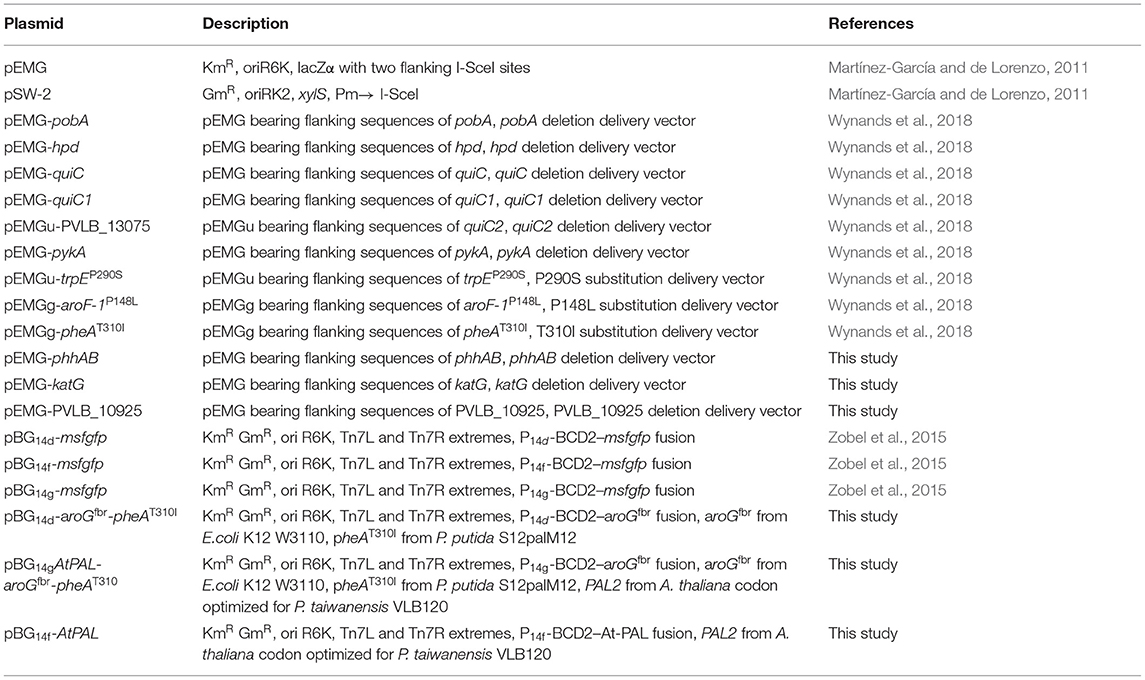
Rational Engineering of Phenylalanine Accumulation in Pseudomonas taiwanensis to Enable High-Yield Production of Trans-Cinnamate
- 1Institute of Bio- and Geosciences (IBG-1: Biotechnology), Forschungszentrum Jülich GmbH, Jülich, Germany
- 2Institute of Applied Microbiology, Rheinisch-Westfälische Technische Hochschule (RWTH) Aachen University, Aachen, Germany
Microbial biocatalysis represents a promising alternative for the production of a variety of aromatic chemicals, where microorganisms are engineered to convert a renewable feedstock under mild production conditions into a valuable chemical building block. This study describes the rational engineering of the solvent-tolerant bacterium Pseudomonas taiwanensis VLB120 toward accumulation of L-phenylalanine and its conversion into the chemical building block t-cinnamate. We recently reported rational engineering of Pseudomonas toward L-tyrosine accumulation by the insertion of genetic modifications that allow both enhanced flux and prevent aromatics degradation. Building on this knowledge, three genes encoding for enzymes involved in the degradation of L-phenylalanine were deleted to allow accumulation of 2.6 mM of L-phenylalanine from 20 mM glucose. The amino acid was subsequently converted into the aromatic model compound t-cinnamate by the expression of a phenylalanine ammonia-lyase (PAL) from Arabidopsis thaliana. The engineered strains produced t-cinnamate with yields of 23 and 39% Cmol Cmol−1 from glucose and glycerol, respectively. Yields were improved up to 48% Cmol Cmol−1 from glycerol when two enzymes involved in the shikimate pathway were additionally overexpressed, however with negative impact on strain performance and reproducibility. Production titers were increased in fed-batch fermentations, in which 33.5 mM t-cinnamate were produced solely from glycerol, in a mineral medium without additional complex supplements. The aspect of product toxicity was targeted by the utilization of a streamlined, genome-reduced strain, which improves upon the already high tolerance of P. taiwanensis VLB120 toward t-cinnamate.
Introduction
Trans-cinnamate is an aromatic compound naturally occurring in plants, where it serves as central intermediate for the biosynthesis of a large number of substances, including coumarins, flavonoids, and phenylpropanoids (Chemler and Koffas, 2008; Vogt, 2010). It is widely used in industry for flavoring, pharmaceuticals, and cosmetics (Fausta et al., 1999; De et al., 2011; Vargas-Tah and Gosset, 2015) and can serve as precursor for bio-based drop-in chemicals such as styrene (McKenna and Nielsen, 2011) or for value-added chemicals such as the stilbene pinosylvin (van Summeren-Wesenhagen and Marienhagen, 2015). Commercial production currently happens from petroleum-based, non-renewable feedstocks in processes that demand high amounts of energy and release toxic by-products (Bruckner, 2010; Tietze et al., 2015). This comes along with increasing apprehension on global climate change and depleting aromatic fossil resources. Hence, there are both environmental and economic drivers for alternative synthesis routes.
Microbial production by whole-cell biocatalysis represents a less environmentally demanding alternative to the common and well-established petrochemical processes (Hatti-Kaul et al., 2007; Becker and Wittmann, 2012; Cho et al., 2015; Kallscheuer et al., 2019). Herein, microbes can be supplied with renewable feedstocks (e.g., glucose, glycerol, or lignin, Kohlstedt et al., 2018; Johnson et al., 2019), and metabolic conversion enables the synthesis of diverse products under mild production conditions, either via native enzymes (Hosseinpour Tehrani et al., 2019) or by heterologous expression of foreign genes (Kallscheuer et al., 2019). In plants, t-cinnamate is formed through deamination of the amino acid L-phenylalanine by phenylalanine ammonia-lyase (PAL) (Cochrane et al., 2004; Huang et al., 2010), and can be further converted into the cis-isoform by photoisomerization (Salum and Erra-Balsells, 2013). While only the trans-isoform is involved in biosynthesis pathways in plants (Salum and Erra-Balsells, 2013), cis-cinnamate displays higher anti-bacterial activities (Chen et al., 2011). The heterologous synthesis of this enzyme in various microorganisms, including Escherichia coli (Vargas-Tah et al., 2015; Bang et al., 2018) or Streptomyces lividans (Noda et al., 2011) enabled t-cinnamate production. Product toxicity is a limiting factor for the efficiency of many microbial production process with these hosts (McKenna and Nielsen, 2011). From a biochemical engineering perspective, it is important to “begin with the end in mind” when developing microbial production strains (Straathof et al., 2019), and product toxicity is one important aspect for this.
Bacteria of the genus Pseudomonas are considered as promising alternative host to produce aromatics such as t-cinnamate. Their robust growth behavior and metabolic versatility recently enabled the synthesis of many different industrially relevant compounds, including a variety of aromatics (Nijkamp et al., 2007; Kuepper et al., 2015; Wynands et al., 2018), rhamnolipids (Tiso et al., 2017), terpenes (Mi et al., 2014), or prodiginines (Domröse et al., 2015). This is in particular due to their high stress tolerance, which has been extensively investigated in the past (Kusumawardhani et al., 2018). Pseudomonads are able to thrive under both endogenous and exogenous oxidative stress, enabled by their particular central carbon metabolism architecture (Chavarría et al., 2013; Nikel et al., 2015). Furthermore, they are equipped with a variety of native tolerance mechanisms that allow growth in the presence of highly toxic compounds (Sardessai and Bhosle, 2002; Segura et al., 2012). For this, Pseudomonads are able to adapt the composition of the inner and outer membrane to lower the permeability for substrates and they can increase their production of energy to fuel energy-consuming tolerance mechanisms (Isken and de Bont, 1998; Ramos et al., 2002; Belda et al., 2016). Some strains of Pseudomonas additionally express solvent efflux pumps that can actively extrude toxic compounds from the inner membrane (Kieboom et al., 1998) and degrade solvents such as toluene (Ramos et al., 2002) or styrene (Velasco et al., 1998). Additional transporters confer higher resistance to further aromatic molecules like p-hydroxybenzoate or p-coumarate (Verhoef et al., 2010; Calero et al., 2018). The native capabilities of Pseudomonas species thus form a strong basis for a biocatalytic process for t-cinnamate synthesis by expanding process options due to reduced product toxicity.
To further exploit its potential as industrial production strain, we recently enhanced the bioprocess features of the solvent-tolerant strain Pseudomonas taiwanensis VLB120 by successive feature reduction (Wynands et al., 2019). The deletion of redundant genomic elements such as proviral segments, genes for biofilm formation and flagella expression and the megaplasmid pSTY resulted in strains with 15% enhanced growth rates and increased biomass yields, thereby improving the overall performance of the strain under bioprocess conditions. In addition, genes encoding for the efflux pump TtgGHI located on the pSTY plasmid were re-integrated into the chromosome of the genome-reduced chassis (GRC) strains to maintain tolerance enabled by this efflux pump. These modifications, in combination with inherent tolerance traits, make the engineered strains promising hosts to efficiently synthesize aromatic compounds like t-cinnamate, as well as potential hydrophobic derivatives such as styrene or stilbenes.
The potential of Pseudomonas species for t-cinnamate production has been demonstrated in a strain of P. putida S12 by the expression of PAL from Rhodosporidium toruloides (Nijkamp et al., 2005). Here, enhanced precursor supply was realized by mutagenesis and subsequent selection on the toxic analog m-fluoro-phenylalanine, leading to the production of 5 mM t-cinnamate with a yield of 6.7% Cmol Cmol−1. However, although genomic analysis of this strain provided leads to enhance the flux through the shikimate pathway in P. taiwanensis VLB120 (Wynands et al., 2018), the genetic rearrangements enabling the enhanced flux specifically toward L-phenylalanine have not been determined. We recently reported a study on phenol production from L-tyrosine in P. taiwanensis VLB120, where 22 mutations in 50 different strains delivered a comprehensive insight on the rational engineering of L-tyrosine accumulation (Wynands et al., 2018). In the most productive strain, five genes involved in the degradation of aromatic intermediates and the gene pykA were deleted to increase the precursor supply for the synthesis of aromatic amino acids. The flux through the shikimate pathway was further enhanced by genomic modifications of genes at their native chromosomal locus resulting in a strain able to accumulate up to 2.8 mM of L-tyrosine from 20 mM glucose.
The aim of this study was to build on this knowledge to redirect the enhanced flux to L-tyrosine in P. taiwanensis GRC3 Δ5ΔpykA-tap (Wynands, 2018) toward L-phenylalanine, given the close relation of their biosynthesis pathways, and thereby expanding the potential product range. Furthermore, the superior tolerance of the streamlined P. taiwanensis GRC toward high concentrations of t-cinnamate was assessed. Routes involved in L-phenylalanine degradation were deleted, leading to L-phenylalanine accumulation. Titers were further increased by an additional overexpression of feedback-resistant versions of the bottleneck enzymes AroG and PheA. Heterologous expression of a PAL from Arabidopsis thaliana in the engineered strains then enabled the production of t-cinnamate. In a completely minimal medium, the plasmid-free chassis strains produced up to 6.3 mM of t-cinnamate from glycerol, which corresponds to a yield of 48% Cmol Cmol−1. Cultivation in fed-batch fermentations resulted in titers of around 33.5 mM, which is the highest reported titer of microbially produced t-cinnamate in a cultivation medium without complex supplements. The engineered strains can further serve as efficient platform to produce various products from L-phenylalanine and t-cinnamate, including styrene, benzoate, or plant polyphenols such as pinosylvin.
Materials and Methods
Bacterial Strains, Plasmids, and Cultivation Conditions
Strains and plasmids used in this study can be found in Tables 1, 2. For cloning purposes, E. coli and Pseudomonas cells were cultivated at 37 or 30°C, respectively, either in liquid LB medium containing 5 g L−1 sodium chloride or on solid LB agar plates [additionally containing 1.5% (w/v) agar]. After tri- or four-parental mating procedures, Pseudomonads were isolated on cetrimide agar (Sigma Aldrich) plates supplemented with 10 mL L−1 glycerol. Kanamycin (50 μg mL−1) or gentamicin (20 μg mL−1) was added to cultures or plates when necessary.
During production and toxicity experiments in shake flasks and well plates, liquid cultures of P. taiwanensis were grown in mineral salt medium (MSM) adapted from Hartmans et al. (1989) at pH 7.0 without the addition of antibiotics. The medium's standard phosphate buffer capacity was increased to 5-fold (111.5 mM K2HPO4 and 68 mM NaH2PO4). Glucose or glycerol were added as sole carbon source at indicated concentrations. Main cultures were inoculated at an OD600 of 0.2, from seed cultures grown in MSM containing glucose as carbon source. Production experiments were performed in 500 mL Erlenmeyer flasks with a culture volume of 50 mL, which were cultivated in a rotary shaker with a frequency of 200 rpm and a throw of 50 mm. Toxicity assays were performed in 96-round low-well plates with a filling volume of 200 μL in the System Duetz® cultivation system (EnzyScreen, Leiden, Netherlands) with a shaking frequency of 225 rpm. OD600 values were calculated from the measured green values using a calibration. Phenylalanine accumulation was analyzed in 24-square deep-well plates with a filling volume of 5 mL and a shaking frequency of 225 rpm, and growth was monitored in the Growth profiler® system.
Plasmid Construction and Genomic Modification
For plasmid construction, DNA fragments were PCR amplified using the Q5 high-fidelity polymerase (New England Biolabs, New Ipswich, USA) with corresponding overhangs to enable subsequent Gibson assembly (Gibson et al., 2009) with the HiFi DNA assembly master mix (New England Biolabs, New Ipswich, USA). Primers were ordered from Eurofins Genomics (Ebersberg, Germany). The gene sequences for aroGfbr (from E. coli K12 W3110) and AtPAL (from A. thaliana) were codon-optimized for P. taiwanensis VLB120 using the online tool OPTIMIZER (Puigbo et al., 2007) and ordered as synthetic DNA fragments from Thermo Fisher Scientific (Waltham, USA). The gene pheAT310I was amplified from genomic DNA of strain P. putida S12palM12. pEMG-based plasmids were transformed into E. coli DH5α λpir cells, pBG-based plasmids were transformed into E. coli PIR2. Correct plasmid assembly was verified by Sanger sequencing performed by Eurofins Genomics (Ebersberg, Germany). Integration at the attTn7-site was achieved by patch-mating of the E. coli donor strain holding the respective pBG-plasmid, the helper strain E. coli HB101 pRK2013, DH5α λpir pTNS1 providing the required transposase and the recipient Pseudomonas taiwanensis strain as described by Wynands et al. (2018). After the mating procedure, Pseudomonas were isolated on Cetrimide agar containing gentamicin and correct integration was confirmed by colony PCR using the OneTaq Quick-Load Master Mix (New England Biolabs, New Ipswich, USA).
Genomic deletions and point mutations were realized using the I-SceI-based method developed by Martínez-García and de Lorenzo (2011) using a streamlined protocol adapted by Wynands et al. (2018). Successful deletions were verified by colony PCR using the OneTaq quick-Load Master Mix (New England Biolabs, New Ipswich, USA).
Fed-Batch in Controlled Bioreactors
Fermentations were carried out in fed-batch operation mode in DASbox® mini-bioreactors using the DASware control software (Eppendorf, Hamburg, Germany). The reactors were set up of 385 ml glass vessels, two Rushton-type impellers driven by direct overhead drives, feeding lines for acid, base, and carbon source, a temperature sensor, an EasyFerm Plus K8 120 pH-sensor (Hamilton Company, Reno, NV, USA) and an InPro® 6800 series O2 sensor (Mettler-Toledo International Inc., Columbus, OH, USA). The starting volume was 100 mL and the temperature was maintained at 30°C. Gas supply was provided via headspace with a starting gas flow rate of 6 sL h−1. The initial agitation frequency was 500 rpm. The dO2 was controlled with a cascade, keeping the concentration of diluted oxygen at 35% by first increasing the agitation speed up to 1,200 rpm, then increasing the oxygen concentration in the air supply to a maximum of 80% and subsequently increasing the gas flow rate. pH 7 was maintained by automatic addition of 5% NH3 or 1 M H2SO4. Cells were grown in MSM according to Hartmans et al. (1989), where the addition of mineral salts stock was increased by 2-fold (MgCl2.6H2O 0.2 g L−1, ZnSO4.7H2O 0.004 g L−1, CaCl2.2H2O 0.002 g L−1, FeSO4.7H2O 0.01 g L−1, Na2MoO4.2H2O 0.0004 g L−1, CuSO4.5H2O 0.0004 g L−1, CoCl2.6H2O 0.0008 g L−1, MnCL2.2H2O 0.002 g L−1). The initial batch medium contained either 20 mM of glucose or 40 mM of glycerol as sole carbon source. The reactors were operated in a dO2-controlled fed-batch mode. When glucose or glycerol was depleted, the dO2 signal rapidly increased as a result of metabolic arrest. A dO2 signal >70 triggered a feed pump, resulting in a feed shot of 5 mM of glucose or 10 mM of glycerol per trigger initiation.
Analytical Methods
Optical densities of liquid cultures were measured at 600 nm using an Ultrospec 10 Cell Density Meter (GE Healthcare, Illinois, USA).
Samples taken from the cultures were centrifuged at 13,000 rpm for 2–5 min and the supernatant was analyzed by HPLC. Aromatics quantification was performed using a Beckman System Gold 126 Solvent Module with a 168 diode 201 array detector (Beckman Coulter, Brea, USA) and an ISAspher 100-5 C18 BDS reversed-phase 202 column (ISERA, Düren, Germany) at 30°C and a flow rate of 0.8 mL min−1. Elution took place with a gradient starting at 90% H2O containing 0.1% (v/v) TFA and 10% methanol. This ratio was held for 2 min, followed by gradual increase to 100% methanol over the course of 8 min. After 2 min at 100% methanol, initial ratios were reached again within 1 min and held constant for further 2 min. UV detection was conducted at a wavelength of 245 nm. L-phenylalanine and L-tyrosine quantification shown in Figure 3 was performed with the same column and elution setup as mentioned before, in a Dionex Ultimate 3000 HPLC system, where detection took place with a Corona Veo charged aerosol detector (Thermo Fisher Scientific, Waltham, MA, USA). Glucose, gluconate and glycerol analysis was performed using a Beckman System Gold 126 Solvent Module with a System Gold 166 UV-detector (Beckman Coulter, Brea, USA) and Smartline RI Detector 2300 (KNAUER, Berlin, Germany) on a MetabAAC column (ISERA, Düren, Germany). Elution took place with 5 mM H2SO4 at an isocratic flow of 0.5 mL min−1 and a temperature of 30°C for 20 min. Glucose and glycerol were analyzed using the RI detector, gluconate concentrations were determined with the UV detector at a wavelength of 210 nm. Due to co-elution of glucose and gluconate, glucose concentrations were determined by subtraction of the gluconate concentration.
Results and Discussion
Enhanced Trans-Cinnamate Tolerance of Streamlined P. taiwanensis Chassis Strains
Product toxicity vastly influences the efficiency of a microbial production process. Cellular stress induced by toxic compounds leads to decreased production rates or complete growth arrest, thereby limiting feasible product titers and yields (Sardessai and Bhosle, 2002; McKenna and Nielsen, 2011; Kusumawardhani et al., 2018). Compared to hydrophobic aromatics such as styrene, t-cinnamate is mildly toxic at neutral pH and it exhibits a different mode of action. Increasing concentrations of t-cinnamate lead to enhanced osmotic stress in cell cultures, but will also cause the inhibition of specific enzymes and disturb cellular processes, resulting in growth defects (Olasupo et al., 2003; Guzman, 2014). Growth of E. coli, for example, is heavily impaired at concentrations above 2.7 mM (400 mg L−1) and completely inhibited at >5.4 mM (800 mg L−1) when t-cinnamate is added at the beginning of cultivation (Olasupo et al., 2003; McKenna and Nielsen, 2011). This necessitates the addition of expensive complex supplements such as yeast extract or casamino acids or a biotransformation approach with late PAL induction at high cell densities to allow for sufficient accumulation of the precursor L-phenylalanine (Noda et al., 2011; Bang et al., 2018).
These problems can be avoided by the utilization of robust, natively stress-resistant host strains such as Pseudomonas taiwanensis VLB120. As shown in Figure 1A, the wildtype can grow in the presence of 30 mM (4.4 g L−1) t-cinnamate in MSM with a rate of 0.24 ± 0.01 h−1. P. taiwanensis VLB120 is equipped with particular mechanisms that enable this superior tolerance toward a variety of toxic compounds. While RND-type efflux pumps mainly act on hydrophobic aromatic solvents and a number of antibiotics (Terán et al., 2003; Köhler et al., 2013; Volmer et al., 2014), an ABC transporter (Ttg2ABC) was recently identified as crucial for p-coumarate tolerance, the hydroxylated derivative of t-cinnamate (Calero et al., 2018). Furthermore, chaperone upregulation is observed as response to protein misfolding (Segura et al., 2012).
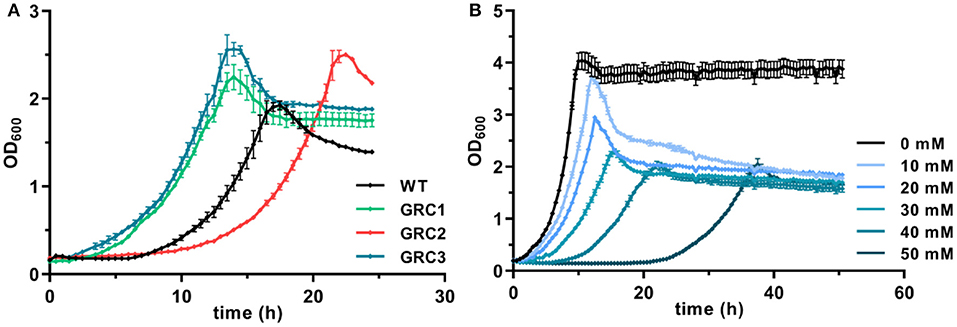
Figure 1. Enhanced growth of genome-reduced Pseudomonas taiwanensis chassis strains in the presence of t-cinnamate. (A) Growth of the P. taiwanensis VLB120 wildtype and the three genome-reduced chassis strains in the presence of 30 mM of t-cinnamate in mineral salts medium (MSM) containing 20 mM of glucose. (B) Growth of P. taiwanensis GRC3 at increasing t-cinnamate concentrations (0–50 mM) in MSM. Growth was monitored in the Growth Profiler®, error bars represent the standard error of the mean (n = 3).
To further exploit the native potential of P. taiwanensis VLB120 as microbial cell factory, we recently engineered a genome-reduced variant of this strain which exhibited higher growth rates and enhanced biomass formation (Wynands et al., 2019). The benefit of genome reduction for potential aromatics production strains is underlined by the tolerance test in the presence of 30 mM t-cinnamate (Figure 1A). The streamlined strains GRC1 and GRC3 had a reduced lag phase in the presence of high t-cinnamate concentrations, while the growth rate remains similar to the wildtype. In addition, the GRC strains reached higher final OD600 values compared to the wildtype. This is particularly important to improve the efficiency of a microbial production process by reducing the overall process time and lowering the amount of substrate needed to generated and maintain the biomass. The only exception is strain P. taiwanensis GRC2, which grew slower and with a longer lag phase than the wildtype. In contrast to GRC1 and 3, GRC2 constitutively expresses ttgGHI genes that encode a solvent-efflux pump. Although this constitutive expression greatly increases the fitness of GRC2 in the presence of hydrophobic solvents such as toluene, it causes a fitness reduction in the absence of solvents (Wynands et al., 2019). This drawback is absent in the other two strains, where the whole ttg operon is absent (GRC1), or the regulatory genes ttgVW are included to induce the pump in response to solvents (GRC3). The similar performance of GRC1 and GRC3 in the presence of t-cinnamate indicates that this aromatic acid does not induce expression of the ttgGHI, nor does the TtgGHI pump contribute to cinnamate tolerance. Given this similar performance, and in light of the potential to use a t-cinnamate-producing strain as platform for a variety of further industrially relevant hydrophobic aromatics, the strain GRC3 was chosen as host to maintain the possibility of solvent efflux by TtgGHI. As shown in Figure 1B, P. taiwanensis GRC3 can grow in the presence of t-cinnamate concentrations of up to 50 mM (7.4 g L−1), which is 10-fold higher than MIC for E. coli. The native tolerance potential of P. taiwanensis VLB120 in combination with improved bioprocess features obtained by streamlining of this strain hence set an ideal starting point for an efficient microbial production process of t-cinnamate.
Rational Engineering of L-phenylalanine Production
t-cinnamate is the deamination product of L-phenylalanine and the enhanced supply of this precursor in microbial chassis strains is thus crucial to allow efficient production. A variety of approaches toward L-phenylalanine overproduction were reported over the last years, delivering highly productive strains of e.g., E. coli (63 g L−1 from glucose) (Ding et al., 2016) or Corynebacterium glutamicum (16 g L−1 from glucose) (Zhang et al., 2015). Biosynthesis of L-phenylalanine and L-tyrosine both starts with the conversion of chorismate into prephenate, which then branches via phenylpyruvate to L-phenylalanine or via 4-hydroxyphenylpyruvate to L-tyrosine. Popular strategies aim to deregulate a 3-deoxy-D-arabino-heptolusonate-7-phosphate (DAHP) synthase (e.g., AroG, AroF, or AroH) or the chorismate mutase/prephenate dehydratase (PheA) activity and to increase the availability of the precursors phosphoenolpyruvate (PEP) and erythrose-4-phosphate (E4P) (Rodriguez et al., 2014; Ding et al., 2016; Huccetogullari et al., 2019). Pseudomonads have the catabolic potential to degrade a high number of aromatic compounds. This ability is a key feature applied for many biotechnological aspects such as the metabolism of complex substrates (Xu et al., 2018), but it also increases the difficulty of manufacturing aromatic amino acid accumulation in this species. We recently reported a study describing the rational engineering of P. taiwanensis VLB120 to produce phenol via the aromatic amino acid L-tyrosine (Wynands et al., 2018). The deletion of five genes (pobA, hpd, quiC, quiC1, quiC2) associated to the degradation of shikimate pathway-derived compounds resulted in strains unable to catabolize L-tyrosine, L-phenylalanine, p-hydroxybenzoate, and 3-dehydroshikimate to ensure aromatics accumulation. The flux toward L-tyrosine was subsequently enhanced by the insertion of point mutations into the native locus of genes in the shikimate pathway, resulting in enzymes with amino acid substitutions (TrpEP290S, AroF-1P148L, PheAT310I). The deletion of pykA (encoding for a pyruvate kinase) additionally reduced the flux of PEP toward pyruvate, thereby increasing the precursor pool for the shikimate pathway. An introduction of these mutations in the genome-reduced strains of P. taiwanensis VLB120 led to the efficient production of phenol from L-tyrosine (Wynands et al., 2019). As shown in Figure 3, the strain P. taiwanensis GRC3 Δ5ΔpykA-tap accumulates 2.85 ± 0.02 mM of tyrosine from 20 mM of glucose, which is comparable to the non-genome-reduced strain. Three genes involved in phenylalanine catabolism were subsequently deleted from the chromosome of P. taiwanensis GRC3 Δ5ΔpykA-tap: (i) phhAB encoding for phenylalanine-4-monooxygenase involved in the conversion of phenylalanine to tyrosine (Herrera et al., 2010), (ii) PVLB_10925, putatively encoding an aromatic-L-amino-acid decarboxylase responsible for the decarboxylation into phenylethylamine, and (iii) katG, putatively coding for a catalase-peroxidase which converts phenylalanine into phenyl acetamide (Figure 2).
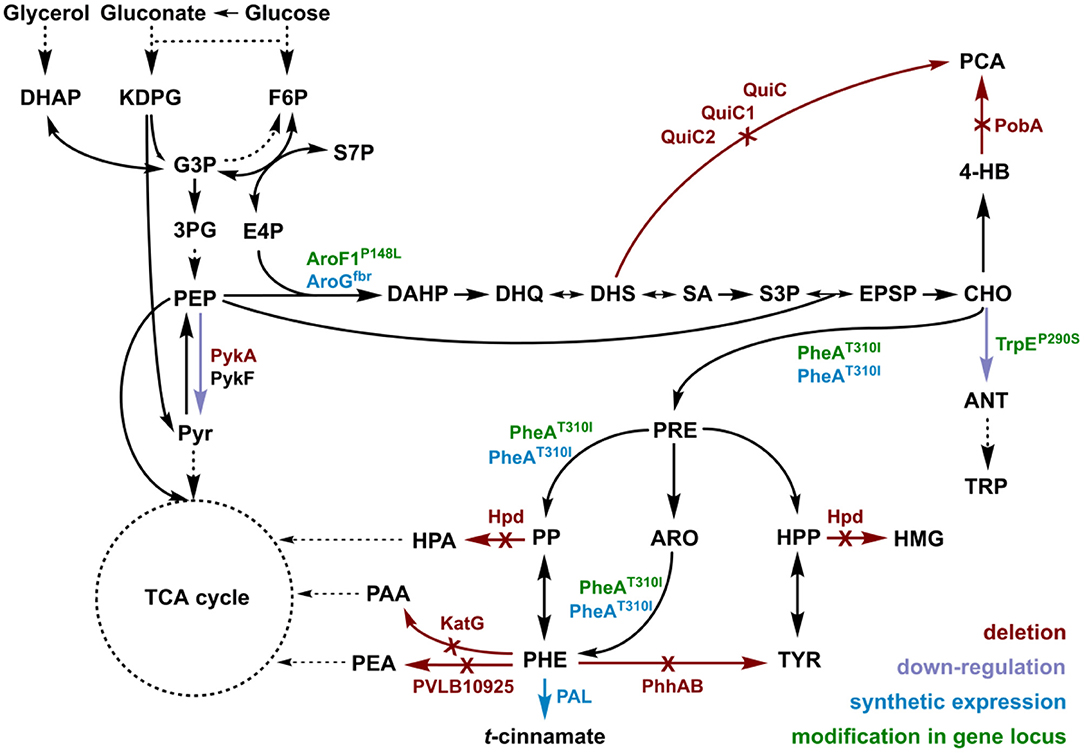
Figure 2. Genomic modifications introduced into strain P. taiwanensis GRC3 to enable accumulation of L-phenylalanine and subsequent deamination to t-cinnamate. Red arrows and annotations indicate gene deletions, purple arrows represent enzymatic downregulation, green annotations highlight point mutations introduced into the native gene locus, and blue tags and arrows represent the overexpression of heterologous genes. DAHP, dihydroxyacetone phosphate; KDPG, 2-keto-3-deoxy-6-phosphogluconate; F6P, fructose-6-phosphate; G3P, glyceraldehyde-3-phosphate; 3PG, 3-phosphoglycerat; S7P, seduheptulose-7-phosphate; PEP, phosphoenolpyruvate; PYR, pyruvate; E4P, erythrose-4-phosphate; DAHP, 3-deoxy-D-arabinoheptulosonate-7-phosphate; DHQ, 3-dehydroquinate; DHS, 3-dehydroshikimate; SA, shikimate; S3P, shikimate-3-phosphate; EPSP, 5-enolpyruvyl-shikimate-5-phosphate; CHO, chorismate; 4-HB, 4-hydroxybenzoate; PCA, protocatechuate; ANT, anthranilate; TRP, tryptophan; PRE, prephenate; PP, phenylpyruvate; HPP, 4-hydroxyphenylpyruvate; ARO, arogenate; TYR, tyrosine; PHE, phenylalanine; HMG, homogentisate; PAA, 2-phenylacetamide; PEA, phenylethylamine; PykA/PykF, pyruvate kinase isozymes; QuiC/QuiC1/QuiC2, 3-dehydroshikimate dehydratase isozymes; PobA, 4-hydroxybenzoate 3-monooxygenase; Hpd, 4-hydroxyphenylpyruvate dioxygenase; AroF-1P148L/AroGfbr, DAHP synthase isozymes; TrpEP290S, anthranilate synthase (component I); PheAT310I, bi-functional chorismate mutase/prephenate dehydratase; PhhAB, phenylalanine 4-monooxygenase; KatG, catalase-peroxidase; PVLB_10925, aromatic-L-amino-acid decarboxylase; PAL, phenylalanine ammonia-lyase.
Pseudomonads possess a variety of transporters that allow export and import of aromatic amino acids. Transcriptome analysis of a Pseudomonas putida strain with increased flux toward tyrosine revealed that upon increased intracellular aromatic amino acid levels, amino acid exporters are upregulated, while uptake systems are downregulated (Wierckx et al., 2008). Aromatics accumulation shown in Figure 3 displays extracellular concentrations, indicating enhanced efflux of both tyrosine and phenylalanine.
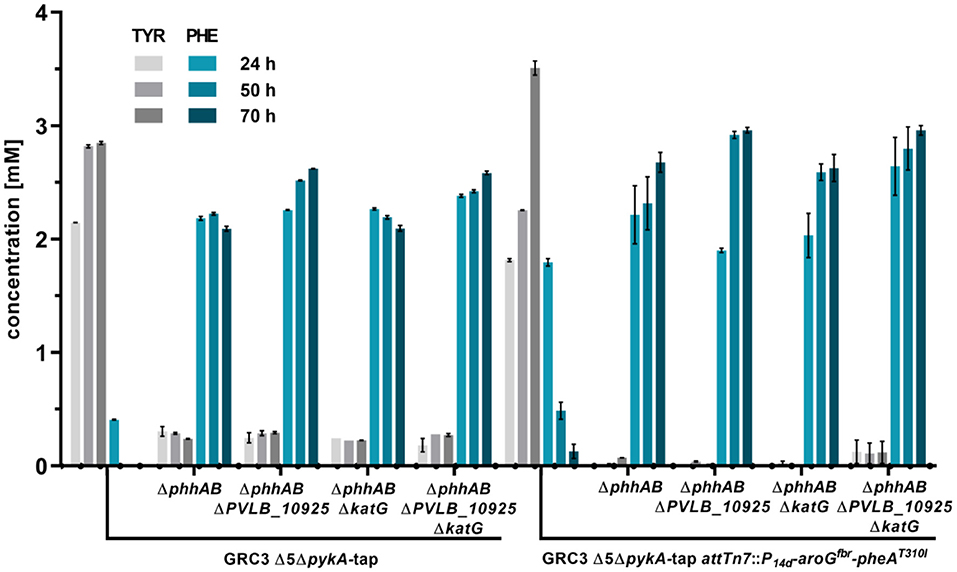
Figure 3. L-phenylalanine and L-tyrosine accumulation of P. taiwanensis GRC3 Δ5ΔpykA-tap and subsequent mutants with deletions to prevent L-phenylalanine degradation and additional overexpression of AroGfbr and PheAT310I. The strains were cultivated in MSM containing 20 mM glucose in a System Duetz® shaker. Error bars represent the standard error of the mean (n = 3).
The deletion of phhAB led to the accumulation of 2.22 ± 0.02 mM of phenylalanine and 0.29 ± 0.01 mM tyrosine after 50 h (Figure 3). This indicates that about 88% of L-tyrosine accumulating due to the enhanced flux toward prephenate in strain P. taiwanensis GRC3 Δ5ΔpykA-tap is stemming from L-phenylalanine. In most microorganisms, L-phenylalanine cannot be converted into L-tyrosine in which case it is solely synthesized from prephenate (Guroff and Ito, 1965). In Pseudomonas however, the pathway via L-phenylalanine seems to be a major route for L-tyrosine formation under the given conditions (Wierckx et al., 2009). As described for other Pseudomonas species, Xanthomonads and Alcalignes (Ahmad et al., 1990), this diversity of aromatic amino acid metabolism account for the flexibility of the organism to cope with a variety of end product analogs (Fiske et al., 1983). The concentration of L-phenylalanine in the culture of P. taiwanensis GRC3 Δ5ΔpykA-tapΔphhAB was slightly reduced after 70 h, indicating that other pathways involved in L-phenylalanine degradation were still active in this strain. The subsequent deletion of gene PVLB_10925 prevented this degradation, resulting instead in a further increase of the final titer to 2.62 ± 0.00 mM L-phenylalanine. The deletion of katG had no influence on phenylalanine accumulation and the strain shows a similar production pattern as the strain without a deletion. Indeed, as the progenitor strain P. taiwanensis GRC3 Δ5ΔpykA-tap was already not able to grow on L-phenylalanine and L-tyrosine (Wynands et al., 2018), this confirms that catabolic pathways are either not active (katG) or not connected to the central carbon metabolism (PVLB_10925) under the applied conditions, even though this strain contains the genetic inventory for the full degradation of the resulting 2-phenylacetamide and phenylethylamine. This is underlined by similar observations in an L-phenylalanine overproducing strain of P. putida DOT-T1E (Molina-Santiago et al., 2016).
L-Tyrosine and L-phenylalanine production could be increased further by the additional overexpression of AroGfbr, a feedback inhibition resistant versions of the DAHP synthase from E. coli K12 W3110 (Kikuchi et al., 1997) and PheAT310I from P. putida S12palM12 (Nijkamp et al., 2005). The coding genes were integrated chromosomally at the attTn7-site and expressed under the control of the constitutive promoter P14g (Zobel et al., 2015). Overexpression in the tyrosine-accumulating strain P. taiwanensis GRC3 Δ5ΔpykA-tap led to tyrosine titers of about 3.5 ± 0.11 mM after 70 h. Transient L-phenylalanine accumulation was observed with this strain, further underlining the high flux through PhhAB. Upon overexpression of aroGfbr and pheAT310I in strains with deletions of phhAB, katG and PVLB_10925, the resulting strain GRC3 Δ8ΔpykA-tap attTn7::P14d-aroGfbr-pheAAT31I (Δ8 = eight deletions of pathways involved in aromatics degradation) accumulated 3.0 ± 0.07 mM of L-phenylalanine after 70 h. Also, L-tyrosine production was reduced, likely due to PheAT310I increasing the flux of prephenate toward L-phenylalanine.
Production of t-cinnamate From Glucose and Glycerol in Shake Flasks
The L-phenylalanine-accumulating strain P. taiwanensis GRC3 Δ8ΔpykA-tap was subsequently used as host for the production of t-cinnamate. The deamination of L-phenylalanine into t-cinnamate was achieved using the PAL2 enzyme from A. thaliana. While four genes encoding PAL have been identified in A. thaliana (Cochrane et al., 2004), PAL2 displayed the highest specific activity when expressed in E. coli (McKenna and Nielsen, 2011). In addition, it has no activity on L-tyrosine as a substrate, in contrast to a variety of other yeast PALs (Cui et al., 2008). As overexpression of aroGfbr and pheAT310I improved L-phenylalanine yields by 15% (Figure 3), combinatorial expression with PAL on the Tn7 transposon under the control of P14g was applied for t-cinnamate production in the strain P. taiwanensis GRC3 Δ8ΔpykA-tap attTn7::P14gAtPAL- aroGfbr-pheAT310. The strain was cultivated in shake flasks in MSM containing either 20 mM of glucose or 40 mM of glycerol as sole carbon source. Glycerol is a promising alternative as feedstock for microbial production processes as abundant by-product of the bio-diesel production (Zambanini et al., 2016). When cultivated on glucose, the strain accumulated 3.80 ± 0.20 mM t-cinnamate, corresponding to a yield of 25.9 ± 0.1% Cmol Cmol−1, with a volumetric productivity of 0.10 ± 0.00 mM h−1 (Figure 4). From glycerol, the t-cinnamate titer was further increased to 6.33 ± 0.12 mM, corresponding to a yield of 47.5 ± 0.9% Cmol Cmol−1. As observed for the production of phenol and p-hydroxybenzoate (Wynands et al., 2018; Lenzen et al., 2019), metabolism of glycerol is highly beneficial for enhanced titers and yields of compounds derived from aromatic amino acids, likely due to metabolic rearrangements favoring the supply of PEP and E4P (Nikel et al., 2014; Poblete-Castro et al., 2019). The volumetric productivity on glycerol is slightly lower compared to glucose (0.09 ± 0.01 mM h−1), as a result of the reduced growth rate. While this strain has a very high yield, its reliable application as a production host was problematic. When the experiment described above was repeated, the cultures displayed varying growth behavior in terms of lag-phase, growth rates, and productivity (Figure S1), even though the cultivation procedure remained identical. Re-transformation of the Tn7-transposon bearing the overexpression constructs and culturing of single colonies yielded the same unstable phenotype. Sequencing of the attTn7-regions of theses strains revealed no mutations in this sequence and verified correct integration. Likely, a combination of the extreme drain on carbon posed by the high t-cinnamate yield, the burden of heterologous overexpression of genes, and the production of a mildly toxic compound provide a strong selection for suppressor mutations. This impairs the reproducibility and renders the strain P. taiwanensis GRC3 Δ8ΔpykA-tap attTn7::P14gAtPAL- aroGfbr-pheAT310 unsuitable as reliable production host in its current form.
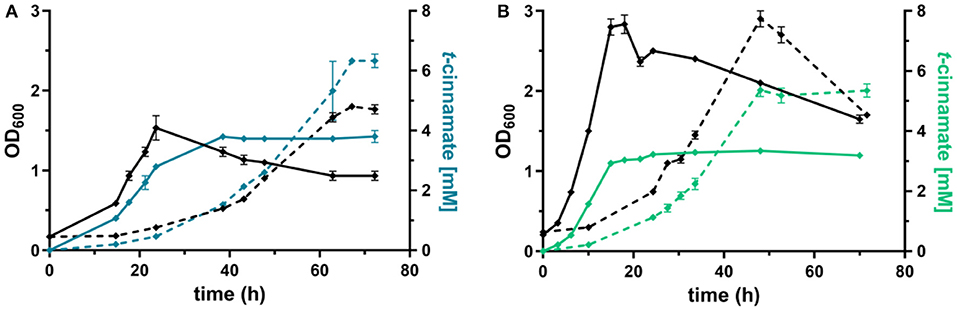
Figure 4. Shake flask cultivations of P. taiwanensis GRC3 Δ8ΔpykA-tap with varying heterologous expression modules. (A) Growth (black lines) and t-cinnamate production (blue lines) by P. taiwanensis GRC3 Δ8ΔpykA-tap attTn7::P14gAtPAL- aroGfbr-pheAT310 in MSM containing 20 mM glucose (solid line) or 40 mM of glycerol (dotted line). (B) Growth (black lines) and t-cinnamate production (green lines) by P. taiwanensis GRC3 Δ8ΔpykA-tap attTn7::P14fAtPAL in MSM containing 20 mM glucose (solid line) or 40 mM of glycerol (dotted line). Error bars represent the standard error of the mean (n = 3).
In contrast, P. taiwanensis GRC3 Δ8ΔpykA-tap attTn7::P14fAtPAL, harboring only the PAL for L-phenylalanine deamination under the control of the weaker P14f promoter, showed reliable growth and production patterns. This strain still carries feedback-resistant aroF-1P148L, pheAT310I, but only in their native context. The lack of in trans overexpression of aroGfbr and pheAT310I reduced t-cinnamate titers and yields slightly, with 3.34 ± 0.07 mM produced from glucose and 5.25 ± 0.22 mM from glycerol, which corresponds to yields of 22.8 ± 0.5 and 38.9 ± 1.6% Cmol Cmol−1, respectively. To the best of our knowledge, these are still the highest yields of t-cinnamate produced in a microbial process using a mineral medium without the addition of complex supplements. This strain furthermore reached higher final biomass and exhibited improved volumetric productivity on glucose (0.13 ± 0.00 mM h−1) and glycerol (0.11 ± 0.00 mM h−1) compared to the strain additionally expressing aroGfbr and pheAT310I. In the course of the cultivations, no accumulation of L-phenylalanine was observed. This indicates that PAL activity is not inhibited by the concentrations of t-cinnamate reached in these cultures, in contrast to observations in other hosts and with PALs from different organisms (Nijkamp et al., 2005; McKenna and Nielsen, 2011; Molina-Santiago et al., 2016). Furthermore, no L-tyrosine accumulation was observed, in contrast to the phenylalanine-producing equivalent that does not express PAL (Figure 3), likely due to a reduction of product inhibition of the upstream metabolic enzymes as a result of efficient conversion to t-cinnamate.
Production in Controlled Bioreactors
In order to assess higher-level t-cinnamate production under more industrially relevant conditions, the strains were cultivated in fed-batch fermentations in controlled bioreactors. The fermenters were operated in a dO2-stat fed-batch mode where carbon depletion and the resulting increase of the dO2 due to metabolic arrest triggered the initiation of a pulse feed of either 5 mM glucose or 10 mM glycerol (Johnson et al., 2016, 2019). This feeding protocol allows controlled addition of carbon source throughout the whole fermentation process, independent from time-varying carbon demands of the cell, thereby preventing excess carbon surplus (Johnson et al., 2016). The resulting feeding trends of single reactors are exemplarily shown in Figure S3. Cultivation took place at pH 7 in a mineral medium without the addition of complex supplements.
Under these conditions, the strain P. taiwanensis GRC3 Δ8ΔpykA-tap attTn7::P14gAtPAL-aroGfbr-pheAT310 accumulated 17.2 ± 0.28 mM of t-cinnamate from glucose, corresponding to a yield of 11.2 ± 0.6% (Cmol Cmol−1) and a volumetric productivity of 0.19 ± 0.20 mM h−1. As shown in Figure 5A, the culture reaches a final OD600 of 12.7 after around 65 h. At this point, a decrease in biomass concentration was observed. Furthermore, there was no significant increase in t-cinnamate titers from this point on, in spite of the fact that the base strain GRC3 tolerated higher concentrations of t-cinnamate up to 50 mM (Figure 1). It is possible that several factors impact the lowered growth performance of the production strains in the presence of t-cinnamate. One factor is the overexpression of heterologous genes which leads to lowered growth rates and impaired fitness as observed in shake flask experiments (Figure 4). Especially the overexpression of AroG and PheA appears to lead to increased cellular stress, which in turn also lowers the growth performance in the presence of toxic compounds. Furthermore, experiments regarding styrene toxicity have demonstrated a crucial difference in tolerance depending on whether a compound is added exogenously to the medium or produced intracellularly (Lian et al., 2016).
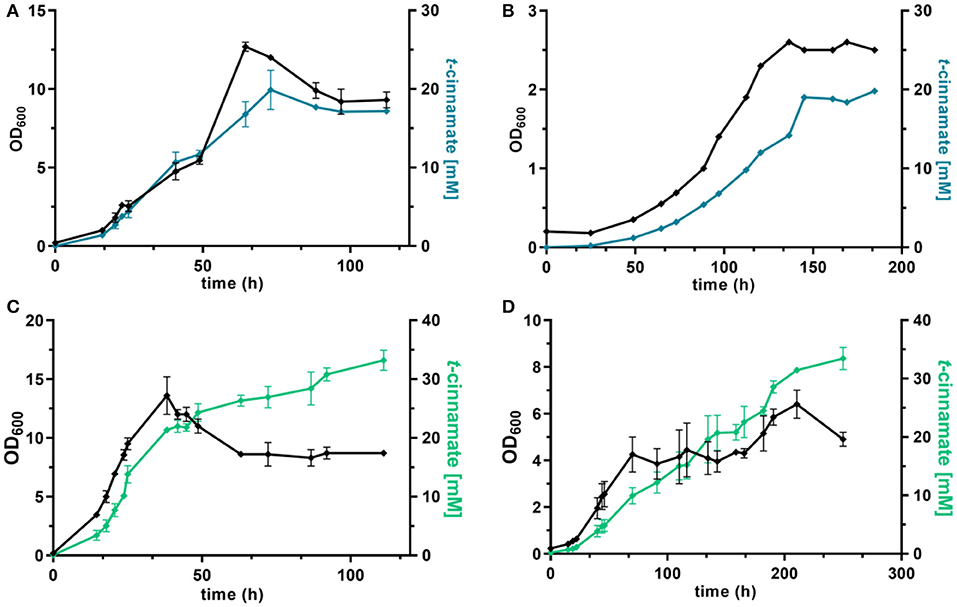
Figure 5. dO2-stat fed-batch fermentations of t-cinnamate producing strains of P. taiwanensis. Cultivations were performed in MSM, where the initial batch medium contained either glucose or glycerol as sole carbon source and the subsequent feeding solution contained solely the respective carbon source. The top graphs show growth (black lines) and t-cinnamate accumulation (blue lines) during fermentation of P. taiwanensis GRC3 Δ8ΔpykA-tap attTn7::P14gAtPAL- aroGfbr-pheAT310 on glucose (A) and glycerol (B). The glycerol figure (B) displays data of a single reactor. The bottom graphs represent growth (black lines) and t-cinnamate accumulation (green lines) during fermentation of P. taiwanensis GRC3 Δ8ΔpykA-tap attTn7::P14fAtPAL on glucose (C) and glycerol (D). The error bars represent the standard error (n = 2).
Fed-batch fermentations using glycerol as carbon source further underline complications using the phenotypically unstable strain P. taiwanensis GRC3 Δ8ΔpykA-tap attTn7::P14gAtPAL-aroGfbr-pheAT310 (Figure 5B; Figure S2). During fermentations, cultures in duplicate reactors showed different growth behavior while stemming from the same pre-culture. The cultures showed differences in lag-phase, growth rate and productivity. The final titers remained comparable, likely as a result of product inhibition as described before. Figure 5B shows a single fermentation on glycerol as an example. Here, the strain produced up to 19.8 mM t-cinnamate, with a yield of 47.8% Cmol and a volumetric productivity of 0.13 mM h−1. The extended lag phase observed during glycerol assimilation (Poblete-Castro et al., 2019) might even increase the occurrence of suppressor mutations by induction of the SOS response.
As observed for shake flask cultivations, reproducibility and stable production was restored in a strain overexpressing solely PAL for L-phenylalanine conversion. The strain P. taiwanensis GRC3 Δ8ΔpykA-tap attTn7::P14fAtPAL produced up to 33.2 ± 2.4 mM t-cinnamate from glucose (Figure 5C) and 33.5 ± 2.7 mM from glycerol (Figure 5D) in a fed-batch fermentation. While the productivity throughout the whole cultivation of P. taiwanensis GRC3 Δ8ΔpykA-tap attTn7::P14fAtPAL on glucose was 0.30 ± 0.02 mM h−1, increasing concentrations of t-cinnamate impaired the strain's fitness and productivity. The volumetric productivity within the first 25 h is 0.55 ± 0.08 mM h−1, which is reduced after 38 h to 0.16 ± 0.04 mM h−1. At the same time, a drop in OD600 was observed at concentrations above 20 mM t-cinnamate. As production continued and cells were still viable at this point as indicated by the ongoing substrate consumption, this drop in OD600 values hints toward an increased cumulative burden on the cell. The drop in OD600 was not caused by biofilm formation, likely due to the deletion of the biofilm-associated lap genes in the GRC3 strain. Membrane adaptation is a key mechanisms of Pseudomonas to cope with environmental stress. Membrane active substances and certain environmental conditions cause adaptations such as cis- to trans-isomerization of fatty acids to increase membrane rigidity and lead to the formation of outer membrane vesicles to facilitate biofilm formation (Eberlein et al., 2018). In addition, many porins as well as import and export pumps are differentially expressed under stress, altering membrane permeability (Volkers et al., 2006; Ramos et al., 2015). The importance of membrane structure on similar compounds such as p-coumarate has recently been demonstrated (Calero et al., 2018) and can likely be partially be transferred to t-cinnamate tolerance.
As enzyme inhibition of different PALs by t-cinnamate has been observed in various studies (Nijkamp et al., 2007; McKenna and Nielsen, 2011), the decrease in productivity could also be linked to this effect. However, no L-phenylalanine accumulation was observed over the course of the fermentations, indicating that the precursor was completely converted by the PAL. Furthermore, no L-tyrosine accumulation was observed during fermentations.
In contrast to the strain overexpressing AroGfbr and PheAT310I, experiments with strain P. taiwanensis GCR3 Δ8ΔpykA-tap attTn7::P14fAtPAL are highly reproducible. While an additional expression of AroGfbr and PheAT310I led to higher yields in shake flasks, solely PAL expression in the L-phenylalanine-overproducing chassis strains enables both higher titers and volumetric productivities in fed-batch fermentations (Table 3). To the best of our knowledge, these are the highest t-cinnamate yields reported for a microbial production process. While titers of 6.5 g L−1 (43.9 mM) have been reported in fed-batch fermentations of t-cinnamate-producing strains of E. coli (Bang et al., 2018), these processes required the addition of yeast extract in the initial batch medium and casamino acid in the feeding solution. The utilization of these engineered P. taiwanensis GRC strains enables growth without the addition of complex supplements. However, tolerance mechanisms for t-cinnamate remain to be further investigated and exploited to enhance titers and thus the feasibility of microbial t-cinnamate synthesis.
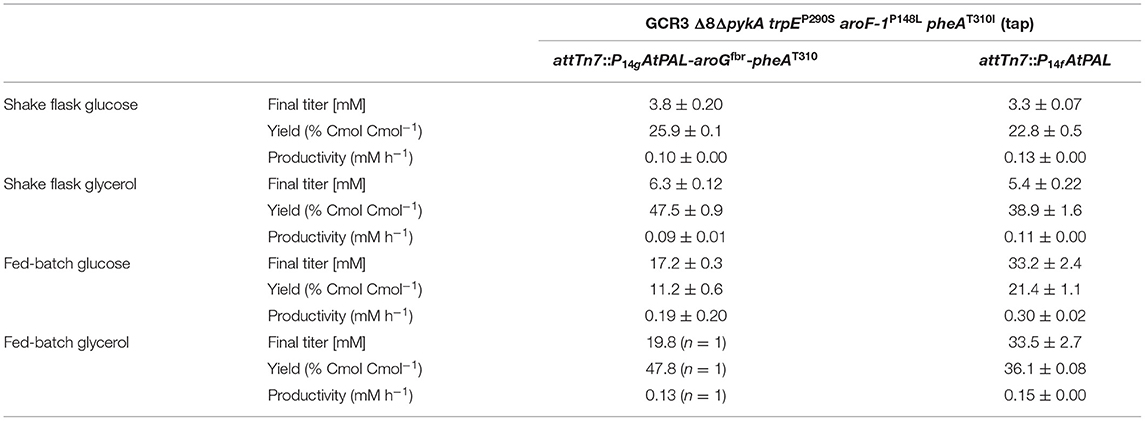
Table 3. Comparison of t-cinnamate titer, yield, and volumetric productivity of strains P. taiwanensis GRC3 Δ8ΔpykA-tap attTn7::P14gAtPAL- aroGfbr-pheAT310 and GRC3 Δ8ΔpykA-tap attTn7::P14fAtPAL in shake flask cultivations and fed-batch fermentations.
Conclusion
In this study, we describe the rational engineering of P. taiwanensis VLB120 towards efficient t-cinnamate production. The plasmid-free strain bearing no auxotrophies synthesized t-cinnamate from glucose or glycerol, with yields of up to 48% Cmol Cmol−1 in shake flask cultivations. Titers were increased up to 33.35 mM in fed-batch fermentations using glycerol as sole carbon source. As product titers achieved in this study impair fitness and productivity of the chassis strains, the native tolerance features of Pseudomonas allowing enhanced t-cinnamate tolerance require further investigation and enhancement to increase process efficiency. One promising target is the ABC transporter Ttg2ABC, an extrusion pump involved in p-coumarate tolerance (Calero et al., 2018). An overexpression of this pump might deliver enhanced tolerance toward t-cinnamate, its regulation however remains to be investigated. Fine-tuning of additional aroG and pheA overexpression, e.g., by using weaker or inducible promoters, could furthermore avoid the observed growth defects while still maintaining the high yields achieved with this setup. Overall, the results underline the high potential of Pseudomonas species to produce chemical building blocks using aromatic amino acids as precursors. The establishment of efficient microbial production of the model compound t-cinnamate will in future serve as foundation to expand the product collection of this versatile species, ranging from bulk chemicals such as styrene (Lee et al., 2019) to specialty compounds such as stilbenes (van Summeren-Wesenhagen and Marienhagen, 2015).
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
NW conceived and supervised the study with the help of LB. MO and MF performed the experiments with support of BW and CL. BW provided the GRC strains with deletions for L-tyrosine accumulation and deletion vectors. MO wrote the manuscript with the help of BW and NW. All authors read and approved the manuscript.
Funding
MO, BW, CL, and NW were funded by the German Research Foundation (DFG) through the Emmy Noether program (WI 4255/1-1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We kindly acknowledge Eppendorf AG for assistance regarding scripting of automated feed protocols.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2019.00312/full#supplementary-material
References
Ahmad, S., Weisburg, W. G., and Jensen, R. A. (1990). Evolution of aromatic amino acid biosynthesis and application to the fine-tuned phylogenetic positioning of enteric bacteria. J. Bacteriol. 172, 1051–1061. doi: 10.1128/jb.172.2.1051-1061.1990
Bang, H. B., Lee, K., Lee, Y. J., and Jeong, K. J. (2018). High-level production of trans-cinnamic acid by fed-batch cultivation of Escherichia coli. Process Biochem. 68, 30–36. doi: 10.1016/j.procbio.2018.01.026
Becker, J., and Wittmann, C. (2012). Bio-based production of chemicals, materials and fuels – Corynebacterium glutamicum as versatile cell factory. Curr. Opin. Biotechnol. 23, 631–640. doi: 10.1016/j.copbio.2011.11.012
Belda, E., van Heck, R. G. A., José Lopez-Sanchez, M., Cruveiller, S., Barbe, V., Fraser, C., et al. (2016). The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ. Microbiol. 18, 3403–3424. doi: 10.1111/1462-2920.13230
Boyer, H. W., and Roulland-Dussoix, D. (1969). A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41, 459–472. doi: 10.1016/0022-2836(69)90288-5
Bruckner, R. (2010). Organic Mechanisms: Reactions, Stereochemistry and Synthesis, 3rd Edn, ed M. Harmata. Berlin: Springer. doi: 10.1007/978-3-642-03651-4
Calero, P., Jensen, S. I., Bojanovič, K., Lennen, R. M., Koza, A., and Nielsen, A. T. (2018). Genome-wide identification of tolerance mechanisms toward p-coumaric acid in Pseudomonas putida. Biotechnol. Bioeng. 115, 762–774. doi: 10.1002/bit.26495
Chavarría, M., Nikel, P. I., Pérez-Pantoja, D., and de Lorenzo, V. (2013). The Entner-Doudoroff pathway empowers Pseudomonas putida KT2440 with a high tolerance to oxidative stress. Environ. Microbiol. 15, 1772–1785. doi: 10.1111/1462-2920.12069
Chemler, J. A., and Koffas, M. A. (2008). Metabolic engineering for plant natural product biosynthesis in microbes. Curr. Opin. Biotechnol. 19, 597–605. doi: 10.1016/j.copbio.2008.10.011
Chen, Y.-L., Huang, S.-T., Sun, F.-M., Chiang, Y.-L., Chiang, C.-J., Tsai, C.-M., et al. (2011). Transformation of cinnamic acid from trans- to cis-form raises a notable bactericidal and synergistic activity against multiple-drug resistant Mycobacterium tuberculosis. Eur. J. Pharm. Sci. 43, 188–194. doi: 10.1016/j.ejps.2011.04.012
Cho, C., Choi, S. Y., Luo, Z. W., and Lee, S. Y. (2015). Recent advances in microbial production of fuels and chemicals using tools and strategies of systems metabolic engineering. Biotechnol. Adv. 33, 1455–1466. doi: 10.1016/j.biotechadv.2014.11.006
Cochrane, F. C., Davin, L. B., and Lewis, N. G. (2004). The Arabidopsis phenylalanine ammonia lyase gene family: kinetic characterization of the four PAL isoforms. Phytochemistry 65, 1557–1564. doi: 10.1016/j.phytochem.2004.05.006
Cui, J. D., Jia, S. R., and Sun, A. Y. (2008). Influence of amino acids, organic solvents and surfactants for phenylalanine ammonia lyase activity in recombinant Escherichia coli. Lett. Appl. Microbiol. 46, 631–635. doi: 10.1111/j.1472-765X.2008.02364.x
de Lorenzo, V., and Timmis, K. N. (1994). [31] Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235, 386–405. doi: 10.1016/0076-6879(94)35157-0
De, P., Baltas, M., and Bedos-Belval, F. (2011). Cinnamic acid derivatives as anticancer agents - a review. Curr. Med. Chem. 18, 1672–1703. doi: 10.2174/092986711795471347
Ding, D., Liu, Y., Xu, Y., Zheng, P., Li, H., Zhang, D., et al. (2016). Improving the production of L-phenylalanine by identifying key enzymes through multi-enzyme reaction system in vitro. Sci. Rep. 6:32208. doi: 10.1038/srep32208
Domröse, A., Klein, A. S., Hage-Hülsmann, J., Thies, S., Svensson, V., Classen, T., et al. (2015). Efficient recombinant production of prodigiosin in Pseudomonas putida. Front. Microbiol. 6:972. doi: 10.3389/fmicb.2015.00972
Eberlein, C., Baumgarten, T., Starke, S., and Heipieper, H. J. (2018). Immediate response mechanisms of Gram-negative solvent-tolerant bacteria to cope with environmental stress: cis-trans isomerization of unsaturated fatty acids and outer membrane vesicle secretion. Appl. Microbiol. Biotechnol. 102, 2583–2593. doi: 10.1007/s00253-018-8832-9
Fausta, N., Mirella, N., di Felice, M., and Scaccini, C. (1999). Benzoic and cinnamic acid derivatives as antioxidants: structure–activity relation. J. Agric. Food Chem. 47, 1453–1459. doi: 10.1021/jf980737w
Fiske, M. J., Whitaker, R. J., and Jensen, R. A. (1983). Hidden overflow pathway to L-phenylalanine in Pseudomonas aeruginosa. J. Bacteriol. 154, 623–631.
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Guroff, G., and Ito, T. (1965). Phenylalanine hydroxylation by Pseudomonas species (ATCC 11299a). J. Biol. Chem. 240, 142–146.
Guzman, J. D. (2014). Natural cinnamic acids, synthetic derivatives and hybrids with antimicrobial activity. Molecules 19, 19292–19349. doi: 10.3390/molecules191219292
Hartmans, S., Smits, J. P., Van Der Werf, M. J., Volkering, F., and De Bont, J. A. M. (1989). 2-Phenylethanol in the styrene-degrading metabolism of styrene oxide and 2-phenylethanol in the styrene-degrading Xanthobacter strain 124X. Appl. Environ. Microbiol. 55, 2850–2855.
Hatti-Kaul, R., Törnvall, U., Gustafsson, L., and Börjesson, P. (2007). Industrial biotechnology for the production of bio-based chemicals - a cradle-to-grave perspective. Trends Biotechnol. 25, 119–124. doi: 10.1016/j.tibtech.2007.01.001
Herrera, M. C., Duque, E., RodrÃ-guez-Herva, J. J., Fernández-Escamilla, A. M., and Ramos, J. L. (2010). Identification and characterization of the PhhR regulon in Pseudomonas putida. Environ. Microbiol. 12, 1427–1438. doi: 10.1111/j.1462-2920.2009.02124.x
Hosseinpour Tehrani, H., Geiser, E., Engel, M., Hartmann, S. K., Hossain, A. H., Punt, P. J., et al. (2019). The interplay between transport and metabolism in fungal itaconic acid production. Fungal Genet. Biol. 125, 45–52. doi: 10.1016/j.fgb.2019.01.011
Huang, J., Gu, M., Lai, Z., Fan, B., Shi, K., Zhou, Y.-H., et al. (2010). Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 153, 1526–1538. doi: 10.1104/pp.110.157370
Huccetogullari, D., Luo, Z. W., and Lee, S. Y. (2019). Metabolic engineering of microorganisms for production of aromatic compounds. Microb. Cell Fact. 18:41. doi: 10.1186/s12934-019-1090-4
Isken, S., and de Bont, J. A. M. (1998). Bacteria tolerant to organic solvents. Extremophiles 2, 229–238. doi: 10.1007/s007920050065
Johnson, C. W., Salvachúa, D., Khanna, P., Smith, H., Peterson, D. J., and Beckham, G. T. (2016). Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab. Eng. Commun. 3, 111–119. doi: 10.1016/j.meteno.2016.04.002
Johnson, C. W., Salvachúa, D., Rorrer, N. A., Black, B. A., Vardon, D. R., St. John, P. C., et al. (2019). Innovative chemicals and materials from bacterial aromatic catabolic pathways. Joule 3, 1523–1537. doi: 10.1016/j.joule.2019.05.011
Kallscheuer, N., Classen, T., Drepper, T., and Marienhagen, J. (2019). Production of plant metabolites with applications in the food industry using engineered microorganisms. Curr. Opin. Biotechnol. 56, 7–17. doi: 10.1016/j.copbio.2018.07.008
Kieboom, J., Dennis, J. J., Zylstra, G. J., and de Bont, J. A. (1998). Active efflux of organic solvents by Pseudomonas putida S12 is induced by solvents. J. Bacteriol. 180, 6769–6772.
Kikuchi, Y., Tsujimoto, K., and Kurahashi, O. (1997). Mutational analysis of the feedback sites of phenylalanine-sensitive 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase of Escherichia coli. Appl. Environ. Microbiol. 63, 761–762.
Köhler, K. A., Rückert, C., Schatschneider, S., Vorhölter, F. J., Szczepanowski, R., Blank, L. M., et al. (2013). Complete genome sequence of Pseudomonas sp. strain VLB120 a solvent tolerant, styrene degrading bacterium, isolated from forest soil. J. Biotechnol. 168, 729–730. doi: 10.1016/j.jbiotec.2013.10.016
Kohlstedt, M., Starck, S., Barton, N., Stolzenberger, J., Selzer, M., Mehlmann, K., et al. (2018). From lignin to nylon: cascaded chemical and biochemical conversion using metabolically engineered Pseudomonas putida. Metab. Eng. 47, 279–293. doi: 10.1016/j.ymben.2018.03.003
Kuepper, J., Dickler, J., Biggel, M., Behnken, S., Jäger, G., Wierckx, N., et al. (2015). Metabolic engineering of Pseudomonas putida KT2440 to produce anthranilate from glucose. Front. Microbiol. 6:1310. doi: 10.3389/fmicb.2015.01310
Kusumawardhani, H., Hosseini, R., and de Winde, J. H. (2018). Solvent tolerance in bacteria: fulfilling the promise of the biotech era? Trends Biotechnol. 36, 1025–1039. doi: 10.1016/j.tibtech.2018.04.007
Lee, K., Bang, H. B., Lee, Y. H., and Jeong, K. J. (2019). Enhanced production of styrene by engineered Escherichia coli and in situ product recovery (ISPR) with an organic solvent. Microb. Cell Fact. 18:79. doi: 10.1186/s12934-019-1129-6
Lenzen, C., Wynands, B., Otto, M., Bolzenius, J., Mennicken, P., Wierckx, N., et al. (2019). High-yield production of 4-hydroxybenzoate from glucose or glycerol by an engineered Pseudomonas taiwanensis VLB120. Front. Bioeng. Biotechnol. 7:130. doi: 10.3389/fbioe.2019.00130
Lian, J., McKenna, R., Rover, M. R., Nielsen, D. R., Wen, Z., and Jarboe, L. R. (2016). Production of biorenewable styrene: utilization of biomass-derived sugars and insights into toxicity. J. Ind. Microbiol. Biotechnol. 43, 595–604. doi: 10.1007/s10295-016-1734-x
Martínez-García, E., and de Lorenzo, V. (2011). Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ. Microbiol. 13, 2702–2716. doi: 10.1111/j.1462-2920.2011.02538.x
McKenna, R., and Nielsen, D. R. (2011). Styrene biosynthesis from glucose by engineered E. coli. Metab. Eng. 13, 544–554. doi: 10.1016/j.ymben.2011.06.005
Mi, J., Becher, D., Lubuta, P., Dany, S., Tusch, K., Schewe, H., et al. (2014). De novo production of the monoterpenoid geranic acid by metabolically engineered Pseudomonas putida. Microb. Cell Fact. 13:170. doi: 10.1186/s12934-014-0170-8
Molina-Santiago, C., Cordero, B. F., Daddaoua, A., Udaondo, Z., Manzano, J., Valdivia, M., et al. (2016). Pseudomonas putida as a platform for the synthesis of aromatic compounds. Mircobiology 162, 1535–1543. doi: 10.1099/mic.0.000333
Nijkamp, K., van Luijk, N., de Bont, J. A. M., and Wery, J. (2005). The solvent-tolerant Pseudomonas putida S12 as host for the production of cinnamic acid from glucose. Appl. Microbiol. Biotechnol. 69, 170–177. doi: 10.1007/s00253-005-1973-7
Nijkamp, K., Westerhof, R. G., Ballerstedt, H., De Bont, J. A., and Wery, J. (2007). Optimization of the solvent-tolerant Pseudomonas putida S12 as host for the production of p-coumarate from glucose. Appl. Microbiol. Biotechnol. 74, 617–624. doi: 10.1007/s00253-006-0703-0
Nikel, P. I., Chavarría, M., Fuhrer, T., Sauer, U., and de Lorenzo, V. (2015). Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the Entner-Doudoroff, Embden-Meyerhof-Parnas, and pentose phosphate pathways. J. Biol. Chem. 290, 25920–25932. doi: 10.1074/jbc.M115.687749
Nikel, P. I., Kim, J., and de Lorenzo, V. (2014). Metabolic and regulatory rearrangements underlying glycerol metabolism in Pseudomonas putida KT2440. Environ. Microbiol. 16, 239–254. doi: 10.1111/1462-2920.12224
Noda, S., Miyazaki, T., Miyoshi, T., Miyake, M., Okai, N., Tanaka, T., et al. (2011). Cinnamic acid production using Streptomyces lividans expressing phenylalanine ammonia lyase. J. Ind. Microbiol. Biotechnol. 38, 643–648. doi: 10.1007/s10295-011-0955-2
Olasupo, N. A., Fitzgerald, D. J., Gasson, M. J., and Narbad, A. (2003). Activity of natural antimicrobial compounds against Escherichia coli and Salmonella enterica serovar Typhimurium. Lett. Appl. Microbiol. 37, 448–451. doi: 10.1046/j.1472-765X.2003.01427.x
Panke, S., Witholt, B., Schmid, A., and Wubbolts, M. G. (1998). Towards a biocatalyst for (S)-styrene oxide production: characterization of the styrene degradation pathway of Pseudomonas sp. strain VLB120. Appl. Environ. Microbiol. 64, 2032–2043.
Poblete-Castro, I., Wittmann, C., and Nikel, P. I. (2019). Biochemistry, genetics and biotechnology of glycerol utilization in Pseudomonas species. Microb. Biotechnol. 1–22. doi: 10.1111/1751-7915.13400
Puigbo, P., Guzman, E., Romeu, A., and Garcia-Vallve, S. (2007). OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 35, W126–W131. doi: 10.1093/nar/gkm219
Ramos, J.-L., Sol Cuenca, M., Molina-Santiago, C., Segura, A., Duque, E., Gómez-García, M. R., et al. (2015). Mechanisms of solvent resistance mediated by interplay of cellular factors in Pseudomonas putida. FEMS Microbiol. Rev. 39, 555–566. doi: 10.1093/femsre/fuv006
Ramos, J. L., Duque, E., Gallegos, M.-T., Godoy, P., Ramos-Gonzalez, M. I., Rojas, A., et al. (2002). Mechanisms of solvent tolerance in Gram-negative bacteria. Annu. Rev. Microbiol. 56, 743–768. doi: 10.1146/annurev.micro.56.012302.161038
Rodriguez, A., Martínez, J. A., Flores, N., Escalante, A., Gosset, G., and Bolivar, F. (2014). Engineering Escherichia coli to overproduce aromatic amino acids and derived compounds. Microb. Cell Fact. 13:126. doi: 10.1186/s12934-014-0126-z
Salum, M. L., and Erra-Balsells, R. (2013). High purity cis-cinnamic acid preparation for studying physiological role of trans-cinnamic and cis-cinnamic acids in higher plants. Environ. Control Biol. 51, 1–10. doi: 10.2525/ecb.51.1
Sardessai, Y., and Bhosle, S. (2002). Tolerance of bacteria to organic solvents. Res. Microbiol. 153, 263–268. doi: 10.1016/S0923-2508(02)01319-0
Segura, A., Molina, L., Fillet, S., Krell, T., Bernal, P., Muñoz-Rojas, J., et al. (2012). Solvent tolerance in Gram-negative bacteria. Curr. Opin. Biotechnol. 23, 415–421. doi: 10.1016/j.copbio.2011.11.015
Straathof, A. J. J., Wahl, S. A., Benjamin, K. R., Takors, R., Wierckx, N., and Noorman, H. J. (2019). Grand research challenges for sustainable industrial biotechnology. Trends Biotechnol. 37, 1042–1050. doi: 10.1016/j.tibtech.2019.04.002
Terán, W., Felipe, A., Segura, A., Rojas, A., Ramos, J.-L., and Gallegos, M.-T. (2003). Antibiotic-dependent induction of Pseudomonas putida DOT-T1E TtgABC efflux pump is mediated by the drug binding repressor TtgR. Antimicrob. Agents Chemother. 47, 3067–3072. doi: 10.1128/AAC.47.10.3067-3072.2003
Tietze, L.-F., Eicher, T., Diederichsen, U., Speicher, A., and Schützenmeister, N. (2015). Reactions and Syntheses : In the Organic Chemistry Laboratory, 1st Edn, eds J. Fischer and D. P. Rotella. Weinheim: Wiley VCH.
Tiso, T., Zauter, R., Tulke, H., Leuchtle, B., Li, W.-J., Behrens, B., et al. (2017). Designer rhamnolipids by reduction of congener diversity: production and characterization. Microb. Cell Fact. 16:225. doi: 10.1186/s12934-017-0838-y
van Summeren-Wesenhagen, P. V., and Marienhagen, J. (2015). Metabolic engineering of Escherichia coli for the synthesis of the plant polyphenol pinosylvin. Appl. Environ. Microbiol. 81, 840–849. doi: 10.1128/AEM.02966-14
Vargas-Tah, A., and Gosset, G. (2015). Production of cinnamic and p-hydroxycinnamic acids in engineered microbes. Front. Bioeng. Biotechnol. 3:116. doi: 10.3389/fbioe.2015.00116
Vargas-Tah, A., Martínez, L. M., Hernández-Chávez, G., Rocha, M., Martínez, A., Bolívar, F., et al. (2015). Production of cinnamic and p-hydroxycinnamic acid from sugar mixtures with engineered Escherichia coli. Microb. Cell Fact. 14:6. doi: 10.1186/s12934-014-0185-1
Velasco, A., Alonso, S., García, J. L., Perera, J., and Díaz, E. (1998). Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J. Bacteriol. 180, 1063–1071.
Verhoef, S., Ballerstedt, H., Volkers, R. J. M., de Winde, J. H., and Ruijssenaars, H. J. (2010). Comparative transcriptomics and proteomics of p-hydroxybenzoate producing Pseudomonas putida S12: novel responses and implications for strain improvement. Appl. Microbiol. Biotechnol. 87, 679–690. doi: 10.1007/s00253-010-2626-z
Volkers, R. J. M., de Jong, A. L., Hulst, A. G., van Baar, B. L. M., de Bont, J. A. M., and Wery, J. (2006). Chemostat-based proteomic analysis of toluene-affected Pseudomonas putida S12. Environ. Microbiol. 8, 1674–1679. doi: 10.1111/j.1462-2920.2006.01056.x
Volmer, J., Neumann, C., Bühler, B., and Schmid, A. (2014). Engineering of Pseudomonas taiwanensis VLB120 for constitutive solvent tolerance and increased specific styrene epoxidation activity. Appl. Environ. Microbiol. 80, 6539–6548. doi: 10.1128/AEM.01940-14
Wierckx, N., Ruijssenaars, H. J., de Winde, J. H., Schmid, A., and Blank, L. M. (2009). Metabolic flux analysis of a phenol producing mutant of Pseudomonas putida S12: verification and complementation of hypotheses derived from transcriptomics. J. Biotechnol. 143, 124–129. doi: 10.1016/j.jbiotec.2009.06.023
Wierckx, N. J., Ballerstedt, H., de Bont, J. A., de Winde, J. H., Ruijssenaars, H. J., and Wery, J. (2008). Transcriptome analysis of a phenol-producing Pseudomonas putida S12 construct: genetic and physiological basis for improved production. J. Bacteriol. 190, 2822–2830. doi: 10.1128/JB.01379-07
Wynands, B. (2018). Engineering of Pseudomonas taiwanensis VLB120 for the sustainable production of hydroxylated aromatics (dissertation). RWTH Aachen University, Aachen, Germany.
Wynands, B., Lenzen, C., Otto, M., Koch, F., Blank, L. M., and Wierckx, N. (2018). Metabolic engineering of Pseudomonas taiwanensis VLB120 with minimal genomic modifications for high-yield phenol production. Metab. Eng. 47, 121–133. doi: 10.1016/j.ymben.2018.03.011
Wynands, B., Otto, M., Runge, N., Preckel, S., Polen, T., Blank, L. M., et al. (2019). Streamlined Pseudomonas taiwanensis VLB120 chassis strains with improved bioprocess features. ACS Synth. Biol. 8, 2036–2050. doi: 10.1021/acssynbio.9b00108
Xu, R., Zhang, K., Liu, P., Han, H., Zhao, S., Kakade, A., et al. (2018). Lignin depolymerization and utilization by bacteria. Bioresour. Technol. 269, 557–566. doi: 10.1016/j.biortech.2018.08.118
Zambanini, T., Sarikaya, E., Kleineberg, W., Buescher, J. M., Meurer, G., Wierckx, N., et al. (2016). Efficient malic acid production from glycerol with Ustilago trichophora TZ1. Biotechnol. Biofuels 9:67. doi: 10.1186/s13068-016-0483-4
Zhang, C., Zhang, J., Kang, Z., Du, G., and Chen, J. (2015). Rational engineering of multiple module pathways for the production of L-phenylalanine in Corynebacterium glutamicum. J. Ind. Microbiol. Biotechnol. 42, 787–797. doi: 10.1007/s10295-015-1593-x
Keywords: Pseudomonas, metabolic engineering, trans-cinnamic acid, L-phenylalanine, rational engineering, glycerol, glucose
Citation: Otto M, Wynands B, Lenzen C, Filbig M, Blank LM and Wierckx N (2019) Rational Engineering of Phenylalanine Accumulation in Pseudomonas taiwanensis to Enable High-Yield Production of Trans-Cinnamate. Front. Bioeng. Biotechnol. 7:312. doi: 10.3389/fbioe.2019.00312
Received: 11 September 2019; Accepted: 23 October 2019;
Published: 20 November 2019.
Edited by:
Jean Marie François, UMS3582 Toulouse White Biotechnology (TWB), FranceReviewed by:
Jian-Ming Liu, Technical University of Denmark, DenmarkJohn A. Morgan, Purdue University, United States
Copyright © 2019 Otto, Wynands, Lenzen, Filbig, Blank and Wierckx. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nick Wierckx, n.wierckx@fz-juelich.de
 Maike Otto
Maike Otto Benedikt Wynands
Benedikt Wynands Christoph Lenzen
Christoph Lenzen Melanie Filbig2
Melanie Filbig2  Nick Wierckx
Nick Wierckx