Editorial: Advances in genomic and genetic tools, and their applications for understanding embryonic development and human diseases
- 1Center of Excellence in Genomic Medicine Research, King Abdulaziz University, Jeddah, Saudi Arabia
- 2Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
- 3Department of Biology, Jamoum University College, Umm Al-Qura University, Mecca, Saudi Arabia
- 4Centre of Biotechnology of Sfax, University of Sfax, Sfax, Tunisia
Significant advances have been recently made in the development of the genetic and genomic platforms. This has greatly contributed to a better understanding of gene expression and regulation machinery. Consequently, this led to considerable progress in unraveling evidence of the genotype-phenotype correlation between normal/abnormal embryonic development and human disease complexity. For example, advanced genomic tools such as next-generation sequencing, and microarray-based CGH have substantially helped in the identification of gene and copy number variants associated with diseases as well as in the discovery of causal gene mutations. In addition, bioinformatic analysis tools of genome annotation and comparison have greatly aided in data analysis for the interpretation of the genetic variants at the individual level. This has unlocked potential possibilities for real advances toward new therapies in personalized medicine for the targeted treatment of human diseases. However, each of these genomic and bioinformatics tools has its limitations and hence further efforts are required to implement novel approaches to overcome these limitations. It could be possible that the use of more than one platform for genotype-phenotype deep analysis is an effective approach to disentangling the cause and treatment of the disease complexities. Our research topic aimed at deciphering these complexities by shedding some light on the recent applications of the basic and advanced genetic/genomic and bioinformatics approaches. These include studying gene-gene, protein-protein, and gene-environment interactions. We, in addition, aimed at a better understanding of the link between normal/abnormal embryonic development and the cause of human disease induction.
Editorial on the Research Topic
Advances in genomic and genetic tools, and their applications for understanding embryonic development and human diseases
Introduction
The current Research Topic (RT) was aimed at deciphering the molecular signaling pathways’ interactions that orchestrate the complexity of embryonic development and human disease. Understanding these pathways should aid in developing targeted therapies. Several reports showed that many genes and/or microRNAs have a dual functional role during normal embryonic development and in human disease conditions (Miyaki et al., 2010; Ulveling et al., 2011; Hosseini-Farahabadi et al., 2013; Lan et al., 2019; Mahajan et al., 2021). Hence, discovering the missing links driving changes in gene expression and regulation during embryonic development to cause human disease induction has been a hot topic for many years (Loch-Caruso and Trosko, 1985; Nunes et al., 2003; Taylor et al., 2004; Lees et al., 2005; Kloosterman and Plasterk, 2006; Hashimoto et al., 2014; Degrauwe et al., 2016; Mazzoccoli et al., 2016; Tatetsu et al., 2016; Shah et al., 2018; Chahal et al., 2019; Prieto-Colomina et al., 2021). The themes of the RT attracted 61 research groups around the world to submit their work among which 32 manuscripts were peer-reviewed and accepted https://www.frontiersin.org/research-topics/10510/advances-in-genomic-and-genetic-tools-and-their-applications-for-understanding-embryonic-development. These included 26 original research articles, 3 case reports, 1 brief research report, 1 systematic review, and 1 opinion article (Table 1). The findings of these articles have certainly added to our knowledge and understanding of the field and have widely opened the door for more extended research to be carried out by other research groups worldwide. Without a doubt, several accepted manuscripts have achieved the main goals of the Research Topic. The main findings of these articles were classified into 3 main research themes and highlighted as follows:
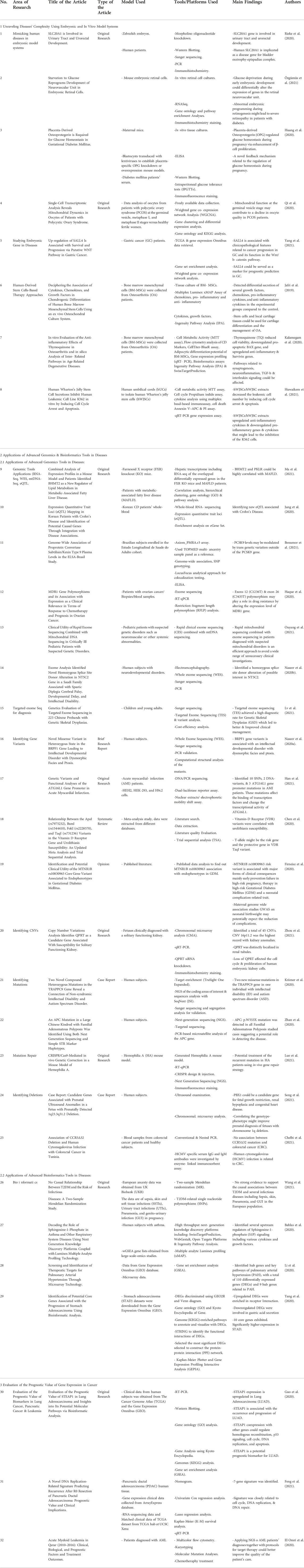
TABLE 1. A list of the accepted articles of the research topic summarizing the research area of each study, article’s title, article’s type, the model used, tool/platform used, main findings and the reference to each study.
Unraveling disease complexity using embryonic and in vitro model systems
The RT included eight articles under this category in which either a developmental gene was analyzed during embryonic development and in disease, a known developmental gene analyzed in human disease or stem cells were used as an in vitro experimental model. In a consortium study, Rieke and his colleagues Rieke et al., showed that SLC20A1 is involved in urinary tract and urorectal development. The study used morpholino to manipulate SLC20A1 function in the zebrafish embryo and the gene was analyzed in patients. The study showed that SLC20A1 was associated with bladder exstrophy-epispadias complex (BEEC, genitourinary malformations). Another research group in the RT used mouse embryonic retinal cells as their model and RNA-seq analysis to study perinatal glucose deprivation during early embryonic development and showed changes in gene expression during retinal neurogenesis. The study concluded that abnormal embryonic programming during retinogenesis could lead to retinopathy in patients with diabetes Özgümüs et al. Mice blastocysts transfected with lentivirus aiming at establishing placenta-specific knockdown or overexpression mouse model were used to study the role of Osteoprotegerin (OPG) in gestational diabetes mellitus (GDM). The study showed that OPG regulates glucose homeostasis during pregnancy through a negative feedback mechanism Huang et al. Polycystic ovary syndrome (PCOS) is another important disease studied in the RT using mitochondrial sequencing of patient oocytes. Transcriptomic dynamics analysis at the single-cell level using the weighted gene co-expression network analysis (WGCNA) approach was applied to show that abnormal mitochondrial function may contribute to a deteriorated oocyte quality in patients diagnosed with polycystic ovary syndrome (PCOS). The authors concluded that the oocyte quality might be affected by mitochondrial function Qi et al. Sall4 is a developmental gene that plays a crucial role in the pluripotency of embryonic stem cells (Chen et al., 2022). In this RT, SALL4 expression was analyzed in gastric cancer patients using gene set enrichment and found to be associated with the cancer progression through Wnt/β-catenin signaling pathway Yang et al. This study reiterated previously reported findings that there is a direct link between development and cancer (Papaioannou et al., 1984; Cheng et al., 2004; Tzukerman et al., 2006; Ma et al., 2010).
Human stem cells are an in vitro model system that offers great promise for developmental studies, disease research, and the emergence of targeted therapies. In our RT, mesenchymal stem cells (BM-MSCs) derived from the bone marrow of osteoarthritis (OA) patients were used to show that the expression of several chemokines, and pro- and anti-inflammatory factors were upregulated. The authors concluded that BM-MSCs could be used for cartilage differentiation and repair Jafri et al. Another study showed that Thymoquinone natural product has anti-inflammatory properties in BM-MSCs derived from OA patients suggesting a therapeutic potential of this natural product Kalamegam et al. Moreover, Wharton’s Jelly stem cell secretion was used in vitro to suppress human leukemia cells through cell cycle arrest and apoptosis induction Huwaikem et al.
In summary, the research theme discussed above showed that analyzing gene expression and regulation during embryonic development could give clues to human disease induction and progression. Several developmental genes have a functional dual role in development and disease including SLC20A1, Osteoprotegerin, and SALL4. Stem cell-based therapies or regenerative medicine represent a highly promising alternative route for disease cure.
Applications of advanced genomics and bioinformatics tools in disease
A considerable number of articles in the RT aimed at using a wide range of genomic/genetic and/or bioinformatics tools to decode the biological complexity and identify causal genes in human diseases were published (Table 1). These articles fall, in general, into the following areas:
Applications of advanced genomic tools in disease
Authors of articles published in this RT applied several advanced genomic tools including RNA-sequencing Jung et al. and Rieke et al., genome-wide association Bensenor et al., and whole exome-sequencing Naseer et al., Haque et al., and Ouyang et al. There was a tendency in several studies for using a combination of more than one genomic tool to critically discover the gene/marker/factor associated with the disease. For example, BHMT2 was identified as a new regulator for lipid metabolism in metabolic-associated fatty liver disease by combining RNA-seq analysis of the expression profile both in a mouse model and human patients Ma et al. In another study, RNA-seq was combined with the eQTL analysis to show the association of causal genes in Crohn’s disease Jung et al. Moreover, a combined exome and mitochondrial sequencing were successfully used for the rapid diagnosis of suspected pediatric mitochondrial disorders Ouyang et al. Next-generation sequencing (NGS) combined with simple short tandem repeat (STR) marker haplotypes were both used to identify a mutation in the adenomatous polyposis coli (APC) of Chinese kindred diagnosed with familial adenomatous polyposis (FAP) Zhan et al.
Targeted exome sequence (TES) was recommended in one of the RT studies as an effective approach for the genetic skeletal dysplasias (GSD) diagnosis Lv et al. Additionally, several studies identified novel genetic variants associated with many disorders and/or diseases such as developmental disorders Naseer et al., acute myocardial infarction Han et al., urolithiasis Chen et al., and endophenotypes in gestational diabetes Firneisz et al. Other RT studies identified copy number variants (CNVs) Zhou et al., gene mutations Krämer et al. and Zhan et al., and gene deletions Chelbi et al. and Song et al., with all, were shown to be associated with different human diseases.
An interesting RT study used a genomic CRISPR/Cas9 mutation repair approach and showed for the first time successful treatment of Hemophilia A in a mouse model Luo et al. This with no doubt gives hope to follow this approach for treatment in humans for Hemophilia A or any other mutation associated with other diseases.
Applications of advanced bioinformatics tools in disease
There have been significant advances in the use of bioinformatics tools and datasets available online that could be used for deciphering the complexity of human disease (Pereira et al., 2020). In our RT, these freely available online resources were used to decode the functional role of genes in human disease. For example, knowledge-based bioinformatics tools such as SwissTargetPrediction, WebGestalt, Open Targets Platforms and Ingenuity Pathway Analysis (IPA) were used to analyze the role of Sphingosine-1-phosphate in respiratory system diseases Bahlas et al., and to identify therapeutic targets for pulmonary arterial hypertension Li et al. Other tools such as Gene Expression Omnibus (GEO), Gene ontology (GO), Kyoto Encyclopedia of Gene, Genome (KEGG) enriched pathways, and STRING online database were applied to identify the functional interactions of the differentially expressed genes (DEGs) associated with the progression of stomach adenocarcinoma Yang et al.
In summary, the research theme described above focused on identifying causal genes, gene variants, copy number variants, mutations, or deletions associated with human diseases. A wide range of advanced bioinformatics tools was used to analyze the genomic data. Combined genomic/genetic/bioinformatics tools and model systems were applied to decipher the mechanistic complexity of the disease.
Evaluation of the prognostic value of gene expression
Gene ontology (GO), gene analysis using Kyoto Encyclopedia, genomes (KEGG) analysis, and gene enrichment analysis (GSEA) were all used to show that STEAP1 is a potential prognostic biomarker for lung adenocarcinoma (LAUD) Guo et al. Feng and his team used Cox regression analysis and LASSO algorithm on a considerable number of DNA replication-related genes to stratify the postoperative tumor-free margin (R0) treated pancreatic ductal adenocarcinoma (PDAC) patients with different recurrence risks. This study resulted in identifying novel DNA replication-related gene signatures Feng et al. A combined approach using NGS data, karyotyping, mutation detection analysis, and flow cytometry was shown to be a better protocol for targeted therapy of acute myeloid leukemia (AML) patients El Omri et al. In summary, this research theme showed that data collected from genomic and bioinformatics analysis were effectively used to evaluate the prognostic value of biomarkers in cancer.
Discussion
We had two aims with this RT: the first was to study genes and/or microRNAs that play a dual function in embryonic development and disease and to deliver an update about the applications of the genomics and bioinformatics advanced tools for understanding the complexity of the disease. To a large extent, these aims were met and achieved in the RT. All accepted articles were divided into 3 categories. The results published in the first category of the RT supported the notion of previous reports showing that many genes function in a dual role manner in development and disease (Gong et al., 2011; Teven et al., 2014; Sohn et al., 2016; Stepicheva and Song, 2016; Mahajan et al., 2021; McMillen et al., 2021; Assidi et al., 2022). Other reports showed that during development components of the FGF, Wnt, Notch, BPM, and Hedgehog signaling orchestrate crucial cellular events including proliferation, migration, differentiation, epithelial-mesenchymal transition (EMT), morphogenesis and somite myogenesis patterning. When gene expression controlling these events is dysregulated, it leads to disease induction (Hansson et al., 2004; Logan and Nusse, 2004; Ma et al., 2010; Teven et al., 2014; Xiao et al., 2017; Abreu de Oliveira et al., 2022; Alrefaei and Abu-Elmagd, 2022; Towler and Shore, 2022). Although our RT did not receive microRNA-related manuscripts, it is well-established that microRNAs play an important role in physiology and disease (Sayed and Abdellatif, 2011; Bhaskaran and Mohan, 2014; Kalayinia et al., 2021).
Stem cell medical research aiming at developing therapies has been recently the promise for chronic disease treatment (Mousaei Ghasroldasht et al., 2022). The current RT has contributed to this important line of research through three studies in which human BM-MSCs and Wharton’s Jelly cells were used to treat osteoarthritis and leukemia respectively Jafri et al., Kalamegam et al., and Huwaikem et al. In addition, stem cells can be also used for cell transplantation methods to cure diseases. For example, in this RT, Luo et al., showed successful CRISPR-mediated targeting of a Hemophilia A mutation, which may eventually one day be offered in the clinic as a treatment option.
For the second aim/category of the RT, a considerable number of published studies applied a wide range of genomic tools to unravel the causal gene(s) in disease Naseer et al., Naseer et al., Haque et al., Jung et al., Krämer et al., Zhan et al., Luo et al., Lv et al., Ma et al., Ouyang et al., and Özgümüs et al.
There was, in addition, substantial use in these studies of many advanced bioinformatics tools aimed at analyzing the involved molecular signaling pathways controlling the disease induction and progression Jafri et al., Kalamegam et al., and Huwaikem et al. In several studies, there was a good use of the freely available gene expression datasets for disease analysis Chen et al., Firneisz et al., Guo et al., Li et al., Qi et al., Yang et al., Feng et al., Yang et al., and Bahlas et al. However, the use of these data presents many ethical challenges (Lathe et al., 2008; Shabani and Borry, 2015). For the most part, these efforts intend to improve and identify early disease prediction, diagnosis, progression, and prognosis.
Conclusion
Our Research Topic aimed at expanding our knowledge of the use of advanced genomic and bioinformatics tools. Results from several articles showed that genes could play a dual functional role in embryonic development and human disease. Stem cell-based therapy and CRISPR/cas9 mutation repair approaches are promising strategies for therapy. Several combined genomic approaches were successfully used to decipher disease induction mechanisms. Advances made in genomic and bioinformatic applications for understanding the complexities of human disease should be of great help to clinicians in personalized therapy.
Author contributions
MA-E: Research Topic idea, write up the Research Topic description, and establish the reviewing panel. AA: Discussion of the Research Topic idea/theme and execution. AR, MA-E, MA: Reviewing process includes appointing the reviewing panel of each manuscript and making provisional acceptance/rejection decisions.
Funding
The authors extend their appreciation to the Deputyship for Research and Innovation, Ministry of Education in Saudi Arabia, for funding this research work through the project number (1033).
Acknowledgments
Guest editors would like to thank all authors who contributed their precious work to our Research Topic. Also, the guest editors would like to thank all reviewers who devoted their time and efforts to producing professional reviewing of the submitted manuscripts. We would like to thank all our colleagues for their support in spreading awareness among other colleagues about our Research Topic. The efforts and commitments of all mentioned here made this Research Topic a very successful one.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abreu De Oliveira, W. A., El Laithy, Y., Bruna, A., Annibali, D., and Lluis, F. (2022). Wnt signaling in the breast: From development to disease. Front. Cell Dev. Biol. 10, 884467. doi:10.3389/fcell.2022.884467
Alrefaei, A. F., and Abu-Elmagd, M. (2022). LRP6 receptor plays essential functions in development and human diseases. Genes (Basel) 13, 120. doi:10.3390/genes13010120
Assidi, M., Buhmeida, A., Al-Zahrani, M. H., Al-Maghrabi, J., Rasool, M., Naseer, M. I., et al. (2022). The prognostic value of the developmental gene FZD6 in young Saudi breast cancer patients: A biomarkers discovery and cancer inducers OncoScreen approach. Front. Mol. Biosci. 9, 783735. doi:10.3389/fmolb.2022.783735
Bhaskaran, M., and Mohan, M. (2014). MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 51, 759–774. doi:10.1177/0300985813502820
Chahal, G., Tyagi, S., and Ramialison, M. (2019). Navigating the non-coding genome in heart development and Congenital Heart Disease. Differentiation. 107, 11–23. doi:10.1016/j.diff.2019.05.001
Chen, K. Q., Anderson, A., Kawakami, H., Kim, J., Barrett, J., and Kawakami, Y. (2022). Normal embryonic development and neonatal digit regeneration in mice overexpressing a stem cell factor, Sall4. PLoS One 17, e0267273. doi:10.1371/journal.pone.0267273
Cheng, T. C., Huang, C. C., Chen, C. I., Liu, C. H., Hsieh, Y. S., Huang, C. Y., et al. (2004). Leukemia inhibitory factor antisense oligonucleotide inhibits the development of murine embryos at preimplantation stages. Biol. Reprod. 70, 1270–1276. doi:10.1095/biolreprod.103.023283
Degrauwe, N., Suvà, M. L., Janiszewska, M., Riggi, N., and Stamenkovic, I. (2016). IMPs: An RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev. 30, 2459–2474. doi:10.1101/gad.287540.116
Gong, B., Liu, T., Zhang, X., Chen, X., Li, J., Lv, H., et al. (2011). Disease embryo development network reveals the relationship between disease genes and embryo development genes. J. Theor. Biol. 287, 100–108. doi:10.1016/j.jtbi.2011.07.018
Hansson, E. M., Lendahl, U., and Chapman, G. (2004). Notch signaling in development and disease. Semin. Cancer Biol. 14, 320–328. doi:10.1016/j.semcancer.2004.04.011
Hashimoto, M., Morita, H., and Ueno, N. (2014). Molecular and cellular mechanisms of development underlying congenital diseases. Congenit. Anom. 54, 1–7. doi:10.1111/cga.12039
Hosseini-Farahabadi, S., Geetha-Loganathan, P., Fu, K., Nimmagadda, S., Yang, H. J., and Richman, J. M. (2013). Dual functions for WNT5A during cartilage development and in disease. Matrix Biol. 32, 252–264. doi:10.1016/j.matbio.2013.02.005
Kalayinia, S., Arjmand, F., Maleki, M., Malakootian, M., and Singh, C. P. (2021). MicroRNAs: Roles in cardiovascular development and disease. Cardiovasc. Pathol. 50, 107296. doi:10.1016/j.carpath.2020.107296
Kloosterman, W. P., and Plasterk, R. H. (2006). The diverse functions of microRNAs in animal development and disease. Dev. Cell 11, 441–450. doi:10.1016/j.devcel.2006.09.009
Lan, L., Wang, W., Huang, Y., Bu, X., and Zhao, C. (2019). Roles of Wnt7a in embryo development, tissue homeostasis, and human diseases. J. Cell. Biochem. 120, 18588–18598. doi:10.1002/jcb.29217
Lathe, W., Williams, J., Mangan, M., and Karolchik, D. (2008). Genomic data resources: Challenges and promises. Nat. Educ. 1, 2.
Lees, C., Howie, S., Sartor, R. B., and Satsangi, J. (2005). The hedgehog signalling pathway in the gastrointestinal tract: Implications for development, homeostasis, and disease. Gastroenterology 129, 1696–1710. doi:10.1053/j.gastro.2005.05.010
Loch-Caruso, R., and Trosko, J. E. (1985). Inhibited intercellular communication as a mechanistic link between teratogenesis and carcinogenesis. Crit. Rev. Toxicol. 16, 157–183. doi:10.3109/10408448509056269
Logan, C. Y., and Nusse, R. (2004). The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810. doi:10.1146/annurev.cellbio.20.010403.113126
Ma, Y., Zhang, P., Wang, F., Yang, J., Yang, Z., and Qin, H. (2010). The relationship between early embryo development and tumourigenesis. J. Cell. Mol. Med. 14, 2697–2701. doi:10.1111/j.1582-4934.2010.01191.x
Mahajan, D., Kancharla, S., Kolli, P., Sharma, A. K., Singh, S., Kumar, S., et al. (2021). Role of fibulins in embryonic stage development and their involvement in various diseases. Biomolecules 11, 685. doi:10.3390/biom11050685
Mazzoccoli, G., Laukkanen, M. O., Vinciguerra, M., Colangelo, T., and Colantuoni, V. (2016). A timeless link between circadian patterns and disease. Trends Mol. Med. 22, 68–81. doi:10.1016/j.molmed.2015.11.007
Mcmillen, P., Oudin, M. J., Levin, M., and Payne, S. L. (2021). Beyond neurons: Long distance communication in development and cancer. Front. Cell Dev. Biol. 9, 739024. doi:10.3389/fcell.2021.739024
Miyaki, S., Sato, T., Inoue, A., Otsuki, S., Ito, Y., Yokoyama, S., et al. (2010). MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 24, 1173–1185. doi:10.1101/gad.1915510
Mousaei Ghasroldasht, M., Seok, J., Park, H.-S., Liakath Ali, F. B., and Al-Hendy, A. (2022). Stem cell therapy: From idea to clinical practice. Int. J. Mol. Sci. 23, 2850. doi:10.3390/ijms23052850
Nunes, F. D., De Almeida, F. C., Tucci, R., and De Sousa, S. C. (2003). Homeobox genes: A molecular link between development and cancer. Pesqui. Odontol. Bras. 17, 94–98. doi:10.1590/s1517-74912003000100018
Papaioannou, V. E., Waters, B. K., and Rossant, J. (1984). Interactions between diploid embryonal carcinoma cells and early embryonic cells. Cell Differ. 15, 175–179. doi:10.1016/0045-6039(84)90071-x
Pereira, R., Oliveira, J., and Sousa, M. (2020). Bioinformatics and computational tools for next-generation sequencing analysis in clinical genetics. J. Clin. Med. 9, E132. doi:10.3390/jcm9010132
Prieto-Colomina, A., Fernández, V., Chinnappa, K., and Borrell, V. (2021). MiRNAs in early brain development and pediatric cancer: At the intersection between healthy and diseased embryonic development. Bioessays. 43, e2100073. doi:10.1002/bies.202100073
Sayed, D., and Abdellatif, M. (2011). MicroRNAs in development and disease. Physiol. Rev. 91, 827–887. doi:10.1152/physrev.00006.2010
Shabani, M., and Borry, P. (2015). Challenges of web-based personal genomic data sharing. Life Sci. Soc. Policy 11, 3. doi:10.1186/s40504-014-0022-7
Shah, K., Patel, S., Mirza, S., and Rawal, R. M. (2018). Unravelling the link between embryogenesis and cancer metastasis. Gene 642, 447–452. doi:10.1016/j.gene.2017.11.056
Sohn, J., Brick, R. M., and Tuan, R. S. (2016). From embryonic development to human diseases: The functional role of caveolae/caveolin. Birth Defects Res. C Embryo Today 108, 45–64. doi:10.1002/bdrc.21121
Stepicheva, N. A., and Song, J. L. (2016). Function and regulation of microRNA-31 in development and disease. Mol. Reprod. Dev. 83, 654–674. doi:10.1002/mrd.22678
Tatetsu, H., Kong, N. R., Chong, G., Amabile, G., Tenen, D. G., and Chai, L. (2016). SALL4, the missing link between stem cells, development and cancer. Gene 584, 111–119. doi:10.1016/j.gene.2016.02.019
Taylor, K. M., Hiscox, S., and Nicholson, R. I. (2004). Zinc transporter LIV-1: A link between cellular development and cancer progression. Trends Endocrinol. Metab. 15, 461–463. doi:10.1016/j.tem.2004.10.003
Teven, C. M., Farina, E. M., Rivas, J., and Reid, R. R. (2014). Fibroblast growth factor (FGF) signaling in development and skeletal diseases. Genes Dis. 1, 199–213. doi:10.1016/j.gendis.2014.09.005
Towler, O. W., and Shore, E. M. (2022). BMP signaling and skeletal development in fibrodysplasia ossificans progressiva (FOP). Dev. Dyn. 251, 164–177. doi:10.1002/dvdy.387
Tzukerman, M., Rosenberg, T., Reiter, I., Ben-Eliezer, S., Denkberg, G., Coleman, R., et al. (2006). The influence of a human embryonic stem cell-derived microenvironment on targeting of human solid tumor xenografts. Cancer Res. 66, 3792–3801. doi:10.1158/0008-5472.CAN-05-3467
Ulveling, D., Francastel, C., and Hubé, F. (2011). When one is better than two: RNA with dual functions. Biochimie 93, 633–644. doi:10.1016/j.biochi.2010.11.004
Wang, H., Guo, Z., Zheng, Y., Yu, C., Hou, H., and Chen, B. (2021). No casual relationship between T2DM and the risk of infectious diseases: A two-sample mendelian randomization study. Front. Genet. 12, 720874. doi:10.3389/fgene.2021.720874
Keywords: genetic/genomic tools, gene expression and regulation, embryonic development, human diseases, Bioinformatics tools, Dual function genes, Molecular signaling
Citation: Abu-Elmagd M, Assidi M, Alrefaei AF and Rebai A (2022) Editorial: Advances in genomic and genetic tools, and their applications for understanding embryonic development and human diseases. Front. Cell Dev. Biol. 10:1016400. doi: 10.3389/fcell.2022.1016400
Received: 10 August 2022; Accepted: 04 October 2022;
Published: 21 November 2022.
Edited and reviewed by:
Ramani Ramchandran, Medical College of Wisconsin, United StatesCopyright © 2022 Abu-Elmagd, Assidi, Alrefaei and Rebai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Abu-Elmagd, mabuelmagd@kau.edu.sa
 Muhammad Abu-Elmagd
Muhammad Abu-Elmagd Mourad Assidi
Mourad Assidi Abdulmajeed F. Alrefaei
Abdulmajeed F. Alrefaei Ahmed Rebai
Ahmed Rebai