- 1Faculty of Bioscience and Bioindustry, Tokushima University, Tokushima, Japan
- 2School of Life Science and Technology, Tokyo Institute of Technology, Yokohama, Japan
Tomato INDOLE-3-ACETIC ACID9 (SlIAA9) is a transcriptional repressor in auxin signal transduction, and SlIAA9 knockout tomato plants develop parthenocarpic fruits without fertilization. We generated sliaa9 mutants with parthenocarpy in several commercial tomato cultivars (Moneymaker, Rio Grande, and Ailsa Craig) using CRISPR-Cas9, and null-segregant lines in the T1 generation were isolated by self-pollination, which was confirmed by PCR and Southern blot analysis. We then estimated shoot growth phenotypes of the mutant plants under different light (low and normal) conditions. The shoot length of sliaa9 plants in Moneymaker and Rio Grande was smaller than those of wild-type cultivars in low light conditions, whereas there was not clear difference between the mutant of Ailsa Craig and the wild-type under both light conditions. Furthermore, young seedlings in Rio Grande exhibited shade avoidance response in hypocotyl growth, in which the hypocotyl lengths were increased in low light conditions, and sliaa9 mutant seedlings of Ailsa Craig exhibited enhanced responses in this phenotype. Fruit production and growth rates were similar among the sliaa9 mutant tomato cultivars. These results suggest that control mechanisms involved in the interaction of AUX/IAA9 and lights condition in elongation growth differ among commercial tomato cultivars.
Introduction
Tomato (Solanum lycopersicum) – a model horticultural crop – is one of the most important vegetable and fruit crops worldwide. In wild-type tomato plants, fruiting can be induced by artificial pollination and the application of exogenous auxin. Parthenocarpy, which allows fruit production without fertilization, is a hugely important agricultural trait and has been produced by traditional breeding in limited species and cultivars. The fruiting of tomato is controlled by auxin signaling factors, tomato Auxin Responsive Factor 8 (SlARF8), and the auxin responsive transcription repressor INDOLE-3-ACETIC ACID 9 (SlIAA9; Wang et al., 2005; Goetz et al., 2006). SlIAA9 has four conserved domains and two functional motifs (Wang et al., 2005; Saito et al., 2011). Loss-of-function mutants of SlIAA9 in tomato exhibit parthenocarpy – a desirable trait in tomato – where fruit set and growth are stimulated without fertilization; parthenocarpy can overcome problems with fruit setting under unfavorable environmental conditions. The sliaa9 mutant tomatoes generated by ethyl methanesulfonate (EMS) treatment of several cultivars, such as Micro-Tom, Ailsa Craig, M82, and Red Setter, showed altered leaf morphology (Zhang et al., 2007; Berger et al., 2009; Saito et al., 2011; Mazzucato et al., 2015). In Micro-Tom, the AS-IAA9 line (SlIAA9 downregulated-line) exhibited phenotypic changes in leaf morphology and parthenocarpy (Wang et al., 2005). Hypersensitivity to auxin, enhanced hypocotyl/stem elongation, reduced apical dominance, and upregulation of SlIAA3 expression were also detected in the AS-IAA9 line (Wang et al., 2005). We have previously reported the generation of sliaa9 knockout mutants using the clustered regularly interspaced short palindromic repeat (CRISPR)-CRISPR-associated protein-9 nuclease (Cas9) system; the sliaa9 mutant showed altered leaf morphology and parthenocarpy (Ueta et al., 2017). In the CRISPR-Cas9 system, the Cas9 nuclease and a guide RNA (gRNA) form a complex and induce a double-strand break (DSB) at the target site. The CRISPR-Cas9 system, which is used to generate target gene-specific mutations, has been applied in many plant species to create novel useful crops by molecular breeding (Osakabe and Osakabe, 2015).
Auxin affects various growth and developmental processes in plants. The relationship between the auxin signal transduction pathway and other environmental factors, such as light conditions, has not been well studied in commercial tomato cultivars. Light conditions lead to various effects in auxin responses (Halliday et al., 2009). When young plants grow in shade, hypocotyl elongation known as the shade avoidance response is induced, in which auxin synthesis is enhanced by PHYTOCHROME B (PHYB) under low red: far red (R:FR; shade) or low light conditions (Trupkin et al., 2014; Ballaré and Pierik, 2017). On the contrary, phytochrome A (PHYA) suppresses shade avoidance responses in plants (Ganesan et al., 2012). It has been suggested that several AUX/IAAs interact with, and are phosphorylated by, PHYA, which inhibits AUX/IAA degradation. As a result, the negative regulation of shade avoidance responses (i.e., inhibition of hypocotyl elongation) is induced under shade (Colón-Carmona et al., 2000; Yang et al., 2018; Ma and Li, 2019). Cryptochrome 1 (CRY1) also interacts with AUX/IAA and negatively regulates auxin signaling in Arabidopsis (Xu et al., 2018).
It is important to clarify the effects of light environments on crop growth for agricultural applications. In this study, we analyzed shoot growth of sliaa9 knockout mutants of three commercial tomato cultivars (Ailsa Craig, Moneymaker, and Rio Grande), which were generated using CRISPR-Cas9, under different light intensity. Null-segregants in next generation progenies of the mutants were isolated, and we demonstrated an effect of normal or low light conditions on shoot growth of sliaa9 mutants. They exhibited different shoot growth phenotypes and photomorphogenesis, although they showed parthenocarpy in common. These results suggest that various control mechanisms involved in tomato shoot growth in commercial cultivars are mediated by AUX/IAA9 and light.
Materials and Methods
Plant Materials and Growth Conditions
Three varieties of Solanum lycopersicum L., i.e., cv. Moneymaker, Rio Grande, and Ailsa Craig, were used in this study. Moneymaker and Ailsa Craig are indeterminate type and Rio Grande is a determinate type tomato. Tomato seeds were sterilized by 70% EtOH and 10% sodium hypochlorite solution (Antiformin, Wako, Japan) including 0.005% Triton X-100 (octylphenol ethoxylate, Wako, Japan). Sterilized seeds imbibed water overnight and were sown on germination medium [1.5% sucrose, 1/2 × MS (pH 5.7)]. Expanded cotyledons 7 days after sowing were used for transformation. Tomato calli and shoots were grown in tissue culture in a growth chamber under conditions of 21–25°C with 15 h light at 30–50 μmol m−2 s−1/9 h dark. T1 or T2 tomato plants imbibed water overnight, germinated in Jiffy-7 (Jiffy), and grown in a growth chamber (CLE-305, TOMY, Japan) or plant growing rack (CR-400L, TOMY, Japan) under conditions of 21–25°C with 15 h light at 40–70 μmol m−2 s−1 (low light conditions) or 150–250 μmol m−2 s−1 (normal light condition)/9 h dark.
CRISPR-Cas9 Vector
In our previous study (Ueta et al., 2017), the SlIAA9 (Solyc04g076850) target sequence, 5'-GAGCTCAGGCTCGGTCTACC-3' (gRNA2) was used for genome editing of all four tomato cultivars. The CRISPR-Cas9 expression vector, pEgP237-2A-GFP (Ueta et al., 2017; Supplementary Figure S1A), was used, which comprises a gRNA expression cassette under the control of the Arabidopsis U6 snRNA-26 (AtU6-26) promoter, and a Cas9 expression cassette for an Arabidopsis codon-optimized spCas9 (AtCas9) fused to GFP via a 2A peptide, which is driven by the 2 × CaMV35SΩ promoter. The AtCas9 contains the 3 × NLS at its C-terminus. In this vector, a kanamycin-resistance marker was used for selection of transgenic plants.
Transformation of Tomato Plants
The CRISPR-Cas9 expression vector was introduced into Agrobacterium tumefaciens (GV2260) and used to transform tomato as described previously (Ueta et al., 2017; Hashimoto et al., 2018). Briefly, Agrobacterium harboring CRISPR-Cas9 vector was cultured on LB medium or AB-MES medium. Leaf discs prepared from cotyledons were subjected to Agrobacterium infection. After co-culture for several days in the dark, the leaf discs were transferred to callus induction medium (CIM) 1 (3% sucrose, 1 × MS, 100 mg/L kanamycin, 1.5 mg/L trans-zeatin, and 25 mg/L meropenem) and cultured for 4 weeks for Moneymaker and Ailsa Craig. After the appearance of calli, shoot induction medium (SIM) 1 (3% sucrose, 1 x MS, 100 mg/L kanamycin, 1.0 mg/L trans-zeatin, and 25 mg/L meropenem) was used for further incubation. Leaf discs from Rio Grande were incubated in CIM2 (3% sucrose, 1 × MS, 50 mg/L kanamycin, 1 mg/L BA, 0.1 mg/L IAA, and 25 mg/L meropenem) for 4–5 weeks after co-culture, then calli were transferred to SIM2 (3% sucrose, 1 × MS, 50 mg/L kanamycin, 1.0 mg/L trans-zeatin, 0.5 mg/L IAA, and 25 mg/L meropenem) and incubate for at least a further 5 weeks. Transgenic shoots were transferred to root induction medium (1.5% sucrose, 0.5 × MS, 50 mg/L kanamycin, and 25 mg/L meropenem) for 2–4 weeks and then to soil pots.
Mutation Analyses in CRISPR-Cas9 Target Sites
Genomic DNA was isolated from tomato leaves using Nucleospin Plant II (TaKaRa, Japan), and used for amplification of target sequences by PCR using PrimeSTAR GXL DNA Polymerase (TaKaRa, Japan). An AccI site is located on the SlIAA9 target sequence, and PCR-RFLP was used for detection of the mutation. In PCR-RFLP, PCR fragments were digested with AccI (NEB, Japan) and analyzed by agarose gel electrophoresis. For Sanger sequencing analysis, PCR fragments purified from agarose gels were cloned by the Seamless ligation cloning extract (SLiCE) method (Motohashi, 2015) into cloning vector pNEB193 (NEB, Japan). All primers used for PCR are listed in Supplementary Table S1.
PCR for Detection and Southern Blot Analysis of Null-Segregant Plants
Around 20–50 plants of the T1 generation and eight plants of the T2 generation were selected randomly for PCR-based detection of T-DNA regions. Preparation of total DNA was as described above. Nine regions in the T-DNA in the CRISPR-Cas9 vector were selected and amplified using PrimeSTAR GXL polymerase (TaKaRa, Japan). All primers used for PCR and the corresponding amplicon sizes are indicated in Supplementary Table S1. PCR experiments were replicated three times for each line.
For Southern blot, total DNA was extracted from leaves of WT and sliaa9 mutants by the modified cetyltrimethylammonium bromide (CTAB)-based DNA extraction method (Murray and Thompson, 1980). Briefly, tomato leaves were ground in liquid nitrogen and incubated in 2% CTAB solution [100 mM Tris-HCl (pH8.0), 20 mM EDTA (pH8.0), 1.4 M NaCl, and 2% (w/v) CTAB]. The lysates were extracted with chloroform:isoamyl alcohol (24:1), and then 1% CTAB solution [50 mM Tris-HCl (pH8.0), 10 mM EDTA (pH8.0), 1% (w/v) CTAB] was added to the water layer. Total DNAs were obtained by CsCl-EtOH precipitation. Total DNA was digested with HpaI, which has a single restriction recognition site in the vector (Supplementary Figure S1A), fractionated on 1% agarose gels, and transferred to nylon membrane (Zeta-Probe, Bio-Rad, United States) by capillary transfer. After UV cross-linking (UVP CL-1000 crosslinker, Analytik Jena, Upland, CA, United States), the membranes were hybridized with DIG-labeled probes for gRNA, Cas9, and LHCB (Solyc02g070970) genes. Probes were amplified using pEgP237-2A-GFP (for gRNA and Cas9) or tomato genomic DNA (for LHCB) using the primers listed in Supplementary Table S1 and labeled with DIG using a DIG High Prime DNA Labeling and Detection Kit (Roche, Basel, Switzerland). Probe labeling, membrane hybridization, and detection were carried out according to the manufacturer’s instructions. Hybridization signals were detected and imaged with PXi (SYNGENE, Bangalore, India).
Off-Target Analysis by Amplicon Sequencing Using MiSeq
Amplicon sequencing was performed using MiSeq (Illumina, Japan) and MiSeq Reagent Kit v2 Nano (Illumina, Japan) was used for analysis. Genomic DNAs were used to amplify the region of the CRISPR-Cas9 target sites or off-target sequences by PCR. One off-target candidate sequence (SL2.50 ch6:26946923-26946946) was selected by Cas-OT to examine the mutation. PCR products were separated by electrophoresis, purified from the agarose gel using the Wizard SV Gel and PCR Clean-Up System (Promega, Japan), and used as templates for a second round of PCR. Second PCR was performed using TruSeq HT primer (Illumina, Japan). All primers used for PCR are listed in Supplementary Table S1. MiSeq data were analyzed using CLC Genomics Workbench software version 7.5.1 (CLC bio, Japan) and mapped on the off-target candidate using Integrative Genomics Viewer (IGV; Broad Institute).
Plant Growth and Leaf Measurements
Shoot lengths of wild-type and sliaa9 mutants under low or normal light conditions were measured until 48 days after germination. Hypocotyl length in young seedlings of the sliaa9 mutants and wild-type were measured at 21 days after germination. The third to tenth internode length of the sliaa9 mutants and the wild-type were measured at 48 days after germination. Several parameters of the fifth to tenth matured leaves from each plant of the sliaa9 mutants and the wild-type were measured 48 days after germination. The parts of leaves measured are shown in Supplementary Figure S4A. Leaf area was measured using ImageJ. Average values were determined using 2–6 lines of the same mutant allele. All experiments repeated 2–3 times.
Measuring Leaf Chlorophyll Concentration in Tomato
SPAD-502Plus (Konica Minolta, Japan) was used to measure leaf chlorophyll concentration in leaves. SPAD values were measured at 3–8 sites near the principal vein on the wide part of the tomato leaf on the third or higher fully elongated leaves. In total, 20–30 SPAD values in a plant were analyzed; average values were determined using 3–6 lines of the same mutant allele. All experiments repeated three times.
qRT-PCR
Hypocotyls of 21-days-old seedlings and the third internodes of 35-days-old seedlings grown under low or normal light intensity were harvested, respectively. Total RNAs were extracted using RNeasy Plant Mini Kit on the QIAcube automated system (Qiagen), followed by reverse-transcribed using a ReverTra Ace qPCR RT Master Mix with gDNA remover (TOYOBO) following the manufacturer’s instructions. Quantitative PCR was performed with GoTaq qPCR Mater Mix (Promega) on LightCycler 96 (Roche). The expression levels of SlIAA3 (5'-GCAATTCAAATCAATCATTCTTTCT-3' and 5'-GTTTATTATCCCAGGCAAACCTAAT-3') were normalized relative to those of a reference gene, Slactin7-like (5'-TGTCCCTATCTACGAGGGTTATGC-3' and 5'-AGTTAAATCACGACCAGCAAGAT-3'; Hashimoto et al., 2018). Relative expression was calculated by two to the power of relative expression (log2), calculated by subtracting Ct values of IAA3 from those of Slactin7-like.
Results
Detection of SlIAA9 Mutations in Commercial Tomato Cultivars
A CRISPR-Cas9 vector targeting exon 2 of the SlIAA9 gene (+217 to +236 from ATG, 5'-GAGCTCAGGCTCGGTCTACC-3'), which was used to generate mutants in tomato cultivars, Micro-Tom and Ailsa Craig, in a previous study (Ueta et al., 2017), was introduced into Moneymaker and Rio Grande via Agrobacterium transformation. PCR-RFLP analysis using an AccI restriction enzyme site located at the CRISPR-Cas9 target was performed to detect CRISPR-Cas9-induced mutations in the T0 regenerated tomato shoots. Non-digested fragments in PCR-RFLP analysis indicated 100% somatic mutations in all cultivars (Figures 1A,B). The progenies of sliaa9 T0 100% mutant lines (T1 lines) were isolated by self-pollination from T0 mutant lines. Sanger sequencing analysis of cloned PCR fragments at the target sites in the T1 mutant lines revealed only one or two types of mutation, which were found at similar levels in all the sequenced clones in these lines (Figure 1C; Supplementary Figure S1). These results suggest the generation of bi-allelic mutants, in which the SlIAA9 mutations introduced a frame shift just after domain I, and a stop codon was generated to produce truncated SlIAA9. The progenies of sliaa9 knockout mutant lines (T1 mutants) were isolated by self-pollination from T0 mutant lines, and T1 mutants showed homozygotes or heterozygotes that were segregated from the parental T0 generation (Supplementary Figure S1). Since T1 mutants were generated from T0 lines by self-pollination, the T-DNA, including the CRISPR-Cas9 cassette, can be segregated out. To detect T-DNA insertion in the genomic DNA of sliaa9 T1 progenies, PCR analysis designed to amplify nine T-DNA regions and Southern blot analysis were performed using specific primers and DIG-labeled probes, respectively (Supplementary Table S1; Supplementary Figure S1A). Several null-segregant mutants were isolated, in which no T-DNA regions were detected by PCR or Southern blot analysis (Supplementary Figures S1B,C). T-DNA regions were also undetected in the T2 progenies of MM#2, i.e., MM#2-13 (Supplementary Figure S1B). An off-target candidate sequence was analyzed using next generation sequencing, Miseq, and the results suggest that off-targets were not detected in mutant plants (Supplementary Figure S2). The T2 progenies of these null-segregant sliaa9 T1 mutant lines MM#2-13, RG#4-1, and AC#1-1 were named as MM-sliaa9,” “RG-sliaa9,” and “AC-sliaa9,” respectively, and used for subsequent experiments.
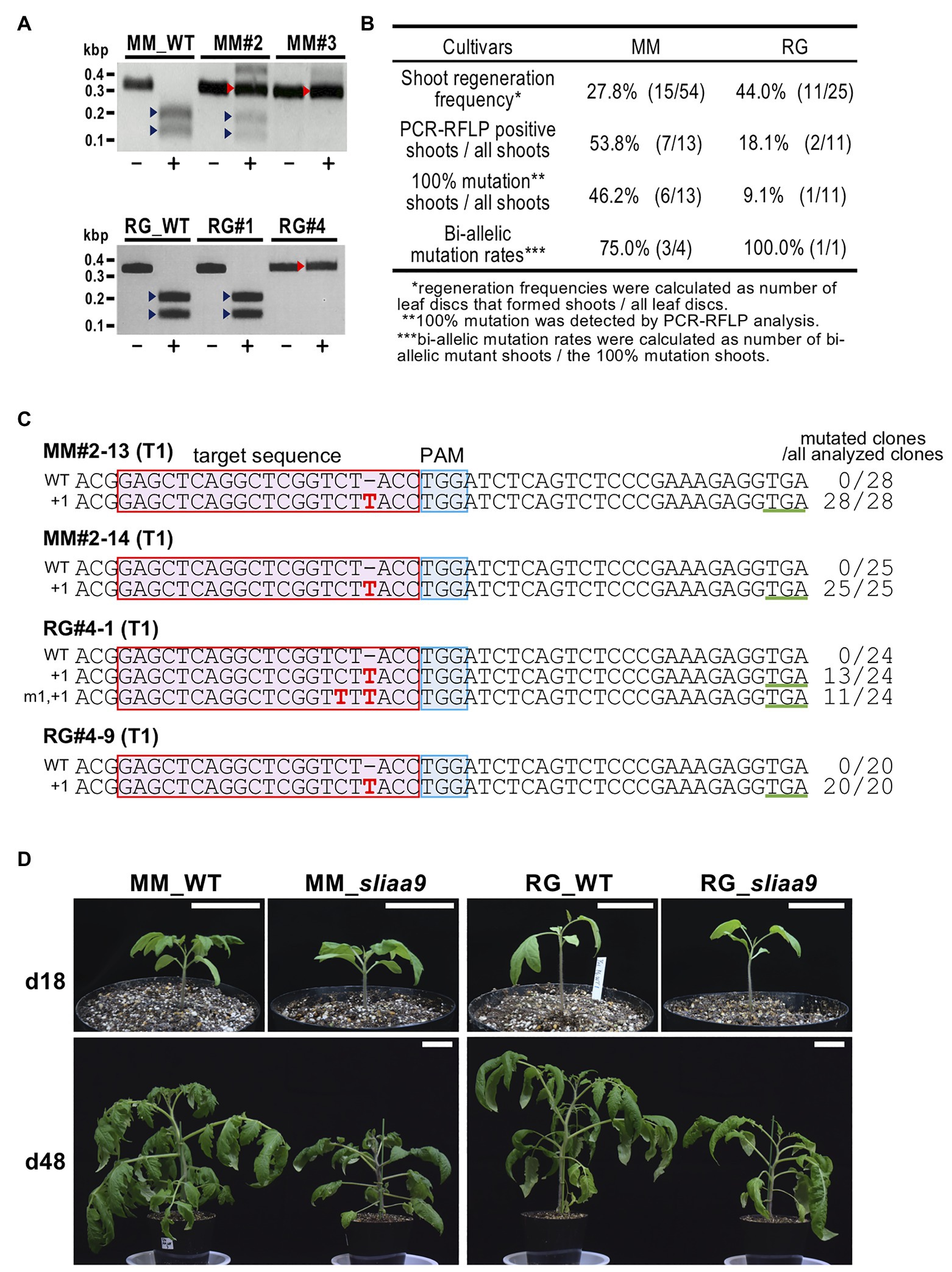
Figure 1. Isolation of sliaa9 Knockout Mutants in Various Tomato Cultivars. (A) PCR-RFLP analysis of SlIAA9 in CRISPR-Cas9 transgenic T0 lines (MM#2, MM#3, RG#1, and RG#4). Target sites in the SlIAA9 gene were amplified by PCR using total DNA isolated from T0 shoots of cultivars Moneymaker and Rio Grande. PCR products were digested with AccI, which has a recognition site at the CRISPR-Cas9 target site. MM; Moneymaker, RG; Rio Grande, −; untreated, +; treated with AccI, red arrowheads; mutated bands, blue arrowheads; wild-type fragments digested with AccI. (B) Mutation efficiency in regenerated shoots (T0). (C) SlIAA9 mutation sequences in sliaa9 T1 lines (MM#2-13, MM#2-14, RG#4-1, and RG#4-9) were analyzed by Sanger sequencing. MM; Moneymaker, RG; Rio Grande, red box; target sequence, blue box; PAM sequence, green underline; newly generated stop codon following CRISPR-Cas9 editing, red characters; and mutated nucleotides. The wild-type (WT) sequences are on top in each alignment. The numbers on the left of the sequences indicate mutated nucleotides. The number of clones analyzed is shown to the right of the sequences. (D) Wild-type and the representative sliaa9 T2 lines 18 days and 48 days after germination. The plants were grown under low light conditions. Left; MM (Moneymaker; T2 line, MM#2-13-4), right; RG (Rio Grande; T2 line, RG#4-1-6). Bars = 5cm.
Altered Elongation Growth in SlIAA9 Knockout Mutants
We next analyzed the effects of SlIAA9 disruption on plant growth among cultivars under different light conditions: low (40–70 μmol m−2 s−1) or normal (150–250 μmol m−2 s−1) light intensities. MM-sliaa9 and RG-sliaa9 showed slower rates of shoot elongation growth in the early growth stages compared with wild-type under low light intensity, whereas AC-sliaa9 did not show clear difference in growth rates (Figures 1D, 2A). Shoot lengths of wild-type and sliaa9 mutants in Moneymaker and Rio Grande grown under normal light intensity were mostly longer than those grown under low light intensity, on the contrary, Ailsa Craig plants did not show large differences between light conditions (Figures 2A,B). Moneymaker sliaa9 showed a strong phenotype in the internode elongation, and this phenotype of Rio Grande mutant was apparent in low light intensity (Figure 2C). On the contrary, there were no significant differences in the internode length of Ailsa Craig sliaa9 and wild-type under the different light intensity (Figure 2C). These results suggest that sliaa9 mutants showed different shoot elongation growth phenotypes among cultivars.
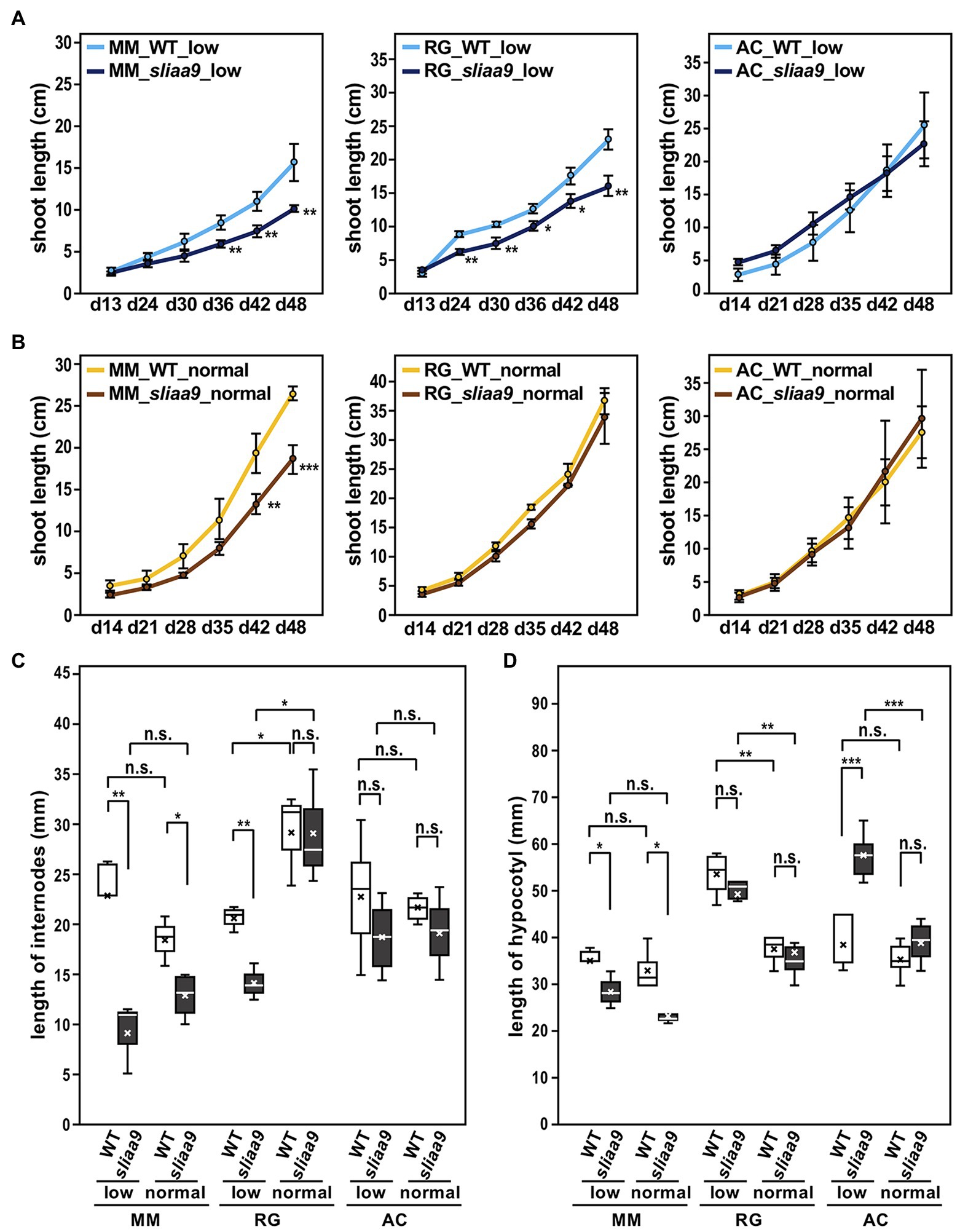
Figure 2. Elongation growth of stem/hypocotyl in the sliaa9 mutants under different light conditions. (A) Shoot growth rates of sliaa9 mutants and the wild-type under low light conditions. (B) Shoot growth rates of sliaa9 mutants and the wild-type under normal light conditions. (C) Length of 3rd–10th internodes of the sliaa9 mutants and the wild-type at day 48. (D) Hypocotyl length of young seedlings of sliaa9 mutants and the wild-type at day 21. MM; Moneymaker, RG; Rio Grande, AC, Ailsa Craig. T2 lines from MM#2-13, RG#4-1, and AC#1-1 (T1 lines) were analyzed. Data are means ± S.D. of independent T2 plant lines (n = 3–6). low; grown in low light conditions, normal; grown in normal light conditions. *p < 0.05, **p < 0.01, and ***p < 0.001 are determined by Student’s t tests. n.s., not significant.
Next, we analyzed the effect of light intensity on hypocotyl growth of young seedlings of tomato sliaa9 mutants grown in different light intensity to identify the AUX/IAA9 function in light response in the tomato cultivars. The results indicated a different response to light conditions among mutants of the three cultivars (Figure 2D). Thus, the hypocotyl length of MM-sliaa9 was smaller than that of wild-type under both light intensity, whereas, AC-sliaa9 showed a longer hypocotyl than that of wild-type in low light intensity; however, the effect on hypocotyl growth was rescued in normal light, and hypocotyl length of mutant and wild-type were similarly decreased (Figure 2D). There was no clear differences between RG-sliaa9 and wild-type; hypocotyl length of mutant and wild-type were similarly elongated under low light intensity (Figure 2D). Furthermore, hypocotyl elongation in Moneymaker is strongly affected by SlIAA9 knockout and did not show any further effects of light intensity. These results might suggest that photomorphogenesis in the hypocotyl elongation of young seedlings differs in different tomato cultivars, and the relationship of the auxin signaling pathway mediated by SlIAA9 and photomorphogenesis might be clearer in Ailsa Craig than in Moneymaker and Rio Grande.
To understand molecular mechanisms of elongation growth controlled by SlIAA9 and the downstream pathway, we focused on the expression of SlIAA3 gene. The SlIAA3 expression levels were analyzed in the third internodes and hypocotyls of young seedlings grown under low or normal light intensity (Supplementary Figure S3). The results indicated that there were not close relationship of the SlIAA3 expression levels, elongation growth mediated by SlIAA9, and the light intensity among cultivars, although weak downregulation of the SlIAA3 expression was observed in the SlIAA9 Rio Grande mutants. The results suggest that complicated pathways might be existed in the downstream of SlIAA9 in elongation growth of the tomato cultivars.
Leaf Morphology of Tomato sliaa9 Mutants
We next analyzed the leaf morphology of tomato sliaa9 mutants. Previous studies had shown that disruption of SlIAA9 in tomato led to altered leaf morphology, with the compound leaf changing to a single leaf (Wang et al., 2005; Berger et al., 2009; Saito et al., 2011; Ueta et al., 2017). In the present study, the sliaa9 mutants in the three cultivars exhibited morphological changes to the single leaf (Figure 3A). To analyze the phenotypes in elongation growth in leaves, we measured several parameters of fully expanded leaves in young plants (Supplementary Figures S4, S5; Figures 3B–E). The length of sliaa9 mutant leaves in Moneymaker and Rio Grande showed no significant differences when compared with those of wild-type in normal lights, whereas those of the Moneymaker mutant in low light conditions and the Ailsa Craig mutant in both light conditions were shorter than those of wild-type (Figures 3A,B), indicating different mechanisms related to elongation in leaves and shoots between cultivars. When the ratio of petiole length to full length of leaves was compared among the mutants, all three cultivars exhibited a higher ratio in sliaa9 mutants compared with wild-type in both light conditions (Figure 3C). Leaf width in sliaa9 mutants of all three cultivars was narrower than that of wild-type, and the mutants of Moneymaker and Ailsa Craig also showed a smaller leaf area than wild-type especially in low light conditions (Figures 3D,E). These results suggest that SlIAA9 knockout affected the several parameters in leaf shapes that might be differentially controlled in various tomato cultivars.
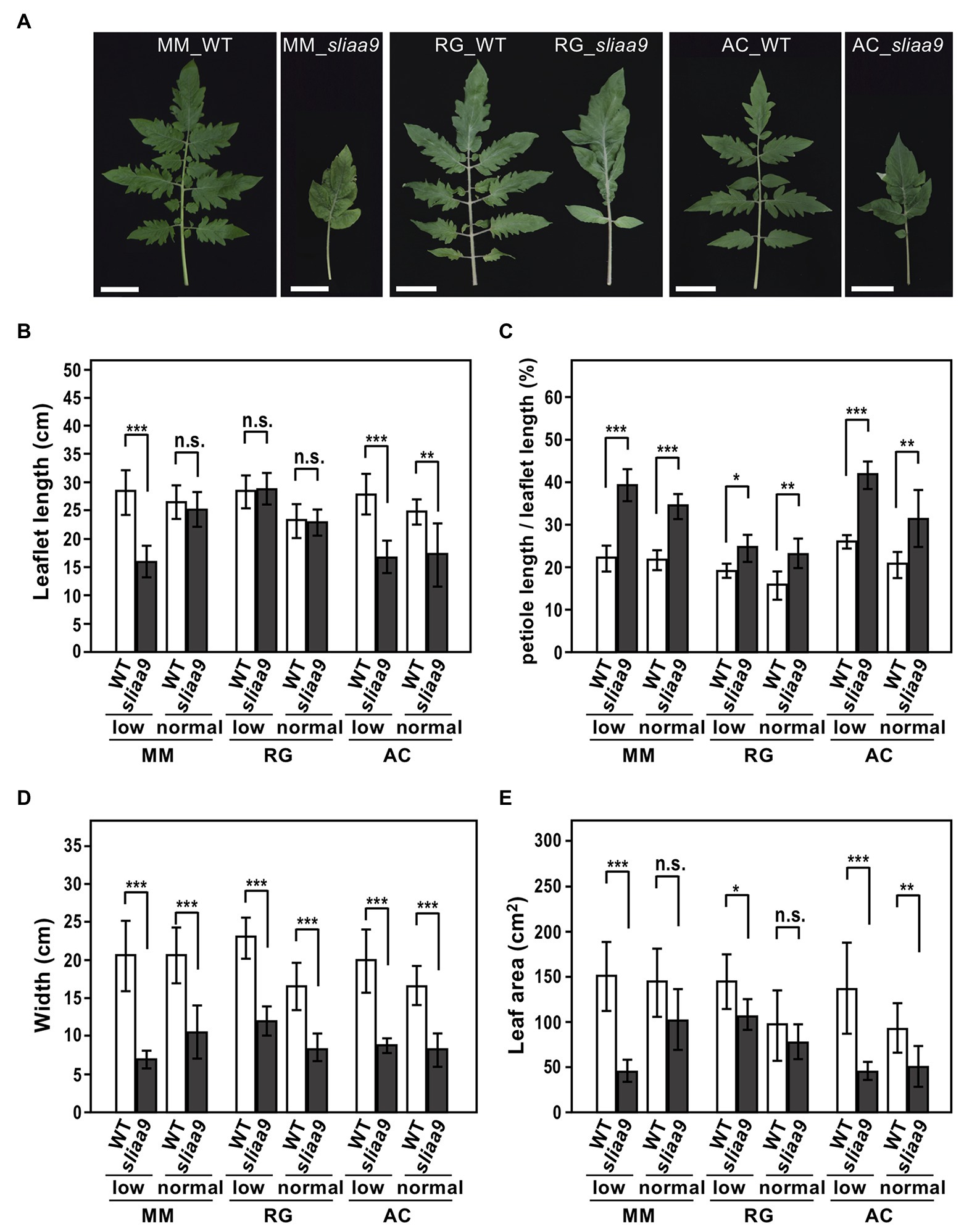
Figure 3. Altered leaf morphology of sliaa9 mutants in three tomato cultivars. (A) Leaf morphology of sliaa9 mutants and wild-type plants grown under low light conditions. Bars = 5cm. (B–E) Leaflet length (B), the ratio of petioles to total leaf length (C), leaf width (D), and leaf area (E) in sliaa9 plants and the wild-type. T2 lines from MM#2-13, RG#4-1, and AC#1-1 (T1 lines) were analyzed. Data are means ± S.D. of 2–5 leaves of individual T2 plant line (total 2–4 lines; n = 6–10). *p < 0.05, **p < 0.01, and ***p < 0.001 are determined by Student’s t tests. n.s., not significant.
To analyze any further effects of SlIAA9 disruption on plant growth, we measured leaf numbers and chlorophyll content in mature leaves of mutant plants using a Soil Plant Analysis Development (SPAD) chlorophyll meter. Leaf numbers increased similarly in sliaa9 mutants and the wild-type of all three cultivars growing under both light conditions (Supplementary Figure S5). There were no significant differences in SPAD values between sliaa9 mutants and the wild-type plants, except that Moneymaker plants showed larger values under normal light than in low light conditions (Supplementary Figure S4E).
Parthenocarpy Phenotypes in SlIAA9 Mutants
We next analyzed flower and fruit phenotypes of the sliaa9 mutants of Moneymaker and Rio Grande. Flower development in all these mutants showed no differences when compared with wild-type plants (Figure 4A). Fruit shape in sliaa9 mutants was also similar to that of the wild-type; on the contrary, several fruits had no, or only few, seeds, suggesting a parthenocarpic phenotype as shown in previous studies using cultivars Micro-Tom and Ailsa Craig (Figure 4A; Ueta et al., 2017). The total number of fruit, and seed numbers, were measured 2 months after the first fruit was fully ripened, indicating that parthenocarpic fruits were generated only in sliaa9 mutants (Figures 4B,C). These results indicate that the role of SlIAA9 in fruit development is common to the three cultivars, and that parthenocarpic phenotypes can be efficiently introduced into various cultivars by CRISPR-Cas9 technology.
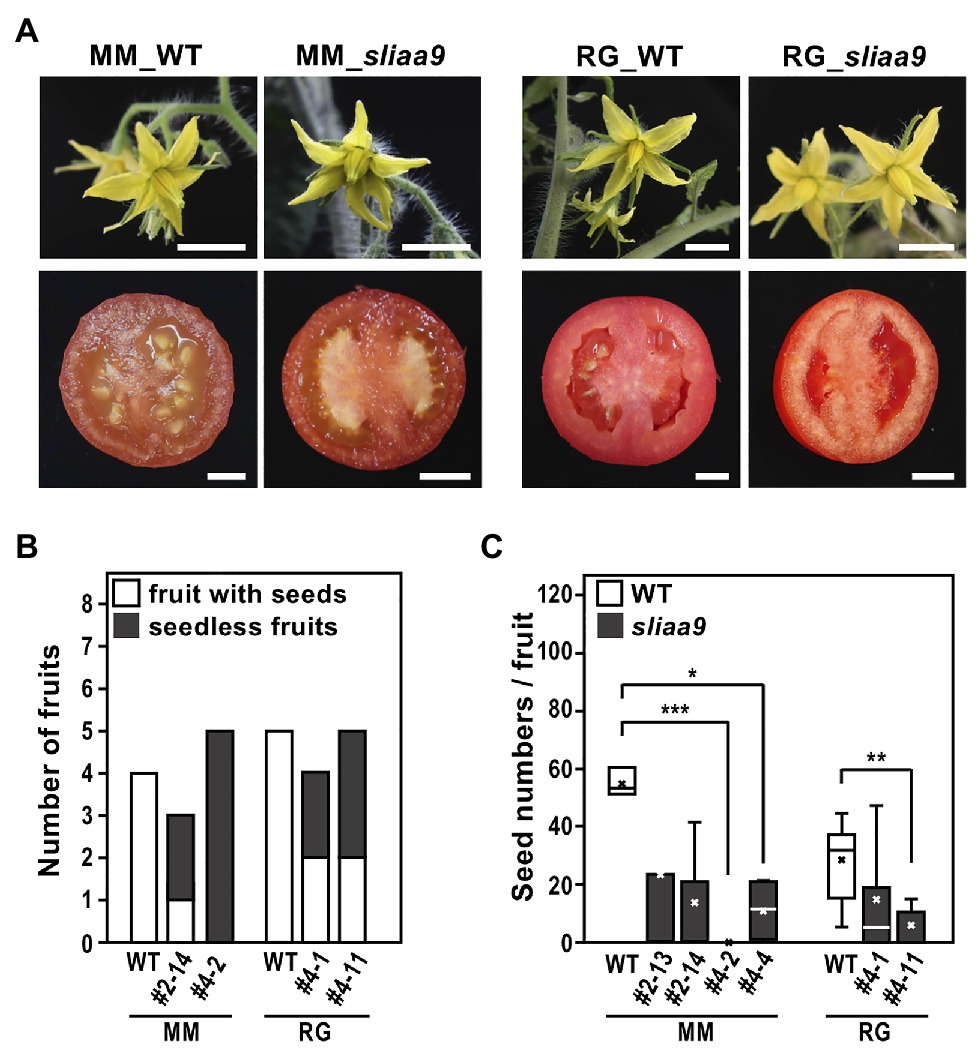
Figure 4. Flowers and fruits phenotypes of sliaa9 mutants. (A) The representative T2 flower (upper, MM_sliaa9; MM#2-13-1, RG_sliaa9; and RG#4-1-1) and T0 fruit (bottom, MM_sliaa9; MM#4, RG_sliaa9; and RG#4) phenotypes of the sliaa9 mutants of Moneymaker (left) and Rio Grande (Right). Bars = 1 cm. (B) Number of seedless mature fruits developed without pollination (parthenocarpic fruits) and all the fruits obtained from sliaa9 T1 mutant lines (MM#2-14, MM#4-2, and RG#4-1, and RG#4-11) grown under low light for 5 months. (C) Seed numbers of sliaa9 T1 mutant lines (MM#2-13, #2-14, #4-2, and #4-4) of Moneymaker and Rio Grande (RG#4-1, and #4-11). Average seed numbers were calculated in the mutant fruits (n = 3–12) without pollination and in wild-type (WT) fruits (n = 5–12) with pollination. *p < 0.05, **p < 0.01, and ***p < 0.001 are determined by Student’s t tests.
Discussion
Disruption and repression of SlIAA9 – a negative regulator of the Aux/IAA transcription factor – has been reported to induce parthenocarpy in tomato cultivars Micro-Tom and Ailsa Craig and also changes in leaf morphology and plant form in several more cultivars, including Micro-Tom, Ailsa Craig, M82, and Red Setter (Wang et al., 2005; Zhang et al., 2007; Berger et al., 2009; Saito et al., 2011; Mazzucato et al., 2015; Ueta et al., 2017). Several other phenotypes in SlIAA9 antisense lines of Micro-Tom (AS-IAA9) have also been reported, e.g., hypersensitivity to auxin, enhanced hypocotyl/stem elongation, and reduced apical dominance were observed in AS-IAA9 plants (Wang et al., 2005); however, few studies have compared SlIAA9 knockout phenotypes in various tomato cultivars. In this study, sliaa9 mutant lines of various commercial tomato cultivars were generated using CRISPR-Cas9 technology. Null-segregants of sliaa9 knockout mutants (progenies of MM#2, RG#4, and AC#1), were isolated and plant phenotypes – shoot elongation, leaf morphology, and parthenocarpy – were analyzed using the mutants grown under the different light conditions.
The elongation of shoots and internodes in cultivars Moneymaker and Rio Grande were commonly decreased in sliaa9 mutants in low light conditions. Although elongation growth were increased through enhanced auxin signaling following SlIAA9 disruption in the previous studies (Wang et al., 2005), the opposite effect on shoot elongation was detected in these cultivars. Elongation growth patterns in leaves of mutants were also different among these two cultivars, as similar to shoot elongation phenotypes. Previous studies had shown that AS-IAA9 (SlIAA9 antisense line) in Micro-Tom exhibited an enhanced stem/hypocotyl elongation growth phenotype (Wang et al., 2005), and the gain-of-function mutant of SlIAA9 with a single amino acid substitution reduced the elongation of internodes and showed a strong dwarf phenotype in cv. VF36 (Koenig et al., 2009). These results suggested the existence of different mechanisms of auxin signaling among tomato cultivars and leaf and shoots under different environmental conditions (Figure 5). Double knockout mutants of Arabidopsis IAA8 and IAA9 genes, which belong to the same clade as SlIAA9, exhibited no differences other than leaf morphology between wild-type plants (Overvoorde et al., 2005; Koenig et al., 2009), suggesting that SlIAA9 function in leaf morphogenesis also varied among plant species.
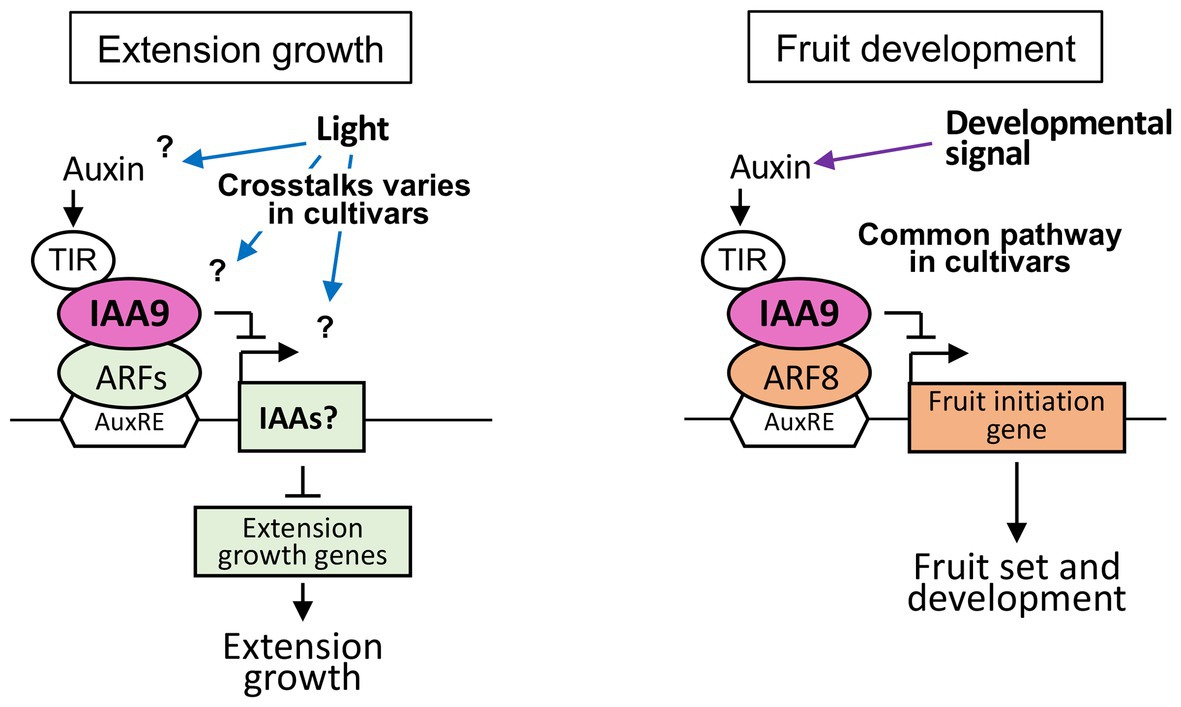
Figure 5. Control mechanisms of elongation growth and fruit development mediated by SlIAA9 in various tomato cultivars. (Left) A hypothetical model of shoot elongation controlled by SlIAA9 in tomato cultivars, Moneymaker, Rio Grande, and Ailsa Craig. (Right) A model of fruit development commonly controlled by SlIAA9 in three tomato cultivars.
We analyzed the effects of light intensity on plant stem and leaf growth of the various tomato cultivars and the relationship with SlIAA9 disruption. Interestingly, the results indicated that SlIAA9 function in stem elongation growth varied among cultivars and light conditions, with Rio Grande showing the enhanced growth of young seedlings; this was even more apparent in the sliaa9 mutant of Aila Craig. In Arabidopsis, it has been shown that light conditions affect the auxin signal pathway, biosynthesis, and transport system (Halliday et al., 2009). Under high light conditions, PHYB and CRY1 are activated and inhibit the AUX/IAA degradation, resulting in the inhibition of expression of auxin responsive genes for elongation growth (Ma and Li, 2019). Under low light or low F:FR conditions, transcription of auxin responsive genes is upregulated through the activation of PIF4/5/7, and elongation growth is enhanced in Arabidopsis (Ma and Li, 2019). Tomato cultivar M82 and wild relatives of tomato (e.g., Solanum pennellii) exhibited shade avoidance responses (e.g., hypocotyl and petiole elongation) under shade or dim-light conditions (Chitwood et al., 2015), suggesting that the mechanisms of elongation growth under light conditions of these tomatoes might be similar with those in Rio Grande.
The upregulation of SlIAA3 – an auxin responsive gene (Chaabouni et al., 2009) – has been reported in SlIAA9 knockout plants, Ailsa Craig entire LA2922 and Micro-Tom AS-IAA9 (Wang et al., 2005; Zhang et al., 2007). The Micro-Tom AS-IAA3 line (SlIAA3 antisense line) showed decreased auxin responses and reduced hypocotyl elongation (Chaabouni et al., 2009). In the present study, AC-sliaa9 hypocotyls were longer than those of wild-type under low light conditions, suggesting that upregulation of SlIAA3 might have occurred in this mutant (Wang et al., 2005). In this study, the SlIAA3 expression levels is not strongly related to the mutant phenotypes for elongation growth and response to light intensity except Rio Grande and more complicated pathways might be existed in the downstream of IAA9 in elongation growth of the tomato cultivars (Figure 5). Further detailed studies at the molecular level are needed to clarify the regulatory mechanisms involved, including the crosstalk of auxin and light signal transduction in tomato cultivars.
The sliaa9 mutants generated by CRISPR-Cas9 in the various commercial cultivars used here showed similar parthenocarpic phenotypes without any changes in fruit yield, suggesting the usefulness of these mutant lines for agricultural applications. Our results suggest that the function of SlIAA9 in fruit development is conserved among tomato varieties, although shoot growth varied, and that these phenotypic changes are also affected by environmental factors, including light conditions. Our results add to the accumulating studies using novel mutants created by genome editing in useful cultivars that will further contribute to new findings in plant functions for further application of environmental control in agriculture.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
YO, KO, and CA-H wrote the manuscript. CA-H performed most of the experiments and analyzed the data. KY and NW performed the genetic analysis. RU and RH designed the gRNA and transformation. KO supervised the research. YO designed, led, and coordinated the overall study. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Program on Open Innovation Platform with Enterprises, Research Institute and Academia (OPERA). This work was also supported by JSPS KAKENHI Grant Number JP19H02932.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank N. Nakashima, M. Fukuhara, and N. Kira for their technical assistance. We also thank T. Miyaji for her critical discussion of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.627832/full#supplementary-material
References
Ballaré, C. L., and Pierik, R. (2017). The shade-avoidance syndrome: multiple signals and ecological consequences. Plant Cell Environ. 40, 2530–2543. doi: 10.1111/pce.12914
Berger, Y., Harpaz-Saad, S., Brand, A., Melnik, H., Sirding, N., Alvarez, J. P., et al. (2009). The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136, 823–832. doi: 10.1242/dev.031625
Chaabouni, S., Jones, B., Delalande, C., Wang, H., Li, Z., Mila, I., et al. (2009). Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. J. Exp. Bot. 60, 1349–1362. doi: 10.1093/jxb/erp009
Chitwood, D. H., Kumar, R., Ranjan, A., Pelletier, J. M., Townsley, B. T., Ichihashi, Y., et al. (2015). Light-induced indeterminacy alters shade-avoiding tomato leaf morphology. Plant Physiol. 169, 2030–2047. doi: 10.1104/pp.15.01229
Colón-Carmona, A., Chen, D. L., Yeh, K. C., and Abel, S. (2000). Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 124, 1728–1738. doi: 10.1104/pp.124.4.1728
Ganesan, M., Han, Y. J., Bae, T. W., Hwang, O. J., Chandrasekkhar, T., Shin, A. Y., et al. (2012). Overexpression of phytochrome A and its hyperactive mutant improves shade tolerance and turf quality in creeping bentgrass and zoysiagrass. Planta 236, 1135–1150. doi: 10.1007/s00425-012-1662-6
Goetz, M., Vivian-Smith, A., Johnson, S. D., and Koltunow, A. M. (2006). AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 18, 1873–1886. doi: 10.1105/tpc.105.037192
Halliday, K. J., Martínez-García, J. F., and Josse, E. M. (2009). Integration of light and auxin signaling. Cold Spring Harb. Perspect. Biol. 1:a001586. doi: 10.1101/cshperspect.a001586
Hashimoto, R., Ueta, R., Abe, C., Osakabe, Y., and Osakabe, K. (2018). Efficient multiplex genome editing induces precise, and self-ligated type mutations in tomato plants. Front. Plant Sci. 9:916. doi: 10.3389/fpls.2018.00916
Koenig, D., Bayer, E., Kang, J., Kuhlemeier, C., and Sinha, N. (2009). Auxin patterns Solanum lycopersicum leaf morphogenesis. Development 136, 2997–3006. doi: 10.1242/dev.033811
Ma, L., and Li, G. (2019). Auxin-dependent cell elongation during the shade avoidance response. Front. Plant Sci. 10:914. doi: 10.3389/fpls.2019.00914
Mazzucato, A., Cellini, F., Bouzayen, M., Zouine, M., Mila, I., Minoia, S., et al. (2015). A TILLING allele of the tomato Aux/IAA9 gene offers new insights into fruit set mechanisms and perspectives for breeding seedless tomatoes. Mol. Breed. 35:22. doi: 10.1007/s11032-015-0222-8
Motohashi, K. (2015). A simple and efficient seamless DNA cloning method using SLiCE from Escherichia coli laboratory strains and its application to SLiP site-directed mutagenesis. BMC Biotechnol. 15:47. doi: 10.1186/s12896-015-0162-8
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. doi: 10.1093/nar/8.19.4321
Osakabe, Y., and Osakabe, K. (2015). Genome editing with engineered nucleases in plants. Plant Cell Physiol. 56, 389–400. doi: 10.1093/pcp/pcu170
Overvoorde, P. J., Okushima, Y., Alonso, J. M., Chan, A., Chang, C., Ecker, J. R., et al. (2005). Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17, 3282–3300. doi: 10.1105/tpc.105.036723
Saito, T., Ariizumi, T., Okabe, Y., Asamizu, E., Hiwasa-Tanase, K., Fukuda, N., et al. (2011). TOMATOMA: a novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol. 52, 283–296. doi: 10.1093/pcp/pcr004
Trupkin, S. A., Legris, M., Buchovsky, A. S., Rivero, M. B. T., and Casal, J. J. (2014). Phytochrome B nuclear bodies respond to the low red to far - red ratio and to the reduced irradiance of canopy shade in Arabidopsis. Plant Physiol. 165, 1698–1708. doi: 10.1104/pp.114.242438
Ueta, R., Abe, C., Watanabe, T., Sugano, S. S., Ishihara, R., Ezura, H., et al. (2017). Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 7:507. doi: 10.1038/s41598-017-00501-4
Wang, H., Jones, B., Li, Z., Frasse, P., Delalande, C., Regad, F., et al. (2005). The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17, 2676–2692. doi: 10.1105/tpc.105.033415
Xu, F., He, S., Zhang, J., Mao, Z., Wang, W., Li, T., et al. (2018). Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit auxin signaling in Arabidopsis. Mol. Plant 11, 523–541. doi: 10.1016/j.molp.2017.12.003
Yang, C., Xie, F., Jiang, Y., Li, Z., Huang, X., and Li, L. (2018). Phytochrome A negatively regulates the shade avoidance response by increasing Auxin/Indole acidic acid protein stability. Dev. Cell 44, 29–41. doi: 10.1016/j.devcel.2017.11.017
Keywords: parthenocarpy, photomorphogenesis, elongation growth, shoot growth, auxin, SlIAA9, CRISPR-Cas9, tomato
Citation: Abe-Hara C, Yamada K, Wada N, Ueta R, Hashimoto R, Osakabe K and Osakabe Y (2021) Effects of the sliaa9 Mutation on Shoot Elongation Growth of Tomato Cultivars. Front. Plant Sci. 12:627832. doi: 10.3389/fpls.2021.627832
Edited by:
Antonio Ferrante, University of Milan, ItalyReviewed by:
Luigi Lucini, Catholic University of the Sacred Heart, ItalyGabriela Toledo-Ortiz, Lancaster University, United Kingdom
Junhong Zhang, Huazhong Agricultural University, China
Copyright © 2021 Abe-Hara, Yamada, Wada, Ueta, Hashimoto, Osakabe and Osakabe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuriko Osakabe, osakabe.y.ab@m.titech.ac.jp
 Chihiro Abe-Hara
Chihiro Abe-Hara Kohji Yamada
Kohji Yamada Naoki Wada
Naoki Wada Risa Ueta
Risa Ueta Ryosuke Hashimoto
Ryosuke Hashimoto Keishi Osakabe
Keishi Osakabe Yuriko Osakabe
Yuriko Osakabe