Deep learning for prediction of isocitrate dehydrogenase mutation in gliomas: a critical approach, systematic review and meta-analysis of the diagnostic test performance using a Bayesian approach
Introduction
Gliomas with isocitrate dehydrogenase (IDH) mutation show a better prognosis, overall survival, and response to chemotherapy than IDH wild-type gliomas, independent of histological grade (1-4). On the other hand, IDH wild-type gliomas may show comparable survival outcomes to glioblastomas with similar molecular and clinical characteristics (5). At present, identification of IDH mutation is based on histopathological analysis of tissue specimens acquired via stereotactic biopsy or definitive resection. Immunohistochemistry and deoxyribonucleic acid (DNA) sequencing techniques have been used to identify IDH genotype (6,7). Nevertheless, the procedures for obtaining tumor tissue are invasive and have potential risks (8). Additionally, spatial and temporal alterations in genetic expression cause intratumoral heterogeneity of gliomas, creating a possibility for non-representative tissue samples (9). This possibility of sampling error may lead to pitfalls in determining tumor grade and tumor mutation status. Furthermore, molecular genetic testing has other impracticalities. Immunohistochemistry misses about 15% of all IDH mutations, whereas numerous alterations which have no impact on IDH enzyme activity may be detected by sequencing (10).
Considering the beforementioned disadvantages of conventional methods for determining the IDH mutation status, research has been done to predict the IDH mutation status of gliomas noninvasively with imaging studies (11,12). The emerging field of radiology that works on obtaining molecular and genetic information from imaging studies is known as radiogenomics. It involves a series of qualitative and quantitative analyses to predict genetic and molecular properties (13).
The treatment regimen differs based on IDH mutation status in gliomas (14). Therefore, an accurate pre-treatment prediction with radiogenomics of IDH mutation status can guide clinical decision-making. Magnetic resonance imaging (MRI) is done routinely in glioma workup. It helps to assess the entire tumor and the surrounding brain tissue noninvasively. Considering the intratumoral heterogeneity and the possibility of the inadequate sampling of tumor specimens, radiogenomic information from MRI studies holds the potential to be integrated into routine clinical practice with the superiority of obtaining information from the entire tumor.
Artificial intelligence (AI) may help extracting imaging features that are incomprehensible to the human eye and associate them with outcomes. Machine learning (ML), a subgroup method of AI, is used for analyzing the correlation between radiomic features and genetic information. Predictive performance can further be enhanced, including clinical and demographic information, such as age, sex, and performance status (15). ML was reported to have a pooled sensitivity and specificity of 88% and 87%, respectively, for IDH mutation prediction in a recent meta-analysis by Zhao et al. (15). ML methods used in classical radiomic approaches utilize the extraction of predefined features, selection of these features, and application of ML techniques for outcome association and molecular and genetic status prediction (16). This pipeline is simplified and, at the same time, enhanced by using deep learning (DL) (17). In DL, radiomic features are extracted without human predefinition, unlike ML. After each round of training, the model’s internal parameters are recalibrated by the back-propagation algorithm (18).
Although several reviews and meta-analyses were recently published on the diagnostic performance of ML and DL algorithms in IDH mutation prediction, none of them has quantitatively evaluated the diagnostic performance of DL methods with the Bayes theorem (12,15,19,20). With this study, we aimed to perform a systematic review and a meta-analysis of DL algorithms’ diagnostic performance in predicting the IDH mutation status of gliomas. We also utilized the Bayes theorem to calculate the posttest probability using likelihood ratios and predetermined pretest probabilities. We present the following article in accordance with the PRISMA-DTA reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-34/rc) (21).
Methods
The present meta-analysis is exempt from ethical approval of the Institutional Review Board since the analysis only involves de-identified data and all the included prospective studies have received local ethics approval. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Literature search
A comprehensive literature search was performed based on the following combination of Medical Subject Headings (MeSH) terms and keywords for a PubMed database search: ((“deep learning” [tw] OR “neural network*” [tw] OR “Deep Learning”[Mesh]) AND (radiogen* [tw] OR radiomic*[tw]) AND (IDH [tw] OR “isocitrate dehydrogenase” OR “Isocitrate Dehydrogenase” [Mesh]) AND (glioma [tw] OR “Glioma” [Mesh])). Corresponding keywords were used for Cochrane Library, Web of Science, Medline, and Scopus data search. The last search was conducted on August 1, 2021. The reference lists of all included studies were checked manually in order to identify other relevant papers.
Study selection
Two authors [M Karabacak and BB Ozkara (3 and 1 year of experience in performing systematic reviews and meta-analyses, respectively)] independently evaluated the eligibility of the articles, and any disagreement was resolved via discussion with a third author (S Bisdas, 18 years of experience in neuroradiology).
Articles were included based on the fulfillment of all the following criteria: (I) patients with histopathologically confirmed World Health Organization (WHO) grade II, III, or IV gliomas; (II) histopathological examination with the IDH mutation; (III) DL was applied to predict the IDH mutation status; (IV) sufficient data for reconstruction of confusion matrices (2×2 tables) in terms of the diagnostic performance of the DL algorithms; (V) original research articles. Corresponding authors of the studies that did not include sufficient data for reconstruction of confusion matrices but fulfilled the rest of the inclusion criteria were contacted via email to inquire if their study originally contained sufficient data. Studies were included if their corresponding author supplied us with the data via email within two weeks of receiving our email.
Articles were excluded if they fulfilled any of the following criteria: (I) reviews, letters, guidelines, editorials, or errata; (II) ML was applied to predict the IDH mutation status; (III) IDH mutation status was not predicted; (IV) insufficient data for the reconstruction of confusion matrices; (V) studies with overlapping cohorts.
Data extraction
Data were collected by the two authors (M Karabacak and BB Ozkara) for the following variables: (I) study characteristics (author, year, country, number of patients, age, sex, and distribution of tumor WHO grades); (II) MRI sequences used in DL algorithms; (III) information included in DL algorithms; (IV) number of patients whose IDH mutation status predicted with DL; and (V) number of patients who had molecular analysis results that revealed IDH mutation.
Quality assessment
The quality assessments were conducted by two authors (M Karabacak and BB Ozkara) independently according to the Checklist for Artificial Intelligence in Medical Imaging (CLAIM) and Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) (22,23). The CLAIM is a new checklist with 42 items for assessing AI studies in medical imaging. Studies were scored 0 or 1 on a 2-point scale for each item. The CLAIM score was calculated by summing the scores of each item per study. An overall CLAIM compliance score was defined for each article by the ratio of applicable items fulfilled. The items were weighted equally.
Four domains were evaluated with QUADAS-2: (I) patient selection; (II) index test; (III) reference standard; and (IV) flow and timing. The patient selection domain provides questions concerning methods of patient selection. The index test domain includes questions about the index test and how it was conducted and interpreted. The reference standard domain provides questions regarding the reference standard and how it was performed and analyzed. The flow and timing domain investigate whether any patients did not receive the index test or reference standard or were excluded from the confusion matrices. Concerns about applicability and bias risk were rated as low, high, or unclear on a 3-point scale.
Any disagreements during the quality assessment were resolved through discussion or with the assistance of a third reviewer.
Statistical analysis
The primary outcome was to evaluate the diagnostic performance of DL algorithms for predicting IDH mutation status in patients with gliomas. Confusion matrices with true-positive (TP), false-positive (FP), true-negative (TN), and false-negative (FN) values were separately calculated for training and validation sets within each study. Zero-point-five was added to prevent the zero cell count problem if any TP, FP, TN, or FN value was 0 (24). Separate meta-analyses were computed for training and validation sets. All statistical analyses were conducted using R version 3.4.1 (R Foundation for Statistical Computing), implementing R packages mada and meta (25). An alpha level of 0.05 was considered statistically significant.
A bivariate random-effects model was used to obtain the pooled sensitivity and specificity with their 95% confidence intervals (CIs) (26). The diagnostic odds ratio (DOR), defined as the odds of having a positive IDH mutation status prediction by DL algorithms in patients with IDH mutant gliomas compared to the odds of having a positive IDH mutation status prediction by DL algorithms in patients with IDH-wild type gliomas, was also obtained. Sensitivity, specificity, and DOR are expressed by forest plots. Summary receiver operating characteristics (SROC) curves were generated for the overall diagnostic accuracy, and the area under the curve (AUC) was calculated (27). Test performance accuracy was categorized as low (AUC, 0.50–0.69), moderate (AUC, 0.70–0.89), or high (AUC, 0.90–1.00) (28).
It becomes less likely that someone with a positive test will actually have the tested condition and more likely that someone with a positive test will not actually have the tested condition as the prevalence of a specific disease decreases (29). To address this problem, we utilized the Bayes’ theorem in our meta-analysis based on the concept that diagnostic accuracy of a test depends on the prevalence of the tested condition in the test population, as well as the test characteristics such as sensitivity and specificity. Probabilities were calculated as pretest odds × likelihood ratio +/− = posttest odds, where odds were substituted with probabilities as odds = probability/(1 – probability) (30). For pretest probabilities, previous genome-wide association studies were used to calculate the overall percentage of IDH mutant gliomas with an overall 80.2% (5,31). Fagan nomograms were plotted to visualize the relationship between pretest probability, the likelihood ratio, and the posttest probability.
Heterogeneity across all eligible studies was estimated using Q-test with P<0.05, indicating the presence of study heterogeneity and I2 statistics. I2 values were defined as follows: heterogeneity that might not be important (0–25%), low heterogeneity (26–50%), moderate heterogeneity (51–75%), and high heterogeneity (76–100%) (32). Publication bias was not assessed in our analysis, as the small number of studies included in our meta-analysis (n=4) may lead to inconclusive funnel plots and regression tests for detecting publication bias (33).
Results
Literature search
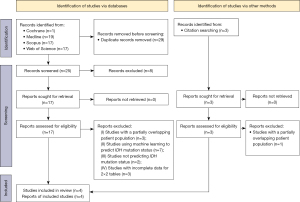
The study selection process is illustrated in Figure 1. The initial literature search yielded 54 articles: 19 from Medline, 17 from Scopus, 17 from Web of Science, and one from Cochrane library, databases, respectively. After removing 29 duplicate articles, the remaining 25 were screened based on their title and abstract, and eight were excluded. Full texts of the remaining 17 articles were obtained and reviewed.
Fifteen articles were excluded because; they used ML to predict the IDH mutation status (n=7), did not predict IDH mutation status (n=2), had an overlapping patient population with one of the other included articles (n=3), and had insufficient data for the reconstruction of confusion matrices (n=3).
Three articles were obtained and reviewed when the references provided in the selected articles were also screened. One article was again excluded because it had an overlapping patient population with one of the other included articles.
Finally, four original articles that included 1,295 patients with glioma were eventually included and analyzed in this study. Among the four studies, three (34-36) included data for validation and training sets, while the remaining one (37) only had data for the validation set. The sample sizes for the training and validation sets were 1,007 and 437, respectively.
Quality assessment
A quality assessment summary of the included studies using the CLAIM is shown in Table 1. Three studies had an item marked as “not applicable” (35-37). The mean CLAIM score of the four studies was 25.25 with a standard deviation of 5.56 (range, 20.00–31.00). The mean CLAIM compliance score of the four studies was 0.61 with a standard deviation of 0.13 (range, 0.49–0.74).
Table 1
| Study | Title/abstract (N=2) | Introduction (N=2) | Methods (N=28) | Results (N=5) | Discussion (N=2) | Other information (N=3) | Total (N=42) | CLAIM compliance |
|---|---|---|---|---|---|---|---|---|
| Chang et al. | 2 | 2 | 22 | 2 | 2 | 1 | 31 | 0.74 |
| Choi et al. | 2 | 2 | 20 (1 N/A) | 2 | 2 | 1 | 29 (1 N/A) | 0.71 |
| Li et al. | 1 | 2 | 15 (1 N/A) | 0 | 1 | 1 | 20 (1 N/A) | 0.49 |
| Matsui et al. | 2 | 2 | 12 (1 N/A) | 2 | 2 | 1 | 21 (1 N/A) | 0.51 |
CLAIM, Checklist for Artificial Intelligence in Medical Imaging; N, number; N/A, not applicable.
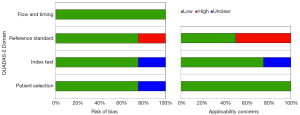
A quality assessment summary of the included studies using the QUADAS-2 tool is shown in Figure 2. Regarding patient selection, one study indicated an unclear risk of bias as they failed to mention the inclusion criteria of patient enrollment (37). Regarding the index test, one study was considered to have an unclear risk of bias, as they did not mention the details of the DL algorithm (36). One study presented a high risk of bias regarding reference standards (34). Chang et al. combined three different cohorts in their study (34). In one cohort, only the IDH 1 R132H mutation was detected in the reference test, while both IDH 1 and 2 mutations were detected in the other two cohorts. One study was assessed to have unclear concerns regarding the applicability of the index test since the details of the DL algorithm were not mentioned (36). Two studies were considered to have high concerns regarding the applicability of the reference standards (34,36). Matsui et al. and Chang et al. detected only IDH R132 and IDH R132H, respectively, which did not match the review question exactly (34,36).
Characteristics of included studies
The patient and study characteristics are described in Table 2. In terms of the applied MRI sequences, three studies used conventional [T1-weighted imaging (T1WI), contrast-enhanced T1WI (T1CE), T2-weighted imaging (T2WI), fluid-attenuated inversion recovery (FLAIR)] MRI sequences (34,36,37), one used dynamic susceptibility contrast-MRI (DSC-MRI) (35). For DL analysis, two studies included only DL extracted radiomics information (35,37), and two studies used radiomics and clinical information (34,36). One study included only age (34), and one included age, sex, and tumor location (36).
Table 2
| Authors | Year | N of Pts | Mean age in years | Sex (M:F) | WHO grade | MRI sequence | DL algorithm | Information used in DL | IDH mutation ratio (%) |
|---|---|---|---|---|---|---|---|---|---|
| Li et al. (37) | 2017 | 119 | 40.7 | 89:30 | Grade II | T1CE, FLAIR | Convolutional neural network | MRI data | 74.0 |
| Chang et al. (34) | 2018 | 496 | 52.0 | 259:237 | Grade II, III, IV | T1, T1CE, T2, FLAIR | Residual neural network | MRI data, age | 47.2 |
| Choi et al. (35) | 2019 | 463 | 52.2 | 272:191 | Grade II, III, IV | DSC perfusion MRI | Recurrent neural network | MRI data | 27.0 |
| Matsui et al. (36) | 2020 | 217 | 42.0 | 131:86 | Grade II, III | T1, T2, FLAIR | Residual neural network | MRI data, age, sex, and tumor position | 77.0 |
| Total | 2017–2019 | 1,295 | 49.4 | 751:544 | Grade II, III, IV | – | – | – | 47.4 |
N, number; Pts, patients; M, male; F, female; WHO, World Health Organization; MRI, magnetic resonance imaging; DL, deep learning; IDH, isocitrate dehydrogenase; T1CE, T1-weighted contrast-enhanced; FLAIR, fluid attenuated inversion recovery; DSC, dynamic susceptibility contrast.
Assessment of diagnostic performance
Training sets
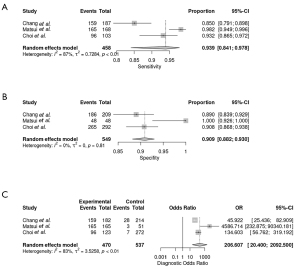
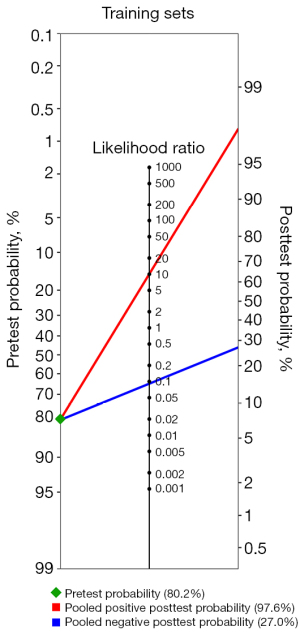
The sensitivities and specificities in the training sets of the individual included studies ranged from 85.1% to 98% and from 88.9% to 100%, respectively. The pooled sensitivity and specificity for the diagnostic performance of DL algorithms for prediction of IDH mutation status were 93.9% (95% CI: 84.1–97.8%; Figure 3A) and 90.9% (95% CI: 88.2–93.0%; Figure 3B), respectively. A DOR of 206.607 (95% CI: 20.400–2,092.500; Figure 3C) was recorded; this value indicates that the likelihood of the distinction of IDH-mutant gliomas from IDH-wild type gliomas was approximately 207 times higher in training sets using DL. The large CI observed was the result of the small number of included studies. The area under the SROC curve was 0.958, showing high test performance accuracy. Figure 4 depicts the SROC curve with the point estimate and associated 95% confidence region for pooled sensitivity/specificity pairs. When the pretest probability was set at 80.2%, the positive posttest probability was 97.6% and the negative posttest probability was 27.0% (Figure 5). The Q-test demonstrated that heterogeneity was present across the studies (Q=11.71, P=0.0029), and the Higgins I2 statistic demonstrated the presence of high heterogeneity in sensitivity (I2=87.2%, 95% CI: 63.6–95.5%) and low heterogeneity in specificity (I2=36.8%, 95% CI: 0.0–79.9%).

Validation sets
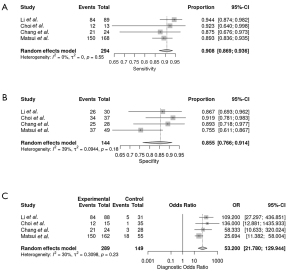
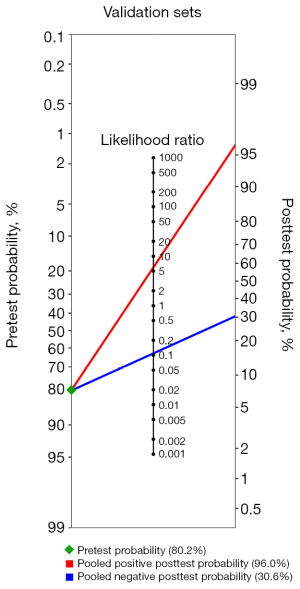
The sensitivities and specificities in the validation sets of the individual included studies ranged from 75.6% to 94.4% and from 67.3% to 93.1%, respectively. The pooled sensitivity and specificity for the diagnostic performance of DL algorithms for prediction of IDH mutation status were 90.8% (95% CI: 86.9–93.6%; Figure 6A) and 85.5% (95% CI: 76.6–91.4%; Figure 6B), respectively. A DOR of 53.200 (95% CI: 21.780–129.944; Figure 6C) was recorded; this value indicates that the likelihood of the distinction of IDH-mutant gliomas from IDH-wild type gliomas was approximately 53 times higher in validation sets using DL. Figure 7 depicts the SROC curve with the point estimate and associated 95% confidence region for pooled sensitivity/specificity pairs. When the pretest probability was set at 80.2%, the positive posttest probability was 96.0% and the negative posttest probability was 30.6% (Figure 8). The Q-test demonstrated that heterogeneity was not present across the studies (Q=4.31, P=0.2297), and the Higgins I2 statistic demonstrated the presence of heterogeneity that might not be important in sensitivity (I2=0.0%, 95% CI: 0.0–84.7%) and low heterogeneity in specificity (I2=38.8%, 95% CI: 0.0–79.1%).

Discussion
To our knowledge, this is the first systematic review and meta-analysis that summarizes the diagnostic performance of DL in the prediction of IDH mutation status in gliomas via the Bayes theorem. Our study revealed that DL models perform well in predicting IDH mutation in gliomas, with a pooled sensitivity and specificity of 93.9% (95% CI: 84.1–97.8%) and 90.9% (95% CI: 88.2–93.0%) in the training sets, and 90.8% (95% CI: 86.9–93.6%) and 85.5% (95% CI: 76.6–91.4%) in the validation sets, respectively. The AUC value was reported in three studies for validation sets and ranged from 0.95 to 0.98. The area under the SROC curve is a diagnostic accuracy index. The diagnostic results are better when it is closer to 1. The area under the SROC curve for training and validation sets was 0.958 and 0.939, indicating high test performance accuracy. Furthermore, more cautious estimates were derived from the Bayes theorem with a pre-established pretest probability of 80.2% and still demonstrated a high positive posttest probability in the training (97.6%) and validation sets (96.0%). This means that the probability of the patient having the disease increases from 80.2% to 97.7% with a positive test result based on the results of the training sets and to 96.0% based on the results of the validation sets. Again, calculations were derived from the Bayes theorem with a pre-established pretest probability of 80.2%, revealing a negative posttest probability of 27.0% and 30.6% in the training and validation sets, respectively. This means that the probability of the patient having the disease decreases from 80.2% to 27.0% with a negative test result based on the results of the training sets and to 30.6% based on the results of the validation sets. The Bayesian method for meta-analyses has the advantage of accounting for the uncertainty around the heterogeneity variance, making it a key strength of our work (38).
Among the four studies included in this meta-analysis, Choi et al. demonstrated the highest positive posttest probability (98.2%) (35). This could be partly due to the utilization of DSC-MRI in the model. Compared to wild-type tumors, hypoxia-inducible-factor 1-alpha activity is lower in IDH mutant tumors. This contributes to a distinct transcriptome signature associated with vasculogenesis and angiogenesis-related signaling pathways, which leads to increased proangiogenic molecules in IDH wild-type tumors (39). Perhaps, the model of Choi et al. achieved a great diagnostic performance by utilizing DSC perfusion-weighted MRI, which is more suitable than conventional MRI for demonstrating the tumor vasculature of gliomas. Prior multi-centered studies have also shown the diagnostic accuracy of ML-assisted DSC-MRI radiomics (40,41). However, the lack of broad clinical dissemination of advanced MRI sequences in most hospital imaging protocols and the needed expertise for the data post-processing and interpretation makes this approach difficult to be widely implemented in clinical practice.
It is worth noting that, because the majority of glioblastomas are IDH wild type, datasets containing a mix of low-grade glioma (LGG) and glioblastomas may suggest a higher level of accuracy than the true accuracy in purely LGG cohorts. The reason is that when the model detects a glioblastoma, it may conclude that it is an IDH wild-type tumor even if the IDH genotype has not been predicted (36). Glioblastoma patients were included in two studies. Choi et al. included 289 glioblastoma patients (62.4%), and Chang et al. included 233 glioblastoma patients (47%) in their studies (34,35).
Several noninvasive methods, both utilizing and not utilizing DL/ML techniques, have been evaluated as predictors of IDH mutation in gliomas. Goyal et al. demonstrated that the T2-FLAIR-mismatch sign was not sensitive but very specific in predicting IDH mutation in gliomas (pooled sensitivity, 32.1%; pooled specificity, 100%) (42). T2-FLAIR mismatch sign is a radiologic signature in which a hyperintense signal on T2WI and a relatively suppressed signal on FLAIR imaging is combined. It does not involve any ML or DL technique, and it can be detected by the naked eye. Goyal et al. also used the Bayes theorem with a pretest probability of 80.2% to generate more conservative estimates, a positive posttest probability of 99.2%, and a negative posttest probability of 73.5% (42). In another meta-analysis, Suh et al. showed that the pooled sensitivity and specificity of 2-hydroxyglutarate magnetic resonance spectroscopy in predicting IDH mutation, without any ML or DL techniques, was 95% and 91%, respectively (43). In another study, Suh et al. also assessed the performances of various conventional and advanced imaging modalities in the prediction of IDH mutation, which consisted of conventional MRI, diffusion-weighted imaging/perfusion-weighted imaging, magnetic resonance spectroscopy, 2-hydroxyglutarate magnetic resonance spectroscopy, and radiomics methods (11). Pooled sensitivity was 86%, and the pooled specificity was 87%. Furthermore, Zhao et al. evaluated the diagnostic accuracy of ML in predicting IDH mutations in a recent meta-analysis (15). The pooled sensitivity and specificity were 87% and 88% in the training set, and 87% and 90% in the validation set, respectively.
DL is a growing field of study that focuses on creating artificial neural networks (ANN) inspired by biological neural networks in the human brain (44). ANN are used in some of the most successful DL approaches. ANNs, by learning and capturing the information contained in the data, recognize complicated nonlinear correlations between dependent and independent variables (45). Choi et al. used a recurrent neural network that can process sequence inputs and take past outputs as inputs using their internal memory (35). Li et al. used a convolutional neural network (CNN), which evaluates the 2D structure of input data using local connections and weights, followed by pooling techniques to derive spatial invariant features (37). In the other two studies, Matsui et al. and Chang et al. used residual neural networks, which employ identical shortcut connections to let information flow between layers without the attenuation that several stacked nonlinear transformations would produce (34,36).
DL algorithms do not require an intermediary feature extraction or an engineering phase to learn the relationship between the input and the appropriate labels. DL combines the processes of data representation and prediction. These are possibly the most significant advantages of DL algorithms compared to conventional ML algorithms (44). However, in medicine, there are some concerns and challenges with DL algorithms. DL models are often referred to as “black boxes” because they are very recursive. According to Rudin, this is causing interpretability issues, which makes clinicians hesitant (46). However, significant progress has been made in developing algorithms that may be able to open the “black box” of DL for a range of deep neural networks (44). Another concern with DL models, when combined with radiomics, is that current models still lack repeatability and validation. There are currently no guidelines for radiomic features or the construction of clinical models using these features (47). There are no evaluation criteria or standardized data collection. Perhaps, these are some of the challenges that need to be resolved to appreciate DL’s potential and value in medicine fully.
The method of identifying the region of interest (ROI) on MRI scans is known as segmentation or labeling (37). Tumor tissue is differentiated from normal surrounding tissues during glioma segmentation. The segmented ROI can then be used to extract radiomic data, which is very time-consuming and highly variable between operators when done manually (48). Automatic tumor segmentation methods have been developed to improve tumor segmentation and overcome these difficulties. In a meta-analysis, van Kempen et al. demonstrated that tumor segmentation based on ML algorithms showed an overall dice similarity coefficient score of 0.84, with high heterogeneity (80.4%) (48). In the included studies in our review, several segmentation methods were used. Li et al. used a CNN (37). They used it to compete in the Brain Tumor Segmentation Challenge (BRATS) (49), where it was placed first and second in BRATS 2013 and BRATS 2015, respectively (37). Choi et al. also used a fully automatic segmentation method based on a CNN algorithm with a manual correction that was the second-placed method in the international BRATS 2017 (35). Matsui et al. and Chang et al. manually segmented the ROIs in their studies (34,36).
This meta-analysis demonstrated that DL is promising in detecting IDH mutations in gliomas. Even though an increasing number of papers being published in recent years, little of them has been translated into clinical practice, indicating the need for several improvements in study design before these methods can be implemented in clinical practice (50). Firstly, standardized data collection, evaluation criteria, and reporting guidelines are required to establish the generalizability and validation of the findings (44). More research for developing clinically oriented DL models with high interpretability is needed to alleviate clinicians’ concerns. According to some authors, current model interpretation methods such as saliency mapping and class activation are unreliable (46,51). As a result, Rudin et al. recommended developing and applying inherently interpretable models (46). Chen et al. proposed a method in which their model distinguishes several parts of an image where it thinks a part of the image mimics a prototypical part of a bird species and then identifies the species of birds based on a weighted combination of the similarity scores between parts of the image and the trained prototypes (52). As a result, their model is interpretable, with a transparent reasoning process similar to human reasoning when making classifications. We believe that inherently interpretable models, such as the one mentioned above, are required to implement DL models into clinical practice by increasing clinician trust in the model. Furthermore, to integrate radiomic models into clinical practice, clinical trials must demonstrate improvements in patient management and decision making (47). Nonetheless, our study revealed that DL models have a high potential for predicting IDH mutations in gliomas. Upon successful improvements, they may be integrated into clinical practice at a low cost compared to conventional methods for detecting IDH mutation and a reliable diagnostic tool.
Our study is not without limitations. Some study data were obtained via email, which reduces their reliability. However, the main limitation is the lack of statistics for the training and test sets. As a result, the statistics of the validation sets were the primary focus of our research. Finally, three studies in the literature were excluded due to a lack of data required to reconstruct a confusion matrix. It is worth noting that, even though it is not a limitation of our study, all the included studies had the limitation of being conducted retrospectively, necessitating the need for additional large-scale prospective studies to validate their findings.
Conclusions
This study revealed that DL algorithms demonstrate excellent diagnostic performance in predicting IDH mutation in gliomas, with an overall 96.0% positive posttest probability in validation sets. Radiomic features related to IDH mutation, and its underlying pathophysiology extracted from advanced MRI, such as perfusion-weighted sequences, may further increase the prediction probability and support large-scale, prospective trials to distill the diagnostic and clinical added value of such DL models in neuro-oncology.
Acknowledgments
We state that the abstract of our study has been published at the 2022 Annual Meeting of the American Academy of Neurology.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the PRISMA-DTA reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-34/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-34/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321:1807-12. [Crossref] [PubMed]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360:765-73. [Crossref] [PubMed]
- Hartmann C, Hentschel B, Wick W, Capper D, Felsberg J, Simon M, Westphal M, Schackert G, Meyermann R, Pietsch T, Reifenberger G, Weller M, Loeffler M, von Deimling A. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol 2010;120:707-18. [Crossref] [PubMed]
- Houillier C, Wang X, Kaloshi G, Mokhtari K, Guillevin R, Laffaire J, Paris S, Boisselier B, Idbaih A, Laigle-Donadey F, Hoang-Xuan K, Sanson M, Delattre JY. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology 2010;75:1560-6. [Crossref] [PubMed]
- Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med 2015;372:2481-98. [Crossref] [PubMed]
- Takano S, Tian W, Matsuda M, Yamamoto T, Ishikawa E, Kaneko MK, Yamazaki K, Kato Y, Matsumura A. Detection of IDH1 mutation in human gliomas: comparison of immunohistochemistry and sequencing. Brain Tumor Pathol 2011;28:115-23. [Crossref] [PubMed]
- Zou Q, Hu Q, Guo M, Wang G. HAlign: Fast multiple similar DNA/RNA sequence alignment based on the centre star strategy. Bioinformatics 2015;31:2475-81. [Crossref] [PubMed]
- Akay A, Rüksen M, Islekel S. Magnetic Resonance Imaging-guided Stereotactic Biopsy: A Review of 83 Cases with Outcomes. Asian J Neurosurg 2019;14:90-5. [Crossref] [PubMed]
- Belden CJ, Valdes PA, Ran C, Pastel DA, Harris BT, Fadul CE, Israel MA, Paulsen K, Roberts DW. Genetics of glioblastoma: a window into its imaging and histopathologic variability. Radiographics 2011;31:1717-40. [Crossref] [PubMed]
- Andronesi OC, Rapalino O, Gerstner E, Chi A, Batchelor TT, Cahill DP, Sorensen AG, Rosen BR. Detection of oncogenic IDH1 mutations using magnetic resonance spectroscopy of 2-hydroxyglutarate. J Clin Invest 2013;123:3659-63. [Crossref] [PubMed]
- Suh CH, Kim HS, Jung SC, Choi CG, Kim SJ. Imaging prediction of isocitrate dehydrogenase (IDH) mutation in patients with glioma: a systemic review and meta-analysis. Eur Radiol 2019;29:745-58. [Crossref] [PubMed]
- Bhandari AP, Liong R, Koppen J, Murthy SV, Lasocki A. Noninvasive Determination of IDH and 1p19q Status of Lower-grade Gliomas Using MRI Radiomics: A Systematic Review. AJNR Am J Neuroradiol 2021;42:94-101. [Crossref] [PubMed]
- Bourgier C, Colinge J, Aillères N, Fenoglietto P, Brengues M, Pèlegrin A, Azria D. Radiomics: Definition and clinical development. Cancer Radiother 2015;19:532-7. [Crossref] [PubMed]
- Wang K, Wang Y, Fan X, Wang J, Li G, Ma J, Ma J, Jiang T, Dai J. Radiological features combined with IDH1 status for predicting the survival outcome of glioblastoma patients. Neuro Oncol 2016;18:589-97. [Crossref] [PubMed]
- Zhao J, Huang Y, Song Y, Xie D, Hu M, Qiu H, Chu J. Diagnostic accuracy and potential covariates for machine learning to identify IDH mutations in glioma patients: evidence from a meta-analysis. Eur Radiol 2020;30:4664-74. [Crossref] [PubMed]
- Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJWL. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci Rep 2015;5:13087. [Crossref] [PubMed]
- Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Communications of the ACM 2017;60:84-90. [Crossref]
- Chang P, Grinband J, Weinberg BD, Bardis M, Khy M, Cadena G, Su MY, Cha S, Filippi CG, Bota D, Baldi P, Poisson LM, Jain R, Chow D. Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas. AJNR Am J Neuroradiol 2018;39:1201-7. [Crossref] [PubMed]
- Jian A, Jang K, Manuguerra M, Liu S, Magnussen J, Di Ieva A. Machine Learning for the Prediction of Molecular Markers in Glioma on Magnetic Resonance Imaging: A Systematic Review and Meta-Analysis. Neurosurgery 2021;89:31-44. [Crossref] [PubMed]
- van Kempen EJ, Post M, Mannil M, Kusters B, Ter Laan M, Meijer FJA, Henssen DJHA. Accuracy of Machine Learning Algorithms for the Classification of Molecular Features of Gliomas on MRI: A Systematic Literature Review and Meta-Analysis. Cancers (Basel) 2021;13:2606. [Crossref] [PubMed]
- Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 2021;372: [PubMed]
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM. QUADAS-2 Group. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011;155:529-36. [Crossref] [PubMed]
- Mongan J, Moy L, Kahn CE Jr. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiol Artif Intell 2020;2:e200029. [Crossref] [PubMed]
- Sweeting MJ, Sutton AJ, Lambert PC. What to add to nothing? Use and avoidance of continuity corrections in meta-analysis of sparse data. Stat Med 2004;23:1351-75. [Crossref] [PubMed]
- mada.pdf [Internet]. [cited 2022 Apr 17]. Available online: https://cran.r-project.org/web/packages/mada/mada.pdf
- Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol 2005;58:982-90. [Crossref] [PubMed]
- Kumar R, Indrayan A. Receiver operating characteristic (ROC) curve for medical researchers. Indian Pediatr 2011;48:277-87. [Crossref] [PubMed]
- Swets JA. Measuring the accuracy of diagnostic systems. Science 1988;240:1285-93. [Crossref] [PubMed]
- Medina LS, Blackmore CC, Applegate KE. Principles of Evidence-Based Imaging. In: Medina L, Applegate K, Blackmore C. editors. Evidence-Based Imaging in Pediatrics. New York, NY: Springer, 2010:3-16.
- Choi BC. Slopes of a receiver operating characteristic curve and likelihood ratios for a diagnostic test. Am J Epidemiol 1998;148:1127-32. [Crossref] [PubMed]
- Labreche K, Kinnersley B, Berzero G, Di Stefano AL, Rahimian A, Detrait I, Marie Y, Grenier-Boley B, Hoang-Xuan K, Delattre JY, Idbaih A, Houlston RS, Sanson M. Diffuse gliomas classified by 1p/19q co-deletion, TERT promoter and IDH mutation status are associated with specific genetic risk loci. Acta Neuropathol 2018;135:743-55. [Crossref] [PubMed]
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ 2003;327:557-60. [Crossref] [PubMed]
- Terrin N, Schmid CH, Lau J. In an empirical evaluation of the funnel plot, researchers could not visually identify publication bias. J Clin Epidemiol 2005;58:894-901. [Crossref] [PubMed]
- Chang K, Bai HX, Zhou H, Su C, Bi WL, Agbodza E, et al. Residual Convolutional Neural Network for the Determination of IDH Status in Low- and High-Grade Gliomas from MR Imaging. Clin Cancer Res 2018;24:1073-81. [Crossref] [PubMed]
- Choi KS, Choi SH, Jeong B. Prediction of IDH genotype in gliomas with dynamic susceptibility contrast perfusion MR imaging using an explainable recurrent neural network. Neuro Oncol 2019;21:1197-209. [Crossref] [PubMed]
- Matsui Y, Maruyama T, Nitta M, Saito T, Tsuzuki S, Tamura M, Kusuda K, Fukuya Y, Asano H, Kawamata T, Masamune K, Muragaki Y. Prediction of lower-grade glioma molecular subtypes using deep learning. J Neurooncol 2020;146:321-7. [Crossref] [PubMed]
- Li Z, Wang Y, Yu J, Guo Y, Cao W. Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci Rep 2017;7:5467. [Crossref] [PubMed]
- Hackenberger BK. Bayesian meta-analysis now - let's do it. Croat Med J 2020;61:564-8. [Crossref] [PubMed]
- Kickingereder P, Sahm F, Radbruch A, Wick W, Heiland S. Deimling Av, Bendszus M, Wiestler B. IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci Rep 2015;5:16238. [Crossref] [PubMed]
- Sudre CH, Panovska-Griffiths J, Sanverdi E, Brandner S, Katsaros VK, Stranjalis G, et al. Machine learning assisted DSC-MRI radiomics as a tool for glioma classification by grade and mutation status. BMC Med Inform Decis Mak 2020;20:149. [Crossref] [PubMed]
- Manikis GC, Ioannidis GS, Siakallis L, Nikiforaki K, Iv M, Vozlic D, Surlan-Popovic K, Wintermark M, Bisdas S, Marias K. Multicenter DSC-MRI-Based Radiomics Predict IDH Mutation in Gliomas. Cancers (Basel) 2021;13:3965. [Crossref] [PubMed]
- Goyal A, Yolcu YU, Goyal A, Kerezoudis P, Brown DA, Graffeo CS, Goncalves S, Burns TC, Parney IF. The T2-FLAIR-mismatch sign as an imaging biomarker for IDH and 1p/19q status in diffuse low-grade gliomas: a systematic review with a Bayesian approach to evaluation of diagnostic test performance. Neurosurg Focus 2019;47:E13. [Crossref] [PubMed]
- Suh CH, Kim HS, Jung SC, Choi CG, Kim SJ. 2-Hydroxyglutarate MR spectroscopy for prediction of isocitrate dehydrogenase mutant glioma: a systemic review and meta-analysis using individual patient data. Neuro Oncol 2018;20:1573-83. [Crossref] [PubMed]
- Parekh VS, Jacobs MA. Deep learning and radiomics in precision medicine. Expert Rev Precis Med Drug Dev 2019;4:59-72. [Crossref] [PubMed]
- Patel JL, Goyal RK. Applications of artificial neural networks in medical science. Curr Clin Pharmacol 2007;2:217-26. [Crossref] [PubMed]
- Rudin C. Stop Explaining Black Box Machine Learning Models for High Stakes Decisions and Use Interpretable Models Instead. Nat Mach Intell 2019;1:206-15. [Crossref] [PubMed]
- Avanzo M, Wei L, Stancanello J, Vallières M, Rao A, Morin O, Mattonen SA, El Naqa I. Machine and deep learning methods for radiomics. Med Phys 2020;47:e185-202. [Crossref] [PubMed]
- van Kempen EJ, Post M, Mannil M, Witkam RL, Ter Laan M, Patel A, Meijer FJA, Henssen D. Performance of machine learning algorithms for glioma segmentation of brain MRI: a systematic literature review and meta-analysis. Eur Radiol 2021;31:9638-53. [Crossref] [PubMed]
- Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging 2015;34:1993-2024. [Crossref] [PubMed]
- Pinto Dos Santos D, Dietzel M, Baessler B. A decade of radiomics research: are images really data or just patterns in the noise? Eur Radiol 2021;31:1-4. [Crossref] [PubMed]
- Seah JCY, Tang JSN, Kitchen A, Gaillard F, Dixon AF. Chest Radiographs in Congestive Heart Failure: Visualizing Neural Network Learning. Radiology 2019;290:514-22. [Crossref] [PubMed]
- Chen C, Li O, Tao C, Barnett AJ, Su J, Rudin C. This Looks Like That: Deep Learning for Interpretable Image Recognition. ArXiv180610574 Cs Stat [Internet] 2019 Dec 28 [cited 2021 Aug 26]. Available online: http://arxiv.org/abs/1806.10574