 11 (10), 889–896. doi:10.1038/ni.1937
Blass, E., and Ott, P. A. (2021). Advances in the development of personalized neoantigen-based therapeutic cancer vaccines. Nat. Rev. Clin. Oncol. 18 (4), 215–229. doi:10.1038/s41571-020-00460-2
Blencowe, B. J. (2006). Alternative splicing: new insights from global analyses. Cell. 126 (1), 37–47. doi:10.1016/j.cell.2006.06.023
Coyne, A. N., Baskerville, V., Zaepfel, B. L., Dickson, D. W., Rigo, F., Bennett, F., et al. (2021). Nuclear accumulation of CHMP7 initiates nuclear pore complex injury and subsequent TDP-43 dysfunction in sporadic and familial ALS. Sci. Transl. Med. 13 (604), eabe1923. doi:10.1126/scitranslmed.abe1923
Coyne, A. N., and Rothstein, J. D. (2022). Nuclear pore complexes - a doorway to neural injury in neurodegeneration. Nat. Rev. Neurol. 18 (6), 348–362. doi:10.1038/s41582-022-00653-6
Gatta, A. T., and Carlton, J. G. (2019). The ESCRT-machinery: closing holes and expanding roles. Curr. Opin. Cell. Biol. 59, 121–132. doi:10.1016/j.ceb.2019.04.005
Genin, M., Clement, F., Fattaccioli, A., Raes, M., and Michiels, C. (2015). M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer 15, 577. doi:10.1186/s12885-015-1546-9
Han, Q., Lv, L., Wei, J., Lei, X., Lin, H., Li, G., et al. (2019). Vps4A mediates the localization and exosome release of β-catenin to inhibit epithelial-mesenchymal transition in hepatocellular carcinoma. Cancer Lett. 457, 47–59. doi:10.1016/j.canlet.2019.04.035
Hausman, D. M. (2019). What is cancer? Perspect. Biol. Med. 62 (4), 778–784. doi:10.1353/pbm.2019.0046
Iranzo, J., Martincorena, I., and Koonin, E. V. (2018). Cancer-mutation network and the number and specificity of driver mutations. Proc. Natl. Acad. Sci. U. S. A. 115 (26), E6010–e9. doi:10.1073/pnas.1803155115
Isono, E. (2021). ESCRT is a great sealer: non-endosomal function of the ESCRT machinery in membrane repair and autophagy. Plant Cell. Physiol. 62 (5), 766–774. doi:10.1093/pcp/pcab045
Jackson, S. P., and Bartek, J. (2009). The DNA-damage response in human biology and disease. Nature 461 (7267), 1071–1078. doi:10.1038/nature08467
Kocaturk, N. M., Akkoc, Y., Kig, C., Bayraktar, O., Gozuacik, D., and Kutlu, O. (2019). Autophagy as a molecular target for cancer treatment. Eur. J. Pharm. Sci. 134, 116–137. doi:10.1016/j.ejps.2019.04.011
Korbei, B. (2022). Ubiquitination of the ubiquitin-binding machinery: how early ESCRT components are controlled. Essays Biochem. 66 (2), 169–177. doi:10.1042/EBC20210042
Ledermann, J. A., Drew, Y., and Kristeleit, R. S. (2016). Homologous recombination deficiency and ovarian cancer. Eur. J. Cancer 60, 49–58. doi:10.1016/j.ejca.2016.03.005
Lee, S. S., and Cheah, Y. K. (2019). The interplay between MicroRNAs and cellular components of tumour microenvironment (TME) on non-small-cell lung cancer (NSCLC) progression. J. Immunol. Res. 2019, 3046379. doi:10.1155/2019/3046379
Levy, J. M. M., Towers, C. G., and Thorburn, A. (2017). Targeting autophagy in cancer. Nat. Rev. Cancer 17 (9), 528–542. doi:10.1038/nrc.2017.53
Lewandowska, A. M., Rudzki, M., Rudzki, S., Lewandowski, T., and Laskowska, B. (2019). Environmental risk factors for cancer - review paper. Ann. Agric. Environ. Med. 26 (1), 1–7. doi:10.26444/aaem/94299
Li, Y., Sun, N., Lu, Z., Sun, S., Huang, J., Chen, Z., et al. (2017). Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett. 393, 40–51. doi:10.1016/j.canlet.2017.02.016
Li, Z., Pearlman, A. H., and Hsieh, P. (2016). DNA mismatch repair and the DNA damage response. DNA Repair (Amst) 38, 94–101. doi:10.1016/j.dnarep.2015.11.019
Liang, X., Zhang, H., Wang, Z., Zhang, X., Dai, Z., Zhang, J., et al. (2022). JMJD8 is an M2 macrophage biomarker, and it associates with DNA damage repair to facilitate stemness maintenance, chemoresistance, and immunosuppression in pan-cancer. Front. Immunol. 13, 875786. doi:10.3389/fimmu.2022.875786
Lim, Z. F., and Ma, P. C. (2019). Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J. Hematol. Oncol. 12 (1), 134. doi:10.1186/s13045-019-0818-2
Malta, T. M., Sokolov, A., Gentles, A. J., Burzykowski, T., Poisson, L., Weinstein, J. N., et al. (2018). Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 173 (2), 338–354.e15. doi:10.1016/j.cell.2018.03.034
Mami-Chouaib, F., Blanc, C., Corgnac, S., Hans, S., Malenica, I., Granier, C., et al. (2018). Resident memory T cells, critical components in tumor immunology. J. Immunother. Cancer 6 (1), 87. doi:10.1186/s40425-018-0399-6
Müller, M., May, S., and Bird, T. G. (2021). Ploidy dynamics increase the risk of liver cancer initiation. Nat. Commun. 12 (1), 1896. doi:10.1038/s41467-021-21897-8
Nakagawa, H., and Fujita, M. (2018). Whole genome sequencing analysis for cancer genomics and precision medicine. Cancer Sci. 109 (3), 513–522. doi:10.1111/cas.13505
Padma, S., Patra, R., Sen Gupta, P. S., Panda, S. K., Rana, M. K., and Mukherjee, S. (2023). Cell surface fibroblast activation protein-2 (Fap2) of fusobacterium nucleatum as a vaccine candidate for therapeutic intervention of human colorectal cancer: an immunoinformatics approach. Vaccines (Basel). 11 (3), 525. doi:10.3390/vaccines11030525
Patra, R., Chakraborty, J., Das, N. C., and Mukherjee, S. (2023). An integrated omics study on the role of HDAC9 gene in the oncogenic events of human gastrointestinal-tract associated cancers. Hum. Gene 37, 201189. doi:10.1016/j.humgen.2023.201189
Patra, R., Das, N. C., and Mukherjee, S. (2021). Exploring the differential expression and prognostic significance of the COL11A1 gene in human colorectal carcinoma: an integrated bioinformatics approach. Front. Genet. 12, 608313. doi:10.3389/fgene.2021.608313
Ritter, A. T., Shtengel, G., Xu, C. S., Weigel, A., Hoffman, D. P., Freeman, M., et al. (2022). ESCRT-mediated membrane repair protects tumor-derived cells against T cell attack. Science 376 (6591), 377–382. doi:10.1126/science.abl3855
Slade, D. (2020). PARP and PARG inhibitors in cancer treatment. Genes. Dev. 34 (5-6), 360–394. doi:10.1101/gad.334516.119
Sturm, G., Finotello, F., Petitprez, F., Zhang, J. D., Baumbach, J., Fridman, W. H., et al. (2019). Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics 35 (14), i436–i445. doi:10.1093/bioinformatics/btz363
Sun, D., Li, H., Cao, M., He, S., Lei, L., Peng, J., et al. (2020). Cancer burden in China: trends, risk factors and prevention. Cancer Biol. Med. 17 (4), 879–895. doi:10.20892/j.issn.2095-3941.2020.0387
Thorsson, V., Gibbs, D. L., Brown, S. D., Wolf, D., Bortone, D. S., Ou Yang, T. H., et al. (2018). The immune landscape of cancer. Immunity 48 (4), 812–830.e14. doi:10.1016/j.immuni.2018.03.023
Ule, J., and Blencowe, B. J. (2019). Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol. Cell. 76 (2), 329–345. doi:10.1016/j.molcel.2019.09.017
Vietri, M., Radulovic, M., and Stenmark, H. (2020). The many functions of ESCRTs. Nat. Rev. Mol. Cell. Biol. 21 (1), 25–42. doi:10.1038/s41580-019-0177-4
Vogelstein, B., Papadopoulos, N., Velculescu, V. E., Zhou, S., Diaz, L. A., and Kinzler, K. W. (2013). Cancer genome landscapes. Science 339 (6127), 1546–1558. doi:10.1126/science.1235122
Wei, D., Zhan, W., Gao, Y., Huang, L., Gong, R., Wang, W., et al. (2021). RAB31 marks and controls an ESCRT-independent exosome pathway. Cell. Res. 31 (2), 157–177. doi:10.1038/s41422-020-00409-1
Wei, J. X., Lv, L. H., Wan, Y. L., Cao, Y., Li, G. L., Lin, H. M., et al. (2015). Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology 61 (4), 1284–1294. doi:10.1002/hep.27660
Xie, J., Zhang, J., Tian, W., Zou, Y., Tang, Y., Zheng, S., et al. (2022). The pan-cancer multi-omics landscape of FOXO family relevant to clinical outcome and drug resistance. Int. J. Mol. Sci. 23 (24), 15647. doi:10.3390/ijms232415647
Yoshihara, K., Shahmoradgoli, M., Martínez, E., Vegesna, R., Kim, H., Torres-Garcia, W., et al. (2013). Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612. doi:10.1038/ncomms3612
Zhang, Z., Wang, Z. X., Chen, Y. X., Wu, H. X., Yin, L., Zhao, Q., et al. (2022). Integrated analysis of single-cell and bulk RNA sequencing data reveals a pan-cancer stemness signature predicting immunotherapy response. Genome Med. 14 (1), 45. doi:10.1186/s13073-022-01050-w
Zheng, S., Liang, J. Y., Tang, Y., Xie, J., Zou, Y., Yang, A., et al. (2023). Dissecting the role of cancer-associated fibroblast-derived biglycan as a potential therapeutic target in immunotherapy resistance: A tumor bulk and single-cell transcriptomic study. Clin. Transl. Med. 13 (2), e1189. doi:10.1002/ctm2.1189
Keywords: pan-cancer, CHMP7, endosomal sorting complex required for transport, immunosuppression, CTL dysfunction, M2 macrophages infiltration
Citation: Guo Y, Wang S, Liang F and Wang M (2023) Identification of CHMP7 as a promising immunobiomarker for immunotherapy and chemotherapy and impact on prognosis of colorectal cancer patients. Front. Cell Dev. Biol. 11:1211843. doi: 10.3389/fcell.2023.1211843
Received: 05 May 2023; Accepted: 09 August 2023;
Published: 30 August 2023.
Edited by:
Subhadip Mukhopadhyay, New York University, United StatesReviewed by:
Jindong Xie, Sun Yat-sen University Cancer Center (SYSUCC), ChinaSuprabhat Mukherjee, Kazi Nazrul University, India
Copyright © 2023 Guo, Wang, Liang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Wang, jdeywangmin@163.com
†These authors have contributed equally to this work
 Yu Guo
Yu Guo Shu Wang2†
Shu Wang2†  Min Wang
Min Wang