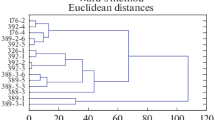
Abstract
Gibberellin signaling pathway genes encoding the DELLA protein and gibberellin receptor GID1 were sequenced in several varieties (Alpha, Valdai, Orlovskaya 9, Pracha) and one line of rye (EM-1) using next generation methods. The revealed multiple alleles of these genes differ mainly in single-nucleotide polymorphisms and less frequently in insertions and deletions. Most of the detected mutations turned out to be synonymous or located in the noncoding regions of the genes. Changes in the amino acid sequences of proteins associated with other mutations are probably functionally neutral. Mutations similar to wheat reduced-height gibberellin-insensitive alleles were not detected.

Similar content being viewed by others
REFERENCES
Schlegel, R.H.J., Rye: Genetics, Breeding, and Cultivation, CRC Press, 2013.
Davière, J.-M. and Achard, P., Gibberellin signaling in plants, Development, 2013, vol. 140, no. 6, pp. 1147–1151.
Peng, J., Richards, D.E., Hartley, N.M., Murphy, G.P., Devos, K.M., Flintham, J.E., Beales, J., Fish, L.J., Worland, A.J., Pelica, F., Sudhakar, D., Christou, P., Snape, J.W., Gale, M.D., and Harberd, N.P., “Green revolution” genes encode mutant gibberellin response modulators, Nature, 1999, vol. 400, no. 6741, pp. 256–261.
Pearce, S., Saville, R., Vaughan, S.P., Chandler, P.M., Wilhelm, E.P., Sparks, C.A., Al-Kaff, N., Korolev, A., Boulton, M.I., Phillips, A.L., Hedden, P., Nicholson, P., and Thomas, S.G., Molecular characterization of Rht-1 dwarfing genes in hexaploid wheat, Plant Physiol., 2011, vol. 157, no. 4, pp. 1820–1831.
Yoshida, H., Hirano, K., Sato, T., Mitsuda, N., Nomoto, M., Maeo, K., Koketsu, E., Mitani, R., Kawamura, M., Ishiguro, S., Tada, Y., Ohme-Takagi, M., Matsuoka, M., and Ueguchi-Tanaka, M., DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins, Proc. Natl. Acad. Sci. U.S.A., 2014, vol. 111, no. 21, pp. 7861–7866.
Li, S., Tian, Y., Wu, K., Ye, Y., Yu, J., Zhang, J., Liu, Q., Hu, M., Li, H., Tong, Y., Harberd, N.P., and Fu, X., Modulating plant growth-metabolism coordination for sustainable agriculture, Nature, 2018, vol. 560, no. 7720, pp. 595–600.
Hirsch, S. and Oldroyd, G.E., GRAS-domain transcription factors that regulate plant development, Plant Signal Behav., 2009, vol. 4, no. 8, pp. 698–700.
Ueguchi-Tanaka, M., Nakajima, M., Katoh, E., Ohmiya, H., Asano, K., Saji, S., Hongyu, X., Ashikari, M., Kitano, H., Yamaguchi, I., and Matsuoka, M., Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin, Plant Cell, 2007, vol. 19, no. 7, pp. 2140–2155.
Fu, X., Richards, D.E., Ait-Ali, T., Hynes, L.W., Ougham, H., Peng, J., and Harberd, N.P., Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor, Plant Cell, 2002, vol. 14, no. 12, pp. 3191–3200.
Börner, A., Plaschke, J., Korzun, V., and Worland, A.J., The relationships between the dwarfing genes of wheat and rye, Euphytica, 1996, vol. 89, no. 1, pp. 69–75.
Braun, E.-M., Tsvetkova, N., Rotter, B., Siekmann, D., Schwefel, K., Krezdorn, N., Plieske, J., Winter, P., Melz, G., Voylokov, A.V., and Hackauf, B., Gene expression profiling and fine mapping identifies a gibberellin 2-oxidase gene co-segregating with the dominant dwarfing gene Ddw1 in rye (Secale cereale L.), Front. Plant Sci., 2019, vol. 10, p. 857.
Kantarek, Z., Masojć, P., Bienias, A., and Milczarski, P., Identification of a novel, dominant dwarfing gene (Ddw4) and its effect on morphological traits of rye, PLoS ONE, 2018, vol. 13.
Grądzielewska, A., Milczarski, P., Molik, K., and Pawłowska, E., Identification and mapping of a new recessive dwarfing gene dw9 on the 6RL rye chromosome and its phenotypic effects, PLOS ONE, 2020, vol. 15, no. 3.
Doyle, P.J., DNA protocols for plants, in Molecular Techniques in Taxonomy, Hewitt, G.M., Johnston, A.W.B., Young, J.P.W., Eds., Berlin–Heidelberg: Springer, 1991, pp. 283–293.
Bauer, E., Schmutzer, T., Barilar, I., et al., Towards a whole-genome sequence for rye (Secale cereale L.), Plant J., 2017, vol. 89, no. 5, pp. 853–869.
IPK Rye blast server. https://webblast.ipk-gatersleben.de/ryeselect/. Accessed June 27, 2020.
Razumova, O.V., Bazhenov, M.S., Nikitina, E.A., Nazarova, L.A., Romanov, D.V., Chernook, A.G., Sokolov, P.A., Kuznetsova, V.M., Semenov, O.G., Karlov, G.I., Kharchenko, P.N., and Divashuk, M.G., Molecular analysis of gibberellin receptor gene GID1 in Dasypyrum villosum and development of DNA marker for its identification, RUDN J. Agron. Anim. Ind., 2020, vol. 15, no. 1, pp. 62–85.
Lischer, H.E.L. and Shimizu, K.K., Reference-guided de novo assembly approach improves genome reconstruction for related species, BMC Bioinf., 2017, vol. 18, no. 1, p. 474.
Bankevich, A., Nurk, S., Antipov, D., et al., SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing, J. Comput. Biol., 2012, vol. 19, no. 5, pp. 455–477.
Huang, X. and Madan, A., CAP3: A DNA sequence assembly program, Genome Res., 1999, vol. 9, no. 9, pp. 868–877.
Zaharia, M., Bolosky, W.J., Curtis, K., Fox, A., Patterson, D., Shenker, S., Stoica, I., Karp, R.M., and Sittler, T., Faster and more accurate sequence alignment with SNAP, 2011. arXiv:1111.5572 [cs, q-bio]
Garrison, E. and Marth, G., Haplotype-based variant detection from short-read sequencing, 2012. arXiv:1207.3907 [q-bio]
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G., Durbin, R., and 1000 Genome Project Data Processing Subgroup, The sequence alignment/map format and SAMtools, Bioinformatics, 2009, vol. 25, no. 16, pp. 2078–2079.
Nicholas, K.B. and Nikolas, H.B., Jr. GeneDoc: A Tool for Editing and Annotating Multiple Sequence Alignments, Pittsburgh Supercomputing Center’s National Resource for Biomedical Supercomputing, 1997.
Choi, Y. and Chan, A.P., PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels, Bioinformatics, 2015, vol. 31, no. 16, pp. 2745–2747.
NCBI Conserved Domain Search. https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi. Accessed June 29, 2020.
ACKNOWLEDGMENTS
We thank Dr. I.G. Loskutov, Doctor of Science, Head of the Department of Genetic Resources of Oat, Rye, and Barley of the Vavilov All-Russia Institute for Plant Genetic Resources; Dr. V.D. Kobylyansky, Doctor of Science, Honorary Professor of Vavilov All-Russia Institute for Plant Genetic Resources; and Dr. V.S. Rubetz, Doctor of Science, Professor of the Department of Genetics, Plant Breeding, and Seed Farming of the Russian State Agrarian University–Moscow Timiryazev Agricultural Academy for kindly providing plant material.
Funding
This work was supported by the Russian Scientific Foundation, project no. 17-76-20023.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
COMPLIANCE WITH ETHICAL STANDARDS
This study does not contain any experiments carried out with animals or people as objects.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Additional information
Translated by Mikhail Bibov
About this article
Cite this article
Bazhenov, M.S., Chernook, A.G. & Divashuk, M.G. Molecular Analysis of the Gibberellin Signaling Pathway Genes in Cultivated Rye (Secale cereale L.). Moscow Univ. Biol.Sci. Bull. 75, 125–129 (2020). https://doi.org/10.3103/S0096392520030025
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0096392520030025