Potential Novel Genes for Late-Onset Alzheimer’s Disease in East-Asian Descent Identified by APOE-Stratified Genome-Wide Association Study
Abstract
The present study reports two novel genome-wide significant loci for late-onset Alzheimer’s disease (LOAD) identified from APOE ε4 non-carrier subjects of East Asian origin. A genome-wide association study of Alzheimer’s disease was performed in 2,291 Korean seniors in the discovery phase, from the Gwangju Alzheimer’ and Related Dementias (GARD) cohort study. The study was replicated in a Japanese cohort of 1,956 subjects that suggested two novel susceptible SNPs in two genes: LRIG1 and CACNA1A. This study demonstrates that the discovery of AD-associated variants is feasible in non-European ethnic groups using samples comprising fewer subjects from the more homogeneous genetic background.
INTRODUCTION
As a genetic component that has been consistently identified as an important antecedent factor in Alzheimer’s disease (AD), Apolipoprotein E (APOE) is the most prevalent risk factor of the disease [1, 2]. The effect of the APOE genotype, however, varies greatly among different ethnicities [3]. For instance, the ɛ4 allele of APOE, which is the primary genetic risk factor for AD, is absent among Arabs from the northern Israeli community residing in Wadi Ara village, even though the prevalence of dementia is approximately twice than that reported in Europeans [4]. Since the identification of the only locus in APOE using a sample consisting of 1,086 individuals in 2007 [5], genome-wide association studies (GWASs) have increased the sample size to improve the statistical power in identifying the missing causal variants for late-onset AD (LOAD) [6, 7]. The latest and largest GWAS includes 1,126,563 cases and controls (including 364,859 proxies) [8]. The variants and associated genes from these GWASs were mostly identified in individuals of European ancestry and found enriched in various biological pathways, including immune responses and endocytosis. These studies, thus suggest the genetic heterogeneity or the complex nature of the LOAD.
Genetic studies in diverse populations have increased our understanding of the genetic architecture of LOAD [9–14]. Studies involving subjects from population with non-European ancestry might be useful in the discovery of variants that are rare or absent, thus showing a smaller effect size in European ancestry [15]. Notably, the discovery of AD-susceptible loci that display allelic heterogeneity among diverse population, has been accomplished using samples from a more homogeneous non-European genetic background with several thousand or much fewer subjects [11, 12].
The East Asian population with Korean ethnicity is a homogeneous population that maintains a distinct genetic profile with a high prevalence of AD among seniors (65 years and older). In the present study, we leverage this distinct genetic architecture of Korean population to discover AD-associated variants. For this purpose, we first performed a GWAS and a post-hoc analysis by performing the APOE stratification of Koreans using the Korea Biobank SNP Array (referred to as Korean Chip) designed for Koreans [16]. The findings were further replicated by combining genotype level data using the findings from another East Asian population [12].
MATERIALS AND METHODS
Study sample
We analyzed the genotype level data of 2,291 subjects (1,119 AD cases and 1,172 controls) for the discovery stage, and 1,956 subjects (980 AD cases and 976 controls) for the replication stage (Table 1). Participants for the discovery stage were sampled from the Gwangju Alzheimer’s & Related Dementias (GARD) cohort, a longitudinal single-center study designed to develop clinical, imaging, genetic, and other biomarkers for early detection and tracking of dementia (mostly AD) from local senior citizens aged 60 years or older in Gwangju, Republic of Korea (Supplementary Material). The study protocol was approved by the Institutional Review Board of Chosun University Hospital, Gwangju, Republic of Korea (CHOSUN 2013-12-018-070). Written informed consents were obtained from all the volunteers or authorized guardians for cognitively impaired subjects before participation.
Table 1
Summary of demographic information for each group in GWAS analyses
| Discovery (n = 2,291) | p | Replication (n = 1,956) | p | |||
| CN | AD | CN | AD | |||
| All samples, n | 1,172 | 1,119 | 976 | 980 | ||
| Female, n (%) | 661 (56.4) | 715 (63.9) | <0.001 | 564 (57.8) | 702 (71.6) | <0.001 |
| Age at exam, m (s.d.) | 76.03 (8.9) | 74.60 (8.9) | <0.001 | 76.94 (5.9) | 72.99 (4.3) | <0.001 |
| APOE ɛ4 non-carrier, n | 976 | 621 | 815 | 435 | ||
| Female, n (%) | 544 (55.7) | 393 (63.3) | 0.003 | 470 (57.7) | 31 (71.3) | <0.001 |
| Age, m (s.d.) | 76.10 (8.9) | 75.43 (9.1) | 0.029 | 77.11 (5.9) | 73.42 (4.3) | <0.001 |
| APOE ɛ4 carrier | 196 | 498 | 161 | 545 | ||
| Female, n (%) | 117 (59.7) | 322 (64.7) | 0.222 | 94 (58.4) | 392 (68.3) | 0.001 |
| Age, m (s.d.) | 95.45 (8.9) | 73.32 (9.0) | <0.001 | 76.06 (5.9) | 72.65 (4.3) | <0.001 |
CN, cognitive normal; AD, Alzheimer’s disease; s.d., standard deviation.
Clinical evaluation was performed through brain imaging of the subjects using the Seoul Neuropsychological Screening Battery (SNSB) [17] test and 3T MRI (Skyra, Siemens Healthineers Ltd., Seoul, Republic of Korea) single scanner. 18F-Florbetaben (FBB) amyloid-beta positron emission tomography (PET) was performed for those who agreed. All diagnoses were evaluated by dementia specialists in neurology and psychiatry at Chosun University Hospital and Chonnam National University Hospital, Gwangju, Republic of Korea. Cognitively normal (CN) subjects exhibited no evidence of neurological disorders and impairment in cognitive function or routine activities. Excluded subjects included those with less than three years of education, a history of brain disease, and poor mental health in addition to those who were on related medications, consuming high levels of alcohol, or diagnosed with depression. All cases were at least 60 years old and fulfilled the NINCDS-ADRDA criteria for AD [18]. The healthy controls were at least 70 years old and either judged to be cognitively normal or did not meet the pathological criteria. The same inclusion criteria of age used in the discovery stage (case over 60 years and control over 70 years) was identically applied in the replication study. Demographic information about the samples used in the replication analysis is summarized in Table 1. Further details about the replication cohort can be found in the study by Miyashita et al. [12].
Genotyping, imputation, and gene mapping
Extensive quality control measures used in the standard protocol [19] were employed and subjects with a low genotyping success rate (95%), heterozygosity outliers, and those that presented cryptic relatedness were excluded. The study population heterogeneity was assessed by means of principal component analysis (PCA) and multidimensional scaling (MDS) and the subjects that did not meet the criteria were eliminated before proceeding with further analyses. Subjects with SNPs with a minor allele frequency < 1%, a genotyping success rate < 95%, or a deviation of the genotype distribution from Hardy-Weinberg equilibrium in the control group (p < 1×10-6) were also excluded. To increase the genotyping coverage between the two datasets, missing genotypes and SNPs were imputed using pre-phased reference haplotypes from the Haplotype Reference Consortium (HRC) panel version 1.1 for each dataset. All the imputed SNPs were subjected to the same quality criteria described above in addition to the requirement of imputation quality with an info score > 0.5, resulting in 35,685,761 and 39,044,005 SNPs for discovery and replication analysis, respectively. Functional mapping of SNPs to genes was conducted using a gene annotation file from Affymetrix [16] and the SNPnexus web tool [20].
Statistical analysis
Power calculations for our genome-wide study were performed using Genetic Association Study (GAS) Power Calculator [21]. Demographic information with the statistical significance was determined by t-test or Chi-square test. A set of GWAS analyses for AD status were performed in three models of a two-stage framework (discovery and replication): 1) all samples; 2) APOE ɛ4 carriers; and 3) APOE ɛ4 non-carriers. The GWAS was performed separately in each model for both the discovery and replication stages. The association of genotype dosages of each SNP in the additive component with the AD case-control status was estimated using logistic regression analysis adjusting for age, sex, and first four principal components using PLINK and EIGENSTRAT software [22–24]. Only the SNPs that attained a suggestively significant association level (p < 5×10-5) were tested in the replication stage using a Japanese dataset. A cohort of 4,247 total subjects was assembled for meta-analysis and SNP effect estimates along with their standard errors (SEs) were combined by a fixed effect model with inverse variance weighted method using the METAL software [25]. The SNP heritability, which is defined as the fraction of the phenotypic variance explained by additive effects of a given set of genetic variants (or SNPs), was estimated using the GCTA software [26, 27].
Gene expression analysis
The mRNA and protein expression in normal human tissues were reviewed via using data generated by Genotype-Tissue Expression (GTEx, release v8) (https://gtexportal.org/home/) [28] and Human Protein Atlas (HPA) (http://www.proteinatlas.org/) [29, 30], and Functional Annotation of Mammalian Genomes 5 (FANTOM5) [31]. We evaluated the genotype-specific expression of rs2280575 via expression quantitative trait loci (eQTL) analysis through the GTEx. In addition, differential expression (DE) analysis of CACNA1A and LRIG1 was performed using a publicly available expression dataset (GEO accession number: GSE118553) [32]. The dataset consists of various brain tissues from 167 AD cases (including both asymptomatic and dementia stage) and 100 CN controls. We used a linear regression model adjusting for age and sex to examine the differences among disease status.
RESULTS
Genome-wide inference for AD status
For discovery, we first conducted GWAS with the GARD sample, which included 2,291 subjects and identified 54 genome-wide significant (p < 5×10-8) SNPs across three genes namely APOE, PVRL2, and TOMM40 (genomic inflation = 1.03), which were found to be associated with LOAD in previous studies (Table 2, Supplementary Figure 1A, B). The heritability of LOAD was estimated without these genes using a liability-threshold model [27, 33] with the set of SNPs excluding the loci annotated in these genes; and a relatively large estimate (0.566±0.152) was obtained. In addition, a portion of SNPs in other previously known genes, such as ABCA7 and BIN1, was replicated at a genome-wide suggestive level (p < 5×10-5) (Supplementary Figure 1A). These results suggest that unreported additional common variants with smaller effects (or possibly rarer variants with larger effects) may be identified through a post-hoc analysis with a less stringent statistical significance. Furthermore, the rs11218343 SNP of SORL1, which was previously identified in a Japanese cohort [12], has been confirmed in this study through replication analysis with a significance of p = 5.9×10-4.
Table 2
Top-ranked genome-wide association results in the Korean discovery sample (p < 5×10–5) and their replication in Japanese and meta-analyses
| Gene | Chr | Lead SNP (type)* | BP | m/M | MAF | Discovery | Replication | Meta-analysis | Previously reported GWAS | ||||||
| OR (95%CI) | p | OR (95%CI) | p | OR | p | GENE (SNP) | P | ref | Alt Allele Frq† | ||||||
| All samples (CN/AD) | (1,172/1,119) | (976/980) | |||||||||||||
| APOE | 19 | rs429358 (m) | 45411941 | C/T | 0.169 | 3.637 (3.03–4.37) | 3.74×10–43 | 2.608 (2.04–3.33) | 1.89× 10–14 | 3.227 | 5.90×10–55 | rs429358 | 1.4×10–546 | [9] | 0.036 |
| TOMM40 | 19 | rs10119 (3’) | 45406673 | A/G | 0.180 | 3.378 (2.83–4.03) | 9.58×10–42 | 2.230 (1.88–2.65) | 7.49×10–20 | 2.731 | 1.59×10–57 | rs10119 | 1.2×10–342 | [46] | 0.279 |
| PVRL2 | 19 | rs12972156 (i) | 45387459 | G/C | 0.151 | 2.942 (2.45–3.54) | 1.52×10–30 | 2.656 (2.06–3.43) | 5.42×10–14 | 2.841 | 8.30×10–43 | PVRL2 (rs412776) | 2.2×10–21 | [9] | 0.001 |
| APOC1 | 19 | rs10414043 (g) | 45415713 | A/G | 0.143 | 3.082 (2.55–3.73) | 3.92×10–31 | 2.629 (2.02–3.43) | 8.76×10–13 | 2.920 | 4.29×10–42 | rs10414043 | 1.2×10–522 | [46] | 0.109 |
| APOE ɛ4 non- carrier (CN/AD) | (976/621) | (815/435) | |||||||||||||
| CACNA1A | 19 | rs189753894 (i) | 13624489 | A/C | 0.082 | 1.726 (1.34–2.23) | 2.68×10–5 | 1.900 (1.35–2.67) | 2.22×10–4 | 1.787 | 2.49 × 10–8 | – | – | – | 0.007 |
| LRIG1 | 3 | rs2280575 (i) | 66542863 | G/A | 0.092 | 0.539 (0.41–0.71) | 1.15×10–5 | 0.551 (0.40–0.76) | 3.46×10–4 | 0.544 | 1.51 × 10 –8 | – | – | – | 0.267 |
| APOE ɛ4 carrier (CN/AD) | (196/498) | (161/545) | |||||||||||||
*m, missense; I, intronic; g, intergenic; 3’, 3’ UTR; †SNP alternative allele frequencies of Europeans reported in the allele frequency aggregator (ALFA) [47]. Chr, chromosome; BP, base-pair position; m/M, minor allele/major allele; MAF, minor allele frequency; OR, odds ratio; CI, confidence interval; ref, reference.
We carried out APOE-stratified GWAS using subgroups of subjects on the basis of APOE ɛ4 carrier status. A total of 219 variants (annotated in 61 genes) and 306 variants (annotated in 82 genes) were identified as ɛ4 carriers (genomic inflation = 1.02) and non-carriers (genomic inflation = 1.03) with statistical significance respectively. Relaxing statistical significance might result in increases false discoveries. Accordingly, an independent cohort dataset was incorporated in our study for replication. For valid novel discovery, a meta-analysis was performed with the replication dataset in which previous GWAS signals were filtered. This analysis resulted in the examination of 7 out of 61 and 15 out of 82 genes for APOE ɛ4 carriers and non-carriers, respectively. Considering the effect direction and statistical significance of the variants in both the discovery and replication datasets, CACNA1A (OR = 1.726, lead SNP rs189753894) and LRIG1 (OR = 0.539, lead SNP rs2280575) SNPs from the ɛ4 non-carrier association result were selected for further functional annotation (Table 2). Additionally, we evaluated the expected power of the ɛ4 non-carrier findings in our discovery study. Given the prevalence of dementia at 0.087 and a SNP in LD (D’ = 1) with a risk allele frequency 0.082, the power was estimated at 85%with significant association at p = 5×10-5 under an additive model with strong effect size of OR 1.8.
Functional annotation of CACNA1A and LRIG1
Tissue specificity analysis with publicly available databases (GTEx, FANTOM5, and HPA) shows that CACNA1A is expressed at both the transcript and protein levels in brain tissues, particularly in the cerebellum (Supplementary Figure 1C, Fig. 1A). The CACNA1A gene plays an important role in neuronal cell death and codes subunits of neuronal calcium channels, which are involved in the neuronal cell death and amyloid-beta toxicity [34, 35].
Fig. 1
The expression level of two genes that show significant differences by tissue, genotype, and diagnosis. A) The CACNA1A is expressed at a high level in the cerebellum (median TPM: 304.8) and in the cerebellar hemisphere (median TPM: 275.0). B) Violin plot showing the effect of the eQTL rs2280575 on LRIG1 expression in the hypothalamus (p = 1.13×10-2). C) The CACNA1A gene showed significant difference in expression between CN and AD in the entorhinal cortex (p = 0.0019). D) The LRIG1 gene showed significant differences in expression between the pairs among CN, aAD, and AD in the entorhinal cortex, frontal cortex, and temporal cortex. The significance was labeled above the box plots (*p < 0.05, **p < 0.01, ***p < 0.001). CN, cognitive normal; AD, Alzheimer’s disease dementia; aAD, asymptomatic AD; n.s, not significant.
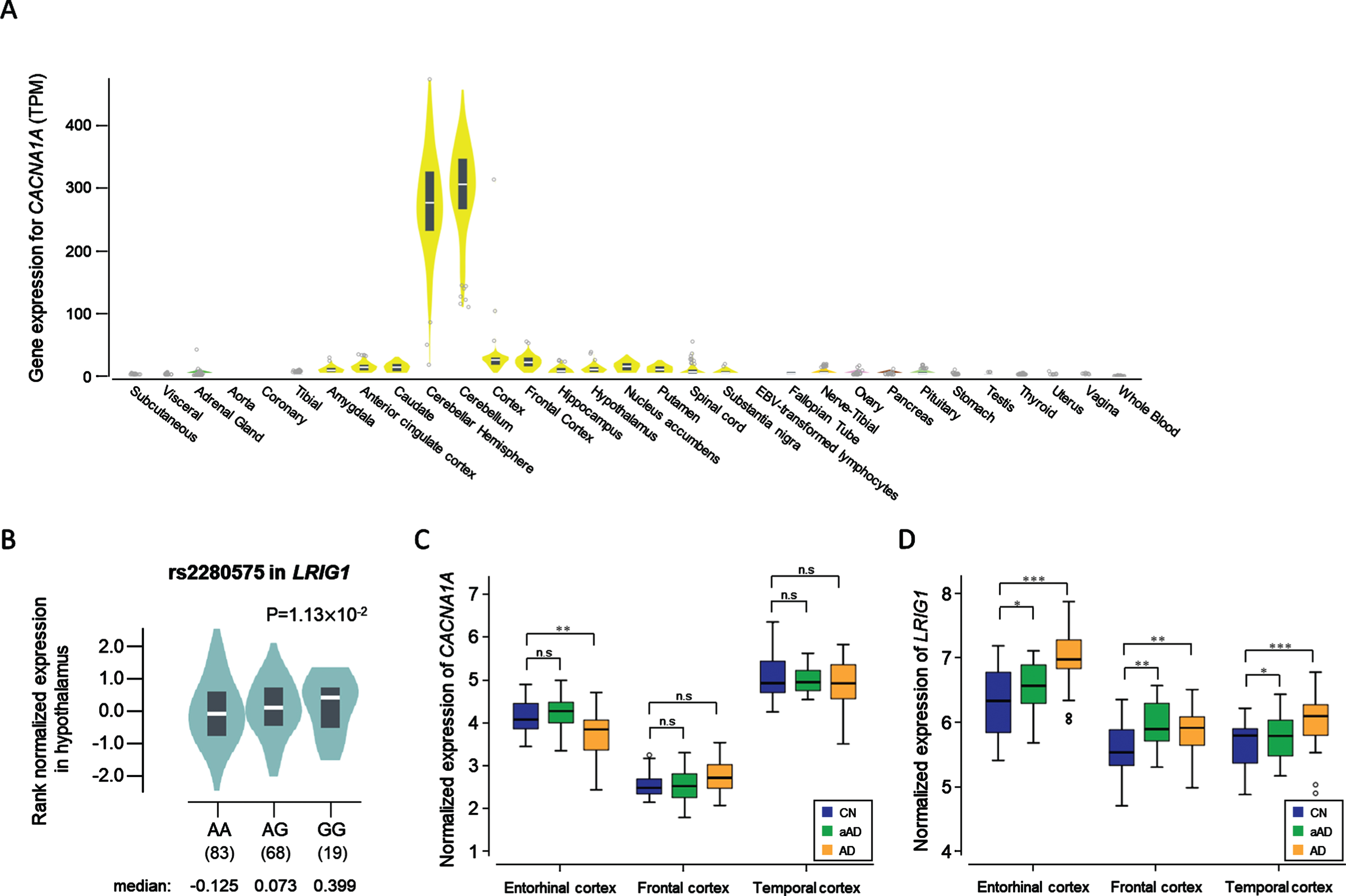
Expression levels of LRIG1 were high in the cerebral cortex at 74.1 protein-coding transcripts per million (pTPM) in HPA while 16.0 LRIG1 pTPM were reported by GTEx (Supplementary Figure 1D). In the FANTOM5 dataset, the RNA expression levels of LRIG1 were higher in the brain tissues than in other tissues, especially for the hippocampal formation, which was found to be 179.9 scaled-tags per million. eQTL analysis with lead SNP rs2280575 in LRIG1 showed significant results in the hypothalamus (Fig. 1B). The association between LRIG1 and the hippocampus was evaluated using in vitro and in vivo analyses from the previous study by Alsina et al. [36].
From the DE analysis with a public dataset (GSE118553), the gene expression level of CACNA1A presented a significant difference (p = 0.0019) only in the entorhinal cortex between CN and AD (Fig. 1C), while the expression of LRIG1 is significantly increased even in cases of asymptomatic stage AD (denoted aAD) for all three tissues analyzed (Fig. 1D).
DISCUSSION
In the present study, we have conducted genome-wide analysis with a Korean sample to identify AD-associated loci. Our study validated the association between AD status and APOE locus in chromosome 19 with very strong genome-wide significance. In addition to APOE, SORL1 has also been identified as a potential risk factor for AD with highly significant association in certain East Asian population [12, 37]. However, this highly significant association of SORL1 with AD is greatly influenced by the presence or absence of ɛ4 allele of APOE. To mask the effect of APOE and identify novel candidate loci in East Asian population, we further carried out an APOE ɛ4-stratified GWAS with two independent East Asian ethnic populations. Our data identified CACAN1A (lead SNP rs189753894) and LRIG1 (lead SNP rs2280575) among APOE ɛ4 non-carriers as novel candidate loci associated with AD.
From the functional annotation and literature survey, we found that the two genes play an essential role in neurobiological function. CACNA1A is a candidate risk gene located on chromosome 19. However, it is not likely to be influenced by the APOE signal because CACNA1A is located far (more than 32 Mb) from the APOE locus and is identified with APOE non-carriers. CACNA1A, a subunit of the neuronal calcium channel, is predominantly and specifically expressed in brain tissue and is found to be involved in neuronal cell death and toxicity of amyloid-beta, suggesting a plausible risk gene for AD [34, 35]. The genetic function of LRIG1 is associated with the dendritic formation of neurons as well as affecting the hippocampus neural cell functions [36]. Abnormal expression and dysfunction of LRIG1 lead to dendritic abnormalities, which are involved in morphogenesis of hippocampal dendrites by brain-derived neurotrophic factor (BDNF) signaling. BDNF plays a major role in the growth, development, and survival of neurons. It is also known to be involved in regulating synaptic plasticity and synaptogenesis for learning and memory in the adult brain [38–40]. BDNF is expressed in low levels in patients with AD [41]. Several studies have shown that high expression of BDNF could slow down cognitive decline in the elderly and in patients with AD [38, 42–44].
This study paved the way for discovery of the novel loci in CACNA1A and LRIG1 genes that could not be revealed in large-scale GWASs using Caucasian subjects. The respective 0.082 and 0.007 allele frequencies of rs18973894 SNP of CACNA1A in Korean and European populations indicate that there is a significant difference in the association of this SNP with AD among these populations. A meta-analysis with a far larger sample size of Caucasians from Alzheimer’s Disease Sequencing Project (ADSP) in NIAGADS demonstrated significant genome-wide difference in the signals for AD risk (3:66455703 in LRIG1 and 19:13395952 in CACNA1A showed p = 6.2×10-12 and p = 3.8×10-27, respectively) of both genes [45].
There are certain limitations to this study that need to be considered. The sample size in this study is relatively smaller than those in other GWASs for AD. Also, this study used only Korean and Japanese population and does not include other East Asian ethnic groups. However, the findings of this study are promising due to the high reliability of accurate diagnostic information based on pathophysiology available in the GARD as well as Japanese datasets. Another limitation of this study is that the results are based on the imputed SNPs. Although the genotype imputation is likely to be inaccurate for the rare variants, the top findings in this work are common variants. Moreover, we have also conducted a replication analysis with an independent sample to improve the accuracy of these findings.
Although the previous studies about the function of CACNA1A and LRIG1 suggest their role in the development of LOAD, a conclusion whether each gene has an impact on the progression of dementia or protecting neurons from degeneration requires further biological validation with an AD model system. We also acknowledge that the conclusions drawn in this study are based on the analysis of two East Asian populations (Korean and Japanese), and therefore, one is required to be cautious during the interpretation of the results to conclude whether our findings are specific to the East Asian or Korean and Japanese populations. A further replication analysis or meta- or mega-analysis with a larger sample size of other East Asian individuals is strongly recommended to conclusively arrive at such an interpretation.
In conclusion, we validated a portion of previously reported LOAD-associated genes, identified from mostly the Caucasian subjects, in the Korean population. We have also identified and replicated East Asian-specific novel loci in CACNA1A and LRIG1 through a post-hoc GWAS with APOE stratification. Our findings thus, provide an improved understanding about the complex genetic signatures associated with LOAD.
ACKNOWLEDGMENTS
The authors greatly appreciate the reviewers’ invaluable comments that helped improving the work. This research was supported by the Original Technology Research Program for Brain Science of the National Research Foundation (NRF) funded by the Korean government, MSIT (NRF-2014M3C7A1046041 and NRF-2014M3C7A1046042); by KBRI basic research program through Korea Brain Research Institute funded by Ministry of Science and ICT (21-BR-03-05); by Healthcare AI Convergence Research & Development Program through the National IT Industry Promotion Agency of Korea (NIPA) funded by the Ministry of Science and ICT (No.1711120216); and by Japan Agency for Medical Research and Development (AMED) (JP18kk0205009 and JP21dk0207045). This study was also supported by the National Institute on Aging of the National Institutes of Health under Award Number U01AG062602. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Authors’ disclosures available online (https://www.j-alz.com/manuscript-disclosures/21-0145r2).
SUPPLEMENTARY MATERIAL
[1] The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/JAD-210145.
REFERENCES
[1] | Farrer LA , Myers RH , Connor L , Cupples LA , Growdon JH ((1991) ) Segregation analysis reveals evidence of a major gene for Alzheimer disease. Am J Hum Genet 48: , 1026–1033. |
[2] | Strittmatter WJ , Weisgraber KH , Huang DY , Dong LM , Salvesen GS , Pericak-Vance M , Schmechel D , Saunders AM , Goldgaber D , Roses AD ((1993) ) Binding of human apolipoprotein E to synthetic amyloid beta peptide: Isoform-specific effects and implications for late-onset Alzheimer disease. Proc Natl Acad Sci U S A 90: , 8098–8102. |
[3] | Singh PP , Singh M , Mastana SS ((2006) ) APOE distribution in world populations with new data from India and the UK. Ann Hum Biol 33: , 279–308. |
[4] | Sherva R , Baldwin CT , Inzelberg R , Vardarajan B , Cupples LA , Lunetta K , Bowirrat A , Naj A , Pericak-Vance M , Friedland RP , Farrer LA ((2011) ) Identification of novel candidate genes for Alzheimer’s disease by autozygosity mapping using genome wide SNP data. J Alzheimers Dis 23: , 349–359. |
[5] | Coon KD MA , Craig DW , Webster JA , Pearson JV , Lince DH , Zismann VL , Beach TG , Leung D , Bryden L , Halperin RF , Marlowe L , Kaleem M , Walker DG , Ravid R , Heward CB , Rogers J , Papassotiropoulos A , Reiman EM , Hardy J , Stephan DA ((2007) ) A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer’s disease. J Clin Psychiatry 68: , 613–618. |
[6] | Ma Y , Jun GR , Zhang X , Chung J , Naj AC , Chen Y , Bellenguez C , Hamilton-Nelson K , Martin ER , Kunkle BW , Bis JC , Debette S , DeStefano AL , Fornage M , Nicolas G , van Duijn C , Bennett DA , De Jager PL , Mayeux R , Haines JL , Pericak-Vance MA , Seshadri S , Lambert JC , Schellenberg GD , Lunetta KL , Farrer LA , Alzheimer’s Disease Sequencing Project and Alzheimer’s Disease Exome Sequencing–France Project ((2019) ) Analysis of whole-exome sequencing data for Alzheimer disease stratified by APOE genotype. JAMA Neurol 76: , 1099–1108. |
[7] | Jansen IE , Savage JE , Watanabe K , Bryois J , Williams DM , Steinberg S , Sealock J , Karlsson IK , Hagg S , Athanasiu L , Voyle N , Proitsi P , Witoelar A , Stringer S , Aarsland D , Almdahl IS , Andersen F , Bergh S , Bettella F , Bjornsson S , Braekhus A , Brathen G , de Leeuw C , Desikan RS , Djurovic S , Dumitrescu L , Fladby T , Hohman TJ , Jonsson PV , Kiddle SJ , Rongve A , Saltvedt I , Sando SB , Selbaek G , Shoai M , Skene NG , Snaedal J , Stordal E , Ulstein ID , Wang Y , White LR , Hardy J , Hjerling-Leffler J , Sullivan PF , van der Flier WM , Dobson R , Davis LK , Stefansson H , Stefansson K , Pedersen NL , Ripke S , Andreassen OA , Posthuma D ((2019) ) Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet 51: , 404–413. |
[8] | Wightman DP , Jansen IE , Savage JE , Shadrin AA , Bahrami S , Rongve A , Børte S , Winsvold BS , Drange OK , Martinsen AE , et al. (2020) Largest GWAS (N = 1,126,563) of Alzheimer’s disease implicates microglia and immune cells. medRxiv 2020.11.20.20235275; doi: https://doi.org/10.1101/2020.11.20.20235275. |
[9] | Ghani M , Reitz C , Cheng R , Vardarajan BN , Jun G , Sato C , Naj A , Rajbhandary R , Wang LS , Valladares O , Lin CF , Larson EB , Graff-Radford NR , Evans D , De Jager PL , Crane PK , Buxbaum JD , Murrell JR , Raj T , Ertekin-Taner N , Logue M , Baldwin CT , Green RC , Barnes LL , Cantwell LB , Fallin MD , Go RC , Griffith PA , Obisesan TO , Manly JJ , Lunetta KL , Kamboh MI , Lopez OL , Bennett DA , Hendrie H , Hall KS , Goate AM , Byrd GS , Kukull WA , Foroud TM , Haines JL , Farrer LA , Pericak-Vance MA , Lee JH , Schellenberg GD , St George-Hyslop P , Mayeux R , Rogaeva E , Alzheimer’s Disease Genetics Consortium ((2015) ) Association of long runs of homozygosity with Alzheimer disease among African American individuals. JAMA Neurol 72: , 1313–1323. |
[10] | Jun GR , Chung J , Mez J , Barber R , Beecham GW , Bennett DA , Buxbaum JD , Byrd GS , Carrasquillo MM , Crane PK , Cruchaga C , De Jager P , Ertekin-Taner N , Evans D , Fallin MD , Foroud TM , Friedland RP , Goate AM , Graff-Radford NR , Hendrie H , Hall KS , Hamilton-Nelson KL , Inzelberg R , Kamboh MI , Kauwe JSK , Kukull WA , Kunkle BW , Kuwano R , Larson EB , Logue MW , Manly JJ , Martin ER , Montine TJ , Mukherjee S , Naj A , Reiman EM , Reitz C , Sherva R , St George-Hyslop PH , Thornton T , Younkin SG , Vardarajan BN , Wang LS , Wendlund JR , Winslow AR , Alzheimer’s Disease Genetics Consortium, Haines J , Mayeux R , Pericak-Vance MA , Schellenberg G , Lunetta KL , Farrer LA ((2017) ) Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimers Dement 13: , 727–738. |
[11] | Meng Y , Baldwin CT , Bowirrat A , Waraska K , Inzelberg R , Friedland RP , Farrer LA ((2006) ) Association of polymorphisms in the Angiotensin-converting enzyme gene with Alzheimer disease in an Israeli Arab community. Am J Hum Genet 78: , 871–877. |
[12] | Miyashita A , Koike A , Jun G , Wang LS , Takahashi S , Matsubara E , Kawarabayashi T , Shoji M , Tomita N , Arai H , Asada T , Harigaya Y , Ikeda M , Amari M , Hanyu H , Higuchi S , Ikeuchi T , Nishizawa M , Suga M , Kawase Y , Akatsu H , Kosaka K , Yamamoto T , Imagawa M , Hamaguchi T , Yamada M , Morihara T , Takeda M , Takao T , Nakata K , Fujisawa Y , Sasaki K , Watanabe K , Nakashima K , Urakami K , Ooya T , Takahashi M , Yuzuriha T , Serikawa K , Yoshimoto S , Nakagawa R , Kim JW , Ki CS , Won HH , Na DL , Seo SW , Mook-Jung I , Alzheimer Disease Genetics C, St George-Hyslop P , Mayeux R , Haines JL , Pericak-Vance MA , Yoshida M , Nishida N , Tokunaga K , Yamamoto K , Tsuji S , Kanazawa I , Ihara Y , Schellenberg GD , Farrer LA , Kuwano R ((2013) ) SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PLoS One 8: , e58618. |
[13] | Rogaeva E , Meng Y , Lee JH , Gu Y , Kawarai T , Zou F , Katayama T , Baldwin CT , Cheng R , Hasegawa H , Chen F , Shibata N , Lunetta KL , Pardossi-Piquard R , Bohm C , Wakutani Y , Cupples LA , Cuenco KT , Green RC , Pinessi L , Rainero I , Sorbi S , Bruni A , Duara R , Friedland RP , Inzelberg R , Hampe W , Bujo H , Song YQ , Andersen OM , Willnow TE , Graff-Radford N , Petersen RC , Dickson D , Der SD , Fraser PE , Schmitt-Ulms G , Younkin S , Mayeux R , Farrer LA , St George-Hyslop P ((2007) ) The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet 39: , 168–177. |
[14] | Jia L , Li F , Wei C , Zhu M , Qu Q , Qin W , Tang Y , Shen L , Wang Y , Shen L , Li H , Peng D , Tan L , Luo B , Guo Q , Tang M , Du Y , Zhang J , Zhang J , Lyu J , Li Y , Zhou A , Wang F , Chu C , Song H , Wu L , Zuo X , Han Y , Liang J , Wang Q , Jin H , Wang W , Lu Y , Li F , Zhou Y , Zhang W , Liao Z , Qiu Q , Li Y , Kong C , Li Y , Jiao H , Lu J , Jia J ((2021) ) Prediction of Alzheimer’s disease using multi-variants from a Chinese genome-wide association study. Brain 144: , 924–937. |
[15] | Kunkle BW , Schmidt M , Klein HU , Naj AC , Hamilton-Nelson KL , Larson EB , Evans DA , De Jager PL , Crane PK , Buxbaum JD , Ertekin-Taner N , Barnes LL , Fallin MD , Manly JJ , Go RCP , Obisesan TO , Kamboh MI , Bennett DA , Hall KS , Goate AM , Foroud TM , Martin ER , Wang LS , Byrd GS , Farrer LA , Haines JL , Schellenberg GD , Mayeux R , Pericak-Vance MA , Reitz C , Writing Group for the Alzheimer’s Disease Genetics Consortium, Graff-Radford NR , Martinez I , Ayodele T , Logue MW , Cantwell LB , Jean-Francois M , Kuzma AB , Adams LD , Vance JM , Cuccaro ML , Chung J , Mez J , Lunetta KL , Jun GR , Lopez OL , Hendrie HC , Reiman EM , Kowall NW , Leverenz JB , Small SA , Levey AI , Golde TE , Saykin AJ , Starks TD , Albert MS , Hyman BT , Petersen RC , Sano M , Wisniewski T , Vassar R , Kaye JA , Henderson VW , DeCarli C , LaFerla FM , Brewer JB , Miller BL , Swerdlow RH , Van Eldik LJ , Paulson HL , Trojanowski JQ , Chui HC , Rosenberg RN , Craft S , Grabowski TJ , Asthana S , Morris JC , Strittmatter SM , Kukull WA ((2021) ) Novel Alzheimer disease risk loci and pathways in African American individuals using the African genome resources panel: A meta-analysis. JAMA Neurol 78: , 102–113. |
[16] | Moon S , Kim YJ , Han S , Hwang MY , Shin DM , Park MY , Lu Y , Yoon K , Jang HM , Kim YK , Park TJ , Song DS , Park JK , Lee JE , Kim BJ ((2019) ) The Korea Biobank Array: Design and identification of coding variants associated with blood biochemical traits. Sci Rep 9: , 1382. |
[17] | Kang Y , Na D , Hahn S ((2003) ) Seoul Neuropsychological Screening Battery. Human Brain Research & Consulting Co, Incheon. |
[18] | McKhann G , Drachman D , Folstein M , Katzman R , Price D , Stadlan EM ((1984) ) Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group* under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 34: , 939–939. |
[19] | Anderson CA , Pettersson FH , Clarke GM , Cardon LR , Morris AP , Zondervan KT ((2010) ) Data quality control in genetic case-control association studies. Nat Protoc 5: , 1564–1573. |
[20] | Dayem Ullah AZ , Oscanoa J , Wang J , Nagano A , Lemoine NR , Chelala C ((2018) ) SNPnexus: Assessing the functional relevance of genetic variation to facilitate the promise of precision medicine. Nucleic Acids Res 46: , W109–W113. |
[21] | Johnson JL , Abecasis GR (2017) GAS Power Calculator: Web-based power calculator for genetic association studies. bioRxiv 164343; doi: https://doi.org/10.1101/164343. |
[22] | Purcell S , Neale B , Todd-Brown K , Thomas L , Ferreira MA , Bender D , Maller J , Sklar P , de Bakker PI , Daly MJ , Sham PC ((2007) ) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: , 559–575. |
[23] | Patterson N , Price AL , Reich D ((2006) ) Population structure and eigenanalysis. PLoS Genet 2: , e190. |
[24] | Price AL , Patterson NJ , Plenge RM , Weinblatt ME , Shadick NA , Reich D ((2006) ) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: , 904–909. |
[25] | Willer CJ , Li Y , Abecasis GR ((2010) ) METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: , 2190–2191. |
[26] | Yang J , Lee SH , Goddard ME , Visscher PM ((2011) ) GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet 88: , 76–82. |
[27] | Yang J , Zeng J , Goddard ME , Wray NR , Visscher PM ((2017) ) Concepts, estimation and interpretation of SNP-based heritability. Nat Genet 49: , 1304–1310. |
[28] | GTEx Consortium ((2015) ) The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348: , 648–660. |
[29] | Uhlen M , Bjorling E , Agaton C , Szigyarto CA , Amini B , Andersen E , Andersson AC , Angelidou P , Asplund A , Asplund C , Berglund L , Bergstrom K , Brumer H , Cerjan D , Ekstrom M , Elobeid A , Eriksson C , Fagerberg L , Falk R , Fall J , Forsberg M , Bjorklund MG , Gumbel K , Halimi A , Hallin I , Hamsten C , Hansson M , Hedhammar M , Hercules G , Kampf C , Larsson K , Lindskog M , Lodewyckx W , Lund J , Lundeberg J , Magnusson K , Malm E , Nilsson P , Odling J , Oksvold P , Olsson I , Oster E , Ottosson J , Paavilainen L , Persson A , Rimini R , Rockberg J , Runeson M , Sivertsson A , Skollermo A , Steen J , Stenvall M , Sterky F , Stromberg S , Sundberg M , Tegel H , Tourle S , Wahlund E , Walden A , Wan J , Wernerus H , Westberg J , Wester K , Wrethagen U , Xu LL , Hober S , Ponten F ((2005) ) A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics 4: , 1920–1932. |
[30] | Uhlen M , Fagerberg L , Hallstrom BM , Lindskog C , Oksvold P , Mardinoglu A , Sivertsson A , Kampf C , Sjostedt E , Asplund A , Olsson I , Edlund K , Lundberg E , Navani S , Szigyarto CA , Odeberg J , Djureinovic D , Takanen JO , Hober S , Alm T , Edqvist PH , Berling H , Tegel H , Mulder J , Rockberg J , Nilsson P , Schwenk JM , Hamsten M , von Feilitzen K , Forsberg M , Persson L , Johansson F , Zwahlen M , von Heijne G , Nielsen J , Ponten F ((2015) ) Proteomics. Tissue-based map of the human proteome. Science 347: , 1260419. |
[31] | Lizio M , Harshbarger J , Shimoji H , Severin J , Kasukawa T , Sahin S , Abugessaisa I , Fukuda S , Hori F , Ishikawa-Kato S , Mungall CJ , Arner E , Baillie JK , Bertin N , Bono H , de Hoon M , Diehl AD , Dimont E , Freeman TC , Fujieda K , Hide W , Kaliyaperumal R , Katayama T , Lassmann T , Meehan TF , Nishikata K , Ono H , Rehli M , Sandelin A , Schultes EA , t Hoen PA , Tatum Z , Thompson M , Toyoda T , Wright DW , Daub CO , Itoh M , Carninci P , Hayashizaki Y , Forrest AR , Kawaji H , FANTOM consortium ((2015) ) Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol 16: , 22. |
[32] | Patel H , Hodges AK , Curtis C , Lee SH , Troakes C , Dobson RJB , Newhouse SJ ((2019) ) Transcriptomic analysis of probable asymptomatic and symptomatic alzheimer brains. Brain Behav Immun 80: , 644–656. |
[33] | Gim J , Kim W , Kwak SH , Choi H , Park C , Park KS , Kwon S , Park T , Won S ((2017) ) Improving disease prediction by incorporating family disease history in risk prediction models with large-scale genetic data. Genetics 207: , 1147–1155. |
[34] | Supnet C , Bezprozvanny I ((2010) ) The dysregulation of intracellular calcium in Alzheimer disease. Cell Calcium 47: , 183–189. |
[35] | Demuro A , Parker I , Stutzmann GE ((2010) ) Calcium signaling and amyloid toxicity in Alzheimer disease. J Biol Chem 285: , 12463–12468. |
[36] | Alsina FC , Hita FJ , Fontanet PA , Irala D , Hedman H , Ledda F , Paratcha G ((2016) ) Lrig1 is a cell-intrinsic modulator of hippocampal dendrite complexity and BDNF signaling. EMBO Rep 17: , 601–616. |
[37] | Zhang CC , Wang HF , Tan MS , Wan Y , Zhang W , Zheng ZJ , Kong LL , Wang ZX , Tan L , Jiang T , Tan L , Yu JT ((2017) ) SORL1 is associated with the risk of late-onset Alzheimer’s disease: A replication study and meta-analyses. Mol Neurobiol 54: , 1725–1732. |
[38] | Lu B , Nagappan G , Guan X , Nathan PJ , Wren P ((2013) ) BDNF-based synaptic repair as a disease-modifying strategy for neurodegenerative diseases. Nat Rev Neurosci 14: , 401–416. |
[39] | Lu B , Nagappan G , Lu Y ((2014) ) BDNF and synaptic plasticity, cognitive function, and dysfunction. Handb Exp Pharmacol 220: , 223–250. |
[40] | Bramham CR , Messaoudi E ((2005) ) BDNF function in adult synaptic plasticity: The synaptic consolidation hypothesis. Prog Neurobiol 76: , 99–125. |
[41] | Ng TKS , Ho CSH , Tam WWS , Kua EH , Ho RC ((2019) ) Decreased serum brain-derived neurotrophic factor (BDNF) levels in patients with Alzheimer’s disease (AD): A systematic review and meta-analysis. Int J Mol Sci 20: , 257. |
[42] | Buchman AS , Yu L , Boyle PA , Schneider JA , De Jager PL , Bennett DA ((2016) ) Higher brain BDNF gene expression is associated with slower cognitive decline in older adults. Neurology 86: , 735–741. |
[43] | Laske C , Stellos K , Hoffmann N , Stransky E , Straten G , Eschweiler GW , Leyhe T ((2011) ) Higher BDNF serum levels predict slower cognitive decline in Alzheimer’s disease patients. Int J Neuropsychopharmacol 14: , 399–404. |
[44] | Jiao SS , Shen LL , Zhu C , Bu XL , Liu YH , Liu CH , Yao XQ , Zhang LL , Zhou HD , Walker DG , Tan J , Gotz J , Zhou XF , Wang YJ ((2016) ) Brain-derived neurotrophic factor protects against tau-related neurodegeneration of Alzheimer’s disease. Transl Psychiatry 6: , e907. |
[45] | Kuzma A , Valladares O , Cweibel R , Greenfest-Allen E , Childress DM , Malamon J , Gangadharan P , Zhao Y , Qu L , Leung YY , Naj AC , Stoeckert CJ , Schellenberg GD , Wang L-S ((2016) ) NIAGADS: The NIA Genetics of Alzheimer’s Disease Data Storage Site. Alzheimers Dement 12: , 1200–1203. |
[46] | Lambert JC , Ibrahim-Verbaas CA , Harold D , Naj AC , Sims R , Bellenguez C , DeStafano AL , Bis JC , Beecham GW , Grenier-Boley B , et al. ((2013) ) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45: , 1452–1458. |
[47] | Phan L , Jin Y , Zhang H , Qiang W , Shekhtman E , Shao D , Revoe D , Villamarin R , Ivanchenko E , Kimura M , Wang ZY , Hao L , Sharopova N , Bihan M , Sturcke A , Lee M , Popova N , Wu W , Bastiani C , Ward M , Holmes JB , Lyoshin V , Kaur K , Moyer E , Feolo M , Kattman BL (2020) ALFA: Allele Frequency Aggregator. National Center for Biotechnology Information, U.S. National Library of Medicine, http://www.ncbi.nlm.nih.gov/snp/docs/gsr/alfa/. |



