An Assessment of LRRK2 Serine 935 Phosphorylation in Human Peripheral Blood Mononuclear Cells in Idiopathic Parkinson’s Disease and G2019S LRRK2 Cohorts
Abstract
The phosphorylated form of LRRK2, pS935 LRRK2, has been proposed as a target modulation biomarker for LRRK2 inhibitors. The primary aim of the study was to characterize and qualify this biomarker for therapeutic trials of LRRK2 inhibitors in Parkinson’s disease (PD). To this end, analytically validated assays were used to monitor levels of pS935 LRRK2 and total LRRK2 in peripheral blood mononuclear cells (PBMCs) from the following donor groups: healthy controls, idiopathic PD, and G2019S carriers with and without PD. Neither analyte correlated with age, gender, or disease severity. While total LRRK2 levels were similar across the four groups, there was a significant reduction in pS935 LRRK2 levels in disease-manifesting G2019S carriers compared to idiopathic PD. In aggregate, these data indicate that phosphorylation of LRRK2 at S935 may reflect a state marker for G2019S LRRK2-driven PD, the underlying biology for which requires investigation in future studies. This study also provides critical foundational data to inform the integration of pS935 and total LRRK2 levels as biomarkers in therapeutic trials of LRRK2 kinase inhibitors.
INTRODUCTION
Missense mutations in the gene encoding leucine-rich repeat kinase 2 (LRRK2) have been associated with autosomal dominant Parkinson’s disease (PD) [1, 2]. The G2019S mutation in the activation loop of LRRK2 is the most common genetic cause of PD [3] and increases LRRK2 kinase activity, which appears to be obligatory for the toxic effects [4]. Thus, discovery and development of LRRK2 kinase inhibitors is an actively pursued therapeutic strategy.
Direct assessment of LRRK2 activity has relied upon autophosphorylation of serine 1292 (S1292) [5] or phosphorylation of its substrates, Rab8 and Rab10 [6]. However, the low stoichiometry of S1292 phosphorylation [5, 7] and non-specificity of Rab phosphorylation by LRRK2 [6] have hampered their utility as pharmacodynamic biomarkers. An alternative robust, but indirect, marker of LRRK2 activity is phosphorylation of serine 935 (pS935). Although the kinase responsible for phosphorylation of S935 remains an area of conjecture [8], all known LRRK2 kinase inhibitors reduce pS935 in cellular and animal studies [9, 10] indicating its usefulness as a pharmacodynamic biomarker of LRRK2 kinase inhibitors. However, since complex biological mechanisms appear to regulate phosphorylation/de-phosphorylation of S935 [11, 12], it is critical to qualify its utility in clinical samples from intended patient populations.
To this end, the Michael J. Fox Foundation for Parkinson’s research (MJFF) established the “LRRK2 Detection in PBMC Consortium”, a precompetitive alliance among MJFF, scientists from five pharmaceutical companies and Columbia University Irving Medical Center (CUIMC). As a prerequisite to join the alliance, each pharmaceutical company presented their in-house assays for pS935 LRRK2 and total LRRK2 developed on a clinically viable platform and characterized for dynamic range, accuracy, precision, test-retest reliability using their preferred peripheral blood mononuclear cell (PBMC) collection protocols. As a first step of this study, a head-to-head comparison of analytical performance of the assays representing three different platforms (Meso Scale Discovery, Quanterix SIMOA single molecule array and CisBio homogeneous time-resolved fluorescence) was performed by each member using PBMCs collected under a standardized protocol at CUIMC. The outcomes of this pilot study led to the selection of the SIMOA assays on the basis of two factors: (a) incompatibility of the PBMC collection protocol with the Meso Scale Discovery platform assays and (b) superiority of the SIMOA platform over CisBio assays regarding sensitivity, dynamic range and test/re-test reliability. However, note that after the start of the current study, one of the consortium members modified their Meso Scale Discovery assay conditions to be able to measure total and pS935 levels in CUIMC-collected PBMCs (Dr. Huntwork-Rodriguez, Denali Therapeutics, personal communication).
This report describes the assessment of total LRRK2 and pS935 LRRK2 in PBMCs collected from healthy controls (PD-G2019S-), idiopathic PD (iPD) (PD+G2019S-) patients and G2019S LRRK2 mutation carriers with (PD+G2019S+) and without PD (PD-G2019S+).
METHODS
Subjects
The CUIMC cohort has been described previously [13]. In brief, healthy controls, iPD patients and LRRK2 G2019S carriers with and without PD were recruited. PD was diagnosed based on the UK PD brain bank criteria [14] except that family history was not an exclusion for recruitment. The study was approved by CUIMC institutional review board, and all participants signed informed consents. Genotyping for G2019S LRRK2 was conducted as previously described [15]. The demographics and clinical summary data on 117 study participants are presented in Table 1.
Table 1
Summary of demographic and clinical characteristics of the PBMC donors. The study cohort consisted of Controls (PD-) and PD subjects (PD+) with (+) and without (–) G2019S LRRK2 mutation. SD, standard deviation; IQR, inter-quartile range; UPDRS, Unified Parkinson’s Disease Rating Scale
| Characteristic | PD-G2019S- | PD-G2019S+ | PD+G2019S- | PD+G2019S+ |
| (n = 22) | (n = 16) | (n = 46) | (n = 33) | |
| Age (years) | ||||
| Mean (SD) | 69.0 (7.1) | 57.9 (11.6) | 64.9 (9.4) | 71.7 (8.9) |
| Range | 57–85 | 37–83 | 41–82 | 56–91 |
| Sex | ||||
| 13 M, 9 F | 6 M, 10 F | 29 M, 17 F | 19 M, 14 F | |
| Disease Duration (Years) | ||||
| Median (IQR) | 7 (2.25–10) | 11 (7–15) | ||
| Range | 0–25 | 0–26 | ||
| Total UPDRS* | ||||
| Mean (SD) | 28.1 (16.4) | 34.4 (18.7) | ||
| Range | 8–94 | 4–81 | ||
| UPDRS Part 3* | ||||
| Mean (SD) | 19.6 (12.3) | 22.2 (11.8) | ||
| Range | 5–64 | 1–53 | ||
| MoCA** | ||||
| Mean (SD) | 27.4 (2.0) | 28.9 (1.1) | 26.6 (3.0) | 26.1 (3.3) |
| Range | 23–30 | 27–30 | 12–30 | 18–30 |
*UPDRS data available for only 64 PD subjects (81%). Total UPDRS includes UPDRS I-III. **MoCA data available for only 58 PD subjects (73%), and missing from 1 PD-G2019S-.
PBMC collection
Whole blood collected during the normal business hours of 9 am to 5 pm in PBS-diluted heparin was transferred into a Ficoll-filled Leucosep tube, centrifuged, upper phase extracted and centrifuged again. CBC data were collected and reviewed to ensure that no samples had abnormal profiles to minimize the variability in the endpoints. The supernatant was then decanted and cells washed. Cell counts were measured by a Countess Automated Cell Counter prior to resuspension and final centrifugation. The cell pellet was stored at –80°C until shipment.
LRRK2 assays
PBMCs were lysed in the following buffer: 50 mM Tris-HCL, 150 mM NaCl, 1% Triton x-100, 2% Glycerol, 10 mM PPA, 20 mM NaF, 2 mM Na3VO4, 2 mM EGTA, and 2 mM EDTA at pH 7.5, with HALT protease and phosphatase inhibitors added fresh. 1 mL of cold lysis buffer was added to frozen aliquots of 3 million cells, and samples were sonicated for 5 s on ice. Lysates were centrifuged at 14,000 x g for 3 min, supernatant was collected and stored at –80°C until day of assay. Samples were randomized on assay plates and blinded to group identity until statistical analysis. Total and pS935 LRRK2 assays developed on the Quanterix Simoa platform [16] used Neuromab N241A/34 for capture and biotinylated detection antibodies (Abcam clone UDD2 for pS935, and Cell Signaling clone D18E12 for total LRRK2). Specificity of the pS935 antibody has been shown previously by depletion of signal following treatment with a LRRK2 inhibitor [17, 18]. Three technical replicates were run, and sample concentrations were interpolated from a standard curve ranging from 9.76 to 40,000 pg/mL of full length recombinant LRRK2 (Life Technologies A15197) in a buffered solution (12 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, 1.25 mM sodium phosphate, 2 mM CaCl2, 25 mM NaHCO3, 25 mM mannose at pH 8.13). While the exact stoichiometry for the phosphorylation state of this recombinant protein has not been extensively characterized, multiple lots tested showed similar performance in both total and pS935 assays. Nonetheless, absolute values generated in this pS935 assay have the caveat that the standard being used is not 100% phosphorylated, so the assay is currently best suited for measuring relative changes. Samples were diluted in the above buffer at 1:10 (total LRRK2) or 1:20 (pS935 LRRK2). The lower limit of detection (defined as 2.5X the standard deviation of the blank) for the assays was 4.1 pg/mL for pS935 (82.8 pg/mL in undiluted lysate) and 12.1 pg/mL for total (121.1 pg/mL in undiluted lysate), and median % CV for standard curves was 5% (pS935) and 19% (total). Assay precision was evaluated using repeat analysis of pilot PBMC samples and spike-in controls. Human PBMC lysates from 6 donors were split into multiple aliquots, as were various concentrations of human recombinant LRRK2 diluted in assay buffer to serve as QC controls. Intra-assay precision was evaluated for each sample across technical replicates within each assay. The samples were run in 7 separate assays, each on a different day, to generate data to test inter-assay variability. Both pS935 and total LRRK2 assays showed robust performance for both intra- and inter-assay precision (Table 2).
Table 2
Precision of the human LRRK2 assays was assessed using 6 pilot PBMC lysate samples and 5 QC samples spiked with recombinant LRRK2 or in vitro phosphorylated LRRK2. Interpolated values normalized to total protein are shown for repeated analysis within the same assay run (intra-assay) or across assays run on separate days (inter-assay) to assess technical variability
| Sample | pS935 LRRK2 | Total LRRK2 | ||||
| pg/mg* | Intra-assay %CV | Inter-assay %CV | pg/mg* | Intra-assay %CV | Inter-assay %CV | |
| Lysate 1 | 40164 | 5.0% | 9.0% | 67422 | 6.0% | 9.0% |
| Lysate 2 | 9734 | 7.0% | 8.0% | 14646 | 15.0% | 17.0% |
| Lysate 3 | 7120 | 8.0% | 8.0% | 14525 | 7.0% | 12.0% |
| Lysate 4 | 29966 | 7.0% | 8.0% | 109675 | 8.0% | 11.0% |
| Lysate 5 | 23133 | 5.0% | 9.0% | 103502 | 5.0% | 15.0% |
| Lysate 6 | 25502 | 4.0% | 10.0% | 62736 | 7.0% | 13.0% |
| pg/mL | Intra-assay %CV | Inter-assay %CV | pg/mL | Intra-assay %CV | Inter-assay %CV | |
| QC 1 | 8669 | 3.0% | 25.0% | 16589 | 7.0% | 21.0% |
| QC 2 | 4035 | 4.0% | 14.0% | 2520 | 6.0% | 19.0% |
| QC 3 | 395 | 3.0% | 8.0% | 390 | 8.0% | 11.0% |
| QC 4 | 135 | 4.0% | 8.0% | 49 | 6.0% | 12.0% |
| QC 5 | 23 | 7.0% | 6.0% | 9 | 15.0% | 18.0% |
*LRRK2 levels were normalized for total protein (pg LRRK2/mg total protein).
Statistical analysis
Data were analyzed by one-way Analysis of variance (ANOVA) using either the two-level factor (all PD versus all controls) or four-level factor (PD+G2019S+, PD+G2019S-, PD-G2019S+, PD-G2019S-) that was applied to each measurement separately. Individual group differences were then assessed using Tukey’s post-hoc tests to account for multiple comparisons within each ANOVA. Sensitivity analysis adjusting for age and sex were conducted using linear regression. Correlation coefficients were based on Pearson correlation coefficients that included all the data.
RESULTS
Total and pS935 LRRK2 levels were first compared for all PD versus all control subjects regardless of G2019S LRRK2 mutation status. While the variances appeared to be higher for the PD subjects (standard deviation = 4,863 vs. 3,084), there were no significant differences between PD and control groups for either total LRRK2 (p = 0.18) or pS935 LRRK2 levels (p = 0.92). When controls and PD subjects were segregated by the G2019S mutation status, there were no difference in total LRRK2 levels among the 4 groups (p = 0.13, Fig. 1A). However, ANOVA indicated a significant group effect for pS935 LRRK2 (p = 0.028), with PD+G2019S+ subjects showing ∼32% lower pS935 LRRK2 levels than PD+G2019S- subjects (p < 0.05, Fig. 1B).
Fig.1
Total and pS935 LRRK2 levels in PBMCs. Box and whisker plots of absolute levels of total LRRK2 and pS935 LRRK2 are shown in panels (A) and (B), respectively. The line through the middle of the boxes corresponds to the median and the lower and the upper lines represent the 25th and 75th percentiles, respectively. The whiskers extend from the 5th percentile on the bottom to the 95th percentile on top. Differences between groups were assessed by ANOVA followed by Tukey’s post-hoc test. *p < 0.05. SD, standard deviation; IQR, inter-quartile range; UPDRS, Unified Parkinson’s Disease Rating Scale.
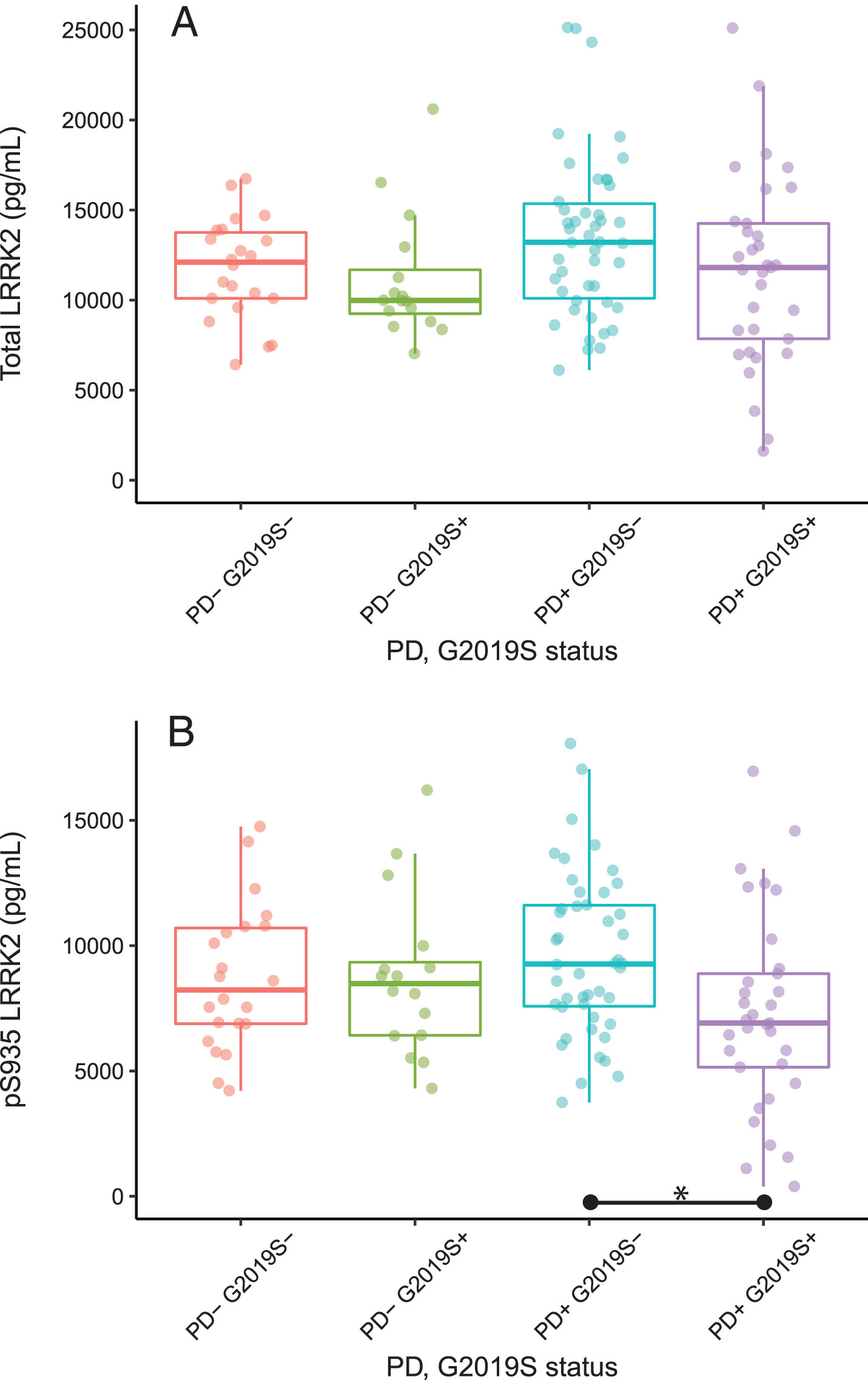
The two LRRK2 measures were compared to Montreal Cognitive Assessment (MoCA) data and UPDRS scores (available for 94% and 83% of subjects, respectively) as well as to the sex of participants, age at donation of the biospecimens and disease duration (where applicable). Although there was a strong correlation between total and pS935 LRRK2 levels (r = 0.89), neither analyte correlated with any demographic or clinical factors (–0.25 < r <0.25, data not shown).
DISCUSSION
In this study, we demonstrate a statistically significant decrease in pS935 LRRK2, but not total LRRK2, levels in human PBMCs from PD subjects with the G2019S LRRK2 mutation compared to iPD subjects, using a sensitive and robust digital immunoassay. Since G2019S mutation results in higher LRRK2 kinase activity [4], the decrease in pS935 LRRK2 levels in carriers with PD was unexpected and indicates the presence of distinct biological mechanisms in LRRK2 G2019S mutation-driven PD compared to iPD or non-manifesting carriers. However, pS935 LRRK2 levels did not correlate with disease severity (based on UPDRS or MoCA), disease duration, age at biospecimen donation, or other demographic factors.
There are three previous reports [19–21] on levels of pS935 and total LRRK2 in PBMCs, but these assessments were conducted using western blotting and restricted to iPD and healthy controls. These studies showed no differences in pS935 or total LRRK2 levels in iPD subjects compared to healthy controls. We have confirmed these findings using a quantitative assay and expanded the investigation to include LRRK2 manifesting and non-manifesting groups. It is noteworthy that the reduction if pS935 LRRK2 levels in PD-manifesting LRRK2 G2019S donors was also observed by a member of the “LRRK2 Detection in PBMC Consortium” using a Meso Scale Discovery assay (Dr. Huntwork-Rodriguez, Denali Therapeutics, personal communication; manuscript in progress). On the other hand, Atashrazm et al. [19] reported higher levels of total LRRK2 levels in neutrophils from iPD cases compared to healthy controls. The apparent discrepancy between our study and [19] may be due to previously reported cell-specific regulation of LRRK2 expression [22].
The S935 residue of LRRK2 is proposed to be a constitutive phosphorylation site that is amenable to regulation by LRRK2 kinase activity through other unidentified kinases and inflammation-related signaling pathways [23]. Interestingly, pathogenic mutations in the GTPase domain of LRRK2, such as the R1441G/C/H, Y1699C, as well as another kinase domain mutation, I2020T, have consistently been associated with reduced phosphorylation of S935, which has been attributed to the disruption in 14-3-3 binding to LRRK2 [24]. However, this effect is spared in G2019S LRRK2 in cellular models as well as in aged G2019S knock-in mice as assessed by immunoblot assays [25]. The current results showing the lack of a reduction in pS935 levels in non-manifesting carriers of G2019S are consistent with these non-clinical findings. On the other hand, our observation of a decrease in p935 levels in the PD-manifesting LRRK2 G2019S compared to non-manifesting carriers indicates that disease manifestation triggers molecular mechanisms that alter the regulation of S935 phosphorylation. Future investigations of such mechanisms are required to garner insights into pathogenic mechanisms of the various LRRK2 mutations.
Although PBMCs represent a relatively simple biospecimen collection method, studies of total and pS935 levels in distinct cell types such as monocytes and neutrophils will be critical since LRRK2 expression is dramatically higher in these cells than in total PBMC population [26] and the cellular heterogeneity of PBMCs may have masked alterations in total or pS935 LRRK2 in the present study. Moreover, in light of the emerging data on LRRK2 expression in specific sub-types of immune cells, it will be critical to demonstrate that the reduction in pS935 LRRK2 levels observed here is not due to differences in the immune cell repertoire between groups as a result of a recent infections or other stresses. Future blood cell collections would also benefit from the inclusion of a questionnaire that captures the immune status of the donors. Additionally, a comparison of pS935 LRRK2 levels with direct markers of LRRK2 kinase activity such as pS1292 LRRK2 or pT73 Rab10 [6] will be important to determine whether despite higher kinase activity of G2019S LRRK2, compensatory biological mechanisms lead to de-phosphorylation of the S935 residue in disease manifesting carriers.
In summary, the data presented here indicate that pS935 LRRK2 level has the potential to be a disease state biomarker of G2019S LRRK2-associated PD but the results need to be replicated and extended in an independent and larger cohort. The decrease in pS935 LRRK2 levels in G2019S-positive PD is indicative of distinct biological mechanism(s) at play in this patient population. Overall, the current study underscores the importance of collaborative consortia for the development and analytical qualification of biomarker assays on standardized human biospecimen collections to enable future studies of not only LRRK2 kinase activity modulation but also spur studies of disease state mechanisms.
CONFLICT OF INTEREST
The following are financial disclosures but none constitute a conflict of interest with the subject matter of this publication. CL, NJ, TCM, NL, SN, SP, SJH, and MB report no conflict of interest. TL, DG, AJ, CC, and RLH are employees of Pfizer Inc. MW is an employee of Quanterix. RA receives consultation fees from ResTORbio, Genzyme/Sanofi, and Roche. KM is paid as the Chief Scientific Officer at Vincere Biosciences, Inc. and receives consulting fees and/or equity from Lysosomal Therapeutics, Sinopia Biosciences and Origami Therapeutics, and the Michael J Fox Foundation for Parkinson’s research.
ACKNOWLEDGMENTS
The Michael J. Fox Foundation would like to thank all the members of the Industry LRRK2 Detection Consortium (Omar Mabrouk from Biogen, Sarah Huntwork-Rodriguez from Denali Therapeutics, Matthew Fell, Julie Lee and Payal Sheth from Merck & Co., Inc., Thomas Lanz from Pfizer Inc., Nathalie Schussler from Sanofi and their respective teams) for their participation and engagement in the Consortium initiative.
SIMOA assay development and testing was funded by Pfizer Inc. Roy N. Alcalay received research funding from the NIH, Parkinson’s Foundation and the Michael J Fox Foundation.
REFERENCES
[1] | Paisan-Ruiz C , Jain S , Evans EW , Gilks WP , Simon J , van der Brug M , Lopez de Munain A , Aparicio S , Gil AM , Khan N , Johnson J , Martinez JR , Nicholl D , Carrera IM , Pena AS , de Silva R , Lees A , Marti-Masso JF , Perez-Tur J , Wood NW , Singleton AB ((2004) ) Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron 44: , 595–600. |
[2] | Zimprich A , Biskup S , Leitner P , Lichtner P , Farrer M , Lincoln S , Kachergus J , Hulihan M , Uitti RJ , Calne DB , Stoessl AJ , Pfeiffer RF , Patenge N , Carbajal IC , Vieregge P , Asmus F , Muller-Myhsok B , Dickson DW , Meitinger T , Strom TM , Wszolek ZK , Gasser T ((2004) ) Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44: , 601–607. |
[3] | Domingo A , Klein C ((2018) ) Genetics of Parkinson disease. Handb Clin Neurol 147: , 211–227. |
[4] | Greggio E , Jain S , Kingsbury A , Bandopadhyay R , Lewis P , Kaganovich A , van der Brug MP , Beilina A , Blackinton J , Thomas KJ , Ahmad R , Miller DW , Kesavapany S , Singleton A , Lees A , Harvey RJ , Harvey K , Cookson MR ((2006) ) Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis 23: , 329–341. |
[5] | Sheng Z , Zhang S , Bustos D , Kleinheinz T , Le Pichon CE , Dominguez SL , Solanoy HO , Drummond J , Zhang X , Ding X , Cai F , Song Q , Li X , Yue Z , van der Brug MP , Burdick DJ , Gunzner-Toste J , Chen H , Liu X , Estrada AA , Sweeney ZK , Scearce-Levie K , Moffat JG , Kirkpatrick DS , Zhu H ((2012) ) Ser1292 autophosphorylation is an indicator of LRRK2 kinase activity and contributes to the cellular effects of PD mutations. Sci Transl Med 4: , 164ra161. |
[6] | Steger M , Tonelli F , Ito G , Davies P , Trost M , Vetter M , Wachter S , Lorentzen E , Duddy G , Wilson S , Baptista MA , Fiske BK , Fell MJ , Morrow JA , Reith AD , Alessi DR , Mann M ((2016) ) Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. Elife 5: , e12813. |
[7] | Christensen KV , Hentzer M , Oppermann FS , Elschenbroich S , Dossang P , Thirstrup K , Egebjerg J , Williamson DS , Smith GP ((2018) ) LRRK2 exonic variants associated with Parkinson’s disease augment phosphorylation levels for LRRK2-Ser1292 and Rab10-Thr73. bioRxiv, doi: 10.1101/447946. |
[8] | Nichols RJ ((2017) ) LRRK2 phosphorylation. Adv Neurobiol 14: , 51–70. |
[9] | Fuji RN , Flagella M , Baca M , Baptista MA , Brodbeck J , Chan BK , Fiske BK , Honigberg L , Jubb AM , Katavolos P , Lee DW , Lewin-Koh SC , Lin T , Liu X , Liu S , Lyssikatos JP , O’Mahony J , Reichelt M , Roose-Girma M , Sheng Z , Sherer T , Smith A , Solon M , Sweeney ZK , Tarrant J , Urkowitz A , Warming S , Yaylaoglu M , Zhang S , Zhu H , Estrada AA , Watts RJ ((2015) ) Effect of selective LRRK2 kinase inhibition on nonhuman primate lung. Sci Transl Med 7: , 273ra215. |
[10] | Baptista M , Merchant K , Barrett T , Bryce D , Ellis M , Estrada A , Fell M , Fiske B , Fuji R , Galatsis P ((2018) ) LRRK2 kinase inhibitors induce a reversible effect in the lungs of non-human primates with no measurable pulmonary deficits. bioRxiv, doi: 10.1101/390815. |
[11] | Dzamko N , Gysbers AM , Bandopadhyay R , Bolliger MF , Uchino A , Zhao Y , Takao M , Wauters S , van de Berg WD , Takahashi-Fujigasaki J , Nichols RJ , Holton JL , Murayama S , Halliday GM ((2017) ) LRRK2 levels and phosphorylation in Parkinson’s disease brain and cases with restricted Lewy bodies. Mov Disord 32: , 423–432. |
[12] | Reynolds A , Doggett EA , Riddle SM , Lebakken CS , Nichols RJ ((2014) ) LRRK2 kinase activity and biology are not uniformly predicted by its autophosphorylation and cellular phosphorylation site status. Front Mol Neurosci 7: , 54. |
[13] | Alcalay RN , Hsieh F , Tengstrand E , Padmanabhan S , Baptista M , Kehoe C , Narayan S , Boehme AK , Merchant K ((2019) ) Higher urine bis(monoacylglycerol)phosphate levels in LRRK2 G2019S mutation carriers: Implications for therapeutic development. Mov Disord, doi: 10.1002/mds.27818 |
[14] | Hughes AJ , Daniel SE , Kilford L , Lees AJ ((1992) ) Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: A clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry 55: , 181–184. |
[15] | Alcalay RN , Levy OA , Waters CC , Fahn S , Ford B , Kuo SH , Mazzoni P , Pauciulo MW , Nichols WC , Gan-Or Z , Rouleau GA , Chung WK , Wolf P , Oliva P , Keutzer J , Marder K , Zhang X ((2015) ) Glucocerebrosidase activity in Parkinson’s disease with and without GBA mutations. Brain 138: , 2648–2658. |
[16] | Wilson DH , Rissin DM , Kan CW , Fournier DR , Piech T , Campbell TG , Meyer RE , Fishburn MW , Cabrera C , Patel PP , Frew E , Chen Y , Chang L , Ferrell EP , von Einem V , McGuigan W , Reinhardt M , Sayer H , Vielsack C , Duffy DC ((2016) ) The Simoa HD-1 Analyzer: A novel fully automated digital immunoassay analyzer with single-molecule sensitivity and multiplexing. J Lab Autom 21: , 533–547. |
[17] | Fell MJ , Mirescu C , Basu K , Cheewatrakoolpong B , DeMong DE , Ellis JM , Hyde LA , Lin Y , Markgraf CG , Mei H , Miller M , Poulet FM , Scott JD , Smith MD , Yin Z , Zhou X , Parker EM , Kennedy ME , Morrow JA ((2015) ) MLi-2, a potent, selective, and centrally active compound for exploring the therapeutic potential and safety of LRRK2 kinase inhibition. J Pharmacol Exp Ther 355: , 397–409. |
[18] | Henderson MX , Sengupta M , McGeary I , Zhang B , Olufemi MF , Brown H , Trojanowski JQ , Lee VMY ((2019) ) LRRK2 inhibition does not impart protection from alpha-synuclein pathology and neuron death in non-transgenic mice. Acta Neuropathol Commun 7: , 28. |
[19] | Atashrazm F , Hammond D , Perera G , Bolliger MF , Matar E , Halliday GM , Schule B , Lewis SJG , Nichols RJ , Dzamko N ((2019) ) LRRK2-mediated Rab10 phosphorylation in immune cells from Parkinson’s disease patients. Mov Disord 34: , 406–415. |
[20] | Dzamko N , Chua G , Ranola M , Rowe DB , Halliday GM ((2013) ) Measurement of LRRK2 and Ser910/935 phosphorylated LRRK2 in peripheral blood mononuclear cells from idiopathic Parkinson’s disease patients. J Parkinsons Dis 3: , 145–152. |
[21] | Perera G , Ranola M , Rowe DB , Halliday GM , Dzamko N ((2016) ) Inhibitor treatment of peripheral mononuclear cells from Parkinson’s disease patients further validates LRRK2 dephosphorylation as a pharmacodynamic biomarker. Sci Rep 6: , 31391. |
[22] | Cook DA , Kannarkat GT , Cintron AF , Butkovich LM , Fraser KB , Chang J , Grigoryan N , Factor SA , West AB , Boss JM , Tansey MG ((2017) ) LRRK2 levels in immune cells are increased in Parkinson’s disease. NPJ Parkinsons Dis 3: , 11. |
[23] | Zhao J , Hermanson SB , Carlson CB , Riddle SM , Vogel KW , Bi K , Nichols RJ ((2012) ) Pharmacological inhibition of LRRK2 cellular phosphorylation sites provides insight into LRRK2 biology. Biochem Soc Trans 40: , 1158–1162. |
[24] | Nichols RJ , Dzamko N , Morrice NA , Campbell DG , Deak M , Ordureau A , Macartney T , Tong Y , Shen J , Prescott AR , Alessi DR ((2010) ) 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson’s disease-associated mutations and regulates cytoplasmic localization. Biochem J 430: , 393–404. |
[25] | Longo F , Russo I , Shimshek DR , Greggio E , Morari M ((2014) ) Genetic and pharmacological evidence that G2019S LRRK2 confers a hyperkinetic phenotype, resistant to motor decline associated with aging. Neurobiol Dis 71: , 62–73. |
[26] | Fan Y , Howden AJM , Sarhan AR , Lis P , Ito G , Martinez TN , Brockmann K , Gasser T , Alessi DR , Sammler EM ((2018) ) Interrogating Parkinson’s disease LRRK2 kinase pathway activity by assessing Rab10 phosphorylation in human neutrophils. Biochem J 475: , 23–44. |



