Biological properties of the BCL-2 family protein BCL-RAMBO, which regulates apoptosis, mitochondrial fragmentation, and mitophagy
- 1Department of Applied Biology, Kyoto Institute of Technology, Kyoto, Japan
- 2Biomedical Research Center, Kyoto Institute of Technology, Kyoto, Japan
Mitochondria play an essential role in the regulation of cellular stress responses, including cell death. Damaged mitochondria are removed by fission and fusion cycles and mitophagy, which counteract cell death. BCL-2 family proteins possess one to four BCL-2 homology domains and regulate apoptosis signaling at mitochondria. BCL-RAMBO, also known as BCL2-like 13 (BCL2L13), was initially identified as one of the BCL-2 family proteins inducing apoptosis. Mitophagy receptors recruit the ATG8 family proteins MAP1LC3/GABARAP via the MAP1LC3-interacting region (LIR) motif to initiate mitophagy. In addition to apoptosis, BCL-RAMBO has recently been identified as a mitophagy receptor that possesses the LIR motif and regulates mitochondrial fragmentation and mitophagy. In the 20 years since its discovery, many important findings on BCL-RAMBO have been increasingly reported. The biological properties of BCL-RAMBO are reviewed herein.
Introduction
Mitochondria play an essential role in energy metabolism by producing ATP and many other metabolites, and also regulate cellular stress responses, such as cell death, autophagy, and inflammation (Green et al., 2011). Mitochondria harbor multiple components that transmit cell death signals and inflammatory signals (Gurung et al., 2015; Bock and Tait, 2020). Damaged mitochondria are disposed by a mitochondrial autophagy process (mitophagy), which counteracts cell death or inflammation (Green et al., 2011). Mitochondrial fission and fusion cycles play a critical role in quality control, which is linked to mitophagy (Youle and van der Bliek, 2012). Mitochondrial dysfunction causes many diseases, including neurodegenerative, cardiovascular, and metabolic disorders (Nunnari and Suomalainen, 2012). Mitochondria also regulate the initiation, growth, survival, and metastasis stages of tumorigenesis (Vyas et al., 2016). Therefore, a detailed understanding of the molecular mechanisms underlying mitophagy is important for therapeutic interventions. Mitophagy is initiated by two major pathways: the ubiquitin-dependent pathway and ubiquitin-independent pathway. PTEN-induced putative kinase 1 (PINK1) and the E3 ubiquitin ligase Parkin play a primary role in the regulation of ubiquitin-dependent mitophagy (Pickrell and Youle, 2015; McWilliams and Muqit, 2017). In contrast, ubiquitin-independent mitophagy is mediated by at least five mitophagy receptors anchored to mitochondrial outer membranes (MOM). In addition to apoptosis, the BCL-2 family protein BCL-RAMBO, also known as BCL2-like 13 (BCL2L13), has recently been identified as a mitophagy receptor that regulates mitochondrial fragmentation and mitophagy. The biological properties of BCL-RAMBO are reviewed herein.
Regulation of cell death
Cell death occurs in multiple forms, including apoptosis, autophagy-dependent cell death, and necroptosis (Tang et al., 2019). Apoptosis is mediated by caspases, which are divided into initiator, effector, and inflammatory caspases (Mcllwain et al., 2013; Man and Kanneganti, 2016). The cascade of caspase activation is triggered by the ligation of cell-surface death receptors and mitochondrial activation resulting in the release of their components (Green et al., 2014). Death receptors and mitochondria trigger signaling pathways that are mediated by different initiator caspases (Figure 1). In the extrinsic pathway, death receptors recruit multiple adaptor proteins and procaspases 8 and 10 in the death-inducing signaling complex, and this is followed by their activation (Lavrik and Krammer, 2012; Siegmund et al., 2017). Initiator active caspases 8 and 10 convert procaspases 3, 6, and 7 to effector active caspases by cleavage, leading to the limited proteolysis of many substrates and the execution of apoptosis (Julien and Wells, 2017). In addition to energy metabolism, mitochondria play an essential role in programmed cell death (Bock and Tait, 2020). In the intrinsic pathway upon apoptotic stimuli, mitochondrial outer membrane permeability (MOMP) causes the release of intermembrane space proteins, two of which are cytochrome c (CYCS) and second mitochondria-derived activator of caspase (SMAC) (Cosentino and García-Sáez, 2017; Kale et al., 2018; Kalkavan and Green, 2018). In mammals, CYCS induces the activation of procaspase 9 through the formation of apoptosomes in the cytosol (Bratton and Salvesen, 2010; Yuan and Akey, 2013). Inhibitor of apoptosis (IAP) proteins target caspases via multiple mechanisms, while SMAC antagonizes IAP proteins (Fulda and Vucic, 2012). In a similar manner to caspases 8 and 10, initiator active caspase 9 cleaves downstream procaspases 3, 6, and 7 into their active forms (Mcllwain et al., 2013). BCL-2 family proteins possess one to four BCL-2 homology (BH) domains (BH1, BH2, BH3, and BH4) and are divided into three groups: anti-apoptotic, pro-apoptotic multi-domain, and pro-apoptotic BH3-only proteins (Adams and Cory, 2018; Singh et al., 2019; Warren et al., 2019). BCL-2 family proteins regulate apoptosis signaling by interacting with pro- and anti-apoptotic proteins at mitochondria (Adams and Cory, 2018; Singh et al., 2019; Warren et al., 2019). Crosstalk exists between extrinsic and intrinsic pathways; BH3-interacting domain death agonist (BID) is cleaved into truncated BID (tBID) by active caspases 8 and 10, translocates to mitochondria, and then interacts with BCL-2 family proteins, which leads to the initiation of apoptosis signaling (Kantari and Walczak, 2011; Kaufmann et al., 2012).
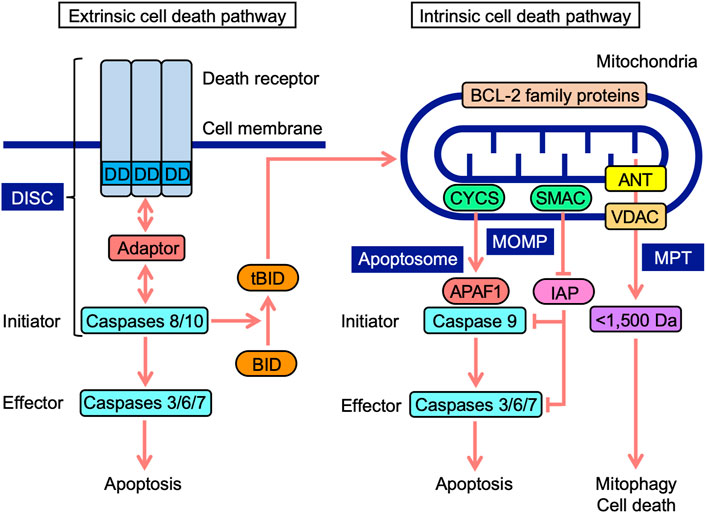
FIGURE 1. Extrinsic and intrinsic cell death signaling pathways. In the extrinsic pathway, death receptors, upon engagement with ligands, recruit adaptor proteins to their death domain (DD) and procaspases 8 and 10, leading to the formation of a death-inducing signaling complex (DISC). In the DISC, procaspases 8 and 10 are activated into initiator active caspases 8 and 10, which cleave procaspases 3, 6, and 7 to effector active caspases 3, 6, and 7, leading to apoptosis. Mitochondria harbor BCL-2 family proteins and VDAC on mitochondrial outer membranes, ANT on mitochondrial inner membranes, and CYCS and SMAC at the intermembrane space. In the intrinsic pathway, mitochondrial outer membrane permeability (MOMP) causes the release of CYCS and SMAC into the cytosol. CYCS induces complex formation consisting of APAF1 and procaspase 9 (apoptosome). In the apoptosome, procaspase 9 is activated to initiator active caspase 9, which cleaves procaspases 3, 6, and 7 to active caspases 3, 6, and 7, leading to apoptosis. SMAC antagonizes IAP proteins, which target caspases. Mitochondrial permeability transition (MPT) releases <1,500-Da solutes, which cause mitophagy and cell death. BH3-interacting domain death agonist (BID) is cleaved into truncated BID (tBID) by active caspases 8 and 10. tBID translocates to mitochondria and interacts with BCL-2 family proteins, triggering apoptosis signaling.
Mitochondrial permeability transition (MPT) promotes the release of <1,500-Da solutes from the matrix via the mitochondrial permeability transition pore (MPTP), which ultimately activates cellular responses ranging from mitophagy to cell death (Bonora et al., 2022). Voltage-dependent anion channel (VDAC), adenine nucleotide translocase (ANT), mitochondrial phosphate carrier, and F1F0-ATPase were initially identified as components of MPTP (Kwong and Molkentin, 2015; Javadov et al., 2017). ANT proteins have been reported to regulate MPTP, but are not essential (Kokoszka et al., 2004; Urbani et al., 2019). VDAC proteins are also dispensable for MPTP (Krauskopf et al., 2006;; Baines et al., 2007; Urbani et al., 2019). Although cyclophilin D is not a component of MPTP, it is a critical modulator of MPTP, desensitizing them to stress (Kwong and Molkentin, 2015; Javadov et al., 2017). VDAC proteins form major channels across MOM, allowing the free passage of small solutes (Maurya and Mahalakshmi, 2016; Mazure, 2017; Magrí et al., 2018). ANT proteins localize to mitochondrial inner membranes (MIM), and mediate the exchange activity of ADP and ATP (Zhivotovsky et al., 2009; Brenner et al., 2011). ANT proteins were recently shown to mediate mitophagy (Hoshino et al., 2019). Therefore, MOMP and MPT regulate mitochondrial stress responses, including cell death and mitophagy.
Regulation of mitochondrial fragmentation
Mitochondrial fission and fusion processes play an essential role in the compartmentalization of damaged and healthy mitochondria, and are linked to mitophagy (Youle and van der Bliek, 2012). The large GTPase dynamin-related protein 1 (DRP1) mediates mitochondrial fission (Otera et al., 2013; Kraus and Ryan, 2017). Mitochondrial fission 1 (FIS1), mitochondrial fission factor (MFF), and mitochondrial dynamics proteins of 49 and 51 kDa (MiD49 and MiD51) are mitochondrial receptor proteins anchored to MOM and mediate mitochondrial fragmentation (Schrader et al., 2012; Rovira-Llopis et al., 2017). DRP1 is translocated from the cytosol to MOM by binding to mitochondrial receptor proteins, and promotes mitochondrial fission through GTPase activity (Otera et al., 2013; Kraus and Ryan, 2017). In contrast, mitofusins 1 and 2 are GTPases involved in the fusion of MOM, while OPA1 mitochondrial dynamin like GTPase mediates the fusion of MIM (Schrepfer and Scorrano, 2016; Tilokani et al., 2018). During fission and fusion cycles, healthy daughter mitochondria undergo the fusion process, while damaged daughter mitochondria are destined for degradation by mitophagy (Youle and van der Bliek, 2012). The dysfunction of mitochondrial dynamics is associated with cardiovascular diseases and neurodegenerative disorders (Burté et al., 2015; Vásquez-Trincado et al., 2016; Murphy and Hartley, 2018).
Regulation of mitophagy
Mitophagy plays an essential role in mitochondrial quality control and cellular homeostasis (Ashrafi and Schwartz, 2013; Palikaras et al., 2018; Pickles et al., 2018). When mitochondria are damaged in response to various stresses, they are enclosed by the elongation of phagophores, which leads to the formation of autophagosomes (Lahiri et al., 2019). Autophagosomes then fuse with lysosomes and their contents are destined for degradation (Lahiri et al., 2019). Autophagy-related protein 8 (ATG8) family proteins are required for the formation of autophagosomes, and are divided into microtubule-associated protein 1 light chain 3 (MAP1LC3) and GABA type A receptor-associated protein (GABARAP) family proteins in multicellular animals, which consist of MAP1LC3A, MAP1LC3B, MAP1LC3C, GABARAP, GABARAPL1, and GABARAPL2 (Schaaf et al., 2016; Johansen and Lamark, 2020). Upon the activation of mitophagy by a number of stimuli, MAP1LC3 family proteins are linked to phosphatidylethanolamine and anchored to phagophore membranes, which is an essential step in the formation of autophagosomes (Schaaf et al., 2016; Johansen and Lamark, 2020).
Ubiquitin-dependent and-independent mitophagy both mediate the recruitment of MAP1LC3-associated phagophores to MOM. Ubiquitin-dependent mitophagy is regulated by PINK1 and Parkin (Figure 2A) (Pickrell and Youle, 2015; McWilliams and Muqit, 2017). PINK1 stability is regulated by mitochondrial membrane depolarization (Pickrell and Youle, 2015; McWilliams and Muqit, 2017). PINK1 is stabilized on MOM and phosphorylates various proteins, such as Parkin, MOM substrates, and ubiquitin, while Parkin ubiquitinates MOM proteins (Pickrell and Youle, 2015; McWilliams and Muqit, 2017). Their polyubiquitin chains are phosphorylated by PINK1 and recruit adaptor proteins, which interact with MAP1LC3 family proteins via their MAP1LC3-interacting region (LIR) motifs comprising the core consensus sequence of [W/F/Y] XX [L/I/V] (where X is any amino acid) (Birgisdottir et al., 2013). NDP52, SQSTM1, NBR1, TAXBP1, and TOLLIP are adaptor proteins that promote the recruitment of phagophores (McWilliams and Muqit, 2017; De et al., 2021).
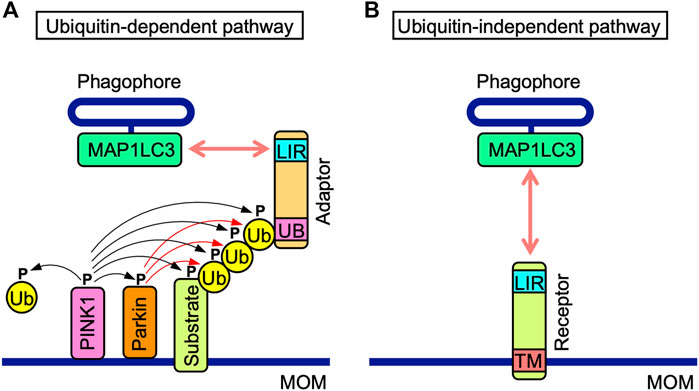
FIGURE 2. Ubiquitin-dependent and-independent mitophagy pathways. (A) In the ubiquitin-dependent pathway, PINK1 is stabilized on mitochondrial outer membranes (MOM) and phosphorylates PINK1 itself, the E3 ubiquitin (Ub) ligase Parkin, MOM-associated substrates, and free and polymerized Ub (black arrows). Parkin ubiquitinates MOM-associated substrates (red arrows). Poly-Ub chains of MOM-associated substrates interact with the ubiquitin-binding domain (UB) of adaptor proteins, which also possess the MAP1LC3-interacting region (LIR) motif. The LIR motif interacts with MAP1LC3 proteins anchored to phagophores. (B) Mitophagy receptors harbor the LIR motif and transmembrane (TM) domain anchored to MOM. In the ubiquitin-independent pathway, mitophagy receptors interact with MAP1LC3 proteins anchored to phagophores.
In addition to Parkin, ubiquitin-dependent mitophagy is mediated by other E3 ubiquitin ligases, such as mitochondrial E3 ubiquitin protein ligase 1 (MUL1), siah E3 ubiquitin protein ligase 1 (SIAH1), and ariadne RBR E3 ubiquitin protein ligase 1 (ARIH1) (Terešak et al., 2022). MUL1 is anchored to MOM and synergistically promotes mitophagy with PINK1 (Calle et al., 2022). SIAH1 forms a complex with PINK1 and synphilin-1, and mediates mitophagy that does not require Parkin activity (Szargel et al., 2016). ARIH1 triggers PINK1-dependent mitophagy in cancer cells in a Parkin-independent manner (Villa et al., 2017). Therefore, multiple E3 ubiquitin ligases, other than Parkin, mediate PINK1-dependent mitophagy.
The ubiquitin-independent pathway is mediated by MOM-anchored mitophagy receptors, all of which possess LIR motifs (Figure 2B). MOM-anchored mitophagy receptors consist of at least five proteins, i.e., BCL-2/adenovirus E1B 19 kDa protein-interacting protein 3 (BNIP3), BCL-2/adenovirus E1B 19 kDa protein-interacting protein 3-like (BNIP3L), also known as Nip3-like protein X (NIX), FUN14 domain-containing 1 (FUNDC1), FK506-bining protein prolyl isomerase 8 (FKBP8), and BCL-RAMBO. The direct interactions of mitophagy receptors with MAP1LC3 family proteins are a prerequisite for the recognition of mitochondria by autophagosomes.
In addition to MOM-anchored mitophagy receptors, prohibitin 2 (PHB2), cardiolipin, and Autophagy/bectin-1 regulator 1 (AMBRA1) mediate the ubiquitin-independent pathway. PHB2 localizes to MIM and mediates Parkin-induced mitophagy by binding to MAP1LC3 proteins upon mitochondrial depolarization and MOM rupture (Wei et al., 2017). Cardiolipin is a phospholipid that localizes to MIM and acts as a receptor for MAP1LC3 proteins when translocated from MIM to MOM in response to stress (Dudek, 2017; Paradies et al., 2019). AMBRA1 is a mitophagy receptor possessing a LIR motif and mediates Parkin-dependent and -independent mitophagy when it localizes to mitochondria (Strappazzon et al., 2011; Strappazzon et al., 2015). HECT, UBA, and WWE domain containing 1 (HUWE1) belongs to the HECT E3 ubiquitin ligase family and possesses the BH3 domain (Kao et al., 2018). HUWE1 interacts with MCL1 and promotes AMBRA1-dependent mitophagy by mediating MCL1 degradation (Strappazzon et al., 2020). The E3 ubiquitin ligase membrane-associated ring-CH-type finger 5 (MARCHF5) mediates the ubiquitination of FUNDC1 at Lys119 for degradation, and MARCHF5 finely regulates hypoxia-induced mitophagy (Chen et al., 2017). Therefore, ubiquitin-dependent protein degradation by E3 ubiquitin ligases plays a regulatory role in the initiation of mitophagy.
Structure of BCL-RAMBO
We initially identified BCL-RAMBO as a unique member of the BCL-2 family that induces apoptosis through its overexpression (Kataoka et al., 2001). Approximately 15 years later, BCL-RAMBO was shown to regulate mitophagy and mitochondrial fragmentation (Murakawa et al., 2015). Research conducted in the last 2 decades has revealed that BCL-RAMBO regulates various cellular processes, including apoptosis, mitochondrial fragmentation, and mitophagy.
BCL-2 family proteins are characterized by one to four BH motifs (Adams and Cory 2018; Singh et al., 2019; Warren et al., 2019). Human BCL-RAMBO was initially identified by the screening of public databases for homology with BCL-2 family proteins (Kataoka et al., 2001). Human BCL-RAMBO consists of 485 amino acids and is composed of four N-terminal BH motifs (BH1, BH2, BH3, and BH4), a No BH motif (BHNo) domain, C-terminal transmembrane (TM) domain, and short cytoplasmic tail (Figures 3A, B). Mouse BCL-RAMBO consists of 434 amino acids, which are highly conserved with human BCL-RAMBO, except for the BHNo domain being approximately 50 amino acids shorter than that of human BCL-RAMBO (Figures 3A, B). The BHNo domain is enriched with glutamic acid and serine. Although the predicted molecular mass of human BCL-RAMBO is approximately 55 kDa, the apparent molecular mass of BCL-RAMBO on SDS-PAGE is approximately 85 kDa, which is attributed to the BHNo domain (Kataoka et al., 2001; Yang et al., 2002). In the BHNo domain, 276–279 amino acids of human BCL-RAMBO (WQQI) and 273–276 amino acids of mouse BCL-RAMBO (WQQI) are LIR motifs required for mitophagy (Murakawa et al., 2015). It currently remains unclear whether the difference in the BHNo domain between human BCL-RAMBO and mouse BCL-RAMBO manifests different functions.
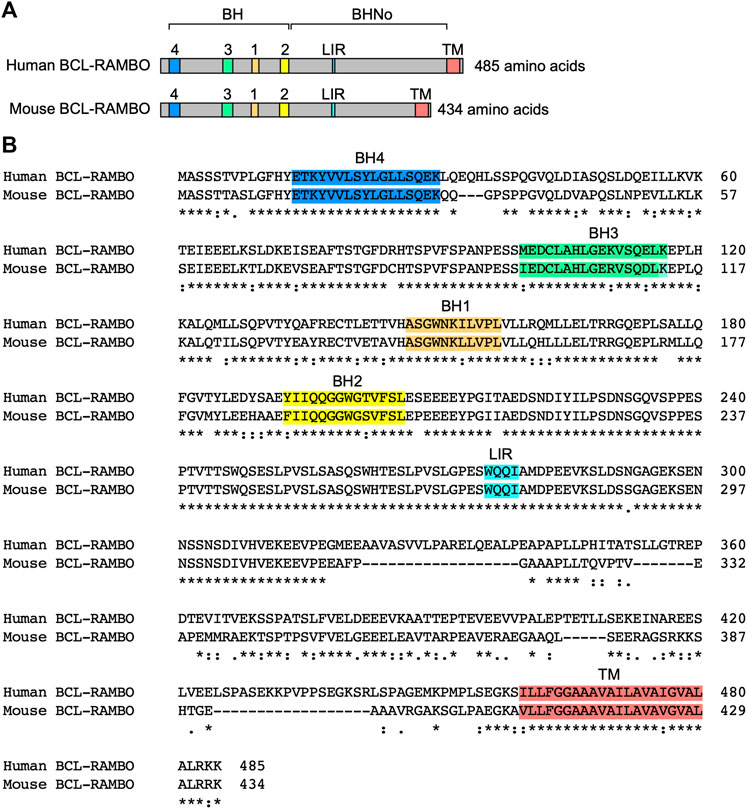
FIGURE 3. Structures of human and mouse BCL-RAMBO. (A) Human BCL-RAMBO consists of 485 amino acids, while mouse BCL-RAMBO consists of 434 amino acids. The BCL-2 homology (BH) domain, No BH motif (BHNo) domain, MAP1LC3-interacting region (LIR) motif, and transmembrane (TM) domain are shown. (B) The amino acid sequences of human BCL-RAMBO and mouse BCL-RAMBO are aligned using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Comparison between BCL-RAMBO and MOM-anchored mitophagy receptors
The structures of five MOM-anchored mitophagy receptors, i.e., BNIP3, BNIP3L, FUNDC1, FKBP8, and BCL-RAMBO, are shown in Figure 4A (Otsu et al., 2015; Hamacher-Brady and Brady, 2016; Tan et al., 2016; Yamaguchi et al., 2016; Rodger et al., 2018; Yoo and Jung, 2018; Zhang et al., 2018; Gustafsson and Dorn, 2019; Ma et al., 2020a; Xie et al., 2020; Xu et al., 2020; Meng et al., 2021a; De et al., 2021; Onishi et al., 2021; Onishi and Okamoto, 2021; Poole and Macleod, 2021). In addition to the LIR motif, BCL-RAMBO, BNIP3, BNIP3L, FUNDC1, and FKBP8 commonly possess a C-terminal TM domain followed by a short cytoplasmic tail (1–11 amino acids) (Figure 4B). The C-terminal TM domain flanked at both ends by at least two basic amino acids is required for the mitochondrial targeting of BCL-xL (Kaufmann et al., 2003). Human and mouse BCL-RAMBO both possess a single C-terminal TM domain flanked by two basic amino acids and three basic amino acids (Figure 3B, Figure 4B). The LIR motifs of these mitophagy receptors are present at regions close to the N terminus, except for BCL-RAMBO (Figure 4C). Other regions in the amino acid sequences of mitophagy receptors are not well conserved.
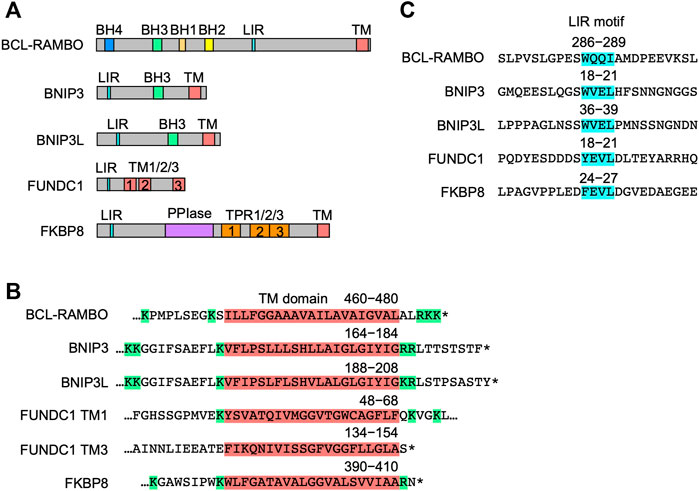
FIGURE 4. Structures of mitophagy receptors anchored to mitochondrial outer membranes (MOM). (A) Mitophagy receptors possess the MAP1LC3-interacting region (LIR) motif and transmembrane (TM) domain. Abbreviations: BCL-2 homology (BH); peptidyl prolyl cis-trans isomerase (PPI); tetratricopeptide repeats (TPR). (B) Structures of the TM domain of human mitophagy receptors. The TM domain (magenta) is shown with numbers referring to positions in the amino acid sequence. Basic amino acids are shown in green. Asterisks indicate the stop codon. (C) Structures of the LIR motif. The LIR motifs (blue) of human mitophagy receptors are shown with numbers referring to positions in the amino acid sequence.
BNIP3 and BNIP3L possess a BH3 domain and are pro-apoptotic proteins (Zhang and Ney, 2009). BCL-2 and BCL-xL interact with BNIP3 and BNIP3L, thereby inhibiting their apoptotic activities (Zhang and Ney, 2009). AMBRA1 is a mitophagy receptor possessing a BH3 domain (Strappazzon et al., 2011; Strappazzon et al., 2015). The C-terminal part of AMBRA1 interacts with BCL-2 and acts as a pro-apoptotic protein upstream of MOMP (Strappazzon et al., 2016). BCL-2 interacts with AMBRA1 and inhibits AMBRA1-induced mitophagy (Strappazzon et al., 2011; Strappazzon et al., 2015). HUWE1 possesses the BH3 domain and mediates MCL1 degradation, while MCL1 delays AMBRA1-dependent mitophagy (Strappazzon et al., 2020). In contrast to these mitophagy receptors harboring the BH3 domain, BCL-RAMBO has not interacted with any of the anti- or pro-apoptotic BCL-2 family proteins examined to date.
Subcellular localization of BCL-RAMBO
BCL-2 family proteins normally reside in or translocate to mitochondria upon apoptotic stimuli. In addition to mitochondria, BCL-2 family proteins manifest different subcellular localizations to the cytosol, endoplasmic reticulum (ER), nuclear outer membrane, nucleus, Golgi apparatus, and peroxisomes (Popgeorgiev et al., 2018). Previous immunostaining studies showed that endogenous BCL-RAMBO co-localizes with CYCS in human glioma LN235 cells (Jensen et al., 2014) and mitochondrial HSP70 in day 3 human embryos (Boumela et al., 2014). The mitochondrial localization of transfected BCL-RAMBO has been revealed by confocal microscopy (Kataoka et al., 2001; Yi et al., 2003; Kim et al., 2012; Murakawa et al., 2015; Nakazawa et al., 2016) and transmission electron microscopy (Jensen et al., 2014). The C-terminal TM domain of BCL-RAMBO is essential for its mitochondrial localization because BCL-RAMBO lacking the TM domain does not co-localize with mitochondrial marker proteins (e.g. SMAC, HSP60, and ATP synthase) or MitoTracker Red in various cell lines (Kataoka et al., 2001; Yi et al., 2003; Kim et al., 2012; Murakawa et al., 2015; Nakazawa et al., 2016). A mitochondrial fractionation study identified BCL-RAMBO as an integral outer membrane protein with N-terminal BH and BHNo domains exposed to the cytosol and a C-terminal TM domain anchored to MOM (Murakawa et al., 2015). Therefore, BCL-RAMBO mainly localizes to MOM with BH and BHNo domains in the cytosol and a C-terminal short tail in the intermembrane space.
BCL-RAMBO-binding proteins
BH domains mediate the interactions of BCL-2 family proteins (Adams and Cory, 2018; Singh et al., 2019; Warren et al., 2019). Immunoprecipitation assays showed that BCL-RAMBO does not interact with seven anti-apoptotic proteins (e.g., BCL-2, BCL-xL, and MCL1) or six pro-apoptotic proteins (e.g., BAX, BAK, and BIM), but possesses four conserved BH motifs (Kataoka et al., 2001). Drosophila possesses two BCL-2 family proteins (Drob-1 and Buffy) (Clavier et al., 2016). Furthermore, BCL-RAMBO does not co-immunoprecipitate with Drob-1 or Buffy (Nakazawa et al., 2016). These findings indicate that BCL-RAMBO does not bind to many BCL-2 family proteins. BCL-RAMBO has so far been shown to bind to ANT, ceramide synthases (CERSs), MAP1LC3/GABARAP, VDAC, unc51-like autophagy-activating kinase 1 (ULK1), phosphoglycerate mutase family member 5 (PGAM5), and SidF (Table 1). These interactions between BCL-RAMBO and its partners appear to regulate apoptosis and mitophagy in mammalian cells (Figures 5A–D).
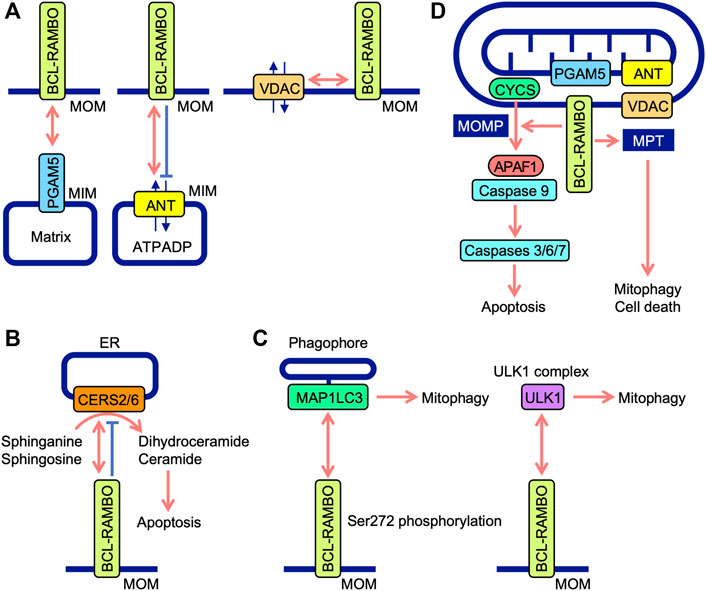
FIGURE 5. Interaction between BCL-RAMBO and BCL-RAMBO-binding proteins. (A) BCL-RAMBO is expressed on mitochondrial outer membranes (MOM). BCL-RAMBO interacts with PGAM5 expressed on the mitochondrial inner membranes (MIM), which is dependent on a transmembrane domain of BCL-RAMBO, but not its C-terminal intermembrane tail. BCL-RAMBO interacts with ANT expressed on the MIM and inhibits ANT activity, which exchanges ATP and ADP. BCL-RAMBO interacts with VDAC expressed on MOM. (B) BCL-RAMBO interacts with CERS2 and CERS6, and inhibits their catalytic activities, thereby inhibiting apoptosis induced by dihydroceramide and ceramide. (C) BCL-RAMBO interacts with MAP1LC3 proteins via its LIR motif. The phosphorylation of BCL-RAMBO at Ser272 is necessary for the interaction with MAP1LC3. BCL-RAMBO interacts with the ULK1 complex, which promotes mitophagy. (D) The BCL-RAMBO-dependent activation of caspases is promoted by PGAM5, ANT, and VDAC. BCL-RAMBO induces the mitochondrial release of CYCS via mitochondrial outer membrane permeability (MOMP). CYCS initiates the APAF1-dependent activation of caspase-9, and subsequent activation of caspases 3, 6, and 7, leading to apoptosis. BCL-RAMBO induces mitochondrial permeability transition (MPT), which causes mitophagy and cell death.
Regulation of apoptosis by BCL-RAMBO
The overexpression of BCL-RAMBO induces apoptosis in the human cancer cell lines, 293T (Kataoka et al., 2001), MCF-7 (Banga et al., 2007), and PC-3 (Kim et al., 2012). BCL-RAMBO promotes taxol-induced cell death in 293A cells and etoposide-induced cell death in HeLa cells (Yi et al., 2003). Wild-type BCL-RAMBO and BCL-RAMBO lacking the N-terminal BH domain, but not BCL-RAMBO lacking the C-terminal TM domain, similarly induces caspase activity by their overexpression in 293T cells (Kataoka et al., 2001). The overexpression of BCL-RAMBO, but not BCL-RAMBO lacking the TM domain, induces apoptosis in PC-3 cells (Kim et al., 2012). These findings indicate that the BHNo domain, but not the BH domain, is required for caspase activation by BCL-RAMBO. This is consistent with BCL-RAMBO not interacting with many anti-apoptotic BCL-2 family members or pro-apoptotic BCL-2 family members (Kataoka et al., 2001). BCL-RAMBO-induced caspase activation occurs independently of death receptor signaling, and, thus, is insensitive to a dominant-negative version of the adaptor protein FADD or the caspase 8 modulator c-FLIPL (Kataoka et al., 2001). Therefore, the BHNo domain of BCL-RAMBO is indispensable for inducing apoptosis signaling in mitochondria.
Previous study demonstrated that BCL-RAMBO induces the release of CYCS from mitochondria, which is not prevented by the caspase inhibitor Z-VAD-fluoromethylketone (Kataoka et al., 2001). Caspase activity induced by BCL-RAMBO is inhibited by IAP proteins (i.e., cIAP1, cIAP2, and XIAP) (Kataoka et al., 2001). These findings suggest that BCL-RAMBO induces MOMP to release CYCS, which triggers the APAF1-dependent activation of caspase 9, leading to caspases 3, 6, and 7 activation and the execution of apoptosis (Figure 5D).
The ANT family of proteins localize to mitochondria, mediate translocation, and exchange ATP and ADP (Zhivotovsky et al., 2009; Brenner et al., 2011). Four ANT isoforms (ANT1, ANT2, ANT3, and ANT4) have different tissue distributions (Brenner et al., 2011). ANT1 and ANT3 are suggested to be pro-apoptotic, while ANT2 and ANT4 are anti-apoptotic (Brenner et al., 2011). These findings indicate that ANT proteins are involved in the regulation of apoptosis. BCL-RAMBO, but not BCL-RAMBO lacking a TM domain, interacts with ANT1 and inhibits ADP/ATP translocation activity in isolated mitochondria (Kim et al., 2012). The knockdown of ANT1 and ANT2 by siRNAs reduces cell death induced by the overexpression of BCL-RAMBO in PC-3 cells (Kim et al., 2012). Consistent with this finding, we showed that BCL-RAMBO physically interacts with ANT1 and ANT2 (Matsubara et al., 2019). BCL-RAMBO induces MPT in isolated mitochondria, which is prevented by bongkrekic acid (Kim et al., 2012). Bongkrekic acid inhibits ANT activity rather than the electron transport chain (Anwar et al., 2017). In addition to MOMP, BCL-RAMBO has been proposed to induce MPT in a manner that is dependent on ANT proteins, leading to the initiation of mitophagy and cell death (Figure 5D).
VDAC proteins, also known as mitochondrial porin, form major channels for small metabolites at MOM, and consist of three isoforms, VDAC1, VDAC2, and VDAC3, in mammals (Mazure, 2017; Magrí et al., 2018). VDAC1 mainly exhibits pro-apoptotic activity, while VDAC2 shows anti-apoptotic activity (Maurya and Mahalakshmi, 2016; Magrí et al., 2018). In Drosophila, the gene porin is widely expressed in adult tissues, while porin2 is exclusively expressed in spermatozoa (Graham and Craigen, 2005; Guarino et al., 2006). Drosophila porin affects the morphology of mitochondria (Park et al., 2010). Comprehensive genetic screening using Drosophila fly lines deficient for the second chromosome showed that human BCL-RAMBO genetically interacts with porin (Matsubara et al., 2019). Glutathione S-transferase-tagged human BCL-RAMBO interacts with VDAC1, albeit to a lesser extent than ANT2 (Matsubara et al., 2019). The co-transfection of VDAC1 or VDAC2 with BCL-RAMBO promotes the cleavage of procaspase 7 and DEVD-hydrolyzing activity in HEK293T cells (Matsubara et al., 2019). These findings indicate that BCL-RAMBO and VDAC cooperatively promote the activation of effector caspases in mammalian cell lines.
Human CERSs comprise six isoforms and catalyze the acylation of sphinganine and sphingosine to convert dihydroceramide and ceramide, respectively (Pewzner-Jung et al., 2006; Wegner et al., 2016). CERS activity mainly localizes to the cytoplasmic leaflet of the ER (Pewzner-Jung et al., 2006). Ceramides are necessary for MOMP and CYCS release (Ueda, 2015). Yeast two-hybrid screening revealed that CERS2 is an interacting partner of BCL-RAMBO (Jensen et al., 2014). BCL-RAMBO binds to CERS2 and CERS6 via the BHNo domain and inhibits their homo- and hetero-dimerization and enzyme activity (Jensen et al., 2014). The overexpression of BCL-RAMBO decreases staurosporine-induced CYCS release, caspase activation, and apoptosis, while its knockdown enhances staurosporine-induced caspase activation and apoptosis (Jensen et al., 2014). Based on these findings, BCL-RAMBO has been proposed to inhibit ceramide-dependent apoptosis by preventing CERS2 and CERS6 and exhibiting anti-apoptotic activity (Figure 5B). The progression of glioblastoma is impaired by the depletion of BCL-RAMBO and enhanced by its overexpression (Jensen et al., 2014). In mouse 3T3-L1 cells cultured under adipogenic and non-differentiation conditions, the knockdown of BCL-RAMBO decreases apoptosis (Fujiwara et al., 2019). In addition to pro-apoptotic activities, BCL-RAMBO exhibits anti-apoptotic activity. Collectively, these findings indicate that BCL-RAMBO exhibits both pro- and anti-apoptotic activities in cellular context-dependent manners.
Regulation of mitophagy by BCL-RAMBO
The screening of the Uniprot database identified BCL-RAMBO as a mammalian Atg32 homologue that shares the WXXI motif, acidic amino acid clusters, and a single TM domain for mitochondrial localization (Murakawa et al., 2015). A yeast two-hybrid assay and pulldown assay revealed that BCL-RAMBO binds to MAP1LC3B via the LIR motif (Murakawa et al., 2015). BCL-RAMBO augments the expression of MAP1LC3B-II, a lipidated form of MAP1LC3B required for the recruitment of phagophores, and promotes mitophagy in a Parkin-independent manner (Murakawa et al., 2015). Carbonyl cyanide 3-chlorophenylhydrazone (CCCP)-induced mitophagy is attenuated by the knockdown of BCL-RAMBO in HEK293A cells (Murakawa et al., 2015). In the Drosophila genetic complementation assay, Atg8a genetically interacts with human BCL-RAMBO (Nakazawa et al., 2016). A subsequent study showed the more selective binding of BCL-RAMBO to MAP1LC3C, GABARAP, and GABARAPL1 than to other MAP1LC3/GABARAP family proteins (Li et al., 2020). Upon transfection in 293A and 293T cells, BCL-RAMBO increases MAP1LC3B-II levels (Murakawa et al., 2015; Hashino et al., 2022). These findings indicate that BCL-RAMBO promotes mitophagy via interactions with MAP1LC3 family proteins. The Ser272 residue of mouse BCL-RAMBO is phosphorylated by its overexpression, while S272A mutants show a reduction in MAP1LC3 binding and MAP1LC3B-II protein levels (Murakawa et al., 2015). BCL-RAMBO is currently proposed to be phosphorylated at Ser272 proximal to the LIR motif and promotes the interaction with MAP1LC3, recruiting phagophores (Figure 5C).
The ULK1 complex consists of ULK1, FAK family kinase-interacting of 200 kDa, ATG13, and ATG101 and initiates the formation of autophagosomes (Lin and Hurley, 2016; Zachari and Ganley, 2017). A recent study showed that BCL-RAMBO co-immunoprecipitates with endogenous ULK1 when transfected in 293 A cells (Murakawa et al., 2019). Endogenous BCL-RAMBO also immunoprecipitates with endogenous ULK1 in CCCP-treated cells (Murakawa et al., 2019). Therefore, BCL-RAMBO appears to recruit the ULK1 complex to MOM for the formation of autophagosomes. In yeast, the interaction of BCL-RAMBO with Atg1 and Atg8 is important for the initiation of BCL-RAMBO-mediated mitophagy (Murakawa et al., 2019). Collectively, these findings demonstrate that Atg8/MAP1LC3 and Atg1/ULK1 proteins are necessary for BCL-RAMBO-mediated mitophagy. BCL-RAMBO interacts with the ULK1 complex and promotes mitophagy (Figure 5C).
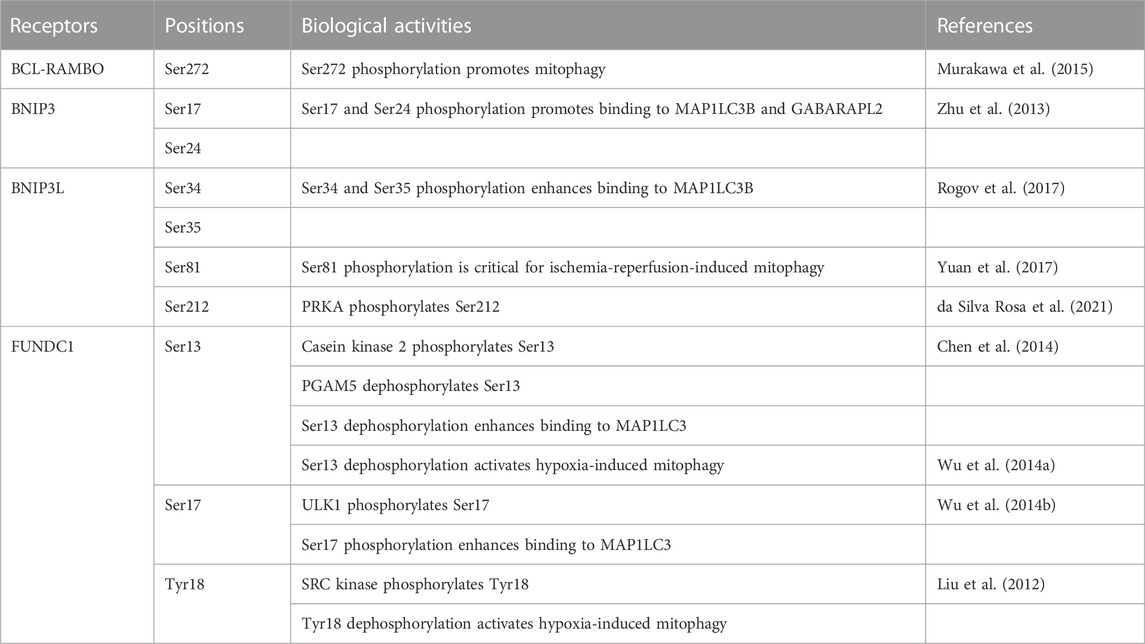
Mitophagy receptors anchored to MOM are regulated by phosphorylation and dephosphorylation (Table 2). Binding to MAP1LC3/GABARAP family proteins is promoted by the phosphorylation of serine residues adjacent to the LIR motif at Ser272 in BCL-RAMBO, Ser17 and Ser24 in BNIP3, Ser34 and Ser35 in BNIP3L, and Ser17 in FUNDC1 (Zhu et al., 2013; Wu et al., 2014b; Murakawa et al., 2015; Rogov et al., 2017), whereas FKBP8 does not possess potential phosphorylated amino acid residues close to the LIR motif (Figure 4C). In contrast, the dephosphorylation of Ser13 and Tyr18 activates FUNDC1-mediated mitophagy in response to hypoxia (Liu et al., 2012; Wu et al., 2014a; Chen et al., 2014). Several protein kinases have been reported to target FUNDC1: casein kinase 2 (CK2) phosphorylates Ser13 (Chen et al., 2014), ULK1 phosphorylates Ser17 (Wu et al., 2014b), and SRC proto-oncogene, non-receptor tyrosine kinase phosphorylates Tyr18 (Liu et al., 2012). Other than MOM-anchored mitophagy receptors, AMBRA1 is phosphorylated by inhibitor of nuclear factor κB kinase subunit α (IKK-α) at Ser1014, which is proximal to the LIR motif WEQL at 1,020–1,023 amino acids (Di Rita et al., 2018). IKK-α-dependent phosphorylation promotes structural changes in AMBRA1 and promotes its interaction with MAP1LC3 (Di Rita et al., 2018). In contrast, PGAM5 dephosphorylates FUNDC1 at Ser13 (Chen et al., 2014). Therefore, the interactions between mitophagy receptors and MAP1LC3/GABARAP family proteins are regulated by phosphorylation and dephosphorylation at LIR and its adjacent regions.
In contrast to mammalian mitophagy receptors, the LIR motif WQAI at 86–89 amino acids of yeast Atg32 does not appear to play a major role in mitophagy (Liu et al., 2014; Furukawa et al., 2019). Ser114 and Ser119 are phosphorylated in yeast Atg32, which mediates the interaction between Atg11 and Atg32 and subsequent mitophagy (Aoki et al., 2011). CK2 phosphorylates Ser114 and Ser119 (Kanki et al., 2013), while the protein phosphatase 2A-like protein phosphatase Ppg1 dephosphorylates Atg32, thereby preventing mitophagy (Furukawa et al., 2018). In the case of BNIP3L, the phosphorylation of Ser81, which is distant from the LIR motif, is essential for BNIP3L-dependent mitophagy (Yuan et al., 2017). Protein kinase cAMP-activated (PRKA) phosphorylates BNIP3L at Ser212 within the C terminus of the TM domain and distal from the LIR motif WVEL at amino acids 36–39 (da Silva Rosa et al., 2021). Other than MOM-anchored mitophagy receptors, PHB2 is phosphorylated by Aurora kinase A at Ser39 distant from the LIR motif YPII at amino acids 121–124, which promotes the interaction between PHB2 and MAP1LC3 (Bertolin et al., 2021). In addition to the LIR motif and its surroundings, mammalian mitophagy receptors are regulated by phosphorylation at other regions distant from the LIR motif to initiate mitophagy.
Regulation of mitochondrial fragmentation by BCL-RAMBO
The BH domain of BCL-RAMBO is responsible for mitochondrial fragmentation in 293A cells, while DRP1 is not required for BCL-RAMBO-induced mitochondrial fragmentation (Murakawa et al., 2015). The overexpression of the BCL-RAMBO S272A mutant induces mitochondrial fragmentation and also reduces MAP1LC3 binding (Murakawa et al., 2015). We showed that the ectopic expression of BCL-RAMBO induces the fragmentation of mitochondria and their perinuclear accumulation in A549 cells (Matsubara et al., 2019). These findings clearly indicate that BCL-RAMBO induces mitochondrial fragmentation via the BH domain, but not the LIR motif. The BH domain of BCL-RAMBO possesses the BH domain consisting of BH1, BH2, BH3, and BH4, in contrast to other mitophagy receptors (BNIP3, BNIP3L, and AMBRA1), which only possess the BH3 domain. Moreover, BCL-RAMBO does not interact with BCL-2 or BCL-xL (Kataoka et al., 2001). Therefore, unlike BNIP3, BNIP3L, and AMBRA1, the BH domain of BCL-RAMBO is unique for the regulation of mitochondrial fragmentation.
Human lung adenocarcinoma A549 cells are more susceptible to the accumulation of fragmented mitochondria by BCL-RAMBO transfection than other cultured cell lines used for subcellular localization (e.g., HeLa, 293A, and PC-3 cells). In A549 cells, the knockdown of three VDAC isoforms (VDAC1, VDAC2, and VDAC3) induces mitochondrial fragmentation, but not their perinuclear accumulation (Matsubara et al., 2019). In Drosophila, porin mutations cause elongated mitochondria, while porin overexpression results in fragmented mitochondria (Park et al., 2010). These findings clearly indicate that VDAC proteins are involved in mitochondrial fragmentation; however, the outcome of the knockdown of VDAC in mammalian cell lines differs from that of VDAC mutations in Drosophila. In mammalian cells, a reduction in VDAC proteins may lead to mitochondrial dysfunction, thereby promoting mitochondrial fragmentation. The MOM-anchored receptors, FIS1, MFF, MiD49, and MiD51 recruit DRP1 to mitochondrial fission sites (Schrader et al., 2012; Rovira-Llopis et al., 2017). Since BCL-RAMBO mediates DRP1-independent mitochondrial fragmentation (Murakawa et al., 2015), it may also promote mitochondrial fragmentation by regulating FIS1, MFF, MiD49, and MiD51 at a step downstream of DRP1.
Regulation of BCL-RAMBO-induced apoptosis and mitophagy by PGAM5
PGAM family proteins possess a conserved PGAM domain that catalyzes mutase and/or phosphatase activities (Sadatomi et al., 2013; Cheng et al., 2021; Liang et al., 2021). PGAM5 lacks mutase activity, but mediates phosphatase activity and plays a regulatory role in mitochondrial dynamics and cell death (Sadatomi et al., 2013; Cheng et al., 2021; Liang et al., 2021). PGAM5 localizes to mitochondria via its N-terminal TM domain (Lo and Hannink, 2008). In mitochondrial compartments, PGAM5 localizes to MIM (Sekine et al., 2012; Lu et al., 2014).
Phenotypic screening using Drosophila deficiency lines identified candidate genes interacting genetically with human BCL-RAMBO. The Pgam5-2 mutant fly line and Pgam 5-2 knockdown fly line suppress the rough eye phenotype induced by BCL-RAMBO (Hashino et al., 2022). Furthermore, PGAM5 modulates the PINK1-mediated pathway in Drosophila (Imai et al., 2010). In human 293T cells, BCL-RAMBO binds to PGAM5, and the BCL-RAMBO TM domain is required for their interaction (Hashino et al., 2022). PGAM5 promotes BCL-RAMBO-induced caspase 3/7 activity, but suppresses the increase of MAP1LC3B-II levels induced by BCL-RAMBO (Hashino et al., 2022). These findings reveal that PGAM5 promotes BCL-RAMBO-induced apoptosis and conversely suppresses BCL-RAMBO-induced mitophagy. BCL-RAMBO localizes to MOM and does not appear to dynamically alter subcellular localization. Phosphorylation and dephosphorylation processes are proposed to regulate BCL-RAMBO-induced apoptosis and mitophagy.
PGAM5 regulates mitochondrial processes by interactions with and the dephosphorylation of multiple proteins. PGAM5 dephosphorylates DRP1 at Ser637 and promotes mitochondrial fragmentation and necrosis (Wang et al., 2012). In contrast, PGAM5 confers protection against necroptosis (Lu et al., 2016). It also interacts with BCL-xL (Lo and Hannick, 2006; Wu et al., 2014a). PGAM5 dephosphorylates BCL-xL at Ser62, restores its binding to BAX and BAK, and provides protection against apoptosis in vinblastine-treated cells (Ma et al., 2020b). Furthermore, PGAM5 regulates Keap1-dependent BCL-xL degradation and exerts anti-apoptotic effects in cardiomyocytes (Yang et al., 2017). In contrast, PGAM5 is required for the activation of BAX and DRP1 dephosphorylation in arenobufagin- and staurosporine-induced apoptosis (Xu et al., 2015). PGAM5 interacts with apoptosis inducing factor and triggers mitophagic cell death, which is inhibited by XIAP (Lenhausen et al., 2016). Therefore, PGAM5 promotes or prevents cell death, including apoptosis and necroptosis, by different mechanisms. In addition to cell death, PGAM5 plays a regulatory role in mitophagy. It dephosphorylates FUNDC1 at Ser13, which promotes mitophagy (Chen et al., 2014). It also interacts with FUNDC1, which requires Syntaxin 17 (Sugo et al., 2018). Moreover, PGAM5 regulates PINK1- and Parkin-mediated mitophagy via DRP1 in CCCP-treated cells (Park et al., 2018). These findings reveal that PGAM5 promotes FUNDC1-dependent mitophagy and PINK1- and Parkin-dependent mitophagy. Ser272 adjacent to the LIR motif of mouse BCL-RAMBO is phosphorylated, which promotes mitophagy (Murakawa et al., 2015). These findings suggest that PGAM5 dephosphorylates BCL-RAMBO at Ser272 to prevent mitophagy; however, PGAM5 may also prevent BCL-RAMBO-mediated mitophagy via other dephosphorylated sites and/or proteins. The protein kinases and protein phosphatases targeting BCL-RAMBO have not yet been identified.
Regulation of adipogenesis by BCL-RAMBO
Brown and beige adipocytes share similar morphological and biochemical characteristics and control systemic energy homeostasis through their thermogenic activity (Harms and Seale, 2013; Rosen and Spiegeman, 2014; Ikeda et al., 2018). In contrast to classical brown adipocytes, beige adipocytes are induced to emerge within white adipose tissues and harbor uncoupling protein 1 (UCP1) (Chouchani et al., 2019). BCL-RAMBO and UCP1 are both increasingly expressed during beige and white adipocyte differentiation (Ju et al., 2018). In response to cold exposure or the β3 adrenergic agonist CL-316,243, the expression of BCL-RAMBO and UCP1 is up-regulated in inguinal white adipose tissue (Ju et al., 2018). The knockdown of BCL-RAMBO inhibits the expression of several genes that are necessary for the browning program during the differentiation of inguinal white adipose tissue (Ju et al., 2018). Therefore, BCL-RAMBO appears to play a novel role in promoting beige adipocyte biogenesis.
Pluripotent mesenchymal stem cells are present in bone marrow stromal cells (BMSCs) and differentiate into different cell types, such as osteoblasts and adipocytes (Chen et al., 2016). BCL-RAMBO expression in BMSCs is higher in an adipogenic culture than in an osteogenic culture (Fujiwara et al., 2019). The knockdown of BCL-RAMBO inhibits the differentiation of adipocytes, oxidative phosphorylation, and apoptosis in mouse 3T3-L1 cells (Fujiwara et al., 2019). These findings indicate that BCL-RAMBO promotes adipogenesis through the regulation of oxidative phosphorylation and apoptosis.
Regulation of bacterial infection by BCL-RAMBO
Programmed cell death plays an essential role in the elimination of cells infected with bacterial pathogens, which target many cellular proteins (Böhme and Rudel, 2009; Ashida et al., 2011). SidF, a substrate of the Dot/Icm transporter system in Legionella pneumophila, confers resistance to apoptosis in L. pneumophila-infected macrophages (Banga et al., 2007). Yeast two-hybrid assays showed that BCL-RAMBO and BNIP3 interact with SidF (Banga et al., 2007). SidF inhibits BCL-RAMBO-induced apoptosis in human breast cancer MCF-7 cells (Banga et al., 2007). In contrast, the L. pneumophila-induced cell death of macrophages is independent of SidF and unaffected by a deficiency in BCL-RAMBO and BNIP3 (Speir et al., 2017). Therefore, the role of BCL-RAMBO in bacterial infection remains controversial and warrants further study.
Expression of BCL-RAMBO in normal cells and cancer cells
Northern blots of normal human tissues showed that human BCL-RAMBO mRNA has multiple splicing forms and is broadly expressed in all tissues tested, while higher expression is observed in the heart, skeletal muscle, and pancreas (Kataoka et al., 2001). Furthermore, human BCL-RAMBO is constitutively expressed during early embryonic development (Boumela et al., 2014). BCL-RAMBO is highly expressed in skeletal muscle (Ávila-Arcos et al., 2019). The content of BCL-RAMBO in skeletal muscle is higher in controls than in sedentary adults, suggesting that chronic exercise maintains BCL-RAMBO levels in skeletal muscle (Arribat et al., 2018). Mitophagy receptors, including BNIP3, BNIP3L, FUNDC1, and BCL-RAMBO, are expressed in adult cardiac progenitor cells, and their expression remains unchanged during differentiation (Lampert et al., 2019). Proteomic profiling showed that BCL-RAMBO is one of the common proteins in human and mouse lens-enriched membrane proteins (Aryal et al., 2020). BCL-RAMBO expression is up-regulated during the browning process of white adipose tissue (Ju et al., 2018) and adipogenic differentiation in mouse ear mesenchymal stem cells and the mouse 3T3-L1 preadipocyte cell line (Fujiwara et al., 2019). Therefore, BCL-RAMBO is widely expressed in adult tissues, and its expression is more abundant in specific tissues, such as skeletal muscle.
BCL-RAMBO protein expression levels differ in human and mouse cancer cell lines (Kataoka et al., 2001). BCL-RAMBO is expressed in various cancers, including childhood acute lymphoblastic leukemia (ALL), gastric cancer, glioblastoma, and acute myeloblastic leukemia (Holleman et al., 2006; Tahir et al., 2010; Yang et al., 2010; Wang et al., 2013; Jensen et al., 2014; Handschuh et al., 2021). In ALL patients, BCL-RAMBO is involved in resistance to L-asparaginase, which is widely used to treat ALL (Holleman et al., 2006). BCL-RAMBO is an independent prognostic factor for ALL (Holleman et al., 2006; Yang et al., 2010) and is associated with a poor clinical outcome for acute myeloid leukemia (Handschuh et al., 2021). In contrast, in a cohort analysis of node-negative breast cancer, BCL-RAMBO expression is associated with a better prognosis (Petry et al., 2010). The BCL-RAMBO transcript is down-regulated in locally advanced rectal cancer that does not respond to chemotherapy (Millino et al., 2017). The down-regulation of BCL-RAMBO is associated with a poor prognosis in clear cell and papillary renal cell carcinoma (Meng et al., 2021b). Collectively, these findings indicate that BCL-RAMBO promotes or prevents the progression of different types of cancers. These opposing roles of BCL-RAMBO are attributed to the different contributions of the pro-apoptotic activity of BCL-RAMBO and its mitophagy-inducing activity counteracting cell death.
Transcriptional regulation of BCL-RAMBO
Several transcription factors have been shown to regulate the transcription of the BCL-RAMBO gene. The Zinc-finger of the cerebellum gene (ZIC) was discovered as the Drosophila melanogaster pair rule gene odd-paired, and ZIC proteins consisting of five tandem C2H2-type zinc fingers regulate gene expression by classical transcription factors and co-factors in mammalian cells (Houtmeyers et al., 2013). Four target genes (i.e. BCL-RAMBO, JunD, Fam57A, and EIF3M) are down-regulated by the knockdown of ZIC1 in liposarcoma cell lines (Brill et al., 2010). The Forkhead box O (FOXO) transcription factor family of proteins are regulated by the insulin signaling pathway and promote target genes involved in various processes, such as cell death and cell cycle arrest (Brown and Webb, 2018; Yadav et al., 2018). Runt-related transcription factor (RUNX) family proteins play a role in development, such as hematopoiesis (Mevel et al., 2019). In a 3D breast epithelial culture, the expression of BCL-RAMBO correlates with FOXO1 at the single-cell level and is controlled by RUNX1 (Wang et al., 2011). A chromatin immunoprecipitation assay revealed that FOXO and RUNX consensus binding sites are present in the promoter region of the BCL-RAMBO gene (Wang et al., 2011). Additional sex comb-like 1 (ASXL1) is a member of the Polycomb group of proteins and mediates transcriptional repression (Katoh, 2013). BCL-RAMBO expression is up-regulated and accompanied by reduced global levels of H3K27me3 and H3K4me3 in mouse Asxl1 −/− hematopoietic stem/precursor cells (Wang et al., 2014). BCL-RAMBO has been identified as one of the 179 genes repressed by the overexpression of anti-silencing function 1B, a conserved histone chaperone, in human MIN6 pancreatic islets (Paul et al., 2016). Limited information is currently available on the transcriptional regulation of BCL-RAMBO; however, the transcription factors responsible for the BCL-RAMBO gene are very important for elucidating the physiological and pathological roles of BCL-RAMBO.
Transcriptional regulation of BCL-RAMBO by miscellaneous experimental conditions
The mRNA expression of BCL-RAMBO has been associated with one of many fluctuating genes by miscellaneous agents. BCL-RAMBO is one of the seven pro-apoptotic genes with expression levels that decrease in human LU-HNSCC-7 squamous cell carcinoma xenografted in nude mice receiving erythropoietin, which is used to treat anemia in cancer patients (Lindgren et al., 2014). These findings indicate that erythropoietin manifests an adverse effect by inhibiting apoptosis in cancer cells. BCL-RAMBO is one of the four apoptotic genes up-regulated by lithocholic acid and bilirubin, which exert deleterious effects on osteoblast functions, in human Saos-2 osteosarcoma cells (Ruiz-Gaspà et al., 2020). BCL-RAMBO is also one of the 13 key genes up-regulated by Periplaneta americana-derived small peptides, which confer resistance to hydrogen peroxide-induced apoptosis in human ovarian granular KGN cells (Wang et al., 2022). TaqMan arrays on apoptotic gene expression revealed that BCL-RAMBO, in addition to BCL-2-binding component 3, is expressed at higher levels in euploid embryos than in aneuploid embryos (Lal et al., 2022). A bioinformatics analysis identified BCL-RAMBO as one of the seven top hub genes down-regulated by an influenza A virus infection (Chen et al., 2022), indicating that BCL-RAMBO is involved in influenza A virus-induced mitophagy. Collectively, these findings show that BCL-RAMBO expression fluctuates to control its pro-apoptotic or mitophagy-inducing activity.
Small-molecule compounds or therapeutic drugs have been reported to affect the transcription of BCL-RAMBO. Cardamonin is a natural compound belonging to the flavonoid family and exhibits anti-cancer activity (Nawaz et al., 2020). BCL-RAMBO is one of the 43 genes down-regulated by cardamonin (1.66 to 6.18-fold) in mouse WEHI-3 leukemia cells (Liao et al., 2019). Cardamonin possesses an α,β-unsaturated carbonyl moiety, which reacts with cysteine residues. Cardamonin targets various cellular proteins, including signaling molecules and transcription factors (Nawaz et al., 2020). Due to the diverse effects of cardamonin, the mechanisms underlying the down-regulation of BCL-RAMBO by cardamonin currently remain unclear. The role of BCL-RAMBO expression in retinal diseases has been examined. Ciprofloxacin is one of the fluoroquinolones currently used as bactericidal antibiotics. It up-regulates BCL-RAMBO expression at the transcriptional level, and this is accompanied by an increase in caspases 3 and 9 in human adult retinal pigment epithelial-19 (ARPE-19) cells and human Müller MIO-M1 cells (Salimiaghdam et al., 2020; Salimiaghdam et al., 2022). Anti-VEGF drugs (i.e., afibercept, ranibizumab, and bevacizumab) also up-regulate the mRNA expression of BCL-RAMBO, together with caspases 3 and 9, in ARPE-19 cells and MIO-M1 cells (Cáceres-del-Carpio et al., 2020; Hamid et al., 2021). These findings suggest that antibiotics or anti-VEGF drugs have a negative impact on retinal diseases by modulating BCL-RAMBO expression.
Regulation of BCL-RAMBO by alternative splicing
Alternative splicing generates multiple transcript variants, which augments the functional diversity of translation products. The human BCL-RAMBO gene is located on chromosome 22q11.21 and is transcribed into many splicing variants. The longest human BCL-RAMBO transcript variant 1 (NM_015367.4) is composed of seven exons and encodes a polypeptide of 485 amino acids (isoform a), which contains all four BH motifs. Human BCL-RAMBO transcript variant 2 (NM_001270726.1) encodes the longest isoform b (a polypeptide of 509 amino acids), which contains the BH1, BH2, and BH3 motifs, but not the BH4 motif. Other transcript variants are shorter than isoform a, mostly due to the lack of exons or alternate exons.
In addition to these transcript variants, transcript variant BCL-RAMBO β, which contains a 98-bp Alu-like sequence, is detected in human adult tissues (e.g., the heart, lymph nodes, and cervix) (Yi et al., 2003). Alu elements are short and repetitive transposable elements that are found in the human genome (Deininger, 2011). The BCL-RAMBO β variant encodes a polypeptide of 104 amino acids, which possesses the BH4 motif, but lacks the BH1, BH2, and BH3 motifs due to the stop codon in the inserted sequence (Yi et al., 2003). The transfection of BCL-RAMBO β and BCL-RAMBO promote etoposide- and taxol-induced cell death (Yi et al., 2003).
Plant homeodomain-finger domain protein 5A (PHF5A) is a component of the splicing factor 3B (SF3B) complex and is involved in the regulation of pluripotency, cell differentiation, and tumorigenesis (Strikoudis et al., 2016; Wang et al., 2019). In human lung adenocarcinoma H1299 and A549 cells, the knockdown of PHF5A or a small molecule inhibitor of PHF5A (pladienolide) induces the alternative splicing of many cell cycle-regulated genes and apoptosis-associated genes, including BCL-RAMBO (Mao et al., 2019).
Circular RNAs (circRNAs) are a new class of RNA molecules that are generated by 3’ to 5’ end-joining as a novel type of alternative splicing, and regulate gene expression by multiple molecular mechanisms (Li et al., 2018). RNA-binding motif protein 20 (RBM20) is an RNA-binding protein with two zinc finger domains that regulates heart-specific alternative splicing (Watanabe et al., 2018). Thirty-eight circRNAs are differently expressed, 26 of which are down-regulated and 12 are up-regulated, in the hearts of RBM20-deficient mice and wild-type mice, and circBcl2l13 is one of the up-regulated circRNAs (Aufiero et al., 2018). The regulation of BCL-RAMBO expression by circRNAs is discussed in the next section.
Regulation of BCL-RAMBO expression by microRNAs (miRNAs)
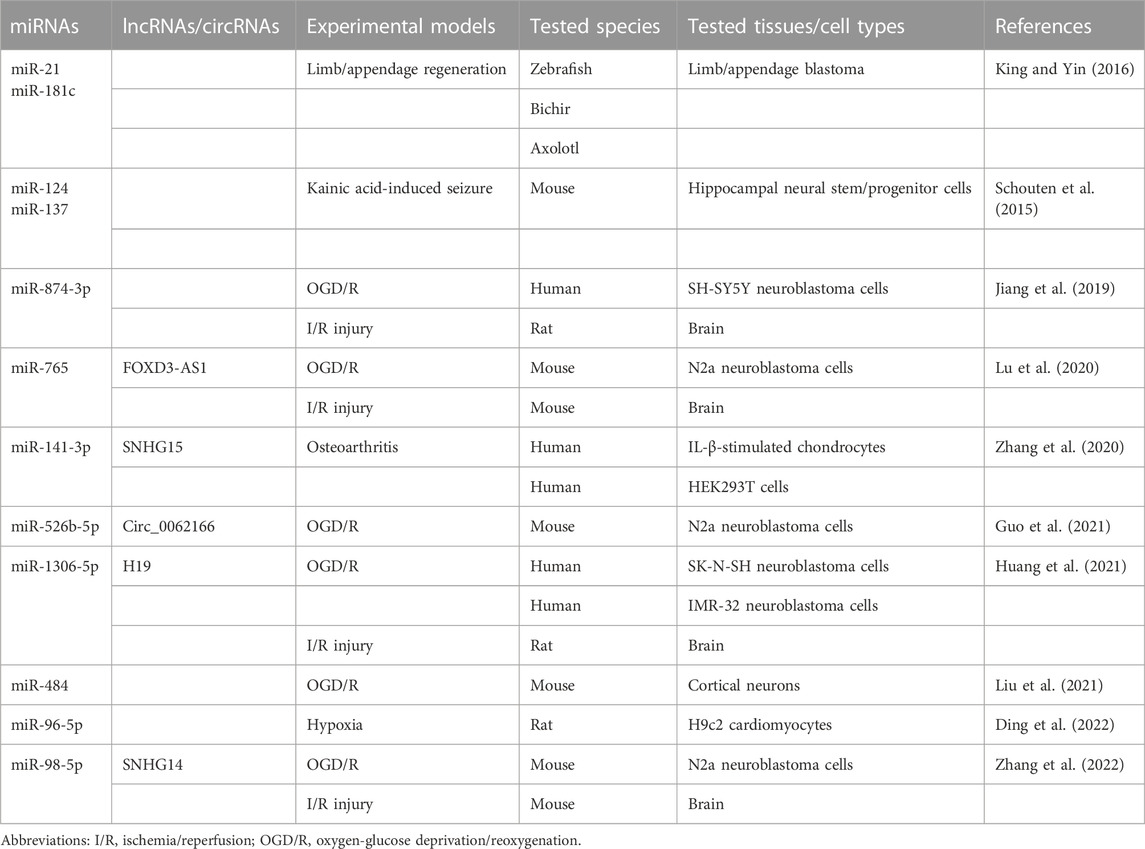
miRNAs are approximately 22-nt RNAs that play a critical role in the repression of their targets (Bartel, 2018). In several experimental models, miRNAs have been reported to module the expression of BCL-RAMBO (Table 3). The up-regulation of miR-21 down-regulates three putative novel target genes (bcl2l13, rgs5, and chka) in addition to two known target genes (pdcd4 and tgfbr2) during limp/appendage blastema formation in Zebrafish, bichir, and axolotl, which provide highly regenerative model systems (King and Yin, 2016). These studies indicate that BCL-RAMBO expression is negatively regulated by multiple miRNAs in vertebrates, including mammals.
The regulation of BCL-RAMBO expression by miRNAs plays a critical role in several experimental disease models. BCL-RAMBO promotes caspase 3 activity and CYCS release in mouse neural stem/progenitor cells (Schouten et al., 2015). The expression of miR-124 and miR-137 is up-regulated in a kainic acid-induced seizure model, which cooperatively reduces endogenous BCL-RAMBO expression and controls caspase 3 activity (Schouten et al., 2015). Cerebral ischemia/reperfusion (I/R) injury is a serious clinical condition that induces the production of reactive oxygen species and ultimately triggers cell death pathways (Wu et al., 2018). In addition to in vivo I/R injury animal models, the oxygen-glucose deprivation and reoxygenation (OGD/R) assay system has been used to examine the effects of miRNAs on BCL-RAMBO expression in in vitro cell cultures. The expression levels of miR-874-3p, miR-765, mi-526b-5p, miR-1306-5p, and miR-98-5p are reduced in OGD/R-induced neuroblastoma cells, which conversely up-regulates BCL-RAMBO expression, thereby promoting pathogenic changes, including apoptosis (Jiang et al., 2019; Lu et al., 2020; Guo et al., 2021; Huang et al., 2021; Zhang et al., 2022). The expression of miR-484 is down-regulated in mouse cortical neurons exposed to OGD/R, which promotes the expression of BCL-RAMBO (Liu et al., 2021). The expression of miR-141-3p down-regulates that of BCL-RAMBO in human IL-1β-stimulated chondrocytes (Zhang et al., 2020). In acute myocardial infarction associated with coronary artery disease, the expression of miR-96-5p is down-regulated, whereas that of BCL-RAMBO is up-regulated (Ding et al., 2022). Based on the pro-apoptotic effects of BCL-RAMBO, these findings show that the suppression of BCL-RAMBO expression is beneficial for therapeutic interventions.
Long non-coding RNAs (lncRNAs) and/or circRNAs have been reported to regulate BCL-RAMBO expression by targeting miRNAs. SNHG15 promotes BCL-RAMBO expression by sponging miR-141-3p in osteoarthritis cartilage tissue (Zhang et al., 2020). In I/R injury and ODG/R, lncRNAs (i.e., FOXD3-AS1, H19, and SNHG14) are up-regulated and promote the expression of BCL-RAMBO by sponging their target miRNAs (Lu et al., 2020; Huang et al., 2021; Zhang et al., 2022). Circ_0062166, generated from exon 3 of BCL-RAMBO, is up-regulated following OGD/R and regulates BCL-RAMBO expression by sequestering miR-526b-5p (Guo et al., 2021). These studies revealed that BCL-RAMBO expression is mainly regulated by the lncRNA/miRNA axis or circRNA/miRNA axis in I/R injury and osteoarthritis.
A Drosophila model expressing human BCL-RAMBO
To the best of our knowledge, BCL-RAMBO homologues have not yet been reported in Drosophila. Human BCL-RAMBO induces apoptosis in Drosophila Schneider 2 cells by triggering the release of CYCS and caspase activation through its overexpression (Nakazawa et al., 2016). The ectopic expression of human BCL-RAMBO in Drosophila by the GAL4-UAS system manifests aberrant morphological changes, which are accompanied by the atrophied wing, split thorax, and rough eye phenotypes (Nakazawa et al., 2016). Among these changes, the rough eye phenotype induced by BCL-RAMBO is severe and characterized with a reduction in eye size, the loss of ommatidia and bristles, and the loss of pigmentation (Nakazawa et al., 2016). Caspase activation also occurs in eye imaginal discs in Drosophila expressing BCL-RAMBO (Nakazawa et al., 2016). The loss of pigmentation induced by the ectopic expression of BCL-RAMBO is partly suppressed by the co-expression of Diap1 and Diap2 as well as ANT mutations (Nakazawa et al., 2016). This is consistent with previous findings showing that BCL-RAMBO-induced caspase activity is suppressed by IAP proteins in 293T cells (Kataoka et al., 2001) and also that ANT is necessary for BCL-RAMBO-induced cell death (Kim et al., 2012). To identify BCL-RAMBO-interacting partner genes, human BCL-RAMBO-expressing fly lines have been crossed with fly lines with the deletion of or mutations in specific genes. Comprehensive screening using fly lines deficient for the second chromosome revealed that the BCL-RAMBO-induced rough eye phenotype is also partly suppressed by mutations in or the knockdown of porin and pgam5-2 (Nakazawa et al., 2016; Matsubara et al., 2019; Hashino et al., 2022). The genetic interaction of BCL-RAMBO with Porin or Pgam5 has been confirmed in human cultured cell lines. Therefore, the humanized Drosophila model expressing BCL-RAMBO may be a useful tool for identifying unknown partner genes.
Conclusion and perspectives
BCL-RAMBO was initially identified as one of the BCL-2 family proteins inducing apoptosis (Kataoka et al., 2001). Approximately 15 years later, BCL-RAMBO has been shown to function as a mammalian Atg32 homologue that regulates mitophagy and mitochondrial fragmentation (Murakawa et al., 2015). In the 20 years since its first discovery, many important findings on the biological properties of BCL-RAMBO have been increasingly reported. BCL-RAMBO is characterized by one to four BH motifs and the LIR motif, and is anchored to MOM. BCL-RAMBO interacts with multiple proteins, including ANT, VDAC, MAP1LC3/GABARAP, ULK1, PGAM5, and CERS, thereby regulating cellular processes, including apoptosis, mitochondrial fragmentation, and mitophagy. BCL-RAMBO expression is regulated by multiple transcription factors, miRNAs, and lncRNAs. BCL-RAMBO is expressed in normal and cancer cells and has been implicated in pathogenic conditions, such as tumorigenesis and I/R injury. Nevertheless, the molecular mechanisms that link BCL-RAMBO to its binding partners, the regulation of its expression, and its physiological and pathological roles have not yet been elucidated in detail. Therefore, future studies are needed to address unanswered questions on BCL-RAMBO and other mitophagy receptors.
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
This work was partly supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number 19H02885.
Acknowledgments
The author is grateful to Takuya Hashino and Jinghong Xu for the critical reading of this review.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adams, J. M., and Cory, S. (2018). The BCL-2 arbiters of apoptosis and their growing role as cancer targets. Cell Death Differ. 25, 27–36. doi:10.1038/cdd.2017.161
Anwar, M., Kasper, A., Steck, A. R., and Schier, J. G. (2017). Bongkrekic acid–a review of a lesser-known mitochondrial toxin. J. Med. Toxicol. 13, 173–179. doi:10.1007/s13181-016-0577-1
Aoki, Y., Kanki, T., Hirota, Y., Kurihara, Y., Saigusa, T., Uchiumi, T., et al. (2011). Phosphorylation of serine 114 on Atg32 mediates mitophagy. Mol. Biol. Cell 22, 3206–3217. doi:10.1091/mbc.E11-02-0145
Arribat, Y., Broskey, N. T., Greggio, C., Boutant, M., Alonso, S. C., Kulkarni, S. S., et al. (2018). Distinct patterns of skeletal muscle mitochondria fusion, fission and mitophagy upon duration of exercise training. Acta Physiol. 225, e13179. doi:10.1111/apha.13179
Aryal, S., Anand, D., Hernandez, F. G., Weaterbee, B. A., Huang, H., Reddy, A. P., et al. (2020). MS/MS in silico subtraction-based proteomic profiling as an approach to facilitate disease gene discovery: application to lens development and cataract. Hum. Genet. 139, 151–184. doi:10.1007/s00439-019-02095-5
Ashida, H., Mimuro, H., Ogawa, M., Kobayashi, T., Sanada, T., Kim, M., et al. (2011). Cell death and infection: A double-edged sword for host and pathogen survival. J. Cell Biol. 195, 931–942. doi:10.1083/jcb.201108081
Ashrafi, G., and Schwarz, T. L. (2013). The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 20, 31–42. doi:10.1038/cdd.2012.81
Aufiero, S., van den Hoogenhof, M. M. G., Reckman, Y. J., Beqqali, A., van der Made, I., Kluin, J., et al. (2018). Cardiac circRNAs arise mainly from constitutive exons rather than alternatively spliced exons. RNA 24, 815–827. doi:10.1261/rna.064394.117
Ávila-Arcos, M., McManus, K. F., Sandoval, K., Rodríguez-Rodríguez, J. E., Villa-Islas, V., Martin, A. R., et al. (2019). Population history and gene divergence in native Mexicans inferred from 76 human exomes. Mol. Biol. Evol. 37, 994–1006. doi:10.1093/molbev/msz282
Baines, C. P., Kaiser, R. A., Sheiko, T., Craigen, W. J., and Molkentin, J. D. (2007). Voltage-dependent anion channels are dispensable for mitochondrial-dependent cell death. Nat. Cell Biol. 9, 550–555. doi:10.1038/ncb1575
Banga, S., Gao, P., Shen, X., Fiscus, V., Zong, W. X., Chen, L., et al. (2007). Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc. Natl. Acad. Sci. U. S. A. 104, 5121–5126. doi:10.1073/pnas.0611030104
Bertolin, G., Alves-Guerra, M. C., Cheron, A., Burel, A., Prigent, C., Le Borgne, R., et al. (2021). Mitochondrial Aurora kinase A induces mitophagy by interacting with MAP1LC3 and Prohibitin 2. Life Sci. Alliance 4, e202000806. doi:10.26508/lsa.202000806
Birgisdottir, Å. B., Lamark, T., and Johansen, T. (2013). The LIR motif – crucial for selective autophagy. J. Cell Sci. 126, 3237–3247. doi:10.1242/jcs.126128
Bock, F. J., and Tait, S. W. G. (2020). Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol. 21, 85–100. doi:10.1038/s41580-019-0173-8
Böhme, L., and Rudel, T. (2009). Host cell death machinery as a target for bacterial pathogens. Microbes Infect. 11, 1063–1070. doi:10.1016/j.micinf.2009.08.014
Bonora, M., Giorgi, C., and Pinton, P. (2022). Molecular mechanisms and consequences of mitochondrial permeability transition. Nat. Rev. Mol. Cell Biol. 23, 266–285. doi:10.1038/s41580-021-00433-y
Boumela, I., Assou, S., Haouzi, D., Déchaud, H., Aït-Ahmed, O., and Hamamah, S. (2014). Developmental regulated expression of anti- and pro-apoptotic BCL-2 family genes during human early embryonic development. Curr. Med. Chem. 21, 1361–1369. doi:10.2174/09298673113206660278
Bratton, S. B., and Salvesen, G. S. (2010). Regulation of the Apaf-1–caspase-9 apoptosome. J. Cell Sci. 123, 3209–3214. doi:10.1242/jcs.073643
Brenner, C., Subramaniam, K., Pertuiset, C., and Pervaiz, S. (2011). Adenine nucleotide translocase family: Four isoforms for apoptosis modulation in cancer. Oncogene 30, 883–895. doi:10.1038/onc.2010.501
Brill, E., Gobble, R., Angeles, C., Lagos-Quintana, M., Crago, A., Laxa, B., et al. (2010). ZIC1 overexpression is oncogenic in liposarcoma. Cancer Res. 70, 6891–6901. doi:10.1158/0008-5472.CAN-10-0745
Brown, A. K., and Webb, A. E. (2018). Regulation of FOXO factors in mammalian cells. Curr. Top. Dev. Biol. 127, 165–192. doi:10.1016/bs.ctdb.2017.10.006
Burté, F., Carelli, V., Chinnery, P. F., and Yu-Wai-Man, P. (2015). Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat. Rev. Neurol. 11, 11–24. doi:10.1038/nrneurol.2014.228
Cáceres-del-Carpio, J., Moustafa, M. T., Toledo-Corral, J., Hamid, M. A., Atilano, S. R., Schneider, K., et al. (2020). In vitro response and gene expression of human retinal Müller cells treated with different anti-VEGF drugs. Exp. Eye Res. 191, 107903. doi:10.1016/j.exer.2019.107903
Calle, X., Garrido-Moreno, V., Lopez-Gallardo, E., Norambuena-Soto, I., Martínez, D., Peñaloza-Otárola, A., et al. (2022). Mitochondrial E3 ubiquitin ligase 1 (MUL1) as a novel therapeutic target for diseases associated with mitochondrial dysfunction. IUBMB Life 74, 850–865. doi:10.1002/iub.2657
Chen, G., Han, Z., Feng, D., Chen, Y., Chen, L., Wu, H., et al. (2014). A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell 54, 362–377. doi:10.1016/j.molcel.2014.02.034
Chen, G., Li, H., Hao, M., Li, X., Dong, Y., Zhang, Y., et al. (2022). Identification of critical genes and pathways for influenza A virus infections via bioinformatics analysis. Viruses 14, 1625. doi:10.3390/v14081625
Chen, Q., Shou, P., Zheng, C., Jiang, M., Cao, G., Yang, Q., et al. (2016). Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 23, 1128–1139. doi:10.1038/cdd.2015.168
Chen, Z., Liu, L., Cheng, Q., Li, Y., Wu, H., Zhang, W., et al. (2017). Mitochondrial E3 ligase MARCH5 regulates FUNDC1 to fine-tune hypoxic mitophagy. EMBO Rep. 18, 495–509. doi:10.15252/embr.201643309
Cheng, M., Lin, N., Dong, D., Ma, J., Su, J., and Sun, L. (2021). PGAM5: A crucial role in mitochondrial dynamics and programmed cell death. Eur. J. Cell Biol. 100, 151144. doi:10.1016/j.ejcb.2020.151144
Chouchani, E. T., Kazak, L., and Spiegelman, B. M. (2019). New advances in adaptive thermogenesis UCP1 and beyond. Cell Metab. 29, 27–37. doi:10.1016/j.cmet.2018.11.002
Clavier, A., Rincheval-Arnold, A., Colin, J., Mignotte, B., and Guénal, I. (2016). Apoptosis in Drosophila: which role for mitochondrial? Apoptosis 21, 239–251. doi:10.1007/s10495-015-1209-y
Cosentino, K., and García-Sáez, A. J. (2017). Bax and Bak pores: Are we closing the circle? Trends Cell Biol. 27, 266–275. doi:10.1016/j.tcb.2016.11.004
da Silva Rosa, S. C., Martens, M. D., Field, J. T., Nguyen, L., Kereliuk, S. M., Hai, Y., et al. (2021). BNIP3L/Nix-induced mitochondrial fission, mitophagy, and impaired myocyte glucose uptake are abrogated by PRKA/PKA phosphorylation. Autophagy 17, 2257–2272. doi:10.1080/15548627.2020.1821548
De, R., Mazumder, S., and Bandyopadhyay, U. (2021). Mediators of mitophagy that regulate mitochondrial quality control play crucial role in diverse pathophysiology. Cell Biol. Toxicol. 37, 333–366. doi:10.1007/s10565-020-09561-1
Deininger, P. (2011). Alu elements: know the SINEs. Genome Biol. 12, 236. doi:10.1186/gb-2011-12-12-236
Di Rita, A., Peschiaroli, A., D´Acunzo, P., Strobbe, D., Hu, Z., Gruber, J., et al. (2018). HUWE1 E3 ligase promotes PINK1/PARKIN-independent mitophagy by regulating AMBRA1 activation via IKKα. Nat. Commun. 9, 3755. doi:10.1038/s41467-018-05722-3
Ding, H., Chen, W., and Chen, X. (2022). Serum miR-96-5p is a novel and non-invasive marker of acute myocardial infarction associated with coronary artery disease. Bioengineered 13, 3930–3943. doi:10.1080/21655979.2022.2031392
Dudek, J. (2017). Role of cardiolipin in mitochondrial signaling pathways. Front. Cell Dev. Biol. 5, 90. doi:10.3389/fcell.2017.00090
Fujiwara, M., Tian, L., Le, P. T., DeMambro, V. E., Becker, K. A., Rosen, C. J., et al. (2019). The mitophagy receptor Bcl-2-like protein 13 stimulates adipogenesis by regulating mitochondrial oxidative phosphorylation and apoptosis in mice. J. Biol. Chem. 294, 12683–12694. doi:10.1074/jbc.RA119.008630
Fulda, S., and Vucic, D. (2012). Targeting IAP proteins for therapeutic intervention in cancer. Nat. Rev. Drug Discov. 11, 109–124. doi:10.1038/nrd3627
Furukawa, K., Fukuda, T., Yamashita, S., Saigusa, T., Kurihara, Y., Yoshida, Y., et al. (2018). The PP2A-like protein phosphatase Ppg1 and the Far complex cooperatively counteract CK2-mediated phosphorylation of Atg32 to inhibit mitophagy. Cell Rep. 23, 3579–3590. doi:10.1016/j.celrep.2018.05.064
Furukawa, K., Innokentev, A., and Kanki, T. (2019). Regulatory mechanisms of mitochondrial autophagy: Lessons from yeast. Front. Plant Sci. 10, 1479. doi:10.3389/fpls.2019.01479
Graham, B. H., and Craigen, W. J. (2005). Mitochondrial voltage-dependent anion channel gene family in Drosophila melanogaster: Complex patterns of evolution, genomic organization, and developmental expression. Mol. Genet. Metab. 85, 308–317. doi:10.1016/j.ymgme.2005.03.009
Green, D. R., Galluzzi, L., and Kroemer, G. (2014). Cell biology. Metabolic control of cell death. Science 345, 1250256. doi:10.1126/science.1250256
Green, D. R., Galluzzi, L., and Kroemer, G. (2011). Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science 333, 1109–1112. doi:10.1126/science.1201940
Guarino, F., Specchia, V., Zapparoli, G., Messina, A., Aiello, R., Bozzetti, M. P., et al. (2006). Expression and localization in spermatozoa of the mitochondrial porin isoform 2 in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 346, 665–670. doi:10.1016/j.bbrc.2006.05.172
Guo, Z., Zhang, G., Xu, B., Zhang, C., Tao, A., He, M., et al. (2021). Circ_0062166 aggravates cerebral ischemia-reperfusion injury through targeting miR-526b-5p/BCL2L13 axis. Brain Inj. 35, 1245–1253. doi:10.1080/02699052.2021.1972143
Gurung, P., Lukens, J. R., and Kanneganti, T. D. (2015). Mitochondria: Diversity in the regulation of the NLRP3 inflammasome. Trends Mol. Med. 21, 193–201. doi:10.1016/j.molmed.2014.11.008
Gustafsson, Å. B., and Dorn, G. W. (2019). Evolving and expanding the roles of mitophagy as a homeostatic and pathogenic process. Physiol. Rev. 99, 853–892. doi:10.1152/physrev.00005.2018
Hamacher-Brady, A., and Brady, N. R. (2016). Mitophagy programs: Mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell. Mol. Life Sci. 73, 775–795. doi:10.1007/s00018-015-2087-8
Hamid, M. A., Moustafa, M. T., Nashine, S., Costa, R. D., Schneider, K., Atilano, S. R., et al. (2021). Anti-VEGF drugs influence epigenetic regulation and AMD-specific molecular markers in ARPE-19 cells. Cells 10, 878. doi:10.3390/cells10040878
Handschuh, L., Wojciechowski, P., Kazmierczak, M., and Lewandowski, K. (2021). Transcript-level dysregulation of BCL2 family genes in acute myeloblastic leukemia. Cancers 13, 3175. doi:10.3390/cancers13133175
Harms, M., and Seale, P. (2013). Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19, 1252–1263. doi:10.1038/nm.3361
Hashino, T., Matsubara, H., Xu, J., Tanaka, R., Kusagawa, E., Ueda, Y., et al. (2022). PGAM5 interacts with Bcl-rambo and regulates apoptosis and mitophagy. Exp. Cell Res. 420, 113342. doi:10.1016/j.yexcr.2022.113342
Holleman, A., den Boer, M. L., de Menezes, R. X., Cheok, M. H., Cheng, C., Kazemier, K. M., et al. (2006). The expression of 70 apoptosis genes in relation to lineage, genetic subtype, cellular drug resistance, and outcome in childhood acute lymphoblastic leukemia. Blood 107, 769–776. doi:10.1182/blood-2005-07-2930
Hoshino, A., Wang, W., Wada, S., McDermott-Roe, C., Evans, C. S., Gosis, B., et al. (2019). The ADP/ATP translocase drives mitophagy independent of nucleotide exchange. Nature 575, 375–379. doi:10.1038/s41586-019-1667-4
Houtmeyers, R., Souopgui, J., Tejpar, S., and Arkell, R. (2013). The ZIC gene family encodes multi-functional proteins essential for patterning and morphogenesis. Cell. Mol. Life Sci. 70, 3791–3811. doi:10.1007/s00018-013-1285-5
Huang, Y., Deng, L., Zeng, L., Bao, S., Ye, K., Li, C., et al. (2021). Silencing of H19 alleviates oxygen–glucose deprivation/reoxygenation-triggered injury through the regulation of the miR-1306-5p/BCL2L13 axis. Metab. Brain Dis. 36, 2461–2472. doi:10.1007/s11011-021-00822-4
Ikeda, K., Maretich, P., and Kajimura, S. (2018). The common and distinct features of Brown and beige adipocytes. Trends Endocrinol. Metab. 29, 191–200. doi:10.1016/j.tem.2018.01.001
Imai, Y., Kanao, T., Sawada, T., Kobayashi, Y., Moriwaki, Y., Ishida, Y., et al. (2010). The loss of PGAM5 suppresses the mitochondrial degeneration caused by inactivation of PINK1 in Drosophila. PLoS Genet. 6, e1001229. doi:10.1371/journal.pgen.1001229
Javadov, S., Jang, S., Parodi-Rullán, R., Khuchua, Z., and Kuznetsov, A. (2017). Mitochondrial permeability transition in cardiac ischemia–reperfusion: whether cyclophilin D is a viable target for cardioprotection? Cell. Mol. Life Sci. 74, 2795–2813. doi:10.1007/s00018-017-2502-4
Jensen, S. A., Calvert, A. E., Volpert, G., Kouri, F. M., Hurley, L. A., Luciano, J. P., et al. (2014). Bcl2L13 is a ceramide synthase inhibitor in glioblastoma. Proc. Natl. Acad. Sci. U. S. A. 111, 5682–5687. doi:10.1073/pnas.1316700111
Jiang, D., Sun, X., Wang, S., and Man, H. (2019). Upregulation of miR-874-3p decreases cerebral ischemia/reperfusion injury by directly targeting BMF and BCL2L13. Biomed. Pharmacother. 117, 108941. doi:10.1016/j.biopha.2019.108941
Johansen, T., and Lamark, T. (2020). Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J. Mol. Biol. 432, 80–103. doi:10.1016/j.jmb.2019.07.016
Ju, L., Chen, S., Alimujiang, M., Bai, N., Yan, H., Fang, Q., et al. (2018). A novel role for Bcl2l13 in promoting beige adipocyte biogenesis. Biochem. Biophys. Res. Commun. 506, 485–491. doi:10.1016/j.bbrc.2018.10.034
Julien, O., and Wells, J. A. (2017). Caspases and their substrates. Cell Death Differ. 24, 1380–1389. doi:10.1038/cdd.2017.44
Kale, J., Osterlund, E. J., and Andrews, D. W. (2018). BCL-2 family proteins: changing partners in the dance towards death. Cell Death Differ. 25, 65–80. doi:10.1038/cdd.2017.186
Kalkavan, H., and Green, D. R. (2018). MOMP, cell suicide as a BCL-2 family business. Cell Death Differ. 25, 46–55. doi:10.1038/cdd.2017.179
Kanki, T., Kurihara, Y., Jin, X., Goda, T., Ono, Y., Aihara, M., et al. (2013). Casein kinase 2 is essential for mitophagy. EMBO Rep. 14, 788–794. doi:10.1038/embor.2013.114
Kantari, C., and Walczak, H. (2011). Caspase-8 and Bid: Caught in the act between death receptors and mitochondria. Biochim. Biophys. Acta 1813, 558–563. doi:10.1016/j.bbamcr.2011.01.026
Kao, S. H., Wu, H. T., and Wu, K. J. (2018). Ubiquitination by HUWE1 in tumorigenesis and beyond. J. Biomed. Sci. 25, 67. doi:10.1186/s12929-018-0470-0
Kataoka, T., Holler, N., Micheau, O., Martinon, F., Tinel, A., Hofmann, K., et al. (2001). Bcl-rambo, a novel Bcl-2 homologue that induces apoptosis via its unique C-terminal extension. J. Biol. Chem. 276, 19548–19554. doi:10.1074/jbc.M010520200
Katoh, M. (2013). Functional and cancer genomics of ASXL family members. Br. J. Cancer 109, 299–306. doi:10.1038/bjc.2013.281
Kaufmann, T., Schlipf, S., Sanz, J., Neubert, K., Stein, R., and Borner, C. (2003). Characterization of the signal that directs Bcl-xL, but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol. 160, 53–64. doi:10.1083/jcb.200210084
Kaufmann, T., Strasser, A., and Jost, P. J. (2012). Fas death receptor signalling: Roles of Bid and XIAP. Cell Death Differ. 19, 42–50. doi:10.1038/cdd.2011.121
Kim, J. Y., So, K. J., Lee, S., and Park, J. H. (2012). Bcl-rambo induces apoptosis via interaction with the adenine nucleotide translocator. FEBS Lett. 586, 3142–3149. doi:10.1016/j.febslet.2012.08.015
King, B. L., and Yin, V. P. (2016). A conserved microRNA regulatory circuit is differentially controlled during limb/appendage regeneration. PLoS ONE 11, e0157106. doi:10.1371/journal.pone.0157106
Kokoszka, J. E., Waymire, K. G., Levy, S. E., Sligh, J. E., Cai, J., Jones, D. P., et al. (2004). The ADP/ATP translocator is not essential for the mitochondrial permeability transition pore. Nature 427, 461–465. doi:10.1038/nature02229
Kraus, F., and Ryan, M. T. (2017). The constriction and scission machineries involved in mitochondrial fission. J. Cell Sci. 130, 2953–2960. doi:10.1242/jcs.199562
Krauskopf, A., Eriksson, O., Craigen, W. J., Forte, M. A., and Bernardi, P. (2006). Properties of the permeability transition in VDAC1-/- mitochondria. Biochim. Biophys. Acta 1757, 590–595. doi:10.1016/j.bbabio.2006.02.007
Kwong, J. Q., and Molkentin, J. D. (2015). Physiological and pathological roles of the mitochondrial permeability transition pore in the heart. Cell Metab. 21, 206–214. doi:10.1016/j.cmet.2014.12.001
Lahiri, V., Hawkins, W. D., and Klionsky, D. J. (2019). Watch what you (self-) eat: Autophagic mechanisms that modulate metabolism. Cell Metab. 29, 803–826. doi:10.1016/j.cmet.2019.03.003
Lal, A., Kranyak, A., Blalock, J., Athavale, D., Barré, A., Doran, A., et al. (2022). Apoptotic qPCR gene expression array analysis demonstrates proof-of-concept for rapid blastocoel fluid-conditioned media molecular prediction. J. Assist. Reprod. Genet. 39, 1515–1522. doi:10.1007/s10815-022-02510-3
Lampert, M. A., Orogo, A. M., Najor, R. H., Hammerling, B. C., Jeon, L. J., Wang, B. J., et al. (2019). BNIP3L/NIX and FUNDC1-mediated mitophagy is required for mitochondrial network remodeling during cardiac progenitor cell differentiation. Autophagy 7, 1182–1198. doi:10.1080/15548627.2019.1580095
Lavrik, I. N., and Krammer, P. H. (2012). Regulation of CD95/Fas signaling at the DISC. Cell Death Differ. 19, 36–41. doi:10.1038/cdd.2011.155
Lenhausen, A. M., Wilkinson, A. S., Lewis, E. M., Dailey, K. M., Scott, A. J., Khan, S., et al. (2016). Apoptosis inducing factor binding protein PGAM5 triggers mitophagic cell death that is inhibited by the ubiquitin ligase activity of X-linked inhibitor of apoptosis. Biochemistry 55, 3285–3302. doi:10.1021/acs.biochem.6b00306
Li, M., Jia, J., Zhang, X., and Dai, H. (2020). Selective binding of mitophagy receptor protein Bcl-rambo to LC3/GABARAP family proteins. Biochem. Biophys. Res. Commun. 530, 292–300. doi:10.1016/j.bbrc.2020.07.039
Li, X., Yang, L., and Chen, L. L. (2018). The biogenesis, functions, and challenges of circular RNAs. Mol. Cell 71, 428–442. doi:10.1016/j.molcel.2018.06.034
Liang, M. Z., Ke, T. L., and Chen, L. (2021). Mitochondrial protein PGAM5 emerges as a new regulator in neurological diseases. Front. Mol. Neurosci. 14, 730604. doi:10.3389/fnmol.2021.730604
Liao, N. C., Shih, Y. L., Chou, J. S., Chen, K. W., Chen, Y. L., Lee, M. H., et al. (2019). Cardamonin induces cell cycle arrest, apoptosis and alters apoptosis associated gene expression in WEHI-3 mouse leukemia cells. Am. J. Chin. Med. 47, 635–656. doi:10.1142/S0192415X19500332
Lin, M. G., and Hurley, J. H. (2016). Structure and function of the ULK1 complex in autophagy. Curr. Opin. Cell Biol. 39, 61–68. doi:10.1016/j.ceb.2016.02.010
Lindgren, G., Ekblad, L., Vallon-Christersson, J., Kjellén, E., Gebre-Medhin, M., and Wennerberg, J. (2014). Erythropoietin suppresses the activation of pro-apoptotic genes in head and neck squamous cell carcinoma xenografts exposed to surgical trauma. BMC Cancer 14, 648. doi:10.1186/1471-2407-14-648
Liu, L., Feng, D., Chen, G., Chen, M., Zheng, Q., Song, P., et al. (2012). Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 14, 177–185. doi:10.1038/ncb2422
Liu, L., Sakakibara, K., Chen, Q., and Okamoto, K. (2014). Receptor-mediated mitophagy in yeast and mammalian systems. Cell Res. 24, 787–795. doi:10.1038/cr.2014.75
Liu, X., Wang, X., Zhang, L., Zhou, Y., Yang, L., and Yang, M. (2021). By targeting apoptosis facilitator BCL2L13, microRNA miR-484 alleviates cerebral ischemia/reperfusion injury-induced neuronal apoptosis in mice. Bioengineered 12, 948–959. doi:10.1080/21655979.2021.1898134
Lo, S. C., and Hannink, M. (2008). PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp. Cell Res. 314, 1789–1803. doi:10.1016/j.yexcr.2008.02.014
Lo, S. C., and Hannink, M. (2006). PGAM5, a Bcl-XL-interacting protein, is a novel substrate for the redox-regulated Keap1-dependent ubiquitin ligase complex. J. Biol. Chem. 281, 37893–37903. doi:10.1074/jbc.M606539200
Lu, W., Karuppagounder, S. S., Springer, D. A., Allen, M. D., Zheng, L., Chao, B., et al. (2014). Genetic deficiency of the mitochondrial protein PGAM5 causes a Parkinson’s-like movement disorder. Nat. Commun. 5, 4930. doi:10.1038/ncomms5930
Lu, W., Sun, J., Yoon, J. S., Zhang, Y., Zheng, L., Murphy, E., et al. (2016). Mitochondrial protein PGAM5 regulates mitophagic protection against cell necroptosis. PLoS ONE 11, e0147792. doi:10.1371/journal.pone.0147792
Lu, Y., Han, Y., He, J., Zhou, B., Fang, P., and Li, X. (2020). LncRNA FOXD3-AS1 knockdown protects against cerebral ischemia/reperfusion injury via miR-765/BCL2L13 axis. Biomed. Pharmacother. 132, 110778. doi:10.1016/j.biopha.2020.110778
Ma, K., Chen, G., Li, W., Kepp, O., Zhu, Y., and Chen, Q. (2020a). Mitophagy, mitochondrial homeostasis, and cell fate. Front. Cell Dev. Biol. 8, 467. doi:10.3389/fcell.2020.00467
Ma, K., Zhang, Z., Chang, R., Cheng, H., Mu, C., Zhao, T., et al. (2020b). Dynamic PGAM5 multimers dephosphorylate BCL-xL or FUNDC1 to regulate mitochondrial and cellular fate. Cell Death Differ. 27, 1036–1051. doi:10.1038/s41418-019-0396-4
Magrí, A., Reina, S., and de Pinto, V. (2018). VDAC1 as pharmacological target in cancer and neurodegeneration: Focus on its role in apoptosis. Front. Chem. 6, 108. doi:10.3389/fchem.2018.00108
Man, S. M., and Kanneganti, T. D. (2016). Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat. Rev. Immunol. 16, 7–21. doi:10.1038/nri.2015.7
Mao, S., Li, Y., Lu, Z., Che, Y., Huang, J., Lei, Y., et al. (2019). PHD finger protein 5A promoted lung adenocarcinoma progression via alternative splicing. Cancer Med. 8, 2429–2441. doi:10.1002/cam4.2115
Matsubara, H., Tanaka, R., Tateishi, T., Yoshida, H., Yamaguchi, M., and Kataoka, T. (2019). The human Bcl-2 family member Bcl-rambo and voltage-dependent anion channels manifests a genetic interaction in Drosophila and cooperatively promote the activation of effector caspases in human cultured cells. Exp. Cell Res. 381, 223–234. doi:10.1016/j.yexcr.2019.05.015
Maurya, S. R., and Mahalakshmi, R. (2016). VDAC-2: Mitochondrial outer membrane regulator masquerading as channel? FEBS J. 283, 1831–1836. doi:10.1111/febs.13637
Mazure, N. M. (2017). VDAC in cancer. Biochim. Biophys. Acta. Bioenerg. 1858, 665–673. doi:10.1016/j.bbabio.2017.03.002
Mcllwain, D. R., Berger, T., and Mak, T. W. (2013). Caspase functions in cell death and disease. Cold Spring Harb. Perspect. Biol. 5, a008656. doi:10.1101/cshperspect.a008656
McWilliams, T. G., and Muqit, M. M. (2017). PINK1 and Parkin: emerging themes in mitochondrial homeostasis. Curr. Opin. Cell Biol. 45, 83–91. doi:10.1016/j.ceb.2017.03.013
Meng, F., Sun, N., Liu, D., Jia, J., Xiao, J., and Dai, H. (2021a). BCL2L13: Physiological and pathological meanings. Cell. Mol. Life Sci. 78, 2419–2428. doi:10.1007/s00018-020-03702-9
Meng, F., Zhang, L., Zhang, M., Ye, K., Guo, W., Liu, Y., et al. (2021b). Down-regulation of BCL2L13 renders poor prognosis in clear cell and papillary renal cell carcinoma. Cancer Cell Int. 21, 332. doi:10.1186/s12935-021-02039-y
Mevel, R., Draper, J. E., Lie-a-Ling, M., Kouskoff, V., and Lacaud, G. (2019). RUNX transcription factors: Orchestrators of development. Development 146, dev148296. doi:10.1242/dev.148296
Millino, C., Maretto, I., Pacchioni, B., Digito, M., De Paoli, A., Canzonieri, V., et al. (2017). Gene and microRNA expression are predictive of tumor response in rectal adenocarcinoma patients treated with preoperative chemoradiotherapy. J. Cell. Physiol. 232, 426–435. doi:10.1002/jcp.25441
Murakawa, T., Okamoto, K., Omiya, S., Taneike, M., Yamaguchi, O., and Otsu, K. (2019). A mammalian mitophagy receptor, Bcl2-L-13, recruits the ULK1 complex to induce mitophagy. Cell Rep. 26, 338–345. doi:10.1016/j.celrep.2018.12.050
Murakawa, T., Yamaguchi, O., Hashimoto, A., Hikoso, S., Takeda, T., Oka, T., et al. (2015). Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat. Commun. 6, 7527. doi:10.1038/ncomms8527
Murphy, M. P., and Hartley, R. C. (2018). Mitochondria as a therapeutic target for common pathologies. Nat. Rev. Drug Discov. 17, 865–886. doi:10.1038/nrd.2018.174
Nakazawa, M., Matsubara, H., Matsushita, Y., Watanabe, M., Vo, N., Yoshida, H., et al. (2016). The human Bcl-2 family member Bcl-rambo localizes to mitochondria and induces apoptosis and morphological aberrations in Drosophila. PLoS ONE 11, e0157823. doi:10.1371/journal.pone.0157823
Nawaz, J., Rasul, A., Shah, M. A., Hussain, G., Riaz, A., Sarfraz, I., et al. (2020). Cardamonin: A new player to fight cancer via multiple cancer signaling pathways. Life Sci. 250, 117591. doi:10.1016/j.lfs.2020.117591
Nunnari, J., and Suomalainen, A. (2012). Mitochondria: In sickness and in health. Cell 148, 1145–1159. doi:10.1016/j.cell.2012.02.035
Onishi, M., and Okamoto, K. (2021). Mitochondrial clearance: Mechanisms and roles in cellular fitness. FEBS Lett. 595, 1239–1263. doi:10.1002/1873-3468.14060
Onishi, M., Yamano, K., Sato, M., Matsuda, N., and Okamoto, K. (2021). Molecular mechanisms and physiological functions of mitophagy. EMBO J. 40, e104705. doi:10.15252/embj.2020104705
Otera, H., Ishihara, N., and Mihara, K. (2013). New insights into the function and regulation of mitochondrial fission. Biochim. Biophys. Acta 1833, 1256–1268. doi:10.1016/j.bbamcr.2013.02.002
Otsu, K., Murakawa, T., and Yamaguchi, O. (2015). BCL2L13 is a mammalian homolog of the yeast mitophagy receptor Atg32. Autophagy 11, 1932–1933. doi:10.1080/15548627.2015.1084459
Palikaras, K., Lionaki, E., and Tavernarakis, N. (2018). Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 20, 1013–1022. doi:10.1038/s41556-018-0176-2
Paradies, G., Paradies, V., Ruggiero, F. M., and Petrosillo, G. (2019). Role of cardiolipin in mitochondrial function and dynamics in health and disease: Molecular and pharmacological aspects. Cells 8, 728. doi:10.3390/cells8070728
Park, J., Kim, Y., Choi, S., Koh, H., Lee, S. H., Kim, J. M., et al. (2010). Drosophila porin/VDAC affects mitochondrial morphology. PLoS ONE 5, e13151. doi:10.1371/journal.pone.0013151
Park, Y. S., Choi, S. E., and Koh, H. C. (2018). PGAM5 regulates PINK1/Parkin-mediated mitophagy via DRP1 in CCCP-induced mitochondrial dysfunction. Toxicol. Lett. 284, 120–128. doi:10.1016/j.toxlet.2017.12.004
Paul, P. K., Rabaglia, M. E., Wang, C. Y., Stapleton, D. S., Leng, N., Kendziorski, C., et al. (2016). Histone chaperone ASF1B promotes human β-cell proliferation via recruitment of histone H3.3. Cell Cycle 15, 3191–3202. doi:10.1080/15384101.2016.1241914
Petry, I. B., Fieber, E., Schmidt, M., Gehrmann, M., Gebhard, S., Hermes, M., et al. (2010). ERBB2 induces an antiapoptotic expression pattern of Bcl-2 family members in node-negative breast cancer. Clin. Cancer Res. 16, 451–460. doi:10.1158/1078-0432.CCR-09-1617
Pewzner-Jung, Y., Ben-Dor, S., and Futerman, A. H. (2006). When do lasses (longevity assurance genes) become CerS (ceramide synthases)? Insights into the regulation of ceramide synthesis. J. Biol. Chem. 281, 25001–25005. doi:10.1074/jbc.R600010200
Pickles, S., Vigié, P., and Youle, R. J. (2018). Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28, R170–R185. doi:10.1016/j.cub.2018.01.004
Pickrell, A. M., and Youle, R. J. (2015). The roles of PINK1, Parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 85, 257–273. doi:10.1016/j.neuron.2014.12.007
Poole, L. P., and Macleod, K. F. (2021). Mitophagy in tumorigenesis and metastasis. Cell. Mol. Life Sci. 78, 3817–3851. doi:10.1007/s00018-021-03774-1
Popgeorgiev, N., Jabbour, L., and Gillet, G. (2018). Subcellular localization and dynamics of the Bcl-2 family of proteins. Front. Cell Dev. Biol. 6, 13. doi:10.3389/fcell.2018.00013
Rodger, C. E., McWilliams, T. G., and Ganley, I. G. (2018). Mammalian mitophagy – from in vitro molecules to in vivo models. FEBS J. 285, 1185–1202. doi:10.1111/febs.14336
Rogov, V. V., Suzuki, H., Marinković, M., Lang, V., Kato, R., Kawasaki, M., et al. (2017). Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins. Sci. Rep. 7, 1131. doi:10.1038/s41598-017-01258-6
Rosen, E. D., and Spiegelman, B. M. (2014). What we talk about when we talk about fat. Cell 156, 20–44. doi:10.1016/j.cell.2013.12.012
Rovira-Llopis, S., Bañuls, C., Diaz-Morales, N., Hernandez-Mijares, A., Rocha, M., and Victor, V. M. (2017). Mitochondrial dynamics in type 2 diabetes: Pathophysiological implications. Redox Biol. 11, 637–645. doi:10.1016/j.redox.2017.01.013
Ruiz-Gaspà, S., Guañabens, N., Jurado, S., Dubreuil, M., Combalia, A., Peris, P., et al. (2020). Bile acids and bilirubin effects on osteoblastic gene profile. Implications in the pathogenesis of osteoporosis in liver diseases. Gene 725, 144167. doi:10.1016/j.gene.2019.144167
Sadatomi, D., Tanimura, S., Ozaki, K., and Takeda, K. (2013). Atypical protein phosphatases: Emerging players in cellular signaling. Int. J. Mol. Sci. 14, 4596–4612. doi:10.3390/ijms14034596
Salimiaghdam, N., Singh, L., Schneider, K., Chwa, M., Atilano, S. R., Nalbandian, A., et al. (2022). Effects of fluoroquinolones and tetracyclines on mitochondria of human retinal MIO-M1 cells. Exp. Eye Res. 214, 108857. doi:10.1016/j.exer.2021.108857
Salimiaghdam, N., Singh, L., Schneider, K., Nalbandian, A., Chwa, M., Atilano, S. R., et al. (2020). Potential adverse effects of ciprofloxacin and tetracycline on ARPE-19 cell lines. BMJ Open Ophthalmol. 5, e000458. doi:10.1136/bmjophth-2020-000458
Schaaf, M. B. E., Keulers, T. G., Vooijs, M. A., and Rouschop, K. M. A. (2016). LC3/GABARAP family proteins: autophagy-(un)related functions. FASEB J. 30, 3961–3978. doi:10.1096/fj.201600698R
Schouten, M., Fratantoni, S. A., Hubens, C. J., Piersma, S. R., Pham, T. V., Bielefeld, P., et al. (2015). MicroRNA-124 and -137 cooperativity controls caspase-3 activity through BCL2L13 in hippocampal neural stem cells. Sci. Rep. 5, 12448. doi:10.1038/srep12448
Schrader, M., Bonekamp, N. A., and Islinger, M. (2012). Fission and proliferation of peroxisomes. Biochim. Biophys. Acta 1822, 1343–1357. doi:10.1016/j.bbadis.2011.12.014
Schrepfer, E., and Scorrano, L. (2016). Mitofusins, from mitochondria to metabolism. Mol. Cell 61, 683–694. doi:10.1016/j.molcel.2016.02.022
Sekine, S., Kanamaru, Y., Koike, M., Nishihara, A., Okada, M., Kinoshita, H., et al. (2012). Rhomboid protease PARL mediates the mitochondrial membrane potential loss-induced cleavage of PGAM5. J. Biol. Chem. 287, 34635–34645. doi:10.1074/jbc.M112.357509
Siegmund, D., Lang, I., and Wajant, H. (2017). Cell death-independent activities of the death receptors CD95, TRAILR1, and TRAILR2. FEBS J. 284, 1131–1159. doi:10.1111/febs.13968
Singh, R., Letai, A., and Sarosiek, K. (2019). Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 20, 175–193. doi:10.1038/s41580-018-0089-8
Speir, M., Vogrin, A., Seidi, A., Abraham, G., Hunot, S., Han, Q., et al. (2017). Legionella pneumophila strain 130b evades macrophage cell death independent of the effector SidF in the absence of flagellin. Front. Cell. Infect. Microbiol. 7, 35. doi:10.3389/fcimb.2017.00035
Strappazzon, F., Di Rita, A., Cianfanelli, V., D´Orazio, M., Nazio, F., Fimia, G. M., et al. (2016). Prosurvival AMBRA1 turns into a proapoptotic BH3-like protein during mitochondrial apoptosis. Autophagy 12, 963–975. doi:10.1080/15548627.2016.1164359
Strappazzon, F., Di Rita, A., Peschiaroli, A., Leoncini, P. P., Locatelli, F., Melino, G., et al. (2020). HUWE1 controls MCL1 stability to unleash AMBRA1-induced mitophagy. Cell Death Differ. 27, 1155–1168. doi:10.1038/s41418-019-0404-8
Strappazzon, F., Nazio, F., Corrado, M., Cianfanelli, V., Romagnoli, A., Fimia, G. M., et al. (2015). AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 22, 419–432. doi:10.1038/cdd.2014.139
Strappazzon, F., Vietri-Rudan, M., Campello, S., Nazio, F., Florenzano, F., Fimia, G. M., et al. (2011). Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J. 30, 1195–1208. doi:10.1038/emboj.2011.49
Strikoudis, A., Lazaris, C., Trimarchi, T., Neto, A. L. G., Yang, Y., Ntziachristos, P., et al. (2016). Regulation of transcriptional elongation in pluripotency and cell differentiation by the PHD-finger protein Phf5a. Nat. Cell Biol. 18, 1127–1138. doi:10.1038/ncb3424
Sugo, M., Kimura, H., Arasaki, K., Amemiya, T., Hirota, N., Dohmae, N., et al. (2018). Syntaxin 17 regulates the localization and function of PGAM5 in mitochondrial division and mitophagy. EMBO J. 37, e98899. doi:10.15252/embj.201798899
Szargel, R., Shani, V., Elghani, F. A., Mekies, L. N., Liani, E., Rott, R., et al. (2016). The PINK1, synphilin-1 and SIAH-1 complex constitutes a novel mitophagy pathway. Hum. Mol. Genet. 25, 3476–3490. doi:10.1093/hmg/ddw189
Tahir, S. K., Wass, J., Joseph, M. K., Devanarayan, V., Hessler, P., Zhang, H., et al. (2010). Identification of expression signatures predictive of sensitivity to the Bcl-2 family member inhibitor ABT-263 in small cell lung carcinoma and leukemia/lymphoma cell lines. Mol. Cancer Ther. 9, 545–557. doi:10.1158/1535-7163.MCT-09-0651
Tan, T., Zimmermann, M., and Reichert, A. S. (2016). Controlling quality and amount of mitochondria by mitophagy: insights into the role of ubiquitination and deubiquitination. Biol. Chem. 397, 637–647. doi:10.1515/hsz-2016-0125
Tang, D., Kang, R., Berghe, T. V., Vandernabeele, P., and Kroemer, G. (2019). The molecular machinery of regulated cell death. Cell Res. 29, 347–364. doi:10.1038/s41422-019-0164-5
Terešak, P., Lapao, A., Subic, N., Boya, P., Elazar, Z., and Simonsen, A. (2022). Regulation of PRKN-independent mitophagy. Autophagy 18, 24–39. doi:10.1080/15548627.2021.1888244
Tilokani, L., Nagashima, S., Paupe, V., and Prudent, J. (2018). Mitochondrial dynamics: overview of molecular mechanisms. Essays Biochem. 62, 341–360. doi:10.1042/EBC20170104
Ueda, N. (2015). Ceramide-induced apoptosis in renal tubular cells: A role of mitochondria and sphingosine-1-phoshate. Int. J. Mol. Sci. 16, 5076–5124. doi:10.3390/ijms16035076
Urbani, A., Giorgio, V., Carrer, A., Franchin, C., Arrigoni, G., Jiko, C., et al. (2019). Purified F-ATP synthase forms a Ca2+-dependent high-conductance channel matching the mitochondrial permeability transition pore. Nat. Commun. 10, 4341. doi:10.1038/s41467-019-12331-1
Vásquez-Trincado, C., García-Carvajal, I., Pennanen, C., Parra, V., Hill, J. A., Rothermel, B. A., et al. (2016). Mitochondrial dynamics, mitophagy and cardiovascular disease. J. Physiol. 594, 509–525. doi:10.1113/JP271301
Villa, E., Proïcs, E., Rubio-Patño, C., Obba, S., Zunino, B., Bossowski, J. P., et al. (2017). Parkin-independent mitophagy controls chemotherapeutic response in cancer cells. Cell Rep. 20, 2846–2859. doi:10.1016/j.celrep.2017.08.087
Vyas, S., Zaganjor, E., and Haigis, M. C. (2016). Mitochondria and cancer. Cell 166, 555–566. doi:10.1016/j.cell.2016.07.002
Wang, J., Li, Z., He, Y., Pan, F., Chen, S., Rhodes, S., et al. (2014). Loss of Asxl1 leads to myelodysplastic syndrome-like disease in mice. Blood 123, 541–553. doi:10.1182/blood-2013-05-500272
Wang, L., Brugge, J. S., and Jones, K. A. (2011). Intersection of FOXO- and RUNX1-mediated gene expression programs in single breast epithelial cells during morphogenesis and tumor progression. Proc. Natl. Acad. Sci. U. S. A. 108, E803–E812. doi:10.1073/pnas.1103423108
Wang, Q., Fu, R., Cheng, H., Li, Y., and Sui, S. (2022). Analysis of the resistance of small peptides from Periplaneta amricana to hydrogen peroxidase-induced apoptosis in human ovarian granular cells based on RNA-seq. Gene 813, 146120. doi:10.1016/j.gene.2021.146120
Wang, Z., Jiang, H., Chen, S., Du, F., and Wang, X. (2012). The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 148, 228–243. doi:10.1016/j.cell.2011.11.030
Wang, Z., Yan, Z., Zhang, B., Rao, Z., Zhang, Y., Liu, J., et al. (2013). Identification of a 5-gene signature for clinical and prognostic prediction in gastric cancer patients upon microarray data. Med. Oncol. 30, 678. doi:10.1007/s12032-013-0678-5
Wang, Z., Yang, X., Liu, C., Li, X., Zhang, B., Wang, B., et al. (2019). Acetylation of PHF5A modulates stress responses and colorectal carcinogenesis through alternative splicing-mediated upregulation of KDM3A. Mol. Cell 74, 1250–1263. doi:10.1016/j.molcel.2019.04.009
Warren, C. F. A., Wong-Brown, M. W., and Bowden, N. A. (2019). BCL-2 family isoforms in apoptosis and cancer. Cell Death Dis. 10, 177. doi:10.1038/s41419-019-1407-6
Watanabe, T., Kimura, A., and Kuroyanagi, H. (2018). Alternative splicing regulator RBM20 and cardiomyopathy. Front. Mol. Biosci. 5, 105. doi:10.3389/fmolb.2018.00105
Wegner, M. S., Schiffmann, S., Parnham, M. J., Geisslinger, G., and Grösch, S. (2016). The enigma of ceramide synthase regulation in mammalian cells. Prog. Lipid Res. 63, 93–119. doi:10.1016/j.plipres.2016.03.006
Wei, Y., Chiang, W. C., Sumpter, R., Mishra, P., and Levine, B. (2017). Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell 168, 224–238. doi:10.1016/j.cell.2016.11.042
Wu, H., Xue, D., Chen, G., Han, Z., Huang, L., Zhu, C., et al. (2014a). The BCL2L1 and PGAM5 axis defines hypoxia-induced receptor-mediated mitophagy. Autophagy 10, 1712–1725. doi:10.4161/auto.29568
Wu, M. Y., Yiang, G. T., Liao, W. T., Tsai, A. P. Y., Cheng, Y. L., Cheng, P. W., et al. (2018). Current mechanistic concepts in ischemia and reperfusion injury. Cell. Physiol. Biochem. 46, 1650–1667. doi:10.1159/000489241
Wu, W., Tian, W., Hu, Z., Chen, G., Huang, L., Li, W., et al. (2014b). ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 15, 566–575. doi:10.1002/embr.201438501
Xie, Y., Liu, J., Kang, R., and Tang, D. (2020). Mitophagy receptors in tumor biology. Front. Cell Dev. Biol. 8, 594203. doi:10.3389/fcell.2020.594203
Xu, W., Jing, L., Wang, Q., Lin, C. C., Chen, X., Diao, J., et al. (2015). Bax-PGAM5L-Drp1 complex is required for intrinsic apoptosis execution. Oncotarget 6, 30017–30034. doi:10.18632/oncotarget.5013
Xu, Y., Shen, J., and Ran, Z. (2020). Emerging views of mitophagy in immunity and autoimmune diseases. Autophagy 16, 3–17. doi:10.1080/15548627.2019.1603547
Yadav, R. K., Chauhan, A. S., Zhuang, L., and Gan, B. (2018). FoxO transcription factors in cancer metabolism. Semin. Cancer Biol. 50, 65–76. doi:10.1016/j.semcancer.2018.01.004
Yamaguchi, O., Murakawa, T., Nishida, K., and Otsu, K. (2016). Receptor-mediated mitophagy. J. Mol. Cell. Cardiol. 95, 50–56. doi:10.1016/j.yjmcc.2016.03.010
Yang, C., Liu, X., Yang, F., Zhang, W., Chen, Z., Yan, D., et al. (2017). Mitochondrial phosphatase PGAM5 regulates Keap1-mediated Bcl-xL degradation and controls cardiomyocyte apoptosis driven by myocardial ischemia/reperfusion injury. In Vitro Cell. Dev. Biol. Anim. 53, 248–257. doi:10.1007/s11626-016-0105-2
Yang, Y. L., Lin, S. R., Chen, J. S., Lin, S. W., Yu, S. L., Chen, H. Y., et al. (2010). Expression and prognostic significance of the apoptotic gene BCL2L13, Livin, and CASP8AP2 in childhood acute lymphoblastic leukemia. Leuk. Res. 34, 18–23. doi:10.1016/j.leukres.2009.07.023
Yang, Y., Ma, J., Song, Z., and Wu, M. (2002). HIV-1 TAT-mediated protein transduction and subcellular localization using novel expression vectors. FEBS Lett. 532, 36–44. doi:10.1016/s0014-5793(02)03624-4
Yi, P., Zhang, W., Zhai, Z., Miao, L., Wang, Y., and Wu, M. (2003). Bcl-rambo beta, a special splicing variant with an insertion of an Alu-like cassette, promotes etoposide- and Taxol-induced cell death. FEBS Lett. 534, 61–68. doi:10.1016/s0014-5793(02)03778-x
Yoo, S. M., and Jung, Y. K. (2018). A molecular approach to mitophagy and mitochondrial dynamics. Mol. Cells 41, 18–26. doi:10.14348/molcells.2018.2277
Youle, R. J., and van der Bliek, A. M. (2012). Mitochondrial fission, fusion, and stress. Science 337, 1062–1065. doi:10.1126/science.1219855
Yuan, S., and Akey, C. W. (2013). Apoptosome structure, assembly, and procaspase activation. Structure 21, 501–515. doi:10.1016/j.str.2013.02.024
Yuan, Y., Zheng, Y., Zhang, X., Chen, Y., Wu, X., Wu, J., et al. (2017). BNIP3L/NIX-mediated mitophagy protects against ischemic brain injury independent of PARK2. Autophagy 13, 1754–1766. doi:10.1080/15548627.2017.1357792
Zachari, M., and Ganley, I. G. (2017). The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 61, 585–596. doi:10.1042/EBC20170021
Zhang, G., Guo, J., Zeng, J., Zhang, X., Chen, R., Wang, G., et al. (2022). LncRNA SNHG14 is beneficial to oxygen glucose deprivation/reoxygenation-induced neuro-2a cell injury via mir-98-5p sequestration-caused BCL2L13 upregulation. Metab. Brain Dis. 37, 2005–2016. doi:10.1007/s11011-022-01009-1
Zhang, J., and Ney, P. A. (2009). Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 16, 939–946. doi:10.1038/cdd.2009.16
Zhang, L., Qin, Y., and Chen, M. (2018). Viral strategies for triggering and manipulating mitophagy. Autophagy 14, 1665–1673. doi:10.1080/15548627.2018.1466014
Zhang, X., Huang, C. R., Pan, S., Pang, Y., Chen, Y. S., Zha, G. C., et al. (2020). Long non-coding RNA SNHG15 is a competing endogenous RNA of miR-141-3p that prevents osteoarthritis progression by upregulating BCL2L13 expression. Int. Immunopharmacol. 83, 106425. doi:10.1016/j.intimp.2020.106425
Zhivotovsky, B., Galluzzi, L., Kepp, O., and Kroemer, G. (2009). Adenine nucleotide translocase: a component of the phylogenetically conserved cell death machinery. Cell Death Differ. 16, 1419–1425. doi:10.1038/cdd.2009.118
Keywords: BCL-RAMBO, BCL2L13, apoptosis, mitochondrial fragmentation, mitophagy, cell death, phosphorylation, miRNAs
Citation: Kataoka T (2022) Biological properties of the BCL-2 family protein BCL-RAMBO, which regulates apoptosis, mitochondrial fragmentation, and mitophagy. Front. Cell Dev. Biol. 10:1065702. doi: 10.3389/fcell.2022.1065702
Received: 10 October 2022; Accepted: 05 December 2022;
Published: 16 December 2022.
Edited by:
Ozgur Kutuk, Başkent University, TurkeyReviewed by:
Yih-Cherng Liou, National University of Singapore, SingaporeAnthea Di Rita, Independent Researcher, Rome, Italy
Copyright © 2022 Kataoka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takao Kataoka, takao.kataoka@kit.ac.jp
 Takao Kataoka
Takao Kataoka