
Relationships of kin17 protein expression with clinical features and prognosis of colorectal cancer
Introduction
Colorectal cancer (CRC) is the most common gastrointestinal tumor and the third most frequently diagnosed malignancy worldwide. In 2012, there were approximately 1.36 million new CRC cases diagnosed and 690,000 deaths, ranking the 4th among malignant tumors. In 2010, the number of new CRC cases in China exceeded 270,000, and 130,000 deaths were reported (1). Although multidisciplinary treatments including surgery, radiotherapy, and chemotherapy have been continuously improved, 40% to 50% of CRC patients eventually suffer from local recurrence and/or distant metastasis, which remain the main causes of treatment failures (2). While many clinicopathological indicators have been developed for disease treatment and prognosis prediction, the mechanisms of CRC recurrence and metastasis remain unclear. Deeper investigations on these mechanisms and more effective prevention and treatment measures are therefore required to further prolong the postoperative survival of CRC patients (3).
Recent studies have found that kin17 gene has extensive biological functions such as DNA-damage repair (4), DNA replication (5), cell proliferation (6), and cell cycle regulation (7). Kin17 is overexpressed in breast cancer (8), liver cancer (9), and chronic myelogenous leukemia cell line K562 (10) and can stimulate tumor cell proliferation. CRC cell lines highly expressing kin17 have shown resistance to chemotherapeutic drugs such as oxaliplatin and 5-fluorouracil that act on DNA and make tumor cells acquire stem-like attributes (11). Therefore, it is of great significance to further explore the expression of kin17 in CRC and its relationships with clinical pathological parameters.
Methods
General data
The clinicopathological data and paraffin specimens of 135 CRC patients in the First Affiliated Hospital of Nanchang University from November 2008 to November 2009 were collected. Among them there were 77 males and 58 females aged 19−88 years (mean: 57 years); 77 patients were younger than 60 years and 58 older than 60 years. There were 20 cases with TNM stage I, 44 cases with stage II, 55 cases with stage III, and 16 cases with stage IV. The lesion types included ulcerative in 79 cases and fungating in 49 cases. Lymph node metastases were noted in 65 cases, and distant metastases occurred in 119 cases. By the end of the follow-up, 66 patients survived and 69 had died. The cancer was low grade in 48 cases and high grade in 87 cases. The tumor types included tubular adenocarcinomas (n=102; highly differentiated in 3 cases, moderately differentiated in 80 cases, and poorly differentiated in 19 cases), mucinous adenocarcinoma (n=28), and signet-ring cell carcinoma (n=3).
Immunohistochemical examinations
The reagent kin17 mouse antibody was purchased from Abcam, and was used at a dilution of 1:200. The paraffin-embedded tissue was cut into 4-µm thick serial sections and then mounted. The sections were de-waxed in xylene for two times with 10 minutes each. Then, the de-waxed sections were re-hydrated in 100–80% alcohol and dis-tilled water. The dried sections were inoculated in 3% H2O2 deionized water at room temperature for 15 minutes to eliminate endogenous peroxidase activity. The sections were then put into a microwave oven with EDTA restoration solution; after restoration under high temperature for 5 min, the sections were thawed for 2 min, followed by intermediate- and low-temperature treatment for 20 min; finally, they were naturally cooled to room temperature. The slides were rinsed with PBS for 3 min and then dried; after having been added with reagent solution A (blocking buffer) that could cover the tissue sections, the sections were incubated for at 37 degrees for 15 minutes. After the solution A was discarded, the primary antibody that could cover the whole tissue section was added and inoculated at 4 °C overnight; in the control group, the primary antibody was replaced by PBS. After the primary antibody was discarded, the sections were rinsed three times with PBS in the shaker; then, the solution B (biotinylated goat anti-mouse secondary antibody) was added and inoculated at 37 °C for 30 min. After the solution B was discarded, the sections were soaked in PBS for 5 min (repeated 3 times), and then the solution C (horseradish peroxidase-labeled streptavidin) was added and incubated at 37 °C for 15 min. After the solution C was discarded, the sections were soaked with PBS for 5 minutes (repeated 3 times) and then the color was developed by incubation with DAB for 10 minutes at room temperature. Color development was terminated by the adding of tap water, and then the sections were counterstained with hematoxylin and differentiated in HCl-alcohol. After gradient dehydration with 30% alcohol, 70% alcohol, and 100% alcohol, the sections were dried in an oven at 37 degrees for 15−20 minutes. Finally, the sections were mounted with neutral balsam for observation under a light microscope.
Judgment of the results
Ki67 was scored according to the percentage of nuclear-stained cells and the staining intensity. Two hundred cells were counted in each visual field to calculate the percentage of positive cells. The percentage of positive cells (defined as nuclear/plasma staining) was scored using the following numeric system: 0 (0−10% positive cells); 1 (10−19%); 2 (20−50%); and 3 (>50%). The staining intensity was scored as either 0 (negative), 1 (yellowish), 2 (brownish yellow), or 3 (brown). For the immunoreactive score, the percentage of positive cells and the staining intensity were multiplied, which produced a score value between 0 and 9, among which 0−2 represents low expression and 3−9 represents high expression.
Statistical analysis
Statistical analysis was implemented by using SPSS 22.0 software package. Chi-square test was used to analyze the relationships between the kin17 protein expression and the clinicopathological features, and a P value of <0.05 was considered statistically significant. Kaplan-Meier univariate survival analysis was performed, along with Log-rank test. A Cox proportional hazards model was used for multivariate survival analysis of CRC, a P value of <0.05 was considered statistically significant. The follow-up ended on November 17, 2013. The follow-up period lasted 3−84 months (mean: 47 months).
Results
Expression of kin17 in CRC tissue and paracancerous normal tissue
The kin17 protein was mainly expressed in the nucleus in the paracancerous tissue but mainly in the cytoplasm and nucleus in the cancer tissues (Figure 1). The number of patients with high kin17 protein expression was 76 in cancer tissue and 20 in paracancerous tissue (49.6% vs. 14.8%, P<0.01). Thus, the kin17 protein expression significantly differed between cancer tissue and paracancerous tissue (P<0.0001) (Table 1).
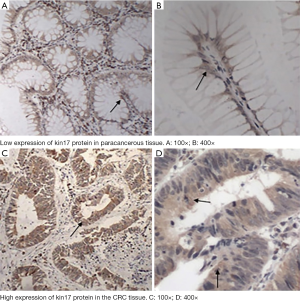
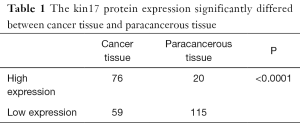
Full table
Relationships of kin17 protein expression with different clinicopathological parameters in CRC patients
High kin17 protein expression was detected in 9 of 20 cases in T1 + T2 group and in 60 cases in T3 + T4 group (31% vs. 60.6%, P<0.05). High kin17 protein expression was detected in 6 patients without lymph node metastasis and 63 patients with lymph node metastasis (9.7% vs. 100%, P<0.01); also, kin17 protein were highly expressed in 63 patients without distant metastasis and in 13 patients with distant metastasis (52.9% vs. 81.25%, P<0.05). Finally, kin17 protein expression showed no significant difference in term of age (P=0.602), gender (P=0.484), and gross pathology (P=0.068) (all P>0.05) (Table 2).
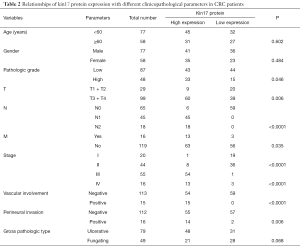
Full table
Results of univariate and multivariate analyses
To further determine the significance of kin17 in CRC, 13 variables including gender, age, tumor invasion depth, lymph node metastasis and distant metastasis, clinical stage, histological grade, vascular invasion, neural invasion, gross pathological type, kin17 protein expression, CEA, and CA199 were analyzed in the univariate analysis and Kaplan-Meier survival analysis, the results showed that kin17 protein expression was a prognostic factor for CRC (P<0.001) (Figure 2).
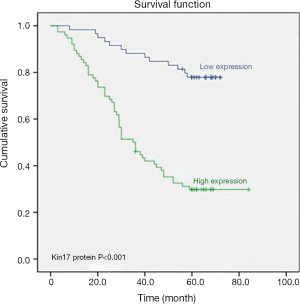
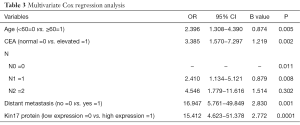
Multivariate Cox regression analysis showed that age, kin17 protein expression, CEA, lymph node metastasis, and distant metastasis were independent prognostic factors for CRC (Table 3).
Full table
Discussion
The kin17 gene is located on human chromosome 10 at p15-p14 or on band A of mouse chromosome 2. Kin17 gene encodes a 45-kDa protein in human, with a C2H2 zinc finger motif on its N-terminal. While there is a 30-kD conserved region in all species, a 15-kD conserved region can be found in higher eukaryotes. Kin17 protein consists of four motifs (12), among which the zinc finger motif is composed of amino acids 28−50 and can bind to DNA; the bipartite nuclear localization signal motif is composed of amino acids 236−266; the region homologous to RccA contains amino acids 163−201; and the KOW motif consists of amino acids 330−363, which mainly bind to RNA (13,14). Research has found that the expression of kin17 protein is associated with the lymph node metastasis of cervical squamous cell carcinoma, and kin17 protein may be related to the malignant biological behavior of the tumor (15). In our current study, we detected the expression of kin17 protein in CRC tissue and paracancerous normal tissue and found the high expression of kin17 protein might be associated with poor prognosis. Thus, kin17 protein may be a prognostic factor for CRC.
A large number of studies have shown that the pathogenesis of tumors has a complex molecular mechanism including activation of proto-oncogenes, inactivation and loss of tumor suppressor genes, and disorders in apoptosis-regulating genes and DNA repair genes (16-19). Tumorigenesis is a multi-step process. Briefly, (I) tumorigenic factors can cause gene damage, activate proto-oncogenes, and/or inactivate tumor suppressor genes and may also induce disorders in apoptosis-regulating genes, DNA repair genes, and other key regulatory genes, leading to polyclonal proliferation of cells; (II) further gene damage leads to clonal proliferation; and (III) after evolution, subclones with different biological characteristics (especially the abilities of infiltration and metastasis) are formed.
Experiments show that kin17 protein is involved in the process of cell proliferation. However, in some experiments, the effects of high kin17 protein expression on cell proliferation were not consistent. Overexpression of kin17 protein can inhibit DNA synthesis in cells and arrest cells in S phase, which can suppress the growth of mammalian cells, especially the proliferation of tumor cells. It may be due to the massive expression of kin17 protein contributes to the forming of a small intranuclear particle-like structure that disturbs cell division. In HEK293 cells transfected with the kin17 plasmid, the number of kin17 protein-expressing cell lines decreased over time. The expression of kin17 protein is low in normal proliferating human fibroblasts but is high in non-small cell adenocarcinoma cell line H1299 and colorectal adenocarcinoma cell line RKO, suggesting high kin17 expression may be needed during tumor development. A recent study has shown that kin17 expression was higher in hepatocellular adenocarcinoma tissue than in normal tissues. In vitro and in vivo experiments showed the abnormally high expression of kin17 protein promotes the growth of hepatocellular adenocarcinoma cells (9). Another study found that the expression of kin17 protein in lung cancer was elevated, which was associated with tumor grade, ki-67/p53 mutation status, and progesterone receptor. Silencing the kin17 gene can suppress epidermal growth factor-dependent cell growth, and high kin17 protein expression promotes the DNA replication and thus the proliferation of immortal MCF-10A cells (8).
In conclusion, it is speculated that kin17 protein plays an important role in the development and progression of CRC and may be a promising biomarker for prognosis prediction and targeted treatment of this malignancy. However, the exact mechanisms underlying its role in these processes still warrant further investigations.
Acknowledgments
Funding: This work is funded by Jiangxi Province Health Department Science and Technology Fund.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.08.16). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Ethics Committee of the First Affiliated Hospital of Nanchang University (ID: 20130809). Informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- International Agency for Research on Cancer. Colorectal cancer estimated incidence, mortality and prevalence worldwide in 2012(EB/OL). Available online: http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx
- Chen W, Zheng R, Zhang S, et al. Report of cancer incidence and mortality in China, 2010. Ann Transl Med 2014;2:61. [PubMed]
- Benefield HC, Baron JA, Stürmer T, et al. Moving from immune phenotyping of colorectal cancer to mechanistic insights on aspirin use. Transl Cancer Res 2016;5:S1131-S1134. [Crossref]
- Masson C, Menaa F, Pinon-Lataillade G, et al. Global genome repair is required to activate KIN17, a UVC-responsive gene involved in DNA replication. Proc Natl Acad Sci U S A 2003;100:616-21. [Crossref] [PubMed]
- Miccoli L, Frouin I, Novac O, et al. The human stress-activated protein kin17 belongs to the multiprotein DNA replication complex and associates in vivo with mammalian replication origins. Mol Cell Biol 2005;25:3814-30. [Crossref] [PubMed]
- Pinon-Lataillade G, Masson C, Bernardino-Sgherri J, et al. KIN17 encodes an RNA-binding protein and is expressed during mouse spermatogenesis. J Cell Sci 2004;117:3691-702. [Crossref] [PubMed]
- Miccoli L, Biard DS, Frouin I, et al. Selective interactions of human kin17 and RPA proteins with chromatin and the nuclear matrix in a DNA damage and cell cycle-regulated manner. Nucleic Acids Res 2003;31:4162-75. [Crossref] [PubMed]
- Zeng T, Gao H, Yu P, et al. Up-regulation of kin17 is essential for proliferation of breast cancer. PLoS One 2011;6:e25343 [Crossref] [PubMed]
- Kou WZ, Xu SL, Wang Y, et al. Expression of Kin17 promotes the proliferation of hepatocellular carcinoma cells in vitro and in vivo. Oncol Lett 2014;8:1190-4. [Crossref] [PubMed]
- Despras E, Miccoli L, Créminon C, et al. Depletion of KIN17, a human DNA replication protein, increases the radio sensitivity of RKO cells. Radiat Res 2003;159:748-58. [Crossref] [PubMed]
- Yu M, Zhang Z, Yu H, et al. KIN enhances stem cell-like properties to promote chemoresistance in colorectal carcinoma. Biochem Biophys Res Commun 2014;448:63-9. [Crossref] [PubMed]
- Kannouche P, Mauffrey P, Pinon-Lataillade G, et al. Molecular cloning and characterization of the human KIN17 cDNA encoding a component of the UVC response that is conserved among metazoans. Carcinogenesis 2000;21:1701-10. [Crossref] [PubMed]
- Tissier A, Kannouche P, Biard DS, et al. The mouse Kin-17 gene codes for a new protein involved in DNA transactions and is akin to the bacterial RecA protein. Biochimie 1995;77:854-60. [Crossref] [PubMed]
- Angulo JF, Rouer E, Mazin A, et al. Identification and expression of the cDNA of KIN17, a zinc-finger gene located on mouse chromosome 2, encoding a new DNA-binding protein. Nucleic Acids Res 1991;19:5117-23. [Crossref] [PubMed]
- Chen N, Wu KH. The expression and significance of kin17 in cervical carcinoma . J Trop Med 2016;16:25-6.
- Nearchou A, Pentheroudakis G. The significance of tumor-associated immune response in molecular taxonomy, prognosis and therapy of colorectal cancer patients. Ann Transl Med 2016;4:271. [Crossref] [PubMed]
- Hur K, Cejas P, Feliu J, et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 2014;63:635-46. [Crossref] [PubMed]
- Chen G, Lin MT. Impact of genomic heterogeneity associated with acquired anti-EGFR resistance in colorectal cancers. Transl Cancer Res 2016;5:S95-S98. [Crossref]
- Mondaca S, Yaeger R. Colorectal cancer genomics and designing rational trials. Ann Transl Med 2018;6:159. [Crossref] [PubMed]


