- 1 Department of Microbiology, Montana State University, Bozeman, MT, USA
- 2 Center for Biofilm Engineering, Montana State University, Bozeman, MT, USA
- 3 Department of Food Science, Purdue University, West Lafayette, IN, USA
- 4 Program in Molecular Structure and Function, The Hospital for Sick Children, Toronto, ON, Canada
- 5 Department of Biochemistry, University of Toronto, Toronto, ON, Canada
Pseudomonas aeruginosa thrives in many aqueous environments and is an opportunistic pathogen that can cause both acute and chronic infections. Environmental conditions and host defenses cause differing stresses on the bacteria, and to survive in vastly different environments, P. aeruginosa must be able to adapt to its surroundings. One strategy for bacterial adaptation is to self-encapsulate with matrix material, primarily composed of secreted extracellular polysaccharides. P. aeruginosa has the genetic capacity to produce at least three secreted polysaccharides; alginate, Psl, and Pel. These polysaccharides differ in chemical structure and in their biosynthetic mechanisms. Since alginate is often associated with chronic pulmonary infections, its biosynthetic pathway is the best characterized. However, alginate is only produced by a subset of P. aeruginosa strains. Most environmental and other clinical isolates secrete either Pel or Psl. Little information is available on the biosynthesis of these polysaccharides. Here, we review the literature on the alginate biosynthetic pathway, with emphasis on recent findings describing the structure of alginate biosynthetic proteins. This information combined with the characterization of the domain architecture of proteins encoded on the Psl and Pel operons allowed us to make predictive models for the biosynthesis of these two polysaccharides. The results indicate that alginate and Pel share certain features, including some biosynthetic proteins with structurally or functionally similar properties. In contrast, Psl biosynthesis resembles the EPS/CPS capsular biosynthesis pathway of Escherichia coli, where the Psl pentameric subunits are assembled in association with an isoprenoid lipid carrier. These models and the environmental cues that cause the cells to produce predominantly one polysaccharide over the others are subjects of current investigation.
Introduction
Pseudomonas aeruginosa is a versatile organism that is able to adapt to a wide range of environments. The bacterium grows in aqueous environments but can also colonize and cause opportunistic infections in both plants and mammals (Lyczak et al., 2000, 2002; Silo-Suh et al., 2002). With its repertoire of virulence factors, P. aeruginosa can even cause death to worms and insects (Tan et al., 1999; Hendrickson et al., 2001; Sibley et al., 2008). The ability of P. aeruginosa to adapt to different environments is due, at least in part, to the high percentage of transcriptional regulators that allow the cells to adapt rapidly to changing environmental conditions (Stover et al., 2000). One strategy that P. aeruginosa possesses that provides the organism with additional survival advantages during changing environmental conditions is the production of several different types of extracellular polysaccharides. The polysaccharides provide the cells with enhanced tolerance to desiccation, oxidizing agents, and host defensive processes (Berry et al., 1989; DeVault et al., 1990; Govan and Deretic, 1996; Pier et al., 2001; Friedman and Kolter, 2004b; Jackson et al., 2004). Production of extracellular polysaccharides during growth on laboratory media provides a visually striking demonstration of the capacity of P. aeruginosa to adapt to its environment. For example, P. aeruginosa isolates obtained from chronic pulmonary infections of patients with cystic fibrosis (CF) are often mucoid. These isolates secrete large amounts of the polysaccharide, alginate. In contrast, some P. aeruginosa CF isolates have a rugose, small colony variant (RSCV) phenotype (Starkey et al., 2009). RSCV isolates autoaggregate during culture in liquid media due to the production of exopolysaccharides that have been termed Pel and Psl. The O-antigen of lipopolysaccharides (LPS) also provides many other adaptive advantages to P. aeruginosa. As LPS has been extensively reviewed elsewhere (Kintz and Goldberg, 2008; King et al., 2009), this review will discuss only the secreted exopolysaccharides, alginate, Pel, and Psl.
The importance of alginate in the pathology of CF disease led to it being the first exopolysaccharide described and it remains the best characterized P. aeruginosa exopolysaccharide in terms of our understanding of its biosynthesis and regulatory mechanisms (Hay et al., 2009). However, alginate is generally not produced by strains isolated from environments other than the CF lung. Isolates obtained from environments other than CF pulmonary tissue produce two different exopolysaccharides, which were identified as a consequence of the complete P. aeruginosa genome sequence becoming available, and through the use of high throughput transposon screening. The Psl polysaccharide was identified using reverse genetics of a putative polysaccharide gene cluster (PA2231–PA2245 of the P. aeruginosa PAO1 genome; Jackson et al., 2004). Mutation of the various Psl genes resulted in clones that were impaired in their ability to form surface-attached communities of microorganisms, termed biofilms (Ma et al., 2006). Pel was identified by screening transposon mutant libraries of the P. aeruginosa strain PA14 for the lack of structured microbial mats that form at the air-water interface of stagnant cultures, termed pellicles (Friedman and Kolter, 2004a).
Alginate and Psl exopolysaccharides have distinct chemical structures (Figure 1). While the structure of Pel has not been fully characterized, it likely has a structure that differs from alginate and Psl (Coulon et al., 2010). Each exopolysaccharide provides differing physiological properties to the cells and the biofilm matrix. In general, individual P. aeruginosa strains, produce predominantly one secreted polysaccharide at any given time (Ohman, 1986; Friedman and Kolter, 2004b; Jackson et al., 2004), although the sequenced strains have the genetic capacity to produce all three polysaccharides (Stover et al., 2000). RSCV isolates may be an exception to this rule, since they have been shown by transcriptional analysis to express both the Psl and Pel operons simultaneously (Starkey et al., 2009). The structure of the alginate and Psl exopolysaccharides have been determined (Evans and Linker, 1973; Byrd et al., 2009). Alginate is a high molecular weight acidic polysaccharide composed of non-repeating subunits of selectively O-acetylated D-mannuronic acid and its C5′ epimer L-guluronic acid (Figure 1A; Linker and Jones, 1964; Chitnis and Ohman, 1990; Franklin and Ohman, 1993). In mucoid strains, alginate is secreted into the surrounding medium and it is not covalently linked to the cell surface. Alginate produced by the mucoid CF P. aeruginosa FRD1 isolate, results in a highly viscous appearance of the colonies on agar medium that can be visualized by atomic force microscopy as a relatively soft but dense gelatinous structure that surrounds the cells (Figure 2A). The Psl polysaccharide is composed of a repeating pentamer containing D-mannose, L-rhamnose, and D-glucose (Figure 1B). Lectin staining of Psl indicated that it forms a helical distribution that surround the cell surface of P. aeruginosa PAO1, thereby facilitating its ability to play a role in cell–cell and cell–surface interactions during biofilm formation (Ma et al., 2009). Fluorescent staining and confocal laser scanning microscopy of P. aeruginosa PAO1 biofilms suggest that Psl forms a fabric-like matrix connecting the biofilm cells (Figure 2B). At present, the structure of Pel has not been determined, but it is proposed to be a glucose-rich polysaccharide that is distinct from cellulose (Friedman and Kolter, 2004b). Scanning electron microscopy of a P. aeruginosa PA14 pellicle shows matrix that connects the cells, allowing them to form a structured assembly at the air-liquid interface (Figure 2C). The connecting matrix is likely composed of Pel polysaccharide, but may contain other secreted polymers including the O-antigen of LPS, and cyclic glucans (Coulon et al., 2010). Diversification of subpopulations of bacteria that produce different polysaccharides has been observed in environmental and CF biofilms, particularly during the production of RSCV. This diversification is hypothesized to be a survival strategy providing members of the biofilm community with a means to withstand changes in environmental conditions (Boles et al., 2004).
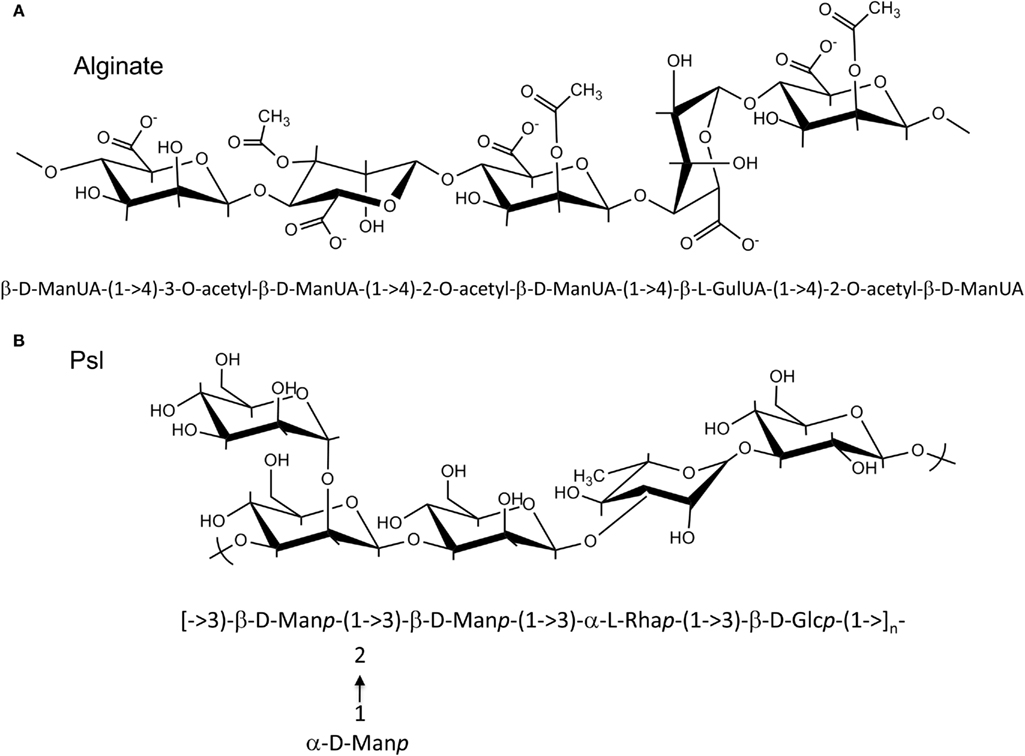
Figure 1. Structures of Alginate and Psl polysaccharide. (A) P. aeruginosa alginate is composed of D-mannuronic acid residues interspersed with L-guluronic acid residues. The hydroxyl groups of the D-mannuronic acid residues may be O-acetylated at the C2′ and/or C3′ positions. Since modification of the D-mannuronate residues occurs at the polymer level, alginate has a random structure. Shown is an example of a possible alginate subunits arrangement, where D-mannuronate subunits are epimerized to L-guluronate or decorated with O-acetyl groups. (B) Psl polysaccharide is composed of a repeating pentamer consisting of D-mannose, L-rhamnose, and D-glucose residues.
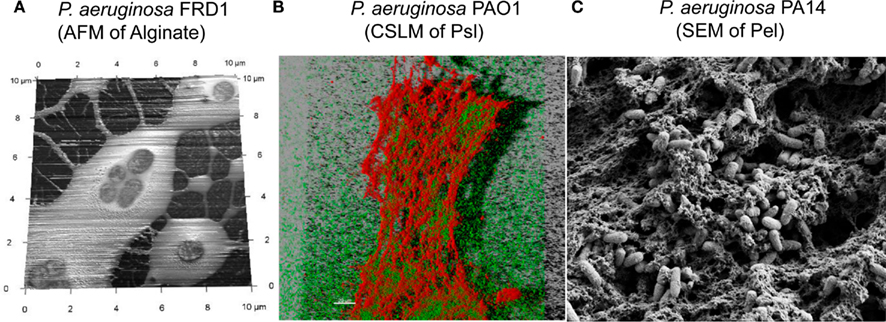
Figure 2. Extracellular polysaccharides of P. aeruginosa, visualized using three approaches. (A) Atomic Force Microscopy image of P. aeruginosa FRD1, showing alginate as a soft loosely adhered polymer that surrounds the cells. (B) Confocal laser scanning microscopy (CLSM) image of hydrated P. aeruginosa PAO1, cultured as a pellicle. The CLSM image shows a three-dimensional reconstruction of the pellicle, with the P. aeruginosa cells expressing the green fluorescent protein and the Psl polysaccharide counterstained with CellMask Orange (Invitrogen Corp.). (C) Scanning Electron Microscopy image of P. aeruginosa PA14, cultured as a pellicle. The image shows the extracellular matrix, which includes Pel, as a fabric-like matrix that surrounds and connects the cells that form a microbial mat at the air-water interface.
Since Psl and Pel were discovered fairly recently, the biosynthetic mechanisms of these two exopolysaccharides are not well established and many aspects of alginate biosynthesis still remain unclear. However, the functional characterization and structures of several proteins involved in alginate production have recently been determined (Regni et al., 2002; Snook et al., 2003; Keiski et al., 2010; Whitney et al., 2011). Using this information and the predicted structures of other alginate, Psl and Pel proteins, herein we describe predictive models for the biosynthetic complexes of Psl and Pel. We predict that the biosynthetic mechanisms for the three P. aeruginosa exopolysaccharides fall into two general classes – an isoprenoid lipid carrier dependent mechanism and most likely a lipid carrier independent mechanism. Biosynthesis of many bacterial exopolysaccharides requires oligosaccharide subunit assembly on a lipid carrier prior to transport across the inner membrane and polymerization of full-length polymer in the periplasm (Whitfield, 2006). As described below, Psl is likely synthesized using this strategy based on the predicted structural similarities to proteins involved in group 1 capsular polysaccharide production in Escherichia coli, such as the K30 antigen (Whitfield, 2006). No isoprenoid lipid carrier has been observed as an intermediate for alginate biosynthesis. Rather, alginate is believed to be polymerized and directly transported across the inner membrane using its glycosyltransferase, Alg8, in conjunction with Alg44, a bis-(3′–5′)-cyclic-dimeric guanosine monophosphate (c-di-GMP) binding protein which has been shown to be required for alginate polymerization (Remminghorst and Rehm, 2006b; Merighi et al., 2007; Oglesby et al., 2008). The mechanism for alginate biosynthesis appears to be similar to that of bacterial cellulose biosynthesis, as this exopolysaccharide similarly does not require a lipid carrier (Romling, 2002). Pel biosynthesis is the least characterized P. aeruginosa exopolysaccharide, but our analysis suggests that it may not require a lipid carrier. Structural modeling of the Pel proteins suggest that its biosynthesis is more similar to that of alginate and cellulose, than Psl, but significant differences also exist (see below).
Each P. aeruginosa exopolysaccharide is encoded on distinct regions of the P. aeruginosa genome (Stover et al., 2000; Figure 3). Alginate is encoded on a 12 gene operon, from PA3540–PA3551 (on the PAO1 genome), Psl on a 12 gene operon from PA2231–PA2242, and Pel are on a seven gene operon from PA3058–PA3064. It should be noted that while PA2243–PA2245 have been termed Psl genes, the results of mutagenesis studies indicate that they are not required for Psl synthesis (Byrd et al., 2009). Figure 3 shows the organization of the exopolysaccharide biosynthetic operons, with the genes color coded by the proposed role(s) that each protein product plays in the biosynthetic process. The biosynthesis of the polymer can be divided into the following distinct steps (Hay et al., 2009): (i) production of the activated sugar-nucleotide precursor; (ii) polymerization and transport across the inner membrane; (iii) post-polymer modification of the polymer in, and passage through the periplasm and; (iv) export from the cell. In the sections below, we combine available published data with the results of our protein modeling studies to describe the four steps in the biosynthetic pathways for each of the three P. aeruginosa extracellular polysaccharides.
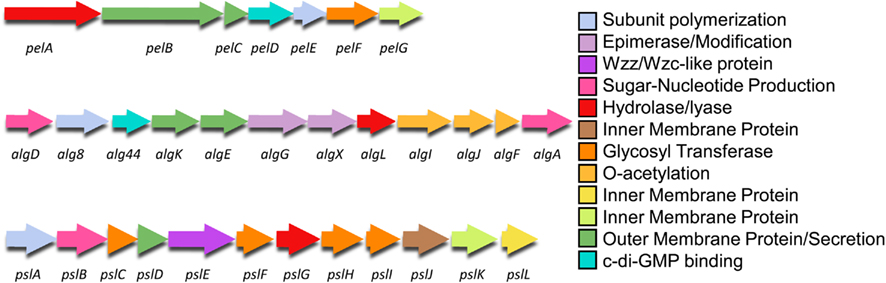
Figure 3. Genetic structure of the Pel, alg, and Psl operons. The genes are color coded by proposed function as shown in Figures 4–6.
Alginate Biosynthesis
Alginate is first synthesized as a linear homopolymer of D-mannuronic acid residues. The polymer is then modified in the periplasm through selective O-acetylation by the concerted action of AlgI, AlgJ, and Alg,F and epimerized by AlgG (Franklin and Ohman, 1993, 2002; Franklin et al., 1994). Since AlgG converts the D-mannuronic acid residues to L-guluronic acid at the polymer level and acetylation can occur at the hydroxyl groups of either the C2′ and/or C3′ positions, alginate has a somewhat random structure (Figure 1A). This distinguishes alginate from many of the E. coli capsular polysaccharides and from Psl as these polymers are composed of regular repeating subunits (Figure 1B).
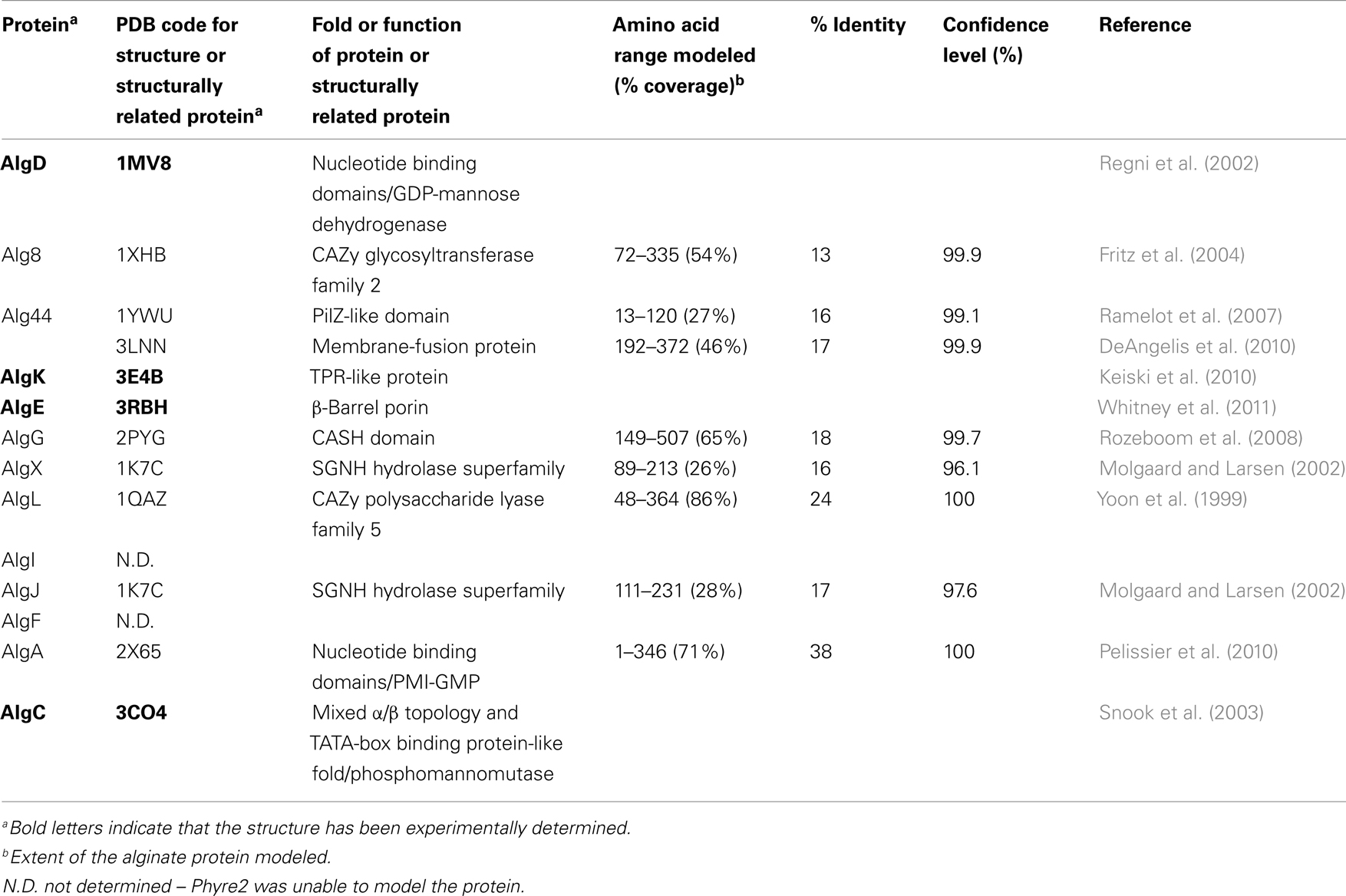
Although direct protein–protein interactions of the 13 alginate proteins have not yet been demonstrated experimentally, genetic and physiological evidence suggest that most of the proteins form a complex that spans the inner membrane, periplasm, and outer membrane (Figure 4 and Table 1). Evidence for a multi-protein biosynthetic apparatus comes from in vitro assays, which found that the cell envelope, including both inner and outer membranes, were required for alginate polymerization (Remminghorst and Rehm, 2006b). The structure of the periplasmic/outer membrane lipoprotein AlgK (PDB ID:3E4B; Keiski et al., 2010), which contains multiple copies of the protein–protein interaction tetratricopeptide-like repeat (TPR), reinforces this hypothesis, as TPR proteins often act as scaffolds for the assembly of multi-protein complexes (Blatch and Lassle, 1999; D’Andrea and Regan, 2003). Further evidence of a multi-protein biosynthetic complex was found when examining the phenotypes of algK, algX, and algG deletion mutants (Jain and Ohman, 1998; Jain et al., 2003; Robles-Price et al., 2004). Deletion of any one of these genes results in the secretion of depolymerized uronic acid subunits, presumably degraded by the periplasmic alginate lyase, AlgL (Jain and Ohman, 1998). These results suggest that each of these proteins is part of the biosynthetic complex and that when this is not formed, the synthesized polymer is exposed to AlgL and degraded. Given its structure, AlgK is likely a scaffold protein, which plays a key role in the assembly of the other periplasmic proteins that form the multi-protein biosynthetic complex (Keiski et al., 2010).
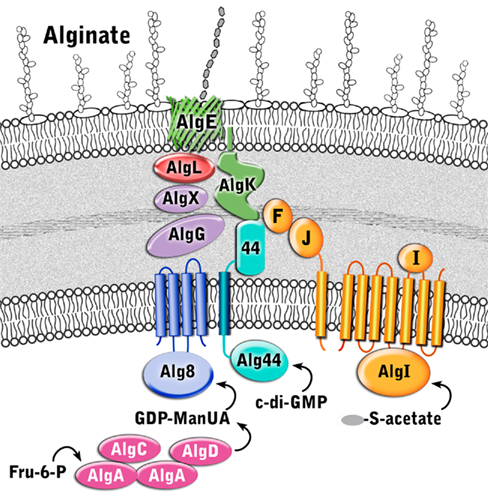
Figure 4. Proposed structure of the alginate biosynthetic complex. The proteins are color coded according to proposed function as described in Figure 3.
Alginate Precursor Synthesis
Exopolysaccharide biosynthesis requires sugar-nucleotide precursors. These are sugar subunits that have been activated with a high-energy bond. For alginate, the sugar subunits are activated with GDP, in a pathway where fructose-6-phosphate, derived from central carbon metabolism (Hay et al., 2009), is converted into GDP-mannuronate in a four-step process:
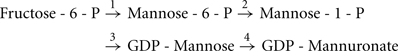
The enzymes required for GDP-mannuronate production include: (i) the bifunctional enzyme, AlgA which exhibits phosphomannose isomerase (PMI) and GDP-mannose pyrophosphorylase (GMP) activity (reactions 1 and 3); (ii) AlgC, a phosphomannomutase (reaction 2); and (iii) AlgD, which is a GDP-mannose dehydrogenase (reaction 4; Darzins et al., 1986; Deretic et al., 1987; Zielinski et al., 1991). Two of the genes required for this process are found on the alginate operon, algA and algD. However, algC is located elsewhere in the genome at PA5322 (Zielinski et al., 1991). AlgC appears to be crucial for general exopolysaccharide biosynthesis, not just alginate, as it is also required for precursor synthesis of Psl, as well as LPS and rhamnolipids (Goldberg et al., 1993; Olvera et al., 1999).
The crystal structures of two of these enzymes, AlgD and AlgC, have been determined (Regni et al., 2002; Snook et al., 2003). A common structural feature of enzymes involved in nucleotide binding, such as in the generation of activated sugars, is the presence of at least one β/α/β nucleotide binding domain. This domain, often referred to as a Rossmann fold, has a secondary structure consisting of alternating β-strands and α-helices arranged such that they form a central six-stranded parallel β-sheet linked to five surrounding α-helices. Many variations of the “classical” Rossmann fold or nucleotide binding domain have been observed in literature and AlgD (PDB ID:1MV8) is one such example (Snook et al., 2003). This protein forms a dimer with each individual subunit containing one complete N-terminal nucleotide binding domain and a C-terminal nucleotide-like binding domain, which lacks the third β-strand and final α-helix of this motif (Snook et al., 2003). The two nucleotide binding domains are separated by a long 33 residue α-helix, that has been termed the spacer-helix. Interestingly, the protein forms a domain-swapped dimer, whereby the N-terminal nucleotide binding domain of one subunit interacts with the C-terminal nucleotide binding domain of the second subunit. The interface of these two domains forms the active site; the location of which was verified by the structure of AlgD in complex with its substrate, NAD(H) and product GDP-mannuronate (ManUA). Two dimers of AlgD likely interact to form a tetrameric structure in the cell cytoplasm, creating the GDP-ManUA product, which is the irreversible step in alginate precursor formation.
The second enzyme involved in alginate precursor production whose structure has been determined is the magnesium dependent mutase, AlgC (PDB ID:3CO4), which exhibits specificity for both phosphomannose and phosphoglucose substrates (Regni et al., 2002). This protein contains four domains of approximately equal size. The first three domains have a common topological core consisting of a four-stranded β-sheet sandwiched between two α-helices, while the fourth domain is unrelated structurally to the first three, and is a member of the TATA-box binding protein-like fold superfamily. This domain consists of a four-stranded anti-parallel β-sheet, flanked by two α-helices and two short β-strands. Residues from all four domains contribute to the formation of a large active site cleft at the center of this “heart” shaped molecule. The specificity (or lack thereof) for glucose versus mannose in this class of enzymes is thought to be determined, at least in part, by a conserved sequence motif GEMS(G/A) found in domain three, which has been postulated to act as the sugar binding loop.
While the structure for AlgA has not been determined, it is predicted by structural modeling to have extensive similarity to other proteins with GDP-mannose pyrophosphorylase activity, such as the Thermotoga maritima guanosine-diphospho-D-mannose pyrophophorylase (PDB ID:2X65; Pelissier et al., 2010; Table 1) and a putative mannose-1-phosphate guanyltransferase from Thermus thermophilus (PDB ID:2CU2). Both of these proteins contain Rossmann-like β/α/β nucleotide binding domains characteristic of proteins that generate or bind sugar-nucleotide precursors.
Polymannuronate Polymerization
Deletion mutations of the genes for the cytoplasmic membrane proteins Alg8 and Alg44 result in no alginate production (Maharaj et al., 1993; Remminghorst and Rehm, 2006a; Oglesby et al., 2008). This is in contrast to deletions of the periplasmic proteins, which result in secretion of degraded polymer and therefore suggests that Alg8 and Alg44 play a key role in alginate polymerization. Alg8 is predicted to have four transmembrane (TM) domains and a large cytoplasmic glycosyltransferase (GT) domain (Remminghorst and Rehm, 2006a; Oglesby et al., 2008). The protein has been classified as a member of the GT-2 family, a large family of inverting glycosyltransferases that include cellulose, chitin, and hyaluronan synthases (Carbohydrate Active Enzymes database, CAZy: http://www.cazy.org/; Coutinho et al., 2003; Cantarel et al., 2009). The cytoplasmic domain is predicted by Phyre2 (Kelley and Sternberg, 2009) to be structurally homologous to UDP-GalNAc:polypeptide α-N-acetylgalactosaminyl transferase-T1 (PDB ID:1XHB; Fritz et al., 2004). As seen above for AlgD, Alg8 is predicted to contain two closely abutting β/α/β Rossmann-like nucleotide binding domains or a GT-A fold. Interestingly, Remminghorst and Rehm (2006b) have demonstrated using an in vitro assay for alginate polymerization with 14C labeled GDP-mannuronic acid as a precursor that alginate chain extension only occurs in the cell envelope fraction. As in vitro polymerization does not occur in the absence of Alg8, this suggests that polymerization requires an association between both the cytoplasmic and outer membranes, and as mentioned above, is one of the primary lines of evidence that an alginate multi-protein biosynthetic complex exists. In complementation studies using a P. aeruginosa PAO1 strain that was engineered to produce alginate (termed P. aeruginosa PDO300), alg8 overexpression in trans resulted in up to a 20-fold increase in alginate production compared to the wild-type strain, and an altered acetylation and epimerization profile (Remminghorst and Rehm, 2006b). This result is surprising, if it is assumed that the alginate biosynthetic proteins form a multi-protein complex; why should extra copies of one component cause an increase in polymer production? Thus, the investigators speculate that Alg8 may be the bottleneck for alginate biosynthesis (Remminghorst and Rehm, 2006b).
The second cytoplasmic membrane protein that has been shown to be absolutely required for alginate production is Alg44 (Merighi et al., 2007). Alg44 has a single TM domain located near the middle of the protein. The protein is predicted to contain a cytoplasmic N-terminal PilZ domain (Merighi et al., 2007). PilZ domains play a role in binding the secondary messenger c-di-GMP, and point mutations in this domain that result in loss of c-di-GMP binding lead to loss of alginate production. While it is clear that binding of c-di-GMP by Alg44 is required for alginate production, the exact molecular mechanism by which this interaction regulates polymerization is not known. It is interesting to note, that Pel biosynthesis also requires c-di-GMP binding to one of its biosynthetic proteins; making c-di-GMP binding a potentially common post-translational regulator of exopolysaccharide production. The C-terminal periplasmic domain of Alg44 is predicted to resemble the membrane-fusion protein MexA from the MexAB-OprM multidrug efflux pump (Remminghorst and Rehm, 2006a). This structural similarity and the requirement for a complete envelope fraction for in vitro polymerization suggest that this domain of Alg44 may play a role in the assembly of the multi-protein complex.
Polymannuronate Modification
In the periplasm, polymannuronic acid is modified to the mature alginate polymer by a series of enzymes that include the O-acetylation complex, AlgI/AlgJ/AlgF, the polymannuronan epimerase, AlgG (Chitnis and Ohman, 1990; Franklin and Ohman, 1993, 2002); Franklin et al., 1994), and possibly, AlgX (Weadge et al., 2010). AlgX does not have a known function, but it likely associates with the multi-protein complex, since removal of AlgX by deletion mutagenesis results in the secretion of depolymerized alginate (Robles-Price et al., 2004). Interestingly, AlgX exhibits 69% sequence similarity to one of the O-acetylation enzymes, AlgJ. Homology modeling suggests that the N-terminal region, exhibits structural similarity to members of the SGNH hydrolase superfamily of enzymes, which typically remove acyl groups from carbohydrates and other compounds (PDB ID:1K7C). This finding suggests that AlgX could have a potential role in polymer modification (Weadge et al., 2010).
Deletion of any of the O-acetylation proteins, AlgI/AlgJ/AlgF, does not impair polymer synthesis or lead to the secretion of depolymerized alginate, indicating that removal of the O-acetylation proteins does not disrupt the formation of the biosynthetic complex and/or allow AlgL access to the polymer (Franklin and Ohman, 2002). AlgI/AlgJ/AlgF O-acetylate mannuronate residues on the hydroxyl groups of the C2′ and/or C3′ positions (Franklin and Ohman, 2002; Figure 1A). The membrane topology of AlgI was mapped using alkaline phosphatase fusion proteins and found to contain seven TM domains, and two large cytoplasmic domains (Franklin and Ohman, 2002). AlgF and AlgJ both localize to the periplasm, where AlgJ is anchored in the cytoplasmic membrane by an uncleaved signal peptide. We have been unable with any confidence to find structural homologs for AlgI or AlgF, but AlgJ shares sequence similarity to the N-terminal region of AlgX and is also predicted to be structurally similar to members of the SGNH hydrolase superfamily (Table 1). The cellular location of AlgI/AlgJ/AlgF suggests a model for alginate O-acetylation, where the acetyl group contained on a donor molecule (possibly an acyl carrier protein or Coenzyme A) is transferred across the membrane by AlgI, then transferred to AlgJ or AlgF for O-acetylation of the mannuronate residues at the polymer level. This seems like an unusual strategy for polysaccharide modification, particularly when compared to N-acetylation of capsular polysaccharides where modification occurs on the sugar precursor prior to polymerization. However, it appears that the AlgI/AlgJ/AlgF esterification mechanism evolved as an ancient strategy to modify a wide variety of exopolysaccharides (Franklin et al., 2004). A similar strategy is used in O-alanylation of lipoteichoic acids in Gram-positive bacteria (via the DltB/DltD complex; Heaton and Neuhaus, 1992), and O-acetylation of cellulose in P. fluorescens (Spiers et al., 2002). AlgI and AlgJ homologs are also found in a wide variety of distantly related bacteria that do not produce alginate, but almost certainly produce other types of exopolysaccharides. The algI, algJ, and algF genes were likely incorporated into the alginate biosynthetic operon by an ancient lateral gene transfer event, as determined by phylogenetic analysis of AlgI and its homologs (Franklin et al., 2004).
AlgG catalyzes the epimerization of the D-mannuronate to L-guluronate at the polymer level (Franklin et al., 1994). This epimerization process alters the structural properties of alginate, including its gelling ability and its ability to bind divalent ions such as calcium (Gacesa, 1988). The structure of the C-terminal domain of AlgG has been modeled and is predicted to contain a right-handed β-helix (RHβH) fold, characteristic of proteins with carbohydrate-binding and sugar hydrolase (CASH) domains (Douthit et al., 2005). The structure of the extracellular alginate epimerase, AlgE4 (PDB ID:2PYG), from Azotobacter vinelandii reinforces this prediction (Rozeboom et al., 2008). CASH domain proteins include the pectate lyase of Erwinia chrysanthemi and several other proteins with glycosidase activity (Jenkins and Pickersgill, 2001; Jenkins et al., 2001). AlgG is the first known CASH domain protein with epimerase rather than lyase activity. The RHβH fold of AlgG includes a shallow cleft that contains the epimerase active site (Douthit et al., 2005). Point mutations of conserved residues in this cleft result in the production of alginate lacking L-guluronate (i.e., O-acetylated polymannuronic acid). Therefore, point mutations in AlgG differ from the algG deletion mutants, which cause secretion of depolymerized alginate (Jain et al., 2003). These results suggest that as long as AlgG maintains its proper tertiary structure it can associate with its protein partner(s) in the multi-protein complex, even if it is enzymatically inactive. It also suggests that alginate may slide through the biosynthetic complex, including the active cleft of AlgG, where it epimerizes approximately every other mannuronate residue.
Some exopolysaccharide biosynthetic operons contain a gene for an enzyme that degrades the polymer. In the case of alginate, AlgL fulfills this function (Boyd et al., 1993; Schiller et al., 1993). Initial attempts at constructing algL deletion mutations in a mucoid strain were unsuccessful, suggesting that AlgL is an essential protein. In a strain where alginate production was regulated by isopropyl β-D-thiogalactopyranoside (IPTG) addition, an algL deletion mutant could be constructed as long as the alg operon was down regulated by lack of IPTG (Jain and Ohman, 2005). Induction of the algL mutant strain with IPTG resulted in cell death and electron micrographs indicated that the periplasm of the mutant strain filled up with a polymer, likely alginate (Jain and Ohman, 2005). This result suggested a role for AlgL in periplasmic processing of alginate or secretion of the polymer, although a role as part of the secretion complex cannot be ruled out. The AlgL protein differs structurally from CASH domain lyases described above and predicted for AlgG. Instead, AlgL is a member of polysaccharide lyase family 5, which is predicted by homology modeling to be an α6α5 barrel and structurally similar to the alginate lyase A1–III from Sphingomonas spp. (PDB ID:1QAZ; Yoon et al., 1999).
Alginate Periplasmic Translocation and Secretion
The structure of the putative scaffold protein, AlgK was recently determined (PDB ID:3E4B) and shown to contain at least 9.5 TPR-like repeats (Keiski et al., 2010). The TPR repeat consists of a pair of anti-parallel α-helices, which are typically found in multiple copies where they pack together to form superhelical structures. This motif and the closely related SEL-1 motif have been implicated in complex formation in a variety of different systems (Blatch and Lassle, 1999; D’Andrea and Regan, 2003). AlgK also contains a lipid moiety that anchors the protein to the outer membrane (Keiski et al., 2010). Deletion mutants of algK suggest that this protein may play a role in the localization of the outer membrane porin protein, AlgE, which led to the hypothesis that AlgK/AlgE interact to form a novel type of secretin that differs structurally from other bacterial capsular polysaccharide secretion systems. AlgE is an outer membrane protein that is capable of spontaneously incorporating into planar lipid bilayers and forming a highly anion specific channel (Rehm et al., 1994). Epitope tagging suggests that it is a monomeric 18-stranded β-barrel porin (Hay et al., 2010), a prediction that the recent structure of AlgE has confirmed (PDB ID:3RBH; Whitney et al., 2009, 2011). AlgE is characterized by a highly electropositive pore constriction formed by an arginine-rich conduit that is occluded on either side by an extracellular loop and an unusually long periplasmic loop, T8. Characterization in vitro and in vivo of a AlgE-T8 loop detection mutant suggests that this loop may play a role in regulating the transport of alginate across the outer membrane.
Pel Biosynthesis
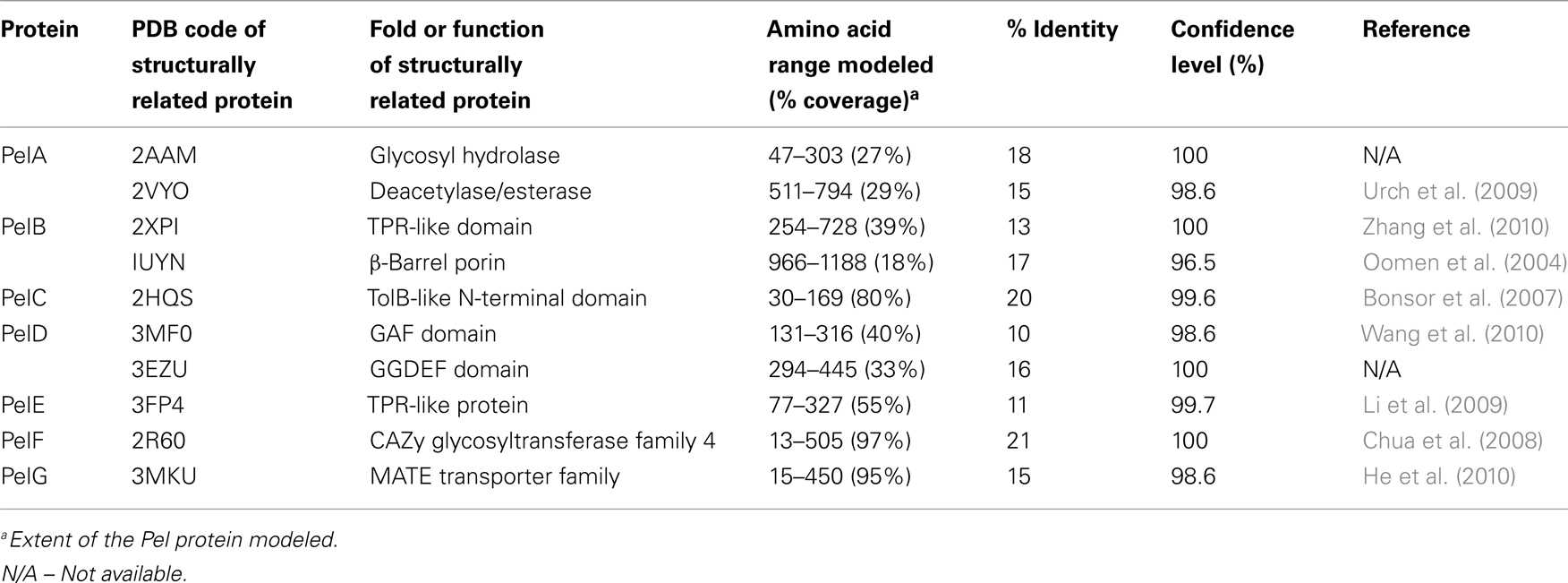
Little is known about Pel biosynthesis, as to date only two of the seven Pel gene products, PelC, and PelD, have been individually examined experimentally (Lee et al., 2007; Vasseur et al., 2007; Ueda and Wood, 2009; Kowalska et al., 2010), and a model of one, PelB, proposed (Keiski et al., 2010). Using these data and our structural homology analysis of the Pel proteins we have generated a preliminary model for Pel biosynthesis and export (Figure 5 and Table 2). Our model suggests that Pel biosynthesis is more closely related to the pathways for alginate and bacterial cellulose biosynthesis (Romling, 2002; Saxena and Brown, 2005), than for capsule production (Whitfield, 2006), but notable differences appear to exist in Pel that would distinguish it from the currently proposed mechanisms for these other secretion systems.
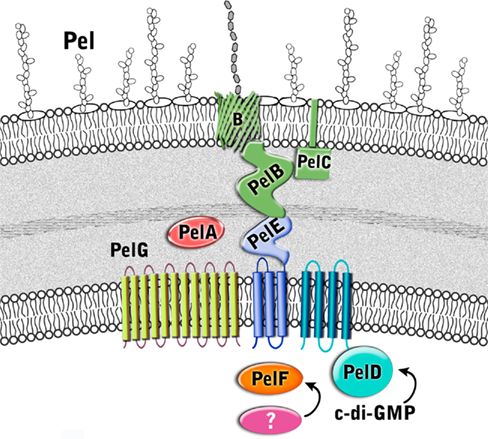
Figure 5. Proposed structure of the Pel biosynthetic complex. The enzymes are color coded according to proposed function as described in Figure 3.
Pel Precursor Production
Unlike alginate, which contains algA and algD on the biosynthetic operon, the Pel operon does not appear to carry any genes for the synthesis of its sugar-nucleotide precursor. This suggests that the biosynthesis of Pel sugar-nucleotide precursors is derived from central carbon metabolism and uses enzymes from other carbohydrate synthesis pathways, similar to the observation that AlgC is required for precursor production for alginate, Psl polysaccharide, rhamnolipid, and LPS production (Goldberg et al., 1993; Coyne et al., 1994; Olvera et al., 1999; Byrd et al., 2009).
Pel Polymerization
Predictions based on the sequences of the Pel proteins suggest that only PelF encodes a protein that localizes to the cytoplasm. PelF is predicted to be a glycosyltransferase that resembles enzymes of the CAZy GT-4 family (Coutinho et al., 2003). Structural homology modeling suggests that PelF has a classical GT-B fold with two Rossmann-like domains and therefore it is likely that this protein is involved in polymerization. Unlike Alg8, which contains multiple TM domains that are thought to aid in polymer export across the IM (Remminghorst and Rehm, 2006b; Oglesby et al., 2008), the cytoplasmic location of PelF and lack of TM domains suggests that Pel export across the inner membrane occurs by a different mechanism. Given that the Pel operon contains only one glycosyltransferase, we anticipate that Pel is a linear homopolymer, and therefore, like alginate, it may not require a lipid carrier for transport across the inner membrane. In contrast to the Psl polysaccharide, no gene for an undecaprenyl-phosphate glycosylphosphotransferase is found on the Pel operon. While it is not clear how the polymer is transported across the inner membrane, the most likely candidates involved in this process are PelD, PelE, and PelG, all of which are located in the inner membrane and are predicted to contain multiple TM domains. The predictions that Pel is a homopolymer and that an isoprenoid lipid does not act as a carrier assumes that there are no Pel genes that are unlinked to PelA-G, that have yet to be identified.
PelD is an inner membrane protein that is essential for Pel polysaccharide production (Lee et al., 2007). PelD binds c-di-GMP and point mutants that abrogate this binding prevent polymer formation. Similarly, manipulation of the cellular levels of c-di-GMP, through a deletion of the tyrosine phosphatase, TpbA, also influence the amount of Pel production and pellicle formation (Ueda and Wood, 2009). Therefore, PelD is functionally similar to Alg44 with respect to its c-di-GMP mediated regulation of polysaccharide production. However, in contrast to Alg44, which has one central TM domain that positions approximately half of Alg44 in the periplasm and the c-di-GMP binding PilZ domain in the cytoplasm (Merighi et al., 2007; Oglesby et al., 2008), the N-terminus of PelD is predicted to contain four TM domains. The presence of these TM domains results in the N-terminus of PelD being positioned in the inner membrane and the C-terminal region being in the cytoplasm where it interacts with c-di-GMP and regulates Pel production (Lee et al., 2007). Also in contrast to Alg44, structural homology predictions using Phyre2 (Kelley and Sternberg, 2009) suggest that the C-terminal domain, encompassing residues 111–455 contains tandemly arranged GAF and GGDEF domains (PDB ID:3MFO and 3EZU), despite the fact that PelD lacks the GG(D/E)EF consensus motif (Lee et al., 2007). How PelD regulates polymer production and why this protein requires four TM domains are questions that remain to be answered.
PelG is an integral inner membrane protein predicted to contain 12 TM domains (Krogh et al., 2001). PelG has structural similarity to NorM from Vibrio cholerae (PDB ID:3MKU), a member of the multidrug and toxic compound extrusion (MATE) family of proteins (He et al., 2010). MATE proteins couple electrochemical gradients to extrusion of molecules across the inner membrane. The similarity of PelG to this family of proteins, suggests that PelG may play a role in export of Pel polysaccharide across the inner membrane or alternatively, like AlgI, it may transport a small molecule substrate that is subsequently used, perhaps by PelA, to modify the Pel polymer.
The third inner membrane protein, PelE, appears to be highly unique. PelE contains a type 1 export signal and two predicted TM domains. This arrangement positions most of the C-terminus of PelE, residues 90–320, in the periplasm. The C-terminal region is predicted to be all α-helical with at least four to five and possibly as many as six copies of the TPR protein–protein interaction motif. Given the role of TPR motifs in the assembly of large protein complexes (Blatch and Lassle, 1999; D’Andrea and Regan, 2003), PelE, may function as a scaffold protein in an analogous manner to AlgK. PelE, may help in the assembly of a secretion complex through interactions possibly with PelA, and PelB. A role in complex assembly at the inner membrane has also been suggested for the periplasmic domain of Alg44 (Hay et al., 2009), indicating that the periplasmic domains of Alg44 and PelE may be functionally equivalent. Multimerization of PelE and/or its association with the four TM domains of PelD could potentially provide the required portal in the inner membrane for the export of the polymer.
Pel Translocation and Export
A second Pel protein with extensive predicted TPR structure is PelB (Keiski et al., 2010). PelB is likely localized to the periplasm and/or outer membrane. In addition to a large TPR containing domain at the N-terminus which homology modeling suggests is similar to the TPR containing anaphase-promoting complex/cyclosome subunit Cdc 16/Cut9 (PDB ID:2XPI; Zhang et al., 2010), PelB is also predicted to be rich in β-sheet structure at its C-terminus, a property that is characteristic of outer membrane porins. PelB may be functionally similar to the alginate secretin, AlgK/AlgE, but rather than two separate proteins, PelB appears to be a fusion protein containing both periplasmic and outer membrane domains in a single polypeptide. Keiski et al. (2010) suggested that this domain structure may be conserved in several other polysaccharide secretion complexes, including the bacterial cellulose and poly-β-1,6-N-acetylglucosamine (PAG) export proteins which contain the TPR/β-barrel fusion proteins, BcsC and PgaA (Romling, 2002; Wang et al., 2004). As in the case of AlgK, the periplasmic domain of PelB is anticipated to serve as a scaffold for the assembly of the secretion complex, and to protect the polysaccharide from degradation as it moves through the periplasm.
The Pel operon contains a second outer membrane protein, the lipoprotein PelC (Vasseur et al., 2007; Kowalska et al., 2010). PelC is lipidated at cysteine 19, and is predicted to contain a short stretch of ∼15 disordered residues followed by a single domain that has mixed α/β topology. Modeling results suggest that residues ∼43 to the C-terminus are structurally similar to the periplasmic N-terminal domain of E. coli TolB (PDB ID:2HQS; Bonsor et al., 2007), a protein that interacts with the peptidoglycan associated lipoprotein, Pal, and required for maintaining outer membrane integrity. TolB contains a three stranded β-sheet flanked by two α-helices. This model is somewhat different from the model proposed by Vasseur et al. (2007), who have suggested that the C-terminal amphipathic α-helix of PelC, which is required for Pel production, is inserted into the membrane in a Wza-like manner (Dong et al., 2006). In this model, PelC would be anchored to the OM by both its lipid anchor and C-terminal α-helix. Until the structure of PelC has been determined experimentally, it is not possible to resolve this discrepancy, but it is clear from the current functional characterization that PelC is essential for polymer production and that the C-terminal α-helix will play a key role either in protein folding and/or in the insertion of the protein into the OM.
The final protein of the Pel operon is PelA, a large 105 kDa protein that is predicted to contain an N-terminal TIM α/β-barrel domain with structural similarity to glycoside hydrolases (PDB ID:2AAM) and a C-terminal carbohydrate esterase domain (PDB ID:2VYO; Urch et al., 2009). Since the chemical structure of the Pel exopolysaccharide is unknown, it is difficult to predict the role of PelA in polymer production, but our model would suggest that the polymer, like alginate, may be modified after polymerization in the periplasm. Deacetylation of exopolysaccharides is not unprecedented as it is well established that N-deacetylation of the PGA polymer is required for biofilm formation in both Gram-negative and Gram-positive bacteria (Agladze et al., 2003). Similarly, if PelA also exhibits hydrolase activity, it may be functionally similar to AlgL, and thus required for proper processing and/or clearance of the Pel polysaccharide in the periplasm (Jain and Ohman, 2005).
Psl Biosynthesis
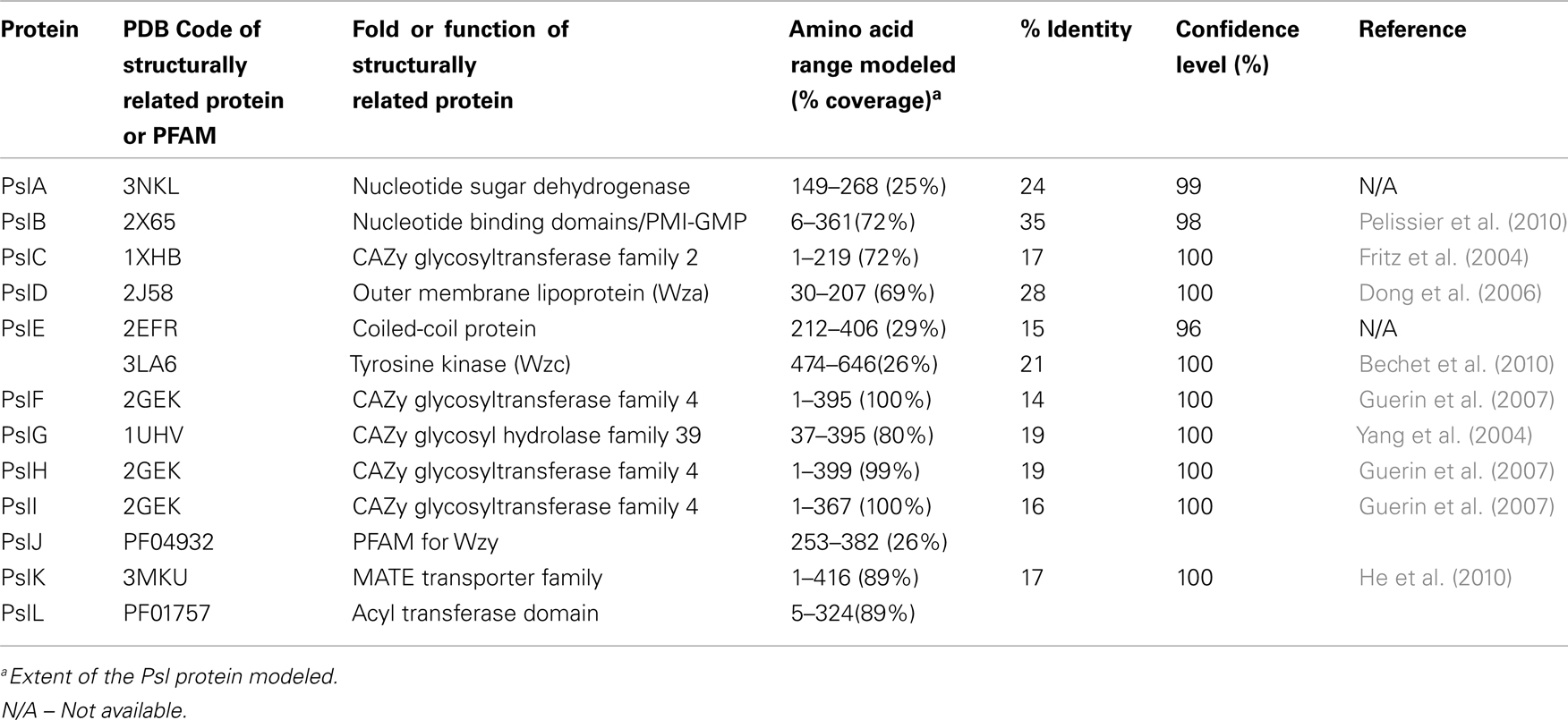
Although the Psl pathway has not been characterized, there is evidence to suggest that its biosynthetic mechanism functionally resembles the isoprenoid lipid-based biosynthesis of E. coli group 1 capsular and extracellular polysaccharides (CPS and EPS; Figure 6 and Table 3). First, Psl has a repeating oligosaccharide structure characteristic of CPS, as opposed to the homomeric (prior to epimerization) structure of alginate, which is polymerized a single sugar at a time. Second, the Psl operon does not encode a TPR protein which is characteristic of alginate and Pel polysaccharide export scaffolds, but does encode several proteins that have structural similarity to group 1 CPS/EPS proteins. These include PslA, PslE, and PslD, which are similar to WbaP, Wzc, and Wza, respectively. Therefore, the Psl biosynthetic pathway appears to be a Wzy-dependent pathway where polymers are built on an isoprenoid lipid carrier (undecaprenyl diphosphate) and then assembled and exported through the action of the group 1 CPS/EPS export process that requires polysaccharide copolymerases (PCP) and outer membrane polysaccharide export (OPX) proteins (Cuthbertson et al., 2009).
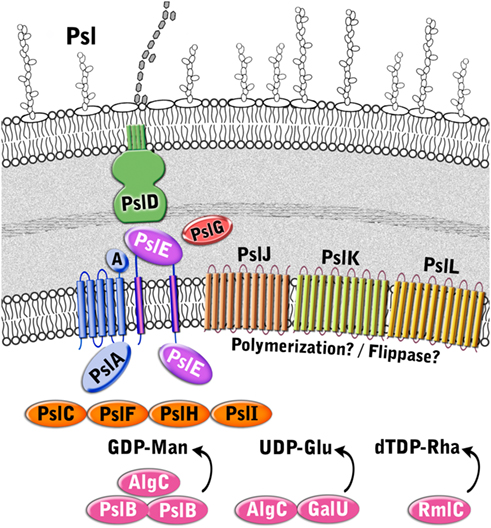
Figure 6. Proposed structure of the Psl biosynthetic complex. The enzymes are color coded according to proposed function as described in Figure 3.
Psl Precursor Production
Since Psl is composed of D-mannose, L-rhamnose, and D-glucose (Figure 1), activated sugar-nucleotide precursors are required for each of these subunits. Only one enzyme from the Psl operon, PslB, is involved in sugar-nucleotide precursor production. The remaining enzymes for precursor activation (AlgC, RmlC, and GalU) are encoded by genes associated with other polysaccharide biosynthetic pathways (Byrd et al., 2009). Byrd et al. (2009) constructed non-polar deletion mutations of each of the Psl genes (PslA-L) and demonstrated that of these mutations only the PslB deletion was still capable of producing Psl polysaccharide. PslB is a bifunctional enzyme that is similar to AlgA, as it is predicted to possess an N-terminal GMP domain and C-terminal PMI domain. P. aeruginosa PAO1 encodes three PMI/GMPs; PslB, WbpW (PA5452 on an LPS biosynthetic operon), and AlgA. A double mutant of PslB and wbpW resulted in loss of Psl production suggesting that the different PMI/GMPs are functionally interchangeable for the production of the GDP-mannose precursor (an algA/PslB double mutant was not tested for Psl production, since algA is not expressed in P. aeruginosa PAO1). As described for AlgA in the alginate pathway shown above, PslB provides the first and third steps (PMI/GMP) for converting fructose-6-phosphate from central metabolism into GDP mannose. AlgC provides the phosphomannomutase activity for the second step (Byrd et al., 2009). Since AlgC also has phosphoglucomutase activity, it is also proposed to be involved in the production of glucose-1-phosphate, which is the substrate for GalU to convert into the UDP-glucose precursor. Byrd et al. (2009) also demonstrated that the L-rhamnose precursor is a product of the Rml pathway, and that a deletion of rmlC results in a defect in Psl production. Taken together, the results indicate that one enzyme for Psl precursor production (PslB) is encoded on the Psl operon, while the other enzymes overlap with other polysaccharide biosynthetic pathways.
Four enzymes with GT domains are encoded on the Psl operon (PslF, PslH, PslI, and PslC). Three of these, PslF, PslH, and PslI, are predicted to belong to the CAZy GT-4 family of enzymes that transfer glycosyl residues by a retaining mechanism and adopt a GT-B fold (Coutinho and Henrissat, 1999). Structural homology predictions using Phyre2 (Kelley and Sternberg, 2009) suggest that PslF, PslH, and PslI all have structural relatedness to the GT-4 enzyme, PimA, a phosphatidylinositol mannosyltransferase from Mycobacteria (PDB ID:2GEK; Guerin et al., 2007). Therefore, these three enzymes may be responsible for the incorporation of the activated sugar subunits into the Psl repeating structure. The remaining soluble GT, PslC, is predicted to belong to the CAZy GT-2 family, which adopt a GT-A fold and catalyze glycosyl transfer by an inverting mechanism (Coutinho et al., 2003). GT-A folded enzymes have a single Rossmann-like fold for nucleotide binding and also have a characteristic Asp-Xxx-Asp (DXD) or equivalent motif (EAD or TDD) that is responsible for coordination of the phosphates of the nucleotide donor via a divalent cation (Coutinho and Henrissat, 1999). The Phyre2 (Kelley and Sternberg, 2009) structural homology predictions for PslC suggest that it is similar to the murine polypeptide α-N-acetylgalactosaminyltransferase-T1 (PDB ID:1XHB; Fritz et al., 2004) and to a putative Bacteriodes fragilis glycosyltransferase (PDB ID:3BCV). However, unlike these proteins, PslC lacks a clear DXD motif, but does possess an EXD sequence that may act as a substitute. Enzymes in the GT-2 family transfer a wide variety of nucleotide activated sugars (including glucose, rhamnose, and mannose), so it is difficult to speculate which of these substrates PslC specifically recognizes. Based on the predicted structures of PslF, PslH, PslI, and PslC, these enzymes likely participate in the transfer of the precursor sugars from activated nucleotide donors to form the repeating unit of the Psl polysaccharide, which is then recognized by the membrane-associated Psl biosynthetic complex.
Psl Polymerization
Five Psl proteins (PslA/PslE/PslJ/PslK/PslL) have inner membrane spanning domains and therefore likely make up the Psl polymerization complex. Two of these proteins, PslA and PslE, have structural similarities to the Wzy-dependent capsule synthesis pathway (Whitfield, 2006). PslA is predicted to contain four TM domains at its N-terminus and one at the C-terminus. Between these transmembrane segments is a cytoplasmically located region that is predicted to have an EPS biosynthesis polyprenyl glycosylphosphotransferase activity and a Rossmann fold. PslA likely plays a similar role to WbaP in providing a site for the assembly of the oligosaccharide repeating unit onto the isoprenoid lipid at the cytoplasmic face of the inner membrane (Whitfield, 2006). PslE has characteristic domains of a Wzz (or Wzc) homolog and is therefore predicted to act as the PCP component in this system. PslE, like other PCPs, possesses two TM domains that flank a large periplasmic domain and a cytoplasmically located C-terminal tail. The large periplasmic domain is predicted to adopt a coiled-coil structure, similar to that seen for Wzz where monomers assemble into a hexameric (possibly octameric) structure with a central pore (Larue et al., 2009). This periplasmic domain is proposed to affect polysaccharide chain length (Tocilj et al., 2008) and is thought to form critical interactions with the CPS/EPS export component (PslD) thereby completing a complex that facilitates transfer of the polymer through the periplasm (Cuthbertson et al., 2009). The C-terminal cytoplasmic domain of PslE has structural similarity to Wzc, another subclass of PCP proteins. Wzc homologs are related to Wzz, but are distinguished by the presence of a C-terminally located tyrosine autokinase domain (Grangeasse et al., 2007). Interestingly, Wzc contains 6 tyrosines in the final 17 amino acids of its C-terminus, while PslE contains only one (Y658). In the case of Wzc, the phosphorylation of all six tyrosines contributes to the export of CPS and no single tyrosine was found to have a dominant phenotype. Thus, it will be interesting to see if the phophorylation of the single tyrosine in PslE affects this process.
The function of the other three inner membrane proteins (PslJ/PslK/PslL) is more difficult to predict. Each of these has extensive membrane topology throughout the protein, with PslJ predicted to have 11 TM domains, PslK, 12 TM domains, and PslL, 11 TM domains. No structural homologs are apparent for PslJ, but sequence similarity suggests that it may have an O-antigen ligase motif (Pfam 04932) that is also found in Wzy.
Similar to PelG, PslK resembles NorM from V. cholerae (PDB ID:3MKU), which is a member of the MATE family (He et al., 2010). Given that group 1 CPS/EPS biosynthesis typically couples Wzy and Wzz proteins to a flippase (Wzx), it is possible that PslJ fills the role of Wzy in the Psl system, and PslK acts as the flippase. However, this remains to be determined. Sequence similarity of PslL suggests that it has an Acyltransferase_3 (Pfam 1757) domain. Since the Psl polysaccharide is not known to be modified by small functional groups it is difficult to speculate what the role of PslL is in the Psl biosynthetic system. However, it is not unreasonable to hypothesize that a modification could be lost during the extraction and analysis of the structure of Psl.
Psl Translocation and Export
PslD is a periplasmic (and possibly outer membrane) protein with structural similarity to the E. coli K30 capsule translocase, Wza (PDB ID:2J58; Dong et al., 2006). Wza belongs to the OPX family of proteins that are responsible for the final stage of CPS/EPS export (Cuthbertson et al., 2009). Based on the structure of Wza, the OPX family of proteins are predicted to be lipoproteins that adopt an octameric configuration with a large central cavity that facilitates CPS export through the periplasm and across the outer membrane. The large periplasmic domain of Wza forms three discrete rings layered on top of each other, which extend the protein toward the inner membrane and thereby facilitate interaction with Wzc to form a molecular scaffold that spans the periplasm. The extreme C-terminus of Wza folds into a final domain that has a novel α-helical barrel structure that forms a pore in the outer membrane (Dong et al., 2006). While most of PslD can be structurally modeled onto Wza, there are some clear differences. PslD possess only two of the three discrete ring domains of Wza and appears to lack the α-helical barrel domain located in the outer membrane. Therefore, it is difficult to understand how the Psl polymer would pass through the outer membrane if the pore is lacking and if there is enough protein present to form a continuous structure with PslE (the Wzc homolog) in order to span the periplasm. One hypothesis is that this may indicate a requirement for additional periplasmic components. Alternatively, the answer may lie in the inherent sequence differences between Wza/Wzc and PslD/PslE. For example, the C-terminal 49 amino acids of PslD that can not be modeled using Wza may still come together in a manner that facilitates the formation of an outer membrane pore. As for the shorter periplasmic portion of PslD, it is interesting to note that the periplasmic domain of PslE is 49 amino acids larger than the same domain of Wzc, so perhaps this provides the additional periplasmic coverage that is needed. These hypotheses will require further experimental evidence.
Structural homology modeling with Phyre2 (Kelley and Sternberg, 2009) suggests that the periplasmic protein PslG, most closely resembles β-D-xylosidases (e.g., PDB ID:1UHV) from the CAZy glycosyl hydrolase family 39 (Cantarel et al., 2009). These enzymes have a (β/α)8 TIM barrel fold that typically consists of an inner ring of eight β-strands surrounded by an outer ring of eight α-helices and a deep active site cleft along the top of the barrel. Since these enzymes hydrolyze glycosidic linkages, it is likely that PslG performs a similar function to AlgL and PelA from the alginate and Pel systems, respectively. Thus, PslG may be responsible for processing the growing polysaccharide for export and/or act as a failsafe enzyme that rids the periplasm of aberrant polymer when the export process goes awry. Indirect evidence for the second function can be implied from wza or wzc mutants that produce short oligosaccharides of CPS instead of the large molecules formed during proper CPS export (Drummelsmith and Whitfield, 1999).
Summary and Future Perspectives
Pseudomonas aeruginosa has the genetic capacity to produce three different secreted extracellular polysaccharides. The biosynthetic mechanism of alginate has been well studied. However, little has been determined experimentally for Psl and Pel biosynthesis. With the ability to predict domain structures of the Psl and Pel biosynthetic proteins, it was possible to construct models for the synthesis of these secreted extracellular polysaccharides (Figures 4–6). The analyses suggest that P. aeruginosa extracellular polysaccharides are synthesized using two general biosynthetic strategies, an isoprenoid lipid carrier dependent mechanism (for Psl) and a lipid carrier independent mechanism (for alginate and likely for Pel). The evidence for a lipid carrier dependent mechanism for Psl include the presence of several proteins with structural similarity to the EPS/CPS Wzy-dependent pathway of E. coli capsule biosynthesis, and the presence of a repeating oligosaccharide structure, that is characteristic of polysaccharides that are first assembled in association with the isoprenoid lipid. The lipid independent mechanism for alginate and possibly for Pel is based on structural similarities of proteins for these two biosynthetic complexes to enzymes involved in bacterial cellulose and Pga biosynthesis (Romling, 2002; Wang et al., 2004) thereby forming a new class of exopolysaccharide biosynthesis/export systems. Hallmarks of this new class are found in both the Pel and alginate biosynthetic pathways. First, both pathways contain a c-di-GMP binding protein that is essential for polysaccharide polymerization. Second, they both appear to utilize TPR-like proteins to form the biosynthetic scaffolds. Finally, the presence of only one glycosyltransferase on the Pel operon, like alginate, suggests that these polysaccharides are first synthesized as a homopolymer. While the models shown for Pel and Psl (Figures 5 and 6) are based primarily on protein structural predictions, they provide a framework and starting point for testing experimentally the structure and function of these biosynthetic proteins and pathways.
Pseudomonas aeruginosa encoding three different polysaccharide biosynthetic operons raises the following questions: (1) Why maintain the genetic potential to synthesize several different polysaccharides? (2) Do environmental signals induce production of one polysaccharide compared to another? and (3) What survival advantages are imparted by the different polysaccharides under different conditions? The structure of a polysaccharide is important for its function, and production of several structurally different polysaccharides may allow cells to adapt to certain environments. Alginate, Pel, and Psl have all been shown to provide cohesive and adhesive properties that allow cells to form pellicles, microcolonies, or biofilms (Nivens et al., 2001; Ma et al., 2009; Colvin et al., 2011). However, the proper structure of the polysaccharide is likely required for formation of these structured communities. For example, a mutant strain that is impaired in the O-acetylation of alginate was unable to form biofilms (Nivens et al., 2001), the predominant phenotype exhibited by strains isolated from chronic disease in the CF lung.
Switching between production of different P. aeruginosa polysaccharides may be induced by specific stress responses (Wood et al., 2006). P. aeruginosa strains in the CF lung are initially derived from environmental reservoirs (Burns et al., 2001). Therefore, these strains likely produce Pel or Psl. Perhaps due to the selective pressures of the CF lung, P. aeruginosa develops alternate phenotypes, including mucoidy associated with alginate production (Lyczak et al., 2002). Alginate production by pulmonary isolates helps protect the cells from host defensive processes, including opsonic phagocytosis (Pier et al., 2001). Interestingly, regulation of both alginate and Psl require alternative sigma factors that are generally associated with stress responses – σE for alginate and σS for Psl (Flynn and Ohman, 1988; Martin et al., 1994; Ramsey and Wozniak, 2005; Irie et al., 2010).
The production of extracellular polymers may temper conditions found in inhospitable environments. Alginate has been shown to play a role in protecting cells from desiccation (Berry et al., 1989; DeVault et al., 1989). Characterization of P. aeruginosa by AFM shows that when an alginate producing strain of P. aeruginosa is transferred from an aqueous environment to a high humidity condition, the strain maintains its shape (Figure 7A). In contrast, when an algD null mutant strain is transferred to equivalent dehydrating conditions, the cells show a collapsed structure (Figure 7B), suggesting that the extracellular polysaccharide may allow cell survival under certain inhospitable conditions. Having the potential to secrete three different polymers with varying hydrophobicity, viscoelastic properties, and charge may provide P. aeruginosa with the flexibility to adjust the physicochemical nature of its microenvironment, and survive in different niches.
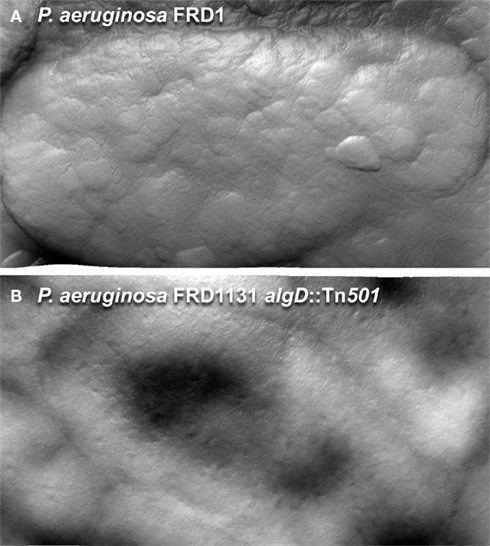
Figure 7. Atomic force microscopy images of hydrated P. aeruginosa FRD1 and FRD11131 algD::Tn501 biofilm cells, collected at 90% humidity. (A) When living at an air/water interface, FRD1 is able to maintain its shape during dehydration. (B) FRD1131 biofilm cells dry out quickly and collapses under the same conditions. Once dry, the extracellular polymer is removed from FRD1 by imaging, but the surface of the FRD1 cells is distinct from FRD1131.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Peg Dirckx for her work with the graphics, Kerry Williamson and Betsey Pitts for generating the TEM and CSLM images, John Whitney, Dustin Little, Laura Riley, Francis Wolfram, Carrie Keiski, Mirela Neculai, and Yura Lobsanov for help with the bioinformatics analyses, and countless discussions that have lead to the generation of these models. This work was supported by grants from the Canadian Institutes of Health Research (CIHR) to P. Lynne Howell (nos. 13337 and 43998); Public Health Service Grant AI065906 to Michael J. Franklin. P. Lynne Howell and Joel T. Weadge are the recipients of a Canada Research Chair, and a Natural Sciences and Engineering Research Council of Canada postdoctoral fellowship, respectively.
References
Agladze, K., Jackson, D., and Romeo, T. (2003). Periodicity of cell attachment patterns during Escherichia coli biofilm development. J. Bacteriol. 185, 5632–5638.
Bechet, E., Gruszczyk, J., Terreux, R., Gueguen-Chaignon, V., Vigouroux, A., Obadia, B., Cozzone, A. J., Nessler, S., and Grangeasse, C. (2010). Identification of structural and molecular determinants of the tyrosine-kinase Wzc and implications in capsular polysaccharide export. Mol. Microbiol. 77, 1315–1325.
Berry, A., Devault, J. D., and Chakrabarty, A. M. (1989). High osmolarity is a signal for enhanced algD transcription in mucoid and non-mucoid Pseudomonas aeruginosa strains. J. Bacteriol. 171, 2312–2317.
Blatch, G. L., and Lassle, M. (1999). The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21, 932–939.
Boles, B. R., Thoendel, M., and Singh, P. K. (2004). Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. U.S.A. 101, 16630–16635.
Bonsor, D. A., Grishkovskaya, I., Dodson, E. J., and Kleanthous, C. (2007). Molecular mimicry enables competitive recruitment by a natively disordered protein. J. Am. Chem. Soc. 129, 4800–4807.
Boyd, A., Ghosh, M., May, T. B., Shinabarger, D., Keogh, R., and Chakrabarty, A. M. (1993). Sequence of the algL gene of Pseudomonas aeruginosa and purification of its alginate lyase product. Gene 131, 1–8.
Burns, J. L., Gibson, R. L., Mcnamara, S., Yim, D., Emerson, J., Rosenfeld, M., Hiatt, P., Mccoy, K., Castile, R., Smith, A. L., and Ramsey, B. W. (2001). Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 183, 444–452.
Byrd, M. S., Sadovskaya, I., Vinogradov, E., Lu, H., Sprinkle, A. B., Richardson, S. H., Ma, L., Ralston, B., Parsek, M. R., Anderson, E. M., Lam, J. S., and Wozniak, D. J. (2009). Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 73, 622–638.
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238.
Chitnis, C. E., and Ohman, D. E. (1990). Cloning of Pseudomonas aeruginosa algG, which controls alginate structure. J. Bacteriol. 172, 2894–2900.
Chua, T. K., Bujnicki, J. M., Tan, T. C., Huynh, F., Patel, B. K., and Sivaraman, J. (2008). The structure of sucrose phosphate synthase from Halothermothrix orenii reveals its mechanism of action and binding mode. Plant Cell 20, 1059–1072.
Colvin, K. M., Gordon, V. D., Murakami, K., Borlee, B. R., Wozniak, D. J., Wong, G. C., and Parsek, M. R. (2011). The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7, e1001264. doi: 10.1371/journal.ppat.1001264
Coulon, C., Vinogradov, E., Filloux, A., and Sadovskaya, I. (2010). Chemical analysis of cellular and extracellular carbohydrates of a biofilm-forming strain Pseudomonas aeruginosa PA14. PLoS ONE 5, e14220. doi: 10.1371/journal.pone.0014220
Coutinho, P. M., Deleury, E., Davies, G. J., and Henrissat, B. (2003). An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328, 307–317.
Coutinho, P. M., and Henrissat, B. (1999). Carbohydrate-Active Enzymes. Avialable at: http://afmb.cnrs-mrs.fr/(cazy/CAZY/index.html
Coyne, M. J. Jr., Russell, K. S., Coyle, C. L., and Goldberg, J. B. (1994). The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. J. Bacteriol. 176, 3500–3507.
Cuthbertson, L., Mainprize, I. L., Naismith, J. H., and Whitfield, C. (2009). Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in gram-negative bacteria. Microbiol. Mol. Biol. Rev. 73, 155–177.
D’Andrea, L. D., and Regan, L. (2003). TPR proteins: the versatile helix. Trends Biochem. Sci. 28, 655–662.
Darzins, A., Frantz, B., Vanags, R. I., and Chakrabarty, A. M. (1986). Nucleotide sequence analysis of the phosphomannose isomerase gene (pmi) of Pseudomonas aeruginosa and comparison with the corresponding Escherichia coli gene manA. Gene 42, 293–302.
De Angelis, F., Lee, J. K., O’connell, J. D. III, Miercke, L. J., Verschueren, K. H., Srinivasan, V., Bauvois, C., Govaerts, C., Robbins, R. A., Ruysschaert, J. M., Stroud, R. M., and Vandenbussche, G. (2010). Metal-induced conformational changes in ZneB suggest an active role of membrane fusion proteins in efflux resistance systems. Proc. Natl. Acad. Sci. U.S.A. 107, 11038–11043.
Deretic, V., Gill, J. F., and Chakrabarty, A. M. (1987). Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J. Bacteriol. 169, 351–358.
DeVault, J. D., Berry, A., Misra, T. K., Darzins, A., and Chakrabarty, A. M. (1989). Environmental sensory signals and microbial pathogenesis: Pseudomonas aeruginosa infection in cystic fibrosis. Biotechnology (N.Y.) 7, 352–357.
DeVault, J. D., Kimbara, K., and Chakrabarty, A. M. (1990). Pulmonary dehydration and infection in cystic fibrosis: evidence that ethanol activates gene expression and induction of mucoidy in Pseudomonas aeruginosa. Mol. Microbiol. 4, 737–745.
Dong, C., Beis, K., Nesper, J., Brunkan-Lamontagne, A. L., Clarke, B. R., Whitfield, C., and Naismith, J. H. (2006). Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature 444, 226–229.
Douthit, S. A., Dlakic, M., Ohman, D. E., and Franklin, M. J. (2005). Epimerase active domain of Pseudomonas aeruginosa AlgG, a protein that contains a right-handed beta-helix. J. Bacteriol. 187, 4573–4583.
Drummelsmith, J., and Whitfield, C. (1999). Gene products required for surface expression of the capsular form of the group 1 K antigen in Escherichia coli (O9a:K30). Mol. Microbiol. 31, 1321–1332.
Evans, L. R., and Linker, A. (1973). Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J. Bacteriol. 116, 915–924.
Flynn, J. L., and Ohman, D. E. (1988). Cloning of genes from mucoid Pseudomonas aeruginosa which control spontaneous conversion to the alginate production phenotype. J. Bacteriol. 170, 1452–1460.
Franklin, M. J., Chitnis, C. E., Gacesa, P., Sonesson, A., White, D. C., and Ohman, D. E. (1994). Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J. Bacteriol. 176, 1821–1830.
Franklin, M. J., Douthit, S. A., and Mcclure, M. A. (2004). Evidence that the algI/algJ gene cassette, required for O acetylation of Pseudomonas aeruginosa alginate, evolved by lateral gene transfer. J. Bacteriol. 186, 4759–4773.
Franklin, M. J., and Ohman, D. E. (1993). Identification of algF in the alginate biosynthetic gene cluster of Pseudomonas aeruginosa which is required for alginate acetylation. J. Bacteriol. 175, 5057–5065.
Franklin, M. J., and Ohman, D. E. (2002). Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 184, 3000–3007.
Friedman, L., and Kolter, R. (2004a). Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690.
Friedman, L., and Kolter, R. (2004b). Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186, 4457–4465.
Fritz, T. A., Hurley, J. H., Trinh, L. B., Shiloach, J., and Tabak, L. A. (2004). The beginnings of mucin biosynthesis: the crystal structure of UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferase-T1. Proc. Natl. Acad. Sci. U.S.A. 101, 15307–15312.
Goldberg, J. B., Hatano, K., and Pier, G. B. (1993). Synthesis of lipopolysaccharide O side chains by Pseudomonas aeruginosa PAO1 requires the enzyme phosphomannomutase. J. Bacteriol. 175, 1605–1611.
Govan, J. R., and Deretic, V. (1996). Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60, 539–574.
Grangeasse, C., Cozzone, A. J., Deutscher, J., and Mijakovic, I. (2007). Tyrosine phosphorylation: an emerging regulatory device of bacterial physiology. Trends Biochem. Sci. 32, 86–94.
Guerin, M. E., Kordulakova, J., Schaeffer, F., Svetlikova, Z., Buschiazzo, A., Giganti, D., Gicquel, B., Mikusova, K., Jackson, M., and Alzari, P. M. (2007). Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria. J. Biol. Chem. 282, 20705–20714.
Hay, I. D., Rehman, Z. U., Ghafoor, A., and Rehm, B. H. (2009). Bacterial biosynthesis of alginates. J. Chem. Technol. Biotechnol. 85, 752–759.
Hay, I. D., Rehman, Z. U., and Rehm, B. H. (2010). Membrane topology of outer membrane protein AlgE, which is required for alginate production in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 76, 1806–1812.
He, X., Szewczyk, P., Karyakin, A., Evin, M., Hong, W. X., Zhang, Q., and Chang, G. (2010). Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 467, 991–994.
Heaton, M. P., and Neuhaus, F. C. (1992). Biosynthesis of D-alanyl-lipoteichoic acid: cloning, nucleotide sequence, and expression of the Lactobacillus casei gene for the D-alanine-activating enzyme. J. Bacteriol. 174, 4707–4717.
Hendrickson, E. L., Plotnikova, J., Mahajan-Miklos, S., Rahme, L. G., and Ausubel, F. M. (2001). Differential roles of the Pseudomonas aeruginosa PA14 rpoN gene in pathogenicity in plants, nematodes, insects, and mice. J. Bacteriol. 183, 7126–7134.
Irie, Y., Starkey, M., Edwards, A. N., Wozniak, D. J., Romeo, T., and Parsek, M. R. (2010). Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 78, 158–172.
Jackson, K. D., Starkey, M., Kremer, S., Parsek, M. R., and Wozniak, D. J. (2004). Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186, 4466–4475.
Jain, S., Franklin, M. J., Ertesvag, H., Valla, S., and Ohman, D. E. (2003). The dual roles of AlgG in C-5-epimerization and secretion of alginate polymers in Pseudomonas aeruginosa. Mol. Microbiol. 47, 1123–1133.
Jain, S., and Ohman, D. E. (1998). Deletion of algK in mucoid Pseudomonas aeruginosa blocks alginate polymer formation and results in uronic acid secretion. J. Bacteriol. 180, 634–641.
Jain, S., and Ohman, D. E. (2005). Role of an alginate lyase for alginate transport in mucoid Pseudomonas aeruginosa. Infect. Immun. 73, 6429–6436.
Jenkins, J., Mayans, O., Smith, D., Worboys, K., and Pickersgill, R. W. (2001). Three-dimensional structure of Erwinia chrysanthemi pectin methylesterase reveals a novel esterase active site. J. Mol. Biol. 305, 951–960.
Jenkins, J., and Pickersgill, R. (2001). The architecture of parallel beta-helices and related folds. Prog. Biophys. Mol. Biol. 77, 111–175.
Keiski, C. L., Harwich, M., Jain, S., Neculai, A. M., Yip, P., Robinson, H., Whitney, J. C., Riley, L., Burrows, L. L., Ohman, D. E., and Howell, P. L. (2010). AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin. Structure 18, 265–273.
Kelley, L. A., and Sternberg, M. J. (2009). Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371.
King, J. D., Kocincova, D., Westman, E. L., and Lam, J. S. (2009). Review: lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun. 15, 261–312.
Kintz, E., and Goldberg, J. B. (2008). Regulation of lipopolysaccharide O antigen expression in Pseudomonas aeruginosa. Future Microbiol. 3, 191–203.
Kowalska, K., Soscia, C., Combe, H., Vasseur, P., Voulhoux, R., and Filloux, A. (2010). The C-terminal amphipathic alpha-helix of Pseudomonas aeruginosa PelC outer membrane protein is required for its function. Biochimie 92, 33–40.
Krogh, A., Larsson, B., Von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580.
Larue, K., Kimber, M. S., Ford, R., and Whitfield, C. (2009). Biochemical and structural analysis of bacterial O-antigen chain length regulator proteins reveals a conserved quaternary structure. J. Biol. Chem. 284, 7395–7403.
Lee, V. T., Matewish, J. M., Kessler, J. L., Hyodo, M., Hayakawa, Y., and Lory, S. (2007). A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65, 1474–1484.
Li, J., Qian, X., Hu, J., and Sha, B. (2009). Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J. Biol. Chem. 284, 23852–23859.
Linker, A., and Jones, R. S. (1964). A polysaccharide resembling alginic acid from a Pseudomonas micro-organism. Nature 204, 187–188.
Lyczak, J. B., Cannon, C. L., and Pier, G. B. (2000). Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2, 1051–1060.
Lyczak, J. B., Cannon, C. L., and Pier, G. B. (2002). Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15, 194–222.
Ma, L., Conover, M., Lu, H., Parsek, M. R., Bayles, K., and Wozniak, D. J. (2009). Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 5, e1000354. doi: 10.1371/journal.ppat.1000354
Ma, L., Jackson, K. D., Landry, R. M., Parsek, M. R., and Wozniak, D. J. (2006). Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 188, 8213–8221.
Maharaj, R., May, T. B., Wang, S. K., and Chakrabarty, A. M. (1993). Sequence of the alg8 and alg44 genes involved in the synthesis of alginate by Pseudomonas aeruginosa. Gene 136, 267–269.
Martin, D. W., Schurr, M. J., Yu, H., and Deretic, V. (1994). Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to sigma E and stress response. J. Bacteriol. 176, 6688–6696.
Merighi, M., Lee, V. T., Hyodo, M., Hayakawa, Y., and Lory, S. (2007). The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65, 876–895.
Molgaard, A., and Larsen, S. (2002). A branched N-linked glycan at atomic resolution in the 1.12 A structure of rhamnogalacturonan acetylesterase. Acta Crystallogr. D Biol. Crystallogr. 58, 111–119.
Nivens, D. E., Ohman, D. E., Williams, J., and Franklin, M. J. (2001). Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183, 1047–1057.
Oglesby, L. L., Jain, S., and Ohman, D. E. (2008). Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology 154, 1605–1615.
Ohman, D. E. (1986). Molecular genetics of exopolysaccharide production by mucoid Pseudomonas aeruginosa. J. Clin. Microbiol. 5, 6–10.
Olvera, C., Goldberg, J. B., Sanchez, R., and Soberon-Chavez, G. (1999). The Pseudomonas aeruginosa algC gene product participates in rhamnolipid biosynthesis. FEMS Microbiol. Lett. 179, 85–90.
Oomen, C. J., Van Ulsen, P., Van Gelder, P., Feijen, M., Tommassen, J., and Gros, P. (2004). Structure of the translocator domain of a bacterial autotransporter. EMBO J. 23, 1257–1266.
Pelissier, M. C., Lesley, S. A., Kuhn, P., and Bourne, Y. (2010). Structural insights into the catalytic mechanism of bacterial guanosine-diphospho-D-mannose pyrophosphorylase and its regulation by divalent ions. J. Biol. Chem. 285, 27468–27476.
Pier, G. B., Coleman, F., Grout, M., Franklin, M., and Ohman, D. E. (2001). Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect. Immun. 69, 1895–1901.
Ramelot, T. A., Yee, A., Cort, J. R., Semesi, A., Arrowsmith, C. H., and Kennedy, M. A. (2007). NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein. Proteins 66, 266–271.
Ramsey, D. M., and Wozniak, D. J. (2005). Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 56, 309–322.
Regni, C., Tipton, P. A., and Beamer, L. J. (2002). Crystal structure of PMM/PGM: an enzyme in the biosynthetic pathway of P. aeruginosa virulence factors. Structure 10, 269–279.
Rehm, B. H., Boheim, G., Tommassen, J., and Winkler, U. K. (1994). Overexpression of algE in Escherichia coli: subcellular localization, purification, and ion channel properties. J. Bacteriol. 176, 5639–5647.
Remminghorst, U., and Rehm, B. H. (2006a). Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett. 580, 3883–3888.
Remminghorst, U., and Rehm, B. H. (2006b). In vitro alginate polymerization and the functional role of Alg8 in alginate production by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 72, 298–305.
Robles-Price, A., Wong, T. Y., Sletta, H., Valla, S., and Schiller, N. L. (2004). AlgX is a periplasmic protein required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 186, 7369–7377.
Romling, U. (2002). Molecular biology of cellulose production in bacteria. Res. Microbiol. 153, 205–212.
Rozeboom, H. J., Bjerkan, T. M., Kalk, K. H., Ertesvag, H., Holtan, S., Aachmann, F. L., Valla, S., and Dijkstra, B. W. (2008). Structural and mutational characterization of the catalytic A-module of the mannuronan C-5-epimerase AlgE4 from Azotobacter vinelandii. J. Biol. Chem. 283, 23819–23828.
Saxena, I. M., and Brown, R. M. Jr. (2005). Cellulose biosynthesis: current views and evolving concepts. Ann. Bot. 96, 9–21.
Schiller, N. L., Monday, S. R., Boyd, C. M., Keen, N. T., and Ohman, D. E. (1993). Characterization of the Pseudomonas aeruginosa alginate lyase gene (algL): cloning, sequencing, and expression in Escherichia coli. J. Bacteriol. 175, 4780–4789.
Sibley, C. D., Duan, K., Fischer, C., Parkins, M. D., Storey, D. G., Rabin, H. R., and Surette, M. G. (2008). Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 4, e1000184. doi: 10.1371/journal.ppat.1000184
Silo-Suh, L., Suh, S. J., Sokol, P. A., and Ohman, D. E. (2002). A simple alfalfa seedling infection model for Pseudomonas aeruginosa strains associated with cystic fibrosis shows AlgT (sigma-22) and RhlR contribute to pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 99, 15699–15704.
Snook, C. F., Tipton, P. A., and Beamer, L. J. (2003). Crystal structure of GDP-mannose dehydrogenase: a key enzyme of alginate biosynthesis in P. aeruginosa. Biochemistry 42, 4658–4668.
Spiers, A., Kahn, S., Bohannon, J., Travisano, M., and Rainey, P. (2002). Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics 16, 33–46.
Starkey, M., Hickman, J. H., Ma, L., Zhang, N., De Long, S., Hinz, A., Palacios, S., Manoil, C., Kirisits, M. J., Starner, T. D., Wozniak, D. J., Harwood, C. S., and Parsek, M. R. (2009). Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J. Bacteriol. 191, 3492–3503.
Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., Brinkman, F. S., Hufnagle, W. O., Kowalik, D. J., Lagrou, M., Garber, R. L., Goltry, L., Tolentino, E., Westbrock-Wadman, S., Yuan, Y., Brody, L. L., Coulter, S. N., Folger, K. R., Kas, A., Larbig, K., Lim, R., Smith, K., Spencer, D., Wong, G. K., Wu, Z., Paulsen, I. T., Reizer, J., Saier, M. H., Hancock, R. E., Lory, S., and Olson, M. V. (2000). Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406, 959–964.
Tan, M. W., Rahme, L. G., Sternberg, J. A., Tompkins, R. G., and Ausubel, F. M. (1999). Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. U.S.A. 96, 2408–2413.
Tocilj, A., Munger, C., Proteau, A., Morona, R., Purins, L., Ajamian, E., Wagner, J., Papadopoulos, M., Van Den Bosch, L., Rubinstein, J. L., Fethiere, J., Matte, A., and Cygler, M. (2008). Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat. Struct. Mol. Biol. 15, 130–138.
Ueda, A., and Wood, T. K. (2009). Connecting quorum sensing, c-di-GMP, Pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog. 5, e1000483. doi: 10.1371/journal.ppat.1000483
Urch, J. E., Hurtado-Guerrero, R., Brosson, D., Liu, Z., Eijsink, V. G., Texier, C., and Van Aalten, D. M. (2009). Structural and functional characterization of a putative polysaccharide deacetylase of the human parasite Encephalitozoon cuniculi. Protein Sci. 18, 1197–1209.
Vasseur, P., Soscia, C., Voulhoux, R., and Filloux, A. (2007). PelC is a Pseudomonas aeruginosa outer membrane lipoprotein of the OMA family of proteins involved in exopolysaccharide transport. Biochimie 89, 903–915.
Wang, H., Robinson, H., and Ke, H. (2010). Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain. J. Biol. Chem. 285, 38149–38156.
Wang, X., Preston, J. F. III, and Romeo, T. (2004). The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186, 2724–2734.
Weadge, J. T., Yip, P. P., Robinson, H., Arnett, K., Tipton, P. A., and Howell, P. L. (2010). Expression, purification, crystallization and preliminary X-ray analysis of Pseudomonas aeruginosa AlgX. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 588–591.
Whitfield, C. (2006). Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75, 39–68.
Whitney, J. C., Hay, I. D., Li, C., Eckford, P. D. W., Robinson, H., Amaya, M. F., Wood, L. F., Ohman, D. E., Bear, C. E., Rehm, B. H., and Howell, P. L. (2011). Structural basis for alginate secretion across the bacterial outer membrane. Proc. Natl. Acad. Sci. U.S.A. 108, 13083�13088.
Whitney, J. C., Neculai, A. M., Ohman, D. E., and Howell, P. L. (2009). Expression, refolding, crystallization and preliminary X-ray analysis of Pseudomonas aeruginosa AlgE. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 463–466.
Wood, L. F., Leech, A. J., and Ohman, D. E. (2006). Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: roles of sigma (AlgT) and the AlgW and Prc proteases. Mol. Microbiol. 62, 412–426.
Yang, J. K., Yoon, H. J., Ahn, H. J., Lee, B. I., Pedelacq, J. D., Liong, E. C., Berendzen, J., Laivenieks, M., Vieille, C., Zeikus, G. J., Vocadlo, D. J., Withers, S. G., and Suh, S. W. (2004). Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase. J. Mol. Biol. 335, 155–165.
Yoon, H. J., Mikami, B., Hashimoto, W., and Murata, K. (1999). Crystal structure of alginate lyase A1-III from Sphingomonas species A1 at 1.78 A resolution. J. Mol. Biol. 290, 505–514.
Zhang, Z., Kulkarni, K., Hanrahan, S. J., Thompson, A. J., and Barford, D. (2010). The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homo-dimer interface similar to Cdc27. EMBO J. 29, 3733–3744.
Keywords: Pseudomonas aeruginosa, alginate, Psl polysaccharide, Pel polysaccharide, Rossmann fold, glycosyltransferase
Citation: Franklin MJ, Nivens DE, Weadge JT and Howell PL (2011) Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front. Microbio. 2:167. doi: 10.3389/fmicb.2011.00167
Received: 15 April 2011;
Paper pending published: 31 May 2011;
Accepted: 19 July 2011;
Published online: 22 August 2011.
Edited by:
Dara Frank, Medical College of Wisconsin, USAReviewed by:
Daniel Wozniak, The Ohio State University, USAChris Whitfield, University of Guelph, Canada
Copyright: © 2011 Franklin, Nivens, Weadge and Howell. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Michael J. Franklin, Department of Microbiology, Montana State University, 109 Lewis Hall, Bozeman, MT 59717, USA. e-mail: franklin@montana.edu; P. Lynne Howell, Molecular Structure and Function, The Hospital for Sick Children, 555 University, 16 Avenue, Toronto, ON, Canada M5G 1X8. e-mail: howell@sickkids.ca