Isolation and identification of protease-producing Bacillus amyloliquefaciens LX-6 and its application in the solid fermentation of soybean meal
- 1Zhejiang Provincial Key Laboratory of Biometrology and Inspection and Quarantine, College of Life Sciences, China Jiliang University, Hangzhou, Zhejiang, China
- 2Department of Aquaculture, Zhejiang Ocean University, Zhoushan, Zhejiang, China
Soybean meal (SM) is considered an ideal substitute for fish meal; however, its application is mainly limited because of its antigen proteins, glycinin and β-conglycinin. To improve the value of SM in the aquaculture industry, we employed an aerobic bacterial strain (LX-6) with protease activity of 1,390.6 ± 12.5 U/mL. This strain was isolated from soil samples and identified as Bacillus amyloliquefaciens based on morphological and physiological biochemical characteristics and 16S rDNA gene sequence analyses. Subsequently, we quantified the extent of glycinin and β-conglycinin degradation and the total protein and water-soluble protein content after SM fermentation with B. amyloliquefaciens LX-6. At 24 h of fermentation, the macromolecular antigen proteins of SM were almost completely degraded; the maximum degradation rates of glycinin and β-conglycinin reached 77.9% and 57.1%, respectively. Accordingly, not only did the concentration of water-soluble proteins increase from 5.74% to 44.45% after 48 h of fermentation but so did the concentrations of total protein and amino acids compared to those of unfermented SM. Field emission scanning electron microscopy revealed that the LX-6 strain gradually disrupted the surface structure of SM during the fermentation process. In addition, B. amyloliquefaciens LX-6 exhibited broad-spectrum antagonistic activity and a wide pH tolerance, suggesting its application in SM fermentation for fish meal replacement.
1 Introduction
Soybean meal (SM) is a by-product of soybean oil extraction, a process involving drying, dehulling, flaking, expanding, and extraction (Fischer et al., 2001; Van Eenennaam and Young, 2014). SM not only contains high protein content and excellent amino acid (AA) quality but also large amounts of antioxidant compounds such as flavonoids and phenols (Lee et al., 2022). SM is often used as a cheap and high-quality protein raw material for poultry feeding and in livestock industries (Qi et al., 2023). The recent development of the aquaculture industry has resulted in a lack of protein sources for aquaculture feed and more expensive conventional raw materials, such as fish meal (Caimi et al., 2021). To reduce aquaculture costs, SM could serve as an ideal substitute for fish meal. However, its application is limited by its relatively high level of antinutritional factors (ANF) (Karr-Lilienthal et al., 2005), especially the antigen proteins glycinin and β-conglycinin. Known food allergens interfere with digestion, absorption, and nutrient use (Peng et al., 2018), causing hypersensitivity in animals (Hotz and Gibson, 2007; Cai et al., 2015). The presence of ANF seriously affects the use value and application scope of SM.
Microbial fermentation is an effective biotechnological approach to improving the nutritional value of SM by degrading macromolecular proteins and ANF compounds (Frias et al., 2008; Sun et al., 2020). Numerous microorganisms can be used in soybean and soy-based product fermentation and for improving the nutritional quality of SM, including Rhizopus oligosporus (Zhang et al., 2022), Aspergillus oryzae (Handa et al., 2019), Lactobacillus (Sun et al., 2022), and yeast (Luján-Rhenals et al., 2015). Owing to its short growth cycle and safety, Bacillus is considered an ideal strain in the fermentation industry. Moreover, SM fermentation with Bacillus produces an end product of high nutritional value and low ANF, which has fostered research in this area (Yao et al., 2021). For example, Zheng et al. (2017) fermented SM with Bacillus siamese, which degraded the allergic protein and loosened its structure, improving its solubility and animal absorption. Moreover, Jones et al. (2010) investigated the effects of fermentation using several strains, discovering that B. subtilis could best degrade antigen proteins and increase the crude protein content. Antigen protein degradation depends on protease hydrolysis; therefore, high protease activity is key for microbial SM fermentation (Yao et al., 2021).
In this study, we screened and identified Bacillus amyloliquefaciens LX-6, a strain with high protease activity, which we used to ferment SM. Later, we quantified the degradation of glycinin and β-conglycinin as well as the water-soluble protein and AA contents in SM after fermentation.
2 Materials and methods
2.1 Source of SM and strains
SM was obtained from Zhejiang Ocean University. Soil samples were collected from different ecological areas in China, such as wetland, forest, waterfall, and lakeshore. Soil samples were dried and stored at 4°C.
2.2 Media and cultural conditions
The medium for isolating protease-producing strains comprised (L−1 distilled water) 50-g skim milk powder and 20-g agar powder. The medium for determining protease activity comprised the following (L-1 distilled water): 40-g SM, 1.5-g CaCl2, 4.1-g glucose, 3-g NaCl, and 9.4-g yeast extract. The solid-state fermentation medium comprised the following (L−1 distilled water): 30-g SM and 30-mL distilled water in a 250-mL conical flask. The SM was sieved through 20 meshes and added to a solid-state fermentation medium.
2.3 Isolation and screening of protease-producing strain
Five grams of soil samples were weighed and added to a 250-mL conical flask containing 45-mL sterile water. Then, dozens of glass beads (4 mm) were added, incubated at 28°C for 2 h at 180 rpm, and immersed in an 80°C water bath for 20 min. Target strains were isolated by serial dilution (diluted gradient to 10–1, 10–2, and 10–3), spread on a skimmed milk plate, and cultured at 37°C for 24 h. Various individual colonies forming hydrolysis zones were streaked onto a skimmed milk plate for further verification and purification. The diameter of the hydrolysis zone in the skimmed milk plate was measured as an indication of the protease activity of the isolated strains. All strains were tested in two independent experiments run in triplicate.
2.4 Determination of enzyme activity
Strains with larger hydrolysis circles were cultured in protease fermentation medium with 4% inoculum at 37°C for 48 h at 180 rpm. The protease activity was determined using the Folin-phenol method of the People’s Republic of China (GB/T 23527–2009). The reaction mixture contained 1.0 mL of 2.0% casein, and 1.0 mL of the supernatant was incubated at 40°C for 20 min. After that, the reaction was terminated by adding 2.0 mL of 0.4 mol/mL trichloroacetic acid to the mixture, followed by incubation at 40°C for 20 min. Then, 1 mL of the supernatant of this mixture was mixed with 5 mL of 0.4 mol/mL Na2CO3 and 1 mL of Folin’s reagent. Finally, the mixture was incubated for another 20 min at 40°C. Tyrosine content was quantified at 660 nm. One unit (U) was defined as the amount of enzyme required to release l-μg tyrosine from the casein per minute under assay conditions.
2.5 LX-6 identification
2.5.1 Morphological observation of the LX-6 strain
The protease-producing strain LX-6 was streaked on Luria-Bertani (LB) solid medium and cultured at 37°C for 24 h to observe colony growth. Spores were stained with malachite green using a spore staining kit (Beijing Sorabio Technology Co., Ltd.).
An appropriate 0.05 g strain LX-6 bacterial solution was fixed with 2.5% glutaraldehyde overnight at 4°C. The strain LX-6 suspension was centrifuged at 11,000 × g for 5 min to obtain the cells, which were subsequently washed three times for 15 min with 1 × phosphate buffered saline buffer. Then, cells were fixed in 1% osmic acid for 1 h and gradually dehydrated with 30%, 50%, 75%, 90%, and 100% ethanol for 15 min (Din et al., 2013). Next, samples were dried with a Hitachi HCP-2 critical point dryer and plated in Hitachi B-1010 ion plating units. Strain LX-6 morphology was observed using a Hitachi SU-8010 scanning electron microscope at 6,000× magnification.
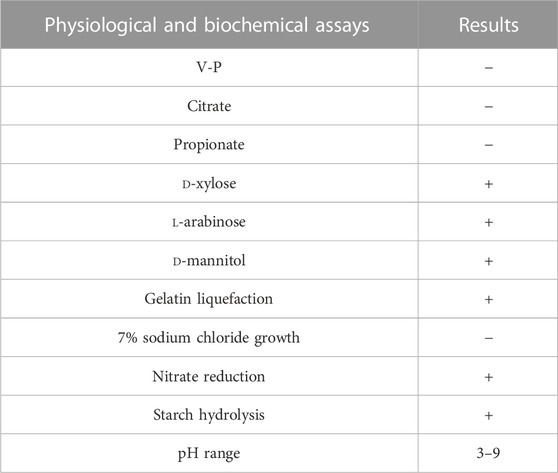
2.5.2 Physiological and biochemical characteristics of strain LX-6
The physiological and biochemical characteristics of strain LX-6 were analyzed by 7% sodium chloride growth, Voges-Proskauer (V-P) determination, nitrate reduction, D-mannitol fermentation, and other tests (Pereira et al., 2020; Ma et al., 2022).
2.5.3 Identification of strain LX-6 by 16S rDNA analysis and phylogenetic analysis
The 16S rDNA of the isolated strain LX-6 was amplified using the primers 16S-F (5′-AGAGTTTGATCATGGCTCAG-3′) and 16S-R (5′-CTACGGCTACCTTGTTACGA-3′). The PCR mixture comprised 1 μL 16S-F, 1 μL 16S-R, 1 μL template DNA, 12.5 μL GC buffer I (2×), 4 μL dNTPs, 0.25 μL Ex taq DNA polymerase, and added sterile water to 25 μL. The amplification procedure was conducted at 94 °C for 5 min, followed by 30 cycles at 94°C for 45 s, 57°C for 30 s, and 72°C for 45 s, with a final elongation step at 72°C for 10 min. The PCR product of the 16S rDNA was purified, sequenced (Tsingke Biotechnology, Hangzhou, China), and deposited in the NCBI database. The sequence was aligned using BLAST analysis, and the molecular phylogenetic tree (neighbor-joining likelihood) was constructed using MEGA 11.
2.6 SM fermentation by the isolated strain LX-6
The isolated strain was grown in LB liquid medium at 37°C and 180 rpm. This liquid culture of the isolated strain (108 CFU/mL) was then inoculated into a fermentation medium containing 30-g SM and 30-mL sterile water (1:1 solid-liquid (g/mL) ratio) in a 500-mL Erlenmeyer flask and fermented at 37 C for 60 h. Samples were collected at 12 h intervals until the end of fermentation. Finally, the fermented SM was freeze-dried, ground, screened through a 60-mesh sieve, and used for further analysis.
2.7 SDS-PAGE and determination of total protein and water-soluble protein contents
To determine the degradation of macromolecular proteins in the fermented SM at different times, 10% Tris sodium-dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) run at 100 V for 120 min was used (Li et al., 2020). The total protein and water-soluble protein content of the samples were determined by the Kjeldahl method (Rizvi et al., 2022) according to GB T6432-2018 and NY/T 1205–2006, respectively. Glycinin and β-conglycinin were analyzed using a commercially available ELISA kit (Hengyuan BioTechnology Co., Ltd., Shanghai, China).
2.8 Analysis of AA composition
The AA composition was measured using an automated AA analyzer (Hitachi L8900, Japan).
2.9 Structural analysis of SM and fermented SM by field emission scanning electron microscopy
Microstructural observation of fermented SM was performed using a FE-SEM (SU-8010, China) at 3,000×magnification according to the protocol of the Electronic Microscopy Center of China Jiliang University.
2.10 Determination of strain LX-6’s antibacterial activity
The antibacterial activity of LX-6 was explored using Serratia, Staphylococcus aureus, Candida albicans, Escherichia coli, and Fusarium oxysporum f. sp. cucumerinum as indicators. Strains were incubated under suitable conditions until the cell/spore concentrations reached around 107 CFU/mL. Indicator plates were prepared by mixing 200 μL of each indicator solution with agar media. The diameter of the inhibition zone was observed after 24–48 h of incubation.
2.11 Statistical analysis
All experiments were performed ≥3 times; the results are expressed as the mean ± standard deviation (SD). The students’ t-test was used for statistical analysis.
3 Results
3.1 Isolation of a strain with high protease activity, LX-6
Bacterial strains from various samples were directly isolated and screened on a skimmed milk powder plate. A total of 28 isolate-forming hydrolysis zones (diameter > 10 mm) were picked and purified again to verify their protease activity. Among them, 16 strains that exhibited large hydrolysis circles (diameter > 15 mm; Figure 1) were then inoculated into protease fermentation medium for enzymatic activity determination. Among the 16 isolated strains, the strain LX-6 exhibited the highest protease activity, 1,390.6 ± 12.5 U/mL (Figure 1). Because we used different proteins as substrates, the growth status and decomposition efficiency of these strains differed. Therefore, the hydrolytic circle values of 16 isolated strains on skim milk plates were not completely consistent with their protease activity in other media containing different substrates. Finally, strain LX-6 was therefore selected for further experiments.
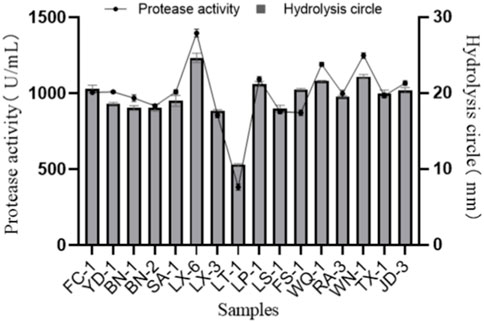
FIGURE 1. Diameter of the hydrolysis zone and protease activity of 16 representative protease-producing strains. The error bars were calculated from three independent experiments.
3.2 Strain LX-6 was identified as Bacillus amyloliquefaciens
The LX-6 colony exhibited a milky white color, a smooth surface, and high viscosity on LB solid plates (Figure 2A). In addition, under SEM, strain LX-6 showed a short rod-shaped morphology (Figure 2B). After staining, spores and cells showed a clear green and red color, respectively (Figure 2C). According to physiological and biochemical assays, strain LX-6 was positive in the V-P, gelatin liquefaction, nitrate reduction, L-arabinose, D-mannitol, and starch hydrolysis tests (Table 1). Therefore, based on its morphological and physiological characteristics, we hypothesized that strain LX-6 belongs to the genus Bacillus. Based on 16S rDNA alignment of strain LX-6 and type Bacillus strains and their phylogenic evolution analyses, strain LX-6 exhibited the highest similarity (98.34%) with B. amyloliquefaciens ATCC 23350 (Figure 3). Accordingly, strain LX-6 was finally identified, named Bacillus amyloliquefaciens LX-6, and deposited in the China Center for Type Culture Collection (No. 20211,457). A 1450-bp 16S rDNA fragment of strain LX-6 was cloned, sequenced, and submitted to the GenBank databases under the accession number OL614753.1.
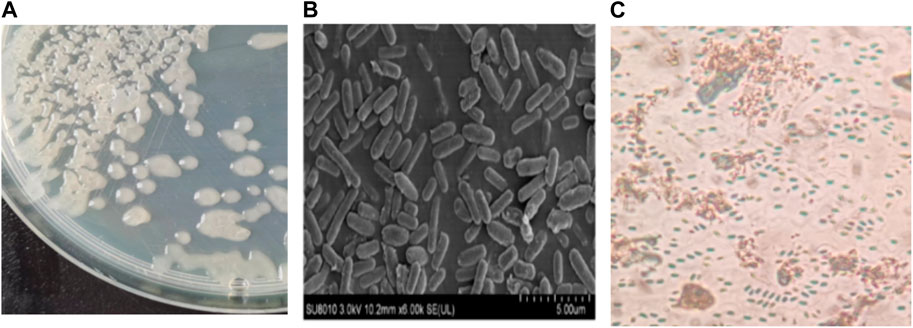
FIGURE 2. Morphological characteristics of the LX-6 strain. To assess colony morphology, the LX-6 strain was streaked and grown on an LB agar plate, which was incubated at 37°C for 2–3 days (A). Scanning electron microscopy of LX-6 grown in LB medium for 24 h (magnification, ×6,000) (B). Spores were stained with malachite green using a spore staining kit (Beijing Sorabio Technology Co., Ltd.) (C).
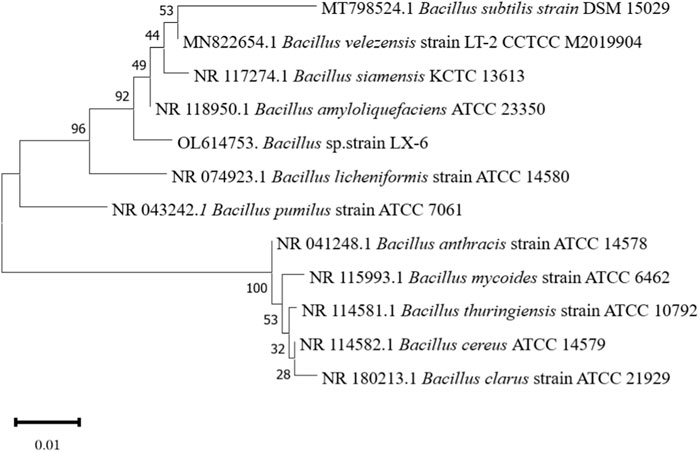
FIGURE 3. Phylogenetic tree analysis based on the 16S rDNA sequence of the LX-6 strain. Phylogenetic analysis was performed with MEGA 11 using the neighbor-joining method in the Jukes-Cantor model. Bootstrap values (>50%) based on 1,000 replicates are shown at the branch nodes. Bar, 0.01 substitutions per nucleotide position.
3.3 SM fermentation with B. amyloliquefaciens LX-6 effectively degraded macromolecular antigen proteins
SM contains mainly two types of allergenic proteins: conglycinin (68 kDa α, 72 kDa α’ subunit, and 52 kDa β subunit) and glycinin (37 kDa acidic subunit and 15 kDa basic subunit). We investigated the effects of B. amyloliquefaciens LX-6 fermentation on the degradation of glycinin and conglycinin in SM by SDS-PAGE (Figure 4). Some macromolecular antigen proteins started to degrade in the initial stage of fermentation (0–12 h), as indicated by the weaker protein bands (33–90 kDa) of fermented SM than those of unfermented SM. The macromolecular proteins in SM were gradually degraded during fermentation, resulting in successively thinner bands. After 24 h of SM fermentation by B. amyloliquefaciens LX-6, macromolecular proteins >20 kDa were almost completely degraded into small molecular products.
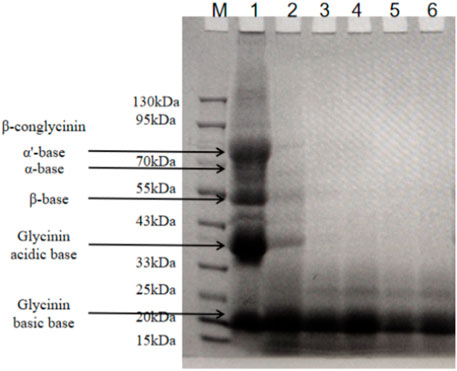
FIGURE 4. SDS-PAGE analyses of the macromolecular antigen proteins of SM at different fermentation times. Lane M, standard protein molecular weight markers; lane 1, unfermented SM; lanes 2-6, protein profiling of fermented SM with B. amyloliquefaciens LX-6 after 12 (lane 2), 24 (lane 3), 36 (lane 4), 48 (lane 5), and 60 h (lane 6) of fermentation at 37°C.
3.4 Increased total and water-soluble protein contents in fermented SM
To investigate the effects of B. amyloliquefaciens LX-6 fermentation on the total protein and water-soluble protein content of SM, we determined the protein contents of unfermented and fermented SM. The total protein content of unfermented SM was 48.87% ± 1.35%, whereas during fermentation by B. amyloliquefaciens LX-6, the total protein content gradually increased, reaching a maximum of 64.58% ± 3.07% at 48 h of fermentation (Figure 5A). Similarly, the water-soluble protein content of fermented SM increased significantly, reaching a maximum of 44.45% after 48 h of fermentation, 7-fold higher than that of unfermented SM (Figure 5A). Moreover, after 60 h of SM fermentation by B. amyloliquefaciens LX-6, the maximum degradation rates of glycinin and β-conglycinin in SM reached 77.9% and 57.1%, respectively (Figure 5B).
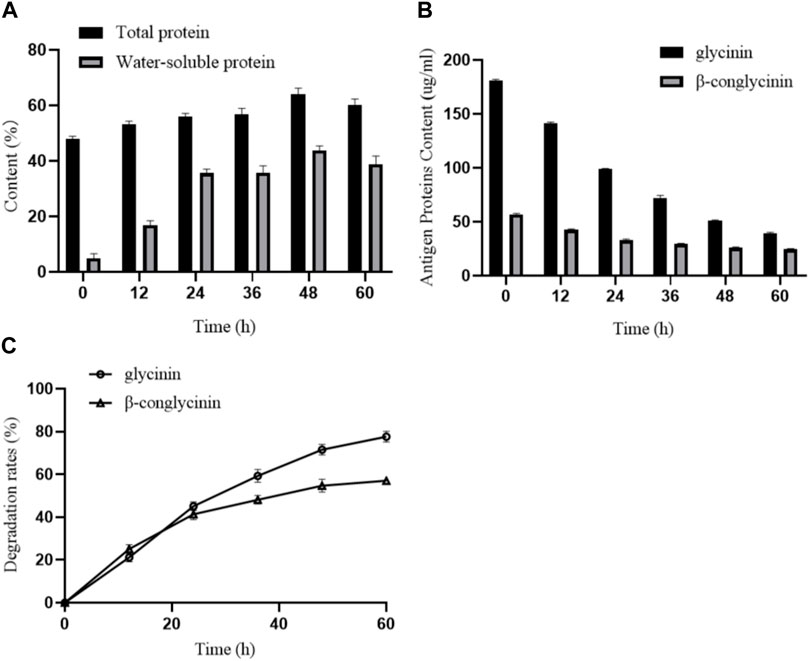
FIGURE 5. Effect of SM fermentation with B. amyloliquefaciens LX-6 on the levels of proteins and degradation of antigenic proteins. SM was inoculated with B. amyloliquefaciens LX-6 and fermented at 37°C. The levels of total proteins and water-soluble proteins (A), antigenic protein levels (glycinin and β-glycinin) (B), and degradation rates of glycinin and β-glycinin (C) were determined after different periods of fermentation.
3.5 The AA content in SM increased after fermentation
The total AA content increased from 148.4 ± 17.39 mg/mL to 1909.43 ± 387.86 mg/mL after SM fermentation with B. amyloliquefaciens LX-6. The levels of essential AA increased more remarkably, from 85.29 ± 8.61 mg/mL to 1,537.72 ± 334.86 mg/mL, than those of nonessential AA. Among essential AAs, the highest increase rate (70-fold) was observed for valine (Val), whose content increased from 15.38 ± 1.26 mg/mL to 1,061.44 ± 220.56 mg/mL, while that of arginine (Arg) decreased after fermentation (Table 2).
3.6 Fermentation by B. amyloliquefaciens LX-6 altered SM’s microstructure
FE-SEM was employed to explore the effect of B. amyloliquefaciens LX-6 fermentation on SM structure. The structure of the unfermented SM was dense and smooth (Figure 6A). However, after 12 h of fermentation (Figure 6B), the SM microstructure started to show loose and broken elements. After 48 h (Figure 6C), the fermented SM was even more fragmentated and had increased porosity.

FIGURE 6. Field emission SEM of unfermented and fermented SM. Unfermented SM (A) was inoculated with B. amyloliquefaciens LX-6 and fermented at 37°C for 12 (B) or 48 h (C). magnification, ×3,000.
4 Discussion
We also tested the viable cell count (Figure 7A) and protease activity (Figure 7B) of LX-6 during different time periods of fermentation SM. Because of B. amyloliquefaciens LX-6’s high protease activity, after 24 h of SM fermentation, glycinin and β-conglycinin were effectively degraded as their corresponding bands were almost impossible to detect via SDS-PAGE. After 48 h of fermentation, we observed the highest degradation rates of glycinin and β-conglycinin using ELISA. The degradation efficiency of glycinin and β-conglycinin in SM fermented by strain LX-6 obtained in the present study is better than that of previous studies using native Bacillus strains (Shikha and Darmwal, 2007; Baweja et al., 2016; Huang et al., 2022), in which the whole fermentation process for full glycinin and β-conglycinin degradation required ≥60 h. The increase in low-molecular protein levels indicated decreased potential allergenicity of SM proteins.
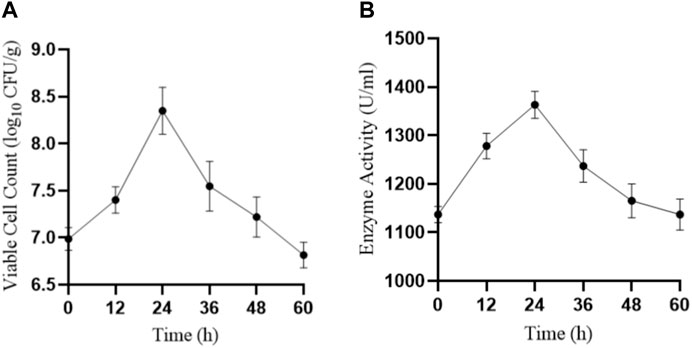
FIGURE 7. SM was inoculated with B. amyloliquefaciens LX-6 and fermented at 37°C. The viable cell count (A) and enzymatic activity (B) were determined after different durations of fermentation.
The degradation of SM antigenic proteins increased total protein, water-soluble protein, and AA contents. In comparison, the soluble protein in SM increased only by 10.14% after fermentation with B. siamensis (Zheng et al., 2017) and by 8.42% and 7.43% when using B. sanfenicol SQVG18 and SQVG22 (Gao et al., 2020). It has been reported that essential AAs are the main limiting factors for SBM replacement of fish meal in aquaculture (Lin and Mui, 2017; Gao et al., 2020). After fermentation with LX-6, the total essential AA levels of SM were 17-fold higher than those of unfermented SM; this increased bio-availability has great potential for replacing fish meal. Excessive SM fermentation causes not only a pungent smell but can also decrease the nutritional value of SM (Medeiros et al., 2018). Accordingly, after 48 h of SM fermentation with B. amyloliquefaciens LX-6, the total and water-soluble protein contents decreased slightly, possibly because small proteins were degraded to ammonia or NH3 due to excessive protease hydrolysis during the fermentation process. Therefore, future research should focus on the optimization of fermentation conditions for improving SM fermentation efficiency with strain B. amyloliquefaciens LX-6. Moreover, SEM revealed that B. amyloliquefaciens LX-6 effectively destroyed the SM microstructure, degrading internal macromolecules and increasing nutritional value and digestibility.
Previous phylogenetic analyses revealed that Bacillus velezensis, Bacillus amyloliquefaciens, and Bacillus siamensis clustered tightly to form the “operational group Bacillus amyloliquefaciens” (Fan et al., 2017), widely used in the food industry and SM fermentation because of their safety and ability to produce proteases. For example, B. amyloliquefaciens ZB has 694 U/g of protease activity and has thus been used for SM fermentation (Wattanakul et al., 2021). The B. siamensis isolate JL8 (protease activity 519·1 U/g) was used in the laboratory production of fermented SM, which showed better properties than unfermented SM (Zheng et al., 2017). B. velezensis 157, exhibiting 236.94 U/g of protease activity, was mixed with Lactiplantibacillus plantarum to degrade ANFs and improve SM nutrition for animal feed via two-stage solid-state fermentation. The protease activity of LX-6 observed in this study (1,390 ± 12.5 U/mL) is much higher than that of the above native or wild-type Bacillus strains. Nevertheless, the protease activity of B. amyloliquefaciens/B. velezensis (Li et al., 2023) could be increased to 20000–30000 U/mL by strategies such as protease purification (Yu et al., 2019), modification of the native strain (Zhou et al., 2020), overexpression or heterologous expression of the npr gene encoding protease (Wang et al., 2016), and optimization of fermentation conditions (Guo et al., 2022). Therefore, SM fermentation with B. amyloliquefaciens LX-6, a strain with high protease production, can improve its efficiency through future genetic modification, mixed fermentation, and medium optimization.
In addition, the antibacterial activity of strain LX-6 was explored using the following five pathogens as indicators: Fusarium oxysporum f. sp. cucumerinum, Escherichia coli, Candida albicans, Serratia, and Staphylococcus aureus. Strain LX-6 had broad-spectrum inhibitory effects on the five tested indicators, exhibiting the strongest inhibition activity against C. albicans (inhibition zone reaching 20 ± 1.7 mm; Figure 8), suggesting a certain degree of safety for the use of strain LX-6 in SM fermentation. However, as we did not characterize the presence of the antifungal or antibacterial metabolite substances of LX-6, this aspect warrants further research. In addition, we investigated the feasibility of using B. amyloliquefaciens LX-6 under various fermentation conditions and found that it had a broad growth range of pH 3–9 (Table 1), suggesting good suitability for SM fermentation.
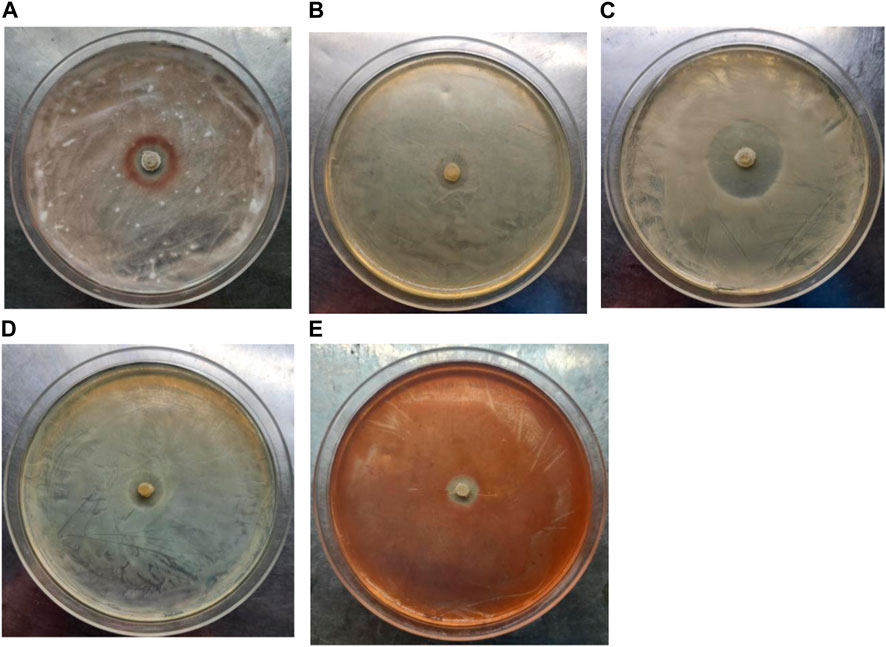
FIGURE 8. Determination of the antibacterial activity of B. amyloliquefaciens LX-6. F. oxysporum f. sp. cucumerinum (A), E. coli (B), C. albicans (C), S. aureus (D), and Serratia (E) were used as indicators.
In conclusion, an efficient protease producer, B. amyloliquefaciens strain LX-6, was isolated and identified. SM fermentation using B. amyloliquefaciens strain LX-6 resulted in effective degradation of the antigenic proteins glycinin and β-conglycinin in SM, which increases digestibility while inhibiting pathogenic bacteria and has broad application in aquaculture.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/nuccore/OL614753.1.
Author contributions
XH and HL conducted experiments. XH and ZM wrote the original draft. TH and JW designed the experiments. ZM revised this article. XY polished English and checked the final version. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Key Research and Development Program of Zhejiang Province (2021C04016), Pioneer and Leading Goose R&D Program of Zhejiang (2022C04020).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Baweja, M., Tiwari, R., Singh, P. K., Nain, L., and Shukla, P. (2016). An alkaline protease from Bacillus pumilus MP 27: Functional analysis of its binding model toward its applications as detergent additive. Front. Microbiol. 7, 1195. doi:10.3389/fmicb.2016.01195
Cai, L., Indrakumar, S., Kiarie, E., and Kim, I. H. (2015). Effects of a multi-strain Bacillus species-based direct-fed microbial on growth performance, nutrient digestibility, blood profile, and gut health in nursery pigs fed corn-soybean meal-based diets. J. Animal Sci. 93 (9), 4336–4342. doi:10.2527/jas.2015-9056
Caimi, C., Biasato, I., Chemello, G., Oddon, S. B., Lussiana, C., Malfatto, V. M., et al. (2021). Dietary inclusion of a partially defatted black soldier fly (Hermetia illucens) larva meal in low fishmeal-based diets for rainbow trout(Oncorhynchus mykiss). J. Animal Sci. Biotechnol. 12, 50–1413. doi:10.1186/s40104-021-00575-1
Din, W. M., Jin, K. T., Ramli, R., Khaithir, T. M., and Wiart, C. (2013). Antibacterial effects of ellagitannins from Acalypha wilkesiana var. macafeana hort.: Surface morphology analysis with environmental scanning electron microscopy and synergy with antibiotics. Phytotherapy Res. PTR 27 (9), 1313–1320. doi:10.1002/ptr.4876
Fan, B., Blom, J., Klenk, H. P., and Borriss, R. (2017). Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an Operational Group B. amyloliquefaciens within the B. subtilis species complex. Front. Microbiol. 8, 22. doi:10.3389/fmicb.2017.00022
Fischer, M., Kofod, L. V., Schols, H. A., Piersma, S. R., Gruppen, H., and Voragen, A. G. (2001). Enzymatic extractability of soybean meal proteins and carbohydrates: Heat and humidity effects. J. Agric. Food Chem. 49 (9), 4463–4469. doi:10.1021/jf010061w
Frias, J., Song, Y. S., Martínez-Villaluenga, C., González de Mejia, E., and Vidal-Valverde, C. (2008). Immunoreactivity and amino acid content of fermented soybean products. J. Agric. Food Chem. 56 (1), 99–105. doi:10.1021/jf072177j
Gao, S., Pan, L., Zhang, M., Huang, F., Zhang, M., and He, Z. (2020). Screening of bacterial strains from the gut of Pacific white shrimp (Litopenaeus vannamei) and their efficiencies in improving the fermentation of soybean meal. FEMS Microbiol. Lett. 367 (2), fnaa017. doi:10.1093/femsle/fnaa017
Guo, L., Guo, Y., Wu, P., Liu, S., Gu, C., YolandaniWu, M., et al. (2022). Enhancement of polypeptide yield derived from rapeseed meal with low-intensity alternating magnetic field. Foods (Basel, Switz. 11 (19), 2952. doi:10.3390/foods11192952
Handa, C. L., de Lima, F. S., Guelfi, M. F. G., Fernandes, M. D. S., Georgetti, S. R., and Ida, E. I. (2019). Parameters of the fermentation of soybean flour by Monascus purpureus or Aspergillus oryzae on the production of bioactive compounds and antioxidant activity. Food Chem. 271, 274–283. doi:10.1016/j.foodchem.2018.07.188
Hotz, C., and Gibson, R. S. (2007). Traditional food-processing and preparation practices to enhance the bioavailability of micronutrients in plant-based diets. J. Nutr. 137 (4), 1097–1100. doi:10.1093/jn/137.4.1097
Huang, C. H., Chen, C. L., Shieh, C. C., Chang, S. H., and Tsai, G. J. (2022). Evaluation of hypoglycemic and antioxidant activities of soybean meal products fermented by Lactobacillus plantarum FPS 2520 and Bacillus subtilis N1 in rats fed with high-fat diet. Metabolites 12 (5), 442. doi:10.3390/metabo12050442
Jones, C. K., DeRouchey, J. M., Nelssen, J. L., Tokach, M. D., Dritz, S. S., and Goodband, R. D. (2010). Effects of fermented soybean meal and specialty animal protein sources on nursery pig performance. J. Animal Sci. 88 (5), 1725–1732. doi:10.2527/jas.2009-2110
Karr-Lilienthal, L. K., Grieshop, C. M., Spears, J. K., and Fahey, G. C. (2005). Amino acid, carbohydrate, and fat composition of soybean meals prepared at 55 commercial U.S. soybean processing plants. J. Agric. Food Chem. 53 (6), 2146–2150. doi:10.1021/jf048385i
Lee, T. Y., Lee, Y. S., Yeh, R. H., Chen, K. H., and Chen, K. L. (2022). Bacillus amyloliquefaciens CU33 fermented feather meal-soybean meal product improves the intestinal morphology to promote the growth performance of broilers. Poult. Sci. 101 (9), 102027. doi:10.1016/j.psj.2022.102027
Li, N., Shen, B., Liu, Y., Weng, P., and Wu, Z. (2023). Heterologous expression and characterization of Bacillus velezensis SW5 serine protease involved in the hydrolysis of anchovy protein. J. Sci. Food Agric. 103 (7), 3468–3478. doi:10.1002/jsfa.12514
Li, Y., Cheng, Y., Zhang, Z., Wang, Y., Mintah, B. K., Dabbour, M., et al. (2020). Modification of rapeseed protein by ultrasound-assisted pH shift treatment: Ultrasonic mode and frequency screening, changes in protein solubility and structural characteristics. Ultrason. Sonochemistry 69, 105240. doi:10.1016/j.ultsonch.2020.105240
Lin, Y. H., and Mui, J. J. (2017). Comparison of dietary inclusion of commercial and fermented soybean meal on oxidative status and nonspecific immune responses in white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 63, 208–212. doi:10.1016/j.fsi.2017.02.011
Luján-Rhenals, D. E., Morawicki, R. O., Gbur, E. E., and Ricke, S. C. (2015). Fermentation of soybean meal hydrolyzates with Saccharomyces cerevisiae and Zymomonas mobilis for ethanol production. J. Food Sci. 80 (7), E1512–E1518. doi:10.1111/1750-3841.12907
Ma, M., Wei, C., Chen, M., Wang, H., Gong, Y., and Hu, Q. (2022). Effects of nutritional mode on the physiological and biochemical characteristics of the mixotrophic flagellate Poterioochromonas malhamensis and the potential ecological implications. Microorganisms 10 (5), 852. doi:10.3390/microorganisms10050852
Medeiros, S., Xie, J., Dyce, P. W., Cai, H. Y., DeLange, K., Zhang, H., et al. (2018). Isolation of bacteria from fermented food and grass carp intestine and their efficiencies in improving nutrient value of soybean meal in solid state fermentation. J. Animal Sci. Biotechnol. 9, 29. doi:10.1186/s40104-018-0245-1
Peng, C., Cao, C., He, M., Shu, Y., Tang, X., Wang, Y., et al. (2018). Soybean glycinin- and β-conglycinin-induced intestinal damage in piglets via the p38/JNK/NF-κB signaling pathway. J. Agric. Food Chem. 66 (36), 9534–9541. doi:10.1021/acs.jafc.8b03641
Pereira, P. P., Torres Tejerizo, G. A., Fernandez, M., Blanch, A. R., Gonzalez, P. S., and Agostini, E. (2020). Polyphasic characterization and identification of the bioremediation agent Bacillus sp. sfc 500-1e. Genomics 112 (6), 4525–4535. doi:10.1016/j.ygeno.2020.08.008
Qi, N., Zhan, X., Milmine, J., Sahar, M., Chang, K. H., and Li, J. (2023). Isolation and characterization of a novel hydrolase-producing probioticBacillus licheniformis and its application in the fermentation of soybean meal. Front. Nutr. 10, 1123422. doi:10.3389/fnut.2023.1123422
Rizvi, N. B., Aleem, S., Khan, M. R., Ashraf, S., and Busquets, R. (2022). Quantitative estimation of protein in sprouts of Vigna radiate (mung beans), Lens culinaris (lentils), and Cicer arietinum (chickpeas) by Kjeldahl and lowry methods. Molecules 27 (3), 814. doi:10.3390/molecules27030814
Shikha, S. A., and Darmwal, N. S. (2007). Improved production of alkaline protease from a mutant of alkalophilic Bacillus pantotheneticus using molasses as a substrate. Bioresour. Technol. 98, 881–885. doi:10.1016/j.biortech.2006.03.023
Sun, T., Wang, R., Sun, D., Li, S., Xu, H., Qiu, Y., et al. (2020). Antibacterial effects of ellagitannins from Acalypha wilkesiana var. macafeana hort.: Surface morphology analysis with environmental scanning electron microscopy and synergy with antibiotics. Bioresour. Technol. 318, 124268. doi:10.1016/j.biortech.2020.124268
Sun, X. X., Chen, D. D., Deng, S. Q., Zhang, G. M., Peng, X., and Sa, R. N. (2022). Using combined Lactobacillus and quorum quenching enzyme supplementation as an antibiotic alternative to improve broiler growth performance, antioxidative status, immune response, and gut microbiota. Poult. Sci. 101 (9), 101997. doi:10.1016/j.psj.2022.101997
Van Eenennaam, A. L., and Young, A. E. (2014). Prevalence and impacts of genetically engineered feedstuffs on livestock populations. J. Animal Sci. 92, 4255–4278. doi:10.2527/jas.2014-8124
Wang, H., Yang, L., Ping, Y., Bai, Y., Luo, H., Huang, H., et al. (2016). Engineering of a Bacillus amyloliquefaciens strain with high neutral protease producing capacity and optimization of its fermentation conditions. PLoS One 11 (1), e0146373. doi:10.1371/journal.pone.0146373
Wattanakul, W., Thongprajukaew, K., Hahor, W., and Suanyuk, N. (2021). Optimal replacement of soybean meal with fermented palm kernel meal as protein source in a fish meal-soybean meal-based diet of sex reversed red tilapia (Oreochromis niloticus × O. mossambicus). Anim. (Basel) 11 (8), 2287. doi:10.3390/ani11082287
Yao, Y., Li, H., Li, J., Zhu, B., and Gao, T. (2021). Anaerobic solid-state fermentation of soybean meal with Bacillus sp. to improve nutritional quality. Front. Nutr. 8, 706977. doi:10.3389/fnut.2021.706977
Yu, P., Wang, X., Huang, X., Ren, Q., and Yan, T. (2019). Purification and characterization of a propanol-tolerant neutral protease fromBacillus sp. ZG20. Prep. Biochem. Biotechnol. 49 (7), 718–726. doi:10.1080/10826068.2019.1605526
Zhang, Y., Wei, R., Azi, F., Jiao, L., Wang, H., He, T., et al. (2022). Solid-state fermentation withRhizopus oligosporusRT-3 enhanced the nutritional properties of soybeans. Front. Nutr. 9, 972860. doi:10.3389/fnut.2022.972860
Zheng, L., Li, D., Li, Z. L., Kang, L. N., Jiang, Y. Y., Liu, X. Y., et al. (2017). Effects of Bacillus fermentation on the protein microstructure and anti-nutritional factors of soybean meal. Lett. Appl. Microbiol. 65 (6), 520–526. doi:10.1111/lam.12806
Keywords: Bacillus amyloliquefaciens, protease, solid fermentation, soybean meal, glycinin
Citation: Huang X, Li H, Han T, Wang J, Ma Z and Yu X (2023) Isolation and identification of protease-producing Bacillus amyloliquefaciens LX-6 and its application in the solid fermentation of soybean meal. Front. Bioeng. Biotechnol. 11:1226988. doi: 10.3389/fbioe.2023.1226988
Received: 22 May 2023; Accepted: 05 July 2023;
Published: 13 July 2023.
Edited by:
Zhi-Qiang Liu, Zhejiang University of Technology, ChinaReviewed by:
Kridsa Unban, Chiang Mai University, ThailandZhenshang Xu, Qilu University of Technology, China
Copyright © 2023 Huang, Li, Han, Wang, Ma and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zheng Ma, mazheng520@163.com
†These authors have contributed equally to this work
 Xinyi Huang
Xinyi Huang Huijie Li1†
Huijie Li1†  Tao Han
Tao Han Jiteng Wang
Jiteng Wang Zheng Ma
Zheng Ma Xiaoping Yu
Xiaoping Yu