
Genomic evolutionary trajectory of metastatic squamous cell carcinoma of the lung
Introduction
Distant metastases are more common in lung adenocarcinoma (LUAD) than in lung squamous cell carcinoma (LUSC). This is one reason why metastatic LUSC has been understudied (1). Most published studies on metastatic LUSC are single case reports, often without genomic analysis of the metastasis due to a lack of sufficient material, or with a limited approach as panel sequencing or FISH (2-5). Advanced LUSC tends to affect older patients with more comorbidities than LUAD, which makes tissue sampling for research studies challenging (6). In daily practice, the biopsies of LUSC metastases are often not large enough for whole exome or whole genome sequencing. Therefore, the mechanisms and evolutionary trajectories of metastasis in LUSC remain poorly understood.
Multi-region sampling and comprehensive molecular profiling of resected tumors have greatly advanced our understanding of intratumoral heterogeneity (ITH) in lung and other cancer types (7). However, the high costs and the diagnostic reality of mostly small biopsy specimens from patients with advanced LUSC make such multi-region analyses difficult if not impossible in clinical practice. Molecular profiling of LUSC is further challenged by the often low tumor cell content of LUSC tissue (8). To overcome this hurdle, we previously showed that the tumor purity of LUAD can be greatly enhanced by an advanced nuclei flow-sorting technique (9). Briefly, cells were stained with 4’,6-Diamin-2-phenylindol (DAPI) and an antibody for TTF-1 to isolate LUAD specific cells. This flow-sorting approach allowed us to distinguish and isolate aneuploid and diploid populations enabling highly informative genomic profiling (9).
Here, we applied a modified nuclei flow-sorting approach to enrich for cytokeratin-labelled tumor nuclei from tumor tissue specimens. An antibody was used for labelling pan-cytokeratins that are highly expressed in LUSC but not in admixed benign mesenchymal cells. Whole exome sequencing (WES) was performed to investigate ITH and clonal evolution from five patients with LUSC and matched spatiotemporal metastases. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-21-48).
Methods
Patient cohort
The retrospective study comprised the following criteria for the inclusion of patients: (I) histologically confirmed LUSC with regional or distant metastases; (II) large enough specimens (i.e., surgical specimens or large biopsies) to allow for nuclei flow-sorting and WES; (III) at least two available tumor samples, fresh-frozen (FF) or formalin-fixed and paraffin-embedded (FFPE), one of which had to be the primary tumor (primary LUSC) and the other the matched metastasis; (IV) tumor samples that had been taken from different time points or body sites of the patient. Multiple regions within the primary tumor were not sampled. Five patients with primary LUSC and six matched metastases were investigated. Tumor samples, FF (n=10) or FFPE (n=6, Table 1), were obtained from the archive of the Institute of Medical Genetics and Pathology of the University Hospital Basel, Switzerland. Histological slides were reviewed by a pulmonary pathologist (LB). The areas with highest tumor cell density were marked for subsequent dissection, avoiding areas rich in stromal cells or benign epithelial cells from entrapped bronchoalveolar tissue. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the local Ethics Committee of Both Basels (EKBB) and Ethics Committee Northwest and Central Switzerland (EKNZ) (NO.: EKBB/EKNZ 31/12) and individual consent for this retrospective analysis was waived.
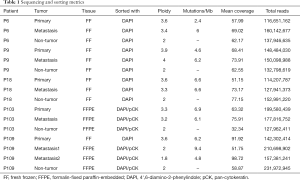
Full table
Nuclei isolation and multiparameter flow-sorting
Nuclei isolation from FF and FFPE samples was conducted as described previously (9-12). All nuclei were DAPI stained for sorting, DNA quantification and ploidy analysis. FF tumor nuclei were only sorted with DAPI (n=7, four patients). Pan-Cytokeratin (pCK, clone MNF116, Dako, Code-Nr. M0821) was used for FFPE tumors as an additional marker for flow-sorting. The applied pCK antibody is specific against CK5, CK6, CK8, CK17 and CK19 and highly expressed in LUSC (13,14). Cytokeratin positive cytoplasmic remnants seem to stay attached to the nuclei of FFPE tissue but are washed away in FF tissue during the procedure. Therefore, only FFPE samples were stained with pCK. This step was crucial to enable refined separation of diploid from aneuploid tumor populations and benign diploid epithelial and stromal cell nuclei in FFPE tissue (n=2, P109, Figure S1). Previously, we described the utility of sorted diploid population as germline (9). In DAPI-sorted aneuploid tumors, the matched diploid populations were used as germline for WES (P6, P9, P18, Figure S2). In samples sorted by DAPI and pCK, diploid pCK-negative populations were used as germline (P103, P109) (9). Initial purity was estimated after sorting by treating all sorted nuclei as 100% and calculating all sorted populations according to their proportions.
DNA extraction and whole exome sequencing
DNA from FFPE isolated nuclei was directly extracted using DNeasy blood and tissue kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s instructions and quantified using the Qubit Fluorometer assay (Life Technologies, Carlsbad, CA, USA), whereas DNA from FF samples was first whole genome amplified using the illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare Life Sciences, Little Chalfont, Buckinghamshire, UK; product: 25660031) as previously described (9,10). In samples that were exclusively sorted with DAPI, the diploid population was used as germline, whereas diploid pCK-negative populations were considered as germline when applicable (9). Flat genomes were considered as a germline (Figure S2). According to manufacturer’s guidelines SureSelect Human All Exon V6 Kit (Agilent) was utilized for the whole exome capturing. Paired-end 100-bp reads were generated on the Illumina NovaSeq 6000. Sequencing was conducted by CeGaT (Tübingen, Germany).
Human genome GRCh37 was used as a reference for aligning the reads. Whole exome sequencing (WES) analysis, comprising sequence processing, calling of somatic single nucleotide variants (SNVs), small insertions and deletions (indels), and allele-specific copy number variations (CNAs), detecting mutational signatures and identifying clonality, was performed using state-of-the-art bioinformatic methods, as previously described with modifications (15,16).
Briefly, SNVs and indels were called using MuTect v1.1.7 and strelka v2.9.10, respectively (17,18). To normalize the FF and FFPE samples we used following approach: All SNVs C:G>T:A with variant allelic fractions (VAFs) less than 10% were discarded to reduce false positive results. Otherwise, SNVs or indels with VAFs <1% or that were supported by fewer than 3 reads were not considered. If SNVs and indels were found in one tumor of a patient, a cut-off of two reads was applied to the remaining tumors. We further excluded variants identified in at least two of a panel of 123 non-tumor samples, including the non-tumor samples included in our study. Variant annotation was performed using SnpEff software v4.1 (19).
The heatmap of non-synonymous mutations was generated using the R package maftools v.2.0.16 by selecting the genes in the Bailey et al. dataset (20) and the significant mutated genes in TCGA dataset (MutSig Q-value <0.05 based on 511 LUSC samples; TCGA, Firehose Legacy, http://www.cbioportal.org) that represents the significantly mutated genes in LUSC.
Analysis of copy number aberrations and clonality
FACETS v0.5.14 enabled the identification of allele-specific CNAs (21). Cancer cell fraction (CCF) of each mutation was inferred using ABSOLUTE v1.0.6 (22). A mutation was classified as clonal, if its probability of being clonal was >50% or if the lower bound of the 95% confidence interval of its CCF was >90%. Mutations were regarded as subclonal if they did not meet the mentioned criteria (23,24).
Phylogenetic analysis and mutational signatures
As described by Murugaesu et al., a maximum parsimony tree was built for each case using binary presence/absence matrices based on the repertoire of non-synonymous and synonymous somatic mutations in the biopsies of the tumors (25-27).
A mutation that was found in both tumor manifestations of one patient was considered as ‘trunk’. Mutations that were detected in only one tumor of the patient were considered as ‘branch’ or ‘private’.
Decomposition of mutational signatures was conducted using the R package deconstructSigs software by selecting the mutational signatures 1, 2, 4, 5 and 13, which are based on the set of 30 mutational signatures (COSMIC) that were observed in LUSC (28). To have a significant result, the amount of mutations must be >50 for the algorithm. For P6 (primary LUSC and metastasis), P109 primary LUSC and P18 primary LUSC it was <50. However, these results can serve as hints to further understand the mechanism of tumor evolution. Phylogenetic trees were drawn using FigTree v1.4.4.
Statistical analysis
All statistical analyses were performed using R software v3.6.3 (R Core Team, 2020). A Wilcoxon test was performed using the R function “wilcox.test” to check whether the primary LUSCs have a significantly different number of non-synonymous mutations than the metastases.
Results
Nuclei flow-sorting achieves high purity of tumor DNA
In this study, we investigated five patients with primary LUSC and their metastases from different sites to explore ITH and clonal evolution (Table 2). All patients were male, and three were former or current smokers, whereas the smoking status was unknown in two patients. Our tissue material cohort consisted of ten FF (Fresh Frozen) and six FFPE (Formalin-fixed paraffin-embedded) samples (Table 1). Although flow-sorting by DAPI allows separating aneuploid tumor nuclei from benign stromal tissue and epithelial cells, it does not detect diploid tumor cells (9,10). To enable genomic profiling of diploid tumor populations in LUSC, we implemented, in addition to DAPI, pCK staining to separately select diploid (2N±0.2) and/or aneuploid (>2.2N) tumor cell populations and remove pCK-negative diploid and near tetraploid benign cells. Across the five patients, we isolated nine aneuploid tumor cell populations using either DAPI alone or DAPI/pCK (Table 1). Diploid tumor cell populations were identified only by DAPI/pCK sorting (Table 1). Our flow-sorting approach enabled us to increase the initial mean purity from 26% (range, 12–50%) of unsorted tumor to a mean of 73% (range, 42–93%) after sorting (Figure S3).
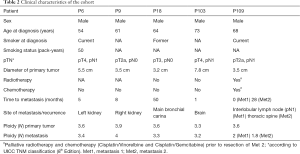
Full table
Mutational landscape of primary-metastatic pairs of LUSC
We performed WES of the sorted tumor cell nuclei and normal samples to a median depth of 69x (range, 58×–99×) and 62× (range, 32×–77×) respectively (Table 1). Median somatic mutation rate was 6.2 mutations/Mb., primary LUSCs and metastases mutation rates were similar (primary LUSC median 6.2; range 2.4–6.9 vs. metastases median 6.1, range 4.8–9.4, Table 1). The five primary LUSCs do not have fewer non-synonymous mutations than the metastases (average 325; range 144–414 vs. average 393.3; range 290–568, Wilcoxon test P value =0.79). A median of 74% (range, 65–94%) of non-synonymous mutations in the primary LUSC were also found in the corresponding metastases (Figure 1A and https://cdn.amegroups.cn/static/public/tlcr-21-48-1.xlsx). On the other hand, a median of 69.7% (range, 35–93%) of the non-synonymous mutations found in the metastases were found in the matched primaries. The most commonly mutated cancer-related genes known to be mutated LUSC included TP53 (9 samples, 4 patients), FAT1 [5, 2], FBXW7 [5, 2], KMT2D [5, 2] and ARID1A [2, 2] (Figure 1B) (29-32).
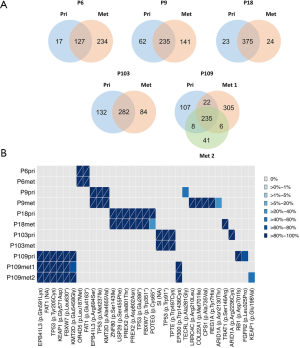
PT and metastatic samples of LUSC shared common mutations known to drive tumorigenesis and tumor progression. Clonality analysis of primary LUSCs and matched metastases revealed that mutations in driver genes, such as the tumor suppressor genes TP53, FBXW7 and KMT2D, were predominantly clonal (Figure 1B). Clonal mutations between the primary LUSCs and metastases were found in each patient. Primary LUSC of P103 and metastases of P9 and P109 displayed private mutations in cancer-related gene.
In all five patients, the trunks were much longer than branches (Figure 2). P109 harbored a long mutational trunk and similar number of mutations in Met1. Private mutations of P109 primary LUSC and Met2 had a much shorter branch compared to Met1. Notably, P109 indicated a common clonal trunk with its two metastases, a lymph node (Met1) and a thoracic spine metastasis (Met2).
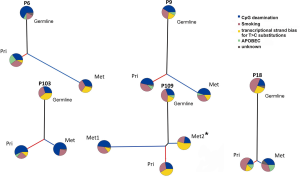
Analysis of mutational signatures revealed that COSMIC signature 1 (CpG deamination, related to aging) and COSMIC signature 4 (related to smoking) were most common being present in all samples across all of our patients (33). COSMIC signature 5 (transcriptional strand bias, related to aging) was present in every primary-metastasis pair but not in all trunks and branches. The APOPEC signature was variably distributed. It was identified in trunk and both branches separately of primary LUSC and metastasis in P9. In contrast it was truncal in P109 but restricted to the branches in P18, and only found in the primary LUSC branch of P103. In P6, it was present in the trunk and the primary LUSC branch but not in the branch of the metastasis.
Taken together, the mutational profiles between the primary LUSCs and their metastasis are closely related as previously seen in LUAD (9). The long trunks of mutations can be primarily attributed to the effects of aging and smoking. The pattern of mutational signatures varied between patients and during the evolution of individual tumors.
Chromosomal aberrations are truncal events
CNAs of the primary LUSCs were highly concordant with those in matched metastases (Figure 3A, Figure S4). In primary LUSCs, a median of 79% (range, 67–85%) of CNAs were also observed in the matched metastases, while a median of 79% (range, 70–99%) of the aberrations in the metastases were also seen in the matched primaries. All tumor samples, primary LUSCs and metastases, shared amplified regions in 3q26.1-33 including the genes PIK3CA, SOX2 and TERC). In addition, there was truncal TERT amplification at 5p15.33 in three of the five patients. We found the following tumor suppressor genes to be lost in our cohort: ATM (2 samples, 2 patients), ARID1A [3, 2], APC [7, 4], CDKN2A [9, 4], CSMD1 [9, 4], FAT1 [3, 2], FOXP1 [8, 4], PTEN [5, 2], SETD2 [8, 4] and amplifications were detected in SOX2 [10, 5], MECOM [10, 5], TERC [10, 5], MYC [4, 2] and CCND1 [4, 2].
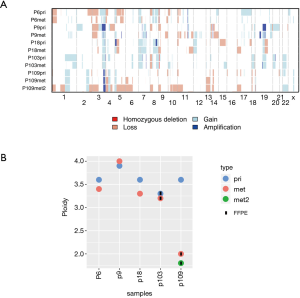
Both chromosomal profiles and ploidy of primary LUSCs and matched metastases were highly similar in most patients (Figure 3B, Figure S4). All primary LUSCs were aneuploid, including primary LUSC of P109 (3.6 N). Interestingly, both metastases of P109 were diploid (2N±0.2N).
Taken together, CNAs and ploidy status were very similar in the primary LUSCs and their metastases suggesting CNAs as early events with limited heterogeneity between primary LUSC and metastasis.
Discussion
In this study, we explored the spatiotemporal heterogeneity and evolution of LUSC by WES using highly enriched nuclei from five primary LUSCs and matched metastases. Intratumoral heterogeneity (ITH) and clonal evolution of primary NSCLC using comprehensive genomic profiling strategies have been analyzed in several previous studies (7,34-36). Although metastasis is the leading cause of death in NSCLC, data on matched pairs of primary LUSCs and metastases remain scarce (9,37,38).
Two other studies have recently been published on spatial and temporal heterogeneity of metastatic LUSC from a small number of patients (39,40). Leong et al. found that the interactions of the DNA repair genes with the tumor microenvironment may play a role in the acquisition of somatic mutations following the metastasis during lung cancer progression (39). Another recent study on metastatic LUSC, demonstrated that large structural variants may play a crucial role in intratumoral genetic heterogeneity (40).
Here, we demonstrate high concordance between the primary-metastasis pairs in terms of SNVs and CNAs in most of the patients. Most aberrations in LUSC were truncal indicating only limited heterogeneity between primary LUSC and metastasis and low subclonal diversity (9). This is in line with the data of a recent pan-cancer study on metastatic solid tumors that included treatment-naive and extensively treated tumors (41). This study revealed that 96% of driver mutations within metastases were clonal with only 4% of subclonal mutations (41). The potential clinical impact of subclonal diversity remains poorly studied. We could adequately address the question whether treatment might affect subclonal diversity in our study since four of the five patients had not received systemic treatment prior to tissue sampling (34).
The high concordance between primary-metastasis pairs in our study corroborates our previous data on a cohort of LUAD (9). In our previous study, more than 80% of CNAs and cancer-related gene mutations were shared between the primary-metastatic LUAD specimens. Ploidy remained stable and did not change over time (9). Here, we found that CNAs and ploidy were equally stable during the metastatic spread of LUSC. This stable aneuploidy in most LUSC and LUAD patients highlights the previously proposed critical importance of a whole genome doubling (WGD) event that might select for fitness to progress and metastasize (42,43). The only discrepant ploidy was found in P109 revealing an aneuploid primary LUSC (3.6N) and two diploid/near-diploid metastases (2N and 1.8N, respectively). Reversion from an aneuploid to a diploid state during progression is unlikely, since WGD is considered irreversible (44). Therefore, we hypothesize that the diploid metastases had emerged from a diploid population prior to WGD of the primary LUSC in this patient. We do not know whether the diploid population had been fully replaced by the aneuploid population after WGD, or whether we have missed a coexistent diploid population due to limited sampling of the primary LUSC. Although most LUSC and LUAD are aneuploid and metastasize after WGD, this example illustrates that stably diploid tumor cell populations can occasionally serve as the backbone of progression and metastasis in NSCLC (9). We have also previously observed this phenomenon in prostate cancer, where a diploid tumor cell population was consistently detectable in repeated biopsies during progression giving rise to temporary aneuploid populations (10).
Somatic mutations in cancers are caused by different mutational processes that generate a characteristic mutational signature (45). The pathogenesis of the vast majority of NSCLC is driven by the accumulation of smoking-induced alterations (7,46). This is in line with the large proportion of signature 4 in our tumor samples reflecting the contribution of smoking-related DNA damage for early acquisition and accumulation of truncal mutations as previously proposed. The smoking status of P9 and P103 was not known. However, the signature profiles showed a significant amount of smoking signature 4, suggesting that these patients were former or current smokers. Mutational signatures were previously inferred to track the evolutionary differences between the primary LUSCs and the metastases (32). In breast cancer, Ng et al. found significant differences in mutational signatures comparing private mutations of the primary tumor and metastasis (27). The discrepancy might contribute to spatial intratumoral heterogeneity. Our study could support these previous results. Mutational signatures changed during metastasis and over time revealing no obvious pattern. Despite the common trunk, the mutational signatures differed in the private branches of the primary LUSCs and metastases. Furthermore, the long trunk of shared genomic alterations in most samples indicates a linear progression model in the metastatic dissemination caused by one major subclone (41,43). In fact, a common ancestor acquired genomic alterations and remained genetically stable even after late dissemination. Nevertheless, there were substantial differences between the private branches of primary LUSCs and metastases. This mutational heterogeneity might be relevant in the context of specific treatments. Notably, Richard et al. have recently reported that different mutational signatures had an impact on the efficacy of immune checkpoint inhibitors (47).
Our study provides new insight into the genomic trajectories of genomic evolution of metastatic LUSC but did not reveal novel specific alterations beyond those that have already been reported by large-scale sequencing consortia (48). Among CNAs, it was most striking that all tumors shared amplifications at 3q26.1-29 [30–40% in cBioPortal (48)] indicating an important role of these CNAs for tumor initiation (49,50). SOX2 and TP63 are among the genes in the amplified 3p26.1-29 region and associated with squamous differentiation (51-53). Notably, SOX2 and p63 are known to be amplified in at least 40% and 30%, respectively, of all LUSC but almost never in LUAD (46).
Our study has several limitations. The low number of patients precludes strong general statements on the genomic trajectories of LUSC. Further confounding factors are the different states of tissue fixation (FF vs. FFPE) and sorting strategies (DAPI vs. DAPI/pCK). The variability in sample quality and the small cohort size is a major limitation but reflects the general difficulties to access appropriate samples from matched primary LUSCs and metastases. Nevertheless, our collection of LUSC is unique being the largest series undergoing WES with matched primary LUSC and metastasis pairs of LUSC so far. Due to incomplete clinical documentation of archived cases, we were unable to reconstruct the full clinicopathological history including smoking history (54). Using WES, we were bound to coding regions preventing any observations on translocations. Furthermore, limited by the sequencing depth and filtering, it is possible that subclonal variants were missed due to detection limits. In fact, one of the limitations of this study, due the possible presence of sequencing and FFPE artefacts, is to remove the mutations with an allele frequency lower then 1% and/or validated by at least 3 reads. This means, especially for the samples with a mean coverage of ~50–70x, failing to detect any subclone with an allele frequency lower than 4–6% even in the ideal circumstances.
Conclusions
Taken together, our study provides new insights into the intratumoral heterogeneity and genomic patterns of primary LUSC and matched metastases using purified tumor populations after nuclei flow-sorting. A close clonal relationship with only limited heterogeneity was revealed in the primary-metastatic pairs, similar to what was seen in LUAD, suggesting late dissemination of the metastases from the primary tumors during tumor evolution. Further studies with larger matched cohorts are required to better elucidate the critical genomic players and related molecular pathways of metastatic disease in NSCLC.
Acknowledgments
We acknowledge CeGaT (Tübingen, Germany) for the technical expertise in sequencing.
Funding: This work was supported by: Swiss National Foundation [SNF 320030_162781 to LB, CR, PZ00P3_168165 to SP]; the Swiss Cancer League [KFS-3995-08-2016 to SP, KFS-4543-08-2018 to CKYN].
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-21-48
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-21-48
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-21-48
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-21-48). LB reports personal fees from MSD, BMS, Roche, Bayer, Takeda, Astra Zeneca and Boehringer Ingelheim outside the submitted work. TL reports grants from Swiss National Science Foundation during the conduct of the study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the local
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhou H, Dong D, Chen B, et al. Diagnosis of Distant Metastasis of Lung Cancer: Based on Clinical and Radiomic Features. Transl Oncol 2018;11:31-6. [Crossref] [PubMed]
- Guo Y, Wang X, Xiao J, et al. Lung squamous cell carcinoma with solitary ocular metastasis and its successful treatment with thoracic surgery and chemotherapy: an interesting and rare case report. BMC Cancer 2018;18:1004. [Crossref] [PubMed]
- Talbot SG, Estilo C, Maghami E, et al. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res 2005;65:3063-71. [Crossref] [PubMed]
- Qiong Z, Na WY, Bo W, et al. Alterations of a spectrum of driver genes in female Chinese patients with advanced or metastatic squamous cell carcinoma of the lung. Lung Cancer 2015;87:117-21. [Crossref] [PubMed]
- Paik PK, Shen R, Won H, et al. Next-Generation Sequencing of Stage IV Squamous Cell Lung Cancers Reveals an Association of PI3K Aberrations and Evidence of Clonal Heterogeneity in Patients with Brain Metastases. Cancer Discov 2015;5:610-21. [Crossref] [PubMed]
- Daaboul N, Nicholas G, Laurie SA. Algorithm for the treatment of advanced or metastatic squamous non-small-cell lung cancer: an evidence-based overview. Curr Oncol 2018;25:S77. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun 2015;6:8971. [Crossref] [PubMed]
- Lorber T, Andor N, Dietsche T, et al. Exploring the spatiotemporal genetic heterogeneity in metastatic lung adenocarcinoma using a nuclei flow-sorting approach. J Pathol 2019;247:199-213. [Crossref] [PubMed]
- Ruiz C, Lenkiewicz E, Evers L, et al. Advancing a clinically relevant perspective of the clonal nature of cancer. PNAS 2011;108:12054-9. [Crossref] [PubMed]
- Krishan A, Dandekar PD. DAPI fluorescence in nuclei isolated from tumors. J Histochem Cytochem 2005;53:1033-6. [Crossref] [PubMed]
- Corver WE, Ter Haar NT. High-resolution multiparameter DNA flow cytometry for the detection and sorting of tumor and stromal subpopulations from paraffin-embedded tissues. Curr Protoc Cytom 2011;7:Unit 737.
- Blobel GA, Moll R, Franke WW, et al. Cytokeratins in normal lung and lung carcinomas. Virchows Arch B Cell Pathol Incl Mol Pathol 1984;45:407-29. [Crossref] [PubMed]
- Quinlan RA, Schiller DL, Hatzfeld M, et al. Patterns of expression and organization of cytokeratin intermediate filaments. Ann N Y Acad Sci 1985;455:282-306. [Crossref] [PubMed]
- Nuciforo S, Fofana I, Matter MS, et al. Organoid Models of Human Liver Cancers Derived from Tumor Needle Biopsies. Cell Rep 2018;24:1363-76. [Crossref] [PubMed]
- Bertucci F, Ng CKY, Patsouris A, et al. Genomic characterization of metastatic breast cancers. Nature 2019;569:560-4. [Crossref] [PubMed]
- Saunders CT, Wong WS, Swamy S, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012;28:1811-7. [Crossref] [PubMed]
- Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213-9. [Crossref] [PubMed]
- Cingolani P, Platts A. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118 iso-2 iso-3. Fly 2012;6:80-92. [Crossref] [PubMed]
- Bailey MH, Tokheim C, Porta-Pardo E, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018;174:1034-5. [Crossref] [PubMed]
- Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44:e131 [Crossref] [PubMed]
- Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012;30:413-21. [Crossref] [PubMed]
- Landau DA, Carter SL, Stojanov P, et al. Evolution and Impact of Subclonal Mutations in Chronic Lymphocytic Leukemia. Cell 2013;152:714-26. [Crossref] [PubMed]
- Guerini-Rocco E, Piscuoglio S, Ng CK. Microglandular adenosis associated with triple-negative breast cancer is a neoplastic lesion of triple-negative phenotype harbouring TP53 somatic mutations. J Pathol 2016;238:677-88. [Crossref] [PubMed]
- Murugaesu N, Wilson GA, Birkbak NJ, et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov 2015;5:821-31. [Crossref] [PubMed]
- Krause A, Roma L, Lorber T, et al. Deciphering the clonal relationship between glandular and squamous components in adenosquamous carcinoma of the lung using whole exome sequencing. Lung Cancer 2020;150:132-8. [Crossref] [PubMed]
- Ng CKY, Bidard FC, Piscuoglio S, et al. Genetic Heterogeneity in Therapy-Naïve Synchronous Primary Breast Cancers and Their Metastases. Clin Cancer Res 2017;23:4402-15. [Crossref] [PubMed]
- Rosenthal R, McGranahan N, Herrero J, et al. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 2016;17:31. [Crossref] [PubMed]
- Zhang XC, Wang J, Shao GG, et al. Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat Commun 2019;10:1772. [Crossref] [PubMed]
- Li Q, Hou J, Hu Z, et al. Multiple mutations of lung squamous cell carcinoma shared common mechanisms. Oncotarget 2016;7:79629-36. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. Corrected in Nature 2013;502:258. [Crossref] [PubMed]
- de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014;346:251-6. [Crossref] [PubMed]
- Zhang LL, Kan M, Zhang MM, et al. Multiregion sequencing reveals the intratumor heterogeneity of driver mutations in TP53-driven non-small cell lung cancer. Int J Cancer 2017;140:103-8. [Crossref] [PubMed]
- Sharma A, Merritt E, Hu X, et al. Non-Genetic Intra-Tumor Heterogeneity Is a Major Predictor of Phenotypic Heterogeneity and Ongoing Evolutionary Dynamics in Lung Tumors. Cell Rep 2019;29:2164-2174.e5. [Crossref] [PubMed]
- Suda K, Suda K, Kim J, et al. Innate Genetic Evolution of Lung Cancers and Spatial Heterogeneity: Analysis of Treatment-Naïve Lesions. J Thorac Oncol 2018;13:1496-1507. [Crossref] [PubMed]
- Roper N, Gao S, Maity TK, et al. APOBEC Mutagenesis and Copy-Number Alterations Are Drivers of Proteogenomic Tumor Evolution and Heterogeneity in Metastatic Thoracic Tumors. Cell Rep 2019;26:2651-2666.e6. [Crossref] [PubMed]
- Leong TL, Gayevskiy V, Steinfort DP, et al. Deep multi-region whole-genome sequencing reveals heterogeneity and gene-by-environment interactions in treatment-naive, metastatic lung cancer. Oncogene 2019;38:1661-75. [Crossref] [PubMed]
- Peng Y, Yuan C, Tao X, et al. Integrated analysis of optical mapping and whole-genome sequencing reveals intratumoral genetic heterogeneity in metastatic lung squamous cell carcinoma. Transl Lung Cancer Res 2020;9:670-81. [Crossref] [PubMed]
- Priestley P, Baber J, Lolkema MP, et al. Pan-cancer whole genome analyses of metastatic solid tumors. Nature 2019;575:210-6. [Crossref] [PubMed]
- López S, Lim EL, Horswell S, et al. Interplay between whole-genome doubling and the accumulation of deleterious alterations in cancer evolution. Nat Genet 2020;52:283-93. [Crossref] [PubMed]
- Turajlic S, Swanton C. Metastasis as an evolutionary process. Science 2016;352:169-75. [Crossref] [PubMed]
- Lens SMA, Medema RH. Cytokinesis defects and cancer. Nat Rev Cancer 2019;19:32-45. [Crossref] [PubMed]
- Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev 2014;24:52-60. [Crossref] [PubMed]
- Alexandrov LB, Ju YS, Haase K, et al. Mutational signatures associated with tobacco smoking in human cancer. Science 2016;354:618-22. [Crossref] [PubMed]
- Richard C, Fumet JD, Chevrier S, et al. Exome Analysis Reveals Genomic Markers Associated with Better Efficacy of Nivolumab in Lung Cancer Patients. Clin Cancer Res 2019;25:957-66. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Teixeira VH, Pipinikas CP, Pennycuick A, et al. Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat Med 2019;25:517-25. [Crossref] [PubMed]
- Chujo M, Noguchi T, Miura T, et al. Comparative genomic hybridization analysis detected frequent overrepresentation of chromosome 3q in squamous cell carcinoma of the lung. Lung Cancer 2002;38:23-9. [Crossref] [PubMed]
- Wilbertz T, Wagner P, Petersen K, et al. SOX2 gene amplification and protein overexpression are associated with better outcome in squamous cell lung cancer. Mod Pathol 2011;24:944-53. [Crossref] [PubMed]
- Tata PR, Chow RD, Saladi SV, et al. Developmental History Provides a Roadmap for the Emergence of Tumor Plasticity. Developmental Cell 2018;44:679-693.e5. [Crossref] [PubMed]
- Mollaoglu G, Jones A, Wait SJ, et al. The Lineage-Defining Transcription Factors SOX2 and NKX2-1 Determine Lung Cancer Cell Fate and Shape the Tumor Immune Microenvironment. Immunity 2018;49:764-779.e9. [Crossref] [PubMed]
- Kenfield SA, Wei EK, Stampfer MJ, et al. Comparison of aspects of smoking among the four histological types of lung cancer. Tob Control 2008;17:198-204. [Crossref] [PubMed]


