PRDM14 Is a Unique Epigenetic Regulator Stabilizing Transcriptional Networks for Pluripotency
- Department of Biomedical Chemistry, School of Science and Technology, Kwansei Gakuin University, Hyogo, Japan
PR-domain containing protein 14 (PRDM14) is a site-specific DNA-binding protein and is required for establishment of pluripotency in embryonic stem cells (ESCs) and primordial germ cells (PGCs) in mice. DNA methylation status is regulated by the balance between de novo methylation and passive/active demethylation, and global DNA hypomethylation is closely associated with cellular pluripotency and totipotency. PRDM14 ensures hypomethylation in mouse ESCs and PGCs through two distinct layers, transcriptional repression of the DNA methyltransferases Dnmt3a/b/l and active demethylation by recruitment of TET proteins. However, the function of PRDM14 remains unclear in other species including humans. Hence, here we focus on the unique characteristics of mouse PRDM14 in the epigenetic regulation of pluripotent cells and primordial germ cells. In addition, we discuss the expression regulation and function of PRDM14 in other species compared with those in mice.
Role of PRDM14 in DNA Demethylation
PRDM14 functions as a site-specific transcriptional activator and repressor through the recruitment of transcriptional regulators in mouse ESCs (mESCs) (Yamaji et al., 2008; Chia et al., 2010; Ma et al., 2011). Prdm14 is transiently expressed in the inner cell mass (ICM) of the blastocyst, followed by rapid downregulation in the epiblast at the post-implantation stage in mice (Yamaji et al., 2008). Primordial germ cells (PGCs) are specified from the most proximal and posterior epiblast cells through BMP4 signaling approximately at embryonic day (E) 6.25 (Lawson et al., 1999). Nascent PGCs enter the embryo proper approximately at E8.0 and migrate through the hindgut, eventually colonizing the embryonic gonad, which constitutes the future testis or ovary. Prdm14 is specifically upregulated during PGC specification from the epiblast and is required for early development of PGCs (Yamaji et al., 2008). In developing PGCs, DNA demethylation occurs in a stepwise manner at the migrating and arriving phases (Seki et al., 2005). DNA demethylation is regulated by two pathways: replication-dependent mechanisms and replication-independent mechanisms (Wu and Zhang, 2017). Soon after PGC specification, the essential factors for epigenomic imprinting, Dnmt3a, Dnmt3b, and Uhrf1, are downregulated (Seisenberger et al., 2012; Kagiwada et al., 2013; Ohno et al., 2013; Kawasaki et al., 2014), which contributes to global DNA hypomethylation in developing PGCs. Prdm14-deficient PGCs fail to repress Dnmt3b and Uhrf1 expression, thereby resulting in stalling of global DNA demethylation (Shirane et al., 2016).
Mouse embryonic stem cells (mESCs) retain metastable pluripotency in serum containing leukemia inhibitory factor (LIF), whereas the transfer of culture conditions from serum plus LIF to the presence of two pharmacological inhibitors for ERK and GSK3β, called 2i plus LIF, leads to ground-state pluripotency associated with pronounced reduction in genome-wide DNA demethylation (Figure 1A) (Ying et al., 2008; Leitch et al., 2013). The resulting global hypomethylation status ensures the activation of pluripotency-associated genes and germline-specific genes. In the 2i plus LIF condition, Dnmt3a/b/l is downregulated, whereas Prdm14 is upregulated (Leitch et al., 2013). Furthermore, DNA methylation levels and Dnmt3a/b/l expression are consistently maintained at high levels in Prdm14-deficient ESCs even in the presence of 2i plus LIF, and PRDM14 directly binds to the upstream region of Dnmt3a/b/l and represses the transcription of these genes (Yamaji et al., 2013; Okashita et al., 2014). These findings indicate that PRDM14 is responsible for global hypomethylation through transcriptional repression of Dnmt3a/b/l in ground-state ESCs (Leitch et al., 2013). A recent study has shown that PRDM14 forms a complex with G9a, a histone methyltransferase, and this complex degrades DNMT3A/B proteins via lysine methylation-dependent polyubiquitination (Figure 1C) (Sim et al., 2017). Together, transcriptional repression of Dnmt3a/b/l and DNMT3A/B/L degradation via PRDM14, along with rapid proliferation, leads to global DNA hypomethylation in ground-state ESCs. Furthermore, DNA methylation of pluripotency-associated genes, germline-specific genes, and imprinted loci was rapidly diminished by Prdm14 induction through the ten-eleven translocation (TET)-thymine DNA glycosylase (TDG)-base excision repair (BER) pathway in ESCs with serum containing LIF (Figures 1B, 2A) (Okashita et al., 2014). Thus, PRDM14 regulates two parallel pathways, DNMT3A/B/L repression and TET recruitment at target loci, to ensure global DNA hypomethylation in ground-state ESCs and PGCs in mice.
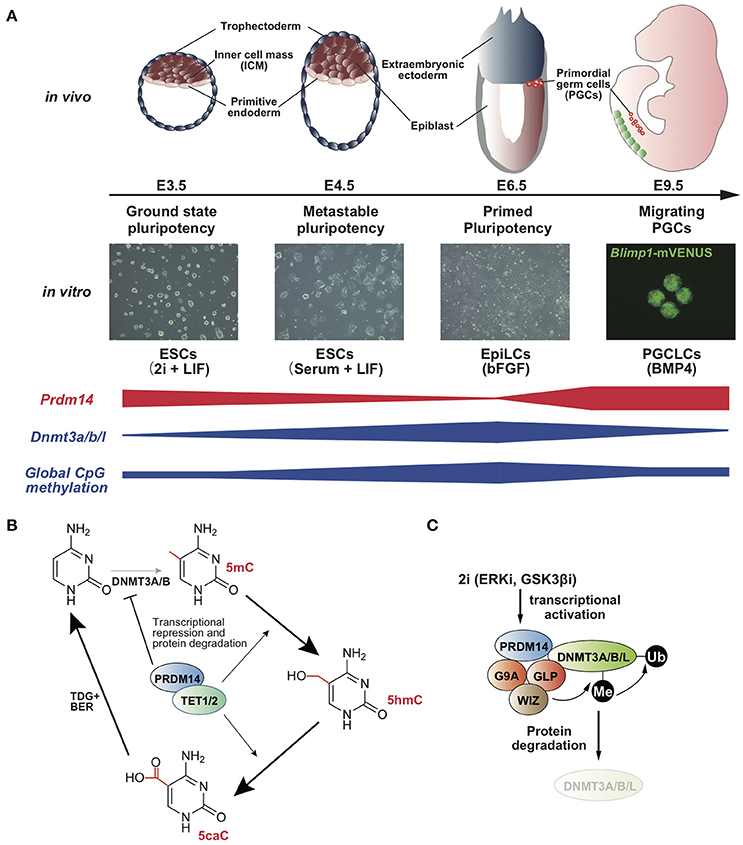
Figure 1. (A) Prdm14 expression negatively correlates with global CpG methylation level in the states of pluripotency and PGC development (Yamaji et al., 2008, 2013). (B) Model for the acceleration of TET-TDG-BER-mediated DNA demethylation by PRDM14 (Okashita et al., 2014). (C) Model for protein degradation of DNMT3A/B/L by PRDM14 (Sim et al., 2017).
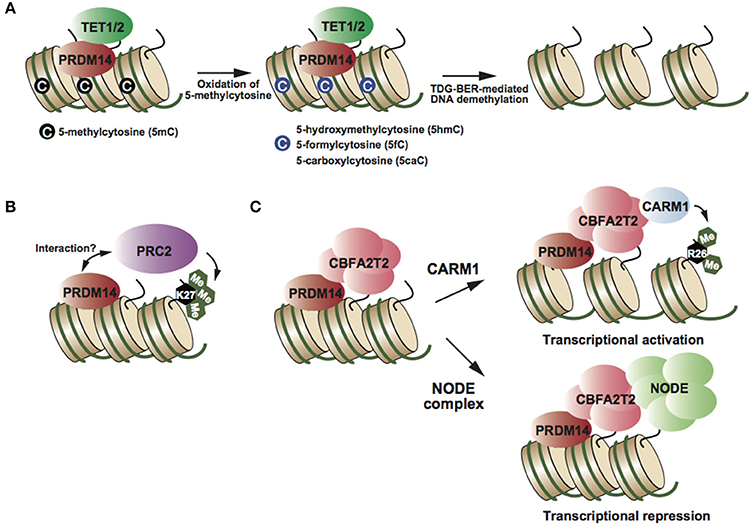
Figure 2. (A) PRDM14 recruits TET proteins at target genes, resulting in oxidation of 5mC and TDG-BER-mediated demethylation (Okashita et al., 2014). (B) PRDM14 might interact with PRC2 and its complex deposits H3K27me3, resulting in transcriptional repression (Yamaji et al., 2013). (C) CBFA2T2 is an essential partner of PRDM14 in both transcriptional activation and repression (Nady et al., 2015; Tu et al., 2016). Partner switching of the PRDM14-CBFA2T2 complex depending on target genes (Burton et al., 2013; Nady et al., 2015).
Role of the PRDM14 Complex in Transcriptional Regulation
PRDM14 has multiple functions in transcriptional regulation, e.g., DNA demethylation and transcriptional activation and repression, depending on the target genes. Biochemical studies have identified CBFA2T2, NODE complex, and BRG1 complex as PRDM14-containing complexes in mESCs (Figure 2) (Nady et al., 2015; Tu et al., 2016). PRDM14 interacts with CBFA2T2 stoichiometrically; this interaction is required to maintain pluripotency in mESCs and early differentiation of PGCs (Nady et al., 2015; Tu et al., 2016). Comparison of global gene expression patterns between Prdm14 knockout (KO) and Cbfa2t2 KO ESCs, revealed that CBFA2T2 was involved in both transcriptional activation and repression via PRDM14 in mESCs (Tu et al., 2016). Therefore, the switching mechanism in transcriptional regulation by PRDM14 complexes might be controlled by another component of the PRDM14 complex. Some studies have reported that PRDM14 interacts with the polycomb repressor complex 2 (PRC2) (Figure 2B) (Payer et al., 2013; Yamaji et al., 2013), whereas other studies failed to reproduce this finding (Nady et al., 2015; Tu et al., 2016). Although PRC2 and H3K27me3 enrichment at PRDM14-binding regions, including Dnmt3b and Xist loci, decreases in Prdm14-deficient mESCs compared with that in wild-type ESCs (Payer et al., 2013; Yamaji et al., 2013), it is largely unknown whether PRDM14 directly recruits the PRC2 complex at these regions. PRDM14 has also been shown to interact with CARM1, a histone arginine methyltransferase (H3R26me2), in mESCs (Figure 2C) (Burton et al., 2013). Prdm14 is expressed heterogeneously at the 4-cell stage, and PRDM14 overexpression in one blastomere at the 2-cell stage drives the predominant contribution of the inner cell mass (ICM) of the blastocyst, similar to the phenotype of CARM1 overexpression. Several studies have reported the functional correlation between PRDM14 and its binding partners; however, the direct functional interaction of PRDM14 with its binding partners in transcriptional regulation has not been investigated.
Diverse Functions and PRDM14 Expression Pattern in Mice and Humans
mESC self-renewal is maintained by the LIF signal, whereas self-renewal of human embryonic stem cells (hESCs) is sustained through bFGF and ACTIVIN signaling. Epiblast stem cells (EpiSCs) derived from epiblast cells at the post-implantation stage of mouse embryos can be expanded and maintained through bFGF and ACTIVIN signaling (Tesar et al., 2007). Therefore, hESCs and EpiSCs are considered to display common features corresponding with the post-implantation epiblast. The expression level of the genes associated with naïve pluripotency including Esrrb, Tcl1, and Klf4 is lower in hESCs and EpiSCs than in mESCs (Sasaki et al., 2015). Recent single-cell transcriptome analyses comparing human and mouse epiblasts derived from the same stage of the developing embryo showed distinct gene expression profiles of epiblasts in humans and mice (Blakeley et al., 2015). Interestingly, Prdm14 expression is closely associated with naïve pluripotency in mESCs and Prdm14 is not expressed in EpiSCs, whereas hESCs showing primed-state pluripotency express PRDM14, and this expression is required for the maintenance of hESC pluripotency but not for EpiSC pluripotency (Chia et al., 2010). Furthermore, single-cell transcriptome analysis of pre- and post-implantation epiblasts in monkeys showed that Prdm14 expression is retained in epiblast cells until soon before gastrulation (Nakamura et al., 2016). In the case of mice, the gene expression profile of epiblast cells dramatically changes after implantation, and it includes the downregulation of naïve pluripotency markers such as Prdm14, Nanog, and Sox2 (Kurimoto et al., 2008). In contrast to mice, however, the post-implantation epiblast relatively retains its gene expression profile for several days, suggesting that primate epiblasts persist for self-renewal of pluripotency (Nakamura et al., 2016). Sustainable Prdm14 expression results in self-renewal of mESCs and hESCs during embryoid body formation (Tsuneyoshi et al., 2008; Okashita et al., 2015). Furthermore, induction of Prdm14 expression in epiblast-like cells (EpiLCs) differentiated from mESCs induces the conversion of EpiLCs into mESCs through activation of pluripotent markers and the repression of differentiation markers (Okashita et al., 2016). These findings clearly indicate that PRDM14 stabilizes the transcriptional network for pluripotency, and that PRDM14 downregulation is necessary for exit from pluripotency. Therefore, the elucidation of distinct regulatory mechanisms for Prdm14 expression in humans and mice is an essential challenge to understand the differences in epiblast maturation between humans and mice.
Mouse and rat embryos display unique epiblast morphology, referred to as an egg cylinder, whereas epiblasts form discs in non-rodent mammals. Such morphological differences in epiblasts between the embryonic disc and egg cylinder correlate with the differences in gene expression profiles and morphology of embryonic stem cells. Lagostomus maximus (plains vizcacha), which is a fossorial rodent, develops flat embryonic discs, as observed in non-rodent mammals (Leopardo and Vitullo, 2017). SOX17 is a critical determinant of germ cell generation in humans but not in mice, and it is not expressed in mouse PGCs. Interestingly, primordial germ cells of Lagostomus maximus embryo express SOX17, as observed in human PGCs. These findings suggest that the embryonic disc constitutes the ancestral morphology of epiblasts in the common ancestor of rodents and muroidea including mice, rats, and hamsters, and might have acquired muroidea-specific mechanisms to change epiblast morphology during evolution from the rodent common ancestor.
Functional and Expression Diversity of PRDM14 in Deuterostomes
Prdm14 is widely distributed in metazoans but not in ecdysozoans including Drosophila and Caenorhabditis elegans (Vervoort et al., 2016). However, limited information is available regarding the expression pattern and function of PRDM14 in non-mammalian deuterostomes. Prdm14 is expressed in pluripotent cells and primordial germ cells in mammals; in contrast, Prdm14 is expressed in motor neurons but not in primordial germ cells in the zebrafish embryo (Liu et al., 2012). In Prdm14-knock down zebrafish embryos, islet2, a critical transcription factor for motor neuron development, is downregulated in motor neurons, and PRM14 binds to the islet2 locus, indicating that PRDM14 regulates motor neuron maturation through islet2 activation. It is unclear whether PRDM14 function in motor neurons is conserved among non-mammalian deuterostomes; therefore, the expression dynamics of Prdm14 in non-mammalian deuterostomes warrant further investigation. Interestingly, isl2, a mouse homolog of zebrafish islet2, is also upregulated soon after Prdm14 upregulation in mouse PGCs (Kurimoto et al., 2008), suggesting that the PRDM14-ISL2 axis might be co-opted from motor neurons to PGCs.
Conclusion and Further Perspectives
Findings regarding the role of Prdm14 expression in mouse germ cell specification led to the consideration that Prdm14 is a critical determinant of germ cell fate in mammals. However, because the expression pattern of Prdm14 differs among deuterostomes, the function of PRDM14 in deuterostomes could be reconsidered. I believe that this diversity of Prdm14 expression depends on the mode of germ cell specification. Germ cell specification comprises two modes: preformation and epigenesis. During preformation, germ cell determinants are asymmetrically distributed in the oocyte, referred to as the “germplasm,” whereas germ cells are induced from pluripotent cells receiving extrinsic signals through epigenesis. It has been proposed that the early segregation of germ cells from somatic lineages might drive the diversity of somatic cells (Johnson and Alberio, 2015). Classical “model” organisms including zebrafish, Xenopus, and chicken employ preformation and early germ cell restriction mediated by BLIMP1, and this the phenomenon, referred to as “Blimping,” even takes place in mice, which undergo epigenesis. Therefore, comparison of the expression and function of Prdm14 in organisms in which germ cell specification occurs in the late stage including urodele amphibians (axolotl), reptiles (gecko), and humans is necessary to uncover the evolutionarily conserved and diverse function of PRDM14 in pluripotent cells and primordial germ cells in deuterostomes.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Funding
This study was supported by JSPS KAKENHI Grant Number 16H01223, 16H01258.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Blakeley, P., Fogarty, N. M., Del Valle, I., Wamaitha, S. E., Hu, T. X., Elder, K., et al. (2015). Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development 142, 3151–3165. doi: 10.1242/dev.123547
Burton, A., Muller, J., Tu, S., Padilla-Longoria, P., Guccione, E., and Torres-Padilla, M. E. (2013). Single-cell profiling of epigenetic modifiers identifies PRDM14 as an inducer of cell fate in the mammalian embryo. Cell Rep. 5, 687–701. doi: 10.1016/j.celrep.2013.09.044
Chia, N. Y., Chan, Y. S., Feng, B., Lu, X., Orlov, Y. L., Moreau, D., et al. (2010). A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 468, 316–320. doi: 10.1038/nature09531
Johnson, A. D., and Alberio, R. (2015). Primordial germ cells: the first cell lineage or the last cells standing? Development 142, 2730–2739. doi: 10.1242/dev.113993
Kagiwada, S., Kurimoto, K., Hirota, T., Yamaji, M., and Saitou, M. (2013). Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J. 32, 340–353. doi: 10.1038/emboj.2012.331
Kawasaki, Y., Lee, J., Matsuzawa, A., Kohda, T., Kaneko-Ishino, T., and Ishino, F. (2014). Active DNA demethylation is required for complete imprint erasure in primordial germ cells. Sci. Rep. 4:3658. doi: 10.1038/srep03658
Kurimoto, K., Yabuta, Y., Ohinata, Y., Shigeta, M., Yamanaka, K., and Saitou, M. (2008). Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 22, 1617–1635. doi: 10.1101/gad.1649908
Lawson, K. A., Dunn, N. R., Roelen, B. A., Zeinstra, L. M., Davis, A. M., Wright, C. V., et al. (1999). Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 13, 424–436. doi: 10.1101/gad.13.4.424
Leitch, H. G., McEwen, K. R., Turp, A., Encheva, V., Carroll, T., Grabole, N., et al. (2013). Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 20, 311–316. doi: 10.1038/nsmb.2510
Leopardo, N. P., and Vitullo, A. D. (2017). Early embryonic development and spatiotemporal localization of mammalian primordial germ cell-associated proteins in the basal rodent Lagostomus maximus. Sci. Rep. 7:594. doi: 10.1038/s41598-017-00723-6
Liu, C., Ma, W., Su, W., and Zhang, J. (2012). Prdm14 acts upstream of islet2 transcription to regulate axon growth of primary motoneurons in zebrafish. Development 139, 4591–4600. doi: 10.1242/dev.083055
Ma, Z., Swigut, T., Valouev, A., Rada-Iglesias, A., and Wysocka, J. (2011). Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat. Struct. Mol. Biol. 18, 120–127. doi: 10.1038/nsmb.2000
Nady, N., Gupta, A., Ma, Z., Swigut, T., Koide, A., Koide, S., et al. (2015). ETO family protein Mtgr1 mediates Prdm14 functions in stem cell maintenance and primordial germ cell formation. Elife 4:e10150. doi: 10.7554/eLife.10150
Nakamura, T., Okamoto, I., Sasaki, K., Yabuta, Y., Iwatani, C., Tsuchiya, H., et al. (2016). A developmental coordinate of pluripotency among mice, monkeys and humans. Nature 537, 57–62. doi: 10.1038/nature19096
Ohno, R., Nakayama, M., Naruse, C., Okashita, N., Takano, O., Tachibana, M., et al. (2013). A replication-dependent passive mechanism modulates DNA demethylation in mouse primordial germ cells. Development 140, 2892–2903. doi: 10.1242/dev.093229
Okashita, N., Kumaki, Y., Ebi, K., Nishi, M., Okamoto, Y., Nakayama, M., et al. (2014). PRDM14 promotes active DNA demethylation through the ten-eleven translocation (TET)-mediated base excision repair pathway in embryonic stem cells. Development 141, 269–280. doi: 10.1242/dev.099622
Okashita, N., Sakashita, N., Ito, K., Mitsuya, A., Suwa, Y., and Seki, Y. (2015). PRDM14 maintains pluripotency of embryonic stem cells through TET-mediated active DNA demethylation. Biochem. Biophys. Res. Commun. 466, 138–145. doi: 10.1016/j.bbrc.2015.08.122
Okashita, N., Suwa, Y., Nishimura, O., Sakashita, N., Kadota, M., Nagamatsu, G., et al. (2016). PRDM14 Drives OCT3/4 recruitment via active demethylationin the transition from primed to naive pluripotency. Stem Cell Rep. 7, 1072–1086. doi: 10.1016/j.stemcr.2016.10.007
Payer, B., Rosenberg, M., Yamaji, M., Yabuta, Y., Koyanagi-Aoi, M., Hayashi, K., et al. (2013). Tsix RNA and the germline factor, PRDM14, link X reactivation and stem cell reprogramming. Mol. Cell 52, 805–818. doi: 10.1016/j.molcel.2013.10.023
Sasaki, K., Yokobayashi, S., Nakamura, T., Okamoto, I., Yabuta, Y., Kurimoto, K., et al. (2015). Robust in vitro induction of human germ cell fate from Pluripotent Stem Cells. Cell Stem Cell 17, 178–194. doi: 10.1016/j.stem.2015.06.014
Seisenberger, S., Andrews, S., Krueger, F., Arand, J., Walter, J., Santos, F., et al. (2012). The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48, 849–862. doi: 10.1016/j.molcel.2012.11.001
Seki, Y., Hayashi, K., Itoh, K., Mizugaki, M., Saitou, M., and Matsui, Y. (2005). Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev. Biol. 278, 440–458. doi: 10.1016/j.ydbio.2004.11.025
Shirane, K., Kurimoto, K., Yabuta, Y., Yamaji, M., Satoh, J., Ito, S., et al. (2016). Global landscape and regulatory principles of DNA methylation reprogramming for germ cell specification by mouse pluripotent stem cells. Dev. Cell 39, 87–103. doi: 10.1016/j.devcel.2016.08.008
Sim, Y. J., Kim, M. S., Nayfeh, A., Yun, Y. J., Kim, S. J., Park, K. T., et al. (2017). 2i Maintains a naive ground state in ESCs through two distinct epigenetic mechanisms. Stem Cell Rep. 8, 1312–1328. doi: 10.1016/j.stemcr.2017.04.001
Tesar, P. J., Chenoweth, J. G., Brook, F. A., Davies, T. J., Evans, E. P., Mack, D. L., et al. (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199. doi: 10.1038/nature05972
Tsuneyoshi, N., Sumi, T., Onda, H., Nojima, H., Nakatsuji, N., and Suemori, H. (2008). PRDM14 suppresses expression of differentiation marker genes in human embryonic stem cells. Biochem. Biophys. Res. Commun. 367, 899–905. doi: 10.1016/j.bbrc.2007.12.189
Tu, S., Narendra, V., Yamaji, M., Vidal, S. E., Rojas, L. A., Wang, X., et al. (2016). Co-repressor CBFA2T2 regulates pluripotency and germline development. Nature 534, 387–390. doi: 10.1038/nature18004
Vervoort, M., Meulemeester, D., Béhague, J., and Kerner, P. (2016). Evolution of Prdm genes in animals: insights from comparative genomics. Mol. Biol. Evol. 33, 679–696. doi: 10.1093/molbev/msv260
Wu, X., and Zhang, Y. (2017). TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet. 18, 517–534. doi: 10.1038/nrg.2017.33
Yamaji, M., Seki, Y., Kurimoto, K., Yabuta, Y., Yuasa, M., Shigeta, M., et al. (2008). Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 40, 1016–1022. doi: 10.1038/ng.186
Yamaji, M., Ueda, J., Hayashi, K., Ohta, H., Yabuta, Y., Kurimoto, K., et al. (2013). PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell 12, 368–382. doi: 10.1016/j.stem.2012.12.012
Keywords: Prdm14, epigenetics, embryonic stem cells (ESCs), primordial germ cells, DNA Methylation
Citation: Seki Y (2018) PRDM14 Is a Unique Epigenetic Regulator Stabilizing Transcriptional Networks for Pluripotency. Front. Cell Dev. Biol. 6:12. doi: 10.3389/fcell.2018.00012
Received: 30 November 2017; Accepted: 29 January 2018;
Published: 13 February 2018.
Edited by:
Alexey Ruzov, University of Nottingham, United KingdomReviewed by:
Ian C. G. Weaver, Dalhousie University, CanadaVladimir B. Teif, University of Essex, United Kingdom
Copyright © 2018 Seki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoshiyuki Seki, yseki@kwansei.ac.jp
 Yoshiyuki Seki
Yoshiyuki Seki