Abstract
Background
Quantitative predictions of in vivo drug-drug interactions (DDIs) resulting from metabolic inhibition are commonly made based upon the inhibitor concentration at the enzyme active site [I] and the in vitro inhibition constant (Ki). Previous studies have involved the use of various plasma inhibitor concentrations as surrogates for [I] along with Ki values obtained from published literature. Although this approach has resulted in a high proportion of successful predictions, a number of falsely predicted interactions are also observed.
Objectives
To focus on three issues that may influence the predictive value of the [I]/Ki ratio approach: (i) the use of unbound Ki (Ki,u) values generated from standardised in vitro experiments compared with literature values; (ii) the selection of an appropriate [I]; and (iii) incorporation of the impact of intestinal metabolic inhibition for cytochrome P450 (CYP) 3A4 predictions. To this end we have selected eight inhibitors of CYP2C9, CYP2D6 and CYP3A4 and 18 victim drugs from a previous database analysis to allow prediction of 45 clinical DDI studies.
Methods
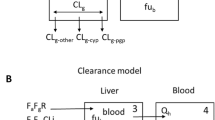
In vitro kinetic and inhibition studies were performed in human liver microsomes using prototypic probe substrates of CYP2C9 and CYP2D6, with various inhibitors (miconazole, sulfaphenazole, fluconazole, ketoconazole, quinidine, fluoxetine, fluvoxamine). The Ki estimates obtained were corrected for non-specific microsomal binding, and the Ki,u was incorporated into in vivo predictions using various [I] values. Predictions for CYP3A4 were based upon in vitro data obtained from a previous publication within our laboratory, and an assessment of the impact of the interaction in the gut wall is included. Predictions were validated against 45 in vivo studies and those within 2-fold of the in vivo ratio of area under the plasma concentration-time curve of the substrate, in the presence and absence of the inhibitor (AUCi/AUC) were considered successful.
Results
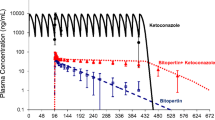
Predictions based upon the average systemic total plasma drug concentration ([I]av) [incorporating the effects of parallel drug elimination pathways] and the Ki,u value resulted in 91% of studies predicted to within 2-fold of the in vivo AUCi/AUC. This represents a 35% improvement in prediction accuracy compared with predictions based upon total Ki values obtained from various published literature sources. A corresponding reduction in bias and an increase in precision were also observed compared with the use of other [I] surrogates (e.g. the total and new unbound maximum hepatic input plasma concentrations). No significant improvement in prediction accuracy was observed by incorporating consideration of gut wall inhibition for CYP3A4.
Conclusion
DDI predictions based upon the use of Ki,u data obtained under a set of optimal standardised conditions were significantly improved compared with predictions using in vitro data collated from various sources. The use of [I]av as the [I] surrogate generated the most successful predictions as judged by several criteria. Incorporation of either plasma protein binding of inhibitor or gut wall CYP3A4 inhibition did not result in a general improvement of DDI predictions.







Similar content being viewed by others
References
Tucker GT, Houston JB, Huang S-M. Optimising drug development: strategies to assess drug metabolism/transporter interaction potential: toward a consensus. Br J Clin Pharmacol 2001 Jul; 52(1): 107–17
Yao C, Levy RH. Inhibition-based metabolic drug-drug interactions: predictions from in vitro data. J Pharm Sci 2002 Sep; 91(9): 1923–35
Ito K, Brown HS, Houston JB. Database analyses for the prediction of in vivo drug-drug interactions from in vitro data [published erratum appears in Br J Clin Pharmacol 2004 Nov; 58 (5): 565-8]. Br J Clin Pharmacol 2004 Apr; 57(4): 473–86
Lin JH, Pearson PG. Prediction of metabolic drug interactions: quantitative or qualitative. In: Rodrigues D, editor. Drug-drug interactions. New York: Marcel Dekker Inc., 2002: 415–438
Ito K, Chiba K, Horikawa M, et al. Which concentration of the inhibitor should be used to predict in vivo drug interactions from in vitro data? AAPS Pharm Sci 2002; 4(4): E25
Rostami-Hodjegan A, Tucker GT. ’In silico’ simulations to assess the ‘in vivo’ consequences of ‘in vitro’ metabolic drug-drug interactions. Drug Discov Today 2004; 1(4): 441–8
Ito K, Hallifax D, Obach RS, et al. Impact of parallel pathways of drug elimination and multiple CYP involvement on drug-drug interactions: CYP2D6 paradigm. Drug Metab Dispos 2005 Jun; 33(6): 837–44
Brown HS, Ito K, Galetin A, et al. Prediction of in vivo drug-drug interactions from in vitro data: impact of incorporating parallel pathways of drug elimination and inhibitor absorption rate constant. Br J Clin Pharmacol 2005 Nov; 60(5): 508–18
Bjornsson TD, Callaghan JT, Einolf HJ, et al. The conduct of in vitro and in vivo drug-drug interaction studies: a pharmaceutical research and manufacturers of America (PhRMA) perspective. Drug Metab Dispos 2003 Jul; 31(7): 815–32
Galetin A, Burt H, Gibbons L, et al. Prediction of time-dependent CYP3A4 drug-drug interactions: impact of enzyme degradation, parallel elimination pathways and intestinal inhibition. Drug Metab Dispos 2006; 34(1): 166–75
Obach RS, Walsky RL, Venkatakrishnan K, et al. In vitro cytochrome P450 inhibition data and the prediction of drug-drug interactions: qualitative relationships, quantitative predictions, and the rank order approach. J Pharmacol Exp Ther. Epub 2005 Sep 28
Hall SD, Thummel KE, Watkins PB, et al. Molecular and physical mechanisms of first-pass extraction. Drug Metab Dispos 1999 Feb; 27(2): 161–6
Ito K, Iwatsubo T, Kanamitsu S, et al. Quantitative prediction of in vivo drug clearance and drug interactions from in vitro data on metabolism, together with binding and transport. Ann Rev Pharmacol Toxicol 1998 Apr; 38: 461–99
Ito K, Iwatsubo T, Kanamitsu S, et al. Prediction of pharmacokinetic alterations caused by drug-drug interactions: metabolic interaction in the liver. Pharmacol Rev 1998 Sep; 50(3): 387–411
Blanchard N, Richert L, Coassolo P, et al. Qualitative and quantitative assessment of drug-drug interaction potential in man, based on Ki, IC50 and inhibitor concentration. Curr Drug Metab 2004 Apr; 5(2): 147–56
Yuan R, Parmelee T, Balian JD, et al. In vitro metabolic interaction studies: experience of the food and drug administration. Clin Pharmacol Ther 1999 Jul; 66(1): 9–15
Huang S-M. Drug interaction studies: study design, data analysis, and implications for dosing and labelling [online]. Available from URL: http://www.fda.gov/ohrms/dockets/ac/04/briefing/2004-4079B1_04_Topic2-TabA.htm [Accessed 2004]
Houston JB, Kenworthy KE, Galetin A. Typical and atypical enzyme kinetics. In: Lee JS, Obach RS, Fisher MB, editors. Drug metabolizing enzymes: cytochrome P450 and other enzymes in drug discovery and development. New York: Fontis Media, Marcel Dekker, 2003
Galetin A, Ito K, Hallifax D, et al. CYP3A4 substrate selection and substitution in the prediction of potential drug-drug interaction. J Pharmacol Exp Ther 2005 Jul; 314(1): 180–90
Kenworthy KE, Bloomer JC, Clarke SE, et al. CYP3A4 drug interactions: correlation of 10 in vitro probe substrates. Br J Clin Pharmacol 1999 Nov; 48(5): 716–27
Debruyne D, Ryckelynck JP. Clinical pharmacokinetics of fluconazole. Clin Pharmacokinet 1993 Jan; 24(1): 10–27
Barriere SL. Pharmacology and pharmacokinetics of traditional systemic antifungal agents. Pharmacother 1990, 40S
Preskorn SH. Clinically relevant pharmacology of selective serotonin reuptake inhibitors: an overview with emphasis on pharmacokinetics and effects on oxidative drug metabolism. Clin Pharmacokinet 1997; 32 Suppl. 1: 1–21
Edwards DJ, Axelson JE, Slaughter RL, et al. Factors affecting quinidine protein binding in humans. J Pharm Sci 1984 Sep; 73(9): 1264–7
Wang Y-H, Jones DR, Hall SD. Prediction of cytochrome P450 3A inhibition by verapamil enantiomers and their metabolites. Drug Metab Dispos 2004 Feb; 32(2): 259–66
Olkkola KT, Ahonen J, Neuvonen PJ. The effects of the systemic antimycotics, itraconazole and fluconazole, on the pharmacokinetics and pharmacodynamics of intravenous and oral midazolam. Anesth Analg 1996 Mar; 82(3): 511–6
Floren LC, Bekersky I, Benet LZ, et al. Tacrolimus oral bioavailability doubles with coadministration of ketoconazole. Clin Pharmacol Ther 1997 Jul; 62(1): 41–9
Lennernas H. Clinical pharmacokinetics of atorvastatin. Clin Pharmacokinet 2003; 42(13): 1141–60
Sheiner BL, Beal SL. Evaluation of methods for estimating pharmacokinetic parameters: II. bioexponential model and experimental pharmacokinetic data. J Pharmacokinet Biopharm 1981 Oct; 9(5): 635–51
Obach RS, Baxter JG, Liston TE, et al. The prediction of human pharmacokinetic parameters from preclinical and in vitro metabolism data. J Pharmacol Exp Ther 1997 Oct; 283(1): 46–58
Venkatakrishnan K, von Moltke LL, Obach RS, et al. Drug metabolism and drug interactions: application and clinical value of in vitro models. Curr Drug Metab 2003 Oct; 4(5): 423–59
Hickman D, Wang JP, Wang Y, et al. Evaluation of the selectivity of in vitro probes and suitability of organic solvents for the measurement of human cytochrome P450 monooxygenase activities. Drug Metab Dispos 1998 Mar; 26(3): 207–15
Chauret N, Gauthier A, Nicoll-Griffith DA. Effect of common organic solvents on in vitro CYP450 mediated metabolic activities in human liver microsomes. Drug Metab Dispos 1998 Jan; 26(1): 1–4
Tran TH, von Moltke LL, Venkatakrishnan K, et al. Microsomal protein concentration modifies the apparent inhibitory potency of CYP3A inhibitors. Drug Metab Dispos 2002 Dec; 30(12): 1441–5
Margolis JM, Obach RS. Impact of non-specific binding to microsomes and phospholipid on the inhibition of cytochrome P4502D6: Implications for relating in vitro inhibition data to in vivo drug interactions. Drug Metab Dispos 2003 May; 31(5): 606–11
von Moltke LL, Greenblatt DJ, Schmider J, et al. In vitro approaches to predicting drug interactions in vivo. Biochem Pharmacol 1998 Jan; 55(2): 113–22
McGinnity DF, Tucker J, Trigg S, et al. Prediction of CYP2C9 mediated drug-drug interactions: a comparison using data from recombinant enzymes and human hepatocytes. Drug Metab Dispos 2005 Nov; 33(11): 1700–7
Grime K, Riley RJ. The impact of in vitro binding on in vitro-in vivo extrapolations, projections of metabolic clearance and clinical drug-drug interactions. Curr Drug Metab 2006; 7(3): 251–64
Tsunoda SM, Velez RL, von Moltke LL, et al. Differentiation of intestinal and hepatic cytochrome P450 3A activity with the use of midazolam as an in vivo probe: effect of ketoconazole. Clin Pharmacol Ther 1999 Nov; 66(5): 461–71
Acknowledgements
This work was funded by a consortium of pharmaceutical companies (AstraZeneca, GlaxoSmithKline, Bristol Myers Squibb, F. Hoffmann La Roche, Novartis, Pfizer and Servier) within the Centre for Applied Pharmacokinetic Research at the University of Manchester. H.S. Brown was financially supported by a Bristol Myers Squibb studentship. The authors wish to thank Kiyomi Ito and Susan Murby for valuable contributions to this work.
The authors have no conflict of interests directly relevant to the content of this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Brown, H.S., Galetin, A., Hallifax, D. et al. Prediction of In Vivo Drug-Drug Interactions from In Vitro Data. Clin Pharmacokinet 45, 1035–1050 (2006). https://doi.org/10.2165/00003088-200645100-00006
Published:
Issue Date:
DOI: https://doi.org/10.2165/00003088-200645100-00006