
Utility of high-throughput sequencing of T-cell receptor rearrangements in assessing treatment response in patients with mature T-cell lymphoma
T-cell lymphomas (TCL) represent a heterogeneous group of non-Hodgkin lymphomas frequently characterized by refractoriness to standard chemotherapy even in the first-line setting (1). Relapsed/refractory (RR) disease is associated with poor outcomes, and overall survival usually less than 9 months (2). The current standard of care in the evaluation of treatment responses in lymphoid malignancies is centered primarily on imaging studies such as conventional computed tomography (CT) and positron emission tomography-computerized tomography (PET/CT) scans, which are costly and require routinely exposing patients to radiation. Furthermore, despite favorable responses on interim or end of treatment imaging, many patients eventually relapse, indicative of their limited predictiveness for long term survival (3). Identifying methods that achieve earlier identification of patients with suboptimal treatment responses may allow for more prompt treatment intervention and improved outcomes. For this reason, there has been growing interest in utilizing non-invasive disease monitoring for evaluating response to treatment and detecting early relapse in lymphoid malignancies, including blood-based high-throughput sequencing/next-generation sequencing (HTS/NGS) that allows for identification of clonal sequences in both circulating tumor cells as well as cell-free DNA (cfDNA) (4). Indeed, HTS/NGS of cfDNA or peripheral blood mononuclear cells (PBMC) can detect a lower burden of disease with higher sensitivity and specificity than that afforded by standard imaging techniques, and can also serve as an independent prognostic variable in various B-cell lymphoid malignancies (4,5). However, there is currently limited data available regarding the role of this technology in the setting of TCL.
Herein, we provide a preliminary report on the potential utility of HTS of T-cell receptors (TCR) to evaluate disease response to treatment in patients with RR TCL in a subgroup of patients participating in a previously published clinical trial evaluating treatment of patients with RR TCL with the PD-1 inhibitor pembrolizumab (6). Detailed inclusion criteria were previously published. The observed overall response rate was 33% with a complete response (CR) rate of 27%. HTS was performed on the variable regions of the TCR beta (TCRβ) and gamma (TCRγ) loci of 6 out of 18 patients as a planned correlative study. Samples for HTS assessment were collected at diagnosis and pre-specified time points on treatment and at time of progression. Baseline clinical patient characteristics and available samples are outlined in Figure 1.
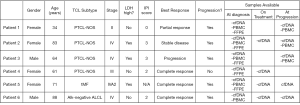
Immunosequencing of the CDR3 regions of human TCRβ and TCRγ chains was performed using the TCRβ and TCRγ Assay (Adaptive Biotechnologies, Seattle, USA). Plasma was collected in EDTA tubes which were processed within 2 hours to extract cfDNA using a Qiagen kit. Similar techniques were employed for the formalin-fixed, paraffin embedded (FFPE) and PBMC sample collections. Extracted genomic DNA was amplified in a bias-controlled multiplex PCR, followed by high-throughput sequencing. Sequences were collapsed and filtered in order to identify and quantitate the absolute abundance of each unique TCRβ and TCRγ CDR3 region for further analysis as previously described (7-9). To be classified as potentially associated with malignant cells, a calibrating sequence had to be present at >3% frequency in the repertoire with >40 templates present, had to make up ≥0.2% of the nucleated cell count in PBMC specimens or ≥0.1% of the nucleated cell count in FFPE specimens, and had to have a sufficiently high frequency such that there were <6 clones within a decade below. Calibrating clones were identified from PBMC and FFPE specimens as there was insufficient material to determine repertoire dominance in the cfDNA samples. Dynamics in changes of identified T-cell clones over time in responders versus non-responders were assessed.
We found that 4 of the 6 included patients had calibrating clonal sequences at baseline; all 4 had ≥2 identifiable clonal sequences. Calibrating clonal sequences amongst these 4 patients made up 5–50% of the repertoire in the baseline PBMCs or FFPE samples, and were identified in both TCRβ and TCRγ loci. These sequences were detectable in cfDNA specimens even when only 100 T-cell templates were available for analysis, and the baseline calibrating clonal sequences were identified as present in all of the cfDNA samples in the patients who progressed. The two patients without calibrating sequences at baseline achieved CR to therapy on clinical trial.
The cfDNA specimens were quantitatively insufficient to analyze statistical significance of clonal frequency variation over time, which was likely due to inadequate specimen collection. At baseline, only 1 (25%) of the CR patient’s (patient 4) 4 baseline calibrating sequences was not detectable in the cfDNA. Unfortunately, there was insufficient material to obtain PBMC samples from this patient. In the 3 progression patients (patients 1–3), all calibrating clonal sequences were detectable in all cfDNA specimens. Amongst these patients, all had a PBMC sample taken at time of progression. Patients 2 and 3 had both TCRβ and TCRBγ sequencing at progression, while patient 1 had sufficient material only for TCRβ. Five of the 6 calibrating sequences observed in a progression PBMC sample showed a significant increase in frequency relative to baseline. The specific HTS findings of each patient are outlined in Figures 2,3.
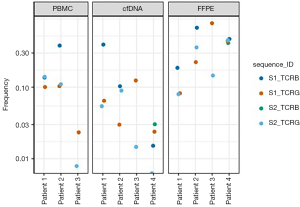
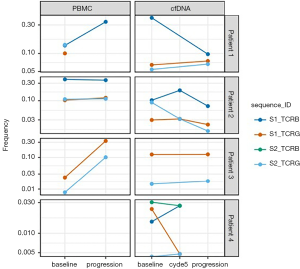
Our pilot project demonstrates that HTS assessment is able to identify a malignant clone that can be followed over time in patients with TCL. This study also suggests that baseline calibrating clones derived from most FFPE and PBMC samples can be identified and tracked in cfDNA specimens. However, our results are very preliminary and have several limitations. These include the failure to identify a trackable clonotype in all patients, which may be related to low frequency clones in patients with low tumor burden, as well as discordance of trackable TCR rearrangements, which may be related to presence of potential non-disease associated clones. While larger investigations will be needed, these preliminary data suggest that frequency of the tumor clone at baseline and reduction in frequency may be predictive of response to treatment. Another study that investigated the role of HTS of the TCR locus to track circulating tumor DNA (ctDNA) in patients with lymphoma treated with allogeneic hematopoietic stem cell transplantation found that the presence of detectable ctDNA was associated with disease relapse or progression (10). If HTS is confirmed to be an accurate means of tracking disease burden in TCL in larger studies, this would provide a more efficient and noninvasive method for disease monitoring, and may allow an opportunity to modify therapy earlier thereby resulting in more optimal care of patients with aggressive TCL.
Acknowledgments
The authors would like to thank the research support received from Merck. They would also like to thank the biostatisticians at Adaptive Biotechnologies for their collaboration on this project.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Precision Cancer Medicine. The article has undergone external peer review.
Peer Review File: Available at http://dx.doi.org/10.21037/pcm-20-45
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm-20-45). HCF reports personal fees from Genetech, personal fees from Sandofi, personal fees from Astrazeneca, personal fees from Abbvie, personal fees from Amgen, personal fees from Takada, personal fees from Jansen, personal fees from BMS, personal fees from Karyopharm, outside the submitted work. PAF is an employee at Adaptive Biotechnologies. JZ reports personal fees from Seattle genetics, verastem, kiyowa Kirin, and Mundi pahrma, outside the submitted work. NHF reports personal fees from Bostongene, during the conduct of the study; grants from Roche, grants and personal fees from Gilead, grants and personal fees from TG Therapeutics, outside the submitted work. YO reports other support from Genentech, outside the submitted work. SKB reports grants from Merck, during the conduct of the study; grants and personal fees from Seattle Genetics, grants from Celgene, grants from Bayer, personal fees from Atara, personal fees from Mundipharma, personal fees from Pfizer, outside the submitted work. And SKB serves as an unpaid editorial board member of Precision Cancer Medicine from Jun 2020 to May 2022. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Teras LR, DeSantis CE, Cerhan JR, et al. 2016 US lymphoid malignancy statistics by World Health Organization subtypes. CA Cancer J Clin 2016;66:443-59. [Crossref] [PubMed]
- Mak V, Hamm J, Chhanabhai M, et al. Survival of patients with peripheral T-cell lymphoma after first relapse or progression: spectrum of disease and rare long-term survivors. J Clin Oncol 2013;31:1970-6. [Crossref] [PubMed]
- Mehta-Shah N, Ito K, Bantilan K, et al. Baseline and interim functional imaging with PET effectively risk stratifies patients with peripheral T-cell lymphoma. Blood Adv 2019;3:187-97. [Crossref] [PubMed]
- Kurtz DM, Green MR, Bratman SV, et al. Noninvasive monitoring of diffuse large B-cell lymphoma by immunoglobulin high-throughput sequencing. Blood 2015;125:3679-87. [Crossref] [PubMed]
- Ladetto M, Bruggemann M, Monitillo L, et al. Next-generation sequencing and real-time quantitative PCR for minimal residual disease detection in B-cell disorders. Leukemia 2014;28:1299-307. [Crossref] [PubMed]
- Barta SK, Zain J. Phase II Study of the PD-1 Inhibitor Pembrolizumab for the Treatment of Relapsed or Refractory Mature T-cell Lymphoma. Clin Lymphoma Myeloma Leuk 2019;19:356-64.e3. [Crossref] [PubMed]
- Robins HS, Campregher PV, Srivastava SK, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood 2009;114:4099-107. [Crossref] [PubMed]
- Carlson CS, Emerson RO, Sherwood AM, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun 2013;4:2680. [Crossref] [PubMed]
- Robins H, Desmarais C, Matthis J, et al. Ultra-sensitive detection of rare T cell clones. J Immunol Methods 2012;375:14-9. [Crossref] [PubMed]
- Herrera AF, Kim HT, Kong KA, et al. Next-generation sequencing-based detection of circulating tumour DNA After allogeneic stem cell transplantation for lymphoma. Br J Haematol 2016;175:841-50. [Crossref] [PubMed]
Cite this article as: Phull PM, Tan CR, Fung HC, Fields PA, Liu Y, Zain J, Fowler NH, Alpaugh RK, Oki Y, Barta SK. Utility of high-throughput sequencing of T-cell receptor rearrangements in assessing treatment response in patients with mature T-cell lymphoma. Precis Cancer Med 2020;3:31.

